Abstract
Various factors modulate the risk of hepatoblastoma. In this study, we aimed to investigate whether single nucleotide polymorphisms (SNPs) in the YTHDF1 gene could predispose to hepatoblastoma. We used TaqMan assay to genotype two YTHDF1 SNPs (rs6011668 C>T and rs6090311 A>G) in a Chinese population composed of 313 subjects with hepatoblastoma and 1446 controls from seven hospitals. We then evaluated the associations of these two SNPs with hepatoblastoma risk using unconditional logistic regression. We found that rs6090311 G allele exhibited a significant association with decreased hepatoblastoma risk [AG vs. AA: adjusted odds ratio (OR)=0.75; 95% confidence interval (CI)=0.58-0.98, P=0.033; AG/GG vs. AA: adjusted OR=0.76, 95% CI=0.59-0.97, P=0.029]. Furthermore, the combined analysis of protective genotypes revealed that subjects carrying two protective genotypes were less likely to have hepatoblastoma than those with 0-1 protective genotypes (adjusted OR=0.75, 95% CI=0.59-0.96, P=0.022). Subjects ≥17 months of age had decreased hepatoblastoma risk, in case that they carried rs6090311 AG/GG (adjusted OR=0.63, 95% CI=0.44-0.91, P=0.012), or two protective genotypes (adjusted OR=0.63, 95% CI=0.44-0.91, P=0.012). False-positive report probability analysis validated the reliability of the significant results. Preliminary functional annotations revealed that rs6090311 G was correlated with decreased expression of its surrounding genes in the expression quantitative trait locus (eQTL) analysis. In conclusion, our results indicate that the rs6090311 A>G in the YTHDF1 gene is related to decreased hepatoblastoma risk.
Keywords: hepatoblastoma, risk, YTHDF1, polymorphism, case-control study
Introduction
Hepatoblastoma is the most prevalent liver tumor in childhood, accounting for up to 80% of all pediatric liver tumor cases 1, 2. The incidence of hepatoblastoma is low, with about 1.5 cases per million annually 3. The occurrence rate of hepatoblastoma is about 1.1 cases in a million children in China 4. The five-year survival rate for hepatoblastoma is about 80% 5. However, the survival rate may sharply decrease to 30-40% in the high-risk hepatoblastoma 6-8.
Hepatoblastoma mainly derives from abnormal differentiation of hepatocyte precursors of an epithelial lineage 9. Fibrosing diseases, hepatitis B virus, or cirrhosis are tightly associated with hepatocellular carcinoma, but not hepatoblastoma 10-12. Alternatively, it has been well documented that hepatoblastoma is associated with prematurity, low birth weight, trisomy 18, familial adenomatous polyposis, and Beckwith-Wiedemann syndrome 13-16. Moreover, parental tobacco use, loss of heterozygosity (LOH) at 11p15, and mutations in genes (such as FGFR3, APC, and β-catenin) have also been observed to significantly associate with hepatoblastoma risk in some studies, despite conflicting reports 17-22. Advances in the etiology of hepatoblastoma have been far lagged behind, when compared to those more common pediatric cancers. Only a few case-control studies have investigated hepatoblastoma susceptibility genetic variants to date. Besides, the sample size of these studies was relatively small, with less than 200 cases 23, 24. Previously, we conducted three extensive epidemiology investigations to identify the genetic variants for hepatoblastoma 25-27.
N6-methyladenosine (m6A) modification is the most recurrent internal modification for mRNAs/ncRNAs 28, 29. m6A modification is dynamically deposited, removed, and recognized by m6A methyltransferases (“writers”), demethylases (“erasers”), and m6A-specific binding proteins (“readers”), respectively 30. m6A “readers” include the YTH-family proteins YTHDF1-3, YTHDC1-2, and insulin-like growth factor 2 mRNA-binding proteins IGF2BP1-3. YTHDF1 interacts with translation initiation factors eIF3 and eIF4A3 to facilitate the translation process of m6A-modified mRNAs in the cytosol 31.
Multiple evidence has suggested the implicated of m6A regulators in the carcinogenesis, including METTL3, METTL14, ALKBH5, FTO, and YTHDF2 32-40. However, the significance of YTHDF1 in cancer is largely unknown. Given the rarity of hepatoblastoma, most of the existing epidemiology studies suffer from small sample size. With this in mind, we performed a relatively large case-control study to identify more hepatoblastoma susceptibility genetic variants in the YTHDF1 gene.
Materials and Methods
Sample selection
The characteristics information of subjects were listed in Table S1. Overall, 313 patients with histologically confirmed hepatoblastoma were recruited from seven hospitals located in seven cities of China (Guangzhou, Xi'an, Zhengzhou, Changsha, Kunming, Shenyang, Taiyuan). Meanwhile, we recruited 1446 geographically matched control participants without a history of cancer or other diseases 41. Blood samples were obtained from all participants, along with informed consents. The study was approved by the institutional review board of all the participating hospitals.
Polymorphism selection and genotyping
Through extensively mining the dbSNP and SNPinfo databases, we identified two potentially functional polymorphisms (rs6011668 C>T and rs6090311 A>G) in the YTHDF1 gene. To be specific, the following criteria were adopted to choose potentially functional polymorphisms: SNPs located in the 5' flanking region, 5' untranslated region, 3' untranslated region, and exon of YTHDF1 gene. Both the two selected SNPs (rs6011668 C>T and rs6090311 A>G) are located in the transcription factor binding sites (TFBS). There is no significant linkage disequilibrium (LD) (R2<0.8) among these two selected SNPs of YTHDF1 gene (R2=0.094 between rs6011668 C>T and rs6090311 A>G) (Figure S1). Genomic DNA was prepared from peripheral blood, as previously described 42. SNP genotyping was implemented using the TaqMan assay according to the manufacturer's instructions (Applied Biosystems) 43. Internal negative controls (water) were used to ensure genotyping accuracy. About 10% of all samples were randomly selected to be genotyped again. The concordant rate of these duplicate samples was 100%.
Statistical analysis
The χ2 test and the student's t-test was used to assess the differences in the distribution of gender and age, respectively, between cases and controls. Each SNP was tested in controls to ensure the fitting with Hardy-Weinberg equilibrium (HWE) using a χ2 test. Multiple logistic regression analysis was performed to evaluate an association between SNPs and hepatoblastoma risk. The odds ratios (ORs), 95% confidence intervals (CIs), and P values were calculated. Data were further stratified by age, gender, and clinical stages. False-positive report probability (FPRP) analysis was conducted to assess noteworthy associations by the means as described elsewhere 44, 45. Expression quantitative trait loci (eQTL) analysis using GTEx portal web site (http://www.gtexportal.org/home/) was conducted to determine the correlation between the SNPs and expression levels of nearby genes 46. The level of significance was set at 0.05. All statistics were performed using SAS 9.1 (SAS Institute, Cary, NC).
Results
Association between YTHDF1 polymorphisms and hepatoblastoma susceptibility
We successfully genotyped the two polymorphisms in 313 cases and 1444 controls. The genotype frequencies of the two SNPs are listed in Table 1. Both of the two SNPs were in agreement with the HWE in controls (PHWE=0.882 for rs6011668 C>T; PHWE=0.240 for rs6090311 A>G). No significant association was found between the rs6011668 C>T polymorphism and hepatoblastoma susceptibility. We found that the rs6090311 G allele was significantly associated with protection against hepatoblastoma (AG vs. AA: adjusted OR=0.75, 95% CI=0.58-0.98, P=0.033; AG/GG vs. AA: adjusted OR=0.76, 95% CI=0.59-0.97, P=0.029). We next sought to determine whether the combination of rs6011668 C>T and rs6090311 A>G have a more powerful effect on hepatoblastoma risk than either one alone. The rs6011668 CC/CT and rs6090311 AG/GG were treated as protective genotypes. Subjects carrying two protective genotypes showed significantly decreased hepatoblastoma risk when compared with those with 0-1 protective genotypes (adjusted OR=0.75, 95% CI=0.59-0.96, P=0.022).
Table 1.
Association between YTHDF1 polymorphisms and hepatoblastoma risk
| Genotype | Cases (N=313) | Controls (N=1444) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs6011668 C>T (HWE=0.882) | |||||||
| CC | 221 (70.61) | 1047 (72.51) | 1.00 | 1.00 | |||
| CT | 83 (26.52) | 366 (25.35) | 1.07 (0.81-1.42) | 0.614 | 1.08 (0.82-1.42) | 0.603 | |
| TT | 9 (2.88) | 31 (2.15) | 1.38 (0.65-2.93) | 0.408 | 1.35 (0.63-2.87) | 0.441 | |
| Additive | 0.405 | 1.11 (0.87-1.40) | 0.405 | 1.10 (0.87-1.40) | 0.413 | ||
| Dominant | 92 (29.39) | 397 (27.49) | 0.497 | 1.10 (0.84-1.44) | 0.497 | 1.10 (0.84-1.44) | 0.495 |
| Recessive | 304 (97.12) | 1413 (97.85) | 0.433 | 1.35 (0.64-2.86) | 0.435 | 1.32 (0.62-2.81) | 0.469 |
| rs6090311 A>G (HWE=0.240) | |||||||
| AA | 142 (45.37) | 558 (38.64) | 1.00 | 1.00 | |||
| AG | 133 (42.49) | 696 (48.20) | 0.75 (0.58-0.98) | 0.032 | 0.75 (0.58-0.98) | 0.033 | |
| GG | 38 (12.14) | 190 (13.16) | 0.79 (0.53-1.17) | 0.231 | 0.79 (0.53-1.17) | 0.240 | |
| Additive | 0.066 | 0.84 (0.70-1.01) | 0.066 | 0.84 (0.70-1.01) | 0.070 | ||
| Dominant | 171 (54.63) | 886 (61.36) | 0.028 | 0.76 (0.59-0.97) | 0.028 | 0.76 (0.59-0.97) | 0.029 |
| Recessive | 275 (87.86) | 1254 (86.84) | 0.627 | 0.91 (0.63-1.32) | 0.627 | 0.92 (0.63-1.33) | 0.643 |
| Combined effect of protective genotypes c | |||||||
| 0 | 8 (2.56) | 31 (2.15) | 1.00 | 1.00 | |||
| 1 | 135 (43.13) | 527 (36.50) | 0.99 (0.45-2.21) | 0.986 | 1.01 (0.45-2.26) | 0.976 | |
| 2 | 170 (54.31) | 886 (61.36) | 0.026 | 0.74 (0.34-1.65) | 0.465 | 0.76 (0.34-1.68) | 0.497 |
| 0-1 | 143 (45.69) | 558 (38.64) | 1.00 | 1.00 | |||
| 2 | 170 (54.31) | 886 (61.36) | 0.021 | 0.75 (0.59-0.96) | 0.021 | 0.75 (0.59-0.96) | 0.022 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
a χ2 test for genotype distributions between hepatoblastoma patients and cancer-free controls.
b Adjusted for age and gender.
c Protective genotypes were carriers with rs6011668 CC/CT, and rs6090311 AG/GG genotypes.
Stratification analysis
To assess whether these variants influence susceptibility in specific subgroups, we performed a stratification analysis regarding age, gender, and clinical stages (Table 2). When compared to the AA genotype, the protective effect of rs6090311 AG/GG genotypes was more evident among children ≥17 months of age (adjusted OR=0.63, 95% CI=0.44-0.91, P=0.012). Besides, the combined analysis also demonstrated that the carriers with two protective genotypes had a significantly decreased hepatoblastoma risk among children aged 17 months and older (adjusted OR=0.63, 95% CI=0.44-0.91, P=0.012).
Table 2.
Stratification analysis for association between YTHDF1 genotypes and hepatoblastoma susceptibility
| Variables | rs6011668 (case/control) | AOR (95% CI) a | P a | rs6090311 (case/control) |
AOR (95% CI) a | P a | Combine genotypes (case/control) | AOR (95% CI) a | P a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT/TT | AA | AG/GG | 0-1 | 2 | |||||||
| Age, month | ||||||||||||
| <17 | 121/465 | 47/176 | 1.02 (0.70-1.49) | 0.933 | 71/255 | 97/386 | 0.90 (0.64-1.28) | 0.564 | 72/255 | 96/386 | 0.88 (0.63-1.25) | 0.476 |
| ≥17 | 100/582 | 45/221 | 1.18 (0.80-1.74) | 0.395 | 71/303 | 74/500 | 0.63 (0.44-0.91) | 0.012 | 71/303 | 74/500 | 0.63 (0.44-0.91) | 0.012 |
| Gender | ||||||||||||
| Female | 99/427 | 30/168 | 0.77 (0.49-1.21) | 0.255 | 60/236 | 69/359 | 0.76 (0.52-1.11) | 0.151 | 60/236 | 69/359 | 0.76 (0.52-1.11) | 0.151 |
| Male | 122/620 | 62/229 | 1.37 (0.97-1.93) | 0.072 | 82/322 | 102/527 | 0.77 (0.56-1.06) | 0.105 | 83/322 | 101/527 | 0.75 (0.54-1.03) | 0.080 |
| Clinical stages | ||||||||||||
| I+II | 116/1047 | 44/397 | 1.00 (0.69-1.44) | 0.997 | 73/558 | 87/886 | 0.75 (0.54-1.05) | 0.092 | 73/558 | 87/886 | 0.75 (0.54-1.05) | 0.092 |
| III+IV | 63/1047 | 28/397 | 1.17 (0.74-1.86) | 0.496 | 41/558 | 50/886 | 0.77 (0.50-1.17) | 0.218 | 42/558 | 49/886 | 0.73 (0.48-1.12) | 0.150 |
AOR, adjusted odds ratio; CI, confidence interval;
a Adjusted for age and gender, omitting the corresponding stratify factor.
False-positive report probability (FPRP) analysis
FPRP analysis was carried out to interrogate our significant findings (Table 3). The threshold for FPRP was preset as 0.2. At the prior probability level of 0.1, findings for 2 vs. 0-1 protective genotypes remained noteworthy.
Table 3.
False-positive report probability values for the associations between YTHDF1 gene polymorphisms and hepatoblastoma susceptibility
| Genotype | Crude OR (95% CI) | P a | Statistical Power b | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs6090311 A>G | ||||||||
| AG vs. AA | 0.75 (0.58-0.98) | 0.032 | 0.847 | 0.101 | 0.252 | 0.787 | 0.974 | 0.997 |
| AG/GG vs. AA | 0.76 (0.59-0.97) | 0.028 | 0.840 | 0.091 | 0.230 | 0.767 | 0.971 | 0.997 |
| ≥17 | 0.63 (0.44-0.90) | 0.011 | 0.377 | 0.082 | 0.212 | 0.748 | 0.968 | 0.997 |
| Protective genotypes | ||||||||
| 2 vs. 0-1 | 0.75 (0.59-0.96) | 0.021 | 0.814 | 0.073 | 0.191 | 0.722 | 0.963 | 0.996 |
| ≥17 | 0.63 (0.44-0.90) | 0.011 | 0.377 | 0.082 | 0.212 | 0.748 | 0.968 | 0.997 |
OR, odds ratio; CI, confidence interval.
a Chi-square test was used to calculate the genotype frequency distributions.
b Statistical power was calculated using the number of observations in the subgroup and the OR and P values in this table.
Expression quantitative trait loci (eQTL) analyses
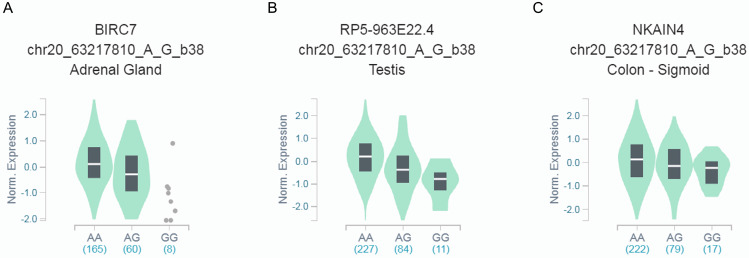
We further explored whether rs6090311 A>G could influence the mRNA level of neighboring genes, using released data from GTEx. We found that rs6090311 G genotype was significantly associated with lower BIRC7 (Figure 1A), RP5-963E22.4 (Figure 1B), and NKAIN4 (Figure 1C) mRNA levels when compared to rs6090311 A genotype.
Figure 1.
Functional implication of rs6090311 polymorphism based on the public database GTEx Portal. (A) The genotype of rs6090311 and expression of BIRC7 gene in adrenal gland; (B) The genotype of rs6090311 and expression of RP5-963E22.4 gene in testis; (C) The genotype of rs6090311 and expression of NKAIN4 gene in colon-sigmoid.
Discussion
The etiology of hepatoblastoma remains obscure. To identify the genetic profile and elucidate the pathogenesis of hepatoblastoma, we performed this case-control study including 313 cases and 1446 controls. Overall, we identified that the YTHDF1 gene rs6090311 A>G polymorphism was associated with a decreased hepatoblastoma risk.
YTHDF1 gene resides in chromosome 20q11. Several lines of evidence suggest the implication of YTHDF1 in tumorigenesis. Shi et al. 47 showed that YTHDF1 deficiency suppressed non-small cell lung cancer cell proliferation and xenograft tumor formation. However, they found that high expression of YTHDF1 was associated with better clinical outcomes. Zhao et al. 48 observed that upregulation of YTHDF1 was correlated with poor prognosis in patients with hepatocellular carcinoma. Han et al. 49 demonstrated that YTHDF1 regulated the anti-tumor immunity response via RNA m6A modification. Nishizawa et al. 50 reported that colorectal cancer patients with high YTHDF1 expression had significantly more reduced overall survival. Molecular mechanism experiments showed that c-Myc could drive YTHDF1 to promote cancer proliferation. The oncogenic role of YTHDF1 in colorectal cancer was also observed by Bai et al. 51.
Only one study has been conducted so far, regarding the epidemiology assessment of YTHDF1 gene SNPs and cancer risk. Meng et al. 52 carried out a two-stage association study on colorectal cancer in China, consisting of the discovery stage with 1150 cases and 1342 controls, and the replication stage with 932 cases and 966 controls. They extensively genotyped 240 SNPs in 20 genes involved in m6A modification. Finally, the SNP rs118049207 located in the SND1 gene was found to associate with colorectal cancer risk in the Chinese population. Further functional experiments suggested that rs118049207 could regulate mRNA expression of the SND1 gene and then alter m6A level. Nevertheless, they failed to detect the association of SNPs rs2024768 and rs6090289 in the YTHDF1 gene with colorectal cancer risk. In the current study, we assayed the genotype frequencies of two YTHDF1 SNPs in a representative set of 313 hepatoblastoma cases and 1446 controls. No correlation between rs6011668 C>T and hepatoblastoma risk was observed, but hepatoblastoma risk was significantly decreased in participants with the rs6090311 G allele. Moreover, subjects carrying two protective genotypes showed significantly reduced hepatoblastoma risk when compared with those with 0-1 protective genotypes. FPRP analysis further validated the strength of the significant findings. These data indicate that YTHDF1 gene SNPs are associated with hepatoblastoma susceptibility. We further attempted to interpret the possible mechanism of YTHDF1 gene rs6090311 A>G-mediated hepatoblastoma risk. eQTL evidence suggested that the rs6090311 G allele is significantly associated with decreased BIRC7, RP5-963E22.4, and NKAIN4 levels. Further functional experiments are needed to elucidate this mechanism.
Our study is strengthened by its size, multiple-center participants, and inclusion of strict genotyping quality control; however, some limitations should be addressed. Firstly, the sample size of our study was moderate. There is a possibility that some of the stratification analysis results might be merely fortuitous events. Secondly, all participants here are restricted to children of Chinese descent. Further studies are therefore warranted to delineate genetic contributions of YTHDF1 to hepatoblastoma susceptibility in populations of non-Chinese ancestries. Thirdly, only two SNPs were analyzed in the current study, more SNPs are needed to be investigated in the near future. Finally, the current study only focused on genetic factors, and environmental factors were not considered in this study. The impact of YTHDF1 gene SNPs is weak. This finding reinforces the notion that hepatoblastoma is a polygenic process and thus a combined analysis of multiple factors may have a greater ability to characterize this disease.
In all, we present the first epidemiology evidence supporting the involvement of YTHDF1 gene polymorphisms in hepatoblastoma risk. Intensive future research will be needed to extend the discovery of hepatoblastoma susceptibility loci to individuals of non-Chinese ancestries.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Guangdong Province (No: 2019A1515010360), Science and Technology Project of Guangzhou (No: 201904010046), and Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (No: 2019B030301004).
References
- 1.Sharma D, Subbarao G, Saxena R. Hepatoblastoma. Semin Diagn Pathol. 2017;34:192–200. doi: 10.1053/j.semdp.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Haas JE, Muczynski KA, Krailo M, Ablin A, Land V, Vietti TJ. et al. Histopathology and prognosis in childhood hepatoblastoma and hepatocarcinoma. Cancer. 1989;64:1082–95. doi: 10.1002/1097-0142(19890901)64:5<1082::aid-cncr2820640520>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 3.Stiller CA, Pritchard J, Steliarova-Foucher E. Liver cancer in European children: incidence and survival, 1978-1997. Report from the Automated Childhood Cancer Information System project. Eur J Cancer. 2006;42:2115–23. doi: 10.1016/j.ejca.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Bao PP, Li K, Wu CX, Huang ZZ, Wang CF, Xiang YM. et al. [Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010] Zhonghua Er Ke Za Zhi. 2013;51:288–94. [PubMed] [Google Scholar]
- 5.Horton JD, Lee S, Brown SR, Bader J, Meier DE. Survival trends in children with hepatoblastoma. Pediatr Surg Int. 2009;25:407–12. doi: 10.1007/s00383-009-2349-3. [DOI] [PubMed] [Google Scholar]
- 6.Meyers RL, Maibach R, Hiyama E, Haberle B, Krailo M, Rangaswami A. et al. Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the Children's Hepatic tumors International Collaboration. Lancet Oncol. 2017;18:122–31. doi: 10.1016/S1470-2045(16)30598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hafberg E, Borinstein SC, Alexopoulos SP. Contemporary management of hepatoblastoma. Curr Opin Organ Transplant. 2019;24:113–7. doi: 10.1097/MOT.0000000000000618. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Polychronidis G, Heger U, Frongia G, Mehrabi A, Hoffmann K. Incidence trends and survival prediction of hepatoblastoma in children: a population-based study. Cancer Commun (Lond) 2019;39:62. doi: 10.1186/s40880-019-0411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czauderna P, Lopez-Terrada D, Hiyama E, Haberle B, Malogolowkin MH, Meyers RL. Hepatoblastoma state of the art: pathology, genetics, risk stratification, and chemotherapy. Curr Opin Pediatr. 2014;26:19–28. doi: 10.1097/MOP.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 10.Wiwanitkit V. Hepatitis virus B is not a risk factor in hepatoblastoma patients. Asian Pac J Cancer Prev. 2005;6:213–4. [PubMed] [Google Scholar]
- 11.Spector LG, Birch J. The epidemiology of hepatoblastoma. Pediatr Blood Cancer. 2012;59:776–9. doi: 10.1002/pbc.24215. [DOI] [PubMed] [Google Scholar]
- 12.Ranganathan S, Lopez-Terrada D, Alaggio R. Hepatoblastoma and Pediatric Hepatocellular Carcinoma: An Update. Pediatr Dev Pathol. 2020;23:79–95. doi: 10.1177/1093526619875228. [DOI] [PubMed] [Google Scholar]
- 13.Mussa A, Molinatto C, Baldassarre G, Riberi E, Russo S, Larizza L. et al. Cancer Risk in Beckwith-Wiedemann Syndrome: A Systematic Review and Meta-Analysis Outlining a Novel (Epi)Genotype Specific Histotype Targeted Screening Protocol. J Pediatr. 2016;176:142–9. doi: 10.1016/j.jpeds.2016.05.038. e1. [DOI] [PubMed] [Google Scholar]
- 14.Oue T, Kubota A, Okuyama H, Kawahara H, Nara K, Kawa K. et al. Hepatoblastoma in children of extremely low birth weight: a report from a single perinatal center. J Pediatr Surg. 2003;38:134–7. doi: 10.1053/jpsu.2003.50027. discussion 134-7. [DOI] [PubMed] [Google Scholar]
- 15.Trobaugh-Lotrario AD, Lopez-Terrada D, Li P, Feusner JH. Hepatoblastoma in patients with molecularly proven familial adenomatous polyposis: Clinical characteristics and rationale for surveillance screening. Pediatr Blood Cancer. 2018;65:e27103. doi: 10.1002/pbc.27103. [DOI] [PubMed] [Google Scholar]
- 16.Valentin LI, Perez L, Masand P. Hepatoblastoma Associated with Trisomy 18. J Pediatr Genet. 2015;4:204–6. doi: 10.1055/s-0035-1565265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chitragar S, Iyer VK, Agarwala S, Gupta SD, Sharma A, Wari MN. Loss of heterozygosity on chromosome 11p15.5 and relapse in hepatoblastomas. Eur J Pediatr Surg. 2011;21:50–3. doi: 10.1055/s-0030-1267208. [DOI] [PubMed] [Google Scholar]
- 18.Sorahan T, Lancashire RJ. Parental cigarette smoking and childhood risks of hepatoblastoma: OSCC data. Br J Cancer. 2004;90:1016–8. doi: 10.1038/sj.bjc.6601651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang A, Sisson R, Gupta A, Tiao G, Geller JI. Germline APC mutations in hepatoblastoma. Pediatr Blood Cancer. 2018;65:e26892. doi: 10.1002/pbc.26892. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Meyfeldt J, Wang H, Kulkarni S, Lu J, Mandel JA. et al. beta-Catenin mutations as determinants of hepatoblastoma phenotypes in mice. J Biol Chem. 2019;294:17524–42. doi: 10.1074/jbc.RA119.009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heck JE, Meyers TJ, Lombardi C, Park AS, Cockburn M, Reynolds P. et al. Case-control study of birth characteristics and the risk of hepatoblastoma. Cancer Epidemiol. 2013;37:390–5. doi: 10.1016/j.canep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baynam GS, Goldblatt J. A child with an FGFR3 mutation, a laterality disorder and an hepatoblastoma: novel associations and possible gene-environment interactions. Twin Res Hum Genet. 2010;13:297–300. doi: 10.1375/twin.13.4.297. [DOI] [PubMed] [Google Scholar]
- 23.Pakakasama S, Chen TT, Frawley W, Muller C, Douglass EC, Tomlinson GE. Myeloperoxidase promotor polymorphism and risk of hepatoblastoma. Int J Cancer. 2003;106:205–7. doi: 10.1002/ijc.11191. [DOI] [PubMed] [Google Scholar]
- 24.Pakakasama S, Chen TT, Frawley W, Muller CY, Douglass EC, Lee R. et al. CCND1 polymorphism and age of onset of hepatoblastoma. Oncogene. 2004;23:4789–92. doi: 10.1038/sj.onc.1207499. [DOI] [PubMed] [Google Scholar]
- 25.Yang T, Li J, Wen Y, Tan T, Yang J, Pan J. et al. LINC00673 rs11655237 C>T Polymorphism Impacts Hepatoblastoma Susceptibility in Chinese Children. Front Genet. 2019;10:506. doi: 10.3389/fgene.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T, Wen Y, Li J, Tan T, Yang J, Pan J. et al. NRAS and KRAS polymorphisms are not associated with hepatoblastoma susceptibility in Chinese children. Exp Hematol Oncol. 2019;8:11. doi: 10.1186/s40164-019-0135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang T, Wen Y, Li J, Yang J, Tan T, Pan J. et al. Association of the TP53 rs1042522 C>G polymorphism and hepatoblastoma risk in Chinese children. J Cancer. 2019;10:3444–9. doi: 10.7150/jca.33063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balacco DL, Soller M. The m(6)A Writer: Rise of a Machine for Growing Tasks. Biochemistry. 2019;58:363–78. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 29.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4:379–86. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 30.Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019;79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y. et al. RNA m(6)A Methyltransferase METTL3 Promotes The Growth Of Prostate Cancer By Regulating Hedgehog Pathway. Onco Targets Ther. 2019;12:9143–52. doi: 10.2147/OTT.S226796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao Y, Shang J, Ji W. ALKBH5-m(6)A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Biophys Res Commun. 2020;521:499–506. doi: 10.1016/j.bbrc.2019.10.145. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L. et al. METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther. 2020;28:599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M. et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene. 2019;38:3667–80. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 36.Dahal U, Le K, Gupta M. RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res. 2019;29:382–9. doi: 10.1097/CMR.0000000000000580. [DOI] [PubMed] [Google Scholar]
- 37.Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z. et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18:163. doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y. et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512:479–85. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 39.Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D. et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/beta-catenin signalling pathway. Cell Prolif. 2020;53:e12768. doi: 10.1111/cpr.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Peng C, Chen J, Chen D, Yang B, He B. et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P, Zhuo ZJ, Zhu J, Yang Z, Xin Y, Li S, Association of TP53 rs1042522 C>G and miR-34b/c rs4938723 T>C polymorphisms with hepatoblastoma susceptibility: a seven-center case-control study. J Gene Med. 2020. p: e3182. DOI: 10.1002/jgm.3182. [DOI] [PubMed]
- 42.Zhuo ZJ, Liu W, Zhang J, Zhu J, Zhang R, Tang J. et al. Functional Polymorphisms at ERCC1/XPF Genes Confer Neuroblastoma Risk in Chinese Children. EBioMedicine. 2018;30:113–9. doi: 10.1016/j.ebiom.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y. et al. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20:1481–90. doi: 10.1111/jcmm.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He J, Wang MY, Qiu LX, Zhu ML, Shi TY, Zhou XY. et al. Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol Carcinog. 2013;52(Suppl 1):E70–9. doi: 10.1002/mc.22013. [DOI] [PubMed] [Google Scholar]
- 45.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank. 2015;13:307–8. doi: 10.1089/bio.2015.29031.hmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L. et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J. et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–68. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 49.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R. et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–4. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N. et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476–86. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai Y, Yang C, Wu R, Huang L, Song S, Li W. et al. YTHDF1 Regulates Tumorigenicity and Cancer Stem Cell-Like Activity in Human Colorectal Carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng Y, Li S, Gu D, Xu K, Du M, Zhu L. et al. Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis. 2020;41:8–17. doi: 10.1093/carcin/bgz165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.