Abstract
The outbreak of SARS-CoV-2 in December 2019 in China subsequently lead to a pandemic. Lack of vaccine and specific anti-viral drugs started a global health disaster. For a sustained control and protection, development of potential anti-viral drugs is one of the targeted approach. Although, designing and developing a panel of new drugs molecules are always encouraged. However, in the current emergency, drug repurposing study is one of the most effective and fast track option. The crystal structure of a SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) RNA Dependent RNA Polymerase (RdRp) has recently been deciphered through X-ray crystallography. The single-chain of core RNA Dependent RNA Polymerase relies on virus-encoded cofactors nsp7 and two units of nsp8 for its optimum function. This study explored the FDA approved database of 7922 molecules and screened against the core polymerase along with cofactors. Here we report a panel of FDA approved drugs that show substantial interactions with key amino acid residues of the active site. Interestingly, some of the identified drugs (Ornipressin, Lypressin, Examorelin, Polymyxin B1) bind strongly within the binding pockets of both forms of RdRp. Besides, we found strong candidates for the complex form as well which include Nacortocin, Cistinexine, Cisatracurium (among others). These drugs have the potential to be considered while contriving therapeutic options.
Keywords: Molecular docking simulation, Drug repurposing, SARS-CoV-2, RNA Dependent RNA polymerase, COVID-19, Biological sciences, Bioinformatics, Pharmaceutical science, Biochemistry, Molecular biology
Molecular docking simulation; Drug repurposing; SARS-CoV-2; RNA Dependent RNA polymerase; COVID-19; Biological Sciences; Bioinformatics; Pharmaceutical Science; Biochemistry; Molecular Biology.
1. Introduction
COVID-19, an infectious disease caused by a novel strain of coronavirus SARS-CoV-2 (Severe Acute Respiratory Syndrome). It caught the attention of the world community after its outbreak in December 2019 from the city of Wuhan, China. Since then it has spread globally, WHO declared COVID-19 as pandemic on 11 March 2020 [1,2]. The emergence of this new strain of coronavirus (SARS-CoV-2) which is highly contagious and caused the deaths of 444,813 (as of 17th, June 2020, WHO) individuals worldwide, making it as one of the biggest challenge for the scientific community worldwide. In this global health emergency, drug repurposing (or repositioning) is one of the fast track option that involves screening of existing FDA approved drugs for the identification of potential molecules that can disrupt the function of key proteins of the SARS-CoV-2 and can be used for treatment against COVID-19.
Coronaviruses are enveloped viruses belonging to family Coronoviridae, which is further subdivided into four genera i.e. alpha, beta, gamma, and delta coronavirus [3]. The members of the alpha and beta coronavirus causes infections in mammals however gamma and delta coronavirus causes disease in birds [4, 5].
SARS-CoV-2 belongs to genus beta-coronavirus. Other members of the same genus that are known to infect humans include HCoV-OC43, HCoV-HKU1, SARS-CoV, and MERS-CoV [4, 5].
SARS-CoV-2 genome is a single strand positive-sense RNA. The size of the genome is 29.8kb and has 14 Open Reading Frames (ORFs) which encode information for 27 structural and non-structural proteins [4]. At the 5′-end of the genome, there are two ORFs (ORF-1a and ORF-1ab) encoding two long stretches of poly-proteins, pp-1a and pp-1ab. These encode information for 15 non-structural proteins (nsp 1–10 and nsp 12–16) [4]. The vital non-structural proteins are; nsp3 (multi-domain protein including PL-pro domain), nsp5 (3CL chymotrypsin-like), nsp9 (helicase, may participate in viral replication), nsp12 (RNA dependant RNA polymerase) and nsp13 (helicase). The 3′-end of the genome encodes information for four structural and eight accessory proteins. The structural proteins are; Spike surface glycoproteins (S), Envelope (E), Matrix (M), and Nucleocapsid (N) proteins. The accessory proteins are; 3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14 [4].
1.1. RNA dependent RNA polymerase (RdRp)
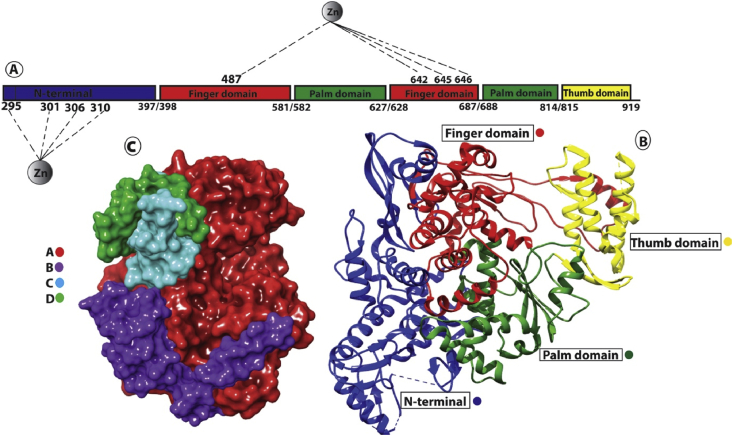
RNA dependent RNA polymerase (RdRp) is involved in the replication and transcription of the SARS-CoV-2 genome. It is the cleavage product of the polyproteins 1a and 1ab from ORF1a and ORF1ab [6]. There is a high degree of conservation among RNA dependent RNA polymerases of different RNA viruses. The Core protein is a single chain of approximately 900 amino acids, showing minimal activity. However, enhanced activity is attained with the attachment of other key subunits [6, 7, 8]. The minimum complex for the proper activity of SARS-CoV-2 requires an attachment of nsp7/nsp8 and an additional nsp8 protein to the core protein [6]. The attachment site for the second additional nsp8 is different. Also, other non-structural proteins (nsp (s)) are involved in this replication and transcription cascade [6]. The core protein looks like a cupped right hand which is further sub-divided into subdomain including finger domain (amino acid residues 398–581, 628–687), palm domain (582–627, 688–815), and thumb domain (816–919) [9] (Figure 1).
Figure 1.
A) Linear structure of RNA Dependent RNA Polymerase (RdRp) showing the N-terminal domain and the main polymerase domain showing sub-domains (Finger, Palm, Thumb). Two metal ion (Zn) are shown in coordination with respective amino acid residues. B) Ribbon diagram showing important domain in RdRp. C) Surface representation of RNA Dependent RNA Polymerase along with its cofactors.
Two additional Zn ions are also required for the structural stability of the RdRp. One of the Zn ion is attached to four amino acids residue (His295, Cys301, Cys306, and Cys310) in the N-terminal domain while the second Zn ion is attached to four amino acid residues (Cys487, His642, Cys645, Cys646) located in finger domain [6]. The presence of Zn in this location shows that it plays an essential role in stabilizing the overall three-dimensional structure of the protein. It has no direct role in the activity of the polymerase as both are quite distal to the active site. All the important sites including, template entry and binding, polymerase activity reaction site followed by the exit through the tunnel (thumb) remain highly conserved among coronaviruses including SARS-CoV-2 [6] (Figure 1). To the extent charge potential is concerned, the palm (RNA template and NTP binding site) has positive electrostatic potential. However, the other sites including attachment of nsp7/8 complex, nsp8 site, and the template exit site (thumb) is neutral [6]. The key residues that are involved in the interaction include; Tyr618, Cys622, Asn691, Asn695, Met755, Ile756, Leu757, Leu758, Ser759, Asp760, Asp761, Ala762, Val763, Glu811, Phe812, Cys813 and Ser814 (numbering about the recently solved structure PDB ID 6M71) [10, 11]. The active site key residues are adjacent aspartates i.e. Asp761 and Asp762, which are involved in the actual reaction of the RdRp enzyme [10, 12].
1.2. Repositioned antiviral drugs and their protein targets
The panel of different antiviral molecules has already been identified by targeting key proteins involved in different stages of the SARS-CoV-2 life cycle. Some of them are in a clinical trial stage. Through repurposing study, the existing antiviral drugs (Ribavirin, Sofosbuvir, Remdesiver, Tenofovir) have already been screened [10, 12]. Those in a clinical trial stage include; ASC09 and Ritonavir (both are HIV inhibitor), GS-5734 (a broad-spectrum investigational antiviral), and Kaletra (inhibitor of 3-CL of SARS and MERS) [13]. Similarly, other compounds in a clinical trial (Favipiravir, used against influenza) didn't show strong activity in clinical isolates [13], while some others with positive results need further investigations [14, 15].
Owing to its significance, the current study encompasses screening and identifying novel FDA approved drugs against the recently elucidated structure of SARS-CoV RNA Dependent RNA Polymerase. The possible lead molecules can further be explored to combat this disease.
2. Materials and methods
2.1. Protein structure and preparation
The crystal structure of RdRp was downloaded from PDB (ID 6M71) with a reported resolution of 2.90Å. RNA Dependent RNA polymerase is a multimeric protein. The minimum complex required for its proper functioning is completed by attachment of three additional protein peptides (nsp7-nsp8, and one additional nsp8) to the core polymerase which is chain A and contains 851 amino acids residues. The current structure is unraveled in complex with all the required cofactors. To investigate the binding affinity of the drugs, we prepared both the structures of RdRp i.e. with and without cofactors. Protein preparation wizard of Maestro was used for protein preparation. Hydrogen atoms, missing residues, and loops were added. The side chains were fixed during protein preparation of both forms of RdRp (with and without cofactors). Besides, disulfide bonds were also created. Protonation states of amino acids were generated at pH 7.4, by using PROPKA to simulate physiological conditions [16, 17]. For the protein structures minimization, the OPLS-2005 force field was used [18]. Grid box was generated by selecting the active site residues as mentioned [10]. Active site residues Asp760 and 761 (DD) were treated as flexible during docking. Grid box was generated as 30, 30, 30 centred (x, y, z) of (114.52, 114.11, 122.91) Å as reported [10]. Receptor grid was generated using Maestro tools and the flexible rotation of certain amino acids residues (Tyr455, Tyr456, Thr462, Cys482, Tyr483, Ser549, Thr586, Thr591, Thr604, Tyr619, Cys622, Thr680, Ser681, Ser682, Thr686, Thr687, Tyr689, Ser692, Cys697, Thr701, Ser754, Ser759, Cys765, Ser778, Tyr788, Ser795, Cys799, Thr801, Cys813, Ser814, Thr817) were allowed.
FDA approved drug database (~7922) was downloaded from the NIH chemical genomics Centre (NCGC) pharmaceutical collection (NPC) database. The database was prepared using the lig-prep module of Maestro [18]. During protein preparation, different combinations of enantiomers and tautomers were generated, which ultimately enhanced the total number of drug molecules. For the protonation states of the ligand at pH 7.4, Epik was used [19]. To check the binding efficacy of the FDA approved drugs, accurate and fast Glide SP- protocol was used for the molecular docking simulations [20, 21]. Glide Induced Fit Docking (IFD) was used to re-dock the top ten molecules in the binding pocket of RdRp [20]. IFD was performed in various steps. Initially, ligands were docked to rigid protein employing potential softened-potential glide docking with the van dar Waals radii scaling of 0.50/0.50, for the receptor and ligand respectively. In the following stage receptor, sampling and refinement were carried out. Residues with minimum one atom inside 5 Å of any of the 10 ligand poses were selected to conformational search and minimization whereas residues lying outside this range were fixed. In this manner, the protein flexibility was considered in the process. Additional redocking of the ligands were performed. For all docking calculations, Glide-XP was analyzed. 3D structure of the protein model, 2D structures of molecules and interaction maps were generated using Maestro (Schrodinger). For other illustration, Adobe Illustrator was used.
3. Results and discussion
For the screening of novel potential inhibitors against RdRp, we selected FDA approved drugs database. A similar type of repurposing study has already been performed, in which the homology model of RdRp was generated, validated and various existing antiviral drugs were investigated for their binding potential within the active site of RdRp without cofactors [10, 12]. Furthermore, other proteins of the SARS-CoV-2 for which X-ray crystal structures were available have also been used as a potential drug target [22]. The current study is the first of its type in which recently determined X-Ray crystal structure of RNA Dependent RNA polymerase was used for the screening of potential inhibitors from the FDA approved drugs database. In the homology model, two aspartates at position 255 and 256 were reported as active site residues [12]. However, in the crystal structure, these similar active site aspartates are represented as Asp760 and Asp761. We have used both forms of the polymerase, a single-core chain, and along with its cofactors (holoenzyme, replication/transcription complex). Initially, a single chain of the RdRp polymerase was selected for the screening, followed by further screening for the holoenzyme.
3.1. Screened compounds against RdRp (Single chain)
Based on docking scores (Glide SP, Glide IFD) and interactions with crucial residues and ligand efficiencies, the top-ranked molecules were selected and analyzed. Table 1 explains the review of the top ten screened molecules and the existing usage of these drugs.
Table 1.
2D structures of top 10 drugs along with their interacting residues, ligand efficiency, docking scores Glide/SP and Glide/IFD (kcal/mol) and existing usage of the drugs.
| FDA Approved Drugs | 2D Structures | Docking Score Kcal/mol | IFD kcal/mol | Glide Ligand Efficiency (SP) | Interacting Residues (SP) | Treatment (Existing usage) |
|---|---|---|---|---|---|---|
| Ornipressin |  |
-10.376 | -10.15 | -0.144 | ASP760, THR591, ASP865, GLN815, SER814, CYS813, GLU811, TYR619 | Vasoconstrictor, Renal failure during decompensated liver cirrhosis [23, 24] |
| Atosiban |  |
-10.096 | -11.11 | -0.148 | ASP760, GLU811, SER814, TYR619, ASP833 | Inhibitor of oxytocin and vasopressin [25] |
| Lanreotide |  |
-10.059 | -9.81 | -0.131 | ASP761, ASP760, ASN691, SER682 | Analogue of Somatostatin involved in suppressing growth hormones, glucagon, and insulin [26] |
| Argiprestocin |  |
-9.874 | -11.68 | -0.137 | ASP761, TRP617 SER814, CYS813, GLU811 | Septic shock [27] |
| Demoxytocin |  |
-9.689 | -8.75 | -0.142 | TRP617, TYR619, LYS621, SER682, ASP760, ASP761, GLU811 | Oxytocin analogue [28] |
| Carbetocin |  |
-9.479 | -9.61 | -0.137 | ALA550, ARG553, THR556, ASP623, ASP760, TYR619, TRP617, TRP800 | Synthetic analogue Oxytocin, postpartum hemorrhage [29] |
| Lypressin |  |
-9.413 | -9.61 | -0.129 | ARG624, VAL557, ARG555, ARG553, ASP760, ASP761, ALA762, GLU811, TRP617, TYR619 | Vasopressins (Lysine vasopressin), diabetes insipidus, anti-diuretic [30] |
| Examorelin |  |
-9.308 | -10.45 | -0.143 | GLU811, ASP760, SER814, GLN815, ARG555 | Synthetic peptide increases the plasma level of growth hormones [31, 32] |
| Colistin |  |
-9.241 | No pose | -0.113 | ASN691, ASP684, SER682, ASP760, TRP617, ASP618, TYR619, LYS551 | Lung infection [33], Cystic fibrosis [34, 35, 36], Ventilator-associated pneumonia [37], nosocomial pneumonia [38], antibiotic against multidrug-resistant gram-negative bacterial infections [39] |
| Polymyxin B1 |  |
-9.228 | -11.92 | -0.109 | ARG555, ASN691, ASP618, ASP761, ASP760, CYS813 | Antibiotic used against multidrug-resistant gram-negative bacterial infection [39, 40] |
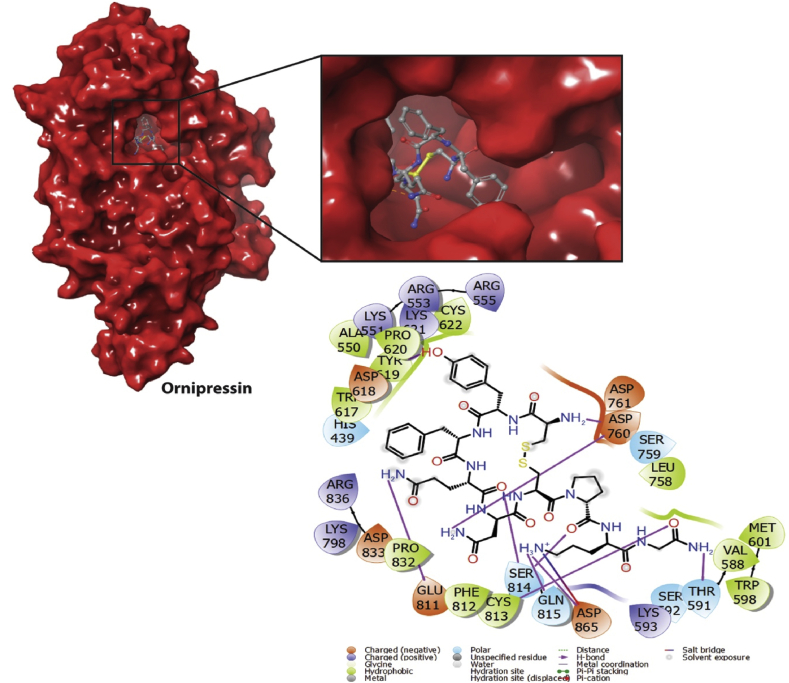
Based on the maximum occupancy of compounds bound to the reported binding tunnel of RdRp, top-ranked conformations were selected considering the hydrogen bonds and other non-covalent interactions. Ornipressin binds into the RdRp binding tunnel with a docking score of -10.37 kcal/mol and the ligand efficiency of -0.14. Ligand interaction analysis of the Ornipressin/RdRp complex shows that ligand mostly made H-bonds and salt bridges. Asp865 is involved in H-bond and salt bridge while Asp760, Thr591, Gln815, Ser814, Cys813, Glu811, Tyr619 formed H-bonds. Ornipressin also depicted some interactions with the hydrophobic and polar residues (Figure 2).
Figure 2.
2D and 3D interaction maps of Ornipression. Surface representation shows the binding mode of drug molecule in the binding tunnel.
In Atosiban complex seven H-bonds, two salt bridges and non-covalent interactions were observed with a docking score of -10.09 kcal/mol and the ligand efficiency of -0.148. Significant interactions were observed with catalytic residues and surface accessible residues. Asp760, Tyr619, and Ser814 made H-bonds while Glu811 and Asp833 formed H-bond as well as salt bridges (Supplementary Figure S1).
Four H-bonds and one salt bridge were noticed in Lareotide with active site residues i.e. Asp760 and Asp761 and other surfaces accessible amino acid residues i.e. Asn691 and Ser682 having a docking score of -10.05 kcal/mol and ligand efficiency of -0.131 (Supplementary Figure S2). The binding potential of Argiprespocin with RdRp was mainly governed by H-bonds. Argiprespocin made six H-bonds with Lys798, Glu811, Leu758, and Ser814. While two H-bonds and one salt bridge were observed for Asp761, one π-cation and H-bond was observed for Trp617 with a docking score of -9.87 kcal/mol and ligand efficiency of -0.137 (Supplementary Figure S2). Whereas, Demoxytocin showed ten H-bonds with both active site Asp760 and Asp761 and other key residues e.g. Trp617, Tyr619, Lys621, Ser682, Glu811, Lys621, Tyr619, Trp617, Ser682 and Glu811 with dock score -9.68 kcal/mol and ligand efficiency of -0.142 (Supplementary Figure S3). Carbetocin, a synthetic analogue of oxytocin, also has the potential to inhibit RdRp (single chain) and showed stable interaction with a docking score of -9.479 kcal/mol and ligand efficiency of -0.137. Eight H-bonds were observed with Lys551, Arg553, Thr556, Trp617, Tyr619, Asp623, and Asp760 (Supplementary Figure S3).
Another screened drug Lypressin showed a docking score of -9.413 kcal/mol and ligand efficiency of -0.129. Ligand interactions showed salt bridge and H-bonding with Asp761 and only H-bonding with other residues including Arg553, Arg555, Thr556, Trp617, Tyr619, Arg624, Asp760, Asp761, Ala762 and Glu811 (Supplementary Figure S4).
Examorelin, a screened inhibitor of this study showed most of H-bonds with Asp760, Cys813, Ser814, Gln815, and Glu811. Salt bridge was also observed with Glu811. Arg555 showed π-cation interaction with a docking score of -9.30 kcal/mol and ligand efficiency of -0.143 (Supplementary Figure S4). Colistin (polymyxin E, polypeptide antibiotics) showed most of the H-bonding with Lys551, Trp617, Tyr619, Asp618, Ser682, Asp684, Asn691, and both catalytic residues i.e. Asp760, Asp761, with a docking score of -9.24 kcal/mol and ligand efficiency of -0.113 (Supplementary Figure S5). Polymyxin B1 also bound tightly with RdRp core protein through salt bridges and H-bonding. Catalytic Asp760 and Asp761 made four salt bridges and three H-bonds with RdRp which shows its strong binding affinity. Moreover, H-bonds were formed with Arg-555, Trp-617, Asp681, Asn691, Cys813, and Ser814 with a docking score of -9.22 kcal/mol and ligand efficiency of -0.109 (Supplementary Figure S5).
To investigate the difference in binding energies, molecules were re-docked in the binding pocket of RdRp by using the Glide-Induced Fit Docking protocol. No significant changes were observed for drugs (except for Colistin which didn't show any pose in IFD) in both the protocols. A similar type of interaction was observed in Glide-IFD as found in Glide-SP (Table 1). Glide-IFD docking poses of selected top 10 molecules are given in Supplementary figures (Figures S6 and S7). All complexes showed promising binding and maximum occupancy of crucial binding pocket residues. It has been observed that the top ten screened drugs binding affinity was stronger with RdRp core protein.
As previously reported, Sofosbuvir, Ribavirin, Tenofovir, Setrobuvir, Guanosine derivative (IDX-184) were shown with a docking score of -7.5, -7.8, -6.9, -9.3, -9.0 kcal/mol respectively against RdRp single-chain [12]. In current study, we have also docked Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, Tenofovir, Guanosine derivative (IDX-184) and Setrobuvir with a docking score of -5.94, -5.57, -4.94, -7.20, -4.52, -8.57, -4.45 kcal/mol respectively (Supplementry Figure S8). The difference in the docking score may be due to the use of different docking algorithms. These antivirals were analysed on the basis of interactions with crucial residues and compared the interactions with screened molecules. Our identified molecules showed more stable H-bonds, salt bridges, and ionic interactions as compared to reported antivirals.
For a potent drug, it is essential to interact with key amino acid residues in the active site pocket of the polymerase. Once the drug binds to the active site of RdRP, the access of positive sense RNA of the virus which serves as a template for anti-genome is blocked. No new viral genomes are synthesized and mature virus particles cannot be assembled and propagated further. In this study, all the identified FDA approved drugs have shown stable interaction with key residues, along with lowest binding energies which indicates the potential for these drugs to inhibit the activity of RdRp, therefore contributing to stop viral proliferation.
The URL link of additional top 90 molecules including 2D structures, docking scores, ligand efficiencies and glide energies are given in the supplementary data.
3.2. Screened compounds against RdRp (Complex)
The minimal complex of RdRp along with essential cofactors (nsp7-nsp8, nsp8) required for enhanced activity is used for this drug repurposing study. We found a panel of molecules that showed significant binding with the RdRp complex. Molecules were analyzed based on docking energies, ligand efficiencies, and other protein-ligand interactions. Some of the screened molecules remained similar as found for single chain of RdRP, they therefore can be taken as common inhibitors for both single chain and in complex form of RdRp. The results of top 14 selected molecules from Glide/SP along with their 2D structures, ligand efficiency, interacting residues and existing usage of the drugs are shown in Table 2. Top 14 molecules including common molecules among both structures were redocked through Glide IFD. Docking score of Gide/IFD are shown in Table 2.
Table 2.
The 2D structures of top 14 drugs along with their interacting residues, ligand efficiency, docking score (Glide/SP and Glide/IFD) kcal/mol against RdRp complex and the existing usage of the durgs.
| Title | 2D Structure | Docking Score Glide/SP | Docking score IFD | Glide Ligand Efficiency (SP) | Interacting Residues (SP) | Treatment (Existing usage) |
|---|---|---|---|---|---|---|
| Nacartocin |  |
-13.943 | -11.15 | -0.202 | LYS798 | Oxytocin analogue having natriuretic activity [41] |
| Cistinexine |  |
-13.687 | -7.26 | -0.201 | ARG553 | Mucolytic Expectorant drug used in chronic bronchitis [42] |
| Cisatracurium |  |
-13.064 | No pose | -0.195 | ARG553, ARG555, GLU811 | Neuromuscular blocking agent [43] |
| Pegamotecan |  |
-12.782 | No pose | -0.178 | LYS621 | Advanced and Metastatic gastric and Gastro-esophageal junction adenocarcinoma [44] |
| Polymyxin B1 | 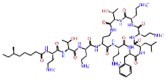 |
-12.646 | -10.99 | -0.149 | ARG555, ASP760, ASP761 | Antibiotic used against multidrug-resistant gram-negative bacterial infection [39, 40] |
| Ebiratide | 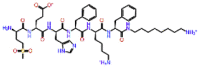 |
-12.628 | -9.50 | -0.18 | TRP800 | Neurotrophic effect (Alzheimer diseases) [45] |
| Sulfomyxine | 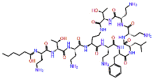 |
-12.468 | -10.25 | -0.152 | ASP760, GLU811 | Sulphur containing Antibiotic [46] |
| Diagastrin | 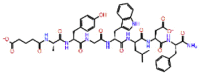 |
-12.313 | -8.88 | -0.173 | ARG553, TYR619, ASP833, ASP836 | Peptide analog of Gastrin, stimulants of gastric acid secretion [47] |
| Ditercalinium chloride |  |
-12.308 | -8.27 | -0.228 | ASP623, ASP760 | Pro-anticancer drug [48] |
| Benzquercin |  |
-12.174 | -9.71 | -0.214 | ARG553, LYS798 | Flavonoid drug [49] |
| Examorelin |  |
-12.139 | -8.69 | -0.187 | ASN691, HIS810 | Synthetic peptide increases the plasma level of growth hormones [31, 32] |
| Lypressin |  |
-11.923 | -9.16 | -0.163 | ARG555, CYS622, LYS621 | Vasopressins (Lysine vasopressin), diabetes insipidus, anti-diuretic [30] |
| Ornipressin |  |
-11.717 | -9.23 | -0.163 | ASP623, LYS621 | Vasoconstrictor, Renal failure during decompensated liver cirrhosis [23, 24] |
| Colistin | 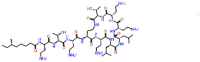 |
-11.077 | No pose | -0.135 | ILE548 | Lung infection [33], Cystic fibrosis [34, 35, 36], Ventilator-associated pneumonia [37], nosocomial pneumonia [38], antibiotic against |
For a complex, increased docking score was observed in Glide/SP. This phenomenon might be due to the allosteric behaviour of the polymerase, as it is a common observation that upon binding of the molecules on an allosteric site, it brings conformational changes in the active site. This then facilitates the efficient binding of the substrate within the active site. The attachment of cofactors (nsp8/nsp7, nsp8) with a core protein (nsp12) might have similar effect on the activity of RdRp. Additionally, the binding of some of the molecules in both the forms depend on the chemical structures of the screened molecules, changed interacting residues, steric hindrance, and stereo chemistry of the active site.
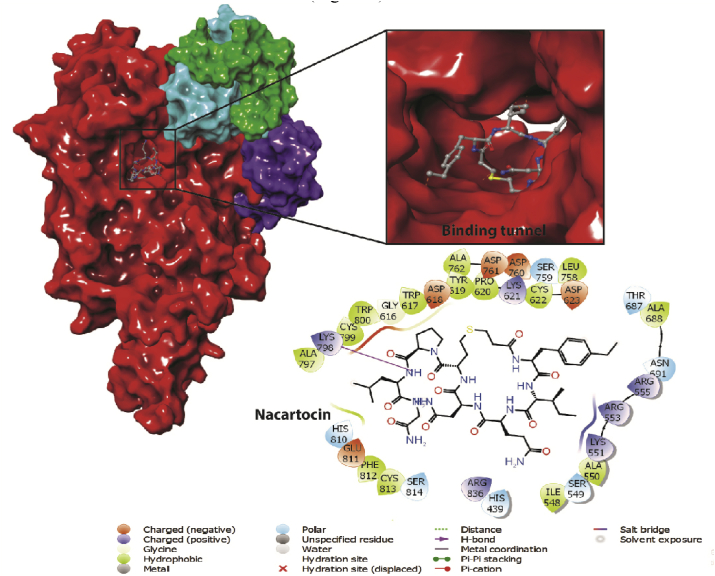
Nacartocin is one of the lead molecules with a docking score of -13.943 kcal/mol and ligand efficiency of -0.202. Nacartocin showed H-bonds and non-covalent interactions with key residues and surface accessible residues (Figure 3).
Figure 3.
2D and 3D interaction map of top lead molecule Nacartocin.
π -cation interaction with Arg-553 and non-covalent interactions with crucial residues were observed for Cistinexine, with a docking score of -13.687 kcal/mol and a ligand efficiency of -0.201. Whereas a neurotransmitter blocking agent Cisatracurium showed two H-bonds with Arg553 and one salt bridge with Glu811 with a docking score of -13.064, and a ligand efficiency of -0.195. Other non-covalent interactions were also observed with polar, non-polar, and hydrophobic residues (Supplementary Figure S9).
Pegamotecan is another lead molecule that showed a strong binding affinity towards RdRp with their essential cofactors. H-bond with Lys621 and other non-covalent interactions with key residues were observed with a docking score of -12.782 kcal/mol and ligand efficiency of -0.178 respectively. Among the top 14 screened drugs Polymyxin B1 is one of the common drugs, identified in both forms of RdRp. Almost similar types of interactions were observed for Polymyxin B1 in the catalytic site of the RdRP complex as found during core protein interactions. Polymyxin B1 showed two salt bridges and one H-bond with Asp760 and Asp761 and two H-bonds with Arg-555 with a docking score of -12.646 kcal/mol and ligand efficiency of -0.149 (Supplementary Figure S9).
Non-covalent interactions and one H-bond with Trp800 were observed for Ediratide with a docking score of -12.682 kcal/mol and with ligand efficiency of -0.18. Asp760, Ser759, and Glu811 made H-bonds with Sulfomyxin through docking score of -12.468 kcal/mol and ligand efficiency of -0.152 (Supplementary Figure S10). π-cation with Arg553 and H-bonds with Tyr619, Arg86, and Asp833 were observed for Diagastrin, which is a peptide analogue of gastrin showed a docking score of -12.313 kcal/mol and ligand efficiency of -0.173. Two salt bridges with Asp760 and Asp623 were observed for Ditercalinium chloride with a docking score of -12.308 kcal/mol and ligand efficiency of -0.228 (Supplementary Figure S10).
Flavonoid drug, Benzquercin showed two π-cation interactions with Arg553 and Lys798 and other non-covalent interactions with polar, non-polar and hydrophobic residues with docking score of -12.174 kcal/mol and ligand efficiency of -0.214 (Supplementary Figure S11).
Examorelin, Lypressin, Ornipressin, and Colistin are also common drugs in both form of RdRp. Only one H-bond with His810 and other non-covalent interactions were observed for Examorelin showed a docking score of -12.139 kcal/mol and ligand efficiency of -0.187. Lypressin made three H-bonds with Arg555, Lys621, and Cys622 with a docking score of -11.923 kcal/mol and ligand efficiency of -0.163 followed by Ornipressin, showed a docking score of -11.717 kcal/mol (ligand efficiency of -0.163). Salt bridge with Asp623 and H-bond with Lys621 is observed during interactions however, Colistin showed two H-bonds with Ile548 and Ser814 with a docking score of -11.077 kcal/mol (-0.135). Colistin is also known as Polymyxin E (Supplementary Figure S11).
IFD poses of top 14 molecules are given in supplementary figures S12A and S12B. Pegamotecan, Cisatracurium, and colistin didn't show any pose during IFD docking simulation. Colistin showed similar results in both structures with and without co-factors.
Based on docking analysis and its reconfirmation, we didn't find any significant difference in the docking energies, therefore our identified molecules have the potential to strongly bind in the binding tunnel of both single chain and complex from of the RdRp. The URL link of 3D structures, docking scores, ligand efficiencies, and Glide energies of additional top 100 molecules against RdRp complex are given in supplementary data.
In a drug repurposing study, the aim is to screen existing approved drugs and suggest their reuse for new medical complications, being the most feasible approach as compared to the de-novo drug designing and development. The benefits associated with repurposed drugs include; safety for human use, no escalating cost, and reduced timeline for its development. For instance, Zidovudine, which was originally used for the treatment in cancer, was repurposed as the first anti-HIV drug, approved by the FDA in 1987 [50,51]. Similarly, Rituximab, originally used in the treatment of various cancers was approved by the FDA in 2006 as a repurposed drug against rheumatoid arthritis [52]. Raloxifene, another drug used for osteoporosis, has also been approved for the treatment of breast cancer [52]. Aspirin, which was primarily used as an analgesic is now recommended and approved by the FDA in 2015 against colorectal cancer and in cardiovascular diseases [52]. In the last few years, about one-third of the FDA approved drugs relate to repurposed drugs [52, 53].
All of the screened molecules identified in this study are FDA approved drugs. We used fast and accurate docking algorithms Glide-SP to screen potential hits against RdRp (core and complex). Glide-IFD was used to compare and confirm the difference between the binding energies of top screened molecules. We have identified molecules for two forms of the RdRp individually. Some of the molecules have shown promising interactions with both forms (core and holoenzyme) of the RdRp, e.g. Polymyxin B1, Ornipressin, Lypressin, and Exemorelin.
Although all the top ten screened molecules for both the forms showed strong binding. Polymyxin B1 shows strong binding with key amino acid residues in core as well as in the holoenzyme form. Polymyxin B1 is currently used as an antibiotic for the treatment of respiratory tract infections i.e. pneumonia (nosocomial, healthcare, or ventilator-associated), due to the multidrug-resistant gram-negative bacteria. For the treatment of respiratory tract infection, it is generally used as oral inhalation via nebulization. The general mechanism of action of Polymyxin involves the destabilization of the integrity of the bacterial membrane, thereby killing gram-negative bacterial cells through membrane lysis. It interacts with the lipopolysaccharides of the Gram-negative bacteria and disrupts the outer membrane. The other known mechanism is by vesicle-vesicle contact pathway, in which Polymyxin induces an osmotic imbalance, that ultimately leads to membrane lysis. Based on the findings of the current study, Polymyxin B1 can be used for the treatment of COVID-19 by blocking the active site of RdRp.
4. Conclusion
Due to the current COVID-19 pandemic, to date, 444,813 people have lost their lives. SARS-CoV-2A is spreading worldwide at an alarming rate. To overcome this infection, the current study was designed to screen all those drugs which are available in the market or currently in a clinical trial. For a potent drug, it is essential to interact with key amino acid residues in the active site pocket of the polymerase. Once the drug binds to the active sites of RdRp, the access of positive sense RNA of the virus which serves as a template for anti-genome is blocked. No new viral genomes are synthesized and mature virus particles cannot be assembled and propagated further. In this study, all the identified FDA approved drugs have shown stable interaction with key residues, along with lowest binding energies which indicates the potential for these drugs to inhibit the activity of RdRp, therefore contributing to stop viral proliferation.
The top candidate drugs we found that interact with a single chain core RdRp include; Ornipressin, Otosiban, Lanreotide, Argiprestocin, Demoxytocin, Carbetocin, Lypressin, Examorelin, Colistin, and Polymyxin B1. Additionally, we also identified compounds that interact with the complex form of RdRp (holoenzyme). It includes; Nacartocin, Cistinexine, Cisatracurium, Pegamopecan, Ebiratide, Sulfomyxine, Diagastrin, Ditercalinium chloride, and Benzquercin. Among these lead molecules, Ornipressin, Lypressin, Examorelin, Polymyxin B1 showed strong binding efficacy with both core and holoenzyme. Although all of the screened molecules showed strong binding affinity towards RdRp (core and holoenzyme), however Polymyxin B1 and Colistin are already in use against respiratory tract infections. Based on our findings, we, therefore, suggest that these screened drugs can be clinically tested to handle this intricate infection.
Declarations
Author contribution statement
J. Ahmad: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
S. Ikram: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
F. Ahmad: Performed the experiments; Analyzed and interpreted the data.
M. Mushtaq: Analyzed and interpreted the data.
Irshad Ur Rehman: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We are thankful to Shelvia Malik and other Schrodinger team members for facilitating us with the licence to use Schrodinger Suite which enabled us to successfully complete this study.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary_Data_RdRp
References
- 1.Wu D. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F. Structure, function, and evolution of coronavirus Spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu A. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Q. Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition. Sci. Rep. 2015;5:17155. doi: 10.1038/srep17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn D.G. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157(11):2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subissi L. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald S.M. RNA synthetic mechanisms employed by diverse families of RNA viruses. Wiley Interdiscip. Rev. RNA. 2013;4(4):351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublie S., Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8(6):704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 12.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 14.Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antivir. Res. 2020;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yazdany J., Kim A.H.J. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: what every clinician should know. Ann. Intern. Med. 2020 doi: 10.7326/M20-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sastry G.M. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 17.Bas D.C., Rogers D.M., Jensen J.H. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73(3):765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen W.L., Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 19.Shelley J.C. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21(12):681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 20.Friesner R.A. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 21.Halgren T.A. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 22.Khan S.A. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 23.Kam P., Tay T. The pharmacology of ornipressin (POR–8): a local vasoconstrictor used in surgery. European journal of anaesthesiology. 1998;15(2):133–139. doi: 10.1111/j.0265-0215.1998.00247.x. [DOI] [PubMed] [Google Scholar]
- 24.Lenz K. Ornipressin in the treatment of functional renal failure in decompensated liver cirrhosis: effects on renal hemodynamics and atrial natriuretic factor. Gastroenterology. 1991;101(4):1060–1067. doi: 10.1016/0016-5085(91)90734-3. [DOI] [PubMed] [Google Scholar]
- 25.de Heus R., Mulder E.J., Visser G.H. Management of preterm labor: atosiban or nifedipine? International journal of women's health. 2010;2:137. doi: 10.2147/ijwh.s7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godara A. The safety of lanreotide for neuroendocrine tumor. Expet Opin. Drug Saf. 2019;18(1):1–10. doi: 10.1080/14740338.2019.1559294. [DOI] [PubMed] [Google Scholar]
- 27.Demiselle J. Vasopressin and its analogues in shock states: a review. Ann. Intensive Care. 2020;10(1):9. doi: 10.1186/s13613-020-0628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westergaard J. Use of oral oxytocics for stimulation of labor in cases of premature rupture of the membranes at term: a randomized comparative study of prostaglandin E2 tablets and demoxytocin resoriblets. Acta obstetricia et gynecologica Scandinavica. 1983;62(2):111–116. doi: 10.3109/00016348309155773. [DOI] [PubMed] [Google Scholar]
- 29.Larciprete G. Carbetocin versus oxytocin in caesarean section with high risk of post-partum haemorrhage. J. Prenat. Med. 2013;7(1):12. [PMC free article] [PubMed] [Google Scholar]
- 30.Mimica N., Wegienka L.C., Forsham P.H. Lypressin nasal spray. Usefulness in patients who manifest allergies to other antidiuretic hormone preparations. JAMA. 1968;203(9):802–803. doi: 10.1001/jama.203.9.802. [DOI] [PubMed] [Google Scholar]
- 31.Imbimbo B.P. Growth hormone-releasing activity of hexarelin in humans. A dose-response study. Eur. J. Clin. Pharmacol. 1994;46(5):421–425. doi: 10.1007/BF00191904. [DOI] [PubMed] [Google Scholar]
- 32.Arvat E. Effects of GHRP-2 and hexarelin, two synthetic GH-releasing peptides, on GH, prolactin, ACTH and cortisol levels in man. Comparison with the effects of GHRH, TRH and hCRH. Peptides. 1997;18(6):885–891. doi: 10.1016/s0196-9781(97)00016-8. [DOI] [PubMed] [Google Scholar]
- 33.Gurjar M. Colistin for lung infection: an update. J. Intensiv. Care. 2015;3(1):3. doi: 10.1186/s40560-015-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beringer P. The clinical use of colistin in patients with cystic fibrosis. Curr. Opin. Pulm. Med. 2001;7(6):434–440. doi: 10.1097/00063198-200111000-00013. [DOI] [PubMed] [Google Scholar]
- 35.Horianopoulou M. Effect of aerosolized colistin on multidrug-resistant Pseudomonas aeruginosa in bronchial secretions of patients without cystic fibrosis. J. Chemother. 2005;17(5):536–538. doi: 10.1179/joc.2005.17.5.536. [DOI] [PubMed] [Google Scholar]
- 36.Kasiakou S.K. Combination therapy with intravenous colistin for management of infections due to multidrug-resistant Gram-negative bacteria in patients without cystic fibrosis. Antimicrob. Agents Chemother. 2005;49(8):3136–3146. doi: 10.1128/AAC.49.8.3136-3146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garnacho-Montero J. Treatment of multidrug-resistant Acinetobacter baumannii ventilator-associated pneumonia (VAP) with intravenous colistin: a comparison with imipenem-susceptible VAP. Clin. Infect. Dis. 2003;36(9):1111–1118. doi: 10.1086/374337. [DOI] [PubMed] [Google Scholar]
- 38.Michalopoulos A. Aerosolized colistin for the treatment of nosocomial pneumonia due to multidrug-resistant Gram-negative bacteria in patients without cystic fibrosis. Crit. Care. 2005;9(1):R53–R59. doi: 10.1186/cc3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falagas M.E., Kasiakou S.K. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005;40(9):1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 40.Zavascki A.P. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 2007;60(6):1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 41.Hrbas P. Nacartocin–analogue of oxytocin with enhanced natriuretic properties: natriuretic and hemodynamic characteristics. Endocrinol. Exp. 1984;18(2):117–124. [PubMed] [Google Scholar]
- 42.Santolicandro A., Baldi S., Giuntini C. The effect of a new expectorant drug on mucus transport in chronic bronchitis. J. Aerosol Med. 1995;8(1):33–42. doi: 10.1089/jam.1995.8.33. [DOI] [PubMed] [Google Scholar]
- 43.Szakmany T., Woodhouse T. Use of cisatracurium in critical care: a review of the literature. Minerva Anestesiol. 2015;81(4):450–460. [PubMed] [Google Scholar]
- 44.Scott L.C. A phase II study of pegylated-camptothecin (pegamotecan) in the treatment of locally advanced and metastatic gastric and gastro-oesophageal junction adenocarcinoma. Canc. Chemother. Pharmacol. 2009;63(2):363–370. doi: 10.1007/s00280-008-0746-2. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto T., Tsuda S., Nakamura S. The neurotrophic effects of ebiratide, an analog of ACTH4-9, on cultured septal cells and aged rats. J. Neural Transm. Gen. Sect. 1995;100(1):1–15. doi: 10.1007/BF01276861. [DOI] [PubMed] [Google Scholar]
- 46.Egawa Y. Sulfomycins, a series of new sulfur-containing antibiotics. I. Isolation, purification and properties. J. Antibiot. (Tokyo) 1969;22(1):12–17. doi: 10.7164/antibiotics.22.12. [DOI] [PubMed] [Google Scholar]
- 47.Schally A.V. Effect of somatostatin analogs on gastric acid secretion in dogs and rats. Int. J. Pept. Protein Res. 1990;36(3):267–274. doi: 10.1111/j.1399-3011.1990.tb00977.x. [DOI] [PubMed] [Google Scholar]
- 48.Okamaoto M. Ditercalinium chloride, a pro-anticancer drug, intimately associates with mammalian mitochondrial DNA and inhibits its replication. Curr. Genet. 2003;43(5):364–370. doi: 10.1007/s00294-003-0393-4. [DOI] [PubMed] [Google Scholar]
- 49.Qamar M.T. In-silico identification and evaluation of plant flavonoids as dengue NS2B/NS3 protease inhibitors using molecular docking and simulation approach. Pak. J. Pharm. Sci. 2017;30(6):2119–2137. [PubMed] [Google Scholar]
- 50.Mitsuya H. 3'-Azido-3'-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc. Natl. Acad. Sci. U. S. A. 1985;82(20):7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarchoan R. Administration of 3'-azido-3'-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. Lancet. 1986;1(8481):575–580. doi: 10.1016/s0140-6736(86)92808-4. [DOI] [PubMed] [Google Scholar]
- 52.Pushpakom S. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 53.Talevi A., Bellera C.L. Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expet Opin. Drug Discov. 2020;15(4):397–401. doi: 10.1080/17460441.2020.1704729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Data_RdRp