Abstract
Objectives
Studies are needed to better understand the genomic evolution of the recently emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This study aimed to describe genomic diversity of SARS-CoV-2 by next-generation sequencing (NGS) in a patient with longitudinal follow-up for SARS-CoV-2 infection.
Methods
Sequential samples collected between January 29th and February 4th, 2020, from a patient infected by SARS-CoV-2 were used to perform amplification of two genome fragments—including genes encoding spike, envelope, membrane and nucleocapsid proteins—and NGS was carried out with Illumina® technology. Phylogenetic analysis was performed with PhyML and viral variant identification with VarScan.
Results
Majority consensus sequences were identical in most of the samples (5/7) and differed in one synonymous mutation from the Wuhan reference sequence. We identified 233 variants; each sample harboured in median 38 different minority variants, and only four were shared by different samples. The frequency of mutation was similar between genes and correlated with the length of the gene (r = 0.93, p = 0.0002). Most of mutations were substitution variations (n = 217, 93.1%) and about 50% had moderate or high impact on gene expression. Viral variants also differed between lower and upper respiratory tract samples collected on the same day, suggesting independent sites of replication of SARS-CoV-2.
Conclusions
We report for the first time minority viral populations representing up to 1% during the course of SARS-CoV-2 infection. Quasispecies were different from one day to the next, as well as between anatomical sites, suggesting that in vivo this new coronavirus appears as a complex and dynamic distributions of variants.
Keywords: Infection follow-up, Minority variants, NGS, Quasispecies, SARS-CoV-2
Introduction
The genome organization in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is similar to that in the other betacoronaviruses, with the open reading frame (ORF) 1a/b encoding non-structural proteins at the 5′-end, and structural proteins as follows: spike (S)– envelope (E)–membrane (M)–nucleocapsid (NC)–3′-end [1]. Since the spike surface glycoprotein plays a major role in infection of the host cell, genomic variations may impact the interaction with the host receptor but also viral pathogenesis, transmissibility and infectivity [2].
As intra-host variants in a transversal study or from the same patient by nanopore sequencing have already been reported [3,4], this study aimed to describe genomic diversity of SARS-CoV-2 by next-generation sequencing (NGS) in a patient with longitudinal follow-up for SARS-CoV-2 infection.
Methods
The first patient diagnosed with SARS-CoV-2 infection in Pitié-Salpêtrière Hospital, Paris, France, was followed daily for SARS-CoV-2 by RT-PCR of respiratory samples; viral genome could be detected between January 29th and February 10th, 2020 [5]. This patient, hospitalized on day 2 of a mild form of coronavirus disease 2019 (Covid-19), did not receive any antiviral or immunomodulation treatment during the entire study period.
Two fragments of about 4000 nucleotides (nt) were amplified by nested PCR (Supplementary Material Table S1), and NGS was performed with paired-end reads (MiSeq v3, 2 x 300 bp) on the MiSeq Illumina® system. Reads were trimmed using Trimmomatic, then mapped on SARS-CoV-2 reference sequence (NC_045512.2) with Geneious Prime software and finally assembled de novo with SPAdes 3.12.0 [6] to generate majority consensus sequences.
Multiple alignment was performed with Mafft7 [7] and phylogenetic analysis of S, E, M and NC genes with PhyML3.0 [8] and GTR substitution model with 1000 bootstraps resampling.
Intra-host variants were called using VarScan [9] with the following requirements: sequencing depth ≥1000, minor allele frequency ≥1% and found at least 100 times. Intra-sample viral variants were studied by comparing each consensus sequence with all cleaned reads generated from the same sample and viral variants during follow-up by comparing consensus sequence of the first nasopharyngeal sample (01292020_NP) with all reads generated from the different samples. Synonymous mutations were identified as having a low impact, missense mutations and insertions with conservative inframe as having a moderate impact, and acquisition or loss of stop codon as well as frameshift as having a high impact on gene expression.
The Spearman rank correlation test was performed on GraphPad.
Results
The sequencing was effective for the first seven samples (one induced sputum and six nasopharyngeal swabs) from January 29th to February 4th, 2020, with a Ct value of SARS-CoV-2 RT-PCR <30. A full-length fragment of 8257 nt was generated with a median (IQR) of 45 523 (41 014–46 023) depth sequencing per sample (Supplementary Material Table S2).
Phylogenetic analysis
Compared to the NC_045512.2 reference sequence, our majority consensus sequences differed in the S gene by only four variations. They all harboured the synonymous mutation 3591T > C, whereas a non-synonymous mutation (859G > A) was found only in sample 02012020_NP. In sample 01312020_NP, a deletion of one nucleotide led to the appearance of a premature stop codon and a non-synonymous substitution at position 3554.
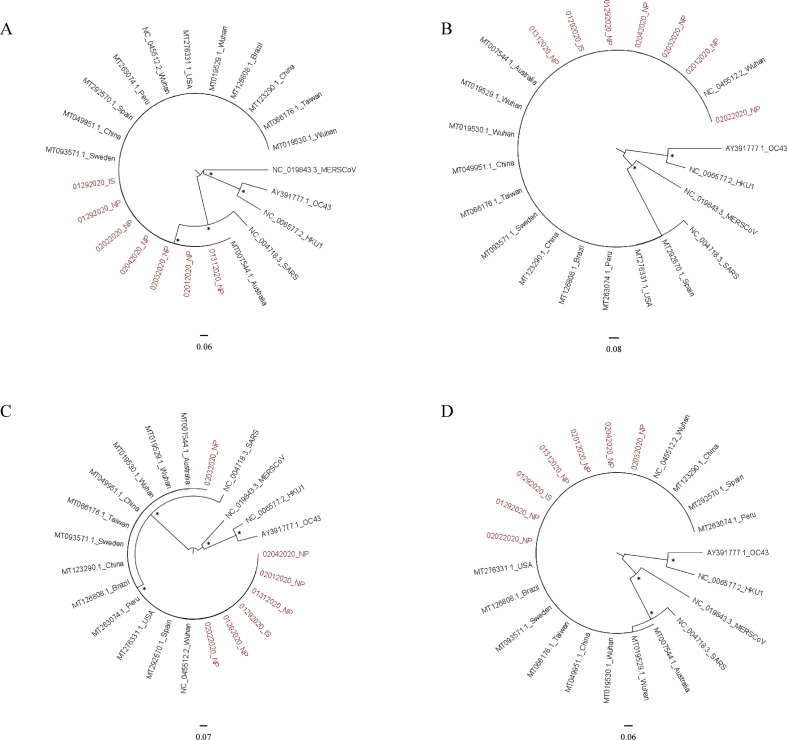
By phylogenetic analysis of the four structural genes, our sequences clustered with all the SARS-CoV-2 reference sequences issued from the NCBI database, and were distinct from the other human coronaviruses (Supplementary Material Fig. S1).
Intra-sample viral variant diversity
We identified 233 viral variants, and the number of variants per sample was not correlated with the depth sequencing (r = 0.23, p = 0.28).
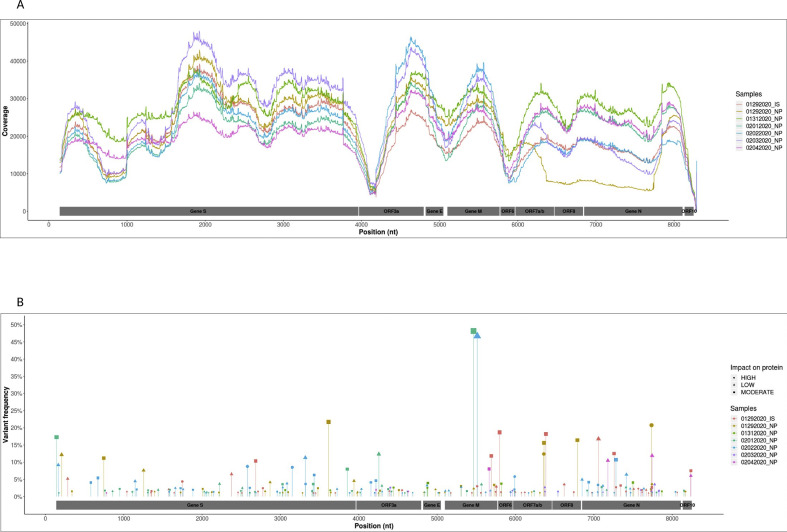
Each sample harboured in median 38 (11–51.5) minority variants (<20%). Only 4/233 identical minority variants were common between two specimens, and 6/233 other mutations were identified at the same position in two samples but induced different variants (Supplementary Material Table S3). Although majority consensus sequences of the two specimens collected on January 29th were strictly identical, each one harboured their specific viral population with 59 variants identified in the induced sputum and 40 in the nasopharyngeal specimens (Fig. 1 ).
Fig. 1.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome diversity during infection. (A) Genome coverage (y axis) according to nucleotide position (x axis). (B) Distribution (x axis) and frequency (y axis) of the 233 intra-sample viral variants identified. Each sample is represented by the same colour in (A) and (B), and the impact of mutations on gene expression is represented by a different symbol (low: a rhombus, moderate: a square, high: a circle).
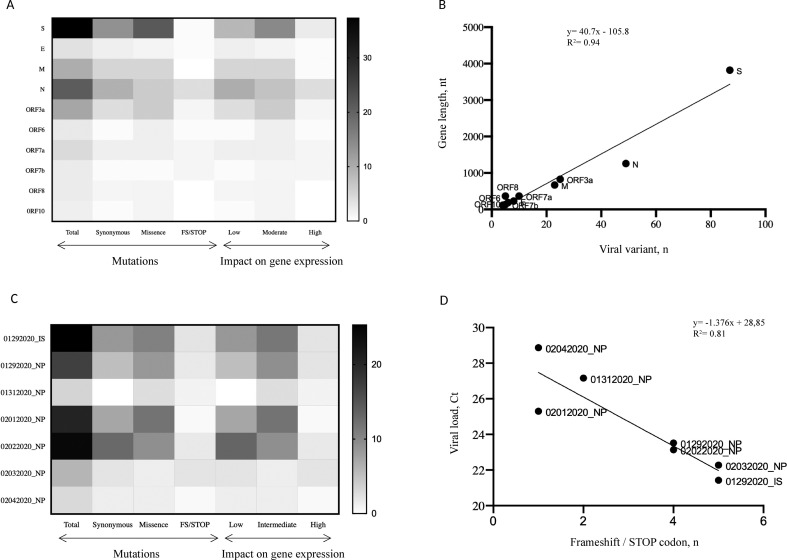
Nucleotide variations occurred in decreasing order in the S gene, N gene, ORF3a, M gene, ORF7a, E gene, ORF6, ORF7b and ORF8, and finally ORF10 (Fig. 2 A). However, according to gene length, the frequency of mutation was similar and correlated with the length of the gene (r = 0.93, p = 0.0002) (Fig. 2B).
Fig. 2.
Distribution of mutation frequency and correlation with gene length or viral load. (A) HeatMap representing the frequency and distribution of the mutations and their impact on gene expression between the different genes. (B) Linear regression line between the number of viral variant (x axis) and the gene length (y axis). (C) HeatMap representing the frequency and distribution of the mutations and their impact on gene expression between the different samples. (D) Linear regression line between the number of frameshift and stop codons (x axis) and the viral load expressed in cycle threshold (Ct) value (y axis). Viral variants by gene: S, n = 87; N, n = 49; ORF3a, n = 25; M, n = 23; ORF7a, n = 10; E, n = 8; ORF6, n = 6; ORF7b, n = 5; ORF8, n = 5; ORF10, n = 4. Scale on the right of (A) and (C) represents the frequency in percentages, with the largest value in dark and the lowest value in light.
Most of the mutations were substitution variations (217/233), including 87/233 synonymous mutations and 107/233 missense mutations. According to gene expression, 88/233 variants had a low impact, 111/233 an intermediate impact, and 23/233 a high impact (Fig. 1). Between samples, only the frequencies of frameshift and stop codons were significantly and strongly correlated with the viral load (r = 0.92, p = 0.0095) (Fig. 2D).
Follow-up of viral variant diversity
By comparing with the consensus sequence collected on January 29th from the nasopharyngeal site, we found the same viral quasispecies in each sample as reported above. However, three majority variants emerged in the S gene obtained from the nasopharyngeal samples collected on January 31st and February 1st, corresponding to the three mutations described previously in the consensus sequences. None of them were found in the previous and following samples as majority or minority variants (Supplementary Material Table S4).
Discussion
The virus identified in this patient was almost identical to the reference sequence from Wuhan [1]. This result was expected, as the patient was a general practitioner presumably infected by tourists from Wuhan and their guide who was later diagnosed SARS-CoV-2-positive [5].
Quasispecies in RNA viruses have previously been reported for SARS-CoV and MERS-CoV [10,11], as well as within individuals during SARS-CoV-2 infection [3,12]. The present study, allowing the analysis of SARS-CoV-2 minority variants at 1%, supports the previous finding. Indeed, we found a median of 38 different viral variants per sample during the follow-up of a single patient, with almost no common variant from one day to the next. More than half of the variants had an intermediate or high impact on gene expression and may explain the lack of persistence over time. Among the different types of mutations, the number of mutations inducing frameshift and stop codons were highly correlated with the viral load, reflecting the loss of fitness in variants harbouring deleterious mutations during intensive viral replication [13]. Otherwise, the viral variant population was also different between samples from the lower (induced sputum) and upper (nasopharyngeal swab) respiratory tract collected on the same day, suggesting independent replication of SARS-CoV-2, as previously reported [14].
Contrary to a previous study which identified a hotspot in ORF8 [15], the mutations identified in this study appeared to be spread fairly evenly throughout the sequenced fragment. Indeed, a limited number of viral variants was shared by two samples, the remainder (97%) being specific to each sample and occurring in different genomic sites, and a strong correlation was found between the number of variants and the length of each gene.
The main limitation of this study is that a fragment of only about 8000 nt was studied, in only one patient, and during a short period of follow-up because of low viral load in samples collected after February 5th. However, our results highlighted that during the first week of infection the major viral population remained identical (5/7), with several specific minority variants which did not seem to persist over time. Larger studies are needed to explore the entire intra-patient variability during the course of the infection, and in different clinical situations, to better understand the impact of the minority viral population on SARS-CoV-2 evolution, physiopathology and transmission.
Author contributions
DB, AGM, SB, VC planned the research; EKF, VP and EC collected the clinical data; VL, IM, ET and CS performed the experiments; AJ, VL, BA and MW analysed the data; AJ, SM, SB and DB wrote the paper. All the authors read and corrected the manuscript and approved the final version.
Transparency declaration
All the authors declare no competing interests. This study was funded by the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS, AC43), the Agence National de la Recherche (ANR) and Sorbonne Université.
Ethics
The study was carried out in accordance with the Declaration of Helsinki. It was a retrospective non-interventional study with no addition to standard care procedures. Reclassification of biological remnants into research material after completion of the ordered virological tests was approved by the local interventional review board of Pitié-Salpêtrière Hospital. According to the French Public Health Code (CSP Article L.1121-1.1) such protocols are exempted from individual informed consent.
Acknowledgements
We thank the SMIT PSL COVID cohort Team for its support.
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.07.032.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- 1.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbe. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung T.S., Liu D.X. Human coronavirus: host–pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 3.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., et al. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klement E., Godefroy N., Burrel S., Kornblum D., Monsel G., Bleibtreu A., et al. The first locally acquired novel case of 2019-nCoV infection in a healthcare worker in the Paris area. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 9.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu D., Zhang Z., Wang F.-S. SARS-associated coronavirus quasispecies in individual patients. N Engl J Med. 2004;350:1366–1367. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- 11.Park D., Huh H.J., Kim Y.J., Son D.-S., Jeon H.-J., Im E.-H., et al. Analysis of intrapatient heterogeneity uncovers the microevolution of Middle East respiratory syndrome coronavirus. Cold Spring Harb Mol Case Stud. 2016;2:a001214. doi: 10.1101/mcs.a001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capobianchi M.R., Rueca M., Messina F., Giombini E., Carletti F., Colavita F., et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin Microbiol Infect. 2020;26:954–956. doi: 10.1016/j.cmi.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 14.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 15.Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.