Abstract
In contrast to Eurasia and North America, powdery mildews (Ascomycota, Erysiphales) are understudied in Australia. There are over 900 species known globally, with fewer than currently 60 recorded from Australia. Some of the Australian records are doubtful as the identifications were presumptive, being based on host plant-pathogen lists from overseas. The goal of this study was to provide the first comprehensive catalog of all powdery mildew species present in Australia. The project resulted in (i) an up-to-date list of all the taxa that have been identified in Australia based on published DNA barcode sequences prior to this study; (ii) the precise identification of 117 specimens freshly collected from across the country; and (iii) the precise identification of 30 herbarium specimens collected between 1975 and 2013. This study confirmed 42 species representing 10 genera, including two genera and 13 species recorded for the first time in Australia. In Eurasia and North America, the number of powdery mildew species is much higher. Phylogenetic analyses of powdery mildews collected from Acalypha spp. resulted in the transfer of Erysiphe acalyphae to Salmonomyces, a resurrected genus. Salmonomyces acalyphae comb. nov. represents a newly discovered lineage of the Erysiphales. Another taxonomic change is the transfer of Oidium ixodiae to Golovinomyces. Powdery mildew infections have been confirmed on 13 native Australian plant species in the genera Acacia, Acalypha, Cephalotus, Convolvulus, Eucalyptus, Hardenbergia, Ixodia, Jagera, Senecio, and Trema. Most of the causal agents were polyphagous species that infect many other host plants both overseas and in Australia. All powdery mildews infecting native plants in Australia were phylogenetically closely related to species known overseas. The data indicate that Australia is a continent without native powdery mildews, and most, if not all, species have been introduced since the European colonization of the continent.
Keywords: host jumps, host-pathogen interactions, invasive species, obligate biotrophs, plant-microbe interactions, rapid evolution
Introduction
Powdery mildews (Ascomycota, Erysiphales) are common obligate biotrophic fungal plant pathogens, comprising ~900 species that infect more than 10,000 dicot and monocot plant species globally (Braun and Cook, 2012). Many powdery mildew species cause economically important diseases of agricultural and horticultural crops, including wheat, barley, grapevine, fruit, and vegetable species (Glawe, 2008). Others are forest pathogens (Marçais and Desprez-Loustau, 2014). Some powdery mildews have become invasive in different parts of the world (Kiss, 2005; Desprez-Loustau et al., 2010), posing plant health biosecurity risks (Jones and Baker, 2007; Brasier, 2008; Biosecurity Australia, 2010; Desprez-Loustau et al., 2010). Two species, Blumeria graminis infecting cereals, and Erysiphe necator infecting grapevine, have become model species in plant pathology research (Gadoury et al., 2012; Bindschedler et al., 2016), while the interactions between Podosphaera plantaginis and its host Plantago lanceolata have long been in focus in the study of wild plant pathosystems (Susi et al., 2015).
Since the early 2000s, molecular phylogenetic analyses have shown that the traditional generic concept of the Erysiphales does not mirror their phylogeny based on the morphological characteristics of the sexual morphs (chasmothecia) (Braun et al., 2002). Surprisingly, grouping species according to the characteristics of their asexual morphs reflect their molecular phylogeny (for reviews, see Takamatsu, 2004, 2013a). This discovery triggered major taxonomic revisions (Braun et al., 2002), and the currently accepted generic concept is now based on a combination of the morphological characteristics of the asexual and sexual morphs (Braun, 2011; Takamatsu, 2013b). The most recent monograph of powdery mildews recognized 17 genera (Braun and Cook, 2012). A further genus, Bulbomicroidium, was described from Mexico (Marmolejo et al., 2018).
In Australia, despite their importance, the biodiversity of the Erysiphales has received little attention. Based on morphological observations and field data accumulated until the early 1980s, Walker (1983) noted that powdery mildews were not recorded on native Australian plants under natural conditions. Braun and Cook (2012) listed ~50 powdery mildew species for Australia. However, some of these records are doubtful as the original identifications were presumptive, being based only on the morphology of the asexual morph, in the absence of the sexual morphs, and/or plant disease lists from overseas. It is difficult, and sometimes impossible, to distinguish powdery mildew species belonging to the same genus by morphological characteristics of the asexual morph (Takamatsu et al., 2002; Jankovics et al., 2008; Bereczky et al., 2015; Desprez-Loustau et al., 2018).
Like in other fungal groups, the advent of molecular biology has facilitated a vastly more precise dissection of the relationships among taxa. DNA barcodes, particularly sequences of the internal transcribed spacer (ITS) region of the nuclear ribosomal DNA (nrDNA), have been most useful for the identification of powdery mildews at the species level, while sequences of the nrDNA 18S and 28S region have supported the identification and classification of the genera (Braun et al., 2002; Takamatsu, 2004, 2013a; Takamatsu et al., 2015a,b; Cabrera et al., 2018; Marmolejo et al., 2018). Other DNA regions have also been tested as molecular barcodes for some groups of the powdery mildews (Inuma et al., 2007; Desprez-Loustau et al., 2017; Ellingham et al., 2019; Qiu et al., 2020), but these studies have not yet resulted in the development of new DNA barcodes that are useful to distinguish species across the Erysiphales. To date, ITS sequences is still the sole species-level DNA barcode available for this fungal group, despite its limitations. These include intragenomic variations within some powdery mildew species on one hand (Kovács et al., 2011), and no differences between some other, morphologically distinguishable species on the other hand (Braun et al., 2019; Qiu et al., 2020). ITS sequences as species barcodes have limitations in other fungal groups, as well (Kiss, 2012; Stadler et al., 2020).
Most of the species listed by Braun and Cook (2012) as present in Australia have not been supported by DNA sequence data. Molecular studies of Australian powdery mildews (e.g., Cunnington et al., 2003, 2004a,b, 2005a,b; Liberato and Cunnington, 2006, 2007; McTaggart et al., 2012) identified common exotic species on crops and wild plants that were introduced to Australia during European colonization, and had major impacts on the entire vegetation of the continent (Kirkpatrick, 1999; Cook and Dias, 2006; Randall, 2007; Fensham and Laffineur, 2019). A comprehensive molecular study of species of Golovinomyces collected from non-native asteraceous hosts concluded that all these powdery mildews were introduced to Australia from the northern hemisphere (Cunnington et al., 2010). Only three powdery mildew taxa have been considered native to Australia based on molecular studies, specifically, Oidium ixodiae on Ixodia achillaeoides (Cunnington et al., 2005a); Neoerysiphe kerribeeensis, on the indigenous Senecio glossanthus, and several other Senecio spp. introduced to Australia (Beilharz et al., 2010); and Pseudoidium hardenbergiae on Hardenbergia spp. (Cunnington et al., 2004b). However, Cunnington et al. (2004b) did not exclude the possibility that Ps. hardenbergiae was introduced to Australia from New Zealand, where it was originally described (Boesewinkel, 1977), while Braun and Cook (2012) noted that the asexual morphs on Hardenbergia spp. in Australia and North America may differ from Ps. hardenbergiae. Neither O. ixodiae nor N. kerribeeensis have been reported outside Australia. Curiously, another powdery mildew species known only from Australia is Golovinomyces lycopersici on tomato, a vegetable introduced from overseas (Kiss et al., 2001, 2005; Braun et al., 2019). This tomato pathogen is closely related to the Australian O. ixodiae (Cunnington et al., 2005a), and also to G. longipes that causes epidemics on solanaceous vegetables and ornamentals in Europe and the USA (Kiss et al., 2008; Kovács et al., 2011).
The distinction between native vs. introduced powdery mildews has special relevance for Australia, an island continent where the native vegetation has largely evolved in isolation until the start of European settlement in 1788 (Kirkpatrick, 1999; Randall, 2007; Fensham and Laffineur, 2019). That particular year is considered as a sharp biogeographic landmark in the history of the Australian flora of vascular plants (Fensham and Laffineur, 2019) as it marked the commencement of deliberate and accidental human-assisted introductions of altogether over 28,000 plant species from overseas, including agricultural and horticultural crops, ornamentals, and pasture species (Randall, 2007). Walker (1983) noted that until the early 1980s, most reports of powdery mildews in Australia were on dicots and monocots introduced from overseas after 1788, and only a few on natives. Walker (1983) also observed that all of the reports of powdery mildew infections on native plants in Australia were observed in artificial conditions, particularly glasshouses or nurseries. Clearly, the distribution and host range of powdery mildews in Australian vegetation needs to be considered in the light of the massive introduction of exotic plant species since 1788.
This study provides the first comprehensive database of powdery mildew species identified in Australia. Our objectives were to (i) compile an up-to-date list of all the powdery mildew taxa identified in Australia based on published DNA barcode sequences; and (ii) identify as many powdery mildew species as possible from across the country based on morphological characteristics and nrDNA ITS sequences. Special attention was paid to the identification of powdery mildews infecting native Australian plants to better understand the host ranges and global distribution of the Erysiphales.
Materials and Methods
Compiling a List of Powdery Mildews Identified in Australia Based on DNA Barcode Sequences Published Prior This Study
Searches were conducted in the NCBI nucleotide database, GenBank, with each genus name of the Erysiphales listed by Braun and Cook (2012), supplemented with “AND Australia” (e.g., “Oidium AND Australia”), to find all the publicly available DNA sequences for powdery mildew specimens collected in Australia. All the old and the currently accepted names of anamorphic and teleomorphic genera were included in these searches. Due to a number of reasons, some ITS sequences deposited in GenBank as fungal entries are inaccurate (Nilsson et al., 2006; Bidartondo, 2008; Irinyi et al., 2015; Hibbett et al., 2016; Selosse et al., 2016). Therefore, BLASTn searches were carried out with the retrieved entries to ensure that these represented powdery mildews.
When necessary, powdery mildew taxon names retrieved from GenBank were updated to correspond to the currently accepted nomenclature, e.g., for taxa belonging to the well-known species complex G. orontii (Braun et al., 2019). Records representing other well-known species complexes that have not been resolved from a taxonomic perspective, e.g. B. graminis (Braun and Cook, 2012), were considered as a single taxon. Those powdery mildew records in GenBank that have only been determined at the generic level were considered as representative of different species if their ITS sequences differed in more than 3 nucleotide positions based on the results obtained by Kovács et al. (2011).
Field Collection, DNA Extraction, and Microscopic Examination of Specimens
Starting from 2017, powdery mildew-infected plant samples were collected from across Australia. Systematic surveys were conducted by four plant pathologists in Queensland and New South Wales, and ad-hoc surveys by 20 members of the National Plant Biosecurity Diagnostic Network. Powdery mildew mycelia were removed from the host plant surfaces with 1–1.5 cm2 pieces of cellotape, and genomic DNA was extracted using the buffers from an Extract-N-Amp Plant PCR kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. A part of the powdery mildew material was examined and photographed under a light microscope whilst still fresh. All plant samples were dried and pressed as herbarium specimens, and then rehydrated in the laboratory for comprehensive morphological examinations if required. Their rehydration was conducted by boiling small pieces of infected plant materials in lactic acid (100% v/v) on a microscope slide, as described by Shin and La (1993). During microscopy with bright field, phase contrast, and differential interference contrast (DIC) optics, the following information was noted: shape and size of conidia, based on measurements of 25 conidia per specimen, presence or absence of fibrosin bodies in fresh conidia, nature of conidiogenesis, characteristics of the conidiophore, e.g., size and shape of foot cell, position of the basal septum, shape and position of hyphal appressoria, position of germ tubes of conidia, when found, and shape of appressoria on germ tubes of conidia. Droplets in conidia that appeared to be made of lipids were stained with 0.3% v/v alcoholic Sudan Black B solution by slightly warming the microscope slides over a flame before microscopic examination. Conidial germination in fresh powdery mildew samples collected from Acalypha spp. was studied on cellophane placed on 1.5% water agar (Szentiványi and Kiss, 2003), and the epidermis of onion scale (To-anun et al., 2005), after incubation at 25°C for 24, 48, 72, and 96 h. Representative specimens of each host-pathogen combination were deposited at the Queensland Plant Pathology Herbarium (BRIP), the New South Wales Plant Pathology and Mycology Herbarium (DAR), and the Victorian Plant Pathology Herbarium (VPRI).
PCR Amplification of the ITS Region From Fresh Specimens
Amplification of the ITS region followed the nested PCR method described by Cunnington et al. (2003), with some modifications, and using the Extract-N-Amp Plant PCR kit. The first PCRs were carried out in 20 μL final volumes, consisting of 10 μL hot start PCR ReadyMix from the Extract-N-Amp Plant PCR kit, 1 μL of each powdery mildew-specific primers PMITS1 and PMITS2 (Table 1) at 10 μM, 6 μL ultrapure water, and 2 μL total genomic DNA. The nested reactions were also carried out in 20 μL final volumes, and consisted of 10 μL ReadyMix, 1 μL of each universal fungal primers ITS1-F (Gardes and Bruns, 1993) and ITS4 (White et al., 1990) at 10 μM, 7 μL ultrapure water, and 1 μL of the product of the first PCRs. The conditions for the first PCRs were as follows: 94°C for 10 min; 35 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C; and finally 10 min at 72°C. The nested PCRs consisted of 94°C for 5 min; 35 cycles of 45 s at 94°C, 45 s at 55°C, and 1 min at 72°C; and finally 10 min at 72°C. PCR products of the nested reactions were purified and sequenced by Macrogen Inc. (Seoul, Korea) with primers ITS1-F and ITS4.
Table 1.
The powdery mildew-specific, and other specific rDNA primers used in this work.
| Primer designation | Orientation | Sequence (5′ → 3′) | Purpose | References |
|---|---|---|---|---|
| PMITS1 | Forward | TCG GAC TGG CCY AGG GAG A | Amplification of the ITS region and a part of the flanking 18S and 28S rDNA regions in most powdery mildews | Cunnington et al., 2003 |
| PMITS2 | Reverse | TCA CTC GCC GTT ACT GAG GT | Cunnington et al., 2003 | |
| PM-Ei-F1 | Forward | CCG TGT CGA TTT GTA TCG TG | Amplification of a large part of the entire ITS region in Erysiphe izuensis | This paper |
| PM-Ei-R1 | Reverse | ACT CTG TCG CGA GAA GCA AG | This paper | |
| T3 | Reverse | ACG CTC GAA CAG GCA TGC CC | Binding site at the 3′-end of the 5.8S rDNA region of most powdery mildews and some other fungi | Hirata and Takamatsu, 1996 |
| PM5 | Forward | TTG CTT TGG CGG GCC GGG | Binding site in the ITS1 region of most powdery mildews | Takamatsu and Kano, 2001 |
| TW14 | Reverse | GCT ATC CTG AGG GAA ACT TC | Binding site at the 3′-end of the D1-D2 domain of the 28S rDNA region in most powdery mildews and some other fungi | https://nature.berkeley.edu/brunslab/tour/primers.html#28s |
| T4 | Forward | TCA ACA ACG GAT CTC TTG GC | Binding site at the 5′-end of the 5.8S rDNA region in most powdery mildews and some other fungi | Hirata and Takamatsu, 1996 |
| T2 | Forward | GGG CAT GCC TGT TCG AGC GT | Binding site at the 3′-end of the 5.8S rDNA region in most powdery mildews and some other fungi | Hirata and Takamatsu, 1996 |
| Pm-LSU-R2 | Reverse | ACT CCA AGG GAG CCA GAT TT | Amplification of the D1-D2 domain of the 28S rDNA in Salmonomyces, in combination with the universal primer LR3 | This paper |
| Pm-LSU-R3 | Reverse | GCT TTA CAT AGG CGC AGG TC | This paper |
PCR Amplification of an ITS Fragment of E. izuensis With Specific Primers Designed in This Study
Amplifications from fresh powdery mildew colonies sampled from the leaves of Rhododendron spp. as described above, consistently yielded ITS sequences that were 98–99% similar to those of a number of uncultured fungi reported from environmental samples. As the powdery mildew species infecting the examined Rhododendron leaves was identified as E. izuensis based on the morphology of the asexual and sexual morphs, species-specific primers PM-Ei-F1 and PM-Ei-R1 (Table 1) were designed based on the ITS sequence of this species available in GenBank under accession number LC009975 using the software Primer3 (Untergasser et al., 2012). Instead of ITS1-F/ITS4, the E. izuensis-specific PM-Ei-F1/PM-Ei-R1 primer pair was used to amplify an ~450 bp long fragment, including the partial ITS1, complete 5.8S, and partial ITS2 regions in the powdery mildew samples from Rhododendron spp. using the nested PCR protocol described above. PCR products were sequenced by Macrogen Inc. with primers PM-Ei-F1 and PM-Ei-R1.
DNA Extraction and PCR Amplification of the ITS Region From Herbarium Specimens Collected Between 1975 and 2013 in Australia
During a review of older powdery mildew specimens available at BRIP, collected between 1975 and 2013, 30 specimens were selected based on the amount and quality of the powdery mildew mycelium available on plant tissues, assessed under a dissecting microscope. Approximately 20 mg of infected plant tissue was taken from dried herbarium specimens, and disrupted in a Tissue Lyser (QIAGEN) with 2.5 mm glass beads (Daintree Scientific). Genomic DNA was extracted with the DNeasy Plant Mini Kit (QIAGEN) according to the manufacturer's instructions, with the modification that N-phenacylthiazolium bromide (PTB; Epichem, Perth) was added to the lysis buffer (final concentration of 2.5 mM). PTB is known to enhance DNA purification efficiency from herbarium specimens (Lister et al., 2008; Telle and Thines, 2008). Amplification of the ITS region was as described above for fresh specimens, with the exception that the PCR was performed with Phusion® High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs), supplemented with Bovine Serum Album (New England Biolabs; final concentration of 1 μg/μL). Purification of the nested PCR products and sequencing was as described above for fresh specimens.
Study of Herbarium Specimens From China and Argentina
Powdery mildew specimens collected from Acalypha spp. in China and Argentina (Supplementary Table 1) were rehydrated during boiling in lactic acid and observed under a light microscope as described above. PCR amplifications of the ITS region were carried out according to the protocol described above.
Sequencing the ITS Region in Powdery Mildew Specimens Collected From Acalypha spp. Following Cloning the PCR Product
Chromatograms obtained from direct sequencing of the ITS region in all the fresh powdery mildew specimens collected from Acalypha spp. in Australia contained double peaks at certain positions. Since intra-sample variation in ITS sequences of powdery mildew species has been reported previously (Kovács et al., 2011), the ITS region of two specimens, one from a young powdery mildew colony infecting A. nemorum, and the other from a young colony from A. wilkesiana, were sequenced after cloning, to reveal potential intra-sample ITS variants. The nested PCR products from these two specimens were cloned at Macrogen Inc. using TA cloning. Ten recombinant plasmids per PCR product were sequenced with the primer M13F.
PCR Amplification of the 28S and 18S nrDNA Regions in Powdery Mildew Specimens Collected From Acalypha spp.
The 18S nrDNA region was amplified in two steps: the first PCR was carried out with the universal fungal primer NS1 (White et al., 1990) and the powdery mildew-specific primer T3 (Hirata and Takamatsu, 1996), followed by two sets of semi-nested PCRs, one with the universal fungal primers NS1 and ITS2, and another one with NS1 and NS8 (White et al., 1990). The 5′-end of the 28S rDNA (including domains D1 and D2) was also amplified in two steps, the first being PCRs with primers PM5 and TW14, followed by two sets of semi-nested PCRs, one with primers T4 and TW14, and another one with T2 (Hirata and Takamatsu, 1996) and TW14. All the specific primers used in this work are listed in Table 1.
The composition of the PCR mixes was as described above for the ITS PCRs from fresh powdery mildew specimens. The parameters for the first PCRs were as follows: 94°C for 10 min; 35 cycles of 1 min at 94°C, 1 min at 52°C, and 2 min at 72°C; and finally 10 min at 72°C. The semi-nested PCRs consisted of 94°C for 5 min; 35 cycles of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C; and finally 10 min at 72°C. All the products of the semi-nested PCRs were purified and sequenced at Macrogen Inc., using primers NS1, NS8, and ITS2 for the 18S products, and primers T2, T4, and TW14 for the 28S products.
A poly-A/T motif in the ITS2 region made all the chromatograms of the 28S products unreliable starting from that motif; therefore, two new nested reverse primers, Pm-LSU-R2 and Pm-LSU-R3 (Table 1) were designed at ~180 and 300 bp downstream of the repeat region, using Primer3, to enable re-sequencing of the repetitive regions to achieve reliable consensus sequences. Each of these new reverse primers was used in combination with the universal fungal primer LR3 (Vilgalys and Hester, 1990) in nested PCRs, using the PCR products from the first 28S PCRs as templates. The products of these nested PCRs were purified and sequenced at Macrogen Inc. with primers LR3, Pm-LSU-R2, and Pm-LSU-R3.
Initial Sequence Analyses
Sequences were compiled from chromatograms, following visual inspections for potential polymorphisms. In order to minimize false positives arising from potential sequencing errors, stringent quality control was conducted. To obtain a reliable consensus sequence, single nucleotide polymorphisms were only accepted if the base call quality was at least 30, and also the polymorphism occurred in more than one sequence (James et al., 2009). Consensus sequences were produced by trimming and assembling the forward and reverse sequences using Geneious Prime 2019.1.3 (Biomatters Ltd.).
Phylogenetic Analyses of 18S, 28S, and 5.8S Sequences to Investigate the Generic Placement of the Powdery Mildews Infecting Acalypha spp. Within the Erysiphales
To reveal the generic placement of the powdery mildews collected from Acalypha spp. within the Erysiphales, a three-locus phylogenetic analysis was conducted using sequences of the nrDNA 28S, 18S, and 5.8S regions. Sequences of representative specimens of all but one of the 18 powdery mildew genera accepted to date were retrieved from GenBank (Supplementary Table 2). The genus Takamatsuella, for which no authentic sequence data is available, could not be included in this analysis. In Brasiliomyces, there are only two specimens of Br. malachrae that have been included in molecular studies so far; a large part of 18S, together with the ITS region was sequenced from specimen MUMH 3119, while the ITS and a large part of 28S was determined from specimen MUMH 3093 (Cabrera et al., 2018). The sequence available for the latter specimen was included in the analysis, and the nucleotides for the 18S sequence were coded as missing. This was also done in the case of all the four powdery mildew specimens known from Acalypha spp. to date, and described as Ps. javanicum (Meeboon and Takamatsu, 2015), as the 18S sequences are not available for these specimens (Supplementary Table 2).
All sequence alignments were generated using MAFFT v. 7.388 (Katoh and Standley, 2013), and visually inspected for potential misalignments or ambiguously-aligned regions. Due to the variable size of the 28S and 18S sequences retrieved from GenBank, alignments for these loci were trimmed to the length of the shortest sequence. For the multi-locus phylogenetic analysis, congruency of loci was tested through incongruence-length difference (ILD) partition homogeneity test (Farris et al., 1995) implemented in PAUP v. 4.0b10 (Swofford, 2002), which detected no significant conflict in tree topologies (P-value = 1.0), and allowed concatenation of loci.
Phylogenetic analyses were conducted using two methods; Bayesian Inference (BI) and Maximum likelihood (ML). For BI, the best-fit nucleotide substitution model for each locus was determined by comparing the Akaike Information Criterion using PAUP v. 4.0 (Swofford, 2002) and MrModeltest v. 2.3 (Nylander, 2004). Two Markov Chain Monte Carlo (MCMC) chains were run using MrBayes v. 3.2.4 (Ronquist and Huelsenbeck, 2003). One tree per 100 generations was saved, and the runs were ended when the standard deviation of split frequencies reached below 0.01. The 50% majority rule consensus tree was estimated after a 25% burn-in of the saved trees. Maximum likelihood analysis was conducted in RAxML v. 8.2.11 (Stamatakis, 2014) using the GTRGAMMA model applied to the individual partitions, with 1,000 bootstrap replicates. Alignments and trees were deposited in TreeBASE (submission no. 25949).
Phylogenetic Analyses of ITS Sequences of Specimens Belonging to Different Genera
ITS sequences of powdery mildew species belonging to distantly related genera are too divergent to allow for non-ambiguous alignment of all sequences (Takamatsu et al., 1998, 2015a; Mori et al., 2000). Therefore, the ITS sequences determined in this study were grouped in the following four subsets of closely related taxa: (i) the Microsphaera lineage of the genus Erysiphe; (ii) Podosphaera and Sawadaea species; (iii) Golovinomyces, Neoerysiphe, Arthrocladiella and Microidium species; and (iv) Leveillula species. At least 95% similar ITS sequences coming from similar taxa collected overseas were added to each of these four subsets based on BLAST searches (Supplementary Table 3), and each subset of sequences was analyzed separately. No ITS analysis was performed for Blumeria as this genus was represented by only three specimens collected in Australia, and Blumeria is only distantly related to all the other genera of the Erysiphales. Similarly, sequences of E. necator and E. australiana were not included in our phylogenetic analyses because these taxa belong to the Uncinula lineage of the genus Erysiphe as defined by Takamatsu et al. (2015b), and their ITS sequences are too divergent to be analyzed together with those of the Microsphaera lineage of Erysiphe (Takamatsu et al., 2015a).
Alignments of the four subsets of ITS sequences were generated using MAFFT v. 7.388 (Katoh and Standley, 2013), and visually inspected for potential misalignments or ambiguously-aligned regions. Phylogenetic analyses were conducted using BI and ML methods as described above. Alignments and trees were deposited in TreeBASE (submission no. 25949).
Results
Powdery Mildews Identified in Australia Based on Their ITS Sequences Published Prior This Study
As of February 2020, the identity of 32 powdery mildew taxa collected from over 60 host plant species in Australia was supported by partial or complete ITS sequences deposited in GenBank (Table 2). DNA barcodes other than the ITS sequences were not available in GenBank for any of these specimens. Those 32 taxa belonged to eight genera, and included 30 well-defined species, and two more taxa identified only to genus level, namely, Pseudoidium sp. on Convolvulus erubescens, closely related to E. heraclei and E. convolvuli based on ITS analyses (Cunnington et al., 2003), and Euoidium sp. on Nicotiana alata and Solanum melongena, with ITS sequences identical to those of G. lycopersici and G. longipes (Cunnington et al., 2005b). Therefore, we considered that 30 powdery mildew species were reliably identified based on their ITS sequences in Australia prior to our study.
Table 2.
List of powdery mildews identified in Australia based on nrDNA ITS sequences and morphological characteristics.
| Powdery mildew genera/species | Host plant | Herbarium accession number | Place and date of collection | ITS GenBank accession number | References |
|---|---|---|---|---|---|
| ARTHROCLADIELLA | |||||
| A. mougeotii | Lycium barbarum | VPRI 18039 | Bridgewater, VIC, July 1992 | AF073358 | Cunnington et al., 2003 |
| Lycium barbarum | BRIP 66057 | Killarney, QLD, April 2017 | MF496139 | Kiss et al., 2018a | |
| Lycium barbarum | BRIP 68795 | Killarney, QLD, October 2017 | MT174179 | This paper | |
| BLUMERIA | |||||
| Blumeria graminis | Hordeum vulgare | VPRI 18279 | Horsham, VIC, September 1992 | AF073352 | Cunnington et al., 2003 |
| Hordeum vulgare | BRIP 68826 | Gatton, QLD, June 2017 | MT174180 | This paper | |
| Phalaris canariensis | BRIP 52847 | Condamine Plains, QLD, September 2009 | MT174181 | This paper | |
| ERYSIPHE | |||||
| E. alphitoides | Jagera pseudorhus | BRIP 68798 | Mt. Tamborine, QLD, December 2017 | MT174182 | This paper |
| Jagera pseudorhus | BRIP 68799 | Burleigh Heads, QLD, July 2018 | MT174183 | This paper | |
| Mangifera indica | VPRI 20379 | Tweed Valley, NSW, October 1994 | AB237799 | Limkaisang et al., 2006 | |
| Mangifera indica | VPRI 20364 | Hopkins Creek, QLD, October 1994 | AB237798 | Limkaisang et al., 2006 | |
| Mangifera indica | VPRI 18420 | Indooroopilly, QLD, October 1992 | AB237795 | Limkaisang et al., 2006 | |
| Quercus robur | BRIP 68796 | Toowoomba, QLD, October 2017 | MT174184 | This paper | |
| Quercus sp. | VPRI 18763 | Mitcham, VIC, January 1993 | AB292705 | Takamatsu et al., 2007 | |
| Quercus sp. | VPRI 20423 | The Basin, VIC, December 1994 | AB292704 | Takamatsu et al., 2007 | |
| E. aquilegiae | Aquilegia sp. | VPRI 20820 | Parkville, VIC, March 1996 | AY452800 | Cunnington et al., 2004a |
| Catharanthus roseus | BRIP 46649, VPRI 32380 | Brisbane, QLD, July 2005 | DQ335569 | Liberato and Cunnington, 2006 | |
| Delphinium sp. | VPRI 19613 | Panton Hill, VIC, November 1993 | AY452802 | Cunnington et al., 2004a | |
| Nigella damascena | VPRI 18533 | Burnley, VIC, December 1992 | AY452804 | Cunnington et al., 2004a | |
| Ranunculus sp. | VPRI 18740 | Pheasant Creek, VIC, January 1993 | AY452805 | Cunnington et al., 2004a | |
| E. australiana | Lagerstroemia indica | VPRI 21732 | Hawthorn East, VIC, February 1998 | AF073347 | Cunnington et al., 2003 |
| Lagerstroemia indica | BRIP 68797 | Geebung, QLD, March 2017 | MT174185 | This paper | |
| Lagerstroemia indica | BRIP 48722 | Sherwood, December 2006 | MT174186 | This paper | |
| E. cruciferarum | Brassica juncea | BRIP 69033 | Jondaryan, QLD, November 2017 | MT174187 | This paper |
| E. diffusa | Glycine clandestina | BRIP 68827 | Rangeville, QLD, June 2018 | MT174188 | This paper |
| Glycine max | BRIP 55388 | QLD, 2012 | JX136797 | McTaggart et al., 2012 | |
| Glycine max | BRIP 58458 | Nangwee, QLD, 2013 | MT174189 | This paper | |
| Glycine max | BRIP 62030 | Giru, QLD, 2014 | MT174190 | This paper | |
| Glycine max cv. Jackson | BRIP 68997 | Grafton, NSW, April 2018 | MT174191 | This paper | |
| E. euonymicola | Euonymus japonicus | BRIP 69034 | Toowoomba, QLD, November 2017 | MT174192 | This paper |
| E. heraclei | Anethum graveolens | BRIP 59438 | Mareeba, QLD, July 2013 | MT174193 | This paper |
| Anethum graveolens | BRIP 68828 | Toowoomba, QLD, December 2017 | MT174194 | This paper | |
| Daucus carota | BRIP 68829 | Toowoomba, QLD, December 2017 | MT174195 | This paper | |
| Daucus carota | VPRI 41227 | Murrumbidgee, NSW, August 2007 | EU371725 | Cunnington et al., 2008b | |
| E. izuensis | Rhododendron sp. | BRIP 69724 | Toowoomba, QLD, June 2018 | MT174196 | This paper |
| Rhododendron indicum | BRIP 68833 | Toowoomba, QLD, August 2018 | MT174197 | This paper | |
| Rhododendron sp. | BRIP 70493 | Mt. Coot-tha, QLD, July 2018 | MT174198 | This paper | |
| E. necator | Vitis vinifera | VPRI 19719 | Dandenong, VIC, December 1993 | AF073346 | Cunnington et al., 2003 |
| Vitis vinifera cv. Fiano | BRIP 69725 | Stanthorpe, QLD, December 2018 | MT174232 | This paper | |
| E. pisi | Pisum sativum | VPRI 19688 | Hopetoun, VIC, November 1993 | AF073348 | Cunnington et al., 2003 |
| E. platani | Platanus x hybrida | BRIP 68800 | Adelaide, SA, March 2018 | MT174199 | This paper |
| Platanus x hybrida | DAR 83490 | Adelaide, SA, 2018 | MT174200 | This paper | |
| Platanus occidentalis | VPRI 21733 | Melbourne, VIC, February 1998 | AF073349 | Cunnington et al., 2003 | |
| E. quercicola | Acacia holosericea | VPRI 20468 | Humpty Doo, NT, February 1995 | AB237806 | Limkaisang et al., 2006 |
| Acacia mangium | VPRI 20374 | Tully, QLD, October 1994 | AB237807 | Limkaisang et al., 2006 | |
| Acacia mangium | VPRI 20907 | Mission Beach, QLD, June 1993 | AB237808 | Limkaisang et al., 2006 | |
| Eucalyptus camaldulensis | VPRI 19251 | West Pennant Hills, NSW, May 1993 | AB237796 | Limkaisang et al., 2006 | |
| Mangifera indica | VPRI 20332 | Douglas, QLD, August 1994 | AB237797 | Limkaisang et al., 2006 | |
| Quercus robur | VPRI 19013 | Stirling, SA, March 1993 | AB295454 | Takamatsu et al., 2007 | |
| Quercus sp. | VPRI 20422 | The Basin, VIC, December 1994 | AB295455 | Takamatsu et al., 2007 | |
| E. syringae | Syringa vulgaris | VPRI 41368 | Ormond, VIC, March 2008 | FJ755790 | Cunnington and Brett, 2009 |
| Syringa vulgaris | VPRI 43062 | VIC, February 2018 | MH368484 | GenBank | |
| E. cf. trifoliorum | Acacia orites | BRIP 70580 | Canungra, QLD, April 2019 | MT174201 | This paper |
| Pisum sativum | BRIP 68831 | Killarney, QLD, November 2017 | MT174202 | This paper | |
| Vicia tetrasperma | BRIP 68838 | Tipton, QLD, Oct 2017 | MT174203 | This paper | |
| Erysiphe sp. | Araujia sericifera | BRIP 66128 | Rosebery, NSW, July 2017 | MG551720 | Southwell et al., 2018 |
| Araujia sericifera | BRIP 66129 | Chiefley, NSW, July 2017 | MG551721 | Southwell et al., 2018 | |
| Araujia sericifera | BRIP 68832 | Rangeville, QLD, March 2018 | MT174204 | This paper | |
| Cassia fistula | BRIP 68834 | Toowoomba, QLD, June 2018 | MT174205 | This paper | |
| Pseudoidium hardenbergiae | Hardenbergia sp. | VPRI 19879 | Devenport, TAS, March 1994 | AY450959 | Cunnington et al., 2004b |
| Pseudoidium hortensiae | Hydrangea macrophylla | BRIP 68852 | Toowoomba, QLD, January 2018 | MT174206 | This paper |
| Pseudoidium sp. | Convolvulus erubescens | VPRI 20708 | Flinders ranges, SA, July 1987 | AF154328 | Cunnington et al., 2003 |
| GOLOVINOMYCES | |||||
| Euoidium sp. | Nicotiana alata | VPRI 19196 | Burnley, VIC, May 1993 | AY683041 | Cunnington et al., 2005b |
| Solanum melongena | VPRI 17907 | Adelaide, SA, May 1992 | AY683038 | Cunnington et al., 2005b | |
| G. ambrosiae | Persicaria decipiens | HAL 3274 F, BRIP 69035 | Redwood Park, QLD, November 2017 | MH745099 | Braun et al., 2019 |
| G. biocellatus | Mentha sp. | BRIP 46650, VPRI 32381 | Brisbane, QLD, July 2005 | EU035602 | Liberato and Cunnington, 2007 |
| Salvia sp. | VPRI 18671 | Stanthorpe, QLD, January 1993 | AF154323 | Cunnington et al., 2003 | |
| Solanum lycopersicum | VPRI 19373 | St. Lucia, QLD, July 1993 | AY683040 | Cunnington et al., 2005b | |
| G. bolayi | Capsella bursa-pastoris | BRIP 68842 | Toowoomba, QLD, June 2018 | MT174213 | This paper |
| Solanum tuberosum | BRIP 50498 | Gatton, QLD, September 2007 | MT174214 | This paper | |
| G. cichoracearum | Aster subulatus | VPRI 17692 | Ardmona, VIC, October 1991 | GQ183937 | Cunnington et al., 2010 |
| Leucanthemum paludosum (syn. Chrysanthemum paludosum) | VPRI 20465 | Brisbane, QLD, January 1995 | GQ183947 | Cunnington et al., 2010 | |
| Dahlia sp. | VPRI 21385 | Burwood, VIC, April 1997 | GQ183949 | Cunnington et al., 2010 | |
| Solidago sp. | VPRI 19021 | Burnley, VIC, March 1993 | GQ183940 | Cunnington et al., 2010 | |
| Tanacetum balsamita | VPRI 19067 | Olinda, VIC, March 1993 | GQ183942 | Cunnington et al., 2010 | |
| Tanacetum parthenium | VPRI 18942 | Dromana, VIC, March 1993 | GQ183939 | Cunnington et al., 2010 | |
| Tanacetum parthenium | BRIP 68841 | Sydney, NSW, September 2017 | MT174215 | This paper | |
| Tanacetum vulgare | VPRI 20223 | Mt. Coot-tha, QLD, August 1994 | GQ183945 | Cunnington et al., 2010 | |
| Zinnia sp. | VPRI 19824 | Toowoomba, VIC, February 1994 | GQ183944 | Cunnington et al., 2010 | |
| G. glandulariae | Glandularia aristigera | BRIP 70490 | Bunya Mountains, QLD, July 2019 | MN190239 | Crous et al., 2019 |
| Glandularia aristigera | BRIP 70491 | Bunya Mountains - Maclagan Road, QLD, June 2019 | MN190241 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 70492 | Bunya Mountains, QLD, February 2017 | MN190240 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 68801 | Bunya Mountains, QLD, March 2018 | MN190242 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 70531 | Bunya Mountains, QLD, June 2019 | MN190243 | Crous et al., 2019 | |
| G. ixodiae | Ixodia achillaeoides | VPRI 20703 | Lenswood, SA, September 1995 | AY769954 | Cunnington et al., 2005a |
| G. latisporus | Aster subulatus | BRIP 68839 | Jondaryan, QLD, November 2017 | MT174207 | This paper |
| Helianthus annuus | BRIP 68840 | Toowoomba, QLD, April 2018 | MT174208 | This paper | |
| Helianthus annuus | BRIP 28996 | Toowoomba, QLD, | MT174209 | This paper | |
| Helianthus annuus | BRIP 49653 | Gatton, QLD, May 2007 | MT174210 | This paper | |
| Helianthus sp. | BRIP 66781 | Kununurra, WA, July 2017 | MT174211 | This paper | |
| Xanthium strumarium | BRIP 11128 | Nebo, QLD, July 1975 | MT174212 | This paper | |
| G. lycopersici | Solanum lycopersicum | VPRI 19847 | Timmering, VIC, February 1994 | AF229021 | Kiss et al., 2001 |
| Solanum lycopersicum | DAR 70008 | Lauderdale, TAS, 1994 | HQ286673 | Kovács et al., 2011 | |
| Solanum lycopersicum | DAR 35763 | White Hills, Bendigo, VIC, 1980 | EU327330 | Kiss et al., 2008 | |
| Solanum lycopersicum | DAR 71625 | Unknown locality, SA, 1996 | EU327332 | Kiss et al., 2008 | |
| Solanum lycopersicum | BRIP 68830 | Maclean, NSW, December 2017 | MT174216 | This paper | |
| G. orontii | Penstemon serrulatus | VPRI 19066 | Olinda, VIC, March 1993 | GQ183941 | Cunnington et al., 2010 |
| Phyla nodiflora | VPRI 20467 | Brisbane, QLD, February 1995 | GQ183948 | Cunnington et al., 2010 | |
| Solanum tuberosum | VPRI 20740 | Toolangi, VIC, October 1995 | AY683039 | Cunnington et al., 2005b | |
| Taraxacum officinale | VPRI 19707 | Kingswood, SA, November 1993 | GQ183943 | Cunnington et al., 2010 | |
| Viola sp. | VPRI 18422 | Parkville, VIC, November 1992 | GQ183938 | Cunnington et al., 2010 | |
| LEVEILLULA | |||||
| L. taurica | Capsicum annuum | VPRI 20146 | Adelaide, SA, June 1994 | AF073351 | Cunnington et al., 2003 |
| Capsicum annuum | BRIP 63342 | Gumlu, QLD, September 2015 | MT174217 | This paper | |
| Euphorbia cyathophora | BRIP 68802 | Iluka, NSW, December 2017 | MT174218 | This paper | |
| Euphorbia heterophylla | BRIP 66778 | Iama, Torres Strait, May 2017 | MT174219 | This paper | |
| Solanum lycopersicum | BRIP 68843 | Carnarvon, WA, May 2018 | MT174220 | This paper | |
| MICROIDIUM | |||||
| M. phyllanthi | Phyllanthus debilis | BRIP 68803 | Cairns, QLD, July 2018 | MT174221 | This paper |
| NEOERYSIPHE | |||||
| N. galeopsidis | Ajuga reptans | VPRI 19007 | Melbourne, VIC, 23 March 1993 | AF073357 | Cunnington et al., 2003 |
| N. kerribeeensis | Senecio glossanthus | DAR 33493 | Kerribee Station, NSW, September 1978 | GU356546 | Beilharz et al., 2010 |
| Senecio glossanthus | VPRI 21786 | Natimuk, VIC, October 1995 | GU356545 | Beilharz et al., 2010 | |
| Senecio minimus (syn. Erechtites minimus) | VPRI 19119 | Kinglake N.P., VIC, April 1993 | GU356541 | Beilharz et al., 2010 | |
| Senecio sp. | VPRI 19944 | Hobart, TAS, March 1994 | GU356544 | Beilharz et al., 2010 | |
| Senecio sp. | VPRI 19929 | Devonport, TAS, March 1994 | GU356543 | Beilharz et al., 2010 | |
| Senecio sp. | VPRI 19834 | Olinda, VIC, February 1994 | GU356542 | Beilharz et al., 2010 | |
| Senecio vulgaris | VPRI 18591 | Burnley, VIC, December 1992 | GU356540 | Beilharz et al., 2010 | |
| PODOSPHAERA | |||||
| P. aphanis | Fragaria x ananassa | VPRI 19031 | Toolangi, VIC, March 1993 | AF073355 | Cunnington et al., 2003 |
| P. fusca | Calendula officinalis | VPRI 20625 | North Carlton, VIC, July 1995 | AF154324 | Cunnington et al., 2003 |
| P. leucotricha | Malus domestica | VPRI 17729 | Lake Eucumbene, NSW, Nov 1991 | AF073353 | Cunnington et al., 2003 |
| Malus pumila | VPRI 18381 | Applethorpe, QLD, October 1992 | e.g., MT178379 | Smith et al., 2020 | |
| Malus domestica | VPRI 18536 | Taroona, TAS, November 1992 | e.g., MT178355 | Smith et al., 2020 | |
| Malus domestica | VPRI 18575 | Beaconsfield, TAS, December 1992 | e.g., MT178381 | Smith et al., 2020 | |
| Malus sylvestris | VPRI 19785 | Kingswood, SA, January 1994 | e.g., MT178378 | Smith et al., 2020 | |
| Malus sp. | VPRI 19947 | Hobart, TAS, March 1994 | e.g., MT178367 | Smith et al., 2020 | |
| P. pannosa | Rosa sp. | BRIP 68844 | Highfields, QLD, June 2017 | MT174222 | This paper |
| P. plantaginis | Plantago lanceolata | BRIP 68845 | Toowoomba, QLD, May 2018 | MT174223 | This paper |
| Lactuca serriola | BRIP 68851 | Carnarvon, WA, May 2018 | MT174224 | This paper | |
| P. tridactyla | Prunus armeniaca | VPRI 19864 | Knoxfield, VIC, March 1994 | AY833657 | Cunnington et al., 2005c |
| Prunus cerasifera | VPRI 19238 | Taroona, TAS, May 1993 | AY833656 | Cunnington et al., 2005c | |
| Prunus persica | VPRI 19866 | Knoxfield, VIC, March 1994 | AY833651 | Cunnington et al., 2005c | |
| Prunus persica | VPRI 19591 | Burnley, VIC, November 1982 | AY833653 | Cunnington et al., 2005c | |
| Prunus sp. | VPRI 19006 | Burnley, VIC, March 1993 | AF154321 | Cunnington et al., 2003 | |
| P. xanthii | Cephalotus follicularis | VPRI 41238 | Keysborough, VIC, September 2007 | EU367960 | Cunnington et al., 2008a |
| Citrullus lanatus | DAR 83497 | Mascot, NSW, 2017 | MT174225 | This paper | |
| Cucurbita maxima | BRIP 69723 | Unknown locality, NSW, 2017 | MT174226 | This paper | |
| Dahlia pinnata | BRIP 68846 | Preston, QLD, May 2018 | MT174227 | This paper | |
| Euryops chrysanthemoides | BRIP 46314 | Unknown locality, QLD, 2005 | DQ205330 | Liberato et al., 2006 | |
| Glandularia aristigera | BRIP 70490 | Bunya Mountains, QLD, July 2019 | MN190026 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 70491 | Bunya Mountains - Maclagan Road, QLD, June 2019 | MN190028 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 70492 | Bunya Mountains, QLD, February 2017 | MN190027 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 68801 | Bunya Mountains, QLD, March 2018 | MN190029 | Crous et al., 2019 | |
| Glandularia aristigera | BRIP 70531 | Bunya Mountains, QLD, June 2019 | MN190244 | Crous et al., 2019 | |
| Trema tomentosa | BRIP 70494 | Canungra, QLD, 8 July 2018 | MT174228 | This paper | |
| Trema tomentosa | BRIP 70495 | Canungra, QLD, 25 April 2019 | MT174229 | This paper | |
| Vigna radiata | VPRI 19910 | Formartin, QDL, March 1994 | AY450961 | Cunnington et al., 2004b | |
| Vigna radiata | BRIP 68847 | Warwick, QLD, April 2017 | MT174230 | This paper | |
| Vigna unguiculata | VPRI 18815 | Berrimah, NT, February 1993 | AY450960 | Cunnington et al., 2004b | |
| SALMONOMYCES | |||||
| S. acalyphae | Acalypha nemorum | BRIP 68805 | Ravensbourne, QLD, June 2018 | MT133545 | This paper |
| Acalypha wilkesiana | BRIP 68804 | Brisbane, QLD, July 2018 | MT133544 | This paper | |
| SAWADAEA | |||||
| S. bicornis | Acer negundo | VPRI 19684 | Duffy, ACT, November 1993 | AF073356 | Cunnington et al., 2003 |
| S. polyfida | Acer palmatum | BRIP 68806 | Toowoomba, QLD, May 2018 | MT174231 | This paper |
Powdery mildew species/genera, and/or their host plants that were recorded for the first time in Australia based on open access molecular data are shown in bold.
Powdery Mildews Identified in This Study
Our field surveys resulted in the collection of over 300 fresh powdery mildew specimens from 35 host plant species from across Australia. In addition, 30 herbarium specimens from BRIP, collected from 24 host plant species between 1975 and 2013, were also included in this study. All powdery mildew specimens were identified based on their morphology and ITS sequences. Altogether, these represented 25 species from nine genera (Table 2). Some of these powdery mildew species have already been reported from Australia based on their ITS sequences; another 12 species have not been identified with publicly available DNA sequences in Australia prior to our study.
In total, this study found 42 reliably identified powdery mildew species in 10 genera on over 90 host plant species across 68 plant genera in Australia (Table 2). Amongst these, a total of 26 plant species are reported here for the first time as hosts of several powdery mildews in Australia. Four of these newly reported host plants are native, i.e., Acacia orites, Acalypha nemorum, Jagera pseudorhus, and Trema tomentosa. Earlier studies have identified powdery mildews on other native hosts, i.e., Acacia spp., Eucalyptus camaldulensis (Limkaisang et al., 2006), Cephalotus follicularis (Cunnington et al., 2008a), Convolvulus erubescens (Cunnington et al., 2003), Hardenbergia sp. (Cunnington et al., 2004b), Ixodia achilleoides (Cunnington et al., 2005a), and Senecio glossanthus and S. minimus (Beilharz et al., 2010) (Table 2). A number of powdery mildews that are common on their host plants in many parts of the world, such as E. cruciferarum on oilseed brassicas (e.g., B. juncea), E. heraclei on dill (Anethum graveolens), G. latisporus on sunflower (Helianthus annuus), and P. pannosa on rose (Rosa sp.), are reported here for the first time based on molecular identifications, although these species could be identical to those recorded in Australia much earlier, under various names (Goss, 1964; Simmonds, 1966; Walker, 1983; Uloth et al., 2016).
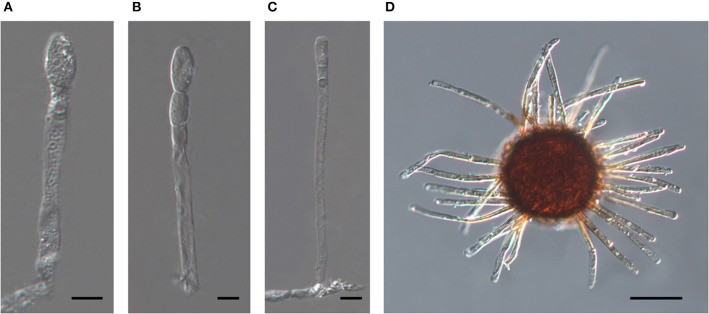
The sexual morphs of three powdery mildew species, i.e., E. izuensis infecting Rhododendron sp., G. latisporus on Helianthus annuus, and P. plantaginis on Plantago lanceolata (Figure 1) were documented for the first time in Australia. Earlier, chasmothecia were reported for E. necator (Wicks and Magarey, 1985; Magarey et al., 1997), and Neoerysiphe kerribeeensis (Beilharz et al., 2010). Walker (1983) listed a few more powdery mildew species producing chasmothecia in Australia, including B. graminis, E. australiana, and P. leucotricha, without details of the host plant species, collection dates, and localities.
Figure 1.
Chasmothecia of three powdery mildew species observed in this study. (A) Erysiphe izuensis on Rhododendron sp. (BRIP 69724). Bar = 100 μm. (B) Podosphaera plantaginis on Plantago lanceolata (BRIP 68845) Note the production of a single ascus in the chasmothecium. Bar = 25 μm. (C) Golovinomyces latisporus on Helianthus annuus (BRIP 28996) Bar = 20 μm. Inset: a chasmothecium with two asci stained with cotton blue. Bar = 50 μm.
Description of a Newly Recognized Lineage of the Erysiphales as a Reintroduced Genus
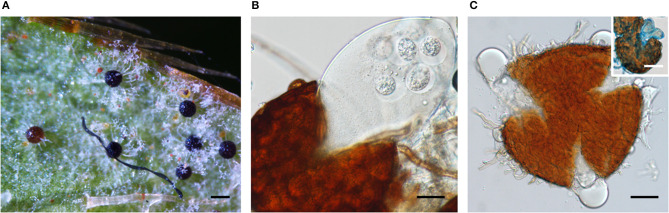
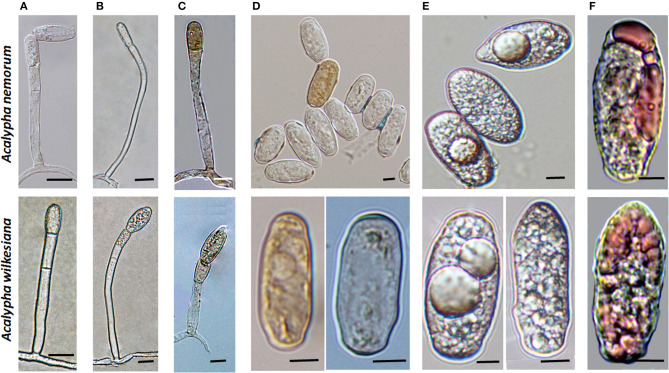
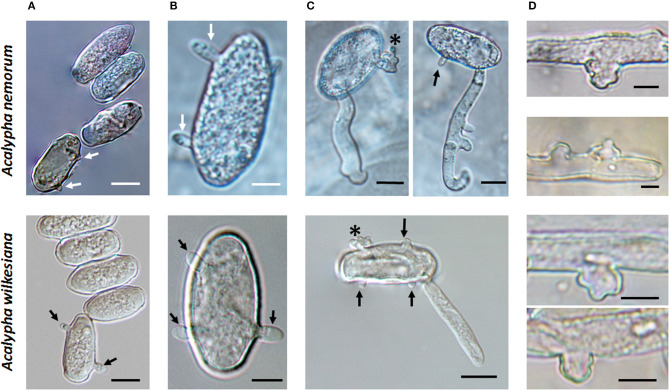
During the field surveys, powdery mildews with very unusual morphological characteristics were collected from Acalypha nemorum and A. wilkesiana. Acalypha nemorum is a native Australian shrub (Sagun et al., 2010), endemic to central and southern Queensland and north-eastern New South Wales, mostly in vine and open eucalypt forests (Forster, 1994). Acalypha wilkesiana is an ornamental shrub widely planted in subtropical and tropical regions of Australia and South East Asia (Sagun et al., 2010), and is native to Fiji (Sanz and Rodríguez, 2012). The symptoms of powdery mildew infections were unusual on A. nemorum, and mainly consisted of yellow spots on the upper leaf surfaces (Figure 2A), and intensively sporulating powdery mildew colonies on the corresponding lower leaf surfaces. The central, older parts of these colonies were brownish (Figures 2B,C). Occasionally, small, sporulating colonies were also observed on the upper surfaces of a few leaves. On A. wilkesiana var. macrophylla (Figure 3A) and cv. Inferno (Figures 3B,C), powdery mildew colonies covered large parts of both the upper and the lower leaf surfaces, causing typical symptoms of powdery mildew infections. The asexual morph exhibited some unusual characteristics on both A. nemorum and A. wilkesiana: (i) conidiophores were dimorphic, either short, Pseudoidium-like, that resembled common Erysiphe anamorphs (Figure 4A), or very long (Figure 4B); (ii) up to 3% of conidiophores and conidia were melanized (Figures 4C,D); (iii) conidia were filled with highly granular cytoplasm that appeared as a shiny, well-structured material, occasionally with voluminous oil droplets, when microscopically examined as fresh materials in water or lactic acid (Figure 4E); the lipid content of the granular cytoplasm and the droplets was confirmed by staining with Sudan Black B (Figure 4F); and (iv) germination of conidia, examined experimentally on sterile cellophane placed on water agar, and also on onion scales, differed from any conidial germination patterns described to date in the Erysiphales (Cook and Braun, 2009; Braun and Cook, 2012). In 24 to 48 h, germinated conidia, including the melanized ones, first produced short, thin, simple germ tubes (Figures 5A,B), up to four per conidium. Once developed, these primary germ tubes remained unchanged for 96 h. In 48 to 96 h, some conidia developed new, thicker secondary germ tubes that terminated in simple or lobed apices (Figure 5C). Some secondary germ tubes that had lobed to multi-lobed apices remained short, while others elongated, and became as long, or longer, than the conidium, ending in simple or lobed apices in 96 h (Figure 5C). Germination characteristics were similar on cellophane and onion scale surfaces. Hyphal appressoria were mostly lobed (Figure 5D). The unusual conidiophore and conidial morphology as well as the unusual conidial germination patterns indicated that this powdery mildew represents a genus on its own. Its molecular phylogenetic relationships were investigated based on nrDNA ITS, 18S and 28S sequences.
Figure 2.
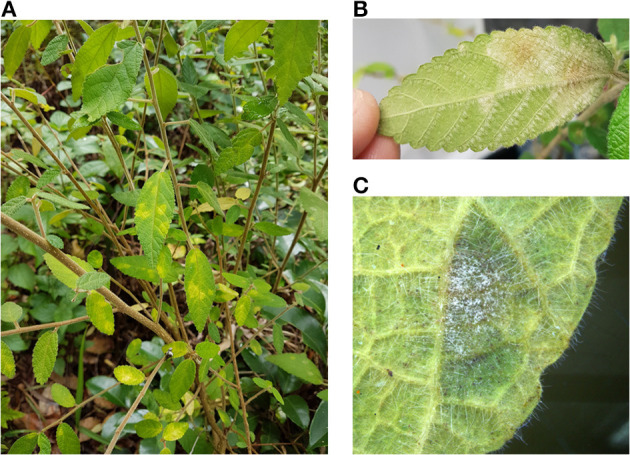
Acalypha nemorum infected with Salmonomyces acalyphae. (A) Symptoms on the upper leaf surfaces. (B) Symptoms on the lower leaf surfaces. (C) Sporulating mycelium on the lower leaf surface.
Figure 3.
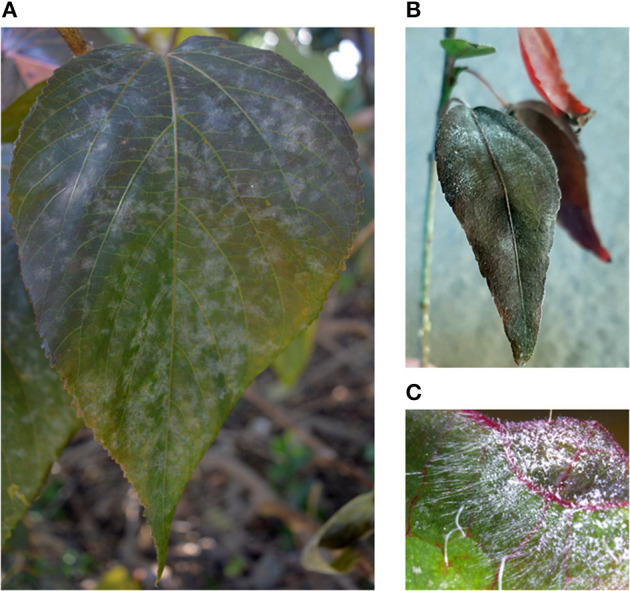
Acalypha wilkesiana infected with Salmonomyces acalyphae. (A) Symptoms on A. wilkesiana var. macrophylla. (B) Symptoms on A. wilkesiana cv. Inferno. (C) Sporulating mycelium on the upper leaf surface.
Figure 4.
Conidiophores and conidia of Salmonomyces acalyphae on Acalypha nemorum and A. wilkesiana collected in Australia. (A) Short conidiophores. Bar = 15 μm. (B) Long conidiophores. Bar = 10 μm. (C) Melanized conidiophores. Bar = 10 μm. (D) Melanized and hyaline conidia after boiling in lactic acid. Note the disappearance of lipid inclusions due to boiling. Bar = 5 μm. (E) Fresh conidia filled with highly granular cytoplasm, occasionally containing voluminous oil droplets. Bar = 5 μm. (F) Conidia stained with Sudan Black B during gentle warming of the samples. A part of the lipid inclusions were stained, and deformed during warming. Bar = 5 μm.
Figure 5.
Germination of conidia of Salmonomyces acalyphae collected from Acalypha nemorum and A. wilkesiana on cellophane placed on Water Agar (WA) and onion scales (OS), respectively. Arrows indicate primary germ tubes, asterisks lobed secondary germ tubes. (A) Development of the first primary germ tube initials on WA. Bar = 10 μm. (B) More developed primary germ tubes on OS (above) and WA (below). Bar = 5 μm. (C) Secondary germ tubes with both simple and lobed ends developed on OS (above) and WA (below). Note the first hyphal elongation with lateral branching on OS. Bar = 10 μm. (D) Hyphal appressoria. Bar = 5 μm.
To obtain unambiguous ITS sequences of the powdery mildew on both Acalypha spp., the ITS regions of specimens BRIP 68804 and BRIP 68805, from A. wilkesiana and A. nemorum, respectively, were re-sequenced following cloning of the PCR products. This resulted in a total of 20 identical sequences which differed by a single nucleotide in the ITS2 region from those determined in four specimens (MUMH5149, MUMH5150, MUMH5152, and MUMH5559) collected from A. wilkesiana and A. argentea in Java, and described as Pseudoidium javanicum by Meeboon et al. (2013). The morphology (Figure 6) and the ITS sequence of a specimen collected from A. wilkesiana in Argentina (HMJAU-PM91866) were also identical to those of BRIP 68804 and BRIP 68805. However, according to Meeboon et al. (2013) the morphology of Ps. javanicum is very different from what we observed in our specimens from A. nemorum and A. wilkesiana. Most importantly, Meeboon et al. (2013) reported that all the conidiophores of Ps. javanicum were short and hyaline, typical of the asexual morphs of the genus Erysiphe (Pseudoidium), and its conidia were also described as typical of Pseudoidium. The conidial germination patterns were not described for Ps. javanicum.
Figure 6.
Conidiophores and conidia of Salmonomyces acalyphae on Acalypha wilkesiana collected in Argentina (HMJAU-PM91866), after boiling in lactic acid. (A) Long conidiophore producing conidia singly. Bar = 10 μm. (B) Conidiophore producing melanized conidia in a short chain. Bar = 10 μm. (C) Short conidiophores. Bars = 5 μm. (D) Conidia. Bar = 10 μm.
The nrDNA 18S and 28S sequences were also determined in specimens BRIP 68804 and BRIP 68805, and analyzed together with the 18S, 28S, and 5.8S sequences of representative specimens of all but one powdery mildew genera accepted to date (Supplementary Table 1). Takamatsuella was the only genus of the Erysiphales that could not be included in this study as no authentic sequence data are available for this monotypic genus. The 28S and 5.8S sequences of P. javanicum determined by Meeboon et al. (2013) were included in the analysis, and the 18S sequences coded as missing, just like for Brasiliomyces (Supplementary Table 1). The dataset consisted of concatenated sequences of 36 taxa, the aligned matrix had a total length of 1,397 characters (28S: 663, 18S: 578, 5.8S: 156). The consensus phylogeny obtained using BI analysis supported the tree topology obtained with ML, therefore, only the best scoring ML phylogram is shown (Figure 7).
Figure 7.
Maximum likelihood phylogram based on the concatenated sequences of the 28S, 5.8S, and 18S regions of the nuclear ribosomal DNA of representatives of all genera of the Erysiphales, except Takamatsuella. Salmonomyces acalyphae comb. nov. is shown in bold blue. The tree is rooted to Byssoascus striatosporus CBS 642.66. Maximum Likelihood bootstrap values >80% and Bayesian Posterior Probability values >0.80 are shown above or below the branches. Thickened branches represent Maximum Likelihood bootstrap value of 100% and Bayesian Posterior Probability of 1.00. The scale bar represents nucleotide substitutions per site.
The Australian powdery mildew specimens from A. nemorum and A. wilkesiana had identical sequences at all the three loci. Their 28S and 5.8S sequences were also identical to those of MUMH5149, MUMH5150, MUMH5152, and MUMH5559, described as P. javanicum infecting Acalypha spp. (Meeboon et al., 2013). The clade consisting of the six specimens from Acalypha spp. had 100% Maximum Likelihood bootstrap (BS) value, and Bayesian Posterior Probability (PP) value of 1.0, being sister to a clade consisting of Brasiliomyces malachrae and Bulbomicroidium bauhiniicola (Figure 7). As both the morphological characteristics and the phylogenetic analysis have clearly indicated that the powdery mildew from Acalypha spp. is different from all the other powdery mildews, its allocation to a genus of its own was warranted.
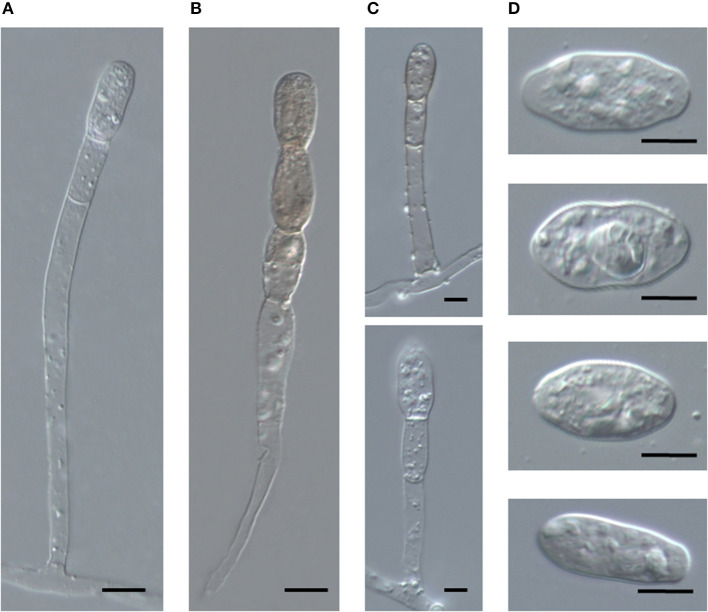
Attempts to identify this powdery mildew led to the genus Salmonomyces, described by Chiddarwar (1959), as well as to Erysiphe acalyphae (≡ Uncinula acalyphae), a widespread species reported from various Acalypha spp. (Braun and Cook, 2012). The history of the taxonomic treatment of the Acalypha powdery mildew is summarized as follows. The first description of this powdery mildew dates back to Sawada (1943). He introduced the genus name Orthochaeta and the species name O. acalyphae, based on type material on A. australis from Taiwan, but both names are invalid, as they were published without Latin description or diagnosis (Art. 39.1). Tai (1946) published Uncinula acalyphae, based on a Chinese collection on A. superba, the first valid species name for this powdery mildew, which was later transferred to Erysiphe (Zheng and Chen, 1981). Doidge (1948) described Uncinula eylesii on A. ciliata from South Africa. Later, Chiddarwar (1959) introduced the new genus and species, Salmonomyces kamatii, for a powdery mildew found on A. ciliata in India. Chiddarwar (1959) treated this species as a member of a new genus, Salmonomyces, based on the unusual characteristics of the sexual morph, without being aware of the previous publications of Tai (1946) and Doidge (1948). Later, Pirozynski (1965) discussed the taxonomy of this Acalypha powdery mildew in detail, and reduced Sa. kamatii and U. eylesii to synonymy with U. acalyphae, based on re-examinations of type material of the synonyms involved, except for type material of U. acalyphae, which was not available for that study. Sathe (1969) considered Salmonomyces as a synonym of Erysiphopsis (Halsted, 1899), a North American genus with Er. parnassiae on Parnassia caroliniana as type species. Zheng and Chen (1981), Braun (1987), and Zheng and Yu (1987) followed Pirozynski (1965) taxonomic conclusions and synonymy. Braun and Cook (2012) treated this species under the name Erysiphe acalyphae. Recently, Meeboon et al. (2013) described Pseudoidium javanicum based on specimens collected from Acalypha spp. in Indonesia, and considered Ps. javanicum to be different from E. acalyphae by having shorter conidiophores.
The oldest validly introduced species name for the powdery mildew on Acalypha is U. acalyphae. The type specimen of U. acalyphae collected in South China (Tai, 1946), HMAS 153, and additional collections from China deposited at HMAS (Supplementary Table 1) were re-examined in this work. The characteristics of the asexual morph (Figures 8A–C) agreed with those of the specimens collected in Australia (Figures 4, 5) and Argentina (Figure 6); importantly, pigmented conidiophores and conidia have been observed in the type specimen and other Chinese specimens, as well. The sexual morph was also present in most of the HMAS specimens examined in this work (Figure 8D; Supplementary Table 1). Attempts to amplify nrDNA regions from the type specimen failed, and a trip to the type collection site in Southern China did not locate powdery mildew-infected Acalypha plants. The morphological agreement of the asexual morph of the type specimen of U. acalyphae with the specimens from Argentina and Australia, together with our phylogenetic analyses, warrant the introduction of the following new combination to accommodate U. acalyphae:
Figure 8.
The asexual and the sexual morphs of Salmonomyces acalyphae on Acalypha brachystachya collected in China (HMAS 152), after boiling in lactic acid. (A) Short conidiophore producing conidia singly. Bar = 10 μm. (B) Conidiophore producing conidia in a short chain. Bar = 10 μm. (C) Long conidiophore producing conidia singly. Bar = 20 μm. (D) Chasmothecium. Bar = 50 μm.
Salmonomyces acalyphae(F.L. Tai) L. Kiss, D.N. Jin and S.Y. Liu, comb. nov.
MycoBank, MB834807
Basionym: Uncinula acalyphae F.L. Tai, Bull. Torrey Bot. Club 73: 123, 1946.
≡ Erysiphe acalyphae (F.L. Tai) R.Y. Zheng and G.Q. Chen, Sydowia 34: 261, 1981.
= Orthochaeta acalyphae Sawada, Rep. Gov. Res. Inst. Formosa 85: 23, 1943, nom. inval. (Art. 39.1).
= Uncinula eylesii Doidge, Bothalia 4: 844, 1948 [type: South Africa, Zimbabwe, Harare (former Salisbury), on Acalypha ciliata, Feb. 1920, F. Eyles 2071 (PREM 13992, holotype)].
= Salmonomyces kamatii Chidd., Sydowia 13: 56, 1959 [type: India, Poona, Purander Fort, on Acalypha ciliata, Sep. 1957, P.P. Chiddarwar 38 (HCIO, holotype; IMI83199, isotype)].
≡ Erysiphopsis kamatii (Chidd.) Sathe, Bull. Torrey Bot. Club 96: 102, 1969.
= Pseudoidium javanicum Meeboon and S. Takam., in Meeboon, Hidayat and Takamatsu, Mycoscience 54(3): 184, 2013 [types: Indonesia, West Java province, Bogor, Cibodas Botanical Garden, 7 Mar. 2012, on Acalypha wilkesiana, J. Meeboon et al. (TNS-F-46915, holotype; MUMH 5559, isotype)].
Illustrations: Tai (1946, p. 116, Figure 4), Zheng and Chen (198, p. 262, Figure 22), Braun (1987, p. 457, Pl. 207), Zheng and Yu (1987, p. 50, Figure 8), Braun and Cook (2012, p. 359, Figure 396).
References (descriptions and taxonomic discussions): Pirozynski (1965, p. 4–6), Braun (1987a, p. 234), Zheng and Yu (1987, p. 49).
Exsiccatae: Fungi Sinensis Exsiccati 4.
Holotype: on Acalypha superba [= A. brachystachya], China, Yunnan, Shishan, Kunming, Nov. 1938, F.L. Tai (HMAS 153). Isotypes: BPI 559117, HMAS 152.
Host range and distribution: on Acalypha australis [Taiwan (Amano, 1986), type of Orthochaeta acalyphae], A. ×cristata [Indonesia (Meeboon et al., 2013)], A. indica [India (Amano, 1986; Pande, 2008); Mauritius (Pirozynski, 1965; Amano, 1986), voucher specimen – IMI 102713], A. ciliata [India (Amano, 1986; Pande, 2008), voucher specimens – IMI 104267, 230417, 264545, type of Salmonomyces kamatii; South Africa (Pirozynski, 1965; Amano, 1986), type of Uncinula eylesii; Sudan (Amano, 1986); Zambia (Amano, 1986); Zimbabwe (Amano, 1986)], A. lanceolata [India (Amano, 1986), voucher specimen – IMI 90203], A. nemorum [Australia, this publication], A. sinensis [Tanzania (Amano, 1986)], A. superba [China (Tai, 1946; Amano, 1986; Zheng and Yu, 1987), type of Uncinula acalyphae; India (Amano, 1986; Pande, 2008); Uganda (Amano, 1986)], A. wilkesiana [Argentina (this publication); Australia (this publication); Indonesia (Meeboon et al., 2013), type of Pseudoidium javanicum], Acalypha sp. [Sudan, voucher specimen – IMI 45223; Zambia (Amano, 1986), voucher specimen – IMI 87889, 95790], Euphorbiaceae.
Notes: Meeboon et al. (2013) reported Pseudoidium javanicum on “Acalypha argentea hort.,” which does not exist as a validly published name, therefore, this host can only be referred to as Acalypha sp.
Morphology
Mycelium and asexual morph (based on the examination of collections from Australia, Argentina, and Indonesia and a re-examination of type material of Uncinula acalyphae): Mycelium amphigenous, thin, effuse, persistent, rarely evanescent; hyphae branched, straight to sinuous, thin-walled, smooth, hyaline; hyphal appressoria nipple-shaped or slightly to distinctly lobed, single or in opposite pairs; conidiophores dimorphic, short, and Pseudoidium-like, consisting of a 25–75 × 5–7 μm foot-cell, followed by 0–2 shorter cells, each (9–)12–20(−24) × 6–8 μm, and a single conidium or conidium initial, and/or long and slender, consisting of a very long foot-cell, 50–150(−243) × 4.5–7 μm, followed by 0–3 shorter cells, each (11–)16–23(−29) × 6–8 μm, and a single conidium or conidium initial or conidia formed in short chains, most conidiophores colorless, but up to three percent becoming pigmented; conidia ellipsoid-ovoid, subcylindrical to doliiform, occasionally with irregular margins, or pointed at one end, 16–32 × 8–16 μm, hyaline or up to three percent becoming pigmented, brownish, content (cytoplasm) granular, fresh conidia with voluminous, up to 9 μm diameter oil droplets.
Conidial germination (based on Australian collections): Primary germ tubes short, thin, simple germ tubes, up to four per conidium. Secondary germ tubes thicker, very short, or as long, or longer, than the conidium, ending in simple or lobed apices.
Sexual morph (based on the holotype, HMAS 153, and the isotype, HMAS 152): Chasmothecia scattered to gregarious, (70–)80–145 μm diam.; peridium cells irregularly polygonal to rounded, not very conspicuous, 5–20 μm diam.; appendages numerous, (10–)20–50, arising equatorially or from the upper half of the ascomata, rather stiff, setiform, 0.5–1.5 times as long as the chasmothecial diam., 4–9 μm wide, width uniform or decreasing from base to top, apex straight, pointed to rounded, rarely somewhat inflated, somewhat recurved or irregular, rarely abruptly constricted just below the apex or at the base, 0–1-septate, occasionally few septum-like lines in the upper half, pigmented, brown throughout or only below, paler toward the tip, apex hyaline or yellowish, walls thin to moderately thick, rarely thin above and thick toward the base, smooth to verrucose; asci 3–12, ellipsoid-obovoid, 30–60 × 20–35 μm, sessile or short-stalked, mostly immature with undeveloped ascospores (4 per ascus, 11–17 × 8–14 μm, according to Tai, 1946).
Detection of an Uncultured Fungus From Rhododendron Powdery Mildew Samples
In most cases, the nested PCRs that included the powdery mildew-specific PMITS1/PMITS2 primer pair (Cunnington et al., 2003) resulted in ITS sequences that were identical, or at least 98% similar to those of different powdery mildews as revealed by BLAST searches in GenBank. Surprisingly, these PCRs, when performed with DNA samples from any powdery mildew colonies sampled from Rhododendron spp., consistently yielded ITS sequences that were identical or over 97% similar to those of several other, unidentified and uncultured fungi. Those fungal ITS sequences were reported from Holcus lanatus roots collected in Germany (JF755916, Kreyling et al., 2012), house dust and indoor air samples collected in the USA (KF800177, Rittenour et al., 2014), Fagus sylvatica leaf litter collected in Austria (JF495199), and rainwater in China (KX516515, KX516323, and KX516237). These results indicate that some ubiquitous, cosmopolitan, but so far unidentified fungi were always associated with the powdery mildew colonies sampled from Rhododendron spp. in different places in this study (Table 2), and their ITS region was amplified consistently with the PMITS1/PMITS2 primer pair. These were the only powdery mildew samples in this work where the nested PCR protocol (Cunnington et al., 2003) has never amplified the ITS region of the respective powdery mildew fungus. A representative sequence obtained from Rhododendron samples was deposited in GenBank as the ITS sequence of an uncultured fungus under accession number MT152274.
Sequencing of an ITS Fragment of E. izuensis
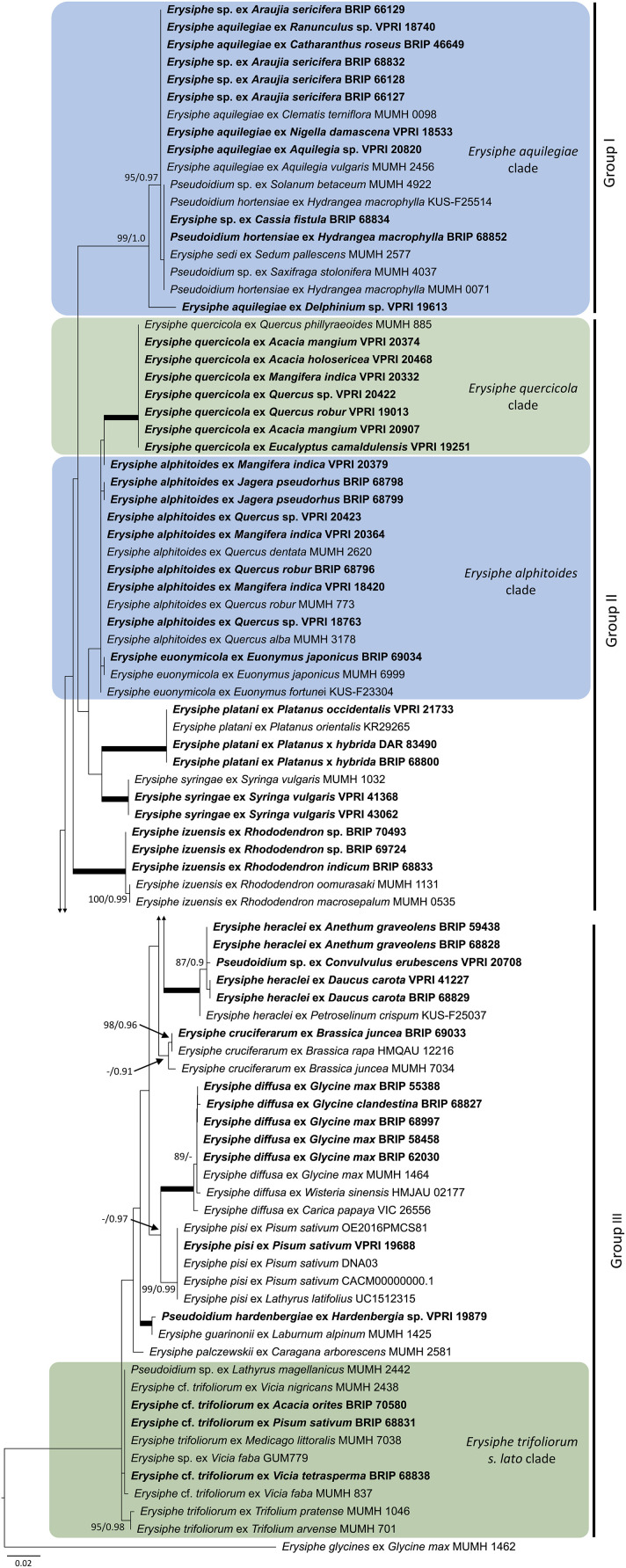
As the powdery mildew species collected from Rhododendron was identified as E. izuensis based on the morphology of the asexual and sexual morphs (Figure 1A), a species-specific primer pair was designed (Table 1) and used to amplify and sequence an ~450 bp long fragment of the ITS region of E. izuensis in six specimens. These were all identical to the corresponding ITS region of an E. izuensis specimen from Japan (LC009975), and were included in the phylogenetic analysis of the Microsphaera lineage of Erysiphe (Figure 9).
Figure 9.
Maximum likelihood phylogram based on the internal transcribed spacers of the nuclear ribosomal DNA and the intervening 5.8S region for powdery mildew species belonging to the Microsphaera lineage of the genus Erysiphe (Takamatsu et al., 2015a). Groups I to III represent the clades defined by Takamatsu et al. (2015a). The tip labels in bold represent specimens collected in Australia (supporting data in Table 2). All the other specimens were collected overseas (supporting data in Supplementary Table 3). The tree is rooted to Erysiphe glycines MUMH 1462. Maximum Likelihood bootstrap values >80% and Bayesian Posterior Probability values >0.80 are shown above or below the branches. Thickened branches represent Maximum Likelihood bootstrap value of 100% and Bayesian Posterior Probability of 1.00. The scale bar represents nucleotide substitutions per site.
Species Belonging to the Uncinula Lineage of the Genus Erysiphe
The genus Erysiphe includes the highest number of species within the Erysiphales (Braun and Cook, 2012). Two major lineages, Microsphaera and Uncinula, have been repeatedly identified within the genus (Takamatsu et al., 2015a,b). The ITS sequences of these two groups are too divergent to be included in a single analysis (Takamatsu et al., 2015a,b). As the Uncinula lineage is only represented by two species in this study, E. necator and E. australiana (Table 2), no phylogenetic analysis was performed for this group.
Species Belonging to the Microsphaera Lineage of the Genus Erysiphe
Based on morphological examinations and ITS sequence analyses, a total of 51 specimens collected in this study or reported from Australia earlier (Table 2) belonged to the Microsphaera lineage. These specimens represented 15 species. Their ITS sequences were analyzed together with 31 ITS sequences from similar taxa collected overseas (Supplementary Table 3). The consensus phylogeny (Figure 9) revealed that powdery mildew infections of a number of native plants were caused by common species known from overseas. For example, the severe powdery mildew infections of young individuals of a native rainforest tree, Jagera pseudorhus (Sapindaceae) (Supplementary Figure 1A), recorded in two distant locations in Queensland (Table 2), was caused by E. alphitoides. This powdery mildew is a well-known pathogen of oaks (Quercus spp.), and has also been found on other woody hosts such as mango (Mangifera indica), and on herbaceous hosts, such as Oenothera spp., in different parts of the world (Limkaisang et al., 2006; Takamatsu et al., 2007, 2015a; Bereczky et al., 2015; Desprez-Loustau et al., 2017, 2018). In Australia, E. alphitoides was detected earlier on Quercus spp. and M. indica (Table 2). The ITS sequences determined in both specimens collected from J. pseudorhus were identical, and differed in one nucleotide in the ITS2 region from E. alphitoides specimens collected in Australia (Figure 9). Erysiphe euonymicola, recorded for the first time in Australia based on molecular evidence, is closely related to E. alphitoides (Figure 9).
Erysiphe quercicola is another species that infects Quercus spp., and also a number of distantly related woody plants, such as Citrus spp., M. indica, and rubber tree (Hevea brasiliensis), in different parts of the world (Takamatsu et al., 2007; Desprez-Loustau et al., 2017, 2018). It was recorded earlier in Australia on M. indica, and also on the native Acacia holosericea, A. mangium, and Eucalyptus camaldulensis (Table 2). No information is available about the severity of the disease caused by E. quercicola on any of these plants in Australia. During our study, powdery mildew infection was observed on another native Acacia species, A. orites, where the mycelium was restricted to the bipinnate leaves at the tips of the phyllodes of young plants (Supplementary Figure 1B). Based on the ITS sequence analysis, the causal agent belonged to the E. trifoliorum s. lato group (Figure 9). Specimens with ITS sequences identical to those of the A. orites powdery mildew were also collected from pea (Pisum sativum) and Vicia tetrasperma in this study, and the same ITS sequences were available in GenBank for a number of overseas powdery mildews (Figure 9). The taxonomy of the E. trifoliorum s. lato group is still unresolved.
Powdery mildews belonging to the E. aquilegiae group, defined by Takamatsu et al. (2015a), have been reported from plant species of at least 15 families in different parts of the world (Jankovics et al., 2008; Meeboon and Takamatsu, 2015), including Australia (Cunnington et al., 2004a; Liberato and Cunnington, 2006; Southwell et al., 2018). This study confirmed that a powdery mildew on Araujia sericifera reported recently from Australia by Southwell et al. (2018) belongs to this group based on its ITS sequence. A similar powdery mildew infecting Cassia fistula has also been identified in this work (Figure 9). To date, Ps. cassiae-siameae is the only known Erysiphe species from C. fistula, reported from Africa and Asia (Braun and Cook, 2012), but ITS sequences are not available for this taxon. The ITS sequences determined in our specimen from C. fistula were identical to a number of other powdery mildews reported from overseas, and also to P. hortensiae infecting Hydrangea macrophylla (Figure 9). This is the first confirmation of the latter species in Australia. Clearly, the taxa belonging to the E. aquilegiae group based on their ITS sequences need taxonomic treatment (Shin et al., 2019).
Golovinomyces Species
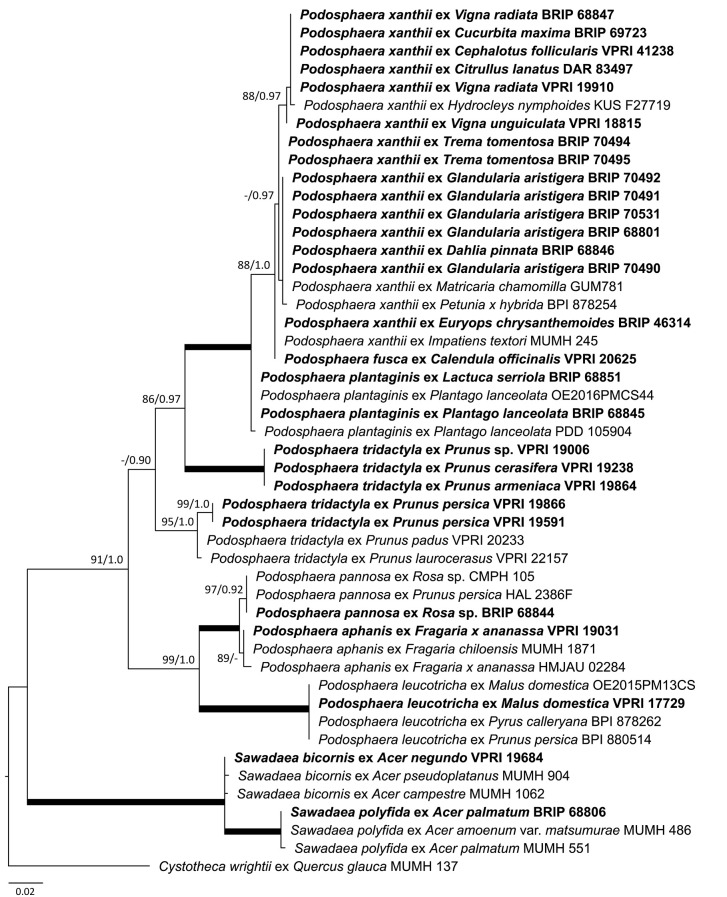
Based on morphology and ITS sequence analyses, a total of 10 Golovinomyces species have been identified in Australia (Table 2; Figure 10). Two of these, G. bolayi and G. glandulariae have only been recently described. Golovinomyces bolayi was delimited from the G. orontii species complex, and recognized as being globally distributed on a wide range of host plant families (Braun et al., 2019). This newly described species was represented by two specimens examined in this study, collected from Capsella bursa-pastoris and Solanum tuberosum. Other Australian specimens from S. tuberosum, Taraxacum officinale, and Phyla nodiflora, identified earlier as G. orontii (Cunnington et al., 2010), had ITS sequences identical to the Australian and overseas G. bolayi specimens (Figure 10). Many other G. orontii specimens with ITS sequences identical to all these powdery mildews have been reported earlier from a wide range of host plants across the globe; these data may indicate the polyphagous nature of these fungi (Matsuda and Takamatsu, 2003; Takamatsu et al., 2013; Braun et al., 2019).
Figure 10.
Maximum likelihood phylogram based on the internal transcribed spacers of the nuclear ribosomal DNA and the intervening 5.8S region for powdery mildew species belonging to Arthrocladiella, Golovinomyces, Microidium, and Neoerysiphe genera. The tip labels in bold represent specimens collected in Australia (supporting data in Table 2). All the other specimens were collected overseas (supporting data in Supplementary Table 3). Golovinomyces ixodiae comb. nov. is shown in bold blue. The tree is rooted to Phyllactinia adesmiae MUMH 1938. Maximum Likelihood bootstrap values >80% and Bayesian Posterior Probability values >0.80 are shown above or below the branches. Thickened branches represent Maximum Likelihood bootstrap value of 100% and Bayesian Posterior Probability of 1.00. The scale bar represents nucleotide substitutions per site.
The other recently recognized species, G. glandulariae, was described from Australia as a pathogen of Glandularia aristigera [= Verbena aristigera] (Crous et al., 2019), a verbenaceous species native to South America (O'Leary and Thode, 2016). It seems that this is not an endemic Australian powdery mildew species either because the ITS sequences determined in all the G. glandulariae specimens were identical to two other Golovinomyces specimens, KR-M-43410 and KR-M-43411 (GenBank acc. nos.: LC076839 and LC076840, respectively), collected from Verbena in Germany without being identified at the species level (Scholler et al., 2016). In addition, a powdery mildew identified as G. verbenae without DNA sequence data has been recently recorded in Europe on G. aristigera (Kruse et al., 2018).
Ixodia achillaeoides was the only Australian native hosting a Golovinomyces species. This was described as Oidium ixodiae by Cunnington et al. (2005a), and later transferred to Euoidium by Braun and Cook (2012). As of February 2020, there are no powdery mildew records in GenBank with ITS sequences identical to that of E. ixodiae. Based on the current “International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code, 2018),” and the “One fungus one name” principle, Euoidium being a heterotypic synonym of Golovinomyces, all phylogenetically and taxonomically confirmed Euoidium spp. have to be transferred to Golovinomyces (Braun, 2012, 2013):
Golovinomyces ixodiae(Cunningt., Beilharz and Pascoe) U. Braun and L. Kiss, comb. nov.
MycoBank, MB834808
Basionym: Oidium ixodiae Cunningt., Beilharz and Pascoe, Australas. Pl. Pathol. 34(1): 92, 2005.
= Euoidium ixodiae (Cunningt., Beilharz and Pascoe) U. Braun and R.T.A. Cook, Taxonomic Manual of the Erysiphales (Powdery Mildews): 334, 2012.
Golovinomyces ixodiae is sister to the clade containing G. longipes, G. lycopersici and Euoidium sp. on Solanum melongena and Nicotiana alata with identical ITS sequences (Cunnington et al., 2005b) (Figure 10). Amongst these taxa, G. lycopersici on tomato has never been recorded outside Australia (Kiss et al., 2005; Braun et al., 2019), although tomato is a crop introduced from overseas. In Australia, G. lycopersici has been reported since 1980 (Kiss et al., 2001), including in this study (Table 2). This species is morphologically and molecularly similar to G. longipes (Cunnington et al., 2005a; Kiss et al., 2008; Kovács et al., 2011) that has caused epidemics on solanaceous vegetables and ornamentals in Europe and the USA (Kiss et al., 2008).
All the powdery mildew specimens collected from sunflower (Helianthus annuus) in this study were identified as G. latisporus based on both ITS sequence analyses (Figure 10) and morphological characteristics, particularly the unique conidial germination patterns (Cook and Braun, 2009; Braun and Cook, 2012). This species was delimited from the G. cichoracearum complex based on its morphology (Cook and Braun, 2009). A number of Australian and overseas specimens identified earlier as G. cichoracearum had ITS sequences identical to G. latisporus (Figure 10). Another globally distributed species separated from the G. cichoracearum complex is G. ambrosiae, characterized with an ITS sequence that is identical to that of G. latisporus, but with morphological characteristics that readily distinguish it from the latter species (Braun et al., 2019). A recent multi-locus analysis distinguished G. latisporus and G. ambrosiae based on non-nrDNA sequences (Qiu et al., 2020). In Australia, G. ambrosiae was identified on Persicaria decipiens, the first member of the Polygonaceae that has been reported as a host of a Golovinomyces species worldwide (Braun et al., 2019).
Neoerysiphe, Arthrocladiella, and Microidium Species
As the alignments of the ITS sequences of the species of Golovinomyces, Neoerysiphe, Arthrocladiella, and Microidium were non-ambiguous, these were analyzed together. The results for the latter three genera are shown in Figure 10, and are discussed below, separately from Golovinomyces, for clarity purposes.
Powdery mildews representing the genus Neoerysiphe were not found in this study, but have been collected and intensively studied earlier in Australia (Cunnington et al., 2003; Beilharz et al., 2010). One of these fungi, named as N. kerribeeensis, produced chasmothecia on S. glossanthus, and was described as the first teleomorphic powdery mildew recognized on a native host in Australia (Beilharz et al., 2010). Its asexual morph has also been identified on several other Senecio spp. introduced to Australia (Beilharz et al., 2010). As of February 2020, there are no powdery mildew records in GenBank with ITS sequences identical or at least 98% similar to that of N. kerribeeensis. The closest taxon is N. galeopsidis (Figure 10), which has been recorded in Australia (Cunnington et al., 2003), and is known from other parts of the world as well (Takamatsu et al., 2008; Heluta et al., 2010).
Arthrocladiella mougeotii, representing a monotypic genus, has been consistently found on its host, Lycium barbarum, an introduced solanaceous species, in Australia (Cunnington et al., 2003; Kiss et al., 2018a). This species is found globally on Lycium spp. (Braun and Cook, 2012). In contrast, Microidium phyllanthi, one of the four described Microidium species (To-anun et al., 2005; Meeboon and Takamatsu, 2017), is reported here for the first time from Australia (Table 2).
Podosphaera and Sawadaea species
This study confirmed the presence of seven Podosphaera species in Australia (Figure 11; Table 2), all known to be globally distributed on their hosts (Braun and Cook, 2012). Amongst these taxa, P. xanthii is regarded as a heterogeneous species complex of many races that could each be specialized to distinct hosts or have broad host ranges that include cucurbits, composites, legumes, and several other plant families (Hirata et al., 2000; Braun et al., 2001). In Australia, P. xanthii has been identified on nine plant species (Table 2); some of these hosts are only distantly related. For example, powdery mildew infections observed in this study on young leaves of Trema tomentosa (Cannabaceae), a native rainforest tree species, were caused by P. xanthii (Supplementary Figure 1C). These infections, observed on a single site in Queensland, in 2 consecutive years (Table 2), are of interest because T. tomentosa is reported for the first time as a powdery mildew host, and also because P. xanthii, despite its broad host range, is uncommon on woody species. Earlier, another interesting Australian record of P. xanthii came from a nursery where potted Cephalotus follicularis (Cephalotaceae) plants became heavily infected with this powdery mildew (Cunnington et al., 2008a). This unusual host of P. xanthii is an attractive insectivorous plant native, and endemic to Western Australia. A number of its clones have been propagated commercially in different parts of the world for decades (Ko et al., 2010); therefore, these may have been repeatedly exposed to powdery mildews under various conditions. However, to our knowledge, the only powdery mildew that has been recorded on this native plant so far was P. xanthii in a nursery in Melbourne (Cunnington et al., 2008a). In nurseries, greenhouses and other artificial environments, P. xanthii has caused a number of unusual and unique powdery mildew infections, such as a serious disease of jellyfish tree (Medusagyne oppositifolia), imported from the Seychelles to the United Kingdom (Pettitt et al., 2010), or an infection of waterpoppy (Hydrocleys nymphoides), a monocot grown in pots in an urban area in Korea (Cho et al., 2018).
Figure 11.
Maximum likelihood phylogram based on the internal transcribed spacers of the nuclear ribosomal DNA and the intervening 5.8S region for powdery mildew species belonging to Podosphaera and Sawadaea genera. The tip labels in bold represent specimens collected in Australia (supporting data in Table 2). All the other specimens were collected overseas (supporting data in Supplementary Table 3). The tree is rooted to Cystotheca wrightii MUMH 137. Maximum Likelihood bootstrap values >80% and Bayesian Posterior Probability values >0.80 are shown above or below the branches. Thickened branches represent Maximum Likelihood bootstrap value of 100% and Bayesian Posterior Probability of 1.00. The scale bar represents nucleotide substitutions per site.
Podosphaera plantaginis infecting Plantago lanceolata in many parts of the world has become a model species in evolutionary and ecological studies of wild plant-pathogen interactions (Numminen et al., 2019). Its presence in Australia is confirmed in this study. Mating assays and molecular evidence have demonstrated that P. plantaginis is homothallic (Tollenaere and Laine, 2013), and therefore it is not surprising that its chasmothecia have been found on Pl. lanceolata (Figure 1B). Interestingly, P. plantaginis was also identified on Lactuca serriola in Western Australia (Figure 11; Table 2). This is the first record of P. plantaginis on this host. Another powdery mildew species, G. bolayi (earlier known as G. cichoracearum s. lat.), has been commonly found on L. serriola in many parts of the globe (Lebeda and Mieslerová, 2011; Braun et al., 2019), but was not recorded on L. serriola in Australia.
Powdery mildews infecting Prunus spp. were intensively studied by Cunnington et al. (2005c) based on specimens collected in Australia and elsewhere. Recently, the taxonomy of this group was revised (Moparthi et al., 2019; Meeboon et al., 2020), therefore the identity of the Australian specimens should be revisited.
Powdery mildew infections of maples (Acer spp.) are caused by species belonging to the genus Sawadaea (Hirose et al., 2005). So far, the presence of two species, S. bicornis and S. polyfida, have been confirmed in Australia (Figure 11; Table 2).
Blumeria and Leveillula Species
Both genera include a number of distinct phylogenetic lineages (Inuma et al., 2007; Khodaparast et al., 2007, 2012; Kusch et al., 2020) that have not been taxonomically resolved. These powdery mildew lineages represent globally distributed species complexes, e.g., B. graminis on cereals and wild grasses; and L. taurica on a wide range of vegetables, ornamentals, and wild plant species. Blumeria graminis occurs on all major cereal species in Australia (Simmonds, 1966; Golzar et al., 2016), but has been confirmed molecularly only on barley (Hordeum vulgare) and Phalaris canariensis (Table 2). Leveillula taurica has been identified on Capsicum (Cunnington et al., 2003), tomato (Solanum lycopersicum) and two species of Euphorbia in Australia by morphology and ITS sequence analyses (Figure 12; Table 2).
Figure 12.
Maximum likelihood phylogram based on the internal transcribed spacers of the nuclear ribosomal DNA and the intervening 5.8S region for powdery mildew species belonging to the genus Leveillula. The tip labels in bold represent specimens collected in Australia (supporting data in Table 2). All the other specimens were collected overseas (supporting data in Supplementary Table 3). The tree is rooted to Phyllactinia adesmiae MUMH 1938. Maximum Likelihood bootstrap values >80% and Bayesian Posterior Probability values >0.80 are shown above or below the branches. Thickened branches represent Maximum Likelihood bootstrap value of 100% and Bayesian Posterior Probability of 1.00. The scale bar represents nucleotide substitutions per site.
Number of Powdery Mildew Species in Australia and Overseas
The number of powdery mildew species in the genera Erysiphe, Golovinomyces, Neoerysiphe, and Podosphaera, reported for Asia, Europe, and North America (Braun and Cook, 2012), were compared with those recorded in Australia in this study. The data in Braun and Cook (2012) may be biased by doubtful species, species complexes, collection intensity, and other factors, whereas the data from this study were similarly biased by collection intensity. The comparison showed that the number of species of Erysiphe, Golovinomyces, Neoerysiphe, and Podosphaera, were much lower in Australia than the other continents (Table 3).
Table 3.
Number of powdery mildew species belonging to four genera in Australia, based on this paper, and in Asia, Europe and North America, compiled from Braun and Cook (2012).
| Genus | Australia | Asia | Europe | North America |
|---|---|---|---|---|
| Erysiphe | 17 | 340 | 103 | 100 |
| Podosphaera | 7 | 85 | 50 | 40 |
| Golovinomyces | 10 | 37 | 33 | 34 |
| Neoerysiphe | 2 | 7 | 5 | 3 |
Discussion
The establishment of the first European settlement in Australia in 1788 represents a sharp biogeographic watershed in the history of the entire Australian flora. There is no other continent where the introduction and spread of European agricultural and forestry practices, including the cultivation of crops imported from overseas, had happened as suddenly, and as recently as in Australia (Fensham and Laffineur, 2019). Almost all broad-acre, fruit, and vegetable crops currently grown in Australia were recently introduced from overseas (Kirkpatrick, 1999; Randall, 2007). In addition, the Acclimatization Societies founded in different part of Australia in the nineteenth and early twentieth century to “enrich” the flora and fauna in Australia, and create landscapes that were more familiar for the European settlers (Cook and Dias, 2006) had an enormous contribution to the introduction and establishment of a high number of ornamental and other plant species from all over the world, together with the associated microbiota. Currently, the number of vascular plant species introduced to Australia since 1788 outnumbers the native species; the entire native Australian vegetation has been deeply transformed by massive land management practices and human-mediated plant introductions (Kirkpatrick, 1999; Randall, 2007).
During our investigation started in 2017, most powdery mildews were collected from introduced hosts. In this study, powdery mildew was found on only four native plants, i.e., Acacia orites, Acalypha nemorum, Jagera pseudorhus, and Trema tomentosa. Each of these native species was infected with a single powdery mildew species, i.e., Erysiphe cf. trifoliorum, Salmonomyces acalyphae, E. alphitoides, and Podosphaera xanthii, respectively, known to infect a number of other plant species overseas (Braun and Cook, 2012; Takamatsu et al., 2015a; Desprez-Loustau et al., 2017). These four powdery mildew species were also identified in this study on introduced hosts; for example on Pisum sativum, Acalypha wilkesiana, Quercus robur, and Vigna spp., respectively. All powdery mildew infections of the four natives were collected in dry rainforests. On A. orites, the infection was limited to the bipinnate leaves of young plants. As well, only the young leaves of T. tomentosa were infected with powdery mildew, while on the other two natives, A. nemorum and J. pseudorhus, the infection was widespread on older leaf tissues. These four native plants may be considered as “accidental” hosts of the respective powdery mildews, although E. alphitoides was found in two distant locations on J. pseudorhus, and both P. xanthii and S. acalyphae have been repeatedly identified in the same site, but on different plants during surveys in 2018 and 2019. These observations suggest that the respective powdery mildews have been established on their native hosts. Less is known about powdery mildew infections of other Australian native species belonging to the genera Acacia, Cephalotus, Convolvulus, Eucalyptus, Hardenbergia, Ixodia, and Senecio, reported prior to this study (Cunnington et al., 2003, 2004b, 2005a, 2008a; Limkaisang et al., 2006; Beilharz et al., 2010). With the notable exception of G. ixodiae (= Oidium ixodiae) on I. achillaeoides (Cunnington et al., 2005a), and N. kerribeeensis on S. glossanthus (Beilharz et al., 2010), to be discussed below, all the other powdery mildews recorded on these natives are known as polyphagous species, i.e., infecting diverse other plant species, both in Australia and overseas. Therefore, in this paper we hypothesize that all powdery mildews have been introduced to Australia, since 1788, and some of these have adapted to become pathogens of a few native plant species, as a result of host range expansion events. Our arguments for this hypothesis are presented below.
Host range expansions appear to be frequent events in the evolution of some powdery mildews (Vági et al., 2007; Kirschner, 2010; Jankovics et al., 2011; Takamatsu, 2013a; Menardo et al., 2016; Beenken, 2017; Frantzeskakis et al., 2019a), and these could explain a number of “accidental” hosts of these fungi in different parts of the world. The genetic and genomic background of such rapid evolutionary processes has been partly revealed in obligate biotrophic plant pathogens (for reviews, see Frantzeskakis et al., 2019a; Thines, 2019). These events may explain the quick establishment of some polyphagous species, notably P. xanthii, E. alphitoides, E. quercicola, E. cf. trifoliorum, and P. xanthii, on Australian native plants. A number of unexpected records of this study, i.e., powdery mildew species on introduced plants that have not been known as their hosts overseas, including Erysiphe sp. (in the E. aquilegiae complex) on C. fistula, G. ambrosiae on Pe. decipiens, or P. plantaginis on La. serriola, may have resulted from rapid host range expansion events that have recently happened in the deeply disturbed Australian vegetation.
In this study, all powdery mildews were identified by morphology and ITS sequences, supplemented with 18S and 28S sequence analyses in the case of Salmonomyces. The use of ITS as the single DNA barcode for species identifications is clearly a bias in powdery mildew research (Kovács et al., 2011; Ellingham et al., 2019), and the limits of the routine fungal identification procedures based on ITS sequences are well-known in mycology (Nilsson et al., 2006; Schoch et al., 2012; Kiss, 2012; Nilsson et al., 2014; Irinyi et al., 2015; Hibbett et al., 2016; Selosse et al., 2016). ITS sequences are currently still the only reliable molecular tool for powdery mildew species identifications, despite recent advancements toward new DNA barcodes (Desprez-Loustau et al., 2017; Ellingham et al., 2019). These could be boosted by the recently determined genomes of distinct lineages (Bindschedler et al., 2016; Frantzeskakis et al., 2019b; Bradshaw and Tobin, 2020). However, closely related powdery mildew species cannot be reliably distinguished based on their ITS sequences because these are sometimes identical (Takamatsu et al., 2015a), or the intra-specific, and intra-sample variation is sometimes higher than the differences between the ITS sequences reported for closely related taxa (Kovács et al., 2011; Ellingham et al., 2019).
Another fundamental bias of the molecular identification of powdery mildews is the lack of DNA sequence information from the type specimens of hundreds of species described and named several decades ago. All those species are currently identified by their morphologies and ITS sequences determined in fresh specimens collected from the respective host plant species. Epitypification (ICN, Art. 9.9) is an established and useful tool to overcome problems in the genetic characterization of ascomycete species, including powdery mildews, when type specimens cannot be used for genetic analyses due to their age, state of conservation, or sparseness. Furthermore, many plant species, and even single leaves of the same host plants, are sometimes infected with more than one powdery mildew species (Chen et al., 2008; Kiss et al., 2008; Krístková et al., 2009), and these could be morphologically indistinguishable (Takamatsu et al., 2002; Jankovics et al., 2008; Bereczky et al., 2015; Desprez-Loustau et al., 2018). Such mixed infections may complicate the identification of powdery mildews. In obligate biotrophs, the host range is decisive in speciation as growth and asexual and sexual reproduction of these pathogens take place while structurally and nutritionally linked to the living host tissues. Gene flow cannot happen between obligate biotrophic pathogens which do not share the same hosts. However, still little is known about the host ranges of morphologically similar powdery mildews that exhibit identical, or highly similar ITS sequences. To date, only a few experimental studies have determined the host ranges of these powdery mildews (Hirata et al., 2000; Jankovics et al., 2008; Pastirčáková et al., 2016). Although phylogenies based on DNA sequences should not be regarded as species phylogenies (Doyle, 1992), the current practice is to apply the same taxon names for powdery mildews that share identical, or highly similar ITS sequences (e.g., Takamatsu et al., 2015a,b; Braun et al., 2019). This approach has been widely applied even in the absence of experimental evidence for an overlap in the host plant ranges of the respective pathogens, i.e., whether powdery mildews with identical, or highly similar ITS sequences can actually meet and recombine in the field. Our study has followed this current practice in powdery mildew research, although we recognize that many more experimental host range studies are needed to reliably delimit closely related taxa based on their potential constraints in gene flow, i.e., what can reasonably be considered true biological species.
There are two powdery mildew taxa, namely G. ixodiae on the native I. achillaeoides (Cunnington et al., 2005a), and G. lycopersici infecting tomato (Kiss et al., 2001) that have never been identified outside Australia. Based on their morphologies and ITS sequences, both taxa are closely related to G. longipes, a well-known pathogen of solanaceous vegetables and ornamentals in Europe and the USA (Kiss et al., 2008; Kovács et al., 2011). Also, an Euoidium sp. recorded on N. alata and S. melongena in Australia has identical ITS sequences to those of G. longipes and G. lycopersici (Cunnington et al., 2005b). More work, including experimental host range studies are needed to determine whether all these taxa in fact represent the same globally distributed species.
Neoerysiphe kerribeeensis is the only taxon which has been described as a native Australian teleomorphic powdery mildew species producing chasmothecia on the native S. glossanthus. Its asexual morph was identified on several Senecio spp. introduced to Australia (Beilharz et al., 2010). There are no closely related Neoerysiphe species known overseas; therefore, the origin of N. kerribeeensis needs further investigation. Beilharz et al. (2010) suggested that N. kerribeeensis was native to Australia, and had become a pathogen of introduced Senecio spp. A contrasting explanation is that this species reached Australia as a pathogen of introduced Senecio spp., and its current occurrence on the native S. glossanthus is the result of its rapid host range expansion. If N. kerribeeensis was found overseas, this would support the second hypothesis.
Some powdery mildews become invasive when introduced to new geographic regions where their host plant species are also present (Kiss, 2005; Jones and Baker, 2007; Brasier, 2008; Desprez-Loustau et al., 2010, 2018). The introduction of E. necator from North America to Europe with infected grapevines is a textbook example (Brewer and Milgroom, 2010; Gadoury et al., 2012). Amongst all crop pathogenic fungi, a number of powdery mildews appear to be spreading most rapidly on a global scale, and are predicted to reach all available hosts by 2050 in many countries of the world (Bebber et al., 2014, 2019; Bebber and Gurr, 2015). Most probably, their introduction and spread in Australia has happened as part of the fundamental changes in the entire vegetation and landscape since 1788 (Kirkpatrick, 1999; Cook and Dias, 2006; Fensham and Laffineur, 2019). As conidia of powdery mildews are short-lived, and their long-distance dispersal is debated (Glawe, 2008), it is likely that all the species identified in this study had reached Australia on their living host plants.
Molecular clock studies indicated that the Erysiphales may have emerged in the late Cretaceous (~100–80 million years ago), and the first radiation of the major lineages occurred in the Cretaceous/Paleogene boundary (70–58 million years ago), in the Northern Hemisphere (Takamatsu, 2013a). By that time the Australian continent may have already been separated from other large pieces of land, so its terrestrial flora continued to evolve in isolation, without being exposed to those plant pathogenic fungi that could not reach the continent from overseas.
It is often difficult to explain how plant pathogenic fungi, especially plant-associated obligate biotrophs, reach new and remote geographic regions, particularly if these pathogens infect weeds and other wild plant species (Fontaine et al., 2013; Kiss et al., 2018b). The possibility that some powdery mildew species arrived in Australia prior to the first European settlement in 1788 cannot be excluded, particularly given Dutch exploration of the Australian continent from the early 1600s. Indigenous Australians arrived at least 50,000 years prior to the beginning of modern agriculture, and may have introduced a number of plant species to the Australian continent. Most of the taxa confirmed by this study are linked to the modern agricultural and horticultural trade, and recent, accidental, human-mediated introductions. The number of powdery mildew species in Australia compared to other parts of the world is low. The close phylogenetic relationships of all Australian powdery mildews to those known overseas, including the specimens collected from Australian native plants, indicate that Australia is a continent without native powdery mildews. Most, if not all, powdery mildew species in Australia appear to have been introduced since the European colonization of the continent.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s (Vilgalys, 2003).
Author Contributions
LKi designed and coordinated the research. LKi, NV, and UB wrote the paper. LKi, JD, ST, KB, JB, MC, AD, RGS, PD, LJ, TN, JE, WH, LKe, SM, KS, JM, SP, S-YL, RSm, RSo, UT, KV, AB, DW, and AY collected and examined the fresh specimens and determined some of the ITS sequences. CM, JD, and KB provided botanical expertise and UB and RGS taxonomic expertise. YT examined the old herbarium specimens and sequenced the ITS region in those specimens. LKi, KB, KS, LKe, D-NJ, and MN determined the rest of the nrDNA sequences. NV carried out the phylogenetic analyses. LKi and ST contributed to those analyses. All authors commented on the manuscript and read and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by University of Southern Queensland and a grant of the Australian Department of Agriculture, Water and the Environment. Preliminary results of this work were published in an abstract of the Australasian Plant Pathology Society Conference APPS 2019, held in Melbourne in November 2019.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01571/full#supplementary-material
References
- Amano K. (1986). Host Range and Geographical Distribution of the Powdery Mildew Fungi. Tokyo: Japan Scientific Societies Press. [Google Scholar]
- Bebber D. P., Field E., Gui H., Mortimer P., Holmes T., Gurr S. J. (2019). Many unreported crop pests and pathogens are probably already present. Global Change Biol. 25, 2703–2713. 10.1111/gcb.14698 [DOI] [PubMed] [Google Scholar]
- Bebber D. P., Gurr S. J. (2015). Crop-destroying fungal and oomycete pathogens challenge food security. Fungal Genet. Biol. 74, 62–64. 10.1016/j.fgb.2014.10.012 [DOI] [PubMed] [Google Scholar]
- Bebber D. P., Holmes T., Gurr S. J. (2014). The global spread of crop pests and pathogens. Global Ecol. Biogeogr. 23, 1398–1407. 10.1111/geb.12214 [DOI] [Google Scholar]
- Beenken L. (2017). First records of the powdery mildews Erysiphe platani and E. alphitoides on Ailanthus altissima reveal host jumps independent of host phylogeny. Mycol. Prog. 16, 135–143. 10.1007/s11557-016-1260-2 [DOI] [Google Scholar]
- Beilharz V., Cunnington J. H., Pascoe I. G. (2010). Neoerysiphe kerribeeensis sp. nov. (Ascomycota: Erysiphales), a new species of Neoerysiphe on native and introduced species of Senecio (Asteraceae) in Australia. Fungal Biol. 114, 340–344. 10.1016/j.funbio.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Bereczky Z., Pintye A., Csontos P., Braun U., Kiss L. (2015). Does the parasite follow its host? Occurrence of morphologically barely distinguishable powdery mildew anamorphs on Oenothera spp. in different parts of the world. Mycoscience 56, 267–272. 10.1016/j.myc.2014.08.001 [DOI] [Google Scholar]
- Bidartondo M. I. (2008). Preserving accuracy in GenBank. Science 319:1616. 10.1126/science.319.5870.1616a [DOI] [PubMed] [Google Scholar]
- Bindschedler L. V., Panstruga R., Spanu P. D. (2016). Mildew-Omics: how global analyses aid the understanding of life and evolution of powdery mildews. Front. Plant Sci. 7:123. 10.3389/fpls.2016.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosecurity Australia (2010). Final Review of Policy: Importation Of Hop (Humulus Species) Propagative Material Into Australia. Canberra, ACT: Biosecurity Australia. [Google Scholar]
- Boesewinkel H. (1977). Identification of Erysiphaceae by conidial characteristics. Rev. Mycol. 41, 493–507. [Google Scholar]
- Bradshaw M., Tobin P. C. (2020). Sequencing herbarium specimens of a common detrimental plant disease (powdery mildew). Phytopathology (in press). 10.1094/PHYTO-04-20-0139-PER [DOI] [PubMed] [Google Scholar]
- Brasier C. (2008). The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 57, 792–808. 10.1111/j.1365-3059.2008.01886.x [DOI] [Google Scholar]
- Braun U. (1987). A monograph of the Erysiphales (powdery mildews). Beih. Nova Hedwigia 89, 1–700. [Google Scholar]
- Braun U. (2011). The current systematics and taxonomy of the powdery mildews (Erysiphales): an overview. Mycoscience 52, 210–212. 10.1007/s10267-010-0092-1 [DOI] [Google Scholar]
- Braun U. (2012). The impacts of the discontinuation of dual nomenclature of pleomorphic fungi: the trivial facts, problems, and strategies. IMA Fungus 3, 81–86. 10.5598/imafungus.2012.03.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U. (2013). (2210–2232) Proposals to conserve the teleomorph-typified name Blumeria against the anamorph-typified name Oidium and twenty-two teleomorph-typified powdery mildew species names against competing anamorph-typified names (Ascomycota: Erysiphaceae). Taxon 62, 1328–1331. 10.12705/626.20 [DOI] [Google Scholar]
- Braun U., Cook R. T. A. (2012). Taxonomic Manual of the Erysiphales (Powdery Mildews). Utrecht: CBS-KNAW Fungal Biodiversity Centre. [Google Scholar]
- Braun U., Cook R. T. A., Inman A. J., Shin H. D. (2002). “The taxonomy of the powdery mildew fungi,” in The Powdery Mildews: a Comprehensive Treatise, eds R. Bélanger, A. Dik, and W. Bushnell (St. Paul, MN: APS Press; ), 13–54. [Google Scholar]
- Braun U., Shin H.-D., Takamatsu S., Meeboon J., Kiss L., Lebeda A., et al. (2019). Phylogeny and taxonomy of Golovinomyces orontii revisited. Mycol. Prog. 18, 335–357. 10.1007/s11557-018-1453-y [DOI] [Google Scholar]
- Braun U., Shishkoff N., Takamatsu S. (2001). Phylogeny of Podosphaera sect. Sphaerotheca subsect. Magnicellulatae (Sphaerotheca fuliginea auct. s. lat.) inferred from rDNA ITS sequences - a taxonomic interpretation. Schlechtendalia 7, 45–52. Available online at: https://www.zobodat.at/pdf/Schlechtendalia_7_0045-0052.pdf [Google Scholar]
- Brewer M. T., Milgroom M. G. (2010). Phylogeography and population structure of the grape powdery mildew fungus, Erysiphe necator, from diverse Vitis species. BMC Evol. Biol. 10:268. 10.1186/1471-2148-10-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M. G., Alvarez R. E., Takamatsu S. (2018). Morphology and molecular phylogeny of Brasiliomyces malachrae, a unique powdery mildew distributed in Central and South America. Mycoscience 59, 461–466. 10.1016/j.myc.2018.04.003 [DOI] [Google Scholar]
- Chen R.-S., Chu C., Cheng C.-W., Chen W.-Y., Tsay J.-G. (2008). Differentiation of two powdery mildews of sunflower (Helianthus annuus) by a PCR-mediated method based on ITS sequences. Eur. J. Plant Pathol. 121, 1–8. 10.1007/s10658-007-9234-5 [DOI] [Google Scholar]
- Chiddarwar P. (1959). Salmonomyces—a new member of the Erysiphaceae. Sydowia 13, 55–56. [Google Scholar]
- Cho S., Han K., Choi I., Shin H.-D. (2018). First Report of Powdery Mildew Caused by Podosphaera xanthii on Hydrocleys nymphoides in Korea. Plant Dis. 102, 247–247. 10.1094/PDIS-06-17-0920-PDN30722488 [DOI] [Google Scholar]
- Cook G. D., Dias L. (2006). It was no accident: deliberate plant introductions by Australian government agencies during the 20th century. Aust. J. Bot. 54, 601–625. 10.1071/BT05157 [DOI] [Google Scholar]
- Cook R. T. A., Braun U. (2009). Conidial germination patterns in powdery mildews. Mycol. Res. 113, 616–636. 10.1016/j.mycres.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Crous P. W., Wingfield M. J., Lombard L., Roets F., Swart W. J., Alvarado P., et al. (2019). Fungal Planet description sheets: 951–1041. Persoonia 43, 223–425. 10.3767/persoonia.2019.43.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnington J. H., Beilharz V. C., Pascoe I. G., Lawrie A. C. (2005a). Oidium ixodiae sp. nov. on Ixodia achilleoides in Australia. Australas. Plant Pathol. 34, 91–94. 10.1071/AP04090 [DOI] [Google Scholar]
- Cunnington J. H., Brett R. W. (2009). S-type powdery mildew on lilac in Australia. Australas. Plant Dis. Notes 4, 21–22. 10.1071/DN09009 [DOI] [Google Scholar]
- Cunnington J. H., Jones R. H., De Alwis S. K. (2008a). First record of powdery mildew on the Cephalotaceae. Australas. Plant Dis. Notes 3, 51–52. 10.1007/BF03211237 [DOI] [Google Scholar]
- Cunnington J. H., Lawrie A. C., Pascoe I. G. (2004a). Unexpected ribosomal DNA internal transcribed spacer sequence variation within Erysiphe aquilegiae sensu lato. Fungal Divers. 16, 1–10. Available online at: http://www.fungaldiversity.org/fdp/sfdp/16-1.pdf [Google Scholar]
- Cunnington J. H., Lawrie A. C., Pascoe I. G. (2004b). Molecular determination of anamorphic powdery mildew fungi on the Fabaceae in Australia. Austral. Plant Pathol. 33, 281–284. 10.1071/AP04017 [DOI] [Google Scholar]
- Cunnington J. H., Lawrie A. C., Pascoe I. G. (2005b). Molecular identification of Golovinomyces (Ascomycota: Erysiphales) anamorphs on the Solanaceae in Australia. Austral. Plant Pathol. 34, 51–55. 10.1071/AP04082 [DOI] [Google Scholar]
- Cunnington J. H., Lawrie A. C., Pascoe I. G. (2005c). Genetic variation within Podosphaera tridactyla reveals a paraphyletic species complex with biological specialization towards specific Prunus subgenera. Mycol. Res. 109, 357–362. 10.1017/S0953756204002072 [DOI] [PubMed] [Google Scholar]
- Cunnington J. H., Lawrie A. C., Pascoe I. G. (2010). Genetic characterization of the Golovinomyces cichoracearum complex in Australia. Plant Pathol. 59, 158–164. 10.1111/j.1365-3059.2009.02175.x [DOI] [Google Scholar]
- Cunnington J. H., Takamatsu S., Lawrie A. C., Pascoe I. G. (2003). Molecular identification of anamorphic powdery mildews (Erysiphales). Austral. Plant Pathol. 32, 421–428. 10.1071/ap03045 [DOI] [Google Scholar]
- Cunnington J. H., Watson A., Liberato J. R., Jones R. H. (2008b). First record of powdery mildew on carrots in Australia. Austral. Plant Dis. Notes 3, 38–41. 10.1007/BF03211231 [DOI] [Google Scholar]
- Desprez-Loustau M.-L., Courtecuisse R., Robin C., Husson C., Moreau P.-A., Blancard D., et al. (2010). Species diversity and drivers of spread of alien fungi (sensu lato) in Europe with a particular focus on France. Biol. Invas. 12, 157–172. 10.1007/s10530-009-9439-y [DOI] [Google Scholar]
- Desprez-Loustau M.-L., Massot M., Feau N., Fort T., De Vicente A., Torés J. A., et al. (2017). Further support of conspecificity of oak and mango powdery mildew and first report of Erysiphe quercicola and Erysiphe alphitoides on mango in mainland Europe. Plant Dis. 101, 1086–1093. 10.1094/PDIS-01-17-0116-RE [DOI] [PubMed] [Google Scholar]
- Desprez-Loustau M.-L., Massot M., Toïgo M., Fort T., Kaya A. G. A., Boberg J., et al. (2018). From leaf to continent: the multi-scale distribution of an invasive cryptic pathogen complex on oak. Fungal Ecol. 36, 39–50. 10.1016/j.funeco.2018.08.001 [DOI] [Google Scholar]
- Doidge E. M. (1948). South African Ascomycetes in the National Herbarium, Part IV, Nos 196–254. Bothalia 4, 837–878. [Google Scholar]
- Doyle J. J. (1992). Gene trees and species trees: molecular systematics as one-character taxonomy. Syst. Bot. 17, 144–163 [Google Scholar]
- Ellingham O., David J., Culham A. (2019). Enhancing identification accuracy for powdery mildews using previously underexploited DNA loci. Mycologia 111, 798–812. 10.1080/00275514.2019.1643644 [DOI] [PubMed] [Google Scholar]
- Farris J. S., Källersjö M., Kluge A. G., Bult C. (1995). Testing significance of incongruence. Cladistics 10, 315–319. 10.1111/j.1096-0031.1994.tb00181.x [DOI] [Google Scholar]
- Fensham R. J., Laffineur B. (2019). Defining the native and naturalised flora for the Australian continent. Aust. J. Bot. 67, 55–69. 10.1071/BT18168 [DOI] [Google Scholar]
- Fontaine M. C., Gladieux P., Hood M. E., Giraud T. (2013). History of the invasion of the anther smut pathogen on Silene latifolia in North America. New Phytol. 198, 946–956. 10.1111/nph.12177 [DOI] [PubMed] [Google Scholar]
- Forster P. I. (1994). A taxonomic revision of Acalypha L. (Euphorbiaceae) in Australia. Austrobaileya 4, 209–226. [Google Scholar]
- Frantzeskakis L., Di Pietro A., Rep M., Schirawski J., Wu C. H., Panstruga R. (2019a). Rapid evolution in plant–microbe interactions – a molecular genomics perspective. New Phytol. 225, 1134–1142. 10.1111/nph.15966 [DOI] [PubMed] [Google Scholar]
- Frantzeskakis L., Németh M. Z., Barsoum M., Kusch S., Kiss L., Takamatsu S., et al. (2019b). The Parauncinula polyspora draft genome provides insights into patterns of gene erosion and genome expansion in powdery mildew fungi. mBio 10, e01692–e01619. 10.1128/mBio.01692-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadoury D. M., Cadle-Davidson L., Wilcox W. F., Dry I. B., Seem R. C., Milgroom M. G. (2012). Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol. Plant Pathol. 13, 1–16. 10.1111/j.1364-3703.2011.00728.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M., Bruns T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Glawe D. A. (2008). The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 46, 27–51. 10.1146/annurev.phyto.46.081407.104740 [DOI] [PubMed] [Google Scholar]
- Golzar H., Shankar M., D'Antuono M. (2016). Responses of commercial wheat varieties and differential lines to western Australian powdery mildew (Blumeria graminis f. sp. tritici) populations. Austral. Plant Pathol. 45, 347–355. 10.1007/s13313-016-0420-9 [DOI] [Google Scholar]
- Goss O. (1964). A revised list of diseases of ornamental plants recorded in Western Australia. J. Depart. Agric. Western Aust. Ser. 4, 589–603. [Google Scholar]
- Halsted B. D. (1899). A new genus of powdery mildews - erysiphopsis. Bull. Torrey Bot. Club 26, 594–595. [Google Scholar]
- Heluta V., Takamatsu S., Harada M., Voytyuk S. (2010). Molecular phylogeny and taxonomy of Eurasian Neoerysiphe species infecting Asteraceae and Geranium. Persoonia 24, 81. 10.3767/003158510X501696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett D., Abarenkov K., Kõljalg U., Öpik M., Chai B., Cole J., et al. (2016). Sequence-based classification and identification of Fungi. Mycologia 108, 1049–1068. 10.3852/16-130 [DOI] [PubMed] [Google Scholar]
- Hirata T., Cunnington J. H., Paksiri U., Limkaisang S., Shishkoff N., Grigaliunaite B., et al. (2000). Evolutionary analysis of subsection Magnicellulatae of Podosphaera section Sphaerotheca (Erysiphales) based on the rDNA internal transcribed spacer sequences with special reference to host plants. Can. J. Bot. 78, 1521–1530. 10.1139/cjb-78-12-1521 [DOI] [Google Scholar]
- Hirata T., Takamatsu S. (1996). Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience 37, 283–288. 10.1007/BF02461299 [DOI] [Google Scholar]
- Hirose S., Tanda S., Levente K., Grigaliunaite B., Havrylenko M., Takamatsu S. (2005). Molecular phylogeny and evolution of the maple powdery mildew (Sawadaea, Erysiphaceae) inferred from nuclear rDNA sequences. Mycol. Res. 109, 912–922. 10.1017/S0953756205003527 [DOI] [PubMed] [Google Scholar]
- Inuma T., Khodaparast S. A., Takamatsu S. (2007). Multilocus phylogenetic analyses within Blumeria graminis, a powdery mildew fungus of cereals. Mol. Phylogenet. Evol. 44, 741–751. 10.1016/j.ympev.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Irinyi L., Serena C., Garcia-Hermoso D., Arabatzis M., Desnos-Ollivier M., Vu D., et al. (2015). International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 53, 313–337. 10.1093/mmy/myv008 [DOI] [PubMed] [Google Scholar]
- James S. A., O'Kelly M. J., Carter D. M., Davey R. P., Van Oudenaarden A., Roberts I. N. (2009). Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res. 19, 626–635. 10.1101/gr.084517.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovics T., Bai Y., Kovács M. G., Kiss L. (2008). Oidium neolycopersici: intraspecific variability inferred from AFLP analysis and relationship with closely related powdery mildew fungi infecting various plant species. Phytopathology 98, 529–540. 10.1094/PHYTO-98-5-0529 [DOI] [PubMed] [Google Scholar]
- Jankovics T., Dolovac N., Bulajić A., Krstić B., Pascal T., Bardin M., et al. (2011). Peach rusty spot is caused by the apple powdery mildew fungus, Podosphaera leucotricha. Plant Dis. 95, 719–724. 10.1094/pdis-10-10-0711 [DOI] [PubMed] [Google Scholar]
- Jones D., Baker R. (2007). Introductions of non-native plant pathogens into Great Britain, 1970–2004. Plant Pathol. 56, 891–910. 10.1111/j.1365-3059.2007.01619.x [DOI] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodaparast S. A., Niinomi S., Takamatsu S. (2007). Molecular and morphological characterization of Leveillula (Ascomycota: Erysiphales) on monocotyledonous plants. Mycol. Res. 111, 673–679. 10.1016/j.mycres.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Khodaparast S. A., Takamatsu S., Harada M., Abbasi M., Samadi S. (2012). Additional rDNA ITS sequences and its phylogenetic consequences for the genus Leveillula with emphasis on conidium morphology. Mycol. Prog. 11, 741–752. 10.1007/s11557-011-0785-7 [DOI] [Google Scholar]
- Kirkpatrick J. B. (1999). A Continent Transformed: Human Impact on the Natural Vegetation of Australia. Oxford: Oxford University Press. [Google Scholar]
- Kirschner R. (2010). First record of Erysiphe magnifica on lotus, a host outside the Magnoliales. Mycol. Prog. 9, 417–424. 10.1007/s11557-009-0651-z [DOI] [Google Scholar]
- Kiss L. (2005). Powdery mildew as invasive plant pathogens: new epidemics caused by two North American species in Europe. Mycol. Res. 109, 259–260. 10.1017/S0953756205232793 [DOI] [PubMed] [Google Scholar]
- Kiss L. (2012). Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109:E1811. 10.1073/pnas.1207143109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss L., Cook R. T. A., Saenz G. S., Cunnington J. H., Takamatsu S., Pascoe I., et al. (2001). Identification of two powdery mildew fungi, Oidium neolycopersici sp. nov. and O. lycopersici, infecting tomato in different parts of the world. Mycol. Res. 105, 684–697. 10.1017/S0953756201004105 [DOI] [Google Scholar]
- Kiss L., Jankovics T., Kovács G. M., Daughtrey M. L. (2008). Oidium longipes, a new powdery mildew fungus on Petunia in the USA: a potential threat to ornamental and vegetable solanaceous crops. Plant Dis. 92, 818–825. 10.1094/pdis-92-5-0818 [DOI] [PubMed] [Google Scholar]
- Kiss L., Kovács G. M., Bóka K., Bohár G., Bohárné K. V., Németh M. Z., et al. (2018b). Deciphering the biology of Cryptophyllachora eurasiatica gen. et sp. nov., an often cryptic pathogen of an allergenic weed, Ambrosia artemisiifolia. Sci. Rep. 8:10806. 10.1038/s41598-018-29102-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss L., Stuart K., Grigg J., Calvert J., Dearnaley J. (2018a). First report of powdery mildew on goji berry (Lycium barbarum) caused by Arthrocladiella mougeotii in Queensland, Australia. Plant Dis. 102, 446–446. 10.1094/PDIS-07-17-1055-PDN [DOI] [Google Scholar]
- Kiss L., Takamatsu S., Cunnington J. H. (2005). Molecular identification of Oidium neolycopersici as the causal agent of the recent tomato powdery mildew epidemics in North America. Plant Dis. 89, 491–496. 10.1094/PD-89-0491 [DOI] [PubMed] [Google Scholar]
- Ko C.-Y., Lin T.-Y., Ho C.-W., Shaw J.-F. (2010). In vitro regeneration of Cephalotus follicularis. Hortscience 45, 260–264. 10.21273/HORTSCI.45.2.260 [DOI] [Google Scholar]
- Kovács G. M., Jankovics T., Kiss L. (2011). Variation in the nrDNA ITS sequences of some powdery mildew species: do routine molecular identification procedures hide valuable information? Eur. J. Plant Pathol. 131, 135–141. 10.1007/s10658-011-9793-3 [DOI] [Google Scholar]
- Kreyling J., Peršoh D., Werner S., Benzenberg M., Wöllecke J. (2012). Short-term impacts of soil freeze-thaw cycles on roots and root-associated fungi of Holcus lanatus and Calluna vulgaris. Plant Soil 353, 19–31. 10.1007/s11104-011-0970-0 [DOI] [Google Scholar]
- Krístková E., Lebeda A., Sedláková B. (2009). Species spectra, distribution and host range of cucurbit powdery mildews in the Czech Republic, and in some other European and Middle Eastern countries. Phytoparasitica 37, 337–350. 10.1007/s12600-009-0045-4 [DOI] [Google Scholar]
- Kruse J., Thiel H., Beenken L., Bender H., Braun U., Ecker J., et al. (2018). Bemerkenswerte Funde phytoparasitischer Kleinpilze (9). Z. Mykol. 84, 87–135. Available online at: https://www.semanticscholar.org/paper/Bemerkenswerte-Funde-phytoparasitischer-Kleinpilze-Kruse-Thiel/aec03bc861d93c86564f4cafbcb2416865a8accc
- Kusch A., Németh M. Z., Vaghefi N., Ibrahim H. M. M., Panstruga R., Kiss L. (2020). A short-read genome assembly resource for Leveillula taurica causing powdery mildew disease of sweet pepper (Capsicum annuum). Mol. Plant-Microbe Interact. 33, 782–786. 10.1094/MPMI-02-20-0029-A [DOI] [PubMed] [Google Scholar]
- Lebeda A., Mieslerová B. (2011). Taxonomy, distribution and biology of lettuce powdery mildew (Golovinomyces cichoracearum sensu stricto). Plant Pathol. 60, 400–415. 10.1111/j.1365-3059.2010.02399.x [DOI] [Google Scholar]
- Liberato J., Cunnington J. (2006). First record of Erysiphe aquilegiae on a host outside the Ranunculaceae. Austral. Plant Pathol. 35, 291–292. 10.1071/AP06002 [DOI] [Google Scholar]
- Liberato J., Cunnington J. (2007). Powdery mildew on Mentha in Australia. Austral. Plant Dis. Notes 2, 83–86. 10.1071/DN07034 [DOI] [Google Scholar]
- Liberato J., Shivas R., Cunnington J. (2006). Podosphaera xanthii on Euryops chrysanthemoides in Australia. Austral. Plant Pathol. 35, 739–741. 10.1071/AP06067 [DOI] [Google Scholar]
- Limkaisang S., Cunnington J. H., Wui L. K., Salleh B., Sato Y., Divarangkoon R., et al. (2006). Molecular phylogenetic analyses reveal a close relationship between powdery mildew fungi on some tropical trees and Erysiphe alphitoides, an oak powdery mildew. Mycoscience 47, 327–335. 10.1007/s10267-006-0311-y [DOI] [Google Scholar]
- Lister D. L., Bower M. A., Howe C. J., Jones M. K. (2008). Extraction and amplification of nuclear DNA from herbarium specimens of emmer wheat: a method for assessing DNA preservation by maximum amplicon length recovery. Taxon 57, 254–258. 10.2307/25065966 [DOI] [Google Scholar]
- Magarey P., Gadoury D., Emmett R., Biggins L., Clarke K., Wachtel M., et al. (1997). Cleistothecia of Uncinula necator in Australia. Vitic. Enol. Sci. 52, 210–218. [Google Scholar]
- Marçais B., Desprez-Loustau M.-L. (2014). European oak powdery mildew: impact on trees, effects of environmental factors, and potential effects of climate change. Ann. For. Sci. 71, 633–642. 10.1007/s13595-012-0252-x [DOI] [Google Scholar]
- Marmolejo J., Siahaan S. A., Takamatsu S., Braun U. (2018). Three new records of powdery mildews found in Mexico with one genus and one new species proposed. Mycoscience 59, 1–7. 10.1016/j.myc.2017.06.010 [DOI] [Google Scholar]
- Matsuda S., Takamatsu S. (2003). Evolution of host–parasite relationships of Golovinomyces (Ascomycete: Erysiphaceae) inferred from nuclear rDNA sequences. Mol. Phylogenet. Evol. 27, 314–327. 10.1016/S1055-7903(02)00401-3 [DOI] [PubMed] [Google Scholar]
- McTaggart A., Ryley M., Shivas R. (2012). First report of the powdery mildew Erysiphe diffusa on soybean in Australia. Austral. Plant Dis. Notes 7, 127–129. 10.1007/s13314-012-0065-7 [DOI] [Google Scholar]
- Meeboon J., Hidayat I., Takamatsu S. (2013). Pseudoidium javanicum, a new species of powdery mildew on Acalypha spp. from Indonesia. Mycoscience 54, 183–187. 10.1016/j.myc.2012.08.006 [DOI] [Google Scholar]
- Meeboon J., Takamatsu S. (2015). Erysiphe takamatsui, a powdery mildew of lotus: rediscovery of teleomorph after 40 years, morphology and phylogeny. Mycoscience 56, 159–167. 10.1016/j.myc.2014.05.002 [DOI] [Google Scholar]
- Meeboon J., Takamatsu S. (2017). Microidium phyllanthi-reticulati sp. nov. on Phyllanthus reticulatus. Mycotaxon 132, 289–297. 10.5248/132.289 [DOI] [Google Scholar]
- Meeboon J., Takamatsu S., Braun U. (2020). Morphophylogenetic analyses revealed that Podosphaera tridactyla constitutes a species complex. Mycologia 112, 244–266. 10.1080/00275514.2019.1698924 [DOI] [PubMed] [Google Scholar]
- Menardo F., Praz C. R., Wyder S., Ben-David R., Bourras S., Matsumae H., et al. (2016). Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nat. Genet. 48, 201–205. 10.1038/ng.3485 [DOI] [PubMed] [Google Scholar]
- Moparthi S., Grove G. G., Pandey B., Bradshaw M., Latham S. R., Braun U., et al. (2019). Phylogeny and taxonomy of Podosphaera cerasi, sp. nov., and Podosphaera prunicola sensu lato. Mycologia 111, 647–659. 10.1080/00275514.2019.1611316 [DOI] [PubMed] [Google Scholar]
- Mori Y., Sato Y., Takamatsu S. (2000). Evolutionary analysis of the powdery mildew fungi using nucleotide sequences of the nuclear ribosomal DNA. Mycologia 92, 74–93. 10.2307/3761452 [DOI] [Google Scholar]
- Nilsson R. H., Hyde K. D., Pawłowska J., Ryberg M., Tedersoo L., Aas A. B., et al. (2014). Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Divers. 67, 11–19. 10.1007/s13225-014-0291-8 [DOI] [Google Scholar]
- Nilsson R. H., Ryberg M., Kristiansson E., Abarenkov K., Larsson K.-H., Kõljalg U. (2006). Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1:e59. 10.1371/journal.pone.0000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numminen E., Vaumourin E., Parratt S. R., Poulin L., Laine A.-L. (2019). Variation and correlations between sexual, asexual and natural enemy resistance life-history traits in a natural plant pathogen population. BMC Evol. Biol. 19:142. 10.1186/s12862-019-1468-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest Version 2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden. [Google Scholar]
- O'Leary N., Thode V. (2016). The Genus Glandularia (Verbenaceae) in Brazil. Ann. Missouri Bot. Gard. 101, 699–749. 10.3417/2014008 [DOI] [Google Scholar]
- Pande A. (2008). Ascomycetes of Peninsular India. Jodhpur: Scientific Publishers. [Google Scholar]
- Pastirčáková K., Jankovics T., Komáromi J., Pintye A., Pastirčák M. (2016). Genetic diversity and host range of powdery mildews on Papaveraceae. Mycol. Prog. 15:36 10.1007/s11557-016-1178-8 [DOI] [Google Scholar]
- Pettitt T., Henricot B., Matatiken D., Cook R. T. A. (2010). First record of Oidium anamorph of Podosphaera xanthii on Medusagyne oppositifolia. Plant Pathol. 59:1168 10.1111/j.1365-3059.2010.02333.x [DOI] [Google Scholar]
- Pirozynski K. (1965). African species of Uncinula. Myc. Papers 101, 2–23. [Google Scholar]
- Qiu P. L., Liu S. Y., Bradshaw M., Rooney-Latham S., Takamatsu S., Bulgakov T. S., et al. (2020). Multi-locus phylogeny and taxonomy of an unresolved, heterogeneous species complex within the genus Golovinomyces (Ascomycota, Erysiphales), including G. ambrosiae, G. circumfusus and G. spadiceus. BMC Microbiol. 20:51 10.1186/s12866-020-01731-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R. P. (2007). The Introduced Flora of Australia and Its Weed Status. Adelaide, SA: CRC for Australian Weed Management. [Google Scholar]
- Rittenour W. R., Ciaccio C. E., Barnes C. S., Kashon M. L., Lemons A. R., Beezhold D. H., et al. (2014). Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environ. Sci. Process. Impacts 16, 33–43. 10.1039/c3em00441d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sagun V., Levin G., Van Welzen P. (2010). Revision and phylogeny of Acalypha (Euphorbiaceae) in Malesia. Blumea 55, 21–60. 10.3767/000651910X499141 [DOI] [Google Scholar]
- Sanz J. M. C., Rodríguez P. M. (2012). Synopsis of Acalypha (Euphorbiaceae) of continental Ecuador. Phytokeys 17, 1–17. 10.3897/phytokeys.17.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe A. V. (1969). Erysiphopsis replaces Salmonomyces. Bull. Torrey Bot. Club 96, 101–102. 10.2307/2484013 [DOI] [Google Scholar]
- Sawada K. (1943). Descriptive catalogue of the Formosan fungi. Part VIII. Rept. Dep. Agric. Gov. Res. Inst. Formosa 85, 1–130. [Google Scholar]
- Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler M., Schmidt A., Siahaan S., Takamatsu S., Braun U. (2016). A taxonomic and phylogenetic study of the Golovinomyces biocellatus complex (Erysiphales, Ascomycota) using asexual state morphology and rDNA sequence data. Mycol. Prog. 15, 56 10.1007/s11557-016-1197-5 [DOI] [Google Scholar]
- Selosse M. A., Vincenot L., Öpik M. (2016). Data processing can mask biology: towards better reporting of fungal barcoding data? New Phytol. 210, 1159–1164. 10.1111/nph.13851 [DOI] [PubMed] [Google Scholar]
- Shin H.-D., La Y.-J. (1993). Morphology of edge lines of chained immature conidia on conidiophores in powdery mildew fungi and their taxonomic significance. Mycotaxon 46, 445–451. [Google Scholar]
- Shin H.-D., Meeboon J., Takamatsu S., Adhikari M. K., Braun U. (2019). Phylogeny and taxonomy of Pseudoidium pedaliacearum. Mycol. Prog. 18, 237–246. 10.1007/s11557-018-1429-y [DOI] [Google Scholar]
- Simmonds J. H. (1966). Host Index of Plant Diseases in Queensland. Brisbane, QLD: Queensland Department of Primary Industries. [Google Scholar]
- Smith R. L., Sawbridge T., Mann R., Kaur J., May T. W., Edwards J. (2020). Rediscovering an old foe: optimised molecular methods for DNA extraction and sequencing applications for fungarium specimens of powdery mildew (Erysiphales). PLos ONE 15:e0232535. 10.1371/journal.pone.0232535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell R. J., Stuart K., Kiss L. (2018). First conclusive report of an Erysiphe sp. causing powdery mildew on moth vine (Araujia sericifera) in Australia and worldwide. Plant Dis. 102:1452 10.1094/PDIS-12-17-1880-PDN [DOI] [Google Scholar]
- Stadler M., Lambert C., Wibberg D., Kalinowski J., Cox R. J., Kolarík M., et al. (2020). Intragenomic polymorphisms in the ITS region of high-quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycol. Prog. 19, 235–245. 10.1007/s11557-019-01552-9 [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susi H., Barrès B., Vale P. F., Laine A.-L. (2015). Co-infection alters population dynamics of infectious disease. Nat. Commun. 6:5975. 10.1038/ncomms6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Szentiványi O., Kiss L. (2003). Overwintering of Ampelomyces mycoparasites on apple trees and other plants infected with powdery mildews. Plant Pathol. 52, 737–746. 10.1111/j.1365-3059.2003.00937.x [DOI] [Google Scholar]
- Tai F. L. (1946). Further studies on the Erysiphaceae of China. Bull. Torrey Bot. Club 73, 108–130. 10.2307/2481610 [DOI] [Google Scholar]
- Takamatsu S. (2004). Phylogeny and evolution of the powdery mildew fungi (Erysiphales, Ascomycota) inferred from nuclear ribosomal DNA sequences. Mycoscience 45, 147–157. 10.1007/s10267-003-0159-316175793 [DOI] [Google Scholar]
- Takamatsu S. (2013a). Origin and evolution of the powdery mildews (Ascomycota, Erysiphales). Mycoscience 54, 75–86. 10.1016/j.myc.2012.08.004 [DOI] [Google Scholar]
- Takamatsu S. (2013b). Molecular phylogeny reveals phenotypic evolution of powdery mildews (Erysiphales, Ascomycota). J. Gen. Plant Pathol. 79, 218–226. 10.1007/s10327-013-0447-5 [DOI] [Google Scholar]
- Takamatsu S., Braun U., Limkaisang S., Kom-Un S., Sato Y., Cunnington J. H. (2007). Phylogeny and taxonomy of the oak powdery mildew Erysiphe alphitoides sensu lato. Mycol. Res. 111, 809–826. 10.1016/j.mycres.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Takamatsu S., Havrylenko M., Wolcan S. M., Matsuda S., Niinomi S. (2008). Molecular phylogeny and evolution of the genus Neoerysiphe (Erysiphaceae, Ascomycota). Mycol. Res. 112, 639–649. 10.1016/j.mycres.2008.01.004 [DOI] [PubMed] [Google Scholar]
- Takamatsu S., Hirata T., Sato Y. (1998). Phylogenetic analysis and predicted secondary structures of the rDNA internal transcribed spacers of the powdery mildew fungi (Erysiphaceae). Mycoscience 39, 441–453. 10.1007/BF02460905 [DOI] [Google Scholar]
- Takamatsu S., Ito H., Shiroya Y., Kiss L., Heluta V. (2015a). First comprehensive phylogenetic analysis of the genus Erysiphe (Erysiphales, Erysiphaceae) I. The Microsphaera lineage. Mycologia 107, 475–489. 10.3852/15-007 [DOI] [PubMed] [Google Scholar]
- Takamatsu S., Ito H., Shiroya Y., Kiss L., Heluta V. (2015b). First comprehensive phylogenetic analysis of the genus Erysiphe (Erysiphales, Erysiphaceae) II: the Uncinula lineage. Mycologia 107, 903–914. 10.3852/15-062 [DOI] [PubMed] [Google Scholar]
- Takamatsu S., Kano Y. (2001). PCR primers useful for nucleotide sequencing of rDNA of the powdery mildew fungi. Mycoscience 42, 135–139. 10.1007/BF02463987 [DOI] [Google Scholar]
- Takamatsu S., Matsuda S., Grigaliunaite B. (2013). Comprehensive phylogenetic analysis of the genus Golovinomyces (Ascomycota: Erysiphales) reveals close evolutionary relationships with its host plants. Mycologia 105, 1135–1152. 10.3852/13-046 [DOI] [PubMed] [Google Scholar]
- Takamatsu S., Shin H.-D., Paksiri U., Limkaisang S., Taguchi Y., Binh N. T., et al. (2002). Two Erysiphe species associated with recent outbreak of soybean powdery mildew: results of molecular phylogenetic analysis based on nuclear rDNA sequences. Mycoscience 43, 333–341. 10.1007/s102670200049 [DOI] [Google Scholar]
- Telle S., Thines M. (2008). Amplification of cox2 (~620 bp) from 2 mg of up to 129 years old herbarium specimens, comparing 19 extraction methods and 15 polymerases. PLoS ONE 3:e3584. 10.1371/journal.pone.0003584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines M. (2019). An evolutionary framework for host shifts – jumping ships for survival. New Phytol. 224, 605–617. 10.1111/nph.16092 [DOI] [PubMed] [Google Scholar]
- To-anun C., Kom-Un S., Sunawan A., Fangfuk W., Sato Y., Takamatsu S. (2005). A new subgenus, Microidium, of Oidium (Erysiphaceae) on Phyllanthus spp. Mycoscience 46, 1–8. 10.1007/s10267-004-0202-z [DOI] [Google Scholar]
- Tollenaere C., Laine A.-L. (2013). Investigating the production of sexual resting structures in a plant pathogen reveals unexpected self-fertility and genotype-by-environment effects. J. Evol. Biol. 26, 1716–1726. 10.1111/jeb.12169 [DOI] [PubMed] [Google Scholar]
- Uloth M. B., You M. P., Barbetti M. J. (2016). Cultivar resistance offers the first opportunity for effective management of the emerging powdery mildew (Erysiphe cruciferarum) threat to oilseed brassicas in Australia. Crop Pasture Sci. 67, 1179–1187. 10.1071/CP16182 [DOI] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40:e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vági P., Kovács G. M., Kiss L. (2007). Host range expansion in a powdery mildew fungus (Golovinomyces sp.) infecting Arabidopsis thaliana: Torenia fournieri as a new host. Eur. J. Plant Pathol. 117, 89–93. 10.1007/s10658-006-9072-x [DOI] [Google Scholar]
- Vilgalys R. (2003). Taxonomic misidentification in public DNA databases. New Phytol. 160, 4–5. 10.1046/j.1469-8137.2003.00894.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. (1983). Pacific mycogeography: deficiencies and irregularities in the distribution of plant parasitic fungi. Aust. J. Bot. Suppl. Ser. 13, 89–136. 10.1071/BT8310089 [DOI] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White (San Diego, CA: Academic Press; ), 315–322. [Google Scholar]
- Wicks T. J., Magarey P. (1985). First report of Uncinula necator cleistothecia on grapevines in Australia. Plant Dis. 69:727 10.1094/PD-69-727d [DOI] [Google Scholar]
- Zheng R., Chen G. (1981). The genus Erysiphe in China. Sydowia 34, 214–327. [Google Scholar]
- Zheng R.-Y., Yu Y.-N. (1987). Flora Fungorum Sinicorum. Vol. 1, Erysiphales. Beijing: Science Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s (Vilgalys, 2003).