Abstract
Background/aim
We aimed to develop a rapid method to enumerate Listeria monocytogenes (L. monocytogenes) utilizing magnetic nanoparticle based preconcentration and surface-enhanced Raman spectroscopy measurements.
Materials and methods
Biological activities of magnetic Au-nanoparticles have been observed to have the high biocompatibility, and a sample immunosensor model has been designed to use avidin attached Au-nanoparticles for L. monocytogenes detection. Staphylococcus aureus (S. aureus) and Salmonella typhimurium (S. typhimurium) bacteria cultures were chosen for control studies. Antimicrobial activity studies have been done to identify bio-compatibility and bio-characterization of the Au-nanoparticles in our previous study and capturing efficiencies to bacterial surfaces have been also investigated.
Results
We constructed the calibration graphs in various population density of L. monocytogenes as 2.2 × 101 to 2.2 × 106 cfu/mL and the capture efficiency was found to be 75%. After the optimization procedures, population density of L. monocytogenes and Raman signal intensity showed a good linear correlation (R2 = 0.991) between 102 to 106 cfu/mL L. monocytogenes. The presented sandwich assay provides low detection limits and limit of quantification as 12 cfu/mL and 37 cfu/mL, respectively. We also compared the experimental results with reference plate-counting methods and the practical utility of the proposed assay is demonstrated using milk samples.
Conclusion
It is focused on the enumeration of L. monocytogenes in milk samples and the comparision of results of milk analysis obtained by the proposed SERS method and by plate counting method stay in food agreement. In the present study, all parameters were optimized to select SERS-based immunoassay method for L. monocytogenes bacteria to ensure LOD, selectivity, precision and repeatablity.
Keywords: Immunomagnetic separation (IMS), surface-enhanced Raman scattering (SERS), Listeria monocytogenes(L. monocytogenes)
1. Introduction
Listeria monocytogenes (L. monocytogenes) is a crucial foodborne pathogenentailing disease. L. monocytogenes can grow and develop even at refrigerator temperatures and is a major problem, especially in ready-to-eat foods. Listeriosis illness is caused by contaminated foods with L. monocytogenes [1]. Raw milk is known as an important source of L. monocytogenes. In 1986, Hayes et al. isolated this bacterium from 12 samples from 100 raw milk samples in USA [2].
Rapid pathogenic bacterial diagnosis has been applied to conduct measurements in biological and food matrix [3]. Up to date, different method has been applied by several research group for enumeration of pathogenic bacteriaespecially L. monocytogenes using polymerase chain reaction immunoassay [4,5], electrochemical sensors [6–8], bioluminescence [9,10], DNA-based sensors [u2935], ELISA [u2936], surface plasmon resonance [u2937], fluorescence [u2938], surface-enhanced Raman scattering (SERS) [19–21]. It was indicated that the reported methods were optimized to select proper system usage to obtain selectivity and precision, there were some problems such as poor sensitivity and long experimental procedures. Also, the enumeration of pathogenin food matrix is problematic [22]. Therefore, new analytical methods are required for the detecting of pathogens and other biomolecules in food matrix. Recently, immunomagnetic separation (IMS) overcomes the matrix effect and is used for the enumeration of bacteria. IMS can eliminate the potential interferences and it has been applied to conduct measurements in food matrix, thereby bacteria can be captured easily [23,24].
In recent years, SERS is commonly used due to its high sensitivity (single molecules can be detected), ability to analyse multiple analytes in one sample, small sample volume, selective to target molecule signal [25–27]. More target molecule can be detected with using the combination of SERS and IMS techniques. Furthermore, the usage of a SERS tag as 5,5’-dithiobis(2-nitrobenzoic acid [28–30], rhodamine dye [31], Texas red [32] enhances the SERS signal and can reach low detection limits compared to label-free detection methods [33,34].
The biocompatibility of nanomaterials in biological systems was characterized and thus, it was aimed to increase the usage possibilities of these nanoparticles. In this study, biological characterization studies such as antimicrobial, antioxidant activities, cytotoxic and anticarcinogenic effects, genotoxicity tests and capturing efficiencies of nanoparticles which would be used as immunoassay design were conducted. In the first part, some parameters (antioxidant activities, cytotoxic, anticarcinogenic effects and genotoxicity tests) of this study were given in our previous study [35]. As a continuation study, antimicrobial characterization and capturing efficiency studies of nanoparticles were performed and the bioassay design of L. monocytogenes was developed. In order to determine the antimicrobial effects of nanoparticles, the studies were performed with L. monocytogenes, S. aureus, S. typhimurium bacteria and a liveness rate of approximately 96% was reached on each bacterium and thus, the antimicrobial effects of the magnetic Au-nanospheres were shown to be quite low. The competitive and noncompetitive capturing amount of nanoparticles on bacteria were also studied. The competitive capturing efficiency of magnetic-Au-nanoparticles was found as 75% in immunoassay model. In the noncompetitive studies, the attachment ratio of L. monocytogenes was found as higher than the attachment of S. aureus and S. typhimurium. Then, SERS-based immunoassay method was developed using Au-nanorods (for SERS labeling) and magnetic Au-nanospheres (for IMS). A calibration curve was constracted for the enumaration of L. monocytogenes in a model system. The present paper is focused on the enumeration of L. monocytogenes in milk samples and the comparision of results of milk analysis obtained by the proposed SERS method and by plate counting method stay in food agreement. In the present study, all parameters were optimized to select SERS-based immunoassay method for L. monocytogenes bacteria to ensure LOD, selectivity, precision and repeatablity.
2. Experimental
2.1. Materials
Disodium hydrogen phosphate (Na2HPO4), silver nitrate (AgNO3), sodium borohydride (NaBH4), solution (30%), absolute ethanol, perchloric acid, ethanolamine, iron (II) sulfate heptahydrate were purchased from Merck KGaA (Darmstadt, Germany). N-Hydroxysulphosuccinimide sodium salt (NHS) was purchased from Pierce Biotechnology (Bonn, Germany). NaCl, Na2HPO4, and KH2PO4 were purchased from J.T. Baker (Deventer, Netherlands). Hydrogen tetrachloroaurate (HAuCl4), was purchased from Sigma-Aldrich Chemie GmbH (Steinheim, Germany). Other chemicals are analytical grade.
2.2. Buffers
Physiological saline (PS) (0.875g/100mL) was prepared by NaCl and distilled water. Na2HPO4, KH2PO4, and NaCl were used for the preparation of PBS buffers (0.1 M, pH 7.4) and adjusted the pH with HCl or NaOH. To adjust the pH of MES buffer (0.05 M, pH 6.5), 0.1 N NaOH was used. The same buffer was also used for the preparation of avidin (0.5 mg/mL). Gluteraldehyde (2.5%) and Osmium tetraoxide (0.1%) were prepared with PS solution (0.875g/100mL). Milli-Q quality water (18 MΩ cm) was used throughout the study.
2.3. Microorganisms
Staphylococcus aureus (S. aureus), Listeria monocytogenes (L. monocytogenes), Salmonella typhimurium (S. typhimurium) bacteria cultures were received from Biotechnology Laboratory at Gazi University, Ankara, Turkey. For L. monocytogenes, S. aureus, S. typhimurium detection nutrient broth was purchased from Merck KGaA (Darmstadt, Germany). L. monocytogenes colonies were selected easily by using CHROMagarTM Listeria culture medium (CHROMagar Microbiology, Paris, France Listeria). We diluted cultures serially (10-fold steps) with PS buffer and plated with 100 μL diluted solution of the culture. We counted colonies after incubation at 37 °C for 24 h.
2.4. Instrumentation
Absorbance measurements of nanoparticles were obtained with an UV-Visible spectrophotometer (Agilent Technologies, Inc., Santa Clara, CA, USA). The Tecnai G2 F30 instrument (FEI Company, Hillsboro, OR, USA) was used to capture TEM images at operated 120 kV. For TEM measurements, 10 μL of nanoparticle solution was dropped and waited for 10 min. FEI Nova NanoSEM 430 microscope (FEI, Eindhoven, Netherlands) was used to get SEM images. Bacteria concentrations were adjusted using a Densitometer (Grant Instruments Ltd., Cambridge, UK). Raman measurements were performed using a Raman Microscopy (Deltanu Inc., Laramie, WY, USA). In the present study, laser source is 785 nm and 20x objective, 30 mm laser spot size, 0.15 W laser power, and 20 s acquisition time.
2.5. Fabrication of Au-coated magnetic spherical nanoparticles
In our previous work, we synthesized a core-shell Au@Fe3O4 nanoparticles. Here, with a brief modification, FeCl3 (1.28 M) and FeSO4.7H2O (0.64 M) were prepared and a solution of 1 M NaOH was added dropwise into the mixture with stirring for 40 min. After addition of 1M NaOH, black participate was obtained. This participate was removed from the reaction chamber via simple magnet and washed 3 times. To coat gold layer onto the iron nanoparticles, we performed the same procedure as reported our previous report (37).
2.6. Fabrication of Au-nanorods
For the SERS tag, we synthesized rod shaped Au nanoparticles based on our previous report. Briefly, we prepared a seed solution mixing CTAB (7.5 mL, 0.1 M) and HAuCl4 (250 µL, 0.01 M) solution. Then, we added NaBH4 (ice-cold, 600µL, 0.01 M) to the resulting solution. After waiting for 5 min, CTAB (4.75 mL, 0.1 M), HAuCl4 (1.0 mL, 0.01 M) and AgNO3 (60µL, 0.004 M) were mixed and the orange colour solution was observed. After adding of ascorbic acid (250µL, 0.01 M), the colour turned colourless. Finally, 5 µL seed solution was added to the resulting solution and waited for 3 h.
2.7. Immunomagnetic separation (IMS)with modified magnetic nanoparticles
We modified the gold coated magnetic nanoparticles using 0.15 M 11-MUA to form a SAM in ethanol overnight. Then, we collected the nanoparticles using a permanent magnet. EDC/NHS (1 mL) was added to the nanoparticle solution and waited for 40 min. After washing steps (2 times), 50 mM MES buffer solution was added. To modify with avidin, the resulting nanoparticles were incubated with avidin solution for 40 min. To eliminate the nonspecific interactions, we used 1% (v/v) ethanolamine for 1 hour. Then, the biotin-labeled L. monocytogenes antibody was added to the avidin modified nanoparticle solution. Then, washing procedure was carried out using PBS to remove unconjugated biotinylated antibodies. All washing procedures have been conducted in an ultrasound bath for 10 s.
2.8. Determination of nanoparticles’ antimicrobial activities
Two different methods were used to determine the antimicrobial activity. In the first (direct) method, antimicrobial activities of magnetic Au-nanoparticles on L. monocytogenes, S. aureus and S. typhimurium strains were tested directly. Each bacteria culture was activated twice in nutrient broth before use. All activated bacteria (L. monocytogenes, 6.8 × 107 cfu/mL; S. aureus, 10.4 × 1010 cfu/mL; S. typhimurium, 7.2 × 107 cfu/mL) concentrations were adjusted to 0.5 McFarland scale using McFarland device and next prepared sterile, nutrient broth was injected with 100 µL bacteria, and this was used as control. In another nutrient broth 100 µL bacterial solution was added with nanoparticle solution having 1 mg/mL in 100 µL. All these mixtures were treated at 37°C for 24 h. The liveness was indicated with inoculation the bacteria on the nutrient agar and the results were also compared with control cultures.
In the second (indirect) method L. monocytogenes, S. aureus and S. typhimurium strains were activated twice and later their concentrations were adjusted separately using McFarland device (Grant-bio, DEN1) to 0.5 McFarland scale using McFarland device (L. monocytogenes, 6.8 × 107 cfu/mL; S. aureus, 10.4 × 1010 cfu/mL; S. typhimurium, 7.2 × 107cfu/mL). 1% (v/v) of bacteria cultures was inoculated into the nutrient broths which contain 1.5% agar. Agar was used as a solidifying agent. After solidifying the medium, the holes were punched with a cork borer in plates of nutrient agar. The holes were then filled with a solution of 25 µL of nanoparticle solution having of 1 mg/mL concentration. The incubation was applied for 24 h at 37 °C and the diameter of clear zones surrounding the wells were determined and indicated the antibacterial activity [36]. All antimicrobial studies were performed with 5 parallel and 2 replicates.
2.9. Determination of nanoparticles’ capturing efficiencies
The capturing efficiency studies were performed with avidin coated nanoparticles. Each of the bacteria was activated twice and used in these experiments.The experiments were conducted in mixed culture media including the control medium in order to both determine the adhesion of various pathogenic microorganisms on the avidin coated nanoparticle surfaces and the success of the immunoassay which is specific for L. monocytogenes antibody bound nano surfaces. For this purpose, studies were conducted to determine the nanoparticles’ capturing efficiencies of each bacterium in competitive and noncompetitive systems.
2.9.1. Determination of noncompetitive capturing efficiencies
In all the capturing efficiency studies, the concentration of nanoparticles was adjusted to 0.5 mg/mL in sterile PS solution. The bacteria were activated twice, and the active cultures were obtained after centrifuge at 10,000 rpm for 15 min and washed and resuspended in PS solution. All activated bacteria (L. monocytogenes, 4.6 × 107 cfu/mL; S. aureus, 9.6 × 1010 cfu/mL; S. typhimurium, 4.4 × 107 cfu/mL) concentrations were adjusted to 0.5 McFarland scale using McFarland device. Then, 0.5 mg/mL nanoparticles were transferred to the bacteria medium and waited for 30 min for incubation. After incubation period, a magnet was used to collect the modified nanoparticles and washing procedure was applied 2 times with PS solution. In the present study, we performed a plate counting methodin the supernatant to determine the capture efficiency by plating the unbound bacteria.
2.9.2. Determination of competitive capturing efficiencies
In order to determination of the competitive capturing efficiencies, 2 experiments were performed. In the first study, the capturing amounts of avidin modified nanoparticles (unmodified with L. monocytogenes antibody) of mixed cultures where L. monocytogenes and S. typhimurium were present in the A medium and L. monocytogenes and S. aureus were in the B medium were investigated. In another study, it was designed to test the success of immunoassay detection of L. monocytogenes and the capturing amounts of L. monocytogenes antibody modified nanoparticles of mixed cultures where L. monocytogenes and S. typhimurium were present in the A medium and L. monocytogenes and S. aureus were in the B medium were also investigated.
The concentration of avidin modified nanoparticles was mixed medium containing L. monocytogenes and S. typhimurium and L. monocytogenes and S. aureus in sterile PS solution. The bacteria were activated twice, and centrifugation procedure was applied at 10,000 rpm for 15 min. Then, resulting cultures were washed and resuspended in PS solution. All activated bacteria concentrations were adjusted to 1 McFarland scale. 1 mL of each culture was added in a sterile tube to form a new mixed culture. Then, 2 mL of this mixed bacterial culture and 2 mL of 0.5 mg/mL nanoparticle concentration were taken into a new sterile tube and placed in a dark medium for 30 min. Afterwards, a permanent magnet was used to collect nanoparticles and nanoparticles were washed twice with PS solution. Thus, the liveness values of the bacteria that the only attached to the nanoparticle surfaces were calculated in cfu/mL using the CHROMagar Listeria.
In the developed immunosensor, we treated nanoparticles with bacterial cells and the capturing amount of L. monocytogenes on the magnetic Au-nanoparticles was shown using SEM and TEM images. For this purpose, 2% of L. monocytogenes cultures were inoculated into nutrient medium and incubation was performed at 37 °C for 24 h. After being activated twice, and centrifugation was performed at 5,000 rpm for 10 min and washed and transformed to the PS. All activated bacteria concentrations were adjusted to 0.5 McFarland scale. 100 µL of each culture was added in a 900 sterile µL PS. Here, 0.5 mg/mL nanoparticle solution was transferred to the diluted bacteria medium and incubated for 30 min at room temperature. Then, a magnet was used to obtain bacteria bounded nanoparticles and washing procedures were applied twice with PS solution. TEM images were captured by dropping nanoparticle-bacteria complex (10 µL) using formvar–carbon coated cupper grids and waited for 10 min.
After adjusting to 0.5 McFarland scale, SEM images were captured to obtain control (L. monocytogenes without nanoparticle) and immunoassay model with L. monocytogenes.
Briefly, we applied IMS and collected all bacterial cells interacted with nanoparticles. Then, glutaraldehyde (2.5%) was added to the cell suspensions for fixation procedure at 4 ℃ and waited overnight. After fixation procedure, the cells were pelleted and washed in PBS buffer. Then, we immersed the pellet in osmium tetroxide (1%) in buffer for postfixation procedure. After washing steps with PBS and water for 10 min each, different ethanol concentrations (initial value from 30 mL/100 mL to 100 mL/100 mL) were used for dehydration during 15 min. After applying three 10 min washing procedure with ethanol (100 g/100 g), dehydration process was achieved. To capture SEM images, air-dried SEM stubs were used to form a layer using gold sputter. Here, 10 µL sample was transferred on SEM stubs. In the present study, SEM was used with an acceleration voltage of 10 kV.
2.10. Preparation of SEM tag
We performed SERS measurements based on labelled sandwich immunoassay. For this purpose, we synthesized goldnanorods modified with DTNB. Here, 50 mM DTNB was dissolved in ethanol and interacted with gold nanorod for 18 h at room temperature. After washing step with MES buffer (50 mM) for 3 times, centrifugation was applied at 7000 rpm for 5 min. Subsequently, the labelled nanoparticles were taken into 1 mL of MES buffer.
2.11. Detection of L. monocytogenes
A sandwich complex was obtained in a solution phase by interacted with magnetic gold nanospheres with L. monocytogenes and DTNB modified gold nanorods. The resulting sandwich complex was interacted for half an hour. Then, a permanent magnet was used to collect the complex. To gain SERS signals from the resulting sandwich complex, we dropped it onto chromatography paper and SERS measurements were conducted 3 times. The SERS spectra corresponding to L. monocytogenes were collected. The calibration curve was constructed by obtaining the average SERS reading of L. monocytogenes (101–107 cfu/mL). The enumeration was completed by counting the number of colonies plating on CHROMagar Listeria agar subsequent incubation at 37 °C for 24 h. The peak signal intensity at 1336 cm−1 was selected for the SERS measurements and we calculated the coefficient of determination (R2) and linearity from the constructed calibration curve.
The sandwich assay for L. monocytogenes cells in milk samples was applied after careful optimization of experimental parameters. Furthermore, comparison of results obtained from SERS method and the counting was made. Dilutions of samples were conducted in buffer (PBS) for the plate-counting method, and a 100 μL sample was plated on CHROMagar Listeria agar and incubated at 37 °C for 24 h.
3. Results and discussion
3.1. Fabrication of the nanoparticles
In our previous work, we constructed the sandwich immunoassay concerning bacteria enumeration with DTNB-labelled rod-shaped gold nanoparticles. The rod-shaped gold nanoparticles are commonly used to conduct SERS measurements. Interaction of gold nanoparticles with target analyte resulted in increasing of sensitivity in SERS measurement [37].
3.2. Determination of nanoparticles’ antimicrobial activities
The determination of nanoparticles’ antimicrobial activities was given in Table 1. The liveness rates were found about 95% for L. monocytogenes, S. aureus and S. typhimurium strains.
Table 1.
Antimicrobial activities of magnetic Au-nanospheres on L. monocytogenes, S. typhimurium and S. aureus.
| Bacteria strains | Live bacteria amount (cfu/mL) | Live bacteria amount afternanoparticle interaction (cfu/mL) | Liveness (%) |
|---|---|---|---|
| L. monocytogenes ATCC 7644 | 6.8 × 107 | 6.5 × 107 | 96 |
| S. typhimurium BAST01 | 7.5 × 107 | 7.2 × 107 | 96 |
| S. aureus ATCC 25923 | 10.4 × 1010 | 9.9 × 1010 | 95 |
3.3. Determination of nanoparticles’ noncompetitive capturing efficiencies
In order to determination of the noncompetitive capturing efficiencies of nanoparticles were performed as shown in Table 2.
Table 2.
The noncompetitive capturing efficiencies of magnetic Au-nanospheres on L. monocytogenes, S. typhimurium and S. aureus.
| Noncompetitive attachment of avidin modificated magnetic Au-nanospheres | |||
|---|---|---|---|
| Bacteria strains | Live bacteria amount(cfu/mL) | Capturingefficiency (%) | |
| L. monocytogenesATCC 7644 | a | 2.8 × 107 | 20 |
| b | 5.7 × 106 | ||
| S. typhimuriumBAST01 | a | 4.4 × 107 | 2 |
| b | 8.0 × 105 | ||
| S. aureusATCC 25923 | a | 9.6 × 1010 | 0.7 |
| b | 6.9 × 108 | ||
| Noncompetitive attachment on immunoassay model | |||
| Bacteria strains | Live bacteria amount(cfu/mL) | ||
| L. monocytogenesATCC 7644 | a | 2.8 × 107 | 75 |
| b | 2.1 × 107 | ||
The noncompetitive attachment of avidin bound magnetic Au-nanospheres’ capturing efficiencies were found as 20%, 2% and 0.7% for L. monocytogenes, S. aureus and S. typhimurium strains, respectively. Noncompetitive attachment on immunoassay model for L. monocytogenes was found as 75%.
3.4. Determination of nanoparticles’ competitive capturing efficiencies
In order to determination of the competitive capturing efficiencies, 2 experiments were performed, and the obtained results were given in Table 3.
Table 3.
The competitive capturing efficiencies of magnetic Au-nanospheres in the A medium. (L. monocytogenes and S. typhimurium) and B medium (L. monocytogenes and S. aureus).
| Competitive attachment of avidin modificated magnetic Au-nanospheres | |||
|---|---|---|---|
| Bacteria strains | Live bacteria amount(cfu/mL) | Liveness ratio ( L. monocytogenes /other bacteria) | |
| L. monocytogenesATCC 7644S. aureusATCC 25923 | a | 5.8 × 107 | 2.1 |
| b | 2.7 × 107 | ||
| L. monocytogenesATCC 7644S. typhimuriumBAST01 | a | 4.5 × 107 | 1.9 |
| b | 2.4 × 107 | ||
| Competitive attachment on immunoassay model | |||
| Bacteria strains | Live bacteria amount(cfu/mL) | Liveness ratio (L. monocytogenes /other bacteria) | |
| L. monocytogenesATCC 7644S. aureusATCC 25923 | a | 6.8 × 107 | 2.3 |
| b | 2.9 × 107 | ||
| L. monocytogenesATCC 7644S. typhimuriumBAST01 | ab | 5.4 × 1072.1 × 107 | 2.6 |
In the first study, the capturing amounts of avidin coated nanoparticles (uncoated with L. monocytogenes antibody) of mixed cultures where L. monocytogenes and S. typhimurium were present in the A medium and L. monocytogenes and S. aureus were in the B medium were investigated. The liveness ratios (L. monocytogenes/other bacteria) were found as 2.1 in medium A and 1.9 in medium B.
In the other study, the capturing amounts of L. monocytogenes antibody modified nanoparticles of mixed cultures where L. monocytogenes and S. typhimurium were present in the A medium and L. monocytogenes and S. aureus were in the B medium were investigated. The liveness ratios were found as 2.3 in medium A and 2.6 in medium B.
3.5. Detection of bacteria capturing on the magnetic Au nanoparticles with SEM and TEM measurements
In the present study, SEM and TEM imaging of (a) control (L. monocytogenes without nanoparticle), (b) immunoassay model with L. monocytogenes were taken to verify and confirm the interactions between bacteria and nanoparticles as shown in Figures 1A and B.
Figure 1.
(A) Scanning electron micrograms of (a) control (L. monocytogenes without nanoparticle), (b) immunoassay model with L. monocytogenes, (B) transmission electron micrograms of (a) control (L. monocytogenes without nanoparticle) (b) immunoassay model with L. monocytogenes.
3.6. Enumeration of L. monocytogenes using SERS
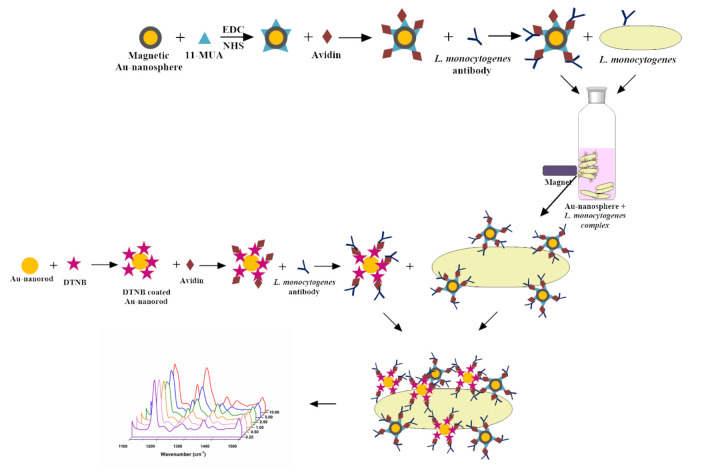
Shown in Figure 2, the stepwise immunoassay strategy was proposed in this study and we focused on the selective detection of L. monocytogenesas SERS based diagnostic test. The proposed method was evaluated in terms of analytical performance. The presented solution is similar to the SERS assay described in our previous studies, but the main advantages of the present assay for enumeration of L. monocytogenesis simplicity due to the elimination of sophisticated sample preparation procedures especially in milk samples. It is also provided that there is no interference from sophisticated milk matrix on L. monocytogenes enumeration using SERS based assay.
Figure 2.
Schematic illustration of SERS-based immunoassay for L. monocytogenes detection.
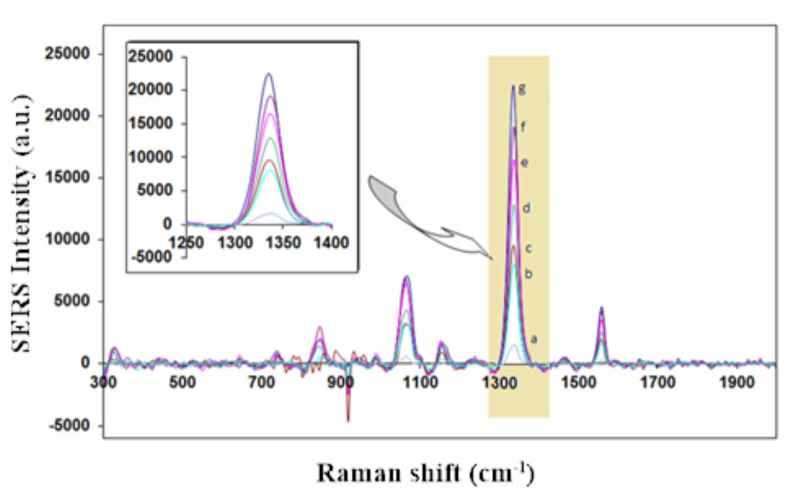
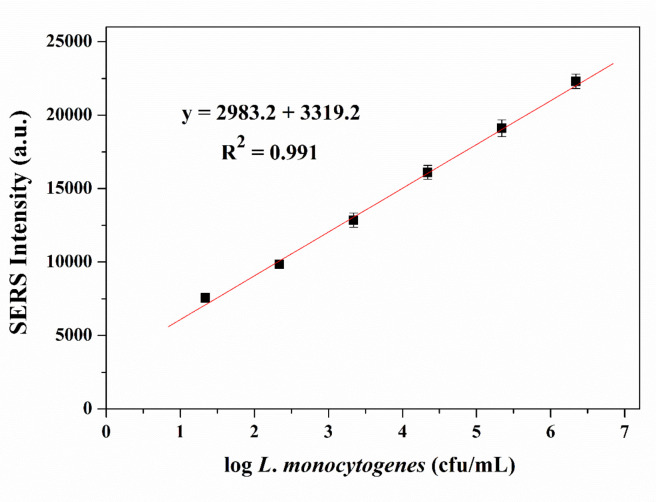
The SERS spectra for L. monocytogenes assays detection method conducted by using gold nanorod as a Raman tag label shows good response with the addition of various concentrations of L. monocytogenes as shown in Figure 3. We observed the symmetric nitro group stretching at 1336 cm-1which is attributable to the DTNB as reporter molecule.SERS peak intensity were measured to quantify L. monocytogenes. We performed to construct the calibration curve with the various concentrations of L. monocytogenes (from 2.2 × 101 to 2.2 × 106 cfu/mL). As shown in Figure 4, with the increase of different concentrations of L. monocytogenes, we observed the increase of SERS signal intensity. The SERS signal tracks with L. monocytogenes population density and becomes distinguishable in the presence of 101 bacteria. It is also mentioned that a good linear correlation (R2 = 0.991) was obtained between 102–106 cfu/mL L. monocytogenes concentration. We calculated the limit of detection and limit of quantification values as 12 cfu/mL and 37 cfu/mL, respectively.
Figure 3.
Symmetric NO2 stretching bands of DTNB range from 2.2 × 101 to 2.2 × 106 cfu/mL L. monocytogenes concentration in SERS-based sandwich immunoassay; L. monocytogenes concentrations of a) no Listeria monocytogenes, b) 2.2 × 101 cfu/mL, c) 2.2 × 102 cfu/mL, d) 2.2 × 103 cfu/mL, e) 2.2 × 104 cfu/mL, f ) 2.2 × 105 cfu/mL, g) 2.2 × 106 cfu/mL.
Figure 4.
Calibration curve for target oligonucleotide sequence in a range from 2.2 × 101 to 2.2 × 106 cfu/mL in SERS-based immunoassay.
The accuracy of the proposed assay was obtained using milk samples by the SERS-immunoassay and compared with plate-counting methods as shown in Table 4.
Table 4.
Comparison of the results obtained for the analysis of milk samples by the SERS-immunoassay and classical counting methods.MethodsConcentrations (cfu/mL)SERS-immunoassay3.6 × 1013.9 × 1024.5 × 1036.0 × 1044.8 × 105Classical counting3.8 × 1014.0 × 1024.2 × 1035.2 × 1044.5 × 105
| Methods | Concentrations (cfu/mL) | ||||
|---|---|---|---|---|---|
| SERS-immunoassay | 3.6 × 101 | 3.9 × 102 | 4.5 × 103 | 6.0 × 104 | 4.8 × 105 |
| Classical counting | 3.8 × 101 | 4.0 × 102 | 4.2 × 103 | 5.2 × 104 | 4.5 × 105 |
It was found that the results procured by the presented immunoassay method and the plate counting method were very similar.
4. Discussion
L. monocytogenes species are among the crucial foodborne pathogens that cause disease in humans and animals. This species can be found especially in milk and dairy products. In this study, the immunosensor model was developed to detect L. monocytogenes in mixed culture media. In literature, many rapid analysis methods were developed for L. monocytogenes detection. Alhogail et al. designed colorimetric biosensor to detect rapidly the amidolytic activity of Listeria protease [38]. The detection limit was found to be 2.17 × 102 cfu/mL in milk and meat samples. Another study was performed from Zhang et al. using Fe3O4 nanoparticle cluster which possesses high efficient peroxidase-like activity with a 5.2 × 103 cfu/mL detection limit [39]. The other study was based on fluorescence assay using aptamer-conjugated magnetic nanoparticles [40]. The detection limit of 102 cfu/mL of L. monocytogenes was obtained. To the best of our knowledge, this is the first report in which a IMS-SERS based assay was utilized to detect L. monocytogenes. The analytical parameters of nanoparticle coated L. monocytogenes sensor were investigated and the developed immunosensor was found as quite selective for L. monocytogenes. A linear correlation between population density of L. monocytogenes and SERS signal intensity was found from 2.2 × 101 to 2.2 × 106 cfu/mL and LOD was found to be 12 cfu/mL. Also, L. monocytogenes was detected easily in milk samples and the results generated by the SERS-immunoassay were comparable with the reference plate-counting methods. Thereby, the assay was very promising for monitoring and enumeration of bacteria in complex matrices such as milk.
Acknowledgment
The authors gratefully acknowledge the financial support from the Gazi University Research Fund through Grant No. 46/2010-02.
Conflict of interest
All authors disclose no conflict of interest that may have influenced either the conduct or the presentation of the research.
Informed consent
Manuscripts reporting the results of experimental investigations did not conduct with humans.
This study was presented at the Taiwan-Turkey Science Summit entitled “Translation of Cells, Nanomaterials and Signaling Molecules into Regenerative Medicine” between April 1 to 3, 2018.
References
- Farber JM Peterkin PI Listeria-Monocytogenes, a food-borne pathogen. Microbiological Reviews. 1991;55:476. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes PS Feeley JC Graves LM Ajello GW Fleming DW Isolation of Listeria-monocytogenes from raw-milk. Applied and Environmental Microbiology. 1986;51:438. doi: 10.1128/aem.51.2.438-440.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naja G Bouvrette P Hrapovic S Luong JHT Raman-based detection of bacteria using silver nanoparticles conjugated with antibodies. Analyst. 2007;132:679. doi: 10.1039/b701160a. [DOI] [PubMed] [Google Scholar]
- Aznar R Alarcon B. PCR detection of Listeria monocytogenes: a study of multiple factors affecting sensitivity. Journal of Applied Microbiology. 2003;95:958. doi: 10.1046/j.1365-2672.2003.02066.x. [DOI] [PubMed] [Google Scholar]
- Carloni E Rotundo L Brandi G Amagliani G Rapid and simultaneous detection of Salmonella spp., Escherichia coli O157, and Listeria monocytogenes by magnetic capture hybridization and multiplex real-time PCR. Folia Microbiologica. 2018;63:735. doi: 10.1007/s12223-018-0617-0. [DOI] [PubMed] [Google Scholar]
- Vizzini P Braidot M Vidic J Manzano M. Electrochemical and optical biosensors for the detection of Campylobacter and Listeria: an update look. Micromachines. 2019;10 doi: 10.3390/mi10080500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebana S Brandao D Cortes P Campoy S Alegret S Electrochemical genosensing of Salmonella, Listeria and Escherichia coli on silica magnetic particles. Analytica Chimica Acta. 2016;904:1. doi: 10.1016/j.aca.2015.09.044. [DOI] [PubMed] [Google Scholar]
- Yan LJ Zhao WS Wen ZR Li XY Niu XL Electrochemical DNA sensor for hly gene of Listeria monocytogenes by three-dimensional graphene and gold nanocomposite modified electrode. International Journal of Electrochemical Science. 2017;12:4086. [Google Scholar]
- Rahman SU Stanton M Casey PG Spagnuolo A Bensi G Development of a click beetle luciferase reporter system for enhanced bioluminescence imaging of Listeria monocytogenes: Analysis in cell culture and murine infection models. Frontiers in Microbiology. 2017;10 doi: 10.3389/fmicb.2017.01797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kheir SM Cherrat L Awussi AA Ramia NE Taha S High-throughput identification of candidate strains for biopreservation by using bioluminescent Listeria monocytogenes. Frontiers in Microbiology. 2018;10 doi: 10.3389/fmicb.2018.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu XL Zheng W Yin CX Weng WJ Li GJ Electrochemical DNA biosensor based on gold nanoparticles and partially reduced graphene oxide modified electrode for the detection of Listeria monocytogenes hly gene sequence. Journal of Electroanalytical Chemistry. 2017;806:116. [Google Scholar]
- Amagliani G Brandi G Omiccioli E Casiere A Bruce IJ Direct detection of Listeria monocytogenes from milk by magnetic based DNA isolation. Food Microbiology. 2004;21:597. [Google Scholar]
- Scheu P Gasch A Berghof K. Rapid detection of Listeria monocytogenes by PCR-ELISA. Letters in Applied Microbiology. 1999;29:416. doi: 10.1046/j.1472-765x.1999.00667.x. [DOI] [PubMed] [Google Scholar]
- Curiale MS Lepper W Robison B Enzyme-Linked Immunoassay for detection of Listeria-monocytogenes in dairy-products, seafoods, and meats - collaborative study. Journal of Aoac International. 1994;77:1472. [PubMed] [Google Scholar]
- Zhang XG Tsuji S Kitaoka H Kobayashi H Tamai M Simultaneous detection of Escherichia coli O157:H7, Salmonella enteritidis, and Listeria monocytogenes at a very low level using simultaneous enrichment broth and multichannel SPR Biosensor. Journal of Food Science. 2017;82:2357. doi: 10.1111/1750-3841.13843. [DOI] [PubMed] [Google Scholar]
- Boulade M Morlay A Piat F Roupioz Y Livache T Early detection of bacteria using SPR imaging and event counting: experiments with Listeria monocytogenes and Listeria innocua. Rsc Advances. 2019;9:15554. doi: 10.1039/c9ra01466g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QR Zhang S Cai YX Yang YX Hu F Rapid detection of Listeria monocytogenes using fluorescence immunochromatographic assay combined with immunomagnetic separation technique. International Journal of Food Science and Technology. 2017;52:1559. [Google Scholar]
- Radhakrishnan R Fluorescence-free biosensor methods in detection of food pathogens with a special focus on Listeria monocytogenes. Biosensors-Basel. 2017;7 doi: 10.3390/bios7040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HB Du XJ Zang YX Li P Wang S. SERS-based lateral flow strip biosensor for simultaneous detection of Listeria monocytogenes and Salmonella enterica Serotype enteritidis. Journal of Agricultural and Food Chemistry. 2017;65:10290. doi: 10.1021/acs.jafc.7b03957. [DOI] [PubMed] [Google Scholar]
- Uusitalo S Kogler M Valimaa AL Popov A Ryabchikov Y Detection of Listeria innocua on roll-to-roll produced SERS substrates with gold nanoparticles. Rsc Advances. 2016;6:62981. [Google Scholar]
- Stambach NR Carr SA Cox CR Voorhees KJ Rapid detection of Listeria by bacteriophage amplification and SERS-Lateral flow immunochromatography. Viruses-Basel. 2015;7:6631. doi: 10.3390/v7122962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya S Chen W Mulchandani A Molecular beacons: a real-time polymerase chain reaction assay for detecting Escherichia coli from fresh produce and water. Analytica Chimica Acta. 2008;614:208. doi: 10.1016/j.aca.2008.03.026. [DOI] [PubMed] [Google Scholar]
- Brandao D Liebana S Campoy S Alegret S Pividori MI Immunomagnetic separation of Salmonella with tailored magnetic micro and nanocarriers. A comparative study. Talanta. 2015;143:198. doi: 10.1016/j.talanta.2015.05.035. [DOI] [PubMed] [Google Scholar]
- Chen J Park B Effect of immunomagnetic bead size on recovery of foodborne pathogenic bacteria. International Journal of Food Microbiology. 2018;267:1. doi: 10.1016/j.ijfoodmicro.2017.11.022. [DOI] [PubMed] [Google Scholar]
- Shanmukh S Jones L Driskell J Zhao YP Dluhy R Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Letters. 2006;6:2630. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- Etchegoin P Maher RC Cohen LF Hartigan H Brown RJC New limits in ultrasensitive trace detection by surface enhanced Raman scattering (SERS) Chemical Physics Letters. 2003;375:84. [Google Scholar]
- Brown RJC Milton MJT Nanostructures and nanostructured substrates for surface-enhanced Raman scattering (SERS) Journal of Raman Spectroscopy. 2008;39:1313. [Google Scholar]
- Guven B Boyaci IH Tamer U Acar-Soykut E Dogan U Development of rolling circle amplification based surface-enhanced Raman spectroscopy method for 35S promoter gene detection. Talanta. 2015;136:68. doi: 10.1016/j.talanta.2014.11.051. [DOI] [PubMed] [Google Scholar]
- Wang JF Wu XZ Wang CW Rong Z Ding HM Facile synthesis of Au-coated magnetic nanoparticles and their application in bacteria detection via a SERS method. Acs Applied Materials & Interfaces. 2016;8:19958. doi: 10.1021/acsami.6b07528. [DOI] [PubMed] [Google Scholar]
- Song D Yang R Fang SY Liu YP Long F SERS based aptasensor for ochratoxin A by combining Fe3O4@Au magnetic nanoparticles and Au-DTNB@Ag nanoprobes with multiple signal enhancement. Microchimica Acta. 2018;185 doi: 10.1007/s00604-018-3020-2. [DOI] [PubMed] [Google Scholar]
- Sharma HSS Carmichael E McCall D Fabrication of SERS substrate for the detection of rhodamine 6G, glyphosate, melamine and salicylic acid. Vibrational Spectroscopy. 2016;83:159. [Google Scholar]
- Liu GK Ren B Wu DY Lin TM Gu RA Electrochemical polymerization of acetylene on Rh electrodes probed by surface-enhanced Raman spectroscopy. Journal of Electroanalytical Chemistry. 2006;594:73. [Google Scholar]
- Wang P Xia M Liang O Sun K Cipriano AF Label-Free SERS selective detection of dopamine and serotonin using graphene-Au nanopyramid heterostructure. Analytical Chemistry. 2015;87:10255. doi: 10.1021/acs.analchem.5b01560. [DOI] [PubMed] [Google Scholar]
- Wang CW Wang JF Li M Qu XY Zhang KH A rapid SERS method for label-free bacteria detection using polyethylenimine-modified Au-coated magnetic microspheres and Au@Ag nanoparticles. Analyst. 2016;141:6226. doi: 10.1039/c6an01105e. [DOI] [PubMed] [Google Scholar]
- Yegenoglu H Aslim B Guven B Zengin A Boyaci IH The comparison of antioxidant capacity and cytotoxic, anticarcinogenic, and genotoxic effects of Fe@Au nanosphere magnetic nanoparticles. Turkish Journal of Biology. 2017;41:302. [Google Scholar]
- Reinheimer JA Demkow MR Candioti MC Inhibition of coliform bacteria by lactic cultures. Australian Journal of Dairy Technology. 1990;45:5. [Google Scholar]
- Wang YL Lee K Irudayaraj J SERS aptasensor from nanorod-nanoparticle junction for protein detection. Chemical Communications. 2010;46:613. doi: 10.1039/b919607b. [DOI] [PubMed] [Google Scholar]
- Alhogail S Suaifan GARY Zourob M. Rapid colorimetric sensing platform for the detection of Listeria monocytogenes foodborne pathogen. Biosensors & Bioelectronics. 2016;86:1061. doi: 10.1016/j.bios.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Zhang LS Huang R Liu WP Liu HX Zhou XM Rapid and visual detection of Listeria monocytogenes based on nanoparticle cluster catalyzed signal amplification. Biosensors & Bioelectronics. 2016;86:1. doi: 10.1016/j.bios.2016.05.100. [DOI] [PubMed] [Google Scholar]