ABSTRACT
MicroRNAs (miRNAs) are active regulators of numerous biological and physiological processes including most of the events of mammalian reproduction. Understanding the biological functions of miRNAs in the context of mammalian reproduction will allow a better and comparative understanding of fertility and sterility in male and female mammals. Herein, we summarize recent progress in miRNA‐mediated regulation of mammalian reproduction and highlight the significance of miRNAs in different aspects of mammalian reproduction including the biogenesis of germ cells, the functionality of reproductive organs, and the development of early embryos. Furthermore, we focus on the gene expression regulatory feedback loops involving hormones and miRNA expression to increase our understanding of germ cell commitment and the functioning of reproductive organs. Finally, we discuss the influence of miRNAs on male and female reproductive failure, and provide perspectives for future studies on this topic.
Keywords: miRNAs, mammalian reproduction, gametogenesis, hormonal balance, peri‐implantation, germ layer specification
I. INTRODUCTION
Mammalian microRNAs (miRNAs) are non‐coding RNAs that can regulate post‐transcriptional gene expression by targeting the 3′‐untranslated region and/or coding region of messenger RNAs (mRNAs) (Hausser et al., 2013), and are involved in the regulation of approximately one‐third of all mammalian genes (Lewis, Burge, & Bartel, 2005). While a single miRNA may target many genes, the expression of a particular gene can be modulated by multiple miRNAs (Sood et al., 2006). Hence, miRNAs are involved in the active regulation of biological and physiological processes in both normal and disease conditions (Reza et al., 2016, 2017, 2018a; Reza, Choi, & Kim, 2018b). In mammalian reproduction, miRNAs are widely involved in sex differentiation (Cook & Blelloch, 2013), gametogenesis (Hayashi et al., 2008), fertilization (Hong et al., 2008), zygotic genome activation (ZGA) and early development (Suh et al., 2010), implantation (Liu et al., 2016), germ layer specification (Vidigal & Ventura, 2012), and pregnancy (Otsuka et al., 2008). Understanding the functions of miRNAs in mammalian reproduction will therefore improve our understanding of the biological challenges of mammalian reproduction.
Several reviews have focused on the event‐specific involvement of non‐coding RNAs (miRNAs, endogenous siRNAs, piwi‐interacting RNAs) in the reproduction of different species. However, those reviews included all types of non‐coding RNAs and a wide range of species (insects, non‐vertebrates, fishes, and birds), in order to develop an understanding of their roles in reproductive biology. To comprehend specifically the roles of miRNAs in mammalian reproduction requires an in‐depth review focusing on mammalian species, as mammals are relatively unique in their structure, physiology, and psychology. Furthermore, the available reviews mainly focused on particular aspects of reproduction such as oogenesis (Donadeu, Schauer, & Sontakke, 2012; Hossain et al., 2012a), spermatogenesis (Kotaja, 2014; Yadav & Kotaja, 2014; Hilz et al., 2016; Chen et al., 2017), early embryonic development (Suh & Blelloch, 2011; Cook & Blelloch, 2013; Dallaire & Simard, 2016), or implantation (Liu & Yang, 2011; Dior et al., 2014; Galliano & Pellicer, 2014; Liu et al., 2016), with a holistic review focusing on all processes of mammalian reproduction lacking. Such a holistic review focusing on miRNA involvement in all aspects of mammalian reproduction is necessary to improve our understanding of reproductive biology and to mitigate mammalian reproductive challenges.
Herein, we summarize recent progress in miRNA‐mediated regulation of mammalian reproduction, and highlight the significance of miRNAs in different aspects of mammalian reproduction including the biogenesis of germ cells, functionality of reproductive organs, and development of early embryos, as well as covering infertility and sterility derived from miRNA deregulation in males and females. First, we discuss impairments and abnormalities caused by depletion of miRNA‐processing machinery during germ cell commitment and the functioning of reproductive organs. Second, we discuss how miRNAs interfere at different stages of male and female reproduction. Furthermore, we focus on the distribution and dynamics of miRNAs during gametogenesis, fertilization, embryo development, and implantation, as well as specification to three different germ layers. Finally, we discuss the influence of miRNAs on male and female reproductive failure, and provide a set of conclusive remarks to guide future studies in this topic.
II. LOSS OF miRNA‐PROCESSING MACHINERY IMPAIRS MAMMALIAN REPRODUCTION
Loss of one or both of the components of the miRNA‐processing machinery (Dicer and Drosha) causes severe impairment during gametogenesis, leading to infertility in both males and females (Fig. 1). Especially, Dicer and Drosha are indispensable during the early stages of development such as before sex differentiation (Bernstein et al., 2003), while in the later stages, their loss causes impairment to different extents depending on the stage at which Dicer or Drosha is deleted (Hong et al., 2008; Liu et al., 2010; Liu et al., 2012). However, it is clear that this impairment does not arise from the absence of the proteins themselves, but rather from the associated failure of biogenesis of small‐RNAs (miRNAs and endogenous siRNAs) (Papaioannou et al., 2009; Hossain et al., 2012b). The severity of Dicer1 deletion is highly stage and organ specific, with other organs or processes often remaining functional. Interestingly, sexual behaviour, mounting ability, external genitalia, and oestrous are not affected by the depletion of Dicer (Papaioannou et al., 2009).
Figure 1.
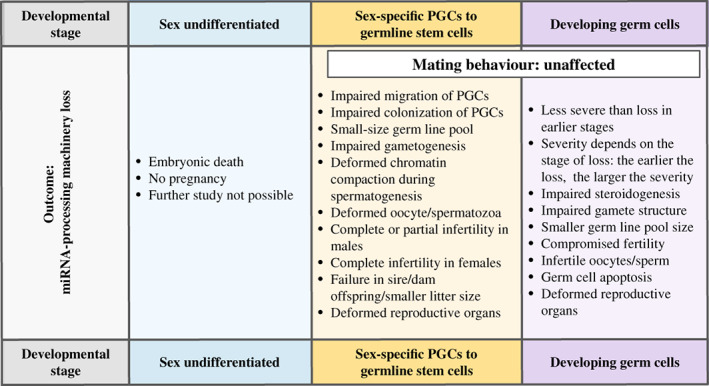
Adverse effects of the loss of microRNA (miRNA)‐processing machinery during mammalian reproduction. The loss of miRNA‐processing machinery can cause reproductive impairment to different extents, including embryonic death, based on the stage at which it is lost. However, mating behaviour of both males and females remains intact. PGC, primordial germ cell.
In the male, primordial germ cell (PGC)‐specific deletion of Dicer and Drosha restricts the propagation, differentiation, and maturation of male PGCs (Hayashi et al., 2008) and causes defective spermatozoa and infertility (Maatouk et al., 2008). Spermatogonia‐specific ablation of Dicer1 impairs the leptotene to zygotene/pachytene transition during prophase I with an increase in apoptotic pachytene spermatocytes, deformed chromatin compaction, acrosome formation, and sperm‐head structure (Romero et al., 2011). Round spermatid‐specific deletion of Dicer1 deregulated actin related protein 2/3 complex subunit 5 (Arpc5), leading to abnormal chromatin compaction and causing deformed acrosomes and sperm heads, as well as apoptosis of spermatids in mice (Chang et al., 2012). Sertoli cell‐specific deletion of Dicer1 caused apoptosis of Sertoli cells after birth leading to degenerated testes, defective prepubertal spermatogenesis, and infertility (Papaioannou et al., 2009). Epididymis‐specific deletion of Dicer1 resulted in imbalanced lipid homeostasis and instability of the sperm membrane (Bjorkgren et al., 2015) as well as leakage of immature spermatocytes to the epididymis and a thick blood–testis barrier with a multilayered organization (Korhonen et al., 2015). Hence, Dicer/Drosha safeguard meiotic progression and the production of spermatozoa, as well as the development of accessory organs, by supplying functional miRNAs.
In females, Dicer1 loss causes abnormal spindle formation and chromosomal misalignment (Liu et al., 2010), failures in meiotic maturation and polar body formation (Murchison et al., 2007; Liu et al., 2010) and functional defects including impaired granulosa cell (GC) proliferation and steroidogenesis, recruitment of immature follicles, follicular atresia, a smaller pool size of pre‐ovulatory follicles, and ovulatory dysfunction followed by infertility (Murchison et al., 2007; Hong et al., 2008; Lei et al., 2010). Dicer1 deficiency impairs the biogenesis of small‐RNAs altering gene expression and signalling networks: Dicer1‐ablated ovaries show deregulation of miR‐503 and its targets including anti‐mullerian hormone (Amh), inhibin beta A subunit (Inhba), cytochrome P450 family 17 subfamily A member 1 (Cyp17a1), cytochrome P450 family 19 subfamily A member 1 (Cyp19a1), zona pellucida glycoprotein (Zp), growth differentiation factor 9 (Gdf9), and bone morphogenetic protein 15 (Bmp15), leading to impairment of folliculogenesis in mice (Lei et al., 2010). Similarly, inhibition of Dicer1 reduced the expression of polo like kinase 1 (Plk1), aurora kinase A (Aurka), BUB1 mitotic checkpoint serine/threonine kinase (Bub1), and BUB1 mitotic checkpoint serine/threonine kinase B (Bub1b), enhanced abnormal spindle formation and chromosomal misalignment, and lowered the maturation rate of oocytes (Murchison et al., 2007; Tang et al., 2007; Liu et al., 2010).
The replacement of Dicer1‐ablated ovaries with wild‐type (WT) ovaries enable the mice to produce viable offspring, while implantation of Dicer1‐ablated ovaries into WT mice results in failure to produce offspring (Otsuka et al., 2008). The failure is prominent during post‐implantation development, while ovulation, fertilization, and first embryonic cell division appear normal (Otsuka et al., 2008) and may be due to disintegration of the corpus luteum (CL) and lower progesterone levels (Otsuka et al., 2008), which may derive from impairment to miRNA regulation required for the angiogenesis, maintenance, and functioning of the CL (Yang et al., 2005; Otsuka et al., 2008). Moreover, depletion of Dicer1 causes disintegration of the structural, mechanical, and functional ability of the fallopian tube including loss of the smooth muscle layer, disorganized epithelium, and deregulation of signalling networks (Luense, Carletti & Christenson, 2009): this might also cause failures in the transportation and implantation of embryos to the uterus (Luense, Carletti, & Christenson, 2009), and the resulting offspring may have impaired reproductive organs because of maternal loss of oviductal Dicer1 (Hong et al., 2008). Fallopian tube‐specific loss of Dicer1 results in shorter tubule length, abnormal coil formation and the presence of fluid‐filled sacs (Hong et al., 2008; Gonzalez & Behringer, 2009). Taken together, the observations suggest that Dicer1 in the female safeguards molecular and signalling networks related to gametogenesis, steroidogenesis, the genomic and structural architecture of the oocytes as well as the development and implantation of embryos.
III. miRNAs ARE INVOLVED IN SEX‐SPECIFICATION AND COMMITMENT OF MAMMALIAN GONADS
Prior to sex determination PGCs carry a unique miRNA profile (Hayashi et al., 2008) that does not match sex‐specific PGCs (Fig. 2), and PGCs undergoing sex determination display dimorphic expression patterns of miRNAs (see online Supporting information, Table S1). For example, the let‐7 family, miR‐125a, and miR‐9 are overexpressed in PGCs for male germline commitment (Hayashi et al., 2008). By contrast, female germline‐committing PGCs show overexpression of miR‐29b, which targets DNA methyltransferase 3 alpha (Dnmt3a) and DNA methyltransferase 3 beta (Dnmt3b), and epigenetically regulates gene expression during oogenesis in mice (Takada et al., 2009). Embryonic loss of miRNAs resulting from Dicer1 deletion is associated with embryonic lethality in mice near the time of PGC specification (Bernstein et al., 2003). Dicer1 depletion just before PGC migration impaired the proliferation and gonad‐specific colonization of PGCs in mice (Hayashi et al., 2008), and oocytes originating from Dicer1‐ablated mouse germline cells failed to establish functional zygotes and commence first zygotic cell division (Tang et al., 2007). These findings indicate that sex differentiation and embryonic development require parentally derived miRNAs.
Figure 2.
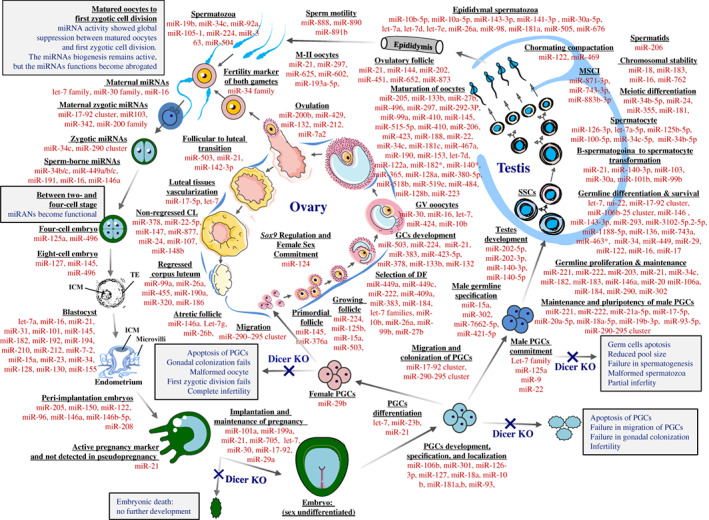
MicroRNA (miRNA) regulation at different stages of sex specification and gametogenesis. The miRNAs that are listed for different stages of oogenesis, spermatogenesis and early embryonic development are the predominantly expressed miRNAs of these stages. miRNAs are involved in every stage of early embryonic development and gametogenesis, except the period between the mature oocyte and first zygotic cell division, where the functions of miRNAs are abrogated (reported in mouse, but yet to be verified in other mammals), and then re‐established between two‐ and four‐cell stage pluripotent embryos. Loss of Dicer during the sex‐undifferentiated stage typically causes embryonic death, while primordial germ cell (PGC)‐specific loss causes the impairment and failure of migration, colonization, and commitment of sex‐specific gonads. miRNAs are highly expressed during gametogenesis, and some miRNAs have sex‐ and purpose‐specific roles; for example, miR‐124 plays an important role in female gonad commitment by suppressing Sox‐9 expression, and the loss of miR‐124 before sex commitment can cause sex reversal in females. Other miRNAs are equally important in nearly all stages; for example, miR‐21 is important in all stages of gametogenesis and is responsible for the viability and survival of cells. The expression of miR‐21 during pregnancy indicates the presence of activated and live embryo (s). CL, corpus luteum; TE, trophectoderm; KO, knockout; GV, germinal vesicle; GC, granulosa cell; DF, dominant follicle; MSCI, meiotic sex chromosome inactivation.
B‐lymphocyte‐induced maturation protein 1 (Blimp1) regulates PGC specification by repressing Hox genes (Ohinata et al., 2005; Vincent et al., 2005; Robertson et al., 2007) and is predominantly targeted by let‐7 (West et al., 2009). It is thought that suppression of Blimp1 by let‐7 upregulates Hox genes, leading totipotent PGCs towards male germline specification. Migration of PGCs to the female gonad is mediated by the miR‐17‐92 and miR‐290‐295 cluster, and the loss of this cluster disrupts PGC colonization in the female gonad, resulting in premature ovarian failure (POF) and sterility (Hayashi et al., 2008; Medeiros et al., 2011). The impact of PGC loss is much more severe in females than in males as female germline stem cells cannot propagate and self‐renew during adulthood, while spermatogonial stem cells (SSCs) successfully propagate and self‐renew throughout the lifetime, thus the loss of these miRNAs results in infertility in females but only subfertility in males.
miR‐22 suppresses oestrogen receptor 1 (ESR1)‐mediated oestrogen signalling in ovine foetal testes, which is necessary for male gonad establishment (Torley et al., 2011). Expression of miR‐202‐5p, miR‐202‐3p, miR‐140‐3p, and miR‐140‐5p is upregulated in committing testes and may mediate testis development as well as Leydig cell formation (Eggers, Ohnesorg, & Sinclair, 2014). By contrast, SRY‐box 9 (Sox9) suppression is required for female gonad commencement (Eggers et al., 2014); in its absence, sex reversal can occur (Eggers et al., 2014). Thus, Sox9 regulatory miR‐124 plays a key role in female gonad establishment (Eggers et al., 2014). let‐7, miR‐23b, and miR‐21 regulate male PGC differentiation by mediating Lin‐28 homolog A (Lin28)/let‐7/Blimp1 signalling (Brieno‐Enriquez et al., 2015). In mice, the abundance of miR‐485 and miR‐878‐5p changes in a time‐dependent manner in the ovary and testes and may be crucial for sex specification (Rakoczy et al., 2013). Similarly unique and differential expression of miRNAs is reported in the testes (Watanabe et al., 2006; Luo et al., 2010; Yang et al., 2013a; Luo et al., 2015b) and ovaries (Choi et al., 2007; Liang et al., 2007; Ahn et al., 2010; Nothnick, 2012; Xiao et al., 2014; Li et al., 2015) as well as other reproductive organs and tissues including testicular somatic cells (Rakoczy et al., 2013), epididymis (Belleannee et al., 2012; Chu et al., 2015; Nixon et al., 2015a, 2015b), fallopian tube (Nothnick, 2012), uterus (Nothnick, 2012) and cervix (Nothnick, 2012) of different mammalian species. Several miRNAs (such as let‐7, miR‐503, miR‐672, miR‐465, miR‐21‐5p and miR‐143‐3p) could play a housekeeping role (Ahn et al., 2010; Li et al., 2015) and the miR‐34 family can be considered as a fertility biomarker (Tscherner et al., 2014) in both sexes. Clearly, imbalances in miRNA regulation at any stage can disrupt the whole process of gonad specification and sex commitment and may cause sexual disorders as well as sex reversal of embryos.
IV. ROLES OF miRNA IN MALE REPRODUCTION
A range of miRNAs are male germ cell stage‐specific and are predominantly detected in male reproductive tissues (Fig. 2), with impaired miRNA expression causing reduced testes size, degenerated seminiferous tubules, impaired spermatogenesis, increased germ cell apoptosis, and the production of abnormal spermatozoa (fragmented acrosomes, impaired chromatin compaction, deformed sperm head, folded tail, immotile sperm), and a decrease in the number of mature spermatozoa, leading to infertility (Table S2).
(1). The formation of male gonads
Most miRNAs that are highly expressed in male PGCs are also detected in spermatogonia cells. Validated targets of these miRNAs are involved in mitogen‐activated protein kinase (MAPK), janus kinase (JAK)‐signal transducer and activator of transcription proteins (STAT), and transforming growth factor beta (TGFβ) signalling pathways (Wu et al., 2014; Garcia‐Lopez et al., 2015). A panel of miRNAs was detected in human and mouse testes (Watanabe et al., 2006). Among these miRNAs, miR‐449a is abundant in both mouse and human testes (Luo et al., 2015b). In boar testes, a subset of miRNAs are expressed differentially between mature and immature testes (Luo et al., 2010) and regulates stage‐specific testicular gene expression. For example, miR‐34b and miR‐34c regulate the differential expression of AQN‐1 protein (AQN‐1), hyaluronan synthase 3 (HAS3), ring finger protein 4 (RNF4), sperm mitochondria‐associated cysteine‐rich protein (SMCP), and sperm adhesion molecule 1 (SPAM1) between mature and immature porcine testes (Luo et al., 2010), indicating that these miRNAs play important roles in species‐ and stage‐specific male germ cell development. Similarly, the testicular somatic cells of fertile and infertile males express miRNAs differentially, for example miR‐202‐5p, a member of the let‐7 family, is selectively expressed in Sertoli cells of fertile males with normal germ cells, but not in the Sertoli cells of infertile males (Dabaja et al., 2015). Previous studies also showed that miR‐202‐5p, which is a downstream mediator of the testis‐determining factor SOX9 (Wainwright et al., 2013), is expressed in Sertoli cells in the early XY gonad (Rakoczy et al., 2013) and its expression is up‐regulated again in the testis at later developmental stages (Ro et al., 2007). These data indicate that miR‐202‐5p is closely associated with male gonad development.
(2). The commitment and maintenance of SSCs
Differential expression of miRNAs is observed at different stages of development and maturation of male germline cells (Fig. 2) (Hayashi et al., 2008). A group of these miRNAs regulating phosphatase and tensin homolog (Pten) and Stat signalling is predominantly downregulated in spermatogonia compared to PGCs, and thus enhances the expression of Pten and Stat levels in spermatogonia compared to PGCs (Garcia‐Lopez et al., 2015). Likewise, members of the miR‐290‐295 cluster are pluripotency related, and are downregulated in spermatogonia compared to PGCs and gonocytes (Zheng et al., 2011; McIver et al., 2012b), the proliferation‐ and differentiation‐regulatory miR‐143‐3p is upregulated in mice spermatogonia compared to in PGCs (Garcia‐Lopez et al., 2015), and miR‐293 is expressed differentially between gonocytes and spermatogonia (McIver et al., 2012b). This indicates that these miRNAs may specifically participate in the commitment of PGCs to SSCs. Among the miRNAs involved in SSC maintenance, miR‐21 regulates the stemness, proliferation, and survival of SSCs by regulating ETS variant 5 (ETV5)/glial cell‐derived neurotrophic factor (GDNF) signalling (Niu et al., 2011). GDNF is a well‐defined paracrine factor that promotes SSCs self‐renewal (Yang & Oatley, 2014), and is closely connected to the ret proto‐oncogene (RET) that is required for GDNF family receptor alpha 1 (GFRα1) signalling in mouse SSCs (Jing et al., 1996; Naughton et al., 2006). With the assistance of the phosphoinositide 3‐kinase (PI3K)/AKT serine/threonine kinase (AKT)‐dependent pathway or sarcoma (Src) family kinase pathway, GDNF regulates the expression of ETV5, inhibitor of DNA binding 4 (ID4), LIM homeobox 1 (LHX1), and POU class 3 homeobox 1 (POU3F1) to drive SSC self‐renewal (Lee et al., 2007; Oatley, Avarbock, & Brinster, 2007). Thus, miR‐21 appears to have a very important role in PGCs self‐renewal and maintenance.
Similarly, miR‐221/222 promotes the stemness characteristics of SSCs by downregulating KIT proto‐oncogene receptor tyrosine kinase (c‐Kit) expression in mice (Yang et al., 2013b), and both miR‐20 and miR‐106a directly target signal transducer and activator of transcription 3 (Stat3) and cyclin D1 (Ccnd1), and play critical roles in maintenance and renewal of SSCs by increasing proliferating cell nuclear antigen (Pcna) and promyelocytic leukaemia zinc finger (Plzf), and reducing c‐Kit expression in mice (He et al., 2013). miR‐290 and miR‐302 are also involved in the maintenance and pluripotency of SSCs in mice (Zovoilis et al., 2008). It is well known that miR‐135a and miR‐544 are essential factors for the regulation of self‐renewal of SSCs in rat and goat, respectively (Moritoki et al., 2014; Song et al., 2015). In addition, miR‐184 and miR‐204 are involved in mouse and goat SSC proliferation (Wu et al., 2011), and protocadherins, which are also important cell‐adhesion molecules, are also targeted by miR‐320 during germ cell biogenesis (Marcon et al., 2008). Differential expression of numerous miRNAs occurs between early and late stages of mammalian spermatogonia; these miRNAs are involved in either the maintenance or differentiation of SSCs (Niu et al., 2011; Tong et al., 2011, 2012; He et al., 2013; Yang et al., 2013b). Thus, growing evidence strongly suggests that many miRNAs are closely linked to the post‐transcriptional regulation of SSCs self‐renewal and maintenance.
(3). The differentiation of male SSCs to spermatocytes
A global shift in testicular miRNA expression is observed during the very early stages of spermatogenesis and differentiation of spermatogonia (Golestaneh et al., 2009). For example, miR‐146 downregulates mediator complex subunit 1 (Med1), c‐Kit, and spermatogenesis and oogenesis specific basic helix‐loop‐helix 2 (Sohlh2), and mediates the differentiation of SSCs (Huszar & Payne, 2013). c‐Kit is required for the differentiation of SSCs and is a target of miR‐221 and miR‐222 (Yang et al., 2013b). Thus, suppression of miR‐221 and miR‐222 may promote the differentiation of SSCs. Also, miR‐34c promotes SSC differentiation in mice by suppressing nanos C2HC‐type zinc finger 2 (Nanos2) and simultaneously upregulating stimulated by retinoic acid 8 (Stra8) and synaptonemal complex protein 3 (Scp3) (Yu et al., 2014). In addition, members of the miR‐34 family regulate notch 1 (NOTCH1), notch 2 (NOTCH2), TGFB‐induced factor homeobox 2 (TGIF2), cyclin dependent kinase 4 (CDK4), and MYC proto‐oncogene, BHLH transcription factor (c‐MYC) (Yan et al., 2009; Bouhallier et al., 2010), and miR‐449 regulates NOTCH1 and B‐cell lymphoma 2 (BCL2) (Yan et al., 2009); the genes together regulate the survival and differentiation of SSCs through Notch and Tgfβ signalling in mammals (Damestoy et al., 2005; Kostereva & Hofmann, 2008; Yan et al., 2009). High mobility group AT‐hook 2 (Hmga2) and outer dense fiber of sperm tails 1 (ODF1) are involved in the structural organization and differentiation of germ cells in mice and human, respectively (Chieffi et al., 2002; Zhao et al., 2007), and are targeted by the let‐7 family and miR‐22, respectively (Curry, Safranski, & Pratt, 2011). The let‐7 family also targets insulin like growth factor 1 (Igf1) and may control spermatogonial differentiation in mice through extracellular signal‐regulated kinase 1/2 (Erk1/2) and PI3K signalling (Shen et al., 2014). miR‐383 regulates mitotic division in spermatogonia and primary spermatocytes by targeting interferon regulatory factor 1 (IRF1), and subsequently downregulating CCND1, cyclin dependent kinase 2 (CDK2), and cyclin dependent kinase inhibitor 1A (p21) (Lian et al., 2010; Luo, Hou, & Yang, 2016). Additionally, miR‐294 and miR‐295 are specific to SSCs, while miR‐201 and miR‐547 are specific to premeiotic male germ cells (Smorag et al., 2012). This indicates that these miRNAs regulate differentiation of SSCs by controlling mainly NOTCH, Erk1/2 and PI3K signalling.
Transition of B‐spermatogonia to spermatocytes accompanies major alterations in miRNA profiles (Liu et al., 2015), and the deregulated miRNAs regulate the differential expression of E2F transcription factor 1 (E2F1), ETS variant 1 (ETV1), TNF alpha‐induced protein 8 like 2 (TNFAIP8l2), target of Myb1 membrane trafficking protein (TOM1), transforming growth factor beta receptor 1 (TGFBR1), and bone morphogenetic protein receptor type 2 (BMPR2) between spermatogonia and spermatocytes (Liu et al., 2015), potentially regulating apoptosis, proliferation, differentiation, junctional assembly, and cell cycle during spermatogenesis (Luo et al., 2015a). For example, methyl‐CpG binding domain protein 6 (MBD6) and H2A histone family member X (H2AX) are important for meiotic division during spermatogenesis and are putative targets of miR‐24 (McIver et al., 2012a). c‐Kit, RNA binding motif protein 44 (Rbm44), and cyclin dependent kinase 6 (Cdk6) play roles in SSC maintenance and meiotic differentiation in mice and are regulated by miR‐221, miR‐203, and miR‐34b‐5p, respectively (Smorag et al., 2012). In the early stages of male germ cell specification in mice, cyclin T2 (Ccnt2) is regulated by miR‐15a, which is expressed stage specifically and time dependently during spermatogenesis (Teng et al., 2011). miR‐15b targets GDP dissociation inhibitor 1 (GDI1) and isocitrate dehydrogenase 3 (NAD (+)) alpha (IDH3A), and regulates TCA‐cycle mediated energy metabolism (Curry et al., 2011). Heat shock transcription factor 2 (Hsf2), an important molecule required for mammalian spermatogenesis, is regulated by miR‐18 during spermatogenesis in mice (Bjork et al., 2010). miR‐18, which belongs to the miR‐17‐92 cluster, is abundantly expressed in spermatocytes. Finally, miR‐34b‐5p regulates meiotic progression by targeting Cdk6 (Smorag et al., 2012 ). Taken together, miRNAs are important regulators of cell cycle progression and differentiation during the transition of B‐spermatogonia to spermatocytes.
(4). Meiotic sex chromosome inactivation (MSCI) and chromosomal stability during spermatogenesis
MSCI refers to the transcriptional silencing of genes located on the X‐chromosome at the pachytene spermatocyte stage. While most X‐chromosomal genes are silenced, X‐chromosome linked miRNAs including miR‐718‐3p, miR‐883a‐3p, and miR‐883a‐5p can escape MSCI‐mediated silencing in mice (Song et al., 2009). This exceptional event allows these miRNAs to remain active throughout spermatogenesis and play a crucial role in the differentiation of SSCs (Song et al., 2009). miRNAs are fine‐tuners of many important genes, and thus their absence can cause overexpression of X‐linked genes that are normally silenced by MSCI (Greenlee et al., 2012), ultimately leading to the impairment of meiotic progress, acrosomal defects, abnormal chromatin condensation and nuclear shaping, degenerated seminiferous tubules, mixing of different stages of haploid spermatogonia, and the presence of exfoliated and immature germ cells in the epididymis (Korhonen et al., 2011), followed by infertility in males (Greenlee et al., 2012). Although miRNAs such as miR‐871‐3p, miR‐743‐3p, and miR‐883b‐3p may regulate MSCI‐mediated gene expression (Song et al., 2009), but there is not yet direct evidence for this. miRNAs are also important for maintaining chromosomal stability; the loss of several miRNAs such as miR‐16, miR‐18, miR‐183, and miR‐762 caused abundant activation of ATM serine/threonine kinase (ATM) and deregulation of genes including mediator of DNA damage checkpoint 1 (Mdc1), Cdk2, trimethylation of lysine 9 on histone H3 (H3K9me3), and ring finger protein 8 (Rnf8), which are involved in chromosomal stability and DNA repair during spermatogenesis in mice (Modzelewski et al., 2015). miR‐762 downregulates DNA damage‐related gene H2A histone family member X (γ‐H2AX) and upregulates the DNA damage repair‐related genes RNF8, ring finger protein 168 (RNF168), DNA repair associated (BRCA1), tumor protein P53 binding protein 1 (TP53BP1), and poly (ADP‐ribose) polymerase 1 (PARP), as well as promoting the growth of immature Sertoli cells in pigs (Ma et al., 2016a), indicating that both miRNA‐mediated gene regulation and fine‐tuning of gene expression are critical factors for MSCI and chromosomal stability during spermatogenesis.
(5). miRNAs as regulators of spermiogenesis and chromatin body compaction
A wide range of miRNAs are expressed differentially during the transition of pachytene spermatocytes to round spermatids and might be important in the spermiogenesis process. For example, round spermatid basic protein 1 (Rsbn1) is an important molecule for the transcriptional regulation of haploid germ cell formation and is a target gene of miR‐355, miR‐181b, and miR‐181c (Yan et al., 2007; McIver et al., 2012a). In addition, miR‐122 downregulates transition protein 2 (Tnp2) (Yu, Raabe, & Hecht, 2005), which replaces protamine, an essential component for the compaction and patterning of chromatin bodies into small sperm heads (Krawetz et al., 2011); testis‐specific miR‐469 also plays an important role in chromatin remodelling and elongation of spermatids by repressing Tnp2 and protamine 2 (Prm2) (de Mateo & Sassone‐Corsi, 2014). Similarly, loss of miRNA‐dependent regulation of Arpc5 impairs translational activation of protamine 1 (Prm1)/Prm2 by blocking 80S ribosome formation and facilitating mRNA transportation to chromatoid/P bodies in mice (Chang et al., 2012). miRNA‐dependent regulation of tudor domain containing 6 (Tdrd6) during spermiogenesis of mice is essential for chromatin body formation (Vasileva et al., 2009). However, unlike the early stages of spermatogenesis, most of these miRNAs disappear during spermiogenesis, leaving a few sperm‐borne miRNAs such as miR‐34 to regulate fertilization and early zygote development (Liu et al., 2012). Therefore, some of the sperm‐borne miRNAs are important epigenetic contributors to the embryo. However, the roles of miRNAs in chromatin compaction require further investigation to determine whether the impairments caused by deregulated miRNAs are permanent or recoverable.
(6). miRNAs regulate maturation and viability of spermatozoa during storage and transportation through the epididymis
Epididymal spermatozoa are enriched with distinctive miRNAs (Fig. 2), which are uncommon in other spermatozoal stages (Nixon et al., 2015b; Reilly et al., 2016). Additionally, epididymal epithelial cell‐derived miRNAs may influence the post‐testicular progression of sperm to their destination and mediate cell‐to‐cell communication (Belleannee et al., 2013a; Belleannee, 2015; Qing et al., 2017); vasectomy results in alteration of miRNAs in both epididymal and seminal microvesicles (Belleannee et al., 2013b). Differential expression of miRNAs is also found between epididymal and freshly ejaculated sperm, as well as among the segments (caput, corpus, cauda) of the epididymis in different species including human (Belleannee et al., 2012), horse (Twenter et al., 2017), mouse (Nixon et al., 2015a, b ), pig (Chang et al., 2016) and bull (Belleannee et al., 2013a). These miRNAs might be involved in spermatozoa maturation (Twenter et al., 2017), intracellular trafficking signalling (Nixon et al., 2015a), death signalling (Chu et al., 2015), and epididymal cell proliferation (Ma et al., 2012). The differential expression of miRNAs among the epididymal segments is common to various mammalian species, thus it could be assumed that the contribution of epididymal miRNAs is an essential part of mammalian sperm maturation and concentration, and that these epididymal miRNAs potentially contribute to the motility of spermatozoa as well as to the content of the seminal fluid.
(7). miRNAs are potent regulators of male germline cell apoptosis in mammals
Apoptosis in spermatocytes is a common event as meiotic cells have to undergo homologous recombination through double‐strand breaks (Hamer et al., 2003). Several miRNAs such as the miR‐449 cluster and miR‐34b/c, regulate male germ cells by targeting the E2F‐retinoblastoma protein (pRb) pathway in the early meiotic phase (Bao et al., 2012). In male sheep with disrupted spermatogenesis, the expression of miR‐144 (homolog miR‐98) was upregulated; miR‐144 regulates the apoptosis‐related genes fas ligand (FASL), caspase 3 (CAS3), tumor protein 53 (TP53), and BCL2‐like 1 (BCL2L1) (Guan et al., 2015). Furthermore, miR‐26a induces G1 arrest by regulating cyclin E1 (CCNE1), cyclin E2 (CCNE2), cyclin D2 (CCND2), and CDK6 (Kota et al., 2009) and then induces apoptosis in parallel with the direct effects of miRNA‐144. Similarly, changes in the expression of miRNA‐99a seem to affect the organization of Sertoli‐cell tight junctions by targeting zona occludens 1 (ZO‐1) (Turcatel et al., 2012), whereas miR‐34c enhances germinal phenotypes in late spermatogenesis (Bouhallier et al., 2010).
In oestradiol benzoate‐treated rats, the miR‐29 family resulted in extensive apoptosis of male germ cells by downregulating Dnmt and myeloid cell leukemia 1 (Mcl‐1) (Meunier et al., 2012). Ochratoxin‐A treatment induces extensive apoptosis of male germ cells, while inhibition of miR‐122 minimizes this Ochratoxin‐A‐mediated toxicity (Chen et al., 2015a), indicating that miR‐122 is a critical regulator for the germ cell apoptosis‐induction pathway. In pigs, expression of miR‐16 and miR‐34 regulates the cell cycle progression during spermatogenesis and senescence of sperm (Luo et al., 2015b). miR‐16 promotes the apoptosis of immortalized GC‐1 spermatogonia cells (GC‐1 SPG) by downregulating Ccnd1 (Li et al., 2016b). Deletion of the miR‐17‐92 cluster, which is potentially regulated by transcriptional regulation of c‐MYC and E2F1 (Novotny et al., 2007), results in testicular atrophy and empty seminiferous tubules by reducing SSCs and increasing germ cell apoptosis (Xie et al., 2016). Taken together, these observations suggest that deregulation in miRNA expression is an important mechanism behind mammalian male germline cell apoptosis.
V. ROLES OF miRNAs IN FEMALE REPRODUCTION
Sex‐specific and unique miRNA profiles are observed at every step of female reproduction (Fig. 2): female‐specific miR‐124 promotes the commitment of female gonads and prevents sex reversal by silencing Sox9 in the mouse ovary. Similarly, many miRNAs play substantial roles throughout folliculogenesis and oogenesis by regulating expression of important genes (see Table S3), and many are specific to particular developmental stages of the follicles, as well as segments of the reproductive tract.
(1). Growth and maturation of ovarian follicles
Folliculogenesis refers to the progression of small primordial follicles into large preovulatory follicles that occurs in part during the oestrus cycle. During folliculogenesis, the majority of follicles commit to atresia, and a few develop into Graafian follicles. miR‐145 regulates initiation of the growth, development, and maintenance of mouse primordial follicles (Fig. 2) by regulating Tgfβ signalling including transforming growth factor beta receptor 2 (Tgfbr2), activin A receptor type 1B (Acvr1b), SMAD family member 3 (Smad3), and SMAD family member 5 (Smad5) (Yang et al., 2013c), while loss of miR‐145 over‐activates primordial follicles and deregulates zona pellucida in growing follicles (Yang et al., 2013c). Similarly, miR‐224 enhances folliculogenesis in mice by targeting SMAD family member 4 (Smad4) and regulating transforming growth factor beta 1 (Tgfβ1)‐induced proliferation of granulosa cells (GCs) (Yao et al., 2010a), and miR‐133b regulates the development of ovaries and maturation of oocytes by targeting transgelin 2 (TAGLN2), an actin protein‐encoding gene (Xiao et al., 2014). In porcine oocytes, miR‐205 participates in brain‐derived neurotrophic factor (BDNF)‐induced maturation by targeting pentraxin 3 (PTX3) (Li et al., 2016a) and miR‐29 has a role during the development of bovine follicles (Hossain et al., 2009).
Numerous miRNAs play a role in growing follicles (Fig. 2) (Lei et al., 2010; Yao et al., 2010a; Xu et al., 2011b; Sen et al., 2014), selection of dominant follicles (Salilew‐Wondim et al., 2014), formation of germinal vesicle (GV) oocytes (Murchison et al., 2007; Tripurani et al., 2010), maturation of oocytes (Tesfaye et al., 2009; Abd El Naby et al., 2013; Xiao et al., 2014; Song et al., 2016; Li et al., 2016a), ovulatory follicles (Carletti, Fiedler, & Christenson, 2010; Sontakke et al., 2014; Wright et al., 2016), the ovulation process (Fiedler et al., 2008; Hasuwa et al., 2013; Ahmed et al., 2017), and development of metaphase II (M‐II) oocytes (Xu et al., 2011b; Wright et al., 2016). Among these miRNAs, miR‐202 is gonad‐specific and might be important in the prevention of premature ovarian failure (POF) (Sontakke et al., 2014), and miR‐27b and its target, peroxisome proliferator activated receptor gamma (PPARγ), is crucial for the maturation of porcine oocytes (Song et al., 2016). In humans, miR‐15a potentially regulates the growth and maturation of oocytes by regulating BCL2 and cell division cycle 25A (CDC25A) (Xu et al., 2011b), while miR‐335‐5p regulates spindle formation and cytoskeleton dynamics via MAPK signalling in mouse oocytes (Cui et al., 2013). These data indicate that each stage of folliculogenesis has a unique profile of miRNA expression and that miRNAs are involved in the selection, growth, development, and maturation of ovarian follicles.
(2). miRNA expressions varies in the granulosa cells (GCs) and thecal cells (TCs) of the dominant follicles (DFs) and subordinate follicles (SFs)
DFs and SFs show differential expression of some miRNAs, which may regulate gonadotropin‐releasing hormone (GnRH) signalling (Salilew‐Wondim et al., 2014). These miRNAs are expressed differentially among different subgroups of GCs (Tesfaye et al., 2009), TCs (Zielak‐Steciwko et al., 2014), and between mural and cumulus cells (Velthut‐Meikas et al., 2013). These miRNAs are also potential regulators of numerous signalling pathways including meiosis, wingless‐type MMTV integration site family (WNT), TGFβ, ErbB receptor tyrosine kinases (ERBB), insulin, P13K–AKT, and MAPK (Zielak‐Steciwko et al., 2014). Another panel of miRNAs including miR‐125b, miR‐145, and miR‐199a‐3p target leukemia inhibitory factor (LIF), cyclin dependent kinase inhibitor 1A (CDKN1A), and prostaglandin‐endoperoxide synthase 2 (PTGS2), respectively, in GCs, but not in TCs of bovine follicles (Donadeu et al., 2012). Furthermore, a panel of miRNAs is highly expressed in atretic follicles compared to healthy follicles, with all miRNAs except miR‐21‐3p and miR‐378 showing higher expression in TCs compared to GCs (Donadeu, Mohammed, & Ioannidis, 2017). This suggests that these miRNAs might regulate the phenotypic characters of different cell types by controlling the expression of important signalling molecules (Donadeu et al., 2017). Additionally, miR‐708, miR‐221, miR‐21‐3p, miR‐335, and miR‐214 are expressed abundantly in the follicular fluid of SFs compared to that of DFs (Gebremedhn et al., 2015) indicating a potential role in the selection of follicles. Hence, differential expression of miRNAs between TCs and GCs may play an important role in the selection and recruitment of DFs, as well as during the follicular wave (in different mammalian species the recruitment, growth and development of several waves of follicles is possible before follicle rupture and ovulation occur; during this process two or more follicles might develop, but only one follicle successfully releases the egg) and atresia.
(3). Atresia and degeneration of ovarian follicles
miRNAs are potent regulators of follicular atresia and degeneration (Fig. 2) (Lin et al., 2012; Cao et al., 2015; Chen et al., 2015b). In brief, the apoptosis of human GCs is regulated by miR‐146a, which directly targets interleukin 1 receptor associated kinase 1 (IRAK1) and TNF receptor associated factor 6 (TRAF6), and indirectly regulates nuclear factor kappa B (NFκB), phospho‐inhibitory subunit of NF kappa B alpha (pIκBα), caspase 8 (CAS8), caspase 9 (CAS9), CAS3, and PARP (Chen et al., 2015b). Follicular atresia in pigs is regulated by members of the let‐7 family, which regulate MAPK, TGFβ, and p53 signalling pathways; all other members are downregulated in atretic and progressively atretic follicles except let‐7 g (Cao et al., 2015). In addition, miR‐26b targets ATM and enhances GC apoptosis during follicular atresia in pigs (Lin et al., 2012). miR‐181a, which is highly expressed in primary follicles over preantral and antral follicles and leads to POF (Zhang et al., 2013b) inhibits the production of GCs and follicular development in mice by suppressing Acvr2a and decreasing phosphorylation of Activin‐signalling. Similarly, miR‐143 inhibits the proliferation of pregranulosa cells in mice by regulating Cdks, Ccnb1, Ccnd2, and Ccne2, and thus inhibits the formation of primordial follicles (Zhang et al., 2013a). In cattle, miR‐21‐5p, miR‐21‐3p, miR‐222, miR‐155, and miR‐199a‐5p are upregulated in atretic follicles compared to healthy follicles (Donadeu et al., 2017). Another two miRNAs, miR‐378 and miR‐21‐5p, were increased in subordinate and anovulatory follicles in horses (Donadeu & Schauer, 2013; Schauer et al., 2013). Of note, miR‐21 prevents follicular atresia and regulates follicle survival, follicular to luteal transition, and ovulation (Carletti et al., 2010). Androgen‐induced miR‐125b suppresses the pro‐apoptotic genes BCL2 antagonist/killer 1 (Bak), BCL2 associated X protein (Bax), Bcl2 modifying factor (Bmf), and TP53 in mouse GCs and promotes follicular survival by preventing atresia (Sen et al., 2014). miR‐376a reduces oocyte apoptosis, increases primordial follicle number, and regulates follicular assembly by modulating Pcna in mice (Zhang et al., 2014). Therefore, miRNAs appear to have a very important regulatory role during apoptosis of GCs and follicular atresia.
(4). Regulation of luteinization and formation of the CL
The importance of miRNA signalling for regulation of the CL has been demonstrated using hypomorphic Dicer1 mutant mice, which fail to implant embryos and maintain pregnancy due to impaired growth of new capillary vessels in the CL (Otsuka et al., 2008). The study demonstrated that the impaired CL angiogenesis in Dicer1 (d/d) mice was associated with a lack of miR‐17‐5p and let‐7b, which target tissue inhibitor of metalloproteinases 1 (Timp1). These data suggest that the pro‐angiogenic miR‐17‐5p and let‐7b are critical factors for the normal development of the CL. miRNAs are known to be involved in the process of CL formation (Fig. 2) such as during follicular to luteal transition (McBride et al., 2012), vascularization of luteal tissue (Otsuka et al., 2008), and in non‐regressed CL (Ma et al., 2011), and regressed CL (Ma et al., 2011). In particular, reduction of miR‐503 expression in pre‐ovulatory follicles, signals the onset of luteinization, with its expression subsequently restored during CL development (Lei et al., 2010; McBride et al., 2012). miR‐503 directly regulates Gdf9, follicle stimulating hormone Receptor (Fshr), oestrogen receptor 2 (Esr2), activin A receptor type 2A (ActRIIa), Ccnd2, and activin A receptor type 2B (ActRIIb) and indirectly regulates inhibin alpha subunit (Inha), Inhba, Inhbb, Cyp19a1, luteinizing hormone/choriogonadotropin receptor (Lhcgr), Esr2, androgen receptor (Ar), Cdkn1b, and Cas3 (Lei et al., 2010). Thus, miR‐503 could have a direct involvement in CL formation and maintenance. Another study showed that miR‐378 plays an anti‐apoptotic role in luteal cells of non‐regressed CL by targeting interferon gamma receptor 1 (IFNGR1) (Ma et al., 2011). These observations provide evidence to explain the physiological requirement for miRNAs during luteal development.
(5). The prediction of oocyte quality
miRNAs are expressed abundantly in follicular fluid, cumulus cells, and oocytes. Therefore, it may be possible to use miRNAs to evaluate the quality of oocytes. Uhde et al. (2017) attempted to assess the quality of bovine oocytes based on the miRNA expression of the surrounding cumulus cells although they concluded that the quality of pre‐fertilization‐stage oocytes could not be determined in this way. As highlighted in Fig. 2, the functions of miRNAs are globally suppressed between matured oocytes and first zygotic cell division stages (Suh et al., 2010). As a result, the functional activity of miRNAs are generally abrogated (although the expression of miRNAs remains abundant) during those stages (Tang et al., 2007; Suh et al., 2010) representing an obstacle to the use of miRNAs as biomarkers to assess the quality of pre‐fertilization‐stage oocytes. However, the roles of maternal miRNAs (oocyte‐derived miRNAs) are indispensable during the post‐fertilization stages (Tang et al., 2007; Karakaya et al., 2015), especially during and after zygotic genome activation. It may be that the roles of oocyte‐derived miRNAs will be understood only by monitoring the post‐fertilization development potential of oocytes.
(6). miRNAs and extracellular vesicles within follicular fluid
Extracellular vesicles including exosomes are tiny particles of biological origin, contain RNAs, proteins, lipids and other small molecules, and are transported by the circulatory system to neighbouring or distant organs (da Silveira et al., 2012; Reza et al., 2016). These particles function as a cargo transport system and participate in cell–cell communication, embryo–maternal crosstalk (Saadeldin, Oh, & Lee, 2015) and communication among ovarian follicles (da Silveira et al., 2012). Vesicular miRNAs derived from follicular fluid are being investigated for their suitability as biomarkers for oocyte quality in matured and immature oocytes (Sohel et al., 2013; Santonocito et al., 2014; da Silveira et al., 2015), as well as in healthy and unhealthy follicles (Sang et al., 2013; Roth et al., 2014). For example, matured follicle‐derived microvesicles are enriched with a panel of miRNAs including miR‐99a, miR‐100, miR‐132 and miR‐218 (Santonocito et al., 2014), among which miR‐132, miR‐212 and miR‐214 negatively regulate the factors related to the inhibition of follicular maturation while miR‐29a is involved in epigenetic changes (Santonocito et al., 2014). In addition, follicular‐fluid‐derived circulatory miRNAs are potentially involved in steroidogenesis processes (Sang et al., 2013), for example, miR‐132, miR‐320, miR‐520c‐3p, miR‐24 and miR‐222 regulate the concentration of oestradiol, while miR‐24, miR‐193b and miR‐483‐5p regulate the concentration of progesterone (Sang et al., 2013). These data indicate that follicular‐fluid‐derived extracellular miRNAs could be an important tool to assess oocyte quality and to differentiate between healthy and unhealthy follicles.
VI. miRNAs are potent regulators of the steroidogenic process during gametogenesis
miRNAs are potent regulators of testicular, ovarian, hypothalamic and pituitary hormones (Figs 3 and 4) (Messina & Prevot, 2017), and regulate the expression and functions of numerous steroidogenic genes (see Table S4). Thus miRNAs act as an essential mediator to maintain interactions among the hypothalamus, pituitary, and gonad.
Figure 3.
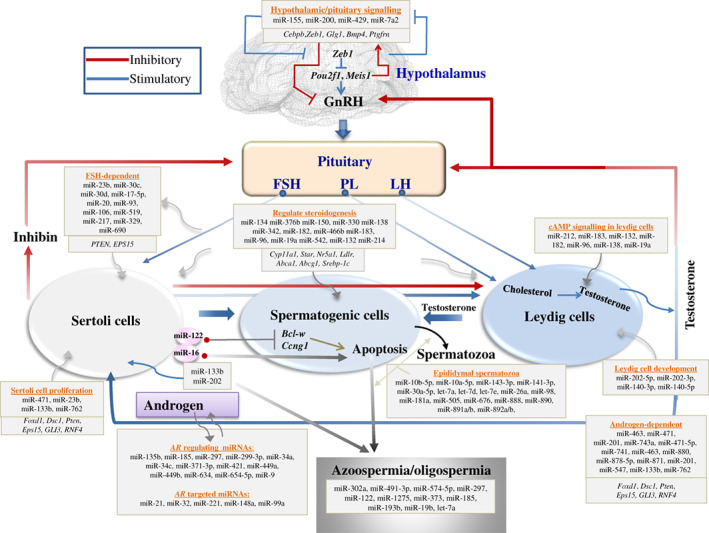
MicroRNA (miRNA) regulation in the maintenance of male hormonal balance. A panel of miRNAs regulates Pou2f1‐ and Meis1‐mediated gonadotropin‐releasing hormone signalling by targeting Zeb1, and thus these miRNAs may play important roles in the maintenance of hormonal balance. The expression of miRNAs changes with levels of different reproductive hormones; additionally, hormonal balance is reported to be miRNA‐dependent, as miRNAs regulate the proliferation, differentiation, function, and apoptosis of male steroidogenic cells. Thus, miRNAs, genes, and hormones develop a feedback loop that optimizes spermatogenesis; any imbalance in the feedback loop can result in azoospermia, oligospermia, infertility, and reproductive failure in males. FSH, follicle‐stimulating hormone; LH, luteneizing hormone; PL, prolactin; GnRH, gonadotropin‐releasing hormone; PTEN, phosphatase and tensin homolog; EPS15, epidermal growth factor receptor pathway substrate 15; AR, androgen receptor; Cebpb, CCAAT/enhancer binding protein beta; Zeb1, zinc finger E‐box binding homeobox 1; Glg1, golgi glycoprotein 1; BMP4, bone morphogenetic protein 4; Ptgfrn, prostaglandin F2 receptor inhibitor; Pou2f1, POU class 2 homeobox 1; Meis1, meis homeobox 1; Cyp11a1, cytochrome P450 family 11 subfamily A member 1; Star, steroidogenic acute regulatory protein; Nr5a1, nuclear receptor subfamily 5 group A member 1; Ldlr, low density lipoprotein receptor; Abca1, ATP binding cassette subfamily A Member 1; Abcg1, ATP binding cassette subfamily G member 1; Srebp‐1c, sterol regulatory element binding transcription factor 1; Bcl‐w, BCL2 like 2; Ccng1, cyclin G1; Foxd1, forkhead box D1; Dsc1, desmocollin 1; GLI3, GLI family zinc finger 3; RNF4, ring finger protein 4.
Figure 4.
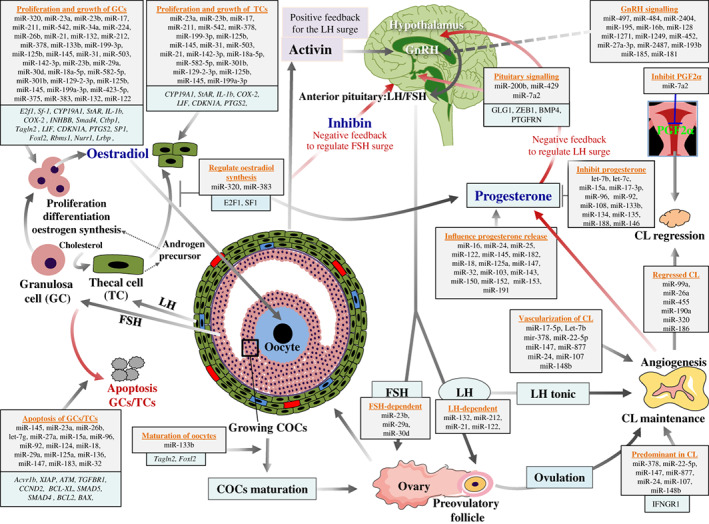
MicroRNA (miRNA) regulation in the maintenance of female hormonal balance. The proliferation, growth, differentiation, functioning, and apoptosis of female steroidogenic cells [granulosa cell (GC) and thecal cell (TC)] are highly regulated by miRNAs. miRNAs are also active regulators of female steroidogenic genes including cytochrome P450 family 19 subfamily A member 1 (Cyp19a1); steroidogenic acute regulatory protein (Star), and prostaglandin‐endoperoxide synthase 2 (Ptgs2); thus, miRNAs directly regulate the steroidogenic process in females through gonadotropin‐releasing hormone (GnRH), follicle‐stimulating hormone (FSH), and luteinizing hormone (LH) signalling. The vasculogenesis and angiogenesis of progesterone‐producing corpus lutem (CL) is also related to the expression of miRNAs, making miRNAs important regulatory molecules during the female steroidogenic processes. miRNAs are involved in both follicular steroidogenesis and luteal steroidogenesis. PGF2α, prostaglandin; COC, cumulus‐oocyte complex; E2f1, E2F transcription factor 1; Sf‐1, steroidogenic factor 1 nuclear receptor; IL‐1b, interleukin 1 beta; COX‐2, cyclooxygenase 2; INHBB, inhibin beta B subunit; Ctbp1, C‐terminal binding protein 1; Tagln2, transgelin 2; LIF, leukemia inhibitory factor; CDKN1A, cyclin dependent kinase inhibitor 1A; SP1, specificity protein 1; Foxl2, forkhead box L2; Rbms1, RNA binding motif single stranded interacting protein 1; Nurr1, nuclear receptor subfamily 4 group A member 2; Lrbp, LH receptor mRNA‐binding protein; Acvr1b, activin A receptor type 1B; XIAP, X‐linked inhibitor of apoptosis; ATM, ATM serine/threonine kinase; TGFBR1, transforming growth factor beta receptor 1; CCND2, cyclin D2; BCL‐XL, BCL2 like 1; SMAD5, SMAD family member 5; SMAD4, SMAD family member 4; BCL2, B‐cell lymphoma 2; BAX, BCL2 associated X protein; GLG1, Golgi glycoprotein 1; ZEB1, zinc finger E‐box binding homeobox 1; BMP4, bone morphogenetic protein 4; PTGFRN, prostaglandin F2 receptor inhibitor; IFNGR1, interferon gamma receptor 1.
(1). Hormonal balance and steroidogenesis in males
miRNAs and hormones together with genes develop an effective feedback loop to coordinate successful spermatogenesis (Fig. 3). The genes in the hypothalamus that regulate GnRH release and pituitary functions including zinc finger E‐box binding homeobox 1 (Zeb1), POU class 2 homeobox (Pou2f), meis homeobox 1 (Meis1), golgi glycoprotein 1 (Glg1), bone morphogenetic protein 4 (Bmp4) and prostaglandin F2 receptor inhibitor (Ptgfrn), are effectively controlled by a panel of miRNAs including miR‐155, miR‐200, miR‐429 and miR‐7a2 (Hasuwa et al., 2013; Ahmed et al., 2017; Messina & Prevot, 2017). Therefore, these miRNAs might have direct or indirect control over the hormones released by the pituitary. Following downstream signalling from the pituitary gland, follicle‐stimulating hormone (FSH), luteinizing hormone (LH) and androgens become functional or inactivated, and this could stimulate the expression of different miRNAs in the testes (Panneerdoss et al., 2012; Ma et al., 2016a), leading to miRNA‐mediated regulation of growth, proliferation, maintenance and apoptosis of testicular cells (Nicholls et al., 2011; Panneerdoss et al., 2012; Shih et al., 2015; Yao et al., 2016; Ma et al., 2016a). Alternatively, any deregulation of miRNAs in the testes could alter the expression of important genes that could disrupt the hormone balance, resulting in spermatogenesis failure, reproductive diseases and infertility such as azoospermia or oligospermia in males (Eggers et al., 2014; Hu et al., 2013; Shih et al., 2015; Ghorbian, 2012).
In human, miR‐133b regulates Sertoli cell proliferation by directly targeting GLI family zinc finger 3 (GLI3) following the upregulation of CCNB1 and CCND1 (Yao et al., 2016). In pigs, miR‐762 promotes the growth and proliferation of immature Sertoli cells by targeting RNF4 (Ma et al., 2016a). In the mouse, miR‐471 directly participates in steroidogenesis by regulating forkhead box D1 (Foxd1) and desmocollin 1 (Dsc1): these genes regulate the metabolism of Sertoli cells and cell–cell adhesion in the testis, respectively (Panneerdoss et al., 2012). In the rat, miR‐132 and miR‐214 repress sterol regulatory element binding transcription factor 1 (Srebp‐1c) and low density lipoprotein receptor (Ldlr), respectively, and both regulate steroidogenesis (Hu et al., 2013). miRNA‐23b regulates Pten and epidermal growth factor receptor pathway substrate 15 (Eps15) in an FSH‐dependent manner, and its function is implicated in endocytosis, focal adhesion, and actin cytoskeleton regulation during spermatogenesis (Nicholls et al., 2011). By contrast, numerous miRNAs in steroidogenic cells are expressed in an FSH‐ and androgen‐dependent manner (Nicholls et al., 2011; Panneerdoss et al., 2012; Shih et al., 2015). This indicates that the expression of miRNAs and hormones are interdependent; a change in one of them can alter the expression of the other. It has been reported that patients suffering from azoospermia and oligospermia showed deregulated miRNA expression profiles (Ghorbian, 2012). Therefore, miRNAs and hormones potentially develop a feedback loop in the testes, and hormone‐mediated gene expression in the testes is regulated in a miRNA‐dependent manner.
(2). Hormonal balance and steroidogenesis in females
miRNAs mainly regulate steroidogenesis in females by regulating the growth, proliferation, and apoptosis of female steroidogenic cells, GCs and TCs. In addition, hypothalamic GnRH signalling following changes in levels of FSH and LH is also regulated by a panel of miRNAs (Hasuwa et al., 2013; Ahmed et al., 2017; Messina & Prevot, 2017). The synthesis and inhibition of steroidogenic hormones including oestrogen, progesterone, and prostaglandin (PGF2α) are also regulated in a miRNA‐dependent manner. In humans, miR‐23a/b targets CYP19A1 (Alford et al., 2007), and miR‐17, miR‐211, and miR‐542 target steroidogenic acute regulatory protein (StAR), interleukin 1 beta (IL‐1b), and cyclooxygenase 2 (COX‐2), respectively. Thus, these miRNAs regulate the steroidogenesis process in follicular cells (Toloubeydokhti, Bukulmez, & Chegini, 2008). In pigs, miR‐34a and miR‐26b target INHBB and SMAD4, respectively, and negatively regulate the steroidogenic process by promoting the apoptosis of GCs (Liu et al., 2014; Tu et al., 2014). Similarly, miR‐423‐5p and miR‐378 post‐transcriptionally downregulate CYP19A1 (Xu et al., 2011a; Sui et al., 2014) and miR‐375 binds to the 3′ untranslated region of specificity protein 1 (SP1) and mediates corticotropin releasing hormone (CRH) signalling (Yu et al., 2017): these miRNAs directly regulate oestradiol synthesis in porcine GCs. In mice, miR‐224 enhances Tgfβ‐induced proliferation of GCs by targeting Smad4 and promotes oestradiol biogenesis by increasing Cyp19a1 (Yao et al., 2010a). miR‐21 plays anti‐apoptotic roles and facilitates the maintenance of GCs, both in vitro and in vivo (Carletti et al., 2010), while miR‐132 and miR‐212 regulate the differentiation of GCs by targeting C‐terminal binding protein 1 (Ctbp1) (Fiedler et al., 2008). miR‐132 regulates cyclic adenosine monophosphate (cAMP) signalling and promotes oestradiol production in mouse GCs through translational repression of nuclear receptor related 1 (Nurr1) following upregulation of Cyp19a1 (Wu et al., 2015), and miR‐122 increases LHCGR mRNA‐binding protein (Lrbp) expression by activating Srebp in rat ovaries (Menon et al., 2013).
Steroid‐producing GCs are also regulated by TAGLN2‐ and forkhead box L2 (FOXL2)‐targeting miR‐133b (Dai et al., 2013; Xiao et al., 2014), which enhances StAR and CYP19A1, as well as oestradiol production (Dai et al., 2013). Similarly, miR‐383 facilitates oestradiol biosynthesis by inhibiting RNA binding motif single‐stranded interacting protein 1 (Rbms1), a DNA‐binding protein that activates c‐Myc in mouse GCs (Yin et al., 2012), while the expression of miR‐383 is regulated by equine chorionic gonadotropin (eCG) and human chorionic gonadotropin (hCG) (Donadeu et al., 2012). Additionally, FSH alters the expression of 31 miRNAs in rat GCs. Among them, miR‐23b is increased, whereas miR‐29a and miR‐30d are decreased after 12 h of exposure; however, both miR‐29a and miR‐30d were increased after 48 h of exposure (Yao et al., 2010b), indicating that the length of hormonal exposure influences miRNA expression. Similarly, exposure of GCs to LH/hCG enhances the expression of miR‐132, miR‐212, miR‐21, and miR‐122 (Fiedler et al., 2008; Carletti et al., 2010; Menon et al., 2013). In addition, 57 miRNAs including members of the let‐7 family, miR‐16, and miR‐122 play a role in female reproduction by inhibiting testosterone biosynthesis (Sirotkin et al., 2009). miR‐320 also regulates the proliferation and functions of GCs by targeting E2f1 and splicing factor 1 (Sf1), thereby promoting progesterone and testosterone levels and inhibiting E2 biosynthesis in mice (Yin et al., 2014). Similarly, 36 miRNAs including the let‐7 family, miR‐15a, and miR‐17‐3p inhibit progesterone biosynthesis, and 16 miRNAs including miR‐16, miR‐24, and miR‐191 promote progesterone synthesis in GCs (Sirotkin et al., 2009). By contrast, miR‐107 enhances testosterone biosynthesis in female gonads (Sirotkin et al., 2009). In addition, prenatal exposure to testosterone greatly influences the expression of miR‐497 and miR‐15b in sheep ovaries (Luense et al., 2011).
By contrast, many miRNAs play anti‐steroidogenic roles by inhibiting growth and proliferation, and increasing apoptosis of GCs and TCs: miR‐145 attenuates mouse GC proliferation by targeting Acvr1b (Yan et al., 2012), miR‐23a plays a pro‐apoptotic role in luteinized human GCs by reducing expression of X‐linked inhibitor of apoptosis (XIAP) and increasing CAS3 (Yang et al., 2012), and miR‐23a and miR‐27a also increase the apoptosis of human GCs by targeting SMAD5 and positively regulating Fas‐induced death signals (Nie et al., 2015). Expression of miR‐26b induces apoptosis of GCs in pigs by directly targeting ATM, a gene involved in DNA repair (Lin et al., 2012), and by suppressing SMAD4 signalling and BCL2 (Liu et al., 2014); let‐7 g promotes GC apoptosis in pigs by targeting TGFBR1 and downregulating the TGFβ signalling pathway (Zhou et al., 2015) as well as by targeting CCND2 and BCL2L1 and regulating MAPK and p53 signalling (Cao et al., 2015). Additionally, a group of miRNAs including miR‐15a, and miR‐29a promote the accumulation of BAX, thus enhancing the apoptosis of human GCs (Sirotkin et al., 2010). In addition, the follicular to luteal transition process and ovulation are stimulated by the presence of steroidogenic genes that are also regulated by miRNAs (Donadeu et al., 2012; Iwamune et al., 2014). Taken together, miRNAs control the biosynthesis of female steroidogenic hormones including oestrogen, progesterone, and androgens by regulating the proliferation, growth, and apoptosis of ovarian GCs and TCs.
VII. miRNAs AS REGULATORS OF ZYGOTIC AND EARLY EMBRYO REPROGRAMMING
Endometrial miRNAs along with parental miRNAs from the germline or uterine fluid could participate in maternal–embryo crosstalk (Saadeldin et al., 2015) and could play important roles during epigenetic reprogramming of the embryonic cells by regulating the expression and functions of genes related to early embryonic development (see Table S5).
(1). Roles during fertilization, ZGA, and epigenetic reprogramming of preimplantation embryos
Recent data suggested that sperm‐borne miRNAs must be transferred into a fertilized embryo to maintain maternal and paternal communication. For example, Yuan et al. (2016) produced zygotes using Drosha conditional knockout (cKO) mouse‐derived miRNA‐depleted sperm and found the resulting zygotes to have reduced developmental potential. Of note, the developmental competence of these zygotes could be recovered through the injection of small miRNAs derived from WT sperm. Furthermore, they found 14 sperm‐borne miRNAs to be present in both WT sperm and the zygote, but not in oocytes, indicating that sperm‐borne miRNAs can affect the development potential of fertilized embryos. By contrast, most zygotic miRNAs are derived from maternal oocytes (Tang et al., 2007). Altered miRNA expression in cumulus cells could result in a poor ovarian response in women undergoing in vitro fertilization (Karakaya et al., 2015), and some miRNAs are highly specific to the blastocyst and are not expressed in the oocytes (Tulay et al., 2015). In addition, sperm‐borne miR‐34c‐5p is thought to initiate the first cleavage divisions in the mouse (Liu et al., 2012), and is positively correlated with intracytoplasmic sperm injection outcomes, as well as high‐quality embryos, implantation, pregnancy and live birth in humans (Cui et al., 2015), and the presence of miR‐34c‐5p indicates viability and high‐quality spermatozoa in humans (Abu‐Halima et al., 2014). Similarly, sperm‐borne miR‐34b/c and mir‐449a/b/c play a critical role in the in vitro first cleavage division of mouse embryos (Liu et al., 2012; Yuan et al., 2015). Based on the above findings, the presence of miR‐34c‐5p can be considered a non‐invasive biomarker for viable sperm and embryos in the mouse and human. In future, miR‐34c‐5p could be a universal biomarker for viable mammalian sperm and embryos; however, it requires intensive validation using other species.
Another study suggested that miR‐34b/c‐ and miR‐449‐null male mice were fertile, whereas miR‐34b/c and miR‐449 double‐knockout (miR‐dKO) male mice were infertile due to severe spermatogenic disruptions and oligoasthenoteratozoospermia (Yuan et al., 2015). However, injection of miR‐dKO round spermatids into fertile mouse oocyte successfully gave rise to healthy offspring (Yuan et al., 2015): indicating that this miRNA family is essential for normal spermatogenesis, but not for fertilization and embryonic development (Liu et al., 2016). Sperm‐derived miR‐191, miR‐16, and miR‐146a are the most abundant miRNAs in early zygotes of mice (Yang et al., 2016), but their roles are not clear. Despite being silenced functionally, the biogenesis of miRNAs is not abrogated during fertilization to ZGA in mice (Tang et al., 2007; Suh et al., 2010), suggesting that miRNAs might have important roles during transformation of zygotes to pluripotent blastocysts (Tang et al., 2007; Suh et al., 2010). Some miRNAs are stage‐specific: miR‐125a and miR‐496 are specific to four‐cell bovine embryos, while miR‐127, miR‐145, and miR‐496 are specific to eight‐cell bovine embryos. However, most miRNAs showed fluctuating expression patterns according to the stage of bovine embryonic development (Tesfaye et al., 2009). This switch in miRNA expression patterns between ZGA and pluripotent blastocyst involves massive molecular rewiring (Hemberger, Dean, & Reik, 2009), and thus miRNAs might play a vital role during the transition of fertilized oocytes to pluripotent blastocysts (Svoboda & Flemr, 2010). Based on these findings, we can conclude that parental miRNAs might have important but limited roles during fertilization and the ZGA; however, miRNAs might be a key regulatory player during the developmental transition from the ZGA to pluripotent blastocyst.
miR‐29b has been suggested to target de novo DNA methyltransferases (Dnmt3a and Dnmt3b) transcripts in PGCs (Takada et al., 2009). The suppression of Dnmt by miRNAs safeguards proper DNA demethylation and establishes the new imprinting of genes. Similarly, Dnmt3a/3b expression was negatively regulated by miR‐29b at the time of mouse ZGA (Zhang et al., 2015). Given that DNA methylation plays a critical role in the morula‐to‐blastocyst transition, these data suggest a feedback relationship between the regulation of miRNA expression and DNA methylation. In addition, pluripotency markers including kruppel like factor 4 (Klf4) and nanog homeobox (Nanog) were downregulated in the morula stage upon miR‐29b inhibition (Wang et al., 2016), indicating the involvement of miR‐29b in the regulation of pluripotency. Similarly, the miR‐290/302 cluster is involved with embryonic pluripotency, although an opposing role of this cluster was shown in one study (Gu et al., 2016). Again, differential expression of miRNAs subsets during trophectoderm specification (Viswanathan et al., 2009) or severe preeclamptic placentas (Pineles et al., 2007) evidence a role for miRNAs during the implantation process. The endometrial miRNAs along with miRNAs from the uterine fluid also could participate during maternal–embryo crosstalk and play an important role during the epigenetic reprogramming of the embryonic cells (Fig. 5). Thus, accumulating evidence suggests that miRNAs are at least indirectly engaged in the epigenetic reprogramming process of early embryos.
Figure 5.
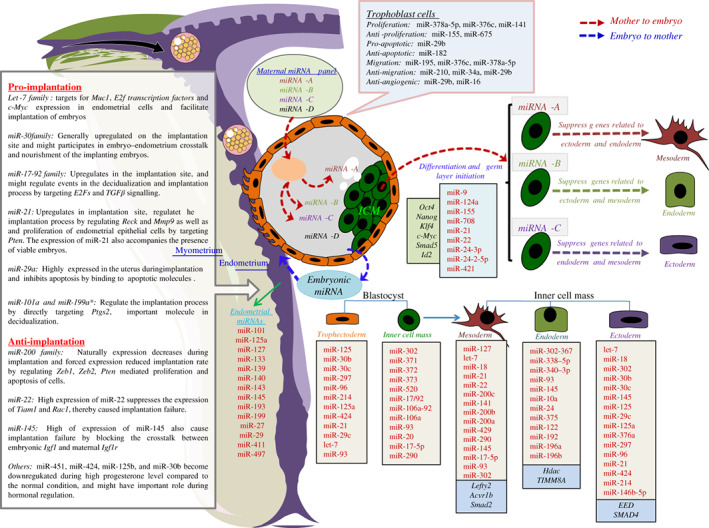
MicroRNA (miRNA) regulation during implantation and germ layer specification. Based on their functions, miRNAs are categorized as anti‐ or pro‐implantation miRNAs, pluripotency miRNAs, or miRNAs that promote differentiation and commitment of mesoderm, endoderm, ectoderm, and trophectoderm lineages. Parental miRNAs might be important regulators during embryonic germ layer specification; variation in the absorption of parental miRNAs could initiate variation in differentiation potential of the inner cell mass (ICM), and thus could direct the ICM towards different germ layers. For example, absorption of miRNA‐A by an ICM cell could suppress the endoderm and ectoderm lineage in that particular cell and thus facilitate mesoderm commitment. Similarly, the absorption of miRNA‐B by an ICM cell could suppress the mesoderm and ectoderm lineage in that respective cell and could direct the cell towards the endoderm lineage; the absorption of miRNA‐C by an ICM cell might facilitate ectoderm commitment by suppressing the mesoderm and endoderm lineages. Muc1, mucin 1, cell surface associated; c‐Myc, MYC proto‐oncogene, BHLH transcription factor; TGFβ, transforming growth factor beta; Reck, reversion inducing cysteine‐rich protein with kazal motifs; Mmp9, matrix metallopeptidase 9; Pten, phosphatase and tensin homolog; Ptgs2, prostaglandin‐endoperoxide synthase 2; Zeb1, zinc finger E‐box binding homeobox 1; Zeb2, zinc finger E‐box binding homeobox 2; Tiam1, T‐cell lymphoma invasion and metastasis 1; Rac1, Rac family small GTPase 1; Igf1, insulin like growth factor 1; Igf1r, insulin like growth factor 1 receptor; Oct4, octamer‐binding protein 4; Nanog, nanog homeobox; Klf4, kruppel like factor 4; Smad5, SMAD family member 5; Id2, inhibitor of DNA binding 2; Lefty2, left–right determination factor 2; Acvr1b, activin A receptor type 1B; Smad2, SMAD family member 2; Hdac, histone deacetylase; TIMM8A, translocase of inner mitochondrial membrane 8A; EED, embryonic ectoderm development; SMAD4, SMAD family member 4.
(2). miRNAs regulate germ‐layer specification in early embryos
During the gastrulation stage, embryonic stem cells (ESCs) differentiate to epiblast stem cells (EpSCs) and then EpSCs begin to differentiate into the endoderm, mesoderm and ectoderm lineages. Differentiation starts with the downregulation of pluripotency markers including octamer‐binding protein 4 (Oct4), Nanog, Klf4, and c‐Myc, which are regulated by miR‐24‐3p and miR‐24‐2‐5p, while expression of protein arginine methyltransferase 7 (Prmt7) inhibits miR‐24‐3p and miR‐24‐2‐5p. Thus, miR‐24‐3p and miR‐24‐2‐5p create a feedback loop with Prmt7 and regulate the differentiation of mouse ESCs (Lee et al., 2016). Furthermore, miR‐124a in humans, and miR‐200a/b/c, miR‐141 and miR‐429 in the mouse regulate early gastrulation‐stage embryos (Berardi et al., 2012). In ESCs, miR‐191 and miR‐16‐1 are abundant, and inhibit the formation of mesoderm by downregulating Smad2, an essential component of Activin–Nodal signalling (Hadjimichael et al., 2016), while miR‐23a may suppress the differentiation of ESCs towards the endoderm and ectoderm lineages (Hadjimichael et al., 2016). By contrast, expression of miR‐9, miR‐124a, miR‐155, and miR‐708 facilitate the differentiation of mouse ESCs (Marson et al., 2008), and the overexpression of miR‐21 facilitates the commitment of mesodermal tissues such as adipose and bone (Eguchi et al., 2013; Kang et al., 2013). Similarly, miR‐303‐367 and miR‐290‐295 clusters may play an important role during the patterning and specification of embryonic germ layers in humans and mice, respectively, by regulating Nodal signalling (Vidigal & Ventura, 2012). During this stage, BMPs, Activin/Nodal, and canonical Wnts signals play critical roles for the transition of inner cell mass (ICM) to mesoderm or endoderm, whereas non‐canonical Wnts, fibroblast growth factors (FGFs), Notch, and hedgehog (Hh) signalling are closely involved in the transition of mesoderm posterior 1 homolog (Mesp1)‐specified mesoderm to cardiac mesoderm.
Embryonic mesoderm shows predominant expression of miR‐145 and miR‐302 in humans and miR‐290, miR‐93, and miR‐17‐5p in mice (Berardi et al., 2012). Similarly, the miR‐200 family is known for its role in early mesoderm commitment by regulating snail family transcriptional repressor 1 (Snail) (Gill et al., 2011). miR‐145 accelerates mesoderm and ectoderm commitment by targeting OCT4, sex determining region Y‐box 2 (SOX2), and KLF4 in human ESCs (Xu et al., 2009), and miR‐127 induces differentiation of mouse ESCs to the mesendoderm lineage by targeting left–right determination factor 2 (Lefty2) and regulating Nodal signalling (Ma et al., 2016b). let‐7 and miR‐18 target Acvr1b and Smad2, respectively, and may attenuate Nodal responsiveness as well as bias the blastomeres towards the ectoderm and mesoderm (Colas et al., 2012). Thus, in vivo blocking of let‐7 and miR‐18 activity may divert the presumptive mesoderm and ectoderm towards the endoderm (Colas et al., 2012).
Embryonic endoderm expresses miR‐145, miR‐10a, miR‐24, miR‐375, miR‐122, miR‐192, miR‐196a, and miR‐196b in humans and miR‐93, miR‐338‐5p, and miR‐340‐3p in mice (Berardi et al., 2012). miR‐421 represses Oct4 and BMP signalling components such as Smad5 and inhibitor of DNA binding 2 (Id2) in mouse ESCs (Hadjimichael et al., 2016), while miR‐338‐5p and miR‐340‐3p promote endodermal differentiation of mouse ESCs by regulating histone deacetylase (Hdac) activity (Fu et al., 2011), and miR‐375 plays important roles in endoderm commitment in human ESCs by targeting translocase of inner mitochondrial membrane 8A (TIMM8A) (Hinton et al., 2010).
Embryonic ectoderm also represents a unique miRNA profile, and predominant expression of miR‐125, miR‐30b, and miR‐30c have been observed in embryonic human ectoderm, while miR‐29c, miR‐125a, miR‐376a, miR‐297, miR‐96, miR‐21, let‐7, miR‐424, and miR‐214 were predominant in the embryonic ectoderm of mice (Berardi et al., 2012). In addition, miR‐30b modulates formation of early human ectoderm by regulating embryonic ectoderm development (EED) (Song et al., 2011), miR‐125 and miR‐146b‐5p regulate the commitment of the neural lineage in humans by targeting SMAD4 (Boissart et al., 2012; Zhang et al., 2017), miR‐17‐5p promotes proliferation and inhibits the differentiation of lung epithelial progenitor cells in mice by targeting RB transcriptional corepressor like 2 (Rbl2) (Lu et al., 2007), and overexpression of miR‐125b‐2 represses the differentiation of mouse ESCs to the ectodermal lineage by regulating Lin28 and differentiation of embryonic stem cells 1 (Dies1) (Deng et al., 2015).
miR‐125, miR‐30b, and miR‐30c are specific to human embryonic trophectoderm, while the expression of miR‐297, miR‐214, miR‐96, miR‐125a, miR‐21, miR‐93, miR‐424, miR‐29c, let‐7, and miR‐376a was detected in mouse trophectoderm (Berardi et al., 2012). Additionally, mouse blastocysts showed upregulated expression of a panel of miRNAs and downregulated expression of miR‐376a compared to the morula stage, indicating the involvement of related miRNAs during trophectoderm commitment (Viswanathan et al., 2009).
Finally, early embryos may be enriched with a panel of parental miRNAs (Fig. 5), either through gametes (Tang et al., 2007; Yuan et al., 2016) or embryo–maternal crosstalk (Saadeldin et al., 2015; Bidarimath et al., 2017). Exosomes and cell‐membrane‐derived micro‐ and nano‐vesicles could be the main medium for this embryo–maternal crosstalk (Diehl et al., 2012; Saadeldin et al., 2015), however, miRNAs could also be carried through the circulatory system by apoptotic bodies, RNA‐binding lipoproteins and Argonaute protein complex (Ling, Bao‐feng, & Jing, 2013; Sohel, 2016). These circulating miRNAs originating from one cell or organ could be transported to and absorbed by neighbouring or distant cells, for example, miRNAs originating from tumour cells could be absorbed by other healthy cells leading to their transformation to malignancy (Guo et al., 2017). Thus, it could be assumed that each cell of the embryonic ICM randomly absorbs different maternal miRNAs delivered through the circulatory system; this variation in miRNA intake among the cells of the embryonic ICM may lead the cells towards different lineages. For example, one cell may absorb a miRNA‐A panel, leading to the mesoderm lineage, another may absorb a miRNA‐B panel and commit to the endoderm lineage, while a third cell may absorb a miRNA‐C panel to specify the ectoderm lineages (Fig. 5). However, further in‐depth experiments, both in vitro and in vivo, are required to support these hypotheses.
(3). miRNAs regulate pregnancy by mediating adhesion molecules, vasculature and angiogenesis
miRNAs involved in the implantation process can be categorized into two groups, anti‐ or pro‐implantation miRNAs (Fig. 5); these miRNAs are potent regulators of many genes related to adhesion, angiogenesis, and vasculature (see Table S5), as well as foetal–maternal immune tolerance following endometrial reception of the conceptus (Bidarimath et al., 2014, 2015). Alterations in expression of these miRNAs in the uterine endometrium could cause implantation failure as well as disease (Hiroki et al., 2010). For example, the let‐7 family regulates the expression of mucin 1, cell surface associated (Muc1) and facilitates implantation of mouse embryos (Inyawilert et al., 2015), the miR‐30 family is generally upregulated at the implantation site and may participate in embryo–endometrium crosstalk and nourishment of the implanting embryo (Moreno‐Moya et al., 2014), the miR‐17‐92 cluster is upregulated at the implantation site, and may regulate the decidualization (morphological and functional changes of the endometrium in preparation for pregnancy) and implantation process by targeting E2F and TGFβ signalling (Mogilyansky & Rigoutsos, 2013). Similarly, miR‐21 is upregulated at the implantation site and regulates matrix metallopeptidase 2 (Mmp2) and matrix metallopeptidase 9 (Mmp9) expression by targeting reversion inducing cysteine‐rich protein with kazal motifs (Reck) during implantation in mice (Hu et al., 2008; Carletti & Christenson, 2009) and PTEN during the proliferation of endometrial epithelial cells (Qin et al., 2012). miR‐29a is highly expressed in the uterus during implantation and inhibits apoptotic molecules (Xia et al., 2014), and miR‐101a and miR‐199a* regulate the implantation process by directly targeting Cox‐2 (Ptgs), an important molecule for decidualization (Chakrabarty et al., 2007). A panel of miRNAs including miR‐10a, miR‐27a, miR‐29c, miR‐323, miR‐331‐5p, miR‐339‐3p, miR‐374‐5p, and miR‐935 are reported to be associated with spontaneous foetal arrest in pigs (Wessels et al., 2013), while miR‐296‐5P, miR‐150, miR‐17P‐5P, miR‐18a, and miR‐19 are associated with angiogenesis of the endometrial tissues in pigs (Bidarimath et al., 2015), and are differentially expressed between healthy and arresting conceptus attachment site (Bidarimath et al., 2015). Thus these miRNAs could be used to assess foetal health, although this requires validation with other mammalian species. Additionally, some miRNAs participate in the implantation process indirectly, such as by promoting the proliferation of trophoblast cells (Luo et al., 2012; Fu et al., 2013), by facilitating trophoblast migration (Bai et al., 2012; Luo et al., 2012; Fu et al., 2013), by inhibiting the apoptosis of trophoblasts (Pineles et al., 2007), and by participating in conceptus–endometrial crosstalk (Bidarimath et al., 2017).
By contrast, the expression of oestradiol‐ and progesterone‐regulatory miR‐200 family members decreases naturally, while its targets Zeb1 and zinc finger E‐box binding homeobox 2 (Zeb2) are upregulated during implantation in mice (Jimenez et al., 2016), and forced expression of miR‐200a reduced the implantation rate of mouse embryos by regulating Pten‐mediated proliferation and apoptosis of cells (Shen et al., 2013). Overexpression of miR‐22 suppresses T‐cell lymphoma invasion and metastasis 1 (Tiam1) and Rac family small GTPase 1 (Rac1), causing implantation failure (Ma et al., 2015), while overexpression of miR‐145 also causes implantation failure in mice by blocking crosstalk between embryonic Igf1 and maternal insulin like growth factor 1 receptor (Igf1r) (Kang et al., 2015). Similarly, miR‐451, miR‐424, miR‐125b, and miR‐30b are downregulated when progesterone is elevated compared to that under normal conditions (Li et al., 2011). Oestrogen‐mediated inhibition of miR‐705 facilitates Mmp9 translation (Carletti & Christenson, 2009), which may be important for proper implantation of embryos and thus pregnancy. Some other miRNAs play anti‐implantation roles by inhibiting proliferation (Dai et al., 2012; Gao et al., 2012), inducing apoptosis (Li et al., 2013), and inhibiting migration (Zhang et al., 2012; Umemura et al., 2013) of the trophoblasts, as well as by suppressing vascularization and angiogenesis during implantation of embryos (Wang et al., 2012).
Active or pseudopregnancy could be confirmed by the expression of miR‐21, which is expressed only in the presence of active embryos (Hu et al., 2008; Luense et al., 2009). As discussed earlier, miRNAs also participate in the maintenance of pregnancy by regulating angiogenesis and vasculogenesis of luteal tissues. In brief, miR‐125b, miR‐145, mir‐31, miR‐503 and miR‐21 regulate interferon‐mediated cell death, and vascularization and angiogenesis in ruminant CL (Donadeu et al., 2012), and miR‐378 regulates the maintenance of the bovine CL by regulating its target IFNGR1 (Ma et al., 2011). miR‐17‐5p and let‐7 play important roles in CL formation by targeting the anti‐angiogenic gene Timp1 (Otsuka et al., 2008). In sheep, miR‐199a‐3p, miR‐145 and miR‐503 are expressed differentially at different stages of the CL and are involved in fibrosis, and immune and inflammatory responses (Donadeu et al., 2012). Taken together, miRNAs mediate implantation and pregnancy in mammals by regulating hormone levels throughout pregnancy, maintaining crosstalk between the embryo and mother, and facilitating angiogenesis and vascularization of the endometrium and luteal tissues.
VIII. CONCLUSIONS
(1) Loss of miRNA processing machinery could impair reproduction at the molecular level and causes infertility or sterility to different extents depending on sex, age and stage of loss, but the external sex characteristics along with mating behaviour remain normal in both males and females.
(2) miRNAs are mostly indispensable during pre‐ and post‐fertilization stages (such as sex differentiation, germline establishment, gametogenesis, preimplantation development and implantation of embryos) but might have very limited roles during the fertilization event. miRNA‐loss‐mediated infertility therefore derives from the malformation of gametes, not because of its absence during the fertilization event itself.
(3) miRNAs could have a direct involvement in the global silencing of X‐linked gene expression during the MSCI process of spermatogenesis and in the initiation of ZGA and the first embryonic cell division.
(4) Steroidogenesis in both sexes is regulated by potent feedback loops comprising miRNAs, genes, and hormones; deregulation in any of which could impair the whole process of mammalian reproduction.
(5) miRNAs could be used as fertility biomarkers for both gametes, and also to differentiate between pseudo and viable pregnancy.
(6) Parental miRNAs (derived from gametes or embryo–maternal crosstalk) could play very important roles in epigenetic modifications and germ‐layer specification of embryos. They may also be important in the implantation and maintenance of pregnancy by regulating angiogenesis and vascularization of the endometrium tissues and CL.
(7) Much work remains to be done to understand the roles of miRNAs in mammalian reproduction, especially in MSCI, the first embryonic cell division and ZGA, germ‐layer specification and embryo–maternal crosstalk.
Supporting information
Table S1. Functions of microRNAs (miRNAs) in sex specification and commitment of the mammalian reproductive system.
Table S2. Functions of microRNAs (miRNAs) in spermatogenesis and spermiogenesis.
Table S3. Functions of microRNAs (miRNAs) during folliculogenesis and oogenesis.
Table S4. Functions of microRNAs (miRNAs) in the regulation of steroidogenic cells and hormonal balance.
Table S5. Functions of microRNAs (miRNAs) in fertilization, implantation and germ‐layer specification.
IX. ACKNOWLEDGMENTS
This work was supported by the Science Research Center (2015R1A5A1009701) from the National Research Foundation of Korea. A.M.M.T.R., K.H., and J.‐H.K. wrote the manuscript and designed the figs A.M.M.T.R. performed the literature search and analysed the information. A.M.M.T.R., S.G.H., and H.S. prepared the figs Y.‐J.C. and C.P. assisted with the literature search.
Contributor Information
Kwonho Hong, Email: hongk@konkuk.ac.kr.
Jin‐Hoi Kim, Email: jhkim541@konkuk.ac.kr.
X. REFERENCES
References marked with asterisk have been cited within the supporting information.
- Abd El Naby, W. S. , Hagos, T. H. , Hossain, M. M. , Salilew‐Wondim, D. , Gad, A. Y. , Rings, F. , Cinar, M. U. , Tholen, E. , Looft, C. , Schellander, K. , Hoelker, M. & Tesfaye, D. (2013). Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote 21, 31–51. [DOI] [PubMed] [Google Scholar]
- Abu‐Halima, M. , Hammadeh, M. , Backes, C. , Fischer, U. , Leidinger, P. , Lubbad, A. M. , Keller, A. & Meese, E. (2014). Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertility and Sterility 102, 989–997.e1. [DOI] [PubMed] [Google Scholar]
- Ahmed, K. , LaPierre, M. P. , Gasser, E. , Denzler, R. , Yang, Y. , Rulicke, T. , Kero, J. , Latreille, M. & Stoffel, M. (2017). Loss of microRNA‐7a2 induces hypogonadotropic hypogonadism and infertility. Journal of Clinical Investigation 127, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, H. W. , Morin, R. D. , Zhao, H. , Harris, R. A. , Coarfa, C. , Chen, Z. J. , Milosavljevic, A. , Marra, M. A. & Rajkovic, A. (2010). MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Molecular Human Reproduction 16, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford, C. , Toloubeydokhti, T. , Al‐Katanani, Y. , Drury, K. C. , Williams, R. & Chenini, N. (2007). The expression of microRNA (miRNA) mir‐23a and 23b and their target gene, CYP19A1 (aromatase) in follicular cells obtained from women undergoing ART. Fertility and Sterility 88, S166–S167. [Google Scholar]
- Bai, Y. , Yang, W. , Yang, H. X. , Liao, Q. , Ye, G. , Fu, G. , Ji, L. , Xu, P. , Wang, H. , Li, Y. X. , Peng, C. & Wang, Y. L. (2012). Downregulated miR‐195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS One 7, e38875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, J. , Li, D. , Wang, L. , Wu, J. , Hu, Y. , Wang, Z. , Chen, Y. , Cao, X. , Jiang, C. , Yan, W. & Xu, C. (2012). MicroRNA‐449 and microRNA‐34b/c function redundantly in murine testes by targeting E2F transcription factor‐retinoblastoma protein (E2F‐pRb) pathway. Journal Biological Chemistry 287, 21686–21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannee, C. (2015). Extracellular microRNAs from the epididymis as potential mediators of cell‐to‐cell communication. Asian Journal of Andrology 17, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannee, C. , Calvo, É. , Caballero, J. & Sullivan, R. (2013a). Epididymosomes convey different repertoires of microRNAs throughout the bovine epididymis. Biology of Reproduction 89(30), 1–11. [DOI] [PubMed] [Google Scholar]
- Belleannee, C. , Calvo, E. , Thimon, V. , Cyr, D. G. , Legare, C. , Garneau, L. & Sullivan, R. (2012). Role of microRNAs in controlling gene expression in different segments of the human epididymis. PLoS One 7, e34996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannee, C. , Legare, C. , Calvo, E. , Thimon, V. & Sullivan, R. (2013b). microRNA signature is altered in both human epididymis and seminal microvesicles following vasectomy. Human Reproduction 28, 1455–1467. [DOI] [PubMed] [Google Scholar]
- Berardi, E. , Pues, M. , Thorrez, L. & Sampaolesi, M. (2012). miRNAs in ESC differentiation. American Journal of Physiology‐Heart and Circulatory Physiology 303, H931–H939. [DOI] [PubMed] [Google Scholar]
- Bernstein, E. , Kim, S. Y. , Carmell, M. A. , Murchison, E. P. , Alcorn, H. , Li, M. Z. , Mills, A. A. , Elledge, S. J. , Anderson, K. V. & Hannon, G. J. (2003). Dicer is essential for mouse development. Nature Genetics 35, 215–217. [DOI] [PubMed] [Google Scholar]
- Bidarimath, M. , Edwards, A. K. , Wessels, J. M. , Khalaj, K. , Kridli, R. T. & Tayade, C. (2015). Distinct microRNA expression in endometrial lymphocytes, endometrium, and trophoblast during spontaneous porcine fetal loss. Journal of Reproductive Immunology 107, 64–79. [DOI] [PubMed] [Google Scholar]
- Bidarimath, M. , Khalaj, K. , Kridli, R. T. , Kan, F. W. , Koti, M. & Tayade, C. (2017). Extracellular vesicle mediated intercellular communication at the porcine maternal‐fetal interface: a new paradigm for conceptus‐endometrial cross‐talk. Scientific Reports 7, 40476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidarimath, M. , Khalaj, K. , Wessels, J. M. & Tayade, C. (2014). MicroRNAs, immune cells and pregnancy. Cellular and Molecular Immunology 11, 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork, J. K. , Sandqvist, A. , Elsing, A. N. , Kotaja, N. & Sistonen, L. (2010). miR‐18, a member of Oncomir‐1, targets heat shock transcription factor 2 in spermatogenesis. Development 137, 3177–3184. [DOI] [PubMed] [Google Scholar]
- Bjorkgren, I. , Gylling, H. , Turunen, H. , Huhtaniemi, I. , Strauss, L. , Poutanen, M. & Sipila, P. (2015). Imbalanced lipid homeostasis in the conditional Dicer1 knockout mouse epididymis causes instability of the sperm membrane. The FASEB Journal 29, 433–442. [DOI] [PubMed] [Google Scholar]
- Boissart, C. , Nissan, X. , Giraud‐Triboult, K. , Peschanski, M. & Benchoua, A. (2012). miR‐125 potentiates early neural specification of human embryonic stem cells. Development 139, 1247–1257. [DOI] [PubMed] [Google Scholar]
- Bouhallier, F. , Allioli, N. , Lavial, F. , Chalmel, F. , Perrard, M. H. , Durand, P. , Samarut, J. , Pain, B. & Rouault, J. P. (2010). Role of miR‐34c microRNA in the late steps of spermatogenesis. RNA 16, 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieno‐Enriquez, M. A. , Garcia‐Lopez, J. , Cardenas, D. B. , Guibert, S. , Cleroux, E. , Ded, L. , Hourcade Jde, D. , Peknicova, J. , Weber, M. & Del Mazo, J. (2015). Exposure to endocrine disruptor induces transgenerational epigenetic deregulation of microRNAs in primordial germ cells. PLoS One 10, e0124296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, R. , Wu, W. J. , Zhou, X. L. , Xiao, P. , Wang, Y. & Liu, H. L. (2015). Expression and preliminary functional profiling of the let‐7 family during porcine ovary follicle atresia. Molecules and Cells 38, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti, M. Z. & Christenson, L. K. (2009). MicroRNA in the ovary and female reproductive tract. Journal of Animal Science 87, E29–E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti, M. Z. , Fiedler, S. D. & Christenson, L. K. (2010). MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biology of Reproduction 83, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty, A. , Tranguch, S. , Daikoku, T. , Jensen, K. , Furneaux, H. & Dey, S. K. (2007). MicroRNA regulation of cyclooxygenase‐2 during embryo implantation. Proceedings of the National Academy of Sciences of the United States of America 104, 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. , Dai, D. H. , Li, Y. , Zhang, Y. , Zhang, M. , Zhou, G. B. & Zeng, C. J. (2016). Differences in the expression of microRNAs and their predicted gene targets between cauda epididymal and ejaculated boar sperm. Theriogenology 86, 2162–2171. [DOI] [PubMed] [Google Scholar]
- Chang, Y. F. , Lee‐Chang, J. S. , Imam, J. S. , Buddavarapu, K. C. , Subaran, S. S. , Sinha‐Hikim, A. P. , Gorospe, M. & Rao, M. K. (2012). Interaction between microRNAs and actin‐associated protein Arpc5 regulates translational suppression during male germ cell differentiation. Proceedings of the National Academy of Sciences of the United States of America 109, 5750–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. , Deng, L. , Yu, X. , Wang, X. , Zhu, L. , Yu, T. , Zhang, Y. , Zhou, B. , Xu, W. , Chen, L. & Luo, H. (2015a). MiR‐122 partly mediates the ochratoxin A‐induced GC‐2 cell apoptosis. Toxicology In Vitro 30, 264–273. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Li, X. , Guo, J. , Zhang, P. & Zeng, W. (2017). The roles of microRNAs in regulation of mammalian spermatogenesis. Journal of Animal Science and Biotechnology 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Xie, M. , Liu, D. & Shi, K. (2015b). Downregulation of microRNA146a inhibits ovarian granulosa cell apoptosis by simultaneously targeting interleukin1 receptorassociated kinase and tumor necrosis factor receptorassociated factor 6. Molecular Medicine Reports 12, 5155–5162. [DOI] [PubMed] [Google Scholar]
- Chieffi, P. , Battista, S. , Barchi, M. , Di Agostino, S. , Pierantoni, G. M. , Fedele, M. , Chiariotti, L. , Tramontano, D. & Fusco, A. (2002). HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene 21, 3644–3650. [DOI] [PubMed] [Google Scholar]
- Choi, Y. , Qin, Y. , Berger, M. F. , Ballow, D. J. , Bulyk, M. L. & Rajkovic, A. (2007). Microarray analyses of newborn mouse ovaries lacking Nobox. Biology of Reproduction 77, 312–319. [DOI] [PubMed] [Google Scholar]
- Chu, C. , Zheng, G. , Hu, S. , Zhang, J. , Xie, S. , Ma, W. , Ni, M. , Tang, C. , Zhou, L. , Zhou, Y. , Liu, M. , Li, Y. & Zhang, Y. (2015). Epididymal region‐specific miRNA expression and DNA methylation and their roles in controlling gene expression in rats. PLoS One 10, e0124450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas, A. R. , McKeithan, W. L. , Cunningham, T. J. , Bushway, P. J. , Garmire, L. X. , Duester, G. , Subramaniam, S. & Mercola, M. (2012). Whole‐genome microRNA screening identifies let‐7 and mir‐18 as regulators of germ layer formation during early embryogenesis. Genes & Development 26, 2567–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, M. S. & Blelloch, R. (2013). Small RNAs in germline development. Current Topics in Developmental Biology 102, 159–205. [DOI] [PubMed] [Google Scholar]
- Cui, L. , Fang, L. , Shi, B. , Qiu, S. & Ye, Y. (2015). Spermatozoa micro ribonucleic acid‐34c level is correlated with intracytoplasmic sperm injection outcomes. Fertility and Sterility 104, 312–317.e1. [DOI] [PubMed] [Google Scholar]
- Cui, X. S. , Sun, S. C. , Kang, Y. K. & Kim, N. H. (2013). Involvement of microRNA‐335‐5p in cytoskeleton dynamics in mouse oocytes. Reproduction, Fertility and Development 25, 691–699. [DOI] [PubMed] [Google Scholar]
- Curry, E. , Safranski, T. J. & Pratt, S. L. (2011). Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology 76, 1532–1539. [DOI] [PubMed] [Google Scholar]
- da Silveira, J. C. , de Andrade, G. M. , Nogueira, M. F. , Meirelles, F. V. & Perecin, F. (2015). Involvement of miRNAs and cell‐secreted vesicles in mammalian ovarian antral follicle development. Reproductive Sciences 22, 1474–1483. [DOI] [PubMed] [Google Scholar]
- da Silveira, J. C. , Veeramachaneni, D. N. , Winger, Q. A. , Carnevale, E. M. & Bouma, G. J. (2012). Cell‐secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biology of Reproduction 86(71), 1–10. [DOI] [PubMed] [Google Scholar]
- Dabaja, A. A. , Mielnik, A. , Robinson, B. D. , Wosnitzer, M. S. , Schlegel, P. N. & Paduch, D. A. (2015). Possible germ cell‐Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell‐specific MicroRNA, miR‐202‐5p, in human testis. Basic and Clinical Andrology 25, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, A. , Sun, H. , Fang, T. , Zhang, Q. , Wu, S. , Jiang, Y. , Ding, L. , Yan, G. & Hu, Y. (2013). MicroRNA‐133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Letters 587, 2474–2482. [DOI] [PubMed] [Google Scholar]
- Dai, Y. , Qiu, Z. , Diao, Z. , Shen, L. , Xue, P. , Sun, H. & Hu, Y. (2012). MicroRNA‐155 inhibits proliferation and migration of human extravillous trophoblast derived HTR‐8/SVneo cells via down‐regulating cyclin D1. Placenta 33, 824–829. [DOI] [PubMed] [Google Scholar]
- Dallaire, A. & Simard, M. J. (2016). The implication of microRNAs and endo‐siRNAs in animal germline and early development. Developmental Biology 416, 18–25. [DOI] [PubMed] [Google Scholar]
- Damestoy, A. , Perrard, M. H. , Vigier, M. , Sabido, O. & Durand, P. (2005). Transforming growth factor beta‐1 decreases the yield of the second meiotic division of rat pachytene spermatocytes in vitro. Reproductive Biology and Endocrinology 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mateo, S. & Sassone‐Corsi, P. (2014). Regulation of spermatogenesis by small non‐coding RNAs: role of the germ granule. Seminars in Cell & Developmental Biology 29, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, S. , Zhang, Y. , Xu, C. & Ma, D. (2015). MicroRNA‐125b‐2 overexpression represses ectodermal differentiation of mouse embryonic stem cells. International Journal of Molecular Medicine 36, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, P. , Fricke, A. , Sander, L. , Stamm, J. , Bassler, N. , Htun, N. , Ziemann, M. , Helbing, T. , El‐Osta, A. , Jowett, J. B. M. & Peter, K. (2012). Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovascular Research 93, 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dior, U. P. , Kogan, L. , Chill, H. H. , Eizenberg, N. , Simon, A. & Revel, A. (2014). Emerging roles of microRNA in the embryo‐endometrium cross talk. Seminars in Reproductive Medicine 32, 402–409. [DOI] [PubMed] [Google Scholar]
- Donadeu, F. X. , Mohammed, B. T. & Ioannidis, J. (2017). A miRNA target network putatively involved in follicular atresia. Domestic Animal Endocrinology 58, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadeu, F. X. & Schauer, S. N. (2013). Differential miRNA expression between equine ovulatory and anovulatory follicles. Domestic Animal Endocrinology 45, 122–125. [DOI] [PubMed] [Google Scholar]
- Donadeu, F. X. , Schauer, S. N. & Sontakke, S. D. (2012). Involvement of miRNAs in ovarian follicular and luteal development. Journal of Endocrinology 215, 323–334. [DOI] [PubMed] [Google Scholar]
- Eggers, S. , Ohnesorg, T. & Sinclair, A. (2014). Genetic regulation of mammalian gonad development. Nature Reviews Endocrinology 10, 673–683. [DOI] [PubMed] [Google Scholar]
- Eguchi, T. , Watanabe, K. , Hara, E. S. , Ono, M. , Kuboki, T. & Calderwood, S. K. (2013). OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One 8, e58796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler, S. D. , Carletti, M. Z. , Hong, X. & Christenson, L. K. (2008). Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biology of Reproduction 79, 1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G. , Ye, G. , Nadeem, L. , Ji, L. , Manchanda, T. , Wang, Y. , Zhao, Y. , Qiao, J. , Wang, Y. L. , Lye, S. , Yang, B. B. & Peng, C. (2013). MicroRNA‐376c impairs transforming growth factor‐beta and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension 61, 864–872. [DOI] [PubMed] [Google Scholar]
- Fu, S. , Fei, Q. , Jiang, H. , Chuai, S. , Shi, S. , Xiong, W. , Jiang, L. , Lu, C. , Atadja, P. , Li, E. & Shou, J. (2011). Involvement of histone acetylation of Sox17 and Foxa2 promoters during mouse definitive endoderm differentiation revealed by microRNA profiling. PLoS One 6, e27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliano, D. & Pellicer, A. (2014). MicroRNA and implantation. Fertility and Sterility 101, 1531–1544. [DOI] [PubMed] [Google Scholar]
- Gao, W. L. , Liu, M. , Yang, Y. , Yang, H. , Liao, Q. , Bai, Y. , Li, Y. X. , Li, D. , Peng, C. & Wang, Y. L. (2012). The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR‐675 that targets nodal modulator 1 (NOMO1). RNA Biology 9, 1002–1010. [DOI] [PubMed] [Google Scholar]
- Garcia‐Lopez, J. , Alonso, L. , Cardenas, D. B. , Artaza‐Alvarez, H. , Hourcade Jde, D. , Martinez, S. , Brieno‐Enriquez, M. A. & Del Mazo, J. (2015). Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA 21, 946–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebremedhn, S. , Salilew‐Wondim, D. , Ahmad, I. , Sahadevan, S. , Hossain, M. M. , Hoelker, M. , Rings, F. , Neuhoff, C. , Tholen, E. , Looft, C. , Schellander, K. & Tesfaye, D. (2015). MicroRNA expression profile in bovine granulosa cells of Preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. PLoS One 10, e0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbian, S. (2012). Micro‐RNAs, next‐generation molecular markers in male infertility field. Translational Andrology and Urology 1, 245–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, J. G. , Langer, E. M. , Lindsley, R. C. , Cai, M. , Murphy, T. L. , Kyba, M. & Murphy, K. M. (2011). Snail and the microRNA‐200 family act in opposition to regulate epithelial‐to‐mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells 29, 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestaneh, N. , Beauchamp, E. , Fallen, S. , Kokkinaki, M. , Uren, A. & Dym, M. (2009). Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 138, 151–162. [DOI] [PubMed] [Google Scholar]
- Gonzalez, G. & Behringer, R. R. (2009). Dicer is required for female reproductive tract development and fertility in the mouse. Molecular Reproduction & Development 76, 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee, A. R. , Shiao, M. S. , Snyder, E. , Buaas, F. W. , Gu, T. , Stearns, T. M. , Sharma, M. , Murchison, E. P. , Puente, G. C. & Braun, R. E. (2012). Deregulated sex chromosome gene expression with male germ cell‐specific loss of Dicer1. PLoS One 7, e46359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, K.‐L. , Zhang, Q. , Yan, Y. , Li, T.‐T. , Duan, F.‐F. , Hao, J. , Wang, X.‐W. , Shi, M. , Wu, D.‐R. , Guo, W.‐T. & Wang, Y. (2016). Pluripotency‐associated miR‐290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Research 26, 350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y. , Liang, G. , Hawken, P. A. R. , Malecki, I. A. , Cozens, G. , Vercoe, P. E. , Martin, G. B. & Guan, L. L. (2015). Roles of small RNAs in the effects of nutrition on apoptosis and spermatogenesis in the adult testis. Scientific Reports 5, 10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Gao, Y. , Li, N. , Shao, F. , Wang, C. , Wang, P. , Yang, Z. , Li, R. & He, J. (2017). Exosomes: new players in cancer. Oncology Reports 38, 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjimichael, C. , Nikolaou, C. , Papamatheakis, J. & Kretsovali, A. (2016). MicroRNAs for fine‐tuning of mouse embryonic stem cell fate decision through regulation of TGF‐β signaling. Stem Cell Reports 6, 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer, G. , Roepers‐Gajadien, H. L. , van Duyn‐Goedhart, A. , Gademan, I. S. , Kal, H. B. , van Buul, P. P. & de Rooij, D. G. (2003). DNA double‐strand breaks and gamma‐H2AX signaling in the testis. Biology of Reproduction 68, 628–634. [DOI] [PubMed] [Google Scholar]
- Hasuwa, H. , Ueda, J. , Ikawa, M. & Okabe, M. (2013). miR‐200b and miR‐429 function in mouse ovulation and are essential for female fertility. Science 341, 71–73. [DOI] [PubMed] [Google Scholar]
- Hausser, J. , Syed, A. P. , Bilen, B. & Zavolan, M. (2013). Analysis of CDS‐located miRNA target sites suggests that they can effectively inhibit translation. Genome Research 23, 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K. , Chuva de Sousa Lopes, S. M. , Kaneda, M. , Tang, F. , Hajkova, P. , Lao, K. , O'Carroll, D. , Das, P. P. , Tarakhovsky, A. , Miska, E. A. & Surani, M. A. (2008). MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3, e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z. , Jiang, J. , Kokkinaki, M. , Tang, L. , Zeng, W. , Gallicano, I. , Dobrinski, I. & Dym, M. (2013). MiRNA‐20 and mirna‐106a regulate spermatogonial stem cell renewal at the post‐transcriptional level via targeting STAT3 and Ccnd1. Stem Cells 31, 2205–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger, M. , Dean, W. & Reik, W. (2009). Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nature Reviews Molecular Cell Biology 10, 526–537. [DOI] [PubMed] [Google Scholar]
- Hilz, S. , Modzelewski, A. J. , Cohen, P. E. & Grimson, A. (2016). The roles of microRNAs and siRNAs in mammalian spermatogenesis. Development 143, 3061–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton, A. , Afrikanova, I. , Wilson, M. , King, C. C. , Maurer, B. , Yeo, G. W. , Hayek, A. & Pasquinelli, A. E. (2010). A distinct microRNA signature for definitive endoderm derived from human embryonic stem cells. Stem Cells and Development 19, 797–807. [DOI] [PubMed] [Google Scholar]
- Hiroki, E. , Akahira, J. , Suzuki, F. , Nagase, S. , Ito, K. , Suzuki, T. , Sasano, H. & Yaegashi, N. (2010). Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Science 101, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, X. , Luense, L. J. , McGinnis, L. K. , Nothnick, W. B. & Christenson, L. K. (2008). Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 149, 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M. M. , Ghanem, N. , Hoelker, M. , Rings, F. , Phatsara, C. , Tholen, E. , Schellander, K. & Tesfaye, D. (2009). Identification and characterization of miRNAs expressed in the bovine ovary. BMC Genomics 10, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M. M. , Salilew‐Wondim, D. , Schellander, K. & Tesfaye, D. (2012a). The role of microRNAs in mammalian oocytes and embryos. Animal Reproduction Science 134, 36–44. [DOI] [PubMed] [Google Scholar]
- Hossain, M. M. , Sohel, M. M. , Schellander, K. & Tesfaye, D. (2012b). Characterization and importance of microRNAs in mammalian gonadal functions. Cell and Tissue Research 349, 679–690. [DOI] [PubMed] [Google Scholar]
- Hu, S. J. , Ren, G. , Liu, J. L. , Zhao, Z. A. , Yu, Y. S. , Su, R. W. , Ma, X. H. , Ni, H. , Lei, W. & Yang, Z. M. (2008). MicroRNA expression and regulation in mouse uterus during embryo implantation. Journal of Biological Chemistry 283, 23473–23484. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Shen, W. J. , Cortez, Y. , Tang, X. , Liu, L. F. , Kraemer, F. B. & Azhar, S. (2013). Hormonal regulation of microRNA expression in steroid producing cells of the ovary, testis and adrenal gland. PLoS One 8, e78040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar, J. M. & Payne, C. J. (2013). MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in mice. Biology of Reproduction 88(15), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inyawilert, W. , Fu, T. Y. , Lin, C. T. & Tang, P. C. (2015). Let‐7‐mediated suppression of mucin 1 expression in the mouse uterus during embryo implantation. Journal of Reproduction and Development 61, 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamune, M. , Nakamura, K. , Kitahara, Y. & Minegishi, T. (2014). MicroRNA‐376a regulates 78‐kilodalton glucose‐regulated protein expression in rat granulosa cells. PLoS One 9, e108997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, P. T. , Mainigi, M. A. , Word, R. A. , Kraus, W. L. & Mendelson, C. R. (2016). miR‐200 regulates endometrial development during early pregnancy. Molecular Endocrinology 30, 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, S. , Wen, D. , Yu, Y. , Holst, P. L. , Luo, Y. , Fang, M. , Tamir, R. , Antonio, L. , Hu, Z. , Cupples, R. , Louis, J. C. , Hu, S. , Altrock, B. W. & Fox, G. M. (1996). GDNF‐induced activation of the ret protein tyrosine kinase is mediated by GDNFR‐alpha, a novel receptor for GDNF. Cell 85, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Kang, M. , Yan, L. M. , Zhang, W. Y. , Li, Y. M. , Tang, A. Z. & Ou, H. S. (2013). Role of microRNA‐21 in regulating 3T3‐L1 adipocyte differentiation and adiponectin expression. Molecular Biology Reports 40, 5027–5034. [DOI] [PubMed] [Google Scholar]
- Kang, Y. J. , Lees, M. , Matthews, L. C. , Kimber, S. J. , Forbes, K. & Aplin, J. D. (2015). MiR‐145 suppresses embryo‐epithelial juxtacrine communication at implantation by modulating maternal IGF1R. Journal of Cell Science 128, 804–814. [DOI] [PubMed] [Google Scholar]
- Karakaya, C. , Guzeloglu‐Kayisli, O. , Uyar, A. , Kallen, A. N. , Babayev, E. , Bozkurt, N. , Unsal, E. , Karabacak, O. & Seli, E. (2015). Poor ovarian response in women undergoing in vitro fertilization is associated with altered microRNA expression in cumulus cells. Fertility and Sterility 103, 1469–1476.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen, H. M. , Meikar, O. , Yadav, R. P. , Papaioannou, M. D. , Romero, Y. , Da Ros, M. , Herrera, P. L. , Toppari, J. , Nef, S. & Kotaja, N. (2011). Dicer is required for haploid male germ cell differentiation in mice. PLoS One 6, e24821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen, H. M. , Yadav, R. P. , Da Ros, M. , Chalmel, F. , Zimmermann, C. , Toppari, J. , Nef, S. & Kotaja, N. (2015). DICER regulates the formation and maintenance of cell‐cell junctions in the mouse seminiferous epithelium. Biology of Reproduction 93, 1391–1313. [DOI] [PubMed] [Google Scholar]
- Kostereva, N. & Hofmann, M. C. (2008). Regulation of the Spermatogonial stem cell niche. Reproduction in Domestic Animal 43, 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota, J. , Chivukula, R. R. , O'Donnell, K. A. , Wentzel, E. A. , Montgomery, C. L. , Hwang, H. W. , Chang, T. C. , Vivekanandan, P. , Torbenson, M. , Clark, K. R. , Mendell, J. R. & Mendell, J. T. (2009). Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja, N. (2014). MicroRNAs and spermatogenesis. Fertility and Sterility 101, 1552–1562. [DOI] [PubMed] [Google Scholar]
- Krawetz, S. A. , Kruger, A. , Lalancette, C. , Tagett, R. , Anton, E. , Draghici, S. & Diamond, M. P. (2011). A survey of small RNAs in human sperm. Human Reproduction 26, 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Kanatsu‐Shinohara, M. , Inoue, K. , Ogonuki, N. , Miki, H. , Toyokuni, S. , Kimura, T. , Nakano, T. , Ogura, A. & Shinohara, T. (2007). Akt mediates self‐renewal division of mouse spermatogonial stem cells. Development 134, 1853–1859. [DOI] [PubMed] [Google Scholar]
- Lee, S. H. , Chen, T. Y. , Dhar, S. S. , Gu, B. , Chen, K. , Kim, Y. Z. , Li, W. & Lee, M. G. (2016). A feedback loop comprising PRMT7 and miR‐24‐2 interplays with Oct4, Nanog, Klf4 and c‐Myc to regulate stemness. Nucleic Acids Research 44, 10603–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, L. , Jin, S. , Gonzalez, G. , Behringer, R. R. & Woodruff, T. K. (2010). The regulatory role of Dicer in folliculogenesis in mice. Molecular and Cellular Endocrinology 315, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B. P. , Burge, C. B. & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Li, C. , Chen, C. , Chen, L. , Chen, S. , Li, H. , Zhao, Y. , Rao, J. & Zhou, X. (2016a). BDNF‐induced expansion of cumulus‐oocyte complexes in pigs was mediated by microRNA‐205. Theriogenology 85, 1476–1482. [DOI] [PubMed] [Google Scholar]
- Li, C. , Chen, S. , Li, H. , Chen, L. , Zhao, Y. , Jiang, Y. , Liu, Z. , Liu, Y. , Gao, S. , Wang, F. , Yu, J. , Wang, H. , Rao, J. & Zhou, X. (2016b). MicroRNA‐16 modulates melatonin‐induced cell growth in the mouse‐derived spermatogonia cell line GC‐1 spg cells by targeting Ccnd1. Biology of Reproduction 95(57), 1–10. [DOI] [PubMed] [Google Scholar]
- Li, P. , Guo, W. , Du, L. , Zhao, J. , Wang, Y. , Liu, L. , Hu, Y. & Hou, Y. (2013). microRNA‐29b contributes to pre‐eclampsia through its effects on apoptosis, invasion and angiogenesis of trophoblast cells. Clinical Science 124, 27–40. [DOI] [PubMed] [Google Scholar]
- Li, R. , Qiao, J. , Wang, L. , Li, L. , Zhen, X. , Liu, P. & Zheng, X. (2011). MicroRNA array and microarray evaluation of endometrial receptivity in patients with high serum progesterone levels on the day of hCG administration. Reproductive Biology and Endocrinology 9, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Fang, Y. , Liu, Y. & Yang, X. (2015). MicroRNAs in ovarian function and disorders. Journal of Ovarian Research 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, J. , Tian, H. , Liu, L. , Zhang, X. S. , Li, W. Q. , Deng, Y. M. , Yao, G. D. , Yin, M. M. & Sun, F. (2010). Downregulation of microRNA‐383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death and Disease 1, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Ridzon, D. , Wong, L. & Chen, C. (2007). Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. , Li, R. , Pan, Z. X. , Zhou, B. , Yu, D. B. , Wang, X. G. , Ma, X. S. , Han, J. , Shen, M. & Liu, H. L. (2012). miR‐26b promotes granulosa cell apoptosis by targeting ATM during follicular atresia in porcine ovary. PLoS One 7, e38640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, X. , Bao‐feng, Y. & Jing, A. (2013). MicroRNA transport: a new way in cell communication. Journal of Cellular Physiology 228, 1713–1719. [DOI] [PubMed] [Google Scholar]
- Liu, H. C. , Tang, Y. , He, Z. & Rosenwaks, Z. (2010). Dicer is a key player in oocyte maturation. Journal of Assisted Reproduction and Genetics 27, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Du, X. , Zhou, J. , Pan, Z. , Liu, H. & Li, Q. (2014). MicroRNA‐26b functions as a proapoptotic factor in porcine follicular granulosa cells by targeting Sma‐and mad‐related protein 4. Biology of Reproduction 91(146), 1–12. [DOI] [PubMed] [Google Scholar]
- Liu, J. L. & Yang, Z. M. (2011). Non‐coding RNAs and embryo implantation. Frontiers in Bioscience (Elite Ed) 3, 1092–1099. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Niu, Z. , Li, Q. , Pang, R. T. , Chiu, P. C. & Yeung, W. S. (2016). MicroRNA and embryo implantation. American Journal of Reproductive Immunology 75, 263–271. [DOI] [PubMed] [Google Scholar]
- Liu, W. M. , Pang, R. T. , Chiu, P. C. , Wong, B. P. , Lao, K. , Lee, K. F. & Yeung, W. S. (2012). Sperm‐borne microRNA‐34c is required for the first cleavage division in mouse. Proceedings of the National Academy of Scienes ot the United States of America 109, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Niu, M. , Yao, C. , Hai, Y. , Yuan, Q. , Liu, Y. , Guo, Y. , Li, Z. & He, Z. (2015). Fractionation of human spermatogenic cells using STA‐PUT gravity sedimentation and their miRNA profiling. Scientific Reports 5, 8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , Thomson, J. M. , Wong, H. Y. , Hammond, S. M. & Hogan, B. L. (2007). Transgenic over‐expression of the microRNA miR‐17‐92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Developmental Biology 310, 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luense, L. J. , Carletti, M. Z. & Christenson, L. K. (2009). Role of Dicer in female fertility. Trends in Endocrinology and Metabolism 20, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luense, L. J. , Veiga‐Lopez, A. , Padmanabhan, V. & Christenson, L. K. (2011). Developmental programming: gestational testosterone treatment alters fetal ovarian gene expression. Endocrinology 152, 4974–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. , Ye, G. , Nadeem, L. , Fu, G. , Yang, B. B. , Honarparvar, E. , Dunk, C. , Lye, S. & Peng, C. (2012). MicroRNA‐378a‐5p promotes trophoblast cell survival, migration and invasion by targeting nodal. Journal of Cell Science 125, 3124–3132. [DOI] [PubMed] [Google Scholar]
- Luo, L. , Ye, L. , Liu, G. , Shao, G. , Zheng, R. , Ren, Z. , Zuo, B. , Xu, D. , Lei, M. , Jiang, S. , Deng, C. , Xiong, Y. & Li, F. (2010). Microarray‐based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS One 5, e11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. F. , Hou, C. C. & Yang, W. X. (2016). Small non‐coding RNAs and their associated proteins in spermatogenesis. Gene 578, 141–157. [DOI] [PubMed] [Google Scholar]
- Luo, M. , Hao, L. , Hu, F. , Dong, Y. , Gou, L. , Zhang, W. , Wang, X. , Zhao, Y. , Jia, M. , Hu, S. & Zhang, X. (2015a). MicroRNA profiles and potential regulatory pattern during the early stage of spermatogenesis in mice. Science China Life Sciences 58, 442–450. [DOI] [PubMed] [Google Scholar]
- Luo, Z. , Liu, Y. , Chen, L. , Ellis, M. , Li, M. , Wang, J. , Zhang, Y. , Fu, P. , Wang, K. , Li, X. & Wang, L. (2015b). microRNA profiling in three main stages during porcine spermatogenesis. Journal of Assisted Reproduction and Genetics 32, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C. , Song, H. , Yu, L. , Guan, K. , Hu, P. , Li, Y. , Xia, X. , Li, J. , Jiang, S. & Li, F. (2016a). miR‐762 promotes porcine immature Sertoli cell growth via the ring finger protein 4 (RNF4) gene. Scientific Reports 6, 32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. , Lin, Y. , Zhao, Z. A. , Lu, X. , Yu, Y. , Zhang, X. , Wang, Q. & Li, L. (2016b). MicroRNA‐127 promotes mesendoderm differentiation of mouse embryonic stem cells by targeting left‐right determination factor 2. Journal of Biological Chemistry 291, 12126–12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. L. , Gong, F. , Tang, Y. , Li, X. , Li, X. , Yang, X. & Lu, G. (2015). Inhibition of endometrial Tiam1/Rac1 signals induced by miR‐22 up‐regulation leads to the failure of embryo implantation during the implantation window in pregnant mice. Biology of Reproduction 92(152), 1–13. [DOI] [PubMed] [Google Scholar]
- Ma, T. , Jiang, H. , Gao, Y. , Zhao, Y. , Dai, L. , Xiong, Q. , Xu, Y. , Zhao, Z. & Zhang, J. (2011). Microarray analysis of differentially expressed microRNAs in non‐regressed and regressed bovine corpus luteum tissue; microRNA‐378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene. Journal of Applied Genetics 52, 481–486. [DOI] [PubMed] [Google Scholar]
- Ma, W. , Xie, S. , Ni, M. , Huang, X. , Hu, S. , Liu, Q. , Liu, A. , Zhang, J. & Zhang, Y. (2012). MicroRNA‐29a inhibited epididymal epithelial cell proliferation by targeting nuclear autoantigenic sperm protein (NASP). Journal of Biological Chemistry 287, 10189–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk, D. M. , Loveland, K. L. , McManus, M. T. , Moore, K. & Harfe, B. D. (2008). Dicer1 is required for differentiation of the mouse male germline. Biology of Reproduction 79, 696–703. [DOI] [PubMed] [Google Scholar]
- Marcon, E. , Babak, T. , Chua, G. , Hughes, T. & Moens, P. B. (2008). miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Research 16, 243–260. [DOI] [PubMed] [Google Scholar]
- Marson, A. , Levine, S. S. , Cole, M. F. , Frampton, G. M. , Brambrink, T. , Johnstone, S. , Guenther, M. G. , Johnston, W. K. , Wernig, M. , Newman, J. , Calabrese, J. M. , Dennis, L. M. , Volkert, T. L. , Gupta, S. , Love, J. , Hannett, N. , Sharp, P. A. , Bartel, D. P. , Jaenisch, R. & Young, R. A. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, D. , Carre, W. , Sontakke, S. D. , Hogg, C. O. , Law, A. , Donadeu, F. X. & Clinton, M. (2012). Identification of miRNAs associated with the follicular‐luteal transition in the ruminant ovary. Reproduction 144, 221–233. [DOI] [PubMed] [Google Scholar]
- McIver, S. C. , Roman, S. D. , Nixon, B. & McLaughlin, E. A. (2012a). miRNA and mammalian male germ cells. Human Reproduction Update 18, 44–59. [DOI] [PubMed] [Google Scholar]
- McIver, S. C. , Stanger, S. J. , Santarelli, D. M. , Roman, S. D. , Nixon, B. & McLaughlin, E. A. (2012b). A unique combination of male germ cell miRNAs coordinates gonocyte differentiation. PLoS One 7, e35553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros, L. A. , Dennis, L. M. , Gill, M. E. , Houbaviy, H. , Markoulaki, S. , Fu, D. , White, A. C. , Kirak, O. , Sharp, P. A. , Page, D. C. & Jaenisch, R. (2011). Mir‐290‐295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proceedings of the National Academy of Sciences of the United States of America 108, 14163–14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, B. , Sinden, J. , Franzo‐Romain, M. , Botta, R. B. & Menon, K. M. (2013). Regulation of LH receptor mRNA binding protein by miR‐122 in rat ovaries. Endocrinology 154, 4826–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina, A. & Prevot, V. (2017). Hypothalamic microRNAs flip the switch for fertility. Oncotarget 8, 8993–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier, L. , Siddeek, B. , Vega, A. , Lakhdari, N. , Inoubli, L. , Bellon, R. P. , Lemaire, G. , Mauduit, C. & Benahmed, M. (2012). Perinatal programming of adult rat germ cell death after exposure to xenoestrogens: role of microRNA miR‐29 family in the down‐regulation of DNA methyltransferases and Mcl‐1. Endocrinology 153, 1936–1947. [DOI] [PubMed] [Google Scholar]
- Modzelewski, A. J. , Hilz, S. , Crate, E. A. , Schweidenback, C. T. , Fogarty, E. A. , Grenier, J. K. , Freire, R. , Cohen, P. E. & Grimson, A. (2015). Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. Journal of Cell Science 128, 2314–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky, E. & Rigoutsos, I. (2013). The miR‐17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death and Differentiation 20, 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Moya, J. M. , Vilella, F. , Martinez, S. , Pellicer, A. & Simon, C. (2014). The transcriptomic and proteomic effects of ectopic overexpression of miR‐30d in human endometrial epithelial cells. Molecular Human Reproduction 20, 550–566. [DOI] [PubMed] [Google Scholar]
- Moritoki, Y. , Hayashi, Y. , Mizuno, K. , Kamisawa, H. , Nishio, H. , Kurokawa, S. , Ugawa, S. , Kojima, Y. & Kohri, K. (2014). Expression profiling of microRNA in cryptorchid testes: miR‐135a contributes to the maintenance of spermatogonial stem cells by regulating FoxO1. The Journal of Urology 191, 1174–1180. [DOI] [PubMed] [Google Scholar]
- Murchison, E. P. , Stein, P. , Xuan, Z. , Pan, H. , Zhang, M. Q. , Schultz, R. M. & Hannon, G. J. (2007). Critical roles for Dicer in the female germline. Genes & Development 21, 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton, C. K. , Jain, S. , Strickland, A. M. , Gupta, A. & Milbrandt, J. (2006). Glial cell‐line derived neurotrophic factor‐mediated RET signaling regulates spermatogonial stem cell fate. Biology of Reproduction 74, 314–321. [DOI] [PubMed] [Google Scholar]
- Nicholls, P. K. , Harrison, C. A. , Walton, K. L. , McLachlan, R. I. , O'Donnell, L. & Stanton, P. G. (2011). Hormonal regulation of sertoli cell micro‐RNAs at spermiation. Endocrinology 152, 1670–1683. [DOI] [PubMed] [Google Scholar]
- Nie, M. , Yu, S. , Peng, S. , Fang, Y. , Wang, H. & Yang, X. (2015). miR‐23a and miR‐27a promote human granulosa cell apoptosis by targeting SMAD5. Biology of Reproduction 93, 98, 1–10. [DOI] [PubMed] [Google Scholar]
- Niu, Z. , Goodyear, S. M. , Rao, S. , Wu, X. , Tobias, J. W. , Avarbock, M. R. & Brinster, R. L. (2011). MicroRNA‐21 regulates the self‐renewal of mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America 108, 12740–12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, B. , Stanger, S. J. , Mihalas, B. P. , Reilly, J. N. , Anderson, A. L. , Dun, M. D. , Tyagi, S. , Holt, J. E. & McLaughlin, E. A. (2015a). Next generation sequencing analysis reveals segmental patterns of microRNA expression in mouse epididymal epithelial cells. PLoS One 10, e0135605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, B. , Stanger, S. J. , Mihalas, B. P. , Reilly, J. N. , Anderson, A. L. , Tyagi, S. , Holt, J. E. & McLaughlin, E. A. (2015b). The microRNA signature of mouse spermatozoa is substantially modified during epididymal maturation. Biology of Reproduction 93(91), 1–20. [DOI] [PubMed] [Google Scholar]
- Nothnick, W. B. (2012). The role of micro‐RNAs in the female reproductive tract. Reproduction 143, 559–576. [DOI] [PubMed] [Google Scholar]
- Novotny, G. W. , Sonne, S. B. , Nielsen, J. E. , Jonstrup, S. P. , Hansen, M. A. , Skakkebaek, N. E. , Rajpert‐De Meyts, E. , Kjems, J. & Leffers, H. (2007). Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR‐17‐92 cluster. Cell Death and Differentiation 14, 879–882. [DOI] [PubMed] [Google Scholar]
- Oatley, J. M. , Avarbock, M. R. & Brinster, R. L. (2007). Glial cell line‐derived neurotrophic factor regulation of genes essential for self‐renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. Journal of Biological Chemistry 282, 25842–25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata, Y. , Payer, B. , O'Carroll, D. , Ancelin, K. , Ono, Y. , Sano, M. , Barton, S. C. , Obukhanych, T. , Nussenzweig, M. , Tarakhovsky, A. , Saitou, M. & Surani, M. A. (2005). Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213. [DOI] [PubMed] [Google Scholar]
- Otsuka, M. , Zheng, M. , Hayashi, M. , Lee, J. D. , Yoshino, O. , Lin, S. & Han, J. (2008). Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. The Journal of Clinical Investigation 118, 1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerdoss, S. , Chang, Y. F. , Buddavarapu, K. C. , Chen, H. I. , Shetty, G. , Wang, H. , Chen, Y. , Kumar, T. R. & Rao, M. K. (2012). Androgen‐responsive microRNAs in mouse Sertoli cells. PLoS One 7, e41146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou, M. D. , Pitetti, J. L. , Ro, S. , Park, C. , Aubry, F. , Schaad, O. , Vejnar, C. E. , Kuhne, F. , Descombes, P. , Zdobnov, E. M. , McManus, M. T. , Guillou, F. , Harfe, B. D. , Yan, W. , Jegou, B. & Nef, S. (2009). Sertoli cell Dicer is essential for spermatogenesis in mice. Developmental Biology 326, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineles, B. L. , Romero, R. , Montenegro, D. , Tarca, A. L. , Han, Y. M. , Kim, Y. M. , Draghici, S. , Espinoza, J. , Kusanovic, J. P. , Mittal, P. , Hassan, S. S. & Kim, C. J. (2007). Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. American Journal of Obstetrics and Gynecology 196, 261.e1–261.e6. [DOI] [PubMed] [Google Scholar]
- Qin, X. , Yan, L. , Zhao, X. , Li, C. & Fu, Y. (2012). microRNA‐21 overexpression contributes to cell proliferation by targeting PTEN in endometrioid endometrial cancer. Oncology Letters 4, 1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing, X. , Shi, J. , Dong, T. , Wu, C. , Hu, L. & Li, H. (2017). Dysregulation of an X‐linked primate‐specific epididymal microRNA cluster in unexplained asthenozoospermia. Oncotarget 8, 56839–56849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoczy, J. , Fernandez‐Valverde, S. L. , Glazov, E. A. , Wainwright, E. N. , Sato, T. , Takada, S. , Combes, A. N. , Korbie, D. J. , Miller, D. , Grimmond, S. M. , Little, M. H. , Asahara, H. , Mattick, J. S. , Taft, R. J. & Wilhelm, D. (2013). MicroRNAs‐140‐5p/140‐3p modulate Leydig cell numbers in the developing mouse testis. Biology of Reproduction 88(143), 1–11. [DOI] [PubMed] [Google Scholar]
- Reilly, J. N. , McLaughlin, E. A. , Stanger, S. J. , Anderson, A. L. , Hutcheon, K. , Church, K. , Mihalas, B. P. , Tyagi, S. , Holt, J. E. , Eamens, A. L. & Nixon, B. (2016). Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Scientific Reports 6, 31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza, A. , Cho, S. K. , Choi, Y. J. , Hong, K. & Kim, J. H. (2018a). Microarray profiling of miRNA and mRNA expression in Rag2 knockout and wild‐type mouse spleens. Scientific Data 5, 170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza, A. , Choi, Y.‐J. & Kim, J.‐H. (2018b). MicroRNA and transcriptomic profiling showed miRNA‐dependent impairment of systemic regulation and synthesis of biomolecules in Rag2 KO mice. Molecules 23, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza, A. , Choi, Y. J. , Yuan, Y. G. , Das, J. , Yasuda, H. & Kim, J. H. (2017). MicroRNA‐7641 is a regulator of ribosomal proteins and a promising targeting factor to improve the efficacy of cancer therapy. Scientific Reports 7, 8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza, A. M. , Choi, Y. J. , Yasuda, H. & Kim, J. H. (2016). Human adipose mesenchymal stem cell‐derived exosomal‐miRNAs are critical factors for inducing anti‐proliferation signalling to A2780 and SKOV‐3 ovarian cancer cells. Scientific Reports 6, 38498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro, S. , Park, C. , Sanders, K. M. , McCarrey, J. R. & Yan, W. (2007). Cloning and expression profiling of testis‐expressed microRNAs. Developmental Biology 311, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, E. J. , Charatsi, I. , Joyner, C. J. , Koonce, C. H. , Morgan, M. , Islam, A. , Paterson, C. , Lejsek, E. , Arnold, S. J. , Kallies, A. , Nutt, S. L. & Bikoff, E. K. (2007). Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development 134, 4335–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, Y. , Meikar, O. , Papaioannou, M. D. , Conne, B. , Grey, C. , Weier, M. , Pralong, F. , De Massy, B. , Kaessmann, H. , Vassalli, J. D. , Kotaja, N. & Nef, S. (2011). Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One 6, e25241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, L. W. , McCallie, B. , Alvero, R. , Schoolcraft, W. B. , Minjarez, D. & Katz‐Jaffe, M. G. (2014). Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. Journal of Assisted Reproduction and Genetics 31, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadeldin, I. M. , Oh, H. J. & Lee, B. C. (2015). Embryonic–maternal cross‐talk via exosomes: potential implications. Stem Cells and Cloning : Advances and Applications 8, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Salas‐Huetos, A. , Blanco, J. , Vidal, F. , Mercader, J. M. , Garrido, N. & Anton, E. (2014). New insights into the expression profile and function of micro‐ribonucleic acid in human spermatozoa. Fertility and Sterility 102, 213–222.e4. [DOI] [PubMed] [Google Scholar]
- Salilew‐Wondim, D. , Ahmad, I. , Gebremedhn, S. , Sahadevan, S. , Hossain, M. D. , Rings, F. , Hoelker, M. , Tholen, E. , Neuhoff, C. , Looft, C. , Schellander, K. & Tesfaye, D. (2014). The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS One 9, e106795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Q. , Yao, Z. , Wang, H. , Feng, R. , Wang, H. , Zhao, X. , Xing, Q. , Jin, L. , He, L. , Wu, L. & Wang, L. (2013). Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. The Journal of Clinical Endocrinology & Metabolism 98, 3068–3079. [DOI] [PubMed] [Google Scholar]
- Santonocito, M. , Vento, M. , Guglielmino, M. R. , Battaglia, R. , Wahlgren, J. , Ragusa, M. , Barbagallo, D. , Borzi, P. , Rizzari, S. , Maugeri, M. , Scollo, P. , Tatone, C. , Valadi, H. , Purrello, M. & Di Pietro, C. (2014). Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertility and Sterility 102, 1751–1761.e1. [DOI] [PubMed] [Google Scholar]
- Schauer, S. N. , Sontakke, S. D. , Watson, E. D. , Esteves, C. L. & Donadeu, F. X. (2013). Involvement of miRNAs in equine follicle development. Reproduction 146, 273–282. [DOI] [PubMed] [Google Scholar]
- Sen, A. , Prizant, H. , Light, A. , Biswas, A. , Hayes, E. , Lee, H. J. , Barad, D. , Gleicher, N. & Hammes, S. R. (2014). Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA‐125b expression. Proceedings of the National Academy of Sciences of the United States of America 111, 3008–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, G. , Wu, R. , Liu, B. , Dong, W. , Tu, Z. , Yang, J. , Xu, Z. & Pan, T. (2014). Upstream and downstream mechanisms for the promoting effects of IGF‐1 on differentiation of spermatogonia to primary spermatocytes. Life Sciences 101, 49–55. [DOI] [PubMed] [Google Scholar]
- Shen, L. J. , He, J. L. , Yang, D. H. , Ding, Y. B. , Chen, X. M. , Geng, Y. Q. , Liu, S. J. , Liu, X. Q. & Wang, Y. X. (2013). Mmu‐microRNA‐200a overexpression leads to implantation defect by targeting phosphatase and tensin homolog in mouse uterus. Reproductive Sciences 20, 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, J. W. , Wang, L. Y. , Hung, C. L. , Kung, H. J. & Hsieh, C. L. (2015). Non‐coding RNAs in castration‐resistant prostate cancer: regulation of androgen receptor signaling and cancer metabolism. International Journal of Molecular Sciences 16, 28943–28978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin, A. V. , Laukova, M. , Ovcharenko, D. , Brenaut, P. & Mlyncek, M. (2010). Identification of microRNAs controlling human ovarian cell proliferation and apoptosis. Journal of Cellular Physiology 223, 49–56. [DOI] [PubMed] [Google Scholar]
- Sirotkin, A. V. , Ovcharenko, D. , Grossmann, R. , Laukova, M. & Mlyncek, M. (2009). Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome‐scale screen. Journal of Cellular Physiology 219, 415–420. [DOI] [PubMed] [Google Scholar]
- Smorag, L. , Zheng, Y. , Nolte, J. , Zechner, U. , Engel, W. & Pantakani, D. V. (2012). MicroRNA signature in various cell types of mouse spermatogenesis: evidence for stage‐specifically expressed miRNA‐221, −203 and ‐34b‐5p mediated spermatogenesis regulation. Biology of the Cell 104, 677–692. [DOI] [PubMed] [Google Scholar]
- Sohel, M. H. (2016). Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achievements in the Life Sciences 10, 175–186. [Google Scholar]
- Sohel, M. M. , Hoelker, M. , Noferesti, S. S. , Salilew‐Wondim, D. , Tholen, E. , Looft, C. , Rings, F. , Uddin, M. J. , Spencer, T. E. , Schellander, K. & Tesfaye, D. (2013). Exosomal and non‐exosomal transport of extra‐cellular microRNAs in follicular fluid: implications for bovine oocyte developmental competence. PLoS One 8, e78505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C. , Yao, J. , Cao, C. , Liang, X. , Huang, J. , Han, Z. , Zhang, Y. , Qin, G. , Tao, C. , Li, C. , Yang, H. , Zhao, J. , Li, K. & Wang, Y. (2016). PPARgamma is regulated by miR‐27b‐3p negatively and plays an important role in porcine oocyte maturation. Biochemical and Biophysical Research Communications 479, 224–230. [DOI] [PubMed] [Google Scholar]
- Song, P. P. , Hu, Y. , Liu, C. M. , Yan, M. J. , Song, G. , Cui, Y. , Xia, H. F. & Ma, X. (2011). Embryonic ectoderm development protein is regulated by microRNAs in human neural tube defects. American Journal of Obstetrics and Gynecology 204, 544.e9–544.e17. [DOI] [PubMed] [Google Scholar]
- Song, R. , Ro, S. , Michaels, J. D. , Park, C. , McCarrey, J. R. & Yan, W. (2009). Many X‐linked microRNAs escape meiotic sex chromosome inactivation. Nature Genetics 41, 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W. , Mu, H. , Wu, J. , Liao, M. , Zhu, H. , Zheng, L. , He, X. , Niu, B. , Zhai, Y. , Bai, C. , Lei, A. , Li, G. & Hua, J. (2015). miR‐544 regulates dairy goat male germline stem cell self‐renewal via targeting PLZF. Journal of Cellular Biochemistry 116, 2155–2165. [DOI] [PubMed] [Google Scholar]
- Sontakke, S. D. , Mohammed, B. T. , McNeilly, A. S. & Donadeu, F. X. (2014). Characterization of microRNAs differentially expressed during bovine follicle development. Reproduction 148, 271–283. [DOI] [PubMed] [Google Scholar]
- Sood, P. , Krek, A. , Zavolan, M. , Macino, G. & Rajewsky, N. (2006). Cell‐type‐specific signatures of microRNAs on target mRNA expression. Proceedings of the National Academy of Sciences of the United States of America 103, 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, N. , Baehner, L. , Moltzahn, F. , Melton, C. , Shenoy, A. , Chen, J. & Blelloch, R. (2010). MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current Biology 20, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, N. & Blelloch, R. (2011). Small RNAs in early mammalian development: from gametes to gastrulation. Development 138, 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, S. , Jia, Y. , He, B. , Li, R. , Li, X. , Cai, D. , Song, H. , Zhang, R. & Zhao, R. (2014). Maternal low‐protein diet alters ovarian expression of folliculogenic and steroidogenic genes and their regulatory microRNAs in neonatal piglets. Asian‐Australasian Journal of Animal Science 27, 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda, P. & Flemr, M. (2010). The role of miRNAs and endogenous siRNAs in maternal‐to‐zygotic reprogramming and the establishment of pluripotency. EMBO Reports 11, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S. , Berezikov, E. , Choi, Y. L. , Yamashita, Y. & Mano, H. (2009). Potential role of miR‐29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA 15, 1507–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F. , Kaneda, M. , O'Carroll, D. , Hajkova, P. , Barton, S. C. , Sun, Y. A. , Lee, C. , Tarakhovsky, A. , Lao, K. & Surani, M. A. (2007). Maternal microRNAs are essential for mouse zygotic development. Genes & Development 21, 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Y. , Wang, Y. , Fu, J. , Cheng, X. , Miao, S. & Wang, L. (2011). Cyclin T2: a novel miR‐15a target gene involved in early spermatogenesis. FEBS Letters 585, 2493–2500. [DOI] [PubMed] [Google Scholar]
- Tesfaye, D. , Worku, D. , Rings, F. , Phatsara, C. , Tholen, E. , Schellander, K. & Hoelker, M. (2009). Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Molecular Reproduction & Development 76, 665–677. [DOI] [PubMed] [Google Scholar]
- Toloubeydokhti, T. , Bukulmez, O. & Chegini, N. (2008). Potential regulatory functions of microRNAs in the ovary. Seminars in Reproductive Medicine 26, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, M. H. , Mitchell, D. , Evanoff, R. & Griswold, M. D. (2011). Expression of Mirlet7 family microRNAs in response to retinoic acid‐induced spermatogonial differentiation in mice. Biology of Reproduction 85, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, M. H. , Mitchell, D. A. , McGowan, S. D. , Evanoff, R. & Griswold, M. D. (2012). Two miRNA clusters, Mir‐17‐92 (Mirc1) and Mir‐106b‐25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biology of Reproduction 86(72), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torley, K. J. , da Silveira, J. C. , Smith, P. , Anthony, R. V. , Veeramachaneni, D. N. , Winger, Q. A. & Bouma, G. J. (2011). Expression of miRNAs in ovine fetal gonads: potential role in gonadal differentiation. Reproductive Biology and Endocrinology 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripurani, S. K. , Xiao, C. , Salem, M. & Yao, J. (2010). Cloning and analysis of fetal ovary microRNAs in cattle. Animal Reproduction Science 120, 16–22. [DOI] [PubMed] [Google Scholar]
- Tscherner, A. , Gilchrist, G. , Smith, N. , Blondin, P. , Gillis, D. & LaMarre, J. (2014). MicroRNA‐34 family expression in bovine gametes and preimplantation embryos. Reproductive Biology and Endocrinology 12, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, F. , Pan, Z. X. , Yao, Y. , Liu, H. L. , Liu, S. R. , Xie, Z. & Li, Q. F. (2014). miR‐34a targets the inhibin beta B gene, promoting granulosa cell apoptosis in the porcine ovary. Genetic and Molecular Research 13, 2504–2512. [DOI] [PubMed] [Google Scholar]
- Tulay, P. , Naja, R. P. , Cascales‐Roman, O. , Doshi, A. , Serhal, P. & SenGupta, S. B. (2015). Investigation of microRNA expression and DNA repair gene transcripts in human oocytes and blastocysts. Journal of Assisted Reproduction and Genetics 32, 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcatel, G. , Rubin, N. , El‐Hashash, A. & Warburton, D. (2012). MIR‐99a and MIR‐99b modulate TGF‐β induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS One 7, e31032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twenter, H. M. , Belk, A. D. , Klohonatz, K. M. , Bass, L. D. , Bouma, G. J. & Bruemmer, J. E. (2017). An investigation into miRNAs in the equine epididymis as potential regulators of Spermatozoal maturation. Journal of Equine Veterinary Science 48, 61–68. [Google Scholar]
- Uhde, K. , van Tol, H. , Stout, T. & Roelen, B. (2017). MicroRNA expression in bovine cumulus cells in relation to oocyte quality. Non‐Coding RNA 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura, K. , Ishioka, S. , Endo, T. , Ezaka, Y. , Takahashi, M. & Saito, T. (2013). Roles of microRNA‐34a in the pathogenesis of placenta accreta. The Journal of Obstetrics and Gynaecology Research 39, 67–74. [DOI] [PubMed] [Google Scholar]
- Vasileva, A. , Tiedau, D. , Firooznia, A. , Muller‐Reichert, T. & Jessberger, R. (2009). Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Current Biology 19, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velthut‐Meikas, A. , Simm, J. , Tuuri, T. , Tapanainen, J. S. , Metsis, M. & Salumets, A. (2013). Research resource: small RNA‐seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Molecular Endocrinology 27, 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal, J. A. & Ventura, A. (2012). Embryonic stem cell miRNAs and their roles in development and disease. Seminars in Cancer Biology 22, 428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, S. D. , Dunn, N. R. , Sciammas, R. , Shapiro‐Shalef, M. , Davis, M. M. , Calame, K. , Bikoff, E. K. & Robertson, E. J. (2005). The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132, 1315–1325. [DOI] [PubMed] [Google Scholar]
- Viswanathan, S. R. , Mermel, C. H. , Lu, J. , Lu, C. W. , Golub, T. R. & Daley, G. Q. (2009). microRNA expression during trophectoderm specification. PLoS One 4, e6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright, E. N. , Jorgensen, J. S. , Kim, Y. , Truong, V. , Bagheri‐Fam, S. , Davidson, T. , Svingen, T. , Fernandez‐Valverde, S. L. , McClelland, K. S. , Taft, R. J. , Harley, V. R. , Koopman, P. & Wilhelm, D. (2013). SOX9 regulates microRNA miR‐202‐5p/3p expression during mouse testis differentiation. Biology of Reproduction 89(34), 1–12. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Fan, H. , Zhao, G. , Liu, D. , Du, L. , Wang, Z. , Hu, Y. & Hou, Y. (2012). miR‐16 inhibits the proliferation and angiogenesis‐regulating potential of mesenchymal stem cells in severe pre‐eclampsia. The FEBS Journal 279, 4510–4524. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhou, T. , Wan, J. , Yang, Y. , Chen, X. , Wang, J. , Zhou, C. , Liu, M. , Ling, X. & Zhang, J. (2016). Comparative transcriptome analysis reveals a regulatory network of microRNA‐29b during mouse early embryonic development. Oncotarget 7, 53772–53782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T. , Takeda, A. , Tsukiyama, T. , Mise, K. , Okuno, T. , Sasaki, H. , Minami, N. & Imai, H. (2006). Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon‐derived siRNAs in oocytes and germline small RNAs in testes. Genes & Development 20, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, J. M. , Edwards, A. K. , Khalaj, K. , Kridli, R. T. , Bidarimath, M. & Tayade, C. (2013). The microRNAome of pregnancy: deciphering miRNA networks at the maternal‐fetal interface. PLoS One 8, e72264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, J. A. , Viswanathan, S. R. , Yabuuchi, A. , Cunniff, K. , Takeuchi, A. , Park, I. H. , Sero, J. E. , Zhu, H. , Perez‐Atayde, A. , Frazier, A. L. , Surani, M. A. & Daley, G. Q. (2009). A role for Lin28 in primordial germ‐cell development and germ‐cell malignancy. Nature 460, 909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, E. C. , Hale, B. J. , Yang, C. X. , Njoka, J. G. & Ross, J. W. (2016). MicroRNA‐21 and PDCD4 expression during in vitro oocyte maturation in pigs. Reproductive Biology and Endocrinology 14, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Bao, J. , Wang, L. , Hu, Y. & Xu, C. (2011). MicroRNA‐184 downregulates nuclear receptor corepressor 2 in mouse spermatogenesis. BMC Developmental Biology 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Zhu, H. , Song, W. , Li, M. , Liu, C. , Li, N. , Tang, F. , Mu, H. , Liao, M. , Li, X. , Guan, W. , Li, X. & Hua, J. (2014). Identification of conservative microRNAs in Saanen dairy goat testis through deep sequencing. Reproduction in Domestic Animal 49, 32–40. [DOI] [PubMed] [Google Scholar]
- Wu, S. , Sun, H. , Zhang, Q. , Jiang, Y. , Fang, T. , Cui, I. , Yan, G. & Hu, Y. (2015). MicroRNA‐132 promotes estradiol synthesis in ovarian granulosa cells via translational repression of Nurr1. Reproductive Biology and Endocrinology 13, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H. F. , Jin, X. H. , Cao, Z. F. , Hu, Y. & Ma, X. (2014). MicroRNA expression and regulation in the uterus during embryo implantation in rat. The FEBS Journal 281, 1872–1891. [DOI] [PubMed] [Google Scholar]
- Xiao, G. , Xia, C. , Yang, J. , Liu, J. , Du, H. , Kang, X. , Lin, Y. , Guan, R. , Yan, P. & Tang, S. (2014). MiR‐133b regulates the expression of the actin protein TAGLN2 during oocyte growth and maturation: a potential target for infertility therapy. PLoS One 9, e100751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, R. , Lin, X. , Du, T. , Xu, K. , Shen, H. , Wei, F. , Hao, W. , Lin, T. , Lin, X. , Qin, Y. , Wang, H. , Chen, L. , Yang, S. , Yang, J. , Rong, X. , Yao, K. , Xiao, D. , Jia, J. & Sun, Y. (2016). Targeted disruption of miR‐17‐92 impairs mouse spermatogenesis by activating mTOR signaling pathway. Medicine (Baltimore) 95, e2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N. , Papagiannakopoulos, T. , Pan, G. , Thomson, J. A. & Kosik, K. S. (2009). MicroRNA‐145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658. [DOI] [PubMed] [Google Scholar]
- Xu, S. , Linher‐Melville, K. , Yang, B. B. , Wu, D. & Li, J. (2011a). Micro‐RNA378 (miR‐378) regulates ovarian estradiol production by targeting aromatase. Endocrinology 152, 3941–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. W. , Wang, B. , Ding, C. H. , Li, T. , Gu, F. & Zhou, C. (2011b). Differentially expressed micoRNAs in human oocytes. Journal of Assisted Reproduction and Genetics 28, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, R. P. & Kotaja, N. (2014). Small RNAs in spermatogenesis. Molecular and Cellular Endocrinology 382, 498–508. [DOI] [PubMed] [Google Scholar]
- Yan, G. , Zhang, L. , Fang, T. , Zhang, Q. , Wu, S. , Jiang, Y. , Sun, H. & Hu, Y. (2012). MicroRNA‐145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB. FEBS Letters 586, 3263–3270. [DOI] [PubMed] [Google Scholar]
- Yan, N. , Lu, Y. , Sun, H. , Qiu, W. , Tao, D. , Liu, Y. , Chen, H. , Yang, Y. , Zhang, S. , Li, X. & Ma, Y. (2009). Microarray profiling of microRNAs expressed in testis tissues of developing primates. Journal of Assisted Reproduction and Genetics 26, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, N. , Lu, Y. , Sun, H. , Tao, D. , Zhang, S. , Liu, W. & Ma, Y. (2007). A microarray for microRNA profiling in mouse testis tissues. Reproduction 134, 73–79. [DOI] [PubMed] [Google Scholar]
- Yang, Q. , Hua, J. , Wang, L. , Xu, B. , Zhang, H. , Ye, N. , Zhang, Z. , Yu, D. , Cooke, H. J. , Zhang, Y. & Shi, Q. (2013a). MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS One 8, e66809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q. , Lin, J. , Liu, M. , Li, R. , Tian, B. , Zhang, X. , Xu, B. , Liu, M. , Zhang, X. , Li, Y. , Shi, H. & Wu, L. (2016). Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Molecular Biology 2, e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q. E. & Oatley, J. M. (2014). Spermatogonial stem cell functions in physiological and pathological conditions. Current Topics in Developmental Biology 107, 235–267. [DOI] [PubMed] [Google Scholar]
- Yang, Q. E. , Racicot, K. E. , Kaucher, A. V. , Oatley, M. J. & Oatley, J. M. (2013b). MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development 140, 280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Wang, S. , Luo, A. , Ding, T. , Lai, Z. , Shen, W. , Ma, X. , Cao, C. , Shi, L. , Jiang, J. , Rong, F. , Ma, L. , Tian, Y. , Du, X. , Lu, Y. , Li, Y. & Wang, S. (2013c). Expression patterns and regulatory functions of microRNAs during the initiation of primordial follicle development in the neonatal mouse ovary. Biology of Reproduction 89(126), 1–11. [DOI] [PubMed] [Google Scholar]
- Yang, W. J. , Yang, D. D. , Na, S. , Sandusky, G. E. , Zhang, Q. & Zhao, G. (2005). Dicer is required for embryonic angiogenesis during mouse development. Journal of Biological Chemistry 280, 9330–9335. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Zhou, Y. , Peng, S. , Wu, L. , Lin, H. Y. , Wang, S. & Wang, H. (2012). Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir‐23a in granulosa cell apoptosis. Reproduction 144, 235–244. [DOI] [PubMed] [Google Scholar]
- Yao, C. , Sun, M. , Yuan, Q. , Niu, M. , Chen, Z. , Hou, J. , Wang, H. , Wen, L. , Liu, Y. , Li, Z. & He, Z. (2016). MiRNA‐133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget 7, 2201–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, G. , Yin, M. , Lian, J. , Tian, H. , Liu, L. , Li, X. & Sun, F. (2010a). MicroRNA‐224 is involved in transforming growth factor‐beta‐mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Molecular Endocrinology 24, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, N. , Yang, B. Q. , Liu, Y. , Tan, X. Y. , Lu, C. L. , Yuan, X. H. & Ma, X. (2010b). Follicle‐stimulating hormone regulation of microRNA expression on progesterone production in cultured rat granulosa cells. Endocrine 38, 158–166. [DOI] [PubMed] [Google Scholar]
- Yin, M. , Lu, M. , Yao, G. , Tian, H. , Lian, J. , Liu, L. , Liang, M. , Wang, Y. & Sun, F. (2012). Transactivation of microRNA‐383 by steroidogenic factor‐1 promotes estradiol release from mouse ovarian granulosa cells by targeting RBMS1. Molecular Endocrinology 26, 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, M. , Wang, X. , Yao, G. , Lu, M. , Liang, M. , Sun, Y. & Sun, F. (2014). Transactivation of micrornA‐320 by microRNA‐383 regulates granulosa cell functions by targeting E2F1 and SF‐1 proteins. Journal of Biological Chemistry 289, 18239–18257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C. , Li, M. , Wang, Y. , Liu, Y. , Yan, C. , Pan, J. , Liu, J. & Cui, S. (2017). MiR‐375 mediates CRH signaling pathway in inhibiting E2 synthesis in porcine ovary. Reproduction 153, 63–73. [DOI] [PubMed] [Google Scholar]
- Yu, M. , Mu, H. , Niu, Z. , Chu, Z. , Zhu, H. & Hua, J. (2014). miR‐34c enhances mouse spermatogonial stem cells differentiation by targeting Nanos2. Journal of Cellular Biochemistry 115, 232–242. [DOI] [PubMed] [Google Scholar]
- Yu, Z. , Raabe, T. & Hecht, N. B. (2005). MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biology of Reproduction 73, 427–433. [DOI] [PubMed] [Google Scholar]
- Yuan, S. , Schuster, A. , Tang, C. , Yu, T. , Ortogero, N. , Bao, J. , Zheng, H. & Yan, W. (2016). Sperm‐borne miRNAs and endo‐siRNAs are important for fertilization and preimplantation embryonic development. Development 143, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, S. , Tang, C. , Zhang, Y. , Wu, J. , Bao, J. , Zheng, H. , Xu, C. & Yan, W. (2015). Mir‐34b/c and mir‐449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biology Open 4, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Jiang, X. , Zhang, Y. , Xu, B. , Hua, J. , Ma, T. , Zheng, W. , Sun, R. , Shen, W. , Cooke, H. J. , Hao, Q. , Qiao, J. & Shi, Q. (2014). microRNA 376a regulates follicle assembly by targeting Pcna in fetal and neonatal mouse ovaries. Reproduction 148, 43–54. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Ji, X. , Zhou, D. , Li, Y. , Lin, J. , Liu, J. , Luo, H. & Cui, S. (2013a). miR‐143 is critical for the formation of primordial follicles in mice. Frontiers in Bioscience (Landmark Ed) 18, 588–597. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Wang, Y. , Liu, X. , Jiang, S. , Zhao, C. , Shen, R. , Guo, X. , Ling, X. & Liu, C. (2015). Expression and potential role of microRNA‐29b in mouse early embryo development. Cellular Physiology and Biochemistry 35, 1178–1187. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Lyu, Y. , Pan, X. , Xu, L. , Xuan, A. , He, X. , Huang, W. & Long, D. (2017). miR146b5p promotes the neural conversion of pluripotent stem cells by targeting Smad4. International Journal of Molecular Medicine 40, 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Sun, H. , Jiang, Y. , Ding, L. , Wu, S. , Fang, T. , Yan, G. & Hu, Y. (2013b). MicroRNA‐181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS One 8, e59667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Fei, M. , Xue, G. , Zhou, Q. , Jia, Y. , Li, L. , Xin, H. & Sun, S. (2012). Elevated levels of hypoxia‐inducible microRNA‐210 in pre‐eclampsia: new insights into molecular mechanisms for the disease. Journal of Cellular and Molecular Medicine 16, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Huo, R. , Wang, F. Q. , Lin, M. , Zhou, Z. M. & Sha, J. H. (2007). Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertility and Sterility 87, 436–438. [DOI] [PubMed] [Google Scholar]
- Zheng, G. X. , Ravi, A. , Calabrese, J. M. , Medeiros, L. A. , Kirak, O. , Dennis, L. M. , Jaenisch, R. , Burge, C. B. & Sharp, P. A. (2011). A latent pro‐survival function for the mir‐290‐295 cluster in mouse embryonic stem cells. PLoS Genetics 7, e1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Liu, J. , Pan, Z. , Du, X. , Li, X. , Ma, B. , Yao, W. , Li, Q. & Liu, H. (2015). The let‐7g microRNA promotes follicular granulosa cell apoptosis by targeting transforming growth factor‐beta type 1 receptor. Molecular and Cellular Endocrinology 409, 103–112. [DOI] [PubMed] [Google Scholar]
- Zielak‐Steciwko, A. E. , Browne, J. A. , McGettigan, P. A. , Gajewska, M. , Dzieciol, M. , Szulc, T. & Evans, A. C. (2014). Expression of microRNAs and their target genes and pathways associated with ovarian follicle development in cattle. Physiological Genomics 46, 735–745. [DOI] [PubMed] [Google Scholar]
- Zovoilis, A. , Nolte, J. , Drusenheimer, N. , Zechner, U. , Hada, H. , Guan, K. , Hasenfuss, G. , Nayernia, K. & Engel, W. (2008). Multipotent adult germline stem cells and embryonic stem cells have similar microRNA profiles. Molecular Human Reproduction 14, 521–529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Functions of microRNAs (miRNAs) in sex specification and commitment of the mammalian reproductive system.
Table S2. Functions of microRNAs (miRNAs) in spermatogenesis and spermiogenesis.
Table S3. Functions of microRNAs (miRNAs) during folliculogenesis and oogenesis.
Table S4. Functions of microRNAs (miRNAs) in the regulation of steroidogenic cells and hormonal balance.
Table S5. Functions of microRNAs (miRNAs) in fertilization, implantation and germ‐layer specification.


