Abstract
Chronic hepatitis B virus (HBV) infection is a major cause of liver disease and cancer worldwide. While current therapeutic approaches can efficiently control viral infection, efficient curative antivirals are absent. The understanding of virus–hepatocyte interactions and sensing of viral infection is an important prerequisite for the development of novel antiviral therapies for cure. Hepatocyte intrinsic innate immunity provides a rapid first line of defense to combat viral infection through the upregulation of antiviral and inflammatory genes. However, the functional relevance of many of these antiviral signaling pathways in the liver and their role in HBV pathogenesis is still only partially understood. The recent identification of intracellular RNA and DNA sensing pathways and their involvement in disease biology, including viral pathogenesis and carcinogenesis, is currently transforming our understanding of virus–host interactions. Here the authors review the current knowledge on intrinsic antiviral innate immune responses including the role of viral nucleic acid sensing pathways in the liver. Since HBV has been designated as a “stealth virus,” the study of the impact of HBV on signaling pathways in the hepatocyte is of significant interest to understand viral pathogenesis. Characterizing the mechanism underlying these HBV–host interactions and targeting related pathways to enhance antiviral innate responses may open new strategies to trigger noncytopathic clearance of covalently closed circular DNA to ultimately cure patients with chronic HBV infection.
Keywords: hepatitis b virus, chemokines, cytokines, antiviral immune responses, IRF3, nuclear factor-kappaB, pattern recognition receptor, host defense, intrinsic innate responses
Hepatitis B virus (HBV) infection is a significant cause of morbidity that includes the development of cirrhosis, hepatic decompensation, and hepatocellular carcinoma (HCC).1 Furthermore, death from HBV-related liver disease remains one of the highest causes of mortality worldwide even though a functional vaccine has existed for decades.2 Given that there are more than 240 million individuals with chronic HBV infection globally, the mechanisms underlying disease pathogenesis are of significant impact.3
Hepatitis B virus pathogenesis involves the activation of the immune system as a host cytolytic response generated to clear infected hepatocytes. In addition, virus control can also be achieved through noncytolytic mechanisms. Failure of these usually potent antiviral immune responses can lead to chronic HBV infection and subsequently clinically significant hepatitis and liver disease.4 Due to the existence of a robust and specific host immune response to HBV in humans, manipulation through delivery of targeted and nontargeted therapies represents a viable approach for the development of a “sterilizing” cure.5 With implications for both our understanding of pathogenesis and toward the realization of curative therapies, studying antiviral immune mechanisms is of paramount importance.6
Initial breakthroughs to generate an effective vaccine and to understand HBV pathogenesis leveraged our understanding of the adaptive immune response.7 More recently, the role of innate responses, which are not pathogen specific, has garnered significant attention.8 With regard to host defense in general, the liver is enriched with immune cells, particularly, cells of the innate immune system including myeloid cells.9 Furthermore, intrinsic innate responses to hepatitis viruses, within the hepatocyte, have been implicated in both protective direct antiviral and inflammatory responses.10 In the case of the hepatitis C virus (HCV), which also causes significant liver disease and is generally considered a noncytopathic virus, many studies have demonstrated that the virus both activates and blocks intrinsic innate antiviral responses within infected cells.11 However, HCV can also evade host immunity, given its ability to bypass the adaptive immune response, making it very challenging to develop an effective vaccine as was developed for HBV several decades ago.12 Although early success with the generation of a vaccine for HBV was realized, HBV cure will be a challenging future endeavor.13 In the case of HBV, which may be considered as a more complex virus than HCV given the presence of both DNA and RNA viral products and the existence of replication intermediates in multiple cellular compartments (e.g., nuclear and cytoplasmic),14 the data are much less clear regarding interactions with the hepatocyte host defense machinery.15 This review summarizes the current literature on cell culture model systems and the absence or presence of functional cell-intrinsic host defense mechanisms. The focus here is on antiviral and hepatocyte responses within HBV-infected cells, while other aspects of innate immunity including natural killer (NK) cells and toll-like receptors (TLRs) are not discussed in detail. In addition, published data are summarized from reports that provide experimental insight, from the characterization of innate responses within hepatocytes, to address the following questions. (1) Does the hepatocyte contain the molecular machinery that would be needed to detect or sense HBV or isolated components of the virion and are the associated signaling pathways functional? (2) Does HBV inhibit functional intrinsic innate immune signaling pathways? (3) Does HBV or isolated components of the virion activate innate signaling?
Hepatitis B Virus Virology and Life Cycle
Hepatitis B virus is a member of the hepadnaviridae family with a partially double-stranded genome that infects hepatocytes. After entry and following uncoating, the viral nucleocapsid is released into the cytoplasm and its relaxed circular DNA (rcDNA) genome is subsequently released at which point its genome translocates to the nucleus. The viral genome is imported into the nucleus through mechanisms that are still only partially understood. In the nucleus, its genome is modified to the covalently closed circular DNA (cccDNA) form that exists stably as an extrachromosomal viral genome. This “minichromosome” serves as a template for both pregenomic RNA (pgRNA) and viral messenger RNA (mRNA) transcription. Viral RNA is transcribed from the DNA genome through an RNA polymerase II-dependent mechanism and is translated into the major proteins that make up the virion.14 Reverse transcription of the pgRNA by the viral reverse transcriptase, within the nucleocapsid with subsequent envelopment, results in newly formed virions (►Fig. 1). HBV proteins including the HBsAg are extremely potent antigens that have facilitated the development of effective vaccines.7 Currently licensed nucleos(t)ide analogues efficiently inhibit viral replication but fail to stop pgRNA and viral protein production and they do not facilitate clearance of cccDNA from infected hepatocytes.
Fig. 1. Model of the HBV life cycle.
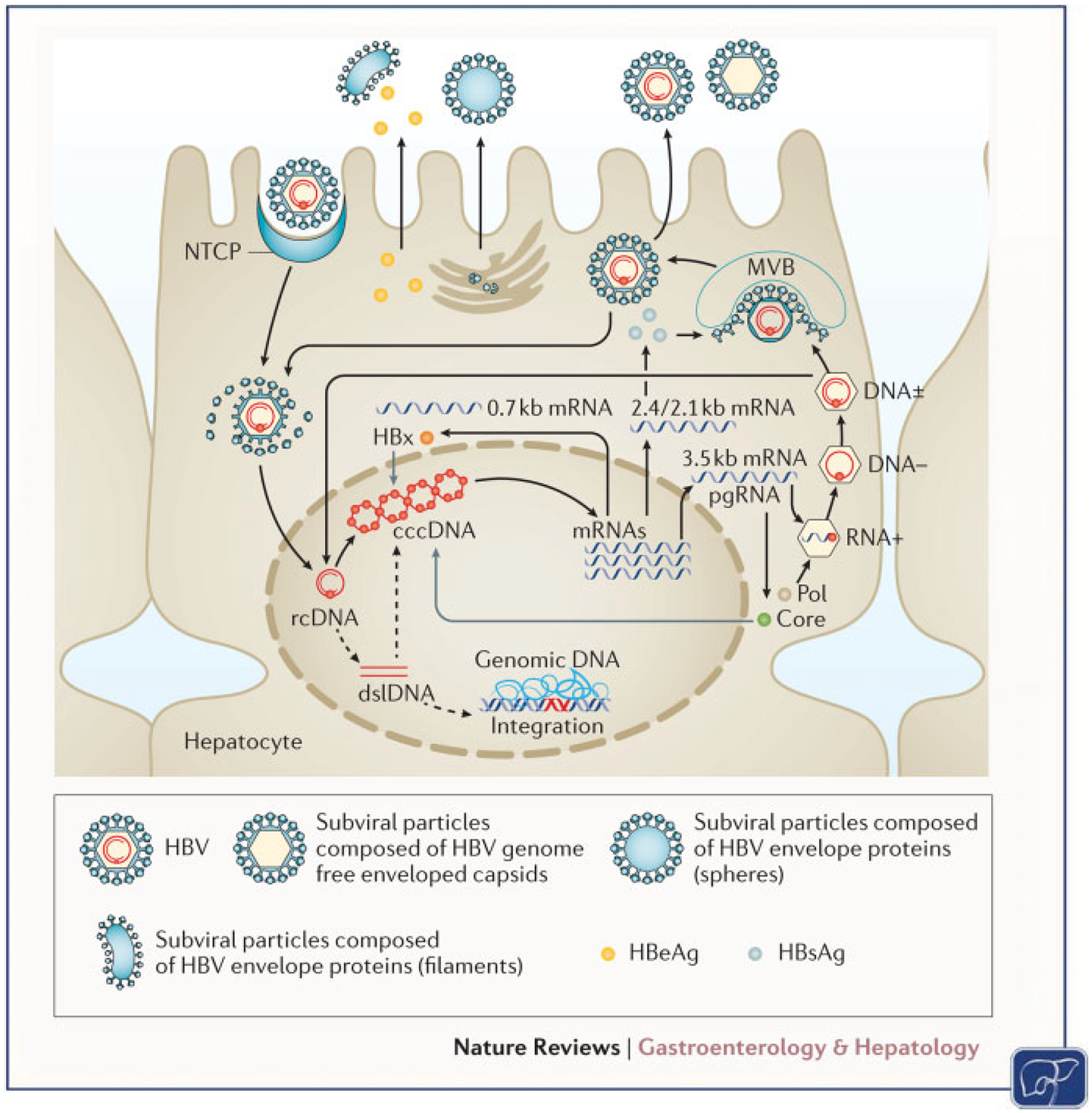
Schematic model depicting the major steps in the HBV life cycle in infected human hepatocytes. The life cycle of HBV, including attachment, entry, uncoating, trafficking to nucleus, cccDNA formation, integration, transcription, translation, encapsidation, and secretion, is depicted. Initially, HBV particles attach to the cell membrane and enter the hepatocytes through mechanisms that involve NTCP. After internalization, viral capsids are released and subsequently directed to the nucleus where the HBV genomes are liberated. In the nucleus, rcDNA genomes are converted into cccDNA that persists in the nucleus of infected cells as a “minichromosome,” which serves as a template for viral RNA transcription. dsDNA is also produced that can be integrated into the cellular genome or also converted into cccDNA. Viral mRNAs are transported to the cytoplasm where they are translated into viral proteins and together with the viral polymerase, the pgRNA is encapsidated and reverse transcribed within the nucleocapsid into progeny rcDNA. Mature nucleocapsids are then either directed to the multivesicular body pathway for envelopment with HBVenvelope proteins or redirected to the nucleus to establish a cccDNA pool. cccDNA, covalently closed circular DNA; dsDNA, double-stranded DNA; HBV, hepatitis B virus; mRNAs, messenger RNAs; NTCP, sodium-taurocholate cotransporting polypeptide; pgRNA, pregenomic RNA; rcDNA, relaxed circular DNA. (Reproduced with permission from Nature Publishing Group, Thomas, E. et al Nat. Rev. Gastroenterol. Hepatol. https://www.nature.com/articles/nrgastro.2016.37.)
Hepatocyte Cell-Intrinsic Innate Immunity and HBV
Hepatocyte cell-intrinsic innate immunity provides a first line of defense to thwart invading pathogens including hepatitis viruses.11,16 Production of type I and III interferon (IFN) are potent effectors of this initial response with type III/IFN-λ (IFNL) being a significant component of the antiviral response in the hepatocyte; however, the production of IFN is only one component of a multipronged cell-intrinsic antiviral response.10
Significant progress has been made recently in our understanding of how cells trigger host defense responses through recognition of pathogen-associated molecular patterns (PAMPs). Of the many PAMPs that have been identified, viral RNA has emerged as a major stimulator of intrinsic cellular defense mechanisms. In the case of an RNA virus like HCV, the viral genome itself is recognized through several distinct pathways that will be discussed in upcoming sections. Briefly, once the virus uncoats and the RNA is released into the cytoplasm, significant secondary structure in the viral genome, particularly in the untranslated region, or double-stranded RNA (dsRNA) intermediates, produced during viral replication, are potent activators of pattern recognition receptors (PRRs) including the retinoic acid-inducible gene (RIG-I) like receptors (RLRs) and TLR3.17–21 However, it has been less clear how cells in the liver trigger innate immune signaling in response to DNA species originating from hepatitis viruses.22
Recent breakthroughs in our understanding of RNA- and DNA-dependent signaling processes arose through the discovery of proteins that have been implicated in the sensing of these nucleic acids.23 DNA has been shown to directly or indirectly, through RNA intermediates arising from RNA polymerase III activity, induce cytokines through the activation of transcription factors including IFN regulatory factor 3 (IRF3) and nuclear factor-kappa B (NF-κB).24,25 However, these DNA sensing mechanisms which can be tissue specific have yet to be fully characterized regarding their functional role in hepatocytes, contribution to HBV antiviral immune responses, and the development of hepatitis.26–29
Hepatitis B virus has a DNA genome that is converted to RNA intermediates through the activity of cellular RNA polymerases.30–32 Although HBV can replicate to high titers in hepatocytes, which can appear as “ground-glass” with hematoxylin and eosin staining due to large load of viral proteins in infected cells, HBV is not a strongly cytopathic virus.33 Rather, HBV-associated liver damage is thought to be the consequence of chronic cytolytic immune responses, targeting infected hepatocytes, by liver-infiltrating immune cells.34
Using experimentally infected chimpanzees, microarray analyses, performed at early time points, suggested that HBV does not activate innate antiviral responses in hepatocytes nor inhibit other intrahepatic innate immune responses.35 After this study, HBV was designated as a “stealth virus.”36 Interestingly, another study demonstrated that HBV might be cleared from infected hepatocytes before any detectable adaptive immune response is mounted,37 thus suggesting that innate immunity or antiviral responses at the level of infected cells could play an important role in viral clearance. In addition, inflammatory chemokines can be upregulated to detectable levels in HBV-infected patients. Initial studies demonstrated that these chemokines were mainly produced and detected only after an adaptive immune response.38–40 However, more recent studies have shown that HBV can also stimulate production of chemokines at earlier time points.41–43 Recent publications have also demonstrated that hepatocytes and macrophages, stimulated with HBV in vitro, can produce inflammatory cytokines eluding to the possibility that HBV does not completely evade immune recognition and that the virus may directly modulate cell-intrinsic innate immune pathways that are involved in the production of inflammatory cytokines and chemokines.26–28,44 PRRs that have been implicated in HBV sensing are discussed in upcoming sections and include cyclic GMP-AMP (cGAMP) synthase (cGAS) and RLRs.17,26,45
Overall, antiviral innate immunity against HBV, which occurs very early after viruses contact with hepatocytes, is an area that has been less studied until recently and necessitates further investigation in appropriate models. In addition, as compared with RNA viruses such as HCV, less is known about how human hepatocytes recognize DNA viruses such as HBV and their replicative RNA intermediates. Given that HBV replication, in humans, is generally not detectable until about 1 month after HBV infection,1,46,47 cell-intrinsic innate immunity may be important for controlling early virus replication.
Cell Culture Models to Study Hepatocyte Cell-Intrinsic Innate Immunity to HBV
The understanding of HBV–host interactions, including cell-intrinsic innate immune responses after infection, has been hampered for many years by the paucity of robust and physiologic cell culture model systems47,48. This is specifically true for the study of HBV infection in hepatocyte models possessing functional host defense pathways that may also require the inclusion of nonparenchymal liver cells to be fully biologically relevant.27,48 As for most viruses, tumor-derived cells lines have been useful in increasing our understanding of HBV biology. Gene-editing approaches have been used to overexpress viral proteins and to generate cell lines that continuously express hepatitis viral genomes. Specifically, HBV DNA genomes and plasmids encoding viral proteins have been delivered intracellularly through various transfection techniques and viral transduction systems.47 Overall, the use of transformed cell lines has relied on the fact that several of them harbor defects in intrinsic innate immune antiviral pathways in cells that would normally restrict expression of viral genomes and proteins.20,49 These cell lines, although abnormal, enabled these viruses to be studied without major interference from host defense signaling pathways. Specifically, a distinguishing characteristic of hepatocyte derived cell lines is the ability to detect extracellular and endosomal pathogen-derived RNAs through the TLR3 pathway that is important for antiviral responses.10 However, the use of transformed cells may be suboptimal for studies focused on understanding innate hepatocyte responses to HBV given the variance in functional TLR3 signaling or other components of intracellular host defense.28
To investigate the pathways involved in sensing of HBV, including its DNA and associated RNA intermediates in human hepatocytes within minutes to hours after viral cell entry, several models have been described.47,48 These include primary human hepatocytes (PHHs), which have functional intrinsic innate immune responses, and the HepaRG cell line, which is immortalized with some host defense pathways intact, as opposed to other transformed hepatoma cell lines.50,51 Interestingly, owing to the differences in antiviral innate immunity within the HepG2 cell line when compared with Huh7 cells, this model has proven to be useful in the study of host defense pathways and to validate results obtained in PHHs. Specifically, the genetically altered HepG2 cell line was capable of producing large amounts of type III IFNL in response to HCV infection, which also has been observed in PHHs.10,52
With regard to the hepatoma cell lines that can support HBV infection, the development of sodium-taurocholate cotransporting polypeptide (NTCP)-overexpressing hepatoma cells, such as HepG2-NTCP cells, facilitates the study of the full HBV life cycle in a robust and easy-to-use cell culture model.47 As previously mentioned, HepG2 cells are capable of mounting efficient innate immune responses after infection by HCV52; however, the level and breadth of activation of antiviral responses are far less than those observed in PHHs.10 Another study utilized the HBV-infected HepG2-NTCP cells for studying the interaction between RIG-I and HBV RNA, suggesting that this cell line can be useful for the study of innate immune responses after HBV infection.17 The PH5CH8 cell line has also been utilized for related studies, albeit less frequently, because it has a functional TLR3 system.19,53,54
These data suggest that results from immortalized and transformed cell lines should be validated in HepaRGs, PHHs, as well as in vivo models, if available, that may include humanized mice.26,48,55 Unfortunately, even these models also have limitations. HepaRG cells are bipotent hepatic progenitor cells that can differentiate into both biliary and hepatocyte-like cells and can divide indefinitely.56 To be fully capable of supporting infection with HBV, these cells must also be treated with dimethyl sulfoxide to foster additional maturation into more differentiated hepatocyte-like cells. According to head-to-head comparisons, HepaRG cells have many similarities to PHHs that are considered to be the gold standard for cell-based models.57,58 Initially, the HepaRG cell line was demonstrated to be capable of supporting HBV infection and replication.56 Unfortunately, HBV infection is optimized through the use of polyethylene glycol (PEG) that is employed during the infection to enhance virion uptake. Infection may be further optimized through continuous administration of PEG to facilitate viral entry by increasing interactions between the HBV virion and the cell membrane59 but the overall infection rate is low with minimal cell-to-cell virus spread.60 HepaRGs may best support HBV infection when they are engineered to overexpress NTCP.61 However, the use of overexpression models has several important caveats: first, it is clear that introduction of the DNA plasmid itself to deliver the target protein will activate cell-intrinsic innate immunity. Second, cell lines that are able to produce high levels of the target protein will rely on a blunted antiviral response given the propensity for these host defense pathways to shut down the translation of foreign proteins, which is a major mechanism of pathogen resistance.62,63
As previously mentioned, PHHs are considered the gold standard for laboratory studies of hepatocyte function. PHHs are obtained from patients undergoing liver resection (usually for a metastasis from a nonliver cancer) and they are isolated from the adjacent “healthy” parenchymal tissue. In addition, PHH can also be obtained from the fetal livers of aborted embryos and serve as a substrate for hepatitis virus infection.64 However, since these fetal hepatocytes, often a combination of both hepatocytes and hepatoblasts depending on the gestational age of the donor fetus, are utilized and discarded, their use is much less controversial than that of embryonic stem cells, which offer the prospect for long-term biomedical applications that include cloning.65 Unlike HepaRG cells, PHHs once plated do not replicate and therefore have a limited life span in tissue culture, usually between 1 and 2 weeks although fetal hepatocytes can be stable for several more weeks.64 Once in culture, these cells rapidly dedifferentiate, concomitantly downregulating biological characteristics found in mature hepatocytes.66 Although these cells have a limited life span in culture, they readily support infection by HBV. However, viral replication is usually limited when compared with levels seen in cell lines since these PHHs presumably have intact host defense pathways that combat the infection.10,26 Many studies have also demonstrated productive infection of PHHs with serum-derived and cell-culture-derived HBV.33,67–69 A challenge with using PHHs is the presence of contaminating cells from lymphoid and myeloid lineages that also have functional and distinct cell-intrinsic innate immune pathways.27 Ideally, single-cell analysis or studies using immunofluorescence are optimal to ensure that any innate immune responses that are observed are arising from an infected hepatocyte. These techniques may be dispensable for experiments with HepaRG cells were there are no contaminating immune cells.
Induced pluripotent stem cells are a newly developed source of hepatocytes named hepatocyte like cells (HLCs) or induced hepatocytes (iHeps) that can be used for studies on viral hepatitis.70,71 These cells, once generated, can be a reliable source of cells that can be differentiated into partially mature hepatocytes. The advantages of stem-cell-derived hepatocytes over PHHs include the ability to obtain an unlimited supply of pure normal hepatocytes and these cells would be less variable when compared with PHHs that are obtained from different donors that can vary by gender, age, exposure to medications/chemicals, genetic polymorphisms, and the presence or absence of underlying liver disease.72,73 These human stem-cell-derived hepatocytes have proven to be a useful substrate to successfully support infection by HBV.70 In addition, HLCs/iHeps would offer the benefit of not containing contaminating white blood cells that are often present in PHH cultures. Unfortunately, stem-cell-derived hepatocytes do not fully differentiate into functional mature hepatocytes. However, various approaches have been taken to overcome this limitation through the use of small molecules that promote differentiation and manipulation of the microenvironment used in the in vitro culture systems.74–76 Since HBV has a DNA genome that is transcribed into viral RNAs,30–32 careful characterization of nucleic acid sensing pathways is needed in more advanced models including HepaRG cells, PHHs, and HLCs/iHeps. This would facilitate additional insight into the mechanisms by which HBV can possibly regulate innate responses in human hepatocytes to promote the liver inflammation that is observed in infected patients.
Does the Hepatocyte Contain the Molecular Machinery to Sense HBV and are the Associated Signaling Pathways Functional?
The liver is a major immunologic organ in the human body9 being a site of initial hematopoiesis in fetal development and also a site where viruses and associated particles may accumulate.77 As a consequence of these observations, it would not be unexpected that hepatocytes, the major parenchymal cell of the liver, have developed adequate machinery to detect and combat foreign pathogens. Furthermore, as epithelial cells, hepatocytes can respond to the major IFN proteins including IFN-α, β, γ, and λ (IFNL).10,78 This opens up the exciting possibility that pretreatment of hepatocytes first with IFN, to upregulate levels of proteins involved in innate immune signaling, may facilitate the detection and characterization of pathways that can sense HBV and components of the virion.28 These pathways may subsequently be amenable to therapeutic interventions in efforts to achieve a sterilizing cure for HBV. One drawback of this approach would be with regard to HBV infection in vitro where pretreatment with IFN may render cells less likely to be infected due to the upregulated host defense mechanism.
Given their earlier initial discovery and importance in immune responses that are conserved throughout multicellular organisms, TLR expression and functionality have been extensively studied in hepatocytes. Hepatocytes express TLRs and upon stimulation with their cognate ligands, they activate downstream antiviral and inflammatory pathways.79 These responses can be broadly classified as those involving foreign nucleic acids versus other types of molecular patterns.80 Since HBV has a DNA genome and produces RNA intermediates, these nucleic acid detection pathways are pertinent. For nucleic acid detection, hepatocytes express TLR3, 7, 8, and 9 and these pathways are functionally activated upon ligand stimulation.81 Specifically polyinosinic:polycytidylic acid (poly(I:C)) that is added to the cell culture media directly, not delivered intracellularly, is able to upregulate well-characterized viral stimulated genes as previously described.10,21. With regard to nonnucleic acid detection, both the TLR 2 and 4 pathways are functional.82 In addition, hepatocytes express the major signal adaptor molecules for TLRs, which are TRIF/TICAM1 (TIR-domain-containing adapter-inducing interferon-β/TIRdomain-containing adaptor molecule 1) and MYD88 (Myeloid Differentiation Primary Response 88). Given that the expression of TLRs is mainly on membranes associated with the extracellular space including endocytic vesicles, they represent an important surveillance pathway for the presence of components of foreign pathogens.83
More recently, the identification of intracellular cytoplasmic signaling pathways that recognize nucleic acids has yielded great insight into intrinsic cellular innate immune responses. In regard to RNA species, clearly both the RIG-I/melanoma differentiation-associated gene 5 (MDA5) pathways are present and functional in hepatocytes as has been demonstrated through studies involving HCV.10 In addition, targeted analysis of the presence and functionality of these pathways has been demonstrated through the use of specific viral RNA mimetics. Transfection of poly(I:C), typically performed using lipofection, to deliver the molecule to the cytoplasmic compartment, is able to upregulate well-characterized viral stimulated genes as previously described.10
Since HBV has a DNA genome, hepatocyte defense pathways that can detect this form of nucleic acid are of paramount interest. However, there has been much more variance in reported data regarding the presence and functionality of innate immune pathways within the hepatocyte that sense DNA species. Recently, cGAS was identified and characterized as a DNA sensor exhibiting antiviral activity against a broad range of DNA and RNA viruses.84,85 cGAS is encoded by the MB21D1 gene and directly binds to cytoplasmic double-stranded DNAs (dsDNAs) inducing the production of cGAMP that is an intracellular, second messenger to activate cell-intrinsic innate immunity. Importantly, cGAS is an IFN stimulated gene (ISG) that is significantly induced following exposure of the cell to IFN.63 A recent report has demonstrated that cGAS is expressed in both human liver and PHH and is also able to activate host defense pathways in hepatocytes after stimulation with DNA.26 Furthermore, it would be expected that cGAS-dependent pathways would be more active following IFN treatment given that cGAS itself is an ISG.
To further understand the recognition of foreign DNA in hepatocytes, studies have been performed focusing on the stimulator of interferon genes (STING) pathway that has been shown to be important for DNA responses and innate responses to HBV.23,86,87 cGAMP, which is produced by cGAS, is subsequently recognized by STING that is encoded by the TMEM173 gene. This triggers the expression of antiviral and inflammatory genes through TANK-binding kinase 1 (TBK1) activation and subsequent nuclear translocation of IRF3 and NF-κB. Recent reports have demonstrated that STING is expressed in both the human liver and PHHs and is also able to activate host defense pathways in hepatocytes.26,28,88 In contrast to cGAS, one recent study has reported that hepatocytes lack the DNA signaling molecule STING.29 In addition, STING itself may not be significantly induced by IFN giving more importance to baseline expression of this molecule. Ultimately, STING is highly expressed in macrophages/Kupffer cells and hepatocytes may express lower levels in comparison.88
Given the likelihood that hepatocytes possess functional intracellular sensing pathways for pathogen-derived DNA that can also be further upregulated by IFN, studies have sought to characterize responses to diverse species of DNA molecules. The innate immune response mounted following stimulation with dsDNA in human hepatocytes has been studied using dsDNA viral mimetics to stimulate in vitro cell culture models including PHHs and HepaRG cells. Transfection of DNA species, typically done through lipofection, including poly(deoxyadenylic-deoxythymidylic) acid (poly (dA:dT)) (b-DNA) and poly(deoxyguanylic-deoxycytidylic) acid (poly(dG:dC)) (z-DNA) has been shown to produce robust antiviral responses in hepatocytes. poly(dG:dC) and poly(dA:dT) have also been shown to activate the downstream signaling proteins TBK1 and IRF3 to stimulate IRF3 nuclear translocation that can be blocked using selective TBK1 inhibitors (e.g., BX795).28
Additional pathogen DNA mimetics including IFN stimulatory DNA (ISD) and cyclic [G(2′,5′)pA(3′,5′)p] = (2′3′-cGAMP) have been shown to stimulate robust innate immune responses in transfected cells.89,90 ISD is a 90-base pair non-CpG oligomer that can be easily synthesized. In both PHHs and HepaRG, both ISD and 2′3′-cGAMP can activate intracellular sensing pathways similar to cells treated with poly(dA:dT) and poly(dG:dC). However, transfected ISD and 2′3′-cGAMP may not be as potent stimulators as poly (dA:dT) and poly(dG:dC).28 Certain DNA mimetics including poly(dG:dC) and poly(dA:dT), when transfected, are potent stimulators of IFN production in hepatocytes; however, this is thought to occur through the production of an RNA intermediary species that is generated from intracellular RNA polymerase III activity. These RNA species would subsequently activate RIG-I/MDA5 signaling.25
Interestingly, DNA mimetics that stimulate antiviral responses independent of an RNA intermediary, including ISD and 2′3′-cGAMP, resulted in significant upregulation of antiviral and proinflammatory genes at both the mRNA and protein level. However, IFNL2/3 was not detected at the protein level suggesting that RNA-independent DNA sensing pathways do not result in secretion of IFNL2/3 at early time points. Interestingly knockdown of cGAS, STING, and downstream signaling proteins TBK1 and IRF3 had dramatic effects on gene induction by ISD demonstrating almost complete inhibition.28 Overall, through the utilization of small interfering RNAs (siRNAs) targeting several DNA sensors, the importance of the cGAS–STING–TBK1–IRF3 pathway in the recognition of DNA in hepatocytes has been demonstrated.26 In addition, the NF-κB pathway has also been demonstrated to play an important role in the upregulation of proinflammatory cytokines following stimulation with DNA in hepatocytes.28
As previously mentioned, recent data have demonstrated that the hepatocyte cGAS–STING pathway is functionally active even without first pretreating cells with IFN. With regard to HBV infection, this was demonstrated in hepatocytes by the reduction of viral cccDNA levels in gain-of-function studies. In addition, CRISPR/Cas9-mediated downregulation of cGAS resulted in a marked increase, whereas overexpression of cGAS resulted in a marked decrease in HBV infection and HBV cccDNA levels.26 The importance of these findings is underscored by the fact that cccDNA is the key viral nucleic acid species responsible for HBV persistence. Furthermore, silencing of cGAS, STING, and TBK1 expression significantly increased HBV infection, demonstrating their importance as viral restriction factors in hepatocytes.26
To further address the mechanisms by which DNA can activate host defense pathways in hepatocytes, other proteins that have been implicated in foreign DNA sensing have been studied. Since some DNA sensors can demonstrate tissue-specific expression, there has been some variance in data from published reports. Gamma IFN inducible protein 16 (IFI16), a known DNA sensor,91 was found to be absent in hepatoma cell lines except HepaRG cells. After inducing a marked decrease in IFI16 protein levels using siRNA, there was no detectable role for this protein in the upregulation of proinflammatory genes by transfected DNA.28 Also, the role of absent in melanoma 2 (AIM2), another known DNA signaling molecule,92 was studied and similar results were obtained with minimal impact being observed in hepatocyte DNA responses. Overall, with regard to DNA sensors that are expressed in hepatocytes, in contrast to studies performed with cGAS and STING, the silencing of IFI16 or AIM2 had minimal impact on the sensing of DNA and HBV infection.26,28 A list of PRR ligands that have been used to study the sensing of nucleic acids in hepatocytes is presented in ►Table 1. Overall, there is experimental evidence to support the possibility that hepatocytes do contain the molecular machinery needed to sense HBV and mount a subsequent antiviral response.
Table 1.
Examples of pathogen-associated molecular patterns (PAMPs) with potential relevance for hepatocyte host defense signaling pathways and HBV control
| PAMPsa | DNA26,28 | RNA19,21,116 | Hepatocyte recognition19,20,26,28,44 |
|---|---|---|---|
| Poly dAdT | +CDS | +RLR | Yes |
| Poly dGdC | +CDS | − | Yes |
| ISD | +CDS | − | Yes |
| 2’3′cGAMP | +STING | Yes | |
| 5′ppp-dsRNA | − | +RLR | Yes |
| PolyI:C | − | +RLR | Yes |
| ssPolyU | − | +TLR8 | Yes |
Abbreviations: CDS: cytosolic DNA sensor; HBV, hepatitis B virus; ISD, IFN stimulatory DNA; RLR, RIG-I like receptors; STING, stimulator of interferon genes; TLR8, toll-like receptor 8.
PAMPs may require transfection for stimulation.
Does HBV Inhibit Functional Intrinsic Innate Immune Signaling Pathways?
Hepatitis B virus has been described as a “stealth virus.” As a consequence, there has been a lot of interest in studying the intriguing possibility that HBV actively blocks antiviral and inflammatory signaling within hepatocytes. Clearly, HCV has the ability to block antiviral signaling pathways. Strong experimental evidence has been published that the HCV NS3/4A protease cleaves the cytoplasmic adaptor protein mitochondrial antiviral signaling protein (MAVS) as a major viral mechanism for blocking RIG-I/MDA5 signaling.11 With regard to HBV, the data have been less clear and many genes have been reported to be regulated by this virus (►Table 2).
Table 2.
HBV-regulated genes
| Host defense genes inhibiteda or activatedb or bothc by HBV | ||
|---|---|---|
| APOBEC3, Lymphotoxin Ba | IRF3c | RIG-Ic |
| cGASc | IRF7c | RUBICONa |
| CUL4-DDB1a | MAVSc | STINGc |
| DDX3a | MDA5c | TBK1c |
| Interferonc | MxAa | TLR2b |
| HSP90a | NFKBc | TRIFa |
| IKKc | Parkina | TRIM22a |
Abbreviations: cGAS, cyclic GMP-AMP synthase; HBV, hepatitis B virus; IKK, IκB kinases; IRF, interferon regulatory factor; MAVS, mitochondrial antiviral signaling protein; MDA5, melanoma differentiation-associated gene 5; RIG-I, retinoic acid-inducible gene; RUBICON, RUN domain Beclin-1-interacting and cysteine-rich domain-containing protein; STING, stimulator of interferon genes; TBK1, TANK-binding kinase 1; TLR2, toll-like receptor 2.
Hepatitis B virus has been reported to specifically block the downstream IFN signaling pathway93 but others have failed to replicate this result.94 IFN itself has been used as a therapeutic agent for chronic HBV infection for decades. Unfortunately, due to its pleiotropic effects as a cytokine, the precise mechanistic understanding of the antiviral actions of IFN, which specifically impact HBV infection, remains to befullycharacterized.95 IFN may contribute to the stimulation of cytotoxic T-lymphocyte and NK cell-mediated killing of HBV infected hepatocytes.96 In addition, there is evidence that IFN has significant direct antiviral effects in hepatocytes and that this activity may depend on the upregulation of ISGs in hepatocytes.95,96 Treatment with IFN-α was able to decrease levels of both HBV DNA and cccDNA.28 Furthermore, activation of the lymphotoxin-β receptor was reported to alter the specific degradation of cccDNA within the nucleus through a cytidine-deamination mechanism via APOBEC3B upregulation. This resembles activation of APOBEC3A by IFN that has been extensively studied.97 However, even though several cellular mechanisms have been proposed that could limit the number of cccDNA molecules, including the deaminase function of APOBEC3 family members, their role in actually affecting cccDNA quantity in vivo is still being investigated.98 Given the evidence of the efficacy of therapeutic IFN on HBV infection and recent data from mechanistic studies involving the APOBEC family of proteins, the possibility that endogenous IFN is released by resident hepatic cells or infiltrating immune cells is of interest. Endogenous IFN-mediated induction of ISGs may result in subtle antiviral activity; however, it remains to be determined how much ISGs directly contribute to viral clearance in humans or if they slightly decrease virus replication or viral protein translation until more robust adaptive immune responses take effect.98 This is in contrast to HCV infection, where IFN responses have been strongly implicated in viral clearance.99
Currently, it is unclear whether HBV possesses molecular mechanisms to directly block the direct sensing of PAMPs or suppress host defense responses once activated.26,27,94 Recent studies have demonstrated that HBV has the ability to downregulate early antiviral responses44 and that HBV proteins and replicative intermediates interfere with innate immune responses in different in vitro models.15 Other studies suggest an active inhibition of innate immune responses by HBV.26 Furthermore, regions of the HBV pgRNA are double stranded and may activate dsRNA-dependent host defense mechanisms.17 To block this host defense pathway, HBV has been reported to induce Parkin-dependent recruitment of the linear ubiquitin assembly complex (LUBAC). The downstream consequence of this was attenuation of signal transduction through MAVS-dependent pathways.100 More recently, HBV infection has been shown to decrease the levels of cGAS as a potential mechanism for interfering with innate signaling. Specifically, HBV infection suppressed cGAS expression and function in cell culture models and humanized liver chimeric mice.26 In addition, the authors reported that HBV represses both expression of cGAS and its effector genes in humanized mice and studies are being conducted to further characterize the underlying mechanisms.
Hepatitis B virus has also been reported to possess numerous other additional strategies to counteract host innate immune responses. It was reported that the HBV polymerase protein, in addition to functioning as reverse transcriptase for pgRNA, inhibits host defense to foreign nucleic acids by interfering with STING-mediated IFN-β induction and also by preventing the activation of IRF3.101 The HBV polymerase has also been shown to directly interfere with PRR signaling via interaction with DDX3 and subsequent inhibition of downstream signaling through TBK1.102 Additionally, the enzyme was shown to prevent the activation of IκB kinases (IKKs) through interaction with HSP90. Furthermore, the polymerase can mask the antigenic step of reverse transcription by delaying reverse transcription until the polymerase/pgRNA complex is successfully encapsidated.98
With a more undefined role in the viral life cycle, the HBV X protein (HBx) has been reported to negatively regulate multiple aspects of host defense in hepatocytes. HBx has been shown to specifically block innate immune signaling by MAVS and TRIF and subsequent IRF3-induced gene expression.98,103 In addition, data have been generated that suggest that HBx specifically inhibits the transcription of TRIM22 that has been shown to be a host restriction factor for HBV.104 HBx has also been shown to target multiple downstream signaling pathways including IRF3 and IRF7 nuclear translocation through the NF-κB essential modulator (NEMO) by the RUN domain Beclin-1-interacting and cysteine-rich domain-containing (RUBICON) protein.105 The HBx protein was also reported to interact with damaged DNA binding protein 1 (DDB1), which is a component of the cullin 4 (CUL4)-DDB1 ubiquitin ligase complex.106 Building on these initial studies, it was subsequently demonstrated that HBx facilitates the CUL4-DDB1 E3 ligase to degrade the structural maintenance of chromosome (Smc) 5/6 complex. The Smc 5/6 complex appears to bind to only episomal HBV DNA and thereby inhibits transcription. However, it does not appear to bind to chromosomally integrated HBV DNA. Thus, Smc 5/6 acts as a host restriction factor and HBx targets the host ubiquitin–proteasome system to degrade the Smc 5/6 complex, thereby enabling HBV gene expression from cccDNA.107,108 Lastly, the HBV protein core (HBVc) and pre-core (HBVpc) can inhibit the expression of MxA at the transcriptional level based on in vitro experiments.109
It is important to point out that many of these mechanisms of innate immune evasion by HBV encoded proteins stem from overexpression studies that require artificial introduction into hepatoma cell lines. In general, overexpression of viral proteins may not provide physiologically relevant data when considering natural infection in humans with HBV infection as previously mentioned. Indeed, new studies utilizing more efficient in vitro HBV infection models demonstrate that HBV may not interfere with PRR-mediated activation of innate responses in the hepatocyte.27,94 Furthermore, using ex vivo cultured human liver biopsies, PRR-mediated IFN and ISG induction was not suppressed in HBV-infected hepatocytes.110 Overall, HBV may employ multiple strategies to evade sensing and antiviral activity of PRRs and their effector pathways and it may also block IFN production and signaling. However, additional data from in vivo and in vitro models suggest that HBV has a mild or even undetectable effect on the activation and downstream signaling of cell-intrinsic innate immune pathways.
Does HBV or Isolated Components of the Virion Activate Innate Signaling?
Given that hepatocytes have numerous mechanisms to sense multiple components of invading pathogens and there have been numerous reports of inhibition of these pathways by HBV, the dynamic interplay between HBV and its host cell has been intriguing. HBV can replicate to high titers, produce large amounts of viral antigens, but still not cause overt cytopathicity to infected hepatocytes. Published reports are presented here to discuss nuances within experimental findings that characterize this complex interaction.
Components of the HBV Virion
The HBV virion is a complex macromolecular structure composed of several distinct components. Purification of distinct components with subsequent stimulation of hepatocytes is a routinely used strategy to determine if there is functional innate immune sensing of HBV in the liver. In regard to its nucleic acid products, since HBV utilizes the hepatocyte RNA polymerase II enzyme to complete its life cycle, its mRNA transcripts resemble cellular mRNAs reducing the likelihood of immune recognition of these viral transcripts. In addition, regulation of viral transcription by the HBx and core proteins may also play a role in shaping the transcriptional activity of the HBV genome from cccDNA to avoid triggering innate responses. This would be accomplished by allowing the viral gene expression to proceed in a controlled manner that is more similar to endogenous gene expression.98 However, recent studies have suggested direct sensing of the pgRNA or other HBV RNAs by either MDA5111 or RIG-I.17 Naked rcDNA has also recently been reported to be sensed in a cGAS-dependent manner in hepatoma cell lines and PHHs.26 The study concluded that in human hepatocytes, exposure to naked HBV genomes leads to the activation of innate antiviral immune responses. Collectively, these data suggest that HBV-derived nucleic acid, including nonencapsidated HBV DNA and pgRNA, is sensed by cGAS and other PRRs; however, this sensing is minimized or absent during natural HBV infection when HBV DNA is encapsidated.
Hepatitis B virus viral proteins have also been reported to stimulate cell-intrinsic innate immunity. Specifically, the HBV nucleocapsid protein may be a ligand for TLR2 that can stimulate proinflammatory chemokine production PHHs. However, there may not be high levels of nonenveloped HBV nucleocapsid structures in patients, as may be found in cell-culture-derived HBV isolates, so the biologic significance of this remains to be clarified.15 The use of HBV virions purified from patients would support the relevance of these experiments70; however, many laboratories continue to use cell-culture-derived virions. Ultraviolet (UV)-inactivated HBV has also been used to stimulate HepaRG cells and PHHs. UV-irradiated virus can typically be inactivated following a standard dose of ~12 J/cm.44 Upregulation of inflammatory chemokines has been reported using UV-inactivated virus28 demonstrating that components of the intact virion may contribute to activation of host defense pathways in hepatocytes.
Hepatitis B Virus Infectious Virions and Innate Immunity
As mentioned previously, physiologically relevant studies of direct HBV infection and cell-intrinsic innate immunity are challenging. This comes from the fact that in vitro models used to study the HBV life cycle are suboptimal arising from either the virions employed or the cells that are infected. Specifically, to generate a strong and productive infection in PHHs, up to 1,000 virus-genome-equivalents (GEq) per cell of recombinant HBV virions are needed.56,60,68 This may stem from hepatocyte host defense pathways that limit virus replication in vitro. Moreover, HBV inefficiently spreads through a monolayer of hepatocytes in vitro, unless additional modifications are made to standard infection protocols.59 The prevention of viral spread may be due to the activity of unknown host defense pathways or mechanisms that do not lead to global changes in gene expression as is observed with HCV infection in PHHs.10
Another challenge is that the HBV inoculum, used for in vitro experiments, may not resemble virus obtained from patients with chronic infection. Most researchers use HBV produced by HepG2.2.15 or HepAD38 cells, HBV serotype ayw, genotype D, to easily infect PHHs, differentiated HepaRG cells, or NTCP overexpressing hepatoma cells. It is possible that these viral preparations may contain contaminants that are not found in normal HBV containing patient sera that can contribute to nonphysiologic results.15 The use of functional HBV virions purified from a patient may be the most biologically relevant70; however, this continues to be a challenge for the field.
Overall, HBV appears to only marginally activate and/or regulate innate immune responses in cell culture hepatocyte models most likely without the production of IFN from hepatocytes. These results have also been supported by experiments from in vivo models.112 Thus HBV behaves like a “stealth virus” avoiding strong viral DNA and RNA sensing and also the activation of pathways involved with recognition of viral proteins except when considering HBsAg that drives antiviral responses from the adaptive immune response. In further support of this concept, the amount of intracellular HBV DNA generated in cell culture models can approach approximately more than 10 million copies, which is robust, significant, and relevant. These data confirm that the levels of HBV DNA in HBV-infected cells are most likely sufficient to activate host defense pathways and the absence of HBV sensing in infected cells is not due to low multiplicity of infections (MOIs).26 The ability of the virus genome to be packaged in a viral capsid, subside in the nucleus, and the generation of viral RNAs that are very similar to cellular RNAs may contribute to evasion of host defense pathways in this study. This is also possibly due to the fact that most host defense pathways are based in the cytoplasm and not the nuclear compartment.113–115
However, studies have reported that HBV can be sensed by hepatocytes at early time points postinfection and even later. Although there was only moderate elevation of some genes involved in inflammation, the results have been confirmed in independent studies.28,44,70 Given that this response was minimal and possibly even transient, it was hypothesized that HBV subsequently blocks this stimulation of gene expression.26,44 Microarray data from published studies have also demonstrated that HBV causes changes in the expression of genes involved in the inflammatory response; however, the level of induction was much less than that observed with HCV from experiments in similar cell culture models.10 ►Fig. 2 highlights possible stimulation of cytoplasmic DNA and RNA sensing pathways by HBV-generated nucleic acids. More recently, several well-executed and controlled studies have been unable to detect any effect of HBV infection on gene expression in cultured PHHs and other relevant models such as ex vivo liver tissue.27,94,110
Fig. 2. Proposed model that highlights the role of nucleic acid signaling in intrinsic innate immunity to HBV.

First, HBV pgRNA may be sensed by the IFN-induced genes MDA5 and RIG-I, leading to activation of NF-kB and IRF3. Second, transfected poly(dG:dC) and poly(dA:dT) activate TBK1 leading to activation of IRF3, resulting in production of inflammatory cytokines. Third, DNA mimetics, transfected into cells, such as ISD and 2’3′cGAMP and HBV rcDNA bind cGAS resulting in activation of STING dependent activation of IRF3 and NF-kB producing inflammatory cytokines. cGAMP, cyclic GMP-AMP; cGAS, cyclic GMP-AMP synthase; HBV, hepatitis B virus; IFN, interferon; IRF3, IFN regulatory factor 3; ISD, IFN stimulatory DNA; MDA5, melanoma differentiation-associated gene 5; NF-kB, nuclear factor-kappa B; pgRNA, pregenomic RNA; rcDNA, relaxed circular DNA; RIG-I, retinoic acid-inducible gene; STING, stimulator of interferon genes; TBK1, TANK-binding kinase 1.
Conclusion
In patients and chimpanzees infected with HBV, previous studies have been unable to demonstrate a robust intrinsic innate immune response35 that can be seen with other hepatitis viruses such as HCV10 even though HBV can replicate to very high levels in hepatocytes. Recent studies taking advantage of improved cell culture models for HBV infection have provided evidence for activation of distinct components of innate immune signaling.17,44 Intriguingly, reports have been published that demonstrate that foreign DNA and HBV components can stimulate innate immune responses in hepatocytes in multiple models. These models have utilized controls to rule out the possibility of stimulation arising from contaminants in the viral preparation. In addition, data have been generated demonstrating that HBV blocks innate immune signaling pathways in hepatocytes. These observations have been studied in patient liver biopsies, where new hepatocytes were constantly being infected. It is therefore possible that the HBV virion possesses the ability to downregulate inflammatory gene induction through experimentally validated mechanisms and also through mechanisms that have yet to be characterized. This ability to downregulate innate response would support the characterization of HBV as a “stealth” virus when compared with other viruses such as HCV.
We also considered studies that have reported changes in the transcriptome following stimulation with HBV in in vitro models. When compared with cells stimulated with HCV, it is apparent that HBV stimulates a much weaker response that may be difficult to detect. It is evident that inflammatory pathways may be significantly stimulated by HBV to cause hepatitis in patients but this is in contrast to that observed following HCV infection where IFN pathways are most activated.10 Comparative microarray results of HBV- and HCV-infected PHHs demonstrate a much more robust and broad stimulation of the innate immune responses by HCV which may also support the notion that HBV is a “stealth” virus when compared with HCV.
Overall, HBV may be able to stimulate innate immunity through NF-κB-dependent pathways, whereas HCV mainly activates both IRF3 and NF-κB.10 In addition, other recent studies have demonstrated a role for the cGAS–STING–TBK1–IRF3 pathway in the detection of HBV.86,87 More recent data have provided evidence that in human hepatocytes, naked HBV genomic rcDNA is sensed in a cGAS-dependent manner, whereas the incorporation into nucleocapsids appears to shield the ability of this host defense pathway to sense this viral component. Furthermore, the cGAS–STING pathway has been shown to exhibit antiviral activity against HBV infection including reduction of viral cccDNA levels.26 The lack of strong IRF3 activation in hepatocytes could explain the difficulty in detecting a functional IFN response in humans. However, in humans, activation of an IFN-independent innate response in hepatocytes could drive the subsequent inflammatory responses that are responsible for recruitment of immune cells into the liver in HBV infected patients.
Further characterization of chemokine and cytokine responses to HBV using appropriate models will add crucial insight into the pathogenesis of HBV infection and possibly unravel novel innovative targets for curative strategies for HBV including the degradation of cccDNA. Specifically, targeted activation of functional antiviral pathways in hepatocytes may aid in the inactivation or degradation of nuclear cccDNA. Additionally, clearance of infected cells through adaptive responses may be facilitated through stimulation of cell-intrinsic inflammatory pathways that would expose infected hepatocytes as specific targets for elimination. Given the effectiveness of current suppressive antiviral therapy, it would be prudent to ensure that stimulation of host defense pathways in hepatocytes circumvents overt liver toxicity that can be seen during a “flare” that can result in a cure. It is anticipated that an increased understanding of host defense in hepatocytes may not only facilitate HBV cure but may also yield new preventive or therapeutic options for virus-induced HCC where perturbation of innate immune response has been shown to play a pathogenic role.
Main Concepts and Learning Points.
Hepatitis B infects hepatocytes and drives liver inflammation that can result in cirrhosis and hepatocellular carcinoma.
Current efforts to study hepatocyte cell-intrinsic innate immune responses are limited by the lack of a robust cell culture model with limitations from both the HBV viruses utilized and the cellular systems that are available.
Results from published data are highly variable spanning from studies reporting complete lack of detection and influence of HBV on antiviral host defenses to detectable and robust responses.
Hepatocytes express pathogen recognition receptors that can recognize components of HBV but their expression levels and functional significance in viral clearance and disease progression in patients with chronic infection are unknown.
A better understanding of HBV sensing and interference with innate immune responses may contribute to the discovery and development of novel antiviral strategies for viral cure.
Funding
E.T. acknowledges support from the NIH (R35GM124915) and Florida Department of Health Bankhead-Coley Cancer Research Program (7BC03). T.F.B. acknowledges support from the European Union (ERC-AdG-HEPCIR, ERC-PoC-2016-PRELICAN, and EU H2020-667273-HEPCAR) ARC, Paris and Institut Hospitalo-Universitaire, Strasbourg (TheraHCC IHUARC IHU201301187), U Strasbourg Foundation HEPKIN, the National Institutes of Health (NCI 1R21CA209940-01A1, NIAID R03AI131066, and NIAID 5U19AI123862-02), and the Institut Universitaire de France (IUF). This work has been published under the framework of the LABEX ANR-10-LABX-0028_HEPSYS and benefits from funding from the state managed by the French National Research Agency as part of the Investments for the Future Program.
Footnotes
Conflicts of Interest
The authors have no conflicts of interests to disclose.
References
- 1.McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology 2009;49(5, Suppl):S45–S55 [DOI] [PubMed] [Google Scholar]
- 2.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol 2014;28(05): 753–770 [DOI] [PubMed] [Google Scholar]
- 3.Burns GS, Thompson AJ. Viral hepatitis B: clinical and epidemiological characteristics. Cold Spring Harb Perspect Med 2014;4(12):a024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai KN, Kuo CF, Ou JJ. Mechanisms of hepatitis B virus persistence. Trends Microbiol 2018;26(01):33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gehring AJ. New treatments to reach functional cure: rationale and challenges for emerging immune-based therapies. Best Pract Res Clin Gastroenterol 2017;31(03):337–345 [DOI] [PubMed] [Google Scholar]
- 6.Chang J, Block TM, Guo JT. The innate immune response to hepatitis B virus infection: implications for pathogenesis and therapy. Antiviral Res 2012;96(03):405–413 [DOI] [PubMed] [Google Scholar]
- 7.Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J 2013;10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb C, Arbuthnot P. Activating the innate immune response to counter chronic hepatitis B virus infection. Expert Opin Biol Ther 2016;16(12):1517–1527 [DOI] [PubMed] [Google Scholar]
- 9.Gao B, Jeong WI, Tian Z. Liver: an organ with predominant innate immunity. Hepatology 2008;47(02):729–736 [DOI] [PubMed] [Google Scholar]
- 10.Thomas E, Gonzalez VD, Li Q, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology 2012;142(04):978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen HR. Emerging concepts in immunity to hepatitis C virus infection. J Clin Invest 2013;123(10):4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoukry NH. Hepatitis C vaccines, antibodies, and T cells. Front Immunol 2018;9:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Testoni B, Durantel D, Zoulim F. Novel targets for hepatitis B virus therapy. Liver Int 2017;37(Suppl 1):33–39 [DOI] [PubMed] [Google Scholar]
- 14.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology 2015. 479–480:672–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faure-Dupuy S, Lucifora J, Durantel D. Interplay between the hepatitis B virus and innate immunity: from an understanding to the development of therapeutic concepts. Viruses 2017;9(05): E95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol 2009;21(04):317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato S, Li K, Kameyama T, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 2015;42(01):123–132 [DOI] [PubMed] [Google Scholar]
- 18.Foy E, Li K, Sumpter R Jr, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A 2005;102(08):2986–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li K, Chen Z, Kato N, Gale M Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem 2005;280(17):16739–16747 [DOI] [PubMed] [Google Scholar]
- 20.Sumpter R Jr, Loo YM, Foy E, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 2005;79(05):2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol 2009;83(19):9824–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Q, Wang Q, Scott MJ, Billiar TR. Immune activation in the liver by nucleic acids. J Clin Transl Hepatol 2016;4(02):151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008;455(7213):674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii KJ, Coban C, Kato H, et al. A toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 2006;7(01):40–48 [DOI] [PubMed] [Google Scholar]
- 25.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009;138(03):576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verrier ER, Yim SA, Heydmann L, et al. Hepatitis B virus evasion from cyclic guanosine monophosphate-adenosine monophosphate synthase sensing in human hepatocytes. Hepatology 2018;68(05):1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng X, Xia Y, Serti E, et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology 2017;66(06):1779–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoneda M, Hyun J, Jakubski S, et al. Hepatitis B virus and DNA stimulation trigger a rapid innate immune responsethrough NF-κB. J Immunol 2016;197(02):630–643 [DOI] [PubMed] [Google Scholar]
- 29.Thomsen MK, Nandakumar R, Stadler D, et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 2016;64(03):746–759 [DOI] [PubMed] [Google Scholar]
- 30.Rall LB, Standring DN, Laub O, Rutter WJ. Transcription of hepatitis B virus by RNA polymerase II. Mol Cell Biol 1983;3(10):1766–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Standring DN, Rall LB, Laub O, Rutter WJ. Hepatitis B virus encodes an RNA polymerase III transcript. Mol Cell Biol 1983;3(10):1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa-2a and nucleos(t)ide analogues. J Infect Dis 2016; 213(02):224–232 [DOI] [PubMed] [Google Scholar]
- 33.Galle PR, Hagelstein J, Kommerell B, Volkmann M, Schranz P, Zentgraf H. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology 1994;106(03):664–673 [DOI] [PubMed] [Google Scholar]
- 34.Seeger C, Zoulim F, Mason WS. Hepadnaviruses In: Knipe DM, Howley PM, eds. Field Virology. 5th ed., Vol. 2 Philadelphia, PA: Lippincott Williams & Wilkins; 2007:2977–3027 [Google Scholar]
- 35.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A 2004;101(17):6669–6674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol 2005;79(15):9369–9380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science 1999;284(5415):825–829 [DOI] [PubMed] [Google Scholar]
- 38.Stacey AR, Norris PJ, Qin L, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol 2009;83(08):3719–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn C, Peppa D, Khanna P, et al. Temporal analysis of early immune responses in patients with acute hepatitis B virus infection. Gastroenterology 2009;137(04):1289–1300 [DOI] [PubMed] [Google Scholar]
- 40.Tan AT, Koh S, Goh W, et al. A longitudinal analysis of innate and adaptive immune profile during hepatic flares in chronic hepatitis B. J Hepatol 2010;52(03):330–339 [DOI] [PubMed] [Google Scholar]
- 41.Luangsay S, Ait-Goughoulte M, Michelet M, et al. Expression and functionality of Toll- and RIG-like receptors in HepaRG cells. J Hepatol 2015;63(05):1077–1085 [DOI] [PubMed] [Google Scholar]
- 42.Giersch K, Allweiss L, Volz T, et al. Hepatitis Delta co-infection in humanized mice leads to pronounced induction of innate immune responses in comparison to HBV mono-infection. J Hepatol 2015;63(02):346–353 [DOI] [PubMed] [Google Scholar]
- 43.Papatheodoridis G, Goulis J, Manolakopoulos S, et al. Changes of HBsAg and interferon-inducible protein 10 serum levels in naive HBeAg-negative chronic hepatitis B patients under 4-year entecavir therapy. J Hepatol 2014;60(01):62–68 [DOI] [PubMed] [Google Scholar]
- 44.Luangsay S, Gruffaz M, Isorce N, et al. Early inhibition of hepatocyte innate responses by hepatitis B virus. J Hepatol 2015;63(06):1314–1322 [DOI] [PubMed] [Google Scholar]
- 45.Bauer T, Sprinzl M, Protzer U. Immune control of hepatitis B virus. Dig Dis 2011;29(04):423–433 [DOI] [PubMed] [Google Scholar]
- 46.Villeneuve JP. The natural history of chronic hepatitis B virus infection. J Clin Virol 2005;34(Suppl 1):S139–S142 [DOI] [PubMed] [Google Scholar]
- 47.Verrier ER, Colpitts CC, Schuster C, Zeisel MB, Baumert TF. Cell culture models for the investigation of hepatitis B and D virus infection. Viruses 2016;8(09):E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas E, Liang TJ. Experimental models of hepatitis B and C – new insights and progress. Nat Rev Gastroenterol Hepatol 2016;13(06):362–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naik S, Russell SJ. Engineering oncolytic viruses to exploit tumor specific defects in innate immune signaling pathways. Expert Opin Biol Ther 2009;9(09):1163–1176 [DOI] [PubMed] [Google Scholar]
- 50.Keskinen P, Nyqvist M, Sareneva T, Pirhonen J, Melén K, Julkunen I. Impaired antiviral response in human hepatoma cells. Virology 1999;263(02):364–375 [DOI] [PubMed] [Google Scholar]
- 51.Melén K, Keskinen P, Lehtonen A, Julkunen I. Interferon-induced gene expression and signaling in human hepatoma cell lines. J Hepatol 2000;33(05):764–772 [DOI] [PubMed] [Google Scholar]
- 52.Israelow B, Narbus CM, Sourisseau M, Evans MJ. HepG2 cells mount an effective antiviral interferon-lambda based innate immune response to hepatitis C virus infection. Hepatology 2014;60(04):1170–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naka K, Dansako H, Kobayashi N, Ikeda M, Kato N. Hepatitis C virus NS5B delays cell cycle progression by inducing interferon-beta via toll-like receptor 3 signaling pathway without replicating viral genomes. Virology 2006;346(02):348–362 [DOI] [PubMed] [Google Scholar]
- 54.Yu S, Chen J, Wu M, Chen H, Kato N, Yuan Z. Hepatitis B virus polymerase inhibits RIG-I- and toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3. J Gen Virol 2010;91(Pt 8):2080–2090 [DOI] [PubMed] [Google Scholar]
- 55.Bility MT, Cheng L, Zhang Z, et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog 2014;10(03):e1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gripon P, Rumin S, Urban S, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A 2002;99(24):15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 2007;168(01):66–73 [DOI] [PubMed] [Google Scholar]
- 58.Andersson TB, Kanebratt KP, Kenna JG. The HepaRG cell line: a unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin Drug Metab Toxicol 2012;8(07):909–920 [DOI] [PubMed] [Google Scholar]
- 59.Michailidis E, Pabon J, Xiang K, et al. A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci Rep 2017;7(01):16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007;46(06):1759–1768 [DOI] [PubMed] [Google Scholar]
- 61.Tu T, Budzinska MA, Vondran FWR, Shackel NA, Urban S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via NTCP-dependent uptake of enveloped virus particles. J Virol 2018:JVI.02007–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li MMH, MacDonald MR, Rice CM. To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol 2015;25(06):320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011;472(7344):481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrus L, Marukian S, Jones CT, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology 2011;54(06):1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyun I. Policy: Regulate embryos made for research. Nature 2014;509(7498):27–28 [DOI] [PubMed] [Google Scholar]
- 66.Elaut G, Henkens T, Papeleu P, et al. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab 2006;7(06): 629–660 [DOI] [PubMed] [Google Scholar]
- 67.Rijntjes PJM, Moshage HJ, Yap SH. In vitro infection of primary cultures of cryopreserved adult human hepatocytes with hepatitis B virus. Virus Res 1988;10(01):95–109 [DOI] [PubMed] [Google Scholar]
- 68.Gripon P, Diot C, Thézé N, et al. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol 1988;62(11):4136–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ochiya T, Tsurimoto T, Ueda K, Okubo K, Shiozawa M, Matsubara K. An in vitro system for infection with hepatitis B virus that uses primary human fetal hepatocytes. Proc Natl Acad Sci U S A 1989; 86(06):1875–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shlomai A, Schwartz RE, Ramanan V, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A 2014;111(33):12193–12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X, Robotham JM, Lee E, et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog 2012;8(04):e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gómez-Lechón MJ, Donato MT, Castell JV, Jover R. Human hepatocytes in primary culture: the choice to investigate drug metabolism in man. Curr Drug Metab 2004;5(05):443–462 [DOI] [PubMed] [Google Scholar]
- 73.Sheahan T, Imanaka N, Marukian S, et al. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe 2014;15(02):190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwartz RE, Shan J, Duncan SA, Goessling W, Bhatia S. Engineering the microenvironment of differentiating IPS derived hepatocyte-like cells enables acquisition of an adult phenotype. Gastroenterology 2013;144(05):S1025 [Google Scholar]
- 75.Shan J, Schwartz RE, Ross NT, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol 2013;9(08):514–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Asumda FZ, Hatzistergos KE, Dykxhoorn DM, et al. Differentiation of hepatocyte-like cells from human pluripotent stem cells using small molecules. Differentiation 2018;101:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L, Dailey PJ, Gettie A, Blanchard J, Ho DD. The liver is a major organ for clearing simian immunodeficiency virus in rhesus monkeys. J Virol 2002;76(10):5271–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He XS, Nanda S, Ji X, Calderon-Rodriguez GM, Greenberg HB, Liang TJ. Differential transcriptional responses to interferon-alpha and interferon-gamma in primary human hepatocytes. J Interferon Cytokine Res 2010;30(05):311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakamoto N, Kanai T. Role of toll-like receptors in immune activation and tolerance in the liver. Front Immunol 2014;5:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol 2005;17(01):1–14 [DOI] [PubMed] [Google Scholar]
- 81.Pei RJ, Chen XW, Lu MJ. Control of hepatitis B virus replication by interferons and toll-like receptor signaling pathways. World J Gastroenterol 2014;20(33):11618–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang E, Lu M. Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis B virus (HBV) infection. Med Microbiol Immunol (Berl) 2015;204(01):11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J 2009;420(01):1–16 [DOI] [PubMed] [Google Scholar]
- 84.Gao D, Wu J, Wu YT, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013;341 (6148):903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schoggins JW, MacDuff DA, Imanaka N, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014;505(7485):691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cui X, Clark DN, Liu K, Xu XD, Guo JT, Hu J. Viral DNA-dependent induction of innate immune response to hepatitis B virus in immortalized mouse hepatocytes. J Virol 2015;90(01):486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dansako H, Ueda Y, Okumura N, et al. The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS J 2016;283(01):144–156 [DOI] [PubMed] [Google Scholar]
- 88.Guo F, Han Y, Zhao X, et al. STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob Agents Chemother 2015;59(02):1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity 2013; 38(05):870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Shi H, Wu J, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 2013;51(02):226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dempsey A, Bowie AG. Innate immune recognition of DNA: a recent history. Virology 2015;479–480:146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bürckstümmer T, Baumann C, Blüml S, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 2009;10(03):266–272 [DOI] [PubMed] [Google Scholar]
- 93.Chen J, Wu M, Zhang X, et al. Hepatitis B virus polymerase impairs interferon-α-induced STA T activation through inhibition of importin-α5 and protein kinase C-δ. Hepatology 2013;57(02):470–482 [DOI] [PubMed] [Google Scholar]
- 94.Mutz P, Metz P, Lempp FA, et al. HBV bypasses the innate immune response and does not protect HCV from antiviral activity of interferon. Gastroenterology 2018;154(06):1791.e22–1804.e22 [DOI] [PubMed] [Google Scholar]
- 95.Konerman MA, Lok AS. Interferon treatment for hepatitis B. Clin Liver Dis 2016;20(04):645–665 [DOI] [PubMed] [Google Scholar]
- 96.Rehermann B, Bertoletti A. Immunological aspects of antiviral therapy of chronic hepatitis B virus and hepatitis C virus infections. Hepatology 2015;61(02):712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014; 343(6176):1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ortega-Prieto AM, Dorner M. Immune evasion strategies during chronic hepatitis B and C virus infection. Vaccines (Basel) 2017;5(03):E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009;461 (7265):798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khan M, Syed GH, Kim SJ, Siddiqui A. Hepatitis B virus-induced Parkin-dependent recruitment of linear ubiquitin assembly complex (LUBAC) to mitochondria and attenuation of innate immunity. PLoS Pathog 2016;12(06):e1005693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suslov A, Wieland S, Menne S. Modulators of innate immunity as novel therapeutics for treatment of chronic hepatitis B. Curr Opin Virol 2018;30:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H, Ryu WS. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion. PLoS Pathog 2010;6(07):e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morikawa K, Shimazaki T, Takeda R, Izumi T, Umumura M, Sakamoto N. Hepatitis B: progress in understanding chronicity, the innate immune response, and cccDNA protection. Ann Transl Med 2016;4(18):337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lim KH, Park ES, Kim DH, et al. Suppression of interferon-mediated anti-HBV response by single CpG methylation in the 5′-UTR of TRIM22. Gut 2018;67(01):166–178 [DOI] [PubMed] [Google Scholar]
- 105.Wan Y, Cao W, Han T, et al. Inducible Rubicon facilitates viral replication by antagonizing interferon production. Cell Mol Immunol 2017;14(07):607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wentz MJ, Becker SA, Slagle BL. Dissociation of DDB1-binding and transactivation properties of the hepatitis B virus X protein. Virus Res 2000;68(01):87–92 [DOI] [PubMed] [Google Scholar]
- 107.Murphy CM, Xu Y, Li F, et al. Hepatitis B virus x protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Reports 2016;16(11):2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Decorsière A, Mueller H, van Breugel PC, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016;531(7594):386–389 [DOI] [PubMed] [Google Scholar]
- 109.Fernández M, Quiroga JA, Carreño V. Hepatitis B virus downregulates the human interferon-inducible MxA promoter through direct interaction of precore/core proteins. J Gen Virol 2003;84(Pt 8):2073–2082 [DOI] [PubMed] [Google Scholar]
- 110.Suslov A, Boldanova T, Wang X, Wieland S, Heim MH. Hepatitis B virus does not interfere with innate immune responses in the human liver. Gastroenterology 2018;154(06):1778–1790 [DOI] [PubMed] [Google Scholar]
- 111.Lu HL, Liao F. Melanoma differentiation-associated gene 5 senses hepatitis B virus and activates innate immune signaling to suppress virus replication. J Immunol 2013;191(06):3264–3276 [DOI] [PubMed] [Google Scholar]
- 112.Fletcher SP, Chin DJ, Ji Y, et al. Transcriptomic analysis of the woodchuck model of chronic hepatitis B. Hepatology 2012;56(03):820–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev 2011;243(01):99–108 [DOI] [PubMed] [Google Scholar]
- 114.Takeda K. Evolution and integration of innate immune recognition systems: the toll-like receptors. J Endotoxin Res 2005;11 (01):51–55 [DOI] [PubMed] [Google Scholar]
- 115.Kondo T, Kobayashi J, Saitoh T, et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A 2013;110(08):2969–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Du K, Liu J, Broering R, et al. Recent advances in the discovery and development of TLR ligands as novel therapeutics for chronic HBV and HIV infections. Expert Opin Drug Discov 2018;13(07): 661–670 [DOI] [PubMed] [Google Scholar]


