Abstract
Background
The prognostic significance of focal adhesion kinase (FAK) in breast cancer remains controversial. Here, we conducted a meta-analysis to explore the prognostic value of FAK expression in breast cancer.
Materials and methods
Possible prognostic significance of protein or mRNA expression of FAK in breast cancer was investigated with searches of electronic databases for relevant publications. Pooled hazard ratios (HRs) and odds ratios (ORs) with 95% confidence intervals (CIs) were extracted from eligible studies.
Results
A total of eight eligible studies which included 2604 participants were analyzed in this meta-analysis. Increased expression of FAK protein was found to significantly correlate with shorter overall survival (OS) (HR = 1.43, 95% CI: 1.12–1.83; P = 0.004), and not with disease-free survival (HR = 1.31, 95% CI: 0.92–1.85; P = 0.14). Elevated FAK protein expression was also associated with negative estrogen receptor (ER) expression (OR, 1.34; 95% CI, 1.06–1.68; P = 0.01), negative progesterone receptor (PR) expression (OR, 1.54; 95% CI, 1.22–1.93; P < 0.001), positive human epidermal growth factor receptor 2 (HER2) expression (OR, 1.64; 95% CI, 1.28–2.09; P < 0.001), triple-negative breast cancer (TNBC) (OR, 1.57; 95% CI, 1.14–2.17; P = 0.006), high nuclear grade (OR, 1.70; 95% CI, 1.05–2.78; P = 0.03), high Ki-67 expression level (OR, 2.87; 95% CI, 1.94–4.24; P < 0.001), and positive p53 status (OR, 2.28; 95% CI, 1.58–3.29; P < 0.001).
Conclusion
Our meta-analysis identifies an association between increased FAK protein expression and worse OS among breast cancer patients. Moreover, enhanced FAK expression is associated with negative ER expression, negative PR expression, positive HER2 expression, TNBC, high nuclear grade, high Ki-67 expression level, and positive p53 status in breast carcinoma.
Keywords: Breast cancer, FAK, Survival, Prognosis, Meta-analysis
Introduction
Breast cancer is becoming an increasingly serious public health concern. Worldwide, it is predicted that approximately 2.1 million cases of breast cancer among females will have been diagnosed in 2018 [1]. Furthermore, despite the reduced mortality of breast cancer due to individualized treatment encompassing early diagnosis, surgery, chemotherapy, radiotherapy, endocrine, and targeted therapy [2], distant metastasis remains one of the major challenges in the treatment of breast cancer cases. It is recognized that the mechanism(s) mediating metastasis are complex. Thus, it has been proposed that novel prognostic markers are needed to provide insight into these molecular mechanisms and improve treatment management of breast cancer cases.
Focal adhesion kinase (FAK) is a non-receptor tyrosine kinase which contributes to cellular physiological processes through the activation of integrins and focal adhesions of cell receptors [3,4]. Previous studies have demonstrated that FAK promotes malignancy by regulating numerous cellular processes, including adhesion, motility, proliferation, migration, invasion, and angiogenesis. Both highly coordinated signaling networks and activated cancer stem cells mediate the roles of FAK [5,6]. For example, Rigiracciolo et al. [7] have demonstrated that FAK promotes the migration of triple-negative breast cancer (TNBC) cells by activating signaling via the estrogenic G-protein coupled estrogen receptor pathway. Bianchi-Smiraglia and coworkers [8] have also reported that integrin β5 promotes breast cancer cell migration via the Src-FAK and MEK-ERK signaling pathways. Correspondingly, FAK expression has been detected in various cancers, including breast cancer [9], pancreatic cancer [10], head and neck squamous cell carcinoma [11], rectal cancer [12], hepatocellular carcinoma [13], and lung cancer [14].
Previous studies have revealed that up-regulation of FAK expression is associated with poor survival outcome in breast carcinoma [15,16], while other studies have reported no significance [17,18]. Therefore, we performed a pooled meta-analysis in order to investigate a possible correlation between FAK expression and its prognostic value in breast cancer.
Materials and methods
Search strategy
Searches were conducted of the PubMed, Embase, Cochrane Library, and Web of Science databases through August 12, 2019 with the following keywords: “breast neoplasms”/“breast cancer” and “focal adhesion kinase”/“FAK” and “prognosis”/“survival”.
Study inclusion/exclusion criteria
Inclusion criteria were: (1) research focused on breast cancer patients, (2) investigations of associations between FAK protein or gene expression and clinical parameters and prognosis, and (3) articles with sufficient information for extraction of hazard ratio (HR) or odds ratio (OR) with 95% confidence interval (CI) data. Exclusion criteria were: non-human studies, studies of cell lines or animals, conference reports, letters, reviews, and studies lacking sufficient information to estimate associations.
Data extraction
Two authors independently extracted baseline characteristics from the studies selected. Author surname, year of publication, country where study conducted, number of patients, outcomes, detection method, staining location, cut-off score, and proportion of up-regulated FAK expression were recorded for each study. From eligible studies, information focused on prognosis and clinicopathologic features were also extracted. The Newcastle Ottawa Scale (NOS) was applied to assess quality of the included studies [19]. Studies with a NOS score ≥ 7 were considered to be of high quality.
Statistical analysis
This meta-analysis was conducted based on Preferred Reporting Items for Systematic Reviews and Meta-analyses guidelines [20]. All analyses were performed with Review Manager version 5.3 (Cochrane Collaboration, Copenhagen, Denmark) and STATA version 12.0 (Stata Corporation, TX, USA) software programs. Heterogeneity was evaluated by using the Chi-squared test and I2 statistics [21]. I2 values >50% and P-values <0.05 indicated significant heterogeneity. A fixed effects model or random effects model was applied according to heterogeneity [22]. Sensitivity analysis was used to estimate stability of the pooled results. Publication bias was assessed with Begg's and Egger's tests [23,24]. Triple sequential analysis (TSA) was performed to estimate required sample information [25]. False-positive report probability (FPRP) analysis was performed to verify true correlations [26]. P-values less than 0.05 were considered statistically significant.
Results
Search results
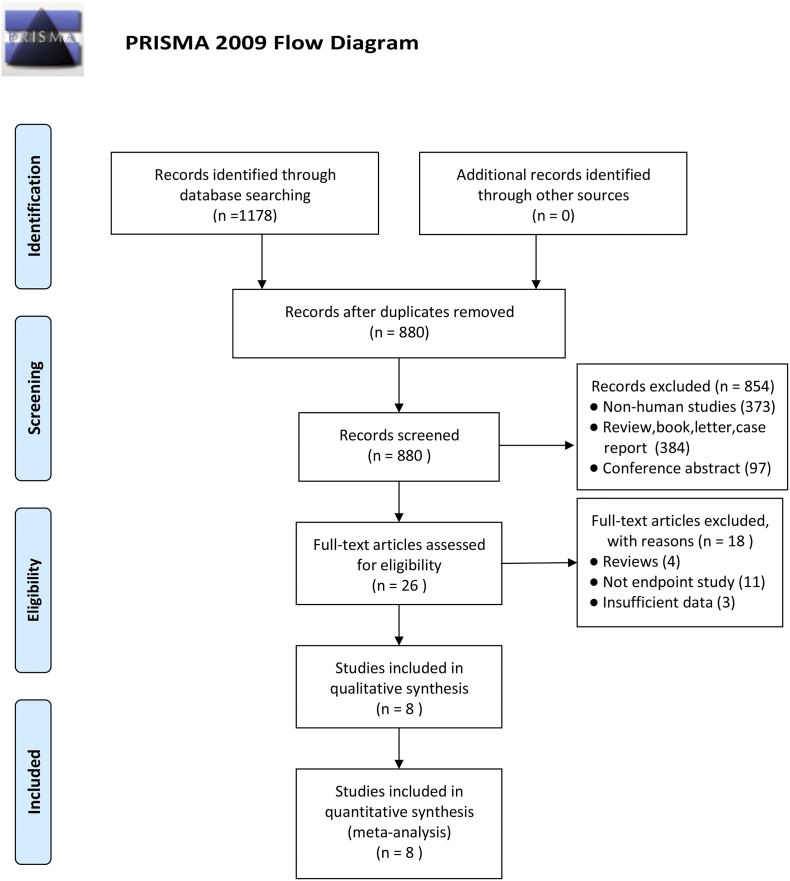
As shown in Fig. 1, a total of 1178 articles were identified from our search strategy (S1 Table). After removing duplicates (n = 298), titles and abstracts were screened. A total of 854 studies were excluded because they either involved cell culture or animal model studies, or were reviews, case reports, or meeting abstracts. Overall text of the remaining 26 studies were then reviewed. Four reviews, eleven no endpoint studies, and three studies lacking relevant data were excluded. Finally, a total of 8 eligible studies involving 2604 participants were selected for a meta-analysis [[15], [16], [17], [18],[27], [28], [29], [30]].
Fig. 1.
Flow chart of the process used to select eligible studies.
Search characteristics
Baseline characteristics of the qualifying studies are summarized in Table 1, Table 2. The included studies were retrospective or observational in a nature, and were of high quality according to NOS criteria (score ≥ 7) (S2 Table). For the integrity of the publication data, we considered relapse-free survival (RFS) equivalent to disease-free survival (DFS) [18], and cancer-specific survival (CSS) equivalent to overall survival (OS) [15] in the eligible studies. In addition, FAK mRNA expression studies were excluded due to insufficient data. Therefore, our eligible studies included detection of FAK expression by immunohistochemistry. However, the antibodies used, the staining locations, and the cut-off values used for FAK expression varied in the eligible studies. Correspondingly, the reported proportion of increased FAK expression ranged from 18.5% to 88%.
Table 1.
Characteristics of the eligible studies examined.
| Publication | Year | Country | Cancer subtype | No. of patients | Age, years (median, range) | Follow-up time, months (median, range) | Outcome | Survival analysis | NOS (score) |
|---|---|---|---|---|---|---|---|---|---|
| Almstedt | 2017 | Germany | Node-negative BC | 335 | 58 (range, 32–90) | 183 (range, 0–348) | DFS,OS | Multivariate | 8 |
| Andisha | 2019 | UK | Primary BC | 474 | 50 | 150 | OS | Multivariate | 8 |
| Golubovskaya | 2014 | USA | Stage II-IV BC | 196 | 56 (range, 27–91) | NR | NR | NR | 7 |
| Guo | 2017 | China | BC | 300 | 56.9 (range, 29–88) | NR | OS | Multivariate | 7 |
| Lark | 2005 | USA | Invasive BC | 629 | 48 (range, 23–74) | NR | NR | NR | 7 |
| Schmitz | 2005 | Germany | BC | 162 | 59 | 89.8 | NR | NR | 7 |
| Theocharis | 2009 | France | BC | 73 | 59 (range, 31–85) | NR | NR | NR | 7 |
| Yom | 2011 | Korea | Invasive BC | 435 | 46 (range, 25–79) | 53 (range, 7–85) | DFS, 0S | Multivariate | 8 |
BC, breast cancer; NR, not reported; DFS, disease-free survival; OS, overall survival; NOS, Newcastle Ottawa Scale.
Table 2.
Methods of quantitative FAK measurement of eligible studies.
| Publication | Year | FAK phenotype | Detection method | FAK expression | Staining location | Antibody | Cut-off value (low/high level) | High FAK expression |
|---|---|---|---|---|---|---|---|---|
| Almstedt | 2017 | FAK | IHC | protein | cytoplasmic and membranous | anti-FAK (1:100,Dako,Germany) | high (IHC score ≥ 6) | 45.1%(151/335) |
| Andisha | 2019 | nuclear ph-FAK Y397 | IHC | protein | nuclear Ph-FAK Y397 | anti-FAK (1:200,ab39967,Abcam) | high (stained ≥ + 2x%) | 50.8%(213/419) |
| Golubovskaya | 2014 | FAK | IHC | protein | NR | anti-FAK (Millipore #05–537) | high (stained score > 4) | 27%(53/196) |
| Guo | 2017 | FAK | IHC | protein | membranous | anti-FAK (1:50,#3285,USA) | high (grade 4–7) | 74.7%(215/288) |
| Lark | 2005 | FAK | IHC | protein | cytoplasmic | anti-FAK (1:250,4.47,USA) | high (stained ≥3+) | 25%(154/629) |
| Schmitz | 2005 | FAK | IHC | protein | cytoplasmic and membranous | anti-FAK (1:100,Polyclonal) | high (stained ≥3+) | 18.5%(30/162) |
| Theocharis | 2009 | FAK | IHC | protein | cytoplasmic and membranous | anti-FAK (sc-1688,USA) | high (stained ≥5%) | 88%(64/73) |
| Yom | 2011 | FAK | IHC | protein | cytoplasmic | anti-FAK (monoclonal,4.47) | high (stained score > 3) | 27.5%(108/393) |
IHC, immunohistochemistry; NR, not reported.
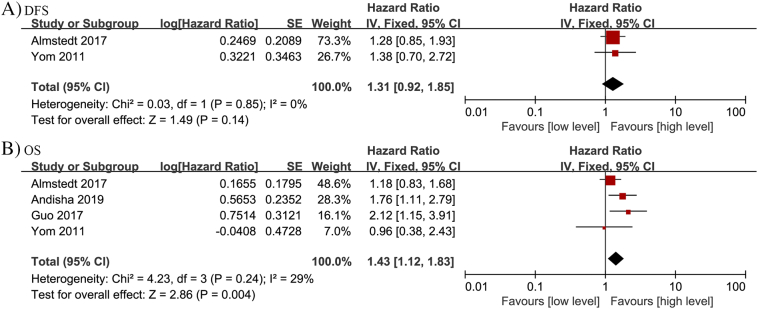
Association of FAK protein expression and DFS and OS
Two studies assessed the relationship between FAK protein expression and DFS in breast cancer, and no significant correlation was observed (HR = 1.31, 95% CI: 0.92–1.85; P = 0.14) (Fig. 2A). Therefore, a fixed effects model was applied according to an absence of heterogeneity (P = 0.85, I2 = 0%). Importantly, increased FAK protein expression was found to significantly correlate with worse OS (n = 4; HR = 1.43, 95% CI: 1.12–1.83; P = 0.004) in the populations of breast cancer patients analyzed (Fig. 2B). Therefore, a fixed effects model was applied based on insignificant heterogeneity (P = 0.24, I2 = 29%).
Fig. 2.
Forest plots depicting correlations between FAK protein expression and (A) DFS, and (B) OS among the breast cancer patients examined.
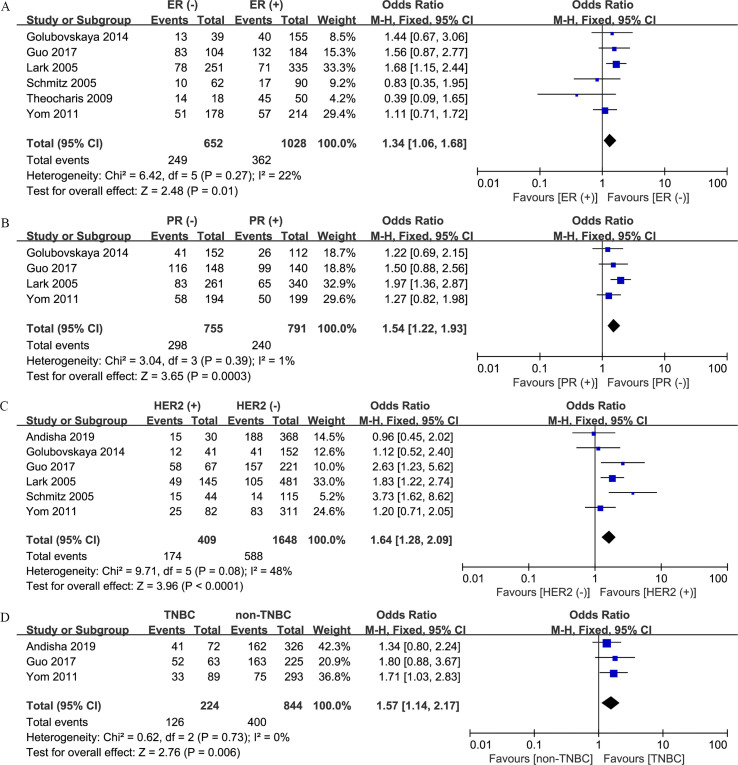
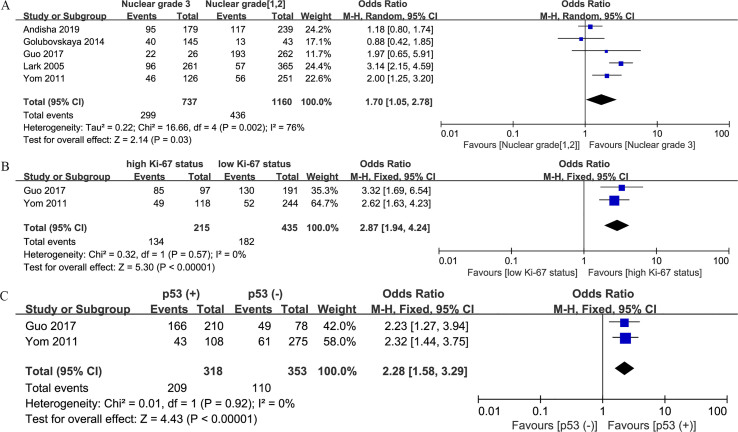
Association between FAK protein expression and clinicopathologic characteristics
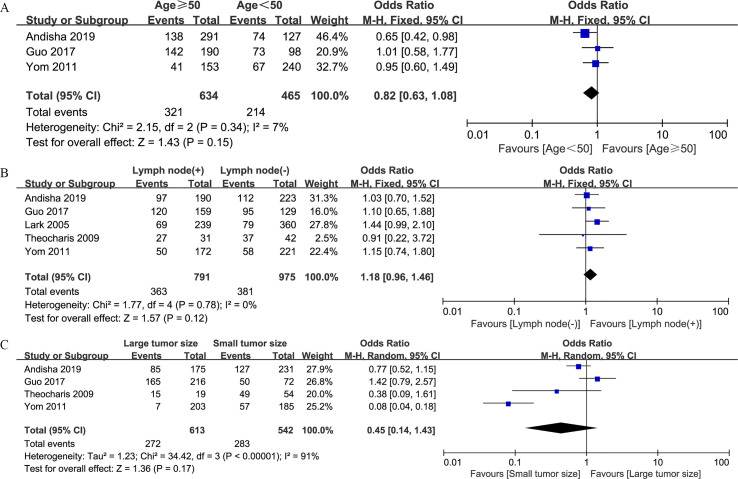
Correlations between increased FAK protein expression and clinicopathologic factors are summarized in Table 3. Overexpression of FAK protein was found to be related to negative estrogen receptor (ER) expression (n = 6; OR, 1.34; 95% CI, 1.06–1.68; P = 0.01) (S1 Fig. A), negative progesterone receptor (PR) expression (n = 4; OR, 1.54; 95% CI, 1.22–1.93; P < 0.001) (S1 Fig. B), positive human epidermal growth factor receptor 2 (HER2) expression (n = 6; OR, 1.64; 95% CI, 1.28–2.09; P < 0.001) (S1 Fig. C), TNBC (n = 3; OR, 1.57; 95% CI, 1.14–2.17; P = 0.006) (S1 Fig. D), high nuclear grade (n = 5; OR, 1.70; 95% CI, 1.05–2.78; P = 0.03) (S2 Fig. A), high Ki-67 expression level (n = 2; OR, 2.87; 95% CI, 1.94–4.24; P < 0.001) (S2 Fig. B), and positive p53 status (n = 2; OR, 2.28; 95% CI, 1.58–3.29; P < 0.001) (S2 Fig. C). However, no significant associations between FAK protein expression and other features, including age (n = 3; OR, 0.82; 95% CI, 0.63–1.08; P = 0.15) (S3 Fig. A), lymph node involvement (n = 5; OR, 1.18; 95% CI, 0.96–1.46; P = 0.12) (S3 Fig. B), and tumor size (n = 4; OR, 0.45; 95% CI, 0.14–1.43; P = 0.17) (S3 Fig. C), were identified.
Table 3.
Meta-analysis of the correlation between FAK expression and clinicopathological factors of breast cancer.
| Clinicopathological parameter | No. of studies | No. of patients | OR (95% CI) | P-value | Heterogeneity |
|
|---|---|---|---|---|---|---|
| I2 (%) | P-value | |||||
| ER (− vs. +) | 6 | 1680 | 1.34 (1.06–1.68) | 0.01 | 22 | 0.27 |
| PR (− vs. +) | 4 | 1546 | 1.54 (1.22–1.93) | <0.001 | 1 | 0.39 |
| HER2 (+ vs. −) | 6 | 2057 | 1.64 (1.28–2.09) | <0.001 | 48 | 0.08 |
| TNBC (TNBC vs. non-TNBC) | 3 | 1068 | 1.57 (1.14–2.17) | 0.006 | 0 | 0.73 |
| Nuclear grade (3 vs. 1 and 2) | 5 | 1897 | 1.70 (1.05–2.78) | 0.03 | 76 | 0.002 |
| Ki-67 (high vs. low) | 2 | 650 | 2.87 (1.94–4.24) | <0.001 | 0 | 0.57 |
| p53 (+ vs. −) | 2 | 671 | 2.28 (1.58–3.29) | <0.001 | 0 | 0.92 |
| Age (≥50 vs.<50) | 3 | 1099 | 0.82 (0.63–1.08) | 0.15 | 7 | 0.34 |
| Lymph node (+ vs. −) | 5 | 1766 | 1.18 (0.96–1.46) | 0.12 | 0 | 0.78 |
| Tumor size (large vs. small) | 4 | 1155 | 0.45 (0.14–1.43) | 0.17 | 91 | <0.001 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer; OR, odds ratio.
S1 Fig.
Forest plots depicting correlations between FAK protein expression and (A) ER status (negative vs. positive), (B) PR status (negative vs. positive), (C) HER2 status (positive vs. negative), and (D) TNBC (TNBC vs. non-TNBC).
S2 Fig.
Forest plots depicting correlations between FAK protein expression and (A) nuclear grade (3 vs. 1 and 2), (B) Ki-67 status (high vs. low), and (C) p53 status (positive vs. negative).
S3 Fig.
Forest plots depicting correlations between FAK protein expression and (A) age (≥50 vs. <50), (B) lymph node involvement (positive vs. negative), and (C) tumor size (large vs. small).
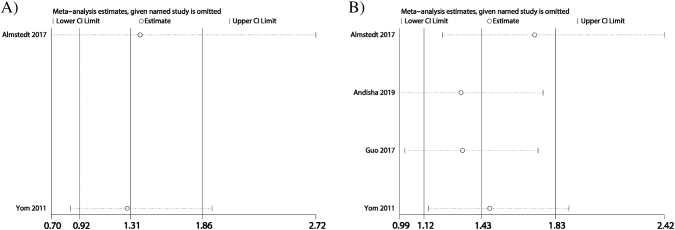
Sensitivity analysis and publication bias
Sensitivity analysis was conducted for DFS (Fig. 3A) and OS (Fig. 3B) data, and pooled HRs were stable. Application of Begg's test (P = 0.734) and Egger's test (P = 0.836) to the OS data further revealed no evidence for publication bias.
Fig. 3.
Sensitivity analysis of FAK protein expression in relation to (A) DFS and (B) OS.
Trial sequential and FPRP analyses
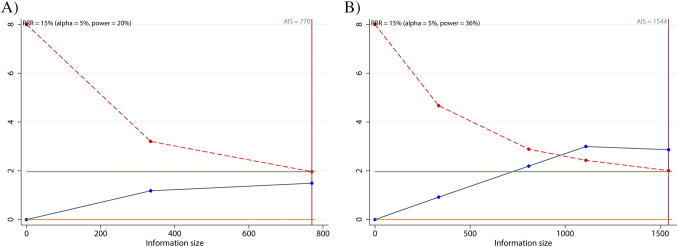
Trial sequential analysis was conducted to evaluate the required sample information of the selected studies. The cumulative Z-curve (blue line) does not cross either the traditional boundary line or the trial sequential monitoring boundary (red line) for DFS (Fig. 4A). However, the cumulative Z-curve (blue line) crosses both the traditional boundary line and the trial sequential monitoring boundary (red line) for OS (Fig. 4B). In addition, the cumulative information reaches the required information size for OS. Therefore, the involved sample size is sufficient for OS, yet insufficient for DFS, in the meta-analysis.
Fig. 4.
Trial sequential analysis (TSA) assessing the required sample information in (A) DFS and (B) OS.
Prognosis information was also subjected to FPRP analysis. With a prior probability of 0.01, the FPRP values for DFS and OS were both >0.2, indicating that the pooled HRs were not truly significant (Table 4).
Table 4.
False-positive report probability analysis values for DFS and OS.
| Survival outcome | Crude HR (95% CI) | P-value | Statistical power | Prior probability |
||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| DFS | 1.31 (0.92–1.85) | 0.125 | 0.779 | 0.325 | 0.591 | 0.941 | 0.994 | 0.999 |
| OS | 1.43 (1.12–1.83) | 0.004 | 0.648 | 0.020 | 0.059 | 0.406 | 0.873 | 0.986 |
DFS, disease-free survival; OS, overall survival.
Discussion
We performed a meta-analysis to clarify a potential association between FAK expression and prognosis of breast cancer. In a previous meta-analysis, a correlation between FAK expression and OS in various types of cancer was investigated. No significant correlation was observed, although this meta-analysis only included two studies which focused on breast cancer cases [31]. In contrast, the present meta-analysis included a total of eight breast cancer articles to investigate an association between FAK protein expression and clinicopathologic factors of breast cancer. In the latter, higher FAK protein expression was found to be related to worse OS (HR, 1.43; 95% CI, 1.12–1.83; P = 0.004). However, the high probability of false-positive reports in the FPRP analysis performed indicates that additional research is needed to confirm this relationship. Meanwhile, no significant correlation was observed between DFS and FAK protein expression (HR, 1.31; 95% CI, 0.92–1.85; P = 0.14). It is possible that this insignificant correlation to DFS may be due to an insufficient sample size was verified with the TSA analysis.
As indicated above, enhanced FAK expression was linked to shorter OS, yet not to DFS, in the breast cancer cases examined. Aglan et al. [32] previously demonstrated that increased total FAK (tFAK) expression correlates with poor DFS in breast cancer. Similarly, Charpin et al. [33] reported that high FAK expression is related to worse DFS in breast cancer. These results, in combination with those of the present study, prove that FAK may represent a valuable prognostic marker of shorter DFS and OS in patients with breast cancer. Indeed, previous studies have reported a relationship between FAK expression and survival outcome for various types of tumors. For example, Thanapprapasr et al. [34] observed that elevated pFAKY397 expression is associated with poor OS in metastatic osteosarcoma. Similarly, de Vicente et al. [35] observed that enhanced expression of FAK correlates with adverse disease-specific survival (DSS) in oral squamous cell carcinoma (OSCC). Min et al. [36] also demonstrated that FAK regulates increased migration and invasion by carcinoma-associated fibroblasts (CAFs) in OSCC via an increase in MCP-1/CCL2 expression. In hepatocellular carcinoma, Ko et al. [37] revealed that elevated expression of FAK is associated with reduced OS. Ko et al. [37] further indicated that FAK may collaborate or crosstalk with 14–3-3ε to promote hepatocellular cancer progression by activating the NFκB signaling pathway. Albasri et al. [38] found that positive nuclear P-FAK expression correlates with poor DSS in colorectal cancer. Meanwhile, Bian et al. [39] further demonstrated that ACP5 promotes colorectal cancer cell proliferation via the FAK/PI3K/AKT signaling pathway. However, Giaginis et al. [40] reported that enhanced FAK expression correlates with favorable OS in patients with diffuse-type gastric cancer. In non-small cell lung cancer, Dy et al. [14] did not find a significant association between FAK expression and OS. Thus, in various tumor types, FAK expression has been associated with variable prognosis. This variability potentially contributes to the value of FAK expression for clinical pathological and cellular characteristics in different carcinomas. Thus, FAK represents a novel and pivotal indicator for evaluating the biological behavior and prognosis of carcinomas. Moreover, FAK represents an effective therapeutic target for regulating the expression and activity of FAK, and for controlling and treating cancer by promoting apoptosis and inhibiting the proliferation of tumor cells.
Numerous studies have demonstrated that certain molecular markers, including negative ER expression, negative PR expression, positive HER2 expression, TNBC, and a high Ki-67 expression level, indicate poor prognosis in breast cancer [41,42]. In the present study, all of these factors were associated with increased FAK expression, and those results support our finding that increased FAK expression was correlated with worse OS in breast cancer. Moreover, Abubakar et al. [43] revealed that up-regulation of the proliferating cell nuclear antigen, Ki-67, is associated with an adverse prognosis in breast cancer. The present results also demonstrate that increased FAK protein expression correlates with a high Ki-67 expression level, thereby suggesting that elevated FAK expression promotes the proliferation of breast cancer cells. Kaur et al. [44] have revealed that p53 also contributes to the pathogenesis of breast cancer via its roles in various cellular processes, including DNA damage repair, cell cycle arrest, and apoptosis. The present results demonstrate that enhanced FAK expression correlates with positive p53 status. Therefore, these results support the hypothesis that FAK plays a role in regulating p53 to affect tumor growth and proliferation.
There were limitations associated with the present meta-analysis. First, the numbers of included studies and patients were relatively small. Thus, further studies involving more samples are needed to confirm our results. Second, the antibodies used, staining locations, and cut-off values for FAK expression varied among the eligible studies. Therefore, it is possible that these factors contributed to the heterogeneity observed among the eligible studies examined.
In conclusion, the present meta-analysis demonstrates that increased FAK protein expression is associated with worse OS in breast cancer. Moreover, elevated FAK protein expression correlates with negative ER expression, negative PR expression, positive HER2 expression, TNBC, high nuclear grade, high Ki-67 expression level, and positive p53 status in breast cancer. Thus, support is provided for FAK to serve as an indicator of the biological behavior and prognosis of carcinomas, while also serving as an effective therapeutic target.
The following are the supplementary data related to this article.
Search strategy.
Quality assessment of the included studies using the Newcastle-Ottawa Scale.
Author contributions
Miao Deng: Conceptualization, Methodology, Software. Weiqiang Qiao: Data curation, Writing - Original draft preparation. Wenhui Wang: Visualization, Investigation. Heyang Liu: Supervision. Wanying Guo: Software, Validation. Peng Li: Writing - Reviewing and Editing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Saini G., Mittal K., Rida P., Janssen E., Gogineni K., Aneja R. Panoptic view of prognostic models for personalized breast cancer management. Cancers (Basel) 2019;11 doi: 10.3390/cancers11091325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J., Yi Q., Tang L. The roles of nuclear focal adhesion kinase (FAK) on cancer: a focused review. J. Exp. Clin. Cancer Res. 2019;38:250. doi: 10.1186/s13046-019-1265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naser R., Aldehaiman A., Diaz-Galicia E., Arold S.T. Endogenous control mechanisms of FAK and PYK2 and their relevance to cancer development. Cancers (Basel) 2018;10 doi: 10.3390/cancers10060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee B.Y., Timpson P., Horvath L.G., Daly R.J. FAK signaling in human cancer as a target for therapeutics. Pharmacol. Ther. 2015;146:132–149. doi: 10.1016/j.pharmthera.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Golubovskaya V.M. Survival signaling through focal adhesion kinase in tumors. Anti Cancer Agents Med. Chem. 2013;13:531. doi: 10.2174/1871520611313040001. [DOI] [PubMed] [Google Scholar]
- 7.Rigiracciolo D.C., Santolla M.F., Lappano R. Focal adhesion kinase (FAK) activation by estrogens involves GPER in triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. 2019;38:58. doi: 10.1186/s13046-019-1056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi-Smiraglia A., Paesante S., Bakin A.V. Integrin beta5 contributes to the tumorigenic potential of breast cancer cells through the Src-FAK and MEK-ERK signaling pathways. ONCOGENE. 2013;32:3049–3058. doi: 10.1038/onc.2012.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berdan C.A., Ho R., Lehtola H.S. Parthenolide covalently targets and inhibits focal adhesion kinase in breast cancer cells. Cell Chem. Biol. 2019;26:1027–1035. doi: 10.1016/j.chembiol.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanteti R., Mirzapoiazova T., Riehm J.J. Focal adhesion kinase a potential therapeutic target for pancreatic cancer and malignant pleural mesothelioma. Cancer Biol. Ther. 2018;19:316–327. doi: 10.1080/15384047.2017.1416937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores A., Dias K.B., Hildebrand L.C., Oliveira M.G., Lamers M.L., Sant'Ana F.M. Focal adhesion kinases in head and neck squamous cell carcinoma. J. Oral Pathol. Med. 2018;47:246–252. doi: 10.1111/jop.12674. [DOI] [PubMed] [Google Scholar]
- 12.Gomez D.P.T., Cebrian A., Fernandez-Acenero M.J. Focal adhesion kinase: predictor of tumour response and risk factor for recurrence after neoadjuvant chemoradiation in rectal cancer. J. Cell. Mol. Med. 2016;20:1729–1736. doi: 10.1111/jcmm.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang N., Arteaga M., Zaidi A. FAK kinase activity is required for the progression of c-MET/beta-catenin-driven hepataocellular carcinoma. Gene Expr. 2016;17:79–88. doi: 10.3727/105221616X691604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dy G.K., Ylagan L., Pokharel S. The prognostic significance of focal adhesion kinase expression in stage I non-small-cell lung cancer. J. Thorac. Oncol. 2014;9:1278–1284. doi: 10.1097/JTO.0000000000000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andisha N.M., McMillan D.C., Gujam F., Roseweir A., Edwards J. The relationship between phosphorylation status of focal adhesion kinases, molecular subtypes, tumour microenvironment and survival in patients with primary operable ductal breast cancer. Cell. Signal. 2019;60:91–99. doi: 10.1016/j.cellsig.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y., Maimaiti Y., Yu P. Overexpression of enhancer of zeste homolog 2 (EZH2) and focal adhesion kinase (FAK) is associated with cancer metastasis and poor prognosis in breast cancer. Int. J. Clin. Exp. Med. 2017;10:2672–2683. [Google Scholar]
- 17.Almstedt K., Sicking I., Battista M.J. Prognostic significance of focal adhesion kinase in node-negative breast cancer. Breast Care (Basel) 2017;12:329–333. doi: 10.1159/000477895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yom C.K., Noh D.Y., Kim W.H., Kim H.S. Clinical significance of high focal adhesion kinase gene copy number and overexpression in invasive breast cancer. Breast Cancer Res. Treat. 2011;128:647–655. doi: 10.1007/s10549-010-1150-2. [DOI] [PubMed] [Google Scholar]
- 19.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 20.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 2010;8:336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DerSimonian R., Laird N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials. 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 24.Egger M., Davey S.G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wetterslev J., Thorlund K., Brok J., Gluud C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J. Clin. Epidemiol. 2008;61:64–75. doi: 10.1016/j.jclinepi.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Wacholder S., Chanock S., Garcia-Closas M., El G.L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golubovskaya V.M., Ylagan L., Miller A. High focal adhesion kinase expression in breast carcinoma is associated with lymphovascular invasion and triple-negative phenotype. BMC Cancer. 2014;14:769. doi: 10.1186/1471-2407-14-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lark A.L., Livasy C.A., Dressler L. High focal adhesion kinase expression in invasive breast carcinomas is associated with an aggressive phenotype. Mod. Pathol. 2005;18:1289–1294. doi: 10.1038/modpathol.3800424. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz K.J., Grabellus F., Callies R. High expression of focal adhesion kinase (p125FAK) in node-negative breast cancer is related to overexpression of HER-2/neu and activated Akt kinase but does not predict outcome. Breast Cancer Res. 2005;7:R194–R203. doi: 10.1186/bcr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theocharis S.E., Klijanienko J.T., Padoy E., Athanassiou S., Sastre-Garau X.X. Focal adhesion kinase (FAK) immunocytochemical expression in breast ductal invasive carcinoma (DIC): correlation with clinicopathological parameters and tumor proliferative capacity. Med. Sci. Monit. 2009;15:R221–R226. [PubMed] [Google Scholar]
- 31.Zeng X.Q., Li N., Ma L.L., Tseng Y.J., Zhao N.Q., Chen S.Y. Prognostic value of focal adhesion kinase (FAK) in human solid carcinomas: a meta-analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0162666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aglan H., Timbrell S., Cramer A., Foden P., Clarke R., Bundred N. Focal adhesion kinase and aldehyde dehydrogenase 1 are prognostic cancer stem cell markers in invasive ductal carcinoma. Breast Cancer Res. Tr. 2018;167:330. [Google Scholar]
- 33.Charpin C., Secq V., Giusiano S. A signature predictive of disease outcome in breast carcinomas, identified by quantitative immunocytochemical assays. Int. J. Cancer. 2009;124:2124–2134. doi: 10.1002/ijc.24177. [DOI] [PubMed] [Google Scholar]
- 34.Thanapprapasr K., Nartthanarung A., Thanapprapasr D., Jinawath A. pFAK-Y397 overexpression as both a prognostic and a predictive biomarker for patients with metastatic osteosarcoma. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Vicente J.C., Rosado P., Lequerica-Fernandez P., Allonca E., Villallain L., Hernandez-Vallejo G. Focal adhesion kinase overexpression: correlation with lymph node metastasis and shorter survival in oral squamous cell carcinoma. Head Neck. 2013;35:826–830. doi: 10.1002/hed.23038. [DOI] [PubMed] [Google Scholar]
- 36.Min A., Zhu C., Wang J. Focal adhesion kinase knockdown in carcinoma-associated fibroblasts inhibits oral squamous cell carcinoma metastasis via downregulating MCP-1/CCL2 expression. J. Biochem. Mol. Toxicol. 2015;29:70–76. doi: 10.1002/jbt.21669. [DOI] [PubMed] [Google Scholar]
- 37.Ko B.S., Jan Y.J., Chang T.C. Upregulation of focal adhesion kinase by 14-3-3epsilon via NFkappaB activation in hepatocellular carcinoma. Anti Cancer Agents Med. Chem. 2013;13:555–562. doi: 10.2174/1871520611313040004. [DOI] [PubMed] [Google Scholar]
- 38.Albasri A., Fadhil W., Scholefield J.H., Durrant L.G., Ilyas M. Nuclear expression of phosphorylated focal adhesion kinase is associated with poor prognosis in human colorectal cancer. Anticancer Res. 2014;34:3969–3974. [PubMed] [Google Scholar]
- 39.Bian Z.Q., Luo Y., Guo F., Huang Y.Z., Zhong M., Cao H. Overexpressed ACP5 has prognostic value in colorectal cancer and promotes cell proliferation and tumorigenesis via FAK/PI3K/AKT signaling pathway. Am. J. Cancer Res. 2019;9:22–35. [PMC free article] [PubMed] [Google Scholar]
- 40.Giaginis C.T., Vgenopoulou S., Tsourouflis G.S., Politi E.N., Kouraklis G.P., Theocharis S.E. Expression and clinical significance of focal adhesion kinase in the two distinct histological types, intestinal and diffuse, of human gastric adenocarcinoma. Pathol. Oncol. Res. 2009;15:173–181. doi: 10.1007/s12253-008-9120-2. [DOI] [PubMed] [Google Scholar]
- 41.Lee K.L., Kuo Y.C., Ho Y.S., Huang Y.H. Triple-negative breast cancer: current understanding and future therapeutic breakthrough targeting cancer stemness. Cancers (Basel) 2019;11 doi: 10.3390/cancers11091334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicolini A., Ferrari P., Duffy M.J. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin. Cancer Biol. 2018;52:56–73. doi: 10.1016/j.semcancer.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Abubakar M., Figueroa J., Ali H.R. Combined quantitative measures of ER, PR, HER2, and KI67 provide more prognostic information than categorical combinations in luminal breast cancer. Mod. Pathol. 2019;32:1244–1256. doi: 10.1038/s41379-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur R.P., Vasudeva K., Kumar R., Munshi A. Role of p53 gene in breast cancer: focus on mutation spectrum and therapeutic strategies. Curr. Pharm. Des. 2018;24:3566–3575. doi: 10.2174/1381612824666180926095709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Search strategy.
Quality assessment of the included studies using the Newcastle-Ottawa Scale.