Abstract
Attention deficit hyperactivity disorder (ADHD) is one of the most common neurobehavioural in the children. Genetic factor is known one of the factors which contributed in ADHD development. VNTR polymorphism in 3’UTR exon 15 of DAT1 gene and exon 3 of DRD4 gene are reported to be associated in ADHD. In this study we examine the association of ADHD with VNTR polymorphism of DAT1 and DRD4 gene in Indonesian children. Sixty-five ADHD children and 70 normal children (6-13 years of age), were included in the study, we matched by age and gender. ADHD was diagnosed by DSM-IV. We performed a casecontrol study to found the association between ADHD and VNTR polymorphism of DAT1 and DRD4 genes. The 10-repeat allele of DAT1 and 2-repeat allele of DRD4 were higher in Indonesian children. Although the frequency of these allele was higher, but it was similar both in ADHD and control groups. Neither DAT1 nor DRD4 gene showed showed significant difference in genotype distribution and frequency allele between both groups (p > 0.05). No association between ADHD and VNTR polymorphism of DAT1 and DRD4 genes found in Indonesian children. This data suggest that DAT1 and DRD4 do not contribute to etiology of ADHD in Indonesian children. Further studies are needed to clarify association between VNTR polymorphism of DAT1 and DRD4 genetic with ADHD of Indonesian children in larger sample size and family based study.
Key words: Children, ADHD, DAT, DRD4, Indonesian, case-control study
Introduction
Attention deficit hyperactivity disorder (ADHD) is one of the most common neurobehavioural disorders in the children which is characterized by inattention, overactivity, and impulsivity.1 Beside found in childhood, ADHD sometimes persist into adulthood in worldwide.2 Recent studies reported that ADHD commonly found in the boys than girls with the prevalence was around 5.29% in the world.3,4
Studies in ADHD showed many factors contributed to ADHD development. The development was affected by enviromental and genetic factors.5 Environmental factor reported having correlation with ADHD as prenatal exposure of alcohol, tobacco, food additives and prematurity.6,7 Besides enviromental factor, there was also genetic factor which reported having correlation with ADHD. A hereditary studies reported that increased risk of ADHD was found in children with hyperactive parents.8Moreover, Morrison in 1973 showed that ADHD parents were more likely to have ADHD children to their own biological children than their adoptive children. This study revealed that there was association between genetic factor with ADHD.
Several genes were known strongly contributed to ADHD development.9 Some studies reported that ADHD correlated with genes in the dopamine system. This system regulated the reinforcement learning structures in brain which associated with ADHD.10 Barr and Misener in 2008 showed that receptors and transporter of dopamine which encoding protein that regulate synthesis, release and degradation of dopamine played role in ADHD. Molecular genetic studies explained that the dopamine transporter 1 (DAT1) and dopamine D4 receptor (DRD4) genes plays a role in the development of ADHD.11-13 Studies of ADHD-related genes have focused on variable number of tandem repeats (VNTR) polymorphism of DAT1 and DRD4 gene.14 VNTR is a repetitive DNA sequence which is repeated several times and continuously in the genome.15VNTR is a marker which plays role to regulate gene expression.15,16 DAT1 gene, located on chromosome 5p15.3, contains a 40 bp VNTR in its 3’-untranslated region (UTR) which length is varying from 3-13 repeat.17 DRD4 gene, G protein-coupled receptor located on chromosome 11p15.5,18 contains a 48-base-pair VNTR in the third exon.19 Association between a VNTR polymorphism in 3’UTR of DAT1, and third axon of DRD4 genes with ADHD have been reported in recent studies.12 Study in Jordan children showed that the 10-repeat allele of DAT1 associated with ADHD and the 7-repeat allele of DRD4 associated with ADHD in the children in European countries.12,20 Instead of meta analysis studies in ADHD showed that a VNTR polymorphism in DAT1 and DRD4 gene had a small association to ADHD, Faraone et al. and Yang et al. suggested that these gene may be had role in the risk of ADHD.21,22 On the other hand, conflicting results were reported in the studies in the Omani children, Han Chinese children, Iranian population and Turkish population which showed no association significant between a VNTR polymorphism with ADHD.13,23-25 However ADHD study among Indonesian population has not been performed yet. To know the association between DAT1 and DRD4 gene with ADHD development, we conducted this study.
Materials and Methods
Subjects
Our study included 65 children with ADHD (58 boys = 89.2% and 7 girls = 10.8%) and 70 children as healthy control (62 boys = 88.5% and 8 girls = 11.4%). ADHD and healthy control participants were Javanese population aged 6-12 years old (mean ±SD: 9.3±1.4 and mean ± SD: 8.78±1.1). They were selected from the Elementary School in Yogyakarta, Indonesia. Final diagnosis of ADHD were made by clinician by Diagnostic and Statistical Manual of Mental Disorder, 4th Edition, Text Revision (DSM IV TR). Participants were excluded if there were any evidence of conduct disorder, neurological conditions and have chronic disease, and mental retardation (IQ≤70). A full scale IQ of participants were above 70 (mean±SD: 98.53±14.6 for ADHD, and mean ± SD: 110±15.6 for healthy control). Written informed consent was obtained from all participants. This study was approved by the Ethical Committee of Faculty of Medicine, Universitas Gadjah Mada, Yogyakarta, Indonesia.
Preparation of DNA Samples
Genomic DNA was extracted from whole blood using Wizard-Genomic DNA Purification Kit (Promega, USA).
PCR Amplification of genomic DNA
The primer sequence was used to amplify the DAT1 40-bp VNTR polymorphisms in exon 15. PCR amplification of the DAT1 in exon 15 used pair of DAT1F 5’TGTGGTGTAGG GAACGGCCTGAG-3’, and DAT1R 5’CTTCCTGGA GGTCACGGCTCAAGG-3’ (Demiralp et al., 2007). Thirty-five cycles were conducted consisting of denaturation 94oC for 1 minute, annealing 62oC for 1 minute, elongation 72oC for 1 minute. We also added initial denaturation 94oC for 7 minutes and final elongation 72oC for 7 minutes. Total volume of each PCR reaction was 30 μl which consisted of distilled water, genomic DNA template (100 ng), 20 pmol/μl of each primer, 10 mM dNTP, 10 x Buffer with MgCl2, Fast Start Taq polymerase, and G-C Rich. The products of PCR (5 μl) were separated on the agarose gel 3% with 1x TBE buffer and visualized by staining with ethidium bromide. 100 bp DNA Ladder (New England Biolabs® Inc, UK) was used as marker to identify the various repeat alleles by the size of the DNA fragment. Repeat alleles variation from the result were determined as follows: 315 bp (6-repeat), 355 bp (7-repeat), 435 bp (9-repeat), 475 bp (10-repeat), 515 bp (11—repeat), and 555 bp (12-repeat). PCR amplification of the DRD4 48-bp VNTR in exon 3 was carried out using the following pair of DRD4F 5`-GCGACTACG TGGTCTA CTCG-3` and DRD4R 5`-AGGACCCT CATGGCCTTG-3`(Demiralp et al., 2007). Thirty-five cycles were conducted consisting of denaturation 94oC for 1 minute, annealing 62oC for 1 minute, elongation 72oC for 1 minute. We also added initial denaturation 94oC for 7 minutes and final elongation 72oC for 7 minutes. Total volume of each PCR reaction was 30 μl which consisted of distilled water, genomic DNA template (100 ng), 20pmol/μl of each primer, 10 mM dNTP, 10 x Buffer with MgCl2, Fast Start Taq polymerase, and G-C Rich. The products of PCR (5 μl) were separated on the agarose gel 3% with 1x TBE buffer and visualized by staining with ethidium bromide. 100 bp DNA Ladder (New England Biolabs® Inc, UK) was used as marker to identify the various repeat alleles by the size of the amplicon. Repeat alleles from the result were determined as follows: 366 bp (2-repeat), 414 bp (3-repeat), 462 bp (4-repeat), 510 bp (5-repeat), 588 bp (6—repeat), and 606 bp (7-repeat).
GeneScanTM analysis
To determine the exact size of several DNA fragments, we performed GeneScanTM analysis. PCR products were used as template which its DAT1F and DRD4F primer was labelled with a flourescent 5-Flourescein Amidite (5-FAM). PCR product was incubated with HiDi Formamide (Applied Biosystems) and GenescanTM TamraTMdye size standard (Applied Biosystems, Foster City, CA, USA) at 95oC for 5 minutes and immediately cooled for 3 minutes. Electrophoresed the mixture on an ABI PRISM 3100 Genetic Analyser (Applied Biosystems, Foster City, CA, USA) for 45 minutes. To analysis the data we used Gene Mapper V4.0 Software (Applied Biosystems).
Statistics
Statistical significant of allele and genotype frequencies in ADHD and control group was computed with analysis of variants (ANOVA) test after observing the pattern of electrophoretic band from Indonesian subjects. A p value < 0.05 was considered statistically significant.
Results
DAT 1
First, we measured the exact size of a DNA fragment using GeneScan analysis. Figure 1 shows that the DNA fragment (REF 9/10) had 9- repeat (435 bp) and 10- repeat (475 bp) of allele of the DAT1 gene (Figure 1). Next, we checked the electrophoresis pattern of the DNA fragment including REF (9/10). Then, we determined the genotype and repeat allele of the samples (65 ADHD and 70 control samples) based on the pattern of DNA fragment on the gel electrophoresis (Figure 2).
Tables 1 and 2 showed the distribution of DAT1 genotypes and the frequency of the allele of DAT1 in the 3’UTR both in the control and ADHD. The genotype distributions between ADHD and control groups were not significantly different (p=0.187). The genotype 10/10 was the most common between both groups in our study: the frequency of the 10-repeat allele was higher and similar in ADHD (89.8%) and control (84.7%). The alleles frequencies between both groups were also no significantly different (p=0.263).
Table 1.
Distribution of the DAT1 genotypes.
| Genotype frequency | ||||
|---|---|---|---|---|
| 9/9 | 9/10 | 10/10 | 10/11 | |
| ADHD (N 61) | 0 | 4 (9.1%) | 39 (88.6%) | 1 (2.3%) |
| Control (N=44) | 1 (14.8%) | 9 (14.8%) | 50 (82%) | 1 (2.6%) |
| Total (N=105) | 1 (0.9%) | 13 (12.4%) | 89 (84.8%) | 2 (1.9%) |
Table 2.
Frequency of the DAT1 alleles.
| Alleles | Control | ADHD | Total |
|---|---|---|---|
| (N = 72) | (N = 49) | (N = 121) | |
| DAT1*9 | 10 (13.9%) | 4 (8.2%) | 14 (11.6%) |
| DAT1*10 | 61 (84.7%) | 44 (89.8%) | 105 (86.8%) |
| DAT1*11 | 1 (1.4%) | 1 (2%) | 2 (1.6%) |
DRD4
To measure the exact size of a DNA fragment, we used GeneScan analysis. GeneScan analysis showed two types of DNA fragments (REF 2/4): 2- repeat (366 bp) and 4- repeat (462 bp) (Figure 3). Next, we checked the electrophoresis pattern of the DNA fragment including REF (2/4).
We determined the genotype and repeat allele of the samples (65 ADHD and 70 control samples) based on the pattern of DNA fragment on the electrophoresis gel (Figure 4). Genotypes distribution and repeat allele frequency of DRD4 VNTR polymorphism in the third exon are shown in the Tables 3 and 4. Distribution of DRD4 genotypes showed that the genotype 2/2 was the most common in our study (60.6%): the 2-repeat allele was the predominant polymorphism both in ADHD (72.8%) and control group (61.7%). The results showed no significant differences in terms of genotype distribution (p=0.334) and allele frequency (p=0.198) between the groups.
Discussion and Conclusions
The present study showed no association between ADHD and a particular VNTR allele of the DAT1 and DRD4 gene in Indonesian sample by the case-control design. In Indonesian children, we found the most common allele in DAT1 gene was 10-repeat allele and in DRD4 gene was 2-repeat allele. Athough frequency of the 10-repeat and 2- repeat alleles were higher in our study, the distribution of these alleles were similar between both in ADHD and control groups (Table 1 and 4). No statistically significant different was found for genotype distribution and allele frequency of DAT1 and DRD4 polymorphism in ADHD and control groups (p>0.05). Notably, we did not found 7-repeat alele for DRD4 gene in ADHD chidren, although that was reported that 7-repeat allele in DRD4 gene may be associated in the etiology of ADHD (Gornick et al. 2007).
Previous genetic studies for ADHD found that VNTR allele of DAT1 gene contributed in the ADHD development.22 The 10-repeat allele in DAT1 was reported contributed for ADHD and played role in the ADHD development.12,26 In family based study, using the haplotype-based haplotype relative risk (HHRR) analysis, Cook and colleague in 1995 revealed significant association between ADHD and 10-repeat allele of DAT1 gene (p=0.06). They showed the 10-repeat allele was higher in the total parent (76.2%). In addition, child’s genotype distribution also showed the 10-repeat allele was common in their study. Despite the association was small, a meta-analysis of association studies was suggestive significant association between ADHD and this allele.22 In Jordan children, using case-control-based association studies, Gharaibeh et al.12 in 2010 also displayed that frequency of 10-repeat allele was higher in ADHD (50%) compared to controls group (24%). They revealed that 10-repeat allele significantly associated with ADHD (p<0.05). Some studies reported that this polymorphism contributed in the treatment response in ADHD children.27-29
Figure 1.
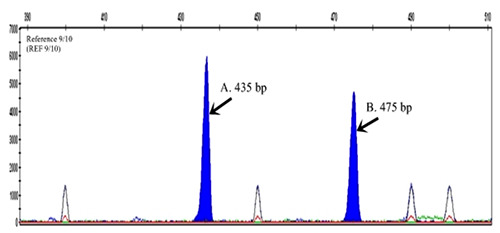
Electrophoregram of the Genescan Analysis of the DAT1 repeat polymorphism. The Genescan™ data represents a heterozygous DAT1 gene with VNTR sizes 435 bp and 475 bp (black arrow).
Figure 2.
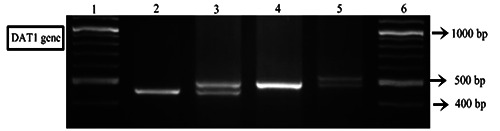
PCR Analysis of the DAT1 repeat polymorphism. Lane 1: DNA Marker (100 bp). Lane 2: homozygote, 9- repeat. Lane 3: heterozygote, 9- and 10- repeat (REF (9/10)). Lane 4: homozygote, 10- repeat. Lane 5: heterozygote, 10- and 11- repeat. Lane 6: DNA Marker. PCR products were run on a 3 % Agarose gel/1x TBE Buffer.
Figure 3.
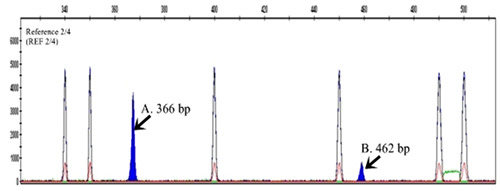
Electrophoregram of the Genescan Analysis of the DRD4 repeat polymorphism. The gene scan analysis of the band contain: a) 2-repeat (366 bp), b) 4-repeat (462 bp) (black arrow).
Figure 4.
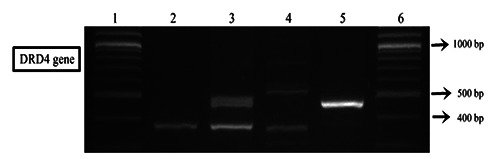
PCR Analysis of the DRD4 repeat polymorphism. PCR Analysis of the DRD4 repeat polymorphism. Lane 1: DNA Marker (100 bp). Lane 2: homozygote, 2- repeat. Lane 3: heterozygote, 2- and 4- repeat (REF 2/4). Lane 4: heterozygote, 2- and 5- repeat. Lane 5: homozygote, 4- repeat. Lane 6: DNA Marker. PCR products were run on a 3 % Agarose gel/1x TBE Buffer.
A number of other studies have found conflicting results. Family-based of association study of Langley et al. in 2005 using the haplotype analysis reported no significant association between the-10 repeat allele and ADHD (p=0.85). They also had similar result for case-control analysis (p=0.91). Allele frequency of 10-repeat allele was similar between ADHD and control groups in their study. Case-control study in Omani children found no association between the 10-repeat allele and ADHD while this allele was higher in the study.25 Their results showed the distribution of this allele was similar in ADHD (64,6%) and control group (60.9%). Banoei et al. in 2008 also reported no association between the 10-repeat allele and ADHD in Iranian population.24
Another gene which known associated with ADHD was DRD4 gene. A meta-analysis of the association studies found association between ADHD and the 7-repeat allele. Although their results showed small association, Faraone et al.21 suggest there was association between ADHD and the 7-repeat allele. Association between ADHD and the 7- repeat allele also found in the case-control study of Gornick et al.32 However there are several conflicting findings. Several studies have demonstrated different results. Familybased study using FBAT analysis and case-control study found no association between ADHD and the 7-repeat allele of DRD4 gene.31 Study in Dutch, Taiwanese and other population families also showed similar results.. Bidwell et al. in family-based of association studies, showed significant association between ADHD and the 4-repeat allele of DRD4 based on FBAT analysis (p<0.07). But, Cheuk et al. in 2006 reported no association between ADHD and 4- repeat allele of DRD4 gene in their study (p>0.05).
Genotyping results in our study for DRD4 gene showed different result. We found no 7- repeat allele in our Indonesian samples. In agreement with our result, in the several Asian studies, the 7-repeat allele was low or not shown.22,23 We found the 2-repeat allele was the most common allele in Indonesian samples. Study of Chang et al. in 1996 reported that the 2-repeat allele was reported quite frequently found in East and South Asia. Recent studies revealed association between ADHD and the 2-repeat allele in Han Chinese children (Leung et al., 2005). Their result was different with our result which showed no association between ADHD and 2-repeat allele, although the 2-repeat allele was higher in our study. There are several limitations of this study. First, the sample size is small for genetic study which may not have enough power to identify a small association between the DAT1, DRD4 VNTR polymorphism and ADHD. Second, because the data of the ADHD families is not available in our study, we just observed an association using the case-control design. Third, our study found different result with previous. It is possibly due to population stratification in ADHD children is needed for genetic studies.
In conclusion, there is no association between a particular VNTR polymophism of DAT1 and DRD4 gene and ADHD in Indonesian children. We suggest further independent analysis of genetic association studies of Indonesian children in larger sample size and family based study.
Table 3.
Distributionof the DRD4 genotypes.
| Genotype frequency | ||||
|---|---|---|---|---|
| 2/2 | 2/4 | 2/5 | 4/4 | |
| ADHD (N = 45) | 31 (68.9%) | 8 (17.8%) | 2 (4.4%) | 4 (8.9%) |
| Control (N = 59) | 32 (54.2%) | 18 (30.5%) | 1 (1.7%) | 8(13.6%) |
| Total (N = 104) | 63 (60.6%) | 26 (25%) | 3 (2.9%) | 12 (11.5%) |
Table 4.
Frequency of the DRD4 alleles.
| Alleles | Control | ADHD | Total |
|---|---|---|---|
| (N = 81) | (N = 59) | (N = 140) | |
| DRD4*2 | 50 (61.7%) | 43 (72.8%) | 93 (66.4%) |
| DRD4*4 | 26 (32.1%) | 12 (20.3%) | 38 (27.2%) |
| DRD4*5 | 1 (1.2%) | 2 (3.4%) | 3 (2.1%) |
Acknowledgments
The authors would like to thank the children, parents, teachers, and school officers in Cangkringan District, Yogyakarta who are willing to participate in this study. We would like to express our highest gratitude also to Dr. Indra Sari Kusuma Harahap, Dr. Nur Imma Fatimah Harahap, Nihayatus Sa’adah, M.Sc, Dr. Sari Kusumaningrum, member of Biochemistry laboratory and resident doctors of Neurology Department UGM for their technical assistance.
References
- 1.National Collaborating Centre for Mental Health. Attention Deficit Hyperactivity Disorder. The Nice Guideline on Diagnosis and Management of ADHD in Children, Young People and Adults. National Clinical Practice Guideline Number 72. London: The British Psychological Society & The Royal College of Psychiatrists; 2009. [Google Scholar]
- 2.Faraone SV, Sergeant J, Gillberg C, Biederman J. The worldwide prevalence of ADHD: is it an American condition?. World Psychiatry 2003;2:104-13. [PMC free article] [PubMed] [Google Scholar]
- 3.Polanczyk G, Silva de Lima M, Horta BL, et al. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am J Psychiatry 2007;164:942-8. [DOI] [PubMed] [Google Scholar]
- 4.Pastor PN, Reuben CA. Diagnosed attention deficit hyperactivity disorder and learning disability: United States, 2004-2006. National Center for Health Statistics. Vital Health Stat 2008;10:1-22. [PubMed] [Google Scholar]
- 5.Sharp SI, McQuillin A, Gurling HMD. Genetics of attention-deficit hyperac tivity disorder (ADHD). Neuropharmocol 2009;57:590-600. [DOI] [PubMed] [Google Scholar]
- 6.Rowland AS, Lesesne CA, Abramowitz AJ. The epidemiology of attentiondeficit/ hyperactivity disorder (ADHD): a public health view. MRDD Res Rev 2002;8:162-70. [DOI] [PubMed] [Google Scholar]
- 7.Biederman J. Attention-deficit/ hyperactivity disorder: a selective overview. Biol Psychiatry 2005;57: 1215-20. [DOI] [PubMed] [Google Scholar]
- 8.Hechtman L. Families of children with attention deficit hyperactivity disorder: a review. Can J Psychiatry 1996;41:350-60. [DOI] [PubMed] [Google Scholar]
- 9.Faraone SV, Khan SA. Candidate gene studies of attention-deficit/hyperactivity disorder. J Clin Psychiatry 2006;67:13-20. [PubMed] [Google Scholar]
- 10.Tripp G, Wickens JR. Neurobiology of ADHD. Neuropharmacology 2009;57: 579-89. [DOI] [PubMed] [Google Scholar]
- 11.Barkley RA, Smith KM, Fischer M, Navia B. An examination of the behavioral and neuropsychological correlates of three ADHD candidate gene polymorphisms (DRD4 7+, DBH Taql A2, and DAT1 40bp VNTR) in hyperactive and normal children followed to adulthood. Am J Med Genet B Neuropsychiatr Genet 2006;141B:487-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gharaibeh MY, Batayneh S, Khabour OF, Daoud A. Association between polymorphisms of the DBH and DAT1 genes and attention deficit hyperactivity disorder in children from Jordan. Exp Therapeut Med 2010;1:701-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guney E, Iseri E, Ergun SG, et al. The correlation of attention deficit hyperactivity disorder with DRD4 gene polymorphism in Turkey. Int J Hum Genet 2013;13:145-52. [Google Scholar]
- 14.Fuke S, Suo S, Takahashi N, et al. The VNTR polymorphism of the human dopamine transporter (DAT1) gene affects gene expression. Pharmacogenom J 2001;1:152-6. [DOI] [PubMed] [Google Scholar]
- 15.Brookes KJ. The VNTR in complex disorders; the forgotten polymorphisms? A functional way forward?. Genomics 2013;101:173-281. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura Y, Koyama K, Matsushima M. VNTR (variable number of tandem repeat) sequences as transcriptional, transtional or functional regulators. J Hum Genet 1998;43:149-52. [DOI] [PubMed] [Google Scholar]
- 17.Vandenbergh DJ, Persico AM, Hawkins AL, et al. Human doopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics 1992;14:1104-6. [DOI] [PubMed] [Google Scholar]
- 18.Gelernter J, Kennedy JL, Van Tol HHM, et al. The D4 dopamine receptor (DRD4) maps to distal 11p close to HRAS. Genomics 1992;13:208-10. [DOI] [PubMed] [Google Scholar]
- 19.Van Tol HHM, Wu CM, Guan HC, et al. Multiple dopamine D4 receptor variants in the human population. Nature 1992;358:149-52. [DOI] [PubMed] [Google Scholar]
- 20.Altink ME, Rommelse NNJ, Slaats- Willemse DIE, et al. The dopamine receptor D4 7-repeat allele influences neurocognitive functioning, but this effect is moderated by age and ADHD status: an exploratory study. World J Biol Psychiatry 2011;1:1-13. [DOI] [PubMed] [Google Scholar]
- 21.Faraone SV, Doyle AE, Mick E, Biederman J. Meta-analysis of the association between the 7-repeat allele of the dopamine D4 receptor gene and attention deficit hyperactivity disorder. Am J Psychiatry 2001;158:1052-7. [DOI] [PubMed] [Google Scholar]
- 22.Yang B, Chan RCK, Jing J, et al. A metaanalysis of association studies between the 10-repeat allele of a VNTR polymorphism in the 3’-UTR of dopamine transporter gene and attention deficit hyperactivity disorder. Am J Med Genet Part B 2007;144B:541-50. [DOI] [PubMed] [Google Scholar]
- 23.Qian Q, Wang Y, Zhou R, et al. Familybased and case-control association studies of DRD4 and DAT1 polymorphisms in Chinese attention deficit hyperactivity disorder patients suggest long repeats contribute to genetic risk for the disorder. Am J Med Genet Part B 2004;128B:84-9. [DOI] [PubMed] [Google Scholar]
- 24.Banoei MM, Majidizadeh T, Shirazi E, et al. No association between the DAT1 10- repeat allele and ADHD in the Iranian population. Am J Med Genet Part B 2008;147B:110-1. [DOI] [PubMed] [Google Scholar]
- 25.Simsek M, Al-Sharbati M, Al-Adawi S, et al. Association of the risk allele of dopamine transporter gene (DAT1*10) in Omani male children with attentiondeficit hyperactivity disorder. Clin Biochem 2005;38:739-42. [DOI] [PubMed] [Google Scholar]
- 26.VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats elements in DAT1 regulates in vitro dopamine transporter density. BMC Genetics 2005;6:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellgrove MA, Hawi Z, Kirley A, et al. Association between dopamine transporter (DAT1) genotype, left-sided inattention, and an enhanced response to methylphenidate in attention-deficit hyperactivity disorder. Neuropsychopharmacology 2005;30:2290-7. [DOI] [PubMed] [Google Scholar]
- 28.Cheon KA, Ryu YH, Kim JW, Cho DY. The homozygosity for 10-repeat allele at dopamine transporter gene and dopamine transporter density in Korean children with attention deficit hyperactivity disorder: relating to treatment response to methylphenidate. Eur Neuropsychopaharmacol 2005;15: 95-101. [DOI] [PubMed] [Google Scholar]
- 29.Pasini A, Sinibaldi L, Paloscia C, et al. Neurocognitive effects of methylphenidate on ADHD children with different DAT genotypes: A longitudinal open label trial. Eur J Paediatr Neurol 2013;17:407-14. [DOI] [PubMed] [Google Scholar]
- 30.Cheuk DKL, Li SYH, Wong V. Exon 3 polymorphisms of dopamine D4 receptor (DRD4) gene and attention deficit hyperactivity disorder in Chinese children. Am J Med Genet Part B 2006:141B:907-11. [DOI] [PubMed] [Google Scholar]
- 31.Hawi Z, McCarron M, Kirley A, et al. No association of the dopamine DRD4 receptor (DRD4) gene polymorphism with attention deficit hyperactivity disorder (ADHD) in the Irish population. Am J Med Genet 2000;96:268-72. [DOI] [PubMed] [Google Scholar]
- 32.Gornick MC, Addington A, Shaw P, et al. Association of the dopamine receptor D4 (DRD4) gene 7-repeat allele with chidren with attention-deficit/hyperactivity disorder (ADHD): an update. Am J Med Genet Part B 2007;144B:379-82. [DOI] [PubMed] [Google Scholar]


