Abstract
Interleukin-31 (IL-31) is a major protein involved in severe inflammatory skin disorders. Its signaling pathway is mediated through two type I cytokine receptors, IL-31RA (also known as gp130-like receptor) and oncostatin M receptor (OSMR). Understanding molecular details in these interactions would be helpful for developing antagonist anti-IL-31 monoclonal antibodies (mAbs) as potential therapies. Previous studies suggest that human IL-31 binds to IL-31RA and then recruits OSMR to form a ternary complex. In this model, OSMR cannot interact with IL-31 in the absence of IL-31RA. In this work we show that feline IL-31 (fIL-31) binds independently with feline OSMR using surface plasmon resonance, ELISA, and yeast surface display. Moreover, competition experiments suggest that OSMR shares a partially overlapping epitope with IL-31RA. We then used deep mutational scanning to map the binding sites of both receptors on fIL-31. In agreement with previous studies on the human homolog, the binding site for IL31-RA contains fIL-31 positions E20 and K82, while the binding site for OSMR comprises the “PADNFERK” motif (P103-K110) and positions G38. However, our results also revealed a new overlapping site, composed of positions R69, R72, P73, D76, D81, and E97, between both receptors which we called the “shared site”. The conformational epitope of an anti-feline IL-31 mAb that inhibits both OSMR and IL-31RA also mapped to this shared site. Combined, our results show that fIL-31 binds IL-31RA and OSMR independently through a partially shared epitope. These results suggest reexamination of the putative canonical mechanisms for IL-31 signaling in higher animals.
Keywords: IL-31, conformational epitope mapping, OSMR, IL-31RA, deep mutational scanning, monoclonal antibodies
Graphical Abstract
INTRODUCTION
Cytokines comprise a large family of small proteins that play a critical role in the development and control of the immune response. Certain cytokines are associated with the initiation and the persistence of pathological pain behavior including nerve and skin injuries. A more recently discovered cytokine, interleukin-31 (IL-31), has been linked to the induction of chronic skin inflammation1. Human and murine data have shown high expression of IL-31 associated with severe inflammatory skin disorders including pruritis, alopecia, skin lesion, and atopic dermatitis (AD) and with other regulated allergic diseases such as asthma1–7. An experimental animal model for human AD reported a strong correlation between itch-associated scratching behavior in NC/Nga mice and expression of IL-31 mRNA5. Elevated IL-31 serum levels were found in adult patients with AD compare to healthy control subjects8 and in pediatric patients during AD flare and quiescence9. Together, these data suggest that IL-31 represents an important target for the development of treatments against such skin inflammatory diseases in humans. Antagonist anti-IL-31 mAbs are currently in development for human health10,11 and have already been developed in animal health12. For example, an anti-hIL-31RA mAb, CIM331, binds to IL-31RA, inhibits IL-31 signaling, and reduces severe pruritus11. In veterinary medicine a “caninized” anti-IL-31 mAb, Lokivetmab, showed efficacy in clinical trials for canine pruritis and is currently approved as an AD therapy for dogs12.
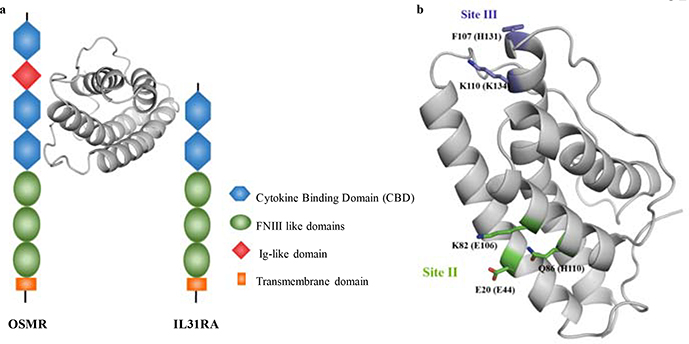
IL-31 is a member of the IL-6 cytokine superfamily produced preferentially by T helper type 2 cells1. Mature human IL-31 (hIL-31) is composed of 141 amino acids1 with a predicted topology of four antiparallel helices13. The IL-31 signaling pathway is thought to be mediated through a gp130-like type 1 cytokine receptor (IL-31RA, also known as GPL) and oncostatin M receptor (OSMR)1,13–15. Both receptors belong to the type I cytokine receptor family which share a common cytokine binding domain (CBD) formed by two fibronectin type III-like domains16. Previous studies supplied immunoprecipitation evidence that human IL-31RA (hIL-31RA) binds directly to hIL-31. In these same studies, immunoprecipitation results failed to detect direct human OSMR (hOSMR) binding to hIL-3113,14. However, an increase in binding was distinguished when hIL-31RA and hOMSR were combined, suggesting that hIL-31 binds first to hIL-31RA, at which time hOMSR is recruited to form the ternary complex13,14. In this model, the ternary complex activates numerous downstream signaling pathways1,13,14,17,18 (Figure 1a).
Figure 1.
Interactions between IL-31 and its receptors. The cartoon model of IL-31 structure was generated using the feline sequence. (a.) The ternary complex formed between IL-31, IL-31RA, and OSMR. Different domains of both receptors are indicated in the figure legend. (b.) Binding sites for the interaction between human IL-31 (hIL-31) and its receptors as described by Le Saux et at13. Residues corresponding to the hIL-31 sequence are indicated in parenthesis.
Specific atomistic knowledge of the binding interactions between IL-31 and its receptors would be helpful for the development of antagonist therapeutic mAbs. Based on the structure of IL-6/IL-6 α-Receptor/gp130 complex19, the IL-6 cytokine superfamily is thought to interact with their receptors through three different contact binding sites (sites I, II, and III). Le Saux et al. used computational analysis and sparse alanine scanning to delineate sites II and III only as critical binding sites for the interaction between hIL-31 and its receptors13. In particular, Glu44, Glu106, and His110 were identified as critical residues for binding site II while Lys134 was identified within the binding site III (Figure 1b). However, this original study tested only 11 point mutants in total: 10 predicted to be at site II or site III and one irrelevant mutation distal to these sites. Thus, it remains unclear whether more a comprehensive mutational study could uncover additional protein interaction surfaces.
The aim of this study is to gain insights into the interactions between feline IL-31 (fIL-31) and its feline receptors fOSMR and fIL-31RA and to map the conformational epitope for an anti-fIL-31 mAb (mAb #1). In contrast to previous studies conducted with homologs, we show through multiple biophysical methods that fOSMR directly binds fIL-31 and partially interferes with fIL-31RA binding. We identified the potential fIL-31 binding sites for fOSMR, fIL31-RA, and an anti-fIL-31 mAb (mAb#1) using a predicted structural model combined with fine epitope mapping20 using yeast surface display21, nicking mutagenesis22, and deep sequencing23. The constructed binding sites agreed with the sites previously found by Le Saux et al13 and showed an additional overlapping site between both receptors to which we termed the “shared site”. Finally, mAb#1 also bound this shared site between both receptors demonstrating its ability to inhibit the signaling pathway. Together, these results suggest that the current model for IL-31 mediated signaling in higher mammals is incomplete and suggest efficient therapeutic strategies to antagonize IL-31-mediated signaling.
MATERIALS AND METHODS
Strains
The Escherichia coli strain used in this study was XL1-Blue (Agilent, Santa Clara, CA) endA1 supE44 thi-1 hsdR17 recA1 gyrA96 relA1 lac [F’ proAB lacI1qZLiM15 Tn10 (Tetr)]. The Saccharomyces cerevisiae strain used in this study was EBY100 (American Type Culture Collection, Manassas, VA) MATa AGA1::GAL1-AGA1::URA3 ura3–52 trp1 leu2-delta200 his3-delta200 pep4::HIS3 prb11.6R can1 GAL.
Plasmid Constructs
pETconNK_fIL-31 was created by inserting a codon-optimized gene encoding the Ser1 – Gln136 from the mature portion of feline interleukin-31 (fIL-31; NCBI XP_011286140.1) (GenScript, Piscataway, NJ) into yeast display vector pETconNK22 (Addgene plasmid #81169) using standard restriction cloning. Sequences were verified by Sanger sequencing (Genewiz, South Plainfield, NJ). The full nucleotide and amino acid sequence is listed in Supporting Note S1, and a sequence alignment to other IL-31 homologs is given in Supporting Note S2.
Preparation of IL-31, OSMR, Il-31RA constructs, and mAb#1
fIL-31 was produced recombinantly in E. coli and CHO cells with a C-terminal His-tag and purified by Ni-NTA affinity chromatography. Other proteins were prepared as human IgG1 Fc fusions and produced in mammalian cell culture and purified by protein A affinity chromatography. Each protein was prepared in phosphate-buffered saline (PBS) at a concentration of at least 0.082 mg/mL (Zoetis Global Therapeutic Research, Kalamazoo, MI). Proteins were biotinylated at a molar ratio of 1:20 protein: biotin using the EZ-link NHS-biotin kit following the manufacturer’s instruction (Life Technologies, Carlsbad, CA). Biotinylated proteins were desalted using Zeba Spin desalting columns (Thermo Fisher, Waltham, MA) and stored at 4°C.
Analytical Size Exclusion Chromatography
Size exclusion chromatography was performed using a TSK SuperSW3000 4.6 × 30 mm gel permeation column. 50 μg of fIL-31 was injected at a flow rate of 0.25 mL/min for 25 minutes with a mobile phase of 200 mM sodium phosphate, pH 7.2.
Surface Plasmon Resonance binding assays
Surface Plasmon Resonance was performed on a Biacore T200 (GE Healthcare, Pittsburgh, PA) to measure binding affinities of fOSMR-ECD and mAb#1 to fIL-31. fIL-31 immobilization on CM5 sensor and the binding measurements were conducted as previously described24. Data was analyzed with Biacore T200 Evaluation software by using the method of double referencing. The resulting curve was fitted with the 1:1 binding model.
ELISA
The plate was coated with fIL-31 overnight at 4°C in carbonate-bicarbonate buffer, pH 9.6 (Sigma-Aldrich, St. Louis, MO) follow by a blocking step with 5% skim milk in PBS with 0.05% TWEEN 20 for 1 hour at room temp. Individual proteins were diluted at different concentrations in blocking buffer and were added to the coated plate for 2 hrs at room temp. Next, HRP conjugated secondary antibodies were added to appropriate wells at 1:10000 in blocking buffer for 1 hr at room temp (goat anti-human Fc for receptors, jackson goat anti-cat Fc for mAb#1, KPL anti-mouse). Finally, KPL SureBlue TMB (VWR) was added to develop absorbance at 650 nm.
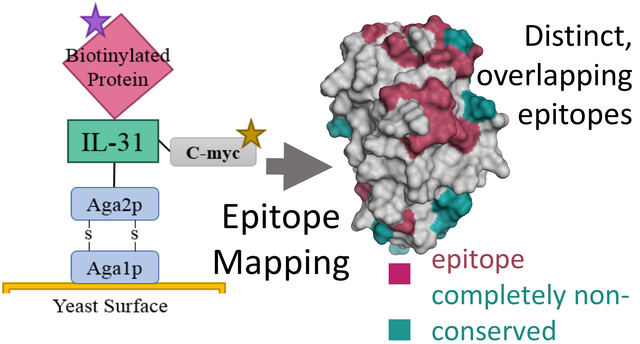
Yeast Surface Display Expression and Binding Activity
Mean fluorescence intensities (MFI) were measured using a BD Accuri C6 flow cytometer. The expression of fIL-31 on the yeast surface was detected using anti-c-myc-FITC (Miltenyi Biotec, San Diego, CA) and the binding interaction with biotinylated proteins was detected using streptavidin- R- phycoerythrin conjugate (Thermo Fisher). Dissociation constant (KD) values were determined according to Chao et al.21 Titrations were performed at triplicates on at least two separate days and MFI values were used to calculate the experimental KD using one site–specific binding equation (Hill coefficient of 1) in Graph Pad Prism software. Labeling concentrations tested vary from 0.064 nM to 262.1 nM for fOSMR and mAb#1, and from 2.05 nM to 524.3 nM for fIL31RA-1FNIII.
Competition Binding Assays
For YSD, 1×105 yeast cells were labeled at twelve times their KD values with either non- biotinylated fOMSR or non-biotinylated mAb#1 for 30 mins at room temperature in PBS containing 1g/liter of bovine serum albumin (PBS-BSA). After spinning down and washing with 200 μl of PBS-BSA, cells were labeled at 1 times their KD values with either biotinylated mAb#1, fOSMR, or fIL31RA-1FNIII for 30 mins at room temperature in PBS-BSA. Then, after a second step of centrifugation and washing, cells were labeled with 0.6 μl of anti-cymc-FITC and 0.25 μl of streptavidin- R- phycoerythrin conjugate in 49.15 μl of PBS-BSA for 10 mins at 4°C. Cells were washed thoroughly with PBS-BSA and read on a flow cytometer. Feline IL 31-VTM-238.A4 was immobilized on CM5 sensor by amine coupling and reached 100 RU. Kinetic binding was measured by inject 50 nM + 0 nM feline OSMR and mAb1 for 250 seconds at flow rate 30 ul/min with 500 seconds dissociation on feline IL 31 directly, and competition binding was measured the same as kinetic binding but on either 500 nM feline OSMR or mAb1 caputured on feline IL 31 surface. Data was analyzed with Biacore T200 Evaluation software by using the method of double referencing and fitted with the 1:1 binding model.
Cell-based assays
E. coli-derived or CHO-derived fIL-31 at various concentrations was incubated with the adherent feline macrophage cell line FCWF-4 (ATCC CRL-2787) for 5 minutes at 37oC. Activation of pSTAT signaling was determined using an AlphaLISA SureFire Ultra pSTAT3 kit (PerkinElmer). For inhibition experiments, fOSMR-ECD was pre-incubated with 0.2 μg/mL fIL-31 for 1 hr at 37oC prior to testing for activation of pSTAT signaling.
Preparation of Mutagenesis Libraries
Two comprehensive single-site saturation mutagenesis libraries were constructed using nicking mutagenesis as described22. All “NNK” mutagenesis oligos were designed using Quick Change Primer Design Program (www.agilent.com) and were ordered from Integrated DNA Technologies (Coralville, IA). Library 1 covered residues Ser1 – Phe68 and library 2 covered Arg69 – Gln136 of the mature fIL-31. 5 μg of library DNA plasmid was transformed into chemically competent Saccharomyces cerevisiae EBY100 cells, and cells were grown and stored at a concentration of 1×107 cells per ml in yeast storage buffer at −80°C according to published protocols20.
Determination of receptor binding sites and conformational epitope of mAb#1
The library screening through FACS and deep sequencing preparation was performed exactly as previously described20. The sorting was done on an Influx Cell Sorter (Michigan State University Flow facility). After preparing the plasmid DNA for deep sequencing, libraries were pooled and sequenced on an Illumina MiSeq using 2 × 250 bp pair-end reads at the Michigan State University Genomic Sequencing Core facility or the University of Illinois at Chicago DNA Service facility. The supplementary material contains the sorting conditions (Table S1), the primers used for deep sequencing (Table S2), and the summary table of statistics (Table S3).
Data Analysis
A modified version of Enrich 0.2 software as described in Kowalsky et al.25 was used to compute enrichment ratios from the raw sequencing files. The relative fluorescence (ζi) for variant i was defined as:
| (1) |
where is the mean fluorescence of variant i and is the mean fluorescence of wild type fIL-31. This equation can be written in terms of experimental observables according to:
| (2) |
where is the log normal fluorescence standard deviation of the clonal population, ε is the enrichment ratio and Φ is the percentage of cells collected by the sorting gate on the flow cytometer. Custom python scripts available at Github (user: JKlesmith) were used to normalize the relative fluorescence, and to calculate the Shannon Entropy and overall statistics26.
Data Availability
Full datasets including normalized relative fluorescence, pre- and post-selection read counts, and raw log base two enrichment scores for each variant can be found in Supplementary Data S1–S3. Raw sequencing reads for this work have been deposited in the SRA (SAMN11289369–72, 79, 81–83, 90–91, SAMN11289422–23).
Results
fOSMR and fIL31-RA bind independently to fIL-31
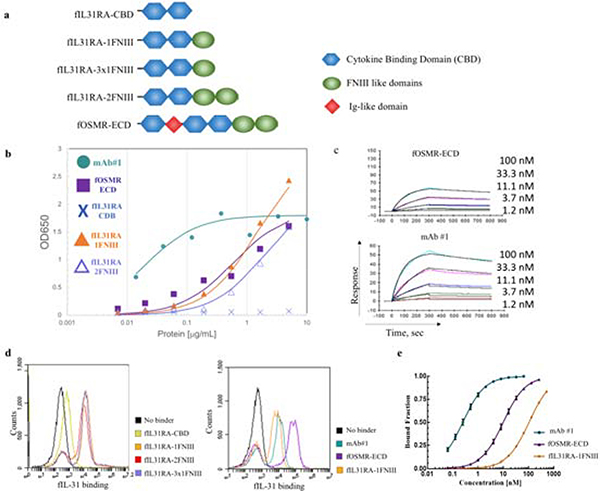
We assessed the binding activity of fIL-31 to soluble fOSMR extracellular domain (fOSMR-ECD), four versions of soluble fIL-31RA (fIL31RA-CBD comprising the CBD only, two fIL31RA-1FNIII isoforms comprising CBD with 1 FNIII domain, and fIL31RA-2FNIII comprising CBD with two FNIII domains (Figure 2a), and to an anti-fIL-31 mAb (mAb#1) (Figure 2). All receptors were expressed as fusion proteins with a C-terminal Fc. Recombinant proteins were produced by transient expression in CHO cells and purified by affinity chromatography using Protein A resin. The expected molecular weight of fIL-31 was determined using mass spectrometry (Figure S1a) following PNGase treatment. fIL-31 was monomeric with a small dimeric peak as judged by analytical gel permeation chromatography (Figure S1b).
Figure 2.
Binding profiles of feline IL-31 (fIL31) with all different constructs of fIL-31RA, fOMSR-ECD, and mAb#1. (a.) fIL-31RA and fOSMR-ECD constructs tested in this work. (b.) ELISA plots and (c.) Surface plasmon resonance sensorgrams showing the binding of fIL-31 with all constructs. For both datasets best-fit curves are shown assuming 1:1 binding kinetics. (d.) Flow cytograms showing the increase in fluorescence in the fIL-31 binding channel with all constructs. (e.) Binding titrations curves for yeast surface-displayed fIL-31 with mAb#1, fOSMR-ECD, and fIL31RA-1FNIII. Titrations were performed at least in triplicates on different days. (error bars, 1 s.d., n≥3)
We first assessed the ability of fOSMR-ECD and fIL31-RA to independently recognize soluble fIL-31 using ELISAs. fIL-31 was coated and then receptors or mAb#1 were incubated at indicated concentrations. After washing, receptors were detected either with an anti-human Fc or an anti-feline Fc (see Methods). fOSMR-ECD bound fIL-31 in a dose dependent fashion, whereas the fIL-31RA CBD alone did not recognize fIL-31 at any concentration tested (Figure 2b). However, both fIL31RA-FNIII isoforms recognized fIL-31. Anti fIL-31 mAb#1 also bound fIL-31 using this ELISA format (Figure 2b). Surface plasmon resonance (SPR) measurements using immobilized fIL-31 showed similar binding results for fOSMR-ECD and mAb#1. (Figure 2c). Binding equilibrium measurements were determined using kinetic fitting with 1:1 binding mode, revealing high-pM and low-nM affinities of fIL-31-fOSMR-ECD and anti fIL-31mAb#1 to fIL-31 (Table 1).
Table 1.
Summary of experimental determined dissociation constants (KD) using yeast surface display and surface plasmon resonance. Error bars represent the standard error of the mean of at least 3 independent measurements. ND, not determined.
| Average KD values [nM] | |||||
|---|---|---|---|---|---|
| Name | Yeast Surface Display | Hill Coefficient | Surface Plasmon Resonance | Fit | Chi2 (RU2) |
| fOSMR-ECD | 10.3 ± 0.5 | 0.99 ± 0.04 | 3.8 | 1:1 | 0.84 |
| fIL31RA-1FNIII | 106 ± 2.7 | 1.00 ± 0.01 | ND | 1:1 | ND |
| mAb #1 | 0.26 ± 0.01 | 0.99 ± 0.04 | 0.3 | 1:1 | 3.17 |
We also measured these intermolecular interactions using yeast surface display (YSD), where fIL-31 was displayed on the yeast surface with an N-terminal yeast surface protein Aga2p and a C-terminal c-myc epitope tag. Saturating amounts of biotinylated soluble fIL-31RA constructs, fOSMR-ECD, and mAb#1 were incubated with yeast cells displaying fIL-31, followed by secondary labeling (Figure 2d). fOSMR-ECD bound fIL-31 with nanomolar affinity (Figure 2d–e). While fIL31RA-CBD did not bind, both fIL31RA-1FNIII isoforms and fIL31RA-2FNIII were bound fIL-31, consistent with ELISA and SPR data. Given that the fIL31RA-1FNIII isoform obtained the highest mean fluorescence intensity (MFI) under saturating amounts of receptor, we used this receptor for the remainder of this work. Dissociation constants ranged from 0.26 ± 0.01 nM for mAb#1 to 106 ± 2.7 nM for fIL31RA-1FNIII (Table 1). For all interactions including mAb #1, the binding on the yeast surface could be modeled using 1:1 binding kinetics (best fits of Hill coefficient – 1.0 ± 0.04), consistent with fIL-31 and receptors being mostly monomeric.
fOSMR partially, but not completely, inhibits binding of fIL31RA-1FNIII to fIL-31
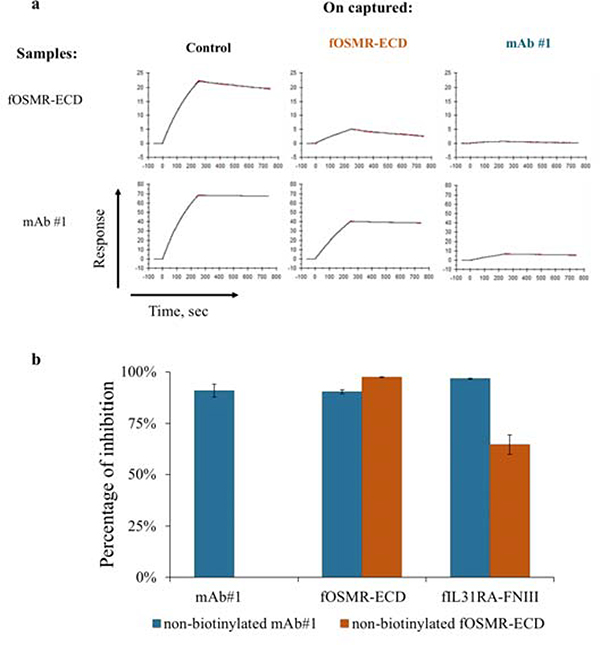
To determine whether the receptors bind to independent sites on fIL-31 or share overlapping sites we performed competition binding assays using SPR and YSD. First, we set-up an SPR-based competitive binding assay where fIL-31 is immobilized to the surface, and then either fOSMR-ECD or mAb#1 were captured at saturating amounts. Next, either fOSMR-ECD or mAb#1 were passed over the captured surface. If receptor and mAb bind at different epitopes there should be no difference in response. When fOSMR-ECD was captured on the surface, mAb#1 binding signal decreased relative to the control indicating some degree of competition. Similarly, the captured mAb#1 decreased the binding signal of fOSMR-ECD by approximately 50% (Figure 3a) relative to the control, indicating at least partially overlapping binding footprints.
Figure 3.
Competition binding assays demonstrated that fOSMR-ECD, fIL31RA-1FNIII, and mAb#1 share an overlapping epitope using SPR and YSD. (a.) SPR sensorgrams showing a decrease in the binding signal when fOMSR-ECD and mAb#1 were captured on the surface. The control was in the presence of 1x HBS-EP buffer. (b.) Percentage of inhibition for yeast cells labeled with either non-biotinylated mAb#1 or non-biotinylated fOSMR-ECD at twelve times their respective Kd values and subsequently labeled with mAb#1, fOSMR-ECD, and fIL31RA-1FNIII at Kd values (error bars, standard error of the mean, n≥3). Inhibition of mAb#1 by non-biotinylated OSMR was not determined.
Next, using YSD, fIL-31 yeast cells were labeled with either non-biotinylated fOMSR-ECD or mAb#1 at 12x their experimentally determined KD values, washed, and subsequently labeled with either biotinylated fOSMR-ECD, fIL31RA-1FNIII or mAb #1 at 1 times their KD. At such labeling concentrations, and considering 1:1 binding kinetics, it was expected that the non-biotinylated proteins inhibit the binding signal of the biotinylated proteins by 0% if binding sites are completely non-overlapping and approximately 92% if proteins occupy overlapping binding sites (fractional occupancy by competitive inhibitor I = I/(KD + I) = 12KD/(KD + 12KD) = 92%). Non-biotinylated mAb #1 decreased binding signal of itself, fOSMR-ECD, and fIL31RA-1FNIII by at least 90% (Figure 3b). These results demonstrated that mAb#1 binds an overlapping epitope or otherwise sterically prevents binding to the two receptors. Non-biotinylated fOMSR-ECD decreases binding to fOSMR-ECD and fIL31RA-1FNIII by 97% and 65%, respectively (Figure 3b). Data from both measurements suggests that fOSMR-ECD and fIL31RA-1FNIII share partially overlapping binding epitopes on fIL-31 and that mAb#1 interferes with binding to both receptors to a different extent.
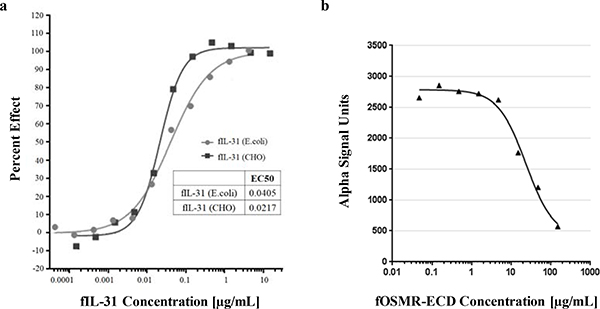
fIL-31 can be specifically inhibited by fOSMR-CBD in cell-based assays
To determine whether recombinant fIL-31 could function in cell-based signaling, we incubated varying amounts of either E. coli-derived or CHO-derived fIL-31 with the feline macrophage cell line FCWF-4 and determined pSTAT signaling using an AlphaLISA pSTAT3 kit. Both recombinant fIL-31s activated pSTAT signaling with an EC50 < 0.1 μg/mL (Figure 4a), indicating comparable bioactivity. To determine whether fOSMR-CBD can inhibit fIL-31 mediated pSTAT3 signaling, we pre-incubated the soluble receptor with 10 μg/mL fIL-31 for 1 hr at 37oC before adding to the cells in culture. Increasing amounts of fOSMR-CBD could directly block fIL-31-mediated pSTAT signaling (Figure 4b), indicating that blocking the IL-31 OSMR-binding epitope is sufficient to block signaling.
Figure 4.
Effect of (a.) fIL-31 to induce STAT3 phosphorylation in macrophage cell line FCWF-4 and (b.) fOSMR-ECD inhibition of pSTAT signaling induced by fIL-31.
Structural homology model for fIL-31
Mapping the fIL-31 binding sites and the conformational epitope requires a reasonably accurate homology model. Given that there is no IL-31 crystal structure available, we generated our initial models using I-TASSER27,28. While all models contained the predicted four helix up-up-down-down topology consistent with a previous structural model from Le Saux et al.13, none were likely to be completely accurate. All models contained several buried hydrophilic loop residues and surface exposed hydrophobic residues, and only some models properly paired the disulfide bond between Cys49-Cys132 (data not shown). Based on the initial set of models, we re-ran I-TASSER using additional restraints based on putative hydrophobic contacts (Contact required between residues 64 – 120; 68 – 120; 14 – 90; 90 – 127) and requiring a distance constraint of 2Å for Cys49-Cys123. Then, we used Rosetta to identify the lowest-scoring variant to refine all atoms within the structure. The resulting homology model of the mature protein has a mostly hydrophobic core formed from the four antiparallel helices (Figure S2; model coordinates in Text S1). Molprobity analysis29 shows a structure with minimal clashes, over 99% favored rotamers, and only 2 Ramachandran outliers (Glu41, Ser42) on a loop covering the interface between the A and D helix. While there will certainly be deviations between this model and the true structure, we judged this model sufficient to evaluate epitope mapping experiments.
YSD and saturation mutagenesis reveals partially overlapping binding sites for fOSMR and fIL31RA
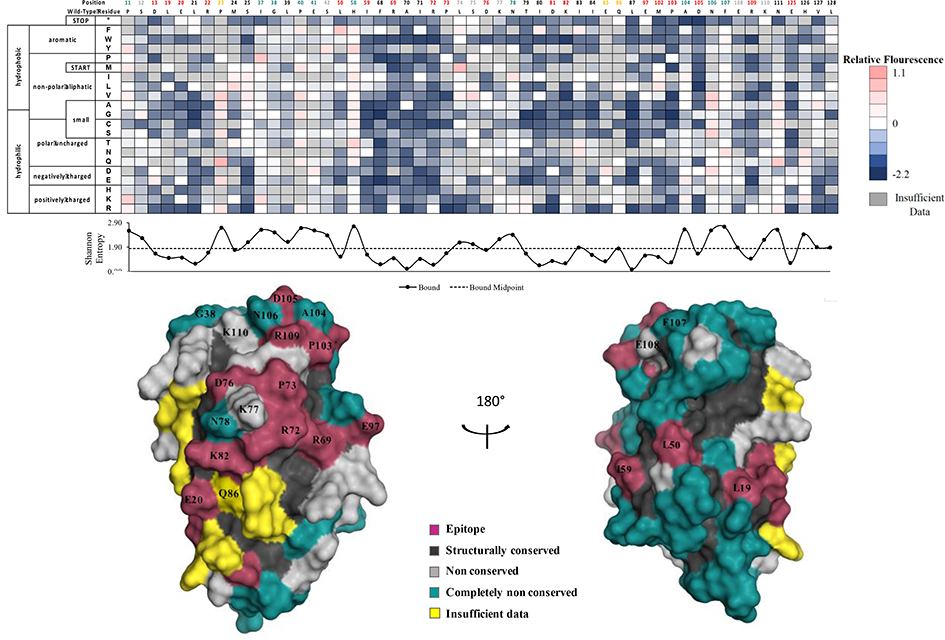
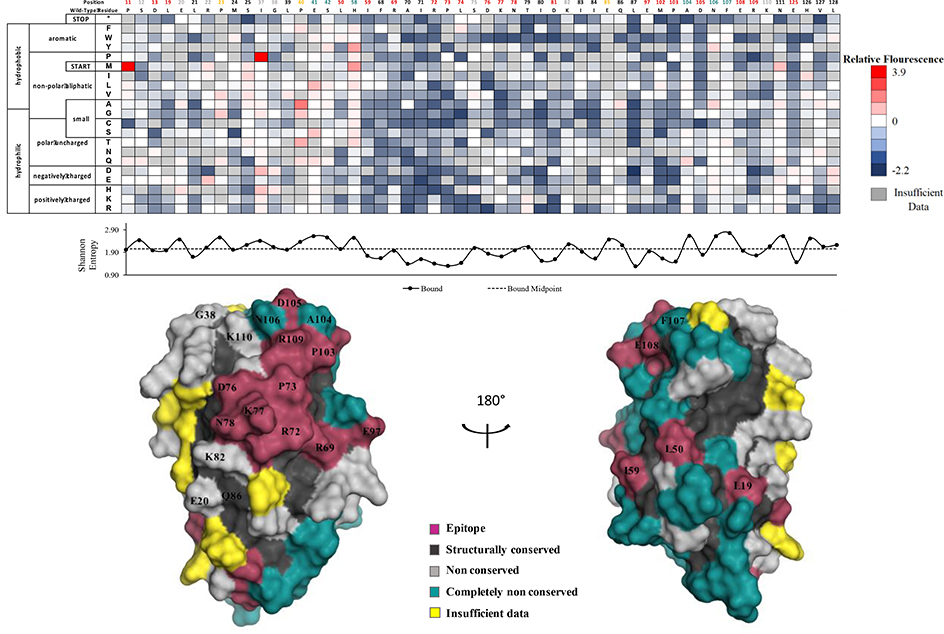
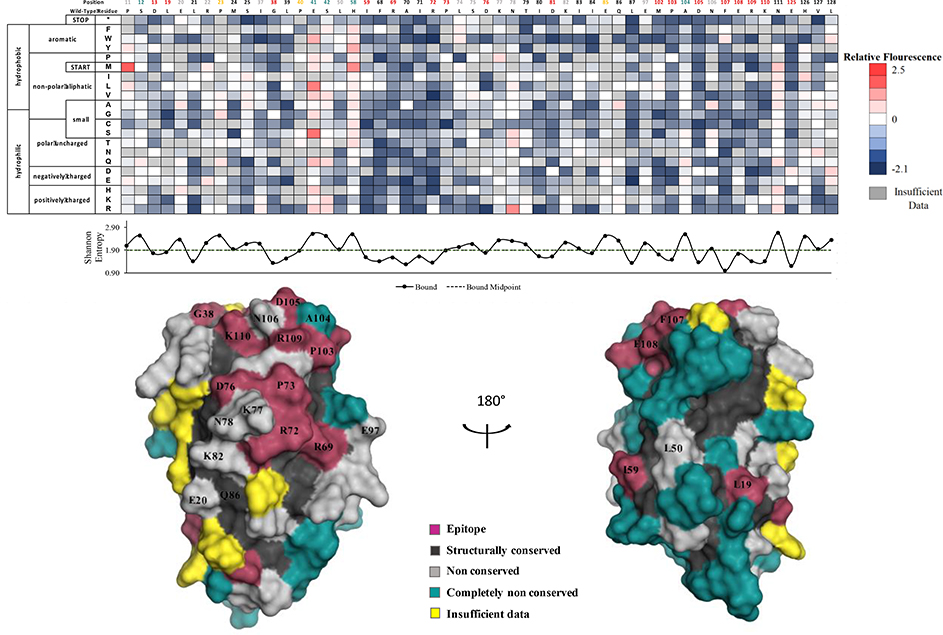
We determined comformational epitopes on fIL-31 for its binding partners using our previously published method combining yeast surface display, nicking mutagenesis, and deep sequencing20,26. Fine epitope mapping using yeast surface display has been established for over 15 years30. The general concept behind our implementation of this method is that mutations that result in loss of binding will predominantly map to the epitope. To that end, we created two comprehensive single-site saturation mutagenesis (SSM) libraries for fIL-31 using nicking mutagenesis22, transformed these libraries into S. cerevisiae EBY100, and deep sequenced the population. We observed an average coverage of 79.5% for every possible single missense and nonsense nonsynonymous substitution (2,161 out of 2,720 mutations; Table S3). The two SSM fIL-31 libraries were expressed on the surface of yeast and each labeled with biotinylated fOSMR-ECD, fIL31RA-1FNIII or mAb #1 at half of the experimentally determined dissociation constant (Table S1). Libraries were sorted by FACS into two distinct populations: one population corresponding to approximately the top 7% by fluorescence for the channel corresponding to the biotinylated protein (bound population), and a reference population of yeast cells that passed through the cell sorter. Plasmid DNA was extracted and deep sequenced. For each variant we calculated the relative fluorescence values based on the change in frequency between the bound and reference populations26. We also determined the per-position Shannon entropy (SE), a measure of sequence conservation, in order to determine the epitope. Summary statistics and complete per-position fIL-31 heatmaps are given in Table S3 and Figures S3–S5, respectively. A subset of the heatmaps for both receptors and mAb#1 are shown in Figure 5–7.
Figure 5.
Determination of the fIL31RA-1FNIII binding site using deep sequencing. Shannon entropy with its respective cut-off (dashed lines) is plotted below the heatmap as well as the structural homology model with the determined binding footprint.
Figure 7.
Determination of the mAb#1 conformational epitope using deep sequencing. Shannon entropy with its respective cut-off (dashed lines) is plotted below the heatmap as well as the structural homology model with the determined binding footprint.
Positions with less than 25% accessible surface area (ASA) are removed from analysis, as mutations at these positions often result in misfolded protein. Applied to the structural model of fIL-31, this analysis removed 60/136 positions; we define these positions as structurally conserved. The SE values for these positions are significantly lower than for surface exposed positions (2.17 vs. 1.78 for fOSMR dataset, 2.24 vs, 1.93 for fIL31RA dataset, and 2.30 vs. 2.00 for mAb #1, p-values for all < 10−5). Positions with insufficient data for more than ten mutations were also excluded from the analysis, leaving 68 fIL-31 positions for further analysis. Surface exposed residues with lower then midpoint SE values were deemed epitope positions. Of the remaining non-conserved positions, ones with the highest 10% SE were classified as completely non-conserved.
Figure 5 shows a restricted per-position heatmap and fIL-31annotated structural model for the fIL-31RA interaction. 16 fIL-31 residues were identified as belonging to the fIL-31RA epitope. These residues form a semi-contiguous patch surrounding the previously described site II from Le Saux et al. for hIL-3113. In particular, E20 on helix A and K82 on helix C were two of three previously identified epitope positions on Site II from alanine scanning experiments by Le Saux et al. The third identified residue, Q86, does show reduced binding upon alanine mutation, although there are insufficient data for other mutations at that positions to make a definitive epitope determination. Unexpectedly, an adjacent contiguous patch on helix B (R69, R72, P73), BC loop (D76), helix C (D81), and CD loop (E97) also appears part of the fIL-31RA epitope, as most mutations at these positions were strongly depleted in the bound library. Other positions identified as belonging to the epitope (L19, L50, and I59) are discontinuous with the rest of the epitope positions and may represent structural conserved positions that reflect inadequacies with the structural model.
Figure 6 shows the fOSMR-fIL31 heatmap and structural model. Overall, 17 positions mapped to the binding site while 28 were completely non-conserved. The epitope covers the expected patch on site III, including G38 and K110 previously identified by Le Saux et al.13 for the hIL31-hOSMR interaction. This epitope is characterized by strong binding at the “PADNFERK” motif (P103-K110) at the beginning of the D helix. However, we also observed low SE and thus strong conservation for the same contiguous patch (R69, R72, P73, D76, D81, E97) as observed with the fIL-31RA interaction. We deemed this region starting with R69 the shared site. Although the sequence entropy of L50 is slightly above the cutoff, the discontinuous L19, L50, and I59 residues observed in the fIL-31RA binding maps are still conserved in this structure and most likely represent structurally conserved residues.
Figure 6.
Determination of the fOSMR-ECD binding site using deep sequencing. Shannon entropy with its respective cut-off (dashed lines) is plotted below the heatmap as well as the structural homology model with the determined binding footprint.
mAb#1 conformational epitope reveals nature of inhibition
In our final approach, we determined the conformational epitope for the anti-fIL-31 mAb #1 using the same procedure. 20 positions mapped to the epitope (Fig 7, Fig S4). mAb#1 binds to all the positions within the shared site between fOSMR and fIL-31RA as well as adjacent positions K77 and N78. However, high SE was observed at both canonical site II (E20, K82) and site III (G38, K110) positions, suggesting that mAb#1 does not directly inhibit either canonical IL6-like receptor binding sites. Based on these results, mAb#1 potentially inhibits the signaling of both receptors through the shared site.
Mutations conferring increases in relative fluorescence are distal from inferred epitopes
There are a few mutations conferring large increases in the relative fluorescence of the yeast display measurements for binding against all three proteins. It is important to note that it is difficult to deconvolute the biophysical interpretation of such individual mutations in terms of binding affinity, relative expression on the yeast surface, and fraction of folded proteins that display on the yeast surface. Nevertheless, for the fIL31RA epitope map only position 46 present on the loop between helix A and B shows greatly improved relative fluorescence, particularly S46F. For OSMR and mAB#1, which share a common epitope, mutations that show increased fluorescence map to the loop between helix A and B (for OSMR: mutations between E41-S46; for mAb#1: I37P and mutations at P40). Both proteins also share a P11M mutation that improves relative fluorescence. The number of gain of function mutations, including mutations to and away from proline, suggests that localized loop restructuring can improve recognition of the binding partners for IL31.
DISCUSSION AND CONCLUSION
In this study we have explored the biochemistry for intermolecular interactions between fIL-31, its receptors, and an anti-fIL-31 mAb. Additionally, we used our established deep mutational scanning pipeline with an improved fIL-31 structural model to map the binding sites of fIL-31 receptors and the conformational epitope of a mAb known to inhibit the bioactivity of fIL-31. We found that Site II residues contributed to fIL-31RA recognition while Site III residues were important in fOSMR binding. However, there were two relative surprises resulting from the current work.
First, in contrary to previous results with human orthologs13, 14, our work shows that fOSMR-ECD can directly recognize fIL-31 without the presence of fIL-31RA. Importantly, we used three different independent biophysical methods for assessing binding. We also show that fIL-31 used in binding experiments is functional in cell-based assays and can be specifically inhibited by fOSMR-ECD. Taken together, the biochemical evidence for this interaction is unambiguous. However, we note that our experiments were all performed with feline IL-31 and receptors; thus, it remains to be seen whether there are species-specific differences between IL-31 signaling pathways (see sequence alignment of IL-31 in Note S2). In particular, given that the lack of interaction between human OSMR and IL-31 was determined by immunoprecipitation, it would be of interest to perform more stringent biochemical validation on the human orthologs to assess whether the mechanism of interaction is conserved across higher animals. Also importantly, we did not establish in the present work exactly how signaling occurs – are both OSMR and IL-31RA required in felines, or is binding to one receptor sufficient?
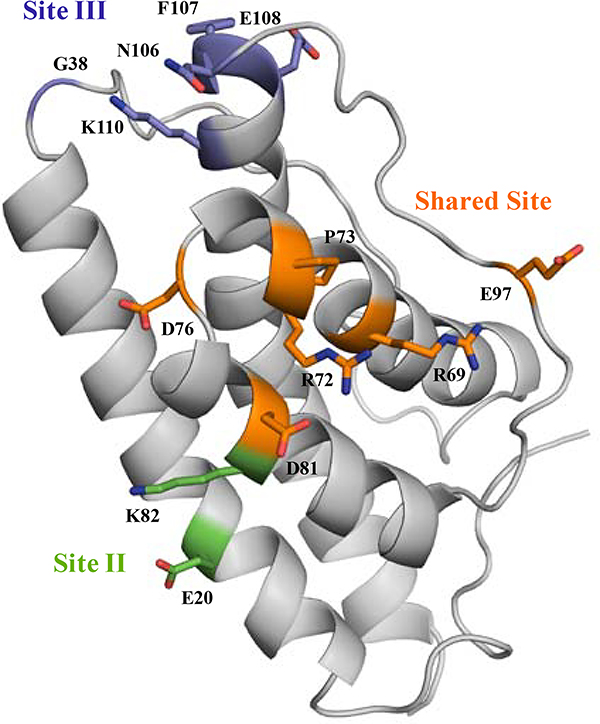
Second, we found that fOSMR and fIL-31RA could compete for binding on fIL-31, and fine epitope mapping using our yeast display pipeline supported the epitope location as a contiguous patch we deemed the “shared site” between both receptors (Figure 8). There are two caveats with an unambiguous determination of this shared site as an epitope for OSMR and IL-31RA recognition. First, we relied on a homology model of IL-31. Thus, it is possible that these shared site positions may not be surface exposed in the monomer. However, we think this unlikely given the contiguous patch revealed by the epitope mapping and the relative hydrophilicity of the involved side-chains. Second, although fIL-31 was mainly produced as a monomeric protein, size exclusion chromatography shows a small percentage of recombinant fIL-31 is dimeric. Such results lead us to ask whether this shared site is exposed on the surface or if it is buried in the interface between each monomer. While unlikely given Hill coefficients of 1 determined through kinetic assays on YSD, this is a real possibility and points to the limitations of fine epitope mapping without a high resolution experimentally determined structure. In particular, mutations that alter the structural integrity of the protein may impact the readout of relative fluorescence from the fine epitope mapping just the same as mutations that alter the binding affinity at the epitope. Since the “shared site” mutants result in lessened binding for all targets, the data is consistent with those sites being epitope sites for all three binding partners but also consistent with those sites being structurally important. In the absence of atomic resolution of the IL31/receptor complex, a key control would be to identify antibodies or other binding proteins that target distal epitopes and ask whether mutations at the “shared site” impact binding. With the pace of antibody development for the IL31 pathway, these experiments should be forthcoming.
Figure 8.
Specific binding sites revealed for fIL31RA-1FNIII and fOSMR-ECD. The shared site (orange) is composed of B-helix, BC loop, C-helix, and CD loop. The binding site for fIL31RA-1FNIII (green), site II, includes A helix and C helix. The binding site for fOSMR-ECD (purple), site III, comprises the N-terminal of AB loop and D-helix.
For over the past few years, IL-31 and its signaling pathway have been identified as one of the central causes of severe inflammatory skin disorders. Fine epitope information presented here may facilitate to the development of antagonist mAbs that inhibit the downstream signaling pathway.
Supplementary Material
Supporting Note S1: Feline IL-31 DNA and Amino acid sequences; mAb #1 VH and VL amino acid sequences.
Supporting Note S2: Sequence alignment of Feline, Human, Mouse, and Canine IL-31.
Supplementary Figure S1: Mass spectrometry and analytical SEC for feline IL-31 construct.
Supplementary Figure S2: Structural model of feline IL-31.
Supplementary Figures S3-S5: Per-position heatmaps for epitope mappings.
Supplementary Table S1: Sorting condition and FACS collection statistics for fIL-31 libraries.
Supplementary Table S2: Primer sets used for deep sequencing analysis.
Supplementary Table S3: fIL-31 library statistics.
SupportingTextS1-fIL31model.cif – homology model of fIL-31
Supplementary_DataS1_fIL-31_IL31RA-1FNIII_FitnessMetric.txt – Textfiles of processed deep sequencing dataset.
Supplementary_DataS2_fIL-31_OSMR_FitnessMetric.txt – Textfiles of processed deep sequencing dataset.
Supplementary_DataS3_fIL-31_mAb#1_FitnessMetric.txt – Textfiles of processed deep sequencing dataset.
ACKNOWLEDGEMENTS
This work was supported by a Zoetis research grant, by a NIH T32 Biotechnology Training Grant (Award # T32-GM110523) to A.M.C, and by Johansen Crosby endowed chair to T.A.W.
Footnotes
NCBI Accession Codes.
fIL-31 XP_011286140.1
Conflict of Interest
T.A.W., A.V.M.C., and C.A.S. have no competing interests. All other authors are employees of Zoetis, Inc. Zoetis has filed patent applications relating to monoclonal antibodies targeting different portions of the IL-31 signaling pathway.
References
- (1).Dillon SR, Sprecher C, Hammond A, Bilsborough J, Rosenfeld-Franklin M, Presnell SR, Haugen HS, Maurer M, Harder B, Johnston J, Bort S, Mudri S, Kuijper JL, Bukowski T, Shea P, Dong DL, Dasovich M, Grant FJ, Lockwood L, Levin SD, LeCiel C, Waggie K, Day H, Topouzis S, Kramer J, Kuestner R, Chen Z, Foster D, Parrish-Novak J, and Gross JA (2004) Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat. Immunol. 5, 752–760. [DOI] [PubMed] [Google Scholar]
- (2).Neis MM, Peters B, Dreuw A, Wenzel J, Bieber T, Mauch C, Krieg T, Stanzel S, Heinrich PC, Merk HF, Bosio A, Baron JM, and Hermanns HM (2006) Enhanced expression levels of IL-31 correlate with IL-4 and IL-13 in atopic and allergic contact dermatitis. J. Allergy Clin. Immunol. 118, 930–937. [DOI] [PubMed] [Google Scholar]
- (3).Rabenhorst A, and Hartmann K (2014) Interleukin-31: A Novel Diagnostic Marker of Allergic Diseases. Curr. Allergy Asthma Rep. 14, 423. [DOI] [PubMed] [Google Scholar]
- (4).Cornelissen C, Lüscher-Firzlaff J, Baron JM, and Lüscher B (2012) Signaling by IL-31 and functional consequences. Eur. J. Cell Biol. 91, 552–566. [DOI] [PubMed] [Google Scholar]
- (5).Takaoka A, Arai I, Sugimoto M, Honma Y, Futaki N, Nakamura A, and Nakaike S (2006) Involvement of IL-31 on scratching behavior in NC/Nga mice with atopic-like dermatitis. Exp. Dermatol. 15, 161–167. [DOI] [PubMed] [Google Scholar]
- (6).Sonkoly E, Muller A, Lauerma AI, Pivarcsi A, Soto H, Kemeny L, Alenius H, Dieu-Nosjean M-C, Meller S, Rieker J, Steinhoff M, Hoffmann TK, Ruzicka T, Zlotnik A, and Homey B (2006) IL-31: A new link between T cells and pruritus in atopic skin inflammation. J. Allergy Clin. Immunol. 117, 411–417. [DOI] [PubMed] [Google Scholar]
- (7).Lewis KE, Holdren MS, Maurer MF, Underwood S, Meengs B, Julien SH, Byrnes-Blake KA, Freeman JA, Bukowski TR, Wolf AC, Hamacher NB, Rixon MW, and Dillon SR (2017) Interleukin (IL) 31 induces in cynomolgus monkeys a rapid and intense itch response that can be inhibited by an IL-31 neutralizing antibody. J. Eur. Acad. Dermatology Venereol. 31, 142–150. [DOI] [PubMed] [Google Scholar]
- (8).Raap U, Wichmann K, Bruder M, Ständer S, Wedi B, Kapp A, and Werfel T (2008) Correlation of IL-31 serum levels with severity of atopic dermatitis. J. Allergy Clin. Immunol. 122, 421–423. [DOI] [PubMed] [Google Scholar]
- (9).Ezzat M, Hasan Z, and Shaheen K (2011) Serum measurement of interleukin-31 (IL-31) in paediatric atopic dermatitis: elevated levels correlate with severity scoring. J. Eur. Acad. Dermatology Venereol. 25, 334–339. [DOI] [PubMed] [Google Scholar]
- (10).Pantazi E, Valenza G, Hess M, and Hamad B (2017) The atopic dermatitis market. Nat. Rev. Drug Discov. 17, 237–238. [DOI] [PubMed] [Google Scholar]
- (11).Nemoto O, Furue M, Nakagawa H, Shiramoto M, Hanada R, Matsuki S, Imayama S, Kato M, Hasebe I, Taira K, Yamamoto M, Mihara R, Kabashima K, Ruzicka T, Hanifin J, and Kumagai Y (2016) The first trial of CIM331, a humanized antihuman interleukin-31 receptor A antibody, in healthy volunteers and patients with atopic dermatitis to evaluate safety, tolerability and pharmacokinetics of a single dose in a randomized, double-blind, placebo-co. Br. J. Dermatol. 174, 296–304. [DOI] [PubMed] [Google Scholar]
- (12).Michels GM, Ramsey DS, Walsh KF, Martinon OM, Mahabir SP, Hoevers JD, Walters RR, and Dunham SA (2016) A blinded, randomized, placebo-controlled, dose determination trial of lokivetmab (ZTS-00103289), a caninized, anti-canine IL-31 monoclonal antibody in client owned dogs with atopic dermatitis. Vet. Dermatol. 27, 478–e129. [DOI] [PubMed] [Google Scholar]
- (13).Le Saux S, Rousseau F, Barbier F, Ravon E, Grimaud L, Danger Y, Froger J, Chevalier S, and Gascan H (2010) Molecular dissection of human interleukin-31-mediated signal transduction through site-directed mutagenesis. J. Biol. Chem. 285, 3470–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Diveu C, Lak-Hal A-HL, Froger J, Ravon E, Grimaud L, Barbier F, Hermann J, Gascan H, and Chevalier S (2004) Predominant expression of the long isoform of GP130-like (GPL) receptor is required for interleukin-31 signaling. Eur. Cytokine Netw. 15, 291–302. [PubMed] [Google Scholar]
- (15).Zhang Q, Putheti P, Zhou Q, Liu Q, and Gao W (2008) Structures and biological functions of IL-31 and IL-31 receptors. Cytokine Growth Factor Rev. 19, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Diveu C, Lelièvre E, Perret D, Lak-Hal A-HL, Froger J, Guillet C, Chevalier S, Rousseau F, Wesa A, Preisser L, Chabbert M, Gauchat J-F, Galy A, Gascan H, and Morel A (2003) GPL, a novel cytokine receptor related to GP130 and leukemia inhibitory factor receptor. J. Biol. Chem. 278, 49850–49859. [DOI] [PubMed] [Google Scholar]
- (17).Dambacher J, Beigel F, Seiderer J, Haller D, Göke B, Auernhammer CJ, and Brand S (2007) Interleukin 31 mediates MAP kinase and STAT1/3 activation in intestinal epithelial cells and its expression is upregulated in inflammatory bowel disease. Gut 56, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dreuw A, Radtke S, Pflanz S, Lippok BE, Heinrich PC, and Hermanns HM (2004) Characterization of the signaling capacities of the novel gp130-like cytokine receptor. J. Biol. Chem. 279, 36112–36120. [DOI] [PubMed] [Google Scholar]
- (19).Boulanger MJ, Chow D, Brevnova EE, and Garcia KC (2003) Hexameric Structure and Assembly of the Interleukin-6/IL-6 α-Receptor/gp130 Complex. Science. 300, 2101–2104. [DOI] [PubMed] [Google Scholar]
- (20).Medina-Cucurella AV, and Whitehead TA (2019) Characterizing Protein-Protein Interactions Using Deep Sequencing Coupled to Yeast Surface Display. Methods Mol. Biol. 1764, 101–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, and Wittrup KD (2006) Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768. [DOI] [PubMed] [Google Scholar]
- (22).Wrenbeck EE, Klesmith JR, Stapleton JA, Adeniran A, Tyo KEJ, and Whitehead TA (2016) Plasmid-based one-pot saturation mutagenesis. Nat. Methods 13, 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Araya CL, and Fowler DM (2011) Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 29, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Medina-Cucurella AV, Zhu Y, Bowen SJ, Bergeron LM, and Whitehead TA (2018) Pro region engineering of nerve growth factor by deep mutational scanning enables a yeast platform for conformational epitope mapping of anti-NGF monoclonal antibodies. Biotechnol. Bioeng. 115, 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kowalsky CA, Klesmith JR, Stapleton JA, Kelly V, Reichkitzer N, and Whitehead TA (2015) High-Resolution Sequence-Function Mapping of Full-Length Proteins. PLoS One 10, e0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kowalsky CA, Faber MS, Nath A, Dann HE, Kelly VW, Liu L, Shanker P, Wagner EK, Maynard JA, Chan C, and Whitehead TA (2015) Rapid fine conformational epitope mapping using comprehensive mutagenesis and deep sequencing. J. Biol. Chem. 290, 26457–26470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Roy A, Kucukural A, and Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen VB, Bryan W, Arendall III W, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chao G, Cochran JR, and Dane Wittrup K (2004) Fine Epitope Mapping of anti-Epidermal Growth Factor Receptor Antibodies Through Random Mutagenesis and Yeast Surface Display. J. Mol. Biol. 342, 539–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Note S1: Feline IL-31 DNA and Amino acid sequences; mAb #1 VH and VL amino acid sequences.
Supporting Note S2: Sequence alignment of Feline, Human, Mouse, and Canine IL-31.
Supplementary Figure S1: Mass spectrometry and analytical SEC for feline IL-31 construct.
Supplementary Figure S2: Structural model of feline IL-31.
Supplementary Figures S3-S5: Per-position heatmaps for epitope mappings.
Supplementary Table S1: Sorting condition and FACS collection statistics for fIL-31 libraries.
Supplementary Table S2: Primer sets used for deep sequencing analysis.
Supplementary Table S3: fIL-31 library statistics.
SupportingTextS1-fIL31model.cif – homology model of fIL-31
Supplementary_DataS1_fIL-31_IL31RA-1FNIII_FitnessMetric.txt – Textfiles of processed deep sequencing dataset.
Supplementary_DataS2_fIL-31_OSMR_FitnessMetric.txt – Textfiles of processed deep sequencing dataset.
Supplementary_DataS3_fIL-31_mAb#1_FitnessMetric.txt – Textfiles of processed deep sequencing dataset.
Data Availability Statement
Full datasets including normalized relative fluorescence, pre- and post-selection read counts, and raw log base two enrichment scores for each variant can be found in Supplementary Data S1–S3. Raw sequencing reads for this work have been deposited in the SRA (SAMN11289369–72, 79, 81–83, 90–91, SAMN11289422–23).