Abstract
The world is facing the third coronavirus caused pandemic in less than twenty years. The SARS-CoV-2 virus not only affects the human respiratory system, but also the gastrointestinal tract. The virus has been found in human feces, in sewage and in wastewater treatment plants. It has the potential to become a panzootic disease, as it is now proven that several mammalian species become infected. Since it has been shown that the virus can be detected in sewage even before the onset of symptoms in the local population, Wastewater Based Epidemiology should be developed not only to localize infection clusters of the primary wave but also to detect a potential second, or subsequent, wave. To prevent a panzootic, virus removal techniques from wastewater need to be implemented to prevent the virus dissemination into the environment. In that context, this review presents recent improvements in all the fields of wastewater treatment from treatment ponds to the use of algae or nanomaterials with a particular emphasis on membrane-based techniques.
Keywords: Wastewater, SARS-CoV-2, Virus, COVID-19, Membranes, Wastewater-based epidemiology
1. Introduction
SARS-CoV-2 is the third coronavirus to spread worldwide in the last two decades [1]. As of July 9th 2020, the virus has infected more than 12 million people with the COVID-19 disease and claimed the lives of 552,000 people. The Severe Acute Respiratory Syndrome (SARS), as its given name indicates, primarily affects the respiratory system of the patients. However, COVID-19 has a broader range of symptoms, with digestive tract symptoms widely spread amongst patients. Moreover, a substantial portion of infected people are asymptomatic or have mild symptoms but still can transmit the virus. Lastly, SARS-CoV-2 is not only supposed to have been transmitted to humans through animal carriers, but humans can also transmit the disease to other animals.
In this review, the literature concerning the presence of the disease in the digestive tract of patients and its dissemination in the wastewater network will be detailed. The present literature concerning which animals have been identified as potential carriers of SARS-CoV-2 will then be summarized.
To limit the propagation of CoVid-19 and to avoid the possibility of a pan-zoonotic disease, wastewater based epidemiology that allows rapid detection of the presence of the disease in an area and the tools available to remove viruses from the wastewater at the wastewater treatment plant to reduce the risk of spreading the disease further within the environment, either by discharge into water systems or by the production of reclaimed water for irrigation or human consumption will be presented.
2. SARS-CoV-2 (2019-nCoV) information
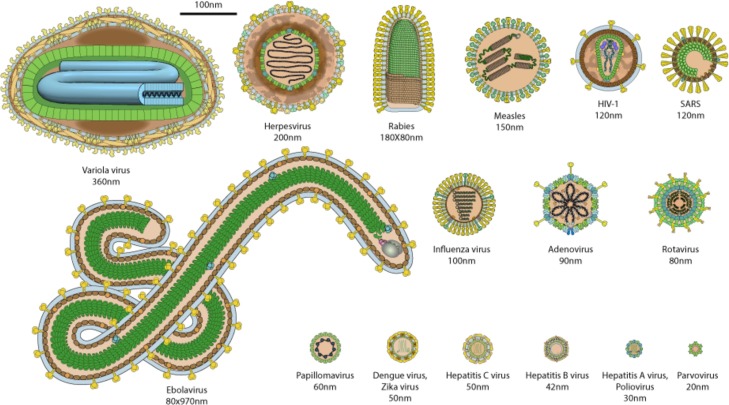
SARS-CoV-2 is an enveloped single-stranded RNA virus which belongs to the Coronaviridae family. This family is subdivided into four subfamilies ranging from alpha to delta. Subfamilies alpha and beta can affect humans. SARS-CoV-2 is from the subfamily Coronavirinae genera β [2]sub-genus Sarbecovirus [3] Since 1960, there have been seven coronaviruses from these two subfamilies that have been reported to have affected humans [4,5]: 229E, OC43, SARS-CoV, NL63, MERS, HKU1 and SARS-CoV-2.- 229E, OC43, NL63, HKU1 are involved in 15 % of the common colds [5] whereas SARS-CoV, MERS and SARS-CoV-2 viruses are more virulent and they cause a severe acute respiratory syndrome(SARS). SARS-CoV outbreak was in 2002–2003 and the Middle East Respiratory Syndrome coronavirus (MERS-CoV) broke out in the Arabian Peninsula in 2012 [1]. Bar-on et al. [6] provide regularly updated data in which can be found the SARS-CoV-2 size, which is around 100 nm. For membrane-based treatment processes, size is important as it determines the maximum pore size of the membrane which needs to be selected to remove the viral particles. Fig. 1 provides size and shape comparisons of several viruses. In this figure, virus sizes range from 20 nm to 970 nm. With 100 nm, SARS-CoV-2 is slightly smaller than SARS-CoV but is bigger than most common viruses.
Fig. 1.
Human virus relative size from https://viralzone.expasy.org/5216 [7].
3. Spread of SARS-CoV-2 in the environment
The two main ways which were initially reported for SARS-CoV-2 transmission are through direct contact and through aerosols produced by contaminated people when sneezing or coughing. SARS-CoV-2 is able to survive on surfaces, with survival duration highly dependent on surface type [[8], [9], [10]], although it has not as yet been been demonstrated as a route for transmission.
However, SARS-CoV-2 RNA has also been found in human feces [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]] of at least 39 % of tested patients [22] and therefore the question of the possibility of fecal-oral transmission is raised [15,17].
Zang et al. [23] studied the infection of human small intestinal enterocytes and reported that two mucosa-specific serine proteases promoted virus entry in enterocytes. Mart et al. [24] also demonstrated that enterocytes are easily infected and can actually be used as experimental models. But regarding the possibility of fecal oral transmission, it was indicated that the SARS-CoV-2 was inactivated by simulated human colonic fluid [23]. These data are in vitro data and it is also reported that live SARS-CoV-2 virus was found in patient’s stool samples [25,26].
Detection of SARS-CoV-2 virus was obtained in both nasopharyngeal and rectal swabs in a pediatrical study of 10 children [27]. Moreover, from the day of admission, the rectal swabs returned positive for 27 days, whereas no virus was detected in any nasopharyngeal swabs after 15 days [19,27]. However, virus replication tests realized on fecal swabs returned negative. Jiang et al. [28] even detected the virus in the stools of patients for as many as 42 days.
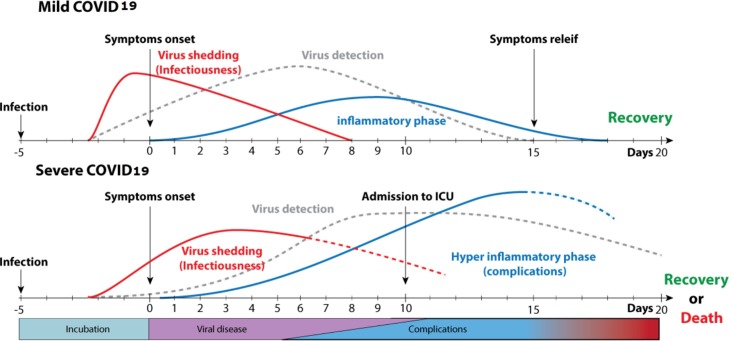
Numerous teams have indicated that COVID-19 could be transmissible by the fecal-oral route [[11], [12], [13], [14], [15], [16], [17],19,[25], [26], [27], [28], [29], [30], [31]], but there is no demonstrated case at this point. However, caution should be taken to limit the possibilities of virus shedding in the environment for both human and animals. He et al. [32] provided data about when and the duration that infected people can shed virus and for how long the RNA virus can be detected [33]. Those data were compiled by ViralZone [34] and are presented in Fig. 2 .
Fig. 2.
Timeline for SARS-CoV-2 shedding and detection https://viralzone.expasy.org/9116 [34].
Since the virus is present in feces, is it found in municipal wastewater and this raises the possibility of spread to the wider environment from insufficiently treated effluent. Several articles have been published indicating detection of SARS-CoV-2 [[35], [36], [37], [38], [39], [40]] in sewage.
Wurtzer et al. [36] were able to correlate the results of the quantitative monitoring of SARS-CoV-2 genome in Paris sewage to the number of people infected. They even suggested that sewage water samples should be stored in a databank to allow them to be investigated retrospectively when an outbreak is starting. The importance of this archiving was equally outlined by Dolfing [41].
Medema et al. [39] report a study in the Netherlands where they started looking for the presence of COVID-19 RNA in 6 locations on February 6th 2020. The detection started on March 4/5, six days before the first cases were reported. Similarly, Randazzo et al. [42] in Spain and Wu et al. [43] in the USA detected COVID-19 RNA in wastewater, even in low prevalence area in Spain. La Rosa et al. [44] reported the detection of COVID-19 in untreated wastewater in Italy.
This raises the question of the presence of SARS-CoV-2 in the aquatic environment. Since COVID19 is a pandemic spread in 215 countries, it covers countries with wastewater treatment plants and other developing countries where wastewater is directly sent to the aquatic environment untreated. Thus, the presence of viable SARS-CoV-2 virus or RNA in aquatic environment is reported by several authors [[45], [46], [47], [48], [49], [50], [51], [52]].
4. SARS-CoV-2 zoonosis
COVID19 disease was first detected in the city of Wuhan in the Hubei province of China in December 2019. The corona virus that causes the disease is now named SARS-CoV-2. Since it shares 96 % whole genome identity to a bat coronavirus: BatCoV RaTG13 [53], the bat Rhinolophus affinis from Yunnan province is suspected to be the original mammal hosting the precursor of SARS-CoV-2. However, the two provinces are distant from each other and the contacts between bats and humans are limited. It is possible that an intermediate animal host is involved in the transmission from animals to humans. The presence of pangolins in the putative starting point of the pandemic, which is the seafood market in Wuhan China, and the 91 % nucleotide identity of the Sars-CoV-2 with a pangolin CoV [6,53] makes it a conjectured intermediate host [54], although this is far from certain and efforts are ongoing to find other candidate intermediate hosts.
Apart from the search for the intermediate host, studies have been made to identify animal species that could have been infected. A Chinese study examined antibody levels in pigs, sheep, horses, chicken and ducks [55] and the results were negative.
However, it is reported that diseased humans have transmitted the virus naturally to ferrets, cats, dogs, tigers, lion and minks [3,[56], [57], [58], [59]].Apart from minks farms, there is no evidence so far that livestock for human consumption such as cows, sheep and pigs have been infected in farms [60]. This last point is rather important if it is considered that in the case of MERS-CoV, which belongs to Coronaviridae beta subfamily like SARS-CoV-2 but not to the subgenus Sarbecovirus, camels [61,62] were suspected to be possible reservoirs or intermediate hosts for successive outbreaks. Even raw milk and improperly cooked meat were discussed as potential sources of infection.
Apart from the natural transmission of SARS-CoV-2 virus from human to animals, studies have reported of experimental inoculation of the virus to animals and study the possibility of transmission from animals and the transmission route by putting in the same cage infected and non-infected animals and having other animals in another cage close to the first [[63], [64], [65], [66]]. This has demonstrated that ferrets and cats are highly susceptible to the virus, whist dogs are less susceptible and livestock (pigs, sheep and ducks) studied were not found to be susceptible It is to be noted that those tests were done on a limited number of animals, so how representative they are of the species as a whole is not entirely known.
Transmission back to humans was discussed by Franklin et al. [67] who raised the possibility of wild hosts in North America as a potential source of perpetuation of the pathogen in the environment. Damas et al. [68] studied in-silico the probabilities of a wide range of animals to get infected and ranked them in five levels from very low to very high based on the conservation of genes for 25 amino acids important in the binding of the virus to a putatively important receptor, ACE2. The ranking used the fact that the main receptor of SARS-CoV-2 is the angiotensin I converting enzyme 2 (ACE2). Their study on 410 vertebrates, including 252 mammals could help identify potential animals that could be viral host. From this they judged that only mammals fell into the categories of medium or higher risk. In the meantime, in the Nederland on mink farms where minks had been found to be contaminated, sequencing was done on samples taken from diseased farmers and, while the topic is still under investigation, the transmission from minks to human seems to have occurred [56].
Gollakner et al. [69] underlined that although the data gathered so far does not allow definite conclusion of the possibility of a panzootic aspect for COVID-19, allowance should be made for the possibility. Since there is shedding of the virus to the environment through sewage, the question has been raised about the possibility of aquatic mammals to become contaminated [70].
5. Wastewater based epidemiology
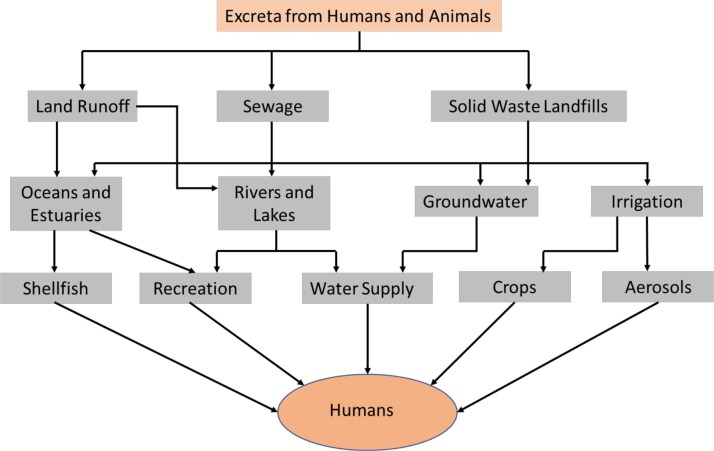
From the previous sections it was shown that SARS-CoV-2 is found live in human feces and that it can be transmitted to animals which can also excrete it. Although the fecal oral transmission has still not been demonstrated, precaution measures should be taken to minimize this possibility. Fig. 3 summarizes the pathways that excreta from both human and animals can follow to potentially transmit the virus back to humans [71]. Sewage and then Wastewater Treatment Plant (WTT) are at the center of this potential transmission route and WTT can play an important role in detection and control of SARS-CoV-2 spread. For the detection part, Wastewater Based Epidemiology (WBE) could be used.
Fig. 3.
Transmission routes for pathogens in human and animals excreta. Adapted with permission from ref. [71].
WBE is a relatively new tool whose idea comes from the fact that relatively stable environmental contaminants that are released in the sewage system can be sampled and quantified. It potentially provides real time information on wastewater contaminants [72] and was first used in 2001 to evaluate illicit drug usage [73]. More recently, WBE has been proposed for the early detection of viral outbreaks [[74], [75], [76], [77], [78], [79], [80], [81], [82]] as well as for the persistence of viral indicators in the aquatic environment [75,83]. The methods used are high throughput RNA sequencing (RNA-Seq) more commonly than PCR (qRT-PCR) [75,84]. In 2019, O’Brien and Xagoraraki [74,85] underlined the importance of wastewater data collection to understand spatial and temporal evolution of viral diseases.
In the case of SARS-CoV-2, considering that the virus was found both in stool samples and wastewater as previously described, several authors have proposed to use WBE as a tool to monitor the pandemic [41,[76], [77], [78], [79], [80],[86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]]. Thus, Medema et al. [38] reported that sewage surveillance could be a sensitive tool to monitor SARS-CoV-2 circulation in the population since it could be detected prior to symptoms being reported in the local population. Moreover, monitoring of international airport sewage would allow very early detection of the entrance of the virus into a country.
Wurtzer et al. [36] even suggested that wastewater sample storage would allow detection of an outbreak retroactively to provide historic information of the spread of the virus. In the present SARS-CoVid-2 outbreak, it could have allowed the localization of the first patients and define which cities should be on lockdown instead of putting the lockdown on a whole country. For example, until now it was believed that the SARS-CoV-2 outbreak in France started late January 2020 but SARS-CoV-2 was detected retrospectively by RT-PCR in samples from a patient admitted in intensive care in late December 2019 [101].
Mao et al. [102] proposed a small, portable paper-based device for the detection of SARS-CoV-2 and Hata and Honda [91] provided information on the potential sensitivity of wastewater monitoring for SARS-CoV-2 which is one diseased person in 100 000.
6. Wastewater treatment for virus removal
The WHO [103] underlined in its interim guidance of April 23rd 2020 the importance of safely managing wastewater. Limiting the dissemination of the SARS-CoV-2 into the environment is of great importance considering that several animals have already been identified as potential hosts and that reclaimed water could be used in irrigation. Gerba et al. [104] reported the virus removal reduction factor that wastewater plants need to provide depending on the reclaimed water use. For irrigation of edible crops, they indicate a 6 log10 removal is sufficient, whereas a 12 log10 removal would be needed to make the water potable. The same removal targets have been reported by Ahmed et al. [105] in their 2020 article on recycled water safety.
Wastewater treatment is a wide field and its full description is not the target of this review. This section will focus on the literature containing techniques used for virus removal. After presenting the model viruses that are used to evaluate the efficiency of wastewater virus removal treatment, recently developed methods will be presented.
6.1. Model virus
SARS-CoV-2 is a potentially lethal virus to humans and some precautions need to be taken for scientific testing to be made. Thus, the Center for disease Control and Prevention (CDC) provides guidelines for Biosafety and COVID-19 [106]. For environment specimen testing, the CDC defines that ‘Procedures that concentrate viruses, such as precipitation or membrane filtration, can be performed in a BSL-2 laboratory with unidirectional airflow and BSL-3 precautions’. As a result, in general, virus surrogates are used instead of dangerous pathogens. The surrogate to be used depends on the experiment type to be handled. For example, to compare the virus concentration methods for wastewater testing, murine hepatitis virus (MHV) has been used as a SARS-CoV-2 surrogate [84]. Other animal coronaviruses such as the transmissible gastroenteritis virus (TGEV) and the feline infectious peritonitis virus (FIPV) have also been used for testing the persistence and disinfection of human coronaviruses in water and wastewater [107]. For safety reasons some laboratories are using the enveloped bacteriophage φ6 [[107], [108], [109]], whose size is 85 nm [110], or the non-enveloped Escherichia virus MS2 [109,[111], [112], [113], [114], [115], [116], [117], [118], [119], [120], [121], [122]], whose size is 26 nm [123,124].
However, Shirasaki et al. [125] studied different virus models and they concluded that the Pepper mild mottle virus (PMMoV), which is a plant virus, appears to be a suitable surrogate for human enteric virus whereas MS2 and φX174 are not. PMMoV has also been validated together with AiV-1 and TMV as surrogates of human enteric viruses for testing wastewater virus reduction [126].
6.2. Wastewater treatment pond systems
Wastewater treatment ponds are very common worldwide as they are the technically simplest way to handle wastewater. Verbyla and Mihelcic [120] reviewed 71 different wastewater treatment pond systems for virus removal and concluded that one log10 reduction of virus content in water was achieved between 14.5–20.9 days. Considering the World Health Organization guidelines for wastewater reuse for irrigation, a pathogen reduction of 6–7 log10 units is required [127]. This target can be reached only by the combination of several methods. Ponds are used as primary treatment and there can be up to three treatment steps.
Virus removal in pond treatment systems is not very well understood and depends on the particular characteristics of the pond, including chemical composition and optical properties, but the process seems to be a combination of sunlight-mediated mechanisms and interactions of virus particles with other particulates including other microorganisms and sedimentation [120].
6.3. UV inactivation and chlorine treatment
Qiu et al. [128] reported the inactivation of human infectious virus at two wastewater plants. They measured the reduction using qPCR on different virus types, the mean log10 reduction they obtained was 1.2 and 1.8. Simon et al. [129] reported the use of chlorine and ultraviolet light disinfection against different viruses. They obtained up to 2.5–3 log10 reduction(LR) on coliphage and E. coli, otherwise the LRs were in the range of 0.3 to 1.3. They observed that results are better on sand-filtered samples. UV inactivation and treatment with hypochlorite generally renders viruses inactive by damaging replicative machinery through oxidation processes [130].
6.4. Ozone treatment
Ozone is highly effective for the inactivation of many viruses. A statement by the International Ozone Association (IOA) indicated that it was not aware of any research and testing that had been conducted specifically on the SARS-CoV-2 coronavirus. Peer Reviewed Research has not yet been completed and therefore definitive conclusions cannot be made regarding ozone inactivation of SARS-CoV-2 (IOA Statement on COVID-19, 2020 [131]).
The US EPA has published disinfection contact time (CT) values for virus inactivation by ozone in the surface water treatment rule (SWTR) (US EPA 1989 [132]). To assess the disinfection efficiency in water treatment, the CT-concept is applied. C represents the concentration of disinfectant (ozone) and T the contact time; CT is the product of the disinfectant concentration and contact time. Sigmon et al. [133] calculated CT values for specified log inactivation levels of a number of viruses and surrogates in wastewater at pH 7.6 and 16 °C, with log reduction values (LRV) for all viruses ranging from 0.56 for an LRV of 1–1.32 for an LRV of 1.32 [133]. These data indicated that all organisms were generally inactivated to 4-log at a CT of no more than 1.0 mg min/L.
Enteric viruses do not get proper inactivation in wastewater effluent discharged to the environment [134]. As such, effluent discharged from sewage treatment plants will have an elevated number of viruses [135]. Ozone has been used as a disinfectant instead of chlorine for the disinfection of sewage effluents used for irrigation of crops, and for discharge to surface water, due to the issue of high chlorine demand and problematic disinfectant byproducts [136]. A study by Perez-Rey et al. [137], reported that selected microorganisms including four viruses, three non-sporulated bacteria, two sporulated bacteria and one fungus, were employed in vaccine preparation for animals. Viruses were the most readily-inactivated microorganisms and sporulated bacteria the most resistant strains. The inactivation followed a second order kinetic law, depending on both ozone and microorganism concentration. The conclusion of that study was total inactivation of microorganisms in wastewater is possible by ozone application. A study by Aydogan et al. [138], indicated the effectiveness of ozone gas to inactivate Bacillus subtilis spores, which share the same physiological characteristics as Bacillus anthracis spores that cause the anthrax disease. Increased humidity during the ozone application increased the rate of inactivation.
The City of Montreal, Québec, Canada, investigated three disinfection processes (Ozone, UV, and Performic Acid) to improve wastewater treatment to reduce pharmaceuticals and endocrine chemicals in one of the primary-treated effluents (high flow system) [139]. The City of Montreal municipal wastewater has the target criteria for coliforms of 9000 UFC/100 mL that needs to be met. Ozone treatment was found to be the most efficient of the three disinfection processes investigated in removing selected pharmaceutical products from lightly treated effluents [139].
6.5. Sand filters for virus removal
Sand filters are commonly used in wastewater treatment, but they usually have less than one-unit log10 virus removal, they are used more for particle removal. However, Samineni et al. [112] developed functionalized sand filters using a water extract of Moringa Oleifera seeds. Then they tested the efficiency of the obtained sand using MS2 bacteriophage virus, obtaining an impressive 7 log10 reduction in virus content. The mechanism implies that MS2 has affinity for some components from the seeds and binds to it. It also implies that the functionalized sand will progressively get saturated and will eventually have to be replaced or regenerated in some way.
6.6. Algal systems for virus removal
Delanka-Pedige et al. [[140], [141], [142]] recently published three articles dealing with the use of algae for removing viruses in wastewater. Results obtained are in the same range as for activated sludge with log removal values in the range of 1–3 depending on the virus type. They seem, however, to reduce the number of species more efficiently and thus reduce the need for chlorination. Mechanisms for viral removal in algae based wastewater treatment systems include sedimentation after viral particles have attached to algal biomass, increased temperatures denaturing algal nucleic acids and proteins and sunlight mediated degradation [140].
6.7. Membranes use in wastewater treatment for virus removal
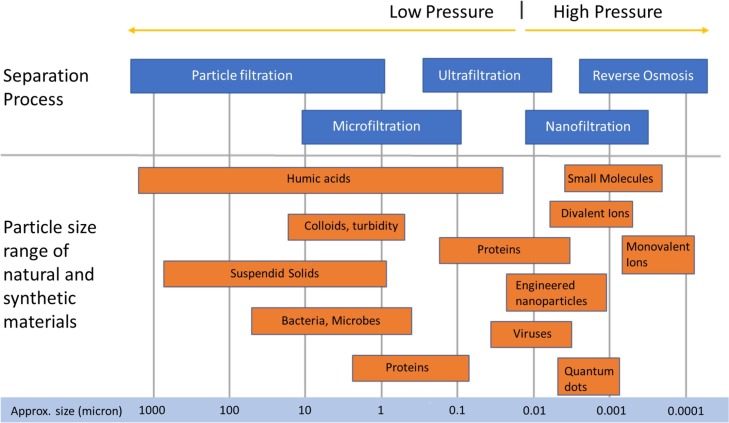
Water filtration membranes are categorized by their pore size and their distributions in four categories: Reverse Osmosis, Nanofiltration, Ultrafiltration and Microfiltration. Fig. 4 gives a representation of those pore sizes together with the main contaminants, including viruses, that can be found in water.
Fig. 4.
Comparison of water filtration membranes pore sizes with water contaminant sizes, reproduced from open source [144].
Considering that SARS-CoV-2 size is 100μm, membranes such as Reverse Osmosis, Nanofiltration and Ultrafiltration should be able to remove it. Goswani et al. [143] reviewed the use of polymeric and ceramic membranes for virus removal from water. The reported removal efficiencies were highly variable since the range reported was 0.2–7. This section will outline very recent literature showing the potential use of those membranes for SARS-CoV-2 before considering microfiltration and membrane bioreactor technology.
6.7.1. Reverse osmosis
Wlodarczyk and Kwarciak-Kozlowska [71] recently reviewed the treatment of waterborne pathogens by reverse osmosis. They provide complete description of the type of pathogens that are waterborne which are divided in three different groups: protozoans (5–100 μm), bacteria (0.5–1.0 μm) and viruses (0.01-0.1 μm); as well as requirements for Reverse Osmosis membrane materials. They indicated that Reverse Osmosis is seldom used to remove pathogens from water even if it is one of the techniques reported by the EPA with indicative log removals above 6. This is because reverse osmosis is typically coupled with a pre-treatment system, often ultrafiltration, to reduce foulants which may interfere with the reverse osmosis process. However, RO can be used in combination with such a suitable a pretreatment, to remove particulate matter, and post treatment, to complete the removal of any remaining contaminants [145].
Prado et al. [146] reported that Membrane Bioreactor/Reverse Osmosis systems were providing virus removal efficiencies in the range of 2.3–2.9 log10 which is rather lower than what was reported by Vickers et al. [147] who used reverse osmosis for the removal of MS2-coliphage. They obtained a nomimal 5 log10 removal without the MBR. Considering that MS2 is 27 μm in size, which is roughly 60 times smaller than SARS-CoV-2, the removal efficiency on the latter would be expected to be higher. The same authors [148] also reported the use of sand-anthracite filters and a membrane bioreactor/reverse osmosis system for the removal of noroviruses, which are non-enveloped capsids of about 38−40 nm in diameter [149], in raw sewage. The efficiency of the system led to a virus concentration below the detection level, which would mean, considering the initial virus concentration and the detection limit of the analysis, a log10 removal higher than 6 or 7.
6.7.2. Nanofiltration
Pore sizes of nanofiltration membranes are generally less than 10 nm which is smaller than any virus presented in Fig. 1 [150]. Although the literature on nanofiltration removal of viruses is not extensive, several studies have been published which generate log removal rates for various viruses at between 3 and 8 [[151], [152], [153], [154], [155]].
6.7.3. Ultrafiltration
As ultrafiltration is often used as a pre-treatment step prior to RO treatment, its efficacy in removal of viruses is more widely reported in the scientific literature than for other membrane-based technologies. Recently Al Aani et al. [156] reviewed the use of Ultrafiltration Membranes for wastewater and water process engineering and report their use for bacteria and virus removal.
Lee et al. [117] used a combination of coagulation and ultrafiltration(UF) on a pilot scale for wastewater reclamation. They improved removal of MS2 bacteriophage by optimizing the pH of the secondary effluent to a range of 5–6 depending on the effluent. Once improved, the virus removal factor was in the 6.8–7.5 log10 range. Additionally, to obtain a more stable transmembrane pressure, an additional sedimentation step was required. The ideal combination was a hybrid coagulation-sedimentation-UF system.
Other researchers [157] used a polyethersulfone Ultrafiltration membrane with average membrane pore sizes of 67 nm for the removal of the bacteriophage PP7. They studied the effect of pH in the range of 5–8 and modelled virus and membrane electrostatic interaction forces. The Log10 removal range obtained was in the 1.5–2.8 range depending on ionic strength. Divalent cations present in feed solutions were found to have a negative impact on the viral removal effectiveness compared with monovalent cations due to their modification of electrostatic interactions.
Grafted zwitterionic polymer hydrogels were used by Lu et al. [158] for the modification of a 150 kDa polyethersulfone membrane. They studied the membrane efficiency on MS2 and HAdV-2 human viruses. The grafting reduced water permeability but provided a Log10 removal in the 3–4 range.
A pore interconnectivity method of evaluation of asymmetric ultrafiltration membranes has been developed commercially specifically for virus removal [159]. Gold nanoparticles of different sizes were used and their capture by the membranes was visualized using SEM. It was found that membranes shown by this method to have poor pore interconnectivity were also membranes which had been previously shown to have significant loss of viral retention after process recovery, whereas membranes shown to have better interconnectivity were more robust in terms of virus retention over a range of operating conditions.
6.7.4. Microfiltration
Since microfiltration membrane average pore sizes are larger than 100 nm, they are more suited to the removal of protozoa and bacteria rather than the smaller viruses [160]. However, this only considers separation by sieving mechanisms. Sinclair et al. [161] modified a microfiltration membranes using a cationic polymer. They reached a 3 log10 MS2 reduction with the resulting membrane with only 22 % reduction in flux. They proposed such membranes for low pressure point of use water filtration, due to the high-water permeability of such membranes.
6.7.5. Membrane bioreactor
O’Brien and Xagoraraki reported that membrane bioreactors, under the correct conditions are capable of a 7log10 reduction in virus concentration [162]. From a comparison of a number of studies it was found that the presence of a biofilm on the membrane was important in virus removal efficiency, where a biofilm, whilst reducing water flux also restricted viral penetration of the membrane. It was concluded that there must be a trade-off between maintaining a biofilm to keep viral removal efficiency high with backwashing of the membrane to remove biofilm to maintain acceptable water fluxes. They also concluded that model viruses do not necessarily show the same removal efficiency as viruses which are pathogenic to humans, with data obtained using harmless bacteriophages likely to lead to overestimation of viral removal efficiencies for pathogenic viruses.
Miura et al. [163] investigated the removal of three virus families in a full scale membrane bioreactor plant. The best results were obtained at pH4 with a log10 reduction range of 2.3–4.5
6.7.6. Ceramic membranes
Im et al. [122,164] first compared ozonation and coagulation as pretreatments for virus removal with a ceramic membrane. They then used a combination of ozonation, coagulation and ceramic membrane filtration for water reclamation. They obtained on a pilot plant a 12 log10 removal factor of MS2.
The use of hydrophobic ceramic capillary membranes for virus removal was reported by Bartels et al. [165]. They functionalized ceramic membranes with hexyl or octyl triethoxysilanes in order to increase hydrophobic surface properties. Without functionalization, they obtained log10 removal in the range of 0.3–3.4 for the bacteriophages MS2 and PhiX174, whereas after functionalization the log10 removal was around 9. This increase in removal efficiency was due to favorable virus- treated membrane surface interactions, primarily due to hydrophobic interactions, which lead to adsorption of viruses to the membrane material. Presumably, such membranes would potentially be subjected to a sufficiently high saturation of viral concentration, thus necessitating cleaning.
In-line coagulation with ceramic membrane filtration for the removal of the bacteriophage Qβ was investigated by Wattanachira et al. [166]. The coagulant they used is polyaluminum chloride. They underlined that they had to adjust coagulant dosage with virus concentration. Overall, they obtained 6.7 log10 virus removal under optimal conditions.
6.7.7. Nanomaterials
The use of nanomaterials to remove and de-activate viruses in wastewater is a broad field and includes the use of fullerenes, photocatalysts and membranes incorporating nanomaterials, such as CNTs, TiO2 and ZVIs [[167], [168], [169]]. For instance, Kim et al. [170] reported that Silver Multiwall nanotubes (Ag-MWCNT) used in a nanocomposite filter system reached a 100 % removal for several different viruses [170]. Domaga et al. [113] studied Cu2O/MWCNTs filters for MS2 virus removal from water. One of their three samples reached a 7 log10 reduction in MS2 at pH5. Nemeth et al. [171] used a Cu2O-coated MWCNT membrane and obtained a 4 log10 MS2 removal in a pH range of 5–9. Kuo et al. [115] obtained a 7 log10 reduction on three viruses, Qβ bacteriophage, MS2 bacteriophage, and Aichi virus, with smectic liquid-crystalline ionic membranes.
6.8. Integrity testing techniques and imperfection fixing
For the Reverse Osmosis, Nano-Filtration and Ultra-Filtration membranes to efficiently remove viruses, their integrity has to be optimal. For instance, membranes may contain defect obtained during manufacture or may suffer mechanical damage during use from high hydraulic operating pressures or scouring by particulates in the feed water. A number of reviews are found in the literature for techniques to assess the integrity and structure property relationships of separation membranes including by Wang et al. [172], Ostarcevic et al. [144] and Pype et al. [173]. High-pressure membranes are only credited at 2 log10 virus removal by regulatory agencies, whereas in the sections above articles were presented where it reached as high as 7 log10. This difference is due to a lack of sufficiently sensitive detection technologies available industrially [174].
Frenkel and Cohen [175] proposed new techniques for real-time monitoring of reverse osmosis membrane integrity for virus removal. They proposed a pulsed marker membrane integrity monitoring system. The potentials and limitations of this type of fluorescent-based technique was then investigated by Yoon [174], who managed to obtain a sensitivity level at the 4 log10 level.
Then, Niewersch et al. [176] proposed a solution-diffusion-imperfection model with a decision tree for diagnosis to help monitor the integrity of membranes, which was capable of identifying the types of imperfections present in a particular membrane under study.
For ultrafiltration, Khahnstover et al. [177] published a comparison of different methods that can be used to assess integrity and separation efficiency. Nanoparticle counting was found to be the most sensitive method overall in terms of lowest LRV values reported.
As a solution to imperfections in nanoscale membranes, Suzuki et al. [178] proposed to use a small volume of aqueous polyvinyl alcohol to selectively plug imperfections. These plugs were then stabilized through exposures to citric acid, sodium hydroxide, and ethylenediaminetetraacetic acid.
7. Conclusions
SARS-CoV-2 is a virus that emerged at the end of 2019 in China and generated a pandemic of COVID-19 within a few months. Initially thought to affect only the respiratory system, it was soon proven to be also gastrointestinal. Then, the environmental implication is that SARS-CoV-2 is shed into the sewage system and thus reaches Wastewater Treatment Plants when existing or more globally the aquatic environment.
SARS-CoV-2 has also been proven to be transmitted from humans to animals, which can potentially be a virus reservoir. Considering the possibility of SARS-CoV-2 being a panzoonotic, it becomes even more important to limit SARS-CoV-2 spread into the environment. Wastewater Treatment Plants could be a solution for the early detection of the disease in an area through the development of a Wastewater Based Epidemiology plan. It could also be used as a decision helper for lockdown application or removal.
After reporting some of the virus models that can be used to characterize virus removal efficiency, a wide range of recently developed wastewater treatments were presented. This include some classical and advanced technologies treatments (Ozone, UV, etc.,) and numerous membrane-based systems. As a general rule, it is the combination of different techniques in primary, secondary and tertiary treatments that would allow enough virus removal to limit the possibility of environment contamination.
The SARS-CoV-2 pandemic should develop a global awareness that solutions have to be found for improving the wastewater treatment even in developing countries, due to the possibility of its spread through the aquatic environment from inadequately treated waste water. SARS-CoV-2 transmission through the fecal oral route is not predominant, but considering that it is the third new coronavirus to evolve to a pandemic in less than twenty years, preparations should be made for the possibility of a virus pandemic more readily transmissible through this route. Considering the economic costs of the present pandemic, greater efforts to improve wastewater treatment specifically with the removal or inactivation of viral contaminants on a global scale should be an urgent priority.
Declaration of Competing Interest
The authors of this manuscript declare that there is no conflict of interest.
References
- 1.Guarner J. Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19. Am. J. Clinical Pathol. 2020;153(4):420–421. doi: 10.1093/ajcp/aqaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30(3):e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khamisse E., et al. Opinion paper: severe acute respiratory syndrome coronavirus 2 and domestic animals: what relation? Animal. 2020:1–4. doi: 10.1017/S1751731120001639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulse J.D. Human coronaviruses: the deadly seven. ACTA Sci. Microbiol. 2020;3(6):86–89. [Google Scholar]
- 5.Yang Y., et al. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar-On Y.M., et al. SARS-CoV-2 (COVID-19) by the numbers. eLife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viralzone S.Io.B.S. 2020. Human Virus Relative Size.https://viralzone.expasy.org/5216 [Google Scholar]
- 8.van Doremalen N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New England J. Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin A., et al. Stability of SARS-CoV-2 in different environmental conditions. medRxiv. 2020 doi: 10.1016/S2666-5247(20)30003-3. p. 2020.03.15.20036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santarpia J.L., et al. Transmission potential of SARS-CoV-2 in viral shedding observed at the University of Nebraska medical center. medRxiv. 2020 p. 2020.03.23.20039446. [Google Scholar]
- 11.Ding S., Liang T.J. Is SARS-CoV-2 also an enteric pathogen with potential fecal-oral transmission: a COVID-19 virological and clinical review. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infectious Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nouri-Vaskeh M., Alizadeh L. Fecal transmission in COVID-19: a potential shedding route. J. Med. Virol. 2020 doi: 10.1002/jmv.25816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quilliam R.S., et al. COVID-19: the environmental implications of shedding SARS-CoV-2 in human faeces. Environ. Int. 2020;140 doi: 10.1016/j.envint.2020.105790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heller L., Mota C.R., Greco D.B. COVID-19 faecal-oral transmission: are we asking the right questions? Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng S.C., Tilg H. COVID-19 and the gastrointestinal tract: more than meets the eye. Gut. 2020 doi: 10.1136/gutjnl-2020-321195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foladori P., et al. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao F., et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimolai N. Features of enteric disease from human coronaviruses: implications for COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.26066. n/a(n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 23.Zang R., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. p. eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamers M.M., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindson J. COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17(5) doi: 10.1038/s41575-020-0295-7. p. 259-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao F., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X., et al. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020 doi: 10.1002/jmv.25941. n/a(n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25825. n/a(n/a) [DOI] [PubMed] [Google Scholar]
- 30.Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal– ;oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Amico F., et al. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention, and management. Clinical Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 33.Wölfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 34.Viralzone S.Io.B.S. 2020. COVID-19 and Treatment.https://viralzone.expasy.org/9116 [Google Scholar]
- 35.Ahmed W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sc. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurtzer S., et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. p. 2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kocamemi B.A. 2020. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters inTurkey. [Google Scholar]
- 38.Medema G., et al. Presence of SARS-coronavirus-2 in sewage. medRxiv. 2020 doi: 10.1021/acs.estlett.0c00357. p. 2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- 39.Medema G., et al. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 40.Sayess R., Hychka K.C., Rahm B.G. NEW YORK STATE WATER RESOURCES INSTITUTE; 2020. Presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the Virus Causing COVID-19, in Raw and Partially-Treated Sewage. [Google Scholar]
- 41.Dolfing J. The importance of sewage archiving in coronavirus epidemiology and beyond. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c02972. [DOI] [PubMed] [Google Scholar]
- 42.Randazzo W., et al. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1128/mSystems.00614-20. p. 2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Rosa G., et al. FIRST DETECTION OF SARS-COV-2 IN UNTREATED WASTEWATERS IN ITALY. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.139652. p. 2020.04.25.20079830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerrero-Latorre L., et al. First SARS-CoV-2 detection in river water: implications in low sanitation countries. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shutler J., et al. Risk of SARS-CoV-2 infection from contaminated water systems. medRxiv. 2020 p. 2020.06.17.20133504. [Google Scholar]
- 47.Kassem I.I., Jaafar H. The potential impact of water quality on the spread and control of COVID-19 in Syrian refugee camps in Lebanon. Water Int. 2020:1–7. [Google Scholar]
- 48.Carducci A., et al. Making waves: coronavirus detection, presence and persistence in the water environment: State of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rimoldi S.G., et al. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.140911. p. 2020.05.01.20086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wartecki A., Rzymski P. On the coronaviruses and their associations with the aquatic environment and wastewater. Water. 2020;12(6):1598. [Google Scholar]
- 51.Carraturo F., et al. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cahill N., Morris D. Recreational waters – a potential transmission route for SARS-CoV-2 to humans? Sci. Total Environ. 2020;740 doi: 10.1016/j.scitotenv.2020.140122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biol. 2020;30(7):1346–1351. doi: 10.1016/j.cub.2020.03.022. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P., et al. Are pangolins the intermediate host of the 2019 novel coronavirus (2019-nCoV)? PLoS Pathog. 2020;16(5) doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Opriessnig T., Huang Y.-W. Update on possible animal sources for COVID-19 in humans. Xenotransplantation. 2020;27(3) doi: 10.1111/xen.12621. p. e12621-e12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oreshkova N., et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance. 2020;25(23) doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hossain M.G., Akter S., Saha S. SARS-CoV-2 host diversity: an update of natural infections and experimental evidences. J. Microbiol. Immunol. Infection. 2020 doi: 10.1016/j.jmii.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiwari R., et al. COVID-19: animals, veterinary and zoonotic links. Vet. Quart. 2020:1–22. doi: 10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sit T.H.C., et al. Infection of dogs with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hernández M., et al. Are animals a neglected transmission route of SARS-CoV-2? Pathogens. 2020;9(6):480. doi: 10.3390/pathogens9060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus: transmission and phylogenetic evolution. Trends Microbiol. 2014;22(10):573–579. doi: 10.1016/j.tim.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonilla-Aldana D.K., et al. 2020. MERS-CoV and SARS-CoV Infections in Animals: A Systematic Review and Meta-Analysis of Prevalence Studies. [PubMed] [Google Scholar]
- 63.Shi J., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. p. eabb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q., et al. SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. bioRxiv. 2020 p. 2020.04.01.021196. [Google Scholar]
- 65.Halfmann P.J., et al. Transmission of SARS-CoV-2 in domestic cats. New England J. Med. 2020 doi: 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dung N.T., et al. Formation of carbonate phases and their effect on the performance of reactive MgO cement formulations. Cement Concrete Res. 2019;125 [Google Scholar]
- 67.Franklin A.B., Bevins S.N. Spillover of SARS-CoV-2 into novel wild hosts in North America: a conceptual model for perpetuation of the pathogen. Sci. Total Environ. 2020;733 doi: 10.1016/j.scitotenv.2020.139358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Damas J., et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. bioRxiv. 2020 doi: 10.1073/pnas.2010146117. p. 2020.04.16.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gollakner R., Capua I. Is COVID-19 the first pandemic that evolves into a panzootic? Veterinaria Italiana. 2020 doi: 10.12834/VetIt.2246.12523.1. [DOI] [PubMed] [Google Scholar]
- 70.Nabi G., Khan S. Risk of COVID-19 pneumonia in aquatic mammals. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wlodarczyk R., Kwarciak-Kozlowska A. In: Waterborne Pathogens. Vara Prasad M.N., Grobelak A., editors. Butterworth-Heinemann; 2020. Chapter 4 - treatment of waterborne pathogens by reverse osmosis; pp. 57–80. [Google Scholar]
- 72.Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Current Opinion Environ. Sci. Health. 2019;9:77–84. [Google Scholar]
- 73.Daughton C.G. Pharmaceuticals and Care Products in the Environment. American Chemical Society; 2001. Illicit drugs in municipal sewage; pp. 348–364. [Google Scholar]
- 74.Xagoraraki I., O’Brien E. In: Women in Water Quality: Investigations by Prominent Female Engineers. O’Bannon D.J., editor. Springer International Publishing; Cham: 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [Google Scholar]
- 75.Adriaenssens E.M., et al. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. mSystems. 2018;3(3):e00025–18. doi: 10.1128/mSystems.00025-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mao K., et al. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Current Opinion Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hrudey Steve E., et al. Wastewater-based epidemiology for SARS-CoV-2. RSC. 2020 [Google Scholar]
- 78.Daughton C.G. Wastewater surveillance for population-wide covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farkas K., et al. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Current Opinion Environ. Sci. Health. 2020;17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chavarria-Miró G., et al. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. medRxiv. 2020 p. 2020.06.13.20129627. [Google Scholar]
- 81.Kitajima M., et al. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Collivignarelli M.C., et al. SARS-CoV-2 in wastewater treatment plants. medRxiv. 2020 p. 2020.06.04.20122218. [Google Scholar]
- 83.Farkas K., et al. Viral indicators for tracking domestic wastewater contamination in the aquatic environment. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmed W., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Brien E., Xagoraraki I. A water-focused one-health approach for early detection and prevention of viral outbreaks. One Health. 2019;7 doi: 10.1016/j.onehlt.2019.100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghernaout D., Ghernaout B. Controlling COVID-19 pandemic through wastewater monitoring. Open Access Library J. 2020;7(5):1–20. [Google Scholar]
- 87.Orive G., Lertxundi U., Barceló D. Do we really need to invoke heroic measures for early SARS-CoV-2 outbreak detection? Eur. J. Epidemiol. 2020:1–2. doi: 10.1007/s10654-020-00654-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar M., et al. The first proof of the capability of wastewater surveillance for COVID-19 in India through the detection of the genetic material of SARS-CoV-2. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bonilla-Aldana D.K., et al. Importance of the one health approach to study the SARS-CoV-2 in Latin America. One Health. 2020 doi: 10.1016/j.onehlt.2020.100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Venugopal A., et al. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Current Opinion Environ. Sci. Health. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hata A., Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with norovirus cases. Environ. Sci. Technol. 2020;54(11):6451–6452. doi: 10.1021/acs.est.0c02271. [DOI] [PubMed] [Google Scholar]
- 92.Bar Or I., et al. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.3389/fpubh.2021.561710. p. 2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Randazzo W., et al. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. medRxiv. 2020 doi: 10.1016/j.ijheh.2020.113621. p. 2020.04.23.20076679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murakami M., et al. Letter to the editor: wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- 95.Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580(7802):176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- 96.Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma V.K., Jinadatha C., Lichtfouse E. Environmental chemistry is most relevant to study coronavirus pandemics. Environ. Chem. Lett. 2020 doi: 10.1007/s10311-020-01017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson N., et al. Detecting the emergent or re-emergent COVID-19 pandemic in a country: modelling study of combined primary care and hospital surveillance. medRxiv. 2020 p. 2020.05.13.20100743. [PubMed] [Google Scholar]
- 99.Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Venugopal A., et al. Novel wastewater surveillance strategy for early detection of COVID – 19 hotspots. Current Opinion Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deslandes A., et al. SARS-CoV-2 was already spreading in France in late December 2019. Int. J. Antimicrobial Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54(7):3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- 103.Organization W.H. World Health Organization; 2020. Water, Sanitation, Hygiene, and Waste Management for the COVID-19 Virus: Interim Guidance, 23 April 2020. [Google Scholar]
- 104.Gerba C.P., Betancourt W.Q., Kitajima M. How much reduction of virus is needed for recycled water: a continuous changing need for assessment? Water Res. 2017;108:25–31. doi: 10.1016/j.watres.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahmed W., et al. Recycled water safety: current status of traditional and emerging viral indicators. Current Opinion Environ. Sci. Health. 2020;16:62–72. [Google Scholar]
- 106.Center for Disease Control C. 2020. Interim Guidelines for Biosafety and COVID-19. [Google Scholar]
- 107.Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- 108.Aquino De Carvalho N., et al. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environ. Sci. Technol. 2017;51(15):8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- 109.Zielińska M., Galik M. Use of ceramic membranes in a membrane filtration supported by coagulation for the treatment of dairy wastewater. Water Air Soil Pollut. 2017;228(5):173. doi: 10.1007/s11270-017-3365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Viralzone S.Io.B.S. 2020. Cystoviridae Phi6.https://viralzone.expasy.org/165 [Google Scholar]
- 111.Huang H., et al. Mechanisms of virus removal from secondary wastewater effluent by low pressure membrane filtration. J. Membrane Sci. 2012;409-410:1–8. [Google Scholar]
- 112.Samineni L., et al. 7 log virus removal in a simple functionalized sand filter. Environ. Sci. Technol. 2019;53(21):12706–12714. doi: 10.1021/acs.est.9b03734. [DOI] [PubMed] [Google Scholar]
- 113.Domagała K., et al. Efficiency and stability evaluation of Cu2O/MWCNTs filters for virus removal from water. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115879. [DOI] [PubMed] [Google Scholar]
- 114.Lee S., et al. Evaluation of virus reduction at a large-scale wastewater reclamation plant by detection of indigenous F-specific RNA bacteriophage genotypes. Environ. Technol. 2019;40(19):2527–2537. doi: 10.1080/09593330.2018.1444675. [DOI] [PubMed] [Google Scholar]
- 115.Kuo D., et al. High virus removal by self-organized nanostructured 2D liquid-crystalline smectic membranes for water treatment. Small. 2020 doi: 10.1002/smll.202001721. n/a(n/a) [DOI] [PubMed] [Google Scholar]
- 116.Fievre A., et al. High-Resolution fourier transform ion cyclotron resonance mass spectrometry of humic and fulvic acids by laser desorption/ionization and electrospray ionization. Energy Fuels. 1997;11(3):554–560. [Google Scholar]
- 117.Lee S., et al. Improvement of virus removal by pilot-scale coagulation-ultrafiltration process for wastewater reclamation: effect of optimization of pH in secondary effluent. Water Res. 2017;114:23–30. doi: 10.1016/j.watres.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 118.Mazurkow J.M., et al. Nano-sized copper (oxide) on alumina granules for water filtration: effect of copper oxidation state on virus removal performance. Environ. Sci. Technol. 2020;54(2):1214–1222. doi: 10.1021/acs.est.9b05211. [DOI] [PubMed] [Google Scholar]
- 119.Torii S., et al. Repeated pressurization as a potential cause of deterioration in virus removal by aged reverse osmosis membrane used in households. Sci. Total Environ. 2019;695 doi: 10.1016/j.scitotenv.2019.133814. [DOI] [PubMed] [Google Scholar]
- 120.Verbyla M.E., Mihelcic J.R. A review of virus removal in wastewater treatment pond systems. Water Res. 2015;71:107–124. doi: 10.1016/j.watres.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 121.Ye Y., et al. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 122.Im D., et al. Performance of combined ozonation, coagulation and ceramic membrane process for water reclamation: effects and mechanism of ozonation on virus coagulation. Separation Purification Technol. 2018;192:429–434. [Google Scholar]
- 123.Viralzone S.Io.B.S. 2020. Levivirus M.S.2.https://viralzone.expasy.org/291 [Google Scholar]
- 124.Wu B., Wang R., Fane A.G. The roles of bacteriophages in membrane-based water and wastewater treatment processes: a review. Water Res. 2017;110:120–132. doi: 10.1016/j.watres.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 125.Shirasaki N., et al. Assessment of the efficacy of membrane filtration processes to remove human enteric viruses and the suitability of bacteriophages and a plant virus as surrogates for those viruses. Water Res. 2017;115:29–39. doi: 10.1016/j.watres.2017.02.054. [DOI] [PubMed] [Google Scholar]
- 126.Tandukar S., Sherchan S.P., Haramoto E. Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Reports. 2020;10(1):3616. doi: 10.1038/s41598-020-60547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.World Health O. WHO guidelines for the safe use of wastewater. Excreta Greywater. 2006;2 [Google Scholar]
- 128.Qiu Y., et al. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 129.Simhon A., et al. Enteric viruses in municipal wastewater effluent before and after disinfection with chlorine and ultraviolet light. J. Water Health. 2019;17(5):670–682. doi: 10.2166/wh.2019.111. [DOI] [PubMed] [Google Scholar]
- 130.Wigginton K.R., et al. Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity. Environ. Sci. Technol. 2012;46(21):12069–12078. doi: 10.1021/es3029473. [DOI] [PubMed] [Google Scholar]
- 131.Jassim S.Y. 2020. International Ozoine Association Statement on COVID-19.https://www.ioa-pag.org/resources/Documents/EOC%20Files/IOA%20Coronavirus%20Statement.pdf Available from: [Google Scholar]
- 132.EPA . 1989. Surface Water Treatment Rule 1989.https://www.epa.gov/dwreginfo/surface-water-treatment-rules Available from: [Google Scholar]
- 133.Sigmon C., et al. Establishing surrogate–virus relationships for ozone disinfection of wastewater. Environ. Eng. Sci. 2015;32(6):451–460. [Google Scholar]
- 134.Harakeh M.S., Butler M. Factors increasing the ozone inactivation of enteric viruses in effluent. Ozone: Sci. Eng. 1984;6(4):235–243. [Google Scholar]
- 135.Irving L.G., Smith F.A. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 1981;41(1):51–59. doi: 10.1128/aem.41.1.51-59.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.White G.C. Van Nostrand Reinhold; 1978. Disinfection of Wastewater and Water for Reuse. [Google Scholar]
- 137.Perez-Rey R., Cháez H., Baluja C. Ozone inactivation of biologically-risky wastewaters. Ozone: Sci. Eng. 1995;17(5):499–509. [Google Scholar]
- 138.Aydogan A., Gurol M.D. Application of gaseous ozone for inactivation of bacillus subtilis spores. J. Air Waste Manage. Assoc. 2006;56(2):179–185. doi: 10.1080/10473289.2006.10464443. [DOI] [PubMed] [Google Scholar]
- 139.Gagnon C., et al. Degradation of selected acidic and neutral pharmaceutical products in a primary-treated wastewater by disinfection processes. Ozone: Sci. Eng. 2008;30(5):387–392. [Google Scholar]
- 140.Delanka-Pedige H.M.K., et al. Metagenomic insights into virus removal performance of an algal-based wastewater treatment system utilizing galdieria sulphuraria. Algal Res. 2020;47 [Google Scholar]
- 141.Delanka-Pedige H.M.K., et al. Algal pathway towards meeting United nation’s sustainable development goal 6. Int. J. Sustainable Dev. World Ecol. 2020:1–9. [Google Scholar]
- 142.Delanka-Pedige H.M.K., et al. Bacteria and virus reduction in secondary treatment: potential for minimizing post disinfectant demand. Water Res. 2020;177 doi: 10.1016/j.watres.2020.115802. [DOI] [PubMed] [Google Scholar]
- 143.Goswami K.P., Pugazhenthi G. Credibility of polymeric and ceramic membrane filtration in the removal of bacteria and virus from water: a review. J. Environ. Manage. 2020;268 doi: 10.1016/j.jenvman.2020.110583. [DOI] [PubMed] [Google Scholar]
- 144.Ostarcevic E.R., et al. Current and emerging techniques for high-pressure membrane integrity testing. Membranes. 2018;8(3):60. doi: 10.3390/membranes8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tang C.Y., et al. Potable water reuse through advanced membrane technology. Environ. Sci. Technol. 2018;52(18):10215–10223. doi: 10.1021/acs.est.8b00562. [DOI] [PubMed] [Google Scholar]
- 146.Prado T., et al. Performance of wastewater reclamation systems in enteric virus removal. Sci. Total Environ. 2019;678:33–42. doi: 10.1016/j.scitotenv.2019.04.435. [DOI] [PubMed] [Google Scholar]
- 147.Vickers J.C., et al. Removal of MS-2 coliphage in full-scale reverse osmosis systems. AWWA Water Sci. 2019;1(6):e1158. [Google Scholar]
- 148.Prado T., et al. Noroviruses in raw sewage, secondary effluents and reclaimed water produced by sand-anthracite filters and membrane bioreactor/reverse osmosis system. Sci. Total Environ. 2019;646:427–437. doi: 10.1016/j.scitotenv.2018.07.301. [DOI] [PubMed] [Google Scholar]
- 149.Viralzone S.Io.B.S. 2020. Norovirus.https://viralzone.expasy.org/194 [Google Scholar]
- 150.Singh R., et al. In: Waterborne Pathogens. Vara Prasad M.N., Grobelak A., editors. Butterworth-Heinemann; 2020. Nanofiltration technology for removal of pathogens present in drinking water; pp. 463–489. Chapter 21. [Google Scholar]
- 151.Zhao X., et al. Evaluation of viral removal by nanofiltration using real‐time quantitative polymerase chain reaction. Biotechnol. Appl. Biochem. 2007;47(2):97–104. doi: 10.1042/BA20060195. [DOI] [PubMed] [Google Scholar]
- 152.Burnouf‐Radosevich M., et al. Nanofiltration, a new specific virus elimination method applied to high‐purity factor IX and factor XI concentrates. Vox sanguinis. 1994;67(2):132–138. doi: 10.1111/j.1423-0410.1994.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 153.Burnouf T., Radosevich M. Nanofiltration of plasma‐derived biopharmaceutical products. Haemophilia. 2003;9(1):24–37. doi: 10.1046/j.1365-2516.2003.00701.x. [DOI] [PubMed] [Google Scholar]
- 154.O’Grady J., et al. Virus removal studies using nanofiltration membranes. Dev. Boil. standardization. 1996;88:319–326. [PubMed] [Google Scholar]
- 155.Zheng X., Liu J. Virus rejection with two model human enteric viruses in membrane bioreactor system. Sc. China Series B: Chem. 2007;50(3):397–404. doi: 10.1007/s11426-007-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Al Aani S., Mustafa T.N., Hilal N. Ultrafiltration membranes for wastewater and water process engineering: a comprehensive statistical review over the past decade. J. Water Process Eng. 2020;35 [Google Scholar]
- 157.Gentile G.J., et al. Electrostatic interactions in virus removal by ultrafiltration membranes. J. Environ. Chem. Eng. 2018;6(1):1314–1321. [Google Scholar]
- 158.Lu R., et al. Improvement of virus removal using ultrafiltration membranes modified with grafted zwitterionic polymer hydrogels. Water Res. 2017;116:86–94. doi: 10.1016/j.watres.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 159.Fallahianbijan F., et al. Quantitative analysis of internal flow distribution and pore interconnectivity within asymmetric virus filtration membranes. J. Membrane Sci. 2020;595 [Google Scholar]
- 160.Kwarciak-Kozlowska A., Wlodarczyk R. In: Waterborne Pathogens. Vara Prasad M.N., Grobelak A., editors. Butterworth-Heinemann; 2020. Chapter 5 - treatment of waterborne pathogens by microfiltration; pp. 81–103. [Google Scholar]
- 161.Sinclair T.R., et al. Virus reduction through microfiltration membranes modified with a cationic polymer for drinking water applications. Colloids and Surfaces A. 2018;551:33–41. [Google Scholar]
- 162.O’Brien E., Xagoraraki I. Removal of viruses in membrane bioreactors. J. Environ. Eng. 2020;146(7) [Google Scholar]
- 163.Miura T., et al. Virus type-specific removal in a full-scale membrane bioreactor treatment process. Food Environ. Virol. 2018;10(2):176–186. doi: 10.1007/s12560-017-9330-4. [DOI] [PubMed] [Google Scholar]
- 164.Im D., et al. Pretreatment of ceramic membrane microfiltration in wastewater reuse: a comparison between ozonation and coagulation. J. Environ. Manage. 2019;251 doi: 10.1016/j.jenvman.2019.109555. [DOI] [PubMed] [Google Scholar]
- 165.Bartels J., et al. Hydrophobic ceramic capillary membranes for versatile virus filtration. J. Membrane Sci. 2019;570-571:85–92. [Google Scholar]
- 166.Wattanachira L., et al. Bacteriophage Removal Efficiency of In-line Coagulation with Ceramic Membrane Filtration. Eng. J. 2017;21(4):1–9. [Google Scholar]
- 167.Indarto A., Ikhsan N.A., Wibowo I. In: Waterborne Pathogens. Vara Prasad M.N., Grobelak A., editors. Butterworth-Heinemann; 2020. Chapter 20 - applications of carbon nanotubes for controlling waterborne pathogens; pp. 433–461. [Google Scholar]
- 168.Ojha A. In: Waterborne Pathogens. Vara Prasad M.N., Grobelak A., editors. Butterworth-Heinemann; 2020. Chapter 19 - nanomaterials for removal of waterborne pathogens: opportunities and challenges; pp. 385–432. [Google Scholar]
- 169.Sahu J.N., et al. Current perspectives and future prospects of nano-biotechnology in wastewater treatment. Separation Purification Rev. 2019:1–20. [Google Scholar]
- 170.Kim J.P., et al. A nanofilter composed of carbon nanotube-silver composites for virus removal and antibacterial activity improvement. J. Environ. Sci. 2016;42:275–283. doi: 10.1016/j.jes.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 171.Németh Z., et al. Enhanced virus filtration in hybrid membranes with MWCNT nanocomposite. Royal Soc. Open Sci. 2019;6(1) doi: 10.1098/rsos.181294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang K., et al. Mechanical properties of water desalination and wastewater treatment membranes. Desalination. 2017;401:190–205. [Google Scholar]
- 173.Pype M.L., et al. Reverse osmosis integrity monitoring in water reuse: the challenge to verify virus removal - a review. Water Res. 2016;98:384–395. doi: 10.1016/j.watres.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 174.Yoon S.-H. Potential and limitation of fluorescence-based membrane integrity monitoring (FMIM) for reverse osmosis membranes. Water Res. 2019;154:287–297. doi: 10.1016/j.watres.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 175.Frenkel V.S., Cohen Y. New techniques for real-time monitoring of reverse osmosis membrane integrity for virus removal. Water Practice Technol. 2018;13(4):947–957. [Google Scholar]
- 176.Niewersch C., et al. Reverse osmosis membrane element integrity evaluation using imperfection model. Desalination. 2020;476 [Google Scholar]
- 177.Krahnstöver T., Hochstrat R., Wintgens T. Comparison of methods to assess the integrity and separation efficiency of ultrafiltration membranes in wastewater reclamation processes. J. Water Process Eng. 2019;30 [Google Scholar]
- 178.Suzuki T., Okamura M., Niinae M. Plugging nanoscale imperfections in the polyamide active layer of thin-film composite reverse osmosis membrane to inhibit advective solute transport. Desalination. 2020;487 [Google Scholar]