Abstract
Aims
To investigate whether metabolic signature composed of multiple plasma metabolites can be used to characterize adherence and metabolic response to the Mediterranean diet and whether such a metabolic signature is associated with cardiovascular disease (CVD) risk.
Methods and results
Our primary study cohort included 1859 participants from the Spanish PREDIMED trial, and validation cohorts included 6868 participants from the US Nurses’ Health Studies I and II, and Health Professionals Follow-up Study (NHS/HPFS). Adherence to the Mediterranean diet was assessed using a validated Mediterranean Diet Adherence Screener (MEDAS), and plasma metabolome was profiled by liquid chromatography-tandem mass spectrometry. We observed substantial metabolomic variation with respect to Mediterranean diet adherence, with nearly one-third of the assayed metabolites significantly associated with MEDAS (false discovery rate < 0.05). Using elastic net regularized regressions, we identified a metabolic signature, comprised of 67 metabolites, robustly correlated with Mediterranean diet adherence in both PREDIMED and NHS/HPFS (r = 0.28–0.37 between the signature and MEDAS; P = 3 × 10−35 to 4 × 10−118). In multivariable Cox regressions, the metabolic signature showed a significant inverse association with CVD incidence after adjusting for known risk factors (PREDIMED: hazard ratio [HR] per standard deviation increment in the signature = 0.71, P < 0.001; NHS/HPFS: HR = 0.85, P = 0.001), and the association persisted after further adjustment for MEDAS scores (PREDIMED: HR = 0.73, P = 0.004; NHS/HPFS: HR = 0.85, P = 0.004). Further genome-wide association analysis revealed that the metabolic signature was significantly associated with genetic loci involved in fatty acids and amino acids metabolism. Mendelian randomization analyses showed that the genetically inferred metabolic signature was significantly associated with risk of coronary heart disease (CHD) and stroke (odds ratios per SD increment in the genetically inferred metabolic signature = 0.92 for CHD and 0.91 for stroke; P < 0.001).
Conclusions
We identified a metabolic signature that robustly reflects adherence and metabolic response to a Mediterranean diet, and predicts future CVD risk independent of traditional risk factors, in Spanish and US cohorts.
Keywords: Mediterranean diet, Metabolomics, Dietary metabolic response, Cardiovascular disease, Mendelian randomization analysis, Risk prediction
See page 2657 for the editorial comment on this article (doi: 10.1093/eurheartj/ehaa260)
Introduction
The 2015–20 Dietary Guidelines for Americans1 recommend the Mediterranean diet as an important and cost-effective strategy for the prevention of cardiovascular disease (CVD)—the cause of one-third of deaths globally.2 The Mediterranean diet is characterized by high consumption of fruits, vegetables, seafood, nuts, legumes, whole grains, and olive oil, moderate intake of wine mainly within meals, and lower intake of red/processed meats, saturated fat, and sugary desserts and beverages.3 Compared with individual foods/nutrients, dietary patterns reflect a person’s habitual diet, and incorporate the synergistic and accumulative effects of various dietary components, thereby minimizing confounding by individual dietary factors. Although the health benefits of adhering to the Mediterranean diet have been demonstrated by intervention trials and prospective cohort studies,3–6 individuals’ metabolic responses to the same diet may vary, and the underlying mechanisms are not completely understood.
Diet may contribute to disease risk through modulation of metabolic pathways and homeostasis7 , 8; therefore, differences in metabolic responses to diet may explain some individual variations in diet-disease associations. However, systematic evaluations of adherence and metabolic responses to complex dietary patterns have been challenging. Traditional dietary surveys, such as food frequency questionnaires (FFQs) and dietary recalls, focus on assessing dietary intakes and are prone to measurement or reporting errors.9 Validated biological markers may represent an alternative approach for measuring dietary intakes (e.g. fatty acids in biospecimens for measuring fat intake), but they are only known for a few dietary factors.
Recent advances in high-throughput metabolomics profiling open a new avenue for developing complementary strategies to evaluate diet and its clinical relevance.10 By measuring intermediate molecules and products of metabolism, metabolomic profiles reflect individuals’ dietary intakes and other sources of variability in metabolism (e.g. genetic variations11 and diet-gut microbiome interplay7) and may, therefore, objectively assess the overall adherence and metabolic responses to complex dietary patterns.10 , 12 Prior studies have identified some metabolites associated with intakes of specific foods/nutrients,13–15 adherence to a few dietary patterns,10 , 13 , 16–18 and CVD risk.19 , 20 However, it remains unknown whether there are inter-individual variations in the metabolome for individuals consuming the Mediterranean diet and whether such metabolomic variations are associated with disease risk.
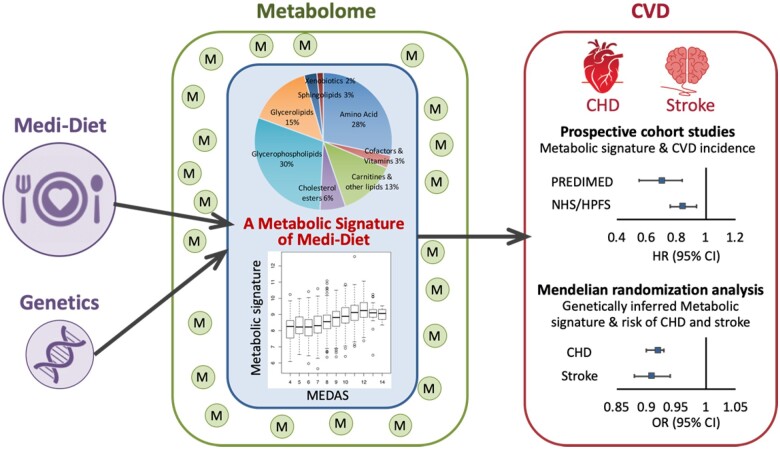
In the present study, leveraging dietary, metabolomic, and clinical data in the Spanish PREDIMED trial (Prevención con Dieta Mediterránea)3 and three US prospective cohorts (the Nurses’ Health Study [NHS], NHSII, and Health Professionals Follow-Up Study [HPFS]) (Figure 1), we examined metabolomic variations in relation to Mediterranean diet adherence, and identified and validated a metabolic signature reflecting adherence to a Mediterranean diet. We then assessed whether this signature was associated with CVD risk independently of known cardiovascular risk factors. In secondary analyses, we further examined genetic variants associated with the metabolic signature and whether such a genetic component of the signature was associated with CVD risk (Take home figure; Figure 2 and Supplementary material online, Figure S1).
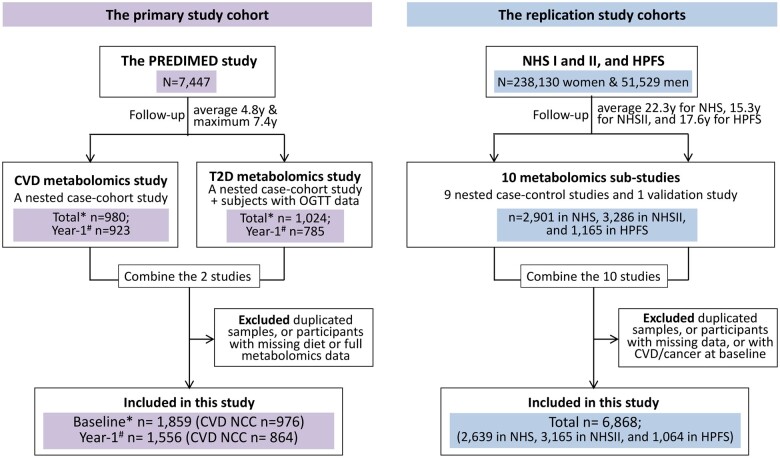
Figure 1.
Flowchart of study cohorts. For PREDIMED study, * indicates the total number of participants with complete diet and metabolomics data at baseline, and # is the number of participants with repeated measurements at year-1 visit. CVD NCC, the nested case-cohort study for incident cardiovascular disease.
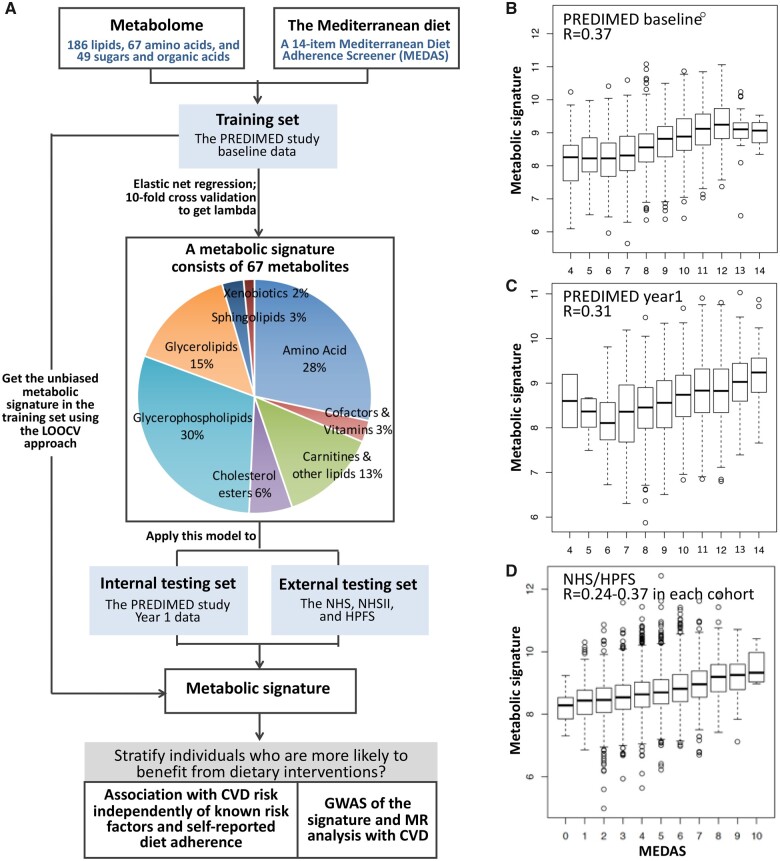
Figure 2.
The metabolic signature for adherence to the Mediterranean diet: flow chart for analytic approach and validation. (A) The training and testing procedures of a metabolic signature for the Mediterranean diet adherence screener (MEDAS). (B) The metabolic signature was trained using PREDIMED baseline measurements and tested with PREDIMED year-1 (interval validation) and the NHS/HPFS baseline measurements (external validation). The correlations between MEDAS and the 67-metabolite signature in the two validation sets are shown in (C) and (D). In the training set, in order to include the training dataset in association analyses, we used the leave-one-out-cross validation (LOOCV) approach to acquire an unbiased metabolic signature score; and the correlation between MEDAS and the LOOCV-derived score is presented in (B).
Methods
Study participants
Our primary study cohort was PREDIMED, a multicentre trial examining the efficacy of two Mediterranean diet interventions over a control diet, for primary prevention of CVD (primary outcome)3 and type 2 diabetes (T2D; secondary outcome of the trial).21 Two nested case-cohort studies were designed for metabolomics profiling22 , 23: the PREDIMED-CVD study that consisted of 229 incident CVD cases and 788 sub-cohort participants (overlapping n = 37),22 and the PREDIMED-T2D study containing 251 incident T2D cases and 641 sub-cohort participants (overlapping n = 53) without T2D at baseline.23 Participants with complete data on oral glucose tolerance (n = 132) were also included in PREDIMED-T2D. Data from PREDIMED-CVD and PREDIMED-T2D studies were combined for developing the metabolic signature model. We excluded individuals with missing data on baseline diet or named metabolites, and the PREDIMED-T2D record if the same individual was also included in the PREDIMED-CVD. A total of 1859 participants remained in the analyses. Of these, 1556 had repeated measurements of diet and metabolomics at year 1 of intervention (Figure 1 and Supplementary material online, Figure S2). The protocol was approved by the Institutional Review Boards at all PREDIMED study locations, and all participants provided written informed consent.
Replication studies were performed in three prospective cohorts: NHS (started in 1976 with 121 701 female nurses aged 30–55 years24), NHSII (started in 1989 with 116 429 female nurses aged 25–42 years24), and HPFS (started in 1986 with 51 529 male health professionals aged 40–75 years25). Participants in these cohorts completed a baseline questionnaire regarding lifestyle, medical history, and health-related questions, and were followed up biennially. Blood samples were collected from subsamples of the NHS during 1989–90,26 NHSII during 1996–99,26 and HPFS during 1993–9527 (follow-up rate >95%26). Participants in our study (referred to as NHS/HPFS) were combined from 10 prior metabolomics sub-studies, after quality controls and data standardization in each sub-study (Supplementary material online, Table S1). We excluded duplicates across sub-studies, individuals missing dietary data or whole sets of named metabolites, and those having CVD or cancer at study baseline. Finally, 6868 participants were included in the replication analyses (Figure 1). The Institutional Review Boards at Brigham and Women’s Hospital and at Harvard T.H. Chan School of Public Health approved the study.
Metabolomics profiling and genotyping
The plasma metabolomics profiling for the PREDIMED and NHS/HPFS were performed in the same laboratory at the Broad Institute of Harvard University and M.I.T. (Cambridge, MA), using high-throughput liquid chromatography-tandem mass spectrometry techniques.28 After quality filtering and standardization (Supplementary material online, Supplementary Methods), 302 named metabolites were qualified for primary analyses in the PREDIMED (Supplementary material online, Figure S3A), with a majority of them also being qualified for replication studies in the NHS/HPFS (Supplementary material online, Table S1). Genotyping in NHS/HPFS was performed using six whole-genome arrays and was merged and imputed based on the 1000 Genomes Project Phase 1 Integrated Release Version 3 (ph1v3, hg19) (Supplementary material online, Supplementary Method).29 , 30 Genetic variants with a minor allele frequency >1% and imputation quality R 2 > 0.3 were used in analyses.
Adherence to the Mediterranean dietary pattern
We used a validated 14-item Mediterranean Diet Adherence Screener (MEDAS, range 0–14) to assess adherence to a traditional Mediterranean diet.31 , 32 The screener includes 14 dichotomous questions on habitual intakes of several food items (Supplementary material online, Table S2). In PREDIMED, trained dieticians completed the screener during in-person dietary-training sessions at baseline and year 1.3 Habitual diet in NHS/HPFS was assessed using a validated FFQ every 4 years.33 The MEDAS score was estimated using average dietary intakes from two FFQs closest to the time of blood draw (NHS: 1986 and 1990; NHSII: 1995 and 1999; HPFS: 1994 and 1998). Because information on olive oil use was as main culinary fat and sofrito intake (Supplementary material online, Table S2) was not available from FFQs, these two items were not included in MEDAS calculation in NHS/HPFS.
Ascertainment of cardiovascular disease
Incident CVD was defined as the composite of non-fatal myocardial infarction, non-fatal stroke, and cardiovascular death, occurring from study baseline (i.e. time of blood collection) through end of follow-up (PREDIMED: December 2010, median follow-up = 4.8 years; NHS: June 2012, median follow-up = 22.3 years; NHSII: June 2013, median follow-up = 15.3 years; HPFS: January 2012, median follow-up = 17.6 years). In PREDIMED, CVD cases were confirmed by an adjudication committee based on information collected from contact with participants and family physicians, ad hoc review of medical charts, and consultation of National Death Index.3 In NHS/HPFS, an adjudication committee confirmed non-fatal cases according to standard criteria.34 , 35 Fatal cases were confirmed by autopsy records or death certificate with evidence of prior CVD. Deaths (follow-up rate > 98%) were identified from next of kin, postal authorities, or the National Death Index.36
Assessment of cardiovascular risk factors and covariates
In PREDIMED, medical history and risk factors were collected through questionnaires. Anthropometric traits were measured by trained personnel. Blood lipids, fasting glucose, and other blood biochemistry markers were assayed using plasma collected at baseline and year 1. Two propensity scores were estimated based on 30 baseline variables to account for the probability of assignment to each intervention group.3 In NHS/HPFS, medical history, risk factors, and blood draw characteristics were collected from biennial questionnaires preceding blood collections and questionnaires completed at blood draw.
Statistical methods
Associations between individual metabolites (inverse-normal transformed) and MEDAS (and secondarily, its components) was assessed using multivariable linear regression. A false discovery rate (FDR) <0.05 was considered as statistically significant. To identify a metabolic signature for adherence to the Mediterranean diet, we used MEDAS and the metabolome from the PREDIMED baseline as the training set (i.e. the combined data of PREDIMED CVD and T2D nested case-control studies, to increase sample size, and ensure sufficient statistical power and model precision). Data from PREDIMED year 1 (for internal replication) and NHS/HPFS baseline (for external validation) were used as the testing sets (Figure 1). We first standardized all metabolites to the same scale; we then used elastic net37 to regress MEDAS on the 302 named metabolites (Supplementary material online, Figure S1B formula #3) and then applied the trained model to the testing sets to calculate the metabolic signature for the PREDIMED year 1 and NHS/HPFS baseline samples. The metabolic signature was calculated as the weighted sum of the selected metabolites with weights equal to coefficients from the elastic net regression (Supplementary material online, Figure S1). In NHS/HPFS, we calculated the signature separately in each sub-study (based on all metabolites available in each sub-study, after standardization) and we combined data from all sub-studies for subsequent analyses (Supplementary material online, Table S1; Supplementary methods). In order to avoid overfitting, the metabolic signature in the training set was obtained using a leave-one-out cross-validation approach.
The metabolic signature and MEDAS were standardized by z-score (mean = 0, SD = 1) before analyses to ensure that hazard ratios (HRs) and confidence interval (CI) for CVD risk were comparable between study cohorts. In PREDIMED, we used weighted Cox regressions with Barlow weights and robust variance estimator22 , 38 to assess associations of MEDAS and the metabolic signatures at baseline and year 1 with incident CVD risk (277 incident events from baseline; 143 incident events from year 1), within the CVD nested case-cohort study (Supplementary material online, Figure S2). Multivariable models were stratified by intervention groups and study centres, adjusting for age, sex, body mass index (BMI), smoking status, diabetes, dyslipidaemia, hypertension, and family history of premature coronary heart disease (CHD). In sensitivity analyses, we further adjusted for blood lipids, alanine aminotransferase, aspartate aminotransferase, estimated glomerular filtration rate, or propensity scores.3 In NHS/HPFS, associations of MEDAS and metabolic signature with CVD risk were estimated using Cox regressions, stratifying by cohort, original sub-studies, and original case-control status, and adjusting for age, fasting status, aspirin use, BMI, smoking status, physical activity, diabetes, dyslipidaemia, hypertension, and family history of premature CHD. As a sensitivity analysis, we further adjusted for all MEDAS food components instead of the total MEDAS score. A P < 0.05 was considered as statistically significant. Associations between individual metabolites (inverse-normal transformed) and CVD risk were assessed by the same model. Associations between MEDAS/metabolic signature and cardiovascular risk factors were analysed using multivariable linear regressions, and Bonferroni corrections were used to define statistical significance.
We performed a genome-wide association study (GWAS) of the metabolic signature in 1925 NHS/HPFS participants with both metabolomics and genotype data (Supplementary material online, Table S3), adjusting for age, sex, MEDAS, the first five genetic principle components, and/or interactions between genetic variants and MEDAS. A P < 5 × 10−8 was considered genome-wide significant and a P < 1 × 10−6 as marginally significant. We applied the mode-based estimate of Hartwig (MBE), a Mendelian randomization (MR)39 approach, to examine the potential causal associations of the metabolic signature on risk of CHD and stroke, based on GWAS summary statistics obtained separately for the metabolic signature, CHD, and stroke. Variants associated with the signature at P < 1 × 10−6 were used as instrumental variables. To maximize the statistical power, we used the most recent consortia GWAS summary statistics for CHD (122 733 cases, 424 528 controls)40 and stroke (40 585 cases, 406 111 controls of Europeans).41 Furthermore, we examined the role of potential mediators (i.e. BMI, lipid, systolic blood pressure, and diabetes) using a two-step MR analysis.
Detailed descriptions of all study methods are provided in Supplementary material online, Supplementary Methods.
Results
Characteristics of the study participants and adherence to the Mediterranean diet
Our primary study cohort included 1859 participants (1556 of whom had year-1 repeated measurements) from PREDIMED (58% women, mean age = 67 years). Baseline characteristics of participants in the sub-cohort were similar to those in the whole trial,3 with an improved lipid profile and a lower prevalence of hypertension and dyslipidaemia after 1 year of dietary intervention (Table 1A). The replication study included 6868 participants from the NHS (all women, mean age = 56 years), NHSII (all women, mean age = 45 years), and HPFS (all men, mean age = 64 years) (Table 1B). Compared with participants from PREDIMED, those from NHS/HPFS were younger and less likely to have obesity, diabetes, dyslipidaemia, and hypertension at baseline (Table 1).
Table 1.
Characteristics of the study participants from baseline and year-1 of the PREDIMED study and at time of blood collection in the NHS, NHS II, and HPFS
| (A) Study participants from the PREDIMED trial |
||||||
|---|---|---|---|---|---|---|
| Characteristics | The PREDIMED-CVD studya
|
The PREDIMED-T2D study |
||||
| At baseline |
At year-1 |
At baseline | At year-1 | |||
| Incident CVD | Sub-cohort | Incident CVD | Sub-cohort | N = 1017 | N = 779 | |
| N = 227 | N = 785 | N = 143 | N = 731 | |||
| Female, n (%) | 91 (40.1) | 449 (57.2) | 61 (42.7) | 416 (56.9) | 621 (61.1) | 474 (60.8) |
| Age, years | 69.4 (6.5) | 67.1 (5.9) | 70.3 (6.6) | 68.2 (5.9) | 66.6 (5.9) | 67.6 (5.8) |
| Body mass index, kg/m2 | 29.6 (3.8) | 29.7 (3.6) | 29.8 (4.0) | 29.7 (3.7) | 30 (3.5) | 30 (3.7) |
| MEDASb | 8.4 (1.8) | 8.9 (1.8) | 10.1 (1.9) | 10 (1.9) | 8.6 (1.9) | 10.1 (1.9) |
| Smoking status, n (%) | ||||||
| Never | 104 (45.8) | 490 (62.4) | 67 (46.9) | 467 (63.9) | 607 (59.7) | 485 (62.3) |
| Former | 77 (33.9) | 198 (25.2) | 43 (30.1) | 182 (24.9) | 228 (22.4) | 172 (22.1) |
| Current | 46 (20.3) | 97 (12.4) | 33 (23.1) | 82 (11.2) | 182 (17.9) | 122 (15.7) |
| Prevalence of diseases/conditions, n (%) | ||||||
| Hypertension | 187 (82.4) | 655 (83.4) | 69 (48.3) | 404 (55.3) | 935 (91.9) | 503 (64.6) |
| Dyslipidaemia | 132 (58.1) | 576 (73.4) | 103 (72.0) | 469 (64.2) | 853 (83.9) | 539 (69.2) |
| Diabetes | 146 (64.3) | 370 (47.1) | 91 (63.6) | 342 (46.8) | 0 | 0 |
| Family history of premature CHD | 44 (19.4) | 198 (25.2) | 29 (20.3) | 185 (25.3) | 268 (26.4) | 208 (26.7) |
| Fasting glucose, mg/dL | 126.7 (41.0) | 116.1 (32.7) | 123.3 (37.2) | 114.5 (31.0) | 102.5 (16.2) | 101.4 (16.7) |
| LDL cholesterol, mg/dL | 130.8 (33.8) | 130.5 (33.2) | 127.2 (31.4) | 128.4 (32.5) | 138.3 (33.4) | 134.2 (33.8) |
| HDL cholesterol, mg/dL | 49.8 (14.3) | 51.2 (12.9) | 51 (16.7) | 52.8 (13.8) | 53 (12.4) | 55 (14.1) |
| Total cholesterol, mg/dL | 208.3 (33.8) | 207.7 (36.8) | 205.7 (35.5) | 205.7 (35.8) | 217.2 (36.5) | 214.4 (37.8) |
| Triglycerides, mg/dL | 147.8 (78) | 132.9 (81.6) | 146.1 (79.2) | 130.3 (73.9) | 133.3 (74.9) | 131.4 (70.1) |
| (B) Study participants from the NHS, NHSII, and HPFS | ||||||
|---|---|---|---|---|---|---|
| Characteristics | NHS (women) |
NHSII (women) |
HPFS (men) |
|||
| Incident CVD | Non-case | Incident CVD | Non-case | Incident CVD | Non-case | |
| N = 192 | N = 2447 | N = 23 | N = 3142 | N = 136 | N = 928 | |
| Age, years | 59.7 (6.1) | 55.7 (6.7) | 45.9 (4.9) | 44.5 (4.5) | 68.3 (7.0) | 62.8 (8.3) |
| Body mass index, kg/m2 | 27.2 (5.6) | 26.1 (4.7) | 27.1 (6.0) | 25.8 (5.8) | 25.8 (2.9) | 25.6 (3.0) |
| MEDASb | 4.1 (1.7) | 4.2 (1.7) | 4.1 (1.6) | 4.2 (1.7) | 4.5 (1.7) | 4.5 (1.8) |
| Smoking status, n (%) | ||||||
| Never | 88 (45.8) | 1187 (48.5) | 9 (39.1) | 2113 (67.3) | 62 (45.6) | 484 (52.2) |
| Former | 75 (39.1) | 1003 (41.0) | 10 (43.5) | 779 (24.8) | 65 (47.8) | 411 (44.3) |
| Current | 29 (15.1) | 257 (10.5) | 4 (17.4) | 250 (8.0) | 9 (6.6) | 33 (3.6) |
| Prevalence of diseases/conditions, n (%) | ||||||
| Hypertension | 94 (49.0) | 697 (28.5) | 2 (8.7) | 368 (11.7) | 61 (44.9) | 261 (28.1) |
| Dyslipidaemia | 93 (48.4) | 999 (40.8) | 8 (34.8) | 801 (25.5) | 63 (46.3) | 391 (42.1) |
| Diabetes | 14 (7.3) | 44 (1.8) | 0 | 41 (1.3) | 9 (6.6) | 37 (4.0) |
| Family history of premature CHD | 58 (30.2) | 551 (22.5) | 6 (26.1) | 668 (21.3) | 28 (20.6) | 142 (15.3) |
| Aspirin use, n (%) | 94 (49.0) | 1156 (47.2) | 4 (17.4) | 456 (14.5) | 61 (4.9) | 349 (37.6) |
| Postmenopausal, n (%) | 160 (83.3) | 1688 (69.0) | 9 (39.1) | 718 (22.9) | — | — |
| Postmenopausal HT,c n (%) | 50 (31.2) | 707 (41.9) | 5 (55.6) | 504 (70.2) | — | — |
| Total calorie, kcal/day | 1724 (474) | 1771 (515) | 1953 (671) | 1830 (534) | 2096 (671) | 2056 (623) |
| Physical activity, MET-hours/week | 15.1 (17.4) | 15.5 (24) | 18.6 (18.8) | 17.9 (21.7) | 29.1 (25.6) | 33.6 (29.9) |
CHD, coronary heart disease; CVD, cardiovascular disease; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MEDAS, Mediterranean Diet Adherence Screener.
Incident CVD cases and the sub-cohort shared 36 overlapping samples at baseline and 26 at year 1, excluding participants missing data on metabolomics or diet.
In PREDIMED, 14 items were used in the calculation of MEDAS and the range of MEDAS was 4–14; in NHS/HPFS, 12 items were used in the calculation of MEDAS and the range of MEDAS was 0–10.
Among post-menopausal women.
The mean MEDAS was 8.7 in the PREDIMED at baseline and 10.1 at year-1 (range: 4–14) (Table 1A); the improvement in the score was due to increased intakes of extra-virgin olive oil (EVOO), fish/seafood, nuts, and legumes resulting from the dietary interventions (Supplementary material online, Figure S4). The mean MEDAS was 4.2 in NHS, 4.2 in NHSII, and 4.5 in HPFS (range: 0–10), lower than that in PREDIMED partially because two items (olive oil as main culinary fat and sofrito intake) were not available for MEDAS calculation (Table 1B and Supplementary material online, Table S2). In addition, compared with those in the PREDIMED, participants from NHS/HPFS reported lower intakes of olive oil, fruit, wine, nuts, fish/seafood, and poultry, but higher intakes of red meat, butter/margarine, and sweets (Supplementary material online, Figure S4).
Variation of the plasma metabolome in response to the Mediterranean diet
Of the 302 named metabolites used in the analyses (Supplementary material online, Figure S3A), we identified 97 (32.1%) metabolites significantly associated with MEDAS at baseline in the PREDIMED (FDR < 0.05) (Supplementary material online, Figure S3D). These metabolites were primarily lipids (n = 82; accounting for 37% of all assayed lipids), and also included amino acids (n = 9; 21% of assayed amino acids) and metabolites of other categories (n = 6) (Supplementary material online, Figure S3B). Results from the PREDIMED year-1 data were highly concordant (Supplementary material online, Figure S3E). The enrichment of significant associations in lipid categories was likely due to strong correlations among lipid species (Supplementary material online, Figure S5).
We then applied elastic net regressions on the 302 named metabolites in PREDIMED baseline data (training set) to identify a metabolic signature for Mediterranean diet adherence. The model selected a combination of 67 metabolites most significantly associated with MEDAS while robust to the effects of collinearity between metabolites (Figure 2 and Supplementary material online, Figure S1). At PREDIMED year-1 (internal testing set) and NHS/HPFS baseline (external testing set), we found that the metabolic signature was significantly correlated with MEDAS (PREDIMED year-1: r = 0.31, P = 3 × 10−35; NHS/HPFS: 34–62 metabolites were available for signature calculation in various sub-studies; overall r = 0.28, P = 4 × 10−118) (Figure 2 and Supplementary material online, Table S1). In the training set, the unbiased metabolic signature acquired using the leave-one-out cross-validation approach was significantly correlated with MEDAS with a similar magnitude (r = 0.37, P = 2 × 10−62) (Figure 2). Further stratification analyses suggested that correlations between MEDAS and the metabolic signature in both PREDIMED and NHS/HPFS were consistent across categories of study-design characteristics (e.g. sub-studies and case-control status) (Supplementary material online, Figures S6 and S7). Secondary analysis in PREDIMED showed a weak, but statistically significant (P = 0.006) positive association between 1-year changes in MEDAS and concurrent changes in the metabolic signature (Supplementary material online, Figure S8).
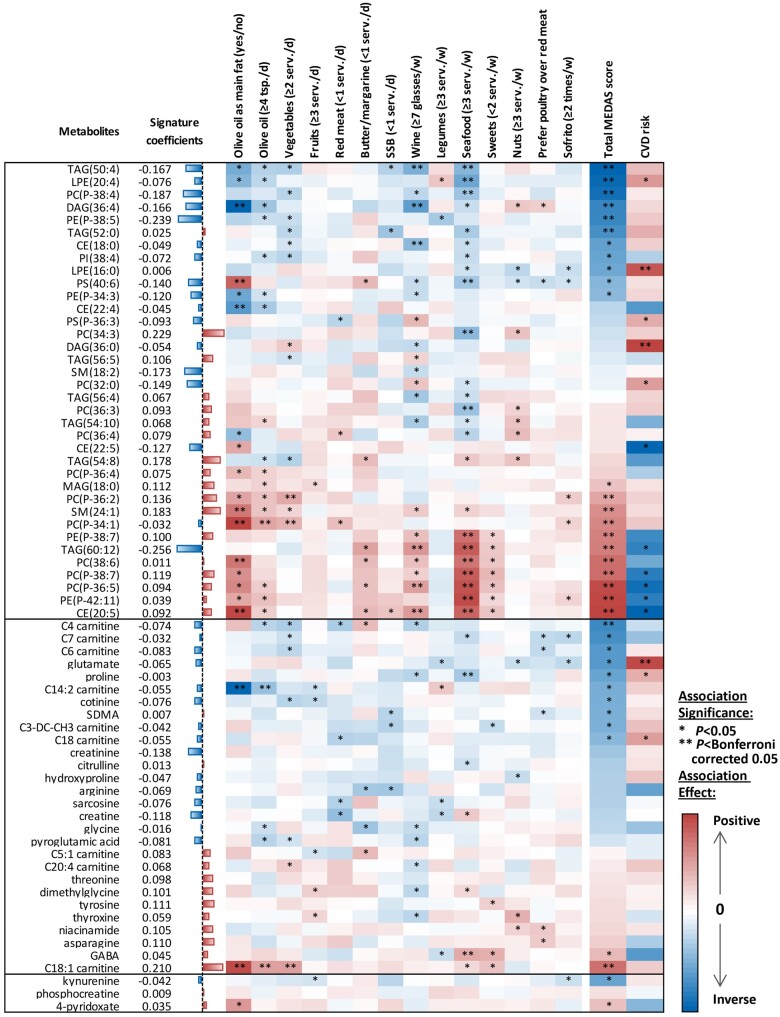
Metabolites in the signature included 45 lipid species and acylcarnitines (20% of all assayed lipids), 19 amino acids (an enriched set of 44% of assayed amino acids), 2 vitamins (29% of assayed vitamins/cofactors), and 1 xenobiotic (Figures 2 and 3 and Supplementary material online, Figure S3C). The selected metabolites showed extensive associations with MEDAS that were highly concordant between PREDIMED and NHS/HPFS. Different MEDAS components were associated with various sub-sets of selected metabolites, but the associations appeared to be stronger and most reproducible for intakes of olive oil, wine, fish/seafood, and sweets (Figure 3 and Supplementary material online, Figure S9–S12). As a notable example, associations between the 67 metabolites constituting the metabolic signature and fish/seafood intake were most concordant across datasets (PREDIMED baseline vs. year 1, r = 0.78; PREDIMED baseline vs. NHS/HPFS, r = 0.72); higher fish/seafood intake was significantly and reproducibly associated with higher levels of highly unsaturated lipid metabolites containing ≥1 chain(s) of eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), and/or docosapentaenoic acid (DPA) (Figure 3 and Supplementary material online, Figure S9–S12).
Figure 3.
Associations of the 67 metabolites constituting the metabolic signature with MEDAS components, total MEDAS score, and subsequent cardiovascular disease risk. Data were based on measurements at PREDIMED baseline (replications were provided in the supplementary material). Presented from left to right are the metabolites’ coefficients (weights) in the signature, and associations with each food component, MEDAS, and subsequent cardiovascular disease risk. Coefficients for associations with food items and MEDAS indicate the SD changes in metabolites per dietary score increment. Coefficients for cardiovascular disease risk indicate ln (hazard ratio) of cardiovascular disease risk per SD increment in metabolites. Colours denote the association directions (red-positive and blue-inverse) and magnitudes (the darker the colour, the stronger the magnitude); asterisks represent association significance (*P < 0.05 and **P < Bonferroni corrected 0.05; for associations with total MEDAS score and cardiovascular disease risk, we Bonferroni corrected for 67 metabolites; for associations with each of the food items, we Bonferroni corrected for 67 metabolites × 14 food items). CVD, cardiovascular disease; MEDAS, the Mediterranean Diet Adherence Screener.
We observed substantial variations in the metabolic signature among participants reporting the same MEDAS (Figure 2 and Supplementary material online, Figure S1C). Secondary analyses suggested that differences in dietary components explained a small proportion of such variation (Supplementary material online, Figure S13), suggesting the influences of other factors (Supplementary material online, Figure S1).
Associations with risk of incident cardiovascular disease events
In multivariable analyses of the PREDIMED study, we observed a significant inverse association between baseline MEDAS with incident CVD [cases n = 227; HR per SD increment in MEDAS =0.77 (95% CI: 0.64–0.93)], and a statistically non-significant inverse association between year-1 MEDAS with risk of subsequent CVD events [cases n = 143; HR = 0.87 (0.71–1.05)]. The metabolic signatures at baseline and year 1 were both inversely associated with incident CVD at a similar magnitude [baseline signature: HR = 0.71 (0.58–0.87); year-1 signature: HR = 0.72 (0.57–0.92)]; notably, these associations remained significant after further adjustment for MEDAS [baseline signature, HR = 0.73 (0.59–0.91); year-1 signature, HR = 0.74 (0.58-0.94)] or MEDAS components (Table 2 and Supplementary material online, Table S4). In contrast, the association between MEDAS and CVD was 37.2% (95% CI: 13.1–70.1%) mediated by the metabolic signature (Supplementary material online, Figure S1D) and, after further adjustment for the metabolic signature, the association was attenuated and became non-significant (Table 2 and Supplementary material online, Table S4). Further adjustment for blood lipids, biomarkers indicating liver/kidney functions, or propensity scores did not change the results (Supplementary material online, Table S4).
Table 2.
Associations of MEDAS and the metabolic signature with CVD risk in the PREDIMED study and the NHS/HPFS
| Analysis model | MEDAS |
Metabolic signature |
||
|---|---|---|---|---|
| HR (95% CI)a | P | HR (95% CI)a | P | |
| PREDIMED baseline MEDAS/signature and 227 incident CVD eventsb | ||||
| Age, sex-adjusted Model | 0.77 (0.65–0.93) | 0.006 | 0.69 (0.57–0.84) | <0.001 |
| Multivariable Model | 0.77 (0.64–0.93) | 0.008 | 0.71 (0.58–0.87) | <0.001 |
| MV + mutual adjustmentc | 0.86 (0.70–1.07) | 0.17 | 0.73 (0.59–0.91) | 0.004 |
| PREDIMED year 1 MEDAS/signature and 143 subsequent CVD eventsd | ||||
| Age, sex-adjusted Model | 0.84 (0.70–1.01) | 0.07 | 0.68 (0.55–0.84) | <0.001 |
| Multivariable Model | 0.87 (0.71–1.05) | 0.15 | 0.72 (0.57–0.92) | 0.008 |
| MV + mutual adjustmentc | 0.95 (0.78–1.15) | 0.59 | 0.74 (0.58–0.94) | 0.01 |
| NHS/HPFS baseline MEDAS/signature and 351 incident CVD eventse | ||||
| Age, sex-adjusted Model | 0.90 (0.81–1.00) | 0.05 | 0.81 (0.74–0.90) | <0.001 |
| Multivariable Model | 0.92 (0.82–1.03) | 0.13 | 0.85 (0.76–0.94) | 0.001 |
| MV + mutual adjustmentc | 0.97 (0.86–1.09) | 0.56 | 0.85 (0.77–0.95) | 0.004 |
BMI, body mass index; CHD, coronary heart disease; CVD, cardiovascular disease; MEDAS, Mediterranean Diet Adherence Screener.
Hazard ratio (HR) and 95% confidence interval (CI) of CVD risk per standard deviation increment in MEDAS or the metabolic signature.
MEDAS, metabolic signature, and covariates were assessed at baseline, and outcome was incident CVD events occurred from baseline through end of follow-up. The basic model was stratified by study centres and intervention arms and adjusted for age and sex. The multivariable (MV) model further adjusted for BMI, smoking status, diabetes, dyslipidaemia, hypertension, and family history of premature CHD.
We included both MEDAS and the metabolic signature of MEDAS simultaneously in the MV model to examine association independence.
MEDAS, metabolic signature, and covariates were assessed at year 1, and outcome was the incident CVD events occurred after year-1 visit through end of follow-up. The analytic models were the same as in (b) except not stratifying by intervention arms because of high co-linearity with MEDAS.
The basic model was stratified by study cohorts (NHS/NHSII: women; HPFS: men), original sub-studies, and the case-control status in the original sub-study, and was adjusted for age in years. MV Model further adjusted for fasting status and aspirin use at blood draw, BMI, smoking status, physical activity, diabetes, dyslipidaemia, hypertension, and family history of premature CHD.
Similar findings were observed in the NHS/HPFS. We documented 351 incident CVD events during up to 22 years of follow-up. Baseline metabolic signature was significantly associated with CVD risk after adjusting for known risk factors and potential confounders [HR = 0.85 (0.76–0.94)] and after further controlling for MEDAS [HR = 0.85 (0.77–0.95)] or MEDAS components. The marginal association between MEDAS and CVD risk [HR =0.90 (0.81–1.00) in age and sex-adjusted model and 0.92 (0.82–1.03) in the multivariable model] was attenuated after further adjusting for the metabolic signature [the associated was 64.5% (95% CI, 1.6–99.5%) mediated by the metabolic signature; HR after adjustment = 0.97 (0.86–1.09)] (Table 2 and Supplementary material online, Table S4 and Figure S1D).
In secondary analyses, we noted significant associations between several metabolites constituting the metabolic signature and CVD risk and the overall association pattern was concordant across PREDIMED and NHS/HPFS (Figure 3 and Supplementary material online, Figures S9–S12). Of note, metabolites associated with higher MEDAS (especially, higher intakes of olive oil, wine, and fish/seafood, and lower intakes of sugar sweetened beverages and sweets) were more likely to be associated with a lower CVD risk; this was exemplified by highly unsaturated lipid metabolites, which showed positive associations with intakes of olive oil, wine, and/or fish/seafood, and a beneficial association with incident CVD. In contrast, metabolites associated with lower MEDAS were more likely to be associated with higher CVD risk (e.g. glutamate) (Figure 3). In sensitivity analyses, the association between the metabolic signature and CVD did not change after excluding any individual metabolite, or unsaturated lipids containing EPA/DHA/DPA, from the signature (Supplementary material online, Table S5), suggesting that such association represented cumulative effects from many metabolites.
Associations with cardiovascular traits/risk factors
In multivariable analyses, MEDAS and the metabolic signature showed a similar association pattern with cardiovascular traits/risk factors, including an inverse association with BMI, waist circumference, and current smoking, and a positive association with physical activity. Compared with MEDAS, the metabolic signature showed stronger associations with metabolic disease/traits, including an inverse association with prevalent diabetes, triglycerides, and total to high-density lipoprotein (HDL) cholesterol ratio, and a positive association with HDL cholesterol (Supplementary material online, Figure S14).
The genetic determinants of the metabolic signature and cardiovascular disease risk
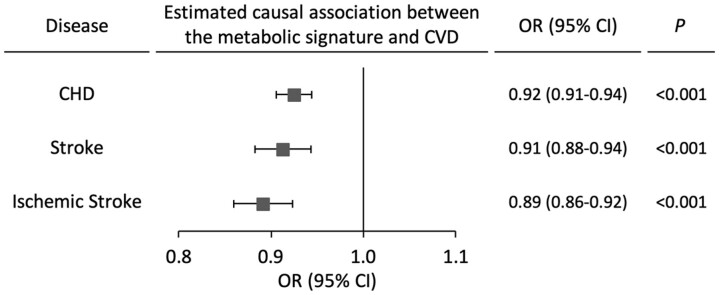
As secondary analyses, we included genetic analyses to facilitate the interpretation of the identified metabolic signature. The estimated genetic heritability of the metabolic signature was 12% (h 2 = 0.12, SE = 0.26). We identified three loci associated with the metabolic signature after adjusting for MEDAS (FADS1-3 and DAOA, P < 5 × 10−8; LINC01187, P = 2 × 10−7) and two additional loci influencing the signature through interactions with MEDAS (ABCC1-6 and LOC101928516, P interaction < 5 × 10−8) (Supplementary material online, Table S6). Mendelian randomization analyses showed that the genetic component of the signature was inversely associated with risk of CHD (OR per SD increment in the genetically inferred metabolic signature = 0.92, 95% CI, 0.91–0.94, P < 0.001), total stroke (OR = 0.91, 95% CI, 0.88–0.94, P < 0.001), and ischaemic stroke (OR = 0.89, 95% CI, 0.86–0.92, P < 0.001) (Figure 4). Several risk factors (including blood lipids, systolic blood pressure, and diabetes) showed weak albeit statistically significant mediating effects in the MR analyses (Supplementary material online, Figure S15).
Figure 4.
Mendelian Randomization analyses of the metabolic signature with risk of coronary heart disease and stroke. In mode-based estimate of Hartwig (MBE) Mendelian randomization analyses, 58 genetic variants associated with the metabolic signature at P < 1 × 10−6 were used as the instrument variable. CHD, coronary heart disease; CVD, cardiovascular disease; MR, Mendelian randomization; OR, odds ratio; CI, confidence interval.
Take home figure.
Integrating clinical, metabolomic, and genetic data from one Spanish and three US prospective cohorts, this study identified and validated a metabolic signature that robustly reflects adherence and metabolic response to a Mediterranean diet and predicts future cardiovascular disease risk independent of traditional risk factors. CHD, coronary heart disease; CI, confidence interval; CVD, cardiovascular disease; HR, hazards ratio; M, metabolites; MEDAS, a 14-item Mediterranean Diet Adherence Screener; Medi-Diet, Mediterranean diet; OR, odds ratio.
Discussion
Leveraging data from a large intervention trial and three prospective cohorts, this study is the first to identify a metabolic signature that measures adherence to the Mediterranean diet and importantly, predicts future CVD risk independently of known CVD risk factors in both Spanish and US populations (Take home figure). Given its ability to identify CVD risk independent of self-reported dietary measures, the metabolic signature holds promise in complementing traditional dietary assessments, stratifying individuals with different dietary response and disease risk, and potentially facilitating personalized nutrition interventions.
Previous studies have identified many individual metabolites associated with some dietary patterns, and intakes of foods such as fish, nuts, and vegetables,12 , 13 , 15–17 , 42 indicating that metabolites in biospecimens may serve as biomarkers for assessing diet. In our study, the 67 metabolites that comprise the metabolic signature showed reproducible associations with MEDAS and its food components. Given that the Mediterranean diet is high in unsaturated fats, it is not surprising that a large proportion of these metabolites are involved in polyunsaturated fatty acid and lipid metabolic pathways, a finding that is also consistent with prior metabolomics studies.16 The positive associations between fish/seafood intake and lipid species containing long-chain n-3 fatty acids are consistent with the dietary source of these fatty acids and prior metabolomic studies of fish intake.16 , 42 Extra-virgin olive oil is rich in monounsaturated fat, and both EVOO and red/white wine are high in anti-oxidative polyphenols.43 Concordantly in our study, intakes of EVOO and wine were positively associated with unsaturated lipids, especially plasmalogen phospholipids that have anti-oxidative properties and are protective for CVD.19 It is worth noting that, in line with the beneficial effect of fish/seafood, olive oil, and wine in CVD prevention,44 , 45 metabolites associated with higher intakes of these dietary factors were more likely to be associated with a lower CVD risk, indicating potential pathways through which these dietary factors are associated with CVD outcomes. Our findings suggested that greater adherence to the Mediterranean diet may lead to profound changes in the metabolome that are associated with favourable cardiometabolic health.
The plasma metabolome reflects the overall metabolic homeostasis resulting from the interactive effects of all metabolism-influencing factors, including diet,13 genetic variabilities,11 the microbiome,7 and health status.46 In our study, self-reported MEDAS was a combination of true dietary intakes and reporting errors. The metabolome, while can be changed by diet, is expected to be independent of reporting errors. By regressing MEDAS on metabolites (Supplementary material online, Figure S1B formula #3), the resulting metabolic signature captured cumulative changes in the metabolome that are correlated with Mediterranean dietary adherence, incorporated individual metabolic variations from other factors that influence dietary metabolism, while also minimized measurement errors which are inherent to self-reported dietary assessments (Supplementary material online, Figure S1A). Consistent with this notion, besides a moderate but robust positive correlation between the metabolic signature and MEDAS, we also observed substantial variation in the metabolic signature among participants with the same MEDAS scores (Supplementary material online, Figure S1C). Some of the variation could be attributed to differences in dietary components and between-person variabilities in responses to diet as a result of genetic variations. Indeed, our genetic analyses found that the metabolic signature was significantly influenced by genetic loci involved in metabolism of fatty acids (e.g. the fatty acid desaturase gene cluster FADS1-3)47 and amino acids (e.g. D-amino acid oxidase activator DAOA). The metabolic signature was also associated with several risk factors known to influence dietary metabolism including physical activity, smoking, and metabolic conditions. Importantly, the metabolic signature was able to identify individuals at different CVD risk among participants with similar CVD risk factors and reported the same MEDAS and dietary components in both PREDIMED and NHS/HPFS. Our MR and mediation analyses suggested the metabolic signature as a potential causal mediator on the path from the Mediterranean diet to risk of CHD and stroke (Supplementary material online, Figure S1D). Taken together, the metabolic signature may represent an integrated picture of the metabolic homeostasis resulting from adherence to the Mediterranean diet and individual metabolic responses to diet.
In the PREDIMED, baseline MEDAS was significantly associated with CVD risk whereas the association for year-1 MEDAS was marginal. This may be due to a reduced sample size at year 1, and that short-term dietary changes immediately after the intervention were not representative of dietary adherence during the entire intervention. As expected, we found, in the whole PREDIMED study, a significant association for year-1 MEDAS (with a similar HR as in our study) and a stronger association for cumulative average of MEDAS since year 1 with CVD risk (Supplementary material online, Table S7).
Cotinine, a metabolite of nicotine, was selected into the metabolic signature by elastic net regression possibly due to the strong correlation between lower MEDAS and smoking. Still, the correlations between MEDAS and metabolic signature were consistent across smoking status categories. Furthermore, the association between the metabolic signature with CVD risk did not change after excluding cotinine from the signature (Supplementary material online, Figure S16).
Although previous studies have identified many diet-associated metabolites,12 , 13 , 15–17 , 42 our study is the first to identify a metabolic signature for the evaluation of adherence/metabolic response to the Mediterranean diet and examined its association with long-term CVD risk. Our findings were reproducible among Spanish and US participants with different food environments and dietary habits. The signature was derived by machine-learning models using well-characterized metabolites and is robust to a few missing predictor metabolites. This allows possible applications by future studies to measure adherence/metabolic response to the Mediterranean diet in other cohorts and intervention studies. The well-characterized data in PREDIMED and NHS/HPFS allowed us to control for confounders/risk factors, and to evaluate genetic determinants of the metabolic signature and the potential causal association between the signature and CVD risk, highlighting the value of the metabolic signature in evaluating diet and disease risks.
Several limitations warrant discussion. First, the metabolic signature was developed based on 302 named metabolites. While the interplay between metabolites may be non-linear, elastic net regression constructed the signature using a linear combination of metabolites. Although the metabolic signature performed robustly, our approach might benefit from the inclusion of additional metabolites, and advanced machine-learning methods that also consider non-linear relations/interactions between metabolites. Second, the Mediterranean diet shares many foods/nutrients with other healthy eating patterns and the metabolic signature was associated with other factors influencing dietary metabolism; this may limit the specificity of the signature in assessing the Mediterranean diet. Future studies are warranted to examine potential metabolic differences across different healthy eating patterns. Third, due to the observational nature, our study was unable to confirm causality; however, we used a prospective cohort design and applied MR analyses, thus reducing concerns of reverse causation. Finally, although we evaluated the cross-population reproducibility of the signature, it should be validated in other independent populations, and examined in associations with other chronic diseases.
In summary, based on reproducible findings in Spanish and three US cohorts, the present study identified a metabolic signature that measures adherence and metabolic response to the Mediterranean diet, and predicts future CVD risk. Further examinations of the metabolic signature and biological pathways of the constituting metabolites may improve our understanding of biological mechanisms through which diet impacts health. Metabolomics profiling holds the promise for an objective and more comprehensive evaluation of adherence and metabolic response to diet, stratifying individuals based on dietary response and disease risk, and facilitating more effective and individualized approaches for dietary interventions.
Supplementary Material
Acknowledgements
The authors thank all of the PREDIMED, NHS, NHSII, and HPFS participants for their participation, as well as the personnel of PREDIMED and affiliated primary care centres, and staff of the NHS, NHSII, and HPFS for their contribution.
Funding
The PREDIMED metabolomics studies were funded by National Institutes of Health (NIH) grants R01 HL118264 and R01 DK102896. The PREDIMED trial was funded by the Spanish Ministry of Health (Instituto de Salud Carlos III, The PREDIMED Network grant RD 06/0045, 2006–13, coordinated by M.A. Martínez-González; and a previous network grant RTIC-G03/140, 2003–05, coordinated R. Estruch). Additional grants were received from the Ministerio de Economía y Competitividad-Fondo Europeo de Desarrollo Regional (Projects CNIC-06/2007, CIBER 06/03, PI06-1326, PI07-0954, PI11/02505, SAF2009-12304, and AGL2010–22319-C03-03) and by the Generalitat Valenciana (PROMETEO17/2017, ACOMP2010-181, AP111/10, AP-042/11, ACOM2011/145, ACOMP/2012/190, ACOMP/2013/159 and ACOMP/213/165). The NHS, NHSII, and HPFS, and their metabolomics studies were supported by NIH grants U01 HL145386, UM1 CA186107, R01 CA49449, R01 HL034594, R01 HL088521, UM1 CA176726, R01 CA67262, UM1 CA167552, R01 HL35464, HL60712, R01 CA50385, P01 CA87969, and R01 AR049880. J.L. was supported by 9-17-CMF-011 from the American Diabetes Association (ADA), K99 DK122128 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and a pilot and feasibility grant from the NIDDK-funded Boston Nutrition Obesity Research Center (P30 DK046200). M.G.-F. was supported by 1-18-PMF-029 from ADA. F.K.T was supported by R00 CA207736 from the National Cancer Institute. D.K.T is supported by 1-18-JDF-104977 from ADA. Role of sponsor: the funders had no role in study design, data collection, data analysis, data interpretation, the writing of the report, or decision of publication of the results.
Conflict of interest: B.M.W. reported grant funding from Celgene and Eli Lilly, and consulting at BioLineRx, Celgene, G1 Therapeutics, and GRAIL. No other co-authors reported COI related to this study.
Contributor Information
Jun Li, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA.
Marta Guasch-Ferré, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA.
Wonil Chung, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Department of Biostatistics, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 4th Floor, Boston, MA 02115, USA.
Miguel Ruiz-Canela, Department of Preventive Medicine and Public Health, University of Navarra, Irunlarrea 1, Pamplona 31008, Spain; IdiSNA (Instituto de Investigación Sanitaria de Navarra), Edificio LUNA-Navarrabiomed, Irunlarrea 3, Pamplona 31008, Spain; CIBER Fisiopatología de la Obesidad y Nutrición (CIBERObn), Instituto de Salud Carlos III, C/Monforte de Lemos 3-5, Pabellón 11, Madrid 28029, Spain.
Estefanía Toledo, Department of Preventive Medicine and Public Health, University of Navarra, Irunlarrea 1, Pamplona 31008, Spain; IdiSNA (Instituto de Investigación Sanitaria de Navarra), Edificio LUNA-Navarrabiomed, Irunlarrea 3, Pamplona 31008, Spain; CIBER Fisiopatología de la Obesidad y Nutrición (CIBERObn), Instituto de Salud Carlos III, C/Monforte de Lemos 3-5, Pabellón 11, Madrid 28029, Spain.
Dolores Corella, CIBER Fisiopatología de la Obesidad y Nutrición (CIBERObn), Instituto de Salud Carlos III, C/Monforte de Lemos 3-5, Pabellón 11, Madrid 28029, Spain; Department of Preventive Medicine, University of Valencia, Valencia 46010, Spain.
Shilpa N Bhupathiraju, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA.
Deirdre K Tobias, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Division of Preventive Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 900 Commonwealth Avenue, Boston, MA 02115, USA.
Fred K Tabung, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Division of Medical Oncology, Department of Internal Medicine, The Ohio State University College of Medicine and Comprehensive Cancer Center – James Cancer Hospital and Solove Research Institute, 410 W 12th Ave Columbus, OH 43210, USA.
Jie Hu, Division of Women’s Health, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 1620 Tremont St, 3rd floor, Boston, MA 02120, USA.
Tong Zhao, Department of Biostatistics, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 4th Floor, Boston, MA 02115, USA.
Constance Turman, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA.
Yen-Chen Anne Feng, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Metabolomics Platform,Broad Institute of Harvard and MIT, 415 Main St, Cambridge, MA 02142, USA.
Clary B Clish, Metabolomics Platform,Broad Institute of Harvard and MIT, 415 Main St, Cambridge, MA 02142, USA.
Lorelei Mucci, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA.
A Heather Eliassen, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA.
Karen H Costenbader, Division of Rheumatology, Inflammation and Immunity, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 75 Francis St, Boston, MA 02115, USA.
Elizabeth W Karlson, Division of Rheumatology, Inflammation and Immunity, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 75 Francis St, Boston, MA 02115, USA.
Brian M Wolpin, Department of Medical Oncology, Dana-Farber Cancer Institute, 450 Brookline Avenue, Boston, MA 02215, USA.
Alberto Ascherio, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA.
Eric B Rimm, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA.
JoAnn E Manson, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Division of Preventive Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 900 Commonwealth Avenue, Boston, MA 02115, USA; Mary Horrigan Connors Center for Women’s Health and Gender Biology, Brigham and Women’s Hospital and Harvard Medical School, 75 Francis Street, Boston, MA 02115, USA.
Lu Qi, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA; Department of Epidemiology, School of Public Health and Tropical Medicine, Tulane University, 1440 Canal Street, New Orleans, LA 70112, USA.
Miguel Ángel Martínez-González, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Department of Preventive Medicine and Public Health, University of Navarra, Irunlarrea 1, Pamplona 31008, Spain; IdiSNA (Instituto de Investigación Sanitaria de Navarra), Edificio LUNA-Navarrabiomed, Irunlarrea 3, Pamplona 31008, Spain; CIBER Fisiopatología de la Obesidad y Nutrición (CIBERObn), Instituto de Salud Carlos III, C/Monforte de Lemos 3-5, Pabellón 11, Madrid 28029, Spain.
Jordi Salas-Salvadó, CIBER Fisiopatología de la Obesidad y Nutrición (CIBERObn), Instituto de Salud Carlos III, C/Monforte de Lemos 3-5, Pabellón 11, Madrid 28029, Spain; Human Nutrition Unit, Faculty of Medicine and Health Sciences, Institut d’Investigació Sanitària Pere Virgili, Universitat Rovira i Virgili, C/Sant Llorenç 21, Reus 43201, Spain.
Frank B Hu, Department of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 3rd Floor, Boston, MA 02115, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, 181 Longwood Avenue, Boston, MA 02115, USA.
Liming Liang, Department of Epidemiology, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 2nd Floor, Boston, MA 02115, USA; Department of Biostatistics, Harvard T.H. Chan School of Public Health, 655 Huntington Avenue, Building II 4th Floor, Boston, MA 02115, USA.
References
- 1.US Department of Agriculture and US Department of Health and Human Services. 2015–2020 Dietary Guidelines for Americans. Washington, DC: US Department of Health and Human Services; 2015. https://health.gov/dietaryguidelines/2015/ (15 January 2019).
- 2. Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P; American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 2017;135:e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Estruch R, Ros E, Salas-Salvadó J, Covas M-I, Corella D, Arós F, Gómez-Gracia E, Ruiz-Gutiérrez V, Fiol M, Lapetra J, Lamuela-Raventos RM, Serra-Majem L, Pintó X, Basora J, Muñoz MA, Sorlí JV, Martínez JA, Fitó M, Gea A, Hernán MA, Martínez-González MA; PREDIMED Study Investigators. Primary prevention of cardiovascular disease with a Mediterranean diet supplemented with extra-virgin olive oil or nuts. N Engl J Med 2018;378:e34. [DOI] [PubMed] [Google Scholar]
- 4. Fung TT, Rexrode KM, Mantzoros CS, Manson JE, Willett WC, Hu FB. Mediterranean diet and incidence of and mortality from coronary heart disease and stroke in women. Circulation 2009;119:1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salas-Salvado J, Bullo M, Babio N, Martinez-Gonzalez MA, Ibarrola-Jurado N, Basora J, Estruch R, Covas MI, Corella D, Aros F, Ruiz-Gutierrez V, Ros E; PREDIMED Study Investigators. Reduction in the incidence of type 2 diabetes with the Mediterranean diet: results of the PREDIMED-Reus nutrition intervention randomized trial. Diabetes Care 2011;34:14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martínez-González MA, Gea A, Ruiz-Canela M. The Mediterranean diet and cardiovascular health. Circ Res 2019;124:779–798. [DOI] [PubMed] [Google Scholar]
- 7. Shoaie S, Ghaffari P, Kovatcheva-Datchary P, Mardinoglu A, Sen P, Pujos-Guillot E, de Wouters T, Juste C, Rizkalla S, Chilloux J, Hoyles L, Nicholson Jeremy K, Dore J, Dumas Marc E, Clement K, Bäckhed F, Nielsen J. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab 2015; 22:320–331. [DOI] [PubMed] [Google Scholar]
- 8. Widmer RJ, Flammer AJ, Lerman LO, Lerman A. The Mediterranean diet, its components, and cardiovascular disease. Am J Med 2015;128:229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu FB, Satija A, Rimm EB, Spiegelman D, Sampson L, Rosner B, Camargo CA Jr, Stampfer M, Willett WC. Diet assessment methods in the nurses’ health studies and contribution to evidence-based nutritional policies and guidelines. Am J Public Health 2016;106:1567–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guasch-Ferré M, Bhupathiraju SN, Hu FB. Use of metabolomics in improving assessment of dietary intake. Clin Chem 2018;64:82–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Illig T, Gieger C, Zhai G, Römisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmüller G, Kato BS, Mewes H-W, Meitinger T, de Angelis MH, Kronenberg F, Soranzo N, Wichmann HE, Spector TD, Adamski J, Suhre K. A genome-wide perspective of genetic variation in human metabolism. Nat Genet 2010;42:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guertin KA, Moore SC, Sampson JN, Huang WY, Xiao Q, Stolzenberg-Solomon RZ, Sinha R, Cross AJ. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr 2014;100:208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esko T, Hirschhorn JN, Feldman HA, Hsu YH, Deik AA, Clish CB, Ebbeling CB, Ludwig DS. Metabolomic profiles as reliable biomarkers of dietary composition. Am J Clin Nutr 2017;105:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibbons H, McNulty BA, Nugent AP, Walton J, Flynn A, Gibney MJ, Brennan L. A metabolomics approach to the identification of biomarkers of sugar-sweetened beverage intake. Am J Clin Nutr 2015;101:471–477. [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Aloy M, Llorach R, Urpi-Sarda M, Tulipani S, Estruch R, Martínez-González MA, Corella D, Fitó M, Ros E, Salas-Salvadó J, Andres-Lacueva C. Novel multimetabolite prediction of walnut consumption by a urinary biomarker model in a free-living population: the PREDIMED study. J Proteome Res 2014;13:3476–3483. [DOI] [PubMed] [Google Scholar]
- 16. Playdon MC, Moore SC, Derkach A, Reedy J, Subar AF, Sampson JN, Albanes D, Gu F, Kontto J, Lassale C, Liao LM, Männistö S, Mondul AM, Weinstein SJ, Irwin ML, Mayne ST, Stolzenberg-Solomon R. Identifying biomarkers of dietary patterns by using metabolomics. Am J Clin Nutr 2017;105:450–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Sullivan A, Gibney MJ, Brennan L. Dietary intake patterns are reflected in metabolomic profiles: potential role in dietary assessment studies. Am J Clin Nutr 2011;93:314–321. [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Perez I, Posma JM, Gibson R, Chambers ES, Hansen TH, Vestergaard H, Hansen T, Beckmann M, Pedersen O, Elliott P, Stamler J, Nicholson JK, Draper J, Mathers JC, Holmes E, Frost G. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol 2017;5:184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alshehry ZH, Mundra PA, Barlow CK, Mellett NA, Wong G, McConville MJ, Simes J, Tonkin AM, Sullivan DR, Barnes EH, Nestel PJ, Kingwell BA, Marre M, Neal B, Poulter NR, Rodgers A, Williams B, Zoungas S, Hillis GS, Chalmers J, Woodward M, Meikle PJ. Plasma lipidomic profiles improve on traditional risk factors for the prediction of cardiovascular events in type 2 diabetes mellitus. Circulation 2016;134:1637–1650. [DOI] [PubMed] [Google Scholar]
- 20. Ussher JR, Elmariah S, Gerszten RE, Dyck JR. The emerging role of metabolomics in the diagnosis and prognosis of cardiovascular disease. J Am Coll Cardiol 2016;68:2850–2870. [DOI] [PubMed] [Google Scholar]
- 21. Salas-Salvado J, Bullo M, Estruch R, Ros E, Covas MI, Ibarrola-Jurado N, Corella D, Aros F, Gomez-Gracia E, Ruiz-Gutierrez V, Romaguera D, Lapetra J, Lamuela-Raventos RM, Serra-Majem L, Pinto X, Basora J, Munoz MA, Sorli JV, Martinez-Gonzalez MA. Prevention of diabetes with Mediterranean diets: a subgroup analysis of a randomized trial. Ann Intern Med 2014;160:1–10. [DOI] [PubMed] [Google Scholar]
- 22. Guasch-Ferré M, Zheng Y, Ruiz-Canela M, Hruby A, Martínez-González MA, Clish CB, Corella D, Estruch R, Ros E, Fitó M, Dennis C, Morales-Gil IM, Arós F, Fiol M, Lapetra J, Serra-Majem L, Hu FB, Salas-Salvadó J. Plasma acylcarnitines and risk of cardiovascular disease: effect of Mediterranean diet interventions. Am J Clin Nutr 2016;103:1408–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruiz-Canela M, Guasch-Ferré M, Toledo E, Clish CB, Razquin C, Liang L, Wang DD, Corella D, Estruch R, Hernáez Á, Yu E, Gómez-Gracia E, Zheng Y, Arós F, Romaguera D, Dennis C, Ros E, Lapetra J, Serra-Majem L, Papandreou C, Portoles O, Fitó M, Salas-Salvadó J, Hu FB, Martínez-González MA. Plasma branched chain/aromatic amino acids, enriched Mediterranean diet and risk of type 2 diabetes: case-cohort study within the PREDIMED Trial. Diabetologia 2018;61:1560–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Colditz GA, Manson JE, Hankinson SE. The Nurses’ Health Study: 20-year contribution to the understanding of health among women. J Womens Health 1997;6:49–62. [DOI] [PubMed] [Google Scholar]
- 25. Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, Stampfer MJ. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet 1991;338:464–468. [DOI] [PubMed] [Google Scholar]
- 26. Tworoger SS, Eliassen AH, Zhang X, Qian J, Sluss PM, Rosner BA, Hankinson SE. A 20-year prospective study of plasma prolactin as a risk marker of breast cancer development. Cancer Res 2013;73:4810–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Platz EA, Clinton SK, Giovannucci E. Association between plasma cholesterol and prostate cancer in the PSA era. Int J Cancer 2008;123:1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paynter NP, Balasubramanian R, Giulianini F, Wang DD, Tinker LF, Gopal S, Deik AA, Bullock K, Pierce KA, Scott J, Martinez-Gonzalez MA, Estruch R, Manson JE, Cook NR, Albert CM, Clish CB, Rexrode KM. Metabolic predictors of incident coronary heart disease in women. Circulation 2018;137:841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindstrom S, Loomis S, Turman C, Huang H, Huang J, Aschard H, Chan AT, Choi H, Cornelis M, Curhan G, De Vivo I, Eliassen AH, Fuchs C, Gaziano M, Hankinson SE, Hu F, Jensen M, Kang JH, Kabrhel C, Liang L, Pasquale LR, Rimm E, Stampfer MJ, Tamimi RM, Tworoger SS, Wiggs JL, Hunter DJ, Kraft P. A comprehensive survey of genetic variation in 20,691 subjects from four large cohorts. PLoS One 2017;12:e0173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, Amin N, Buchkovich ML, Croteau-Chonka DC, Day FR, Duan Y, Fall T, Fehrmann R, Ferreira T, Jackson AU, Karjalainen J, Lo KS, Locke AE, Magi R, Mihailov E, Porcu E, Randall JC, Scherag A, Vinkhuyzen AA, Westra HJ, Winkler TW, Workalemahu T, Zhao JH, Absher D, Albrecht E, Anderson D, Baron J, Beekman M, Demirkan A, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Fraser RM, Goel A, Gong J, Justice AE, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Lui JC, Mangino M, Mateo Leach I, Medina-Gomez C, Nalls MA, Nyholt DR, Palmer CD, Pasko D, Pechlivanis S, Prokopenko I, Ried JS, Ripke S, Shungin D, Stancakova A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Afzal U, Arnlov J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Bluher M, Bolton JL, Bottcher Y, Boyd HA, Bruinenberg M, Buckley BM, Buyske S, Caspersen IH, Chines PS, Clarke R, Claudi-Boehm S, Cooper M, Daw EW, De Jong PA, Deelen J, Delgado G, Denny JC, Dhonukshe-Rutten R, Dimitriou M, Doney AS, Dorr M, Eklund N, Eury E, Folkersen L, Garcia ME, Geller F, Giedraitis V, Go AS, Grallert H, Grammer TB, Grassler J, Gronberg H, de Groot LC, Groves CJ, Haessler J, Hall P, Haller T, Hallmans G, Hannemann A, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hemani G, Henders AK, Hillege HL, Hlatky MA, Hoffmann W, Hoffmann P, Holmen O, Houwing-Duistermaat JJ, Illig T, Isaacs A, James AL, Jeff J, Johansen B, Johansson A, Jolley J, Juliusdottir T, Junttila J, Kho AN, Kinnunen L, Klopp N, Kocher T, Kratzer W, Lichtner P, Lind L, Lindstrom J, Lobbens S, Lorentzon M, Lu Y, Lyssenko V, Magnusson PK, Mahajan A, Maillard M, McArdle WL, McKenzie CA, McLachlan S, McLaren PJ, Menni C, Merger S, Milani L, Moayyeri A, Monda KL, Morken MA, Muller G, Muller-Nurasyid M, Musk AW, Narisu N, Nauck M, Nolte IM, Nothen MM, Oozageer L, Pilz S, Rayner NW, Renstrom F, Robertson NR, Rose LM, Roussel R, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Schunkert H, Scott RA, Sehmi J, Seufferlein T, Shi J, Silventoinen K, Smit JH, Smith AV, Smolonska J, Stanton AV, Stirrups K, Stott DJ, Stringham HM, Sundstrom J, Swertz MA, Syvanen AC, Tayo BO, Thorleifsson G, Tyrer JP, van Dijk S, van Schoor NM, van der Velde N, van Heemst D, van Oort FV, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Waldenberger M, Wennauer R, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, Arveiler D, Bakker SJ, Beilby J, Bergman RN, Bergmann S, Biffar R, Blangero J, Boomsma DI, Bornstein SR, Bovet P, Brambilla P, Brown MJ, Campbell H, Caulfield MJ, Chakravarti A, Collins R, Collins FS, Crawford DC, Cupples LA, Danesh J, de Faire U, den Ruijter HM, Erbel R, Erdmann J, Eriksson JG, Farrall M, Ferrannini E, Ferrieres J, Ford I, Forouhi NG, Forrester T, Gansevoort RT, Gejman PV, Gieger C, Golay A, Gottesman O, Gudnason V, Gyllensten U, Haas DW, Hall AS, Harris TB, Hattersley AT, Heath AC, Hengstenberg C, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Hovingh GK, Humphries SE, Hunt SC, Hypponen E, Jacobs KB, Jarvelin MR, Jousilahti P, Jula AM, Kaprio J, Kastelein JJ, Kayser M, Kee F, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kooner JS, Kooperberg C, Koskinen S, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimaki T, Lupoli S, Madden PA, Mannisto S, Manunta P, Marette A, Matise TC, McKnight B, Meitinger T, Moll FL, Montgomery GW, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Ouwehand WH, Pasterkamp G, Peters A, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ritchie M, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schwarz PE, Sebert S, Sever P, Shuldiner AR, Sinisalo J, Steinthorsdottir V, Stolk RP, Tardif JC, Tonjes A, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Electronic Medical R, Genomics C, Consortium MI, Consortium P, LifeLines Cohort S, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PI, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hayes MG, Hui J, Hunter DJ, Hveem K, Jukema JW, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, Marz W, Melbye M, Moebus S, Munroe PB, Njolstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Perusse L, Peters U, Powell JE, Power C, Quertermous T, Rauramaa R, Reinmaa E, Ridker PM, Rivadeneira F, Rotter JI, Saaristo TE, Saleheen D, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Strauch K, Stumvoll M, Tuomilehto J, Uusitupa M, van der Harst P, Volzke H, Walker M, Wareham NJ, Watkins H, Wichmann HE, Wilson JF, Zanen P, Deloukas P, Heid IM, Lindgren CM, Mohlke KL, Speliotes EK, Thorsteinsdottir U, Barroso I, Fox CS, North KE, Strachan DP, Beckmann JS, Berndt SI, Boehnke M, Borecki IB, McCarthy MI, Metspalu A, Stefansson K, Uitterlinden AG, van Duijn CM, Franke L, Willer CJ, Price AL, Lettre G, Loos RJ, Weedon MN, Ingelsson E, O’Connell JR, Abecasis GR, Chasman DI, Goddard ME, Visscher PM, Hirschhorn JN, Frayling TM. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014;46:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Estruch R, Martínez-González MA, Corella D, Salas-Salvadó J, Ruiz-Gutiérrez V, Covas MI, Fiol M, Gómez-Gracia E, López-Sabater MC, Vinyoles E, Arós F, Conde M, Lahoz C, Lapetra J, Sáez G, Ros E; PREDIMED Study Investigators. Effects of a Mediterranean-style diet on cardiovascular risk factors: a randomized trial. Ann Intern Med 2006;145:1–11. [DOI] [PubMed] [Google Scholar]
- 32. Schröder H, Fitó M, Estruch R, Martínez‐González MA, Corella D, Salas‐Salvadó J, Lamuela‐Raventós R, Ros E, Salaverría I, Fiol M, Lapetra J, Vinyoles E, Gómez‐Gracia E, Lahoz C, Serra‐Majem L, Pintó X, Ruiz‐Gutierrez V, Covas MI. A short screener is valid for assessing mediterranean diet adherence among older Spanish men and women. J Nutr 2011;141:1140–1145. [DOI] [PubMed] [Google Scholar]
- 33. Willett WC, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol 1985;122:51–65. [DOI] [PubMed] [Google Scholar]
- 34. Mendis S, Thygesen K, Kuulasmaa K, Giampaoli S, Mahonen M, Ngu Blackett K, Lisheng L; . Writing group on behalf of the participating experts of the WHO consultation for revision of WHO definition of myocardial infarction. World Health Organization definition of myocardial infarction: 2008-09 revision. Int J Epidemiol 2011;40:139–146. [DOI] [PubMed] [Google Scholar]
- 35. Walker AE, Robins M, Weinfeld FD. The National Survey of Stroke. Clinical findings. Stroke 1981;12:I13–I44. [PubMed] [Google Scholar]
- 36. Stampfer MJ, Willett WC, Speizer FE, Dysert DC, Lipnick R, Rosner B, Hennekens CH. Test of the National Death Index. Am J Epidemiol 1984;119:837–839. [DOI] [PubMed] [Google Scholar]
- 37. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 38. Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. J Clin Epidemiol 1999;52:1165–1172. [DOI] [PubMed] [Google Scholar]
- 39. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017;46:1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pvd H, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD Jr, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen WM, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu FC, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jimenez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee JM, Lemmens R, Leys D, Lewis CM, Lin WY, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmae K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf SAFGen Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; International Genomics of Blood Pressure (iGEN-BP) Consortium; INVENT Consortium; STARNETBis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Bjorkegren JLM, Codoni V, Civelek M, Smith NL, Tregouet DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD; BioBank Japan Cooperative Hospital Group; COMPASS Consortium; EPIC-CVD Consortium; EPIC-InterAct Consortium; International Stroke Genetics Consortium (ISGC); METASTROKE Consortium; Neurology Working Group of the CHARGE Consortium; NINDS Stroke Genetics Network (SiGN); UK Young Lacunar DNA Study; MEGASTROKE Consortium, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT Jr., Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–537.29531354 [Google Scholar]
- 42. Lu Y, Zou L, Su J, Tai E, Whitton C, van Dam R, Ong C. Meat and seafood consumption in relation to plasma metabolic profiles in a Chinese population: a combined untargeted and targeted metabolomics study. Nutrients 2017;9:683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cazzola R, Cestaro B. Red wine polyphenols protect n−3 more than n−6 polyunsaturated fatty acid from lipid peroxidation. Food Res Int 2011;44:3065–3071. [Google Scholar]
- 44. Chowdhury R, Stevens S, Gorman D, Pan A, Warnakula S, Chowdhury S, Ward H, Johnson L, Crowe F, Hu FB, Franco OH. Association between fish consumption, long chain omega 3 fatty acids, and risk of cerebrovascular disease: systematic review and meta-analysis. BMJ 2012;345:e6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guasch-Ferré M, Hu FB, Martínez-González MA, Fitó M, Bulló M, Estruch R, Ros E, Corella D, Recondo J, Gómez-Gracia E, Fiol M, Lapetra J, Serra-Majem L, Muñoz MA, Pintó X, Lamuela-Raventós RM, Basora J, Buil-Cosiales P, Sorlí JV, Ruiz-Gutiérrez V, Martínez JA, Salas-Salvadó J. Olive oil intake and risk of cardiovascular disease and mortality in the PREDIMED Study. BMC Med 2014;12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Newgard CB. Metabolomics and metabolic diseases: where do we stand? Cell Metab 2017;25:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism 2010;59:993–999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.