Abstract
Proper neural function depends on the correct specification of individual neural fates, controlled by combinations of neuronal transcription factors. Different neural types are sequentially generated by neural progenitors in a defined order, and this temporal patterning process can be controlled by Temporal Transcription Factors (TTFs) that form temporal cascades in neural progenitors. The Drosophila medulla, part of the visual processing center of the brain, contains more than 70 neural types generated by medulla neuroblasts which sequentially express several TTFs, including Homothorax (Hth), eyeless (Ey), Sloppy paired 1 and 2 (Slp), Dichaete (D) and Tailless (Tll). However, it is not clear how such a small number of TTFs could give rise to diverse combinations of neuronal transcription factors that specify a large number of medulla neuron types. Here we report how temporal patterning specifies one neural type, the T1 neuron. We show that the T1 neuron is the only medulla neuron type that expresses the combination of three transcription factors Ocelliless (Oc or Otd), Sox102F and Ets65A. Using CRISPR-Cas9 system, we show that each transcription factor is required for the correct morphogenesis of T1 neurons. Interestingly, Oc, Sox102F and Ets65A initiate expression in neurons beginning at different temporal stages and last in a few subsequent temporal stages. Oc expressing neurons are generated in the Ey, Slp and D stages; Sox102F expressing neurons are produced in the Slp and D stages; while Ets65A is expressed in subsets of medulla neurons born in the D and later stages. The TTF Ey, Slp or D is required to initiate the expression of Oc, Sox102F or Ets65A in neurons, respectively. Thus, the neurons expressing all three transcription factors are born in the D stage and become T1 neurons. In neurons where the three transcription factors do not overlap, each of the three transcription factors can act in combination with other neuronal transcription factors to specify different neural fates. We show that this way of expression regulation of neuronal transcription factors by temporal patterning can generate more possible combinations of transcription factors in neural progeny to diversify neural fates.
Keywords: temporal patterning, neuroblasts, Temporal Transcription Factors, neuronal differentiation, neuronal morphogenesis
Introduction
A great diversity of neurons and glia constitute the complex nervous system and form the basis of neural function. Understanding the specification of different neural types during development is a question of great interest. Studies in C. elegans, Drosophila and vertebrates have shown that in post-mitotic neurons, neuronal transcription factors (TFs) act in combinations to control the expression of neuron type-specific effector genes to define functional and structural identities of neurons (Allan and Thor, 2015; Hobert and Kratsios, 2019). These neuronal TFs include those that control functional identities such as neurotransmitter / receptor expression and electrophysiological properties, as well as those TFs that control neuronal morphogenesis during the development, specifically termed morphology TFs (Allan and Thor, 2015; Enriquez et al., 2015; Hobert and Kratsios, 2019). In different neural types, the same phenotypic traits can be controlled by distinct combinations of TFs (Serrano-Saiz et al., 2013; Konstantinides et al., 2018). Neurons are generated from a small pool of neural progenitors during development. Temporal patterning and spatial patterning of neural progenitors have been shown to contribute to the generation of neural diversity in both vertebrates and invertebrates (Guillemot, 2007; Lin and Lee, 2012; Allan and Thor, 2015; Holguera and Desplan, 2018). There have been great advances in the study of temporal patterning of neuroblasts in Drosophila (Doe, 2017; Miyares and Lee, 2019). Drosophila embryonic ventral nerve cord neuroblasts, larval medulla neuroblasts, and the Intermediate Neural Progenitor (INPs) of central brain type II neuroblasts are temporally patterned by the sequential expression of Temporal Transcription Factors (TTFs), although different sequences of TTFs are used in different regions (Isshiki et al., 2001; Bayraktar and Doe, 2013; Li et al., 2013; Suzuki et al., 2013; Bertet et al., 2014). In the larval central brain and nerve cord neuroblasts, temporal patterning is controlled by complementary gradients of two RNA binding proteins: IGF-II mRNA-binding protein (Imp) and Syncrip (Syp), as well as candidate TTFs such as D, Cas, Svp, Broad and Eip93 (Maurange, Cheng and Gould, 2008; Liu et al., 2015; Ren et al., 2017; Syed, Mark and Doe, 2017; Pahl, Doyle and Siegrist, 2019).
What is the relationship between TTFs expressed in neuroblasts and neuronal TFs that specify neural fates? How does temporal patterning of neuroblasts generate combinatorial codes of TFs in neurons to determine different neural types? In many cases, the number of TTFs is much less than the number of neural fates specified. Studies in the Drosophila ventral nerve cord and central brain have defined regulatory logics by which the expression of neuronal TFs are controlled by integration of temporal, sub-temporal and spatial cues (Baumgardt et al., 2009; Karlsson, Baumgardt and Thor, 2010; Liu et al., 2019; Sen et al., 2019). In the Drosophila medulla, the largest region of the optic lobe, more than 70 neural types are generated by medulla neuroblasts, which sequentially express TTFs including Homothorax (Hth), eyeless (Ey), Sloppy paired 1 and 2 (Slp), Dichaete (D) and Tailless (Tll) as they age (Li et al., 2013; Suzuki et al., 2013). One possible way for how TTFs in neuroblasts control neuronal TFs is that one neuronal TF can only be activated by one TTF and expressed in only one temporal stage. There are examples for this kind of regulation in the Drosophila medulla: Bsh is only expressed in the Notch-on neuronal progeny of GMCs derived from Hth neuroblasts, and Hth is required for Bsh expression (Li et al., 2013; Suzuki et al., 2013). Bsh is required and sufficient for the Mi1 neuron fate (Hasegawa et al., 2013). However, through this mechanism, the number of possible combinations of transcription factors generated by temporal patterning of neuroblasts will be greatly limited. In this work, we studied how the temporal patterning of medulla neuroblasts generates the TF combinatorial code that controls the morphogenesis of T1 neurons.
T1 neurons are a class of mysterious neurons that connect the lamina and the medulla part of the optic lobe. They are uni-columnar neurons with one in each of the 800 columns of the medulla (Fischbach and Dittrich, 1989). The cell body of the T1 neuron is found in the medulla cortex, and its axon branches in a characteristic “T” shape on the distal surface of the medulla. One branch projects through the outer optic chiasm back to the lamina and then forms a basket like structure of processes surrounding each lamina cartridge. The other branch arborizes in the M2 layer of the medulla with a dense bush like structure (Fischbach and Dittrich, 1989) (Fig. 1A). T1 neuron is post-synaptic to amc (lamina amacrine cells), and amc/T1 pathway was shown to enhance the lamina neuron L1 motion detection pathway at intermediate contrast (Rister et al., 2007). Depolarizing T1 neurons affected the flight steering responses to visual stimuli (Tuthill et al., 2013).
Figure 1. Oc, Ets65A and Sox102F are expressed in T1 neurons.
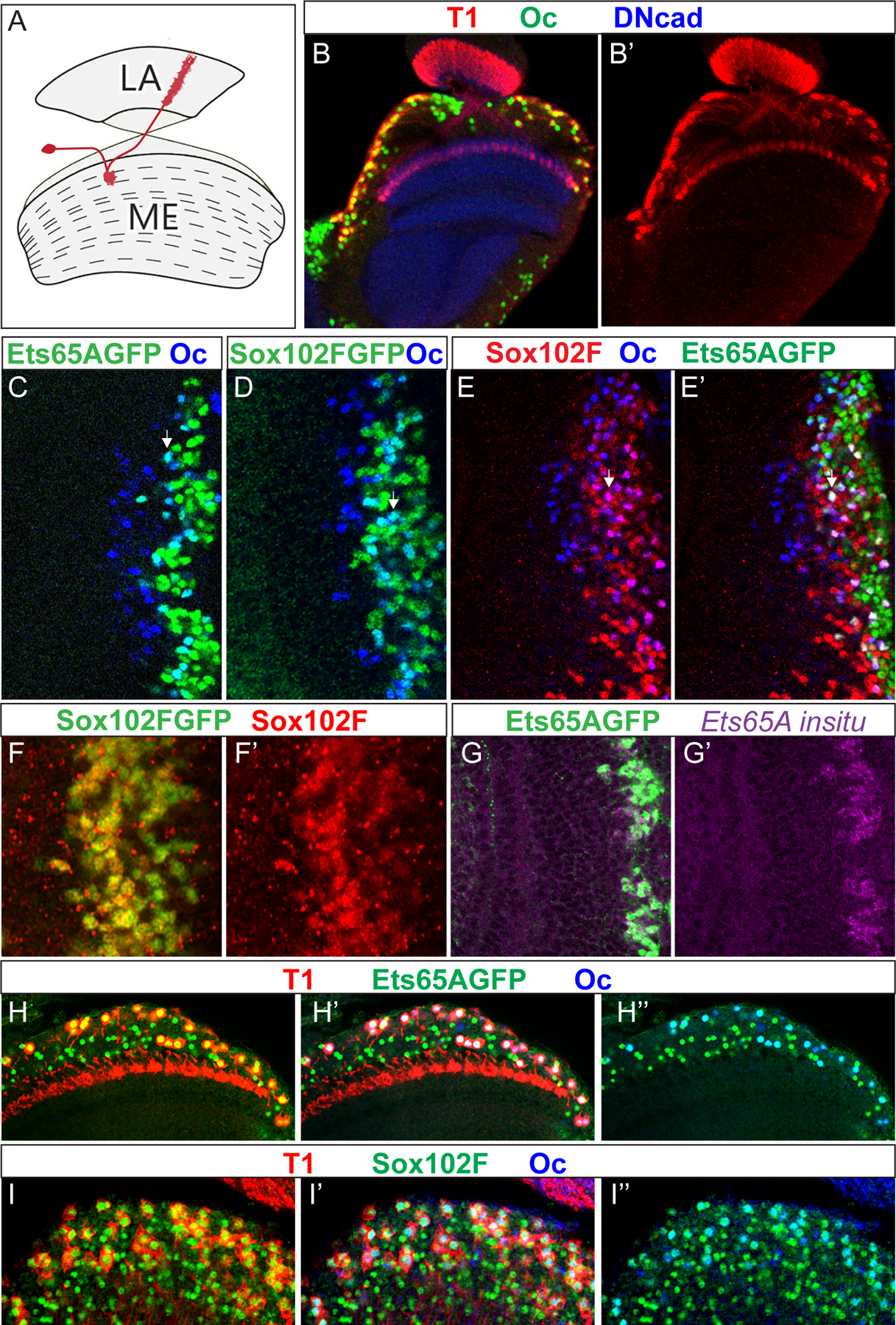
(A) A schematic drawing of the normal T1 morphology in red. LA: lamina. ME: Medulla. (B-B’) The expression of Oc (green) in the adult medulla carrying T1LexA>LexopRFP (red). (C,D) The expression of Ets65A::GFP (green in C) or Sox102F::GFP (green in D) partially overlaps with that of Oc (blue) in the 3rd instar larval medulla. The early-born (deeper layer) neurons are to the left, and later born (more superficial layers) neurons are to the right. Neurons expressing both appear cyan in the overlay. White arrow indicates one example. (E,E’) Neurons that express both Sox102F (red) and Oc (blue) also express Ets65A::GFP (green). (F,F’) Sox102F::GFP (green) is expressed in the same neurons as Sox102F immunostaining (red) in the 3rd instar larval medulla. (G,G’) In the 3rd instar larval medulla, the expression of Ets65A::GFP (green) is in the nucleus of the same cells that express Ets65A mRNA as shown by in situ hybridization against all isoforms (purple). (H–H”) T1 neurons (red) in the adult medulla express both Ets65A::GFP (green) and Oc (blue). (I,I’) T1 neurons (red) in the adult medulla express both Sox102F (green) and Oc (blue).
Through screening antibodies and GFP fusion lines, we found that Ocelliless (oc), Sox102F and Ets65A are expressed in T1 neurons, and the combination of these three TFs can distinguish T1 neurons from all other medulla neurons. Using CRISPR-Cas9 system, we generated bi-allelic somatic mutations of each of the three TFs in T1 neurons, and showed that knock-down of each one affected different aspects of the T1 neuron morphology. We next examined how the expression of each TF is controlled by temporal patterning to generate the combination code. We found that Oc expression in neurons starts in the Ey temporal stage, and continues in the Slp and D temporal stages, and Ey is required for the initiation of Oc expression in neurons; while Sox102F expression in neurons starts in the Slp temporal stage, and continues in the D temporal stage, and Slp is required for initiating the expression of Sox102F in neurons; finally, Ets65A is expressed in subsets of medulla neurons born in the D and later temporal stages, and D is required for the expression of Ets65A. Thus, the three TFs that control T1 neuron morphology initiate their expression in neurons beginning at different temporal stages controlled by different TTFs, but each of them spans a few temporal stages, and the neurons expressing all three TFs are born in the D stage and become T1 neurons. In neurons where the three transcription factors do not overlap, each of the three TFs could also act with other neuronal TFs to specify different neural fates. In this way, more combinations of TFs can be generated through temporal patterning.
Results
A combination of three transcription factors specifically marks T1 neurons
We screened for transcription factors expressed in T1 neurons by staining a T1-LexA lexopRFP line (Pfeiffer et al., 2010) with available antibodies and found that Oc is expressed in T1 neurons (Fig. 1B–B’). However, not all Oc expressing neurons become T1 neurons. In parallel, in a screen of a collection of GFP tagged TFs expressed under native control (created by Spokony, R. and White, K), we found that Oc expression partially overlaps with that of three transcription factors fused to GFP: Ets65A, Sox102F and Fork head (Fkh) in the larval medulla (Fig. 1C,D, Sup. Fig. S1C). Among them, Fkh::GFP only overlaps with Oc expression in the posterior Dpp domains of the medulla, and is not expressed in T1 neurons (Sup. Fig. S1A–D). We confirmed that Sox102F::GFP and Ets65A::GFP are expressed in the same cells as the endogenous gene in the larval medulla, by Immunostaining (Sox102F) or In situ hybridization (Ets65A) (Fig. 1 F–G’). Triple labeling of Sox102F, Oc and Ets65A::GFP in the larval medulla showed that some medulla neurons express all three transcription factors (Fig. 1 E,E’). In the adult medulla, both Ets65A::GFP and Sox102F are expressed in T1 neurons together with Oc (Fig. 1H–I”). All neurons expressing both Ets65A::GFP and Oc are T1 neurons, and vice versa (Fig. 1H–H”). Similarly, all neurons expressing both Sox102F and Oc are T1 neurons, and vice versa (Fig 1I–I”). Thus, T1 neurons are the only cells in the adult medulla that express Oc, Ets65A and Sox102F together.
Oc, Sox102F and Ets65A are each required for T1 neuron morphogenesis
To examine the function of each of the three TFs in the specification and morphogenesis of T1 neurons, we used the CRISPR-Cas9 system to generate bi-allelic somatic mutations of each TF gene in medulla neuroblasts and neurons. The three transcription factors are not expressed in neuroblasts (Sup. Fig. S1 E–F”‘), therefore the phenotype will be specifically post-mitotic phenotype in neurons. We designed two gRNAs for each gene and used ey>Gal4 that is expressed in early stage medulla neuroblasts to drive the two gRNAs and UAS-Cas9. In subsets of medulla neuroblasts, bi-allelic somatic mutations of the corresponding TF gene will be generated, so that later stage neuroblasts and their progeny will be mutant for each of the three TFs. CRISPR against Sox102F or oc removed the expression of the corresponding TF in a large subset of T1 neurons as assessed by immunostaining. In the adult medulla (n=8) of oc-CRISPR flies, Oc expression is lost in ~59(±13) percent of T1 neurons (Fig. 2B,C, compare to no-gRNA control in A). In Sox102F-CRISPR adult medulla (n=9), ~52(±9) percent of T1 neurons lose Sox102F expression (Fig. 2E,F, compare to no-gRNA control in D). For Ets65A, because the antibody is not available, and in situ hybridization in combination with RFP staining didn’t work well in adult brains, we used a different method to examine whether Ets65A can be knocked down by CRISPR. We generated GFP marked clones in which Gal4 drives UASCas9 and UASgRNA against Ets65A. In such CRISPR clones (14 clones in 12 lobes), Ets65A expression was partially knocked down as assessed by in situ hybridization against all isoforms in larval brains (Fig. 2G–G”).
Figure 2. Knocking down of Oc, Sox102F and Ets65A using CRISPR-Cas9 system.
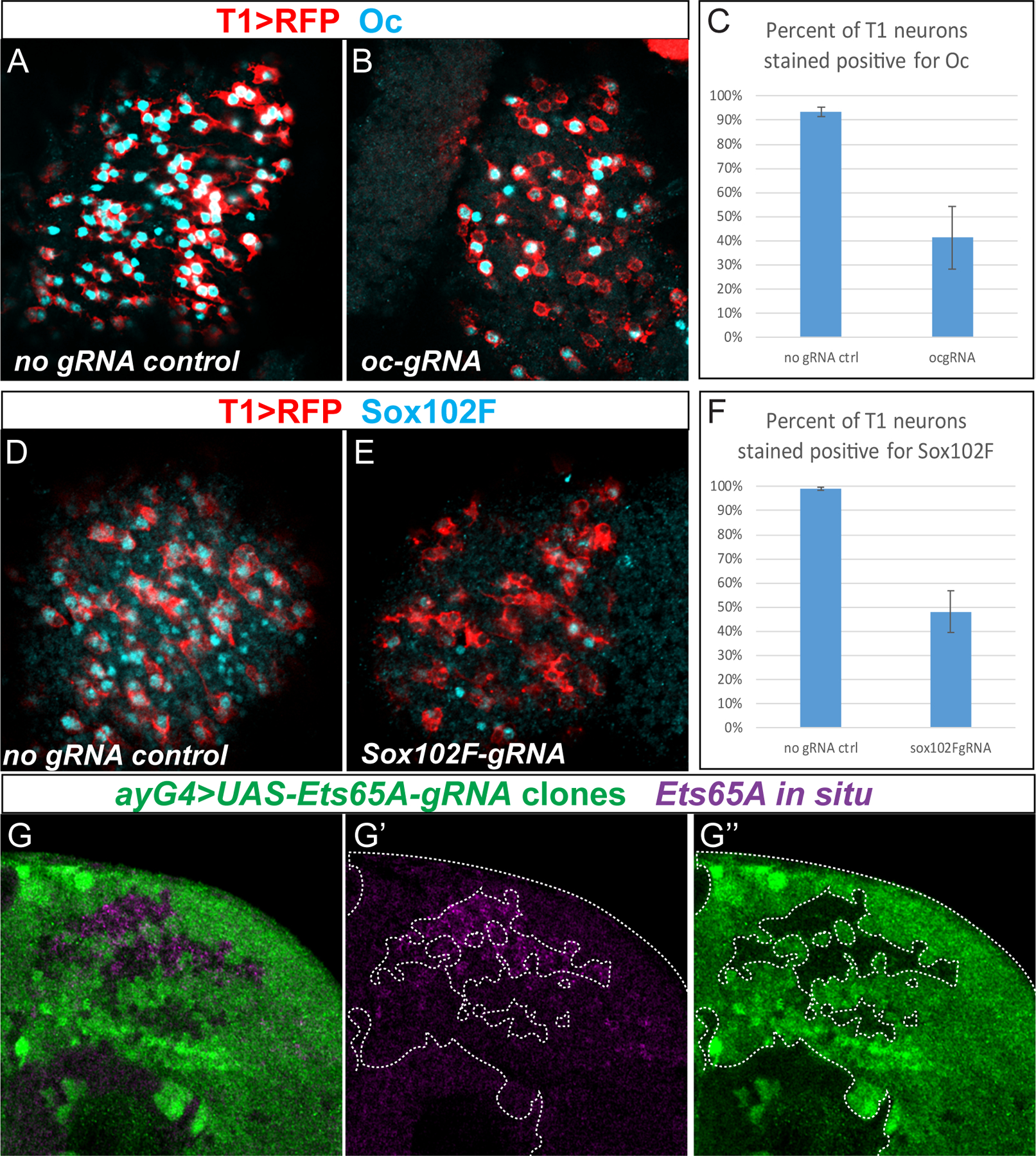
(A) Oc expression (cyan) in control medulla (genotype T1LexA>LexopRFP/ UASCas9; eyGal4/+) with T1 labeled in red. (B) Oc expression (cyan) in ocgRNA medulla (genotype T1LexA>LexopRFP /UASCas9; eyGal4/ocgRNA) with T1 labeled in red. (C) Quantification of percentage of T1 neurons stained positive for Oc in control and ocgRNA brains, n=4 for control, n=8 for ocgRNA. Less than 100% ( ~93±1.8 %) of wild types T1s are stained positive for Oc, possibly due to focal planes or weak antibody signals, etc. Oc is lost in ~59±13 % of T1 neurons when Oc is knocked down with ocgRNA (t-test: p=6.24×10−6 ). (D) Sox102F expression (cyan) in control medulla (genotype T1LexA>LexopRFP/ UASCas9; eyGal4/+) with T1 labeled in red. (E) Sox102F expression (cyan) in Sox102FgRNA medulla (genotype T1LexA>LexopRFP/ UASCas9; eyGal4/Sox102FgRNA) with T1 labeled in red. (F) Quantification of percentage of T1 neurons stained positive for Sox102F in control and Sox102FgRNA brains. n=4 for control, n=9 for Sox102F-gRNA. Sox102F is lost in ~52±9% of T1 neurons in Sox102FgRNA brains. (t-test: p=8.39×10−8 ). (G–G”) In clones (marked in green) where Cas9 and Ets65AgRNA are expressed (genotype yw hs FLP; act>y+>Gal4 UAS GFP / UASCas9; UASEts65AgRNA/+), in situ hybridization of Ets65A (purple) in 3rd instar larval medulla showed that Ets65A mRNA is partially knocked down in the clones. Note the clone margin perfectly correlates with that of higher Ets65A expression.
Next we observed how loss of each of the three TFs affects T1 neuron morphology. We used the MultiColor FlpOut (MCFO) approach (Nern, Pfeiffer and Rubin, 2015) to densely (Fig. 3 A,E,I,M) or sparsely label (Fig. 3B,F,J,N) T1 neurons in either no gRNA control or CRISPR mosaic mutant flies, and used immunostaining to identify the mutant neurons. All T1 neurons in the no gRNA control optic lobes (4 densely labeled and 10 sparsely labeled) exhibit normal morphology: they form a bush like dense structure of arborizations in the M2 layer, and also extend an axon back into the lamina neuropil (Fig. 3A,B,D). In oc-CRISPR densely labeled optic lobes (n=8), some T1 neurons exhibit abnormal morphology: their arborizations in the medulla appear dis-organized, often go beyond M2, and do not form the characteristic dense bush like structure (Fig. 3E,H). Their axons still project back to the lamina. To reconstruct the whole neuron morphology and track the TF expression in the cell body, we examined sparsely labeled optic lobes (n=10) where only a few neurons were labeled per brain. In these brains, 11 out of 13 T1 neurons that lose the Oc expression exhibit the abnormal medulla arborizations (Fig. 3F, G–G”,H). These neurons still retain Sox102F expression (Fig. 3G’). In Sox102F-CRISPR densely (n=5) or sparsely labeled optic lobes (n=11): neurons marked by T1 LexA that lose the expression of Sox102F (15 out of 15 such neurons reconstructed), have large and loose medulla arborizations resembling that of multi-columnar medulla intrinsic neurons (Fig. 3I–L). Some mutant neurons (3 out of 15) lost their projection to the lamina. These neurons still express Oc (Fig. 3K”). In Ets65A-CRISPR optic lobes (n=16 in total for two experiments), since Ets65A antibody is not available, we were not able to determine the percentage of mutant neurons, so we quantified the number of T1 neurons that showed a phenotype versus the total T1 neurons examined. All T1 neurons still form a bush shape of arborizations at M2, but 32±8% of them (42 out of 103 T1s in experiment 1, and 24 out of 98 T1s in experiment 2) extend a single long branch that reaches M6 or deeper layer. They still have an axon that projects back to the lamina (Fig. 3M,N,P). These neurons still express Sox102F and Oc (Fig. 3O–O”). These data suggest that each of the three transcription factors is required for certain aspects of the T1 neuron morphology.
Figure 3. Oc, Ets65A and Sox102F are each required for T1 neuron morphogenesis.
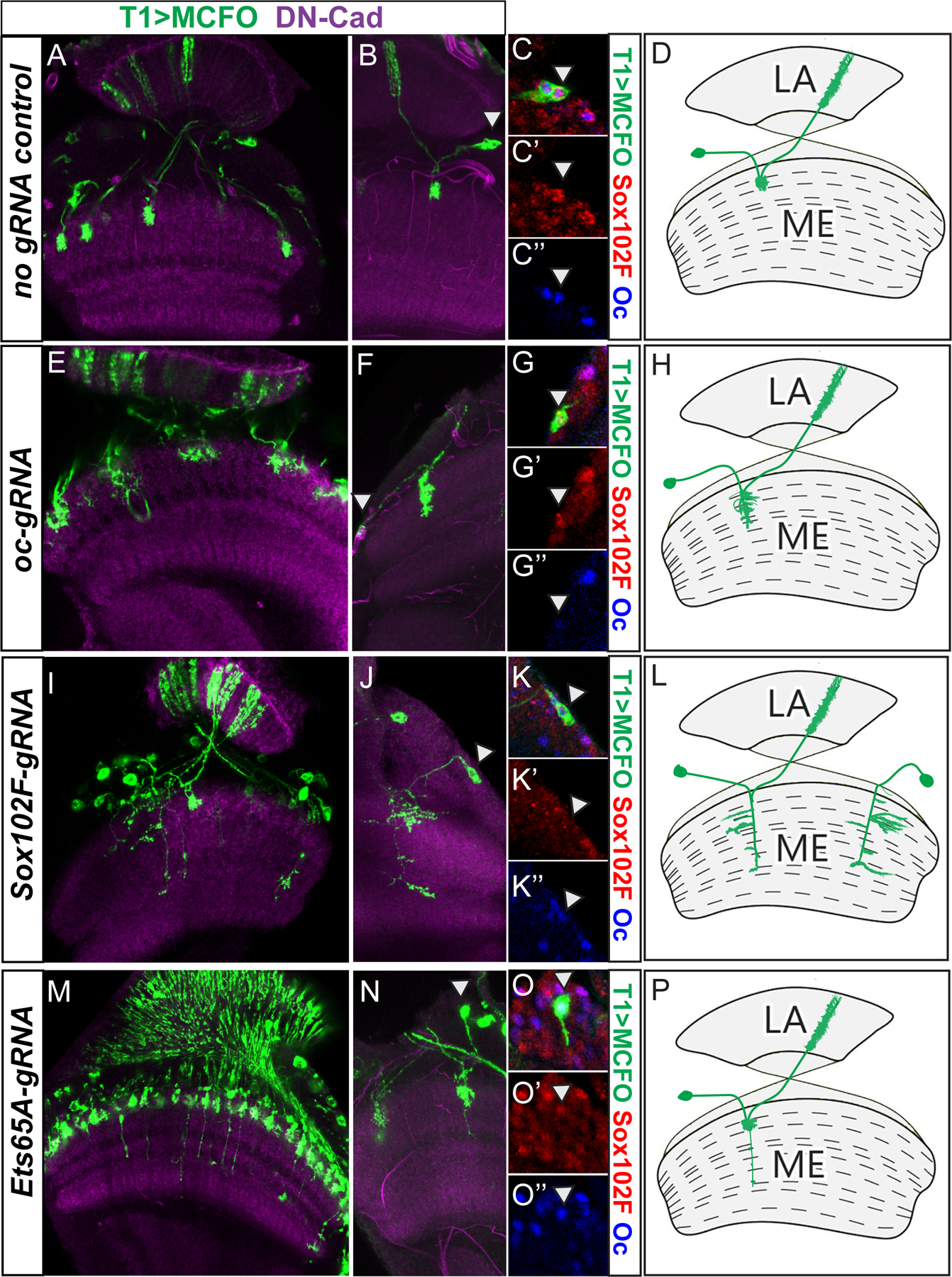
MCFO is used to visualize wild type or mutant T1 morphology with HA staining in green. In all panels, white arrowhead indicates the location of the cell body. (A–D) no gRNA wild type control (genotype: hsFLP/+; T1lexA /UASCas9; ey>Gal416F10, 13xLexAop2->dSTOP>-myr::smGFP-HA /+). (A) Densely labeled. (B) Sparsely labeled. (C–C”) Magnified view of the cell body marked with an arrowhead in (B) showing Sox102F (red) and Oc (blue) staining. (D) Drawing of the wild type T1 morphology. (E–H) oc is knocked down using ocgRNA (Genotype: hsFLP/+; T1lexA /UASCas9; ey>Gal416F10, 13xLexAop2->dSTOP>-myr::smGFP-HA / UAS-ocgRNA). (E) Densely labeled. (F) Sparsely labeled. (G-G”) Magnified view of the cell body marked with arrowhead in (F) showing Sox102F (red) and Oc (blue) staining. (H) Drawing of typical oc mutant T1 morphology. (I–L) Sox102F is knocked down using Sox102FgRNA (Genotype: hsFLP/+; T1lexA /UASCas9; ey>Gal416F10, 13xLexAop2->dSTOP>-myr::smGFP-HA / UAS-Sox102FgRNA). (I) Densely labeled. (J) Sparsely labeled. (K–K”) Magnified view of the cell body marked with arrowhead in (J) showing Sox102F (red) and Oc (blue) staining. (L) Drawing of typical Sox102F mutant T1 morphology. (M–P) Ets65A is knocked down using Ets65AgRNA (Genotype: hsFLP/+; T1lexA /UASCas9; ey>Gal416F10, 13xLexAop2->dSTOP>-myr::smGFP-HA / UAS-Ets65AgRNA). (M) Densely labeled. (N) Sparsely labeled. (O–O”) Magnified view of the cell body marked with arrowhead in (N) showing Sox102F (red) and Oc (blue) staining. (P) Drawing of typical Ets65A mutant T1 morphology.
The three transcription factors initiate expression in neurons beginning at different temporal stages
To understand how the combinatorial code of the three transcription factors is generated, we examined the expression pattern of the three transcription factors in the larval medulla. In larval brain, Oc and Ets65A::GFP are expressed in populations of medulla neurons that partially overlap: Oc is expressed in early-born to mid-born layers of medulla neurons, and Ets65A::GFP is expressed in mid to late-born neurons at more superficial layers, and there are one layer of neurons that express both in the middle (Fig. 1C). Similarly for Oc and Sox102F: there are some neurons that express Oc but not Sox102F, some neurons that express Sox102F but not Oc, and some neurons that express both TFs (Fig. 1D,E). To determine which temporal stage(s) the transcription factors are expressed in, we first characterized three Gal4 lines that can mark neurons born at different temporal stages. Through screening Flylight Gal4 lines (Jenett et al., 2012) (Li et al., 2014) generated using genomic DNA fragments from the temporal genes ey, slp, and D, we identified three Gal4 lines that initiate lacZ reporter expression at the same time as the corresponding temporal gene (ey-R16F10>Gal4, slp-R35H02>Gal4, and D-R12G08>Gal4) (Fig. 4A–C’). Among them, the D-R12G08>Gal4 is less than ideal, and includes the majority but not all of the D expressing neuroblasts. Due to the persistence of the LacZ protein, LacZ reporter is maintained in all progeny of the corresponding temporal stage, as well as in all later stage neuroblasts and progeny (Fig. 4A–C, Fig. 5F). However, through the nested expression patterns of the three Gal4 lines, we can deduce which temporal stage(s) the neurons are generated in. Neurons that express Ets65A::GFP are largely contained within the window of D>Gal4 driving UAS-NuLacZ (Fig. 4D, Fig. 5F), suggesting that they are born in the D (and possible later) stage(s). All Ets65A::GFP expressing neurons also express slp>>LacZ due to the LacZ persistence, but Ets65A::GFP is not expressed at the time when slp>Gal4 initiates its expression (Sup. Fig. S2 D,D’). In contrast, only some of the Sox102F::GFP expressing neurons express D>Gal4 driving UAS-NuLacZ, but all of them express slp>Gal4 driving UAS-NuLacZ or ey>Gal4 driving UAS-NuLacZ (Fig. 4 E,G, Sup. Fig. S2A–A’, E–E’, Fig. 5F). Similarly, only some of the Oc expressing neurons express D>Gal4 driving UAS-NuLacZ, but all of them express slp>Gal4 driving UAS-NuLacZ, or ey>Gal4 driving UAS-NuLacZ (Fig. 4F, H, I, Sup. Fig. S2B–C’, Fig. 5F). These results would suggest that the generation of Sox102F or Oc expressing neurons started in the Slp stage, and continued in the D stage. However, there is a significant overlap between the expression of Ey and Slp in neuroblasts, and the transition stage (late Ey stage) neuroblasts have already started a low level of Slp expression (Li et al., 2013), so the slp>Gal4 is already turned on in the transition stage neuroblasts and thus their progeny (Fig. 5F). Therefore, it is not clear whether Sox102F and Oc started their expression in neurons born in the Slp stage or the transition stage (late Ey stage). To address this question, we tested which TTF is required for the expression of each of the three transcription factors.
Figure 4. The three transcription factors initiate expression in neurons born at different temporal stages.
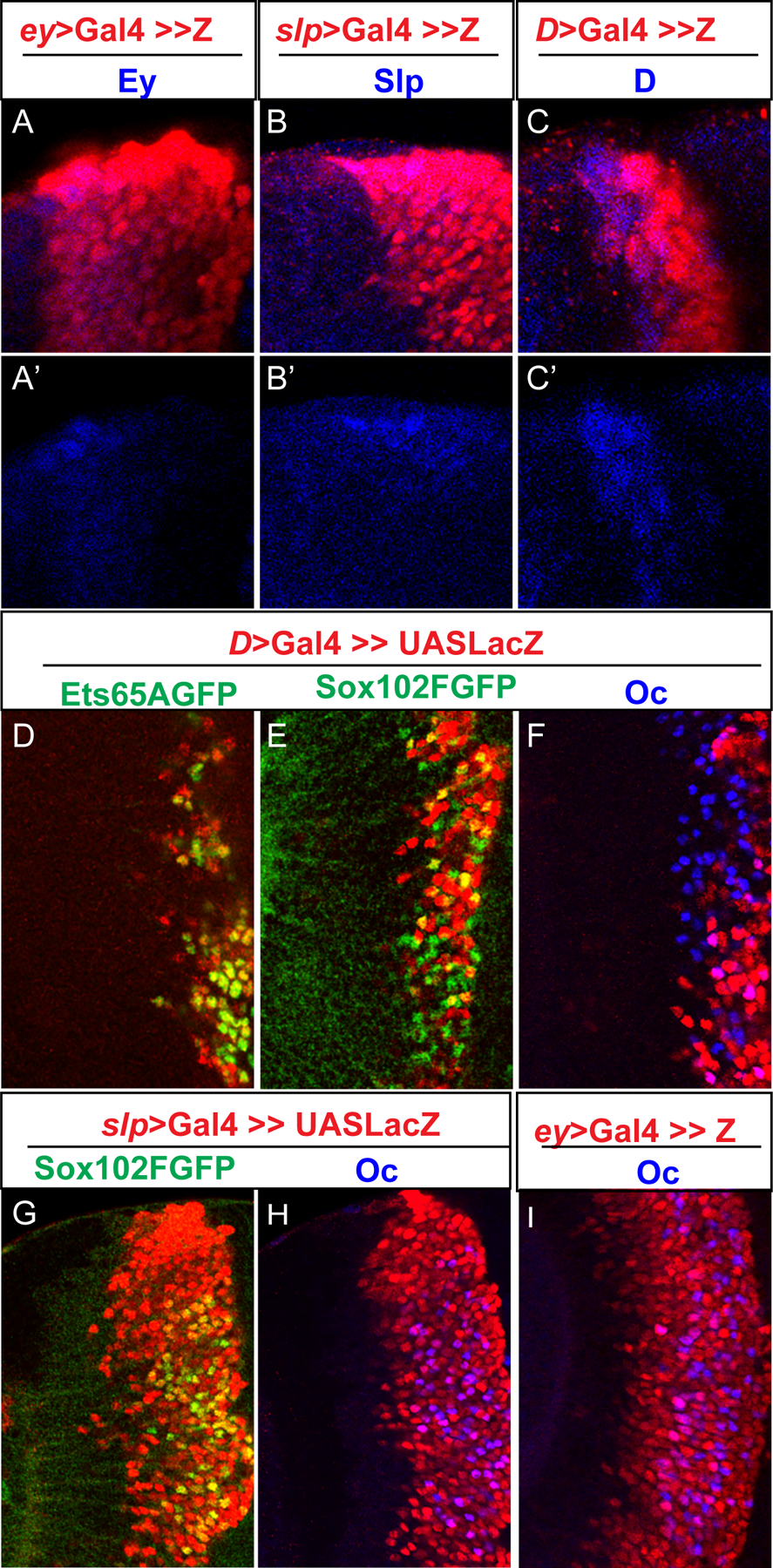
All images shown are of 3rd instar larval medulla. (A–A’) UAS-NuLacZ reporter (red) driven by an ey> Gal4 initiates expression at the same time as endogenous Ey expression (blue). (B–B’) UAS-NuLacZ reporter (red) driven by a slp> Gal4 initiates expression at the same time as endogenous Slp expression (blue). (C–C’) UAS-NuLacZ reporter (red) driven by a D> Gal4 initiates expression at the same time as endogenous D expression (blue). (D–F) D>Gal4 is used to drive UAS-NuLacZ (red) in D stage (and all later stage) neuroblasts and progeny. (D) Almost all Ets65A::GFP (green) expressing cells express D>> LacZ. (E) Some Sox102F::GFP (green) expressing cells express D>> LacZ (yellow cells), while others do not (green cells). (F) Some Oc (blue) expressing cells express D>>LacZ (purple cells), while others do not (blue cells). (G-H) slp>Gal4 is used to drive UAS-NuLacZ (red) expression. (G) All neurons expressing Sox102F::GFP (green) also express slp>>LacZ. (H) All neurons expressing Oc (blue) also express slp>>LacZ. (I) ey>Gal4 is used to drive UAS-NuLacZ (red) in Ey stage (and all later stage) neuroblasts and progeny. All Oc (blue) expressing neurons also express LacZ(red).
Figure 5. D, Slp and Ey are required for initiation of Ets65A, Sox102F and Oc expression in neurons, respectively.
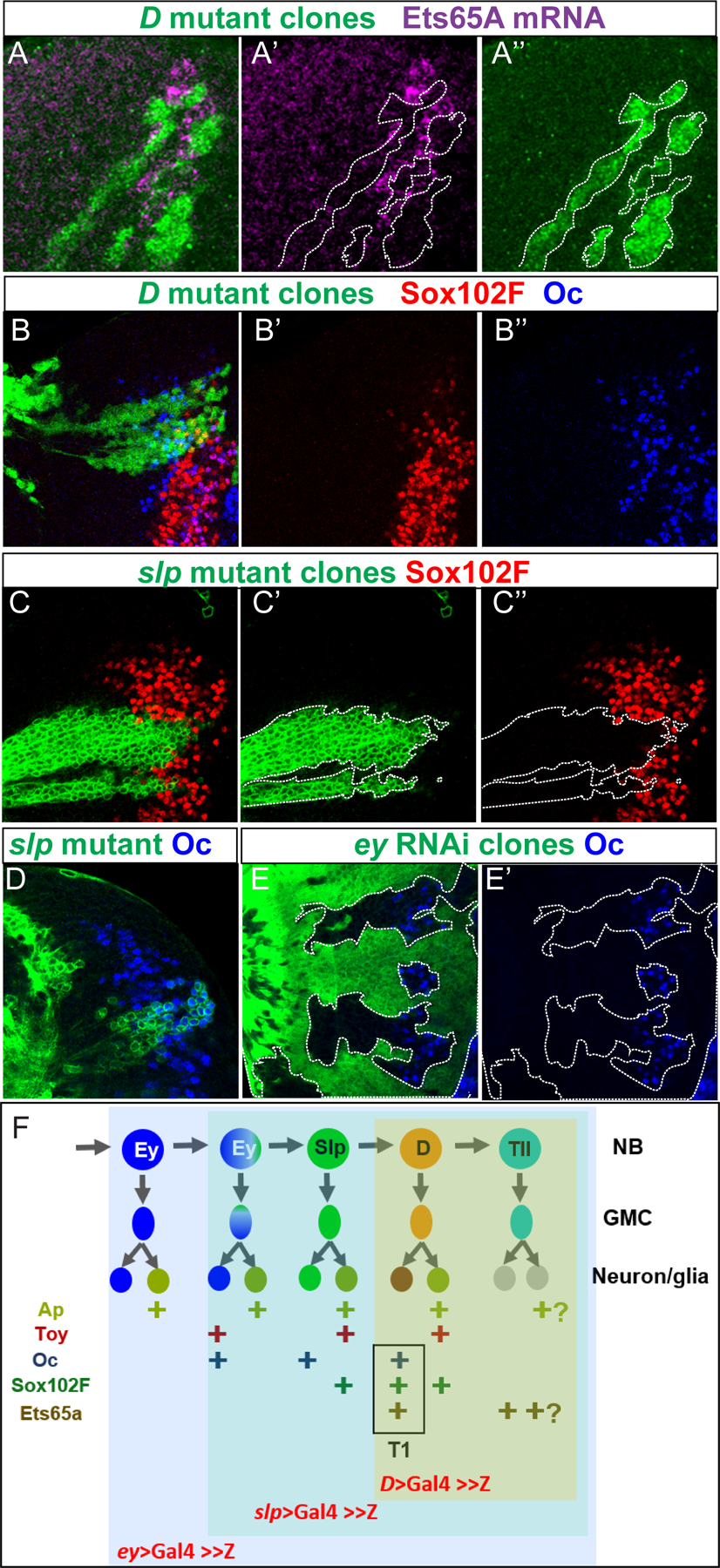
All images shown are of 3rd instar larval medulla. (A–A”) In situ hybridization of Ets65A mRNA (purple) in brains with D mutant clones marked by GFP (green). (B–B”) Immunostaining of Sox102F (red) and Oc (blue) in brains with D mutant clones marked by GFP (green). (C–C”) Immunostaining of Sox102F (red) in brains with slp mutant clones marked by GFP (green). (D) Immunostaining of Oc (blue) in brains with slp mutant clones marked by GFP (green). (E,E’) Immunostaining of Oc (blue) in brains with clones expressing eyRNAi driven by actin>Gal4 marked by GFP (green). (F) A model summarizing how temporal patterning generate different combinations of transcription factor expression in medulla neurons. Oc expression is started in late Ey stage, and is expressed in Notch-off progeny of Ey, Slp and D stages; Sox102F is expressed in Notch-on neurons born in the Slp and D temporal stages, and also in Notch-off neurons born in the D stage; Ets65A is expressed in Notch-off neurons born in the D stage, and continue to be expressed in progeny born at later stages. The Ap and Toy expression pattern are based on (Li et al., 2013).
The three transcription factors require different neuroblast TTFs to initiate expression in neurons
To examine whether and how the expression of the three T1 transcription factors are controlled by neuroblast TTFs, we examined their expression in mutant clones of temporal genes encoding TTFs. The expression of Ets65A mRNA is lost in D mutant clones (12 out of 12 clones in 6 lobes examined), confirming that Ets65A expressing neurons are born in the D (or later) temporal stage, and the D TTF is required for the Ets65A expression (Fig. 5 A–A”, Sup. Fig. S3A–A”). Interestingly, the expression of Ets65A in lobula plug neurons is also lost in D mutant clones (Sup. Fig. S3B–B”). In contrast, in D mutant clones (25 out of 25 clones in 12 lobes examined), Sox102F or Oc expression is not lost (Fig. 5 B–B”). Expression of Sox102F is lost in 18 out of 18 slp mutant clones in 6 brain lobes (Fig. 5 C–C”). These results suggest that the generation of Sox102F expressing neurons starts in the Slp stage, and the TTF Slp is required for initiating the Sox102F expression in neuronal progeny (Fig. 5F). Finally, the expression of Oc is not lost in 11 out of 18 slp mutant clones (Fig. 5D), but is lost in 25 out of 25 clones expressing RNAi against ey (Fig. 5E–E’). These results suggest that the generation of Oc expressing neurons starts in the late Ey stage, and continues in the Slp and D temporal stages, and the TTF Ey is required for initiating the expression of Oc in neuronal progeny (Fig. 5F). Although Ey is required for the expression of Oc in neuronal progeny, Oc is only expressed in progeny born from the late stage Ey neuroblasts, suggesting possibly some sub-temporal regulatory logic similar to those described in (Baumgardt et al., 2009) might be in action to start Oc expression only in the late Ey stage progeny. For example, it is possible that a sub-temporal factor is only activated by a high level of Ey or by the prolonged expression of Ey in the late Ey stage NBs, and this sub-temporal factor is responsible to turn on the Oc expression.
T1 neurons are the Notch-off neuronal progeny derived from D stage neuroblasts in all spatial domains of the main medulla.
Since Ets65A starts its expression in neurons born in the D stage, neurons expressing all three transcription factors, which will become T1 neurons should be born in the D temporal stage. It has been shown that the Notch pathway is involved in the binary fate choices between the two neuronal progeny of one medulla GMC. Cells with the Notch signaling pathway turned on will express the Apterous (Ap) transcription factor, and Ap is lost in SuH mutant clones that lose Notch activity (Li et al., 2013). Using an ap>lacZ reporter that is expressed in exactly the same pattern as Ap (Li et al., 2013), we examined whether each of the three transcription factors is expressed in the Notch-on or Notch-off hemi-lineages. All cells expressing Oc do not express Ap, and the majority of the cells that express Ets65A do not express Ap except a very small population at the most superficial layer that have weak Ap staining (Fig. 6A–A”) Thus, the cells that express Oc and Ets65A together do not express Ap (white arrows in Fig. 6A–A”), suggesting that T1 neurons are from the Notch-off hemi-lineage (Fig. 5F). Similarly, Oc and Sox102F are expressed together in neurons that do not express Ap in the medulla (Fig. 6B–B” cyan arrows). However, the cells that express Sox102F but not Oc are expressing Ap (Fig. 6B–B” yellow arrows). Thus, there are at least three populations of neurons that express Sox102F: N-on neurons born in the Slp stage that do not overlap with Oc, because Oc is in the N-off neurons in the Slp stage; N-on neurons in the D stage that also inherit D (Li et al. 2013); and N-off neurons born in the D stage that also express Ets65A and Oc, and this population of neurons will become T1 neurons (Fig. 5F). Previously it was shown that the N-on neuronal progeny born in the Slp and D temporal stages express Ap and Twin of Eyeless (Toy), and they become specified as Tm5 and Tm20 neurons labeled by OrtC1Gal4 (Li et al., 2013). Indeed, a subset of Sox102F neurons co-express Toy and Ap (Fig. 6C–C” white arrows). Together these suggest that a combination of Sox102F, Ap and Toy are expressed in Tm5 and/or Tm20 neurons. A search in the sequencing data published for all medulla neurons (Konstantinides et al., 2018) confirmed that Tm5a/b neurons express Sox102F, Toy and Ap.
Figure 6. T1 neurons are derived from the Notch-off progeny and present in all spatial domains of the main medulla.
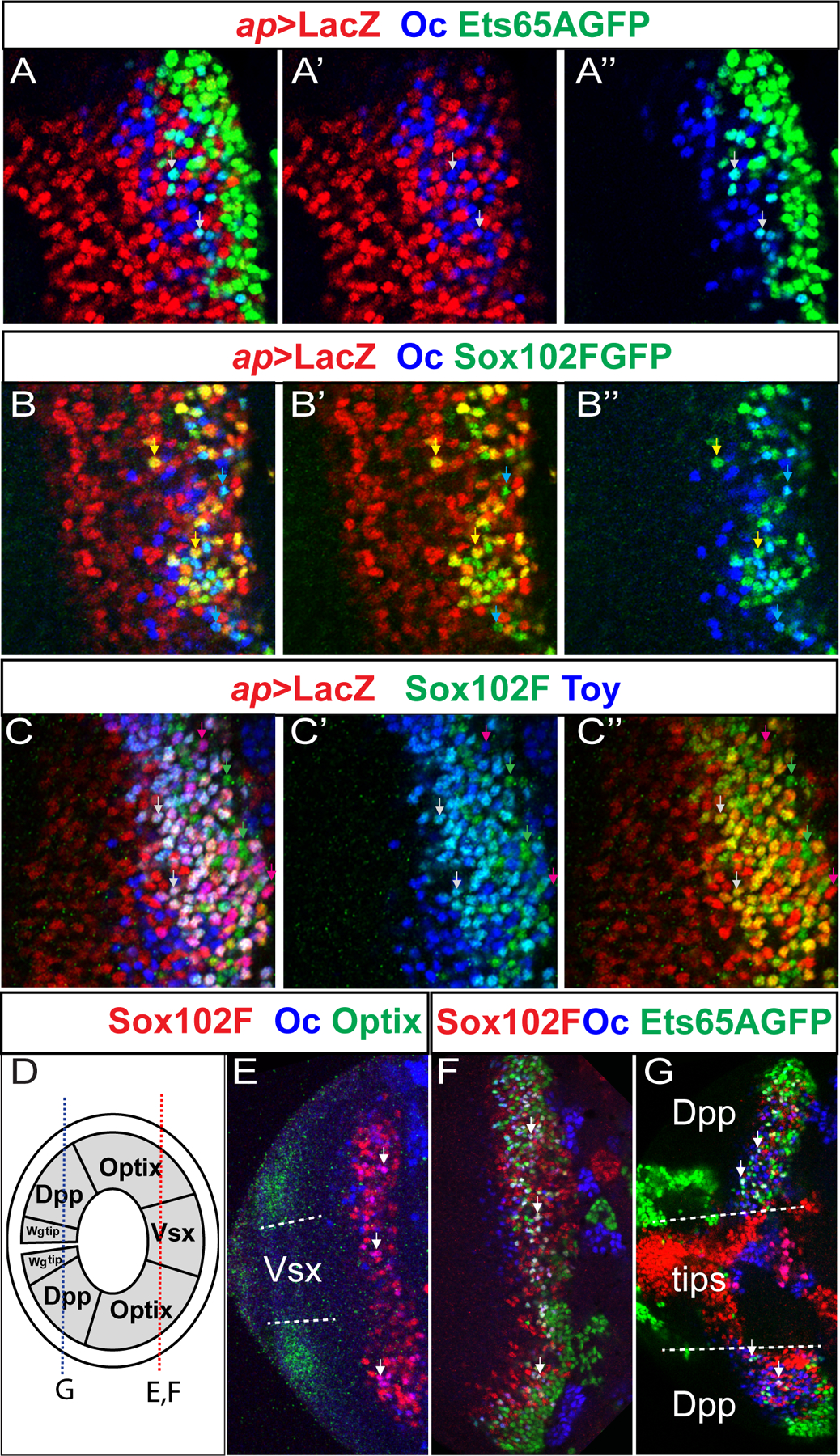
All images shown are of 3rd instar larval medulla. (A–A”) The expression of Ets65A::GFP (green), Oc (blue) and ap>LacZ (red) in larval medulla. Cells expressing both Oc and Ets65A appear cyan in the overlay (A), and two examples are indicated by white arrows. (B–B”) The expression of Sox102F::GFP (green), Oc (blue) and ap>LacZ (red) in larval medulla. Cells expressing both Oc and Sox102F appear cyan in the overlay (B,B”), and two examples are indicated by cyan arrows. Cells expressing both Ap and Sox102F appear yellow in the overlay (B,B’) and two examples are indicated by yellow arrows. (C-C”) The expression of Sox102F (green), Toy (blue) and ap>LacZ (red) in larval medulla. Examples of cells expressing Sox102F but not Toy or ap>LacZ are indicated by green arrows; Examples of cells expressing all three are indicated by white arrows; there are also cells that express Toy and ap>Lac but not Sox102F, and examples are indicated by purple arrows. (D) A schematic drawing showing the different spatial domains of the main medulla (the center Vsx domain, Optix domains, Dpp domains) and the two tips that express Wg. Drawing not to scale. The red dashed line indicates the location of the focal planes for panels E and F, and the blue dashed line indicates the location of the focal plane for the panel G. (E) Cells expressing Oc (blue) and Sox102F (red) are present in the center Vsx domain and the two neighboring Optix (green) domains. Cells expressing both TFs appear purple in the overlay and examples are indicated by white arrows. (F) Cells expressing all three TFs: Oc (blue), Sox102F (red) and Ets65A::GFP (green) are present in the center Vsx domain and the two neighboring Optix domains. Cells expressing all three TFs appear white in the overlay and examples are indicated by white arrows. (G) Cells expressing all three TFs: Oc (blue), Sox102F (red) and Ets65A::GFP (green) are also present in the two Dpp domains (dorsal and ventral), but not present in the two tips (Wg domains).
We also examined whether T1 neurons are generated in all spatial domains of the main medulla crescent, which include the center domain that expresses Vsx, the neighboring two domains that express Optix and the two posterior Dpp-expressing domains (Fig. 6 D) (Erclik et al., 2017). The two tips expressing Wg are special domains in the optic lobe that generate specialized cell types (Bertet et al., 2014). T1 neuron is a uni-columnar neural type, i.e. there is one T1 in every column of the medulla. Uni-columnar neurons are suggested to be generated in all of the main spatial domains of the main medulla (Erclik et al., 2017) . Consistent with this, we observed neurons expressing all three TFs in all domains of the main medulla region (Fig. 6 D–G).
Discussion
In this study, we identified a combination of three transcription factors that control T1 neuron morphology, and examined how the expression of these three transcription factors are controlled by temporal patterning of medulla neuroblasts. Oc is turned on in neurons starting in the late Ey stage, and Oc expressing neurons continue to be generated in the Slp and D stages, although the fates will be different, possibly dependent on the co-expression with other neuronal TFs. Sox102F expressing neurons start to be generated in the Slp stage and continue in the D stage. Ets65A expressing neurons are generated in the D and later temporal stages. Thus, the three TFs that control T1 neuron morphology start their expression in neurons born at different temporal stages, and require the corresponding TTF for initiation of their expression, and each neuronal TF is expressed in neurons spanning a few temporal stages. One advantage for such temporal control of neuronal TFs is that more combinations of TFs can be generated to specify different fates. For example, Toy is expressed in the N-on neuronal progeny born from the Slp and D stages, and also in some N-off progeny born from the late Ey stage neuroblasts in some regions of the medulla (Li et al., 2013). Results from our study and others suggest that the subset of Sox102F neurons that do not express Oc, express Toy and Ap instead (Fig. 5F, Fig. 6C–C”), and they are specified as Tm5 neurons. In addition, the neurons that express both Toy and Oc in the N-off progeny of some late Ey stage neuroblasts could determine another unknown neural type (Fig. 5F). Although it remains to be determined whether these TF combinations are indeed required for the corresponding neural fates, these examples do suggest that different combinations of neuronal TFs can be created that might determine different fates.
We showed that mutation of each of the three TFs expressed in T1 caused a certain morphological defect, similar to the morphology TFs that act in combinations to determine motor neuron morphology (Enriquez et al., 2015). For oc and Ets65A mutant neurons, it appeared that they still maintained the T1 fate, but the morphology was abnormal. Some Sox102F mutant neurons resembled medulla intrinsic neurons, but without functional assay, it was not clear whether they were fully transformed to a normal Mi neuron fate, or they were still T1 neurons that underwent dramatic morphological changes. One question is whether the combination of TFs regulate neuron morphology by simple addition (each TF determines one feature, and the simple addition of these features determines one neural type), or in a synergistic way (three TFs together can determine features not determined by either TF alone). In the case of T1, when we removed Sox102F from T1 neurons, the driver we used (T1-LexA) was still expressed in the mutant neurons, but the neurons became more like medulla intrinsic neurons, and some neurons lost the projection back to the lamina. However, Sox102F is not expressed in other neurons that project back to the lamina like lamina wide field neurons (lawf 1/2) which express Hth and Eya (Chen et al., 2016). Instead, Sox102F is also expressed in a Transmedulla neural type (Tm5) which do not resemble T1 neurons. Thus, our results favor the synergistic action model of neuronal TFs to control neuron morphology.
Our results are consistent with the principle that integration between temporal /spatial patterning of neuroblasts and the Notch-dependent binary neuron fate choice further diversifies neural fates (Baumgardt et al., 2009; Karlsson, Baumgardt and Thor, 2010; Lin et al., 2010; Erclik et al., 2017; Liu et al., 2019; Sen et al., 2019). We found that T1 neurons are derived from the Notch-off hemilineage of D stage neuroblasts. In addition, although T1 neurons are uni-columnar neurons that are generated throughout the main medulla region, there is a spatial component that regulates Oc expression and neural fate specification. Neurons that co-express Oc and Forkhead are only localized in the Dpp domains (Fig. 1E). Through analyzing the sequencing data published for all medulla neurons (Konstantinides et al., 2018), the neurons expressing both Oc and Fkh should become the Dm12 neuron, a multi-columnar neuron with arborizations spanning several columns. Thus, our results support the conclusion that uni-columnar neurons are generated throughout the medulla main region, while multi-columnar neurons are generated in special spatial domains determined by spatial patterning (Erclik et al., 2017).
In summary, our study of T1 neuron specification illustrated an example how temporal patterning of neuroblasts sequentially turns on the expression of three TFs in neuronal progeny, and generates different combinational codes to determine neural fates. In the future it will be interesting to examine how TTFs in neuroblasts regulate the expression of neuronal TFs in neurons that often span a few temporal stages. Only a subset of neurons maintain the expression of TTFs, while other neurons do not. Thus the TTFs should determine the expression of neuronal TFs already in neuroblasts. It is possible that the TTF promotes epigenetic modifications in the neuronal TF gene locus, so that the TF will be turned on in its progeny as well as in neurons born in subsequent temporal stages. It is also possible that the expression of the same neuronal TF in two subsequent temporal stages are controlled by two separate enhancers that respond to different TTFs. Addressing these questions will further advance our understanding of the link between neuroblast temporal patterning and neural fate specification.
Materials and Methods:
Fly stocks and genetic crosses:
The following stocks were obtained from Bloomington Drosophila Stock Center: ywhsflp, T1lexA/CyO, Tm2/Tm6 (BL 52753); Ets65A::GFP (BL38640), Sox102F::GFP (BL 42288), Fkh::GFP(BL 43951) (generated by Spokony, R. and White, K.); UAS-Cas9 (BL 54594), ey-R16F10>Gal4 (BL 48737), slp-R35H02>Gal4 (BL 49923), and D-R12G08>Gal4 (BL47855); UAS-eyRNAi (BL32486). The 13xLexAop2->dSTOP>-myr::smGFP-HA fly stock is a gift from Aljoscha Nern and Gerald Rubin (Nern, Pfeiffer and Rubin, 2015). To generate slp mutant MARCM clones, male flies of FRT40A slp[S37A] /SM6-TM6B (gift from Andrew Tomlinson) were crossed with virgin female flies of y,w, hsFLP, UASCD8GFP; FRT40A tubGal80; tubGal4/TM6B, and the progeny were heat shocked once at 37 °C for 40min at 1st instar larvae stage, and dissected at wandering 3rd instar stage or white pupae stage. To generate D mutant clones, males of yw; D[d23] FRT2A / TM6B were crossed with virgin females of yw hs FLP; act GAL4 UASGFP / CyO; tub GAL80 FRT2A / TM6B (gifts from Makoto Sato), and the progeny were heat shocked twice at early larvae stage at 37 °C for 1.5hr each, and dissected at wandering 3rd instar stage or white pupae stage. To generate ey RNAi clones, males of UAS-eyRNAi (BL32486) were crossed with virgin females of yw hs FLP act>y+>Gal4 UAS GFP / CyO, and progeny were heat shocked at 37 °C for 8 min at 1st instar larvae stage, and dissected at wandering 3rd instar stage or white pupae stage.
To examine the efficiency of bi-allelic somatic mutations generated using gRNAs against oc, or Sox102F in T1 neurons, flies carrying yw; T1lexA loxopRFP/CyO; ey>Gal416F10/TM6B were crossed with each of the two stocks carrying UAS-Cas9 and UASgRNA against each gene. Crosses were kept at 18 °C for 8 days, and then transferred to 29 °C until adult eclosion. The immunostaining results are quantified with Fiji. To examine the efficiency of bi-allelic somatic mutations generated using gRNAs against Ets65A, virgin females of yw hs FLP; act>y+>Gal4 UAS GFP / CyO were crossed to males of yw; UASCas9;UASgRNA-Ets65A, progeny were heat shocked at 37 °C for 8 min at 1st instar larval stage, and dissected at wandering 3rd instar larval stage for in situ hybridization.
To examine the single mutant neuron morphology, virgin females of yw hsFLP; T1lexA /CyO; ey>Gal416F10, 13xLexAop2->dSTOP>-myr::smGFP-HA /TM6B were crossed with each of these three stocks carrying UAS-Cas9 and UASgRNA against each gene. Crosses were kept at 18 °C for 8 days, and then transferred to 29 °C until adult eclosed. The eclosed adult flies were heat shocked at 37 °C for 15min to 2hrs to generate sparsely or densely labeled T1 neurons, and dissected 2 days later.
Antibody Staining
Antibodies used in this work include Guinea pig anti Oc (1:750, gift from Tiffany Cook) (Xie et al., 2007); Rabbit anti Sox102F and Rabbit anti Slp1 (1:500 for both, Gifts from Claude Desplan); Rabbit anti-D (1:200) (from John R. Nambu), mouse anti-Ey (1:10, DSHB), sheep anti-GFP (1:500, AbD Serotec), Goat anti anti-beta-gal (Abcam 1:1000), rabbit anti-RFP (Abcam 1:1000), Rabbit anti HA (Cell Signaling Technology, 1:1000). Secondary antibodies are from Jackson or Invitrogen. Immunostaining was done as described (Li et al., 2013) with a few modifications: 3rd instar Larval brains or adult brains were dissected in 1XPBS, and fixed in 4% Formaldehyde for 30 minutes on ice (larval) or 45min at RT (adult). Brains were incubated in primary antibody solution overnight at 4°C, washed three times and incubated in secondary antibody solution overnight at 4°C, washed three times and mounted in Slowfade. Images are acquired using a Zeiss Confocal Microscope. Figures are assembled using Photoshop and Illustrator.
In situ hybridization
In situ hybridization for Ets65A was done according to a published protocol (Long et al., 2017) with minor modifications (The step of treatment with NaBH4 to reduce background was skipped). Probes were designed by entering transcript sequences of common exons for all Ets65A isoforms in the online Stellaris Designer and 48 of 20nt probes were purchased from Biosearch Technologies.
CRISPR/CAS9 gRNA Design, Cloning and transgenic animals
CRISPR/Cas9 system were used to generate loss of function somatic mutations for Oc, Ets65A, and Sox102F genes. We used the flyCRISPR Optimal Target Finder tool to select guide RNAs with PAM sequence NGG. Two guide RNAs for each gene were cloned into a pCFD6 vector (Port and Bullock, 2016). The sequences of the primers (ordered from Sigma) used for incorporating two gRNAs for each gene into pCFD6 vector are listed below, with the gRNA sequence being lowercase (for the right primer, the lowercase sequence is the reverse complement of the gRNA sequence). Cloning was done per the “pCFD6 cloning protocol” (www.crisprflydesign.org).
Oc-gRNA1, 2
Left primer: GCGGCCCGGGTTCGATTCCCGGCCGATGCAggcggcgggcttcctcaaatGTTTCAGAGCTATGCTGGAAAC
Right primer: ATTTTAACTTGCTATTTCTAGCTCTAAAACagcgatggcatgggcatgccTGCACCAGCCGGGAATCGAACCC
Ets65A-gRNA1, 2
Left primer: GCGGCCCGGGTTCGATTCCCGGCCGATGCAggtgatgcagctggcgttatGTTTCAGAGCTATGCTGGAAAC
Right primer: ATTTTAACTTGCTATTTCTAGCTCTAAAACcaaggcgcgactgagcttatTGCACCAGCCGGGAATCGAACCC
Sox102F-gRNA1,2
Left primer: GCGGCCCGGGTTCGATTCCCGGCCGATGCAttctccaggaggcttcatatGTTTCAGAGCTATGCTGGAAAC
Right primer: ATTTTAACTTGCTATTTCTAGCTCTAAAACcgtactggctgctcagatgaTGCACCAGCCGGGAATCGAACCC
Supplementary Material
Supplementary Figure S1. The expression pattern of Fkh, Oc, Ets65A and Sox102F. (A) A schematic drawing showing the different spatial domains of the main medulla (including the center Vsx domain, Optix domains and Dpp domains) and the two tips that express Wg. Drawing not to scale. The red dashed line indicates the location of the focal plane of panel B, and the blue dashed line indicates the location of the focal plane of panel C. (B) The expression of Fkh::GFP (green) and Oc (blue) in the anterior domains of the 3rd instar larval medulla do not overlap. (C) The expression of Fkh::GFP (green) and Oc (blue) overlap in the posterior Dpp domains. White arrow indicates one example. (D) Fkh::GFP (green) is not expressed in T1 neurons (red). (E-E’”) Ets65AGFP (green) and Oc (blue) are not expressed in medulla neuroblasts marked by Dpn (red). (F-F’”) Sox102F (green) and Oc (blue) are not expressed in medulla neuroblasts marked by Dpn (red).
Supplementary Figure S2. The three transcription factors initiate expression in neurons born at different temporal stages. (A-B’, D-D”) slp>Gal4 is used to drive UAS-NuLacZ (red) expression. (C,C’,E,E’) ey>Gal4 is used to drive UAS-NuLacZ (red) expression. (A,A’) Split channels are shown for Figure 4G. Sox102F::GFP is in green. (B,B’) Split channels are shown for Figure 4H. Oc is in blue. (C,C’) Split channels are shown for Figure 4I. Oc is in blue. (D,D’) All neurons expressing Ets65A::GFP (green) also express slp>>LacZ(red). Ets65A-GFP is not expressed at the time when slp>Gal4 initiates its expression. (E,E’) All neurons expressing Sox102F (green) also express ey>>LacZ(red). Sox102F is not expressed at the time when ey>Gal4 initiates its expression.
Supplementary Figure S3. D is required for Ets65A expression in both the medulla and the lobula plug. All images shown are of 3rd instar larval brain. (A-B”) In situ hybridization of Ets65A mRNA (purple) in brains with D mutant clones marked by GFP (green). (A-A”) A clone in the medulla region. (B-B”) Clones in the lobula plug region.
Highlights:
A combination of three transcription factors controls T1 neuron morphogenesis;
Temporal Transcription Factors in neuroblasts control expression of neuronal TFs;
Temporal progression of neuroblasts generates different transcription factor codes.
Acknowledgements:
We thank the fly community, especially Aljoscha Nern, Gerald Rubin, Andrew Tomlinson, Makoto Sato, Tiffany Cook, John R. Nambu and Claude Desplan for generous gifts of antibodies and fly stocks. We thank Fillip Port for help with the pCFD6 vector. We thank the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, the Developmental Studies Hybridoma Bank, and TriP at Harvard Medical School (NIH/NIGMS R01-GM084947) for fly stocks and reagents. We thank Zhenqing Chen for critically reading the manuscript and for help with the Fiji software. This work was supported by National Institutes of Health (Grant 1 R01 EY026965-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- Allan DW and Thor S (2015) ‘Transcriptional selectors, masters, and combinatorial codes: Regulatory principles of neural subtype specification’, Wiley Interdisciplinary Reviews: Developmental Biology. doi: 10.1002/wdev.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M et al. (2009) ‘Neuronal Subtype Specification within a Lineage by Opposing Temporal Feed-Forward Loops’, Cell. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA and Doe CQ (2013) ‘Combinatorial temporal patterning in progenitors expands neural diversity’, Nature. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C et al. (2014) ‘Temporal patterning of neuroblasts controls notch-mediated cell survival through regulation of hid or reaper’, Cell. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z et al. (2016) ‘A Unique Class of Neural Progenitors in the Drosophila Optic Lobe Generates Both Migrating Neurons and Glia’, Cell Reports. doi: 10.1016/j.celrep.2016.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ (2017) ‘ Temporal Patterning in the Drosophila CNS ‘, Annual Review of Cell and Developmental Biology. doi: 10.1146/annurev-cellbio-111315-125210. [DOI] [PubMed] [Google Scholar]
- Enriquez J et al. (2015) ‘Specification of Individual Adult Motor Neuron Morphologies by Combinatorial Transcription Factor Codes’, Neuron. doi: 10.1016/j.neuron.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erclik T et al. (2017) ‘Integration of temporal and spatial patterning generates neural diversity’, Nature. doi: 10.1038/nature20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KF and Dittrich APM (1989) ‘The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure’, Cell and Tissue Research. doi: 10.1007/BF00218858. [DOI] [PubMed] [Google Scholar]
- Guillemot F (2007) ‘Spatial and temporal specification of neural fates by transcription factor codes’, Development. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Hasegawa E et al. (2013) ‘Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe’, Developmental Biology. doi: 10.1016/j.ydbio.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Hobert O and Kratsios P (2019) ‘Neuronal identity control by terminal selectors in worms, flies, and chordates’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Holguera I and Desplan C (2018) ‘Neuronal specification in space and time’, Science. doi: 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T et al. (2001) ‘Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny’, Cell. doi: 10.1016/S0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jenett A et al. (2012) ‘A GAL4-Driver Line Resource for Drosophila Neurobiology’, Cell Reports. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson D, Baumgardt M and Thor S (2010) ‘Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues’, PLoS Biology. doi: 10.1371/journal.pbio.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N et al. (2018) ‘Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features’, Cell. doi: 10.1016/j.cell.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH et al. (2014) ‘A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila’, Cell Reports. doi: 10.1016/j.celrep.2014.06.065. [DOI] [PubMed] [Google Scholar]
- Li X et al. (2013) ‘Temporal patterning of Drosophila medulla neuroblasts controls neural fates’, Nature. Nature Publishing Group, 498(7455), pp. 456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S et al. (2010) ‘Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain’, Development. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S and Lee T (2012) ‘Generating neuronal diversity in the Drosophila central nervous system’, Developmental Dynamics. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- Liu LY et al. (2019) ‘Mamo decodes hierarchical temporal gradients into terminal neuronal fate’, eLife. doi: 10.7554/eLife.48056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z et al. (2015) ‘Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates’, Science. doi: 10.1126/science.aad1886. [DOI] [PubMed] [Google Scholar]
- Long X et al. (2017) ‘Quantitative mRNA imaging throughout the entire Drosophila brain’, Nature Methods. doi: 10.1038/nmeth.4309. [DOI] [PubMed] [Google Scholar]
- Maurange C, Cheng L and Gould AP (2008) ‘Temporal Transcription Factors and Their Targets Schedule the End of Neural Proliferation in Drosophila’, Cell. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Miyares RL and Lee T (2019) ‘Temporal control of Drosophila central nervous system development’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2018.10.016. [DOI] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD and Rubin GM (2015) ‘Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl MC, Doyle SE and Siegrist SE (2019) ‘E93 Integrates Neuroblast Intrinsic State with Developmental Time to Terminate MB Neurogenesis via Autophagy’, Current Biology. doi: 10.1016/j.cub.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD et al. (2010) ‘Refinement of tools for targeted gene expression in Drosophila’, Genetics. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F and Bullock SL (2016) ‘Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs’, Nature Methods. doi: 10.1038/nmeth.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q et al. (2017) ‘Stem Cell-Intrinsic, Seven-up-Triggered Temporal Factor Gradients Diversify Intermediate Neural Progenitors’, Current Biology. doi: 10.1016/j.cub.2017.03.047. [DOI] [PubMed] [Google Scholar]
- Rister J et al. (2007) ‘Dissection of the Peripheral Motion Channel in the Visual System of Drosophila melanogaster’, Neuron. doi: 10.1016/j.neuron.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Sen SQ et al. (2019) ‘Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci’, eLife. doi: 10.7554/eLife.44036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E et al. (2013) ‘XModular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins’, Cell. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T et al. (2013) ‘A temporal mechanism that produces neuronal diversity in the Drosophila visual center’, Developmental Biology. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Syed MH, Mark B and Doe CQ (2017) ‘Steroid hormone induction of temporal gene expression in drosophila brain neuroblasts generates neuronal and glial diversity’, eLife. doi: 10.7554/eLife.26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC et al. (2013) ‘Contributions of the 12 Neuron Classes in the Fly Lamina to Motion Vision’, Neuron. doi: 10.1016/j.neuron.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B et al. (2007) ‘Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila’, Development. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. The expression pattern of Fkh, Oc, Ets65A and Sox102F. (A) A schematic drawing showing the different spatial domains of the main medulla (including the center Vsx domain, Optix domains and Dpp domains) and the two tips that express Wg. Drawing not to scale. The red dashed line indicates the location of the focal plane of panel B, and the blue dashed line indicates the location of the focal plane of panel C. (B) The expression of Fkh::GFP (green) and Oc (blue) in the anterior domains of the 3rd instar larval medulla do not overlap. (C) The expression of Fkh::GFP (green) and Oc (blue) overlap in the posterior Dpp domains. White arrow indicates one example. (D) Fkh::GFP (green) is not expressed in T1 neurons (red). (E-E’”) Ets65AGFP (green) and Oc (blue) are not expressed in medulla neuroblasts marked by Dpn (red). (F-F’”) Sox102F (green) and Oc (blue) are not expressed in medulla neuroblasts marked by Dpn (red).
Supplementary Figure S2. The three transcription factors initiate expression in neurons born at different temporal stages. (A-B’, D-D”) slp>Gal4 is used to drive UAS-NuLacZ (red) expression. (C,C’,E,E’) ey>Gal4 is used to drive UAS-NuLacZ (red) expression. (A,A’) Split channels are shown for Figure 4G. Sox102F::GFP is in green. (B,B’) Split channels are shown for Figure 4H. Oc is in blue. (C,C’) Split channels are shown for Figure 4I. Oc is in blue. (D,D’) All neurons expressing Ets65A::GFP (green) also express slp>>LacZ(red). Ets65A-GFP is not expressed at the time when slp>Gal4 initiates its expression. (E,E’) All neurons expressing Sox102F (green) also express ey>>LacZ(red). Sox102F is not expressed at the time when ey>Gal4 initiates its expression.
Supplementary Figure S3. D is required for Ets65A expression in both the medulla and the lobula plug. All images shown are of 3rd instar larval brain. (A-B”) In situ hybridization of Ets65A mRNA (purple) in brains with D mutant clones marked by GFP (green). (A-A”) A clone in the medulla region. (B-B”) Clones in the lobula plug region.


