Abstract
A total of seven yeast strains from 18 xylanolytic and/or xylose-fermenting yeast species isolated from the wood-feeding termite Reticulitermes chinenesis could efficiently decolorize various azo dyes under high-salt conditions. Of these strains, a novel and unique azo-degrading and halotolerant yeast, Sterigmatomyces halophilus SSA1575, has been investigated in this study. This strain could significantly decolorize four combinations of a mixture of dyes. It showed a high capability for decolorizing Reactive Black 5 (RB5) even at 1,500 mg L−1. The strain SSA1575 still showed a high capability for decolorizing a 50 mg L−1 RB5 with a salt mixing at a NaCl concentration of up to 80 g L−1. It also exhibited significant ability to decolorize repeated additions of dye aliquots, with a reduction in time of up to 18 h. Most of the tested carbon and nitrogen sources could significantly enhance a RB5 decolorization. However, this process was inhibited by the addition of sucrose and sodium nitrate. NADH-dichlorophenol indophenol (NADH-DCIP) reductase and lignin peroxidase were determined as the key reductase and oxidase of S. halophilus SSA1575. Finally, strain SSA1575, can effectively detoxify RB5 into non-toxic products. Overall, S. halophilus SSA1575, might be a promising halotolerant yeast valued for the treatment of various textile effluents with high salinity.
Subject terms: Biological techniques, Microbiology, Environmental sciences
Introduction
A variety of textile dyestuffs (approximately 10,000 synthetic dyes; 7 × 107 metric tons) produced every year worldwide, posing a threat to the environmental safety1,2. Azo dyes represent the largest and most common used class of synthetic dyes in textile industry. However, the effluents from these industries have proven toxic, mutagenic and carcinogenic activities due to the presence of one or more azo bond3,4. Recently, the discharge of dye effluents into natural streams has gained great public attention owing to the serious ecological risks. Therefore, more effective and ecofriendly methods to treat industrial wastewater effluents are urgent prior to their discharge into the natural environment.
Despite the removal of azo dyes from effluents using conventional treatments, such as flocculation, membrane filtration, photocatalysis and adsorption5–8, microbial decolorization is a cheaper and eco-friendly accepted technique for the removal of dyes compared to conventional physical and chemical methods3,9. Hence, most current studies on the degradation of azo dyes are mainly focused on microbial degradation using filamentous fungi and bacteria. However, the production of aromatic amines has adverse impact on bacterial activity10. In comparison, fungi have been found to decolorize dyes without production of aromatic amines based on their extracellular lignin-modifying enzymes, including lignin peroxidase (LiP), manganese peroxidase (MnP) and laccase (Lac) along with other supporting enzymes6. However, filamentous fungi (molds) are sometimes poorly adapted in the industrial effluents due to their mycelial ageing, the large amounts of sludge produced, as well as their growth rates are usually slow when compared to most yeast species11. Yeasts, by contrast, have many advantages over bacteria and filamentous fungi, which present a potential capability to deal with some stringent wastewater conditions, such as a salty or acidic condition, and particularly in regard to their unique non-pathogenicity and fast growing nature12–14. As far as it is known, relatively limited studies are available in literature about yeast based azo dye decolorization under hypersaline conditions15–17.
The effluents of the wastewater textile industry are composed of a high concentration of dyes and other pollutant substances, such as a high salt concentration. For the treatment of textile effluents, it is important to use microbial strains capable of tolerating high salt concentrations greater than 3%6. Halophilic and salt-tolerant microorganisms have biological advantages compared to other organisms that could die or become dormant when survive under the same extreme conditions. For example, many salt-tolerant or halophilic microorganisms can keep a relatively high growth rate and metabolic efficiency under high salt conditions, since these organisms do not permit salt into their cell, while they apply the accumulation mechanism of solutes or osmolytes for living in saline habitats6, 18. Some salt-tolerant yeast strains, Pichia occidentalis G1, Galactomyces geotrichum GG and Scheffersomyces spartinae TLHS-SF1, have been confirmed as efficient decolorizing and detoxifying alternatives of azo dyes under high salinity15–17.
As lignin is a phenol aromatic polymer, the presence of azo dyes may induce the microbial ligninases, which have great potential in processes related to the degradation of wastewater aromatic dyes. However, the actual pathways for the biodegradation of azo dyes are still not fully understood, since the involvement of ligninase enzymes in dye degradation is "non-specific" in nature19. The biodegradation of azo dye Reactive Black 5 (RB5) led to the formation of dissimilar metabolic end products in two independent studies. For example, the mechanism of RB5 degradation by a yeast strain, Trichosporon akiyoshidainum HP-2023 led to the formation of Quinone B and phenildiazene, which were driven via phenol oxidase and peroxidase enzymes20. However, oxalic acid was produced as the end metabolic product of the RB5 degradation by Aeromonas hydrophila isolated from textile azo dye wastewater, via the cleavage of azo (–N=N–) bonds21. On the other hand, the application of the purified enzymes in textile dye effluents is an expensive treatment method. These enzymes often lose their catalytic potential under extreme conditions and are difficult to recover6,22,23. Hence, catalytically efficient ligninases-producing salt-tolerant or halophilic yeasts are required to overcome these challenges. Recently, some strains of Sterigmatomyces halophilus, capable of tolerating various extreme conditions, such as salt concentration, were successfully isolated and identified from the gut symbionts of a wood-feeding termite species, Reticulitermes chinenesis by Ali et al.13. This promising model insect has been reported for its capability in digesting lignin through in-situ gut symbionts that are responsible for lignin degradation24. These symbionts exhibited potential for some industrial applications, especially in lignin removal from lignocellulosic biomass-based biorefinery3,24.
Exploration of ligninase-producing yeast strains from termite gut symbionts on their capability to degrade aromatic compounds, as well as to survive under extreme conditions, such as high salt concentration is gaining attention, making the treatment of textile dyes by halotolerant yeasts a novel biodegradation approach. Therefore, this study was undertaken to identify and characterize novel halotolerant yeast species or strains isolated from the gut of R. chinenesis and capable of decolorizing azo dyes efficiently. Various physcio-chemical and nutritional parameters were evaluated to achieve maximum dye decolorization efficiency by S. halophilus SSA1575. Activities of some enzymes which might participate in dye degradation were also determined. Phytotoxicity and microbial toxicity tests were performed for estimating the toxicity of the metabolites after dye decolorization. To well understand possible degradation pathways of the target dye, the enzymatic system involved and possible metabolic intermediates analyzed, as well as related literature were studied.
Results and discussion
Identification and characteristics of halotolerant azo dye-decolorizing yeasts
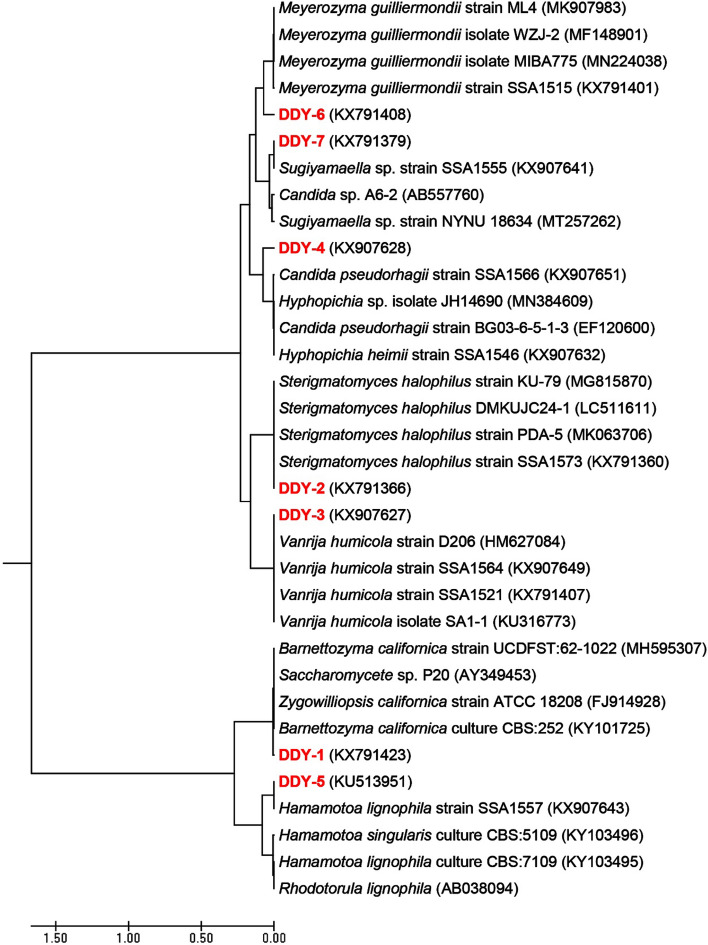
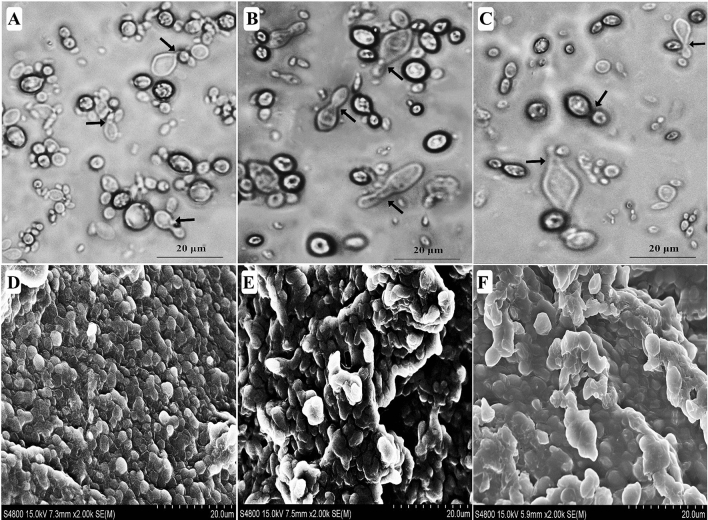
Among 18 xylanolytic and/or xylose-fermenting yeast species isolated from the wood-feeding termite R. chinenesis13, seven yeast strains could efficiently decolorize various azo dyes as confirmed from the decolorized halos observed in the Minimal Saline (MS) agar plates containing 50 mg L−1 of the dye tested. These yeast strains were also examined similarly in MS broth medium supplemented with 50 mg L−1 of the dye and incubated at 30 °C for 24 h. As given in Table 1, all yeast strains successfully decolorized RB5 with a decolorization efficiency of over 90% within 24 h of static incubation. Figure 1 depicts the neighbor-joining tree showing the phylogenetic relationship of the 26S rDNA gene sequences obtained from the seven dye decolorizing yeast strains (DDY-1 to DDY-7) alongside GenBank organisms closely related to them at NCBI (Table 2). Clearly, the strain DDY-2 presented an impressive decolorization performance of various azo dyes tested over other yeast strains (Table 1). The yeast strain DDY-2 exhibited high identity (99.34%) to S. halophilus strain SSA1573 (with accession number KX791360). Hence, DDY-2 was identified as S. halophilus strain SSA1575 with the GeneBank accession number KX791366 (Table 2 and Fig. 1). Morphologically, the cells of the newly isolated deuteromycete S. halophilus strain SSA1575, isolated from R. chinenesis, were mostly spherical (Fig. 2A) to ovoid (Fig. 2B) and produced conidia on interconnecting sterigmata (Fig. 2C). These cells did not have mycelial elements or undergo sexual recombination.
Table 1.
Performance of tested yeast strains on decolorizing various azo dyes.
| Isolate code | RB5 | RR120 | RB19 | AzB | AS-GR | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MD (%) | T (h) | MD (%) | T (h) | MD (%) | T (h) | MD (%) | T (h) | MD (%) | T (h) | |
| DDY-1 | 93.52 | 24 | 78.23 | 18 | 93.75 | 18 | 90.80 | 18 | 81.98 | 15 |
| DDY-2 | 98.71 | 12 | 93.46 | 18 | 94.80 | 15 | 95.88 | 18 | 96.56 | 21 |
| DDY-3 | 95.78 | 21 | 91. 10 | 21 | 80.72 | 12 | 78.76 | 21 | 90.11 | 18 |
| DDY-4 | 90. 82 | 18 | 88.34 | 24 | 69.12 | 24 | 91.70 | 24 | 77.98 | 24 |
| DDY-5 | 90.76 | 24 | 75.16 | 24 | 55.89 | 24 | 88. 64 | 21 | 89.71 | 18 |
| DDY-6 | 98.12 | 24 | 90.87 | 18 | 79. 34 | 18 | 91.09 | 24 | 69.89 | 24 |
| DDY-7 | 93.45 | 18 | 90.06 | 18 | 92.51 | 24 | 87. 83 | 15 | 88.03 | 24 |
RB5 Reactive Black 5, RR120 Reactive Red 120, RB19 Reactive Blue 19, AzB Azure B, AS-GR Acid Scarlet GR, MD maximum decolorization, T time.
Figure 1.
Evolutionary relationships of the seven azo-decolorizing yeast strains based on the UPGMA method as conducted by using MEGA7.
Table 2.
Molecular identification of azo-degrading yeast strains based on BLAST comparison to the GeneBank database.
| Isolate code | Yeast species | Strain | Accession no | Closest relative (accession no.) | Sequence identity (%) |
|---|---|---|---|---|---|
| DDY-1 | Barnettozyma californica | SSA1537 | KX791423 | Barnettozyma californica culture CBS:252 (KY101725) | 99.45 |
| DDY-2 | Sterigmatomyces halophilus | SSA1575 | KX791366 | Sterigmatomyces halophilus strain SSA1573 (KX791360) | 99.34 |
| DDY-3 | Vanrija humicola | SSA1541 | KX907627 | Vanrija humicola strain D206 (HM627084) | 99.34 |
| DDY-4 | [Candida] pseudorhagii | SSA1542 | KX907628 | [Candida] pseudorhagii strain SSA1566 (KX907651) | 88.07 |
| DDY-5 | Hamamotoa lignophila | SSA1567 | KU513951 | Hamamotoa lignophila strain SSA1557 (KX907643) | 100 |
| DDY-6 | Meyerozyma guilliermondii | SSA1522 | KX791408 | Meyerozyma guilliermondii strain SSA1515 (KX791401) | 87.94 |
| DDY-7 | Sugiyamaella sp. | SSA1592 | KX791379 | Sugiyamaella sp. strain SSA1555 (KX907641) | 99.64 |
Figure 2.
Sterigmatomyces halophilus SSA1575 from the wood-feeding termite R. chinenesis. Cell morphology of the strain SSA1575 showing spherical forms (A), ovoid forms (B) and conidia formation on interconnecting sterigmata (arrows) (C). Scanning electron micrographs of S. halophilus SSA1575 under a high salt concentration; 20 g L−1 NaCl (D), 30 g L−1 NaCl (E) and 50 g L−1 NaCl (F).
The majority of the yeast species S. halophilus was commonly isolated from marine environments25. However, the xylanase-producing and D-xylose-fermenting yeast S. halophilus was first identified from an insect gut system as per our previous study13. To date, to the authors’knowledge, no report has investigated the performance of S. halophilus on dye decolorization under high salinity. Therefore, changes in the morphological structure of strain SSA1575 under the stress of high salt conditions were observed by scanning electron microscopy (Fig. 2D–F). The growing cells with 20 g L−1 NaCl remained relatively normal with oval/spherical shapes. However, increasing NaCl concentration up to 50 g L−1 revealed obvious metamorphosis including massive compact clumps with distorted edges and irregular clumps in some yeast cells. The increasing need for bioremediation of hypersaline habitats and for biological control agents that can be used in agriculture irrigated by saline water, stimulate the search for halophilic/salt-tolerant yeasts. The cellular stress response in yeasts has a defined set of targeted cellular functions including DNA/chromatin stabilization and repair, protein chaperoning, removal of damaged proteins, and cell cycle control26.
As a novel yeast species that was first identified from an insect gut system, S. halophilus strain SSA1575 has exhibited some unique and impressive properties that are potentially valuable for biorefinery and bioremediation purposes. No report has been published on the dye decolorization using S. halophilus. In addition, the S. halophilus strain SSA1575, showed significantly higher capability on decolorizing RB5 when compared with the other yeasts that were isolated from R. chinenesis. Simultaneously, SSA1575 showed a high tolerance under hypersaline conditions, which suggested its valuable potential in the treatment of industrial effluents containing a wide variety of azo dyes at a high salt environment. Based on these reasons, the S. halophilus strain SSA1575 and RB5 were actually selected for further investigation.
Decolorization of mixture of azo dyes
The performance of different microorganisms on the decolorization of the mixture of azo dyes was previously studied in a few reports27–29. While evaluating the decolorization of the mixtures of azo dyes by the S. halophilus strain SSA1575, it was observed that this halotolerant yeast strain was effectively decolorized mixture of azo dyes. Based on the values obtained from the American Dye Manufactuers’ Institute (ADMI) calculations, it was found that mixture I (RB5 and Reactive Black 19; RB19) was decolorized more effectively than three other mixtures (II, III and IV) at 24 h of dye addition. Mixture I showed 83.17% ADMI removal which was found to be higher than mixture II (RB5, RB19 and Reactive Red 120; RR120), mixture III (RB5, RB19, RR120 and Azure B; AzB) and mixture IV (RB5, RB19, RR120, AzB and Acid Scarlet GR; AS-GR) by 72.56%, 58.42% and 51.21%, respectively. Tamboli et al.29 reported that the structure variability of dye mixtures may induce a maximum production of the polyhydroxyalkanoate (PHA) synthase enzyme. PHAs are the cytoplasmic microbial inclusions, which formed in stressed conditions, i.e. in the presence of excess carbon source and when growth is restricted by the lack of other nutrients such as nitrogen, phosphorus or sulphur30. As the nutritional supply is reestablished, the PHA can be degraded by intracellular polymerase and subsequently metabolized to a carbon and energy source31.
Dye concentration
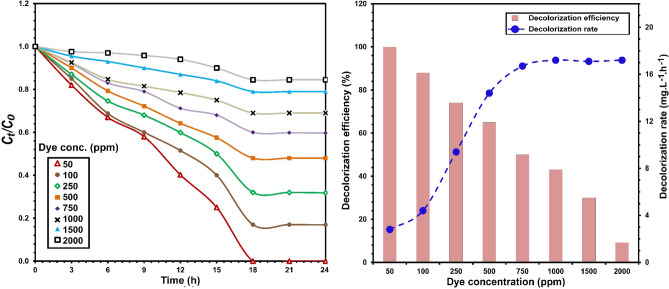
The effects of initial dye concentration on the decolorization efficiency of RB5 by S. halophilus SSA1575 are shown in Fig. 3. Clearly, the decolorization efficiency reached 100% at 50 mg L−1 RB5 after incubation for up to 18 h. Beside dye removal, the decolorization rate was also calculated to understand the effect of the initial dye concentration on decolorization. It has been reported that the decolorization rate may be increased with increasing dye concentration32. In this study, although the decolorization efficiencies decreased with increasing RB5 concentrations, the decolorization rate increased with increasing dye concentration. At a concentration of 50 mg L−1, the maximum decolorization rate of RB5 was obtained at 2.8 mg L−1 h−1, while it reached 17.2 mg L−1 h−1) at 2000 mg L−1 after 24 h. Meanwhile, maximum decolorization efficiency (> 70%) was observed at 100–500 mg L−1 RB5 within 24 h. Then, decolorization efficiency of RB5 decreased significantly (p < 0.001) with increasing dye concentration, reaching 45% at 1,000 mg L−1, which was probably due to the toxicity of dye at high concentrations, hence inhibiting microbial growth33. Dye decolorization of RB5 by S. halophilus SSA1575 was compared with other strains reported in literature (Table 3) and it showed a competitive decolorization performance. As the highest concentrations tested in this study are much above the dye levels in wastewater and aquatic environments, the newly isolated yeast S. halophilus SSA1575, could potentially be utilized for bioremediation of azo dye wastewaters at a high concentration of azo dyes. Singh34 reported that the dye concentrations in textile wastewater generally range between 10 and 200 mg L−1.
Figure 3.
Influence of the initial concentration of RB5 on the dye decolorization performance of S. halophilus SSA-1575, inset with the corresponding decolorization efficiency and decolorization rate.
Table 3.
Comparison of the promising feasibility of S. halophilus SSA1575 on decolorization efficiency with other halotolerant yeast strains reported in literature.
| Strain | Dye concentration (mg L−1) | NaCl concentration (g L−1) | Maximum decolorization (%) | Time (h) | References |
|---|---|---|---|---|---|
| Pichia occidentalis G1 | 50 | 30 | 98 | 16 | 15 |
| Scheffersomyces spartinae TLHS-SF1 | 80 | ⩽ 30 | 90 | 16 | 16 |
| Galactomyces geotrichum GG | 100 | ⩽ 40 | 92 | 10 | 17 |
| Cyberlindnera samutprakarnensis S4 | 50 | 30 | 97 | 18 | 18 |
| S. halophilus SSA1575 | 50 | 5 | 100 | 6 | This study |
| S. halophilus SSA1575 | 50 | 20 | 98.71 | 12 | This study |
| S. halophilus SSA1575 | 50 | 40 | 98 | 24 | This study |
| S. halophilus SSA1575 | 50 | 50 | 92 | 24 | This study |
Salt concentration
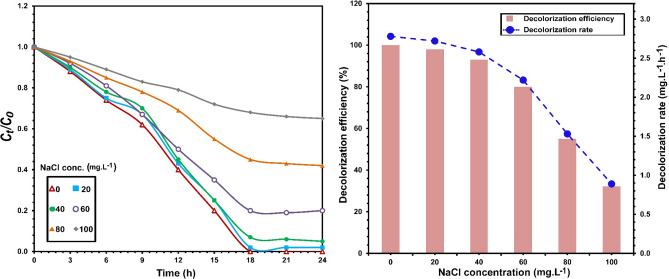
The efficiency of S. halophilus SSA1575 to decolorize 50 mg L−1 RB5 in the presence of a high salt concentration was evaluated (Fig. 4). After incubation for 12 h, nearly complete decolorization of RB5 by the SSA1575 strain was observed under the effect of 20 g L−1 NaCl. However, the RB5 decolorization efficiency reached above 90% at NaCl concentration of up to 50 g L−1 after 18–24 h. A further increase in the salt concentration above 50 g/L showed significant decrease in the decolorization efficiency of RB5 by the yeast strain SSA1575, reaching less than 29% when the concentration NaCl was 100 g L−1. On the other hand, the growth rate of strain SSA1575 at salt concentration (0–50 g L−1 NaCl) was significantly better than any higher salinity when beyond 50 g L−1 (data not shown). As a result, the newly isolated yeast strain S. halophilus SSA-1575 is identified as a halotolerant yeast instead of a halophilic one35. Our findings are in agreement with halotolerant yeasts that have been reported in references for azo dye decolorization15–18. Thus, it could be concluded that S. halophilus SSA1575 could be an efficient halotolerant azo-degrading yeast with great potential for disposal of azo dye wastewaters with the salt contaminant at a high concentration.
Figure 4.
Influence of salt concentration on the decolorization of RB5 by S. halophilus SSA-1575, inset with the corresponding decolorization efficiency and decolorization rate.
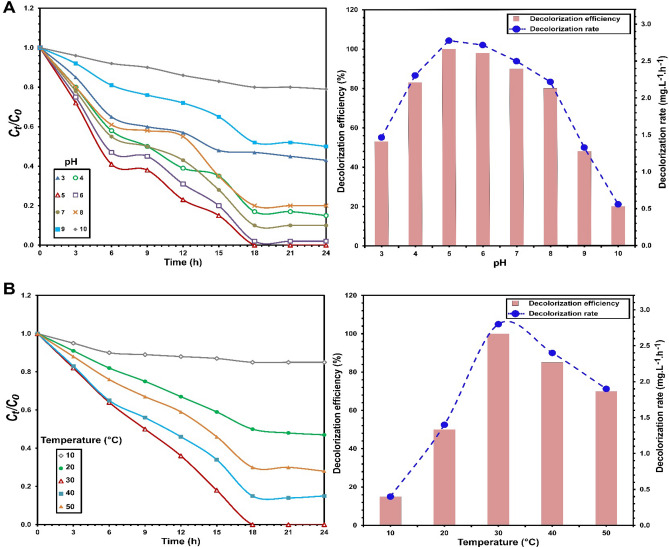
pH and temperature
With regard to pH and temperature parameters, their effects on dye decolorization performance driven by S. halophilus SSA1575 were evaluated (Fig. 5). As depicted in Fig. 5A, the influence of pH variation differed. The decolorization efficiency of RB5 reached 100% at pH 5 after incubation for 18 h with a maximum dye removal rate of 2.78 mg L−1 h−1, whereas a significant decrease in decolorization efficiency was observed at strongly acidic or alkaline pH conditions (p = 0.003). Hsueh and Chen36 reported that the diminishment of dye decolorization at acidic pH, was probably due to the formation of protonated azo dyes that lead to change in the chemical structure of the dye. Hence, microorganism could not recognize the modified dye. However, over 50% RB5 decolorization by S. halophilus SSA1575 was still observed at a strongly acidic pH value (pH 3.0) with a maximum dye removal rate of 1.47 mg L−1 h−1, which might be meaningful in the treatment of azo dyes at acidic conditions. Our findings are in agreement with Du et al.37.
Figure 5.
Influence of pH (A) and environmental temperature (B) on the decolorization of RB5 by S. halophilus SSA-1575, inset with the corresponding decolorization efficiency and decolorization rate.
To evaluate the influence of environmental temperature, which has an effect on enzymatic activities related to dye decolorization, the effect of temperature on RB5 decolorization by S. halophilus SSA1575 was investigated (Fig. 5B). Complete decolorization was observed at 30 °C after incubation for 18 h with a maximum dye decolorization rate of 2.8 mg L−1 h−1, but nearly 100% and over 85% decolorization efficiency of RB5 were achieved at 40 and 50 °C after 24 h. Significant sharp decline in decolorization efficiency was also observed at 10 °C. Our findings are in agreement with Du et al.37 and Tan et al.38 who found that low temperatures could significantly inhibit dye decolorization. Comparatively, in this study, S. halophilus SSA1575 still showed a potential decolorizing capability at higher temperatures up to 50 °C, suggesting its valuable potential in the bioremediation of azo dyes.
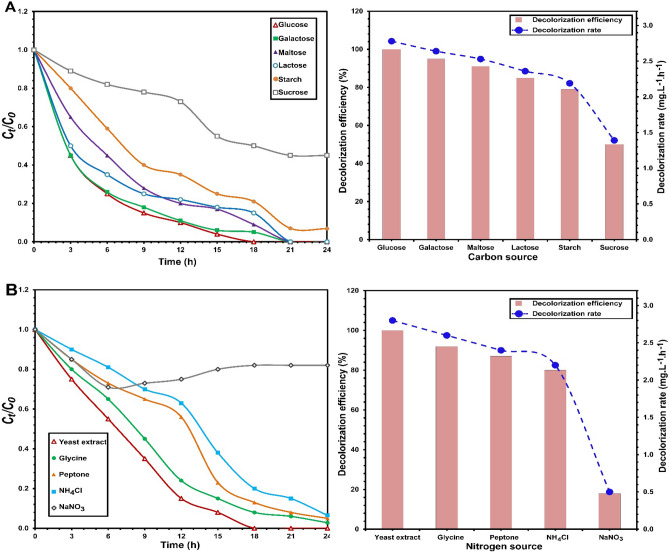
Carbon and nitrogen sources
The performance of S. halophilus SSA1575 on decolorization efficiency of RB5 in the presence of various carbon and nitrogen sources was also studied (Fig. 6). Typically, azo dyes have low carbon content, which makes biodegradation of azo dyes extremely difficult without any supplemental sources of carbon or nitrogen37,39,40. Except sucrose, which was slightly inhibited dye decolorization, other added carbon sources (glucose, galactose, maltose, lactose and starch) could significantly (p < 0.001) enhance RB5 decolorization by S. halophilus SSA1575 (Fig. 6A). Of these, glucose was the optimal carbon source, showing complete decolorization of RB5 after 18 h with a maximum dye decolorization rate of 2.78 mg L−1 h−1, which was probably due to the easy metabolism of glucose that was taken up inside the yeast cells41. Similar to the carbon sources, most tested nitrogen sources could also significantly (p < 0.001) enhance RB5 decolorization (Fig. 6B). Yeast extract was the best nitrogen source, showing complete decolorization of RB5 after 18 h with a maximum dye decolorization rate of 2.8 mg L−1 h−1. However, the addition of sodium nitrate (NaNO3) strongly inhibited RB5 decolorization. Similar results were reported previously37.
Figure 6.
Influence of various carbon sources (A) and nitrogen sources (B) on the decolorization of RB5 by S. halophilus SSA-1575, inset with the corresponding decolorization efficiency and decolorization rate.
Repeated additions of dye aliquots
To understand the capability of S. halophilus SSA1575 to decolorize repeated additions of dye aliquots well, 50 mg L−1 RB5 was tested in decolorization experiments. The decolorization of the first dye aliquot reached 100% within 42 h. The second dye aliquot was also decolorized (100%) by S. halophilus SSA1575 within the next 36 h. However, the third and fourth RB5 aliquots, which were subsequently added, showed complete decolorization within 30 and 24 h of addition, respectively. Further added dye aliquots (fifth, sixth and seventh) revealed also complete decolorization of RB5 only within 18 h. Clearly, the reduction in time required for decolorization of RB5 by S. halophilus SSA1575 in the next cycle was probably due to acclimatization of this yeast strain to the particular azo dye11. Therefore, the ability of S. halophilus SSA1575 to decolorize repeated additions of dye aliquots within a short time is significant for its commercial application.
Toxicity assessment
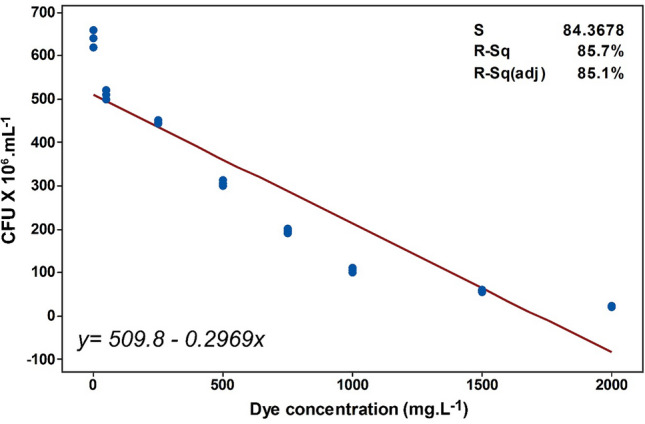
Some of the intermediate metabolites, produced from dye degradation, could accumulate in the environment and might be more toxic than the original dye42. Hence, phytotoxicity and microbial toxicity assays were conducted finally, to evaluate the safety of the metabolic intermediates produced after the decolorization of RB5 by S. halophilus SSA1575. Phytotoxicity, which becomes a more prevalent assay due to being less expensive and easier than other methods, was also performed to evaluate the toxicity of the untreated and treated dye. Therefore, phytotoxicity of RB5 and its extracted metabolites formed after degradation by S. halophilus SSA1575, was evaluated using Sorghum vulgare and Phaseolus mungo seeds (Table 4). The RB5 (100 ppm) showed a higher inhibitory effect on the plumule and radicle lengths of both plant seeds when compared to extracted metabolites obtained after dye degradation. The RB5 solution exhibited 50 and 60% germination inhibition in the case of S. vulgare and P. mungo seeds respectively, when compared with metabolites (Table 4). These results are in agreement with Guo et al.17 and Saratale et al.39 who reported that the metabolites of azo dyes exhibited lower toxicity when compared with the original dyes. On the other hand, the microbial toxicity based on the number of viable cells of Sinorhizobium meliloti was also performed (Fig. 7). The viability of S. meliloti cells was significantly decreased along with an increased RB5 concentration, as revealed from the negative linear correlation between the number of colonies and the dye concentration (p < 0.001; r =− 0.9). The linear regression model indicated that the RB5 dye concentration has a significant predicted impact on cell viability that can account for 85.7% of explained variability in the living cell count. This finding was also consistent with previous reports43. It can be concluded that the results of toxicity evaluations indicate a capability of the newly isolated halotolerant S. halophilus SSA1575, in converting the recalcitrant azo dye RB5 into some non-toxic metabolites. Hence, this yeast strain can be safely implemented in a bioremediation process, particularly for industrial wastewater containing high-salt azo dyes.
Table 4.
Phytotoxicity assessment of RB5 and its metabolites formed after degradation by S. halophilus SSA-1575.
| Parameters | Distilled water | RB5 | Extracted metabolites |
|---|---|---|---|
| Sorghum vulgare | |||
| Germination (%) | 100 | 50 | 100 |
| Plumule (cm) | 7.4 ± 0.11 | 3.1 ± 0.13* | 6.2 ± 0.09** |
| Radicle (cm) | 3.5 ± 0.09 | 1.2 ± 0.07* | 2.4 ± 0.07** |
| Phaseolus mungo | |||
| Germination (%) | 100 | 40 | 100 |
| Plumule (cm) | 10.7 ± 0.40 | 2.3 ± 0.33* | 7.5 ± 0.35** |
| Radicle (cm) | 5.3 ± 0.11 | 0.7 ± 0.15* | 3.1 ± 0.15** |
Values are mean of triplicate ± SD.
*Significantly different from the seeds germinated in distilled water at p < 0.05 by one-way ANOVA with Tukey–Kramer multiple comparisons test.
**Significantly different from the seeds germinated in RB5 dye at p < 0.05 by one-way ANOVA with Tukey–Kramer multiple comparisons test.
Figure 7.
Toxicity assessment. Microbial toxicity of biotransformation metabolites based on the viable colony counting bacterial cells, Sinorhizobium meliloti. p-value < 0.05 is considered significant.
Decolorization mechanism in terms of enzymatic analysis and byproducts detection
As depicted in Table 5, the activities of three oxidases (Lac, LiP and MnP) and two reductases (NADH-DCIP reductase and azoreductase) were analyzed over a period of RB5 decolorization by S. halophilus SSA1575. Song et al.18 reported that NADH-DCIP reductase and azoreductase enzymes were primarily responsible for the cleavage of –N=N– of azo dyes, while Lac, MnP and LiP were involved in further steps of the biodegradation intermediates, such as amines. In this study, the oxidases and reductases were measured under both low (0 g L−1 NaCl) and high (40 g L−1 NaCl) salt concentrations. The activity of NADH-DCIP reductase was significantly induced (p < 0.005) intracellularly and extracellularly under high-salt concentration by 102% and 94%, respectively over the control (Table 5). On the other hand, the activity azoreductase was not detected under high salinity, which suggested that the NADH-DCIP reductase was probably involved in the first step of RB5 degradation. The activities of LiP, MnP and Lac, which were probably involved in further biodegradation steps of RB5, were detected only intracellularly under both low- and high-salt concentrations. However, these activities were significantly decreased under high salinity during the degradation processing when compared with their controls (p < 0.001). As shown in Table 5, the activities of LiP and Lac were detected as crucial intracellular oxidases of the yeast S. halophilus SSA1575, with relatively higher activities when compared with the activity of MnP. Song et al.15 reported the possible inhibitory effect at a high salt concentration on ligninases (oxidases). Clearly, S. halophilus SSA1575 could efficiently decolorize RB5 due to the unique enzymatic system involved.
Table 5.
Extracellular and intracellular enzymes’ activities produced after decolorizing 50 mg L−1 RB5 by the halotolerant yeast strain SSA-1575.
| Enzyme | Control | High salinity | ||
|---|---|---|---|---|
| Intracellular | Extracellular | Intracellular | Extracellular | |
| Laca | 0.367 ± 0.002 | ND | 0.083 ± 0.001* | ND |
| LiPa | 0.452 ± 0.008 | ND | 0.283 ± 0.001* | ND |
| MnPa | 0.171 ± 0.006 | ND | 0.023 ± 0.0002* | ND |
| NADH-DCIP reductaseb | 20.7 ± 0.04 | 12.93 ± 0.05* | 41.74 ± 0.67* | 25.03 ± 0.11* |
| Azoreductasec | 2.83 ± 0.63 | 0.11 ± 0.015 | ND | ND |
Values are mean of triplicates ± SD. Significantly of enzyme activities after dye decoloration differ from target dye (control) at * p < 0.05 using one-way (ANOVA) with Tukey–Kramer comparison test. ND not detected.
aActivity measured by U/min/mg protein.
bActivity measured by µg of DCIP reduced/min/mg protein.
cActivity measured by µg of methyl red reduced/min/mg protein.
To understand well the possible mechanism of dye decolorization, the byproducts made by the halotolerant yeast strain S. halophilus SSA1575 during RB5 degradation were analyzed via UV–Vis spectroscopy, Fourier transformed infrared spectroscopy (FTIR) and Mass Spectrometry techniques. The changes in the UV–vis absorption spectra of RB5 solution (with an initial dye concentration of 50 mg L−1 and salt concentration of 40 g L−1 NaCl) treated with S. halophilus SSA1575 within 24 h were presented (Fig. 8). It has been reported that the intensity of –N=N– bond is proportional to the concentration of azo dye in solution1. UV–vis absorption spectra of RB5 exhibited a strong absorption at λmax = 595 nm, which originated from the conjugated aromatic rings connected by the –N=N– bond. The other observed two peaks at 203 and 310 nm were attributed to the benzene and naphthalene rings of the dye44. Azo dye could be decolorized by microbial cells through biodegradation or adsorption. In the biodegradation process, either the major absorption peaks in the visible light region of the spectrum disappears completely or a new peak appears. However, in the case of the adsorption process, the UV–vis absorption peaks decrease approximately in proportion to each other45,46. As depicted in Fig. 8, the absorbance observed at 595 nm (at the beginning of the dye decolorization process) was decreased with a small shift towards a shorter wavelength, where two peaks at 562 and 530 nm were observed and probably attributed to the formation of other metabolites, such as quinone and benzene molecules11,20,43. On the other hand, the peaks at 211, 230 and 278 nm disappeared, while a new peak (206 nm) was observed. As the reaction progressed, the intensity of the absorption peak (595 nm) decreased to almost zero after 24 h of RB5 decolorization by S. halophilus SSA1575, which was verified from the fully transparent solution (Fig. 8). Clearly, such changes in the absorbance were probably attributed to the changes in the molecular structure of RB5 (deconstruction of its primary chromatophores), indicating degradation of the –N=N– bond, formation of amino groups and other intermediate metabolites, and deconstruction of RB5 during the dye decolorization process7,47.
Figure 8.
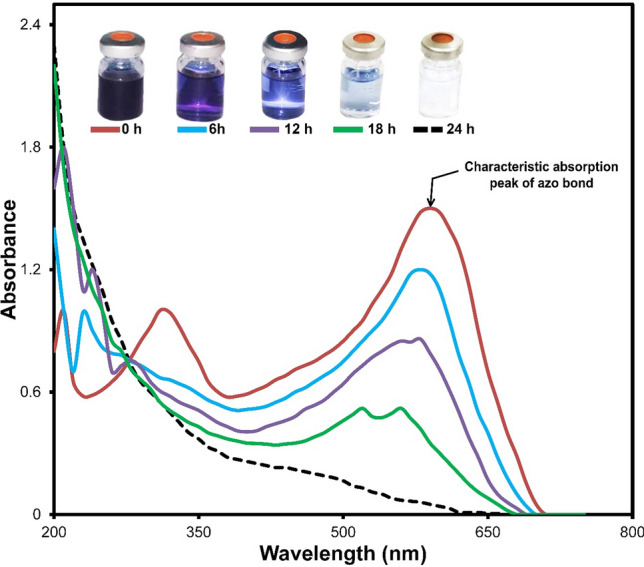
UV–vis spectra of 50 mg L−1 RB5 before and after decolorization by S. halophilus SSA1575.
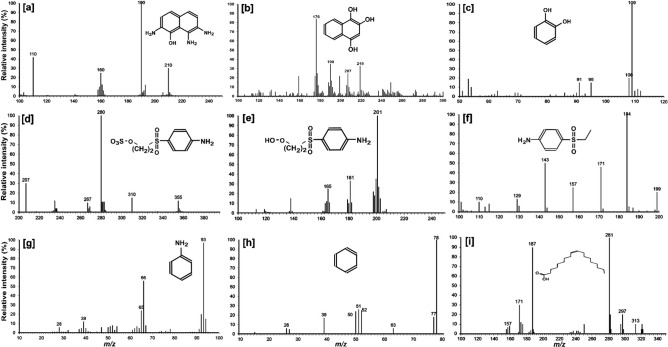
The nature of the degradation product of RB5 by S. halophilus SSA1575 was verified from FT-IR analysis. Comparison of the FT-IR spectrum of the original dye molecule (RB5) with metabolic products extracted after the complete decolorization process clearly indicated significant changes and biodegradation of the RB5 molecule, which were evidenced from the formation of some new peaks as well as the disappearance of some initial peaks (Table 6). In fact, –N=N–, NH2, aromatic amines, C–C linkages showed a cleavage with a prolonged decolorization reaction time and cleavage of azo bonds in RB548. The decrease in the intensities of the peaks at the low-frequency region of spectra (620–850 cm−1) suggested the fission of aromatic rings49. As a result, it is very clear that the molecular structure of RB5 was significantly deconstructed during the decolorization process, by the newly isolated yeast strain S. halophilus SSA1575. To further propose the possible degradation pathway of RB5 by SSA1575, possible metabolites were detected using Mass Spectrometry technique. The structures of nine possible decolorization byproducts could be drawn from the mass spectra and m/z values as depicted in Fig. 9.
Table 6.
Absorption spectra of RB5 before and after its decolorization.
| Wave number (cm−1) | Assignment | Before decolorization | After decolorization |
|---|---|---|---|
| 3,497 | N–H stretching vibration in amides | + | − |
| 1,730 | COOH stretching | − | + |
| 1,670 | C = C aromatic stretching vibration | + | − |
| 1,609, 1,551 | N = N stretching | + | − |
| 1,487 | Ring vibrations | + | − |
| 1,422 | Benzene ring | + | + |
| 1,335 | NO2 stretching | + | − |
| 1,170 | Sulfone SO2 stretching | + | + |
| 1,045, 1,086, 1,202 | C–N stretching confirm the azo nature of dye | + | − |
| 845 | C–H deformation | + | − |
| 783 | N–O stretching | + | − |
| 699, 623 | –C–S-stretching vibration | + | − |
+ : present, −: absent.
Figure 9.
The intermediate metabolites of RB5 degradation identified by GC–MS analysis.
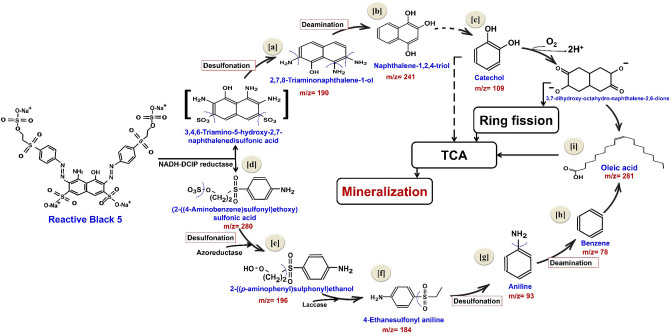
Figure 10 depicts the possible pathway of RB5 biodegradation by S. halophilus SSA1575. The asymmetrical reduction of azo bonds of RB5 by the action of NADH-DCIP reductase was considered as the first step towards bye degradation, resulting in the formation of amines (2-((4-aminobenezene)sulfonyl)ethoxy)sulfonic acid [d] and 1,2,7-triamino-8-hydroxy-3,6-naphthalinedisulfonate (TAHNDS). However, TAHNDS is an unstable compound and it might be rapidly transformed to other smaller intermediate metabolites16,18,50,51, such as 2,7,8-Triaminonaphthalen-1-ol [a]. Subsequently, both of the compounds [a] and [d] were probably further oxidatively transformed into smaller compounds and may eventually be mineralized as depicted in Fig. 10. The intermediate metabolite 2,7,8-Triaminonaphthalen-1-ol [a] was probably deaminated into naphthalene-1,2,4-triol [b], then transformed to catechol [c], which might be cleaved oxidatively into aliphatic metabolites via the cis-muconic acid pathway52 followed by the TCA cycle and finally leading to complete mineralization of RB5 by S. halophilus SSA1575. On the other hand, (2-((4-aminobenezene)sulfonyl)ethoxy)sulfonic acid [d] was probably further oxidatively transformed into 2-((4-aminophenyl)sulfonyl)ethanol [e], 4-ethanesulfonyl aniline [f] and aniline [g] through desulfonation. The later metabolite was probably then deaminated into benzene [h]. According to Gutiérrez et al.53, benzene was successfully used as a precursor for synthesizing a saturated fatty acid by Rhodococcus sp. strain 33. Finally, cis-9-octadecenoic acid [i] was probably mineralized. Overall findings suggested the efficiency of S. halophilus SSA1575 to decolorize and degrade RB5 under high salinity, suggesting that this newly isolated halotolerant yeast strain would have a high potential in the bioremediation of azo dye wastewater with a high load of salts.
Figure 10.
Proposed pathway for the degradation of RB5 by S. halophilus SSA1575.
Methods
Reagents and culture media
Five Azo dyes (Sigma-Aldrich, St. Louis, USA) were used in this study. The molecular structures and absorption wavelength of these dyes were shown in Fig. S1. Other chemical reagents were purchased from Sinopharm Chemical Reagent Co., Ltd. (China). Biochemical reagents and Dr. GenTLER (from Yeast) High Recovery were purchased from TaKaRa Biotechnology Co., Ltd. (Japan). Yeast Malt Extract (YME) consisted of (g/L): 3.0 yeast extract; 5.0 peptone 3.0 malt extract and 10 glucose. Minimal Saline (MS) medium consisted of (g/L): 0.05 CaCl2, 0.05 MgSO4, 1.0 KH2PO4, 0.2 (NH4)2SO4 and 20 NaCl was used, containing 10 glucose as carbon source.
Isolation and identification of azo-decolorizing yeasts
A total of seven novel yeast species or strains which could efficiently decolorize azo dyes under high salt conditions were successfully isolated from the gut of R. chinenesis as per our previously described method13. Phenotypic characterization of the highest decolorization efficiency performed by a yeast strain in a relatively short period of time was first examined following standard methods54,55. To identify the yeast genetically, genomic DNA was extracted using Dr. GenTLER (from Yeast) High Recovery according to the manufacturer's instructions. The 26S rDNA gene was amplified by PCR with the primers NL1/NL43,13. Then the purified PCR products were sequenced by Sangon Biotech (Shanghai, China). All sequences were submitted to GenBank (accession numbers in Table 2) and aligned with sequences available at BLAST-n (https://www.ncbi.nlm.nih.gov/BLAST/). The evolutionary history was inferred using the UPGMA method56. The evolutionary distances were computed using the Kimura 2-parameter method57 and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 2). All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in Evolutionary Genetics Analysis (MEGA) version 758.
Decolorization experiments
The Aqueous solutions (0.1 g) of RB5 as well as other azo dyes were prepared by dissolving the dye tested at the desired concentration in distilled water to prepare 500 mL of solution. Decolorization experiments were conducted in an Erlenmeyer flask (100 mL) containing a volume (5%) of yeast suspension (OD600 of 0.2). Before incubation at 30 °C for 24 h under static condition, the dye was added to flasks at a concentration of 50 mg L−1. Periodically, a 2 mL sample was withdrawn and centrifuged at 12,000 rpm for 3 min. The clear supernatant was used to measure the concentration of the dye by UV–vis spectrometer (Model Shimadzu-UV2600, Japan) at the absorbance maximum of 650 nm (AzB), 595 nm (RB5), 537 nm (RR120), 592 nm (RB19) and 511 nm (AS-GR). Un-inoculated control was included to compare color loss during the decolorization experiments.
The fastest decolorized dye in a relatively short period of time was selected as the target dye for further investigation of the optimization of dye decolorization. The decolorization performance of RB5 (50 mg L−1) by S. halophilus SSA1575, was evaluated at varying salt concentrations (10–100 g L−1 NaCl with an interval of 10 g L−1), dye concentrations (50, 100, 250, 500, 750, 1,000, 1,500 and 2000 mg L−1), temperatures (5–50 °C with an interval of 5 °C) and initial pH (3.0–10.0 with an interval of 1.0). The effect of different carbon and nitrogen sources (2% each) on decolorization efficiency of RB5 by S. halophilus SSA1575 was also studied using glucose, galactose, sucrose, maltose, lactose and starch as carbon sources, while yeast extract, peptone, NaNO3, NH4Cl and glycine were used as nitrogen sources. Further experiments were also performed to evaluate the performance of S. halophilus SSA1575 to decolorize repeated additions of 50 mg L−1 RB5 aliquots.
Decolorization of mixture of azo dyes were calculated following the ADMI protocol59. ADMI removal percentage is defined as the ratio between the ADMI removal value after a particular contacting time (t) and the ADMI value at initial concentration. Four combination mixtures containing two dyes (RB5 and RB19), three dyes (RB5, RB19 and RR120), four dyes (RB5, RB19, RR120 and AzB) and five dyes (RB5, RB19, RR120, AzB and AS-GR) were used at a concentration of 0.2 g L−1. All decolorization experiments were performed in triplicates.
Analytical methods
The percentage of decolorization of dye mixtures was calculated following the Standard Method60. To identify possible metabolites as well as to predict possible mechanisms during decolorization process of the target dye by S. halophilus SSA1575, UV–vis spectroscopy in the region spectrum of 200–700 nm, FTIR in the IR region of 400–4,000 cm−1, and Mass Spectrometry were used under the operation conditions reported earlier61,62.
Toxicity assessment
Phytotoxicity assay
Phytotoxicity was determined in this study based on the inhibition of seed germination using the most-used species S. vulgare (monocot seeds) and P. mungo (dicot seeds)63. The ethyl acetate products of RB5 degradation were dissolved in sterile distilled water to make a final concentration of 100 mg/L. Twenty seeds for each plant set were separately watered with a 5 mL solution of RB5 (100 ppm) or the decolorization metabolites. The seeds watered by distilled water were used as a control set. The changes in radical and plumule lengths were measured after 7 days of germination.
Microbial toxicity assay
This assay was performed using the model soil bacterium S. meliloti that establishes nitrogen-fixing symbiosis with alfalfa64. In this test, the products of dye biodegradation were assessed based on the viable colony counting of S. meliloti cells. A volume of 5% freshly cell suspension (OD600 of 0.2) was inoculated into 5 mL of sterilized yeast mannitol broth at different concentrations of the dye decolorized medium which was previously filtered (0.2 µm). The viable bacterial colonies were counted using the pour plate technique.
Enzyme assay
Extracellular and intracellular enzyme solutions of S. halophilus SSA1575 cells before and after the target dye decolorization for 24 h in a liquid medium containing 50 mg L−1 RB5 and 40 g L−1 NaCl were prepared following the method described previously65. Activities of reductase enzymes including azoreductase and nicotinamide adenine dinucleotide-dichlorophenol indophenol (NADH-DCIP) reductase were monitored as described by Song et al.15. Activities of oxidative enzymes such as LiP, MnP and Lac were also determined as reported earlier by Ali et al.65 . All enzyme assays were carried out spectrophotometrically at room temperature. A blank contained all components except the enzyme. One unit of enzyme activity was defined as the amount of substrate consumed or product generated per milligram protein per minute. Protein concentration of the enzyme solution was monitored using the Bradford method66.
Statistical analysis
Results were statistically analyzed using Minitab 17.1.0.0 and SigmaPlot Software 12.5.0.38. The normality of data was estimated by the Shapiro Wilk test. One-, two- or three-way ANOVA test used with multiple comparisons by Tukey methods. Simple linear regression analysis was performed to estimate the effect of the dye and its biotransformed metabolites on the viability of cells using a regression equation for prediction. The p-value < 0.05 is considered significant.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31772529), the National Key R&D Program of China (2018YFE0107100), and the Project funded by the Priority of Academic Program Development of Jiangsu Higher Education Institutions (PAPD 4013000011).
Author contributions
R.A.-T.: Methodology, Writing-Original Draft preparation, Software. S.A.A.: Conceptualization, Data curation, Methodology, Writing-Reviewing and Editing. M.F.F.: Ecotoxicology assessment, Investigation. E.-R.K.: Analyzed chemical data, Writing-Reviewing and Editing. J.S.: Funding acquisition, Validation, Project administration, Writing-review and editing. All the authors agree to be accountable for the content of the work.
Data availability
All data are available upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianzhong Sun, Email: jzsun1002@ujs.edu.cn.
Sameh S. Ali, Email: samh@ujs.edu.cn
Supplementary information
is available for this paper at 10.1038/s41598-020-69304-4.
References
- 1.Qin X, Zhu Z, Liu, Fu H, Zhang H, Wang A, Li H, Zhang H. Ultrafast degradation of azo dyes catalyzed by cobalt-based metallic glass. Sci. Rep. 2016;5:18226. doi: 10.1038/srep18226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sampaio AC, Bezerra RMF, Dias AA. Mediterranean forested wetlands are yeast hotspots for bioremediation: a case study using azo dyes. Sci. Rep. 2018;8:15943. doi: 10.1038/s41598-018-34325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali SS, Al-Tohamy R, Sun J, Wu J, Huizi L. Screening and construction of a novel microbial consortium SSA-6 enriched from the gut symbionts of wood-feeding termite, Coptotermes formosanus and its biomass-based biorefineries. Fuel. 2019;236:1128–1145. doi: 10.1016/j.fuel.2018.08.117. [DOI] [Google Scholar]
- 4.Droguett T, Mora-Gómez J, García-Gabaldón M, Ortega E, Mestre S, Cifuentes G, Pérez-Herranz V. Electrochemical degradation of Reactive Black 5 using two-different reactor configuration. Sci. Rep. 2020;10:4482. doi: 10.1038/s41598-020-61501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali SS, Al-Tohamy R, Xie R, El-Sheekh, Sun J. Construction of a new lipase- and xylanase-producing oleaginous yeast consortium capable of reactive azo dye degradation and detoxification. Bioresour. Technol. 2020;313:123631. doi: 10.1016/j.biortech.2020.123631. [DOI] [PubMed] [Google Scholar]
- 6.Giovanella P, Vieira GAL, Otero IVR, Pellizzer EP, Fontes BJ, Sette LD. Metal and organic pollutants bioremediation by extremophile Microorganisms. J. Hazard. Materi. 2020;382:121024. doi: 10.1016/j.jhazmat.2019.121024. [DOI] [PubMed] [Google Scholar]
- 7.Al-Tohamy R, Kenawy E, Sun J, Ali SS. Performance of a newly isolated salt-tolerant yeast strain Sterigmatomyces halophilus SSA-1575 for azo dye decolorization and detoxification. Front. Microbiol. 2020;11:1163. doi: 10.3389/fmicb.2020.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Amato CA, Giovannetti R, Zannotti M, Rommozzi E, Minicucci M, Gunnella R, Cicco A. Band gap implications on nano-TiO2 surface modification with ascorbic acid for visible light-active polypropylene coated photocatalyst. Nanomater. 2018;8:599. doi: 10.3390/nano8080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zamel D, Hassanin AH, Ellethy R, Singer G, Abdelmoneim A. Novel bacteria-immobilized cellulose acetate/poly(ethylene oxide) nanofibrous membrane for wastewater treatment. Sci. Rep. 2019;9:18994. doi: 10.1038/s41598-019-55265-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan S, Sadasivam K, Gunalan S, Shanmugam G, Kothandan G. Application of docking and active site analysis for enzyme linked biodegradation of textile dyes. Environ. Pollut. 2019;248:599–608. doi: 10.1016/j.envpol.2019.02.080. [DOI] [PubMed] [Google Scholar]
- 11.Parshetti GK, Telke AA, Kalyani DC, Govindwar SP. Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J. Hazard. Materi. 2010;176:503–509. doi: 10.1016/j.jhazmat.2009.11.058. [DOI] [PubMed] [Google Scholar]
- 12.Martorell MM, Pajot HF, Defigueroa LIC. Dye-decolorization yeasts isolated from Las Yungas rain forest. Dye assimilation and removal used as selection criteria. Int. Biodeter. Biodegr. 2012;66:25–32. doi: 10.1016/j.ibiod.2011.10.005. [DOI] [Google Scholar]
- 13.Ali SS, Wu J, Xie R, Zhou F, Sun J, Huang M. Screening and characterizing of xylanolytic and xylose-fermenting yeasts isolated from the wood-feeding termite, Reticulitermes chinenesis. PLoS ONE. 2017;12(7):e0181141. doi: 10.1371/journal.pone.0181141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali SS, Al-Tohamy R, Sun J, Wu J, Huang M. The role of gut symbionts from termites: a unique hidden player from yeasts (Review) Acta Microbiol. Sin. 2018;58(6):1004–1015. doi: 10.13343/j.cnki.wsxb.20170610. [DOI] [Google Scholar]
- 15.Song L, Shao Y, Ning S, Tan L. Performance of a newly isolated salt-tolerant yeast strain Pichia occidentalis G1 for degrading and detoxifying azo dyes. Bioresour. Technol. 2017;233:21–29. doi: 10.1016/j.biortech.2017.02.065. [DOI] [PubMed] [Google Scholar]
- 16.Tan L, He M, Song L, Fu X, Shi S. Aerobic decolorization, degradation and detoxification of azo dyes by a newly isolated salt-tolerant yeast Scheffersomyces spartinae TLHS-SF1. Bioresour. Technol. 2016;203:287–294. doi: 10.1016/j.biortech.2015.12.058. [DOI] [PubMed] [Google Scholar]
- 17.Guo G, Tian F, Zhao Y, Tang M, Liu W, Liu C, Xue S, Kong W, Sun Y, Wang S. Aerobic decolorization and detoxification of Acid Scarlet GR by a newly isolated salt-tolerant yeast strain Galactomyces geotrichum GG. Int. Biodeter. Biodegr. 2019;145:104818. doi: 10.1016/j.ibiod.2019.104818. [DOI] [Google Scholar]
- 18.Song Z, Song L, Shao Y, Tan L. Degradation and detoxification of azo dyes by a salt-tolerant yeast Cyberlindnera samutprakarnensis S4 under high-salt conditions. World J. Microbiol. Biot. 2018;34:131. doi: 10.1007/s11274-018-2515-7. [DOI] [PubMed] [Google Scholar]
- 19.Rawat D, Mishra V, Sharma RS. Detoxification of azo dyes in the context of environmental processes. Chemosphere. 2016;155:591–605. doi: 10.1016/j.chemosphere.2016.04.068. [DOI] [PubMed] [Google Scholar]
- 20.Martorell MM, Pajot HF, de Figueroa IC. Biological degradation of Reactive Black 5 dye by yeast Trichosporon akiyoshidainum. J. Environ. Chem. Engin. 2017;5:5987–5993. doi: 10.1016/j.jece.2017.11.012. [DOI] [Google Scholar]
- 21.El Bouraie M, El Din W. Biodegradation of reactive balck 5 by Aeromonas hydrophila strain isolated from dye-contaminated textile wastewater. Sustain. Environ. Res. 2016;26:209–216. doi: 10.1016/j.serj.2016.04.014. [DOI] [Google Scholar]
- 22.Taguchi T, Ebihara K, Yanagisaki C, Yoshikawa J, Horiguchi H, Amachi S. Decolorization of recalcitrant dyes by a multicopper oxidase produced by Iodidimonas sp. Q-1 with iodide as a novel inorganic natural redox mediator. Sci. Rep. 2018;8:6717. doi: 10.1038/s41598-018-25043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Díez-Méndez A, García-Fraile P, Solano F, Rivas R. The ant Lasius niger is a new source of bacterial enzymes with biotechnological potential for bleaching dye. Sci. Rep. 2019;9:15217. doi: 10.1038/s41598-019-51669-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun JZ, Ding SY, Peterson DJ. Biological conversion of biomass for fuels and chemicals: explorations from natural biomass utilization systems. R. Soc. Chem. (RSC) 2014 doi: 10.1039/9781849734738. [DOI] [Google Scholar]
- 25.Fell JW. Sterigmatomyces Fell (1966) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts: A Taxonomic Study. 5. Berlin: Elsevier; 2011. pp. 1991–1994. [Google Scholar]
- 26.Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- 27.Zahran SA, Ali-Tammam M, Hashem AM, Aziz RK, Ali AE. Azoreductase activity of dye-decolorizing bacteria isolated from the human gut microbiota. Sci. Rep. 2019;9:5508. doi: 10.1038/s41598-019-41894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado K, Compart L, Morais R, Rosa L, Santos M. Biodegradation of reactive textile dyes by Basidiomycetous fungi from Brazelian ecosystems. Braz. J. Microbiol. 2006;37:481–487. doi: 10.1590/S1517-83822006000400015. [DOI] [Google Scholar]
- 29.Tamboli DP, Kurade MB, Waghmode TR, Joshi SM, Govindwar SP. Exploring the ability of Sphingobacterium sp. ATM to degrade textile dye Direct Blue GLL, mixture of dyes and textile effluent and production of polyhydroxyhexadecanoic acid using waste biomass generated after dye degradation. J. Hazard. Mater. 2010;182:169–176. doi: 10.1016/j.jhazmat.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Hai T, Lange D, Rabus R, Steinbüchel A. Polyhydroxyalkanoates (PHA) accumulation in sulfate-reducing bacteria and identification of a class III PHA synthase (PhaEC) in Desulfococcus multivorans. Appl. Environ. Microbiol. 2004;70:4440–4448. doi: 10.1128/AEM.70.8.4440-4448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonartseva G, Myshkina V, Nikolaeva D, Rebrov A, Gerasin V, Makhin T. The biodegradation of polyhydroxybutyrate (PHB) by a model soil community: the effect of cultivation conditions on the degradation rate and the physicochemical characteristics of PHB. Mikrobiologiia. 2002;71:221–226. doi: 10.1023/A:1015162608031. [DOI] [PubMed] [Google Scholar]
- 32.Cruz A, Buitrón G. Biodegradation of disperse blue 79 using sequenced anaerobic/aerobic biofilters. Water Sci. Technol. 2001;44:159–166. doi: 10.2166/wst.2001.0209. [DOI] [PubMed] [Google Scholar]
- 33.Singh, S. N. (ed.) Microbial Degradation of Xenobiotics, Environmental Science and Engineering XIV, Springer, Berlin, Heidelberg (2012). 10.1007/978-981-13-7462-3
- 34.Singh, S. N. (ed.) 2014. Microbial Degradation of Synthetic Dyes in Wastewaters, Environmental Science XIV, Springer International Publishing, Switzerland. (2014). 10.1007/978-3-319-10942-8
- 35.Woolard CR, Irvine RL. Treatment of hypersaline wastewater in the sequencing batch reactor. Water Res. 1995;29:1159–1168. doi: 10.1016/0043-1354(94)00239-4. [DOI] [Google Scholar]
- 36.Hsueh C-C, Chen B-Y. Comparative study on reaction selectivity of azo dye decolorization by Pseudomonas luteola. J. Hazard. Mater. 2007;141:842–849. doi: 10.1016/j.jhazmat.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 37.Du L-N, Li G, Zhao YH, Xu H-K, Wang Y, Zhou Y, Wang L. Efficient metabolism of the azo dye methyl orange by Aeromonas sp. strain DH-6: Characteristics and partial mechanism. Int. Biodeter. Biodegr. 2015;105:66–72. doi: 10.1016/j.ibiod.2015.08.019. [DOI] [Google Scholar]
- 38.Tan L, Ning SX, Xia HZ, Sun J. Aerobic decolorization and mineralization of azo dyes by a microbial community in the absence of an external carbon source. Int. Biodeterior. Biodegrad. 2013;85:210–216. doi: 10.1016/j.ibiod.2013.02.018. [DOI] [Google Scholar]
- 39.Saratale RG, Saratale GD, Chang JS, Govindwar SP. Ecofriendly degradation of sulfonated diazo dye C.I. Reactive Green 19A using Micrococcus glutamicus NCIM-2168. Bioresour. Technol. 2009;100:3897–3905. doi: 10.1016/j.biortech.2009.03.051. [DOI] [PubMed] [Google Scholar]
- 40.Jamee R, Siddique R. Biodegradation of synthetic dyes of textile effluent by microorganisms: an environmentally and economically sustainable approach. Eur. J. Microbiol. Immunol. 2019;9(4):114–118. doi: 10.1556/1886.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Zhu Y, He XB, Han GM, Tian XJ. Evaluation of a new fungus Ceriporia lacerate strain P2-its ability to decolorize Alizarin Red and Methyl Orange. World J. Microbiol. Biotechnol. 2008;24:3097–3104. doi: 10.1007/s11274-008-9822-3. [DOI] [Google Scholar]
- 42.Heo N, Jun Y, Park J. Dye molecules in electrolytes: new approach for suppression of dye-desorption in dye-sensitized solar cells. Sci. Rep. 2013;3:1712. doi: 10.1038/srep01712. [DOI] [Google Scholar]
- 43.Enayatizamir N, Tabandeh F, Rodríguez-Couto S, Yakhchali B, Alikhani HA, Mohammadi L. Biodegradation pathway and detoxification of the diazo dye Reactive Black 5 by Phanerochaete chrysosporium. Bioresour. Technol. 2011;102:10359–10362. doi: 10.1016/j.biortech.2011.08.130. [DOI] [PubMed] [Google Scholar]
- 44.Cao J, Wei L, Huang Q, Wang L, Han S. Reducing degradation of azo dye by zero-valent iron in aqueous solution. Chemosphere. 1999;38:565–571. doi: 10.1016/S0045-6535(98)00201-X. [DOI] [PubMed] [Google Scholar]
- 45.Asad S, Amoozegar MA, Pourbabaee AA, Sarbolouki MN, Dastgheib SMM. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour. Technol. 2007;98:2082–2088. doi: 10.1016/j.biortech.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 46.Bheemaraddi MC, Patil S, Shivannavar CT, Gaddad SM. Isolation and characterization of Paracoccus sp. GSM2 capable of degrading textile azo dye Reactive Violet 5. Sci. World J. 2014;2014:410704. doi: 10.1155/2014/410704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dafale N, Rao NN, Meshram SU, Wate SR. Decolorization of azo dyes and simulated dye bath wastewater using acclimatized microbial consortium biostimulation and halo tolerance. Bioresour. Technol. 2008;99(7):2552–2558. doi: 10.1016/j.biortech.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 48.Yang H, Liu J, Wang Y-X, He C-S, Zhang L-S, Mu Y, Li W. Bioelectrochemical decolorization of a reactive diazo dye: Kinetics, optimization with a response surface methodology, and proposed degradation pathway. Bioelectrochemistry. 2019;128:9–16. doi: 10.1016/j.bioelechem.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Kolekar YM, Kodam KM. Decolorization of textile dyes by Alishewanella sp KMK6. Appl. Microbiol. Biotechnol. 2012;95:521–529. doi: 10.1007/s00253-011-3698-0. [DOI] [PubMed] [Google Scholar]
- 50.Jain K, Shah V, Chapla D, Madamwar D. Decolorization and degradation of azo dye—Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. J. Hazard. Mater. 2012;213–214:378–386. doi: 10.1016/j.jhazmat.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Qu Y, Cao X, Ma Q, Shi S, Tan L, Li X, Zhou H, Zhang X, Zhou J. Aerobic decolorization and degradation of Acid Red B by a newly isolated Pichia sp. TCL. J. Hazard. Mater. 2012;223–224:31–38. doi: 10.1016/j.jhazmat.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 52.Oturkar CC, Nemade HN, Mulik PM, Patole MS, Hawaldar RR, Gawai KR. Mechanistic investigation of decolorization and degradation of Reactive Red 120 by Bacillus lentus BI377. Bioresour. Technol. 2011;102:758–764. doi: 10.1016/j.biortech.2010.08.094. [DOI] [PubMed] [Google Scholar]
- 53.Gutiérrez J, Nichols P, Couperwhite I. Changes in whole cell-derived fatty acids induced by benzene and occurrence of the unusual 16:1g6c in Rhodococcus sp. 33. FEMS Microbiol. Lett. 1999;176:213–218. doi: 10.1111/j.1574-6968.1999.tb13664.x. [DOI] [Google Scholar]
- 54.Kurtzman CP. Hyphopichia von Arx & van der Walt. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5. Amsterdam: Elsevier; 2011. pp. 435–438. [Google Scholar]
- 55.Yarrow D. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW, editors. The Yeasts, a Taxonomic Study. 4. Amsterdam: Elsevier; 1998. pp. 77–100. [Google Scholar]
- 56.Sneath PHA, Sokal RR. Numerical Taxonomy. San Francisco: Freeman; 1973. [Google Scholar]
- 57.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 58.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen K, Wu J, Yang W, Hwang S. Evaluation of effective diffusion coefficient and intrinsic kinetic parameters on azo dye biodegradation using PVA-immobilized cell beads. Biotechnol. Bioeng. 2003;83:821–832. doi: 10.1002/bit.10730. [DOI] [PubMed] [Google Scholar]
- 60.APHA, Standard Method for the Examination of Water and Wastewater, 20th ed., American Public Health Association, 2120E, Washington, DC, USA. (1998).
- 61.Kenawy E, Ali SS, Al-Etewy M, Sun J, Wu J, El-Zawawy N. Synthesis, characterization and biomedical applications of a novel Schiff base on methyl acrylate-functionalized chitosan bearing p-nitrobenzaldehyde groups. Int. J. Biol. Macromol. 2019;122:833–843. doi: 10.1016/j.ijbiomac.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Ali SS, El-Zawawy NA, Al-Tohamy R, El-Sapagh S, Mustafa AM, Sun J. Lycium shawii Roem. & Schult.: A new bioactive antimicrobial and antioxidant agent to combat multi-drug/pan-drug resistant pathogens of wound burn infections. J Tradit. Complement. Med. 2019;10:13–25. doi: 10.1016/j.jtcme.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khandare RV, Kabra AN, Kurade MB, Govindwar SP. Phytoremediation potential of Portulaca grandiflora Hook. (Moss-Rose) in degrading a sulfonated diazo reactive dye Navy Blue HE2R (Reactive Blue 172) Bioresour. Technol. 2011;102:6774–6777. doi: 10.1016/j.biortech.2011.03.094. [DOI] [PubMed] [Google Scholar]
- 64.Cosquer A, Pichereau V, Le Mee D, Le Roch M, Renault J, Carboni B, Uriac P, Bernard T. Toxicity and osmoprotective activities of analogues of glycine betaine obtained by solid phase organic synthesis towards Sinorhizobium meliloti. Bioorg. Med. Chem. Lett. 1999;9:49–54. doi: 10.1016/S0960-894X(98)00679-9. [DOI] [PubMed] [Google Scholar]
- 65.Ali SS, Kornaros M, Manni A, Sun J, El-Shanshoury AER, Kenawy E-R, Khalil MA. Enhanced anaerobic digestion performance by two artificially constructed microbial consortia capable of woody biomass degradation and chlorophenols detoxification. J. Hazard. Mater. 2020;389:122076. doi: 10.1016/j.jhazmat.2020.122076. [DOI] [PubMed] [Google Scholar]
- 66.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request.