Summary
Recent data suggest that HLA epitope matching is beneficial for the prevention of de novo donor specific antibody (DSA) formation after transplantation. In this review, different approaches to predict the immunogenicity of an HLA mismatch will be discussed. The parameters used in these models are often called epitopes but the actual antibody epitope is far more complex. Exact knowledge of the antibody epitope is crucial if epitope matching is also used as a tool to select compatible donors for (highly) sensitized patients. Evidence is provided that it is not always possible to give an exact definition of an antibody epitope. We conclude that HLA “epitope” matching is superior over HLA antigen matching with respect to the prevention of de novo DSA formation and will enhance the prediction of acceptable HLA mismatches for sensitized patients. However, epitope matching at our current level of knowledge will not solve all histocompatibility problems as unexpected antibody reactivity still may occur.
Keywords: antibodies, epitopes, HLA matching, immunogenicity, virtual crossmatch
Introduction
After the discovery that blood transfusion and pregnancy can lead to the induction of leucocyte‐reactive antibodies 1, 2, 3, it soon became clear that the antigens recognized by these antibodies played a pivotal role in the immune response leading to graft rejection after kidney transplantation. When donors and recipients had the same HLA type, graft and patient survival was significantly better than in case of HLA mismatched transplants 4. These first results were obtained with transplants using living related donors, often siblings of the patient, which made the chance of transplanting an HLA identical graft relatively high. Selection of an HLA identical unrelated donor is far more difficult, due to the enormous polymorphism of the HLA system. Only in case a large pool of unrelated donors is available, full HLA matching on the serological level might become feasible. In order to reach this goal, the international organ exchange organization “Eurotransplant” was founded in 1967 by Jon van Rood 5. By creating a common waiting list and a common donor pool of several countries in Europe, HLA matching became a realistic option, at least for a subpopulation of patients. It appeared that this initiative was successful, as about 20% of the patients within Eurotransplant were transplanted with an HLA‐A, ‐B, ‐DR identical donor, which was associated with superior graft survival compared to HLA mismatched transplants 6. Unfortunately, still the majority of patients were transplanted with a (partially) HLA mismatched graft. Nonetheless, it appeared that decreasing the number of HLA mismatches was already a tool to minimize sensitization and prevent early graft loss. During the years following, more efficient immunosuppression became available leading to prolonged graft survival, also in HLA mismatched transplants. Nevertheless, sensitization towards HLA still occurs, and those patients are at risk for early graft loss 7. Moreover, once sensitized the chance of finding a suitable donor organ for a patient becomes far more difficult. Recently, several approaches of alternative HLA matching have been described to prevent the induction of donor specific antibody (DSA) in order to improve graft survival. Knowledge of the exact amino acid sequence of the different HLA antigens appears to be crucial for the selection of an optimally HLA mismatched donor 8. In this review, we will focus on the differential immunogenicity of epitopes, the requirement for T cell help and the difficulties in determining the exact binding determinants of HLA antibodies.
From HLA antigens towards HLA epitopes
Soon after the serological identification of HLA antigens it became clear that HLA molecules belonged to a highly polymorphic system. While the introduction of molecular typing was crucial for a more accurate definition of HLA antigens, it also resulted in an enormous increase of the number of HLA alleles reported. At the moment, more than 15 000 alleles are known and it is to be expected that this number will increase far more by broader application of next generation sequencing as a tool for HLA typing. At first glance, these developments make the selection of an HLA mismatched donor organ that will not induce DSA a mission impossible.
Fortunately, knowledge on the exact amino acid sequence of the different HLA alleles has given more insight in the crucial positions on the HLA molecules for the induction and reactivity of allo‐antibodies. Already in the early days of HLA, when the HLA antigens were still serologically characterized with allo‐antisera, it became clear that the different HLA molecules share antigenic determinants that we now call epitopes 9. Several CREGs (Cross REactive Groups) could be identified based on their shared reactivity with the same allo‐antibodies 10. However, for an optimal characterization of the epitopes expressed by the different HLA antigens, molecularly HLA typing at the allele level is crucial 8. Nowadays it is clear that every HLA antigen consists of a unique set of antibody epitopes, while the individual epitopes can be shared by multiple HLA antigens 11. The consequence is that the number of foreign antibody epitopes varies within the same level of HLA antigen mismatches (Fig. 1). Some HLA mismatches have many epitopes not shared by the HLA antigens of the patient. These mismatches are likely to be more immunogenic compared to an HLA mismatch which shares most epitopes with the patient. Several tools have been developed to determine the relative immunogenicity of an HLA mismatch on basis of this principle.
Figure 1.
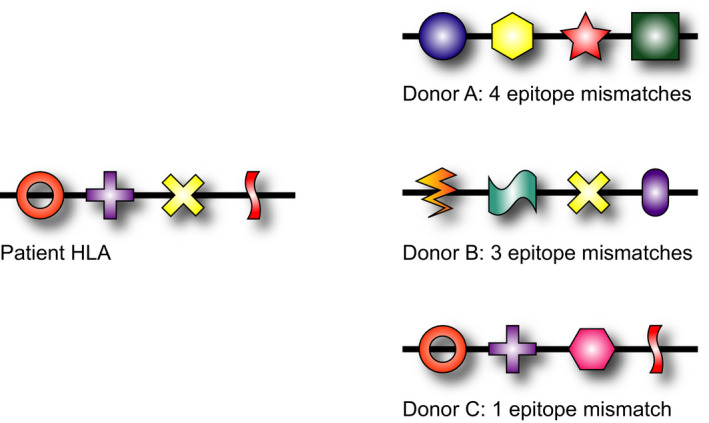
HLA alleles can be considered as a string of potential antibody epitopes. A specific HLA allele consists of a unique set of epitopes while the individual epitopes can be shared with other alleles. The consequence is that the number of foreign epitopes on an individual HLA mismatch can differ and depends on the HLA type of the potential antibody producer. It is to be expected that the HLA mismatch of donor A (four foreign epitopes for the patient) is more immunogenic than the one of donor C (only one nonself epitope).
Tools to define the immunogenicity of an HLA mismatch
The first individual who realized that the patchwork character of HLA molecules with respect to antibody epitopes could provide a basis to determine the immunogenicity of foreign HLA molecules, was Rene Duquesnoy in Pittsburgh. He defined HLA molecules as strings of crucial amino acid polymorphisms, which potentially can trigger an antibody response 12. Originally these polymorphisms were defined as consisting of three amino acids, adjacent to each other on antibody accessible sites of the molecule, called triplets. Later, Duquesnoy redefined the crucial polymorphism as amino acids within 3 Ångstrom radius, structurally near to each other on the tertiary conformation of the HLA molecule, called eplets 13. The computer algorithm HLAMatchmaker made it possible to compare the triplets or eplets on a foreign HLA molecule with those present on the patient's own HLA molecules 14. During the past years, several studies demonstrated clear associations between the number of triplets or eplets on a mismatched HLA antigen and the chance that a patient develops de novo DSA 15, 16, 17, 18. Both in case of mismatched HLA class I and HLA class II antigens, the incidence of antibody formation increases with the number of foreign triplets or eplets. Some studies even suggest that it is possible to define a threshold of a certain number of eplets, which predicts whether antibody production will occur 19. However, even a very low number of mismatched eplets can already give rise to DSA formation as was already shown by the studies of Dankers et al. 16, which demonstrated a clear association between the number of mismatched triplets and the chance that a patient will develop de novo DSA. In case of 12 or more triplet mismatches 100% of the patients, who rejected their graft, had developed DSA but also in case of one or two triplet mismatches respectively 10% and 22% of the patients produced DSA. Furthermore, it remains to be established whether the definition of the number of foreign eplets is the optimal way to predict the immunogenicity of an HLA antigen. A similar predictive value has been demonstrated if one considers the total number of antibody‐accessible amino acid substitutions of the mismatched HLA antigen in comparison with the own HLA antigens of the patient 20.
A completely different approach has been developed by the Cambridge group. Their studies show a clear role of the physiochemical properties of mismatched amino acids. If the physiochemical properties of mismatched amino acids are very different from those of the own HLA antigens of the patients, induction of donor specific antibodies is far more likely than in case of similar properties 20, 21, 22. They have validated a score system (EMS‐2D), which determines the degree of foreignness of the physiochemical characteristics, both for the induction of DSA after renal transplantation and after blood transfusion. The higher the score, the more likely it is that a patient will make antibodies.
The different approaches, which consider polymorphisms on antibody accessible sites of the donor HLA molecule, all suggest that quantitative aspects i.e. the number of foreign triplets, eplets, amino acids play a crucial role but do not consider a potential difference in the immunogenic properties of the individual polymorphisms 23.
Differential immunogenicity of individual “epitopes”
Although the relationship between the number of triplet/eplet mismatches and the chance to develop DSA is well established, the immunogenicity of the individual triplets/eplets may vary. Not every polymorphic site has the same immunogenic potential and one could hypothesize that a higher number of triplet/eplet mismatches makes it likely that one of these is particularly immunogenic. This is also suggested by a study on the development of DSA associated with graft loss in a cohort of 1311 previously nonimmunized males, who returned on the waiting list of Eurotransplant after failure of their first transplant. This study, which focused on the development of antibodies against HLA‐A and ‐B showed that some mismatched triplets led to antibody formation in about 50% of the cases whereas others led to a lower incidence of DSA or hardly induced any antibodies (Table 1). Similarly to the frequencies of HLA alleles, which differ amongst populations, the frequency of potential antibody epitopes will also be different between populations in the world (www.allelefrequencies.net). The chance that a particular HLA allele mismatch will induce antibodies depends amongst others on the frequency of its most immunogenic epitope in the population. If this frequency is high, it is more likely that donor and recipient share this epitope, resulting in a low number of patients with that antibody specificity. If the frequency of the immunogenic epitope is low in a population, the incidence of antibody formation against that HLA allele in case that it is mismatched will be high. A preliminary study comparing the incidence of antibody formation in transplant recipients in Israel versus Eurotransplant confirms this hypothesis (Israeli et al. manuscript in preparation). These data suggest that future matching strategies should not only focus on the number of mismatched eplets, triplets or amino acids but, especially, on prevention of mismatches for highly immunogenic polymorphisms. In order to be able to reach this goal, it is crucial to identify the immunogenicity of the different polymorphisms with respect to the induction of allo‐antibodies. This is one of the aims of the 18th International Histocompatibility workshop, which will take place in Amsterdam in 2021. By collecting information on the incidence of de novo DSA in a large group of high resolution typed donor‐recipient combinations from different populations, it should be feasible to identify the most immunogenic antibody epitopes. In future allocation, these should be avoided as a mismatch in order to prevent the induction of DSA. The other side of the coin is the identification of polymorphisms, who do not lead to allo‐antibodies in an individual patient and are likely to serve as acceptable mismatches.
Table 1.
The immunogenicity of individual epitopes differs as demonstrated by analyzing the epitope specificity of donor specific antibody (DSA) developed in previously nonsensitized males, who returned on the waiting list after failure of their first kidney transplant
| Positions + amino acids | Yes DSA | No DSA | % DSA | Donor mismatches |
|---|---|---|---|---|
| 79G 80T 81L | 30 | 23 | 56.6 | A23 A24 A25 A32 |
| 11A 12M 13S | 55 | 49 | 52.9 | B7 B18 B27 B37 B38 B39 B46 B48 B61 |
| 150A 151H 152V | 104 | 95 | 52.3 | A1 A3 A11 A23 A25 A26 A29 A30 A31 A32 A33 |
| 126L 127K 128E | 102 | 94 | 52 | A1 A3 A11 A25 A26 A29 A30 A31 A32 A33 |
| 130L 131S 132S | 30 | 30 | 50 | B7 B8 |
| 73I 74D 75R | 14 | 47 | 23 | A31 A3375 |
| 73R 76E 77D | 20 | 69 | 22.5 | B27, B37, B47 |
| 185P 186R 187T | 1 | 15 | 6.3 | A33 |
Very immunogenic epitopes are associated with specific antibodies in more than 50% of the cases at risk whereas other epitopes are less immunogenic.
An additional role for T cell epitopes
The majority of the donor specific antibodies that develop after transplantation belong to the IgG class. Production of IgG antibodies is only feasible if CD4+ T cells provide help to the B cells, as without such an interaction activation of B cells will only lead to IgM antibodies. T cell help is based on the recognition of peptides derived from allogeneic HLA molecules presented by the HLA class II molecules on the B cell (Fig. 2). Whereas, B cell epitopes solely reside on antibody accessible locations, polymorphisms throughout the HLA molecule can theoretically give rise to T cell epitopes. Indeed, studies on the location of B cell and T cell epitopes showed overlapping regions, as one would expect, but also the presence of T cell epitopes in areas where no B cell epitopes were present 24. Identification of the actual peptides, which function as targets for this so called indirect allorecognition by CD4+ T cells, has shown to be very difficult due to the low frequency of indirectly recognizing T cells 25. Nonetheless, a computer algorithm has been developed, which calculates the number of potential allogeneic peptides derived from the mismatched HLA molecule that are able to bind to the HLA‐DR molecules present on the B cells of the recipient 26. This PIRCHE approach (Predictable Indirectly ReCognizable HLA Epitopes) has shown to be of additional value for the prediction of the chance that donor specific antibodies will be produced 26, 27, 28. An increased number of theoretical T cell epitopes is associated with a higher chance that a patient will start to make antibodies to a mismatched HLA antigen. A recent publication showed that the number of PIRCHE mismatches and the number of eplet mismatches were independent risk factors for both the development of DSA after transplantation, and graft outcome 18. Similar to B cell epitopes, the challenge is to discriminate the actual immunogenic PIRCHEs from the nonimmunogenic theoretical ones. It is to be expected that a combinatorial approach, focussing on the identification of both B cell and T cell epitopes, will be the optimal basis of future matching strategies.
Figure 2.
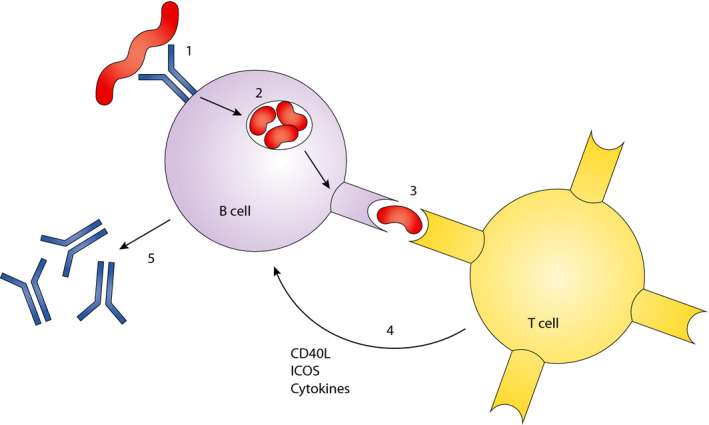
The production of IgG antibodies depends on a specific interaction between CD4 + T cells and B cells. 1: The B cell receptor recognizes an epitope on a foreign HLA molecule. 2: This leads to internalization of the target antigen, which is then degraded into peptides. 3: Some of these peptides bind to the HLA class II molecules on the B cell and the foreign (nonself) peptides are recognized by CD4 + T cells. 4: his leads to activation of the T cells associated with the production of immunoregulatory molecules. 5: These molecules trigger a class switch of the antibodies produced.
Immunogenic determinants versus actual antibody epitopes
So far, the immunogenicity of an HLA mismatch for the humoral immune response has been discussed with a special emphasis on the predictive value of polymorphic structures which serve as a trigger for, or are associated with, the induction of an antibody response. As discussed above, these structures are rather simple and consist of one or a few amino acids. However, the actual antibody epitope, which consists of the crucial contact sites on the HLA molecule necessary for the interaction with antibody molecule is far more complex. In principle, an antibody molecule has a paratope, consisting of six so called complementary determining regions (CDRs), three on the heavy chain and three on the light chain, which interact with the HLA antigen recognized. The interaction of the epitope with these CDRs leads to a kind of footprint on the HLA molecule (Fig. 3). The specificity of the antibody is determined by the amino acids on the HLA molecule, which interact with the CDR3 on the heavy chain of the immunoglobulin molecule. These amino acids include the triplets/eplets, which were responsible for the induction of the antibody response. The other contact sites contribute to the stability and avidity of the interaction between antibody and antigen. In order to be able to predict the HLA antigen reactivity pattern of an antibody, it is essential to determine which of these interactions are crucial for proper binding of the antibody. In some cases, antigens only sharing the polymorphism interacting with the CDR3 with the immunizing antigen, are already targets for an alloantibody. In other cases an additional requirement is that one or more other CDRs bind exactly the same amino acids as present on the immunizing antigen. The use of human HLA‐specific monoclonal antibodies (mAbs) has been instrumental for the definition of actual antibody epitopes, while also absorption elution experiments are suitable to define the crucial polymorphisms defining an antibody epitope. A dedicated website has been developed to register the actual antibody epitopes on the different HLA antigens 29, 30.
Figure 3.
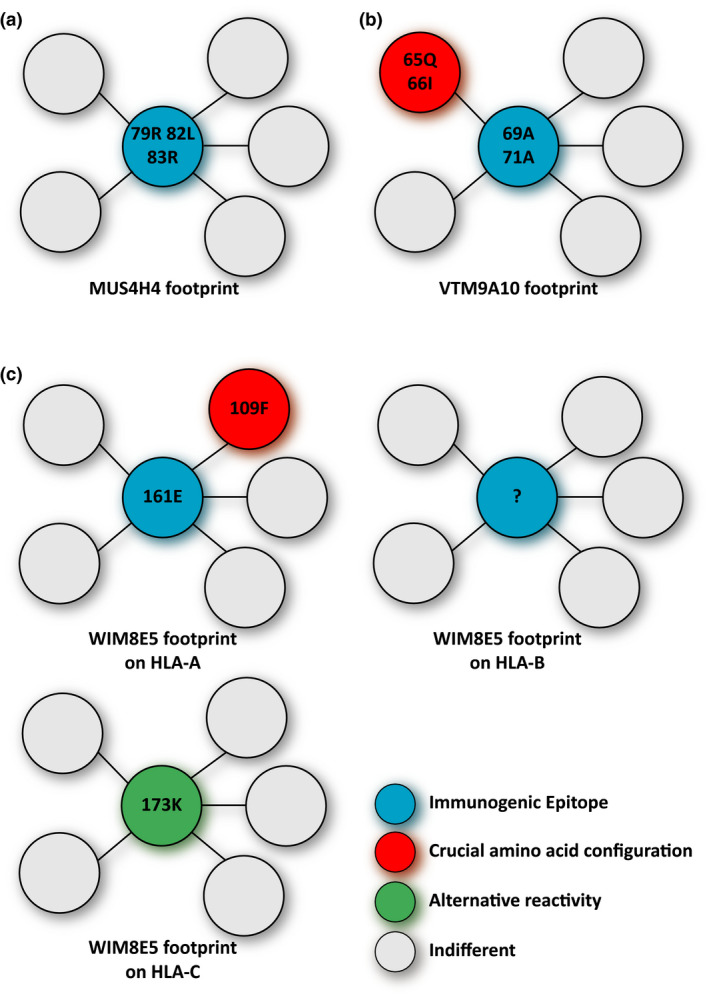
Foot prints of three human monoclonal antibodies on their target antigens. (a) The reactivity of monoclonal MUS4H4 depends only on sharing of the immunogenic epitope with the HLA molecule, which has triggered the production of this antibody. (b) For the reactivity of monoclonal antibody VTM9A10 sharing of both the immunogenic epitope and an additional contact site with the immunizing antigen is crucial. (c) The reactivity of monoclonal antibody WIM8E5 is very complex. It appears that the reactivity with HLA‐A antigens depends on sharing of the immunogenic epitope and an additional contact site. The observed cross‐reactivity with HLA‐B and –C antigens has completely different requirements. The reactive HLA‐C alleles have one particular polymorphic position in common (in green) whereas the basis of the reactivity with HLA‐B targets remains unclear. Note: more details on the immunizing effect leading to the production of these antibodies is given in table 2.
It is clear that some of the epitopes can be easily defined, while others are more complex, as shown below.
Definition of the actual HLA antibody epitope can be difficult, even with monoclonal antibodies
Both human mAbs and absorption/elution studies with alloantisera have been very instrumental for the definition of the actual antibody epitopes on the HLA molecules 31, 32, 33. These verified epitopes are registered at the epitope registry (http://www.epregistry.com.br) As mentioned above, antigen‐antibody interaction involves six CDRs of which the CDR3 binds to the immunogenic epitope and determines the specificity of the antibody. In most cases, the presence of this immunogenic epitope, alone or in combination with one or two other polymorphic sites, can explain the antibody reactivity pattern with the different HLA alleles (Table 2, Fig. 3a,b, monoclonal antibodies MUS4H4 and VTM9A10). However, sometimes very complicated reaction patterns are observed, which make it virtually impossible to define an epitope that explains all antibody reactivities.
Table 2.
HLA types of the antibody producer and immunizer and epitope specificity of the three monoclonal antibodies depicted in Fig. 3
| Human mAb | HLA antibody producer | HLA immunizer | Epitopes on reactive HLA class I allele | ||
|---|---|---|---|---|---|
| HLA‐A | HLA‐B | HLA‐C | |||
| MUS4H4 (IgG) | A*02:01, A*26:01, B*39:01, B*41:01, C*12:03, C*17:01 | A*24:02 | 79R82L83R | 79R82L83R | – |
| VTM9A10 (IgG) | A*25:01, A*29:02, B*44:03, B*15:01, C*05:01, C*16:01 | B*07:02 | – | 69A71A(65Q66I) | – |
| WIM8E5 (IgG) | A*03:01, A*03:02, B*47:01, B*51:01, C*06:02, C*15:02 | A*11:01 | 161E(109F) | ? | 173K |
? indicates the epitope recognized on HLA‐B is unclear.
Here we describe an example of an HLA‐A*11:01 induced mAb, WIM8E5, derived from a woman who became sensitized during pregnancy by paternal antigens. The trigger of the antibody response can be any epitope on the mismatched HLA, dependent on the phenotype of the mother. In this case, comparison of all the HLA class I molecules of the mother with the immunising HLA‐A*11:01, reveals that there are only two amino acid differences, which may have triggered the antibody response. However, if we compare only the HLA‐A molecules of the mother with HLA‐A*11:01, there are six amino acid differences.
When WIM8E5 was screened with single antigen bead (SAB) assays of two independent vendors, a broad HLA antigen reactivity was observed, including specificities of all three classical HLA class I loci. In contrast to other mAbs, the reactivity of WIM8E5 cannot be explained by the fact that the reactive alleles share the immunogenic epitope and some other crucial amino acid configurations, which are absent on the nonreactive antigens. Almost all HLA‐A antigens are reactive, except for three, including the self‐antigens. In addition, some HLA‐B and HLA‐C antigens were reactive. Extensive comparisons of the amino acid composition of the different reactive HLA antigens did not lead to a clear definition of an antibody epitope (Fig. 3c).
Others have shown that dilution of serum samples can clarify the patterns observed in SAB assays 34. Dilution of WIM8E5 led to a decrease in the breadth of reactive HLA antigens. One group of HLA‐A antigens, including the immunising HLA‐A*11:01, remained highly reactive in SAB assays upon dilution. The reactivity of the other HLA‐A antigens was less consistent and decreased with each dilution step, while the reactivity of the HLA‐B and HLA‐C antigens decreased rapidly upon dilution.
Most of the HLA class I epitopes have been defined on the basis of interlocus comparisons of amino‐acid sequences. However, for WIM8E5 the reactive HLA‐C antigens do not have any amino acid configuration in common with the reactive HLA‐A antigens. However, they do share a unique amino acid that is absent on the nonreactive HLA‐C antigens. These data suggest that the induction of the WIM8E5 antibody was induced by an epitope mismatch specific for HLA‐A and that the crossreactivity with HLA‐B and HLA‐C is not based on reactivity of the antibody with the same, HLA‐A specific epitope.
This example shows that verification of HLA epitopes using mAbs involves more than screening the mAb with SAB assay, and just identifying the shared amino acid configurations of the reactive antigens. The different reactivity patterns upon dilution of WIM8E5 do support the conclusion that different epitopes are involved and that the affinity of the antibody for the different target antigens is different. An alternative explanation for the observed cross‐reactivity might be that the reactive antigens have similar physiochemical properties, despite amino acid variation within the critical contact site.
Concluding remarks
For many years, HLA matching strategies have focused on the selection of donors with a minimal number of HLA antigen mismatches. However, it is clear that the immunogenicity of mismatched HLA antigens can differ. If one would like to prevent the induction of de novo DSA, which is known to be associated with a poor outcome, then limiting the number of HLA antigen mismatches is not the optimal strategy. Our view on HLA antigens as possible targets for antibodies has been changed considerably over time. Antibodies are not specific for an HLA antigen but for an epitope present on the HLA molecule. Every HLA antigen can be considered as a string of antibody epitopes. A particular HLA antigen consists of a unique combination of epitopes while the individual epitopes can also be expressed on other HLA antigens. This is the reason why the number of foreign epitopes present on a mismatched HLA antigen will differ and depends on the HLA type of the recipient. Some HLA mismatches express many epitopes whereas others have only a few or even no foreign epitope. As a consequence, the presence of two HLA antigen mismatches on a donor organ may, for some recipients, be associated with fewer epitope mismatches than the presence of a single antigen mismatch. Therefore, novel matching strategies aim at the limitation of the number of foreign epitopes rather than the number of HLA antigen mismatches.
Current activities in the field of epitope matching can be divided in two categories. The first one is aiming at the prevention of de novo DSA formation by limiting the number of potentially antibody inducing polymorphisms on the mismatched HLA molecules. Indeed, several tools have been described, which can successfully predict the chance that DSA will be induced 23. These include the number of foreign eplets/triplet, amino‐acids or PIRCHE's, and the physicochemical properties of the mismatched HLA molecule. Such parameters are already excellent predictors of the immunogenicity of an HLA mismatch and as soon as we are able to distinguish the very immunogenic polymorphisms from the nonimmunogenic ones, their prediction will be even better. However, none of these parameters can be considered as the actual antibody epitope. Exact knowledge of the antibody epitope is crucial if one would like to introduce epitope matching as a tool for virtual crossmatching of (highly) sensitized patients. Human monoclonal antibodies and absorption/elution studies have been very helpful for the definition of several antibody epitopes, which are described in the HLA epitope registry. At the moment, this collection is far from complete and the example described in this review suggests that it will not always be feasible to explain the possible reactivity of an HLA antibody based on the knowledge of the amino acid sequence of the immunizing antigen.
In conclusion, “epitope” matching is superior over antigen matching with respect to the prevention of de novo DSA formation and will enhance the prediction of acceptable HLA mismatches for sensitized patients. However, one should realize that epitope matching at our current level of understanding will not solve all histocompatibility problems, as unexpected antibody reactivity still may occur.
Funding
The authors have declared no funding.
Conflicts of interest
The authors have declared no conflicts of interest.
References
- 1. Dausset J, Brecy H. Identical nature of the leucocyte antigens detectable in monozygotic twins by means of immune iso‐leuco‐agglutinins. Nature 1957; 180: 1430. [DOI] [PubMed] [Google Scholar]
- 2. Payne R, Rolfs MR. Fetomaternal leukocyte incompatibility. J Clin Invest 1958; 37: 1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Rood JJ, Eernisse JG, Van Leeuwen A. Leucocyte antibodies in sera from pregnant women. Nature 1958; 181: 1735. [DOI] [PubMed] [Google Scholar]
- 4. Van Rood JJ, Van Leeuwen A, Eernisse JG, Frederiks E, Bosch LJ. Relationship of leukocyte groups to tissue transplantation compatibility. Ann N Y Acad Sci 1964; 120: 285. [DOI] [PubMed] [Google Scholar]
- 5. van Rood JJ. A proposal for international cooperation in organ transplantation: eurotransplant In: Curtoni ES, Mattiuz PL, Tosi RM, eds. Histocompatibility Testing. Baltimore, MD: Williams and Wilkins, 1967: 451. [Google Scholar]
- 6. Persijn GG, D'Amaro J, De Lange P, et al The effect of mismatching and sharing of HLA‐A, ‐B, and ‐DR antigens on kidney graft survival in Eurotransplant 1982 to 1988. Clin Transpl 1988; 237. [PubMed] [Google Scholar]
- 7. Lachmann N, Terasaki PI, Budde K, et al Anti‐human leukocyte antigen and donor‐specific antibodies detected by luminex posttransplant serve as biomarkers for chronic rejection of renal allografts. Transplantation 2009; 87: 1505. [DOI] [PubMed] [Google Scholar]
- 8. Duquesnoy RJ, Kamoun M, Baxter‐Lowe LA, et al Should HLA mismatch acceptability for sensitized transplant candidates be determined at the high‐resolution rather than the antigen level? Am J Transplant 2015; 15: 923. [DOI] [PubMed] [Google Scholar]
- 9. Claas F, Castelli‐Visser R, Schreuder I, van Rood J. Allo‐antibodies to an antigenic determinant shared by HLA‐A2 and B17. Tissue Antigens 1982; 19: 388. [DOI] [PubMed] [Google Scholar]
- 10. Rodey GE, Fuller TC. Public epitopes and the antigenic structure of the HLA molecules. Crit Rev Immunol 1987; 7: 229. [PubMed] [Google Scholar]
- 11. Tambur AR, Claas FH. HLA epitopes as viewed by antibodies: what is it all about? Am J Transplant 2015; 15: 1148. [DOI] [PubMed] [Google Scholar]
- 12. Duquesnoy RJ. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. I. Description of the algorithm. Hum Immunol 2002; 63: 339. [DOI] [PubMed] [Google Scholar]
- 13. Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol 2006; 67: 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Claas FH, Dankers MK, Oudshoorn M, et al Differential immunogenicity of HLA mismatches in clinical transplantation. Transpl Immunol 2005; 14: 187. [DOI] [PubMed] [Google Scholar]
- 15. Wiebe C, Pochinco D, Blydt‐Hansen TD, et al Class II HLA epitope matching‐A strategy to minimize de novo donor‐specific antibody development and improve outcomes. Am J Transplant 2013; 13: 3114. [DOI] [PubMed] [Google Scholar]
- 16. Dankers MK, Witvliet MD, Roelen DL, et al The number of amino acid triplet differences between patient and donor is predictive for the antibody reactivity against mismatched human leukocyte antigens. Transplantation 2004; 77: 1236. [DOI] [PubMed] [Google Scholar]
- 17. Kosmoliaptsis V, Bradley JA, Sharples LD, et al Predicting the immunogenicity of human leukocyte antigen class I alloantigens using structural epitope analysis determined by HLAMatchmaker. Transplantation 2008; 85: 1817. [DOI] [PubMed] [Google Scholar]
- 18. Lachmann N, Niemann M, Reinke P, et al Donor‐recipient matching based on predicted indirectly recognizable HLA epitopes independently predicts the incidence of de novo donor‐specific HLA antibodies following renal transplantation. Am J Transplant 2017; 17: 3076. [DOI] [PubMed] [Google Scholar]
- 19. Wiebe C, Nickerson P. Strategic use of epitope matching to improve outcomes. Transplantation 2016; 100: 2048. [DOI] [PubMed] [Google Scholar]
- 20. Kosmoliaptsis V, Mallon DH, Chen Y, Bolton EM, Bradley JA, Taylor CJ. Alloantibody responses after renal transplant failure can be better predicted by donor‐recipient HLA amino acid sequence and physicochemical disparities than conventional HLA matching. Am J Transplant 2016; 16: 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kosmoliaptsis V, Dafforn TR, Chaudhry AN, Halsall DJ, Bradley JA, Taylor CJ. High‐resolution, three‐dimensional modeling of human leukocyte antigen class I structure and surface electrostatic potential reveals the molecular basis for alloantibody binding epitopes. Hum Immunol 2011; 72: 1049. [DOI] [PubMed] [Google Scholar]
- 22. Mallon DH, Bradley JA, Taylor CJ, Kosmoliaptsis V. Structural and electrostatic analysis of HLA B‐cell epitopes: inference on immunogenicity and prediction of humoral alloresponses. Curr Opin Organ Transplant 2014; 19: 420. [DOI] [PubMed] [Google Scholar]
- 23. Wiebe C, Kosmoliaptsis V, Pochinco D, Taylor C, Nickerson P. A comparison of HLA molecular mismatch methods to determine HLA immunogenicity. Transplantation 2018; 102: 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otten HG, Calis JJ, Kesmir C, van Zuilen AD, Spierings E. Predicted indirectly recognizable HLA epitopes presented by HLA‐DR correlate with the de novo development of donor‐specific HLA IgG antibodies after kidney transplantation. Hum Immunol 2013; 74: 290. [DOI] [PubMed] [Google Scholar]
- 25. Waanders MM, Heidt S, Koekkoek KM, et al Monitoring of indirect allorecognition: wishful thinking or solid data? Tissue Antigens 2008; 71: 1. [DOI] [PubMed] [Google Scholar]
- 26. Geneugelijk K, Honger G, van Deutekom HW, et al Predicted indirectly recognizable HLA epitopes presented by HLA‐DRB1 are related to HLA antibody formation during pregnancy. Am J Transplant 2015; 15: 3112. [DOI] [PubMed] [Google Scholar]
- 27. Geneugelijk K, Thus KA, Spierings E. Predicting alloreactivity in transplantation. J Immunol Res 2014; 2014: 159479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geneugelijk K, Spierings E. Matching donor and recipient based on predicted indirectly recognizable human leucocyte antigen epitopes. Int J Immunogenet 2018; 45: 41. [DOI] [PubMed] [Google Scholar]
- 29. Duquesnoy RJ, Marrari M, da M Sousa LC, et al 16th IHIW: a website for antibody‐defined HLA epitope Registry. Int J Immunogenet 2013; 40: 54. [DOI] [PubMed] [Google Scholar]
- 30. Duquesnoy RJ, Marrari M, Mulder A, Sousa LC, da Silva AS, do Monte SJ. First report on the antibody verification of HLA‐ABC epitopes recorded in the website‐based HLA Epitope Registry. Tissue Antigens 2014; 83: 391. [DOI] [PubMed] [Google Scholar]
- 31. Mulder A, Kardol M, Regan J, Buelow R, Claas F. Reactivity of twenty‐two cytotoxic human monoclonal HLA antibodies towards soluble HLA class I in an enzyme‐linked immunosorbent assay (PRA‐STAT). Hum Immunol 1997; 56: 106. [DOI] [PubMed] [Google Scholar]
- 32. Duquesnoy RJ, Marrari M, Jelenik L, Zeevi A, Claas FH, Mulder A. Structural aspects of HLA class I epitopes reacting with human monoclonal antibodies in Ig‐binding, C1q‐binding and lymphocytotoxicity assays. Hum Immunol 2013; 74: 1271. [DOI] [PubMed] [Google Scholar]
- 33. El‐Awar N, Lee JH, Tarsitani C, Terasaki PI. HLA class I epitopes: recognition of binding sites by mAbs or eluted alloantibody confirmed with single recombinant antigens. Hum Immunol 2007; 68: 170. [DOI] [PubMed] [Google Scholar]
- 34. Tambur AR. Hiding in plain sight‐a new look at HLA epitopes: a case report. Am J Transplant 2016; 16: 3286. [DOI] [PubMed] [Google Scholar]


