Abstract
Apoptosis is an important contributing factor in spinal cord injury (SCI). ZBTB38 is involved in the transcriptional regulation of multiple signaling pathways, is differentially expressed at different SCI stages, and may provide a therapeutic strategy for the treatment of patients with SCI. In this study, we found that autophagy is blocked in ZBTB38 knockdown SH-SY5Y cells and that the expression levels of LC3B II/I decreased and P62 increased. We used transcriptome high-throughput sequencing to identify the target in ZBTB38 knockdown cells. From the transcriptome profile, RB1CC1 (i.e., FIP200), a key component of the initiation machinery of autophagy (FIP200-ATG13-ULK1-ATG101), was found to decrease 4.2-fold following ZBTB38 knockdown. When RB1CC1-overexpressed plasmids were transfected into ZBTB38 knockdown cells, they rescued the phenotype of ZBTB38 knockdown cells. Cell proliferation and viability were significantly enhanced by RB1CC1 overexpression, and LC3B and P62 expression returned to their original levels. We also injected ZBTB38-overexpressed lentivirus into the injured center of the spinal cord and detected significant upregulation of RB1CC1 in the spinal cord. ZBTB38 overexpression can promote autophagy and partly rescue the secondary damage of SCI. Therefore, our findings provide a new strategy for the treatment of SCI.
Keywords: ZBTB38, RNA sequencing, Spinal Cord Injury, RB1CC1, Autophagy
1. Introduction
ZBTB38 encodes a protein called CIBZ, a member of the C2H2 zinc finger protein family, which typically contains the BTB domain (Oikawa et al., 2011). The BTB domain is often contained in transcription factors, which regulate the transcription of multiple target genes and participate in several signaling pathways. ZBTB38 also plays an important role in gene expression regulation, cell differentiation, and embryo development (Sasai et al., 2005; Stogios et al., 2005). Although hundreds of BTB/POZ genes have been identified in the human genome, few have been functionally characterized.
Spinal cord injury (SCI) is characterized by spinal cord damage resulting from a blunt or penetrating trauma and constitutes a severe disease of the central nervous system, with a high disablement rate. SCI still lacks effective clinical treatment (Furlan and Fehlings, 2008; Mountney et al., 2010; Nakamura and Okano, 2013; Wright et al., 2011; Zurita et al., 2012). Scientists have suggested that apoptosis is an important contributing factor in spinal cord damage in animal models and in human tissues (Penas et al., 2007). However, the signaling pathways involved in SCI remain unexplored. Our previous study in an SCI mice model showed that the ZBTB38/CIBZ protein is differentially expressed at different SCI stages (Cai et al., 2017). In addition, cell apoptosis was induced by endoplasmic reticulum stress (Cai et al., 2017). We therefore believe that functionally restored ZBTB38 can provide a therapeutic strategy for the treatment of patients with SCI.
Autophagy, a highly conserved lysosome-dependent degradation pathway, is widely present in eukaryotic cells and serves as an important alternative defense and protection mechanism complementary to endoplasmic reticulum stress (Klionsky and Emr, 2000; Yang and Klionsky, 2010). The organism enhances autophagy to eliminate non-functional organelles and degraded proteins, thereby enabling cell recycling and reuse. However, excessive autophagy induction may trigger autophagic cell death, a programmed cell death that differs from apoptosis (Korkmaz et al., 2012; Menghini et al., 2014; Mikhaylova et al., 2012; Ucar et al., 2012; Wang et al., 2013a). Recently, it was found that autophagic cell death plays an important role in tumor formation and infection (Kanzawa et al., 2004; Suzuki et al., 2001). In a previous study, we found that ZBTB38 expression can be regulated by the binding of the endoplasmic reticulum stress-responsive transcription factor ATF4 to its promoter region (Cai et al., 2017). Whether ZBTB38 plays an important role in the autophagic regulation pathway of SCI remains unknown.
RB1CC1, also known as the focal adhesion kinase family-interacting protein of 200 kDa (FIP200), is a potential target for the RB1 gene (Melkoumian et al., 2005). It is widely expressed in human and mouse tissues and is particularly abundant in the heart, testes, and musculoskeletal tissues. FIP200 is involved in various cellular regulatory processes, including cell proliferation, differentiation, apoptosis, and migration (Melkoumian et al., 2005; Wei et al., 2009). FIP200 also serves as a key factor in the initiation of antophagic synthesis in the mammalian target of rapamycin (mTOR) signaling pathway and participates in the formation of the autophagy initiation complex, comprising ULK1 (unc-51-like kinase 1), ATG13 (autophagy-related protein 13), FIP200, and ATG101. In FIP200-deficient cells, mTOR autophagy induction is inhibited, irreparable DNA damage occurs, and cell death is enhanced (Abbi et al., 2002; Wang et al., 2011).
To investigate the possible function of ZBTB38 in the spinal cord, we knocked down the ZBTB38 gene using small interfering RNA (siRNA) in SH-SY5Y cells. Transcriptome high-throughput sequencing was also performed in the control and siZBTB38 groups to assess the role of ZBTB38 in the autophagy pathway. To determine whether restoring RB1CC1 expression can rescue autophagy-mediated death after ZBTB38 knockdown, an RB1CC1-overexpressed plasmid was transfected into siZBTB38 SH-SY5Y cells. We then generated a traumatic SCI mouse model by constructing a lentivirus carrying full-length ZBTB38 cDNA and injecting it into the lesions of SCI mice.
2. Materials and methods
2.1. Cell culture and standard assays
SH-SY5Y cells were purchased from American Type Culture Collection (Rockville, MD, USA) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin–streptomycin. Transient transfections, lentiviral transduction and infection, quantitative real-time polymerase chain reaction (qRT-PCR), Western blot, and immunofluorescence were performed as described previously (Cai et al., 2011; Cai et al., 2012; Cai et al., 2017). The primers used in qRT-PCR and siRNA suppression assays are listed in Supplemental Table 1.
2.2. Constructs and reagents
The RB1CC1 (NM_014781)-overexpressed plasmid vector (Genechem, Shanghai, China) containing a full-length human RB1CC1 (RB1CC1-F: CGAGCTCAAGCTTCGAATTCCGCCACCATGAAGTTATATGTATTTCTGGTTAACACTG; RB1CC1-R: TGGTGGCGACCGGTGGATCCCGTACTTTCTTATTCCATGATACGGCTTTC) was used to overexpress the RB1CC1 gene in SH-SY5Y cells. Null vector transfection was used as control. To construct the ZBTB38 (NM_175537) lentiviral expression vector, a full-length mouse ZBTB38 (ZBTB38-F: GAGGATCCCCGGGTACCGGTCGCCACCATGACAGTCATGTCCCTCTCCAGGGAC; ZBTB38-R: TCCTTGTAGTCCATACCAAGGACGTTTTCAGCAAAGGCTTTTATG) was cloned into the GV358 vector (Genechem, Shanghai, China) according to manufacturer’s instructions. The construction of the RB1CC1-overexpressed plasmid vector and the ZBTB38 lentiviral expression vector was performed as described previously (Cai et al., 2011; Cai et al., 2012; Cai et al., 2017; Wang et al., 2013b). RB1CC1/FIP200, LC3B, p62, and GAPDH antibodies were obtained from Abcam (Cambridge, MA).
2.3. Cell counting kit-8 experiments
Cell counting kit-8 (CCK-8) experiments were conducted according to manufacturer’s instructions. The CCK-8 was purchased from Cell Signaling (Beverly, MA). The experimental groups were the following: the null vector transfection control group, the siZBTB38 SH-SY5Y group (SH-SY5Y cells transfected with ZBTB38 siRNA), and the RB1CC1/siZBTB38 group (i.e., a plasmid with overexpressing RB1CC1 was transfected into siZBTB38 SH-SY5Y cells). Absorbance at 450 nm was measured 0, 12, 18, 24, 36, and 72 h after transient transfection.
2.4. Library construction for RNA sequencing
Transcriptome high-throughput sequencing was performed in the control group (SH-SY5Y cells transfected with liposome alone) and the siZBTB38 group (SH-SY5Y cells transfected with ZBTB38 siRNA). Total RNA was isolated from SH-SY5Y cells using TRIzol and the pure-link RNA mini kit (ThermoFisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using the Qubit RNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
In total, 2 μg RNA per sample was used as input material for RNA sample preparations. This study included two groups of three biological replicates. Sequencing libraries were generated using a NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, USA), and index codes were added to attribute sequences to each sample. Fragmentation was performed using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After the adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with a hairpin loop structure was ligated to prepare for hybridization. The library fragments were purified using AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min, followed by 5 min at 95 °C before PCR. Following this, PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers, and index (X) Primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed using the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v4-cBot-HS (Illumia). Following cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform, and paired-end reads were generated. Staff at Beijing Biomarker Technologies (Beijing, China) performed isolation of mRNA, fragment interruption, cDNA synthesis, addition of adapters, PCR amplification and RNA-Seq.
2.5. Traumatic SCI animal model and immunofluorescence analysis
SCI was performed as described previously (Cai et al., 2011; Cai et al., 2012). Eight-week-old male Kunming mice weighing 25–30 g were purchased from the Jiangning Qinglongshan Animal Cultivation Farm (Nanjing, China). All experimental procedures were approved by the Institutional Animal Care and Use Committee at Nanjing Agricultural University, China (Cai et al., 2012).
The control group underwent sham injuries wherein only vertebral plates were cut off without causing spinal injuries. The spinal processes from Th7–Th9 were exposed, and SCI success was confirmed by quick jerks of the hind limbs observed in trauma-surgery animals. To prevent urinary retention, we squeezed the bladders of SCI mice three times a day to assist with urination until the mice regained the automatic micturition reflex. SCI mice were euthanized by cervical vertebra dislocation 2, 4 and 8 days after trauma, and spinal cord samples were collected and processed. The spinal cords of Th7–Th9 were fixed in PFA overnight and embedded in paraffin wax. For immunofluorescence, sections were stained with the RB1CC1/FIP200 antibody (Abcam, Cambridge, MA) as described previously (Cai et al., 2011; Cai et al., 2012; Cai et al., 2017).
2.6. Injection of the ZBTB38 lentivirus
Mice were divided into two groups (n = 10 each) at random 24 h after SCI model success. Into the injury center, SCI groups were injected with 50 μl of empty lentivirus and treatment groups were injected with 50 μl of ZBTB38 lentivirus (titer: 108 TU/ml). Spinal cord samples were collected and processed 2, 4, and 8 days after treatment.
2.7. Data and statistical analysis
2.7.1. Gene functional annotation
Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), KO (KEGG Ortholog database), and GO (Gene Ontology).
2.7.2. Differential expression analysis
Differential expression analysis of the two conditions was performed using the DEGseq (2010) R package. The p-values obtained from a negative binomial model of gene expression were adjusted using Benjamini and Hochberg corrections to control for false discovery rates. Genes with an adjusted p-value < 0.05 were considered to be differently expressed between groups.
2.7.3. GO enrichment and KEGG pathway enrichment analysis
GO enrichment analysis of the differently expressed genes (DEGs) was implemented in the “GO seq” package in R based on a Wallenius non-central hyper-geometric distribution, which can adjust for gene length bias in DEGs (Young et al., 2010).
KEGG is a database for understanding high-level functions and utilities of biological systems through large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/) (Kanehisa et al., 2008). We used the KOBAS (Mao et al., 2005) software to test for the statistical enrichment of differentially expressed genes in KEGG pathways.
2.7.4. Statistical analysis
All data were reported as mean ± standard deviation and analyzed using one-way analysis of variance in SPSS v.17.0. Statistical tests were performed with the Kruskal–Wallis and Mann–Whitney U tests. A least significant difference test was used for comparisons between groups. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. ZBTB38 knockdown significantly inhibits autophagic expression in SH-SY5Y
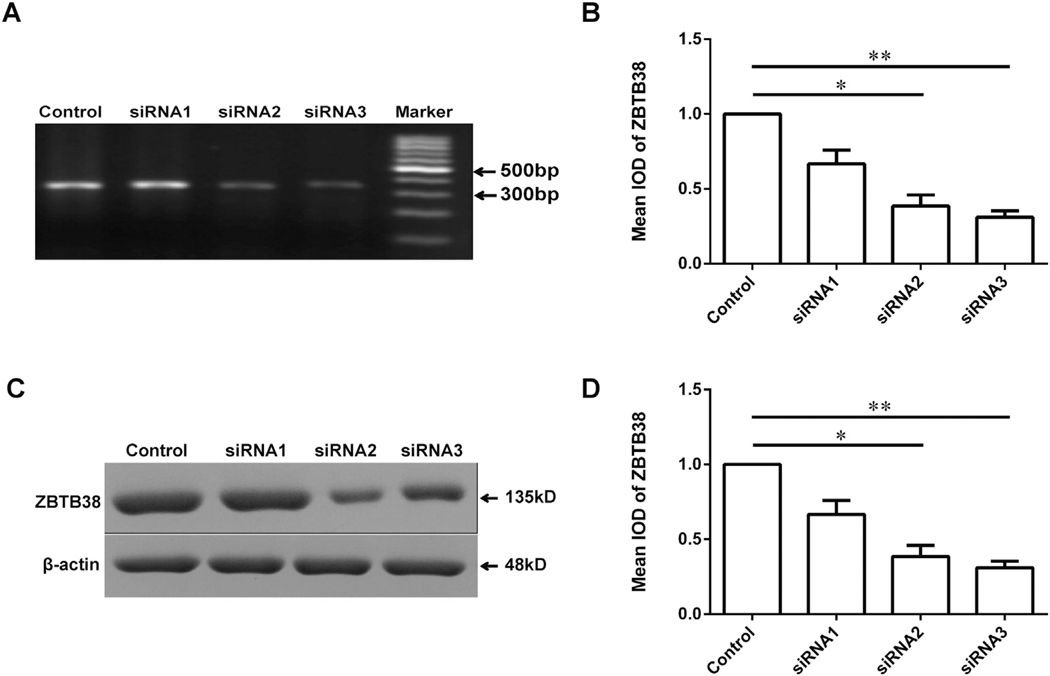
The ZBTB38 expression levels significantly decreased in the knockdown condition in SH-SY5Y cells (Fig. 1), indicating that ZBTB38 knockdown cells could be used for following experiments.
Fig. 1.
Down regulations of ZBTB38 by siRNA in SH-SY5Y cells. (A, B) SH-SY5Y cells were divided into two groups underwent control group and siRNA, respectively. cDNA was harvested from these cells on the 4 h after transfecting different sequences of ZBTB38 siRNAs (siRNA1, siRNA2 and siRNA3 indicate three different siRNA primers). Representative images of these assays are shown in (A) and quantitative data are shown in (B); (C, D) cell lysates were collected for Western blot analysis. Representative images of these assays are shown in (C) and quantitative data are shown in (D), β-actin was used as an internal control. *p < 0.05; **p < 0.01.
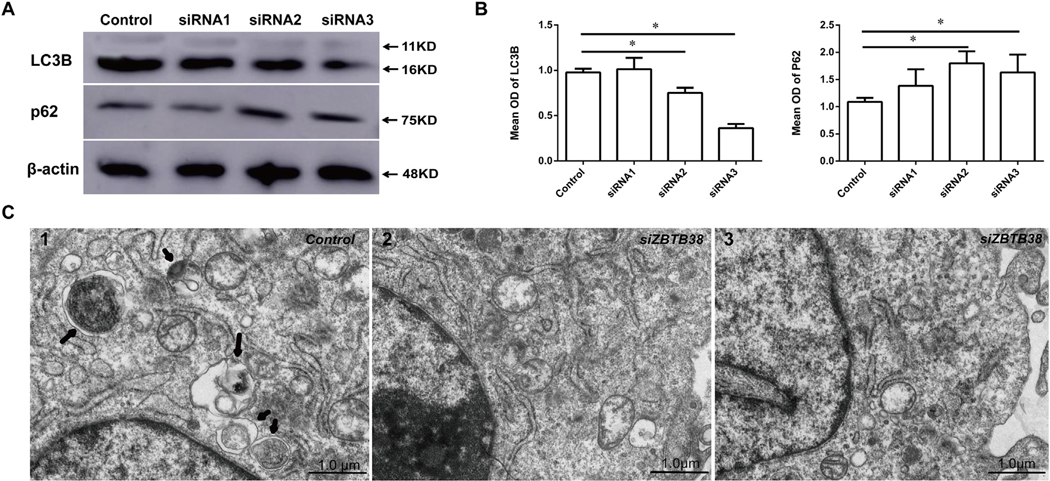
LC3B and P62 are biomarkers that reflect the level of autophagy. Western blot analysis showed that the LC3B expression level decreased in the ZBTB38 knockdown groups compared with the control group. The P62 expression level increased in the ZBTB38 knockdown groups compared with the control group. The differences were statistically significant (p < 0.05) (Fig. 2A and B).
Fig. 2.
Reduction of ZBTB38 by siRNA induces down-regulation of two autophagy-related genes in SH-SY5Y cells. (A) Expression of LC3B (LC3-I:11kD, LC3-II:16kD) and p62 (75 kD) were detected was detected by Western blotting with corresponding antibodies. β-actin were used as internal controls for Western blotting. SH-SY5Y cells were transfected with three ZBTB38 siRNA duplexes. (B) Mean OD of LC3B and p62 was compared by histogram. *p < 0.05. (C) Transmission electron microscopy showed that there were significantly less phagosomes present in the siZBTB38 SH-SY5Y cells (SH-SY5Y cells transfected with ZBTB38 siRNA), as compared to the control group treated with liposome alone. The black arrow shows the autophagosome.
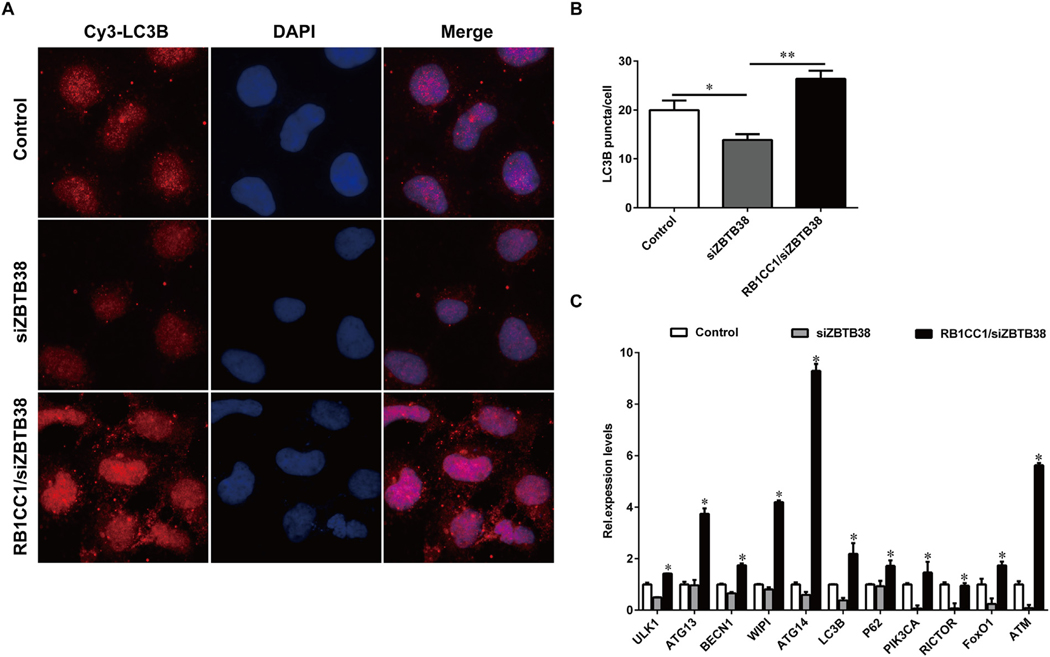
Transmission electron microscopy showed a few vacuolar structures surrounded by bilayer membranes (i.e., typical autophagosomes or autolysosome structures) in the cytoplasm of the ZBTB38 knockdown cells, suggesting that autophagosomes were not significantly induced by ZBTB38 knockdown (Fig. 2C). In addition, the autophagic membrane protein LC3B was labeled with Cy3 and the red fluorescence intensity of autophagosomes in each group was measured by laser confocal microscopy (Fig. 3A and B). The autophagic expression in ZBTB38 knockdown cells was significantly reduced compared with that in the control group treated with liposome alone (p < 0.05).
Fig. 3.
The RB1CC1 overexpression plasmid was transfected into SH-SY5Y cells treated with ZBTB38 siRNA and the autophagy was restarted. (A) Immunofluorescence assays showed ZBTB38 knockdown SH-SY5Y cells have a significantly reduced punctate pattern of LC3B staining (red), consistent with the formation of fewer autophagosomes. The LC3B immunofluorescence of SH-SY5Y cells transfected with RB1CC1 overexpression plasmid was stronger. DAPI (blue) was used for nuclear staining. (B) Quantification of the number of autophagosomes per cell. (C) Transfected cells were processed for qRT-PCR analysis. The mRNA levels of autophagic marker genes were quantified and normalized relative to GAPDH. *p < 0.05 and **p < 0.01 vs. control group treated with liposome alone. Data are presented as means ± SEM from at least 3 independent experiments. Control, SH-SY5Y cells treated with liposome alone; siZBTB38, SH-SY5Y cells transfected with ZBTB38 siRNA; RB1CC1/siZBTB38, siZBTB38 cells transfected with plasmid overexpressing RB1CC1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. ZBTB38 knockdown downregulates RB1CC1 expression in SH-SY5Y cells
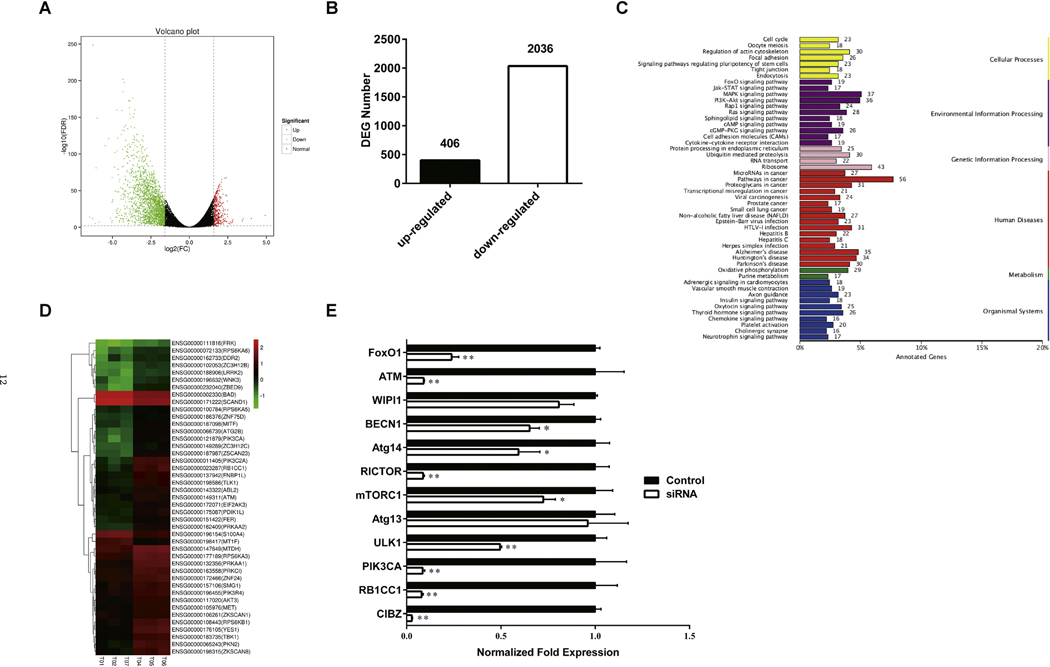
Using transcriptome high-throughput sequencing, three parallel experiments were repeated and 47.05 GB of clean data were obtained with 6.12 GB of clean data for each sample on average. Clean reads of each sample were aligned with the designated reference genome. Based on the comparison results, differentially expressed genes in the two groups of samples were easily identified with a volcano plot (Fig. 4A). The number of differentially expressed genes is summarized in Fig. 4B. KEGG annotation of differentially expressed genes was ranked according to pathway type in the KEGG database, and environmental information processing results showed that differentially expressed genes are mainly enriched in the MAPK and PI3K/Akt signaling pathways after ZBTB38 knockdown (Fig. 4C). Genes with similar expression patterns were clustered, and RB1CC1 expression was significantly decreased in SH-SY5Y cells in the siZBTB38 group (p < 0.01) (Fig. 4D and E).
Fig. 4.
ZBTB38 knockdown SH-SY5Y cells were analyzed by RNA-seq. (A) Distribution of the differentially expressed genes (DEGs) shown as a volcano plot. Each point represents a gene. The green dots represent the down-regulated differentially expressed genes, red dots represent the up-regulated differentially expressed genes, and black dots represent non-differentially expressed genes. (B) Statistical analysis of differentially expressed genes number. (C) DEG KEGG classification. The vertical axis lists the various metabolic pathways, and horizontal axis gives the percent of annotated genes in the pathways. (D) The heatmap of the subset DEGs in different samples (T04, T05, T06, T01, T02, T07). The significance of the difference in the gene expression between the patient groups were assessed using the t-test at p < 0.05 and Log2 fold change > 2 (LFC > 2) was set as the cut off. (E) RNA was isolated from the SH-SY5Y cells treated with Liposomes or ZBTB38 siRNA for 48 h and analyzed by qPCR. Values for each mRNA are normalized to GAPDH and are relative to levels in untreated wild-type cells (n = 3, error bars are SD). *p < 0.05 vs control group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To verify the reliability of RNA-Seq data, 12 genes closely related to autophagic synthesis were detected by qRT-PCR, and the results showed that the expression of these genes in the siZBTB38 group was significantly downregulated (p < 0.05). The differential expression of the RB1CC1, PIK3C2A, RICTOR, ATM, FoxO1, and ULK1 genes was highly significant (p < 0.01) (Fig. 4E).
3.3. Autophagy, cell proliferation, and cell viability after recovery of RB1CC1 expression
To determine whether restoring RB1CC1 expression can rescue autophagy-mediated death after ZBTB38 knockdown, an RB1CC1-overexpressed plasmid was transfected into siZBTB38 SH-SY5Y cells. The expression levels of all genes examined in this study (e.g., PIK3C2A, RICTOR, ATM, FoxO1, BECN1, ULK1, ATG13, and ATG14) sharply increased in the co-transfection group compared with the other two groups (Fig. 3C). ATG14 and ATM expression increased almost 10 times in the co-transfection group compared with the ZBTB38 knockdown groups (Fig. 3C).
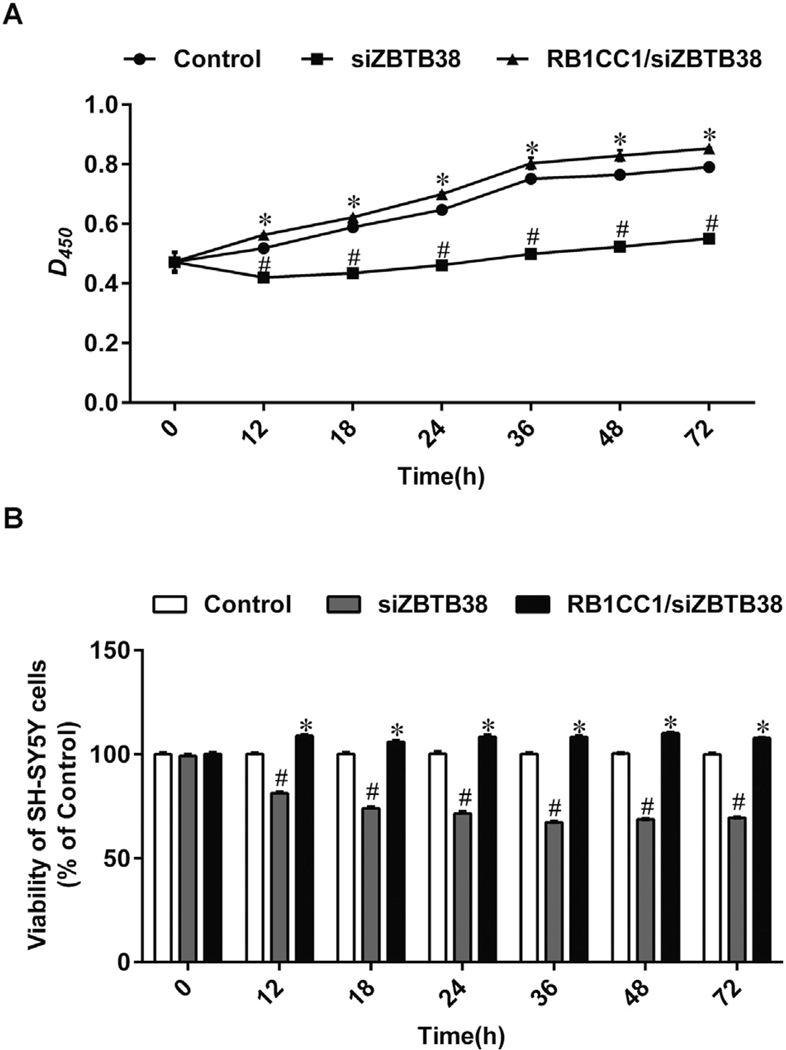
No significant difference in cell proliferation was observed among the initial phases of each group after culture by transient transfection (p > 0.05). From 12 to 72 h, cell proliferation in the RB1CC1/siZBTB38 group was significantly higher than that in the siZBTB38 SH-SY5Y group (p < 0.05). The siZBTB38 SH-SY5Y group showed significantly lower cell proliferation than the control group (p < 0.05) (Fig. 5A). These results indicate that ZBTB38 siRNA inhibits the proliferation of SH-SY5Y cells (Fig. 5B).
Fig. 5.
Proliferation and viability of ZBTB38 knockdown SH-SY5Y cells were promoted by RB1CC1. SH-SY5Y cell proliferation (A) and viability (B) in different groups. No significant difference was detected in each group initial stage after culture by transient transfections. SH-SY5Y cell proliferation and viability was significantly greater in the RB1CC1/siZBTB38 group than in siZBTB38 group from the 12 h to the 72 h after culture. The results also showed a significant difference in SH-SY5Y cell proliferation between control group and siZBTB38 group, *p < 0.05 compared with siZBTB38 group; #p < 0.05 compared with the control group. Data are presented as means ± SEM from at least 3 independent experiments. Control, SH-SY5Y cells treated with liposome alone; siZBTB38, SH-SY5Y cells transfected with ZBTB38 siRNA; RB1CC1/siZBTB38, siZBTB38 cells transfected with plasmid overexpressing RB1CC1.
3.4. ZBTB38 overexpression upregulates RB1CC1 expression and enhances autophagy in SCI mice
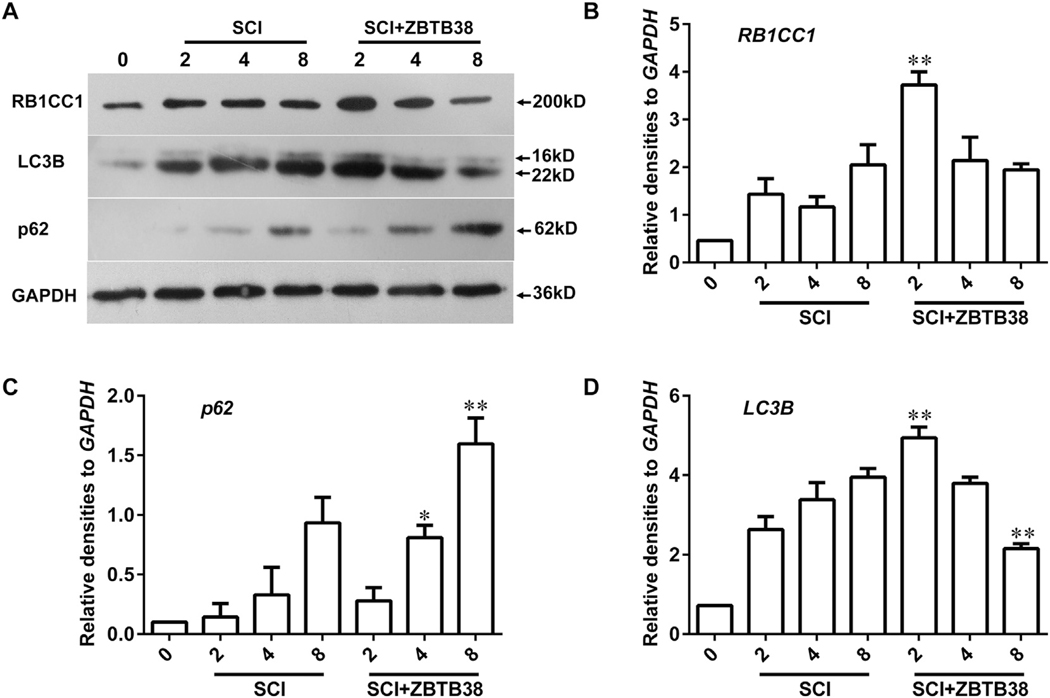
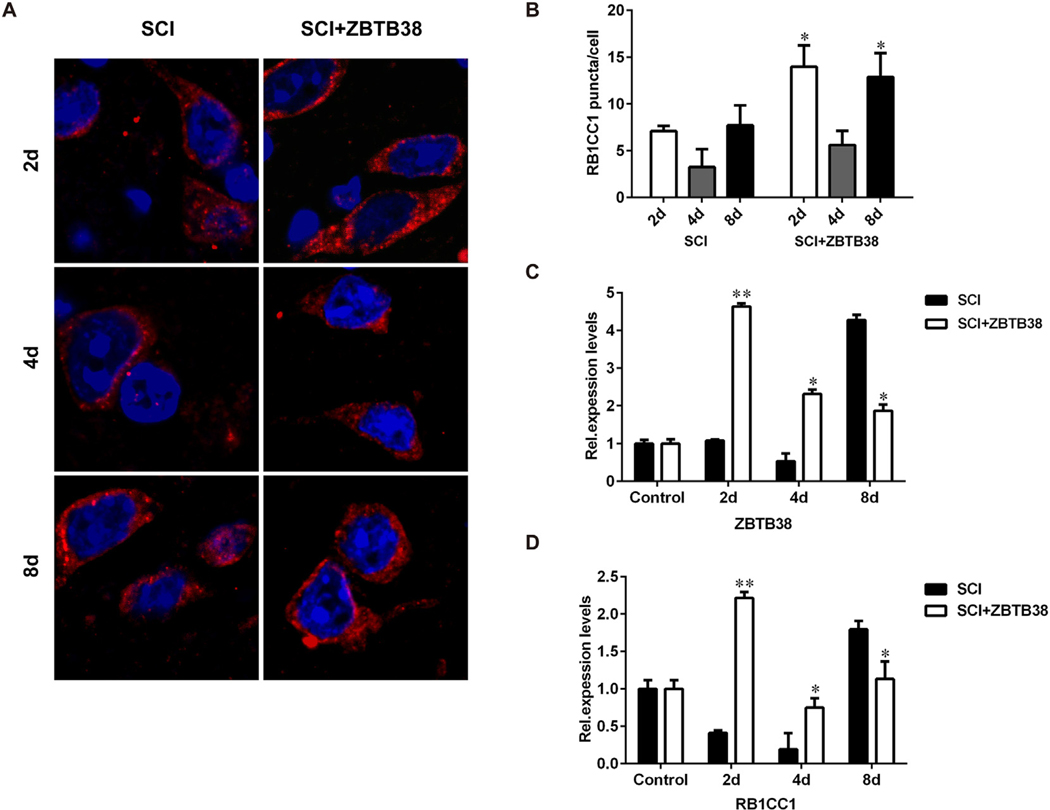
Exogenous ZBTB38 was stably expressed in the injured spinal cord of mice within 8 days after injection. No toxicity effects were observed in SCI mice after the lentivirus injection. Autophagic expression peaked 2 days after the lentivirus injection, and RB1CC1 and LC3B expression was significantly upregulated compared with that in the SCI control group (p < 0.05), but it decreased to a normal level on the eight day after the lentivirus injection P62 expression in the treatment group was significantly reduced compared with that in the SCI control group (Fig. 6). RB1CC1 reached its highest value on the second day and a normal level on the eight day (Fig. 7). Our data indicate that the synthesis of autophagosomes significantly increased with ZBTB38 overexpression and autophagy as programmed cell death promoted SCI repair.
Fig. 6.
Overexpression of ZBTB38 up-regulated the expression of RB1CC1 and promoted the occurrence of autophagy in vivo. (A) The spinal cord samples were harvested from the SCI mice with or without ZBTB38 lentivirus injection on days 0, 2, 4 and 8 post treatment (n = 10), and cell lysates were collected for Western blot analysis, and quantitative data are shown in (B, C, D). 0, healthy mice; SCI, Mice with spinal cord injury; SCI + ZBTB38, The constructed lentivirus containing full-length ZBTB38 cDNA was injected into the lesions of SCI mice.*p < 0.05; **p < 0.01, compared with SCI mice in the same period.
Fig. 7.
Expression of RB1CC1 is consistent with ZBTB38 in the ZBTB38 lentivirus-injected SCI mice. (A) The tissue sections from the same samples were used for immunofluorescence assays of RB1CC1 (red). DAPI (blue) was used for nuclear staining. Representative images of these assays are shown in (A) and quantitative data are shown in (B). (C, D) The RNA from the samples was collected for qRT-PCR analysis to determine the expression levels of ZBTB38 and RB1CC1 genes. Control, healthy mice; SCI, Mice with spinal cord injury; SCI + ZBTB38, The constructed lentivirus containing full-length ZBTB38 cDNA was injected into the lesions of SCI mice.*p < 0.05; **p < 0.01, compared with SCI mice in the same period. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Autophagy is an important defense and protection mechanism in eukaryotic cells. Damaged and degenerated proteins and non-functional organelles in the body are eliminated by enhanced autophagy, and cell recycling and reuse are ultimately promoted. Autophagy therefore plays an essential role in the processes of cell homeostasis, growth, and tumor formation and infection and has become a subject of great concern in recent years. Autophagy exerts a protective function on nerve cells during brain injury and neurodegenerative diseases, which partly prevents cell death (Chen et al., 2013; Hou et al., 2014; Sekiguchi et al., 2012).
We found that in the SCI model, early treatment with rapamycin (i.e., an enhanced autophagy inducer that inhibits mTOR) after injury can increase Beclin1 and LC3 expression in injured spinal cord tissue and reduce apoptosis, indicating that enhanced autophagy after SCI can protect nerve cells, reduce nerve tissue injury, and promote nerve function recovery (Chen et al., 2013; Hou et al., 2014; Sekiguchi et al., 2012; Walker et al., 2012; Zhang et al., 2013). Our previous study showed that endoplasmic reticulum stress-mediated apoptosis is effectively inhibited and SCI repair is promoted in model mice injected with a ZBTB38-expressing lentivirus (Cai et al., 2017).
The downregulation of ZBTB38 sharply decreased the autophagy gene expression levels (e.g., RB1CC1, PIK3C2A, RICTOR, ATM, FoxO1, and ULK1) in cultured SH-SY5Y cells. We further demonstrated that RB1CC1, a key gene for autophagy initiation, and LC3B II/I, the biomarkers for autophagosomes, significantly increased in SCI mice after injection of the ZBTB38 lentivirus, indicating that the restoration of ZBTB38 can promote autophagy and play an active role in the repair of secondary damage in traumatic SCI.
Previously, endoplasmic reticulum stress was induced to regulate cellular homeostasis in ZBTB38 knockdown SH-SY5Y cells (Cai et al., 2017). In the condition of endoplasmic reticulum stress, the unfolded protein response reduces protein production, increases the ability of the endoplasmic reticulum to treat proteins, and contributes to cell survival. When a large number of unfolded and/or misfolded proteins accumulate in the endoplasmic reticulum and the proteasome cannot be fully degraded, the unfolded protein response upregulates autophagy. Autophagy initiation also provides additional protection. However, in the present study, we found that autophagy in ZBTB38 knockdown SH-SY5Y cells was suppressed, and this conditional suppression was likely caused by the regulation of an autophagy-related gene.
The RB1CC1/FIP200 protein is a focal adhesion kinase family-interacting protein of 200 kD and is involved in the formation of the autophagy initiation complex (FIP200-ATG13-ULK1-ATG101) (Hara et al., 2008). KEGG pathway classification analysis of the differentially expressed genes identified by transcriptome sequencing showed that the differentially expressed genes after ZBTB38 knockdown were mainly enriched in the PI3K/Akt signaling pathway, a key pathway for autophagy. When the transcription factor ZBTB38 was knocked down, the PI3K/Akt signaling pathway was inhibited, resulting in significant downregulation of downstream RB1CC1.
Phosphorylation of the ULK (FIP200-ATG13-ULK1-ATG101) complex by downstream mTORC1 may inactivate the pathway and negatively regulate the formation of autophagosomes (Ganley et al., 2009; Kamada et al., 2010; Morselli et al., 2011). When amino acids are deficient or mTORC1 is inhibited, ULK1 is activated to interact with RB1CC1, and ATG13 is recruited to strengthen the interaction. The ULK complex is then formed. ULK1 enhances the activity of class III phosphoinositide 3-kinases by phosphorylating the Ser14 in BECN 1 (i.e., the homologue of yeast ATG6) (Russell et al., 2013). The activated ULK complex along with the PI3K complex targets autophagy-specific pools of phosphatidyl inositol-3-phosphoric acid (PtdIns3P) generated after being recruited to autophagy initiation sites (Chen et al., 2014). These processes facilitate the nucleation of isolated membranes and the recruitment of other ATG proteins and autophagy-specific PtdIns3P effectors, such as the WD-repeat domain phosphoinositide-interacting (WIPI) protein (Russell et al., 2013; Chen et al., 2014; Simonsen and Tooze, 2009).
PtdIns3P interacts with the WIPI protein and works synergistically with ATG2 to participate in autophagic nucleation. Gene expression in the downstream mTOR signaling pathway (e.g., ULK1, ATG13, RB1CC1, BECN1, and WIPI expression) was significantly downregulated in ZBTB38 knockdown SH-SY5Y cells. Our results indicate that the autophagy regulation mechanism of the mTOR signaling pathway was also inhibited, which further proves that ZBTB38 gene deficiency can block autophagy. ZBTB38 can therefore be used as a novel regulator of autophagy initiation targeted by RB1CC1.
The expression of downstream genes closely related to RB1CC1 was significantly upregulated in ZBTB38 knockdown SH-SY5Y cells transfected with RB1CC1-overexpressing plasmid. The intensity of LC3B was higher than that in the other two groups in the immunofluorescence staining assay, indicating that autophagy in ZBTB38 knockdown SH-SY5Y cells was reinitiated, and its proliferation and viability also markedly increased.
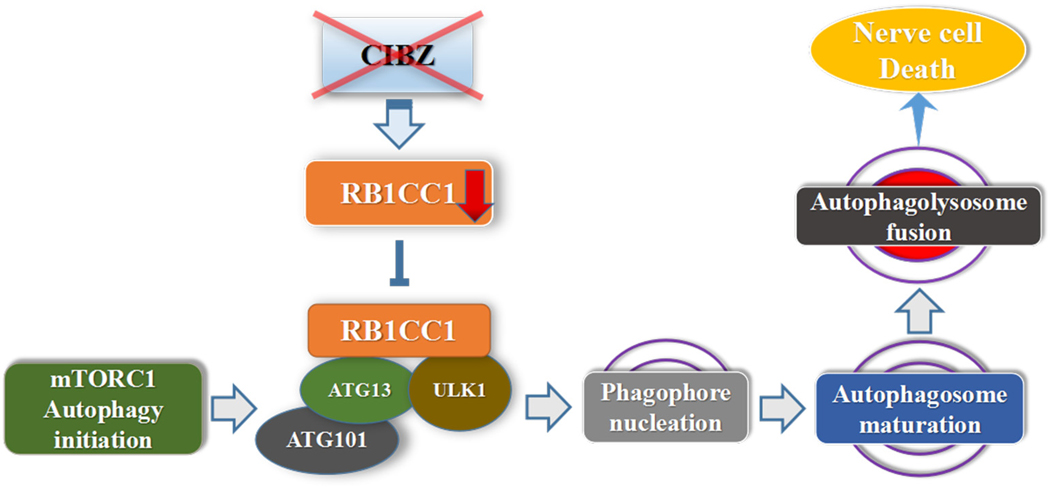
In conclusion, we believe that ZBTB38 can serve as a positive regulator for autophagy initiation and the outcome of this regulation can be negatively regulated by RB1CC1. When ZBTB38 is knocked down, RB1CC1 is intensely downregulated and triggers the switch to secondary SCI. This causes the inhibition of the initial stage of cell autophagosome biosynthesis, eventually blocking autophagy. Nerve cells lose their autophagy repair mechanism, accelerating the apoptosis of nerve cells (Fig. 8). Autophagy-related RB1CC1/FIP200 may be another novel target for SCI, which could provide new strategies for the treatment of SCI.
Fig. 8.
The model depicting the relationship between CIBZ, RB1CC1 and autophagy. RB1CC1, the key mediator of mTORC1 signaling to autophagy, regulates early initial stages of autophagosome formation in response to starvation or mTORC1 inhibition, and forms a multiprotein complex by interacting with ATG13, ULK1 and ATG101. While knockdown of ZBTB38 decrease the RB1CC1 level, inhibit initiation of the mTORC1 autophagy cascade, the autophagy pathway was blocked, and finally induces the death of neural cells.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (NSFC31372207 and 81570094), the Innovation Team of Scientific Research Platform in Anhui Province, and a start-up grant from Nanjing Agricultural University (804090).
Abbreviations:
- ATG2
autophagy-related protein 2
- ATG13
autophagy-related protein 13
- ATG14
autophagy-related protein 14
- ATG101
autophagy-related protein 101
- ATM
ataxia telangiectasia-mutated gene
- BECN1
the homologue of yeast autophagy-related protein 6
- CCK-8
cell counting kit-8
- Cy3
Alexa Fluor fluorescent dye
- DEGs
differently expressed genes
- DEGseq
differential expression gene sequence
- FoxO1
forkhead box protein O1
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KOBAS
KEGG Orthology Based Annotation System
- LC3B
microtubule-associated proteins 1A/1B light chain 3B
- RB1
retinoblestoma 1
- RB1CC1/FIP200
the focal adhesion kinase family-interacting protein of 200 kDa
- RICTOR
rapamycin-insensitive companion of mTOR
- MAPK
mitogen-activated protein kinase
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- PI3K/Akt
class III phosphoinositide 3-kinase/Protein Kinase B
- P62
sequestosome 1
- PIK3C2A
phosphoinositide-3-Kinase Class-2-Alpha Polypeptide
- qRT-PCR
quantitative real-time polymerase chain reaction
- SCI
spinal cord injury
- SH-SY5Y
human neuroblastoma cells
- siRNA
Small interfering RNA
- Th7
seventh
- Th9
ninth
- ULK1
unc-51 like autophagy activating kinase 1
- WIPI
WD repeat domain phosphoinositide-interacting protein
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2018.07.073.
Declaration of interest
The authors declare no competing financial interests.
Compliance with ethical standards
All animal experiments complied with the ARRIVE guidelines and were carried out according to the National Institutes of Health guide for the care and use of laboratory animals (NIH Publications No. 8023, revised 1978). All animal experiments were approved by the Anhui Normal University Academic Ethics Committee.
References
- Abbi S, Ueda H, Zheng C, et al. , 2002. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol. Biol. Cell 13, 3178–3191. 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Fan R, Hua T, Liu H, Li J, 2011. Nimodipine alleviates apoptosis-mediated impairments through the mitochondrial pathway after spinal cord injury. Curr. Zool 57, 340–349. 10.1093/czoolo/57.3.340. [DOI] [Google Scholar]
- Cai Y, Li J, Yang S, Li P, Zhang X, Liu H, 2012. CIBZ, a novel BTB domain-containing protein, is involved in mouse spinal cord injury via mitochondrial pathway independent of p53 gene. PLoS One 7, e33156 10.1371/journal.pone.0033156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Li J, Zhang Z, et al. , 2017. ZBTB38 is a novel target for spinal cord injury. Oncotarget 8, 45356–45366. 10.18632/oncotarget.17487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Fong TH, Hsu PW, Chiu WT, 2013. Multifaceted effects of rapamycin on functional recovery after spinal cord injury in rats through autophagy promotion, anti-inflammation, and neuroprotection. J. Surg. Res 179, e203–e210. 10.1371/journal.pone.0030012. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu XR, Yin YQ, et al. , 2014. Unravelling the multifaceted roles of Atg proteins to improve cancer therapy. Cell Prolif. 47, 105–112. 10.1111/cpr.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan JC, Fehlings MG, 2008. Cardiovascular complications after acute spinal cord injury: pathophysiology, diagnosis, and management. Neurosurg. Focus 25, E13 10.3171/FOC.2008.25.11.E13. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X, 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem 284, 12297–12305. 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, et al. , 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol 181, 497–510. 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Zhang L, Zhang L, et al. , 2014. Acute spinal cord injury in rats induces autophagy activation. Turk. Neurosurg 24, 369–373. 10.5137/1019-5149.JTN.8623-13.0. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, et al. , 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol 30, 1049–1058. 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, et al. , 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S, 2004. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 11, 448–457. 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD, 2000. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D, 2012. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 8, 165–176. 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L, 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Melkoumian ZK, Peng X, Gan B, Wu X, Guan JL, 2005. Mechanism of cell cycle regulation by FIP200 in human breast cancer cells. Cancer Res. 65, 6676–6684. 10.1158/0008-5472.CAN-04-4142. [DOI] [PubMed] [Google Scholar]
- Menghini R, Casagrande V, Marino A, et al. , 2014. MiR-216a: a link between endothelial dysfunction and autophagy. Cell Death Dis. 5, e1029 10.1038/cddis.2013.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova O, Stratton Y, Hall D, et al. , 2012. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell 21, 532–546. 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Shen S, Ruckenstuhl C, et al. , 2011. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle 10, 2763–2769. 10.4161/cc.10.16.16868. [DOI] [PubMed] [Google Scholar]
- Mountney A, Zahner MR, Lorenzini I, Oudega M, Schramm LP, Schnaar RL, 2010. Sialidase enhances recovery from spinal cord contusion injury. Proc. Natl. Acad. Sci. U. S. A 107, 11561–11566. 10.1073/pnas.1006683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Okano H, 2013. Cell transplantation therapies for spinal cord injury focusing on induced pluripotent stem cells. Cell Res. 23, 70–80. 10.1038/cr.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa Y, Omori R, Nishii T, Ishida Y, Kawaichi M, Matsuda E, 2011. The methyl-CpG-binding protein CIBZ suppresses myogenic differentiation by directly inhibiting myogenin expression. Cell Res. 21, 1578–1590. 10.1038/cr.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penas C, Guzmán MS, Verdú E, Forés J, Navarro X, Casas C, 2007. Spinal cord injury induces endoplasmic reticulum stress with different cell-type dependent response. J. Neurochem 102, 1242–1255. 10.1111/j.1471-4159.2007.04671.x. [DOI] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, et al. , 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol 15, 741–750. 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Matsuda E, Sarashina E, Ishida Y, Kawaichi M, 2005. Identification of a novel BTB-zinc finger transcriptional repressor, CIBZ, that interacts with CtBP corepressor. Genes Cells 10, 871–885. 10.1111/j.1365-2443.2005.00885.x. [DOI] [PubMed] [Google Scholar]
- Sekiguchi A, Kanno H, Ozawa H, Yamaya S, Itoi E, 2012. Rapamycin promotes autophagy and reduces neural tissue damage and locomotor impairment after spinal cord injury in mice. J. Neurotrauma 29, 946–956. 10.1089/neu.2011.1919. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA, 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol 186, 773–782. 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG, 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y, 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, Gupta SK, Fiedler J, et al. , 2012. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun 3, 1078 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CL, Walker MJ, Liu NK, et al. , 2012. Systemic bisperoxovanadium activates Akt/mTOR, reduces autophagy, and enhances recovery following cervical spinal cord injury. PLoS One 7, e30012 10.1371/journal.pone.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Olman MA, Stewart J Jr., et al. , 2011. Downregulation of FIP200 induces apoptosis of glioblastoma cells and microvascular endothelial cells by enhancing Pyk2 activity. PLoS One 6, e19629 10.1371/journal.pone.0019629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhang J, Zhang L, et al. , 2013a. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 145, 1133–1143. 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang C, Hong Z, Chen H, Chen W, Chen G, 2013b. C/EBP homologous protein (CHOP) mediates neuronal apoptosis in rats with spinal cord injury. Exp. Ther. Med 5, 107–111. 10.3892/etm.2012.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Gan B, Wu X, Guan JL, 2009. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J. Biol. Chem 284, 6004–6013. 10.1074/jbc.M806375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KT, El Masri W, Osman A, Chowdhury J, Johnson WE, 2011. Concise review: bone marrow for the treatment of spinal cord injury: mechanisms and clinical applications. Stem Cells 29, 169–178. 10.1002/stem.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ, 2010. Eaten alive: a history of macroautophagy. Nat. Cell Biol 12, 814–822. 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A, 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Wang ZG, Wu FZ, et al. , 2013. Regulation of autophagy and ubiquitinated protein accumulation by bFGF promotes functional recovery and neural protection in a rat model of spinal cord injury. Mol. Neurobiol 48, 452–464. 10.1007/s12035-013-8432-8. [DOI] [PubMed] [Google Scholar]
- Zurita M, Aguayo C, Bonilla C, et al. , 2012. The pig model of chronic paraplegia: a challenge for experimental studies in spinal cord injury. Prog. Neurobiol 97, 288–303. 10.1016/j.pneurobio.2012.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.