Abstract
Synthetic or natural derived cell-penetrating peptides (CPPs) are vastly investigated as tools for the intracellular delivery of membrane-impermeable molecules. As viruses are intracellular obligate parasites, viral originated CPPs have been considered as suitable intracellular shuttling vectors for cargo transportation. A total of 310 CPPs were identified in the proteome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Screening the proteome of the cause of COVID-19 reveals that SARS-CoV-2 CPPs (SCV2-CPPs) span the regions involved in replication, protein-nucleotide and protein-protein interaction, protein-metal ion interaction, and stabilization of homo/hetero-oligomers. However, to find the most appropriate peptides as drug delivery vectors, one might face several hurdles. Computational analyses showed that 94.3% of the identified SCV2-CPPs are non-toxins, and 38% are neither antigenic nor allergenic. Interestingly, 36.70% of SCV2-CPPs were resistant to all four groups of protease families. Nearly 1/3 of SCV2-CPPs had sufficient inherent or induced helix and sheet conformation leading to increased uptake efficiency. Heliquest lipid-binding discrimination factor revealed that 44.30% of the helical SCV2-CPPs are lipid-binding helices. Although Cys-rich derived CPPs of helicase (NSP13) can potentially fold into a cyclic conformation in endosomes with a higher rate of endosomal release, the most optimal SCV2-CPP candidates as vectors for drug delivery were SCV2-CPP118, SCV2-CPP119, SCV2-CPP122, and SCV2-CPP129 of NSP12 (RdRp). Ten experimentally validated viral-derived CPPs were also used as the positive control to check the scalability and reliability of our protocol in SCV2-CPP retrieval. Some peptides with a cell-penetration ability known as bioactive peptides are adopted as biotherapeutics themselves. Therefore, 59.60%, 29.63%, and 32.32% of SCV2-CPPs were identified as potential antibacterial, antiviral, and antifungals, respectively. While 63.64% of SCV2-CPPs had immuno-modulatory properties, 21.89% were recognized as anti-cancers. Conclusively, the workflow of this study provides a platform for profound screening of viral proteomes as a rich source of biotherapeutics or drug delivery carriers.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, Cell-penetrating peptide, Drug delivery, Bioactive peptide
Abbreviations: a.a, amino acid; ACP, anticancer peptide; AH, amphipathic helix; AIP, anti-inflammatory peptide; AMP, antimicrobial peptide; CoV, coronavirus; COVID, coronavirus disease; CPP, cell-penetrating peptide; ERGIC, endoplasmic reticulum-Golgi intermediate compartment; NSP, nonstructural protein; RTC, replication transcription complex; SCV, severe acute respiratory syndrome coronavirus; VLP, virus-like particle
Graphical abstract
1. Introduction
The origin of the first introduced cell-penetrating peptide (CPP) is back to the crucial role of the TAT protein of HIV-1 as a trans-activator for transcription, which stimulates viral infection and immunosuppression (Frankel and Pabo, 1988). Thereafter, CPPs were defined as 4–40 amino acid short peptides, some of which are involved in the pathogenesis of viruses. For example, the presence of a conserved cationic CPP at the C-terminus of human papillomavirus (HPV) L2 protein participates in the intracellular viral trafficking (Zhang et al., 2018).
The cell membrane acts as a hydrophobic obstacle for drug delivery; hence, a meaningful number of viral derived CPPs have been used to facilitate the internalization of cargos through the host cell membrane. Cargos are defined as conjugated or non-conjugated macromolecules such as proteins, small organic molecules, and nucleic acids. Peptides including pepR and pepM derived from dengue virus capsid protein (Freire et al., 2014), Xentry and X-pep acquired from the X-protein of hepatitis B virus (Montrose et al., 2014), FHV originated peptide from flock house virus coat protein (Nakase et al., 2009), and VG-21 from vesicular stomatitis virus glycoprotein (Tiwari et al., 2014) are some of the examples of viral derived CPPs providing cell permeability for the added cargo molecule. Since viruses are intracellular obligate parasites, some CPPs have also been used for intracellular shuttling of antiviral medications such as the conjugated form of an antisense peptide nucleic acid with the TAT peptide applied for suppressing SARS-CoV-1 replication (Ahn et al., 2011). The other notable aspect is the biological properties of CPPs as bioactive peptides, including antimicrobial, anti-inflammatory, and anticancer properties (Langel, 2019).
Despite the aforementioned applications, proteolytic cleavage of peptides with proteases, metabolic instability, adverse off-target side effects, probable allergenicity, immunogenicity or toxicity (at high concentrations or during prolonged administration) are potential drawbacks of some CPPs. Endosomal entrapment or degradation in the process of endosome maturation is another interruption during the administration of some other CPPs (Reissmann, 2014). Since safety, stability, and specificity of peptides for various utilizations is of great value, there is always the need to introduce novel natural or synthetic CPPs. As protein derived penetrating peptides constitute about 80–90% of CPPs, it is arguable that huge quantities of CPPs are hidden in protein sequences (Fu et al., 2019). Screening the proteome of the Coronaviridae family for this purpose was somehow neglected until the recent pandemic. In parallel to the conventional high throughput screening for CPP discovery by wet-lab methods, the primary prediction of CPPs using validated computational analyses is growing research focus, especially when we encounter high numbers of proteins or mining proteome resources. The latter approach would lead to high authentic hits, which can be proved with an increased success rate than the former strategy (Gautam et al., 2015). Machine-learning based prediction methods are amenable to scan protein sequences for the presence of CPPs. Some of the most frequent models of the current prediction methods are support vector machine (SVM), neural network (NN), random forest (RF), kernel extreme learning machine (KELM), and extremely randomized tree (ERT). However, few methods are provided as freely available web interfaces. Different predictions based on SVM, NN, RF, KELM, and ERT models have accuracies up to 97.4%, 83% 90.6%, 83.1%, and 89.6%, respectively (Su et al., 2020). There are various indicators such as sensitivity, specificity, accuracy, receiver operating characteristic, and Matthew's correlation coefficient to assess the performance of the models. Then statistical methods such as jackknife test, k-fold cross validation, and independent test are used to verify indicators in different predictor models (Fu et al., 2019).
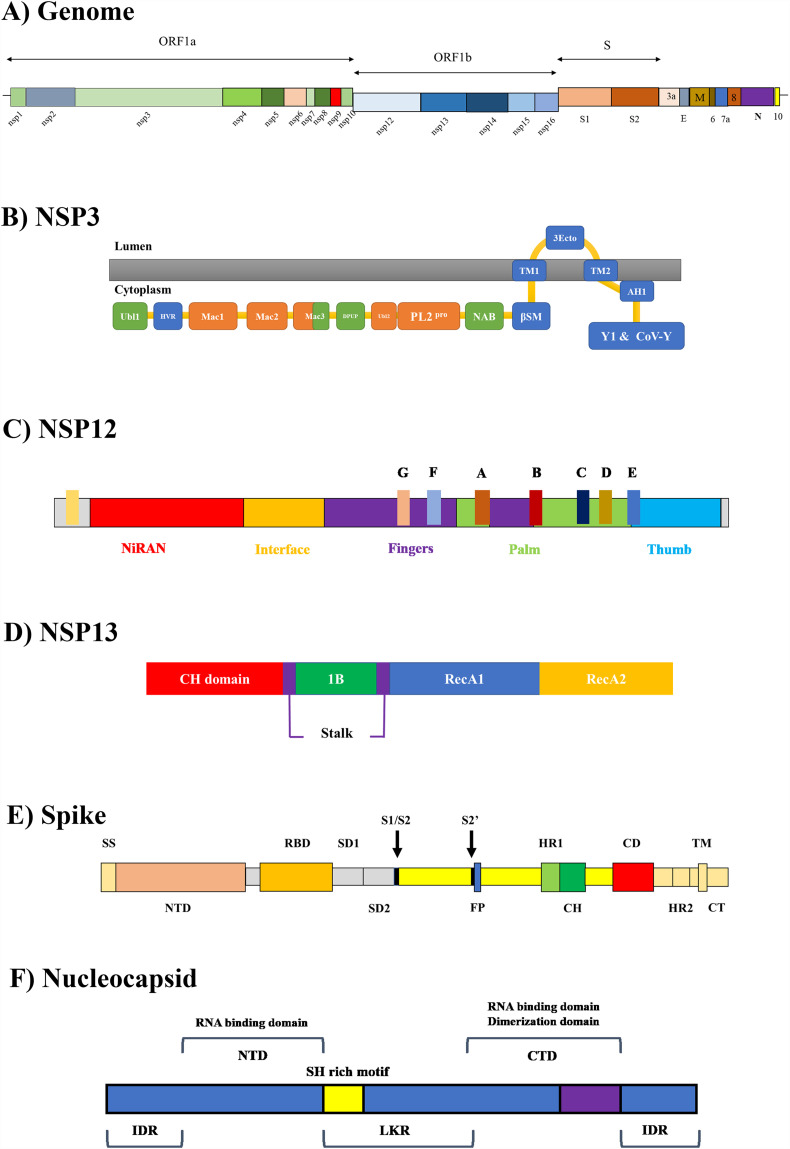
The 2019-novel coronavirus or 2019-nCoV latter annotated as severe acute respiratory syndrome coronavirus (SARS-CoV-2) belongs to the Coronaviridae family. SARS-CoV-2 led to a pandemic known as COVID-19 infecting the upper respiratory system with pneumonia-like symptoms (Chan et al., 2020). SARS-CoV-2 has a 5′ cap structure and 3′ polyA tail in its positive-sense single-stranded RNA (+ssRNA). The longer part of the genome proximal to the 5′ end (replicase gene) with two ORFs (ORF1a and ORF1b) encodes two overlapping polyproteins (pp) known as pp1a and pp1ab due to a (−1) ribosomal frameshifting (Fig. 1A). While pp1a harbors ten nonstructural proteins (NSP) known as NSP1–10, the longer pp1ab contains 15 NSPs displaying as NSP1–10 and NSP12–NSP16 (Wu et al., 2020). The formation of mature NSPs is achieved mainly through the processing of pp1a/pp1ab by two viral proteases encoded by NSP3 and NSP5 known as papain-like protease (PLpro) and chymotrypsin-like cysteine protease (3CLpro), which process N- and C-terminal regions of pps, respectively (Chen et al., 2020). The 1/3 of the coronavirus virion proximal to the 3′ region contains ORFs leastwise encoding four structural proteins in S-E-M-N order, which are Spike, Envelope, Membrane, and Nucleocapsid, respectively. Other accessory and structural proteins are also translated from subgenomic RNAs (sgRNA) of SARS-CoV-2. Spike as a fusion protein is associated with the cellular entrance of the virus and consists of two subunits, namely S1 and S2 for the attachment to the cellular receptor and viral fusion to the membrane, respectively (Coutard et al., 2020). The cleavage of S protein is initiated by a host cellular serine protease (TMPRSS2). It seems that in SARS-CoV-2 acquisition of a new multi-basic arginine-rich cleavage site at the S1/S2 boundary —absent in other SARS-CoVs— has facilitated the S protein processing for entrance into the host cells (Hoffmann et al., 2020). Such a furin-like cleavage site was also admitted in the H5N1 hemagglutinin HA cleavage site (addition of a multibasic motif “RERRRKKR↓GL”) throughout the Hong Kong 1997 outbreak (Braun and Sauter, 2019).
Fig. 1.
A) A schematic model of the SARS-CoV-2 genome (Chen et al., 2020; Coutard et al., 2020; Wu et al., 2020). B–D) Domain organization of nonstructural proteins including, NSP3, NSP12, and NSP13, respectively. E–F) The domain organization of spike and nucleocapsid as structural proteins.
The emergence of SARS-CoV-2 with a distressing transmission and mortality needs to shed light on all aspects at the molecular level. Various studies have performed rigorous comparative genomic studies to reveal the ancestral origin and unique mutations in SARS-CoV-2 compared with the other members of the family (Lam et al., 2020). Enveloped viruses sequester their virion into the cytoplasm through membrane fusion (White and Whittaker, 2016). Peptides derived from such proteins imitate the role of their parent proteins for cargo delivery. The presence of an arginine-rich peptide at the N-terminal of S2 subunit (Arg-rich peptides are efficient CPPs), has urged us to screen the proteome of SARS-CoV-2 for penetrating peptides. In the current study, finding and characterization of CPPs not only in novel insertions but also in the whole proteome of the virus was of paramount concern. To introduce the good out of the bad and the ugly, such CPPs could be mediators for potential drug delivery like the well-known TAT (Sadeghian et al., 2018) or act as a bioactive peptide by themselves. This investigation might also improve our understanding of replication, transcription, and pathogenesis of the virus.
2. Material and methods
2.1. Determination of potential cell-penetrating sequences in the proteome of SARS-CoV-2
Twenty-four proteins from SARS-CoV-2 proteome including nonstructural proteins 1–16, four major structural proteins (S, E, M, N), and accessory proteins encoded by the genome of SARS- CoV-2 isolate Wuhan-Hu-1 (GenBank ID# MN908947.3) were submitted to the protein scanning tool in CellPPD web server (http://crdd.osdd.net/raghava/cellppd/) (Gautam et al., 2015). Prediction models on various CellPPD modules are either SVM-based or SVM-Motif-based. The confidence of CPP prediction in these models can be easily determined by employing filters such as threshold and e-value. Higher threshold and/or lower e-values increase the confidence of CPP prediction in protein sequences. The SVM prediction method was selected to mine the SARS-CoV-2 proteome with a motif e-value of 10 and an SVM score threshold of 0.0. CellPPD web server is unique among other CPP identification web servers since it can predict all possible overlapping peptides with a specified length in a protein sequence and evaluate each sequence separately to verify whether it has cell penetration ability or not.
Furthermore, several experimentally validated viral-derived CPPs including NLS-A from porcine circovirus 2 (PCV2) (Yu et al., 2018), FHV coat (35–49) from flock house virus (FHV) (Futaki et al., 2001), pepR from dengue virus capsid protein (DENVC) (Freire et al., 2014), VP22 from herpes simplex virus 1 (HSV-1) (Elliott and O'Hare, 1997), TAT and REV from human immunodeficiency virus 1 (HIV-1) (Futaki et al., 2001), Erns from classical swine fever virus (CSFV) (Langedijk, 2002), HPV-WT from human papillomavirus (HPV) (Zhang et al., 2018), chimeric Pep1 originated from simian virus (SV40) (Morris et al., 2001), and VG-21 from vesicular stomatitis virus (VSV) (Tiwari et al., 2014) were analyzed by CellPPD webserver as positive controls using the above-mentioned settings. The cell permeability of most of these CPPs has been validated using at least one cell line, and in some instances such as TAT and Pep1 by in vivo models as well.
2.2. Evaluation of uptake efficiency of SARS-CoV-2 CPPs (SCV2-CPPs)
The MLCPP web server (http://www.thegleelab.org/MLCPP/) was applied to predict the uptake efficiency of 310 potential cell-penetrating sequences in the proteome of SARS-CoV-2 (Manavalan et al., 2018b). This web server implements a two-layered prediction framework, which in its first layer the submitted peptides are classified into two groups of CPPs and non-CPPs. In the next layer, peptides are divided into CPPs with high and low uptake efficiencies. A prediction confidence score between 0.0 and 1.0 is provided for each layer. Zero is accounting for the least confident, and one is considering for the most confident predictions. The output depends on characteristics such as amino acid, dipeptide, and atomic compositions, as well as the physiochemical properties of the peptide sequence. The uptake efficiency of experimentally validated viral-derived CPPs as positive controls also evaluated using the MLCPP server.
2.3. Calculation of physiochemical parameters of SCV2-CPPs
Several physiochemical parameters such as molecular weight (Mw), theoretical isoelectric point (pI), instability index, and Grand average of hydropathicity (GRAVY) were computed for the CPP sequences using ProtParam tool on the EXPASY server (https://web.expasy.org/protparam/). The sequences were then submitted to the Pepcalc tool (https://pepcalc.com/) to estimate their water solubility. Pepcalc server classifies peptides as either good or poor water-soluble.
2.4. Analyses regarding in vivo administration of SCV2-CPPs
To investigate in vivo behaviors of the identified peptides, several web servers such as Toxinpred (Gupta et al., 2015) (https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php), Plifepred (Mathur et al., 2018) (https://webs.iiitd.edu.in/raghava/plifepred/batch.php), Vaxijen V 2.0 (Zaharieva et al., 2017) (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), HemoPI (Chaudhary et al., 2016) (https://webs.iiitd.edu.in/raghava/hemopi/multiple_test.php), Allertop v.2 (Dimitrov et al., 2014) (https://www.ddg-pharmfac.net/AllerTOP/), and Prosper (Song et al., 2012) (https://prosper.erc.monash.edu.au/) were used to predict the toxicity, half-life, antigenicity, hemolytic potency, allergenicity, and protease susceptibility of SCV2-CPPs, respectively. The most probable disulfide bonds were predicted using DiANNA web server (http://bioinformatics.bc.edu/clotelab/DiANNA/) (Ferrè and Clote, 2005).
2.5. Secondary structure and membrane interaction prediction of SCV2-CPPs
Amphipathic CPPs form α-helices and/or β-sheets in solution or after interaction with membrane. The formation of these secondary structures enhances membrane transduction efficiency. Investigation of the CPPs' secondary structures was conducted using the PEP2D web server (http://crdd.osdd.net/raghava/pep2d/submit2.html). To predict the effect of membrane on the folding of CPPs, sequences were further submitted to the fmap server (https://membranome.org/fmap) using peptide in the membrane model (Lomize et al., 2017). This model predicts the formation of individual stable α-helical regions of peptides in the lipid bilayer. Experimental conditions were set to a temperature of 310 K and a pH of 7.4.
2.6. Determination of lipid-binding potential of helical SCV2-CPPs
SARS-CoV-2 CPPs with more than 50% α-helical conformation as well as several experimentally validated viral-derived CPPs mentioned in section 2.1 were submitted to the Heliquest web server (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py). For peptides shorter and longer than 18 amino acids in length, the FULL and 1-TURN window size was adjusted, respectively. Using Heliquest several attributes such as hydrophobic moment (μH) and net charge (z) were calculated per peptide (Gautier et al., 2008). These values were applied to calculate the discrimination factor (D) for each peptide. Peptides with discrimination factors above 0.68 are expected to have membrane-binding potential. Since the Heliquest server considers any submitted sequence as helical, only SCV2-CPPs with at least 50% helicity were used for this analysis. All the helical wheel illustrations of this study are drawn using Heliquest.
2.7. Further potential bioactivities of SCV2-CPPs
Predicted SCV2-CPPs were studied for other bioactivities using bioinformatics tools. The SVM based iAMPpred server (http://cabgrid.res.in:8080/amppred/) was used to predict antibacterial, antifungal, and antiviral probabilities for each peptide. If a peptide has a score above 0.5 in a category, it can be considered to potentially have the respective bioactivity. The potential anticancer activity prediction of SCV2-CPPs was conducted using the iACP server (http://server.malab.cn/ACPred-FL/ProcessServlet) (Chen et al., 2016). Furthermore, the AIPpred server (http://www.thegleelab.org/AIPpred/) was used to determine the anti-inflammatory effects of the peptides (Manavalan et al., 2018a).
3. Results and discussion
3.1. Determination of potential cell-penetrating regions in the proteome of SARS-CoV-2
The proteome of SARS-CoV-2 was analyzed using the protein scanning tool of the CellPPD server. A total of 310 potential CPPs were distinguished in the proteome named SCV2-CPP 1-310 (Table 1 , Table 2 , Supplementary material 1). The number of CPPs (with a length of 10 amino acid residues) in each protein ranges from 0 to 54. Protein N (nucleocapsid protein) and NSP3 with 54 and 51 potential CPPs had the highest number of identified CPPs among all SARS-CoV-2 proteins. No CPPs were identified in NSP7, protein E, and the accessory protein translated from ORF10. For a deeper insight into the distribution of CPPs within each protein, the number of CPPs per 100 amino acids of length was calculated for each protein. Protein N with 12.9 CPP per 100 residues had the most uniform distribution of CPPs among all SARS-CoV-2 proteins. This observation seems reasonable for nucleic acid-binding proteins in viruses to be rich in positively charged residues. These basic residues are usually part of a cationic or amphipathic CPP and interact efficiently with negatively charged groups of RNA (Krüger et al., 2018).
Table 1.
Nonstructural (NSP), structural, and accessory proteins of SARS-CoV-2 and the number of CPPs found in each protein.
| Gene/Protein name | Length | Number of potential CPPs | CPPs per 100 amino acid residues of protein |
|---|---|---|---|
| Nsp1 | 180 | 8 | 4.4 |
| Nsp2 | 638 | 18 | 2.8 |
| Nsp3 (PLpro) | 1945 | 51 | 2.6 |
| Nsp4 | 500 | 11 | 2.2 |
| Nsp5 (3CLpro) | 306 | 6 | 2.0 |
| Nsp6 | 290 | 1 | 0.3 |
| Nsp7 | 83 | 0 | 0.0 |
| Nsp8 | 198 | 11 | 5.6 |
| Nsp9 | 113 | 3 | 2.7 |
| Nsp10 | 139 | 1 | 0.7 |
| Nsp12 (RdRP) | 932 | 23 | 2.5 |
| Nsp13 (Helicase) | 601 | 32 | 5.3 |
| Nsp14 (N7-MTase) | 527 | 6 | 1.1 |
| Nsp15 (NendoU) | 346 | 8 | 2.3 |
| Nsp16 (2’-O MTase) | 298 | 10 | 3.4 |
| S (Surface glycoprotein) | 1273 | 24 | 1.9 |
| ORF3a | 275 | 18 | 6.5 |
| E (Envelope protein) | 75 | 0 | 0.0 |
| M (Membrane protein) | 222 | 14 | 6.3 |
| ORF6 | 61 | 1 | 1.6 |
| ORF7a | 121 | 7 | 5.8 |
| ORF8 | 121 | 3 | 2.5 |
| N (Nucleocapsid protein) | 419 | 54 | 12.9 |
| ORF10 | 38 | 0 | 0.0 |
Table 2.
Predicted CPPs defined by CellPPD server and the uptake efficiency (UE) of SARS-CoV-2 CPPs (SCV2-CPP) using the MLCPP server.
| Peptide name | Sequence | UE | Peptide name | Sequence | UE | Peptide name | Sequence | UE | Peptide name | Sequence | UE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SCV2-CPP1 | IKRSDARTAP | Low | SCV2-CPP79 | LAYYFMRFRR | Low | SCV2-CPP157 | NARLRAKHYV | Low | SCV2-CPP235 | KLIFLWLLWP | Low |
| SCV2-CPP2 | KRSDARTAPH | Low | SCV2-CPP80 | AYYFMRFRRA | Low | SCV2-CPP158 | PAPRTLLTKG | Low | SCV2-CPP236 | FIASFRLFAR | Low |
| SCV2-CPP3 | PVAYRKVLLR | High | SCV2-CPP81 | YYFMRFRRAF | High | SCV2-CPP159 | APRTLLTKGT | Low | SCV2-CPP237 | ASFRLFARTR | Low |
| SCV2-CPP4 | VAYRKVLLRK | High | SCV2-CPP82 | FMRFRRAFGE | High | SCV2-CPP160 | FNSVCRLMKT | Low | SCV2-CPP238 | SFRLFARTRS | Low |
| SCV2-CPP5 | AYRKVLLRKN | Low | SCV2-CPP83 | MRFRRAFGEY | High | SCV2-CPP161 | FLGTCRRCPA | High | SCV2-CPP239 | FRLFARTRSM | Low |
| SCV2-CPP6 | YRKVLLRKNG | Low | SCV2-CPP84 | WFFSNYLKRR | High | SCV2-CPP162 | DNKLKAHKDK | Low | SCV2-CPP240 | RLFARTRSMW | Low |
| SCV2-CPP7 | RKVLLRKNGN | Low | SCV2-CPP85 | FSNYLKRRVV | Low | SCV2-CPP163 | KLKAHKDKSA | Low | SCV2-CPP241 | FARTRSMWSF | Low |
| SCV2-CPP8 | KVLLRKNGNK | Low | SCV2-CPP86 | KEMYLKLRSD | – | SCV2-CPP164 | FLTRNPAWRK | Low | SCV2-CPP242 | HGTILTRPLL | Low |
| SCV2-CPP9 | KDLLARAGKA | Low | SCV2-CPP87 | YNRYLALYNK | High | SCV2-CPP165 | RNPAWRKAVF | Low | SCV2-CPP243 | GAVILRGHLR | High |
| SCV2-CPP10 | FEIKLAKKFD | – | SCV2-CPP88 | RYLALYNKYK | High | SCV2-CPP166 | GIPKDMTYRR | Low | SCV2-CPP244 | RIAGHHLGRC | Low |
| SCV2-CPP11 | SIIKTIQPRV | – | SCV2-CPP89 | FRKMAFPSGK | Low | SCV2-CPP167 | DMTYRRLISM | – | SCV2-CPP245 | YSRYRIGNYK | Low |
| SCV2-CPP12 | KTIQPRVEKK | Low | SCV2-CPP90 | TANPKTPKYK | Low | SCV2-CPP168 | FSRVSAKPPP | Low | SCV2-CPP246 | LIIKNLSKSL | Low |
| SCV2-CPP13 | TIQPRVEKKK | Low | SCV2-CPP91 | ANPKTPKYKF | Low | SCV2-CPP169 | SRVSAKPPPG | Low | SCV2-CPP247 | HVYQLRARSV | Low |
| SCV2-CPP14 | IQPRVEKKKL | Low | SCV2-CPP92 | PKTPKYKFVR | Low | SCV2-CPP170 | KINAACRKVQ | Low | SCV2-CPP248 | QLRARSVSPK | Low |
| SCV2-CPP15 | SGLKTILRKG | Low | SCV2-CPP93 | KTPKYKFVRI | Low | SCV2-CPP171 | GNPKAIKCVP | Low | SCV2-CPP249 | RARSVSPKLF | Low |
| SCV2-CPP16 | LKTILRKGGR | Low | SCV2-CPP94 | RWFLNRFTTT | Low | SCV2-CPP172 | ELWAKRNIKP | Low | SCV2-CPP250 | RSVSPKLFIR | Low |
| SCV2-CPP17 | KTILRKGGRT | Low | SCV2-CPP95 | MNSQGLLPPK | Low | SCV2-CPP173 | LWAKRNIKPV | Low | SCV2-CPP251 | ITLCFTLKRK | High |
| SCV2-CPP18 | GNFKVTKGKA | Low | SCV2-CPP96 | SEVVLKKLKK | Low | SCV2-CPP174 | WAKRNIKPVP | Low | SCV2-CPP252 | TLCFTLKRKT | Low |
| SCV2-CPP19 | FKVTKGKAKK | Low | SCV2-CPP97 | VVLKKLKKSL | Low | SCV2-CPP175 | RNIKPVPEVK | Low | SCV2-CPP253 | LCFTLKRKTE | Low |
| SCV2-CPP20 | KVTKGKAKKG | Low | SCV2-CPP98 | VLKKLKKSLN | Low | SCV2-CPP176 | KTQFNYYKKV | Low | SCV2-CPP254 | SKWYIRVGAR | Low |
| SCV2-CPP21 | KGKAKKGAWN | Low | SCV2-CPP99 | KKLKKSLNVA | Low | SCV2-CPP177 | SRNLQEFKPR | Low | SCV2-CPP255 | KWYIRVGARK | Low |
| SCV2-CPP22 | GGAKLKALNL | Low | SCV2-CPP100 | DAAMQRKLEK | – | SCV2-CPP178 | RNLQEFKPRS | Low | SCV2-CPP256 | YIRVGARKSA | Low |
| SCV2-CPP23 | SKGLYRKCVK | Low | SCV2-CPP101 | AAMQRKLEKM | Low | SCV2-CPP179 | GLAKRFKESP | Low | SCV2-CPP257 | PQNQRNAPRI | Low |
| SCV2-CPP24 | KGLYRKCVKS | Low | SCV2-CPP102 | MQRKLEKMAD | – | SCV2-CPP180 | KMQRMLLEKC | High | SCV2-CPP258 | ERSGARSKQR | Low |
| SCV2-CPP25 | GLYRKCVKSR | Low | SCV2-CPP103 | YKQARSEDKR | Low | SCV2-CPP181 | VLRQWLPTGT | Low | SCV2-CPP259 | RSGARSKQRR | Low |
| SCV2-CPP26 | GLLMPLKAPK | Low | SCV2-CPP104 | KQARSEDKRA | Low | SCV2-CPP182 | DMSKFPLKLR | High | SCV2-CPP260 | SGARSKQRRP | Low |
| SCV2-CPP27 | QRKQDDKKIK | Low | SCV2-CPP105 | QARSEDKRAK | Low | SCV2-CPP183 | MSKFPLKLRG | Low | SCV2-CPP261 | GARSKQRRPQ | Low |
| SCV2-CPP28 | RKQDDKKIKA | Low | SCV2-CPP106 | MLFTMLRKLD | Low | SCV2-CPP184 | SKFPLKLRGT | Low | SCV2-CPP262 | ARSKQRRPQG | Low |
| SCV2-CPP29 | KQDDKKIKAC | Low | SCV2-CPP107 | QDLKWARFPK | Low | SCV2-CPP185 | KFPLKLRGTA | Low | SCV2-CPP263 | RSKQRRPQGL | Low |
| SCV2-CPP30 | DITFLKKDAP | – | SCV2-CPP108 | DLKWARFPKS | Low | SCV2-CPP186 | MILSLLSKGR | Low | SCV2-CPP264 | SKQRRPQGLP | Low |
| SCV2-CPP31 | MLAKALRKVP | High | SCV2-CPP109 | LKWARFPKSD | Low | SCV2-CPP187 | LLSKGRLIIR | High | SCV2-CPP265 | KQRRPQGLPN | Low |
| SCV2-CPP32 | LAKALRKVPT | Low | SCV2-CPP110 | RCHIDHPNPK | Low | SCV2-CPP188 | GRLIIRENNR | Low | SCV2-CPP266 | RRPQGLPNNT | Low |
| SCV2-CPP33 | EAKTVLKKCK | Low | SCV2-CPP111 | GVSAARLTPC | – | SCV2-CPP189 | RLIIRENNRV | Low | SCV2-CPP267 | QIGYYRRATR | Low |
| SCV2-CPP34 | AKTVLKKCKS | Low | SCV2-CPP112 | GFAKFLKTNC | Low | SCV2-CPP190 | NLTTRTQLPP | Low | SCV2-CPP268 | IGYYRRATRR | Low |
| SCV2-CPP35 | KTVLKKCKSA | Low | SCV2-CPP113 | PHISRQRLTK | Low | SCV2-CPP191 | RFQTLLALHR | Low | SCV2-CPP269 | GYYRRATRRI | Low |
| SCV2-CPP36 | KSAFYILPSI | Low | SCV2-CPP114 | HISRQRLTKY | Low | SCV2-CPP192 | YLQPRTFLLK | Low | SCV2-CPP270 | YYRRATRRIR | Low |
| SCV2-CPP37 | KAIVSTIQRK | Low | SCV2-CPP115 | ISRQRLTKYT | Low | SCV2-CPP193 | SVYAWNRKRI | Low | SCV2-CPP271 | YRRATRRIRG | Low |
| SCV2-CPP38 | STIQRKYKGI | Low | SCV2-CPP116 | SRQRLTKYTM | Low | SCV2-CPP194 | YAWNRKRISN | Low | SCV2-CPP272 | RRATRRIRGG | Low |
| SCV2-CPP39 | TIQRKYKGIK | Low | SCV2-CPP117 | RQRLTKYTMA | Low | SCV2-CPP195 | AWNRKRISNC | Low | SCV2-CPP273 | RATRRIRGGD | Low |
| SCV2-CPP40 | IQRKYKGIKI | Low | SCV2-CPP118 | GERVRQALLK | Low | SCV2-CPP196 | WNRKRISNCV | Low | SCV2-CPP274 | TRRIRGGDGK | Low |
| SCV2-CPP41 | GARFYFYTSK | Low | SCV2-CPP119 | RVRQALLKTV | High | SCV2-CPP197 | RQIAPGQTGK | – | SCV2-CPP275 | RIRGGDGKMK | Low |
| SCV2-CPP42 | ARYMRSLKVP | Low | SCV2-CPP120 | KPYIKWDLLK | Low | SCV2-CPP198 | YNYLYRLFRK | High | SCV2-CPP276 | GKMKDLSPRW | Low |
| SCV2-CPP43 | GIEFLKRGDK | – | SCV2-CPP121 | RLKLFDRYFK | Low | SCV2-CPP199 | YLYRLFRKSN | High | SCV2-CPP277 | SQASSRSSSR | Low |
| SCV2-CPP44 | DNLKTLLSLR | High | SCV2-CPP122 | KLFDRYFKYW | High | SCV2-CPP200 | YRLFRKSNLK | Low | SCV2-CPP278 | ASSRSSSRSR | Low |
| SCV2-CPP45 | YMSALNHTKK | Low | SCV2-CPP123 | FPFNKWGKAR | Low | SCV2-CPP201 | RLFRKSNLKP | Low | SCV2-CPP279 | SSRSSSRSRN | Low |
| SCV2-CPP46 | SALNHTKKWK | Low | SCV2-CPP124 | KWGKARLYYD | Low | SCV2-CPP202 | RKSNLKPFER | Low | SCV2-CPP280 | SRSSSRSRNS | Low |
| SCV2-CPP47 | ALNHTKKWKY | Low | SCV2-CPP125 | YAISAKNRAR | Low | SCV2-CPP203 | KKSTNLVKNK | Low | SCV2-CPP281 | RSSSRSRNSS | Low |
| SCV2-CPP48 | LNHTKKWKYP | Low | SCV2-CPP126 | AISAKNRART | Low | SCV2-CPP204 | KSTNLVKNKC | Low | SCV2-CPP282 | SSSRSRNSSR | Low |
| SCV2-CPP49 | NHTKKWKYPQ | Low | SCV2-CPP127 | KNRARTVAGV | Low | SCV2-CPP205 | HADQLTPTWR | Low | SCV2-CPP283 | SSRSRNSSRN | Low |
| SCV2-CPP50 | HTKKWKYPQV | Low | SCV2-CPP128 | NRQFHQKLLK | Low | SCV2-CPP206 | YQTQTNSPRR | Low | SCV2-CPP284 | RSRNSSRNST | Low |
| SCV2-CPP51 | KKPASRELKV | Low | SCV2-CPP129 | RQFHQKLLKS | Low | SCV2-CPP207 | TQTNSPRRAR | Low | SCV2-CPP285 | GSSRGTSPAR | Low |
| SCV2-CPP52 | KPASRELKVT | Low | SCV2-CPP130 | KSIAATRGAT | Low | SCV2-CPP208 | TNSPRRARSV | Low | SCV2-CPP286 | AALALLLLDR | Low |
| SCV2-CPP53 | KHYTPSFKKG | Low | SCV2-CPP131 | RIMASLVLAR | High | SCV2-CPP209 | NSPRRARSVA | Low | SCV2-CPP287 | ALALLLLDRL | Low |
| SCV2-CPP54 | YTPSFKKGAK | Low | SCV2-CPP132 | RNLQHRLYEC | Low | SCV2-CPP210 | PRRARSVASQ | Low | SCV2-CPP288 | KKSAAEASKK | Low |
| SCV2-CPP55 | PSFKKGAKLL | Low | SCV2-CPP133 | RLYECLYRNR | Low | SCV2-CPP211 | KQIYKTPPIK | Low | SCV2-CPP289 | KSAAEASKKP | Low |
| SCV2-CPP56 | FKKGAKLLHK | Low | SCV2-CPP134 | SLRCGACIRR | High | SCV2-CPP212 | SQILPDPSKP | Low | SCV2-CPP290 | SAAEASKKPR | Low |
| SCV2-CPP57 | KKGAKLLHKP | Low | SCV2-CPP135 | RCGACIRRPF | High | SCV2-CPP213 | RLITGRLQSL | Low | SCV2-CPP291 | AAEASKKPRQ | Low |
| SCV2-CPP58 | KGAKLLHKPI | Low | SCV2-CPP136 | CGACIRRPFL | High | SCV2-CPP214 | SASKIITLKK | Low | SCV2-CPP292 | AEASKKPRQK | Low |
| SCV2-CPP59 | WCIRCLWSTK | High | SCV2-CPP137 | GACIRRPFLC | High | SCV2-CPP215 | ASKIITLKKR | Low | SCV2-CPP293 | EASKKPRQKR | Low |
| SCV2-CPP60 | CIRCLWSTKP | Low | SCV2-CPP138 | ACIRRPFLCC | High | SCV2-CPP216 | SKIITLKKRW | Low | SCV2-CPP294 | ASKKPRQKRT | Low |
| SCV2-CPP61 | ANYAKPFLNK | Low | SCV2-CPP139 | CIRRPFLCCK | High | SCV2-CPP217 | KIITLKKRWQ | Low | SCV2-CPP295 | SKKPRQKRTA | Low |
| SCV2-CPP62 | TNIVTRCLNR | Low | SCV2-CPP140 | IRRPFLCCKC | High | SCV2-CPP218 | IITLKKRWQL | Low | SCV2-CPP296 | KKPRQKRTAT | Low |
| SCV2-CPP63 | CTFTRSTNSR | Low | SCV2-CPP141 | RRPFLCCKCC | High | SCV2-CPP219 | KKRWQLALSK | Low | SCV2-CPP297 | KPRQKRTATK | Low |
| SCV2-CPP64 | TCMMCYKRNR | High | SCV2-CPP142 | MSYYCKSHKP | Low | SCV2-CPP220 | KRWQLALSKG | Low | SCV2-CPP298 | PRQKRTATKA | Low |
| SCV2-CPP65 | MCYKRNRATR | Low | SCV2-CPP143 | ANTCTERLKL | – | SCV2-CPP221 | VRIIMRLWLC | Low | SCV2-CPP299 | RQKRTATKAY | Low |
| SCV2-CPP66 | CYKRNRATRV | Low | SCV2-CPP144 | KLFAAETLKA | Low | SCV2-CPP222 | RIIMRLWLCW | Low | SCV2-CPP300 | RQGTDYKHWP | Low |
| SCV2-CPP67 | YKRNRATRVE | Low | SCV2-CPP145 | SWEVGKPRPP | Low | SCV2-CPP223 | IIMRLWLCWK | Low | SCV2-CPP301 | KTFPPTEPKK | Low |
| SCV2-CPP68 | KRNRATRVEC | Low | SCV2-CPP146 | VGKPRPPLNR | Low | SCV2-CPP224 | IMRLWLCWKC | High | SCV2-CPP302 | FPPTEPKKDK | Low |
| SCV2-CPP69 | RDLSLQFKRP | Low | SCV2-CPP147 | GKPRPPLNRN | Low | SCV2-CPP225 | MRLWLCWKCR | High | SCV2-CPP303 | PPTEPKKDKK | Low |
| SCV2-CPP70 | SLQFKRPINP | Low | SCV2-CPP148 | KALKYLPIDK | Low | SCV2-CPP226 | RLWLCWKCRS | High | SCV2-CPP304 | PTEPKKDKKK | Low |
| SCV2-CPP71 | HNIALIWNVK | – | SCV2-CPP149 | DKCSRIIPAR | – | SCV2-CPP227 | LWLCWKCRSK | High | SCV2-CPP305 | TEPKKDKKKK | Low |
| SCV2-CPP72 | LSEQLRKQIR | Low | SCV2-CPP150 | KCSRIIPARA | Low | SCV2-CPP228 | WLCWKCRSKN | High | SCV2-CPP306 | EPKKDKKKKA | Low |
| SCV2-CPP73 | QLRKQIRSAA | Low | SCV2-CPP151 | CSRIIPARAR | Low | SCV2-CPP229 | LCWKCRSKNP | High | SCV2-CPP307 | PKKDKKKKAD | Low |
| SCV2-CPP74 | LRKQIRSAAK | Low | SCV2-CPP152 | SRIIPARARV | Low | SCV2-CPP230 | CWKCRSKNPL | High | SCV2-CPP308 | KKDKKKKADE | Low |
| SCV2-CPP75 | RKQIRSAAKK | Low | SCV2-CPP153 | RIIPARARVE | Low | SCV2-CPP231 | KCRSKNPLLY | Low | SCV2-CPP309 | TQALPQRQKK | Low |
| SCV2-CPP76 | KQIRSAAKKN | Low | SCV2-CPP154 | SVVNARLRAK | Low | SCV2-CPP232 | NRNRFLYIIK | Low | SCV2-CPP310 | ALPQRQKKQQ | Low |
| SCV2-CPP77 | QIRSAAKKNN | Low | SCV2-CPP155 | VVNARLRAKH | Low | SCV2-CPP233 | RNRFLYIIKL | Low | |||
| SCV2-CPP78 | NNWLKQLIKV | High | SCV2-CPP156 | VNARLRAKHY | Low | SCV2-CPP234 | YIIKLIFLWL | Low | |||
To develop a reliable predicting model, candidate peptides are obtained from confirmed databases. Then, practice and test sets for model training and validating the authenticity of the trained model are considered, respectively (Su et al., 2020). However, in the current study, we have analyzed some experimentally validated viral-derived CPPs to check the scalability and reliability of CPPs originated from SARS-CoV-2. Although all of the experimentally validated viral-derived CPPs considering as the positive control (Table 3 ) were identified as CPP in the “multiple peptides” module of CellPPD using the SVM + Motif hybrid model, the SVM-based model could not detect VG-21 as a CPP. This observation is due to the prediction state in the SVM-based model. As the threshold of prediction rate varying from −1 to +1, the sensitivity and specificity alter by displacing from the default threshold (Gautam et al., 2015). Selecting a higher threshold value results in high specificity (high confidence) with less positive outputs. However, a lower threshold increases the sensitivity and covers a higher number of CPPs, though with an increased number of false-positive results. Therefore, some few CPPs such as VG21 might not be detected as positive in SVM-based model. As the specificity correlates to the capacity to predict non-CPPs (negatives), it is of prominent significance for wet-lab investigations. The low specificity of a model provides a high amount of false-positive peptides when implemented for proteome mining. These false-positives boost the cost of laboratory validation by itself.
Table 3.
The predictions of CellPPD and MLCPP servers regarding ten viral-derived CPPs as positive controls compared to the experimentally validated reports.
| Name of the peptide | Sequence | Origin | CPP prediction (CellPPD) | SVM score⁎ | Uptake efficiency (MLCPP) | Prediction confidenc# (MLCPP) | Uptake efficiency (literature) | References |
|---|---|---|---|---|---|---|---|---|
| NLS-A | MTYPRRRFRRRRHRPRS | PCV2 | CPP | 0.80 | High | 0.81 | High | (Yu et al., 2018) |
| FHV coat (35–49) | RRRRNRTRRNRRRVR | FHV | CPP | 1.13 | High | 0.74 | High, Comparable to the TAT peptide | (Futaki et al., 2001) |
| PepR | LKRWGTIKKSKAINVLRGF RKEIGRMLNILNRRRR |
DENVC | CPP | 0.26 | High | 0.50 | High (uptake by 96% of the cells) | (Freire et al., 2014) |
| VP22 | DAATATRGRSAASRPTQRP RAPARSASRPRRPVE |
HSV-1 | CPP | 1.00 | Low | 0.36 | controversial (cell type & cargo dependent) | (Hakkarainen et al., 2005) (Carnevale et al., 2018) |
| TAT | YGRKKRRQRRR | HIV-1 | CPP | 1.07 | High | 0.84 | High | (Duong and Yung, 2013) (Shin et al., 2014) (Li et al., 2013) |
| Erns | RQGAARVTSWLGRQLRIA GKRLEGRSK |
CSFV | CPP | 1.00 | Low | 0.38 | Higher uptake than penetratin, MAP, and pVEC in yeast cells | (Parenteau et al., 2005) |
| Pep1 | KETWWETWWTEWSQPK KKRKV |
SV40 | CPP | 1.00 | Low | 0.49 | Lower than the penetratin analog EB1 | (Akahoshi et al., 2016) |
| VG-21 | VTPHHVLVDEYTGEWVDSQFK | VSV | – | −0.72 | – | – | Higher uptake of nanoparticles in HEp-2 cells | (Tiwari et al., 2014) |
| REV | TRQARRNRRRRWRERQR | HIV-1 | CPP | 1.14 | High | 0.87 | High, comparable to the TAT peptide | (Futaki et al., 2001) |
| HPV-WT | CSPQYTIIADAGDFYLHPSYY MLRKRRKR |
HPV | CPP | 0.09 | – | – | Uptake by HeLa and HaCat cell line | (Zhang et al., 2018) |
The SVM score provided by CellPPD.
The prediction confidence of uptake efficiency provided by MLCPP.
Cell-penetrating peptides are categorized into various groups, namely cationic, amphipathic, hydrophobic, and anionic, with distinct physiochemical characteristics (Langel, 2019). Among the 310 predicted SCV2-CPP sequences, there are cationic, amphipathic, and hydrophobic peptides. Some highly cationic SCV2-CPPs including, SCV2-CPP270, SCV2-CPP271, and SCV2-CPP272 (with a total net charge of +5) derived from the Arg-rich region of nucleoprotein showed the highest peptide sequence identity to cationic positive controls such as TAT, NLS-A, and FHV coat (35–49). Some other SCV2-CPPs are highly amphipathic, including SCV2-CPP304, SCV2-CPP305, SCV2-CPP306, and SCV2-CPP307 with the highest degree of peptide sequence identity to Pep1 as an amphipathic CPP according to alignment of peptides in UniProt.
3.2. The role of SCV2-CPPs in the replication-transcription machinery and pathogenesis
It is noteworthy that several viral proteins, such as RNA binding proteins, viral particle envelope proteins, and trans-activators of gene transcription are proper candidates for the discovery of novel CPPs. Within the following subsections, the distribution of SCV2-CPPs in nonstructural, structural, and accessory proteins of SARS-CoV-2 and their potential roles in protein-nucleic acid association, and protein-protein or protein-cofactor interaction identified in SARS-CoV-2 so far or according to SARS-CoV-1 homologs are discussed.
3.2.1. Nonstructural protein 1 (NSP1) and SCV2-CPPs
NSP1 is the N-terminus cleavage outcome of the proteolytic action of PLpro on the ORF1a at the consensus splitting site “LXGG”. The cytoplasmic NSP1 protein of SARS-CoVs disrupts the host translational machinery by binding to the 40S ribosomal subunit; hence, it acts as the main virulence factor (Narayanan et al., 2015). The host mRNAs that are not entailed in the operative process of translation are conveyed into the host mRNA degradation pathways. Therefore, the NSP1-40S complex triggers the endolytic cleavage of the host mRNAs. Coronaviruses display divergent NSP1 protein sequences; however, an almost conserved functional domain defined as “LLRKXGXKG” has been detected among coronaviruses, including SARS-CoV-1 (Lei et al., 2013). This domain corresponds to the residues detected in SCV2-CPP8 of SARS-CoV-2 NSP1. A significant attenuation was observed in a mutant with 27 nucleotide deletion (encoding the above domain) in the mouse hepatitis virus. Substitution of “Arg” and “Lys” in the NSP1 of SARS-CoV-1 mutants (corresponding to R124 and K125 in SCV2-CPP4–8 of SARS-CoV-2 homolog) by “Ser” and “Gln” residues was not able to defeat antiviral pathways of the host and displayed damaged replication. The two above positively charged regions are proposed as potential sites for binding to RNA (Tanaka et al., 2012). Although R124/K125 mutants were still able to inhibit translation, they had lost the ability to perform the mRNA cleavage in the host. Therefore, it can be suggested that the presence of positive residues at R124/K125 positions of SARS-CoV-2 NSP1 now observed in SCV2-CPP4-8 is essential for the replication process.
3.2.2. Nonstructural protein 2 (NSP2) and SCV2-CPPs
The functional role of NSP2 containing SCV2-CPP9–26 has not been explained precisely. The deletion of NSP2 in SARS-CoV-1 ends in a moderate decline in viral titers. Hence, NSP2 might be nonessential for replication. However, its linkage to NSP3 before processing assigns a potential regulative role to NSP2. It is of note that NSP2 interacts with prohibins 1 and 2, which are connected to cell cycle progression and apoptosis. Thus, NSP2 might affect changes in the host cell environment (Cornillez-Ty et al., 2009).
3.2.3. Nonstructural protein 3 (NSP3) and SCV2-CPPs
Amid all RNA viruses, coronavirus has the largest genome identified thus far; NSP3 is the largest encoded protein therein. As a multi-domain protein, NSP3 is organized as follows in β-CoVs: a ubiquitin-like domain 1 (Ubl1), a hypervariable region (HVR), three macrodomains (Mac), a domain preceding Ubl2 and PLpro (DPUP), a ubiquitin-like domain 2 (Ubl2), the PLpro, a nucleic acid-binding domain (NAB), a beta-coronavirus specific marker domain (βSM), transmembrane domain 1 (TM1), NSP3 ectodomain (3Ecto), transmembrane domain 2 (TM2), an amphipathic helix (AH1), a conserved domain with the undiscovered function (Y1), and a specific domain found only in coronaviruses (CoV-Y) (Fig. 1B) (Lei et al., 2018). According to the annotation of NCBI conserved domain search service, and alignment with SARS-CoV-1 NSP3, the Ubl1 domain in SARS-CoV-2 spanning residues 1–111 with the core residues 20–106 might form a typical ubiquitin-like fold. CoV Ubl1s are committed to ssRNA binding and interaction with the “N” protein (Keane and Giedroc, 2013). Therefore, Ubl1 is ascribed to the replication and induction of viral infection. The main interaction of the Ubl1-N complex includes the acidic residues of Ubl1 (E52 and D59) and the serine-arginine (SR)-rich region of the structural nucleocapsid (N) protein present in the penetrating peptide SCV2-CPP278 (ASSRSSSRSR). It is of note that no CPP could be identified in the first two domains of NSP3, including the Ubl1 + HVR (known as NSP3a), which exists in all CoVs. Mac1 or X-domain displays phosphatase activity and removes the 1″-phosphate from ADP-ribose-1″-phosphate. Mac1 is a conserved domain forming residues 206–387 in SARS-CoV-2 without any CPP. However, three subsequent unique domains in the SARS-CoV known as the “SARS-unique domain” (SUD) or NSP3c all contain CPPs. Mac2 (SUD-N), Mac3 (SUD-M), and DPUP (SUD-C) are located at the N-terminus, middle, and C-terminus of this domain, respectively. Mac2 (spanning residues 413–548) harbors a part of SCV2-CPP27–29 and the penetrating peptides SCV2-CPP30–36. Mac3 that flanks residues 549–678 of NSP3 protein contains SCV2-CPP37–42. DPUP that constructs residues 679–743 of NSP3 comprises SCV2-CPP43–44. The dominant established function of Mac2–3 (SUD-NM) is related to RNA binding. Mac2–3 favorably interacts with oligo(G) that can generate G-quadruplexes. In the Mac2 domain of SARS-CoV-1, there are two positively charged “Lys” spots participating in oligo(G) attachment (Tan et al., 2009). The corresponding positive residues in SARS-CoV-2 are R500K501 represented in SCV2-CPP31–32 (MLAKALR 500 K 501VPT) and K529K530 in SCV2-CPP33–35 (EAKTVLK 529 K 530CKSA). Although one of the lysine residues (K476 in SARS-CoV-1) is replaced with R500 in SARS-CoV-2, the positive charge of arginine still plays the role. Another “Lys” patch in Mac3, which is conserved in SARS-CoVs corresponds to K587K589K592 in SARS-CoV-2. This could be found in SCV2-CPP39–40 (XXK 587YK 589GIK 592XX). In SARS-CoV-1 mutation of the above-mentioned “Lys” residues has eliminated virus replication (Kusov et al., 2015). This indicates that the G-quadruplex binding is an indispensable factor for the activity of replication transcription complex (RTC). Ubl2-PLpro of SARS-CoV-2 spans residues 746–1059. Despite several other CoVs with two paralogs of PLpro, SARS-CoVs have just one copy of the gene. The development of an oxyanion hole to maintain the negative charge throughout peptide hydrolysis is a noteworthy property of serine and cysteine proteases (Ratia et al., 2006). As in SARS-CoV-1, an essential hydrogen bond interaction between the side chain of D853 and W838 is required to retain a reachable active site. The corresponding “Trp” residue resides in SCV2-CPP46–50. NAB (residues 1089–1203 of SARS-CoV-2 NSP3) and βSM (residues 1226–1341 of SARS-CoV-2 NSP3) domains are inclusively known as NSP3e and could be found only in βCoVs. Similar to Mac3, the NAB domain also shows RNA binding capacity for both oligo(G) and oligo(A) and interacts with ssRNA (Johnson et al., 2010). A positively charged fragment in SARS-CoV-1 corresponding to K1162, K1163, and R1193 of the SARS-CoV-2 NAB domain could be involved in RNA binding. The first two “Lys” residues reside in SCV2-CPP54–57, and the “Arg” residue lies in SCV2-CPP59–60. Although SCV2-CPP61 and a segment of SCV2-CPP62 are in the βSM region, the exact function of this unit is not yet defined. The TM domains of NSP3 promote the construction of the membrane-related replicase complex. Neither TMs (TM1 flanks a.a 1414–1436 and TM2 is situated at residues 1519–1541) nor 3Ecto (residues 1437–1518) display any CPP. No CPP could be detected in the AH1 region (a.a 1546–1568). AH1 does not span the membrane and has an amphipathic character. Located at the cytosolic side of the ER, the Y1 + CoV-Y domains flank a region between residues 1569–1945. However, the border between the two is not exactly determined. While the Y1 domain is conserved in the order Nidovirales, the CoV-Y is just preserved in CoVs. SCV2-CPP64-77 resides in the last two domains. It has been demonstrated that in the absence of Y1 and CoV-Y domains the effectiveness of NSP3-NSP4 is diminished (Neuman, 2016). Conclusively, and in comparison to the SARS-CoV-1 homolog, it seems that the positively charged residues of CPPs in NSP3 are vital for replication via interaction with oligo(G) and RNA binding.
3.2.4. Nonstructural protein 4 (NSP4) and SCV2-CPPs
NSP4 is a transmembrane protein with four TM helices and a cytosolic C-terminal domain. Hence, the presence of multiple hydrophobic and membrane spanning domains are indicative of the involvement of NSP4 in the formation of double-membrane vesicles (DMVs) (Sakai et al., 2017). NSP4 harbors SCV2-CPP78–88, none of which locate in the TM helices.
3.2.5. Nonstructural protein 5 (NSP5) and SCV2-CPPs
The NSP5 protein of coronavirus is a chymotrypsin-like protease perceived as the main protease (Mpro). Belonging to the C30 family of endopeptidases, NSP5 protein is in charge of the cleavage of NSP4-NSP16 to assemble into the RTC (Yang et al., 2003). NSP5 is well conserved in Nidovirales with three domains known as DomI, DomII, and DomIII corresponding to residues 8–101, 102–184, and 201–303 in SARS-CoV-2 NSP5, respectively. DomI and DomII are catalytic domains, and DomIII is the dimerization domain. The dimerization of NSP5 is between E290 of one protomer and R4 of the other (Zhang et al., 2020). The “Arg” residue involved in this interaction is found in SCV2-CPP89 (FR ⁎KMAFPSGK).
3.2.6. Nonstructural protein 6 (NSP6) and SCV2-CPPs
NSP6 is indispensable for the formation of DMVs that are representative of CoV replicative organelles. Although the TMHMM server predicts that NSP6 comprises seven TMs, only six of these serve as membrane-spanning helices. The appearance of extra non-transmembrane hydrophobic domains adjacent to the actual TM domain is typical for double-membrane organelles (Neuman, 2016). NSP6 contains only one CPP named as SCV2-CPP95.
3.2.7. Nonstructural protein 8 (NSP8) and SCV2-CPPs
NSP7 does not have any CPP. While the second subunit of NSP8 involves in the polymerase activity of NSP12, NSP7 and NSP8 heterodimers stabilize NSP12 regions requires for RNA binding. NSP8 is constituted of an N-terminal shaft domain containing all identified CPPs in SCV2-NSP8 and a C-terminal head domain (Xiao et al., 2012). SCV2-CPP103–106 are in a region of NSP8 containing some basic amino acids committing to the positive electrostatic interactions in the template-binding channel. To verify if positive residues are associated with nucleic acid attachment, various basic amino acids were mutated. Accordingly, the nucleic acid binding affinity of NSP8 mutants in SARS-CoV-1 equivalent to K72A, R75A, and R80A in SCV2-CPP103 (YK 72QAR 75SEDKR 80AK) of SARS-CoV-2 was much weaker than that of the wild-type (Zhai et al., 2005). “Ala” mutation of “Asp” in SARS-CoV-1 corresponding to D99A of SARS-CoV-2 in SCV2-CPP106 also disturbed the electrostatic interaction of NSP8 to the K332 residue of NSP12 (Subissi et al., 2014).
Interactions between NSP7 and NSP8 reveal that the binding region for NSP7-NSP8 heterodimer is entirely conserved (Zhai et al., 2005). Homologous residues of SARS-CoV-2 NSP8 (M87, M90, L91, M94, and L98), including SCV2-CPP106 (M 90 L 91FTM 94LRKL 98D) and NSP7 (V6, C8, V12, and V16) can form a hydrophobic core like the observed core in SARS-CoV-1 NSP8. Similarly, the side chains of residues of NSP8 (F92, L95, L103, I106, I107, and A110) wherein the first two residues reside in SCV2-CPP106 could potentially be involved in hydrophobic interactions with M52, V53, L56, L59, L60, and I68 of NSP7. The empty barrel-shaped structure of the hexadecamer infers that its purpose is to surround and stabilize RNA, thus retaining the RNA to expedite adequate replication and transcription. Conclusively, NSP8 CPPs are involved in the interaction with NSP7 and NSP12.
3.2.8. Nonstructural protein 9 (NSP9) and SCV2-CPPs
The NSP9 of CoVs is perceived as an indispensable ssRNA binding protein in the replication complex. Despite the obscure exact mechanism, NSP9 dimerization is critical for replication. However, dimerization is not necessary for RNA binding. It has been demonstrated that in SARS-CoV-1, K52, and R55 are decisive residues for RNA binding (Egloff et al., 2004). The replacement of K52 decreased the capacity of RNA binding in other members of the family. Some aromatic residues, such as W53 might also contribute a stacking interaction with nucleotides. The corresponding residues in SARS-CoV-2 are SCV2-CPP107–109 (QDLK 52 W 53AR 55FPKSD). Collectively, it seems that a pack of positively charged and an aromatic residue in NSP9 are implicated in RNA binding.
3.2.9. Nonstructural protein 10 (NSP10) and SCV2-CPPs
To cap the SARS-CoV viral mRNA several NSPs are involved. NSP13, NSP14, and the complex of NSP10/16 are committed to the capping process in a rigid order. Capping of the 5′ end of viral mRNA is a shield for the virus not to be identified as a pathogen by the host immune system. When NSP13 discards the phosphate group from the mRNA to produce ppRNA the next step is catalyzed by a yet unidentified guanylyl transferase to form GMP from GTP. The resulted GMP binds to the ppRNA. Then, a methyl group is added to the N7 atom of guanine by NSP14 (N7-MTase) for the production of the Cap0 structure. The activated NSP16 (2’-OMTase) by NSP10 is responsible for methylation of the 2’-O position of ribose for the construction of protective Cap1 (Menachery et al., 2014).
NSP10 is known as a conserved small non-enzymatic protein with 139 residues in SARS-CoV-2 that acts as a transcription factor in replication and transcription of SARS-CoV. NSP10 also binds to two Zn2+ ions with high affinity (Su et al., 2006). His83 in SARS-CoV-1 is tetrahedrally chelated with one of the zinc ions. NSP10 binds and triggers the activity of 3′–5′ ExoN and 2′-O-MTase activity of NSP14 and NSP16, respectively. Replacement of His83R abolished the interaction with NSP14. Lys87 has a detrimental effect on the binding of NSP10 to NSP14. Mutation studies have shown that the substitution of R78G or R78A in NSP10 results in a diminished ExoN activity of NSP14 (Bouvet et al., 2014). It seems that the key residues of NSP10 associates with the interaction and activity of NSP14 correspond to the homologous position in SARS-CoV-2 and reside in the single identified CPP named SCV2-CPP110 (R 78CH 80IDH 83PNPK 87). In an equivalent manner, residues 77–80 of NSP10 inside SCV2-CPP110 are also involved in the interaction with NSP16 (Decroly et al., 2011). It is of note that NSP10 is usually produced in a 3–6 fold ratio than NSP14 and NSP16. Consequently, the NSP14–10 and NSP16–10 complex can exist at the same time (Bouvet et al., 2014). Collectively, some positively charged residues in NSP10 as a transcription factor are connected to the interaction with NSP14 and NSP16, while His residue coordinates with Zn2+ ion.
3.2.10. Nonstructural protein 12 (NSP12) and SCV2-CPPs
RNA-dependent RNA polymerase (RdRp) is a protein with multiple domains and invokes the formation of phosphodiester bonds between ribonucleotides (Yin et al., 2020). The RdRp is a complex with several NSPs, including NSP12, NSP8, and NSP7. In this section, only NSP12 of SARS-CoV-2 RdRp is described (Fig. 1C). The SARS-CoV-2 NSP12 with 932 amino acid residues is composed of an N-terminal nidovirus-unique extension domain (residues 1–396 containing SCV2-CPP111–122) and a polymerase domain (residues 397–932 harboring SCV2-CPP123–133). The nidovirus-unique extension is separated into two discrete regions: the NiRAN domain (a.a. 117–250 including SCV2-CPP113–119) and an interface domain (a.a. 251–365 with SCV2-CPP120–122). It seems that the NiRAN does not have a functional role in polymerase activity. The polymerase domain at the C-terminal of NSP12 is constituted of a finger (residues 397–581 and 621–679), a palm (residues 582–620 and 680–815), and a thumb subdomain (a.a. 816–932). Viral polymerases also have conserved motif regions (A–G). These subdomains and motifs are correlated to catalysis and interaction with the template and incoming nucleotides. SCV2-CPP123–124 and SCV2-CPP125–127 reside in motif G and motif F of the finger subdomain, respectively. The rest of the NSP12 CPPs including SCV2-CPP128–131 and SCV2-CPP132–133 lie in the finger and palm subdomains, sequentially.
The RdRps have entry channels or tunnels lined with positively charged residues; hence, favor the entry of NTPs and the template RNA into the active site (Venkataraman et al., 2018). The triphosphate part of ATP mostly interacts with positively charged residues, such as K551, R553, and R555 in SCV2-CPP125–127 and hydrophilic S549 in SCV2-CPP125–126. The nucleoside part of ATP (adenosine) interacts with more diverse residues such as hydrophilic T556, and hydrophobic residues including V557 and A558 in SCV2-CPP127. As a result, the ATP binding site resides inclusively in SCV2-CPP127 (K 551NR 553AR 555 T 556 V 557 A 558GV) in motif F (Zhang and Zhou, 2020). It has been designated that the V557 in SCV2-CPP127 regulates polymerase accuracy. The valine side chain encounters the template RNA and associates with the base to be matched with the arriving NTP. It is of note that ExoN enhances the accuracy of RNA synthesis by altering mistakes caused by RdRp (Shannon et al., 2020).
Out of the residues that are involved in recognition and interaction with RNA by RdRp complex the followings are notable in SARS-CoV-2; A580 of SCV2-CPP130 (KSIA 580ATRGAT) in the finger subdomain, A558 and G559 in SCV2-CPP127 (KNRARTVA 558 G 559V) of motif F, and N507 in SCV2-CPP123 (FPFN 507KWGKAR) of motif G. All the above-mentioned residues are in the finger subdomain (Yin et al., 2020). This is compatible with the evidence that no particular sequence is needed for NSP12 activity at the elongation level. The finger subdomain acts as a holder of the template RNA in the right geometry and promotes polymerization (Yin et al., 2020). In summation, CPPs of NSP12 are associated with the interaction with NTP as well as recognition and binding to the template RNA.
3.2.11. Nonstructural protein 13 (NSP13) and SCV2-CPPs
The NSP13 (helicase) in coronavirus has various catalytic activities including, 1) hydrolysis of NTPs and dNTPs, 2) unwinding nucleic acid duplexes, and 3) RNA 5′-triphosphatase activity. In other words, helicases break hydrogen bonds in DNA/RNA duplexes in an NTP/metal-dependent mode with conserved motifs associated with NTP and nucleotide-binding. NSP13 has core and accessory domains (Lehmann et al., 2015). While RecA-like domains form the core of the NSP13 enzyme to hydrolyze NTPs, accessory domains support the catalytic activity and interact with other proteins (Fig. 1D). NSP13 in SARS-CoV-2 has 601 amino acid residues. The N-terminal “Cys-His-rich” domain, known as the CH domain (residues 1–113) correlates with three zinc atoms. Cys72 and His75 residues participating in the third zinc coordination in the CH domain of other coronaviruses are correlated to residues situated in SCV2-CPP142 (MSYYC 72KSH 75KP). An equivalent to P408 of RecA1 (residues 241–443) in SCV2-CPP158–159 (AP 408RTLLTKGT) is committed to the recognition and attachment to RNA. In a homologous comparison, R443 of RecA2 (residues 444–596) in SCV2-CPP161 (FLGTCRR 443CPA) plays a role in NTP hydrolysis. Mutation of “Ala” to “Val” in murine coronavirus corresponding to A336V in SCV2-CPP149–153 led to the diminished virus growth (Zhang et al., 2015). R337 and R339 in SCV2-CPP151–153 are positively charged residues located on the entry of nucleotide-binding groove and drag negatively charged nucleic acids. Physical interaction has been claimed between NSP12 and NSP13, wherein the RdRP elevates the unwinding of the helicase (Jia et al., 2019). Homologous to SARS-CoV-1, the “LKA” residues in the third zinc finger located in SCV2-CPP144 (KLFAAETLKA), and the complete sequence of SCV2-CPP148 “KALKYLPIDK” in RecA1 domain are both potentially assigned in this interaction. Therefore, some positively charged residues in NSP13 are involved in recognition and attachment to RNA nucleotides, while “His” and “Cys” residues coordinate with Zn2+ ion.
3.2.12. Nonstructural protein 14 (NSP14) and SCV2-CPPs
As a bifunctional protein, NSP14 plays a vital role in RNA synthesis. SARS-CoV-2 NSP14 protein with 527 residues has dual enzymatic properties. One is the exoribonuclease (ExoN) activity that resides in residues 1–287 at the N-terminal domain. The other is a guanine-N7 methyltransferase (N7-MTase) activity spanning residues 288–527 at the C-terminal domain. The ExoN is essential for proofreading to avoid lethal mutations during RNA synthesis, and the SAM-dependent N7-MTase activity is required for viral mRNA capping (Snijder et al., 2016). SCV2-CPP166–169 is located in the ExoN, and SCV2-CPP170–171 is based in the N7-MTase domain. As mentioned earlier, NSP10 interacts with the ExoN domain and is the activator of NSP14 in the complex (Ma et al., 2015). A1-R76 of NSP14 is a flexible region known to interact with NSP10 in SARS-CoV-1. Identically, SCV2-CPP166–167 is situated in this region of NSP14 of SARS-CoV-2. It has been demonstrated that “DXGXPXA” is a SAM or AdoMet-binding site in NSP14, which aligns with SCV2-CPP171 in SARS CoV-2 (Snijder et al., 2016). The tyrosine residue at position 51 in SCV2-CPP166 of NSP14 (GIPKDMTY 51RR) is equivalently connected to the formation of hydrogen bonds with NSP10. The structure flanking A119-D144 also links to NSP14-NSP10 interaction and locates in SCV2-CPP168–169. Conclusively, NSP14 CPPs are involved in SAM binding. Any disruption in the NSP14-NSP10 interaction results in a significant reduction in the replication constancy.
3.2.13. Nonstructural protein 15 (NSP15) and SCV2-CPPs
NSP15 of SARS-CoVs is a uridylate-specific Mn2+-dependent endoribonuclease known as NendoU. NSP15 cleaves 3′ uridylates and generates 2′-3′ cyclic phosphodiester (Bhardwaj et al., 2008). NSP15 is an essential part of the RTC required for the synthesis of RNA. Nonetheless, there is little information about the characteristics of NSP15 in vivo. For example, arterivirus and mouse hepatitis virus displayed diminished sub-genomic RNA levels correlated to NSP15 mutations. In mouse hepatitis virus, NSP15 was connected to the RTC-associated proteins such as viral primase NSP8 and NSP12 during infection (Athmer et al., 2017). NSP15 of SARS-CoV-2 is composed of 346 a.a residues and in the monomer state is composed of three domains, including an N-terminal domain (a.a 1–62), a central domain (a.a 63–191), and a catalytic long C-terminal domain (residues 192–346) compared with SARS-CoV-1 (Ricagno et al., 2006). While SCV2-CPP172–175 is in the border of N- and central domain, SCV2-CPP176 and SCV2-CPP177–179 lie in the central and C-terminal domains, respectively. NSP15 is enzymatically active in the hexameric state (Joseph et al., 2007). Several interactions stabilize the hexadecamer in SARS-CoV-1, such as the interaction of R61 with E266. R61 is situated in SCV2-CPP172–175. The H1‑nitrogen atom of R61 interacts with the ε1 oxygen atom of E266 in SARS-CoV-1 (Bhardwaj et al., 2008). The Mn2+ ions provoke conformational variations in the protein such as persuading a switch to the active conformation. Coordination between Mn2+ and the hydroxyl group of S261, the main-chain carbonyl oxygen atom of P262, and the nitrogen of guanidinium moiety of R257 are described in SARS-CoV-2 (Kim et al., 2020) and appears in SCV2-CPP179 (GLAKR 257FKES 261 P 262). Collectively, CPPs of NSP15 are involved in protein-protein interaction to stabilize NendoU hexadecamers and interact with Mn2+ ions.
3.2.14. Nonstructural protein 16 (NSP16) and SCV2-CPPs
NSP16 of SARS-CoV-2 with 298 residues is classified in the S-adenosylmethionine-dependent methyltransferase family. The 2’-O methyltransferase (2’-O MTase) activity of NSP16 is required for thriving coronavirus infection. The 2’-O methylation averts virus detection by the host immune system (Menachery et al., 2014). This 2’-O MTase activity is regulated by NSP10. Four patches, known as I to IV in NSP16 are in contact with NSP10 (Decroly et al., 2011). In line with SARS-CoV-1, SCV2-CPP181 is part of patch II (KGVAPGTAVLRQWLPT), and some residues of SCV2-CPP182–185 exist in patch IV (SLFDMSK). Mutation of R86A in patch II of SARS-CoV-1 NSP16 diminished its interaction with NSP10 and abolished the MTase activity completely. This residue can be found in SCV2-CPP181. Mutation of Q87A in SARS-CoV-1 equivalent to the similar residue in SCV2-CPP-181 has still some degree of interaction with NSP10 with a significant reduction in 2’-O-MTase activity. Mutation of M247A in the patch IV of SARS-CoV-1 (appears in SCV2-CPP182–183) distorted the complex of the NSP10-NSP16 heterodimer and eliminated 2’-O-MTase activity. Totally, in addition to MTase activity, NSP16 CPPs are implicated in the interaction with NSP10.
3.2.15. Spike protein (S) and SCV2-CPPs
Protein “S” is a surface-placed trimeric spike glycoprotein, and as the second most abounding protein in the envelope, it defines host cell propensity, virus-cell, and cell-cell fusion. Posttranslational cleavage of S results into two chains known as S1 and S2. The N-terminal S1 surface unit contains the receptor-binding domain (RBD) and the C-terminus transmembrane unit (S2) comprising the merge of the viral envelope and host cell membrane. S2 protein is classified as the class I fusion proteins. As shown in Fig. 1E (Wrapp et al., 2020), the detailed order of domains in the S protein of SARS-CoV-2 are residues 1–15 known as the signal sequence (SS) followed by an N-terminal domain (NTD: residues 16–305) with SCV2-CPP190–192. RBD flanks the region between residues 330–521 with SCV2-CPP193–202. Subdomain regions SD1 (a.a. 522–589) and SD2 (a.a. 590–676) harbor SCV2-CPP203–204 and SCV2-CPP205 peptides, respectively. Residues that are spanning the regions 816–833, and 908–985 form the flanking peptide (FP) and heptad-repeat 1 (HR1), respectively, without any CPP. While the central helix (CH1: residues 986–1035) contains SCV2-CPP213, the connector domain (CD: residues 1076–1141) lacks any CPP. The rest of the sequence, including heptad-repeat 2 (HR2), transmembrane domain (TM2), and cytoplasmic tail (CT) with no exact defined border, do not have any CPPs. The cleavage of S1 and S2 is essential for the initiation of viral pathogenesis by a protease. Serine proteases split the protein within the motif (R/K)-(2X)n-(R/K)↓ wherein n = 0,1,2,3 amino acid spacer. The S1/S2 cleavage site in SARS-CoV-2 adopts the “RRAR↓S" motif between residues R685/S686 (Coutard et al., 2020). The S1/S2 cleavage site locates in an Arg-rich region of SCV2-CPP208-210. After viral endocytosis, a second cleavage occurs via endolysosomal proteases at S2' cleavage site (R815 ↓), supporting membrane fusion. There are residues such as L455 and F456 in RBD that greatly enhance the affinity of S1 to the receptor (Spinello et al., 2020). These residues are detected in SCV2-CPP198-201.
3.2.16. Membrane protein (M) and SCV2-CPPs
“M” as a type III transmembrane (TM) protein is the most abundant glycoprotein in coronavirus particles. M protein with 222 residues involves in virus assembly and budding via M-M, M-S, and M-N interactions. From its synthesis compartment (ER), the M protein is conveyed to the Golgi complex. It has been demonstrated that the SARS-CoV M protein accumulates at the ERGIC (mostly Golgi complex) then is disseminated to the plasma membrane (Tseng et al., 2013). The M protein is composed of three parts including 1) a short outside N-terminal domain, 2) three TM domains, and 3) a long carboxy-terminal (CT) domain inside the particle. M protein is organized in the following order in SARS-CoV-2: virion surface (residues 1–18), helical TM1 (residues 19–39), cytoplasmic intravirion (residues 40–49), helical TM2 (residues 50–70) containing SCV2-CPP235, virion surface (residues 71–78), TM3 region (residues 79–99) with four residues of SCV2-CPP236, and intravirion region (residues 100–221) including SCV2-CPP239-245. It is of note that the TM1 of SARS-CoV M protein has a “KKXX” Golgi retention signal. Several studies have shown the most important motifs or residues of the M protein involved in virus assembly. Replacement of P59A in SARS-CoV-1 (equivalently reside in SCV2-CPP235) resulted in incomplete virus assembly (Tseng et al., 2013). Mutation of W58F (identically locates in SCV2-CPP235) in TM2 did not influence assembly; however, preservation of an aromatic residue was decisive for SARS-CoV virus-like particle (VLP) assembly. This tryptophan residue might stabilize TM helices. It has been also demonstrated that the conservation of an aromatic residue such as F96 in TM3 of SCV2-CPP236 is also essential for VLP formation. It is known that “Aromatic-X-X-Aromatic” regions such as “WXXW” in TM2 are crucial for stabilizing the M protein dimerization. The corresponding residues are observed in SCV2-CPP235 (KLIFLWLLWP). Replacement of aromatic residues with “Ala” or “Leu” decreases VLP formation. A conserved motif defined as SMWSFNPETNIL within the CT domain is functionally essential for the assembly (Arndt et al., 2010). The five first residues of this motif are found in SCV2-CPP241. Homologous C159 at the CT domain of M protein in SCV2-CPP244 potentially can affect VLP formation through M-M interaction to form homo-oligomers. Y196 found in SCV2-CPP245 (Y 196SRYRIGNYK) of the M protein in the CT domain is discussed as a critical residue for the interaction with spike protein (M-S interaction) (Ujike and Taguchi, 2015). Altogether, it seems that aromatic residues such as “Phe” and “Trp” in a structural protein like M are significantly crucial for VLP formation or viral assembly and stabilizing M protein dimerization. The “Cys” residue is involved in M-M and “Tyr” in M-S interaction.
3.2.17. Nucleocapsid protein (N) and SCV2-CPPs
The large genome (about 30 kb) of coronavirus should be inserted into virions with about 100 nm diameter; hence, considerable supercoiling of the genome is needed in a well-packed ribonucleoprotein (RNP) complex. The most crucial role of N protein is the development of a tight bond with the viral RNA (McBride et al., 2014). As displayed in Fig. 1F, the N protein is organized in the following order; 1) an intrinsically disordered region (IDR) flanking residues 1–44 of SARS-CoV-2 N protein, 2) residues of about 45–181 forms the N-terminal domain (NTD), 3) a middle IDR linker region spanning residues 182–247 called LKR, 4) and a C-terminal domain (CTD) lies in residues 248–365 of the N protein (Chang et al., 2016). SCV2-CPP257–262 is located in IDR1. The NTD has peptides, including SCV2-CPP267–276, and the LKR region consists of SCV2-CPP278–287. The CTD region harbors SCV2-CPP288–299. Finally, SCV2-CPP309–310 is situated in the last IDR domain. NTD is an RNA binding domain, and CTD is an RNA binding and oligomerization domain. The middle IDR (LKR) and the C-terminal IDR have also been connected to the oligomerization of N protein. To pack the genome into RNP, the N protein should have a tight bond with RNA. On the other hand, for adequate expression, the N protein needs a simple dissociation to present the viral RNA. To achieve a dynamic between the above-mentioned opposite roles, two strategies are adopted by the N protein. One is the extra positive charge in the N protein, which is critical for interaction with RNA and repulsive for self-dimerization. The N protein of SARS-CoV is profoundly basic. The prevailing presence of positively charged residues (Arg and Lys) in NTD renders a positively charged pocket for electrostatic interaction with the RNA phosphate group. IDRs are the other strategy for the dynamic activity of N protein as a modulator for RNA binding and oligomerization. IDRs do not have a fixed 3D structure in the native state and links in protein-protein interaction. The LKR has a phosphorylated Ser/Arg-rich region such as (XSSRSSSRSRX) in SCV2-CPP278–279 involved in N-M interaction (Chang et al., 2014). It has been revealed that the absence of a region equivalent to SCV2-CPP294 (ASKKPRQKRT) in the CTD of SARS-CoV-1 reduced protein-protein and protein-nucleic acid interaction substantially. The N protein of SARS-CoV basic charges are mainly distributed in three regions including the SR-rich region of LKR, CTD's N-terminus with SCV2-CPP288 (KKSAAEASKK), and the C-terminus disordered region (IDR3) with SCV2-CPP305–308 rich in lysine residues “KKDKKKK”.
3.2.18. Accessory proteins and SCV2-CPPs
3.2.18.1. ORF3a
ORF3a (X1) with 275 amino acid residues is an ion channel protein, encoded by a section linking S and E proteins. ORF3a has three TM domains and is localized to the ERGIC complex. ORF3a is correlated to virulence through the management of the cytokine and chemokine expression. It also triggers necrotic cell death (Siu et al., 2019). Nineteen different non-synonymous a.a exchanges among 537 SARS-CoV-2 strains have been clarified recently (Issa et al., 2020). Some mutations such as K61N in SCV2-CPP214–217 and W128L in SCV2-CPP221–228 were functionally deleterious.
3.2.18.2. ORF6
With a hydrophobic N- and hydrophilic C-terminus, ORF6 is localized to the ERGIC membrane of infected cells (Frieman et al., 2007). With only one SCV2-CPP246, ORF6 disrupts innate immunity responses.
3.2.18.3. ORF7a
ORF7a involves in viral replication cycle and induction of inflammatory responses. ORF7a is a type I TM protein. Residues 1–96 are in the ectodomain containing SCV2-CPP247-250. Residues 97–116 situated in the TM domain, and a.a 117–121 constitutes a short cytoplasmic tail. SCV2-CPP251-253 is in the border of TM and the cytoplasmic domain. The C-terminus contains a “KRKTE” motif with three positively charged residues that act as a signal for the export of protein from ER to the Golgi apparatus (McBride and Fielding, 2012). Positively charged residues in this motif are represented in SCV2-CPP253 (LCFTLKRKTE).
3.2.18.4. ORF8
SCV2-CPP254–256 exists in ORF8, which can be found only in lineage B of β-CoVs. The exact function of ORF8 in replication or pathogenesis is not yet clarified. Nevertheless, there are reports on the step by step deletion of ORF8 during the epidemic of SARS-CoV-1 (Muth et al., 2018).
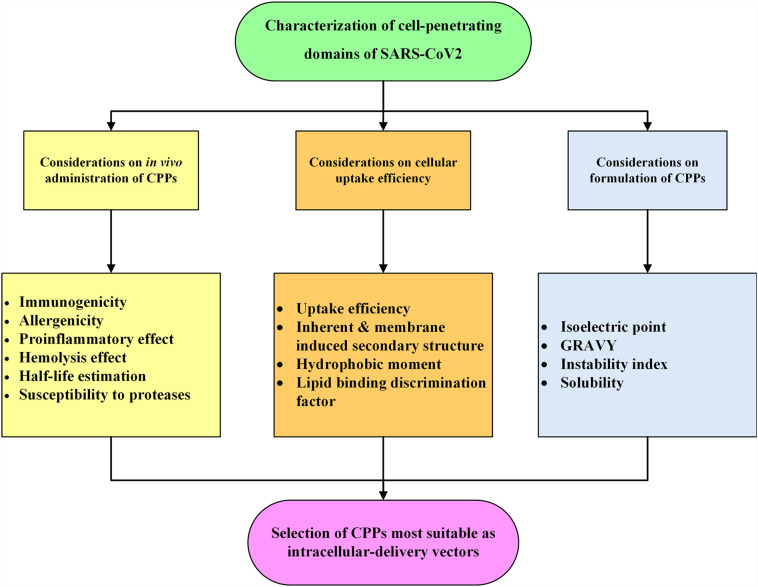
3.3. Characterization of SCV2-CPPs as drug delivery vectors
SARS-CoV-2 provides an opportunity for the discovery of novel CPPs. In the following section, SCV2-CPPs were verified for critical characteristics to be developed as drug carriers or as peptide biotherapeutics. Therefore, SCV2-CPPs were evaluated for their uptake efficiency, physiochemical properties, solubility, half-life, toxicity, immunogenicity, RBC lysis potential, protease susceptibility, inherent and membrane induced secondary structure, amphipathicity, and lipid binding potential.
3.3.1. Uptake efficiency of SCV2-CPPs
Out of 310 potential CPPs recognized by CellPPD web server, 297 were confirmed as confident CPPs using MLCPP web server. Thirteen peptides that were predicted as non-CPPs by MLCPP were excluded from further analyses as drug delivery vectors. CPPs are generally used for the intracellular delivery of therapeutic macromolecules. However, not all CPPs have adequate cellular uptake for sufficient therapeutic response (Keller et al., 2013). Higher cellular uptake of CPPs makes them promising as tools for drug delivery. Besides cell penetration ability, MLCPP predicts the uptake efficiency of submitted peptides. Thirteen percent of the SARS-CoV-2 CPPs were estimated to have high uptake efficiencies (Table 2). NSP4 and ORF3a had the highest number of CPPs with high uptake efficiencies. Prediction confidence of cell penetration and uptake efficiency for all the peptides is available at Supplementary material 2.
For some of the selected viral-derived CPPs as positive controls, the uptake efficiency has been reported individually or in comparison to other well-known CPPs (Table 3). The literature description on the uptake efficiency of the positive control CPPs was compared to the predicted uptake efficiency using web servers (Table 3). Out of ten viral-derived CPPs as positive controls, five of them were predicted to have high uptake efficiency (NLS-A, FHV coat (35–49), PepR, TAT, and REV) and three peptides were identified as CPPs with low uptake efficiency (VP-22, Erns, and Pep1) using the MLCPP server. This observation is in agreement with the experimentally validated uptake efficiency reported in the literature. It is of note that despite the prediction of CellPPD, MLCPP could not recognize VG-21 and HPV-WT as CPPs. There are two highly confident available webservers (CPPred-RF and MLCPP) that can predict not only CPPs but also their uptake efficiency. Of these two servers, MLCPP has been authenticated using an independent dataset (Manavalan et al., 2018b). An empirical study on the web-based CPP prediction tools showed that CPPred-RF has higher sensitivity, whereas MLCPP displays higher specificity and accuracy (Su et al., 2020). While the lower sensitivity implies that some CPP sequences might be missed by the MLCPP server (for example, VG-21 and HPV-WT), the higher accuracy and specificity lower the risk of false-positives among the recognized SCV2-CPPs. In other words, the retrieval of eight positive control CPPs that are confirmed empirically proves the robustness of our procedure to recognizing confident hits. Consideration of stringent conditions in our analyses in a way that two positive controls were not recognized by MLCPP diminishes the possibility of false-positives for further in vitro and in vivo experiments, which in turn saves time and expenses.
3.3.2. Physiochemical properties of SCV2-CPPs
Several physiochemical properties of SCV2-CPPs were calculated using the ProtParam tool (Supplementary material 3). The Mw of CPPs affects their transmembrane transport and protease degradation. Higher Mw corresponds to lower uptake efficiency and higher degradation propensity of peptides (Wang and Li, 2017). The Mw of the identified CPPs ranged from 975 kDa (SCV2-CPP285) to 1466 kDa (SCV2-CPP121). Nevertheless, the differences observed in the Mw of SCV2-CPPs are not as high to be the sole reason for variations regarding cellular uptake or protease susceptibility.
Another determining factor to consider a peptide as a suitable drug delivery vector is pI. pI is defined as a pH at which a peptide or protein is neutral and carries no net electrical charge. The pI of a peptide is calculated using pKa values of its amino acid residues and greatly affects the pH-dependent behavior of the peptide. Furthermore, therapeutic CPPs should not have a pI similar to the pH of their delivery route or target organ. According to our analyses, except for SCV2-CPP212, SCV2-CPP286, and SCV2-CPP287 with an acidic theoretical pI of 6.2 (negative net charge) and SCV2-CPP205 with a near-neutral pI of 7.1 (a net charge of about zero), the rest of SCV2-CPPs (98.65%) displayed basic isoelectric points between 8.6 and 12.6 and are considered as positively charged (Supplementary material 3).
The stability of SCV2-CPPs was estimated by analyzing their instability index values. Proteins and peptides with instability index values lower than 40 are expected to be stable in test tubes (Behzadipour and Hemmati, 2019). According to the calculated data, 47.8% of SCV2-CPPs would be stable in vitro.
The GRAVY value for a peptide is defined by adding the hydropathy index of each residue divided by the sequence length. A more negative value indicates that the peptide is more hydrophilic (Owji and Hemmati, 2018). GRAVY values for SCV2s-CPPs range from −3.3 to 2.2. Only 13.8% of SCV2-CPPs had GRAVY values >0. Therefore, most of the SCV2-CPPs (86.2%) are expected to be fairly hydrophilic with suitable water solubility.
Peptide aggregation is a major stability concern. For pharmaceutical purposes usually concentrated solutions of peptides are required. Therefore, having a highly soluble therapeutic peptide is favorable (Kramer et al., 2012). Pepcalc web server presents an estimation of water solubility of peptides based on their pI, the number of charged residues, and peptide length. According to the estimation, only a small number of peptides (9.8%) were predicted to have poor water solubility, which is in line with previous results based on GRAVY values (supplementary material 3).
3.3.3. Analyses of SCV2-CPPs for in vivo applications
SCV2-CPPs were verified for various important biological properties affecting their in vivo administration (Supplementary material 4). In addition to in vitro stability during the manufacturing process, peptides should have appropriate stability in vivo as well. Suboptimal in vivo half-life of peptides is one of the major challenges in therapeutic peptide development (Mathur et al., 2016). Peptides with low half-lives are rapidly cleared from the circulation and will not achieve adequate biological responses (Böttger et al., 2017). Plifepred web server calculates half-life of peptides in blood medium. The calculated half-lives are in the range of 529 s for SCV2-CPP160 to 2057 s for SCV2-CPP107. This seems an acceptable range compared with the control penetrating peptides such as poly-Arginine (R8) and TAT (vastly used as drug delivery vectors) with calculated half-lives of about 834 and 2329 s, respectively. It seems that the charge and size of amino acid residues influence the half-life of peptides. Peptides consisted of large aromatic and positively charged amino acids displayed lower half-lives in comparison to that of smaller negatively charged residues (Mathur et al., 2018).
One of the hurdles to find an optimum CPP is cytotoxicity. Toxicity might be due to their effect on the cellular and organelle membrane integrity, production of free radicals, or aggregation (Cieślik et al., 2015; Derakhshankhah and Jafari, 2018). According to the Toxinpred server, 94.3% of the identified SCV2-CPPs were recognized as non-toxins (Supplementary material 4).
Hemolysis is one of the concerns during the in vivo administration of peptides and is considered as an index of peptide toxicity (Win et al., 2017). HemoPI server is able to evaluate hemolytic potency of submitted peptides by providing a PROB score. PROBE score ranges between 1 (very likely to be hemolytic) and 0 (very unlikely to be hemolytic). We considered peptides with a PROBE score of 0.50 and higher to be potentially hemolytic. Only 11.11% of SCV2-CPPs were classified as hemolytic (Supplementary material 4).
Despite less immunogenicity than proteins and antibodies, peptides can exert immunologic responses after in vivo administration. Immunogenicity of peptides can lead to allergic reactions or affect their activity (Shankar et al., 2014). Hypersensitivity reactions caused by biologicals result in mild to life-threatening symptoms (Deptuła et al., 2018). The presence of peptides in the body might induce the production of antibodies that can lead to the neutralization of therapeutic and loss of efficacy (Kuriakose et al., 2016). Vaxijen web server predicted that 47.47% of SCV2-CPPs are antigenic peptides. These peptides may stimulate the production of anti-drug antibodies in recipients. On the other hand, 34.68% of the identified CPPs were predicted as probable allergens by the Allertop web server. These peptides have a risk of inducing hypersensitivity reactions after administration (Supplementary material 4). However, 38% of SCV2-CPPs were neither antigenic nor allergenic.
3.3.4. Protease susceptibility of SCV2-CPPs
A protease or peptidase is an enzyme that hydrolyzes proteins and peptides to polypeptides or single amino acids. Proteases are categorized into four families as follows: 1) aspartic proteases such as pepsins, 2) cysteine proteases namely caspases, 3) metalloproteases like matrix metallopeptidase, and 4) serine proteases such as plasmin and thrombin (Varkhede et al., 2020). Inherent susceptibility of peptides to protease degradation and their further inactivation would significantly prevent their development as therapeutic agents (Heard et al., 2013). An efficient CPP should have relative resistance to proteases for intact delivery to target cells. Therefore, CPPs were evaluated for their susceptibility to different classes of proteases using the Prosper server. The obtained results are reported based on the protease family (Supplementary material 5). Only SCV2-CPP36 and SCV2-CPP83 were predicted to be cleaved by aspartic proteases. A total of 22, 107, and 125 SCV2-CPPs were predicted to be cleaved by cysteine proteases, metalloproteases, and serine proteases, respectively. Conclusively, the identified SCV2-CPPs were most susceptible to serine and metalloproteases with the highest resistance to aspartic proteases. Among the identified CPPs, 8 peptides (SCV2-CPP26, SCV2-CPP144, SCV2-CPP177, SCV2-CPP178, SCV2-CPP181, SCV2-CPP207, SCV2-CPP242, and SCV2-CPP243) were predicted to be lysed by three different protease families and 50 peptides were predicted to be cleaved by two different protease families. Interestingly, 36.70% of SCV2-CPPs were resistant to all four groups of protease families.
3.3.5. Secondary structure and membrane interaction of SCV2-CPPs
It has been defined experimentally while some CPPs do not show any secondary structure in aqueous solution, some others display a very low or a mixed α-helix and β-sheet structure. However, those CPPs that are amphipathic in nature exist as α-helical or β-sheet forms (Kalafatovic and Giralt, 2017). The inherent secondary structure of SARS-CoV-2 CPPs was predicted using PEP2D web server. On average, coiled conformations were the most abundant predicted form (76.46% of residues) as anticipated. Following coiled conformation, 16.81% and 6.74% of SCV2-CPP residues had α-helical and β-sheet structures (Supplementary material 6). Amphipathic CPPs are additionally classified into primary and secondary amphipathic peptides. In primary amphipathic peptides, the cationic and hydrophobic zone of the CPP is defined based on the primary structure and at the sequence scale. In secondary amphipathic peptides, the division of cationic and hydrophobic regions occurs after folding of the peptide (Di Pisa et al., 2015). While some amphipathic CPPs can form secondary structures or reveal conformational changes in buffered solutions, others only fold after interaction with the plasma membrane (Lee et al., 2010). Some purely cationic CPPs such as “Tat” or “R9” have random coil conformations, and their translocation across the plasma membrane does not rely on the secondary structure. However, for amphipathic peptides, cellular uptake efficiency is highly influenced by the ability to fold into α-helical or β-sheet structures (Eiríksdóttir et al., 2010; Futaki et al., 2007). The effect of the helical structure of CPPs (whether inherent, membrane or artificially induced) on cellular internalization has been discussed by Kalafatovic and Giralt (Kalafatovic and Giralt, 2017).
To evaluate the influence of peptide-membrane interactions on the secondary structure, SCV2-CPPs were further submitted to the fmap server. Fmap predicts the stable helical regions of peptides in association with the lipid membrane. Among all the identified SCV2-CPPs, 78 CPPs were predicted to fold partially or completely into stable α-helices upon contact with the membrane (Supplementary material 6 and Fig. 2 ).
Fig. 2.
Eight representative SCV2-CPPs with more than 90% helical conformation in contact with the membrane. The helices of various CPPs display different tilt angles. Blue and red surfaces represent the outer and inner layers of the bilayer membrane, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.6. Determination of lipid-binding potential for helical SCV2-CPPs
A notable motif observed in some proteins and peptides is the membrane-binding amphipathic helix (AH). These regions play a prominent role in lipid-protein interplays. (Drin and Antonny, 2010). For example, they are required in membrane remodeling machinery, membrane fission, or can be served as membrane anchors (Zhukovsky et al., 2019). The unique feature of the AH region, which is the separation of the lipophilic and hydrophilic side of an α-helix enables proteins to interact with lipids. The AH motifs are potentially attractive CPP candidates since they are able to have an efficacious interaction with phospholipids of the membrane (Kim et al., 2015). The polar side of the helical peptide has electrostatic interactions with the polar head of phospholipids, whereas the hydrophobic side of the helix is inserted between the acyl-chains. (Seelig, 2004). Numerous studies have been conducted regarding the role, structure, and identification of these domains in proteins and peptides (Keller, 2011). Heliquest web server is applicable to identify AH regions according to the methodology introduced by Eisenberg et al. known as the hydrophobic moment (Eisenberg et al., 1984). Heliquest calculates several parameters for a sequence including μH (a measure of the sequence amphipathicity) and z (the net charge of the sequence). μH and z are used to determine Heliquest lipid-binding discrimination factor (D). Based on the calculated discrimination factor the peptides can be sorted as a lipid-binding helix (D > 1.34), possible lipid-binding helix (0.68 < D < 1.34), and non-lipid binding peptides (D < 0.68) (Gautier et al., 2008). The calculated hydrophobic moment of SARS-CoV-2 helical CPPs was in the range of 0.06 to 0.89 (Table 4 ). The higher value of μH means that the helix has a higher degree of amphipathicity. The lipid-binding discrimination factor of the identified helical SCV2-CPPs ranged from 0.12 to 2.08. Based on the D factor, 44.30% and 48.10% of helical SCV2-CPPs were identified as lipid-binding helices and possible lipid-binding helices, respectively (Table 4).
Table 4.
Hydrophobic moments (μH) and lipid-binding discrimination factors (D) of SARS-CoV-2 helical CPPs.
| Peptide name | μH | D | Interpretation | Peptide name | μH | D | Interpretation |
|---|---|---|---|---|---|---|---|
| SCV2-CPP3 | 0.31 | 1.29 | Possible lipid-binding helix | SCV2-CPP129 | 0.50 | 1.46 | Lipid-binding helix |
| SCV2-CPP4 | 0.21 | 1.52 | Lipid-binding helix | SCV2-CPP131 | 0.18 | 0.83 | Possible lipid-binding helix |
| SCV2-CPP7 | 0.20 | 1.51 | Lipid-binding helix | SCV2-CPP144 | 0.16 | 0.48 | Non lipid-binding helix |
| SCV2-CPP9 | 0.47 | 1.10 | Possible lipid-binding helix | SCV2-CPP151 | 0.43 | 1.39 | Lipid-binding helix |
| SCV2-CPP15 | 0.53 | 1.49 | Lipid-binding helix | SCV2-CPP154 | 0.14 | 1.13 | Possible lipid-binding helix |
| SCV2-CPP16 | 0.63 | 1.91 | Lipid-binding helix | SCV2-CPP155 | 0.14 | 1.12 | Possible lipid-binding helix |
| SCV2-CPP22 | 0.09 | 0.75 | Possible lipid-binding helix | SCV2-CPP156 | 0.10 | 1.08 | Possible lipid-binding helix |
| SCV2-CPP26 | 0.31 | 0.95 | Possible lipid-binding helix | SCV2-CPP157 | 0.06 | 1.04 | Possible lipid-binding helix |
| SCV2-CPP31 | 0.64 | 1.60 | Lipid-binding helix | SCV2-CPP159 | 0.31 | 0.96 | Possible lipid-binding helix |
| SCV2-CPP32 | 0.66 | 1.61 | Lipid-binding helix | SCV2-CPP160 | 0.60 | 1.23 | Possible lipid-binding helix |
| SCV2-CPP33 | 0.51 | 1.47 | Lipid-binding helix | SCV2-CPP179 | 0.57 | 1.20 | Possible lipid-binding helix |
| SCV2-CPP34 | 0.53 | 1.82 | Lipid-binding helix | SCV2-CPP181 | 0.44 | 0.75 | Possible lipid-binding helix |
| SCV2-CPP36 | 0.34 | 0.65 | Non lipid-binding helix | SCV2-CPP182 | 0.44 | 1.07 | Possible lipid-binding helix |
| SCV2-CPP37 | 0.46 | 1.42 | Lipid-binding helix | SCV2-CPP183 | 0.43 | 1.39 | Lipid-binding helix |
| SCV2-CPP39 | 0.53 | 1.82 | Lipid-binding helix | SCV2-CPP185 | 0.34 | 1.31 | Possible lipid-binding helix |
| SCV2-CPP41 | 0.19 | 0.84 | Possible lipid-binding helix | SCV2-CPP186 | 0.38 | 1.02 | Possible lipid-binding helix |
| SCV2-CPP42 | 0.41 | 1.38 | Lipid-binding helix | SCV2-CPP191 | 0.34 | 0.98 | Possible lipid-binding helix |
| SCV2-CPP44 | 0.36 | 0.67 | Non lipid-binding helix | SCV2-CPP192 | 0.23 | 0.87 | Possible lipid-binding helix |
| SCV2-CPP72 | 0.69 | 1.31 | Possible lipid-binding helix | SCV2-CPP198 | 0.69 | 1.64 | Lipid-binding helix |
| SCV2-CPP73 | 0.53 | 1.49 | Lipid-binding helix | SCV2-CPP214 | 0.24 | 1.22 | Possible lipid-binding helix |
| SCV2-CPP74 | 0.62 | 1.91 | Lipid-binding helix | SCV2-CPP215 | 0.31 | 1.62 | Lipid-binding helix |
| SCV2-CPP75 | 0.45 | 2.08 | Lipid-binding helix | SCV2-CPP217 | 0.49 | 1.78 | Lipid-binding helix |
| SCV2-CPP77 | 0.32 | 1.29 | Possible lipid-binding helix | SCV2-CPP219 | 0.34 | 1.64 | Lipid-binding helix |
| SCV2-CPP78 | 0.73 | 1.35 | Lipid-binding helix | SCV2-CPP220 | 0.41 | 1.37 | Lipid-binding helix |
| SCV2-CPP79 | 0.47 | 1.43 | Lipid-binding helix | SCV2-CPP221 | 0.43 | 1.06 | Possible lipid-binding helix |
| SCV2-CPP80 | 0.30 | 1.28 | Possible lipid-binding helix | SCV2-CPP223 | 0.52 | 1.15 | Possible lipid-binding helix |
| SCV2-CPP81 | 0.48 | 1.44 | Lipid-binding helix | SCV2-CPP233 | 0.33 | 1.30 | Possible lipid-binding helix |
| SCV2-CPP82 | 0.60 | 1.23 | Possible lipid-binding helix | SCV2-CPP234 | 0.49 | 0.79 | Possible lipid-binding helix |
| SCV2-CPP87 | 0.50 | 1.13 | Possible lipid-binding helix | SCV2-CPP235 | 0.30 | 0.62 | Non lipid-binding helix |
| SCV2-CPP88 | 0.39 | 1.36 | Lipid-binding helix | SCV2-CPP236 | 0.52 | 1.15 | Possible lipid-binding helix |
| SCV2-CPP96 | 0.51 | 1.47 | Lipid-binding helix | SCV2-CPP237 | 0.34 | 1.31 | Possible lipid-binding helix |
| SCV2-CPP98 | 0.76 | 2.03 | Lipid-binding helix | SCV2-CPP238 | 0.37 | 1.34 | Lipid-binding helix |
| SCV2-CPP101 | 0.52 | 1.15 | Possible lipid-binding helix | SCV2-CPP243 | 0.32 | 0.96 | Possible lipid-binding helix |
| SCV2-CPP106 | 0.56 | 0.86 | Possible lipid-binding helix | SCV2-CPP250 | 0.16 | 1.14 | Possible lipid-binding helix |
| SCV2-CPP112 | 0.53 | 1.16 | Possible lipid-binding helix | SCV2-CPP267 | 0.38 | 1.35 | Lipid-binding helix |
| SCV2-CPP118 | 0.40 | 1.04 | Possible lipid-binding helix | SCV2-CPP268 | 0.45 | 1.74 | Lipid-binding helix |
| SCV2-CPP119 | 0.49 | 1.45 | Lipid-binding helix | SCV2-CPP269 | 0.56 | 1.85 | Lipid-binding helix |
| SCV2-CPP120 | 0.14 | 0.79 | Possible lipid-binding helix | SCV2-CPP286 | 0.13 | 0.12 | Non lipid-binding helix |
| SCV2-CPP121 | 0.62 | 1.58 | Lipid-binding helix | SCV2-CPP287 | 0.29 | 0.27 | Non lipid-binding helix |
| SCV2-CPP122 | 0.89 | 1.50 | Lipid-binding helix | ||||
The lipid-binding discrimination factor for ten positive viral-derived control CPPs was also calculated using μH and z value provided by Heliquest (Table 5 ). The viral-derived CPPs as positive controls were amphipathic, cationic, and negatively-charged in nature. As expected, all of the cationic and amphipathic CPPs were evaluated as lipid-binding peptides (D > 1.34). The negatively charged VG-21 however was recognized as a non-lipid binding peptide (D < 0.68). It seems that the Heliquest method works best with neutral to positively charged helices. Most negatively charged peptides are expected to be repulsed by a negatively charged membrane unless they are highly amphipathic. As VG-21 has increased the uptake of gold nanoparticles experimentally, it has been argued that due to its negative charge, instead of electrostatic interaction, VG-21 might have non-specific or other uptake mechanisms such as receptor-mediated endocytosis or direct penetration (Tiwari et al., 2014).
Table 5.
The nature, hydrophobic moment (μH), and lipid-binding discrimination factor (D) of several experimentally validated viral-derived CPPs as positive controls.
| Peptide name | Nature | μH | D | Interpretation |
|---|---|---|---|---|
| NLS-A | Cationic | 0.25 | 3.21 | Lipid binding |
| FHV coat (35–49) | Cationic | 0.25 | 3.86 | Lipid binding |
| PepR | Cationic | 0.69 | 1.97 | Lipid binding |
| VP22 | Amphipathic | 0.34 | 1.97 | Lipid binding |
| TAT | Cationic | 0.23 | 2.86 | Lipid binding |
| Erns | Amphipathic | 0.47 | 1.76 | Lipid binding |
| Pep1 | Amphipathic | 0.43 | 1.39 | Lipid binding |
| VG-21 | Anionic | 0.30 | −1.04 | Non-lipid binding |
| REV | Cationic | 0.25 | 3.20 | Lipid binding |
| HPV-WT | Amphipathic | 0.16 | 1.47 | Lipid binding |
3.3.7. The best SCV2-CPP candidates as drug delivery vectors
To introduce the most optimal SCV2-CPP candidates for further in vitro and in vivo studies, peptides were assorted based on the above-mentioned analyses (Fig. 3 ). Peptides with the highest sum score are potentially considered as suitable candidates for drug delivery applications. The SCV2-CPPs were classified according to various predicted attributes as follows:
-
•
Uptake efficiency: peptides were scored “0” for low and “+1” for high uptake efficiency.
-
•
Isoelectric point: peptides got a rank of “0” for a theoretical pI within ±0.4 range of the blood pH (7.0–7.8). The rest of the peptides had a score of “+1”.
-
•
Instability index: peptides with an instability index higher and lower than 40 were graded as “0” and “+1”, respectively.
-
•
Water solubility: peptides with poor and good water solubility got a degree of “0” and “+1”, respectively.
-
•
Half-life in blood: peptides with half-lives longer and shorter than 900 s (15 min) were scored “0” and “+1”, respectively.
-
•
Toxicity: toxic and non-toxic peptides got a score of “0” and “+1”, respectively.
-
•
Hemolytic activity: peptides with a probe score equal or higher than 0.50 were defined by “0”, while peptides with a probe score less than 0.50 assigned “+1”.
-
•
Antigenicity: antigenic and non-antigenic peptides were displayed as “0” and “+1”, respectively.
-
•
Allergenicity: probable and non-probable allergic peptides were designated as “0” and “+1”, respectively.
-
•
Protease susceptibility: peptides that were predicted to be cleaved by one or more protease families scored “0”, while peptides that were not cleaved by any of the protease families were defined as “+1”.
-
•
Secondary structure: peptides with 50% or higher helical or sheet secondary structure got a score of “+1”. The rest of the peptides were scored “0” (Supplementary material 7).
Fig. 3.
The proposed workflow to select the most proper SARS-CoV-2 derived CPP candidates as intracellular delivery vectors.
Among all the analyzed SCV2-CPPs, SCV2-CPP118, SCV2-CPP119, SCV2-CPP122, and SCV2-CPP129 had the highest sum-score. These four CPPs all reside in NSP12. These peptides are assumed to have an amphipathic nature after folding into an α-helix and can be regarded as secondary amphipathic. In between, SCV2-CPP122 has the highest hydrophobic moment; as a result, the highest degree of amphiphilicity. The hydrophilic and hydrophobic residues of SCV2-CPP122 are completely separated in the α-helical structure and can fold into a nearly complete α-helix after interaction with the lipid membrane (Fig. 4 ). SCV2-CPP122 had the highest hydrophobic moment value (0.89) even in comparison to the viral-derived CPPs as the positive control, which means a higher degree of amphipathicity. The hydrophobic moment values for the other three selected peptides were higher or comparable to most positive controls with a μH value of 0.40, 0.49, and 0.50 for SCV2-CPP118, SCV2-CPP119, and SCV2-CPP129, respectively (Table 4, Table 5). The lipid-binding discrimination factor for the four selected NSP12-derived CPPs, including SCV2-CPP118, SCV2-CPP119, SCV2-CPP-122, and SCV2-CPP129 are 1.04, 1.45, 1.50, and 1.46, respectively; which is comparable to the D value of the positive control amphipathic peptides such as Pep1 (D = 1.39) and Erns (D = 1.76) (Table 5).
Fig. 4.
Segregation of hydrophilic and hydrophobic residues of SCV2-CPP118, SCV2-CPP119, SCV2-CPP122, and SCV2-CPP129 in their primary sequences and helical structure.
3.4. SCV2-CPPs as bioactive peptides
While CPPs are used as drug delivery vectors, some peptides with cell-penetration ability are adopted as bio-therapeutics themselves, such as antimicrobial and anticancer peptides. There are also reports on the immunomodulatory effects of some CPPs. In addition to the drug delivery capacity, CPPs were further analyzed to verify if SCV2-CPPs can be exploited as agents against bacterial, viral, and fungal infections. Their potential against cancer and chronic inflammatory diseases was also calculated.
3.4.1. Antimicrobial peptides
Antimicrobial peptides (AMPs) are short cationic peptides with high affinity to the membrane, some of which exist naturally as modulators of the eukaryotic immune system (Boman, 2000). However, many AMPs have been artificially designed and synthesized as well. Due to growing antibiotic resistance, these peptides are emerging as a promising alternate to the regular antibiotic therapy. Some AMPs enter cells without perpetual membrane destruction; therefore, they can be used as vectors for intracellular delivery of bioactive macromolecules (Splith and Neundorf, 2011). On the other hand, some CPPs have potent antimicrobial activity (Gaspar et al., 2013).
iAMPpred server categorizes the antimicrobial activity into three classes, including antibacterial, antiviral, and antifungal. A peptide with a probability score equal to or higher than 0.5 is considered as a positive AMP. If a peptide belongs to all the classes mentioned above, the likelihood of that peptide as an AMP is higher. About 59.60%, 29.63%, and 32.32% of SCV2-CPPs were identified as potential antibacterial, antiviral, and antifungal peptides, respectively (Table 6). Moreover, 16.50% of peptides associated with all three classes of AMPs. SCV2-CPP139, SCV2-CPP140, SCV2-CPP187, and SCV2-CPP246 were identified as antibacterial, antiviral, and antimicrobial AMPs with a probability higher than 0.90. Therefore, these four peptides are the most promising potential AMPs in the proteome of SARS-CoV-2.
Table 6.
Potential antimicrobial, anticancer, and anti-inflammatory activities of SARS-CoV-2 CPPs.
| Peptide name | Antimicrobial potential |
Anticancer potential | Anti-inflammatory potential | Peptide name | Antimicrobial potential |
Anticancer potential | Anti-inflammatory potential | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibacterial score | Antiviral score | Antifungal score | Antibacterial score | Antiviral score | Antifungal score | ||||||
| SCV2-CPP1 | 0.02 | 0.02 | 0.07 | x | ✓ | SCV2-CPP161 | 0.49 | 0.53 | 0.54 | ✓ | ✓ |
| SCV2-CPP2 | 0.04 | 0.02 | 0.04 | x | ✓ | SCV2-CPP162 | 0.7 | 0.15 | 0.38 | x | x |
| SCV2-CPP3 | 0.77 | 0.43 | 0.37 | x | x | SCV2-CPP163 | 0.92 | 0.26 | 0.7 | ✓ | x |
| SCV2-CPP4 | 0.91 | 0.59 | 0.7 | x | ✓ | SCV2-CPP164 | 0.25 | 0.41 | 0.09 | x | ✓ |
| SCV2-CPP5 | 0.79 | 0.36 | 0.38 | x | x | SCV2-CPP165 | 0.46 | 0.26 | 0.15 | x | ✓ |
| SCV2-CPP6 | 0.88 | 0.38 | 0.63 | x | ✓ | SCV2-CPP166 | 0.6 | 0.19 | 0.49 | ✓ | x |
| SCV2-CPP7 | 0.46 | 0.15 | 0.25 | x | ✓ | SCV2-CPP168 | 0.17 | 0.17 | 0.16 | x | x |
| SCV2-CPP8 | 0.64 | 0.13 | 0.34 | x | ✓ | SCV2-CPP169 | 0.06 | 0.04 | 0.04 | x | x |
| SCV2-CPP9 | 0.85 | 0.49 | 0.53 | x | ✓ | SCV2-CPP170 | 0.96 | 0.35 | 0.68 | ✓ | ✓ |
| SCV2-CPP12 | 0.27 | 0.21 | 0.18 | x | ✓ | SCV2-CPP171 | 0.92 | 0.39 | 0.54 | x | ✓ |
| SCV2-CPP13 | 0.27 | 0.21 | 0.18 | x | ✓ | SCV2-CPP172 | 0.18 | 0.44 | 0.06 | x | ✓ |
| SCV2-CPP14 | 0.56 | 0.37 | 0.35 | x | ✓ | SCV2-CPP173 | 0.57 | 0.42 | 0.22 | x | x |
| SCV2-CPP15 | 0.98 | 0.64 | 0.97 | ✓ | x | SCV2-CPP174 | 0.48 | 0.19 | 0.15 | x | ✓ |
| SCV2-CPP16 | 0.96 | 0.85 | 0.96 | x | ✓ | SCV2-CPP175 | 0.14 | 0.1 | 0.04 | x | x |
| SCV2-CPP17 | 0.86 | 0.61 | 0.74 | x | x | SCV2-CPP176 | 0.63 | 0.36 | 0.18 | ✓ | ✓ |
| SCV2-CPP18 | 0.97 | 0.38 | 0.94 | x | x | SCV2-CPP177 | 0.12 | 0.34 | 0.12 | x | x |
| SCV2-CPP19 | 0.97 | 0.34 | 0.78 | x | x | SCV2-CPP178 | 0.11 | 0.34 | 0.12 | x | x |
| SCV2-CPP20 | 0.96 | 0.35 | 0.75 | x | x | SCV2-CPP179 | 0.32 | 0.22 | 0.45 | x | x |
| SCV2-CPP21 | 0.97 | 0.39 | 0.64 | ✓ | x | SCV2-CPP180 | 0.6 | 0.5 | 0.63 | x | ✓ |
| SCV2-CPP22 | 0.99 | 0.67 | 0.94 | x | ✓ | SCV2-CPP181 | 0.04 | 0.05 | 0.04 | x | x |
| SCV2-CPP23 | 0.99 | 0.55 | 0.97 | x | ✓ | SCV2-CPP182 | 0.71 | 0.42 | 0.86 | x | x |
| SCV2-CPP24 | 0.99 | 0.57 | 0.97 | x | ✓ | SCV2-CPP183 | 0.9 | 0.64 | 0.97 | x | ✓ |
| SCV2-CPP25 | 0.98 | 0.58 | 0.96 | x | ✓ | SCV2-CPP184 | 0.81 | 0.12 | 0.78 | x | x |
| SCV2-CPP26 | 0.74 | 0.64 | 0.51 | x | ✓ | SCV2-CPP185 | 0.87 | 0.38 | 0.82 | x | ✓ |
| SCV2-CPP27 | 0.69 | 0.47 | 0.47 | ✓ | x | SCV2-CPP186 | 0.85 | 0.56 | 0.92 | ✓ | ✓ |
| SCV2-CPP28 | 0.79 | 0.3 | 0.55 | ✓ | x | SCV2-CPP187 | 0.98 | 0.93 | 0.99 | ✓ | ✓ |
| SCV2-CPP29 | 0.86 | 0.3 | 0.62 | ✓ | x | SCV2-CPP188 | 0.6 | 0.32 | 0.41 | x | ✓ |
| SCV2-CPP31 | 0.8 | 0.79 | 0.59 | x | x | SCV2-CPP189 | 0.48 | 0.16 | 0.24 | x | ✓ |
| SCV2-CPP32 | 0.69 | 0.39 | 0.43 | x | x | SCV2-CPP190 | 0.35 | 0.1 | 0.08 | x | x |
| SCV2-CPP33 | 0.92 | 0.1 | 0.58 | x | x | SCV2-CPP191 | 0.04 | 0.17 | 0.03 | x | ✓ |
| SCV2-CPP34 | 0.99 | 0.23 | 0.91 | x | ✓ | SCV2-CPP192 | 0.06 | 0.06 | 0.05 | x | ✓ |
| SCV2-CPP35 | 0.99 | 0.23 | 0.91 | x | x | SCV2-CPP193 | 0.63 | 0.32 | 0.17 | ✓ | ✓ |
| SCV2-CPP36 | 0.35 | 0.68 | 0.49 | ✓ | x | SCV2-CPP194 | 0.62 | 0.18 | 0.12 | x | ✓ |
| SCV2-CPP37 | 0.43 | 0.58 | 0.5 | x | x | SCV2-CPP195 | 0.53 | 0.12 | 0.11 | x | ✓ |
| SCV2-CPP38 | 0.66 | 0.77 | 0.66 | x | x | SCV2-CPP196 | 0.58 | 0.19 | 0.09 | x | ✓ |
| SCV2-CPP39 | 0.9 | 0.8 | 0.85 | x | ✓ | SCV2-CPP198 | 0.41 | 0.46 | 0.3 | x | ✓ |
| SCV2-CPP40 | 0.94 | 0.89 | 0.9 | ✓ | ✓ | SCV2-CPP199 | 0.32 | 0.36 | 0.25 | x | ✓ |
| SCV2-CPP41 | 0.16 | 0.38 | 0.09 | x | x | SCV2-CPP200 | 0.46 | 0.44 | 0.51 | x | ✓ |
| SCV2-CPP42 | 0.35 | 0.16 | 0.26 | x | ✓ | SCV2-CPP201 | 0.5 | 0.4 | 0.47 | x | ✓ |
| SCV2-CPP44 | 0.13 | 0.22 | 0.38 | x | ✓ | SCV2-CPP202 | 0.16 | 0.22 | 0.16 | x | ✓ |
| SCV2-CPP45 | 0.32 | 0.18 | 0.2 | x | x | SCV2-CPP203 | 0.47 | 0.09 | 0.12 | x | x |
| SCV2-CPP46 | 0.88 | 0.13 | 0.46 | ✓ | x | SCV2-CPP204 | 0.67 | 0.1 | 0.19 | x | ✓ |
| SCV2-CPP47 | 0.88 | 0.19 | 0.54 | ✓ | x | SCV2-CPP205 | 0.12 | 0.1 | 0.05 | x | x |
| SCV2-CPP48 | 0.78 | 0.12 | 0.32 | ✓ | x | SCV2-CPP206 | 0.51 | 0.26 | 0.15 | x | x |
| SCV2-CPP49 | 0.71 | 0.11 | 0.21 | ✓ | ✓ | SCV2-CPP207 | 0.19 | 0.11 | 0.03 | x | x |
| SCV2-CPP50 | 0.74 | 0.25 | 0.43 | ✓ | ✓ | SCV2-CPP208 | 0.04 | 0.02 | 0.03 | x | x |
| SCV2-CPP51 | 0.36 | 0.1 | 0.12 | x | x | SCV2-CPP209 | 0.03 | 0.02 | 0.03 | x | x |
| SCV2-CPP52 | 0.03 | 0.03 | 0.03 | x | x | SCV2-CPP210 | 0.1 | 0.04 | 0.07 | x | x |
| SCV2-CPP53 | 0.85 | 0.24 | 0.73 | x | x | SCV2-CPP211 | 0.67 | 0.39 | 0.56 | x | x |
| SCV2-CPP54 | 0.89 | 0.21 | 0.76 | x | ✓ | SCV2-CPP212 | 0.06 | 0.26 | 0.05 | x | x |
| SCV2-CPP55 | 0.99 | 0.52 | 0.96 | x | ✓ | SCV2-CPP213 | 0.04 | 0.18 | 0.38 | ✓ | ✓ |
| SCV2-CPP56 | 0.99 | 0.59 | 0.94 | ✓ | ✓ | SCV2-CPP214 | 0.92 | 0.63 | 0.82 | ✓ | x |
| SCV2-CPP57 | 0.96 | 0.3 | 0.62 | ✓ | ✓ | SCV2-CPP215 | 0.94 | 0.59 | 0.94 | ✓ | ✓ |
| SCV2-CPP58 | 0.98 | 0.58 | 0.94 | ✓ | x | SCV2-CPP216 | 0.78 | 0.73 | 0.83 | ✓ | ✓ |
| SCV2-CPP59 | 0.74 | 0.92 | 0.43 | x | ✓ | SCV2-CPP217 | 0.66 | 0.83 | 0.85 | ✓ | ✓ |
| SCV2-CPP60 | 0.74 | 0.74 | 0.8 | x | ✓ | SCV2-CPP218 | 0.35 | 0.8 | 0.77 | ✓ | ✓ |
| SCV2-CPP61 | 0.57 | 0.21 | 0.3 | ✓ | x | SCV2-CPP219 | 0.69 | 0.52 | 0.56 | x | ✓ |
| SCV2-CPP62 | 0.45 | 0.07 | 0.18 | x | ✓ | SCV2-CPP220 | 0.55 | 0.4 | 0.45 | x | ✓ |
| SCV2-CPP63 | 0.82 | 0.26 | 0.22 | x | ✓ | SCV2-CPP221 | 0.26 | 0.58 | 0.12 | x | ✓ |
| SCV2-CPP64 | 0.69 | 0.52 | 0.44 | ✓ | ✓ | SCV2-CPP222 | 0.55 | 0.74 | 0.22 | x | ✓ |
| SCV2-CPP65 | 0.67 | 0.22 | 0.14 | x | ✓ | SCV2-CPP223 | 0.62 | 0.83 | 0.26 | ✓ | ✓ |
| SCV2-CPP66 | 0.8 | 0.16 | 0.35 | x | ✓ | SCV2-CPP224 | 0.59 | 0.85 | 0.27 | ✓ | ✓ |
| SCV2-CPP67 | 0.22 | 0.06 | 0.06 | x | x | SCV2-CPP225 | 0.57 | 0.85 | 0.24 | ✓ | ✓ |
| SCV2-CPP68 | 0.19 | 0.05 | 0.04 | x | ✓ | SCV2-CPP226 | 0.52 | 0.86 | 0.26 | ✓ | ✓ |
| SCV2-CPP69 | 0.14 | 0.14 | 0.2 | x | ✓ | SCV2-CPP227 | 0.65 | 0.9 | 0.32 | ✓ | ✓ |
| SCV2-CPP70 | 0.22 | 0.52 | 0.25 | x | x | SCV2-CPP228 | 0.73 | 0.83 | 0.23 | ✓ | ✓ |
| SCV2-CPP72 | 0.31 | 0.72 | 0.69 | x | ✓ | SCV2-CPP229 | 0.81 | 0.66 | 0.4 | ✓ | ✓ |
| SCV2-CPP73 | 0.32 | 0.55 | 0.39 | x | ✓ | SCV2-CPP230 | 0.81 | 0.66 | 0.4 | x | ✓ |
| SCV2-CPP74 | 0.74 | 0.74 | 0.78 | x | ✓ | SCV2-CPP231 | 0.88 | 0.26 | 0.28 | x | ✓ |
| SCV2-CPP75 | 0.88 | 0.67 | 0.75 | x | ✓ | SCV2-CPP232 | 0.6 | 0.59 | 0.35 | x | ✓ |
| SCV2-CPP76 | 0.76 | 0.59 | 0.67 | x | x | SCV2-CPP233 | 0.69 | 0.91 | 0.58 | x | ✓ |
| SCV2-CPP77 | 0.35 | 0.35 | 0.32 | x | ✓ | SCV2-CPP234 | 0.5 | 0.84 | 0.38 | x | ✓ |
| SCV2-CPP78 | 0.51 | 0.26 | 0.2 | x | ✓ | SCV2-CPP235 | 0.69 | 0.88 | 0.25 | x | ✓ |
| SCV2-CPP79 | 0.5 | 0.56 | 0.24 | ✓ | ✓ | SCV2-CPP236 | 0.35 | 0.69 | 0.35 | x | ✓ |
| SCV2-CPP80 | 0.5 | 0.54 | 0.28 | x | ✓ | SCV2-CPP237 | 0.07 | 0.18 | 0.05 | x | ✓ |
| SCV2-CPP81 | 0.54 | 0.54 | 0.28 | x | ✓ | SCV2-CPP238 | 0.14 | 0.13 | 0.12 | x | ✓ |
| SCV2-CPP82 | 0.55 | 0.53 | 0.43 | x | ✓ | SCV2-CPP239 | 0.23 | 0.27 | 0.19 | x | ✓ |
| SCV2-CPP83 | 0.44 | 0.43 | 0.27 | x | ✓ | SCV2-CPP240 | 0.18 | 0.26 | 0.08 | x | ✓ |
| SCV2-CPP84 | 0.37 | 0.56 | 0.14 | x | ✓ | SCV2-CPP241 | 0.31 | 0.23 | 0.18 | x | ✓ |
| SCV2-CPP85 | 0.43 | 0.33 | 0.27 | x | ✓ | SCV2-CPP242 | 0.08 | 0.13 | 0.09 | x | ✓ |
| SCV2-CPP87 | 0.34 | 0.34 | 0.24 | x | ✓ | SCV2-CPP243 | 0.64 | 0.73 | 0.66 | x | ✓ |
| SCV2-CPP88 | 0.46 | 0.43 | 0.35 | x | ✓ | SCV2-CPP244 | 0.83 | 0.74 | 0.66 | x | ✓ |
| SCV2-CPP89 | 0.88 | 0.81 | 0.97 | x | ✓ | SCV2-CPP245 | 0.45 | 0.47 | 0.48 | ✓ | x |
| SCV2-CPP90 | 0.8 | 0.11 | 0.29 | x | x | SCV2-CPP246 | 0.98 | 0.93 | 0.98 | x | ✓ |
| SCV2-CPP91 | 0.73 | 0.15 | 0.26 | x | x | SCV2-CPP247 | 0.25 | 0.28 | 0.15 | x | ✓ |
| SCV2-CPP92 | 0.94 | 0.33 | 0.72 | ✓ | ✓ | SCV2-CPP248 | 0.06 | 0.11 | 0.05 | x | x |
| SCV2-CPP93 | 0.91 | 0.47 | 0.79 | ✓ | ✓ | SCV2-CPP249 | 0.1 | 0.1 | 0.15 | x | x |
| SCV2-CPP94 | 0.53 | 0.49 | 0.16 | x | ✓ | SCV2-CPP250 | 0.32 | 0.37 | 0.59 | x | x |
| SCV2-CPP95 | 0.13 | 0.1 | 0.08 | x | x | SCV2-CPP251 | 0.88 | 0.33 | 0.95 | x | ✓ |
| SCV2-CPP96 | 0.87 | 0.3 | 0.33 | x | ✓ | SCV2-CPP252 | 0.86 | 0.22 | 0.61 | x | ✓ |
| SCV2-CPP97 | 0.98 | 0.57 | 0.73 | x | ✓ | SCV2-CPP253 | 0.63 | 0.18 | 0.59 | x | ✓ |
| SCV2-CPP98 | 0.96 | 0.27 | 0.49 | ✓ | ✓ | SCV2-CPP254 | 0.61 | 0.44 | 0.53 | x | ✓ |
| SCV2-CPP99 | 0.97 | 0.31 | 0.59 | x | ✓ | SCV2-CPP255 | 0.89 | 0.79 | 0.79 | x | ✓ |
| SCV2-CPP101 | 0.55 | 0.59 | 0.64 | x | ✓ | SCV2-CPP256 | 0.59 | 0.25 | 0.55 | x | x |
| SCV2-CPP103 | 0.34 | 0.17 | 0.63 | x | x | SCV2-CPP257 | 0.45 | 0.2 | 0.16 | x | ✓ |
| SCV2-CPP104 | 0.18 | 0.17 | 0.27 | x | x | SCV2-CPP258 | 0.28 | 0.15 | 0.43 | x | x |
| SCV2-CPP105 | 0.18 | 0.17 | 0.26 | x | ✓ | SCV2-CPP259 | 0.75 | 0.24 | 0.53 | x | ✓ |
| SCV2-CPP106 | 0.74 | 0.58 | 0.72 | x | ✓ | SCV2-CPP260 | 0.38 | 0.09 | 0.24 | x | x |
| SCV2-CPP107 | 0.28 | 0.27 | 0.16 | ✓ | ✓ | SCV2-CPP261 | 0.58 | 0.19 | 0.27 | x | ✓ |
| SCV2-CPP108 | 0.28 | 0.26 | 0.2 | ✓ | ✓ | SCV2-CPP262 | 0.58 | 0.19 | 0.28 | x | ✓ |
| SCV2-CPP109 | 0.29 | 0.26 | 0.21 | ✓ | ✓ | SCV2-CPP263 | 0.67 | 0.24 | 0.37 | x | ✓ |
| SCV2-CPP110 | 0.57 | 0.28 | 0.32 | x | ✓ | SCV2-CPP264 | 0.25 | 0.12 | 0.13 | x | x |
| SCV2-CPP112 | 0.9 | 0.65 | 0.95 | ✓ | x | SCV2-CPP265 | 0.43 | 0.18 | 0.09 | x | x |
| SCV2-CPP113 | 0.08 | 0.08 | 0.31 | x | ✓ | SCV2-CPP266 | 0.33 | 0.07 | 0.05 | x | x |
| SCV2-CPP114 | 0.22 | 0.17 | 0.4 | x | ✓ | SCV2-CPP267 | 0.62 | 0.5 | 0.46 | x | ✓ |
| SCV2-CPP115 | 0.31 | 0.27 | 0.21 | x | ✓ | SCV2-CPP268 | 0.74 | 0.59 | 0.58 | x | ✓ |
| SCV2-CPP116 | 0.37 | 0.16 | 0.2 | x | ✓ | SCV2-CPP269 | 0.74 | 0.58 | 0.58 | x | ✓ |
| SCV2-CPP117 | 0.43 | 0.28 | 0.22 | x | ✓ | SCV2-CPP270 | 0.74 | 0.61 | 0.54 | x | ✓ |
| SCV2-CPP118 | 0.16 | 0.14 | 0.06 | x | ✓ | SCV2-CPP271 | 0.79 | 0.58 | 0.45 | x | ✓ |
| SCV2-CPP119 | 0.19 | 0.17 | 0.1 | x | ✓ | SCV2-CPP272 | 0.73 | 0.54 | 0.34 | x | ✓ |
| SCV2-CPP120 | 0.8 | 0.4 | 0.48 | ✓ | ✓ | SCV2-CPP273 | 0.37 | 0.16 | 0.21 | x | ✓ |
| SCV2-CPP121 | 0.49 | 0.55 | 0.45 | ✓ | ✓ | SCV2-CPP274 | 0.69 | 0.35 | 0.54 | x | ✓ |
| SCV2-CPP122 | 0.42 | 0.62 | 0.23 | ✓ | ✓ | SCV2-CPP275 | 0.94 | 0.65 | 0.86 | x | ✓ |
| SCV2-CPP123 | 0.65 | 0.81 | 0.36 | x | ✓ | SCV2-CPP276 | 0.46 | 0.28 | 0.37 | x | ✓ |
| SCV2-CPP124 | 0.7 | 0.39 | 0.66 | ✓ | x | SCV2-CPP277 | 0.47 | 0.34 | 0.33 | x | x |
| SCV2-CPP125 | 0.43 | 0.29 | 0.31 | x | x | SCV2-CPP278 | 0.51 | 0.35 | 0.33 | x | x |
| SCV2-CPP126 | 0.14 | 0.1 | 0.15 | x | x | SCV2-CPP279 | 0.47 | 0.35 | 0.31 | x | x |
| SCV2-CPP127 | 0.35 | 0.04 | 0.15 | ✓ | x | SCV2-CPP280 | 0.47 | 0.35 | 0.31 | x | x |
| SCV2-CPP128 | 0.53 | 0.28 | 0.44 | ✓ | ✓ | SCV2-CPP281 | 0.48 | 0.36 | 0.32 | x | x |
| SCV2-CPP129 | 0.55 | 0.3 | 0.65 | ✓ | ✓ | SCV2-CPP282 | 0.48 | 0.36 | 0.32 | x | x |
| SCV2-CPP130 | 0.62 | 0.19 | 0.19 | ✓ | x | SCV2-CPP283 | 0.36 | 0.3 | 0.25 | x | x |
| SCV2-CPP131 | 0.08 | 0.28 | 0.1 | x | ✓ | SCV2-CPP284 | 0.24 | 0.2 | 0.15 | x | x |
| SCV2-CPP132 | 0.58 | 0.22 | 0.25 | x | ✓ | SCV2-CPP285 | 0.07 | 0.02 | 0.08 | x | ✓ |
| SCV2-CPP133 | 0.7 | 0.53 | 0.46 | x | ✓ | SCV2-CPP286 | 0.13 | 0.48 | 0.05 | x | ✓ |
| SCV2-CPP134 | 0.7 | 0.57 | 0.67 | ✓ | ✓ | SCV2-CPP287 | 0.18 | 0.5 | 0.08 | x | ✓ |
| SCV2-CPP135 | 0.92 | 0.76 | 0.84 | ✓ | ✓ | SCV2-CPP288 | 0.77 | 0.38 | 0.53 | x | x |
| SCV2-CPP136 | 0.86 | 0.79 | 0.83 | ✓ | ✓ | SCV2-CPP289 | 0.5 | 0.28 | 0.45 | x | x |
| SCV2-CPP137 | 0.86 | 0.79 | 0.83 | ✓ | ✓ | SCV2-CPP290 | 0.11 | 0.15 | 0.18 | x | x |
| SCV2-CPP138 | 0.92 | 0.85 | 0.85 | ✓ | ✓ | SCV2-CPP291 | 0.17 | 0.2 | 0.22 | x | x |
| SCV2-CPP139 | 0.99 | 0.96 | 0.96 | x | ✓ | SCV2-CPP292 | 0.42 | 0.19 | 0.34 | x | x |
| SCV2-CPP140 | 0.99 | 0.95 | 0.96 | x | ✓ | SCV2-CPP293 | 0.51 | 0.22 | 0.3 | x | x |
| SCV2-CPP141 | 0.94 | 0.92 | 0.87 | x | ✓ | SCV2-CPP294 | 0.8 | 0.25 | 0.39 | x | x |
| SCV2-CPP142 | 0.59 | 0.41 | 0.45 | x | x | SCV2-CPP295 | 0.8 | 0.25 | 0.39 | x | x |
| SCV2-CPP144 | 0.66 | 0.68 | 0.54 | x | ✓ | SCV2-CPP296 | 0.83 | 0.3 | 0.38 | x | ✓ |
| SCV2-CPP145 | 0.09 | 0.04 | 0.06 | x | x | SCV2-CPP297 | 0.83 | 0.3 | 0.38 | x | x |
| SCV2-CPP146 | 0.79 | 0.16 | 0.11 | x | ✓ | SCV2-CPP298 | 0.62 | 0.2 | 0.16 | x | x |
| SCV2-CPP147 | 0.8 | 0.13 | 0.09 | x | ✓ | SCV2-CPP299 | 0.7 | 0.3 | 0.27 | x | ✓ |
| SCV2-CPP148 | 0.94 | 0.34 | 0.74 | ✓ | ✓ | SCV2-CPP300 | 0.35 | 0.06 | 0.21 | x | x |
| SCV2-CPP150 | 0.61 | 0.24 | 0.75 | ✓ | ✓ | SCV2-CPP301 | 0.76 | 0.28 | 0.36 | x | ✓ |
| SCV2-CPP151 | 0.46 | 0.15 | 0.46 | ✓ | ✓ | SCV2-CPP302 | 0.66 | 0.18 | 0.34 | x | x |
| SCV2-CPP152 | 0.31 | 0.07 | 0.32 | ✓ | ✓ | SCV2-CPP303 | 0.75 | 0.16 | 0.3 | x | x |
| SCV2-CPP153 | 0.31 | 0.13 | 0.25 | ✓ | x | SCV2-CPP304 | 0.8 | 0.16 | 0.39 | x | x |
| SCV2-CPP154 | 0.2 | 0.18 | 0.08 | x | ✓ | SCV2-CPP305 | 0.8 | 0.25 | 0.4 | x | x |
| SCV2-CPP155 | 0.5 | 0.37 | 0.11 | x | ✓ | SCV2-CPP306 | 0.8 | 0.24 | 0.36 | x | x |
| SCV2-CPP156 | 0.43 | 0.28 | 0.15 | x | x | SCV2-CPP307 | 0.77 | 0.34 | 0.45 | x | x |
| SCV2-CPP157 | 0.44 | 0.29 | 0.15 | x | ✓ | SCV2-CPP308 | 0.72 | 0.33 | 0.41 | x | x |
| SCV2-CPP158 | 0.24 | 0.09 | 0.07 | x | x | SCV2-CPP309 | 0.37 | 0.24 | 0.18 | x | ✓ |
| SCV2-CPP159 | 0.59 | 0.11 | 0.11 | x | x | SCV2-CPP310 | 0.52 | 0.29 | 0.26 | x | ✓ |
| SCV2-CPP160 | 0.43 | 0.13 | 0.25 | x | ✓ | ||||||
Anticancer potential: anticancer (✓), non-anticancer (x),
Anti-inflammatory potential: Anti-inflammatory (✓), non-anti-inflammatory (x).
3.4.2. Anticancer peptides
Anticancer peptides (ACPs) are perceived as a branch of AMPs manifesting antitumor traits (Schaduangrat et al., 2019). Some ACPs are virulent toward microbes, tumor cells, and healthy mammalian cells, while others are solely lethal to tumor cells (Gaspar et al., 2013). The antitumor activity of an ACP is generally accomplished through the interaction with cellular membranes using membranolytic or non-membranolytic mechanisms. There are several variances between cellular membranes of cancerous and normal mammalian cells. For instance, the membrane of cancerous cells usually has a larger surface area because of microvilli formation, higher negative net charge, and increased fluidity due to lower cholesterol content (Schweizer, 2009). These variations are the basis of the selectivity of some ACPs facing tumor cells. The iACP server predicts whether a peptide has the potential to be an ACP or not using the g-gap dipeptide identification method. About 21.89% of SCV2-CPPs were recognized as ACPs by the iACP web server. As expected a dominant number of the identified ACPs (88.36%) were identified earlier as antimicrobial peptides (Table 6).
3.4.3. Anti-inflammatory peptides
Inflammation is the natural response to situations such as physical injury or infections and has a significant role in wound healing and microbial resistance. However, prolonged, uncontrolled inflammation can lead to various chronic diseases (Dadar et al., 2019). Several natural and synthetic peptides with immune-modulatory properties known as anti-inflammatory peptides (AIPs) have emerged as anti-inflammatory agents. These peptides can modulate the differentiation of immune cells or inhibit the signal transduction pathway of inflammatory cytokines (Gupta et al., 2017). Recently, several CPPs and AMPs with anti-inflammatory properties have been reported (Fu et al., 2020; Steel et al., 2012; Wang et al., 2011). AIPpred server determines anti-inflammatory epitopes and categorizes the submitted peptides into high, medium, and low confidence AIP or non-AIPs. In this study, more than half of the SCV2-CPPs (63.64%) were predicted as AIPs (Table 6).
3.5. The endosomal entrapment and endosomal escape of SCV2-CPPs
A determinative step in the process of peptide delivery is endosomal entrapment. To our knowledge, there is no tool to predict the potentials of peptides to escape the endosome. One of the strategies to overcome endosomal entrapment is inspired by viruses. Viruses can destabilize the lipid bilayer through the insertion of hydrophobic domain side chains. Covalent attachment of these natural or modified domains to CPPs can enhance their translocation and endosomal escape (Han et al., 2001; Kalafatovic and Giralt, 2017; Lonn et al., 2016). Hydrophobic regions of the spike protein S2 unit responsible for the fusion of SARS-CoV-2 to the membrane are promising as endosomal escape domains.
The other approach is the incorporation of glutamate residues into the hydrophobic segment of amphipathic helices. “Glu” has a negative charge in the extracellular environment and is neutral inside the slightly acidic endosome, thus promotes the formation of an α-helix capable of lysing the endosomes (Kobayashi et al., 2009; Li et al., 2004). SCV2-CPP51, SCV2-CPP52, SCV2-CPP188, and SCV2-CPP189 have the helical structure and a “Glu” residue in the hydrophobic face of the helix. These peptides are expected to have a higher amount of release from endosomes (Fig. 5 ).
Fig. 5.
A) SCV2-CPPs expected to achieve cyclization due to an intramolecular disulfide bond formation after entering the endosome. The most probable disulfide bonds are predicted using DiANNA web server. B) Helical wheel illustrations of CPPs with glutamate residues in the hydrophobic side of the helix structure. Hydrophilic and hydrophobic sides of the helices are roughly divided by the red line. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Cysteine residues are also effective amino acids in a peptide sequence to endorse endosomal release capability. In the oxidative conditions of endosomes, “Cys” harboring peptides form membranolytic oligomers by intermolecular disulfide bonds. Controlling this process so that it eventuates to the establishment of dimers has shown to improve peptide uptake (Jha et al., 2011). About 18.18% of SCV2-CPPs had “Cys” residues in their sequences. Cyclization of CPPs is one of the tacts to boost their cellular uptake and protection against proteases (Kalafatovic and Giralt, 2017). The consequence of intra-molecular “Cys” interactions can give rise to the cyclization of peptides. Cyclization via disulfide bonds is reversible upon delivery to the reductive environment. SCV2-CPP137, SCV2-CPP138, and SCV2-CPP139 originated from the Cys-rich N-terminal region of NSP13 (helicase) can potentially fold into a cyclic conformation in endosomes and results in the higher profile of endosomal release (Fig. 5).
4. Conclusion
According to the obtained data, one can conclude that SARS-CoV-2 RNA interacting proteins have the highest number of cell-penetrating domains. In contrast, proteins containing membrane interacting transduction domains had overall higher uptake efficiency. To be applied as a drug delivery vector, most of the identified SCV2-CPPs had suitable physiochemical characteristics and water solubility for formulation purposes. SCV2-CPPs were predicted to have acceptable half-lives in comparison to the control, low toxicity, and low hemolysis potential. More than half of SCV2-CPPs had a very low probability of cleavage by major protease families. Although the shortcoming of SCV2-CPPs is their uptake efficiency, we could describe that one of the most crucial factors is their secondary structure. Alpha-helices and β-sheets in the secondary structure of SCV2-CPPs positively affect uptake efficiency. Based on the performed analyses, SCV2-CPPs were scored, and four peptides with the most positive attributes were selected as candidates for further experimentation. It is of special note that antimicrobial, anticancer, and cell-penetrating peptides have similar sequential characteristics. A significant number of SCV2-CPPs were identified as antimicrobial, anticancer, and anti-inflammatory peptides. Among the selected candidate as drug delivery vectors, SCV2-CPP122 and SCV2-CPP129 are expected to have antimicrobial, anticancer, and anti-inflammatory activity and can be considered as bioactive CPPs. However, it should be declared that based on the identity of the cargo, route of administration, and the target of delivery, other SCV2-CPPs might also be proper candidates, which has to be resolved by further in vitro and in vivo experiments. However, adjustment of stringent accuracy conditions in our analyses increased the specificity to avoid suggestion of false-positives in introducing novel CPPs. Conclusively, this study provides a platform for screening viral proteomes as a rich source of biotherapeutics or drug delivery carriers.
Funding information
This work was supported by Shiraz University of Medical Sciences, Shiraz, Iran; project number 99-01-106-22161.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgements
Authors would like to thank Shiraz University of Medical Sciences, Shiraz, Iran. Yasaman Behzadipour appreciates her mother –Farkhondeh Gholamzadeh– the nursing supervisor of the Persian Gulf Martyrs Hospital, Bushehr, Iran, for the struggle in the front-line during the COVID-19 pandemic.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104474.
Appendix A. Supplementary data
Supplementary material 1. Proteins of SARS-CoV-2. Cell-penetrating peptides with the highest prediction confidence according to the SkipCPP-Pred server are highlighted in each region.
Supplementary material 2. SARS-CoV2 proteins were scanned by CellPPD amino acid sequences to find CPPs. Confident CPPs were determined by MLCPP. *Prediction confidence of cell penetration. **Prediction confidence of uptake efficiency.
Supplementary material 3. Physiochemical properties of SCV2-CPPs calculated by ProtParam and Pepcalc.
Supplementary material 4. Prediction of half-life, toxicity, hemolytic potency, antigenicity, and allergenicity of SCV2-CPPs.
Supplementary material 5. Evaluation of the susceptibility of SCV2-CPPs to different protease superfamilies, including aspartic proteases, cysteine proteases, metalloproteases, and serine proteases.
Supplementary material 6. Secondary structure prediction of SCV2-CPPs in solution predicted by PEP2D server (C: coil, H: helix, E: sheet). The membrane induced helical region column is defined as predicted by the fmap server.
Supplementary material 7. Scoring SCV2-CPPs for drug delivery applications. Peptides with the highest sum score are highlighted in green
References
- Ahn D.G., Lee W., Choi J.K., Kim S.J., Plant E.P., Almazán F., Taylor D.R., Enjuanes L., Oh J.W. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 2011;91:1–10. doi: 10.1016/j.antiviral.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahoshi A., Matsuura E., Ozeki E., Matsui H., Watanabe K., Ohtsuki T. Enhanced cellular uptake of lactosomes using cell-penetrating peptides. Sci. Technol. Adv. Mater. 2016;17:245–252. doi: 10.1080/14686996.2016.1178056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athmer J., Fehr A.R., Grunewald M., Smith E.C., Denison M.R., Perlman S. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. mBio. 2017;8 doi: 10.1128/mBio.02320-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadipour Y., Hemmati S. Considerations on the rational design of covalently conjugated cell-penetrating peptides (CPPs) for intracellular delivery of proteins: a guide to CPP selection using glucarpidase as the model cargo molecule. Molecules. 2019;24:4318. doi: 10.3390/molecules24234318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J.M., Yi L.L., Guarino L., Sacchettini J.C., Kao C.C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 2008;283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman H.G. Innate immunity and the normal microflora. Immunol. Rev. 2000;173:5–16. doi: 10.1034/j.1600-065x.2000.917301.x. [DOI] [PubMed] [Google Scholar]
- Böttger R., Hoffmann R., Knappe D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178943. e0178943-e0178943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lécine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunology. 2019;8 doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale K.J., Muroski M.E., Vakil P.N., Foley M.E., Laufersky G., Kenworthy R.…Strouse G.F. Selective uptake into drug resistant mammalian cancer by cell penetrating peptide-mediated delivery. Bioconjug. Chem. 2018;29(10):3273–3284. doi: 10.1021/acs.bioconjchem.8b00429. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H., To, K.K, Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Lo S.C., Wang Y.S., Hou M.H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today. 2016;21:562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K., Kumar R., Singh S., Tuknait A., Gautam A., Mathur D., Anand P., Varshney G.C., Raghava G.P. A web server and mobile app for computing hemolytic potency of peptides. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Ding, H., Feng, P., Lin, H., Chou, K.-C., 2016. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget 7, 16895. Doi: 10.18632/oncotarget.7815. [DOI] [PMC free article] [PubMed]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieślik M., Czapski G.A., Strosznajder J.B. The molecular mechanism of amyloid β42 peptide toxicity: the role of sphingosine kinase-1 and mitochondrial sirtuins. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., 3rd, Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar M., Shahali Y., Chakraborty S., Prasad M., Tahoori F., Tiwari R., Dhama K. Antiinflammatory peptides: current knowledge and promising prospects. Inflamm. Res. 2019;68:125–145. doi: 10.1007/s00011-018-1208-x. [DOI] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2'-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deptuła M., Wardowska A., Dzierżyńska M., Rodziewicz-Motowidło S., Pikuła M. Antibacterial peptides in dermatology-strategies for evaluation of allergic potential. Molecules. 2018;23:414. doi: 10.3390/molecules23020414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshankhah H., Jafari S. Cell penetrating peptides: a concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018;108:1090–1096. doi: 10.1016/j.biopha.2018.09.097. [DOI] [PubMed] [Google Scholar]
- Di Pisa M., Chassaing G., Swiecicki J.-M. Translocation mechanism (s) of cell-penetrating peptides: biophysical studies using artificial membrane bilayers. Biochemistry. 2015;54:194–207. doi: 10.1021/bi501392n. [DOI] [PubMed] [Google Scholar]
- Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP v. 2—a server for in silico prediction of allergens. J. Mol. Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- Drin G., Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Duong H.H., Yung L.Y. Synergistic co-delivery of doxorubicin and paclitaxel using multi-functional micelles for cancer treatment. Int. J. Pharm. 2013;454:486–495. doi: 10.1016/j.ijpharm.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiríksdóttir E., Konate K., Langel Ü., Divita G., Deshayes S. Secondary structure of cell-penetrating peptides controls membrane interaction and insertion. Biochim. Biophys. Acta (BBA)-Biomembranes. 2010;1798:1119–1128. doi: 10.1016/j.bbamem.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Elliott G., O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- Ferrè F., Clote P. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 2005;33:W230–W232. doi: 10.1093/nar/gki412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A.D., Pabo C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- Freire J.M., Veiga A.S., Rego de Figueiredo I., de la Torre B.G., Santos N.C., Andreu D., Da Poian A.T., Castanho M.A. Nucleic acid delivery by cell penetrating peptides derived from dengue virus capsid protein: design and mechanism of action. FEBS J. 2014;281:191–215. doi: 10.1111/febs.12587. [DOI] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Ke L., Cai L., Chen X., Ren X., Gao M. Improved prediction of cell-penetrating peptides via effective orchestrating amino acid composition feature representation. IEEE Access. 2019;7:163547–163555. doi: 10.1109/ACCESS.2019.2952738. [DOI] [Google Scholar]
- Fu T.-K., Kuo P.-H., Lu Y.-C., Lin H.-N., Wang L.H.-C., Lin Y.-C., Kao Y.-C., Lai H.-M., Chang M.D.-T. Cell penetrating peptide as a high safety anti-inflammation ingredient for cosmetic applications. Biomolecules. 2020;10:101. doi: 10.3390/biom10010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- Futaki S., Nakase I., Tadokoro A., Takeuchi T., Jones A.T. Arginine-rich peptides and their internalization mechanisms. Biochem. Soc. Trans. 2007;35:784–787. doi: 10.1042/BST0350784. [DOI] [PubMed] [Google Scholar]
- Gaspar D., Veiga A.S., Castanho M.A.R.B. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013;4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam A., Chaudhary K., Kumar R., Raghava G.P.S. In: Cell-Penetrating Peptides. Langel Ü., editor. Humana Press; New York, NY: 2015. Computer-aided virtual screening and designing of cell-penetrating peptides; pp. 59–69. [DOI] [PubMed] [Google Scholar]
- Gautier R., Douguet D., Antonny B., Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P. In: Computational Peptidology. Zhou P., Huang J., editors. Humana Press; New York, NY: 2015. Peptide toxicity prediction; pp. 143–157. [DOI] [Google Scholar]
- Gupta S., Sharma A.K., Shastri V., Madhu M.K., Sharma V.K. Prediction of anti-inflammatory proteins/peptides: an insilico approach. J. Transl. Med. 2017;15:7. doi: 10.1186/s12967-016-1103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkarainen T., Wahlfors T., Meriläinen O., Loimas S., Hemminki A., Wahlfors J. VP22 does not significantly enhance enzyme prodrug cancer gene therapy as a part of a VP22-HSVTk-GFP triple fusion construct. J. Gene Med. 2005;7:898–907. doi: 10.1002/jgm.737. [DOI] [PubMed] [Google Scholar]
- Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- Heard K.R., Wu W., Li Y., Zhao P., Woznica I., Lai J.H., Beinborn M., Sanford D.G., Dimare M.T., Chiluwal A.K., Peters D.E., Whicher D., Sudmeier J.L., Bachovchin W.W. A general method for making peptide therapeutics resistant to serine protease degradation: application to dipeptidyl peptidase IV substrates. J. Med. Chem. 2013;56:8339–8351. doi: 10.1021/jm400423p. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271-280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S.T. SARS-CoV-2 and ORF3a: non-synonymous mutations and polyproline regions. BioRxiv. 2020 doi: 10.1101/2020.03.27.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha D., Mishra R., Gottschalk S., Wiesmuller K.H., Ugurbil K., Maier M.E., Engelmann J. CyLoP-1: a novel cysteine-rich cell-penetrating peptide for cytosolic delivery of cargoes. Bioconjug. Chem. 2011;22:319–328. doi: 10.1021/bc100045s. [DOI] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Chatterjee A., Neuman B.W., Wüthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 2010;400:724–742. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81:6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafatovic D., Giralt E. Cell-penetrating peptides: design strategies beyond primary structure and amphipathicity. Molecules. 2017;22:1929. doi: 10.3390/molecules22111929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Giedroc D.P. Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 2013;87:3502–3515. doi: 10.1128/JVI.03112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. New user-friendly approach to obtain an Eisenberg plot and its use as a practical tool in protein sequence analysis. Int. J. Mol. Sci. 2011;12:5577–5591. doi: 10.3390/ijms12095577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A.-A., Mussbach F., Breitling R., Hemmerich P., Schaefer B., Lorkowski S., Reissmann S. Relationships between cargo, cell penetrating peptides and cell type for uptake of non-covalent complexes into live cells. Pharmaceuticals. 2013;6:184–203. doi: 10.3390/ph6020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., Yum S.Y., Jang G., Ahn D.-R. Discovery of a non-cationic cell penetrating peptide derived from membrane-interacting human proteins and its potential as a protein delivery carrier. Sci. Rep. 2015;5:11719. doi: 10.1038/srep11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Nakase I., Kawabata N., Yu H.H., Pujals S., Imanishi M., Giralt E., Futaki S. Cytosolic targeting of macromolecules using a pH-dependent fusogenic peptide in combination with cationic liposomes. Bioconjug. Chem. 2009;20:953–959. doi: 10.1021/bc800530v. [DOI] [PubMed] [Google Scholar]
- Kramer R.M., Shende V.R., Motl N., Pace C.N., Scholtz J.M. Toward a molecular understanding of protein solubility: increased negative surface charge correlates with increased solubility. Biophys. J. 2012;102:1907–1915. doi: 10.1016/j.bpj.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D.M., Neubacher S., Grossmann T.N. Protein-RNA interactions: structural characteristics and hotspot amino acids. RNA. 2018;24:1457–1465. doi: 10.1261/rna.066464.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose A., Chirmule N., Nair P. Immunogenicity of biotherapeutics: causes and association with posttranslational modifications. J Immunol Res. 2016;2016:1298473. doi: 10.1155/2016/1298473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., Leung G.M., Holmes E.C., Hu Y.L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Langedijk J.P. Translocation activity of C-terminal domain of pestivirus erns and ribotoxin L3 loop. J. Biol. Chem. 2002;277:5308–5314. doi: 10.1074/jbc.M104147200. [DOI] [PubMed] [Google Scholar]
- Langel Ü. first ed. Springer Singapore; 2019. CPP, Cell-Penetrating Peptides. [DOI] [Google Scholar]
- Lee C.-C., Sun Y., Huang H.W. Membrane-mediated peptide conformation change from α-monomers to β-aggregates. Biophys. J. 2010;98:2236–2245. doi: 10.1016/j.bpj.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Ying S., Baojun L., Yi Y., Xiang H., Wenli S., Zounan S., Deyin G., Qingyu Z., Jingmei L., Guohui C. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Nicol F., Szoka F.C., Jr. GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv. Drug Deliv. Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- Li K., Qin W., Ding D., Tomczak N., Geng J., Liu R., Liu J., Zhang X., Liu H., Liu B., Tang B.Z. Photostable fluorescent organic dots with aggregation-induced emission (AIE dots) for noninvasive long-term cell tracing. Sci. Rep. 2013;3:1150. doi: 10.1038/srep01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize A.L., Lomize M.A., Krolicki S.R., Pogozheva I.D. Membranome: a database for proteome-wide analysis of single-pass membrane proteins. Nucleic Acids Res. 2017;45:D250–D255. doi: 10.1093/nar/gkw712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn P., Kacsinta A.D., Cui X.S., Hamil A.S., Kaulich M., Gogoi K., Dowdy S.F. Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 2016;6:32301. doi: 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan B., Shin T.H., Kim M.O., Lee G. AIPpred: sequence-based prediction of anti-inflammatory peptides using random forest. Front. Pharmacol. 2018;9:276. doi: 10.3389/fphar.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan B., Subramaniyam S., Shin T.-H., Kim M., Lee G. Machine-learning-based prediction of cell-penetrating peptides and their uptake efficiency with improved accuracy. J. Proteome Res. 2018;17:2715–2726. doi: 10.1021/acs.jproteome.8b00148. [DOI] [PubMed] [Google Scholar]
- Mathur D., Prakash S., Anand P., Kaur H., Agrawal P., Mehta A., Kumar R., Singh S., Raghava G.P. PEPlife: a repository of the half-life of peptides. Sci. Rep. 2016;6:1–7. doi: 10.1038/srep36617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D., Singh S., Mehta A., Agrawal P., Raghava G.P. In silico approaches for predicting the half-life of natural and modified peptides in blood. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Debbink K., Baric R.S. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrose K., Yang Y., Krissansen G.W. X-pep, a novel cell-penetrating peptide motif derived from the hepatitis B virus. Biochem. Biophys. Res. Commun. 2014;453:64–68. doi: 10.1016/j.bbrc.2014.09.057. [DOI] [PubMed] [Google Scholar]
- Morris M.C., Depollier J., Mery J., Heitz F., Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat. Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtarič D., Toplak I., Ameneiros R.S., Pfeifer A., Thiel V., Drexler J.F., Müller M.A., Drosten C. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase I., Hirose H., Tanaka G., Tadokoro A., Kobayashi S., Takeuchi T., Futaki S. Cell-surface accumulation of flock house virus-derived peptide leads to efficient internalization via macropinocytosis. Mol. Ther. 2009;17:1868–1876. doi: 10.1038/mt.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antivir. Res. 2016;135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owji H., Hemmati S. A comprehensive in silico characterization of bacterial signal peptides for the excretory production of Anabaena variabilis phenylalanine ammonia lyase in Escherichia coli. 3. Biotech. 2018;8:488. doi: 10.1007/s13205-018-1517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J., Klinck R., Good L., Langel Ü., Wellinger R.J., Abou Elela S. Free uptake of cell-penetrating peptides by fission yeast. FEBS Lett. 2005;579(21):4873–4878. doi: 10.1016/j.febslet.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U S A. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S. Cell penetration: scope and limitations by the application of cell-penetrating peptides. J. Pept. Sci. 2014;20:760–784. doi: 10.1002/psc.2672. [DOI] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghian I., Khalvati B., Ghasemi Y., Hemmati S. TAT-mediated intracellular delivery of carboxypeptidase G2 protects against methotrexate-induced cell death in HepG2 cells. Toxicol. Appl. Pharmacol. 2018;346:9–18. doi: 10.1016/j.taap.2018.03.023. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Kawachi K., Terada Y., Omori H., Matsuura Y., Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaduangrat N., Nantasenamat C., Prachayasittikul V., Shoombuatong W. ACPred: a computational tool for the prediction and analysis of anticancer peptides. Molecules. 2019;24:1973. doi: 10.3390/molecules24101973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009;625:190–194. doi: 10.1016/j.ejphar.2009.08.043. [DOI] [PubMed] [Google Scholar]
- Seelig J. Thermodynamics of lipid–peptide interactions. Biochim. Biophys. Acta. 2004;1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Shankar G., Arkin S., Cocea L., Devanarayan V., Kirshner S., Kromminga A., Quarmby V., Richards S., Schneider C.K., Subramanyam M., Swanson S., Verthelyi D., Yim S. Assessment and reporting of the clinical immunogenicity of therapeutic proteins and peptides-harmonized terminology and tactical recommendations. AAPS J. 2014;16:658–673. doi: 10.1208/s12248-014-9599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antivir. Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M.C., Zhang J., Ah Min K., Lee K., Moon C., Balthasar J.P., Yang V.C. Combination of antibody targeting and PTD-mediated intracellular toxin delivery for colorectal cancer therapy. J. Control Release. 2014;194:197–210. doi: 10.1016/j.jconrel.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Yuen K.S., Castaño-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Tan H., Perry A.J., Akutsu T., Webb G.I., Whisstock J.C., Pike R.N. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? An answer from all-atoms simulations. ChemRxivTM. 2020 doi: 10.26434/chemrxiv.12091260.v3. [DOI] [PubMed] [Google Scholar]
- Splith K., Neundorf I. Antimicrobial peptides with cell-penetrating peptide properties and vice versa. Eur. Biophys. J. 2011;40:387–397. doi: 10.1007/s00249-011-0682-7. [DOI] [PubMed] [Google Scholar]
- Steel R., Cowan J., Payerne E., O’Connell M.A., Searcey M. Anti-inflammatory effect of a cell-penetrating peptide targeting the Nrf2/Keap1 interaction. ACS Med. Chem. Lett. 2012;3:407–410. doi: 10.1021/ml300041g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 2006;80:7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Hu J., Zou Q., Manavalan B., Wei L. Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools. Brief. Bioinform. 2020;21:408–420. doi: 10.1093/bib/bby124. [DOI] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U S A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P.M., Eroglu E., Bawage S.S., Vig K., Miller M.E., Pillai S., Dennis V.A., Singh S.R. Enhanced intracellular translocation and biodistribution of gold nanoparticles functionalized with a cell-penetrating peptide (VG-21) from vesicular stomatitis virus. Biomaterials. 2014;35:9484–9494. doi: 10.1016/j.biomaterials.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Tseng Y.T., Chang C.H., Wang S.M., Huang K.J., Wang C.T. Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkhede N., Bommana R., Schöneich C., Forrest M.L. Proteolysis and oxidation of therapeutic proteins after intradermal or subcutaneous administration. J. Pharm. Sci. 2020;109:191–205. doi: 10.1016/j.xphs.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S., Prasad B., Selvarajan R. RNA dependent RNA polymerases: insights from structure. Function and Evolution. Viruses. 2018;10 doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Li B. Effect of molecular weight on the transepithelial transport and peptidase degradation of casein-derived peptides by using Caco-2 cell model. Food Chem. 2017;218:1–8. doi: 10.1016/j.foodchem.2016.08.106. [DOI] [PubMed] [Google Scholar]
- Wang Y.F., Xu X., Fan X., Zhang C., Wei Q., Wang X., Guo W., Xing W., Yu J., Yan J.-L., Liang H.-P. A cell-penetrating peptide suppresses inflammation by inhibiting NF-κB signaling. Mol. Ther. 2011;19:1849–1857. doi: 10.1038/mt.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win T.S., Malik A.A., Prachayasittikul V.S., Wikberg J.E., Nantasenamat C., Shoombuatong W. HemoPred: a web server for predicting the hemolytic activity of peptides. Future Med. Chem. 2017;9:275–291. doi: 10.4155/fmc-2016-0188. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci U S A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Zhan Y., Xue B., Dong Y., Wang Y., Jiang P., Wang A., Sun Y., Yang Y. Highly efficient cellular uptake of a cell-penetrating peptide (CPP) derived from the capsid protein of porcine circovirus type 2. J. Biol. Chem. 2018;293:15221–15232. doi: 10.1074/jbc.RA118.004823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharieva N., Dimitrov I., Flower D., Doytchinova I. Immunogenicity prediction by VaxiJen: a ten year overview. J Proteomics Bioinformatics. 2017;10:298–310. doi: 10.4172/jpb.1000454. [DOI] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou R. 2020. Binding Mechanism of Remdesivir to SARS-CoV-2 RNA Dependent RNA Polymerase. Published online ahead of print. [DOI] [PubMed] [Google Scholar]
- Zhang R., Li Y., Cowley T.J., Steinbrenner A.D., Phillips J.M., Yount B.L., Baric R.S., Weiss S.R. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J. Virol. 2015;89:3598–3609. doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., da Silva G.M., Deatherage C., Burd C., DiMaio D. Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 protein to trigger retrograde trafficking. Cell. 2018;174:1465–1476. doi: 10.1016/j.cell.2018.07.031. e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovsky M.A., Filograna A., Luini A., Corda D., Valente C. Protein amphipathic Helix insertion: a mechanism to induce membrane fission. Front. Cell Dev. Biol. 2019;7 doi: 10.3389/fcell.2019.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1. Proteins of SARS-CoV-2. Cell-penetrating peptides with the highest prediction confidence according to the SkipCPP-Pred server are highlighted in each region.
Supplementary material 2. SARS-CoV2 proteins were scanned by CellPPD amino acid sequences to find CPPs. Confident CPPs were determined by MLCPP. *Prediction confidence of cell penetration. **Prediction confidence of uptake efficiency.
Supplementary material 3. Physiochemical properties of SCV2-CPPs calculated by ProtParam and Pepcalc.
Supplementary material 4. Prediction of half-life, toxicity, hemolytic potency, antigenicity, and allergenicity of SCV2-CPPs.
Supplementary material 5. Evaluation of the susceptibility of SCV2-CPPs to different protease superfamilies, including aspartic proteases, cysteine proteases, metalloproteases, and serine proteases.
Supplementary material 6. Secondary structure prediction of SCV2-CPPs in solution predicted by PEP2D server (C: coil, H: helix, E: sheet). The membrane induced helical region column is defined as predicted by the fmap server.
Supplementary material 7. Scoring SCV2-CPPs for drug delivery applications. Peptides with the highest sum score are highlighted in green