SUMMARY
The PD-1 pathway regulates dysfunctional T cells in chronic infection and cancer, but the role of this pathway during acute infection remains less clear. Here, we demonstrate that PD-1 signals are needed for optimal memory. Mice deficient in the PD-1 pathway exhibit impaired CD8+ T cell memory following acute influenza infection, including reduced virus-specific CD8+ T cell numbers and compromised recall responses. PD-1 blockade during priming leads to similar differences early post-infection but without the defect in memory formation, suggesting that timing and/or duration of PD-1 blockade could be tailored to modulate host responses. Our studies reveal a role for PD-1 as an integrator of CD8+ T cell signals that promotes CD8+ T cell memory formation and suggest PD-1 continues to fine-tune CD8+ T cells after they migrate into nonlymphoid tissues. These findings have important implications for PD-1-based immunotherapy, in which PD-1 inhibition may influence memory responses in patients.
Graphical Abstract
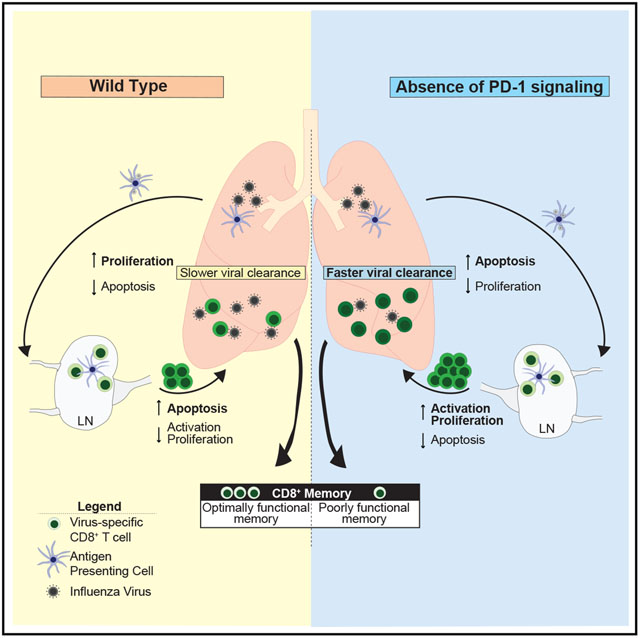
In Brief
The role of PD-1 in memory development is poorly understood. Here, Pauken et al. show that constitutive loss of PD-1 during acute infection causes overactivation of CD8+ T cells during the effector phase and impairs memory and recall responses. These data indicate PD-1 is required for optimal memory.
INTRODUCTION
The development of effector and memory CD8+ T cells requires coordinated signals from the T cell receptor (TCR) (signal 1), costimulation (signal 2), and inflammation (signal 3) (Curtsinger et al., 1999). The quantity and quality of the three signals can affect CD8+ T cell activation, but how such signals regulate memory CD8+ T cell differentiation remains incompletely understood (Chang et al., 2014). Signal 2 encompasses many costimulatory and coinhibitory pathways. Costimulatory signals such as CD28 and inducible T cell costimulator (ICOS or CD278) augment T cell survival, function, and metabolic activity and sustain T cell responses (Francisco et al., 2010; Chen and Flies, 2013). Conversely, coinhibitory receptors such as cytotoxic T lymphocyte associated protein-4 (CTLA-4 or CD152) and programmed death-1 (PD-1 or CD279) dampen these positive signals. The importance of signal 2 has been highlighted by the application of antibodies blocking coinhibitory receptors for treating cancer and chronic infections (Barber et al., 2006; Day et al., 2006; Brahmer et al., 2012; Topalian et al., 2012, 2015; Page et al., 2014; Sharpe and Pauken, 2018). PD-1 pathway blockade has been approved by the U.S. Food and Drug Administration (FDA) for at least 20 types of tumors, including melanoma, non-small cell lung cancer, renal cell carcinoma, Hodgkin’s lymphoma, bladder cancer, and microsatellite instability high or mismatch-repair-deficient solid tumors, and this number continues to grow (Sharpe and Pauken, 2018; Pardoll, 2012; Topalian et al., 2015). Considering the increasing use of PD-1 checkpoint blockade alone or in combination with other therapies (Chen and Mellman, 2017), an understanding of how the PD-1 pathway regulates immunological memory has significant therapeutic relevance. However, how this pathway regulates CD8+ memory T cell differentiation, function, and survival remains poorly understood.
In addition to the well-established role of the PD-1 pathway in regulating exhausted CD8+ T cells, PD-1 is expressed by all T cells during activation (Sharpe and Pauken, 2018). Consequently, PD-1 is critically positioned to shape the ensuing effector response and, by extension, the memory response. Previous work showed that a lack of PD-1:programmed death ligand (PD-L) signals during some primary infections resulted in more robust effector T cell responses (Frebel et al., 2012; Odorizzi et al., 2015; Ahn et al., 2018) and enhanced CD8+ T cell memory and/or skewed T cells toward a central memory phenotype (Allie et al., 2011; Ahn et al., 2018). In addition, the secondary expansion of unhelped memory CD8+ T cells was increased by PD-1 blockade (Fuse et al., 2009). However, these studies focused mainly on early time points during memory development, and further work is needed to fully understand how the timing and/or duration of loss of PD-1 signals affect memory responses. For example, other studies have shown that loss of PD-1 signals during acute infection can reduce, rather than augment, effector and/or memory T cell responses (Rowe et al., 2008; Talay et al., 2009; Yao et al., 2009; Xu et al., 2013). Consequently, additional studies are needed to clarify how the PD-1 pathway shapes the development of effector and memory CD8+ T cells following acute infections.
Here we examined the role of the PD-1 pathway in effector and memory CD8+ T cell differentiation during influenza virus infection using mice lacking PD-1 (PD-1 knockout [KO]) or both ligands, PD-L1 (B7-H1) and PD-L2 (B7-DC) (PD-L1/L2 double knockout [DKO]), or experiencing PD-1 pathway blockade. Lack of PD-1:PD-L signals in the whole animal led to compromised CD8+ T cell memory, including reduced cell numbers and impaired secondary responses. There were major cell-intrinsic alterations in CD8+ T cell memory, because PD-1 KO CD8+ T cells transferred into wild-type (WT) mice exhibited similar defects in memory formation. PD-1 mediates these effects by controlling key transcriptional pathways involved in the durability of CD8+ T cell memory, signaling, and cell cycle. Importantly, blocking the PD-1 pathway only during the priming and effector phase of infection induced phenotypic changes in CD8+ T cells similar to those seen in PD-L1/L2 DKO mice at day 8 post-infection (p.i.) but did not result in the severe memory defect observed in mice with constitutive PD-1 deficiency. These data demonstrate a role for PD-1 in tempering the strength of initial activation to promote optimal CD8+ T cell memory formation and highlight the importance of timing and/or duration of loss of PD-1 signals in regulating memory formation.
RESULTS
Increased Proliferation and Death of CD8+ T Cells without PD-1 Signaling
To investigate the role of the PD-1 pathway during acute H3N2 influenza virus X31 infection in mice, we first assessed the expression of PD-1, PD-L1, and PD-L2 on both hematopoietic and non-hematopoietic cells in the lung during the first 12 days p.i. We used a strain of X31 engineered to express the GP33–41 epitope from lymphocytic choriomeningitis virus (LCMV) (called X31-GP33) to allow tracking of DbGP33–41+ CD8+ T cells (Laidlaw et al., 2013). PD-L1 and PD-L2 expression increased substantially during X31-GP33 infection on hematopoietic antigen-presenting cells (APCs), including dendritic cells (DCs), macro phages, and B cells, and PD-L1 and PD-L2 were expressed on some non-hematopoietic cells, such as epithelial cells (Figure S1). Expression of PD-L1 and PD-L2 peaked between day 3 and day 6 p.i. on lung APCs and EpCAM+ lung epithelial cells (Figures S1B–S1F). PD-L1, but not PD-L2, was expressed on CD31+ lung endothelial cells and peaked between day 3 and day 8 p.i.; PD-L1 remained significantly elevated even at day 12 p.i. on alveolar macrophages and lung endothelial cells (Figures S1B, S1D, and S1E). PD-1 was rapidly upregulated on CD4+ and CD8+ T cells in the lung, with high levels of expression maintained at day 12 p.i. (Figure S2A). Upregulation of PD-1 was also observed on splenic CD4+ and CD8+ T cells, albeit to a lesser extent than in the lung (Figures S2A and S2B). Thus, dynamic changes in PD-1, PD-L1, and PD-L2 expression occur in the respiratory tract during influenza infection, suggesting that the PD-1 pathway may regulate CD8+ T cell effector and/or memory responses in this setting.
To investigate the impact of the PD-1 pathway on the generation of effector T cell responses, WT or PD-L1/L2 DKO mice were infected with X31-GP33 and virus-specific CD8+ T cells were examined from day 8 to day 10 p.i. Although all mice had similar frequencies of influenza-specific CD8+ T cells in the lung, there was a significant increase in the absolute number of virus-specific CD8+ T cells in PD-L1/L2 DKO mice for all three epitopes examined (DbNP366–374, DbPA224–223, and DbGP33–41) (Figure 1A). This finding suggested that loss of PD-1 did not alter immunodominance during the effector phase of the response but rather affected the magnitude of the CD8+ T cell response in the lung. At day 10 p.i., there were no significant differences in CD8+ T cell functionality or quality of cytokine-producing cells in the lungs following ex vivo stimulation with NP366–374 peptide (Figure S3A). However, the increase in absolute number of virus-specific CD8+ T cells at these early time points corresponded with better viral control: PD-1 KO and PD-L1/L2 DKO mice lost less weight during primary X31-GP33 infection and contained less viral RNA per nanogram (ng) of lung than WT control mice (Figures S3B and S3C). Collectively, these data suggest that effector responses were enhanced during primary X31-GP33 infection in PD-1 pathway-deficient mice.
Figure 1. Altered Effector CD8+ T Cell Expansion and Contraction in the Absence of PD-1.

(A) Frequencies (left) and numbers (right) of indicated tetramer+ CD8+ T cells in the lungs of WT and PD-L1/L2 DKO mice at day 10 after X31-GP33 infection. (B) Significantly enriched (FDR < 0.01) gene sets in WT CD8+ T cells clustered based on gene member overlap and annotated for the biological states/processes they represent. (C) Representative GO terms enriched in microarray data from DbGP33–41+ and DbPA224–233+ CD8+ T cells from lungs of WT versus PD-L1/L2 DKO mice at day 8 p.i. (D) Flow cytometric analysis of Ki-67 expression by DbNP366–374+ CD8+ T cells from the spleen (top) and lung (middle) of WT and PD-L1/L2 DKO mice at day 8 p.i. Numbers indicate the fraction of Ki-67+ DbNP366–374+ CD8+ T cells. Representative plots are shown on the left, and a summary of Ki-67+ cells is shown on the right. Bottom, FLICA staining on DbNP366–374+ lung CD8+ T cells from WT and PD-L1/L2 DKO mice at day 8 p.i. Numbers indicate the fraction of FLICA+ DbNP366–374+ CD8+ T cells based on unstained controls. Representative plots are shown on the left, and a summary of numbers is shown on the right. (E) Frequencies (left) and numbers (right) of indicated tetramer+ CD8+ T cells in the lungs of WT and PD-L1/L2 DKO mice at day 15 after X31-GP33 infection. Data are representative of 2–3 independent experiments with 4–5 mice per group and represented as mean ± SEM. Significance was assessed using Student’s t test; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001. GP, DbGP33–41 tetramer; PA, DbPA224–233 tetramer; NP, DbNP366–374 tetramer. See Figures S1–S3 for supporting data.
To begin to investigate the mechanisms responsible for the quantitative differences in effector CD8+ T cells generated in the absence of PD-1 pathway signals, we performed microarray analysis of DbPA224–233 and DbGP33–41 CD8+ T cells from the lungs of WT and PD-L1/L2 DKO mice at day 8 p.i. Gene set enrichment analysis (GSEA) using Gene Ontology (GO) (Ashburner et al., 2000; Subramanian et al., 2005) revealed that most significantly enriched pathways (p < 0.001, false discovery rate [FDR] < 0.001) were related to cell division (Figures 1B and 1C; Figure S2C). Unexpectedly, WT cells were enriched for cell-cycle pathways, whereas PD-L1/L2 DKO cells appeared to be exiting cell cycle (Figure 1C). We further analyzed three representative pathways enriched in the WT cells: mitosis, spindle assembly, and DNA replication. Among those genes upregulated in WT relative to PD-L1/L2 DKO CD8+ T cells, most were members of these pathways (Figure S2C). Flow cytometric analysis supported this phenotype. At day 8 p.i., a lower frequency of virus-specific CD8+ T cells expressed Ki-67 in the lung of PD-L1/L2 DKO mice compared with WT mice, but a higher frequency expressed Ki-67 in the spleen in PD-L1/L2 DKO mice (Figure 1D). These data suggest that virus-specific CD8+ T cells in PD-L1/L2 DKO mice had started to exit the cell cycle in the lung and proliferated more in the secondary lymphoid organs, whereas the WT CD8+ T cells were at an earlier phase of the response in which the cells that had proliferated recently trafficked to the lung and the cells were still entering the cycle in the spleen. This finding was consistent with the increased numbers of virus-specific CD8+ T cells observed in PD-L1/L2 DKO in the lung compared with WT mice at day 10 (Figure 1A). Concomitantly, a higher frequency of virus-specific CD8+ T cells in the lung also showed active caspase staining (with Fluorochrome Inhibitor of Caspases [FLICA] reagent) in PD-L1/L2 DKO mice (Figure 1D), suggesting that these cells were more susceptible to death than their WT counterparts. The decrease in proliferation and increase in cell death in the absence of PD-1 signals in the lung at day 8 p.i. led to a more significant contraction of influenza-specific CD8+ T cells at day 15 p.i. for the subdominant epitopes (DbPA224–233 and DbGP33–41) (Figure 1E). These data suggest a link between PD-1:PD-L deficiency and cell cycle during early activation of virus-specific CD8+ T cells in the lung during influenza infection, and early excessive proliferation correlated with increased cell death during the contraction phase in PD-L1/L2 DKO mice.
Impaired Virus-Specific CD8+ T Cell Memory in Mice Lacking PD-1 Signaling
We hypothesized that these early changes in PD-L1/L2 DKO mice would affect the formation of CD8+ T cell memory responses. To test this hypothesis, we first assessed the ratio of PD-L1/L2 DKO:WT virus-specific CD8+ T cells in the lung over time following X31-GP33 infection (Figure 2A). From day 8 to day 10 p.i., all three influenza epitopes showed a higher number in PD-L1/L2 DKO compared with WT, consistent with increased effector expansion (Figures 1A and 2A). However, at day 15 and later, PD-L1/L2 DKO showed a greater contraction of the DbPA224–233 and DbGP33–41 epitopes (Figure 2A) compared with WT control mice. The immunodominant (DbNP366–374) CD8+ T cell response displayed slower kinetics in this deterioration (Figure 2A) but eventually (day 35+ p.i.) showed the same trend as the subdominant epitopes, with WT outnumbering the DKO CD8+ T cells (Figure 2A). Thus, PD-1 pathway deficiency led to an initial increase in virus-specific CD8+ T cells that eroded over time.
Figure 2. CD8+ T Cell Memory Is Impaired in PD-1 Pathway-Deficient Mice.
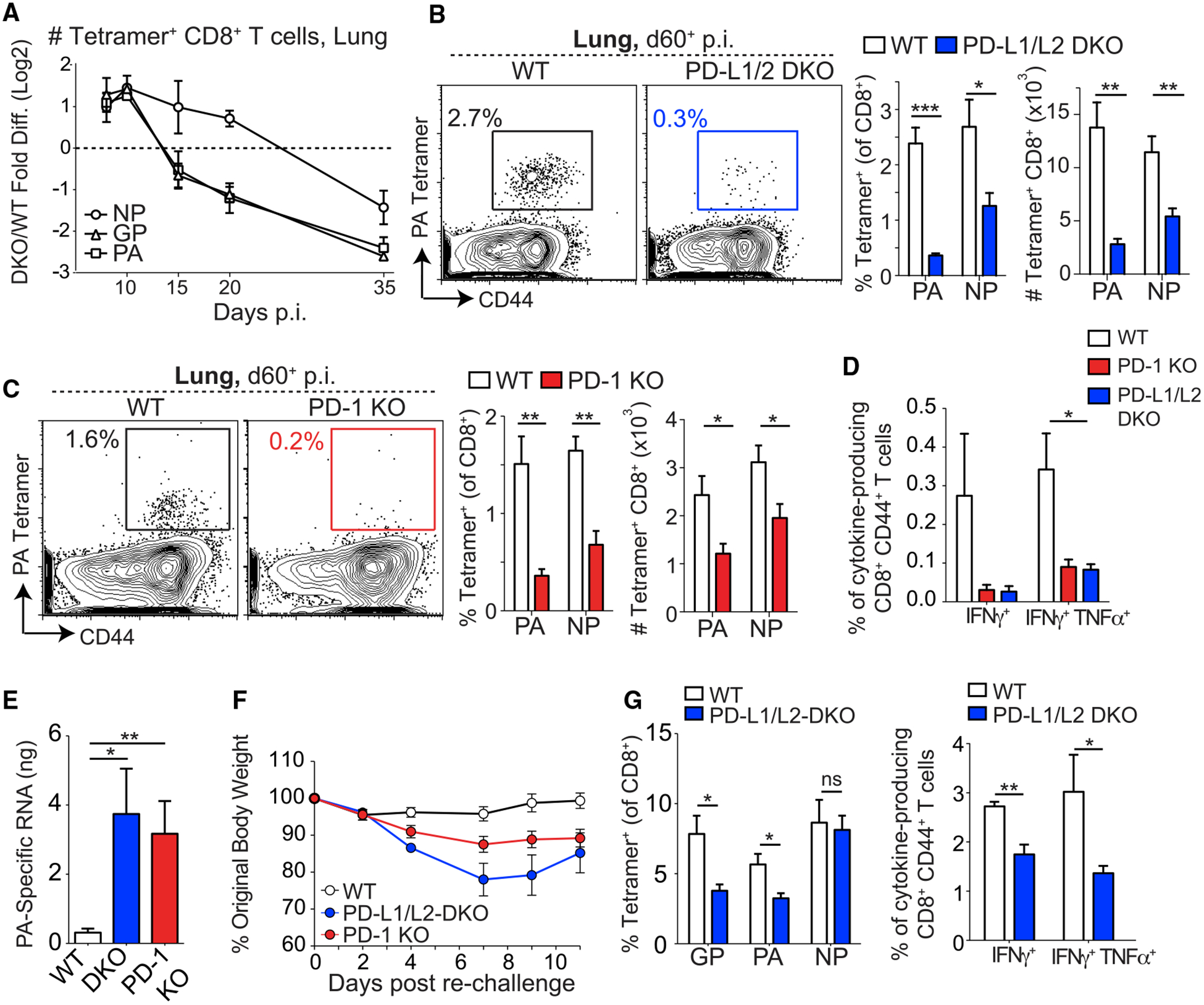
(A) Ratio of numbers of influenza-specific CD8+ T cells in the lungs of PD-L1/L2 DKO and WT mice over time after X31-GP33 infection. The log2 fold change of the ratio of DKO to WT cells was plotted over time. Here, a value of zero indicates a 1:1 ratio of DKO to WT, a value greater than zero indicates more DKO than WT, and a value less than zero indicates more WT than DKO. (B and C) Representative plots (left) show the frequency of DbPA224–233+ CD8+ T cells in the lungs of X31-GP33-infected WT or PD-L1/L2 DKO mice (B) and WT or PD-1 KO mice (C) at day 60+ p.i. Numbers on plots indicate the percentages of DbPA224–233+ cells of the CD8+ population. Percentages and numbers of DbPA224–233+ and DbNP366–374+ T cells are summarized on the right. (D) Summary of frequencies of cytokine-producing CD8+ CD44+ T cells from the lungs of WT, PD-L1/L2 DKO, and PD-1 KO mice at day 60+ p.i. stimulated ex vivo with NP366–374 peptide. (E) Influenza viral titers in the lung at day 3.5 p.i. after PR8-GP33 challenge in mice that had been infected with X31-GP33 at least 60 days before challenge. (F) Weight loss in WT, PD-L1/L2 DKO, and PD-1 KO X31-GP33-immune mice (day 60+) following rechallenge with PR8-GP33. (G) Frequencies of tetramer+ CD8+ T cells (left) and cytokine-producing CD8+ CD44+ T cells (right) following stimulation ex vivo with NP366–374 peptide in lungs of X31-GP33-immune WT and PD-L1/L2 DKO mice at day 3.5 following rechallenge with PR8-GP33. Data are representative of 3–5 independent experiments with 4–5 mice per group and represented as mean ± SEM. Significance was assessed using Student’s t test; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001. See Figure S4 for supporting data.
Consistent with the steeper decline of influenza-specific CD8+ T cells in DKO during the contraction phase of influenza, the frequencies and numbers of virus-specific CD8+ T cells were reduced in the lungs of PD-L1/L2 DKO and PD-1 KO mice compared with WT at day 60+ p.i. (Figures 2B and 2C). Although the magnitude of the difference was greater for the subdominant (DbPA224–233) than the immunodominant (DbNP366–374) response, both responses showed impaired memory formation (Figures 2B and 2C). A similar decrease in CD8+ T cell frequencies was observed in the spleen, mediastinal (lung draining) lymph node (dLN), and bone marrow (Figure S4A). In addition, lower frequencies of cytokine-producing CD8+ T cells were detected at day 60+ p.i. following ex vivo peptide stimulation in PD-L1/L2 DKO and PD-1 KO mice (Figure 2D), indicating a qualitative defect in the memory response in these mice.
To test whether PD-1:PD-L interactions during primary viral infection affected secondary responses, we rechallenged X31-GP33-immune mice with the heterotypic strain of influenza PR8, which also expressed the DbGP33–41 epitope from LCMV (called PR8-GP33) (Mueller et al., 2010). Because neutralizing antibodies do not cross-react between these viruses, memory CD8+ T cells play a central role in protective immunity (Liang et al., 1994; Flynn et al., 1998; Mueller et al., 2010). PD-L1/L2 DKO mice had significantly increased viral titers in the lung 3.5 days following rechallenge (Figure 2E) and lost more weight than WT mice (Figure 2F). Lower frequencies and total numbers of influenza-specific, as well as cytokine-producing, CD8+ T cells (Figure 2G; Figure S4B) were also observed in the lung at day 3.5 post-rechallenge in PD-L1/L2 DKO mice compared with WT mice. However, by day 7 after rechallenge, CD8+ T cell responses in the lungs were comparable in WT and PD-L1/L2 DKO mice and the virus was cleared in the PD-L1/L2 DKO mice (Figures S4C–S4F). Similar defects in viral control were observed in PD-1 KO mice (Figures 2E and 2F). Thus, PD-1 pathway deficiency resulted in compromised memory CD8+ T cells that failed to mount optimal recall responses. This defect manifested as delayed viral clearance and increased weight loss (Figures 2E and 2F; Figure S4C). Altogether, these data indicate a key role for the PD-1 pathway in regulating the development of optimal CD8+ T cell memory.
Cell-Intrinsic Regulation of CD8+ T Cell Memory Differentiation by PD-1
The compromised CD8+ T cell memory responses in PD-1 pathway-deficient mice could result from CD8+ T cell-intrinsic functions of PD-1 or extrinsic effects. Because PD-1 pathway-deficient mice cleared primary X31-GP33 virus infection more rapidly than WT mice (Figure S3C), and previous work identified a critical role for antigen in the development and maintenance of CD8+ T cell memory in the lung (Jelley-Gibbs et al., 2005; Zammit et al., 2006; Lee et al., 2011; McMaster et al., 2018), the altered CD8+ T cell dynamics in this setting could be related to enhanced clearance of influenza infection during primary infection. To circumvent these issues and to interrogate CD8+ T cell-intrinsic mechanisms of PD-1-mediated regulation, we cotransferred equal numbers of WT and PD-1 KO P14 TCR transgenic CD8+ T cells (recognizing the DbGP33–41 epitope) into WT mice followed by infection with X31-GP33 virus. This approach did not alter viral load at day 8 p.i. compared with mice without P14 cells (Figure S5A) and enabled direct comparison of WT and PD-1 KO CD8+ T cells in the same environment while controlling for precursor frequency, TCR repertoire, viral control, and influenza pathogenesis. Consistent with findings in germline KO mice, the frequency and numbers of PD-1 KO P14 cells were significantly higher than WT P14 cells in the lung and spleen at day 7 p.i. (Figure 3A), with similar trends in the blood and dLN (Figure S5B). The higher frequency of PD-1 KO P14 cells during primary X31-GP33 virus infection was associated with increased bromodeoxyuridine (BrdU) incorporation in the spleen and dLN, but not the lung (Figure 3B), indicating that PD-1 regulates CD8+ T cell proliferation in a cell-intrinsic manner in secondary lymphoid organs.
Figure 3. Cell-Intrinsic Defect in CD8+ T Cell Memory in the Absence of PD-1.
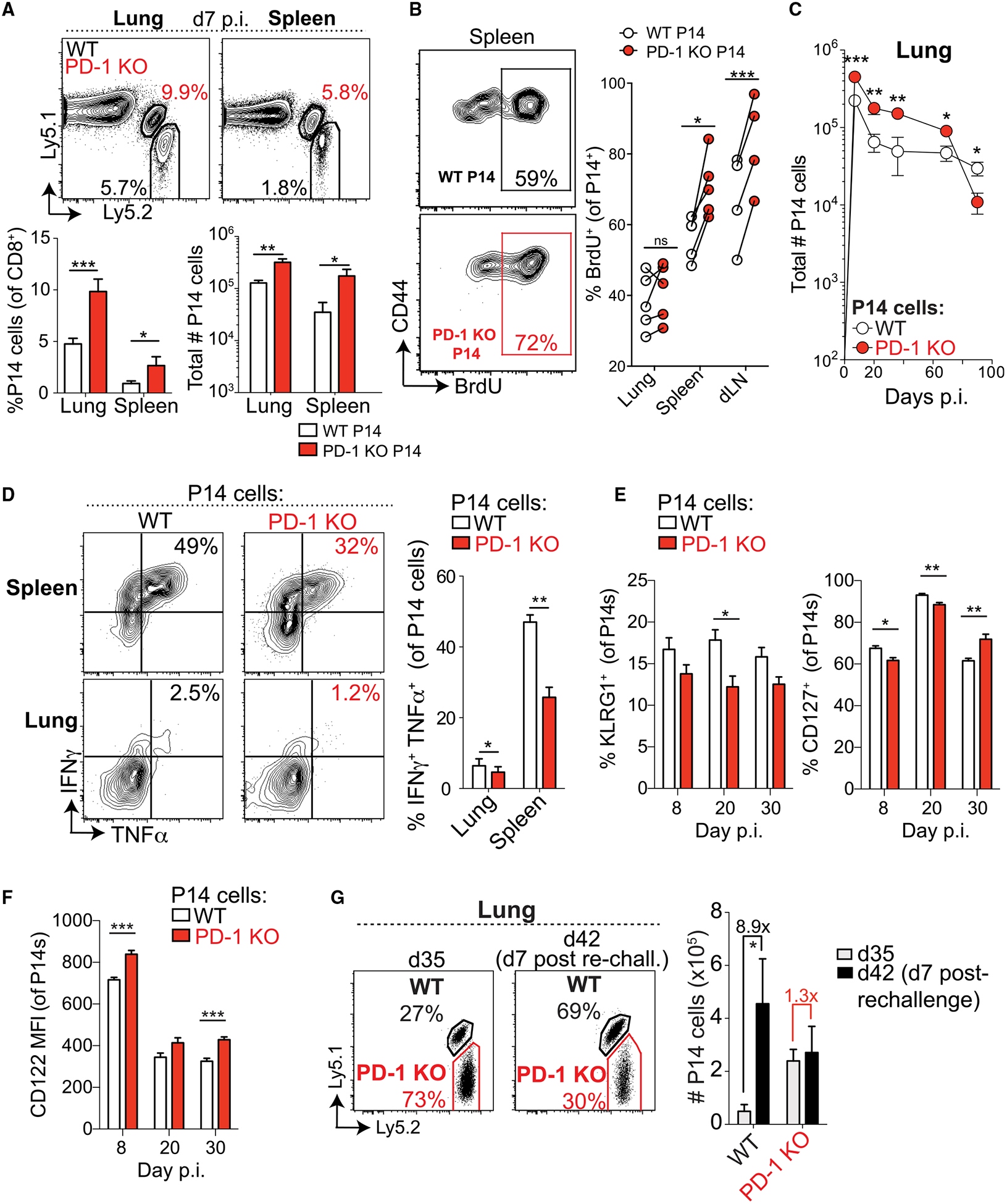
(A) (top) Representative plots of WT (black) and PD-1 KO (red) P14 cells in the lung and spleen 7 days after X31-GP33 infection. Numbers indicate frequencies of P14 cells as percentages of CD8+ T cells (top). Summaries of frequencies and numbers of P14 cells (bottom). (B) Flow cytometric analysis of BrdU incorporation by WT and PD-1 KO P14 cells at day 7 p.i. Numbers indicate the fraction of BrdU+ P14 cells. Representative plots from the spleen (left). Summary of BrdU+ P14 cells in the indicated organs (right). (C) Longitudinal analysis of WT and PD-1 KO P14 cell numbers in the lung during primary X31-GP33 infection. (D) Representative plots of intracellular cytokine staining for IFNγ and TNF-α (left) in WT and PD-1 KO P14 cells from spleen (top) and lung (bottom) at day 47+ p.i. and quantification (right). (E and F) Longitudinal analysis of KLRG1 and CD127 (E) and CD122 (F) expression on transferred PD-1 KO and WT P14 cells on days 8, 20, and 30 p.i. in the lung. (G) Representative plots of WT and PD-1 KO P14 cells at day 35 p.i. with X31-GP33 (day 35, left) and 7 days after rechallenge with PR8-GP33 (day 42 after primary infection, right). Numbers indicate frequencies of P14 cells as percentages of CD8+ T cells. A summary of numbers of P14 cells pre- and post-rechallenge is shown (right). Numbers above the bars indicate the fold change between day 35 and day 42 (7 days after rechallenge). Data are representative of 3–4 independent experiments with 4–6 mice per group and represented as mean ± SEM. Significance was assessed using Student’s t test; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001. See Figures S5 and S6 for supporting data.
Similar to global PD-1 pathway KO mice, PD-1 KO P14 cells in WT mice were more prone to cell death in the lung, as shown by increased FLICA staining at day 10 p.i. (Figure S5C). PD-1 KO P14 cells also underwent a more progressive contraction and were less abundant than WT P14 cells in the lung and spleen after 3 months (Figure 3C; Figure S5D). Moreover, PD-1 KO memory P14 cells showed significantly reduced interferon gamma (IFNγ) and tumor necrosis factor alpha (TNF-α) coproduction and a decreased amount of IFNγ made per cell compared with WT cells (Figure 3D; Figure S5E). Hence, although the kinetics differed, these results were consistent with the quantitative and qualitative memory attrition observed in the global KO mice. There were significant, albeit small, differences in KLRG1, CD127, and CD122 expression in lung at days 8, 20, and 30 p.i. (Figures 3E and 3F). However, these changes were subtle compared with the quantitative (Figure 3C) and qualitative (Figure 3D) defects in memory formation observed. These data highlight the importance of functional readouts to determine residual memory capacity during acute infection. Considering this, we assessed the per-cell capacity of WT versus PD-1 KO P14 memory cells to mount a productive recall response following re-challenge with PR8-GP33. Here, PD-1 KO P14 memory cells displayed substantially impaired secondary expansion compared with WT P14 cells in the same mice (Figure 3G). These results point to a key cell-intrinsic function for PD-1 signals in regulating CD8+ T cell proliferation and survival during the acute phase of infection and subsequent optimal CD8+ T cell memory formation.
Although PD-1 KO P14 cells showed a similar memory defect in WT hosts as virus-specific CD8+ T cells in intact PD-1 pathway KO mice, we did note a difference in the kinetics of the quantitative memory defect. This difference could result from variations in cell-extrinsic factors such as viral control (Figures S3C and S5A) or cell-intrinsic factors such as the resting state of naive T cell populations in these mice. To evaluate this issue, we examined CD8+ T cell function in the steady state of unmanipulated PD-L1/L2 DKO and PD-1 KO mice and compared this to PD-1 KO P14 cells (Figure S6). There were no significant differences in the numbers of CD8+ T cells in the spleen or dLNs of WT versus PD-L1/L2 DKO or PD-1 KO mice, and most T cells displayed a naive phenotype (Figures S6A and S6B; data not shown). Both PD-L1/L2 DKO and PD-1 KO mice showed a subtle but significant increase in frequencies of CD44high CD8+ T cells, and this effect was more pronounced in PD-1 KO mice (Figures S6A and S6B). Moreover, most markers of recent activation (including CD69, CD25, Ki-67, and KLRG1) were uniformly low in both PD-L1/L2 DKO and PD-1 KO mice and comparable to WT mice, suggesting no ongoing activation of CD8+ T cells in the steady state (Figures S6C and S6D). However, in unimmunized PD-1 KO mice (but not PD-L1/L2 DKO mice), both CD44low and CD44high CD8+ T cells showed increased CXCR3 expression compared with WT mice (Figures S6E–S6G). Restricting TCR specificity eliminated this effect, because CXCR3 expression was similar in PD-1 KO P14 cells and WT P14 cells, as were levels of other activation markers, including Ki-67, Tim-3, and CD160 (Figure S6H). Considering the similar defects in memory CD8+ T cell responses in intact PD-1 KO and PD-L1/L2 DKO mice, and in PD-1 KO P14 CD8+ T cells transferred into WT mice, and that elevated CXCR3 expression only is seen in non-TCR transgenic PD-1 KO mice, we think it is unlikely that elevated CXCR3 expression in the PD-1 KO mouse influences the memory phenotype observed during acute influenza infection.
PD-1 Restrains CD8+ T Cell Effector and Memory Responses to Multiple Acute Respiratory Infections
To investigate whether PD-1 plays a similar role on CD8+ T cells during other acute respiratory infections, we cotransferred equal numbers of WT and PD-1 KO P14 cells into WT mice followed by intranasal infection with LCMV Armstrong, vaccinia virus-GP33 (VV-GP33) or influenza virus (PR8-GP33). All viruses were administered intranasally to investigate the role of PD-1 in regulating memory CD8+ T cell formation in the lung. PD-1 deficiency resulted in increased frequency of P14 cells in the blood during the effector phase in all infections (Figure 4A). However, substantial attrition of virus-specific CD8+ T cell responses occurred over time (Figure 4A). The fold contraction of the PD-1 KO P14 population after the effector phase was substantially greater in each infection, indicating less efficient per-cell memory formation in the absence of PD-1 (Figure 4B). At day 100+ p.i., PD-1 KO P14 cells produced less IFNγ and TNF-α compared with WT P14 cells, with the most significant results observed during VV-GP33 and PR8-GP33 infections (Figures 4C and 4D). Thus, PD-1 can restrain early expansion of virus-specific CD8+ T cells during multiple acute respiratory infections, profoundly affecting memory responses.
Figure 4. Cell-Intrinsic Defect in PD-1-Deficient CD8+ T Cell Memory Is Observed in Multiple Intranasal Infections.
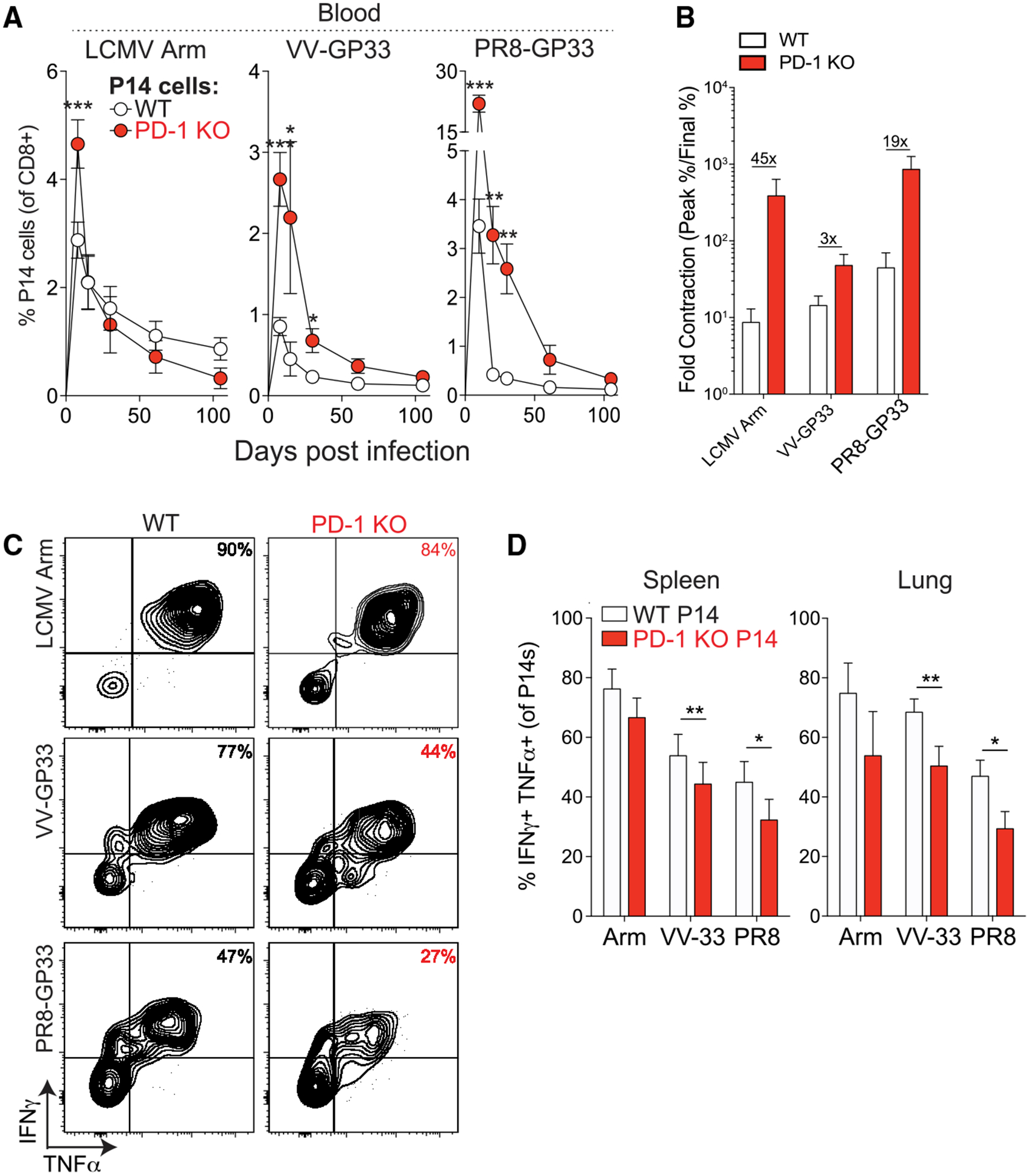
(A) Frequencies of WT and PD-1 KO P14 cells in mixed chimeras in the peripheral blood following primary intranasal infections with LCMV Armstrong, VV-GP33, or PR8-GP33. (B) Individual bars show fold changes in frequencies of WT and PD-1 KO P14 cells between day 8–10 and day 100+ from mice in (A). Numbers above bar graphs indicate the difference in ratios observed in WT versus PD-1 KO P14 cells. (C and D) Representative plots (C) and summary (D) of frequencies of IFN-γ+ TNF-α+ P14 cells at day 100+ p.i. Summary data (D) in the spleen (left) and lung (right) are shown. Data are representative of 2 independent experiments with 4–5 mice per group and represented as mean ± SEM. Significance was assessed using Student’s t test; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001. Arm, LCMV Armstrong; PR8, PR8-GP33.
Temporally Restricting PD-1 Pathway Deficiency to the Priming Phase of Infection Does Not Result in the Memory Defect Observed in Constitutive PD-1 Pathway-Deficient Mice
The results described earlier with constitutive PD-1 pathway deficiency suggested that absence of PD-1 signaling impaired memory formation. We next asked whether temporal loss of PD-1 signaling during the priming and effector phase of infection resulted in a similar memory defect. We administered anti-PD-1 blocking antibody to WT B6 mice early during X31-GP33 infection (day −1 through 8), and compared effector (day 8 p.i.) and memory (day 60+ p.i.) responses to isotype control (Rat Immunoglobulin (Ig)G2a)-treated WT B6 mice or PD-L1/L2 DKO mice as controls. During the effector phase (day 8 p.i.), significant differences in the frequency and number of DbGP33–41+ CD8+ T cells in the lung were not observed (data not shown). Phenotypically, both PD-L1/L2 DKO mice and anti-PD-1-treated mice showed a significant decrease in the frequency of KLRG1+ CD127− DbGP33–41+ T cells in the lung at day 8 p.i. (Figure 5A), suggesting that blockade of PD-1 had effects similar to genetic PD-1 pathway deficiency during the effector phase. These phenotypic differences were not sustained at the memory time point (Figure 5B). There were no differences in CD127+ KLRG1− (Figures 5A and 5B) DbGP33–41+ CD8+ T cells at either time point. At day 60+, there was a quantitative defect in memory observed in PD-L1/L2 DKO that was not seen in anti-PD-1-treated mice (Figure 5C). Collectively, these data suggest that restricting PD-1 pathway deficiency to the priming stage of influenza led to similar phenotypic changes during the effector phase but did not result in the same memory defect observed in the absence of PD-1 signals.
Figure 5. Transient Early Blockade of the PD-1 Pathway Does Not Result in the Same Memory Defects as Permanent PD-1 Loss.
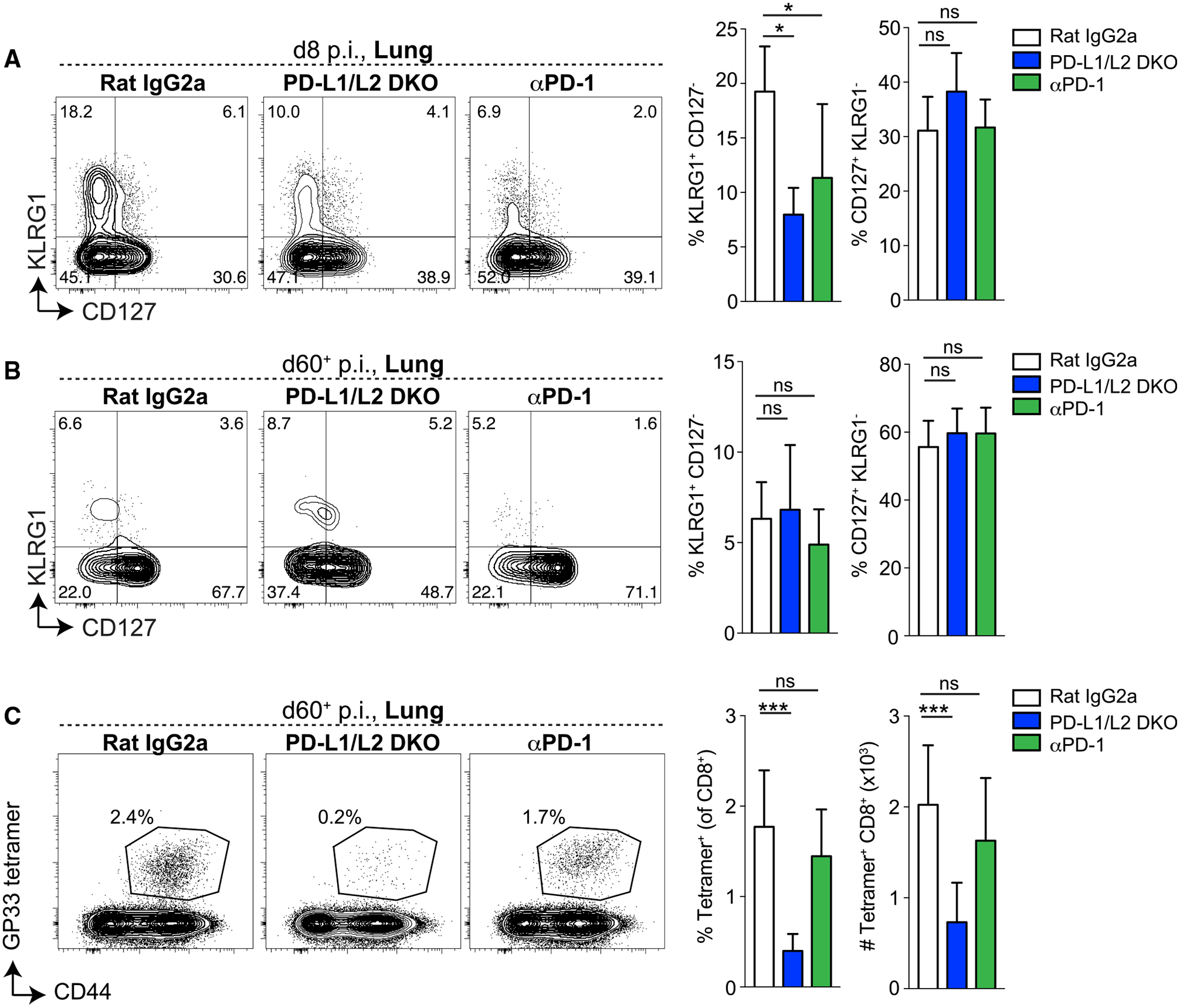
WT B6 mice were treated with anti-PD-1 antibody (clone 29F.1A12) or isotype control antibody (rat IgG2a) on days −1, 2, and 5 and harvested on day 8 p.i., or treated on days −1, 2, 5, and 8 and harvested on day 60+ p.i. PD-L1/L2 DKO mice were included as a control. (A and B) Representative plots (left) and summary (right) of KLRG1 and CD127 expression on DbGP33–41+ CD8+ T cells in the lung at day 8 (A) or day 60+ (B) p.i. (C) Representative plots showing GP and CD44 staining in the lung on day 60+ p.i. Numbers on plots indicate the frequencies of DbGP33–41+ cells of the CD8+ population (left). Summary of frequencies (of the CD8+ population, middle) and numbers (right) of DbGP33–41+ T cells in the lung. Data from day 8 p.i. are representative of 2 independent experiments, and data from day 60+ are pooled from 2 independent experiments with 3–8 mice per group. Data are represented as mean ± SEM. Significance was assessed using ANOVA (Kruskal-Wallis test) followed by Dunn’s multiple comparison test; ns, not significant; *p < 0.05, ***p < 0.001.
DISCUSSION
PD-1 is expressed by all T cells during activation, positioning this pathway to play an essential role in shaping quantitative and qualitative aspects of CD8+ T cell responses early during differentiation. It is well established that TCR signal strength in the acute phase of an immune response and the formation of self-renewing central memory T cells are inversely correlated, with stronger TCR signals leading to less central memory formation (Wherry et al., 2003; Badovinac et al., 2007; Sarkar et al., 2007; Buchholz et al., 2013; Gerlach et al., 2013; Knudson et al., 2013). Because PD-1 can modulate TCR (Yokosuka et al., 2012; Zinselmeyer et al., 2013; Freeman et al., 2000; Parry et al., 2005; Butte et al., 2007) and CD28 signaling (Hui et al., 2017; Kamphorst et al., 2017), with PD-1 loss resulting in stronger TCR stimulation, we hypothesized that PD-1 KO mice would have defects in the formation of durable memory responses during acute infection. Here we found that complete PD-1 deficiency indeed resulted in the development of suboptimal memory CD8+ T cells that lacked long-term stability and responded poorly to rechallenge. These defects were associated with early excessive proliferation of effector CD8+ T cells and increased cell death. The PD-1 pathway regulated this process, at least partly, by controlling expression of cell-cycle genes and other pathways. In contrast, transient PD-1 blockade during the priming and effector stage failed to result in the same memory defect as constitutive PD-1 pathway deficiency, suggesting that the timing and/or duration of PD-1 loss was critical for determining the effects on memory CD8+ T cells. Collectively, these findings suggest the PD-1 pathway acts as a key regulator of effector and memory CD8+ T cell responses during acute viral infection.
The complex interplay between TCR signaling, costimulation, and coinhibition at different stages of differentiation (e.g., effector, memory, and exhaustion) and in different anatomical locations (e.g., secondary lymphoid organs versus non lymphoid tissues) likely influences the effects of PD-1 on T cell responses (Sharpe and Pauken, 2018). Influenza virus infection provides an excellent model for studying the role of PD-1 in regulating CD8+ T cell differentiation, because this model shares key features with common human infections, including localized rather than systemic antigen and viral replication in a mucosal tissue. One interesting aspect of influenza infection is that although replicating virus is only detectable for ~7–10 days, complexes of peptide and major histocompatibility complex (MHC) can persist for ~30 days or more (Zammit et al., 2006; Kim et al., 2010). Although antigen presentation in this setting may be more prolonged than during other acute infection models like LCMV Armstrong, influenza virus infection still results in the development of bona fide memory CD8+ T cells that mount effective secondary responses that are protective upon rechallenge (Wu et al., 2014; Laidlaw et al., 2014; Kreijtz et al., 2007), a feature that distinguishes influenza from chronic infections like LCMV clone 13 (Blackburn et al., 2008). Similar to influenza infection, numerous other models used for studying memory T cells have some degree of prolonged antigen presentation, including intranasal infection with vesicular stomatitis virus (VSV) (Turner et al., 2007) or Sendai virus (Takamura et al., 2010), implying that persistent antigen presentation may be a feature of intranasal infections in general. Moreover, local antigen recognition is critical for the generation of lung memory CD8+ T cells (Lee et al., 2011; McMaster et al., 2018; Zammit et al., 2006; Jelley-Gibbs et al., 2005). Considering that the lung is a site of many clinically relevant human infections, as well as tumors, it is important to understand the dynamics of CD8+ T cell biology, antigen persistence, and the role of PD-1 in this location.
Previous work showed that PD-1 inhibits cell-cycle progression through the G1 phase in vitro by modulating key cell-cycle regulators in CD4+ T cells (Latchman et al., 2001; Patsoukis et al., 2012). PD-1 also can inhibit phosphatidylinositol 3-kinase (PI3K)/Akt signaling (Parry et al., 2005; Yokosuka et al., 2012). Our transcriptional profiling data showed less T cell proliferation in the lungs of a whole-body KO setting compared with the WT setting at day 8 p.i. We speculate that differences in the kinetics of expansion contribute to the transcriptional profiles observed here; CD8+ T cell proliferation had already peaked in the lungs of the PD-1 pathway KO mice and these cells were beginning to exit cell cycle, whereas the WT CD8+ T cells were still in the peak proliferative phase at day 8 p.i. In the P14 chimera setting, PD-1 KO P14 cells incorporated significantly more BrdU within the first 7 days p.i. than WT P14 cells, suggesting that the PD-1 KO cells were undergoing more or faster cell cycles during that early phase. Similarly, the CD8+ T cells in the spleen of PD-L1/L2 DKO showed a higher frequency of Ki-67+ cells at day 8 p.i. compared with WT CD8+ T cells. Our transcriptional data suggest that PD-1 pathway deficiency resulted in T cells that were responding with faster kinetics and that had already proliferated at the time of analysis, whereas control WT T cells were still proliferating. Thus, our data indicate that CD8+ T cells responding in the genetic absence of PD-1 pathway signals were able to rapidly proliferate but that this occurs earlier compared with WT T cells.
PD-1 deficiency resulted in impaired virus-specific memory CD8+ T cell responses for all epitopes examined, but there were temporal differences in the erosion of the responses to dominant and subdominant epitopes. During influenza virus infection, CD8+ T cells specific for different epitopes receive qualitatively different signals that affect memory CD8+ T cell responses (Ballesteros-Tato et al., 2014). DbNP366–374+ CD8+ T cells encounter antigen on CD40-licensed DCs later than DbPA224–233+ CD8+ T cells, leading to sustained CD25 expression, better responses to interleukin-2 (IL-2), and the ability to dominate secondary responses. The differences in these responses may explain temporal differences in the effects of PD-1 deficiency on CD8+ T cell differentiation of the immunodominant CD8+ T cell response (DbNP366–374) compared with the subdominant responses (DbPA224–233+ and DbGP33–41+). Although the subdominant responses showed more rapid contraction in PD-1 pathway-deficient mice, ultimately the DbNP366–374+ CD8+ T cell response contracted more compared with WT mice, consistent with the notion of PD-1 signals modulating CD8+ T cell memory formation. However, the timing of contraction and the severity of the memory defect following PD-1 loss may be multifactorial depending on TCR affinity, access to antigen, costimulation, and/or growth factors in the tissue microenvironment. Consistent with this notion, the virus-specific CD8+ T cell population in lung contracted more dramatically in intact PD-1 pathway KO mice compared with the P14 mixed chimera setting in which WT and PD-1 KO CD8+ T cells were present in the same WT mice. These differences between the two settings may reflect changes in cell-extrinsic factors such as viral burden between the two models.
Our studies suggest that the PD-1 pathway regulates CD8+ T cell responses to infection in several ways. Previous work showed that antibody blockade or genetic deletion of the PD-1 pathway enhanced early virus-specific effector CD8+ T cell responses to respiratory infection (Erickson et al., 2012), consistent with the enhanced effector responses observed here. Our work extends these findings by showing that early augmented effector responses without PD-1 signals result in subsequent defects in CD8+ T cell memory in the absence of PD-1 signals. The continued contraction of PD-1 KO CD8+ T cells likely points to a defect in memory maintenance, which would be consistent with the role of antigen in the long-term maintenance of memory T cells in the lung (Jelley-Gibbs et al., 2005; Zammit et al., 2006; Lee et al., 2011; McMaster et al., 2018) and continued regulation by PD-1 during later stages of the memory response. Moreover, selective PD-1 blockade only during the early stages of infection (day −1 to 8 p.i.) did not result in the same memory defect observed in the genetic KO setting, suggesting that the PD-1 pathway may continue to function as an integrator of signals required for memory T cell maintenance after the initial priming and effector stages of the immune response. Additional studies are needed to define how the timing and/or duration of PD-1 loss affect the effector and memory stages of CD8+ T cell differentiation. Lastly, further studies are needed to determine whether the results obtained here using respiratory infections are broadly applicable to other types of infections, as well as tumors, or whether these findings are specific to the lung microenvironment.
In summary, our findings reveal that a key function of PD-1 is to temper excessive T cell proliferation and facilitate the development of optimal CD8+ T cell memory. Although our studies have defined a CD8+ T cell memory defect in global PD-1 pathway KO mice, selective PD-1 pathway disruption only during the priming and effector phase did not show the same memory defect, suggesting that the timing and/or duration of PD-1 blockade during CD8+ T cell differentiation can critically influence outcomes. Thus, targeting the PD-1 pathway during different stages of an immune response may result in distinct effects on CD8+ T cell differentiation, recall responses, and/or memory maintenance. These concepts may be of interest in the cancer setting, in which the timing, duration, and/or sequence of PD-1 blockade relative to other therapies may influence patient outcomes. Control of cell cycle and apoptosis likely contributes to how PD-1:PD-L signals influence CD8+ T cell memory. Future work clarifying the mechanisms by which PD-1 contributes to effector versus memory differentiation during both infection and cancer will be important for determining how to optimally administer PD-1 blocking agents alone and with other therapies to achieve durable improved immunity.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arlene H. Sharpe (Arlene_Sharpe@hms.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The microarray data generated during this study are available on GEO. The accession number for the microarray data reported in this paper is GEO: GSE149425. This study did not generate unique code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
For primary infections, 6–12 week old mice were utilized. Wild-type (WT) C57BL/6 mice were purchased from the Jackson Laboratory (stock number 000664). PD-1 KO and PD-L1/L2 DKO have been described (Keir et al., 2006, 2007), and are available from the Jackson Laboratory (stock numbers 028276 and 32239-JAX, respectively). For primary infections of intact WT, PD-1 KO, and PD-L1/L2 DKO mice male or female mice were utilized. Age and gender matched mice were used for all experiments. To generate PD-1 KO P14 mice, PD-1 KO mice were crossed to P14 TCR transgenic mice (Pircher et al., 1989). Ly5.2+ or Ly5.2+Ly5.1+ PD-1 KO and WT P14 cells were isolated from the blood and transferred i.v. into WT C57BL/6 mice (Ly5.2+) at a 1:1 ratio (500 cells each) at least one day prior to infection. Female P14 TCR transgenic mice were used as donors, and WT female mice were used for recipients. For naive mouse studies, female WT, PD-1 KO, and PD-L1/L2 DKO mice between 8–12 weeks of age and female WT and PD-1 KO P14 between 16–21 weeks of age were utilized. All mice were maintained in specific pathogen-free facilities at Harvard Medical School or the University of Pennsylvania and maintained under standard housing, husbandry, and diet conditions according to Institutional Animal Care and Use Committee and NIH guidelines. All experimental procedures performed had been approved by the Institutional Animal Care and Use Committee at each institution.
Viral infections and antibody treatments
Viruses were administered via the intranasal (i.n.) route to generate respiratory infections. Most studies utilized primary influenza X31 expressing the GP33–41 epitope from LCMV (X31-GP33, 1.6×105 TCID50) (Laidlaw et al., 2013). In some studies, primary LCMV Armstrong (104 PFU), vaccinia virus-expressing GP33–41 from LCMV (VV-GP33, 104 PFU) (Rodriguez et al., 1998), or influenza strain PR8 expressing the GP33–41 epitope from LCMV (PR8-GP33, 0.3 LD50) (Mueller et al., 2010) were used and delivered via the i.n. route. For re-challenge experiments (secondary PR8-GP33 challenge), mice were infected i.n. with PR8-GP33 (10 LD50) at > 35 days after primary infection. Recombinant influenza strains containing the LCMV GP33–41 epitope were provided by Dr. Richard Webby (St. Jude Children’s Research Hospital, Memphis, TN) (Laidlaw et al., 2013; Mueller et al., 2010). Viral titers in lungs were determined by quantitative real-time PCR as described (Laidlaw et al., 2013). For in vivo antibody blockade experiments, WT C57BL/6 mice were given 200 μg per mouse per injection of rat anti-PD-1 (clone 29F.1A12) or isotype control antibody (Rat IgG2a, clone 2A3 from BioXCell) on days −1, 2, and 5 p.i. and analyzed at day 8 p.i., or on days −1, 2, 5, and 8 p.i. and analyzed at day 60+ p.i.
METHOD DETAILS
Isolation of Lymphocytes and Flow Cytometry
Lymphocytes were isolated from spleen, lung and lung draining LNs as described (Laidlaw et al., 2013). Single cell suspensions were stained with fluorescently labeled antibodies specific to: CD8α (clone 53–6.7), CD44 (clone IM7), IFNγ (clone XMG1.2), TNFα (clone MP6-XT22), CD127 (clone SB/199 or A7r34), CD122 (clone TM-β1), CXCR3 (clone CXCR3–173), Tim-3 (clone RMT3–23), CD160 (clone 7H1), CD25 (clone PC61), CD69 (clone H1.2F3), CD62L (MEL-14), TCR Vα2 (B20.1), CD45.1 (clone A20), CD45.2 (clone 104), PD-1 (clone RMP1–30), PD-L1 (clone 10F.9G2), PD-L2 (clone TY25), CD11b (clone M1/70), CD11c (clone N418), CD19 (clone 6D5), CD4 (clone GK1.5), CD326 or Ep-CAM (clone G8.8), and CD31 (clone 390) purchased from BioLegend, Ki-67 (clone B56), CD3 (clone 145–2C11), CD8α (clone 53–6.7), and CD62L (clone MEL-14) from BD Biosciences, CD4 (RM4–5) and KLRG1 (clone 2F1) from Thermo Fisher Scientific, and KLRG1 (clone 2F1) from Abcam and Southern Biotech. Intracellular staining for Ki-67 was performed following permeabilization with the eBioscience Foxp3/transcription factor fixation/permeabilization kit according to manufacturer’s instructions (Thermo Fisher Scientific). Poly-caspase analysis was performed with the FLICA Vybrant FAM Assay Kit according to manufacturer’s instructions (Thermo Fisher Scientific). Dead cell exclusion was performed by live/dead fluorescent reactive dye (Thermo Fisher Scientific) staining according to manufacturer’s instructions. H-2 DbGP33–41, DbNP366–374, and DbPA224–233 biotinylated monomers were either made as described (Wherry et al., 2003) or obtained from the National Institutes of Health (NIH) tetramer core facility, conjugated to Streptavidin-R-Phycoerythrin or Streptavidin-Allophycocyanin (Thermo Fisher Scientific) in house to make tetramers as recommended by the NIH tetramer core facility, and used as recommended. For intracellular cytokine staining (ICS), single cell suspensions from spleens or lungs, which included T cells as well as APCs from the same animal, were incubated with 0.5 μg/ml influenza NP366–374 or GP33–41 peptide (Genscript) or no peptide for 5 hours at 37°C in the presence of GolgiPlug (BD Biosciences), surface stained, fixed/permeabilized, and intracellularly stained using the Cytofix/Cytoperm kit (BD Biosciences) as directed by the manufacturer. For BrdU detection, animals were treated with 2 mg of BrdU (Sigma-Aldrich) i.p. 12–24 hours prior to analysis. BrdU incorporation was assessed by the BrdU Flow Kit per manufacturer’s instructions (BD Biosciences). Data were acquired on a LSR II or Symphony flow cytometer (BD) and analyzed using FlowJo software (Tree Star).
Gene Expression Microarray
CD8+ T cells specific for the subdominant (GP33–41, PA224–233) influenza epitopes were sorted from the lungs of WT and PD-L1/L2 DKO mice on d8 following X31-GP33 infection. For the microarray, RNA was extracted using RNAdvance tissue isolation kit (Agencourt) and amplified using the WT-Ovation One Direct System (NuGEN). Fragmented and labeled cDNA was hybridized to Affymetrix Mouse430_2 microarray. Microarray data were processed and analyzed as described previously (Haining et al., 2008; Subramanian et al., 2005).
QUANTIFICATION AND STATISTICAL ANALYSIS
All non-microarray data were analyzed for statistical significance using GraphPad Prism software. Statistical details of experiments (including the statistical tests used, number of animals in each experiment, number of times experiments have been replicated, and precision measures) can be found in the Figure Legends. Statistical tests performed included Student’s t tests, one way ANOVA, or two way ANOVA. P values < 0.05 were considered significant. Asterisks in the figure legends indicating significance correspond to: * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | ||
|---|---|---|---|---|
| Antibodies | ||||
| Rat anti-mouse PD-1 clone 29F.1A12 (used for in vivo treatments) | BioXCell | Cat# BE0273; RRID:AB_2687796 | ||
| Rat IgG2a clone 2A3 (used for in vivo treatments) | BioXCell | Cat#BE0089; RRID:AB_1107769 | ||
| Rat anti-mouse CD8α (53–6.7)(used for flow cytometry) | BioLegend | Cat#100723; RRID:AB_389304 | ||
| Rat anti-mouse CD8α (53–6.7)(used for flow cytometry) | BioLegend | Cat#100712; RRID:AB_312751 | ||
| Rat anti-mouse CD8α (53–6.7)(used for flow cytometry) | BD Biosciences | Cat#564297; RRID:AB_2722580 | ||
| Rat anti-mouse CD8α (53–6.7)(used for flow cytometry) | BD Biosciences | Cat#612898; N/A | ||
| Rat anti-mouse CD8α (53–6.7)(used for flow cytometry) | Thermo Fisher Scientific | Cat#61–0081–82; RRID:AB_2574524 | ||
| Rat anti-mouse/human CD44 (IM7)(used for flow cytometry) | BioLegend | Cat#103059; RRID:AB_2571953 | ||
| Rat anti-mouse/human CD44 (IM7)(used for flow cytometry) | BioLegend | Cat#103020; RRID:AB_493683 | ||
| Rat anti-mouse/human CD44 (IM7)(used for flow cytometry) | BioLegend | Cat#103026; RRID:AB_493713 | ||
| Rat anti-mouse/human CD44 (IM7)(used for flow cytometry) | BioLegend | Cat#103030; RRID:AB_830787 | ||
| Rat anti-mouse IFNγ (XMG1.2)(used for flow cytometry) | BioLegend | Cat#505822; RRID:AB_961359 | ||
| Rat anti-mouse IFNγ (XMG1.2)(used for flow cytometry) | BioLegend | Cat#505810; RRID:AB_315404 | ||
| Rat anti-mouse TNFα (MP6–XT22)(used for flow cytometry) | BioLegend | Cat#506322; RRID:AB_961434 | ||
| Rat anti-mouse TNFα (MP6–XT22)(used for flow cytometry) | BioLegend | Cat#506314; RRID:AB_493330 | ||
| Rat anti-mouse TNFα (MP6–XT22)(used for flow cytometry) | BioLegend | Cat#506328; RRID:AB_2562902 | ||
| Rat anti-mouse CD127 (SB/199)(used for flow cytometry) | BioLegend | Cat#121112; RRID:AB_493509 | ||
| Rat anti-mouse CD127 (A7r34)(used for flow cytometry) | BioLegend | Cat#135024; RRID:AB_11218800 | ||
| Rat anti-mouse CD127 (A7r34)(used for flow cytometry) | BioLegend | Cat#135010; RRID:AB_1937251 | ||
| Rat anti-mouse CD122 (TM–β1)(used for flow cytometry) | BioLegend | Cat#123208; RRID:AB_940613 | ||
| Rat anti-mouse CD122 (TM–β1)(used for flow cytometry) | Thermo Fisher Scientific | Cat#48–1222–82; RRID:AB_2016697 | ||
| Armenian hamster anti-mouse CXCR3 (CXCR3–173) (used for flow cytometry) | BioLegend | Cat#126529; RRID:AB_2563100 | ||
| Armenian hamster anti-mouse CXCR3 (CXCR3–173) (used for flow cytometry) | BioLegend | Cat#126522; RRID:AB_2562205 | ||
| Rat anti-mouse Tim-3 or CD366 (RMT3–23)(used for flow cytometry) | BioLegend | Cat#119721; RRID:AB_2616907 | ||
| Rat anti-mouse CD62L (MEL-14)(used for flow cytometry) | BioLegend | Cat#104445; RRID:AB_2564215 | ||
| Rat anti-mouse CD62L (MEL-14)(used for flow cytometry) | BD Biosciences | Cat#562404; RRID:AB_11154046 | ||
| Rat anti-mouse CD160 (7H1) (used for flow cytometry) | BioLegend | Cat#143008; RRID:AB_2562676 | ||
| Rat anti-mouse CD160 (7H1) (used for flow cytometry) | BioLegend | Cat#143004; RRID:AB_10960743 | ||
| Rat anti-mouse CD25 (PC61) (used for flow cytometry) | BioLegend | Cat#102008; RRID:AB_312857 | ||
| Rat anti-mouse TCR Vα2 (B20.1) (used for flow cytometry) | BioLegend | Cat#127814; RRID:AB_1186116 | ||
| Armenian hamster anti-mouse CD69 (H1.2F3) (used for flow cytometry) | BioLegend | Cat#104514; RRID:AB_492843 | ||
| Mouse anti-mouse CD45.1 (A20)(used for flow cytometry) | BioLegend | Cat#110708; RRID:AB_313497 | ||
| Mouse anti-mouse CD45.1 (A20)(used for flow cytometry) | BioLegend | Cat #110714; RRID:AB_313503 | ||
| Mouse anti-mouse CD45.1 (A20)(used for flow cytometry) | BioLegend | Cat#110724; RRID:AB_493733 | ||
| Mouse anti-mouse CD45.2 (104)(used for flow cytometry) | BioLegend | Cat#109841; RRID:AB_2563485 | ||
| Mouse anti-mouse CD45.2 (104)(used for flow cytometry) | BioLegend | Cat#109828; RRID:AB_893350 | ||
| Mouse anti-mouse CD45.2 (104)(used for flow cytometry) | BioLegend | Cat#109806; RRID:AB_313443 | ||
| Rat anti-mouse PD-1 (RMP1–30)(used for flow cytometry) | BioLegend | Cat#109110; RRID:AB_572017 | ||
| Rat anti-mouse PD-L1 (10F.9G2)(used for flow cytometry) | BioLegend | Cat#124312; RRID:AB_10612741 | ||
| Rat anti-mouse PD-L1 (10F.9G2)(used for flow cytometry) | BioLegend | Cat# 124308; RRID:AB_2073556 | ||
| Rat anti-mouse PD-L2 (TY25)(used for flow cytometry) | BioLegend | Cat#107206; RRID:AB_2162011 | ||
| Rat anti-mouse/human CD11b (M1/70) (used for flow cytometry) | BioLegend | Cat#101228; RRID:AB_893232 | ||
| Armenian hamster anti-mouse CD11c (N418)(used for flow cytometry) | BioLegend | Cat#117322; RRID:AB_755988 | ||
| Rat anti-mouse CD19 (6D5)(used for flow cytometry) | BioLegend | Cat#115530; RRID:AB_830707 | ||
| Rat anti-mouse CD4 (GK1.5)(used for flow cytometry) | BioLegend | Cat#100428; RRID:AB_493647 | ||
| Rat anti-mouse CD4 (GK1.5)(used for flow cytometry) | BioLegend | Cat#100412; RRID:AB_312697 | ||
| Rat anti-mouse Ep-CAM or CD326 (G8.8) (used for flow cytometry) | BioLegend | Cat#118218; RRID:AB_2098648 | ||
| Rat anti-mouse CD31 (390) (used for flow cytometry) | BioLegend | Cat#102416; RRID:AB_493410 | ||
| Rat anti-mouse CD4 (RM4–5) (used for flow cytometry) | Thermo Fisher Scientific | Cat#47–0042–82; RRID:AB_1272183 | ||
| Mouse anti-human/mouse Ki-67 (B56) (used for flow cytometry) | BD Biosciences | Cat#561277; RRID:AB_10611571 | ||
| Mouse anti-human/mouse Ki-67 (B56) (used for flow cytometry) | BD Biosciences | Cat#556026; RRID:AB_396302 | ||
| Armenian hamster anti-mouse CD3 (145–2C11) | BD Biosciences | Cat#563565; RRID:AB_2738278 | ||
| Syrian hamster anti-mouse KLRG1 (2F1) (used for flow cytometry) | Southern Biotech | Cat#1807–02; RRID:AB_2795367 | ||
| Syrian hamster anti-mouse KLRG1 (2F1) (used for flow cytometry) | Abcam | Cat#ab25205; RRID:AB_448711 | ||
| Syrian hamster anti-mouse KLRG1, eBioscience (2F1) (used for flow cytometry) | Thermo Fisher Scientific | Cat#61–5893–82; RRID:AB_2574630 | ||
| Bacterial and Virus Strains | ||||
| Influenza strain X31 expressing the GP33–41 epitope from LCMV | Richard Webby (Laidlaw et al., 2013) | Grown up in house | ||
| Influenza strain PR8 expressing the GP33–41 epitope from LCMV | Richard Webby (Mueller et al., 2010) | Grown up in house | ||
| LCMV Armstrong strain | Rafi Ahmed | Grown up in house | ||
| Vaccinia virus expressing the GP33–41 epitope from LCMV | Rodriguez et al., 1998 | N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||||
| NP366–374 peptide (sequence ASNENMETM) | GenScript | Cat#RP20267; N/A | ||
| GP33–41 peptide (sequence KAVYNFATM) | GenScript | Cat#RP20257; N/A | ||
| 5-bromo-2’-deoxyuridine (BrdU) | Sigma-Aldrich | Cat#19–160; N/A | ||
| Deposited Data | ||||
| Raw and analyzed microarray data | This paper | GEO: GSE149425 | ||
| Experimental Models: Organisms/Strains | ||||
| Mouse: WT C57BL/6J strain | The Jackson Lab | Stock No. 000664; RRID:IMSR_JAX:000664 | ||
| Mouse: PD-1 KO C57BL/6 | Originally described in Keir et al., 2007 | Mouse line developed in house, available from the Jackson lab as Stock No. 028276; RRID:IMSR_JAX:028276 | ||
| Mouse: PD-L1/L2 DKO C57BL/6 mice | Originally described Keir et al., 2006 | Mouse line developed in house, available from the Jackson lab as MMRRC Stock No. 32239-JAX; N/A | ||
| Mouse: P14 TCR transgenic mice | Originally described in Pircher etal., 1989 | N/A | ||
| Software and Algorithms | ||||
| GraphPad Prism | GraphPad Software | N/A | ||
| Flow Jo | Tree Star | N/A | ||
| Other | ||||
| BD GolgiPlug Protein Transport Inhibitor (containing Brefeldin A) | BD Biosciences | Cat#555029; N/A | ||
| Vybrant FAM Poly Caspases Assay Kit (FLICA) | Thermo Fisher Scientific | Cat#V35117; N/A | ||
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34966; N/A | ||
| LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L10119; N/A | ||
| eBioscience Foxp3/transcription factor fixation/permeabilization kit | Thermo Fisher Scientific | Cat#00–5521–00; N/A | ||
| BD Cytofix/Cytoperm fixation/permeabilization kit | BD Biosciences | Cat#554714; N/A | ||
| DbGP33–41 biotinylated monomer, conjugated to Steptavidin R-Phycoerythrin (Cat#S866) or Allophycocyanin (Cat#S868) to make tetramer in house | NIH Tetramer Core Facility or prepared in house as described (Wherry et al., 2003) | N/A | ||
| DbPA224–233 biotinylated monomer, conjugated to Steptavidin R-Phycoerythrin (Cat#S866) or Allophycocyanin (Cat#S868) to make tetramer in house | NIH Tetramer Core Facility or prepared in house as described (Wherry et al., 2003) | N/A | ||
| DbNP366–374 biotinylated monomer, conjugated to Steptavidin R-Phycoerythrin (Cat#S866) or Allophycocyanin (Cat#S868) to make tetramer in house | NIH Tetramer Core Facility or prepared in house as described (Wherry et al., 2003) | N/A | ||
| Steptavidin, R-Phycoerythrin Conjugate (SA-PE) | Thermo Fisher Scientific | Cat#S866; N/A | ||
| Streptavidin, Allophycocyanin crosslinked, conjugate | Thermo Fisher Scientific | Cat#S868; N/A | ||
| RNAdvance tissue isolation kit (Agencourt) (for microarray profiling) | Agencourt Bioscience, Beckman Coulter | Cat#A32645; N/A | ||
| WT-Ovation One Direct System (NuGEN) (for microarray profiling) | NuGEN | Cat#3500–12; N/A | ||
| Affymetrix Mouse 430_2 microarray (for microarray profiling) | Thermo Fisher Scientific | Cat#900495; N/A | ||
Highlights.
Early loss of PD-1 leads to overactivation of CD8+ T cells during acute infection
Mice constitutively lacking PD-1 or PD-L develop impaired CD8+ T cell memory
Cell-intrinsic PD-1 signals suppress effector cell expansion and promote memory
Timing of PD-1 blockade determines impact on memory generation
ACKNOWLEDGMENTS
We thank Dr. Jason Schenkel, Dr. Alison Ringel, and members of the Sharpe and Wherry labs for discussions. This work was funded by NIH grants P01 AI078897, P01 AI112521, and P01 AI108545 and BAA HHSN266200500030C (to A.H.S. and E.J.W.), R01 AI105343 and U19 AI082630 (to E.J.W.), P01 AI56299, P01 AI039671, and R01 CA229851 (to A.H.S.), and the Cancer Research Institute Predoctoral Emphasis Pathway in Tumor Immunology (to J.G.). E.J.W. is supported by the Parker Institute for Cancer Immunotherapy.
DECLARATION OF INTERESTS
A.H.S. and G.J.F. have patents/pending royalties on the PD-1 pathway from Roche, Merck, Bristol-Myers-Squibb, EMD-Serono, Boehringer-Ingelheim, AstraZeneca, Dako, and Novartis. G.J.F. has equity in Nextpoint, Triursus, and Xios. G.J.F. has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, and Origimed. A.H.S. is on advisory boards for Surface Oncology, Elstar, SQZ Biotechnologies, Selecta, Elpiscience, and Monopteros and has research funding from Novartis, Roche, Ipsen, Quark, and Merck. E.J.W. receives honoraria, consulting fees, and/or research support from BMS, Celgene, Dynavax, Eli Lilly, Elstar, Merck, MedImmune, Pieris, Roche, Surface Oncology, and KyMab. E.J.W. is a founder of Arsenal Biosciences. E.J.W. has a patent licensing agreement for the PD-1 pathway licensed by Genentech/Roche. W.N.H. is a founder of and equity holder at Arsenal Biosciences, equity holder at Tango therapeutics, and employee of Merck and Co.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107827.
REFERENCES
- Ahn E, Araki K, Hashimoto M, Li W, Riley JL, Cheung J, Sharpe AH, Freeman GJ, Irving BA, and Ahmed R (2018). Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. USA 115, 4749–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allie SR, Zhang W, Fuse S, and Usherwood EJ (2011). Programmed death 1 regulates development of central memory CD8 T cells after acute viral infection. J. Immunol 186, 6280–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. ; The Gene Ontology Consortium (2000). Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Haring JS, and Harty JT (2007). Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity 26, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, León B, Lee BO, Lund FE, and Randall TD (2014). Epitope-specific regulation of memory programming by differential duration of antigen presentation to influenza-specific CD8(+) T cells. Immunity 41, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, and Ahmed R (2006). Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687. [DOI] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Freeman GJ, and Wherry EJ (2008). Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc. Natl. Acad. Sci. USA 105, 15016–15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. (2012). Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med 366, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Gräf P, Verschoor A, Schiemann M, Höfer T, and Busch DH (2013). Disparate individual fates compose robust CD8+ T cell immunity. Science 340, 630–635. [DOI] [PubMed] [Google Scholar]
- Butte MJ, Keir ME, Phamduy TB, Sharpe AH, and Freeman GJ (2007). Programmed death-1 ligand 1 interacts specifically with the B7–1 costimulatory molecule to inhibit T cell responses. Immunity 27, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Wherry EJ, and Goldrath AW (2014). Molecular regulation of effector and memory T cell differentiation. Nat. Immunol 15, 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, and Flies DB (2013). Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol 13, 227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, and Mellman I (2017). Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330. [DOI] [PubMed] [Google Scholar]
- Curtsinger JM, Schmidt CS, Mondino A, Lins DC, Kedl RM, Jenkins MK, and Mescher MF (1999). Inflammatory cytokines provide a third signal for activation of naive CD4+ and CD8+ T cells. J. Immunol 162, 3256–3262. [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. (2006). PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354. [DOI] [PubMed] [Google Scholar]
- Erickson JJ, Gilchuk P, Hastings AK, Tollefson SJ, Johnson M, Downing MB, Boyd KL, Johnson JE, Kim AS, Joyce S, and Williams JV (2012). Viral acute lower respiratory infections impair CD8+ T cells through PD-1. J. Clin. Invest 122, 2967–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, and Doherty PC (1998). Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 8, 683–691. [DOI] [PubMed] [Google Scholar]
- Francisco LM, Sage PT, and Sharpe AH (2010). The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev 236, 219–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebel H, Nindl V, Schuepbach RA, Braunschweiler T, Richter K, Vogel J, Wagner CA, Loffing-Cueni D, Kurrer M, Ludewig B, and Oxenius A (2012). Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J. Exp. Med 209, 2485–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. (2000). Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med 192, 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse S, Tsai CY, Molloy MJ, Allie SR, Zhang W, Yagita H, and Usherwood EJ (2009). Recall responses by helpless memory CD8+ T cells are restricted by the up-regulation of PD-1. J. Immunol 182, 4244–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Rohr JC, Perié L, van Rooij N, van Heijst JW, Velds A, Urbanus J, Naik SH, Jacobs H, Beltman JB, et al. (2013). Heterogeneous differentiation patterns of individual CD8+ T cells. Science 340, 635–639. [DOI] [PubMed] [Google Scholar]
- Haining WN, Ebert BL, Subrmanian A, Wherry EJ, Eichbaum Q, Evans JW, Mak R, Rivoli S, Pretz J, Angelosanto J, et al. (2008). Identification of an evolutionarily conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells. J. Immunol 181, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, and Vale RD (2017). T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355, 1428–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelley-Gibbs DM, Brown DM, Dibble JP, Haynes L, Eaton SM, and Swain SL (2005). Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J. Exp. Med 202, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, et al. (2017). Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Liang SC, Guleria I, Latchman YE, Qipo A, Albacker LA, Koulmanda M, Freeman GJ, Sayegh MH, and Sharpe AH (2006). Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med 203, 883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Freeman GJ, and Sharpe AH (2007). PD-1 regulates self-reactive CD8+ T cell responses to antigen in lymph nodes and tissues. J. Immunol 179, 5064–5070. [DOI] [PubMed] [Google Scholar]
- Kim TS, Hufford MM, Sun J, Fu YX, and Braciale TJ (2010). Antigen persistence and the control of local T cell memory by migrant respiratory dendritic cells after acute virus infection. J. Exp. Med 207, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson KM, Goplen NP, Cunningham CA, Daniels MA, and Teixeiro E (2013). Low-affinity T cells are programmed to maintain normal primary responses but are impaired in their recall to low-affinity ligands. Cell Rep. 4, 554–565. [DOI] [PubMed] [Google Scholar]
- Kreijtz JH, Bodewes R, van Amerongen G, Kuiken T, Fouchier RA, Osterhaus AD, and Rimmelzwaan GF (2007). Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25, 612–620. [DOI] [PubMed] [Google Scholar]
- Laidlaw BJ, Decman V, Ali MA, Abt MC, Wolf AI, Monticelli LA, Mozdzanowska K, Angelosanto JM, Artis D, Erikson J, and Wherry EJ (2013). Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog. 9, e1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, and Kaech SM (2014). CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, et al. (2001). PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol 2, 261–268. [DOI] [PubMed] [Google Scholar]
- Lee YT, Suarez-Ramirez JE, Wu T, Redman JM, Bouchard K, Hadley GA, and Cauley LS (2011). Environmental and antigen receptor-derived signals support sustained surveillance of the lungs by pathogen-specific cytotoxic T lymphocytes. J. Virol 85, 4085–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Mozdzanowska K, Palladino G, and Gerhard W (1994). Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol 152, 1653–1661. [PubMed] [Google Scholar]
- McMaster SR, Wein AN, Dunbar PR, Hayward SL, Cartwright EK, Denning TL, and Kohlmeier JE (2018). Pulmonary antigen encounter regulates the establishment of tissue-resident CD8 memory T cells in the lung airways and parenchyma. Mucosal Immunol. 11, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, Langley WA, Li G, García-Sastre A, Webby RJ, and Ahmed R (2010). Qualitatively different memory CD8+ T cells are generated after lymphocytic choriomeningitis virus and influenza virus infections. J. Immunol 185, 2182–2190. [DOI] [PubMed] [Google Scholar]
- Odorizzi PM, Pauken KE, Paley MA, Sharpe A, and Wherry EJ (2015). Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med 212, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DB, Postow MA, Callahan MK, Allison JP, and Wolchok JD (2014). Immune modulation in cancer with antibodies. Annu. Rev. Med 65, 185–202. [DOI] [PubMed] [Google Scholar]
- Pardoll DM (2012). The blockade of immune checkpoints in cancer immuno-therapy. Nat. Rev. Cancer 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, and Riley JL (2005). CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol 25, 9543–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsoukis N, Brown J, Petkova V, Liu F, Li L, and Boussiotis VA (2012). Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci. Signal 5, ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher H, Bürki K, Lang R, Hengartner H, and Zinkernagel RM (1989). Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature 342, 559–561. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, An LL, Harkins S, Zhang J, Yokoyama M, Widera G, Fuller JT, Kincaid C, Campbell IL, and Whitton JL (1998). DNA immunization with minigenes: low frequency of memory cytotoxic T lymphocytes and inefficient antiviral protection are rectified by ubiquitination. J. Virol 72, 5174–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JH, Johanns TM, Ertelt JM, and Way SS (2008). PDL-1 blockade impedes T cell expansion and protective immunity primed by attenuated Listeria monocytogenes. J. Immunol 180, 7553–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Teichgräber V, Kalia V, Polley A, Masopust D, Harrington LE, Ahmed R, and Wherry EJ (2007). Strength of stimulus and clonal competition impact the rate of memory CD8 T cell differentiation. J. Immunol 179, 6704–6714. [DOI] [PubMed] [Google Scholar]
- Sharpe AH, and Pauken KE (2018). The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol 18, 153–167. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S, Roberts AD, Jelley-Gibbs DM, Wittmer ST, Kohlmeier JE, and Woodland DL (2010). The route of priming influences the ability of respiratory virus-specific memory CD8+ T cells to be activated by residual antigen. J. Exp. Med 207, 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talay O, Shen CH, Chen L, and Chen J (2009). B7-H1 (PD-L1) on T cells is required for T-cell-mediated conditioning of dendritic cell maturation. Proc. Natl. Acad. Sci. USA 106, 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, and Pardoll DM (2015). Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Cauley LS, Khanna KM, and Lefrançois L (2007). Persistent antigen presentation after acute vesicular stomatitis virus infection. J. Virol 81, 2039–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, and Ahmed R (2003). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, and Cauley LS (2014). Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol 95, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Fu HH, Obar JJ, Park JJ, Tamada K, Yagita H, and Lefrançois L (2013). A potential new pathway for PD-L1 costimulation of the CD8-T cell response to Listeria monocytogenes infection. PLoS ONE 8, e56539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Wang S, Zhu Y, Luo L, Zhu G, Flies S, Xu H, Ruff W, Broad-water M, Choi IH, et al. (2009). PD-1 on dendritic cells impedes innate immunity against bacterial infection. Blood 113, 5811–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, and Saito T (2012). Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med 209, 1201–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit DJ, Turner DL, Klonowski KD, Lefrançois L, and Cauley LS (2006). Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity 24, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeyer BH, Heydari S, Sacristán C, Nayak D, Cammer M, Herz J, Cheng X, Davis SJ, Dustin ML, and McGavern DB (2013). PD-1 promotes immune exhaustion by inducing antiviral T cell motility paralysis. J. Exp. Med 210, 757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data generated during this study are available on GEO. The accession number for the microarray data reported in this paper is GEO: GSE149425. This study did not generate unique code.


