Abstract
Renal cell carcinoma (RCC) is one of most common cancers with gradually increasing incidence and high mortality. Chromogenic RCC (chRCC) is the third most common histological subtype of RCC, accounting for approximately 5–7% of RCC. In our study, the transcriptome expression profile data (n=89) of chRCC, corresponding clinical data (n=113) and the somatic mutation data (n=66) were obtained from the TCGA database. We first analyzed the mutation data of chRCC patients and divided chRCC patients into high and low tumor mutation burden (TMB) groups based on the median TMB. We found that high TMB was significantly associated with worse prognosis and could promote tumor metastasis and development. Moreover, four different immune-related genes (BIRC5, PDGFRL, INHBE, IL20RB) were also identified. We found that BIRC5 was significantly overexpressed in the high TMB group and correlated with worse prognosis. The results of univariate and multivariate COX analyses demonstrated that BIRC5 (hazard ratio (HR) = 2.094) may serve as a prognostic indicator for patients with chRCC with high TMB. In addition, we identified the possible functional pathways of BIRC5 through gene set enrichment analysis (GSEA) enrichment. A positive correlation was obtained between BIRC5 and the abundance of CD4+ T cells. The results of our study revealed their correlation between the immune-related genes and clinicopathologic features as well as potential functional pathways as well as immune infiltrating cells, which may provide more data about the development of chRCC immunotherapy.
Keywords: chromophobe renal cell carcinoma, immunotherapy, TCGA, tumor mutation burden
Introduction
Kidney cancer is well known as the third most common malignant tumor in the urinary system after prostate cancer and bladder cancer [1–3]. According to the literature, the globally estimated new cases and deaths of kidney cancer were 403262 and 175098 in 2018, respectively [4]. The prognosis of RCC is poor, with the overall survival of stage IV RCC patients to be 10–15 months. Approximately 90% of kidney cancers are renal cell carcinoma (RCC), including clear cell RCC (ccRCC), chromogenic RCC (chRCC) and papillary RCC (pRCC). chRCC is the third most common histological subtype of RCC, accounting for approximately 5–7% of RCC [5–7]. There are many treatments for chRCC, but immunotherapy is considered to be the most promising and immune checkpoint inhibitor proven useful for RCC [8,9]. However, the role of immunotherapy in chRCC is far from fully clarified.
Increasing evidences revealed that immunotherapy is the most effective way to treat advanced or aggressive cancer [10–13]. The blockade of these immune checkpoints has translated into effective strategies for cancer immunotherapy. Immune checkpoints have achieved great success in suppressing lung cancer, breast cancer and melanoma [14–17]. Tumor mutation burden (TMB) is referred to the number of mutations that exist within a megabase of genomic territory [18]. Previous studies suggest that the TMB is closely related to the efficacy of immunotherapy in most cancer types [19–22]. Moreover, high TMB predicts immunotherapy benefit and TMB can predict survival after immunotherapy across multiple cancer types [20,23]. However, only approximately 20% of cancer patients can benefit from immunotherapy [24]. What is worse is that few related studies have focused on primary chRCC in the TMB subgroup. These sobering data illustrate a critical need to determine the immunotherapy response mechanism for primary chRCC.
In the present study, we analyzed significantly different immune-related genes in the high and low TMB subgroups, explored the prognostic role of immune-related genes in chRCC and its potential correlation with immune-infiltrating cells. The results of our study may provide sufficient information for patient prognosis prediction and additional choice for the immunotherapy of primary chRCC.
Materials and methods
Datasets
The primary chRCC transcriptome expression profile (n=89) and corresponding clinical data (n=113) including age, gender, tumor grade, pathological stage, etc. were downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/). Somatic mutation data (n=66) were also obtained from the TCGA database with data type of ‘Masked Somatic Mutation’. Meanwhile, we obtained a list of the immune-related genes from the ImmPort database (https://www.immport.org/) [25].
Calculation of TMB scores and prognostic analysis
TMB is defined as the total number of mutations per megabase in tumor tissue. ‘maftools’ R package was used to calculate the mutations of each sample in chRCC, and all the chRCC samples were divided into low and high TMB groups based on median data. It was combined with clinical information to analyze the relationship between TMB and Stage and tumor metastasis. In addition, we used the ‘survival’ R package to analyze the relationship between high/low TMB groups and prognosis.
Identification of differentially expressed genes and immune-related genes
We used the ‘Limma’ R package to identify differentially expressed genes in the high and low TMB groups with a fold change (FC) of 2 and P-value of 0.05. And the result was visualized with the ‘pheatmap’ package. Compared with a list of immune-related genes from the immunology database (Immport), we determine the immune-related genes from all differentially expressed genes.
Functional enrichment analysis
In order to identify potential functions and approaches of differentially expressed genes, enrichment analysis including GO (Gene Ontology) function and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analyses were performed using ‘clusterProfiler’ R package.
Analysis of immune-related genes in high and low TMB groups
The immune-related genes were selected to further evaluate the prognostic value of differential immune-related genes in patients with low and high TMB levels. In addition, we compared the expression of four immune-related genes in the high and low TMB groups through the ‘beeswarm’ package, and then further evaluated their prognostic value using univariate and multivariate factor Cox regress analyses.
Gene Set Enrichment Analysis
We performed a Gene Set Enrichment Analysis (GSEA) analysis using the GSEA_4.0.3 software in order to further understand the BIRC5-related pathways. We divided the patients in the TCGA cohort into two groups based on TMB score for GSEA of BIRC5 and selected the c2.cp.kegg.v7.0.symbols.gmt gene set as the reference gene set, with a nominal P-value of <5% as a standard [26].
Statistical analysis
All analyses were performed using R software (version 3.6.2). The Cox regression analysis was performed based on the ‘survival’ R package. The ‘Limma’ package was mainly used for the analysis of differences. A P-value <0.05 was considered significant.
Results
The landscape of mutation profiles in chRCC
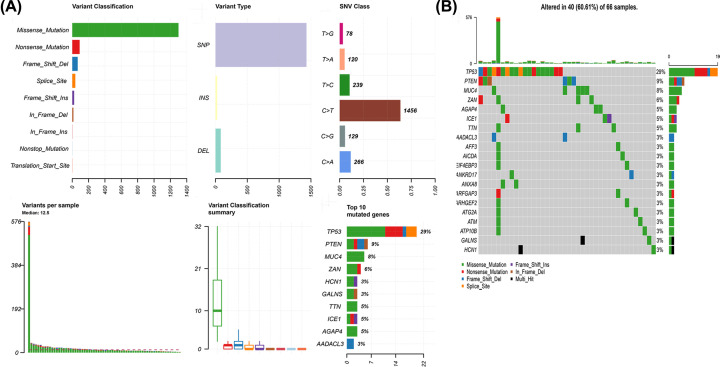
Obtained from the TCGA database, somatic mutation data (n=66) were analyzed with R, and the results were visualized with ‘maftools’ package. We further classified these mutations according to different categories. As shown in Figure 1A, missense mutation was the most common type of variant classification, and single nucleotide polymorphisms occurred more frequently than insertions or deletions. C > T was the most common single-nucleotide variation (SNV) in chRCC (Figure 1A). Then we calculated the number of base changes in each sample, with different colors representing different mutation types (Figure 1A). The waterfall chart revealed the top 20 mutant genes of the mutation profile in chRCC, including TP53 (29%), PTEN (9%), MUC4 (8%), ZAN (6%), HCN1 (3%), TTN (5%), ICE1 (5%), AGAP4 (5%), and AADACL3 (3%) et al. (Figure 1B).
Figure 1. TCGA chRCC mutation cohort.
(A) Overview of TCGA chRCC cohort mutations. (B) Waterfall of the top 20 mutated genes in the TCGA chRCC cohort.
Correlation of TMB with prognosis, clinicopathological characteristics, and tumor grades of chRCC patients, and functional enrichment analysis
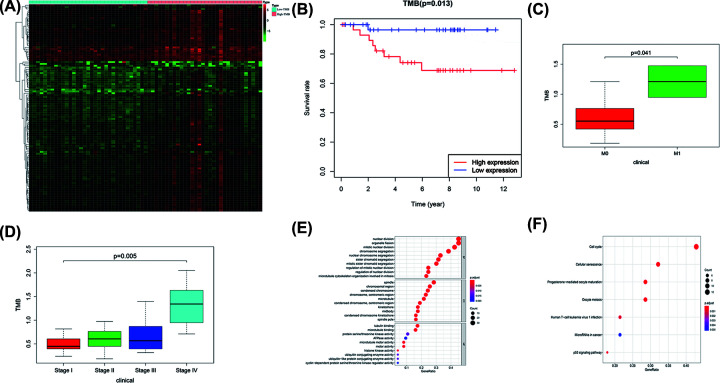
Based on the median TMB, we divided chRCC into high TMB group (n=32) and low TMB group (n=33), and analyzed its gene expression profile to identify DEGs with a false discovery rate (FDR) of 0.05 and an FC of 1.5, and the result was visualized with a heatmap (Figure 2A). Kaplan–Meier survival analysis revealed that patients with high TMB were associated with worse prognosis (P=0.013, Figure 2B). Next, we analyzed the relationship between TMB and tumor metastasis, and the results showed that high TMB may promote tumor metastasis (P=0.041, Figure 2C). And TMB levels correlate with advanced tumor grade (P=0.005, Figure 2D). Finally, we used the ‘clusterProfiler’ R package to explore the potential functions and pathways of these genes. A total of 291 GO terms and 7 pathways were identified (P<0.05, enrichment score of >1.5). The results showed that the top cancer-related biological processes were associated with the regulation of mitotic nuclear division, nuclear division, and organelle fission (Figure 2E), and the results suggested that these genes were mainly enriched in the cancer‐related signaling pathway, such as cell cycle, progesterone-mediated oocyte maturation, cellular senescence, oocyte meiosis, et al. (Figure 2F).
Figure 2. TMB correlation analysis.
(A) Heatmap of differentially expressed genes in high and low TMB groups. (B) Kaplan–Meier survival analysis. (C) Relationship between TMB and tumor metastasis. (D) Relationship between TMB and tumor stage. (E,F) GO and KEGG results.
Identification of immune-related genes
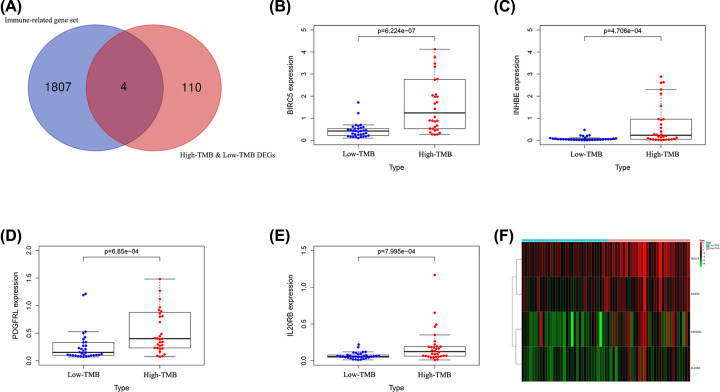
A total of 114 DEGs were identified by comparing the high and low TMB groups (Supplementary Table S1). We also downloaded a list of immune-related genes from the immunology database (Immport), from which we identified four immune-related genes, including BIRC5, PDGFRL, INHBE, and IL20RB (Figure 3A).
Figure 3. Identification of immune-related genes in high and low TMB groups.
(A) Four immune-related genes. (B–E) Expression levels of four immune-related genes in high and low TMB groups. (F) Heatmap of differences in four immune-related genes.
Expression levels of four immune-related genes in high and low TMB groups
We extracted the expression data of four immune-related genes from the expression file, then compared their expression in the high and low TMB groups, and visualized them through the ‘beeswarm’ package. From the results, we found that BIRC5, PDGFRL, INHBE, and IL20RB were highly expressed in the high TMB group (Figure 3B–E). The heatmap in Figure 3F showed the expression of four immune-related genes in the high and low TMB groups.
The correlation between four immune-related genes with TMB, tumor metastasis, patients’ survival, and immune checkpoint genes
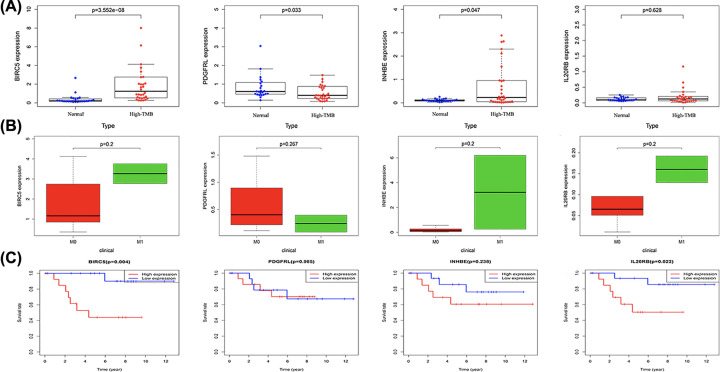
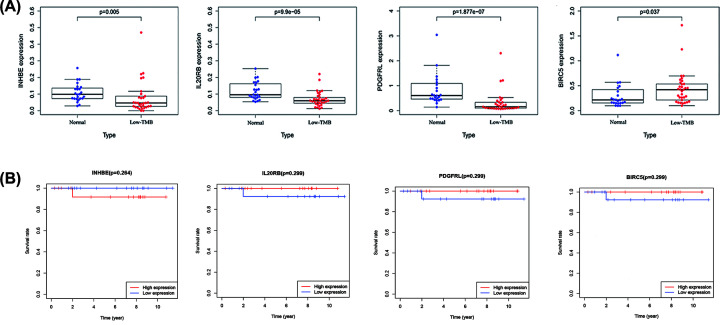
In order to better understand the role of these four immune-related genes in chRCC, we divided chRCC patients into high and low TMB groups, and analyzed the expression levels of these four immune-related genes in high and low TMB groups, and then analyzed their correlation with tumor metastasis and patients’ prognosis. In the results of the high TMB group (Figure 4A), we found that BIRC5 (P=3.552e-08) and INHBE (P=0.047) were significantly up-regulated in the high TMB group, while PDGFRL (P=0.033) was significantly down-regulated in the high TMB group. However, all these four immune-related genes were not related to tumor metastasis (Figure 4B). We then found that high TMB chRCC patients with BIRC5 (P=0.004) and IL20RB (P=0.022) were correlated with poor prognosis (Figure 4C). As a result in low TMB chRCC group, we found that INHBE (P=0.005), IL20RB (P=9.9e-05), and PDGFRL (P=1.877e-07) were down-regulated in the low TMB group (Figure 5A). In contrast, BIRC5 (P=0.037) was up-regulated in the low TMB group (Figure 5A). Moreover, we revealed that the expression of these four immune-related genes would not affect the prognosis of chRCC patients with low TMB (Figure 5B). The correlation between the expression of four immune-related genes and the reported immune checkpoint genes were also analyzed, which revealed that PDGFRL was certainly positively correlated with the immune checkpoint genes (Supplementary Table S2).
Figure 4. Correlation of four immune-related genes with high TMB group.
(A) Expression levels of four immune-related genes in normal group and high TMB groups. (B) Expression levels of four immune-related genes in high TMB group patients with and without tumor metastasis. (C) The overall survival of high TMB group patients with high and low expression levels of four immune-related genes.
Figure 5. Correlation of four immune-related genes with low TMB group.
(A) Expression levels of four immune-related genes in normal group and low TMB groups. (B) The overall survival of low TMB group patients with high and low expression levels of four immune-related genes.
Prognosis of BIRC5 in high TMB group
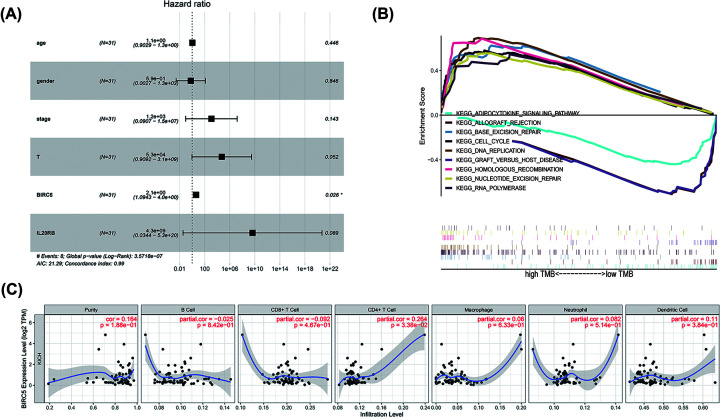
Previous results indicated that BIRC5 and IL20RB were significantly associated with the prognosis of patients with high TMB. In order to further study the prognosis in the chRCC high TMB group, we further performed a COX regression analysis in the high TMB group (Table 1). First, we found that stage (hazard ratio (HR) = 9.985, 95% confidence interval (CI) = 2.481–40.187, P=0.001), T (HR = 19.829, 95% CI = 2.487–158.045, P=0.005), and BIRC5 (HR = 1.177, 95% CI = 1.062–1.303, P=0.002) were significantly correlated with prognosis in univariate COX analysis. After that, we performed a multivariate COX analysis, which revealed that BIRC5 (HR = 2.094; 95% CI = 1.094–4.010; P=0.026) was independently associated with a worse prognosis (Figure 6A, Table 1).
Table 1. Univariate and multivariate Cox regression analyses.
| Parameter | Univariate Cox analysis | Multivariate Cox analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.040 | 0.970–1.116076 | 0.262 | 1.067 | 0.970–1.261 | 0.445 |
| Gender | 2.428 | 0.488–12.0696 | 0.277 | 0.587 | 0.488–125.377 | 0.845 |
| Stage | 9.985 | 2.481–40.187 | 0.001 | 1175.509 | 2.481–15228757 | 0.143 |
| T | 19.829 | 2.487–158.045 | 0.005 | 52662.11 | 2.487–3.05E+09 | 0.052 |
| BIRC5 | 1.177 | 1.062–1.303 | 0.002 | 2.094934 | 1.062–4.010 | 0.025 |
| IL20RB | 4.66 | 0.603–36.015 | 0.140 | 4.27E+09 | 0.603–5.30E+20 | 0.088 |
P<0.05 has statistical significance.
Figure 6. Cox analysis, GSEA, and tumor-infiltrating immune cells analysis of BIRC5.
(A) Cox analysis of BIRC5. (B) GSEA of BIRC5 based on based on TMB score. (C) Tumor-infiltrating immune cells analysis of BIRC5.
GSEA and immune infiltration of BIRC5
GSEA was performed to explored the BIRC5 associated-functions in chRCC. The results revealed prominent enrichment of signatures related in the base excision repair, cell cycle, DNA replication, homologous reorganization, nucleotide excision repair, and RNA polymerase in the high TMB group (Figure 6B, Table 2). In addition, adipocytokine signaling pathway, allograft rejection, and graft versus host disease were enriched in the low TMB group group (Figure 6B, Table 2).
Table 2. GSEA.
| Name | ES | NES | NOM P-val | FDR q-val |
|---|---|---|---|---|
| KEGG_ADIPOCYTOKINE_SIGNALING_PATHWAY | −0.45 | −1.64 | 0.017 | 0.323 |
| KEGG_ALLOGRAFT_REJECTION | −0.72 | −1.68 | 0.031 | 0.481 |
| KEGG_BASE_EXCISION_REPAIR | 0.62 | 1.75 | 0.014 | 0.765 |
| KEGG_CELL_CYCLE | 0.58 | 1.74 | 0.010 | 0.283 |
| KEGG_DNA_REPLICATION | 0.69 | 1.60 | 0.024 | 0.331 |
| KEGG_GRAFT_VERSUS_HOST_DISEASE | −0.72 | −1.65 | 0.036 | 0.421 |
| KEGG_HOMOLOGOUS_RECOMBINATION | 0.69 | 1.73 | 0.014 | 0.233 |
| KEGG_NUCLEOTIDE_EXCISION_REPAIR | 0.55 | 1.60 | 0.041 | 0.375 |
| KEGG_RNA_POLYMERASE | 0.55 | 1.68 | 0.017 | 0.288 |
NOM P-val <0.05 has statistical significance.
To further analyze the function of BIRC5 in the high TMB group, we used TIMER to verify the correlation between BIRC5 and immune cell infiltration levels (Figure 6C). We found a positive correlation between BIRC5 expression and the abundance of CD4 + T cells (Cor = 0.264; P=3.38e-02).
Discussion
At present, the immunotherapy for RCC is mainly based on immune checkpoints of PD-1 and PD-L1 inhibitors [27–29]. However, the immunotherapy of chRCC is still insufficient. TMB has proven to be a determinant of immune-related survival in many tumor patients and is extremely important in the treatment of tumors [30–33]. TMB can also be used as one of the indicators for the treatment of RCC patients, but the current TMB research on chRCC patients is still insufficient [34–36]. The prognostic role of immune-related genes in high and low TMB groups and their relevance to immunotherapy have not yet been explored. Thus, this research investigated the prognostic role of immune-related genes and the potential association with immune infiltrate cells in chRCC.
In the current study, chRCC somatic mutation data were obtained from the TCGA database and then divided into high TMB and low TMB groups based on the median. We then analyzed the immune gene differences in the high and low TMB groups and analyzed the correlation of immune-related gene function and prognosis. As a result, it was found through research that mutations in chRCC are also common, and their mutations are mainly missense mutations. Moreover, SNP was the most common variant type. Actually, somatic missense mutations strongly contribute to the generation of novel tumor epitopes [37] Previous studies have demonstrated the significance of missense mutation and SNP in tumorigenesis, progression, and prognosis in various cancer types, including bladder cancer [38–41]. The three most frequently mutated genes were TP53, PTEN, and MUC4. TP53 is one of the famous tumor suppressor genes reported to regulate the cell cycle thus inhibits the development of cancerous cells [42]. P53 protein maintains genome stability and prevents the occurrence of genomic mutation [43]. PTEN was referred as a dormant tumor suppressor in RCC and associated with associated with patients’ prognosis [44].
We divided chRCC patients into high and low TMB groups based on TMB levels and analyzed the differences between high and low TMB groups. We found that there was a significant difference between high TMB and low TMB groups. High TMB was correlated with a worse prognosis and would promote tumor metastasis and development. In fact, another study about ccRCC also suggested that patients with higher TMB tended to be with a worse prognosis [45]. Moreover, Chuanjie et al. found that ccRCC with higher TMB levels are associated with higher tumor grades and advanced pathological stages [46].
Another important finding of our study was that four differentially expressed immune-related genes (BIRC5, INHBE, PDGFRL, and IL20RB) were identified in the high and low TMB groups. BIRC5 and IL20RB were correlated with the worse prognosis in high TMB groups analysis. Previous studies revealed that BIRC5 could promote tumor cell proliferation in RCC [46]. Moreover, BIRC5 was found to be a biomarker for prognosis and therapy in other types of cancers, including lung cancer and pancreatic cancer [47,48]. Interestingly, IL20RB was also suggested as a prognosis biomarker in pRCC, and promoted cell proliferation, invasion and migration [49]. Our study further highlighted the significance of BIRC5 and IL20RB in the tumorigenesis and progress of RCC.
BIRC5, selected for further study and univariate as well as multivariate Cox analyses, demonstrated that it can be used as an independent prognostic factor for chRCC. Moreover, the result of GSEA revealed that BIRC5 was involved in base excision repair, cell cycle, DNA replication, homologous reorganization, nucleotide excision repair, RNA polymerase, and adipocytokine signaling pathway in chRCC. DNA replication is one of the fundamental biological processes in which dysregulation can cause genome instability [50]. And DNA replication errors are the main drivers of cancers, including RCC [51]. The normal process of cell division occurs via the cell cycle, and dysregulation of cell cycle would result in sustained unscheduled cell growth, proliferation, a hallmark of cancer [52]. In hepatocellular carcinoma, down-regulation of BIRC5 could induce cancer cell apoptosis and cell cycle arrest [53]. Thus, BIRC5 may regulate RCC development via cell cycle.
In our study, we also found that BIRC5 in chRCC was involved in immune cell infiltration. Actually, another study also found that BIRC5 was associated with immune cell infiltration and served as a prognostic biomarker and therapeutic target for HCC [54]. In chRCC, BIRC5 showed positive correlation with CD4+ T cells. Previous study revealed that CD4+ T cells could promote tumor cell proliferation in RCC [55]. Moreover, another study found that CD4+ T cells in RCC patients were associated with favorable prognosis [56]. Therefore, BIRC5 may also regulate tumor cell biological process, thus affecting the prognosis of chRCC via CD4+ T cells. And further studies should test this hypothesis.
Admittedly, our research also had some limitations. First of all, the results of the current study were not validated using another independent patient cohort. Moreover, it would be better if in vitro or in vivo experiments were performed to validate our findings.
In summary, our study shows that high TMB in chRCC patients correlated with worse prognosis, while BIRC5 in the high TMB group can be used as an independent prognostic indicator of chRCC. We also predict that BIRC5-enriched functional pathways and potentially affected immune-infiltrating cells, and this information may contribute to the development of chRCC therapy.
Supplementary Material
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- chRCC
chromogenic renal cell carcinoma
- CI
confidence interval
- DEG
differentially expressed genes
- FC
fold change
- GO
gene ontology
- GSEA
gene set enrichment analysis
- HR
hazard ratio
- PD-L1
programmed death ligand 1
- PD-1
programmed cell death protein 1
- pRCC
papillary renal cell carcinoma
- RCC
renal cell carcinoma
- SNV
single-nucleotide variation
- TCGA
the cancer genome atlas
- TIMER
tumor immune estimation resource
- TMB
tumor mutation burden
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China [grant number 81703062].
Author Contribution
Lei Li and Xi Chen performed data analysis work and aided in writing the manuscript. Qing Zhou designed the study, assisted in writing the manuscript. Lu Hao, Qiuyan Chen and Haosheng Liu edited the manuscript. All authors read and approved the final manuscript.
References
- 1.Delahunt B. and Eble J.N. (1997) Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod. Pathol. 10, 537–544 [PubMed] [Google Scholar]
- 2.Delahunt B., Eble J.N., McCredie M.R., Bethwaite P.B., Stewart J.H. and Bilous A.M. (2001) Morphologic typing of papillary renal cell carcinoma: comparison of growth kinetics and patient survival in 66 cases. Hum. Pathol. 32, 590–595 10.1053/hupa.2001.24984 [DOI] [PubMed] [Google Scholar]
- 3.Meskawi M., Sun M., Trinh Q.D., Bianchi M., Hansen J., Tian Z. et al. (2012) A review of integrated staging systems for renal cell carcinoma. Eur. Urol. 62, 303–314 10.1016/j.eururo.2012.04.049 [DOI] [PubMed] [Google Scholar]
- 4.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. and Jemal A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 5.Kim M., Joo J.W., Lee S.J., Cho Y.A., Park C.K. and Cho N.H. (2020) Comprehensive immunoprofiles of renal cell carcinoma subtypes. Cancers (Basel) 12, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volpe A., Novara G., Antonelli A., Bertini R., Billia M., Carmignani G. et al. (2012) Chromophobe renal cell carcinoma (RCC): oncological outcomes and prognostic factors in a large multicentre series. BJU Int. 110, 76–83 10.1111/j.1464-410X.2011.10690.x [DOI] [PubMed] [Google Scholar]
- 7.Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H. et al. (2014) The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330 10.1016/j.ccr.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(2017) Checkpoint inhibitor combo effective for RCC. Cancer Discov. 7, Of5 10.1158/2159-8290.CD-NB2017-133 [DOI] [PubMed] [Google Scholar]
- 9.Linehan W.M. and Ricketts C.J. (2017) Kidney cancer in 2016: RCC - advances in targeted therapeutics and genomics. Nat. Rev. Urol. 14, 76–78 10.1038/nrurol.2016.260 [DOI] [PubMed] [Google Scholar]
- 10.Topalian S.L., Wolchok J.D., Chan T.A., Mellman I., Palucka K., Banchereau J. et al. (2015) Immunotherapy: the path to win the war on cancer? Cell 161, 185–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasser S., Lim L.H.K. and Cheung F.S.G. (2017) The role of the tumour microenvironment in immunotherapy. Endocr. Relat. Cancer 24, T283–T295 10.1530/ERC-17-0146 [DOI] [PubMed] [Google Scholar]
- 12.Yang Y. (2015) Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Invest. 125, 3335–3337 10.1172/JCI83871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steven A., Fisher S.A. and Robinson B.W. (2016) Immunotherapy for lung cancer. Respirology 21, 821–833 10.1111/resp.12789 [DOI] [PubMed] [Google Scholar]
- 14.Di Tucci C., Schiavi M.C., Faiano P., D'Oria O., Prata G., Sciuga V. et al. (2018) Therapeutic vaccines and immune checkpoints inhibition options for gynecological cancers. Crit. Rev. Oncol. Hematol. 128, 30–42 10.1016/j.critrevonc.2018.05.011 [DOI] [PubMed] [Google Scholar]
- 15.Postow M.A., Sidlow R. and Hellmann M.D. (2018) Immune-related adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 378, 158–168 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- 16.Bednarski J.J. and Sleckman B.P. (2019) At the intersection of DNA damage and immune responses. Nat. Rev. Immunol. 19, 231–242 10.1038/s41577-019-0135-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topalian S.L. (2017) Targeting immune checkpoints in cancer therapy. JAMA 318, 1647–1648 10.1001/jama.2017.14155 [DOI] [PubMed] [Google Scholar]
- 18.Ritterhouse L.L. (2019) Tumor mutational burden. Cancer Cytopathol. 127, 735–736 10.1002/cncy.22174 [DOI] [PubMed] [Google Scholar]
- 19.Dyck L. and Mills K.H.G. (2017) Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 47, 765–779 10.1002/eji.201646875 [DOI] [PubMed] [Google Scholar]
- 20.(2018) High TMB predicts immunotherapy benefit. Cancer Discov. 8, 668. [DOI] [PubMed] [Google Scholar]
- 21.Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A. et al. (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 30, 44–56 10.1093/annonc/mdy495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y., Feng X.-F., Yang W.-X. and You C.-G. (2019) Exploring the pathological mechanism of bladder cancer based on tumor mutational burden analysis. Biomed Res. Int. 2019, 1093815 10.1155/2019/1093815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y. et al. (2019) Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun D.A., Burke K.P. and Van Allen E.M. (2016) Genomic approaches to understanding response and resistance to immunotherapy. Clin. Cancer Res. 22, 5642–5650 10.1158/1078-0432.CCR-16-0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalocusky K.A., Kan M.J., Hu Z., Dunn P., Thomson E., Wiser J. et al. (2018) The 10,000 Immunomes Project: building a resource for human immunology. Cell Rep. 25, 513.e513–522.e513 10.1016/j.celrep.2018.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damian D. and Gorfine M. (2004) Statistical concerns about the GSEA procedure. Nat. Genet. 36, 663, author reply 663 10.1038/ng0704-663a [DOI] [PubMed] [Google Scholar]
- 27.Ohaegbulam K.C., Assal A., Lazar-Molnar E., Yao Y. and Zang X. (2015) Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 21, 24–33 10.1016/j.molmed.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelsey R. (2018) Kidney cancer: PDL1 as a biomarker in high-risk RCC. Nat. Rev. Urol. 15, 202. [DOI] [PubMed] [Google Scholar]
- 29.Sunela K.L., Lehtinen E.T., Kataja M.J., Kujala P.M., Soimakallio S. and Kellokumpu-Lehtinen P.L. (2014) Development of renal cell carcinoma (RCC) diagnostics and impact on prognosis. BJU Int. 113, 228–235 10.1111/bju.12242 [DOI] [PubMed] [Google Scholar]
- 30.Boumber Y. (2018) Tumor mutational burden (TMB) as a biomarker of response to immunotherapy in small cell lung cancer. J. Thorac. Dis. 10, 4689–4693 10.21037/jtd.2018.07.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladányi A. and Tímár J. (2020) Immunologic and immunogenomic aspects of tumor progression. Semin. Cancer Biol. 60, 249–261 10.1016/j.semcancer.2019.08.011 [DOI] [PubMed] [Google Scholar]
- 32.Greillier L., Tomasini P. and Barlesi F. (2018) The clinical utility of tumor mutational burden in non-small cell lung cancer. Transl. Lung Cancer Res. 7, 639–646 10.21037/tlcr.2018.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Xu J., Du C., Wu Y., Xia D., Lv W. et al. (2019) The predictive value of tumor mutation burden on efficacy of immune checkpoint inhibitors in cancers: a systematic review and meta-analysis. Front. Oncol. 9, 1161 10.3389/fonc.2019.01161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azzi S., Gallerne C., Romei C., Le Coz V., Gangemi R., Khawam K. et al. (2015) Human renal normal, tumoral, and cancer stem cells express membrane-bound interleukin-15 isoforms displaying different functions. Neoplasia 17, 509–517 10.1016/j.neo.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergerot P.G., Hahn A.W., Bergerot C.D., Jones J. and Pal S.K. (2018) The role of circulating tumor DNA in renal cell carcinoma. Curr. Treat. Option Oncol. 19, 10 10.1007/s11864-018-0530-4 [DOI] [PubMed] [Google Scholar]
- 36.Guo W., Fu Y., Jin L., Song K., Yu R., Li T. et al. (2020) An exon signature to estimate the tumor mutational burden of right-sided colon cancer patients. J. Cancer 11, 883–892 10.7150/jca.34363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klebanov N., Artomov M., Goggins W.B., Daly E., Daly M.J. and Tsao H. (2019) Burden of unique and low prevalence somatic mutations correlates with cancer survival. Sci. Rep. 9, 4848 10.1038/s41598-019-41015-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korphaisarn K., Morris V.K., Overman M.J., Fogelman D.R., Kee B.K., Raghav K.P.S. et al. (2017) FBXW7 missense mutation: a novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget 8, 39268–39279 10.18632/oncotarget.16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIntyre J.B., Nelson G.S., Ghatage P., Morris D., Duggan M.A., Lee C.H. et al. (2014) PIK3CA missense mutation is associated with unfavorable outcome in grade 3 endometrioid carcinoma but not in serous endometrial carcinoma. Gynecol. Oncol. 132, 188–193 10.1016/j.ygyno.2013.11.015 [DOI] [PubMed] [Google Scholar]
- 40.Yuan Y., Wang W., Li H., Yu Y., Tao J., Huang S. et al. (2015) Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer 15, 346 10.1186/s12885-015-1349-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo G., Sun X., Chen C., Wu S., Huang P., Li Z. et al. (2013) Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat. Genet. 45, 1459–1463 10.1038/ng.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer M., Grossmann P., Padi M. and DeCaprio J.A. (2016) Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 44, 6070–6086 10.1093/nar/gkw523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negrini S., Gorgoulis V.G. and Halazonetis T.D. (2010) Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- 44.Tang L., Li X., Gao Y., Chen L., Gu L., Chen J. et al. (2017) Phosphatase and tensin homolog (PTEN) expression on oncologic outcome in renal cell carcinoma: a systematic review and meta-analysis. PLoS ONE 12, e0179437 10.1371/journal.pone.0179437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C., He H., Hu X., Liu A., Huang D., Xu Y. et al. (2019) Development and validation of a metastasis-associated prognostic signature based on single-cell RNA-seq in clear cell renal cell carcinoma. Aging (Albany N.Y.) 11, 10183–10202 10.18632/aging.102434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Z., Chen Y., Yang C., Zhang M., Chen A., Yang J. et al. (2019) STAT gene family mRNA expression and prognostic value in hepatocellular carcinoma. Onco Targets Ther. 12, 7175–7191 10.2147/OTT.S202122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y., Zhu W., Chen W., Wu J., Hou G. and Li Y. (2019) Prognostic value of BIRC5 in lung adenocarcinoma lacking EGFR, KRAS, and ALK mutations by integrated bioinformatics analysis. Dis. Markers 2019, 5451290 10.1155/2019/5451290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S.H., Hong Y., Markowiak S., Sanchez R., Creeden J., Nemunaitis J. et al. (2019) BIRC5 is a target for molecular imaging and detection of human pancreatic cancer. Cancer Lett. 457, 10–19 10.1016/j.canlet.2019.04.036 [DOI] [PubMed] [Google Scholar]
- 49.Cui X.F., Cui X.G. and Leng N. (2019) Overexpression of interleukin-20 receptor subunit beta (IL20RB) correlates with cell proliferation, invasion and migration enhancement and poor prognosis in papillary renal cell carcinoma. J. Toxicol. Pathol. 32, 245–251 10.1293/tox.2019-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitao H., Iimori M., Kataoka Y., Wakasa T., Tokunaga E., Saeki H. et al. (2018) DNA replication stress and cancer chemotherapy. Cancer Sci. 109, 264–271 10.1111/cas.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomasetti C., Li L. and Vogelstein B. (2017) Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 10.1126/science.aaf9011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingham M. and Schwartz G.K. (2017) Cell-cycle therapeutics come of age. J. Clin. Oncol. 35, 2949–2959 10.1200/JCO.2016.69.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao L., Li C., Shen S., Yan Y., Ji W., Wang J. et al. (2013) OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer 13, 82 10.1186/1471-2407-13-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ali H.R., Provenzano E., Dawson S.J., Blows F.M., Liu B., Shah M. et al. (2014) Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 25, 1536–1543 10.1093/annonc/mdu191 [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Wang Y., Xu L., Lu X., Fu D., Su J. et al. (2018) CD4+ T cells promote renal cell carcinoma proliferation via modulating YBX1. Exp. Cell Res. 363, 95–101 10.1016/j.yexcr.2017.12.026 [DOI] [PubMed] [Google Scholar]
- 56.Nishida K., Kawashima A., Kanazawa T., Kidani Y., Yoshida T., Hirata M. et al. (2020) Clinical importance of the expression of CD4+CD8+ T cells in renal cell carcinoma. Int. Immunol. 32, 347–357 10.1093/intimm/dxaa004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.