Abstract
Pollen lipids are essential for sexual reproduction, but our current knowledge regarding lipid dynamics in growing pollen tubes is still very scarce. Here, we report unique lipid composition and associated gene expression patterns during olive pollen germination. Up to 376 genes involved in the biosynthesis of all lipid classes, except suberin, cutin and lipopolysaccharides, are expressed in olive pollen. The fatty acid profile of olive pollen is markedly different compared with other plant organs. Triacylglycerol (TAG), containing mostly C12–C16 saturated fatty acids, constitutes the bulk of olive pollen lipids. These compounds are partially mobilized, and the released fatty acids enter the β-oxidation pathway to yield acetyl-CoA, which is converted into sugars through the glyoxylate cycle during the course of pollen germination. Our data suggest that fatty acids are synthesized de novo and incorporated into glycerolipids by the ‘eukaryotic pathway’ in elongating pollen tubes. Phosphatidic acid is synthesized de novo in the endomembrane system during pollen germination and seems to have a central role in pollen tube lipid metabolism. The coordinated action of fatty acid desaturases FAD2–3 and FAD3B might explain the increase in linoleic and alpha-linolenic acids observed in germinating pollen. Continuous synthesis of TAG by the action of diacylglycerol acyltransferase 1 (DGAT1) enzyme, but not phosphoplipid:diacylglycerol acyltransferase (PDAT), also seems plausible. All these data allow for a better understanding of lipid metabolism during the olive reproductive process, which can impact, in the future, on the increase in olive fruit yield and, therefore, olive oil production.
Keywords: Lipid composition, Lipid droplet, Olea europaea, Olive, Pollen tube, Transcriptomics
Introduction
The male gametophyte (i.e. the pollen grain) of higher plants comprises a walled vegetative cell that encloses a smaller generative cell, which splits into two sperm cells. When a compatible pollen grain lands on the appropriate stigma, the vegetative cell extrudes an outgrowth to form a pollen tube, which delivers the male gametes to the female gametophyte to achieve fertilization (Higashiyama and Takeuchi 2015). This process is of major importance from an agronomical viewpoint since a substantial fraction of the plant-derived food for human and animal consumption (i.e. seeds and fruits) typically requires a preceding fertilization step.
The pollen grain contains four structures of lipid nature, which differ in their origin, composition and function (Piffanelli et al. 1998). The vegetative cell of the male gametophyte is surrounded by a durable exine wall that is mostly made up of an amalgam of biopolymers termed sporopollenin (Ariizumi and Toriyama 2011). This barrier provides structural support to the male gametophyte and protects it from pathogen attacks. The exine also likely mediates the initial adhesion of pollen to the stigma surface (Zinkl et al. 1999). The sporopollenin is built up in the diploid tapetal cells from saturated precursors, such as long-chain fatty acids or long aliphatic chains (Domínguez et al. 1999) and, to a lesser extent, phenolic compounds (Osthoff and Wiermann 1987). The exine precursors are released to the anther lumen and assembled onto the microspore surface, likely with the guidance of the primexine wall that would act as a template (Blackmore and Barnes 1990). Lipid metabolism in the tapetum is somehow linked to proper exine wall formation (Ariizumi et al. 2004).
After apoptosis, the tapetal remnants are retained by the exine and form an extracellular matrix known as ‘pollen coat’ or ‘pollenkitt’ (Murphy 2006). The nature of the pollen coat is mainly lipid, being neutral lipids esterified by saturated fatty acids (SFA) the most abundant constituents (Hernández-Pinzón et al. 1999, Murphy 2006). This sticky coating aids the male gametophyte to preserve its water status after leaving the anther (Elleman and Dickinson 1986, Preuss et al. 1993), protects it against UV radiation and attack from pathogens (Hsieh and Huang 2007) and mediates pollen–pollinator interactions (Pacini and Franchi 1994). In plants with dry stigmas, pollen coat lipids and proteins are mobilized and form a foot at the contact point that contributes to adhesion and promotes pollen–stigma recognition and water transport into the grain (Preuss et al. 1993, Mayfield and Preuss 2000). The pollen coat is left behind on the stigma, so its influence seems to be restricted to the early steps of germination.
The haploid vegetative cell of the developing male gametophyte also synthesizes and accumulates a large amount of neutral lipids in specialized organelles called lipid droplets (LD) (Zienkiewicz et al. 2011). These storage lipids appear densely packed and surrounded by a single layer of polar lipids containing a few unique proteins (Huang 2018). These reserves are critical for normal pollen development, as knockout mutations in the two triacylglycerol (TAG) synthesizing enzymes phosphatidyl:diacylglycerol acyltransferase 1 (PDAT1) and diacylglycerol acyltransferase 1 (DGAT1) of Arabidopsis lead to microspore abortion after first mitosis (Zhang et al. 2009). During pollen germination, LD polarize near the germinative aperture and move to the emerging pollen tube (Rodríguez-García et al. 2003). Then, storage lipids are partially mobilized during the early stages of pollen tube growth (Zienkiewicz et al. 2010), likely through the action of lipase and lipoxygenase (LOX) enzymes (Zienkiewicz et al. 2013, Müller and Ischebeck 2018), as observed in seeds (Feussner et al. 1995, Eastmond 2006). Interestingly, the absence of sugars in the culture medium does not affect olive pollen germination and pollen tube growth rates (Zienkiewicz et al. 2013). Therefore, soluble sugars (e.g. glucose) resulting from starch mobilization and lipid reserves may be sufficient for the onset of germination. Moreover, LD mobilization is sped up in a medium lacking a carbon source, suggesting that sugars may modulate the activity of lipid-degrading enzymes. For a long time, LD were regarded as a mere energy source. However, continuous pollen tube growth also demands a vast amount of membrane precursors. That need could be initially covered by stored TAG, which may serve as an inert intermediate product for the synthesis of membrane lipids (Ischebeck 2016). The endoplasmic reticulum (ER) network surrounding LD may facilitate their rapid incorporation (Piffanelli et al. 1998). Intracellular lipids also play key roles as signaling molecules in processes that are essential for pollen tube growth, such as signal transduction, cytoskeleton dynamics and vesicle trafficking (Murphy 2012).
A comprehensive lipid profiling of the mature male gametophyte was previously reported in a number of species including rapeseed (Piffanelli et al. 1997), maize (Bianchi et al. 1990), tobacco (Dorne et al. 1988), sunflower (Schulz et al. 2000), Aleppo pine (Andrikopoulos et al. 1985) and olive (Rodríguez-Rosales and Donaire 1988) among others. Different lipid classes, such as sterols (Villette et al. 2015), galactolipids (Nakamura et al. 2009), sphingolipids (Luttgeharm et al. 2015) and free fatty acids (FFA) and alkanes (Bashir et al. 2013), were also deeply analyzed in the mature pollen of several species. To date, a single study describes the kinetics of neutral and polar lipids during rapeseed microspore/pollen development (Piffanelli et al. 1997). These authors also depicted the expression patterns of several lipid biosynthetic genes that were differentially regulated in developing microspores/pollen grains and tapetal cells. A pioneer comparative study reported the lipid composition of the olive pollen grain at maturity and after 24 h of germination (Rodríguez-Rosales and Donaire 1988). Transcriptomic data from olive pollen are also publicly available (Carmona et al. 2015, Iaria et al. 2016). More recently, Rotsch et al. (2017) outlined the sterol and fatty acid profile at 13 stages of tobacco pollen development, from Te to germinated pollen. Therefore, our current knowledge regarding how lipid metabolism is adjusted in growing pollen tubes is still very scarce.
The olive tree (Olea europaea L.) is a typical oil-storing crop, and its cultivation has a great agronomical, ecological and social impact in the Mediterranean basin. Olive oil is the cornerstone of the Mediterranean diet, and its consumption also provides health benefits. The annual world production of olive oil ranges from about 2.8 to 3.3 million t per year on nearly 10 million ha of land (Vossen 2013). For this reason, the olive is a strategic crop for the agrifood industry. Plant lipid compounds supply 25% of dietary calories to the developed countries, as well as a source for renewable biomaterials and fuel (Chapman and Ohlrogge 2012). Moreover, fruit set and olive oil production highly depends on the success of pollination. In this work, we carried out for the first time a comprehensive study of the lipid composition and associated quantitative gene expression in the olive pollen during germination and early pollen tube growth. This information will allow us a better understanding of lipid metabolism during plant reproduction in an oil crop of great agronomical relevance as olive.
Results and Discussion
The lipid composition of olive pollen is unique
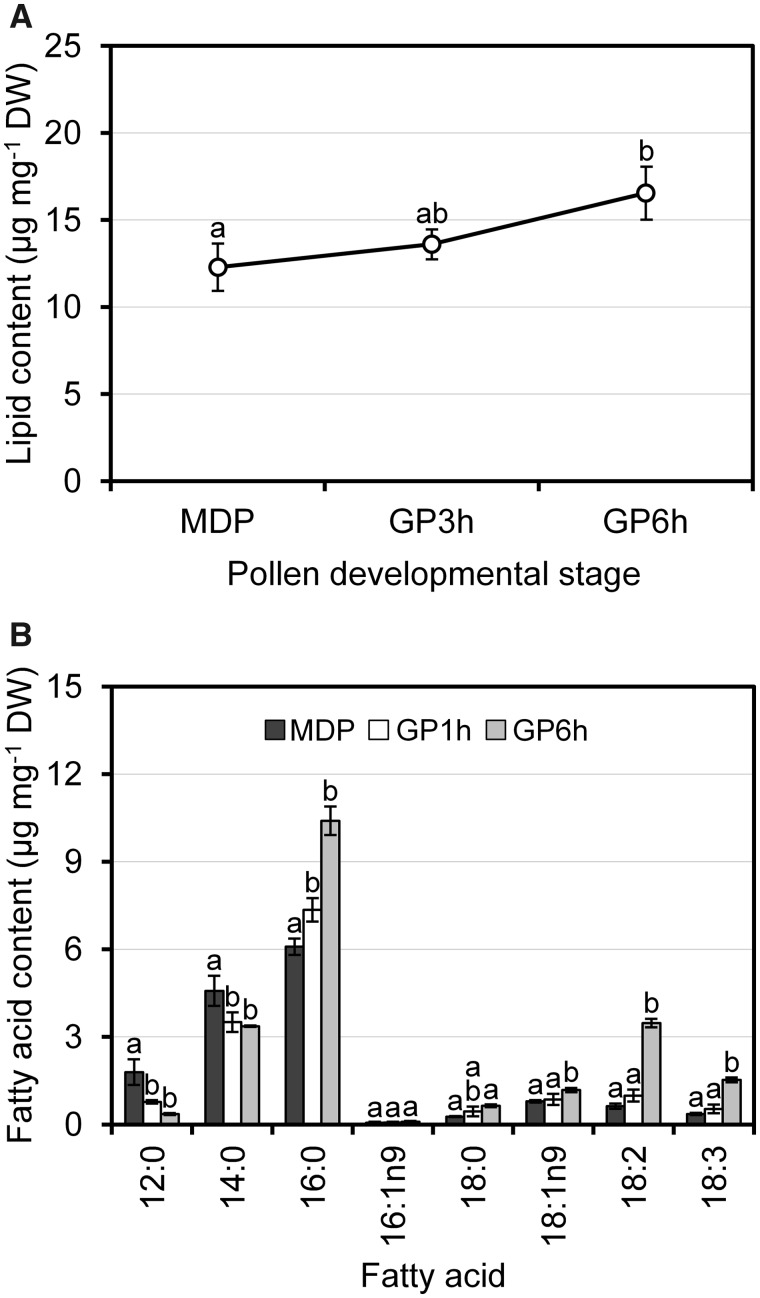
The mature pollen of olive (cv. Picual) contained about 12.3 ± 1.4 µg of total lipids per mg of DW. This amount was increased by one-third after the first 6 h of in vitro culture (Fig. 1A). These values were similar to those previously reported in olive pollen (Rodríguez-Rosales and Donaire 1988) but differed quantitatively from the average amount of acyl lipids present in other tissues, such as leaves, seeds and mesocarp (Donaire et al. 1975, Bianchi and Blahov 1994, Hernández et al. 2016). In addition, the total amount of lipids in the pollen grain greatly varies depending on the plant family (Evans et al. 1987, Thompson et al. 1993). These discrepancies can be explained in the framework of pollination. Thus, it has been shown convincingly that entomophilous pollen grains accumulate relatively more storage lipids than wind-pollinated ones (Stanley 1971, Baker and Baker 1979). Accordingly, we observed that the content of lipids in the olive pollen, a wind-pollinated species, was significantly lower than in the entomophilous pollen of other oleaginous plants, such as oil palm, rapeseed and sunflower (Opute 1975, Evans et al. 1987, Shakya and Bhatla 2010).
Fig. 1.
Analysis of total lipid content (A) and fatty acid composition (B) in olive pollen during in vitro germination. Different letters (a, b) denote statistically significant differences at P < 0.05 level. Error bars of three analytical replicates are shown. GPXh, germinated pollen for a period of X hours; MDP, mature dehydrated pollen; lauric acid (12:0); myristic acid (14:0); palmitic acid (16:0); palmitoleic acid (16:1); stearic acid (18:0); oleic acid (18:1n9); linoleic acid (18:2); α-linolenic acid (18:3).
The mature olive pollen grain displays a range of fatty acids from dodecanoic acid (C12) to octadecanoic acids (C18) (Fig. 1B). One notable feature is that SFA made up about 87.4% of the total fatty acid content. In fact, palmitic acid was the most abundant fatty acid (41.8%), followed by myristic and lauric acids (Fig. 1B). Interestingly, the fatty acid composition of the olive pollen grain was markedly different from those of leaves, and oil-accumulating tissues like seeds and mesocarp (Hernández et al. 2009). All these tissues are enriched in unsaturated fatty acids (UFA), such as oleic and linoleic acids in the case of seeds and mesocarp, and α-linolenic acid in leaves. The olive pollen grain fatty acid profile was also different from those reported in other oleaginous species, which contain larger amounts of linoleic or α-linolenic polyunsaturated fatty acids (PUFA) (Opute 1975, Evans et al. 1987, Brown et al. 2012). Moreover, we did not identify very-long-chain fatty acids (C20–C22), which are commonly detected in the pollen grain of other oleaginous species (Evans et al. 1987, Shakya and Bhatla 2010, Brown et al. 2012). PUFA could serve as precursors for the synthesis of volatile compounds, which might act as attractants of insect pollinators (Hopkins et al. 1969). This fact is in agreement with the low degree of unsaturation in olive pollen since this species is wind-pollinated. However, all these results should be taken with precaution since the pollen fatty acid content and composition greatly fluctuate depending on the genotype, as shown in maize (Kostić et al. 2017).
The variation in fatty acid quantities as a function of time during germination was also studied. Thus, among the SFA pool, palmitic and stearic acids significantly increased their levels during the first 6 h of germination, while the amount of myristic and lauric acids diminished. On the other hand, PUFA levels increased up to 5-fold and oleic acid also slightly increased its content in the 6-h germinated pollen (Fig. 1B). These PUFA may act as signaling molecules in processes that are essential for pollen growth as signal transduction or vesicle trafficking (Murphy 2012).
Transcriptomic analysis of lipid metabolism reveals the central role of glycerolipids during olive pollen germination and early pollen tube growth
Analysis of an olive pollen transcriptome available at the ReprOlive database allowed us to retrieve up to 735 tentative transcripts (TTs) associated with lipid metabolism (Supplementary Tables S1, S2), which represent about 2.64% of total TTs. After in silico sequence reconstruction and validation, we were able to identify up to 376 unique genes encoding for proteins/enzymes involved in the biosynthesis of all lipid classes, except those responsible for the production of lipopolysaccharides, suberin and cutin. These 376 olive pollen gene sequences were orthologs of 267 Arabidopsis proteins/enzymes (Supplementary Tables S1, S2). Among gene families, a total of 51 olive pollen proteins/enzymes displayed two distinct isoforms, whereas 19 of them showed three isoforms. In addition, we reported four different FAD2, FAH1 and CGI58 proteins, and one kinase, annotated as PIP5K6, exhibited up to seven unique isoforms. This transcriptomic dataset compiled here can be used in further functional comparisons across other oil-accumulating tissues and developmental stages in this species and also expands the transcriptomic resources to non-model species, thus also allowing comparative studies.
A comprehensive analysis of the lipid metabolism-related pathways pointed out that overall transcript levels remained steady or slightly decreased during pollen germination and pollen tube growth for all the biochemical routes dissected (Supplementary Table S2). A similar temporal expression pattern was previously reported in developing oil-storing seeds from several species (Troncoso-Ponce et al. 2011). However, this decline was not a general feature that could be applied to all lipid-related genes (Supplementary Table S2). Our data here highlight the importance of phospholipid-based signaling processes in pollen tubes, which accounted for ∼43% of the total mapped reads (Fig. 2A) and ∼23% of expressed genes (Fig. 2B). Accordingly, phosphatidylinositol (PI)-derived phosphoinositides and their associated enzymes have been shown to be essential for suitable pollen development and pollen tube apical growth, acting as secondary messengers in distinct signaling pathways and regulating key processes, such as endocytic trafficking, ionic transport and cell volume among others (Heilmann and Ischebeck 2016). The transcriptomic analysis also revealed that glycerolipid metabolism represented as much as 28% and 36% of total mapped reads and expressed genes, respectively (Fig. 2). Remarkably, the synthesis of TAG and phospholipids accounted for about 22% of mapped reads and proteins/enzymes (Fig. 2). These numbers may reflect the high requirements of membrane lipids for the rapid apical growth of pollen tubes on their way to the megagametophyte. Thus, the amount of membrane lipids needed was estimated as roughly 1.8 ng per cm of length for a 10-µm-width pollen tube (Kucerka et al. 2006). Interestingly, our data suggested that sphingolipid metabolism may also be important for pollen tube elongation since the associated biosynthetic genes made up ∼16% of mapped reads (Fig. 2). The first step in the synthesis of these lipid compounds is catalyzed by a serine palmitoyltransferase. This enzyme regulates the process of apoptosis, which is of vital importance for the male gametophyte development (Teng et al. 2008). To go in-depth in our study, we selected specific key genes of each lipid pathway for further quantitative expression studies, whose results will be discussed in parallel with lipidomic data below.
Fig. 2.
Transcriptomic analysis of lipid metabolism in the olive pollen during in vitro germination. A) Total number of RPMMs for each lipid metabolic pathway. B) Number of genes expressed in the olive pollen for each lipid metabolic route. GPXh, germinated pollen for a period of X hours; MDP, mature dehydrated pollen; RPMMs, reads per million mapped.
Storage lipids are mobilized during the early stages of olive pollen germination
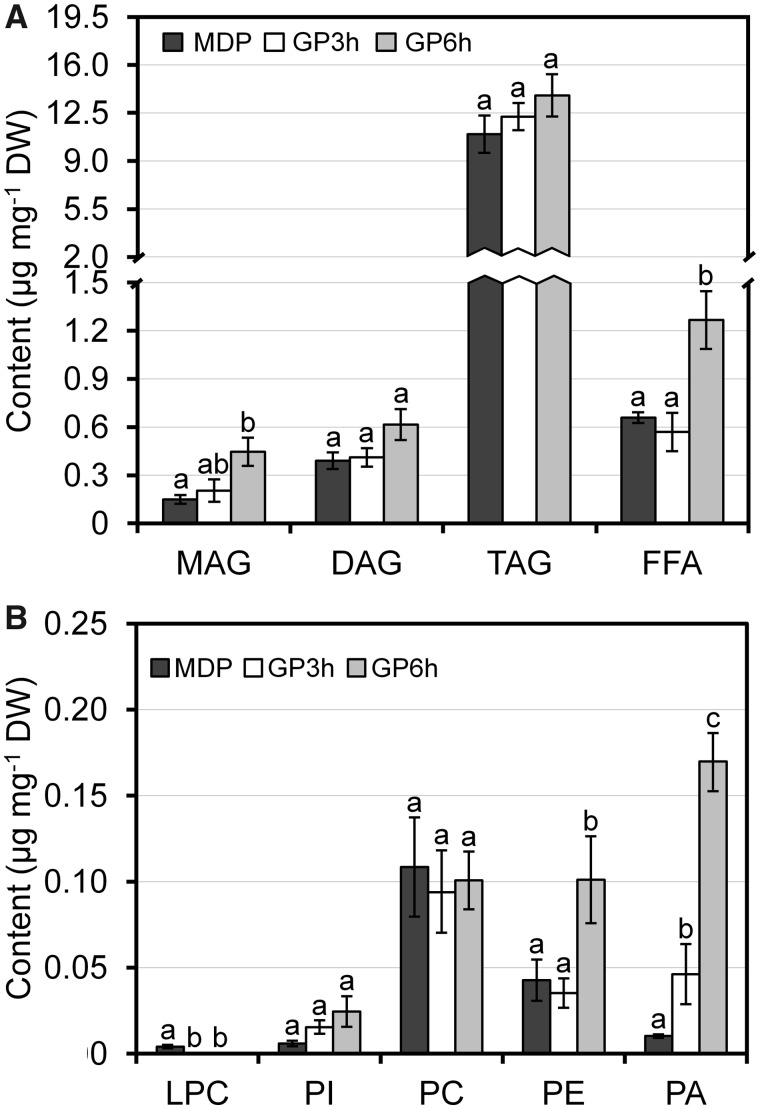
Neutral lipids constitute the bulk of lipids at the mature pollen stage (Fig. 3). The predominant component in the neutral lipid fraction was the TAG pool (11.0 ± 1.4 µg mg−1 DW) (Fig. 3A). TAG synthesis begins at the Te stage, continues during microspore development and speeds up after pollen first mitosis (Zienkiewicz et al. 2011). At pollen maturity, LD fill up most of the cytoplasm of the vegetative cell. During rehydration, these organelles polarize near the aperture through which the pollen tube emerges (Rodríguez-García et al. 2003). We observed that the TAG content remained unchanged during the first 6 h of germination (Fig. 3A). However, the fatty acid composition of TAG significantly changed during in vitro germination, with a significant drop in myristic and lauric acids, as well as an increase in linoleic and α-linolenic acids (Table 1). These data suggest that there is a simultaneous and continuous degradation and synthesis of TAG during the early stages of olive pollen in vitro germination.
Fig. 3.
Analysis of neutral (A) and polar (B) lipids in the olive pollen during in vitro germination. Different letters (a, b, c) denote statistically significant differences at P < 0.05 level. Error bars of three analytical replicates are shown. DAG, diacylglycerol; FFA, free fatty acid; DW, dry weigh; GPXh, germinated pollen for a period of X hours; LPC, lysophosphatidylcholine; MAG, monoacylglycerol; MDP, mature dehydrated pollen; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; TAG, triacylglycerol.
Table 1.
Fatty acid composition of major lipids during olive (Olea europaea L.) pollen in vitro germination
| Lipid | Fatty acid (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| 12:0 | 14:0 | 16:0 | 16:1 | 18:0 | 18:1 | 18:2 | 18:3 | |
| MAG | ||||||||
| MDP | 0.8 ± 0.8a | 7.6 ± 1.5a | 41.5 ± 1.8a | nd | 32.3 ± 4.7a | 6.7 ± 0.6a | 7.8 ± 1.8a | 3.7 ± 0.3a |
| GP3h | nd | 3.1 ± 1.7b | 41.3 ± 5.7a | 1.1 ± 1.9 | 25.0 ± 2.8a | 9.7 ± 1.5b | 12.6 ± 1.9b | 6.8 ± 1.0ab |
| GP6h | 0.2 ± 0.3a | 4.3 ± 0.1ab | 45.7 ± 0.9a | nd | 12.2 ± 0.5b | 4.6 ± 0.2a | 22.6 ± 1.5c | 10.4 ± 2.7b |
| DAG | ||||||||
| MDP | 3.1 ± 0.4a | 29.3 ± 0.4a | 53.1 ± 0.2a | 0.5 ± 0.2a | 3.4 ± 0.1a | 8.8 ± 0.1a | 0.6 ± 0.1a | 1.4 ± 1.0a |
| GP3h | 0.5 ± 0.5b | 21.9 ± 3.1b | 57.7 ± 2.3a | 0.2 ± 0.4a | 5.7 ± 0.8b | 10.5 ± 0.1b | 2.2 ± 0.9b | 1.3 ± 0.7a |
| GP6h | 0.2 ± 0.4b | 28.1 ± 2.4a | 55.2 ± 3.5a | 0.7 ± 0.2a | 5.2 ± 0.9b | 6.8 ± 0.2c | 2.5 ± 0.4b | 1.8 ± 0.7a |
| TAG | ||||||||
| MDP | 22.4 ± 0.3a | 32.6 ± 0.3a | 36.8 ± 0.1a | 0.2 ± 0.1a | 1.3 ± 0.1a | 4.4 ± 0.2a | 1.7 ± 0.3a | 0.7 ± 0.1a |
| GP3h | 18.8 ± 2.2b | 31.2 ± 0.3b | 38.4 ± 1.5ab | 0.3 ± 0.1a | 1.6 ± 0.1b | 4.3 ± 0.1a | 3.9 ± 0.1b | 1.6 ± 0.1b |
| GP6h | 17.0 ± 1.1b | 29.5 ± 0.2c | 39.1 ± 0.1b | 0.3 ± 0.0a | 1.7 ± 0.1c | 3.6 ± 0.0b | 6.4 ± 0.9c | 2.4 ± 0.3c |
| FFA | ||||||||
| MDP | 5.8 ± 0.8a | 25.4 ± 0.2a | 40.1 ± 1.1a | 0.7 ± 0.1a | 2.3 ± 0.1a | 6.2 ± 0.5a | 5.6 ± 0.4a | 14.0 ± 0.4a |
| GP3h | 1.8 ± 0.6b | 19.7 ± 0.9b | 42.7 ± 0.2b | 0.2 ± 0.3a | 4.6 ± 0.4b | 6.9 ± 0.4a | 9.6 ± 0.3b | 14.7 ± 0.9a |
| GP6h | 4.0 ± 1.5ab | 14.8 ± 0.6c | 39.2 ± 0.9a | 0.7 ± 0.4a | 3.7 ± 0.2c | 4.6 ± 0.1b | 21.7 ± 0.6c | 11.4 ± 0.3b |
| LPC | ||||||||
| MDP | nd | nd | 35.9 ± 5.1 | nd | 20.3 ± 0.3 | 14.0 ± 1.5 | 19.9 ± 1.2 | 9.8 ± 2.4 |
| GP3h | nd | nd | nd | nd | nd | nd | nd | nd |
| GP6h | nd | nd | nd | nd | nd | nd | nd | nd |
| PI | ||||||||
| MDP | nd | nd | 29.6 ± 6.8a | nd | 21.1 ± 5.0a | 13.3 ± 5.5a | 25.0 ± 4.6a | 11.3 ± 0.6a |
| GP3h | nd | nd | 38.1 ± 2.9a | nd | 18.2 ± 2.4a | 6.6 ± 3.7a | 23.8 ± 3.6a | 13.4 ± 3.4a |
| GP6h | nd | nd | 39.6 ± 7.2a | nd | 9.2 ± 2.1b | 8.0 ± 3.1a | 30.8 ± 2.1a | 12.5 ± 2.7a |
| PC | ||||||||
| MDP | nd | 0.1 ± 0.1 | 34.7±6.1a | 0.2 ± 0.2a | 2.7 ± 0.3a | 2.9 ± 0.2a | 43.4 ± 3.0a | 16.4 ± 2.1a |
| GP3h | nd | nd | 34.3 ± 3.0a | 0.1 ± 0.2a | 5.4 ± 0.3b | 7.3 ± 2.5ab | 37.7 ± 1.5b | 15.6 ± 1.2ab |
| GP6h | nd | nd | 36.9 ± 3.8a | 0.2 ± 0.1a | 5.1 ± 0.8b | 9.5 ± 2.6b | 36.2 ± 1.6b | 12.1 ± 1.5b |
| PE | ||||||||
| MDP | nd | 0.2 ± 0.3 | 27.8 ± 6.4a | 0.2 ± 0.3a | 4.9 ± 0.2a | 2.4 ± 0.5a | 49.2 ± 3.9a | 15.4 ± 1.4a |
| GP3h | nd | nd | 30.9±6.5a | nd | 9.5 ± 0.9b | 7.0 ± 0.7b | 38.8 ± 3.2b | 13.8 ± 2.3a |
| GP6h | nd | nd | 34.5 ± 3.5a | 0.3 ± 0.3a | 4.9 ± 0.6a | 4.3 ± 0.9c | 43.9 ± 1.3ab | 12.4 ± 0.8a |
| PA | ||||||||
| MDP | nd | nd | 27.7 ± 7.6a | nd | 18.9 ± 4.3a | 9.4 ± 2.5a | 28.9 ± 1.8a | 15.2 ± 1.6a |
| GP3h | nd | nd | 27.4 ± 5.9a | nd | 10.3 ± 1.8b | 7.0 ± 0.9a | 40.9 ± 5.5b | 14.4 ± 2.2a |
| GP6h | nd | nd | 31.5 ± 2.7a | 0.1 ± 0.2 | 4.0 ± 0.9b | 5.6 ± 0.8a | 43.9 ± 1.7b | 14.8 ± 0.9a |
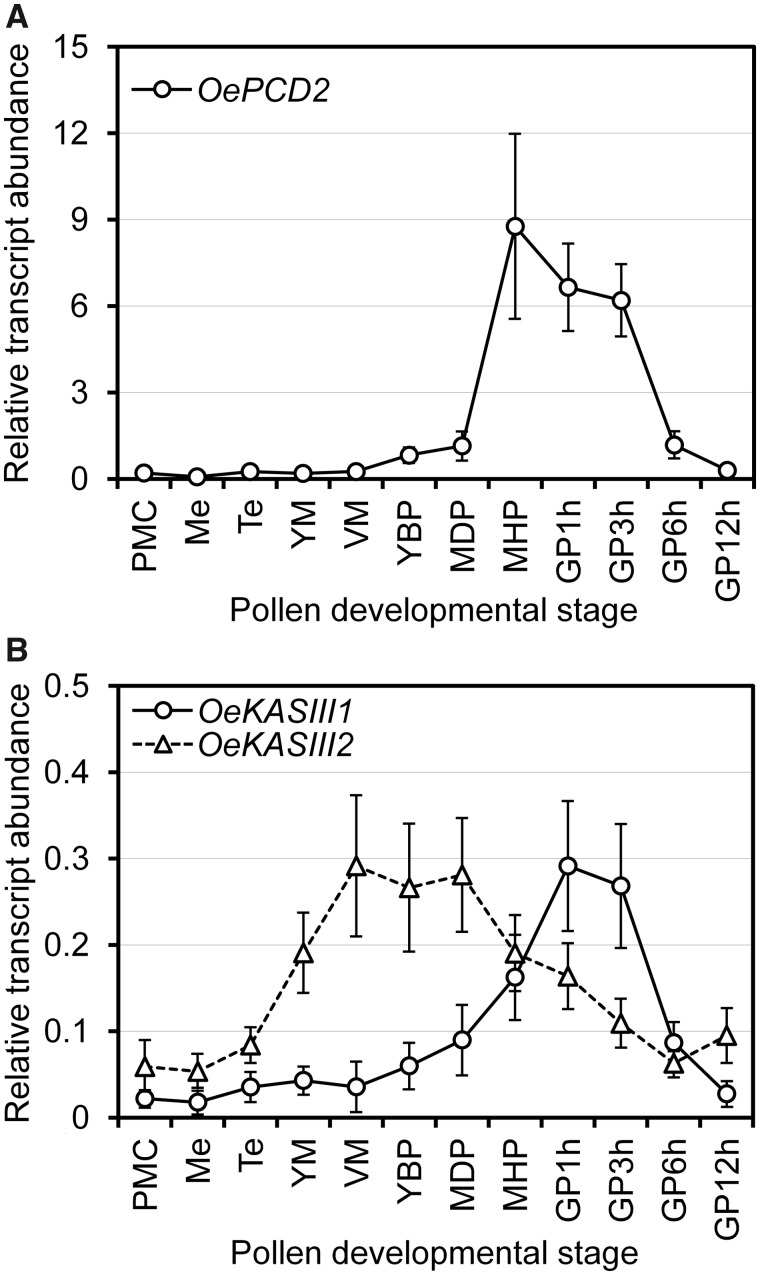
We previously reported that a phospholipase A enzyme may promote the access of hydrolytic enzymes to the storage lipids in olive pollen grains (Zienkiewicz et al. 2013). This enzyme may be active and stably associated with LD at the mature pollen stage. This is in good agreement with the idea that phospholipid breakdown is a prerequisite for TAG mobilization (Matsui et al. 1999). Accordingly, atomic force microscopy experiments demonstrated that a phospholipase A2 (PLA2) enzyme effectively produces holes in the phospholipid monolayer of cucumber seed LD (Noll et al. 2000). These holes are wide enough (∼80 nm) to provide access to TAG lipases (TAGL) and LOX enzymes. The survey of the olive pollen transcriptome revealed a single full-length cDNA, annotated as a ‘probable phospholipase A2 homolog 1’ (Supplementary Table S2). The protein displays 59.8% identity to an Arabidopsis phospholipase A2-gamma (At4g29460). Remarkably, this ER- and Golgi-localized PLA2γ enzyme plays critical roles in Arabidopsis pollen development and germination (Kim et al. 2011). Therefore, we analyzed the expression pattern of the OePLA2γ gene at the same well-defined stages of pollen development and germination. We observed that OePLA2γ began to increase its expression level just after meiosis (Me) of pollen mother cells (PMCs). Thus, OePLA2γ transcripts augmented almost 4-fold after first pollen mitosis (Fig. 4A). Expression levels remained constant during pollen maturation and the first 6 h of germination and then started to come down until 12 h of culture, which is in agreement with its mentioned putative function. According to the transcriptomic data, this is the sole PLA2 gene to be expressed in olive pollen, so its participation in the mobilization of storage TAG is quite likely. However, further investigations will be necessary to confirm this hypothesis.
Fig. 4.
Expression analysis by qPCR of selected key genes involved in storage TAG mobilization (A–C), fatty acid degradation by β-oxidation (D–F) and succinate production from acyl-CoA by the glyoxylate cycle (G–I) throughout olive pollen ontogeny, germination and pollen tube growth. The following genes were analyzed: ACX, CSY, ECI, GLK, ICL, LACS, MS, PLA2γ, and TAGL. GPXh, germinated pollen for a period of X hours; MDH, mature dehydrated pollen; Me, meiosis; MHP, mature rehydrated pollen; PMC, pollen mother cell; Te, tetrad; VM, vacuolated microspore; YBP, young and mid bicellular pollen; YM, young microspore.
TAG degradation involved both TAGL and LOX activities. These enzymes are likely recruited at the onset of germination as lipolytic activity was not detected on LD in the mature pollen grain (Zienkiewicz et al. 2013). Ebelactone B effectively inhibited olive pollen lipases, hampering the germination of pollen (Zienkiewicz et al. 2013). Up to 10 putative TAGL genes were expressed in olive pollen (Supplementary Fig. S1A), which correlates well with the high number of active lipases previously reported (Rejón et al. 2012). Most of these lipases were predicted to be secreted and located in the extracellular space, but a few are associated with LD (Supplementary Table S2). Two of them, named OeTGL5 and OeTGL6, were annotated as ‘TAG lipase SDP1-like’. OeTGL6 gene showed a high similarity with Arabidopsis SDP1 (SUGAR DEPENDENT PROTEIN 1, At5g04040). This gene encodes a TAG lipase with a patatin-like domain (Pfam01734) that catalyzes the initial step of breakdown of storage TAG in germinating seeds (Eastmond 2006). On the other hand, OeTGL5 showed high identity to an SDP1-like gene (At3g57140), which is strongly expressed in pollen tubes (Zou et al. 2009). The OeTGL5 protein showed a 90% identity to OeTGL6, and both proteins contain the conserved lipase motif (GXSXG) found in the patatin-like domain. Therefore, OeTGL5 and OeTGL6 genes may be good candidates to play a role in TAG degradation at the onset of pollen germination. To evaluate this possibility, we carried out quantitative expression analysis using specific primers to discriminate between these two SDP1-like genes. Hence, OeTGL5 showed low levels of transcription from PMCs to microspore stages (Fig. 4B). The expression of this gene was upregulated after vacuolated microspores (VM) underwent the first mitosis, reaching a peak after 3 h of germination, and then mRNA levels gradually decreased (Fig. 4B). On the other hand, OeTGL6 began to increase its expression after Me, at the Te stage, and then transcript levels remained high during pollen maturation (Fig. 4B). One hour after the emergence of the pollen tube, the transcription of OeTGL6 was downregulated and mRNA levels dropped. Moreover, a good temporal correlation was observed between OePLA2γ and OeTAGL genes during germination (Fig. 4A, B).
The contribution of other TAG lipases to TAG degradation cannot be ruled out. Thus, recently, an LD-located GXSXG lipase gene (OIL BODY LIPASE 1, OBL1, At3g14360) was functionally characterized in Arabidopsis and tobacco pollen tubes (Müller and Ischebeck 2018). OBL1 enzyme displayed lipase activity on TAG, diacylglycerol (DAG) and monoacylglycerol (MAG), and the obl1 null mutant was hindered in pollen tube growth. A partial sequence that showed homology to AtOBL1 was also identified in the olive pollen transcriptome (Supplementary Table S2). However, its expression level was much lower compared with the SDP1-like lipases, so its contribution to TAG mobilization in olive pollen tubes is not clear. Further experiments will be necessary to shed light on this question.
The amount of DAG followed a similar pattern to TAG (Fig. 3A). The SDP1 lipase is also able to hydrolyze DAG, although the activity rate was lower compared to TAG (Eastmond 2006), making additional DAG lipases likely. The olive pollen transcriptome also showed up to eight putative MAG lipase (MAGL) genes (Supplementary Fig. S1B). These enzymes catalyze the hydrolysis of MAG to fatty acid and glycerol (Kim et al. 2016). Indeed, an increase in glycerol levels was observed in tobacco pollen grains just after rehydration and pollen tube protrusion (Rotsch et al. 2017). Glycerol utilization in plants likely involves the production of glycerol-3-phosphate (G3P) by glycerol kinase (GLK) enzymes (Huang 1975). The olive pollen expressed a single GLK gene (Supplementary Table S2), which is orthologous to Arabidopsis GLI1 (At1g80460). This gene is upregulated during early postgerminative seedling growth (Eastmond 2004). Quantitative expression analysis showed two waves of transcription of OeGLK in the olive pollen, the first just after Me and the second after pollen rehydration (Fig. 4C). These data suggest that MAG is being degraded during pollen germination. However, despite the content of FFA increasing 2-fold after 6 h of germination, MAG levels also slightly increased (Fig. 3A). A putative explanation for this could be that MAG is being simultaneously degraded and synthesized during pollen in vitro germination, as reported for TAG. The presence of FFA in pollen grains has been reported previously in several plant species (Andrikopoulos et al. 1985, Bianchi et al. 1990, Piffanelli et al. 1997). Despite FFA being usually negligible in healthy plant cells, their content is increased in response to various stresses and they play a pivotal role in plant–microbe interactions (Walley et al. 2013). However, the origin and physiological role of these compounds in pollen grains is still unknown.
Fatty acids released from storage TAG may fuel early pollen tube growth
The fate of fatty acids released from TAG in the pollen tube is still unclear. It has been suggested that TAG may serve as a source of carbon since fast pollen tube growth is highly energy-consuming (Zienkiewicz et al. 2013). The drop in myristic and lauric acid levels observed during olive pollen germination is consistent with this hypothesis (Table 1). For this purpose, fatty acids should be channeled into β-oxidation within glyoxysomes in a similar way to that reported in germinating seeds (Graham 2008). Indeed, these organelles frequently localize in contact with LD (Hayashi et al. 2001). Typically, the vegetative cell of the olive pollen grain contains numerous peroxisomes scattered through its cytoplasm, which later appear in the pollen tube cytoplasm (Zafra et al. 2012). In addition, the olive pollen grain contains the full set of enzymes that constitute the core of β-oxidation (Supplementary Table S2). To address this issue, we analyzed the expression profile of several key enzymes that participate in fatty acid catabolism.
First, we studied long-chain acyl-CoA synthetases (LACS), which activate released FFA for feeding β-oxidation cycle (Graham 2008). The Arabidopsis genome contains nine LACS genes (Shockey et al. 2002) but only two, namely AtLACS6 and AtLACS7, were found to be involved in peroxisomal β-oxidation (Fulda et al. 2002). A single AtLACS7 ortholog, called OeLACS7, was retrieved from the olive pollen transcriptome. We found that the amount of OeLACS7 transcripts gradually increased during pollen ontogeny and was further doubled during rehydration before gradually returned to basal levels (Fig. 4D).
The first step in the β-oxidation pathway involves the participation of acyl-CoA oxidase (ACX) enzymes, which catalyze the desaturation of acyl-CoAs to 2-trans-enoyl-CoAs. Up to five different partial gene sequences, called OeACX1-1, OeACX1-2, OeACX3-1, OeACX3-2 and OeACX4, were identified in the olive pollen transcriptome (Supplementary Table S2). OeACX1-1, OeACX3-2, OeACX3-1 and OeACX4, in this order, showed the highest transcriptional activity in the olive pollen (Fig. 4E), while the contribution of OeACX1-2 was negligible. OeACX1-1 and OeACX3-2 genes exhibited similar patterns of temporal expression. Thus, mRNAs accumulated during pollen maturation, and expression peaked after 3 h of pollen tube growth followed by a decline. Transcripts of OeACX3-1 began to increase later in pollen maturation and its reduction started earlier, just after pollen rehydration. OeACX4 transcripts were only detected at a very low level in the mature pollen. The presence of multiple ACX proteins may be explained by their different substrate specificities. Thus, the Arabidopsis ACX1, ACX3 and ACX4 enzymes act on long- (C16–C18), medium- (C10–C14) and short-chain (C4–C8) acyl-CoAs, respectively (Hayashi et al. 1999, Hooks et al. 1999, Froman et al. 2000).
We further explored the expression profile of enoyl-CoA isomerase (ECI) genes. These enzymes are able to isomerize both 3-cis and 3-trans double bonds into the 2-trans form, being essential for the β-oxidation of UFA, such as oleic acid (Goepfert et al. 2008). The olive pollen expressed two distinct ECI genes, which were called OeECI1 and OeECI2 (Supplementary Table S2). These two genes displayed similar patterns of temporal expression, although OeECI2 transcript levels were significantly higher (Fig. 4F). Expression of both ECI genes reached a maximum in pollen grains germinated for 1 h and then gradually decreased, although at a higher rate in the case of OeECI1. Together, our data support the idea that fatty acids released from TAG might be degraded to acetyl-CoA during the early steps of olive pollen germination.
The glyoxylate cycle function is restricted to the early stages of pollen germination
In germinating oilseeds, the acetyl-CoA produced from β-oxidation of fatty acids can enter the glyoxylate cycle, resulting in the net production of four-carbon compounds, which can be used as precursors for the synthesis of sugars in the cytosol (Kornberg and Beevers 1957). Alternatively, these compounds can move to the mitochondria and enter the TCA cycle, being used for respiration (Pracharoenwattana et al. 2005). It was previously reported that two specific markers of the glyoxylate cycle, the isocitrate lyase (ICL) and malate synthase (MS) enzymes, were also fully active in rapeseed pollen (Zhang et al. 1994). These authors concluded that the glyoxysomal function was induced during pollen development. Similarly, the olive pollen transcriptome exhibits all the genes required for the glyoxylate cycle, including single ICL and MS genes (Supplementary Table S3). To explore their expression pattern during pollen germination and pollen tube growth, we performed qPCR expression analyses. Thus, we found that OeICL transcripts gradually accumulated during microspore/pollen development, reaching a maximum after pollen rehydration, and then declined (Fig. 4G). On the other hand, OeMS transcript levels triplicated at the bicellular pollen stage (Fig. 4H). The second wave of expression was further observed at short times of germination (1 h). Remarkably, the peak of expression of OeICL preceded that of OeMS, as previously reported in rapeseed pollen (Zhang et al. 1994). The glyoxylate cycle may not be functional at longer times of germination since MS activity could not be detected in tobacco pollen tubes after 4 h of growth (Mellema et al. 2002). Peroxisomal citrate synthase (CSY) is another key enzyme of the glyoxylate cycle that catalyzes an essential step in the respiration of fatty acids (Pracharoenwattana et al. 2005). We identified two members of the CSY gene family in olive pollen that were annotated as OeCSY2 and OeCSY3 (Supplementary Table S3). Both sequences exhibited high identity but a fragment of 37 nt in length was deleted in OeCSY3, so we could design specific primers for OeCSY2 but not for OeCSY3. Expression analysis showed that CSY2 was highly upregulated during pollen maturation, as well as in early growing pollen tubes (Fig. 4I). Overall, our data suggest that the glyoxylate cycle is active during olive pollen germination. Yet, its function may be restricted to the early stages, when the vegetative cell forms a nascent pollen tube.
Fatty acids are synthesized de novo in olive pollen tubes
The acetyl-CoA pool needed for fatty acid production is likely generated from pyruvate by a plastidial pyruvate dehydrogenase (PDH) complex. However, in germinating tobacco pollen, 50% of sugars are fermented through a PDH bypass. This pathway involves a pollen-specific pyruvate decarboxylase (PDC) that produces acetaldehyde, which is further converted into ethanol in a reversible reaction by alcohol dehydrogenase (Bucher et al. 1995). Alternatively, acetaldehyde may be oxidized to acetate by an aldehyde dehydrogenase (ALDH), which serves as a substrate of an acetyl-CoA synthase enzyme for the synthesis of acetyl-CoA (Mellema et al. 2002). The PDH bypass is essential for pollen development and fertility. Thus, in maize, the lack of RF2-encoded ALDH activity causes anther development arrest and rf2 plants fail to shed pollen (Liu et al. 2001). Interestingly, all the genes that participate in the so-called PDH bypass were identified in the olive pollen transcriptome (Supplementary Table S2), suggesting that this pathway may be active in growing olive pollen tubes as previously described for tobacco (Mellema et al. 2002). To investigate this hypothesis, we carried out an expression analysis of the PDC gene. A single PDC gene sequence, called OePDC2, was obtained from the olive pollen transcriptome database (Supplementary Table S2). When this enzyme is impaired, pollen tube growth through the style is severely reduced, as demonstrated in null pdc2 mutant plants of Petunia hybrida (Gass et al. 2005). The OePDC2 gene showed basal expression levels at the early stages of pollen ontogeny and was upregulated during pollen maturation (Fig. 5A). Transcript levels further increased up to 8-fold after pollen rehydrates and remained high and relatively constant during the first 3 h of germination. Finally, OePDC2 expression declined to basal levels at later times of culture. The expression pattern of the OePDC2 gene closely resembles that of the pollen-specific PDC2 gene described in Solanaceae species (Bucher et al. 1995, Gass et al. 2005). Therefore, our data suggest that the PDH bypass may be a common feature in the pollen tube of Angiosperms. The ethanolic fermentation pathway in pollen is primarily controlled by sugar supply rather than oxygen (Bucher et al. 1995), a reminiscent of what occurs in the yeast Saccharomyces cerevisiae (Pronk et al. 1996). The synthetic culture medium mimics the olive stigmatic/stylar exudate, which contains a high amount of sugars (Suárez et al. 2012). Therefore, at high sugar levels, respiration and fermentation likely take place concurrently in olive pollen tubes as previously reported in tobacco (Tadege and Kuhlemeier 1997).
Fig. 5.
Expression analysis by qPCR of fatty acid synthesis-associated genes PDC (A) and KASIII (B) throughout olive pollen ontogeny, germination and pollen tube growth. Pollen developmental stages are abbreviated as in Fig. 4.
The vegetative cell of the olive mature pollen contains plastids that are later localized at the organelle-rich subapical region of the pollen tube (Alché et al. 2004). Moreover, the olive pollen grain contains the full set of enzymes that constitute the fatty acid synthase complex (Supplementary Table S2). Therefore, fatty acids may be de novo synthesized in the olive pollen tube, as previously reported in tobacco (Mellema et al. 2002). To test this hypothesis, we carried out the qPCR expression analysis of two gene sequences retrieved from the olive pollen transcriptome, called OeKASIII1 and OeKASIII2, that exhibit a high identity to Arabidopsis β-ketoacyl-ACP synthase III (AtKASIII) gene (At1g62640) (Supplementary Table S2). This KAS enzyme catalyzes the first condensation reaction that starts fatty acid synthesis and therefore likely regulates its production. Olive pollen KASIII1 and KASIII2 genes showed different expression patterns during pollen development and germination. Thus, OeKASIII1 transcript levels stayed basal during microspore/pollen development and a rise in transcription occurred just after pollen rehydration, with a peak of expression at the interval between 1 and 3 h of germination (Fig. 5B). In contrast, OeKASIII2 began its transcription after Me and reached a maximum at the VM stage. Then, mRNA levels remained steady during pollen maturation and declined after pollen rehydration (Fig. 5B). These data suggest that both KASIII genes are developmentally regulated: OeKAIII2 might have a role in fatty acid synthesis during pollen development within the anther, while OeKAIII1 might be involved in the synthesis of fatty acids in elongating pollen tubes.
Plastidial glycerolipid biosynthesis through the ‘prokaryotic pathway’ is not active in olive pollen tubes
In higher plants, fatty acids are incorporated into glycerolipids through two different pathways, the ‘prokaryotic pathway’ that takes place in the chloroplast and the ‘eukaryotic pathway’, which is located in the ER (Browse and Somerville 1991). In the ‘prokaryotic pathway’, the G3P is converted to phosphatidic acid (PA) by acylation at the sn-1 and sn-2 positions, with fatty acids being transferred directly from the acyl–acyl carrier protein pool. These two metabolic steps are catalyzed by the plastidial G3P acyltransferase (ACT1/ATS1) and lysophosphatidic acid acyltransferase (LPAAT1) enzymes, respectively. However, ACT1/ATS1 and LPAAT1 genes were not identified in the olive pollen transcriptome (Supplementary Table S2). In addition, we did not detect the presence of palmitolinolenic acid (16:3) either in mature or germinated pollen grains (Fig. 1B), which is in agreement with the fact that the olive is an ‘18:3 plant’ (Hernández et al. 2016). Finally, our analysis showed that olive pollen, either mature or germinated, does not contain phosphatidylglycerol (PG) (Fig. 3B). PG is present at very low amounts in rapeseed, tobacco and lily pollen grains, while it has not been detected in Arabidopsis pollen. In this last species, PG seems to be dispensable for the pollen as the ACT1/ATS1, LPAAT1 and phosphatidylglycerol-phosphate synthase (PGP1) genes are not essential for pollen fertility (Hagio et al. 2002, Yu et al. 2004, Xu et al. 2006). Altogether, our data point out that the ‘prokaryotic pathway’ is not active in olive pollen tubes.
On the other hand, we did not observe the presence of monogaloctosyldiacylglycerol, digalactosyldiacylglycerol and sulfoquinovosyldiacylglycerol in olive pollen (Fig. 3). This fact suggests that these galactolipids and sulfolipids are not synthesized in plastids from DAG moieties synthesized in the ER through ‘the eukaryotic pathway’. This feature seems to be specific of pollen because the olive synthesizes plastidial glycerolipids in other plant organs and tissues (Hernández et al. 2008, 2016).
PA is synthesized de novo and plays a central role in glycerolipid metabolism in olive pollen tubes
The microsomal GPAT and LPAAT enzymes incorporate fatty acids activated with CoA to G3P. The ER-located GPAT enzyme (GPAT9) is essential for pollen fertility since the gpat9 pollen is unable to germinate in vitro (Shockey et al. 2016). Moreover, two ER-located LPAAT proteins (LPAAT2 and LPAAT3) seem to function redundantly in Arabidopsis pollen (Kim et al. 2005). A close inspection of the olive pollen transcriptome allowed us to identify the corresponding GPAT9 gene ortholog (Supplementary Table S2). This gene, called OeGPAT9, began its expression after Me and transcripts gradually accumulated during pollen maturation (Fig. 6A). A second peak was reached later, just after pollen rehydration, and was extended until the third hour of germination (Fig. 6A). On the other hand, three LPAAT-like partial transcripts, termed OeLPAAT4, OeLPAAT2-1 and OeLPAAT2-2, were also identified (Supplementary Table S2). Expression analyses demonstrated that only OeLPAAT2-1 and OeLPAAT4 genes were expressed in pollen tubes, although their temporal profiles clearly differed (Fig. 6B). Notably, OeLPAAT2-1 expression was upregulated in growing pollen tubes, with maximum and constant values during the first 6 h of culture (Fig. 6B). OeLPAAT4 transcript levels remained low and unchanged during microspore/pollen development but increased 10-fold at pollen maturity and then gradually declined during germination. Therefore, these data indicate that the ‘eukaryotic pathway’ of glycerolipid synthesis is likely activated during olive pollen maturation and continues operating during germination and pollen tube growth.
Fig. 6.
Expression analysis by qPCR of GPAT (A) and LPAAT (B) genes throughout olive pollen ontogeny, germination and pollen tube growth. Pollen developmental stages are abbreviated as in Fig. 4.
Polar lipids are essential constituents of biological membranes. Thus, tip growth requires the continuous maintenance of a plasma membrane domain to which Golgi-derived vesicles are targeted and fused. Phosphatidylcholine (PC) was the most abundant phospholipid in the olive mature pollen and doubled the content of phosphatidylethanolamine (PE) (Fig. 3B). These two phospholipids accounted for three-quarters of total polar lipids. The mature pollen grain also contained PI, lysophosphatidylcholine (LPC) and PA at lower amounts (Fig. 3B). A similar phospholipid distribution was reported in olive mesocarp (Hernández et al. 2016) and leaves (Guerfel et al. 2008), although no PA was detected in this latter tissue. Rapeseed, tobacco and lily pollen grains also contain similar ratios of PC, PE and PI (Dorne et al. 1988, Evans et al. 1990, Nakamura et al. 2009). By contrast, in Arabidopsis pollen, PA, PC and PI each represents ∼25% of total polar lipids (McDowell et al. 2013). Mutations disrupting genes involved in PC and PE production in Arabidopsis cause male infertility (Liu et al. 2015, Chen et al. 2018) or embryonic lethality (Mizoi et al. 2006).
The content of PE remained steady during the first 3 h of culture and then increased 2-fold in the next 3 h, while PC levels did not change during the process (Fig. 3B). By contrast, the amount of PA notably increased during germination to reach a peak of concentration in the interval between 3 and 6 h of culture (Fig. 3B). Similarly, PA levels transiently increased during tobacco pollen germination (Dorne et al. 1988, Potocký et al. 2003). PA and the enzymes involved in its synthesis are essential for regulating the pollen tube apical growth (Potocký et al. 2003, Monteiro et al. 2005, Pleskot et al. 2012, Vaz Dias et al. 2019). Moreover, a reduction in PA levels was associated with a lower capacity of pollen to germinate under high-temperature stress (Djanaguiraman et al. 2013). The hydrolysis of PC and PE by phospholipase D (PLD) enzymes generates PA (Potocký et al. 2003). However, the fatty acid composition of PA was very different from that of PC and PE (Table 1). Therefore, PA production directly from PC and PE through the action of PLD enzymes does not look plausible. Instead, the reassembling of preexisting membrane fragments into functional membranes may explain the behavior of PC and PE at the onset of germination (Elleman and Dickinson 1986). The increase in PA in olive pollen tubes correlates well with the expression pattern of its biosynthetic genes GPAT and LPAAT as shown in Fig. 6. These data suggest that part of the PA pool may be synthesized de novo in the ER via G3P. Another important route for PA production is by the sequential action of phospholipase C (PLC) and diacylglycerol kinase (DGK) enzymes on membrane inositol phospholipids (Vaz Dias et al. 2019). Accordingly, we identified two PI-PLC and seven DGK genes that are expressed at different levels in the olive pollen transcriptome (Supplementary Table S2). Whether these enzymes contribute to PA increase in olive pollen tubes requires more investigation.
Fatty acid desaturation in olive pollen tubes takes place in the ER by the coordinated action of FAD2-3 and FAD3B enzymes
Palmitic acid together with other SFA, such as lauric and myristic acids, was the most abundant acyl groups in TAG, DAG and FFA pools, while palmitic and stearic acids stand out in MAG (Table 1). This high content in SFA of TAG certainly influences the high proportion of SFA observed in total lipids, as TAG is the most abundant lipid in the olive pollen grain (Fig. 3). SFA do not have double bonds, so they can be more tightly packed in LD. A similar fatty acid composition was reported in maize pollen TAG, where palmitic acid accounts for about 85% of total fatty acids (Bianchi et al. 1990). However, this is not a common feature since UFA are usually more abundant in pollen TAG (Andrikopoulos et al. 1985, Dorne et al. 1988, Brown et al. 2012, Rotsch et al. 2017). The fatty acid composition of olive pollen TAG was also markedly different from those of leaves (Guerfel et al. 2008) and oil-accumulating tissues like oilseeds and mesocarp (Hernández et al. 2016, Parvini et al. 2016), which mainly contain TAG species enriched in oleic acid and also contain a high proportion of linoleic acid. The fatty acid composition followed a similar pattern in all neutral lipids during pollen germination, with minor differences (Table 1). Thus, lauric, myristic and oleic acids mostly diminished in parallel to a rise in stearic acid and PUFA. In addition, the amount of palmitic acid remained unchanged in MAG, DAG and FFA and slightly augmented in TAG. In the phospholipid pool, palmitic and linoleic acids were the most abundant fatty acids in LPC, PI, PC, PE and PA at the mature pollen stage (Table 1). In PC and PE pools, linoleic and α-linolenic acid levels came down after pollen germination, whereas the amount of stearic and oleic acids significantly augmented (Table 1). The opposite pattern was true for PA and PI. Palmitic acid levels remained constant during germination in all phospholipid classes.
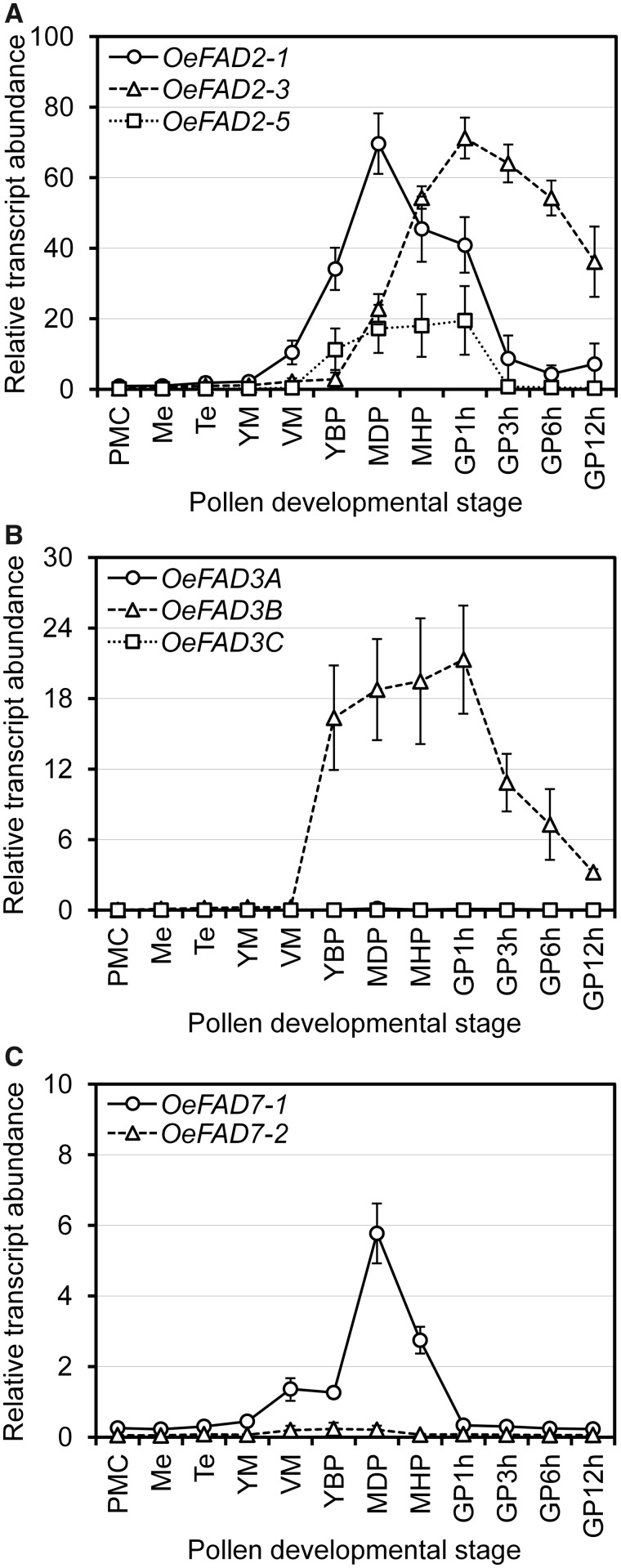
The increase in PUFA levels in TAG and FFA could explain the rise of the total linoleic and α-linolenic acid contents observed in the 6-h germinated pollen (Fig. 1B). Consequently, we retrieved four FAD2 genes from the olive pollen transcriptome (Supplementary Table S2). Two of these microsomal oleate desaturase genes were previously characterized in olive mesocarp and annotated as FAD2-1 and FAD2-2 (Hernández et al. 2005). FAD2-1 is strongly expressed in very young seeds and in leaves (Hernández et al. 2005), while FAD2-2 is responsible for the linoleic acid content in the olive fruit mesocarp (Hernández et al. 2009). Two additional FAD2 genes, named OeFAD2-3 and OeFAD2-5, were also recently identified and characterized (Hernández et al. 2020). To determine the contribution of each FAD2 gene to oleic acid desaturation in olive developing and germinating pollen grains, we carried out qPCR expression analysis using specific probes. All the four FAD2 genes differed in their expression levels. Thus, they could be grouped into highly (OeFAD2-1 and OeFAD2-3) and moderately (OeFAD2-5) expressed (Fig. 7A), while the expression of OeFAD2-2 was negligible (data not shown). OeFAD2-1 and OeFAD2-3 seemed to be the major contributors in quantitative terms to oleic acid desaturation in olive pollen, but they exhibited distinct temporal regulation patterns. Thus, OeFAD2-1 began its expression much earlier, just after microspores were released from Te. OeFAD2-1 transcripts accumulated during pollen maturation and then decreased (Fig. 7A). On the other hand, OeFAD2-3 was upregulated at the latest stage of pollen maturation and reached a peak of expression during the first 3 h of pollen tube growth. Later, transcript levels gradually declined but still remained high after 12 h of culture (Fig. 7A). Therefore, most of the oleate desaturase activity in olive pollen tubes could be mainly attributed to OeFAD2-3 gene.
Fig. 7.
Expression analysis by qPCR of fatty acid desaturases FAD2 (A), FAD3 (B) and FAD7 (C) throughout olive pollen ontogeny, germination and pollen tube growth. Pollen developmental stages are abbreviated as in Fig. 4.
Concerning the desaturation of linoleic acid to α-linolenic acid, up to three different FAD3 genes were identified in the olive pollen transcriptome (Supplementary Table S2). Two of them, namely OeFAD3A and OeFAD3B, were previously cloned and functionally characterized (Banilas et al. 2007, Hernández et al. 2016). OeFAD3A is expressed in olive seeds and leaves and is responsible for linoleic acid desaturation in the seed, while OeFAD3B gene did not show any expression in the vegetative tissues (Hernández et al. 2016). We retrieved a third FAD3 partial gene sequence from the olive pollen transcriptome, called OeFAD3C (Supplementary Table S2). However, OeFAD3A and OeFAD3C showed very low basal transcriptional activity in olive developing and germinating pollen grains (Fig. 7B). By contrast, OeFAD3B was highly expressed in olive pollen. OeFAD3B gene transcription started after the first pollen mitosis, mRNA levels remained quite high and constant until the first hour of tube growth was completed and then gradually declined (Fig. 7B). These data suggest that this microsomal linoleate desaturase may be pollen specific since OeFAD3A was suggested to be the main gene responsible for the production of α-linolenic acid in olive seeds (Hernández et al. 2016). On the other hand, in the olive fruit mesocarp, desaturation of linoleic acid is carried out by two plastidial linoleate desaturases, called OeFAD7-1 and OeFAD7-2 (Poghosyan et al. 1999, Hernández et al. 2016). A single FAD7-1 orthologous gene was identified after surveying the olive pollen transcriptome (Supplementary Table S2). OeFAD7-1 showed high expression levels in developing olive pollen grains, but transcription was downregulated in pollen tubes (Fig. 7C), suggesting a low contribution of these genes to the synthesis of α-linolenic acid in the olive pollen grain. Moreover, no FAD8 genes were expressed in either mature or germinating pollen grains. Interestingly, the triple fad3-2 fad7-2 fad8 mutation in Arabidopsis led to male sterility, but exogenous applications of α-linolenic acid or jasmonate restored fertility (McConn and Browse 1996). Actually, jasmonic acid synchronizes pollen maturation and anther dehiscence, being essential for male fertility (Ishiguro et al. 2001). However, the contribution of each desaturase to the fad3-2 fad7-2 fad8 pollen phenotype was not determined in Arabidopsis. Here, our expression data strongly suggest that only OeFAD3B contributes to the α-linolenic acid content in olive pollen tubes.
TAG is mainly synthesized de novo through the action of DGAT1 enzyme but not PDAT activity in olive pollen tubes
Germinating pollen grains are expected to degrade rather to synthesize storage lipids. However, a pioneering study in tobacco showed that all lipid classes, including storage TAG, are synthesized de novo in growing pollen tubes (Dorne et al. 1988). In plants, TAG is synthesized from DAG by DGAT, which catalyzes the last step of the so-called Kennedy pathway. TAG is also produced by an acyl-CoA-independent pathway that involves PDAT activity. These two enzymes are essential for normal pollen development as the knockout dgat1-1 pdat1-2 double mutation in Arabidopsis resulted in sterile pollen that lacked LD (Zhang et al. 2009). To address whether TAG is also newly synthesized in olive pollen tubes, we first surveyed the olive pollen transcriptome to search for DGAT and PDAT orthologous genes. Thus, two DGAT genes, called OeDGAT1 (Giannoulia et al. 2000) and OeDGAT2 (Banilas et al. 2011), and two PDAT genes, named OePDAT1-1 and OePDAT1-2, were identified (Supplementary Table S2). Subsequent qPCR analysis revealed that both DGAT genes began to be transcribed after the first mitosis but their expression patterns diverged at this point (Fig. 8A). Thus, the transcriptional activity of OeDGAT2 stayed constant during pollen maturation and the first hour of germination and was strongly downregulated afterward. Yet, OeDGAT1 transcripts gradually accumulated in growing pollen tubes until reaching a maximum at the 3 h of growth (Fig. 8A). On the other hand, only the OePDAT1-1 gene was expressed at high levels in the olive pollen grain (Fig. 8B). This gene was induced at the late stages of pollen maturation. A peak of expression was observed after rehydration, but levels decreased during germination. So, the expression profile of these two biosynthetic genes points to DGAT1 as the enzyme involved in TAG de novo biosynthesis during olive pollen germination and pollen tube growth. Its physiological meaning is not yet clear, but some authors have speculated that packed-TAG may serve as an inert intermediate sink to finely adjust lipid synthesis/turnover or be involved in acyl editing during pollen tube growth (Ischebeck 2016).
Fig. 8.
Expression analysis by qPCR of DGAT (A) and PDAT (B) genes throughout olive pollen ontogeny, germination and pollen tube growth. Pollen developmental stages are abbreviated as in Fig. 4.
Materials and Methods
Plant material collection
Olive (Olea europaea L. cv. Picual) pollen grains were collected at the time of anthesis by shaking the panicles inside large paper bags. Samples were further sieved through a set of nylon meshes to remove flower remnants. Olive developing anthers at the stages of PMC, Me, Te, young microspore (YM), VM, young-mid bicellular pollen (YBP) and mature dehydrated pollen (MDP) were also dissected under a stereomicroscope. Developmental stages were determined by correlating the size (i.e. length and width) of floral buds with the morphological and cellular traits of developing microspores/pollen grains (Supplementary Table S4). Microspore/pollen staging was carried out by squashing and DAPI staining under an Eclipse Ti fluorescence microscope (Nikon, Tokyo, Japan) (Supplementary Fig. S2). All samples were harvested from genotyped olive trees at three different locations in Andalusia (Spain): Granada (sample # 1; GPS coordinates: S37°11′17.5″, W3°36′24.1″, Armilla (sample # 2; S37°8′27.6″, W3°37′6.6″) and Mengíbar (sample # 3; S37°58′42.6″, W3°47′52.9″). Collected samples were immediately processed as described below, or frozen in liquid N2 and properly stored at −80°C until use.
In vitro pollen germination
Freshly collected pollen grains were rehydrated in a humid chamber for 30 min at 30°C in the dark and germinated at room temperature in a synthetic growth medium [0.03% (w/v) Ca(NO3)2, 0.01% (w/v) KNO3, 0.02% (w/v) MgSO4 and 0.01% (w/v) H3BO3] supplemented with 5% (w/v) sucrose. Pollen grains were sampled just after rehydration, and at several time intervals (1, 3, 6 and 12 h) of in vitro germination. Germinated pollen-enriched fractions were prepared by passing the culture through a nylon mesh (ca. 25 μm of pore size).
Lipid extraction and analysis
Total lipids were extracted from mature pollen and germinated pollen samples following the method of Folch et al. (1957), with minor modifications (Piffanelli et al. 1997). Briefly, pollen grain samples (∼0.5 g each) were osmotically shocked in 20 mM phosphate buffer (pH 7.2), sonicated three times for 10 s each and incubated in a chloroform:methanol (2:1) mixture for 30 min. Then, 0.88% (w/v) KCl was added and samples were centrifuged at 5,000 × g for 5 min to separate the two phases. The lower phase was collected, and an equal volume of a mixture of methanol:water (1:1) was added. After centrifugation at 5,000× g for 5 min, the lower phase was collected, the solvent was completely removed under a stream of N2 and total lipids were dissolved in chloroform for further analysis.
For polar lipids analysis, total lipids were fractionated in a Lichrolut 0.5 g silica gel cartridge (Merck, Kenilworth, NJ, USA) equilibrated with chloroform, using a vacuum manifold. The solution of total lipids was loaded on the column, which was then washed with hexane:diethyl ether (1:1) to elute neutral lipids from the column. Subsequently, the column was washed with methanol to recover the polar lipids fractions, which was evaporated to dryness under an N2 stream, and dissolved in chloroform for the separation of different polar lipid classes.
Neutral and polar lipids separation was carried out by thin-layer chromatography according to Hernández et al. (2008). Individual lipids were visualized under iodine vapor and identification was made by reference to authentic standards. Fatty acid methyl esters (FAMEs) of the total lipid fraction and the individual lipid classes were produced by acid-catalyzed transmethylation (Garcés and Mancha 1993) and analyzed using a 7890A gas chromatograph (Agilent Technologies, Santa Clara, CA, USA), according to Román et al. (2015). Heptadecanoic acid was used as an internal standard to calculate the lipid and fatty acid contents in the samples. Results are presented as means (μg per mg of DW) ± SD of three independent biological replicas (i.e. samples # 1–3 described above).
Transcriptomic analysis of lipid metabolism in olive pollen
Lipid metabolism-related transcripts were identified and retrieved from a de novo assembled and automatically annotated olive pollen transcriptome that is publicly available at the ‘ReprOlive’ website (http://reprolive.eez.csic.es/olivodb, last accessed 15/05/2020), which was previously generated in our laboratory from pollen cDNA libraries subjected to 454 sequencing (Carmona et al. 2015). This transcriptome comprises three different developmental stages, namely MDP and germinated pollen at two different times (1 and 6 h). To manually annotate the olive pollen genes related to lipid metabolism, we performed sequence similarity searches using the BLAST+ algorithm (Camacho et al. 2009) of the ReprOlive toolbar with a cutoff E-value of <1e−10, and selecting the option ‘pollen_transcriptome_v1.1’ from the list of assemblies. For this purpose, the Arabidopsis acyl-lipid metabolism gene sequences depicted at the ARALIP website (http://aralip.plantbiology.msu.edu, last accessed 15/05/2020; Li-Beisson et al. 2013) were used as query sequences. In addition, we performed text-based searches using the ‘Annotation Search’ tool by combining Gene Ontology (GO) terms, EC numbers, descriptions and gene names. We also surveyed the olive pollen transcriptome to search for genes/enzymes associated with the glyoxylate cycle. All retrieved TTs were further manually verified using the BLAST algorithm against the Viridiplantae database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Chimeric sequences were discarded, and those transcripts annotated as ‘Unknown protein’ or ‘Uncharacterized protein’ were suitably re-annotated. When an Arabidopsis query gene blasted to two or more partial TT at the ‘ReprOlive’ record, the corresponding sequences were aligned using the Clustal Omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/) and blasted against the olive (cv. Farga) and oleaster (i.e. wild olive; Olea europaea var. sylvestris) genomes (Cruz et al. 2016, Unver et al. 2017), to distinguish and annotate putative distinct protein/enzyme isoforms.
All retrieved olive pollen gene sequences are listed in Supplementary Tables S2, S3. These tables also incorporate useful information about the subcellular localization (only when inferred by direct assay), EC number, ReprOlive accession number, sequence status, AA sequence identity (%) with the corresponding Arabidopsis gene orthologs and abbreviation of each reported gene. Supplementary Tables S2, S3 also provide expression raw data (i.e. number of mapped reads) for each TT. Quantitative data were expressed and plotted as reads per million mapped to enable comparison across the olive pollen developmental stages. Some key genes were subsequently selected for expression analysis by qPCR (Supplementary Table S5).
RNA isolation and cDNA synthesis
Total RNA was extracted from developing anthers and germinated pollen samples (∼1.5 g each) by using the RNeasy Tissue RNA isolation kit (Qiagen, Hilden, Germany). The removal of gDNA was carried out by RNase-free DNase treatment (Qiagen) as described by the manufacturer. First-strand cDNA was synthesized with 2 μg of total RNA using a High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions.
qPCR analysis
Expression analysis of lipid metabolism-related genes was performed by qPCR using a LightCycler® 480 system (Roche, Basel, Switzerland). Partial gene sequences were blasted against the olive genome to recover the full mRNA sequences to design specific probes. Primers for gene-specific amplification (Supplementary Table S6) were designed using the Primer3 version 4.1.0 software (http://bioinfo.ut.ee/primer3). Target and housekeeping (olive phosphoenolpyruvate carboxylase, PEPC; ReprOlive database accession # po11_olive_008366) genes were subjected to qPCR in 96-well optical reaction plates in 20 μl mixtures per well, using a LightCycler® 480 SYBR Green I Master mix (Roche). Cycle threshold (Ct) values were obtained with the LightCycler® 480 software (Roche). The relative expression level of each gene was calculated using the equation 2−ΔCt, where ΔCt = CtTARGET − CtPEPC (Livak and Schmittgen 2001, Pfaffl 2004). Data are presented as means ± SD of three independent biological experiments (i.e. samples # 1–3 described above) and two technical replicas each.
Statistical analysis
The Shapiro–Wilk test was used to test the normality of data, and the Levene test was applied to assess the equality of variances. Statistical comparisons for each variable were carried out by the one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test for multiple comparisons. All analyses were performed using the SPSS v.23 software (IBM, USA). Different letters in figures and tables denote statistically significant differences at P < 0.05 level.
Supplementary Data
Supplementary data are available at PCP online.
Supplementary Material
Acknowledgment
We thank the CIFA ‘Venta del Llano’ (Mengíbar, Jaén, Spain) for kindly providing us the plant material. We also thank the Open Access Publication Support Initiative of CSIC through its Unit of Information Resources for Research (URICI) for kindly supporting the publication fee.
Funding
Agencia Estatal de Investigación from the Ministerio de Economía y Competitividad (MINECO-AEI) (ERDF co-financed AGL2013-43042P and AGL2017-84298P to A.J.C., AGL2014-55300R and AGL2017-87871R to J.M.M.-R.) and Consejo Superior de Investigaciones Científicas (CSIC) (JAEDOC089 to M.L.H.).
Disclosures
The authors have no conflicts of interest to declare.
References
- Alché J.D.D., M’rani-Alaoui M., Castro A.J., Rodríguez-García M.I. (2004) Ole e 1, the major allergen from olive (Olea europaea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol. 45: 1149–1157. [DOI] [PubMed] [Google Scholar]
- Andrikopoulos N.K., Siafaka-Kapadai A., Demopoulos C.A., Kapoulas V.M. (1985) Lipids of Pinus halepensis pollen. Phytochemistry 24: 2953–2957. [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., et al. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181. [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K. (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460. [DOI] [PubMed] [Google Scholar]
- Baker H.G., Baker I. (1979) Starch in angiosperm pollen grains and its evolutionary significance. Am. J. Bot. 66: 591–600. [Google Scholar]
- Banilas G., Karampelias M., Makariti I., Kourti A., Hatzopoulos P. (2011) The olive DGAT2 gene is developmentally regulated and shares overlapping but distinct expression patterns with DGAT1. J. Exp. Bot .62: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banilas G., Nikiforiadis A., Makariti I., Moressis A., Hatzopoulos P. (2007) Discrete roles of a microsomal linoleate desaturase gene in olive identified by spatiotemporal transcriptional analysis. Tree Physiol. 27: 481–490. [DOI] [PubMed] [Google Scholar]
- Bashir M.E.H., Lui J.H., Palnivelu R., Naclerio R.M., Preuss D. (2013) Pollen lipidomics: lipid profiling exposes a notable diversity in 22 allergenic pollen and potential biomarkers of the allergic immune response. PLoS One 8: e57566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi G., Blahov G. (1994) Composition of lipid classes in the morphologically different parts of the olive fruit, cv. Coratina (Olea europaea L.). Eur. J. Lipid Sci. Technol. 96: 72–77. [Google Scholar]
- Bianchi G., Murelli C., Ottaviano E. (1990) Maize pollen lipids. Phytochemistry 29: 739–744. [Google Scholar]
- Blackmore S., Barnes S.H. (1990) Pollen wall development in angiosperms In Microspores: Evolution and Ontogeny. Edited by Blackmore S., Knox R.B. pp. 173–192. Academic Press, London. [Google Scholar]
- Brown A.P., Kroon J.T.M., Swarbreck D., Febrer M., Larson T.R., Graham I.A., et al. (2012) Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One 7: e30100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J., Somerville C. (1991) Glycerolipid synthesis: biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 467–506. [Google Scholar]
- Bucher M., Brander K.A., Sbicego S., Mandel T., Kuhlemeier C. (1995) Aerobic fermentation in tobacco pollen. Plant Mol. Biol. 28: 739–750. [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009) BLAST+: architecture and applications. BMC Bioinform. 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona R., Zafra A., Seoane P., Castro A.J., Guerrero-Fernández D., Castillo-Castillo T., et al. (2015) ReprOlive: a database with linked data for the olive tree (Olea europaea L.) reproductive transcriptome. Front. Plant Sci. 6: 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.D., Ohlrogge J.B. (2012) Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287: 2288–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Salari H., Taylor M.C., Jost R., Berkowitz O., Barrow R., et al. (2018) NMT1 and NMT3 N-methyltransferase activity is critical to lipid homeostasis, morphogenesis, and reproduction. Plant Physiol. 177: 1605–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F. Julca I. Gómez-Garrido J. Loska D. Marcet-Houben M. Cano E., et al. (2016) Genome sequence of the olive tree, Olea europaea. GigaSci. 5: 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djanaguiraman M., Prasad P.V.V., Schapaugh W.T. (2013) High day- or nighttime temperature alters leaf assimilation, reproductive success, and phosphatidic acid of pollen grain in soybean [Glycine max (L.) Merr.]. Crop Sci. 53: 1594–1604. [Google Scholar]
- Domínguez E., Mercado J.A., Quesada M.A., Heredia A. (1999) Pollen sporopollenin: degradation and structural elucidation. Sex. Plant Reprod. 12: 171–178. [Google Scholar]
- Donaire J.P., Sánchez A.J., López-Gorge J., Recalde L. (1975) Metabolic changes in fruit and leaf during ripening in the olive. Phytochemistry 14: 1167–1169. [Google Scholar]
- Dorne A.J., Kappler R., Kristen U., Heinz E. (1988) Lipid metabolism during germination of tobacco pollen. Phytochemistry 27: 2027–2031. [Google Scholar]
- Eastmond P.J. (2004) Cloning and characterization of the acid lipase from castor beans. J. Biol. Chem. 279: 45540–45545. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J. (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman C.J., Dickinson H.G. (1986) Pollen-stigma interactions in Brassica. IV. Structural reorganization in the pollen grains during hydration. J. Cell Sci. 80: 141–157. [DOI] [PubMed] [Google Scholar]
- Evans D.E., Rothnie N.E., Palmer M.V., Burke D.G., Sang J.P., Hilliard E.P., et al. (1987) Comparative analysis of fatty acids in pollen and seed of rapeseed. Phytochemistry 26: 1895–1897. [Google Scholar]
- Evans D.E., Sang J.P., Cominos X., Rothnie N.E., Knox R.B. (1990) A study of phospholipids and galactolipids in pollen of two lines of Brassica napus L. (rapeseed) with different ratios of linoleic to linolenic acid. Plant Physiol. 93: 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C., Kindl H., Kuhn H. (1995) Lipoxygenase-catalyzed oxygenation of storage lipids is implicated in lipid mobilization during germination. Proc. Natl. Acad. Sci. USA 92: 11849–11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., Stanley G.H.S. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- Froman B.E., Edwards P.C., Bursch A.G., Dehesh K. (2000) ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol. 123: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M., Shockey J., Werber M., Wolter F.P., Heinz E. (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant J. 32: 93–103. [DOI] [PubMed] [Google Scholar]
- Garcés R., Mancha M. (1993) One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal. Biochem. 211: 139–143. [DOI] [PubMed] [Google Scholar]
- Gass N., Glagotskaia T., Mellema S., Stuurman J., Barone M., Mandel T., et al. (2005) Pyruvate decarboxylase provides growing pollen tubes with a competitive advantage in petunia. Plant Cell 17: 2355–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoulia K., Haralampidis K., Poghosyan Z., Murphy D.J., Hatzopoulos P. (2000) Differential expression of diacylglycerol acyltransferase (DGAT) genes in olive tissues. Biochem. Soc. Trans. 28: 695–697. [PubMed] [Google Scholar]
- Goepfert S., Vidoudez C., Tellgren-Roth C., Delessert S., Hiltunen J.K., Poirier Y. (2008) Peroxisomal Δ3,Δ2-enoyl CoA isomerases and evolution of cytosolic paralogues in embryophytes. Plant J. 56: 728–742. [DOI] [PubMed] [Google Scholar]
- Graham I.A. (2008) Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142. [DOI] [PubMed] [Google Scholar]
- Guerfel M., Baccouri O., Boujnah D., Zarrouk M. (2008) Changes in lipid composition, water relations and gas exchange in leaves of two young ‘Chemlali’ and ‘Chetoui’ olive trees in response to water stress. Plant Soil 311: 121–129. [Google Scholar]
- Hagio M., Sakurai I., Sato S., Kato T., Tabata S., Wada H. (2002) Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol. 43: 1456–1464. [DOI] [PubMed] [Google Scholar]
- Hayashi H., De Bellis L., Ciurli A., Kondo M., Hayashi M., Nishimura M. (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J. Biol. Chem. 274: 12715–12721. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M., Hayashi H., Hara-Nishimura I., Nishimura M. (2001) Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218: 83–94. [DOI] [PubMed] [Google Scholar]
- Heilmann I., Ischebeck T. (2016) Male functions and malfunctions: the impact of phosphoinositides on pollen development and pollen tube growth. Plant Reprod. 29: 3–20. [DOI] [PubMed] [Google Scholar]
- Hernández M.L., Guschina I.A., Martínez-Rivas J.M., Mancha M., Harwood J.L. (2008) The utilization and desaturation of oleate and linoleate during glycerolipid biosynthesis in olive (Olea europaea L.) callus cultures. J. Exp. Bot. 59: 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández M.L., Mancha M., Martínez-Rivas J.M. (2005) Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 66: 1417–1426. [DOI] [PubMed] [Google Scholar]
- Hernández M. L. Padilla M. N. Mancha M. and Martínez-Rivas J. M. (2009) Expression Analysis Identifies FAD2-2 as the Olive Oleate Desaturase Gene Mainly Responsible for the Linoleic Acid Content in Virgin Olive Oil. J. Agric. Food Chem. 57: 6199–6206. [DOI] [PubMed] [Google Scholar]
- Hernández M.L., Sicardo M.D., Martínez-Rivas J.M. (2016) Differential contribution of endoplasmic reticulum and chloroplast ω-3 fatty acid desaturase genes to the linolenic acid content of olive (Olea europaea) fruit. Plant Cell Physiol. 57: 138–151. [DOI] [PubMed] [Google Scholar]
- Hernández M.L., Sicardo M.D., Arjona P.M., Martínez-Rivas J.M. (2020) Specialized functions of olive FAD2 gene family members related to fruit development and the abiotic stress response. Plant Cell Physiol. 61: 427–441. [DOI] [PubMed] [Google Scholar]
- Hernández-Pinzón I., Ross J.H., Barnes K.A., Damant A.P., Murphy D.J. (1999) Composition and role of tapetal lipid bodies in the biogenesis of the pollen coat of Brassica napus. Planta 208: 588–598. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Takeuchi H. (2015) The mechanism and key molecules involved in pollen tube guidance. Annu. Rev. Plant Biol. 66: 393–413. [DOI] [PubMed] [Google Scholar]
- Hooks M.A., Kellas F., Graham I.A. (1999) Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 20: 1–13. [DOI] [PubMed] [Google Scholar]
- Hopkins C.Y., Jevans A.W., Boch R. (1969) Occurrence of octadeca-trans-2, cis-9, cis-12-trienoic acid in pollen attractive to the honey bee. Can. J. Biochem. 47: 433–436. [DOI] [PubMed] [Google Scholar]
- Hsieh K., Huang A.H.C. (2007) Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19: 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.H.C. (1975) Enzymes of glycerol metabolism in the storage tissues of fatty seedlings. Plant Physiol. 55: 555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.H.C. (2018) Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol. 176: 1894–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaria D., Chiappetta A., Muzzalupo I. (2016) De novo transcriptome sequencing of Olea europaea L. to identify genes involved in the development of the pollen tube. Sci. World J. 2016: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T. (2016) Lipids in pollen—they are different. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1861: 1315–1328. [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Ok S.H., Bahn S.C., Jang J., Oh S.A., Park S.K., et al. (2011) Endoplasmic reticulum- and Golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. Plant Cell 23: 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.U., Li Y., Huang A.H.C. (2005) Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17: 1073–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R.J., Kim H.J., Shim D., Suh M.C. (2016) Molecular and biochemical characterizations of the monoacylglycerol lipase gene family of Arabidopsis thaliana. Plant J. 85: 758–771. [DOI] [PubMed] [Google Scholar]
- Kornberg H.L., Beevers H. (1957) A mechanism of conversion of fat to carbohydrate in castor beans. Nature 180: 35–36. [DOI] [PubMed] [Google Scholar]
- Kostić A.Ž., Mačukanović-Jocić M.P., Špirović Trifunović B.D., Vukašinović I.Ž., Pavlović V.B., Pešić M.B. (2017) Fatty acids of maize pollen. Quantification, nutritional and morphological evaluation. J. Cereal Sci. 77: 180–185. [Google Scholar]
- Kucerka N., Tristram-Nagle S., Nagle J.F. (2006) Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 208: 193–202. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y., Shorrosh B., Beisson F., Andersson M.X., Arondel V., Bates P.D., et al. (2013) Acyl-lipid metabolism. Arabidopsis Book 11: e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Cui X., Horner H.T., Weiner H., Schnable P.S. (2001) Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell 13: 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang G., Wang X. (2015) Role of aminoalcoholphosphotransferases 1 and 2 in phospholipid homeostasis in Arabidopsis. Plant Cell 27: 1512–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luttgeharm K.D., Kimberlin A.N., Cahoon R.E., Cerny R.L., Napier J.A., Markham J.E., et al. (2015) Sphingolipid metabolism is strikingly different between pollen and leaf in Arabidopsis as revealed by compositional and gene expression profiling. Phytochemistry 115: 121–129. [DOI] [PubMed] [Google Scholar]
- Matsui K., Hijiya K., Tabuchi Y., Kajiwara T. (1999) Cucumber cotyledon lipoxygenase during postgerminative growth. Its expression and action on lipid bodies. Plant Physiol. 119: 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J.A., Preuss D. (2000) Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2: 128–130. [DOI] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell S.C., López-Marques R.L., Poulsen L.R., Palmgren M.G., Harper J.F. (2013) Loss of the Arabidopsis thaliana P(4)-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development. PLoS One 8: e62577.23667493 [Google Scholar]
- Mellema S., Eichenberger W., Rawyler A., Suter M., Tadege M., Kuhlemeier C. (2002) The ethanolic fermentation pathway supports respiration and lipid biosynthesis in tobacco pollen. Plant J. 30: 329–336. [DOI] [PubMed] [Google Scholar]
- Mizoi J., Nakamura M., Nishida I. (2006) Defects in CTP: phosphorylethanolamine cytidylyltransferase affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18: 3370–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro D., Liu Q., Lisboa S., Scherer G.E.F., Quader H., Malhó R. (2005) Phosphoinositides and phosphatidic acid regulate pollen tube growth and reorientation through modulation of [Ca2+]c and membrane secretion. J. Exp. Bot. 56: 1665–1674. [DOI] [PubMed] [Google Scholar]
- Müller A.O., Ischebeck T. (2018) Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol. 217: 1062–1076. [DOI] [PubMed] [Google Scholar]
- Murphy D.J. (2006) The extracellular pollen coat in members of the Brassicaceae: composition, biosynthesis, and functions in pollination. Protoplasma 228: 31–39. [DOI] [PubMed] [Google Scholar]
- Murphy D.J. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249: 541–585. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kobayashi K., Ohta H. (2009) Activation of galactolipid biosynthesis in development of pistils and pollen tubes. Plant Physiol. Biochem. 47: 535–539. [DOI] [PubMed] [Google Scholar]
- Noll F., May C., Kindl H. (2000) Phospholipid monolayer of plant lipid bodies attacked by phospholipase A2 shows 80 nm holes analyzed by atomic force microscopy. Biophys. Chem. 86: 29–35. [DOI] [PubMed] [Google Scholar]
- Opute F.I. (1975) Lipid and sterol composition of the pollen of the west African oilpalm, Elaeis guineensis. Phytochemistry 14: 1023–1026. [Google Scholar]
- Osthoff K.S., Wiermann R. (1987) Phenols as integrated compounds of sporopollenin from Pinus pollen. J. Plant Physiol. 131: 5–15. [Google Scholar]
- Pacini E., Franchi G.G. (1994) Role of the tapetum in pollen and spore dispersal. Plant Syst. Evol. 7: 1–11. [Google Scholar]
- Parvini F., Sicardo M.D., Hosseini-Mazinani M., Martínez-Rivas J.M., Hernández M.L. (2016) Transcriptional analysis of stearoyl-acyl carrier protein desaturase genes from olive (Olea europaea) in relation to the oleic acid content of the virgin olive oil. J. Agric. Food Chem. 64: 7770–7781. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2004) Quantification strategies in real-time PCR InA-Z of Quantitative PCR. Edited by Bustin S.A. pp. 87–112. International University Line, La Jolla, CA. [Google Scholar]
- Piffanelli P., Ross J.H.E., Murphy D.J. (1997) Intra- and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J. 11: 549–562. [DOI] [PubMed] [Google Scholar]
- Piffanelli P., Ross J.H.E., Murphy D.J. (1998) Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11: 65–80. [Google Scholar]
- Pleskot R., Pejchar P., Bezvoda R., Lichtscheidl I.K., Wolters-Arts M., Marc J., et al. (2012) Turnover of phosphatidic acid through distinct signaling pathways affects multiple aspects of pollen tube growth in tobacco. Front. Plant Sci. 3: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poghosyan Z.P., Haralampidis K., Martsinkovskaya A.I., Murphy D.J., Hatzopoulos P. (1999) Developmental regulation and spatial expression of a plastidial fatty acid desaturase from Olea europaea. Plant Physiol. Biochem. 37: 109–119. [Google Scholar]
- Potocký M., Eliáš M., Profotová B., Novotná Z., Valentová O., Žárský V. (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J.E., Smith S.M. (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Lemieux B., Yen G., Davis R.W. (1993) A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7: 974–985. [DOI] [PubMed] [Google Scholar]
- Pronk J.T., Steensma H.Y., van Dijken J.P. (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12: 1607–1633. [DOI] [PubMed] [Google Scholar]
- Rejón J.D., Zienkiewicz A., Rodríguez-García M.I., Castro A.J. (2012) Profiling and functional classification of esterases in olive (Olea europaea) pollen during germination. Ann. Bot. 110: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-García M.I., M'rani-Alaoui M., Fernández M.C. (2003) Behavior of storage lipids during development and germination of olive (Olea europaea L.) pollen. Protoplasma 221: 237–244. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rosales M.P., Donaire J.P. (1988) Germination-induced changes in acyl lipids and free sterols of olive pollen. New Phytol. 108: 509–514. [Google Scholar]
- Román Á., Hernández M.L., Soria-García Á., López-Gomollón S., Lagunas B., Picorel R., et al. (2015) Non-redundant contribution of the plastidial FAD8 ω-3 desaturase to glycerolipid unsaturation at different temperatures in Arabidopsis. Mol. Plant 8: 1599–1611. [DOI] [PubMed] [Google Scholar]
- Rotsch A.H., Kopka J., Feussner I., Ischebeck T. (2017) Central metabolite and sterol profiling divides tobacco male gametophyte development and pollen tube growth into eight metabolic phases. Plant J. 92: 129–146. [DOI] [PubMed] [Google Scholar]
- Schulz S., Arsene C., Tauber M., McNeil J.N. (2000) Composition of lipids from sunflower pollen (Helianthus annuus). Phytochemistry 54: 325–336. [DOI] [PubMed] [Google Scholar]
- Shakya R., Bhatla S.C. (2010) A comparative analysis of the distribution and composition of lipidic constituents and associated enzymes in pollen and stigma of sunflower. Sex. Plant Reprod. 23: 163–172. [DOI] [PubMed] [Google Scholar]
- Shockey J., Regmi A., Cotton K., Adhikari N., Browse J., Bates P.D. (2016) Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 170: 163–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J.M., Fulda M.S., Browse J.A. (2002) Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129: 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley R.G. (1971) Pollen chemistry and tube growth InPollen: development and Physiology. Edited by Heslop-Harrison J. pp. 131–135. Butterworth, London. [Google Scholar]
- Suárez C., Castro A.J., Rapoport H.F., Rodríguez-García M.I. (2012) Morphological, histological and ultrastructural changes in the olive pistil during flowering. Sex. Plant Reprod. 25: 133–146. [DOI] [PubMed] [Google Scholar]
- Tadege M., Kuhlemeier C. (1997) Aerobic fermentation during tobacco pollen development. Plant Mol. Biol. 35: 343–354. [DOI] [PubMed] [Google Scholar]
- Teng C., Dong H., Shi L., Deng Y., Mu J., Zhang J., et al. (2008) Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol. 146: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T.E., Senter S.D., Grauke L.J. (1993) Lipid content and fatty acids of pecan pollen. HortScience 28: 1191–1193. [Google Scholar]
- Troncoso-Ponce M.A., Kilaru A., Cao X., Durrett T.P., Fan J., Jensen J.K., et al. (2011) Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68: 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unver T., Wu Z., Sterck L., Turktas M., Lohaus R., Li Z., et al. (2017) Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA 114: E9413–E9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz Dias F., Serrazina S., Vitorino M., Marchese D., Heilmann I., Godinho M., et al. (2019) A role for diacylglycerol kinase 4 in signalling crosstalk during Arabidopsis pollen tube growth. New Phytol. 222: 1434–1446. [DOI] [PubMed] [Google Scholar]
- Villette C., Berna A., Compagnon V., Schaller H. (2015) Plant sterol diversity in pollen from angiosperms. Lipids 50: 749–760. [DOI] [PubMed] [Google Scholar]
- Vossen P. (2013) Growing olives for oil InHandbook of Olive Oil. Analysis and Properties. Edited by Aparicio R., Harwood J. pp. 19–56. Springer, New York. [Google Scholar]
- Walley J.W., Kliebenstein D.J., Bostock R.M., Dehesh K. (2013) Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 16: 520–526. [DOI] [PubMed] [Google Scholar]
- Xu C., Yu B., Cornish A.J., Froehlich J.E., Benning C. (2006) Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant J. 47: 296–309. [DOI] [PubMed] [Google Scholar]
- Yu B., Wakao S., Fan J., Benning C. (2004) Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 45: 503–510. [DOI] [PubMed] [Google Scholar]
- Zafra A., Jiménez-Quesada M.J., Traverso J.A., Corpas F.J., Rodríguez-García M.I., Alché J.D. (2012) Peroxisomal localization of Cu, Zn-superoxide dismutase in the male reproductive tissues of the olive tree. Microsc. Microanal. 18: 33–34. [Google Scholar]
- Zhang M., Fan J., Taylor D.C., Ohlrogge J.B. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Laudencia-Chingcuanco D., Comai L., Li M., Harada J.J. (1994) Isocitrate lyase and malate synthase genes from Brassica napus L. are active in pollen. Plant Physiol. 104: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz K., Castro A.J., Alché J.D., Zienkiewicz A., Suárez C., Rodríguez-García M.I. (2010) Identification and localization of a caleosin in olive (Olea europaea L.) pollen during in vitro germination. J. Exp. Bot. 61: 1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz A., Zienkiewicz K., Rejón J.D., Rodríguez-García M.I., Castro A.J. (2013) New insights into the early steps of oil body mobilization during pollen germination. J. Exp. Bot. 64: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz K., Zienkiewicz A., Rodríguez-García M.I., Castro A.J. (2011) Characterization of a caleosin expressed during olive (Olea europaea L.) pollen ontogeny. BMC Plant Biol. 11: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl D.J., Zwiebel B.I., Grier D.G., Preuss D. (1999) Pollen-stigma adhesion in Arabidopsis: a species specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440. [DOI] [PubMed] [Google Scholar]
- Zou J., Song L., Zhang W., Wang Y., Ruan S., Wu W.H. (2009) Comparative proteomic analysis of Arabidopsis mature pollen and germinated pollen. J. Integr. Plant Biol. 51: 438–455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.