Summary
Pf filamentous prophages are prevalent among clinical and environmental Pseudomonas aeruginosa isolates. Pf4 and Pf5 prophages are integrated into the host genomes of PAO1 and PA14, respectively, and play an important role in biofilm development. However, the genetic factors that directly control the lysis‐lysogeny switch in Pf prophages remain unclear. Here, we identified and characterized the excisionase genes in Pf4 and Pf5 (named xisF4 and xisF5, respectively). XisF4 and XisF5 represent two major subfamilies of functional excisionases and are commonly found in Pf prophages. While both of them can significantly promote prophage excision, only XisF5 is essential for Pf5 excision. XisF4 activates Pf4 phage replication by upregulating the phage initiator gene (PA0727). In addition, xisF4 and the neighboring phage repressor c gene pf4r are transcribed divergently and their 5′‐untranslated regions overlap. XisF4 and Pf4r not only auto‐activate their own expression but also repress each other. Furthermore, two H‐NS family proteins, MvaT and MvaU, coordinately repress Pf4 production by directly repressing xisF4. Collectively, we reveal that Pf prophage excisionases cooperate in controlling lysogeny and phage production.
A proposed model of the lysis‐lysogeny switch of Pf4 in PAO1. Integration of Pf4 in host genome is maintained by MvaT/MvaU by binding to the excisionase gene xisF4. Inactivation of MvaT/MvaU upregulates xisF4 and subsequently, XisF4 activates the replication initiator gene PA0727, leading to Pf4 phage replication and release.
![]()
Introduction
Filamentous phages, among the simplest biological entities known, were discovered over a half‐century ago. Profoundly different from tailed double‐stranded DNA (dsDNA) phages, filamentous phages feature long, thin filaments and a small, circular single‐stranded DNA (ssDNA) genome (Mai‐Prochnow et al., 2015).The Ff filamentous phages which infect conjugative Escherichia coli strains, including f1, fd and M13, were isolated from sewage systems in the early 1960s (Loeb, 1960; Hofschneider and Preuss, 1963; Marvin and Hoffmann‐Berling, 1963). The Pseudomonas aeruginosa phage Pf1 was described in 1966 and is twice the size of Ff phages (Takeya and Amako, 1966). Subsequently, these phages have been found in a variety of Gram‐negative bacteria and occasionally in Gram‐positive bacteria in diverse habitats (Mai‐Prochnow et al., 2015). Rather than kill host bacteria, filamentous phages generally become lysogens, with the phage genome either being integrated into the bacterial chromosome or remaining extrachromosomal as episomes (Mai‐Prochnow et al., 2015). Increasing evidence demonstrates that they can strongly affect host physiology and virulence expression as lysogens. For example, the filamentous phage CTXφ in Vibrio cholera carries genes encoding the cholera toxin and plays a critical role in the conversion of nontoxigenic strains into pathogens (Waldor and Mekalanos, 1996; Davis et al., 2002). Another filamentous phage, VPIφ in V. cholera, encodes the structural gene for a toxin‐co‐regulated pilus, which is not only the receptor for CTXφ but also a colonization factor (Li et al., 2003). The filamentous phage φRSM3 in Ralstonia solanacearum encodes a transcriptional regulator that represses the expression of the host virulence genes (Addy et al., 2012).
Pf1‐like filamentous phages are prevalent among clinical and environmental P. aeruginosa isolates, and more than half of the 241 clinical isolates were found to harbor at least one Pf1‐like genetic element (Knezevic et al., 2015). The Pf prophage Pf1 infects P. aeruginosa strain K (PAK) without integrating into the host genome and replicates exclusively as an episome. In contrast, Pf4 and Pf5 are integrated into the host genomes of PAO1 and PA14, respectively, and maintained as prophages (Mooij et al., 2007). During P. aeruginosa biofilm development, Pf1‐like genes are among the most strongly induced genes (Whiteley et al., 2001). Pf4 in PAO1 plays an essential role in biofilm development and the structural integrity of the biofilm (Rice et al., 2009). During biofilm formation, Pf4 can develop into mature virus particles and then can convert into a superinfective form that is correlated with cell death and the appearance of small‐colony variants (SCVs) (Webb et al., 2004). Pf4 also contributes to the virulence of PAO1, as shown by the increased survival of the strain without Pf4 using a mice infection model (Rice et al., 2009). More specifically, Pf4 can promote bacterial adhesion to mucin, alter progression of the inflammatory response, and contribute to noninvasive infection in a murine pneumonia model (Secor et al., 2017). Furthermore, the production of Pf4 phage during PAO1 biofilm development is found to be associated with the formation of highly ordered liquid crystals, thus promoting the pathogenic features of biofilms (Secor et al., 2015). In addition, inactivation of Pf5 genes increases biofilm formation in PA14 (Lee et al., 2018). It has become clear that a mutualistic relationship has developed between P. aeruginosa and the Pf prophages.
Phage integration is a crucial step for lysogeny, while prophage excision is a critical step for phage production (Feiner et al., 2015). For tailed dsDNA phages, prophage excision requires integrase and additional recombinase activity normally conferred by a phage‐encoded recombination directionality factor (RDF) or excisionase (Bertani and Bertani, 1971; Gottesman, 1974; Couturier, 1976). Filamentous phages have smaller genomes than tailed dsDNA phages, and many of them do not encode their own recombinases. In some cases, host‐encoded recombinases are needed for prophage excision. For example, excision of filamentous prophages CTXφ and VGJφ in Vibrio cholera and XacF1 in Xanthomonas axonopodis are mediated by the host‐encoded recombinases XerC/D (Huber and Waldor, 2002; Das et al., 2011; Ahmad et al., 2014). For Pf prophages, nonintegrated Pf1 prophage carries a truncated integrase but Pf4 and Pf5 prophages both carry intact integrases. However, it remains unknown whether Pf prophages encode their own excisionases or use the host recombinase to mediate excision.
Prophages can maintain two distinct life forms: a lytic cycle and a lysogenic cycle. Although in most cases, the lysogenic state is relatively stable, alterations in host cell physiology may lead to the lytic cycle which initiates phage production and/or host cell lysis (Ofir and Sorek, 2018). The best‐studied lysis‐lysogeny conversion is between lambda prophage and its E. coli host, and this conversion is regulated by host‐encoded proteins RecA and LexA under the SOS response (Little and Mount, 1982). In recent years, several studies showed that several prophages induced during biofilm development are specifically regulated by the host‐encoded histone‐like nucleoid structuring (H‐NS) family proteins (Wang et al., 2009; Hong et al., 2010; Liu et al., 2015; Zeng et al., 2016). H‐NS family proteins are shown to bind to intrinsically curved DNA to regulate the expression and excision of mobile genetic elements (Dorman, 2007; Singh and Grainger, 2013). For example, in Salmonella enterica, H‐NS binds to AT‐rich sequences and silences the Salmonella pathogenicity islands (Navarre et al., 2006). In E. coli, H‐NS binds to the promoter region of the tra operon of the conjugative F plasmid, which encodes components of the transfer apparatus, resulting in the repression of plasmid transfer (Will and Frost, 2006). We found that CP4‐57 and rac prophages in E. coli K‐12 are excised during E. coli biofilm development and that these two prophages are not regulated by the SOS response (Wang et al., 2010). Indeed, excision of the prophage rac is controlled by H‐NS through its binding to the rac excisionase gene xisR (Hong et al., 2010; Liu et al., 2015), while excision of the P4‐family prophage CP4‐57 in E. coli is controlled by another DNA binding protein, Hha (Wang et al., 2009), a protein known to interact with H‐NS (Nieto et al., 2002) and recently shown to behave as a toxin with antitoxin TomB (Marimon et al., 2016). We recently reported that the host‐encoded H‐NS represses CP4So prophage excision in Shewanella oneidensis by binding to the excisionase gene alpA at warm temperatures, while the de‐repression of alpA by H‐NS induces prophage excision to increase host fitness at cold temperatures (Zeng et al., 2016). In PAO1, two H‐NS family proteins, MvaT and MvaU, function coordinately as xenogeneic silencers and control the activation of Pf4 (Castang et al., 2008; Li et al., 2009). A previous ChIP‐chip assay also suggested that MvaT and MvaU bind to the region upstream of the Pf4 structural genes (Castang and Dove, 2012). However, the underlying mechanism of action of the H‐NS family proteins on Pf prophages remains elusive.
In this study, we identified a new ORF (xisF4) in the prophage Pf4 that encodes a functional excisionase. This is the first report that demonstrates that filamentous prophages carry their own excisionases. Excision of the prophage Pf4 increased 104‐fold upon overexpression of xisF4, and deletion of xisF4 abolished the ability of Pf4 to excise from the host chromosome. XisF4 also activates the replication of phage production by upregulating the recently reported replication initiator gene (PA0727). In addition, MvaT and MvaU coordinately repress Pf4 production by directly repressing the promoter activity of xisF4. Similarly, the excisionase XisF5 in the prophage Pf5 also induces excision of Pf5 in PA14. Taken together, we demonstrate that Pf prophages can encode their own excisionase and that host factors control Pf excision and production through the regulation of this excisionase gene.
Results
Identification of excisionase genes in the prophages Pf4 and Pf5
Unlike Pf1 in strain PAK, Pf4 and Pf5 integrate as prophages in the genomes of PAO1 and PA14, respectively. In PAO1, Pf4 is integrated between PA0714 (encoding a hypothetical protein) and PA0729.1 (encoding tRNA‐gly) (Webb et al., 2004). In PA14, Pf5 is integrated into a different site, between PA14_49040 and PA14_48870 (Mooij et al., 2007). Comparative genomic analysis shows that a majority of the genes encoding structural proteins of the filamentous phages (PA0717‐PA0726) share higher sequence identity (> 90%) than the rest of the regions, except for PA0724 which encodes the minor coat protein (~50%) (Fig. 1). Downstream of the region coding for the structural genes is PA0727, which was recently found to be responsible for phage replication (Martínez and Campos‐Gómez, 2016). Moreover, PA0727 also shares high sequence identity among Pf1, Pf4 and Pf5, and it encodes the phage replication initiator protein in PAO1 (Fig. 1). Downstream of PA0727 is PA0728 (renamed as intF4) which encodes a putative integrase; however, a truncated integrase gene (integrase in Pf1 is 100 aa and the integrase in Pf4 is 328 aa) is present in the phage Pf1.
Figure 1.
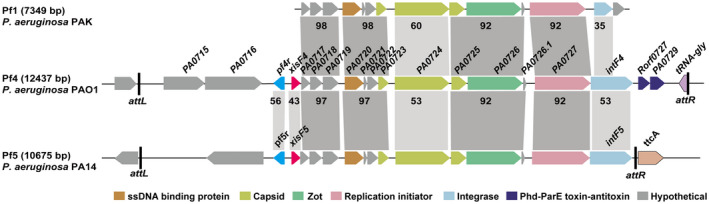
Sequence alignment of prophage genes in Pf1, Pf4 and Pf5. Names of the Pf4 genes in PAO1 are shown to scale. The reannotated genes xisF4 and pf4r in PAO1 and xisF5 and pf5r in PA14 are also shown here. attL and attR represent the left and right attachment sites, respectively. Numbers in the dark gray shaded regions represent the percentage of DNA sequence identity between genes (> 90%), and the numbers in the light gray shaded regions represent the percentage of amino acid sequence identity between proteins (40~60%).
We analyzed the genomic region between PA0716 and PA0717. Although this region is absent in the phage Pf1, moderate sequence identity was found in a region of ~600 bp next to PA0717 in Pf4 and Pf5 prophages. Previous analysis by the Kjelleberg group suggested that this region contained an ORF encoding putative regulatory proteins of 88 aa with 42% sequence identity with the repressor C of the phage P2 (Webb et al., 2004). Thus, we proposed to name the putative repressor protein in the prophage Pf4 of PAO1 as Pf4r (Pf4 repressor C) and in the prophage Pf5 in PA14 as Pf5r (Pf5 repressor C PA14_49020). Pf4r and Pf5r share 56% sequence identity. Between pf4r and PA0717, we found a putative ORF of 216 bp which encodes a protein of 71 aa. At a similar genomic location in the prophage Pf5, we found an ORF sharing 43.5% sequence identity with the one in Pf4. Thus, we named these ORFs as xisF4 in Pf4 (Pf4 excisionase) and xisF5 in Pf5 (Pf5 excisionase PA14_49010). XisF4 has a HTH DNA binding domain predicted by a BlastP search in the NCBI and IMG database, and shares 29% identity with the Xis of mycobacteriophage Pukovnik by structural similarity search using Phyre2 (Kelley et al., 2015) (Fig. S1). Xis is required for Pukovnik prophage excision (Singh et al., 2014), thus we hypothesized that XisF4 may function as an excisionase of Pf4.
XisF4 and XisF5 promote prophage excision
As far as we know, no prophage‐encoded excisionase have been characterized in filamentous prophages; thus, we first checked whether these newly identified genes encode functional excisionases. We cloned the predicted coding region of xisF4 into pHERD20T to overexpress xisF4 using a PBADinducible promoter in the PAO1 wild‐type and the ΔxisF4 strains. qPCR using a forward primer (Pf4‐f) flanking the left attachment site and a reverse primer (Pf4‐r) flanking the right attachment site of the prophage Pf4 were employed to quantify the proportion of cells with Pf4 excised from the host chromosome (Fig. 2A). The frequency of Pf4 excision was very low in the PAO1 wild‐type during planktonic growth (approximately one out of 106 cells), suggesting that the prophage Pf4 resides stably in the host genome (Fig. 2B). The frequency of Pf4 excision in PAO1 increased approximately 105‐fold when xisF4 was overexpressed via pHERD20T‐xisF4 in the PAO1 wild‐type strain, reaching up to ~1%. As expected, excision of Pf4 was undetectable (less than one out of 106 cells) in the ΔxisF4 strain, and it was restored by overexpressing xisF4 (Fig. 2B). These results show that XisF4 can promote prophage excision. To confirm whether XisF4 is required for Pf4 excision, the frequency of Pf4 excision was also detected in the ΔxisF4 strain when the integrase IntF4 was overproduced. Although the ability of IntF4 to induce Pf4 excision in the ΔxisF4 is lower than that of XisF4 under similar conditions, the frequency of Pf4 excision was similar in the PAO1 wild‐type strain and in the ΔxisF4 strain when IntF4 was overproduced, suggesting that XisF4 is not required for Pf4 excision. Furthermore, similar results were obtained using PCR assays to detect Pf4 excision using a different pair of primers (Fig. S2). To further check the excisionase activity of XisF4, we performed electrophoretic mobility shift (EMSA) assays. As shown in Fig. 2C, XisF4 binds and shifts the attachment site of Pf4 prophage, and the presence of XisF4 also enhanced the binding of IntF4 to the attachment site. As a negative control, XisF4 could not bind or shift the bacterial attachment site (attB) once Pf4 is excised (Fig. 2D). These results show that XisF4 functions as an excisionase and can promote Pf4 excision. However, XisF4 is non‐essential for Pf4 excision as the integrase can still induce Pf4 excision in the absence of XisF4.
Figure 2.
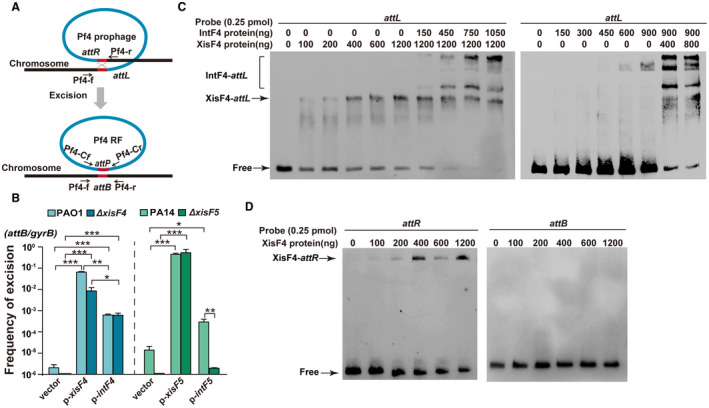
Integrase and excisionase in Pf4 and Pf5 promote prophage excision. A. A schematic diagram illustrates the excision of the prophage Pf4 from the PAO1 host chromosome and the formation of Pf4 replicative form (RF) molecules in the cytoplasm of PAO1. B. The frequency of Pf4 excision was quantified in PAO1 and ΔxisF4 overexpressing intF4 or xisF4 via pHERD20T‐based plasmids, and the frequency of Pf5 excision was quantified in PA14 and ΔxisF5 overexpressing intF5 or xisF5 via pHERD20T‐based plasmids. Three independent cultures of each strain were used, and error bars indicate standard deviation. Unpaired two‐tailed Student’s t tests were used for statistical analysis; p < 0.05 is marked *, p < 0.01 is marked **, and p < 0.001 is marked *** throughout the study. C. EMSA showed that XisF4 and IntF4 bound to attL in a concentration‐dependent manner. D. EMSA showed that XisF4 bound to attR in a concentration‐dependent manner but not to attB.
Next, we also investigated the function of the putative excisionase gene xisF5 and the putative integrase gene intF5 (PA14_48880) in Pf5 prophage in the PA14 wild‐type strain. As expected, the frequency of Pf5 excision greatly increased (approximately 105‐fold) when xisF5 was overexpressed via pHERD20T‐xisF5 in the PA14 wild‐type strain (Fig. 2B). In addition, excision of Pf5 was undetectable in the ΔxisF5 strain, and Pf5 excision can be restored by overexpressing xisF5 in the ΔxisF5 strain (Fig. 2B). To check whether XisF5 is required for Pf5 excision, the frequency of Pf5 excision was also detected in the ΔxisF5 strain when the integrase was overproduced. Different from Pf4, no Pf5 excision was detected when the Pf5 integrase gene intF5 was overexpressed in the ΔxisF5 strain. Additionally, Pf5 excision was also undetectable in the ΔintF5 strain even when xisF5 was overexpressed (Fig. S3A). Furthermore, as expected, XisF5 and IntF5 can bind and shift attL of Pf5 as shown by EMSA (Fig. S3B). Taken together, we demonstrate that both excisionase and integrase in Pf prophages can promote prophage excision, and XisF5 is essential for Pf5 excision while XisF4 is non‐essential for Pf4 excision.
XisF4 and XisF5 activate phage replication
We observed that overexpressing xisF4 in the PAO1 wild‐type strain resulted in a severe growth inhibition. To test whether the growth inhibition is dependent on the presence of Pf4 prophage, we constructed a Pf4 deletion mutant strain (ΔPf4) in which the whole prophage was removed from PAO1 (Table 1, Fig. S5). As expected, growth inhibition was no longer detected when xisF4 was overexpressed in the ΔPf4 strain (Fig. 3A). It has been shown that some excisionases such as Cox protein of P2 phage can function as a transcriptional regulator in addition to as an excisionase or RDF (Saha et al., 1989; Yu and Haggard‐Ljungquist, 1993). Thus, we tested whether XisF4 can regulate the production of Pf4 phage particles. Active Pf4 phage particles produced by PAO1/pHERD20T‐xisF4 were quantified by applying the supernatant collected from this strain onto the bacterial lawn formed by the ΔPf4 strain. Notably, Pf4 phage particles increased approximately 106‐fold when xisF4 was overexpressed, reaching up to 1011 PFU ml–1 (Fig. 3B), indicating that XisF4 increases phage production. Replication of filamentous phages involves the generation of double‐stranded DNA known as replicative form (RF) molecules (Marvin, 1998). Thus, we further explored whether XisF4 can promote the formation of Pf4 RF using qPCR. An inner forward primer near the left attachment site (Pf4‐Cr) and an inner reverse primer near the right attachment site (Pf4‐Cf) of the prophage Pf4 were then employed to quantify the number of Pf4 RF molecules in the mixed population using qPCR (Fig. 3C). Overexpressing xisF4 via pHERD20T‐xisF4 in the PAO1 wild‐type strain greatly increased the number of Pf4 RF molecules by approximately 104‐fold, while Pf4 RF was undetectable in the ΔxisF4 strain (Fig. 3C). In contrast, although IntF4 can also increase prophage excision, overproducing IntF4 via pHERD20T‐intF4 did not affect the number of Pf4 RF (Fig. 3C). Similar results were obtained for xisF5 in Pf5 prophage in PA14 strain. Overexpressing xisF5 via pHERD20T‐xisF5 in the PA14 wild‐type strain greatly increased the number of Pf5 RF molecules by approximately 104‐fold, while overexpressing intF5 did not affect the number of Pf5 RF (Fig. S4).
Table 1.
Bacterial strains and plasmids used in this study.
| 10 | Description | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F– φ80lacZ∆M15 ∆(lacZYA‐argF)U169 recA1 endA1 hsdR17(rk–, mk+)phoA supE44 thi‐1 gyrA96 relA1 tonA | Novagen |
| K‐12 BW25113 | lacI q rrnB T14 ΔlacZ WJ16 hsdR514 ΔaraBAD AH33 ΔrhaBAD LD78 | Baba et al. (2006) |
| BL21(DE3) | F– ompT hsdSB(rB–mB–) gal dcm λ(DE3) Ω PtacUV5::T7 polymerase | Novagen |
| P. aeruginosa | ||
| PAO1 | Wild type | Stover et al. (2000) |
| Δpf4r | pf4r deletion mutant derived from PAO1 | This study |
| ΔxisF4 | xisF4 deletion mutant derived from PAO1 | This study |
| ΔPf4 | whole Pf4 prophage removed from PAO1 host chromosome | This study |
| PA14 | Wild type | Liberati et al. (2006) |
| ΔxisF5 | xisF5 deletion mutant derived from PA14 | This study |
| ΔintF5 | intF5 deletion mutant derived from PA14 | This study |
| ΔmvaT | mvaT deletion mutant derived from PAO1 | This study |
| ΔmvaU | mvaU deletion mutant derived from PAO1 | This study |
| ΔmvaUΔmvaT | mvaT and mvaU double deletion mutant derived from PAO1 | This study |
| PAO1::Ppf4r‐lacZ | LacZ reporter strain | This study |
| ΔPf4::Ppf4r‐lacZ | LacZ reporter strain | This study |
| PAO1:: PxisF4‐lacZ | LacZ reporter strain | This study |
| ΔPf4:: PxisF4‐lacZ | LacZ reporter strain | This study |
| PAO1::PPA0720‐lacZ | LacZ reporter strain | This study |
| PAO1::PPA0724‐lacZ | LacZ reporter strain | This study |
| PAO1::PPA0727‐lacZ | LacZ reporter strain | This study |
| Plasmids | ||
| pHERD20T | ApR, expression vector with araC‐PBAD promoter | Qiu et al., (2008) |
| pHERD20T‐xisF4 | ApR, xisF4 in pHERD20T EcoRI/HindIII | This study |
| pHERD20T‐pf4r | ApR, pf4r in pHERD20T EcoRI/HindIII | This study |
| pHERD20T‐intF4 | ApR, intF4 in pHERD20T EcoRI/XbaI | This study |
| pHERD20T‐xisF5 | ApR, xisF5 in pHERD20T EcoRI/HindIII | This study |
| pHERD20T‐pf5r | ApR, pf5r in pHERD20T EcoRI/HindIII | This study |
| pHERD20T‐intF5 | ApR, intF5 in pHERD20T EcoRI/XbaI | This study |
| pET28b | KmR, expression vector | Novagen |
| pET28b‐intF4 | KmR, intF4 in pET28b NcoI/HindIII | This study |
| pET28b‐xisF5 | KmR, xisF5 in pET28b NcoI/HindIII | This study |
| pET28b‐intF5 | KmR, intF5 in pET28b NcoI/HindIII | This study |
| pEX18AP | ApR, oriT +, sacB +, gene replacement vector | Hoang et al. (1998) |
| pFLP2 | ApR, Flp recombinase‐expressing plasmid | Hoang et al. (1998) |
| pPS856 | ApR, GmR; for amplifying gentamycin resistance cassette | Hoang et al. (1998) |
| pEX18AP‐pf4r‐up‐GM‐down | GmR, CarR, for deleting pf4r | This study |
| pEX18AP‐xisF4‐up‐GM‐down | GmR, CarR, for deleting xisF4 | This study |
| pEX18AP‐Pf4‐up‐GM‐down | GmR, CarR, for deleting Pf4 | This study |
| pEX18Ap‐mvaT‐up‐GM‐down | GmR, CarR, for deleting mvaT | This study |
| pEX18Ap‐mvaU‐up‐GM‐down | GmR, CarR, for deleting mvaU | This study |
| mini‐CTX‐LacZ | TetR, integration vector for single‐copy, chromosomal lacZ fusions; Ω‐FRT‐attP‐MCS, ori, int, and oriT | Becher and Schweizer (2000) |
| pCTX‐Ppf4r‐lacZ | TetR, −313 bp relative to translational start site of pf4r cloned into mini‐CTX‐lacZ | This study |
| pCTX‐ PxisF4‐lacZ | TetR, −300 bp relative to translational start site of xisF4 cloned into mini‐CTX‐lacZ | This study |
| pCTX‐PPA0720‐lacZ | TetR, −345 bp relative to translational start site of PA0720 cloned into mini‐CTX‐lacZ | This study |
| pCTX‐PPA0724‐lacZ | TetR, −334 bp relative to translational start site of PA0724 cloned into mini‐CTX‐lacZ | This study |
| pCTX‐PPA0727‐lacZ | TetR, −360 bp relative to translational start site of PA0727 cloned into mini‐CTX‐lacZ | This study |
Figure 3.
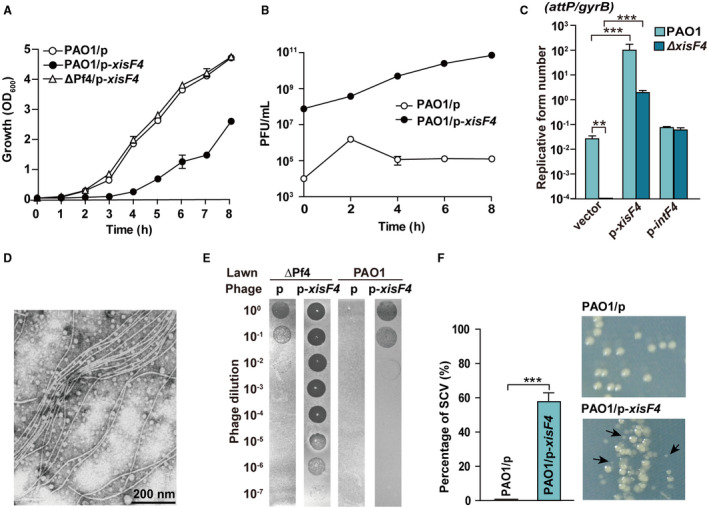
XisF4 activates Pf4 production. A. Growth (OD600) was tested in planktonically growing PAO1 carrying pHERD20T (p), PAO1 carrying pHERD20T‐xisF4 (p‐xisF4) and ΔPf4 carrying pHERD20T‐xisF4; 10 mM arabinose was added at the beginning. B. Pf4 phage titers (PFU ml–1) were quantified on ΔPf4 lawns using supernatant from PAO1 carrying pHERD20T or pHERD20T‐xisF4 under the same condition as shown in A. C. The number of Pf4 RF molecules were quantified in PAO1 and ΔxisF4 overexpressing intF4 or xisF4. D. Transmission electron microscopy (TEM) of Pf4 phage particles collected from the supernatant of PAO1 carrying pHERD20T‐xisF4 at 4 h as shown in B. E. Plaque formation by the phage lysates at 4 h as shown in B. Phage lysates were serially diluted and 10 μl samples were dropped on PAO1 and ΔPf4 lawns, respectively. F. The percentage of SCVs (relatively small colonies formed) was calculated from PAO1 carrying pHERD20T or pHERD20T‐xisF4 at 4 h as shown in B. Three independent cultures of each strain were used, and error bars indicate standard deviation.
A large number of filamentous phage particles were also observed using electron microscopy in the supernatant of PAO1 when xisF4 was overexpressed (Fig. 3D). In addition, we found that high titers of Pf4 (> 108 PFU ml–1) can not only infect the ΔPf4 strain but can also re‐infect the PAO1 wild‐type strain that contains an integrated copy of the same phage (Fig. 3E). Previous work showed, by using the flow‐cell biofilm assay, that the formation of small colony variants (SCVs) was induced at the late stage of PAO1 biofilm development (Rice et al., 2009). Here, the occurrence of SCVs increased up to 55% when xisF4 was overexpressed via PAO1/pHERD20T‐xisF4, while SCVs were rarely detected in PAO1/pHERD20T in planktonic growth conditions (Fig. 3F). These results collectively demonstrate that XisF4 and XisF5 can activate phage replication.
XisF4 promotes Pf4 replication by activating PA0727
To explore how XisF4 regulates phage production, we first performed qRT‐PCR to check the expression of Pf4 genes by overexpressing xisF4 via pHERD20T‐xisF4 with 10 mM arabinose for 30 min. The structural genes conserved in Pf4 and Pf5 (from PA0717 to PA0726) were all upregulated by 10~20‐fold when xisF4 was overexpressed (Fig. 4AB). In addition, PA0727, which encodes the replication initiator protein, was also highly induced. In contrast, the genes upstream and downstream of PA0717‐intF4 were only slightly induced (< 2‐fold), likely due to the presence of more copies of Pf4 RF (Fig. 4B). These results suggest that genes encoding phage structural gene and phage replication initiator were specifically activated when xisF4 was overexpressed. To gain further insights into the regulation of XisF4 on Pf4 genes, four different chromosomal lacZ transcriptional fusions (PxisF4‐lacZ, PPA0720‐lacZ, PPA0724‐lacZ and PPA0727‐lacZ) (Table 1) were constructed to test the promoter activity of the four operons in vivo by measuring β‐galactosidase activity. When xisF4 was overexpressed in PAO1 with the addition of 10 mM arabinose (at OD600 ~ 0.1) for 3 h, the promoter activity of xisF4 (PxisF4‐lacZ) increased 11 ± 1‐fold, and the promoter activity of PA0727 (PPA0727‐lacZ) increased 2.2 ± 0.3‐fold (Fig. 4C). However, overexpressing xisF4 did not affect the promoter activities of PA0720 and PA0724 (Fig. 4C). Furthermore, electrophoretic mobility shift (EMSA) assays showed that XisF4 bound and shifted its own promoter and the promoter region of PA0727 but not the promoter regions of PA0720 or PA0724 (Fig. 4D). Taken together, these results demonstrate that XisF4 positively regulates PA0727 and that overproduction of XisF4 greatly increases Pf4 phage production.
Figure 4.
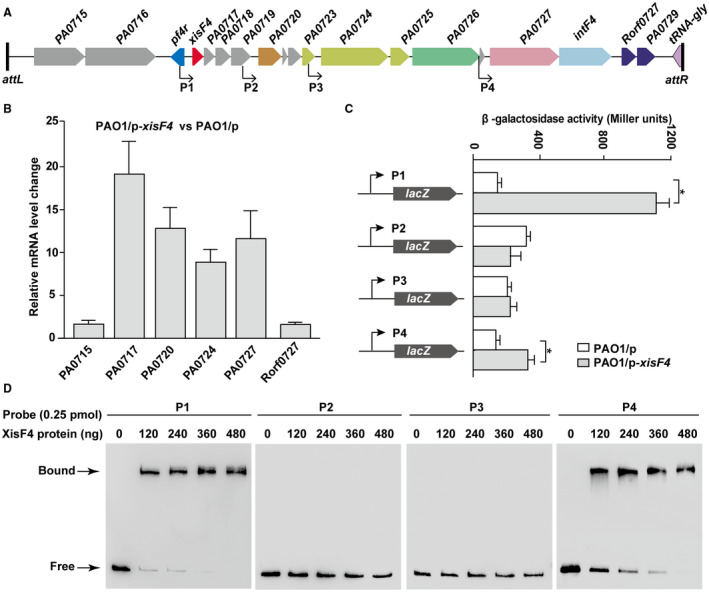
XisF4 induces the expression of PA0727. A. Start sites of four putative promoter regions of the related operons in Pf4 are indicated with arrows, with P1 for xisF4‐PA0719, P2 for PA0720‐0723, P3 for PA0724‐0726 and P4 for PA0727‐0728. B. Relative mRNA levels of the first gene (except PA0717) of the putative operons shown in A in PAO1/pHERD20T‐xisF4 versus PAO1/pHERD20T. C. The lacZ reporter activities were determined in strain PAO1 carrying pHERD20T and pHERD20T‐xisF4. When cells were grown to OD600 ~ 0.1, 10 mM arabinose was added for 3 h of induction. Three independent cultures were used, and error bars indicate standard deviation in B and C. D. EMSA showed that XisF4 bound to the promoter regions of P1 and P4 in a concentration‐dependent manner, but XisF4 did not bind to the promoter regions of P2 or P3.
Pf4r confers immunity to Pf4
Pf4r in Pf4 shares 42% homology with the repressor C of the phage P2 (Webb et al., 2004). Repressor C in P2 controls the lytic conversion of P2 prophage (Saha et al., 1987b); thus, we first tested whether Pf4r can control the lysogenic conversion of Pf4. We constructed a pf4r deletion‐mutant strain (Δpf4r) in PAO1 (Table 1). Pf4 phage production was quantified by applying the supernatant collected from the planktonic culture of the Δpf4r and the PAO1 wild‐type strains to the bacterial lawn formed by the ΔPf4 strain. As expected, deletion of pf4r increased Pf4 production 105‐fold compared to the PAO1 wild‐type strain (Fig. 5A). Furthermore, complementation of pf4r via pHERD20T‐pf4r in the Δpf4r strain greatly reduced Pf4 production (Fig. 5B), suggesting that Pf4r functions as a repressor for Pf4 phage production. In lambdoid, CTXφ and P2 phages, phage repressors are known to mediate phage immunity (Wilgus et al., 1973; Kimsey and Waldor, 1998). To explore the role of Pf4r in Pf4 immunity, we used Pf4 phages to infect the ΔPf4 strain in the absence of pf4r (carrying pHERD20T) and in the presence of pf4r (carrying pHERD20T‐pf4r). As expected, the ability of Pf4 to infect the ΔPf4 strain decreased 106‐fold when Pf4r was overproduced via pHERD20T‐pf4r (Fig. 5C). Additionally, overproduction of Pf4r via pHERD20T‐pf4r in the PAO1 wild‐type strain reduced the ability of Pf4 to re‐infect PAO1 approximately 100‐fold, and overproduction of Pf5r via pHERD20T‐pf5r in the PAO1 wild‐type strain also reduced the ability of Pf4 phages to re‐infect PAO1 approximately 10‐fold (Fig. 5C). These results collectively demonstrate that Pf4r and Pf5r both confer immunity to Pf4 infection.
Figure 5.

Pf4r confers immunity to Pf4. A. Pf4 phage titers (PFU ml–1) were quantified on ΔPf4 lawn using culture supernatant from planktonically growing PAO1 and Δpf4r strains at different times. B. Pf4 phage titers (PFU ml–1) were quantified on ΔPf4 lawns using culture supernatant from planktonically growing PAO1 carrying pHERD20T, Δpf4r carrying pHERD20T and Δpf4r carrying pHERD20T‐xisF4. At the beginning of the culture, 10 mM arabinose was added to induce the expression of xisF4. C. Pf4 phages were collected from planktonic culture supernatant of PAO1 with overexpression of XisF4. Serially diluted phages were then applied to lawns of ΔPf4 carrying pHERD20T, pHERD20T‐pf4r and pHERD20T‐pf5r, and lawns of PAO1 carrying pHERD20T, pHERD20T‐pf4r and pHERD20T‐pf5r. Three independent cultures of each strain were used, and error bars indicate standard deviation in A and B.
Pf4r auto‐activates itself and represses xisF4
Previous analysis predicted that the upstream region of the structural gene PA0717 is the regulatory region of Pf4 (McElroy et al., 2014). Since the new ORF (XisF4) identified in this study is located between PA0717 and pf4r, 5′‐RACE was employed to determine the transcriptional start sites of pf4r and xisF4, respectively. As shown in Fig. 6A, the transcriptional start site of xisF4 is 129 bp upstream of the translational start site of xisF4 and 11 bp downstream of that of pf4r. On the other hand, the transcriptional start site of pf4r is 150 bp upstream of the translational start site of pf4r and 31 bp downstream of that of xisF4. The 5′‐untranslated regions of the pf4r and xisF4 transcripts overlap and contain a directed repeat (DR, 5′‐GGGGAAATA‐3′) and an inverted repeat (IR, 5′‐AATTATTT‐3′) (Fig. 6A).
Figure 6.
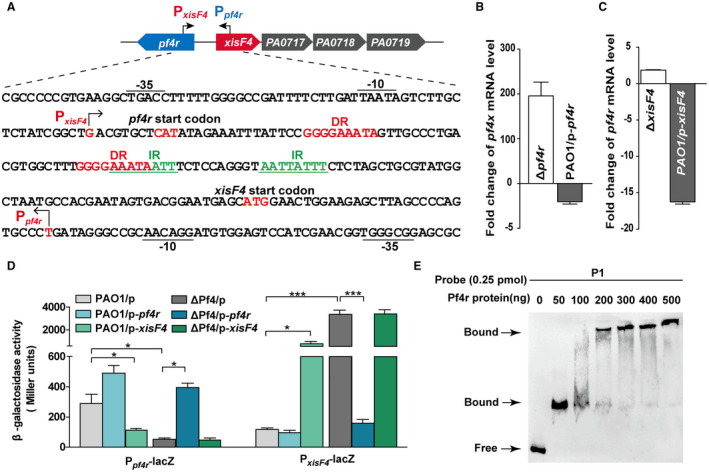
Pf4r represses xisF4 while activating itself. A. A schematic diagram indicates the intergenic region between xisF4 and xisF5. The transcriptional start sites of xisF4 and pf4r were determined by 5′‐RACE, and arrows indicate the direction of transcription. DR and IR indicate directed and inverted repeats, respectively. B. Fold change of mRNA levels of xisF4 in Δpf4r versus PAO1, and in PAO1/pHERD20T‐pf4r versus PAO1/pHERD20T. C. Fold change of mRNA levels of pf4r in ΔxisF4 versus PAO1, and in PAO1/pHERD20T‐xisF4 versus PAO1/pHERD20T. D. The β‐galactosidase activity of Ppf4r – lacZ and PxisF4 – lacZ were determined in PAO1 and ΔPf4 carrying pHERD20T, pHERD20T‐pf4r and pHERD20T‐xisF4, respectively. 10 mM arabinose was added for induction for 3 h at OD600 ~ 0.1. Three independent cultures of each strain were used, and error bars indicate standard deviation in B, C and D. E. EMSA showed that Pf4r bound to the promoter region of P1 (as shown in Fig. A) in a concentration‐dependent manner.
To check the cross‐regulation between Pf4r and XisF4, expression of xisF4 in the presence versus in the absence of pf4r was assessed by qRT‐PCR. The level of xisF4 transcript was induced 196 ± 32‐fold in the Δpf4r strain compared with the PAO1 wild‐type strain, and the level of xisF4 transcript was repressed 2.9 ± 0.9‐fold when pf4r was overexpressed via pHERD20T‐pf4r compared with the empty plasmid in PAO1 (Fig. 6B). Similarly, expression of pf4r in the presence versus in the absence of xisF4 was also assessed. The level of pf4r transcript was repressed 16.2 ± 0.7‐fold when xisF4 was overexpressed via pHERD20T‐xisF4 compared with the empty plasmid in the PAO1 strain (Fig. 6C). However, the level of pf4r transcript was not changed in the ΔxisF4 strain compared with the PAO1 wild‐type strain. These results showed that Pf4r represses the expression of xisF4 and vice versa. Furthermore, these results also suggested that xisF4 was repressed while pf4r was expressed in the PAO1 wild‐type strain to ensure lysogeny under normal conditions. To further determine how Pf4r regulates xisF4 and pf4r, two transcriptional lacZ fusions, one containing the xisF4 promoter (PxisF4‐lacZ) and the other containing the pf4r promoter (Ppf4r‐lacZ), were constructed and integrated into the PAO1 wild‐type strain and the ΔPf4 strain, respectively. Noticeably, the promoter activity of pf4r was 5.2 ± 0.2‐fold higher in the PAO1 wild‐type strain (PAO1/pHERD20T) compared with the ΔPf4 strain (ΔPf4/pHERD20T). In contrast, the promoter activity of xisF4 was 28 ± 1‐fold higher in the ΔPf4 strain (ΔPf4/pHERD20T) compared with the PAO1 wild‐type strain (PAO1/pHERD20T). Corroborating the above qRT‐PCR results, the promoter activity of pf4r decreased 3.0 ± 0.5‐fold when xisF4 was overexpressed in the PAO1 wild‐type strain and increased 7.0 ± 0.3‐fold when pf4r was overexpressed in the ΔPf4 strain (Fig. 6D). These results demonstrate that XisF4 repressed the expression of pf4r and Pf4r activated itself in the absence of xisF4. Similarly, the promoter activity of xisF4 reduced 21 ± 1‐fold when pf4r was overexpressed in the ΔPf4 strain and increased 11 ± 1‐fold when xisF4 was overexpressed in the PAO1 wild‐type strain (Fig. 6D). These results demonstrated that Pf4r repressed the expression of xisF4 and XisF4 can activate itself in the presence of pf4r. Furthermore, EMSA was performed to test the DNA binding activity of Pf4r to the overlapped promoter regions of pf4r and xisF4. As expected, Pf4r bound and shifted this region (Fig. 6E) and XisF4 also bound and shifted the same region as shown above (Fig. 4D). Taken together, these results collectively show that Pf4r represses the transcription of xisF4 and XisF4 represses the expression of pf4r.
MvaT/MvaU control xisF4 transcription
Our earlier work revealed that H‐NS controls P4‐like excisionase gene alpA in Shewanella oneidensis and Hha controls P4‐like excisionase gene alpA in E. coli K‐12 (Wang et al., 2009; Zeng et al., 2016). In PAO1, two H‐NS family proteins, MvaT and MvaU, function coordinately as xenogeneic silencers (Castang et al., 2008). A previous ChIP‐chip assay also suggested that one of the binding regions of MvaT and MvaU is located between PA0715 and PA0717 (Castang and Dove, 2012). Here, to investigate whether the H‐NS family protein regulates xisF4 in PAO1, two single‐deletion strains, each lacking one of the H‐NS family genes, and a double‐deletion strain lacking two H‐NS family genes were constructed (Table 1). Next, a chromosomal lacZ transcriptional fusion of the xisF4 promoter (PxisF4‐lacZ) was integrated into these deletion mutants to test the promoter activity of xisF4 in vivo by measuring β‐galactosidase activity. The promoter activity of xisF4 increased 16 ± 1‐fold in the ΔmvaTΔmvaU double‐mutant strain compared with the wild‐type PAO1 strain (Fig. 7A). However, the promoter activity of xisF4 in the ΔmvaT strain or in the ΔmvaU strain was similar to that in the wild‐type PAO1 strain (Fig. 7A). Furthermore, deleting both H‐NS family proteins also greatly induced Pf4 production (103‐fold), Pf4 excision (105‐fold) and the number of Pf4 RF molecules (102‐fold) (Fig. 7B–D). Taken together, these results suggest that the host proteins MvaT and MvaU coordinately repress the production of Pf4 by directly repressing xisF4 transcription.
Figure 7.
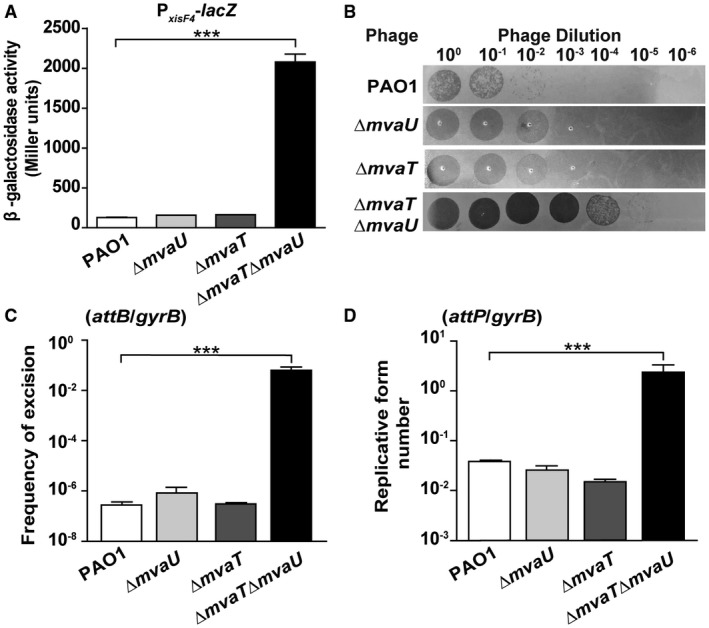
MvaT and MvaU coordinately repress xisF4. A. The β‐galactosidase activity of the PxisF4 – lacZ reporter was determined in PAO1, ΔmvaT, ΔmvaU and ΔmvaTΔmvaU. B. Phages were collected from planktonic culture (OD600 ~ 1.0) supernatants of PAO1, ΔmvaT, ΔmvaU and ΔmvaTΔmvaU. Serial dilutions were applied to lawns of ΔPf4. C. The frequency of Pf4 excision was quantified in PAO1, ΔmvaT and ΔmvaU and ΔmvaTΔmvaU. D. The numbers of Pf4 RF molecules were quantified in PAO1, ΔmvaT, ΔmvaU and ΔmvaTΔmvaU. Three independent cultures were used, and error bars indicate standard deviation.
XisF4 and XisF5 represent two major subfamilies of excisionases in Pf prophages
To investigate the prevalence of XisF4 and XisF5 among Pseudomonas species, we first set a gene profile within the genus Pseudomonas in IMG/M database with the minimal identity of 60% and e‐value of 0.01 (Mukherjee et al., 2017). Among 1999 sequenced Pseudomonas strains, 233 of them carry a XisF4 homologue, 165 of them carry a XisF5 homologue and 62 of them carry both XisF4 and XisF5 homologues (Fig. 8A). Among 696 sequenced P. aeruginosa strains deposited in the IMG/M database, 194 carry a XisF4 homologue, 146 carry a XisF5 homologue and 58 carry homologues of both XisF4 and XisF5 (Fig. 8A). We next investigated the phylogenetic relationships among the XisF4 or XisF5 homologues. Notably, we found that the excisionase genes were clustered into two distinct groups, the XisF4 group and the XisF5 group (Fig. 8B). Importantly, overexpressing xisF5 in PAO1 was unable to induce Pf4 excision, and vice versa (Fig. S6), suggesting these two groups might be functionally divergent. To further determine the prevalence of excisionases among Pf prophages, we first analyzed the abundance and diversity of Pf prophages in P. aeruginosa. Considering that the phylogenetic relationship of phages can be represented by their conserved coat proteins (Kauffman et al., 2018), a tree based on the minor coat protein (PA0724) of Pf prophages (carrying the conserved region PA0720‐PA0727) was constructed. According to the phylogenetic tree, Pf prophages were clustered into three clades, Pf4, Pf5 and Pf‐LES (Fig. 8C), which is similar to the previous phylogenetic analysis of Pf prophages based on whole‐genome analysis (four clades: Pf4, Pf5 Pf7, and Pf‐LES) (Knezevic et al., 2015). The Pf4 group obtained based on minor coat proteins contains the Pf7 clade, including the prophages of strains PA7, PA38182 and MTB‐1t. Among the 433 Pf phages, 247 (57%) contain a XisF4 or XisF5 excisionase (Fig. 8C, Table S2), suggesting that more than half of the Pf phages are integrated into the host chromosome instead of replicating episomally. Our analysis also revealed that many P. aeruginosa strains carry multiple copies of Pf phages belonging to the same or different clades (Fig. 8C). These multiple copies of Pf prophages are integrated into different tRNA genes, which are matched to the different subfamilies of integrase based on phylogenetic analysis (Figs 8C and S7). Taken together, these analyses indicate that XisF4 and XisF5 are widespread among Pf phages in P. aeruginosa.
Figure 8.
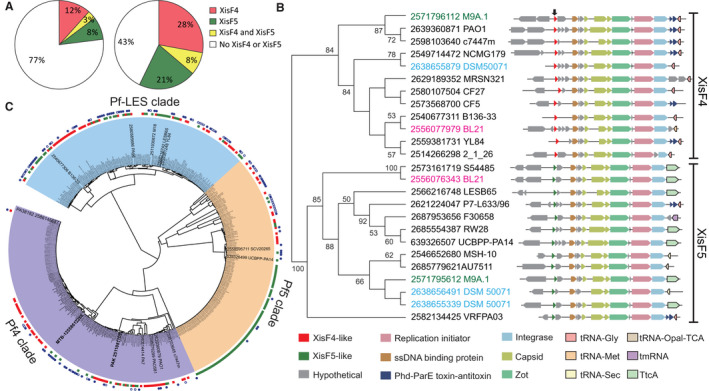
XisF4 and XisF5 are prevalent in Pf Phage. A. The left pie chart shows the proportions of the strains carrying XisF4 (red), XisF5 (green) or both (yellow) among 1999 Pseudomonas strains. The right pie chart shows the proportions of the strains carrying XisF4, XisF5 or both among 696 Pseudomonas aeruginosa strains. B. Neighbor‐joining tree of excisionases showing the phylogenetic relationships of the excisionase homologues in P. aeruginosa strains. Bootstrap values greater than 50% are indicated at the nodes. The name on each node indicates the Gene ID and the strain name in the IMG/M database, and the excisionase homologues from the same strain are marked by the same font color. The arrow on the top shows the position of the excisionase. The regions neighboring these excisionase genes were analyzed and are shown for each node. C. Phylogenetic tree of PA0724 (minor coat protein) homologues indicates the relationship of the Pf prophages. Three clades are indicated by color coding: purple‐Pf4, orange‐Pf5 and blue‐Pf‐LES. The first outer ring represents the presence of XisF4 (red), XisF5 (green) or no XisF4 or XisF5 (blank) in the specific Pf phage carrying the corresponding PA0724 homologue at each node. The second outer ring indicates that the strain at each node has multiple copies of PA0724 homologues located in a complete Pf phage; a filled blue circle indicates two copies, and a blue circle indicates more than two copies.
Discussion
In this study, we report for the first time that filamentous phages encode their own excisionases. More importantly, we demonstrate that the two excisionases (XisF4 in Pf4 and XisF5 in Pf5) not only promote Pf prophage excision but also activate prophage replication. To our knowledge, due to their relatively small genome sizes (< 12 kb), filamentous phages usually do not encode their own excisionases. The identification of excisionases in Pf4 and Pf5 prophages fundamentally extends the current understanding of genome excision and host regulation of filamentous phages. Specifically, we show that high expression of pf4r is needed to ensure the lysogeny of PAO1 under normal conditions while high expression of xisF4 is needed to induce prophage excision and replication under specific conditions. XisF4 and XisF5 share medium sequence identity, and our results demonstrate that XisF5 is essential for Pf5 excision while XisF4 is non‐essential for Pf4 excision. In addition, phylogenetic analysis demonstrated that XisF4 and XisF5 represent the two subfamilies of excisionase of Pf phages in P. aeruginosa strains, and they are highly prevalent in Pf prophages. Thus, prophage excision regulated by prophage‐encoded excisionase should be common for Pf prophages in P. aeruginosa strains. Also, our results suggest that using genetically modified excisionase to induce prophage excision without regulating phage replication would be a feasible way to remove integrated filamentous prophage from the Pseudomonas host genome.
The genome sizes of Pf1, Pf4 and Pf5 are 7.3 kb, 12.4 kb and 10.6 kb, respectively (Hill et al., 1991). Comparative analysis revealed that both Pf4 and Pf5 exhibit mosaic phage genome structures. Most of the structural genes of Pf1, Pf4 and Pf5 are highly conserved, except for the minor coat proteins g3p (PA0724 in Pf4, PA14_48930 in Pf5 and ORF437 in Pf1) which share a lower sequence similarity. The minor coat protein g3p is involved in the initial step of phage infection by interacting with host pilus (Holland et al., 2006), and the low sequence identity in minor coat proteins suggest that these Pf prophages have been divergent and that there has been a long co‐evolution of the bacteria‐host interaction (Marvin et al., 2014). Apart from the backbone shared by these three Pf prophages, Pf4 and Pf5 also carry two regulatory genes, repressor C (pf4r and pf5r) and excisionase (xisF4 and xisF5). In this study, we demonstrate that XisF4 acts in three ways: acting as a transcriptional repressor of pf4r, acting as a transcriptional activator of PA0727 (which is the replication initiator of Pf4), and promoting the excision of Pf4. The Cox protein of P2 also has similar three functions; it represses the promoter of repressor gene, activates the gene which controls phage replication, and promotes the excision of P2 (Saha et al., 1987a; 1989; Eriksson and Haggard‐Ljungquist, 2000). P2 is a major family of temperate dsDNA phages and is different from filamentous phages in gene arrangement and morphology (Bertani and Bertani, 1971; Crowther, 1980). Although they are functionally equivalent, there is no sequence similarity between Cox and XisF4 or XisF5. Instead, XisF4 shares 30% similarity with a putative AlpA excisionase of P4‐like prophage of Pseudomonas mandelii (Fig. S1). P4 is a satellite phage that relies on P2 as a helper to supply the gene products necessary for phage particle assembly and cell lysis (Barrett et al., 1976). Interestingly, repressor C in Pf4 shows 42% sequence identity with the repressor C of phage P2 (Webb et al., 2004). Moreover, apart from the conserved regulatory genes in both Pf4 and Pf5, there are several unique genes in Pf4 and Pf5. For example, the Phd‐ParE toxin‐antitoxin system, a putative ATPase and a reverse transcriptase are found in Pf4 but not in Pf5 or Pf1 (Webb et al., 2004). Pf5 has a unique gene encoding a protein with a predicated ParA nucleotide‐binding domain (Mooij et al., 2007). Further studies are under way to explore the functions of these unique phage genes.
It has been previously reported that Pf4 phages released from PAO1 biofilms can re‐infect the PAO1 wild‐type that contains an integrated copy of the same phage (Rice et al., 2009). Further sequencing results revealed that mutations within or upstream of repressor c gene of Pf4 were accumulated in the ‘superinfective’ Pf4 phages released from PAO1 flow‐cell biofilms (McElroy et al., 2014). Our analysis showed that these mutations are located at the upstream region of the xisF4 gene. Furthermore, we demonstrate that the repressor C in Pf4 and Pf5 confer immunity to Pf4 infection and Pf4 phages released from PAO1 overexpressing xisF4 were also capable of re‐infecting PAO1 wild‐type strain. However, these phages can only re‐infect PAO1 at high titers (> 108 PFU ml–1) and this ability of re‐infecting PAO1 is further reduced by overexpressing pf4r or pf5r in PAO1. These results suggest that a high amount of phage particles might circumvent the phage immunity conferred by the phage repressor protein. Nevertheless, it remains to be determined whether and how the excisionase and/or the repressor C directly contribute to the emergence of superinfective Pf4 phages in PAO1 flow‐cell biofilms.
For lambda phage, it is clear that two host‐encoded proteins, RecA and LexA, play an important role in the control of the lysis‐lysogeny switch. However, very few studies directly focused on the lysis‐lysogeny switch of Pf prophages. Nevertheless, activation of genes in Pf prophages in P. aeruginosa have been reported in several stressed conditions. Pf prophage‐encoded genes were found to be strongly upregulated in P. aeruginosa biofilm cells using DNA microarrays (Whiteley et al., 2001). Pf4 was induced when PAO1 cells encountered oxidative stress, and the primary oxidative stress response protein OxyR is involved in this process (Wei et al., 2012; Hui et al., 2014). It was reported that Pf4 superinfection in P. aeruginosa is regulated by BfmR which is part of a two‐component signal transduction pathway in response to membrane perturbing stress (Petrova et al., 2011). A recent study showed that Pf5 production is activated a million‐fold by inactivation of the substrate binding protein DppA1 through an unknown mechanism linked to Pf5 nutrient sensing; i.e., when nutrients are low, Pf5 lyses the host (Lee et al., 2018). Further studies are needed to explore whether these genetic factors function through the excisionase gene to regulate the Pf4 and Pf5 prophage induction or whether other host factors can directly activate Pf production under specific conditions.
In PAO1, MvaT and MvaU can form homomeric or heteromeric complexes (Castang et al., 2008). Under normal growing conditions, MvaT and MvaU repress the expression of xisF4 by binding to the xisF4 promoter. As a result, Pf4 prophage stably resides in the host genome, and Pf4 replication is repressed. When mvaT and mvaU are repressed, xisF4 will be derepressed. Subsequently, XisF4 will function as a recombination directionality factor and promote the excision of Pf4, along with the presence of integrase. Meanwhile, XisF4 will bind to the promoter region of PA0727, leading to the activation of PA0727. As a replication initiator protein, PA0727 will promote the replication of Pf4 (Fig. 9). In E. coli, H‐NS was stimulated by cold shock through the protein CspA (La Teana et al., 1991). H‐NS is also repressed by an antisense RNA, called DsrA, along with an RNA‐binding protein called Hfq (Lease and Belfort, 2000; Brescia et al., 2003). In Salmonella enterica, H‐NS is an essential component in thermoregulation that responds to the temperature change (Ono et al., 2005). Recently, in Shewanella oneidensis, H‐NS was found to be responsible for the prophage excision during cold adaption (Zeng et al., 2016). However, we found that decreasing the temperature from 37 to 15°C did not change the expression levels of mvaT and mvaU or the production of Pf4. In addition, Pf4 production was induced during biofilm formation (Whiteley et al., 2001); one of the possible causes is that the mvaT‐ and mvaU‐mediated repression of xisF4 was blocked during biofilm formation. However, the stress factors and genes involved in the regulation of mvaT and mvaU in PAO1 have rarely been demonstrated. It is reasonable to speculate that the MvaT and MvaU might be counter‐silenced by other DNA‐binding proteins (Will et al., 2015) or become inactivated in response to environmental stress and consequently increases phage production. Nevertheless, the underlying regulation of MvaT and MvaU in response to environmental stimuli would help understand the trade‐off between Pf filamentous phage and its bacterial host.
Figure 9.
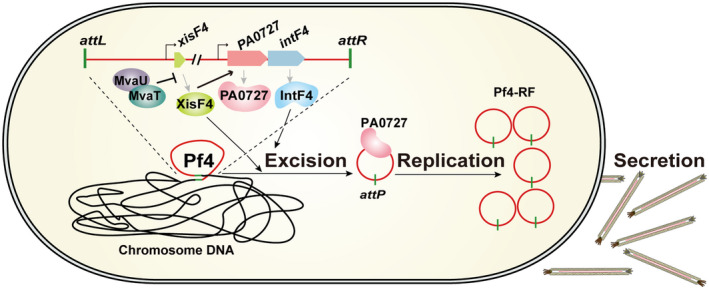
A proposed model of the lysis‐lysogeny switch of Pf4 in PAO1. The stable integration of Pf4 in the host genome is maintained by the host proteins MvaT/MvaU. MvaT and MvaU repress the expression of xisF4 by binding to the promoter of xisF4. When mvaT and/or mvaU are repressed or inactivated, expression of xisF4 will be de‐repressed and induce Pf4 excision. Meanwhile, XisF4 can activate the expression of PA0727 (replication initiator) and produce a large number of circular forms of Pf4. Finally, Pf4 will be assembled and secreted to the outside of the cell.
Materials and methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids are listed in Table 1, and primers are listed in Table S1. E. coli and P. aeruginosa PAO1 strains were grown in Luria‐Bertani (LB) medium at 37°C unless specified otherwise. When necessary, the following antibiotics were added at the indicated concentrations: tetracycline (50 µg ml–1), gentamycin (30 µg ml–1), carbenicillin (100 µg ml–1), kanamycin (50 µg ml–1).
Construction of deletion mutants
The gene deletion method used here in P. aeruginosa was previously reported (Hoang et al., 1998). To delete Pf4, xisF4, xisF5, intF5, pf4r, mvaT and mvaU, upstream and downstream homologous sequences (0.7–1 kb) were amplified through PCR from PAO1 or PA14 genomic DNA. Gentamycin resistance gene was amplified through PCR from the plasmid pPS856. These three amplicons were then ligated into pEX18Ap to produce the deletion plasmids. In‐frame deletion mutants were obtained via homologous recombination using sucrose resistance selection. The GmR selectable marker was removed from the chromosome as described previously (Hoang et al., 1998). For the construction of the double‐deletion mutant ΔmvaTΔmvaU, the mvaT single‐deletion strain was used as the recipient for further deleting mvaU. The final obtained mutants were confirmed by sequencing.
Construction of plasmids
For the construction of the expression plasmid pHERD20T, the full coding regions of pf4r, pf5r, xisF4, xisF5, intF4 and intF5 were amplified through PCR from PAO1 or PA14 genomic DNA, and the PCR products were purified, digested with restriction enzymes and ligated into the vector pHERD20T. For construction of promoter reporter strains, the putative promoter regions of xisF4, pf4r, PA0720, PA0724 and PA0727 were amplified by PCR. Each amplicon was ligated into mini‐CTX‐lacZ using the Vazyme ClonExpress II One Step Cloning Kit. The constructed plasmid was then transformed into PAO1 or ΔPf4 hosts and integrated into the chromosome at the attB site near the tRNASer sequence (Becher and Schweizer, 2000). The tetracycline selection marker carried by the plasmid was removed from the chromosome as described previously (Hoang et al., 1998).
Plaque assay
Pf4 phages were collected from planktonic cultures of PAO1 strains. Two milliliters of culture was collected and centrifuged at 12000 rpm for 2 min, and the supernatant was filtered through a 0.22 μm filter (Millipore Millex GP) to obtain a pure Pf4 solution. Then, the supernatant was 10‐fold serially diluted using LB. For preparation of bacterial lawns, we used the top‐layer agar method as previously described (Eisenstark, 1967). Then, 1 ml of stationary culture OD600 ~ 4 (PAO1 or ΔPf4) was mixed with 4 ml of molten LB‐top agar (8 g–l agar, 0.1% glucose, 5 mM CaCl2) at 50°C and poured over LB‐bottom plates (10 g–l agar, 0.1% glucose, 5 mM CaCl2). Finally, 10 μl of the serially diluted Pf4 solution was applied to bacterial lawns, and the plaques were visualized after 8 h of incubation.
Reporter activity assay
Specific β‐galactosidase activity (U mg–1) of strains harboring the pf4r, xisF4, PA0720, PA0724 and PA0727 promoter reporter constructs was determined using the Miller assay (Miller, 1972). To determine the promoter activity of pf4r, xisF4, PA0720, PA0724 and PA0727 under overexpression of xisF4 or pf4r, pHERD20T‐xisF4 or pHERD20T‐pf4r was transformed into the strain carrying the reporter. Strains were grown overnight in LB supplemented with carbenicillin. The overnight cultures were diluted 1:100 in LB, and 10 mM arabinose was added from OD600 ~ 0.1. After induction for 3 h, cells were collected to determine β‐galactosidase activity. To determine the promoter activity of xisF4 in PAO1, ΔmvaT, ΔmvaU and ΔmvaTΔmvaU, strains were grown overnight in LB. The overnight cultures were diluted 1:100 in LB, and the cells were collected to determine β‐galactosidase activity when the OD600 was ~1.0.
Protein purifications
Protein XisF4 and Pf4r were purified from E.coli BW25113 strain containing plasmid pHERD20T‐xisF4 and pHERD20T‐pf4r, respectively. One liter of LB supplemented with carbenicillin was inoculated with 10 ml of overnight culture, and the bacteria were grown with shaking at 37°C. 10 mM arabinose was added from OD600 ~ 0.5 and all the cells were collected by centrifugation after induction for 6 h. Protein IntF4, XisF5 and IntF5 were extracted from E. coli BL21(DE3) strain containing plasmid pET28b‐intF4, pET28b‐xisF5 and pET28b‐ intF5, respectively. One liter of LB supplemented with kanamycin was inoculated with 10 ml of overnight culture, and the bacteria were grown with shaking at 37°C. 0.5 mM IPTG was added from OD600 ~ 0.5 and all the cells were collected by centrifugation after induction for 6 h. The subsequent steps of extraction protein from the collected pellet were performed as previously described (Liu et al., 2015).
Electrophoretic mobility shift assay (EMSA)
Electrophoretic mobility shift assays were performed as previously described (Liu et al., 2015). DNA fragment AttL (122 bp) and AttR (252 bp) flanking the left or right attachment site of Pf4 were amplified through PCR from the genomic DNA of PAO1 strain, using primer pair probe‐Pf4attL‐F/R or probe‐Pf4attR‐F/R (Table S1), respectively. DNA fragment AttB (184 bp) covering the attachment site of Pf4 were amplified from the genomic DNA of ΔPf4 strain, using primer pair probe‐Pf4attB‐F/R (Table S1). DNA fragment AttL of Pf5 (141 bp) flanking the left attachment site of Pf5 were amplified from the genomic DNA of PA14 wild‐type strain, using primer pair probe‐Pf5attL‐F/R (Table S1). DNA fragments of the intergenic region between xisF4 and pf4r (232 bp), the promoter region of PA0720 (253 bp), PA0724 (282 bp) and PA0727 (229 bp) were amplified from PAO1 using the corresponding primer pair listed in Table S1. All the purified DNA fragments were labeled biotin by using the Biotin 3′ End DNA Labeling Kit (Termo scientific, Rockford, USA). DNA fragments (0.25 pmol) were mixed with the purified proteins and incubated at 25°C for 2 h. The binding reaction components were added following the protocol as described in the LightShiſt Chemiluminescent EMSA kit (Termo scientific, Rockford, USA). Then the binding reaction samples were run on a 6% DNA retardation gel at 100 V in 0.5 × TBE and were then transferred to nylon membranes. The membranes were visualized using the Chemiluminescence Nucleic Acid Detection Module Kit (Termo scientific, Rockford, USA).
5′‐RACE
Mapping of the 5′ transcriptional start site was performed using the SMARTer RACE 5′/3′ Kit (TAKARA) following the manufacturer’s recommendations. First, RNA was extracted using an RNA extraction kit (Promega, Madison, WI, USA). Second, polyA tails were added to RNA prior to first‐strand 3′‐cDNA synthesis using a Poly‐(A) Polymerase enzyme (Takara Bio Cat. No. 2180A). Third, the pf4r‐ and xisF4‐specific primers were designed following the protocol in the SMARTer RACE 5′/3′ Kit user manual. Finally, the inserts were sequenced using the M13R primer.
Quantitative reverse‐transcription real‐time PCR (qRT‐PCR)
Strains were grown overnight and adjusted to an OD600 of 0.05 in LB. Cells were collected at OD600 ~ 1.0 by centrifugation (12000 rpm for 1 min), and used for RNA extraction using an RNA extraction kit (Promega, Madison, WI, USA). The medium was supplemented with carbenicillin for strains carrying pHERD20T‐based plasmids, and 10 mM arabinose was added at OD600 ~ 0.8 for 30 min. Total RNA was extracted from exponential‐phase bacteria using The cDNA synthesis was conducted using reverse transcription (Promega, Madison, WI, USA). Total cDNA (50 ng) was used for qRT‐PCR using the Step One Real‐Time PCR System. The level of the 16S rRNA gene transcript was used to normalize the gene expression data.
Quantification of prophage genome excision and extrachromosomal phage copy number
The frequency of prophage excision and the extrachromosomal phage copy number under different conditions were quantified by quantitative PCR (qPCR). To determine the the frequency of Pf4 or Pf5 excision, the number of chromosomes that are devoid of Pf4 or Pf5 were quantified using primers (Pf4‐f/r or Pf5‐f/r) flanking the reconstituted bacterial attachment site (attB) (Fig. 2A), which only generate PCR products when the prophage is excised because of the size of the prophage (Fig. 1). The numbers of Pf4 or Pf5 RF molecules were quantified using primers (Pf4‐Cf/r or Pf5‐Cf/r) flanking the phage attachment site (attP) (Fig. 2A). The final value of frequency of excision and RF number were normalized by reference gene (gyrB), which is a single‐copy housekeeping gene indicating the number of total chromosomes. Strains were grown overnight and adjusted to an OD600 of 0.05 in LB. Cells were collected at OD600 ~ 1.0 by centrifugation (12000 rpm for 1 min), and used for DNA extraction using DNA Isolation Kit (TIANGEN DP302). The medium was supplemented with carbenicillin for strains carrying pHERD20T‐based plasmids, and 10 mM arabinose was added from OD600 ~ 0.05. Total DNA (50–200 ng) was used as the template for the qPCR reaction using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher). The reaction was conducted using the Step One Real‐Time PCR System.
Phylogenetic analysis
The distributions of XisF4 and XisF5 among a total of 1999 sequenced Pseudomonas strains were obtained by using the gene profile and alignment tool in the IMG/M system, version 4.6, with an e‐value of 0.01 and minimal amino acid identity of 60% (Mukherjee et al., 2017). For phylogenetic analysis of XisF4 and XisF5, 266 homologous sequences were retrieved by BLASTp in the IMG/M system (e‐value of 0.01 and minimal amino acid identity of 35%), excluding XisF4 and XisF5 homologues whose neighboring genes are not from Pf prophages (Table S2) (Mukherjee et al., 2017). A final set of 25 representative sequences (including only homologues with amino acid identity of 100%) was selected for construction of a phylogenetic tree in MEGA 5.0 using the neighbor‐joining (NJ) method with 1000 replications (Tamura et al., 2011). For phylogenetic analysis of Pf prophages based on PA0724 (minor coat protein), 433 sequences were retrieved via BLASTp in the IMG/M system (e‐value of 0.01 and minimal amino acid identity of 35%), excluding PA0724 homologues whose neighboring genes are not from Pf prophages (Table S2) (Mukherjee et al., 2017). The phylogenetic tree of PA0724 homologues was constructed with MAFFT using an averaged linkage (UPGMA) approach (Kuraku et al., 2013). The corresponding neighboring gene, the XisF4 or XisF5 homologue, was displayed on the PA0724 tree based on the phylogenetic analysis of XisF4 and XisF5. A set of 397 homologues of IntF4 and of the neighboring gene, PA0724, were retrieved in the IMG/M system for phylogenetic analysis, and the phylogenetic tree of IntF4 was constructed using the same method as for PA0724 (Table S2). The neighboring genes of tRNA, xisF4 or xisF5 are displayed on the phylogenetic tree for IntF4. The placement of genes on trees was visualized and annotated using the iTOL interface (Letunic and Bork, 2016).
Conflicts of interest
The authors declare no conflict of interests.
Supporting information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31625001, 31500025 and 41706172), the National Basic Research Program of China (2017YFC0506303 and 2018YFC1406500) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA13010500). XW is a 1000‐Youth Elite Program recipient in China.
References
- Addy, H.S. , Askora, A. , Kawasaki, T. , Fujie, M. and Yamada, T. (2012) Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by φRSM filamentous phages. Phytopathology, 102, 469–477. [DOI] [PubMed] [Google Scholar]
- Ahmad, A.A. , Askora, A. , Kawasaki, T. , Fujie, M. and Yamada, T. (2014) The filamentous phage XacF1 causes loss of virulence in Xanthomonas axonopodis pv. citri, the causative agent of citrus canker disease. Frontiers in Microbiology, 5, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, T. , Ara, T. , Hasegawa, M. , Takai, Y. , Okumura, Y. , Baba, M. , et al. (2006) Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Molecular Systems Biology, 2, 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, K.J. , Marsh, M.L. and Calendar, R. (1976) Interactions between a satellite bacteriophage and its helper. Journal of Molecular Biology, 106, 683–707. [DOI] [PubMed] [Google Scholar]
- Becher, A. and Schweizer, H.P. (2000) Integration‐proficient Pseudomonas aeruginosa vectors for isolation of single copy chromosomal lacZ and lux gene fusions. BioTechniques, 29(948–950), 952. [DOI] [PubMed] [Google Scholar]
- Bertani, L.E. and Bertani, G. (1971) Genetics of P2 and related phages In: Caspari, E.W. (Ed.) Advances in Genetics. New York: Elsevier, pp. 199–237. [DOI] [PubMed] [Google Scholar]
- Brescia, C.C. , Mikulecky, P.J. , Feig, A.L. and Sledjeski, D.D. (2003) Identification of the Hfq‐binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA, 9, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang, S. and Dove, S.L. (2012) Basis for the essentiality of H‐NS family members in Pseudomonas aeruginosa . Journal of Bacteriology, 194, 5101–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang, S. , McManus, H.R. , Turner, K.H. and Dove, S.L. (2008) H‐NS family members function coordinately in an opportunistic pathogen. Proceedings of the National Academy of Sciences of the USA, 105, 18947–18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier, M. (1976) Integration and excision of bacteriophage Mu‐1. Cell, 7, 155–163. [DOI] [PubMed] [Google Scholar]
- Crowther, R.A. (1980) Structure of bacteriophage Pf1. Nature, 286, 440–441. [DOI] [PubMed] [Google Scholar]
- Das, B. , Bischerour, J. and Barre, F.‐X. (2011) VGJɸ integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proceedings of the National Academy of Sciences of the USA, 108, 2516–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, B.M. , Kimsey, H.H. , Kane, A.V. and Waldor, M.K. (2002) A satellite phage‐encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. The EMBO Journal, 21, 4240–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman, C.J. (2007) H‐NS, the genome sentinel. Nature Reviews Microbiology, 5, 157–161. [DOI] [PubMed] [Google Scholar]
- Eisenstark, A. (1967) Bacteriophage techniques In: Maramorosch, X. and Koprowski H. (Eds.) Methods in Virology. New York: Academic Press, pp. 449–524. [Google Scholar]
- Eriksson, J.M. and Haggard‐Ljungquist, E. (2000) The multifunctional bacteriophage P2 cox protein requires oligomerization for biological activity. Journal of Bacteriology, 182, 6714–6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner, R. , Argov, T. , Rabinovich, L. , Sigal, N. , Borovok, I. and Herskovits, A.A. (2015) A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nature Reviews Microbiology, 13, 641–650. [DOI] [PubMed] [Google Scholar]
- Gottesman, M.E. (1974) Integration and excision of bacteriophage lambda. Cell, 1, 69–72. [Google Scholar]
- Hill, D.F. , Short, N.J. , Perham, R.N. and Petersen, G.B. (1991) DNA sequence of the filamentous bacteriophage Pf1. Journal of Molecular Biology, 218, 349–364. [DOI] [PubMed] [Google Scholar]
- Hoang, T.T. , Karkhoff‐Schweizer, R.R. , Kutchma, A.J. and Schweizer, H.P. (1998) A broad‐host‐range Flp‐FRT recombination system for site‐specific excision of chromosomally‐located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene, 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Hofschneider, P.H. and Preuss, A. (1963) M 13 bacteriphage liberation from intact bacteria as revealed by electron microscopy. Journal of Molecular Biology, 7, 450–451. [DOI] [PubMed] [Google Scholar]
- Holland, S.J. , Sanz, C. and Perham, R.N. (2006) Identification and specificity of pilus adsorption proteins of filamentous bacteriophages infecting Pseudomonas aeruginosa . Virology, 345, 540–548. [DOI] [PubMed] [Google Scholar]
- Hong, S.H. , Wang, X. and Wood, T.K. (2010) Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H‐NS of Escherichia coli . Microbial Biotechnology, 3, 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, K.E. and Waldor, M.K. (2002) Filamentous phage integration requires the host recombinases XerC and XerD. Nature, 417, 656–659. [DOI] [PubMed] [Google Scholar]
- Hui, J.G.K. , Mai‐Prochnow, A. , Kjelleberg, S. , McDougald, D. and Rice, S.A. (2014) Environmental cues and genes involved in establishment of the superinfective Pf4 phage of Pseudomonas aeruginosa . Frontiers in Microbiology, 5, 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, K.M. , Hussain, F.A. , Yang, J. , Arevalo, P. , Brown, J.M. , Chang, W.K. , et al. (2018) A major lineage of non‐tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature, 554, 118–122. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. , Mezulis, S. , Yates, C.M. , Wass, M.N. and Sternberg, M.J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey, H.H. and Waldor, M.K. (1998) CTXφ immunity: application in the development of cholera vaccines. Proceedings of the National Academy of Sciences of the USA, 95, 7035–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic, P. , Voet, M. and Lavigne, R. (2015) Prevalence of Pf1‐like (pro)phage genetic elements among Pseudomonas aeruginosa isolates. Virology, 483, 64–71. [DOI] [PubMed] [Google Scholar]
- Kuraku, S. , Zmasek, C.M. , Nishimura, O. and Katoh, K. (2013) aLeaves facilitates on‐demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Research, 41, W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana, A. , Brandi, A. , Falconi, M. , Spurio, R. , Pon, C.L. and Gualerzi, C.O. (1991) Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H‐NS. Proceedings of the National Academy of Sciences of the USA, 88, 10907–10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, R.A. and Belfort, M. (2000) Riboregulation by DsrA RNA: trans‐actions for global economy. Molecular Microbiology, 38, 667–672. [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Song, S. , Sheng, L. , Zhu, L. , Kim, J.S. and Wood, T.K. (2018) Substrate binding protein DppA1 of ABC transporter DppBCDF increases biofilm formation in Pseudomonas aeruginosa by inhibiting Pf5 prophage lysis. Frontiers in Microbiology, 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I. and Bork, P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research, 44, W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Wally, H. , Miller, S.J. and Lu, C.D. (2009) The multifaceted proteins MvaT and MvaU, members of the H‐NS family, control arginine metabolism, pyocyanin synthesis, and prophage activation in Pseudomonas aeruginosa PAO1. Journal of Bacteriology, 191, 6211–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Kotetishvili, M. , Chen, Y. and Sozhamannan, S. (2003) Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae . Applied and Environmental Microbiology, 69, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati, N.T. , Urbach, J.M. , Miyata, S. , Lee, D.G. , Drenkard, E. , Wu, G. , et al. (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proceedings of the National Academy of Sciences of the USA, 103, 2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, J.W. and Mount, D.W. (1982) The SOS regulatory system of Escherichia coli . Cell, 29, 11–22. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Li, Y. , Guo, Y. , Zeng, Z. , Li, B. , Wood, T.K. , et al. (2015) Physiological function of rac prophage during biofilm formation and regulation of rac excision in Escherichia coli K‐12. Scientific Reports, 5, 16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, T. (1960) Isolation of a bacteriophage specific for the F plus and Hfr mating types of Escherichia coli K‐12. Science, 131, 932–933. [DOI] [PubMed] [Google Scholar]
- Mai‐Prochnow, A. , Hui, J.G.K. , Kjelleberg, S. , Rakonjac, J. , McDougald, D. and Rice, S.A. (2015) Big things in small packages: the genetics of filamentous phage and effects on fitness of their host. FEMS Microbiology Reviews, 39, 465–487. [DOI] [PubMed] [Google Scholar]
- Marimon, O. , Teixeira, J.M.C. , Cordeiro, T.N. , Soo, V.W.C. , Wood, T.L. , Mayzel, M. , et al. (2016) An oxygen‐sensitive toxin‐antitoxin system. Nature Communications, 7, 13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, E. and Campos‐Gómez, J. (2016) Pf filamentous phage requires UvrD for replication in Pseudomonas aeruginosa . mSphere, 1, e00104–e00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin, D.A. (1998) Filamentous phage structure, infection and assembly. Current Opinion in Structural Biology, 8, 150–158. [DOI] [PubMed] [Google Scholar]
- Marvin, D.A. and Hoffmann‐Berling, H. (1963) Physical and chemical properties of two new small bacteriophages. Nature, 197, 517–518. [Google Scholar]
- Marvin, D.A. , Symmons, M.F. and Straus, S.K. (2014) Structure and assembly of filamentous bacteriophages. Progress in Biophysics and Molecular Biology, 114, 80–122. [DOI] [PubMed] [Google Scholar]
- McElroy, K.E. , Hui, J.G. , Woo, J.K. , Luk, A.W. , Webb, J.S. , Kjelleberg, S. , et al. (2014) Strain‐specific parallel evolution drives short‐term diversification during Pseudomonas aeruginosa biofilm formation. Proceedings of the National Academy of Sciences of the USA, 111, E1419–E1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Mooij, M.J. , Drenkard, E. , Llamas, M.A. , Vandenbroucke‐Grauls, C.M. , Savelkoul, P.H. , Ausubel, F.M. , et al. (2007) Characterization of the integrated filamentous phage Pf5 and its involvement in small‐colony formation. Microbiology, 153, 1790–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S. , Seshadri, R. , Varghese, N.J. , Eloe‐Fadrosh, E.A. , Meier‐Kolthoff, J.P. , Goker, M. , et al. (2017) 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nature Biotechnology, 35, 676–683. [DOI] [PubMed] [Google Scholar]
- Navarre, W.W. , Porwollik, S. , Wang, Y.P. , McClelland, M. , Rosen, H. , Libby, S.J. , et al. (2006) Selective silencing of foreign DNA with low GC content by the H‐NS protein in Salmonella . Science, 313, 236–238. [DOI] [PubMed] [Google Scholar]
- Nieto, J.M. , Madrid, C. , Miquelay, E. , Parra, J.L. , Rodriguez, S. and Juarez, A. (2002) Evidence for direct protein‐protein interaction between members of the Enterobacterial Hha/YmoA and H‐NS families of proteins. Journal of Bacteriology, 184, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir, G. and Sorek, R. (2018) Contemporary phage biology: from classic models to new insights. Cell, 172, 1260–1270. [DOI] [PubMed] [Google Scholar]
- Ono, S. , Goldberg, M.D. , Olsson, T. , Esposito, D. , Hinton, J.C.D. and Ladbury, J.E. (2005) H‐NS is a part of a thermally controlled mechanism for bacterial gene regulation. The Biochemical Journal, 391, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, O.E. , Schurr, J.R. , Schurr, M.J. and Sauer, K. (2011) The novel Pseudomonas aeruginosa two‐component regulator BfmR controls bacteriophage‐mediated lysis and DNA release during biofilm development through PhdA. Molecular Microbiology, 81, 767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, D. , Damron, F.H. , Mima, T. , Schweizer, H.P. and Yu, H.D. (2008) PBAD‐based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Applied and Environmental Microbiology, 74, 7422–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, S.A. , Tan, C.H. , Mikkelsen, P.J. , Kung, V. , Woo, J. , Tay, M. , et al. (2009) The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. The ISME Journal, 3, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S. , Haggard‐Ljungquist, E. and Nordstrom, K. (1987a) The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. The EMBO Journal, 6, 3191–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S. , Haggard‐Ljungquist, E. and Nordstrom, K. (1989) Activation of prophage P4 by the P2 Cox protein and the sites of action of the Cox protein on the two phage genomes. Proceedings of the National Academy of Sciences of the USA, 86, 3973–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S. , Lundqvist, B. and Haggardljungquist, E. (1987b) Autoregulation of bacteriophage‐P2 repressor. The EMBO Journal, 6, 809–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor, P.R. , Michaels, L.A. , Smigiel, K.S. , Rohani, M.G. , Jennings, L.K. , Hisert, K.B. , et al. (2017) Filamentous bacteriophage produced by Pseudomonas aeruginosa alters the inflammatory response and promotes noninvasive infection in vivo . Infection and Immunity, 85, e00648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor, P.R. , Sweere, J.M. , Michaels, L.A. , Malkovskiy, A.V. , Lazzareschi, D. , Katznelson, E. , et al. (2015) Filamentous bacteriophage promote biofilm assembly and function. Cell Host & Microbe, 18, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Plaks, J.G. , Homa, N.J. , Amrich, C.G. , Heroux, A. , Hatfull, G.F. , et al. (2014) The structure of Xis reveals the basis for filament formation and insight into DNA bending within a mycobacteriophage intasome. Journal of Molecular Biology, 426, 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.S. and Grainger, D.C. (2013) H‐NS can facilitate specific DNA‐binding by RNA polymerase in AT‐rich gene regulatory regions. PLoS Genetics, 9, e1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, C.K. , Pham, X.Q. , Erwin, A.L. , Mizoguchi, S.D. , Warrener, P. , Hickey, M.J. , et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- Takeya, K. and Amako, K. (1966) A rod‐shaped Pseudomonas phage. Virology, 28, 163–165. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor, M.K. and Mekalanos, J.J. (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science, 272, 1910–1914. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Kim, Y. , Ma, Q. , Hong, S.H. , Pokusaeva, K. , Sturino, J.M. , et al . (2010) Cryptic prophages help bacteria cope with adverse environments. Nature Communications, 1, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Kim, Y. and Wood, T.K. (2009) Control and benefits of CP4‐57 prophage excision in Escherichia coli biofilms. The ISME Journal, 3, 1164–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, J.S. , Lau, M. and Kjelleberg, S. (2004) Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. Journal of Bacteriology, 186, 8066–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Q. , Minh, P.N. , Dotsch, A. , Hildebrand, F. , Panmanee, W. , Elfarash, A. , et al (2012) Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Research, 40, 4320–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley, M. , Bangera, M.G. , Bumgarner, R.E. , Parsek, M.R. , Teitzel, G.M. , Lory, S. , et al (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature, 413, 860–864. [DOI] [PubMed] [Google Scholar]
- Wilgus, G.S. , Mural, R.J. , Friedman, D.I. , Fiandt, M. and Szybalski, W. (1973) λimmλ·434: A phage with a hybrid immunity region. Virology, 56, 46–53. [DOI] [PubMed] [Google Scholar]
- Will, W.R. and Frost, L.S. (2006) Characterization of the opposing roles of H‐NS and TraJ in transcriptional regulation of the F‐plasmid tra operon. Journal of Bacteriology, 188, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, W.R. , Navarre, W.W. and Fang, F.C. (2015) Integrated circuits: how transcriptional silencing and counter‐silencing facilitate bacterial evolution. Current Opinion in Microbiology, 23, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A. and Haggard‐Ljungquist, E. (1993) The Cox protein is a modulator of directionality in bacteriophage P2 site‐specific recombination. Journal of Bacteriology, 175, 7848–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. , Liu, X. , Yao, J. , Guo, Y. , Li, B. , Li, Y. , et al. (2016) Cold adaptation regulated by cryptic prophage excision in Shewanella oneidensis . The ISME Journal, 10, 2787–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


