Abstract
Purpose
Population-based newborn screening (NBS) allows early detection and treatment of inherited disorders. For certain medically-actionable conditions, however, NBS is limited by the absence of reliable biochemical signatures amenable to detection by current platforms. We sought to assess the analytic validity of an ATP7A targeted next generation DNA sequencing assay as a potential newborn screen for one such disorder, Menkes disease.
Methods
Dried blood spots from control or Menkes disease subjects (n = 22) were blindly analyzed for pathogenic variants in the copper transport gene, ATP7A. The analytical method was optimized to minimize cost and provide rapid turnaround time.
Results
The algorithm correctly identified pathogenic ATP7A variants, including missense, nonsense, small insertions/deletions, and large copy number variants, in 21/22 (95.5%) of subjects, one of whom had inconclusive diagnostic sequencing previously. For one false negative that also had not been detected by commercial molecular laboratories, we identified a deep intronic variant that impaired ATP7A mRNA splicing.
Conclusions
Our results support proof-of-concept that primary DNA-based NBS would accurately detect Menkes disease, a disorder that fulfills Wilson and Jungner screening criteria and for which biochemical NBS is unavailable. Targeted next generation sequencing for NBS would enable improved Menkes disease clinical outcomes, establish a platform for early identification of other unscreened disorders, and complement current NBS by providing immediate data for molecular confirmation of numerous biochemically screened condition.
Keywords: Menkes disease, ATP7A, Molecular genetics, Targeted next generation sequencing, Dried blood spots, Newborn screening
1. Introduction
Government-funded and state-mandated newborn screening (NBS) programs are currently guided by the Recommended Uniform Screening Panel (RUSP) to screen for 35 core and 26 secondary conditions [1]. NBS for these conditions enables early presymptomatic identification and institution of dietary or medical interventions that reduce morbidity and mortality. Twenty-five years ago, tandem mass spectrometry revolutionized NBS by enabling multiplex testing of newborn dried blood spots (DBS) to screen simultaneously for numerous metabolic disorders [[2], [3], [4]]. However, NBS for many other potentially treatable infantile-onset disorders remains a formidable challenge, especially when biochemical analytes are unavailable or unamenable to detection from DBS with current NBS platforms. Deoxyribonucleic acid (DNA)-based NBS approaches exploiting recent advances in sequencing and interpretive technologies appear poised to address this gap. Use of DNA sequencing technology is already being adopted by NBS public health laboratories for second-tier confirmatory testing on samples with positive first-tier biochemical results that may have low specificity [[5], [6], [7], [8]].
The inherent universality of DNA sequence conveys potential to reveal risk for nearly any monogenic inherited disorder. In contrast to techniques such as whole genome and whole exome sequencing, targeted next generation sequencing (tNGS) offers the scalability, speed, and resolution to economically evaluate multiple newborn disease genes of interest, in parallel [8]. Therefore, tNGS may be uniquely suited as a primary modality (first-tier) to detect disorders for which a biochemical analyte is not currently measurable from DBS, or to improve first-tier biochemical tests (e.g. to reduce false positives) by providing supportive confirmatory sequence data relevant to those conditions currently screened [6,[8], [9], [10], [11], [12], [13], [14]].
Menkes disease is a X-linked recessive disorder of copper metabolism with a predicted minimum birth prevalence of 1 in 34,810 live male births based on loss-of-function variant frequencies in the Genome Aggregation Database (gnomAD) [15] for ATP7A, which encodes an essential copper-transporting ATPase [[16], [17], [18]]. While onset of this medically-actionable condition occurs in early infancy, population-based NBS by biochemical analyte testing is not currently available or feasible. Abnormal plasma neurochemical levels due to deficiency of a copper-requiring enzyme, dopamine-β-hydroxylase, are diagnostic in affected Menkes newborns [17], however the need for larger blood volume and an alumina extraction step may render this approach impractical for DBS-based NBS. At birth, affected Menkes infants appear healthy and typically develop normally for 6 to 10 weeks before seizures, failure to thrive, irreversible brain atrophy, and stalled neurodevelopment ensue [16]. However, a brief window of opportunity exists in the newborn period during which medical intervention can prevent the inexorable downhill course otherwise expected for this illness, as demonstrated with subcutaneous Copper Histidinate (CuHis) treatment [17,[19], [20], [21], [22], [23]]. Another emerging approach, adeno-associated virus-mediated ATP7A gene therapy, also appears highly promising as a complementary treatment in combination with CuHis, demonstrating a synergistic treatment effect with subcutaneous copper in the Menkes mouse model [24,25].
In this study, we evaluate the analytic validity of a tNGS assay to detect ATP7A variants in DBS from previously genotyped subjects with Menkes disease. With this example, we aim to assess the potential for utility of tNGS for NBS of any actionable genetic disorder not detectable through current biochemical newborn screening approaches.
2. Materials, patients and methods
2.1. Subjects and DBS specimens
Menkes disease patients were evaluated at the NIH Clinical Center in Bethesda, MD under protocol 09-CH-0059 (Clinical Trials.gov number, NCT00811785) for Copper Histidinate (IND 34,166) treatment. The protocol and a specific amendment for collection and evaluation of DBS for ATP7A variants were approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Institutional Review Board. Informed consent was obtained from the patients' parents for this specific amendment.
Twenty-four coded, de-identified DBS samples were assessed: 23 from male subjects with Menkes disease, and one from a healthy male control evaluated under a screening protocol (02-CH-0023) also approved by the NICHD IRB. Sequencing and interpretation were performed blinded to the disease status and number of affected subjects and normal controls. The samples were collected serially during scheduled protocol visits, between February and November 2016. One Menkes patient (Subject B) contributed two DBS specimens, obtained at separate protocol visits.
2.2. Targeted next generation sequencing (tNGS) panel
We utilized an existing tNGS panel validated for use with DBS and designed as a diagnostic tool in Newborn Intensive Care Units for interrogation of 544 genes (7 Mb) associated with genetic disorders that present with a phenotype in the newborn period and are potentially medically actionable (Table S1) [8]. In addition to ATP7A, the tNGS panel includes the most common genes associated with disorders now listed on the US Recommended Uniform Screening Panel (RUSP) of the Advisory Committee on Heritable Diseases in Newborns and Children (ACHDNC; https://www.hrsa.gov/advisory-committees/heritable-disorders/index.html), and offers a model of broad DNA-based NBS coverage. The entire coding sequences, including first and last exons, adjacent untranslated regions and splice junctions of the 544 genes were sequenced. The clinical test pipeline was optimized for use in neonates, utilizing DNA extracted from small blood volume (DBS punches) to detect single-nucleotide variants (SNVs), small insertions and deletions (INDELs) up to 20 bp in length, and large copy number variants (CNVs) with rapid turnaround time (≤120 h). Over 90% of the panel genes were sequenced to a depth of 20× or greater however only ATP7A sequences were analyzed.
2.3. Sample processing
DNA was isolated from several DBS spot punches and processed in a clinical pipeline, as described previously [8]. Custom oligonucleotide probe libraries (SeqCap EZ Choice, Nimblegen/Roche) were utilized to capture genomic DNA regions and targeted libraries were created as described [8]. Sequencing of the targeted libraries was performed on a MiSeq Sequencer (Illumina, San Diego, CA USA). Sequencing reads were aligned to the reference genome (hg19/GRCH build 37) using Burrows-Wheeler Aligner for short alignments (BWA), followed by a Genome Analysis Toolkit v1.6 (GATK) variant calling pipeline running on the Illumina platform, and analyzed with genome interpretation software (Opal 4.0, Fabric Genomics, Oakland, CA USA).
2.4. Variant identification
2.4.1. Identification of ATP7A SNVs and INDELs
SNVs and INDELs were identified using variant filter tools in Opal 4.0. We first reviewed and removed common polymorphisms (>5% in the general population) by comparison with dbSNP 132, the 1000G, the Exome Variant Server (http://evs.gs.washington.edu), and an in-house variant database to identify both common benign variants and recurrent artifact variant calls.
2.4.2. CNV analysis
For each sample, the mean-normalized coverage at each base in the targeted panel was calculated from an aligned reads file produced by the workflow above. Normalized coverage in a patient sample was compared to an empirical model of expected coverage and an assigned Z-score was calculated, with candidate deletion events having negative Z-scores. Loci with Z-scores exceeding a designated threshold (|Z-score| > 3) were identified and consecutive loci above threshold were merged to form a list of candidate copy number gains and losses. The coverage model generated from a reference set of 20 samples from non-CNV individuals was used to validate the workflow. This consisted of the mean, variance and coefficient of variance in coverage observed at each base in the targeted panel.
2.5. False positive estimates
Because concern about generating false-positive results exists for all NBS algorithms and our study design could not provide such data, we estimated false-positive rates via two methods: 1) review of ATP7A SNVs in the Genome Aggregation Database (gnomAD) at Broad Institute, Massachusetts Institute of Technology, Cambridge, MA that covers 125,748 exomes and 15,708 genomes from 141,456 unrelated individuals (https://gnomad.broadinstitute.org/) and 2) evaluation for the presence of pathogenic or likely pathogenic (as previously defined) ATP7A variants in 163 newborns without Menkes disease who had been screened at birth with whole exome sequencing (Newborn Screening Translational Research Network, Longitudinal Pediatric Data Resource, dbGaP accession nbs000002.v1.p1) [[26], [27], [28]].
2.6. Western blotting
Total protein was extracted from cultured human fibroblasts and western blotting performed using a rabbit amino-terminus anti-ATP7A antibody or an anti-β-actin monoclonal antibody, as previously described [29].
2.7. Confocal imaging
For confocal imaging experiments, cultured human fibroblasts were grown on glass cover slips, stained with a carboxy-terminal anti-ATP7A antibody, examined by confocal microscopy (Zeiss 710), and the images captured using Zen software.
2.8. Reverse transcription-polymerase chain reaction
Total RNA was extracted from cultured fibroblasts using TriPure Isolation Reagent (Roche). First strand cDNA synthesis was performed using the Enhanced Avian TR First Strand Synthesis Kit (Sigma, St Louis, MO). ATP7A cDNA was obtained by reverse transcription and polymerase chain reaction (RT-PCR) using total RNA as template. The ATP7A exon 3/exon 4 region was amplified using primers: 5′-TGGCTCAAGCTGGTGAAGTC-3′ (forward) and 5′-TGAGGTACTTGGGCTGCTTT-3′ (reverse).
2.9. Automated DNA sequencing
DNA sequencing of RT-PCR products was performed as previously described [17].
3. Results
3.1. tNGS findings
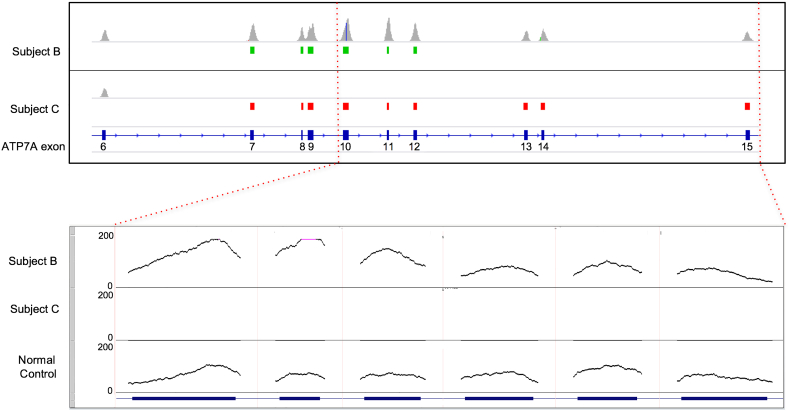
We confirmed successful generation of clinical grade sequence data for the ATP7A sequences analyzed in all 24 blinded DBS specimens. Sequences from either control or Menkes individuals revealed 22 with ATP7A variants consistent with the diagnosis of Menkes disease (Table 1). These included eight copy number variants (six large deletions, two large duplications (Fig. 1), four small insertion/deletions causing translational reading frameshifts, as well as three nonsense, three splice junction, and four missense variants. These results represent the typical ATP7A mutation spectrum associated with this illness [30]. Five benign SNV variants (one intronic, three 3’UTR and one missense variants) were noted in seven subjects. Comparison of study tNGS results on the 21 Menkes disease subjects whose ATP7A genotypes had been previously determined with commercially obtained clinical results showed 100% concordance. DBS sample number 15 was correctly identified as a normal control not affected with Menkes disease. Deletion of exon 1 was found in two samples from unrelated subjects, as was deletion of exons 13–14. Duplication of exon 7–12 was found in two DBS specimens obtained from the same Menkes disease patient (Subject B) at separate protocol visits.
Table 1.
Patient demographics, diagnostics, and results of targeted next Generation sequencing.
| DBS ID | Subject ID | Birth State | Age at diagnosis (months) | Current age | Diagnostic work-up other than ATP7A sequencing | ATP7A Transcript/protein variant detected on diagnostic work-up | ATP7ATranscript/protein variant detected on blinded tNGS | Chromosomal Position/dbSNP [Hg19] |
|---|---|---|---|---|---|---|---|---|
| 1 | A | VA | 14 | 5y, 3 m | Multiple EEGs, brain MRI, serum Cu & Cp, plasma catechols | c.2357 T > G; p.Met786Arga | c.2357 T > G/ p.Met786Arg | ChrX:77268560/ rs797045354 |
| 2 | B | MA | 8.5 | 5y, 4 m | Brain MRI, plasma catechols | dupl exon 7-12b | dupl exon 7–12 | |
| 3 | C | TX | 5 | 3y, 5 m | EEG, brain MRI | del exons 7-23c | del exons 7–23 | |
| 4 | D | AZ | 2.5 | 3y | EEG, brain MRI, plasma catechols | Not performed (no insurance) |
del exon 13–14 | |
| 5 | E | CA | 14 | 3y, 2 m | EEG, brain MRI, serum Cu & Cp, plasma catechols | c.2172 + 6 T > G; (c.IVS9 + 6 T > G)d | c.2172 + 6 T > G (c.IVS9 + 6 T > G)/ n.a. | chrX:77267177/ n.a. |
| 6 | B | MA | 8.5 | 5y, 4 m | Brain MRI, plasma catechols | dupl exon 7-12b | dupl exon 7–12 | |
| 7 | F | IL | 48 | 7y, 4 m | Muscle biopsy, serum Cu&Cp, chromosome microarray, plasma amino acids, urine organic acids, brain MRI/MRS; skin biopsy | c.3014 G > T; p.Gly1005Vale | c.3014G > T/ p.Gly1005Val | ChrX:77284844/ n.a. |
| 8 | G | UT | 0.5⁎ | 1y, 9 m | Plasma catechols | Not performed (Known family variant [16]) |
del exon 13–14 | |
| 9 | H | MI | 4 | 3y, 4 m | Brain and abdomen MRI/MRA | c.2129_2132 del4f p.Ser710LeufsTer2 | c.2129_2132delCTGT/ p.Ser710LeufsTer2 | chrX: 77267128–77,267,131/ n.a. |
| 10 | I | AL | 9 | 2y, 5 m | Karyotype, CNV microarray, Fragile X | Inconclusive resultsd, g | Partial exon 12 del starts at c.2614, ends in IVS12 | |
| 11 | J | GA | 9 | 5y | Brain MRI/MRS, CT head, lysosomal enzyme screen, metabolic LP, chromosomal microarray, dilated eye exam, serum Cu & Cp, amino acids, pyruvate, fatty acid profile, CBC, plasma catechols, urine MPS screen, s-sulfocysteine, oligosaccharides, n-acetyl-aspartic acid and organic acids | None detectedc | None detected/ n.a | n.a./n.a. |
| 12 | K | MA | 12 | 3y, 2 m | Neurological workup, cardiac echo, EKG, EEG, video EEG, cranial ultrasound, brain MRI, plasma catechols, SNP microarray, epilepsy gene panel, PT/OT, blood: folate, vitamin B12, ferritin, thyroid panel, prealbumin, CPK | c.1139_1140delTGd p.Val380AspfsTer4 |
c.1139_1140delTG/ p.Val380AspfsTer4 | chrX:77245255–77,245,256/ n.a. |
| 13 | L | AL | 2.5 | Died at 13 m | Head ultrasound, cardiac echo, upper GI, urine creatinine, serum Cu & Cp, CBC, electrolytes, developmental and speech therapy assessments | c.601C > T p.Arg201Terb | c.601C > T/ p.Arg201Ter | chrX: 77244218/ rs151340633 |
| 14 | M | OK | 7 | 2y, 1 m | Brain MRI, serum Cu & Cp, plasma catechols, CBC, CPK, fatty acid, amino acid and acylcarnitine profiles, comprehensive metabolic panel | c.3526C>Th p.Gln1176Ter | c.3526C > T/ p.Gln1176Ter | chrX: 77294348/ n.a. |
| 15 | NormalControl | – | – | – | – | – | None detected/n.a | n.a./n.a. |
| 16 | N | NY | 0.3⁎ | 1y, 6 m | Prenatal molecular analysis | exon 1 (5’ UTR) deli | exon 1 (5’ UTR) del | |
| 17 | O | LA | 1.5 | 4y, 8 m | Plasma catechols | c.2627-1G > A (c.IVS12-1G > A)b |
c.2627-1G > A (c.IVS12-1G > A)/ n.a. | chrX:77275740/n.a. |
| 18 | P | FL | 0.25 ⁎ | 2y, 3 m | Plasma catechols | exon 1 (5’ UTR) del j | exon1 (5’ UTR) del | |
| 19 | Q | CO | 11 | 2y, 11 m | EEG, skull x-ray, brain MRI, VCUG, serum Cu & Cp | c.4117G>Ck p.Ala1373Pro | c.4117G > C/ p.Ala1373Pro | chrX:77298926/ n.a. |
| 20 | R | PA | 5 | 1y, 8 m | EEG, serum Cu & Cp, plasma catechols | c.3445C>Ta p.Gln1149Ter |
c.3445C > T/ p.Gln1149Ter | chrX:77289253/n.a. |
| 21 | S | MA | 7 | 3y, 2 m | Plasma catechols | c.3071 T>Ab p.Ile1024Lys |
c.3071 T > A/ p.Ile1024Lys | chrX:77284901/ rs797044648 |
| 22 | T | AZ | 9.5 | Died at 2y, 5 m | EEG | c.3496dupAd p.Ile1316AsnfsTer12 | c.3946dupA/ p.Ile1316AsnfsTer12 | chrX:77298226/ n.a. |
| 23 | U | CA | 2 | 1y,1 m | Plasma catechols | c.4006-1G>Al (c.IVS20-1G > A) |
c.4006-1G > A (c.IVS20-1G > A)/ n.a. | chrX:77298814/n.a. |
| 24 | V | TN | 7 | 4y,10 m | EEG, brain MRI, brain and internal auditory canal CT scans, skeletal survey, chest x-ray, serum Cu & Cp, karyotype, microarray, hearing and ophthalmology evaluations, abdominal ultrasound and radiograph, EKG | c.3753delTc p.Leu1252Ter |
c.3753delT/ p.Leu1252Ter | chrX:77296182/n.a. |
ATM = ataxia telangienctasia; Cp = ceruloplasmin; CT = computerized tomography; Cu = copper; EEG = electroencephalogram, EKG = electrocardiogram; MRA = magnetic resonance angiography; MRI = magnetic resonance imaging; MRS = magnetic resonance spectroscopy; NCV = nerve conduction velocity; OT = occupational therapy; PT = physical therapy; SNP = single nucleotide polymorphism; VCUG = voiding cystourethrogram.
Screened early due to known 50% risk (positive family history).
Prevention Genetics.
Emory Genetics Laboratory.
Baylor Medical Genetics Laboratories.
ARUP Laboratories.
Gene Dx.
Connective Tissue Gene Tests.
Quest Diagnostics.
Fulgent Diagnostics.
University of Chicago.
GeneCare Medical Genetics Center (prenatal analysis of subject's older affected brother).
Baylor Miraca.
Invitae.
Fig. 1.
Detection of copy number variants.
Coverage histograms (scale 0–250 read depth) of exons 6–15 of ATP7A (NM_000052) for two samples (Subjects B,C) displaying copy number variants identified by the CNV caller as intervals beneath the coverage track. Duplications are shown in green and deletions in red. Subject B shows a duplication of exons 7–12. Subject C shows a homozygous deletion after exon 6 (the deletion continues for the remaining exons). The inset below shows the above samples and sample 15, the normal control. The control 1,084,460-1shows reduced base coverage at each position on the left three exons than in sample 1,084,460–1 as expected.
3.2. Detection of deletion at exon/intron border by tNGS after prior inconclusive results
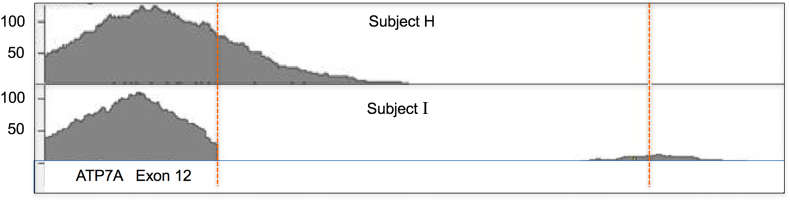
Subject I had inconclusive results from peripheral blood specimens evaluated at two different commercial molecular diagnostic laboratories (Table 1), however the tNGS method detected a deletion involving the final three amino acids of exon 12 and part of intron 12 (Fig. 2). While the capture probes used in our tNGS did not cover intron 12, the sequence data from Subject I contained deep intron 12 reads. The reference map track illustrates the location where capture reads were lost, and the distal intronic region from which sequence reads were evident (Fig. 2). Due to the deletion, deep intron 12 sequence became contiguous with exon 12 sequence, and therefore was read. Loss of the exon 12 splice donor site is predicted to cause exon skipping in this patient. Failure to pick up this variant by commercial laboratories presumably reflects loss of the DNA sequence to which the reverse primer in a polymerase chain reaction (PCR) would normally hybridize.
Fig. 2.
Partial deletion of ATP7A exon 12 detected by tNGS.
Although the tNGS capture probes did not cover the introns for ATP7A (coding + non-coding exons are covered) the deep intronic side of the deletion shows sequencing read coverage (compare 1,084,455–6 (above) vs 1,084,454–7 (below). The RefSeq map of Exon 12 shows the location where the capture reads are lost and the dotted redline runs through the intronic region for which we now see sequencing reads the deep intronic sequence is now so close to the capture region that it is part of the same library fragment and thus sequenced.
3.3. Characterization of false negative result
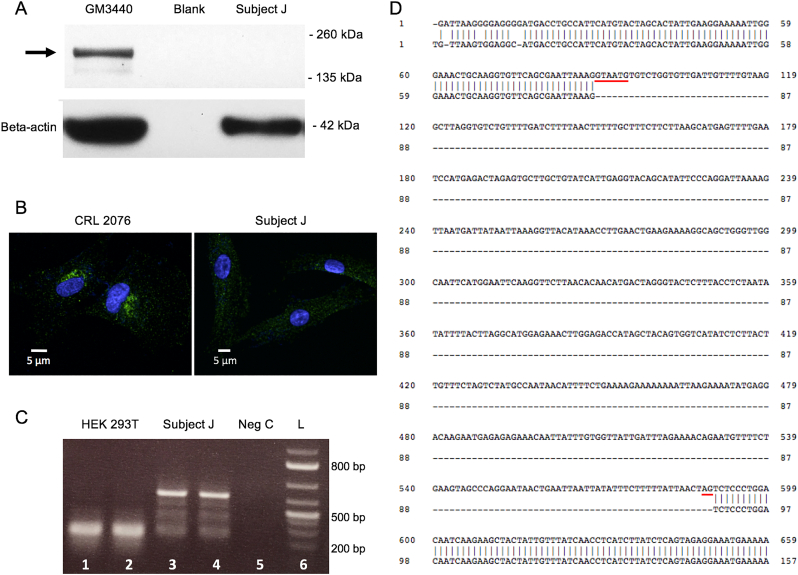
Subject J, like all other protocol 09-CH-0059 enrolled participants, manifested hallmark clinical and biochemical features of Menkes disease, including distinctively abnormal plasma catecholamine levels, a sensitive and specific diagnostic test for this condition [17,31]. No ATP7A variant was identified previously for this subject in commercial testing that included conventional Sanger sequencing of exons and array-based deletion/duplication studies, and neither did tNGS of the subject's DBS in this study. Western blots of fibroblast protein and confocal imaging indicated absence of the ATP7A gene product (Fig. 3a,b). Whole genome sequencing (New York Genome Center, New York, NY) suggested that deep intronic variations in ATP7A intron 2 coincided with an exon splice suppressor signal predicted to disturb mRNA splicing (data not shown). We then documented abnormal splicing in ATP7A transcripts obtained from this subject's fibroblasts, specifically, retention of the 501 bp intron 3 between the exon 3 and exon 4 sequences (Fig. 3c,d). This mutant cDNA sequence predicts insertion of 16 novel amino acids, followed by a nonsense codon and premature termination of translation. These findings suggest the molecular mechanism underlying this subject's Menkes disease phenotype.
Fig. 3.
Characterization of false negative result (Subject J).
A. Western blot of Subject J's cultured fibroblasts indicating absence of the normal ATP7A protein as seen in a normal control cell line (arrow), GM3440. Lower panel: beta-actin control for protein loading. B. Confocal microscopy of cultured fibroblasts from normal control cell line, CRL 2076, (left panel) and Subject J (right panel), stained with an anti-ATP7A antibody. Only the normal control cells show the perinuclear staining (green), consistent with ATP7A protein. C. Reverse transcription- polymerase chain reaction (RT-PCR) of ATP7A exons 3 and 4 shows a much larger transcript in fibroblasts from Subject J (lanes 3,4) compared to that in the normal cell line, CRL2076 (lanes 1,2). The negative control (lane 5) shows no ATP7A transcript, as expected. L = 100 bp DNA ladder (lane 6). D. Alignment of the complementary DNA (cDNA) sequence in Subject J (top) to the normal ATP7A exon 3 and 4 sequence (bottom) disclosed failure to splice exon 3 to exon 4 and insertion of 501 bp. The sequence of this insert matched precisely with that of ATP7A intron 3, including the consensus splice donor (gtaatg) and splice acceptor site (ag) sequences (underlined in red) [32]. Whole genome sequencing of this Subject's DNA (New York Genome Center, New York, NY) identified deep intronic variations in ATP7A intron 2 that coincide with an exon splice suppressor signal predicted to disturb mRNA splicing.
4. Discussion
Annually in the United States, NBS leads to early identification and treatment of 5000–8000 cases of inherited disease based on biochemical analytes recovered from DBS [7,33]. Identification of the residual burden of Mendelian diseases not covered by NBS is addressed ad hoc when phenotypes clinically present. This often involves long times to diagnosis and missed therapeutic opportunities. We have previously reported a 544 gene panel designed for rapid genomic diagnostic evaluation of common phenotypes presenting in the newborn period [8]. In this study, we applied this panel for proof-of-concept that a rapid tNGS based test could have sufficient analytic validity to consider adapting to high-throughput screening for detection of medically actionable genetic disorders. The panel includes conditions for which no suitable NBS analyte currently exists as a primary screening modality. We used a tNGS screening algorithm for Menkes disease on DNA extracted from DBS specimens and were able to assess for multiple mutation types in a single assay while fulfilling the rapid turnaround requirement for NBS [34]. The sensitivity of tNGS for ATP7A abnormalities was significantly increased by adding CNV detection capability since 12–20% of Menkes disease cases involve CNVs [35]. We found CNVs in eight samples, including one deletion missed by two commercial laboratories (Subject I). More than 95% of ATP7A variants in affected individuals were identified and classified correctly. This compares favorably with previous ATP7A variant analyses using more expensive and time-consuming approaches [30,35].
Menkes disease is not included in the current Recommended Uniform Screening Panel (RUSP)1 (Table 2), despite emerging evidence of medical actionability with neonatal diagnosis [17,[19], [20], [21], [22], [23]]. As the untreated illness evolves, abnormal clinical and biochemical phenotypes emerge in infants who are apparently healthy at birth [16]. In the absence of a positive family history, the correct diagnosis is usually not determined until 4–8 months of age. In addition, the rate of disease due to novel variants is high (Table 1). As the probands were not analyzed by segregation analysis and these did not overlap with known family history, we suspect that the mothers of probands are likely obligate carriers or the variant is a de novo variant in the proband. Identification of Menkes disease in the newborn period would not only improve neurodevelopmental outcomes, but also eliminate the need for uncomfortable and costly interventions and diagnostic tests (Table 1, Column 6).
Table 2.
Current use of DNA-based testing in newborn screening algorithms for disorders on 2018 RUSP panel.
| Disorder (RUSP) | 1st Tier Modality | Analyte/Activity | 2nd Tier Modality | Analyte | 3rd Tier Modality | Analyte | Gene |
|---|---|---|---|---|---|---|---|
| 3-Methylcrotonyl-CoA Carboxylase deficiency | MS/MS | C5OH | None | MCCC1MCCC2 | |||
| 3-methylglutaconic Acidemia, Type 1 | MS/MS | C5OH | None | AUH | |||
| aX-linked Adrenoleukodystrophy | MS/MS | C26:0 | MS/MS/HPLC | VLCFA | NGS | DNA | ABCD1 |
| Argininosuccinic aciduria | MS/MS | Citrulline | None | ASL | |||
| Beta-ketothiolase deficiency | MS/MS | C5:1 and C5OH | None | ACAT1 | |||
| Biotinidase deficiency | Enzyme assay | Biotinidase activity | None |
BTD |
|||
| Carnitine Uptake Defect | MS/MS | Free Carnitine, Total Acylcarnitines | None | SLC22A5 | |||
| Citrullinemia | MS/MS | Citrulline | None | ASS1 | |||
| Cobalamin A,B cofactor deficiency | MS/MS | C3 and C4DC | None | MMAA, MMAB | |||
| Cobalamin C, D cofactor deficiency | MS/MS | C3 and methionine | None | MMACHC, MMADHC | |||
| Congenital adrenal hyperplasia | ELISA | 17-OH progesterone | None | CYP21A2 | |||
| Congenital Hypothyroidism | ELISA | T4 | ELISA | TSH | N = 13 SeeTable S2 | ||
| aCystic Fibrosis | ELISA | IRT | PCR | DNA | NGS | DNA | CFTR |
| aGalactosemia | Enzyme assay | Galactose-1-Phosphate Uridyl Transferase | Enzyme assay | Gal—1P | PCR | DNA | GALT |
| Glutaric acidemia, type I | MS/MS | C5DC | None | GCDH | |||
| Homocystinuria | MS/MS | Methionine | None | CBS | |||
| Isovaleric acidemia | MS/MS | C5 | None | IVD | |||
| Long-chain 3-hydroxyacyl CoA dehydrogenase deficiency | MS/MS | C16OH and C18:1OH | None | HADHA | |||
| Maple Syrup Urine Disease | MS/MS | Leucine | None | BCKDHA, BCKDHB, DBT | |||
| aMedium-chain acyl-CoA dehydrogenase deficiency | MS/MS | C6 and C8 | PCR | DNA | ACADM | ||
| Methylmalonyl-CoA mutase deficiency | MS/MS | C3 and C4DC | None | MUT | |||
| Multiple carboxylase deficiency | MS/MS | C3 and C5OH | None | HLCS | |||
| Phenylketonuria | MS/MS | Phenylalanine | None | PAH | |||
| aPompe disease | MS/MS | Acid alpha-glucosidase | NGS | DNA | GAA | ||
| Propionic acidemia | MS/MS | C3 | None | PCCA, PCCB | |||
| aSevere combined immunodeficiency | qPCR | TREC (DNA) | None | N = 16 SeeTable S2 | |||
| Trifunctional protein deficiency | MS/MS | C16OH and C18:1OH | None |
HADHA, HDHB |
|||
| Tyrosinemia type I | MS/MS | Succinylacetone | None | FAH | |||
| Very long-chain acyl-CoA dehydrogenase deficiency | MS/MS | C14 and C14:1 | None | ACADVL | |||
| aMucopolysaccharidosis Type I | MS/MS | IDUA | NGS | DNA | IDUA | ||
| aSickle cell anemia, aHgbS/βeta-thalassemia, aHgbS/HgbC disease | HPLC/IEF | Hemoglobin | PCR/NGS | DNA | HBB, HBA1, HBA2 | ||
| aCongenital hearing loss | Hearing screen | EOAE/BAER | NGS | DNA | N = 106 SeeTable S3 | ||
| bSpinal muscular Atrophy | qPCR | DNA | None | SMN1, SMN2 |
BAER: Brainstem Auditory Evoked Responses; ELISA: Enzyme-linked Immunosorbent Assay; EOAE: Evoked Otoacoustic Emissions; HPLC: High Performance Liquid Chromatography; IEF: Isoelectric Focusing; MS/MS: Tandem Mass Spectroscopy; NGS: Next Generation Sequencing; RUSP: Recommended Uniform Screening Panel; VLCFA: Very Long Chain Fatty Acids; qPCR: quantitative PCR.
Disorders for which DNA-based tests are currently used for 1st tier non-sequencing evaluation (SCID, SMA), 2nd or 3rd tier genotyping, or sequencing of NBS presumptive positives.
Currently in pilot newborn screening evaluation.
Based on the workflow described, we estimate high-throughput tNGS would have a per sample reagent cost ranging from $95–200 for a 200 gene panel. This general cost paradigm meets the desired goals of $1 per NBS test per condition and remains below the population-based screening target of $5 per condition [7]. If, in addition to providing a primary screen for actionable conditions lacking measurable biochemical analytes, a tNGS run can provide data that can be utilized to replace confirmatory second or third-tier DNA testing for the current RUSP conditions, considerable new cost savings could accrue. Eleven of 35 RUSP disorders currently use DNA in their test algorithms (Table 2). We estimate that more than 200 genetic loci may be implicated in the molecular bases of these 35 conditions (Table S2, Table S3).
Cystic fibrosis (CF) NBS that incorporates tNGS has been performed for several years through the New York State CF Newborn Screening Program, providing evidence for technical feasibility of seqeuencing in a public health laboratory setting [36]. All NBS algorithms for CF measure immunoreactive trypsinogen (IRT) levels as a first-tier test. Infants with an elevated IRT level typically undergo second-tier testing with a DNA panel of common variants in CFTR, the CF gene [37,38]. If this second-tier test is inconclusive, sweat chloride testing and/or diagnostic sequencing of the entire CFTR gene is needed. Reagent costs for such tiered testing range from $220 to $1320 per sample [38]. Alternatively, inclusion of CFTR in a tNGS-based NBS panel would enable immediate molecular evaluation to confirm or exclude the diagnosis in infants with elevated IRT, precluding additional test costs. For other conditions, such as severe combined immunodeficiency (SCID), congenital hypothyroidism, and congenital deafness, whose etiologies are heterogeneous (Table S2, Table S3), having tNGS data available immediately also would shorten time to diagnosis and therapy [39]. Associated genes for second-tier testing can be multiplexed in a tNGS panel for essentially the same cost as sequencing for Menkes disease alone, or even Menkes disease plus first-tier tNGS for other treatable unscreened conditions (Table S4).
The specific cost benefit of NBS for Menkes disease includes elimination of the extensive testing often involved in diagnostic odysseys of many affected infants (Table 1, Column 6). Evidentiary principles for clinical validity and clinical utility of genetic testing recently summarized by the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative [40] are fulfilled for ATP7A testing, based on multiple sources of evidence [17,[19], [20], [21], [22], [23],29,31], as well as 2018 FDA designation of Fast Track status for Copper Histidinate treatment for patients diagnosed with classic Menkes disease who have not demonstrated significant clinical progression [41]. All infants found on NBS with ATP7A variants would be evaluated by plasma catecholamine analysis, an inexpensive and highly sensitive and reliable assay [17,31,[42], [43], [44]] to exclude a false positive diagnosis of Menkes disease. The ethical and social aspects (and consequences) of missed early diagnosis of Menkes disease have also been reviewed [45].
Presumptive positive or borderline NBS results for all screened biochemical disorders combined were reported in 3.5% of all Florida newborns screened in both 2016 and 2017 [46]. Using these data as estimates for the entire US, where annual births approximate 3.9 million, nearly 150,000 US newborns annually may require molecular testing for second or third-tier follow-up testing for confirmation of NBS positive results and resolving false positives. The considerable cost of such testing would be averted or reduced by the immediate reflexive availability of molecular information from a tNGS screen performed routinely as a part of newborn screening at birth. Irrespective of whether all public health laboratories currently have sequencing capabilities, NBS may be the only way to provide molecular information in an equitable manner for early determination and treatment.
Concern about generating false-positive results exists for all NBS algorithms. A review of ATP7A SNVs in the gnomAD database of 141,456 unrelated individuals revealed 452 singleton ATP7A missense variants that could be construed as possibly deleterious (VUS) if found during NBS [15,26]. Based on these data, we estimate the maximum probability of a false-positive from an ATP7A singleton variant to be approximately 0.2%, as is supported by the absence of findings in newborns screened with whole exome sequencing in the BabySeq Project (Genomic Sequencing for Childhood Risk and Newborn Illness, U19 HD077671) [27,28]. While plasma catecholamine measurement may not be suitable as a first tier-screen, it provides an ideal potential second-tier follow-up test for presumed positive results for Menkes disease based on NBS tNGS identification of ATP7A variants [17,31,[42], [43], [44]]. For inherited disorders currently screened, false positive rates in the range of 0.1–0.2% are commonly observed [33,34]. Based on the current results, and previous ATP7A variant analyses [35], we also anticipate the false negative rate among true cases of Menkes disease to be less than 5%. Thus, at a population level, the estimated false positive and false negative rates that would add to the burden of overall NBS can be kept within the ranges considered acceptable for NBS.
The one false negative from the Menkes disease cohort (Subject J) required whole genome sequencing and confirmatory analysis of mRNA splicing in the patient's cells to unravel the etiology. Known pathogenic deep intronic variants are relatively rare in most genes, and are not covered by tNGS design, which focuses on coding regions. tNGS reliably detects the most exonic and splice junction variants; therefore, tNGS screen failures (false negatives) are expected to be uncommon. Based on incidence rates and false negative cases in the cohort, we estimate 1 in 1.9 million screens would miss detection for Menkes disease. The finding also underscores the importance of developing other molecular methods such as RNA sequencing, functional studies, or biochemical assays that could reduce false negatives.
Although we used a 544-gene panel for this exercise in evaluating analytic validity, all the conditions may not be appropriate for NBS. In clinical practice, a tailored tNGS panel could range between 30 and 100 conditions. In an evaluation of 14,821 exomes to determine a “hit rate” for 30 heritable conditions for which biochemical screening tests are not available, the hit rate was approximately 0.1% (M. Bainbridge, personal communication). Thus, there is considerable room for expansion of DNA-based screening as a first-tier tNGS NBS test.
This work confirms that targeted NGS as a first-tier DNA based test in NBS represents an approach by which the molecular bases of multiple medically-actionable diseases, including Menkes disease, can be assessed rapidly in a single efficient assay [47]. Reduced health care costs could be realized by avoiding the illnesses and diagnostic odysseys of unrecognized affected infants, as well as removing the need for later molecular testing for diagnostic confirmation of biochemically screened genetic disorders. As such, tNGS can provide a useful supplement to tandem mass spectrometry and enhance current NBS efforts. For Menkes specifically, the ten Wilson and Jungner principles of screening for disease are fulfilled (Table 3) [48].
Table 3.
Evidence that Menkes disease fulfills Wilson and Jungner Principles [48].
| Principle | Evidence |
|---|---|
| 1. The condition represents an important health problem. | Menkes disease represents an important health problem when unrecognized and untreated [16]. |
| 2. There should be an accepted treatment for patients with recognized disease. | Evidence from clinical trials indicate that early (within 28 days of birth, corrected for prematurity) CuHis therapy enhances survival as well as quality of clinical outcome in classic Menkes disease [17,[19], [20], [21], [22], [23]]. Based on these results, the FDA granted Fast Track Designation to CuHis in June 2018. |
| 3. Facilities for diagnosis and treatment should be available. | There is a Menkes Disease Clinic established at Nationwide Children's Hospital, Columbus OH (https://www.nationwidechildrens.org/specialties/menkes-disease-clinic). Other university-based pediatric medical centers are also available for diagnosis and treatment of this illness. |
| 4. There should be a recognizable latent or early symptomatic stage. | Infants affected with Menkes disease are neurologically asymptomatic in the neonatal period, and remain so for six to eight weeks after birth [16]. |
| 5. There should be a suitable test or examination. | Both the plasma neurochemical assay and ATP7A gene sequencing are suitable and available for diagnosis of Menkes disease [30,31,41,42]. |
| 6. The test should be acceptable to the population. | Newborn screening is widely accepted by parents in the US and world-wide as an effective approach to reducing the burden of inherited metabolic diseases [2,4,5,33,46]. |
| 7. The natural history of the condition, including development from latent to declared disease, should be adequately understood. | The medical literature on Menkes disease is quite clear in this regard, based on publications dating to the original description of the phenotype (Menkes JH et al., J Pediatr. 1962). |
| 8. There should be an agreed policy on whom to treat as patients. | There is general acceptance in the Biochemical Genetics provider community that asymptomatic newborns known to have the diagnosis of Menkes disease should be treated with CuHis injections. |
| 9. The cost of case-finding (including diagnosis and treatment of patients diagnosed) should be economically balanced in relation to possible expenditure on medical care as a whole. | While the exact costs of early diagnosis and three years treatment with CuHis are not known, they appear unlikely to exceed the costs of extensive diagnostic odysseys, hospitalizations, anti-convulsive medications, and other health care needs of untreated affected individuals. |
| 10. Case-finding should be a continuing process and not a “once and for all” project. | Newborn screening for Menkes disease would represent a continuing process, not a single occasion. |
Acquisition of DNA sequences at birth may also benefit individuals at-risk for illness later in life and help establish robust genotype-phenotype correlations in other medically actionable newborn diseases yet to be screened.
The following are the supplementary data related to this article.
Complete gene list included in tNGS panel (n = 544)
119 genes from Table S1 associated with disorders currently covered by NBS
106 genes associated with congenital hearing loss from the 544 Gene tNGS Panel.
Genes from 544 Gene tNGS panel associated with actionable newborn disorders not currently screened by NBS
Compliance with ethics guidelines
Informed Consent: All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained for all patients included in the study.
Details of the contributions of individual authors
AB, RBP and SGK contributed to the conception, planning, conduct, and reporting of the overall work described in the article. TS coordinated sample processing and performed variant calling. EM developed the CNV algorithm, managed the interpretation pipeline, and implemented informatics workflows. LY characterized the false negative result. AB, RBP, EM and SGK wrote the manuscript. SGK managed patients, blinded the patient samples, and oversaw call concordance.
Declaration of Competing Interest
Dr. Kaler's laboratory received a Collaborative Research and Development Award (CRADA) entitled “Development of Combination Copper Plus AAV-based ATP7A-targeted Gene Therapy for Menkes Disease” from Cyprium Therapeutics, Inc., New York, NY in 2017. Cyprium is focused on development of novel therapies for the treatment of Menkes disease and related copper metabolism disorders.
Acknowledgements
This work was supported by the National Institutes of Health Intramural Research Program (Z01 HD008768) (SGK, LY), and a grant from The Menkes Foundation (California, MD USA) to AB. RP was supported by a National Institutes of Health Grant (U19HD077671). We thank the BabySeq project Executive Committee and Ozge Ceyhan-Birsoy for review and sharing of ATP7A data.
References
- 1.Urv T.K., Parisi M.A. Newborn screening: beyond the spot. Adv. Exp. Med. Biol. 2017;1031:323–346. doi: 10.1007/978-3-319-67144-4_19. [DOI] [PubMed] [Google Scholar]
- 2.Levy H.L. Newborn screening by tandem mass spectrometry: a new era. Clin. Chem. 1998;44:2401–2402. [PubMed] [Google Scholar]
- 3.American College of Medical Genetics/American Society of Human Genetics Test and Technology Transfer Committee Working Group Tandem mass spectrometry in newborn screening. Genet. Med. 2000;2:267–269. doi: 10.1097/00125817-200007000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Wilcken B., Wiley V., Hammond J., Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N. Engl. J. Med. 2003;348:2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 5.Berg J.S., Agrawal P.B., Bailey D.B., Jr. Newborn sequencing in genomic medicine and public health. Pediatrics. 2017;139 doi: 10.1542/peds.2016-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boemer F., Fasquelle C., d'Otreppe S. A next-generation newborn screening pilot study: NGS on dried blood spots detects causal mutations in patients with inherited metabolic diseases. Sci. Rep. 2017;7:17641. doi: 10.1038/s41598-017-18038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botkin J.R., Rothwell E. Whole genome sequencing and newborn screening. Curr. Genet. Med. Rep. 2016;4:1–6. doi: 10.1007/s40142-016-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharjee A., Sokolsky T., Wyman S.K. Development of DNA confirmatory and high- risk diagnostic testing for newborns using targeted next-generation DNA sequencing. Genet. Med. 2015;17:337–347. doi: 10.1038/gim.2014.117. [DOI] [PubMed] [Google Scholar]
- 9.Peng G., Shen P., Gandotra N., Le A., Fung E., Jelliffe-Pawlowski L., Davis R.W., Enns G.M., Zhao H., Cowan T.M., Scharfe C. Combining newborn metabolic and DNA analysis for second-tier testing of methylmalonic acidemia. Genet. Med. 2019;21(4):896–903. doi: 10.1038/s41436-018-0272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X., Wang R., Fan Y., Gu X., Yu Y. Next-generation sequencing as a second-tier diagnostic test for newborn screening. J. Pediatr. Endocrinol. Metab. 2018;31(8):927–931. doi: 10.1515/jpem-2018-0088. [DOI] [PubMed] [Google Scholar]
- 11.Ko J.M., Park K.S., Kang Y., Nam S.H., Kim Y., Park I., Chae H.W., Lee S.M., Lee K.A., Kim J.W. A new integrated newborn screening workflow can provide a shortcut to differential diagnosis and confirmation of inherited metabolic diseases. Yonsei Med. J. 2018;59(5):652–661. doi: 10.3349/ymj.2018.59.5.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Mousa H., Al-Dakheel G., Jabr A., Elbadaoui F., Abouelhoda M., Baig M., Monies D., Meyer B., Hawwari A., Dasouki M. High incidence of severe combined immunodeficiency disease in Saudi Arabia detected through combined T cell receptor excision circle and next generation sequencing of newborn dried blood spots. Front. Immunol. 2018;9:782. doi: 10.3389/fimmu.2018.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smon A., Repic Lampret B., Groselj U., Zerjav Tansek M., Kovac J., Perko D., Bertok S., Battelino T., Trebusak Podkrajsek K. Next generation sequencing as a follow-up test in an expanded newborn screening programme. Clin. Biochem. 2018;52:48–55. doi: 10.1016/j.clinbiochem.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Qian J., Wang X., Liu J., Zhong J., Le Y., Melchior Tellier L.C.A., Liu C., Jiang P., Gao R., Wang Y. Applying targeted next generation sequencing to dried blood spot specimens from suspicious cases identified by tandem mass spectrometry-based newborn screening. J. Pediatr. Endocrinol. Metab. 2017;30(9):979–988. doi: 10.1515/jpem-2017-0003. [DOI] [PubMed] [Google Scholar]
- 15.Kaler S.G., Ferreira C., Yam L. Estimated birth prevalence of Menkes disease and ATP7A- related disorders based on the Genome Aggregation Database (gnomAD) Mol. Genet. Metab. Rep. 2020;24:100602. doi: 10.1016/j.ymgmr.2020.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaler S.G. Menkes disease. Adv. Pediatr. 1994;41:263–304. [PubMed] [Google Scholar]
- 17.Kaler S.G., Holmes C.S., Goldstein D.S. Neonatal diagnosis and treatment of Menkes disease. N. Engl. J. Med. 2008;358:605–614. doi: 10.1056/NEJMoa070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaler S.G. ATP7A-related copper transport diseases-emerging concepts and future trends. Nat. Rev. Neurol. 2011;7:15–29. doi: 10.1038/nrneurol.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaler S.G. Neurodevelopment and brain growth in classic Menkes disease is influenced by age and symptomatology at initiation of copper treatment. J. Trace Elem. Med. Biol. 2014;28:427–430. doi: 10.1016/j.jtemb.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaler S.G., Buist N.R.M., Holmes C.S., Goldstein D.S., Miller R.C., Gahl W.A. Early copper therapy in classic Menkes disease patients with a novel splicing mutation. Ann. Neurol. 1995;38:921–928. doi: 10.1002/ana.410380613. [DOI] [PubMed] [Google Scholar]
- 21.Kaler S.G., Das S., Levinson B., Goldstein D.S., Holmes C.S., Patronas N.J., Packman S., Gahl W.A. Successful early copper therapy in Menkes disease associated with a mutant transcript containing a small in-frame deletion. Biochem. Mol. Med. 1996;57:37–46. doi: 10.1006/bmme.1996.0007. [DOI] [PubMed] [Google Scholar]
- 22.Tang J., Donsante A., Desai V., Patronas N., Kaler S.G. Clinical outcomes in Menkes disease patients with a copper-responsive ATP7A mutation, G727R. Mol. Genet. Metab. 2008;95:174–181. doi: 10.1016/j.ymgme.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaler S.G., Liew C.J., Donsante A., Hicks J.D., Sato S., Greenfield J.C. Molecular correlates of epilepsy in early diagnosed and treated Menkes disease. J. Inherit. Metab. Dis. 2010;33:583–589. doi: 10.1007/s10545-010-9118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donsante A., Yi L., Zerfas P. ATP7A gene addition to the choroid plexus results in long-term rescue of the lethal copper transport defect in a Menkes disease mouse model. Mol. Ther. 2011;19:2114–2123. doi: 10.1038/mt.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haddad M.R., Choi E.Y., Zerfas P. Cerebrospinal fluid-directed rAAV9-rsATP7A plus subcutaneous copper Histidinate advance survival and outcomes in a Menkes mouse model. Mol. Ther. Methods Clin. Dev. 2018;10:165–178. doi: 10.1016/j.omtm.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski K.J., Francioli L.C., Tiao G. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. (preprint) [DOI] [Google Scholar]
- 27.Ceyhan-Birsoy O., Machini K., Lebo M.S. A curated gene list for reporting results of newborn genomic sequencing. Genet. Med. 2017;19:809–818. doi: 10.1038/gim.2016.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm I.A., Agrawal P.B., Ceyhan-Birsoy O., Christensen K.D., Fayer S., Frankel L.A., Genetti C.A., Krier J.B., RC LaMay, Levy H.L., AL McGuire, Parad R.B., Park P.J., Pereira S., Rehm H.L., Schwartz T.S., Waisbren S.E., Yu T.W., Team BabySeq Project, Green R.C., Beggs A.H. The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr. 2018;18(1):225. doi: 10.1186/s12887-018-1200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaler S.G., Tang J.R., Donsante A., Kaneski C. Translational read-through of a nonsense mutation in ATP7A. Ann. Neurol. 2009;65:108–113. doi: 10.1002/ana.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P.-C., McAndrew P.E., Kaler S.G. Rapid and robust screening of the Menkes disease /occipital horn syndrome gene. Genet. Test. 2002;6:255–260. doi: 10.1089/10906570260471778. [DOI] [PubMed] [Google Scholar]
- 31.Kaler S.G., Gahl W.A., Berry S.A., Holmes C.S., Goldstein D.S. Predictive value of plasma catecholamine levels in neonatal detection of Menkes disease. J. Inherit. Metab. Dis. 1993;16:907–908. doi: 10.1007/BF00714295. [DOI] [PubMed] [Google Scholar]
- 32.Kaler S.G., Gallo L.K., Proud V.K. Occipital horn syndrome and a mild Menkes phenotype associated with splice site mutations at the MNK locus. Nat. Genet. 1994;8:195–202. doi: 10.1038/ng1094-195. [DOI] [PubMed] [Google Scholar]
- 33.Howell R.R., Terry S., Tait V.F. CDC grand rounds: newborn screening and improved outcomes. MMWR Morb. Mortal. Wkly Rep. 2012;61(21):390–393. [PubMed] [Google Scholar]
- 34.Tang H., Feuchtbaum L., Neogi P., Ho T., Gaffney L., Currier R.J. Damaged goods?: an empirical cohort study of blood specimens collected 12 to 23 hours after birth in newborn screening in California. Genet. Med. 2016;18(3):259–264. doi: 10.1038/gim.2015.154. [DOI] [PubMed] [Google Scholar]
- 35.Tümer Z., Birk Møller L., Horn N. Screening of 383 unrelated patients affected with Menkes disease and finding of 57 gross deletions in ATP7A. Hum. Mutat. 2003;22:457–464. doi: 10.1002/humu.10287. [DOI] [PubMed] [Google Scholar]
- 36.Kay D.M., Maloney B., Hamel R., Pearce M., DeMartino L., McMahon R., McGrath E., Krein L., Vogel B., Saavedra-Matiz C.A., Caggana M., Tavakoli N.P. Screening for cystic fibrosis in New York state: considerations for algorithm improvements. Eur. J. Pediatr. 2016 Feb;175(2):181–193. doi: 10.1007/s00431-015-2616-3. (Epub 2015 Aug21. PubMed PMID: 26293390) [DOI] [PubMed] [Google Scholar]
- 37.Baker M.W., Atkins A.E., Cordovado S.K., Hendrix M., Earley M.C., Farrell P.M. Improving newborn screening for cystic fibrosis using next-generation sequencing technology: a technical feasibility study. Genet. Med. 2016;18:231–238. doi: 10.1038/gim.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Currier R.J., Sciortino S., Liu R., Bishop T., Alikhani Koupaei R., Feuchtbaum L. Genomic sequencing in cystic fibrosis newborn screening: what works best, two-tier predefined CFTR mutation panels or second-tier CFTR panel followed by third-tier sequencing? Genet. Med. 2017;19:1159–1163. doi: 10.1038/gim.2017.32. [DOI] [PubMed] [Google Scholar]
- 39.Smith L.D., Bainbridge M.N., Parad R.B., Bhattacharjee A. Second tier molecular genetic testing in newborn screening for Pompe disease: landscape and challenges. Int. J. Neonatal Screen. 2020 Jun;6(2) doi: 10.3390/ijns6020032. pii: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.National Academies of Sciences, Engineering, and Medicine . The National Academies Press; Washington, DC: 2017. An Evidence Framework for Genetic Testing. [DOI] [PubMed] [Google Scholar]
- 41.https://www.fda.gov/drugs/ind-activity/fast-track-designation-requests
- 42.Kaler S.G., Goldstein D.S., Holmes C., Salerno J.A., Gahl W.A. Plasma and cerebrospinal fluid neurochemical pattern in Menkes disease. Ann. Neurol. 1993;33:171–175. doi: 10.1002/ana.410330206. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein D.S., Holmes C.S., Kaler S.G. Relative efficiencies of plasma catechol levels and ratios for neonatal diagnosis of menkes disease. Neurochem. Res. 2009 Aug;34(8):1464–1468. doi: 10.1007/s11064-009-9933-8. (Epub 2009 Feb 21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vairo F.P.E., Chwal B.C., Perini S., Ferreira M.A.P., de Freitas Lopes A.C., Saute J.A.M. A systematic review and evidence-based guideline for diagnosis and treatment of Menkes disease. Mol. Genet. Metab. 2019 Jan;126(1):6–13. doi: 10.1016/j.ymgme.2018.12.005. (Epub 2018 Dec 11) [DOI] [PubMed] [Google Scholar]
- 45.Sheela S.R., Manoj L., Liu P.-C., Lem K.E., Kaler S.G. Copper replacement treatment for symptomatic Menkes disease: ethical considerations. Clin. Genet. 2005;68:278–283. doi: 10.1111/j.1399-0004.2005.00496.x. [DOI] [PubMed] [Google Scholar]
- 46.State of Florida NBS Yearly Statistics, 2013-2017 . February 23, 2018. Florida Genetics and Newborn Screening Advisory Council Public Meeting, Jacksonville, FL. [Google Scholar]
- 47.van Campen J.C., Sollars E.S.A., Thomas R.C., Bartlett C.M., Milano A., Parker M.D., Dawe J., Winship P.R., Peck G., Grafham D., Kirk R.J., Bonham J.R., Goodeve A.C., Dalton A. Next generation sequencing in newborn screening in the United Kingdom National Health Service. Int. J. Neonatal Screen. 2019 Dec;5(4):40. doi: 10.3390/ijns5040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson J.M.G., Jungner G. Public Health Papers No. 34. WHO; Geneva: 1968. Principles and practice of screening for disease. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete gene list included in tNGS panel (n = 544)
119 genes from Table S1 associated with disorders currently covered by NBS
106 genes associated with congenital hearing loss from the 544 Gene tNGS Panel.
Genes from 544 Gene tNGS panel associated with actionable newborn disorders not currently screened by NBS