ABSTRACT
Autophagy is primarily an efficient intracellular catabolic pathway used for degradation of abnormal cellular protein aggregates and damaged organelles. Although autophagy was initially proposed to be a cellular stress responder, increasing evidence suggests that it carries out normal physiological roles in multiple biological processes. To date, autophagy has been identified in most organs and at many different developmental stages, indicating that it is not only essential for cellular homeostasis and renovation, but is also important for organ development. Herein, we summarize our current understanding of the functions of autophagy (which here refers to macroautophagy) in the mammalian life cycle.
Keywords: autophagy, development, life cycle, mammal, physiological roles
I. INTRODUCTION
Autophagy is a catabolic process that is essential for cellular homeostasis and renovation through the elimination of large cytoplasmic components, such as abnormal protein aggregates and damaged organelles, via lysosomal degradation. The term ‘autophagy’ was first coined in 1963 by Christian de Duve, the discoverer of lysosomes. For a long time subsequently, research into autophagy was limited to morphological descriptions, due to the lack of an effective system for genetic analysis (Ohsumi, 2014). In the 1990s, yeast genetic screening was employed to identify the molecules regulating autophagy, and this led to the breakthrough discovery of a series of autophagy‐related (Atg) genes and their signal transduction pathways, which are critical for the induction and maintenance of autophagy (Tsukada & Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1995).
Autophagy has both anti‐stress and physiological functions. On one hand, autophagy generates new building blocks and energy for anabolism upon nutrient deprivation or other stress by degradation of intracellular components, on the other hand, autophagy participates in cell homeostasis regulation and disease prevention by degrading damaged organelles or harmful components under normal conditions. Numerous studies have demonstrated that, under physiological conditions, autophagy takes place in a variety of tissues and organs, and at distinct developmental stages. These results indicate that autophagy is indispensable in the development and functional maintenance of normal tissues. Accordingly, the physiological roles of autophagy can be divided into two aspects: promoting cell remodelling during morphogenesis, and maintaining cellular homeostasis after maturation. As a result, autophagy has an important function in protection from various diseases, such as neurodegeneration, liver disease, and cancer (Kundu & Thompson, 2008; Deretic, Saitoh & Akira, 2013; Santana‐Codina, Mancias & Kimmelman, 2017).
In this review, we firstly outline the molecular mechanisms of autophagy, then summarize progress in understanding basal autophagy in mammals. We also attempt to delineate the physiological roles of autophagy throughout the mammalian life cycle.
II. THE MOLECULAR MECHANISMS OF AUTOPHAGY IN MAMMALS
In mammalian cells, the three primary types of autophagy are defined as macroautophagy, microautophagy, and chaperone‐mediated autophagy. Macroautophagy, the classic type of autophagy, is the most widely studied. Its distinctive feature is the formation of double‐membrane vesicles (autophagosomes), which contain substrates for degradation and eventually fuse with lysosomes. Autophagy can be categorized as non‐selective or selective. Non‐selective macroautophagy randomly engulfs cytoplasmic components into autophagosomes for lysosomal degradation. Selective macroautophagy, however, specifically recognizes and degrades a particular cargo, either a protein complex or an organelle such as a mitochondrion. Mitophagy, which can occur by either macro‐ or microautophagy, refers to selective removal of superfluous or damaged mitochondria with the help of mitochondrial autophagy receptors. Herein, we summarize the roles of macroautophagy (referred as autophagy hereafter) in the mammalian life cycle.
An important function of autophagy is to degrade long‐lived proteins. This process is regulated by a series of protein complexes (Fig. 1). These complexes, which are made up of distinct sets of ATG proteins, form the core autophagy machinery and are responsible for autophagy induction, autophagosome formation, autophagosome trafficking, and fusion of autophagosomes with lysosomes (Fig. 2).
Figure 1.
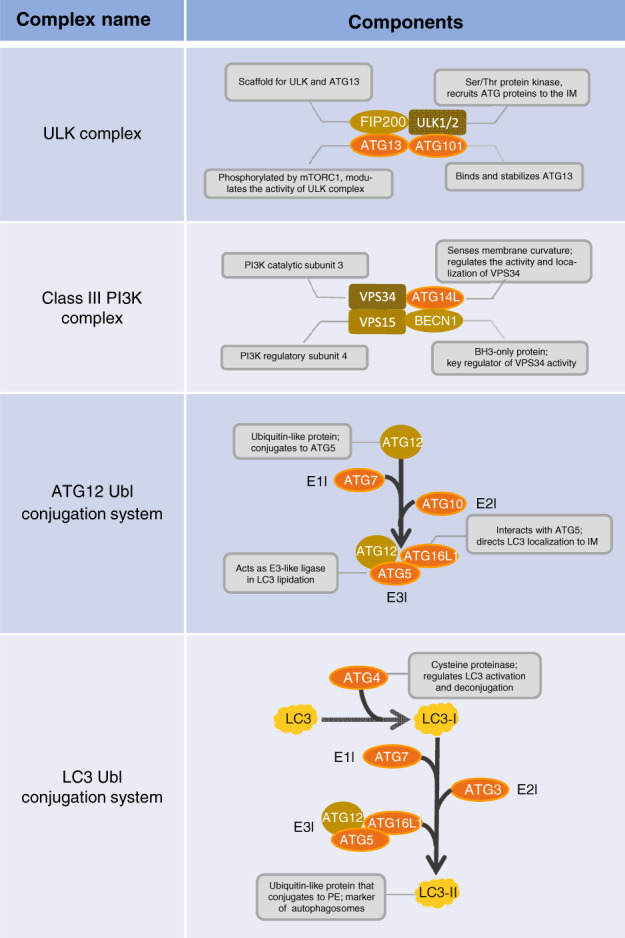
The major complexes and conjugating systems of autophagy in mammals. The Unc‐51‐like kinase (ULK) complex, composed of ULK1/2, the focal adhesion kinase family‐interacting protein of 200 kD (FIP200), autophagy related 13 (ATG13) and ATG101, is crucial for autophagy induction. Class III phosphatidylinositol 3‐kinase (PI3K) complex contains an important catalytic subunit, vacuolar protein sorting 34 (VPS34), which is recruited by the ULK complex and is essential for initiating autophagosome assembly. The ATG12 and microtubule associated protein 1 light chain 3 (LC3) systems are two ubiquitin‐like (Ubl) conjugation systems. Their activation requires different ubiquitin‐activating enzyme (E1), ubiquitin‐conjugating enzyme (E2) and ubiquitin ligase enzyme (E3)‐like enzymes, and they function in autophagosome expansion and maturation. BECN1, beclin 1; BH3, BCL2 homology region 3; IM, isolation membrane; mTORC1, mammalian target of rapamycin complex 1; PE, phosphatidylethanolamine.
Figure 2.
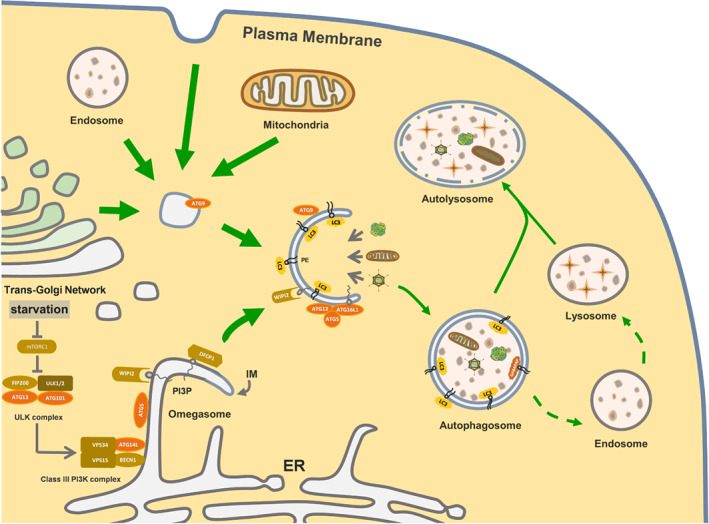
The molecular mechanism of autophagy in mammals. Starvation or other triggers cause inhibition of the mammalian target of rapamycin complex 1 (mTORC1), leading to activation of the Unc‐51‐like kinase (ULK1) complex. This complex then modulates the translocation of Class III phosphatidylinositol 3‐kinase (PI3K) complex to the endoplasmic reticulum (ER), which facilitates nucleation and assembly of the isolation membrane (IM). Autophagy related 9 (ATG9) can carry membranes from other components to promote IM expansion, which plays an important role in the early stages of autophagosome formation. In addition, two ubiquitin‐like conjugation systems, the microtubule associated protein 1 light chain 3 (LC3) system and the autophagy related 12 (ATG12) system, are critical for IM elongation and closure. After the autophagosome forms, it fuses with lysosomes or endosomes to eventually form an autolysosome. The contents of the autophagosome are then degraded and recycled to the cytoplasm. BECN1, beclin 1; DFCP1, double FYVE domain‐containing protein 1; FIP200, the focal adhesion kinase family‐interacting protein of 200 kD; PE, phosphatidylethanolamine; PI3P, phosphatidylinositol 3‐phosphate; VPS34, vacuolar protein sorting 34; WIPI2, WD repeat domain, phosphoinositide interacting 2.
(1). Autophagy induction
Autophagy is induced by many stimuli, including nutrient deficiency (e.g. amino acids, glucose), withdrawal of growth factors (e.g. insulin and insulin‐like growth factors), reduced cellular energy levels (e.g. ATP), extra‐ or intracellular stress [e.g. endoplasmic reticulum (ER) stress, hypoxia, oxidative stress], and pathogenic infections (He & Klionsky, 2009). Amino acid or growth factor deprivation are the most effective stimuli inducing autophagy, and both are linked to the mammalian target of rapamycin (TOR) complex 1 (mTORC1) pathway (Sengupta, Peterson & Sabatini, 2010). TOR is an important serine/threonine protein kinase involved in regulating cellular metabolism by sensing nutrient signals. In nutrient‐sufficient conditions, mTORC1 phosphorylates Unc‐51‐like kinase 1 (ULK1) and ATG13 (Hosokawa et al., 2009; Kim et al., 2011), which are both components of the ULK1 complex, to prevent autophagy initiation. Once the activity of mTORC1 is inhibited by starvation, rapid dephosphorylation of ATG13 may promote the formation of a stable complex between ATG13 and ULK1, focal adhesion kinase family‐interacting protein of 200 kD (FIP200) and ATG101 (Fig. 2). ULK1, the only protein kinase among the core ATG proteins, is crucial for modifying and recruiting downstream ATG proteins, including ATG13 and FIP200, to trigger autophagy (Jung et al., 2009).
Interestingly, recent studies have shown that mTOR and 5'‐AMP‐activated protein kinase (AMPK) coordinate to regulate cellular nutrient and energy signals to maintain cellular homeostasis through phosphorylation of ULK1 at distinct serine residues (Kim et al., 2011). Under conditions of cellular energy deficiency, AMPK phosphorylates ULK1 at Ser (555) to disrupt the interaction between mTOR and ULK1, resulting in activation of autophagy. Accordingly constitutive phosphorylation of ULK1 by AMPK, but not mTOR, has been shown to protect normal autophagy flux in embryonic stem cells (ESCs) to maintain their stemness properties (Gong et al., 2018).
(2). Autophagosome formation
Upon ULK1 complex formation, the class III phosphatidylinositol 3‐kinase (PI3K) complex I is recruited to the isolation membrane (IM), which is the autophagosomal precursor that functions in the initial sequestering of cargos. PI3K complex I is required for IM nucleation and assembly. Its main component vacuolar protein sorting 34 (VPS34), the catalytic subunit of the PI3K complex, is recruited by ULK1 and produces phosphatidylinositol 3‐phosphate (PI3P) at initiation sites. PI3P is critical for autophagosome formation and is considered a marker of autophagosome membranes. Although the origin of the IM is still controversial, several lines of evidence indicate that the ER provides the membrane to initiate IM formation in mammals. Evidence for this is the presence of a PI3P‐rich area in the ER membrane with an Ω‐like shape, termed the omegasome (Mizushima, Yoshimori & Ohsumi, 2011). The omegasome is also the location of the PI3P downstream effector double‐FYVE‐containing protein 1 (DFCP1), another marker of IM initiation sites (Axe et al., 2008; Hayashi‐Nishino et al., 2009). Morphological observations also reveal that most IM initiation sites are located in the close vicinity of the ER microdomains (Hayashi‐Nishino et al., 2009; Yla‐Anttila et al., 2009; Uemura et al., 2014). After the ULK1 and PI3K complexes become localized to the ER‐derived omegasomes, other ATG proteins are recruited and facilitate IM expansion (Fig. 2).
In addition to recruiting the PI3K complex, the ULK1 complex can also recruit ATG9 to the IM. ATG9, the only transmembrane protein among the core ATG proteins, shuttles between the IM and other membrane components (at least the trans‐Golgi network, plasma membrane and endosomes) in the form of ATG9 vesicles (Fig. 2). Its transport seems dependent on WIPI2 (a homolog of yeast ATG18) and the WIPI2‐interacting protein ATG2 (Lu et al., 2011; Orsi et al., 2012). Therefore, ATG9 vesicles are considered to be carriers of membranes to promote IM expansion. The mitochondrial membrane has also been shown to be a major contributor to expansion (Hailey et al., 2010).
During the process of expansion, two ubiquitin‐like (Ubl) conjugation systems are essential: the microtubule associated protein 1 light chain 3 (LC3) system and the ATG12 system (Figs 1 and 2). Both LC3 and ATG12 are Ubl proteins. ATG12 is successively activated by ubiquitin‐activating enzyme (E1)‐like enzyme ATG7 and ubiquitin‐conjugating enzyme (E2)‐like enzyme ATG10, then conjugated to ATG5 to form an ATG12–ATG5–ATG16L1 complex on the IM (Fig. 2). Recent studies have shown that recruitment of ATG12–ATG5–ATG16L1 requires WIPI2 to link LC3 conjugation (see below) with PI3P at the site of autophagosome formation (Dooley et al., 2014), This suggests that the ATG18/WIPI family has an important role in assembling the conjugation system.
Before undergoing conjugation, the LC3 precursor must be cleaved by the cysteine protease ATG4 to reveal the C‐terminal glycine. The product, cytosolic LC3‐I, is then activated by E1‐like enzyme ATG7 and E2‐like enzyme ATG3, and finally conjugated to phosphatidylethanolamine (PE) to yield the membrane‐associated LC3‐PE (LC3‐II) (Kabeya et al., 2004). Unlike ATG12 conjugation, the last step in ATG3 conjugation requires an ubiquitin ligase enzyme (E3)‐ligase, ATG12–ATG5–ATG16L1 complex (Hanada et al., 2007). Studies have revealed that ATG12 only localizes on the IM, whereas LC3‐II is found on the autophagosomal membrane throughout the whole process of autophagosome formation and tethering/fusing with lysosomes (Nakatogawa, Ichimura & Ohsumi, 2007).
(3). Fusion of the autophagosome with lysosomes
The precise mechanism of autophagosome–lysosome fusion is still elusive. Three intracellular membrane‐trafficking‐associated protein families, RAS‐related GTP‐binding protein (RAB) GTPases, membrane‐tethering complexes, and soluble‐N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNARE) proteins, are known to be involved in this process (Bento et al., 2016). In addition, evidence has shown that the microtubule system is essential for trafficking mature autophagosomes from random initiation sites towards the perinuclear area (Jahreiss, Menzies & Rubinsztein, 2008), where they fuse with endosomes or lysosomes. Furthermore, the class III PI3K complex II, in which ATG14L is replaced by UV irradiation resistance‐associated gene (UVRAG), acts in regulating the trafficking of mature autophagosomes to lysosomes (Bento et al., 2016).
In mammals, as well as directly fusing with lysosomes, autophagosomes can first fuse with endosomes and then traffic to lysosomes to form autolysosomes (Berg et al., 1998). Upon autolysosome formation, the cargos are degraded by lysosome‐derived hydrolases, then recycled to the cytoplasm to participate in the next round of biosynthesis or energy generation.
III. THE ROLES OF AUTOPHAGY IN THE MAMMALIAN LIFE CYCLE
As an evolutionarily conserved process, autophagy exists in all eukaryotic organisms, from unicellular yeasts to higher mammals. In lower organisms, such as Saccharomyces cerevisiae, Dictyostelium discoideum, and Caenorhabditis elegans, it is comparatively easy to study the functional involvement of autophagy in developmental processes. For instance, genetic‐screening studies have shown that yeast strains with Atg gene mutations are unable to differentiate into spores under nutrient‐limiting conditions (Tsukada & Ohsumi, 1993). In C. elegans, mitochondrial accumulation was observed to be associated with a decline in mitophagy during aging, indicating that mitophagy is crucial in anti‐aging (Palikaras, Lionaki & Tavernarakis, 2015). However, in mammals, the roles of autophagy are more complicated and relatively hard to delineate. Nevertheless, the physiological roles of autophagy in mammals during different development stages are gradually being uncovered, due to the availability of model systems with conditional loss or gain of autophagy function through modification of Atg genes (Fig. 3).
Figure 3.
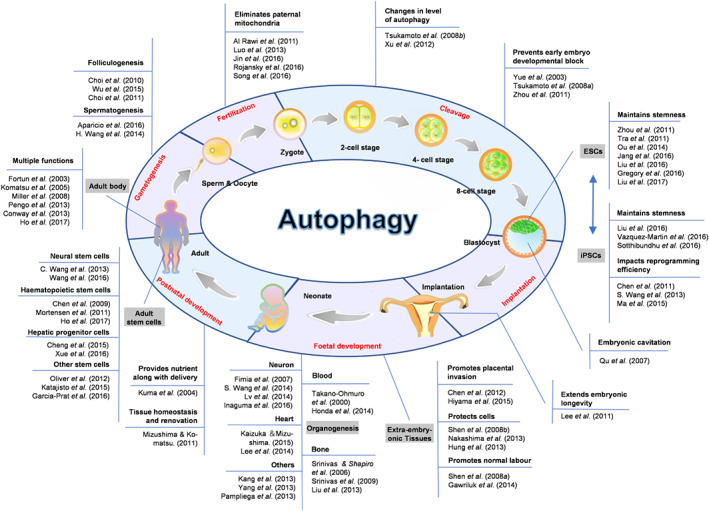
Autophagy plays essential roles during the mammalian life cycle. Representative research publications documenting the physiological roles of autophagy are summarized, according to development stage. These studies span from fertilization, cleavage, implantation, foetal development and postnatal development, to the next generation of gametogenesis, indicating that autophagy has important functions during the whole mammalian life cycle. ESC, embryonic stem cell; iPSC, induced pluripotent stem cell.
(1). Autophagy in the development of preimplantation embryos
Among the characteristics of mammals, viviparity and lactation endow their offspring with a high survival rate. The associated developmental features may be reflected in the functions of autophagy. Below, we discuss the roles of autophagy in the development of mammalian early embryos.
(a). Autophagy in fertilization
Embryonic development begins with the fertilization of the egg by sperm. In mice, autophagy is triggered by calcium oscillation during sperm–oocyte fusion rather than at the ovulation stage (Tsukamoto et al., 2008b). During fertilization and early embryonic development, certain intracellular materials derived from oocytes and sperm must be degraded (Stitzel & Seydoux, 2007). For example, mitochondria in sperm are degraded during fertilization, and this mitochondrial degradation is crucial for maternal cytoplasmic inheritance (a universal principle in animals, in which the offspring inherit only maternal cytoplasm). The elimination of sperm‐borne mitochondria was originally thought to occur through the autophagy pathway because autophagy markers, such as LC3, were detected around the paternal mitochondria (Al Rawi et al., 2011). However, later research suggested that LC3 occurs only transiently and detaches from mitochondria before they are degraded. Furthermore, some undegraded paternal mitochondria are unevenly distributed in blastomeres and converge in several cells (Luo et al., 2013; Jin et al., 2016). Although controversy still exists, some recent studies provide clues about this process. Rojansky, Cha & Chan (2016) carried out gain‐ or loss‐of‐function assays on two non‐lethal mitophagy‐related E3 ubiquitin ligases, parkin RBR E3 ubiquitin protein ligase (PARKIN) and mitochondrial E3 ubiquitin protein ligase 1 (MUL1), revealing that mitophagy is required for elimination of paternal mitochondria. Song et al. (2016) showed that elimination of paternal mitochondria requires both sequestosome 1 (p62/SQSTM1)‐dependent autophagy and the valosin containing protein (VCP)‐associated ubiquitin–proteasome system. Further studies are required to delineate the exact function of autophagy in this process.
(b). Autophagy in cleavage
After fertilization, the level of autophagy in mice is high between the one‐ and the four‐cell stage, although there is a transient depression from the late one‐cell stage to the middle of the two‐cell stage. This transient depression might be to protect newly synthesized nuclear factors that are necessary for reprogramming (Egli et al., 2007; Tsukamoto, Kuma & Mizushima, 2008a). The autophagy level then declines after the four‐cell stage. Unlike in mice, autophagy in porcine parthenotes initiates at the one‐cell stage and then gradually declines through the two‐cell stage to the blastocyst stage (Xu et al., 2012). This implies that autophagy levels are not identical in different species.
Studies have revealed an essential role of ATG proteins in early embryonic development. Mouse embryos generated by fusing Atg5‐null oocytes with Atg5‐null sperm arrest at the four‐ to eight‐cell stage and cannot reach the blastocyst stage (Tsukamoto et al., 2008b). Evidence from protein synthesis analysis suggests that new protein synthesis is blocked by Atg5 deficiency because autophagy is impaired and cannot degrade old proteins to provide amino acid substrates (Tsukamoto et al., 2008b). In support of these findings, beclin1 −/− and vps34 −/− mouse embryos are also unable to accomplish embryogenesis (Yue et al., 2003; Zhou, Takatoh & Wang, 2011). Together, these findings suggest that autophagy prevents a growth block during early embryonic development.
(c). Autophagy in embryonic stem cells
At the blastocyst stage, embryonic cells undergo preliminary differentiation into two types: trophoblast cells and inner cell mass (ICM) cells. Additionally, a cavity appears in the trophoblast, called the blastocoel. Its formation is thought to be associated, at least partially, with autophagic cell death (or autosis, a type of programmed cell death). Deficiency of Atg genes, such as Atg5 or Beclin1, leads to cell corpse accumulation in embryoid bodies (EBs) and failure to cavitate (Qu et al., 2007). Such defects might be due to a shortage of ‘eat‐me’ signals (phosphatidylserine exposure) and ‘come‐get‐me’ signals (lysophosphatidylcholine). These results suggest that there is a relationship between autophagy and apoptosis, in accordance with the findings of Yousefi et al. (2006) that ATG5 can promote apoptosis.
Of the two constituents of the blastocyst, the trophoblast is destined to become the placenta, while the ICM will develop into the foetus. The fact that the ICM forms all the foetal cells and tissues indicates its ability for pluripotency. ICM cells can be isolated and cultured in vitro as a pluripotent stem cell line, named embryonic stem cells (ESCs). ESCs represent an ideal model for the study of embryonic development in vitro. By establishing human ESC lines with stable green fluorescent protein (GFP)‐LC3 expression, Tra et al. (2011) observed that autophagy occurred at basal levels in undifferentiated human ESCs, but was upregulated upon differentiation. Besides differentiation, autophagy can be enhanced by oxidative stress in ESCs. Under oxidative stress, autophagy and mitochondrial function seem to be regulated by sirtuin 1 (SIRT1), partly through the class III PI3K/beclin 1 (BECN1) and mTOR pathways (Ou et al., 2014). Correspondingly, class III PI3K‐deficient ICM cells lose the ability to form colonies in culture (Zhou et al., 2011).
The primary cilium is a sleuder protuberance existing on nearly all cell surfaces, which senses extracellular physical or chemical stimuli and transmits them to the nucleus. Based on the discovery that the primary cilium is expressed in ESCs (Kiprilov et al., 2008) and interacts with autophagy (Pampliega et al., 2013), Gregory et al. (2016) proposed a model in which autophagy interacts with the primary cilium to maintain the function of pluripotent stem cells. Starvation is sensed by the primary cilium to induce autophagy, which results in the development and differentiation of pluripotent stem cells; normal nutrition attenuates autophagy, which results in self‐renewal and reprogramming of pluripotent stem cells. By exploring the role of primary cilium‐induced autophagy in the differentiation of ESCs, Jang et al. (2016) demonstrated that autophagy was elevated once ciliation increased, driving human embryonic stem cells (hESC) commitment to a neuroectoderm but not mesendoderm fate. These data indicate that autophagy plays important roles during the early differentiation of ESCs.
Autophagic flux encompasses the entire dynamic process of autophagy including autophagosome synthesis, delivery of autophagic substrates to the lysosome, and degradation of autophagic substrates inside the lysosome. It precisely reflects the intensity of autophagic degradation activity. Recently, high autophagic flux was identified as an intrinsic characteristic of mouse ESCs for maintenance of pluripotency (Liu et al., 2017). The ESCs harness a substantial infrastructure, which includes enhanced expression of autophagy machinery genes driven by forkhead box O1 (FOXO1), to guard the autophagic flux for maintenance of stemness. Loss of expression of the autophagy genes Atg3 or Ulk1 in ESCs leads to accumulation of damaged mitochondria and reactive oxygen species (ROS), resulting in compromised pluripotency (Liu et al., 2016, 2017; Gong et al., 2018). Consequently, ESC differentiation is also impaired.
Taken together, these studies suggest that autophagy is essential for maintenance of ESC pluripotency. The autophagic flux level in ESCs is higher than in somatic cells. This high autophagic flux protects the stemness of ESCs by efficiently generating building blocks to facilitate rapid cell proliferation. Alternatively, the high autophagic flux may protect ESCs from oxidative damage to the genome by rapidly removing damaged organelles and abnormal protein aggregates.
(d). Autophagy in somatic cell reprogramming
Induced pluripotent stem cells (iPSCs), which are reprogrammed from somatic cells by ectopic expression of transcription factors, have similar characteristics and functional proprieties to ESCs, and can be used as a powerful tool in developmental biology (Takahashi & Yamanaka, 2006; Wernig et al., 2007). The stemness of iPSCs is impaired if the autophagy level becomes too high or too low: treatment with bafilomycin (an inhibitor of autophagy) induces cell death, while adding a high concentration of rapamycin (which activates autophagy by suppressing mTOR) facilitates spontaneous differentiation into EBs (Sotthibundhu et al., 2016). However, treatment with a proper concentration of rapamycin enhances the reprogramming efficiency (Chen et al., 2011). Consistent with these findings, studies have demonstrated that ectopic expression of the transcription factor SRY‐box 2 (Sox2) transiently activates autophagy to induce somatic cell reprogramming by suppressing the activity of mTOR (S. Wang et al., 2013b).
During the reprogramming process, metabolic switching is an important step. Compared to differentiated cells, pluripotent stem cells have fewer mitochondria and lower levels of associated oxidative phosphorylation, and rely more on glycolysis for energy production (Zhang et al., 2012). Liu et al. (2016) found that mitochondrial remodelling occurs during somatic reprogramming, and this remodelling is mediated by ATG3‐dependent autophagy. Atg3 deficiency results in reduced reprogramming efficiency. Although iPSCs can be obtained from atg3 −/− mouse embryonic fibroblasts (MEFs), they are not healthy and contain many abnormal mitochondria. Like atg3 −/− ESCs, atg3 −/− iPSCs accumulate abnormal mitochondria, and show elevated ROS levels and reduced ATP production, leading to impaired self‐renewal and abnormal differentiation. These data clearly show that ATG3‐dependent basal autophagy is crucial for pluripotency induction and maintenance. Consistent with these findings, loss of PTEN‐induced putative kinase 1 (PINK1), a serine–threonine kinase that specifically participates in mitophagy, also perturbs PSC homeostasis (Vazquez‐Martin et al., 2016).
Another recent study showed that an ATG5‐independent non‐canonical autophagic process is important for reprogramming of somatic cells. This non‐canonical pathway seems to be regulated by AMPK as the upstream signal, and mediates mitochondrial clearance to facilitate metabolic switching and iPSC induction (Ma et al., 2015).
Taken together, these studies provide evidence that both canonical and non‐canonical autophagy play crucial roles in mitochondrial clearance during somatic cell reprogramming and thus contribute significantly to pluripotency induction.
(e). Conclusions
Autophagy is carefully adjusted during early embryonic development. It is triggered by calcium oscillation during fertilization to eliminate paternal mitochondria, which is crucial for maternal cytoplasmic inheritance. Subsequently, autophagic levels first rise and then gradually fall (with or without fluctuation), depending on the species. Atg‐deficient mouse models suggest that one of the functions of autophagy in this process is the promotion of new protein synthesis, but the exact mechanisms still await further study.
(2). Autophagy in embryo implantation and embryonic development
Embryo implantation occurs at the blastula stage. At this point, a mature blastocyst breaks out of the zona pellucida, then attaches to and penetrates the endometrium. Embryonic diapause is an interesting phenomenon characterized by a period of delayed implantation. Delayed implantation is considered to be an adaptation to resist nutrient shortage in certain mammals (Lopes, Desmarais & Murphy, 2004). Some studies have shown that autophagy is involved in this complex process. Lee et al. (2011) found that autophagy is an important protector of dormant blastocysts to extend embryonic longevity, although it may lead to reduced developmental competency of the blastocysts.
Organogenesis is a pivotal process that takes place during embryonic development. Different tissues and organs initiate during the embryonic stage, and undergo further development during the foetal period. Increasing evidence has shown that autophagy plays a role in organogenesis during embryonic development.
(a). Autophagy in extra‐embryonic tissues
After implantation, it is necessary for embryos to establish a tight relationship with the maternal uterus in order to obtain nutrients. At this time, the trophoblast differentiates further into the villous trophoblast (VT) and the extravillous trophoblast (EVT). Invasion of the EVT into the uterine wall finally establishes the blood circulation between the foetus and mother. During the early period of pregnancy, the uterus is in a hypoxic and low‐nutrient state, which induces autophagy in the EVT (Nakashima et al., 2013). An elevated level of autophagy is also observed in early placental tissues and is proposed to play a pro‐survival role in resisting apoptotic cell death (Chen, Longtine & Nelson, 2012). Impairment of autophagy by soluble endoglin weakens the invasion and vascular remodelling ability of EVTs, leading to poor placentation. In support of these findings, another study found that autophagy is highly induced in maternal decidual cells and the placenta (Hiyama et al., 2015).
During pregnancy, ATG proteins are continuously expressed in the placenta. There is no obvious difference in expression levels of Atg genes in the early, mid‐, and late gestation stages. Autophagy is thought to protect cells from deficiency of oxygen and glucose (Hung et al., 2013). In amniotic epithelial (AE) cells, endocytosis and autophagic activity are enhanced by nutrient limitation (Shen et al., 2008b). Furthermore, both autophagic and apoptotic cell death can be detected by transmission electron microscopy in AE cells taken from ruptured term amnions, indicating that autophagy contributes to amniotic membrane rupture during labour (Shen et al., 2008a).
Not every fetus is delivered at the end of a full‐term pregnancy, with some pregnancies ending in preterm birth. One of the reasons for this may be BECN1‐dependent autophagy. Evidence has shown that deficiency of Beclin1 impairs accumulation of lipid droplets for progesterone production in luteal cells, eventually leading to a preterm labour phenotype (Gawriluk et al., 2014).
(b). Autophagy in embryonic organogenesis
(i). Neurons
Neurogenesis is the earliest form of organogenesis during embryonic development. Loss of the ATG protein autophagy and beclin 1 regulator 1 (AMBRA1), a positive regulator of BECN1‐dependent autophagy, causes severe neural tube defects including midbrain/hindbrain exencephaly and/or spina bifida at stages E10–E14.5 (embryonic day 10–14.5) (Fimia et al., 2007). Consistent with these findings, ATG5‐dependent autophagy has been shown to regulate neural differentiation. During astrocyte development, ATG5‐mediated autophagy promotes astrocyte differentiation by degrading the Janus kinase 2 (JAK2)‐signal transducer and activator of transcription 3 (STAT3) inhibitor suppressor of cytokine signaling 2 (SOCS2) (S. Wang et al., 2014a). Moreover, ATG5 mediates the differentiation and proliferation of cortical neural progenitor cells (NPCs) via the β‐Catenin signaling pathway (Lv et al., 2014). In support of this finding, the ATG protein VPS34 has been shown to contribute to cortical neurogenesis by regulating excitatory neuron migration and axon extension (Inaguma et al., 2016).
(ii). Blood
In developing embryos, blood is first generated in the yolk sac. The differentiation of yolk sac‐derived embryonic erythroid cells requires elimination of organelles like mitochondria. During this process, mitophagy is upregulated (4.6‐fold increase) and decomposed mitochondria can be detected in lysosomes (Takano‐Ohmuro et al., 2000). As the embryos develop further, blood is generated in the foetal spleen and liver. Recent studies have shown that mitochondria in foetal definitive reticulocytes in the liver are engulfed and digested by ULK1‐dependent ATG5‐independent autophagy (Honda et al., 2014). These studies indicate that autophagy is required for reticulocyte development during early embryonic developmental stages.
(iii). Heart
The heart is considered a critically important organ due to its role in pumping blood through the body. Recent studies have shown that myocardial growth is abnormal at the post‐implantation stage when Atg13 is depleted. Mechanistic studies found that Atg13 deficiency leads to defective autophagosome formation and facilitates tumor necrosis factor alpha (TNF‐α)‐induced apoptosis, implying that both autophagic and non‐autophagic functions of ATG13 contribute to heart development (Kaizuka & Mizushima, 2015). In another study, Lee et al. (2014) revealed that Atg5 deficiency leads to abnormal myocardial development. All pups at postnatal day 0 (P0) exhibit an enlarged right atrium, and almost half also had atrioventricular canal defects and abnormal, thickened valves (Lee et al., 2014). Together, these data provide supporting evidence that autophagy plays pivotal roles in embryonic cardiac development.
(iv). Bone
Endochondral ossification is an essential process during the embryonic development of bone tissues. It requires chondrocytes, which are specialized cartilage‐producing cells. Autophagosomes have been detected during a brief window of terminal chondrocyte differentiation, and autophagy is induced in a hypoxia inducible factor 1 (HIF1)‐dependent manner by regulation of the activities of mTOR and AMPK in a hypoxic environment in the absence of a blood supply (Srinivas, Bohensky & Shapiro, 2009). Evidence has shown that autophagy promotes chondrocyte survival under such conditions (Srinivas & Shapiro, 2006; Srinivas et al., 2009). Furthermore, deletion of the Atg gene Rb1cc1 (encoding FIP200) leads to defective osteoblast terminal differentiation and osteopenia during osteoblast differentiation into bone (Liu et al., 2013).
(v). Other organs
In MEFs and primary lung fibroblasts, autophagy is markedly induced by interleukin‐2 (IL‐2). IL‐2 induces autophagy via an ATG5–BECN1–high mobility group box 1 (HMGB1)‐dependent pathway that can protect fibroblasts from apoptosis (Kang et al., 2013). Autophagic genes are also detected in tooth germs during odontogenesis, and LC3 was found to colocalize with the enamel organ and dental papilla, indicating the involvement of autophagy in tooth development (Yang et al., 2013). Other studies have shown that autophagy participates in ciliogenesis by regulating the proteins associated with intraflagellar transport (Pampliega et al., 2013).
(c). Conclusions
Successful implantation results in an embryo initially experiencing a hypoxic and low‐nutrient state in the uterus, which drives autophagy in EVT and placental tissues and facilitates placentation. During delayed implantation, autophagy may protect dormant blastocysts, extending their longevity. Autophagy promotes embryonic organogenesis, such as neural tube formation, neural differentiation, reticulocyte development, myocardial growth, chondrocyte survival and differentiation, etc., during early embryonic development. Autophagy is also thought to contribute to amniotic membrane rupture during labour. Given that embryogenesis is a long‐term and multi‐lineage developmental process, additional efforts should be made to uncover the diverse functions of autophagy and the underlying regulatory mechanisms.
(3). Autophagy in neonates and adults
The transplacental nutrient supply ceases when the developed fetus is delivered. Such sudden nutrient deprivation will represent an extremely harsh stress, and immediately induces a high level of autophagy for 3–12 h until the neonate receives maternal milk (Kuma et al., 2004). Autophagy is critical for neonatal survival: atg5 −/− mice die within the first day after delivery, even though they appear normal at birth. These atg5 −/− mice are able to survive to birth because they inherit maternal ATG proteins from the oocyte during fertilization. In wild‐type mice, autophagy returns to basal levels around 1–2 days after birth.
In adults, autophagy occurs extensively in various organs and tissues. At any given time point, it has many different roles in regulating a range of developmental and homeostatic processes. Mizushima & Komatsu (2011) comprehensively reviewed the role of autophagy in tissue homeostasis and renovation previously, so we restrict our discussion here to a general description of the potential functions of autophagy in adults, and introduce some recent advances in research into adult stem cells.
(a). The potential functions of autophagy in adults
The primary function of autophagy is to degrade self‐components to supply energy or to provide building blocks for the synthesis of new materials. This process produces a variety of metabolic substances, such as amino acids, glucose, and fatty acids (Singh & Cuervo, 2011). Increasing evidence indicates that autophagy is not only activated upon starvation, but also functions under normal physiological conditions, whether locally (tissue‐specific effects) or globally (endocrine effects) (Kim & Lee, 2014).
Autophagy functions in cellular quality control. Autophagy degrades protein aggresomes or damaged organelles to maintain normal cellular function. In the liver, autophagy catabolizes about 1–1.5% of cellular proteins per hour, independent of nutritional status (Mizushima & Komatsu, 2011). Autophagy deficiency leads to multiple cellular abnormalities, including abnormal mitochondria, accumulated peroxisomes and ubiquitin‐positive aggregates in liver (Komatsu et al., 2005). In Schwann cells, autophagy degrades mutant or misfolded peripheral myelin protein 22 (PMP22) proteins (Fortun et al., 2003), which are thought to form aggresomes and cause demyelinating peripheral neuropathies. Moreover, autophagy can remove invading intracellular pathogens, including bacteria, viruses, and parasites as a defence mechanism.
Autophagy also participates in cell differentiation and cell‐fate determination. For example, autophagy is required for the transition of pro‐B to pre‐B cells (Miller et al., 2008). Autophagy‐deficient B cells have a diminished capacity to differentiate into plasma cells, show deregulated expression of plasma cell transcription factors and decreased antibody secretion (Conway et al., 2013).
The functions of autophagy are widespread and alteration of normal autophagy leads to a plethora of diseases, which have been reviewed elsewhere (Mizushima & Komatsu, 2011; Kim & Lee, 2014). Dysfunction of autophagy in different tissues results in distinct a range of diseases (Saha et al., 2018; Xie & Zhou, 2018), including neurodegeneration (Xie & Zhou, 2018), cardiac diseases (Lavandero et al., 2015), immune diseases (Pengo et al., 2013; Netea‐Maier et al., 2016; Hua et al., 2018), obesity (Zhang, Sowers & Ren, 2018), tumorigenesis (Kimmelman & White, 2017),renal diseases (De Rechter et al., 2016) and oral diseases (Tan, Zhang & Zhou, 2017), among others.
(b). Autophagy in adult stem cells
Adult stem cells exist in various tissues and organs, and function in tissue repair and regeneration. Increasing evidence indicates that autophagy is essential for adult stem cell self‐renewal and differentiation. Autophagy has also been shown to enhance the ability of adult stem cells to resist stress conditions (Salemi et al., 2012).
(i). Neural stem cells
Autophagic protein FIP200, but not ATG5, ATG16L1, or ATG7, has been found to be essential for the maintenance and differentiation of postnatal neural stem cells. Loss of FIP200 leads to reduced numbers of neural stem cells and impaired neuronal differentiation capacity of the brain (C. Wang et al., 2013a). These defects are caused by accumulation of p62/SQSTM1 aggregates, which impairs the function of superoxide dismutase and increases the level of superoxide (Wang et al., 2016).
(ii). Haematopoietic stem cells
Autophagy has been identified as an essential mechanism protecting haematopoietic stem cells (HSCs) from metabolic stress (Warr et al., 2013). Autophagy‐deficient HSCs display impaired self‐renewal activity and other features similar to aged HSCs (Ho et al., 2017). This indicates the involvement of autophagy in maintaining pluripotency and combating aging in HSCs. When the autophagic genes Atg7/Atg12 are conditionally depleted, or mTOR is activated, HSC dysfunction is observed, characterized by mitochondrion accumulation, ROS elevation, DNA damage, etc. (Chen et al., 2009; Mortensen et al., 2011; Ho et al., 2017). Consequently, HSCs tend to proliferate, differentiate and lose the ability to repair the haematopoietic system. Further mechanistic studies showed that autophagy clears active and healthy mitochondria, allowing HSCs to have a low metabolic rate in order to maintain their quiescence. Most intriguingly, about one‐third of old HSCs have a high level of autophagy combined with a low metabolic rate with normal long‐term regeneration potency similar to healthy young HSCs (Ho et al., 2017). These studies suggest that autophagy becomes increasingly necessary with age to maintain the regenerative ability of old HSCs.
(iii). Hepatic progenitor cells
Hepatic progenitor cells (HPCs) have a higher autophagic activity than differentiated hepatocytes. Inhibition of autophagy in mouse HPCs compromises their ability to undergo self‐renewal, proliferation and hepatic differentiation under normal culture conditions, but has little effect on cell viability (Cheng et al., 2015). Consistent with this finding, autophagy has been identified to promote HPC‐mediated regeneration of rat liver in vivo. Damaged mitochondria and ROS accumulate in ATG5‐deficient rat HPCs. Furthermore, homologous recombination, which is one of the methods for repairing damaged DNA, is suppressed upon Atg5 depletion in rat HPCs. These results suggest that autophagy maintains the stemness of HPCs and protects them from DNA damage (Xue et al., 2016).
(iv). Other adult stem cells
Autophagy is constitutively active in human mesenchymal stem cells (MSCs) at a high level. Inhibition of autophagy by knockdown of Bcl2l1 inhibits the survival and promotes the differentiation of MSCs, indicating the importance of autophagy in pluripotency regulation in MSCs (Oliver et al., 2012). In addition, it was reported that more autophagosomes co‐localized with old mitochondria during the division of human mammary stem‐like cells. This demonstrates that mitochondrial quality control, mediated by mitophagy, is crucial for the asymmetrical division of stem cells and for preserving the stemness of daughter cells (Katajisto et al., 2015). In muscle stem cells (satellite cells), autophagy is essential for maintaining their quiescent state and preventing senescence, similar to HSCs. This suggests that autophagy may be part of a common strategy for maintaining adult stem cell pools in vivo (Garcia‐Prat et al., 2016).
(c). Conclusions
Autophagy is important in neonatal survival; it occurs at a high level immediately following birth until the neonate obtains a dietary source of nutrition. In adults, autophagy takes place in various organs with different roles. Its potential functions include the supply of energy or new building blocks, cellular quality control, cell differentiation, and cell fate determination, among others. Autophagy also functions in adult stem cell self‐renewal and differentiation, and may protect adult stem cells from metabolic stress, DNA damage, and help to maintain a quiescent state to prevent senescence. Disruption of the autophagic balance leads to numerous diseases. Thus, an in‐depth understanding of autophagy will help us to understand and fight such diseases.
(4). Autophagy in gametogenesis
Mammalian gametogenesis is an essential process by which diploid gametocytes undergo meiosis to form mature haploid gametes. It is a key process required in mammalian sexual reproduction, as haploid gametes are essential for creating a stable diploid genome upon fertilization.
(a). Autophagy in folliculogenesis
In the mammalian ovary, most follicles undergo atresia and/or apoptosis, and only a small population undergoes maturation and ovulation. A few studies have demonstrated that autophagy is involved in folliculogenesis (Choi et al., 2011), which is vital for oocyte development. During ovarian development, KIT proto‐oncogene receptor tyrosine kinase (KIT) promotes autophagy by activation of the PI3K–AKT serine/threonine kinase (AKT)–mTOR pathway, and inhibition of PI3K–AKT–mTOR results in dramatically decreased generation of germ cells (Adhikari et al., 2010; Gawriluk et al., 2011). Further mechanistic studies have shown that IL‐33, a member of the IL‐1 cytokine family, is responsible for the rapid clearance of cellular waste generated during follicular atresia by inducing autophagy (Wu et al., 2015). Thus, autophagy has been proposed to be essential for maintaining the pool of dormant female germ cells. Consistent with this finding, studies have shown that autophagy maintains the primordial follicles in a dormant state by the suppression of mTORC1 activity (Reddy et al., 2008; Choi et al., 2010).
(b). Autophagy in spermatogenesis
Autophagy components have been detected in rat spermatocytes, indicating the involvement of autophagy in spermatogenesis (Bustamante‐Marin et al., 2012). In support of this, autophagy was reported to be activated during spermatogenesis in the horse (Gallardo Bolanos et al., 2012). Such autophagy was proposed to protect spermatogenesis by improving spermatocyte survival (Kanninen, de Andrade Ramos & Witkin, 2013).
Recent studies have revealed that many ATG proteins like LC3, ATG5, PINK1, and p62 are highly expressed in human spermatozoa, especially in the middle piece of the tail where mitochondria are located (Aparicio et al., 2016). Autophagy is thought to be crucial for sperm survival and motility. Inhibition of autophagy resulted in reduced ATP generation, decreased intracellular calcium concentration, and compromised motility and viability of spermatozoa (Aparicio et al., 2016).
The acrosome is an important structure facilitating penetration of sperm into the egg. Evidence has shown that acrosome biogenesis requires ATG7‐dependent autophagy. Sperm lacking ATG7 fail to form intact acrosomal vesicles, and this results in irregular or nearly round‐headed spermatozoa, and fertilization failure (H. Wang et al., 2014b). These data suggest that autophagy plays a pivotal role in normal acrosome development.
(c). Conclusions
Autophagy contributes to both folliculogenesis and spermatogenesis. Autophagy may function in the rapid removal of cellular waste generated during follicular atresia and help to maintain the pool of dormant female germ cells. It protects spermatogenesis by improving spermatocyte survival and functioning in sperm motility. Autophagy is also required for acrosome biogenesis, which is pivotal for successful fertilization. Future studies should attempt to delineate other contributions of autophagy to spermatogenesis.
IV. CONCLUSIONS
(1) Although autophagy was first identified as a cellular stress responder, increasing evidence suggests that it carries out normal physiological roles during the life of mammals, and participates in precise regulation of the growth, development, and aging of organisms.
(2) Autophagy functions locally and specifically in different tissues, and is likely to influence tissue development in different ways. As a result, when autophagy is impaired, distinct differentiating tissues may appear to display diverse autophagy‐related phenotypes, even though they are at the same developmental stage.
(3) Autophagy plays distinct functions in adult stem cells and pluripotent stem cells. It maintains HSC quiescence by clearing active and healthy mitochondria to keep the cells in a state of reduced metabolism. By contrast, a high autophagic flux in ESCs ensures that they have a high metabolic rate to sustain their self‐renewal properties and pluripotency.
(4) Further efforts are required to examine the roles of autophagy systematically in the mammalian life cycle. The mechanisms underlying differential functions of autophagy during development also require investigation. Such information will provide a deeper understanding of developmental biology, and new targets to treat diseases or to modulate the differentiation of pluripotent stem cells in vitro.
V. ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China 2018YFA0108402, the Strategic Priority Research Program of the Chinese Academy of Sciences XDA16030302, the National Natural Science Foundation of China Program 31720103907, 31621004, 31570995, the Key Research Program of the Chinese Academy of Sciences ZDRW‐ZS‐2017‐5, and National Thousand Young Talents Program to T.Z. The authors declare no conflicts of interest.
VI. REFERENCES
- Adhikari, D. , Zheng, W. , Shen, Y. , Gorre, N. , Hamalainen, T. , Cooney, A. J. , Huhtaniemi, I. , Lan, Z. J. & Liu, K. (2010). Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Human Molecular Genetics 19, 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rawi, S. , Louvet‐Vallee, S. , Djeddi, A. , Sachse, M. , Culetto, E. , Hajjar, C. , Boyd, L. , Legouis, R. & Galy, V. (2011). Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334, 1144–1147. [DOI] [PubMed] [Google Scholar]
- Aparicio, I. M. , Espino, J. , Bejarano, I. , Gallardo‐Soler, A. , Campo, M. L. , Salido, G. M. , Pariente, J. A. , Pena, F. J. & Tapia, J. A. (2016). Autophagy‐related proteins are functionally active in human spermatozoa and may be involved in the regulation of cell survival and motility. Science Reports 6, 33647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe, E. L. , Walker, S. A. , Manifava, M. , Chandra, P. , Roderick, H. L. , Habermann, A. , Griffiths, G. & Ktistakis, N. T. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3‐phosphate and dynamically connected to the endoplasmic reticulum. The Journal of Cell Biology 182, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento, C. F. , Renna, M. , Ghislat, G. , Puri, C. , Ashkenazi, A. , Vicinanza, M. , Menzies, F. M. & Rubinsztein, D. C. (2016). Mammalian autophagy: how does it work? Annual Review of Biochemistry 85, 685–713. [DOI] [PubMed] [Google Scholar]
- Berg, T. O. , Fengsrud, M. , Stromhaug, P. E. , Berg, T. & Seglen, P. O. (1998). Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. The Journal of Biological Chemistry 273, 21883–21892. [DOI] [PubMed] [Google Scholar]
- Bustamante‐Marin, X. , Quiroga, C. , Lavandero, S. , Reyes, J. G. & Moreno, R. D. (2012). Apoptosis, necrosis and autophagy are influenced by metabolic energy sources in cultured rat spermatocytes. Apoptosis 17, 539–550. [DOI] [PubMed] [Google Scholar]
- Chen, B. , Longtine, M. S. & Nelson, D. M. (2012). Hypoxia induces autophagy in primary human trophoblasts. Endocrinology 153, 4946–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Liu, Y. , Liu, Y. & Zheng, P. (2009). mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Science Signaling 2, ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Shen, L. , Yu, J. , Wan, H. , Guo, A. , Chen, J. , Long, Y. , Zhao, J. & Pei, G. (2011). Rapamycin and other longevity‐promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell 10, 908–911. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Wang, B. , Zhou, H. , Dang, S. , Jin, M. , Shi, Y. , Hao, L. , Yang, Z. & Zhang, Y. (2015). Autophagy is required for the maintenance of liver progenitor cell functionality. Cellular Physiology and Biochemistry 36, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Choi, J. Y. , Jo, M. W. , Lee, E. Y. & Choi, D. S. (2011). Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertility and Sterility 95, 1482–1486. [DOI] [PubMed] [Google Scholar]
- Choi, J. Y. , Jo, M. W. , Lee, E. Y. , Yoon, B. K. & Choi, D. S. (2010). The role of autophagy in follicular development and atresia in rat granulosa cells. Fertility and Sterility 93, 2532–2537. [DOI] [PubMed] [Google Scholar]
- Conway, K. L. , Kuballa, P. , Khor, B. , Zhang, M. , Shi, H. N. , Virgin, H. W. & Xavier, R. J. (2013). ATG5 regulates plasma cell differentiation. Autophagy 9, 528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rechter, S. , Decuypere, J. P. , Ivanova, E. , van den Heuvel, L. P. , De Smedt, H. , Levtchenko, E. & Mekahli, D. (2016). Autophagy in renal diseases. Pediatric Nephrology 31, 737–752. [DOI] [PubMed] [Google Scholar]
- Deretic, V. , Saitoh, T. & Akira, S. (2013). Autophagy in infection, inflammation and immunity. Nature Reviews. Immunology 13, 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley, H. C. , Razi, M. , Polson, H. E. , Girardin, S. E. , Wilson, M. I. & Tooze, S. A. (2014). WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12‐5‐16L1. Molecular Cell 55, 238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, D. , Rosains, J. , Birkhoff, G. & Eggan, K. (2007). Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 447, 679–685. [DOI] [PubMed] [Google Scholar]
- Fimia, G. M. , Stoykova, A. , Romagnoli, A. , Giunta, L. , Di Bartolomeo, S. , Nardacci, R. , Corazzari, M. , Fuoco, C. , Ucar, A. , Schwartz, P. , Gruss, P. , Piacentini, M. , Chowdhury, K. & Cecconi, F. (2007). Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121–1125. [DOI] [PubMed] [Google Scholar]
- Fortun, J. , Dunn, W. A. Jr. , Joy, S. , Li, J. & Notterpek, L. (2003). Emerging role for autophagy in the removal of aggresomes in Schwann cells. The Journal of Neuroscience 23, 10672–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo Bolanos, J. M. , Miro Moran, A. , Balao da Silva, C. M. , Morillo Rodriguez, A. , Plaza Davila, M. , Aparicio, I. M. , Tapia, J. A. , Ortega Ferrusola, C. & Pena, F. J. (2012). Autophagy and apoptosis have a role in the survival or death of stallion spermatozoa during conservation in refrigeration. PLoS One 7, e30688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Prat, L. , Martinez‐Vicente, M. , Perdiguero, E. , Ortet, L. , Rodriguez‐Ubreva, J. , Rebollo, E. , Ruiz‐Bonilla, V. , Gutarra, S. , Ballestar, E. , Serrano, A. L. , Sandri, M. & Munoz‐Canoves, P. (2016). Autophagy maintains stemness by preventing senescence. Nature 529, 37–42. [DOI] [PubMed] [Google Scholar]
- Gawriluk, T. R. , Hale, A. N. , Flaws, J. A. , Dillon, C. P. , Green, D. R. & Rucker, E. B. III (2011). Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction 141, 759–765. [DOI] [PubMed] [Google Scholar]
- Gawriluk, T. R. , Ko, C. , Hong, X. , Christenson, L. K. & Rucker, E. B. III (2014). Beclin‐1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proceedings of the National Academy of Sciences of the United States of America 111, E4194–E4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J. , Gu, H. , Zhao, L. , Wang, L. , Liu, P. , Wang, F. , Xu, H. & Zhao, T. (2018). Phosphorylation of ULK1 by AMPK is essential for mouse embryonic stem cell self‐renewal and pluripotency. Cell Death & Disease 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, S. , Swamy, S. , Hewitt, Z. , Wood, A. , Weightman, R. & Moore, H. (2016). Autophagic response to cell culture stress in pluripotent stem cells. Biochemical and Biophysical Research Communications 473, 758–763. [DOI] [PubMed] [Google Scholar]
- Hailey, D. W. , Rambold, A. S. , Satpute‐Krishnan, P. , Mitra, K. , Sougrat, R. , Kim, P. K. & Lippincott‐Schwartz, J. (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada, T. , Noda, N. N. , Satomi, Y. , Ichimura, Y. , Fujioka, Y. , Takao, T. , Inagaki, F. & Ohsumi, Y. (2007). The Atg12‐Atg5 conjugate has a novel E3‐like activity for protein lipidation in autophagy. The Journal of Biological Chemistry 282, 37298–37302. [DOI] [PubMed] [Google Scholar]
- Harding, T. M. , Morano, K. A. , Scott, S. V. & Klionsky, D. J. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. The Journal of Cell Biology 131, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi‐Nishino, M. , Fujita, N. , Noda, T. , Yamaguchi, A. , Yoshimori, T. & Yamamoto, A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature Cell Biology 11, 1433–1437. [DOI] [PubMed] [Google Scholar]
- He, C. & Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annual Review of Genetics 43, 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama, M. , Kusakabe, K. T. , Takeshita, A. , Sugi, S. , Kuniyoshi, N. , Imai, H. , Kano, K. & Kiso, Y. (2015). Nutrient starvation affects expression of LC3 family at the feto‐maternal interface during murine placentation. The Journal of Veterinary Medical Science 77, 305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T. T. , Warr, M. R. , Adelman, E. R. , Lansinger, O. M. , Flach, J. , Verovskaya, E. V. , Figueroa, M. E. & Passegue, E. (2017). Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, S. , Arakawa, S. , Nishida, Y. , Yamaguchi, H. , Ishii, E. & Shimizu, S. (2014). Ulk1‐mediated Atg5‐independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nature Communications 5, 4004. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N. , Hara, T. , Kaizuka, T. , Kishi, C. , Takamura, A. , Miura, Y. , Iemura, S. , Natsume, T. , Takehana, K. , Yamada, N. , Guan, J. L. , Oshiro, N. & Mizushima, N. (2009). Nutrient‐dependent mTORC1 association with the ULK1‐Atg13‐FIP200 complex required for autophagy. Molecular Biology of the Cell 20, 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Y. , Shen, M. , McDonald, C. & Yao, Q. (2018). Autophagy dysfunction in autoinflammatory diseases. Journal of Autoimmunity 88, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, T. H. , Hsieh, T. T. , Chen, S. F. , Li, M. J. & Yeh, Y. L. (2013). Autophagy in the human placenta throughout gestation. PLoS One 8, e83475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaguma, Y. , Matsumoto, A. , Noda, M. , Tabata, H. , Maeda, A. , Goto, M. , Usui, D. , Jimbo, E. F. , Kikkawa, K. , Ohtsuki, M. , Momoi, M. Y. , Osaka, H. , Yamagata, T. & Nagata, K. I. (2016). Role of class III phosphoinositide 3‐kinase in the brain development: possible involvement in specific learning disorders. Journal of Neurochemistry 139, 245–255. [DOI] [PubMed] [Google Scholar]
- Jahreiss, L. , Menzies, F. M. & Rubinsztein, D. C. (2008). The itinerary of autophagosomes: from peripheral formation to kiss‐and‐run fusion with lysosomes. Traffic 9, 574–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J. , Wang, Y. , Lalli, M. A. , Guzman, E. , Godshalk, S. E. , Zhou, H. & Kosik, K. S. (2016). Primary cilium‐autophagy‐Nrf2 (PAN) axis activation commits human embryonic stem cells to a neuroectoderm fate. Cell 165, 410–420. [DOI] [PubMed] [Google Scholar]
- Jin, Y. X. , Zheng, Z. , Yu, X. F. , Zhang, J. B. , Namgoong, S. , Cui, X. S. , Hyun, S. H. & Kim, N. H. (2016). Autophagy and ubiquitin‐mediated proteolysis may not be involved in the degradation of spermatozoon mitochondria in mouse and porcine early embryos. Zygote 24, 31–41. [DOI] [PubMed] [Google Scholar]
- Jung, C. H. , Jun, C. B. , Ro, S. H. , Kim, Y. M. , Otto, N. M. , Cao, J. , Kundu, M. & Kim, D. H. (2009). ULK‐Atg13‐FIP200 complexes mediate mTOR signaling to the autophagy machinery. Molecular Biology of the Cell 20, 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y. , Mizushima, N. , Yamamoto, A. , Oshitani‐Okamoto, S. , Ohsumi, Y. & Yoshimori, T. (2004). LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form‐II formation. Journal of Cell Science 117, 2805–2812. [DOI] [PubMed] [Google Scholar]
- Kaizuka, T. & Mizushima, N. (2015). Atg13 is essential for autophagy and cardiac development in mice. Molecular and Cellular Biology 36, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, R. , Tang, D. , Lotze, M. T. & Zeh Iii, H. J. (2013). Autophagy is required for IL‐2‐mediated fibroblast growth. Experimental Cell Research 319, 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanninen, T. T. , de Andrade Ramos, B. R. & Witkin, S. S. (2013). The role of autophagy in reproduction from gametogenesis to parturition. European Journal of Obstetrics, Gynecology, and Reproductive Biology 171, 3–8. [DOI] [PubMed] [Google Scholar]
- Katajisto, P. , Dohla, J. , Chaffer, C. L. , Pentinmikko, N. , Marjanovic, N. , Iqbal, S. , Zoncu, R. , Chen, W. , Weinberg, R. A. & Sabatini, D. M. (2015). Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. H. & Lee, M. S. (2014). Autophagy‐‐a key player in cellular and body metabolism. Nature Reviews Endocrinology 10, 322–337. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Kundu, M. , Viollet, B. & Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology 13, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman, A. C. & White, E. (2017). Autophagy and tumor metabolism. Cell Metabolism 25, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprilov, E. N. , Awan, A. , Desprat, R. , Velho, M. , Clement, C. A. , Byskov, A. G. , Andersen, C. Y. , Satir, P. , Bouhassira, E. E. , Christensen, S. T. & Hirsch, R. E. (2008). Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. The Journal of Cell Biology 180, 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, M. , Waguri, S. , Ueno, T. , Iwata, J. , Murata, S. , Tanida, I. , Ezaki, J. , Mizushima, N. , Ohsumi, Y. , Uchiyama, Y. , Kominami, E. , Tanaka, K. & Chiba, T. (2005). Impairment of starvation‐induced and constitutive autophagy in Atg7‐deficient mice. The Journal of Cell Biology 169, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma, A. , Hatano, M. , Matsui, M. , Yamamoto, A. , Nakaya, H. , Yoshimori, T. , Ohsumi, Y. , Tokuhisa, T. & Mizushima, N. (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Kundu, M. & Thompson, C. B. (2008). Autophagy: basic principles and relevance to disease. Annual Review of Pathology 3, 427–455. [DOI] [PubMed] [Google Scholar]
- Lavandero, S. , Chiong, M. , Rothermel, B. A. & Hill, J. A. (2015). Autophagy in cardiovascular biology. The Journal of Clinical Investigation 125, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. E. , Oh, H. A. , Song, H. , Jun, J. H. , Roh, C. R. , Xie, H. , Dey, S. K. & Lim, H. J. (2011). Autophagy regulates embryonic survival during delayed implantation. Endocrinology 152, 2067–2075. [DOI] [PubMed] [Google Scholar]
- Lee, E. , Koo, Y. , Ng, A. , Wei, Y. , Luby‐Phelps, K. , Juraszek, A. , Xavier, R. J. , Cleaver, O. , Levine, B. & Amatruda, J. F. (2014). Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy 10, 572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Fang, F. , Yuan, H. , Yang, D. , Chen, Y. , Williams, L. , Goldstein, S. A. , Krebsbach, P. H. & Guan, J. L. (2013). Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. Journal of Bone and Mineral Research 28, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K. , Zhao, Q. , Liu, P. , Cao, J. , Gong, J. , Wang, C. , Wang, W. , Li, X. , Sun, H. , Zhang, C. , Li, Y. , Jiang, M. , Zhu, S. , Sun, Q. , Jiao, J. , Hu, B. , Zhao, X. , Li, W. , Chen, Q. , Zhou, Q. & Zhao, T. (2016). ATG3‐dependent autophagy mediates mitochondrial homeostasis in pluripotency acquirement and maintenance. Autophagy 12, 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P. , Liu, K. , Gu, H. , Wang, W. , Gong, J. , Zhu, Y. , Zhao, Q. , Cao, J. , Han, C. , Gao, F. , Chen, Q. , Li, W. , Jiao, J. , Hu, B. , Zhou, Q. & Zhao, T. (2017). High autophagic flux guards ESC identity through coordinating autophagy machinery gene program by FOXO1. Cell Death and Differentiation 24, 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, F. L. , Desmarais, J. A. & Murphy, B. D. (2004). Embryonic diapause and its regulation. Reproduction 128, 669–678. [DOI] [PubMed] [Google Scholar]
- Lu, Q. , Yang, P. , Huang, X. , Hu, W. , Guo, B. , Wu, F. , Lin, L. , Kovacs, A. L. , Yu, L. & Zhang, H. (2011). The WD40 repeat PtdIns(3)P‐binding protein EPG‐6 regulates progression of omegasomes to autophagosomes. Developmental Cell 21, 343–357. [DOI] [PubMed] [Google Scholar]
- Luo, S. M. , Ge, Z. J. , Wang, Z. W. , Jiang, Z. Z. , Wang, Z. B. , Ouyang, Y. C. , Hou, Y. , Schatten, H. & Sun, Q. Y. (2013). Unique insights into maternal mitochondrial inheritance in mice. Proceedings of the National Academy of Sciences of the United States of America 110, 13038–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, X. , Jiang, H. , Li, B. , Liang, Q. , Wang, S. , Zhao, Q. & Jiao, J. (2014). The crucial role of Atg5 in cortical neurogenesis during early brain development. Scientific Reports 4, 6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, T. , Li, J. , Xu, Y. , Yu, C. , Xu, T. , Wang, H. , Liu, K. , Cao, N. , Nie, B. M. , Zhu, S. Y. , Xu, S. , Li, K. , Wei, W. G. , Wu, Y. , Guan, K. L. & Ding, S. (2015). Atg5‐independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nature Cell Biology 17, 1379–1387. [DOI] [PubMed] [Google Scholar]
- Miller, B. C. , Zhao, Z. , Stephenson, L. M. , Cadwell, K. , Pua, H. H. , Lee, H. K. , Mizushima, N. N. , Iwasaki, A. , He, Y. W. , Swat, W. & Virgin, H. W. T. (2008). The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 4, 309–314. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. & Komatsu, M. (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. , Yoshimori, T. & Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annual Review of Cell and Developmental Biology 27, 107–132. [DOI] [PubMed] [Google Scholar]
- Mortensen, M. , Soilleux, E. J. , Djordjevic, G. , Tripp, R. , Lutteropp, M. , Sadighi‐Akha, E. , Stranks, A. J. , Glanville, J. , Knight, S. , Jacobsen, S. E. , Kranc, K. R. & Simon, A. K. (2011). The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of Experimental Medicine 208, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, A. , Yamanaka‐Tatematsu, M. , Fujita, N. , Koizumi, K. , Shima, T. , Yoshida, T. , Nikaido, T. , Okamoto, A. , Yoshimori, T. & Saito, S. (2013). Impaired autophagy by soluble endoglin, under physiological hypoxia in early pregnant period, is involved in poor placentation in preeclampsia. Autophagy 9, 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa, H. , Ichimura, Y. & Ohsumi, Y. (2007). Atg8, a ubiquitin‐like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. [DOI] [PubMed] [Google Scholar]
- Netea‐Maier, R. T. , Plantinga, T. S. , van de Veerdonk, F. L. , Smit, J. W. & Netea, M. G. (2016). Modulation of inflammation by autophagy: consequences for human disease. Autophagy 12, 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi, Y. (2014). Historical landmarks of autophagy research. Cell Research 24, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, L. , Hue, E. , Priault, M. & Vallette, F. M. (2012). Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells and Development 21, 2779–2788. [DOI] [PubMed] [Google Scholar]
- Orsi, A. , Razi, M. , Dooley, H. C. , Robinson, D. , Weston, A. E. , Collinson, L. M. & Tooze, S. A. (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Molecular Biology of the Cell 23, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, X. , Lee, M. R. , Huang, X. , Messina‐Graham, S. & Broxmeyer, H. E. (2014). SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 32, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras, K. , Lionaki, E. & Tavernarakis, N. (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528. [DOI] [PubMed] [Google Scholar]
- Pampliega, O. , Orhon, I. , Patel, B. , Sridhar, S. , Diaz‐Carretero, A. , Beau, I. , Codogno, P. , Satir, B. H. , Satir, P. & Cuervo, A. M. (2013). Functional interaction between autophagy and ciliogenesis. Nature 502, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengo, N. , Scolari, M. , Oliva, L. , Milan, E. , Mainoldi, F. , Raimondi, A. , Fagioli, C. , Merlini, A. , Mariani, E. , Pasqualetto, E. , Orfanelli, U. , Ponzoni, M. , Sitia, R. , Casola, S. & Cenci, S. (2013). Plasma cells require autophagy for sustainable immunoglobulin production. Nature Immunology 14, 298–305. [DOI] [PubMed] [Google Scholar]
- Qu, X. , Zou, Z. , Sun, Q. , Luby‐Phelps, K. , Cheng, P. , Hogan, R. N. , Gilpin, C. & Levine, B. (2007). Autophagy gene‐dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946. [DOI] [PubMed] [Google Scholar]
- Reddy, P. , Liu, L. , Adhikari, D. , Jagarlamudi, K. , Rajareddy, S. , Shen, Y. , Du, C. , Tang, W. , Hamalainen, T. , Peng, S. L. , Lan, Z. J. , Cooney, A. J. , Huhtaniemi, I. & Liu, K. (2008). Oocyte‐specific deletion of Pten causes premature activation of the primordial follicle pool. Science 319, 611–613. [DOI] [PubMed] [Google Scholar]
- Rojansky, R. , Cha, M. Y. & Chan, D. C. (2016). Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 5, e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S. , Panigrahi, D. P. , Patil, S. & Bhutia, S. K. (2018). Autophagy in health and disease: a comprehensive review. Biomedicine & Pharmacotherapy 104, 485–495. [DOI] [PubMed] [Google Scholar]
- Salemi, S. , Yousefi, S. , Constantinescu, M. A. , Fey, M. F. & Simon, H. U. (2012). Autophagy is required for self‐renewal and differentiation of adult human stem cells. Cell Research 22, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana‐Codina, N. , Mancias, J. D. & Kimmelman, A. C. (2017). The role of autophagy in cancer. Annual Review of Cancer Biology 1, 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, S. , Peterson, T. R. & Sabatini, D. M. (2010). Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Molecular Cell 40, 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Z. Y. , Li, E. M. , Lu, S. Q. , Shen, J. , Cai, Y. M. , Wu, Y. E. , Zheng, R. M. , Tan, L. J. & Xu, L. Y. (2008a). Autophagic and apoptotic cell death in amniotic epithelial cells. Placenta 29, 956–961. [DOI] [PubMed] [Google Scholar]
- Shen, Z. Y. , Xu, L. Y. , Li, E. M. , Zhuang, B. R. , Lu, X. F. , Shen, J. , Wu, X. Y. , Li, Q. S. , Lin, Y. J. , Chen, Y. W. & Tan, L. J. (2008b). Autophagy and endocytosis in the amnion. Journal of Structural Biology 162, 197–204. [DOI] [PubMed] [Google Scholar]
- Singh, R. & Cuervo, A. M. (2011). Autophagy in the cellular energetic balance. Cell Metabolism 13, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W. H. , Yi, Y. J. , Sutovsky, M. , Meyers, S. & Sutovsky, P. (2016). Autophagy and ubiquitin‐proteasome system contribute to sperm mitophagy after mammalian fertilization. Proceedings of the National Academy of Sciences of the United States of America 113, E5261–E5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotthibundhu, A. , McDonagh, K. , von Kriegsheim, A. , Garcia‐Munoz, A. , Klawiter, A. , Thompson, K. , Chauhan, K. D. , Krawczyk, J. , McInerney, V. , Dockery, P. , Devine, M. J. , Kunath, T. , Barry, F. , O'Brien, T. & Shen, S. (2016). Rapamycin regulates autophagy and cell adhesion in induced pluripotent stem cells. Stem Cell Research & Therapy 7, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas, V. , Bohensky, J. & Shapiro, I. M. (2009). Autophagy: a new phase in the maturation of growth plate chondrocytes is regulated by HIF, mTOR and AMP kinase. Cells, Tissues, Organs 189, 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas, V. & Shapiro, I. M. (2006). Chondrocytes embedded in the epiphyseal growth plates of long bones undergo autophagy prior to the induction of osteogenesis. Autophagy 2, 215–216. [DOI] [PubMed] [Google Scholar]
- Stitzel, M. L. & Seydoux, G. (2007). Regulation of the oocyte‐to‐zygote transition. Science 316, 407–408. [DOI] [PubMed] [Google Scholar]
- Takahashi, K. & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takano‐Ohmuro, H. , Mukaida, M. , Kominami, E. & Morioka, K. (2000). Autophagy in embryonic erythroid cells: its role in maturation. European Journal of Cell Biology 79, 759–764. [DOI] [PubMed] [Google Scholar]
- Tan, Y. Q. , Zhang, J. & Zhou, G. (2017). Autophagy and its implication in human oral diseases. Autophagy 13, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm, M. , Egner, R. , Koch, B. , Schlumpberger, M. , Straub, M. , Veenhuis, M. & Wolf, D. H. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Letters 349, 275–280. [DOI] [PubMed] [Google Scholar]
- Tra, T. , Gong, L. , Kao, L. P. , Li, X. L. , Grandela, C. , Devenish, R. J. , Wolvetang, E. & Prescott, M. (2011). Autophagy in human embryonic stem cells. PLoS One 6, e27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M. & Ohsumi, Y. (1993). Isolation and characterization of autophagy‐defective mutants of Saccharomyces cerevisiae. FEBS Letters 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, S. , Kuma, A. & Mizushima, N. (2008a). The role of autophagy during the oocyte‐to‐embryo transition. Autophagy 4, 1076–1078. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, S. , Kuma, A. , Murakami, M. , Kishi, C. , Yamamoto, A. & Mizushima, N. (2008b). Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120. [DOI] [PubMed] [Google Scholar]
- Uemura, T. , Yamamoto, M. , Kametaka, A. , Sou, Y. S. , Yabashi, A. , Yamada, A. , Annoh, H. , Kametaka, S. , Komatsu, M. & Waguri, S. (2014). A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Molecular and Cellular Biology 34, 1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Martin, A. , Van den Haute, C. , Cufi, S. , Corominas‐Faja, B. , Cuyas, E. , Lopez‐Bonet, E. , Rodriguez‐Gallego, E. , Fernandez‐Arroyo, S. , Joven, J. , Baekelandt, V. & Menendez, J. A. (2016). Mitophagy‐driven mitochondrial rejuvenation regulates stem cell fate. Aging (Albany NY) 8, 1330–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Liang, C. C. , Bian, Z. C. , Zhu, Y. & Guan, J. L. (2013a). FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nature Neuroscience 16, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Xia, P. , Ye, B. , Huang, G. , Liu, J. & Fan, Z. (2013b). Transient activation of autophagy via Sox2‐mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell 13, 617–625. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Li, B. , Qiao, H. , Lv, X. , Liang, Q. , Shi, Z. , Xia, W. , Ji, F. & Jiao, J. (2014a). Autophagy‐related gene Atg5 is essential for astrocyte differentiation in the developing mouse cortex. EMBO Reports 15, 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wan, H. , Li, X. , Liu, W. , Chen, Q. , Wang, Y. , Yang, L. , Tang, H. , Zhang, X. , Duan, E. , Zhao, X. , Gao, F. & Li, W. (2014b). Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Research 24, 852–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Chen, S. , Yeo, S. , Karsli‐Uzunbas, G. , White, E. , Mizushima, N. , Virgin, H. W. & Guan, J. L. (2016). Elevated p62/SQSTM1 determines the fate of autophagy‐deficient neural stem cells by increasing superoxide. The Journal of Cell Biology 212, 545–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr, M. R. , Binnewies, M. , Flach, J. , Reynaud, D. , Garg, T. , Malhotra, R. , Debnath, J. & Passegue, E. (2013). FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig, M. , Meissner, A. , Foreman, R. , Brambrink, T. , Ku, M. , Hochedlinger, K. , Bernstein, B. E. & Jaenisch, R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES‐cell‐like state. Nature 448, 318–324. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Carlock, C. , Zhou, C. , Nakae, S. , Hicks, J. , Adams, H. P. & Lou, Y. (2015). IL‐33 is required for disposal of unnecessary cells during ovarian atresia through regulation of autophagy and macrophage migration. Journal of Immunology 194, 2140–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W. & Zhou, J. (2018). Aberrant regulation of autophagy in mammalian diseases. Biology Letters 14, 20170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. N. , Shen, X. H. , Lee, S. E. , Kwon, J. S. , Kim, D. J. , Heo, Y. T. , Cui, X. S. & Kim, N. H. (2012). Autophagy influences maternal mRNA degradation and apoptosis in porcine parthenotes developing in vitro. The Journal of Reproduction and Development 58, 576–584. [DOI] [PubMed] [Google Scholar]
- Xue, F. , Hu, L. , Ge, R. , Yang, L. , Liu, K. , Li, Y. , Sun, Y. & Wang, K. (2016). Autophagy‐deficiency in hepatic progenitor cells leads to the defects of stemness and enhances susceptibility to neoplastic transformation. Cancer Letters 371, 38–47. [DOI] [PubMed] [Google Scholar]
- Yang, J. W. , Zhu, L. X. , Yuan, G. H. , Chen, Y. X. , Zhang, L. , Zhang, L. & Chen, Z. (2013). Autophagy appears during the development of the mouse lower first molar. Histochemistry and Cell Biology 139, 109–118. [DOI] [PubMed] [Google Scholar]
- Yla‐Anttila, P. , Vihinen, H. , Jokitalo, E. & Eskelinen, E. L. (2009). 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185. [DOI] [PubMed] [Google Scholar]
- Yousefi, S. , Perozzo, R. , Schmid, I. , Ziemiecki, A. , Schaffner, T. , Scapozza, L. , Brunner, T. & Simon, H. U. (2006). Calpain‐mediated cleavage of Atg5 switches autophagy to apoptosis. Nature Cell Biology 8, 1124–1132. [DOI] [PubMed] [Google Scholar]
- Yue, Z. , Jin, S. , Yang, C. , Levine, A. J. & Heintz, N. (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America 100, 15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Nuebel, E. , Daley, G. Q. , Koehler, C. M. & Teitell, M. A. (2012). Metabolic regulation in pluripotent stem cells during reprogramming and self‐renewal. Cell Stem Cell 11, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Sowers, J. R. & Ren, J. (2018). Targeting autophagy in obesity: from pathophysiology to management. Nature Reviews. Endocrinology 14, 356–376. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Takatoh, J. & Wang, F. (2011). The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One 6, e16358. [DOI] [PMC free article] [PubMed] [Google Scholar]


