Abstract
The contribution of the nucleotide-binding oligomerization domain protein NOD1 to obesity has been investigated in mice fed a high fat diet (HFD). Absence of NOD1 accelerates obesity as early as 2 weeks after feeding a HFD. The obesity was due to increases in abdominal and inguinal adipose tissues. Analysis of the resting energy expenditure showed an impaired function in NOD1-deficient animals, compatible with an alteration in thyroid hormone homeostasis. Interestingly, free thyroidal T4 increased in NOD1-deficient mice fed a HFD and the expression levels of UCP1 in brown adipose tissue were significantly lower in NOD1-deficient mice than in the wild type animals eating a HFD, thus contributing to the observed adiposity in NOD1-deficient mice. Feeding a HFD resulted in an alteration of the proinflammatory profile of these animals, with an increase in the infiltration of inflammatory cells in the liver and in the white adipose tissue, and an elevation of the circulating levels of TNF-α. In addition, alterations in the gut microbiota in NOD1-deficient mice correlate with increased vulnerability of their ecosystem to the HFD challenge and affect the immune-metabolic phenotype of obese mice. Together, the data are compatible with a protective function of NOD1 against low-grade inflammation and obesity under nutritional conditions enriched in saturated lipids. Moreover, one of the key players of this early obesity onset is a dysregulation in the metabolism and release of thyroid hormones leading to reduced energy expenditure, which represents a new role for these hormones in the metabolic actions controlled by NOD1.
Subject terms: Biochemistry, Immunology, Physiology
Introduction
The nucleotide-binding oligomerization domain (NOD) protein NOD1 belongs to the NOD-like receptor (NLR) family of pathogen recognition receptors. NOD1 is a cytoplasmic protein that recognizes conserved fragments found in the cell wall of many types of Gram-negative or Gram-positive bacteria1. NOD1 resides as an inactive monomer in the cytoplasm and, upon ligand recognition, a conformational change occurs favoring oligomerization and recruitment of the serine/threonine-protein kinase 2 (RIPK2) through homotypic CARD–CARD interactions2,3. RIPK2 mediates the recruitment and activation of the serine/threonine kinase TAK1, which is a prerequisite for activation of the IκB kinase complex, leading to both the canonical NF-κB and MAPK activation pathways4. The bacterial determinants involved in NOD1 activation have not been yet fully elucidated, but several studies have demonstrated that the minimal motif γ-d-glutamyl-meso-diaminopimelic acid (iE-DAP), present in most bacterial peptidoglycans, is sufficient to fully activate NOD11,2. Although NLRs and TLRs are thought to play partial redundant roles, their expression pattern shows certain specificities that can be evidenced in genetically targeted animal models challenged with different pathogens1,5. NOD1 is widely expressed by a variety of cell types, but is significantly represented in epithelia, stromal and endothelial cells, adipocytes and in the thyroid6,7, and to a lesser extent in resting myeloid cells3,4,8–10. Activation of NOD1 in intestinal epithelial cells triggers the production of chemokines and the recruitment of acute inflammatory cells in vivo, playing a role in the immune response against pathogenic microbes4,11,12. Indeed, NOD1 KO mice are more susceptible to multiple bacterial pathogens suggesting a specific role for this receptor in the innate defense mechanisms of the intestinal epithelium against acute infections13–16. Moreover, NOD1 also exerts a protective role against chronic intestinal inflammatory processes, like colorectal cancer as deduced from NOD1 or RIPK2 deficient mice challenged with different colonic tumor inducers13,15–17, proving its role in the maintenance of intestinal homeostasis.
The innate immune system is strongly associated with obesity, insulin resistance, metabolic syndrome and diabetes18. Recent studies suggest that much of this interplay is due to NLR activation, in particular NOD118–20. Moreover, insulin resistance has been associated with both NOD1 and NOD2 activities by eliciting pro-inflammatory signaling pathways in adipocyte cell models and in in vivo studies on human adipose tissue20–23. Indeed, it appears that several fatty acids can moderately activate these NLRs leading to a chronic low-grade inflammatory tone15,24. Several studies suggest that the intestinal barrier becomes compromised during HFD feeding, resulting in translocation of gut bacterial components into systemic circulation18. Therefore, intestinal epithelial NOD1 may play a protective role in strengthening the gut barrier and restricting this bacterial influx in obesity. Accordingly, mice deficient in NOD1 present an exacerbated systemic inflammation under HFD, in addition to alterations in the broad metabolic homeostasis of the animals2,20,24,25. In this work, we have examined the impact of a HFD in NOD1 deficient mice compared to wild-type (WT) mice during the early onset of diet-induced obesity. Our data show that NOD1 deficient mice fed a HFD for six weeks exhibit an exacerbated increase in lipogenesis in the iWAT and eWAT, but this occurs in the absence of an evident insulin resistance phenotype. In addition to this, important alterations in thyroid hormone dynamics were observed when comparing WT vs. NOD1 KO mice fed a HFD, which probably are involved in the progression to further metabolic dysfunctions. The NOD1 deletion also influences the composition of the gut microbiota and the HFD-induced alterations associated with a worse metabolic phenotype.
Materials and methods
Materials
Common reagents were from Sigma-Aldrich-Merck (St Louis, MO, USA) or Roche (Darmstadt, DE). Murine or human cytokines were obtained from PeproTech (London, UK). Antibodies were from Ambion (Austin, TX, USA), Abcam (Cambridge, UK) or Cell Signaling (Danvers, MA, USA). Reagents for electrophoresis were from Bio-Rad (Hercules, CA, USA). Tissue culture dishes were from Falcon (Lincoln Park, NJ, USA), and serum and culture media were from Invitrogen (Life Technologies/Thermo-Fisher, Madrid, ES).
Animal care and tissue preparation
Wild type (WT) and homozygous Nod1−/− (NOD1 KO) mice (both in the mixed C57BL/6 and 129/C57BL/6 background), have been previously described7,26,27. Male NOD1 WT or KO were housed under 12 h light/dark cycle and food and water was provided ad libitum (chow diet: CHD; 2.05% fat, mainly oleic and linoleic acids; SAFE-PANLAB, ES) for 12 weeks. Animals were then maintained in CHD or switched to a high-fat diet (HFD; 10.2% hydrogenated coconut fat and 0.75% cholesterol; Ssniff, Soest, DE) which was provided ad libitum for six weeks. Treatment with the NOD1 activator iE-DAP was achieved as previously described7. Animals were cared for according to the protocol approved by the Ethical Committee of CISC, and in accordance to directive 2010/63/EU of the European Parliament.
Evaluation of animal metabolic profile
Animals under analysis were maintained individually in Phenomaster Metabolic Cages (TSE Systems International Group) from day 33 to day 38 and the evaluation was done during day 35–38. Spontaneous physical movement, drink and food intake, respiratory exchange (O2 and CO2), weight and heat production were monitored as previously described28.
Evaluation of the glucose tolerance test
Mice underwent a glucose tolerance test (GTT) at the beginning of the HFD and at week 6 of treatment. Animals were fasted overnight and injected with 2 g of glucose per kg body weight. Blood glucose was measured with an Accu-Check glucometer (Roche) from the tail vein at 0, 15, 30, 60 and 120 min. Plasma insulin levels were determined using an ELISA kit (Mercodia, Uppsala, SE) and in parallel to the glucose determination (15 min after glucose administration).
Assay of total bile acids (BA)
A fluorimetric assay (Sigma-Aldrich) was used to determine total bile acids in serum and in liver extracts. The assay is based on the use of 3-hydroxysteroid dehydrogenase that reacts with twelve BA, reducing NAD to NADH. Appropriate internal controls were used to ensure the linearity and specificity of the reaction.
1H NMR analysis of liver lipids
HRMAS analysis was run at 4 °C in an 11.7 T Bruker Avance spectrometer at 500.113 MHz, and using 5 kHz spinning rate, following a previously described protocol for lipids29,30. The ex vivo spectra were quantified by using MestReC software (Mestrelab Research, Santiago de Compostela, ES). Validation of 1H MR spectra were obtained from different FA standards and data were generated using heuristic analysis. Results were expressed as: SFA: saturated fatty acids (FFAA); UFA: Unsaturated FFAA; MUFA: monounsaturated FFAA; PUFA: polyunsaturated FFAA; MCL: medium chain length FFAA; NDB: number of double bonds in FFAA; PU/UFA, polyunsaturated/unsaturated fatty acids; saturation index (Sat. Index), as the ratio between stearic and oleic acids, reflecting membrane rigidity.
Quantification of myeloid cell populations
Animals were fed as indicated and the presence of myeloid cells in the eWAT and liver at 24 h was assessed in a FACSCanto II flow cytometer as previously described7. The adipose tissue (eWAT) was mechanically digested using an enzymatic solution (type II collagenase, CaCl2 plus BSA) for 20 min at 37 °C in a rotational shaker (200 rpm). Leukocyte subsets were defined using FlowJo software: neutrophils (CD45+CD115−Ly6G+), inflammatory monocytes (CD45+CD115+) and tissue macrophages (CD45+CD115−F4/80+)-population31.
Preparation of total protein cell extracts
Tissues were homogenized as previously described in the presence of 1 mM β-mercaptoethanol, detergent (0.5% CHAPS) and protease and phosphatase inhibitor cocktail from Sigma. Extracts were centrifuged and protein content was quantified7.
RNA isolation and analysis
RNA was extracted in TRIzol and processed in a Real-time PCR equipment as previously described7,32. Samples were analyzed in duplicate vs. 36B4. Dio2, Ucp1 and Dio1 were assayed using specific Taqman probes (Mm01244861m1 for Ucp1, Mn00515664m1 for Dio2, Mn00839358_m1 for Dio1 (Gene expression assays, Applied Biosystems, Foster City, CA). Results were normalized to cyclophilin labelled with VIC (Ppia, Mm02342429g1, Applied Biosystems) in the same well. The fold-change in mRNA expression was calculated by the 2−ΔΔCt method.
Protein analysis by Western blot
Protein cell extracts were boiled in 250 mM Tris–HCl, pH 6.8; 2% SDS, 10% glycerol and 2% β-mercaptoethanol, and equal amounts of protein were separated by SDS-PAGE electrophoresis in 8–10% gels and visualized by ECL chemiluminescence as described7,32. The antibodies used were from Abcam or Cell Signaling.
Thyroid hormones determinations
The concentrations of T3 and T4 were measured after extraction and purification from plasma, liver, brown adipose tissue (BAT) and thyroids using specific radioimmunoassays. For hormone measurement in thyroids, tissues were homogenized in ethanol, and thyroidal free (F) T4 and T3 were determined in the dried extracts as described33.
Deiodinase activities
D2 in BAT and D1 in liver
125I-T4 and 125I-rT3 were synthetized and purified to reduce iodide to less than 1% of the total 125I. D2 activity was assayed as previously described34,35. D1 activity was determined as previously described36.
Histological analysis and immunostaining
Mouse liver and eWAT tissues were fixed in 4% paraformaldehyde in 0.1 M PBS and embedded in paraffin. 5 μm sections of tissue were deparaffinized sections were stained with hematoxylin/eosin (HE) or Oil-Red O using standard procedures.
Microbiota analysis
Analysis of gut microbiota
Fecal samples from individual mice from each experimental group (n = 10 for control, n = 8 for NOD1 KO) were collected after 12 weeks on a CHD and then at 6 weeks after the switch to the HFD and stored in 2 ml cryovials. Samples were immediately frozen in liquid nitrogen and maintained at − 80 °C. DNA extraction was done using Fast DNA Stool Mini Kit (Qiagen) according to the manufacturer’s instructions with several modifications: Fecal samples (ca. 220 mg) were homogenized with glass beads in the presence of 1 ml of Inhibitex buffer (Qiagen) using a beadbeater for 2 successive rounds for 1 min and then heated to 95 °C for 10 min. DNA was PCR-amplified in triplicate using primers (S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21) that target the V4–V5 variable regions of the 16S rRNA gene, following a previous protocol37. Samples were tagged with barcodes to allow multiplexing during the sequencing process. Sample amplicons were combined and purified using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) according to the manufacturer’s instructions and combined in equimolar concentrations before carrying out sequencing on a MiSeq instrument (Illumina). All raw sequence data has been submitted to ENA-EMBL Accession # PRJEB33681.
Bioinformatic processing of data was carried out using the software QIIME1 (version 1.9.1)38, Mothur39, and UPARSE40. Briefly, using QIIME1, paired-end forward and reverse Illumina reads were joined into contigs (join_paired_ends.py), barcodes were extracted (extract_barcodes.py), and reads were demultiplexed (split_libraries_fastq.py). Sequences smaller than 325 bp or containing homopolymers greater than 7 were removed using the screen.seqs() command from the software program Mothur. Primers were then removed using trim.seqs() from Mothur. Using UPARSE, chimeras were removed and reads were clustered at 97% identity into OTUs using default settings. An OTU abundance table was generated within the UPARSE pipeline by mapping reads to representative sequences for each OTU. Using QIIME, a biom file was created from the OTU table and reads were rarefied (single_rarefaction.py) and singletons were removed (filter_otus_from_otu_table.py). A phylogenetic tree was constructed from representative sequences for each OTU, and aligned (align_seqs.py) using PYNAST41 and filtered (filter_alignment.py) using default settings. Alpha diversity metrics (Shannon’s, Chao1, observed OTUs) were calculated (alpha_diversity.py). Beta diversity analysis was conducted using generalized UniFrac (GUniFrac)42. Taxonomy was assigned to representative OTUs (summarize_taxa.py) using UCLUST against the Greengenes database (version 13.8).
Statistical analyses of microbiota
The biomarker discovery method LDA Effect Size (LEfSe43) was employed to compare the effect of genotype (WT vs. NOD1 KO groups) under a CHD and HFD diet, as well as examine the effects of a HFD after 6 weeks. Comparisons of beta diversity between groups using generalized UniFrac were performed by a PERMANOVA with adonis within the GUniFrac package in R.
Statistical analysis
After normality, the statistical analysis was calculated by D’Agostino-Pearson omnibus test, either a nonparametric test (Mann–Whitney U test), or a normality test (unpaired Student’s t test with Welch’s correction, ordinary 1-way ANOVA) was used as appropriate. Removal of outliers was assessed by the ROUT method. Results were considered statistically significant at P < 0.05.
Results
Loss of NOD1 accelerates diet-induced obesity
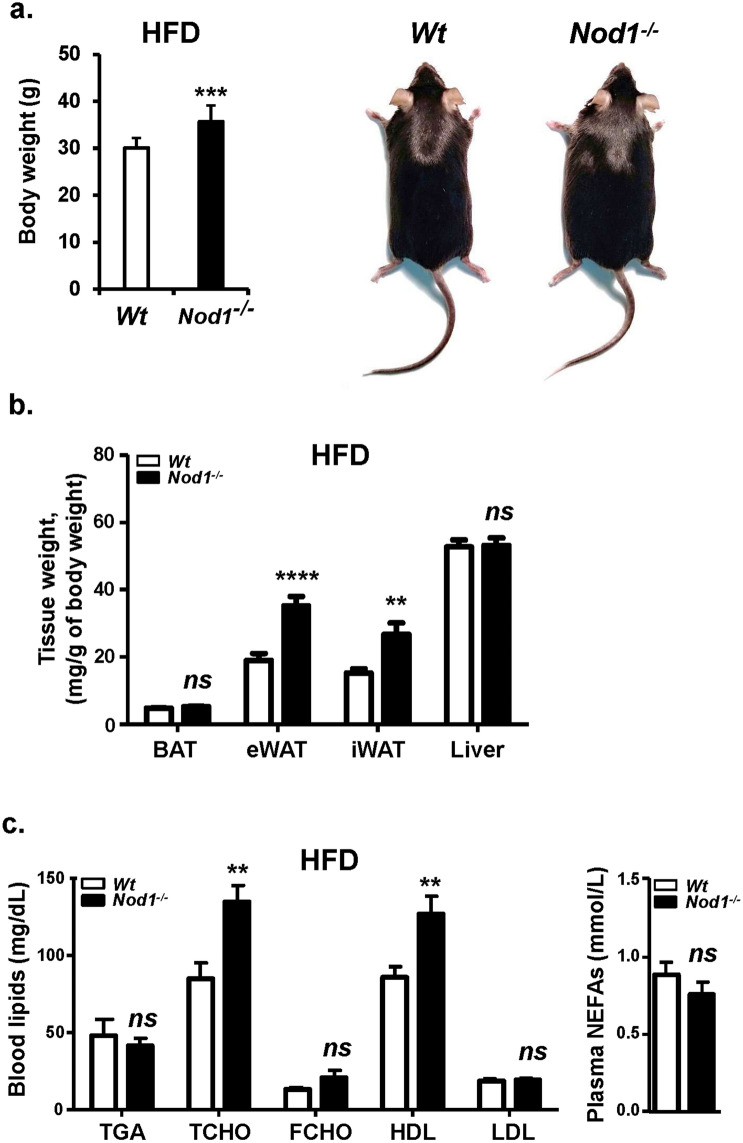
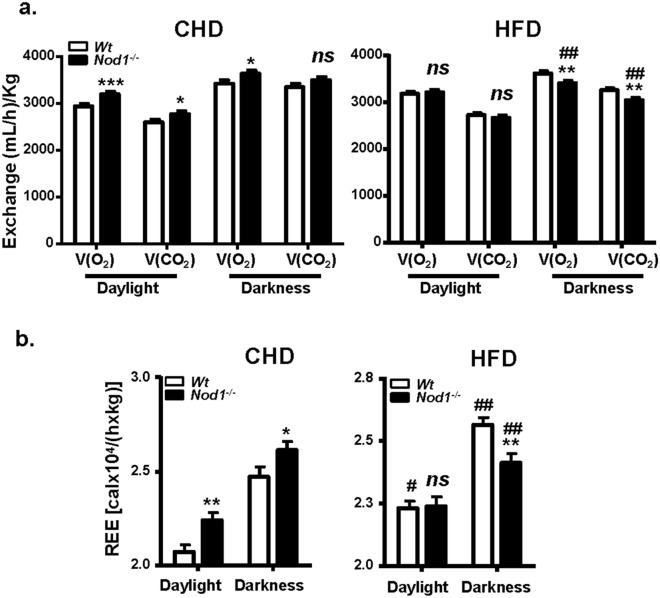
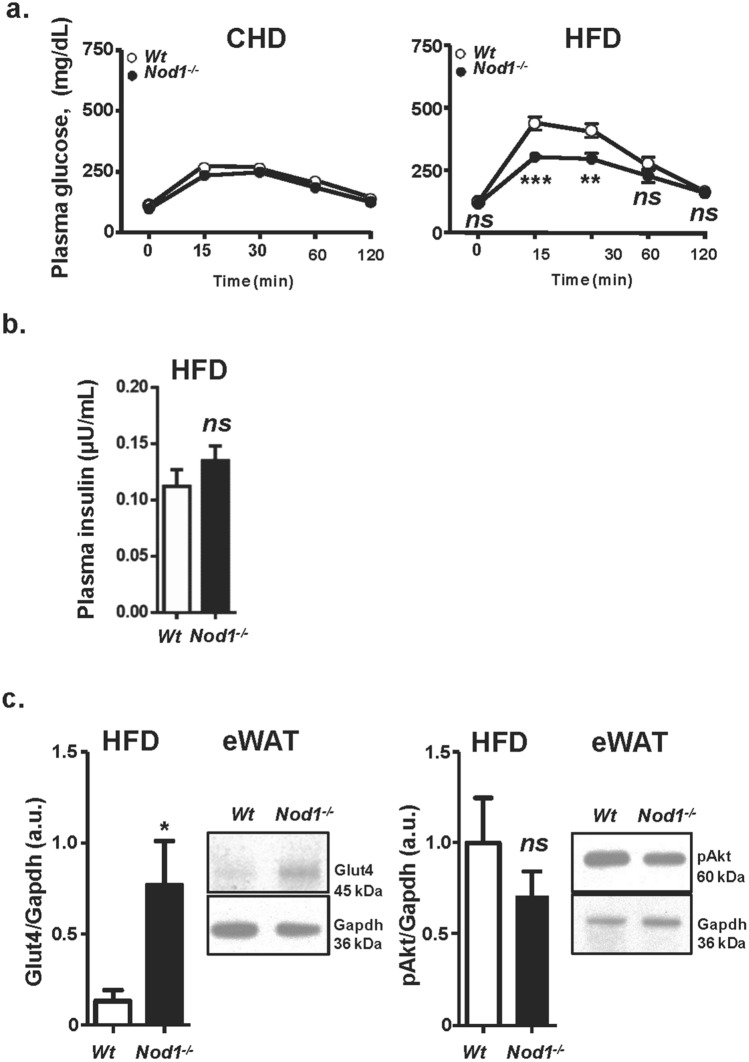
WT and NOD1-deficient mice fed standard CHD did not show differences in body weight across the lifetime of the animals (not shown). However, after 6 weeks on HFD, statistically significant differences in body weight were observed, mainly due to fat accumulation in the eWAT, iWAT and BAT (Supplementary Fig. S1a, Fig. 1a,b), but not in other organs (e.g., liver; Fig. 1b). The increase in body weight of NOD1-deficient mice fed HFD was statistically significant after two weeks of treatment, having both groups the same tibiae size (Supplementary Fig. S1a,b). Regarding blood lipids, HFD-fed mice exhibited an increase in total cholesterol (TCHO) due to an elevation in the HDL fraction, but no differences were observed in other lipids assayed, including triglycerides (TGA) and non-esterified fatty acids (NEFA; Fig. 1c). When animals were analyzed in metabolic cages, NOD1-deficiency in mice fed CHD resulted in higher respiratory activity. However, a depressed respiration during the darkness period was observed in NOD1-deficient mice vs. the WT counterparts when fed a HFD (Fig. 2a). These metabolic differences included a substantial decrease in Resting Energy Expenditure (REE) during darkness and in lesser motion in HFD NOD1 KO animals, a situation opposite to that observed under CHD (Fig. 2b, and Supplementary Fig. S2), suggesting a role for thyroid hormones (a key player in REE) in this differential response44. However, no significant differences in body temperature (not shown), and food and beverage intake were observed between WT and NOD1 KO mice regardless the dietary regimen (Supplementary Fig. S2). To further investigate the mechanisms associated with the observed body weight gain, the glucose tolerance test was evaluated. No differences in the time-dependent glucose drop were observed in animals under CHD; however, NOD1 KO mice fed HFD exhibited an unexpected improvement in the glucose tolerance test that was not due to differences in plasma insulin levels (Fig. 3a,b). These data are compatible with increased GLUT-4 expression from eWAT in NOD1 KO mice, suggesting improved glucose homeostasis (Fig. 3c). Nevertheless, no significant differences in phospho-AKT were observed under these conditions (Fig. 3c).
Figure 1.
NOD1 deficient mice exhibit enhanced body weight and obesity after feeding high-fat diet (HFD). 12-weeks old WT (n = 27) and NOD1 KO mice (n = 26) maintained in chow diet (CHD) were fed for 6 additional weeks with CHD or HFD. (a) Body weight and representative images of animals. (b) Adipose tissues (brown adipose tissue, BAT; epididymal adipose tissue, eWAT; inguinal adipose tissue, iWAT) and liver weight under 6-weeks HFD conditions. (c) Serum lipid profile and non-esterified fatty acids content from these animals (n = 7, for each group). Results show the mean ± SEM of the indicated samples. **P < 0.01; ***P < 0.005; ****P < 0.001 vs. the corresponding condition in WT mice. ns not statistically significant.
Figure 2.
Evaluation of animal metabolic profiles. WT (n = 12) and NOD1 KO (n = 12) animals were fed standard chow diet (CHD) or high-fat diet (HFD) for 6 weeks. The O2 and CO2 exchange (a), the Resting Energy Expenditure (REE) (b) were registered as described in the “Materials and methods” section, during the daylight and darkness periods (12 h). Results show the mean ± SEM of 6 animals per nutritional condition and genetic background. *P < 0.05; **P < 0.01; ***P < 0.005 vs. the corresponding condition in WT mice; #P < 0.05, ##P < 0.01 vs. the corresponding condition under HFD. ns not statistically significant.
Figure 3.
NOD1 KO mice fed HFD exhibit improved glucose tolerance. WT and NOD1 KO mice were fed CHD or HFD for 6 weeks and, at the end of the period were starved and submitted to a glucose tolerance test. Plasma glucose level after i.p. administration of 2 g of glucose per kg body weight (a). Plasma insulin levels from these animals were determined 15 min after glucose administration (b). Immunoblot levels of Glut4 and pAkt in eWAT (c). Results show the mean ± SEM of 6 animals per nutritional condition and genetic background. *P < 0.05; **P < 0.01; ***P < 0.005 vs. the corresponding condition in WT mice. ns not statistically significant. Immunoblot corresponds to a representative assay out of four.
NOD1 deficiency increases white adipocyte lipid content and promotes liver steatosis in HFD-fed mice
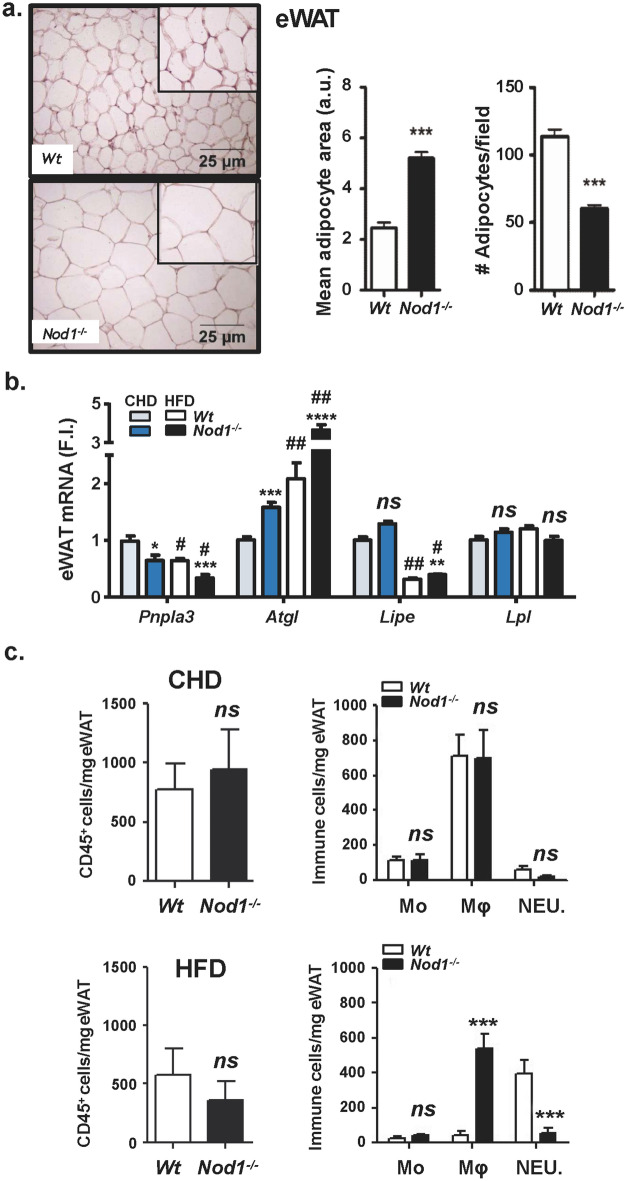
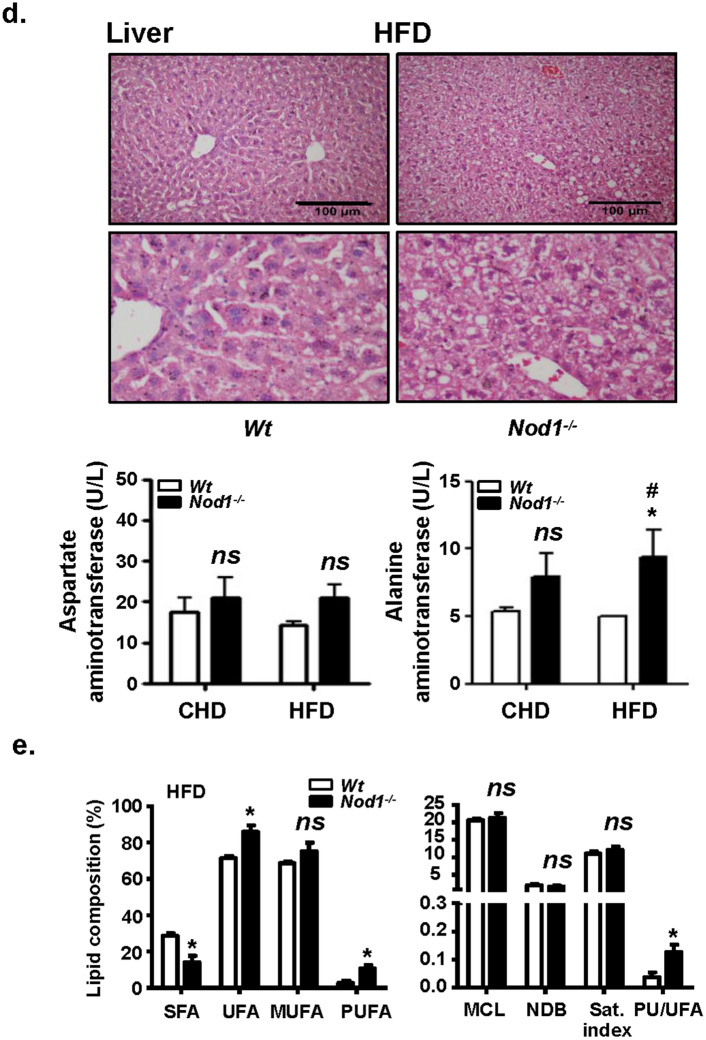
Since eWAT seems to be one of the tissues responsible for the increased body weight in NOD1 KO mice fed HFD, a histological analysis of this tissue was performed. In agreement with the body weight increase of NOD1 KO mice, a very significant enlargement of adipocytes was observed, indicating obesity or at least overweight (Fig. 4a). The RNA levels of key adipocyte lipases and lipid metabolism genes were determined (Fig. 4b). The phospholipase Pnpla3 (patatin-like phospholipase domain-containing protein 3; Pnpla3), which is upregulated by T3 and mediates the hydrolysis of triglycerides in adipocytes playing a role on lipid storage homeostasis45,46, was decreased in NOD1 KO adipose tissue from CHD and HFD mice, a situation compatible with hypothyroidism. The lipase Atgl (patatin-like phospholipase domain-containing protein 2) increased significantly under HFD21,47. Other lipases, such as Lipe (hormone-sensitive lipase), decreased in HFD, in agreement to their role in controlling lipolysis47,48, whereas Lpl (lipoprotein lipase) remained unchanged in all conditions assayed (Fig. 4b). Among other genes involved in lipid homeostasis, a marked increase in Ppara, but not in Pparg was evidenced in the absence of NOD1 (not shown). Because lipid accumulation is evident in eWAT from NOD1 KO animals fed HFD, it can be suggested that lipogenesis is markedly enhanced in this tissue through mechanisms not yet fully established. Since infiltrating immune cells may influence the physiology of adipocytes, eWAT resident immune cell populations were determined by flow cytometry. As Fig. 4c shows, no significant differences in the content of CD45-positive cells or in the tissue distribution of myeloid cells was observed under CHD. However, after HFD, the eWAT tissue from NOD1 KO mice exhibited a large infiltration of macrophages, but with lesser neutrophilia (Fig. 4c, lower panel), despite the larger size of the eWAT adipocytes from NOD1 KO HFD fed mice. In addition to eWAT, the liver from NOD1 KO mice under HFD presented a significant steatosis, without elevation of serum transaminases except for a modest, although statistically significant increase in alanine aminotransferase (ALT; Fig. 4d). This increased lipogenesis in the absence of NOD1 under HFD was confirmed by the fatty acid composition of livers that showed an increase of polyunsaturated fatty acids (PUFA) and a decrease of saturated fatty acids (SFA), a metabolic situation associated with the development of non-alcoholic fatty liver disease (NAFLD)49 (Fig. 4e). Interestingly, WT mice fed a HFD for 4 weeks followed by 2 weeks on CHD fully recovered from the lipid accumulation in liver, whereas NOD1 KO mice under the same nutritional protocol still exhibited clear signs of steatosis and lipid accumulation in the liver (Supplementary Fig. S3). Indeed, HFD significantly activates NOD1 signaling cascade in WT mice through the phosphorylation of RIP2 kinase (Supplementary Fig. S4).
Figure 4.
NOD1 KO mice fed HFD exhibit a significant increase in eWAT size, infiltration of immune cells and hepatic steatosis. Immunohistochemical analysis of eWAT from WT and NOD1 KO mice fed HFD for 6 weeks, and evaluation of adipocyte area and cell density (a). qPCR analysis of eWAT lipases from mice fed CHD or HFD for 6 weeks (b). Infiltration of immune cells in eWAT (c). Accumulation of lipids in liver and serum transaminase levels from WT and NOD1 KO mice fed HFD (d). Hepatic lipid composition from HFD fed animals determined by 1H magnetic resonance spectrum (1H MRS-HRMAS) (e). Results show the mean ± SEM of 4 (b, c) and 6 (a, d, e) animals per nutritional condition and genetic background. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001 vs. the corresponding condition in WT mice. #P < 0.05, ##P < 0.01, ###P < 0.005 vs. the corresponding genetic condition under CHD. Ns not statistically significant. Results were expressed as: SFA: saturated fatty acids (FFAA); UFA: Unsaturated FFAA; MUFA: monounsaturated FFAA; PUFA: polyunsaturated FFAA; MCL: medium chain length FFAA; NDB: number of double bonds in FFAA; PU/UFA, polyunsaturated/unsaturated fatty acids; saturation index (Sat. Index), as the ratio between stearic and oleic acids.
NOD1-deficiency influences bile acid metabolism, inflammation and intestinal lipids in HFD-fed mice
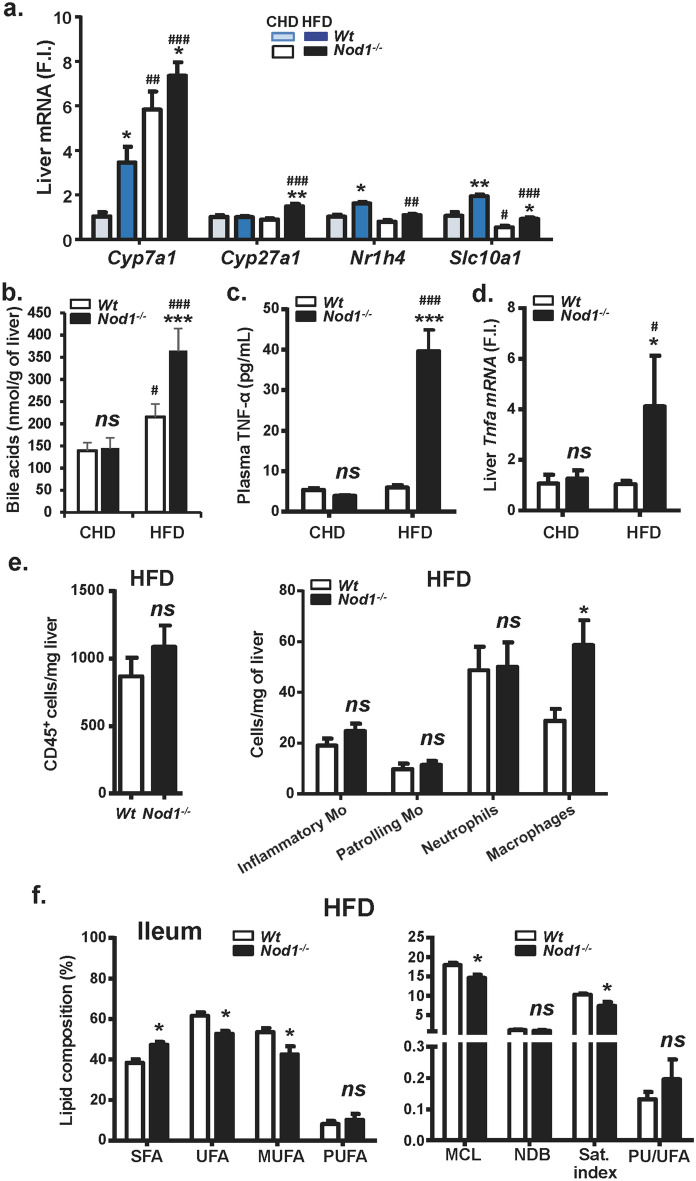
Bile acids (BA), produced in the liver, stimulate energy expenditure through increases in D2 deiodinase in BAT. In addition to this, BA are critically important for balanced lipid metabolism and innate immunity19,50. Thus, we investigated the mRNA levels of relevant genes involved in cholesterol catabolism leading to BA biosynthesis (Fig. 5a, Supplementary Fig. S5). The key sterol hydrolases Cyp7a1 (also increased in NOD1 KO CHD-fed mice) and Cyp27a1 were significantly increased in NOD1 KO HFD fed mice, compatible with an increase in BA synthesis in NOD1 KO liver (Fig. 5b), due to the elevated levels of cholesterol under these conditions. The hepatic levels of the Na+/BA-cotransporter Slc10a1 were significantly higher in NOD1 KO vs. WT mice in CHD regimen, suggesting that the enterohepatic circulation of BA into hepatocytes was reduced upon HFD treatment. Regarding Nr1h4, the farnesoid X receptor that counteracts the expression of Cyp7a1, avoiding excess of BA synthesis, only a minimal but statistically significant decrease was observed in NOD1 KO livers under HFD vs. CHD. Furthermore, in vivo NOD1 activation with iE-DAP induced Cyp7a1 in liver as did the HFD, at the time that decreased the transcription of Slc10a1 (Supplementary Fig. S5), suggesting the existence of alternative mechanisms independent of NOD1 and responsible for the elevation in the levels of both genes under HFD. Moreover, increased serum TNFα levels were observed in NOD1 KO mice fed HFD, probably due to the enhanced liver Tnfa expression measured in these animals (Fig. 5c,d). Interestingly enough, an increase, although not statistically significant, in the liver resident inflammatory monocytes was observed in these animals, that was associated with a statistically significant increase in liver macrophages (Fig. 5e). The lipid composition of intestinal mucosa from NOD1 KO mice fed HFD included a higher proportion of saturated fatty acids and a lower amount of monounsaturated fatty acids vs. their WT counterparts (Fig. 5f).
Figure 5.
NOD1 KO mice fed HFD exhibit an enhanced liver pro-inflammatory profile. WT and NOD1 KO mice were maintained under CHD or HFD for 6 weeks. The hepatic mRNA levels of genes involved in bile acid metabolism were determined (a). Quantification of hepatic bile acids (b). The plasma levels of TNF-α (c) and the hepatic mRNA levels of Tnfa (d) were determined. The infiltration of immune cells in liver were determined by flowcytometry (e). The lipid composition of the ileum was determined by 1H MR spectroscopy (f.). Results show the mean ± SEM of 4 (b–e) and 8 (a, f) animals per nutritional condition and genetic background. *P < 0.05; **P < 0.01; ***P < 0.005 vs. the corresponding condition in WT mice. #P < 0.05, ##P < 0.01, ###P < 0.005 vs. the corresponding genetic condition under CHD. ns not statistically significant. Results were expressed as: SFA: saturated fatty acids (FFAA); UFA: Unsaturated FFAA; MUFA: monounsaturated FFAA; PUFA: polyunsaturated FFAA; MCL: medium chain length FFAA; NDB: number of double bonds in FFAA; PU/UFA, polyunsaturated/unsaturated fatty acids; saturation index (Sat. Index), as the ratio between stearic and oleic acids.
Thyroid function is altered in mice lacking NOD1
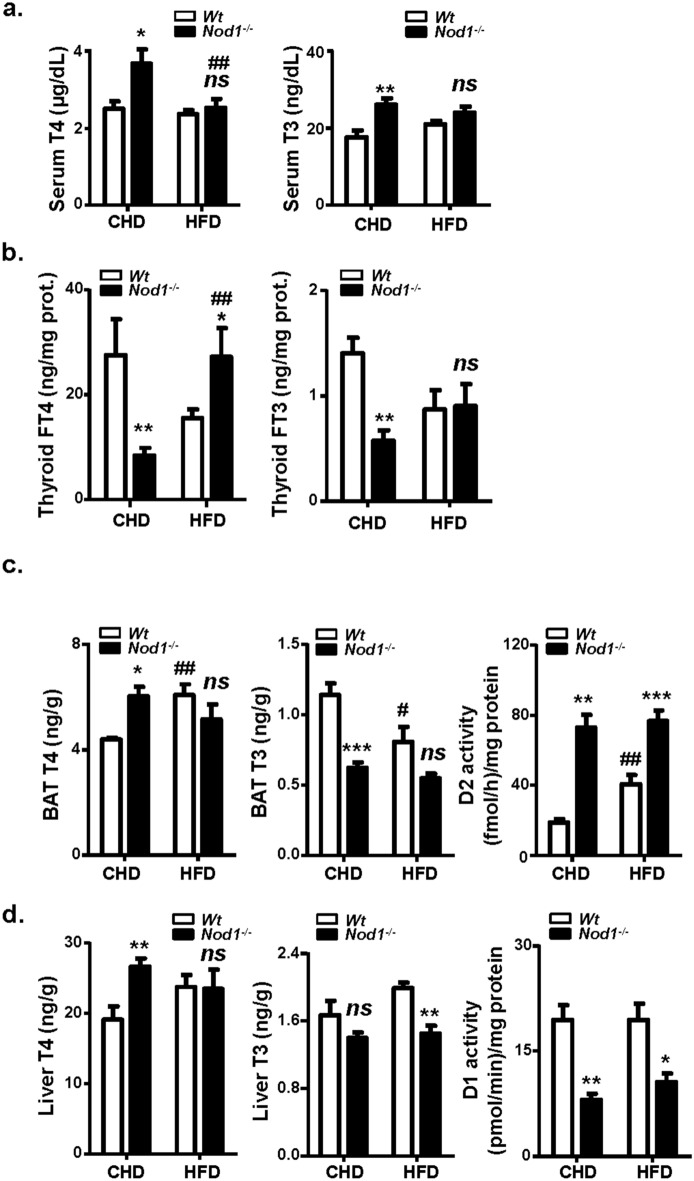
Since thyroid hormones have an impact on tissue homeostasis and lipid metabolism51–53 as well as in energy expenditure44, we investigated their levels in WT and NOD1 KO mice fed CHD or HFD. The serum levels of T4 and T3 were significantly higher in NOD1 KO vs. WT mice under CHD, suggesting increased requirements of T4 and T3 in NOD1 KO mice. However, feeding HFD suppressed the differences between both groups (Fig. 6a). Measurement of free thyroidal T4 and T3 (thyroid FT4 and FT3 ready to be released to serum) in NOD1 KO mice fed CHD showed a strong decrease as compared to the WT, indicating damage in the secretion of T4 and T3 in NOD1 KO mice. However, in HFD, the FT4 is higher in the thyroid of NOD1 KO animals compared with WT counterparts (Fig. 6a,b). Due to the importance of BAT in regulating thermogenesis and energy balance, thyroid hormones levels were assessed in BAT and in liver. We found that T4 concentrations were increased in BAT and liver of NOD1 KO animals fed CHD in parallel to plasma T4, but feeding HFD also suppressed this T4 rise. Regarding T3 levels, absence of NOD1 resulted in equal or low T3 values than in the WT counterparts in BAT and liver, independently of the diet (Fig. 6c,d). However, deficiency in NOD1 results in statistically higher D2 activity in BAT, irrespectively of the nutritional regime of the animals (Fig. 6c) and nine- to ten-fold increases in Dio2 mRNA (Supplementary Fig. S6a), suggesting high activation of BAT machinery to maintain T3 levels in BAT. Of note, that this occurs with a normal or high BAT T4 (low T4 is the main activator of D2 activity), that excludes a role for T4 in the D2 activity and Dio2 mRNA increases, pointing to other activators for D2 deiodinase as BA or fatty acids. Ucp1 mRNA was decreased to 60% in NOD1 mice on HFD, which agrees with the observed obesity under HFD in NOD1 KO mice (Supplementary Fig. S6b). Conversely, D1 activity in liver was significantly lower in NOD1 KO animals, which correlates with the T3 levels measured in this tissue (Fig. 6d), despite to observe similar mRNA levels for Dio1 (Supplementary Fig. S6c).
Figure 6.
Thyroid hormones in plasma, thyroid, BAT and liver; liver D1 and BAT D2 deiodinase activities in WT and NOD1 KO mice fed CHD or HFD. Animals were maintained on CHD or fed HFD for 6 weeks. The serum (a) and free thyroidal concentrations (b) of T4 T3 were determined. The BAT T4, T3 and T4 type 2 deiodinase activity (D2 activity) were determined (c). The liver T4, T3 and type 1 deiodinase activity (D1 activity) were determined (d). Results show the mean ± S.E.M. of 9–15 animals per nutritional condition and genetic background. *P < 0.05; **P < 0.01; ***P < 0.005 vs. the corresponding condition in WT mice. #P < 0.05; ##P < 0.01 vs. the corresponding genetic condition under CHD. ns not statistically significant.
Gut microbiota differences in WT and NOD1 KO mice under different diets
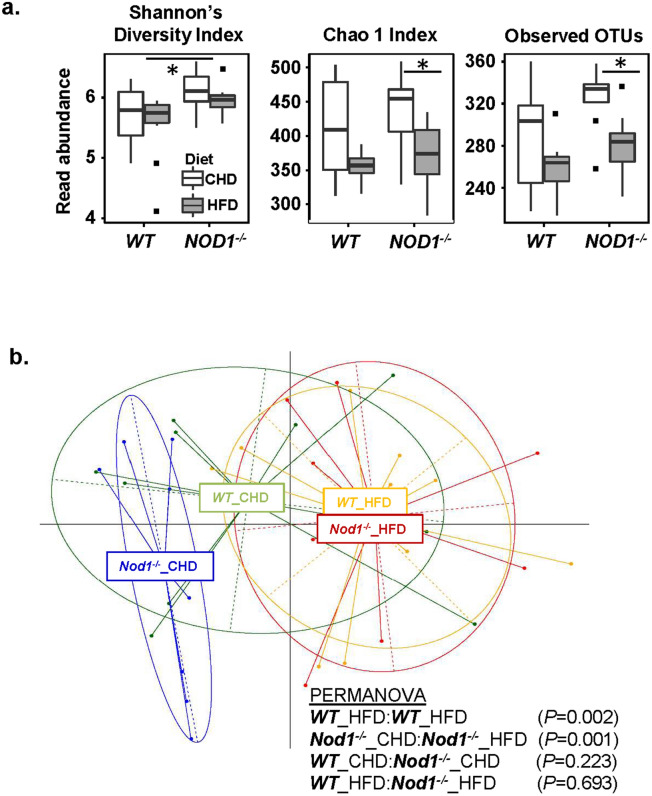
Analyses of the intestinal microbiota were performed in the two experimental groups (NOD1 KO and WT mice) under the CHD and after switching to a HFD. These enabled us to identify differences in microbiota related to both the diet (HFD vs. CHD) and the genotype (NOD1 KO vs. WT). In general, indices of α diversity tended to be reduced by the HFD but the differences varied depending on the genotype (Fig. 7a). As shown in Fig. 7a, the HFD feeding caused decreases in observed OTUs and in the Chao1 index (a measure of species richness) in NOD1 KO mice but not in WT mice, suggesting that HFD exerted stronger adverse effects in the gut microbiota composition of the NOD1 KO. Nonetheless, Shannon’s diversity index (species diversity) was slightly lower in WT than in NOD1 KO under HFD (Fig. 7a). Comparison of β diversity showed that after a HFD, it appears that the microbiota composition changed for both genotype groups when compared to CHD. Analysis using generalized UniFrac (which down-weights abundant or rare taxonomic groups) revealed significant differences between CHD and HFD for WT and NOD1 KO groups, individually. However, no significant differences in β diversity could be attributed to the genotype, when comparing WT vs NOD1 KO groups for CHD and HFD, respectively (Fig. 7b). When differences in specific taxonomic categories were analyzed as a function of the genotype under each specific dietary period, it was observed that the NOD1 KO group was characterized by lower abundances of the family S24-7, as well as the genera Bacteroides and Mucispirillum compared to the WT group under a standard CHD (Supplementary Fig. S7). In contrast, higher abundances of the genera Oscillospira, Prevotella, [Prevotella], Parabacteroides, AF12 (family Rikenellaceae), Rikenella, Dehalobacterium, Helicobacter, Desulfovibrio and rc4_4 (family Peptococcaceae) were observed in the NOD1 KO group compared to the WT under a CHD. After HFD feeding, the NOD1 KO group was characterized by higher abundances of the family Desulfovibrioaceae and the genera Odoribacter, Prevotella, Parabacteroides, [Ruminococcus], Rikenella, Paraprevotella and Helicobacter compared to the WT group. Therefore, four bacterial groups were increased in NOD1 KO mice compared to WT regardless of the diet, including Prevotella, Parabacteroides, Rikenella and Helicobacter. The HFD feeding caused a decrease in several genera (Bifidobacterium, Rikenella, Bilophila, rc4_4 and Enterococcus) and an increase in others (Helicobacter) in both WT and NOD1 KO groups, compared to the groups fed the CHD, revealing the key influence of the diet on the gut microbiota composition above the genetic background. Of these bacterial groups, the initial increased levels of Helicobacter found in NOD1 KO mice could aggravate the effects of the HFD, which also could contribute to their increase. Specific effects of the HFD depending on the genotype were also detected. Indeed, the HFD decreased the abundance of some taxonomic genera (Oscillospira, Coprococcus and Streptococcus) only in the NOD1 KO group. Other taxonomic genera were increased only in the NOD1 KO group (Bacteroides, [Ruminococcus], Dorea) or the WT group (Desulfovibrio, Allobaculum) under HFD feeding. These findings suggest that the genotype influences the susceptibility of the host to diet-induced microbiota changes (Supplementary Fig. S7).
Figure 7.
Characterization of the gut microbiota. (a) Alpha diversity indices (Shannon’s diversity index, Chao1, Observed OTUs) from each diet and genotype treatment group. (b) Principle coordinates analysis (PCoA) plot using generalized UniFrac distances comparing microbial communities from each treatment group. Group means are indicated by the center of each ellipse. Distance-based non-parametric PERMANOVA tests were conducted as pairwise comparisons of treatment groups. P values of all pairwise comparisons were corrected for multiple comparisons using false discovery rate.
Discussion
In this work, we show that under a standard CHD, the absence of NOD1 does not alter lipid metabolism nor promote obesity, as deduced by the presence of normal adipose tissue and complete absence of liver steatosis in these mice. However, HFD favors a rapid and significant development of obesity, or at least overweight, caused by an enlargement of eWAT adipocytes and enhanced hepatic steatosis in NOD1 KO mice, whereas this situation is delayed in the WT counterparts. Blood lipids from NOD1 KO mice fed HFD show an increase in total cholesterol, mainly due to the elevation of the HDL fraction. When animals are analyzed in metabolic cages, the absence of NOD1 results in higher REE values. However, under a HFD regimen, the rise in REE during the darkness period is lesser than in the WT animals, contributing to the increased obesity in NOD1 KO mice vs. the same genotype in CHD. Indeed, the significant decreased levels of Pnpla3 in eWAT suggest a moderate hypothyroidism, as previously described35. In fact, among the targets responsive to thyroid hormones and involved in obesity, Pnpla3 plays a role in the enlargement of eWAT35. However, the situation is more complex in view of the elevation of Atgl levels, a key enzyme in triglyceride lipolysis47, in the absence of NOD1, both under CHD and HFD vs. the corresponding levels in the WT counterparts.
We provide evidence of different mechanisms that could explain this accelerated obesity of NOD1 KO mice fed HFD. According to our data, one of the key players is a significant alteration of thyroid hormone homeostasis at 6 weeks after HFD. Under a CHD, NOD1 deficiency results in higher serum levels of both T3 and T4, accompanied by lower FT4 and FT3 hormones in the thyroid (hormone ready to be released). Thyroidal FT4 and FT3 are strongly decreased in NOD1 KO mice fed CHD, showing a profound effect of thyroid secretion. If similar decreases in NOD1 expression or activity were found in humans, with decreases in thyroidal FT4 and FT3, this would indicate the early involvement of the thyroid in pathological events, such as obesity associated to alterations in this NLR. This thyroid situation is reversed under HFD, which may contribute to the increased obesity of NOD1 deficient mice. The strong response of BAT D2 deiodinase points to higher T3 requirements in NOD1 KO mice. Regarding other alternatives accounting for T3 decreased levels, such as Dio3, it should be mentioned that Dio3 is expressed mainly during fetal life and in proliferative situations, including cells in culture. Dio3 is the main pathway for T3 degradation. It has been reported that Dio3 is not expressed or it is undetectable in murine BAT, thyroid and adult liver. Therefore, thyroid hormone dysfunction appears to be an early event prior to the development of insulin resistance, and related to the gain in body weight mainly through lipid accumulation in fat and liver tissues.
It is known that HFD alters the permeability of the intestinal barrier, and its integrity is compromised by this dietary regimen24,25,54–56. HFD lipids, in combination with bacterial molecules from the gut microbiota (bacterial cell wall components, DNA, etc.) are translocated across this barrier and gain access to systemic circulation and activate pro-inflammatory pathways, not only in immune cells but also in other tissues where NOD1 and NOD2 are expressed11,12,50. Our data suggest that HFD is involved in the rise of circulating NOD1 activatos as deduced by the increase in the phosphorylation of the adaptor RIPK2 in the WT mice. Interestingly, the absence of NOD1 per se has an impact on the gut microbiota composition, which exhibits a higher content in Parabacteroides, Prevotella, Rikenella and Helicobacter regardless of the diet. The differences in microbiota related to the NOD1 deletion seem to influence the subsequent HFD-induced effects on the gut ecosystem, which could contribute to aggravate the immune-metabolic phenotype of the obese mice. In addition to this, NOD1 deficiency in epithelial intestinal cells is likely associated with a higher translocation of gut-derived molecules that contribute to activate immunity.
Exacerbation of the adverse metabolic effects of the HFD in NOD1 KO mice compared to WT mice, including increased adiposity and lipogenesis and changes in ileum lipid composition could be related to dietary-induced alterations in the natural intestinal ecosystem of the NOD1 KO animals. This is characterized by reductions in some diversity indexes and shifts in the abundance of specific bacterial taxonomic groups. A role for dietary-induced changes in the microbiota in dietary lipid absorption and deposition in peripheral tissues has been reported in different study models12,57. Diet-induced increases in the phylum Firmicutes were shown to increased lipid absorption in the intestine through fecal transplantation experiments57. Other studies reported that that lipid absorption could be increased by a microbiota enriched in Clostridiaceae and with reduced abundance of Bacteroidaceae and Bifidobacteriaceae12. In our study, HFD-induced reductions in Bifidobacterium, which could explain increased lipid absorption leading to obesity and adiposity, although these effects were identified in both WT and NOD1 KO and attributed exclusively to the HFD. Specific dietary effects on the intestinal ecosystem of NOD1 KO mice (not detected in WT mice under HF feeding), such as reductions in Oscillospira, Coprococcus or Streptococcus and increases in others (Dorea, [Ruminococcus] or Bacteroides) could account for the stronger adverse impact of the diet on lipid absorption and metabolism than in WT mice. Oscillospira has often been associated with leanness in human microbiota studies58, although opposite associations have also been reported in WT rodents59. This potential benefits could be attributed to the ability of Oscillospira spp. to produce butyrate, which is thought to play positive roles in obesity58. Oscillospira spp. also utilize as a primary energy source host-derived glycans (glucoronate), instead of dietary starches, which will cost energy to the host and could explain its positive association with leanness in humans58. By contrast, increases in Dorea have been positively associated with obesity/adiposity in rodent59,60 and human61 studies.
Specific taxa of the gut microbiota regulate proteins involved in BA metabolism, such as BA transporters, bile salt hydrolase (BSH) and BA 7α-dehydroxylase activities. These are responsible for both BA deconjugation and subsequent dehydroxylation in the distal gut. Their activity could increase the excretion of BAs in feces and reduce their concentrations in the body, leading to induction of BA synthesis from cholesterol in the liver11,12. Therefore, the induction of the expression of Cyp7a1 in the liver of NOD1 KO mice under both the CHD and the HFD could be related to the specific microbiota alterations associated with the mouse genotype and the subsequent effect of the HFD. For example, Parabacteroides spp., which showed higher abundances in NOD1 KO mice, although were not specifically increased by the HFD, produce secondary BAs and, thereby, could contribute to inducing BA synthesis from cholesterol in the liver as well62–64.
Collectively, the low-grade immune response upon HFD is exacerbated in the case of NOD1 deficiency, contributing to dysfunctional lipid synthesis, due to a higher infiltration of pro-inflammatory immune cells in adipose and liver tissues18,20, as confirmed by the quantification of the infiltrated cells and the rise in circulating TNF-α levels. Indeed, experiments using germ-free mice show protection against HFD-induced obesity in NOD1 KO animals, although this phenotype is suppressed when mice are housed under conventional conditions and the gut becomes colonized13,18,65–67. The HFD used promoted a modest increase in body weight of the WT animals (9.8% increase after 6 weeks); however, in the absence of NOD1 the body weight gain was 33.3% during this period, reflecting this obesity in adipose tissue and in the liver. In addition to this, these tissues became more infiltrated by immune cells, such as macrophages and neutrophils, as it has been previously described4,22,24. The idea that lipids and bacterial-derived molecules cooperate in the induction of ‘metabolic inflammation’ and obesity comes from the fact that particularly NOD1 is overexpressed and activated in adipose tissue in response to these challenges10,18,20,23. However, our data provide a different scenario since constitutive deficiency of NOD1 worsens the metabolic dysfunction due to HFD. In fact, NOD1 polymorphisms in humans -associated with differences in the activity of this NLR- appear to play a relevant role in inflammatory-related diseases, including obesity23,68,69.
Regarding insulin resistance and NLR activity, other groups have described that NOD1 activation after iE-DAP administration promotes insulin resistance20,23,24,70. However, in animals lacking NOD1 in the hematopoietic system, Chan et al.18 reported that after 18 weeks of HFD, the WT animals exhibited hyperinsulinemia, insulin resistance and a reduced capacity to decrease glucose overload, a situation that was significantly more benign in the hematopoietic NOD1 deficient mice. Here we provide a new piece of knowledge since our animals deficient in NOD1 and fed a HFD for 6 weeks, despite an evident obesity, are not resistant to insulin and exhibit an improved glucose tolerance test when compared with the WT counterparts. These results suggest that additional dysfunctional changes are required to establish insulin resistance and defective oral glucose tolerance test, compatible with diabetes progression in WT animals. Moreover, reversion to CHD after 6 weeks of HFD shows a significant recovery in the liver steatosis of the NOD1 KO animals and almost complete absence of Oil-red O staining in the WT counterparts, which is in agreement with the failure of the establishment of the insulin-resistance phenotype in these 6 weeks of HFD regimen.
Finally, although less attention has been paid to thyroid metabolism associated with microbial composition it remains a relevant pathophysiological issue as observed from our results in the NOD1 KO mice, and discussed in a recent review71. Indeed, our data suggest an interplay between NOD1 activity, microbiota and thyroid hormone function that might help to unravel unexpected obesity-related pathologies in humans.
Supplementary information
Acknowledgements
This work was supported by Grants SAF2017-82436R, AGL2017-88801-P and SAF2016-75004R from MINECO/AEI/FEDER/EU, S2017/BMD-3686 from Comunidad de Madrid, CIVP18A3864 from Fundación Ramón Areces and CIBERCV and CIBERHED (funded by the Instituto de Salud Carlos III) and Fondos FEDER.
Abbreviations
- HFD
High fat diets
- JNK
C-Jun N-terminal kinase
- KO
Knockout
- MAPK
Mitogen-activated protein kinase
- q(RT)-PCR
Quantitative real-time (RT) polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- WT
Wild-type
Author contributions
S.G.-R. designed the study, performed experiments, analyzed data and contributed to writing the paper. M.P.-G., V.F.-G., K.J.P. and E.F. performed experiments and analyzed data; M.P.-G. contributed to designing the study. M.J.O. performed experiments, analyzed data and provided intellectual input. Y.S. provided intellectual input on microbiota studies and revised the manuscript. A.C. and P.M.-S. provided intellectual input and revised the manuscript. L.B. designed the study, provided funding and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Silvia González-Ramos, Marta Paz-García and Victoria Fernández-García.
Contributor Information
Silvia González-Ramos, Email: sgramos@iib.uam.es.
Lisardo Boscá, Email: lbosca@iib.uam.es.
Supplementary information
is available for this paper at 10.1038/s41598-020-69295-2.
References
- 1.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 3.Park JH, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 4.He S, Wang X. RIP kinases as modulators of inflammation and immunity. Nat. Immunol. 2018;19:912–922. doi: 10.1038/s41590-018-0188-x. [DOI] [PubMed] [Google Scholar]
- 5.Keestra-Gounder AM, Tsolis RM. NOD1 and NOD2: Beyond peptidoglycan sensing. Trends Immunol. 2017;38:758–767. doi: 10.1016/j.it.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human-Protein-Atlas. https://www.proteinatlas.org/ENSG00000106100-NOD1/tissue.
- 7.Gonzalez-Ramos S, et al. Endothelial NOD1 directs myeloid cell recruitment in atherosclerosis through VCAM-1. Faseb J. 2019;33:9. doi: 10.1096/fj.201801231RR. [DOI] [PubMed] [Google Scholar]
- 8.Moreno L, Gatheral T. Therapeutic targeting of NOD1 receptors. Br. J. Pharmacol. 2013;170:475–485. doi: 10.1111/bph.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubino SJ, Selvanantham T, Girardin SE, Philpott DJ. Nod-like receptors in the control of intestinal inflammation. Curr. Opin. Immunol. 2012;24:398–404. doi: 10.1016/j.coi.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Stroh T, et al. Nucleotide oligomerization domains 1 and 2: Regulation of expression and function in preadipocytes. J. Immunol. 2008;181:3620–3627. doi: 10.4049/jimmunol.181.5.3620. [DOI] [PubMed] [Google Scholar]
- 11.Chevre R, Silvestre-Roig C, Soehnlein O. Nutritional modulation of innate immunity: The fat-bile-gut connection. Trends Endocrinol. Metab. 2018;29:686–698. doi: 10.1016/j.tem.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Guryn K, et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469.e455. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadaleta RM, Cariello M, Sabba C, Moschetta A. Tissue-specific actions of FXR in metabolism and cancer. Biochim. Biophys. Acta. 2015;1851:30–39. doi: 10.1016/j.bbalip.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan Y, Seregin SS, Chen J, Chen GY. Nod1 limits colitis-associated tumorigenesis by regulating IFN-gamma production. J. Immunol. 2016;196:5121–5129. doi: 10.4049/jimmunol.1501822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverio R, et al. Lipases and lipid droplet-associated protein expression in subcutaneous white adipose tissue of cachectic patients with cancer. Lipids Health Dis. 2017;16:159. doi: 10.1186/s12944-017-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KL, et al. Circulating NOD1 activators and hematopoietic NOD1 contribute to metabolic inflammation and insulin resistance. Cell Rep. 2017;18:2415–2426. doi: 10.1016/j.celrep.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 19.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou YJ, Zhou H, Li Y, Song YL. NOD1 activation induces innate immune responses and insulin resistance in human adipocytes. Diabetes Metab. 2012;38:538–543. doi: 10.1016/j.diabet.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Roy D, et al. Coordinated transcriptional control of adipocyte triglyceride lipase (Atgl) by transcription factors Sp1 and peroxisome proliferator-activated receptor gamma (PPARgamma) during adipocyte differentiation. J. Biol. Chem. 2017;292:14827–14835. doi: 10.1074/jbc.M117.783043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma A, et al. Nod1-mediated lipolysis promotes diacylglycerol accumulation and successive inflammation via PKCdelta-IRAK axis in adipocytes. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1865:136–146. doi: 10.1016/j.bbadis.2018.10.036. [DOI] [PubMed] [Google Scholar]
- 23.Zhou YJ, et al. Increased NOD1, but not NOD2, activity in subcutaneous adipose tissue from patients with metabolic syndrome. Obesity (Silver Spring) 2015;23:1394–1400. doi: 10.1002/oby.21113. [DOI] [PubMed] [Google Scholar]
- 24.Schertzer JD, et al. NOD1 activators link innate immunity to insulin resistance. Diabetes. 2011;60:2206–2215. doi: 10.2337/db11-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 26.Delgado C, et al. NOD1, a new player in cardiac function and calcium handling. Cardiovasc. Res. 2015;106:375–386. doi: 10.1093/cvr/cvv118. [DOI] [PubMed] [Google Scholar]
- 27.Val-Blasco A, et al. Role of NOD1 in heart failure progression via regulation of Ca(2+) handling. J. Am. Coll. Cardiol. 2017;69:423–433. doi: 10.1016/j.jacc.2016.10.073. [DOI] [PubMed] [Google Scholar]
- 28.Mojena M, et al. Protection against gamma-radiation injury by protein tyrosine phosphatase 1B. Redox Biol. 2018;17:213–223. doi: 10.1016/j.redox.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouhid L, et al. Yarrow supercritical extract ameliorates the metabolic stress in a model of obesity induced by high-fat diet. Nutrients. 2019;12:e72. doi: 10.3390/nu12010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prieto P, et al. Interplay between post-translational cyclooxygenase-2 modifications and the metabolic and proteomic profile in a colorectal cancer cohort. World J. Gastroenterol. 2019;25:433–446. doi: 10.3748/wjg.v25.i4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr JS, Kennedy AJ, Hasty AH, et al. Isolation of adipose tissue immune cells. J. Vis. Exp. 2013;75:e50707–614. doi: 10.3791/50707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Prados JC, et al. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 33.Morreale de Escobar G, Pastor R, Obregon MJ, Escobar del Rey F. Effects of maternal hypothyroidism on the weight and thyroid hormone content of rat embryonic tissues, before and after onset of fetal thyroid function. Endocrinology. 1985;117:1890–1900. doi: 10.1210/endo-117-5-1890. [DOI] [PubMed] [Google Scholar]
- 34.Obregon MJ, et al. Thyroid hormones and 5'-deiodinase in rat brown adipose tissue during fetal life. Am. J. Physiol. 1989;257:E625–631. doi: 10.1152/ajpendo.1989.257.5.E625. [DOI] [PubMed] [Google Scholar]
- 35.Calvo RM, Obregon MJ. Tri-iodothyronine upregulates adiponutrin mRNA expression in rat and human adipocytes. Mol. Cell Endocrinol. 2009;311:39–46. doi: 10.1016/j.mce.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz de Ona C, Morreale de Escobar G, Calvo R, Escobar del Rey F, Obregon MJ. Thyroid hormones and 5'-deiodinase in the rat fetus late in gestation: Effects of maternal hypothyroidism. Endocrinology. 1991;128:422–432. doi: 10.1210/endo-128-1-422. [DOI] [PubMed] [Google Scholar]
- 37.Herlemann DP, et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segata N, Huttenhower C. Toward an efficient method of identifying core genes for evolutionary and functional microbial phylogenies. PLoS ONE. 2011;6:e24704. doi: 10.1371/journal.pone.0024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell CS, et al. Resistance to thyroid hormone is associated with raised energy expenditure, muscle mitochondrial uncoupling, and hyperphagia. J. Clin. Investig. 2010;120:1345–1354. doi: 10.1172/JCI38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chamoun Z, Vacca F, Parton RG, Gruenberg J. PNPLA3/adiponutrin functions in lipid droplet formation. Biol. Cell. 2013;105:219–233. doi: 10.1111/boc.201200036. [DOI] [PubMed] [Google Scholar]
- 46.Kumari M, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab. 2012;15:691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweiger M, et al. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 48.Watt MJ, et al. Regulation of HSL serine phosphorylation in skeletal muscle and adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2006;290:E500–508. doi: 10.1152/ajpendo.00361.2005. [DOI] [PubMed] [Google Scholar]
- 49.Leporq B, Lambert SA, Ronot M, Vilgrain V, Van Beers BE. Quantification of the triglyceride fatty acid composition with 3.0 T MRI. NMR Biomed. 2014;27:1211–1221. doi: 10.1002/nbm.3175. [DOI] [PubMed] [Google Scholar]
- 50.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 51.Mullur R, Liu YY, Brent GA. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014;94:355–382. doi: 10.1152/physrev.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obregon MJ. Adipose tissues and thyroid hormones. Front. Physiol. 2014;5:479. doi: 10.3389/fphys.2014.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinha RA, Singh BK, Yen PM. Thyroid hormone regulation of hepatic lipid and carbohydrate metabolism. Trends Endocrinol. Metab. 2014;25:538–545. doi: 10.1016/j.tem.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Kawano M, Miyoshi M, Ogawa A, Sakai F, Kadooka Y. Lactobacillus gasseri SBT2055 inhibits adipose tissue inflammation and intestinal permeability in mice fed a high-fat diet. J. Nutr. Sci. 2016;5:e23. doi: 10.1017/jns.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 56.Luck H, et al. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab. 2015;21:527–542. doi: 10.1016/j.cmet.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Semova I, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gophna U, Konikoff T, Nielsen HB. Oscillospira and related bacteria: From metagenomic species to metabolic features. Environ. Microbiol. 2017;19:835–841. doi: 10.1111/1462-2920.13658. [DOI] [PubMed] [Google Scholar]
- 59.Jiao N, et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: A meta-analysis. Physiol. Genom. 2018;50:244–254. doi: 10.1152/physiolgenomics.00114.2017. [DOI] [PubMed] [Google Scholar]
- 60.Mulders RJ, et al. Microbiota in obesity: Interactions with enteroendocrine, immune and central nervous systems. Obes. Rev. 2018;19:435–451. doi: 10.1111/obr.12661. [DOI] [PubMed] [Google Scholar]
- 61.Karvonen AM, et al. Gut microbiota and overweight in 3-year old children. Int. J. Obes. (Lond.) 2019;43:713–723. doi: 10.1038/s41366-018-0290-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 2015;11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu Y, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat. Commun. 2017;8:1785. doi: 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26:222–235.e225. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 65.Rabot S, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez-Nunez I, et al. Nod2 and Nod2-regulated microbiota protect BALB/c mice from diet-induced obesity and metabolic dysfunction. Sci. Rep. 2017;7:548. doi: 10.1038/s41598-017-00484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suarez-Zamorano N, et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015;21:1497–1501. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feerick CL, McKernan DP. Understanding the regulation of pattern recognition receptors in inflammatory diseases: A 'Nod' in the right direction. Immunology. 2017;150:237–247. doi: 10.1111/imm.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cuda C, Badawi A, Karmali M, El-Sohemy A. Effects of polymorphisms in nucleotide-binding oligomerization domains 1 and 2 on biomarkers of the metabolic syndrome and type II diabetes. Genes Nutr. 2012;7:427–435. doi: 10.1007/s12263-012-0287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prajapati B, Jena PK, Rajput P, Purandhar K, Seshadri S. Understanding and modulating the Toll like Receptors (TLRs) and NOD like Receptors (NLRs) cross talk in type 2 diabetes. Curr. Diabetes Rev. 2014;10:190–200. doi: 10.2174/1573399810666140515112609. [DOI] [PubMed] [Google Scholar]
- 71.Virili C, Centanni M. "With a little help from my friends": The role of microbiota in thyroid hormone metabolism and enterohepatic recycling. Mol. Cell Endocrinol. 2017;458:39–43. doi: 10.1016/j.mce.2017.01.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.