Abstract
Nitrogen (N) is a major limiting factor affecting crop yield in unfertilized soil. Thus, cultivars with a high N use efficiency (NUE) and good grain protein content (GPC) are needed to fulfill the growing food demand and to reduce environmental burden. This is especially true for rice (Oryza sativa L.) that is cultivated with a high input of N fertilizer and is a primary staple food crop for more than half of the global population. Here, we report that rice asparagine synthetase 1 (OsASN1) is required for grain yield and grain protein contents under both N-sufficient (conventional paddy fields) and N-limiting conditions from analyses of knockout mutant plants. In addition, we show that overexpression (OX) of OsASN1 results in better nitrogen uptake and assimilation, and increased tolerance to N limitation at the seedling stage. Under field conditions, the OsASN1 OX rice plants produced grains with increased N and protein contents without yield reduction compared to wild-type (WT) rice. Under N-limited conditions, the OX plants displayed increased grain yield and protein content with enhanced photosynthetic activity compared to WT rice. Thus, OsASN1 can be an effective target gene for the development of rice cultivars with higher grain protein content, NUE, and grain yield under N-limiting conditions.
Keywords: Asparagine synthetase 1, Grain quality and yield, Knockout, Nitrogen use efficiency, Overexpression, Rice
Introduction
Nitrogen (N) is an essential macronutrient in plants and a fundamental building block of amino acids, nucleic acids and chlorophyll (Fukushima and Kusano 2014, Chen and Ma 2015). N deficiency in soil limits plant productivity, nutritional value and crop yield, in part by reducing the photosynthetic components (Fukushima and Kusano 2014). Application of N fertilizer has been a major factor responsible for the increase in crop yield over the past five decades. However, overuse of N fertilizers has caused adverse environmental and economic consequences (Fox et al. 2017). Hence, reducing the dependency of plants on N fertilizer and improving their N use efficiency (NUE; the total biomass or grain yield produced per unit of N fertilizer applied) are desperately needed for environmental and agricultural sustainability (Masclaux-Daubresse et al. 2010). This is especially important for rice (Oryza sativa L.), which is a major food staple, providing 21% of global human per capita energy and 15% of per capita protein (http://www.knowledgebank.irri.org). Therefore, enhancing the nutritional quality of rice represents an economical strategy for the improvement in human nutrition and health.
The grain protein content (GPC) of cereals is an important component of the total protein in the human diet. The amount of GPC in rice (8.5%) is relatively low compared with that in other cereals like wheat (12.3%), barley (12.8%) and Millet (13.4%); however, net protein utilization (which is an indicator of nutritive value) of the rice grain is highest in rice among cereal grains (Peng et al. 2014, Mahender et al. 2016, Yang et al. 2019). Various approaches have been employed to enhance the GPC and essential amino acid (e.g. lysine and threonine) content of rice grains (Jiang et al. 2016). Overexpression (OX) of aspartate aminotransferase genes resulted in an increase in total amino acid (5.4–2.0%) and protein (up to 22.2%) contents in rice grain (Zhou et al. 2009). A quantitative trait locus in rice qPC1 that controls GPC by regulating the synthesis and accumulation of glutelins, prolamins, globulins, albumins and starch encodes a putative amino acid transporter OsAAP6. The positive allele significantly enhanced the GPC of rice up to 15.3% (Peng et al. 2014). The OX of artificially synthesized fusion proteins containing lysine/threonine motifs significantly increased the content of crude protein (20.45%), total amino acids (19.43%), lysine (33.87%) and threonine (21.21%) in transgenic rice grains (Jiang et al. 2016).
Glutamine and asparagine are the major forms of N in the phloem sap (Hayashi and Chino, 1990), and the synthesis of these two amino acids in source organs is, therefore, critical for N recycling and remobilization at the whole plant level (Lea et al. 2007).
Glutamine synthetase (GS) plays a major role in the condensation of ammonium and glutamate to form the amino group of the glutamine (Kusano et al. 2011). Disruption of OsGS1.1, which is the major component of the cytosolic GS in rice, caused a severe reduction in plant growth and grain filling (Tabuchi et al. 2005), and lack of OsGS1;2 displayed remarkable reduction in spikelet number per panicle with a decrease in yield (Funayama et al. 2013). Glutamate synthase (GOGAT) catalyzes the transamination of glutamine and 2-oxoglutarate to two molecules of glutamate, thus providing glutamate for ammonium assimilation through the GS and for the synthesis of the other amino acids (Bernard and Habash 2009). Disruption of the OsNADH-GOGAT1, OsNADH-GOGAT2 and OsFd-GOGAT1 causes a remarkable decrease in yield (Tamura et al. 2010, Tamura et al. 2011, Sun et al. 2017).
Asparagine synthetase (ASN) that transfers an amide group from glutamine to aspartate, to form asparagine, functions in N assimilation, remobilization and allocation within the plant, and in glutamate and glutamine recovery (Gaufichon et al. 2013, Moison et al. 2018). The ASN genes are regulated by environmental and developmental factors, including the level of N supply in growth media (Lam et al. 1998, Lam et al. 2003, Wong et al. 2004). Among the three ASN genes in Arabidopsis thaliana, ASN1 and ASN2 are antagonistically regulated by light and carbon (C) and N metabolites (Lam et al. 1998). Disruption of ASN2 results in asparagine depletion in phloem saps, lower 15N flux from source leaves to sinks, delayed senescence and abnormal growth phenotypes, suggesting that ASN2 is essential for N assimilation, distribution and remobilization (Gaufichon et al. 2013). The ASN1 gene is preferentially expressed in flowers and reproductive organs compared with the two other ASN genes. Accordingly, the asn1 mutant shows defective embryo development and reduced seed N content, suggesting a critical role for ASN1 in seed N filling (Gaufichon et al. 2017).
The rice genome harbors two ASN genes, OsASN1 and OsASN2 (Ohashi et al. 2015). OsASN1 is mainly expressed in roots when supplied with sufficient amounts of , whereas OsASN2 accumulates to only low levels in roots grown in the presence or absence of 1 mM (Ohashi et al. 2015). Knockout mutants of OsASN1 generated by Tos17 insertion exhibit an approximately 80–90% reduction in free asparagine content in roots and xylem sap at the seedling stage. Hence, OsASN1 is thought to be regulated by N supply in the soil since OsASN1, but not OsASN2 is detected in roots grown with added (Ohashi et al. 2015). Recently, the analysis of osasn1 mutants generated by T-DNA and CRISPR-Cas9 showed a reduced growth compared to wild type (WT), with a notable reduction in tiller numbers, suggesting that OsASN1 has an important role in rice development (Luo et al. 2018). In contrast, no significant differences between WT and osasn1 mutants generated by Tos17 were observed with respect to tiller number and growth pattern under different supply in a hydroponic culture and in a paddy field (Ohashi et al. 2018).
Here, we used both knockout and OX transgenic lines to investigate the role of OsASN1 in plant growth and development in paddy fields and its importance in NUE. Measuring ammonium uptake, yield and grain filling, we show that OsASN1 OX represents a valuable plant breeding strategy for developing rice cultivars with improved NUE and GPC under N-limiting conditions.
Results
Effect of N availability on OsASNs expression
To test that OsASNs expression responds to changes in external N levels, we grew rice seedlings on a modified 1/2 MS medium with increasing concentrations of for 4 d. Significant increases in OsASN1 expression were observed in shoots and roots at concentrations >1 and 10 mM, respectively (Supplementary Fig. S1A, C). However, OsASN2 expression was not induced by (Supplementary Fig. S1B, D). We selected OsASN1 for the further study, since the gene was responsive to the N supply. Murashige and Skoog.
Effect of osasn1 loss-of-function mutation on agronomic traits
To determine how reduced OsASN1 activity affects plant growth and yield, we identified two independent T-DNA knockout mutants, osasn1-1 and osasn1-2, from the rice flanking-sequence tag database (Jeon et al. 2000, An et al. 2003). In the osasn1-1 and osasn1-2 mutant lines, T-DNA was inserted in the first and fourth introns, respectively (Supplementary Fig. S2A). These lines were confirmed as knockout mutants based on the lack of OsASN1 transcripts determined by quantitative real-time (qRT-PCR) analysis, irrespective of external N supply at the seedling stage (Supplementary Fig. S2B). In addition, OsASN1 transcripts were not detected in flag leaves of osasn1 mutants at the flowering stage (Supplementary Fig. S2C). As a result of OsASN1 disruption, mutant lines displayed lower asparagine concentrations compared with the WT in their flag leaves (Supplementary Fig. S2D); however, the level of glutamine, which is a substrate of OsASN1, was similar in WT and mutant plants (Supplementary Fig. S2E).
Next, we investigated the growth and yield of knockout mutant plants in a paddy field using conventional agricultural practices (Fig. 1A). No differences in flowering time were observed between osasn1 mutants and WT plants in the field (Supplementary Fig. S2F). The plant height and aboveground biomass of knockout plants were significantly reduced compared with those of WT controls (Supplementary Fig. S2G, H). However, the number of tillers and spikelet number per panicle were similar between mutant and WT plants (Supplementary Fig. S2I, J). The total grain yield per plant was severely reduced to 59.3% and 60.0% of the WT in osasn1-1 and osasn1-2 mutants, respectively, and fertility of mutant plants was also reduced (Fig. 1B; Supplementary Fig. S2K). In addition, the thickness of the seed of osasn1 mutants was reduced compared with the WT, resulting in reduced 1,000-seed weight (Fig. 1C, D). However, the length and width of mutant seeds were similar to those of WT seeds (Supplementary Fig. S2L, M).
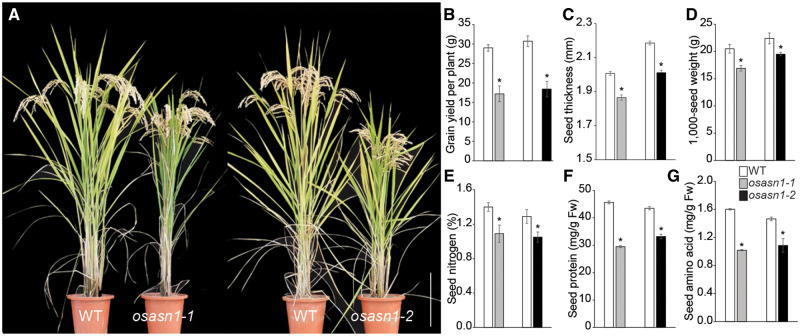
Fig. 1.
Agronomic traits of osasn1 knockout mutants of rice. (A) Comparison of osasn1-1 and osasn1-2 mutant rice plants and WT controls grown in a paddy field using conventional agricultural practices. The two knockout lines are in different genetic backgrounds, osasn1-1 in Hwayoung cultivar and osasn1-2 in Dongjin cultivar. Plants were transferred to pots from paddy fields, and representative photographs were taken before harvest (scale bar = 20 cm). (B) Grain yield per plant grown in paddy fields (n = 5). (C) Grain thickness (n = 20) of seeds from WT and osasn1 mutants. (D) The 1,000-grain weight from the WT and osasn1 mutants (n = 5). Quantification of nitrogen (E), protein (F) and amino acid (G) concentrations in grains of osasn1 and WT plants. Data represent mean ± SE of five replicates. Significant differences between mutant and WT plants are indicated with an asterisk (*P < 0.05).
Because OsASN1 may affect N remobilization during grain filling, we analyzed total N content of the flag leaves and panicles as representative source and sink tissues, respectively (Supplementary Fig. S3A–D). Total N content gradually decreased in the flag leaves but increased in the panicles of the WT plants, suggesting that N was remobilized from the flag leaves to the panicles during the grain-filling period. N levels were higher in the mutant flag leaves than in the WT flag leaves (Supplementary Fig. S3A, B), and lower in the mutant panicles than in the WT panicles (Supplementary Fig. S3C, D). Accordingly, N content of the mutant seeds was lower than that of the WT seeds; N in osasn1-1 and osasn1-2 grains was decreased to 78% and 81% of that in WT grains (Fig. 1E). Disruption of OsASN1 also led to less accumulation of asparagine in the grains of mutant compared to the WT (Supplementary Fig. S3E). Nevertheless, the concentrations of glutamine were similar in the seeds of the osasn1 mutant and of the WT (Supplementary Fig. S3F). The majority of nitrogen is remobilized from source leaves, stems or roots to the reproductive sink organs as amino acid (Masclaux-Daubresse et al. 2010). Nitrogen content in the grain, which is an important nutritional quality trait of rice, relies on protein and amino acid contents (Tang et al. 2007, Huang et al. 2019). The protein and free amino acid contents in the seeds were lower in knockout mutant than in WT (Fig. 1F, G).
osasn1 mutants show reduced tolerance to N limitation at the seedling stage
Next, we compared the seedling growth of osasn1 and WT in N-sufficient and N-limited conditions for 8 d. In N-sufficient media containing 10 mM , the growth of mutant seedlings was similar to that of WT seedlings (Fig. 2A); plant height and fresh weight showed no significant differences between mutant and WT plants (Fig. 2A–C). However, in N-limited medium containing 0.1 mM , osasn1 mutant seedlings were smaller than WT seedlings (Fig. 2A); the height of osasn1-1 and osasn1-2 seedlings was reduced to 83% and 75% compared to that of WT seedlings, respectively (Fig. 2B), and the fresh weight of osasn1-1 and osasn1-2 seedlings was reduced to 89% and 83% compared to that of WT seedlings, respectively (Fig. 2C). A similar response was also observed in seedlings grown in N-sufficient and N-limited media for a longer period of 14 d (Supplementary Fig. S4). The shoots of both mutants accumulated less N (91% of WT), whereas mutant roots contained more N than WT roots, irrespective of the external supply (Fig. 2D).
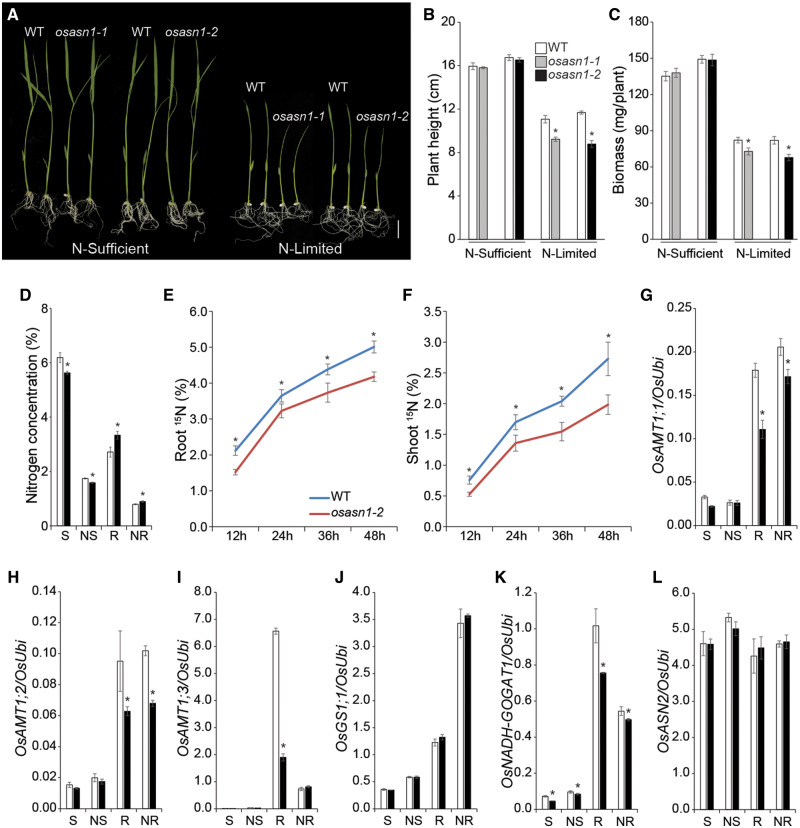
Fig. 2.
The osasn1 knockout mutants showed enhanced sensitivity to N limitation at the seedling stage. (A) Phenotypes of WT and knockout mutant plants. Plants were grown in N-sufficient (10 mM ) or N-limited (0.1 mM ) media for 8 d. Scale bars = 2.5 cm. Measurement of plant height (B) and fresh weight (C) of WT and osasn1 seedlings. (D) Nitrogen (N) concentrations in shoots and roots (n = 4 each) of WT and osasn1-2 mutant plants. (E, F) N uptake analysis in WT and osasn1-2 mutant plants at the seedling stage. Seven-day-old seedlings were treated with 15N-labeled NH4Cl, and 15N contents of roots (E) and shoots (F) were quantified at the indicated time points after treatment. Expression analysis of OsAMT1;1 (G), OsAMT1;2 (H), OsAMT1;3 (I), OsGS1;1 (J), OsNADH-GOGAT1 (K) and OsASN2 (L) in WT and osasn1-2 mutant plants using qRT-PCR. Data represent mean ± SE of four replicates. Significant differences between mutant and WT plants were determined using Student’s t-tests and indicated with an asterisk (*P < 0.05). S, shoots of plants grown in N-sufficient medium; NS, shoots of plants grown in N-limited medium; R, roots of plants grown in N-sufficient medium; NR, roots of plants grown in N-limited medium.
Because disruption of OsASN1 affected the N distribution in seedling shoots and roots, we monitored N influx into the roots and N transport from roots to shoots by performing 15N labeling and tracing assays at the seedling stage (Fig. 2E, F). Results showed that 15N content in roots and shoots of osasn1-2 plants was lower than in the corresponding tissues of WT plants (Fig. 2E, F). This showed that knockout plants had lower ammonium uptake capacities than WT. Next, we tested the transcript levels of ammonium transporters (OsAMTs) to evaluate the effect of OsASN1 disruption on the expression of OsAMTs. Among the 12 genes encoding putative OsAMTs in rice, only OsAMT1 has been characterized so far as a high affinity transporter for ammonium transporter (Bao et al. 2015, Li et al. 2017). The expression of OsAMT1;1, OsAMT1;2 and OsAMT1;3 was lower in the osasn1 mutants than in WT (Fig. 2G–I), coinciding the lower ammonium uptake capacities in mutant as shown by the 15N tracing assay. Next, the expression of genes involved in N assimilation was monitored to evaluate the physiological significance of OsASN1 gene disruption. In the plant, is mainly assimilated via the GS/GOGAT cycle, and in a lower extent by ASN (Masclaux-Daubresse et al. 2010). The expression of OsGS1;1 was not significantly affected (Fig. 2J) while the expression of OsNADH-GOGAT1 was decreased in osasn1 mutant compared to WT irrespective of the amount of supply (Fig. 2K), suggesting that disruption of OsASN1 could disturb the GS/GOGAT cycle. However, the transcript level of OsASN2 remained unchanged in the osasn1 mutants (Fig. 2L).
Characterization of OsASN1 overexpressing transgenic plants in paddy fields
Based on the effects of OsASN1 disruption on N transport and assimilation, we hypothesized that the OX of OsASN1 could improve NUE in rice and, in turn, decrease rice dependency on N availability in the soil. We overexpressed OsASN1 in the rice cultivar Dongjin under the control of the maize ubiquitin promoter. Several transgenic lines were obtained, and we selected the two independent transgenic lines (OX1 and OX2) that displayed elevated expression of OsASN1 in seedling shoots compared to WT when grown on media containing 10 mM (Supplementary Fig. S5A, B). Additional qRT-PCR analyses confirmed the higher expression of OsASN1 in shoots and roots of OX1 and OX2 lines than in those of WT plants under N-sufficient and N-limited conditions (Supplementary Fig. S5C). Transcripts of OsASN1 also accumulated to higher levels in flag leaves of OX lines than in those of WT plants at the flowering stage (Supplementary Fig. S5D). Accordingly, OX lines displayed higher asparagine concentrations than the WT, thus confirming the increased OsASN1 activity; asparagine levels were 3.4- and 2.2-fold higher in leaves of OX1 and OX2 lines, respectively, than in WT leaves (Supplementary Fig. S5E). By contrast, glutamine levels were reduced to 42.9% and 66.4% in leaves of OX1 and OX2 lines, respectively, compared with WT leaves (Supplementary Fig. S5F). This suggests that glutamine serves as an N donor for asparagine synthesis by OsASN1.
To determine the effect of the increase in OsASN1 activity on N utilization and plant growth and morphology, we grew WT and OX plants in a paddy field using conventional agricultural practices. The OX plants were morphologically similar to WT plants (Fig. 3A). OX of OsASN1 did not affect flowering time (Supplementary Fig. S5G). Plant height, aboveground dry matter, tiller number per plant, spikelet number per panicle and grain-filling rates of OX plants were not different in OX and WT plants, leading then to similar grain yield (Fig. 3B; Supplementary Fig. S5H–L). Moreover, the length, width and thickness of OX grains were similar to those of WT grains (Supplementary Fig. S5M–O), resulting in no significant differences in grain weight between the WT and OX plants (Fig. 3C).
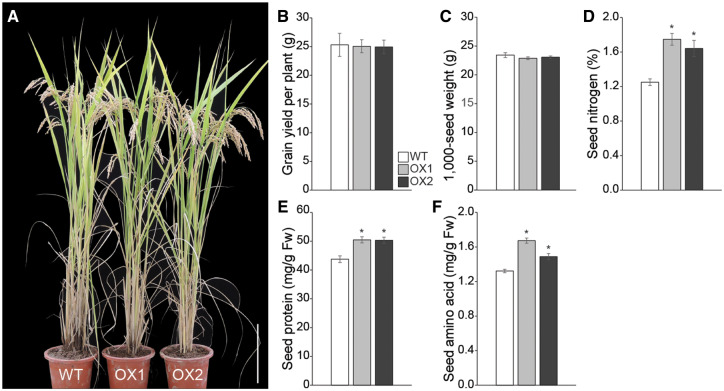
Fig. 3.
Agronomic traits of OsASN1 overexpressing transgenic plants. (A) Morphology of OsASN1 overexpressor lines (OX1 and OX2) and their segregant WT rice plants grown in a conventional paddy field using conventional agricultural practices. Representative photographs were taken before harvest (scale bar = 20 cm). Agronomic traits of WT and OX plants grown in paddy fields, including gran yield per plant grown in paddy fields (n = 5) (B) and 1,000-grain weight (C). Quantification of N (D), protein (E) and amino acid (F) concentrations in grains of WT and OX plants. Data represent mean ± SE of five replicates. Significant differences between OX and WT plants are indicated with an asterisk (*P < 0.05).
Total N content of leaves was higher in OX plants than in WT plants during the early stages of grain filling, but no significant differences were observed between OX and WT plants at later stages (Supplementary Fig. S6A). In panicles, OX plants accumulated significantly more N than WT plants (Supplementary Fig. S6B). Subsequently, we found that the N concentration in the grains of OX plants was higher than in the WT grains; N in OX1 and OX2 grains was increased to 140% and 131% of that in WT grains, respectively (Fig. 3D). The levels of asparagine in the grains of OX1 and OX2 lines were 3.4- and 3.1-fold higher, respectively, than that in WT grains, indicating the enhanced OsASN1 activity in grains (Supplementary Fig. S6C). On the contrary, glutamine levels were reduced to 69.5% and 79.6% in grains of OX1 and OX2 lines, respectively, compared with WT grains (Supplementary Fig. S6D). Grain protein concentration of both OX lines was elevated to 120% of that in WT grains (Fig. 3E), and free amino acid concentrations in grains of OX lines were increased to 127% (OX1) and 113% (OX2) of that in WT grains (Fig. 3F), suggesting that OX of OsASN1 could improve the grain quality.
Transgenic OX lines showed enhanced tolerance to N limitation at the seedling stage
To examine the growth of OX plants under N-limiting conditions, we first compared seedling growth with and without N in the growth media. The growth of OX and WT seedlings showed no significant differences in N-sufficient medium (Fig. 4A–C). However, in N-limited medium, OX seedlings exhibited increased plant height and fresh weight compared with WT seedlings (Fig. 4A–C). The height of OX1 and OX2 seedlings increased to 119% and 113% of that of WT seedlings, respectively (Fig. 4B). Accordingly, the fresh weight of OX1 and OX2 seedlings was 137% and 127% of that of WT seedlings, respectively (Fig. 4C). A similar response was also observed in OX seedlings grown in N-sufficient and N-limited media for a longer period of 14 d (Supplementary Fig. S7). When grown on N-sufficient media for 2 weeks, WT and OX seedlings showed similar plant height and biomass (Supplementary Fig. S7A–C). Nevertheless, under N limitation, the height of OX1 and OX2 seedling displayed increased plant height (113% and 112% of that of WT, respectively; Supplementary Fig. S7A, B) and fresh weight (114% and 115% of that of WT, respectively; Supplementary Fig. S7A, C). When grown under N-sufficient conditions, OX lines accumulated more N, 138% (OX1) and 131% (OX2) in shoots and 125% (OX1) and 119% (OX2) in roots compared with the WT (Fig. 4D). Under N-limiting condition, relative N concentrations in OX plants increased to 125% (OX1) and 121% (OX2) in shoots and 133% (OX1) and 130% (OX2) in roots compared with WT plants (Fig. 4D). We then conducted 15N labeling and tracing assays at the seedling stage to monitor N influx into the root and transport capacity from the root to the shoot (Fig. 4E, F). Consistent with the phenotype of the OX plants that displayed higher N concentrations in their root and shoot than WT (Fig. 4D), 15N uptake in roots (Fig. 4E) and shoots (Fig. 4F) of OX1 plants was higher than that of WT plants.
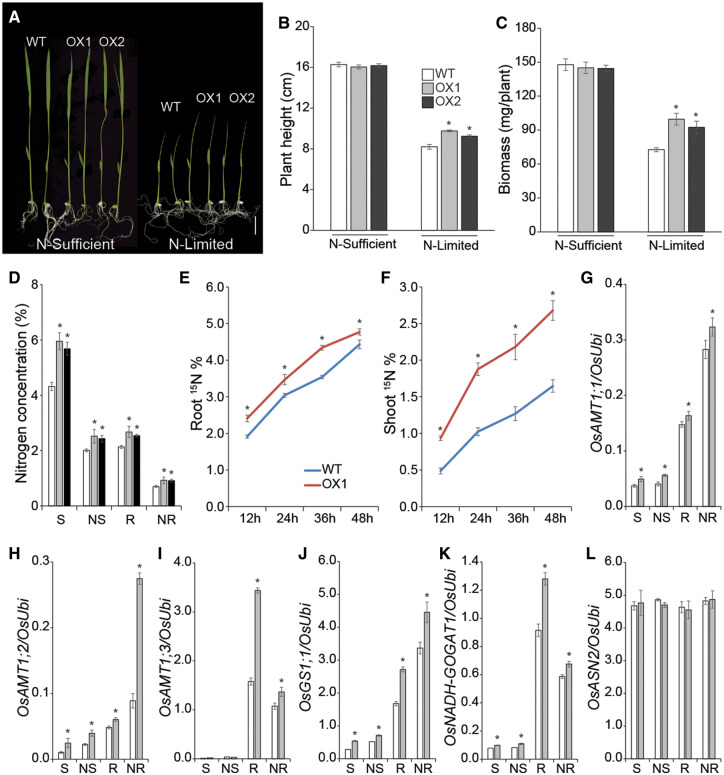
Fig. 4.
Transgenic OsASN1 OX lines showed enhanced tolerance to N limitation at the seedling stage. (A) Phenotypes of OX1, OX2 and WT plants grown. Plants were grown in N-sufficient (10 mM ) or N-limited (0.1 mM ) media for 8 d. Scale bars = 2.5 cm. Measurement of the height (B) and fresh weight (C) of WT and OX seedlings. (D) N concentrations in shoots and roots of WT and OX plants. (E, F) N uptake analysis of WT and OX1 seedlings grown in Yoshida solution. Seven-day-old seedlings were treated with 15N-labeled NH4Cl, and 15N contents of roots (E) and shoots (F) were measured at the indicated time points after treatment. Expression analysis of OsAMT1;1 (G), OsAMT1;2 (H), OsAMT1;3 (I), OsGS1;1 (J), OsNADH-GOGAT1 (K) and OsASN2 (L) in WT and OX1 plants using qRT-PCR. Data represent mean ± SE of five biological replicates. Significant differences between WT and OX plants were determined using Student’s t-test and are indicated with an asterisk (*P < 0.05). S, shoots of plants grown in N-sufficient medium; NS, shoots of plants grown in N-limited medium; R, roots of plants grown in N-sufficient medium; NR, roots of plants grown in N-limited medium.
As OX of OsASN1 enhanced the N uptake and transport activities, expressions of OsAMT1;1, OsAMT1;2 and OsAMT1;3 were higher in the OX seedlings than in WT, accompanying with the elevated levels of N in the OX seedlings under both N-sufficient and N-limited conditions (Fig. 4G–I). Furthermore, the expression of OsGS1;1 and OsNADH-GOGAT1 was higher in OX seedlings compared with WT seedlings irrespective of external N supply (Fig. 4J, K). The expression of OsASN2 was not significantly different between OX and WT under both N-sufficient and N-limiting conditions (Fig. 4L).
Because OsASN1 overexpressors and loss-of-function mutants displayed modified N uptake and translocation, we monitored their sensitivity to N toxicity. When grown on N-rich media (100 mM ) for 8 d, the OX seedlings exhibited a decreased plant height and fresh weight (Supplementary Fig. S8A–C) with higher N concentrations in shoots and roots than WT seedlings (Supplementary Fig. S8D). On the contrary, osasn1 mutant seedlings grown under high N supply showed an increased plant height and fresh weight (Supplementary Fig. S8E–G) with lower N concentrations in shoots and roots than WT seedlings (Supplementary Fig. S8H). When grown under 100 mM for 2 weeks, rice seedlings exhibited clear N toxicity symptoms (Supplementary Fig. S9A, D), which have not been observed in the 8-day-old seedling. N-toxicity symptoms included shortening of plant height, chlorosis and appearance of necrotic spots on leaves. The symptoms were evidently observed from the newly emerging leaves, which finally led to leaf browning (Supplementary Fig. S9A, D). Under this growth condition, OX seedlings exhibited an increased N toxicity as shown by a decreased plant height and fresh weight (Supplementary Fig. S9A–C) than WT seedlings. On the contrary, osasn1 mutants showed an increased plant height and fresh weight than WT seedlings (Supplementary Fig. S9D–F). The chlorosis and necrotic spots appeared similarly in WT, OX and the mutant seedlings.
OX of OsASN1 improves grain yield under N-limiting conditions
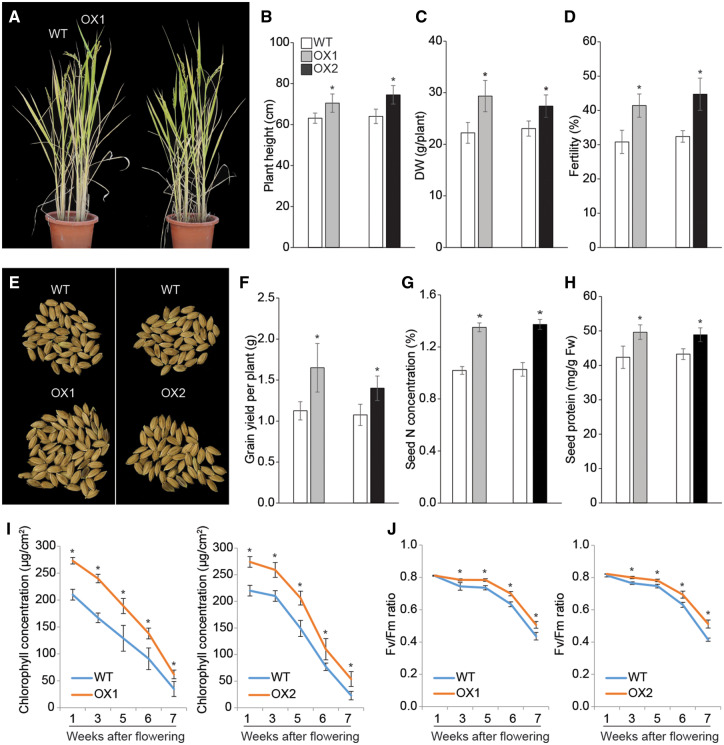
The growth and yield of OX lines were evaluated under N-limiting growth conditions until harvest. Plants were grown in pots containing N-limited nursery soil containing 1/16th of the amount of N in regular N-enriched nursery soil. The OX plant height (112% and 116% of WT for OX1 and OX2, respectively) and dry weight (132% and 119% of WT for OX1 and OX2, respectively) were larger than that of WT (Fig. 5A–C). Fertility of OX plants was also higher than that of the WT (Fig. 5D). However, tiller number per plant, spikelet number per panicle and 1,000-seed weight showed no differences between the OX and WT plants (Supplementary Fig. S10A–C). In addition, grain yield per plant of OX1 and OX2 lines showed an increase of 147% and 130%, respectively, compared with the WT (Fig. 5E, F). N concentrations of OX1 and OX2 plants were also increased to 132% and 133%, respectively, compared with the WT (Fig. 5G). Consequently, grain protein contents were enhanced to 117% in OX1 and 114% in OX2 compared with that in WT (Fig. 5H).
Fig. 5.
Improved tolerance of OsASN1 overexpressing transgenic plants to N limitation at the reproductive stage. (A) Morphological comparison of OX1, OX2 and WT plants grown in N-limiting conditions at the reproductive stage. Plants were grown in an outdoor condition until harvest in pots containing 1/16th of the N amount present in regular N-enriched nursery soil. Representative photographs were taken 5 weeks after flowering. Scale bars = 20 cm. Characteristics of WT and OX plants, including plant height (B), DW of aboveground biomass (C) and fertility (D). (E, F) Total grain yield per plant. N (G) and protein (H) concentrations in mature seeds of WT and OX plants grown in conditions described in (A). Quantification of photosynthesis-related traits in WT and OX plants, including total chlorophyll concentrations (I) and photochemical efficiency (J). Data represent mean ± SE of five replicates. Significant differences between OX and WT plants are indicated with an asterisk (*P < 0.05).
Next, we found that OX plants grown under N-limiting condition retained higher chlorophyll contents than WT plants at the heading stage (Fig. 5I). Although the maximum values for photochemical efficiency (Fv/Fm ratio) were similar for OX and WT plants at the heading stage, the ratio was higher in transgenic leaves than in WT leaves during the ripening stage (Fig. 5J). These data suggest that increased grain yield of OX lines under N-limiting conditions is at least in part due to increased photosynthetic activity. Analysis of OX and WT plants in the following year confirmed higher grain yield and increased grain N and grain protein concentrations in OX plants compared with the WT (Supplementary Fig. S11A–F).
The growth of osasn1 seedlings under the same N-limiting outdoor conditions was also examined until harvest. Knockout osasn1 plants grown in pots containing N-limited nursery soil showed lower plant height, aboveground dry mass, fertility, grain yield per plant and grain N and protein concentrations than the WT plants (Supplementary Figs. S12A–K, S13A–F). However, chlorophyll content and Fv/Fm ratio were not different from those of WT plants (Supplementary Fig. S12L, M).
Discussion
N affects many aspects of plant growth and development. Hence, improvement in NUE contributes to better crop production and grain quality (Kabir 2016). Many reviews have reported the numerous attempts made to modify NUE by manipulating nitrate or transporters (Tegeder and Masclaux-Daubresse 2018) or N assimilation enzymes (Chardon et al. 2012).
Among the enzymes involved in N remobilization during grain filling, the cytosolic GS1 and ASN play essential roles because glutamine and asparagine are central in the transport and storage of nitrogen at different developmental stages, including N mobilization in germinating seeds, N recycling in vegetative cells in response to biotic and abiotic stresses and N remobilization from source to sink organs (Gaufichon et al. 2010). Recently, Moison et al. (2018) showed that induction of ASN2 increases asparagine content in the Arabidopsis triple GS1 mutant to compensate for the N remobilization defects caused by the reduction in the cytosolic glutamine synthesis.
Characterization of OsASN1 knockout mutants (Ohashi et al. 2015, 2018, Luo et al. 2018) were previously reported with some discrepancies. Ohashi et al. (2015) characterized the knockout mutants of OsASN1 generated by Tos17 insertion (as1-m1, as1-m2) and showed that OsASN1 is responsible for the synthesis of asparagine in rice roots when supplied with . Luo et al. characterized a T-DNA mutant (asn1) and the CRISPR/Cas9 mutant lines. The mutant plants showed reduced growth compared to WT with a notable reduction in tiller numbers, suggesting that OsASN1 has an important role in rice development (Luo et al. 2018). In contrast, no significant differences between WT and OsASN1 mutants generated by Tos17 were observed in tiller number and growth pattern under different supply in a hydroponic culture and in a paddy field (Ohashi et al. 2018). In this report, we characterized the same T-DNA mutant (indicated as osasn1-1) line that was used in a previous report (Luo et al. 2018) and another T-DNA mutant line (osasn1-2) that was newly isolated by our group. In our field growth condition, tiller numbers in the mutant plants were indistinguishable from those of WT plants (Supplementary Fig. S2I). In addition, our OsASN1 overexpressing transgenic plants had tiller numbers similar to those in WT (Supplementary Fig. S5J) in a paddy field. Thus, in our field condition, OsASN1 has a limited role in tiller development. In contrast, the mutants showed a reduced growth pattern in consistent with a previous report (Luo et al. 2018).
However, the effect of OsASN1 on grain yield and N remobilization has not yet been demonstrated under field conditions. In this study, we investigated the role of OsASN1 in plant growth and development under field conditions using knockout mutants and OX lines. The osasn1 mutants showed impaired growth with less biomass under N limitation and in conventional field condition (Figs 1, 2, Supplementary Figs. S4, S12, S13). Because disruption of OsASN1 changed the N distribution between seedling shoots and roots (Fig. 2D), we measured (i) N influx into the roots and N translocation from roots to shoots by 15N tracing assay and (ii) the expressions of three representative OsAMTs (OsAMT1;1, OsAMT1;2 and OsAMT1;3), and N assimilation-related genes (Fig. 2E–L). Our results indicate that the disruption of OsASN1 reduced the ammonium uptake capacity by regulating the expression of OsAMTs and disturbed the balance of GS/GOGT cycle. Therefore, without OsASN1, assimilation of ammonium into amino acids was reduced, leading to less need for ammonium and unbalanced GS/GOGT cycle. Consequently, grain N and protein concentrations and grain-filling rate of osasn1 mutant plants grown in the paddy field were lower than those of WT plants. Higher concentration of N in the leaves of knockout mutants was in good accordance with the lower N remobilization to the seeds. Together, these suggest that the OsASN1 gene is required for optimal growth, yield and nutritional quality of rice grown in N-sufficient conventional paddy fields.
In contrast to osasn1 mutants, OsASN1 OX lines displayed increased nitrogen uptake and influx as shown by the 15N tracing assay and expression analyses of OsAMTs (Fig. 4E–I). The enhanced expressions of OsGGS1;1 and OsGOGAT1 suggested that N assimilation by GS/GOGAT cycle was more active in OsASN1 OX than in WT plants and that more ammonium could be assimilated into amino acids (Fig. 4J, K). Consequently, we assume that OX lines increased asparagine synthesis in their leaves and then displayed higher nitrogen and protein allocation to their seeds. However, no change in plant height, biomass or yield could be observed in conventional paddy fields. This demonstrates the potential of OsASN1 OX to improve grain protein content without compromising crop yield, when grown in conventional paddy fields and using conventional agricultural practices. Under N-limiting growth conditions at reproductive stage, the leaves of OsASN1 OX lines displayed higher photochemical efficiency than WT leaves, leading to better agronomic performance including increased biomass, grain filling and yields of OX lines as compared to those of WT plants. OX of OsASN1 leads to practical improvements in grain yield and N and protein concentrations, in plants grown under N-limiting conditions.
Although the labeling of plants with 15N isotope is a valuable strategy to analyze the remobilization of nitrogen from source to sinks (Havé et al. 2017), we did not conduct such labeling experiments at the reproductive stage, but we analyzed nitrogen budget at different time points during seed development. The total N contents of flag leaves and panicles were measured at four time points during the grain-filling stage, which indicated that changes in the expression of OsASN1 affected the remobilization of N from flag leaf to the seeds all along the grain-filling stage (Supplementary Figs. S3, S6). Although several lines of evidence we presented here surely suggested the changes in nitrogen remobilization, a tracer experiment with 15N isotope would have further clarified our conclusion.
Several studies have been investigated the OX of ASN genes. OX of Arabidopsis ASN1 increases seed protein content and free amino acid content allocated to flowers and developing siliques (Lam et al. 2003). Constitutive OX of asparagine synthetase A from Escherichia coli in lettuce has been shown to enhance vegetative growth and leaf N content (Giannino et al. 2008). OX of pepper (Capsicum annuum) asparagine synthetase 1 (CaAS1) in Arabidopsis increases the conversion of aspartate to asparagine, leading to enhanced resistance to pathogens (Hwang et al. 2011). In soybean (Glycine max) seeds, the expression of ASN shows a positive correlation between free asparagine levels in developing seeds and protein concentration at maturity (Pandurangan et al. 2012). Our results also show the clear association between asparagine level and grain protein content in mature seeds by OX of OsASN1.
In cereals, total biomass and N concentration in grains, rather than any other organ, are important indicators of NUE (Pathak et al. 2011). Despite the attempts to improve NUE via transgenic approaches, there have been limited reports of successful increases in grain yield through the manipulation of NUE in cereals (Li et al. 2017). It may be difficult to increase NUE with a single transgene because of the complicated regulation of genes involved in N uptake, assimilation and remobilization. However, we here demonstrate that OX of a single ASN gene, OsASN1, can significantly improve both grain yield and N content in rice under N-limiting field conditions. This increased performance may be partly due to enhanced tolerance to N-limiting conditions at the seedling stage, and partly due to increased photosynthetic activity in mature plants. Even in N-sufficient field conditions, OX of the OsASN1 gene resulted in increased N concentration in grains, and consequently enhanced nutritional value of rice grains. Enhancing the nutrient contents of cereals is important for proper human nutrition and health (Peng et al. 2014). As rice is the most widely consumed staple food crop, feeding more than half of the world’s populations, improving its nutritional quality is critical. Grains from OX plants accumulated more N (31–40% higher than WT), and consequently, protein concentration in grains of OX lines was increased to 120% of that in WT grains. This clearly shows that the genetic manipulation of OsASN1 is a promising strategy to improve the nutritional status. Furthermore, the gain provided by OsASN1 OX regarding the grain protein content (GPC) was comparable to those previously reported on GPC using various different approaches (Zhou et al. 2009, Peng et al. 2014, Jiang et al. 2016). Our approach consisting in OX of a single gene improved GPC in real field practice.
In addition, although the protein content of rice grains is relatively low compared with other cereal crops (Mahender et al. 2016), the protein quality of rice grains is superior due to a better balanced amino acid profile and the presence in high amounts of highly nutritive and digestible glutelins (Chattopadhyay et al. 2018). The protein digestibility and nutritional value of rice grains are higher than those of the other major cereals, such as wheat, maize and barley (Amagliani et al. 2017). Rice proteins are generally regarded as hypoallergenic and suitable for the development of protein-enriched ingredients and products with high nutritional value (Helm and Burks 1996). Our results showed that OX of OsASN1 enhanced seed protein content in plants grown under N-sufficient and N-limiting outdoor conditions, indicating that transgenic OsASN1 seeds may serve as a valuable source of rice protein-containing products.
Here, we present a role of OsASN1 for optimal plant development in both N-sufficient conventional paddy fields and N-limiting conditions. Interestingly, OX of OsASN1 does not alter plant morphology and improves nutritional quality without causing yield reduction in paddy fields. Therefore, rice overexpressing a single gene encoding OsASN1 provides a significant and practical contribution toward the increase in yield and grain protein contents in nitrogen-limited soil as well as in agricultural practice with reduced nitrogen fertilization.
Materials and Methods
Plant growth
Rice seedlings were grown on a modified 1/2 MS medium containing NH4Cl as a sole N source and 5 mM KCl instead of 5 mM KNO3. To test the expression of OsASN genes responding to changes in external N supply, seedlings were grown on the modified 1/2 MS medium with 0, 0.1, 1, 10 or 100 mM NH4Cl as the only N source. To test the effect of N limitation at seedling stage, we germinated and then grew seedlings on the modified 1/2 MS medium containing 0.1 mM NH4Cl as the only N source and 5 mM KCl instead of 5 mM KNO3. For nitrogen sufficiency, 10 mM NH4Cl was added to the modified 1/2 MS medium as the only N source and KCl was substituted for KNO3. For N toxicity test, we added 100 mM NH4Cl to the modified 1/2 MS medium.
RNA preparation and mRNA quantification
Total RNA was isolated with WelPrep Total RNA isolation reagent (WELGENE, Republic of Korea), according to the manufacturer’s instructions, and treated with RNase-free DNase I (Takara Bio, Shiga, Japan) to prevent genomic DNA contamination. The first-strand cDNA was synthesized from 2 μg of total RNA in a 25-μl volume using ImProm II Reverse Transcriptase System (Promega, Madison, WI, USA). To determine the gene expression levels, qRT-PCR was performed on CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA) using a SYBR Premix ExTaq Kit (Takara Bio, Shiga, Japan). Levels of OsUbi mRNA served to normalize the expression ratio for each gene. Changes in gene expression were calculated using the ΔΔCt method. Gene-specific primers used for qRT-PCR are listed in Supplementary Table S1.
Isolation of knockout mutants and generation of OX lines of OsASN1
Two putative OsASN1 mutant lines were isolated from the rice flanking-sequence tag database (Jeon et al. 2000, An et al. 2003). Genotyping was performed using two gene-specific primers and one T-DNA-specific primer (Supplementary Table S1). We did not backcross the T-DNA mutants. Instead, we used the T3/T4 generations to analyze their phenotypes along with the segregant WTs. We originally obtained the two T-DNA mutant lines in a heterozygote state and the T-DNA in these lines segregated with the ratio of 1:2:1 (WT; heterozygotes:homozygotes), suggesting only one copy of T-DNA in both lines. After the isolation of T-DNA homozygote lines and their segregant WT lines harboring no T-DNA, we grew them in a paddy field for the generation progress and seed amplification. Transcript levels of OsASN1 were determined by qRT-PCR using cDNA prepared from shoots harvested from 7-day-old osasn1-1 in Hwayoung cultivar (line 3D-02739), osasn1-2 in Dongjin cultivar (line 3A-05359) and segregated WT seedlings. To construct an OsASN1 OX vector, the full-length OsASN1 cDNA was amplified using OxF and OxR primers (Supplementary Table S1). The PCR products were cloned into pGA1611 under the control of maize (Zea mays) ubiquitin promoter, and the resulting construct was introduced into the rice cultivar Dongjin via Agrobacterium-mediated transformation (Lee et al. 1999, Kim et al. 2009).
Agronomic traits
Knockout mutants and OX lines of OsASN1 were transplanted and grown to maturity in paddy fields located at the Daegu Gyeongbuk Institute of Science and Technology (35.8°N; 128.7°E). Fertilizer was applied as described previously (Oh et al. 2009). To test the effect of N limitation on agronomic traits of plants, WT, OX and osasn1 seedlings at the four-leaf stage were transplanted and grown to maturity in the same pot containing N-limited nursery soil, in which the N level was 1/16th (25 mg/kg) of the regular N level in N-enriched nursery soil (400 mg/kg). To evaluate the yield components under conventional field or N-limiting growth condition, we grew T3 and T4 generations of T-DNA mutants and OX lines along with their segregant WT in 2016 and 2017. Therefore, the growth of the mutants and overexpression lines were mostly synchronized with their respective segregant WT plants. Agronomic traits were evaluated in five plants of each genotype (WT, OX and osasn1 mutants) at the ripening stage. The following agronomic traits were evaluated: heading date, plant height, aboveground mass, tiller number per plant, spikelet number per panicle, filling rate, grain yield per plant, 1,000-grain weight, seed weight, seed length, seed thickness and seed width.
Determination of total N, soluble protein and amino acid content
Tissues from WT, OX and osasn1 plants were dried in an oven at 80°C to a constant weight. Samples were then ground to a fine powder using a mortar and pestle and stored in a desiccator. The total N content was determined using an Elemental Analyzer (Vario MICRI cube; Elementar, Germany), according to the manufacturer’s instructions, with acetanilide as a standard. Soluble protein content was determined as described previously (He et al. 2011). Amino acids were extracted using 2% (w/v) sulfosalicylic acid and quantified as described previously (Rosen 1957). Concentrations of glutamine and asparagine were determined using the l-Asparagine/l-Glutamine/Ammonia Assay Kit (Megazyme International Ireland, Bray, Ireland), according to the manufacturer’s instructions.
N uptake assay
Seeds of WT, OX and osasn1 plants were germinated and seedlings were cultured for 7 d in Yoshida nutrient solution (Yoshida et al. 1976), in which 1.44 mM NH4NO3 was replaced by 2.88 mM NH4Cl. Next, seedlings were treated with 15N-labeled NH4Cl (10% atom 15NH4Cl; Sigma-Aldrich). Roots and shoots were harvested at 12, 24, 36 and 48 h after treatment and dried at 70°C. The dried samples were weighed and ground, and 15N abundance was estimated as described previously (Masclaux-Daubresse and Chardon 2011).
Supplementary Data
Supplementary data are available at PCP online.
Supplementary Material
Acknowledgments
We thank Kyungsook An for generating the transgenic lines and handling the seed stock and Dr. Gi-Gyeong Park for maintaining plants.
Funding
Institute for Basic Science (IBS-R013-D1) (to H.G.N.); PHC STAR (no 34300ZK) to C.M.-D., 2017K1A3A1A21013795 to P.O.L.); and the IJPB benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
Disclosures
The authors have no conflicts of interest to declare.
References
- Amagliani L., O’Regan J., Kelly A.L., O’Mahony J.A. (2017) The composition, extraction, functionality and applications of rice proteins: a review. Trends Food Sci. Technol. 64: 1–12. [Google Scholar]
- An S., Park S., Jeong D.-H., Lee D.-Y., Kang H.-G., Yu J.-H., et al. (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 133: 2040–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao A., Liang Z., Zhao Z., Cai H. (2015) Overexpressing of OsAMT1–3, a high affinity ammonium transporter gene, modifies rice growth and carbon–nitrogen metabolic status. Int. J. Mol. Sci. 16: 9037–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S.M., Habash D.Z. (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182: 608–620. [DOI] [PubMed] [Google Scholar]
- Chardon F., Noël V., Masclaux-Daubresse C. (2012) Manipulating NUE in Arabidopsis and crop plants to improve yield and seed quality. J. Exp. Bot. 63: 3401–3412. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay K., Sharma S.G., Bagchi T.B., Molla K.A., Sarkar S., Marndi B.C., et al. (2018) Development of recombinant high yielding lines with improved protein content in rice (Oryza sativa L.). J. Agric. Sci. 156: 241–257. [Google Scholar]
- Chen Z.C., Ma J.F. (2015) Improving nitrogen use efficiency in rice through enhancing root nitrate uptake mediated by a nitrate transporter, NRT1.1B. J. Genet. Genomics 42: 463–465. [DOI] [PubMed] [Google Scholar]
- Fox T., DeBruin J., Haug Collet K., Trimnell M., Clapp J., Leonard A., et al. (2017) A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 15: 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A., Kusano M. (2014) A network perspective on nitrogen metabolism from model to crop plants using integrated ‘omics’ approaches. J. Exp. Bot. 65: 5619–5630. [DOI] [PubMed] [Google Scholar]
- Funayama K., Kojima S., Tabuchi-Kobayashi M., Sawa Y., Nakayama Y., Hayakawa T., et al. (2013) Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 54: 934–943. [DOI] [PubMed] [Google Scholar]
- Gaufichon L., Marmagne A., Belcram K., Yoneyama T., Sakakibara Y., Hase T., et al. (2017) ASN1-encoded asparagine synthetase in floral organs contributes to nitrogen filling in Arabidopsis seeds. Plant J. 91: 371–393. [DOI] [PubMed] [Google Scholar]
- Gaufichon L., Masclaux-Daubresse C., Tcherkez G., Reisdorf-Cren M., Sakakibara Y., Hase T., et al. (2013) Arabidopsis thaliana ASN2 encoding asparagine synthetase is involved in the control of nitrogen assimilation and export during vegetative growth. Plant Cell Environ. 36: 328–324. [DOI] [PubMed] [Google Scholar]
- Gaufichon L., Reisdorf-Cren M., Rothstein S.J., Chardon F., Suzukia A. (2010) Biological functions of asparagine synthetase in plants. Plant Sci. 179: 141–153. [Google Scholar]
- Giannino D., Nicolodi C., Testone G., Frugis G., Pace E., Santamaria P., et al. (2008) The overexpression of asparagine synthetase A from E. coli affects the nitrogen status in leaves of lettuce (Lactuca sativa L.) and enhances vegetative growth. Euphytica 162: 11–22. [Google Scholar]
- Havé M., Marmagne A., Chardon F., Masclaux-Daubresse C. (2017) Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68: 2513–2529. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Chino M. (1990) Chemical composition of phloem sap from the upper most internode of the rice plant. Plant Cell Physiol. 31: 247–251. [Google Scholar]
- He D., Han C., Yang P. (2011) Gene expression profile changes in germinating rice. J. Integr. Plant Biol. 53: 835–844. [DOI] [PubMed] [Google Scholar]
- Helm R.M., Burks A.W. (1996) Hypoallergenicity of rice protein. Cereal Food World 41: 839–843. [Google Scholar]
- Huang M., Zhang H., Zhao C., Chen G., Zou Y. (2019) Amino acid content in rice grains is affected by high temperature during the early grain-filling period. Sci. Rep. 9: 2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.S., An S.H., Hwang B.K. (2011) Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 67: 749–762. [DOI] [PubMed] [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22: 561–570. [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Ma A., Xie L., Ramachandran S. (2016) Improving protein content and quality by over-expressing artificially synthetic fusion proteins with high lysine and threonine constituent in rice plants. Sci. Rep. 6: 34427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir G. (2016) Genetic approaches of increasing nutrient use efficiency especially in cereal crops—a review. J. Bio-Sci. 22: 111–125. [Google Scholar]
- Kim S.R., Lee D.Y., Yang J.I., Moon S., An G. (2009) Cloning vectors for rice. J. Plant Biol. 52: 73–78. [Google Scholar]
- Kusano M., Tabuchi M., Fukushima A., Funayama K., Diaz C., Kobayashi M., et al. (2011) Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J. 66: 456–466. [DOI] [PubMed] [Google Scholar]
- Lam H.M., Hsieh M.S., Coruzzi G. (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J. 16: 345–353. [DOI] [PubMed] [Google Scholar]
- Lam H.M., Wong P., Chan H.K., Yam K.M., Chen L., Chow C.M., et al. (2003) Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 132: 926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea P.J., Sodek L., Parry M.A.J., Shewry P.R., Halford N.G. (2007) Asparagine in plants. Ann. Appl. Biol. 150: 1–26. [Google Scholar]
- Lee S., Jeon J.S., Jung K.H., An G. (1999) Binary vector for efficient transformation of rice. J. Plant Biol. 42: 310–316. [Google Scholar]
- Li H., Hu B., Chu C. (2017) Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68: 2477–2488. [DOI] [PubMed] [Google Scholar]
- Luo L., Qin R., Liu T., Yu M., Yang T., Xu G. (2018) OsASN1 plays a critical role in asparagine-dependent rice development. Int J Mol Sci. 20: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahender A., Anandan A., Pradhan S.K., Pandit E. (2016) Rice grain nutritional traits and their enhancement using relevant genes and QTLs through advanced approaches. Springerplus 5: 2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Chardon F. (2011) Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J. Exp. Bot. 62: 2131–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Daniel-Vedele F., Dechorgnat J., Chardon F., Gaufichon L., Suzuki A. (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105: 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moison M., Marmagne A., Dinant S., Soulay F., Azzopardi M., Lothier J., et al. (2018) Three cytosolic glutamine synthetase isoforms located in different order veins work together for N remobilization and seed filling in Arabidopsis. J. Exp. Bot. 69: 4379–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., Kim Y.S., Kwon C.W., Park H.K., Jeong J.S., Kim J.K. (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150: 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M., Ishiyama K., Kojima S., Konishi N., Nakano K., Kanno K., et al. (2015) Asparagine synthetase 1, but not asparagine synthetase 2, is responsible for the biosynthesis of asparagine following the supply of ammonium to rice roots. Plant Cell Physiol. 56: 769–778. [DOI] [PubMed] [Google Scholar]
- Ohashi M., Ishiyama K., Kojima S., Konishi N., Sasaki K., Miyao M., et al. (2018) Outgrowth of rice tillers requires availability of glutamine in the basal portions of shoots. Rice 11: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandurangan S., Pajak A., Molnar S.J., Cober E.R., Dhaubhadel S., Hernández-Sebastià C., et al. (2012) Relationship between asparagine metabolism and protein concentration in soybean seed. J. Exp. Bot. 63: 3173–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R.R., Lochab S., Raghuram N. (2011) Improving Plant Nitrogen-Use Efficiency in Comprehensive Biotechnology, Vol. 4, 2nd edn.Elsevier, Oxford: pp. 209–218. [Google Scholar]
- Peng B., Kong H., Li Y., Wang L., Zhong M., Sun L., et al. (2014) OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 5: 4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H. (1957) A modified ninhydrin colorimetric analysis of amino acids. Arch. Biochem. Biophys. 67: 10–15. [DOI] [PubMed] [Google Scholar]
- Sun L., Wang Y., Liu L.L., Wang C., Gan T., Zhang Z., et al. (2017) Isolation and characterization of a spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice. Sci. Rep. 7: 41846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M., Sugiyama K., Ishiyama K., Inoue E., Sato T., Takahashi H., et al. (2005) Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 42: 641–651. [DOI] [PubMed] [Google Scholar]
- Tamura W., Hidaka Y., Tabuchi M., Kojima S., Hayakawa T., Sato T., et al. (2010) Reverse genetics approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids 39: 1003–1012. [DOI] [PubMed] [Google Scholar]
- Tamura W., Kojima S., Toyokawa A., Watanabe H., Tabuchi-Kobayashi M., Hayakawa T., et al. (2011) Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice. Front. Plant Sci. 2: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.L., Huang J.F., Cai S.H., Wang R.C. (2007) Nitrogen contents of rice panicle and paddy by hyperspectral remote sensing. Pak. J. Biol. Sci. 10: 4420–4425. [DOI] [PubMed] [Google Scholar]
- Tegeder M., Masclaux-Daubresse C. (2018) Source and sink mechanisms of nitrogen transport and use. New Phytol. 217: 35–53. [DOI] [PubMed] [Google Scholar]
- Wong H.-K., Chan H.-K., Coruzzi G.M., Lam H.-M. (2004) Correlation of ASN2 gene expression with ammonium metabolisms in Arabidopsis. Plant Physiol. 134: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Guo M., Sun S., Zou Y., Yin S., Liu Y., et al. (2019) Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat. Commun. 10: 1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Foorno D., Cock J., Gomez K. (1976) Laboratory Manual for Physiological Studies of Rice, 3rd edn International Rice Research Institute, Philippines: p. 61. [Google Scholar]
- Zhou Y., Cai H., Xiao J., Li X., Zhang Q., Lian X. (2009) Over‐expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 118: 1381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.