Abstract
Multidrug resistance prompts the search for new sources of antibiotics with new targets at bacteria cell. To investigate the antibacterial activity of Cinnamomum cassia L. essential oil (CCeo) alone and in combination with antibiotics against carbapenemase-producing Klebsiella pneumoniae and Serratia marcescens. The antimicrobial susceptibility of the strains was determined by Vitek® 2 and confirmed by MALDI-TOF/TOF. The antibacterial activity of CCeo and its synergism with antibiotics was determined using agar disk diffusion, broth microdilution, time-kill, and checkboard methods. The integrity of the bacterial cell membrane in S. marcescens was monitored by protein leakage assay. CCeo exhibited inhibitory effects with MIC = 281.25 μg.mL-1. The association between CCeo and polymyxin B showed a decrease in terms of viable cell counts on survival curves over time after a 4 hour-treatment with a FIC index value of 0.006. Protein leakage was observed with increasing concentrations for CCeo and CCeo + polymyxin B treatments. CCeo showed antibacterial activity against the studied strains. When associated with polymyxin B, a synergistic effect was able to inhibit bacterial growth rapidly and consistently, making it a potential candidate for the development of an alternative treatment and drug delivery system for carbapenemase-producing strains.
Introduction
Klebsiella pneumoniae and Serratia marcescens are able to acquire and share their resistance genes, leading to multidrug resistance, which can result in severe infections. One of the most important therapeutic challenges posed by Enterobacterales is their resistance to carbapenems [1]. Carbapenemase encoding-genes are located on self-conjugative plasmids, capable of disseminating among bacteria, resulting in the spread of resistance [2]. Clinical options for hard-to-treat infections include tigecycline, polymyxins, aminoglycosides, and fosfomycin [1]. However, the use of these antibiotics is limited because of their toxicity and pharmacokinetic properties [3].
Serratia is a genus naturally resistant to polymyxins. It has a modified lipopolysaccharide (LPS) where the normally encountered Enterobacterales cationic phosphate groups are substituted for 4-amino-4-deoxy-L-arabinose (L-Ara4N) or phosphoethanolamine [4]. The cationic substitution of the phosphate groups by L-Ara4N [5] is the most common modification. This increases the net negative charge of lipid A of LPS results in polymyxins being repelled by bacterial cells [4].
The operon arnBCADTEF-pmrE (also called pmrHFIJKLM-ugd) mediates the synthesis and transfer of L-Ara4N to lipid A [4–6]. The inactivation of the arnB and arnC genes can result in polymyxin sensitivity in S. marcescens [4]. The arnBCADTEF-pmrE operon appears to be constitutively expressed in intrinsically resistant bacteria [6]. Due to widespread distribution of carbapenemase-producing bacteria across the world and the limitations of current treatments for its infections, new therapeutic alternatives are needed [7]. Thus, novel sources of chemicals with antimicrobial activity against multidrug resistance strains are required.
Essential oils are potential sources of antimicrobial compounds. Several studies have demonstrated the antibacterial, antifungal, antiviral, and antiparasitic properties of essential oils from plants [8, 9]. These essential oils and their components have a variety of cellular targets, particularly the membrane and cytoplasm, and may alter the morphology of cells [9], making them an interesting area of research. Essential oils are promising novel functional excipients in the development of new drug delivery systems [10]. Excipients are considered as essential constituents able to guarantee the performance of a drug and optimize the attainment of its therapeutic effects [11]. Essential oils will need to be studied in order to make the best use of their antibacterial activities and to reduce the concentrations of antibiotics required to achieve an antibacterial effect. The antimicrobial activities of Cinnamomum cassia and its essential oil have been reported in bacteria sensitive to antibiotics [12, 13]. The main compound responsible for antimicrobial activity in cinnamon bark essential oil is cinnamaldehyde, which is also the dominant substance [7].
This study describes the antibacterial efficacy of C. cassia L. essential oil (CCeo) against carbapenemase-producing K. pneumoniae and S. marcescens and its synergistic effects when used in combination with antibiotics.
Materials and methods
Essential oil
The essential oil used in this study was C. cassia, a yellow to yellow brown clear oily liquid, extracted from the leaves, bark, and branches by steam distillation, with a density of 1.053 g/cm3, originating from China and acquired commercially from Ferquima (Vargem Grande Paulista, SP, Brazil) under the product name of “Essential Oil of Cinnamon Cassia” (CAS number: 84961-46-6; batch number: 217). Trans-cinnamaldehyde 99% (CAS number: 14371-10-9) was obtained from Sigma-Aldrich (St. Louis, MO, USA). According to the chromatographic technical report provided by the supplier, cinnamaldehyde was the most prevalent compound (87.6%), followed by α-humulene (3.1%), γ-elemene (2.5%), borneol (1.5%), cinnamic acid (0.7%), benzaldehyde (0.5%), eugenol (0.4%), and other minor components (3.7%) (S1 Fig).
Bacterial strains
Carbapenemase-producing K. pneumoniae (KP-KPC) and S. marcescens (SM-KPC) were collected and isolated from human specimens by rectal swab and urine sample, respectively. Samples were taken from patients over 50 years old, hospitalized in different wards at a tertiary hospital in Brazil’s Midwestern region and are representative of the carbapenemase-producing Gram-negative bacteria group. Bacterial strains evaluated in this study were representative from predominant clonal type (33 KPC-producing K. pneumoniae and 6 KPC-producing S. marcescens), previously characterized [14, 15].
Selected isolates were maintained at –70°C, then sub-cultured on MacConkey agar for 24 h before testing. These isolates were identified and characterized according to their sensibility to antibiotics by Vitek® 2 (bioMérieux, Hazelwood, MO, USA). Their minimal inhibitory concentrations (MIC) were confirmed by broth microdilution. Preliminary screening for the presence of carbapenemase was performed using the modified Hodge test [16] and confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF/TOF) using a Microflex LT spectrometer (Bruker Daltonics, Billerica, MA, USA) [17].
Detection of resistance genes
Polymerase chain reaction (PCR) was performed to confirm the presence of the genes blaCTX-M-1-like, blaCTX-M-2-like, blaCTX-M-8-like, blaCTX-M-14-like, blaGES-like, blaGIM-like, blaIMP-10, blaIMP-like, blaKPC-2, blaNDM-like, blaOXA-23, blaOXA-48-like, blaSHV-like, blaSIM-like, blaSME-like, blaSPM-like, blaTEM-like, and blaVIM-like in KP-KPC and SM-KPC strains. Total DNA of KP-KPC and SM-KPC isolates was extracted using the QIAamp DNA minikit® (Qiagen, Courtaboeuf, France) and used as the template in PCR experiments. Amplification was performed using specific primers and cycling parameters, according to the target sequence. Genomic DNA was quantified using a NanoDrop TM 1000 spectrophotometer (Thermo Fischer Scientific, Waltham, Massachusetts, MA, USA). Polymerase chain reactions (PCRs) were prepared as standard 25 μL volumes comprising 12.5 μL of 2 x Master mix [0.05 U/μL Taq DNA polymerase, reaction buffer, 4 mM MgCl2, 0.4 mM of each dATP, dCTP, dGTP and dTTp], 0.25 μL of each primer (1 μL), 1 μL template DNA (20−30 ng/μL) and 11 μL nuclease free water. PCR was performed using a Thermal cycler (model C1000 Touch) supplied by BIO-RAD, California, USA. Sequences of the primers used to the amplification of bla genes are shown in Table 1. The PCR products were analyzed in horizontal electrophoresis using a 1% agarose gel and a 50-bp DNA ladder (Ludwig, Brazil) as a molecular marker, then PCR products were visualized by UV light at 336 nm.
Table 1. Sequences of the primers used to the amplification of bla genes.
| Primer | Sequence (5'-3') | Reference | Product length |
|---|---|---|---|
| blaCTX-M-1-like | F: CGCTTTGCGATGTGCAG | [18] | 512 bp |
| R: ACCGCGATATCGTTGGT | |||
| blaCTX-M-2-like | F: CGACGCTACCCCTGCTATT | [19] | 552 bp |
| R: CCAGCGTCAGATTTTTCAGG | |||
| blaCTX-M-8-like | F: TCGCGTTAAGCGGATGATGC | [19] | 666 bp |
| R: AACCCACGATGTGGGTAGC | |||
| blaCTX-M-14-like | F: TACAGCCCTTCGGCGATGA | [18] | 874 bp |
| R: GGTGACAAAGAGAGTGCAACGGAT | |||
| blaGES-like | F: GTTTTGCAATGTGCTCAACG | [20] | 371 bp |
| R: TGCCATAGCAATAGGCGTAG | |||
| blaGIM-like | F: TCGACACACCTTGGTCTGAA | [20] | 477 bp |
| R: AACTTCCAACTTTGCCATGC | |||
| blaIMP-10 | F: CCAAACYACTASGTTATC | [18] | 188 bp |
| R: GAATAGRRTGGCTTAAYTCTC | |||
| blaIMP-like | F: GGAATAGAGTGGCTTAA(C/T)TCTC | [20] | 232 bp |
| R: GGTTTAA(C/T)AAAACAACCACC | |||
| blaKPC-2 | F: TCTGGACCGCTGGGAGCTGG | [21] | 399 bp |
| R: TGCCCGTTGACGCCCAATCC | |||
| blaNDM-like | F: GGTTTGGCGATCTGGTTTTC | [20] | 621 bp |
| R: CGGAATGGCTCATCACGATC | |||
| blaOXA-23 | F: GATCGGATTGGAGAACCAGA | [22] | 501 bp |
| R: ATTTCTGACCGCATTTCCAT | |||
| blaOXA-48-like | F: TTGGTGGCATCGATTATCGG | [18] | 798 bp |
| R: GAGCACTTCTTTTGTGATGGC | |||
| blaSHV-like | F: CTTGACCGCTGGGAAACGG | [18] | 200 bp |
| R: AGCACGGAGCGGATCAACGG | |||
| blaSIM-like | F: TACAAGGGATTCGGCATCG | [20] | 570 bp |
| R: TAATGGCCTGTTCCCATGTG | |||
| blaSME-like | F: TATGGAACGATTTCTTGGCG | [23] | 300 bp |
| R: CTCCCAGTTTTGTCACCTAC | |||
| blaSPM-like | F: AAAATCTGGGTACGCAAACG | [20] | 271 bp |
| R: ACATTATCCGCTGGAACAGG | |||
| blaTEM-like | F: CCCTTATTCCCTTTYTTGCGG | [18] | 680 bp |
| R: AACCAGCCAGCCWGAAGG | |||
| blaVIM-like | F: GATGGTGTTTGGTCGCATA | [20] | 390 bp |
| R: CGAATGCGCAGCACCAG |
bp: base pairs; F: forward primer; R: reverse primer.
Antibacterial assays of CCeo
The antibacterial activity of raw CCeo was determined using the standard method of agar disk diffusion. Polymyxin B and tigecycline were used as the positive standards. Filter paper disks of 6 mm diameter, containing 10 μl of the CCeo were evaluated against KP-KPC and SM-KPC. After 30 min at room temperature, the dishes were incubated at 37 ± 1°C, for 24 h and the diameter of the inhibition zone formed was measured in millimeters [24]. The test was performed in duplicate and the means of the values obtained were used to classify the bacterium as sensitive (≥ 10 mm) or resistant (< 10 mm) [25]. The MIC of CCeo and trans-cinnamaldehyde (TC) were determined by broth microdilution, as described by Cavalcanti et al. [26] and was determined by resazurin colorimetric assay. CCeo was diluted with distilled water and 0.5% Tween-80.
Synergy tests
The agar disk diffusion assay was performed using the Kirby–Bauer methods for synergy screening. Here, CCeo was evaluated alone and in combination several antibiotics (amikacin, ciprofloxacin, imipenem (IPM), piperacillin/tazobactam, polymyxin B (POL), and tigecycline (Sigma-Aldrich). Antibiotic disks were placed on a Müller-Hinton plate inoculated with KP-KPC or SM-KPC and 10 μl of the CCeo were added in the antibiotic disk [24]. After incubation, the inhibition zone of the combination was comparatively examined with the independent test samples. Synergistic interactions were evaluated if the inhibition zone in the combination was larger than 2 mm or more in comparison to either antibiotics or CCeo alone [27]. The test was performed in duplicate.
Time-kill
The time-kill method was performed using the broth macrodilution technique [28]. CCeo alone and in combination with antibiotic (imipenem or polymyxin B) was tested against the KP-KPC and SM-KPC strains. The concentration of each antimicrobial agent tested represented the MIC value. The polymyxin B concentration used as MIC for SM-KPC was 1 μg.mL-1, which was the same as the polymyxin B MIC for KP-KPC. The time-kill studies were performed with approximately 1.5 × 106 colony forming units (CFU).mL-1 in a final volume of 3.2 mL, verified with a spectrophotometer by Vitek® 2 (bioMérieux). The tubes were shaken periodically and incubated at 37°C. Samples were obtained after 0, 4, 8, 12, 16, 20, and 24 h of incubation. At each sample time, 1 μl was withdrawn from each tube using a sterile loop and seeded in MacConkey agar plates. The plates were incubated for 24 h at 37°C and the colony counts were determined. The resulting colony count of the samples treated with the combination was compared to the samples treated with the most effective single agent. A decrease in colony count of 100-fold or more was considered synergism, while an increase in colony count of 100-fold or more was considered antagonism. Changes that were limited to 10-fold were defined as additive or indifferent [29]. A negative control of BHI was used for the bacterial strain, and saline was used as a sterility control. Polymyxin B and gentamicin (GEN) were used as the positive controls for KP-KPC and SM-KPC strains, respectively. The test was performed in triplicate.
Checkboard
To evaluate the potentiating effect of CCeo, combinations of CCeo and polymyxin B were assessed against the KP-KPC and SM-KPC strains. The concentration of each antimicrobial agent ranged from 1/32 to 2 × MIC. The polymyxin B concentration used as MIC for SM-KPC was 1 μg.mL-1. Using a microtiter plate, two-fold serial dilutions of the oils were prepared in the horizontal rows and two-fold serial dilutions of the antibiotics were prepared in the vertical rows. The plates were prepared well-by-well to obtain a single plate in which both antimicrobial agents were cross-diluted. The inoculum used was 1.5 × 106 CFU.mL-1. Resazurin was used to indicate viable bacteria. The fractional inhibitory concentration (FIC) of each combination was then calculated as the ratio of MIC of the antimicrobial agent in combination versus the MIC of the antimicrobial agent alone [29]. The ΣFIC is then calculated for each test sample independently.
The FIC index was calculated as follows: ΣFIC = FIC(*i) + FIC(*ii). The results were interpreted as either synergistic (ΣFIC ≤ 0.5), additive (0.5 < ΣFIC < 1.0), noninteractive (1.0 < ΣFIC ≤ 4.0), or antagonistic effects (ΣFIC > 4.0) [29]. All experiments were carried out in duplicate as two independent experiments and the calculated FIC indexes were averaged. A negative control of BHI was used for the bacterial strains and saline was used as a sterility control. The sterility of CCeo, polymyxin B, and water was assessed. A second negative control, in the form of 0.5% Tween-80, was also used.
Determination of cell membrane integrity
The integrity of the bacterial cell membrane was monitored by the release of proteins from the cell into the supernatant. Carbapenemase-producing SM-KPC was incubated in MacConkey agar at 37°C for 24 h. Three groups with bacterial inoculum in the logarithmic growth phase (1.5 × 106 CFU.mL-1) were placed in a 96-well microplate, treated with the combination CCeo + polymyxin B, with 0.5, 1, and 2 × the MIC of each agent. The microplate was incubated at 37°C for 0, 1, 2, and 4 h. Then, 1 μL aliquot of CCeo + polymyxin B treatment (concentration of MIC) were seeded in Müller-Hinton agar to confirm the viability of the strain at each time-point. The content of each microplate well was then centrifuged at 2,500 rpm for 5 min at 4°C. The concentration of the proteins released from the cytoplasm was then assessed in the supernatant using the PierceTM BCA Protein Assay kit. The optical absorbance (OD) was read at 490 nm using the iMarkTM Microplate Absorbance Reader (Bio-Rad, São Paulo, SP, Brazil). The experiment was performed in duplicate. Saline with bacterium was used as negative control. CCeo and polymyxin B were diluted with distilled water.
Statistical analysis
The experimental data derived from time–kill and determination of cell membrane integrity assays were analyzed using linear regression slopes comparison—GraphPad Prism 6.01 (GraphPad Inc., San Diego, CA, USA). For time kill, log CFU values were plotted against time for each antibiotic. The kill rate was determined at different time intervals (0, 4, 8, 12, 16, 20 and 24 h), undertaking a linear regression to find the slope for each antimicrobial to test the difference between the slopes and intercepts. For determination of cell membrane integrity assay, concentration of protein was plotted against antimicrobial agent’s concentration. P value < 0.05 was considered significant.
Molecular analysis of 16S and arnB genes in SM-KPC strain
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) analysis was performed to measure the changes in the expression levels of the small subunit 16S of ribosomal RNA during treatment with CCeo in association with polymyxin B. Total RNA was isolated from SM-KPC before and after treatment (at MIC values), transformed into cDNA using the iScriptTM cDNA Synthesis kit (Bio-Rad). The cDNA was then subjected to RT-qPCR using the primers pairs 16S forward (16SF) and 16S reverse (16SR) (Table 2) and SYBR® Green JumpStartTM Taq ReadyMixTM (Sigma-Aldrich). The reactions were performed using the following reagents: SYBR Green mix, 16SF primer, and 16SR primer (10 μM), and template (20 ng), using double-distilled water to obtain the reaction final volume. RT-qPCR cycling was carried out using the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad) as follows: initial denaturation at 94°C for 2 min, then 40 cycles of 94°C (15 sec) and 60°C (1 min). All PCR assay reactions were performed in triplicate. Negative controls were included with each PCR assay. Following amplification, the results were analysed on the PCR computer using the CFX Manager ver. 3.1.15 software. Once amplification was completed, the level of amplification was reported by the software as the mean cycle threshold (CT) value of the replicate samples. A sample was considered positive by real-time RT-qPCR and expressed the 16S gene if the CT value was above the threshold value before the 40 cycles of the PCR reaction and below the negative control CT.
Table 2. Primers used for amplification of arnB and 16S genes.
| Primer | Sequence 5´-3´ | Nucleotide position | Product length |
|---|---|---|---|
| arnBF | CCAAAGCGATTGTTCCGGTG | 365–384 | 169 bp |
| arnBR | AAAGAGAAAATCGCCGTGCC | 609–590 | |
| 16SF | ATTCCAGGTGTAGCGGTGAA | 646–665 | 238 bp |
| 16SR | TGAGTTTTAACCTTGCGGCC | 883–864 |
16SF: 16S gene forward primer; 16SR: 16S gene reverse primer; arnBF: arnB gene forward primer; arnBR: arnB gene reverse primer; bp: base pairs.
Reverse transcriptase polymerase chain reaction (RT-PCR) was also performed on the arnB genes in the SM-KPC strain before and after treatment with CCeo + polymyxin B or polymyxin B alone [14, 15]. The primers arnBF and arnBR were designed based on the gene sequences of S. marcescens subsp. marcescens Db11 and S. marcescens 2880STDY5682918 (GenBank Accession No. HG326223.1, NZ_FCJC01000002.1) (Table 2). Total RNA was isolated and transformed into cDNA using iScriptTM cDNA Synthesis kit (Bio-Rad) and assessed by RT-PCR (Lin2014). The PCR products were examined by 1% agarose gel electrophoresis and ethidium bromide staining.
Ethical approval
This study was conducted with the approval of the Research Ethics Committee from the Universidade Federal da Grande Dourados (Process No. 877.292/2014).
Results
The isolates showed resistance to carbapenems. Sensitivity was observed only to polymyxin B for KP-KPC and amikacin and gentamicin for SM-KPC (Table 3). The strains were identified as carbapenemase-producers by modified Hodge test and MALDI-TOF/TOF. PCR amplification showed that the blaKPC-2 gene was present in KP-KPC, and blaKPC-2, blaIMP-10, and arnB in SM-KPC. The presence of blaCTX-M-1-like, blaCTX-M-2-like, blaCTX-M-8-like, blaCTX-M-14-like, blaGES-like, blaGIM-like, blaIMP-like, blaNDM-like, blaOXA-23, blaOXA-24, blaOXA-48, blaOXA-48-like, blaOXA-51, blaOXA-58, blaSHV-like, blaSIM-like, blaSME-like, blaSPM-like, blaTEM-like, and blaVIM-like genes was not detected.
Table 3. Sensitivity of bacterial strains to the action of different antibiotics (results expressed in μg.mL-1) assessed by Vitek® 2.
| Antibiotics | MIC (Interpretation) | |
|---|---|---|
| KP-KPC | SM-KPC | |
| Amikacin | > 32 (R) | 16 (S) |
| Ampicillin | > 16 (R) | > 16 (R) |
| Aztreonam | > 16 (R) | > 16 (R) |
| Cefoxitin | > 16 (R) | > 16 (R) |
| Ceftazidime | 16 (R) | 8 (R) |
| Cefepime | > 16 (R) | 8 (R) |
| Ciprofloxacin | > 2(R) | > 2 (R) |
| Ertapenem | > 4 (R) | > 4 (R) |
| Gentamicin | > 8 (R) | ≤ 2 (S) |
| Imipenem | > 8 (R) | > 8 (R) |
| Levofloxacin | > 4 (R) | > 4 (R) |
| Meropenem | > 8 (R) | > 8 (R) |
| Nitrofurantoin | > 64 (R) | > 64 (R) |
| Piperacillin/Tazobactam | > 64/4 (R) | 64/4 (R) |
| Polymyxin B | ≤ 1 (S) | (IR) |
| Tigecycline | 2 (I) | 4 (R) |
| Sulfamethoxazole/Trimethoprim | > 4/76 (R) | ≤ 1/19 (S) |
KP-KPC: carbapenemase-producing K. pneumoniae; MIC: minimal inhibitory concentration; SM-KPC: carbapenemase-producing S. marcescens; (S): susceptibility; (I): intermediate; (R): resistance; (IR): intrinsic resistance.
CCeo exhibited an inhibitory effect against KP-KPC and SM-KPC strains, with MICs of 281.25 μg.mL-1. The MIC of trans-cinnamaldehyde was evaluated and found to be the same as the MIC of CCeo for both strains. The initial synergism trial using the disk diffusion method showed the potentiation of imipenem and polymyxin B when associated with CCeo. The PCR results, the disk diffusion, and the MIC of CCeo against KP-KPC and SM-KPC strains and the synergism effect are shown in Table 4.
Table 4. Antibacterial action of CCeo and trans-cinnamaldehyde using disk-diffusion and broth microdilution and synergism between CCeo and antibiotics using disk-diffusion as screening.
| Method | Antimicrobial agent | Bacteria | |
|---|---|---|---|
| KP-KPC | SM-KPC | ||
| MIC | CCeo | 25 mm (S) | 16 mm (S) |
| TC | 281.25 μg.mL-1 | 281.25 μg.mL-1 | |
| Disk-diffusion | CCeo | 281.25 μg.mL-1 | 281.25 μg.mL-1 |
| IPM | 0 mm | 0 mm | |
| CCeo + IPM | 25 mm * | 20 mm * | |
| POL | 20 mm | 14 mm | |
| CCeo + POL | 2 mm * | 6 mm * | |
CCeo: Cinnamomum cassia essential oil; IPM: imipenem; IR: intrinsic resistance; KP-KPC: carbapenemase-producing K. pneumoniae; MIC: minimum inhibitory concentration; POL: polymyxin B; S: susceptibility; SM-KPC: carbapenemase-producing S. marcescens; TC: trans-cinnamaldehyde
*: diameter of inhibition zone in mm of CCeo + antibiotic minus diameter of inhibition zone of antibiotic.
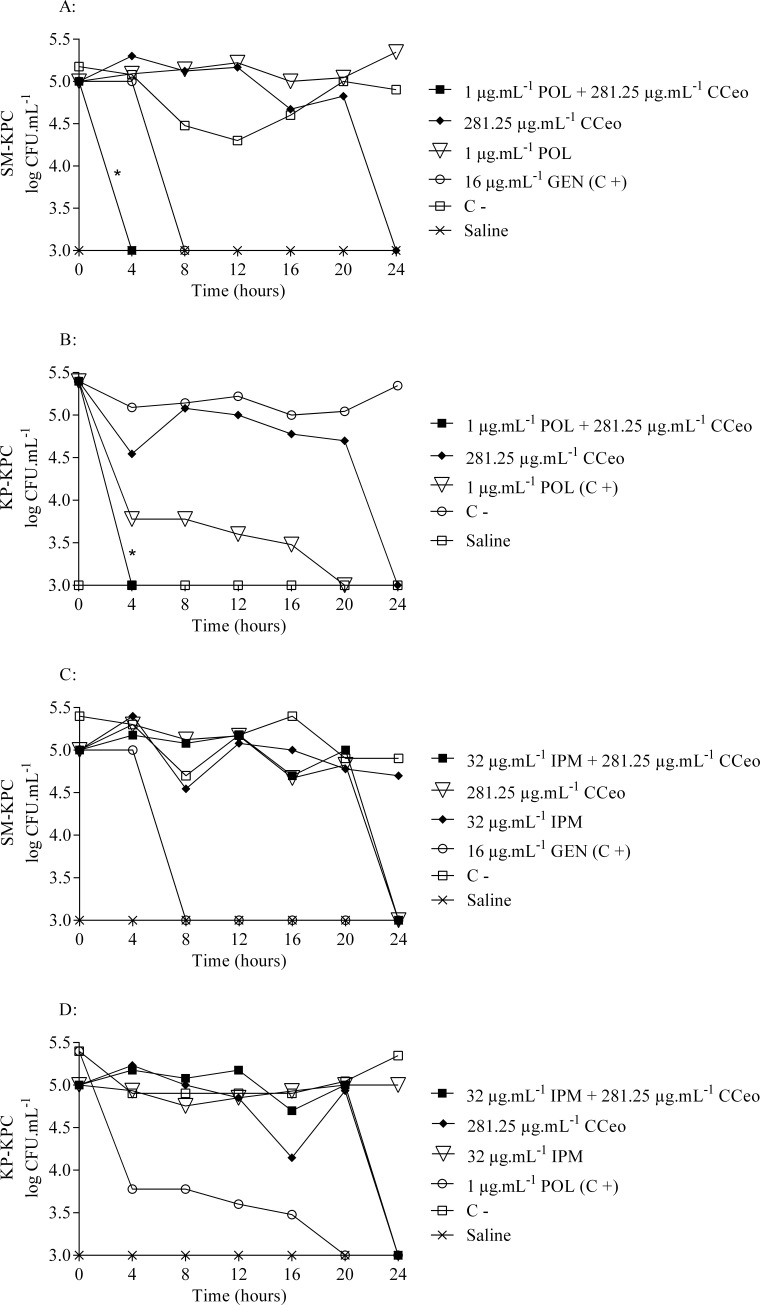
The survival curves of the strains showed a decrease in viable cell counts over time when CCeo was in association with polymyxin B (Fig 1). CCeo + polymyxin B promoted decreases in cellular count in a range of 5 log10 CFU.mL-1. Considering the time of cell death, CCeo inhibited KP-KPC and SM-KPC strains within 24 h. Moreover, CCeo + polymyxin B reduced the bacterial load to undetectable levels within only four hours of treatment, which was faster than polymyxin B (20 h) (p < 0.05) and gentamicin (8 h) (p < 0.05) for KP-KPC and SM-KPC, respectively. For KP-KPC, CCeo + polymyxin B time-kill curve presented statistical difference against the negative control (p < 0.05) but not from POL curve (p > 0.05) showing that the synergy CCeo + polymyxin B is as effective as polymyxin B, an used antibiotic against this carbapenemase-producing strain. For SM-KPC, the CCeo + polymyxin B time-kill curve was significantly different from the polymyxin B curve (p < 0.05). The combination of CCeo and imipenem was unable to reduce the bacterial load (p > 0.05). For all treatments, imipenem had no activity within 24 h. The control antibiotics (polymyxin B and gentamicin) successfully inhibited both strains before 24 h. Saline was used as negative control, for which no growth was observed. The effect of the addition of Tween-80 (0.5%) as an oil solubilizer to the antibacterial assays did not interfere with bacterial growth.
Fig 1. Time-kill curves of the studied carbapenemase-producing strains.
(A) CCeo x polymyxin B against SM-KPC. (B) CCeo x POL against KP-KPC. (C) CCeo x IPM against SM-KPC. (D) CCeo x IPM against KP-KPC. C+: positive control; C-: negative control (SM-KPC or KP-KPC and BHI–Brain Heart Infusion broth); CCeo: C. cassia essential oil; GEN: gentamicin; IPM: imipenem; POL: polymyxin B. Linear regression slopes comparison, *: p < 0.05 comparing to C+.
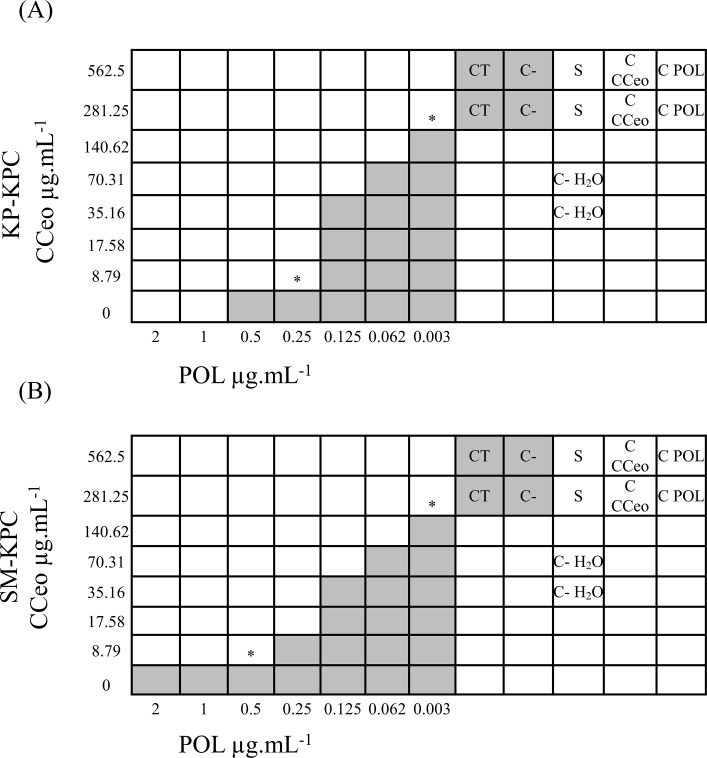
In the checkboard assay, 49 different combinations of CCeo and polymyxin B were tested for each strain, ranging from twice the MIC to several dilutions below the MIC. The FIC index values were calculated by considering the lowest combinations of CCeo and polymyxin B in which there was no visible growth. The FIC index values were 0.006 for both KP-KPC and SM-KPC. A 320-fold reduction of the polymyxin B MIC was able to inhibit the growth of the strains (Fig 2). Checkboard assay was not performed using imipenem because the time-kill curves did not prove synergism.
Fig 2. Checkboard assays with CCeo and POL against the studied carbapenemase-producing strains.
(A) KP-KPC. (B) SM-KPC. Shading: visible growth; *: optimal association concentrations; CT: tween 80 control; C-: negative control; S: sterility control of saline; C- H2O: water control; C CCeo: sterility control of C. cassia essential oil; C POL: sterility control of polymyxin B.
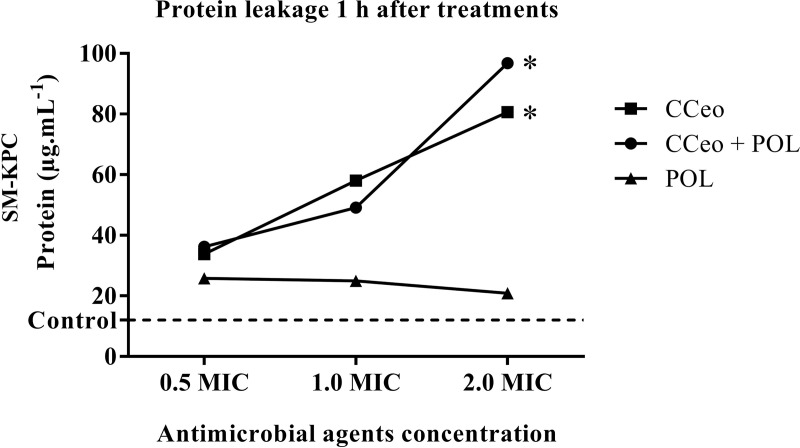
When SM-KPC was treated with increasing concentrations of CCeo and CCeo + polymyxin B, the cells externalized the proteins. The protein leakage from the bacteria in the negative control at 1 h resulted in 12.0 μg.mL-1. At a sub-inhibitory concentration (0.5 × MIC), 33.8 μg.mL-1 of protein was present in the supernatant of CCeo treatment and 36.2 μg.mL-1 for CCeo + polymyxin B, both at 1 h. With CCeo treatment (2 × MIC), 80.6 μg.mL-1 of protein was present in the supernatant. With CCeo + polymyxin B (1 h), 96.8 μg.mL-1 of protein was present in the supernatant, showing an increase in the protein leakage levels. The concentrations of externalized proteins when cells were treated with CCeo and CCeo + polymyxin B differed significantly from cells treated only with polymyxin B (p < 0.05). Polymyxin B alone did not show any leakage of protein (Fig 3). In addition, no difference was observed in the protein leakage between the time-points studied (1, 2 and 4h). Protein leakage after 1 h in all treatments was the time-point chosen to be represented graphically because the bacterial cells are still viable to grow in the culture media at this time.
Fig 3. Protein leakage from SM-KPC after 1 hour of CCeo and CCeo + POL treatments.
CCeo (MIC, 281.25 μg.mL-1), CCeo + POL (MIC CCeo, 281.25 μg.mL-1 + MIC POL, 1 μg.mL-1) and POL (MIC, 1 μg.mL-1). Control: saline with SM-KPC. CCeo: C. cassia essential oil; MIC: minimal inhibitory concentration; POL: polymyxin B. Linear regression slopes comparison, *: p < 0.05 comparing to POL.
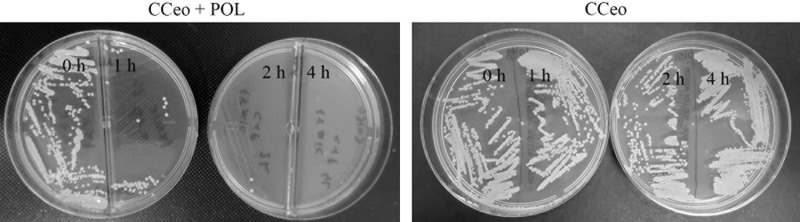
When the cells were treated with CCeo + polymyxin B, Müller-Hinton agar plates showed abundant growth at 0 h, an expressive growth reduction at 1 h, only 2 CFU at 2 h, and a total inhibition at 4 h. No difference in bacterium growth at 0, 1, 2, and 4 h was observed when the cells were treated with CCeo alone (Fig 4), consistent with the time-kill assay results.
Fig 4. SM-KPC growth after CCeo and CCeo + POL treatments.
Müller-Hinton agar plates seeded with 0 h, 1 h, 2 h, and 4 h aliquots of SM-KPC treated with CCeo + POL 1 x MIC and CCeo 1 x MIC.
The 16S rRNA transcripts were reduced in SM-KPC cells after treatment CCeo + polymyxin B, as verified by RT-qPCR, confirming a decrease in the bacterial cell number (S2 Fig). The arnB transcripts were present before and after treatment with CCeo + polymyxin B. Also, arnB gene expression was observed after treatment with polymyxin B (S3 Fig).
Discussion
As far as we know, there are no studies on the antibacterial action of C. cassia alone or in combination with antibiotics against carbapenemase-producing Enterobacterales bacteria. Here, C. cassia was able to inhibit two strains of Gram-negative bacteria resistant to almost all classes of clinical antibiotics, either alone or in combination with polymyxin B. Antibacterial assays demonstrated the potential of action of CCeo against KP-KPC and SM-KPC (MIC of 281.25 μg.mL-1). Trans-cinnamaldehyde is the main component with antibacterial action of CCeo, and showed the same MIC as CCeo. Following a new tendency to study the whole plant instead of single compounds [29], assays were conducted with CCeo as a whole. CCeo + polymyxin B was found to be faster than polymyxin B or gentamicin in inhibiting KP-KPC and SM-KPC. This susceptibility was independent of the acquired resistant gene, since KP-KPC expressed blaKPC-2, and SM-KPC expressed blaKPC-2 and blaIMP-10, indicating a mechanism of action not related to the genus, species, or resistance genes. The FIC index value of CCeo and polymyxin B in combination confirmed the synergistic effects between these two agents for KP-KPC and SM-KPC. This indicated that the best dosages of the antimicrobial agents where obtained when these compounds were used in combination, providing a basis for the use of different combinations in the development of alternative treatments.
Imipenem and polymyxin B were chosen for synergism confirmatory tests because they are used clinically for the therapeutic treatment of Gram-negative infections. The doses of CCeo and antibiotics in this study were selected according to the MIC values presented by the strains studied. For carbapenem-resistant bacteria, polymyxin B alone or in combination with other antibiotics is currently the treatment of choice for these infections [30] and this study´s aim was to reach the lowest doses of the antimicrobial agents tested in synergism for an efficient alternative treatment. As the species Serratia marcescens has intrinsic resistance to polymyxins, the experimental polymyxin B concentration used as MIC for SM-KPC was 1 μg.mL-1, which was the same as the polymyxin B MIC for KP-KPC. The widespread distribution of carbapenemase-producing Enterobacterales has resulted in the increased use of this drug, with the inevitable risk of developing resistance. However, the use of polymyxin is associated with a high incidence of toxicity, such as nephrotoxicity and neurotoxicity [31]. Despite the polymyxins toxicity, when there is no other pharmacological option for treatment, this dosage is administered in patients who present infections with bacteria resistant of carbapenem [32]. The possibility of reducing the necessary dose for treatment with polymyxin B, when associated with CCeo, is a important result since the polymyxin B dose is directly related to its toxicity when used therapeutically. Our study demonstrates that when polymyxin B is associated with CCeo there is a drastic reduction of 320-fold in the dose required for in vitro treatment of the studied multidrug-resistant strains, which brings a benefit to the patient who will not have to receive higher doses of a toxic medication. Moreover, lower doses are expected to reduce the selective pressure upon carbapenemase-producing bacteria, thereby minimizing the appearance of resistance [30]. Concerning CCeo toxicity, the major components of C. cassia are considered to be non-toxic and safe agents with no acute or chronic toxicity, no mutagenicity or genotoxicity, and no carcinogenicity detected in mammalian studies [33].
The synergism results in our study also demonstrate that a dose of 8.79 μg.mL-1 of C. cassia essential oil is able to inhibit the growth of the multi-resistant strains tested. Cinnamaldehyde has been reported as the main component in CCeo and studies showed that Cinnamaldehyde absorption appears to be rapid and complete in mice [34] and in humans. The elimination of cinnamaldehyde by two adult volunteers, who had received a single oral dose of 0.7 mg/kg, was rapid with 100% recovered in the urine within 8 h [35]. Humans also clear systemically available cinnamic acid quickly. Eleven adult human volunteers received single intravenous doses of cinnamic acid, equivalent to 5 mg/kg bodyweight. Plasma was cleared of cinnamic acid (cinnamaldehyde is converted to cinnamic acid in organism) within 20 min [36]. Acute toxicity, oral LD50 values for cinnamaldehyde in rats and guinea pigs have been reported as 2220 mg/kg and 1160 mg/kg, respectively [33]. Mutagenicity and genotoxicity in mammalian, there was no evidence of an increase in unscheduled DNA synthesis when rats were administered up to 500 mg/kg bodyweight by intraperitoneal injection in a micronucleus assay [37]. The CCeo oil are intended to be used at low levels of exposure relative to doses that elicit adverse effects in laboratory animals via systemic exposure. A C. cassia oral safe dose is 0.7 mg/kg/day [33], and the bio-accessibility of C. cassia by oral route was approximately 79% [38]. The minor C. cassia essential oil dose, demonstrated in our study (8.79 μg.mL-1), able to inhibit the growth of the multi-resistant strains tested when associated with polymyxin B is lower than the literature describes as safe concentrations of C. cassia for humans.
In this study, CCeo was found to be efficient as an excipient for polymyxin B, reducing the dose of antibiotic required for treatment. Thus, micro-emulsions and nano-emulsions synthesized using CCeo may be used as drug delivery systems. Due to their potent antibacterial activity and physicochemical properties, these drug delivery systems could be used to improve the treatment options for human diseases and provide better delivery vehicles for drugs with low bioavailability, thereby decreasing drug toxicity and prolonging the usable life of current antimicrobial drugs [39]. Nanostructured systems have been designed intending essential oils encapsulation as approach to enhance their bio-availability and bio-efficacy as a result of high cellular uptake and controlled release delivery. According to the literature, polymer-based nano-carriers are extensively used for this purpose and no significant cytotoxicity on the normal growth and the development of cultured diploid human cells were observed [40].
The concentrations of leaked proteins after sub-inhibitory (0.5 × MIC) treatments were 2.8-fold (CCeo) and 3.0-fold (CCeo + polymyxin B) higher than the negative control at 1 h. These sub-inhibitory treatment results indicated that the bacterial cells start to leak proteins when they are exposed to CCeo. However, the cells are still viable to grow in culture media. The protein leakage indicates an affected membrane [9] is still able to allow the entrance of polymyxin B to conclude the lysis of the bacterial cell membrane. At 1 × MIC and 2 × MIC of the CCeo and CCeo + polymyxin B treatments, protein leakage was higher. However, late intracellular content extravasation may occur due to other apoptosis pathways being activated [7]. It is possible to assume that CCeo induced concentration-dependent damage in cell membranes, destabilized the outer membrane of the SM-KPC, allowing polymyxin B to enter the periplasm of the cell, disrupting outer membrane integrity, with leakage of cellular contents, and cell death [7]. Moreover, it has already been shown that cinnamon reduces the negativity of the electric charge of E. coli cell membranes, allowing antibiotics to access the penicillin-binding proteins to induce of cell death [27].
A low FIC value index indicates the low MICs of CCeo and polymyxin B in the inhibition of SM-KPC growth. The mechanisms of action of this synergistic effect could involve CCeo blocking the SM-KPC strategies for resistance to polymyxin B or increasing the outer membrane permeability, allowing polymyxin B to enter the bacterial periplasmic space. The resistance strategies of S. marcescens to polymyxins include modifications on their LPS, which normally have negative charges and are the initial targets of polymyxins [4]. Several bacterial cell injuries have already been reported for bacteria treated with cinnamon essential oils [27] and cinnamaldehyde [41]. In this study, the expression of the arnB gene was observed in SM-KPC before and after treatment with polymyxin B. It is known that polymyxin B can upregulate the arnB expression in S. marcescens, indicating that the bacterial cell is able to sense the presence of polymyxin B and undergo a positive feedback reaction by enhancing the arnB expression in order to promote better protection against an imminent drug threat [4]. These results confirm the hypothesis that CCeo disturbs the bacterial outer membrane allowing polymyxin B to enter the cell since, when it was associated with polymyxin B, CCeo was able to inhibit arnB overexpression.
The findings presented here are of great significance given the current importance of polymyxin B in clinical practice and an increase in the generalized bacterial resistance to this drug. However, this research was limited to in vitro studies. Since our findings were promising with KP-KPC and SM-KPC, strains classified with “Critical” priority pathogens on the list for the research and development of new antibiotics by The World Health Organization (WHO) [42], the development of research with another clinically relevant bacteria, should be extended to in vivo investigations, including models of infection.
Conclusion
CCeo was able to inhibit KP-KPC and SM-KPC and was able to exert this inhibition in combination with polymyxin B at a reduced antibiotic dose. The results presented here, therefore, provide a basis for further in vivo and clinical studies for the development of novel treatments using a combination of CCeo and polymyxin B for use against multidrug resistant strains.
Supporting information
AGILENT 7820A Gas Chromatograph. Column: HP-5 30 m x 0.32 mm x 0.25 μm (AGILENT). Temp.: Column: 70°C (0 min), 3°C / min at 240°C. Injector: 240°C Split: 1/50. FID detector: 250°C. Vol. injection time: 1 ul (conc 1.0% in chloroform). Vany Ferraz, Chemistry Department–UFMG (Universidade Federal de Minas Gerais). Requested by Ferquima (Vargem Grande Paulista, SP, Brazil).
(TIF)
Green lines: S. marcescens without CCeo + POL treatment; blue lines: S. marcescens after 1 hour with CCeo + POL treatment; red lines: negative control. Electroforese gel: RT-qPCR products. RFU: Relative fluorescence units.
(TIF)
(A) Agarose gel electrophoresis for arnB mRNA (169 bp) stained with GelRed—CCeo + POL treatment. Lane 1: molecular weight marker Ludwig 50 x (50 bp); lane 2: negative control; lane 3: before treatment; lane 4: after 1 hour of treatment. (B) Agarose gel electrophoresis for arnB mRNA (169 bp) stained with GelRed–POL treatment. Lane 1: molecular weight marker Ludwig 50 x (50 bp); lane 2: negative control; lane 3: before treatment; lane 4: after 1 hour of treatment.
(TIF)
Acknowledgments
The authors thank the financial support and infrastructure provided by the Laboratório de Pesquisa em Ciência da Saúde (LPCS) and Universidade Federal da Grande Dourados (UFGD) which made this study possible, thank the Hospital Universitário da Universidade Federal da Grande Dourados (HU-UFGD) for the strains of the study. K.E.S. received a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and J.H.F.S.Q. from Support Foundation for the Development of Education, Science and Technology in the State of Mato Grosso do Sul (FUNDECT).
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was partially supported by the National Council for Scientific and Technological Development (CNPq grants 480949/2013-1 and 407791/2013-2) – www.cnpq.br; the Support Foundation for the Development of Education, Science and Technology in the State of Mato Grosso do Sul (FUNDECT grants 05/2011 and 04/2012) – www.fundect.ms.gov.br; Coordination for the Improvement of Higher Education Personnel (CAPES) and Federal University of Grande Dourados (UFGD). K.E.S. received a scholarship from CAPES and J.H.F.S.Q from FUNDECT. The sponsors had no role in the collection, analysis and interpretation of data or the writing of the manuscript. There was no additional external funding received for this study.
References
- 1.Campos AC, Albiero J, Ecker AB, Kuroda CM, Meirelles LEF, Polato A, et al. Outbreak of Klebsiella pneumoniae carbapenemase–producing K pneumoniae: A systematic review. Am J Infect Control. 2016;44(11):1374–80. 10.1016/j.ajic.2016.03.022 [DOI] [PubMed] [Google Scholar]
- 2.Hammoudi D, Moubareck CA, Sarkis DK. How to detect carbapenemase producers? A literature review of phenotypic and molecular methods. J Microbiol Methods. 2014;107:106–18. 10.1016/j.mimet.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 3.Javan AO, Shokouhi S, Sahraei Z. A review on colistin nephrotoxicity. Eur J Clin Pharmacol. 2015;71:801–10. 10.1007/s00228-015-1865-4 [DOI] [PubMed] [Google Scholar]
- 4.Lin QY, Tsai YL, Liu MC, Lin WC, Hsueh PR, Liaw SJ. Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob Agents Chemother. 2014;58:5181–90. 10.1128/AAC.00013-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. 10.1128/mmbr.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasconcelos NG, Croda J, Simionatto S. Antibacterial mechanisms of cinnamon and its constituents: A review. Microb Pathog. 2018;120:198–203. 10.1016/j.micpath.2018.04.036 [DOI] [PubMed] [Google Scholar]
- 8.Barbosa LN, Probst IS, Andrade BF, Alves FC, Albano M, da Cunha Mde L, et al. In vitro antibacterial and chemical properties of essential oils including native plants from Brazil against pathogenic and resistant bacteria. J Oleo Sci. 2015;64(3):289–98. 10.5650/jos.ess14209 [DOI] [PubMed] [Google Scholar]
- 9.Nazzaro F, Fratianni F, De Martino L, Coppola R, De Feo V. Effect of Essential Oils on Pathogenic Bacteria. Pharmaceuticals. 2013;6:1451–74. 10.3390/ph6121451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marto J, Gouveia LF, Gonçalves LM, Ribeiro HM, Almeida AJ. Design of minocycline-containing starch nanocapsules for topical delivery. J Microencapsul. 2018;11:1–28. 10.1080/02652048.2018.1487472 [DOI] [PubMed] [Google Scholar]
- 11.Dave VS, Saoji SD, Raut NA, Haware RV. Excipient variability and its impact on dosage form functionality. J Pharm Sci. 2015;104(3):906–15. 10.1002/jps.24299 [DOI] [PubMed] [Google Scholar]
- 12.Trinh NT, Dumas E, Thanh ML, Degraeve P, Ben Amara C, Gharsallaoui A, et al. Effect of a Vietnamese Cinnamomum cassia essential oil and its major component trans-cinnamaldehyde on the cell viability, membrane integrity, membrane fluidity, and proton motive force of Listeria innocua. Can J Microbiol. 2015;61(4):263–71. 10.1139/cjm-2014-0481 [DOI] [PubMed] [Google Scholar]
- 13.Andrade BFMT Barbosa LN, Probst IS, Fernandes Júnior A. Antimicrobial activity of essential oils. J Essent Oil Res. 2014;26(1):34–40. 10.1080/10412905.2013.860409 [DOI] [Google Scholar]
- 14.Silva KE, Maciel WG, Sacchi FP, Carvalhaes CG, Rodrigues-Costa F, da Silva AC, et al. Risk factors for KPC-producing Klebsiella pneumoniae: watch out for surgery. J Med Microbiol. 2016;65(6):547–53. 10.1099/jmm.0.000254 [DOI] [PubMed] [Google Scholar]
- 15.Silva KE, Cayô R, Carvalhaes CG, Sachi FPC, Rodrigues-Costa F, Ramos da Silva AC, et al. Coproduction of KPC-2 and IMP-10 in Carbapenem-Resistant Serratia marcescens Isolates from an Outbreak in a Brazilian Teaching Hospital. J Clin Microbiol. 2015;53(7):2324–8. 10.1128/JCM.00727-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Five Informational Supplement. CLSI Document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 17.Carvalhaes CG, Cayô R, Assis DM, Martins ER, Juliano L, Juliano MA, et al. Detection of SPM-1-producing Pseudomonas aeruginosa and class D beta-lactamase-producing Acinetobacter baumannii isolates by use of liquid chromatography-mass spectrometry and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:287–90. 10.1128/JCM.02365-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brasiliense D, Cayô R, Streling AP, Nodari CS, Barata RR, Lemos PS, et al. Diversity of Metallo-β-Lactamase-Encoding Genes Found in Distinct Species of Acinetobacter Isolated From the Brazilian Amazon Region. Mem Inst Oswaldo Cruz. 2019;114:e190020 10.1590/0074-02760190020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum b-lactamases. J Antimicrob Chemother. 2006;57(1):154–5. 10.1093/jac/dki412 [DOI] [PubMed] [Google Scholar]
- 20.Gheorghe I, Czobor I, Chifiriuc MC, Borcan E, Ghiţă C, Banu O, et al. Molecular Screening of Carbapenemase-Producing Gram-negative Strains in Romanian Intensive Care Units During a One Year Survey. J Med Microbiol. 2014;63(Pt 10):1303–10. 10.1099/jmm.0.074039-0 [DOI] [PubMed] [Google Scholar]
- 21.Cole JM, Schuetz AN, Hill CE, Nolte FS. Development and Evaluation of a Real-Time PCR Assay for Detection of Klebsiella pneumoniae Carbapenemase Genes. J Clin Microbiol. 2009;47(2):322–6. 10.1128/JCM.01550-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for Genes Encoding Prevalent OXA Carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–3. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 23.Mlynarcik P, Roderova M, Kolar M. Primer Evaluation for PCR and Its Application for Detection of Carbapenemases in Enterobacteriaceae. Jundishapur J Microbiol. 2016;9(1):e29314 10.5812/jjm.29314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adukwu EC, Bowles M, Edwards-Jones V, Bone H. Antimicrobial Activity, Cytotoxicity and Chemical Analysis of Lemongrass Essential Oil (Cymbopogon flexuosus) and Pure Citral. Appl Microbiol Biotechnol. 2016;100(22):9619–27. 10.1007/s00253-016-7807-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong-Leung YL. Antibacterial activities of some Hong Kong plants used in Chinese medicine. Fitoterapia. 1988;69(1):11–6. [Google Scholar]
- 26.Cavalcanti YW, Almeida LFD, Padilha WWN. Antifungal Activity of Three Essential Oils on Candida Strains. Rev Odontol Bras Central. 2011;20(52):68–73. 10.1155/2017/7158756 [DOI] [Google Scholar]
- 27.Yap PS, Krishnan T, Chan KG, Lim SH. Antibacterial mode of action of Cinnamomum verum bark essential oil, alone and in combination with piperacillin, against a multi-drug-resistant Escherichia coli strain. J Microbiol Biotechnol. 2015;25(8):1299–306. 10.4014/jmb.1407.07054 [DOI] [PubMed] [Google Scholar]
- 28.Morinaka A, Tsutsumi Y, Yamada K, Takayama Y, Sakakibara S, Takata T, et al. In Vitro and In Vivo Activities of OP0595, a New Diazabicyclooctane, against CTX-M-15-Positive Escherichia coli and KPC-Positive Klebsiella pneumoniae. Antimicrob Agents Chemother. 2016;60(5):3001–6. 10.1128/AAC.02704-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Vuuren S, Viljoen A. Plant-Based Antimicrobial Studies–Methods and Approaches to Study the Interaction between Natural Products. Planta Med. 2011;77:1168–82. 10.1055/s-0030-1250736 [DOI] [PubMed] [Google Scholar]
- 30.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 31.Bialvaei AZ, Samadi Kafil H. Colistin, Mechanisms and Prevalence of Resistance. Curr Med Res Opin. 2015;31(4):707–21. 10.1185/03007995.2015.1018989 [DOI] [PubMed] [Google Scholar]
- 32.Liang Q, Huang M, Xu Z. Early use of polymyxin B reduces the mortality of carbapenem-resistant Klebsiella pneumoniae bloodstream infection. Braz J Infect Dis. 2019;23(1):60–5. 10.1016/j.bjid.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bickers D, Calow P, Greim H, Hanifin JM, Rogers AE, Saurat JH, et al. A toxologic and dermatologic assessment of cinnamyl alcohol, cinnamaldehyde and cinnamic acid when used as fragrance ingredients. The RIFM expert panel. Food Chem Toxicol. 2005;43:799–836. 10.1016/j.fct.2004.09.013 [DOI] [PubMed] [Google Scholar]
- 34.Peters M, Caldwell J. Studies on trans-cinnamaldehyde. The influence of dose size and sex on its disposition in the mouse and rat. Food Chem Toxicol. 1994;32:869–76. 10.1016/0278-6915(94)90084-1 [DOI] [PubMed] [Google Scholar]
- 35.Peters M. Metabolic and mechanistic studies in the safety evaluation of trans-cinnamaldehyde. A Thesis submitted for the Degree of Doctor of Philosophy in the University of London, Department of Pharmacology and Toxicology, 1993.
- 36.Quarto di Palo FM, Bertolini AM Cinnamic acid administration to renal patients. Atti Accad Med Lombarada. 1961;16:180–3. [Google Scholar]
- 37.Hayashi M, Kishi M, Sofuni T, Ishidate M Jr. Micronucleus tests in mice on 39 food additives and eight miscellaneous chemicals. Food Chem Toxicol. 1988;26:487–500. 10.1016/0278-6915(88)90001-4 [DOI] [PubMed] [Google Scholar]
- 38.Helal A, Tagliazucchi D, Verzelloni E, Conte A. Bioaccessibility of Polyphenols and Cinnamaldehyde in Cinnamon Beverages Subjected to in Vitro Gastro-Pancreatic Digestion. J Funct Foods. 2014;7:506–16. 10.1016/j.jff.2014.01.005 [DOI] [Google Scholar]
- 39.Franklyne JS, Mukherjee A, Chandrasekaran N. Essential oil micro- and nanoemulsions: promising roles in antimicrobial therapy targeting human pathogens. Lett Appl Microbiol. 2016;63(5):322–34. 10.1111/lam.12631 [DOI] [PubMed] [Google Scholar]
- 40.Lammari N, Louaer O, Meniai AH, Elaissari A. Encapsulation of Essential Oils via Nanoprecipitation Process: Overview, Progress, Challenges and Prospects. Pharmaceutics. 2020;7:12(5). pii: E431 10.3390/pharmaceutics12050431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albano M, Alves FCB, Andrade BFMT, Barbosa LN, Pereira AFM, daCunha MLRS, et al. Antibacterial and anti-staphylococcal enterotoxin activities of phenolic compounds. Innov Food Sci Emerg Technol. 2016;38:83–90. 10.1016/j.ifset.2016.09.003 [DOI] [Google Scholar]
- 42.WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, 2017. http://www.who.int/meedicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed 01st June 2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AGILENT 7820A Gas Chromatograph. Column: HP-5 30 m x 0.32 mm x 0.25 μm (AGILENT). Temp.: Column: 70°C (0 min), 3°C / min at 240°C. Injector: 240°C Split: 1/50. FID detector: 250°C. Vol. injection time: 1 ul (conc 1.0% in chloroform). Vany Ferraz, Chemistry Department–UFMG (Universidade Federal de Minas Gerais). Requested by Ferquima (Vargem Grande Paulista, SP, Brazil).
(TIF)
Green lines: S. marcescens without CCeo + POL treatment; blue lines: S. marcescens after 1 hour with CCeo + POL treatment; red lines: negative control. Electroforese gel: RT-qPCR products. RFU: Relative fluorescence units.
(TIF)
(A) Agarose gel electrophoresis for arnB mRNA (169 bp) stained with GelRed—CCeo + POL treatment. Lane 1: molecular weight marker Ludwig 50 x (50 bp); lane 2: negative control; lane 3: before treatment; lane 4: after 1 hour of treatment. (B) Agarose gel electrophoresis for arnB mRNA (169 bp) stained with GelRed–POL treatment. Lane 1: molecular weight marker Ludwig 50 x (50 bp); lane 2: negative control; lane 3: before treatment; lane 4: after 1 hour of treatment.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.