Abstract
We developed a model to predict the cyclic pattern of porcine reproductive and respiratory syndrome virus (PRRSV) RNA detection by reverse-transcription real-time PCR (RT-rtPCR) from 4 major swine-centric veterinary diagnostic laboratories (VDLs) in the United States and to use historical data to forecast the upcoming year’s weekly percentage of positive submissions and issue outbreak signals when the pattern of detection was not as expected. Standardized submission data and test results were used. Historical data (2015–2017) composed of the weekly percentage of PCR-positive submissions were used to fit a cyclic robust regression model. The findings were used to forecast the expected weekly percentage of PCR-positive submissions, with a 95% confidence interval (CI), for 2018. During 2018, the proportion of PRRSV-positive submissions crossed 95% CI boundaries at week 2, 14–25, and 48. The relatively higher detection on week 2 and 48 were mostly from submissions containing samples from wean-to-market pigs, and for week 14–25 originated mostly from samples from adult/sow farms. There was a recurring yearly pattern of detection, wherein an increased proportion of PRRSV RNA detection in submissions originating from wean-to-finish farms was followed by increased detection in samples from adult/sow farms. Results from the model described herein confirm the seasonal cyclic pattern of PRRSV detection using test results consolidated from 4 VDLs. Wave crests occurred consistently during winter, and wave troughs occurred consistently during the summer months. Our model was able to correctly identify statistically significant outbreak signals in PRRSV RNA detection at 3 instances during 2018.
Keywords: cyclic, outbreak signal, PRRSV, prediction, swine pathogens, veterinary diagnostic laboratories
Introduction
Every day, thousands of porcine samples representing hundreds of submissions are submitted to veterinary diagnostic laboratories (VDLs) for routine testing for porcine reproductive and respiratory syndrome virus 1 and 2 (PRRSV; Betaarterivirus suid 1, Betaarterivirus suid 2). Considering a “case” as the information corresponding to a sample submission to a VDL and identified by a unique accession identifier, 67,039 submissions were sent during 2018 to 4 major swine-centric VDLs in the United States and tested for PRRSV by reverse-transcription real-time PCR (RT-rtPCR), according to the procedures in use and under a fee-for-service basis, which was an average of 1,289 weekly submissions, or 258 submissions per working day.12 Submission information, sample types, and test results are routinely stored on the individual VDLs’ database servers known as laboratory information management systems. The patterns identified using these veterinary laboratory results can provide valuable insights for animal health monitoring and disease surveillance. Three steps should be taken with animal surveillance and disease outbreak investigation4: 1) retrospective evaluation of the data to identify any temporal or spatial patterns, or external covariates, or both; 2) model or remove these explainable patterns from the data (pre-processing); and 3) monitor pre-processed data prospectively to detect unexpected clusters of events (possible outbreak signals) in time or space-time.
Different levels of animal health syndromic surveillance and various statistical approaches have been applied in different situations and scenarios for veterinary surveillance.4 The most commonly reported approaches in veterinary syndromic surveillance include descriptive summaries, control charts, Bayesian belief networks, regression models, and various algorithms from the surveillance package in R.4 Model selection depends on database organization and structural characteristics, and the monitoring or surveillance objective. To use a large database for veterinary surveillance, the ability must exist to process a large volume of data and to efficiently aggregate it in a structured format, in order for the data to be used prospectively.13 However, the use of large datasets in veterinary medicine in a way that generates useful information in a prospective format is challenging.13 Information needs to be analyzed and distributed for the users as quickly as it is generated to allow informed decision-making on control and management of disease.
The Swine Disease Reporting System (SDRS; http://www.fieldepi.org/SDRS) has been aggregating results from swine samples routinely submitted and tested for PRRSV by RT-rtPCR from 4 U.S. VDLs: Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL), University of Minnesota Veterinary Diagnostic Laboratory (UMN-VDL), South Dakota State University Animal Disease Research & Diagnostic Laboratory (SDSU-ADRDL), and Kansas State University Veterinary Diagnostic Laboratory (KSU-VDL).12 This large database containing PRRSV test results can be used for PRRSV surveillance and monitoring at the national level. The 4 VDLs reporting to the SDRS test > 95% of all porcine submissions submitted for the National Animal Health Laboratory Network (NAHLN) in the United States.12 Submissions with the following reasons were removed: submission as “research,” reason for testing as “exporting,” specimen as “vaccine,” or specimen as “environmental sample.” After removing these submissions, SDRS recorded 547,873 PRRSV submissions between 2007 and 2018, collected from 49 of 50 states in the United States and submitted for testing by RT-rtPCR.12
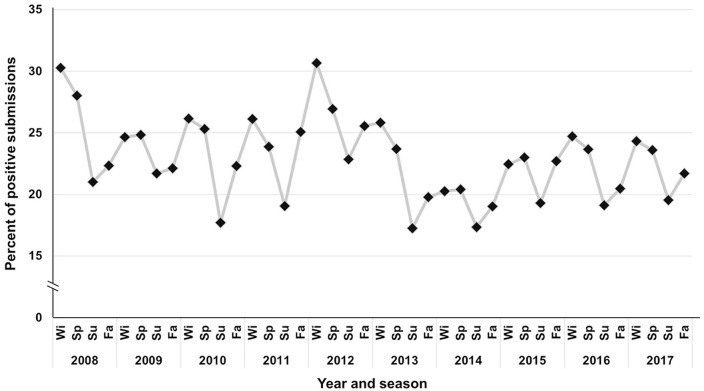
Historical data on PRRSV RNA testing by RT-rtPCR viewed as the “PRRSV-positive submissions” tested divided by total submissions tested for PRRSV (positive, negative, suspect, or inconclusive) revealed an apparent seasonal cyclic pattern of PRRSV detection.10 The highest proportion of PRRSV RT-rtPCR–positive results, organized at a submission level, was consistently detected during winter (December, January, and February) and spring months (March, April, and May), and the lowest during summer (June, July, and August) and fall months (September, October, and November).
In human disease surveillance, the cyclic pattern of disease occurrence and detection has long been explored to monitor disease occurrence and identify outbreak signals. Regression models have been used to predict seasonal changes in human mortality to allow detection of periods with excess mortality caused by influenza epidemics.9 In 2008, a study from Australia described a refinement of this model to monitor the proportion of mortality in humans caused by pneumonia and influenza with a cyclic pattern of occurrence.6 SDRS PRRSV detection information has demonstrated great similarity to human influenza disease occurrence, creating an opportunity to apply a cyclic algorithm to proactively detect PRRSV outbreaks in near-real-time.
The purpose of our study was to develop and apply an algorithm to historical SDRS results for PRRSV RNA detection to characterize the cyclic pattern of detection, construct a capability to forecast the proportion of positive submissions for the upcoming 52-wk period, and thus identify weeks having a significant increase or decrease in detection.
Materials and methods
To confirm the cyclic pattern of PRRSV detection reported in the SDRS database (Fig. 1), we used the following method to address the 3 proposed steps for animal surveillance and disease outbreak investigations.4 An algorithm (SAS v.9.4; SAS Institute, Cary, NC) was created to organize SDRS database results by weekly counts of positive submissions, negative submissions, and total submissions tested. A function to calculate the percentage of positive submissions was applied. Each year was considered and organized in a cycle composed of 52 observations corresponding to 52 wk. Weekly data from 2015, 2016, and 2017 were used to construct the baseline for PRRSV detection. Further description of the data structure used in our work is available in detail in our previous study.12
Figure 1.
Percent of porcine reproductive and respiratory syndrome virus (PRRSV)-positive submissions reported at the Swine Disease Reporting System (SDRS) project over time and apparent seasonal pattern of detection of PRRSV over time. The seasons are organized by months: winter (Wi) = December, January, and February; spring (Sp) = March, April, and May; summer (Su) = June, July, and August; fall (Fa) = September, October, and November. For the SDRS database, a full 4-season cycle starts December 1 and ends November 30 of the subsequent year.
The model to analyze the cyclic pattern of PRRSV detection by VDLs was based on the cyclic regression model used to monitor the proportion of deaths attributable to influenza in humans in Australia.6 We refined the original model used to calculate the 95% confidence interval (95% CI) for predicted values and data visualization. A SAS standard procedure resistant to outliers named PROC ROBUSTREG was used to fit a robust regression model. The M estimation available using the SAS PROC ROBUSTREG procedure was used to estimate the predicted percentage of positive submissions for the upcoming year of 2018. It has been established that the M procedure can be feasibly used when outlier contamination is present mainly in the response direction and produces reasonable estimation.
The cyclic regression model included a linear time term, T, with values 1, 2, 3, . . . 207, 208. Term T is composed of 4 cycles of 52-wk observations corresponding to the first 3 cycles to each individual historic year period used to construct the baseline (January 2015–December 2017), and the last cycle for the sequences time series for the upcoming year (January–December 2018) was filled with blank values during the forecast steps. This step triggers the SAS procedure to insert forecast values for those weeks based on the model fitted to the earlier data. These forecasted results generated by the model were used as the upcoming year’s baseline reference results. The T variable has the function to accommodate the long-term linear and curvilinear changes in the background of the percentage of positive results.6 Annual seasonal harmonic variables were added to describe the cyclic seasonal background pattern. The harmonic variables are functions of the week of the year, t, and the periodicity is in the same units, in this case, yearly (52.18 wk). The 2 harmonic variables, in this case, are sine and cosine of the week of the year, t, and calculated as sine of (2π t/52.18) and cosine of (2π t/52.18),6 where π is the constant “Pi.” Each 52-wk year defines a cycle responsible for forming the wave pattern. Sine and cosine accommodate the curvilinear wave high, amplitude, wave crest, and wave trough. The final model can be written as: predicted percentage of weekly positive results = œ +ß1 T + ß2 sine + ß3 cosine, in which œ is the intercept estimate for the percentage of positive submissions, ß1 is the estimate for the T variable, ß2 is the estimate for sine, and ß3 is the estimate for cosine.
The forecast of the predicted individual results for each of the 52 wk of the upcoming year of 2018 was retrieved from the model. A 95% CI for each predicted weekly value was calculated using the SAS DATA STEP procedure using the predicted standard error value for each individual weekly result captured from the output of the robust regression model. For the purpose of our study and aligned with the World Health Organization’s definition, an outbreak signal was defined as the percentage of observed PCR-positive submissions exceeding the number of expected PCR-positive submissions (i.e., not site-level PRRSV detection). The goodness of fit of the model with relation to leverage and the potential influence of extreme outliers was assessed by the standard procedure available in the PROC ROBUSTREG procedure. The pattern of detection of PRRSV in the United States significantly changed during and following the porcine epidemic diarrhea virus (PEDV) and porcine deltacoronavirus (PDCoV) epidemic years of 2013 and 2014.10,14 Thus, inclusion of historical data was restricted to the period after 2014. Otherwise, the R2 reported by the model output would have been used to select the number of years included in the baseline (historic period), and we would have used the model with the highest R2.
Historical results, 2018 predicted results, and the 95% CI for the predicted weekly result were plotted using the SAS PROC SGPANEL procedure. An algorithm was developed to capture the 2018 weekly results released by participant VDLs and include them in the SDRS database. The algorithm was also used to calculate the percentage of positive submissions, integrate the predicted information generated by the model from historical data, and plot historical and 2018 results. For the purpose of our study, an outbreak signal was defined as the time at which the percentage of positive submissions results for PRRSV detection rate crossed the threshold of 95% CI boundaries.
The use of results from the same dataset to generate a model is the most plausible way to validate an indicator of outbreak signals.4 During 2018, outbreaks signals were validated by comparing monthly changes in the pattern of agent detection by production phase, and by input from a panel of specialists. The original SDRS project described 8 age categories: adult, boar stud, breeding herd, replacement, suckling piglets, nursery, grow-finish, and unknown. To better understand the origin of an outbreak signal, we grouped the age categories in 3 swine production phases: adult/sow farm, wean-to-market, and unknown. Adult/sow farm aggregated the SDRS age categories of adult, boar stud, breeding herd, replacement, and suckling piglets. Wean-to-market aggregated the SDRS age categories of nursery and grow-finish. The unknown category corresponds to the SDRS unspecified age category. Plots of the percentage of positive results by year and month were constructed (Power Business Intelligence; Microsoft, Redmond, WA). Generated outbreak signals and monthly changes in the percentage of positive results by phase were discussed with a panel of specialists comprised of the SDRS advisory council and VDL diagnosticians during 2018. The SDRS advisory council was composed of swine field veterinarians from different production systems distributed across the United States. The major inputs and findings were preliminarily incorporated into the SDRS monthly report released in 2018.
Results
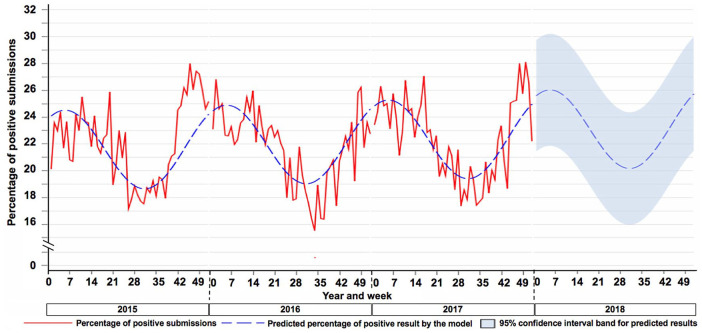
The proposed work was able to develop and apply an algorithm to scan historical SDRS data, forecast the weekly expected percentage of RT-rtPCR–positive submissions for the upcoming year, and characterize a cyclic pattern of PRRSV RNA detection in the United States using VDL results. The weekly predicted percentage of positive results for 2018 followed a consistent pattern for wave crest and wave trough according to the weekly detected proportion of positive submissions in 2015–2017 (Fig. 2). The percent of positive results for PRRSV forecasted by the model was expected to increase over time. The increase in the wave crest was expected to increase by 1.5% from 2015 to the predicted year of 2018, moving from 24.5% in 2015 to 26.0% in 2018 (Fig. 2).
Figure 2.
Predicted value and 95% confidence interval (CI) for the 2018 forecasted percentage of submissions tested positive for porcine reproductive and respiratory syndrome virus. Dashed blue line = predicted weekly percentage of positive results. Continuous red line = observed percentage of weekly positive submissions for 2015–2017. 2018 blue band = 95% CI for the weekly predicted percentage of positive results.
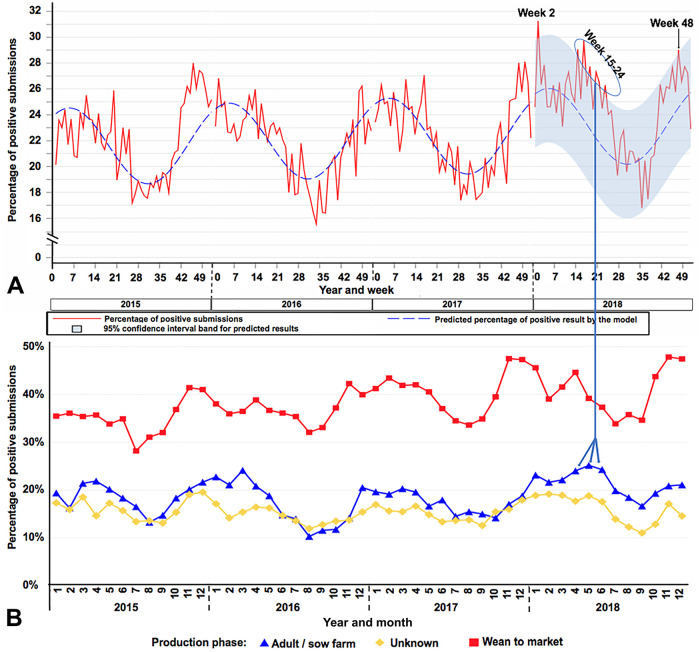
During 2018, 3 time periods generated outbreak signals: the first was on week 2 (January 7–13), the second was week 15–24 (April 8–June 16), and the third was on week 48 (November 25–December 1; Fig. 3A). The first outbreak signal was driven by increased detection in wean-to-market animals. In week 2 of 2018, 185 of 357 (51.8%) submissions coming from wean-to-market pigs tested positive for PRRSV, whereas in the previous week only 128 of 297 (43.1%) tested positive. The second and largest outbreak signal was driven mainly by increased detection of PRRSV in adult/sow farms (Fig. 3B). Data suggest that the second outbreak signal was preceded by an increased percentage of positive submissions detected in the production phase wean-to-market that occurred late in March and April (Fig. 3B). During 2018, a specimen described as “processing fluid”5 was increasingly used for PRRSV testing. Prior to 2018, processing fluid was not consistently used for PRRSV testing. During 2018, processing fluid represented 7,721 of 27,733 (27.8%) submissions tested for adult/sow farms, and 2,513 of 7,721 (32.5%) submissions tested positive; in all other specimens, 3,390 of 20,012 (16.9%) submissions tested positive. The third outbreak signal was also driven by increased detection in the wean-to-market phase. In week 48, 228 of 454 (50.2%) submissions tested positive for PRRSV in the wean-to-market phase, whereas in the previous week 218 of 451 (48.3%) tested positive. Additionally, the average number of weekly submissions tested from wean-to-market moved from 362 in October to 410 in November, with the percentage of positive results from 727 of 1,663 (43.7%) to 889 of 1,859 (47.8%), respectively.
Figure 3.
A. Percentage of positive submissions detected during 2018. Inertia ellipses represent outbreaks signals. Arrows point to the production phase results (in panel B) that were associated with the outbreak signal. B. Percentage of positive submissions by production phase over year and month. Each color and marker format represent a different production phase as indicated at the bottom of the chart.
The use of production phase–grouped results to validate the outbreak signal provided 3 pieces of additional information:
The wean-to-market pattern of the percentage of positive submissions was consistent over the years, with the lowest points of detection between July and September, and an increasing trend starting thereafter (Fig. 3B).
The pattern of the percentage of positive results in adult/sow farms was similar over time, and the increase in the percentage of positive results occurring in the second half of the year was preceded by an increased proportion of positivity in the wean-to-market phase (Fig. 3B).
The overall percentage of PRRSV-positive submissions in wean-to-market is consistently higher than the percentage of PRRSV-positive submissions in adult/sow farms (Fig. 3B).
Discussion
The SDRS project described a seasonal cyclic pattern of PRRSV RT-rtPCR detection. Our goal was to develop an algorithm to scan the SDRS historical database for PRRSV detection, forecast the predicted weekly results for the upcoming year, and to issue early outbreak signals. Our algorithm integrating the cyclic pattern of occurrence was able to confirm the cyclic pattern of PRRSV detection and predict the expected detection for the upcoming year based on historical data. Outbreak signals were detected and further validated with changes in the pattern of detection by phase and with input from a panel of specialists. Outbreaks were defined as a significant increase in PRRSV-positive submissions relative to expected values for that time of the year.
During 2018, 3 outbreak signals were detected and further validated with increased detection in at least 1 phase of production. The first and third outbreaks signals lasted for 1 wk only. In the first outbreak signal, there was an increased proportion of positive submissions in wean-to-market and an increase in the number of submissions submitted for PRRSV testing. In the third outbreak signal, the number of submissions tested coming from wean-to-market only changed by 3 from the previous week, but the proportion of positive submissions increased from 48.3% to 50.2%. In addition, the average number of submissions tested for PRRSV from wean-to-market increased from 362 in October to 410 in November and the proportion of positive submissions increased from 43.7% to 47.8%. When comparing the first and third outbreak signals, the first was more likely to be the consequence of the New Year holiday whereby fewer samples were submitted for testing in the first week of the year. This was reflected by a subsequent high number of submissions in the following week. In comparison, the third outbreak signal was likely a true outbreak signal based on a consistent average weekly increase in the number of submissions tested (48 submissions/week) from October to November. This was accompanied by a 4.1% increase in the percentage of positive submissions.
The longest outbreak signal occurred between April and June and was mainly driven by tested submissions from adult/sow farms. The occurrence of these outbreak signals coincided with a previously increased detection of PRRSV in the age category wean-to-market and the adoption of processing fluid to monitor PRRSV in the piglets during the first week of life. Potential reasons noted by the advisory council for increased detection of PRRSV at that time included 1) likely lateral transmission of PRRSV from wean-to-market farms to adult/sow farms, and 2) the possibility of more piglets sampled by using processing fluid, which is a population-based sampling method. Detection of PRRSV in processing fluids contributed to an increase in the percentage of positive submissions for adult/sow farms by 5.2% and a 1.5% increase in all production phases during spring months (March, April, and May), and by 5.8% in adult/sow farms, and a 1.8% in all production phases during summer months (June, July, and August).
The increased proportion of positive submissions observed in wean-to-market animals before the second outbreak corresponds with the historical recurring pattern of detection wherein the proportion of positive submissions in wean-to-market precedes the increase in adult/sow farms. This finding strongly suggests that wean-to-market animals potentially play a major role in PRRSV macroepidemiologic dynamics and should receive attention. Additionally, this finding perhaps should be taken into consideration in design plans for regional PRRSV control and eradication programs.
Our model was able to scan the PRRSV RT-rtPCR VDL results and then predict the expected proportion of submissions status results. The observed results followed the prediction and crossed the 95% CI when a contributing factor, such as holidays and or the introduction of a new specimen type, was detected (Fig. 3). Our findings strongly suggest that VDL historical results can be used to forecast PRRSV detection. There was a marked seasonal pattern of pathogen detection, with an increased percentage of positive submissions during relatively “cold” months. The relatively higher detection of PRRSV during colder months agrees with the relatively higher incidence of PRRSV outbreaks in sow farms.7,11 The higher detection of PRRSV during winter months may be a result of the virus remaining infectious longer in the environment and allowing an opportunity to be easily mechanically transported during colder weather3 compared with warmer weather.2 The seasonal temperature effect has been previously noted as a major risk factor associated with PRRSV spread in the midwestern United States.1 Additionally, regional effects and clustering of different PRRSV strains may contribute to the spatial and temporal distribution of PRRSV.8
In addition to the macroepidemiologic level of monitoring by our model, veterinarians and producers may use the model to detect changes in disease status early at the farm level. When outbreak signals are issued, reinforcement of farm biosecurity, review of pig and people flow, and truck decontamination audit procedures are measures that could potentially limit further pathogen spread. Finally, our algorithm could be further explored for the detection of other swine disease agents that have a cyclic pattern of occurrence, such as PEDV and PDCoV.
Acknowledgments
We thank VDL clients for submitting samples for testing. We also thank current and former SDRS advisory council members for their valuable input and volunteered time: Drs. Clayton Johnson, Emily Byers, Hans Rotto, Jeremy Pittman, Mark Schwartz, Paul Yeske, Pete Thomas, Rebecca Robbins, Tara Donovan, Matthew Turner, Deborah Murray, Scott Dee, and Melissa Hensch.
Footnotes
Availability of data and materials: The aggregated information of the percentage of positive results by season is publicly available online at http://www.fieldepi.org/SDRS in the PRRSV dashboard. Monthly reports containing the prediction model plots are available at the Swine Health Information Center (SHIC) under the Domestic Disease Monitoring Reports: https://www.swinehealth.org/domestic-disease-surveillance-reports/. Restrictions apply to the availability of additional weekly data, given VDL confidentiality, and are not publicly available. SAS algorithm procedures can be made available upon reasonable request and approval by SDRS principal investigator, SDRS project coordinator, and VDL directors.
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This project was co-funded by the American Association of Swine Veterinarians Foundation award 018743-00001 and by the Swine Health Information Center (SHIC) awards 17-210 and 19-155 SHIC to D.C.L. Linhares.
ORCID iDs: Giovani Trevisan  https://orcid.org/0000-0002-4980-526X
https://orcid.org/0000-0002-4980-526X
Leticia C. M. Linhares  https://orcid.org/0000-0001-5305-7981
https://orcid.org/0000-0001-5305-7981
Eric R. Burrough  https://orcid.org/0000-0003-4747-9189
https://orcid.org/0000-0003-4747-9189
References
- 1. Alkhamis MA, et al. Novel approaches for spatial and molecular surveillance of porcine reproductive and respiratory syndrome virus (PRRSv) in the United States. Sci Rep 2017;7:4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dee S, et al. Mechanical transmission of porcine reproductive and respiratory syndrome virus throughout a coordinated sequence of events during warm weather. Can J Vet Res 2003;67:12–19. [PMC free article] [PubMed] [Google Scholar]
- 3. Dee S, et al. Mechanical transmission of porcine reproductive and respiratory syndrome virus throughout a coordinated sequence of events during cold weather. Can J Vet Res 2002;66:232–239. [PMC free article] [PubMed] [Google Scholar]
- 4. Dórea FC, Vial F. Animal health syndromic surveillance: a systematic literature review of the progress in the last 5 years (2011–2016). Vet Med (Auckl) 2016;7:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lopez WA, et al. Porcine reproductive and respiratory syndrome monitoring in breeding herds using processing fluids. J Swine Health Prod 2018;26:146–150. [Google Scholar]
- 6. Muscatello DJ, et al. Prospective surveillance of excess mortality due to influenza in New South Wales: feasibility and statistical approach. Commun Dis Intell Q Rep 2008;32:435–442. [DOI] [PubMed] [Google Scholar]
- 7. Perez AM, et al. Individual or common good? Voluntary data sharing to inform disease surveillance systems in food animals. Front Vet Sci 2019;6:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosendal T, et al. Spatial and temporal patterns of porcine reproductive and respiratory syndrome virus (PRRSV) genotypes in Ontario, Canada, 2004–2007. BMC Vet Res 2014;10:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serfling RE. Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Rep 1963;78:494–506. [PMC free article] [PubMed] [Google Scholar]
- 10. Stevenson GW, et al. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 2013;25:649–654. [DOI] [PubMed] [Google Scholar]
- 11. Tousignant SJ, et al. Temporal and spatial dynamics of porcine reproductive and respiratory syndrome virus infection in the United States. Am J Vet Res 2015;76:70–76. [DOI] [PubMed] [Google Scholar]
- 12. Trevisan G, et al. Macroepidemiological aspects of porcine reproductive and respiratory syndrome virus detection by major United States veterinary diagnostic laboratories over time, age group, and specimen. PLoS One 2019;14:e0223544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. VanderWaal K, et al. Translating big data into smart data for veterinary epidemiology. Front Vet Sci 2017;4:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang L, et al. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 2014;20:1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]