Abstract
Among the biotic constraints to wheat (Triticum aestivum L.) production, fusarium head blight (FHB), caused by Fusarium graminearum, leaf rust (LR), caused by Puccinia triticina, and stripe rust (SR) caused by Puccinia striiformis are problematic fungal diseases worldwide. Each can significantly reduce grain yield while FHB causes additional food and feed safety concerns due to mycotoxin contamination of grain. Genetic resistance is the most effective and sustainable approach for managing wheat diseases. In the past 20 years, over 500 quantitative trait loci (QTLs) conferring small to moderate effects for the different FHB resistance types have been reported in wheat. Similarly, 79 Lr-genes and more than 200 QTLs and 82 Yr-genes and 140 QTLs have been reported for seedling and adult plant LR and SR resistance, respectively. Most QTLs conferring rust resistance are race-specific generally conforming to a classical gene-for-gene interaction while resistance to FHB exhibits complex polygenic inheritance with several genetic loci contributing to one resistance type. Identification and deployment of additional genes/QTLs associated with FHB and rust resistance can expedite wheat breeding through marker-assisted and/or genomic selection to combine small-effect QTL in the gene pool. LR disease has been present in the southeast United States for decades while SR and FHB have become increasingly problematic in the past 20 years, with FHB arguably due to increased corn acreage in the region. Currently, QTLs on chromosome 1B from Jamestown, 1A, 1B, 2A, 2B, 2D, 4A, 5A, and 6A from W14, Ning7840, Ernie, Bess, Massey, NC-Neuse, and Truman, and 3B (Fhb1) from Sumai 3 for FHB resistance, Lr9, Lr10, Lr18, Lr24, Lr37, LrA2K, and Lr2K38 genes for LR resistance, and Yr17 and YrR61 for SR resistance have been extensively deployed in southeast wheat breeding programs. This review aims to disclose the current status of FHB, LR, and SR diseases, summarize the genetics of resistance and breeding efforts for the deployment of FHB and rust resistance QTL on soft red winter wheat cultivars, and present breeding strategies to achieve sustainable management of these diseases in the southeast US.
Keywords: fusarium head blight, leaf rust, stripe rust, quantitative trait locus mapping, marker-assisted selection, resistance breeding, soft red winter wheat, southeast US
Introduction
Wheat (Triticum aestivum L.) is a staple food crop grown in 17% of the total world cropping area and contributes towards 18.3% of the global human calorie intake just next to rice (Peng et al., 2011; FAOSTAT, 2018). The United States (US) shares 7% of total global wheat production, and the crop ranks third among US field crops in terms of planted area, production, and gross farm income after corn and soybeans (USDA, 2020). Among the five major wheat classes in the US, soft red winter wheat (SRWW) common to the southeast US shares 15–20% of total area and 17% of total production (Vocke and Ali, 2013). However, this major cereal crop is under continuous threat due to several biotic and abiotic constraints resulting in a significant reduction in grain yield and quality (Limbalkar et al., 2018; Ghimire et al., 2020). Savary et al. (2019) reported 31 pests responsible for an estimated 21.5% economic yield loss in wheat. Among these, fusarium head blight (FHB), leaf rust (LR), and stripe rust (SR) are reported as the most problematic fungal diseases both in the southeast US and throughout the world (Chen et al., 2002; Cowger and Sutton, 2005; Milus et al., 2006; Kolmer et al., 2007; Walter et al., 2016; Kolmer et al., 2019). SRWW production in the southeast has decreased by nearly 61% over the past decade (2008 to 2019) due to several factors of which FHB, LR, and SR are the most prominent (https://usda.library.cornell.edu/concern/publications/5t34sj573). While FHB alone was ranked the second most challenging disease in the US midwest, Canada, South Brazil, Paraguay, Uruguay, and Argentina next to tan spot, both FHB and LR were ranked the topmost diseases in China and across the globe (Savary et al., 2019).
FHB, also widely known as wheat scab, head scab or scab, is a floral disease of wheat caused by Fusarium graminearum Schwabe [teleomorph Gibberella zeae (Schwein.) Petch]. FHB is of great concern to wheat producers because of its yield-reducing ability as well as food and feed safety threat associated with harmful mycotoxin contamination mostly by deoxynivalenol (DON) in the infected grain (McMullen et al., 1997; Dweba et al., 2017; ElDoliefy et al., 2020). Smith (1884) first described the disease symptoms and morphological characters of Fusisporium culmorum (present day Fusarium culmorum) causing FHB, and epidemics were also reported from the UK in 1884 (Hao et al., 2020). FHB was reported in the US states of Indiana, Delaware, and Ohio in the 1890s (Chester, 1890; Arthur, 1891). One hundred years later, a major FHB epidemic affected over 10 million acres of wheat in Minnesota, North Dakota, South Dakota, and the Canadian province of Manitoba causing over $1 billion in yield losses in 1993 alone and $7.7 billion gross loss over 1993–2001 (McMullen et al., 1997; Nganje et al., 2004).
FHB has become increasingly problematic in the past 20 years in the southeast US possibly due to corn acreage expansion. Increased corn acreage in the US (6 million hectares more in 2018) including all southeastern states since 1990 along with diminishing wheat acreage (about 12 million hectares less in 2018) also correlates with increased FHB epidemics in the region (USDA, 1990; USDA, 2018; https://www.ers.usda.gov/data-products/chart-gallery/gallery/chart-detail/?chartId=76955). Cowger and Sutton (2005) reported a total of $13.6 million yield loss from five southeastern states due to FHB in 2003. Nearly 50% disease incidence was observed in experimental plots in 2013–2014, while incidence as high as 80% was observed in some commercial wheat fields across Georgia in 2014–2015 and 2018–2019 (Lilleboe, 2014; Buck and Youmans, 2015). As a result, wheat millers have incurred higher costs for mycotoxin testing, additional cleaning and blending, as well as additional shipping costs to access better quality grain.
The ability of the FHB pathogens to produce mycotoxins poses a persistent global threat to human and livestock health. The US Food and Drug Administration (FDA) has restricted DON levels below 1 ppm for finished wheat products such as flour and bran consumed by human and 5–10 ppm for all livestock feed (FDA, 2010). Type-B trichothecenes, such as DON, are acutely phytotoxic and can act as virulence factors on a sensitive host maintaining positive relationship with FHB severity as observed across several studies (O’Donnell et al., 2000; Lemmens et al., 2005; Buerstmayr et al., 2019). Nevertheless, Cowger and Arellano (2010) observed that asymptomatic wheat field with low infected grain might also constitute higher DON due to late infection and rainfall immediately after anthesis. Recently, there have been increased concerns about higher mycotoxin levels in wheat straw for livestock feed due to reduced DON levels observed in grain during grain fill compared to non-grain tissues such as rachises and glumes (Cowger and Arellano, 2013; Bissonnette et al., 2018).
LR and SR, also known as brown rust and yellow rust, caused by Puccinia triticina Eriks. (Pt) and Puccinia striiformis Westend. (Ps) respectively, are the most common rust diseases of wheat; the other being stem rust caused by Puccinia graminis subsp. graminis Pers.:Pers (Kolmer, 2005; Bolton et al., 2008). These widespread and destructive foliar diseases have been closely scrutinized due to the continuous evolution of the novel and more virulent pathogen races which are difficult to manage (Kolmer, 2005; Kolmer et al., 2007). Reports on physiologic races of LR from the former Soviet Union and the US originated as early as 1960 with 5–100% disease severity observed in the field (Herr, 1961). Huerta-Espino et al. (2011) reported an estimated 3 million tons of wheat yield loss worth of $350 million in the US due to LR between 2000 and 2004. LR normally reduces grain yield by 5–15%; however, losses up to 40% have been reported depending on the climatic conditions, time and duration of infection, and resistance levels of wheat cultivars (Samborski, 1985; Marasas et al., 2004; Kolmer et al., 2007). Earlier reports on describing phenotypes of Pt as well as yield loss assessment in SRWW from the south Atlantic States also reflect the importance of LR in this region (Kolmer, 2002; Green et al., 2014). SR is the most frequent disease of wheat in the western US causing up to 70% of yield loss (Chen et al., 2002). Since 2000, it has been widespread in the southeast US and become destructive on SRWW with the emergence of new races (Chen et al., 2002; Milus et al., 2006). Interestingly, Khan et al. (1997) revealed that for every 1% increase in rust severity, there is a 1% reduction in the wheat grain yield. A photosynthetically active wheat flag leaf significantly impacts grain formation, therefore higher yield reduction together with decreased number of kernels per spike, lower kernel weight, plant biomass, and harvest index often occur with an increased infection on flag leaves (Chester, 1946; Bolton et al., 2008; Huerta-Espino et al., 2011; Green et al., 2014).
Reviews at the turn of the twenty-first century included epidemiology (Parry et al., 1995) and conventional breeding of FHB (Mesterhazy, 1995; Miedaner, 1997; Mesterházy et al., 1999). More recently, reviews of the literature have been published on diverse fields of FHB resistance including QTL mapping and marker-assisted selection (MAS) (Buerstmayr et al., 2009; Gupta et al., 2010; Shah et al., 2017), genomic selection (GS) (Larkin et al., 2019), and resistance breeding (Kosová et al., 2009; Buerstmayr et al., 2014; Steiner et al., 2017; Buerstmayr et al., 2019; Zhu et al., 2019). Similarly, review papers on broad aspects of breeding and genomic selection (Todorovska et al., 2009), epidemiology (Eversmeyer and Kramer, 2000; Brown and Hovmøller, 2002; Chen, 2005; Bolton et al., 2008; Jin et al., 2010; Zhao et al., 2016), and host resistance (Kolmer, 1996; Chen, 2007; Kolmer et al., 2007; Singh et al., 2011; Kolmer, 2013; Ellis et al., 2014; Aktar-Uz-Zaman et al., 2017; McIntosh et al., 2017) on LR and SR have been presented. In addition, several QTL meta-analyses on FHB (Liu et al., 2009; Löffler et al., 2009; Venske et al., 2019), LR (Soriano and Royo, 2015), and SR (Cheng et al., 2018) have added additional evidence to the breeding efforts undertaken globally. However, to our knowledge, there is no single review focusing on SRWW breeding for resistance to FHB, LR, and SR in the southeast US.
Epidemiology and Management of FHB
F. graminearum is a homothallic, facultative parasite of wheat that normally overwinters as a saprophyte in the plant debris of small grains and corn which serves as a reservoir for primary inoculum the next season (McMullen et al., 2012; Dweba et al., 2017). The pathogen has a brief biotrophic phase until it colonizes the living tissue to maintain the necrotrophic relation with its host (Goswami and Kistler, 2004). However, it is still unclear whether F. graminearum is a true hemibiotroph or not (Dweba et al., 2017). The fungus has a wide host range and can infect several hosts including wheat, barley, rice, oats and causes Gibberella stalk and ear rot disease on corn (Goswami and Kistler, 2004; McMullen et al., 2012). The FHB pathogens can also cause seedling blight, root and stem rot if infected wheat seeds are planted the following year.
A warm and moist environment with high relative humidity (above 90%) can stimulate ascospore and conidial dispersal by wind, rain or insects to healthy wheat spikes to trigger infections during anthesis (McMullen et al., 1997; Schmale III et al., 2006). Cowger et al. (2009) revealed that post-anthesis moisture is crucial for disease development and contributes to increased FHB severity and DON contamination levels. The fungus colonizes wheat anthers and eventually the middle rachis blocking the vascular cambium; thus the portion of the wheat head above the affected area stops growing and exhibits a typical premature bleaching symptom (Figures 1A, B) (Pritsch et al., 2000; Buerstmayr and Buerstmayr, 2016). The most discernable symptoms later in the season are the formation of pinkish-orange colored asexual fruiting structures (sporodochia) in the spikelet hosting both microconidia and macroconidia (Figure 1B) (Schmale III et al., 2006; McMullen et al., 2012). Infected wheat spikes often harbor shriveled tombstone grains that are discolored with low kernel weight and inferior quality due to high mycotoxin contamination (Figure 1C). It is also common to observe black-colored overwintering perithecia and sporodochia on matured wheat spikes as well as corn debris (Figure 1E) which serve as primary inoculum for next season continuing the disease cycle (McMullen et al., 2012). The pathogens can be easily isolated and cultured on the growth media from infected wheat spikes or corn debris allowing morphological and genetic analyses (Figure 1F).
Figure 1.
Fusarium head blight (FHB) symptom; FHB disease in the field used to assess FHB incidence (type I resistance) (A), infected spike with orange-colored sporodochia used to assess FHB severity (type II resistance) (B), infected seed used to assess Fusarium-damaged kernel (FDK) (type IV resistance) (C) compared to healthy seed lot (D), F. graminearum overwintering in corn stalk (E), and FHB isolate cultured in potato dextrose agar (PDA) plate showing characteristic pinkish-red colony (F). (Source: Figures 1A–F, B. Ghimire).
The filamentous, ascomycetous fungus F. graminearum was perceived as a single panmictic species responsible for FHB until the end of the 20th century (O’Donnell et al., 2000). At present, F. graminearum producing the 15ADON chemotype is considered one of the 16 phylogenetically distinct species within the F. graminearum species complex (FGSC) primarily responsible for FHB in the US including the southeast region (Sarver et al., 2011; Bec et al., 2015; Ghimire et al., 2019a; Ghimire et al., 2019b). There has been a gradual shift from 15ADON to more toxigenic 3ADON population in the US and Canada presumably due to climate change and fitness advantages towards a more aggressive chemotype with exact reason not known yet (Ward et al., 2008; Puri and Zhong, 2010; Schmale et al., 2011). Additionally, the high proportion of recently evolved NX-2 chemotype in the northern US and Canada foretells the possible threat of this novel strain in the southeast US (Kelly and Ward, 2018; Lofgren et al., 2018).
Management of FHB requires an integrated approach incorporating the use of cultural practices, fungicides, and host resistance. Since planting of wheat crop directly on corn stubble is a common practice, interrupting the corn–wheat cropping pattern and adopting tillage practices in no-till cropping systems are the best viable cultural practices (McMullen et al., 1997; McMullen et al., 2012). The adoption of proper crop rotation along with the removal of infected residue could reduce FHB infection and DON by 30% (McMullen et al., 2012). The use of demethylation inhibitor (DMI) fungicides in combination with a moderately resistant cultivar at anthesis was partially successful in managing FHB and DON level (McMullen et al., 2012; Madden et al., 2014; Martinez-Espinoza et al., 2014; Bowen et al., 2016; Paul et al., 2019); however, the effective use of fungicides within the short window of anthesis is challenging per se (Cowger et al., 2016).
The development of FHB resistant wheat cultivars is the single most cost-effective and sustainable approach to manage this disease. Although complete FHB resistance has not been obtained yet (Steiner et al., 2017), identification and validation of major and minor effect QTLs and their introgression through diverse breeding methodologies including recently implemented methods such as marker-assisted and genomic selections are of paramount important. These methods have allowed gene pyramiding for durable resistance across wheat breeding programs (Buerstmayr et al., 2009; Li et al., 2016b; Shah et al., 2017; Zhao et al., 2018; Zhu et al., 2019). Meanwhile, the catastrophic epidemics in the 1990s resulted in the initiation of the US wheat and barley scab initiative (USWBSI; https://scabusa.org/) in 1997 (McMullen et al., 2012). The initiative has used a multi-state, integrated approach to minimize FHB risk through breeding for FHB resistance sharing germplasm through uniform scab nurseries, weather-based forecasting systems (FHB Alerts and ScabSmart), and uniform fungicide trials (McMullen et al., 1997). In the southeast US, SunGrains (http://www.sungrains.lsu.edu/about.shtml) was established in 2005 with currently seven universities collaborating to develop superior wheat germplasm with insect and disease resistance by sharing and screening germplasm.
Epidemiology and Management of LR and SR
Unlike the FHB pathogen, Pt and Ps which cause LR and SR in wheat, respectively, are obligate parasites and cannot be cultured outside the host tissue (Zhao et al., 2016). These biotrophic fungal pathogens interact with host resistance genes in a gene-for-gene manner (Chen, 2007; Kolmer, 2013; Zhao et al., 2016). Pt and Ps are heteroecious and require two genetically unrelated hosts to complete their life cycle (Roelfs et al., 1992; Bolton et al., 2008; Jin et al., 2010; Zhao et al., 2013): the primary host wheat in which it completes its asexual life cycle and alternate hosts, Thalictrum speciosissimum (meadow rue) for LR and Berberis spp. for SR, in which they complete the sexual life cycle. Rust pathogens are macrocyclic and have five different spore stages i.e., teliospores, basidiospores, urediniospores, pycniospores, and aeciospore (Bolton et al., 2008; Jin et al., 2010; Kolmer, 2013). LR and SR are polycyclic diseases; urediniospores can re-infect wheat under conducive climatic conditions leading to disease epidemics (Bolton et al., 2008; Kolmer, 2013; Chen et al., 2014). At later growing stages as temperatures rise, thick-walled black telia are formed which enable the pathogens to survive non-host periods (Chen et al., 2014). With favorable environmental conditions, the haploid basidiospores formed from the telia are wind-dispersed to nearby alternate hosts to form pycnium which later forms aeciospores to infect the primary host thus completing the life cycle (Bolton et al., 2008; Kolmer, 2013; Chen et al., 2014).
The typical symptoms of LR on wheat leaves appear as round lesions with orange-brown urediniospores on the upper leaf surface (Figures 2A, B) (Roelfs et al., 1992; Zhao et al., 2016). SR symptoms appear as yellow to orange uredinial pustules arranged in stripes usually between veins on leaves, glumes, and awns (Figure 2D) (Chen et al., 2014). Usually, LR and SR appear successively in the field during warm and dry, and cool and humid periods, respectively. Pt and Ps can infect wheat leaves if exposed to 4–8 hours of dew period with a temperature of 10–25°C and 5–12°C, respectively (Roelfs et al., 1992; de Vallavieille-Pope et al., 1995). However, new races of SR adapted to high temperature became prevalent worldwide, leading to epidemics in warmer areas where the disease was not problematic (Milus et al., 2008; Mboup et al., 2009; Hovmøller et al., 2010). The occurrence of several identical multilocus genotypes has justified that wheat rust spores can undergo long-distance dispersal of thousands of miles across continents and ocean by wind and intercontinental exchanges (Brown and Hovmøller, 2002; Kolmer, 2005; Bahri et al., 2009; Hovmøller et al., 2010; Walter et al., 2016; Kolmer et al., 2019).
Figure 2.
Wheat rusts; leaf rust (LR) uredinia on wheat seedlings in greenhouse (A), and adult plants in the field (B) used to assess seedling and adult plant resistance, respectively, stripe rust (SR) disease in the field plot compared to healthy wheat (C), and SR uredinia showing the elongated stripes typical of the disease on the flag leaf of adult plants in the field (B). Field photos are taken from southwest Georgia research and education center, Plains, GA (Source: Figures 2A, B, S. Sapkota; 2C, D, A. D. Martinez-Espinoza).
The populations of rust pathogens in the US have been dynamic with the discovery of new rust resistance genes, migration of virulence phenotypes, clonal reproduction, and mutation of rust urediniospores as documented by annual surveys conducted since the 1930s for LR (Mains and Jackson, 1926; Ordoñez and Kolmer, 2009; Kolmer, 2019) and the 1960s for SR (Chen et al., 2002; Line, 2002). Since Johnston (1961) reported a total of 16 physiologic races of LR from 22 US states, new races have appeared while others have disappeared. A collection of rust isolates from Georgia, North Carolina, South Carolina, and Virginia in 1999 revealed that virulence phenotype MBRK (virulence to LR genes Lr1, Lr3, Lr3ka, Lr11, Lr30, Lr10, Lr14a, and Lr18) at 38.7% and TLGF (virulence to Lr1, Lr2a, Lr2c, Lr3, Lr9, Lr11, Lr14a, and Lr18) at 33.8% of isolates were the most common phenotypes suggesting the south Atlantic states were a single epidemiological area for LR (Kolmer, 2002). However, a recent study updated the presence of 40 multilocus genotype groups widely distributed across wheat growing regions worldwide of which six exist in North America (Kolmer et al., 2019). MBTNB and MCTNB phenotypes with virulence to Lr11 and Lr18 are typical to the southeastern states and Ohio valley region. Similarly, Wan and Chen (2014) identified 146 races of stripe rust in 2010 compared to 21 races in 2000 in the US (Chen et al., 2002). Prior to 2009, SR races with virulence against Yr1, Yr3, Yr8, Yr9, Yr10, Yr11, Yr12, Yr16, Yr17, Yr18, Yr19, and Yr20 were frequently reported in the southeast US; whereas, Yr6, Yr7, Yr8, Yr9, Yr27, Yr43, Yr44, and YrExp2 virulence combinations have been identified in the last few years (https://striperust.wsu.edu/races/data/).
Similar to the management of FHB, an integrated management approach is the most effective means of controlling LR and SR (Roelfs et al., 1992). Cultural practices such as eradication of volunteer plants, crop debris, and alternate hosts (if present) can reduce primary inoculum (Roelfs et al., 1992) but do not guarantee complete freedom from rust spores in the field since urediniospores are carried long-distance by wind. Aerial dispersal of rust spores makes quarantine methods ineffective to control this disease. Although an effective management of rust diseases is possible by the use of fungicides, the approach is not always economically feasible (Bowen et al., 1991; Roelfs et al., 1992; Buck et al., 2009; Buck et al., 2011). Genetic resistance is therefore the preferred method to manage rust diseases. To date, 79 Lr genes and more than 200 QTLs, as well as 82 Yr genes and 140 QTLs have been identified for both seedling and adult plant LR and SR resistance, respectively (Rosewarne et al., 2013; McIntosh et al., 2017; Sapkota et al., 2019b).
Genetics of FHB Resistance and Breeding Strategies
Types of FHB Resistance
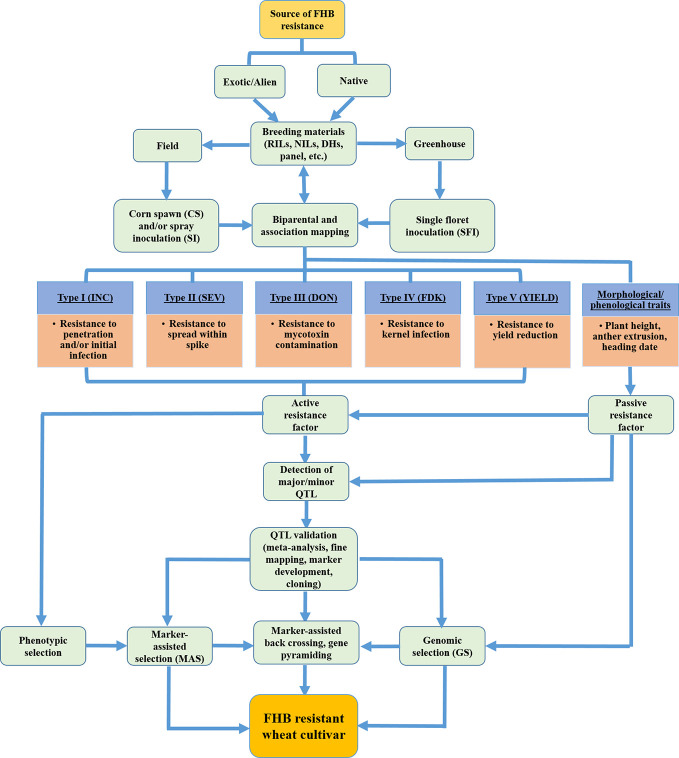
Host plant resistance to FHB is “horizontal” and neither species nor race-specific in nature (Mesterházy et al., 1999); rather resistance has low heritability, is quantitatively inherited and polygenic trait. There are no reports of genetic immunity to FHB; however multiple resistance genes can overlap to confer one or the other types of resistance (Parry et al., 1995; Ma et al., 2020). Type I and type II resistance was presented initially by Schroeder and Christensen (1963). This was later broadened to the five different types of host resistance mechanisms to FHB widely accepted to date (Mesterhazy, 1995; Mesterházy et al., 1999). Type I (FHB incidence) involves resistance to penetration and/or initial infection; type II (disease severity) is the condition when host plants resist spread of infection from one spikelet to another within an infected spike; type III reduces accumulation of mycotoxin(s) in the infected kernel; type IV provides resistance to kernel infection also known as Fusarium-damaged kernel (FDK); and type V provides resistance to yield loss (Figure 3). FHB resistant cultivars typically exhibit more than one resistance type since each QTL/gene can produce several resistance reactions (Dweba et al., 2017). Although type II resistance has been widely studied as the primary resistance type and type V is almost a neglected class, studies on the sources of type III and type IV resistance are becoming of primary interest for most of the wheat breeding programs due to increased health concerns associated with harmful vomitoxins (Venske et al., 2019; Zhu et al., 2019).
Figure 3.
Schematic flow diagram showing overall breeding effort for assessing FHB resistance types, QTL detection and validation, and development of FHB resistant wheat cultivars. (Source: Figure 3, B. Ghimire).
Sources of FHB Resistance
There are two major approaches for introducing FHB resistance in SRWW; deployment of resistant QTL from alien and exotic gene pools such as Fhb1 from Sumai 3 and exploitation of native sources of resistance from local cultivars (Brown-Guedira et al., 2008; Steiner et al., 2017). Despite the complexity of FHB resistance, the larger genetic variation from both exotic and native sources has been harnessed for deploying several resistance traits in modern wheat cultivars.
Exotic and Alien Sources of FHB Resistance
Sumai 3 is a highly resistant spring wheat cultivar developed from a cross between resistant parents Funo and Taiwan Xiaomai and was released by the Suzhou Institute of Agricultural Sciences in 1970 (Cuthbert et al., 2006; Buerstmayr et al., 2009). Sumai 3 is the most widely used donor parent to deploy the major effect QTL Fhb1 in several wheat breeding programs throughout the world (Zhu et al., 2019). While Sumai 3 has been extensively used in North America, other regions are either relying on different sources of Fhb1 such as Shinchunaga in Japan and Norin 129 and Nigmai 9 in China or even dropping Fhb1 due to preferential selection for the stem rust gene Sr2 in International Maize and Wheat Improvement Center (CIMMYT) as both of these genes are linked in repulsion (Buerstmayr et al., 2019). Besides Sumai 3, novel sources of FHB resistance from several other Asian cultivars or landraces such as Chokwang conferring type II resistance from Korea and Wangshuibai, Nyu-Bai and Nobeokabozu and their descendants CM-82036 and Ning7840 from China are widely used in US wheat breeding programs (Yang et al., 2005; Buerstmayr et al., 2009; Buerstmayr et al., 2014; Li et al., 2016b; Buerstmayr et al., 2019). Before the introduction of Sumai 3, US wheat breeding programs primarily used type I resistance sourced from Brazilian donor cultivar Frontana with stable QTL reported on chromosomes 3AL and 5A (Steiner et al., 2004; Buerstmayr et al., 2009). In addition, wheat relatives including wild emmer (T. dicoccoides) were also successfully used in hard red spring wheat (HRSW) to release several adapted cultivars including Steele-ND (Mergoum et al., 2005). Nevertheless, the occurrence of Fhb2 and Fhb5 QTLs in Sumai 3 in addition to Fhb1 has made this spring wheat cultivar a universal donor in FHB resistance breeding (Buerstmayr et al., 2019).
McVey is considered the first Sumai 3-derived US cultivar and was developed in Minnesota in 1999 (Hao et al., 2020). Since then, several top derivatives of Sumai 3 have been developed in the US by University programs: Alsen (2002–2006) (Frohberg et al., 2006), Glenn (2007–2011) (Mergoum et al., 2006), Barlow (2012–2015) (Mergoum et al., 2011), SY Soren (2016), and SY Ingmar (2017–2018) in North Dakota, Faller (2009–2012) (Mergoum et al., 2008) and Prosper (2013–2015) (Mergoum et al., 2013) in Minnesota, Prevail (2016–2017) in South Dakota, and AAC Brandon (2016–2018) in Manitoba, Canada (Zhu et al., 2019; Hao et al., 2020). Private seed companies are also actively involved in developing FHB resistant SRWW. For example, Pioneer Breeding Company developed 25R18 (1999), 25R42 (2001), 25R35 (2003), 25R54 (2003), INW0412 (2004), and 25R51 (2005) exploiting both Asian (3BS and 5AS) and/or European (1B and 3A) sources in the native background (Griffey, 2005; Zhu et al., 2019). Though more than 20 HRSW cultivars with Sumai 3 in their pedigrees are widely grown in the northern US and Canada, a negligible number of SRWW cultivars have been released for the southeast US (Steiner et al., 2017; Zhu et al., 2019).
The Fhb1 locus previously reported as Qfhs.ndsu-3BS was mapped as a single Mendelian gene in a high-resolution mapping population (Cuthbert et al., 2006). Bakhsh et al. (2013) suggested Fhb1 does not have any deleterious effects on agronomic and end-use quality traits in hard red winter wheat (HRWW) and near-isogenic lines (NILs) with Fhb1 locus reduced FHB disease severity and infected kernels by 23 and 27%, respectively (Pumphrey et al., 2007). Further, the unique ability of Fhb1 to detoxify DON to DON-3-O-glucoside can be further explored to reveal the mechanism of detoxification for wider applications (Lemmens et al., 2005).
Oliver et al. (2005) conducted an extensive evaluation of several wheat-alien species derivatives (Roegneria kamoji, R. ciliaris, Leymus racemosus, Thinopyrum ponticum, Th. elongatum, Th. junceum, Th. intermedium, Dasypyrum villosa, Secale cereal, and Avena sativa) and concluded these were an invaluable gene pool and could serve as novel sources for FHB resistance. FHB resistance is also present in more closely related species including Triticum tauschii (Oliver et al., 2005), T. macha (Steed et al., 2005), T. timopheevii (Malihipour et al., 2017), and T. spelta (Góral et al., 2008), and distantly related species Agropyron (Elymus spp.) (Ban, 1997) and Lophopyrum elongatum (Shen and Ohm, 2006). T. dicoccoides and tall wheatgrass have been widely exploited in the southeast US donating the major FHB QTL Qfhs.ndsu-3AS and Qfhs.pur-7EL, respectively (Brown-Guedira et al., 2008).
Native Sources of FHB Resistance
Native sources of resistance are more convenient to use compared to exotic sources due to higher adaptability as well as the presence of good agronomic and post-harvest quality traits. After the identification of the first native source of FHB resistance in SRWW Freedom in 1991 with a QTL mapped to chromosome 2AS, several other resistance sources were explored (Griffey, 2005). The most popular native cultivars documented with FHB resistance include: Massey (3BL) (1981), COKER 9474 (3BS), Ernie (1A/2B/3BL/4A/4BL/5A/6A) (1994), Foster (1995), Patton (Fhb2/3B/6B), Roane, Hondo, Heyne (3AS/4DL/4AL) (1998), Goldfield (2B/7B), Wesley (Fhb1) (1999), McCormick (3BS/2D/5A), Tribute (2002), NC-Neuse (1A/4A/6A), Truman (2ASc/2DS/3DS), INW0304 (2B), IL94-1653 (2B/4B) (2003), Cecil, INW0411 (Fhb1/1B/3A) (2004), Bess (1A/2B/3B), NY88046-8138, COKER 9511, WestBred X00-1079 (2005), and USG3555 (2007) (Van Sanford et al., 1997; Gilsinger et al., 2005; Griffey, 2005; Liu et al., 2007; Brown-Guedira et al., 2008; Griffey et al., 2008; Bonin and Kolb, 2009; Kang et al., 2011; Eckard et al., 2015; Islam et al., 2016; Bai et al., 2018; Murphy et al., 2019; Zhu et al., 2019). The SRWW cultivar Jamestown developed from the cross between Roane and Pioneer Brand 2691 by the Virginia Agricultural Experiment Station in 2007 is considered as a strong native source of FHB resistance (1BL) and is widely used a donor parent and FHB resistance check in the annual Southern Uniform Winter Wheat Scab Nursery (SUWWSN) (Griffey et al., 2010a; Zhu et al., 2019).
QTL Mapping for FHB Resistance and Validation
With the rapid advancements in genotyping along with reduced costs, it has become a routine approach to detect QTL associated with disease resistance mostly using conventional biparental QTL mapping (Buerstmayr et al., 2019; ElDoliefy et al., 2020; Ma et al., 2020). Since the first QTL mapping studies conducted independently by Waldron et al. (1999) and Bai et al. (1999), almost every wheat breeding program has integrated QTL mapping for FHB resistance with other important agronomic and quality traits (Figure 3). In 2003, several research groups initiated MAS (Yang et al., 2003; Zhou et al., 2003) leading to the first MAS-developed wheat cultivar Sabin from the University of Minnesota Agricultural Experimental Station in 2009 (Anderson et al., 2012). To date, more than 500 QTLs conferring resistance to different FHB traits except for grain yield (type V resistance) have been reported (Buerstmayr et al., 2019; Venske et al., 2019). However, only 20% are major effect QTLs (Buerstmayr et al., 2019).
Although hundreds of QTLs have been identified from all 42 wheat chromosomes, few have only been validated and successfully deployed in breeding programs to date (Buerstmayr et al., 2009; Steiner et al., 2017; Buerstmayr et al., 2019; Venske et al., 2019). Several QTL meta-analyses performed for FHB resistance in wheat at different time periods have also cross-validated to ensure the applicability of promising QTL (Liu et al., 2009; Löffler et al., 2009; Mao et al., 2010; Venske et al., 2019). For example, a comprehensive QTL meta-analysis conducted by Venske et al. (2019) utilized 556 QTLs from multiple projects and concluded that only two out of ten FHB-responsive genes recovered from meta-QTL 1/chr. 3B encoding for glycosiltransferase and cytochrome P450 were validated while the rest of the promising loci required further study. The analysis also revealed that the wheat B sub-genome had the largest number of QTL (238 QTLs) mapped further down on the chromosome 3B region (81 QTLs). The highest number of QTLs (41.5%) was reported for type II resistance while the fewest QTLs were for type I resistance (11.5%). The meta-analysis emphasized that future breeding activities should be directed towards fine mapping for QTL validation and the development of diagnostic markers for routine use in MAS and GS.
However, it is often questionable whether markers can be validated in a diverse background to enhance the visibility of the QTL mapping results. Nonetheless, Li et al. (2016a) observed very few markers (~3%) holding reproducible marker-trait association while testing 364 genome-wide informative SSR and sequence-tagged site (STS) markers associated with type II FHB resistance. Additionally, Petersen et al. (2017) found that only one QTL (Qfhb.nc-2B.1) and its associated marker contributed resistance to disease severity and FDK and was identified as the best candidate for use in MAS among twelve genomic loci identified from diverse wheat nurseries. The same research group previously revealed that Kompetitive Allele-Specific PCR (KASP) markers, Qfhb.nc-1A, Qfhb.nc-1B, and Qfhb.nc-6A developed from SRWW NC-Neuse are good candidates for use in MAS (Petersen et al., 2016). With the identification of the Fhb1 locus from Sumai 3 in 1999, several diagnostic markers have been developed allowing the successful introgression of this durable resistance locus in SRWW through MAS (Cuthbert et al., 2006; Bernardo et al., 2012; Steiner et al., 2017; Su et al., 2018).
FHB resistance is partially additive, so gene pyramiding is feasible using marker-assisted backcrossing or using either a single or combination of different selection strategies including phenotypic selection, MAS, and GS (Figure 3) (Shah et al., 2017). Agostinelli et al. (2012) suggested that an initial round of phenotypic selection for FHB traits at a moderate intensity will increase the homozygous resistant alleles at the major locus thus intensifying the MAS in the next round. Zhou et al. (2003) suggested MAS for major QTL during the seedling stage followed by phenotypic selection after anthesis benefited from effective FHB screening. Gupta et al. (2010) reviewed the progress and prospects of several aspects of MAS strategies such as AB-QTL, mapping-as-you-go, marker-assisted recurrent selection, and genome-wide selection. To date, Fhb1, Fhb2, and Qfhs.ifa-5A derived from Sumai 3 are reported the most used/validated and therefore, pyramided type II resistance QTL (Cuthbert et al., 2006; Cuthbert et al., 2007; Xue et al., 2011; Balut et al., 2013; Steiner et al., 2017; Su et al., 2018). Several authors have confirmed an enhanced type III resistance when Fhb1 locus was pyramided with QFhs.nau-2DL (Jiang et al., 2007; Agostinelli et al., 2012; Clark et al., 2016). Some other popular QTL widely used across wheat breeding programs include Fhb3 for type II resistance translocated from chromosome 7Lr#1 of wild grass L. racemosus (Qi et al., 2008), Fhb4 for type I resistance descended from the Chinese landrace Wangshiubai and mapped to 4BL (Xue et al., 2010), Fhb7 from Th. ponticum (Guo et al., 2015), and Fhb7AC on 7A (Jayatilake et al., 2011) most of which are pyramided to Fhb1. A recent study on the Fhb7 gene encoding a glutathione S-transferase reveals that broad resistance conferred to Fusarium species by Fhb7 is due to xenobiotic detoxification of the trichothecene compounds as was observed in Fhb1 (Wang et al., 2020). Further, the occurrence of the Fhb7 gene in the wild wheat relative Th. elongatum genome might be through a “natural” fungus-to-plant horizontal gene transfer event (Wang et al., 2020; Wulff and Jones, 2020). Wheat breeding programs will benefit from successful introgression of Fhb7 in diverse wheat backgrounds through distance hybridization conferring broad resistance to both FHB and crown rot.
Phenological and Morphological Traits Associated With FHB Resistance
Buerstmayr et al. (2019) grouped FHB resistance mechanisms into two classes: one conferred by host plant resistance gene(s) and is widely considered in breeding programs (active resistance factors with five resistance types) and the other due to developmental and morphological traits (passive resistance factors) that mainly constitute plant stature, anther extrusion, and flowering date (Figure 3). Thus, QTL mapping should consider both customary FHB resistance as well as morphological traits associated with FHB resistance to enrich the genetic diversity for durable resistance. The potential of phenological and morphological traits to enhance FHB resistance has been currently deemed as a key to successful strategic breeding (Buerstmayr et al., 2019) although the importance of these traits on resistance expression was previously reported (Mesterhazy, 1995; Miedaner, 1997; Liu et al., 2013). A summary of QTL mapping studies conducted between 2009 and 2019 revealed that nearly 40% of the QTLs for plant height, 60% for anther extrusion, and 25% for flowering date overlapped with FHB resistance QTL (Buerstmayr et al., 2019). This could be due to linkage drag (tightly linked locus) or pleiotropy between morphological and disease traits.
Several studies have claimed that the semi-dwarf plants harboring Rht-D1b and Rht-B1b alleles are more prone to FHB epidemics (Buerstmayr et al., 2009; Mao et al., 2010; Steiner et al., 2017). Mesterhazy (1995) demonstrated that awnless, taller wheat plants are usually less infected compared to shorter plants with awns. A possible explanation could be that rain hitting soil/debris only splashes 40–60 cm high and thus taller plants would be escaped from infection. Wheat awns forming larger head surface could trap and funnel airborne conidia down to open florets and enhance infection. Flower morphology also affects FHB resistance where a narrow flower opening is more prone to FHB infection (Gilsinger et al., 2005). Buerstmayr and Buerstmayr (2016) observed increased FHB severity as an effect of the high proportion of retained anthers and reduced plant height associated with Rht-D1b and Rht-B1b where the former allele had a significantly higher impact compared to the latter. These ‘green revolution’ genes introduced to enhance grain yield were also found to have a significant association with FHB infection counting up to 41% of type I susceptibility and 13–23% of reduced anther extrusion (Miedaner and Voss, 2008; Perovic et al., 2008; He et al., 2016). Interestingly, Yi et al. (2018) underlined the unique genetic relationship of FHB QTL to morphological traits where spike compactness strongly affected FHB in point inoculation while plant height and spike compactness contributed more to FHB severity compared to days to flowering with spray inoculation.
Modern Genomic Approaches for Breeding FHB Resistance
Modern breeding programs utilize novel technologies including transgenesis, gene cloning, and gene editing to complement conventional breeding for incorporation of durable resistance against FHB (Ma et al., 2020). The successful cloning of Fhb1, 2DL, and recently Fhb7 and the development of diagnostic markers have contributed towards deployment of these QTL (Kage et al., 2017; Li et al., 2019; Su et al., 2019; Wang et al., 2020). Lagudah and Krattinger (2019) compared three recent contradictory papers on Fhb1 gene cloning and suggested that both the Pore-forming toxin-like gene (Rawat et al., 2016) and Histidine-rich calcium-binding-protein gene (Li et al., 2019; Su et al., 2019) could possibly be contributing independently towards FHB resistance. The possibility of either dominant-negative model proposed by Lagudah and Krattinger (2019) or some unknown mechanism(s) of spatiotemporal gene function due to differential inoculation time (pre-anthesis, early anthesis, and anthesis) applied in all three studies is a debatable subject. Indeed, it could be a topic of beneficial scientific debate to gain further knowledge on the Fhb1 gene complex and its use in breeding for FHB resistance (Hao et al., 2020).
Genes contributing to FHB resistance have been introduced from alien species using transgenic approaches for over 20 years (Ma et al., 2020). These include the pathogenesis-related (PR) gene, ScNPR1 (Secale cereale-NPR1) from rye (Yu et al., 2017), overexpression of barley HvUGT13248 and Brachypodium distachyon UGT Bradi5gUGT03300 for improved detoxification of DON to D3G and NIV to NIV3G respectively (Li et al., 2015; Gatti et al., 2019), and introduction of barley class II chitinase gene for enhancing Type II and III resistance in wheat (Shin et al., 2008). Despite the daunting efforts, only limited success has been achieved to engineer transgenes in greenhouse environments, which also need further verification under field conditions to validate its functionality and genetic stability. Additionally, novel approaches such as expressed QTL (eQTL) combining transcriptomes data with conventional QTL mapping to identify genomic regions harboring candidate genes for FHB resistance have been initiated (Kazan and Gardiner, 2018). Similarly, host susceptibility genes could be explored to disable these deleterious genes through mutagenesis or editing through CRISPR/Cas9 (Wang et al., 2014). Host-induced gene silencing (HIGS) through the production of small interfering RNA molecules (RNAi) as demonstrated recently in wheat and model organism B. distachyon for FHB resistance could be further utilized (Cheng et al., 2015; He et al., 2019). However, regulatory hurdles have limited success for obtaining improved FHB resistant cultivars via a transgenic approach.
The success in sequencing whole genome of wheat has opened new avenues for screening multiple loci for FHB resistance at a much faster speed (IWGSC, 2014; Appels et al., 2018; Ma et al., 2020). Along with the readily available genomic data in public data repositories, there has been increased use of single nucleotide polymorphism (SNP) and microsatellite markers using modern next-generation sequencing (NGS) platforms such as genotyping-by-sequencing (GBS) and high-density Illumina 90K SNP assay in several QTL and association mapping projects (Deschamps et al., 2012; Liu et al., 2012; Islam et al., 2016; Petersen et al., 2016; Zhao et al., 2018). The sequence availability of both the F. graminearum and the wheat genome will allow greater understanding of the inherent mechanisms and genes underlying host–pathogen interactions contributing to effective management.
Molecular Mechanisms of FHB Resistance
Recent advancement in molecular/genetic analysis along with the advent of genomic tools and modern technologies contributed substantially in partially understanding the mechanism of disease resistance. However, a comprehensive understanding of the complex network of cellular and molecular events and pathway involved in FHB resistance remains to be elucidated. Pritsch et al. (2000) initiated a study on the role of PR genes (PR-1, PR-2, PR-3, PR-4, and PR-5) on the resistance mechanism in response to F. graminearum infection and disease spread in Sumai 3. The study revealed that this basal defense response triggered as early as 6–12 hours after inoculation (hai) and peaked at 36–48 hai, much faster in resistant compared to susceptible cultivars. Later, Golkari et al. (2007) demonstrated that glume and rachis were the most responsive organs with the earliest transcriptome pattern observed at 24 hai. Since then, transcriptomic data has been widely explored to portray the potential mechanisms behind QTL-mediated defense responses against Fusarium species and elucidate the system-level understanding on the biology of the pathogen (Kazan and Gardiner, 2018; Ma et al., 2020). The successful combination of transcriptomics with metabolomics and proteomics approaches identified the defense-response functionality of the FHB resistance QTL 2D, Fhb1 and Qfhs.ifa-5A in wheat NILs (Warth et al., 2015; Kage et al., 2017).
Salicylic acid (SA) inducing systemic acquired resistance (SAR) has been perceived as a key element in early signaling events during FHB infection (Ma et al., 2020). Ding et al. (2011) observed the up-regulation of SA pathways with higher hormone accumulation in the resistant Chinese cultivar Wangshuibai inducing PAL, EDS1, NPR1, and Glu2 genes within 3 hai. The early signaling pathways for SAR, mediated by coordinated and ordered expression of SA, jasmonic acid, ethylene, calcium ions, phosphatidic acid, and phenylpropanoid in addition to reactive oxygen species (ROS) production and scavenging, programmed cell death, cell wall fortification, and lignin biosynthesis have been widely discussed (Ding et al., 2011; Ma et al., 2020). The ability of the host plant to curb F. graminearum infection through the production of detoxification enzymes as observed in recent studies is another important approach to understand FHB resistance mechanisms (Lemmens et al., 2005; Gatti et al., 2019; Wang et al., 2020).
Genetic Sources of FHB Resistance in the Southeast
Although initial efforts were mostly focused on mapping FHB resistance QTL in spring wheat due to the severe epidemics in the Northern Grain Plains in the 1990s (McMullen et al., 1997), increased FHB in the southeast has shifted breeding efforts on SRWW to include this disease. Seven QTL mapping studies consisting of eight mapping populations, recombinant inbred lines (RILs), doubled-haploids (DHs), or mapping panel with 98–256 lines were considered for FHB resistance (Table 1) (Liu et al., 2012; Liu et al., 2013; Islam et al., 2016; Petersen et al., 2016; Petersen et al., 2017; Tessmann and Van Sanford, 2018; Tessmann et al., 2019). The major sources of FHB resistant alleles in those studies were: VA00W-38, Massey, Becker, Ernie, MO 94-317, NC-Neuse, and Truman. All the mapping studies considered type I–IV resistance in both greenhouse and field-based studies. Except for one study (Islam et al., 2016), all other mapping studies assessed morphological and phenological traits to explore their association with FHB traits. A total of 76 QTLs were detected across seven mapping studies of which 28 were considered as major effect QTLs (11, 10, and seven on chromosomes A, B, and D respectively). Since a single QTL was contributing to more than one FHB trait implying that the FHB QTL has a pleiotropic effect, the studies found a total of 10 major QTLs associated with type I, 10 with type II, 6 with FHB Index, 13 with type III, and 17 with type IV resistance. The identification of some major effect QTL for type III and IV resistance observed in recent years in the SRWW region encouraged the wheat breeders in the southeast US to screen wheat varieties with major available QTLs associated with low DON and FDK. Further, the higher number of type III and type IV resistance QTL reported in these studies is in contrast to the highest number of type II QTL reported in a review by Venske et al. (2019) showcasing the interest shift on FHB traits in this region. Furthermore, it was evident that the lower number of QTLs was detected on a small mapping population, while the number of QTLs increased for the large-sized population as mentioned by Buerstmayr et al. (2009). A higher number of FHB traits were associated with QTL when studies were carried out both in the greenhouse and field as compared to field studies alone. This could be possible as low heritable FHB QTLs are only fully expressed when populations are tested across multiple environments for multiple years (Mesterházy et al., 1999). Interestingly, the association of FHB QTL with plant height and heading date observed in most of the studies corroborates the necessity to consider these phenotypes in QTL mapping projects (Miedaner and Voss, 2008; He et al., 2016; Buerstmayr et al., 2019). For instance, out of the four common QTLs associated with FHB variables in both Becker/Massey and Ernie/MO 94–317 mapping populations, three of them overlapped with genes governing plant height (Rht-B1 and Rht-D1) and photoperiod sensitivity (Ppd-D1) (Table 1) (Liu et al., 2013). The pleiotropic effects of these morphological genes suggest that deploying and pyramiding FHB resistance QTL with these important morphological traits could be an effective strategy to improve FHB resistance in SRWW cultivars. Further, the low phenotypic variation (−2.1 to 26.7%) explained by QTL identified in the SRWW panels and mapping population is not unusual for polygenic nature of FHB resistance where multiple small effects QTL contribute to resistance (Petersen et al., 2017; Tessmann et al., 2019).
Table 1.
Summary of QTL mapping studies for FHB resistance in SRWW in the southeast US.
| Mapping Population/panel | Population sizea | Number of experimentsb | Inoculation techniquesc | FHB traits assessedd | Associated morphological/phenological traits assessede | Source of resistance allele | Chromosome allocation of all detected QTL | Major QTL/chromosome with associated traits | Linked markers for major QTL | R2f | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VA00W-38/Pioneer brand ‘26R46’ | 182 RILs | Field (4) | SI | INC, SEV, IND, FDK, DON | FT or HD | VA00W-38 | 1BL, 2AS, 2ASc, 2AL, 2DL, 5B, 6A, 7A | 2DL (INC), 5B (INC, DON), 6A (FDK) | Xgwm349 for 2DL; Xgwm271 for 5B | 5.7–21.3 | Liu et al., 2012 |
| Becker/Massey | 152 RILs | GH (4); Field (7) | SFI (GH); SI (Field) | INC, SEV, IND, FDK, DON | FT (photoperiod sensitivity), PHT | Massey & Becker | 1AS, 1DS, 2DS, 2BL, 3BL, 4BS, 4DS, 4DL, 6BL, 7A | 2DS (INC, IND), 4BS (INC, IND, FDK), 4DS (INC) | Xgwm261, Xgwm484 for 2DS; Xgwm513, Xgwm149 for 4BS; Xbarc334, Xgwm192 for 4DS | – | Liu et al., 2013 |
| Ernie/MO 94-317 | 231 RILs | GH (2); Field (5) | SFI (GH); SI (Field) | INC, SEV, IND, FDK, DON | FT (photoperiod sensitivity), PHT | Ernie & MO 94-317 | 2AL, 2BL, 2DS, 3BL, 4BS, 4DS, 5AL, 6AL | 2DS (SEV, IND), 4BS (INC, SEV, IND, FDK), 4DS (INC, SEV, IND, FDK, DON), 5AL (INC, SEV, IND) | Xgwm261, Xgwm484 for 2DS; Xgwm513, Xgwm149 for 4BS; Xbarc334, Xgwm192 for 4DS; Xgwm291 for 5AL | – | Liu et al., 2013 |
| NC-Neuse/AGS2000 | 170 RILs | Field (7) | CS | INC, SEV, FDK, DON | PHT, HD | NC-Neuse | 1A, 1B, 1D, 2A, 4A, 5B, 6A | 1A (INC, SEV, FDK, DON), 1B (FDK, DON), and 6A (INC, SEV) | KASP markers Qfhb.nc‐1A for 1A; Qfhb.nc‐1B for 1B; Qfhb.nc‐6A for 6A | 5.7–19.5 | Petersen et al., 2016 |
| Truman/MO 94-317 | 167 RILs | GH (2); Field (2) | SFI (GH); SI or CS (Field) | INC, SEV, IND, FDK, DON | – | Truman | 1BSc, 1BL, 1DLc, 2ASc, 2BL, 2DS, 3AL, 3BSc, 3BL, 3DS | 2ASc, 2DS, 3DS (SEV, FDK, DON for all locus) | wPt8826 for 2ASc; wPt666223 for 2DS; wPt5390 for 3DS | 6.7–25.3 | Islam et al., 2016 |
| Truman/NC-Neuse | 98 DHs | Field (7) | CS, SI | INC, SEV, FDK, DON | PHT, HD | NC-Neuse, Truman | 1A, 1B, 2A, 2B, 3B, 4A.2a, 4A.2b, 4D, 5B, 5D, 6A2, 6A.3 | 2B (SEV, FDK, DON) | KASP marker Qfhb.nc-2B.1 | 6.2–26.7 | Petersen et al., 2017 |
| Mapping panel | 236 SRWW cultivars/breeding lines | Field (2) | SI, CS | FHB rating (0-9 scale), FDK, DON | PHT, HD | – | 1A, 1B, 1D, 2A, 3B, 3D, 4A, 5A, 5B, 5D, 6A, 6B, 7A, 7B | 1A, 1B, 2A, 3B, 5A (FDK), 4A, 6A, 6B (DON) | – | 0.08–0.19 | Tessmann and Van Sanford, 2018 |
| Mapping panel | 256 SRWW cultivars/breeding lines | Field (2) | CS | FHB rating (0-9 scale), INC, SEV, IND, FDK, DON | PHT, HD | – | 3A, 4A, 5B, 6A, 6B, 7A, 7B | 4A (DON), 5B (FDK, DON), 6B (FDK) | – | −2.1–4.0 | Tessmann et al., 2019 |
aRILs, recombinant inbred lines; DHs, double-haploids; SRWW, soft red winter wheat.
bGH, greenhouse.
cSI, spray inoculation (conidial); SFI, single floret inoculation; CS, corn spawn.
dINC, disease incidence; SEV, disease severity; IND, FHB Index; FDK, Fusarium-damaged kernel; DON, deoxynivalenol.
eFT, flowering time; HD, heading date; PHT, plant height.
fThe highest amount of phenotypic variation (%) explained by the QTL.
QTL validation is another important step towards the development of resistant cultivars. Four studies on QTL validation were conducted in the southeast US using eleven mapping populations developed as back-crossed lines, NILs or RILs (Table 2) (Chen et al., 2006; Kang et al., 2011; Balut et al., 2013; Clark et al., 2016). These studies had either W14 or Sumai 3 as a donor parent and validated Fhb1 (3BS), QFhs.nau-2DL, and 5AS (Fhb5) QTLs in diverse backgrounds. Most of the lines in these studies developed by marker-assisted backcrossing and selection highlight the wider application of MAS in the southeast for screening and developing wheat cultivars. As an instance, three FHB QTL 3BS (Fhb1), 2D, and 5A from non-adapted Chinese cultivar Ning7840 was pyramided into SRWW cultivar McCormick by marker-assisted backcrossing (Kang et al., 2011). Further, the exotic and native QTLs were deployed from Ning7840 and McCormick, respectively, to develop SRWW germplasm KY06C-11-3-10 using the same approach (Clark et al., 2014). This germplasm has been further used as a donor parent for developing breeding populations. Chen et al. (2006) confirmed the presence of major FHB resistance QTLs on chromosomes 3BS and 5AS in Chinese line W14 contributed to reduced initial infection, disease spread, kernel infection, and DON accumulation. The use of double-haploid populations in this study has shortened the breeding cycle thereby expediting the validation and further QTL deployment processes. Overall, greater emphasis is ongoing to introgress FHB resistance QTL both from exotic and native sources in current SRWW cultivars through efficient breeding approaches such as genomic selection and hastily using DH method.
Table 2.
Summary of main QTL validation studies for FHB resistance in SRWW in the southeast US.
| Mapping Population | Population sizea | Number of experimentsb | Inoculation techniquesc | Targeted FHB traitsd | Associated morphological/phenological traitse | Resistant donor | Validated QTL/chromosome | Markers used for screening | References |
|---|---|---|---|---|---|---|---|---|---|
| W14/Pion2684 | 96 DHs | GH (2); Field (1) | SFI (GH); SI (Field) | INC, SEV, FDK, DON (for both QTL) | – | W14 | 3BS (Fhb1), 5AS (Fhb5) | Xbarc133, Xgwm493 for 3BS; Xbarc117, Xbarc56 for 5AS | Chen et al., 2006 |
| McCormick/Ning7840 | 8 NILs using marker-assisted backcrossing | GH (1); Field (3) | SFI (GH); CS (Field) | INC, SEV, FDK, DON (2DL and 3BS) | – | Ning7840 (Sumai 3) | 3BS (Fhb1), 2D, 5A | Xgwm533, Xcfd79, XUMN10 for Fhb1; Xgwm304, Xbarc186 for 5A; Xgwm539, Xgwm608 for 2D | Kang et al., 2011 |
| 5 different populations | 155 RILs | Field (2) | CS | INC, SEV, IND, FDK, DON | Agronomic, milling, and baking quality | W14 | Fhb1, QFhs.nau-2DL | Xgwm533 for Fhb1; Xcfd233 for QFhs.nau-2DL | Balut et al., 2013 |
| (4 populations) KY97C-0321-05-2/VA01W-476; KY97C-0519-04-05/VA01W-476; KY97C- 0540-01-03/VA01W-476; KY97C-0508-01-01A/VA01W-476 | 379 lines from all 4 crosses (BC1F1:3 and BC1F1:4) | Field (2) | CS | INC, SEV, IND, FDK, DON | PHT, HD, milling, and baking quality | W14 | Fhb1, QFhs-nau-2DL | UMN10, gwm533 for Fhb1; cfd233, gwm539 for QFhs.nau-2DL | Clark et al., 2016 |
aRILs, recombinant inbred lines; DHs, double-haploids; NILs, near isogenic lines; BC, backcrossing.
bGH, greenhouse.
cSI, spray inoculation (conidial); SFI, single floret inoculation; CS, corn spawn.
dINC, disease incidence; SEV, disease severity; IND, FHB Index; FDK, Fusarium-damaged kernel; DON, deoxynivalenol.
eHD, heading date; PHT, plant height.
In the southeast US, each participating program routinely screens for FHB resistance as an important criterion before releasing new wheat cultivar in the region. The elite lines are screened for FHB resistance in the SUWWSN (https://scabusa.org/) and Cooperative SRWW Nurseries (consisting of 6-State Advanced and Preliminary (6ST-ADV & 6ST-PRE), Gulf Atlantic Wheat (GAWN) and Sun Wheat nurseries) for multiple years. These lines are also genotyped in the USDA Eastern Regional Small Grains Genotyping Laboratory at Raleigh, NC to identify robust markers associated with FHB resistance traits. The most common resistance loci found across SRWW and breeding lines in SUWWSN recently include Fhb1, Fhb_3B_Massey, Fhb_5A_Ernie, Fhb_5A_Ning, Fhb_2DL_Wuhan1/W14, Fhb_1B_Jamestown, Fhb_1A_Neuse, Fhb_4A_Neuse, Fhb_6A_Neuse, Fhb_2B_Bess, and Fhb_3B_Bess (Murphy et al., 2019). Breeding materials are also tested in the regional Uniform Eastern & Southern SRWW Nurseries and screened for all these resistance loci except Fhb_2DL_Wuhan1/W14. A detailed list of the SSR and SNP markers used in the genotypic analysis can be found at: https://www.ars.usda.gov/southeast-area/raleigh-nc/plant-science-research/docs/small-grains-genotyping-laboratory/regional-nursery-marker-reports/. Notably, the registration of FHB resistant SRWW germplasm NC-Neuse (Murphy et al., 2004), Jamestown (Griffey et al., 2010a), VA04W-433, VA04W-474 (Chen et al., 2012), KY06C-11-3-10 (Clark et al., 2014), and AGS 3015 (Mergoum et al., 2019) is among the FHB breeding achievements obtained so far in the region (see Table 3 for representative list of SRWW cultivars).
Table 3.
List of representative major FHB and rusts resistant SRWW cultivars/germplasm released in the southeast US.
| SRWW cultivars/germplasm | Pedigree | Resistance against FHBa | Resistance against LRb | Resistance against SRc | QTL/chromosomed | Year of registration | Institution(s) involved | References | |
|---|---|---|---|---|---|---|---|---|---|
| Jaypee | ‘Doublecrop’/AR 39-3 | – | ✓ | ✓ | Lr10, Lr18 | 1995 | Arkansas Agricultural Experiment Station | Bacon et al., 1998 | |
| Roane | VA 71-54-147/’Coker 68-15’//IN65309C1-18-2-3-2 | ✓ | ✓ | – | Lr10, Lr11 | 1999 | Virginia Agricultural Experiment Station | Griffey et al., 2001 | |
| NC-Neuse | Coker 86-29//’Stella’/CHD 756-80/3/’Coker 9907’ | ✓ | ✓ | – | Lr9, Lr10, Lr11 | 2003 | North Carolina Agricultural Research Service | Murphy et al., 2004 | |
| McCormick | VA92-51-39/AL870365 | ✓ | ✓ | ✓ | Lr24 | 2002 | Virginia Agricultural Experiment Station | Griffey et al., 2005a | |
| Tribute | VA92-51-39/AL870365 | ✓ | ✓ | ✓ | Lr9, Lr24 | 2002 | Virginia Agricultural Experiment Station | Griffey et al., 2005b | |
| Allegiance (KY90C-054-6) | Pioneer Brand ‘2548’/’SS 555’ | ✓ | ✓ | ✓ | Lr10 | 2002 | Kentucky Agricultural Experiment Station | Van Sanford et al., 2006 | |
| Chesapeake | VA91-54-222 (‘Roane’”S”)/’FFR555W’//VA93-52-55 | – | ✓ | – | Lr26 | 2005 | Maryland Agricultural Experiment Station and Virginia Agricultural Experiment Station | Costa et al., 2007 | |
| 5205 (VA01W-205) | Pioneer Brand ‘2684’/VA93-54-185//’Pocahontas’ | ✓ | ✓ | ✓ | - | 2008 | Virginia Agricultural Experiment Station | Griffey et al., 2009a | |
| USG 3555 (VA02W-555) | VA94-52-60/Pioneer Brand ‘2643’//’USG 3209’ | ✓ | ✓ | ✓ | Lr11, Lr26 | 2007 | Virginia Agricultural Experiment Station | Griffey et al., 2009b | |
| Jamestown | ‘Roane’/Pioneer Brand ‘2691’ | ✓ | ✓ | ✓ | 1B; Lr10, Lr18 | 2007 | Virginia Agricultural Experiment Station | Griffey et al., 2010a | |
| Shirley (VA03W-409) | VA94-52-25/’Coker 9835’ (PI 548846 PVPO)//VA96–54-234 | ✓ | ✓ | – | - | 2008 | Virginia AgriculturalExperiment Station | Griffey et al., 2010b | |
| SW049029104 | ‘38158’ (PI 619052)/Pioneer brand 2552 (PI 566924)//’Roane’ (PI 612958) | ✓ | ✓ | – | Lr11 | 2009 | Virginia Agricultural Experiment Station | Griffey et al., 2011 | |
| VA04W-433 | Ning7840 (PI 531188)/Pioneer brand ‘2684’ (PI566923)//VA96-54-244 | ✓ | ✓ | – | 3BS | 2009 | Virginia Agricultural Experiment Station | Chen et al., 2012 | |
| VA04W-474 | ‘Roane’ (PI 612958)//W14(PI 641164)/’Coker 9134’ (PI 573034PVPO) | ✓ | ✓ | – | 5AS | 2009 | Virginia Agricultural Experiment Station | Chen et al., 2012 | |
| KY06C-11-3-10 | Ning7840’/’McCormick | ✓ | ✓ | ✓ | Fhb1, 5A, 2DL; Lr34, Lr24; Yr18 | 2013 | Kentucky Agricultural Experiment Station, Maryland Agricultural Experiment Station, Virginia Polytechnic Institute and State University, North Carolina State University, and the USDA–ARS | Clark et al., 2014 | |
| Pembroke 2014 | ‘25R18’/KY92C-0010-17//KY96C-0767-1 | ✓ | – | ✓ | Fhb1 | 2014 | Kentucky Agricultural Experiment Station | Van Sanford et al., 2016 | |
| Hilliard | ‘25R47’/’Jamestown | ✓ | ✓ | ✓ | - | 2015 | Virginia Agricultural Experiment Station | Baik et al., 2017 | |
| GA 03564-12E6 | SS 8641/4/’AGS2000’*3/931433//’2684’/3*AGS 2000 | – | ✓ | ✓ | Lr37; Yr17 | 2015 | UGA small grain breeding program and SUNGRAINS | Johnson et al., 2017 | |
| Pembroke 2016 | ‘Pioneer 25W33’/’Pioneer 25W60’//Pioneer 25W33/KY90C-042-37-1 | ✓ | ✓ | ✓ | - | 2016 | Kentucky Agricultural Experiment Station | Van Sanford et al., 2018 | |
| Savoy | GA 931233/’USG3592’//GA 941208-2E35 | – | ✓ | ✓ | - | 2015 | UGA small grain breeding program and SUNGRAINS | Johnson et al., 2018 | |
| AR11LE24 (AGS 2055) | GA961591-3E42/GA96229-3A41 | – | ✓ | ✓ | - | 2015 | University of Arkansas’s Division of Agriculture, UGA, and SunGrains | Mason et al., 2018 | |
| GA09129-16E55 (AGS 3015) | 991109-6E8/IL00-8530//991109-6E8 | ✓ | ✓ | ✓ | 1A, 1B, 4A, 6A; Lr37; Yr17 | 2019 | University of Georgia small grain breeding program | Mergoum et al., 2019 |
a,b,c(✓) indicates SRWW resistance to FHB, LR, and SR, respectively.
dQTL resistance for FHB, LR, and SR is listed in order and each category is separated by a semi-colon (;).
Genetics of LR and SR Resistance and Breeding Strategies
Types of Resistance
Resistance to rust diseases in wheat is broadly classified into two categories; seedling and adult plant resistance (APR) (Kolmer, 1996; Chen, 2007; Kolmer et al., 2007). Seedling resistance, also known as all-stage resistance, is detected in the seedling stage and remains effective at all stages of plant growth. Seedling resistance is race-specific, controlled by a single gene and mostly effective for a short duration due to the continuous evolution of new races. In contrast, APR is detected at the adult stage, controlled by multiple genes with a small effect, and is typically more durable (Line and Chen, 1995). APR genes are further classified into race-specific and race non-specific resistance genes (Ellis et al., 2014). The majority of the winter wheat cultivars possess high-temperature adult plant (HTAP) resistance typical to SR which is expressed only at the high post-inoculation temperatures (Qayoum and Line, 1985). Wheat cultivars that possess only HTAP resistance are susceptible at the seedling stage when the temperature is low, but as the temperature increases, the level of their resistance also increases (Line and Chen, 1995; Chen, 2007; Ellis et al., 2014). Research evidence proved that race non-specific HTAP resistance mostly triggers at the jointing and later growth stages with flag leaves exhibiting more resistance than the lower leaves within the same plant (Qayoum and Line, 1985). Moreover, SR development was much slower on highly resistant cultivars establishing direct correlation of rust development with HTAP resistance. Apart from seedling and APR, wheat cultivars were also found to have slow rusting or partial resistance to LR and SR (Line and Chen, 1995; Singh et al., 2005; Figlan et al., 2020). Similar to APR, slow rusting resistance is controlled by multiple genes with small effects that are race non-specific and more durable (Kolmer, 1996).
Sources of Resistance
A wild relative of wheat, Sharon goatgrass (Aegilops sharonensis), native to the fertile crescent region, has long been considered a possible source of unique resistance genes to several wheat diseases with the highest resistance frequency (60–77%) observed for LR and intermediate resistance (45%) for SR (Olivera et al., 2007; Olivera et al., 2008; Millet et al., 2014). Today, several other wild relatives of wheat contributing a diverse genetic pool against rust resistance are widely exploited. Some of the successful transformation from donor relatives documented include major resistance genes Lr21, Lr22a, and Lr39 from Tausch’s goatgrass (Ae. tauschii) (Raupp et al., 2001; Kolmer, 2013), Lr24 from wheatgrass (Thinopyrum ponticum) (Li et al., 2003), Lr57 from Ae. geniculate (Kuraparthy et al., 2007), Lr37/Yr17 from Ae. ventricosa (Kolmer, 2013), Lr9 and Lr26/Yr9 from Ae. umbellulata and common rye (Secale cereal) respectively (Browder, 1980), Yr15 from T. dicoccoides (Gerechter-Amitai et al., 1989), and Yr8 from Ae. comosa (Friebe et al., 1996). Cryptic alien introgressions may provide a unique source of resistance but are also likely to bring additional undesirable or even deleterious genes that reduce cultivar fitness, quality, and even grain yield due to linkage drag (Kuraparthy et al., 2007).
QTL Mapping for LR and SR Resistance and Validation
Similar to FHB, biparental QTL mapping using two divergent cultivars was instrumental in identifying rust resistance loci. Currently, 79 Lr-genes (Lr1-Lr79) and 82 Yr-genes have been formally cataloged from common wheat, durum wheat, and other related species (McIntosh et al., 2017; Qureshi et al., 2018; USDA Cereal disease lab, https://www.ars.usda.gov/midwest-area/stpaul/cereal-disease-lab/docs/resistance-genes/resistance-genes/). Of the 79 Lr-genes and 82 Yr-genes, 64 and 15 Lr-genes, and 59 and 19 Yr-genes confer seedling and APR, respectively (McIntosh et al., 2017; Da Silva et al., 2018; Sapkota et al., 2019a). Among the 15 Lr-genes and 19 Yr-genes for APR, seven (Lr12, Lr13, Lr22a/b, Lr35, Lr37, Lr48, and Lr49) and eight (Lr34, Lr46, Lr67, Lr68, Lr74, Lr75, Lr77, and Lr78) Lr-genes and five (Yr11, Yr12, Yr13, Yr14, and Yr34) and fourteen (Yr16, Yr18, Yr29, Yr30, Yr36, Yrns-B, YrA1, YrA2, YrA3, YrA4, YrA5, YrA6, YrA7, and YrA8) Yr-genes are reported to confer race-specific and race non-specific resistance, respectively (McIntosh et al., 2017; Da Silva et al., 2018; Waqar et al., 2018).
QTL mapping of slow rusting genes expressing a prolonged latent period has gained scientific interest to assure durable resistance to wheat LR. For example, the winter wheat line CI13227, well known for slow rusting resistance, has been widely used in mapping studies. Several research groups have demonstrated that CI13227 has both seedling resistance gene Lr3ka as well as APR genes on 1BL likely to be Lr46 along with 2B, 2DS, 7AL, and 7BL (Xu et al., 2005a; Xu et al., 2005b; Kolmer et al., 2012; Kolmer et al., 2015). The seedling resistance genes found in CI13227 might have contributed to the development of small uredinia, lower field infection, and a longer latent period possibly attributing additive effects with the APR genes (Kolmer et al., 2012). Later, Kolmer et al. (2015) found that Lr46 gene additionally confers APR to SR, stem rust, and powdery mildew. Another major effect QTL Lr74 mapped to 3BS region was detected in two SRWW cultivars Clark and Caldwell and is considered an important genomic region that contains stem rust resistance gene Sr2 and FHB QTL Fhb1 (Li et al., 2017; Kolmer et al., 2018). Further studies to determine if Lr74 is linked in coupling phase with Sr2 as in the case for Sr2 and Fhb1 would assist in combining these genes in breeding programs (Li et al., 2017). In addition, the presence of seedling resistance genes Lr9, Lr10, and Lr14a in Coker 9663 and Lr14b and Lr26 in Pioneer 26R61 together with APR gene Lr13 in the latter demonstrates the availability of copious genetic resources across SRWW cultivars (Kolmer, 2010).
With increasing numbers of QTL mapping studies, the identification of closely linked markers and their validation for MAS is indispensable. Validation of molecular markers for some of the LR resistance genes such as Rph5 and Rph7 in barley is noteworthy in terms of its applicability in gene postulation, MAS, and eventual gene pyramiding with other resistance genes (Mammadov et al., 2007). Similarly, several breeding approaches have been carried out to develop diagnostic markers that are tightly linked to LR resistance locus in wheat. Dedryver et al. (1996) developed a sequence characterized amplified region (SCAR) marker converted from a random amplified polymorphic DNA marker, OP-H5, linked to Lr24 locus indicating the possibility of molecular screening of LR resistance through MAS. Gene pyramiding for gaining durable LR resistance is usually effective through MAS once the diagnostic markers are developed and validated in diverse backgrounds. Kloppers and Pretorius (1997) revealed that wheat lines with combinations of Lr34 (race non-specific) and Lr13 (race-specific) as well as Lr34 and Lr37 genes showed improved resistance exhibiting active complementary effect even in the presence of LR races possessing virulence for Lr13 gene. CIMMYT has been working for an extended period of time to pyramid additive slow rusting LR and SR genes (usually four or five) having small to intermediate effects in locally adapted wheat cultivars through backcrossing breeding approach (Singh et al., 2005). In this light, several kinds of PCR-based molecular markers, including SCAR, AFLP, and cleaved amplified polymorphic sequences (CAPS), are available to screen for LR resistance genes Lr1, Lr9, Lr10, Lr19, Lr21, Lr24, Lr25, Lr28, Lr29, Lr34, Lr35, Lr37, Lr39, Lr47, and Lr51 and SR resistance genes Yr10, Yr15, Yr17, Yr18, and Yr32 further assisting in gene pyramiding (Robert et al., 1999; Spielmeyer et al., 2005; Murphy et al., 2009; Todorovska et al., 2009).
Modern Genomic Approaches for Breeding LR and SR Resistance
Limited success has been achieved by using phenotyping screening, physiologic specification of virulence races and conventional QTL mapping studies to breed wheat cultivars with durable LR and SR resistances. Similar to the approach for FHB resistance, a system-based approach understanding the host plant and their obligate parasites can help to develop resilient breeding strategies. Development of transgenic wheat and exploiting HIGS still need to be largely explored (Figlan et al., 2020). Yin et al. (2011) summarized the ability of RNAi technology to suppress the expression pattern of SR fungus gene through the use of barley stripe mosaic virus system. The presence of RNAi triggering the downregulation of protein kinase A gene PsCPK1, an important pathogenicity factor, was found to enhance resistance to wheat SR (Qi et al., 2018). The expression of hairpin RNAi constructs homologous to Pt MAP-kinase (PtMAPK1) encoding genes resulted in the silencing of corresponding genes in LR fungi opening an alternative avenue for developing rust-resistant wheat cultivars (Panwar et al., 2018). Further, gene-deployment strategies to gain durable rust resistance require better understanding of potential pathogen variability. The emergence of new rust races can be constantly scouted by using sophisticated surveillance technology called “field pathogenomics” which uses RNAseq or genomic DNA-based approaches to generate high-resolution NGS data for describing pathogen dynamics (Derevnina and Michelmore, 2015; Figlan et al., 2020). These modern tools show considerable promise to provide insights into the biology, population structure, and pathogenesis of wheat SR and could help formulate breeding decisions while deploying rust resistance genes in SRWW cultivars (Hubbard et al., 2015).
Map-based cloning of rust resistance genes is still an arduous endeavor due to the polyploidy complex, large wheat genome size, and high content of repetitive DNA (Cloutier et al., 2007). Nonetheless, some LR resistance genes have been successfully cloned such as Lr1 (Cloutier et al., 2007), Lr10 (Feuillet et al., 2003), Lr21 (Huang et al., 2003), Lr22a (Thind et al., 2017), Lr34 (Krattinger et al., 2009), and Lr67 (Moore et al., 2015). Similarly, SR resistance genes that have been cloned include Yr5/YrSP and Yr7 (Marchal et al., 2018), Yr10 (Liu et al., 2014), Yr15 (Klymiuk et al., 2018), Yr18 (Krattinger et al., 2009), Yr36 (Fu et al., 2009), and Yr46 (Moore et al., 2015). Yr10 is considered the first seedling resistance gene cloned while other genes such as Yr18/Lr34/Sr57/Pm38, and Yr46/Lr67/Sr55/Pm39 possess multi-pathogen (partial) slow rusting APR functionality against the three wheat rusts and powdery mildew (Spielmeyer et al., 2005; Krattinger et al., 2009; Moore et al., 2015; Klymiuk et al., 2018; Figlan et al., 2020). Despite the paucity of success in gene cloning, pathways to engineer genes of interest via high throughput modern genome editing technology have already been unlocked (Figlan et al., 2020) tracking success in powdery mildew resistance (Wang et al., 2014). Other modern genomic approaches to rust resistance gene isolation such as mutant chromosome sequencing (MutChromSeq), resistance gene enrichment sequencing (RenSeq), and platform combining association genetics (GWAS) with RenSeq (AgRenSeq) have been discussed recently (Dinh et al., 2020). Above all, the molecular and genomic basis of resistance breeding relies on our understanding of host resistance as well as pathogen virulence genes and their interactive mechanisms involved in rust resistance.
Molecular Mechanisms of LR and SR Resistance
Plants defense mechanisms against pathogens are broadly classified into two categories i.e., basal defense and R-gene mediated defense mechanism (Dinh et al., 2020). Basel resistance is effective against a wider range of plant pathogens and includes both host and non-host resistance; whereas, R-gene mediated resistance is effective against specific pathogens (Gururani et al., 2012; Dinh et al., 2020). Overall, rust disease resistance mechanisms in wheat take place in three main steps: recognition of the pathogens, signal transduction, and defense responses (Dinh et al., 2020). When the rust pathogens land on the wheat plant, pattern recognition receptors (PRRs) in the host plant detect the pathogens which is followed by the transduction of signals to the immune system of the plants. Once the pathogen is recognized and the signal is received, different mechanisms are induced in plant cells including programmed cell death (PCD) or sacrificing the infected cells and blocking the nutrient sources to the pathogens, as a disease resistance mechanism (Dinh et al., 2020).
Cereal hosts restrict the invasion and growth of biotrophic rust pathogens using two broad strategies, penetration resistance and PCD-mediated resistance (Gebrie, 2016). Penetration resistance occurs when non-adapted pathogens fail to recognize the plant’s physical and chemical signals necessary for subsequent infection. PCD-mediated resistance occurs inside the penetrated epidermal cell where the plant terminates nutrient supply to the fungal pathogens restricting further development by induction of invaded program cell death. Wheat resistance responses to rust pathogens have been characterized into surface-localized pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI), intracellular effector-triggered immunity (ETI), and broad-spectrum quantitative (partial) disease resistance (Dangl et al., 2013; Gebrie, 2016; Klymiuk et al., 2018). Dinh et al. (2020) discussed the significance of PAMPs conserved across plant pathogens that are recognized by PRRs in host plants. One of the well-known PAMPs present in cereal rust fungi is chitin (Dinh et al., 2020). Recognition of PAMPs triggers multiple defense responses in the host plant including generation of ROS, biosynthesis of defense hormones and phytoalexin, and cell wall strengthening which eventually generate PTI response (Gebrie, 2016; Dinh et al., 2020). The second layer of plant defense response, ETI, uses resistance (R) proteins that are activated upon the recognition of pathogen effectors (Dangl et al., 2013). The association of ETI with a hypersensitive response leading to PCD prevents rust pathogens from acquiring nutrients from cereal hosts (Gebrie, 2016; Dinh et al., 2020). Most of the cloned LR resistance genes, such as Lr1, Lr10, Lr21, and Lr22a, encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins (Feuillet et al., 2003; Huang et al., 2003; Cloutier et al., 2007; Dinh et al., 2020). However, only a few of the SR resistance genes, such as Yr10, Yr5/YrSP, and Yr7 encode this protein (Liu et al., 2014; Marchal et al., 2018). The activation of these NBS-LRR genes normally induces PCD or produces ROS in plants (Qi and Innes, 2013; Dinh et al., 2020). Therefore, the presence of these NBS-LRR genes in wheat is considered to be important to confer resistance against LR and SR with hypersensitive reaction via PCD.
Genetic Sources of LR and SR Resistance in the Southeast
Significant progress has been made on developing SRWW cultivars with varying degrees of rust resistance (Kolmer, 2019). Almost all breeding programs that work on FHB resistance as mentioned before are actively involved in breeding LR and SR resistance SRWW cultivars in the southeast US. The UGA small grain breeding program in collaboration with the USDA-ARS Cereal Rust Laboratory registered six LR resistant SRWW germplasm in 1991 adapted to the southeast US: Ceruga-1, Ceruga-2, Ceruga-3, Ceruga-4, Ceruga-5, and Ceruga-6. This was the result of a cooperative research program initiated in 1982 (Long et al., 1992). Subsequently, most released SRWW cultivars in the southeast US have good resistance to LR and SR. At UGA, LR resistant cultivars recently released include AGS 2027, PIO 26R94, and SS 8629 (2013), AGS 2024, Savoy, and SH 0555 (2014), AGS 2033 (2015), Fury (Progeny 16-1) and AGS 2055 (2016), GW 2032, AGS 3030, and AGS 3040 (2017), USG 3640 (2018), and AGS 3015, Dyna-Gro Rutledge and Blanton (2019) most of which have Lr37 resistance gene in their background (M. Mergoum, personal communication and unpublished data). The most common LR and SR resistance genes reported in SRWW cultivars for the southeastern states include the combination of Lr1, Lr2a, Lr9, Lr10, Lr11, Lr12, Lr14a, Lr18, Lr26, and Lr37 and Yr17 and YrR6, respectively (Kolmer, 2002; Hao et al., 2011; Kolmer, 2013; Kolmer, 2019).
Notably, gene postulation studies in SRWW cultivars and breeding lines also revealed the presence of seedling resistance genes Lr1, Lr2a, Lr9, Lr10, Lr11, Lr18, and Lr26 and APR genes Lr12 and Lr34 when tested in the field of North Carolina (Kolmer, 2003). However, cultivars with Lr2a, Lr9, and Lr26 genes in combination with APR genes Lr12 and Lr34 only exhibited higher resistance to LR. Additionally, gene postulation studies on 116 contemporary SRWW suggested the presence of seedling resistance genes Lr1, Lr2a, Lr2c, Lr3, Lr3ka, Lr9, Lr10, Lr11, Lr14a, Lr18, Lr20, Lr23, Lr24, and Lr26 (Wamishe and Milus, 2004a) as well as the contribution of either Lr12, Lr13, or Lr34 for APR to LR (Wamishe and Milus, 2004b). Recent reports on a genotypic analysis of SRWW breeding lines also revealed the presence of LR and SR resistance genes Lr34/Yr18 (on 7DS), Yr17/Lr37/Sr38 (2NS:2A translocation), Lr9 (on 6BL), Lr18 (from T. timopheevii on 5BL), Sr24/Lr24 (introgressed from Agropyron elongatum on 3DS or 1RS), and Yr_4BL (2019 Uniform Eastern & Southern SRWW Nursery Marker Report). The detailed list on different diagnostic markers used in the analysis can be found at: https://www.ars.usda.gov/southeast-area/raleigh-nc/plant-science-research/docs/small-grains-genotyping-laboratory/regional-nursery-marker-reports/. Some of the most common SRWW cultivars/germplasm lines registered and/or released for the southeast with LR and SR resistance include CK9553 and AGS 2056 (Lr11), AGS 2000 and Arcadia (Lr26), Jamestown and Hilliard (Lr10, Lr18), KY06C-11-3-10 (Lr34/Lr24/Yr18), and GA 03564-12E6 and AGS 3015 (Lr37/Yr17) (Clark et al., 2014; Baik et al., 2017; Johnson et al., 2017; Kolmer, 2019; Mergoum et al., 2019) (see Table 3 for a representative list of cultivars). Despite the fact that Lr34 is present in few breeding lines and germplasm, this most renowned durable LR resistance gene is still absent in all released SRWW cultivars to date (Kolmer et al., 2018). A complete update on wheat rust across the US including available Lr and Yr genes and gene postulations on wheat cultivars can be found at the USDA-ARS Cereal Disease Laboratory (https://www.ars.usda.gov/midwest-area/stpaul/cereal-disease-lab/).
There have been few QTL mapping studies for rust resistance conducted in the southeast region. Three biparental QTL mapping studies were recently conducted for LR: one study looked at accessing seedling resistance (Sapkota et al., 2019b) and two focused on APR (Table 4) (Carpenter et al., 2017; Sapkota, 2019). The studies utilized RIL population (175–225 lines) and detected major QTL for LR resistance in 2BS (seedling), 1AL and 5B (APR) explaining 8.1–75.3% of phenotypic variations which were either validated or under validation for future use. Among those, LrA2k recently mapped on chromosome 2BS from SRWW cultivar AGS 2000 confers a high level of LR resistance at the seedling stage (Sapkota et al., 2019b). Similarly, Lr2K38 has been mapped on 1AL chromosome from SRWW cultivar AGS 2038 and confers a high level of APR to LR (Sapkota et al., 2019). Both of these genes were effective against the current prevalent Pt races MFGKG and MBTNB and are invaluable genetic resources for resistance breeding in the southeast region. In addition, a validation study was conducted in the growth chamber for seedling resistance using RILs (Table 4) (Carpenter et al., 2018). Linkage analysis validated the presence of Lr18 gene linked to SNP marker IWB41960 in Jamestown/VA10W-21 and RL6009/VA10W21 F2 populations which can be successfully deployed in breeding programs.
Table 4.
Summary of recently identified QTL for LR and SR resistances and their validation in SRWW in the southeast US.
| Mapping population | Population sizea | Number of experiments | Traits studiedb | Gene/QTL identified/validated | Linked markers | R2c | References |
|---|---|---|---|---|---|---|---|
| QTL Identification | |||||||
| Pioneer ‘25R47’/Jamestown | 186 RILs | Field (6) | IT (0-9 scale), DS |
QLr.vt-5B1 QLr.vt-5B2 |
IWB7835/IWB24418 IWB32871/IWB26068 |
22.1 8.1 |
Carpenter et al., 2017 |
| AGS 2038/UGA 111729 | 225 RILs | Field (3); greenhouse (1) | IT (0-4 scale), DS | Lr2K38 | IWB20487 | 34.4 | Sapkota, 2019 |
| Pioneer® variety 26R61/AGS 2000 | 175 RILs | Growth chamber (3) | IT (0-4 scale) | LrA2K | Xwmc770 | 75.3 | Sapkota et al., 2019b |
| Pioneer variety® 26R61/AGS 2000 | 178 RILs | Field (3) | IT (0-9 scale) | YrR61 | Xbarc124/Xgwm359 | 56.0 | Hao et al., 2011 |
| VA00W-38/Pioneer Brand 26R46 | 182 RILS | Field (4) | IT (0-9 scale), DS |
QYrva.vt-2AS QYrva.vt-4BL QYrpi.vt-6BL |
Xgwm296b Xbarc163 Xwmc756 |
58.9 19.3 11.7 |
Christopher et al., 2013 |
| VA96W-270/Coker 9835 | 146 RILs | Field (4) | DS and AUDPC |
QYr.ar-3BS QYr.ar-4BL |
IWA6471 IWA3551 |
28.3 26.3 |
Subramanian et al., 2016 |
| Pioneer ‘25R47’/Jamestown | 186 RILs | Field (6) | IT (0-9 scale), DS |
QYr.vt-3B QYr.vt-6A |
IWB60574/IWB23272 IWB5971/IWB63000 |
11.1 14.3 |
Carpenter et al., 2017 |
| QTL Validation | |||||||
| Pioneer ‘25R47’/Jamestown | 186 RILs; 1600 individuals (200 from each fight F2 population) | Growth chamber (1) | IT (0-4 scale) | Lr18 | IWB41960 | – | Carpenter et al., 2018 |
aRILs, recombinant inbred lines.
bIT, infection type; DS, disease severity; AUDPC, area under disease progress curve.
cThe highest amount of phenotypic variation (%) explained by the QTL.
Similarly, four biparental QTL mapping studies were conducted for adult plant SR resistance (Table 4) (Hao et al., 2011; Christopher et al., 2013; Subramanian et al., 2016; Carpenter et al., 2017). The first study utilized a RIL population (178 lines) and detected a major QTL on 2A, contributing up to 56% of phenotypic variation in the SRWW cultivars Pioneer 26R61 (Hao et al., 2011). The second study utilized 182 RILs derived from VA00W-38 (resistant) and Pioneer Brand 26R46 and mapped four QTLs, on chromosomes 2AS, 3BL, 4BL, and 6BL (Christopher et al., 2013). Of these, 2AS QTL explained up to 58.9% of phenotypic variation. The third study utilized a RIL population (146 RILs) and mapped two QTLs on chromosomes 3BS and 4BL (Subramanian et al., 2016). The fourth study utilized a RIL population (186 lines) and detected two QTLs for SR resistance in SRWW cultivar Jamestown in chromosomes 3B and 6A, contributing up to 11.1 and 14.3% of phenotypic variation, respectively (Carpenter et al., 2017). Interestingly, the presence of LR and SR resistance QTL on 5B, 3B, and 6A together with FHB resistance QTL in 1B makes SRWW cultivar Jamestown a good donor parent in the SRWW breeding programs (Carpenter et al., 2017). In addition, donor parents Pioneer26R61 and VA00W-38 in these mapping studies acquiring FHB and LR resistance QTL besides SR resistance hold a good genetic pool for multi-trait resistance.
Challenges and Breeding Strategies for FHB, LR, and SR Resistance
Disease phenotyping in the greenhouse and in particular under field conditions is necessary to validate germplasm selections based on molecular data. However, FHB and rusts differ dramatically on the relative ease of disease phenotyping. Disease ratings for rust are fairly straightforward and consider infection type (either a 0–4 or 0–9 scale) and disease severity to assess both seedling and APR (Figure 2) (Stakman et al., 1962; Qayoum and Line, 1985; Carpenter et al., 2017). Inoculum can be either urediniospore suspensions (greenhouse) or infected spreader plants (field) to initiate localized epidemics. The presence of five resistance types and the need for floral infection for FHB assessments increase the difficulty of phenotyping for this disease (Figures 1 and 3) (Mesterhazy, 1995). Field and greenhouse assessments typically require an exogenous water source (e.g. mist system) that can be programmed to provide moisture throughout the day and night during anthesis. This can often be over several week periods due to differing anthesis across heterogeneous germplasm. Inoculum can be applied as corn spawn and conidial spray inoculation or naturally infected corn debris in the field to assess all resistance types while single floret inoculations of a conidial suspension are typically used in the greenhouse particularly to assess type II, III, and IV resistance. Differences in plant height and flowering date of wheat germplasm may require multiple inoculations creating uneven disease pressure in the field with the likelihood of disease escape on early and later flowering germplasm. High disease incidence resulting in a majority of infected spikelets can also make type II screening challenging (M. Mergoum, personal communication). Additionally, Stagonospora nodorum infection on the wheat spike may add confusion to correctly rate FHB in the field (Brown-Guedira et al., 2008).
The selection of wheat cultivars with reduced FDK and DON levels is becoming a major criterion in the majority of the southeast wheat breeding programs. However, collecting these data requires intensive labor and resources for large and early generation germplasm selections. Several research groups are developing high throughput phenotyping such as detached leaf assays (Browne et al., 2005), air separation and digital photo analysis (Maloney et al., 2014), near-infrared reflectance (Dvorjak, 2014), optical sorter (Carmack et al., 2019) or grain imaging platform (Ward et al., 2019a) and have observed a high level of correlation for FDK and DON level between actual and predicted values. Despite these achievements, there are still some other impediments to successful field phenotyping for both FHB and rust diseases.
Low heritability and high genotype by environment interaction (G×E) are the major bottlenecks to the complexity of FHB resistance breeding as it is usually challenging to detect minor effect QTL both from adapted and exotic sources (Verges et al., 2006). Plant breeders can have hundreds of thousands of breeding lines per season and are limited in the greenhouse for FHB and rust screening due to time and space constraints. High G×E can result in a poor correlation between greenhouse and field screening for quantitative traits such as FHB severity, DON levels, and FDK which suggests that greenhouse test alone would not be sufficient to explore resistance sources which might actually be expressed only in the field (Hall and Van Sanford, 2003). Regardless of the time and effort, phenotypic screening in multiple environments is still indispensable to lay the foundation for successful MAS and GS. A recent study showed that the incorporation of the G×E effect and multiple traits in the GS model increased the prediction accuracy by 9.6% for low-heritability traits across environments (Ward et al., 2019b). Notably, the higher prediction ability of the phenotypic selection over GS with higher selection response vice versa documented by Steiner et al. (2019) underscores that genomics and phenomics should be complementary to further cultivar improvements. Similarly, the higher accuracy of genome-wide maker-based models tested in recent studies to detect even minor effect QTL unchecked by MAS ensures the success of GS for future rusts resistance in SRWW (Todorovska et al., 2009; Bulli et al., 2016; Steiner et al., 2017).
Though significant strides have been made in FHB resistance using universal donor Sumai 3, inadvertent linkage drag associated with Fhb1 has been a problem for its successful deployment in breeding lines (Schweiger et al., 2016). It is often challenging to gain acceptable agronomic and quality traits while introgressing major effect FHB resistance QTL from exotic and alien sources. Benson et al. (2012) studied the genetic diversity and population structure of the eastern winter wheat population and concluded that the short intense breeding history against FHB resistance might have localized effects on linkage drag. However, appropriate breeding methodologies such as repeated backcrossing with the recurrent parent and large population size can help to identify the few recombinants with minimum negative effects (“bad linkages”). More importantly, breeding winter wheat for both FHB and rusts resistance is time consuming (requires vernalization) compared to spring wheat. In addition, the lack of QTL validation in diverse backgrounds and the paucity of diagnostic markers have disadvantaged FHB and rusts breeding programs in the southeast US (Brown-Guedira et al., 2008; Waqar et al., 2018). Low accuracy in QTL size and location along with a low representation of genomic allele distribution often warrants the need for validating QTL and associated markers in a diverse background to ensure wider applicability (Buerstmayr et al., 2019).
Unlike FHB, the emergence of new virulence phenotype groups and races of the LR and SR pathogens is the single most recurring problem for durable rust resistance breeding in the southeast US and elsewhere (Kolmer et al., 2019). The replacement of LR virulence phenotype MBRK and TLGF dominant during the early 2000s with MBTNB and MCTNB phenotypes at present highlights the dynamic nature of LR isolates (Kolmer, 2002; Kolmer et al., 2019). Similarly, new races of SR have been continuously appearing since races virulent to Yr8 and Yr9 started threatening SRWW in the southeast US in 2000 (Hao et al., 2011). Wheat breeding programs in the southeast US should, therefore, continue the rigorous search for novel QTL and slow rusting genes for LR and SR resistance to overcome new races and gain long term genetic resistance in the field. In addition, stewardship and deployment of the few resistance genes for LR and SR available to breeding programs in the southeast are necessary to preserve and prolong the efficacy of these genes.
With the recent achievements on whole-genome sequencing (IWGSC, 2014) and the genome annotation (Appels et al., 2018), the future breeding endeavor of SRWW should orient towards the exploration of high-throughput genomic tools such as field pathogenomics, transgenes, gene cloning, genome editing, NGS, and GBS to gain deeper insights into host and underlying pathogen to achieve durable FHB and rust resistance (Bulli et al., 2016; Dinh et al., 2020; Figlan et al., 2020). Above all, a continued collaboration among wheat breeders for sharing and multi-location screening of the promising germplasm against FHB, LR, and SR resistance will be a key to successful resistance breeding in the southeast US.
Author Contributions
BG, SS, and BB respectively wrote the major sections on FHB, LR, and SR of the manuscript. BG designed the overall outline and prepared the first draft of the manuscript. JB, MM, and AM-E provided guidance, critical suggestions, and feedback on the overall content throughout the manuscript preparation. MM initiated the concept of this chapter and led the group in accomplishing it efficiently and timely. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Benjamin Lopez, Steve Sutton, and John Youmans for providing valuable information on FHB and LR resistance efforts ongoing at the UGA Small Grain Breeding Program. The authors are also thankful for valuable information obtained from the SunGrains, USWBSI, and USDA-ARS websites to prepare this manuscript.
References
- Agostinelli A. M., Clark A. J., Brown-Guedira G., Van Sanford D. A. (2012). Optimizing phenotypic and genotypic selection for Fusarium head blight resistance in wheat. Euphytica 186, 115–126. 10.1007/s10681-011-0499-6 [DOI] [Google Scholar]
- Aktar-Uz-Zaman M., Tuhina-Khatun M., Hanafi M. M., Sahebi M. (2017). Genetic analysis of rust resistance genes in global wheat cultivars: an overview. Biotechnol. Biotechnol. Equip. 31, 431–445. 10.1080/13102818.2017.1304180 [DOI] [Google Scholar]
- Anderson J. A., Wiersma J. J., Linkert G. L., Kolmer J. A., Jin Y., Dill-Macky R., et al. (2012). Registration of ‘Sabin’ Wheat. J. Plant Regist. 6, 174–179. 10.3198/jpr2011.06.0344crc [DOI] [Google Scholar]
- Appels R., Eversole K., Feuillet C., Keller B., Rogers J., et al. (2017). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, Eaar7191. 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Arthur J. C. (1891). Indiana Agricultural Experiment Station Bulletin. 36, 129–132. [Google Scholar]
- Bacon R. K., Kelly J. T., Milus G. A. (1998). Registration of ‘Jaypee’ Wheat. Crop Sci. 38, 1723–1723. 10.2135/cropsci1998.0011183x003800060067x [DOI] [Google Scholar]
- Bahri B., Leconte M., Ouffroukh A., De Vallavieille-Pope C., Enjalbert J. (2009). Geographic limits of a clonal population of wheat yellow rust in the Mediterranean region. Mol. Ecol. 18, 4165–4179. 10.1111/j.1365-294X.2009.04267.x [DOI] [PubMed] [Google Scholar]
- Bai G., Kolb F. L., Shaner G., Domier L. L. (1999). Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathology 89, 343–348. 10.1094/PHYTO.1999.89.4.343 [DOI] [PubMed] [Google Scholar]
- Bai G., Su Z., Cai J. (2018). Wheat resistance to Fusarium head blight. Can. J. Plant Pathol. 40, 336–346. 10.1080/07060661.2018.1476411 [DOI] [Google Scholar]
- Baik B.-K., Penning B., Sturbaum A. (2017). Soft Wheat Quality Laboratory, 63rd Annual Research Review 2017 Annual Report. Wooster, OH: Corn, Soybean and Wheat Quality Research Unit, USDA-ARS; Available at: http://ars.usda.gov/Main/Docs.htm?docid=3032 [Accessed March 4, 2020]. [Google Scholar]
- Bakhsh A., Mengistu N., Baenziger P. S., Dweikat I., Wegulo S. N., Rose D. J., et al. (2013). Effect of fusarium head blight resistance gene Fhb1 on agronomic and end-use quality traits of hard red winter wheat. Crop Sci. 53, 793–801. 10.2135/cropsci2012.06.0364 [DOI] [Google Scholar]
- Balut A. L., Clark A. J., Brown-Guedira G., Souza E., Van Sanford D. A. (2013). Validation of Fhb1 and QFhs.nau-2DL in several soft red winter wheat populations. Crop Sci. 53, 934–945. 10.2135/cropsci2012.09.0550 [DOI] [Google Scholar]
- Ban T. (1997). Evaluation of resistance to fusarium head blight in indigenous Japanese species of Agropyron (Elymus). Euphytica 97, 39–44. 10.1023/A:1003078109694 [DOI] [Google Scholar]
- Bec S., Ward T. J., Farman M., O’Donnell K., Hershman D. E., Van Sanford D. A., et al. (2015). Characterization of Fusarium strains recovered from wheat with symptoms of head blight in Kentucky. Plant Dis. 99, 141105070717008. 10.1094/PDIS-06-14-0610-RE [DOI] [PubMed] [Google Scholar]
- Benson J., Brown-Guedira G., Murphy J. P., Sneller C. (2012). Population structure, linkage disequilibrium, and genetic diversity in soft winter wheat enriched for fusarium head blight resistance. Plant Genome 5, 71–80. 10.3835/plantgenome2011.11.0027 [DOI] [Google Scholar]
- Bernardo A. N., Ma H., Zhang D., Bai G. (2012). Single nucleotide polymorphism in wheat chromosome region harboring Fhb1 for Fusarium head blight resistance. Mol. Breed. 29, 477–488. 10.1007/s11032-011-9565-y [DOI] [Google Scholar]
- Bissonnette K. M., Kolb F. L., Ames K. A., Bradley C. A. (2018). Effect of Fusarium head blight management practices on mycotoxin contamination of wheat straw. Plant Dis. 102, 1141–1147. 10.1094/PDIS-09-17-1385-RE [DOI] [PubMed] [Google Scholar]
- Bolton M. D., Kolmer J. A., Garvin D. F. (2008). Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 9, 563–575. 10.1111/j.1364-3703.2008.00487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin C. M., Kolb F. L. (2009). Resistance to fusarium head blight and kernel damage in a winter wheat recombinant inbred line population. Crop Sci. 49, 1304–1312. 10.2135/cropsci2008.08.0459 [DOI] [Google Scholar]
- Bowen K. L., Everts K. L., Leath S. (1991). Reduction in yield of winter wheat in North Carolina due to powdery mildew and leaf rust. Phytopathology 81, 503. 10.1094/phyto-81-503 [DOI] [Google Scholar]
- Bowen K., Sharma N., McMaster N., Jones J., Pegues M. (2016). Management of Fusarium head blight and DON in Alabama, in: APS Annu. Meet. Available at: https://www.apsnet.org/meetings/Documents/2016_meeting_abstracts/aps2016625.htm (Accessed February 25, 2020).
- Browder L. E. (1980). A compendium of information about named genes for low reaction to Puccinia recondita in wheat. Crop Sci. 20, 775–779. 10.2135/cropsci1980.0011183x002000060024x [DOI] [Google Scholar]
- Brown J. K. M., Hovmøller M. S. (2002). Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541. 10.1126/science.1072678 [DOI] [PubMed] [Google Scholar]
- Browne R. A., Murphy J. P., Cooke B. M., Devaney D., Walsh E. J., Griffey C. A., et al. (2005). Evaluation of components of fusarium head blight resistance in soft red winter wheat germ plasm using a detached leaf assay. Plant Dis. 89, 404–411. 10.1094/PD-89-0404 [DOI] [PubMed] [Google Scholar]
- Brown-Guedira G., Griffey C., Kolb F., McKendry A., Murphy J. P., Van Sanford D. (2008). Breeding FHB-resistant soft winter wheat: Progress and prospects. Cereal Res. Commun. 36, 31–35. 10.1556/CRC.36.2008.Suppl.B.5 [DOI] [Google Scholar]
- Buck J. W., Youmans J. (2015). Georgia 2014-2015 Small Grain Performance Tests -Annual Publication 100-7. Available at: http://www.swvt.uga.edu/canola.html (Accessed February 25, 2020).
- Buck J., Martinez-Espinoza A., Youmans J. (2009). Effect of seed treatment and foliar applications of insecticide or fungicide on soft red winter wheat yield in Georgia. Can. J. Plant Pathol. 31, 478. 10.1080/07060660909507622 [DOI] [Google Scholar]
- Buck J. W., Wise K., Dong W. (2011). Effect of postsymptom application of fungicides on urediniospore production by Puccinia triticina on wheat and P. hemerocallidis on daylily. Plant Dis. 95, 325–330. 10.1094/PDIS-09-10-0646 [DOI] [PubMed] [Google Scholar]
- Buerstmayr M., Buerstmayr H. (2016). The semidwarfing alleles Rht-D1b and Rht-B1b show marked differences in their associations with anther-retention in wheat heads and with fusarium head blight susceptibility. Phytopathology 106, 1544–1552. 10.1094/PHYTO-05-16-0200-R [DOI] [PubMed] [Google Scholar]
- Buerstmayr H., Ban T., Anderson J. (2009). QTL mapping and marker assisted selection for Fusarium head blight resistance in wheat. Cereal Res. Commun. 36, 1–3. 10.1556/CRC.36.2008.Suppl.B.1 [DOI] [Google Scholar]
- Buerstmayr H., Buerstmayr M., Schweiger W., Steiner B. (2014). Breeding for resistance to head blight caused by Fusarium spp. in wheat. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 9, 1–13. 10.1079/PAVSNNR20149007 [DOI] [Google Scholar]
- Buerstmayr M., Steiner B., Buerstmayr H. (2019). Breeding for Fusarium head blight resistance in wheat—Progress and challenges. Plant Breed., 1–26. 10.1111/pbr.12797 [DOI]
- Bulli P., Zhang J., Chao S., Chen X., Pumphrey M. (2016). Genetic architecture of resistance to stripe rust in a global winter wheat germplasm collection. G3 Genes Genomes Genet. 6, 2237–2253. 10.1534/g3.116.028407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmack W. J., Clark A. J., Dong Y., Van Sanford D. A. (2019). Mass selection for reduced deoxynivalenol concentration using an optical sorter in SRW wheat. Agronomy 9, 816. 10.3390/agronomy9120816 [DOI] [Google Scholar]
- Carpenter N. R., Griffey C. A., Malla S., Barnett M., Marshall D., Fountain M. O., et al. (2017). Identification of quantitative resistance to Puccinia striiformis and Puccina triticinia in the soft red winter wheat cultivar ‘Jamestown’. Crop Sci. 57, 2991–3001. 10.2135/cropsci2017.03.0143 [DOI] [Google Scholar]
- Carpenter N. R., Griffey C. A., Rosso L., Malla S., Chao S., Guedira G. L., et al. (2018). Mapping Lr18: A leaf rust resistance gene widely deployed in soft red winter wheat. J. Plant Dis. Biomarkers 1, 4–10. [Google Scholar]
- Chen X., Moore M., Milus E. A., Long D. L., Line R. F., Marshall D., et al. (2002). Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis. 86, 39–46. 10.1094/PDIS.2002.86.1.39 [DOI] [PubMed] [Google Scholar]
- Chen J., Griffey C. A., Saghai Maroof M. A., Stromberg E. L., Biyashev R. M., Zhao W., et al. (2006). Validation of two major quantitative trait loci for fusarium head blight resistance in Chinese wheat line W14. Plant Breed. 125, 99–101. 10.1111/j.1439-0523.2006.01182.x [DOI] [Google Scholar]
- Chen J., Griffey C. A., Liu S., Maroof S. M. A., Murphy J. P., Navarro R. A., et al. (2012). Registration of fusarium head blight-resistant soft red winter wheat germplasm VA04W-433 and VA04W-474. J. Plant Regist. 6, 111–116. 10.3198/jpr2011.07.0397crg [DOI] [Google Scholar]
- Chen W., Wellings C., Chen X., Kang Z., Liu T. (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 15, 433–446. 10.1111/mpp.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. M. (2005). Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 27, 314–337. 10.1080/07060660509507230 [DOI] [Google Scholar]
- Chen X. M. (2007). Challenges and solutions for stripe rust control in the United States. Aust. J. Agric. Res. 58, 648–655. 10.1071/AR07045 [DOI] [Google Scholar]
- Cheng W., Song X. S., Li H. P., Cao L. H., Sun K., Qiu X. L., et al. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. 10.1111/pbi.12352 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Yao F., Ye X., Jiang Q., Li W., Deng M. (2018). Construction of linkage map of the meta quantitative trait loci (MQTL) on stripe rust resistance in Wheat (Triticum aestivum L.). Acta Phytopathol. Sin. 10, 1–23. 10.13926/j.cnki.apps.000292 [DOI] [Google Scholar]
- Chester F. D. (1890). Third Annual Report of the Delaware College Agricultural Experiment Station. Wilmington, DE: Mercantile Printing Company; 89–90. [Google Scholar]
- Chester K. S. (1946). The nature and prevention of the cereal rusts as exemplified in the leaf rust of wheat. Waltham, MA: : Chronica Botanica Company. [Google Scholar]
- Christopher M. D., Liu S., Hall M. D., Marshall D. S., Fountain M. O., Johnson J. W., et al. (2013). Identification and mapping of adult plant stripe rust resistance in soft red winter wheat VA00W-38. Crop Sci. 53, 871–879. 10.2135/cropsci2012.02.0086 [DOI] [Google Scholar]
- Clark A. J., Costa J. M., Griffey C. A., Brown-Guedira G. L., Dong Y., Souza E. J., et al. (2014). Registration of scab-resistant KY06C-11-3-10 soft red winter wheat germplasm. J. Plant Regist. 8, 211–216. 10.3198/jpr2013.07.0039crg [DOI] [Google Scholar]
- Clark A. J., Sarti-Dvorjak D., Brown-Guedira G., Dong Y., Baik B. K., Van Sanford D. A. (2016). Identifying rare FHB-resistant segregants in intransigent backcross and F2 winter wheat populations. Front. Microbiol. 7, 1–14. 10.3389/fmicb.2016.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S., McCallum B. D., Loutre C., Banks T. W., Wicker T., Feuillet C., et al. (2007). Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 65, 93–106. 10.1007/s11103-007-9201-8 [DOI] [PubMed] [Google Scholar]
- Costa J. M., Griffey C. A., Bockelman H. E., Cambron S. E., Cooper A., Gaines C., et al. (2007). Registration of ‘Chesapeake’ Wheat. J. Plant Regist. 1, 26. 10.3198/jpr2006.10.0630crc [DOI] [Google Scholar]
- Cowger C., Arellano C. (2013). Fusarium graminearum infection and deoxynivalenol concentrations during development of wheat spikes. Phytopathology 103, 460–471. 10.1094/PHYTO-03-12-0054-R [DOI] [PubMed] [Google Scholar]
- Cowger C., Arrellano C. (2010). Plump kernels with high deoxynivalenol linked to late Gibberella zeae infection and marginal disease conditions in winter wheat. Phytopathology 100, 719–728. 10.1094/PHYTO-100-7-0719 [DOI] [PubMed] [Google Scholar]
- Cowger C., Sutton A. L. (2005). The southeastern U . S . Fusarium head blight epidemic of 2003. Plant Heal. Prog. 6, 4. 10.1094/PHP-2005-1026-01-RS.Abstract [DOI] [Google Scholar]
- Cowger C., Patton-Ozkurt J., Brown-Guedira G., Perugini L. (2009). Post-anthesis moisture increased Fusarium head blight and deoxynivalenol levels in North Carolina winter wheat. Phytopathology 99, 320–327. 10.1094/PHYTO-99-4-0320 [DOI] [PubMed] [Google Scholar]
- Cowger C., Weisz R., Arellano C., Murphy P. (2016). Profitability of integrated management of fusarium head blight in North Carolina winter wheat. Phytopathology 106, 814–823. 10.1094/PHYTO-10-15-0263-R [DOI] [PubMed] [Google Scholar]
- Cuthbert P. A., Somers D. J., Thomas J., Cloutier S., Brulé-Babel A. (2006). Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 112, 1465–1472. 10.1007/s00122-006-0249-7 [DOI] [PubMed] [Google Scholar]
- Cuthbert P. A., Somers D. J., Brulé-Babel A. (2007). Mapping of Fhb2 on chromosome 6BS: A gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 114, 429–437. 10.1007/s00122-006-0439-3 [DOI] [PubMed] [Google Scholar]
- Da Silva G. B. P., Zanella C. M., Martinelli J. A., Chaves M. S., Hiebert C. W., McCallum B. D., et al. (2018). Quantitative trait loci conferring leaf rust resistance in hexaploid wheat. Phytopathology 108, 1344–1354. 10.1094/PHYTO-06-18-0208-RVW [DOI] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawich B. J. (2013). Pivoting the plant immune system from dissection to deployment. Sci. 80 341, 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vallavieille-Pope C., Huber L., Leconte M., Goyeau H. (1995). Comparative effects of temperature and interrupted wet periods on germination, penetration, and infection of Puccinia recondita f. sp. tritici and P. striiformis on wheat seedlings. Phytopathology 85, 409–415. 10.1094/phyto-85-409 [DOI] [Google Scholar]
- Dedryver F., Jubier M. F., Thouvenin J., Goyeau H. (1996). Molecular markers linked to the leaf rust resistance gene Lr24 in different wheat cultivars. Genome 39, 830–835. 10.1139/g96-105 [DOI] [PubMed] [Google Scholar]
- Derevnina L., Michelmore R. W. (2015). Wheat rusts never sleep but neither do sequencers: will pathogenomics transform the way plant diseases are managed? Genome Biol. 16, 44. 10.1186/s13059-015-0615-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S., Llaca V., May G. D. (2012). Genotyping-by-sequencing in plants. Biology 1, 460–483. 10.3390/biology1030460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Xu H., Yi H., Yang L., Kong Z., Zhang L., et al. (2011). Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PloS One 6, e19008. 10.1371/journal.pone.0019008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh H. X., Singh D., Periyannan S., Park R. F., Pourkheirandish M. (2020). Molecular genetics of leaf rust resistance in wheat and barley. Theor. Appl. Genet., 133, 2035–2050. 10.1007/s00122-020-03570-8 [DOI] [PubMed] [Google Scholar]
- Dvorjak D. S. (2014). Fusarium head blight resistance and agronomic performace in soft red winter wheat populations.[dissertation] (Kentucky (KY): University of Kentucky; ). Available at: https://uknowledge.uky.edu/pssetds/38 (Accessed March 3, 2020). [Google Scholar]
- Dweba C. C., Figlan S., Shimelis H. A., Motaung T. E., Sydenham S., Mwadzingeni L., et al. (2017). Fusarium head blight of wheat: Pathogenesis and control strategies. Crop Prot. 91, 114–122. 10.1016/j.cropro.2016.10.002 [DOI] [Google Scholar]
- Eckard J. T., Gonzalez-Hernandez J. L., Caffe M., Berzonsky W., Bockus W. W., Marais G. F., et al. (2015). Native Fusarium head blight resistance from winter wheat cultivars ‘Lyman,’ ‘Overland,’ ‘Ernie,’ and ‘Freedom’ mapped and pyramided onto ‘Wesley’-Fhb1 backgrounds. Mol. Breed. 35, 6. 10.1007/s11032-015-0200-1 [DOI] [Google Scholar]
- ElDoliefy A. E. F. A., Kumar A., Anderson J. A., Glover K. D., Mamidi S., Elias E. M., et al. (2020). Genetic dissection of Fusarium head blight resistance in spring wheat cv. ‘Glenn.’. Euphytica 216, 71. 10.1007/s10681-020-02610-0 [DOI] [Google Scholar]
- Ellis J. G., Lagudah E. S., Spielmeyer W., Dodds P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5, 1–13. 10.3389/fpls.2014.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eversmeyer M. G., Kramer C. L. (2000). Epidemiology of wheat leaf and stem rust in the central Great Plains of the USA. Annu. Rev. Phytopathol. 38, 491–513. 10.1146/annurev.phyto.38.1.491 [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2018). “Food and Agriculture Organization of the United Nations” in Statistical Database. Available at: https://search.library.wisc.edu/database/UWI12320 (Accessed February 25, 2020).
- FDA (2010). Guidance for Industry and FDA: Advisory Levels for Deoxynivalenol (DON) in Finished Wheat Products for Human Consumption and Grains and Grain By-Products used for Animal Feed. Available at: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/guidance-industry-and-fda-advisory-levels-deoxynivalenol-don-finished-wheat-products-human (Accessed February 25, 2020).
- Feuillet C., Travella S., Stein N., Albar L., Nublat A., Keller B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U. S. A. 100, 15253–15258. 10.1073/pnas.2435133100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlan S., Ntushelo K., Mwadzingeni L., Terefe T., Tsilo T. J., Shimelis H. (2020). Breeding wheat for durable leaf rust resistance in Southern Africa: variability, distribution, current control strategies, challenges and future prospects. Front. Plant Sci. 11, 549. 10.3389/fpls.2020.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe B., Jiang J., Raupp W. J., McIntosh R. A., Gill B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 91, 59–87. 10.1007/BF00035277 [DOI] [Google Scholar]
- Frohberg R. C., Stack R. W., Olson T., Miller J. D., Mergoum M. (2006). Registration of ‘Alsen’ Wheat. Crop Sci. 46, 2311–2312. 10.2135/cropsci2005.12.0501 [DOI] [Google Scholar]
- Fu D., Uauy C., Distelfeld A., Blechl A., Epstein L., Chen X., et al. (2009). A Kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. 10.1126/science.1166289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Góral T., Ochodzki P., Mazurek A., Bulinskaradomska Z. (2008). Resistance of species from genus Triticum to fusarium head blight and accumulation of Fusarium-metabolites in grain. Cereal Res. Commun. 36, 95–97. [Google Scholar]
- Gatti M., Cambon F., Tassy C., Macadre C., Guerard F., Langin T., et al. (2019). The Brachypodium distachyon UGT Bradi5gUGT03300 confers type II fusarium head blight resistance in wheat. Plant Pathol. 68, 334–343. 10.1111/ppa.12941 [DOI] [Google Scholar]
- Gebrie S. A. (2016). Biotrophic fungi infection and plant defense mechanism. J. Plant Pathol. Microbiol. 7, 378. 10.4172/2157-7471.1000378 [DOI] [Google Scholar]
- Gerechter-Amitai Z. K., van Silfhout C. H., Grama A., Kleitman F. (1989). Yr15 - a new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G-25. Euphytica 43, 187–190. 10.1007/BF00037912 [DOI] [Google Scholar]
- Ghimire B., Martinez-Espinoza A. D., Glenn A. E., Mergoum M., Bowen K. L., Buck J. W. (2019. a). A preliminary investigation into the genetic diversity of Fusarium species causing Fusarium head blight (FHB) of wheat in Georgia and Alabama. in. Phytopathology S2, 166. 10.1094/PHYTO-109-10-S2.1 [DOI] [Google Scholar]
- Ghimire B., Mergoum M., Johnson J., Glenn A. E., Bowen K. L., Youmans J., et al. (2019. b). “Understanding the Genetic Diversity of Fusarium Species causing Fusarium Head Blight (FHB) of Wheat in Georgia,” in Proceedings of the 2019 National Fusarium Head Blight Forum, vol. 71 . Eds. Canty S., Hoffstetter A., Wiermer B., Dill-Macky R. (Lexington, KY: East Lansing, MI; ). 10.1017/CBO9781107415324.004 [DOI] [Google Scholar]
- Ghimire B., Hulbert S. H., Steber C. M., Garland-Campbell K., Sanguinet K. A. (2020). Characterization of root traits for improvement of spring wheat in the Pacific Northwest. Agron. J. 112, 228–240. 10.1002/agj2.20040 [DOI] [Google Scholar]
- Gilsinger J., Kong L., Shen X., Ohm H. (2005). DNA markers associated with low Fusarium head blight incidence and narrow flower opening in wheat. Theor. Appl. Genet. 110, 1218–1225. 10.1007/s00122-005-1953-4 [DOI] [PubMed] [Google Scholar]
- Golkari S., Gilbert J., Prashar S., Procunier J. D. (2007). Microarray analysis of Fusarium graminearum-induced wheat genes: Identification of organ-specific and differentially expressed genes. Plant Biotechnol. J. 5, 38–49. 10.1111/j.1467-7652.2006.00213.x [DOI] [PubMed] [Google Scholar]
- Goswami R. S., Kistler H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. 10.1111/J.1364-3703.2004.00252.X [DOI] [PubMed] [Google Scholar]
- Green A. J., Berger G., Griffey C. A., Pitman R., Thomason W., Balota M. (2014). Genetic resistance to and effect of leaf rust and powdery mildew on yield and its components in 50 soft red winter wheat cultivars. Crop Prot. 64, 177–186. 10.1016/j.cropro.2014.06.023 [DOI] [Google Scholar]
- Griffey C. A., Starling T. M., Price A. M., Sisson W. L., Das M. K., Pridgen T. H., et al. (2001). Registration of “Roane” wheat. Crop Sci. 41, 1359–1360. 10.2135/cropsci2001.4141359x [DOI] [Google Scholar]
- Griffey C. A., Rohrer W. L., Pridgen T. H., Brooks W. S., Chen J., Wilson J. A., et al. (2005. a). Registration of ‘McCormick’ wheat. Crop Sci. 45, 417–419. 10.2135/cropsci2005.0417 [DOI] [Google Scholar]
- Griffey C. A., Rohrer W. L., Pridgen T. H., Brooks W. S., Chen J., Wilson J. A. (2005. b). Registration of “Tribute” wheat. Crop Sci. 45, 419–420. 10.2135/cropsci2005.0417 [DOI] [Google Scholar]
- Griffey C. A., Brown-Guedira G., Liu S., Murphy J. P., Sneller C. (2008). “Characterization and development of FHB resistant soft winter wheat cultivars in the eastern U.S,” in Proceedings of the 2008 National Fusarium Head Blight Forum. Eds. Canty S., Clark A., Walton E., Ellis D., Mundell J., Van Sanford D. (Indianapolis, IN. Lexington, KY: University of Kentucky; ), 162–165. [Google Scholar]
- Griffey C. A., Thomason W. E., Pitman R. M., Beahm B. R., Paling J. J., Chen J., et al. (2009. a). Registration of “5205” wheat. J. Plant Regist. 3, 283–288. 10.3198/jpr2009.05.0259crc [DOI] [Google Scholar]
- Griffey C. A., Thomason W. E., Pitman R. M., Beahm B. R., Paling J. J., Chen J., et al. (2009. b). Registration of ‘USG 3555’ wheat. J. Plant Regist. 3, 273–278. 10.3198/jpr2009.05.0258crc [DOI] [Google Scholar]
- Griffey C. A., Thomason W. E., Pitman R. M., Beahm B. R., Paling J. J., Chen J., et al. (2010. a). Registration of “Jamestown” wheat. J. Plant Regist. 4, 28–33. 10.3198/jpr2009.05.0257crc [DOI] [Google Scholar]
- Griffey C. A., Thomason W. E., Pitman R. M., Beahm B. R., Paling J. J., Chen J., et al. (2010. b). Registration of “Shirley” wheat. J. Plant Regist. 4, 38–43. 10.3198/jpr2009.05.0260crc [DOI] [Google Scholar]
- Griffey C. A., Thomason W. E., Pitman R. M., Beahm B. R., Gundrum P. G., Liu S. Y., et al. (2011). Registration of “SW049029104” wheat. J. Plant Regist. 5, 91–97. 10.3198/jpr2010.03.0146crc [DOI] [Google Scholar]
- Griffey C. A. (2005). “Identification and incorporation of FHB resistance in winter wheat: An overview of progress and strategies,” in Proceedings of the 2005 National Fusarium Head Blight Forum, vol. 31 Eds. Canty S. M., Boring T., Wardwell J., Siler L., Ward R. (Milwaukee, WI: Michigan State University; ). 10.1007/s00122-007-0577-2 [DOI] [Google Scholar]
- Guo J., Zhang X., Hou Y., Cai J., Shen X., Zhou T., et al. (2015). High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 128, 2301–2316. 10.1007/s00122-015-2586-x [DOI] [PubMed] [Google Scholar]
- Gupta P. K., Langridge P., Mir R. R. (2010). Marker-assisted wheat breeding: Present status and future possibilities. Mol. Breed. 26, 145–161. 10.1007/s11032-009-9359-7 [DOI] [Google Scholar]
- Gururani M. A., Venkatesh J., Upadhyaya C. P., Nookaraju A., Pandey S. K., Park S. W. (2012). Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 78, 51–65. 10.1016/j.pmpp.2012.01.002 [DOI] [Google Scholar]
- Hall M. D., Van Sanford D. A. (2003). Diallel analysis of Fusarium head blight in soft red winter wheat. Crop Sc 43, 1663–1670. 10.1007/s10681-010-0195-y [DOI] [Google Scholar]
- Hao Y., Chen Z., Wang Y., Bland D., Buck J., Brown-Guedira G., et al. (2011). Characterization of a major QTL for adult plant resistance to stripe rust in US soft red winter wheat. Theor. Appl. Genet. 123, 1401–1411. 10.1007/s00122-011-1675-8 [DOI] [PubMed] [Google Scholar]
- Hao Y., Rasheed A., Zhu Z., Wulff B. B. H., He Z. (2020). Harnessing wheat Fhb1 for fusarium resistance. Trends Plant Sci. 25, 1–3. 10.1016/j.tplants.2019.10.006 [DOI] [PubMed] [Google Scholar]
- He X., Singh P. K., Dreisigacker S., Singh S., Lillemo M., Duveiller E. (2016). Dwarfing genes Rht-B1b and Rht-D1b are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PloS One 11, 1–14. 10.1371/journal.pone.0162499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Zhang R., Zhao J., Qi T., Kang Z., Guo J. (2019). Host-induced silencing of Fusarium graminearum genes enhances the resistance of Brachypodium distachyon to Fusarium head blight. Front. Plant Sci. 10, 1362. 10.3389/fpls.2019.01362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr R. R. (1961). Control of diseases and insect pests of crop plants in the Soviet Union. Plant Dis. Rep. 45, 399–410. [Google Scholar]
- Hovmøller M. S., Walter S., Justesen A. F. (2010). Escalating threat of wheat rusts. Science 329, 369. 10.1126/science.1194925 [DOI] [PubMed] [Google Scholar]
- Huang L., Brooks S. A., Li W., Fellers J. P., Trick H. N., Gill B. S. (2003). Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard A., Lewis C. M., Yoshida K., Ramirez-Gonzalez R. H., de Vallavieille-Pope C., Thomas J., et al. (2015). Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 16, 23. 10.1186/s13059-015-0590-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Espino J., Singh R. P., Germán S., McCallum B. D., Park R. F., Chen W. Q., et al. (2011). Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179, 143–160. 10.1007/s10681-011-0361-x [DOI] [Google Scholar]
- Islam M. S., Brown-Guedira G., Van Sanford D., Ohm H., Dong Y., McKendry A. L. (2016). Novel QTL associated with the Fusarium head blight resistance in Truman soft red winter wheat. Euphytica 207, 571–592. 10.1007/s10681-015-1550-9 [DOI] [Google Scholar]
- IWGSC (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 10.1126/science.1251788 [DOI] [PubMed] [Google Scholar]
- Jayatilake D. V., Bai G. H., Dong Y. H. (2011). A novel quantitative trait locus for Fusarium head blight resistance in chromosome 7A of wheat. Theor. Appl. Genet. 122, 1189–1198. 10.1007/s00122-010-1523-2 [DOI] [PubMed] [Google Scholar]
- Jiang G. L., Dong Y., Shi J. R., Ward R. W. (2007). QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. II. Resistance to deoxynivalenol accumulation and grain yield loss. Theor. Appl. Genet. 115, 1043–1052. 10.1007/s00122-007-0630-1 [DOI] [PubMed] [Google Scholar]
- Jin Y., Szabo L. J., Carson M. (2010). Century-old mystery of Puccinia striiformis life history solved with the identification of berberis as an alternate host. Phytopathology 100, 432–435. 10.1094/PHYTO-100-5-0432 [DOI] [PubMed] [Google Scholar]
- Johnson J. W., Chen Z., Buck J. W., Buntin G. D., Babar M. A., Mason R. E., et al. (2017). ‘GA 03564-12E6’: A high-yielding soft red winter wheat cultivar adapted to Georgia and the southeastern regions of the United States. J. Plant Regist. 11, 159–164. 10.3198/jpr2016.07.0036crc [DOI] [Google Scholar]
- Johnson J. W., Chen Z., Buck J. W., Buntin G. D., Babar M. A., Mason R. E., et al. (2018). ‘Savoy’, an adapted soft red winter wheat cultivar for Georgia and the southeast regions of the United States. J. Plant Regist. 12, 85–89. 10.3198/jpr2017.06.0039crc [DOI] [Google Scholar]
- Johnston C. O. (1961). Distribution and prevalence of physiologic races of leaf rust of wheat, Puccinia recondita rob. ex desm., in the United States in 1960. Plant Dis. Rep. 45, 661–662. [Google Scholar]
- Kage U., Karre S., Kushalappa A. C., McCartney C. (2017). Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant Biotechnol. J. 15, 447–457. 10.1111/pbi.12641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Clark A., Van Sanford D., Griffey C., Brown-Guedira G., Dong Y., et al. (2011). Exotic scab resistance quantitative trait loci effects on soft red winter wheat. Crop Sci. 51, 924–933. 10.2135/cropsci2010.06.0313 [DOI] [Google Scholar]
- Kazan K., Gardiner D. M. (2018). Transcriptomics of cereal–Fusarium graminearum interactions: what we have learned so far. Mol. Plant Pathol. 19, 764–778. 10.1111/mpp.12561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. C., Ward T. J. (2018). Population genomics of Fusarium graminearum reveals signatures of divergent evolution within a major cereal pathogen. PloS One 13, 1–32. 10.1371/journal.pone.0194616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Trevathan L. E., Robbins J. T. (1997). Quantitative relationship between leaf rust and wheat yield in Mississippi. Plant Dis. 81, 769–772. 10.1094/PDIS.1997.81.7.769 [DOI] [PubMed] [Google Scholar]
- Kloppers F. J., Pretorius Z. A. (1997). Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 46, 737–750. 10.1046/j.1365-3059.1997.d01-58.x [DOI] [Google Scholar]
- Klymiuk V., Yaniv E., Huang L., Raats D., Fatiukha A., Chen S., et al. (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9, 3735. 10.1038/s41467-018-06138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmer J. A., Jin Y., Long D. L. (2007). Wheat leaf and stem rust in the United States. Aust. J. Agric. Res. 58, 631–638. 10.1071/AR07057 [DOI] [Google Scholar]
- Kolmer J. A., Lin M., Bai G. (2012). Genetics of leaf rust resistance in the winter wheat line CI13227. Crop Sci. 52, 2166–2172. 10.2135/cropsci2012.02.0136 [DOI] [Google Scholar]
- Kolmer J. A., Lagudah E. S., Lillemo M., Lin M., Bai G. (2015). The Lr46 gene conditions partial adult- plant resistance to stripe rust, stem rust, and powdery mildew in thatcher wheat. Crop Sci. 55, 2557–2565. 10.2135/cropsci2015.02.0082 [DOI] [Google Scholar]
- Kolmer J. A., Chao S., Brown-Guedira G., Bansal U., Bariana H. (2018). Adult plant leaf rust resistance derived from the soft red winter wheat cultivar ‘Caldwell’ maps to chromosome 3BS. Crop Sci. 58, 152–158. 10.2135/cropsci2017.05.0272 [DOI] [Google Scholar]
- Kolmer J. A., Ordoñez M. E., German S., Morgounov A., Pretorius Z., Visser B., et al. (2019). Multilocus genotypes of the wheat leaf rust fungus Puccinia triticina in worldwide regions indicate past and current long-distance migration. Phytopathology 109, 1453–1463. 10.1094/PHYTO-10-18-0411-R [DOI] [PubMed] [Google Scholar]
- Kolmer J. A. (1996). Genetics of resistance to wheat leaf rust. Annu. Rev. Phytopathol. 34, 435–455. 10.1146/annurev.phyto.34.1.435 [DOI] [PubMed] [Google Scholar]
- Kolmer J. A. (2002). Virulence phenotypes of Puccinia triticina in the South Atlantic states in 1999. Plant Dis. 86, 288–291. 10.1094/PDIS.2002.86.3.288 [DOI] [PubMed] [Google Scholar]
- Kolmer J. A. (2003). Postulation of leaf rust resistance genes in selected soft red winter wheats. Crop Sci. 43, 1266–1274. 10.2135/cropsci2003.1266 [DOI] [Google Scholar]
- Kolmer J. A. (2005). Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 8, 441–449. 10.1016/j.pbi.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Kolmer J. A. (2010). Genetics of leaf rust resistance in the soft red winter wheat cultivars Coker 9663 and Pioneer 26R61. Plant Dis. 94, 628–632. 10.2135/cropsci2016.12.1029 [DOI] [PubMed] [Google Scholar]
- Kolmer J. (2013). Leaf rust of wheat: Pathogen biology, variation and host resistance. Forests 4, 70–84. 10.3390/f4010070 [DOI] [Google Scholar]
- Kolmer J. A. (2019). Virulence of Puccinia triticina, the wheat leaf rust fungus, in the United States in 2017. Plant Dis. 103, 2113–2120. 10.1094/PDIS-09-18-1638-SR [DOI] [PubMed] [Google Scholar]
- Kosová K., Chrpová J., Šíp V. (2009). Cereal resistance to Fusarium head blight and possibilities of its improvement through breeding. Czech J. Genet. Plant Breed. 45, 87–105. 10.17221/63/2009-cjgpb [DOI] [Google Scholar]
- Krattinger S. G., Lagudah E. S., Spielmeyer W., Singh R. P., Huerta-Espino J., McFadden H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. 10.1126/science.1166453 [DOI] [PubMed] [Google Scholar]
- Kuraparthy V., Chhuneja P., Dhaliwal H. S., Kaur S., Bowden R. L., Gill B. S. (2007). Characterization and mapping of cryptic alien introgression from Aegilops geniculata with new leaf rust and stripe rust resistance genes Lr57 and Yr40 in wheat. Theor. Appl. Genet. 114, 1379–1389. 10.1007/s00122-007-0524-2 [DOI] [PubMed] [Google Scholar]
- Löffler M., Schön C. C., Miedaner T. (2009). Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol. Breed. 23, 473–488. 10.1007/s11032-008-9250-y [DOI] [Google Scholar]
- Lagudah E. S., Krattinger S. G. (2019). A new player contributing to durable Fusarium resistance. Nat. Genet. 51, 1070–1071. 10.1038/s41588-019-0454-3 [DOI] [PubMed] [Google Scholar]
- Larkin D. L., Lozada D. N., Mason R. E. (2019). Genomic selection—considerations for successful implementation in wheat breeding programs. Agronomy 9, 1–18. 10.3390/agronomy9090479 [DOI] [Google Scholar]
- Lemmens M., Scholz U., Berthiller F., Dall’Asta C., Koutnik A., Schuhmacher R., et al. (2005). The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for fusarium head blight resistance in wheat. Mol. Plant-Microbe Interact. 18, 1318–1324. 10.1094/MPMI-18-1318 [DOI] [PubMed] [Google Scholar]
- Li H., Chen Q., Conner R. L., Guo B., Zhang Y., Graf R. J., et al. (2003). Molecular characterization of a wheat - Thinopyrum ponticum partial amphiploid and its derivatives for resistance to leaf rust. Genome 46, 906–913. 10.1139/g03-053 [DOI] [PubMed] [Google Scholar]
- Li X., Shin S., Heinen S., Dill-Macky R., Berthiller F., Nersesian N., et al. (2015). Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Mol. Plant-Microbe Interact. 28, 1237–1246. 10.1094/MPMI-03-15-0062-R [DOI] [PubMed] [Google Scholar]
- Li T., Luo M., Zhang D., Wu D., Li L., Bai G. (2016. a). Effective marker alleles associated with type 2 resistance to Fusarium head blight infection in fields. Breed. Sci. 66, 350–357. 10.1270/jsbbs.15124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Zhang D., Zhou X., Bai G., Li L., Gu S. (2016. b). Fusarium head blight resistance loci in a stratified population of wheat landraces and varieties. Euphytica 207, 551–561. 10.1007/s10681-015-1539-4 [DOI] [Google Scholar]
- Li C., Wang Z., Li C., Bowden R., Bai G., Li C., et al. (2017). Mapping of quantitative trait loci for leaf rust resistance in the wheat population Ning7840 × Clark. Plant Dis. 101, 1974–1979. 10.1094/PDIS-12-16-1743-RE [DOI] [PubMed] [Google Scholar]
- Li G., Zhou J., Jia H., Gao Z., Fan M., Luo Y., et al. (2019). Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 51, 1106–1112. 10.1038/s41588-019-0426-7 [DOI] [PubMed] [Google Scholar]
- Lilleboe D. (2014). FHB in 2014 : From Nonexistent to Severe. USWBSI, 1–14.
- Limbalkar O. M., Meena K., Singh M., Sunilkumar V. P. (2018). Genetic improvement of wheat for biotic and abiotic stress tolerance. Int. J. Curr. Microbiol. Appl. Sci. 7, 1962–1971. 10.20546/ijcmas.2018.712.226 [DOI] [Google Scholar]
- Line R. F., Chen X. (1995). Successes in breeding for and managing durable resistance to wheat rusts. Plant Dis. 79, 1254–1255. 10.1094/PD-79-1254 [DOI] [Google Scholar]
- Line R. F. (2002). Stripe rust of wheat and barley in North America: A retrospective historical review. Annu. Rev. Phytopathol. 40, 75–118. 10.1146/annurev.phyto.40.020102.111645 [DOI] [PubMed] [Google Scholar]
- Liu S., Abate Z. A., Lu H., Musket T., Davis G. L., McKendry A. L. (2007). QTL associated with Fusarium head blight resistance in the soft red winter wheat Ernie. Theor. Appl. Genet. 115, 417–427. 10.1007/s00122-007-0577-2 [DOI] [PubMed] [Google Scholar]
- Liu S., Hall M. D., Griffey C. A., McKendry A. L. (2009). Meta-analysis of QTL associated with fusarium head blight resistance in wheat. Crop Sci. 49, 1955–1968. 10.2135/cropsci2009.03.0115 [DOI] [Google Scholar]
- Liu S., Christopher M. D., Griffey C. A., Hall M. D., Gundrum P. G., Brooks W. S. (2012). Molecular characterization of resistance to fusarium head blight in U.S. soft red winter wheat breeding line VA00W-38. Crop Sci. 52, 2283–2292. 10.2135/cropsci2012.03.0144 [DOI] [Google Scholar]
- Liu S., Griffey C. A., Hall M. D., McKendry A. L., Chen J., Brooks W. S., et al. (2013). Molecular characterization of field resistance to Fusarium head blight in two US soft red winter wheat cultivars. Theor. Appl. Genet. 126, 2485–2498. 10.1007/s00122-013-2149-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Frick M., Huel R., Nykiforuk C. L., Wang X., Gaudet D. A., et al. (2014). The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol. Plant 7, 1740–1755. 10.1093/mp/ssu112 [DOI] [PubMed] [Google Scholar]
- Lofgren L., Riddle J., Dong Y., Kuhnem P. R., Cummings J. A., Del Ponte E. M., et al. (2018). A high proportion of NX-2 genotype strains are found among Fusarium graminearum isolates from northeastern New York State. Eur. J. Plant Pathol. 150, 791–796. 10.1007/s10658-017-1314-6 [DOI] [Google Scholar]
- Long D. L., Roberts J. J., Schafer J. F., Johnson J. W., Fowler H. A., Cunfer B. M. (1992). Registration of six leaf rust resistant soft red winter wheat germplasm lines. Crop Sci. 32, 1514–1515. 10.2135/cropsci1992.0011183x003200060053x [DOI] [Google Scholar]
- Ma Z., Xie Q., Li G., Jia H., Zhou J., Kong Z., et al. (2020). Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 133, 1541–1568. 10.1007/s00122-019-03525-8 [DOI] [PubMed] [Google Scholar]
- Madden L. V., Bradley C. A., Dalla Lana F., Paul P. A. (2014). “Meta-analysis of 19 years of fungicide trials for the control of fusarium head blight of wheat,” in Proceedings of the 2014 National Fusarium Head Blight Forum. Eds. Canty S., Clark A., Turcott N., Van Sanford D. (Lexington, KY: East Lansing, MI; ), 17–18. [Google Scholar]
- Mains E. B., Jackson H. S. (1926). Physiologic specialisation in the leaf rust of wheat, Puccinia triticina Erikas. Phytopathology 16, 89–120. [Google Scholar]
- Malihipour A., Gilbert J., Fedak G., Brûlé-Babel A., Cao W. (2017). Mapping the A genome for QTL conditioning resistance to Fusarium head blight in a wheat population with Triticum timopheevii background. Plant Dis. 101, 11–19. 10.1094/PDIS-02-16-0144-RE [DOI] [PubMed] [Google Scholar]
- Maloney P. V., Petersen S., Navarro R. A., Marshall D., McKendry A. L., Costa J. M., et al. (2014). Digital image analysis method for estimation of Fusarium-damaged kernels in wheat. Crop Sci. 54, 2077–2083. 10.2135/cropsci2013.07.0432 [DOI] [Google Scholar]
- Mammadov J. A., Brooks W. S., Griffey C. A., Saghai Maroof M. A. (2007). Validating molecular markers for barley leaf rust resistance genes Rph5 and Rph7. Plant Breed. 126, 458–463. 10.1111/j.1439-0523.2007.01387.x [DOI] [Google Scholar]
- Mao S. L., Wei Y. M., Cao W., Lan X. J., Yu M., Chen Z. M., et al. (2010). Confirmation of the relationship between plant height and Fusarium head blight resistance in wheat (Triticum aestivum L.) by QTL meta-analysis. Euphytica 174, 343–356. 10.1007/s10681-010-0128-9 [DOI] [Google Scholar]
- Marasas C. N., Smale M., Singh R. P. (2004). The economic impact in developing countries of leaf rust resistance breeding in CIMMYT-related spring bread wheat. Economics Program Paper 04-01 (Mexico, D.F.: CIMMYT; ). Available at: https://repository.cimmyt.org/handle/10883/1045 (Accessed March 2, 2020). [Google Scholar]
- Marchal C., Zhang J., Zhang P., Fenwick P., Steuernagel B., Adamski N. M., et al. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 4, 662–668. 10.1038/s41477-018-0236-4 [DOI] [PubMed] [Google Scholar]
- Martinez-Espinoza A. D., Youmans J., Vermeer C. B., Buck J. W. (2014). Evaluation of fungicides for control of stripe rust and fusarium head blight of winter wheat in southwest Georgia 2015. Plant Dis. Manage. Rep. 10, CF034. [Google Scholar]
- Mason R. E., Johnson J. W., Mergoum M., Miller R. G., Moon D. E., Carlin J. F., et al. (2018). ‘AR11LE24’, a soft red winter wheat adapted to the Mid-South region of the USA. J. Plant Regist. 12, 357–361. 10.3198/jpr2017.09.0060crc [DOI] [Google Scholar]
- Mboup M., Leconte M., Gautier A., Wan A. M., Chen W., de Vallavieille-Pope C., et al. (2009). Evidence of genetic recombination in wheat yellow rust populations of a Chinese oversummering area. Fungal Genet. Biol. 46, 299–307. 10.1016/j.fgb.2008.12.007 [DOI] [PubMed] [Google Scholar]
- McIntosh R. A., Dubcovsky J., Rogers W. J., Morris C., Xia X. C., Designators L. (2017). “Catalogue of ene Symbols for Wheat: 2017 Supplement,” in Komugi Wheat Genet. Resour. Database (Japan: Comm. Natl, Kyoto; ). updated Jan. 11. [Google Scholar]
- McMullen M., Jones R., Gallenberg D. (1997). Scab of wheat and barley: A re-emerging disease of devastating impact. Plant Dis. 81, 1340–1348. 10.1094/PDIS.1997.81.12.1340 [DOI] [PubMed] [Google Scholar]
- McMullen M., Bergstrom G., De Wolf E., Dill-Macky R., Hershman D., Shaner G., et al. (2012). A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 96, 1712–1728. 10.1094/PDIS-03-12-0291-FE [DOI] [PubMed] [Google Scholar]
- Mergoum M., Frohberg R. C., Miller J. D., Stack R. W. (2005). Registration of “Steele-ND” wheat. Crop Sci. 45, 1163–1164. 10.2135/cropsci2004.308CV [DOI] [Google Scholar]
- Mergoum M., Frohberg R. C., Olson T., Friesen T. L., Rasmussen J. B., Stack R. W. (2006). Registration of “Glenn” wheat. Crop Sci. 46, 473–474. 10.2135/cropsci2005.0287 [DOI] [Google Scholar]
- Mergoum M., Frohberg R. C., Stack R. W., Rasmussen J. W., Friesen T. L. (2008). Registration of ‘Faller’ spring wheat. J. Plant Regist. 2, 224–229. 10.3198/jpr2008.03.0166crc [DOI] [Google Scholar]
- Mergoum M., Simsek S., Frohberg R. C., Rasmussen J. B., Friesen T. L., Adhikari T. (2011). ‘Barlow’: A high-quality and high-yielding hard red spring wheat cultivar adapted to the North Central Plains of the USA. J. Plant Regist. 5, 62–67. 10.3198/jpr2010.05.0259crc [DOI] [Google Scholar]
- Mergoum M., Frohberg R. C., Stack R. W., Simsek S., Adhikari T. B., Rasmussen J. B., et al. (2013). ‘Prosper’: A high-yielding hard red spring wheat cultivar adapted to the North Central Plains of the USA. J. Plant Regist. 7, 75–80. 10.3198/jpr2012.05.0271crc [DOI] [Google Scholar]
- Mergoum M., Johnson J., Buck J., Chen Z., Harrison S. A., Mason R. E., et al. (2019). “GA09129-16E55 (AGS 3015), a new soft red winter wheat cultivar adapted to the US southeast with improved FHB resistance,” in Proceedings of the 2019 National Fusarium Head Blight Forum. Eds. Canty S., Hoffstetter A., Wiermer B., Dill-Macky R. (Lexington, KY: East Lansing, MI; ), 101–102. [Google Scholar]
- Mesterházy Á., Bartók T., Mirocha C. G., Komoróczy R. (1999). Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 118, 97–110. 10.1046/j.1439-0523.1999.118002097.x [DOI] [Google Scholar]
- Mesterhazy A. (1995). Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 114, 377–386. 10.1111/j.1439-0523.1995.tb00816.x [DOI] [Google Scholar]
- Miedaner T., Voss H. H. (2008). Effect of dwarfing Rht genes on fusarium head blight resistance in two sets of near-isogenic lines of wheat and check cultivars. Crop Sci. 48, 2115–2122. 10.2135/cropsci2008.02.0107 [DOI] [Google Scholar]
- Miedaner T. (1997). Breeding wheat and rye for resistance to Fusarium diseases. Plant Breed. 116, 201–220. 10.1111/j.1439-0523.1997.tb00985.x [DOI] [Google Scholar]
- Millet E., Manisterski J., Ben-Yehuda P., Distelfeld A., Deek J., Wan A., et al. (2014). Introgression of leaf rust and stripe rust resistance from Sharon goatgrass (Aegilops sharonensis Eig) into bread wheat (Triticum aestivum L.). Genome 57, 309–316. 10.1139/gen-2014-0004 [DOI] [PubMed] [Google Scholar]
- Milus E. A., Seyran E., Mcnew R. (2006). Aggressiveness of Puccinia striiformis f. sp. tritici isolates in the South-Central United States. Plant Dis. 90, 847–852. 10.1094/PD-90-0847 [DOI] [PubMed] [Google Scholar]
- Milus E. A., Kristensen K., Hovmøller M. S. (2008). Increased aggressiveness of Puccinia striiformis f. sp. tritici at least partially explains recent stripe rust epidemics. Phytopathology 98, S107. 10.1094/PHYTO.2008.98.6.S9 [DOI] [PubMed] [Google Scholar]
- Moore J. W., Herrera-Foessel S., Lan C., Schnippenkoetter W., Ayliffe M., Huerta-Espino J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. 10.1038/ng.3439 [DOI] [PubMed] [Google Scholar]
- Murphy J. P., Navarro R. A., Leath S., Bowman D. T., Weisz P. R., Ambrose L. G., et al. (2004). Registration of ‘NC-Neuse’ wheat. Crop Sci. 44, 1479–1480. 10.2135/cropsci2004.1479 [DOI] [Google Scholar]
- Murphy L. R., Santra D., Kidwell K., Yan G., Chen X., Campbell K. G. (2009). Linkage maps of wheat stripe rust resistance genes YR5 and YR15 for use in marker-assisted selection. Crop Sci. 49, 1786–1790. 10.2135/cropsci2008.10.0621 [DOI] [Google Scholar]
- Murphy J. P., Lyerly J. H., Acharya R., Page J., Ward B., Brown-Guedira G. (2019). Southern Uniform Winter Wheat Scab Nursery. Available at: https://scabusa.org/publications (Accessed May 20, 2020).
- Nganje W. E., Kaitibie S., Wilson W. W., Leistritz F. L., Bangsund D. A. (2004). Economic Impacts of Fusarium Head Blight in Wheat and Barley: 1993-2001. (Agribusiness and Applied Economics Report No 538). North Dakota State University, Fargo, ND: Department of Agribusiness and Applied Economics, Agricultural Experiment Station. [Google Scholar]
- O’Donnell K., Kistler H. C., Tacke B. K., Casper H. H. (2000). Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. 97, 7905–7910. 10.1073/pnas.130193297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver R. E., Cai X., Xu S. S., Chen X., Stack R. W. (2005). Wheat-alien species derivatives: A novel source of resistance to Fusarium head blight in wheat. Crop Sci. 45, 1353–1360. 10.2135/cropsci2004.0503 [DOI] [Google Scholar]
- Olivera P. D., Kolmer J. A., Anikster Y., Steffenson B. J. (2007). Resistance of sharon goatgrass (Aegilops sharonensis) to fungal diseases of wheat. Plant Dis. 91, 942–950. 10.1094/PDIS-91-8-0942 [DOI] [PubMed] [Google Scholar]
- Olivera P. D., Millet E., Anikster Y., Steffenson B. J. (2008). Genetics of resistance to wheat leaf rust, stem rust, and powdery mildew in Aegilops sharonensis. Phytopathology 98, 353–358. 10.1094/PHYTO-98-3-0353 [DOI] [PubMed] [Google Scholar]
- Ordoñez M. E., Kolmer J. A. (2009). Differentiation of molecular genotypes and virulence phenotypes of Puccinia triticina from common wheat in North America. Phytopathology 99, 750–758. 10.1094/PHYTO-99-6-0750 [DOI] [PubMed] [Google Scholar]
- Panwar V., Jordan M., McCallum B., Bakkeren G. (2018). Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol. J. 16, 1013–1023. 10.1111/pbi.12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. W., Jenkinson P., McLeod L. (1995). Fusarium ear blight (scab) in small grain cereals—a review. Plant Pathol. 44, 207–238. 10.1111/j.1365-3059.1995.tb02773.x [DOI] [Google Scholar]
- Paul P. A., Salgado J. D., Bergstrom G., Bradley C. A., Byamukama E., Byrne A. M., et al. (2019). Integrated effects of genetic resistance and prothioconazole + tebuconazole application timing on fusarium head blight in wheat. Plant Dis. 103, 223–237. 10.1094/PDIS-04-18-0565-RE [DOI] [PubMed] [Google Scholar]
- Peng J. H., Sun D., Nevo E. (2011). Domestication evolution, genetics and genomics in wheat. Mol. Breed. 28, 281–301. 10.1007/s11032-011-9608-4 [DOI] [Google Scholar]
- Perovic D., Förester J., Welz G., Kopahnke D., Lein V., Löschenberger F., et al. (2008). Marker-assisted wheat improvement: creating semi-dwarf phenotypes with superior fusarium head blight resistance. Cereal Res. Commun. 36, Suppl. B, 153–155. 10.1556/crc.36.2008.suppl.b.14 [DOI] [Google Scholar]
- Petersen S., Lyerly J. H., Maloney P. V., Brown-Guedira G., Cowger C., Costa J. M., et al. (2016). Mapping of Fusarium head blight resistance quantitative trait loci in winter wheat cultivar NC-Neuse. Crop Sci. 56, 1473–1483. 10.2135/cropsci2015.05.0312 [DOI] [Google Scholar]
- Petersen S., Lyerly J. H., McKendry A. L., Islam M. S., Brown-Guedira G., Cowger C., et al. (2017). Validation of Fusarium head blight resistance QTL in US winter wheat. Crop Sci. 57, 1–12. 10.2135/cropsci2015.07.0415 [DOI] [Google Scholar]
- Pritsch C., Muehlbauer G. J., Bushnell W. R., Somers D. A., Vance C. P. (2000). Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol. Plant-Microbe Interact. 13, 159–169. 10.1094/MPMI.2000.13.2.159 [DOI] [PubMed] [Google Scholar]
- Pumphrey M. O., Bernardo R., Anderson J. A. (2007). Validating the Fhb1 QTL for fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop Sci. 47, 200–206. 10.2135/cropsci2006.03.0206 [DOI] [Google Scholar]
- Puri K. D., Zhong S. (2010). The 3ADON population of Fusarium graminearum found in North Dakota is more aggressive and produces a higher level of DON than the prevalent 15ADON population in spring wheat. Phytopathology 100, 1007–1014. 10.1094/PHYTO-12-09-0332 [DOI] [PubMed] [Google Scholar]
- Qayoum A., Line R. F. (1985). High-temperature, adult-plant resistance to stripe rust of wheat. Phytopathology 75, 1121. 10.1094/phyto-75-1121 [DOI] [Google Scholar]
- Qi D., Innes R. W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4, 348. 10.3389/fimmu.2013.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. L., Pumphrey M. O., Friebe B., Chen P. D., Gill B. S. (2008). Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor. Appl. Genet. 117, 1155–1166. 10.1007/s00122-008-0853-9 [DOI] [PubMed] [Google Scholar]
- Qi T., Zhu X., Tan C., Liu P., Guo J., Kang Z., et al. (2018). Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 16, 797–807. 10.1111/pbi.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N., Bariana H., Kumran V. V., Muruga S., Forrest K. L., Hayden M. J., et al. (2018). A new leaf rust resistance gene Lr79 mapped in chromosome 3BL from the durum wheat landrace Aus26582. Theor. Appl. Genet. 131, 1091–1098. 10.1007/s00122-018-3060-3 [DOI] [PubMed] [Google Scholar]
- Raupp W. J., Singh S., Brown-Guedira G. L., Gill B. S. (2001). Cytogenetic and molecular mapping of the leaf rust resistance gene Lr39 in wheat. Theor. Appl. Genet. 102, 347–352. 10.1007/s001220051652 [DOI] [Google Scholar]
- Rawat N., Pumphrey M. O., Liu S., Zhang X., Tiwari V. K., Ando K., et al. (2016). Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 48, 1576–1580. 10.1038/ng.3706 [DOI] [PubMed] [Google Scholar]
- Robert O., Abelard C., Dedryver F. (1999). Identification of molecular markers for the detection of the yellow rust resistance gene Yr17 in wheat. Mol. Breed. 5, 167–175. 10.1023/A:1009672021411 [DOI] [Google Scholar]
- Roelfs A. P., Singh R. P., Saari E. E. (1992). Rust diseases of wheat: Concepts and methods of disease management (Mexico: CIMMYT; ). [Google Scholar]
- Rosewarne G. M., Herrera-Foessel S. A., Singh R. P., Huerta-Espino J., Lan C. X., He Z. H. (2013). Quantitative trait loci of stripe rust resistance in wheat. Theor. Appl. Genet. 126, 2427–2449. 10.1007/s00122-013-2159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborski D. J. (1985). “The Cereal Rusts Volume II Diseases, Distribution, Epidemiology, and Control,” in Wheat Leaf Rust. Eds. Roelfs A. P., Bushnell W. R. (Orlando, FL: Academic Press; ), 39–59. [Google Scholar]
- Sapkota S., Hao Y., Johnson J., Buck J., Aoun M., Mergoum M. (2019. a). Genome-wide association study of a worldwide collection of wheat genotypes reveals novel quantitative trait loci for leaf rust resistance. Plant Genome 12, 190033. 10.3835/plantgenome2019.05.0033 [DOI] [PubMed] [Google Scholar]
- Sapkota S., Hao Y., Johnson J., Lopez B., Bland D., Chen Z., et al. (2019. b). Genetic mapping of a major gene for leaf rust resistance in soft red winter wheat cultivar AGS 2000. Mol. Breed. 39, 8. 10.1007/s11032-018-0909-8 [DOI] [Google Scholar]
- Sapkota S. (2019). Genetics of leaf rust disease resistance in wheat (Triticum aestivum L.) (Georgia, GA: University of Georgia; ). [Google Scholar]
- Sarver B. A. J., Ward T. J., Gale L. R., Broz K., Corby Kistler H., Aoki T., et al. (2011). Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 48, 1096–1107. 10.1016/j.fgb.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Savary S., Willocquet L., Pethybridge S. J., Esker P., McRoberts N., Nelson A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. 10.1038/s41559-018-0793-y [DOI] [PubMed] [Google Scholar]
- Schmale D. G., Wood-Jones A. K., Cowger C., Bergstrom G. C., Arellano C. (2011). Trichothecene genotypes of Gibberella zeae from winter wheat fields in the eastern USA. Plant Pathol. 60, 909–917. 10.1111/j.1365-3059.2011.02443.x [DOI] [Google Scholar]
- Schmale III D. G., Leslie J. F., Zeller K. A., Saleh A. A., Shields E. J., Bergstrom G. C. (2006). Genetic structure of atmospheric populations of Gibberella zeae. Phytopathology 96, 1021–1026. 10.1094/PHYTO-96-1021 [DOI] [PubMed] [Google Scholar]
- Schroeder H. W., Christensen J. J. (1963). Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 53, 831–838. [Google Scholar]
- Schweiger W., Steiner B., Vautrin S., Nussbaumer T., Siegwart G., Zamini M., et al. (2016). Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 129, 1607–1623. 10.1007/s00122-016-2727-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah L., Ali A., Zhu Y., Wang S., Si H., Ma C. (2017). Wheat resistance to Fusarium head blight and possibilities of its improvement using molecular marker-assisted selection. Czech J. Genet. Plant Breed. 53, 47–54. 10.17221/139/2016-CJGPB [DOI] [Google Scholar]
- Shen X., Ohm H. (2006). Fusarium head blight resistance derived from Lophopyrum elongatum chromosome 7E and its augmentation with Fhb1 in wheat. Plant Breed. 125, 424–429. 10.1111/j.1439-0523.2006.01274.x [DOI] [Google Scholar]
- Shin S., Mackintosh C. A., Lewis J., Heinen S. J., Radmer L., Dill-Macky R., et al. (2008). Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 59, 2371–2378. 10.1093/jxb/ern103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Huerta-Espino J., William H. M. (2005). Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turkish J. Agric. For. 29, 121–127. 10.3906/tar-0402-2 [DOI] [Google Scholar]
- Singh R. P., Huerta-Espino J., Bhavani S., Herrera-Foessel S. A., Singh D., Singh P. K., et al. (2011). Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179, 175–186. 10.1007/s10681-010-0322-9 [DOI] [Google Scholar]
- Smith W. G. (1884). “New Diseases of Wheat, Barley, and Rye-grass, caused by Fusisporium culmorum, horde, and Lolii,” in Diseases of Field and Garden Crops: Chiefly Such as are Caused by Fungi. Ed. Smith W. G. (New London: MacMillan and Co.), 208–213. [Google Scholar]
- Soriano J. M., Royo C. (2015). Dissecting the genetic architecture of leaf rust resistance in wheat by QTL meta-analysis. Phytopathology 105, 1585–1593. 10.1094/PHYTO-05-15-0130-R [DOI] [PubMed] [Google Scholar]
- Spielmeyer W., McIntosh R. A., Kolmer J., Lagudah E. S. (2005). Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor. Appl. Genet. 111, 731–735. 10.1007/s00122-005-2058-9 [DOI] [PubMed] [Google Scholar]
- Stakman E. C., Stewart D. M., Loegering W. Q. (1962). Identification of physiologic races of Puccinia graminis var. tritici (Washington: USDA; ). [Google Scholar]
- Steed A., Chandler E., Thomsett M., Gosman N., Faure S., Nicholson P. (2005). Identification of type I resistance to Fusarium head blight controlled by a major gene located on chromosome 4A of Triticum macha. Theor. Appl. Genet. 111, 521–529. 10.1007/s00122-005-2043-3 [DOI] [PubMed] [Google Scholar]
- Steiner B., Lemmens M., Griesser M., Scholz U., Schondelmaier J., Buerstmayr H. (2004). Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor. Appl. Genet. 109, 215–224. 10.1007/s00122-004-1620-1 [DOI] [PubMed] [Google Scholar]
- Steiner B., Buerstmayr M., Michel S., Schweiger W., Lemmens M., Buerstmayr H. (2017). Breeding strategies and advances in line selection for Fusarium head blight resistance in wheat. Trop. Plant Pathol. 42, 165–174. 10.1007/s40858-017-0127-7 [DOI] [Google Scholar]
- Steiner B., Michel S., Maccaferri M., Lemmens M., Tuberosa R., Buerstmayr H. (2019). Exploring and exploiting the genetic variation of Fusarium head blight resistance for genomic-assisted breeding in the elite durum wheat gene pool. Theor. Appl. Genet. 132, 969–988. 10.1007/s00122-018-3253-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Jin S., Zhang D., Bai G. (2018). Development and validation of diagnostic markers for Fhb1 region, a major QTL for Fusarium head blight resistance in wheat. Theor. Appl. Genet. 131, 2371–2380. 10.1007/s00122-018-3159-6 [DOI] [PubMed] [Google Scholar]
- Su Z., Bernardo A., Tian B., Chen H., Wang S., Ma H., et al. (2019). A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 51, 1099–1105. 10.1038/s41588-019-0425-8 [DOI] [PubMed] [Google Scholar]
- Subramanian N. K., Mason R. E., Milus E. A., Moon D. E., Brown-Guedira G. (2016). Characterization of two adult-plant stripe rust resistance genes on chromosomes 3BS and 4BL in soft red winter wheat. Crop Sci. 56, 143–153. 10.2135/cropsci2015.01.0043 [DOI] [Google Scholar]
- Tessmann E. W., Van Sanford D. A. (2018). GWAS for fusarium head blight related traits in winter wheat (Triticum aestivum L.) in an artificially warmed treatment. Agronomy 8, 68. 10.3390/agronomy8050068 [DOI] [Google Scholar]
- Tessmann E. W., Dong Y., Van Sanford D. A. (2019). GWAS for fusarium head blight traits in a soft red winter wheat mapping panel. Crop Sci. 59, 1823–1837. 10.2135/cropsci2018.08.0492 [DOI] [Google Scholar]
- Thind A. K., Wicker T., Šimková H., Fossati D., Moullet O., Brabant C., et al. (2017). Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat. Biotechnol. 35, 793–796. 10.1038/nbt.3877 [DOI] [PubMed] [Google Scholar]
- Todorovska E., Christov N., Slavov S., Christova P., Vassilev D. (2009). Biotic stress resistance in wheat - Breeding and genomic selection implications. Biotechnol. Biotechnol. Equip. 23, 1417–1426. 10.2478/V10133-009-0006-6 [DOI] [Google Scholar]
- USDA, F. A. S (2020). Grain: World Markets and Trade. Available at: https://public.govdelivery.com/accounts/USDAFAS/subscriber/new (Accessed March 3, 2020).
- USDA, N. A. S. S (1990). Acreage. Available at: https://usda.library.cornell.edu/concern/publications/j098zb09z?locale=en (Accessed March 3, 2020).
- USDA, N. A. S. S (2018). Acreage. Available at: https://usda.library.cornell.edu/concern/publications/j098zb09z?locale=en (Accessed March 3, 2020).
- Van Sanford D. A., Swanson C. S., Pearce W. L., Tutt C. R., Tomes L. J., Hershman D. E. (1997). Registration of ‘Foster’ wheat. Crop Sci. 37, 627. 10.2135/cropsci1997.0011183x003700020054x [DOI] [Google Scholar]
- Van Sanford D. A., Connelley J., Swanson C. S., Kennedy B., Tutt C. R., Tomes L. J., et al. (2006). Registration of ‘Allegiance’ wheat. Crop Sci. 46, 2305. 10.2135/cropsci2005.11.0413 [DOI] [Google Scholar]
- Van Sanford D. A., Clark A. J., Hershman D., Brown-Guedira G. L., Cowger C., Dong Y., et al. (2016). Registration of ‘Pembroke 2014’ soft red winter wheat. J. Plant Regist. 10, 41–46. 10.3198/jpr2015.07.0045crc [DOI] [Google Scholar]
- Van Sanford D. A., Clark A. J., Bradley C., Brown-Guedira G. L., Cowger C., Dong Y., et al. (2018). Registration of ‘Pembroke 2016’ soft red winter wheat. J. Plant Regist. 12, 373–378. 10.3198/jpr2017.12.0089crc [DOI] [Google Scholar]
- Venske E., dos Santos R. S., Farias D., da R., Rother V., da Maia L. C., et al. (2019). Meta-analysis of the QTLome of fusarium head blight resistance in bread wheat: Refining the current puzzle. Front. Plant Sci. 10, 1–19. 10.3389/fpls.2019.00727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges V. L., Van Sanford D., Brown-Guedira G. (2006). Heritability estimates and response to selection for Fusarium head blight resistance in soft red winter wheat. Crop Sci. 46, 1587–1594. 10.2135/cropsci2005.11-0412 [DOI] [Google Scholar]
- Vocke G., Ali M. (2013). U.S. Wheat Production Practices, Costs, and Yields: Variations Across Regions, EIB-116 (U.S. Department of Agriculture, Economic Research Service; ). Available at: https://www.ers.usda.gov/publications/pub-details/?pubid=43786 (Accessed March 3, 2020). [Google Scholar]
- Waldron B. L., Moreno-Sevilla B., Anderson J. A., Stack R. W., Frohberg R. C. (1999). RFLP mapping of QTL for fusarium head blight resistance in wheat. Crop Sci. 39, 805–811. 10.2135/cropsci1999.0011183X003900030032x [DOI] [Google Scholar]
- Walter S., Ali S., Kemen E., Nazari K., Bahri B. A., Enjalbert J., et al. (2016). Molecular markers for tracking the origin and worldwide distribution of invasive strains of Puccinia striiformis. Ecol. Evol. 6, 2790–2804. 10.1002/ece3.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamishe Y. A., Milus E. A. (2004. a). Genes for adult-plant resistance to leaf rust in soft red winter wheat. Plant Dis. 88, 1107–1114. 10.1094/PDIS.2004.88.10.1107 [DOI] [PubMed] [Google Scholar]
- Wamishe Y. A., Milus E. A. (2004. b). Seedling resistance genes to leaf rust in soft red winter wheat. Plant Dis. 88, 136–146. 10.1094/PDIS.2004.88.2.136 [DOI] [PubMed] [Google Scholar]
- Wan A., Chen X. (2014). Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis. 98, 1534–1542. 10.1094/PDIS-01-14-0071-RE [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- Wang H., Sun S., Ge W., Zhao L., Hou B., Wang K., et al. (2020). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435. 10.1126/science.aba5435 [DOI] [PubMed] [Google Scholar]
- Waqar A., Khattak S. H., Begum S., Rehman T., Shehzad A., Ajmal A., et al. (2018). Stripe rust: A 1688 review of the disease, Yr genes and its molecular markers. Sarhad J. Agric. 34, 188–201. 10.17582/journal.sja/2018/34.1.188.201 [DOI] [Google Scholar]
- Ward T. J., Clear R. M., Rooney A. P., O’Donnell K., Gaba D., Patrick S., et al. (2008). An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 45, 473–484. 10.1016/j.fgb.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Ward B. P., Brown-Guedira G., Cowger C., Marshall D., Dong Y. (2019. a). “Machine learning models for predicting deoxynivalenol concentration from grain imaging,” in Proceedings of the 2019 National Fusarium Head Blight Forum, vol. 124 . Eds. Canty S., Hoffstetter A., Wiermer B., Dill-Macky R. (Lexington, KY: East Lansing, MI; ). [Google Scholar]
- Ward B. P., Brown-Guedira G., Tyagi P., Kolb F. L., Van Sanford D. A., Sneller C. H., et al. (2019. b). Multienvironment and multitrait genomic selection models in unbalanced early-generation wheat yield trials. Crop Sci. 59, 491–507. 10.2135/cropsci2018.03.0189 [DOI] [Google Scholar]
- Warth B., Parich A., Bueschl C., Schoefbeck D., Neumann N. K. N., Kluger B., et al. (2015). GC–MS based targeted metabolic profiling identifies changes in the wheat metabolome following deoxynivalenol treatment. Metabolomics 11, 722–738. 10.1007/s11306-014-0731-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B. B. H., Jones J. D. G. (2020). Breeding a fungal gene into wheat. Science 368, 822–823. 10.1126/science.311.5769.1843b [DOI] [PubMed] [Google Scholar]
- Xu X., Bai G., Carver B. F., Shaner G. E., Hunger R. M. (2005. a). Molecular characterization of slow leaf-rusting resistance in wheat. Crop Sci. 45, 758–765. 10.2135/cropsci2005.0758 [DOI] [Google Scholar]
- Xu X. Y., Bai G. H., Carver B. F., Shaner G. E., Hunger R. M. (2005. b). Mapping of QTLs prolonging the latent period of Puccinia triticina infection in wheat. Theor. Appl. Genet. 110, 244–251. 10.1007/s00122-004-1819-1 [DOI] [PubMed] [Google Scholar]
- Xue S., Li G., Jia H., Xu F., Lin F., Tang M., et al. (2010). Fine mapping Fhb4, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 121, 147–156. 10.1007/s00122-010-1298-5 [DOI] [PubMed] [Google Scholar]
- Xue S., Xu F., Tang M., Zhou Y., Li G., An X., et al. (2011). Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 123, 1055–1063. 10.1007/s00122-011-1647-z [DOI] [PubMed] [Google Scholar]
- Yang Z. P., Gilbert J., Somers D. J., Fedak G., Procunier J. D., McKenzie I. H. (2003). Marker assisted selection of Fusarium head blight resistance genes in two doubled haploid populations of wheat. Mol. Breed. 12, 309–317. 10.1023/B:MOLB.0000006834.44201.48 [DOI] [Google Scholar]
- Yang J., Bai G., Shaner G. E. (2005). Novel quantitative trait loci (QTL) for Fusarium head blight resistance in wheat cultivar Chokwang. Theor. Appl. Genet. 111, 1571–1579. 10.1007/s00122-005-0087-z [DOI] [PubMed] [Google Scholar]
- Yi X., Cheng J., Jiang Z., Hu W., Bie T., Gao D., et al. (2018). Genetic analysis of fusarium head blight resistance in CIMMYT bread wheat line C615 using traditional and conditional QTL mapping. Front. Plant Sci. 9, 1–12. 10.3389/fpls.2018.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Jurgenson J. E., Hulbert S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant-Microbe Interact. 24, 554–561. 10.1094/MPMI-10-10-0229 [DOI] [PubMed] [Google Scholar]
- Yu G., Zhang X., Yao J., Zhou M. P., Ma H. (2017). Resistance against Fusarium head blight in transgenic wheat plants expressing the ScNPR1 gene. J. Phytopathol. 165, 223–231. 10.1111/jph.12553 [DOI] [Google Scholar]
- Zhao J., Wang L., Wang Z., Chen X., Zhang H., Yao J., et al. (2013). Identification of eighteen berberis species as alternate hosts of Puccinia striiformis f. sp. tritici and virulence variation in the pathogen isolates from natural infection of barberry plants in China. Phytopathology 103, 927–934. 10.1094/PHYTO-09-12-0249-R [DOI] [PubMed] [Google Scholar]
- Zhao J., Wang M., Chen X., Kang Z. (2016). Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Annu. Rev. Phytopathol. 54, 207–228. 10.1146/annurev-phyto-080615-095851 [DOI] [PubMed] [Google Scholar]
- Zhao M., Wang G., Leng Y., Wanjugi H., Xi P., Grosz M. D., et al. (2018). Molecular mapping of Fusarium head blight resistance in the spring wheat line ND2710. Phytopathology 108, 972–979. 10.1094/PHYTO-12-17-0392-R [DOI] [PubMed] [Google Scholar]
- Zhou W. C., Kolb F. L., Bai G. H., Domier L. L., Boze L. K., Smith N. J. (2003). Validation of a major QTL for scab resistance with SSR markers and use of marker-assisted selection in wheat. Plant Breed. 122, 40–46. 10.1046/j.1439-0523.2003.00802.x [DOI] [Google Scholar]
- Zhu Z., Hao Y., Mergoum M., Bai G., Humphreys G., Cloutier S., et al. (2019). Breeding wheat for resistance to Fusarium head blight in the Global North: China, USA, and Canada. Crop J. 7, 730–738. 10.1016/j.cj.2019.06.003 [DOI] [Google Scholar]