Abstract
Objective
Finding relevant datasets is important for promoting data reuse in the biomedical domain, but it is challenging given the volume and complexity of biomedical data. Here we describe the development of an open source biomedical data discovery system called DataMed, with the goal of promoting the building of additional data indexes in the biomedical domain.
Materials and Methods
DataMed, which can efficiently index and search diverse types of biomedical datasets across repositories, is developed through the National Institutes of Health–funded biomedical and healthCAre Data Discovery Index Ecosystem (bioCADDIE) consortium. It consists of 2 main components: (1) a data ingestion pipeline that collects and transforms original metadata information to a unified metadata model, called DatA Tag Suite (DATS), and (2) a search engine that finds relevant datasets based on user-entered queries. In addition to describing its architecture and techniques, we evaluated individual components within DataMed, including the accuracy of the ingestion pipeline, the prevalence of the DATS model across repositories, and the overall performance of the dataset retrieval engine.
Results and Conclusion
Our manual review shows that the ingestion pipeline could achieve an accuracy of 90% and core elements of DATS had varied frequency across repositories. On a manually curated benchmark dataset, the DataMed search engine achieved an inferred average precision of 0.2033 and a precision at 10 (P@10, the number of relevant results in the top 10 search results) of 0.6022, by implementing advanced natural language processing and terminology services. Currently, we have made the DataMed system publically available as an open source package for the biomedical community.
Keywords: data discovery index, metadata, dataset, information storage and retrieval, information dissemination
INTRODUCTION
With the advances in recent technologies producing large datasets, the bottleneck of biomedical research is shifting from digital data generation to data management and analysis. Large, complex, and diverse data are continually being generated and are accumulating exponentially, becoming valuable sources for biomedical discovery. To take full advantage of existing data, facilitate knowledge discovery, and make scientific discoveries more productive and reproducible, following the widely endorsed FAIR principles (to make data Findable, Accessible, Interoperable, and Reusable)1 is recommended. However, there are unique challenges in collecting and normalizing preexisting experimental data from disparate sources for different purposes.
The biomedical and healthCAre Data Discovery Index Ecosystem (bioCADDIE) project,2 funded by the National Institutes of Health (NIH) via the Big Data to Knowledge program, is focused on the discovery of biomedical datasets. Since its start, researchers, service providers, and knowledge experts around the globe have participated in various aspects of bioCADDIE, such as working groups, pilot projects, and dataset retrieval challenges (https://biocaddie.org/). To instantiate the concepts and recommendations developed by this large community, bioCADDIE developed a prototype data discovery index (DDI) named DataMed, which collects and indexes metadata from broad types of biomedical datasets of interest from heterogeneous sources and makes them searchable through a web-based interface.3 We believe that metadata from diverse datasets can be mapped to a unified representation model, thus enabling more efficient search across domain-specific repositories and making data more discoverable by users. Further details and discussion of the motivations for building DataMed are available here.3 DataMed is available as an open source package, to allow the research community to leverage its technologies to build additional biomedical data indexes. This article describes technical details about developing DataMed, including its metadata ingestion/indexing pipeline and search engine functionalities.
BACKGROUND
To embrace the big data era, the biomedical research community has devoted substantial effort and resources to the goal of enabling biomedical research as a digital enterprise. Making datasets findable is key to promoting the reuse of existing datasets, and major initiatives have been established to build repositories and knowledge bases for specific types of data and domains.4 For example, Gene Expression Omnibus is a public functional genomics data repository for gene expression data.5 Protein Data Bank serves as an information portal for biological macromolecular structures.6 ImmPort is a data repository for public data sharing of immunological studies.7
Such data repositories have greatly improved the discoverability and reuse of datasets, since researchers can easily find datasets from a familiar repository. However, with an increasing number of repositories, search capabilities for different types of data across multiple repositories is needed. Currently, researchers need to search individual repositories, which is time consuming and also limits the ideas that researchers can have when they know the datasets they have access to. An integrated biomedical data retrieval and discovery system across different repositories is a first step toward removing this limitation. A successfully integrated search engine will provide a one-stop shop, where data seekers can quickly access all these resources, improving the community’s capability to utilize existing databases for data query, knowledge dissemination, and integrative analysis.
DDI systems to help users find datasets across multiple repositories exist. The Omics Discovery Index aggregates and indexes “omics” datasets, including 90 729 datasets from 15 repositories.8 Other resources, such as Datacite, provide basic foundations for data discovery for general research data. As of the time of writing, Datacite includes 8 452 860 works and 1287 data centers globally from many different domains.9 The Neuroscience Information Framework10 and the National Institute of Diabetes and Digestive and Kidney Disease Information Network11 are community aggregators focused on specific biomedical domains, assisting researchers in finding data and information such as organisms, reagents, etc. They aggregate information from >230 resources and support efforts such as the Resource Identification Initiative.12 Dataverse, which includes 49 122 datasets, is an open source system for researchers, data authors, publishers, etc., to share, preserve, cite, explore, and analyze research data.13 In the biomedical domain, there is no comprehensive search engine covering a broad spectrum of repositories. Several technical challenges exist when building such an integrated search engine, including extracting and normalizing metadata from different repositories to a unified metadata model,14 as well as finding highly relevant datasets for users in a huge search space.
The mission of DataMed is to provide a DDI to help users efficiently find and access existing datasets that are distributed across a wide range of repositories in the biomedical domain. In DataMed, we developed a metadata ingestion pipeline which extracts, maps, and indexes by following the DatA Tag Suite (DATS)14 based on input from the community and a thorough analysis of existing metadata from popular repositories. We implemented a fully functional search engine for retrieving relevant datasets, with a user-friendly interface and other advanced technologies, such as Elasticsearch,15 natural language processing (NLP), and terminology services. This paper provides a detailed description of DataMed’s architecture and technologies, as well as evaluations of individual components.
METHODS
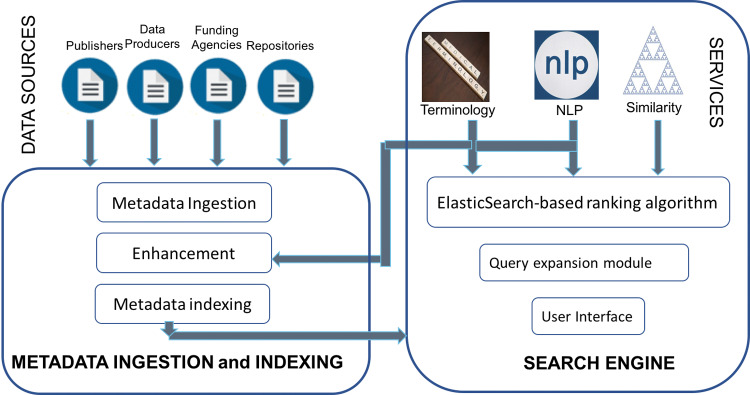
DataMed consists of 2 major components: the ingestion and indexing pipeline and the search engine (Figure 1). The ingestion and indexing pipeline collects metadata from different repositories, maps them to the DATS model, and then indexes them to the Elasticsearch endpoint. The search engine is a web-based application that consists of a user interface and various search functionalities, including the core Elasticsearch-based ranking algorithm, query expansion module utilizing NLP and terminologies, and other advanced services, such as a dataset similarity calculator.
Figure 1.
Architecture of DataMed
Ingestion and indexing pipeline
System architecture
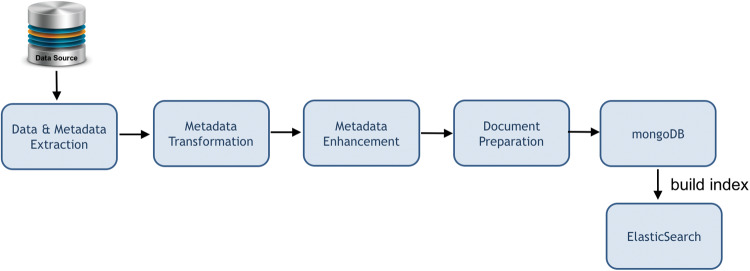
For data ingestion, transformation, and enhancement, DataMed uses a horizontally scalable message oriented extract-transform-load system. As shown inFigure 2, the pipeline is a loosely coupled distributed system consisting of a message dispatcher and one or more data processing components (consumers), with a command line management interface. The dispatcher acts as a hub, orchestrating the data ingestion and processing pipeline using persistent queues. The consumers are managed within a consumer container, wherein each consumer receives a data record wrapper document from the consumer container, does an operation such as a transformation, cleanup, and/or enhancement on the document, and returns the document to the consumer container. The consumer container saves the updated document to the data store (MongoDB), and then places a message in the message queue for the dispatcher. The dispatcher uses the pipeline specification to decide the next step and places a message in the corresponding consumer’s input message queue. All running consumer containers listen to the message queues configured for the consumers they manage and provide the data record wrapper for processing upon receipt of a new message in the input queue in an asynchronous manner.
Figure 2.
Workflow of the ingestion and indexing pipelines
Data ingestion
To handle heterogeneity in the data availability from multiple institutions and laboratories, the pipeline abstracts out retrieval modes (eg, REST API, FTP), data formats (eg, XML, CSV, JSON), and data traversal functionality. Different ingestors for retrieval mode and data format combinations are developed as specific consumers using a specialized plug-in interface. The extent of effort required to develop a new plug-in for a consumer varies. If a new ingest consumer needs to be created from scratch, it takes, with testing, anywhere between half a day (4 h) to multiple days, depending on the complexity. For new enhancement modules that work as consumers, the time required to develop the module also depends on the complexity of the processing required (4 h and up for a simple one). Now, we have developed a number of ingest consumers for different repositories; therefore, for many new sources, we just need to configure a current consumer rather than develop a new ingest consumer for the source. Data traversal is generalized where the iterators allow streaming to retrieve data only when needed, facilitating the processing of datasets much larger than the system memory. Each specific ingestor uses these iterator(s) to retrieve and traverse the records. Each traversed raw data record is converted to JavaScript Object Notation (JSON) format, wrapped in a JSON document with additional pipeline management and provenance information, and stored in a MongoDB database for further processed.
Data transformation
The ingested raw data is transformed into the DATS format using a domain-specific language called JSONTL. The language uses JSONPath, similar to XPath, to specify branches in a JSON object tree. A matching branch from the source JSON document is mapped to a branch in the destination document. Currently, the incoming converted JSON is mapped to the index DATS representation manually for each repository. Thus, the DATS metadata model provides the analogous structures for the curators to map. This mapping can be one-to-one, many-to-one, one-to-many, or many-to-many. To allow all forms of mapping and arbitrary field value manipulation and combination, the language allows embedding scripts written in the Python language to be included in the transformation rules. The language also allows conditional transformations. The transformation engine is integrated into a consumer and run as a part of the processing pipeline by the consumer containers.
Data enhancement
In addition to the original metadata, enhancements to the metadata records are performed by the current DataMed ingestion and indexing pipeline. These include data citation enhancer (which attaches information on citations of the dataset from other resources) and an NLP-based biomedical named entity enhancer (please see the subsection on NLP service for details). Using the NLP enhancement on a metadata record, particularly on longer text descriptions and abstracts, provides a detailed list of semantic concepts contained within the dataset description. This enhanced DATS transformation for each record is stored in the MongoDB and indexed to the Elasticsearch cluster.
Search engine
The DataMed search engine is a PHP-based web application following the MVC (Model, View, and Control) architecture.16 The user interface provides various ways for users to interactively refine their search queries and navigate among returned results. Different search functionalities based on NLP and terminology technologies are implemented to improve the search performance and user experience.
User interface
DataMed provides a Google-like search box query entry as well as an advanced search option allowing expert users to define the search fields and build specialized queries using Boolean operators. The ranked list of relevant results can be further filtered and refined by the user with facets (eg, data types, repository names). Each dataset record provides general information, such as title, description, released data, etc., and link to the dataset in the original data resource for users to access the dataset.
The general view applies across all repositories, while the detailed view for a single repository has additional repository-specific information. The DataMed user interface (UI) also provides many other functions, such as sharing selected datasets via e-mail, downloading them, or storing them as collections using users’ DataMed accounts. We provide additional information, such as similar datasets in DataMed via similar dataset service, and associated publications and grants, by linking to external resources such as PubMed17 and NIH RePORT.18
Search functionalities
Figure 3 shows the workflow of the DataMed search engine. After a user enters a query, the NLP service extracts biomedical concepts that are used to generate synonyms via terminology service. The synonyms are added to the original search query and sent to Elasticsearch to retrieve results. We describe each of the services below.
Figure 3.
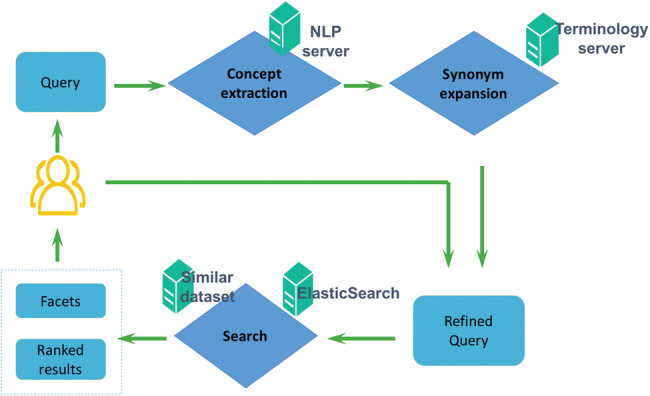
Workflow of the DataMed web application
NLP service: Two different NLP solutions are used to identify biomedical concepts from queries: (1) general Medical Subject Headings concepts are extracted using the existing MetaMap Lite system,19 and (2) specific concepts such as diseases, chemicals, genes, and biological processes are identified using in-house NLP programs. Both rule-based and machine learning-based NLP pipelines developed using the CLAMP NLP toolkit (clamp.uth.edu) are utilized. Identified entities are mapped to Unified Medical Language System (UMLS) Concept Unique Identifiers.20–22 Our NLP service is implemented as (1) web service, used for real-time query expansion, and (2) Java program, used as NLP enhancer in the ingestion pipeline.
Terminology service: The terminology server is based on SciGraph (https://github.com/SciGraph/SciGraph) and Neo4j, via adoption of major ontologies such as Medical Subject Headings,23 SNOMED CT,24 Gene Ontology,25 Foundational Model of Anatomy,26 National Center for Biotechnology Information Taxonomy,27 and Hugo Gene Nomenclature.28 These different terminologies are integrated in the context of the UMLS Metathesaurus to obtain a unified terminology of related terms. A web service supports real-time concept and relationship (eg, synonym and parent-child) identification, which is used along with NLP service for query expansion and metadata enrichment and as a stand-alone component for spelling correction and auto-completion functions.
Ranking algorithm: Currently, DataMed 3.0 uses Elasticsearch’s default ranking algorithm (cosine similarity based on vector space model using TF-IDF weighting29) to retrieve and rank datasets from the entire collection. We are in the process of integrating novel search algorithms from the bioCADDIE dataset retrieval challenge,30 and this should be reflected in future releases.
Similar dataset : The similar dataset service is an iterative variant of the Random Indexing paradigm.31,32 Dataset vectors are composed from word vectors using distributional semantics methods.33 The word vectors undergo transformations prior to being added to the dataset vector, indicating the field in which they occur such that datasets with similar (but not necessarily identical) words in the same field have similar vectors. Numerical fields such as date are encoded into semantic vector representations34,35 such that vectors for datasets published at a similar (but not identical) time are related to one another.36
Evaluation
To evaluate the accuracy of the ingestion pipeline, we randomly collected metadata of 50 datasets and their outputs by the ingestion pipeline – the mapped DATS elements. We manually compared each DATS element with the original metadata records to determine whether it was correctly mapped by the ingestion pipeline. The accuracy, defined as the ratio between the number of correctly mapped DATS elements and the number of total elements, is reported. We also evaluated the scalability of the ingestion pipeline by setting up a test system on the Amazon cloud consisting of 6 EC2 instances: one for the MongoDB database, another for the dispatcher, ActiveMQ messaging server, and consumer container process, and 4 running a consumer container. The consumer containers were serially introduced after at least one hour of processing. We report the number of datasets processed per second after the introduction of each consumer container.
To determine how the DATS elements are represented in biomedical data repositories, we assessed the frequency of each DATS element across repositories. For each DATS element, we manually checked whether a repository contained this element in its metadata records. We then defined the frequency of an element as the ratio between the number of data repositories that contained this element and the number of total repositories that we examined. We report the frequency for each DATS element.
We also evaluated the NLP service using a corpus containing textual description of 700 randomly selected datasets. For each dataset, entities (eg, genes and chemicals) were annotated by domain experts, resulting in a total of 2303 entities. We report performance using precision, recall, and F1-measure. To evaluate the terminology service for generating synonyms, we collected search terms in DataMed with a frequency >10 (156 terms in total) in October 2016 and manually examined the synonyms we obtained from the terminology service. We report the number of terms that have the correct synonyms.
In the bioCADDIE Dataset Retrieval Challenge,30,37 we generated a benchmark dataset, which contains 15 user queries and 20 184 samples with relevance judgments. Using this benchmark dataset, we evaluated DataMed’s ranking algorithm by reporting the inferred average precision (infAP), the inferred normalized discounted cumulative gain (infNDCG), and the precision at 10 (p@10). p@10 is the number of relevant results in the top 10 search results, which is a metric often used in modern information retrieval. We also report infAP, infNDCG, and p@10 for DataMed without synonym expansion of user queries to demonstrate the utility of NLP and terminology services.
RESULTS
As of July 2017, DataMed had ingested 2 336 403 datasets from 74 repositories across 15 data types. Fifteen organizations have submitted requests to DataMed to index their repositories. We are in the process of ingesting more repositories. Regular updates to the index are planned to include recently added datasets and changes of the metadata of datasets (in case of any modifications) in a timely manner. Among different types of ingestors, the Web Ingestor and Database Ingestor are the 2 most widely used across different repositories.
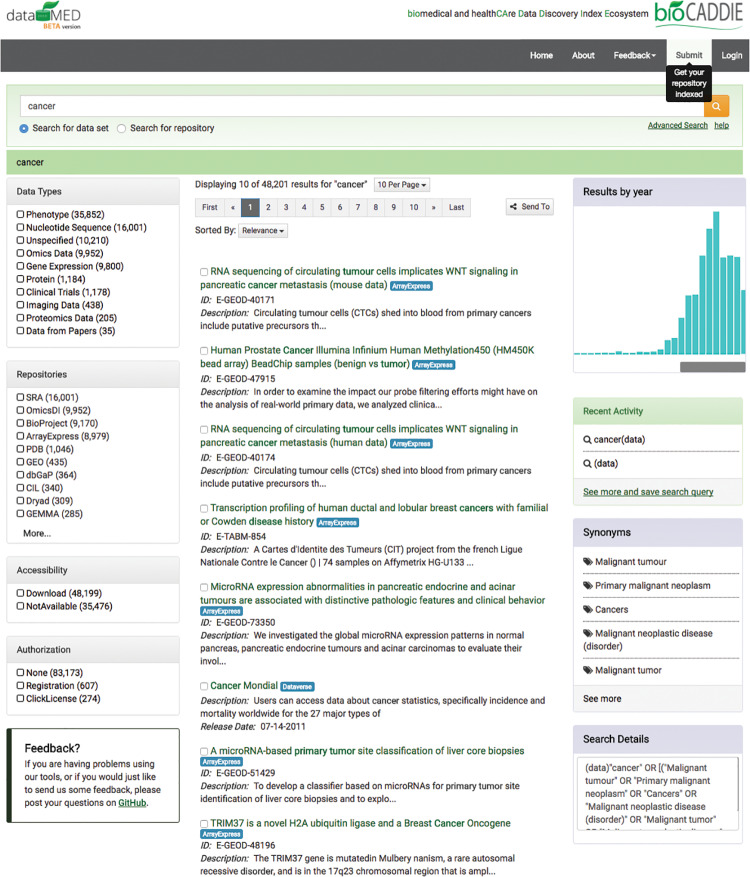
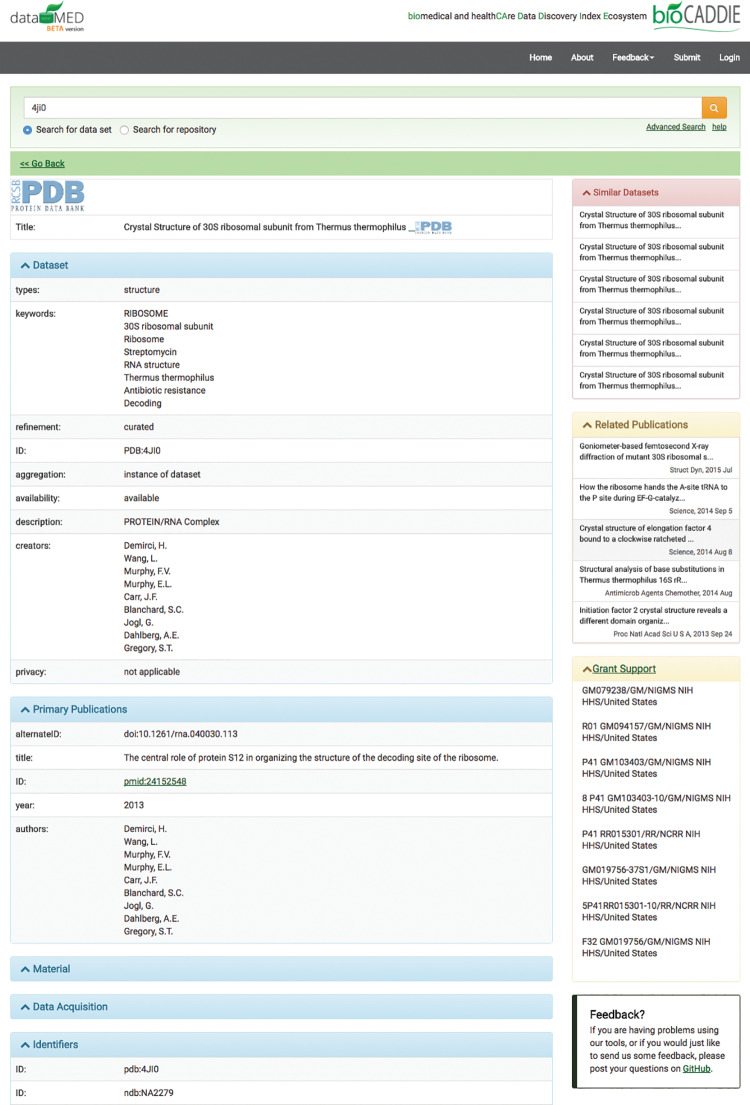
Figure 4 shows a screenshot of the search page in DataMed. In the left column, facets are provided for users to refine search results. The middle column displays the search results. Visualization of dataset release dates via a timeline graph, synonyms, and search query details are shown on the right.Figure 5 shows screenshots of the information page for a selected dataset. Besides the metadata of the dataset, similar datasets, related publications, and grant information are also provided if available.
Figure 4.
Screenshot of search page in DataMed
Figure 5.
Screenshot of a single item page in DataMed
To improve DataMed and monitor usage, a user-activity tracking module is implemented that logs all queries as well as keystrokes and clicks. To further engage users and solicit user input during the development process, DataMed collects feedback (https://datamed.org/feedback.php) via multiple modes: (1) a “Contact Us” form, (2) a System Usability Scale (SUS)-style questionnaire, and (3) an issue-/bug-reporting repository. Feedback from all routes is logged into GitHub, which serves as a central node to track user-reported issues for the development team. To better understand users’ needs, usability studies were also conducted, providing guidance for the iterative development of DataMed.38
The evaluation exercise showed that among 1361 DATS elements from the 50 randomly chosen datasets, 1225 were correctly identified, indicating an accuracy of 90% for the ingestion pipeline. In the scalability testing, the number of datasets processed per second shows a linear increase with each additional consumer container computation unit, demonstrating that the ingestion pipeline is computationally scalable.
Table 1 shows the frequencies of core DATS elements across repositories. Frequencies of all DATS element are provided inSupplementary Table S1. Elements such as Dataset Title and Data Repository Name show high frequencies >85%. However, some data fields, such as Grant Name and Software Name, have very low frequencies, indicating a need to link these external resources.
Table 1.
Frequency of core DATS elements
| DATS legend | Core DATS element | Frequency (%) |
|---|---|---|
| Dataset | Title | 86 |
| Types | 73 | |
| Creators | 52 | |
| Grant | Name | 4 |
| Dimension | Name | 11 |
| Type | 4 | |
| Data repository | Name | 89 |
| Organization | Name | 84 |
| Software | Name | 2 |
Table 2 shows the performance of the NLP engine on recognizing different types of biomedical entities. The overall precision, recall, and F-measures are 91.88%, 71.38%, and 79.66%, respectively, on 700 randomly selected datasets. For synonym generation by the terminology service, out of the total 156 search terms examined, 124 returned the correct synonyms, achieving an accuracy rate of 79.49%.
Table 2.
NER results of the NLP system
| Evaluation metrics | Gene (%) | Disease (%) | Chemical (%) | Biological process (%) | Overall (%) |
|---|---|---|---|---|---|
| Precision | 89.95 | 92.54 | 91.08 | 93.96 | 91.88 |
| Recall | 50.75 | 88.89 | 69.06 | 76.80 | 71.38 |
| F-score | 64.89 | 90.68 | 78.55 | 84.52 | 79.66 |
| No. of Entities | 670 | 684 | 462 | 487 | 2303 |
On the benchmark dataset generated for the bioCADDIE dataset retrieval challenge,30,37 the current ranking algorithm of DataMed (with query expansion by NLP and terminology services) has an infAP of 0.203, an infNDCG of 0.354, and a p@10 of 0.602, which are lower than the best entry in the challenge (infAP, infNDCG, and p@10 of 0.147, 0.513, 0.760), indicating a need to improve the ranking algorithm. When we disabled the query expansion function, infAP, infNDCG, and p@10 dropped to 0.098, 0.259, and 0.468, respectively.
DataMed was first publicly launched on June 30, 2016. Since then, we have been tracking the traffic to DataMed using Google Analytics. About a year later (July 2017), and still in a prototype stage, DataMed had attracted 11 144 users from 126 countries.
DISCUSSION
Increased availability of digital data and growing multidomain research areas in the biomedical field have created a need for users from differing disciplines to find and retrieve datasets that are not from their direct areas of expertise. Therefore, toolsets that provide a coherent presentation of metadata information across repositories housing biomedical datasets are important for data search and access. DataMed is one of the first data discovery indexes that harvest metadata from a broad range of data providers and make it available through a single integrated search system. The proximal goal of DataMed is to develop the capability to search across datasets from different repositories, which it has achieved. The depth in search capabilities achieved by domain-specific search engines forms the next step that the DataMed team would like to pursue, and further evaluation is needed to validate its usefulness.
On account of the diverse nature of metadata fields and repositories, some of the issues arising during the development of DataMed overlap with those of data integration efforts aiming to link information across heterogeneous datasets (for a review, see39). The application of knowledge resources such as ontologies and semantic web technologies to draw connections across disparate datasets has been an active area of research over the past decade (eg, see40–42). The goals of these efforts have generally involved developing an integrated data model to permit reasoning across data drawn from different sources. In contrast, DataMed aims to facilitate search across a broad range of inconsistently indexed data repositories. However, many of the technologies and approaches used to facilitate dataset integration are also pertinent to dataset indexing for information retrieval purposes, as is evident in DataMed’s utilization of a common data model (DATS14) for metadata fields, and the application of NLP and terminology services to attempt to map between different expressions of related concepts. Our project is not the first to apply these technologies to dataset retrieval. One line of related research concerns the mapping of dataset metadata to concepts in the UMLS,43–46 including at times using the resulting annotations to draw novel inferences.47 As the primary goal of the DataMed prototype is to facilitate search and retrieval across a large number of repository types, it differs from these efforts, in both its focus and scope.
DataMed uses a modular architecture and supports use cases from the general, biological, and translational communities with a number of common needs for metadata searches. This makes DataMed broad (rather than deep) in its search capabilities, allowing it to easily span a range of diverse domains and types of data. There are other centralized repositories, such as the Omics Discovery Index,8 developed by the biomedical community that address needs that are specific to one or few of the various data types of biomedicine. DataMed ingests such aggregators (ie, other indices) in addition to the repositories that house the datasets themselves in an effort to provide a comprehensive overview of all the available information for a particular dataset. However, such data aggregators have not yet been developed for the majority of biomedical areas, so future efforts could involve developing a deeper index for each specialized domain.
DataMed is still a prototype and a work in progress. There are limitations to the system. Since DataMed was designed to cover a broad range of repositories, there is no in-depth indexing of the different repositories yet. DATS is also generalized and broad. Therefore, DataMed in its current state cannot address use cases that require detailed, granular metadata to answer queries. We are developing extendable attributes in DATS that can be used for deeper search, eg, finding datasets with certain variables. Furthermore, the current ranking algorithm of DataMed is a relatively baseline system. We are exploring innovative search strategies (such as learning-to-rank) that arose from the Dataset Retrieval Challenge30 for integration into DataMed.
The DataMed team has also worked toward exposing the harvested metadata to other general search applications (eg, Google, Yahoo, Bing, etc.) by providing schema.org markups. We have developed a RESTful API to provide programmatic access to all of DataMed’s harvested metadata and additional user-interface features. Such sharing capabilities make DataMed easily accessible not only to the biomedical community, but also to those outside of the biomedical sphere.
We are developing a proof-of-concept module to link DataMed to external resources. For example, DataMed will use a plug-in to access remotely located databases and tools from the Library of Integrated Network-based Signatures Data Coordination and Integration Center (http://lincs-dcic.org/#/) as a visualization tool for the results of user-requested data. Reusing components like these not only provides additional data usability options, but also significantly reduces duplication of programming effort and development costs.
The consortium approach to development has allowed DataMed to receive input from various community members to produce a reusable, modular, portable, robust, feature-rich application. The DataMed team is continuously improving the quality of the index and expanding the scope of the ingested repositories. Evaluation of the DATS model and other DataMed tools, not only by the bioCADDIE team but also by the community, is critical for the long-term sustenance of the initiative. To this end, the team has made the codes available to the community as an open-source package in GitHub (https://github.com/biocaddie), as well as datasets such as the benchmark dataset from the Dataset Retrieval Challenge.37 We are not only providing information about the existence and usefulness of the tool to the biomedical community, but are also actively pursuing different strategies for long-term sustenance and maintenance of DataMed.
CONCLUSIONS
DataMed leverages scalable technologies to ingest, index, and search diverse biomedical datasets across repositories. It demonstrates a successful prototype for building an integrated dataset search engine for the biomedical domain. Its flexible service oriented architecture and the open source nature make it valuable for building other data discovery indexes in the biomedical domain.
Competing interest
The authors declare no competing financial interests.
Author contributions
LO-M, IF, JG, HX, HK, SS, RM, IF, GA, and TJ supervised the research. XC, BO, AG, TC, LO-M, JG, and HX wrote the manuscript. XC, BO, ES, TC, RL, FT, YL, NZ, AG, PR-S, AG-B, DR, MJ, and CF performed the research and analyzed the data.
SUPPLEMENTARY MATERIAL
Supplementary material is available atJournal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
This project is funded by grant U24AI117966 from the NIH National Institute of Allergy and Infectious Diseases as part of the Big Data to Knowledge program. We thank all members of the bioCADDIE community for their valuable input on the overall project.
References
- 1. Wilkinson MD,Dumontier M,Aalbersberg IJJ,et al. The FAIR Guiding Principles for scientific data management and stewardship.Scientific Data. 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lucila O-M,Alter G,Fore I,Martone M,Sansone S-A,Xu H.. bioCADDIE White Paper – Data Discovery Index.2015. figshare. 10.6084/m9.figshare.1362572. Accessed August 3, 2016. [DOI] [Google Scholar]
- 3. Ohno-Machado L,Sansone S-A,Alter G,et al. DataMed: Finding useful data across multiple biomedical data repositories.Nature Genet. 2017;49:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. NIH Data Sharing Repositories.www.nlm.nih.gov/NIHbmic/nih_data_sharing_repositories.html. Accessed August 15, 2017. [Google Scholar]
- 5. Edgar R,Domrachev M,Lash AE. Gene expression Omnibus: NCBI gene expression and hybridization array data repository.Nucleic Acids Res. 2002;301:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berman HM,Westbrook J,Feng Z,et al. The Protein Data Bank.Nucleic Acids Res. 2000;281:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhattacharya S,Andorf S,Gomes L,et al. ImmPort: disseminating data to the public for the future of immunology.Immunol Res. 2014;58(2–3):234–39. [DOI] [PubMed] [Google Scholar]
- 8. Perez-Riverol Y,Bai M,da Veiga Leprevost F,et al. Discovering and linking public omics data sets using the Omics Discovery Index.Nat Biotechnol. 2017;355:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brase J. DataCite – A Global Registration Agency for Research Data. In2009 Fourth International Conference on Cooperation and Promotion of Information Resources in Science and Technology.2009 10.1109/COINFO.2009.66. [DOI] [Google Scholar]
- 10. Bandrowski AE,Cachat J,Li Y,et al. A hybrid human and machine resource curation pipeline for the Neuroscience Information Framework.Database,2012: bas005. 10.1093/database/bas005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whetzel PL,Grethe JS,Banks DE,et al. The NIDDK information network: a community portal for finding data, materials, and tools for researchers studying diabetes, digestive, and kidney diseases.PLOS ONE. 2015;109 10.1371/journal.pone.0136206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bandrowski A,Brush M,Grethe JS,et al. The resource identification initiative: a cultural shift in publishing.F1000Res. 2015;4:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King G. An Introduction to the Dataverse Network as an Infrastructure for Data Sharing.Soc Methods Res. 2007;36:173–99. [Google Scholar]
- 14. Sansone SA,Gonzalez-Beltran A,Rocca-Serra P,et al. DATS, the data tag suite to enable discoverability of datasets.Sci Data. 2017;4:170059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuć R,Rogozinski MR. ElasticSearch Server. Birmingham, UK: Packt Publishing Ltd.;2013. [Google Scholar]
- 16. Cui W,Huang L,Liang L,Li J,et al. The Research of PHP Development Framework Based on MVC Pattern. In:2009 Fourth International Conference on Computer Sciences and Convergence Information Technology.2009. DOI: 10.1109/ICCIT.2009.130. [DOI] [Google Scholar]
- 17. PubMed Entrez Programming Utilities.2017www.ncbi.nlm.nih.gov/home/develop/api/. Accessed August 15, 2017. [Google Scholar]
- 18. Research Portfolio Online Reporting Tools (RePORT).2015https://exporter.nih.gov/ExPORTER_Catalog.aspx. Accessed August 15, 2016. [Google Scholar]
- 19. Demner-Fushman D,Rogers WJ,Aronson AR. MetaMap Lite: an evaluation of a new Java implementation of MetaMap.J Am Med Inform Assoc. 2017;244:841–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu J,Wu Y,Zhang Y,et al. UTH-CCB@BioCreative V CDR Task: Identifying Chemical-induced Disease Relations in Biomedical Text. In:Fifth BioCreative Challenge Evaluation Workshop.2015:254–259. [Google Scholar]
- 21. Binns D,Dimmer E,Huntley R,et al. QuickGO: a web-based tool for Gene Ontology searching.Bioinformatics. 2009;2522:3045–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaewphan S,Van Landeghem S,Ohta T,et al. Cell line name recognition in support of the identification of synthetic lethality in cancer from text.Bioinformatics. 2016;322:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rogers FB. Medical subject headings.Bull Med Libr Assoc. 1963;51:114–16. [PMC free article] [PubMed] [Google Scholar]
- 24. International Health Terminology Standards Development Organisation.History Of SNOMED CT.www.snomed.org/snomed-ct/what-is-snomed-ct/history-of-snomed-ct. Accessed August 15, 2017. [Google Scholar]
- 25. Harris MA,et al. The Gene Ontology (GO) database and informatics resource.Nucleic Acids Res. 2004;32(Database issue):D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Structural Informatics Group.Foundational Model of Anatomy.2017http://si.washington.edu/projects/fma. Accessed August 10, 2017. [Google Scholar]
- 27. Federhen S.The NCBI Taxonomy database.Nucleic Acids Res. 2012;40(Database issue):D136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gray KA,et al. Genenames.org: the HGNC resources in 2015.Nucleic Acids Res. 2015;43(Database issue):D1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elasticsearch.Theory Behind Relevance Scoring.www.elastic.co/guide/en/elasticsearch/guide/current/scoring-theory.html. Accessed August 14, 2017. [Google Scholar]
- 30. Roberts K,Gururaj AE,Chen X,et al. Information retrieval for biomedical datasets: the 2016 bioCADDIE dataset retrieval challenge.Database 2017:bax068. 10.1093/database/bax068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cohen T,Schvaneveldt R,Widdows D. Reflective Random Indexing and indirect inference: a scalable method for discovery of implicit connections.J Biomed Inform. 2010;432:240–56. [DOI] [PubMed] [Google Scholar]
- 32. Kanerva P,Kristoferson J,Holst A. Random indexing of text samples for latent semantic analysis.Proc 22nd Annual Conf Cogn Sci Soc. 2000;22https://escholarship.org/uc/item/5644k0w6. [Google Scholar]
- 33. Cohen T,Widdows D. Empirical distributional semantics: methods and biomedical applications.J Biomed Inform. 2009;422:390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen T,Widdows D,Wahle M,Schvaneveldt. Orthogonality and Orthography: Introducing Measured Distance into Semantic Space. InQuantum Interaction: 7th International Conference. Leicester, UK,July 25–27, 2013. Selected Papers,Atmanspacher H,et al. , ed. Berlin, Heidelberg: Springer;2014:34–46. [Google Scholar]
- 35. Widdows D,Cohen T. Graded semantic vectors: an approach to representing graded quantities in generalized quantum models. InQuantum Interaction: 9th International Conference, Filzbach, Switzerland,July 15–17, 2015Revised Selected Papers,Atmanspacher H,Filk T,Pothos E, ed. Cham, Switzerland: Springer International Publishing;2016:231–44. [Google Scholar]
- 36. Widdows D,Cohen T. The Semantic Vectors Package: New Algorithms and Public Tools for Distributional Semantics. In2010 IEEE Fourth International Conference on Semantic Computing.2010. DOI: 10.1109/ICSC.2010.94. [DOI] [Google Scholar]
- 37. Cohen T,Roberts K,Gururaj AE,et al. A Publicly Available Benchmark for Biomedical Dataset Retrieval: The Reference Standard for the 2016 bioCADDIE Dataset Retrieval Challenge.Database 2017: bax061. 10.1093/database/bax061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dixit R,Rogith D,Narayana V,et al. User needs analysis and usability assessment of DataMed–a biomedical data discovery index.J Am Med Inform Assoc. 2017;253:337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Louie B,Mork P,Martin-Sanchez F,Halevy A,Tarczy-Hornoch P. Data integration and genomic medicine.J Biomed Inform. 2007;401:5–16. [DOI] [PubMed] [Google Scholar]
- 40. Belleau F,Nolin MA,Tourigny N,Rigault P,Morissette J. Bio2RDF: towards a mashup to build bioinformatics knowledge systems.J Biomed Inform. 2008;415:706–16. [DOI] [PubMed] [Google Scholar]
- 41. Noy NF,Shah NH,Whetzel PL,et al. BioPortal: ontologies and integrated data resources at the click of a mouse.Nucleic Acids Res. 2009;37(Web Server issue):W170–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen B,Dong X,Jiao D,et al. Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data.BMC Bioinformatics. 2010;11:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology.Nucleic Acids Res. 2004;32(Database issue):D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jonquet C,Musen M,Shah N. A system for ontology-based annotation of biomedical data.Proceedings of the 5th International Workshop on Data Integration in the Life Sciences. 2008:144–52. [Google Scholar]
- 45. Shah NH,Jonquet C,Chiang AP,Butte AJ,Chen R,Musen MA. Ontology-driven indexing of public datasets for translational bioinformatics.BMC Bioinformatics. 2009;10(Suppl 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Doan S,Lin KW,Conway M,et al. PhenDisco: phenotype discovery system for the database of genotypes and phenotypes.J Am Med Inform Assoc. 2014;211:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Butte AJ,Kohane IS. Creation and implications of a phenome-genome network.Nat Biotechnol. 2006;241:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.