Abstract
Telomerase is a ribonucleoprotein complex, the catalytic core of which includes the telomerase reverse transcriptase (TERT) and the non-coding human telomerase RNA (hTR), which serves as a template for the addition of telomeric repeats to chromosome ends. Telomerase expression is restricted in humans to certain cell types, and telomerase levels are tightly controlled in normal conditions. Increased levels of telomerase are found in the vast majority of human cancers, and we have recently begun to understand the mechanisms by which cancer cells increase telomerase activity. Conversely, germline mutations in telomerase-relevant genes that decrease telomerase function cause a range of genetic disorders, including dyskeratosis congenita, idiopathic pulmonary fibrosis and bone marrow failure. In this Review, we discuss the transcriptional regulation of human TERT, hTR processing, assembly of the telomerase complex, the cellular localization of telomerase and its recruitment to telomeres, and the regulation of telomerase activity. We also discuss the disease relevance of each of these steps of telomerase biogenesis.
Telomeres comprise repetitive sequences at the ends of chromosomes that maintain the integrity of linear chromosomes. These chromosome ends must be distinguished from other types of linear DNA, such as broken DNA ends, which are destined for repair. Telomeric sequences are specifically bound by a set of proteins that together make up the shelterin complex (FIG. 1a). The shelterin complex serves to solve the end-protection problem, by preventing DNA repair responses generated to free DNA ends. These responses include activation of the DNA damage response and repair by non-homologous end-joining, alternative non-homologous end-joining or homologous recombination1,2. To serve these functions, the protein subunits of shelterin recruit many other protein cofactors and, in recent years, mass spectrometry-based approaches have expanded the landscape of shelterin-associated proteins3. A second critical problem at chromosome ends is telomere shortening, which occurs because the DNA polymerase that synthesizes DNA in the 5'–3' direction incompletely replicates the lagging strand, thereby causing gradual shortening of telomere sequences over time. This end-replication problem was solved by the evolution of telomerase, which is a ribonucleoprotein (RNP) complex with a catalytic core composed of telomerase reverse transcriptase (TERT) and the non-coding RNA, telomerase RNA component (TERC; herein referred to as human telomerase RNA (hTR)), which serves as a template for the addition of telomeric repeats to chromosome ends.
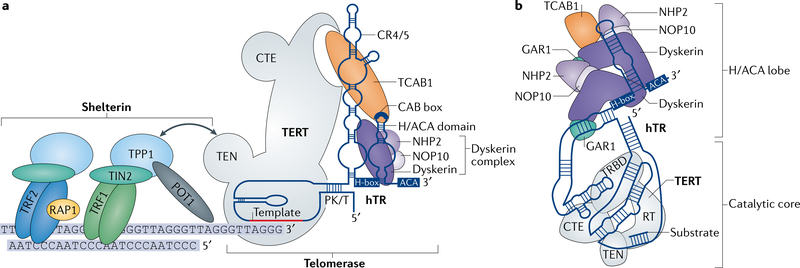
Fig. 1 |. Telomerase interacts with shelterin at telomeres.
a | Telomerase is a ribonucleoprotein complex comprising the scaffolding non-coding human telomerase RNA (hTR), the enzyme telomerase reverse transcriptase (TERT) and associated cofactors. TERT binds to hTR through its telomerase RNA binding domain (not shown) at the pseudoknot/template (PK/T) domain and the conserved region 4/5(CR4/5) domain of hTR. The dyskerin complex comprising dyskerin, NOP10 and NHP2 binds to the H/ACA domain of hTR. The H/ACA domain includes a single-strand hinge (H-box) followed by a stem-loop and the sequence ACA. The H/ACA domain of hTR also contains a conserved four-nucleotide motif called the CAB box, which binds the protein telomerase Cajal body protein 1 (TCAB1). The template region of hTR binds to the telomeric 3'-end strand. The telomere repeats are bound by the protein complex shelterin. Two double-strand DNA binding proteins, telomeric repeat-binding factor 1 (TRF1) and TRF2, bind directly to telomeric DNA. TRF2 interacts with RAP1 (also known as TRF2-interacting telomeric protein 1), and both TRF1 and TRF2 interact with TRF1-interacting nuclear factor 2 (TIN2), which also binds TPP1. TPP1 additionally binds the single-strand DNA binding protein protection of telomeres protein 1 (POT1) and recruits telomerase to telomeres through the N-terminal domain (TEN) of TERT. b | Schematic of telomerase showing the relationship of TERT domains with hTR and telomerase cofactors, adapted from the cryo-electron microscopy structure of human telomerase. Telomerase adopts a flexible, RNA-tethered two-lobed structure. The H/ACA domain of hTR bound by two sets of the dyskerin complex (dyskerin, NHP2, NOP10 and GAR1) and by TCAB1 comprises one lobe. The second lobe contains the catalytic core, where hTR and TERT encircle the telomere substrate. The two lobes are connected by the CR4/5 domain of hTR. CTE, C-terminal extension; RT, reverse transcriptase; TRBD, telomerase RNA binding domain. Adapted from REF.49, Springer Nature Ltd.
In single-cell eukaryotes, telomerase is expressed constitutively to enable long-term cell division and viability. In metazoans, and particularly in humans, the expression of telomerase became restricted to certain cell types or cell states. Human telomerase expression is highest in the germline and in progenitor cell compartments, but is limited in many differentiated cell types. This restricted pattern may have evolved in part to reduce theriskofcancer,asnearly allhumantumoursshow increased telomerase expression. Germline mutations in telomerase-pathway genes that decrease telomerase function cause the stem cell failure diseases dyskeratosis congenita, aplastic anaemia and idiopathic pulmonary fibrosis. These genetic findings indicate that cells must maintain an optimum level of telomerase, which is sufficient to support tissue homeostasis but low enough to decrease the risk of cancer. In addition to tight control of enzyme levels, telomerase is regulated at multiple additional steps.
In this Review, we discuss the latest information regarding how the human telomerase complex is regulated in normal progenitor cells and in cancer cells and how these processes are altered in disease. Transcription of the TERT gene is key in the regulation of telomerase expression and dictates which cell types may express the telomerase enzyme. Processing of the 3' end of hTR is required to make a mature hTR molecule and is subjected to quantitative regulation that dictates steady-state telomerase levels. Telomerase is a large multisubunit RNP, which includes several protein components with distinct roles in telomerase function. Assembly of this complex requires the activity of many additional proteins and movement of the developing complex through different subcompartments of the nucleus. Recruitment of telomerase to telomeres is controlled by specific protein–protein interactions, and the enzyme is further regulated after telomere engagement. During cancer development, TERT is upregulated through diverse mechanisms to support the enhanced cell proliferation associated with tumorigenesis. Impaired maintenance of telomeres underlies a series of tissue phenotypes characterized by cell depletion and end-stage organ dysfunction. Study of these disease states enables the direct investigation of biochemical defects in telomerase function in human tissues.
Telomerase transcription and maturation
Crucial regulation of telomerase activity occurs through the control of TERT transcription, which dictates TERT levels and determines the cell types in which telomerase is expressed. Regulation of hTR is carried out through a post-transcriptional maturation process of its 3' end, which sets the levels of mature hTR and, in turn, the levels of the mature telomerase complex (FIG. 2).
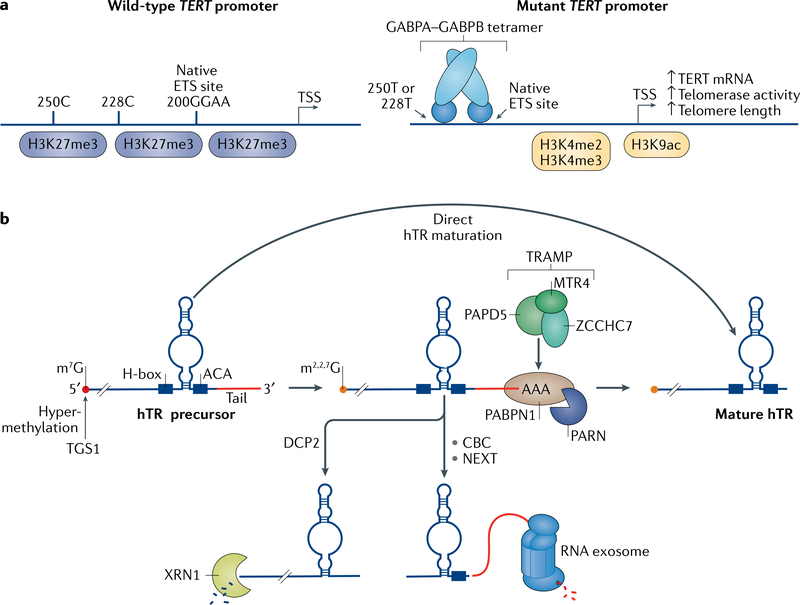
Fig. 2 |. Regulation of the TERT–hTR catalytic core of telomerase.
a | Regulation of telomerase reverse transcriptase (TERT) transcription and the role of mutations in the TERT promoter. The wild-type promoter of the TERT gene is often silenced by the gene-repressive trimethylation of histone H3 Lys27 (H3K27me3) modification. The wild-type promoter contains native binding sites (with the motif GGAA) for the ETS family of transcription factors. In diverse cancer types, C?T transitions occur 124 bp and 146 bp upstream of the translation start codon, which are referred to as C228T and C250T based on their genomic coordinates. These mutations are mutually exclusive and create additional ETS binding sites three to five helical turns (not shown) away from the native ETS sites, which are optimal for recruiting the ETS hetero-tetramer GABPA–GABPB. Mutant TERT alleles more commonly associate with RNA polymerase II and are marked by the gene-activating H3K4me2, H3K4me3 and acetylated histone H3 Lys9 (H3K9ac) modifications. MutantTERT alleles cause upregulation of TERT transcription and increased telomerase activity. TSS, transcription start site. b | Regulation of human telomerase RNA (hTR) biogenesis. hTR is transcribed as a precursor molecule with a 5' methylguanosine cap (m7G). The m7G cap is further methylated by trimethylguanosine synthase (TGS1) into m2,2,7G. The majority of hTR transcripts have a short (<10 nucleotides), genomically encoded tail. These precursors are exonucleolytically trimmed to produce mature hTR. hTR precursors can be oligo-adenylated by non-canonical poly(A) RNA polymerase PAPD5, which is a member of the TRAMP complex along with the RNA-exosome helicase MTR4 and the zinc-finger protein ZCCHC7. A subset of these oligo-adenylated hTR intermediates are substrates for degradation by the RNA exosome, to which they are recruited by the nuclear exosomal targeting (NEXT) complex and the nuclear cap-binding complex (CBC). hTR can also undergo decapping by the cytoplasmic decapping enzyme DCP2 and degradation in the 5'–3 direction by 5'–3' exoribonuclease 1 (XRN1). The remaining oligo-adenylated hTR intermediates are bound by PABPN1 and can be de-adenylated by the disease-associated poly(A) ribonuclease (PARN), which promotes hTR maturation and accumulation. There is also a direct maturation pathway for hTR precursors, which functions in the absence of both PARN and PAPD5; the enzymes involved in this pathway are still unknown. H-box, single-strand hinge in H/ACA domain.
Regulation of TERT transcription
Transcription from the TERT promoter is highly regulated and is the primary means of controlling telomerase levels in diverse cell types and cellular states. TERT is not expressed in human fibroblasts, which accounts for the progressive shortening of telomeres and replicative senescence encountered by fibroblasts after extensive cell division in culture4–6. This ‘Hayflick limit’ (the finite proliferation potential of primary human cells) is overcome by forced overexpression of TERT or through the introduction of transcription-activating, cancer-associated mutations in the TERT promoter4,7. Similarly, cultured human keratinocytes or mammary epithelial cells also lack TERT and encounter telomere shortening-based roadblocks to proliferation8,9. In contrast to the silenced expression of TERT in these cultured human cell types, TERT is strongly upregulated in human T cells during mitogenic stimulation10. This upregulation is presumably important for long-term clonal expansion of lymphocytes during immune responses. Consistent with this notion, telomerase-knockout mouse lymphocytes with short telomeres show impaired proliferative responses when exposed to mitogens11. In contrast to the restricted expression of TERT in certain adult human cells, TERT is expressed in many fetal tissues, but its expression becomes limited in early postnatal life12. TERT is expressed in human embryonic stem cells, human neural stem cells and human bone marrow progenitors13–15. Thus, TERT is expressed in many human stem cell populations, but not in certain cultured cell types.
Analysis of telomerase expression patterns in mouse cells suggested that telomerase is distributed more widely than in human cells. This conclusion is most commonly derived from the observation that telomerase and TERT are detectable in mouse fibroblasts16. Despite the less restricted expression of TERT in mouse fibroblasts compared with human fibroblasts, the advent of transcription reporters embedded within the TERT locus has revealed that the TERT promoter is highly regulated in mouse tissues. Generally, telomerase expression is closely associated with germline tissues in organisms from worm to mammals17–20. Using a mouse knock-in TERT-transcription reporter, TERT levels were found to be highest in undifferentiated spermatogonia, mitotic stem cells in the adult testis19,20. During the differentiation of spermatogonia to committed progenitor cells and subsequent entry into meiosis, telomerase levels diminish in a stepwise fashion. At the spermatid stage, TERT promoter activity and TERT expression was undetected19.
TERT-CreER.
A mouse strain engineered to induce the expression of the recombinase CreERT2, which enables fluorescence labelling of telomerase-expressing cells in vivo.
The development of cell lineage tracing from the TERT promoter using TERT-CreER mouse strains has enabled the analysis of TERT expression patterns in cell subsets within tissues and the assessment of their functional behaviour as stem cells. Analysis of the small intestineusingatransgenic TERT-CreERstrainshowed that TERT is expressed in mouse intestinal crypts and that these TERT-positive cells serve as intestinal stemcellsinrenewingthepopulationofdifferentiated cells of the intestinal villus21. A knock-in TERT-CreER strain revealed the presence of a subset population of hepatocytes in adult liver with high TERT expression and elevated telomerase activity22. These TERThigh hepatocytes function as hepatocyte stem cells, repopulating the liver in homeostatic conditions and after injury22. Together, these results show that, in renewing tissues in adults, TERT expression is enriched in progenitor cell populations to support their long-term proliferation, but that TERT is downregulated or extinguished in more differentiated progeny cells. Terminally differentiated cells, such as skeletal muscle myocytes, cardiac myocytes, neurons and spermatids, lack telomerase expression even in mice, further supporting a link between proliferative capacity and TERT expression19,23. Telomerase enrichment in tis sue progenitor cells is crucial for stem cell function, as inactivation of telomerase genes in mice leads to defects in spermatogonial stem cells, gastrointestinal crypt progenitors and haematopoietic stem cells11,19,24.
Pseudouridylation.
Enzymatic alteration of the linkage between the uracil base and the ribose sugar moiety of uridine to create its isomer pseudouridine.
Studies of TERT expression patterns in human tissues are limited, but there is evidence that TERT is expressed in stem cell and progenitor cell compartments within tissues, including in the crypts of the gastrointestinal tract, the hair follicle bulb, the hair follicle bulge and the interfollicular epidermis25–30. The mechanisms by which TERT is enriched in progenitor cells and silenced during differentiation in both mouse and human remain unknown. A core TERT-promoter construct encompassing the transcription start site is active not only in telomerase-expressing human cancer cells but also in telomerase-negative fibroblasts, suggesting that the information controlling cell-type regulation is not encoded in the proximal promoter31–33 (FIG. 2a). Many studies have dissected the TERT promoter and identified cis-elements bound by general transcription factors. The elements and factors include two E-boxes with the sequence CACGTG, which are bound by the oncoprotein MYC and its interacting proteins MAX and MAD1, and binding sites for the transcription factors SP1, upstream stimulatory factor 1 (USF1) and USF2, ID2 and ETS2 (REFS34–37). Significant advances in understanding the transcriptional regulation of TERT have been recently achieved with the discovery of highly recurrent mutations in the TERT promoter in widespread cancer types (FIG. 2a; see below).
Restriction of TERT expression to progenitor cell compartments and downregulation of TERT during differentiation strongly control telomerase activity, but the mechanisms by which this regulation is achieved in either mice or humans remain unknown. Another important open question is why human keratinocytes lack telomerase activity in culture but exhibit telomerase activity in human skin9,38. Finally, we lack a fundamenta understanding of how TERT is more effectively silenced during differentiation in human tissues compared with differentiation in mouse tissues, or indeed what accounts for the differences in TERT regulation across the many mammalian species that have been studied39.
hTR synthesis and maturation
The non-coding RNA hTR contains the template for the addition of telomeric repeats40–42 (FIG. 1a). In vertebrates, hTR belongs tothe family of small Cajal body RNAs (scaRNAs) and small nucleolar RNAs (snoRNAs). Both scaRNAs and snoRNAs are encoded in introns and transcribed by RNA polymerase II (Pol II) along with their host genes. By contrast,hTR istranscribedfroma dedicatedlocus with its own promoter43,44.
Metazoan telomerase RNAs acquired a box H/ACA domain, which includes a hairpin–hinge–hairpin–tail structure with a single-strand hinge (H-box) of the sequence ANANNA and a short tail containing the sequence ACA (FIG. 1a). This RNA domain is shared with a subset of H/ACA-containing snoRNAs and scaRNAs45,46. H/ACA-containing snoRNAs and scaRNAs guide the pseudouridylation of ribosomal RNA and of small nuclear RNAs, respectively47. A complex comprising dyskerin, NOP10 and NHP2 binds to the H/ACA sequences and executes pseudouridylation on the targets of the RNA guides. The H/ACA sequences are crucial for metazoan telomerase biogenesis, and mutations in the H/ACA domain or in components of the dyskerin complex cause hTR loss and the disease dyskeratosis congenita (see below). Interestingly, although several pseudouridine residues exist in hTR, their relevance to telomerase function remains unclear48. Nevertheless, dyskerin is crucial for telomerase function by supporting hTRstabilityand processing(see below).
The H/ACA domain of hTR contains a three-nucleotide sequence embedded in a terminal stem-loop termed the CAB box, which binds telomerase Cajal body protein 1 (TCAB1; FIG. 1a). TCAB1 is required for both the catalytic activity of telomerase and trafficking of telomerase to Cajal bodies, which are proposed to aid in telomerase RNP assembly and/or in telomerase trafficking to telomeres (see below). Biochemical experiments and a recent cryo-electron microscopy structure revealed that the dyskerin complex and TCAB1 are components of the mature telomerase holoenzyme49,50.
The mechanism of hTR transcription termination in vertebrates is unknown as, unlike the related snoRNAs, which are embedded in introns, mature hTR is not generated by the spliceosome. Consistent with variations in the length and sequence of telomerase RNAs in evolution, the processing of telomerase RNAs is substantially different between species. However, one commonality is the conservation of post-transcriptional modification of telomerase RNAs from single-cell eukaryotes to mammals. In Saccharomyces cerevisiae, transcription termination of the telomerase RNA TLC1 occurs through the Nrd1–Nab3–Sen1 (NNS) pathway51–53. Although no homologue of NNS proteins exists in vertebrates, a proposed functional analogue is the Integrator complex, which is responsible for transcription termination l of some non-coding RNAs (such as small nuclear RNAs) and mRNAs54–56. Following termination, the NNS com plex recruits a complex termed the TRAMP complex that oligo-adenylates TLC1 transcripts and thus primes them for degradation by the RNA exosome, which is a multisubunit RNAse57. TRAMP homologues in verte brates include non-canonical poly(A) RNA polymerase PAPD5 (also known as TENT4B), the AIR2 homologue ZCCHC7 and MTR4 (REFS58,59) (FIG. 2b). Similar tothe yeast complex, vertebrate TRAMP oligo-adenylates precursor telomerase RNA transcripts.
In steady-state conditions, hTR exists as a pool of transcripts, the majority of which end at position 451 and a minority extend past nucleotide 451. These 3'-end extensions are genomically encoded by the hTR locus and, additionally, most of the extended molecules possess short untemplated adenosine tails60. It had been unclear whether extended hTR molecules represented precursors for mature hTR or whether they were aberrant transcripts. To address this question, nascent RNA end-sequencing — a technique for enriching RNA precursors and sequencing their 3' ends — was developed, revealing that hTR is initially transcribed as an extended molecule of approximately 461 nucleotides, and that this precursor is processed to the mature 451-nucleotide RNA61 (FIG. 2b). Longer hTR precursors, exceeding 1,500 nucleotides in length, have also been detected using PCR with reverse transcription and may represent either very early hTR precursors that are rapidly processed into the short forms or unproductive hTR transcripts that are targeted for destruction by nuclear RNA surveillance62,63
hTR precursors are rapidly oligo-adenylated primarily by the poly(A) polymerase PAPD5 (REFS61,63,64) (FIG. 2b). The oligo-adenylated hTR intermediates can be deadenylated by poly(A) ribonuclease (PARN), which was initially discovered as a gene mutated in families with short telomeres and pulmonary fibrosis (see below). In the absence of PARN, hTR intermediates initially accumulate and are subsequently degraded, resulting in diminished hTR levels and reduced telomerase activity (see below). PARN has increased activity towards poly(A) substrates and may preferentially remove untemplated A tails from hTR precursors65,66. Alternatively, PARN could both remove untemplated A tails and trim the genomic extensions of hTR precursors to produce mature hTR. This idea was suggested on the basis of evidence for PARN in vitro activity towards hTR genomic tail sequences62. However, in the absence of both PARN and PAPD5, the extent of oligo-adenylation of hTR precursors is considerably lower, and maturation of hTR transcript proceeds normally61. These observations indicate the presence of additional 3'–5' exonucleases in addition to PARN that are responsible for trimming in vivo the hTR precursor into its mature length directly without the requirement for oligo-adenylation and de-adenylation. One such exonuclease candidate is the Cajal body-localized 3'–5' RNA exonuclease TOE1, which contributes to the trimming of small nuclear RNAs, snoRNAs and scaRNAs as well as hTR precursor molecules67–69.
Oligo-adenylated hTR transcripts that are not processed to mature hTR are degraded by the RNA exosome (FIG. 2b). The poly(A)-binding protein PABPN1 (also known as PABP2) binds oligo-adenylated hTR transcripts, and its loss leads to loss of hTR transcripts70. PABPN1-unbound oligo-adenylated hTR intermediates are subject to degradation by the recruitment of the nuclear RNA exosome by the nuclear exosome targeting (NEXT) complex in conjunction with the cap binding complex (CBC)63,71. Depletion of exosome components, NEXT or CBC can rescue the phenotype of low hTR levels owing to dyskerin loss or hTR point mutation-sin the H/ACA domain, suggesting that the exosome serves also to remove defective or misassembled hTR transcripts71–73. Early during transcription, telomerase RNAs acquire the m7G monomethylguanosine cap, which is hypermethylated to a m2,2,7G trimethylguanosine cap by the enzyme trimethylguanosine synthase (TGS1)74,75 (FIG. 2b). Hypermethylation by TGS1 suppresses hTR accumulation, and TGS1-negative cell lines exhibit increased telomerase activity and telomere lengthening, indicating that TGS1 negatively regulates telomerase76. Cytoplasmic decapping of hTR transcripts by DCP2 (also known as m7GpppN-mRNA hydrolase) and subsequent degradation by 5'–3' exoribonuclease 1 (XRN1) also leads to hTR decay71. Thus, hTR transcripts undergo a multistep process of maturation that includes cycles of adenylation and de-adenylation, which together control the steady-state levels of hTR. These mechanisms arecrucialfor the regulationoftelomeraselevels,and many of these steps are disrupted in disease states.
Assembling the telomerase holoenzyme
Many telomerase-associated proteins have been identified biochemically. These proteins can be divided into two classes: stably associated components of the mature holoenzyme and transiently associated proteins, which typically serve a function in the assembly of the holoenzyme (BOX 1).
Box 1 |. The telomerase holoenzyme and its accessory factors.
Although combining telomerase reverse transcriptase (TERT) and the telomerase RNA (hTR) in the presence of a cell extract yields telomerase activity, such assembly cannot occur inside a living cell without other components of the telomerase holoenzyme and without many accessory proteins. Distinguishing between the components of the mature holoenzyme and the accessory proteins can be achieved through biochemical and genetic studies. Telomerase holoenzyme components are stoichiometrically associated with TERT and hTR, such that immunodepletion of a holoenzyme component depletes telomerase levels and activity from a human cancer cell extract. By contrast, an accessory protein may be transiently associated with telomerase, and its immunodepletion does not quantitatively deplete telomerase levels or activity. Genetic studies are also crucial for determining whether a telomerase-associated factor is required for telomerase function. Depletion of either a holoenzyme component or an accessory factor may diminish telomerase levels or telomerase activity, or impair telomerase function, depending on the role the protein serves in the pathway.
The telomerase core
All telomerase enzymes across evolution share a common catalytic core, comprising TERT and telomerase RNA. These core components were originally identified in single-celled ciliated protozoans using biochemical approaches77,78. Human TERT and hTR were subsequently cloned based, in part, on sequence similarity79. Co-expression of TERT and hTR in vitro is sufficient to reconstitute telomerase activity, but only in the presence of a cell extract that provides additional enzymatic activities that stimulate telomerase assembly80,81. The association of TERT and hTR depends on the binding of RNA secondary structures in hTR by TERT. Interactions between TERT and hTR are mediated by specific domains in hTR and TERT. The human TERT protein contains an N-terminal domain (TEN), a telomerase RNA binding domain (TRBD), the catalytic reverse transcriptase (RT) domain and a C-terminal extension (CTE). Human telomerase RNA contains a 5' pseudoknot/template (PK/T) domain that templates the addition of telomerase repeats, a central domain comprising a three-way junction between stem-loops termed conserved region 4/5 (CR4/5) and the 3' H/ACA domain (FIG. 1a).
Detailed information on the TERT–hTR interaction is derived from cryo-electron microscopy structures of the telomerase complex, in human and Tetrahymena thermophila, and from biochemical analyses49,82,83 (FIG. 1b). Biochemical experiments showed that TERT binds to the PK/T domain and the CR4/5 domains of hTR81,84 (FIG. 1a). Although TERT can bind to hTR though the PK/T domain alone, the additional binding with CR4/5 is required for full telomerase activity84,85. Structura analysis showed that the TERT domains are arranged in a ring structure characteristic of other reverse transcriptases (FIG. 1b). The assembled catalytic core contains a central canal accommodating the template–substrate RNA–DNA hybrid and providing an exit for the newly synthesized telomere DNA49,82,83. Many of the germline mutations that cause dyskeratosis congenita perturb the associations between hTR and hTERT (see below).
Many cofactors are required for the assembly of the telomerase holoenzyme
Although the catalytic core is sufficient for telomerase activity in vitro, the telomerase holoenzyme in human cell extracts comprises a much larger complex of approximately 500 kDa86,87. The holoenzyme has been isolated by affinity purification using tagged TERT proteins, antibodies against endogenous TERT proteins or affinity chromatography with a telomere-DNA oligonucleotide substrate88–90. Telomerase stability, full activation and recruitment to telomeres require the assembled holoenzyme, which includes two dyskerin complexes that bind to hTR— one at each of the two hairpins in the H/ACA domain. Dyskerin and NOP10 interact with the RNA, and in turn GAR1 (which is not part of the dyskerin complex) and NHP2 interact with dyskerin and NOP10 (REFS49,91,92). Dyskerin–NOP10–NHP2 co-localize with the site of H/ACA snoRNA transcription but GAR1 does not, suggesting GAR1 may be a non-core component of telomerase that is assembled at a later stage of maturation. GAR1 is co-localized with the holoenzyme following its recruitment to Cajal bodies93–96. TCAB1 is a stably associated component of the holoenzyme and engages telomerase through an interaction with the hTR CAB box. TCAB1 controls the catalytic activity and nuclear trafficking of telomerase (see below).
In addition to these stably associated constituents of the telomerase holoenzyme, several proteins have been identified that associate with telomerase transiently and are required for its biogenesis. NAF1 binds to dyskerin and is required for the stable association of dyskerin with telomerase96–98. SHQ1 is another dyskerin-interacting protein that facilitates telomerase assembly99. Two AAA+ ATPases, pontin and reptin, bind to TERT and to dyskerin and promote the assembly of telomerase into a stable complex. Pontin and reptin associate with telomerase in S phase, and their depletion leads to loss of hTR and telomerase activity88. TERT binding to hTR is aided by the chaperone heat shock protein 90 (HSP90)100–102, and telomerase assembly with TCAB1 requires the chaperone TriC, which is dedicated to folding certain classes of protein clients, including WD40 repeat-containing proteins such as TCAB1 (REF.103). Thus, although minimal telomerase activity requires only TERT and hTR, telomerase function in vivo requires assembly with stably associated holoenzyme components, such as the dyskerin complex and TCAB1, and with transiently associated proteins, such as pontin, reptin and chaperones like HSP90 and TRiC.
WD40 repeat.
A protein domain of 44–60 amino acids, which often mediates protein–protein or protein–RNA interactions.
Telomerase localization
Elongation of telomeres requires telomerase access to telomeric DNA. However, processing of telomerase components and assembly of the complex likely occur at multiple sites within the nucleus, and defects in these processes result in telomerase mislocalization.
Localization to Cajal bodies
Like other scaRNPs, the telomerasecomplexislocalizedtoCajalbodies,which are nuclear compartments enriched in factors of splicing and of other RNA-related processes93,104. The protein coilin scaffolds Cajal bodies; loss of coilin abrogates Cajal body formation and mislocalizes telomerase105,106. Telomerase localization in Cajal bodies is mediated also by TCAB1, which binds to the CAB box of hTR; in the absence of TCAB1–CAB box interaction, telomerase mislocalizes to the nucleolus50,103,107,108 (FIG. 3). Single-molecule, live cell imaging showed that telomerase exhibits extended residence time in Cajal bodies109. The purpose of telomerase localization in Cajal bodies remains unclear, as telomeres are effectively maintained in coilin-knockout cancer cells despite the disruption of Cajal bodies106,110. Furthermore, telomerase does not localize to Cajal bodies in certain mouse cells111. Nevertheless, it remains possible that Cajal bodies serve an important function in regulating telomerase in cellular contexts of low-level telomerase expression, or in certain cell types that have not yet been studied.Owing to the technical challenges in studying telomerase, its subnuclear localization has only been studied in a few cell types.
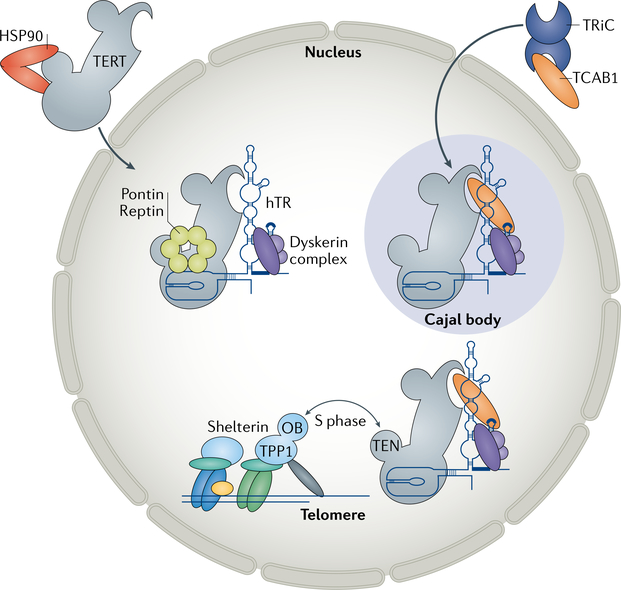
Fig. 3 |. Assembly and trafficking of telomerase within the cell.
Telomerase reverse transcriptase (TERT) is folded with the help of the chaperone heat shock protein 90 (HSP90), whereas human telomerase RNA (hTR) is bound co-transcriptionally by the dyskerin complex, comprising dyskerin, NOP10, NHP2 and the associated factor NAF1. The cofactors pontin and reptin promote assembly of hTR with the dyskerin complex and TERT. Telomerase is localized to Cajal bodies by telomerase Cajal body protein 1 (TCAB1), which is folded by the chaperone TRiC complex. In the absence of TCAB1, telomerase mislocalizes to the nucleolus (not shown) and is not recruited to telomeres. During S phase, the N-terminal domain (TEN) of TERT and the TEL patch (not shown) on the oligosaccharide/oligonucleotide binding (OB) fold domain of TPP1 interact to recruit telomerase to telomeres.
Recruitment to telomeres and association with shelterin
Recruitment of telomerase to telomeres is mediated by an interaction between TERT and the shelterin protein TPP1 (FIG. 1a). TPP1 is connected to the rest of the shelterin complex through TRF1-interacting nuclear factor 2 (TIN2), which also forms contacts with the double-strand DNA-binding proteins telomeric repeat-binding factor 1 (TRF1) and TRF2 (REFS112,113). TPP1 also associates with protection of telomeres protein 1 (POT1), which directly binds the single-strand telomere overhang114. Loss of TPP1 or TIN2 reduces telomerase recruitment to telomeres115. The specific interaction between TERT and TPP1 is mediated by the TEN domain of TERT and a patch of amino acids on the sur-face of the oligosaccharide/oligonucleotide binding fold domain of TPP1, termed the TEL patch87,106,115–117 (FIG. 3). The importance of the TEL patch is underscored by the discovery of mutations in this region of TPP1 that cause telomere shortening and severe dyskeratosis congenita118. Live cell imaging of single telomerase molecules revealed that telomerase RNPs diffuse through the nucleus and form both transient and long-term interactions with telomeres109. LossofTERT–TPP1bindingbymutation oftheTELpatchinhuman embryonicstemcells causes telomere shortening and loss of cell viability, underlining the importance of telomerase recruitment for telomere maintenance117. Interestingly, tethering of TERT to TPP1 in this context could not completely rescue telomere shortening, suggesting that the TEL patch of human TPP1 has additional roles in telomerase activation.
TIN2 is also essential for telomere length maintenance119–122; in its absence, TPP1 and POT1 cannot localize to the telomere123. Owing to this bridging function, TIN2 is required for recruitment of telomerase to telomeres115. Reflecting its importance in telomere length maintenance, TIN2 is mutated in individuals with short-telomere syndromes, with mutations clustering in exon 6, near the C-terminal region of the protein124,125. Although these mutations do not affect TIN2 localization or telomerase activity, there is some evidence that they impair telomerase recruitment to the telomere in human cells126,127. However, a mouse model of the disease-causing TIN2-K280E mutation exhibits telomere shortening through a telomerase-independent mechanism128. More studies are needed to precisely clarify how these TIN2 mutations cause telomere shortening.
Telomeric proteins can also inhibit telomerase activity at the telomere. The CST complex, comprising CTC1, STN1 and TEN1, is a conserved trimeric complex that binds the telomeric 3' overhang129,130 (FIG. 4). The human CST complex assists DNA replication at telomeres by stimulating DNA polymerase α-primase131. Additionally, CST restricts telomere extension by limiting access of telomerase to the telomere and inhibiting telomerase stimulation by TPP1–POT1 (see below)132. Thus, the CST complex may facilitate a switch from telomerase-mediated elongation of telomeres to fill-in synthesis of telomeres.
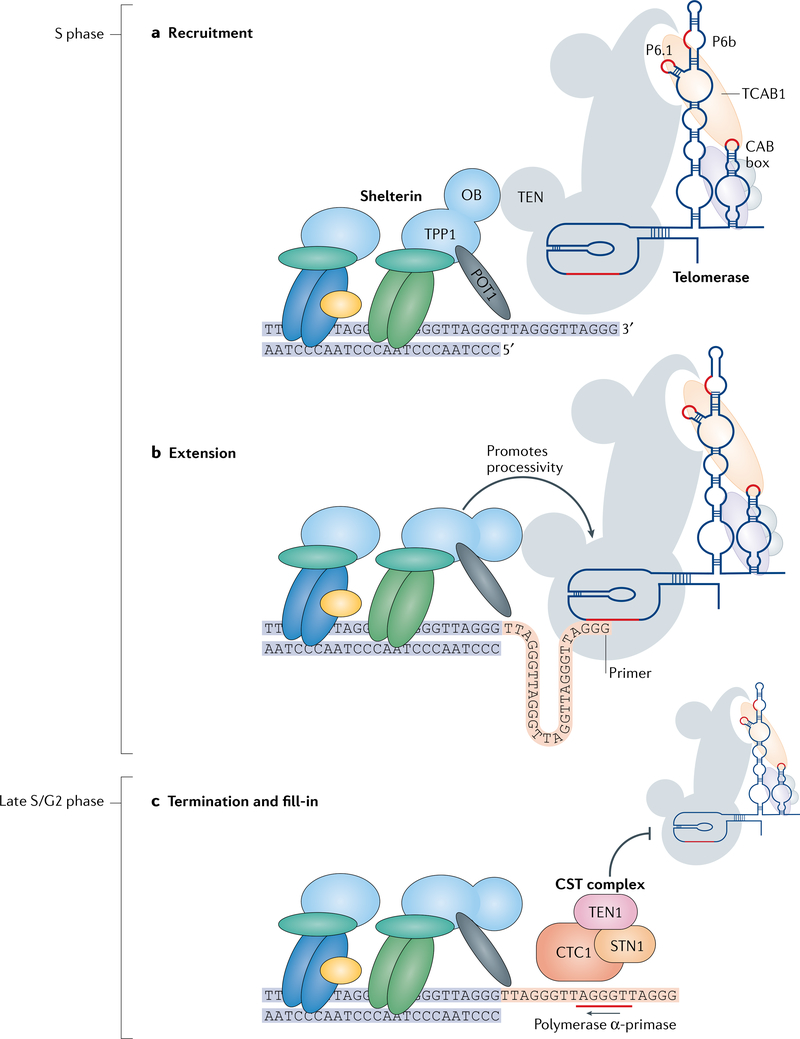
Fig. 4 |. Regulation of telomerase activity at the telomere.
a | Recruitment: telomerase is recruited to the telomere during S phase through contacts between the oligosaccharide/oligonucleotide binding (OB) fold domain of the shelterin complex subunit TPP1 and the N-terminal domain (TEN) of telomerase reverse transcriptase (TERT). Telomerase activity is regulated by telomerase Cajal body protein 1 (TCAB1), which binds to the H/ACA domain of human telomerase RNA (hTR) at a conserved four-nucleotide sequence called the CAB box. TCAB1 binding causes conformational changes in the P6.1 and P6b regions of the hTR conserved region 4/5 (CR4/5) domain, which enhance telomerase activity.TCAB1-interacting sites in hTR are marked in red. b | Extension: telomerase extends the G-rich strand (orange) of the telomere. Telomerase processivity — the number of telomere repeats that telomerase can add in one primer-binding event — is enhanced by the shelterin components TPP1 and protection of telomeres protein 1 (POT1). TPP1–POT1 promote telomerase processivity by encouraging the translocation of the telomerase enzyme along the telomere substrate and/or by preventing dissociation of telomerase from the telomeric primer. c | Termination and fill-in: during late S phase/G2 phase, the CST complex — comprising CTC1, TEN1 and STN1 — binds the newly synthesized G(-rich) strand of the telomere and prevents telomerase from accessing it. CST also impairs the interaction of telomerase with TPP1–POT1 and promotes the activity of DNA polymerase α-primase, which synthesizes the RNA primer (red line) that is necessary for the fill-in of the telomeric C strand.
Recruitment of telomerase to telomeres is regulated in a cell cycle-dependent manner. In both budding yeast and fission yeast, interaction of telomerase with telomeres is restricted to S phase133. Similarly, in human cells, co-localization of telomerase and telomeres occurs primarily during S phase134,135 (FIGS 3 and 4). Cell cycle-regulated assembly of the telomerase enzyme could account for some of the restriction of telomerase recruitment to S phase. Along these lines, the association between TERT and pontin varies during the cell cycle and peaks in S phase. Interestingly, assembly of the telomerase complex with TCAB1 varies during the cell cycle88. The TCAB1–hTR association is lost in mitotic cells, suggesting that some elements of the telomerase enzyme are assembled in a cell cycle-regulated manner136. This observation fits with the previously established importance of TCAB1 in telomerase recruitment to telomeres and suggests that regulation of TCAB1 binding to telomerase accounts for some of the cell cycle-dependent recruitment. Thus, telomerase recruitment to telomeres requires an interaction between TERT and the shelterin protein TPP1 and occurs primarily during S phase; however, the precise mechanisms controlling the timing and regulation of this recruitment remain unknown.
Regulation of telomerase activity
Whether telomerase activity can be regulated by its protein cofactors has been an active area of study. TCAB1 was initially characterized as a telomerase protein required for telomerase trafficking to Cajal bodies and for maintaining telomere length50,107. In human and mouse cells deleted of TCAB1 using genome editing, telomerase catalytic activity was impaired. Diminished telomerase activity in these cells was caused by partial unfolding of crucial RNA helices within the hTR CR4/5domain,whichinturnimpaireditsbinding with TERT110. These findings indicate that binding of TCAB1 promotes a catalytically active state of telomerase by enhancing RNA folding and encouraging proper association of TERT and hTR (FIG. 4a).
Once recruited to the telomere, telomerase must be further activated to efficiently add telomeric repeats. Regulation of telomerase at the telomere occurs through the activity of the shelterin components. As TPP1–POT1 binds to single-strand DNA through POT1, the initial prediction was that telomerase and TPP1–POT1 would compete for telomere binding, and that TPP1–POT1 would inhibit telomerase activity by sequestering the 3' overhang. In support of this hypothesis, overexpression of a POT1 mutant incapable of binding single-strand DNA results in rapid telomere lengthening by enhancing telomerase activity at telomeres137. In contrast to this finding, addition of recombinant TPP1 and POT1 to telomerase in vitro caused a twofold to threefold increase in telomerase processivity (defined as the number of telomere repeats added following a single telomerase–primer binding event)138–140. Further studies showed th TPP1–POT1 improve telomerase processivity in vitro by preventing its dissociation from telomeric DNA or by increasing the efficiency of telomerase translocation after each repeat addition138 (FIG. 4b). Stimulation of telomerase processivity is mediated by the TEL patch of TPP1, which also functions in telomerase recruitment116 In conjunction with recombinant TPP1–POT1, the addition of exogenous TIN2 can also stimulate telomerase processivity in cells141. Although in vivo studies of telomerase processivity are lacking, theoretically this activation mechanism of telomerase allows telomerase to synthesize long tracts of telomere DNA in vitro with only a few primer-binding events. Additional investigation is required to better understand this dual role of TPP1–POT1 in enhancing and inhibiting telomerase function.
Telomerase upregulation is nearly universal in human cancers
Upregulation or reactivation of telomerase occurs in more than 90% of all human tumours142–144. Although telomerase appears to be expressed in most proliferating progenitor cells, the amount of telomerase is insufficient to maintain telomeres during ageing and in homeostasis. Insufficient levels of telomerase may explain the general observation that telomeres shorten in human tissues during ageing. As discussed above, many types of primary human cells grown in the laboratory completely lack telomerase and show progressive telomere shortening with cell division. After a long lag phase of several months, human fibroblasts cease division and enter a state of replicative senescence. We now understand that replicative senescence is induced by a subset of very short telomeres that lose the ability to protect chromosome ends and are recognized as damaged DNA, which induces the DNA damage response, including activation of the p53 and RB tumour suppressor pathways145,146. Inactivation of p53 and RB extends the proliferation of cells with critically short telomeres, but culminates in telomere crisis: a period of genomic instability and cell death. Although telomere crisis impairs cell division, it can also drive malignant transformation by inducing non-reciprocal translocations, aneuploidy and copy number changes147,148. The cell death induced during telomere crisis is driven by autophagy149. In fibroblasts, overexpression of TERT reconstitutes telomerase, prevents cell senescence and crisis, and enables cell immortality4. Upregulation of telomerase in human tumours serves the same function to confer cell immortalization, thereby facilitating tumour invasion and metastasis.
Telomerase is upregulated in cancer through diverse mechanisms, including gene amplification of TERT and of the hTR-encoding gene TERC. The best understood mechanism by which cancers increase TERT transcription is through acquisition of non-coding mutations in the TERT promoter (FIG. 2a). TERT promoter mutations represent the most common mutations in several tumour types, including glioblastoma, melanoma, bladder cancer and hepatocellular carcinoma150–154. Furthermore, they are the most common non-coding mutations in human cancers155. TERT promoter mutations activate TERT transcription, resulting in increased mRNA levels, elehat vated telomerase activity and increased telomere length in urothelial carcinoma cell lines156. The two commonly occurring C→T transition mutations are located in the proximal promoter, 124 bp and 146 bp upstream of the translationstartcodon.Thesemutationsarereferredto as C228T and C250T, based on their hg19 genomic coordinates (chr5, 1,295,228 C>T and chr5, 1,295,25 C>T). The C228T and C250T mutations are heterozygous, are mutually exclusive and create de novo binding sites for ETSfamilytranscriptionfactors(FIG. 2a).
The mutated TERT promoter specifically recruits the ETS family transcription factor GABPA with its cofactor GABPB157. The two transcription factors form a hetero-tetramer that binds to a motif at the TERT promoter as well as to the new motif created by the mutations. In cancer cells harbouring TERT promoter mutations, TERT mRNAs specifically derive from the mutated allele158. Consistent with this finding, the wild-type allele remains marked by trimethylated histone H3 Lys27 (H3K27me3), which is a gene-repressive histone modification, whereas the mutated allele is marked by the gene-active histone modifications H3K4me2, H3K4me3 and acetylated histone H3 Lys9 (H3K9ac)158–160 (FIG. 2a). Thus, the mutations in the TERT promoter may prevent normal silencing of the promoter. Generation of these TERT promoter mutations in telomerase-expressing human embryonic stem cells resulted in a modest increase in TERT mRNA levels. However, upon differentiation into fibroblasts, the mutant cells retained telomerase expression whereas isogenic control cell lines silenced TERT15. The TERT promoter mutations may also alter the long-range contacts of the TERT locus to favour transcription activation160.
The frequency of TERT upregulation in cancer suggests that it is crucial for tumorigenesis. Based on the restriction of TERT promoter mutations to cancers of tissues with typically low rates of self-renewal(such a sthe central nervous system, the liver and melanocytes) and their absence from cancers of more proliferative tissues (such as the colon and blood), it has been proposed that tissues with low basal telomerase activity require activating telomerase mutations early during tumorigenesis161. In favo ur ofthis argument, TERT promoter mutations n occur in early, precursor lesions in cancers, such as hepatocellular carcinoma, bladder cancer, cutaneous melanoma, thyroid carcinoma, squamous cell carcinoma, basal cell carcinoma and oligodendroglioma153,162–164.
In addition to selecting for TERT promoter mutations, cancers upregulate TERT transcription by diverse mechanisms. In some cases of hepatocellular carcinoma, the hepatitis B virus (HBV) can integrate its genome in close proximity to the TERT promoter, thereby placing viral enhancer elements near the promoter without disrupting coding sequences165–168. Chromosom rearrangements in neuroblastoma position the TERT locusnearstrongenhancerelements,thereby upregulating TERT transcription169,170. Furthermore, in a set of nearly 7,000 tumour samples, 4% had amplifications of the TERTgene, most commonly in ovarian cancer and lung adenocarcinoma samples171. Importantly, TERT promoter mutations in glioblastoma and TERT rearrangements are each mutually exclusive with mutations in ATRX or DAXX, which are histone chaperones that are commonly inactivated in cancer cells employing a telomerase-independent mechanism of telomere maintenance, termed alternative lengthening of telomeres (ALT). ALT is based on homologous recombination and uses break-induced replication to synthesize tracks of telomere DNA. ALT in cancer cells results in long and heterogeneous telomeres and a lack of telomerase expression172. ALT is a common mechanism of telomere maintenance in glioblastoma, neuroendocrine tumours and certain sarcomas173.
Upregulation of hTR in cancer is less well studied, but some evidence suggests that, like TERT, hTR is transcriptionally activated in human tumours. An increase in copy number of the hTR-encoding gene TERC has been detected in multiple cancer types, including in cervix, ovary and lung cancers174,175. Finally, although itis estimated that more than 90% of cancers upregulate telomerase activity, most telomerase-positive tumours lack TERT promoter mutations. A study of sequencing data from >18,000 tumour and matched normal tissue samples found that, among TERT-expressing tumours, 32% carried TERT promoter mutations, gene amplifications or chromosomal rearrangements171. These observations suggest that tumours that have detectable telomerase activity but no activating TERT mutations must upregulate telomerase by some other mechanism. One possible means for modulating telomerase activity relates to the telomere position effect, an epigenetic means by which telomere shortening can enhance the expression of telomere-proximal genes, such as TERT176,177. Further work is needed to ascertain whether this is a common mechanism by which TERT transcription is increased during carcinogenesis. It also remains to be seen whether cancer cells increase telomerase activity by other mechanisms, such as increasing the assembly, recruitment to telomeres or catalytic activity of telomerase.
Diminished telomerase precipitates tissue-failure diseases
In stark contrast to telomerase upregulation in cancer, germline reduction in telomerase function precipitates a set of tissue-failure diseases characterized by very short telomeres. These diseases, which are known as telomere biology disorders (TBDs), are remarkably heterogeneous in their severity and can manifest at birth, in childhood or even in middle age. The classic telomerase insufficiency disorder was initially defined as dyskeratosis congenita, which is a multiorgan, systemic disease characterized by abnormal skin pigmentation, dystrophic nails, bone marrow failure and predisposition to specific cancers, including squamous cell carcinoma and leukaemia178. Based on recent genetic and clinical findings, it is now understood that mutations in telomerase-relevant genes can additionally cause idiopathic pulmonary fibrosis, aplastic anaemia, and liver fibrosis and cirrhosis (TABLE 1). Mutations in every component of the telomerase holoenzyme have been identified in individuals with TBDs and have provided insight into telomerase structure and function.
Table 1 |.
Telomerase and telomere-associated genes mutated in tissue-failure disorders
| Gene | Product | Normal function | Diseases associated with loss of function | Refs |
|---|---|---|---|---|
| TERC | hTR | Non-coding RNA template for telomere synthesis | Aplastic anaemia, dyskeratosis congenita, pulmonary fibrosis | 190–192 |
| TERT | TERT | Reverse transcriptase that catalyses telomere synthesis | Aplastic anaemia, dyskeratosis congenita, pulmonary fibrosis | 192–194 |
| DKC1 | Dyskerin | Pseudouridine synthase that controls hTR processing | Dyskeratosis congenita, aplastic anaemia, pulmonary fibrosis | 195,196 |
| TINF2 | TIN2 | Shelterin protein that connects the DNA binding proteins TRF1 and TRF2 with TPP1 | Dyskeratosis congenita, aplastic anaemia | 125,124 |
| PARN | PARN | RNA exonuclease that aids hTR maturation | Dyskeratosis congenita, pulmonary fibrosis | 186 |
| WRAP53 | TCAB1 | WD40 protein that binds hTR and promotes RNA folding and telomerase localization | Dyskeratosis congenita | 107 |
| CTC1 | CTC1 | Component of the CST complex that promotes telomere DNA replication | Dyskeratosis congenita, Coats plus | 188,197 |
| RTEL1 | RTEL1 | DNA helicase that promotes telomere stability during replication | Dyskeratosis congenita, pulmonary fibrosis | 189 |
| Nola2 | NHP2 | Component of the dyskerin complex | Dyskeratosis congenita | 198 |
| Nola3 | NOP10 | Component of the dyskerin complex | Dyskeratosis congenita | 185 |
| Naf1 | NAF1 | Component of the dyskerin complex | Pulmonary fibrosis | 184 |
| ACD | TPP1 | Shelterin protein that recruits TERT to telomeres | Dyskeratosis congenita, aplastic anaemia | 118,199 |
| ZCCHC8 | ZCCHC8 | Component of nuclear exosome targeting complex | Pulmonary fibrosis | 200 |
Telomerase and telomere-associated genes that are mutated in telomere biology disorders are listed from the most common to the least common. CST, CTC1–STN1–TEN1; hTR, human telomerase RNA; PARN, poly(A) ribonuclease; RTEL1, regulator of telomere elongation helicase 1; TCAB1, telomerase Cajal body protein 1; TERC, telomerase RNA component; TERT, telomerase reverse transcriptase; TIN2, TRF1-interacting nuclear factor 2; TRF1, telomeric repeat-binding factor 1.
Heterozygous mutations in the TERC(encoding hTR) or TERT genes are common in TBDs and cause haploinsufficiency (reduced enzyme levels). Mutations in TERC typically reduce telomerase catalysis or decrease hTR levels. Most TERC mutations occur within the PK/T domain of hTR, which binds TERT and contains the template sequence for reverse transcription. Many of these mutations impair TERT binding and reduce telomerase function, but preserve hTR expression85. Mutations in the central CR4/5 domain of hTR also reduce telomerase catalytic activity, by impairing the e hTR–TERT association84,179. Mutations in the 3' H/ACA region of hTR often cause loss of hTR through diminished dyskerin binding180. Germline mutations in the TERT gene in individuals with TBD are distributed throughout the open reading frame, in contrast to the somatic mutations in the TERT promoter in human cancers. Coding mutations in TERT commonly lead to reduced catalytic activity, although many mutations preserve catalytic function181. These findings suggest that TERT mutations may disrupt other aspects of telomerase biology, including protein stability, complex assembly or recruitment to telomeres. In support of this notion, mutations in the TERT TEN domain or TERT C-terminus impair its recruitment to telomeres106. Consistent with the importance of telomerase recruits ment for telomere maintenance, rare mutations have been identified in TPP1 that specifically disrupt its interaction with TERT and impair recruitment of telomerase to telomeres118.
The core dyskerin complex (dyskerin–NHP2–NOP10) recognizes that the hTR H/ACA domain is crucial for hTR stability and processing. Mutations in the geneencodingdyskerinreduce hTRlevelsandcause the X-linked form of dyskeratosis congenita182, and rare mutations in NOP10 and in NHP2 have been identified in autosomal recessive forms of dyskeratosis congenita. Rare mutations have also been found in the gene encoding the dyskerin-associated cofactor NAF1, which lead to diminished hTR levels183–185. A common means by whic hTR levels are reduced in TBDs is through mutations that inactivate PARN186. Such mutations cause autosomal recessive forms of idiopathic pulmonary fibrosis, aplastic anaemia and dyskeratosis congenita and commonly occur in the nuclease or RNA binding domains of PARN. PARN mutations slow hTR maturation rates, leading to a relative accumulation of hTR precursors and global reduction in the levels of mature hTR61. Rare autosomal recessive mutations in the gene encoding TCAB1 have also been identified in TBDs107,187. These mutation reside in the WD40-repeat domain and interfere with TCAB1 protein folding by the TRiC chaperone complex, thereby causing loss of TCAB1 protein103. In addition to these mutations directly impairing mutations in genes encodingcomponentsofthetelomeraseholoenzyme, mutations in individuals with TBD are also found in TIN2, in CTC1 and in regulator of telomere elongation helicase 1 (RTEL1), which is a DNA helicase involved in the maintenance of telomere integrity124,125,188,189. Itis striking that most TBD-causing mutations affect hTR, by reducing its levels or function, thereby emphasizing the central role of hTR in telomerase function.
Conclusions and future perspective
Telomerase evolved to provide a simple solution for replicating chromosome ends in single-cell eukaryotes, but the telomerase complex has assumed greater importance in long-lived, complex metazoans, such as humans. The fundamental challenge of supporting robust cell division for tissue renewal while reducing the likelihood of developing cancer accounts for this elevated importance of telomerase in humans. The requirement for facilitating cell renewal while suppressing cancer, combined with the special characteristics of RNPs, explains the complexity of regulation of the telomerase complex. These features account for exquisite transcriptional control of TERT expression, the need to precisely process and stabilize the telomerase RNA, regulation of assembly of the active complex and regulation of recruitment of the enzyme to telomeres. In organisms and tissues, certain cells are licensed to express telomerase, and the amount of telomerase varies with the differentiation state of the cell. The multiple levels of telomerase control are essential as an increase or a decrease in telomerase activity beyond a narrow range is associated with human diseases. Despite important progress, we continue to lack an understanding of the cis-regulatory elements and the trans-acting factors that activate TERT transcription in normal progenitor cells and that silence TERT during differentiation.
Telomerase RNA was once thought to be unregulated and expressed in a ubiquitous fashion. The study of TBDs has revealed surprising new levels of regulation of hTR, involving the function of several enzymes in processing its 3' end. These enzymes control the rate of hTR maturation and, in turn, dictate its steady-state levels. The maturation of hTR is dictated by the core dyskerin complex, which binds the hTR H/ACA domain. Thus, the evolution of an H/ACA domain in telomerase RNAs in metazoans served to dictate the mechanism of RNA processing, while regulating the levels and protecting the 3' end of hTR. Based on these observations, the dyskerin complex can then be considered a telomerase RNA end-processing and stabilizing complex. This feature is visually evident in the cryo-electron microscopy structure of human telomerase, which shows the core dyskerin complex to be distant from the catalytic site of TERT. The hTR-processing enzymes PAPD5 and PARN can modulate the levels of hTR, which represents a potential therapeutic avenue for individuals with TBD. Other potential means of modulating telomerase activity include telomerase assembly, recruitment to telomeres or using catalytic activators and inhibitors.
Telomerase upregulation is nearly universal in diverse cancer types but is now best understood in cancers with activating mutations in the TERT promoter. These simple non-coding mutations create a new consensus binding site for the transcription factor GABP, and thus targeting GABP has been suggested as a means for inhibiting TERT in this subset of cancers. However, the majority of human cancers lack TERT promoter mutations, and therefore control of TERT transcription in cancer continues to remain poorly understood. Advances in this area could improve our understanding of TERT regulation in cancer and provide new avenues for cancer therapy.
Acknowledgements
This work was supported by NIH grants CA197563 and AG056575 to S.E.A. C.M.R. was supported by MSTP Training Grant GM007365 and by a Gerald J. Lieberman Fellowship.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palm W & de Lange T How shelterin protects mammalian telomeres. Annu. Rev. Genet. 41, 301–334 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Lazzerini-Denchi E & Sfeir A Stop pulling my strings — what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol. 17, 364–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grolimund L et al. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun. 4, 2848 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Bodnar AG et al. Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Hayflick L & Moorhead PS The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961). [DOI] [PubMed] [Google Scholar]
- 6.Allsopp RC et al. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA 89, 10114–10118(1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiba K et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 357, 1416–1420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romanov SR et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409, 633–637 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Kiyono T et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396, 84–88 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Weng NP, Levine BL, June CH & Hodes RJ Regulated expression of telomerase activity in human T lymphocyte development and activation. J. Exp. Med. 183, 2471–2479 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HW et al. Essential role of mouse telomerase in highly proliferative organs. Nature 392, 569–574 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Wright WE, Piatyszek MA, Rainey WE, Byrd W & Shay JW Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Chiu CP et al. Differential expression of telomerase activity in hematopoietic progenitors from adult human bone marrow. Stem Cell 14, 239–248 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Chiba K et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 4, e07918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prowse KR & Greider CW Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl Acad. Sci. USA 92, 4818–4822 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier B et al. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2, e18 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare I I. et al. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell 160, 1013–1026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pech MF et al. High telomerase is a hallmark of undifferentiated spermatogonia and is required for maintenance of male germ line stem cells. Genes Dev. 29, 2420–2434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbuzov A et al. Purification of GFRα1+ and GFRα1spermatogonial stem cells reveals a niche-dependent mechanism for fate determination. Stem Cell Rep. 10, 553–567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery RK et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl Acad. Sci. USA 108, 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Artandi SE et al. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc. Natl Acad. Sci. USA 99, 8191–8196 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allsopp RC, Morin GB, DePinho R, Harley CB & Weissman IL Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood 102, 517–520(2003). [DOI] [PubMed] [Google Scholar]
- 25.Gunes C & Rudolph KL The role of telomeres in stem cells and cancer. Cell 152, 390–393 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Aubert G Telomere dynamics and aging. Prog. Mol. Biol. TransI Sci 12 5, 89–111 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Amit M et al. Clonally derived human embryonic stem cell lines maintain piuripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 227, 271–278 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Yui J, Chiu CP & Lansdorp PM Telomerase activity in candidate stem cells from fetal liver and adult bone marrow. Blood 91,3255–3262 (1998). [PubMed] [Google Scholar]
- 29.Morrison SJ, Prowse KR, Ho P & Weissman IL Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity 5, 207–216(1996). [DOI] [PubMed] [Google Scholar]
- 30.Schepers AG, Vries R, van den Born M, van de Wetering M & Clevers H Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBOJ. 30, 1104–1109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wick M, Zubov D & Hagen G Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 232, 97–106(1999). [DOI] [PubMed] [Google Scholar]
- 32.Takakura M et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 59, 551–557 (1999). [PubMed] [Google Scholar]
- 33.Horikawa I, Cable PL, Afshari C & Barrett JC Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 59, 826–830 (1999). [PubMed] [Google Scholar]
- 34.Wu KJ et al. Direct activation of TERT transcription by c-MYC. Nat. Genet. 21,220–224 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Greenberg RA et al. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 18, 1219–1226(1999). [DOI] [PubMed] [Google Scholar]
- 36.Goueli BS & Janknecht R Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene 22, 8042–8047 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Kyo S et al. Spl cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 28, 669–677 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harle-Bachor C & Boukamp P Telomerase activity in the regenerative basal layer of the epidermis inhuman skin and in immortal and carcinoma-derived skin keratinocytes. Proc. Natl Acad. Sci. USA 93, 6476–6481 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes NM et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell 10, 761–768 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachand F, Kukolj G & Autexier C Expression of hTERT and hTR in cis reconstitutes and active human telomerase ribonucleoprotein. RNA 6, 778–784 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinrich SL et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17, 498–502 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Beattie TL, Zhou W, Robinson MO & Harrington L Reconstitution of human telomerase activity in vitro. Curr. Biol. 8, 177–180 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Feng J et al. The RNA component of human telomerase. Science 269, 1236–1241 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Blasco MA, Funk W, Villeponteau B & Greider CW Functional characterization and developmental regulation of mouse telomerase RNA. Science 269, 1267–1270(1995). [DOI] [PubMed] [Google Scholar]
- 45.Kiss T, Fayet E, Jady BE, Richard P & Weber M Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 71,407–41 7 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Mitchell JR, Cheng J & Collins K A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3' end. Mol. Cell Biol. 19, 567–576 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matera AG, Terns RM & Terns MP Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Kim NK, Theimer CA, Mitchell JR, Collins K & Feigon J Effect of pseudo uridyl ation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 38, 6746–6756(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen THD et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190–195(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venteicher AS et al. A human telomerase holoenzyme protein required forCajal body localization and telomere synthesis. Science 323, 644–648 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamonnak N et al. Yeast Nrd 1, Nab3, and Sen 1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17, 2011–2025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuehner JN, Pearson EL & Moore C Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12, 283–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noel JF, Larose S, Abou Elela S & Wellinger RJ Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res. 40, 5625–5636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baillat D et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Rubtsova MP et al. Integrator is a key component of human telomerase RNA biogenesis. Sci. Rep. 9, 1701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stadelmayer B et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat. Commun. 5, 5531–5531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tudek A et al. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol. Cell 55, 467–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lubas M et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Schilders G, van Dijk E & Pruijn GJ C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 35, 2564–2572 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldfarb KC & Cech TR 3' terminal diversity of MRP RNA and other human noncoding RNAs revealed by deep sequencing. BMC Mol. Biol. 14, 23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roake CM et al. Disruption of telomerase RNA maturation kinetics precipitates disease. Mol. Cell 74, 688–700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tseng CK, Wang HF, Schroeder MR & Baumann P The H/ACA complex disrupts triplex in hTR precursor to permit processing by RRP6 and PARN. Nat. Commun 9, 5430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tseng CK et al. Human telomerase RNA processing and quality control. Cell Rep. 13, 2232–2243 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Moon DH et al. Poly(A)-specific ribonuclease (PARN) mediates 3'-end maturation of the telomerase RNA component. Nat. Genet. 47, 1482–1488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henriksson N, Nilsson P, Wu M, Song H & Virtanen A Recognition of adenosine residues by the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 285, 163–170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu M et al. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 24, 4082–4093 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lardelli RM et al. Biallelic mutations in the 3' exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat. Genet. 49, 457–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Son A, Park JE & Kim VN PARN and TOE1 constitute a 3' end maturation module for nuclear non-coding RNAs. Cell Rep. 23, 888–898 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Deng T et al. TOE1 acts as a 3' exonuclease for telomerase RNA and regulates telomere maintenance. Nucleic Acids Res. 47, 391–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen D et al. A polyadenylation-dependent 3' end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 13, 2244–2257 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Shukla S, Schmidt JC, Goldfarb KC, Cech TR & Parker R Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat. Struct. Mol. Biol. 23, 286–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boyraz B et al. Posttranscriptional manipulation of TERC reverses molecular hallmarks of telomere disease. J. Clin. Invest. 126, 3377–3382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fok WC et al. Posttranscriptional modulation of TERC by PAPD5 inhibition rescues hematopoietic development in dyskeratosis congenita. Blood 133, 1308–1312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mouaikel J, Verheggen C, Bertrand E, Tazi J & Bordonne R Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9, 891–901 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Tang W, Kannan R, Blanchette M & Baumann P Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484, 260–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen L et al. Loss of human TGS1 hypermethylase promotes increased telomerase RNA and telomere elongation. Cell Rep. 30, 1358–1372.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greider CW & Blackburn EH Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell 43, 405–413 (1985). [DOI] [PubMed] [Google Scholar]
- 78.Lingner J & Cech TR Purification of telomerase from euplotes aediculatus: requirement of a primer 3' overhang. Proc. Natl Acad. Sci. USA 93, 10712–10717 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakamura TM et al. Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959 (1997). [DOI] [PubMed] [Google Scholar]
- 80.Tesmer VM et al. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell Biol. 19, 6207–6216 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitchell JR & Collins K Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6, 361–371 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Jiang J et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350, aab4070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang J et al. Structure of telomerase with telomeric DNA. Cell 173, 1179–1190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen JL, Opperman KK & Greider CW A critical stem–loop structure in the CR4–CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 30, 592–597 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robart AR & Collins K Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J. Biol. Chem. 285, 4375–4386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schnapp G, Rodi HP, Rettig WJ, Schnapp A & Damm K One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 26, 3311–3313 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xin H et al. TPP1 is a homologue of ciliate TEBP-ß and interacts with POT1 to recruit telomerase. Nature 445, 559–562 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Venteicher AS, Meng Z, Mason PJ, Veenstra TD & Artandi SE Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cohen SB et al. Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Fu D & Collins K Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Egan ED & Collins K An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol. Cell Biol. 32, 2428–2439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Egan ED & Collins K Biogenesis of telomerase ribonucleoproteins. RNA 18, 1747–1759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jady BE, Bertrand E & Kiss T Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM & Terns MP Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell 15, 81–90 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang BD et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 59, 3761–3767 (1999). [PubMed] [Google Scholar]
- 96.Darzacq X et al. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 173, 207–218 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang C & Meier UT Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23, 1857–1867 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M & Henry Y hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 12, 832–840 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grozdanov PN, Roy S, Kittur N & Meier UT SHQ1 is required prior to NAF1 for assembly of H/ ACA small nucleolar and telomerase RNPs. RNA 15, 1188–1197 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Holt SE et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13, 817–826 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Toogun OA, Dezwaan DC & Freeman BC The hsp90 molecular chaperone modulates multiple telomerase activities. Mol. Cell. Biol. 28, 457–467 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Keppler BR, Grady AT & Jarstfer MB The biochemical role of the heat shock protein 90 chaperone complex in establishing human telomerase activity. J. Biol. Chem. 281, 19840–19848 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Freund A et al. Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 159, 1389–1403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cristofari G et al. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell 27, 882–889 (2007). [DOI] [PubMed] [Google Scholar]
- 105.Stern JL, Zyner KG, Pickett HA, Cohen SB & Bryan TM Telomerase recruitment requires both TCAB1 and cajal bodies independently. Mol. Cell Biol. 32, 2384–2395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhong FL et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhong F et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 25, 11–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tycowski KT, Shu MD, Kukoyi A & Steitz JA A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell 34, 47–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmidt JC, Zaug AJ & Cech TR Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197 e1189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen L et al. An activity switch in human telomerase based on RNA conformation and shaped by TCAB1. Cell 174, 218–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tomlinson RL, Li J, Culp BR, Terns RM & Terns MP A Cajal body-independent pathway for telomerase trafficking in mice. Exp. Cell Res. 316, 2797–2809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Broccoli D, Smogorzewska A, Chong L & de Lange T Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 (1997). [DOI] [PubMed] [Google Scholar]
- 113.Chong L et al. A human telomeric protein. Science 270, 1663–1667 (1995). [DOI] [PubMed] [Google Scholar]
- 114.Baumann P & Cech TR Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 (2001). [DOI] [PubMed] [Google Scholar]
- 115.Abreu E et al. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell Biol. 30, 2971–2982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nandakumar J et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sexton AN et al. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 28, 1885–1899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kocak H et al. Hoyeraal-Hreidarsson syndrome caused by a germline mutation in the TEL patch of the telomere protein TPP1. Genes Dev. 28, 2090–2102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Houghtaling BR, Cuttonaro L, Chang W & Smith S A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631 (2004). [DOI] [PubMed] [Google Scholar]
- 120.Ye JZ et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu D et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680 (2004). [DOI] [PubMed] [Google Scholar]
- 122.Kim SH, Kaminker P & Campisi J TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Takai KK, Kibe T, Donigian JR, Frescas D & de Lange T Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44, 647–659 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Savage SA et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 82, 501–509 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Walne AJ, Vulliamy T, Beswick R, Kirwan M & Dokal I TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 112, 3594–3600 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Frank AK et al. The shelterin TIN2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet. 11, e1005410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang D, He Q, Kim H, Ma W & Songyang Z TIN2 protein dyskeratosis congenita missense mutants are defective in association with telomerase. J. Biol. Chem. 286, 23022–23030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frescas D & de Lange T A TIN2 dyskeratosis congenita mutation causes telomerase-independent telomere shortening in mice. Genes Dev. 28, 153–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miyake Y et al. RPA-like mammalian Ctc1–Stn1–Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206 (2009). [DOI] [PubMed] [Google Scholar]
- 130.Surovtseva YV et al. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Casteel DE et al. A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 284, 5807–5818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen LY, Redon S & Lingner J The human CST complex is a terminator of telomerase activity. Nature 488, 540–544 (2012). [DOI] [PubMed] [Google Scholar]
- 133.Nandakumar J & Cech TR Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14, 69–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jady BE, Richard P, Bertrand E & Kiss T Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 17, 944–954 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM & Terns MP Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vogan JM et al. Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. eLife 5, e18221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Loayza D & de Lange T POT1 as a terminal transducer of TRF1 telomere length control. Nature 424, 1013–1018 (2003). [DOI] [PubMed] [Google Scholar]
- 138.Latrick CM & Cech TR POT1–TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29, 924–933 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang F et al. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 (2007). [DOI] [PubMed] [Google Scholar]
- 140.Zaug AJ, Podell ER, Nandakumar J & Cech TR Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24, 613–622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pike AM, Strong MA, Ouyang JPT & Greider CW TIN2 functions with TPP1/POT1 to stimulate telomerase processivity. Mol. Cell Biol. 39, e00593–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Greider CW & Blackburn EH Telomeres, telomerase and cancer. Sci. Am. 274, 92–97 (1996). [DOI] [PubMed] [Google Scholar]
- 143.Counter CM et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11, 1921–1929 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim NW et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994). [DOI] [PubMed] [Google Scholar]
- 145.Takai H, Smogorzewska A & de Lange T DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 (2003). [DOI] [PubMed] [Google Scholar]
- 146.d’Adda di Fagagna F et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 (2003). [DOI] [PubMed] [Google Scholar]
- 147.Artandi SE et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641–645 (2000). [DOI] [PubMed] [Google Scholar]
- 148.Davoli T, Denchi EL & de Lange T Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141, 81–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nassour J et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565, 659–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Horn S et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013). [DOI] [PubMed] [Google Scholar]
- 151.Killela PJ et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl Acad. Sci. USA 110, 6021–6026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang FW et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kinde I et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 73, 7162–7167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vinagre J et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun 4, 2185 (2013). [DOI] [PubMed] [Google Scholar]
- 155.Weinhold N, Jacobsen A, Schultz N, Sander C & Lee W Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 10.1038/ng.3101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Borah S et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347, 1006–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bell RJ et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N & Cech TR Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 29, 2219–2224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xi L, Schmidt JC, Zaug AJ, Ascarrunz DR & Cech TR A novel two-step genome editing strategy with CRISPR–Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 16, 231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Akincilar SC et al. Long-range chromatin interactions drive mutant TERT promoter activation. Cancer Discov. 6, 1276–1291 (2016). [DOI] [PubMed] [Google Scholar]
- 161.Killela PJ et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl Acad. Sci. USA 110, 6021–6026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nault JC et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 4, 2218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shain AH et al. The genetic evolution of melanoma from precursor lesions. N. Engl. J. Med. 373, 1926–1936 (2015). [DOI] [PubMed] [Google Scholar]
- 164.Heidenreich B, Rachakonda PS, Hemminki K & Kumar R TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 24, 30–37 (2014). [DOI] [PubMed] [Google Scholar]
- 165.Ferber MJ et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 22, 3813–3820 (2003). [DOI] [PubMed] [Google Scholar]
- 166.Paterlini-Bréchot P et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22, 3911–3916 (2003). [DOI] [PubMed] [Google Scholar]
- 167.Bellon M & Nicot C Regulation of telomerase and telomeres: human tumor viruses take control. J. Natl Cancer Inst. 100, 98–108 (2008). [DOI] [PubMed] [Google Scholar]
- 168.Kawai-Kitahata F et al. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J. Gastroenterol. 51, 473–486 (2016). [DOI] [PubMed] [Google Scholar]
- 169.Valentijn LJ et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet. 47, 1411–1414 (2015). [DOI] [PubMed] [Google Scholar]
- 170.Peifer M et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526, 700–704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Barthel FP et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 49, 349–357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pickett HA & Reddel RR Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat. Struct. Mol. Biol. 22, 875–880 (2015). [DOI] [PubMed] [Google Scholar]
- 173.Dilley RL & Greenberg RA ALTernative telomere maintenance and cancer. Trends Cancer 1, 145–156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Soder AI et al. Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene 14, 1013–1021 (1997). [DOI] [PubMed] [Google Scholar]
- 175.Sugita M et al. Molecular definition of a small amplification domain within 3q26 in tumors of cervix, ovary, and lung. Cancer Genet. Cytogenet. 117, 9–18 (2000). [DOI] [PubMed] [Google Scholar]
- 176.Yuan X, Larsson C & Xu D Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 38, 6172–6183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim W et al. Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): implications for aging and cancer. PLoS Biol. 14, e2000016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Armanios M & Blackburn EH The telomere syndromes. Nat. Rev. Genet. 13, 693–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Brown Y et al. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 35, 6280–6289 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Trahan C & Dragon F Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. RNA 15, 235–243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K & Cech TR Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 41, 8969–8978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mitchell JR, Wood E & Collins K A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999). [DOI] [PubMed] [Google Scholar]
- 183.Vulliamy TJ et al. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood 107, 2680–2685 (2006). [DOI] [PubMed] [Google Scholar]
- 184.Stanley SE et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci. Transl Med 8, 351ra107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Walne AJ et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 16, 1619–1629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Stuart BD et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet. 47, 512–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Batista LF et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 474, 399–402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Walne AJ et al. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica 98, 334–338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Walne AJ, Vulliamy T, Kirwan M, Plagnol V & Dokal I Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am. J. Hum. Genet. 92, 448–453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vulliamy T et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413, 432–435 (2001). [DOI] [PubMed] [Google Scholar]
- 191.Vulliamy T, Marrone A, Dokal I & Mason PJ Association between aplastic anaemia and mutations in telomerase RNA. Lancet 359, 2168–2170 (2002). [DOI] [PubMed] [Google Scholar]
- 192.Armanios MY et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 356, 1317–1326 (2007). [DOI] [PubMed] [Google Scholar]
- 193.Yamaguchi H et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 352, 1413–1424 (2005). [DOI] [PubMed] [Google Scholar]
- 194.Armanios M et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl Acad. Sci. USA 102, 15960–15964 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Heiss NS et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19, 32–38 (1998). [DOI] [PubMed] [Google Scholar]
- 196.Alder JK et al. Diagnostic utility of telomere length testing in a hospital-based setting. Proc. Natl Acad. Sci. USA 115, E2358–e2365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Anderson BH et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44, 338–342 (2012). [DOI] [PubMed] [Google Scholar]
- 198.Vulliamy T et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl Acad. Sci. USA 105, 8073–8078 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Guo Y et al. Inherited bone marrow failure associated with germline mutation of ACD, the gene encoding telomere protein TPP1. Blood 124, 2767–2774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gable DL et al. ZCCHC8, the nuclear exosome targeting component, is mutated in familial pulmonary fibrosis and is required for telomerase RNA maturation. Genes Dev. 33, 1381–1396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]