Abstract
Wheat productivity has been significantly improved worldwide through the incorporation of novel genes from various gene pools, not least from wild relatives of wheat, into the commonly cultivated bread and durum wheat. Here, we present and summarize results obtained from a diverse set of wheat-alien introgression lines with mainly introgressions of rye, but also of Leymus spp. and Thinopyrum junceiforme into bread-wheat (Triticum aestivum L.). From this material, lines carrying 2RL were found with good agronomic performance and multiple resistance not least towards several races of powdery mildew. A novel resistance gene, one of few showing resistance towards all today identified stem rust races, designated Sr59, was also found originating from 2RL. Lines with multiple introgressions from 4R, 5R, and 6R were found resistant towards the majority of the stripe rust races known today. Due to lack of agricultural adaptation in these lines, transfer of useful genes into more adapted wheat material is a necessity, work which is also in progress through crosses with the CSph1b mutant, to be able to only transfer small chromosome segments that carry the target gene. Furthermore, resistance towards Russian wheat aphid was found in lines having a substitution of 1R (1D) and translocations of 3DL.3RS and 5AL.5RS. The rye chromosomes 1R, 2R, and 6R were found responsible for resistance towards the Syrian Hessian fly. High levels of especially zinc was found in several lines obtained from crosses with Leymus racemosus and Leymus mollis, while also some lines with 1R, 2R, or 5R showed increased levels of minerals and in particular of iron and zinc. Moreover, lines with 1R, 2R, 3R, and Leymus spp. introgressions were also found to have a combination of high iron and zinc and low cadmium concentrations. High variation was found both in grain protein concentration and gluten strength, measured as %UPP, within the lines, indicating large variation in bread-making quality. Thus, our study emphasizes the impact that wheat-alien introgression lines can contribute to current wheat lines and shows large opportunities both to improve production, resistance, and quality. To obtain such improvements, novel plant breeding tools, as discussed in this paper, opens unique opportunities, to transfer suitable genes into the modern and adapted wheat cultivars.
Keywords: agronomic performance, baking quality, breeding, disease and pest resistance, Leymus spp., Secale cereale L., Triticum aestivum L.
Introduction
Wheat is one of the three major crops of importance for food security worldwide, the other two being rice and maize (FAO, 2016). Bread wheat (Triticum aestivum L.) is a hexaploid and the most commonly cultivated species of wheat (95%), belonging to the tribe Triticeae and the family Poaceae (McFadden and Sears, 1946; Dubcovsky and Dvorak, 2007). The second most commonly cultivated form of wheat is durum wheat (Triticum durum L.), contributing 5% to the total production (Dubcovsky and Dvorak, 2007). In total, wheat contributes 20% of the total calories and proteins consumed by the human population, thereby contributing to a higher total protein intake than the whole total meat consumption summed (Shewry and Hey, 2015).
Due to the high contribution of wheat to the daily human food intake, human food security is highly vulnerable to the increasing threats to wheat production from climate change, including global warming (Steenwerth et al., 2014). Wheat yield is also negatively affected by abiotic and biotic stresses resulting in economic losses to farmers (Husenov et al., 2020). The population growth predicted to be more than 9 billion people worldwide in 2050, result in additional demand on food production, simultaneously bringing an increasing competition for arable land for food production (FAO, 2016). To meet these challenges, novel wheat cultivars are urgently needed, adapted to contribute high yield under sustainable and demanding cultivation conditions (Shiferaw et al., 2013). For this purpose, novel plant breeding methodologies have to be developed in order to most beneficially use available genetic resources and smart and rapid plant development to produce the needed wheat materials in time to cope with needs and challenges.
Plant breeding to obtain sustainable, high resistance and high-quality crops are dependent on suitable genes for the wanted traits. For many traits, such genes are available within the breeding material in on-going breeding programs for the crop and will be easily transferred by breeders through ordinary crossing schemes. However, domestication and breeding practices have reduced the presence of rare and favorable allelic variation to biotic and abiotic stresses and environmental changes originally found in the wild relatives (Tanksley and McCouch, 1997; Singh et al., 2018). Therefore, wild relatives, landraces, and close relatives of wheat are a unique source of novel genetic variations for introgression into modern cultivars (Molnár-Láng et al., 2015). For wheat, several useful transfers of genes from landraces have been reported including e.g. the Rht dwarfing genes, the powdery mildew resistance gene Pm24, and several biotic and abiotic stress resistance genes (Kihara, 1983; Zeven, 1998; Huang and Röder, 2011; Cavanagh et al., 2013; Singh et al., 2018). Also, genes have been transferred to wheat from non-Triticum (alien) species, where transfers from e.g. rye (Secale cereale) have resulted in widely cultivated wheat cultivars (McIntosh et al., 1995; Friebe et al., 1996). The most successful alien transfer into the wheat genome is that of the 1RS chromosome segment, in the form of 1AL.1RS, 1BL.1RS, and 1DL.1RS translocations (Rabinovich, 1998; Mago et al., 2015), contributing several resistance genes for powdery mildew, leaf, stripe and stem rusts. Out of them, the 1BL.1RS wheat-rye translocation has contributed immensely to global wheat production as a source of resistance genes (Sr31/Yr9/Lr26/Pm9) to wheat fungal diseases (Schlegel, 2020), but it is also known to contribute weak and sticky dough (Dhaliwal et al., 1988). Rye is a unique source of many important traits for wheat improvement, e.g. the resistance genes Sr27, Sr50, Sr1RS Amigo, Lr25, Lr45, Pm7, etc. have been identified from rye (The et al., 1991; Marais and Marais, 1994; McIntosh et al., 1995; Friebe et al., 1996), although these genes have contributed limitedly to agricultural production until now. More recently, some novel resistance genes from rye i.e. Sr59, Yr83, and Pm56 have been introgressed into wheat (Rahmatov et al., 2016a; Hao et al., 2018; Li et al., 2020), which may be used as durable sources against fungal diseases. Herbicide-resistant evolution is challenging weed management; therefore, the allelopathic potential is a good solution to mitigate weed management in crop production. Bertholdsson et al. (2012), reported that rye is an excellent source of allelopathic potential that can be used for wheat breeding. In addition, Iron (Fe) and Zinc (Zn) deficiency are severely affecting human health, causing several physiological disorders, symptomatic anemia, stunting, etc., and therefore high content in staple crops such as wheat are of outmost importance (Johansson et al., 2014; Johansson et al., 2020). The recent great advancements in genomic and cytogenetic tools open opportunities to transfer alien resistance genes to wheat, simultaneously avoiding linkage drag issues.
The present paper is focusing on opportunities and challenges of the use of a diverse set of wheat-alien introgression lines with mainly introgressions of rye, but also of Leymus spp. and Thinopyrum junceiforme into bread-wheat (T. aestivum L.). This provides useful insight into the identification and characterization of wheat-alien introgression lines based on several studies through diseases and pests screening, agronomic performances and molecular markers. Resistances and quality characteristics of wheat within this material, connections to introgressed chromosomes, localization of genes, and status for transfer of these genes are described here. Finally, a short overview is given as to the impact of novel breeding strategies for the use of alien germplasm in modern breeding.
Materials and Methods
Plant Materials
A set of winter and spring wheat-alien introgression lines maintained at the Plant Breeding Department at the Swedish University of Agricultural Sciences were used in different part of the hereby presented studies. These lines were developed by crossing and backcrossing strategies during 1980 to 2000 by the late Professor Arnulf Merker at the Swedish University of Agricultural Sciences ( Table 1 ). The wheat-alien introgression lines used for the present paper contained rye chromosomes with 1R, 2R, 3R, 4R, 5R and 6R in the form of a single disomic substitution wheat–rye translocations such as 1DL.1RS, 1BL.1RS, 2BS.2RL, 3DL.3RS and 5AL.5RS, lines with multiple combinations of rye chromosome substitutions such as 1R + 2R, 1R + 3R, 1R + 6R, 5R + 4R + 7R and 1R + 6R + 4R + 7R (Merker, 1979; Merker and Rogalska, 1984), and lines with introgressed chromatin from Leymus mollis, Leymus racemosus, and T. junceiforme (Ellneskog-Staam and Merker, 2001; Ellneskog-Staam and Merker, 2002). The full material used has previously been completely described in Rahmatov (2016) and Rahmatov et al. (2017).
Table 1.
Wheat-alien introgression lines and respective parents evaluated in this study.
| Cross/Pedigree | Plant habit | No. of lines | Type | Reference |
|---|---|---|---|---|
| Triticalea | Spring and winter | 5 | ×Triticosecale | Forsstrom and Merker, 2001 |
| Wheata | Spring and winter | 8 | Triticum aestivum and Tr. carthlicum | Forsstrom et al., 2002 |
| Sv 876012 x H | Winter | 37 | Wheat–rye introgressions | Forsstrom and Merker, 2001 |
| Sv 876032 x H x K | Winter | 54 | Wheat–rye introgressions | Forsstrom and Merker, 2001 |
| Sv 856003 x H | Winter | 6 | Wheat–rye introgressions | Forsstrom and Merker, 2001 |
| Sub 1R + 2R | Winter | 42 | Wheat–rye introgressions | Forsstrom and Merker, 2001 |
| Malysh | Winter | 6 | Wheat–rye introgressions | Merker, 1984 |
| Starke × Otello | Winter | 7 | Wheat–rye introgressions | Merker, 1984 |
| Uno × Holme | Winter | 8 | Wheat–rye introgressions | Merker, 1984 |
| Triticale VT828041 | Spring | 6 | Wheat–rye introgressions | Merker, 1984 |
| Triticale Drira | Spring | 23 | Wheat–rye introgressions | Merker, 1984 |
| Triticale Beagle | Spring | 12 | Wheat–rye introgressions | Merker, 1984 |
| Triticale VT83 615 | Spring | 2 | Wheat–rye introgressions | Merker, 1984 |
| Triticale VT83 591 | Spring | 4 | Wheat–rye introgressions | Merker, 1984 |
| Triticale VT 82 8039 | Spring | 5 | Wheat–rye introgressions | Merker, 1984 |
| 3R BB14 (Cimmyt 1974) | Spring | 4 | Wheat–rye introgressions | Merker, 1984 |
| Leymus mollis | Winter | 42 | Wheat–L. mollisintrogressions | Merker and Lantai, 1997 |
| Leymus racemosus | Spring | 22 | Wheat–L. racemosus introgressions | Merker and Lantai, 1997 |
| Th. junceiforme | Spring | 16 | Wheat–T. junceiforme introgressions | Merker and Lantai, 1997 |
| aParental cultivars and Lines | 309 | TOTAL |
Field Trials
A total of 180 of the winter wheat lines and 57 of the spring wheat lines were evaluated by field trials for multiple resistance and agronomic performance during two executive seasons, 2014 and 2015, in Svalöv, Sweden and in Harzhof and Laberweinting in Germany. During these seasons, the lines were continuously evaluated and scored (scale 1–9) for lodging (winter wheat) and presences of diseases (spring and winter wheat). Comparisons of presence of diseases and alien material were carried out (Andersson et al., 2016).
Diseases Screening
Stem rust seedling resistance assays with ten Pgt races and adult plant responses with three Pgt races (TTKSK + TTKST, TKTTF and MCCFC), were carried out on 185 and 94 of the winter and spring wheat-alien introgression lines under field conditions following the procedure described in Hysing et al. (2007) and Rahmatov et al. (2015); Rahmatov et al. (2016a, b). For the stripe rust evaluations, 189 of the winter and 73 of the spring wheat-alien introgression lines were tested in the seedling and adult plant stages. Twelve stripe rust races with different virulence/avirulence combinations and geographic origins were used for screening at the seedling stage along with adult plant evaluations in the field according to Rahmatov et al. (2017). Hysing et al. (2007), evaluated a set of 2BS.2RL wheat-rye translocation lines against stripe rust, leaf rust, and powdery mildew races.
Hessian Fly and Russian Wheat Aphid Screenings
A total of 57 spring and 185 winter wheat-alien introgression lines were evaluated in 2011 and 2012 at the seedling stage against Hessian fly (HF) and the Russian wheat aphid (RWA) in collaborations with ICARDA. In brief, the rearing rooms for HF experiments were kept at 20°C, Rh 70–80%, and the cycle of 16/8 h light/dark was used. Six or ten seeds per wheat accession were sown in hill plots in metal flats 55 × 45 × 10 cm, in total 48 accessions per box plus controls, in a mixture of soil:sand:peat (2:1:1). After 5–6 days, at the one-leaf stage, infestation by HF was done with about 30 females and 10 males under net for 3–4 days (El Bouhssini et al., 2013). The scoring took place 20 days after infestation, with the number of resistant and susceptible plants per accession. The first screening was conducted in the spring and winter materials in 2011, and a second screening was only conducted in the winter materials in 2012. Based on these two screenings, lines with 100% resistance reaction to HF were selected for further confirmations in four separate screenings.
The RWA biotype was collected from Tel Hadya, Syria, and thereafter reared on the susceptible wheat cultivar (Andersson et al., 2015). The experiments were carried out in a greenhouse at 19–20°C, with light/dark photoperiod 16/8 h and relative humidity of about 60%. The accessions were planted in a randomized (alpha design) order together with susceptible and resistant controls in each planting tray, in a mixture of soil, sand, and peat (2:1:1). An evaluation was done when symptoms were seen on susceptible checks, using the ICARDA RWA damage scale with a 1–3 scale for leaf rolling (LR) and 1–6 scale for leaf chlorosis (LC) (El Bouhssini et al., 2011). In the second advanced screening, selected accessions from the first screening results were repeated at four separate times (Andersson et al., 2015).
Allelopathic Potential of Wheat-Alien Introgression Lines
Allelopathic potential of the wheat–rye introgression lines were tested according to Bertholdsson et al. (2012). In this study, seeds of Chenopodium alba, Lolium perenne, Brassica napus, Lactuca sativa, Eruca sativa, Sinapis indicum and Sinapis alba were used to find high root growth inhibition when grown together with rye. In this investigation, four pregerminated cereal seedlings were planted along the wall of 400-ml Phytotech tissue culture vials (bottom diameter 75 mm) filled with 20 ml 0.35% water agar, and eight pregerminated mustard seedlings (S. alba cv. Medicus) were planted in a circle in the center of the vials. The experiment was tested in four replicates, and the dry weight of the shoot and root were measured (Bertholdsson et al., 2012).
Analysis of Grain Samples for Micronutrients Concentration and Protein Composition
A total of 40 of the lines were evaluated for micronutrients (e.g. Iron, Zinc, and Cadmium) content with Inductively Coupled Plasma Mass Spectrometry (ICPMS) at the University of Minnesota, similarly as described in Hussain et al. (2010) and Moreira-Ascarrunz et al. (2016). Briefly, all samples were ashed in a muffle furnace for 12 h at 485°C. Then, the ash was dissolved in 5 ml of 20% HCl followed by dilution with 5 ml of deionized water. The ICPMS provides concentration assays for several microelements, including zinc, iron, and cadmium in mg/Kg.
The complete set of winter wheat alien translocation lines were analysed with SE-HPLC according to Johansson et al. (2001) to evaluate the total amount of SDS-extractable proteins (TOTE) and percentage of unextractable polymeric protein in total polymeric protein (%UPP). A high correlation is known to exist between TOTE and grain protein concentration and between %UPP and gluten strength (Malik et al., 2011; Malik et al., 2013) and thereby this methodology can be used to understand relationships with bread-making quality (Hussain et al., 2012; Hussain et al., 2013; Vazquez et al., 2019).
Statistical Analyses
The statistical software SAS 9.3 (SAS, 2011) was used for principal component analyses (PCA) calculations to understand relationships between minerals and protein factors with evaluated wheat-alien introgression lines. In order to understand and visualize the distribution and relationship between variables and factors evaluated, principal component analysis (PCA) can be applied to orthogonally represent the variables in a data matrix vector. PCA is known to show the distribution of dependent variables and independent factors, in a loading and score plot, respectively (Wold et al., 1987). Values of content of Iron, Zinc and Cadmium were calculated by mini tab for wheat, Triticale, wheat–rye and wheat-Leymus lines and presented as boxplots with lowest and highest observations as well as lower and upper quartile and median.
Results
Multiple Resistance and Agronomic Performance
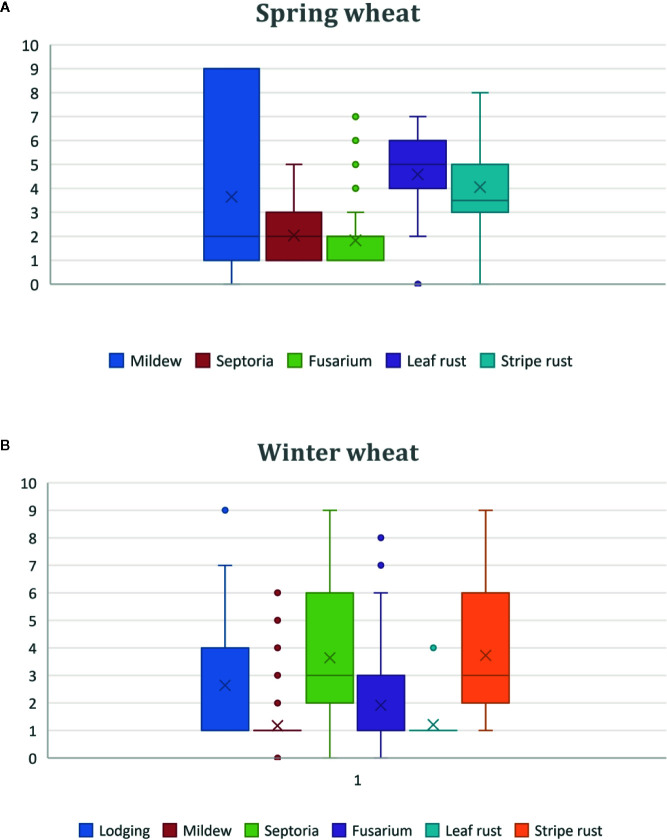
The lines showed varying agronomic performance, with some lines being almost comparable to currently grown wheat in Sweden while others differed largely. Large variation was found in the material both for lodging and presence of diseases ( Figure 1 ). However, the majority of the winter wheat lines had strong stem and with limited lodging, thus making them of interest as a source of lodging resistance ( Figure 1B ). Presence of 1R, 2R, 3R, 5R, 1R + 6R and L. racemosus correlated with decreased levels of infections with powdery mildew, Zymoseptoria tritici (causal agent of Septoria triticae blotch) and Fusarium head blight during field conditions. Lower levels of leaf, stem and stripe rusts infection responses were found in lines with 1R, 2R, 3R, 1R + 3R, 1R + 6R, and L. racemosus, respectively.
Figure 1.
Boxplots showing variation in lodging and various diseases based on scoring of the material from 0 to 9, in wheat alien introgression lines of (A) Spring wheat, and (B) Winter wheat, from field trials during two years in Sweden and Germany. In each boxplot, five bars are represented, indicating smallest observation, lower quartile, median, upper quartile, and largest observation, respectively. X marks the mean value.
Rusts Screenings
From the stem rust seedling evaluation, eleven 2R (2B), three 2R (2D), and three 3R (3D) wheat–rye disomic substitution lines, and seven wheat-T. junceiforme were found to carry potentially new stem rust resistant gene/s ( Table 2 ). Based on the ten Pgt races, known resistance genes could not be postulated because their reactions did not correspond to the avirulence/virulence profile of the races tested. All lines that were resistant at the seedling stage remained resistant at the adult plant stage against races TTKSK + TTKST in Kenya and TKTTF in Turkey. Trace resistance was found in several of the lines tested at St. Paul, Minnesota, against the race MCCFC ( Table 2 ), although only a few number of lines were tested due to winter type of the material and limited seed available.
Table 2.
Stem rust seedling and adult plant resistance tests in the wheat–rye and wheat–T. Junceiforme introgression lines with potential sources of new stem rust resistance gene/s.
| # | Chromosome | Seedling Resistance Test | Adult Plant Resistance | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TTKSK, 1 Rep. | TTKSK, 2 Rep. | TPMKC | TTTTF | QTHJC | RKQQC | TTKST | TRTTF | TTTSK | TKTTF | MCCFC | TTKSK+ TTKST | TKTTF | MCCFC | ||
| SLU73 | 2R susbstituted 2B | 2+ | 2 | 3+ | ;1 | ;01/3+ | 11+ | 2/2+ | 1+2/3+4 | 22+ | 1/2+ | 4 | 20MR | 10MR | – |
| SLU74 | 2R susbstituted 2B | 2+ | 2+ | 1 3+ Z | ;1/1 | ;-1 | ;1 | ;11+ | 1+2/3+ | 22+ | 1/2+ | 4 | 30MR | 10MR | – |
| SLU75 | 2R susbstituted 2B | 2 | 2 | 3+ | 0;/0; 3+ | ;-1 | ;1 | 11+ | 33+ | 22+ | 11+ | 4 | 20MR | 10MR | – |
| SLU76 | 2R susbstituted 2B | 2 | 2 | 3+ | ;1 | ;1/2- | ;11+ | ;11+ | 11+3 | 22+ | 3+ | 4 | 30MRMS | 10MS | – |
| SLU77 | 2R susbstituted 2B | 2 | 2 | 3+ | 2+3/4/;1 | ;1/1+3 | ;11+/2 | ;11+/2/3+ | 33+ | 22+ | 3+ | 4 | 30MRMS | 10MRMS | – |
| SLU78 | 2R susbstituted 2B | 2+ | 2+ | 3+ | ;/3- | 1+3- | ;11+3- | ;11+2+ | 22+3+ | 22+ | 3+ | 4 | 30MRMS | 10MS | – |
| SLU79 | 2R susbstituted 2B | 2 | 2 | 3+ | ;/3- | ;13- | ;13-/3 | ;12+ | 11+/3+ | 22+ | 1 | 4 | 30MRMS | 10MR | – |
| SLU80 | 2R susbstituted 2B | 2+ | 2+ | 33+ | 3+ | 33+/;13- | 33+/;13- | ;11+/3+ | 1+2/3+ | 22+ | 3+ | 4 | 30MRMS | 10MS | – |
| SLU81 | 2R susbstituted 2B | 2+ | 2+ | 3+ | 3-/1; | 11+ | 11+ | 1+3- | 33+ | 22+ | 3+ | 4 | 40MRMS | 10MS | – |
| SLU82 | 2R susbstituted 2B | 2+ | 2+ | 3+ | ; 1 3-/4 | ;13-/3+ | ;13-/3+ | ;1/3 | 3+ | 22+ | 3+ | 4 | 40MRMS | 10MS | – |
| SLU83 | 2R susbstituted 2B | 2+ | 2+ | 4 | ; 1 3- | 13-/3+ | 13-/3+ | 11+/3+ | 33+ | 22+/3+ | 3+ | 4 | 40MRMS | 10MS | – |
| SLU210 | 2R susbstituted 2D | 0; | 0; | 0; | 1+ | 0; | ;1 | 0; | ;12 | ;1 | 1 | 0; | 20RMR | 5RMR | TR |
| SLU214 | 3R susbstituted 3D | 0; | 0; | 11+ | 4 | 0; | 0; | 0; | ;11+ | 3+ | 3+ | 0; | 20R | 10MRMS | TR |
| SLU219 | 3R susbstituted 3D | 0; | 0; | 0; | 0; | 0; | 0; | 0; | 0; | 0; | 0; | 0; | 5TR | 5R | TR |
| SLU222 | 3R susbstituted 3D | 0; | 0; | 33+ | 4 | 1+2/3- | 3 | 0; | 22+ | ; | 33+ | 11+ | 10RMR | 70S | 10R |
| SLU238 | 2R susbstituted 2D | 1 | 1 | 2 | 22- | 12- | ;1 | ;11+ | ;01 | ;1 | 1- | ;01- | 10R | 10RMR | TR |
| SLU239 | 2R susbstituted 2D | 1 | 1- | 2 | 22- | 12- | ;1 | ;01 | ;01 | ;1 | 1- | ;01- | 20RMR | 5RMR | TR |
| SLU251 | Th.-Wheat | 0; | 0; | 3+ | 4 | 33+ | 3+ | 0;1 | 3+ | 0; | 3+ | 4 | – | – | – |
| SLU252 | Th.-Wheat | 0; | 0; | 33+ | 3+ | 33+ | 3+ | 0; | 3+4 | 0; | 3+ | 3+ | – | – | – |
| SLU253 | Th.-Wheat | 1 | 0; | 33+ | 3+ | 33+ | 3+ | 1+3- | 2+3- | 0; | 3+ | 3+ | – | – | – |
| SLU255 | Th.-Wheat | 0; | 0; | 3+ | 4 | 33+ | 3+ | 0,1 | 3+ | 0; | 3+ | 3+ | – | – | – |
| SLU256 | Th.-Wheat | 0; | 0; | 3+ | 3+ | 33+ | 3+ | 0; | 3+4 | 0; | 3+ | 3+ | – | – | – |
| SLU274 | Th.-Wheat | 0; | 0; | 3+ | 4 | 33+ | 3+ | 0,1 | 3+ | 0; | 3+ | 3+ | – | – | 5MS |
| SLU275 | Th.-Wheat | 0; | 0; | 33+ | 3+ | 33+ | 3+ | 0; | 3+4 | 0; | 3+ | 3+ | – | – | 30MS |
Seedling infection types observed based on 0–4 scale (Stakman et al., 1962). The lines with;0–2+ types considered as resistant. The lines with 3–4 types considered as susceptible. Adult plant response was evaluated based on the Cobb Scale (Peterson et al., 1948) and host response to infection based on pustule type and size (Roelfs et al., 1992). TR, Trace Resistance; R, Resistance; MR, Moderately Resistance; MRMS, Moderately Resistance to Moderately Susceptible; and MS, Moderately Susceptible. A total of 94 lines of the total material were screened for adult plant resistance, explaining the lack of data for some of the lines presented here.
The wheat-alien introgression lines showed high variability in resistance/susceptibility reactions against the twelve stripe rust isolates applied to screen for resistance genes ( Table 3 ). The screening resulted in 149 lines (57% of the lines), postulated to contain a combination of known Yr genes e.g. Yr1, Yr2, Yr9, and Yr32. However, six of the multiple wheat–rye introgression lines with 4R, 5R and 6R were identified as highly resistant against a total of 25 stripe rust races, including the twelve used for the full material ( Table 3 ). Thus, these six lines might possess a new stripe rust resistance gene/s. Molecular cytogenetic analysis showed that the 4R, 5R and 6R rye chromosomes substituted 4D, 5D and 6D wheat chromosomes. Further studies are going on for determining the underlying genetic basis of these resistance gene/s.
Table 3.
Resistance(R)/susceptibility(S) of wheat-alien introgression lines to isolates of Puccinia striiformis tritici.
| Isolates | 6 lines fromSv 876012 × H | 256 lines | |||
|---|---|---|---|---|---|
| with 4R + 5R + 6R | R | MR | MS | S | |
| SE 205/12 | R | 12 | 23 | 102 | 119 |
| UK 94/519 | R | 49 | 2 | 24 | 182 |
| DK 66/02 | R | 84 | 4 | 29 | 140 |
| Taj 01a/10 | R | 17 | 34 | 87 | 119 |
| ER 02/03 | R | 174 | 15 | 54 | 14 |
| DK 11/09 | R | 178 | 11 | 46 | 21 |
| DK 71/93 | R | 66 | 34 | 99 | 56 |
| AF 87/12 | R | 207 | 10 | 22 | 16 |
| DK09/11 | R | 39 | 14 | 104 | 98 |
| DK 122/09 | R | 6 | 15 | 126 | 109 |
| SE 100/09 | R | 220 | 8 | 16 | 12 |
| TR 34/11 | R | 170 | 41 | 8 | 32 |
Aphid and Hessian Fly Resistance
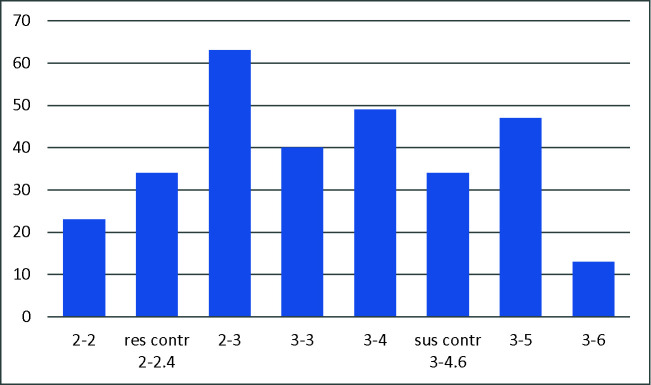
Among the total of 242 evaluated lines, 235 germinated and showed a high variation in resistance to RWA ( Figure 2 ). A total of 23 accessions were identified as resistant against the RWA. Resistance was found to RWA, particularly in accessions having substitutions of 1R instead of 1D [1R (1D) or 1R (1D) + 6R (6D)], in translocations to 3D or 5A (3DL.3RS and 5AL.5RS) and accessions with introgressions of L. mollis.
Figure 2.
Evaluation of 235 wheat alien introgression lines for resistance against Russian wheat aphid (RWA), resulting in number of lines with different scales of resistance. Scale: leaf rolling (LR)-leaf chlorosis (LC); 1-1 and 1-2 = highly resistant, 2-2 = resistant, 2-3 = moderately resistant, 3-3 moderately susceptible, 3-4, 3-5, 3-6 = susceptible. For the susceptible and resistant controls, mean values of 34 lines are used.
The first screening (242 lines) for HF resistance showed 11 winter and two spring wheat accessions with 100% resistance, while in the second screening, nine of the 11 winter wheat accessions were proofed with 100% resistance, which also holds true for the additional four repeated screenings ( Table 4 ). These fully resistant winter wheat accessions contained 1R, 1R + 6R, 1RS + 2RL, 1RL + 2RL, 2RL, and 2R translocations or substitutions. The presence of these genes in our alien wheat material might be one explanation for the HF resistance found although the presence of full resistance in accessions with the substitution 1R.1D in winter wheat and the translocation 1RS.1DL in spring wheat indicate the presence of additional unknown resistance genes in the present material. Besides, high and partial levels of resistance with the presence of 1R, 1RS, 2R, 3R, 3RS, 4R, 5R, 6RL, and L. racemosus and L. mollis substitutions and translocations were found promising sources against HF.
Table 4.
Accessions of Swedish winter wheat with rye substitutions and translocations showing resistance for Hessian fly at separate screenings.
| Acc.No. | Subs./transl. | 1st screen | 2nd screen | Mean 4 screens | |||
|---|---|---|---|---|---|---|---|
| Tot pl | % inf | Tot pl | % inf | Tot pl | % inf | ||
| Kr 08-59 | 1R.1D | 5 | 0 | 10 | 0 | 9.25 | 0 |
| Kr 08-60 | 1R.1D + 6R.6D | 5 | 0 | 10 | 0 | 8.75 | 0 |
| Kr 08-76 | T1RS.1BL + T2BS.2RL | 2 | 0 | 10 | 0 | 9.5 | 0 |
| Kr 08-79 | 2R.2B | 5 | 0 | 10 | 0 | 9.25 | 0 |
| Kr 08-89 | T1RL.1BS + T2BS.2RL | 5 | 0 | 10 | 0 | 9.5 | 0 |
| Kr 08-90 | T1RL.1BS + T1BS.2RL | 2 | 0 | 10 | 0 | 9.5 | 0 |
| Kr 08-91 | T2RL.2BS | 5 | 0 | 10 | 0 | 9 | 0 |
| Kr 08-94 | T2RL.2BS | 5 | 0 | 10 | 0 | 10 | 0 |
| Kr 08-95 | T2RL.2BS | 5 | 0 | 10 | 0 | 9 | 0 |
| Res Cont | 6RL | 5 | 0 | 10 | 0 | 10 | 0 |
| Sus Cont | - | 5 | 100 | 10 | 100 | 10 | 100 |
Nutritional Benefits
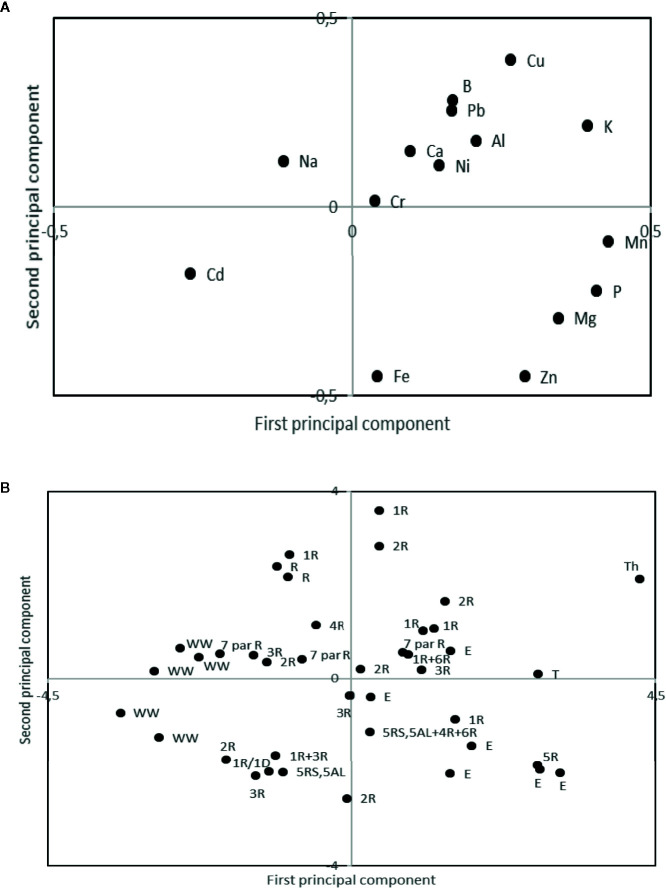
Principal component analyses indicated high levels of Cadmium (Cd) in the winter wheat lines as compared to the rest of the evaluated lines, while Leymus spp. was indicated as containing high levels of Iron (Fe) and Zinc (Zn; Figure 3 ). Mean values of minerals content in the different types of material (Wheat–rye introgressions, Leymus spp. introgressions, wheat, and triticale) verified the high content of Zn in the Leymus spp. introgression lines and the high Cd content in the wheat lines ( Table 5 ). A relatively high Fe content was found in two of the parental wheat lines used in the present study; Sonett (57.0 mg/kg) and Prins (60.6 mg/kg). Furthermore, the triticale parents, Drira (51.8 mg/kg) and Beagle (63.3 mg/kg), were observed to contain a high level of Zn ( Table 5 ). Overall, the minimum 22.7 mg/kg and maximum 64.2 mg/kg for Fe concentrations were observed in the wheat–rye introgression lines with 1R, 2R, 3R, 5R, and 6R rye chromosomes ( Table 5 ). The minimum and maximum Zn concentrations produced by these wheat–rye introgression lines were 32.9 mg/kg and 89.3 mg/kg, respectively. The overall grain Cd concentration ranged from 0.02 to 0.13 mg/kg, in which the lines with low Cd concentration were observed to be 0.015 to 0.017 mg/kg in the wheat–rye introgression 1R (1D), and the lines with L. mollis and L. racemosus chromosomes. Interestingly, nine of the lines with a high combination of Fe (ranged from 47.4 to 64.2 mg/kg) and Zn (ranged from 53.7 to 83.4 mg/kg) concentration and low Cadmium concentration (ranged from 0.02 to 0.07 mg/kg) were detected in the wheat-rye 1R (1D), 2R (2D), 2R (2B), 3R (3B), and L. mollis and L. racemosus intogression lines ( Table 5 ).
Figure 3.
Loading (A) and score (B) plot from principal component analyses of mineral composition in winter wheat (WW), Triticale (T), Rye (R), and alien substitution and translocation lines with rye introgressions (given as R and a what type of), Thinopyrum (Th) and Leymus (E) introgressions. The first and second principal component explained 23.0 and 17.0% of the variation, respectively.
Table 5.
Mean values of zinc, iron and cadmium concentrations in wheat, triticale, Leymus spp., wheat–rye introgression and wheat–Leymus spp. introgression lines.
| Plant lines | Fe (mg/kg) | Zn (mg/kg) | Cd (mg/kg) | |||
|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | |
| Wheat (n = 5) | 45.0 | 31.0–60.6 | 39.5 | 34.5–48.7 | 0.09 | 0.07–0.12 |
| Rye (n = 2) | 39.7 | 38.1–41.2 | 35.2 | 33.8–36.6 | 0.00 | 0.00–0.00 |
| Triticale (n = 5) | 37.9 | 29.5–45.0 | 48.9 | 38.7–63.3 | 0.09 | 0.07–0.15 |
| Leymus spp. (n = 3) | 49.4 | 41.4–59.1 | 75.8 | 62.4–83.4 | 0.02 | 0.00–0.02 |
| Wheat–rye introgression (n = 22) | 38.9 | 22.7–64.2 | 54.8 | 32.9–89.3 | 0.05 | 0.00–0.10 |
| Wheat–Leymus spp. introgression (n = 3) | 47.5 | 43.0–51.9 | 63.6 | 53.1–69.1 | 0.04 | 0.02–0.06 |
Baking Quality
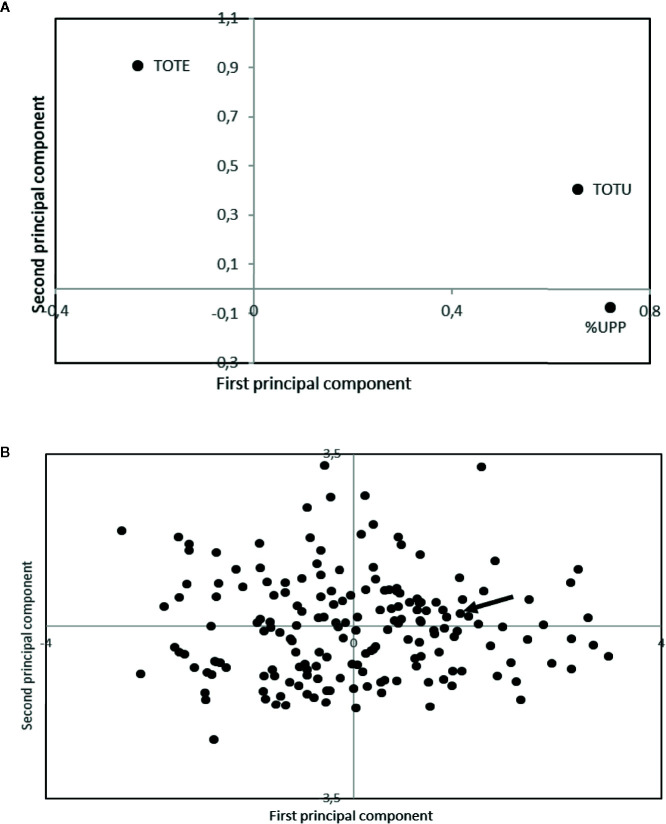
The evaluated alien introgression lines showed a high level of variability both in grain protein concentration and gluten strength ( Figure 4 ). A total of 40% of the lines showed a higher grain protein concentration than the standard cultivar, Dragon, while 8% of the lines showed higher gluten strength than the standard. The 10% of the evaluated lines with the highest grain protein concentration (TOTE), were all found to have either addition of chromosome 1R, 2R, 4R, and 6R or a 1R/1D translocation ( Table 6 ). Several of the high grain protein concentration lines also had additions of 1R and 6R. The lines with high gluten strength (%UPP) were found either to have introgressions from Leymus or additions of either 1R + 2R or 1R + 4R ( Table 6 ).
Figure 4.
Loading (A) and score (B) plot from principal component analyses of storage protein composition from SE-HPLC. The arrow is indicating the Swedish spring wheat line, used as a standard within the analyses. TOTE, total amount of SDS-extractable proteins; TOTU, total amount of SDS-unextractable proteins; and %UPP, percentage of unextractable polymeric protein in total polymeric protein. The first and second principal component explained 58.8 and 35.6% of the variation, respectively.
Table 6.
Accessions of Swedish winter wheat with substitutions and translocations (rye = R, Leymus) showing high (in descending order) total amount of SDS-extractable protein (TOTE—correlating to grain protein concentration) and percentage of unextractable polymeric protein in total polymeric protein (%UPP—correlating with gluten strength).
| TOTE | %UPP | ||||
|---|---|---|---|---|---|
| Acc.No. | Subs./transl. | Rel. values | Acc.No. | Subs./transl. | Values |
| Kr 08-10 | 1R, 4R, 6R, 7R | 1.74 | Kr 08-109 | Leymus | 86.4 |
| Kr 08-54 | 1R/1D | 1.70 | Kr 08-107 | Leymus | 85.7 |
| Kr 08-16 | 1R, 4R, 6R, 7R | 1.64 | Kr 08-111 | Leymus | 85.1 |
| Kr 08-57 | 1R/1D | 1.63 | Kr 08-104 | Leymus | 84.3 |
| Kr 08-15 | 1R, 4R, 6R, 7R | 1.60 | Kr 08-100 | 2RL/2BS | 81.9 |
| Kr 08-08 | 1R, 4R, 6R, 7R | 1.60 | Kr 08-28 | 1R + 6R | 81.6 |
| Kr 08-55 | 1R/1D | 1.59 | Kr 08-79 | 1R + 2R | 81.0 |
| Kr 08-09 | 1R, 4R, 6R, 7R | 1.59 | Kr 08-80 | 1R + 2R | 80.3 |
| Kr 08-53 | 1R/1D | 1.54 | Kr 08-110 | Leymus | 79.3 |
| Kr 08-30 | IR + 6R | 1.53 | Kr 08-04 | 1R + 4R | 79.2 |
| Kr 08-143 | 5R/5A | 1.53 | Kr 08-108 | Leymus | 78.6 |
| Kr 08-63 | 1R + 6R | 1.52 | Kr 08-77 | 1R + 2R | 78.4 |
| Kr 08-75 | 1RS + 2RL | 1.50 | Kr 08-106 | Leymus | 76.6 |
| Kr 08-76 | 1RS + 2RL | 1.50 | Kr 08-01 | 1R + 4R | 75.3 |
| Kr 08-82 | 1R + 2R | 1.47 | Kr 08-95 | 1R + 2R | 75.0 |
| Kr 08-60 | IR + 6R | 1.47 | Dragon | 74.9 | |
| Kr 08-84 | 1R + 2R | 1.45 | |||
| Kr 08-156 | 1BS/1RL | 1.45 | |||
| Kr 08-52 | 1R/1D | 1.45 | |||
| Kr08-20 | IR+6R | 1.44 | |||
| Dragon | 1.22 | ||||
For TOTE the 20 accessions with highest value and their corresponding alien segments are shown while for %UPP, those higher than the standard (Dragon = Swedish winter wheat).
Discussion
New sources of genetic diversity are essential to improve yield, root growth, stand establishment, adaptation to climate change, nitrogen use efficiency, water use efficiency, resistance to abiotic and biotic stresses, biomass, photosynthetic potential, nutritional and end-use quality. In this paper, results from studies over a range of years are compiled to highlight the importance of wheat-alien introgression lines as a potential source of several important traits for wheat improvement. Our studies proved that these wheat-alien introgression lines carry various genetic variation e.g. resistance to diseases (rusts, powdery mildew, S. triticae, and Fusarium head blight), pests (Hessian fly and aphids), agronomic performance, weed competition, yield potential, microelements (Fe, Zn, Cd, etc.), fertility, alpha amylase activity, and positive end-use quality.
The evaluated 2R (2B) and 2R (2D) substitution lines showed resistance to all stem rust races at both the seedling and adult plant stages. Additionally, three of the 3R (3D) (SLU214, SLU219, and SLU222) substitution lines and seven of the wheat-T. junceiforme were found as potential sources of stem rust resistance genes. From the screening of a collection of wheat-alien introgression lines, the line SLU238 [2R (2D) wheat-rye disomic substitution] possessed resistance to many races of Pgt, including the widely virulent race TTKSK (Rahmatov et al., 2016a). In previous studies, Rahmatov et al. (2016b), reported that by the crossing of SLU238 and CS ph1b mutant, a new wheat-rye Robertsonian translocation 2DS·2RL was developed as the source of the gene Sr59. To this date, no stem rust resistance genes have been reported from the 2R chromosome, but chromosome 2R from different rye sources has been described as a source of resistance to various diseases and insects and also various agronomic traits. Previously, the resistance genes to leaf rust Lr25 and Lr45, powdery mildew Pm7 and Hessian fly resistance gene H21 have been reported from the 2R chromosome (Friebe et al., 1996; Friebe et al., 1999). Furthermore, the resistance genes Sr27, Sr31/Yr9/Lr26, Sr50, Sr1RS Amigo and, SrSatu have been described, originating from the rye chromosomes 1R and 3R, and these have been found to be effective against many of all the three rusts races (Marais and Marais, 1994; Mago et al., 2002; Singh et al., 2011; Olivera et al., 2013). Out of these resistance genes, Sr31 has been deployed widely and provided durable resistance against stem rust races for over 30 years in agriculture (Singh et al., 2008).
Agronomic performances of some of the alien-wheat introgression lines were similar to wheat for grain yield, straw length, lodging, grain volume weight, 1000-kernel weight, fertility, grain a-amylase activity, and end-use quality (Hysing et al., 2007; Andersson et al., 2016) while some of the lines showed large variation in agronomic performance. Field studies indicated a correlation between the presence of rye (1R, 2R, 3R, 5R, 1R + 6R) and L. racemosus chromosomes, with low level of powdery mildew, S. triticae and Fusarium head blight infections (Andersson et al., 2016). Previous studies have reported an Fhb3 resistance gene to Fusarium head blight derived from L. racemosus (Qi et al., 2008), which might also be present in our wheat—L. racemosus introgression lines. Therefore, future evaluation of these lines to other powdery mildew and Z. triticae isolates at seedling and adult plant stages are needed. Hysing et al. (2007), reported that red coleoptile color was correlated to the presence of the 2BS.2RL translocation allowing this character to be used as a morphological marker. Furthermore, lines with the 2BS.2RL translocation were demonstrated a high level of resistance against leaf rust and powdery mildew at the seedling stage (Merker and Forsström, 2000; Hysing et al., 2007) and adult plant resistance to TTKSK (Ug99; Rahmatov et al., 2015), thus indicating presence of uncharacterized resistance gene/s. Valuable rye chromosomes harboring beneficial genes from 4R, 5R, and 6R have also been identified (Rahmatov et al., 2017). These lines containing 4R, 5R, and 6R chromosomes are pointed out here as useful due to the fact that they are possessing novel stripe rust resistance genes. Further investigations are needed to understand the underlying genetic basis of this resistance. In various studies, stripe rust and powdery mildew resistance genes have reported on the 4R, 5R, and 6R chromosomes (An et al., 2015; Schneider et al., 2016; Xi et al., 2019), in which Yr83 was mapped on the 6RL (Li et al., 2020). Besides this, chromosomes 4R and 6R have been demonstrated to contribute increased protein content and also to be associated with good pollinator traits (Nguyen et al., 2015; Schneider et al., 2016). Thus, there is a need to further exploit these wheat-alien introgression lines with various chromosome constitutions for wheat improvement.
High levels of resistance were identified in lines with the 1R, 3RS, 1R + 6R, 5R, and L. mollis chromosome introgressions against RWA. Resistances to RWA obtained from the wheat-alien introgression lines particularly lines with the 3R, 5R and L. mollis chromosomes have not previously been reported (Andersson et al., 2015). Previously, Dn7, Gb2, and Gb6 resistance genes to cereal aphids have been reported on chromosome arm 1R (Friebe et al., 1996; Friebe et al., 1999; Anderson et al., 2003). Also, 1RSam.1AL and MA1S.1RLe(1B), 1Re(1D) wheat–rye translocation, and substitution lines were shown with a high level of resistance against HF and RWA, and these lines are now used in the international wheat breeding programs (Crespo-Herrera et al., 2019). The wheat-alien introgression lines with the presence of 1R, 1RS, 2R, 3R, 3RS, 4R, 5R, 6RL, and L. racemosus and L. mollis chromosomes provides resistance to the Syrian HF biotype. Previous studies have verified alien germplasm to contribute HF resistance in wheat through the H21 and H25 resistance genes from rye, located on 2R and 6R, respectively (Friebe et al., 1999). Hysing et al. (2007), reported that lines with the T2BS.2RL were susceptible to the HF biotypes thus this indicates different rye sources used for developing Swedish wheat-alien introgression lines. Host resistance to these insects is the most effective way of control, and various resistance genes have been derived from alien species. The resistances to RWA and HF reported here originating from alien material have not previously been described and can, therefore, be useful to widen the pool of resistance genes in wheat breeding for resistance to RWA and HF.
The wheat-rye introgression lines displayed a good source of allelopathic potential, while lines with L. mollis chromosome showed a low level of allelopathic potential and the bread wheat genotypes showed no allelopathic activity. These wheat-alien lines can be used as a source of allelopathic potential and weed competitiveness in breeding programs to improve weed suppression ability for wheat improvement. Bertholdsson et al. (2012a, b), showed that the highest allelopathic potential was found in lines with 1R and 2R chromosomes. Moreover, some lines with multiple rye chromosomes (1R + 6R and 1R + 4R + 6R + 7R) were also showed high allelopathic activity (Bertholdsson et al., 2012). Previous studies have identified lines with 1R substitution showing early vigour, which can be positive for the root exudation of allelochemicals (Ehdaie et al., 2003). Breeding efforts for the allelopathic potential is considered as a complex trait (Bertholdsson, 2007), although successful examples are present on rice (Kong et al., 2006) and spring wheat (Bertholdsson, 2010). Quantitative trait loci (QTLs) linked to allelopathic traits have found on wheat chromosomes (Wu et al., 2003), thus, this indicates that allelopathic traits inherited quantitatively. The lines with high allelopathic potential identified in this study may be worthwhile for the breeding of allelopathic wheat, particularly for the purpose of organic wheat.
Various zinc, iron, and cadmium concentrations were identified in these lines. Wild relatives of wheat represent a reach source of micronutrient benefits because they have a huge and deep rooting system during its vegetation period that most efficiently uptake micronutrient if they are available in the soil (Borill et al., 2014). This has been proved by using natural genetic diversity for micronutrient uptake that can increase the nutrient content in wheat through genetic improvement (Velu et al., 2014). For instance, studies have indicated high levels of Fe and Zn to be encoded by a Gpc-B1 locus, present in particular in wild emmer wheat (Uauy et al., 2006; Johansson et al., 2020). Thereby, genetic biofortification in wheat can be enhanced using these wheat-alien introgression lines as a source of natural genetic diversity.
Plant breeding is mostly targeting traits that improve yield potential, i.e. resistance to biotic and abiotic stresses, although for wheat improved baking and bread-making quality is also of outmost importance (Helguera et al., 2020). Wheat flour has, in particularly due to its unique protein properties, qualities which makes it outstanding for end-uses for daily food products such as bread, pastries, biscuits, porridge, cookies, etc. (Johansson et al., 2013). The gluten proteins, the gliadins, and the glutenins, encoded on group 1 and group 6 of the wheat chromosomes, are to a high extent responsible for the impact on the baking quality of wheat (Johansson et al., 2013). Alien introgressions into the wheat genome have often resulted in negative effects on the baking quality, e.g. the Sec-1, Sec-2, and Sec-3 genes from rye instead of corresponding wheat genes at the group 1 chromosome of wheat (Kim et al., 2005). However, introgressions of rye from other parts of the genome than from the group 1 chromosomes might have less tremendous effects on the baking quality. Thus, previous results have indicated that 2BS.2RL wheat–rye translocations only had minor effects on baking quality (Hysing et al., 2007). These authors indicated that there were not any significant differences between the translocation and non-translocation groups like for grain a-amylase activity, grain starch, protein content, and other agronomic performances. Bread-making quality is known to be determined to a large extent by the gluten proteins, their amount and distribution (Johansson et al., 2013). Thus, the grain proteins concentration, the specific protein composition, the amount of specific proteins, and the amount and size distribution of polymeric protein are all factors of relevance for the bread-making quality (Finney and Barmore, 1948; Johansson et al., 2002; Johansson et al., 2003; Johansson et al., 2005; Johansson et al., 2008; Johansson et al., 2013). The evaluated alien introgression lines showed a high level of variability in both grain protein concentration and gluten strength. Thus, the alien material evaluated here, seems to have also interesting properties when it comes to specific quality breeding. Introgressions of Leymus seem to be able to contribute both high nutrition and high gluten strength to the material.
Alien Breeding Through Novel Tools
Introgression of desired genes from wild relatives into the bread wheat has become widely recognized as diversifying genetic diversity. However, wheat-alien chromosome additions often contribute negatively to the agricultural value of the line, therefore, desired gene/s has to be transferred into the wheat genome. Such transfers are normally blocked by the presence of a Ph1 (Pairing homoeologous) allele, which strictly controls homologous chromosome pairing across the hexaploid genome to prevent hybridization between wheat and an alien species (Riley and Chapman, 1958). Anyhow, alien chromosome segments carrying gene/s of interest have been widely transferred into the wheat genome using the CS ph1b homoeologous recombination, radiation, and embryo culture techniques (Sears, 1977; Sears, 1993; Chen et al., 1994; Merker and Lantai, 1997). These approaches in a combination of molecular and cytogenetic manipulations were used to facilitate the introgression of Sr26 and Lr19 from Thinopyrum ponticum, Sr39 from Aegilops speltoides, Sr59 from S. cereale, etc. with small alien chromosome segments (Sharma and Knott, 1966; Merker and Lantai, 1997; Niu et al., 2011; Rahmatov et al., 2016a; Rahmatov et al., 2016b). More recently, reference genomes have been made available for wheat, (IWGSC, 2014), rye (Bauer et al., 2017), barley (IBGSC, 2012), rice (IRGSP, 2005), and Brachypodium (IBI, 2010), greatly facilitating the forward and reverse genetics in crops. Various high-throughput genotyping platforms such as the 9K and 90K Illumina Infinium SNP arrays and the 35K and 820K Affymetrix Axiom arrays have been developed for gene and QTL mapping (Wang et al., 2014; Winfield et al., 2016; Allen et al., 2017). In addition, genotyping-by-sequencing and exome capture sequencing opens up more opportunities for markers development and gene isolation (Poland et al., 2012; Krasileva et al., 2017). All these genotyping platforms provide tremendous tools to assess the genetic diversity and allelic variation across plant genomes. However, a low level of SNP polymorphism between hexaploid wheat and wild relatives has been reported which negatively impact the use of the mentioned platforms (Winfield et al., 2016). Therefore, Tiwari et al. (2014) suggested the use of flow cytometric chromosome sorting to develop unique SNP markers for the mapping of alien genes to overcome these challenges. Whole-genome shotgun sequencing is becoming another valuable breeding tool in terms of time and cost, which are already used in major crops such as wheat (Brenchley et al., 2012), maize (Hufford et al., 2012), rice (Huang et al., 2012), and soybean (Fang et al., 2017). However, the transfer of desired alien gene/s remains a challenge, although some advances have been made in transferring resistance genes. Jupe et al. (2013) developed an exome capture and sequencing of nucleotide-binding leucine-rich repeat (NLR) genes in potato. Such resistance gene enrichment sequencing (RenSeq) allowed a rapid cloning of the Sr22 and Sr45 resistance genes through mutational genomics (Steuernagel et al., 2016). Another approach, MutChromSeq, has been applied through mutational genomics, chromosome flow sorting and sequencing that has resulted in the cloning of the Pm2 resistance gene (Sánchez-Martín et al., 2016). Interestingly, another cloning approach suggested a combination of association genetics and R gene enrichment sequencing, which rapidly identified stem rust resistance genes for cloning (Arora et al., 2019). Besides, a combination of cisgenesis and genome editing tools may accelerate the plant breeding process (Cardi, 2016). Also, the use of speed breeding may significantly accelerate the generation times and breeding cycles (Watson et al., 2018). Therefore, integration of high-throughput genotyping and precise phenotyping tools may efficiently assist in transferring the introgression of small alien chromatin segments to develop new genetic diversity for wheat improvement. For example, the development of synthetic wheat and 1RS chromosome arm has made a great contribution to sustainable wheat production. Evidently, for the development of superior crop cultivars requires new genetic variation that meets sustainable agriculture and food security needs.
Conclusions—Alien Genes Into Modern Wheat—Future Perspectives
Every day, the human population is growing, and with that the demand of food from sustainable and healthy crop production. To adequately meet the global food demand required by 2050, there is a need to increase wheat yield annually. These can be achieved through the two unique opportunities; plant breeding and improved agronomic practices. Importantly, to meet projected food demand, the breeding programs need to broaden the existing genetic base, in particular by the use of alien species with the potential to improve yield, resistance to biotic and abiotic stresses and quality. Several of our studies have identified new sources of resistance to fungal diseases and insects in the wheat-alien introgression derivatives from S. cereale, L. mollis, L. racemosus and T. junceiforme. Also, these lines exhibiting good agronomic performances, high allelopathic potential, and superior end-use quality traits. Our results suggest that some of the lines could be used as a source of high Iron and Zinc and low Cadmium concentrations. These findings show that the wheat-alien introgressions are a potentially useful genetic resource for wheat improvement. The introgression of large alien chromosomes usually challenges researchers and breeders by causing linkage drag that can negatively effect on yield and quality properties. Fortunately, with the presence of high-throughput genotyping and phenotyping tools, opportunities increase to transfer desired gene/s with a small alien chromosome segment. Consequently, research is currently underway to transfer stem and stripe rust resistance genes into the elite wheat background to be used by breeders to develop superior wheat cultivars with new resistance genes. Further, additional research is also in progress for characterization and transferring of useful traits such as micronutrients (Zn, Fe, and Cd), allelopathic potential, diseases, and insect resistance as well as stable baking quality.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
EJ, TH, SA and MR planned various parts of the study, the hypothesis, and the objectives. TH, MP-L, SA, RA carried out various parts of the field and lab work. All authors contributed to compiling various parts of the results. EJ and MR planned the writing of this paper and did the first draft. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by Partnership Alnarp grant nos. PA710 and PA1094, the Swedish Research Council VR grant no. 2016-05806 and Formas grant no. 2017-00514.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank our international research collaborators Matthew Rouse at the United States Department of Agriculture, ARS-CDL, Brian Steffenson at the University of Minnesota, Mogens Hovmøller at the GRRC, Aarhus University, Kumarse Nazari and Mustafa El-Bouhssini at the ICARDA. The authors also appreciate Kenyan Agricultural and Livestock Research Organization Food Crops Research Center and CIMMYT.
References
- Allen A. M., Winfield M. O., Burridge A. J., Downie R. C., Benbow H. R., Barker G. L. A., et al. (2017). Characterization of a wheat breeders’ array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 15, 390–401. 10.1111/pbi.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D., Zheng Q., Luo Q., Ma P., Zhang H., Li L., et al. (2015). Molecular cytogenetic identification of a new wheat-rye 6R chromosome disomic addition line with powdery mildew resistance. PloS One 10 (8), e0134534. 10.1371/journal.pone.0134534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G. R., Papa D., Peng J., Tahir M., Lapitan N. L. V. (2003). Genetic mapping of Dn7, a rye gene conferring resistance to the Russian wheat aphid in wheat. Theor. Appl. Genet. 107, 1297–1303. [DOI] [PubMed] [Google Scholar]
- Andersson S. C., Johansson E., Baum M., Rihawi F., El Bouhssini M. (2015). New resistance sources to Russian wheat aphid (Diuraphis noxia) in Swedish wheat substitution and translocation lines with rye (Secale cereale) and Leymus mollis . Czech J. Genet. Plant Breed. 51, 162–165. 10.17221/72/2015-CJGPB [DOI] [Google Scholar]
- Andersson S. C., Johansson E., Henriksson T., Rahmatov M. (2016). Nya resistensgener för framtidens vete. LTV-fakultetens faktablad. Available from: https://pub.epsilon.slu.se/13096/1/andersson_sc_etal_160225.pdf (Accessed March 09, 2020).
- Arora S., Steuernagel B., Gaurav K., Chandramohan S. (2019). Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. 10.1038/s41587-018-0007-9 [DOI] [PubMed] [Google Scholar]
- Bauer E., Schmutzer T., Barilar I., Mascher M., Gundlach H., Martis M. M., et al. (2017). Towards a whole-genome sequence for rye (Secale cereale L.). Plant J. 89, 853–869. 10.1111/tpj.13436 [DOI] [PubMed] [Google Scholar]
- Bertholdsson N.-O., Andersson S. C., Merker A. (2012). Allelopathic potential of Triticum spp., Secale spp. and Triticosecale spp. and use of chromosome substitutions and translocations to improve weed suppression ability in winter wheat. Plant Breed. 131, 75–80. 10.1111/j.1439-0523.2011.01895.x [DOI] [Google Scholar]
- Bertholdsson N.-O. (2007). Varietal variation in allelopathic activity in wheat and barley and possibilities for use in plant breeding. Allelopathy J. 19, 193–201. [Google Scholar]
- Bertholdsson N.-O. (2010). Breeding spring wheat for improved allelopathic potential. Weed Res. 50, 49–57. 10.1111/j.1365-3180.2009.00754.x [DOI] [Google Scholar]
- Borrill P., Connorton J. M., Balk J., Miller A. J., Sanders D., Uauy C. (2014). Biofortification of wheat grain with iron and zinc: integrating novel genomic resources and knowledge from model crops. Front. Plant Sci. 5, 53. 10.3389/fpls.2014.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R., Spannagl M., Pfeifer M., Barker G. L. A., D’Amore R., Allen A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. 10.1038/nature11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardi T. (2016). Cisgenesis and genome editing. Combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed. 135, 139–147. 10.1111/pbr.12345 [DOI] [Google Scholar]
- Cavanagh C. R., Chao S., Wang S., Huang B. E., Stephen S., Kiani S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. 110, 8057–8062. 10.1073/pnas.1217133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. D., Tsujimoto H., Gill B. S. (1994). Transfer of PhI genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theor. Appl. Genet. 88, 97–101. 10.1007/BF00222400 [DOI] [PubMed] [Google Scholar]
- Crespo-Herrera L. A., Singh R. P., Sabraoui A., El-Bouhssini M. (2019). Resistance to insect pests in wheat-rye and Aegilops speltoides Tausch translocation and substitution lines. Euphytica 215, 123 . 10.1007/s10681-019-2449-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal A. S., Mares D. J., Marshall D. R., Skeritt J. H. (1988). Protein composition and pentosan content in relation to dough stickiness of 1B/1R translocation wheats. Cereal Chem. 62, 143–149. [Google Scholar]
- Dubcovsky J., Dvorak J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866. 10.1126/science.1143986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehdaie B., Whitkus R. W., Waines J. G. (2003). Root biomass, water-use efficiency, and performance of wheat-rye translocations of chromosomes 1 and 2 in spring bread wheat ‘‘Pavon’’. Crop Sci. 43, 710–717. 10.2135/cropsci2003.0710 [DOI] [Google Scholar]
- El Bouhssini M., Street K., Amri A., Mackay M., Ogbonnaya F. C., Omran A., et al. (2011). Sources of resistance in berad wheat to Russian wheat aphid (Diuraphis noxia) in Syria identified using the Focused Identification of Germplasm Strategy (FIGS). Plant Breed. 130, 96–97. 10.1111/j.1439-0523.2010.01814.x [DOI] [Google Scholar]
- El Bouhssini M., Ogbonnaya F. C., Chen M., Lhaloui S., Rihawi F., Dabbous A. (2013). Sources of resistance in primary synthetic hexaploid wheat (Triticum aestivum L.) to insect pests: Hessian fly, Russian wheat aphid and Sunn pest in the fertile crescent. Genet. Resour. Crop Evol. 60, 621–627. 10.1007/s10722-012-9861-3 [DOI] [Google Scholar]
- Ellneskog-Staam P., Merker A. (2001). Genome composition, stability and fertility of hexaploid alloploids between Triticum turgidum var. carthlicum and Leymus racemosus . Hereditas 134 79– , 84. 10.1111/j.1601-5223.2001.00079.x [DOI] [PubMed] [Google Scholar]
- Ellneskog-Staam P., Merker A. (2002). Screening for Resistance to Powdery Mildew and Brown Rust in Wheat-Leymus racemosus and Wheat-Thinopyrum junceiforme Alloploids. Acta Agric. Scand. Section B Soil Plant Sci. 52, 158–161. 10.1080/09064710300525 [DOI] [Google Scholar]
- Fang C., Ma Y., Wu S., Liu Z., Wang Z., Yang R., et al. (2017). Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18, 161. 10.1186/s13059-017-1289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2016). Save and Grow in practice: maize, rice, wheat. A guide to sustainable cereal production. Available from: http://www.fao.org/3/a-i4009e.pdf (Accessed March 09, 2020).
- Finney K. F., Barmore M. A. (1948). Loaf volume and protein content of hard winter and spring wheats. Cereal. Chem. 25, 291–312. [Google Scholar]
- Forsström P.-O., Merker A. (2001). Sources of wheat powdery mildew resistance from wheat-rye and wheat-leymus hybrids. Hereditas 134, 115–119. 10.1111/j.1601-5223.2001.00115.x [DOI] [PubMed] [Google Scholar]
- Forsström P., Merker A., Schwarzacher T. (2002). Characterisation of mildew resistant wheat-rye substitution lines and identification of an inverted chromosome by fluorescent in situ hybridisation. Heredity 88, 349–355. 10.1038/sj.hdy.6800051 [DOI] [PubMed] [Google Scholar]
- Friebe B., Jiang J., Raupp W. J., McIntosh R. A., Gill B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. 10.1007/BF00035277 [DOI] [Google Scholar]
- Friebe B., Kynast R. G., Hatchett J. H., Sears R. G., Wilson D. L., Gill B. S. (1999). Transfer of rye-derived Hessian fly resistance genes H21 and H25 from bread wheat into durum wheat. Crop Sci. 3, 1692–1696. 10.2135/cropsci1999.3961692x [DOI] [Google Scholar]
- Hao M., Liu M., Luo J., Fan C., Yi Y., Zhang L., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant Sci. 9, 1040. 10.3389/fpls.2018.01040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helguera M., Abugalieva A., Battenfield S., Békés F., Branlard G., Cuniberti M., et al. (2020). “Grain quality in breeding,” in Wheat quality for improving processing and human health. Eds. Igrejas G. Ikeda T. M., Guzmán C. (Switzerland: Springer; ), pp 273–pp 308. [Google Scholar]
- Huang X., Röder M. S. (2011). High-density genetic and physical bin mapping of wheat chromosome 1D reveals that the powdery mildew resistance gene Pm24 is located in a highly recombinogenic region. Genetica 139, 1179–1187. 10.1007/s10709-011-9620-y [DOI] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wang Z.-X., Wang A., Zhao Q., Zhao Y., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M. B., Xu X., van Heerwaarden J., Pyhäjärvi T., Chia J.-M., Cartwright R. A., et al. (2012). Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811. 10.1038/ng.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husenov B., Otambekova M., Muminjanov H., Morgounov A., Asaad S., Garkava-Gustavsson L., et al. (2020). Constraints and perspectives for sustainable wheat production in Tajikistan. Front. Sustain. Food Syst. 4, 27. 10.3389/fsufs.2020.00027 [DOI] [Google Scholar]
- Hussain A., Larsson H., Kuktaite R., Johansson E. (2010). Mineral composition of organically grown wheat genotypes: contribution to daily minerals intake. Int. J. Environ. Res. Public Health 7, 3442–3456. 10.3390/ijerph7093442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A., Larsson H., Kuktaite R., Prieto-Linde M. L., Johansson E. (2012). Towards the understanding of bread-making quality in organically grown wheat: Dough mixing behaviour, protein polymerisation and structural properties. J. Cereal Sci. 56, 659–666. 10.1016/j.jcs.2012.07.016 [DOI] [Google Scholar]
- Hussain A., Larsson H., Kuktaite R., Prieto-Lindem M. L., Johansson E. (2013). Amount and size distribution of monomeric and polymeric proteins in the grain of organically produced wheat. Cereal Chem. 90, 80–86. 10.1094/CCHEM-05-12-0064-R [DOI] [Google Scholar]
- Hysing S. C., Hsam S. L. K., Singh R. P., Huerta-Espino J., Boyd L. A., Koebner R. M. D., et al. (2007). Agronomic performance and multiple disease resistance in T2BS.2RL wheat-rye translocation lines. Crop Sci. 47, 254–260. 10.2135/cropsci2006.04.0269 [DOI] [Google Scholar]
- IBGSC (The International Barley Genome Sequencing Consortium) (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- IBI (The International Brachypodium Initiative) (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463, 763–768 . 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- IRGSP (International Rice Genome Sequencing Project) (2005). The map-based sequence of the rice genome. Nature 436, 793–800. 10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- IWGSC (The International Wheat Genome Sequencing Consortium) (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum ) genome. Science 345, 1251788. 10.1126/science.1251788 [DOI] [PubMed] [Google Scholar]
- Johansson E., Prieto-Linde M. L., Jönsson J. (2001). Effects of wheat cultivar and nitrogen application on storage protein composition and bread-making quality. Cereal Chem. 78, 19–25. 10.1094/CCHEM.2001.78.1.19 [DOI] [Google Scholar]
- Johansson E., Nilsson H., Mazhar H., Skerritt J., MacRitchie F., Svensson G. (2002). Seasonal effects on storage proteins and gluten strength in four Swedish wheat cultivars. J. Sci. Food Agric. 82, 1305–1311. 10.1002/jsfa.1185 [DOI] [Google Scholar]
- Johansson E., Prieto-Linde M., Svensson G., Jönsson J. (2003). Influences of cultivar, cultivation year and fertilizer rate on amount of protein groups and amount and size distribution of mono- and polymeric proteins in wheat. J. Agr. Sci. 140, 275–284. 10.1017/S0021859603003162 [DOI] [Google Scholar]
- Johansson E., Kuktaite R., Andersson A., Prieto-Linde M. L. (2005). Protein polymer built-up during wheat development: influences of temperature and nitrogen timing. J. Sci. Food Agric. 85, 473–479. 10.1002/jsfa.2006 [DOI] [Google Scholar]
- Johansson E., Prieto-Linde M., Gissén C. (2008). Influences of weather, cultivar and fertiliser rate on grain protein polymer accumulation in field-grown winter wheat, and relations to grain water content and falling number. J. Sci. Food Agric. 88:, 2011–2018. 10.1002/jsfa.3312 [DOI] [Google Scholar]
- Johansson E., Malik A. H., Hussain A., Rasheed F., Newson W. R., Plivelic T., et al. (2013). Wheat gluten polymer structures: the impact of genotype, environment, and processing on their functionality in various applications. Cereal Chem. 90, 367–376. 10.1094/CCHEM-08-12-0105-FI [DOI] [Google Scholar]
- Johansson E., Hussain A., Kuktaite R., Andersson S. C., Olsson M. E. (2014). Contribution of organically grown crops to human health. Int. J. Environ. Res. Public Health 11, 3870–3893. 10.3390/ijerph110403870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E., Branlard G., Cuniberti M., Flagella Z., Hüsken A., Nurit E., et al. (2020). “Genotypic and environmental effect on wheat technological and nutritional quality,” in Wheat quality for improving processing and human health. Eds. Igrejas G. Ikeda T. M., Guzmán C. (Switzerland: Springer; ), pp 171–204. [Google Scholar]
- Jupe F., Witek K., Verweij W., Sliwka J., Pritchard L., Etherington G. J., et al. (2013). Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544. 10.1111/tpj.12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H. (1983). “Origin and history of ‘Daruma’, a parental variety of Norin 10,” in Proceedings of the Sixth International Wheat Genetics Symposium. Ed. Sakamoto S. (Kyoto, Japan, Beijing: Plant Germplasm Institute, University of Kyoto, China Agricultural Scientech Press; ). [Google Scholar]
- Kim W., Johnson J. W., Baenziger P. S., Lukaszewski A. J., Gaines C. S. (2005). Quality effect of wheat-rye (1R) translocation in “Pavon 76”. Plant Breed. 124, 334–337. 10.1111/j.1439 [DOI] [Google Scholar]
- Kong C. H., Li H. B., Hu F., Xu X. H., Wang P. (2006). Allelochemicals released by rice roots and residues in soil. Plant Soil 288, 47–56. 10.1007/s11104-006-9033-3 [DOI] [Google Scholar]
- Krasileva K. V., Vasquez-Gross H. A., Howell T., Bailey P., Paraiso F., Clissold L., et al. (2017). Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. 114, 913–921. 10.1073/pnas.1619268114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Dundas I., Dong C., Li G., Trethowan R., Yang Z., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. 10.1007/s00122-020-03534-y [DOI] [PubMed] [Google Scholar]
- Mago R., Zhang P., Vautrin S., Šimková H., Bansal U., Luo M.-C., et al. (2015). The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants. 1, 15186. 10.1038/NPLANTS.2015.186 [DOI] [PubMed] [Google Scholar]
- Mago R., Spielmeyer W., Lawrence G., Lagudah E., Ellis J., Pryor A. (2002). Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor. Appl. Genet. 104, 1317–1324. [DOI] [PubMed] [Google Scholar]
- Malik A. H., Prieto-Linde M. L., Kuktaite R., Andersson A., Johansson E. (2011). Individual and interactive effects of cultivar maturation time, nitrogen regime and temperature level on accumulation of wheat grain proteins. J. Sci. Food Agric. 91, 2192–2200. 10.1002/jsfa.4439 [DOI] [PubMed] [Google Scholar]
- Malik A. H., Kuktaite R., Johansson E. (2013). Combined effect of genetic and environmental factors on the accumulation of proteins in the wheat grain and their relationship to bread-making quality. J. Cereal Sci. 57, 170–174. 10.1016/j.jcs.2012.09.017 [DOI] [Google Scholar]
- Marais G. F., Marais A. S. (1994). The derivation of compensating translocations involving homoeologous group 3 chromosomes of wheat and rye. Euphytica 79, 75–80. 10.1007/BF00023578 [DOI] [Google Scholar]
- McFadden E. S., Sears E. R. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 107–116. 10.1093/oxfordjournals.jhered.a105594 [DOI] [PubMed] [Google Scholar]
- McIntosh R. A., Wellings C. R., Park R. F. (1995). Wheat rusts: an atlas of resistance genes (Dordrecht: Kluwer Academic Publishers; ). [Google Scholar]
- Merker A., Forsström P. (2000). Isolation of mildew resistant wheat-rye translocation lines from a double substitution line. Euphytica 115, 167–172. 10.1023/A:1004018500970 [DOI] [Google Scholar]
- Merker A., Lantai K. (1997). Hybrids between wheats and perennial Leymus and Thinopyrum species. Acta Agric. Scand. Section B Plant Soil Sci. 47, 48–51. 10.1080/09064719709362438 [DOI] [Google Scholar]
- Merker A., Rogalska S. (1984). The breeding behaviour of a double disomic wheat-rye substitution line. Cereal. Res. Commun. 12, 13–17. [Google Scholar]
- Merker A. (1979). The breeding behaviour of some rye wheat chromosome substitutions. Hereditas 91, 245–255. 10.1111/j.1601-5223.1979.tb01667.x [DOI] [Google Scholar]
- Merker A. (1984). The rye genome in wheat breeding. Hereditas 100, 183–191. 10.1111/j.1601-5223.1984.tb00118.x [DOI] [Google Scholar]
- Molnár-Láng M., Ceoloni C., Doležel J. (2015). Alien Introgression in Wheat. Cytogenetics, Molecular Biology, and Genomics (Berlin/Heidelberg, Germany: Springer; ). [Google Scholar]
- Moreira-Ascarrunz S. D., Larsson H., Prieto-Linde M. L., Johansson E. (2016). Mineral nutritional yield and nutrient density of locally adapted wheat genotypes under organic production. Foods 5, 89. 10.3390/foods5040089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V., Fleury D., Timmins A., Laga H., Hayden M., Mather D., et al. (2015). Addition of rye chromosome 4R to wheat increases anther length and pollen grain number. Theor. Appl. Genet. 128, 953–964. 10.1007/s00122-015-2482-4 [DOI] [PubMed] [Google Scholar]
- Niu Z., Klindworth D. L., Friesen T. L., Chao S., Jin Y., Cai X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. 10.1534/genetics.110.123588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera P. D., Pretorius Z. A., Badebo A., Jin Y. (2013). Identification of resistance to races of Puccinia graminis f. sp. tritici with broad virulence in Triticale (×Triticosecale). Plant Dis. 97, 479–484. [DOI] [PubMed] [Google Scholar]
- Peterson R. F., Campbell A. B., Hannah A. E. (1948). A diagrammatic scale for estimating rust severity on leaves and stems of cereals. Can. J. Res. Section C. 26, 496–500. 10.1139/cjr48c-033 [DOI] [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E., Jannink J. L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PloS One 7, e32253. 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Pumphrey M., Friebe B., Chen P., Gill B. (2008). Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor. Appl. Genet. 117, 1155–1166. 10.1007/s00122-008-0853-9 [DOI] [PubMed] [Google Scholar]
- Rabinovich S. V. (1998). Importance of wheat-rye translocations for breeding modern cultivar of Triticum aestivum L. Euphytica 100, 323–340. 10.1023/A:1018361819215 [DOI] [Google Scholar]
- Rahmatov M., Garkava-Gustavsson L., Wanyera R., Steffenson B., Rouse M., Johansson E. (2015). Stem rust resistance in 1BL.1RS and 2RL.2BS double wheat-rye translocation lines. Czech J. Genet. Plant Breed. 51, 148–154. 10.17221/80/2015-CJGPB [DOI] [Google Scholar]
- Rahmatov M., Rouse M. N., Nirmala J., Danilova T., Friebe B., Steffenson B. J., et al. (2016. a). A new 2DS·2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 129, 1383–1392. 10.1007/s00122-016-2710-6 [DOI] [PubMed] [Google Scholar]
- Rahmatov M., Rouse M. N., Steffenson B. J., Andersson S. C., Wanyera R., Pretorius Z. A., et al. (2016. b). Sources of stem rust resistance in wheat–alien introgression lines. Plant Dis. 100, 1101–1109. 10.1094/PDIS-12-15-1448-RE [DOI] [PubMed] [Google Scholar]
- Rahmatov M., Hovmøller M. S., Nazari K., Andersson S. C., Steffenson B. J., Johansson E. (2017). Seedling and adult plant stripe rust resistance in diverse wheat–alien introgression lines. Crop Sci. 57, 2032–2042. 10.2135/cropsci2016.08.0664 [DOI] [Google Scholar]
- Rahmatov M. (2016). Genetic characterisation of novel resistance alleles to stem rust and stripe rust in wheat-alien introgression lines. Doctoral thesis (SLU Epsilon Open Archive; ). Available from: https://pub.epsilon.slu.se/13585/ (Accessed March 09, 2020). [Google Scholar]
- Riley R., Chapman V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715. 10.1038/182713a0 [DOI] [Google Scholar]
- Roelfs A. P., Singh R. P., Saari E. E. (1992). Rust diseases of wheat: Concepts and methods of disease management (Mexico D.F., Mexico: CIMMYT; ). [Google Scholar]
- Sánchez-Martín J., Steuernagel B., Ghosh S., Herren G., Hurni S., Adamski N. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221. 10.1186/s13059-016-1082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS (2011). SAS Institute Inc 9.3 (NC, USA: SAS Institute Inc.). [Google Scholar]
- Schlegel R. (2020). Current list of wheats with rye and alien introgression 1R.1B - 1R/1B - 1R-1B - 1RS.1BL - 1R(1B) - alien. Available from: http://www.rye-gene-map.de/rye-introgression/index.html (Accessed March 09, 2020).
- Schneider A., Rakszegi M., Molnár-Láng M., Szakács É., et al. (2016). Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor. Appl. Genet. 129, 1045–1059. 10.1007/s00122-016-2682-6 [DOI] [PubMed] [Google Scholar]
- Sears E. R. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. 10.1139/g77-063 [DOI] [Google Scholar]
- Sears E. R. (1993). Use of radiation to transfer alien chromosome segments to wheat. Crop Sci. 33, 897–901. 10.2135/cropsci1993.0011183X003300050004x [DOI] [Google Scholar]
- Sharma D., Knott D. R. (1966). The transfer of leaf rust resistance from Agropyron to Triticum by irradiation. Can. J. Genet. Cytol. 8, 137–143. 10.1139/g66-018 [DOI] [Google Scholar]
- Shewry P. R., Hey S. (2015). The contribution of wheat to human diet and health. Food Energy Secur. 4, 178–202. 10.1002/fes3.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiferaw B., Smale M., Braun H., Duveiller E., Reynolds M., Muricho G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Seurc. 5, 291–317. 10.1007/s12571-013-0263-yn [DOI] [Google Scholar]
- Singh R. P., Hodson D. P., Huerta-espino J., Jin Y., Njau P., Wanyera R., et al. (2008). Will stem rust destroy the world’s wheat crop? Adv. Agron. 98, 271–309. 10.1016/S0065-2113(08)00205-8 [DOI] [Google Scholar]
- Singh R. P., Hodson D. P., Huerta-Espino J., Jin Y., Bhavani S., Njau P., et al. (2011). The emergence of Ug99 races of the stem rust fungus is a threat to the world wheat production. Annu. Rev. Phytopathol. 49, 465–481. 10.1146/annurev-phyto-072910-095423 [DOI] [PubMed] [Google Scholar]
- Singh S., Vikram P., Sehgal D., Burgueño J., Sharma A., Singh S. K., et al. (2018). Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Sci. Rep. 8, 12527. 10.1038/s41598-018-30667-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakman E. C., Stewart D. M., Loegering W. Q. (1962). Identification of physiologic races of Puccinia graminis var. tritici (US Department of Agric., ARS E-617; ). [Google Scholar]
- Steenwerth K. L., Hodson A. K., Bloom A. J., Carter M. R., Cattaneo A., Chartres C. J., et al. (2014). Climate-smart agriculture global research agenda: scientific basis for action. Agric. Food Secur. 3, 11. 10.1186/2048-7010-3-11 [DOI] [Google Scholar]
- Steuernagel B., Periyannan S. K., Hernández-Pinzón I., Witek K., Rouse M. N., Yu G. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. 10.1038/nbt.3543 [DOI] [PubMed] [Google Scholar]
- Tanksley S. D., McCouch S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066. 10.1126/science.277.5329.1063 [DOI] [PubMed] [Google Scholar]
- The T. T., Gupta R. B., Dyck P. L., Appels R., Hohmann U., McIntosh R. A. (1991). Characterization of stem rust resistant derivatives of wheat cultivar Amigo. Euphytica 58, 245–252. 10.1007/BF00025256 [DOI] [Google Scholar]
- Tiwari V. K., Wang S., Sehgal S., Vrana J., Friebe B., Kubalakova M., et al. (2014). SNP discovery for mapping alien introgressions in wheat. BMC Genom. 15, 273. 10.1186/1471-2164-15-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. (2006). A NAC gene regulating senescence improves grain protein, zinc and irons content in wheat. Science 314, 1298–1300. 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D., Berger A., Prieto-Linde M., Johansson E. (2019). Can nitrogen fertilization be used to modulate yield, protein content and bread-making quality in Uruguayan wheat? J. Cereal Sci. 85, 153–161. 10.1016/j.jcs.2018.12.001 [DOI] [Google Scholar]
- Velu G., Ortiz-Monasterio I., Cakmak I., Hao Y., Singh R. P. (2014). Biofortification strategies to increase grain zinc and iron concentrations in wheat. J. Cereal Sci. 59, 365–372. 10.1016/j.jcs.2013.09.001 [DOI] [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., Huang B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A., Ghosh S., Williams M. J., Cuddy S. W., Simmonds J., Rey M.-D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants. 4, 23–29. 10.1038/s41477-017-0083-8 [DOI] [PubMed] [Google Scholar]
- Winfield M. O., Allen A. M., Burridge A. J., Barker G. L. A., Benbow H. R., Wilkinson P. A., et al. (2016). High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206. 10.1111/pbi.12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S., Esbensen K., Geladi P. (1987). Principal component analysis. Chemom. Intell. Lab. Syst. 2 (1-3), 37–52. 10.1016/0169-7439(87)80084-9 [DOI] [Google Scholar]
- Wu H., Pratley J., Ma W., Haig T. (2003). Quantitative trait loci and molecular markers associated with wheat allelopathy. Theor. Appl. Genet. 107, 1477–1481. 10.1007/s00122-003-1394-x [DOI] [PubMed] [Google Scholar]
- Xi W., Tang Z., Luo J., Fu S. (2019). Physical location of new stripe rust resistance gene (s) and PCR-based markers on rye (Secale cereal L.) chromosomes 5 using 5R dissection lines. Agronomy 9, 498. [Google Scholar]
- Zeven A. C. (1998). Landraces: a review of definitions and classifications. Euphytica 104, 127–139. 10.1023/A:1018683119237 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.