Abstract
The intracellular lifestyle of Salmonella enterica is characterized by the formation of a replication-permissive membrane-bound niche, the Salmonella-containing vacuole (SCV). As a further consequence of the massive remodeling of the host cell endosomal system, intracellular Salmonella establish a unique network of various Salmonella-induced tubules (SIT). The bacterial repertoire of effector proteins required for the establishment for one type of these SIT, the Salmonella-induced filaments (SIF), is rather well-defined. However, the corresponding host cell proteins are still poorly understood. To identify host factors required for the formation of SIF, we performed a sub-genomic RNAi screen. The analyses comprised high-resolution live cell imaging to score effects on SIF induction, dynamics and morphology. The hits of our functional RNAi screen comprise: i) The late endo-/lysosomal SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex, consisting of STX7, STX8, VTI1B, and VAMP7 or VAMP8, which is, in conjunction with RAB7 and the homotypic fusion and protein sorting (HOPS) tethering complex, a complete vesicle fusion machinery. ii) Novel interactions with the early secretory GTPases RAB1A and RAB1B, providing a potential link to coat protein complex I (COPI) vesicles and reinforcing recently identified ties to the endoplasmic reticulum. iii) New connections to the late secretory pathway and/or the recycling endosome via the GTPases RAB3A, RAB8A, and RAB8B and the SNAREs VAMP2, VAMP3, and VAMP4. iv) An unprecedented involvement of clathrin-coated structures. The resulting set of hits allowed us to characterize completely new host factor interactions, and to strengthen observations from several previous studies.
Author summary
The facultative intracellular pathogen Salmonella enterica serovar Typhimurium induces the reorganization of the endosomal system of mammalian host cells. This activity is dependent on translocated effector proteins of the pathogen. The host cell factors required for endosomal remodeling are only partially known. To identify such factors for the formation and dynamics of endosomal compartments in Salmonella-infected cells, we performed a live cell imaging-based RNAi screen to investigate the role of 496 mammalian proteins involved in cellular logistics. We identified that endosomal remodeling by intracellular Salmonella is dependent on host factors in the following functional classes: i) the late endo-/lysosomal SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex, ii) the early secretory pathway, represented by regulator GTPases RAB1A and RAB1B, iii) the late secretory pathway and/or recycling endosomes represented by GTPases RAB3A, RAB8A, RAB8B, and the SNAREs VAMP2, VAMP3, and VAMP4, and iv) clathrin-coated structures. The identification of these new host factors provides further evidence for the complex manipulation of host cell transport functions by intracellular Salmonella and should enable detailed follow-up studies on the mechanisms involved.
Introduction
The food-borne, facultative intracellular pathogen Salmonella enterica serovar Typhimurium (STM) is the etiological agent of gastroenteritis in humans or systemic infections in mice [1]. An early step in disease is the active invasion of epithelial cells. This process is dependent on the translocation of effector proteins by STM into the host cell through a type 3 secretion system (T3SS) encoded by genes in Salmonella pathogenicity island 1 (SPI1) [2, 3].
After invasion STM, similar to many other intracellular pathogens, establish a replicative niche in host cells, termed Salmonella-containing vacuole (SCV). This process is dependent on the function of a distinct T3SS, encoded by genes in SPI2 [4, 5] and translocating another set of effectors [6]. Though initially associating with markers of the early endosome (EE) such as EEA1 and the small GTPase RAB5 [7, 8], the SCV finally acquires several markers of the late endosome (LE). These markers include lysosome-associated membrane proteins (LAMPs) [9, 10], the vacuolar ATPase [11], and RAB7 [12, 13]. Concurrently, other canonical organelle markers such as the mannose-6-phosphate receptor are excluded [14].
A unique feature of STM among intravacuolar bacteria is the formation of a diverse array of long tubular structures, Salmonella-induced tubules (SIT) [15]. The first SIT discovered are the LAMP-decorated Salmonella-induced filaments (SIF) [16, 17]. Moreover, SIF have been structurally characterized, revealing the presence of a double membrane tubular network [18, 19]. The host-derived membranes forming SCV, SIF, and other tubular compartments are collectively termed Salmonella-modified membranes (SMM).
The repertoire of bacterial effector proteins necessary for the formation of SMM is quite well-characterized, with the SPI2-T3SS effector protein SifA being the most important factor [20, 21]. However, much less is known about corresponding host factors required for biogenesis of SMM. One crucial factor in SIF biogenesis is the SifA- and kinesin-interacting protein SKIP (a.k.a. PLEKHM2). In conjunction with the effectors SifA and PipB2 [22, 23] and the small GTPase ARL8B [24, 25], SKIP mediates kinesin-1 interaction and thus a link to the microtubule cytoskeleton and organelle motility [26].
Several attempts were made to analyze the interactions of STM with host factors in a systematic manner. These comprise RNA inference (RNAi) screens aiming at different parts of the STM infection process. Two genome-scale screens targeted the invasion [27, 28], while three screens focused on intracellular replication with two sub-genomic screens covering kinases and corresponding phosphatases, respectively [29, 30], and a genome-wide screen [31]. Additionally, two recent proteomic studies also shed light on interactions of intracellular STM with host cells. Vorwerk et al. [32] characterized the proteome of late SMM, while Santos et al. focused on early and maturing SCV [33].
All of these studies identified host factors yet unprecedented in STM pathobiology and showed the general value of such systematic approaches. However, none of these approaches targeted specifically SIF, thus a host-SIF interactome is far from complete. Therefore, we established a targeted RNAi screen comprising 496 human genes mostly involved in cellular logistics to identify host factors involved in the formation of SIF. Using stably LAMP1-GFP-transfected HeLa cells, we performed automated microscopy on a spinning disk confocal microscope (SDCM) system with time-lapse live cell imaging (LCI) of STM infection and scored for altered SIF formation as phenotypic readout. Investigating high-scoring hits of the RNAi screen, we validated several so far unknown host-SIF interactions by LCI: (i) involvement of the late endo-/lysosomal soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex and its interaction partners, (ii) interactions of SIF with early secretory RAB1A/B, (iii) late secretory RAB3A, RAB8A/B, and VAMP2/3/4, and (iv) an interaction of SIF with clathrin-coated structures.
Results
RNAi screen setup and evaluation
We aimed to identify host cell factors that are required for the endosomal remodeling induced by intracellular STM in a SPI2-T3SS-dependent manner. For this, an RNAi screen was performed with siRNAs predominantly targeting mammalian genes involved in cellular logistics and trafficking (75.2% categorized in intracellular transport according to Gene Ontology [GO] terms). This subset of 496 genes was termed ‘trafficome’ and is listed in S1 Table (along with the GO terms from which these genes were selected). Such a screen necessitates specific considerations and controls with the major ones described below, and further experimental issues detailed in Suppl. Materials.
As a phenotypic readout for STM-induced endosomal remodeling, we specifically scored the formation of SIF in infected cells. SIF show a highly dynamic behavior in their early phase after formation, with constant elongation and retraction [34, 35]. Thus, in contrast to previous RNAi screens done by analyzing fixed cells, we decided to perform this screen by LCI to obtain maximal phenotypic information. A previously established HeLa cell line stably transfected with LAMP1-GFP as the marker for SIF [18] was used as the host cell.
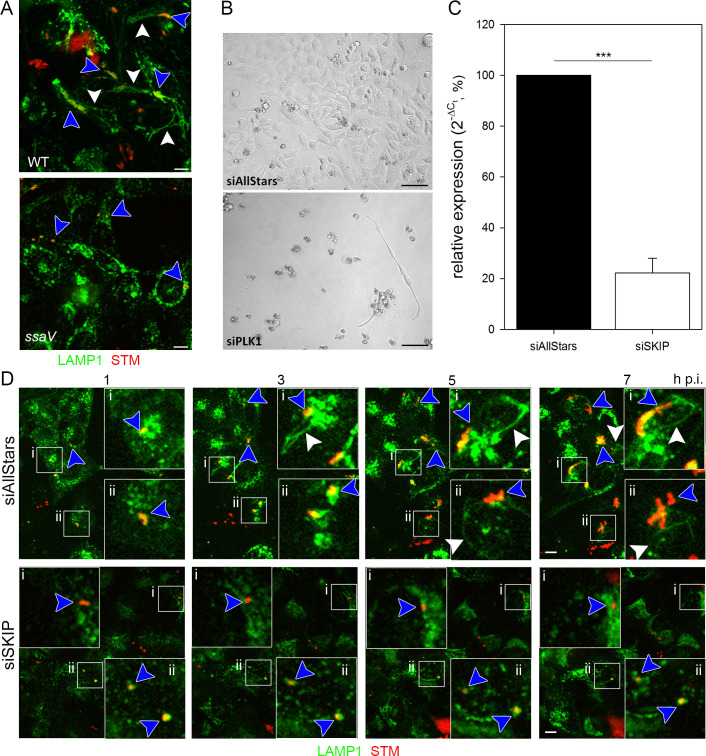
As controls for STM-induced phenotypes, we used STM wild type (WT), capable in SIF induction, and an isogenic strain defective in SsaV, a central component of the SPI2-T3SS, and thus unable to induce SIF formation (Fig 1A). As a control for successful reverse transfection in general, we analyzed the lethal effect of an siRNA directed against polo-like kinase 1 (PLK1), a cell cycle control protein. The knockdown of this protein leads initially to a cell cycle arrest, and ultimately to cell death, as shown in Fig 1B. Besides, a phenotype-related control was established, i.e. a knockdown of a host factor already known to be essential for SIF formation. A host factor directly involved in SIF formation is SKIP [22]. This study already successfully used SKIP silencing, thus we used an siRNA with the same sequence as control. Real-time PCR indicated that the siRNA targeting SKIP yielded not a complete but significant knockdown with a reduction to ca. 22% (Fig 1C). Transfection with AllStars siRNA did not affect SIF formation and dynamics throughout the infection (Fig 1D, S1 Movie), while a SKIP knockdown abolished SIF formation (Fig 1D, S2 Movie) and reduced intracellular replication of STM. Though this outcome does not completely exclude off-target effects, the phenotypic/visual control showed at least the intended purpose of the SKIP siRNA being fulfilled. The partial knockdown explains the rare appearance of SIF. Taken together, the establishment of the proper controls allowed us the execution of a larger scale RNAi screen.
Fig 1. RNAi screen setup and validation.
A) Intracellular phenotypes of STM under screening conditions. HeLa LAMP1-GFP cells were infected with mCherry-labelled STM WT or ssaV strains and imaged live 8 h p.i. by SDCM. Images display the presence of STM in LAMP1-positive SCV (blue arrowhead), the induction of SIF formation by STM WT (white arrowhead), and the lack of SIF formation by the STM ssaV strain. Scale bar, 10 μm. B) Controls for siRNA-mediated knockdown. HeLa LAMP1-GFP cells were reverse transfected with scrambled AllStars siRNA or PLK1 siRNA for 72 h and then imaged. Scale bar, 20 μm. C) Validation of SKIP siRNA knockdown. HeLa LAMP1-GFP cells were reverse transfected with AllStars or SKIP siRNA for 72 h. Then, RT-PCR targeting SKIP was performed. Depicted is the mean with standard deviation of three biological replicates (n = 3) each performed in triplicates. Statistical analysis was performed using Student’s t-test and indicated as: ***, p < 0.001. D) SKIP knockdown as a control for the inhibition of SIF formation. HeLa LAMP1-GFP cells were first reverse transfected with AllStars or SKIP siRNA for 72 h. Then, cells were infected with mCherry-labelled STM WT (MOI = 15) and imaged live by SDCM 1–7 h p.i in hourly intervals. Blue arrowheads indicate SIF-forming or non-SIF-forming single bacteria or microcolonies, white arrowheads indicate SIF. Scale bar, 10 μm.
The RNAi trafficome screen
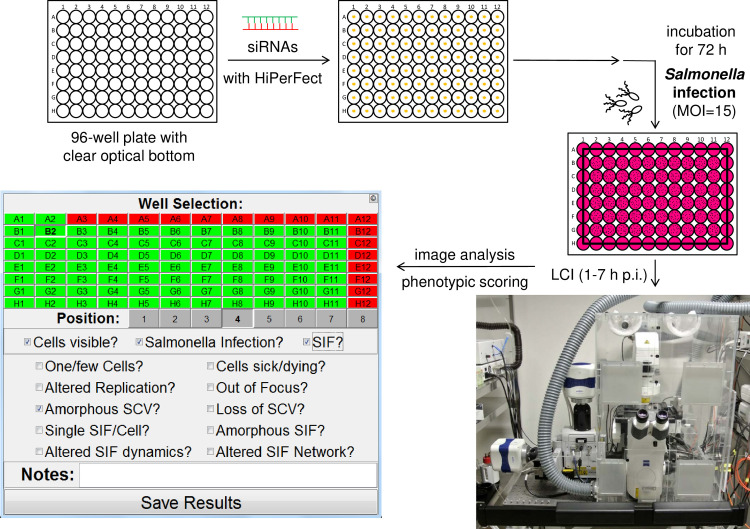
The complete workflow of the RNAi screen executed is summarized in Fig 2. First, siRNAs (with three distinct siRNAs per target) were automatically spotted onto 24 96-well plates per biological replicate (with the total screen being performed in three biological replicates). Additionally, each plate contained siAllStars, siPLK1, and siSKIP as negative and positive siRNA controls, and as a phenotype-specific control, respectively. HeLa LAMP1-GFP cells were seeded onto siRNAs for reverse transfection, incubated for 72 h, and subsequently infected with mCherry-labelled STM WT or ssaV. The formation of SIF was followed by LCI acquiring eight positions per well with single bacteria-focused Z-planes with an SDCM from 1–7 h post infection (p.i.) with hourly intervals.
Fig 2. Basic workflow of the trafficome RNAi screen.
24 96-well plates with clear optical bottoms per biological replicate (with 3 biological replicates in total, n = 3) were automatically spotted with siRNAs. HeLa LAMP1-GFP cells were seeded for reverse transfection with each plate also containing negative, positive, and phenotype-specific controls. After 72 h of incubation, infection with STM WT (and ssaV as control, MOI = 15) was performed, followed by LCI for the acquisition of eight positions per well with single bacteria-focused Z-planes with hourly intervals of imaging from 1–7 h p.i. on an SDCM system. Subsequent phenotypic scoring was performed using the SifScreen utility. The MATLAB-based data input mask allows the entry of well- and position-specific information on general cell behavior and Salmonella/SMM phenotypes and the generation of a results report.
We set out to execute the analysis by visual inspection following the example of Stein et al. [20], who performed a mutant library screen to identify bacterial factors involved in SIF formation. As the dynamic nature of SIF and the phenotypic heterogeneity in the cellular context excluded fully automated analysis, we decided to perform the analysis by visual inspections and used a MATLAB-based tool named SifScreen to support data input and collection (Fig 2). This tool queried the presence of SIF in the examined field of view as the main feature in a binary manner (for detailed information see S1 Text). The scoring was always performed by analyzing the complete time-lapse movies for each position allowing to identify SIF formation due to its dynamic nature more easily.
Since siRNA silencing usually does not yield 100% loss of function, we did not expect a complete lack of SIF in each of the eight images per well. Furthermore, since a considerable number of cells were present per image (roughly 10–30 cells, depending on applied siRNA and position on plate) a single SIF-forming cell would have prompted a SIF-positive scoring, even if generally a SIF-negative knockdown might have occurred. Thus, we decided to define an overall SIF-abolishing hit with a comparably high cutoff of 50%, i.e. if less than 50% of the images showed SIF. However, this cutoff did not take into consideration knockdowns possibly affecting cell viability in general or other circumstances compromising the analysis. Because parameters such as host cell viability were also queried by SifScreen, this assessment allowed us to differentiate between ‘true hits’ and ‘possible hits,’ scoring the former and latter with values of 3 and 1, respectively. This is important as other influences, such as a decreased invasion, could affect the outcome of the scoring, even though they might not have been readily detectable. Additionally, to avoid a possible bias due to visual analysis, each screening plate was analyzed independently by two investigators. With the screen performed in biological triplicates (n = 3), we subsequently compiled all scoring data for each host target from both investigators, also pooling the results of the three individual siRNAs per target. This summary resulted in a list of final hits shown in S2 Table, in which hits with a cumulative scoring of 1–4, 5–7, or ≥8 were classified as low-, mid- and high-ranking hits, respectively. Examples of time-lapse acquisitions of selected siRNAs are given in S1 Fig.
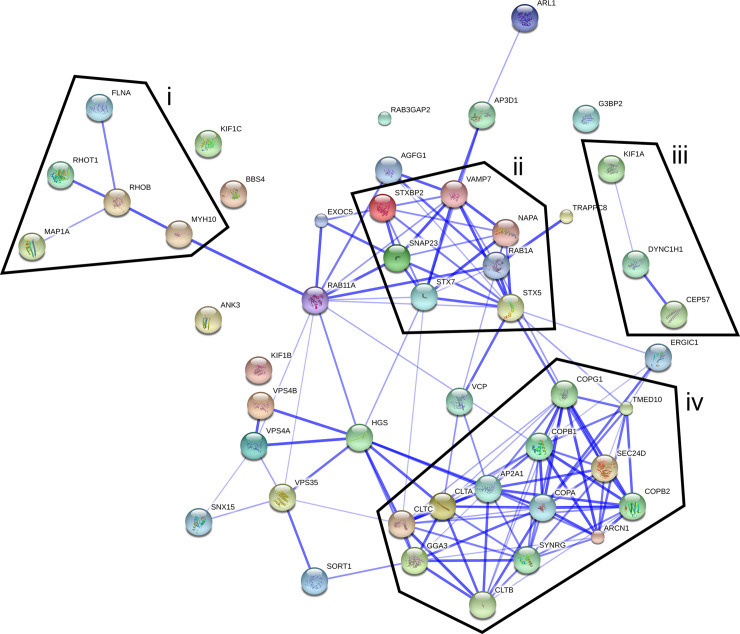
Approximately 81% (404 of 496) of the trafficome targets scored to varying degrees positive, underlining the general importance of trafficking processes for SIF formation. Table 1 shows selected high-ranking hits involved in trafficking and cytoskeleton biology. These hits clearly show the involvement of all protein classes necessary for the vesicle budding and fusion machinery, the core of cellular trafficking. These comprise: (i) small GTPases, especially Rab GTPases, as primary regulators [36–38]; (ii) vesicle coats and their adaptors as cargo and budding mediators [39–43]; (iii) cytoskeleton components as the basis for vesicle motility [44]; (iv) tethering factors as part of the fusion specification [45, 46]; (v) SNAREs as the primary fusion agents [45, 47, 48]. Besides, this list includes hits of diverse subcellular origin, encompassing the complete secretory and endo-/lysosomal system, i.e. endoplasmic reticulum (ER), Golgi apparatus, endo-/lysosomes. Supporting these allocations, the interaction network of the hits from Table 1 shows several distinct clusters (Fig 3). Two of the clusters are connected to cytoskeleton biology (also interconnected if lower-ranking hits are included). Another cluster is SNARE-centered, including RAB1A, with RAB11A as a node between this cluster and one of the cytoskeleton-related clusters. Lastly, one cluster is associated with COPI and clathrin-coated vesicles (CCVs). Collectively, the overall results of the trafficome screen confirm the general importance of host trafficking factors in SIF biogenesis, indicating a crucial role for a plethora of as yet unprecedented factors in STM pathobiology.
Table 1. High-ranking trafficome hits (scoring cutoff of ≥8; see main text for scoring details) involved in trafficking and cytoskeleton biology (see S1 Table for more details on individual host factors).
| Gene symbol | Full name1 | Localization2 |
|---|---|---|
| Small GTPases and interacting proteins | ||
| Arf family | ||
| ARL1 | ADP-ribosylation factor-like GTPase 1 | Golgi |
| Rab family | ||
| RAB1A | RAB1A, member RAS oncogene family | ER, Golgi, EE |
| RAB11A | RAB11A, member RAS oncogene family | RE, vesicle, PM |
| Ras family | ||
| RHOB | ras homolog family member B | nucleus, LE, PM |
| RHOT1 | ras homolog family member T1 | mitochondrion OM |
| Interacting proteins | ||
| G3BP2 | G3BP stress granule assembly factor 2 | cytoplasm |
| RAB3GAP2 | RAB3 GTPase activating non-catalytic protein subunit 2 | cytoplasm |
| Vesicle coats and adaptors | ||
| BBSome | ||
| BBS4 | Bardet-Biedl syndrome 4 | CS/MTOC |
| Clathrin coats | ||
| AGFG1 | ArfGAP with FG repeats 1 | nucleus, vesicle |
| AP2A1 | adaptor related protein complex 2 subunit alpha 1 | CCV, PM |
| AP3D1 | adaptor-related protein complex 3 subunit delta 1 | CCV, Golgi |
| CLTA/B | clathrin light chain A and B | CCV |
| CLTC | clathrin heavy chain | CCV |
| GGA3 | Golgi-associated, gamma adaptin ear containing, ARF binding protein 3 | Golgi, endosome |
| SYNRG | synergin gamma | Golgi |
| COP-I | ||
| ARCN1 | archain 1 | Golgi, vesicle |
| COPA/B1/B2/G1 | COPI coat complex subunit alpha, beta 1, beta 2, and gamma | Golgi, vesicle |
| TMED10 | transmembrane p24 trafficking protein 10 | ER, Golgi, vesicle |
| COP-II | ||
| SEC24D | SEC24 homolog D, COPII coat complex component | ER, Golgi, vesicle |
| Retromer | ||
| VPS35 | VPS35 retromer complex component | EE, LE |
| Tethering factors | ||
| Exocyst complex | ||
| EXOC5 | exocyst complex component 5 | cytoplasm |
| TRAPPIII complex | ||
| TRAPPC8 | trafficking protein particle complex 8 | Golgi |
| SNAREs | ||
| Qa-SNAREs | ||
| STX5 | syntaxin 5 | Golgi |
| STX7 | syntaxin 7 | EE, LE |
| Qb,c-SNAREs | ||
| SNAP23 | synaptosome associated protein 23 | PM |
| R-SNAREs | ||
| SEC22B | SEC22 homolog B, vesicle trafficking protein (gene/pseudogene) | ER, Golgi |
| VAMP7 | vesicle associated membrane protein 7 | ER, Golgi, LE/ lysosome, vesicle, PM |
| Interacting proteins | ||
| NAPA | NSF attachment protein alpha | membrane |
| STXBP2 | syntaxin binding protein 2 | PM |
| Cytoskeleton and motor proteins | ||
| Kinesins | ||
| KIF1A/B/C | kinesin family member 1A, B, and C | CS |
| Dyneins | ||
| DYNC1H1 | dynein cytoplasmic 1 heavy chain 1 | CS |
| Microtubule-associated proteins | ||
| CEP57 | centrosomal protein 57 | CS/MTOC, nucleus |
| MAP1A | microtubule associated protein 1A | CS |
| Myosins | ||
| MYH10 | myosin heavy chain 10 | CS |
| Actin filament membrane linkers | ||
| ANK3 | ankyrin 3 | CS |
| FLNA | filamin A | CS |
| ESCRT complexes | ||
| Adaptors | ||
| HGS | hepatocyte growth factor-regulated tyrosine kinase substrate | EE, MVB |
| AAA ATPase | ||
| VPS4A/B | vacuolar protein sorting 4 homolog A and B | MVB |
| Miscellaneous | ||
| ERGIC1 | endoplasmic reticulum-golgi intermediate compartment 1 | ER, Golgi |
| SNX15 | sorting nexin 15 | vesicle |
| SORT1 | sortilin 1 | nucleus, ER, Golgi, endo-/lysosome |
| VCP | valosin containing protein | nucleus, ER |
1 according to NCBI Gene
2 subcellular localization according to UniProt; CCV = clathrin-coated vesicle, CS = cytoskeleton, EE = early endosome, ER = endoplasmic reticulum, LE = late endosome, MTOC = microtubule-organizing center, MVB = multivesicular body, OM = outer membrane, PM = plasma membrane, RE = recycling endosome
Fig 3. Interaction network of selected trafficome hits.
The interaction of selected high-ranking hits (scoring cutoff of ≥8, see also Table 1) was visualized using the STRING database (confidence view). Borders delineate clusters related to the cytoskeleton (i, iii), SNAREs (ii), or COPI and clathrin-coated vesicles (iv).
Validation of selected hits
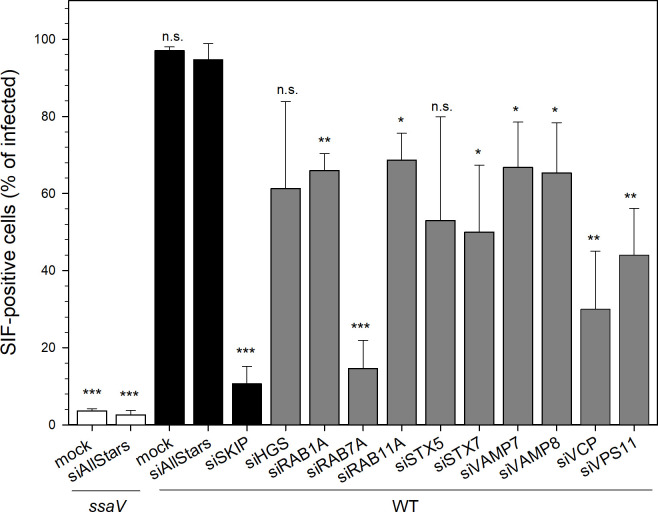
To test the validity of our approach and the resulting hits, we focused on a subset of genes due to their presence in the noticeable interaction clusters depicted in Fig 3, or prior reports on involvement in STM pathobiology. HGS is part of the ‘endosomal sorting complex required for transport’ (ESCRT) complex ESCRT-0. Interaction of SPI1-T3SS effector SopB with HGS was previously reported [49], and HGS was one of the highest-ranking hits (S2 Table). Furthermore, RAB1A and RAB11A were included as highest-ranking Rab GTPases, with RAB11A previously being shown to colocalize with SCV as well as SIF [32, 50]. Consistently, RAB7A served as another well-established SCV- and SIF-localizing control [12, 50–52]. STX5, STX7, VAMP7, and VAMP8 were chosen due to being the highest-ranking SNAREs (except VAMP8 lacking from the trafficome), the previously shown colocalization of STX7 with SIF [50], and the recently reported essential role of VAMP7 in SIF biogenesis [33]. The AAA+ (ATPases associated with diverse cellular activities) protein VCP was included as another of the highest-ranking trafficking-related hits and another host factor already known to be important for proper SCV and SIF biogenesis via the STM effector SptP [53]. Finally, the VPS11 core component of the class C core vacuole/endosome tethering (CORVET) / homotypic fusion and protein sorting (HOPS) group of multisubunit tethering complexes (MTCs) was chosen due to the HOPS complex functionally bridging RAB7 with late endo-/lysosomal SNAREs and the recent recognition of its essential role in STM replication and SCV and SIF biogenesis [54, 55].
The success of the silencing with individually ordered siRNAs for subsequent SIF quantification was first confirmed by RT-PCR with a consistently significant decrease in mRNA in most cases down to 5–10% compared to control siAllStars (S2A Fig, siSKIP served as the screen-inherent phenotype-specific control). As an mRNA reduction does not necessarily affect the actual protein levels due to potential long protein half-lives, we additionally tested the presence of selected targets, i.e. SKIP, RAB7A, and VPS11, by Western blot analysis (S2B and S2C Fig). We observed reduced protein levels for all targets, though the decrease for SKIP and VPS11 was moderate with 82.0% and 69.9%, respectively, compared to siAllstars-treated cells, whereas amounts of RAB7A decreased to 13.8%. Next, we determined the effect of silencing of the selected targets on SIF formation (Fig 4). The ssaV mutant strain and the knockdown with siSKIP served as screen-inherent SIF-abolishing controls. All knockdowns resulted in decreased SIF formation with the reduction being statistically significant except for siHGS and siSTX5. Regarding SKIP and VPS11, this decline also exhibits that moderate silencing on the protein level suffices to exert a biological effect. The siRAB7A had the highest impact that corresponds with the strong reduction of the RAB7A protein level, siVCP was second-highest, and the others ranged similar. Thus, the knockdowns of VAMP7 and VCP meet previous data (even though SIF abolishment regarding VCP depletion here is less pronounced) [33, 53]. Altogether, the effect of silencing various host targets on SIF formation demonstrates that our approach confirms known host factors, but also allows the identification of novel factors to be crucial for SIF biology.
Fig 4. Influence of host factor silencing on SIF formation.
HeLa LAMP1-GFP cells were not transfected (mock), or reverse transfected with siAllStars or the indicated siRNA, infected with STM WT or SPI2-deficient ssaV expressing mCherry as indicated, and SIF-positive infected cells were counted. Depicted are means with standard deviation for three biological replicates (n = 3). Statistical analysis was performed against siAllstars + WT with Student’s t-test and indicated as: n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
STM deploys membranes of early and late secretory, late endo-/lysosomal, and clathrin-coated origin in SIF biogenesis
The fact that host factors appear as hits in our screen clearly indicates a physiologically relevant role in SIF biogenesis. However, whether this role is by direct interaction or an indirect one involving several intermittent steps, remains unclear. Thus, we decided to analyze the localization of selected hits with regard to SIF (Fig 5, Fig 6, Fig 7). Even though mere colocalization in light microscopy is no ultimate proof of direct interaction, it is a first approximation as it potentially allows such a possibility.
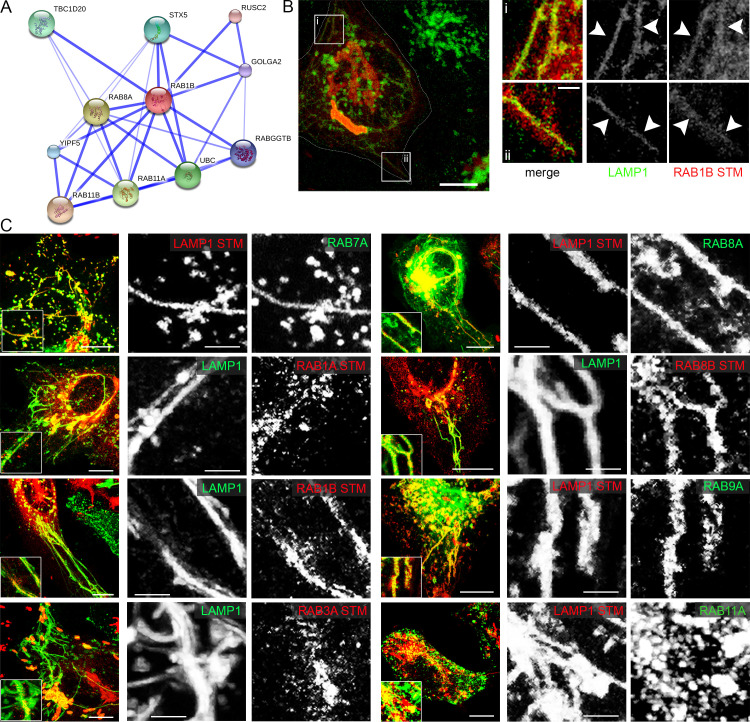
Fig 5. RAB proteins identified by the trafficome screen colocalize with SIF and SCV.
A) Direct interaction network of RAB1B as visualized by STRING. B) and C) HeLa cells either stably transfected with LAMP1-GFP (green) or transiently transfected with LAMP1-mCherry (red) were co-transfected with plasmids encoding various RAB GTPases (RAB7A, RAB1A, RAB1B, RAB3A, RAB8A, RAB8B, RAB9A, RAB11A) fused to GFP (green) or mRuby2 (red) and then infected with STM WT expressing mCherry or GFP. Living cells were imaged from 6–9 h p.i. by CLSM and images are shown as maximum intensity projections (MIP). Insets magnify structures of interest and white arrowheads indicate colocalization with SIF. Scale bars, 10 μm (overviews), 1 μm (details).
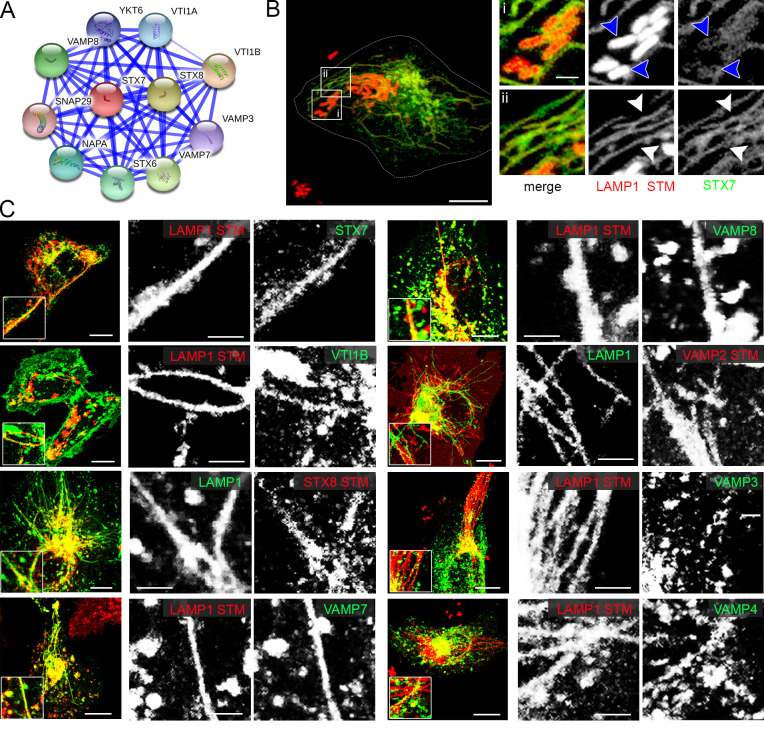
Fig 6. SNARE proteins identified by the trafficome screen colocalize with SIF and SCV.
A) Direct interaction network of STX7 as visualized by STRING. B) and C) HeLa cells either stably transfected with LAMP1-GFP (green), or transiently transfected with LAMP1-mCherry (red) were co-transfected with plasmids encoding various SNAREs (STX7, VTIB, STX8, VAMP7, VAMP8, VAMP2, VAMP3, VAMP4) fused to GFP (green) or mRuby2 (red). Infection and imaging were performed as for Fig 5. Insets magnify structures of interest and white and blue arrowheads indicate colocalization with SIF and SCV, respectively. Scale bars, 10 μm (overviews), 1 μm (details).
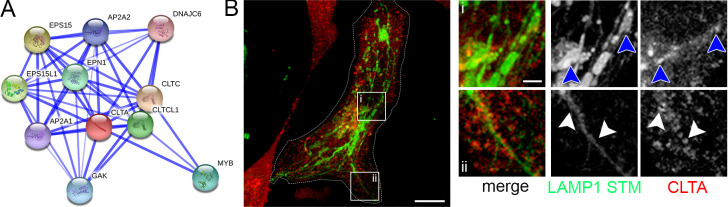
Fig 7. CLTA identified by the trafficome screen colocalizes with SIF and SCV.
A) Direct interaction network of CLTA as visualized by STRING. B) HeLa cells stably transfected with LAMP1-GFP (green) were co-transfected with a plasmid encoding CLTA fused to mRuby2 (red). Infection and imaging were performed as for Fig 5. Insets magnify structures of interest and white and blue arrowheads indicate colocalization with SIF and SCV, respectively. Scale bars, 10 μm (overviews), 1 μm (details).
For analyses of RAB GTPases (Fig 5), we again used RAB7A and RAB9A as positive controls, both showing a clear colocalization with SIF. Of the several Rab GTPases included in the trafficome RAB1A showed the highest score (S2 Table). RAB1 GTPases are responsible for anterograde ER-Golgi trafficking [56–59]. Importantly, RAB1A can be functionally substituted by RAB1B [60, 61] and an STM replication-targeted RNAi screen identified specifically RAB1B as a hit [31]. Hence, we analyzed the infection-related localization of both, RAB1A and RAB1B, and detected a partial and a strong colocalization of RAB1A and RAB1B, respectively, with SIF (Fig 5).
Another high-ranking hit with relation to RAB proteins was RAB3GAP2 (S2 Table), the non-catalytic subunit of the RAB3 inactivating GTPase-activating protein (GAP) complex [62]. RAB3 possesses four isoforms in mammals [63] and is involved in regulated exocytosis [64]. As neither the catalytic GAP subunit, RAB3GAP1, nor one of the four isoforms of RAB3 were present in the trafficome, we decided to analyze the localization of RAB3A and found a partial colocalization with SIF (Fig 5).
Besides, a mid-ranking hit was RAB8A (S2 Table), a Golgi- and endosome-localized RAB likewise involved in exocytic processes [65]. Interestingly, RAB8A isoform RAB8B was observed to be excluded from maturing SCVs (≤ 3 h p.i.) [50]. Therefore, we analyzed the localization of both, RAB8A and RAB8B, and strikingly found a strong colocalization of not only RAB8A but also RAB8B with SIF (Fig 5).
As several RAB proteins participating in the late secretory system/exocytosis seem to play a role in SIF biogenesis, we additionally analyzed three SNAREs with exocytic roles not present in the trafficome: VAMP2, VAMP3, and VAMP4 [66–68], with VAMP2 also shown to be present on early SCV [69]. Apart from that, the presence of the two high-ranking SNARE hits STX7 and VAMP7 (S2 Table) on SIF was previously shown [33, 50]. However, SNAREs, are part of complexes of usually four proteins participating in membrane fusion and consisting of a single v-SNARE (on the vesicle or incoming membrane) and a ternary t-SNARE subcomplex (on the target or accepting membrane). VAMP7 is the v-SNARE in the SNARE complex for heterotypic LE/lysosome fusions with the t-SNAREs STX7, STX8, and VTI1B [70, 71] being replaced by VAMP8 in homotypic LE fusions [72, 73]. In fact, the presence of VTI1B and STX8 on early SCV [69, 74] and their role in STM replication [54], as well as the involvement of VAMP8 in STM invasion were already shown [75]. However, the interaction of VTI1B, STX8, and VAMP8 with SIF remains unclear, except VAMP8 silencing causing SIF reduction identified here (Fig 4). Thus, we analyzed the localization of VTI1B, STX8, and VAMP8 using STX7 and VAMP7 as controls. As shown in Fig 6, we detected a prominent association of STX7 and VAMP7 with SIF, and of VAMP2 and VAMP8. Colocalization of VTIB, STX8, VAMP3, and VAMP4 with SIF was also observed, however, these SNARE subunits showed a more heterogeneous distribution and only a fraction of SIF was positive for these candidates.
Notably, both recent proteomic studies [32, 33] identified AP2A1 as being present on SMM/SCV, and this screen determined AP2A1 as high-ranking hit of host factors involved in endosomal remodeling (S2 Table). AP2A1 represents one of the two core subunit isoforms of the canonical AP-2 adaptor complex usually acting in clathrin-mediated endocytosis (CME) at the plasma membrane [76, 77]. With both clathrin light chains CLTA and CLTB, and the conventional heavy chain CLTC, the main coat components of CCV formation were among the high-scoring hits (S2 Table). We analyzed the localization of CLTA and observed a partial colocalization with SIF (Fig 7).
In conclusion, the colocalization of various host factors involved in cellular transport with SIF validated the results of the RNAi screen. These proteins are components of SIF tubules and, to a variable extent, required for the formation of SIF.
Discussion
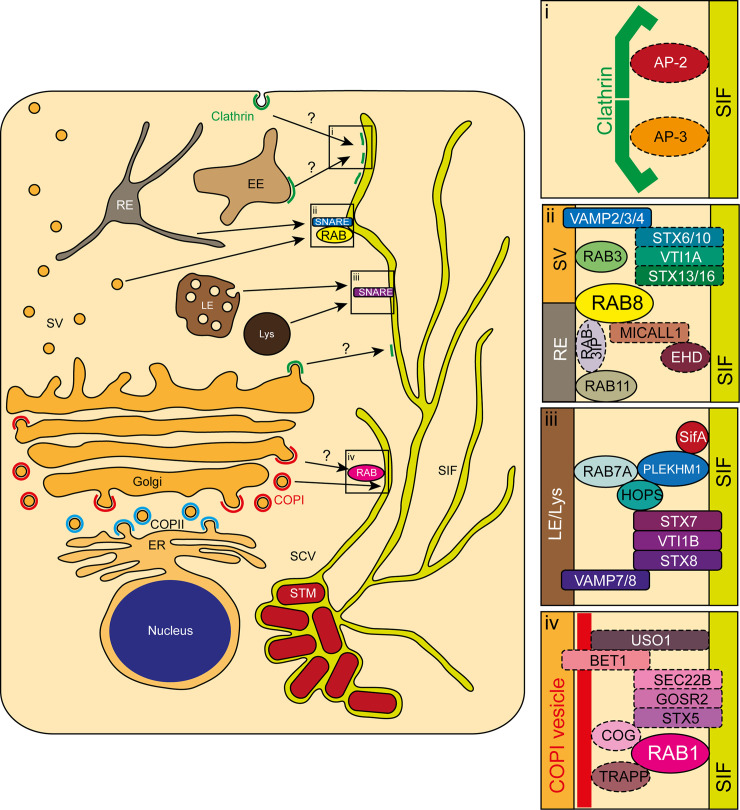
By applying a targeted RNAi screen, we identified several new host factors required for the formation of SIF and partially characterized interactions of host proteins with SMM. Our data strengthen the involvement of the late endo-/lysosomal SNARE complex, and reveal new interactions of SIF with RAB1, RAB3, and RAB8 GTPases, exocytic SNAREs, and clathrin-coated structures. The implications of these findings as discussed below are depicted in Fig 8. Several trafficome targets identified here as high-ranking hits besides those mentioned above were previously shown to be not only involved in infection biology in STM in general, but specifically in SCV and/or SIF biogenesis including: dynein–DYNC1H1 [78–80], filamin–FLNA [81], myosin II–MYH10 [82], VPS4A/B [49]. This also holds true for several mid-ranking hits: kinesin-1 –KIF5A/B [22–25, 83], PIKFYVE [84], RAB9A [85, 86], RAB14 [85], SCAMP3 [87].
Fig 8. Newly identified interactions of intracellular STM with host factors.
Depicted are central eukaryotic endomembrane organelles possibly playing a role in the newly identified interplays of host factors with SIF. Magnifications show the interactions of clathrin (i), late secretory and/or recycling-related RAB3A, RAB8A/B, and VAMP2/3/4 (ii), late endo-/lysosomal VTI1B, STX8, and VAMP8 (iii), and early secretory RAB1A/B (iv) with other host factors added as discussed in the text. Solid lines represent interactions identified here or otherwise known, dashed lines represent putative interactions. COP, coat protein complex; EE, early endosome; ER, endoplasmic reticulum; LE, late endosome; Lys, lysosome; RE, recycling endosome; SCV, Salmonella-containing vacuole; SIF, Salmonella-induced filaments; STM, S. Typhimurium; SV, secretory vesicle.
Data complementary to our screen were recently provided by two proteomic studies. Our group analyzed the SMM proteome in the late phase of infection (8 h p.i.) [32] that contained several host proteins that are mid- or high-ranking hits in this screen (summarized in Table 2, first column). The colocalization of several of these proteins with SMM was shown by immunostaining or LCI in that study. Santos et al. [33] determined the proteomes of early and maturing SCV (30 min p.i. and 3 h p.i., respectively) again identifying proteins appearing as hits in this screen (see Table 2, second and third column). Taken together, these data strongly validate the approach deployed here.
Table 2. Host proteins (gene symbols) identified as hits in the trafficome screen here that are also part of at least one distinct SMM proteome as identified in other studies (see S1 Table for more details on individual host factors).
| 8 h p.i. SMM1 | 30 min p.i. SCV2 | 3 h p.i. SCV2 |
|---|---|---|
| AP2A1 (AP-2) | - | AP2A1 |
| - | - | BET1 (SNARE) |
| - | CLTC (clathrin) | - |
| COPA/G1 (COPI)3 | - | - |
| DYNC1H1 (dynein) | DYNC1H1 | - |
| - | ERGIC1 | ERGIC1 |
| - | ERP29 | - |
| - | - | EXOC5 |
| FLNA (filamin) | FLNA | FLNA |
| G3BP2 | - | G3BP2 |
| - | IQGAP1 | IQGAP1 |
| - | KIF5B (kinesin-1) | KIF5B |
| - | MAP1B | - |
| MYH9/10 (myosin II) | MYH9 | MYH9 |
| NAPA/α-SNAP | - | - |
| RAB2A3 | RAB2A | - |
| - | RAB4A | - |
| RAB7A3 | RAB7A | - |
| RAB11A3 | - | - |
| RAB143 | - | - |
| - | SEC22B (SNARE) | - |
| - | SEC24C (COPII) | - |
| TMED10 (COPI) | - | - |
| VCP | - | - |
The approach reported here has a major advantage compared to studies based on organelle proteomics [32, 33]. Proteomic analyses lead to the identification of the presence or absence of host factors on the organelle of interest, but a particular role in the biogenesis of this organelle cannot be implied directly [32, 33]. In our RNAi approach potentially each, or at least each high-ranking hit, points to a role in STM-induced endosomal remodeling. However, the functional role revealed by RNAi does not necessarily depend on localization of the host factor at SIF and/or SCV. The effect on endosomal remodeling may be mediated indirectly, involving several interacting partners. We analyzed the localization of selected host factors (Fig 5, Fig 6, Fig 7) and found several differences in the host factor sets identified by proteomics or by our approach. Nevertheless, there is a considerable overlap of host factors identified by both approaches, proteomics and RNAi, as represented in Table 2.
The presence on SIF and/or importance for SIF formation of RAB7, the HOPS complex, STX7, and VAMP7, as well as the direct fusion of late endo-/lysosomal-like VAMP7-positive vesicles with the SCV, was shown before [33, 50, 54]. These interactions indicate the involvement of the complete canonical mammalian late endo-/lysosomal vesicle fusion machinery in SIF biogenesis. Whether this interaction cascade also employs the canonical STX7, VTI1B, and STX8 was not fully clarified. Here, we expand this cascade by showing the physiological relevance of STX7 for SIF formation (Fig 4), and the presence of VTI1B and STX8 on SIF (Fig 6) as depicted in Fig 8iii. This cascade is possibly expanded by the host protein PLEKHM1, as the recruitment of RAB7 and the HOPS complex by SifA via the host protein PLEKHM1 and its involvement in SCV biogenesis was recently revealed [55], most likely also being involved in SIF biogenesis. Taken together, SifA seems to recruit the complete late endo-/lysosomal fusion machinery. Thus, SifA performs a dual role besides the binding of SKIP and the SIF mobility connected with it. This is also corroborated by the identification of interactions of SifA with STX7 and VAMP7 by a recent BioID screen [88]. Alternatively or in addition, SopD2 might be likewise involved as it was also shown to interact with STX7 and VAMP7 besides VTI1B in the same study.
The interaction of STM with the early secretory system is poorly characterized. In fact, the involvement of early secretory host factors, e.g. RAB2A, in SMM biology was only recently described by proteomic studies [32, 33]. We now expand this interaction by showing the physiological relevance of RAB1A in SIF formation (Fig 4) and presence of both RAB1A and RAB1B on SIF (Fig 5). The direct association of RAB1A/B with SIF possibly connects several distinct trafficking events. First, RAB1B was shown to be involved in formation of the COPI vesicle coat, which participates in intra-Golgi and retrograde Golgi-to-ER transport [89–91]. Second, the COPI components COPA and COPG1 were shown to partly colocalize with SIF [32]. In accordance, our screen identified the majority of COPI components as mid- or high-ranking hits (ARCN1, COPA/B1/B2/G1, Table 1 and Fig 3). Thus, RAB1A and/or RAB1B might represent a physical link between COPI vesicles and SCV and/or SIF for the redirection of early secretory material as depicted in Fig 8iv.
The physical interaction of SIF with COPI vesicles might be, similar to the late endo-/lysosomal fusion machinery, additionally accompanied by tethering factors and SNAREs. The conserved oligomeric Golgi (COG) tethering complex was shown to be a RAB1 effector and directly bind COPI components [92, 93]. Interestingly, all components of the COG present in the trafficome scored mid- to high-ranking (COG1/2/3/5/7, S2 Table). Additionally, COG binds STX5, a SNARE that is part of several ER-Golgi and intra-Golgi transport-related SNARE complexes [94–97]. Srikingly, only the components of one distinct SNARE complex, comprising STX5, GOSR2/GS27/membrin, BET1, and SEC22B, scored all mid- to high-ranking (Table 1 and S2 Table). STM effectors partaking in this interaction might by SseF and SseG, as they were recently shown in the recent BioID screen [88] to interact with STX5 and SEC22B, besides PipB2 also interacting with SEC22B. However, whether the potential involvements indicated by these collective data expand the potential RAB1/COPI interaction cascade described above remain to be elucidated.
Another tethering factor, the transport protein particle (TRAPP) complexes were identified as RAB1 guanine exchange factors (GEFs) [98–101] and COPI tethers [TRAPPII; 100, 102]. TRAPPI is the core shared by all TRAPP complexes, with II and III possessing unique additional subunits. TRAPPC8, the unique component of TRAPPIII, scored high-ranking. Other components were not present in this trafficome, except the TRAPP core subunit TRAPPC2, which unexpectedly did not score at all (S2 Table). So far TRAPPIII is only characterized to participate in autophagy [101, 103, 104]. Besides, the tethering golgin USO1/p115 scored mid-ranking (S2 Table). USO1 is also a RAB1B effector and COPI tether [91, 105], executing these roles partly in conjunction with COG [106]. Additionally, USO1 is able to bind STX5 [107]. As for the COG complex and ER/Golgi SNAREs, for both, the TRAPP complexes and USO1, a specific role in SIF biogenesis remains to be elucidated.
It has already been described that STM, depending on SseF/SseG, recruits exocytic vesicles from the Golgi apparatus destined to the plasma membrane to the SCV [108]. Which host factors are involved in this process was unclear, and our work now sheds light on this phenotype by showing the presence of exocytic RABs, i.e. RAB3A, RAB8A, and RAB8B (Fig 5), and exocytic SNAREs, i.e. VAMP2, VAMP3, and VAMP4 (Fig 6), on SIF.
Besides their involvement in exocytosis, VAMP4 and VAMP3 are also known to prominently participate in endosome-to-Golgi transport in conjunction with STX16, VTI1A, and STX6 or STX10 for EEs or LEs, respectively [109, 110].This might represent another interaction cascade of SIF as VTI1A was a mid-ranking hit (although STX6 and STX10 were not included in the trafficome and STX16 ranked low, S2 Table). Moreover, the STM-mediated redirection of LAMP1-containing vesicles from the Golgi apparatus to the early SCV was shown to involve recruitment of STX6 and VAMP2 via SPI1-T3SS effector SipC [69]. Alternatively, this might happen via the SPI2-T3SS effector PipB2 that was identified in the recent BioID screen as an interactor of VAMP2 [88]. Furthermore, in homotypic EE fusion STX16 is replaced by STX13 [111], and STX13 was previously shown to be present on early SCV [74, 112]. While the exact role of RAB3A and the identity of SNAREs involved in the processes described above remain to be determined, this might indicate that the interception of secretory vesicles by SMM depends on a SNARE complex comprising a distinct combination of the abovementioned SNAREs as represented in Fig 8ii.
In addition to its exocytic role, the mid-ranking hit RAB8A is involved in recycling processes as indicated by the localization on tubular recycling endosomes (RE) [113]. This localization depends on several factors such as the RAB8 GEF RAB3IP/RABIN8 (which is also part of the trafficome, though it ranked low, S2 Table), concurrently being an effector of the RE master regulator RAB11 [114]. Another factor is the RE-localized MICALL1, which interacts with the dynamin-like ATPases EHD1 and EHD3 [113, 115, 116]. Interestingly, MICALL1 was identified in an RNAi screen with focus on STM intracellular replication [31], EHD1 and EHD2 were present in the proteome of maturing SCV [33], and EHD4 was present in late SMM [32]. Moreover, the association of the maturing SCV and late SMM with RAB11A/B was shown previously [32, 50], with RAB11A following RAB1A as the second-highest-ranking RAB in our screen (S2 Table). The SPI2-T3SS effector SopD2 most likely plays a role in RAB8 recruitment, as it was previously shown to interact with RAB8 [88, 117]. Collectively, these data strongly argue for a continued association of STM not only with exocytic compartments as described above involving a distinct SNARE complex, but also with recycling compartments with RAB8 isoforms at its center at later time points (summarized in Fig 8ii).
Data on the involvement of clathrin-coated structures or adaptor protein complexes in STM pathobiology are scarce. We now show an association of clathrin via one of its light chains, CLTA, with SMM (Fig 7). It is peculiar that two proteomic studies [32, 33], as well as our screen, indicate an involvement of the AP-2 complex in biogenesis of SMM, while the other adaptor complexes were not identified. The presence of the CME-related AP-2 is remarkable as it is primarily plasma membrane-localized, in contrast to the Golgi traffic-related AP-1 and AP-4, or the endo-/lysosomal traffic-related AP-3 and AP-5 [43]. Especially AP-3 deserves detailed analyses in the future since its two core subunit isoforms scored in mid- and high-ranking range (AP3B1 and AP3D1, S2 Table, see Fig 8i). Several SPI2-T3SS effectors, i.e. PipB2, SopD2, and SseG, might participate in such a recruitment because the recent BioID screen revealed the interaction with various AP-2 and AP-3 core subunits [88]. However, the examination of other AP complexes also seems worthwhile, since the latter study indicates interactions with several of them and the trafficome screen did not comprehensively cover AP complexes.
In summary, we successfully employed a sub-genomic RNAi screen to systematically identify new host factors, corresponding protein complexes, and pathways involved in SIF formation. By providing physiologically relevant data regarding SIF formation, this work further corroborates involvements of host factors with SMM indicated by previous proteomics studies [32, 33]. Similar future screens can also reveal the biogenesis of several other SIT [15], and extend to the host cell types important for Salmonella pathogenesis.
Materials and methods
Bacterial strains and growth conditions
For infection STM NCTC 12023 WT and isogenic SPI2-T3SS-defective strain P2D6 harboring plasmid pFPV-mCherry/2 or isogenic GFP-expressing MvP1897 were used (for details see S3 Table). Strains were routinely grown in Luria-Bertani (LB) broth (Difco, BD, Heidelberg, Germany) containing 50 μg/mL carbenicillin for plasmid selection at 37°C with aeration.
Cell lines and cell culture
Experiments were performed using the parental HeLa cell line (ATCC No. CCL-2) or the lentivirus-transfected HeLa cell line stably expressing LAMP1-GFP [18]. Cells were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L glucose, 4 mM stable glutamine, and sodium pyruvate (Biochrom, Berlin, Germany) supplemented with 10% inactivated fetal calf serum (iFCS; Gibco, Darmstadt, Germany) in an atmosphere of 5% CO2 and 90% humidity at 37°C.
siRNA library and individual siRNAs
The siRNAs used here were part of a human whole-genome library obtained from Qiagen (Hilden, Germany) deposited at the Max Planck Institute (MPI) for Infection Biology (Berlin, Germany). The actual siRNA library is a custom-made library similarly built as others from the MPI [118, 119] and comprised siRNAs targeting 496 host proteins with a threefold coverage, i.e. three individual siRNAs per target. The targets are mostly involved in intracellular trafficking as they were all chosen from GO terms associated with trafficking except the terms ‘autophagy’ and ‘canonical glycolysis’ (see S1 Table for a full list of the parental GO terms). A volume of 4 μL of each siRNA (0.2 μM, end concentration of 5.2 nM) was spotted automatically onto 96-well Clear Bottom Black Cell Culture Microplates (Corning, Corning, NY, USA) and frozen at -20°C before transfer. The 1,488 individual siRNAs were distributed on 24 96-well plates in total per biological replicate with three biological replicates performed (n = 3). Additionally, each plate contained the same amount of the following siRNAs from Qiagen as knockdown controls: AllStars as negative and Hs_PLK1_7 as positive controls. A custom siRNA from Qiagen directed against SKIP served as a phenotype-specific control [22] and was spotted on location. Information including target sequences for these siRNAs and those ordered for validation experiments, are listed in S4 Table.
Reverse transfection with siRNA
If not using 96-well screening plates as detailed above, the amount for an end concentration of 5 nM siRNA was spotted onto standard cell culture 6-well plates (for mRNA extraction or Western blot analyses; TPP, Trasadingen, Switzerland) or 8-well polymer bottom chamber slides (for quantification of SIF formation; μ-Slides, ibidi, Martinsried, Germany).
Next, a mixture of the transfection reagent HiPerFect (Qiagen, Hilden, Germany) and serum-free cell culture medium was applied and this was incubated for 5–10 min at room temperature (RT). Subsequently, 5,000, 125,000, or 20,000 cells per well of 96-well plates, 6-well plates, or 8-well chamber slides, respectively, were added in serum-containing medium and incubated for 72 h at 37°C in a humidified atmosphere containing 5% CO2.
Gene expression quantification
After reverse transfection with siRNAs in 6-well plates, total RNA of cells was extracted using the RNeasy Mini Kit following the manufacturer’s instructions (Qiagen, Hilden, Germany). Homogenization during extraction was performed using Qiagen QIAshredder columns. Then, 1 μg of RNA digested with DNaseI (NEB, Frankfurt a. M., Germany) was used for reverse transcription of mRNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Dreieich, Germany) following the manufacturer’s instructions employing the Oligo(dT)18 primer. For RT-PCR, 1 μL of cDNA was used with the Thermo Scientific Maxima SYBR Green/Fluorescein qPCR Master Mix (2x). As reference gene, the housekeeping gene GAPDH was selected [120]. For control of individual host factor knockdowns, primers were used employing the PrimerBank database [121, 122]. Primers for the host factors analyzed and control GAPDH are listed in S5 Table. Primer concentration was 150 nM each, and primer efficiency was determined for each primer pair. RT-PCR was performed in an iCycler instrument (Bio-Rad, Munich, Germany) in triplicates in 96-well plates. Relative expression was determined using the 2-ΔCt method [84, 123] with GAPDH expression set as 100%. Results were plotted using SigmaPlot 11 (Systat Software, Erkrath, Germany).
Western blot analyses
Whole cell lysates were prepared using a lysis buffer (1% Triton X-100, 5% glycerol in phosphate-buffered saline [PBS] with cOmplete, EDTA-free Protease Inhibitor Cocktail; Roche, Mannheim, Germany) from reversely transfected cells in 6-well plates. The resulting extracts were centrifuged at 1,800 x g and the supernatant was quantified for protein content with the Pierce BCA Protein Assay Kit (Thermo Scientific) using BSA as standard following the manufacturer’s instructions. After precipitation by addition of a five-fold volume of acetone and incubation for 1 h at 4°C, pellets were dried and resuspended in SDS-PAGE loading buffer. Of precipitated whole cell protein, 30 μg was loaded onto 10% (for SKIP and VPS11) or 12% (RAB7A) gels and separated by SDS-PAGE. After electrophoresis, samples were blotted onto a 0.45 μm nitrocellulose membrane using a semi-dry electrophoretic transfer unit (Bio-Rad). Blots were incubated with primary antibodies directed against SKIP (dilution 1:1,000; custom), VPS11 (dilution 1:1,000; sc-100893, Santa Cruz Biotechnology, Dallas, TX, USA), or RAB7A (dilution 1:1,000; #9367/clone D95F2, Cell Signaling Technology, Frankfurt a. M., Germany), or γ-tubulin (dilution 1:1,000; T6557/clone GTU-88, Sigma-Aldrich, Taufkirchen, Germany) as loading control. Secondary antibodies coupled to horseradish peroxidase were chosen according to the donor species of the primary antibodies and diluted 1:10,000. Detection was achieved by an ECL detection kit (Thermo Scientific), and blots were visualised with a ChemiDoc imaging system (Bio-Rad). Densitometric analysis was performed with ImageLab (v4.0, Bio-Rad).
Construction of plasmids
Plasmids used in this study were either obtained from Addgene, kind gifts from various laboratories, or cloned by Gibson Assembly or restriction enzyme digests and are listed in S3 Table. Oligonucleotides for the construction of plasmids encoding host proteins fused to mRuby2 or EGFP are listed in S5 Table. First, N- or C-terminal mRuby2 vectors were cloned. For that, the vectors pEGFP-C1 and pEGFP-N1 were amplified and EGFP was exchanged for a fragment encoding mRuby2. Genes encoding host proteins were amplified from vectors obtained from DNASU (S3 Table) and then inserted into mRuby2 vectors by Gibson Assembly. Plasmids encoding host proteins fused to EGFP were constructed using restriction enzyme digests. The vector pEGFP-C3 was digested with KpnI and XbaI or KpnI and BamHI and the larger fragment was recovered. The inserts were treated the same way and fragments were ligated.
Host cell transfection
For LCI for the localization of host factors, HeLa or HeLa LAMP1-GFP cells were seeded 1 d prior to transfection. About 20,000 or 150,000 cells were seeded in 8-well chamber slides (see above) or 3.5 mm glass bottom dishes (FluoroDish, WPI, Berlin, Germany), respectively. For transfection 0.5 or 2 μg of plasmid DNA in 25 or 200 μL serum-free medium were mixed with 1 or 4 μL of FuGENE HD transfection reagent (DNA to reagent ratio of 1:2; Promega, Mannheim, Germany) and incubated for 10 min at RT. Medium on the cells was changed and the transfection mixture applied. Cells were incubated for at least 18 h before infection with medium change during infection. For a complete list of transfection plasmids, see S3 Table.
Infection experiments
Overnight cultures of STM were diluted 1:31 and grown for additional 3.5 h in LB broth in glass test tubes with agitation in a roller drum at 60 rpm. HeLa cells were infected with STM WT or ssaV serving as control for screening approaches in 96-well plates with a multiplicity of infection (MOI) of 15 (OD600 of subcultures ranged from 3.3–4.2 and 3.0–4.3 for WT or ssaV, respectively), otherwise for colocalization analysis or SIF quantification in 8-well chamber slides or FluoroDishes with an MOI of 75 or 50, respectively. Infection only of 96-well plates was synchronized by centrifugation at 500 x g for 5 min, and in all cases proceeded for 25 min at 37°C in a humidified atmosphere containing 5% CO2. Cells were washed thrice with full medium or PBS for screening or non-screen LCI purposes, respectively, and incubated in full medium containing 100 μg/mL gentamicin for 1 h to eliminate extracellular bacteria. Then medium containing 10 μg/mL gentamicin was applied for the remainder of the experiment.
Live cell imaging
For LCI full medium was replaced by imaging medium consisting of Minimal Essential Medium (MEM) with Earle’s salts, but without NaHCO3, L-glutamine and phenol red (Biochrom, Berlin, Germany) and instead supplemented with 30 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (Sigma-Aldrich), pH 7.4, containing 10 μg/mL gentamicin. Fluorescence imaging for screening purposes was performed using a Zeiss Cell Observer microscope with Yokogawa Spinning Disk Unit CSU-X1 (Carl Zeiss, Göttingen, Germany), Evolve 512 x 512 EMCCD camera (Photometrics, Tucson, AZ, USA), automated PZ-2000 stage (Applied Scientific Instrumentation, Eugene, OR, USA), and infrared-based focus system Definite Focus, operated by Zeiss ZEN 2012 software (blue edition). The microscope was equipped with live cell periphery consisting of a custom-made incubation chamber surrounding the microscope body and connected with “The Cube” heating unit (Life Imaging Services, Basel, Switzerland) maintaining 37°C and the Incubation System S for CO2 and humidity supply (PeCon, Erbach, Germany). Images were acquired using the Zeiss LD Plan-Neofluar 40x/0.6 Corr air objective (with bottom thickness correction ring). For acquisition of GFP and mCherry BP 525/50 (Zeiss) and LP 580 (Olympus, Hamburg, Germany) filters, respectively, were applied. Imaging of individual screening plates was executed hourly from 1–7 h p.i. with eight positions per well and a single Z-plane per position (adjusted to the plane with the majority of bacteria). All images obtained were processed by the ZEN software. Non-screen LCI was performed using a Leica SP5 confocal laser-scanning microscope (CLSM) operated by Leica LAS AF software. The microscope was also equipped with live cell periphery consisting of ‘The Box’ incubation chamber (Life Imaging Services, Basel, Switzerland), a custom-made heating unit and a gas supply unit ‘The Brick’. Images were acquired using the HCX PL APO CS 100x/1.4 oil objective (Leica, Wetzlar, Germany), applying the polychroic mirror TD 488/543/633 for acquisition of GFP and mCherry. All images were processed by LAS AF software.
Quantification of SIF formation
After siRNA knockdown and infection as described above, 100 infected HeLa LAMP1-GFP cells per condition as indicated were examined live with a 40x objective from 6–8 h p.i. for the presence of SIF as exhibited by WT-infected cells, and the percentage in relation to siAllstars-treated WT-infected cells calculated. Results from three independent experiments (n = 3) were plotted using SigmaPlot 11.
Data analysis
For the central entry and collection of scoring data, the MATLAB-based utility SifScreen was used. The categorization of targets/hits was executed using the GO classification scheme [124, 125]. For the visualization of protein interactions, the STRING v10 database with default settings was applied [126].
Supporting information
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
HeLa cells expressing LAMP1-GFP (green) were reverse transfected with the indicated siRNAs for 72 h (also corresponding to Fig 4). Then, cells were infected with STM WT expressing mCherry (red) at MOI = 15, and imaged by SDCM. Depicted are representative field of views 7 h p.i. Scale bar, 20 μm.
(TIF)
HeLa LAMP1-GFP cells were left untreated or reverse transfected with siAllStars or the indicated siRNA. A) For RT-PCR, total RNA was extracted, mRNA reverse transcribed, and the generated cDNA was used in RT-PCR. Depicted are means and standard deviation for three biological replicates (n = 3), each performed in triplicates. Statistical analysis was performed against siAllstars with Student’s t-test and indicated as: ***, p < 0.001. B) For Western blot analysis, cell lysates were processed to determine the protein levels of SKIP, RAB7A, and VPS11. Two independent knock-down assays are indicated by k/d 1 and k/d 2. As a loading control, blots were additionally processed for detection of γ-tubulin. C) Densitometry of Western blot signals for the indicated proteins.
(TIF)
The movie corresponds to Fig 1D.
(AVI)
The movie corresponds to Fig 1D.
(AVI)
Acknowledgments
We thank Monika Nietschke and Ursula Krehe for construction of plasmids and technical assistance. Special thanks go to Markus C. Kerr (Brisbane), André P. Mäurer, Peter Braun, Marion Rother, and Thomas F. Meyer (Berlin) for advice and logistics in setting up the screen. We thank Martin Aepfelbacher (Hamburg), Thierry Galli (Paris), Wanjin Hong (Singapore), and Yulong Li (Beijing) for providing transfection vectors and Stéphane Méresse (Marseille) and Christian Ungermann (Osnabrück) for antibodies.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by BMBF grant 0315834D ‘Medizinische Infektionsgenomik’, and the DFG by priority programme SPP 1580 grant HE 1964/18-2 to M.H, the Z project of SFB944. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nat Rev Microbiol. 2015;13(4):191–205. 10.1038/nrmicro3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan JE, Curtiss R. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–7. 10.1073/pnas.86.16.6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramos-Morales F. Impact of Salmonella enterica Type III Secretion System Effectors on the Eukaryotic Host Cell. ISRN Cell Biology. 2012;2012 10.5402/2012/787934 [DOI] [Google Scholar]
- 4.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1996;93(6):2593–7. 10.1073/pnas.93.6.2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A. 1996;93:7800–4. 10.1073/pnas.93.15.7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012;158(Pt 5):1147–61. Epub 2012/03/17. [pii] 10.1099/mic.0.058115-0 . [DOI] [PubMed] [Google Scholar]
- 7.Steele-Mortimer O, Meresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1(1):33–49. 10.1046/j.1462-5822.1999.00003.x [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee K, Siddiqi SA, Hashim S, Raje M, Basu SK, Mukhopadhyay A. Live Salmonella recruits N-ethylmaleimide-sensitive fusion protein on phagosomal membrane and promotes fusion with early endosome. J Cell Biol. 2000;148(4):741–53. 10.1083/jcb.148.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills SD, Finlay BB. Comparison of Salmonella typhi and Salmonella typhimurium invasion, intracellular growth and localization in cultured human epithelial cells. MicrobPathog. 1994;17:409–23. [DOI] [PubMed] [Google Scholar]
- 10.Oh YK, Alpuche-Aranda C, Berthiaume E, Jinks T, Miller SI, Swanson JA. Rapid and complete fusion of macrophage lysosomes with phagosomes containing Salmonella typhimurium. Infect Immun. 1996;64(9):3877–83. Epub 1996/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Intracellular replication of Salmonella within epithelial cells is associated with filamentous structures containing lysosomal membrane glycoproteins. Infect Agents Dis. 1993;2(4):227–31. [PubMed] [Google Scholar]
- 12.Meresse S, Steele-Mortimer O, Finlay BB, Gorvel JP. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 1999;18(16):4394–403. 10.1093/emboj/18.16.4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brumell JH, Tang P, Mills SD, Finlay BB. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic. 2001;2(9):643–53. 10.1034/j.1600-0854.2001.20907.x . [DOI] [PubMed] [Google Scholar]
- 14.Garcia-del Portillo F, Finlay BB. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J Cell Biol. 1995;129(1):81–97. 10.1083/jcb.129.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder N, Mota LJ, Meresse S. Salmonella-induced tubular networks. Trends Microbiol. 2011;19(6):268–77. Epub 2011/03/01. S0966-842X(11)00021-7 [pii] 10.1016/j.tim.2011.01.006 . [DOI] [PubMed] [Google Scholar]
- 16.Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc Natl Acad Sci U S A. 1993;90(22):10544–8. 10.1073/pnas.90.22.10544 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knuff K, Finlay BB. What the SIF is happening—The role of intracellular Salmonella-induced filaments. Front Cell Infect Microbiol. 2017;7:335 10.3389/fcimb.2017.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieger V, Liebl D, Zhang Y, Rajashekar R, Chlanda P, Giesker K, et al. Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLoS Pathog. 2014;10(9):e1004374 10.1371/journal.ppat.1004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liss V, Hensel M. Take the tube: remodelling of the endosomal system by intracellular Salmonella enterica. Cell Microbiol. 2015;17(5):639–47. 10.1111/cmi.12441 . [DOI] [PubMed] [Google Scholar]
- 20.Stein MA, Leung KY, Zwick M, Garcia-del Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20(1):151–64. 10.1111/j.1365-2958.1996.tb02497.x . [DOI] [PubMed] [Google Scholar]
- 21.Zhao W, Moest T, Zhao Y, Guilhon AA, Buffat C, Gorvel JP, et al. The Salmonella effector protein SifA plays a dual role in virulence. Sci Rep. 2015;5:12979 10.1038/srep12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucrot E, Henry T, Borg JP, Gorvel JP, Meresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308(5725):1174–8. 10.1126/science.1110225 [DOI] [PubMed] [Google Scholar]
- 23.Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, Schroeder N, et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc Natl Acad Sci U S A. 2006;103(36):13497–502. 10.1073/pnas.0605443103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaniuk NA, Canadien V, Bagshaw RD, Bakowski M, Braun V, Landekic M, et al. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell Microbiol. 2011;13(11):1812–23. Epub 2011/08/10. 10.1111/j.1462-5822.2011.01663.x . [DOI] [PubMed] [Google Scholar]
- 25.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell. 2011;21(6):1171–8. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leone P, Meresse S. Kinesin regulation by Salmonella. Virulence. 2011;2(1):63–6. Epub 2011/01/11. 14603 [pii]. 10.4161/viru.2.1.14603 . [DOI] [PubMed] [Google Scholar]
- 27.Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, Schlumberger M, et al. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol Syst Biol. 2011;7:474 Epub 2011/03/17. 10.1038/msb.2011.7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornbrough JM, Gopinath A, Hundley T, Worley MJ. Human genome-wide RNAi screen for host factors that facilitate Salmonella invasion reveals a role for potassium secretion in promoting internalization. PLoS One. 2016;11(11):e0166916 10.1371/journal.pone.0166916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450(7170):725–30. Epub 2007/11/30. nature06345 [pii] 10.1038/nature06345 . [DOI] [PubMed] [Google Scholar]
- 30.Albers HM, Kuijl C, Bakker J, Hendrickx L, Wekker S, Farhou N, et al. Integrating chemical and genetic silencing strategies to identify host kinase-phosphatase inhibitor networks that control bacterial infection. ACS Chem Biol. 2014;9(2):414–22. 10.1021/cb400421a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thornbrough JM, Hundley T, Valdivia R, Worley MJ. Human genome-wide RNAi screen for host factors that modulate intracellular Salmonella growth. PLoS One. 2012;7(6):e38097 10.1371/journal.pone.0038097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorwerk S, Krieger V, Deiwick J, Hensel M, Hansmeier N. Proteomes of host cell membranes modified by intracellular activities of Salmonella enterica. Mol Cell Proteomics. 2015;14(1):81–92. 10.1074/mcp.M114.041145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos JC, Duchateau M, Fredlund J, Weiner A, Mallet A, Schmitt C, et al. The COPII complex and lysosomal VAMP7 determine intracellular Salmonella localization and growth. Cell Microbiol. 2015;17(12):1699–720. 10.1111/cmi.12475 . [DOI] [PubMed] [Google Scholar]
- 34.Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, et al. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic. 2008;9(12):2117–29. Epub 2008/09/13. TRA830 [pii] 10.1111/j.1600-0854.2008.00830.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajashekar R, Liebl D, Seitz A, Hensel M. Dynamic remodeling of the endosomal system during formation of Salmonella-induced filaments by intracellular Salmonella enterica. Traffic. 2008;9(12):2100–16. Epub 2008/09/27. TRA821 [pii] 10.1111/j.1600-0854.2008.00821.x . [DOI] [PubMed] [Google Scholar]
- 36.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91(1):119–49. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhuin T, Roy JK. Rab proteins: the key regulators of intracellular vesicle transport. Exp Cell Res. 2014;328(1):1–19. 10.1016/j.yexcr.2014.07.027 . [DOI] [PubMed] [Google Scholar]
- 38.Wandinger-Ness A, Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 2014;6(11):a022616 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brodsky FM. Diversity of clathrin function: new tricks for an old protein. Annu Rev Cell Dev Biol. 2012;28:309–36. 10.1146/annurev-cellbio-101011-155716 . [DOI] [PubMed] [Google Scholar]
- 40.Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6(2). 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popoff V, Adolf F, Brugger B, Wieland F. COPI budding within the Golgi stack. Cold Spring Harb Perspect Biol. 2011;3(11):a005231 10.1101/cshperspect.a005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol. 2013;5(2). 10.1101/cshperspect.a013367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SY, Guo X. Adaptor protein complexes and intracellular transport. Biosci Rep. 2014;34(4). 10.1042/BSR20140069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat Cell Biol. 2012;14(1):11–9. 10.1038/ncb2409 . [DOI] [PubMed] [Google Scholar]
- 45.Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24(1):35–43. 10.1016/j.tcb.2013.09.006 . [DOI] [PubMed] [Google Scholar]
- 46.Chia PZ, Gleeson PA. Membrane tethering. F1000Prime Rep. 2014;6:74 10.12703/P6-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744(3):493–517. . [PubMed] [Google Scholar]
- 48.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–43. 10.1038/nrm2002 . [DOI] [PubMed] [Google Scholar]
- 49.Dukes JD, Lee H, Hagen R, Reaves BJ, Layton AN, Galyov EE, et al. The secreted Salmonella dublin phosphoinositide phosphatase, SopB, localizes to PtdIns(3)P-containing endosomes and perturbs normal endosome to lysosome trafficking. Biochem J. 2006;395(2):239–47. 10.1042/BJ20051451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith AC, Heo WD, Braun V, Jiang X, Macrae C, Casanova JE, et al. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2007;176(3):263–8. Epub 2007/01/31. jcb.200611056 [pii] 10.1083/jcb.200611056 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison RE, Brumell JH, Khandani A, Bucci C, Scott CC, Jiang X, et al. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell. 2004;15:3146–54. 10.1091/mbc.e04-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D'Costa VM, Braun V, Landekic M, Shi R, Proteau A, McDonald L, et al. Salmonella Disrupts Host Endocytic Trafficking by SopD2-Mediated Inhibition of Rab7. Cell reports. 2015;12(9):1508–18. Epub 2015/08/25. 10.1016/j.celrep.2015.07.063 . [DOI] [PubMed] [Google Scholar]
- 53.Humphreys D, Hume PJ, Koronakis V. The Salmonella effector SptP dephosphorylates host AAA+ ATPase VCP to promote development of its intracellular replicative niche. Cell Host Microbe. 2009;5(3):225–33. Epub 2009/03/17. S1931-3128(09)00064-X [pii] 10.1016/j.chom.2009.01.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sindhwani A, Arya SB, Kaur H, Jagga D, Tuli A, Sharma M. Salmonella exploits the host endolysosomal tethering factor HOPS complex to promote its intravacuolar replication. PLoS Pathog. 2017;13(10):e1006700 Epub 2017/10/31. 10.1371/journal.ppat.1006700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McEwan DG, Richter B, Claudi B, Wigge C, Wild P, Farhan H, et al. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015;17(1):58–71. 10.1016/j.chom.2014.11.011 . [DOI] [PubMed] [Google Scholar]
- 56.Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52(6):915–24. 10.1016/0092-8674(88)90433-3 . [DOI] [PubMed] [Google Scholar]
- 57.Bacon RA, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109(3):1015–22. 10.1083/jcb.109.3.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, et al. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115(1):31–43. 10.1083/jcb.115.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119(4):749–61. 10.1083/jcb.119.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukhopadhyay A, Nieves E, Che FY, Wang J, Jin L, Murray JW, et al. Proteomic analysis of endocytic vesicles: Rab1a regulates motility of early endocytic vesicles. J Cell Sci. 2011;124(Pt 5):765–75. 10.1242/jcs.079020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukhopadhyay A, Quiroz JA, Wolkoff AW. Rab1a regulates sorting of early endocytic vesicles. Am J Physiol Gastrointest Liver Physiol. 2014;306(5):G412–24. 10.1152/ajpgi.00118.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagano F, Sasaki T, Fukui K, Asakura T, Imazumi K, Takai Y. Molecular cloning and characterization of the noncatalytic subunit of the Rab3 subfamily-specific GTPase-activating protein. J Biol Chem. 1998;273(38):24781–5. 10.1074/jbc.273.38.24781 . [DOI] [PubMed] [Google Scholar]
- 63.Touchot N, Chardin P, Tavitian A. Four additional members of the ras gene superfamily isolated by an oligonucleotide strategy: molecular cloning of YPT-related cDNAs from a rat brain library. Proc Natl Acad Sci USA. 1987;84(23):8210–4. 10.1073/pnas.84.23.8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlüter OM, Khvotchev M, Jahn R, Südhof TC. Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J Biol Chem. 2002;277(43):40919–29. 10.1074/jbc.M203704200 . [DOI] [PubMed] [Google Scholar]
- 65.Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol. 1993;123(1):35–45. 10.1083/jcb.123.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75(3):409–18. 10.1016/0092-8674(93)90376-2 . [DOI] [PubMed] [Google Scholar]
- 67.Borisovska M, Zhao Y, Tsytsyura Y, Glyvuk N, Takamori S, Matti U, et al. v-SNAREs control exocytosis of vesicles from priming to fusion. EMBO J. 2005;24(12):2114–26. 10.1038/sj.emboj.7600696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cocucci E, Racchetti G, Rupnik M, Meldolesi J. The regulated exocytosis of enlargeosomes is mediated by a SNARE machinery that includes VAMP4. J Cell Sci. 2008;121(Pt 18):2983–91. 10.1242/jcs.032029 . [DOI] [PubMed] [Google Scholar]
- 69.Madan R, Rastogi R, Parashuraman S, Mukhopadhyay A. Salmonella acquires lysosome-associated membrane protein 1 (LAMP1) on phagosomes from Golgi via SipC protein-mediated recruitment of host Syntaxin6. J Biol Chem. 2012;287(8):5574–87. 10.1074/jbc.M111.286120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bogdanovic A, Bennett N, Kieffer S, Louwagie M, Morio T, Garin J, et al. Syntaxin 7, syntaxin 8, Vti1 and VAMP7 (vesicle-associated membrane protein 7) form an active SNARE complex for early macropinocytic compartment fusion in Dictyostelium discoideum. Biochem J. 2002;368(Pt 1):29–39. 10.1042/BJ20020845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SC, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5(6):590–5. 10.1038/sj.embor.7400150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Antonin W, Holroyd C, Fasshauer D, Pabst S, Von Mollard GF, Jahn R. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19(23):6453–64. 10.1093/emboj/19.23.6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antonin W, Holroyd C, Tikkanen R, Honing S, Jahn R. The R-SNARE endobrevin/VAMP-8 mediates homotypic fusion of early endosomes and late endosomes. Mol Biol Cell. 2000;11(10):3289–98. 10.1091/mbc.11.10.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh PK, Kapoor A, Lomash RM, Kumar K, Kamerkar SC, Pucadyil TJ, et al. Salmonella SipA mimics a cognate SNARE for host Syntaxin8 to promote fusion with early endosomes. J Cell Biol. 2018;217(12):4199–214. 10.1083/jcb.201802155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dai S, Zhang Y, Weimbs T, Yaffe MB, Zhou D. Bacteria-generated PtdIns(3)P recruits VAMP8 to facilitate phagocytosis. Traffic. 2007;8(10):1365–74. 10.1111/j.1600-0854.2007.00613.x . [DOI] [PubMed] [Google Scholar]
- 76.Merrifield CJ, Kaksonen M. Endocytic accessory factors and regulation of clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol. 2014;6(11):a016733 10.1101/cshperspect.a016733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Traub LM, Bonifacino JS. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol. 2013;5(11):a016790 10.1101/cshperspect.a016790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marsman M, Jordens I, Kuijl C, Janssen L, Neefjes J. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell. 2004;15(6):2954–64. 10.1091/mbc.e03-08-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abrahams GL, Müller P, Hensel M. Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic. 2006;7(8):950–65. 10.1111/j.1600-0854.2006.00454.x [DOI] [PubMed] [Google Scholar]
- 80.Guignot J, Caron E, Beuzon C, Bucci C, Kagan J, Roy C, et al. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J Cell Sci. 2004;117:1033–45. 10.1242/jcs.00949 [DOI] [PubMed] [Google Scholar]
- 81.Miao EA, Brittnacher M, Haraga A, Jeng RL, Welch MD, Miller SI. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol Microbiol. 2003;48(2):401–15. 10.1046/j.1365-2958.2003.t01-1-03456.x . [DOI] [PubMed] [Google Scholar]
- 82.Wasylnka JA, Bakowski MA, Szeto J, Ohlson MB, Trimble WS, Miller SI, et al. Role for myosin II in regulating positioning of Salmonella-containing vacuoles and intracellular replication. Infect Immun. 2008;76(6):2722–35. Epub 2008/04/16. IAI.00152-08 [pii] 10.1128/IAI.00152-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dumont A, Boucrot E, Drevensek S, Daire V, Gorvel JP, Pous C, et al. SKIP, the host target of the Salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic. 2010;11(7):899–911. Epub 2010/04/22. TRA1069 [pii] 10.1111/j.1600-0854.2010.01069.x . [DOI] [PubMed] [Google Scholar]
- 84.Kerr MC, Wang JT, Castro NA, Hamilton NA, Town L, Brown DL, et al. Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. EMBO J. 2010;29(8):1331–47. 10.1038/emboj.2010.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuijl C, Pilli M, Alahari SK, Janssen H, Khoo PS, Ervin KE, et al. Rac and Rab GTPases dual effector Nischarin regulates vesicle maturation to facilitate survival of intracellular bacteria. EMBO J. 2013;32(5):713–27. 10.1038/emboj.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seixas E, Ramalho JS, Mota LJ, Barral DC, Seabra MC. Bacteria and protozoa differentially modulate the expression of Rab proteins. PLoS One. 2012;7(7):e39858 10.1371/journal.pone.0039858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mota LJ, Ramsden AE, Liu M, Castle JD, Holden DW. SCAMP3 is a component of the Salmonella-induced tubular network and reveals an interaction between bacterial effectors and post-Golgi trafficking. Cell Microbiol. 2009;11(8):1236–53. Epub 2009/05/15. CMI1329 [pii] 10.1111/j.1462-5822.2009.01329.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.D'Costa VM, Coyaud E, Boddy KC, Laurent EMN, St-Germain J, Li T, et al. BioID screen of Salmonella type 3 secreted effectors reveals host factors involved in vacuole positioning and stability during infection. Nat Microbiol. 2019. 10.1038/s41564-019-0580-9 . [DOI] [PubMed] [Google Scholar]
- 89.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14(5):2116–27. 10.1091/mbc.e02-09-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18(7):2400–10. 10.1091/mbc.e06-11-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo Y, Linstedt AD. Binding of the vesicle docking protein p115 to the GTPase Rab1b regulates membrane recruitment of the COPI vesicle coat. Cell Logist. 2013;3:e27687 10.4161/cl.27687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157(4):631–43. 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168(5):747–59. 10.1083/jcb.200412003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J Cell Biol. 2007;179(6):1179–92. 10.1083/jcb.200705145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu D, Joglekar AP, Williams AL, Hay JC. Subunit structure of a mammalian ER/Golgi SNARE complex. J Biol Chem. 2000;275(50):39631–9. 10.1074/jbc.M007684200 . [DOI] [PubMed] [Google Scholar]
- 96.Zhang T, Hong W. Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J Biol Chem. 2001;276(29):27480–7. 10.1074/jbc.M102786200 . [DOI] [PubMed] [Google Scholar]
- 97.Tai G, Lu L, Wang TL, Tang BL, Goud B, Johannes L, et al. Participation of the syntaxin 5/Ykt6/GS28/GS15 SNARE complex in transport from the early/recycling endosome to the trans-Golgi network. Mol Biol Cell. 2004;15(9):4011–22. 10.1091/mbc.e03-12-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, et al. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7(2):433–42. 10.1016/s1097-2765(01)00190-3 . [DOI] [PubMed] [Google Scholar]
- 99.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11(12):4403–11. 10.1091/mbc.11.12.4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20(19):4205–15. 10.1091/mbc.e09-05-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107(17):7811–6. Epub 2010/04/09. 1000063107 [pii] 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cai H, Zhang Y, Pypaert M, Walker L, Ferro-Novick S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J Cell Biol. 2005;171(5):823–33. 10.1083/jcb.200505145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016;35(3):281–301. 10.15252/embj.201592695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shirahama-Noda K, Kira S, Yoshimori T, Noda T. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J Cell Sci. 2013;126(Pt 21):4963–73. 10.1242/jcs.131318 . [DOI] [PubMed] [Google Scholar]
- 105.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17(8):2156–65. 10.1093/emboj/17.8.2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, et al. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8(3):270–84. 10.1111/j.1600-0854.2006.00530.x . [DOI] [PubMed] [Google Scholar]
- 107.Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J Cell Biol. 2002;157(1):45–62. 10.1083/jcb.200112127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuhle V, Abrahams GL, Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic. 2006;7(6):716–30. 10.1111/j.1600-0854.2006.00422.x [DOI] [PubMed] [Google Scholar]
- 109.Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, et al. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156(4):653–64. 10.1083/jcb.200110081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180(1):159–72. 10.1083/jcb.200707136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brandhorst D, Zwilling D, Rizzoli SO, Lippert U, Lang T, Jahn R. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc Natl Acad Sci USA. 2006;103(8):2701–6. 10.1073/pnas.0511138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith AC, Cirulis JT, Casanova JE, Scidmore MA, Brumell JH. Interaction of the Salmonella-containing vacuole with the endocytic recycling system. J Biol Chem. 2005;280(26):24634–41. 10.1074/jbc.M500358200 . [DOI] [PubMed] [Google Scholar]
- 113.Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18(8):2828–37. 10.1091/mbc.e07-02-0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, et al. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci U S A. 2010;107(14):6346–51. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rahajeng J, Giridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic. 2012;13(1):82–93. 10.1111/j.1600-0854.2011.01294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20(24):5181–94. 10.1091/mbc.e09-06-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spano S, Gao X, Hannemann S, Lara-Tejero M, Galan JE. A Bacterial Pathogen Targets a Host Rab-Family GTPase Defense Pathway with a GAP. Cell Host Microbe. 2016;19(2):216–26. 10.1016/j.chom.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gurumurthy RK, Mäurer AP, Machuy N, Hess S, Pleissner KP, Schuchhardt J, et al. A loss-of-function screen reveals Ras- and Raf-independent MEK-ERK signaling during Chlamydia trachomatis infection. Sci Signal. 2010;3(113):ra21 10.1126/scisignal.2000651 . [DOI] [PubMed] [Google Scholar]
- 119.Sharma M, Machuy N, Böhme L, Karunakaran K, Mäurer AP, Meyer TF, et al. HIF-1α is involved in mediating apoptosis resistance to Chlamydia trachomatis-infected cells. Cell Microbiol. 2011;13(10):1573–85. 10.1111/j.1462-5822.2011.01642.x . [DOI] [PubMed] [Google Scholar]
- 120.Vreeburg RA, Bastiaan-Net S, Mes JJ. Normalization genes for quantitative RT-PCR in differentiated Caco-2 cells used for food exposure studies. Food Funct. 2011;2(2):124–9. 10.1039/c0fo00068j . [DOI] [PubMed] [Google Scholar]
- 121.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31(24):e154 10.1093/nar/gng154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40(Database issue):D1144–9. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Town L, McGlinn E, Fiorenza S, Metzis V, Butterfield NC, Richman JM, et al. The metalloendopeptidase gene Pitrm1 is regulated by hedgehog signaling in the developing mouse limb and is expressed in muscle progenitors. Dev Dyn. 2009;238(12):3175–84. 10.1002/dvdy.22126 . [DOI] [PubMed] [Google Scholar]
- 124.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, et al. The GOA database: gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43(Database issue):D1057–63. 10.1093/nar/gku1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]