Abstract
Breast cancer has seriously been threatening physical and mental health of women in the world, and its morbidity and mortality also show clearly upward trend in China over time. Through inquiry, we find that survival rate of patients with early‐stage breast cancer is significantly higher than those with middle‐ and late‐stage breast cancer, hence, it is essential to conduct research to quickly diagnose breast cancer. Until now, many methods for diagnosing breast cancer have been developed, mainly based on imaging and molecular biotechnology examination. These methods have great contributions in screening and confirmation of breast cancer. In this review article, we introduce and elaborate the advances of these methods, and then conclude some gold standard diagnostic methods for certain breast cancer patients. We lastly discuss how to choose the most suitable diagnostic methods for breast cancer patients. In general, this article not only summarizes application and development of these diagnostic methods, but also provides the guidance for researchers who work on diagnosis of breast cancer.
1. INTRODUCTION
Breast cancer (BC) has become one of the most common malignant tumours, and latest dates from CA‐cancer magazine show that the incidence rate is increasing every year. In 2019, approximately 316 700 new cases of BC have been confirmed in US women, and the growth rate is nearly 0.3% per year. 1 The data from China show that the incidence rate of BC also raises per year (272 400 cases in 2015 and 367 900 cases in 2018). 2 , 3 Taking population growth into consideration, experts predict that there will be about 3.2 million new BC cases per year globally by 2050. 4 More notably, not only the number of patients with BC is increasing all over the world, but also the age of affected patients is tending to be younger. 5 There are many factors causing above situation, such as age, family history, lifestyle environments and so on. 4 , 6 , 7 The high incidence rate of BC is unavoidable, but decreasing the mortality of BC is feasible. Early detection and treatment are critical to curing BC, because it tends to metastasize in the middle and last stage. 8 , 9 , 10 Therefore, finding BC is vital in early stage, which can greatly improve the survival rate of patients.
To quickly and accurately screen BC, many diagnostic methods based on imaging and molecular biotechnology have been developed. It is indispensable to summarize and evaluate these methods, to provide value information for clinical diagnosis. Jafari 11 summarized various imaging techniques and biochemical biomarkers used for detection and monitoring BC patients and highlighted that it is helpful to diagnose and treat patients with BC by measuring level of certain biomarkers. Weaver 12 described definitions and applications of imaging “biomarkers,” and thought that they can build the decision support system by these markers to provide help for clinical breast care and BC–related research. Many articles review these methods for diagnosing BC mainly from these aspects, by introducing the contribution of imaging techniques (including molecular imaging markers) in diagnosing BC patients, and summarizing these findings on connection between newly discovered tumour makers and BC patients. 13 , 14 , 15 Many articles describe a large number of diagnostic methods for breast cancer, but few articles introduce how to choose suitable diagnostic methods for different types of BC patients.
In this review, many diagnostic methods are reviewed, such as mammography (MG), ultrasonography (US), magnetic resonance imaging (MRI), nucleic acid hybridization system (NAHS), real‐time fluorescence quantitative PCR system (RT‐qPCR), protein hybridization system (PHS), flow cytometer (FCM) and so on. We herein introduce their development and summarize their advantages and disadvantages and provide some diagnostic schemes for different types of BC patients. The article can help future research and development in diagnosing BC patients and guiding people who are working on BC research, on how to choose the suitable methods for diagnosing BC patients.
2. IMAGING DIAGNOSIS
Utilization of imaging techniques shows clearly the morphology and location of tumour tissues and proves much clinical information that is valuable to doctors. However, imaging techniques may cause harm to patients when using contrast agents and high energy rays. Therefore, we should discuss these imaging techniques and choose the most appropriate diagnostic method for BC patients. These imaging techniques mainly include mammography (MG), ultrasonography (US), magnetic resonance imaging (MRI), positron emission computed tomography (PET), computed tomography (CT) and single‐photon emission computed tomography (SPECT). In Table 1, we list the advantages and disadvantages of these imaging methods. In view of high cost, poor practicability and radiation damage, PET, CT and SPECT are not recommended in diagnosing BC patients. However, these techniques can be used as auxiliary diagnostic methods for diagnosing BC in some special cases, such as screening for metastatic BC, presence of bone and lymphatic metastases. Therefore, we only introduce MG, US and MRI that are preferred methods for screening BC. Summary and evaluation of these common imaging techniques will help doctors to better serve patients and promote the development of clinical diagnosis.
Table 1.
Advantages and disadvantages of imaging techniques
| Imaging techniques | Advantages | Disadvantages |
|---|---|---|
| XRM |
|
|
| US |
|
|
| MRI |
|
|
| PET |
|
|
| CT |
|
|
| SPECT |
|
|
Abbreviations: CT, Computed tomography; MRI, Magnetic resonance imaging; PET, Positron emission tomography; SPECT, Single‐photon emission computed tomography; US, Ultrasonography; XRM, X‐ray mammography.
2.1. Mammography
Mammography (MG) is preferred strategy for screening and diagnosing BC and helps doctors obtain clinic information on BC patients. The evidence suggests that the mortality rate of BC patients could be reduced 30%‐40% though early MG screening. 16 Meanwhile, the diagnostic result of MG is only positive criteria for 4%‐10% of BC patients (eg, patients who exhibited only slight calcification). 17 , 18 MG is developed continuously with passage of time. Contrast‐enhanced mammography (CEM) and digital breast tomosynthesis (DBT) are at present two main strategies that diagnose BC patients in clinic. 19 , 20 Through investigation, CEM is superior to full‐field digital mammography (FFDM), and the value of CEM in diagnostic accuracy and evaluation of disease extent is close to breast MRI. 21 , 22 Similarly, DBT also has good performance, such as higher specificity, when compared with FFDM (96.4%, 57229/59381% vs 97.5%, 23427/24020, P < .001). 23 In 1998, computer‐aided detection (CAD) was developed and it greatly improved the sensitivity of instruments from about 60% to 100%. 24 CEM can be combined with CAD to diagnose BC patients, and it could carry out classification for breast masses, and the ROC curves for patients will be significantly increased to 0.848 ± 0.038 (P < .01). 25 Similarly, the reading time for DBT can be improved to about 29.2%, and the ROC curves for patients will be increased from 0.841 to 0.850 (95% CI, −0.012 to 0.030) when combined with CAD. 26
In general, MG and its derivatives are indispensable part in diagnosis and screening of BC patients. Their advantages are as follows: rapid screening, high accuracy, low cost and suitable for promoted use. Therefore, MG is optimal imaging diagnostic method for patients with low income and eliminates the risk for BC, etc However, these factors may cause MG to not be suitable for everyone. For example, MG needs harmful contrast agent and X‐ray to do imaging, so cannot be used repeatedly in a short period of time, and is not recommended to use for patients under age of 40. 27 In the future, MG will tend to be harmless and with high resolution. Meanwhile, with advancement of artificial intelligence (AI) technique and development of sensors, it is viable to realize automation of detection and analysis of BC.
2.2. Ultrasonography
Ultrasonography (US) is applied in observing morphology and variation condition of tumour tissues, and it can accurately locate the location of lesions. US is not harmful to humans and is suitable for everyone. The development history of US is as follows: the early grayscale US only showed whether the tumour existed at detection site, but it was difficult to distinguish benign and malignant tumours, because its resolution was low. 28 , 29 Surely, the two‐dimensional US only gets some flat images of tumour, and judgement by physicians will be affected. So, three‐dimensional US technology was developed for three‐dimensional imaging of tumour morphology and blood vessel distribution, which are shown when patients are diagnosed. 30 The colour Doppler US is one of many three‐dimensional US and can clearly reflect the situation of tumour and blood flow information and provide doctors with more valuable clinical information, so that it can distinguish benign and malignant tumours. 31 In 1998, Krouskop 32 found that there are elastic differences in different tissues, which provides theoretical foundation for developing elastic US. Moreover, some researches screened the suspected pathological tissues by using elastic US and found that it improves greatly the accuracy for diagnosing BC. 33 , 34 However, when combined with three‐dimensional US, the elastic US can diagnose axillary lymphadenopathy and classify the patient's tumour state. 35 Though MG is optimal method to detect the calcification condition of BC, when the size of calcification is too small, it is difficult to be detected by MG or routine US. 36 A new US image‐processing technique, MicroPure, was therefore developed. This method can reduce speckle by analysing pictures of spatial feature and frequency and create images that have high contrast resolution and high tissue uniformity. 37 Machado et al 38 processed ex vivo surgical breast specimens by using MicroPure examination and found that the MicroPure has high recognition rate to microcalcifications of BC, and conventional US cannot found its situation.
US has many advantages, such as use of few contrast agents, none high energy rays and suitability for all ages. Meanwhile, when MG cannot be used, US can become an alternative diagnostic method for BC. However, the US has limitations that need professional operation and lower definition and resolution than CT. Notably, the people who are obese and those with nodi lymphatici parasternales metastasis are not suitable to use US for diagnosis. In the future, intelligent US detection will be a new tendency, which will greatly reduce errors due to unprofessional judgements, thereby helping doctors to get more accurate diagnostic results.
2.3. Magnetic resonance imaging
Magnetic resonance imaging (MRI) allows early detection of familial BC regardless of patients’ age, breast density or risk status. 39 Figure 1 is schematic diagram of MRI. Water dispersion coefficient of different tissues exists with differences. Magnetic resonance diffusion weighted (MRDW) is a technique that can show clear movement of water molecules in the body. Therefore, MRDW has become a method for diagnosing BC patients. Through literature review, we found that malignant tumours have typical water diffusion‐limited effects in comparison with benign tumours, so researchers can distinguish benign and malignant breast tumours by using MRDW to measure apparent diffusion coefficient (ADC) values (represent diffusion‐limited effects) of tumours (ADC values: normal breast group > benign group > malignant group). 40 , 41 Recently, a review reported that the optimal threshold values for ADC in distinguishing benign and malignant lesions are as follows: 1.06 × 10−3 mm2/s ~ 1.10 × 10−3 mm2/s. 42 Dynamic contrast‐enhanced MRI (DCE‐MRI) has higher resolution of soft tissues than MRDW, and it can clearly show morphological characteristics and haemodynamic characteristics of the lesions in vivo. 43 Researchers found that the positive predictive value (98%) of DCE/MRI is higher than the positive predictive value (77%) of MRI alone, and the specificity points to 97%. 44 Guindalini 45 compared the diagnostic techniques of BC and found that biannual DCE‐MRI and annual MG for BC patients have performed well and low recall rates. Magnetic resonance spectroscopy (MRS) is a non‐invasive method that can also improve diagnostic rate of BC, by evaluating the risk of BC and guiding treatment of BC. 46 , 47
Figure 1.
Schematic diagram of MRI
Magnetic resonance elastography (MRE) is another special magnetic resonance technology that can provide information on tissue elasticity by transmission of mechanical waves in tissues. Bohte et al 48 elaborated MRE’s future tendency that it can delineate pre‐operative tumour and predict response to treatment and metastatic potential of primary tumours. PET/MRI, Positron emission computed tomography (PET) combined with MRI, can display soft tissue structures of the breast and chest wall. PET can provide molecular‐level information in vivo, and PET/MRI can improve the positive predictive rate of patients and has great value in evaluating BC metastasis. 9 , 49 , 50
Magnetic resonance imaging is an auxiliary method that has many advantages in diagnosing BC. However, there are many factors that influence the wide application of MRI, such as long imaging time, high cost, cannot be carried out if has metal material in patient's body, and so on. Therefore, MRI can be used in situations where the primary BC is too small, where all information about tumour needs to be obtained, and for screening of high‐risk groups, etc In the future, MRI will tend to have higher signal‐to‐noise ratio, shorter imaging time and lower cost. Likewise, advancement in MRI should also consider how to reduce the use of contrast agents, so that it serves every stage of BC.
3. MOLECULAR BIOTECHNOLOGY EXAMINATION
Molecular biotechnology examinations can diagnose BC earlier than imaging techniques. Nevertheless, it cannot replace the imaging techniques and become auxiliary methods to diagnose BC. The purposes of molecular biotechnology examination are to analyse specific biomarkers such as nucleic acid, proteins, cells and tissues of patients. These examinations can help doctors obtain much clinical information at the molecular level. At present, these examination techniques mainly include nucleic acid hybridization system, real‐time fluorescence quantitative PCR system, protein hybridization system, flow cytometer, needle biopsy and immunohistochemistry (IHC). These techniques help us analyse BC from the level of nucleic acids, proteins and cells.
3.1. Novel specific biomarkers
Circulating tumour cells (CTCs) enter the blood circulation from primary tumour tissues, and the number of CTCs is about 1 ~ 102/mL in peripheral blood. Jin et al 51 investigated the viability of using CytoSorter®system to detect CTCs and to evaluate the diagnostic value of CTCs in BC. Their results showed that the CTCs can differentiate BC patients from the patients with benign breast diseases or healthy volunteers, as a diagnostic aid for early cancer diagnosis and cancer staging. 51 CTCs could be used as a novel biomarker in assisting BC detection.
Circulating tumour DNA (ctDNA) is fragments of tumour genomic DNA that contains characteristics of gene variations consistent with primary solid tumour. ctDNA is thus very helpful in identifying the DNA from tumour cells or normal cells, as the number of ctDNA is very small in peripheral blood. Thus, the quantitative and qualitative detection methods for ctDNA are based mainly on PCR and next‐generation sequencing (NGS). Ma et al 52 had a longitudinal monitoring of 21 patients during treatment that showed that the molecular tumour burden index (mTBI, a measure of the percentage of ctDNA in samples), positively correlated with tumour size as evaluated by computed tomography (P < .0001, Pearson r = .52), and detected disease progression 8‐16 weeks. 52 Therefore, ctDNA could be used to assess tumour heterogeneity and predict treatment outcomes in metastatic BCs. 52
Exosomes are membrane‐enclosed phospholipid extracellular vesicles with a variety of tumour antigens which can be applied in the diagnosis and treatment of cancer due to their high secretion on the surface of cancer cells. 53 Exosomes have stable chemical properties, and their size is 30‐150 nm. 53 Ni et al 54 investigated whether the enrichment of miRNAs in exosomes reflects the pathogenesis of BC and ductal carcinoma in situ (DCIS). The levels of exosomal miR‐16 were higher in plasma of BC (P = .034) and DCIS (P = .047) patients than healthy women and were associated with oestrogen (P = .004) and progesterone (P = .008) receptor status. Moreover, lower levels of exosomal miR‐30b were associated with recurrence (P = .034), and exosomal miR‐93 was upregulated in DCIS patients (P = .001). 54 Taken together, their result showed that different signatures of miR‐16, miR‐30b and miR‐93 in exosomes from BC and DCIS patients are associated with a particular biology of breast tumours. 54 Therefore, exosomes have become a research hotspot in recent years because of their great diagnostic potential.
Long noncoding RNA (lncRNA) can involve in the regulation of cell cycle of tumour cells and a variety of cell signalling pathways of cancer cell invasion, metastasis, resistance of chemotherapy and so on. Shao et al 55 found two lncRNAs that significantly correlated with outcomes of breast cancer and were regulated by methylation status. Liang et al 56 revealed that RHPN1 antisense RNA 1 (RHPN1‐AS1) was induced by KDM5B and promoted BC via RHPN1‐AS1/miR‐6884‐5p/ANXA11 pathway. Besides, H19, an oestrogen‐inducible lncRNA, was reported to function in the cell survival and proliferation, which was from the oestrogen in breast cancer cells. 57 Therefore, the functions of lncRNAs in initiation, progression and metastasis of breast cancer are emerging and are expected to be a potential new diagnostic marker and therapeutic target for BC. 56
Circular RNAs (circRNAs) were recently discovered as a looped subset of competing endogenous RNAs, with an ability to regulate gene expression by microRNA sponging. 58 Lu et al 59 found that a total of 715 circRNAs were notably overexpressed, and 440 were remarkably downregulated in the BC lesions compared with healthy tissue samples among 1155 differentially expressed circRNAs. In 2019, Yan et al 60 introduced hsa_circ_0072309 as a novel prognostic biomarker in BC, which is a miR‐492 sponge that is downregulated in BC. Dysregulation of this circRNA increases proliferation, migration and invasion in BC cells, and thus, it has a potential role in BC, as it is highly conserved and stable.
In all, these novel biomarkers not only are monitored dynamically, but are also used to judge prognosis. The patient's body fluids are used as samples for CA biopsy.
3.2. Nucleic acid hybridization system
3.2.1. Nucleic acid hybridization
Nucleic acid hybridization techniques mainly include fluorescence in situ hybridization (FISH) and aptamer probe hybridization (APH). They can find special fragments of tumour biomarkers and search new tumour biomarkers when diagnosing BC.
FISH has made huge contributions to the development of molecular biology diagnostics. 61 Its principle follows (Figure 2) base pairing. These data display that approximately 25‐30 per cent of all BC are human epidermal growth factor receptor 2 (HER‐2)–positive BC. 62 , 63 FISH has high response rates (2474 of 2524; 98.0%) to amplify HER‐2 gene and has high HER‐2 copies number per cell (by 2.86; P = .02). 64 FISH detection is an important factor in whether a medication (Herceptin) is needed or not for BC patients. Meanwhile, FISH is considered the “gold standard” for detecting whether the HER‐2 gene is activated. 65 In addition, FISH shows other advantages, including reproducibility, stability and high sensitivity. However, these factors limit its promotion, including the need for complex probes design and special fluorescence detector. In the future, multicolour fluorescence in situ hybridization will be a tendency in greatly improving the throughput when searching genetic sites.
Figure 2.
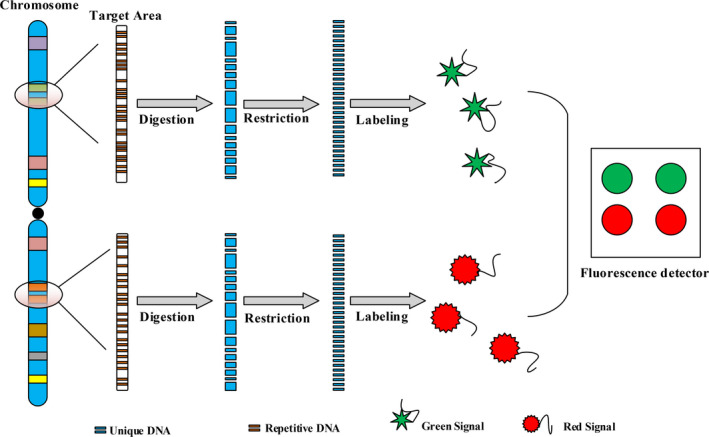
Technical principle of fluorescence in situ hybridization
Aptamer probe hybridization is another highly sensitive and specific technique. Suitable aptamers are key factors in the accuracy of APH. These aptamers mainly are produced by Systematic Evolution of Ligands by Exponential enrichment (SELEX). 66 At present, Cell‐SELEX is one of the most representative of SELEX, and it has become the main method that gets optimal aptamers from tumours. 67 The schematic diagram of Cell‐SELEX is shown in Figure 3. Suitable aptamers can identify some specific fragments that can be used to diagnose diseases. Kim 68 prepared a nucleotide aptamer (SE15‐8‐QDs) for detecting BC and found that it is more sensitive than the common probes. Cai 69 developed a new type of fluorescence aptamer (AAI2‐5) that can detect MCF‐7 BC cells and MDA‐MB‐231 cell lines easily and sensitively from breast cells with an accuracy of 90%. However, the process for obtaining suitable aptamers or probes is complex and difficult, requiring a lot of time and money, and is not suitable for promoting to use in primary hospitals. In the future, APH will have the easy process for screening suitable aptamers and will find more biomarkers of BC.
Figure 3.
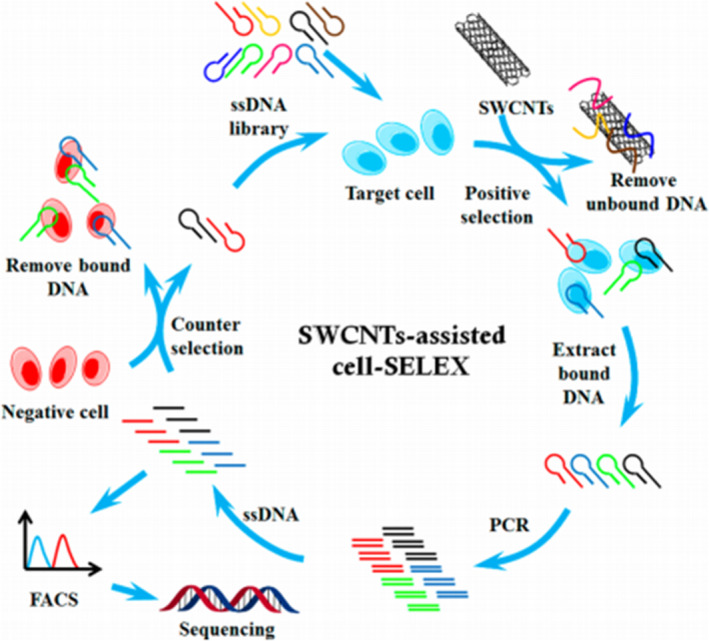
Schematics of cell‐based aptamer selection. 147 (Reproduced with permission from Copyright 2014, American Chemical Society)
3.2.2. Gene chip and next‐generation sequencing
Gene chip can analyse a large number of fragments of nucleic acid simultaneously, and it is applied widely in diagnosing BC. Gene chip is used to observe and analyse the condition of nucleic acids in BC cells or tissues and also can find new diagnostic biomarkers for BC by screening a large number of samples. As well known, gene chip is essentially a high‐density oligonucleotide microarray. 70 , 71 , 72 At present, there are two methods for chip preparation: in situ synthesis and direct point method. 73 However, in situ synthesis is the main method, and its schematic diagram is shown in Figure 4. Using gene chip technology, researchers found mechanisms for doxorubicin resistance in BC and screened these key genes for BC therapy. 74 Jiang et al 75 used LIMMA (Linear Models for Microarray Data) methodology to identify differential expression of lncRNAs between tumour and normal samples, and they identified 26 inter‐genic lncRNAs transcripts that were specifically expressed in tumour cells [P < .005, FDR < 0.15]. There are however limitations of gene chip, such as difficulty in synthesizing probes, easy appearance of positive signals and especially complicated nucleic acid extraction. In the future, with development of nanotechnology, the size of chip will be smaller and the throughput of gene chip will be higher.
Figure 4.
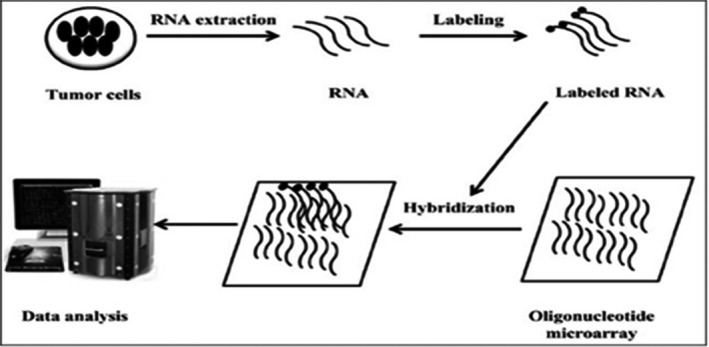
Flow chart of microarray technology. 162 (Reproduced with permission from Copyright 2012, Rajnish Kumar)
Next‐generation sequencing (NGS) was put forward by Metzker. 76 The schematic diagram of NGS is shown in Figure 5. This technique makes great contribution to get the genome sequence information and can help find mutant gene sites. Currently, NGS has been applied widely in diagnosing BC. For example, Dong et al 77 designed targeted NGS platform and found three additional possible disease‐causing mutant genes. Liang et al 78 found twelve common mutant genes by NGS detection, namely TP53, PIK3CA, MYH9, NOTCH2, BRCA2, ERBB4, FGFR3, POLE, LAMA2, ARID1A, NOTCH4 and ROS1, in inflammatory BC. Moreover, Kim et al 79 detected at least one somatic mutation in 44 of 61 tDNA (72.1%) and 29 of 44 (65.9%) and cfDNA, and the overall concordance rate of cfDNA to tDNA was 85.9%, utilizing next‐generation digital sequencing technology. Wu et al 80 used RNA sequencing to detect tumour‑specific miRNAs, and their results showed that the exosome levels of hsa‐miR‐150‐5p, hsa‐miR‐576‐3p and hsa‐miR‐4665‐5p were higher in BC with recurrence compared to those in patients without recurrence. Page et al 81 used a novel targeted NGS panel to examine cfDNA to detect somatic mutations and gene amplification in women with metastatic BC. Their results showed no mutations were identified in cfDNA of healthy controls, whereas exactly half the patients with metastatic BC had at least one mutation or amplification in cfDNA (mean 2, range 1‐6) across a total of 13 genes. 81 Scarpitta et al 82 screened the 24 genes involved in BC predisposition, genome stability maintenance and DNA repair mechanisms by NGS and found that a positive family history is a strong predictor of germline BRCA2 mutations in male BC. Ou‐Yang et al 83 compared differences in gene expressions in parental and CHD4‐deficient cells by NGS and suggested that the chromodomain‐helicase‐DNA‐binding protein 4 regulates β1 integrin in triple‐negative BC. However, the main limitation of NGS is short reads of about 200‐500 bp. Single‐molecule sequencing can offer long read lengths, direct RNA sequencing, direct identification of base modifications and so on, but at present NGS can easily occupy mismatch and is not suitable for analysis of satellite DNA. 84 Therefore, sequencing can help us to analyse the gene mutations in humans and can predict the risk of BC. Research shows that NGS will be main trend of high throughput, high accuracy and fewer mismatch in the future.
Figure 5.
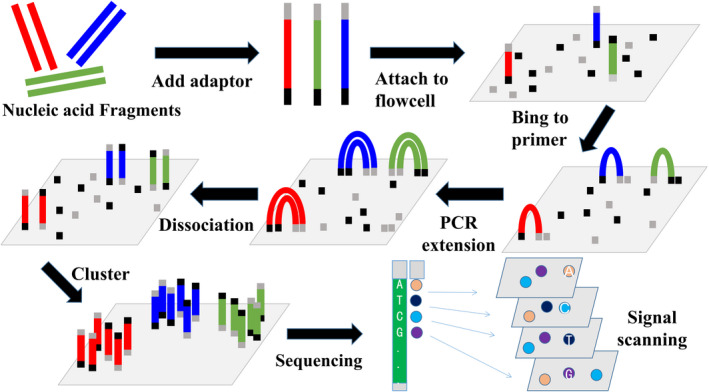
Schematic diagram for NGS
3.3. Real‐time fluorescence quantitative PCR system
Real‐time fluorescence quantitative PCR (RT‐qPCR) system can monitor the process of nucleic acids amplification and predict the protein expression condition. Various biomarkers, such as cfDNA, ctDNA, lncRNA, circRNA, microRNA and so on, have been expressed in BC, but their content is too low to be detected by ordinary instruments. Therefore, RT‐qPCR system is a good choice and can predict risk of BC by analysing the level of mRNA expression. It has some advantages, such as less time consumption, high sensitivity and specificity. In addition, it requires less samples and shorter analysis time compared with other molecular methods. 85 Meanwhile, RT‐qPCR is optimal technology for identifying difference of expression levels of mRNA between malignant tumours and normal tissues. 86 Mansoori et al 87 found that Bach‐1 mRNA was overexpressed, while miR‐142‐3p was downregulated in the BC tumours and then summed up that the expression of miR‐142‐3p has relationship with BC. Moreover, RT‐qPCR system can also guide BC treatment by monitoring specific expression of mRNA. 88 , 89 Matouk et al 90 used the system to analyse the expression condition of H19 gene in BC patients and healthy individual and found the expression difference between them, indicating that the H19 gene is a potential molecular marker for diagnosing BC. However, to obtain satisfactory results, high‐quality mRNA should be extracted. The process for extraction of high‐quality mRNA is difficult because of presence of RNase in the environment. 91 So the full‐automatic nucleic acid extraction device appears and will improve the RNA yield for getting the accurate analysis results. 92
Gene promoter region DNA methylation can also cause cancer, because it can produce similar effects to gene mutations, such as obtaining or losing functions of some specific genes. 93 , 94 Methylation‐based RT‐qPCR system is widely used for analysing genetic methylation. The Table 2 lists part of methylation genes in BC. In order to understand the detection process for methylated genes, we elaborate it by Figure 6. Next, the applications of methylation in BC are expounded. Luo et al 94 identified that these genes, ALDH1L1, HOPX, WNT5A and SOX9, were hypomethylated after neoadjuvant chemotherapy (NAC) treatment by using MethyLight ddPCR and the methylation levels of 4 genes in BC patients after NAC were lower than those before NAC. MethyLight can be used to research expression conditions of methylated silencing genes in cell lines during treatment of BC with drugs. 95 Mastoraki et al 96 considered that methylation of ESR1 gene can become a potential liquid biopsy‐based biomarker to evaluate the risk of BC and ESR1 methylation in CTCs and was associated with response of everolimus/exemestane. In addition, the MethyLight can explore chemo‐resistance to breast tumour by analysing methylation gene. 97 Therefore, MethyLight plays very important part in diagnosing BC. However, MethyLight has some limitations; for example, the nucleic acid needs to be treated (totally methylated or un‐methylated nucleic acid), needs to design complex probes and requires professional operation. In the future, integrating extraction and methylation detection of DNA will be a tendency, which will not only improve the DNA yield, but also the efficiency of methylation.
Table 2.
Partially methylation gene in breast cancer
| Gene | Gene description | References |
|---|---|---|
| BRCA1 | BRCA1 gene is a tumour suppressor, and it can maintain genomic stability. The nuclear phosphoprotein is encoded by BRCA1 gene. Methylation of the BRCA1 gene promoter region can change expression of BRCA1 gene and loss function of tumour suppressor | 148 |
| E2F4 | E2F4 gene is potential basal transcription factor, and it can promote tumour growth. Methylation of E2F4 gene can cause upregulation expression of E2F4 gene and accelerate the development of tumours | 149 |
| PITX2 | PITX2 gene is a prognostic marker for progesterone receptor‐positive patients, and it is closely associated with poor survival and distant metastasis of breast tumours. If PITX 2 gene is methylated, it can be considered low risk of distant metastasis recurrences and need not adjuvant chemotherapy | 150 |
| Hox | The methylation of Hox gene is closely related to the high expression of oestrogen and progesterone receptors, and methylation of HoxD13 gene is closely related to breast tumour size and poor clinical treatment | 151 |
| AKT1 | Methylation of AKT1 gene is observed to be associated with BC, and it affects expression of AKT1 gene. The expression of AKT1 gene has significantly associated with HER‐2 protein status | 152 |
| Soxl7 | Soxl7 gene has significantly associated with breast tumour size and lymphatic metastasis, but un‐methylation of Soxl7 gene is found in normal breast tissue and serum | 153 |
| CDKN2A | The methylation of CDKN2A gene in patients with malignant tumour is found, but un‐methylation of CDKN2A gene is found in patients with benign breast disease. Methylation of CDKN2A gene also is associated with distant metastasis of breast tumours | 154 |
| FHIT | FHIT gene is widely expressed in normal tissues, and methylation of FHIT gene occurs in 31% of patients with primary BC. In particular, after FHIT gene is methylated, its expression quantity is changed in patients with sporadic ductal carcinoma | 155 |
| TIMP‐3 | Methylation of TIMP‐3 gene is found in BC cells, but does not find in normal tissues. The degree of methylation of TIMP‐3 gene is positively correlated with malignancy of BC | 156 |
| MDGI | The MDGI gene is also lowly expression in BC tissues. If promoter region of MDGI gene is methylated in breast cancer patients, methylation of MDGI will be only slightly influenced by surgery, whereas tamoxifen therapy will be a more pronounced effect | 157 |
| RASSF1A | In the patients with sporadic BC, finding 33.3% of RASSF1A gene was deleted or methylated | 158 |
| HSD17B4 | Methylation of HSD17B4 gene is an independent predictive marker for pathological complete response in some studies. If the HSD17B4 is not methylated in patients with BC, these patients will be not benefit from trastuzumab treatment, but will be benefit from lapatinib treatment | 159 |
| ESR1 | Abnormal hyper‐methylation of ESR1 gene is found in BC cells, and it will hope to become a new biomarker of breast tumour | 160 |
| RhoBTB2 | Aberrant methylation of RhoBTB2 gene may affect expression of the RhoBTB2 gene, which influences PR protein status, become the factor that induce BC | 161 |
| NBPF1 | Hypermethylation of promoter region of NBPF1 gene is found in patient's serum or plasma with BC, and thus, the NBPF1 methylated from patient's serum or plasma may become potential tumour biomarker for detection of BC | 160 |
Figure 6.
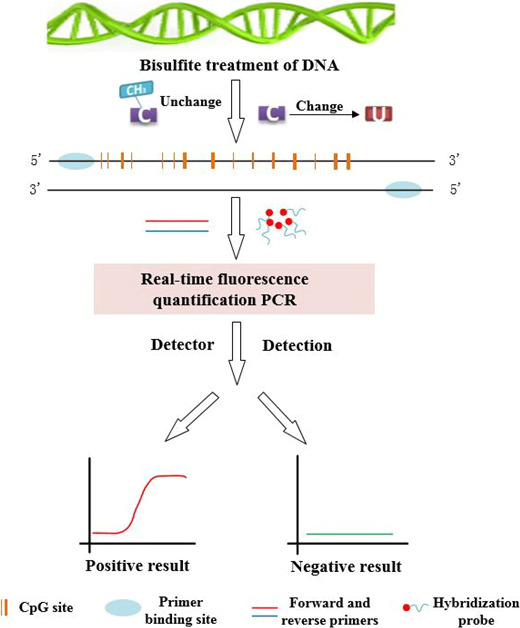
Schematic diagram of MethyLight
3.4. Protein hybridization system
Tumour cells or tissues contain not only the nucleic acids but also many proteins. The “central dogma” of molecular biology shows that proteins are closely associated with nucleic acids. However, if the final protein has no change, the differential expression of nucleic acids may not cause cancer. Therefore, proteins are another important biomarker for diagnosing cancers and analysing the situation of proteins can predict occurrence of cancer. Similarly, proteins, as important biomarker, make great contribution to diagnosis of BC. In Table 3, we introduce the most common and latest oncogene proteins involved in BC. These proteins can be quantitatively evaluated by immunochemistry, RT‐qPCR and Western blot. The difference between them is the different detective object, in which RT‐qRCR is for mRNA and Western blot and immunostaining are for proteins.
Table 3.
Partial oncogene proteins related to breast cancer
| Protein | Protein description | References |
|---|---|---|
| HER‐2 | HER‐2 as therapeutic and prognostic biomarker plays a significant role in Human BC. It is found that adenomas and carcinomas have higher levels of HER‐2 protein than normal mammary glands | 163 |
| CA125 | CA125 as a predictive marker of ovarian/breast carcinoma, it depends on disease nature/stages. CA125 plays an interactive role in the disease processes, and it is closely related to BC | 164 |
| CA19‐9 | Levels of CA19‐9 are correlated with treatment response and survival of BC | 165 |
| MUC1 | MUC1‐MBP is a member of the mucins family, and it is present in normal glandular epithelial cells and tumour cells. MUC1‐MBP consists of a polypeptide core and a side chain sugar chain. MUC1‐MBP widely distributed on the surface of BC cells | 166 |
| ER | ER in the pathophysiology of BC plays an important role, and it as an index can be used to guide pharmacy for BC patients | 167 |
| CypB | BC tissues have higher levels of CypB proteins than para cancerous tissues. Functional study confirms that downregulation levels of CypB may inhibit tumour cell growth, proliferation and migration | 168 |
| CA153 | When the breast becomes cancerous, the activities of protease and salivary enzyme are increased, causing destruction of the cytoskeleton of the gland, causing CA153 saccharide antigen generally separated from the cancer cell membrane and releasing into the blood. It is an important index for screening BC | 169 |
| CEA | CEA is an acidic glycoprotein with a specific determinant of human embryonic antigen. It is a broad‐spectrum tumour marker that can be expressed in a variety of tumours. It is also elevated in the serum of patients with BC, lung cancer and other malignant tumours | 170 |
| PR | Analysis of PR proteins remains controversial in BC. The level of PR + is related to age of BC patients. The deletion of PR proteins might cause BC | 98 |
3.4.1. Immunochemistry
For pathologists, immunostaining (IHC) can accurately locate the site of organization and is an auxiliary method for diagnosing BC. IHC analysis of breast tumours has advantages in the following four aspects: (a) can distinguish between benign and malignant breast tumours; (b) can assess interstitial infiltration; (c) can distinguish between ductal and lobular tumours; and (d) can detect expression of proteins associated with BC treatment and prognosis, to guide endocrine therapy and prognosis. 98 , 99 , 100 , 101 At present, IHC is the best diagnostic method for oestrogen receptor (ER) and progesterone receptor (PR) in BC. 102 The basic principle of IHC (as shown in Figure 7) is antigen‐specific binding of antibodies, and these antibodies are usually labelled with colour reagents (such as fluorescein and metal ion) to detect the antigen, protein, peptides, etc IHC can screen and diagnose BC patients by evaluating the level of marker proteins. 103 HER‐2 gene amplification may cause overexpression of HER‐2, so Suryavanshi et al 104 used IHC to confirm whether HER‐2 gene was amplified abnormally in BC patients by detecting the level of protein and evaluating that the effect of IHC was close to FISH. Surely, IHC also can help researchers to explore the relationship between external factors and BC. For example, Wang et al 105 found the underlying association between alcohol and BC by utilizing IHC. Toomey 106 used IHC to assess the level of PTEN protein and found that 14 of 45 (31.1%) tumours samples had low (absent or weak) PTEN expression and PR‐negative tumour had higher PTEN expression than PR‐positive tumours (37.9% vs 18.8%) in BC. BC formation may cause changes of protein levels, so IHC is able to study the mechanism of breast tumours by analysis of protein levels. However, IHC needs fluorescence labelling which is time‐consuming and difficult to prepare.
Figure 7.
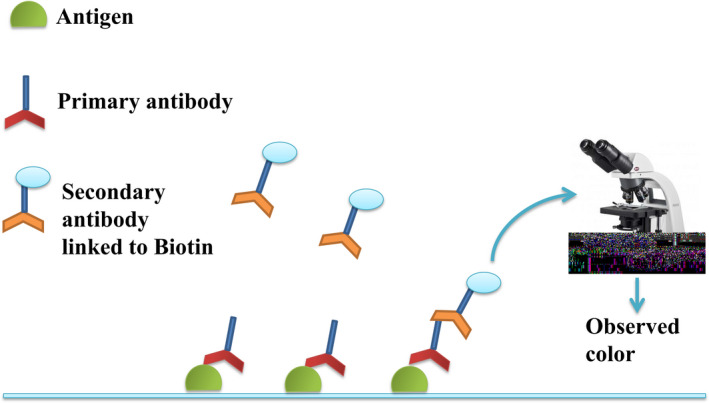
Schematic diagram of immunohistochemical principle
3.4.2. Western blot
Similarly, Western blotting also utilizes the antigen‐antibody binding character that is highly specific. On one hand, the capacity of histological localization of Western blotting is poorer than that of IHC, but the capacity of quantitative protein level is more accurate than that of IHC. On the other hand, for RT‐qPCR, though they all evaluate quantitatively the level of proteins, their detection objects are different, where RT‐qPCR is for nucleic acids, and Western blotting is for proteins. Using exogenous proteins to study proteins’ interaction is a common approach, but the most rigorous approach is to detect interactions between endogenous proteins. 107 Western blotting can satisfy this need and be used in diagnosing BC. For example, Zhou et al 108 used Western blot to investigate the expression of UCA1 and microRNA (miRNA) in BC cells in response to IMP1 expression. Liu used this technique to analyse the relationship between miRNA and IDH1gene. These results showed that Western blot can not only explore whether the proteins are expressed, but also verify whether the protein expression is abnormal. 109 De Francesco et al 110 found by Western blotting analysis that HIF‐1α and GPER expressions increased with time in CAFs cells, but expression decreased over time in SKBR3 cells. Moreover, Ansari 111 utilized Western blot to analyse the level of proteins in BC and found that 191 from 1110 (17%) in the discovery set and 268 from 1554 (17%) in validation sets of cases had positive SLC7A5 expression (>15 H‐score), while 1019 in 1923 (53%) from metastatic BC cases had high mRNA expression (log2 intensity > 8). Surely, Western blot has deficiencies, such as use of expensive agents, easily false positive and needs professional operation. In the future, the decrease in price of Western blot agents will be a tendency and simply the process of Western blot operation.
3.5. Flow cytometer
Flow cytometer (FCM) can reflect multiple physical characteristics of a single cell when the cell flows in suspension, 112 and it has become an indispensable technology in diagnosis of BC. FCM is a high‐tech developed in the 1960s, and it is the combination of many disciplines and technologies, such as cytochemistry, immunology, materials science, molecular biology, spectroscopy, optical system, fluidic system, laser technology and computer technology. 113 , 114 , 115 , 116 Surely, FCM also has sorting function for tumour cells and can rapidly detect cells or biological particles through the one‐by‐one flow state, multi‐parameters or rapidly qualitative and quantitative analysis. 117 , 118 , 119 , 120 Figure 8 shows how marked sites on cell surface are detected by FCM. 121 In the FCM, the cells or biological particles need to firstly be treated and labelled, so that they can be detected by laser.
Figure 8.
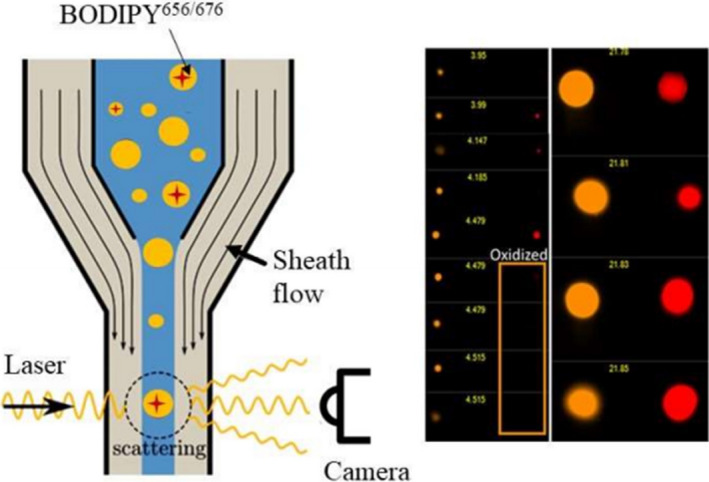
Isolation of individual droplets by flow cytomtry. 121 (Reproduced with permission from Copyright 2020, American Chemical Society)
In recent years, FCM, by combining with other detecting techniques, can achieve quantitative detection of low‐abundance genes. 122 FCM also is excellent method in diagnosing BC and guiding medication. Kim et al 123 used FCM to analyse tumour cell surface markers and found that hypoxic tumour microenvironment may associate with promoting malignant progression and therapy resistance. Chamberlin 124 utilized the FCM to mark different cells and found that the ratio of luminal and basal cells presents a significant increase in obese mammary glands with weight gain and expounded how obesity is linked to BC. Moreover, Tu et al 125 used FCM to trace these tumour cells and found that primary breast tumour growth cannot be affected by an oral administration of an FUT inhibitor (2‐fluorinated‐peracetyl‐fucose), but these medicines greatly reduce the lung metastatic. Through FCM analysis, Xu et al 126 identified the origin of cancer stem cell (CSC)–like cells that would be critical to cancer treatment and found these breast non‐stem cancer cells are transferred to breast CSC‐like cells in apoptosis process. FCM not only can detect the biomarkers of BC cell, but also can also detect BC cells based on morphology. Patel et al 127 utilized digital holographic cytometry (DHC) and found that a special marker, sialic acid‐molecularly imprinted polymers (SA‐MIPs), has impact on different BC cells’ morphology and motility. Similarly, Farghadani et al 128 investigated the mechanism of inhibitory and cytotoxic activity of anticancer agent on BC cells, cell cycle progression using flow cytometry analysis, and found some valuable medicine.
There are some advantages of FCM, including nonspecific binding in antigen antibody may cause the signalling pathway of FCM to be affected. Dyestuff pollution in FCM experiment is also a big trouble, and expensive instruments are required. In the future, it is most important that diagnostic scheme for FCM should be standardized and agents of high efficiency and low cost should be developed.
3.6. Puncture biopsy system
Needle biopsy is a main method to obtain tumours tissue or cells sample for histopathological diagnosis. These puncture biopsies system include fine‐needle aspiration cytology (FNAC), core needle biopsy (CNB) and vacuum‐assisted breast biopsy (VABB). 129 , 130 At present, VABB has excellent effects in the auxiliary diagnosis of BC. Its advantages are as follows: single puncture can accurately and simply collect many samples, accurate positioning, convenient operation, smaller trauma area and so on. In general, obtaining samples (cells or tissues) by puncture needs staining (usually using haematoxylin‐eosin) to easily observe samples under optical microscope, which can rapidly analyse and identify pathological tissue and cell morphology to help doctors make pathological diagnosis. Surely, these samples also are detected by other molecular biology methods. Zhang et al 131 used high‐frequency ultrasound‐guided breast mass biopsy to diagnose BC in two hundred patients. Their results showed that each patient had a successful puncture rate of 100% under the guidance of ultrasound. Moreover, no complications occurred, and 95% (190/200) of the patients were clearly diagnosed, and 5% (10/200) were orientally diagnosed. The biopsy examination results were completely consistent with surgical pathological results in 170 patients, accounting for 85%. Thus, this method can provide strong evidence for diagnosis and identification of benign and malignant breast tumours, and for choosing the correct operation scheme. 131 Hu et al 132 performed US‐guided fine‐needle aspiration biopsy (FNAB) for early‐stage BC and found that its sensitivity and specificity were higher than for US alone, 11.9% and 21.7%, respectively. With development of imaging, the accuracy of puncture biopsy is higher under imaging guidance. Guo et al 133 offered a new integrated precise re‐biopsy algorithm for pathological confirmation and surveillance of recurrent BC. The technology is more sensitive and accurate than conventional imaging technologies in diagnosis of early‐stage BC.
However, there are some disadvantages of needle breast biopsy; for example, it may cause tumours transfer and researchers thought that high‐grade, non‐coaxial biopsies, triple‐negative BCs and multiple insertions may be risk factors for neoplastic seeding. 134 , 135 , 136 , 137 In the future, with development of biopsy needle, the risk of neoplastic seeding will be reduced and the accuracy of diagnosis will be improved. Surely, the latest imaging guidance will also promote the development of puncture biopsy.
4. CONCLUSIONS AND FUTURE PERSPECTIVES
In this review, we mainly introduced the common methods for diagnosis of BC. As the exploration of imaging technology goes deeper, researchers realize that the single imaging technology has lower accuracy and cannot meet the need for BC diagnosis, and the combination of various imaging modalities will be one of the major developing directions. 138 , 139 , 140 Moreover, with development of biosensors, a lot of BC biomarkers have been found. The combination of imaging sensors and biosensors can get unexpected results. 141 , 142 Meanwhile, more and more aptamers are developed, which increases connection between imaging and molecular biology. 143 , 144 These aptamer‐functionalized nano‐composites not only can become indicators for imaging, but also can identify cancer cells, and/or even classify BC cells subsets. In another aspect, screening for new tumour biomarkers is still an important task which can help doctors diagnose BC faster and more accurately. Currently, proteins, nucleic acids and lipids are the main tumour markers in breast cancer, while the question remains whether single markers could not acquire definite diagnosis results. 145 , 146 Hence, multiple tumour markers or screening for a super new marker can greatly improve the positive diagnostic rate for BC and reduce the negative diagnosis rate.
Over the next few years, imaging instruments still will be the routine method for screening BC, because they suit to be widely applied. However, new markers for BC will advance these technologies to higher throughput, faster, higher sensitivity and specificity. In the future, with development and use of these techniques, they not only can diagnose BC from various aspects, but also can evaluate effect of treating BC. Of course, different types of BC also will be evaluated by corresponding diagnostic methods, to get the most accurate results.
CONFLICT OF INTEREST
The authors have no conflict of interest.
AUTHOR CONTRIBUTIONS
The article was written by Ziyu He and Zhu Chen. Miduo Tan, Sauli Elingarami, Yuan Liu, Nongyue He, Taotao Li and Wen Li contributed to conceptualization and revision of the article. The article was funded by Zhu Chen, Yan Deng, Song Li and Juan Fu. All authors reviewed the final manuscript.
ACKNOWLEDGEMENTS
This work was funded by grants from the National Science Foundation of China (Grant No. 61901168, 61971187, 61571187, 61527806 and 61871180), Hunan Provincial Natural Science Foundation of China (Grant No. 2019JJ50122), Hunan Key Research Project (Grant No. 2017SK2174), China Postdoctoral Science Foundation (Grant No. 2018M630498) and Education Department Outstanding Young Project of Hunan (Grant No. 18B299).
He Z, Chen Z, Tan M, et al. A review on methods for diagnosis of breast cancer cells and tissues. Cell Prolif. 2020;53:e12822 10.1111/cpr.12822
Ziyu He and Zhu Chen contributed equally to this work.
Contributor Information
Zhu Chen, Email: chenzhu220@163.com.
Song Li, Email: sosong1980@gmail.com.
Juan Fu, Email: fjuangy@126.com.
DATA AVAILABILITY STATEMENT
Some or all data, models or code generated or used during the study are available from the corresponding author by request.
REFERENCES
- 1. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2018;69(6):438‐451. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐132. [DOI] [PubMed] [Google Scholar]
- 3. Feng R‐M, Zong Y‐N, Cao S‐M, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019;29(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tao ZQ, Shi A, Lu C, et al. Breast cancer: epidemiology and etiology. Cell Biochem Biophys. 2015;72(2):333‐338. [DOI] [PubMed] [Google Scholar]
- 5. DeSantis CE, Ma J, Jemal A. Trends in stage at diagnosis for young breast cancer patients in the United States. Breast Cancer Res Treat. 2019;173(3):743‐747. [DOI] [PubMed] [Google Scholar]
- 6. McPherson K, Steel CM, Dixon JM. ABC of breast diseases: breast cancer–epidemiology, risk factors, and genetics. BMJ. 2000;321(7261):624‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zografos GC, Panou M, Panou N. Common risk factors of breast and ovarian cancer: recent view. Int J Gynecol Cancer. 2004;14(5):721‐740. [DOI] [PubMed] [Google Scholar]
- 8. Robson M, Im S‐A, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523‐533. [DOI] [PubMed] [Google Scholar]
- 9. Tuttle TM, Habermann EB, Grund EH, et al. Increasing use of contralateral prophylactic mastectomy for breast cancer patients: a trend toward more aggressive surgical treatment. J Clin Oncol. 2007;25(33):5203‐5209. [DOI] [PubMed] [Google Scholar]
- 10. Bonneau C, Gurard‐Levin ZA, Andre F, et al. Predictive and prognostic value of the TauProtein in breast cancer. Anticancer Res. 2015;35(10):5179‐5184. [PubMed] [Google Scholar]
- 11. Jafari SH, Saadatpour Z, Salmaninejad A, et al. Breast cancer diagnosis: imaging techniques and biochemical markers. J Cell Physiol. 2018;233(7):5200‐5213. [DOI] [PubMed] [Google Scholar]
- 12. Weaver O, Leung JWT. Biomarkers and imaging of breast cancer. Am J Roentgenol. 2018;210(2):271‐278. [DOI] [PubMed] [Google Scholar]
- 13. Marchiò C, Reis‐Filho JS. Molecular diagnosis in breast cancer. Diagn Pathol. 2008;14(5):202‐213. [Google Scholar]
- 14. Pareja F, Marchiò C, Reis‐Filho JS. Molecular diagnosis in breast cancer. Diagn Histopathol. 2018;24(2):71‐82. [Google Scholar]
- 15. Berse B, Lynch JA. Molecular diagnostic testing in breast cancer. Semin Oncol Nurs. 2015;31(2):108‐121. [DOI] [PubMed] [Google Scholar]
- 16. Ayer T. Inverse optimization for assessing emerging technologies in breast cancer screening. Ann Oper Res. 2015;230(1):57‐85. [Google Scholar]
- 17. Weigel S, Decker T, Korsching E, et al. Calcifications in digital mammographic screening: improvement of early detection of invasive breast cancers? Radiology. 2010;255(3):738‐745. [DOI] [PubMed] [Google Scholar]
- 18. Tse GM, Tan P‐H, Pang ALM, et al. Calcification in breast lesions: pathologists’ perspective. J Clin Pathol. 2007;61(2):145‐151. [DOI] [PubMed] [Google Scholar]
- 19. Covington MF, Pizzitola VJ, Lorans R, et al. The future of contrast‐enhanced mammography. Am J Roentgenol. 2018;210(2):292‐300. [DOI] [PubMed] [Google Scholar]
- 20. Chong A, Weinstein SP, McDonald ES, Conant EF. Digital breast tomosynthesis: concepts and clinical practice. Radiology. 2019;292(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fallenberg EM, Schmitzberger FF, Amer H, et al. Contrast‐enhanced spectral mammography vs. mammography and MRI – clinical performance in a multi‐reader evaluation. Eur Radiol. 2017;27(7):2752‐2764. [DOI] [PubMed] [Google Scholar]
- 22. Sorin V, Yagil Y, Yosepovich A, et al. Contrast‐enhanced spectral mammography in women with intermediate breast cancer risk and dense breasts. Am J Roentgenol. 2018;211(5):W267‐W274. [DOI] [PubMed] [Google Scholar]
- 23. Skaane P, Sebuødegård S, Bandos AI, et al. Performance of breast cancer screening using digital breast tomosynthesis: results from the prospective population‐based Oslo Tomosynthesis Screening Trial. Breast Cancer Res Treat. 2018;169(3):489‐496. [DOI] [PubMed] [Google Scholar]
- 24. Katzen J, Dodelzon K. A review of computer aided detection in mammography. Clin Imaging. 2018;52:305‐309. [DOI] [PubMed] [Google Scholar]
- 25. Danala G, Patel B, Aghaei F, et al. Classification of breast masses using a computer‐aided diagnosis scheme of contrast enhanced digital mammograms. Ann Biomed Eng. 2018;46(9):1419‐1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benedikt RA, Boatsman JE, Swann CA, et al. Concurrent computer‐aided detection improves reading time of digital breast tomosynthesis and maintains interpretation performance in a multireader multicase study. Am J Roentgenol. 2018;210(3):685‐694. [DOI] [PubMed] [Google Scholar]
- 27. Monticciolo DL, Newell MS, Hendrick RE, et al. Breast cancer screening for average‐risk women: recommendations from the ACR commission on breast imaging. J Am Coll Radiol. 2017;14(9):1137‐1143. [DOI] [PubMed] [Google Scholar]
- 28. Ha R, Kim H, Mango V, et al. Ultrasonographic features and clinical implications of benign palpable breast lesions in young women. Ultrasonography. 2014;34(1):66‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H, Shi Q, Gu J, et al. Combined value of virtual touch tissue quantification and conventional sonographic features for differentiating benign and malignant thyroid nodules smaller than 10 mm. J Ultrasound Med. 2014;33(2):257‐264. [DOI] [PubMed] [Google Scholar]
- 30. Helal MH, Mansour SM, Salaleldin LA, et al. The impact of contrast‐enhanced spectral mammogram (CESM) and three‐dimensional breast ultrasound (3DUS) on the characterization of the disease extend in cancer patients. Br J Radiol. 2018;91(1087):20170977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang GCH, Fried KO. Most thyroid cancers detected by sonography lack intranodular vascularity on color doppler imaging: review of the literature and sonographic‐pathologic correlations for 698 thyroid neoplasms. J Ultrasound Med. 2017;36(1):89‐94. [DOI] [PubMed] [Google Scholar]
- 32. Krouskop TA, Wheeler TM, Kallel F, et al. Elastic moduli of breast and prostate tissues under compression. Ultrason Imaging. 1998;20(4):260‐274. [DOI] [PubMed] [Google Scholar]
- 33. Shi X‐Q, Li J, Qian L, et al. Correlation between elastic parameters and collagen fibre features in breast lesions. Clin Radiol. 2018;73(6):595.e1‐595.e7. [DOI] [PubMed] [Google Scholar]
- 34. Signore G, Nifosì R, Albertazzi L, Storti B, Bizzarri R. Polarity‐sensitive coumarins tailored to live cell imaging. J Am Chem Soc. 2010;132(4):1276‐1288. [DOI] [PubMed] [Google Scholar]
- 35. Wang Y, Nasief HG, Kohn S, et al. Three‐dimensional ultrasound elasticity imaging on an automated breast volume scanning system. Ultrason Imaging. 2017;39(6):369‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park AY, Seo BK, Cha SH, et al. An innovative ultrasound technique for evaluation of tumor vascularity in breast cancers: superb micro‐vascular imaging. J Breast Cancer. 2016;19(2):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Machado P, Eisenbrey JR, Stanczak M, et al. Characterization of breast microcalcifications using a new ultrasound image‐processing technique. J Ultrasound Med. 2019;38(7):1733‐1738. [DOI] [PubMed] [Google Scholar]
- 38. Machado P, Eisenbrey JR, Stanczak M, et al. Ultrasound detection of microcalcifications in surgical breast specimens. Ultrasound Med Biol. 2018;44(6):1286‐1290. [DOI] [PubMed] [Google Scholar]
- 39. Riedl CC, Luft N, Bernhart C, et al. Triple‐modality screening trial for familial breast cancer underlines the importance of magnetic resonance imaging and questions the role of mammography and ultrasound regardless of patient mutation status, age, and breast density. J Clin Oncol. 2015;33(10):1128‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orguc S, Basara I, Coskun T. Diffusion‐weighted MR imaging of the breast: comparison of apparent diffusion coefficient values of normal breast tissue with benign and malignant breast lesions. Singapore Med J. 2012;53(11):737‐743. [PubMed] [Google Scholar]
- 41. Bougias H, Ghiatas A, Priovolos D, Veliou K, Christou A. Whole‐lesion apparent diffusion coefficient (ADC) metrics as a marker of breast tumour characterization‐comparison between ADC value and ADC entropy. Br J Radiol. 2016;89(1068):20160304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rabasco P, Caivano R, Simeon V, et al. Can diffusion‐weighted imaging and related apparent diffusion coefficient be a prognostic value in women with breast cancer? Cancer Invest. 2017;35(2):92‐99. [DOI] [PubMed] [Google Scholar]
- 43. Wojcinski S, Farrokh H, Wiskirchen G, et al. The automated breast volume scanner (ABVS): initial experiences in lesion detection compared with conventional handheld B‐mode ultrasound: a pilot study of 50 cases. Int J Womens Health. 2011;3(1);337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moy L, Noz ME, Maguire GQ Jr, et al. Role of fusion of prone FDG‐PET and magnetic resonance imaging of the breasts in the evaluation of breast cancer. Breast J. 2010;16(4):369‐376. [DOI] [PubMed] [Google Scholar]
- 45. Guindalini RSC, Zheng Y, Abe H, et al. Intensive surveillance with biannual dynamic contrast‐enhanced magnetic resonance imaging downstages breast cancer in BRCA1 mutation carriers. Clin Cancer Res. 2019;25(6):1786‐1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clauser P, Marcon M, Dietzel M, et al. A new method to reduce false positive results in breast MRI by evaluation of multiple spectral regions in proton MR‐spectroscopy. Eur J Radiol. 2017;92:51‐57. [DOI] [PubMed] [Google Scholar]
- 47. He D, Mustafi D, Fan X, et al. Magnetic resonance spectroscopy detects differential lipid composition in mammary glands on low fat, high animal fat versus high fructose diets. PLoS ONE. 2018;13(1):e0190929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bohte AE, Nelissen JL, Runge JH, et al. Breast magnetic resonance elastography: a review of clinical work and future perspectives. NMR Biomed. 2018;31(10):e3932. [DOI] [PubMed] [Google Scholar]
- 49. Plecha DM, Faulhaber P. PET/MRI of the breast. Eur J Radiol. 2017;94:A26‐A34. [DOI] [PubMed] [Google Scholar]
- 50. Albano D, Bosio G, Orlando E, et al. Role of fluorine‐18‐fluorodeoxyglucose positron emission tomography/computed tomography in evaluating breast mucosa‐associated lymphoid tissue lymphoma: a case series. Hematol Oncol. 2017;35(4):884‐889. [DOI] [PubMed] [Google Scholar]
- 51. Jin LD, Zhao WH, Zhang J, et al. Evaluation of the diagnostic value of circulating tumor cells with CytoSorter®CTC capture system in patients with breast cancer. Cancer Med. 2020;9:1638‐1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ma F, Guan Y, Yi Z, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer. 2020;146(5):1359‐1368. [DOI] [PubMed] [Google Scholar]
- 53. Zhou YU, Xu H, Wang H, et al. Detection of breast cancer‐derived exosomes using the horseradish peroxidase‐mimicking DNAzyme as an aptasensor. Analyst. 2020;145(1):107‐114. [DOI] [PubMed] [Google Scholar]
- 54. Ni Q, Stevic I, Pan C, et al. Different signatures of miR‐16, miR‐30b and miR‐93 in exosomes from breast cancer and DCIS patients. Sci Rep. 2018;8(1):12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shao M, Ma H, Wan X, et al. Survival analysis for long noncoding RNAs identifies TP53TG1 as an antioncogenic target for the breast cancer. J Cell Physiol. 2020. 10.1002/jcp.29517 [DOI] [PubMed] [Google Scholar]
- 56. Liang D, Liu H, Yang Q, et al. Long noncoding RNA RHPN1‐AS1, induced by KDM5B, is involved in breast cancer via sponging miR‐6884‐5p. J Cell Biochem. 2020. 10.1002/jcb.29645 [DOI] [PubMed] [Google Scholar]
- 57. Sun H, Wang G, Peng Y, et al. H19 lncRNA mediates 17β‐estradiol‐induced cell proliferation in MCF‐7 breast cancer cells. Oncol Rep. 2015;33(6):3045‐3052. [DOI] [PubMed] [Google Scholar]
- 58. Li W, Jia M, Wang J, et al. Association of MMP9‐1562C/T and MMP13‐77A/G polymorphisms with non‐small cell lung cancer in southern Chinese population. Biomolecules. 2019;9(3):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lü L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8(27):44096‐44107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR‐492. Cancer Manag Res. 2019;11:1033‐1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Giovannoni SJ, DeLong EF, Olsen GJ, Pace NR. Phylogenetic group‐specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170(2):720‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Furrer D, Jacob S, Caron C, et al. Concordance of HER2 immunohistochemistry and fluorescence in situ hybridization using tissue microarray in breast cancer. Anticancer Res. 2017;37(6):3323‐3329. [DOI] [PubMed] [Google Scholar]
- 63. Yang W, Klos KS, Zhou X, et al. ErbB2 overexpression in human breast carcinoma is correlated with p21Cip1 up‐regulation and tyrosine‐15 hyperphosphorylation of p34Cdc2. Cancer. 2003;98(6):1123‐1130. [DOI] [PubMed] [Google Scholar]
- 64. Geiersbach KB, Bridge JA, Dolan M, et al. Comparative performance of breast cancer human epidermal growth factor receptor 2 fluorescence in situ hybridization and brightfield in situ hybridization on college of American pathologists proficiency tests. Arch Pathol Lab Med. 2018;142(10):1254‐1259. [DOI] [PubMed] [Google Scholar]
- 65. Bhattacharjee A, Bhattacharyya T, Thomas A. Human epidermal growth factor receptor 2 borderline mortality in breast cancer patients: evidence from surveillance, epidemiology, and end results program population‐based study. Clin Epidemiol Glob Health. 2018;6(2):88‐93. [Google Scholar]
- 66. Kim YS, Song MY, Jurng J, Kim BC. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach. Anal Biochem. 2013;436(1):22‐28. [DOI] [PubMed] [Google Scholar]
- 67. Yuan B, Jiang X, Chen Y, et al. Metastatic cancer cell and tissue‐specific fluorescence imaging using a new DNA aptamer developed by Cell‐SELEX. Talanta. 2017;170(February):56‐62. [DOI] [PubMed] [Google Scholar]
- 68. Kim MY, Jeong S. In vitro selection of RNA Aptamer and specific targeting of ErbB2 in breast cancer cells. Nucleic Acid Ther. 2011;21(3):173‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cai S, Li G, Zhang XI, et al. A signal‐on fluorescent aptasensor based on single‐stranded DNA‐sensitized luminescence of terbium (III) for label‐free detection of breast cancer cells. Talanta. 2015;138:225‐230. [DOI] [PubMed] [Google Scholar]
- 70. Bliss SA, Paul S, Pobiarzyn PW, et al. Evaluation of a developmental hierarchy for breast cancer cells to assess risk‐based patient selection for targeted treatment. Scientific Rep. 2018;8(1):367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Burns MA. An integrated nanoliter DNA analysis device. Science. 1998;282(5388):484‐487. [DOI] [PubMed] [Google Scholar]
- 72. Hao S, Ha L, Cheng G, et al. A spontaneous 3D bone‐on‐a‐chip for bone metastasis study of breast cancer cells. Small. 2018;14(12):1702787. [DOI] [PubMed] [Google Scholar]
- 73. Li C, Ding X, Liu Z, Zhu J. Rapid identification of Candida spp. frequently involved in invasive mycoses by using flow‐through hybridization and Gene Chip (FHGC) technology. J Microbiol Methods. 2017;132:160‐165. [DOI] [PubMed] [Google Scholar]
- 74. Kim M‐H, Kim E‐H, Jung HS, et al. EX4 stabilizes and activates Nrf2 via PKCδ, contributing to the prevention of oxidative stress‐induced pancreatic beta cell damage. Toxicol Appl Pharmacol. 2017;315:60‐69. [DOI] [PubMed] [Google Scholar]
- 75. Jiang Z, Slater CM, Zhou Y, et al. LincIN, a novel NF90‐binding long non‐coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. 2017;19(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Metzker ML. Sequencing technologies‐the next generation. Nat Rev Genet. 2010;11(1):31‐46. [DOI] [PubMed] [Google Scholar]
- 77. Dong LI, Wu N, Wang S, et al. Detection of novel germline mutations in six breast cancer predisposition genes by targeted next‐generation sequencing. Hum Mutat. 2018;39(10):1442‐1455. [DOI] [PubMed] [Google Scholar]
- 78. Liang XU, Vacher S, Boulai A, et al. Targeted next‐generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res. 2018;20(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim ST, Lee WS, Lanman RB, et al. Prospective blinded study of somatic mutation detection in cell‐free DNA utilizing a targeted 54‐gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget. 2015;6(37):40360‐40369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu H, Wang Q, Zhong H, et al. Differentially expressed microRNAs in xosomes of patients with breast cancer revealed by next‐generation sequencing. Oncol Rep. 2020;43:240‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Page K, Guttery DS, Fernandez‐Garcia D, et al. Next Generation sequencing of circulating cell‐free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin Chem. 2017;63(2):532‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Scarpitta R, Zanna I, Aretini P, et al. Germline investigation in male breast cancer of DNA repair genes by next‐generation sequencing. Breast Cancer Res Treat. 2019;178(3):557‐564. [DOI] [PubMed] [Google Scholar]
- 83. Ou‐Yang F, Pan MR, Chang SJ, et al. Identification of CHD4‐beta 1 integrin axis as a prognostic marker in triple‐negative breast cancer using next‐generation sequencing and bioinformatics. Life Sci. 2019;238:116963. [DOI] [PubMed] [Google Scholar]
- 84. Bae JW, Choi KH, Kim HG, et al. The detection of circulating breast cancer cells in peripheral blood by reverse transcriptase‐polymerase chain reaction. J Korean Med Sci. 2000;15(2):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yin W‐B, Yan M‐G, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363‐368. [DOI] [PubMed] [Google Scholar]
- 86. Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17(1):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mansoori B, Mohammadi A, Gjerstorff MF, et al. miR‐142‐3p is a tumor suppressor that inhibits estrogen receptor expression in ER‐positive breast cancer. J Cell Physiol. 2019;234(9):16043‐16053. [DOI] [PubMed] [Google Scholar]
- 88. Ryu TY, Kim K, Kim S‐K, et al. SETDB1 regulates SMAD7 expression for breast cancer metastasis. BMB Rep. 2019;52(2):139‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Geyer CE, Tang G, Mamounas EP, et al. 21‐Gene assay as predictor of chemotherapy benefit in HER2‐negative breast cancer. NPJ Breast Cancer. 2018;4(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Matouk IJ, Raveh E, Abu‐lail R, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta Mol Cell Res. 2014;1843(7):1414‐1426. [DOI] [PubMed] [Google Scholar]
- 91. Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004;68(6):1187‐1197. [DOI] [PubMed] [Google Scholar]
- 92. He Z, Tang C, Chen X, et al. Based on magnetic beads to develop the kit for extraction of high‐quality cell‐free DNA from blood of breast cancer patients. Mater Express. 2019;9(8):956‐961. [Google Scholar]
- 93. Ng J, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015;16(2):2472‐2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Luo Y, Huang J, Tang YI, et al. Regional methylome profiling reveals dynamic epigenetic heterogeneity and convergent hypomethylation of stem cell quiescence‐associated genes in breast cancer following neoadjuvant chemotherapy. Cell Biosci. 2019;9(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McCullough LE, Chen J, Cho YH, et al. DNA methylation modifies the association between obesity and survival after breast cancer diagnosis. Breast Cancer Res Treat. 2016;156(1):183‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mastoraki S, Strati A, Tzanikou E, et al. ESR1 methylation: a liquid biopsy‐based epigenetic assay for the follow‐up of patients with metastatic breast cancer receiving endocrine treatment. Clin Cancer Res. 2018;24(6):1500‐1510. [DOI] [PubMed] [Google Scholar]
- 97. Tuo Y‐L, Ye Y‐F. MGP is downregulated due to promoter methylation in chemoresistant ER+ breast cancer and high MGP expression predicts better survival outcomes. Eur Rev Med Pharmacol Sci. 2017;21(17):3871‐3878. [PubMed] [Google Scholar]
- 98. Yip C‐H, Rhodes A. Estrogen and progesterone receptors in breast cancer. Futur Oncol. 2014;10(14):2293‐2301. [DOI] [PubMed] [Google Scholar]
- 99. Canas‐Marques R, Schnitt SJ. E‐cadherin immunohistochemistry in breast pathology: uses and pitfalls. Histopathology. 2016;68(1):57‐69. [DOI] [PubMed] [Google Scholar]
- 100. Maeda I, Kubota M, Ohta J, et al. Effectiveness of computer‐aided diagnosis (CADx) of breast pathology using immunohistochemistry results of core needle biopsy samples for synaptophysin, oestrogen receptor and CK14/p63 for classification of epithelial proliferative lesions of the breast. J Clin Pathol. 2017;70(12):1057‐1062. [DOI] [PubMed] [Google Scholar]
- 101. Hanley KZ, Birdsong GG, Cohen C, et al. Immunohistochemical detection of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 expression in breast carcinomas. Cancer Cytopathol. 2009;117(4):279‐288. [DOI] [PubMed] [Google Scholar]
- 102. Shen Q, Rao Q, Xia Q‐Y, et al. Perivascular epithelioid cell tumor (PEComa) with TFE3 gene rearrangement: clinicopathological, immunohistochemical, and molecular features. Virchows Arch. 2014;465(5):607‐613. [DOI] [PubMed] [Google Scholar]
- 103. Bogdanovska‐Todorovska M, Kostadinova‐Kunovska S, Jovanovik R, et al. Correlation of immunohistochemistry and fluorescence in situ hybridization for HER‐2 assessment in breast cancer patients: single centre experience. Open Access Maced J Med Sci. 2018;6(4):593‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Suryavanshi M, Mehta A, Jaipuria J, et al. Clinical utility of RT‐PCR in assessing HER 2 gene expression versus traditional IHC and FISH in breast cancer patients. Breast Cancer. 2018;25(4):416‐430. [DOI] [PubMed] [Google Scholar]
- 105. Wang J, Heng YJ, Eliassen AH, et al. Alcohol consumption and breast tumor gene expression. Breast Cancer Res. 2017;19(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Toomey S, Eustace AJ, Fay J, et al. Impact of somatic PI3K pathway and ERBB family mutations on pathological complete response (pCR) in HER2‐positive breast cancer patients who received neoadjuvant HER2‐targeted therapies. Breast Cancer Res. 2017;19(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Moore SC, Matthews CE, Ou Shu X, et al. Estrogen metabolites, and breast cancer risk in postmenopausal Chinese women. J Natl Cancer Inst. 2016;108(10):djw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhou Y, Meng X, Chen S, et al. IMP1 regulates UCA1‐mediated cell invasion through facilitating UCA1 decay and decreasing the sponge effect of UCA1 for miR‐122‐5p. Breast Cancer Res. 2018;20(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Liu W‐S, Chan S‐H, Chang H‐T, et al. Isocitrate dehydrogenase 1–snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res. 2018;20(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. De Francesco EM, Sims AH, Maggiolini M, et al. GPER mediates the angiocrine actions induced by IGF1 through the HIF‐1α/VEGF pathway in the breast tumor microenvironment. Breast Cancer Rese. 2017;19(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. El Ansari R, Craze ML, Miligy I, et al. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 2018;20(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Saeys Y, Van Gassen S, Lambrecht BN. Computational flow cytometry: helping to make sense of high‐dimensional immunology data. Nat Rev Immunol. 2016;16(7):449‐462. [DOI] [PubMed] [Google Scholar]
- 113. Li M, Cui P, Li K, et al. Dual‐site fluorescent probe for highly selective and sensitive detection of sulfite and biothiols. Chinese Chem Lett. 2018;29(6):992‐994. [Google Scholar]
- 114. Tang X, Chen Z, Li Y, et al. Compressive sensing‐based electrostatic sensor array signal processing and exhausted abnormal debris detecting. Mech Syst Signal Process. 2018;105:404‐426. [Google Scholar]
- 115. Darjee SM, Bhatt KD, Panchal US, et al. Scrupulous recongnisation of biologically important acids by Fluorescent “turn off‐on” mechanism of thaicalix reduced silver nanoparticles. Sens Bio‐Sensing Res. 2016;28(2):312‐318. [Google Scholar]
- 116. Guo Y, Luo F, Zhang X, et al. TPPU enhanced exercise‐induced EET concentrations to exert cardioprotection in mice after myocardial infarction. J Cell Mol Med. 2018;22(3):1489‐1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Huang G, Meng Q‐Q, Zhou W, et al. Design and synthesis of biotinylated dimethylation of alkannin oxime derivatives. Chinese Chem Lett. 2017;28(2):453‐457. [Google Scholar]
- 118. Liu B, Dou C, Guerrero JM. Event‐triggered hybrid control based on multi‐agent system for microgrids. IET Gener Transm Distrib. 2014;8(12):1987‐1997. [Google Scholar]
- 119. Long M, Su H, Liu B. Group controllability of two‐time‐scale multi‐agent networks. J Franklin Inst. 2018;355(13):6045‐6061. [Google Scholar]
- 120. Finak G, Jiang W, Krouse K, et al. High‐throughput flow cytometry data normalization for clinical trials. Cytom Part A. 2014;85(3):277‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Li P, McClements DJ, Decker EA. Application of flow cytometry as novel technology in studying the effect of droplet size on lipid oxidation in oil‐in‐water emulsions. J Agric Food Chem. 2019;68(2):567‐573. [DOI] [PubMed] [Google Scholar]
- 122. García‐Foncillas J, Alba E, Aranda E, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol. 2017;28(12):2943‐2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kim H, Lin Q, Glazer PM, et al. The hypoxic tumor microenvironment in vivo selects the cancer stem cell fate of breast cancer cells. Breast Cancer Res. 2018;20(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chamberlin T, D’Amato JV, Arendt LM. Obesity reversibly depletes the basal cell population and enhances mammary epithelial cell estrogen receptor alpha expression and progenitor activity. Breast Cancer Res. 2017;19(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tu C‐F, Wu M‐Y, Lin Y‐C, et al. FUT8 promotes breast cancer cell invasiveness by remodeling TGF‐β receptor core fucosylation. Breast Cancer Res. 2017;19(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xu Y, So C, Lam H‐M, et al. Flow cytometric detection of newly‐formed breast cancer stem cell‐like cells after apoptosis reversal. J Vis Exp. 2019;143:1‐11. [DOI] [PubMed] [Google Scholar]
- 127. Patel M, Feith M, Janicke B, et al. Evaluation of the impact of imprinted polymer particles on morphology and motility of breast cancer cells by using digital holographic cytometry. Appl. Sci. 2020;10(2):750. [Google Scholar]
- 128. Farghadani R, Rajarajeswaran J, Hashim NBM, et al. A novel β‐diiminato manganeseIII complex as the promising anticancer agent induces G0/G1 cell cycle arrest and triggers apoptosis via mitochondrial‐dependent pathways in MCF‐7 and MDA‐MB‐231 human breast cancer cells. RSC Adv. 2017;7(39):24387‐24398. [Google Scholar]
- 129. Zhong J, Sun D‐S, Wei W, et al. Contrast‐enhanced ultrasound‐guided fine‐needle aspiration for sentinel lymph node biopsy in early‐stage breast cancer. Ultrasound Med Biol. 2018;44(7):1371‐1378. [DOI] [PubMed] [Google Scholar]
- 130. Donaldson AR, McCarthy C, Goraya S, et al. Breast cancer risk associated with atypical hyperplasia and lobular carcinoma in situ initially diagnosed on core‐needle biopsy. Cancer. 2018;124(3):459‐465. [DOI] [PubMed] [Google Scholar]
- 131. Zhang JL, Wang JH, Bai AF, et al. The significance of high‐frequency ultrasound‐guided breast mass biopsy in the diagnosis of breast cancer. Eur J Gynaecol Oncol. 2017;38(5):741‐744. [Google Scholar]
- 132. Hu X, Zhou X, Yang H, et al. Axillary ultrasound and fine needle aspiration biopsy in the preoperative diagnosis of axillary metastases in early‐stage breast cancer. Oncol Lett. 2018;15(6):8477‐8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Guo W, Hao B, Luo N, et al. Early re‐staging and molecular subtype shift surveillance of locally recurrent or metastatic breast cancer: a new PET/CT integrated precise algorithm. Cancer Lett. 2018;418:221‐229. [DOI] [PubMed] [Google Scholar]
- 134. Santiago L, Adrada BE, Huang ML, et al. Breast cancer neoplastic seeding in the setting of image‐guided needle biopsies of the breast. Breast Cancer Res Treat. 2017;166(1):29‐39. [DOI] [PubMed] [Google Scholar]
- 135. Jahanbin B, Soleimani V, Azmoudeh‐Ardalan F. Displaced epithelium in breast pathology: a review. Arch Breast Cancer. 2018;5(4):150‐158. [Google Scholar]
- 136. Fan J, Xie Z‐H, Teng X‐X, et al. Determination of methylene blue by resonance light scattering method using silica nanoparticles as probe. Chinese Chem Lett. 2017;28(5):1104‐1110. [Google Scholar]
- 137. Qin W, Xu C, Zhao Y, et al. Recent progress in small molecule fluorescent probes for nitroreductase. Chinese Chem Lett. 2018;29(10):1451‐1455. [Google Scholar]
- 138. Chen H, Gu Z, An H, et al. Precise nanomedicine for intelligent therapy of cancer. Sci China Chem. 2018;61(12):1503‐1552. [Google Scholar]
- 139. Borin TF, Arbab AS, Gelaleti GB, et al. Melatonin decreases breast cancer metastasis by modulating Rho‐associated kinase protein‐1 expression. J Pineal Res. 2016;60(1):3‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Li T, Yang J, Ali Z, et al. Synthesis of aptamer‐functionalized Ag nanoclusters for MCF‐7 breast cancer cells imaging. Sci China Chem. 2017;60(3):370‐376. [Google Scholar]
- 141. Liu M, Wang Z, Tan T, et al. An aptamer‐based probe for molecular subtyping of breast cancer. Theranostics. 2018;8(20):5772‐5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Liu M, Yang T, Chen Z, Wang Z, He N. Differentiating breast cancer molecular subtypes using a DNA aptamer selected against MCF‐7 cells. Biomater Sci. 2018;6(12):3152‐3159. [DOI] [PubMed] [Google Scholar]
- 143. Tan J, Yang N, Zhong L, et al. A new theranostic system based on endoglin aptamer conjugated fluorescent silica nanoparticles. Theranostics. 2017;7(19):4862‐4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liu M, Khan A, Wang Z, et al. Aptasensors for pesticide detection. Biosens Bioelectron. 2019;130:174‐184. [DOI] [PubMed] [Google Scholar]
- 145. Huang R, Chen Z, Liu M, et al. The aptamers generated from HepG2 cells. Sci China Chem. 2017;60(6):786‐792. [Google Scholar]
- 146. Xi Z, Huang R, Li Z, et al. Selection of HBsAg‐specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of hepatitis B virus infection. ACS Appl Mater Interfaces. 2015;7(21):11215‐11223. [DOI] [PubMed] [Google Scholar]
- 147. Tan Y, Guo Q, Xie Q, et al. Single‐walled carbon nanotubes (SWCNTs)‐assisted cell‐systematic evolution of ligands by exponential enrichment (Cell‐SELEX) for improving screening efficiency. Anal Chem. 2014;86(19):9466‐9472. [DOI] [PubMed] [Google Scholar]
- 148. Brianese RC, Nakamura KDdM, Almeida FGDSR, et al. BRCA1 deficiency is a recurrent event in early‐onset triple‐negative breast cancer: a comprehensive analysis of germline mutations and somatic promoter methylation. Breast Cancer Res Treat. 2018;167(3):803‐814. [DOI] [PubMed] [Google Scholar]
- 149. Farman F, Haq F, Muhammad N, et al. Aberrant promoter methylation status is associated with upregulation of the E2F4 gene in breast cancer. Oncol Lett. 2018;15(6):8461‐8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Rahman WFWA, Fauzi MH, Jaafar H. Expression of DNA methylation marker of paired‐like homeodomain transcription factor 2 and growth receptors in invasive ductal carcinoma of the breast. Asian Pacific J Cancer Prev. 2014;15(19):8441‐8445. [DOI] [PubMed] [Google Scholar]
- 151. Pilato B, Pinto R, De Summa S, et al. HOX gene methylation status analysis in patients with hereditary breast cancer. J Hum Genet. 2013;58(1):51‐53. [DOI] [PubMed] [Google Scholar]
- 152. Heng J, Guo X, Wu W, et al. Integrated analysis of promoter mutation, methylation and expression of AKT1 gene in Chinese breast cancer patients. PLoS ONE. 2017;12(3):e0174022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fu D, Ren C, Tan H, et al. Sox17 promoter methylation in plasma DNA is associated with poor survival and can be used as a prognostic factor in breast cancer. Medicine. 2015;94(11):e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Spitzwieser M, Entfellner E, Werner B, et al. Hypermethylation of CDKN2A exon 2 in tumor, tumor‐adjacent and tumor‐distant tissues from breast cancer patients. BMC Cancer. 2017;17(1):260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zaki SM, Abdel‐Azeez HA, El Nagar MR, et al. Analysis of FHIT gene methylation in egyptian breast cancer women: association with clinicopathological features. Asian Pacific J Cancer Prev. 2015;16(3):1235‐1239. [DOI] [PubMed] [Google Scholar]
- 156. Hsu C‐H, Peng K‐L, Kang M‐L, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2(3):568‐579. [DOI] [PubMed] [Google Scholar]
- 157. Liggett TE, Melnikov AA, Marks JR, et al. Methylation patterns in cell‐free plasma DNA reflect removal of the primary tumor and drug treatment of breast cancer patients. Int J Cancer. 2011;128(2):492‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Jezkova E, Zubor P, Kajo K, et al. Impact of RASSF1A gene methylation on the metastatic axillary nodal status in breast cancer patients. Oncol Lett. 2017;14(1):758‐766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Fujii S, Yamashita S, Yamaguchi T, et al. Pathological complete response of HER2‐positive breast cancer to trastuzumab and chemotherapy can be predicted by HSD17B4 methylation. Oncotarget. 2017;8(12):19039‐19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Li D, Li P, Wu J, et al. Methylation of NBPF1 as a novel marker for the detection of plasma cell‐free DNA of breast cancer patients. Clin Chim Acta. 2018;484(7):81‐86. [DOI] [PubMed] [Google Scholar]
- 161. Tang W, Wang C, Fu F, et al. RhoBTB2 gene in breast cancer is silenced by promoter methylation. Int J Mol Med. 2014;33(3):722‐728. [DOI] [PubMed] [Google Scholar]
- 162. Kumar R, Sharma A, Tiwari R. Application of microarray in breast cancer: An overview. J Pharm Bioallied Sci. 2012;4(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Burrai GP, Tanca A, De Miglio MR, et al. Investigation of HER2 expression in canine mammary tumors by antibody‐based, transcriptomic and mass spectrometry analysis: is the dog a suitable animal model for human breast cancer? Tumor Biol. 2015;36(11):9083‐9091. [DOI] [PubMed] [Google Scholar]
- 164. Nazmeen A, Maiti S, Mandal K, et al. Better predictive value of Cancer Antigen125 (CA125) as biomarker in ovary and breast tumors and its correlation with the histopathological type/grade of the disease. Med Chem. 2017;13(8):796‐804. [DOI] [PubMed] [Google Scholar]
- 165. Wang W, Xu X, Tian B, et al. The diagnostic value of serum tumor markers CEA, CA19‐9, CA125, CA15‐3, and TPS in metastatic breast cancer. Clin Chim Acta. 2017;470:51‐55. [DOI] [PubMed] [Google Scholar]
- 166. Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20(6):332‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Park J, Lee Y. Hypoxia induced phosphorylation of estrogen receptor at serine 118 in the absence of ligand. J Steroid Biochem Mol Biol. 2017;174:146‐152. [DOI] [PubMed] [Google Scholar]
- 168. Fang F, Flegler AJ, Du P, et al. Expression of cyclophilin B is associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer. Am J Pathol. 2009;174(1):297‐308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Bjöhle J, Bergqvist J, Gronowitz JS, et al. Serum thymidine kinase activity compared with CA 15–3 in locally advanced and metastatic breast cancer within a randomized trial. Breast Cancer Res Treat. 2013;139(3):751‐758. [DOI] [PubMed] [Google Scholar]
- 170. Yang Z‐M, Ding X‐P, Pen L, et al. Analysis of CEA expression and EGFR mutation status in non‐small cell lung cancers. Asian Pacific J Cancer Prev. 2014;15(8):3451‐3455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data, models or code generated or used during the study are available from the corresponding author by request.


