Abstract
This study detailed the responses of Galleria mellonella larvae to disseminated infection caused by co-infection with Candida albicans and Staphylococcus aureus . Doses of C. albicans (1×105 larva−1) and S. aureus (1×104 larva−1) were non-lethal in mono-infection but when combined significantly (P<0.05) reduced larval survival at 24, 48 and 72 h relative to larvae receiving S. aureus (2×104 larva−1) alone. Co-infected larvae displayed a significantly higher density of S. aureus larva−1 compared to larvae infected solely with S. aureus . Co-infection resulted in dissemination throughout the host and the appearance of large nodules. Co-infection of larvae with C. albicans and S. aureus (2×104 larva−1) resulted in an increase in the density of circulating haemocytes compared to that in larvae infected with only S. aureus . Proteomic analysis of co-infected larval haemolymph revealed increased abundance of proteins associated with immune responses to bacterial and fungal infection such as cecropin-A (+45.4-fold), recognition proteins [e.g. peptidoglycan-recognition protein LB (+14-fold)] and proteins associated with nodule formation [e.g. Hdd11 (+33.3-fold)]. A range of proteins were also decreased in abundance following co-infection, including apolipophorin (−62.4-fold), alpha-esterase 45 (−7.7-fold) and serine proteinase (−6.2-fold). Co-infection of larvae resulted in enhanced proliferation of S. aureus compared to mono-infection and an immune response showing many similarities to the innate immune response of mammals to infection. The utility of G. mellonella larvae for studying polymicrobial infection is highlighted.
Keywords: Galleria, infection, immunity, mini-model, Candida, Staphylococcus, co-infection
Introduction
The human microbiome consists of communities of bacteria, archaea, viruses and fungi that, through their interactions, have both positive and negative effects on each other and on their host [1]. In times of dysbiosis, polymicrobial infections can lead to aggressive forms of diseases that are usually difficult to treat. Co-colonization by Aspergillus fumigatus and Pseudomonas aeruginosa in the cystic fibrosis lung has poorer clinical outcomes compared to colonization by P. aeruginosa alone [2]. A large proportion of infections (30–60 %) caused by Candida albicans are polymicrobial and Staphylococcus aureus is one of the most common microbes isolated during these infections [3–5]. Infections caused by C. albicans affect large numbers of patients and a recent study found that in chemotherapy-immunosuppressed mice C. albicans can influence the resident bacterial community and contribute to mucosal bacterial dysbiosis, facilitating enhanced invasion by Candida [6]. In recent years, advances in sequencing technologies have demonstrated that Candida rarely exists in isolation but rather as a heterogeneous biofilm with other fungal and bacterial species [7]. These types of infections in humans include catheter infections, burn wound infections, septicaemia, ventilator-associated pneumonia, keratitis, denture stomatitis, peritonitis, cystic fibrosis and urinary tract infections [1, 8]. The interactions between S. aureus and C. albicans are synergistic and result in increased mortality in animal models, which is associated with enhanced invasion, biofilm formation, exacerbated inflammatory responses and intrinsic resistance to antimicrobial chemotherapy [8–19]. Proteomic analysis of the interactions between S. aureus and C. albicans has demonstrated an increased abundance of proteins associated with stress and growth responses, and metabolism in the early stages of the interaction, although S. aureus had little effect on the proteome of C. albicans hyphae [20].
C. albicans virulence includes polymorphism and phenotypic switching, production of toxins (e.g. candidalysin), invasins (e.g. Als3p), biofilm formation and metabolic adaption [21, 22]. The virulence of S. aureus is multifaceted and allows the bacterium to interact with the host (fibronectin-binding proteins A and B), evade the immune response (chemotaxis inhibitory protein of staphylococci) and induce tissue damage (alpha toxin) [23–27]. During co-infection S. aureus can bind to Als3p in the C. albicans cell wall and this facilitates disseminated infection in an oral co-colonization model [8]. Interestingly, interactions between C. albicans and S. aureus in mixed species biofilm result in alterations in the proteome, resulting in increased abundance of proteins associated with resistance to oxidative stress and virulence [28]. To date, these pathogens have mostly been studied in isolation in higher vertebrates and there is a lack of a system that is compatible with the 3R policies and enables the generation of rapid results and allows the screening of multiple interactions in a short space of time.
Larvae of the greater wax moth Galleria mellonella are a widely used model to study the virulence of microbial pathogens and to assess the efficacy of antimicrobial agents due to the many advantages associated with their use (e.g. ease of use, cost-effectiveness, lack of legal and ethical restrictions) [29–33]. More recently, larvae have been utilized to study the virulence of bacterial (e.g. S. aureus [34], Streptococcus pneumoniae [35] and Enterococcus faecalis [36, 37]) and fungal (e.g. C. albicans [38, 39], A. fumigatus [40] and Cryptococcus neoformans [41]) pathogens as well as their interactions with the cellular and humoral immune response of larvae. Larvae were also used to assess the interaction between S. aureus and C. albicans and show how co-infection reduced the efficacy of miconazole [14]. The aim of this work was to characterize the development of, and the host immune response to, mixed microbial co-infection in G. mellonella larvae in order to develop a simple but effective system to characterize polymicrobial infections in vivo.
Methods
Strains and culture conditions
C. albicans MEN (a kind gift from Professor D. Kerridge, Cambridge, UK) was cultured in YEPD broth [2 % (w/v) glucose, 2 % (w/v) bactopeptone (Difco Laboratories), 1 % (w/v) yeast extract (Oxoid Ltd, Basingstoke, UK)]. S. aureus (clinical isolate) was cultured in nutrient broth (Oxoid). Cultures were grown overnight at 37 °C and 200 r.p.m. to the early stationary phase as described elsewhere [34]. Cells were harvested by centrifugation (2000 x g) and cell pellets were washed three times with phosphate-buffered saline (PBS) prior to injection into larvae. Stocks of C. albicans and S. aureus were maintained on YEPD agar plates as above but supplemented with 2 % (w/v) agar] or nutrient agar plates, respectively.
Larval culture and inoculation
Sixth instar larvae of the greater wax moth G. mellonella (Livefoods Direct Ltd, UK) were stored in the dark at 15 °C and maintained in wood chippings [38]. Larvae weighing 0.22±0.03 g were selected and used within 2 weeks of receipt. Ten healthy larvae per treatment and controls were placed in sterile 9 cm Petri dishes lined with Whatman filter paper and containing some wood shavings. Larvae were inoculated with S. aureus (enumerated using a spectrophotometer) through the last left pro-leg into the haemocoel with a Myjector U-100 insulin syringe (Terumo Europe NV, Belgium). For co-infection, larvae were administrated 20 µl of a solution containing C. albicans (enumerated using a haemocytometer) and S. aureus at the inoculum stated. Larvae were acclimatized to 37 °C for 1 h prior to all experiments and incubated at 37 °C for all studies. All experiments were performed independently on three separate occasions.
Determination of proliferation of S. aureus in G. mellonella larvae
Larvae were infected with C. albicans or S aureus or co-infected with C. albicans and S aureus , and S. aureus proliferation larva−1 was assessed over 48 h. Infected larvae (n=3) were homogenized using a pestle and mortar with 3 ml of PBS, serially diluted and plated onto nutrient agar ( S. aureus ) plates supplemented with 0.1 g (v/v) amphotericin B and incubated at 37 °C for 24 h. The bacterial load was calculated as the S. aureus colony-forming units (c.f.u.) per larva and was based on the number of colonies that grew at specific dilutions. This protocol is described in detail elsewhere [34, 42].
Cryo-imaging of co-infected G. mellonella larvae
G. mellonella larvae were co-infected with C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) and incubated at 37 °C for 0, 6, 24 and 48 h. Larvae (n=3) were embedded in Cryo-Imaging Embedding Compound (Bioinvision), flash-frozen in liquid nitrogen and mounted on a stage for sectioning [34, 40]. Sectioning and imaging was carried out every 10 µm using a Cryoviz (Bioinvision, Inc., Cleveland, OH, USA) cryo-imaging system.
Determination of haemocyte density in G. mellonella larvae
Changes in haemocyte density was assessed by bleeding 40 µl of haemolymph from G. mellonella larvae (n=5) into a microcentrifuge tube on ice to prevent melanization. Haemolymph was diluted in 0.37 % (v/v) mercaptoethanol-supplemented PBS and cell density was determined using a haemocytometer. Haemocyte density was expressed as haemocyte density per larva. Experiments were performed on three independent occasions and the means±se were determined.
Quantitative proteomics of larval haemolymph
Quantitative proteomics was conducted on haemocyte-free haemolymph from larvae (n=10 with four biological replicates) at 0, 6 and 24 h post-co-infection [C. albicans (1×105 larva−1), S. aureus (2×104 larva−1)]. Protein (75 µg) was prepared according to established protocols [38]. Protein identification from the tandem mass spectrometry (MS/MS) data was performed using the Andromeda search engine in MaxQuant (version 1.2.2.5; http://maxquant.org/) to correlate the data against a six-frame translation of the EST contigs for G. mellonella [43, 44] or the proteome for C. albicans or S. aureus (Uniprot).
Results processing, statistical analyses and graphics generation were conducted using Perseus v 1.5.5.3. Label-free quantification (LFQ) intensities were log2-transformed and analysis of variance (ANOVA) of significance and t-tests between the haemolymph proteomes of 0, 6 and 24 h S . aureus -treated larvae were performed using a P-value of 0.05 and significance was determined using false discovery rate (FDR) correction (Benjamini–Hochberg). The (FDR was set at 1 % for both peptides and proteins. Proteins that had non-existent values (indicative of absence or very low abundance in a sample) were also used in statistical analysis of the total differentially expressed group following imputation of the zero values using a number close to the lowest value of the range of proteins plus or minus the standard deviation. After data imputation these proteins were included in subsequent statistical analysis.
Statistical analysis
All experiments were performed on three independent occasions and the results are expressed as the mean±se. All of the statistical analysis listed was performed using GraphPad Prism v 6.00 (one-way ANOVA; survival, haemocyte density, c.f.u.). Differences were considered significant at P<0.05.
Data availability
The mass spectrometry (MS) proteomics data and MaxQuant search output files have been deposited to the ProteomeXchange Consortium [45] via the PRIDE partner repository with the dataset identifier PXD014273.
Results
Response of G. mellonella larvae to co-infection by C. albicans and S. aureus
Infection of larvae with an inoculum of 1×105 C. albicans larva−1 resulted in no significant decrease in survival over 72 h as compared to non-infected controls. Infection of larvae with S. aureus (2×106 larva−1) resulted in larval survival of 80±10 % at 24 h, 55.93±9.62 % at 48 h and 10±5.15 % at 72 h post-infection (Fig. 1). Co-infection of larvae with C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) resulted in a significant decrease in survival [24 h, 70±3.33 % (P<0.01); 48 h, 10±3.33 % (P<0.0001); 72 h, 5±6.66 % (P<0.0001)] compared to larvae that received S. aureus (2×104 larva−1) alone at 24, 48 (100±0 %) and 72 h (96.66±5.77 %). Co-infection of G. mellonella larvae with C. albicans (1×105 larva−1) and S. aureus (2×106 larva−1) reduced survival to 30±6.66 % (P<0.0001) at 24 h and 0±0 % (P<0.0001) at 48 and 72 h post-infection, relative to mono-infection with S. aureus (2×106 larva−1) (Fig. 1).
Fig. 1.
Survival of G. mellonella larvae following mono- and co- infection with C. albicans and S. aureus . Larvae were infected with 20 µl of C. albicans or S. aureus or co-infected (1×105 C. albicans with varying concentrations of S. aureus ) and survival was assessed over 72 h at 37 °C. Statistical significance was determined by comparing co-infected larvae to the relevant S. aureus mono-infected larvae (**, P<0.01; ****, P<0.0001). Larvae were also injected with 20 µl PBS (PBS-injected). All values are the mean±se of three independent experiments.
Dissemination of infection throughout the host
The microbial burden of co-infected larvae was assessed by measuring S. aureus c.f.u larva−1 in co-infected [C. albicans (1×105 larva−1) with S. aureus (2×104 or 2×106 larva−1)] larvae and comparing these to larvae infected solely with S. aureus (2×104 or 2×106 larva−1). Co-infected larvae displayed a significantly higher S. aureus c.f.u larva−1 at 6 (1.51±0.17×105 c.f.u. larva−1, P<0.05), 24 (1.01±0.38×107 c.f.u. larva−1, P<0.05) and 48 h (2.01±0.23×108 c.f.u larva−1, P<0.001) compared to larvae infected with S. aureus (2×104 larva−1) alone at 6 (4.49±0.5×104 c.f.u larva−1), 24 (4.51±0.37×105 c.f.u larva−1) and 48 h (1.05±0.33×107 c.f.u larva−1). There was a significant increase in S. aureus c.f.u. at 6 (1.55±0.59×106 c.f.u larva−1, P<0.001), 24 (2.58±0.52×108 c.f.u larva−1, P<0.05) and 48 h (6.12±0.73×108, P<0.01) post-infection in co-infected larvae compared to those infected solely with S. aureus (2×106 larva−1) at 6 (9.96±0.68×105 c.f.u. larva−1), 24 (9.41±0.34×107 c.f.u. larva−1) and 48 h (3.21±0.51×108 c.f.u. larva−1) (Fig. 2).
Fig. 2.
S. aureus c.f.u. larva−1 obtained from G. mellonella larvae infected with S. aureus cells (2×106 and 2×106 larva−1) or co-infected larvae over 48 h at 37 °C. Co-infection with C. albicans and S. aureus (C. a+S. a) results in an increase in bacterial load in larvae from 6 to 48 h as determined by plating on nutrient agar plates. Statistical analysis was carried out by comparing co-infected larvae (C. a+S. a) to larvae infected solely with S. aureus (S. a) at their respective timepoints, (*, P<0.05; **, P<0.01). All values are the mean±se of three independent experiments.
Previous work has visualized the dissemination of C. albicans and S. aureus , separately, in larvae [39]. In this case cryo-imaging was employed to assess the development of dual infection within G. mellonella larvae. Larvae were co-infected with C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) and cryo-imaging was performed at 0, 6, 24 and 48 h post-infection (Fig. 3). Six hours post-infection melanized nodules (black arrows) appeared around the site of inoculation (white arrows) and throughout the host. By 24 h post-infection there was extensive melanization (red arrow) of larval tissue, especially around the site of inoculation and nodules were visible throughout the larva. By 48 h post-infection the survival rate for G. mellonella larvae was 10±3.33% and there was extensive melanization throughout the larvae as well as a large number of distinct nodules.
Fig. 3.
CryoViz-imaging of G. mellonella larvae co-infected with C. albicans and S. aureus . Larvae (n=3) were co-infected with C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) for 0, 6, 24 and 48 h at 37 °C. Larvae were embedded, sectioned (10 µm) and visualized using a Cryoviz (Bioinvision, Inc., Cleveland, OH, USA) cryo-imaging system. Point of inoculation, white arrows; nodules, black arrows; extensive melanization, red arrows.
Analysis of larval immune response to C. albicans and S. aureus dual infection
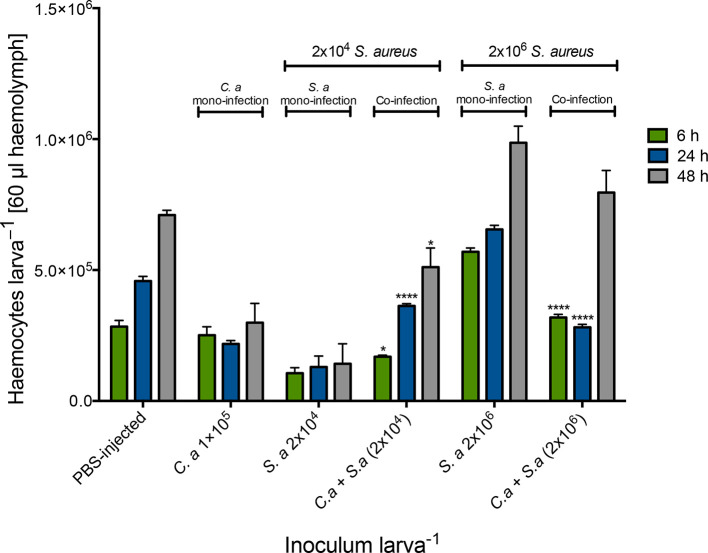
Alterations in circulating haemocyte density in co-infected larvae were determined in order to assess the cellular response to co-infection. Uninfected larvae had a haemocyte density of 2.4±0.17×105 haemocytes larva−1. Mono-infection with C. albicans (1×105 larva−1) resulted in a small alteration in haemocyte density at 24 [2.18±0.22×105 haemocytes larva−1 (60 μl haemolymph)] and 48 h (2.09±0.13×105). Inoculation of larvae with S. aureus (2×104 larva−1) also resulted in a change in haemocyte density at 6 (1.26±0.36×105), 24 (1.30±0.73×105) and 48 h (1.42±0.13×105) post-infection. Mono-infection with S. aureus 2×106 larva−1 produced alterations in circulating haemocyte density at 6 (5.70±0.25×105, P<0.0001) and 24 h (6.55±0.27×105, P<0.01) post-infection (Fig. 4).
Fig. 4.
Alteration in circulating haemocyte density following inoculation with S. aureus cells alone and in combination with C. albicans. G. mellonella larvae were inoculated with 20 µl S. aureus cells (2×104 and 2×106 larva−1) alone or in combination with C. albicans (1×105 larva−1) and haemocytes were extracted and enumerated from 6, 24 and 48 h post-inoculation at 37 °C. Statistical analysis was performed by comparing co-infected larvae (C. a+S.a) to S. aureus -only-treated larvae at their at respective doses and time points (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001). All values are the mean±se of three independent experiments.
Co-infection of larvae with C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) resulted in an increase in the density of circulating haemocytes at 6 (1.69±0.13×105, P<0.05), 24 (3.63±0.14×105, P<0.0001) and 48 h (5.11±0.20×105, P<0.05) post-infection compared to larvae infected solely with S. aureus (2×104 larva−1). Co-infection of C. albicans (1×105 larva−1) with S. aureus at an inoculum of 2×106 larva−1 (3.19±0.31×105, P<0001) resulted in a significant decrease in circulating haemocyte density relative to larvae infected solely with S. aureus (2×106 larva−1) 6 h post-infection (2×105, 3.15±0.17×105; 2×106, 5.70±0.25×105).
Quantitative proteomic analysis was performed on the cell-free haemolymph of G. mellonella larvae following co-infection with C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) for 0, 6 and 24 h post-infection. In total, 2293 peptides were identified, representing 351 proteins with 2 or more peptides and 48 and 83 (6 vs 0 h and 24 vs 0 h, respectively) proteins were determined to be differentially abundant (ANOVA, P<0.05) with a fold change of >1.5. A total of 34, 46 and 84 proteins at 0, 6 and 24 h, respectively, were deemed exclusive (Fig. S1, available in the online version of this article). These proteins were subsequently used to statistically analyse the total differentially expressed group after imputation of the zero values as described and were then included in statistical analysis after data imputation. A principal component analysis (PCA) was performed on all filtered proteins and distinguished the 0, 6 and 24 h C. albicans- and S. aureus -treated samples. A clear difference between the 0, 6 and 24 h proteome was observed (Fig. S2).
The proteins that showed increased relative abundance in the larvae co-infected for 6 h compared to the control were gustatory receptor candidate 25 (+10.5-fold), gloverin-like protein (+7.9-fold), putative defence protein Hdd11 (+7.1-fold), actin 3 (+6.6-fold) and paramyosin (+5-fold). The proteins that showed decreased relative abundance in the larvae co-infected for 6 h compared to the control were cathepsin B-like cysteine proteinase (−4.9-fold), apolipophorins (−2.4-fold), beta-1, 3-glucan-binding protein (−2.3-fold) and methionine-rich storage protein (−2.2-fold) (Fig. 5a , Table S1).
Fig. 5.
Proteomic responses of G. mellonella larval haemolymph following co-infection by C. albicans (1×105 larva−1) and S. aureus (2×104 larva−1) after 6 (a) and 24 (b) h at 37 °C. Volcano plots represent protein intensity difference (log2 mean intensity difference) and significance in differences (- log P-value) based on a two-sided t -test. Proteins above the line are considered statistically significant (P value<0.05) and those to the right and left of the vertical lines indicate relative fold changes>1.5. Annotations are given for the most differentially abundant proteins identified in haemolymph from co-infected larvae 6 and 24 h. These plots are based upon post imputed data.
The proteins that showed increased relative abundance in co-infected larvae at 24 h compared to the control larvae were cecropin-A (+45.4-fold), gustatory receptor candidate 25 (+41.7-fold), cecropin-d-like peptide (+37.8-fold), putative defence protein Hdd11 (+33.3-fold), gloverin (+19.3-fold) peptidoglycan-recognition protein-LB (+14.0-fold), serpin-4B (+12.9-fold) and salivary cysteine-rich peptide precursor (+12.6-fold). The proteins that showed decreased relative abundance in larvae co-infected for 24 h compared to the control larvae were apolipophorins (−62.4-fold), alpha-esterase 45 (−7.7-fold), serine proteinase (−6.2-fold), saposin-related precursor (−5.8-fold), carboxylesterase CarE-11 precursor (−5.6-fold) and cathepsin B-like cysteine proteinase (−4-fold) (Fig. 5b , Table S2). A range of antimicrobial peptides and proteins (Fig. S3a), proteins associated with bacterial and fungal recognition, and proteins involved in nodule formation (Fig. S3b) were identified within the co-infected larval haemolymph.
A range of C. albicans and S. aureus proteins were also detected in co-infected G. mellonella larval haemolymph. A total of 23 C. albicans proteins were identified in co-infected G. mellonella larval haemolymph and included Hsp70 family ATPase, heat shock protein SSA2 and peroxiredoxin TSA1-A (Table S3). In addition, 20 S . aureus proteins such as staphopain B and HTH-type transcriptional regulator SarR were detected in haemolymph 24 h post-infection (Table S4).
Discussion
The incidence of polymicrobial infection is possibly underestimated as current routine clinical microbiological investigation could never encompass the vast number and diversity of microbes present within the human micro- and mycobiome. Advances in technologies associated with the detection of these microbes will eventually enter the clinic and this will require the reclassification of many mono-infections as polymicrobial infections. The work presented here aimed to develop an in vivo system to study the pathogenesis of polymicrobial infection using G. mellonella larvae, which have been widely used for studying the development of single-species infections [46–49].
Inoculation of larvae with C. albicans (1×105 larva−1) or S. aureus (2×104 larva−1) resulted in no reduction in survival, but co-inoculation of larvae with these doses resulted in a large decrease in survival. This suggests that the interactions between C. albicans and S. aureus may be synergistic, which has previously been documented in murine studies [1, 8, 9, 13, 14, 16, 50]. Infection of G. mellonella larvae with S. aureus (2×106 larva−1) or C. albicans (5×105 larva−1) in mono- infection resulted in 70 and 44 % larval mortality after 48 h, respectively [34, 38]. Co-infection of larvae with S. aureus (1×105 larva−1) and C. albicans (2×104 larva−1) produced mortality of 90 %, 48 h post-infection. Interestingly, single doses of C. albicans and S. aureus were non-lethal 5 days post-infection in mice but when combined resulted in 70 % mortality 2 days post-infection [50].
Co-inoculation of larvae resulted in a significant increase in S. aureus c.f.u larva−1 relative to mono-infection. In co-infected mice, the microbial burden was significantly higher in the kidney and spleen compared to mono-infection [50]. Interestingly, the total bacterial c.f.u. in co-infected larvae, regardless of dose, was 10.65±4.40-fold higher than in S. aureus -only-infected larvae.
Cryo-imaging was utilized to assess the dissemination of co-infection throughout the host over 48 h. By 24 h there was significant nodule formation and by 48 h there was extensive melanization and widespread nodule formation. Melanization of co-infected larvae is more systemic than that observed in mono-infection, which appears as localized nodules throughout the host [34, 39]. Previous results indicated that infection of larvae with a higher inoculum of C. albicans (5×105 larva−1) or S. aureus (2×106 larva−1) in mono-infection resulted in more cuticular melanization and larger granuloma (nodule)-type structure formation [34, 39]. This may indicate that co-infection, at a lower cell density, which ultimately produced a larger decrease in survival relative to mono-infection, may have activated a less robust immune response to co-infection (summarized in Table 1) and via less cuticular melanization. This aberrant immune response (e.g. decreased AMP abundance in co-infection relative to mono-infection) could be due to the acute and accelerated proliferation of S. aureus in the presence of C. albicans, which results in an uncontrollable bacterial burden that cannot be cleared in time by the host. Peters and Noverr [50] demonstrated that co-infection was associated with an increased inflammatory response characterized by an up to 100-fold increase in inflammatory cytokines (e.g. IL-6) and neutrophil influx [50].
Table 1.
Comparison of the different proteomic responses between single infection of G. mellonella larvae with C. albicans [30], S. aureus [29] and co-infection (current study). Fold change is 24 h infected larval haemolymph relative to 0 h relevant infected control*
|
Infection and reference |
C. albicans [30] |
Co-infection (C. albicans and S. aureus ) |
||
|---|---|---|---|---|
|
Inoculum size |
5×105 20 µl−1 |
2×106 20 µl−1 |
1×105 (C. albicans) and 2×106 ( S. aureus ) 20 µl−1 |
|
|
Protein name |
Fold change (+/−)* |
Fold change (+/−)* |
Fold change (+/−)* |
|
|
Invasion |
Muscle protein 20-like protein |
+173.3-fold |
nd |
+8.6-fold |
|
Nodulation |
Hdd11 |
+49.4-fold |
+7.24-fold |
+33.3-fold |
|
Antimicrobial peptide |
Gloverin |
+52.5-fold |
+121.69-fold |
+19.3-fold |
|
Cecropin-D |
+22.8-fold |
+73.7-fold |
+37.8-fold |
|
|
Detoxification |
Glutathione s transferase |
+114.1-fold |
nd |
nd |
|
Thioredoxin |
+3.2-fold |
−3.94-fold |
nd |
|
|
Microbial recognition |
PG-RP LB |
+71.6-fold |
+9.53-fold |
+14-fold |
|
PG-RP B |
+7.5-fold |
+17.35-fold |
nd |
|
|
β-glucan RP |
−28.6-fold |
+2.16-fold |
nd |
nd, non detected; PG RP, peptidoglycan recognition protein.
Infection of larvae with a low inoculum of S. aureus (2×104 larva−1) with C. albicans (1×105 larva−1) resulted in a significant increase in haemocyte density, however an inoculum of S. aureus (2×106 larva−1) with C. albicans (1×105 larva−1) produced a decrease in haemocyte density compared to S. aureus mono-infection. The increase in haemocyte density is probably a result of the release of haemocytes usually attached to the linings of internal surface of the cuticle and organs such as the fat body [51], while the decrease in haemocyte density associated with the higher inoculum may be due to the aggregation of haemocytes at the site of inoculation in order to halt the spread of the infection.
The humoral immune response of larvae to co-infection was determined by label-free MS. At 6 h post co-infection antimicrobial peptide gloverin-like protein had increased 7.9-fold. Gloverins are heat-stable antibacterial polypeptides that bind to bacterial lipopolysaccharide and components of the fungal cell wall. It was previously demonstrated that Escherichia coli and S. aureus induce gloverin expression in Bombyx mori and Manduca sexta [52, 53]. Gloverins display activity against yeasts but do not possess anti- S. aureus activity [53]. In C. albicans-infected G. mellonella larvae, gloverin was not detected 6 h post-infection but had increased 52-fold at 24 h post-infection, however in S. aureus -infected larvae gloverin had increased 20-fold at 6 h and 121-fold at 24 h post-infection [34, 38]. Similarly, PGRP-B increased 3-fold in co-infected larvae but 2.3-fold in C. albicans and 6-fold in S. aureus mono-infected larvae [34, 38]. These experiments were performed with five times more C. albicans (5×105 larva−1) and 100 times more S. aureus (2×106 larva−1) compared to co-infected larvae [C. albicans (1×105 larva−1), S. aureus (2×104 larva−1)] in the current study (Table 1). Members of the phenoloxidase cascade (e.g. prophenoloxidase activating factor 3) increased in abundance by 4.2-fold in co-infected larvae 6 h post-co-infection but had only increased 2.4-fold in C. albicans and 2.85-fold in S. aureus at 6 h post-mono-infection (Table 1). The phenoloxidase cascade is a series of enzymatic reactions that result in the formation of phenoloxidase, which participates in melanotic encapsulation, wound healing and cuticle sclerotization, and this reaction is analogous to the mammalian complement cascade in terms of protein structure and function [54].
By 24 h post-co-infection, the abundance of cecropin A had increased 45-fold as compared to control larvae. Cecropin A is a 37 amino acid antimicrobial peptide first isolated from Hyalophora cecropia. It demonstrates antibacterial activity against multidrug-resistant bacteria (e.g. S. aureus ), induces C. albicans apoptosis and has immune-modulatory effects on macrophages [55–57]. Cecropin A had increased in C. albicans mono-infected larvae (4.5-fold) at 6 h post-infection, but increased 10.5-fold 24 h post- S. aureus infection [34, 38]. Moricin-like peptide increased 21- and 20.8-fold during C. albicans and S. aureus mono-infection, respectively, but was not detected in co-infected larvae. Moricins are secreted pro-peptides that are activated via proteolysis and increase the permeability of bacterial and fungal membranes. G. mellonella moricins are highly active against yeasts and S. aureus [58, 59]. Hdd11 had increased by 7.1- and 33.33-fold at 6 and 24 h post-co-infection. However, Hdd11, which shares homology with noduler from the tasar silkworm (Antheraea mylitta) [60, 61], had increased by 49-fold and 7.24-fold at 24 h post-C. albicans and S. aureus infection, respectively. Noduler shares a reeler domain with Hdd11 and binds insect haemocytes, bacterial LPS and fungal beta glucan, is enriched within nodules (G. mellonella nodules are highly enriched for Hdd11 protein; unpublished observation) and is important during the nodulation response [62]. Apolipophorin decreased 1.9-fold in abundance in C. albicans mono-infected larvae and was not detected during S. aureus infection, but was found to be significantly decreased (64.2-fold) during co-infection by S. aureus and C. albicans. Apolipophorin plays a role in lipid transport [63, 64] but has immune-modulatory activity and augments the activity of lysozyme [65], potentiates the activity of antimicrobial peptides [66], regulates phenoloxidase activity [67, 68] and is a pathogen recognition receptor and opsonin of lipopolysaccharide, lipoteichoic acids and fungal β-glucan [69]. Furthermore, lysozyme decreased (4.9-fold) during C. albicans mono-infection of larvae and increased (31.38-fold) during S. aureus mono-infection, but was not detected in co-infected larvae.
It is possible that co-infection with C. albicans and S. aureus activates a less robust immune response in terms of recognition and opsonin proteins (PGRPs), which are essential for early microbial recognition to prevent the spread of infection. Interestingly, the fungicidal AMP gloverin decreased in abundance during co-infection relative to mono-infection by both organisms, which may indicate a defective killing response of larvae to C. albicans, which, in turn, may allow increased proliferation of S. aureus due to increased survival of C. albicans.
In total, 23 C. albicans and 20 S . aureus proteins were found in haemolymph during co-infection of G. mellonella larvae. C. albicans heat shock protein SSA2 and peroxiredoxin TSA1-A and S. aureus staphopain B were found in haemolymph 24 h post-infection. SSA1 can induce host cell endocytosis of C. albicans, which leads to increased virulence [70] and plays a role in resistance to antimicrobial peptides and antifungal agents [71–73]. Peroxiredoxin TSA1-A was also detected in larvae mono-infected with C. albicans [39] and plays a role in cell protection against oxidative stress [74]. S. aureus staphopain B decreased the phagocytic activity of neutrophils and cleaved CD31, resulting in increased clearance by macrophages [75]. It is possible that these proteins display activity against insect haemocytes and antimicrobial peptides or play a role in detoxification of the microbial cell from the host immune response.
Co-infection with C. albicans and S. aureus produced elevated mortality relative to mono-infection and this is associated with increased S. aureus proliferation, alterations in circulating haemocyte densities and systemic dissemination of infection throughout larvae. The immune proteome of co-infected larvae displays an altered immune response relative to infection by C. albicans [38] or S. aureus [34] at a higher inoculum. This study demonstrates that the G. mellonella larvae may be a useful in vivo system to study co-infection by dual-species, inter-kingdom microbial pathogens. This model could be adapted for other common interactions (antagonistic or synergistic) that exist during human infection. This is particularly important as advances in healthcare that ultimately result in inhibition of the host immune response (e.g. SYK inhibitors) will facilitate the emergence of new pathogens and co-infection by diverse pathogens that are well adapted to thrive in the immunocompromised host.
Supplementary Data
Funding information
G. S. is the recipient of a Maynooth University doctoral studentship. The Q-exactive mass spectrometer was funded under the SFI Research Infrastructure Call 2012, grant number 12/RI/2346 (3). This publication emanated from research supported in part by a research grant from Science Foundation Ireland (SFI) and is co-funded under the European Regional Development Fund under grant number 12/RC/2275_P2.
Acknowledgements
The assistance of Dr Ilona Dix with Cryoviz imaging is acknowledged.
Author contributions
G. S. and K. K. designed the experiments. G. S. and L. S. performed the experiments and analysed the results. G. S. and K. K. wrote the manuscript. All authors have read and approved the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMP, antimicrobial peptide; DEP, differentially expressed protein; FDR, false discovery rate; GO, gene ontology; SSDA, statistically significant differentially abundant.
Two supplementary figures and four supplementary tables are available with the online version of this article.
Edited by: J. Cavet and K. Robinson
References
- 1.Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reece E, Segurado R, Jackson A, Mcclean S, Renwick J, et al. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients : an Irish registry analysis. BMC Pulm Med. 2017:1–8. doi: 10.1186/s12890-017-0416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn Microbiol Infect Dis. 2007;59:401–406. doi: 10.1016/j.diagmicrobio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Reno J, Doshi S, Tunali AK, Stein B, Farley MM, et al. Epidemiology of methicillin-resistant Staphylococcus aureus bloodstream coinfection among adults with candidemia in atlanta, GA, 2008–2012. Infect Control Hosp Epidemiol. 2015;36:1298–1304. doi: 10.1017/ice.2015.185. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell LE, Millhouse E, Sherry L, Kean R, Malcolm J, et al. Polymicrobial Candida biofilms: friends and foe in the oral cavity. FEMS Yeast Res. 2015;15:fov077. doi: 10.1093/femsyr/fov077. [DOI] [PubMed] [Google Scholar]
- 6.Bertolini M, Ranjan A, Thompson A, Diaz PI, Sobue T, et al. Candida albicans induces mucosal bacterial dysbiosis that promotes invasive infection. PLoS Pathog. 2019;15:e1007717. doi: 10.1371/journal.ppat.1007717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz PI, Strausbaugh LD, Dongari-Bagtzoglou A. Fungal-bacterial interactions and their relevance to oral health: linking the clinic and the bench. Front Cell Infect Microbiol. 2014;29:101. doi: 10.3389/fcimb.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, et al. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology. 2015;161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong EF, Tsui C, Kucharíková S, Van Dijck P, Jabra-Rizk MA. Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison DL, Scheres N, Willems HME, Bode CS, Krom BP, et al. The host immune system facilitates disseminated Staphylococcus aureus disease due to phagocytic attraction to Candida albicans during coinfection: a case of bait and switch. Infect Immun. 2019;87 doi: 10.1128/IAI.00137-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd OA, Peters BM. Candida albicans and Staphylococcus aureus pathogenicity and polymicrobial interactions: lessons beyond koch’s postulates. Journal of Fungi. 2019;5:81. doi: 10.3390/jof5030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, et al. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. MBio. 2016;7 doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, et al. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Carvalho Dias K, Barbugli PA, de Patto F, Lordello VB, de Aquino Penteado L, et al. Soluble factors from biofilm of Candida albicans and Staphylococcus aureus promote cell death and inflammatory response. BMC Microbiol. 2017;17 doi: 10.1186/s12866-017-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krüger W, Vielreicher S, Kapitan M, Jacobsen ID, Niemiec MJ. Fungal-Bacterial interactions in health and disease. Pathogens. 2019;8:70. doi: 10.3390/pathogens8020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd OA, Fidel PL, Harro JM, Hilliard JJ, Tkaczyk C, et al. Candida albicans augments Staphylococcus aureus virulence by engaging the Staphylococcal agr quorum sensing system. MBio. 2019;10 doi: 10.1128/mBio.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carolus H, Van Dyck K, Van Dijck P, albicans C. Candida albicans and Staphylococcus species: a threatening Twosome. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeh MAC, Fidel PL, Noverr MC. Identification of specific components of the eicosanoid biosynthetic and signaling pathway involved in pathological inflammation during intra-abdominal infection with Candida albicans and Staphylococcus aureus . Infect Immun. 2018;86 doi: 10.1128/IAI.00144-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, et al. Microbial interactions and differential protein expression in Staphylococcus aureus–Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol. 2010;59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasper L, König A, Koenig P-A, Gresnigt MS, Westman J, et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacey K, Geoghegan J, McLoughlin R. The role of Staphylococcus aureus virulence factors in skin infection and their potential as vaccine antigens. Pathogens. 2016;5:22. doi: 10.3390/pathogens5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinji H, Yosizawa Y, Tajima A, Iwase T, Sugimoto S, et al. Role of Fibronectin-Binding Proteins A and B in In Vitro Cellular Infections and In Vivo Septic Infections by Staphylococcus aureus . Infect Immun. 2011;79:2215–2223. doi: 10.1128/IAI.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhakdi S, Tranum-Jensen J. Alpha-Toxin of Staphylococcus aureus . Microbiol Rev. 1991;55:733–751. doi: 10.1128/MMBR.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burman JD, Leung E, Atkins KL, O’Seaghdha MN, Lango L, et al. Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus . J Biol Chem. 2008;283:17579–17593. doi: 10.1074/jbc.M800265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jongerius I, Köhl Jörg, Pandey MK, Ruyken M, van Kessel KPM, et al. Staphylococcal complement evasion by various convertase-blocking molecules. J Exp Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jardeleza C, Jones D, Baker L, Miljkovic D, Boase S, et al. Gene expression differences in nitric oxide and reactive oxygen species regulation point to an altered innate immune response in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2013;3:193–198. doi: 10.1002/alr.21114. [DOI] [PubMed] [Google Scholar]
- 29.Tsai CJ-Y, Loh JMS, Proft T, Jia C, -Yun T, Mei J, Loh S. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senior NJ, Bagnall MC, Champion OL, Reynolds SE, La Ragione RM, et al. Galleria mellonella as an infection model for Campylobacter jejuni virulence. J Med Microbiol. 2011;60:661–669. doi: 10.1099/jmm.0.026658-0. [DOI] [PubMed] [Google Scholar]
- 31.Borman AM, Szekely A, Johnson EM. Comparative Pathogenicity of United Kingdom Isolates of the Emerging Pathogen Candida auris and Other Key Pathogenic Candida Species. mSphere. 2016;1:e00189–16. doi: 10.1128/mSphere.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kavanagh K, Sheehan G. The use of Galleria mellonella larvae to identify novel antimicrobial agents against fungal species of medical interest. JoF. 2018;4:113. doi: 10.3390/jof4030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mylonakis E, Casadevall A, Ausubel FM. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007;3:e101. doi: 10.1371/journal.ppat.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheehan G, Dixon A, Kavanagh K. Utilization of Galleria mellonella larvae to characterize the development of Staphylococcus aureus infection. Microbiology. 2019;165:863–875. doi: 10.1099/mic.0.000813. [DOI] [PubMed] [Google Scholar]
- 35.Evans BA, Rozen DE. A Streptococcus pneumoniae infection model in larvae of the wax moth Galleria mellonella. Eur J Clin Microbiol Infect Dis. 2012;31:2653–2660. doi: 10.1007/s10096-012-1609-7. [DOI] [PubMed] [Google Scholar]
- 36.La Rosa SL, Casey PG, Hill C, Diep DB, Nes IF, et al. In Vivo assessment of growth and virulence gene expression during commensal and pathogenic lifestyles of luxABCDE -Tagged Enterococcus faecalis strains in murine gastrointestinal and intravenous infection models. Appl Environ Microbiol. 2013;79:3986–3997. doi: 10.1128/AEM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leanti La Rosa S, Diep DB, Nes IF, Brede DA. Construction and Application of a luxABCDE reporter system for real-time monitoring of enterococcus faecalis gene expression and growth. Appl Environ Microbiol. 2012;78:7003–7011. doi: 10.1128/AEM.02018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheehan G, Kavanagh K. Analysis of the early cellular and humoral responses of Galleria mellonella larvae to infection by Candida albicans . Virulence. 2018;9:163–172. doi: 10.1080/21505594.2017.1370174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheehan G, Kavanagh K. Proteomic analysis of the responses of Candida albicans during infection of Galleria mellonella larvae. JoF. 2019;5:7. doi: 10.3390/jof5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheehan G, Clarke G, Kavanagh K. Characterisation of the cellular and proteomic response of Galleria mellonella larvae to the development of invasive aspergillosis. BMC Microbiol. 2018;18 doi: 10.1186/s12866-018-1208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.London R, Orozco BS, Mylonakis E. The pursuit of cryptococcal pathogenesis: heterologous hosts and the study of cryptococcal host–pathogen interactions. FEMS Yeast Res. 2006;6:567–573. doi: 10.1111/j.1567-1364.2006.00056.x. [DOI] [PubMed] [Google Scholar]
- 42.Silva LN, Da Hora GCA, Soares TA, Bojer MS, Ingmer H, et al. Myricetin protects Galleria mellonella against Staphylococcus aureus infection and inhibits multiple virulence factors. Sci Rep. 2017;7 doi: 10.1038/s41598-017-02712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox Jürgen, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 44.Vogel H, Altincicek B, Glöckner G, Vilcinskas A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics. 2011;12:308. doi: 10.1186/1471-2164-12-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Côté RG, Griss J, Dianes JA, Wang R, Wright JC, et al. The proteomics identification (pride) converter 2 framework: an improved suite of tools to facilitate data submission to the pride database and the ProteomeXchange Consortium. Mol Cell Proteomics. 2012;11:1682–1689. doi: 10.1074/mcp.O112.021543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maguire R, Kunc M, Hyrsl P, Kavanagh K. Caffeine administration alters the behaviour and development of Galleria mellonella larvae. Neurotoxicol Teratol. 2017;64:37–44. doi: 10.1016/j.ntt.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Gu W, Yu Q, Yu C, Sun S. In vivo activity of fluconazole/tetracycline combinations in Galleria mellonella with resistant Candida albicans infection. J Glob Antimicrob Resist. 2018;13:74–80. doi: 10.1016/j.jgar.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim W, Melse Y, Konings M, Phat Duong H, Eadie K, et al. Addressing the most neglected diseases through an open research model: the discovery of fenarimols as novel drug candidates for eumycetoma. PLoS Negl Trop Dis. 2018;12:e0006437. doi: 10.1371/journal.pntd.0006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters BM, Noverr MC. Candida albicans-staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun. 2013;81:2178–2189. doi: 10.1128/IAI.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratcliffe NA. Invertebrate immunity — a primer for the non-specialist. Immunol Lett. 1985;10:253–270. doi: 10.1016/0165-2478(85)90100-2. [DOI] [PubMed] [Google Scholar]
- 52.Yi HY, Deng XJ, Yang WY, Zhou CZ, Cao Y, et al. Gloverins of the silkworm Bombyx mori: structural and binding properties and activities. Insect Biochem Mol Biol. 2013;43:612–625. doi: 10.1016/j.ibmb.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.XX X, Zhong X, HY Y, XQ Y. Manduca sexta gloverin binds microbial components and is active against bacteria and fungi. Dev Comp Immunol. 2012;38:275–284. doi: 10.1016/j.dci.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheehan G, Garvey A, Croke M, Kavanagh K. Innate humoral immune defences in mammals and insects: The same, with differences ? Virulence. 2018;9:1625–1639. doi: 10.1080/21505594.2018.1526531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yun J, Lee DG. Cecropin A-induced apoptosis is regulated by ion balance and glutathione antioxidant system in Candida albicans . IUBMB Life. 2016;68:652–662. doi: 10.1002/iub.1527. [DOI] [PubMed] [Google Scholar]
- 56.Lee E, Shin A, Kim Y. Anti-Inflammatory activities of cecropin A and its mechanism of action. Arch Insect Biochem Physiol. 2015;88:31–44. doi: 10.1002/arch.21193. [DOI] [PubMed] [Google Scholar]
- 57.Lee E, Jeong K-W, Lee J, Shin A, Kim J-K, et al. Structure-Activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep. 2013;46:282–287. doi: 10.5483/BMBRep.2013.46.5.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hara S, Moricin YM. A novel type of antibacterial peptide isolated from the silkworm, Bombyx mori. J Biol Chem. 1995;270:29923–29927. doi: 10.1074/jbc.270.50.29923. [DOI] [PubMed] [Google Scholar]
- 59.Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. The discovery and analysis of a diverged family of novel antifungal moricin-like peptides in the wax moth Galleria mellonella. Insect Biochem Mol Biol. 2008;38:201–212. doi: 10.1016/j.ibmb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 60.Shin SW, Park SS, Park DS, Kim MG, Kim SC, Woon Shin S, Park D-S, Gwang Kim M, et al. Isolation and characterization of immune-related genes from the fall webworm, Hyphantria cunea, using PCR-based differential display and subtractive cloning. Insect Biochem Mol Biol. 1998;28:827–837. doi: 10.1016/S0965-1748(98)00077-0. [DOI] [PubMed] [Google Scholar]
- 61.Sarauer BL, Gillott C, Hegedus D. Characterization of an intestinal mucin from the peritrophic matrix of the diamondback moth, Plutella xylostella. Insect Mol Biol. 2003;12:333–343. doi: 10.1046/j.1365-2583.2003.00420.x. [DOI] [PubMed] [Google Scholar]
- 62.Gandhe AS, John SH, Nagaraju J, Noduler NJ. Noduler, a novel immune up-regulated protein mediates nodulation response in insects. J Immunol. 2007;179:6943–6951. doi: 10.4049/jimmunol.179.10.6943. [DOI] [PubMed] [Google Scholar]
- 63.Niere M, Meißlitzer C, Dettloff M, Weise C, Ziegler M, et al. Insect immune activation by recombinant Galleria mellonella apolipophorin III. Biochim Biophys Acta. 1999;1433:16–26. doi: 10.1016/S0167-4838(99)00148-X. [DOI] [PubMed] [Google Scholar]
- 64.Niere M, Dettloff M, Maier T, Ziegler M, Wiesner A. Insect immune activation by apolipophorin III is correlated with the lipid-binding properties of this protein. Biochemistry. 2001;40:11502–11508. doi: 10.1021/bi010117f. [DOI] [PubMed] [Google Scholar]
- 65.Zdybicka-Barabas A, Cytryñska M. Apolipophorins and insects immune response. ISJ - Invertebr Surviv J. 2013;10:58–68. [Google Scholar]
- 66.Park SY, Kim CH, Jeong WH, Lee JH, Seo SJ, et al. Effects of two hemolymph proteins on humoral defense reactions in the wax moth, Galleria mellonella. Dev Comp Immunol. 2005 doi: 10.1016/j.dci.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Zdybicka-Barabas A, Cytryńska M. Involvement of apolipophorin III in antibacterial defense of Galleria mellonella larvae. Comp Biochem Physiol B Biochem Mol Biol. 2011;158:90–98. doi: 10.1016/j.cbpb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Zdybicka-Barabas A, Mak P, Jakubowicz T, Cytryńska M. Lysozyme and defense peptides as suppressors of phenoloxidase activity in Galleria mellonella . Arch Insect Biochem Physiol. 2014;87:1–12. doi: 10.1002/arch.21175. [DOI] [PubMed] [Google Scholar]
- 69.Wojda I. Immunity of the greater wax moth Galleria mellonella . Insect Sci. 2017;24:342–357. doi: 10.1111/1744-7917.12325. [DOI] [PubMed] [Google Scholar]
- 70.Sun JN, Solis NV, Phan QT, Bajwa JS, Kashleva H, et al. Host cell invasion and virulence mediated by Candida albicans Ssa1. PLoS Pathog. 2010;6:e1001181. doi: 10.1371/journal.ppat.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho T, Toyoda M, Sudoh M, Nakashima Y, Calderone RA, et al. Isolation and sequencing of the Candida albicans MSI3, a putative novel member of the hsp70 family. Yeast. 2003;20:149–156. doi: 10.1002/yea.952. [DOI] [PubMed] [Google Scholar]
- 72.Li XS, Reddy MS, Baev D, Edgerton M. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J Biol Chem. 2003;278:28553–28561. doi: 10.1074/jbc.M300680200. [DOI] [PubMed] [Google Scholar]
- 73.Nagao J-ichi, Cho T, Uno J, Ueno K, Imayoshi R, et al. Candida albicans Msi3p, a homolog of the Saccharomyces cerevisiae Sse1p of the hsp70 family, is involved in cell growth and fluconazole tolerance. FEMS Yeast Res. 2012;12:728–737. doi: 10.1111/j.1567-1364.2012.00822.x. [DOI] [PubMed] [Google Scholar]
- 74.Urban C, Xiong X, Sohn K, Schröppel K, Brunner H, et al. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans . Mol Microbiol. 2005;57:1318–1341. doi: 10.1111/j.1365-2958.2005.04771.x. [DOI] [PubMed] [Google Scholar]
- 75.Smagur J, Guzik K, Bzowska M, Kuzak M, Zarebski M, et al. Staphylococcal cysteine protease staphopain B (SspB) induces rapid engulfment of human neutrophils and monocytes by macrophages. Biol Chem. 2009;390:361–371. doi: 10.1515/BC.2009.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry (MS) proteomics data and MaxQuant search output files have been deposited to the ProteomeXchange Consortium [45] via the PRIDE partner repository with the dataset identifier PXD014273.