Abstract
Background:
Lysine-specific histone demethylase 1 (LSD1) is a potential target of cancer therapy. In the present study, we aimed to investigate the combined antitumor activity of a novel LSD1 inhibitor (ZY0511) with 5-fluorouracil (5-FU) and elucidate the underlying mechanism in colorectal cancer (CRC).
Methods:
We evaluated LSD1 expression in CRC tissues from patients who received 5-FU treatment. The synergistic antitumor effect of 5-FU with ZY0511 against human CRC cells was detected both in vitro and in vivo. The underlying mechanism was explored based on mRNA sequencing (mRNA-seq) technology.
Results:
Overexpression of LSD1 was observed in human CRC tissues, and correlated with CRC development and 5-FU resistance. ZY0511, a novel LSD1 inhibitor, effectively inhibited CRC cells proliferation, both in vitro and in vivo. Notably, the combination of ZY0511 and 5-FU synergistically reduced CRC cells viability and migration in vitro. It also suppressed Wnt/β-catenin signaling and DNA synthesis pathways, which finally induced apoptosis of CRC cells. In addition, the combination of ZY0511 with 5-FU significantly reduced CRC xenograft tumor growth, along with lung and liver metastases in vivo.
Conclusions:
Our findings identify LSD1 as a potential marker for 5-FU resistance in CRC. ZY0511 is a promising candidate for CRC therapy as it potentiates 5-FU anticancer effects, thereby providing a new combinatorial strategy for treating CRC.
Keywords: colorectal cancer, combination therapy, 5-FU, LSD1 inhibitor, ZY0511
Introduction
Colorectal cancer (CRC) is the third most common malignancy and fourth most common cause of cancer-related deaths worldwide, with annual increases in morbidity and mortality. Over the past decades, 5-fluorouracil (5-FU) has remained the gold standard for CRC treatment.1 5-FU resistance occurs frequently, especially in patients with advanced CRC, which leads to response rates of only 20–25%.2 By combination with other drugs, such as oxaliplatin, irinotecan, and targeted therapies, the response rates for 5-FU-based chemotherapy have increased to 40–50% in advanced CRC, which has improved overall survival.3 However, despite these improvements, the efficiency of the above combinatorial strategies remains far from satisfactory. A better understanding of 5-FU resistance mechanism and better options of combinational therapy strategy are urgently needed to improve the overall survival of patients with advanced CRC.
Epigenetic dysregulation plays crucial roles in development and progression of human CRC. The machinery that controls DNA and histone modification has become a major attraction for targeted CRC therapies.4,5 Lysine-specific histone demethylase 1 (LSD1) is a histone-modifying enzyme responsible for demethylating histone H3 lysine 4 (H3K4) and histone H3 lysine 9 (H3K9) 6,7 ; thus, it regulates the expression of many genes that are important in biological processes. Overexpression of LSD1 is observed in many human cancers, such as acute myeloid leukemia (AML),8 colorectal,9,10 oral,11 breast,12 lung,13,14 and prostate cancer,15 and is associated with a poor survival rate of cancer patients. Increasing evidence suggests that LSD1 is involved in tumorigenesis through regulating cell stemness, motility, epithelial-mesenchymal transition (EMT), and glycolysis metabolism.7 Knockdown or inhibition of LSD1 effectively inhibited cancer cell proliferation, invasion, and migration, supporting LSD1 as a promising cancer therapy target.
Recent studies have shown that LSD1 overexpression is also associated with resistance to chemotherapy drugs such as paclitaxel and cisplatin,16,17 and LSD1 knockdown largely eliminates this resistance. Previous research has shown that LSD1 depletion enhanced the chemosensitivity of human oral squamous cell carcinoma cells to 5-FU,11 suggesting an association between LSD1 overexpression and 5-FU resistance. Moreover, LSD1 could reduce H3K4me2 and H3K9me2 cellular levels, which predicted worse survival outcomes of pancreatic cancer patients who were undergoing adjuvant fluorouracil chemotherapy.18 Therefore, we hypothesized that LSD1 inhibitors could be used for CRC treatment and to enhance the sensitivity to 5-FU. However, the effects of LSD1 inhibitors, alone or in combination with 5-FU, on CRC remains unclear.
Previously, we developed a novel and potent small molecular LSD1 inhibitor, ZY0511, with an IC50 of 1.4 nM. ZY0511 inhibited proliferation of multiple human cancer cell lines, including breast cancer, prostate cancer, and CRC cells in vitro.19,20 Here, our results demonstrate that ZY0511 inhibits growth of human CRC cells both in vitro and in vivo. Notably, LSD1 overexpression is correlated with 5-FU resistance in human CRC specimens, and ZY0511 combined with 5-FU synergistically suppresses CRC cells growth and metastasis, both in vitro and in vivo. Collectively, our findings identify LSD1 as a potential marker for 5-FU resistance in CRC. ZY0511 is a promising candidate for CRC therapy as it potentiates 5-FU anticancer effects, thereby validating a combinatorial strategy for treating CRC.
Materials and methods
Materials
5-FU was purchased from Tianjin Jinyao Amino Acid Co., Ltd. (Tianjin, China). For all in vitro assays, 5-FU was dissolved in dimethyl sulfoxide (DMSO) to a 10-mM stock solution and diluted in the relevant assay medium to provide the final desired concentration. For in vivo experiments, 5-FU was dissolved in 0.9% phosphate-buffered saline (PBS) and dosed at 0.1 ml/10 g of body weight. For all in vitro assays, medium with 0.1% DMSO served as the vehicle control.
Cell lines and cell culture
Human CRC cell lines including SW620, HCT116, SW480, LOVO, DLD-1, SW48, HCT15, HT29, and human normal colonic epithelial cell line NCM460 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in a humidified incubator at 37°C with 5% CO2, and grown in RPMI 1640 or the Dulbecco’s modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Logan, UT, USA) and 1% antibiotics (penicillin and streptomycin).
Synthesis and preparation of ZY0511
ZY0511((E)-N′-(4-chloro-7-hydroxy-2,3-dihydro-1H-inden-1-ylidene)-3-(morpholinosulfonyl) benzohydrazide) was synthesized in the State Key Laboratory of Biotherapy, Sichuan University (Sichuan, China); its structural formula is shown in supplemental Figure S1A. For in vitro assays, ZY0511 was dissolved in DMSO and diluted in the relevant culture medium to a final DMSO concentration of 0.1% (v/v). For in vivo experiments, ZY0511 was suspended in 5% N-methyl-2-pyrrolidone (NMP) and 95% polyethylene glycol (PEG) 400 and administered by oral gavage at volumes of 10 ml/kg/day.
Patient samples
A total of 60 patients with CRC were recruited from the Guizhou Provincial People’s Hospital. This study was conducted with the approval of the Guizhou Provincial People’s Hospital, China (permit number: 2016037), and the patients enrolled in this study did not receive any neoadjuvant chemotherapy or radiation therapy prior to surgical treatment. The clinical information of patients has been summarized in Table 1. The stage of all tissue specimens was determined according to the American Joint Committee on Cancer (AJCC) guidelines for colorectal tumors: stage I, 5 patients; stage II, 27 patients; stage III, 20 patients; stage IV, 8 patients. The tissue samples, dissected by a senior pathologist in the operating room, were frozen immediately in liquid nitrogen and stored at −80°C. The clinical diagnosis, tumor stage, histology differentiation, and resection margin were determined by routine histopathology examination of hematoxylin and eosin (H&E)-stained specimens by a blinded pathologist.
Table 1.
Relationship between expression level of LSD1 and clinical features of CRC patients.
| Characteristics | Number (n) | % | LSD1 |
X 2 | p value | |
|---|---|---|---|---|---|---|
| High | Low | |||||
| Gender | ||||||
| Male | 32 | 53 | 23 | 9 | 1.685 | 0.194 |
| Female | 28 | 47 | 24 | 4 | ||
| Age (years) | ||||||
| <60 | 31 | 52 | 25 | 6 | 0.202 | 0.653 |
| ⩾60 | 29 | 48 | 22 | 7 | ||
| Location | ||||||
| Colon | 34 | 57 | 26 | 8 | 0.090 | 0.764 |
| Rectum | 26 | 43 | 19 | 7 | ||
| TNM stage | ||||||
| I | 5 | 8 | 2 | 3 | 8.843 | 0.031 |
| II | 27 | 45 | 20 | 7 | ||
| III | 20 | 33 | 18 | 2 | ||
| IV | 8 | 13 | 8 | 0 | ||
| Distant | ||||||
| yes | 8 | 25 | 8 | 0 | 3.346 | 0.067 |
| No | 52 | 75 | 39 | 13 | ||
CRC, colorectal cancer; LSD1, lysine-specific histone demethylase 1A; TNM, tumor node metastasis.
MTT assay
Cells (3–5 × 103/well) were plated in 96-well plates and incubated overnight, followed by 5-FU treatment of various concentrations with or without ZY0511. After treatment, 10 μl of MTT (3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide; 5 mg/ml) was added to each well and incubated for an additional 1–4 h. Then, the medium was discarded and 150 μl of DMSO was added to dissolve the formazan. Absorbance was measured at 570 nm using a Microplate Auto reader (Bio-Rad, Hercules, CA, USA). The cell viability was calculated as (OD of drug-treatment) × 100%/(control OD). The IC50 values were calculated using Graph-Pad Prism 6 software (San Diego, CA, USA), using XY modeling.
Combination index score calculation
To evaluate the combination effect of ZY0511 and 5-FU, pre-incubated cells were co-treated with various concentrations of the two drugs for 72 h. Drug synergism was analyzed using the CompuSyn software (Biosoft, Ferguson, MO, USA).21 Following software instructions, drug combinations at non-constant ratios were used to calculate the combination index (CI) values in our study; CI <0.75, CI = 0.75–1.25, and CI >1.25 were considered to reflect synergistic, additive, and antagonistic effects, respectively.
Colony formation assay
For colony formation assays, SW620 and DLD-1 cells were seeded in triplicate into six-well plates (3–5 × 103/well) and incubated overnight. Cells were then cultured in various concentrations of 5-FU, with or without ZY0511 for 10–14 days. Growth medium was replaced every 2 days. Then, cells were fixed with methanol, stained with a 0.5% crystal violet solution for 15−20 min, and the colonies (>50 cells) were counted under a microscope.
Apoptosis assay
For apoptosis assays, cells (1 × 103/well) were seeded into six-well plates and cultured in the presence or absence of drugs, as indicated. After 48 h, the cells were harvested and apoptosis was determined using an Annexin V/PI apoptosis detection kit according to the manufacturer’s instructions using FCM (BD Biosciences, San Jose, CA, USA). The data were analyzed using FlowJo software (Ashland, OR, USA).
Mitochondrial membrane potential (ΔΨm) assay
The ΔΨm of cells treated with ZY0511 and/or 5-FU was measured. Briefly, SW620 and DLD-1 cells (2 × 105/well) were plated in six-well plates, followed by ZY0511 (1.25 and 2.5 μM) and/or 5-FU (10 μM) treatment for 48 h. Then, ΔΨm was determined according to the retention of the rhodamine 123 dye in cells. Cells were harvested, washed with PBS, and then incubated with rhodamine 123 (5 μg/mL) at 37°C for 15 min in the dark. After washing twice, the cells were incubated again in PBS at 37°C for 30 min in the dark and fluorescence was then measured using FCM.
Wound healing and transwell migration assay
SW620 and DLD-1 cells were seeded into six-well plates. When cells grew to 80% confluence, the cell monolayers were damaged with a micropipette to create a 2-mm wide linear wound, followed with various concentrations of 5-FU and/or ZY0511 treatment. After 24 h of incubation, cells were fixed and photographed. Images were acquired using a microscope (Zeiss, Jena, Germany) and the inhibition of migrated cells was expressed using a percentage of the value assigned for the untreated group.
Transwell migration assays were carried out in 24-well plates using Boyden chambers with an 8-μm pore size PET membrane (MCEP24H48, Millipore, Billerica, MA, USA). Briefly, 4–6 × 104 cells in 100 μl of a non-serum culture medium were resuspended in the upper chamber and medium with 10% fetal bovine serum was used as a chemoattractant in the lower chamber. After 24 h of incubation, non-invasive cells on the upper surface membrane were gently removed using a cotton swab. The invasive cells were fixed with paraformaldehyde and stained with 0.2% crystal violet. The stained invasive cells were photographed under an inverted light microscope with a 20X objective and quantified by manual counting in three randomly selected areas.
Immunoblot assay
Western blot analyses were performed as per a previously described method.22 Briefly, cells were harvested, washed twice with chilled PBS, and then lysed with RIPA buffer containing protease inhibitor cocktail. The concentrations of the cell lysate were measured using the Coomassie brilliant blue G-250 method and equalized before loading. Equal amounts of protein from each sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% skim milk and incubated with specific primary antibodies at 4°C overnight. After the incubation with the relevant secondary antibodies, the reactive bands were identified using an enhanced chemiluminescence kit (Amersham Bioscience, Piscataway, NJ, USA), scanned, and analyzed using a Gel-Pro imager and software. Western blot analyses were performed using primary antibodies against LSD1 (CST#2139), Bcl-2 (CST#15071), Bax (CST#2774), caspase-3 (CST#9662), E-cadherin (CST#14472), vimentin (CST#5741), MMP2 (CST#40994), TIMP2 (CST#5738), AKT (CST#4691), GSK3-β (CST#12456), β-catenin (CST#8480), and cyclin D1 (CST#2978). β-Actin (CST#3700) was used as an internal control. All antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA).
mRNA sequencing
mRNA Sequencing (mRNA-Seq) was performed using a profiler service provided by NOVELBIO Corporation (Shanghai, China). Total RNA was purified from SW620 cells after DMSO, 5-FU (10 μM), and/or ZY0511 (1.25 μM) treatment for 48 h and stored in TRIzol (Invitrogen, Carlsbad, CA, USA). Triplicate samples were harvested for each group, and significant probe sets were filtered for detection using a fold-change >1.5, a p < 0.05 (Student’s t test), and a false discovery rate <0.05. The differentially expressed genes were enriched using gene ontology (GO) analysis.
Reverse transcription-polymerase chain reaction
Total RNA was extracted from the eight cell lines using with TRIzol reagent, according to the manufacturer’s instructions. cDNAs was synthesized using a Thermo-Scientific reverse transcription kit (Waltham, MA, USA). Real-time qPCR (Bio-Rad, Hercules, CA, USA) was conducted using the SoAdvancedTM Universal SYBR Green kit (Bio-Rad), according to the manufacturer’s instructions: 95°C pre-denaturation for 30 s, 40 cycles of 95°C denaturation for 5 s, 60°C annealing and extension for 20 s, followed by a dissolution curve analysis (65–95°C in 2–5 s steps of 0.5°C each). The fold change between the target gene mRNA transcripts and the β-actin control was calculated and is shown in the histogram. Primer sequences used were: LSD1: F-GTC TCG TTG GCG TGC TGA TCC; R-GAA GAG TCG TGG AAT CGG CTG TG; β-Actin: F-CAT GTA CGT TGC TAT CCA GGC; R-CTC CTT AAT GTC ACG CAC GAT; CAD: F-CCT ACA CGG ATG GAG ACC TGG AG; R-CAC ATG CTC AGA GAT GGC GAT GG; DHODH: F-TTC TCT TCG CCT CCT ACC TGA TGG; R-GAG GTG AAG CGA ACA GCC AGT C; DCTPP1: F-TCC ATC AGC CTC GGA ATC TCC TC; R-GCT GCT AAT GCC ACC AGG TAG ATG; DUT: F-TCA GGC TTG GCT GCA AAA CA R-CGT TCG CAA ATG AGC TGT GC; TYMS: F-GTG GTG AAC AGT GAG CTG TCC TG; R-TTC AGT GGC TCG ATG TGA TTC AGG; and POLD1: F-CCG CTC CTA CAC GCT CAA TGC; R-GGT CGG TGA TGA TGC TGT GCT G.
Immunohistochemistry analysis
All tissues were fixed in 4% paraformaldehyde for 24–48 h and paraffin-embedded. Then, tissues were cut into 5-μm sections and subjected to standard staining with H&E. The paraffin tumor sections were stained with Ki-67 and cleaved-caspase 3. Images were obtained using a microscope (Leica, DM4000B, Wetzlar, Germany). The intensity of each antigen expression was graded as follows: 0, no staining; 1, weak; 2, moderate; 3, strong. The semi-quantitative evaluation on the basis of staining intensity was scored as the percentage of cells staining positively, as follows: 0, no staining; 1, 1–25% of the cells stained; 2, 26–50% of the cells stained; 3, 51–75% of the cells stained; and 4, >75% of the cells stained. Intensity score was multiplied by the density score to yield an overall score of 0–12 for each specimen. The immunoreactivity of each slide was divided into three groups based on the final scores: 0, negative; 1–4, low expression; 4–12, high expression. Negative controls without the primary antibody incubation were included in each staining run. Each slide was read and scored independently by two pathologists, in a blinded fashion.
Animal studies
All animal experiments were approved by the Institutional Animal Care and Treatment Committee of the State Key Laboratory of Biotherapy, Sichuan University, China (permit number: 20161205-3). Female BALB/C nude mice (18–20 g, 6–8 weeks old) using for animal experiments were obtained from HFK Bioscience Co., Ltd. (Beijing, China). Tumor xenograft models (SW620 and DLD-1) were established by injecting cancer cells subcutaneously into the right flank of mice. Nude mice bearing tumor with an average volume of 100–300 mm3 in size were random divided into four groups (n = 10 per group), the vehicle, 5-FU, ZY0511, and the combination of the two drugs. The dosage and approach of the combination group was given as following: 5-FU: 30 mg/kg, three times per week for three consecutive weeks via intraperitoneal injection (D1/D2/D3, D8/D9/D10, and D16/D17/D18); ZY0511: 50 mg/kg, once daily by oral administration. Tumor growth and mice body weight were regularly measured every 3 days during the treatment. The tumor size was calculated according to the formula: tumor volume (mm3) = (L × W2)/2, where L is the length and W is the width.
Toxicity evaluation
To investigate potential side effects or toxicity on mice during the treatment, mice were observed continuously for relevant indices such as body weight loss, diarrhea, anorexia, and skin ulcer, or toxic death. On the 27th day, all animals were euthanized after blood collection for routine blood analysis and blood chemistry analysis. The organs such as the heart, liver, spleen, lungs, and kidneys were collected and fixed in 4% formaldehyde solution prior to analysis by H&E staining, and observed by two pathologists in a blinded manner.
Mouse models of pulmonary and liver metastasis
Liver metastasis models were established through injecting of SW620 cells (2.5 × 106) to the spleen subcapsular space in BALB/C nude mice. The established metastatic mice were random divided into four groups (n = 10 per group), vehicle, 5-FU, ZY0511 and two drugs combination group. Mice were sacrificed under anesthesia after 9 weeks and the livers were obtained. For the induction of pulmonary metastases, SW620 cells (1 × 106) were delivered by intravenously injection into the tails of mice. Mice were random grouped into four groups (n = 10 per group) as the liver metastasis model. After 10 weeks, mice were sacrificed under anesthesia for collecting the lungs. The tumor metastases were assessed through H&E staining and quantified basing on the visual examination. The data was quantified manually.
Statistical analysis
Data are expressed as the means ± standard error of the mean (SEM). The indicated sample size (n) represents biological replicates. Group allocation and outcome assessment were not performed in a blinded manner. All samples that met proper experimental conditions were included in the analysis. Survival was measured using the Kaplan–Meier method. Statistical significance was determined using Student’s t test, ANOVA, log-rank test, and Pearson’s correlation, using the Prism 6 software (GraphPad Software, La Jolla, CA, USA). Significance was set at p < 0.05.
Results
LSD1 overexpression in human CRC tissues is correlated with CRC development and 5-FU resistance
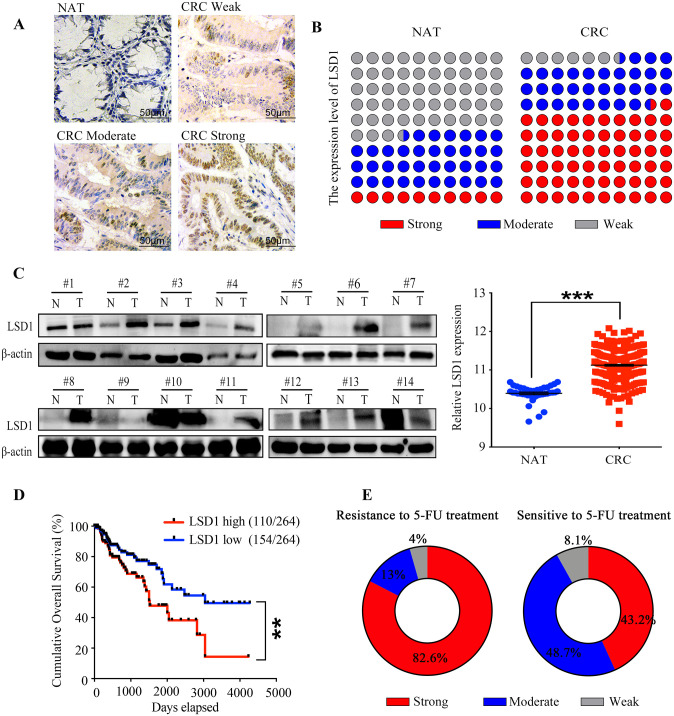
To explore the clinical significance of LSD1 expression in patients’ sensitivity to 5-FU, we detected the expression level of LSD1 in human CRC tissues and paired normal adjacent tissues (NATs) using immunohistochemistry (IHC) staining (the clinical characteristics of patients are provided in Table 1). The relative levels of LSD1 were assessed by staining density scores. Our results showed that, in CRC tissues, 37 cases (62%) exhibited strong, 19 (32%) exhibited moderate, and 4 (6%) exhibited weak or no immunopositivity. In contrast, most NATs (53%) exhibited weak or no LSD1 expression (Figure 1A, 1B). The LSD1 expression on a cohort of 14 CRC tissues selected randomly from these 60 patients was further validated using western blot analysis. Consistently with the results of IHC, we observed elevated protein levels of LSD1 in CRC tissues, compared with NATs (Figure 1C). Statistical analysis revealed that expression levels of LSD1 significantly increased in CRC tissues compared with the corresponding NATs (Figure 1B, 1C). In addition, LSD1 level was negatively correlated with tumor node metastasis (TNM) stage of CRC patients (Table 1). However, no significant association was found between LSD1 expression in CRC tissues and gender, age, or location (p > 0.05, Table 1). In addition, data mining of mRNA expression in human CRC tissues and matched NATs in Gene Expression Omnibus (GEO) database were performed and results showed that evaluated expression of LSD1 in CRC tissues was negatively correlated with the survival rate of CRC patients (Figure 1D). Collectively, these results reveal that LSD1 overexpression is tightly linked to CRC development.
Figure 1.
LSD1 overexpression in human CRC tissues is correlated with CRC development and 5-FU resistance (A) Representative immunohistochemistry images of LSD1 protein in human NATs and CRC tissues with weak, moderate and strong immunopositivity, respectively. Positive cells were stained brown. Scale bar, 50 μM. (B) Quantification of LSD1 expression in CRC tissues based on staining density scores of IHC. (C) Immunoblot analysis of LSD1 expression in 14 pairs of human CRC tissues and NATs. (D) Correlation analysis of LSD1 expression and patient survival by Kaplan–Meier analysis based on the GEO database. (E) Quantification of LSD1 expression in CRC tissues from patients resistance or sensitivity to 5-FU treatment based on IHC staining.
**p < 0.01, ***p < 0.001.
CRC, colorectal cancer; GEO, Gene Expression Omnibus; IHC, immunohistochemistry; LSD1, lysine-specific histone demethylase 1; NAT, normal adjacent tissues; 5-FU, 5-fluorouracil.
Then, we focused on evaluating the correlation between LSD1 overexpression and patient response to 5-FU treatment based on IHC results. Among 60 patients, 23 cases acquired 5-FU resistance within 1 year, and 37 cases were sensitive to 5-FU treatment. For the 23 patients who acquired 5-FU resistance, 19 cases (82.6%) exhibited strong, 3 cases (13%) moderate, and 1 case (4.4%) weak immunopositivity (Figure 1E). Conversely, for the 37 5-FU-sensitive patients, 16 cases (43.2%) exhibited strong, 18 cases (48.7%) exhibited moderate, and 3 cases (8.1%) exhibited weak or no immunopositivity (Figure 1E), These results indicated that LSD1 upregulation in CRC tissues is associated with 5-FU resistance in patients. Taken together, our results suggest that LSD1 overexpression is tightly linked to CRC development and 5-FU resistance.
ZY0511 inhibits human CRC cell proliferation in vitro
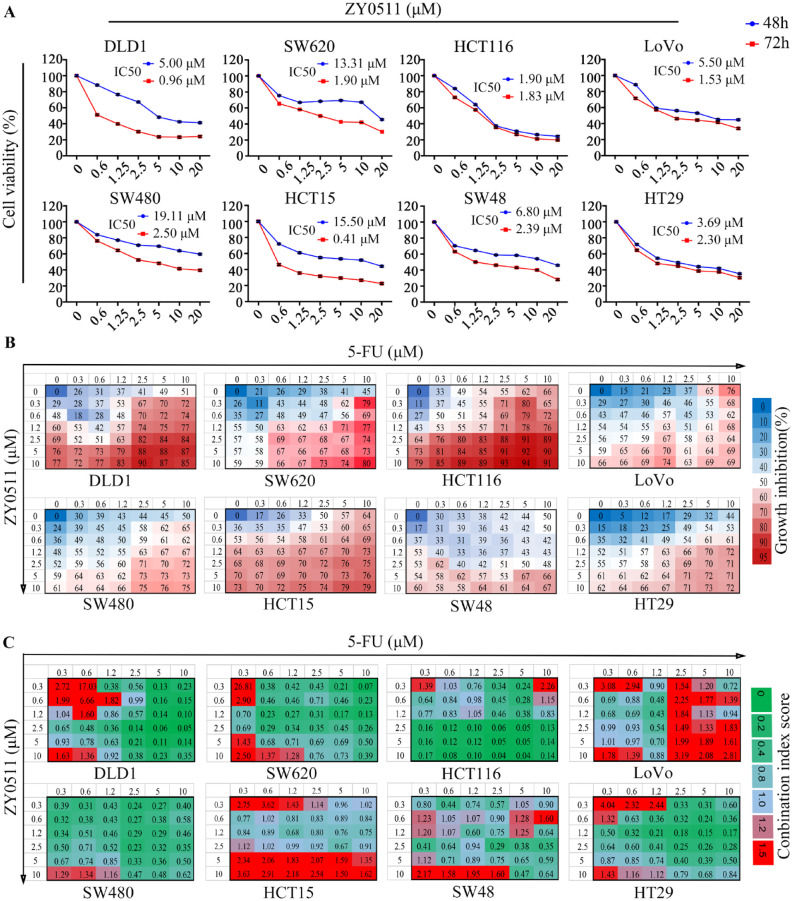
To investigate the potential functional roles of LSD1 in CRC, we profiled mRNA and protein expression of LSD1 in a panel of human CRC cell lines, including SW620, HCT116, SW480, LoVo, DLD-1, SW48, HCT15, and HT29 cells using quantitative reverse transcription-polymerase chain reaction (RT-qPCR) and western blot analysis. The results revealed that all CRC cells expressed a high level of LSD1 compared with normal human colonic epithelial cell NCM 460 (Figure S1B). Specifically, the expression of LSD1 was relatively high in SW620, DLD-1, and HCT116 cells and low in SW48 and HT29 cells (Figure S1B). Our results are consistent with previous studies reporting that LSD1 expression is high in SW620 cells and low in HT29 cells.
Then, we evaluated the antiproliferative activity of ZY0511 using the above cells. CRC cells were treated with various concentrations of ZY0511 for 48 h and 72 h, respectively, followed by MTT assays. The results showed that ZY0511 inhibited CRC cells proliferation in a concentration and time-dependent manner, with IC50 values ranging from 1.84 μM to 19.11 μM for 48 h, and 0.53 μM to 2.5 μM for 72 h, respectively (Figure 2A). In most cells, ZY0511 sensitivity was associated with LSD1 expression level. For example, SW620, HCT116, and DLD-1 cells with high LSD1 expression were more sensitive to ZY0511 treatment, with IC50 values of 1.90 μM, 1.83 μM, and 0.96 μM for 72 h, respectively, whereas SW480 and SW48 cells with low LSD1 expression were less sensitive (IC50 of 2.5 μM and 2.38 μM, respectively). However, the sensitivity of LoVo and HCT15 to ZY0511 is not completely consistent with their LSD1 level. HCT15 with low LSD1 level is highly sensitive to ZY0511 (IC50 of 0.4 μM). Taken together, ZY0511 inhibited human CRC cells proliferation in vitro. Cells sensitivity to ZY0511 was associated mainly with LSD1 level. However, there might be other factors involved in cell sensitivity to ZY0511.
Figure 2.
ZY0511 and 5-FU combination synergistically inhibits CRC cells proliferation in vitro (A) The proliferation of human CRC cells was evaluated by MTT assay after various concentrations of ZY0511 treatment for 48 and 72 h, respectively. The IC50 values were the mean of three individual experiments. (B) Cells were treated with ZY0511 in combination with 5-FU in CRC cells. Percentages of growth inhibition at each concentration of the drugs are presented. Data are show as mean of three independent experiments. (C) CI scores for CRC cells treated with 5-FU in combination with ZY0511 at the indicated concentrations. Each CI score represents data from three independent experiments.
CI, combination index; CRC, colorectal cancer; 5-FU, 5-fluorouracil; MTT, 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide.
ZY0511 and 5-FU combination synergistically inhibits CRC cells proliferation in vitro
Next, we evaluated whether ZY0511 in combination with 5-FU could synergistically inhibit CRC cell proliferation. CRC cells were treated with various concentrations of ZY0511 with or without 5-FU, followed by MTT assays. The results showed that ZY0511 significantly increased 5-FU antiproliferative effect against CRC cells (Figure 2B). For example, in SW620 cells, the inhibition rate for 5-FU (10 μM) and ZY0511 (0.3 μM) treatment alone was 45% and 26%, respectively, whereas 5-FU and ZY0511 combination increased the inhibition rate to 79% (Figure 2B). In DLD-1 cells, the inhibition rate for 5-FU (10 μM) and ZY0511 (0.3 μM) treatment alone was 51% and 29%, respectively, whereas 5-FU and ZY0511 combination increased the inhibition rate to 72% (Figure 2B).
CI scores were analyzed based on inhibition rates, and the CI <0.75, CI = 0.75–1.25, and CI >1.25 indicated synergistic, additive, and antagonistic effects, respectively. In DLD-1 with high LSD1 level, the CI scores for 0.6, 1.2, and 2.5 μM ZY0511 in combination with 10 μM 5-FU were 0.15, 0.1, and 0.05, respectively (Figure 2C). Similarly, in SW620 cells with a high LSD1 level, the CI scores for the above-mentioned combination were 0.23, 0.13, and 0.26, respectively, suggesting strongly synergistic antiproliferative effects (Figure 2C). Notably, most of CI scores in ZY0511 >2.5 μM combinations indicated synergistic effects in all CRC cells, except for HCT15 and LoVo cells. In HCT15 cells with a low LSD1 level, 97% of the CI scores for ZY0511 and 5-FU combination were higher than 0.75, and 50% of the CI scores were higher than 1.25, suggesting additive effects or even antagonistic effects. Collectively, these data suggested that ZY0511 in combination with 5-FU synergistically inhibited proliferation of CRC cells. The synergistic effects were associated with the LSD1 level in cells. Therefore, SW620 and DLD-1 cells with a high LSD1 level and strong synergistic effects were selected for further studies.
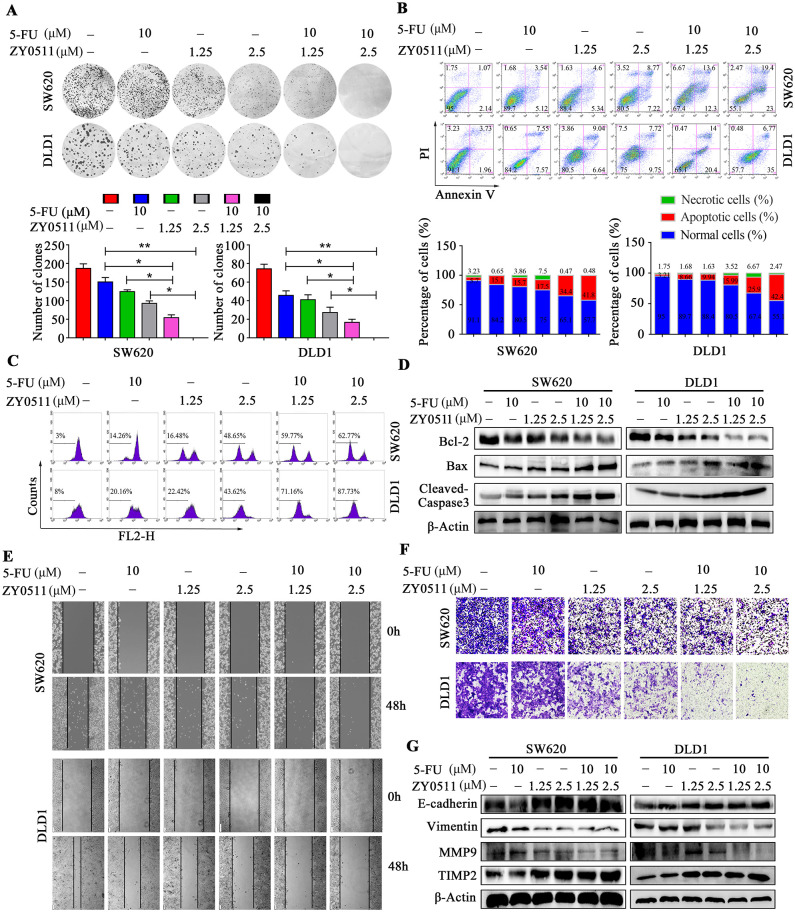
To further assess the antiproliferative effect of ZY0511, colony formation assays were performed. Consistent with MTT results, ZY0511 or 5-FU treatment alone significantly decreased the number and size of colonies, compared with the control. The combination of the two drugs resulted in a more obvious inhibition based on colony number and size (Figure 3A).
Figure 3.
ZY0511 in combination with 5-FU induces apoptosis and inhibits migration of CRC cells (A) Representative images (top) and quantification (bottom) of colony formation assay following ZY0511 and 5-FU treatment in CRC cells. Data are presented as mean ± SD (t test, *, p < 0.05; **, p < 0.01). (B) Representative images (top) and quantification (bottom) of apoptosis induction in CRC cells with ZY0511 and 5-FU treatment. (C) ΔΨm assay in CRC cells with ZY0511 and 5-FU treatment. (D) Western blot analysis of apoptosis proteins in CRC cells with ZY0511 and 5-FU treatment. (E, F) Representative images of wound-healing assay (E) and transwell migration assay (F) in CRC cells treated with 5-FU and ZY0511. (G) Western blot analysis of indicated proteins in CRC cells treated with 5-FU and ZY0511.
CI, combination index; CRC, colorectal cancer; 5-FU, 5-fluorouracil; ΔΨm, mitochondrial membrane potential; SD, standard deviation.
ZY0511 in combination with 5-FU induces apoptosis
Next, we detected apoptosis levels using flow cytometry (FCM) by using Annexin V/PI dual-labeling. SW620 cell with ZY0511 (1.25 μM or 2.5 μM) or 5-FU (10 μM) treatment exhibited 9.94%, 15.99%, and 8.66% apoptosis rates, respectively, whereas the apoptosis rate increased to 25.9% and 42.4%, respectively, when ZY0511 combined with 5-FU treatment was employed (Figure 3B). Similar results were observed in DLD-1 cells. The apoptosis rates were 15.68%, 17.47%, and 15.12% for ZY0511 (1.25 μM or 2.5 μM) or 5-FU (10 μM) alone, 34.4% and 41.77% for ZY0511 and 5-FU combination, respectively (Figure 3B).
The loss of mitochondrial membrane potential (ΔΨm) is an important event in the apoptotic process. Thus, the ΔΨm alteration was detected using the mitochondria-specific and voltage-dependent dye Rh123 in SW620 and DLD-1 cells. In SW620 cells, ZY0511 (1.25 μM and 2.5 μM) and 5-FU (10 μM) led to 16.48%, 48.65%, and 14.26% losses of ΔΨm, respectively, and 59.77% and 62.77% losses of ΔΨm upon the respective combinations (Figure 3C, S2A). Similarly, in DLD-1 cells with the same treatment as SW620, the ΔΨm losses were 22.42%, 43.62%, 20.16%, 71.16%, and 87.73%, respectively (Figure 3C, S2A).
To further confirm the apoptosis induction, Bcl-2, Bax, and cleaved caspase-3 were detected using western blot analysis in SW620 and DLD-1 cells after ZY0511 (1.25 μM and 2.5 μM) and/or 5-FU (10 μM) treatment for 48 h. Unlike 5-FU alone, 5-FU in combination with ZY0511 significantly decreased Bcl-2 expression (Figure 3D). Conversely, cleaved caspase-3 and Bax significantly increased after combination compared with single treatment (Figure 3D). Taken together, ZY0511 in combination with 5-FU significantly induces apoptosis in SW620 and DLD-1 cells.
ZY0511 and 5-FU combination inhibits CRC cells migration in vitro
To assess the effects of ZY0511 and 5-FU combination on cell migration, we performed wound-healing assays. In SW620 and DLD-1 cells, the wound was wider in 5-FU or ZY0511 treatment alone than that in the control group. The wound in the combination group was more obvious compared with the single drug treatment, indicating that ZY0511 in combination with 5-FU synergistically inhibited CRC cells migration (Figure 3E, S2B). Similar results were obtained in the transwell migration assay (Figure 3F, S2C). In SW620 cells, the combination of ZY0511 (1.25 μM and 2.5 μM) and 5-FU (10 μM) significantly inhibited cell migration by 65.9% or 90.2%, respectively, and for DLD1 cells, by 76% and 89%, respectively (Figure 3F, S2C).
As epithelial to mesenchymal transition (EMT) is an important process in the migration and invasion of CRC, we detected the epithelial marker E-cadherin, the mesenchymal markers vimentin, metalloproteinase (MMP9), and metalloproteinase inhibitor (TIMP2) in SW620 and DLD-1 cells after ZY0511 (1.25 μM and 2.5 μM) and/or 5-FU (10 μM) treatment for 48 h, using western blot analysis. 5-FU alone changed vimentin expression only minimally, whereas the combination treatment decreased vimentin and MMP9 expression significantly (Figure 3G). E-cadherin and TIMP2 increased significantly after the combination treatment compared with the single drug treatment. Collectively, ZY0511 in combination with 5-FU significantly inhibits cells migration.
ZY0511 and 5-FU combination suppresses Wnt/β-catenin signaling and pyrimidine metabolism
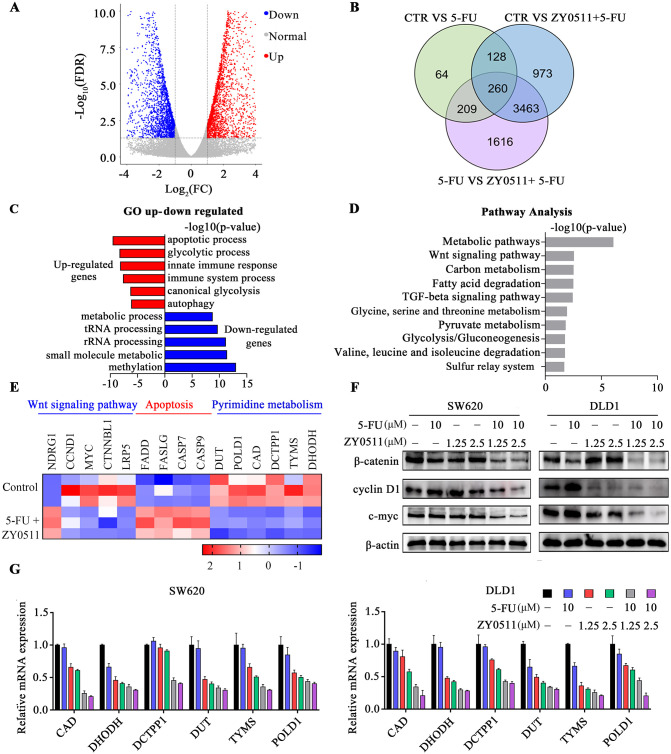
To gain insight into the mechanism of how ZY0511 and 5-FU combination treatment inhibited CRC cells proliferation and migration, we used mRNA sequence (mRNA-Seq) technology to explore the factors or genes involved in the antitumor effects of ZY0511 and 5-FU treatment. Transcriptome analysis results showed that a total of 4824 genes were markedly altered following 5-FU in combination with ZY0511 treatment, compared with 5-FU treatment alone (Figure 4A). Venn analysis revealed that 260 genes changed with significant differences (Figure 4B). GO enrichment analysis revealed that the combination treatment upregulated the expression of genes closely associated with the apoptotic, glycolytic, and immune processes (Figure 4C). Conversely, genes correlated with methylation, metabolic process, and RNA processing decreased (Figure 4C). Further, according to GO analysis, altered gene expression is related with signaling pathways. Pathway analysis showed these were involved in diverse biological processes, including metabolic pathways, the Wnt signaling pathway, fatty acid degradation, and the TGF-beta signaling pathway (Figure 4D). Notably, Wnt signaling, apoptotic, and metabolism pathways were predominantly altered.
Figure 4.
ZY0511 in combination with 5-FU suppresses Wnt/β-catenin signaling and pyrimidine metabolism. (A) Volcano plot of the mRNA-seq analysis of SW620 cells after 5-FU and/or ZY0511 exposure (red represents upregulation, blue represents downregulation, and gray represents no meaningful change). (B) Venn analysis showed significant differences gene expression. (C) Representative GO term analysis of upregulated and downregulated genes after 5-FU and/or ZY0511 exposure. (D) Ingenuity pathway analysis of SW620 cells after 5-FU and/or ZY0511 treatment. Results are expressed as –log (p value). (E) The mRNA-seq representative genes expression in SW620 and DLD1 cells after 5-FU and/or ZY0511 treatment. (F) Western blot analysis of Wnt/β-catenin pathway protein. β-actin was used as the reference protein. (G) mRNA level of pyrimidines metabolic enzymes CAD, DHODH, DCTPP1, DUT, TYMS, and POLD1 in SW620 and DLD1 cells by RT-qPCR.
5-FU, 5-fluorouracil; GO, gene ontology; mRNA-seq, mTNA sequencing; RT-qPCR, quantitative reverse transcription-polymerase chain reaction.
Aberrant activation of the Wnt/β-catenin pathway contributes to the development of various human cancers, including CRC. In CRC, β-catenin accumulates in a free cytosolic form and engages with nuclear T-cell factor (TCF) transcription factors. We further detected the expression of β-catenin and its target proteins cyclin D1 and c-Myc by using western blot analysis. The results showed that ZY0511 and 5-FU combination treatment significantly downregulated the expression of β-catenin, cyclin D1, and c-Myc, which was consistent with mRNA-Seq results, indicating the suppression of β-catenin signaling in the two CRC cell lines (Figure 4E, 4F).
mRNA-Seq results also revealed alterations in the pyrimidine metabolism pathway, a key component of DNA synthesis. In turn, LSD1 plays critical roles in chromatin remodeling via histone demethylation and DNA damage, which are essential for influencing genome DNA repair processes. Consistent with the mRNA-Seq results, RT-qPCR results confirmed the down-regulation of genes in the pyrimidine metabolic pathway upon combination treatment, including carbamoyl-phosphate synthetase (CAD), dihydroorotate dehydrogenase (DHODH), Duchenne deoxyribonucleic acid (DUT), dCTP pyrophosphatase 1 (DCTPP1), TYMS, and DNA polymerase delta catalytic subunit gene 1 (POLD1) (Figure 4E, 4G).
ZY0511 and 5-FU combination synergistically inhibits tumor growth in vivo
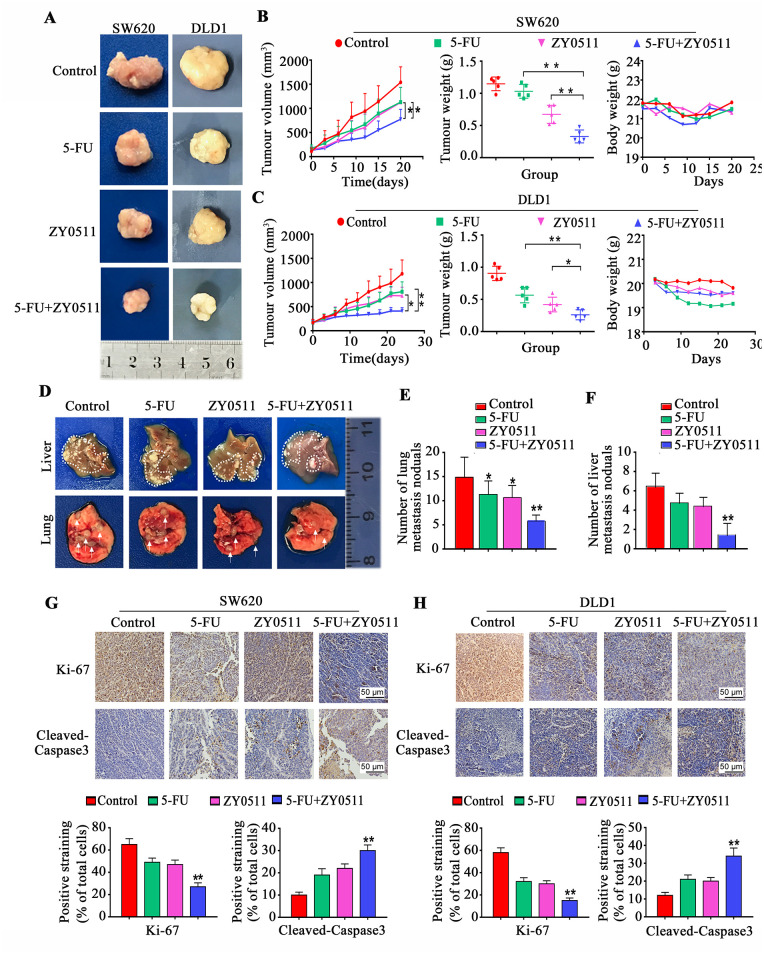
The in vivo antitumor activities of ZY0511 and/or 5-FU were evaluated with mouse subcutaneous xenograft models. Nude mice carrying SW620 and DLD-1 xenograft tumors were administered ZY0511 (50 mg/kg/day, orally), 5-FU (30 mg/kg, thrice weekly, intraperitoneal injection), or both drugs in combination for 3 weeks. Tumor volumes and body weight were measured every 3 days. The results showed ZY0511 or 5-FU treatment inhibited the tumor growth in SW620 xenograft models at an inhibition rate of 30% and 36%, respectively, whereas the combination treatment produced marked tumor regression with an inhibition rate of 66%, despite no apparent toxicity (Figure 5A, 5B). In DLD1 models, the tumor inhibition rates were 43% for ZY0511 and 46% for 5-FU treatment, respectively, whereas the inhibition rate increased to 61% (Figure 5A, 5B). The tumor weights showed a similar pattern (Figure 5B, 5C). No loss of body weight was observed with this dosage (Figure 5B, 5C). We also examined cells proliferation and apoptosis induction in xenograft tumors by IHC staining. Consistent with in vivo results, the combination treatment caused a significant reduction in proliferating cells stained using nuclear Ki-67 in SW620 (Figure 5G) and DLD-1 tumor tissues (Figure 5H), and a significant up-regulation in apoptotic cells stained using cleaved-caspase-3, compared with the single drug treatments (Figure 5G, 5H).
Figure 5.
ZY0511 in combination with 5-FU inhibits CRC growth in vivo (A) Representative images of tumor for SW620 and DLD1 xenografts treated with vehicle, 5-FU (30 mg/kg) and/or ZY0511 (50 mg/kg), respectively. (B–C) Mean tumor volume (mm3) (left), tumor weights (middle) and body weights (right) of SW620 (B) and DLD1 (C) xenografts during treatment with 5-FU and/or ZY0511. Data are shown as mean ± SD. (n = 6 per group). (D) Representative images of live (top) and lung (bottom) metastasis nodules for SW620 metastasis model treated with vehicle, 5-FU (30 mg/kg) and/or ZY0511 (50 mg/kg), respectively. (E–F) Numbers of lung (E) and liver (F) metastasis nodules are shown as mean ± SEM (n = 6 per group). (G–H) Representative images of Ki-67 and cleaved-caspase3 in SW620 (G) and DLD1 (H) xenograft tumor samples.
*p < 0.05, **p < 0.01. Scale bar represents a distance of 50 μm.
5-FU, 5-fluorouracil; SD, standard deviation; SEM, standard error of the mean.
To further evaluate the anti-metastatic effect of ZY0511 in combination with 5-FU in vivo, we established an in vivo metastasis model by intravenously injection of SW620 cells into nude mice. Metastatic nodules of the lungs were quantified after tumor-bearing mice being euthanized at 9 weeks. Multiple large nodules were evident in vehicle-treated groups, whereas the extent of lung metastasis was reduced in ZY0511 and/or 5-FU-treated mice, with a particular reduction of metastatic nodules in the combination treatment group (Figure 5D, 5E).
The mouse liver metastasis model was generated by spleen injection of SW620 cells, which is highly invasive. Mice were euthanized 10 weeks after SW620 injection. ZY0511 or 5-FU treatment alone did not affect the liver metastases or hepatic tumor burden. However, ZY0511 in combination with 5-FU dramatically reduced the number of visible metastatic nodules on the liver surface of tumor-bearing, compared with control mice (Figure 5D, 5F). Thus, ZY0511 in combination with 5-FU significantly inhibits tumor growth and metastasis in vivo.
Safety profile of ZY0511 in combination with 5-FU in vivo
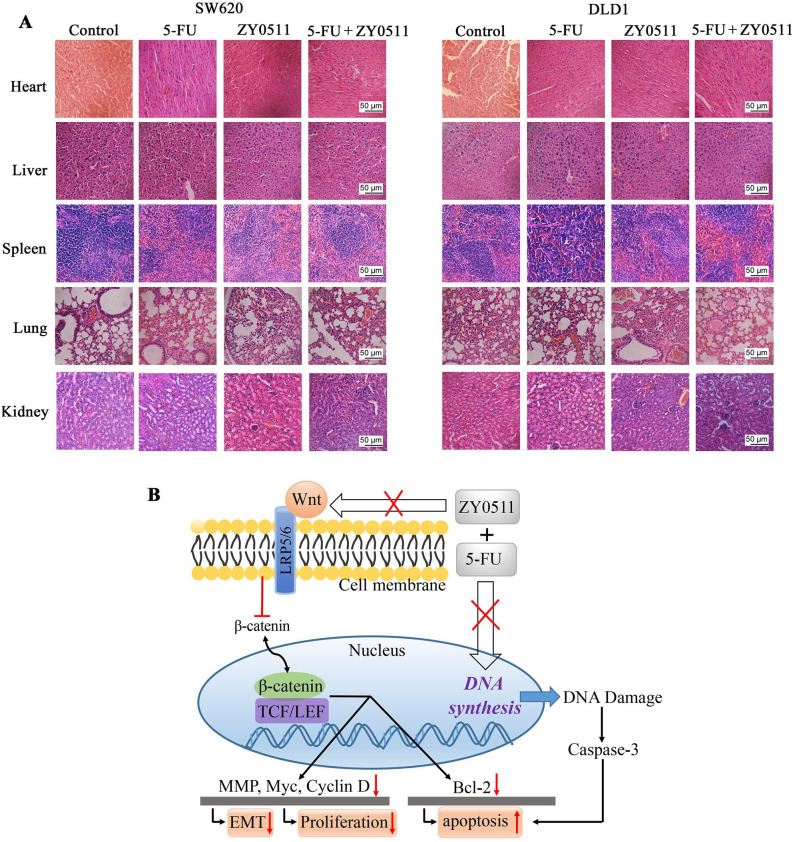
No obvious adverse effects were observed during the ZY0511 and 5-FU treatment, such as toxic death, dermatitis, or body weight loss. Furthermore, microscopic examination showed no pathologic changes in the heart, liver, spleen, lungs, or kidneys of mice after ZY0511 and 5-FU treatment (Figure 6A).
Figure 6.
Safety profile of ZY0511 in combination with 5-FU in vivo and proposed mechanisms. (A) Representative H&E staining images of heart, liver, spleen, lung and kidney of mice after ZY0511 and 5-FU treatment. No obvious pathological changes were observed in the control and treated groups. Scale bar represents a distance of 50 μm. (B) Proposed mechanisms by which 5-FU co-develop hypersensitivity to ZY0511. LSD1 inhibitor ZY0511 in combination with 5-FU synergistically reduced CRC cells proliferation and migration by inhibition Wnt/β-catenin signaling and metabolism enzymes in the DNA synthesis, which finally induced apoptosis and restored sensitivity to 5-FU.
5-FU, 5-fluorouracil; EMT, epithelial-mesenchymal transition; H&E, hematoxylin and eosin; LEF, lymphoid enhancer factor; TCF, T-cell factor.
Discussion
LSD1 plays important roles in tumor progression and drug resistance,23 and LSD1 inhibitors have great potential for cancer therapies.24 In the present study, we demonstrated that ZY0511, a novel and potent LSD1 inhibitor developed by our group, significantly inhibited human CRC cells proliferation and migration both in vitro and in vivo. Importantly, we found that the overexpression of LSD1 was associated with patient sensitivity to 5-FU treatment, and the combination of ZY0511 and 5-FU exerted synergistic anticancer efficacy against CRC cells. Moreover, our studies demonstrated that the combination treatment suppressed the Wnt/β-catenin signaling and pyrimidine metabolic pathways, thus inhibiting tumor proliferation and metastasis. Our results provide a theoretical foundation for the clinical application of LSD1 inhibitors and a new combinatorial strategy for CRC treatment.
The observations of LSD1 overexpression in solid and hematological malignancies have led to the active development of small-molecule LSD1 inhibitors.23,25 Among these inhibitors, irreversible LSD1, such as GSK2879552 and ORY-1001, are currently undergoing clinical studies for cancer therapy.13,26 Though they exhibited good activity against hematologic malignancies, such as AML,26,27 and acute lymphoblastic leukemia (ALL),26,28 their efficiency against solid tumors was not obvious, except against small-cell lung cancer (SCLC).13 Moreover, these LSD1 inhibitors comprise mainly tranylcypromine (TCP) derivatives, which exhibit biological toxicity. Thus, there is a large unmet clinical need to explore novel, potent, and selective LSD1 inhibitors with structural diversity against solid tumors. For example, Sharma et al. developed a potent, specific, and reversible LSD1 inhibitor, HCI-2509, with a benzamide structure. HCI-2509 has remarkable single agent efficacy and in vivo tolerability in poorly differentiated malignancies such as Ewing’s sarcoma, neuroblastoma, and prostate cancer.29–31 We successfully used a conformational strategy to obtain and optimize the novel and potent LSD1 inhibitor ZY0511. It has a benzohydrazide scaffold, unlike the LSD1 inhibitors currently in clinical studies. Our previous study has shown that ZY0511 exhibits significant anticancer effects against a variety of human solid cancer cell lines in vitro, and is well tolerated in vivo, suggesting it is highly safe. We successfully obtained a patent from the State Intellectual Property Office of the People’s Republic of China.
Previous studies have reported that LSD1 is upregulated in human CRC tissues, compared with NATs by IHC staining, western blot analysis, or TCGA database, respectively.32,33 Moreover, the expression of LSD1 is significantly higher in colon cancer with high TNM stages and distant metastasis.9 LSD1 knockdown by siRNA could inhibit the proliferation of CRC cells. CBB1003, a LSD1 inhibitor, suppresses HCT116 cells proliferation with a IC50 value of 250.4 µM.34 These data evoked our interest on the anticancer effect of ZY0511 in CRC. We further confirmed LSD1 overexpression in human CRC specimens using IHC staining, western blot analysis and TCGA database were both used, which is different from the above-mentioned studies, in which only a single assay was used. Our results have shown that ZY0511 exhibits significant anticancer and anti-metastasis effects against human CRC cells, both in vitro and in vivo. Compared with CBB1003 or tranylcypromine, ZY0511 exhibited a 10- to 100-fold stronger antiproliferative effect against CRC cells in vitro. Moreover, to the best of our knowledge, ZY0511 is the first LSD1 inhibitor to exhibit growth inhibition and anti-metastasis efficiency against CRC in vivo, supporting ZY0511 as a promising candidate for CRC treatment. Interestingly, we found that CRC cells with high LSD1 levels (DLD-1, SW620, and HCT116) were sensitive to ZY0511, and the CRC cells with lower LSD1 levels (SW480 and HT29) were less sensitive to ZY0511. These data suggest that ZY0511 sensitivity is associated with LSD1 level, which is consistent with previous studies.13 However, we also found that the sensitivity of small parts of CRC cells (LoVo and HCT15) to ZY0511 was not completely consistent with their LSD1 levels. For example, HCT15 with low LSD1 level has high sensitivity to ZY0511. Thus, there might be other factors involved in sensitivity of LSD1 inhibitors besides the LSD1 level. For example, Zhang et al. found that expression of Sox2, which is a target of LSD1, was associated with sensitivity to LSD1 inhibition in lung, breast, and ovarian cancer cells.35 CBB1007, an LSD1 inhibitor, selectively impaired the proliferation of Sox2-expressing lung squamous cell carcinomas, but not that of Sox2-negative cells.35 Moreover, nonhistone substrates of LSD1 might be involved in LSD1 inhibitor sensitivity. Although LSD1 was originally identified as a histone lysine demethylase, it has been shown to demethylate nonhistone substrates, such as p53, DNMT1, MYPT1, HSP90, STAT3, ERα, and HIF-1α,36,37 which are important in cancer biology. Thus, the basic status of these nonhistone substrates of LSD1 might affect cells sensitivity to LSD1 inhibitors. Pishas et al. found that the induction of KDM1B was strongly associated with hypersensitivity of SP-2509, a LSD1 inhibitor.38 To provide a precision therapy strategy for using LSD1 inhibitor, the factors that are involved in sensitivity of LSD1 inhibitors are worth further investigation.
To date, 5-FU remains a commonly used chemotherapeutic drug for advanced CRC treatments and clinical studies. However, drug resistance remains a critical limitation to the clinical application of 5-FU. Over the past decades, an increased understanding of the 5-FU resistance mechanism has promoted the progress of new strategies that increase antineoplastic activity. Accordingly, 5-FU-based chemotherapy, combined with epigenetic modulatory drugs, is an effective strategy to enhance chemotherapy sensitivity in CRC. A combination of trichosstatin, a histone deacetylase (HDAC) inhibitor, with 5-FU synergistically suppressed CRC viability by downregulating expression of thymidylate synthase (TS) ,39 KRAS, c-Myc,40 or stemness promoting markers, such as LGR5, Nanog, and β-catenin.41 In the present study, we demonstrated that LSD1 overexpression in CRC tissues was associated with patients’ sensitivity to 5-FU treatment. ZY0511 in combination with 5-FU displayed synergistic effects against CRC cell proliferation and migration, which indicated that ZY0511 could enhance chemosensitivity toward 5-FU therapy. Our results are similar to those of a previous study that demonstrated that LSD1 inhibitors pargyline and tranylcypromine enhanced 5-FU sensitivity in oral squamous cell carcinoma cells.11 Thus, LSD1 inhibitors in combination with 5-FU might be an effective strategy for CRC therapy.
However, the underlying mechanism of how LSD1 regulates sensitivity to 5-FU remains unclear. Mechanistically, we found that ZY0511 in combination with 5-FU inhibited the expression of β-catenin and its target genes, which indicated that Wnt signaling was involved in the synergistic anticancer efficiency of ZY0511 and 5-FU. Aberration of the classical Wnt/β-catenin pathway is implicated in the development, occurrence, and metastasis in CRC.42 Overactive Wnt signaling leads to constitutively active β-catenin, which, via lymphoid enhancer factor (LEF)/TCF transcription factors, leads to the inappropriate activation of Wnt target genes that control cells proliferation and migration.43,44 More than 80% of the sporadic CRC tumors exhibit hyperactive Wnt signaling. Activation of the Wnt/β-catenin signaling pathway is an important mechanism of 5-FU resistance, and inhibition of β-catenin and its downstream genes represents an important potential strategy for the clinical reversal of 5-FU resistance. For example, Guo et al. found that HCT-8R, a 5-FU-resistant CRC cell, exhibited high expression of β-catenin and TCF4 by suppressing the checkpoint kinase 1 (CHK1) pathway. Wnt3a stimulation enhanced HCT-8R cells resistance to 5-FU, whereas IWP-2, a Wnt pathway inhibitor, reduced resistance.41 The above-mentioned studies support our results that ZY0511 could enhance 5-FU sensitivity by inhibiting β-catenin. Moreover, there are several studies demonstrating that LSD1 could activate the Wnt/β-catenin pathway, which is consistent with our results. In cancer initiating cells (CICs) of hepatocellular carcinoma (HCC), LSD1 overexpression activated β-catenin by inhibiting the expression of several suppressors of β-catenin signaling, especially Prickle1 and APC. LSD1-associated activation of β-catenin is essential for maintaining the activity of Lgr5+ CICs and drug resistance, such as Cisplatin and sorafenib, in HCC. LSD1 inhibitors, such as pargyline and GSK2879552, suppressed stem-like properties of sorafenib-resistant HCC cells by derepressing the expression of multiple upstream negative regulators of the Wnt pathway.45 In the gastric cancer cell line MKN-28, LSD1 knockdown suppressed the expression of β-catenin, VEGF, and Bcl-2, thus inhibiting cells proliferation and invasion, and increased the activity of cisplatin in vitro.46 Thus, our results demonstrated that LSD1 inhibitor and 5-FU synergistically inhibited CRC proliferation by suppressing the Wnt pathway in CRC.
Apart from the roles in tumor progression and drug resistance, LSD1 also participates in the regulatory network of tumors metastasis.45,47 LSD1 is critical for survival and metastasis in breast cancer.48 We have shown that LSD1 is overexpressed in CRC tissues with distant metastasis, and correlates positively with human CRC cell migration. We found that ZY0511 and 5-FU synergistically inhibited metastasis of CRC both in vitro and in vivo. In metastasis of CRC, E-cadherin epithelial phenotype disappears and vimentin phenotype is up-regulated. E-cadherin is silenced through its promoter methylation or acetylation. For example, treatment with entinostat, a HDACs inhibitor, downregulated E-cadherin expression. LSD1 could be recruited to the promoters of vimentin and E-cadherin, subsequently demethylating H3K4m2 and H3K4m1, activating vimentin, and suppressing E-cadherin transcription, thereby ultimately inducing EMT, which contributes to CRC metastasis.45 These studies support our finding that ZY0511 treatment upregulated E-cadherin and downregulated vimentin, thus suppressed metastasis of CRC. Moreover, β-catenin is a versatile protein that interacts with E-cadherin at the cell junction and participates in the formation of adhesive.7 Overexpression of E-cadherin in cancer cells blocks the transcriptional capacity of β-catenin, effectively shutting down the expression of target genes, thereby preventing cell migration. E-cadherin inhibition by ZY0511 treatment might result in β-catenin suppression. Therefore, β-catenin and E-cadherin interaction are involved in the anti-metastasis efficiency of ZY0511 and 5-FU combination.
TYMS overexpression is one of the most well-established mechanisms responsible for 5-FU resistance.49 Our mRNA-Seq analysis revealed that ZY0511 and 5-FU combination significantly altered the genes of the subordinate pyrimidine metabolic pathway, including TYMS, CAD, DHODH, DUT, DCTPP1, and POLD1, thereby ultimately inhibiting DNA synthesis and leading to apoptosis. TYMS is regulated by DNA methylation and histone modification. It has been reported that HDAC activities are essential for DNA synthesis and expression of repair genes including TYMS. HDAC inhibitors trichostatin and vorinostat, in combination with 5-FU, exhibit synergistic antitumor effects by down-regulating TYMS in CRC cells and enhancing 5-FU cytotoxicity.50 Thus, ZY0511 might increase sensitivity of cells to 5-FU by inhibiting TYMS expression. DCTPP1, a member of the nucleoside triphosphate pyrophosphohydrolase (NTP-PPase) family, maintains the accuracy of DNA replication by cleavage of harmful nucleotides. DCTPP1 can cause DNA promoter hypomethylation, resulting in high MDR1 resistance gene expression and 5-FU resistance, and inhibition of DCTPP1 can enhance 5-FU cytotoxicity and reverse drug resistance.51 DHODH, the limiting enzyme in the pyrimidine synthesis pathway, is a direct molecular target of the β-catenin downstream protein c-Myc.52 POLD1 has proofreading capabilities for exonuclease activity and is important for DNA repair, including nucleotide excision, double-strand break, base excision, and mismatch repairs.53 As these genes are important in nucleotide pool maintenance and DNA repair, their downregulation could impair DNA synthesis and repair, eventually leading to cell death. These findings suggest that ZY0511 may improve 5-FU cytotoxicity by impairing DNA synthesis and repair.
In conclusion, our results provide theoretical evidence for developing a combinatorial strategy of LSD1 inhibitor ZY0511 and 5-FU for treating CRC. Although a systemic toxicity study will be required for the clinical implementation of these findings, our study provides a route map for future personalized combinatorial strategy with epigenetic drugs and traditional chemotherapy.
Supplemental Material
Supplemental material, Figure_legends for Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer by Wen Peng, Huaqing Zhang, Shisheng Tan, Yan Li, Yang Zhou, Liang Wang, Chunqi Liu, Qiu Li, Xiaobo Cen, Shengyong Yang and Yinglan Zhao in Therapeutic Advances in Medical Oncology
Supplemental material, FIGURE_S1 for Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer by Wen Peng, Huaqing Zhang, Shisheng Tan, Yan Li, Yang Zhou, Liang Wang, Chunqi Liu, Qiu Li, Xiaobo Cen, Shengyong Yang and Yinglan Zhao in Therapeutic Advances in Medical Oncology
Supplemental material, Figure_S2 for Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer by Wen Peng, Huaqing Zhang, Shisheng Tan, Yan Li, Yang Zhou, Liang Wang, Chunqi Liu, Qiu Li, Xiaobo Cen, Shengyong Yang and Yinglan Zhao in Therapeutic Advances in Medical Oncology
Acknowledgments
We thank the Guizhou Provincial People’s Hospital Surgical and pathological departments for technical assistance.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Project of the National Natural Sciences Foundation of China (81773198), National S&T Major project (2018ZX09201018) and National Key R&D Program of China (Grant No. 2016YFC1303200). The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Wen Peng, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China; Department of Oncology, The People’s Hospital of Guizhou Province, Guiyang, China.
Huaqing Zhang, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Shisheng Tan, Department of Oncology, The People’s Hospital of Guizhou Province, Guiyang, China.
Yan Li, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Yang Zhou, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Liang Wang, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Chunqi Liu, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Qiu Li, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Xiaobo Cen, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Shengyong Yang, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, and Collaborative Innovation Center for Biotherapy, Sichuan University, Chengdu, China.
Yinglan Zhao, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, West China Medical School, Sichuan University, 17#, 3rd Section, Ren min South Road, Chengdu 610041, China; West China School of Pharmacy, Sichuan University, Chengdu, China.
References
- 1. Fiore D, Proto MC, Pisanti S, et al. Antitumor effect of pyrrolo-1,5-benzoxazepine-15 and its synergistic effect with Oxaliplatin and 5-FU in colorectal cancer cells. Cancer Biol Ther 2015; 17: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carethers JM. Systemic treatment of advanced colorectal cancer: tailoring therapy to the tumor. Therap Adv Gastroenterol 2008; 1: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bossi P, Saba NF, Vermorken JB, et al. The role of systemic therapy in the management of sinonasal cancer: a critical review. Cancer Treat Rev 2015; 41: 836–843. [DOI] [PubMed] [Google Scholar]
- 4. Zamani M, Hosseini SV, Mokarram P. Epigenetic biomarkers in colorectal cancer: premises and prospects. Biomarkers 2018; 23: 105–114. [DOI] [PubMed] [Google Scholar]
- 5. Khare S, Verma M. Epigenetics of colon cancer. Methods Mol Biol 2012; 863: 177–185. [DOI] [PubMed] [Google Scholar]
- 6. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953. [DOI] [PubMed] [Google Scholar]
- 7. Lei ZJ, Wang J, Xiao HL, et al. Lysine-specific demethylase 1 promotes the stemness and chemoresistance of Lgr5+ liver cancer initiating cells by suppressing negative regulators of β-catenin signaling. Oncogene 2015; 34: 3188–3198. [DOI] [PubMed] [Google Scholar]
- 8. Wen S, Wang J, Liu P, et al. Novel combination of histone methylation modulators with therapeutic synergy against acute myeloid leukemia in vitro and in vivo. Cancer Lett 2018; 413: 35–45. [DOI] [PubMed] [Google Scholar]
- 9. Ding J, Zhang ZM, Xia Y, et al. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer 2013; 109: 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramírez-Ramírez R, Gutiérrez-Angulo M, Peregrina-Sandoval J, et al. Somatic deletion of KDM1A/LSD 1 gene is associated to advanced colorectal cancer stages. J Clin Pathol 2020; 73: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Zhu Y, Wang Q, et al. The histone demethylase LSD1 is a novel oncogene and therapeutic target in oral cancer. Cancer Lett 2016; 374: 12–21. [DOI] [PubMed] [Google Scholar]
- 12. Zhu Q, Huang Y, Marton LJ, et al. Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids 2011; 42: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mohammad HP, Smitheman KN, Kamat CD, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 2015; 28: 57–69. [DOI] [PubMed] [Google Scholar]
- 14. Takagi S, Ishikawa Y, Mizutani A, et al. LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res 2017; 77: 4652–4662. [DOI] [PubMed] [Google Scholar]
- 15. Liang Y, Ahmed M, Guo H, et al. LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res 2017; 77: 5479–5490. [DOI] [PubMed] [Google Scholar]
- 16. Kim J, Park UH, Moon M, et al. Negative regulation of ERalpha by a novel protein CAC1 through association with histone demethylase LSD1. FEBS Lett 2013; 587: 17–22. [DOI] [PubMed] [Google Scholar]
- 17. Dalvi MP, Wang L, Zhong R, et al. Taxane-platin-resistant lung cancers co-develop hypersensitivity to jumonjic demethylase inhibitors. Cell Rep 2017; 19: 1669–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manuyakorn A, Paulus R, Farrell J, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol 2010; 28: 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Y, Li Y, Wang W-J, et al. Synthesis and biological evaluation of novel ( E )- N ′-(2,3-dihydro-1 H -inden-1-ylidene) benzohydrazides as potent LSD1 inhibitors. Bioorg Med Chem Lett 2016; 26: 4552–4557. [DOI] [PubMed] [Google Scholar]
- 20. Li Y, Tao L, Zuo Z, et al. ZY0511, a novel, potent and selective LSD1 inhibitor, exhibits anticancer activity against solid tumors via the DDIT4/mTOR pathway. Cancer Lett 2019: 454: 179–190. [DOI] [PubMed] [Google Scholar]
- 21. Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010; 70: 440–446. [DOI] [PubMed] [Google Scholar]
- 22. Shan H, Li X, Xiao X, et al. USP7 deubiquitinates and stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal Transduct Target Ther 2018; 3: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol 2019; 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang G-J, Lei P-M, Wong S-Y, et al. Pharmacological inhibition of LSD1 for cancer treatment. Molecules 2018; 23: 3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng Y-C, Ma J, Wang Z, et al. A systematic review of histone lysine-specific demethylase 1 and its inhibitors. Med Res Rev 2015; 35: 1032–1071. [DOI] [PubMed] [Google Scholar]
- 26. Maes T, Mascaro C, Tirapu I, et al. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell 2018; 33: 495–511.e12. [DOI] [PubMed] [Google Scholar]
- 27. Lokken AA, Zeleznik-Le NJ. Breaking the LSD1/KDM1A addiction: therapeutic targeting of the epigenetic modifier in AML. Cancer Cell 2012; 21: 451–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wada T, Koyama D, Kikuchi J, et al. Overexpression of the shortest isoform of histone demethylase LSD1 primes hematopoietic stem cells for malignant transformation. Blood 2015; 125: 3731–3746. [DOI] [PubMed] [Google Scholar]
- 29. Sankar S, Theisen ER, Bearss J, et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin Cancer Res 2014; 20: 4584–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta S, Doyle K, Mosbruger TL, et al. Reversible LSD1 inhibition with HCI-2509 induces the p53 gene expression signature and disrupts the MYCN signature in highrisk neuroblastoma cells. Oncotarget 2018; 9: 9907–9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta S, Weston A, Bearrs J, et al. Reversible lysine-specific demethylase 1 antagonist HCI-2509 inhibits growth and decreases c-MYC in castration- and docetaxel-resistant prostate cancer cells. Prostate Cancer Prostatic Dis 2016; 19: 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z, Li S, Song W, et al. Lysine-specific demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis via activation of the Wnt/beta-catenin pathway by down-regulating Dickkopf-1 (DKK1). PLoS One 2013; 8: e70077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H-S, Liu H-Y, Zhou Z, et al. TSPAN8 promotes colorectal cancer cell growth and migration in LSD1-dependent manner. Life Sci 2020; 241: 117114. [DOI] [PubMed] [Google Scholar]
- 34. Hsu H-C, Liu Y-S, Tseng K-C. CBB1003, a lysine-specific demethylase 1 inhibitor, suppresses colorectal cancer cells growth through down-regulation of leucine-rich repeat-containing G-protein-coupled receptor 5 expression. J Cancer Res Clin Oncol 2015; 141:11–21. [DOI] [PubMed] [Google Scholar]
- 35. Zhang X, Lu F, Wang J, et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep 2013; 5: 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang J, Sengupta R, Espejo AB, et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007; 449: 105–108. [DOI] [PubMed] [Google Scholar]
- 37. Majello B, Gorini F, Saccà CD, et al. Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers (Basel) 2019; 11: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pishas KI, Drenberg CD, Taslim C, et al. Therapeutic Targeting of KDM1A/LSD1 in Ewing Sarcoma with SP-2509 engages the endoplasmic reticulum stress response. Mol Cancer Ther 2018; 17: 1902–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baretti M, Azad NS. The role of epigenetic therapies in colorectal cancer. Curr Probl Cancer 2018; 42: 530–547. [DOI] [PubMed] [Google Scholar]
- 40. Zhang B, Zhang B, Chen X, et al. Loss of Smad4 in colorectal cancer induces resistance to 5-fluorouracil through activating Akt pathway. Br J Cancer 2014; 110: 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He L, Zhu H, Zhou S, et al. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Exp Mol Med 2018; 50: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng X, Xu X, Chen D, et al. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother 2019; 110: 473–481. [DOI] [PubMed] [Google Scholar]
- 43. Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem 2006; 281: 22429–22433. [DOI] [PubMed] [Google Scholar]
- 44. Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013; 13: 11–26. [DOI] [PubMed] [Google Scholar]
- 45. Huang M, Chen C, Geng J, et al. Targeting KDM1A attenuates Wnt/beta-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett 2017; 398: 12–21. [DOI] [PubMed] [Google Scholar]
- 46. Li Y, Tian X, Sui CG, et al. Interference of lysine-specific demethylase 1 inhibits cellular invasion and proliferation in vivo in gastric cancer MKN-28 cells. Biomed Pharmacother 2016; 82: 498–508. [DOI] [PubMed] [Google Scholar]
- 47. Jiang Chen JD, Ziwei Wang, et al. Identification of downstream metastasis-associated target genes regulated by LSD1 in colon cancer cells. Oncotarget 2017; 8: 19609–19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu J, Feng J, Li L, et al. Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep 2020; 21: e48597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sakatani A, Sonohara F, Goel A. Melatonin-mediated downregulation of thymidylate synthase as a novel mechanism for overcoming 5-fluorouracil associated chemoresistance in colorectal cancer cells. Carcinogenesis 2019; 40: 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okada K, Hakata S, Terashima J, et al. Combination of the histone deacetylase inhibitor depsipeptide and 5-fluorouracil upregulates major histocompatibility complex class II and p21 genes and activates caspase-3/7 in human colon cancer HCT-116 cells. Oncol Rep 2016; 36: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xia LL TY, Song FF, et al. DCTPP1 attenuates the sensitivity of human gastric cancer cells to 5-fluorouracil by up-regulating MDR1 expression epigenetically. Oncotarget 2016; 7: 68623–68637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dorasamy MS, Choudhary B, Nellore K, et al. Dihydroorotate dehydrogenase inhibitors target c-Myc and arrest melanoma, myeloma and lymphoma cells at S-phase. J Cancer 2017; 8: 3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nicolas E, Golemis EA, Arora S. POLD1: central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene 2016; 590: 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Figure_legends for Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer by Wen Peng, Huaqing Zhang, Shisheng Tan, Yan Li, Yang Zhou, Liang Wang, Chunqi Liu, Qiu Li, Xiaobo Cen, Shengyong Yang and Yinglan Zhao in Therapeutic Advances in Medical Oncology
Supplemental material, FIGURE_S1 for Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer by Wen Peng, Huaqing Zhang, Shisheng Tan, Yan Li, Yang Zhou, Liang Wang, Chunqi Liu, Qiu Li, Xiaobo Cen, Shengyong Yang and Yinglan Zhao in Therapeutic Advances in Medical Oncology
Supplemental material, Figure_S2 for Synergistic antitumor effect of 5-fluorouracil with the novel LSD1 inhibitor ZY0511 in colorectal cancer by Wen Peng, Huaqing Zhang, Shisheng Tan, Yan Li, Yang Zhou, Liang Wang, Chunqi Liu, Qiu Li, Xiaobo Cen, Shengyong Yang and Yinglan Zhao in Therapeutic Advances in Medical Oncology