Abstract
Streptococcus suis is one of the most important bacterial swine pathogens worldwide and is an emerging pathogen in humans. There are 29 serotypes, and serotyping, which is based on the antigenicity of the capsular polysaccharide (CPS) or on its coding genes, is often part of routine identification and provides further information regarding S. suis virulence and zoonotic potential. Serotypes 2 and 14 possess high zoonotic potential, and serotype 1/2 is the serotype most frequently isolated from diseased pigs in North America. PCR has replaced antibody-based techniques to perform serotyping. However, traditional PCR is not able to differentiate serotype 2 from 1/2 and serotype 1 from 14, given that the only difference in the cps loci of those serotype pairs is a nonsynonymous single-nucleotide polymorphism. We developed a mismatch amplification mutation assay (MAMA)-PCR that was able to correctly serotype 148 isolates previously known to be serotypes 1, 2, 1/2, or 14. This technique will be highly useful in animal and human health laboratories performing PCR serotyping of S. suis isolates.
Keywords: mismatch amplification mutation assay; PCR identification; Streptococcus suis serotypes 1, 2, 1/2, and 14
Streptococcus suis is one of the most important bacterial pathogens of post-weaned piglets and is responsible for extensive economic losses, with sudden death, meningitis, endocarditis, and arthritis being the most frequent clinical manifestations.6 Moreover, S. suis is a zoonotic agent responsible for meningitis and septic shock in humans.4,17 Serotyping, which is often part of the routine identification of S. suis isolates recovered from diseased pigs and humans, provides further confirmation regarding the identity and virulence potential of this pathogen. A total of 35 serotypes of S. suis serotypes had been originally described based on the antigenicity of their capsular polysaccharide (CPS).6 However, 6 of those serotypes have recently been classified as belonging to another bacterial species.8 Serotypes 20, 22, and 26 have been reclassified as S. parasuis, serotypes 32 and 34 as S. orisratti, and serotype 33 as S. ruminantium.13
Serologic typing, which is one of the most important contributors to the characterization of S. suis infection, has historically been performed with reference antisera using a coagglutination test, capillary precipitation test, or the Neufeld capsular reaction test.6 Some serotypes cross-react, indicating the presence of common antigenic determinants. For example, cross-reactions have been observed between serotype 1/2 and serotypes 1 and 2, and between serotypes 1 and 14.5 These cross-reactions are the result of similar or closely related structural features of the CPS between those serotypes, described in 2016.14 Serotype 2 CPS contains galactose, glucose, N-acetylglucosamine, rhamnose, and sialic acid; serotype 14 CPS possesses galactose, glucose, N-acetylglucosamine, and sialic acid. Serotype 1/2 CPS differs from serotype 2 CPS, and serotype 1 CPS from serotype 14 CPS, by a single substitution of the galactose residue bearing sialic acid in serotypes 2 and 14 CPS side chains by an N-acetylgalactosamine (GalNAc) residue.14–16
Molecular serotyping methods based on CPS-coding genes have been described, including a 2-step multiplex PCR assay.9 However, despite the differences in CPS sugar composition and structure, and the fact that all other serotypes possess a “serotype-specific” gene, serotype pairs 2 and 1/2, and 1 and 14, have identical CPS gene contents.9 Indeed, all PCR tests described so far are unable to differentiate those serotypes.9 It has been reported that the only consistent difference in the cps loci of strains of these serotype pairs was a nonsynonymous single-nucleotide polymorphism (SNP) in codon 161 of gene cpsK, predicted to result in a single amino acid difference in glycosyltransferase CpsK (W161 in serotypes 2 and 14, and C161 in serotypes 1/2 and 1).11 Indeed, it is possible to achieve serotype switching of field strains of serotypes 2 and 1/2, and 14 and 1, solely by replacing the amino acid 161 of CpsK.9 We report herein a new mismatch amplification mutation assay (MAMA) that can correctly serotype strains of S. suis serotypes 1, 2, 1/2, and 14 by PCR.
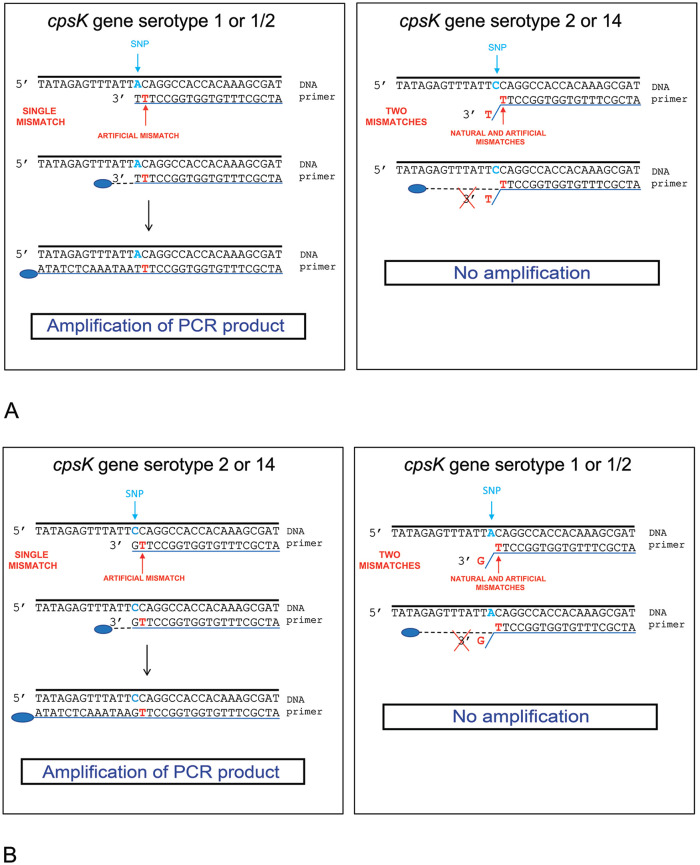
A MAMA is a PCR-based technique for SNP discrimination that uses SNP-specific primers at the 3□-end.2 We selected candidate genes according to a previous publication.1 One forward primer was designed to be complementary to the corresponding allele of the cpsK gene of serotypes 1 and 1/2, with one mismatch introduced at the second nucleotide from the 3□-end of the primer, resulting in 2 mismatches with serotypes 2 and 14 (Fig. 1A). We designed a second forward primer, but this time, complementary to cpsK gene with 1 mismatch for serotypes 2 and 14 and 2 mismatches for serotypes 1 and 1/2 (Fig. 1B). Allele-specific primers carrying a single mismatch had no effect on the overall PCR yield, whereas primers carrying ≥ 2 consecutive mismatches at the 3□-end failed to generate any detectable amplification products.
Figure 1.
Schematic representation of mismatch amplification mutation assay (MAMA) A. PCR1. B. PCR2. SNP = single-nucleotide polymorphism.
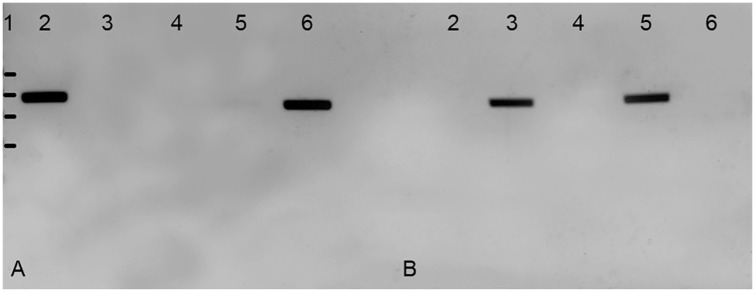
Each 20-μL reaction mixture contained 0.2 μM of each primer (Table 1), 0.2 mM of dNTPs, 1× TopTaq buffer (Qiagen, Toronto, Canada) + 6 mM of MgCl2, 0.1 U of TopTaq DNA polymerase (Qiagen), and 1 µL (corresponding to a mean of 4 ng) of DNA (Instagene matrix; Bio-Rad Laboratories, Mississauga, Canada). Experiments were run on a T-gradient machine (Biometra; Montreal Biotech, Dorval, Canada). The PCR program parameters included an initial denaturation step (94°C, 3 min), followed by 30 cycles of denaturation (94°C, 30 s), annealing (60°C, 30 s), and extension (72°C, 10 s). Electrophoresis was performed in 1.5% agarose gel, and a 100-bp DNA ladder was used as the molecular weight marker. Results were interpreted as follows: serotype 1 and 14 cross-reacting isolates, which were positive by MAMA-PCR1 and negative by MAMA-PCR2, corresponded to serotype 1; those negative by MAMA-PCR1 and positive by MAMA-PCR2 were considered as serotype 14 (Table 2). In the same way, serotype 2 and 1/2 cross-reacting isolates that were positive by MAMA-PCR1 and negative by MAMA-PCR2 corresponded to serotype 1/2; those negative by MAMA-PCR1 and positive by MAMA-PCR2 were considered as serotype 2. Optimization of the thermocycling conditions was carried out with the reference strains of S. suis serotypes 1 (strain 428), 2 (strain S735), 1/2 (strain 2651), and 14 (strain 13730).10 Results obtained were as expected (Fig. 2).
Table 1.
Primers used for mismatch amplification mutation assay (MAMA)-PCR.
| Primer name | Sequence | PCR |
|---|---|---|
| F_MAMA_1 | ATCGCTTTGTGGTGGCCTTT | MAMA-PCR1 |
| R_MAMA_1-2 | AGAAGCTTCTTTTGCTGTTTGC | MAMA-PCR1 and MAMA-PCR2 |
| F_MAMA_2 | ATCGCTTTGTGGTGGCCTTG | MAMA-PCR2 |
Table 2.
Interpretations of mismatch amplification mutation assay (MAMA)-PCR.
| Multiplex traditional PCR | MAMA-PCR1 | MAMA-PCR2 | Serotype |
|---|---|---|---|
| 1, 14 | Positive (367 bp) | Negative | 1 |
| Negative | Positive (367 bp) | 14 | |
| 2, 1/2 | Positive (367 bp) | Negative | 1/2 |
| Negative | Positive (367 bp) | 2 |
Expected product size in parentheses.
Figure 2.
PCR amplification by mismatch amplification mutation assay (MAMA)-PCR of cpsK gene fragments. A. MAMA-PCR1; B. MAMA-PCR2, with serotype 1 (lanes 2), serotype 14 (lanes 3), without DNA (lanes 4), serotype 2 (lanes 5), and serotype 1/2 (lanes 6). Lane 1 = ladder of 200, 300, 400, and 500 bp.
We validated our tests with 148 field isolates recovered from the internal organs of diseased pigs in Canada. S. suis was isolated in pure or predominant cultures as described previously.6 One hundred thirty-two isolates belonging to serotypes 1 (25 isolates), 2 (64 isolates), 1/2 (13 isolates), and 14 (30 isolates) were encapsulated and had been serotyped previously by the coagglutination test using specific antisera.5 The remaining 16 isolates were untypeable by coagglutination but positive with the traditional PCR test for serotypes 2 and 1/2 (12 isolates) or for serotypes 1 and 14 (4 isolates), suggesting that these isolates do not express CPS.9 To confirm the serotype of the coagglutination-negative isolates, the fragment of interest of the cpsK gene was amplified from DNA using primers cpsK-1 (5□-GAGATTCTTCTGGTGAATGACG-3□) and cpsK-2 (5□-CCCCGTTTTCAGAAAGACAC-3□) and the following PCR parameters: an initial denaturation step (95°C, 2 min), followed by 30 cycles of denaturation (95°C, 20 s), annealing (56°C, 10 s), and extension (72°C, 15 s). Sanger sequencing of the generated amplicon using the same primers revealed that 1, 6, 6, and 3 of the coagglutination-negative isolates possess cps loci of serotypes 1, 2, 1/2, and 14, respectively. Results of validation tests using all 152 of these isolates showed that the MAMA-PCR test was able to correctly identify, as a single serotype, all field isolates tested in our study.
The fact that the current PCR-based serotyping system fails to correctly differentiate serotypes 2 from 1/2 and 1 from 14 is a major issue. Serotype 2 is the most widespread virulent S. suis serotype worldwide, and it affects both pigs and humans. In contrast, serotype 1/2 has never been isolated from humans, indicating a different zoonotic potential.7 However, serotype 1/2 is the most prevalent serotype recovered from diseased pigs in the United States.3 Knowing the serotype of an isolate is important, given that there is at least one commercial vaccine available against S. suis serotype 2, and possible cross-protection against serotype 1/2 is unknown. Serotype 14 isolates are also virulent for pigs, and serotype 14 is the second most commonly recovered serotype from humans.7 On the other hand, serotype 1 has been reported in only 3 unconfirmed human cases, suggesting that they were probably not serotype 1 but rather serotype 14.7
Serotyping has traditionally been carried out with antisera, although only a few laboratories in the world possess antisera against all serotypes. In addition, one of the disadvantages of the coagglutination test is that many field isolates do not express CPS after culture. Also, some isolates are unencapsulated in vivo as reported previously in cases of endocarditis.12 One attractive alternative for serotyping is PCR, which is now performed in most laboratories,9 and which can serotype encapsulated isolates as well as those that do not express CPS. However, as mentioned above, isolates belonging to serotypes 2 and 1/2 as well as 1 and 14 cannot be differentiated by existing PCR assays. The importance of MAMA-PCR over gene sequencing can also be justified when a large number of bacterial isolates need to be tested in epidemiologic investigations.1 The MAMA-PCR test developed in our study can be used to clearly differentiate these serotypes.
Acknowledgments
We thank Katerine Aubé for technical assistance.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was supported by the Diagnostic Service, Faculty of Veterinary Medicine, University of Montreal and Natural Sciences and Engineering Research Council of Canada (grant 04435) to M. Gottschalk.
ORCID iDs: Daisuke Takamatsu  https://orcid.org/0000-0001-6875-0498
https://orcid.org/0000-0001-6875-0498
Marcelo Gottschalk  https://orcid.org/0000-0002-2196-2212
https://orcid.org/0000-0002-2196-2212
References
- 1. Athey TB, et al. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol 2016;16:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deekshit VK, et al. Mismatch amplification mutation assay-polymerase chain reaction: a method of detecting fluoroquinolone resistance mechanism in bacterial pathogens. Indian J Med Res 2019;149:146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Estrada AA, et al. Serotype and genotype (multilocus sequence type) of Streptococcus suis isolates from the United States serve as predictors of pathotype. J Clin Microbiol 2019;57:e00377-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gottschalk M, et al. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev 2007;8:29–45. [DOI] [PubMed] [Google Scholar]
- 5. Gottschalk M, et al. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J Clin Microbiol 1993;31:2192–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gottschalk M, Segura M. Streptococcosis. In: Zimmerman J, et al. , eds. Diseases of Swine. 11th ed. Hoboken, NJ: Wiley, 2019;934–950. [Google Scholar]
- 7. Goyette-Desjardins, et al. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 2014;3:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okura M, et al. Current taxonomical situation of Streptococcus suis. Pathogens 2016;5:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okura M, et al. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol 2014;52:1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perch B, et al. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J Clin Microbiol 1983;17:993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roy D, et al. A single amino acid polymorphism in the glycosyltransferase cpsK defines four Streptococcus suis serotypes. Sci Rep 2017;7:4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tohya M, et al. Comparative genome analyses of Streptococcus suis isolates from endocarditis demonstrate persistence of dual phenotypic clones. PLoS One 2016;11:e0159558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tohya M, et al. Defining the taxonomic status of Streptococcus suis serotype 33: the proposal for Streptococcus ruminantium sp. nov. Int J Syst Evol Microbiol 2017;67:3660–3665. [DOI] [PubMed] [Google Scholar]
- 14. Van Calsteren MR, et al. Explaining the serological characteristics of Streptococcus suis serotypes 1 and 1/2 from their capsular polysaccharide structure and biosynthesis. J Biol Chem 2016;291:8387–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Calsteren MR, et al. Structure determination of Streptococcus suis serotype 14 capsular polysaccharide. Biochem Cell Biol 2013;91:49–58. [DOI] [PubMed] [Google Scholar]
- 16. Van Calsteren MR, et al. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem Cell Biol 2010;88:513–525. [DOI] [PubMed] [Google Scholar]
- 17. Wertheim HF, et al. Streptococcus suis: an emerging human pathogen. Clin Infect Dis 2009;48:617–625. [DOI] [PubMed] [Google Scholar]