Summary
Ethylene plays a critical role in many diverse processes in plant development. Recent studies have demonstrated that overexpression of the maize ARGOS8 gene reduces the plant's response to ethylene by decreasing ethylene signaling and enhances grain yield in transgenic maize plants. The objective of this study was to determine the effects of ethylene on the development of nodal roots, which are primarily responsible for root‐lodging resistance in maize. Exogenous application of the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) was found to promote the emergence of nodal roots. Transcriptome analysis of nodal tissues revealed that the expression of genes involved in metabolic processes and cell wall biogenesis was upregulated in response to ACC treatment, supporting the notion that ethylene is a positive regulator for the outgrowth of young root primordia. In BSV::ARGOS8 transgenic plants with reduced ethylene sensitivity due to constitutive overexpression of ARGOS8, nodal root emergence was delayed and the promotional effect of ACC on nodal root emergence decreased. Field tests showed that the BSV::ARGOS8 plants had higher root lodging relative to non‐transgenic controls. When ARGOS8 expression was controlled by the developmentally regulated promoter FTM1, which conferred ARGOS8 overexpression in adult plants but not in the nodal roots and nodes in juvenile plants, the FTM1::ARGOS8 plants had no significant difference in root lodging compared with the wild type but produced a higher grain yield. These results suggest that ethylene has a role in promoting nodal root emergence and that a delay in nodal root development has a negative effect on root‐lodging resistance in maize.
Keywords: ethylene, maize, nodal root, root lodging, ARGOS8, grain yield
Significance Statement
Root‐lodging resistance is a critically important trait for high‐yield maize hybrids and has been selected in maize breeding programs over the past 100 years. In maize, the stem‐borne nodal roots are formed in the normal developmental process and play a major role in root‐lodging resistance. Using the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) and ARGOS8 transgenic plants which have reduced ethylene sensitivity, we found that ACC promotes nodal root emergence and that ARGOS8 transgenic plants have delayed nodal root development and increased root lodging. These results suggest a positive role for ethylene in root‐lodging resistance. These findings have applications in maize breeding for improved agronomic traits.
Introduction
Roots perform functions that are necessary for plant growth and development. Roots absorb water and nutrients and anchor plants in the soil. In maize, the root system consists of embryonic roots (i.e. the primary root and lateral seminal roots), stem‐borne nodal roots and lateral roots that are formed from all roots (Abbe and Stein, 1954; Feldman, 1994). The stem‐borne roots, including the crown roots at the basal nodes of the stem and the brace roots which emerge from the nodes above the soil surface and quickly enter the soil, form the main framework of the underground root system and are the basis for root‐lodging resistance (Hochholdinger, 2009). Root lodging is the inability of plants to stand upright for optimum growth and harvestability (Ennos et al., 1993; Landi et al., 2007). The maize rtcs (rootless concerning crown and seminal roots) mutant deficient in the formation of nodal roots falls over at early developmental stages and cannot survive in typical agronomic environments without human intervention (Hetz et al., 1996). Maize root lodging often occurs during the mid‐growing season when heavy rainfall saturates soils and strong winds displace the root system from its normal vertical axis. In root‐lodged plants, uptake of water and nutrients, as well as interception of light by the canopy, can be reduced and in some cases pollination is delayed, leading to reduced grain yield and poor grain quality (Carter and Hudelson, 1988; Pellerin et al., 1990; Stamp and Kiel, 1992; Guingo and Hébert, 1997; Duvick, 2005; Liu et al., 2012). Harvesting of root‐lodged fields is often difficult and can result in additional yield losses.
Maize nodal roots develop sequentially at each of the seven or eight lowest nodes, beginning at the coleoptile node (Hoppe et al., 1985). These roots originate from cells next to the peripheral cylinder of vascular bundles in the stem, with young root primordia growing through the cortex, penetrating the nodal epidermis and emerging from the stem. Phytohormones play an important role in regulating the development of stem‐borne roots (often referred to as adventitious roots, which include all roots formed from non‐root tissues; for reviews see Orman‐Ligeza et al., 2016; Bellini et al., 2014; Steffens and Rasmussen, 2013). Genetic studies in rice, tomato and Arabidopsis have shown that pin‐formed (PIN) and AUX1 families of auxin efflux and influx carriers are required for the initiation and development of adventitious roots (Tyburski and Tretyn, 2004; Xu et al., 2005; Morita and Kyozuka, 2007; Kitomi et al., 2008; Liu et al., 2009; Negi et al., 2010; Vidoz et al., 2010; Pacurar et al., 2014). Auxin induces the expression of the maize RTCS gene and the rice ortholog ADVENTITIOUS ROOTLESS1 (ARL1; also known as CROWN ROOTLESS1 or CRL1), which are essential for the initiation of stem‐borne roots (Liu et al., 2005; Taramino et al., 2007). The rice ARL1/CRL1 and CROWN ROOTLESS5 (CRL5; an AP2/ERF transcription factor) are targets of the auxin response factor gene family (Inukai et al., 2005; Kitomi et al., 2011). It has been reported that the rice Oscand1 mutant is defective in crown root emergence and the gene responsible for the mutation is involved in auxin signaling to maintain the G2/M cell cycle transition in the crown root meristem (Wang et al., 2011). Cytokinins are antagonistic to auxin and suppress the formation of adventitious roots. In rice, treatment with cytokinin inhibits crown root initiation and loss‐of‐function mutation of WUSCHEL‐related homeobox 11 (WOX11), a repressor of cytokinin signaling, reduces the number and the growth rate of crown roots (Zhao et al., 2009). The rice CRL5 protein promotes crown root initiation through repression of cytokinin signaling by positively regulating type‐A response regulators (Kitomi et al., 2011).
Ethylene is another hormone in the hormonal regulatory network regulating adventitious root development (for review see Steffens and Rasmussen, 2016). In deepwater rice, tomato and Rumex palustris, submergence in flood water increases ethylene biosynthesis and ethylene induces adventitious root formation (Visser et al., 1996; Lorbiecke and Sauter, 1999; Kim et al., 2008; Vidoz et al., 2010). In tomato, the Nr (Never ripe) mutant defective in ethylene signaling has fewer adventitious roots in young seedlings than the wild type (WT), while the epinastic mutant, which has elevated ethylene and constitutive ethylene signaling, has a greater number of adventitious roots (Negi et al., 2010). Exogenous application of the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) promotes adventitious rooting in WT plants. Treatment with ACC was found to negatively regulate transport of indole‐3‐acetic acid (IAA) in hypocotyls, whereas the Nr mutation increased IAA transport. Negi et al. (2010) proposed that ethylene regulation of adventitious root formation in tomato hypocotyls is auxin dependent. Clark et al. (1999) also reported that ACC promotes adventitious root formation in vegetative cuttings taken from older tomato plants, and showed that ethylene signaling is required for auxin‐induced adventitious root formation in tomato and petunia (Clark et al., 1999). These tomato studies, however, did not determine if ethylene promotes the initiation of root primordia or the outgrowth of young root primordia through the cortex (i.e. emergence). In deepwater rice, ethylene enhances adventitious root development during flooding by promoting the emergence of the root primordia which have been formed during normal development (Lorbiecke and Sauter, 1999). However, the role of ethylene in maize nodal root development is far less clear, and that is the focus of this study.
Previously, we have generated transgenic maize plants with reduced ethylene sensitivity by overexpressing the maize ARGOS8 gene (Shi et al., 2015, 2016). ARGOS8 is a member of the ARGOS gene family which contains a conserved TPT domain (Shi et al., 2016). ARGOS8 is expressed in developing kernels, but the transcript abundance in other tissues is very low (Shi et al., 2017). The ARGOS8 protein is a component of the ethylene receptor signaling complex, interacting with the reversion‐to‐ethylene sensitivity1 like (RTL) proteins (Shi et al., 2016). When overexpressed, ARGOS8 reduces the ethylene response in transgenic maize and Arabidopsis plants by reducing ethylene signal transduction (Shi et al., 2015, 2016). Maize hybrids with constitutive ARGOS8 overexpression produce high grain yield under drought‐stressed and well‐watered conditions (Shi et al., 2015, 2017). Because of the constitutive expression patterns of the transgene or the genome‐edited allele, it is expected that ethylene signaling can be downregulated in tissues where reduced ethylene signaling is unnecessary and even harmful to achieving high grain yield. Given the promotional effect of ethylene on adventitious root development in rice and the predominant role of nodal roots in the resistance of maize plants to root lodging, we sought to determine whether ethylene is involved in maize nodal root development and whether reduced ethylene signaling affects root‐lodging resistance.
In this report, ACC treatments and ARGOS8 transgenic plants with reduced ethylene sensitivity were used to determine the effect of ethylene on nodal root emergence. We also examined transcriptional changes in nodal tissues upon ACC treatment and in plants with reduced ethylene sensitivity. Root lodging of transgenic plants with ARGOS8 overexpression driven by constitutive promoters and a developmentally regulated promoter was evaluated in field conditions. The results show that ACC treatments promote nodal root emergence and plants with constitutive ARGOS8 overexpression have delayed nodal root emergence as well as reduced root‐lodging resistance. When the developmentally regulated FTM1 promoter was used to restrict ARGOS8 overexpression to adult plants, the transgenic plants withstood the effects of inclement weather like the non‐transgenic controls but had a higher grain yield.
Results
ACC promotes nodal root emergence in maize
The greenhouse‐grown maize hybrid PHR03/PH12SG developed the first tier of brace roots on node 6, which also bears the fifth leaf (the coleoptile node designated node 1; Hoppe et al., 1985). The brace roots grew into the soil and developed lateral roots. In plants at vegetative stage 8 (V8), the young root primordia had formed on nodes 6 and 7 but were arrested, appearing as a circle of deep green dots and surrounded by a pale green halo underlying the stem epidermis (Figure 1a). The V8 plants were treated with the ethylene precursor ACC by watering them with an aqueous solution of 1 L per plant per day of 0, 50 or 100 μm acc. Brace roots began to emerge on node 6 3 days after initiation of the treatment, and subsequently on node 7 2 days later (Figure 1b). At day 7, the 50 μm ACC‐treated plants had on average 16.2 and 6.1 roots on nodes 6 and 7, respectively, but only 1.4 roots were found on node 6 in water controls (Figure 1c). In the plants treated with 100 μm ACC, almost all primordia on both node 6 and node 7 had broken through the stem surface by day 7. The total number of primordia on node 7, including emerged and unemerged, was not affected by the ACC treatment (Figure 1d). The application of ACC increased ethylene biosynthesis in the plants, as revealed by increased ethylene emission in leaves (Figure 1e), indicating that ethylene is most likely responsible for the promotion of nodal root emergence.
Figure 1.
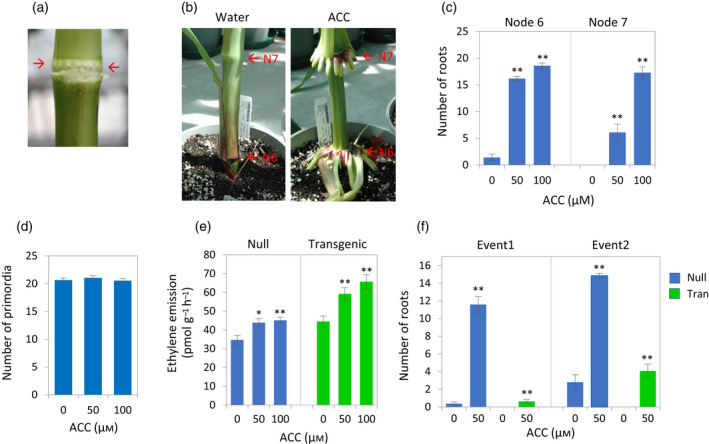
Effects of treatments with the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) on maize brace root emergence and ethylene evolution
(a) Node 6 of a maize hybrid PHR03/PH12SG plant at V10 with the leaf sheath removed showing root primordia (arrows). (b) Images of node 6 (N6) and node 7 (N7) of the plants that were treated with water or 100 μm ACC for 7 days starting at V8. (c) Number of brace roots in the ACC‐treated plants 6 days after initiation of the treatment (n = 10; error bar, SE). The ACC‐treated plants were compared with water controls (Student's t‐test, ** P < 0.01). (d) Number of primordia on node 7, including emerged and unemerged primordia. The plants were treated with ACC for 7 days starting at V9 and scored 8 days after the end of the ACC treatment (n = 20; error bars, SE). (e) Ethylene emission rates in the eighth leaves of ACC‐treated plants and water controls. Samples were taken from the wild‐type (i.e. non‐transgenic segregants; null) and BSV::ARGOS8 transgenic plants at V11 which had been treated with ACC for 5 days (n = 10; error bar, SE). The ACC‐treated plants were compared with water controls (Student's t‐test, *P < 0.05). (f) Number of brace roots on node 6 in the non‐transgenic segregants (null) and BSV::ARGOS8 transgenic plants (Tran) treated with 0 or 50 μm ACC for 7 days starting at V10 (n = 24; error bar, SE). The ACC‐treated plants were compared with water controls (Student's t‐test, **P < 0.01).
To confirm the promotional effect of ACC on nodal root emergence, we tested ARGOS8 transgenic plants, which have reduced ethylene sensitivity (Shi et al., 2015, 2016). In the transgenic plants, maize ARGOS8 was overexpressed under control of the banana streak virus (BSV) promoter which confers strong constitutive expression (Schenk et al., 2001). To determine the brace root response, the BSV::ARGOS8 hybrid PHR03/PH12SG plants were treated at V10 with 50 μm ACC. The ACC treatment did not affect the transcript abundance of the ARGOS8 transgene (Figure S1 in the online Supporting Information), as expected. Like WT plants, the ACC‐treated transgenic plants had increased ethylene emission relative to water controls (Figure 1e). Although the ACC treatment also promoted the emergence of brace roots on node 6 in the transgenic plants, the magnitude of the effect was considerably smaller than that in non‐transgenic segregants (Figure 1f). On node 7, brace roots were observed in some of the non‐transgenic plants 7 days after the initiation of the treatment, but not in any transgenic plants.
Maize seedlings grown in the greenhouse were used to study underground nodal root response. When 17‐day‐old seedlings were treated with ACC, nodal roots began to emerge on node 5 in the treated non‐transgenic plants 6 days after the initiation of treatment. In contrast, there were no roots visible in water (0 μm ACC) controls until 4 days later (Figure 2a). By this time, most of the root primordia on node 5 in 100 μm ACC‐treated plants had penetrated through the node epidermis. Like the brace roots, the underground nodal roots in the BSV::ARGOS8 transgenic plants were not as responsive to ACC as those in the non‐transgenic plants (Figure 2a,b). At 3 days after the end of the 16‐day ACC treatment, the 50 μm ACC‐treated non‐transgenic plants had on average 11.3 roots on node 5 with a mean root length of 152.1 mm, while the transgenic plants had 8.3 nodal roots on the same node and the root length averaged 57.7 mm (Figure 2c,d). For the plants treated with 100 μm ACC, nearly all the root primordia on node 5 had emerged in both transgenic and non‐transgenic plants and the number of nodal roots was comparable, but the non‐transgenic segregants had longer roots (Figure 2c,d). The total number of primordia on node 5, including emerged and unemerged, was not significantly different between the transgenic and non‐transgenic plants or between ACC‐treated and water controls (Figure S2), suggesting that both ACC treatment and ARGOS8 overexpression affected nodal root emergence rather than initiation of root primordia.
Figure 2.
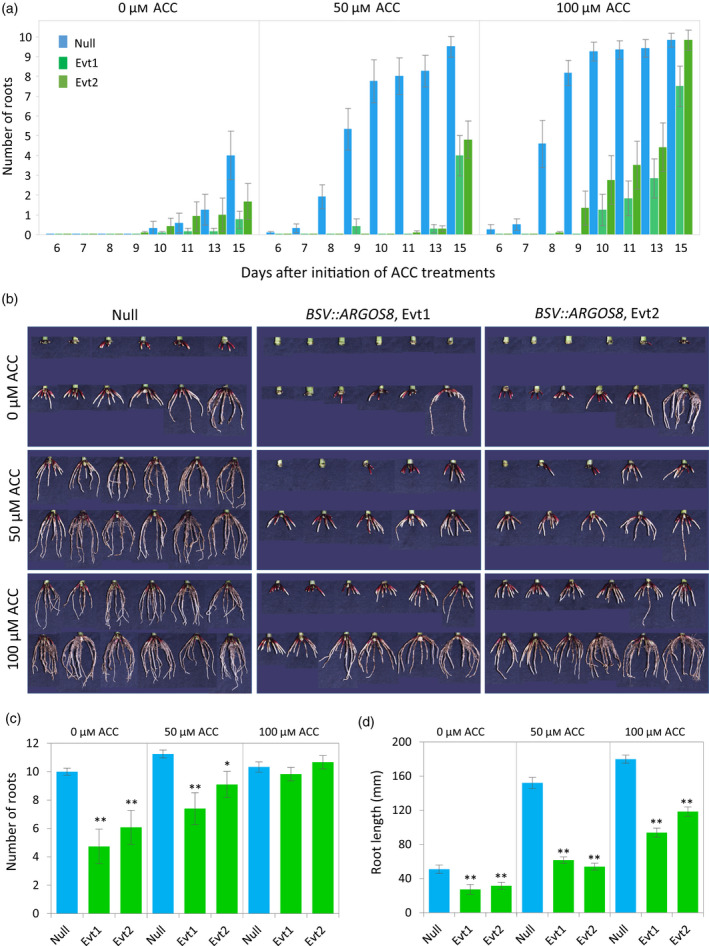
Emergence and growth of underground nodal roots on node 5 in maize plants.
(a) Time courses of nodal root emergence in response to 1‐aminocyclopropane‐1‐carboxylic acid (ACC) treatment in the BSV::ARGOS8 transgenic plants and non‐transgenic controls. Seventeen‐day‐old seedlings of two events (Evt1 and Evt2) and bulked non‐transgenic segregants (Null) were treated with 0, 50 or 100 μm ACC for 16 days (n = 12; error bar, SE).
(b) Images of isolated node 5 with nodal roots from 10 or 12 individual plants per genotype and treatment 3 days after the end of the ACC treatment.
(c) Number of roots on node 5 (n = 12; error bar, SE). Transgenic plants (Evt1 and Evt2) were compared with Null (Student's t‐test, *P < 0.05, **P < 0.01).
(d) Length of roots on node 5. The data represent means of the length of individual roots (n = 61–135; error bar, SE).
Under normal growth conditions without exogenously applied ACC (i.e. water controls), nodal root emergence was delayed in the BSV::ARGOS8 transgenic plants compared with the non‐transgenic segregants (Figure 2a). In the 35‐day‐old plants with overlying leaf sheaths removed, nodal roots were found on node 5 in all non‐transgenic plants examined (Figure 2b). Of the 24 transgenic plants from two events, only 17 had nodal roots. The number of node 5 roots per plant was higher and the root length longer in the non‐transgenic plants (Figure 2c,d). In the V14 plants, brace roots were present on node 6 in 20 out of 28 non‐transgenic plants examined, averaging 8.7 roots per plant, while only one plant with two roots was found from the same number of transgenic plants (Figure S3). The delay of nodal root emergence in the transgenic plants overexpressing ARGOS8 further supports the hypothesis that ethylene is involved in the regulation of nodal root emergence in maize.
Greater root lodging in BSV::ARGOS8 transgenic plants
To determine if a delay in nodal root emergence affects resistance to root lodging, BSV::ARGOS8 transgenic and non‐transgenic hybrids were grown in fields in nine locations across the USA in 2013. Root lodging was evaluated in typical agronomic environments. Out of the nine locations, seven did not have lodging pressure. Only two locations experienced a combination of strong winds and heavy rainfall before flowering, allowing discrimination of root lodging. Compared with non‐transgenic controls, the transgenic plants had a significantly higher frequency of root lodging (Table 1).
Table 1.
Root lodging and grain yield in maize PHR03/PH12SG hybrid overexpressing ARGOS8. Transgenic plants (Trans) of three ARGOS8 constructs (nine to ten events per construct) and non‐transgenic controls (Null) were grown in nine locations across the USA in 2013. Two locations had significant root‐lodging pressures allowing us to evaluate root‐lodging resistance. All analyses were implemented using ASReml with output of the model presented as best linear unbiased predictions
| Trait | Location | Construct | Trans | Null | Difference (95% CI) | n | P‐value |
|---|---|---|---|---|---|---|---|
| Per cent | Per cent | ||||||
| Root‐lodged plants | Location 1 | BSV::ARGOS8 | 48.9 | 13.0 | 35.7 (24.9–46.5) | 27 | 0.00 |
| GOS2::ARGOS8 | 18.2 | 13.0 | 5.2 (−5.4 to 15.7) | 30 | 0.32 | ||
| FTM1::ARGOS8 | 16.8 | 13.0 | 3.6 (−6.9 to 14.0) | 30 | 0.50 | ||
| Location 2 | BSV::ARGOS8 | 6.3 | 0.6 | 5.7 (2.4–9.1) | 26 | 0.00 | |
| GOS2::ARGOS8 | 1.0 | 0.6 | 0.5 (−2.9 to 3.8) | 29 | 0.78 | ||
| FTM1::ARGOS8 | 0.9 | 0.6 | 0.4 (−3.0 to 3.7) | 29 | 0.83 | ||
| bu ac−1 | bu ac−1 | ||||||
| Grain yield | All locations | BSV::ARGOS8 | 144.2 | 143.0 | 1.2 (−0.5 to 2.9) | 733 | 0.17 |
| GOS2::ARGOS8 | 147.8 | 143.0 | 4.8 (3.1–6.4) | 826 | 0.00 | ||
| FTM1::ARGOS8 | 145.8 | 143.0 | 2.8 (1.1–4.4) | 830 | 0.00 |
n = total comparisons: number of locations × number of events × replicates.
A reduction in ethylene sensitivity in the transgenic plants overexpressing ARGOS8 driven by the maize UBIQUITIN1 promoter (UBI1) had been shown to improve grain yield under drought stress conditions and in well‐watered environments (Shi et al., 2015). Relative to the UBI1 promoter, the BSV promoter conferred a considerably higher level of constitutive ARGOS8 overexpression (Figure 3a). In the 2013 field test, the transgenic PHR03/PH12SG hybrid plants gave a yield similar to non‐transgenic controls (Tables 1 and S1). The reason for the lack of increase in yield in the BSV::ARGOS8 plants is that the highly expressed ARGOS8 may have reduced ethylene sensitivity too much (Shi et al., 2015), while high grain yield requires an optimal level of ethylene signaling in transgenic plants. In the same genetic background, moderate expression of ARGOS8 under control of the maize constitutive GOS2 promoter (Shi et al., 2017), which confers weaker expression than UBI1 (Figure 3), was found to increase grain yield by 4.8 bushels per acre (bu ac−1) (Tables 1 and S1). Although the percentage of root‐lodged GOS2::ARGOS8 plants was slightly higher than that in non‐transgenic controls, the difference was not statistically significant (Table 1). However, further testing of the GOS2::ARGOS8 construct in different hybrids revealed that the effects of ARGOS8 overexpression on root lodging are influenced by genetic background. Hybrid plants derived from the GOS2::ARGOS8 transgenic PH184C inbred line developed brace roots later than WT controls in the field (Figure 4a,b). Greenhouse‐grown transgenic PH184C/PH1V69 plants also showed delayed nodal root development and reduced promotional effects of ACC on nodal root emergence relative to WT controls (Figure S4). In a multiple‐location field test in 2016, the GOS2::ARGOS8 transgenic hybrids had a significantly higher percentage of root‐lodged plants than WT controls (Figure 4c). However, these hybrids had a greater grain yield than controls (Figure S5). These results confirmed that constitutively overexpressing ARGOS8 delays nodal root development and negatively affects root‐lodging resistance.
Figure 3.
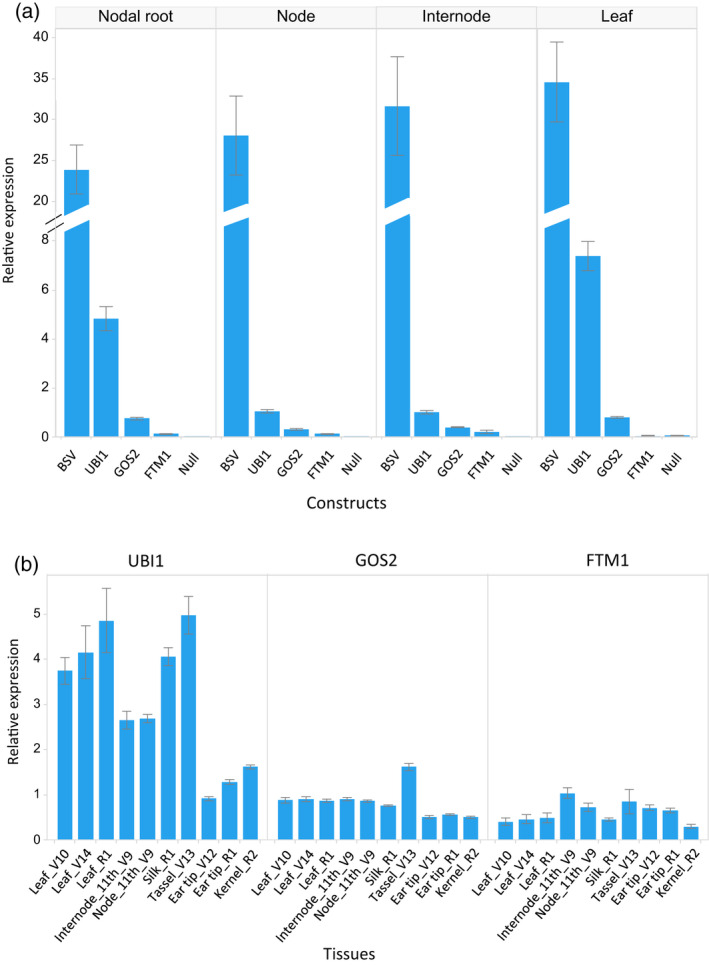
Transcript expression in transgenic ARGOS8 and control plants
(a) Relative expression levels of maize ARGOS8 transcripts in greenhouse‐grown 24‐day‐old transgenic and non‐transgenic plants (Null). Quantitative RT‐PCR was conducted using ARGOS8‐specific primers. Two events per construct (promoter BSV, UBI1, GOS2 or FTM1) and eight replicates per event (error bars, SE).
(b) Relative expression levels of the ARGOS8 transgene in field‐grown transgenic plants. Quantitative RT‐PCR was conducted using primers derived from the OS‐UBIQUITIN terminator used in the constructs. Three events per construct and three replicates per event (error bars, SE).
Figure 4.
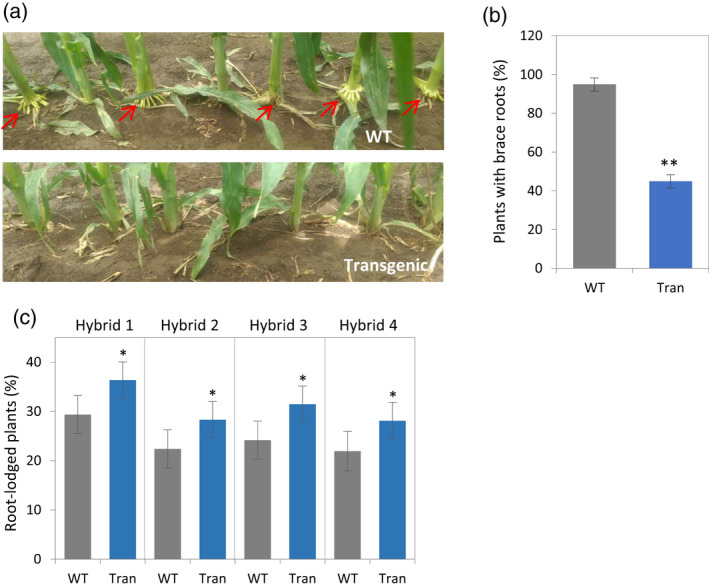
Nodal roots and root lodging of the GOS2::ARGOS8 transgenic plants grown in the field.
(a) Images of the transgenic and wild type (WT) hybrid PH184C/PH1V69 plants 10 days before silking grown in the field in 2017 showing delayed nodal root development in the transgenic plants. Red arrows indicate brace roots on the lowest aboveground node in WT plants.
(b) Percentage of plants with brace roots in the GOS2::ARGOS8 transgenic (Tran) and WT PH184C/PH1V69 plants. Two‐row plots with 57–58 plants per plot (n = 2; error bars, SE; Student's t test, **P < 0.01).
(c) Percentage of root‐lodged plants in the GOS2::ARGOS8 transgenic (Tran) hybrids and WT controls. Four hybrids were produced by crossing the transgenic PH184C inbred with four inbred testers. Transgenic plants and WT controls were planted in seven locations across the USA in 2016 (hybrid 1, PH184C/PH1V69). Transgenic plants were compared with WT. Data analyses were implemented using ASReml with output of the model presented as best linear unbiased predictions (error bars, SE; *P < 0.05).
To determine if increased grain yield and reduced root‐lodging resistance can be decoupled, we tested the transgenic plants which overexpressed ARGOS8 under control of the non‐constitutive FTM1 promoter. The FTM1 promoter was derived from the maize FLORAL TRANSITION MADS1 gene (also known as ZMM4; Danilevskaya et al., 2008). This promoter confers developmentally regulated gene expression, primarily after the transition from vegetative to reproductive growth in developing apical and lateral inflorescences and to a lesser extent in several other tissues in adult plants, but not in embryonic and juvenile tissues (Danilevskaya et al., 2008). For 24‐day‐old seedlings, there were very few transcripts of ARGOS8 in nodal roots and nodes in the FTM1::ARGOS8 transgenic plants, similar to that found in non‐transgenic plants (Figure 3a). Among the four constructs, the highest level of ARGOS8 expression in those tissues was found in the BSV::ARGOS8 plants (Figure 3a). The absence of ARGOS8 overexpression in tissues of the juvenile plants implies unaffected ethylene sensitivity in the FTM1::ARGOS8 transgenic plants at early developmental stages. In adult FTM1::ARGOS8 plants, ARGOS8 was overexpressed in leaves, stalks, tassels, silks, cobs and developing seeds, and the abundance of the transcripts were nearly as high as in the GOS2::ARGOS8 plants (Figure 3b). Field tests showed that the FTM1::ARGOS8 hybrid PHR03/PH12SG yielded 2.8 bu ac−1 more than non‐transgenic plants (P < 0.05; Tables 1 and S1). The percentage of root‐lodged plants was not significantly different between the transgenic plant and controls in the two locations where root‐lodging pressure existed (Table 1). These results indicate that restricting ARGOS8 overexpression to adult plants can minimize the risk of root lodging and increase grain yield in maize.
Transcript profiles of nodal tissues in ACC‐treated plants
Ethylene‐induced transcriptional changes in nodal tissues were investigated with RNA sequencing. Wild‐type plants (21 days old) were treated with 100 μm ACC. The BSV::ARGOS8 transgenic plants which have reduced ethylene sensitivity were included for comparison with untreated WT plants. To identify differentially expressed (DE) genes prior to nodal root emergence, a cross section (about 2 mm thick) of node 5, containing root primordia, was excised 48 and 72 h after the initiation of the ACC treatment (Figure 5a). The differential expression at the gene level was analyzed in pairwise comparisons. The DE genes are defined as those with false discovery‐corrected P < 0.05 and an absolute value of log2(fold change) ≥ 1.
Figure 5.
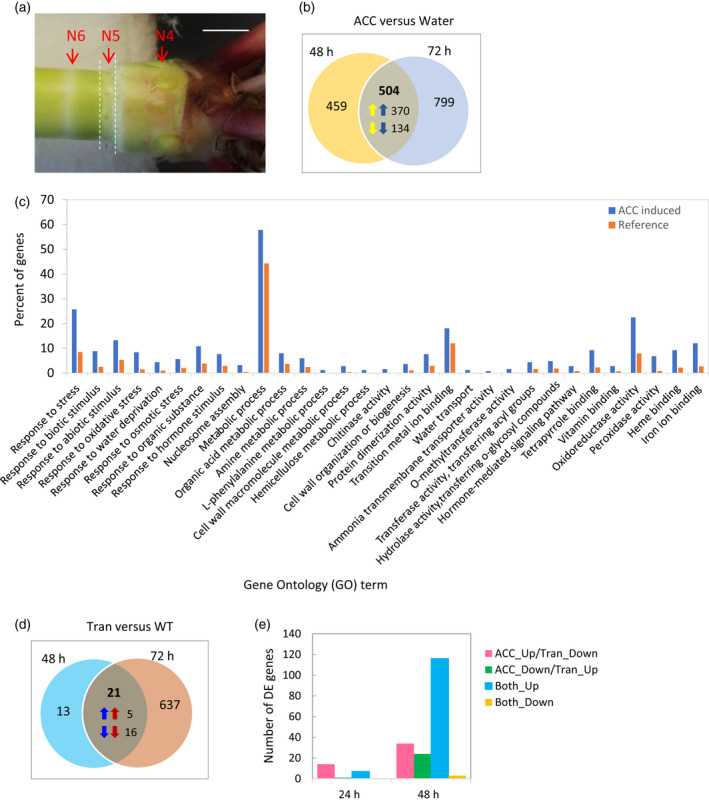
Ethylene‐induced transcriptomic changes in nodes of maize plants
(a) Image of basal nodes of a 21‐day‐old seedling showing nodes 4, 5 and 6 (N4, N5 and N6). Dashed lines indicate the sampling regions. Bar = 5 mm.
(b) Venn diagram showing differentially expressed (DE) genes in nodal tissues treated with 0 or 100 μm ACC at 48 and 72 h. Arrows indicate up‐ or down‐regulation by 1‐aminocyclopropane‐1‐carboxylic acid (ACC).
(c) Overrepresentation of Gene Ontology terms (false discovery‐corrected P < 0.05) among the ACC upregulated DE genes.
(d) Venn diagram showing DE genes between the BSV::ARGOS8 transgenic plants and WT plants.
(e) Number of DE genes in four groups classified based on response to ACC treatments (ACC) in WT and response to overexpression of the BSV::ARGOS8 transgene (Tran) in untreated plants (Up, upregulated; Down, downregulated).
Comparisons of the ACC‐treated WT plants with water controls revealed that 963 and 1303 genes were DE 48 and 72 h after the initiation of the treatment, respectively. There were 504 overlapping DE genes between the two sets (Figure 5b, Table [Link], [Link]). Among those, 370 were upregulated and 134 downregulated by ACC and the direction of change of expression of these genes was consistent between the two time points. The function of the ACC upregulated genes was explored using the Gene Ontology (GO) classification system. The significantly enriched GO categories include metabolic process (e.g. organic acid, amine, l‐phenylalanine, hemicellulose and cell wall macromolecule metabolism; methyltransferase, acyl transferase and hydrolase activity), nucleosome assembly, protein dimerization, transition metal ion binding and cell wall biogenesis (Figure 5c, Table [Link], [Link]). These categories are related to cell growth. Increased expression of these genes is consistent with the faster outgrowth of the young root primordia and emergence of nodal roots in the ACC‐treated plants. Consistent with the well‐known role of ethylene in stress responses and defense to infection, the GO terms response to biotic stimulus, oxidative stress, osmotic stress, water deprivation, organic substance and hormone stimulus were significantly enriched in this set of ACC‐induced genes. In addition, the GO terms oxidoreductase activity (GO 16491) and peroxidase activity (GO 4601) were overrepresented in the ACC‐treated plants (Figure 5c, Table [Link], [Link]). The ACC‐upregulated genes include 17 putative peroxidases. It has been reported that water deficit suppresses the expression of several peroxidase genes in the crown region and significantly reduces the number of nodal roots in the model C4 grass Setaria viridis (Sebastian et al., 2016). In rice, NADPH oxidase was the major enzyme responsible for production of reactive oxygen species (ROS) which mediate the ethylene‐induced growth of the young root primordia (Steffens et al., 2012). For the 134 ACC‐suppressed genes, only two GO terms, transcription factor activity (GO 3700) and O‐acyltransferase activity (GO 8374), were significantly enriched (Table [Link], [Link]).
When the RNA sequencing data from BSV::ARGOS8 plants were compared with those from the WT, 12 genes were found to be upregulated and 22 downregulated at 48 h (i.e. 23‐day‐old plants) in the transgenic plants (Figure 5d, Table [Link], [Link]). A larger number of DE genes were detected at 72 h, 548 being upregulated and 110 downregulated. A comparison of these DE genes with those identified in the ACC treated plants revealed 63 genes that were upregulated by ACC and downregulated in the transgenic plants, or vice versa (Figure 5e, Table [Link], [Link]). One of these genes is the AP2 EREB transcription factor 114 (EREBP114; GRMZM2G008234) which is the maize ortholog of the rice CROWN ROOTLESS5 (Kitomi et al., 2011; Schnable et al., 2012). The others include a receptor protein serine/threonine kinase (GRMZM2G075438), an ethylene signaling component (RTL3, GRMZM2G121208), 10 transcriptional factors [AP2/ERF (1), NAC (1), WUSCHEL‐related homeobox (1), basic helix–loop–helix (2), MYB‐related (3) and C2H2‐type zinc finger (2)], 20 enzymes (e.g. trehalose‐6‐phosphate synthase, GRMZM5G890599; phytosulfokines2, GRMZM2G146222; anionic peroxidase1, GRMZM2G108207; heme peroxidase, AC197758.3_FG004; respiratory burst NADPH oxidase, GRMZM2G300965) and three cortical cell‐delineating proteins (Table [Link], [Link]).
Discussion
The promotion of stem‐borne adventitious root formation by ethylene under stressed conditions is well known in rice and tomato plants (Clark et al., 1999; Lorbiecke and Sauter, 1999; Negi et al., 2010; Vidoz et al., 2010). In maize, nodal roots originate at the stem nodes during the normal developmental process and form the framework of the root system. In this study, BSV::ARGOS8 transgenic plants with reduced ethylene signaling and ACC treatments were used to determine if ethylene plays a role in nodal root development in maize. We found that nodal root emergence was accelerated in maize plants treated with ACC. The transgenic plants with reduced ethylene sensitivity due to ARGOS8 overexpression showed delayed emergence of nodal roots. As expected, the promotional effect of ethylene on nodal roots was less pronounced in the ACC‐treated transgenic plants than in the WT. It has been reported that the ethylene‐producing growth regulator ethephon promoted the development of brace roots in field‐grown maize hybrids at growth stage V10 (Langan and Oplinger, 1987). In another study, maize seedlings growing in liquid media through which ethylene was bubbled showed increased adventitious root formation (Jackson et al., 1981). Taken together, these results demonstrate that ethylene is a positive regulator of nodal root development in maize.
Although nodal roots emerged earlier in the ACC‐treated maize plants, the sequence of nodal root development along the stem remained unchanged. As in the untreated plants, the roots on a node in the ACC‐treated plants did not penetrate the stem epidermis until the roots on the lower node had emerged, indicating that there are other regulators which control the growth of root primordia and certain conditions must be met in the node before ethylene can exert its promotional effect. In deepwater rice, the epidermal cells overlying the tips of the root primordia are a physical barrier restricting the emergence of the stem‐borne roots, and programmed cell death in the epidermal cells is required for the root primordia to emerge from the stem surface (Lorbiecke and Sauter, 1999; Steffens et al., 2012). However, the role of the epidermal cells in restricting root primordia in maize is not clear. For the nodes examined in this study, root primordia had formed within the nodal tissues prior to ACC treatments (Figure 1a). The accelerated emergence of nodal roots was a result of enhanced growth of the young root primordia, which was reflected in the transcriptomic changes in the nodal tissues. A GO analysis of the DE genes revealed that the ACC‐upregulated genes are associated with GO categories nucleosome assembly, protein dimerization, metabolic process, cell wall biogenesis and transporter activities. Increased expression of the genes in these GO categories can support active cell division and expansion, leading to fast tissue growth. It has been shown that accelerated growth of the young root primordia in rice generates a mechanical force which is one of the two signals (the other one being ethylene) that regulate programmed cell death of the epidermal cells overlying the root primordia, facilitating root emergence in rice (Steffens et al., 2012).
Root lodging is one of the major problems in high‐yield maize production. Early establishment of nodal roots and adequate nodal root development can provide plants resistance to root lodging (Pellerin et al., 1990; Stamp and Kiel, 1992; Ennos et al., 1993). Extensive breeding efforts have taken place to develop lodging‐resistant hybrids, but as complex traits, selection for root‐lodging resistance and simultaneous selection for high grain yield is quite challenging. Targeted genetic engineering can be an alternative approach. Two maize mutants with defects in nodal root development have been isolated. The rtcs mutant is devoid of nodal roots (Hetz et al., 1996). The RTCS gene encodes an auxin‐inducible LOB domain protein acting downstream of the auxin response factor ARF34 (Taramino et al., 2007; Majer et al., 2012; Xu et al., 2015). The rootless1 mutant produces fewer crown roots at the first two nodes, and lacks brace roots (Jenkins, 1930). These genes can be overexpressed in transgenic plants to test if they can enhance nodal root formation. In this study, we found that modulation of ethylene signaling also can alter nodal root development, uncovering new targets (i.e. ethylene pathway genes) for regulating root‐lodging resistance. Because reduced ethylene signaling delays nodal root emergence and increases root lodging in the BSV::ARGOS8 transgenic plants and ACC treatments promote nodal root formation, increasing ethylene signaling by altering the expression of ethylene pathway genes in a tissue‐specific fashion could enhance resistance to root lodging. In addition, transcript profiling of the nodal tissues revealed other candidate genes for improving root‐lodging resistance, such as EREBP114, the maize ortholog of the rice CROWN ROOTLESS5 (Kitomi et al., 2011), and WUSCHEL‐RELATED HOMEOBOX 4, which belongs to the same gene family as the rice WOX11 (Zhao et al., 2009), respiratory burst NADPH oxidase and ROS pathway genes.
Ethylene promotes nodal root emergence, which could lead to improved resistance of the plant to root lodging. However, ethylene is involved in many aspects of plant growth and development as well as the response of plants to both biotic and abiotic stresses. Maize can show enhanced grain yield when the sensitivity of the plant to ethylene is reduced or expression of an ethylene biosynthesis gene is silenced (Habben et al., 2014; Shi et al., 2015, 2017). When constitutively overexpressed, ARGOS8 increased kernel number per ear (Shi et al., 2015), but delayed nodal root emergence and reduced plant resistance to root lodging. Using the developmentally regulated FTM1 promoter for ARGOS8 overexpression in adult plants, we obtained transgenic plants which have a greater yield and have minimal root‐lodging problems. The FTM1::ARGOS8 transgene is expressed at a low level in the basal nodes of the stem and nodal roots in young plants; consequently the effect on ethylene signaling in these tissues was negligible. This study shows that selective modification of ethylene signaling at different developmental stages can decouple improved grain yield from root lodging.
Experimental Procedures
Plant materials and growth conditions
The seeds of maize hybrid PHR03/PH12SG and transgenic plants were sown in Ellepot® plugs (Ellepot A/S, https://www.ellepot.com/) in 4‐ by 8‐cell flats and 2‐week‐old seedlings were transferred to 2‐L pots and grown in the greenhouse. The soil mixture consisted of peat (48.7%), Turface (44.6%; https://www.turface.com/), perlite (2.3%), vermiculite (4.4%), lime [3 pounds per cubic yard (lbs yd−3)], Peters’ Professional 11‐5‐11 UniMix Starter (1 lb yd−3; ICL Specialty Fertilizers, https://icl-sf.com/), Suffusion wetting agent (0.5 lbs yd−3; OHP Inc., http://www.ohp.com/) and Osmocote Plus 15‐9‐12 controlled‐release fertilizer (3 lbs yd−3; ICL Specialty Fertilizers). Nutrient was supplemented once a week with water‐soluble fertilizer dissolved in tap water (95 p.p.m.; Peters Excel 15‐5‐15 Cal‐Mag Special; ICL Specialty Fertilizers). To determine the effects of ethylene on nodal root formation, plants at various vegetative growth stages were treated with the ethylene precursor ACC (Accela ChemBio Inc., http://www.accelachem.com/) by watering them daily with an aqueous solution of ACC at three concentrations (0, 50 or 100 μm). Plant vegetative growth stages were determined using the leaf collar method. Stem nodes were numbered according to their order of appearance, the coleoptile node being designated node 1 (Hoppe et al., 1985).
Transgene constructs and plant transformation
To generate constructs overexpressing maize ARGOS8 in maize plants, the coding sequence was PCR‐amplified from the inbred B73, verified by DNA sequencing and integrated between a promoter and the rice UBIQUITIN terminator in a Gateway‐modified derivative of pSB11 (Ishida et al., 1996) using the Invitrogen Gateway system (Thermo Fisher Scientific, https://www.thermofisher.com/). The promoters used in this study include the BSV promoter and the maize UBIQUITIN1 (UBI1), GOS2 and FLORAL TRANSITION MADS1 (FTM1) promoter. The transfer DNA (T‐DNA) region contains the bialaphos resistance gene (phosphinothricin‐N‐acetyl‐transferase) as a selectable marker for transgenic plant selection. A visible marker (red fluorescent protein) was added in some of the constructs to facilitate seed sorting. These plasmids were then co‐integrated into the super binary pSB1 vector in Agrobacterium tumefaciens strain LBA4404 (Komari et al., 1996) by electroporation. Maize transformants were produced in the proprietary inbred lines PHR03 or PH184C by using Agrobacterium‐mediated transformation as described (Cho et al., 2014). Single‐copy T‐DNA integration events that expressed the transgene were selected and advanced for crosses to WT plants and further characterization.
Transgene expression analysis
To determine mRNA expression of the transgene in maize with reverse transcription PCR, cDNA was synthesized with a mixture of oligo(dT) and random hexamer primers using Applied Biosystems HCAP Reverse Transcription Kit (Life Technologies, cat. no. 4368813, https://www.thermofisher.com/). Quantitative RT‐PCR experiments were performed using TaqMan Universal Master Mix (Life Technologies) and a TaqMan probe. Relative quantification values were determined using the difference in Ct from the target gene and internal controls. Maize elF4‐γ was used as an internal control.
Ethylene emission analysis
Ethylene measurements were conducted on leaf disks taken from plants grown in the greenhouse. Thirty leaf disks (1 cm in diameter) were allowed to release wound‐generated ethylene for 2 h, placed in amber glass vials (volume 9.77 ml) containing a disk of filter paper wetted with 50 μl of distilled water and then sealed with aluminum crimp seals. After an incubation period of 24 h, 1‐ml samples were taken from the headspace of each sealed vial. The ethylene content was quantified by gas chromatography as previously described (Habben et al., 2014). The ethylene production rate was expressed as pmol per hour per gram of dry weight.
Gene expression analysis by RNA sequencing
Nodal tissues were sampled from WT plants treated with ACC for 48 and 72 h and water controls as well as BSV::ARGOS8 transgenic plants without ACC treatments. Four biological replicates consisting of four individual plants each were collected for each genotype, treatment and time point. Total RNAs were isolated from the nodal tissues with the Qiagen RNeasy kit for total RNA isolation (Qiagen, http://www.qiagen.com/). Sequencing libraries from the resulting total RNAs were prepared using the TruSeq mRNA‐Seq kit according to the manufacturer's instructions (Illumina, https://www.illumina.com/) and sequenced on the Illumina HiSeq 2500 system with Illumina TruSeq SBS version 3 reagents. Sequences were trimmed based on quality scores, mapped to the maize B73 reference genome sequence v.4 and analyzed via the RSEM pipeline (Li et al., 2010; Li and Dewey, 2011). DESeq2 was used to identify DE genes (Love et al., 2014). The DE genes are defined as those with false discovery‐corrected P < 0.05 and an absolute value of log2(fold change) ≥ 1. The DE genes were analyzed for overrepresentation of GO terms using BiNGO (Maere et al., 2005). Those GO terms with a false discovery‐corrected P < 0.05 were deemed overrepresented.
Maize hybrid field study
To evaluate root‐lodging resistance in hybrids, field trials were conducted in multiple locations across the USA in small plots (approximately 8.4 m2) with two to four replications at each location. Hybrid seed for these trials was generated by crossing heterozygous transgenic inbred lines with inbred testers. Hybrid seed segregated 1:1 for the transgene, and visible selection markers linked to the transgene were used to identify and separate the transgene‐positive F1 seed from the transgene‐negative F1 seed. The non‐transgenic segregants (null) served as controls. For those constructs without visible markers, homozygous transgenic inbred plants and WT were crossed with inbred testers in the same nursery to produce experimental and control hybrid seeds. The field study in 2013 was carried out at research centers in Woodland CA, York NE, Garden City KS, Plainview TX, Miami MO, Marion IA, Johnston IA and Princeton IN. Fertilizer at each location was applied to achieve maximum yields. Weeds and pests were controlled according to local practices. Root‐lodged plants (i.e. plants tilting at more than 30°; Landi et al., 2007) were recorded at the locations where natural lodging due to severe weather conditions occurred before flowering. Grain mass and grain moisture data for all locations were collected using a small plot combine. Grain yield was adjusted to a constant 15% moisture. The root‐lodging study of the GOS2::ARGOS8 transgenic hybrids was performed in 2016 at research centers in Ivesdale IL, San Jose IL, Buda IL, Garden City KS and Marion IA. All locations experienced natural root‐lodging events during the growth season.
The field experimental design was set up as split plot configuration where hybrid background was the whole plot and events were the subplot. Data analysis was performed using ASReml (VSN International Ltd, https://www.vsni.co.uk/), and the values reported are best linear unbiased predictions (BLUPs; Gilmour et al., 2009). A mixed model framework was used to perform the multi‐location analysis. In the analysis, the main effect of constructs and events was considered as a fixed effect whereas hybrids and interactions between event and hybrid were treated as random. The main effect of locations and location interactions with event and hybrid were considered as random effects. The blocking factors such as replicates and spatial variation in the field within each location were considered as random. For grain yield, three components of spatial effects including row, column and autoregressive correlation as AR1 × AR1 were included to reduce noise caused by spatial variation in the field. However, the spatial adjustments were not applied to analysis of lodging data to avoid overfitting. The significance test between event and nulls (or WT) within and across hybrids was performed using a P‐value of 0.05 in a two‐tailed test.
Conflict of interest
The authors are employees of Corteva Agriscience, Agriculture Division of DowDuPont.
Supporting information
Figure S1. Transcript abundance of the ARGOS8 transgene in 1‐aminocyclopropane‐1‐carboxylic acid‐treated BSV::ARGOS8 transgenic plants and non‐transgenic segregants.
Figure S2. Number of primordia on node 5 in BSV::ARGOS8 transgenic and non‐transgenic plants.
Figure S3. Percentage of plants with nodal roots on node 6 at the vegetative stage V14.
Figure S4. Effects of 1‐aminocyclopropane‐1‐carboxylic acid on nodal root emergence in maize PH184C/PH1V69 hybrid plants.
Figure S5. Grain yield of maize GOS2::ARGOS8 transgenic events.
Table S1. Grain yield of maize transgenic events from three ARGOS8 constructs.
Table S2. RNA sequencing analysis of nodal tissues.
Acknowledgements
We thank Rayeann Archibald, Kimberly Glassman, Terry Hu, Carl Simmons, Kellie Reimann, Mei Guo, Mary Trimnell, Jacque Hockenson, John Nau, John Van Hemert, Jesse Ourada and Karen Kratky for excellent assistance with this study. We acknowledge our colleagues at outlying breeding stations who conducted first‐rate yield trials. We thank Tom Greene and Dave Warner for their organizational leadership and helpful input.
References
- Abbe, E.C. and Stein, O.L. (1954) The origin of the shoot apex in maize: embryogeny. Am. J. Bot. 41, 285–293. [Google Scholar]
- Bellini, C. , Pacurar, D.I. and Perrone, I. (2014) Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. [DOI] [PubMed] [Google Scholar]
- Carter, P.R. and Hudelson, K.D. (1988) Influence of simulated wind lodging on corn growth and grain yield. J. Prod. Agric. 1, 295–299. [Google Scholar]
- Cho, M.J. , Wu, E. , Kwan, J. et al. (2014) Agrobacterium‐mediated high‐frequency transformation of an elite commercial maize (Zea mays L.) inbred line. Plant Cell Rep. 33, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Clark, D.G. , Gubrium, E.K. , Barrett, J.E. , Nell, T.A. and Klee, H.J. (1999) Root formation in ethylene‐insensitive plants. Plant Physiol. 121, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya, O.N. , Meng, X. , Selinger, D.A. , Deschamps, S. , Hermon, P. , Vansant, G. , Gupta, R. , Ananiev, E.V. and Muszynski, M.G. (2008) Involvement of the MADS‐box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol. 147, 2054–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick, D.N. (2005) The contribution of breeding to yield advances in maize (Zea mays L.). Adv. Agron. 86, 83–154. [Google Scholar]
- Ennos, A.R. , Crook, M.J. and Grimshaw, C. (1993) The anchorage mechanics of maize, Zea mays. J. Exp. Bot. 44, 147–153. [Google Scholar]
- Feldman, L. (1994) The maize root In The Maize Handbook (Freeling M. and Walbot V., eds). New York: Springer, pp. 29–37. [Google Scholar]
- Gilmour, A.R. , Gogel, B.J. , Cullis, B.R. and Thompson, R. (2009) ASReml User Guide. Release 3.0. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK. [Google Scholar]
- Guingo, E. and Hébert, Y. (1997) Relationships between mechanical resistance of the maize root system and root morphology, and their genotypic and environmental variations. Maydica, 42, 265–274. [Google Scholar]
- Habben, J.E. , Bao, X. , Bate, N.J. et al. (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought‐stress conditions. Plant Biotechnol. J. 12, 685–693. [DOI] [PubMed] [Google Scholar]
- Hetz, W. , Hochholdinger, F. , Schwall, M. and Feix, G. (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J. 10, 845–857. [Google Scholar]
- Hochholdinger, F. (2009) The maize root system: morphology, anatomy, and genetics In Handbook Of Maize: Its Biology. (Bennetzen J. and Hake S., eds). New York: Springer, pp. 145–160. [Google Scholar]
- Hoppe, D.C. , McCully, M.E. and Wenzel, C.L. (1985) The nodal roots of Zea: their development in relation to structural features of the stem. Can. J. Bot. 64, 2524–2537. [Google Scholar]
- Inukai, Y. , Sakamoto, T. , Ueguchi‐Tanaka, M. , Shibata, Y. , Gomi, K. , Umemura, I. , Hasegawa, Y. , Ashikari, M. , Kitano, H. and Matsuoka, M. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell, 17, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, Y. , Saito, H. , Ohta, S. , Hiei, Y. , Komari, T. and Kumashiro, T. (1996) High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14, 745–750. [DOI] [PubMed] [Google Scholar]
- Jackson, M.B. , Drew, M.C. and Giffard, S.C. (1981) Effects of applying ethylene to the root system of Zea mays on growth and nutrient concentration in relation to flooding tolerance. Physiol. Plant. 52, 23–28. [Google Scholar]
- Jenkins, M.T. (1930) Heritable characters of maize XXXIV‐rootless. J. Hered. 21, 79–80. [Google Scholar]
- Kim, H.J. , Lynch, J.P. and Brown, K.M. (2008) Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant, Cell Environ. 31, 1744–1755. [DOI] [PubMed] [Google Scholar]
- Kitomi, Y. , Ogawa, A. , Kitano, H. and Inukai, Y. (2008) CRL4 regulates crown root formation through auxin transport in rice. Plant Root, 2, 19–28. [Google Scholar]
- Kitomi, Y. , Ito, H. , Hobo, T. , Aya, K. , Kitano, H. and Inukai, Y. (2011) The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type‐A response regulator of cytokinin signaling. Plant J. 67, 472–484. [DOI] [PubMed] [Google Scholar]
- Komari, T. , Hiei, Y. , Saito, Y. , Murai, N. and Kumashiro, T. (1996) Vectors carrying two separate T‐DNAs for co‐transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10, 165–174. [DOI] [PubMed] [Google Scholar]
- Landi, P. , Sanguineti, M.C. , Liu, C. , Li, Y. , Wang, T.Y. , Giuliani, S. , Bellotti, M. , Salvi, S. and Tuberosa, R. (2007) Root‐ABA1 QTL affects root lodging, grain yield, and other agronomic traits in maize grown under well‐watered and water‐stressed conditions. J. Exp. Bot. 58, 319–326. [DOI] [PubMed] [Google Scholar]
- Langan, T.D. and Oplinger, E.S. (1987) Growth and yield of ethephon treated maize. Agron. J. 79, 130–134. [Google Scholar]
- Li, B. and Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Ruotti, V. , Stewart, R.M. , Thomson, J.A. and Dewey, C.N. (2010) RNA‐Seq gene expression estimation with read mapping uncertainty. Bioinformatics, 26, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Wang, S. , Yu, X. , Yu, J. , He, X. , Zhang, S. , Shou, H. and Wu, P. (2005) ARL1, a LOB‐domain protein required for adventitious root formation in rice. Plant J. 43, 47–56. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Wang, J. , Wang, L. , Wang, X. , Xue, Y. , Wu, P. and Shou, H. (2009) Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 19, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Song, F. , Liu, F. , Zhu, X. and Xu, H. (2012) Effect of planting density on root lodging resistance and its relationship to nodal root growth characteristics in maize (Zea mays L.). J. Agric. Sci. 4, 182–189. [Google Scholar]
- Lorbiecke, R. and Sauter, M. (1999) Adventitious root growth and cell‐cycle induction in deepwater rice. Plant Physiol. 119, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere, S. , Heymans, K. and Kuiper, M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics, 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Majer, C. , Xu, C. , Berendzen, K.W. and Hochholdinger, F. (2012) Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot‐borne root initiation in maize (Zea mays L.). Philos. Trans. R. Soc. Lond B Biol. Sci. 367, 1542–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, Y. and Kyozuka, J. (2007) Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol. 48, 540–549. [DOI] [PubMed] [Google Scholar]
- Negi, S. , Sukumar, P. , Liu, X. , Cohen, J.D. and Muday, G.K. (2010) Genetic dissection of the role of ethylene in regulating auxin‐dependent lateral and adventitious root formation in tomato. Plant J. 61, 3–15. [DOI] [PubMed] [Google Scholar]
- Orman‐Ligeza, B. , Parizot, B. , Gantet, P.P. , Beeckman, T. , Bennett, M.J. and Draye, X. (2013) Post‐embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci. 18, 459–467. [DOI] [PubMed] [Google Scholar]
- Pacurar, D.I. , Perrone, I. and Bellini, C. (2014) Auxin is a central player in the hormone cross‐talks that control adventitious rooting. Physiol. Plant. 151, 83–96. [DOI] [PubMed] [Google Scholar]
- Pellerin, S. , Trendel, R. and Duparque, A. (1990) Relationship between morphological characteristics and loding susceptibility of maize (Zea mays L.). Agronomie, 10, 439–446. [Google Scholar]
- Schenk, P.M. , Remans, T. , Sági, L. , Elliott, A.R. , Dietzgen, R.G. , Swennen, R. , Ebert, P.R. , Grof, C.P. and Manners, J.M. (2001) Promoters for pregenomic RNA of banana streak badnavirus are active for transgene expression in monocot and dicot plants. Plant Mol. Biol. 47, 399–412. [DOI] [PubMed] [Google Scholar]
- Schnable, J.C. , Freeling, M. and Lyons, E. (2012) Genome‐wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol. 4, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian, J. , Yee, M.C. , Goudinho Viana, W. et al. (2016) Grasses suppress shoot‐borne roots to conserve water during drought. Proc. Natl Acad. Sci. USA, 113, 8861–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Habben, J.E. , Archibald, R.L. , Drummond, B.J. , Chamberlin, M.A. , Williams, R.W. , Lafitte, H.R. and Weers, B.P. (2015) Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 169, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Drummond, B.J. , Wang, H. , Archibald, R.L. and Habben, J.E. (2016) Maize and Arabidopsis ARGOS proteins interact with ethylene receptor signaling complex, supporting a regulatory role for ARGOS in ethylene signal transduction. Plant Physiol. 171, 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Gao, H. , Wang, H. , Lafitte, H.R. , Archibald, R.L. , Yang, M. , Hakimi, S.M. , Mo, H. and Habben, J.E. (2017) ARGOS8 variants generated by CRISPR‐Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamp, P. and Kiel, C. (1992) Root morphology of maize and its relationship to root lodging. J. Agron. Crop Sci. 168, 113–118. [Google Scholar]
- Steffens, B. and Rasmussen, A. (2016) The physiology of adventitious roots. Plant Physiol. 170, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens, B. , Kovalev, A. , Gorb, S.N. and Sauter, M. (2012) Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell, 24, 3296–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino, G. , Sauer, M. , Stauffer, J.L. Jr , Multani, D. , Niu, X. , Sakai, H. and Hochholdinger, F. (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post‐embryonic shoot‐borne root initiation. Plant J. 50, 649–659. [DOI] [PubMed] [Google Scholar]
- Tyburski, J. and Tretyn, A. (2004) The role of light and polar auxin transport in root regeneration from hypocotyls of tomato seedling cuttings. Plant Growth Regul. 42, 39–48. [Google Scholar]
- Vidoz, M.L. , Loreti, E. , Mensuali, A. , Alpi, A. and Perata, P. (2010) Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 63, 551–562. [DOI] [PubMed] [Google Scholar]
- Visser, E. , Cohen, J.D. , Barendse, G. , Blom, C. and Voesenek, L. (1996) An ethylene‐mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palustris Sm. Plant Physiol. 112, 1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.F. , He, F.F. , Ma, X.X. , Mao, C.Z. , Hodgman, C. , Lu, C.G. and Wu, P. (2011) OsCAND1 is required for crown root emergence in rice. Mol. Plant, 4, 289–299. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Zhu, L. , Shou, H. and Wu, P. (2005) A PIN1 family gene, OsPIN1, involved in auxin‐dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. [DOI] [PubMed] [Google Scholar]
- Xu, C. , Tai, H. , Saleem, M. et al. (2015) Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot‐borne root formation. New Phytol. 207, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Hu, Y. , Dai, M. , Huang, L. and Zhou, D.X. (2009) The WUSCHEL‐related homeobox gene WOX11 is required to activate shoot‐borne crown root development in rice. Plant Cell, 21, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Transcript abundance of the ARGOS8 transgene in 1‐aminocyclopropane‐1‐carboxylic acid‐treated BSV::ARGOS8 transgenic plants and non‐transgenic segregants.
Figure S2. Number of primordia on node 5 in BSV::ARGOS8 transgenic and non‐transgenic plants.
Figure S3. Percentage of plants with nodal roots on node 6 at the vegetative stage V14.
Figure S4. Effects of 1‐aminocyclopropane‐1‐carboxylic acid on nodal root emergence in maize PH184C/PH1V69 hybrid plants.
Figure S5. Grain yield of maize GOS2::ARGOS8 transgenic events.
Table S1. Grain yield of maize transgenic events from three ARGOS8 constructs.
Table S2. RNA sequencing analysis of nodal tissues.


