Abstract
RNA granule formation, which can be regulated by RNA-binding proteins (RBPs) such as fragile X mental retardation protein (FMRP), acts as a mechanism to control both the repression and subcellular localization of translation. Dysregulated assembly of RNA granules has been implicated in multiple neurological disorders, such as amyotrophic lateral sclerosis. Thus, it is crucial to understand the cellular pathways impinging upon granule assembly or disassembly. The goal of this review is to summarize recent advances in our understanding of the role of the RBP, FMRP, in translational repression underlying RNA granule dynamics, mRNA transport and localized. We summarize the known mechanisms of translational regulation by FMRP, the role of FMRP in RNA transport granules, fragile X granules and stress granules. Focusing on the emerging link between FMRP and stress granules, we propose a model for how hyperassembly and hypoassembly of RNA granules may contribute to neurological diseases.
Keywords: FMRP, granules, mRNP granules, protein synthesis, RNA, stress granules
1 ∣. INTRODUCTION
The physiologic functioning of neurons relies on the regulation of translation, which underlies critical functions such as the maintenance of neuronal health and synaptic plasticity.1-4 Dysregulation of protein synthesis in neurons therefore leads to severe disruptions in neuronal functioning and can result in impaired cognition. This is exemplified by the hallmark pathology of disrupted protein synthesis in neurodevelopmental disorders such as Angelman syndrome,5-7 autism spectrum disorder (ASD),8-10 and fragile X syndrome (FXS).11-13 FXS is the leading inherited form of intellectual disability and is caused by the loss of fragile X mental retardation protein (FMRP), a translational repressor.14 Studies throughout the past two decades on FMRP function have uncovered several mechanisms by which FMRP represses translation, including stalling polyribosomes and associating with miRNA and related mRNA ribonucleoprotein (mRNP) complexes. These mechanisms are discussed in depth in other reviews.14-17 Additionally, FMRP has been known to regulate the formation of the RNA granules, including mRNP transport granules, fragile X granules (FXGs), P-bodies and stress granules. In addition to regulating translation by multiple mechanisms, FMRP has been well studied to regulate the transport of mRNP granules.16 The regulation of mRNP granule formation is necessary for the transport and localization of specific mRNA species to synaptic sites.18,19 FMRP has also been identified to promote the formation of stress granules20—a membrane-less assembly of RNA-binding proteins (RBPs) and translationally repressed mRNAs formed in response to cellular stress.21 Progress made in studying the regulation of stress granule formation has highlighted their relevance to neurological disorders. Aberrant formation of stress granules, for instance, has been implicated in neurodegenerative22 and neurodevelopmental disorders.23 Here, we review our current understanding of FMRP function in regulating translation and mRNP granule transport, including its role in “FXGs.” Next, we cover the role of FMRP in stress granule dynamics and how posttranslational modifications (PTMs) of FMRP contribute to the regulation of translation and granule dynamics (Figure 1). Further insight into the basic biology of FMRP's role in mRNP and stress granules may help uncover novel points of therapeutic options for neurodevelopmental disorders and perhaps other related neurological disorders.
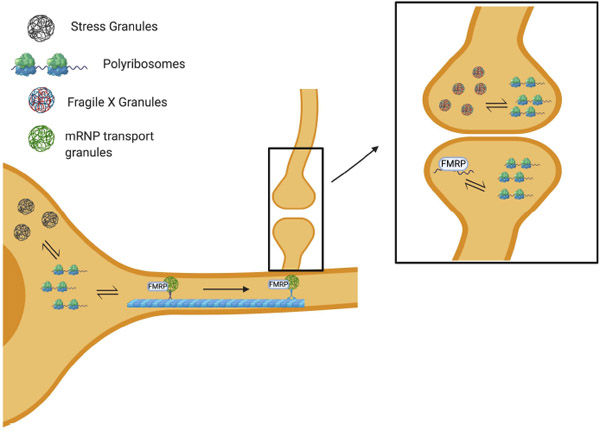
FIGURE 1.
Overview of FMRP function in stress granules, mRNP, local translation at the synapse and FXGs. Stress granules, membrane-less organelles with translationally repressed mRNAs, are in dynamic equilibrium with mRNAs being actively translated. mRNAs can also be sequestered into the mRNP transport granules, through which transcribes can be spatially regulated. For instance, transcripts can be transported to the synapse for localized translation. The formation of FXGs can regulate protein synthesis in the presynaptic terminal. FMRP has been shown to play a role in stress granule dynamics and mRNP granules. FMRP has also been well-studied function as a translational repressor at the synapse, regulating local protein synthesis and synaptic function. Additionally, FMRP homologues FXR1 and FXR2 have been shown to be present in FXGs
2 ∣. FMRP IS A TRANSLATIONAL REPRESSOR
FMRP is an RBP encoded by the FMR1 gene that regulates mRNA translation, localization and stability. FMRP consists of three major RNA-binding domains, including two hnRNP K-homology (KH) domains and an RGG box domain.24 The two KH domains recognize kissing-complex tertiary motifs in RNA,25 whereas the RGG box recognizes G-quadruplex structures.26 The loss of FMRP as a result of a trinucleotide repeat expansion in the 5′ UTR of the FMR1 gene is a hallmark of the monogenic neurodevelopmental disorder FXS.
Following earlier studies on FMRP localization in RNA granules within dendrites and spine synapses,27 subsequent studies provided mechanistic insight into the role of FMRP as a translational repressor at synapses.16 In the absence of FMRP, there is excess basal translation and loss of stimulus-induced translation in dendrites and spines.28 Multiple modes of translational repression have been attributed to FMRP. While several of these mechanisms of translational regulation by FMRP have been reviewed in detail elsewhere,14,15 here we provide a preliminary overview for these major roles of FMRP.
Translation can be evaluated in terms of three steps: initiation, elongation and termination. FMRP regulates both initiation and elongation stages of translation by acting alone29 or through association with factors such as miRNAs and the RNA-induced silencing complex (RISC).30,31 Additionally, FMRP can inhibit cap-dependent translation of mRNAs by forming a repressive mRNP complex with cytoplasmic FMR1-interacting protein 1 (CYFIP1), an eIF4E-binding protein (4EBP).32 Together, the FMRP-CYFIP complex binds eIF4E and inhibits initiation.32 Previous studies have shown that genetic reduction of CYFIP1 levels in neurons in mice increased the level of proteins encoded by FMRP-targeted mRNAs, including Maplb, aCamk2 and App.32 The binding of FMRP to CYFIP1 inhibits interactions of eIF4E with eIF4G. As a result, the formation of FMRP-CYFIP1 complex inhibits cap-dependent translation. This regulatory mechanism is linked to extracellular signaling, as brain-derived neurotrophic factor (BDNF) or glutamate signaling can release CYFIP1 and FMRP from eIF4E to promote translation. In Fmr1-KO mice, both eIF4E-eIF4G binding33 and eIF4E phosphorylation34 are increased. Taken together, these observations suggest that FMRP regulates initiation through both direct and indirect mechanisms.
Previous work has found that FMRP associates directly or indirectly with miRNAs, Argonaut of RISC, Dicer and miRNA precursors. In a Drosophila model, dFMRP modulates miRNA expression to control neuronal development.30,35 In mice, FMRP associates with RISC and/or miRNAs that cooperate to regulate mRNA translation and dendritic spine morphology.31 The stimulation of group 1 metabotropic glutamate receptors (mGluRs) results in the PP2A-mediated dephosphorylation of FMRP, which subsequently promotes the release of RISC from target mRNAs.31 Additionally, this dephosphorylation events also enhance the ubiquitination of FMRP and its degradation by the proteasome.36 Together this pathway serves as a switch for regulating translational activity in response to environmental cues. Through the well-established mechanisms outlined above, FMRP maintains the proper physiologic levels of translation for its target mRNAs.
The majority of FMRP associates with polyribosomes and has been shown to stall polyribosomes at the elongation phase.29 in vivo high-throughput sequencing of RNAs isolated by CLIP showed that FMRP binds most frequently to coding regions of mRNAs, with fewer binding sites within 5′ and 3′ UTRs.24,29 High frequency of binding to coding regions suggests that FMRP acts by physically blocking ribosomes transit. Experiments specifically measuring ribosome transit with and without FMRP showed that polypeptide elongation was 40% to 50% faster in Fmr1-KO than wild-type (WT) brain lysates,29 which further implies a role for FMRP in regulation of ribosomal transit. It remains unclear how FMRP interactions with coding sequences allow for selectivity in FMRP binding. Several studies have worked to identify common sequences on mRNAs to which FMRP binds.24,29,37 While these findings have been reviewed in depth elsewhere,17 it is of note that there seem to be multiple motifs that FMRP binds, including G-quadruplexes often within untranslated regions.29 Bioinformatic overlap of FMRP consensus binding sequences (FCBS) from two published FMRP CLIP sequencing datasets identified the short sequence motifs TGGA and GAC in both coding and untranslated regions.38 Further work is needed to better understand the role of FMRP-binding domains and the motifs recognized on the model of regulation by FMRP. Related to this question, recent work has identified the role of RGG box interactions with Q-quadruplexes in mRNA localization, whereas KH domain interactions regulate translation on distinct FMRP target mRNAs.39
3 ∣. FMRP IN RNA TRANSPORT GRANULES
FMRP is associated with RNA transport granules in neuronal processes, both dendrites27,40 and axons40,41 of cultured neurons. Live cell imaging of FMRP-GFP fusion proteins has revealed dynamic and activity-regulated transport of FMRP.40,42 Several groups have uncovered association of FMRP with molecular motors.42-45 Using the MS2 tagging method for live cell imaging of FMRP target mRNA, several groups have shown that FMRP regulates mRNA transport.42,46,47 One model has emerged that suggests a role for FMRP as an adapter for motors that transport RNA; however, it remains unclear whether FMRP directly binds to motor or adapter proteins. A limitation in the field has been lack of a unifying mechanism for how FMRP may play a role in localization of its target mRNAs. Recent exciting work has uncovered a role for FMRP RGG box interactions with G-quadruplex RNA sequences to localize mRNAs in neurons.39 This study further uncouples the functions of FMRP target mRNA interactions in localization vs translation.
4 ∣. FMRP RECRUITMENT TO FRAGILE X GRANULES
Mammalian cells express two autosomal homologs of FMRP known as FXR1P and FXR2.48,49 Similar to FMRP, FXR1P and FXR2P contain two KH domains and have 86% and 60% amino acid sequence overlap with FMRP, respectively.48,49 Observation of “FXGs” containing FMRP, FXR1P and FXR2P in the presynaptic compartment suggests a physiologic role for the fragile X proteins in regulating the axonal proteome.50,51 While many of the translationally repressive functions of FMRP occur postsynaptically (as discussed above), previous observations of reduced axonal growth cone motility in Fmr1 knockout neurons suggested a potential presynaptic role of FMRP and perhaps its homologs.40
Initial studies on FXG focused on characterizing the protein composition of these granules as well as where in the brain they were localized.51,52 While FXR2P is expressed in essentially every FXG, FXGs are also composed of FMRP and/or FXR1P dependent on the brain region.51,53 For example, the majority of hippocampal FXGs are composed of FXR2P and FMRP, whereas frontal cortex FXGs contain all three fragile X proteins.51 Initially, it was observed in mice that FXGs are present only during developmental periods requiring robust synaptic plasticity.51 A more recent study has identified the persistent expression of FXG in the hippocampus of adult mice and adult humans,50 which has implications for the role of FMRP, FXR1P and FXR2P across the human lifespan.
With the well-described role of FMRP in the regulation of translation, it was hypothesized that FXG may also play a role in the spatiotemporal control of protein synthesis in the presynaptic compartment. Ribosomal subunits were identified to associate with FXG in multiple brain regions including the hippocampus, motor cortex and the olfactory bulb.50 Additionally, several previously identified FMRP mRNA targets including Omp, Map1b and B-catenin29 were observed to colocalize to FXG.50 The association of ribosomal machinery and mRNA with the FXGs is reminiscent of stress granules, another mRNP granule class that represses the translation of associated mRNAs (to be discussed further in depth below).21 Thus, the interaction of ribosomes and mRNAs suggested that FXGs have the capacity to regulate local translation of the associated mRNA. Indeed, OMP expression within the axons of Fmr1 knockout mice was significantly higher compared to expression within the axons of WT mice.50 Similar to how the protein composition of FXG is dependent upon brain region, the specific mRNA transcripts associated with the FXG are differential based on brain region.53 The brain region specificity of FXG protein and mRNA composition support a mechanism by which FXG can tightly regulate the spatiotemporal translation of FMRP targets. Taken together, the studies on FXGs reveal a distinct role for FMRP in the presynaptic compartment in regulating the translation of mRNAs that have an impact on synaptic strength and connectivity. However, it is unclear if other proteins that are involved in other mRNP granules are also involved in the formation of these FXGs. For example, Caprin1 is involved in the formation of both RNA transport granules and stress granules54-57 and interacts with FMRP58; the interaction between Caprin1 and FMRP promotes phase separation, which is an important feature of mRNP granules (discussed further in depth below).59 With its roles in other mRNP granules and its association with FMRP, it is possible that Caprin1 may also be involved in the formation FXGs. To better understand the physiology of FXG, it is necessary to elucidate what other RBPs are involved in the formation and function of FXG.
FMRP is also observed to colocalize to RNA processing bodies (P-bodies),60,61 which are RNA granules that prevent the translation of mRNA and promote the degradation of mRNA via miRNA-guided translational silencing and siRNA-guided RNA degradation.18 The association of FMRP to the RISC,30,31 an integral component of miRNA-guided translational silencing, further supports observations of FMRP localization in P-bodies. The recruitment of FMRP to P-bodies further implies a role of FMRP in the formation of translationally repressive mRNP granules via its interaction with other RBPs.
5 ∣. FMRP REGULATES STRESS GRANULE DYNAMICS
Stress granules are membrane-less ribonucleoprotein assemblies that form during cellular stress.21 mRNAs sequestered into stress granules are translationally repressed at the initiation phase; the precise regulation of stress granules assembly and disassembly is therefore a mechanism for fine-tuning of protein synthesis.
Approximately 50% of the proteome of the “core” of stress granules—the more stable substructure that can be purified—are RBPs.56 Interestingly, the composition of stress granules varies depending on the type of cellular stress used to induce stress granule formation, cell type and disease contexts.57 In the study by Markmiller et al, nearly 25% of the RBPs under study exhibited stress-type-specific stress granule targeting. Just under half of the RBPs tested were found to localize to stress granules in a cell-type-dependent manner. Neuronal stress granules are particularly diverse in their composition, which varies depending on whether they are localized in the dendrites, soma or axon.
Much work has been done on elucidating how cellular processes influence stress granule dynamics.62 Molecular chaperones play a significant role in the dissolution of stress granules during stress recov-ery63-65 and can inhibit the aberrant formation of stress granules in amyotrophic lateral sclerosis (ALS).66 RNA helicases, which regulate many aspects of RNA metabolism, have also been shown to mediate stress granule dissolution. Proteomic analyses of stress granule cores reveal RNA helicases as a conserved component, and abolishing the activities of such RNA helicases reduces stress granule formation.56
Additionally, several PTMs have been found to regulate stress granule dynamics. PTMs can rapidly alter the interactome of target substrates by inducing conformational changes or alteration in charge of the target substrate. Three classes of PTMs have been found to oppose stress granule formation. Glycosylation of α-synuclein and tau have been shown to inhibit their pathologic aggregation into stress granules and other mRNP structures.67,68 Methylation of RBPs, such as the ALS-associated FUS, has been found to inhibit the cation-π interactions–in which a π-bond system stabilizes cationic group–and stress granule assembly.69,70 Similarly, lysine acetylation can disrupt cation-π interactions and is protective against pathologic tau aggregation characteristic of multiple neurodegenerative disorders.71
By far the most well-characterized PTM in regulating stress granule dynamics is phosphorylation. The rapid and reversible conjugation of a charged phosphate moiety serves as an ideal pathway for rapidly altering the RBP interactome in response to environmental cues. Interestingly, phosphorylation can promote or oppose stress granule formation, depending on the kinase involved. Several cases of phosphorylation-mediated inhibition of phase separation have been reported.72(p4),73 Liquid-liquid phase separation (LLPS) occurs when a set of molecules form a dense network of weak interactions, which concentrate these molecules into a separate phase. By altering the conformation and charge of a biomolecule, phosphorylation can switch the ability of a molecule to form the necessary interactions that drive LLPS. For instance, the hyperphosphorylation of tau drastically alters the electrostatic characteristic of the molecule in a way that favors phase separation.74 More recently, phosphorylation of FMRP was observed to result in its phase separation with CAPRIN1,59 a protein that positively regulates and localizes to stress granule assembly. This suggests a mechanism by which FMRP localizes to and promotes the formation of stress granules upon PTM. Other studies have supported this hypothesis. For instance, FMRP and its autosomal homologs are well described to be present in stress granules.23,75 Furthermore, the loss of FMRP results in reduced levels of stress granule formation.20 As described above, it is well established that dephosphorylation of FMRP disinhibits translational repression. This is a critical node downstream of the stimulation of mGluRs, which are critical for protein synthesis-dependent long-term depression (LTD), a form of learning and memory.36 Therefore, the phase separation of FMRP as a result of phosphorylation points to a potential link between the activation of group 1 mGluRs and stress granule dynamics. A better understanding of the mechanistic relationship between extracellular signaling and the role of FMRP in stress granules may uncover novel therapeutic strategies that target stress granule dynamics through receptors such as mGluRs.
6 ∣. FMRP MODIFICATION AS A MOLECULAR SWITCH
As this review has highlighted, FMRP can regulate the translation of mRNA through multiple mechanisms. Interestingly, PTM of FMRP can alter its ability to regulate translation across several of these mechanisms. FMRP has been shown to be phosphorylated on the highly conserved serine 499, which may then promote phosphorylation of nearby serines.76 It was initially concluded that ribosomal protein S6 kinase (S6K1) was primarily responsible for the phosphorylation of FMRP downstream of mGluR signaling.77 However, later work has demonstrated that FMRP is first phosphorylated on serine 499 by casein kinase II (CK2), and then secondary phosphorylation on nearby residues can occur via mGluR-dependent mechanisms.78 FMRP phosphorylated at serine 499 is associated with polyribosomes that are stalled and inhibit translation,76 whereas unphosphorylated FMRP associates with actively translating polyribosomes.76 These observed changes in ribosomal stalling based on FMRP phosphorylation status suggested that the PTM of FMRP serves as a level of regulation on the ability of FMRP to repress translation. Additionally, the phosphorylated form of FMRP acts as a translational repressor through other mechanisms. It has been observed that phosphorylated FMRP forms an inhibitory complex with RISC and miRNAs to repress the translation of FMRP-target mRNAs.31,79 Phosphorylated FMRP represses the translation of HoxB8 mRNA via miR196a-mediated translational repression79 as well as PSD-95 mRNA via miR125a-mediated translational repression.31 Dephosphorylation of FMRP by PP2A downstream of mGluR signaling80 allows for the release of the miRNA-RISC complex from PSD-95 mRNA, thus allowing for its translation.36
While it is evident that the phosphorylation status of FMRP is able to alter its ability to regulate translation, the phosphorylation state of FMRP also has downstream consequence on other PTMs of FMRP. Dephosphorylation of FMRP by PP2A following mGluR signaling triggers the ubiquitination of FMRP.36,81 This ubiquitination event precedes the translation of PSD-95,36 which suggests that ubiquitination of FMRP is a critical step to reverse the repression of translation. Currently, there is an interest to better understand what E3 ubiquitin ligases may be responsible for the ubiquitination of FMRP. Cdh1-APC, an E3 ubiquitin ligase implicated in cell cycle exit, has been demonstrated to ubiquitinate FMRP and trigger its degradation following mGluR stimulation.82 It is currently unclear if ubiquitination of FMRP by Cdh1-APC affects the downstream translation of FMRP-targeted mRNAs. The observations that dephosphorylation and ubiquitination affect the ability of FMRP to stall polyribosomes and cooperate with RISC begs the question if these PTMs are also critical for other functions of FMRP, such as its role in mRNP granules.
As discussed above, a critical step in the formation of mRNP granules is LLPS. Phosphorylated FMRP has the ability to phase separate with RNA into liquid droplets,83 which implies that phosphorylated FMRP is more readily able to form translationally repressive granules, such as FXG or stress granules. Furthermore, the phosphorylation of FMRP promotes its ability to bind to Caprin1,59 another RBP known for its ability to regulate the translation of mRNA via mRNP granules. Given how FXGs, P-bodies and stress granules are able to regulate protein synthesis, it can be concluded that the effects of FMRP phosphorylation on LLPS are another mechanism for regulating protein synthesis. It has yet to be determined whether the dephosphorylation of FMRP by PP2A is able to reverse the ability of FMRP to bind to Caprin1 and phase separate. With the effects that ubiquitination has on RISC-mediated translational repression, it is also of interest to determine whether or not ubiquitination of FMRP also affects LLPS. The ubiquitination proteasome system (UPS) has been previously implicated in stress granule formation,84,85 so the ubiquitination of FMRP may be a step in the regulation of granule formation. A future focus for research may be in the investigation of how dephosphorylation and ubiquitination may regulate the ability of FMRP to phase separate and form mRNP granules that play key physiologic roles in the spatiotemporal regulation of protein synthesis.
A recent study demonstrated that Cdh1-APC, an E3 ubiquitin ligase that ubiquitinates FMRP (as mentioned above), regulates the formation of stress granules in postmitotic neurons.86 In WT cortical neurons, Cdh1-APC activity antagonizes the formation of stress granules, which acts as a mechanism to regulate protein synthesis. In Fmr1 knockout neurons, modulation of Cdh1-APC activity had no effect on the formation of stress granules. Taken together, results from this most recent study suggest that ubiquitination of FMRP by Cdh1-APC antagonizes the formation of stress granules. Elucidation of how ubiquitination of FMRP by Cdh1-APC regulates stress granule formation and downstream protein synthesis brings the field a step closer to better understanding the mechanism by which FMRP itself is regulated and the downstream processes that are affected (Figure 2).
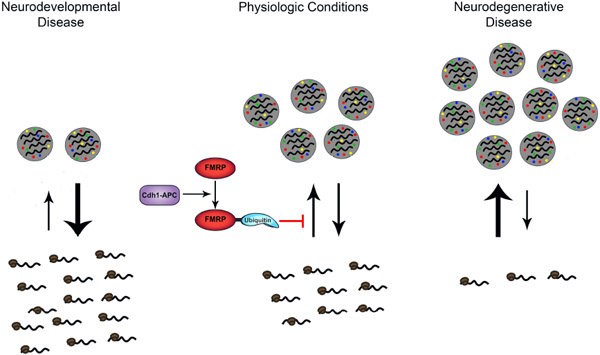
FIGURE 2.
Dynamic equilibrium between stress granules and protein synthesis is regulated by PTMs of RBPs. In physiologic conditions (middle), stress granule assembly and disassembly act as a mechanism to regulate protein synthesis. It has been shown in neurodegenerative diseases (right), such as ALS, that stress granules hyperassemble and lead to reduced protein synthesis.22 Neurodevelopmental disorders (left), such as FXS, are characterized by increased levels of protein synthesis17 and proposed hypoassembly of stress granules.20 Additionally, a recent study86 demonstrates that Cdh1-APC is a complex that regulates the assembly of stress granules through PTM of FMRP, and therefore it may be of interest to investigate manipulation of Cdh1-APC activity as a potential therapeutic opportunity for neurodevelopmental and neurodegenerative diseases
7 ∣. CONCLUSION
The diversity of RNA granules ensures dynamic and spatiotemporal control of RNA localization and translation. Here, we have highlighted these processes through the lens of FMRP, an mRNA-binding protein linked to diverse types of RNA granules including RNA transport granules, stress granules, P-bodies and FXGs. Emerging mechanisms of FMRP-mediated control of RNA localization and translation via RNA granules have provided new insight into the biology of RNA granules in health and disease. FMRP is also a regulator of the dynamics of mRNA fate between RNA granules and polyribosomes. Studies have begun to elucidate the role of PTMs, including phosphorylation and ubiquitination. More recently, the role of phase separation to form biomolecular condensates has emerged for FMRP containing RNA granules, as has been described for diverse types of RNA granules. Here, we have presented a model for how hyperassembly and hypoassembly of RNA granules may contribute to neurological diseases. Further knowledge of the basic regulatory mechanisms of RNA granule dynamics should provide insight into potential therapeutic targets that may correct defects in RNA granule dynamics affecting translational control in neurological disease.
ACKNOWLEDGMENTS
The authors thank members of the Bassell lab for helpful discussions and feedback. They acknowledge support from NIH 1R01MH109026 and F31NS101932 (ANV) (G. J. B.).
Funding information
National Institute of Health, Grant/Award Numbers: 1R01MH109026, NIH F31NS101932 (ANV)
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10(5):587–592. 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 2.Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40(2):347–359. 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- 3.Pfeiffer BE, Huber KM. Current advances in local protein synthesis and synaptic plasticity. J Neurosci. 2006;26(27):7147–7150. 10.1523/JNEUROSCI.1797-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18(1):60–68. 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greer PL, Hanayama R, Bloodgood BL, et al. The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–716. 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15(1):70–73. 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 7.Lee SY, Ramirez J, Franco M, et al. Ube3a, the E3 ubiquitin ligase causing Angelman syndrome and linked to autism, regulates protein homeostasis through the proteasomal shuttle Rpn10. Cell Mol Life Sci. 2014;71(14):2747–2758. 10.1007/s00018-013-1526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tastet J, Decalonne L, Marouillat S, et al. Mutation screening of the ubiquitin ligase gene RNF135 in French patients with autism. Psychiatr Genet. 2015;25(6):263–267. 10.1097/YPG.0000000000000100. [DOI] [PubMed] [Google Scholar]
- 9.Tsai N-P, Wilkerson JR, Guo W, et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151(7):1581–1594. 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi JJ, Berrios J, Newbern JM, et al. An autism-linked mutation disables phosphorylation control of UBE3A. Cell. 2015;162(4):795–807. 10.1016/j.cell.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross C, Bassell GJ. Excess protein synthesis in FXS patient lymphoblastoid cells can be rescued with a p110β-selective inhibitor. Mol Med. 2012;18:336–345. 10.2119/molmed.2011.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross C, Nakamoto M, Yao X, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30(32):10624–10638. 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A, Hoeffer CA, Takayasu Y, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30(2):694–702. 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci. 2012;35:417–443. 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16(10):595–605. 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2): 201–214. 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee A, Ifrim MF, Valdez AN, Raj N, Bassell GJ. Aberrant RNA translation in fragile X syndrome: from FMRP mechanisms to emerging therapeutic strategies. Brain Res. 2018;1693(pt A):24–36. 10.1016/j.brainres.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang HL, Eom T, Oleynikov Y, et al. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31(2):261–275. 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 19.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51(6):685–690. 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Didiot M-C, Subramanian M, Flatter E, Mandel J-L, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell. 2009;20(1):428–437. 10.1091/mbc.e08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668–679. https://doi.org/10.10167j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci. 2019;20(11):649–666. 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gareau C, Houssin E, Martel D, et al. Characterization of fragile X mental retardation protein recruitment and dynamics in Drosophila stress granules. PLoS One. 2013;8(2):e55342 10.1371/journal.pone.0055342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ascano M, Mukherjee N, Bandaru P, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492(7429):382–386. 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell JC, Fraser CE, Mostovetsky O, et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19(8):903–918. 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9(10):1198–1207. 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24(11):2648–2655. 10.1523/JNEUR0SCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ifrim MF, Williams KR, Bassell GJ. Single-molecule imaging of PSD-95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J Neurosci. 2015;35(18):7116–7130. 10.1523/JNEUROSCI.2802-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin P, Zarnescu DC, Ceman S, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the micro-RNA pathway. Nat Neurosci. 2004;7(2):113–117. 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 31.Muddashetty RS, Nalavadi VC, Gross C, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42(5):673–688. 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Napoli I, Mercaldo V, Boyl PP, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134(6):1042–1054. 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Ronesi JA, Collins KA, Hays SA, et al. Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat Neurosci. 2012;15(3):431–440, S1. 10.1038/nn.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gkogkas CG, Khoutorsky A, Cao R, et al. Pharmacogenetic inhibition of eIF4E-dependent Mmp9 mRNA translation reverses fragile X syndrome-like phenotypes. Cell Rep. 2014;9(5):1742–1755. 10.1016/j.celrep.2014.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X-L, Li Y, Wang F, Gao F-B. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci. 2008;28(46):11883–11889. 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012;32(8):2582–2587. 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107(4):489–499. 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 38.Anderson BR, Chopra P, Suhl JA, Warren ST, Bassell GJ. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016;44(14):6649–6659. 10.1093/nar/gkw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goering R, Hudish LI, Guzman BB, et al. FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. bioRxiv. 2019;784728 10.1101/784728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32(1–2): 37–48. 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Bassell GJ, Sasaki Y. Fragile X mental retardation protein is involved in protein synthesis-dependent collapse of growth cones induced by semaphorin-3A. Front Neural Circuits. 2009;3:11 10.3389/neuro.04.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008; 14(6):926–939. 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling S-C, Fahrner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci USA. 2004;101(50):17428–17433. 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bianco A, Dienstbier M, Salter HK, Gatto G, Bullock SL. Bicaudal-D regulates fragile X mental retardation protein levels, motility, and function during neuronal morphogenesis. Curr Biol. 2010;20(16): 1487–1492. 10.1016/j.cub.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindsay AJ, McCaffrey MW. Myosin Va is required for the transport of fragile X mental retardation protein (FMRP) granules. Biol Cell. 2014;106(2):57–71. 10.1111/boc.201200076. [DOI] [PubMed] [Google Scholar]
- 46.Estes PS, O'Shea M, Clasen S, Zarnescu DC. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol Cell Neurosci. 2008;39(2):170–179. 10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Kao D-I, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci U S A. 2010;107(35): 15601–15606. 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, O'Connor JP, Siomi MC, et al. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14(21):5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siomi MC, Siomi H, Sauer WH, Srinivasan S, Nussbaum RL, Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14(11):2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akins MR, Berk-Rauch HE, Kwan KY, et al. Axonal ribosomes and mRNAs associate with fragile X granules in adult rodent and human brains. Hum Mol Genet. 2017;26(1):192–209. 10.1093/hmg/ddw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29(5):1514–1524. 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akins MR, Leblanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. J Comp Neurol. 2012;520(16):3687–3706. 10.1002/cne.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chyung E, LeBlanc HF, Fallon JR, Akins MR. Fragile X granules are a family of axonal ribonucleoprotein particles with circuit-dependent protein composition and mRNA cargos. J Comp Neurol. 2018;526(1): 96–108. 10.1002/cne.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakayama K, Ohashi R, Shinoda Y, et al. RNG105/caprin1, an RNA granule protein for dendritic mRNA localization, is essential for long-term memory formation. eLife. 2017;6:e29677 10.7554/eLife.29677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiina N, Yamaguchi K, Tokunaga M. RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. J Neurosci. 2010;30(38):12816–12830. 10.1523/JNEUROSCI.6386-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164(3):487–498. 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markmiller S, Soltanieh S, Server KL, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172(3):590–604.e13. 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El Fatimy R, Tremblay S, Dury AY, et al. Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes [corrected]. PLoS One. 2012;7(6): e39338 10.1371/journal.pone.0039338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019; 365(6455):825–829. 10.1126/science.aax4240. [DOI] [PubMed] [Google Scholar]
- 60.Barbee SA, Estes PS, Cziko A-M, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52(6):997–1009. 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee EK, Kim HH, Kuwano Y, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010;17(6):732–739. 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snead WT, Gladfelter AS. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol Cell. 2019;76(2):295–305. 10.1016/j.molcel.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cherkasov V, Hofmann S, Druffel-Augustin S, et al. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23(24):2452–2462. 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- 64.Walters RW, Muhlrad D, Garcia J, Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015;21(9):1660–1671. 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroschwald S, Maharana S, Mateju D, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015;4:e06807 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mateju D, Franzmann TM, Patel A, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36(12):1669–1687. 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marotta NP, Lin YH, Lewis YE, et al. O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson's disease. Nat Chem. 2015;7(11):913–920. 10.1038/nchem.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuzwa SA, Shan X, Macauley MS, et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8(4):393–399. 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 69.Qamar S, Wang G, Randle SJ, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173(3):720–734.e15. 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorton BM, Shechter D. Cellular consequences of arginine methylation. Cell Mol Life Sci. 2019;76(15):2933–2956. 10.1007/s00018-019-03140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlomagno Y, Chung DC, Yue M, et al. An acetylation-phosphorylation switch that regulates tau aggregation propensity and function. J Biol Chem. 2017;292(37):15277–15286. 10.1074/jbc.M117.794602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang A, Conicella AE, Schmidt HB, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018;37(5):e97452 10.15252/embj.201797452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monahan Z, Ryan VH, Janke AM, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36(20):2951, 10.15252/embj.201696394-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8:275 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herman AB, Silva Afonso M, Kelemen SE, et al. Regulation of stress granule formation by inflammation, vascular injury, and atherosclerosis. Arterioscler Thromb Vasc Biol. 2019;39(10):2014–2027. 10.1161/ATVBAHA.119.313034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12(24):3295–3305. 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 77.Narayanan U, Nalavadi V, Nakamoto M, et al. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283(27): 18478–18482. 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bartley CM, O'Keefe RA, Blice-Baum A, et al. Mammalian FMRP S499 is phosphorylated by CK2 and promotes secondary phosphorylation of FMRP. eNeuro. 2016;3(6):ENEURO.0092–ENEU16.2016. 10.1523/ENEURO.0092-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Tang W, Zhang L, Zhang C. FMRP regulates miR196a-mediated repression of HOXB8 via interaction with the AGO2 MID domain. Mol Biosyst. 2014;10(7):1757–1764. 10.1039/c4mb00066h. [DOI] [PubMed] [Google Scholar]
- 80.Narayanan U, Nalavadi V, Nakamoto M, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27(52):14349–14357. 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51(4):441–454. 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 82.Huang J, Ikeuchi Y, Malumbres M, Bonni A. A Cdh1-APC/FMRP ubiquitin signaling link drives mGluR-dependent synaptic plasticity in the mammalian brain. Neuron. 2015;86(3):726–739. 10.1016/j.neuron.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsang B, Arsenault J, Vernon RM, et al. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc Natl Acad Sci U S A. 2019;116 (10):4218–4227. 10.1073/pnas.1814385116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi I-E. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18(7):2603–2618. 10.1091/mbc.e06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie X, Matsumoto S, Endo A, et al. Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J Cell Sci. 2018;131(8):jcs210856 10.1242/jcs.210856. [DOI] [PubMed] [Google Scholar]
- 86.Valdez-Sinon AN, Lai A, Shi L, et al. Cdh1-APC regulates protein synthesis and stress granules in neurons through an FMRP-dependent mechanism. iScience. 2020;101132.23(5). 10.1016/j.isci.2020.101132. [DOI] [PMC free article] [PubMed] [Google Scholar]