Abstract
Objective
To assess clinical documentation variations across health care institutions using different electronic medical record systems and investigate how they affect natural language processing (NLP) system portability.
Materials and Methods
Birth cohorts from Mayo Clinic and Sanford Children’s Hospital (SCH) were used in this study (n = 298 for each). Documentation variations regarding asthma between the 2 cohorts were examined in various aspects: (1) overall corpus at the word level (ie, lexical variation), (2) topics and asthma-related concepts (ie, semantic variation), and (3) clinical note types (ie, process variation). We compared those statistics and explored NLP system portability for asthma ascertainment in 2 stages: prototype and refinement.
Results
There exist notable lexical variations (word-level similarity = 0.669) and process variations (differences in major note types containing asthma-related concepts). However, semantic-level corpora were relatively homogeneous (topic similarity = 0.944, asthma-related concept similarity = 0.971). The NLP system for asthma ascertainment had anF-score of 0.937 at Mayo, and produced 0.813 (prototype) and 0.908 (refinement) when applied at SCH.
Discussion
The criteria for asthma ascertainment are largely dependent on asthma-related concepts. Therefore, we believe that semantic similarity is important to estimate NLP system portability. As the Mayo Clinic and SCH corpora were relatively homogeneous at a semantic level, the NLP system, developed at Mayo Clinic, was imported to SCH successfully with proper adjustments to deal with the intrinsic corpus heterogeneity.
Keywords: documentation variation, natural language processing, portability, asthma, electronic medical records
BACKGROUND AND SIGNIFICANCE
The rapid adoption of electronic medical records (EMRs) provides a good opportunity to leverage clinical data for clinical research. Natural language processing (NLP), which can convert unstructured text to a structured format, has been shown to be a promising way to automate chart review to enable large-scale research studies that require information embedded in clinical narratives where labor-intensive manual chart review is infeasible.1–4
NLP techniques have been successfully applied in various clinical applications, including medication information extraction,5 patient medical status identification,6–8 sentiment analysis,9 decision support,10,11 genome-wide association studies,12,13 and diagnosis code assignment.14,15 Over the past decade, multiple clinical NLP systems have been developed and deployed, such as cTAKES,16 YTEX,17 MTERMS,18 HiTEXT,19 MedLEE,20 and MedTagger.21,22 Although clinical NLP systems have proven to be successful at various tasks, their performance often varies across institutions and sources of data.23,24
Clinical practice and workflow vary across institutions, which results in different practice settings and reporting schemes for generating EMRs (ie, process variation). Also, it has been demonstrated that clinical language is not homogeneous, but instead consists of heterogeneous sublanguage characteristics (eg, syntactic variation25–29 or semantic variation30,31). Those variations need to be considered in the tool development process.32 Implementation of an EMR system differs among institutions, and the clinical corpus also differs in its nature. That being said, questions arise whenever an NLP system developed in one corpus is applied to another corpus, such as: How similar are the 2 corpora? If 2 corpora differ, how does the difference affect NLP system portability?
A previous study suggested that the performance of a clinical NLP system may depend on the source of the clinical notes. Thus, the semantic and contextual information of the target notes should be seriously considered in the tool development process.32 Liu et al.33 compared the performance of an existing NLP system for smoking status across institutions and showed that customization was necessary to achieve desirable performance. Similarly, Carroll et al.34 studied the portability of a phenotype algorithm using NLP for medical concept extraction in identifying rheumatoid arthritis. They found that the number of NLP-derived data elements varied among institutions and thus normalized it by International Classification of Diseases, Ninth Revision codes to produce consistent performance. Mehrabi et al.35 developed a rule-based NLP system to identify patients with a family history of pancreatic cancer and showed that a rule-based NLP system relying on specific information extraction was portable across institutions. Additionally, Liu et al.36 emphasized the importance of a semantic lexicon for extracting and encoding clinical information from EMRs to achieve semantic interoperability in developing NLP systems. Most studies of NLP system portability showed the validity of portability by comparing system performance but lacked a systematic analysis of the heterogeneity of the EMR corpus (ie, clinical documentation variations) among institutions and its impact on portability. A systematic comparison of EMR corpus characteristics across institutions would help us better understand the portability of NLP systems.
Mayo Clinic developed an NLP system2 that processes clinical notes and automatically ascertains asthma status based on predetermined asthma criteria (PAC).37 The original system has been restructured as the open source information extraction framework MedTaggerIE,21 one of the 3 components in MedTagger,21,22 and has been significantly refined to improve its performance.1 The current system extracts asthma-related episodes or events from clinical notes and applies expert rules to determine a patient’s asthma status.
In this study, we examined the clinical corpus of asthma birth cohorts using the different EMR systems at Mayo Clinic and Sanford Children’s Hospital (SCH) in 3 aspects: (1) overall corpus at the word level (ie, lexical variation), (2) topics and asthma-related concepts (ie, semantic variation), and (3) clinical note types (ie, process variation). We compared those statistics and explored the NLP system’s portability for asthma ascertainment.
METHODS AND MATERIALS
This study was approved by the Mayo Clinic and SCH institutional review boards.
Measures of clinical documentation variations
Variations in clinical documentation across institutions can be simply examined by basic corpus statistics, such as total number of documents/tokens, frequency of medical concepts, note types, and sections. Further, we explored corpus similarities between Mayo Clinic and SCH in terms of: (1) entire corpus, (2) medical concepts of interest, and (3) note types. For corpus and note similarity, we preprocessed the documents by using tokenization, removing stop words, and stemming. We created a vector space model for each case to compare similarities.
For each case, we calculated the cosine similarity, a measure between 2 vectors that quantifies the angle between them. It measures the orientation of 2 vectors, not magnitude, and is commonly used in high-dimensional positive space (eg, text mining) bounded in [0, 1], with 0 indicating orthogonality (decorrelation) and 1 meaning exactly the same. Thus, cosine similarity is useful to show how 2 documents or corpora are alike in terms of their subject matter.38 The detailed descriptions of a vector representation are as follows:
(1)Corpus: The entire corpus of each institution was compared as a whole using (term frequency–inverse document frequency), (term frequency–inverse patient frequency), and latent topic–based vector. In the setting, each corpus was represented by a normalized vector whose element consisted of the for each termt, defined by the summation of for all documents in the corpus divided by the total number of documents in the corpus, ie,, where
The is a variation oftf − idf, designed to view the word distribution weighted at the patient level instead of the document level, in order to reflect the significance of words across patients. The for the termt is defined by the summation of for all patients in the corpus divided by the total number of patients in the corpus, ie,, where
In order to compare the corpora by topic, we employed latent Dirichlet allocation39,40 to generate the document distributions in the topic space, ie,. The topic for the corpusC is defined as:
(2)Medical concepts: The asthma-related concepts used in the PAC (Figure 1) were extracted and compared. Each concept consists of the corresponding keywords. A vector representation of asthma-related concepts for each corpus was created using the definition of (concept frequency–inverse document frequency) and (concept frequency–inverse patient frequency). The for the conceptc is defined by where
Figure 1.
Predetermined asthma criteria (PAC).
The for the conceptc is defined by, where and are defined as the same as above, but replacing document with patient frequency.
(3)Note types: Clinical documents have various note types based on the event (eg, admission, discharge, progression). Although there is commonality of note types among institutions, the detailed definitions of note types may differ and the same note type may contain heterogeneous topics. Therefore, it is interesting to compare topic distributions of note types between the 2 institutions. Similar to the latent topic vector representations for the corpus, the topic for the clinical note typeT is defined by
where is the number of documents in the note type.
A case study of NLP system portability in asthma birth cohorts
We examined clinical documentation variations in asthma birth cohorts of Mayo Clinic and SCH, which use different EMR systems, and compared the performance of the NLP asthma ascertainment system on the 2 corpora.
Patient cohorts
The patient cohorts used for corpus analysis were randomly selected from the birth cohort at each institution (n = 298 for each); the SCH cohort was born between 2011 and 2012 (male 54%), and the Mayo Clinic cohort was born between 1997 and 2007 (male 50%). As the Mayo Clinic cohort was followed up longer (median = 11.5 years, SD = 3.3) than the SCH cohort (median = 2.3 years, SD = 0.35), we adjusted the last follow-up date of Mayo Clinic subjects to match the same age as the SCH cohort in order to avoid age bias; ie, Mayo clinical notes were selected up to a certain date to match age with SCH.
EMR system at Mayo Clinic vs SCH
Mayo Clinic uses an in-house EMR system originated from GE. Mayo’s clinical notes consist of predefined sections and each section contains specific content. SCH uses an Epic EMR system. Although there are section templates in the SCH EMR system, use of templates is not mandated. Therefore, there are many variations in section names and content. In this study, we used clinical notes exported as plain-text files from both EMR systems.
NLP asthma ascertainment system
The NLP asthma ascertainment system (NLP-PAC system) implements the predetermined asthma criteria, which are based on presence/absence of asthma-related concepts (Figure 1). It has been developed as an alternative to labor-intensive manual chart review and also to overcome noncomprehensive asthma identification by conventional approaches using structured data, such as International Classification of Diseases, Ninth and Tenth Revisions codes.41–43 The NLP-PAC system was implemented in MedTaggerIE,21 a resource-driven open source information extraction framework built under Apache Unstructured Information Management Architecture, which separates domain-specific NLP knowledge engineering from the generic NLP process. Domain-specific knowledge (ie, asthma-related events and episodes; concepts with shade inFigure 1) was defined in the customizable external resources and extracted from clinical notes using regular expression-based pattern match rules, assertion status (eg, nonnegated, associated with patients), and section constraint (eg, diagnosis section for physician-diagnosed asthma). Then, expert rules were implemented in a rule-engine program to ascertain asthma status (ie, asthma vs nonasthma) based on predetermined asthma criteria (seeFigure 1).
The predetermined asthma criteria were originally developed by Yunginger et al.37 and have been used extensively in research on asthma epidemiology.44–48 In our study, probable and definite asthma types were combined, because most probable asthma becomes definite over time.37,49
Comparison of NLP asthma ascertainment
The NLP-PAC system was applied to both Mayo Clinic and SCH corpora, and the performance of asthma ascertainment against the gold standard (ie, manual chart review) was compared in sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), andF-score.
RESULTS
We examined basic corpus statistics and corpus similarities between Mayo Clinic and SCH for a given study cohort (n = 298 for each institution) in terms of: (1) overall corpus at the word level (ie, lexical variation), (2) topics and asthma-related concepts (ie, semantic variations), and (3) clinical note types (ie, process variations). Corpus similarities were measured by the cosine similarity described earlier. The performance of the NLP-PAC system on both cohorts was also reported.
Corpus statistics of Mayo Clinic vs SCH
The basic corpus statistics of Mayo Clinic and SCH are included inTable 1. SCH has a larger number of documents than Mayo Clinic. SCH has certain note types (eg, Care Planning, Patient Instructions, and Clinical Team, consisting of 29% of the total documents) that are not used by Mayo Clinic; however, they are represented as sections within Mayo clinical notes. Also, there are multiple same note type documents in SCH, with a little extra or different text content, created on the same date but at different times, which is not the case at Mayo Clinic. These facts explain the major difference in total number of documents between the 2 institutions. SCH has a greater median number of asthma-related concepts per patient than Mayo Clinic. However, Mayo Clinic has greater median number of tokens and asthma-related concepts per document than SCH.
Table 1.
Corpus statistics of Mayo Clinic and SCH (n = 298 patients each)
| Category | Mayo | SCH |
|---|---|---|
| Total no. of documents | 9604 | 30 589 |
| Total no. of tokens | 2 212 389 | 10 117 963 |
| No. of documents/patient, median (IQR) | 27 (18) | 80 (69.8) |
| No. of tokens/document, median (IQR) | 186 (210) | 103 (331) |
| No. of asthma-related conceptsa/patient, median (IQR) | 19.5 (32.8) | 65.5 (88) |
| No. of asthma-related concepts/document, median (IQR) | 2 (3) | 1 (2) |
| No. of note types | 16 | 32 |
| No. of sections | 17 | 54 |
aEach concept consists of a set of keywords. IQR: interquartile range.
SCH has more segmented note types (eg, Anesthesia Pre-Procedure Evaluation, Anesthesia Post-Procedure Evaluation, Anesthesia Transfer of Care) than Mayo Clinic and thus has a higher number of note types. We were not able to obtain the exact SCH section statistics, since the section is not explicitly defined in the clinical notes. Instead, we examined frequent patterns of potential sections (eg, OBJECTIVE:, SUBJECTIVE:, EXAM:), parsed those sections in clinical notes, and obtained section statistics. It should be noted that these statistics represent only this cohort.
Figure 2 shows the distribution of asthma-related concepts (from PAC inFigure 1 and found in clinical notes in this study) between Mayo Clinic and SCH. The concept “cough” appears most frequently at both institutions. At Mayo Clinic, “asthma,” “dyspnea,” and “wheezing” concepts have similar distribution, but at SCH, “wheezing” is a lot more prevalent.Figures 3 and4 show the distributions of note types and sections that contain any asthma-related concepts, respectively, for the 2 institutions. At Mayo Clinic, the Limited Exam note contains more than half of the asthma-related concepts. At SCH, 3 note types, Progress Note, Telephone Encounter, and Patient Instruction, contain most of the asthma-related concepts. However, it should be noted that asthma-related concepts in Patient Instruction do not represent actual medical conditions a patient presents, but are provided as an educational guide to handle possible events. As shown inFigure 4, major sections containing most asthma-related concepts differ between the 2 institutions, although History of Present Illness and the “plan” section (Impression/Report/Plan at Mayo Clinic and Plan at SCH) are used at both institutions.
Figure 2.
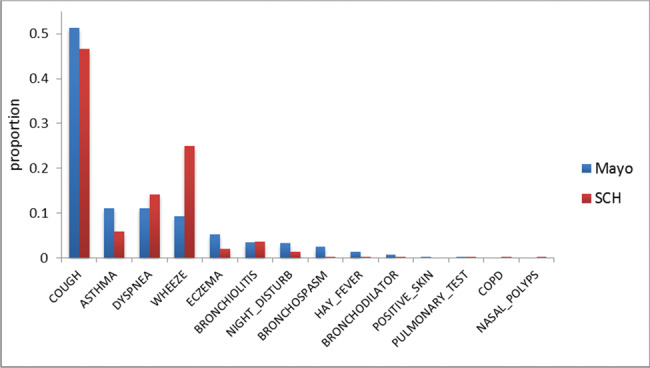
Distribution of asthma-related concepts.
Figure 3.
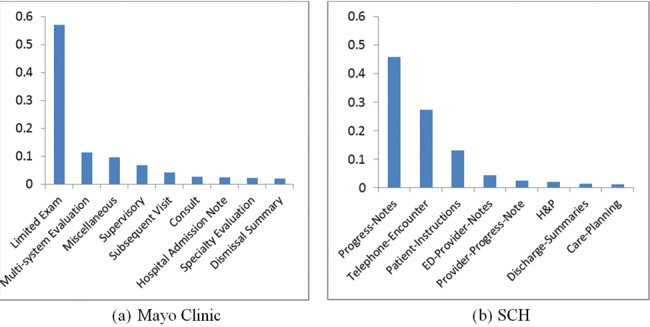
Note types that contain asthma-related concepts (y axis is proportion of asthma-related concepts; includes note types with proportion ≥ 0.01).
Figure 4.
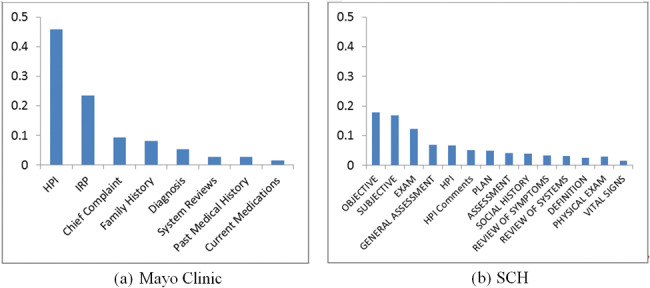
Sections that contain asthma-related concepts (y axis is proportion of asthma-related concepts; includes sections with proportion ≥ 0.01).
Corpus similarities between Mayo Clinic and SCH
The similarities between Mayo Clinic and SCH corpora were examined in terms of the whole corpus and asthma-related concepts (Table 2). Although the word-level similarity (ie, corpustf − idf andtf − ipf) was mediocre, the topic similarity (ie, semantic similarity) of the 2 corpora was high (0.944). The similarity of asthma-related concepts (ie, semantic similarity) between the 2 institutions was 0.971 fortf − idf and 0.855 fortf − ipf. Since asthma-related concepts can be considered as a topic, we did not measure the topic similarity of asthma-related concepts.
Table 2.
The similarities of Mayo Clinic and SCH corpora
| Data source | tf − idf | tf − ipf | Topic |
|---|---|---|---|
| Whole corpus | 0.669 | 0.581 | 0.944 |
| Asthma-related concepts | 0.971 | 0.855 | NA |
NA: not applicable.
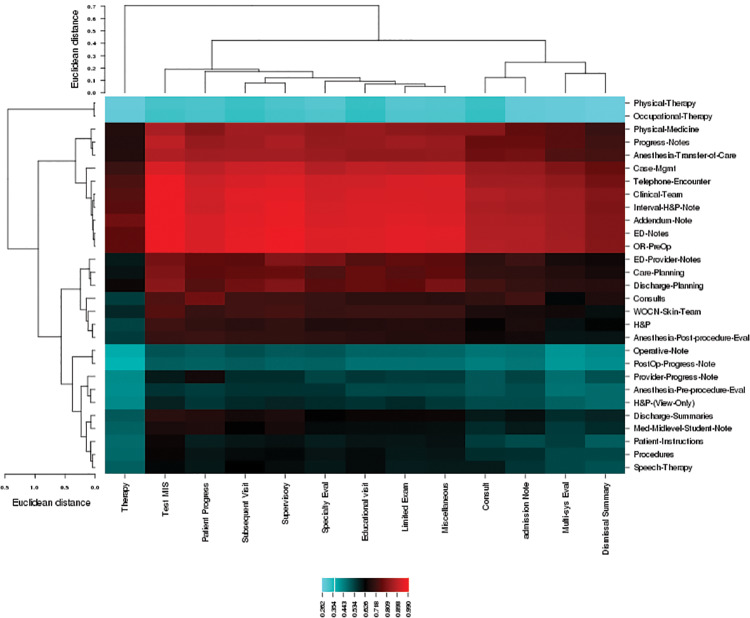
The similarity of note types (ie, process similarity) between Mayo Clinic and SCH corpora was compared based on topics. The note types with frequency ≥20 were compared and a clustered image map (ie, heat map) was plotted. InFigure 5, the color in the cell represents the degree of topic similarity (ie, cosine similarity) of the 2 note types. Euclidean distance was used to cluster rows and/or columns in the heat map. At SCH, Progress Notes and Telephone Encounter are major note types that contain asthma-related concepts. The top 3 notes similar to SCH’s Telephone Encounter are Mayo’s Test MIS, Supervisory, and Miscellaneous notes (similarity = 0.973, 0.948, 0.937, respectively); similar to SCH’s Progress Note are Mayo’s Test MIS, Supervisory, and Limited Exam notes (similarity = 0.897, 0.872, 0.856, respectively).
Figure 5.
A heat map of note type similarity based on topics at Mayo (bottom label) and SCH (right label).
NLP-PAC asthma ascertainment
Portability of the NLP system to SCH has been examined in 2 stages: (1) prototype (stage 1), which required adjustments to be able to run the Mayo NLP-PAC system on the SCH cohort to deal with process variations due to different EMR generation, such as sentence parsing and section segmentation; and (2) refinement (stage 2), which further reduced process variations and refined assertion after error analysis based on the results of the prototype system; ie, selection of practice setting (note type) to be excluded (eg, Psychology, Care Conference, Speech Therapy), adjustment of assertion (eg, negated, possible) to cope with SCH description patterns.
Table 3 contains the NLP-PAC performance of Mayo Clinic and SCH stages 1 and 2. The data used at Mayo Clinic consisted of 497 subjects randomly selected from an Olmsted County birth cohort who had primary care at Mayo Clinic (independent from the data used in NLP-PAC development). The data used in SCH stage 1 are the same cohorts described previously (see Methods and Materials section). SCH stage 2 used another 298 subjects, independent from stage 1. The performance of SCH stage 1 (prototype) was considerably lower than Mayo Clinic, but after moderate refinement, stage 2 produced comparable performance to Mayo Clinic.
Table 3.
NLP-PAC performance for asthma ascertainment
| Metrics | Mayo | SCH Stage 1 (prototype) | SCH Stage 2 (refinement) |
|---|---|---|---|
| Sensitivity | 0.972 | 0.840 | 0.920 |
| Specificity | 0.957 | 0.924 | 0.964 |
| PPV | 0.905 | 0.788 | 0.896 |
| NPV | 0.988 | 0.945 | 0.973 |
| F-score | 0.937 | 0.813 | 0.908 |
DISCUSSIONS
The corpus statistics of asthma birth cohorts between Mayo Clinic and SCH were analyzed and various aspects of their documentation variations were compared. The basic statistics differed in number (ie, number of documents, tokens, asthma-related concepts, note types, and sections), and word-level corpus similarities (ie, corpustf − idf andtf − ipf) were merely mediocre. However, at the concept level, the 2 corpora were relatively homogeneous (corpus topic similarity = 0.944; asthma-related concept similarity = 0.971). This may reflect that clinicians share common semantics to describe asthma episodes and events even though they have heterogeneous clinical sublanguage that shows up in different EMR systems; ie, there exist notable lexical variations to denote semantically similar concepts.
There are process variations across institutions in practice settings and reporting schemes for generating EMR data; ie, the distributions of note types and sections that contain asthma-related concepts differ. Regarding the distribution of asthma-related concepts, “cough,” “asthma,” “dyspnea,” and “wheezing” were dominant, making up 83% and 92% of total concepts at Mayo Clinic and SCH, respectively. Although cough is the most frequent asthma-related symptom, cough alone does not lead to an asthma diagnosis. It should be associated with wheezing in PAC to meet the definition of “asthmatic-cough.” At Mayo Clinic, the “asthma” concept (11%) appears in a similar proportion as “dyspnea” and “wheezing” (11% and 9%, respectively), while at SCH the “asthma” concept (6%) appears much less than “dyspnea” and “wheezing” (14% and 25%, respectively). There is also a 16% difference in the appearance of “wheezing” between the 2 institutions.
The major note types that contain asthma-related concepts were different between the 2 institutions: Limited Exam, Multi-System Evaluation, Miscellaneous (57%, 11%, and 10%, respectively) at Mayo Clinic; Progress Notes and Telephone Encounter (46% and 27%, respectively) at SCH, indicating a difference in the practice of recording asthma-related events and episodes in clinical notes. The major sections that contain asthma-related concepts also differed: 3 sections (History of Present Illness, Impression/Report/Plan, and Chief Complaint) contain almost 80% of asthma-related concepts at Mayo Clinic, but they were spread among many sections at SCH. There were some note types at SCH that were not similar to any of Mayo Clinic’s note types (eg, Physical Therapy, Occupational Therapy, Post-Op Progress Note). However, most of Mayo Clinic’s note types were similar in some degree to SCH notes.
The NLP system first applied to the SCH corpus (stage 1) is the one that dealt with required process variations to be technically operable. It produced anF-score of 0.813, ie, roughly 13% lower than the system’s performance on the Mayo corpus. The error analysis showed that assertion identification (mostly negation detection) was a major source of error and needs to be adjusted accordingly to cope with SCH description patterns (ie, negation sublanguage characteristics). As noted by Wu et al.,50 negation detection suffers when applied to a different corpus and requires domain adaptation. They revealed that negation detection is relatively easy to optimize for a specific corpus but difficult to generalize to an arbitrary clinical corpus. The structured section templates would be helpful to correctly identify sections at SCH, which would eliminate subsequent asthma ascertainment errors due to sectionization. After the refinement process (SCH stage 2) to reduce errors found in stage 1, the NLP-PAC showed comparable performance to Mayo Clinic, except sensitivity (0.972 at Mayo, 0.920 at SCH), suggesting that further effort may be required to analyze SCH data to capture additional asthma cases.
We also reapplied NLP-PAC used in SCH stage 2 on the Mayo data (inTable 3), but using Mayo specifications (ie, section, note type) due to the technical operability. It produced 0.973, 0.963, 0.917, and 0.988 for sensitivity, specificity, PPV, and NPV, respectively, which was a little better but very similar to the performance of the original Mayo NLP-PAC (seeTable 3). Since the major adjustment at SCH is assertion (except required adjustment for technical operability), the revised NLP-PAC on the Mayo data was able to produce comparable asthma ascertainment without loss of performance.
There are limitations in this study. Mayo Clinic and SCH cohorts were matched by age, but the date ranges differ by approximately 10 years. There is potentially clinical practice change related to asthma. However, we believe that it would not have much effect on the outcome, because asthma prevalence did not change considerably over a decade (7.3% in 2001 to 8.4% in 2010).51 The NLP-PAC system we evaluated is an expert system based on information extraction of medical concepts. Other NLP systems using different techniques may not hold the same notions we found. Lastly, this study was conducted on a specific domain, pediatric asthma at 2 institutions. Our findings may not be generalizable to other domains with different patient cohorts at other institutions.
We conducted this study on corpora across institutions using different EMR systems (GE-basedvs Epic). In the future, it would also be interesting to conduct the same study on corpora across institutions using the same EMR system and explore how it differs from the current study in terms of clinical documentation variations and NLP system portability.
CONCLUSION
The different types of clinical documentation variations played different roles in assessing NLP system portability. The NLP system for asthma ascertainment is largely dependent on asthma-related concepts rather than individual word distributions. We believe that concept-wise similarity (ie, semantic similarity) should be emphasized to assess the portability of a knowledge-based NLP system that largely relies on information extraction, such as our NLP-PAC system. The Mayo Clinic and SCH corpora were relatively homogeneous in concepts, which shows good potential for NLP-PAC system portability. However, appropriate adjustments were necessary to deal with the intrinsic corpus heterogeneity, such as process and assertion variations, in order to produce a desirable performance of asthma ascertainment using NLP.
Competing Interests
The authors have no competing interests to declare.
Funding
This work was made possible by National Institute of General Medical Sciences R01GM102282, National Library of Medicine R01LM11934, National Institute of Biomedical Imaging and Bioengineering R01EB19403, National Heart, Lung, and Blood Institute R01HL126667, National Institute of Child Health and Human Development R21Al116839-01, and the T Denny Sanford Pediatric Collaborative Research Fund.
Contributors
SS and HL conceived the study and design. SS and YW acquired the data and implemented the algorithms. SS performed the analysis and drafted the initial manuscript. All authors participated in interpretation of the data and contributed to manuscript revisions.
ACKNOWLEDGMENTS
We gratefully acknowledge Paul R Kingsbury for editorial support.
References
- 1. Wi C-I,Sohn S,Rolfes MC,et al. Application of a natural language processing algorithm to asthma ascertainment: an automated chart review.Am J Respir Crit Care Med. 2017;1964:430–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu ST,Sohn S,Ravikumar K,et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study.Ann Allergy Asthma Immunol. 2013;1115:364–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murff HJ,FitzHenry F,Matheny ME,et al. Automated identification of postoperative complications within an electronic medical record using natural language processing.JAMA. 2011;3068:848–55. [DOI] [PubMed] [Google Scholar]
- 4. Melton GB,Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries.J Am Med Inform Assoc. 2005;124:448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sohn S,Clark C,Halgrim S,Murphy S,Chute C,Liu H. MedXN: an open source medication extraction and normalization tool for clinical text.J Am Med Inform Assoc. 2014;215:858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sohn S,Kocher J-PA,Chute CG,Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients.J Am Med Inform Assoc. 2011;18(Suppl 1):144–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sohn S,Savova GK. Mayo clinic smoking status classification system: extensions and improvements.AMIA Annu Symp. 2009;2009:619–23. [PMC free article] [PubMed] [Google Scholar]
- 8. Sohn S,Ye Z,Liu H,Chute C,Kullo I. Identifying abdominal aortic aneurysm cases and controls using natural language processing of radiology reports.AMIA Jt Summits Transl Sci Proc. 2013;2013:249–53. [PMC free article] [PubMed] [Google Scholar]
- 9. Sohn S,Torii M,Li D,Wagholikar K,Wu S,Liu H. A hybrid approach to sentiment sentence classification in suicide notes.Biomed Inform Insights. 2012;5(Suppl. 1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Demner-Fushman D,Chapman W,McDonald C. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;425:760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aronsky D,Fiszman M,Chapman WW,Haug PJ. Combining decision support methodologies to diagnose pneumonia.J Am Med Inform Assoc. 2001:12–16. [PMC free article] [PubMed] [Google Scholar]
- 12. Kullo IJ,Fan J,Pathak J,Savova GK,Ali Z,Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease.J Am Med Inform Assoc. 2010;175:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kullo IJ,Ding K,Jouni H,Smith CY,Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record.PLoS One. 2010;59:e13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedman C,Shagina L,Lussier Y,Hripcsak G. Automated encoding of clinical documents based on natural language processing.J Am Med Inform Assoc. 2004;115:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pakhomov SVS,Buntrock JD,Chute CG. Automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques.J Am Med Inform Assoc. 2006;135:516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Savova G,Masanz J,Ogren P,et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications.J Am Med Inform Assoc. 2010;175:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garla V,Re VL,Dorey-Stein Z,et al. The Yale cTAKES extensions for document classification: architecture and application.J Am Med Inform Assoc. 2011;185:614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou L,Plasek JM,Mahoney LM,et al. Using medical text extraction, reasoning and mapping system (MTERMS) to process medication information in outpatient clinical notes.AMIA Annu Symp Proc. 2011;2011:1639–48. [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng Q,Goryachev S,Weiss S,Sordo M,Murphy S,Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system.BMC Med Inform Dec Mak. 2006;61:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedman C,Alderson PO,Austin JH,Cimino JJ,Johnson SB. A general natural-language text processor for clinical radiology.J Am Med Inform Assoc. 1994;12:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H,Bielinski S,Sohn S,et al. An information extraction framework for cohort identification using electronic health records.AMIA Jt Summits Transl Sci Proc. 2013;2013:149–53. [PMC free article] [PubMed] [Google Scholar]
- 22. Torii M,Wagholikar K,Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources.J Am Med Inform Assoc. 2011;185:580–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan J,Prasad R,Yabut RM,et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annu Symp Proc. 2011;2011:382–91. [PMC free article] [PubMed] [Google Scholar]
- 24. Wagholikar K,Torii M,Jonnalagadda S,Liu H. Feasibility of pooling annotated corpora for clinical concept extraction.AMIA Jt Summits Transl Sci Proc. 2012;2012:38. [PMC free article] [PubMed] [Google Scholar]
- 25. Friedman C. A broad-coverage natural language processing system.Proc AMIA Symp. 2000:270–74. [PMC free article] [PubMed] [Google Scholar]
- 26. Stetson PD,Johnson SB,Scotch M,Hripcsak G. The sublanguage of cross-coverage.Proc AMIA Symp. 2002:742–46. [PMC free article] [PubMed] [Google Scholar]
- 27. Friedman C,Kra P,Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris.J Biomed Inform. 2002;354:222–35. [DOI] [PubMed] [Google Scholar]
- 28. Harris ZS.A Grammar of English on Mathematical Principles. New York: Wiley;1982. [Google Scholar]
- 29. Harris ZS.A Theory of Language and Information: A Mathematical Approach. Oxford: Clarendon Press;1991. [Google Scholar]
- 30. Wu Y,Denny JC,Rosenbloom ST,et al. A long journey to short abbreviations: developing an open-source framework for clinical abbreviation recognition and disambiguation (CARD).J Am Med Inform Assoc. 2017;24(e1):e79–e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu H,Stetson PD,Friedman C. Methods for building sense inventories of abbreviations in clinical notes.J Am Med Inform Assoc. 2009;161:103–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patterson O,Hurdle JF. Document clustering of clinical narratives: a systematic study of clinical sublanguages.AMIA Annu Symp Proc. 2011;2011:1099–107. [PMC free article] [PubMed] [Google Scholar]
- 33. Liu M,Shah A,Jiang M,et al. A study of transportability of an existing smoking status detection module across institutions.AMIA Annu Symp Proc. 2012;2012:577–86. [PMC free article] [PubMed] [Google Scholar]
- 34. Carroll RJ,Thompson WK,Eyler AE,et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records.J Am Med Inform Assoc. 2012;19(e1):e162–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mehrabi S,Krishnan A,Roch AM,et al. Identification of patients with family history of pancreatic cancer: investigation of an NLP system portability.Stud Health Technol Inform. 2014;216:604–08. [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H,Wu ST,Li D,et al. Towards a semantic lexicon for clinical natural language processing.AMIA Annu Symp Proc. 2012;2012:568–76. [PMC free article] [PubMed] [Google Scholar]
- 37. Yunginger JW,Reed CE,O'Connell EJ,Melton LJ III,O'Fallon WM,Silverstein MD. A community-based study of the epidemiology of asthma: incidence rates, 1964–1983.Am Rev Respir Dis. 1992;1464:888–94. [DOI] [PubMed] [Google Scholar]
- 38. Singhal A. Modern information retrieval: a brief overview.IEEE Data Eng Bull. 2001;244:35–43. [Google Scholar]
- 39. Blei DM,Ng AY,Jordan MI. Latent Dirichlet allocation.J Machine Learn Res. 2003;3:993–1022. [Google Scholar]
- 40. Wang Y,Lee JS,Choi IC. Indexing by latent Dirichlet allocation and an ensemble model.J Assoc Inform Sci Technol. 2016;677:1736–50. [Google Scholar]
- 41. Bisgaard H,Szefler S. Prevalence of asthma-like symptoms in young children.Pediatric Pulmonol. 2007;428:723–28. [DOI] [PubMed] [Google Scholar]
- 42. Molis W,Bagniewski S,Weaver A,Jacobson R,Juhn Y. Timeliness of diagnosis of asthma in children and its predictors.Allergy. 2008;6311:1529–35. [DOI] [PubMed] [Google Scholar]
- 43. Juhn Y,Kung A,Voigt R,Johnson S. Characterisation of children’s asthma status by ICD-9 code and criteria-based medical record review.Prim Care Respir J. 2011;201:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Silverstein MD,Yunginger JW,Reed CE,et al. Attained adult height after childhood asthma: effect of glucocorticoid therapy.J Allergy Clin Immunol. 1997;994:466–74. [DOI] [PubMed] [Google Scholar]
- 45. Yawn BP,Yunginger JW,Wollan PC,Reed CE,Silverstein MD,Harris AG. Allergic rhinitis in Rochester, Minnesota residents with asthma: frequency and impact on health care charges.J Allergy Clin Immunol. 1999;1031:54–59. [DOI] [PubMed] [Google Scholar]
- 46. Bauer BA,Reed CE,Yunginger JW,Wollan PC,Silverstein MD. Incidence and outcomes of asthma in the elderly: a population-based study in Rochester, Minnesota.Chest. 1997;1112:303. [DOI] [PubMed] [Google Scholar]
- 47. Hunt LW,Silverstein MD,Reed CE,O'Connell EJ,O'Fallon WM,Yunginger JW. Accuracy of the death certificate in a population-based study of asthmatic patients.JAMA. 1993;26915:1947–52. [PubMed] [Google Scholar]
- 48. Juhn YJ,Qin R,Urm S,Katusic S,Vargas-Chanes D. The influence of neighborhood environment on the incidence of childhood asthma: a propensity score approach.J Allergy Clin Immunol. 2010;1254:838–43. e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Juhn YJ,Kita H,Lee LA,et al. Childhood asthma and measles vaccine response.Ann Allergy Asthma Immunol. 2006;974:469–76. [DOI] [PubMed] [Google Scholar]
- 50. Wu S,Miller T,Masanz J,et al. Negation’s not solved: generalizability versus optimizability in clinical natural language processing.PLoS One. 2014;911:e112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Akinbami OJ. Trends in asthma prevalence, health care use, and mortality in the United States, 2001-2010.NCHS Data Brief. 2012;(94):1–8. [PubMed] [Google Scholar]