Abstract
When eukaryotic cells encounter double-stranded RNA, genes of matching sequence are silenced through RNA interference. Surprisingly, in some animals and plants, the same gene is specifically silenced even in cells that did not encounter the double-stranded RNA, due to the transport of a gene-specific silencing signal between cells. This silencing signal likely has an RNA component that gives it sequence-specificity, however its precise identity remains unknown. Studies in the worm Caenorhabditis elegans and in plants have revealed parts of a complex protein machinery that transports this silencing signal. Some of these proteins are conserved in vertebrates, including mammals, raising the possibility that higher animals can communicate gene-specific silencing information between cells. Such communication provides antiviral immunity in plants and perhaps in C. elegans. Identifying the transported silencing signal and deciphering the evolutionarily selected role of the transport machinery are some of the key challenges for the future.
Keywords: gene silencing, dsRNA import, siRNA, systemic RNAi, endocytosis, epigenetics
INTRODUCTION
Cells communicate with each other to coordinate development and function in all multicellular organisms. Such communication often involves small molecules or proteins from a signaling cell being recognized by proteins in receiving cells. This recognition is then transduced in the receiving cell and activates specific signaling pathways to generate an appropriate response. Recent analysis of a gene silencing mechanism called RNA interference (RNAi) suggests that cells of a multicellular organism can also communicate with other cells by direct transport of regulatory RNAs.
An early indication that RNA could modulate cell function comes from the observation that double-stranded RNA (dsRNA) could trigger the inactivation of matching sequences through RNAi in the worm Caenorhabditis elegans (35). Such silencing was also soon discovered in plants (144), in trypanosomes (85), and in flies (56). Once dsRNA enters a cell, a member of the Dicer family of RNaseIII-like enzymes cleaves the dsRNA (12) to produce short interfering RNAs (siRNAs) of ~21 nucleotides (32, 45, 155). These processed guide RNAs associate with a protein complex that generally includes a member of the Argonaute family of RNaseH-like enzymes to perform sequence-specific cleavage of RNA (47, 71).
Although first discovered as a response to experimentally delivered RNA triggers, we now know that key steps in RNAi underlie many diverse gene regulatory mechanisms. These include processes that (a) downregulate the expression of endogenous genes, (b) direct transcriptional gene silencing and alter chromatin structure to promote kinetochore function and chromosome segregation (see chapter by K. Ekwall, this issue), and (c) direct elimination of DNA from somatic nuclei in Tetrahymena (150).
RNAi-like mechanisms called quelling in fungi (105) and posttranscriptional gene silencing in plants (136) reduce the expression of genes homologous to DNA introduced as transgenes. Curiously, such silencing in plants is noncell autonomous and can be transmitted between cells (91, 139). A similar spread of silencing between cells is also observed when dsRNA is injected into C. elegans to trigger RNAi (35). However, recent studies in C. elegans suggest that plants and animals use distinct mechanisms to transport silencing information between cells. Here, we review the noncell autonomous effects of RNAi and discuss some of the implications of this novel mode of intercellular signaling.
IN C. ELEGANS
Transport of Silencing Information Between Cells
RNAi (see Sidebar RNAi in C. elegans) can be triggered in C. elegans by injecting dsRNA into worms (35), soaking worms in dsRNA solutions (121), feeding worms bacteria that express dsRNA (127), and using transgenes that express dsRNA in vivo (124) (Figure 1). When worms take up dsRNA from the environment (presumably through the intestinal lumen), the target gene is silenced in both the intestinal cells and in a variety of tissues throughout the animal and its progeny (121, 127). Similar noncell autonomous silencing also occurs when dsRNA is injected into, or expressed in, a specific tissue (35, 146). These observations suggest that cells in C. elegans can transport either dsRNA or a derived silencing signal throughout the animal to cause sequence-specific silencing.
Figure 1.
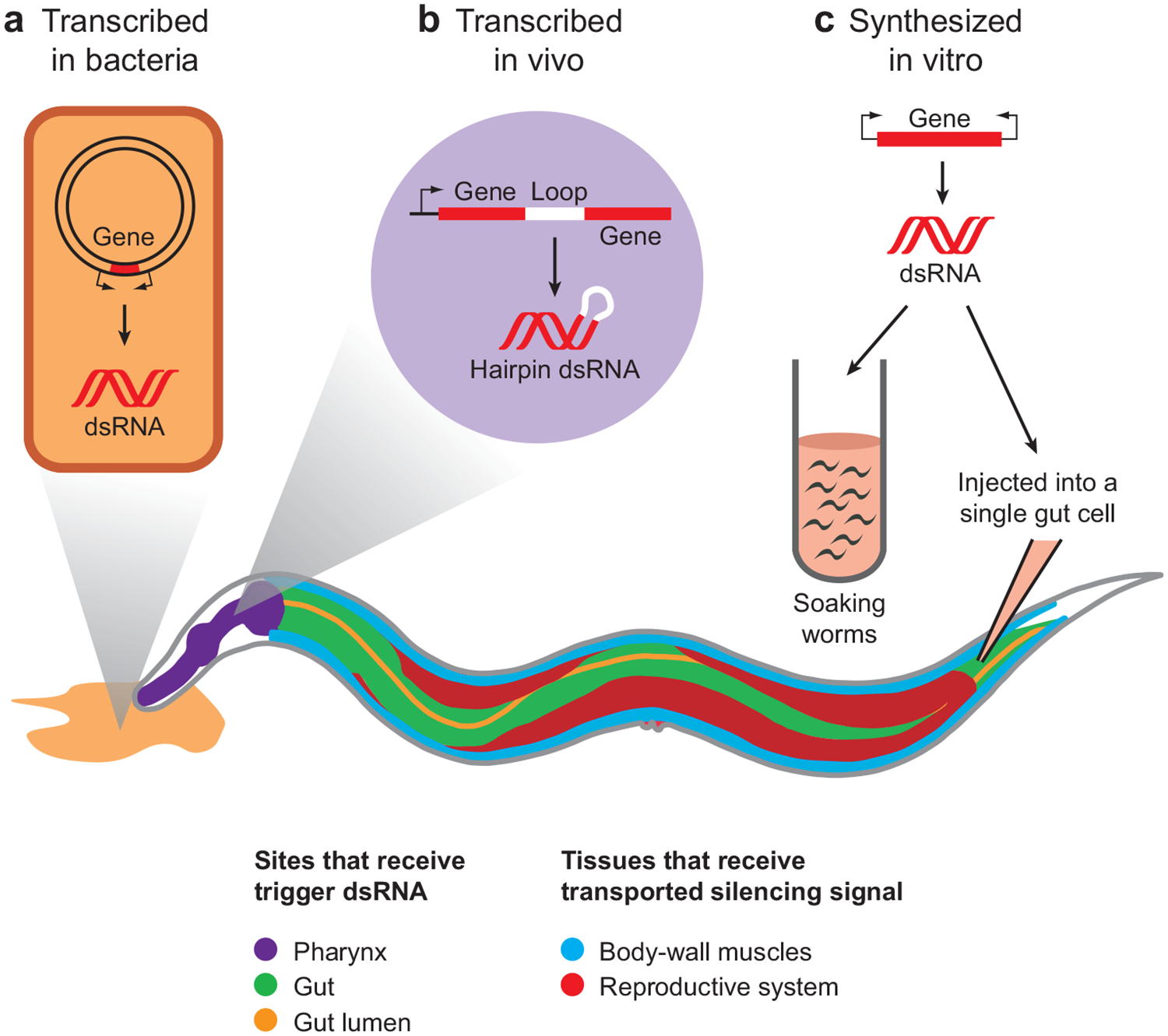
The transport of silencing information between cells can be triggered in many ways in C. elegans. (a) When worms are either fed bacteria that express dsRNA or soaked in a dsRNA solution, the dsRNA is imported from the gut lumen into gut cells, and is disseminated to distant sites such as body-wall muscles and the reproductive system, presumably through the pseudocoelomic fluid (not shown), which bathes all tissues. Transcription start sites (small black arrows) and DNA (bacterial plasmid, PCR product, or inverted-repeat transgene) with target gene are shown. (b) Hairpin dsRNA made in vivo in the pharynx and (c) dsRNA injected into single gut cells likely also cause silencing to spread to other tissues via the pseudocoelom.
The transport of silencing information between cells may be a regulated process. RNAi triggered by transgenes that express gfp hairpin RNA (dsRNA with a connecting loop at one end) in specific tissues leads to detectable transport of silencing information between cells when strong promoters are used to drive the transgene (17, 146). However, when gfp hairpin RNA was expressed in the body-wall muscles using a relatively weak body-wall muscle-specific promoter, GFP was visibly silenced only in body-wall muscle cells (128). Remarkably, introduction of unrelated dsRNA (with no sequences matching gfp) by injection or soaking led to GFP silencing in other tissues. Similar apparent induced transport and/or reception of a hairpin RNA-derived silencing signal was also reported when animals expressing hairpin RNAs were briefly starved (146). Such regulation in response to the environmental or physiological conditions suggests that the transport of sequence-specific silencing information is physiologically important. Notably, there are many sources of dsRNA in the genome of C. elegans (157), and endogenous siRNAs have been detected in vivo (5). However, it is currently unknown whether the silencing information from such endogenous RNAi is transported between cells.
Import of Trigger dsRNA and Silencing Information
To identify genes that control the import of dsRNA and the transport of silencing information between cells during RNAi, three independent genetic screens have been performed in C. elegans (sid screen, fed screen, and rsd screen) and one in Drosophila S2 cells. Analysis of the genes identified in these screens suggests that silencing signals are imported into cells using a dsRNA channel and specific endocytic machinery (Figure 2).
Figure 2.
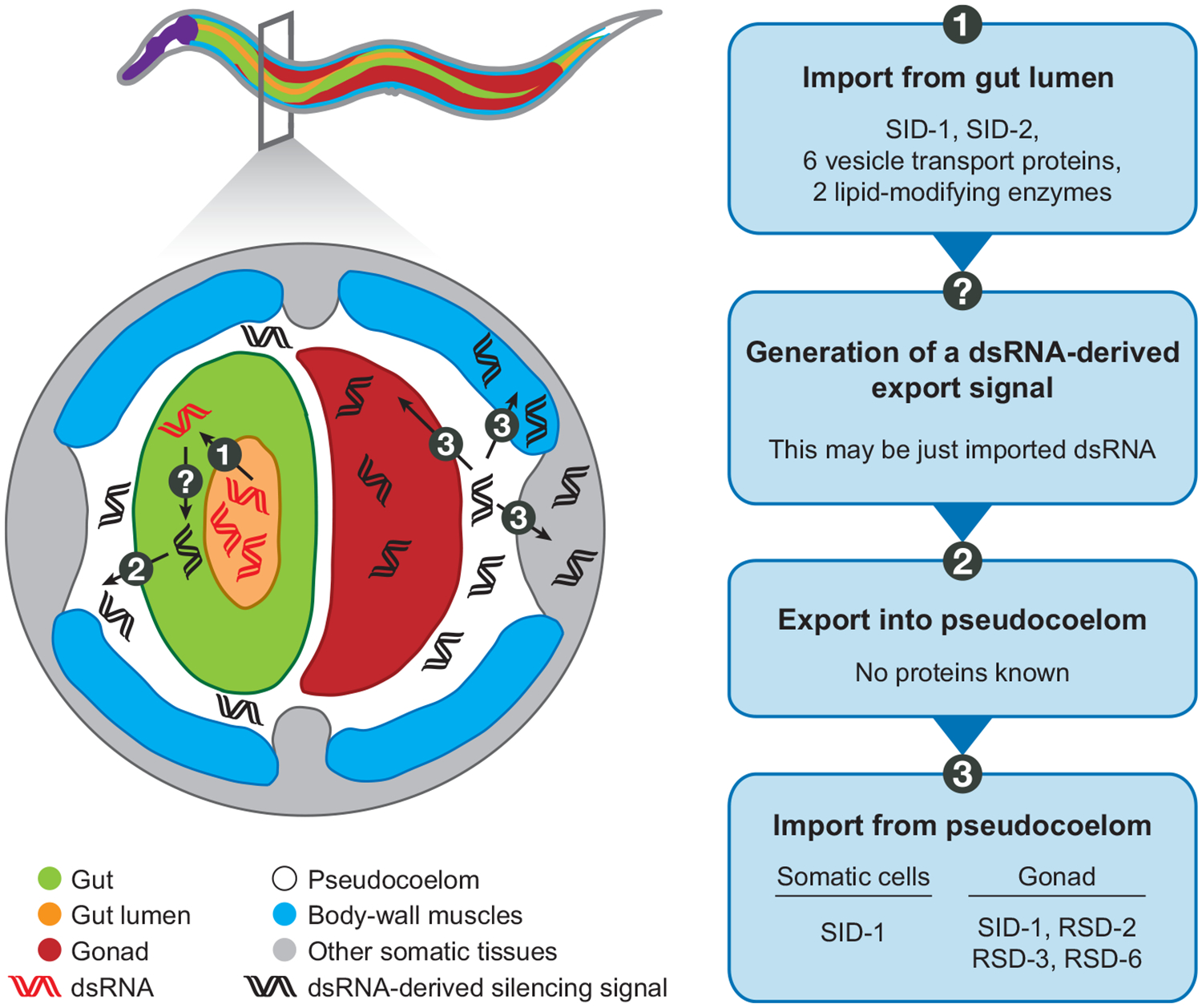
Part of the molecular machinery that controls the uptake of dsRNA and mediates intercellular transport of silencing in C. elegans is known. A cross-section through the worm shows the transport of dsRNA and a dsRNA-derived silencing signal during RNAi. dsRNA is first imported from the gut lumen into gut cells. Imported dsRNA is either converted into a derived silencing signal and then exported or is directly exported out of gut cells into the pseudocoelom. The silencing signal is finally imported into the gonad and into body-wall muscles and other somatic tissues. The flowchart on the right shows the steps and proteins required during the intercellular transport of silencing. SID-1/RSD-8 is a dsRNA channel; SID-2/FED-1/RSD-4 is a transmembrane protein located in the lumenal membrane of the intestine; RSD-2 and RSD-6 form a protein complex that may bind RNA; and RSD-3 may mediate endocytosis. The silencing signal derived from in vivo expression of dsRNA or injection of dsRNA into a single gut cell is also presumably exported into the pseudocoelom from which it is imported into cells as above (not shown).
Sid screen.
Systemic RNAi-defective (sid) mutants were isolated as animals that cannot transport silencing information between cells. The intercellular transport of silencing information was followed by expressing GFP in both pharynx and body-wall muscles, and supplying gfp dsRNA by expressing a transgene specifically in the pharynx while simultaneously feeding the worms bacteria that express gfp dsRNA (146). Wild-type worms silenced GFP expression in the pharynx and due to the transport of silencing information from both the pharynx and the intestinal cells that take up the dsRNA from the food (systemic RNAi), they also silenced GFP in the body-wall muscles. In contrast, sid mutants continued to silence GFP in the pharynx due to autonomous RNAi, but no longer silenced GFP in the body-wall muscles because they lack the ability to transport the silencing information between cells. This screen identified at least five genes (146; C. P. Hunter, unpublished results), two of which, sid-1 and sid-2, have been analyzed in detail.
The sid-1 gene encodes a conserved protein with homologs in most animals including mammals (146), and in C. elegans the SID-1 protein functions as a dsRNA channel for the import of dsRNA and silencing information into most cells (34, 146). When dsRNA that targets a gene expressed in the germline was injected directly into the germline of sid-1 mutants, RNAi was triggered. However, the same dsRNA did not trigger RNAi when it was injected into the body cavity of sid-1 mutants, suggesting that sid-1 is required for the import of exogenously supplied dsRNA. Further, sid-1 mutant worms are also unable to import into body-wall muscles a silencing signal generated by expression of a gfp hairpin RNA in the pharynx. The sid-1 gene is expressed in all non-neuronal cells and encodes a transmembrane protein that accumulates in peripheral cell membranes. Taken together, this suggests that the SID-1 protein may be a channel that imports both dsRNA from the environment and the silencing signal generated by transgenes that express hairpin RNA. To test this hypothesis, SID-1 was expressed in Drosophila cells, which lack both a SID-1 homolog and efficient mechanisms to take up dsRNA from the surrounding medium (34). In this heterologous system, SID-1 enables rapid import of dsRNA even under energy-depleted conditions, consistent with SID-1 acting as a passive dsRNA channel, rather than a pump or receptor. Further, SID-1 shows a strong preference for long dsRNA (≥100 bp) over short dsRNA or siRNAs. Thus, SID-1 likely enables the passive import of dsRNA into cells in which it is expressed.
The finding that sid-1 is not expressed in most neurons may explain why neurons are resistant to silencing triggered by exogenous dsRNA but are susceptible to RNAi triggered by providing dsRNA within neurons using transgene expression (124). However, RNAi does occur in neurons in response to exogenously supplied dsRNA in an enhanced RNAi-1 (eri-1) mutant where the efficiency of RNAi is increased due to the stabilization of siRNAs (55). In these experiments silencing in neurons was scored one generation after dsRNA exposure, thus the silencing signal may gain access into neurons not by transport but by inheritance through the germline. Alternatively, it is possible that a subthreshold level of the silencing signal may be successfully transported into neurons through a SID-1 independent mechanism or that there is undetected SID-1 expression in neurons. Although neurons of C. elegans are refractory to systemic RNAi, this is not a universal property of neurons as neurons of the parasitic nematode G. pallida are susceptible to exogenously supplied dsRNA (58).
The sid-2 gene encodes a single-pass transmembrane protein that is required for the import of dsRNA from the environment into the intestine (147). However, sid-2 is not required for the transport of a silencing signal from either injected dsRNA or endogenously supplied dsRNA. SID-2 is expressed in the intestine, where it localizes to the lumenal membrane. Based on localization and on the defects in animals that lack SID-2, SID-2 may modify SID-1 activity in gut cells to enable efficient dsRNA uptake from the environment. Alternatively, SID-2 may act to bring dsRNA into endocytic vesicles for SID-1-dependent transport into the cytoplasm. The ability to take up dsRNA from the environment is shared by some other Caenorhabditis species, but is notably less widespread among Caenorhabditis species than is the ability to transport a silencing signal between cells. C. briggsae lacks the ability to take up dsRNA from the environment, but transgenic expression of C. elegans SID-2 in C. briggsae is sufficient to confer sensitivity to environmentally supplied dsRNA. Conversely, the C. briggsae ortholog of SID-2 has a highly divergent extracellular domain and is unable to restore import of dsRNA in sid-2 mutant C. elegans. Additional experiments will likely reveal whether SID-2 transports dsRNA into the animal independent of SID-1 or whether SID-2 modifies the import of dsRNA through SID-1.
Fed screen.
Feeding defective (fed) mutants were identified as animals that are insensitive to RNAi triggered by feeding but are sensitive to RNAi triggered by injection (128). This screen was done in two steps. First, mutants were isolated based on their failure to exhibit RNAi when fed bacteria expressing dsRNA against a gene expressed in the germline. Second, the same dsRNA was then injected directly into the germline of the identified mutants and those that exhibited silencing were chosen for further analysis. Thus, fed mutants are defective in the transport of dsRNA-derived silencing information and not in cell-autonomous RNAi. This screen identified two genes (fed-1 and fed-2). Based on the similar phenotype and map position, the fed-1 mutant likely identifies the same gene as sid-2. The gene mutated in fed-2 is yet to be identified.
Rsd screen.
The RNAi spreading defective (rsd) mutants, like the fed mutants, were isolated as animals insensitive to RNAi triggered by feeding but susceptible to RNAi triggered by injection (126). This screen identified at least five genes (rsd-2, −3, −4, −6, and −8).
The rsd-2, rsd-3, and rsd-6 mutants are resistant to RNAi triggered by feeding dsRNA against germline genes but not to RNAi triggered by feeding dsRNA against somatic genes. Thus, these three genes appear to be required for the transport of silencing information into the germline but not into somatic tissues. Although RSD-2 has no detectable homologs in other organisms, it was found to interact with RSD-6, which contains a Tudor domain frequently found in RNA-binding proteins. RSD-3 has an epsin N-terminal homology (ENTH) domain that is often found in vesicle-trafficking proteins (67), and has a human homolog, Enthoprotin, that might also function in vesicle trafficking (143). Thus, endocytosis may play a role in the transport of silencing information into the germline. However, none of the rsd mutants exhibit a generalized defect in endocytosis and mutants defective in endocytosis are not defective in the transport of silencing information into the germline (126). These observations suggest that RSD-3 may control vesicle trafficking in a pathway that is specific for the import of the silencing signal into the germline. Based on the similar phenotype and map position, rsd-4 likely identifies sid-2/fed-1. The rsd-8 mutant identified the same gene as sid-1.
Drosophila screen.
Ten genes required for the endocytic uptake of dsRNA by cultured Drosophila cells have homologs in C. elegans that are required in C. elegans for RNAi triggered by feeding dsRNA (109; also see IN OTHER ANIMALS section). Four of these encode components of intracellular vesicle transport, two encode lipid-modifying enzymes, and four encode genes of unknown function. Worms were fed dsRNA targeting each of the above ten genes and tested four days later for their ability to perform RNAi against a test gene, unc-52, triggered by two days’ supply of unc-52 dsRNA in the food. Silencing any one of these ten genes made worms resistant to RNAi. Although these results suggest that all ten genes are required for the import of dsRNA from the environment, additional experiments are required to determine if they are also required for other steps of silencing transport (Figure 2), such as the export of a silencing signal out of intestinal cells or the passage of dsRNA across the membrane.
The functions of genes identified in all four screens above suggest that the import of dsRNA or a silencing signal involves at least two steps: (a) endocytic uptake into the cell and (b) transport through a channel into the cytoplasm. Further analysis of the import genes identified thus far (Figure 2) and the identification of additional import genes will likely clarify our understanding of import mechanism(s) and may point to pathways that are shared with other intercellular transport mechanisms.
What Is the Exported Silencing Signal?
Although both short and long dsRNAs can trigger RNAi, only long dsRNAs (>50 bp) trigger the detectable transport of silencing information between cells in C. elegans (34). Injection of a 50-bp dsRNA targeting an essential germline gene causes embryonic lethality when injected into the germline. However, when injected into the intestine, the same 50 bp dsRNA requires an additional 50 bp of random dsRNA fused to it (for a total of 100-bp dsRNA) to cause embryonic lethality, suggesting that only long dsRNAs can efficiently initiate the transport of silencing information from intestinal cells to the germline where the target is expressed.
Processing the dsRNA into siRNA, a key step during RNAi, is not required for the transport of silencing information to other cells. When the intestines of rde-4 mutant animals (which do not process long dsRNA into siRNA) are injected with long dsRNA, RNAi is not observed in the injected animal but is observed in heterozygous progeny generated by mating the injected animals with wild-type males (94, 122). Furthermore, target mRNA is not required for the export of a silencing signal (40). Thus, the injected long dsRNA (or a RNAi-independent derived signal) is transported from the intestine into the germline.
During RNAi, RNA-dependent RNA Polymerases (RdRPs) can lead to the generation of dsRNA that includes sequences 5′ of the trigger dsRNA sequence, and these secondary dsRNAs can also act as or generate a transported silencing signal (2, 115). When an unc-22::gfp chimeric mRNA was strongly overexpressed in the intestine and GFP was targeted for RNAi by exposure only to gfp dsRNA, an Unc-22 mutant phenotype was observed although unc-22 functions in body-wall muscles. This was apparently due to the generation of secondary dsRNA homologous to unc-22 in the intestine and the transport of this dsRNA or a derived silencing signal to body-wall muscles.
Further experiments to explore the substrate specificity of the dsRNA channel SID-1 and to characterize the structure of the transported silencing signal will help determine its identity.
Inheritance of Silencing Information
A single encounter with trigger dsRNA can lead to silencing that is inherited for many generations, although this effect is not fully penetrant (138). Several chromatin-remodeling factors are required for this long-term gene silencing, suggesting that the trans-generational inheritance of RNAi may be due to silencing at the transcriptional level, rather than posttranscriptional level. Consistent with this, the RNAi genes rde-1 and rde-4 are only required for the generation of the inherited signal upon initially encountering dsRNA, but not for the subsequent silencing directed by the signal in the next generation (40). Further, rde-1- and rde-4-independent cell-autonomous gene silencing can be triggered by injection of short antisense RNA (125). These considerations suggest that the inherited silencing signal may include secondary siRNAs, which are short and predominantly of antisense polarity (89). Curiously, secondary siRNAs are associated with argonaute-like proteins that lack conserved catalytic residues in the presumed nuclease site (152), suggesting that target cleavage is not important for their activity. Together, these observations indicate that inheritance of silencing information likely involves heterochromatinization in response to a signal that includes secondary siRNAs.
IN OTHER ANIMALS
In addition to C. elegans, many other animals can import silencing information into cells and initiate RNAi (Table 1). Clues to possible mechanisms for the import of dsRNA and, to a lesser extent, the spread of silencing information between cells are available for some of these organisms.
Table 1.
Organisms that can transport silencing information across cell boundaries
| Organism | dsRNA delivered by | Silencing occurs in |
|---|---|---|
| Animals | ||
| Free-living worm (Caenorhabditis) | Injection into gut | Gonad (35) |
| Environment | Gonad (121) | |
| Fed bacteria expressing transgene | Body-wall muscles (127) | |
| Transgene in pharynx | Body-wall muscles (146) | |
| Parasitic worms (Ascaris Brugia Haemonchus Litomosoides Nippostrongylus Onchocerca Ostertagia Schistosoma Trichostrongylus Globodera) | Environment | Skin (52) |
| Environment | Unknown (52) | |
| Environment | Unknown (52) | |
| Fed bacteria expressing transgene | Unknown (52) | |
| Environment | Unknown (52) | |
| Environment | Neurons (52) | |
| Environment | Skin (52) | |
| Environment | Unknown (52) | |
| Environment | Gut (52) | |
| Environment | Unknown (52) | |
| Fed bacteria expressing transgene | Unknown (52) | |
| Environment | Neurons (58) | |
| Planaria (Girardia Schmidtea) | Injection into head cavity | Eye (96) |
| Fed bacteria expressing transgene | Entire animal (84) | |
| Polyp (Hydra) | Fed bacteria expressing transgene | Entire animal (21) |
| Silkworm (Bombyx) | Injection into hemocoel | Midgut, silk glands (123) |
| Fruitfly (Drosophila) | Intra-abdominal injection | Neurons in the head (31) |
| Fleshfly (Sarcophaga) | Injection into larval hemolymph | Pupal hemocytes (86) |
| Mosquito (Aedes Anopheles) | Injection into hemocoel | Gut (11) |
| Thoracic injection | Mid-gut, fat body (14) | |
| Malaria parasite (Plasmodium) | Surrounding medium | Parasites within red-blood cells (77) |
| Beetle (Tribolium) | Intra-abdominal injection of pupae | Next generation (18) |
| Intra-abdominal injection of larvae | Adult bristles (129) | |
| Grasshopper (Schistocerca) | Injection into heart vessel of larvae | Developing eye (27) |
| Cricket (Gryllus) | Intra-abdominal injection | Next generation (82) |
| Locust (Locusta) | Intra-abdominal injection | Next generation (48) |
| Cockroach (Blatella Periplaneta) | Intra-abdominal injection | Ovariole (22) |
| Injection into cercus of larvae | Neurons and skin (79) | |
| Honey bee (Apis) | Injection into hemolymph | Insect fat body (6) |
| Tick (Amblyomma Ixodes) | Injection into hemolymph | Salivary gland (3) |
| Injection into hemolymph | Salivary gland (120) | |
| Feeding in droplet to larvae | Salivary gland (117) | |
| Milkweed bug (Oncopeltus) | Intra-abdominal injection | Next generation (72) |
| Wasp (Nasonia) | Intra-abdominal injection | Next generation (75) |
| Termite (Reticulitermes) | Injection into side of the thorax | Insect fat body (158) |
| Kissing bug (Rhodnius) | Thoracic injection of larvae | Salivary gland (8) |
| Moth (Spodoptera Manduca Epiphyas) | Injection into hemocoel | Midgut (97) |
| Intra-abdominal injection | Insect fat body (33) | |
| Fed droplets to larvae | Antennae of adults (132) | |
| Spider (Cupiennius Achaearanea) | Injection into perivitelline space of embryo | Developing appendages (111) |
| Injection into opisthosoma | Next generation (1) | |
| Mouse (Mus) | Modified siRNA injection into tail-vein | Liver, heart, kidney, adipose, and lung (118) |
| Plants | ||
| Tobacco (Nicotiana) | Silenced rootstock | Grafted scion (91) |
| Infected leaf | Entire plant (139) | |
| Sunflower (Helianthus) | Silenced rootstock | Grafted scion (49) |
| Leaf infiltrated with RNA from silenced plant | New leaves above the infiltrated leaf (49) | |
| Pumpkin, cucumber (Cucurbita, Cucumis) | Silenced pumpkin rootstock | Grafted cucumber scion (153) |
| Thale cress (Arabidopsis) | Induced expression of transgene | 2–3 upper leaves (44) |
| Legume (Medicago) | A. rhizogenes-mediated root transformation | Limited extent in nontransgenic shoot (70) |
| Fern (Ceratopteris) | Transgene bombarded into 1 cell | Entire gametophyte (107) |
Drosophila.
RNAi screens in cultured Drosophila S2 cells, where dsRNA is provided in the growth media, are widely practiced and have been extremely productive (80). These RNAi screens have even been used to identify factors required for the import of dsRNA into these cells, identifying genes that apparently function in endocytosis, that encode scavenger receptors, and that encode lipid-modifying enzymes (109, 134). It is not clear whether any of these genes function, like sid-1, to transport dsRNA across the membrane. However, these genes likely mediate the triggering of RNAi in adult flies by intra-abdominal injection of dsRNA (31). Despite the ability to initiate RNAi in distant tissues by these injections, intercellular transport of silencing information apparently does not occur when RNAi is triggered by transgenes that express dsRNA (104). This might be because intercellular transport of a silencing signal in Drosophila is regulated by environmental or physiological conditions, just as in C. elegans and plants. Alternatively, in contrast to C. elegans and plants, Drosophila lack RdRPs, which can amplify silencing signals, and such amplification of small amounts of transported dsRNA may be required for efficient RNAi and/or for subsequent export of dsRNA (see section below).
Mammals.
Mice and cultured human cells can import siRNA or chemically modified siRNA silencing triggers (87, 118). When overexpressed in cultured human cells, the human ortholog of the dsRNA channel SID-1 improves the ability of those cells to import siRNAs (30). This observation suggests that mammals may also take up dsRNA through the SID-1 channel when physiological conditions activate or cause overexpression of the channel. Additionally, cholesterol-coupled siRNAs as well as antagomirs (single-stranded RNAs antisense to microRNAs, which control gene expression) with stabilizing modifications injected directly into the tail vein of mice trigger detectable silencing of the target in many tissues (63, 118). However, it is currently unknown whether cholesterol-coupled siRNAs enter mouse cells through either of the two mouse homologs of the dsRNA channel SID-1. Analysis of mice lacking SID-1 homologs will likely reveal the physiological role of SID-1 in mice and will also be an invaluable tool to dissect the role of mammalian SID-1 in the transport of trigger dsRNA, siRNA or other silencing signals.
In contrast to RNAi in C. elegans, RNAi in mammals is not typically triggered using long dsRNAs as they can cause a nonsequence-specific innate immune response, called the interferon response, that is associated with viral infections (110). However, some forms of long dsRNA are tolerated by the immune system and do not activate a strong interferon response (103, 119, 131). Further, the potency of the interferon response induced by some long dsRNAs and siRNAs >23 bp is cell-type dependent (100). However, in some cell types such as mouse embryonic stem cells, sequence-specific silencing using long dsRNA is possible (13, 88, 149). These observations suggest that nonspecific and sequence-specific response pathways may be regulated and may coexist in mammals, as is the case in penaeid shrimp (102).
Can All Animals Transport Silencing Information Between Cells?
The lack of RNA-dependent RNA polymerases (RdRPs), which may amplify silencing, is often considered as evidence against the presence of a robust RNAi response and of intercellular transport of silencing information in some insects and vertebrates including mammals (104). However, recent studies suggest that in organisms that do not have an easily identifiable RdRP, DNA-directed RNA polymerases or other enzymes may perform a similar role (20). Both Drosophila and mice lack an identified RdRP, yet small RNAs called repeat-associated siRNA in the Drosophila germline (135) and piwi-interacting RNAs in mice (9, 38, 42, 65, 143a) accumulate in an asymmetric fashion that is reminiscent of synthesis by RdRPs. Furthermore, silencing of unpaired DNA during meiosis, a process that requires RdRP activity in both C. elegans (76) and in N. crassa (114), also occurs in mice (133). Notably, in C. elegans and in plants (see later section) the transport of a silencing signal does not require efficient RNAi. Therefore, in mammals and insects that lack RdRP-based signal amplification mechanisms, transport of silencing information between cells may occur even in the absence of robust cell-autonomous RNAi. Finally, the absence of observed systemic RNAi may reflect a requirement for specific environmental or physiological conditions. In C. elegans, intercellular spread of silencing information from body-wall muscles expressing gfp dsRNA is only detected upon induction by changes in the environment (128) and the extent of silencing in body-wall muscles in response to expression of dsRNA in pharyngeal cells is enhanced by starvation (146). In plants, similar physiological states can determine the extent of spread of silencing information (130).
IN PLANTS
Transport of Silencing Information Between Cells
The transport of silencing information between cells was first discovered in transgenic tobacco plants that spontaneously silenced both the transgene and the homologous endogenous gene (90, 92). In grafting experiments, the silenced state of rootstocks was transmitted to the grafted nonsilenced scions (91). Further, when a leaf of GFP-expressing tobacco plants was infiltrated with Agrobacterium cultures that carry a gfp transgene, GFP expression was silenced throughout the plant (139). Silencing in these cases is likely triggered by improperly processed sense RNAs (aberrant RNAs) made from the transgenes that are converted by cellular RdRPs into dsRNAs (37). For example, silencing by sense-only transgenes required the RdRP RDR6 in Arabidopsis (26, 83). Accordingly, expression of hairpin dsRNA from inverted-repeat transgenes circumvents this requirement for RdRPs in tobacco plants (113). Finally, the transport of silencing can be triggered using either 2025-nt RNAs purified from a silenced transgenic plant (49) or a single synthetic double-stranded siRNA (60), suggesting that the ultimate trigger for the transport of silencing information may be siRNAs in plants.
Plant cells transport silencing information to adjacent cells through intercellular pores called plasmodesmata and to distant cells through the vascular tissue called phloem (Figure 3). Most plant cells are connected with each other through dynamic pores called plasmodesmata (154), thus forming a cytoplasmic continuum. Mature guard cells that regulate gaseous exchange in the leaf are not part of the cytoplasmic continuum and consequently, transport of the silencing signal can cause silencing in guard cells only if the signal is received by immature guard cells that have not yet lost their plasmodesmatal connections (50, 141). Water and minerals are transported through xylem tubes, which are lined with dead cells, while photoassimilates, RNA, proteins, and silencing information are transported through phloem tubes, which are made of living enucleated sieve elements supported by companion cells. Transport through the phloem occurs from mature, photosynthetically autonomous organs (phloem sources) to new growth (phloem sink), and transport of silencing information over long distances also occurs from phloem source to phloem sink (23, 91, 130, 141).
Figure 3.
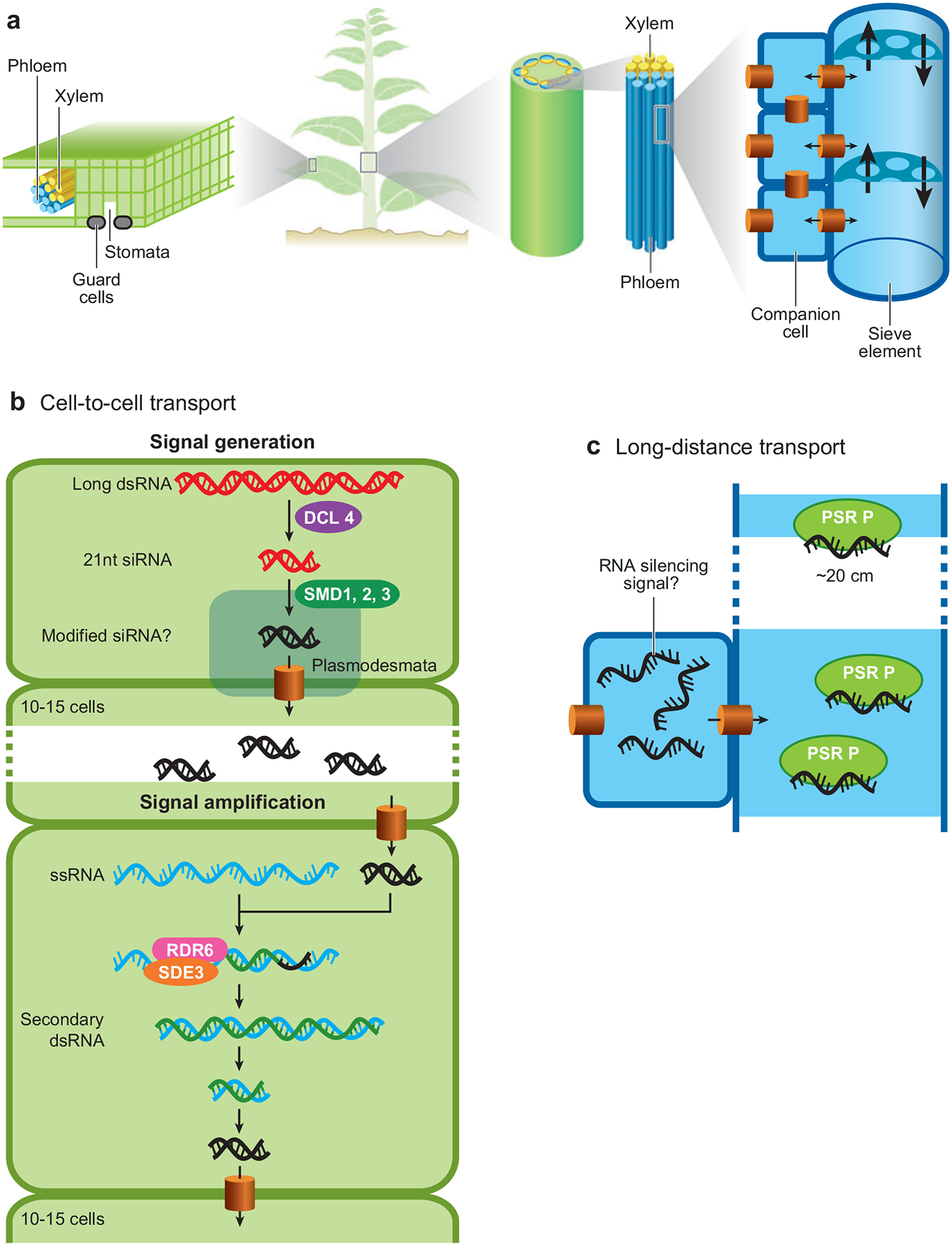
Plants use distinct mechanisms for cell-to-cell and long-distance transport of silencing information. (a) Schematic showing the organization of plant vasculature. Long-distance transport of silencing information occurs through the phloem, comprised of companion cells that support associated sieve elements. Cell-to-cell transport in leaves occurs through plasmodesmal connections, which connect the cytoplasm of all cells except guard cells that surround sites of gaseous exchange (stomata). (b) Model for the generation and amplification of a cell-to-cell silencing signal. The RNAi trigger, long dsRNA, is converted to 21-nt siRNAs by DCL4, and modification of these 21-nt siRNAs for transport and/or their transport through plasmodesmata is controlled by the silencing movement defective genes (SMD1, 2, 3). This signal can travel for ~10–15 cells without any amplification. However, in the presence of target mRNA, a cellular RNA-dependent RNA polymerase (RDR6) and an RNA helicase (SDE3) can direct the synthesis of secondary dsRNA, which may then be processed just as the trigger long dsRNA. (c) Model for the long-distance transport of a silencing signal. The silencing signal (likely RNA) may be transported as single-stranded short RNAs through the phloem since the phloem small RNA-binding protein 1 (PSRP1) of pumpkin specifically binds short single-stranded RNAs. Similar PSRPs may exist in other plants.
The physiological status and surrounding environment of a tissue dictates the extent of transport of silencing information. When silencing is initiated in single cells or small groups of cells, the movement of silencing information is restricted to about 10–15 cells in diameter in phloem source tissues such as mature leaves (50, 90). Conversely, in phloem sink tissues, such as new growth, silencing information progressively spreads from the phloem into the entire lamina of the leaves. Further, when mature leaves are converted into phloem sinks by shading, the transport of silencing information within the leaf becomes more extensive (130). Nutrient depletion may thus increase phloem flow bringing more silencing signals and nutrients to the leaf and/or may induce signal amplifiers such as RdRPs in the leaf.
Cell-to-Cell Transport of Silencing Information
Plant cells appear to use 21-nt siRNAs to transport silencing information through plasmodesmata. In the absence of amplification the spread of silencing is limited to ~10–15 neighboring cells (Figure 3), while more extensive spread of the silencing information can be achieved by a relay mechanism, whereby a silencing signal received by a cell in a leaf is amplified by the action of cellular RdRPs allowing additional cell-to-cell spreading within the leaf.
Limited cell-to-cell transport is observed when the endogenous gene SULPHUR is targeted for silencing by dsRNA expressed in phloem companion cells. Chlorosis (reduction in chlorophyll concentrations) due to RNAi of SULPHUR is observed in the companion cells and in the ~10–15 directly adjacent leaf cells. This limited spreading is unaltered in Arabidopsis mutants that lack the cellular RdRP, rdr6, suggesting signal amplification is not required (29, 50). In contrast, spreading is not observed in Dicer-like-4 (DCL4) mutants, which are deficient in the production of 21-nt siRNAs but not 24-nt siRNAs (29, 45). Furthermore, in an enhanced silencing mutant where the chlorosis spreads beyond 10–15 cells, thelevels of 21-nt siRNA are increased (29). Thus, the RNA component of the silencing signal transported through plasmodesmata is likely 21-nt siRNAs.
Additionally, mutants specifically defective in the transport of a silencing signal would be expected to lose the ability to spread chlorosis without losing the ability to generate 21-nt siRNAs. Screens for such silencing movement defective mutants (29) identified SMD1, SMD2, and SMD3 mutants, and the genes mutated in SMD mutants are currently unknown.
In contrast to the limited transport associated with silencing of endogenous genes, silencing of transgenes often spreads throughout the leaf lamina (presumably due to relay signal amplification). For example, expressing dsRNA matching the 5′ region of gfp in phloem companion cells caused uniform silencing of GFP in the entire leaf of transgenic Arabidopsis plants (50). However, in mutants lacking the cellular RdRP rdr6, GFP silencing (like silencing of endogenous genes) could be detected only in ~10–15 cells adjacent to the veins. This indicates that RdRP activity is required for long-range spread of silencing. Similar experiments implicate the putative RNA helicase SDE3 in the spread of silencing information. In these experiments, wild-type plants as well as rdr6 and sde3 mutant plants accumulated similar amounts of 21-nt and 24-nt siRNA matching the 5′ region of gfp that corresponds to the trigger dsRNA, as is typical for plants undergoing transgene-triggered RNAi (45, 46). However, siRNAs corresponding to the 3′ region of gfp (outside sequences matching trigger dsRNA) were exclusively 21-nt long and more of these 21-nt siRNAs accumulated in wild-type plants than in sde-3 or in rdr-6 mutants (50).
These results suggest a model (Figure 3) whereby long dsRNA is processed into 21-nt siRNAs by DCL4 during silencing. The movement of these 21-nt siRNAs through plasmodesmata is dependent on the silencing movement defective genes SMD1, 2, 3. This results in the spread of silencing to ~10–15 neighboring cells. RDR6 and SDE-3 in these neighboring cells amplify the signal, producing secondary dsRNA beyond the original trigger. These cells then process the secondary dsRNA and relay the silencing information further by spreading the 21-nt siRNAs. The extent of spread depends upon the physiological state of the tissue and this regulation may occur through modulation of RDR6 and SDE3 activity.
Long-Distance Transport of Silencing Information
Plants can also transport silencing information over long distances (~20 cm) through the phloem (91, 141). When a segment of a nontransgenic plant is grafted between silenced transgenic plants and nonsilenced transgenic plants, the silencing information is successfully transmitted through the nontransgenic tissue (91). Thus, aberrant transcripts made from transgenes are not required for propagation of the silencing signal via a relay mechanism as described for cell-to-cell transport through plasmodesmata. Furthermore, abolishing the expression of the tobacco ortholog of the RdRP RDR6 in plant stem sections does not prevent the transmission of the silencing signal through that section, showing that amplification is not required for transport of the silencing signal through the phloem (113). In contrast, reception of the phloem-transported silencing signal requires either abundant target molecules or amplification of the silencing signal (36, 93, 113).
The molecular mechanism that controls phloem-dependent transport of silencing information is largely unknown. Although in one study phloem-dependent transport was found to be strictly correlated with the presence of ~24-nt siRNAs (45), conflicting results suggest that phloem-dependent transport does not correlate with any RNA species (78). However, a phloem small RNA-binding protein, PSRP1, has been isolated biochemically from pumpkin phloem sap and found to bind small RNAs with high affinity (153). Although PSRP1 preferentially binds to small RNAs, the precise nature and size of the RNA bound by PSRP1 during the transport of silencing information is unknown. Due to the low complexity of the PSRP1 sequence, clear homologs have not been identified in Arabidopsis or in tobacco. However, similar small RNA binding proteins were detected in the phloem saps of cucumber and lupine plants, suggesting that small RNAs involved in long-distance silencing may be transported through the phloem as parts of ribonucleoprotein complexes. Thus, the sequence-specific silencing signal that is transported through the phloem as well as the pathway responsible for its biogenesis and transport remain largely unclear.
Lessons From Plant Viruses
RNAi functions in plants as an antiviral defense mechanism (68). Infection with a virus triggers RNA silencing directed against the virus, while plants defective in RNAi show enhanced susceptibility to viral infection. Additionally, and most convincingly, viruses encode virulence factors that inhibit RNAi. Many viral suppressors of RNAi differentially affect cell-autonomous RNAi and the intercellular transport of silencing information providing insight into the regulation of these processes.
The generation and transport of silencing information is not dependent on efficient cell-autonomous RNAi. In grafting experiments, HC-Pro protein of Potato virus Y strongly inhibits cell autonomous RNAi, yet does not affect the long-distance transport of silencing information to grafted scions (78). In contrast, the p25 protein of Potato virus X and the 2b protein of a weak strain of Cucumber mosaic virus do not effectively inhibit cell-autonomous RNAi, yet inhibit long-distance transport of silencing information (43, 140). Similarly, cell-to-cell and long-distance transport can be selectively inhibited by different viral proteins. The AC2 protein of African cassava mosaic virus eliminates cell-to-cell but not long-distance spread of silencing information, whereas the P1 protein of Rice yellow mottle virus eliminates long-distance but not cell-to-cell spread of silencing information (50). Finally, a GFP-tagged Turnip crinckle virus lacking the P38 suppressor protein was restricted to single cells from which the cell-to-cell transport of silencing information occurred but the long-distance transport of silencing information was inhibited (108).
Recent studies suggest that most plant viral suppressors can bind small RNAs and presumably suppress RNAi by sequestering them (64, 81). Similar studies that examine the biochemical functions of viral suppressor proteins may provide a mechanistic understanding of their effects on the intercellular transport of silencing information discussed above.
EVOLUTIONARILY SELECTED ROLE OF SILENCING TRANSPORT MACHINERY?
In plants, the silencing transport machinery appears to be part of a plant-wide antiviral response. In this paradigm, an infected plant cell sends a silencing signal ahead of the viral infection front to enable neighboring cells to mount a strong antiviral RNAi response and thus prevent the establishment of infection. Consistently, some plant viruses encode proteins that suppress the intercellular transport of silencing information during RNAi and such suppression is required for successful infection (68).
In animals, many components of the silencing transport machinery are highly conserved (109, 126, 146), suggesting that this process was selected for over evolutionary time. Although no selected roles for the silencing transport machinery in animals are known, a few speculative possibilities are presented below.
Sharing Silencing RNAs
Antiviral response.
Cell-autonomous RNAi has been suggested to be an antiviral defense mechanism in worms (74, 112, 145), in flies (137, 142), and in mosquitoes (54, 69). Unlike in invertebrates, it is difficult to evaluate whether RNAi is an antiviral mechanism in mammals (25) since dsRNA triggers a nonspecific response called the interferon response that causes degradation of all mRNA in somatic cells. Although several mammalian viruses express proteins that can inhibit RNAi in insect cells (69), some of these same proteins also inhibit the interferon response during viral infection of mammalian cells (53). This suggests that these viral proteins inhibit steps that are common to both the interferon and RNAi responses: e.g., recognition of dsRNA triggers. Furthermore, in mammalian cells, unlike in plants, viral infection does not inhibit RNAi (95). Thus, although the antiviral role (if any) of RNAi in mammals remains unclear, RNAi does appear to be an antiviral response in many invertebrates,
In contrast, it is unknown if the intercellular transport of silencing information during RNAi is part of an antiviral response in any animal, mainly because natural viruses that infect the well-studied animal model C. elegans are unknown and therefore studies akin to those described above in plants are not yet possible. The recent establishment of flock house virus (74), vesicular stomatitis virus (112, 145), and vaccinia virus (73) infection in C. elegans combined with the availability of well-characterized mutants defective in the intercellular transport of silencing such as sid-1 (146) could make it possible to evaluate if, as in plants, the transport of silencing is an antiviral response at least in some animals.
Noncell-autonomous control of gene regulation.
Endogenously made dsRNAs may also trigger the transport of gene regulatory information between cells. Most plants and animals express an abundant class of regulatory RNAs called micro RNAs (miRNAs). These RNAs are ~21 nt long, are made from long dsRNAs, and regulate development and behavior by directing the translational repression or cleavage of cellular mRNAs (61). In addition, there are many sources of sense-antisense dsRNAs in the genome of plants (15), C. elegans (66), mice (59), and humans (101, 151). These dsRNAs may regulate diverse cellular processes: e.g., endogenous siRNAs derived from a natural sense-antisense pair in Arabidopsis controls salt tolerance (15). In all cases examined so far, noncell-autonomous effects have not been uncovered for the action of miRNAs or natural dsRNAs. However, an appealing hypothesis is that dsRNAs and miRNAs that are expressed in cells exposed to the environment are transported in response to specific environmental changes. A rigorous evaluation of this hypothesis requires experiments exploring the expression pattern of miRNAs under various environmental conditions.
Sharing of Nonsilencing RNAs
The transport of silencing information between cells during RNAi may use part of the same machinery as that used by some animal cells to transport proteins, mRNAs, and microRNAs between cells. Cultured cells can control neighboring cells by releasing vesicles called microvesicles (99), in the case of mouse embryonic stem cells, or called exosomes (135a), in the case of mouse and human mast cells, that contain select proteins and RNAs. When neighboring cells import these microvesicles or exosomes, the mRNA enters the cytoplasm through an unknown mechanism and is translated into functional protein. Similar vesicles called argosomes carry signaling molecules to neighboring cells during early development and pattern formation (39). If, as proposed here, the intercellular transport machinery for silencing information is also required for the transport of such essential molecules, why are developmental defects not observed in mutants defective in the intercellular transport of silencing information? All genetic screens performed in C. elegans thus far have been capable of isolating only viable and fertile mutants. Therefore, additional screens designed to isolate lethal or sterile mutants may uncover such essential genes that also mediate the intercellular transport of silencing information.
In recent years, large-scale sequencing of small RNAs has led to the discovery of many different kinds of abundant noncoding RNAs: endogenous secondary siRNAs (89) and 21-U siRNAs (106) in C. elegans, repeat-associated siRNAs (rasiRNAs) (10) and piwi-interacting RNAs (piRNAs) (16) in Drosophila, and piRNAs in mammals (9, 38, 42, 65, 143a). Some of these RNAs appear to arise from dsRNA precursors, while others are found associated with components of the RNAi machinery, but their biogenesis and roles remain unclear. The transport of these endogenous small RNAs or their precursors could potentially be a selected function of the silencing transport machinery.
Experience-Dependent Inheritance
A hen is only an egg’s way of making another egg.
Samuel Butler
RNAi in C. elegans can be transmitted from somatic cells to the germline. Other examples of such epigenetic inheritance are also associated with regulatory double-stranded RNAs (156). For example, DNA methylation and/or heterochromatinization is associated with silenced copies of genes, and in mammals, the sex of the parent providing the chromosome affects which genes are silenced on that chromosome (imprinting) (148). Environmental or dietary changes in pregnant rats or mice, respectively, can also cause changes in DNA methylation and gene expression that are inherited stably through subsequent generations even in the absence of the initial dietary or environmental change (7, 24). Furthermore, in maize and in mice, the heterochromatinized state of certain dominant alleles can be transferred to homologous alleles apparently through the RdRP-dependent production of RNA from the dominant allele (19, 98). Thus, the silencing transport machinery may transport RNAs associated with heterochromatinization from somatic cells to the germline to confer an altered heterochromatin state and therefore altered gene expression in the next generation.
PERSPECTIVE
About a decade ago, Andrew Fire, Craig Mello, and coworkers made the seminal discovery that dsRNA is the trigger for a posttranscriptional gene silencing mechanism called RNAi (35). In addition to RNAi, dsRNA has been subsequently found to be involved in several diverse regulatory processes such as translational repression, transcriptional silencing, and DNA elimination. Similarly, the silencing information transported between cells during RNAi is perhaps just one of many forms of sequence-specific communication between cells that enable multicellular organisms to develop and function coordinately.
RNAi: RNA interference
dsRNA: double-stranded RNA
siRNA: short-interfering RNA
Noncell autonomous effects: effects in naïve cells resulting from processes such as RNAi in other cells
Hairpin RNA: a form of dsRNA typically generated when the mRNA product of an inverted-repeat sequence folds back on itself
RdRP: RNA-directed RNA Polymerase
SID: systemic RNAi defective
Inverted-repeat sequence: a pair of linked DNA sequences that are the reversed complement of each other
Heterochromatinization: the condensation of transcriptionally active euchromatin into silenced heterochromatin
Grafting: the process of joining a stem or a bud of one plant (scion) to the stem of another (rootstock)
Epigenetic inheritance: transmission of information from one generation to the next without encoding it in the DNA sequence
RNAi IN C. ELEGANS.
A current model of cell-autonomous RNAi that incorporates key recent discoveries is presented below (28, 89, 106, 116, 152). When trigger dsRNA enters a cell, it is processed into ~21-nt primary siRNAs by the RNase III-like enzyme DCR-1 (62). Primary siRNAs complementary to target mRNA are bound by the RNase H-like primary Argonaute RDE-1, which is part of the RNA-induced silencing complex (RISC). Subsequent cleavage of target mRNA by the primary siRNA-programmed RISC recruits a cellular RNA-directed RNA Polymerase (RdRP) such as RRF-1 to produce ~21-nt secondary siRNA by target mRNA-directed unprimed synthesis. These secondary siRNAs associate with secondary Argonautes such as SAGO-1, which likely require additional proteins for activity. Subsequent steps that eventually lead to silencing are still unclear. An analogous parallel pathway processes dsRNAs endogenously made within the cell for silencing. For example, endogenous primary siRNAs bind the primary Argonaute ERGO-1 and endogenous secondary siRNAs are likely generated by the RdRP RRF-3. However, secondary siRNAs generated from both endogenous and exogenous pathways compete for association with the same secondary Argonautes. Thus, RNAi triggered by exogenously introduced dsRNA competes with RNAi triggered by endogenous dsRNA. Additional RNAi components (41, 57) have been omitted from the above model for simplicity.
SUMMARY POINTS.
In eukaryotic cells, double-stranded RNA triggers the silencing of genes with matching sequences through a process called RNA interference. In plants and in some animals, this sequence-specific silencing information is transported between cells. As a result, the same genes are silenced even in distant cells that did not initially encounter the double-stranded RNA.
Studies in the animal model C. elegans suggest that a form of long double-stranded RNA likely carries the sequence-specific information between cells during RNAi. Specific endocytosis machinery and a nucleic acid channel, SID-1, control the entry of the silencing signal into cells. The precise identity of the silencing signal and how it is exported out of cells are unknown.
Plants use two different mechanisms, both distinct from that of C. elegans, to transport silencing information. First, the silencing information (likely in the form of 21-nt siRNA) is transported to adjacent cells through dynamic intercellular bridges called plasmodesmata. This form of transport has a range of ~10–15 cells, but can extend further if the transported signal is amplified. In a second mechanism, an unknown silencing signal can be transported over long (~20 cm) distances through the phloem vascular tissue.
RNAi acts as an antiviral mechanism in plants and at least in some animals. Several plant viruses encode proteins that suppress the transport of silencing information between cells, suggesting that intercellular transport of silencing evolved as a defense mechanism against viruses. A similar role for intercellular transport of silencing has not been shown in animals.
In addition to an antiviral role, several speculative roles can be envisioned for the transport of sequence-specific information between animal cells: e.g., noncell-autonomous gene regulation, sharing regulatory RNAs among cells during times of stress, conveying somatic experience to the germline, etc. Further experiments are required to determine which of these, if any, are the evolutionarily selected roles of the silencing transport machinery.
FUTURE ISSUES.
What is the biochemical nature of the silencing signal transported between cells? Is it the RNAi trigger dsRNA or some other modified/derived signal? How is this signal produced and regulated?
Why does the efficiency of transport of silencing information between cells vary with different trigger dsRNAs? Is dsRNA from the environment transported differently from dsRNA made within the cells of an organism? How is the transport of silencing between cells regulated?
What is the evolutionarily selected function of the intercellular transport machinery? Is it to transport silencing information or was this machinery selected for another role?
Do mammals transport RNAs between cells?
ACKNOWLEDGMENTS
We thank Daniel Chase, Edith Myers, Jessica Tanis, and members of the Hunter lab, particularly, Jacqueline Brooks, Daniel Schott, Jessica Smith, and Shai Shen-Orr for critical comments. We apologize for being unable to include the work of some colleagues due to space restrictions. This work was supported by awards from the American Heart Association to A.M.J., and from NSF (MCB 110452) and NIH (GM069891) to C.P.H.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Akiyama-Oda Y, Oda H. 2006. Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development 133:2347–57 [DOI] [PubMed] [Google Scholar]
- 2.Alder MN, Dames S, Gaudet J, Mango SE. 2003. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 9:25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aljamali MN, Bior AD, Sauer JR, Essenberg RC. 2003. RNA interference in ticks: a study using histamine binding protein dsRNA in the female tick Amblyomma americanum. Insect Mol. Biol 12:299–305 [DOI] [PubMed] [Google Scholar]
- 4.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, et al. 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442:295–98 [DOI] [PubMed] [Google Scholar]
- 5.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol 13:807–18 [DOI] [PubMed] [Google Scholar]
- 6.Amdam GV, Simoes ZL, Guidugli KR, Norberg K, Omholt SW. 2003. Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnol 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araujo RN, Santos A, Pinto FS, Gontijo NF, Lehane MJ, Pereira MH. 2006. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol 36:683–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–7 [DOI] [PubMed] [Google Scholar]
- 10.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, et al. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337–50 [DOI] [PubMed] [Google Scholar]
- 11.Bartholomay LC, Fuchs JF, Cheng LL, Beck ET, Vizioli J, et al. 2004. Reassessing the role of defensin in the innate immune response of the mosquito, Aedes aegypti. Insect Mol. Biol 13:125–32 [DOI] [PubMed] [Google Scholar]
- 12.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–66 [DOI] [PubMed] [Google Scholar]
- 13.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 98:14428–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. 2002. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep 3:852–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. 2006. Endogenous siRNAs derived from a pair of natural cis antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–103 [DOI] [PubMed] [Google Scholar]
- 17.Briese M, Esmaeili B, Johnson NM, Sattelle DB. 2006. pWormgatePro enables promoter-driven knockdown by hairpin RNA interference of muscle and neuronal gene products in Caenorhabditis elegans. Invert. Neurosci 6:5–12 [DOI] [PubMed] [Google Scholar]
- 18.Bucher G, Scholten J, Klingler M. 2002. Parental RNAi in Tribolium (Coleoptera). Curr. Biol 12:R85–86 [DOI] [PubMed] [Google Scholar]
- 19.Chandler VL. 2007. Paramutation: from maize to mice. Cell 128:641–45 [DOI] [PubMed] [Google Scholar]
- 20.Chang J, Taylor J. 2002. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J 21:157–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, et al. 2006. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J. Cell Sci 119:846–57 [DOI] [PubMed] [Google Scholar]
- 22.Ciudad L, Piulachs MD, Belles X. 2006. Systemic RNAi of the cockroach vitellogenin receptor results in a phenotype similar to that of the Drosophila yolkless mutant. FEBS J 273:325–35 [DOI] [PubMed] [Google Scholar]
- 23.Crete P, Leuenberger S, Iglesias VA, Suarez V, Schob H, et al. 2001. Graft transmission of induced and spontaneous post-transcriptional silencing of chitinase genes. Plant J 28:493–501 [DOI] [PubMed] [Google Scholar]
- 24.Cropley JE, Suter CM, Beckman KB, Martin DIK. 2006. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc. Natl. Acad. Sci. USA 103:17308–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen BR. 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol 7:563–67 [DOI] [PubMed] [Google Scholar]
- 26.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543–53 [DOI] [PubMed] [Google Scholar]
- 27.Dong Y, Friedrich M. 2005. Nymphal RNAi: systemic RNAi mediated gene knockdown in juvenile grasshopper. BMC Biotechnol 5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124:343–54 [DOI] [PubMed] [Google Scholar]
- 29.Dunoyer P, Himber C, Voinnet O. 2005. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet 37:1356–60 [DOI] [PubMed] [Google Scholar]
- 30.Duxbury MS, Ashley SW, Whang EE. 2005. RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem. Biophys. Res. Commun 331:459–63 [DOI] [PubMed] [Google Scholar]
- 31.Dzitoyeva S, Dimitrijevic N, Manev H. 2003. Gamma-aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: adult RNA interference and pharmacological evidence. Proc. Natl. Acad. Sci. USA 100:5485–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20:6877–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eleftherianos I, Millichap PJ, ffrench-Constant RH, Reynolds SE. 2006. RNAi suppression of recognition protein mediated immune responses in the tobacco hornworm Manduca sexta causes increased susceptibility to the insect pathogen Photorhabdus. Dev. Comp. Immunol 30:1099–107 [DOI] [PubMed] [Google Scholar]
- 34.Feinberg EH, Hunter CP. 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301:1545–47 [DOI] [PubMed] [Google Scholar]
- 35.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–11 [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Perez RD, Houdt HV, Depicker A. 2004. Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J 38:594–602 [DOI] [PubMed] [Google Scholar]
- 37.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. 2004. A link between mRNA turnover and RNA interference in Arabidopsis. Science 306:1046–48 [DOI] [PubMed] [Google Scholar]
- 38.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202 [DOI] [PubMed] [Google Scholar]
- 39.Greco V, Hannus M, Eaton S. 2001. Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell 106:633–45 [DOI] [PubMed] [Google Scholar]
- 40.Grishok A, Tabara H, Mello CC. 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287:2494–97 [DOI] [PubMed] [Google Scholar]
- 41.Grishok A 2005. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett 579:5932–39 [DOI] [PubMed] [Google Scholar]
- 42.Grivna ST, Beyret E, Wang Z, Lin H. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20:1709–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo HS, Ding SW. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J 21:398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo HS, Fei JF, Xie Q, Chua NH. 2003. A chemical-regulated inducible RNAi system in plants. Plant J 34:383–92 [DOI] [PubMed] [Google Scholar]
- 45.Hamilton A, Voinnet O, Chappell L, Baulcombe D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J 21:4671–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamilton AJ, Baulcombe DC. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–52 [DOI] [PubMed] [Google Scholar]
- 47.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146–50 [DOI] [PubMed] [Google Scholar]
- 48.He ZB, Cao YQ, Yin YP, Wang ZK, Chen B, et al. 2006. Role of hunchback in segment patterning of Locusta migratoria manilensis revealed by parental RNAi. Dev. Growth Differ 48:439–45 [DOI] [PubMed] [Google Scholar]
- 49.Hewezi T, Alibert G, Kallerhoff J. 2005. Local infiltration of high- and low-molecular-weight RNA from silenced sunflower (Helianthus annuus L.) plants triggers post-transcriptional gene silencing in nonsilenced plants. Plant Biotechnol. J 3:81–89 [DOI] [PubMed] [Google Scholar]
- 50.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22:4523–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deleted in Proof.
- 52.Kalinna BH, Brindley PJ. 2007. Manipulating the manipulators: advances in parasitic helminth transgenesis and RNAi. Trends Parasitol 23:197–204 [DOI] [PubMed] [Google Scholar]
- 53.Katze MG, He Y, Gale M Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol 2:675–87 [DOI] [PubMed] [Google Scholar]
- 54.Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. 2004. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 101:17240–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy S, Wang D, Ruvkun G. 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427:645–49 [DOI] [PubMed] [Google Scholar]
- 56.Kennerdell JR, Carthew RW. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017–26 [DOI] [PubMed] [Google Scholar]
- 57.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, et al. 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308:1164–67 [DOI] [PubMed] [Google Scholar]
- 58.Kimber MJ, McKinney S, McMaster S, Day TA, Fleming CC, Maule AG. 2007. flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J 21:1233–43 [DOI] [PubMed] [Google Scholar]
- 59.Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. 2005. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res 15:463–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins F Jr. 2002. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA 99:11981–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kloosterman WP, Plasterk RH. 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell 11:441–50 [DOI] [PubMed] [Google Scholar]
- 62.Knight SW, Bass BL. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293:2269–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, et al. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–89 [DOI] [PubMed] [Google Scholar]
- 64.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, et al. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 25:2768–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, et al. 2006. Characterization of the piRNA complex from rat testes. Science 313:363–67 [DOI] [PubMed] [Google Scholar]
- 66.Lee RC, Hammell CM, Ambros V. 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12:589–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. 2004. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci 117:9–18 [DOI] [PubMed] [Google Scholar]
- 68.Li F, Ding SW. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol 60:503–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li WX, Li H, Lu R, Li F, Dus M, et al. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Limpens E, Ramos J, Franken C, Raz V, Compaan B, et al. 2004. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot 55:983–92 [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–41 [DOI] [PubMed] [Google Scholar]
- 72.Liu PZ, Kaufman TC. 2004. hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 131:1515–27 [DOI] [PubMed] [Google Scholar]
- 73.Liu WH, Lin YL, Wang JP, Liou W, Hou RF, et al. 2006. Restriction of vaccinia virus replication by a ced-3 and ced-4-dependent pathway in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103:4174–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, et al. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436:1040–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. 2006. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439:728–32 [DOI] [PubMed] [Google Scholar]
- 76.Maine EM, Hauth J, Ratliff T, Vought VE, She X. 2005. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr. Biol 15:1972–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malhotra P, Dasaradhi PV, Kumar A, Mohmmed A, Agrawal N, et al. 2002. Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and −2) of Plasmodium falciparum. Mol. Microbiol 45:1245–54 [DOI] [PubMed] [Google Scholar]
- 78.Mallory AC, Mlotshwa S, Bowman LH, Vance VB. 2003. The capacity of transgenic tobacco to send a systemic RNA silencing signal depends on the nature of the inducing transgene locus. Plant J 35:82–92 [DOI] [PubMed] [Google Scholar]
- 79.Marie B, Bacon JP, Blagburn JM. 2000. Double-stranded RNA interference shows that Engrailed controls the synaptic specificity of identified sensory neurons. Curr. Biol 10:289–92 [DOI] [PubMed] [Google Scholar]
- 80.Mathey-Prevot B, Perrimon N. 2006. Drosophila genome-wide RNAi screens: Are they delivering the promise? Cold Spring Harbor Symp. Quant. Biol 71:141–48 [DOI] [PubMed] [Google Scholar]
- 81.Merai Z, Kerenyi Z, Kertesz S, Magna M, Lakatos L, Silhavy D. 2006. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol 80:5747–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mito T, Sarashina I, Zhang H, Iwahashi A, Okamoto H, et al. 2005. Non-canonical functions of hunchback in segment patterning of the intermediate germ cricket Gryllus bimaculatus. Development 132:2069–79 [DOI] [PubMed] [Google Scholar]
- 83.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–42 [DOI] [PubMed] [Google Scholar]
- 84.Newmark PA, Reddien PW, Cebria F, Sanchez Alvarado A. 2003. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA 100(Suppl. 1):11861–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ngo H, Tschudi C, Gull K, Ullu E. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishikawa T, Natori S. 2001. Targeted disruption of a pupal hemocyte protein of Sarcophaga by RNA interference. Eur. J. Biochem 268:5295–99 [DOI] [PubMed] [Google Scholar]
- 87.Overhoff M, Sczakiel G. 2005. Phosphorothioate-stimulated uptake of short interfering RNA by human cells. EMBO Rep 6:1176–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paddison PJ, Caudy AA, Hannon GJ. 2002. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 99:1443–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pak J, Fire A. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315:241–44 [DOI] [PubMed] [Google Scholar]
- 90.Palauqui JC, Elmayan T, De Borne FD, Crete P, Charles C, Vaucheret H. 1996. Frequencies, timing, and spatial patterns of co-suppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol 112:1447–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to nonsilenced scions. EMBO J 16:4738–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Palauqui JC, Vaucheret H. 1995. Field trial analysis of nitrate reductase cosuppression: a comparative study of 38 combinations of transgene loci. Plant Mol. Biol 29:149–59 [DOI] [PubMed] [Google Scholar]
- 93.Palauqui JC, Vaucheret H. 1998. Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95:9675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parrish S, Fire A. 2001. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7:1397–402 [PMC free article] [PubMed] [Google Scholar]
- 95.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–76 [DOI] [PubMed] [Google Scholar]
- 96.Pineda D, Gonzalez J, Callaerts P, Ikeo K, Gehring WJ, Salo E. 2000. Searching for the prototypic eye genetic network: Sine oculis is essential for eye regeneration in planarians. Proc. Natl. Acad. Sci. USA 97:4525–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. 2002. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J. Biol. Chem 277:46849–51 [DOI] [PubMed] [Google Scholar]
- 98.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. 2006. RNA-mediated nonmendelian inheritance of an epigenetic change in the mouse. Nature 441:469–74 [DOI] [PubMed] [Google Scholar]
- 99.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, et al. 2006. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20:847–56 [DOI] [PubMed] [Google Scholar]
- 100.Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, et al. 2006. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA 12:988–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.RIKEN Genome Explor. Res. Group, Genome Sci. Group (Genome Network Project Core Group), FANTOM Consort. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564–66 [DOI] [PubMed] [Google Scholar]
- 102.Robalino J, Bartlett T, Shepard E, Prior S, Jaramillo G, et al. 2005. Double-stranded RNA induces sequence-specific antiviral silencing in addition to nonspecific immunity in a marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response? J. Virol 79:13561–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robbins MA, Li M, Leung I, Li H, Boyer DV, et al. 2006. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat. Biotechnol 24:566–71 [DOI] [PubMed] [Google Scholar]
- 104.Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C. 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9:299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Romano N, Macino G. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol 6:3343–53 [DOI] [PubMed] [Google Scholar]
- 106.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127:1193–207 [DOI] [PubMed] [Google Scholar]
- 107.Rutherford G, Tanurdzic M, Hasebe M, Banks JA. 2004. A systemic gene silencing method suitable for high throughput, reverse genetic analyses of gene function in fern gametophytes. BMC Plant Biol 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ryabov EV, van Wezel R, Walsh J, Hong Y. 2004. Cell-to-cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J. Virol 78:3149–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, et al. 2006. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol 8:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev 14:778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schoppmeier M, Damen WGM. 2001. Double-stranded RNA interference (RNAi) in the spider Cupiennius salei: the role of Distal-less is evolutionary conserved. Dev. Genes Evol 211:76–82 [DOI] [PubMed] [Google Scholar]
- 112.Schott DH, Cureton DK, Whelan SP, Hunter CP. 2005. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102:18420–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwach F, Vaistij FE, Jones L, Baulcombe DC. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138:1842–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–16 [DOI] [PubMed] [Google Scholar]
- 115.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, et al. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465–76 [DOI] [PubMed] [Google Scholar]
- 116.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315:244–47 [DOI] [PubMed] [Google Scholar]
- 117.Soares CA, Lima CM, Dolan MC, Piesman J, Beard CB, Zaidner NS. 2005. Capillary feeding of specific dsRNA induces silencing of the isac gene in nymphal Ixodes scapularis ticks. Insect Mol. Biol 14:443–52 [DOI] [PubMed] [Google Scholar]
- 118.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432:173–78 [DOI] [PubMed] [Google Scholar]
- 119.Strat A, Gao L, Utsuki T, Cheng B, Nuthalapaty S, et al. 2006. Specific and nontoxic silencing in mammalian cells with expressed long dsRNAs. Nucleic Acids Res 34:3803–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sukumaran B, Narasimhan S, Anderson JF, DePonte K, Marcantonio N, et al. 2006. An Ixodes scapularis protein required for survival of Anaplasma phagocytophilum in tick salivary glands. J. Exp. Med 203:1507–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tabara H, Grishok A, Mello CC. 1998. RNAi in C. elegans: soaking in the genome sequence. Science 282:430–31 [DOI] [PubMed] [Google Scholar]
- 122.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, et al. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123–32 [DOI] [PubMed] [Google Scholar]
- 123.Tabunoki H, Higurashi S, Ninagi O, Fujii H, Banno Y, et al. 2004. A carotenoid-binding protein (CBP) plays a crucial role in cocoon pigmentation of silkworm (Bombyx mori) larvae. FEBS Lett 567:175–78 [DOI] [PubMed] [Google Scholar]
- 124.Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet 24:180–83 [DOI] [PubMed] [Google Scholar]
- 125.Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RH. 2002. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295:694–97 [DOI] [PubMed] [Google Scholar]
- 126.Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH. 2004. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol 14:111–16 [DOI] [PubMed] [Google Scholar]
- 127.Timmons L, Fire A. 1998. Specific interference by ingested dsRNA. Nature 395:854. [DOI] [PubMed] [Google Scholar]
- 128.Timmons L, Tabara H, Mello CC, Fire AZ. 2003. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell 14:2972–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tomoyasu Y, Denell RE. 2004. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol 214:575–78 [DOI] [PubMed] [Google Scholar]
- 130.Tournier B, Tabler M, Kalantidis K. 2006. Phloem flow strongly influences the systemic spread of silencing in GFP Nicotiana benthamiana plants. Plant J 47:383–94 [DOI] [PubMed] [Google Scholar]
- 131.Tran N, Raponi M, Dawes IW, Arndt GM. 2004. Control of specific gene expression in mammalian cells by coexpression of long complementary RNAs. FEBS Lett 573:127–34 [DOI] [PubMed] [Google Scholar]
- 132.Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, Newcomb RD. 2006. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol 15:383–91 [DOI] [PubMed] [Google Scholar]
- 133.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, et al. 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet 37:41–47 [DOI] [PubMed] [Google Scholar]
- 134.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, et al. 2006. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem 281:14370–75 [DOI] [PubMed] [Google Scholar]
- 135.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313:320–24 [DOI] [PubMed] [Google Scholar]
- 135a.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, et al. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9:654–59 [DOI] [PubMed] [Google Scholar]
- 136.van Blokland N, van der Geest N, Mol JNM, Kooter JM. 1994. Transgenemediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J 6:861–77 [Google Scholar]
- 137.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, et al. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20:2985–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. 2006. Gene expression: long-term gene silencing by RNAi. Nature 442:882. [DOI] [PubMed] [Google Scholar]
- 139.Voinnet O, Baulcombe DC. 1997. Systemic signaling in gene silencing. Nature 389:553. [DOI] [PubMed] [Google Scholar]
- 140.Voinnet O, Lederer C, Baulcombe DC. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157–67 [DOI] [PubMed] [Google Scholar]
- 141.Voinnet O, Vain P, Angell S, Baulcombe DC. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95:177–87 [DOI] [PubMed] [Google Scholar]
- 142.Wang XH, Aliyari R, Li WX, Li HW, Kim K, et al. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312:452–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wasiak S, Legendre-Guillemin V, Puertollano R, Blondeau F, Girard M, et al. 2002. Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol 158:855–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143a.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, et al. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20:1732–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Waterhouse PM, Graham MW, Wang MB. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95:13959–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. 2005. RNA interference is an antiviral defense mechanism in Caenorhabditis elegans. Nature 436:1044–47 [DOI] [PubMed] [Google Scholar]
- 146.Winston WM, Molodowitch C, Hunter CP. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456–59 [DOI] [PubMed] [Google Scholar]
- 147.Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 104:10565–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang PK, Kuroda MI. 2007. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell 128:777–86 [DOI] [PubMed] [Google Scholar]
- 149.Yang S, Tutton S, Pierce E, Yoon K. 2001. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell Biol 21:7807–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yao MC, Chao JL. 2005. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet 39:537–59 [DOI] [PubMed] [Google Scholar]
- 151.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol 21:379–86 [DOI] [PubMed] [Google Scholar]
- 152.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, et al. 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127:747–57 [DOI] [PubMed] [Google Scholar]
- 153.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, et al. 2000. A systemic small RNA signaling system in plants. Plant Cell 16:1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zambryski P 2004. Cell-to-cell transport of proteins and fluorescent tracers via plasmodesmata during plant development. J. Cell. Biol 164:165–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zamore PD, Tuschl T, Sharp PA, Bartel DP. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25–33 [DOI] [PubMed] [Google Scholar]
- 156.Zaratiegui M, Irvine DV, Martienssen RA. 2007. Noncoding RNAs and gene silencing. Cell 128:763–76 [DOI] [PubMed] [Google Scholar]
- 157.Zhao G, Chang KY, Varley K, Stormo GD. 2007. Evidence for active maintenance of inverted repeat structures identified by a comparative genomic approach. PLoS ONE 2:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou X, Oi FM, Scharf ME. 2006. Social exploitation of hexamerin: RNAi reveals a major caste-regulatory factor in termites. Proc. Natl. Acad. Sci. USA 103:4499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]


