Abstract
The insect midgut epithelium is composed of columnar, goblet, and regenerative cells. Columnar epithelial cells are the most abundant and have membrane protrusions that form the brush border membrane (BBM) on their apical side. These increase surface area available for the transport of nutrients, but also provide opportunities for interaction with xenobiotics such as pathogens, toxins and host plant allelochemicals. Recent improvements in proteomic and bioinformatics tools provided an opportunity to determine the proteome of the T. ni BBM in unprecedented detail. This study reports the identification of proteins from BBM vesicles (BBMVs) using single dimension polyacrylamide gel electrophoresis coupled with multi‐dimensional protein identification technology. More than 3000 proteins were associated with the BBMV, of which 697 were predicted to possess either a signal peptide, at least one transmembrane domain or a GPI‐anchor signal. Of these, bioinformatics analysis and manual curation predicted that 185 may be associated with the BBMV or epithelial cell plasma membrane. These are discussed with respect to their predicted functions, namely digestion, nutrient uptake, cell signaling, development, cell–cell interactions, and other functions. We believe this to be the most detailed proteomic analysis of the lepidopteran midgut epithelium membrane to date, which will provide information to better understand the biochemical, physiological and pathological processes taking place in the larval midgut.
Keywords: brush border membrane vesicles, midgut, proteomic analysis; Trichoplusia ni
Introduction
Trichoplusia ni (Lepidoptera: Noctuidae), also known as the cabbage looper moth, is a major agricultural pest in most of the world. T. ni will feed on an exceptionally broad range of plants, including over 160 species in 36 families (Sutherland & Green, 1984). Many of these plants are important crop species, including cruciferous vegetables. As a result, T. ni is a common and devastating pest in vegetable growing areas and greenhouses worldwide.
In lepidopteran larvae, the gut is composed of three sections: the foregut, midgut and hindgut. The foregut is lined with a chitin‐containing cuticle and facilitates physical processing of food. Posterior and anterior sphincters separate the midgut from the foregut and hindgut, respectively. The midgut is lined by the peritrophic matrix (PM), an acellular fibrous sheet composed mainly of chitin, mucopolysaccharides and proteins. The PM separates the food bolus from the midgut epithelial cell layer and prevents access by microorganisms and larger food particles to epithelial cells (Lehane & Billingsley, 1996; Hegedus et al., 2009). In T. ni, the columnar cells are the most common in the midgut epithelium. Underlying the columnar cells are regenerative cells which replenish sloughed columnar cells. Columnar cells have membranous protrusions on the apical surface called the brush border membrane (BBM) which increase the surface area of the epithelium for food absorption. Goblet cells are interspersed among the columnar cells along the midgut epithelium (Braun & Keddie, 1997).
The gut is also the primary site for many physiological, biochemical and biological interactions between the insect and the environment. The main route of entry into the host for most insect pathogens is via the gut. Insect viruses, such as the well‐studied Autographa californica multiple nucleopolyhedrovirus (AcMNPV), cross the PM to infect midgut columnar and regenerative cells (Granados & Lawler, 1981; Keddie et al., 1989; Knebel‐Morsdorf et al., 1996; Javed et al., 2016). Entry of virions into host midgut cells involves binding to cell surface receptors by virus ligands followed by fusion with the BBM (Granados & Lawler, 1981; Horton & Burand, 1993). The viral ligands are believed to be the per os infectivity factors; however, their cognate epithelial cell receptors, responsible for the initial interaction, have yet to be discovered (Peng et al., 2013). The delta‐endotoxin released by Bacillus thuringiensis was first shown to interact with an aminopeptidase‐N that is anchored to the epithelial cell membrane by glycosylphosphatidylinositol (GPI) (Knight et al., 1994; Garczynski & Adang, 1995). Subsequent toxin‐binding guided proteomic analyses revealed additional targets, which included alkaline phosphatase, cadherin, actin and V‐ATP synthase (McNall & Adang, 2003; Krishnamoorthy et al., 2007). The significance of the midgut epithelium extends beyond host–pathogen interactions, for example, insecticidal lectins released as part of the plant defense system interact with specific cell surface glycoproteins (Lagarda‐Diaz et al., 2016).
Despite the importance of the midgut epithelial cell membrane in such interactions, reports on its composition are scarce. Twenty proteins were identified from Manduca sexta larval midgut microvilli (Pauchet et al., 2009), while 74 and 86 proteins were found in BBMVs from Aedes aegypti (Popova‐Bulter & Dean, 2009) and Chilo suppressalis (Ma et al., 2012), respectively. It should be noted that many of the proteins identified in these studies are unlikely to be associated with the outer plasma membrane and are most likely remnants of the cytosolic and endomembrane systems. Recent improvements in protein identification technology from complex protein mixtures and the availability of large genomic and transcriptomic sequence databases provide unparalleled opportunities to determine the identities of the complete cellular and subcellular organelle protein composition. In this report, we present a detailed identification of T. ni BBMV proteins and the most complete set of BBMV proteins reported to date. This will enhance the understanding of interactions between the midgut epithelium and micro‐organisms, host plants, and non‐biological materials.
Materials and methods
Preparation of T. ni BBMVs
T. ni larvae were reared at 27 °C on artificial diet (per L: 128 g Pinto beans, 37 g yeast, 32 g wheat germ, 16 g alfalfa meal, 16 g ascorbic acid, 12 g agar, 11 g Wesson's salts, 4 g corn oil, 3.2 g Tween 80, 1.3 g flax oil, 0.9 g methyl 4‐hydroxybenzoate, 0.8 g sorbic acid, 0.5 g chloramphenicol, 0.5 g tetracycline, and 0.25 g alpha‐tocopherol) based on Diet 7 in Vail et al. (1973). BBMVs were prepared using a modification of the protocols of Wolferberger et al. (1987) and Abdul‐Rauf and Ellar (1999). Actively feeding 4th‐instar larvae (n = 10) were chilled on ice for 15 min, dissected and the PM removed from longitudinally sectioned midguts. The midguts were rinsed three times in buffer A (300 mmol/L mannitol, 5 mmol/L ethylene glycol bis(α‐aminoethyl ether)‐N,N‐tetracetic acid (EGTA), 17 mmol/L Tris‐(hydroxymethyl) aminomethane‐HCl [pH 7.5]) and collected in preweighed 2.0 mL tubes containing 400 μL buffer A supplemented with protease inhibitor cocktail (Roche, Laval, Quebec, Canada). Collection tubes were pre‐cooled on ice and after gut collection each tube was weighed again to determine the net weight of midgut tissue. If necessary, the midgut tissue was stored at –80 °C prior to processing. Frozen gut tissue (0.4 g) was thawed on ice and mixed with buffer A to a total volume of 2 mL and homogenized by 30 strokes of a Dounce homogenizer. An equal volume of 24 mmol/L MgCl2 was added to the homogenized tissue and the mixture was again homogenized (five strokes) and incubated on ice for 20 min. The mixture was centrifuged at 2500 × g for 15 min at 4 °C to separate midgut tissue from the BBMVs. The supernatant containing the BBMVs was removed and saved on ice and the pellet resuspended in 1 mL buffer A supplemented with protease inhibitor cocktail and the process repeated. Finally, supernatants were pooled and centrifuged at 30 000 × g for 30 min at 4 °C. The supernatant was discarded and the pellet containing the BBMVs was resuspended in 100 μL half‐strength ice‐cold buffer A and either used immediately or stored at –80 °C.
Transmission electron microscopy of BBMVs
The BBMVs were immobilized on formvar‐carbon coated copper grids (200 mesh) treated with poly L‐lysine by floating the grids on a drop of BBMVs for 5 min. Excess BBMV suspension was removed by touching the edge of the grid with blotting paper and the grids were negatively stained by floating on a drop of 1% phosphotungstic acid for 30 s. Excess acid was removed with blotting paper and the grids were air‐dried at room temperature overnight. A transmission electron microscope Philips CM10 (Philips Electron Optics, Eindhoven, the Netherlands) was used to observe the BBMVs. Images were captured on a plate film camera, films developed, and scanned.
Proteomics analysis of BBMVs
The concentration of protein in the BBMV preparation and the cell debris fraction was determined using a BCA protein assay kit (Thermo Scientific, Rockford, IL, USA). BBMVs were solubilized in protein loading buffer (2% SDS, 10% glycerol, 0.01% bromophenol blue in 60 mmol/L Tris‐HCl buffer, pH 6.8), proteins separated by electrophoresis on 12% SDS‐PAGE gels and protein bands visualized by staining with Coomassie Blue. A lane of BBMV‐enriched proteins was cut into 27 slices with each slice having about one to two prominent protein bands.
The samples were subjected to LS‐MS/MS analysis at the Genome BC Proteomics Centre, University of Victoria, Canada, as per the following procedure. Trypsin digests were performed as previously described (Parker et al., 2005). Briefly, gel slices were manually cut into 1 mm cubes and transferred to a Genomics Solutions Progest (DigiLab Inc., Holliston, MA, USA) perforated digestion tray. The gel pieces were destained (50/45/5 v/v methanol/water/acetic acid) prior to reduction (10 mmol/L dithiothreitol, MilliporeSigma, Oakville, Ontario, Canada) and alkylation (100 mmol/L iodoacetamide, Sigma). Modified sequencing‐grade porcine trypsin solution (20 ng/μL, Promega, Madison, WI, USA) was added to the gel slices at an enzyme/protein ratio of 1 : 50. Proteins were then digested for 5 h at 37 °C prior to collection of the tryptic digests and acid extraction of the gel slices (50/40/10 v/v acetonitrile/water/formic acid). The samples were then lyophilised and stored at –80 °C prior to analysis.
The peptide digests were separated by online reverse phase chromatography using a Thermo Scientific EASY‐nLC II system with a reverse‐phase Magic C‐18AQ precolumn (100 μm I.D., 2 cm length, 5 μm, 100Å, Michrom BioResources Inc, Auburn, CA, USA) and a reverse phase nanoanalytical column Magic C‐18AQ (75 μm I.D., 15 cm length, 5 μm, 100 Å, Michrom BioResources Inc, Auburn, CA, USA) both prepared in‐house, at a flow rate of 300 nL/min. The chromatography system was coupled online with an LTQ Orbitrap Velos mass spectrometer equipped with a Nanospray II source (Thermo Fisher Scientific, Bremen, Germany). Solvents were A: 2% acetonitrile, 0.1% formic acid; B: 90% acetonitrile, 0.1% formic acid. After a 249 bar (∼10 μL) precolumn equilibration and 249 bar (∼6 μL) nanocolumn equilibration, samples were separated by a 55 min gradient (0 min: 5% B; 45 min: 45% B; 2 min: 80% B; hold 8 min: 80% B).
The LTQ Orbitrap Fusion (Thermo Fisher Scientific) parameters were as follows: Nanoelectrospray ion source with spray voltage 2.1 kV, capillary temperature 225 °C. Survey MS1 scan m/z range 400–2000 profile mode, resolution 60 000 FWHM@400 m/z with AGC target 1E6, and one microscan with maximum inject time of 500 ms. Lock mass Siloxane 445.120024 for internal calibration with preview mode for FTMS master scans: on, injection waveforms: on, monoisotopic precursor selection: on; rejection of charge state: 1. The samples were analyzed by the following methods: (1) top 15 FTMS/IT‐CID method with the 15 most intense ions charge state 2–4 exceeding 5000 counts were selected for CID ion trap MSMS fragmentation (ITMS scans 2–16) with detection in centroid mode. Dynamic exclusion settings were: repeat count: 2; repeat duration: 15 s; exclusion list size: 500; exclusion duration: 60 s with a 10 ppm mass window. The CID activation isolation window was: 2 Da; AGC target: 1E4; maximum inject time: 100 ms; activation time: 10 ms; activation Q: 0.250; and normalized collision energy 35%. Common human keratin and porcine trypsin peptide masses were excluded from MS/MS selection during the analysis.
Data analysis
A MASCOT database was generated by supplementing published transcriptome data with two in‐house libraries. The transcriptome developed by Chen et al. (2014) from T. ni cell line Tnms42 undergoing infection with AcMNPV (NCBI Accession No. PRJNA260558) contributed 70 322 unigenes. In‐house 454‐sequencing libraries from brain (NCBI Accession No. SRX1745247) and midgut (NCBI Accession No. SRX1745246) contributed 141 198 and 146 091 reads, respectively. The sequences from the three databases were combined and assembled de novo using the CLC Genomics Workbench (7.5.1) using a word size of 64 bp resulting in 18 972 contigs (assemblies with two or more sequences). Singletons were added by mapping the published transcriptome to the assembly, collecting the unmapped reads and adding them to the assembly contig sequences to bring the transcriptome to 58 200 sequences. These were translated in all 6 reading frames and filtered for sequences over 50 amino acids resulting in a database of 261 307 hypothetical protein sequences. Common contaminant sequences were downloaded from Maxquant.org and included in the T. ni MASCOT database. MS/MS analysis was performed on each slice and output as an .mgf file. All .mgf files were concatenated into a single file and the mass spectra transformed into data files which were used to search the T. ni MASCOT database. Protein identities were assigned using the Mascot MS/MS Ion search algorithm. Mascot search parameters were as follows: peptide tolerance: 8 ppm, fixed modification: carbamidomethyl, variable modification: oxidation, MS/MS tolerance: 0.6 Da, peptide charge 2+ and 3+, and allowed missed cleavages: 1.
BBMV proteins were annotated using the CLC Genomics Workbench plugin for Blast2GO (refseq May 19, 2015), examined for the presence of signal peptides (SignalP http://www.cbs.dtu.dk/services/SignalP/) if a start codon was present, transmembrane domains (TM) (TMHMMOL/L http://www.cbs.dtu.dk/services/TMHMMOL/L), a glycophosphatidylinositol (GPI) anchor signal (BIG‐PI: http://mendel.imp.ac.at/gpi/gpi_server.html), as well as cellular localization (TargetP: http://www.cbs.dtu.dk/services/TargetP/). Additional structural features were determined using PredictProtein (Yachdav et al., 2014) and HMMOL/LER (Finn et al., 2015).
Results
BBMVs were prepared from midgut tissue isolated from actively feeding 4th instar T. ni larvae. Separation of the T. ni midgut BBMV proteins by SDS‐PAGE yielded a pattern of discreet bands ranging from more than 170 kDa to less than 15 kDa in size (Fig. 1). The pattern of separated proteins in the BBMV fraction was highly distinct from that of the midgut cell debris remaining after BBMV preparation indicating that BBMV purification had been successful.
Figure 1.
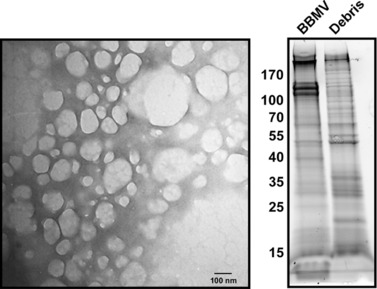
Preparation and analysis of T. ni midgut brush border membrane vesicles (BBMV). (A) Transmission electron microscopic image of negatively stained BBMVs. (B) SDS‐PAGE gel of proteins from 4th‐instar BBMV or cell debris remaining after BBMV isolation. The gel shows enrichment of proteins in the BBMV fraction as reflected by different protein banding patterns in the BBMV and remnant midgut cell debris preparations. The size (kDa) of molecular weight markers is shown in the left hand margin.
The T. ni BBMVs were observed by transmission electron microscopy after negative staining with phosphotungstic acid. The vesicles were circular to oval in shape and of varying sizes (ca. 50–200 nm) (Fig. 1). The morphology of the BBMVs was consistent with the morphology of BBMVs isolated from the midgut of Pieris brassicae, cabbage butterfly (Wolfersberger et al., 1987) and mosquito, Aedes aegypti (Abdul‐Rauf & Ellar, 1999) larvae.
Identification BBMV proteins
A complete genome sequence is not available for T. ni, therefore, virtual protein sequences inferred from contigs compiled from EST sequences from all available database were used. As such, these contigs do not represent sequences from a specific strain/isolate, but rather a composite view. Genebank does not issue accession numbers for virtual genes/proteins used in these types of proteomics studies; however, all of the ESTs which contributed to building the contigs have been deposited in GenBank (see Materials and Methods) with the nucleotide sequences of the contigs and the derived protein sequences provided in Table S1 and Supporting information 1–4.
Proteomic analysis identified 3169 proteins in the BBMV fraction (Table S1; Supporting information 1 and 2). The BBMVs will contain some remnants of the cytosolic components since the process by which they are generated involves disruption of the cell membrane by homogenization followed by spontaneous formation of microvesicles that can recapture such material. To establish which proteins were likely to be associated with the BBMV plasma membrane several criteria were applied. Proteins may either be incorporated within (single and multiple pass transmembrane proteins) or attached to (GPI anchor) the outer plasma membrane. As well, outer membrane proteins often, though not exclusively, begin the trek to this destination in the endoplasmic reticulum which requires a signal peptide for entry. In total, 697 proteins were predicted to possess a signal peptide (333 proteins for which the amino terminus could be inferred from the translated EST contigs), at least one transmembrane domain (484), a GPI‐anchor signal (25), or some combination of these (Table S2; Supporting information 3 and 4). Of these, a few proteins did not show homology to any characterized proteins and, therefore, may represent newly discovered midgut proteins requiring further exploration (Table 1). Of the proteins that could be annotated, many were known or predicted to be localized to subcellular organelles (e.g., mitochondria) and were excluded from the list of putative BBMV plasma membrane proteins, while other proteins were orthologs of known plasma membrane proteins characterized in other insects and/or animals and were included. Manual curation resulted in a final set of 185 putative plasma membrane proteins that were separated into functional categories (Table 1) and are described in more detail below.
Table 1.
Proteins associated with the T. ni brush border or epithelial cell membrane
| Contig name | Protein description† | Signal peptide‡ | Transmembrane domains | GPI signal |
|---|---|---|---|---|
| 1. Enzymes | ||||
| 1.1 Protein metabolism | ||||
| T_ni_cDNA_15821 | Membrane alanyl aminopeptidase | Y | 0 | Y |
| T_ni_cDNA_15865 | Membrane alanyl aminopeptidase | ND | 1 | Y |
| T_ni_cDNA_15923 | Membrane alanyl aminopeptidase | ND | 1 | N |
| T_ni_cDNA_15756 | Membrane alanyl aminopeptidase | ND | 0 | Y |
| T_ni_cDNA_15581 | Aminopeptidase N | Y | 1 | Y |
| T_ni_cDNA_15581 | Aminopeptidase N | Y | 1 | Y |
| T_ni_cDNA_15841 | Zinc carboxypeptidase | ND | 0 | Y |
| T_ni_cDNA_16326 | Zinc carboxypeptidase | ND | 1 | N |
| T_ni_cDNA_11989 | Serine carboxypeptidase | N | 1 | N |
| T_ni_cDNA_9354 | Dipeptidyl peptidase 4 | ND | 1 | N |
| T_ni_cDNA_2674 | Dipeptidyl peptidase 4 | N | 1 | N |
| T_ni_cDNA_11394 | Trypsin‐like protease | ND | 1 | N |
| T_ni_cDNA_16839 | Alkaline c‐like | ND | 1 | N |
| T_ni_cDNA_15309 | Alkaline c‐like | Y | 1 | N |
| T_ni_cDNA_14829 | Gamma‐glutamyltranspeptidase 1 | N | 1 | N |
| T_ni_cDNA_11454 | Angiotensin‐converting enzyme‐like | ND | 2 | N |
| 1.2 Lipid metabolism | ||||
| T_ni_cDNA_15964 | Bile salt‐activated lipase | ND | 1 | N |
| T_ni_cDNA_15786 | Pancreatic triacylglycerol lipase | ND | 1 | N |
| T_ni_cDNA_6516 | Group XV phospholipase A2 | Y | 1 | N |
| 1.3 Alkaline phosphatases | ||||
| T_ni_cDNA_16197 | Membrane‐bound alkaline phosphatase | Y | 0 | Y |
| T_ni_cDNA_7523 | Membrane‐bound alkaline phosphatase | ND | 0 | Y |
| T_ni_cDNA_10461 | Alkaline phosphatase | Y | 1 | N |
| 1.4 Esterases | ||||
| T_ni_cDNA_7581 | FE4 esterase | ND | 0 | Y |
| T_ni_cDNA_5769 | FE4 esterase | ND | 1 | N |
| 2. Transporters | ||||
| 2.1 Nutrient transporters | ||||
| T_ni_cDNA_4704 | Facilitated trehalose transporter TreT1 | N | 12 | N |
| T_ni_cDNA_16594 | Facilitated trehalose transporter TreT1 | ND | 5 | N |
| T_ni_cDNA_16422 | Facilitated trehalose transporter TreT1 | ND | 4 | N |
| T_ni_cDNA_16750 | Facilitated trehalose transporter TreT1 | N | 4 | N |
| T_ni_cDNA_6038 | Facilitated trehalose transporter TreT1 | ND | 7 | N |
| T_ni_cDNA_2642 | Facilitated trehalose transporter TreT1 | ND | 6 | N |
| T_ni_cDNA_8414 | Facilitated trehalose transporter TreT1 | ND | 4 | N |
| T_ni_cDNA_18685 | Facilitated trehalose transporter TreT1 | ND | 4 | N |
| T_ni_cDNA_5339 | Facilitated trehalose transporter TreT1 | N | 12 | N |
| T_ni_cDNA_10089 | Facilitated trehalose transporter TreT1 | Y | 12 | N |
| T_ni_cDNA_6537 | Facilitated trehalose transporter TreT1 | N | 12 | N |
| T_ni_cDNA_1150 | Facilitated trehalose transporter TreT1‐2 | ND | 12 | N |
| T_ni_cDNA_15922 | Facilitated trehalose transporter TreT1‐2 | N | 12 | N |
| T_ni_cDNA_15135 | Facilitated trehalose transporter TreT1‐2 | N | 12 | N |
| T_ni_cDNA_2589 | Facilitated trehalose transporter TreT1‐2 | N | 12 | N |
| T_ni_cDNA_16456 | Monocarboxylate transporter 9 | ND | 7 | N |
| T_ni_cDNA_15428 | Monocarboxylate transporter 12 isoform x1 | ND | 12 | N |
| T_ni_cDNA_7342 | Monocarboxylate transporter 12 | ND | 5 | N |
| T_ni_cDNA_7344 | Monocarboxylate transporter 12 | ND | 5 | N |
| T_ni_cDNA_936 | Monocarboxylate transporter 13 | N | 12 | N |
| T_ni_cDNA_15711 | Peptide transporter family 1 | ND | 10 | N |
| T_ni_cDNA_331 | Sodium‐dependent amino acid transporter 1 | ND | 10 | N |
| T_ni_cDNA_15560 | Sodium‐dependent amino acid transporter 1 | N | 11 | N |
| T_ni_cDNA_268 | Proton‐coupled amino acid transporter 4 | N | 10 | N |
| T_ni_cDNA_10643 | b(+)‐Type amino acid transporter 1 | ND | 12 | N |
| T_ni_cDNA_11849 | Neutral and basic amino acid transport protein RBAT | N | 1 | N |
| T_ni_cDNA_484 | p Protein (tyrosine transport – melanin biosynthesis) | N | 4 | N |
| T_ni_cDNA_16680 | Carnitine transporter (solute carrier family 22 member 5) | ND | 6 | N |
| T_ni_cDNA_15116 | Long‐chain fatty acid transport protein 4 | ND | 4 | N |
| T_ni_cDNA_2856 | Long‐chain fatty acid transport protein 4 | N | 2 | N |
| T_ni_cDNA_16312 | Folate transporter (solute carrier family 46 member 3) | ND | 7 | N |
| T_ni_cDNA_13650 | Folate transporter (solute carrier family 46 member 3) | N | 4 | N |
| T_ni_cDNA_2107 | Folate transporter (solute carrier family 46 member 3) | N | 11 | N |
| T_ni_cDNA_13629 | Folate transporter (solute carrier family 46 member 3) | ND | 5 | N |
| T_ni_cDNA_14535 | Thiamine transporter 1 | N | 9 | N |
| T_ni_cDNA_6134 | Sodium‐dependent multivitamin transporter | N | 13 | N |
| T_ni_cDNA_14772 | Glucose transporter type 1 | N | 10 | N |
| T_ni_cDNA_16283 | sodium‐ and chloride‐dependent glycine transporter 1 | ND | 10 | N |
| T_ni_cDNA_4340 | Equilibrative nucleoside transporter 1 | N | 10 | N |
| T_ni_cDNA_11410 | Nucleoside transporter (solute carrier family 28 member 3) | N | 6 | N |
| 2.2 Ion transporters | ||||
| T_ni_cDNA_7971 | Monoamine transporter (solute carrier family 22 member 3) | N | 10 | N |
| T_ni_cDNA_10297 | Monoamine transporter (solute carrier family 22 member 3) | N | 9 | N |
| T_ni_cDNA_16742 | Organic cation transporter (unknown specificity) | N | 7 | N |
| T_ni_cDNA_16932 | Organic cation transporter (unknown specificity) | ND | 3 | N |
| T_ni_cDNA_15514 | Organic cation transporter (unknown specificity) | ND | 8 | N |
| T_ni_cDNA_16584 | Organic cation transporter (unknown specificity) | ND | 1 | N |
| T_ni_cDNA_16504 | Organic anion transporter (solute carrier family 22 member 6) | N | 12 | N |
| T_ni_cDNA_4393 | Zinc transporter zip1 | N | 8 | N |
| T_ni_cDNA_15008 | Zinc transporter zip1 | N | 7 | N |
| T_ni_cDNA_15544 | Zinc transporter 1 | N | 5 | N |
| T_ni_cDNA_15075 | Zinc transporter 2 | ND | 2 | N |
| T_ni_cDNA_5548 | Zinc transporter 5 | N | 6 | N |
| T_ni_cDNA_4830 | Zinc transporter 8 | N | 6 | N |
| T_ni_cDNA_8634 | Sodium potassium‐transporting ATPase subunit alpha | N | 8 | N |
| T_ni_cDNA_5157 | Sodium potassium‐transporting ATPase subunit beta‐1 | N | 1 | N |
| T_ni_cDNA_15599 | Sodium potassium calcium exchanger 4 | Y | 11 | N |
| T_ni_cDNA_16457 | Otopetrin‐2‐like (calcium) | ND | 2 | N |
| T_ni_cDNA_8680 | Plasma membrane calcium‐transporting ATPase 2 | N | 10 | N |
| T_ni_cDNA_5242 | Membrane magnesium transporter 1 | N | 2 | N |
| T_ni_cDNA_4851 | Metal transporter CNNM2 | ND | 4 | N |
| T_ni_cDNA_2047 | Metal transporter CNNM4 | Y | 5 | N |
| T_ni_cDNA_16108 | High‐affinity copper uptake protein 1 | N | 1 | N |
| T_ni_cDNA_2666 | High‐affinity copper uptake protein 1 | N | 2 | N |
| T_ni_cDNA_1242 | Copper‐transporting ATPase 1 | ND | 8 | N |
| T_ni_cDNA_5956 | Sodium‐independent sulfate anion transporter | ND | 2 | N |
| T_ni_cDNA_2292 | Sodium‐independent sulfate anion transporter | N | 9 | N |
| T_ni_cDNA_17245 | Inorganic phosphate cotransporter | N | 1 | N |
| T_ni_cDNA_9286 | Inorganic phosphate cotransporter | ND | 2 | N |
| T_ni_cDNA_16781 | Inorganic phosphate cotransporter | ND | 5 | N |
| T_ni_cDNA_16392 | Inorganic phosphate cotransporter | ND | 5 | N |
| T_ni_cDNA_11064 | Inorganic phosphate cotransporter | ND | 4 | N |
| T_ni_cDNA_11851 | Inorganic phosphate cotransporter | N | 10 | N |
| T_ni_cDNA_14148 | Inorganic phosphate cotransporter | N | 10 | N |
| T_ni_cDNA_8012 | Band 3 anion transport protein | ND | 3 | N |
| T_ni_cDNA_8012 | Band 3 anion transport protein | ND | 7 | N |
| T_ni_cDNA_11471 | Band 3 anion transport protein | ND | 1 | N |
| T_ni_cDNA_8298 | Bestrophin‐4 (chloride transport) | ND | 5 | N |
| T_ni_cDNA_49 | Transmembrane channel‐like protein 5 | N | 9 | N |
| T_ni_cDNA_6160 | Transmembrane channel‐like protein 7 | ND | 2 | N |
| T_ni_cDNA_5307 | Transmembrane channel‐like protein 7 | ND | 1 | N |
| T_ni_cDNA_3081 | v‐Type proton ATPase subunit s1 | Y | 2 | N |
| T_ni_cDNA_2854 | v‐Type proton ATPase 16 kDa proteolipid subunit | N | 4 | N |
| T_ni_cDNA_16138 | v‐Type proton ATPase 116 kDa subunit a | N | 6 | N |
| T_ni_cDNA_8655 | v‐Type proton ATPase 116 kDa subunit a | N | 7 | N |
| 2.3 ATP‐binding cassette (ABC) and major facilitator (MFS) superfamily transporters | ||||
| T_ni_cDNA_16946 | ABC sub‐family A member 3 | ND | 1 | N |
| T_ni_cDNA_6541 | ABC sub‐family G member 4 | ND | 1 | N |
| T_ni_cDNA_7803 | ABC sub‐family G member 4 | N | 6 | N |
| T_ni_cDNA_12415 | ABC sub‐family G member 4 | N | 7 | N |
| T_ni_cDNA_6541 | ABC sub‐family G member 4 | ND | 5 | N |
| T_ni_cDNA_9811 | Multidrug resistance protein 1a | ND | 2 | N |
| T_ni_cDNA_7617 | Multidrug resistance‐associated protein lethal 03659 | N | 11 | N |
| T_ni_cDNA_16958 | Multidrug resistance‐associated protein lethal 03659 | ND | 2 | N |
| T_ni_cDNA_7620 | Multidrug resistance‐associated protein 4 | N | 6 | N |
| T_ni_cDNA_1434 | Multidrug resistance‐associated protein lethal 03659 | N | 6 | N |
| T_ni_cDNA_9811 | Multidrug resistance protein homolog 49 isoform x1 | ND | 4 | N |
| T_ni_cDNA_17333 | Multidrug resistance protein homolog 49 | ND | 6 | N |
| T_ni_cDNA_14948 | Multidrug resistance‐associated protein 7 | ND | 9 | N |
| T_ni_cDNA_3976 | Multidrug resistance‐associated protein 4 | ND | 4 | N |
| T_ni_cDNA_7619 | Multidrug resistance‐associated protein 4 | ND | 5 | N |
| T_ni_cDNA_3976 | Multidrug resistance‐associated protein 4 | ND | 5 | N |
| T_ni_cDNA_16436 | Multidrug resistance‐associated protein lethal 03659 | ND | 6 | N |
| T_ni_cDNA_11576 | Multidrug resistance‐associated protein lethal 03659 | N | 2 | N |
| T_ni_cDNA_3465 | Major facilitator superfamily domain‐containing protein 6 | N | 10 | N |
| 2.4 Other transporters | ||||
| T_ni_cDNA_16176 | Nose resistant to fluoxetine protein 6 | ND | 5 | N |
| T_ni_cDNA_16598 | Nose resistant to fluoxetine protein 6 | ND | 3 | N |
| T_ni_cDNA_16620 | Nose resistant to fluoxetine protein 6 | ND | 4 | N |
| T_ni_cDNA_16176 | Nose resistant to fluoxetine protein 6 | ND | 4 | N |
| T_ni_cDNA_16467 | Nose resistant to fluoxetine protein 6 | ND | 6 | N |
| T_ni_cDNA_11792 | sid1 Transmembrane family member 1 | Y | 7 | N |
| T_ni_cDNA_1279 | sid1 Transmembrane family member 1 | ND | 4 | N |
| T_ni_cDNA_13757 | sid‐1‐Related gene3 precursor | ND | 3 | N |
| T_ni_cDNA_2198 | Aquaporin | N | 6 | N |
| T_ni_cDNA_16275 | Aquaporin | N | 5 | N |
| T_ni_cDNA_6828 | Cell cycle control protein 50a (phospholipid transport—flippase) | ND | 1 | N |
| T_ni_cDNA_6917 | Ileal sodium bile acid cotransporter‐like | Y | 9 | N |
| 3. Signaling and development | ||||
| T_ni_cDNA_4126 | G‐protein coupled receptor 125 | ND | 5 | N |
| T_ni_cDNA_5794 | G‐protein coupled receptor mth2‐like | ND | 7 | N |
| T_ni_cDNA_10188 | Notch protein isoform X1 | ND | 1 | N |
| T_ni_cDNA_8224 | Dispatched protein (hedgehog signaling) | N | 12 | N |
| T_ni_cDNA_5965 | Patched domain‐containing protein 3‐like (hedgehog receptor) | ND | 9 | N |
| T_ni_cDNA_15629 | Fasciclin‐2 isoform x1 (adhesion protein—inhibitor of EGFR signaling) | ND | 0 | Y |
| T_ni_cDNA_15628 | Fasciclin‐2 isoform x3 | Y | 2 | N |
| T_ni_cDNA_10096 | Fasciclin‐3 isoform x3 | Y | 1 | N |
| T_ni_cDNA_8856 | Plexin‐a4 | ND | 2 | N |
| T_ni_cDNA_6561 | Plexin domain‐containing protein 2‐like | N | 1 | N |
| T_ni_cDNA_16287 | Protein rolling stone‐like | N | 6 | N |
| 4. Cell–cell interaction | ||||
| T_ni_cDNA_1567 | Integrin beta‐nu (midgut cell development) | ND | 1 | N |
| T_ni_cDNA_1572 | Integrin beta‐nu | ND | 1 | N |
| T_ni_cDNA_6146 | Integrin alpha‐ps1 isoform x1 | ND | 1 | N |
| T_ni_cDNA_889 | Integrin alpha‐ps2‐like | Y | 1 | N |
| T_ni_cDNA_1079 | Dystroglycan isoform x1 | ND | 1 | N |
| T_ni_cDNA_16036 | Cadherin‐like membrane protein precursor | ND | 1 | N |
| T_ni_cDNA_15320 | De‐cadherin‐like isoform x2 | ND | 1 | N |
| T_ni_cDNA_13185 | Lachesin‐like isoform x1 | ND | 1 | N |
| T_ni_cDNA_1960 | Innexin inx2 (gap junctions) | N | 4 | N |
| 3.3 Other | 1 | N | ||
| T_ni_cDNA_5683 | 23 kDa integral membrane protein | N | 4 | N |
| T_ni_cDNA_15216 | 23 kDa integral membrane protein | N | 4 | N |
| T_ni_cDNA_5730 | Protein mesh isoform x2 | ND | 1 | N |
| T_ni_cDNA_6939 | Basigin | ND | 2 | N |
| 5. Other proteins | ||||
| 5.1 Immunity/defense | ||||
| T_ni_cDNA_3905 | Peptidoglycan‐recognition protein sc2‐like | N | 1 | N |
| T_ni_cDNA_3908 | Peptidoglycan‐recognition protein 2‐like | N | 1 | N |
| T_ni_cDNA_14656 | Croquemort‐like isoform x3 | N | 2 | N |
| T_ni_cDNA_11150 | Transmembrane protein 120 | ND | 1 | N |
| T_ni_cDNA_6486 | Transmembrane protein 120 | ND | 5 | N |
| 5.2 Chitin metabolism | ||||
| T_ni_cDNA_3621 | Trehalase‐like isoform x1 | Y | 1 | N |
| T_ni_cDNA_8547 | Chitinase‐3‐like protein 2 | N | 1 | N |
| T_ni_cDNA_13856 | Laccase‐1‐like isoform x2 | ND | 0 | Y |
| 5.3 Other | ||||
| T_ni_cDNA_9349 | Protein yellow | Y | 0 | Y |
| T_ni_cDNA_2522 | Protein yellow‐like | ND | 0 | Y |
| T_ni_cDNA_1605 | Transmembrane protein 87a | Y | 7 | N |
| T_ni_cDNA_8393 | Transmembrane protein 177 | ND | 2 | N |
| T_ni_cDNA_5627 | Transmembrane protein 222 | N | 2 | N |
| T_ni_cDNA_7343 | Transmembrane protein 256 homolog isoform x1 | ND | 3 | N |
| T_ni_cDNA_11297 | Leucine‐rich transmembrane protein (aael003720) | ND | 1 | N |
| T_ni_cDNA_9865 | Neutral ceramidase isoform x1 | N | 1 | N |
| T_ni_cDNA_12371 | Leucine‐rich repeat neuronal protein 3‐like isoform x1 | Y | 1 | N |
| T_ni_cDNA_8987 | Platelet glycoprotein v | Y | 1 | N |
| T_ni_cDNA_15370 | Motile sperm domain‐containing protein 2‐like | N | 2 | N |
| T_ni_cDNA_8049 | Cleft lip and palate transmembrane protein 1 homolog | N | 4 | N |
| T_ni_cDNA_13005 | Cklf‐like marvel transmembrane domain‐containing protein 4 | N | 4 | N |
| T_ni_cDNA_3867 | Sel1 (signal transduction) or Skt5p (chitin synthesis) | ND | 1 | N |
| T_ni_cDNA_5900 | TM2‐domain (related to 7 transmembrane GPCR) | ND | 2 | N |
| T_ni_cDNA_14902 | Vacuolar ATPase assembly integral membrane VMA21 | ND | 2 | N |
| T_ni_cDNA_6626 | Uncharacterized insect protein (two protein‐binding domains) | ND | 1 | N |
| T_ni_cDNA_7146 | Uncharacterized insect protein (two protein‐binding domains) | N | 2 | N |
| T_ni_cDNA_11766 | Uncharacterized insect protein (NLS) | ND | 1 | N |
| T_ni_cDNA_16269 | Uncharacterized insect protein | ND | 0 | Y |
†Annotation based on Gene Ontology and BLAST analysis.
‡ND: could not be determined as 5′ open‐reading frame of the contig did not extend to the start codon; N: No; Y: Yes.
Membrane‐associated enzymes
The initial stages of food digestion take place in the endoperitrophic space within the lumen of the lepidopteran midgut as a bolus surrounded by the PM. Here, endoproteases cleave proteins into smaller peptides that can pass through the PM into the ectoperitrophic space adjacent to the midgut epithelium. The peptides are then further processed into component amino acids by membrane‐associated amino‐ and carboxypeptidases so they may be taken up by the midgut cells. Membrane aminopeptidase N (APN) and alanyl peptidases have been isolated from lepidopteran midgut tissue (Wang et al., 2005) and are associated with the brush border membrane of lepidopteran midgut epithelial cells (Yuan et al., 2011). Four isozymes of aminopeptidase N (APN), APN1, APN2, APN3, and APN4, have been identified in T. ni from cDNA sequences (Wang et al., 2005). In this study, six BBMV plasma membrane proteins were identified that were similar to aminopeptidases classified as M1 family zinc metallopeptidases; these included two enzymes classified as APN‐like peptidases and four as alanyl aminopeptidases, although these terms are often used synonymously. In addition, two zinc carboxypeptidase proteins, a venom‐like serine carboxypeptidase and two dipeptidyl peptidases, were identified which would also contribute to the final stages of protein digestion. While endopeptidase activity is generally associated with the initial stages of digestion in the insect gut lumen, an unusual trypsin‐like protease and two alkaline C‐like endopeptidases with single transmembrane domains were identified in the BBMVs which may digest short peptides passing through the peritrophic membrane into the ectoperitrophic space (Bolognesi et al., 2008).
Membrane‐associated enzymes/proteins have been implicated as targets/ligands for insect pathogens or their toxins. Members of the APN family facilitate infection of the lepidopteran midgut by serving as receptors for bacterial toxins, in particular Bacillus thuringiensis delta‐endotoxins (Knight et al., 1994; Sangadala et al., 1994). Insect viruses also attach to, as yet, unidentified ligands on the midgut epithelium (Haas‐Stapleton et al., 2004); a phenomenon that precedes fusion with the epithelial membrane and that may impart host specificity (Horton & Burand, 1993). Interestingly, an alanyl APN was identified as the receptor for an insect‐borne plant virus (Linz et al., 2015) suggesting a role in its transmission. A proteomic study in Plodia interpunctella revealed that resistance to B. thuringiensis Cry1Ab toxin may have arisen from a decrease in the level of chymotrypsin‐like proteases required for its activation in the midgut (Candas et al., 2003); however, enzymes of this type were not predicted to be associated with the T. ni BBMV membrane in this study. Another proteomic study which examined T. ni cells selected in the presence of Cry1Ac toxin identified at least 30 differentially expressed enzymes/proteins; however, most of these were cytosolic or associated with cellular organelles (Gai et al., 2013). Comparison of BBMV proteins from resistant and susceptible insects may uncover additional B. thuringiensis toxin‐binding proteins.
Historically, insect lipases have been annotated according to their most closely related mammalian counterparts. Three enzymes involved in lipid digestion, including a bile salt‐activated lipase, a pancreatic triacylglycerol lipase and a group XV phospholipase A2, were associated with the BBMVs. Interestingly, the T. ni pancreatic lipase associated with the BBMV does not possess the cysteine‐rich C‐terminus domain, which is common to insect midgut pancreatic lipases that are associated with the PM where they may initiate digestion of dietary lipids (Simpson et al., 2007; Campbell et al, 2008; Toprak et al., 2015). One of the enzymes was classified as a bile salt‐activated lipase; however, insects do not produce bile salts (De Veau & Schultz, 1992). Rather, phospholipid surfactants, such as lysolecithin, may be generated by the hydrolysis of phospholipids by phospholipase A (Terra et al., 1994), which was also associated with the BBMVs.
Membrane‐bound alkaline phosphatases localized in the midgut of lepidopteran larvae have been well‐characterized and are used as larval midgut BBMV markers (Wolferberger et al., 1987; Abdul‐Rauf & Ellar, 1999). Similar to APN, midgut alkaline phosphatases are linked to the epithelial cell plasma membrane by a GPI anchor. The oligomeric form of B. thuringiensis delta‐endotoxin has particular affinity for N‐acetylgalactosamine residues on GPI‐anchored proteins (Pardo‐Lopez et al., 2006; Ning et al., 2010). This study identified two membrane alkaline phosphatases associated with the T. ni BBMV; both have a GPI anchor motif and are potential receptors for B. thuringiensis delta‐endotoxins.
Two isoforms of FE4 carboxylesterase were associated with the BBMV. In addition to their role in digestion, these enzymes are capable of cleaving ester bonds in other organic molecules, including certain classes of insecticides. Indeed, amplification of genes encoding insecticide detoxifying enzymes, namely glutathione‐S‐transferase, cytochrome P450 monooxygenases and esterases, is a major cause of insecticide resistance (Bass & Field, 2011). In Myzus persicae, gene amplification leading to increased expression of FE4 esterase and the closely related E4 esterase confers resistance of the insect to organophosphate insecticides (Field & Devonshire, 1998). While insecticide resistance arising from increased esterase activity has been commonly reported in hemipterans and dipterans, the presence of these enzymes in T. ni suggests that this mechanism may also available to lepidopteran pests.
Transporters
Nutrient transporters
The BBM is the principal site for nutrient acquisition and many of the proteins that were found to be associated with the T. ni BBMVs are likely involved in some aspect of nutrient molecule transport. According to the protein family database (Pfam; www.sangar.ac.uk/Software/Pfam/), the Drosophila melanogaster genome encodes 44 proteins with sugar transporter motifs. This study found 15 proteins associated with the T. ni BBMVs with similarity to the facilitated trehalose transporter 1 (TreT1). TreT1 transporters are highly specific for trehalose and do not recognize other disaccharides. Moreover, these transporters facilitate trehalose transport independent of membrane potential (Kikawada et al., 2007). Trehalose is the major hemolymph disaccharide sugar in most insects. It serves as a readily accessible energy source, such as during flight (Shulka et al., 2015), but is also used as a desiccation and cryoprotectant (Kikawada et al., 2007). Trehalose is synthesized in the insect fat body; however, it may also be present in the diet as plants use trehalose as an osmoprotectant in response to salt and drought stress (Grennan, 2007). The association of trehalose transporters with the T. ni midgut epithelial membrane indicate that both de novo synthesis and acquisition from external sources may contribute to the trehalose pool within the hemolymph. The hydrolysis of trehalose by trehalase to produce to two glucose molecules is an early and important regulatory step in the chitin biosynthetic pathway (Shen et al., 2017). While the midgut is not covered in a cuticle, it does secrete chitin, which forms a scaffold that supports the peritrophic matrix (Hegedus et al., 2009). Three membrane‐associated enzymes were identified in the BBMV fraction that are involved in some aspect of trehalose, chitin or cuticle metabolism; trehalase‐like isoform x1 (chitin synthesis), chitinase‐3‐like protein 2 (hydrolysis), and laccase‐1‐like isoform x2 (sclerotization).
Transporters for other carbohydrates, such as glucose, were also associated with the BBMV, as were five monocarboxylate transporters. The latter transporters are members of a family of multiple pass transmembrane proteins, which carry molecules with a single carboxylate group, for example pyruvate and lactate, across the plasma membrane in a proton‐dependent manner (Halestrap, 2012). Several amino acid transporters, as well as two long chain fatty acid transporters and a nucleoside transporter were also identified. Membrane transporters were also present to facilitate the acquisition of folate and thiamine from the diet, and a sodium‐dependent multi‐vitamin transporter may be capable of transporting a variety of vitamins and other cofactors, such as biotin, pantothenic acid and lipoic acid (Vadlapudi et al., 2012).
Ion transporters
The cell membrane is not permeable to most ions so their movement into or out of the cell requires specific transporter proteins. Seven proteins similar to organic cation transporters were associated with the T. ni BBMVs. Organic cationic transporters translocate cations, such as monoamines, coenzymes, and other molecules across the cell membrane (Koepsell et al., 2003). Many transporters capable of transporting inorganic cations were also identified. These included six zinc transporters, several potassium and/or calcium transporters and a bevy of copper and other metal transporters. The importance of phosphate is exemplified by the presence of seven inorganic phosphate transporters. Transporters for other anions were also present, including three band 3 anion transport proteins (solute carrier family 4, member 1), which exchange chloride with bicarbonate across plasma membranes. A bestrophin 4 protein was also identified that allows for calcium‐dependent transport of chloride ions (Milenkovic et al., 2008).
Four proteins were identified as being similar to the vacuolar (V‐type) proton ATPase. V‐type proton ATPases have previously been reported in midgut tissue from Heliothis virescens (Krishnamoorthy et al., 2007), H. armigera (Yuan et al., 2011), Bombyx mori (Kajiwara et al., 2005), and Manduca sexta (Pauchet et al., 2009). V‐type proton ATPases are localized on the apical surface of midgut cells and use energy from ATP hydrolysis to produce a proton gradient across the insect midgut epithelium, which facilitates the activity of other molecule transporters (Wieczorek et al., 1999). It is also possible that proton ATPases help to regulate gut pH as the midgut of T. ni larvae, like other lepidopterans, is highly basic (Braun & Keddie, 1997) and the digestive enzyme complement of lepidopteran larvae is most active at elevated pH (Hegedus et al., 2003). V‐type proton ATPase subunits in Heliothis virescens midgut were shown to interact with the B. thuringiensis toxin Cry1Ac (Krishnamoorthy et al., 2007); however, P. interpunctella larvae resistant to Cry1Ab toxin had higher levels of V‐ATPase subunit B in the midgut epithelium (Candas et al., 2003).
ATP‐binding cassette (ABC) and major facilitator superfamily (MFS) transporters
ABC and MFS transporters exhibit a wide range of specificities including polysaccharides, drugs, sugars, heavy metals, peptides, amino acids, and inorganic ions (Perlin et al., 2014). ABC transporters are the largest and most widely expressed family of transporters and hydrolyze ATP to energize the transport of molecules across the cell membrane (Leslie et al., 2005). Eighteen proteins associated with the T. ni BBMVs were members of the ABC family of transporters. Thirteen of the ABC transporters were similar to multidrug resistance associated proteins (MDRP), which have been implicated in the efflux of host plant phytoalexins in phytopathogens (Perlin et al., 2014). The number of such transporters in the T. ni midgut epithelium may be reflective of its cosmopolitan, oligophagous nature compared to insects with more restricted host specificities.
Other transporters
Five proteins identified in the BBMVs had similarity to, nose‐resistant to fluoxetine protein‐6 (NRF‐6). The NRF‐6 protein is a multipass membrane protein and has been best studied in the invertebrate model organism Caenorhabditis elegans where, among other functions, it is involved in the uptake of a range of molecules, including lipids and xenobiotic compounds from the intestine (Choy et al., 2006; Watts & Browse, 2006).
Three SID‐1‐like double stranded RNA (dsRNA) transporters were associated with the BBMVs. SID‐1 is required for the passive uptake of dsRNA from surrounding medium and for the movement of RNA interference signals between cells (Shih & Hunter, 2011). This is interesting since experiments to induce systemic RNAi in lepidopteran insects has met with limited success (Terenius et al., 2010), while feeding of dsRNA to larvae results in effective silencing of midgut genes (Toprak et al., 2013). It remains to be determined what might be the significance of dsRNA uptake by the midgut epithelium in insect biology and/or insect–host plant interactions.
This study identified two aquaporins that were associated with the BBMV. Aquaporins are membrane spanning proteins that form water channels through the cell membrane. They are an integral component of the system that maintains water homeostasis which is critical for the establishment of concentration gradients and osmotic balance. Aquaporins have also been implicated in freezing and desiccation tolerance in some insects (Cohen, 2012).
Proteins involved in cell signaling and development
The BBMV‐enriched fraction will contain some remnants of the columnar cell membrane, and several proteins involved in cell signaling, development or cell–cell association were identified and are described below. The midgut epithelium is constantly being sloughed and renewed. In addition, the cellular composition (columnar cells, goblet cells, and underlying regenerative cells) of the T. ni midgut changes along its length (Engelhard et al., 1991; Braun, 1996). As such, some of the proteins associated with the BBMV fraction were involved in aspects of signaling or cell–cell interaction related to cellular development and morphogenesis. A G‐protein coupled receptor (GPCR) 125 was identified that was highly similar to the human adhesion GPCR which stimulates tumor angiogenesis and is involved in other aspects of metazoan development (Weis & Cheresh, 2011). A BBMV GPCR similar to Methuselah (mth2) was identified which may regulate cell longevity in response to stress (InterPro: IPR010596); this would be an important consideration for midgut epithelial cell turnover. Notch, a cell surface transmembrane protein that regulates cellular events, such as differentiation, proliferation, and apoptosis (Guruharsha et al., 2012), was found in the BBMV enriched fraction. The Hedgehog signaling pathway is also important for determining cell fate and morphogenesis (Ingham et al., 2011). Hedgehog secretion is dependent upon the membrane protein Dispatched (Tukachinsky et al., 2012), which was found in the BBMV fraction, as was the Hedgehog receptor Patched. Independent of the Notch and Hedgehog signaling pathways, Fascilin 2, the neural cell‐adhesion molecule, specifically inhibits epidermal growth factor‐mediated signaling during development (Mao & Freeman, 2009). Three Fascilin 2 proteins were found in the BBMVs. In Drosophila, the semaphorin receptor Plexin A is involved in axon development and other aspects related to development (Xu et al., 2000). Plexin A and the Plexin A domain‐containing protein 2‐like, which has also been implicated in cell morphogenesis, were associated with the BBMVs. A rolling stone‐like transmembrane protein was also identified, which in Drosophila, is involved in myogenesis (Paululat et al., 1997).
Proteins involved in cell–cell interactions
Two integrin beta‐nu proteins and two integrin alpha isoforms were found in the BBMV fraction. Integrins are transmembrane proteins that connect the extracellular matrix to the cytoskeleton. In insects, integrin beta‐nu is required for migration of primordial midgut cells and for maintaining cell polarity in the midgut epithelium. It is also involved in maintaining endodermal integrity and adhesion of the midgut epithelium to the surrounding muscle (Devenport & Brown, 2004). A dystroglycan was also identified. Dystroglycan is the transmembrane component of the dystroglycan complex that, like integrins, links the extracellular matrix to the actin cytoskeleton (Adams & Brancaccio, 2015). Two cadherins were found in the BBMV fraction. Cadherins are calcium‐dependent cell adhesion transmembrane proteins. The extracellular domains interact to form “adherens” junctions which bind cells within tissues together, while the intracellular domain interacts with other partners to anchor the adherens to actin (Harris & Tepass, 2010). Binding of B. thuringiensis delta‐endotoxin to cadherin is thought to facilitate interaction with GPI‐anchored membrane proteins, such as APN and alkaline phosphatase (Pardo‐Lopez et al., 2006). A lachesin‐like isoform x1 protein was found in the BBMV fraction. Lachesins are another type of cell adhesion protein and in Drosophila regulate cell size and cell adhesion during morphogenesis (Llimargas et al., 2004). An innexin was found in the BBMV fraction. Tight association of cells is necessary not only for maintaining tissue integrity, but also to allow for cell‐to‐cell communication. In this regard, innexins form gap junctions between cells that permit the exchange of ions and small molecules (Bao et al., 2007).
Other BBMV proteins
Other proteins associated with the BBMV plasma membrane may be involved in some aspect of defense against enteric pathogens, these included two peptidoglycan‐recognition proteins, which are expressed in response to bacterial infections and induce the production of antimicrobial peptides, promote phagocytosis or act directly to hydrolyze peptidoglycan (Dziarski & Gupta, 2006). An isoform of Croquemort was associated with the BBMVs. Croquemort is a type of CD36 receptor which in macrophages is required for phagocytosis of apoptotic cells. Although it is not required for engulfment of bacteria, members of this family are part of the ancient innate immune system common to vertebrates and invertebrates (France et al., 1999). Two proteins similar to transmembrane protein 120 were associated with the BBMVs, which are described as also being involved in the innate immune response against Gram‐negative bacteria (www.uniprot.org/uniprot/Q9U1M2).
Discussion
The midgut epithelium's primary role is for the uptake and acquisition of nutrients from the lumen. The analysis of the T. ni BBM presented here identified several membrane‐associated enzymes involved in the terminal stages of macromolecule digestion, as well as a plethora of transporters needed to translocate small molecules across the epithelial cell membrane. The midgut epithelium is also the principal site for interaction with host‐derived anti‐nutritional molecules and toxins. Indeed, transporters were identified that have been implicated in the efflux and metabolism of host plant phytoalexins and other xenobiotics. Importantly, the midgut epithelium is a target for pathogens and several membrane proteins corresponding to known receptors for B. thuringiensis toxin (aminopeptidase‐N, cadherin, V‐type proton ATPase) were found. Finally, the insect virus per os infectivity (pif) complex is believed to interact with a ligand present on the epithelial cell membrane; however, this has yet to be identified for any baculovirus system. Should this model hold true, it is likely that the ligand(s) recognized by the pif complex is one of the membrane proteins described in this study. This is the most detailed proteomic analysis of the lepidopteran midgut epithelial cell membrane to date and will provide information and opportunities to better understand the biochemical, physiological, and pathological processes occurring in the larval midgut.
Supporting information
Supporting Information 1 BBMV protein sequences.
Supporting Information 2 BBMV protein nucleotide sequences.
Supporting Information 3 BBMV membrane protein sequences.
Supporting Information 4 BBMV membrane protein nucleotide sequences.
Table S1 T. ni BBMV proteins.
Table S2 Putative T. ni BBMV and epithelial membrane proteins.
Acknowledgments
These studies were funded by the Canadian Crop Genomics Initiative. MA Javed is the recipient of an NSERC Visiting Research Fellowship from the Government of Canada.
Disclosure
The authors have no conflict of interest.
The copyright line for this article was changed on 18 March 2019 after original online publication.
Contributor Information
Martin A. Erlandson, Email: martin.erlandson@agr.gc.ca.
Dwayne D. Hegedus, Email: dwayne.hegedus@agr.gc.ca.
References
- Abdul‐Rauf, M. and Ellar, D.J. (1999) Isolation and characterization of brush border membrane vesicles from whole Aedes aegypti larvae. Journal of Invertebrate Pathology, 73, 45–51. [DOI] [PubMed] [Google Scholar]
- Adams, J.C. and Brancaccio, A. (2015) The evolution of the dystroglycan complex, a major mediator of muscle integrity. Biology Open, 4, 1163–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, L. , Samuels, S. , Locovei, S. , MacAgno, E. , Muller, K. and Dahl, G. (2007) Innexins form two types of channels. FEBS Letters, 581, 5703–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, C. and Field, L.M. (2011) Gene amplification and insecticide resistance. Pest Management Science, 67, 886–890. [DOI] [PubMed] [Google Scholar]
- Bolognesi, R. , Terra, W. and Ferreira, C. (2008) Peritrophic membrane role in enhancing digestive efficiency. Journal of Insect Physiology, 54, 1413–1422. [DOI] [PubMed] [Google Scholar]
- Braun, L. (1996) A new tissue model for evaluating effects of Bacillus thuringiensis toxins on insect midgut epithelium. Ph.D. Thesis, University of Alberta, Canada. [DOI] [PubMed] [Google Scholar]
- Braun, L. and Keddie, B.A. (1997) A new tissue technique for evaluating effects of Bacillus thuringiensis toxins on insect midgut epithelium. Journal of Invertebrate Pathology, 69, 92–104. [DOI] [PubMed] [Google Scholar]
- Campbell, P.M. , Cao, A.T. , Hines, E.R. , East, P.D. and Gordon, K.H.J. (2008) Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera . Insect Biochemistry and Molecular Biology, 38, 950–958. [DOI] [PubMed] [Google Scholar]
- Candas, M. , Loseva, O. , Oppert, B. , Kosaraju, P. and Bulla, L.A. (2003) Insect resistance to Bacillus thuringiensis . Molecular Cellular Proteomics, 2, 19–28. [DOI] [PubMed] [Google Scholar]
- Chen, Y.R. , Zhong, S.L. , Fei, Z.J. , Gao, S. , Zhang, S.Y. , Li, Z.F. et al (2014) Transcriptome responses of the host Trichoplusia ni to infection by the baculovirus Autographa californica multiple nucleopolyhedrovirus. Journal of Virology, 88, 13781–13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, R.K.M. , Kemner, J.M. , Thomas, J.H. (2006) Fluoxetine‐resistance genes in Caenorhabditis elegans function in the intestine and may act in drug transport. Genetics, 172, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, E. (2012) Roles of aquaporins in osmoregulation, desiccation and cold hardiness in insects. Entomology, Ornithology and Herpertology, S1, 001 [Google Scholar]
- De Veau, E.J.I. and Schultz, J.C. (1992) Reassessment of interaction between gut detergents and tannins in Lepidoptera and significance for gypsy moth larvae. Journal of Chemical Ecology, 18, 1437–1453. [DOI] [PubMed] [Google Scholar]
- Devenport, D. and Brown, N.H. (2004) Morphogenesis in the absence of integrins: mutation of both Drosophila beta subunits prevents midgut migration. Development, 131, 5405–5415. [DOI] [PubMed] [Google Scholar]
- Dziarski, R. and Gupta, D. (2006) The peptidolycan recognition proteins (PGRPs). Genome Biology, 7, e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhard, E.K. , Keddie, B.A. and Volkman, L.E. (1991) Isolation of third, fourth, and fifth instar larval midgut epithelia of the moth, Trichoplusia ni . Tissue and Cell, 23, 917–928. [DOI] [PubMed] [Google Scholar]
- Field, L.M. and Devonshire, A.L. (1998) Evidence that the E4 and FE4 esterase genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer) are part of a gene family. Biochemical Journal, 330, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D. , Clements, J. , Arndt, W. , Miller, B.L. , Wheeler, T.J. , Schreiber, F. et al (2015) HMMER web server: 2015 update. Nucleic Acids Research Web Server, 43, W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc, N.C. , Heitzler, P. , Ezekowitz, R.A. and White, K. (1999) Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila . Science, 284, 1991–1994. [DOI] [PubMed] [Google Scholar]
- Harris, T.J. and Tepass, U. (2010) Adherens junctions: from molecules to morphogenesis. Nature Reviews in Molecular and Cell Biology, 11, 502–514. [DOI] [PubMed] [Google Scholar]
- Gai, Z.C. , Zhang, X.J. , Wang, X. , Peng, J.X. , Li, Y. , Liu, K.Y. et al (2013) Differential proteomic analysis of Trichoplusia ni cells after continuous selection with activated Cry1Ac toxin. Cytotechnology, 65, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczynski, S.F. and Adang, M.J. (1995) Bacillus thuringiensis Cry1Ac delta‐endotoxin binding aminopeptidase in the Manduca sexta midgut has a glycosylphosphatidylinositol anchor. Insect Biochemistry and Molecular Biology, 25, 409–415 [Google Scholar]
- Granados, R.R. and Lawler, K.A. (1981) In vivo pathway of Autographa californica baculovirus invasion and infection. Virology, 108, 297–308. [DOI] [PubMed] [Google Scholar]
- Grennan, A.K. (2007) The role of trehalose biosynthesis in plants. Plant Physiology, 144, 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha, K.G. , Kankel, M.W. and Artavanis‐Tsakonas, S. (2012) The Notch signalling system: recent insights into the complexity of a conserved pathway. Nature Reviews in Genetics, 13, 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap, A.P. (2012) The monocarboxylate transporter family—structure and functional characterization. IUBMB Life, 64, 1–9. [DOI] [PubMed] [Google Scholar]
- Haas‐Stapleton, E.J. , Washburn, J.O. and Volkman, L.E. (2004) P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion‐derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. Journal of Virology, 78, 6786–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus, D.D. , Baldwin, D. , O'Grady, M. , Braun, L. , Gleddie, S. , Sharpe, A. et al (2003) Midgut proteases from Mamestra configurata (Lepidoptera: Noctuidae) larvae: characterization, cDNA cloning and expressed sequence tag analysis. Archives of Insect Biocheistry and Physiology, 53, 30–47. [DOI] [PubMed] [Google Scholar]
- Hegedus, D.D. , Erlandson, M. , Gillott, C. and Toprak, U. (2009) New insights into peritrophic matrix synthesis, architecture and function. Annual Reviews of Entomology, 54, 285–302. [DOI] [PubMed] [Google Scholar]
- Horton, H.M. and Burand, J.P. (1993) Saturable attachment sites for polyhedron‐derived baculovirus on insect cells and evidence for entry via direct membrane fusion. Journal of Virology, 67, 1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham, P.W. , Nakano, Y. and Seger, C. (2011) Mechanisms and functions of Hedgehog signalling across the metazoa. Nature Reviews in Genetics, 12, 393–406. [DOI] [PubMed] [Google Scholar]
- Javed, M.A. , Harris, S. , Willis, L. , Theilmann, D. , Donly, C. , Hegedus, D.D. and Erlandson, M. (2016) Microscopic investigation of AcMNPV infection in the Trichoplusia ni midgut. Journal of Invertebrate Pathology, 141, 24–33. [DOI] [PubMed] [Google Scholar]
- Kajiwara, H. , Ito, Y. , Imamaki, A. , Nakamura, M. , Mita, K. and Ishizaka, M. (2005) Protein profile of silkworm midgut of fifth‐instar day‐3 larvae. Journal of Electrophoresis, 49, 61–69. [Google Scholar]
- Keddie, B.A. , Aponte, G.W. and Volkman, L.E. (1989) The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science, 243, 1728–1730. [DOI] [PubMed] [Google Scholar]
- Kikawada, T. , Saito, A. , Kanamori, Y. , Nakahara, Y. , Iwata, K. , Tanaka, D. et al (2007) Trehalose transporter 1, a facilitated and high‐capacity trehalose transporter, allows exogenous trehalose uptake into cells. Proceedings of the National Academy of Sciences USA, 104, 11585–11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel‐Morsdorf, D. , Flipsen, J.T. , Roncarati, R. , Jahnel, F. , Kleefsman, A.W. and Vlak, J.M. (1996) Baculovirus infection of Spodoptera exigua larvae: lacZ expression driven by promoters of early genes pe38 and me53 in larval tissue. Journal of General Virology, 77, 815–824. [DOI] [PubMed] [Google Scholar]
- Knight, P.J. , Knowles, B.H. and Ellar, D.J. (1994) The receptor for Bacillus thuringiensis CrylA(c) delta‐endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Molecular Microbiology, 11, 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsell, H. , Schmitt, B.M. and Gorboulev, V. (2003) Organic cation transporters. Reviews of Physiology, Biochemistry and Pharmacology, 150, 36–90. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy, M. , Jurat‐Fuentes, J.L. , McNall, R.J. , Andacht, T. and Adang, M.J. (2007) Identification of novel Cry1Ac binding proteins in midgut membranes from Heliothis virescens using proteomic analyses. Insect Biochemistry and Molecular Biology, 37, 189–201. [DOI] [PubMed] [Google Scholar]
- Lagarda‐Diaz, I. , Guzman‐Partida, A.M. , Huerta‐Ocampo, J.A. , Winzerling, J. and Vazquez‐Moreno, L. (2016) Identification of membrane proteins of the midgut of Zabrotes subfasciatus larvae associated with the insecticidal mechanism of PF2 lectin. Journal of Asia‐Pacific Entomology, 19, 677–682. [Google Scholar]
- Leslie, E.M. , Deeley, R.G. and Cole, S.P. (2005) Multidrug resistance proteins: role of P‐glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicology and Applied Pharmacology, 204, 216–237. [DOI] [PubMed] [Google Scholar]
- Lehane, M.J. and Billingsley, P.F. (1996) Biology of the Insect Midgut. Chapman and Hall, London. [Google Scholar]
- Linz, L.B. , Liu, S. , Chougule, N.P. and Bonning, B.C. (2015) In vitro evidence supports membrane alanyl aminopeptidase N as a receptor for a plant virus in the pea aphid vector. Journal of Virology, 89, 11203–11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llimargas, M. , Strigini, M. , Katidou, M. , Karagogeos, D. and Casanova, J. (2004) Lachesin is a component of a septate junction‐based mechanism that controls tube size and epithelial integrity in the Drosophila tracheal system. Development, 131, 181–190. [DOI] [PubMed] [Google Scholar]
- Ma, W.H. , Zhang, Z. , Peng, C.H. , Wang, X.P. , Li, F. and Lin, Y.J. (2012) Exploring the midgut transcriptome and brush border membrane vesicle proteome of the rice stem borer, Chilo suppressalis (Walker). PLoS ONE, 7, e38151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y. and Freeman, M. (2009) Fascilin 2, the Drosophila orthologue of neural cell‐adhesion molecule, inhibits EGF receptor signalling. Development, 136, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNall, R.J. and Adang, M.J. (2003) Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochemistry and Molecular Biology, 33, 999–1010. [DOI] [PubMed] [Google Scholar]
- Milenkovic, V.M. , Langmann, T. , Schreiber, R. , Kunzelmann, K. and Weber, B.H. (2008) Molecular evolution and functional divergence of the bestrophin protein family. BMC Evolutionary Biology, 8, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, C. , Wu, K. , Liu, C. , Gao, Y. , Jurat‐Fuentes, J.L. and Go, X. (2010) Characterization of a Cry1Ac toxin‐binding alkaline phosphatase in the midgut from Helicoverpa armigera (Hübner) larvae. Journal of Insect Physiology, 56, 666–672. [DOI] [PubMed] [Google Scholar]
- Pauchet, Y. , Muck, A. , Svatoš, A. and Heckel, D.G. (2009) Chromatographic and electrophoretic resolution of proteins and protein complexes from the larval midgut microvilli of Manduca sexta . Insect Biochemistry and Molecular Biology, 39, 467–474. [DOI] [PubMed] [Google Scholar]
- Paululat, A. , Goubeaud, A. , Damm, C. , Knirr, S. , Burchard, S. and Renkawitz‐Pohl, R. (1997) The mesodermal expression of rolling stone (rost) is essential for myoblast fusion in Drosophila and encodes a potential transmembrane protein. Journal of Cell Biology, 138, 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, C.E. , Warren, M.R. , Loiselle, D.R. , Dicheva, N.N. , Scarlett, C.O. and Borchers, C.H. (2005) Identification of components of protein complexes. Methods in Molecular Biology, 301, 117–151. [DOI] [PubMed] [Google Scholar]
- Pardo‐Lopez, L. , Gomez, I. , Rausell, C. , Sanchez, J. , Soberon, M. and Bravo, A. (2006) Structural changes of the Cry1Ac oligomeric pre‐pore from Bacillus thuringiensis induced by N‐acetylgalactosamine facilitates toxin membrane insertion. Biochemistry, 45, 10329–10336. [DOI] [PubMed] [Google Scholar]
- Peng, K. , van Lent, J.W.M. , Boeren, S. , Fang, M. , Theilmann, D.A. , Erlandson, M.A. et al (2013) Characterization of novel components of the baculovirus per os infectivity factor complex. Journal of Virology, 86, 4981–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin, M.H. , Andrews, J. and Toh, S.S. (2014) Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Advances in Genetics, 85, 201–253. [DOI] [PubMed] [Google Scholar]
- Popova‐Butler, A. and Dean, D.H. (2009) Proteomic analysis of the mosquito Aedes aegypti midgut brush border membrane vesicles. Journal of Insect Physiology, 55, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangadala, S. , Walters, F.S. , English, L.H. and Adang, M.J. (1994) A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)‐K+ efflux in vitro . Journal of Biological Chemistry, 269, 10088–10092. [PubMed] [Google Scholar]
- Shen, Q.D. , Yang, M.M. , Xie, G.Q. , Wang, H.J. , Zhang, L. , Qiu, L.Y. et al (2017) Excess trehalose and glucose affects chitin metabolism in brown planthopper (Nilaparvata lugens). Journal of Asia‐Pacific Entomology, 20, 449–455. [Google Scholar]
- Shih, J.D. and Hunter, C.P. (2011) SID‐1 is a dsRNA‐selective dsRNA‐gated channel. RNA, 17, 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulka, E. , Thorat, L.J. , Nath, B.B. and Gaikwad, S.M. (2015) Insect trehalase: physiological significance and potential applications. Glycobiology, 25, 357–367. [DOI] [PubMed] [Google Scholar]
- Simpson, R.M. , Newcomb, R.D. , Gatehouse, H.S. , Crowhurst, R.N. , Chagné, D. , Gatehouse, L.N. et al (2007) Expressed sequence tags from the midgut of Epiphyas postvittana (Walker) (Lepidoptera: Tortricidae). Insect Molecular Biology, 16, 675–690. [DOI] [PubMed] [Google Scholar]
- Sutherland, D.W.S. and Greene, G.L. (1984) Cultivated and wild host plants Suppression and Management of Cabbage Looper Populations (eds. Lingren P.D. & Greene G.L.), pp. 1–13. U.S. Department of Agriculture, Technical Bulletin no. 1684, U.S.A. [Google Scholar]
- Terenius, O. , Papanicolaou, A. , Garbutt, J.S. , Eleftherianos, I. , Huvenne, H. , Kanginakudru, S. et al (2010) RNA interference in Lepidoptera—successes and failures. Insect Biochemistry and Molecular Biology, 57, 231–245. [Google Scholar]
- Terra, W.R , Ferreira, C. , Jordao, B.P. and Dillon, R.J. (1994) Digestive enzymes Biology of the Insect Midgut (eds. Lehane M.J. & Billingsely P.F.). Chapman and Hall, London. [Google Scholar]
- Toprak, U. , Erlandson, M. , Baldwin, D. , Karcz, S. , Wan, L. , Coutu, C. et al (2015) Identification of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix proteins and enzymes involved in peritrophic matrix chitin metabolism. Insect Science, 23, 656–674. [DOI] [PubMed] [Google Scholar]
- Toprak, U. , Baldwin, D. , Erlandson, M. , Gillott, C. , Harris, S. and Hegedus, D.D. (2013) In vitro and in vivo application of RNA interference for targeting genes involved in peritrophic matrix synthesis in a lepidopteran system. Insect Science, 20, 92–100. [DOI] [PubMed] [Google Scholar]
- Tukachinsky, H. , Kuzmickas, R.P. , Jao, C.Y. , Liu, J. and Salic, A. (2012) Dispatched and Scube mediate the efficient secretion of the cholesterol‐modified Hedgehog ligand. Cell, 30, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlapudi, A.D. , Vadlapatla, R.K. and Mitra, A.K. (2012) Sodium dependent multivitamin transporter (SMVT): a potential target for drug delivery. Current Drug Targets, 13, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vail, P.V. , Anderson, S.J. and Jay, D.L. (1973) New procedures for rearing cabbage loopers and other lepidopterous larvae for progagation of nuclear polyhedrosis viruses. Environmental Entomology, 2, 339–334. [Google Scholar]
- Wang, P. , Zhang, X. and Zhang, J. (2005) Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni. Insect Biochemistry and Molecular Biology, 35, 611–620. [DOI] [PubMed] [Google Scholar]
- Watts, J.L. and Browse, J. (2006) Dietary manipulation implicates lipid signaling in the regulation of germ cell maintenance in C. elegans . Developmental Biology, 292, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, S.M. and Cheresh, D.A. (2011) Tumor angiogenesis: molecular pathways and therapeutic targets. Nature Medicine, 17, 1359–1370. [DOI] [PubMed] [Google Scholar]
- Wieczorek, H. , Grube, G. , Harvey, W.R. , Huss, M. and Merzendorfer, H. (1999) The plasma membrane H+‐V‐ATPase from tobacco hornworm midgut. Journal of Bioenergetics and Biomembranes, 31, 67–74. [DOI] [PubMed] [Google Scholar]
- Wolfersberger, M. , Luethy, P. , Maurer, A. , Parenti, P. , Sacchi, F.V. , Giordana, B. et al (1987) Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comparative Biochemistry and Physiology, 86, 301–308. [Google Scholar]
- Xu, X.M. , Fisher, D.A. , Zhou, L. , White, F.A. , Ng, S. , Snider, W.D. et al (2000) The transmembrane protein Semaphorin 6A repels embryonic sympathetic axons. Journal of Neuroscience, 20, 2638–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachdav, G. , Kloppmann, E. , Kajan, L. , Hecht, M. , Goldberg, T. , Hamp, T. et al (2014) PredictProtein: an open resource for online prediction of protein structural and functional features. Nucleic Acids Research, 42, W337–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C. , Ding, X.Z. , Xia, L.Q. , Yin, J. , Huang, S.Y. and Huang, F. (2011) Proteomic analysis of BBMV in Helicoverpa armigera midgut with and without Cry1Ac toxin treatment. Biocontrol Science and Technology, 21, 139–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1 BBMV protein sequences.
Supporting Information 2 BBMV protein nucleotide sequences.
Supporting Information 3 BBMV membrane protein sequences.
Supporting Information 4 BBMV membrane protein nucleotide sequences.
Table S1 T. ni BBMV proteins.
Table S2 Putative T. ni BBMV and epithelial membrane proteins.


