Dear Editor,
COVID-19, caused by the SARS-CoV-2 virus, has now spread worldwide with catastrophic human and economic impacts and currently has infected over 10 million people and killed over 500,0001. In an effort to mitigate disease symptoms and impede viral spread, efforts in vaccine development and drug discovery are being conducted at a rapid pace2. Recently, we showed that the well-known anticoagulant heparin has exceptional binding affinity to the spike protein (S-protein) of SARS-CoV-23. The S-protein of SARS-CoV-2 bound more tightly to immobilized heparin (KD = ~10−11 M) than the S-proteins of either SARS-CoV (KD = ~10−7 M) or MERS-CoV (KD = ~10-9 M). However, it is not known whether the tight binding of heparin to the SARS-CoV-2 S-protein translates into potent antiviral activity. In the current study, we evaluated the in vitro antiviral properties of heparin and other closely related polysaccharides to assess the relevance of heparin-related GAGs and other sulfated polysaccharides as part of the pharmacopeia of potential therapeutics that target SARS-CoV-2. Vero-CCL81, which expresses both ACE2 and TMPRSS24, were used for viral replication at high titer5 for use in antiviral assays.
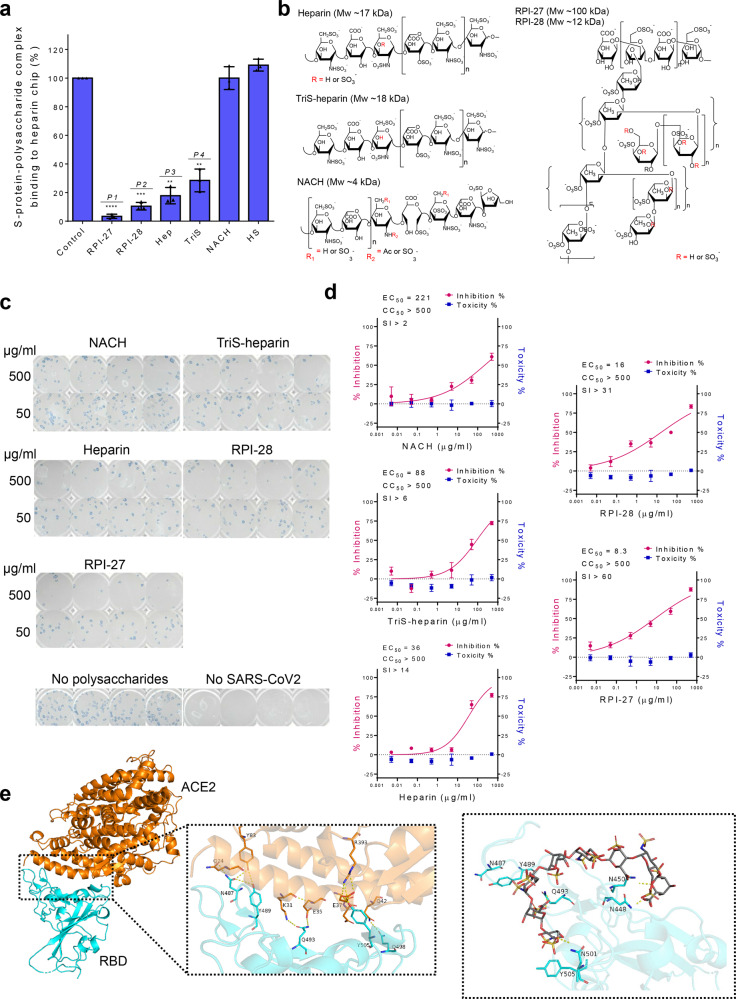
Heparin, heparan sulfates, other glycosaminoglycans (GAGs)3, and fucoidan and other highly sulfated polysaccharides were screened using surface plasmon resonance (SPR) to measure binding affinity to the SARS-CoV-2 S-protein (Fig. 1a). Briefly, solution competition studies between surface immobilized heparin and other sulfated polysaccharides were evaluated by injecting SARS-CoV-2 S-protein (50 nM) alone or mixed with 1 µM of an indicated polysaccharide in SPR buffer at a flow rate of 30 μL/min. After each run, dissociation and regeneration were performed. For each set of competition experiments, a control experiment (S-protein without polysaccharide) was performed to ensure the surface was fully regenerated. Among the tested polysaccharides, RPI-27 and RPI-28, complex sulfated polysaccharides (fucoidans) extracted from the seaweed Saccharina japonica6, chemo-enzymatically synthesized trisulfated (TriS) heparin7, and unfractionated USP-heparin itself were able to compete with heparin for S-protein binding. We selected these compounds along with a non-anticoagulant low molecular weight heparin (NACH)8 for further study (Fig. 1b). The other GAGs including heparan sulfate, the chondroitin sulfates, and keratan sulfate show no competitive binding when compared to the control.
Fig. 1. Assessment of antiviral activities of certain sulfated polysaccharides.
a Surface plasmon resonance (SPR) experiments were used to screen polysaccharides that outcompete immobilized heparin binding to SARS-CoV-2 S-protein. Data are presented as mean±s.d., n=3 biologically independent samples. A two-sided t-test was performed to test significance against the control (P1<0.0001, P2=0.0003, P3=0.0016, P4=0.0041). b Structural units comprising polysaccharides used for in vitro antiviral studies. c Focus reduction assay images of virus infection on treatment of indicated polysaccharides. At 48h after infection, Vero cells were fixed and probed with SARS-CoV-2 spike primary antibody (1:10000, Sino Bio Inc.) and HRP-conjugated goat rabbit (1:10000, Abcam) secondary antibody. d Vero cells were infected with SARS-CoV-2 at a MOI of 2.5×10−3 at different doses of each polysaccharide for 48h. The viral yield was quantified using a focus reduction assay. Cytotoxicity in Vero cells was measured using a WST-1 assay. The left and right y-axis of the graphs represent mean % inhibition of virus yield and cytotoxicity of the polysaccharides, respectively. Cytotoxicity experiments were performed in duplicate with n=3 biologically independent samples. Focus reduction assay experiments were performed in mean±s.d. (quadruplicate measurements) with n=3 biologically independent samples. e The RBD-ACE2-binding interface is stabilized by an extensive hydrogen bonding network involving sidechains of several residues on both RBD and ACE2. Polar sidechains of N487, Y489, Q493, Q498, and Y505 on the spike protein RBD along with other residues would be able to bind to heparin and inhibit RBD-ACE2 interaction. Heparin (here an octasaccharide) forms a hydrogen bond network with N448, N450, Q493, and N501 that aids in its occupancy of this binding regions and sterically restrict access to Q498, Y489, and Y505 necessary for ACE2 receptor binding.
Standard assays were performed to quantify potential cytotoxicity and antiviral activity. Cytotoxicity determination of the polysaccharides was performed using Vero cells and the standard water-soluble tetrazolium salt-1 (WST-1) assay (Takara Bio Inc., Japan). None of the tested polysaccharides showed toxicity even at the highest concentrations tested. Vero cells were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 2.5 × 10−3 with varying dosages of polysaccharide to confirm antiviral activity. A focus reduction assay was performed 48 h post infection to determine efficacy. Antiviral activities correlated with the SPR results. The most potent compound tested, RPI-27, is a high molecular weight, branched polysaccharide related to the known compound fucoidan, and had an EC50 of 8.3 ± 4.6 μg/mL, which corresponds to ~83 nM (Fig. 1c, d and Supplementary Table S1). This is substantially more potent than remdesivir having a reported in vitro EC50 value of 770 nM in Vero-E6 cells9 and 11.4 µM in Vero-CCL81 cells10, currently approved for emergency use for severe COVID-19 infections. The smaller RPI-28 has the same basic structure as RPI-27 but a lower molecular weight and, thus, a lower activity (EC50 = 1.2 μM, Supplementary Table S1). Heparin and the TriS-heparin (an intermediate in the bioengineered heparin synthesis pathway3) also have potent antiviral activity with EC50 values of ~2.1 and 5.0 μM, while the lower molecular weight NACH had an approx. EC50 of 55 μM. Similar antiviral activity of heparin has also been demonstrated recently11. Heparin and TriS-heparin are similar, with the latter devoid of the relatively small fraction of 3-O-sulfate groups present on heparin (Fig. 1b). Thus, their similar activity is expected. However, the low molecular weight NACH had far lower antiviral activity. Less sulfated GAGs, such as heparan sulfate and various chondroitin sulfates, because of their very low S-protein binding were not tested in the antiviral assay.
The high activity of RPI-27 and RPI-28 relative to the other polysaccharides tested may be a result of multivalent interactions between the polysaccharide and viral particle12. While heparin, TriS-heparin, and NACH are linear polysaccharides, RPI-27 and RPI-28 are both highly branched (Fig. 1b), possibly conferring added points of interaction in 3-dimensional space. The higher affinity of RPI-27 compared to RPI-28, and hence its more potent antiviral activity, may be due to the far higher molecular weight of the former providing greater opportunity for multipoint binding to the S-protein of SARS-CoV-2. The non-anticoagulant TriS-heparin may be more desirable in some applications than the potent anticoagulant heparin.
Our results reveal that specific sulfated polysaccharides bind tightly to the S-protein of SARS-CoV-2 in vitro, which suggests that they can act as decoys to interfere with S-protein binding to the heparan sulfate co-receptor in host tissues3,11, inhibiting viral infection. To model this, we constructed a docking model between heparin and the S-protein receptor-binding site (RBD) using the crystal structure of the chimeric RBD-ACE2 complex (PDB ID: 6VW1)13 (Fig. 1e and detailed docking model described in supplementary information). The RBD’s amino acid residues involved in binding the ACE2 (angiotensin-converting enzyme 2) receptor also participated in heparin binding, suggesting a mechanism of viral entry inhibition by heparin. Moreover, the larger the oligosaccharide model used in docking studies, the tighter the binding. Specifically, the octasaccharide binds tighter than the tetrasaccharide (–7.3 vs. –6.1 kcal/mol).
Since these polysaccharides show promising antiviral activity in vitro and low cytotoxicity, we suggest that they may have promising clinical use. Along these lines, SARS-CoV-2 has been found to infect a wide range of tissues that possess sufficient ACE2 levels14, including the nose and the gastrointestinal tract15. Potential routes of delivery of these non-anticoagulant polysaccharide candidates, including the fucoidans (RPI-27, and RPI-28) and the TriS-heparin, could be through a nasal spray, metered dose inhaler, or oral delivery. This is distinct from remdesivir, which must be delivered intravenously16. Indeed, when taken orally, the fucoidans, isolated from edible sulfated seaweed polysaccharides, are considered as “Generally Recognized as Safe” and heparin, an approved drug, is not orally bioavailable. Interestingly, a retrospective clinical study suggests that the administration of anticoagulants, such as heparin, may provide better outcomes for patients hospitalized with COVID-19, including a dramatic reduction in mortality of intubated patients17. It is unknown whether this is a result of heparin’s anticoagulation alone, or to some degree is an effect of its anti-SARS-CoV-2 activity. Inhaled heparin has additional benefits such as reducing pulmonary coagulopathy and inflammation without producing systemic bleeding18. To this end, we suggest that treatment of fucoidans, nebulized heparin, or possibly TriS-heparin in combination with or without current antiviral therapies, should be assessed first in human primary epithelial cells and then in human patients suffering from COVID-19.
Supplementary information
Supplementary information, Materials and Table
Acknowledgements
This work was supported by the Korea Research Institute of Bioscience and Biotechnology Research Initiative Program (KGM4572013) to J.J.H and H.O., and the Global Research Laboratory Program through the National Research Foundation of Korea (2014K1A1A2043032) to J.S.D. and S.-J.K.
Author contributions
S.-J.K., R.J.L., J.S.D, and J.J.H. conceived and designed the experiments. P.S.K, H.O., W.J., and F.Z. performed multiple experiments. K.F. performed the heparin docking model. P.S.K., H.O., F.Z., J.J.H., and S.-J.K. analyzed the data. P.S.K., S.-J.K., J.S.D., and R.J.L. led the manuscript preparation with contributions from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Paul S. Kwon, Hanseul Oh, Seok-Joon Kwon
Contributor Information
Jung Joo Hong, Email: hong75@kribb.re.kr.
Robert J. Linhardt, Email: linhar@rpi.edu
Jonathan S. Dordick, Email: dordick@rpi.edu
Supplementary information
Supplementary Information accompanies the paper at (10.1038/s41421-020-00192-8).
References
- 1.World Health Organization. Coronavirus disease (COVID-199) situation reports. WHO. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (2020).
- 2.Wu D, et al. Vaccines and therapies in development for SARS-CoV-2 infections. J. Clin. Med. 2020;9:E1885. doi: 10.3390/jcm9061885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, S. Y. et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res.181, 104873. 10.1016/j.antiviral.2020.104873 (2020). [DOI] [PMC free article] [PubMed]
- 4.Matsuyama S, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harcourt J, et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin W, Wang J, Ren S, Song N, Zhang Q. Structural analysis of a heteropolysaccharide from Saccharina japonica by electrospray mass spectrometry in tandem with collision-induced dissociation tandem mass spectrometry (ESI-CID-MS/MS) Mar. Drugs. 2012;10:2138–2152. doi: 10.3390/md10102138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cress BF, et al. Heavy heparin: a stable isotope-enriched, chemoenzymatically-synthesized, poly-component drug. Angew. Chem. Int. Ed. Engl. 2019;58:5962–5966. doi: 10.1002/anie.201900768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YP, et al. Non-anticoagulant heparin as a pre-exposure prophylaxis prevents lyme disease infection. ACS Infect. Dis. 2020;6:503–514. doi: 10.1021/acsinfecdis.9b00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon S, et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64:e00819–e00820. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mycroft-West, C. J. et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. BioRxiv10.1101/2020.04.28.066761 (2020). [DOI] [PMC free article] [PubMed]
- 12.Nemanichvili N, et al. Fluorescent trimeric hemagglutinins reveal multivalent receptor binding properties. J. Mol. Biol. 2019;431:842–856. doi: 10.1016/j.jmb.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sungnak W, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grein J, et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paranjpe I, et al. Association of treatment dose anticoagulation with in-hospital survival among hospitalized patients with COVID-19. J. Am. Coll. Cardiol. 2020;76:122–124. doi: 10.1016/j.jacc.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chimenti L, et al. Nebulized heparin attenuates pulmonary coagulopathy and inflammation through alveolar macrophages in a rat model of acute lung injury. Thromb. Haemost. 2017;117:2125–2134. doi: 10.1160/TH17-05-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Materials and Table