Summary
Coral‐associated microorganisms are thought to play a fundamental role in the health and ecology of corals, but understanding of specific coral–microbial interactions are lacking. In order to create a framework to examine coral–microbial specificity, we integrated and phylogenetically compared 21,100 SSU rRNA gene Sanger‐produced sequences from bacteria and archaea associated with corals from previous studies, and accompanying host, location and publication metadata, to produce the Coral Microbiome Database. From this database, we identified 39 described and candidate phyla of Bacteria and two Archaea phyla associated with corals, demonstrating that corals are one of the most phylogenetically diverse animal microbiomes. Secondly, this new phylogenetic resource shows that certain microorganisms are indeed specific to corals, including evolutionary distinct hosts. Specifically, we identified 2–37 putative monophyletic, coral‐specific sequence clusters within bacterial genera associated with the greatest number of coral species (Vibrio, Endozoicomonas and Ruegeria) as well as functionally relevant microbial taxa (“Candidatus Amoebophilus”, “Candidatus Nitrosopumilus” and under recognized cyanobacteria). This phylogenetic resource provides a framework for more targeted studies of corals and their specific microbial associates, which is timely given the escalated need to understand the role of the coral microbiome and its adaptability to changing ocean and reef conditions.
Introduction
Corals are complex ‘holobiont’ organisms (Knowlton and Rohwer, 2003), hosting multi‐species microbial interactions which sustain their productivity and growth in oligotrophic reef waters. It is well known that corals rely on endosymbiotic microalgae (Symbiodinium) to supply sugars, amino acids, lipids and other nutrients (Trench, 1971), but corals also host an assemblage of other microorganisms including endolithic algae, bacteria, archaea, fungi and viruses (Rohwer et al., 2002; Knowlton and Rohwer, 2003; Ainsworth et al., 2017). Of these microorganisms, bacteria and archaea are thought to play a critical role in the cycling and regeneration of nutrients for the coral holobiont (Rohwer et al., 2001, 2002; Beman et al., 2007; Kelly et al., 2014). There is also evidence that coral‐associated bacteria are capable of producing antibiotic and other secondary metabolite compounds, which could provide protection to the coral (Kelman, 2004; Ritchie, 2006; Raina et al., 2009). Understanding the individual contribution of these disparate organisms to the coral holobiont, and if and how these microorganisms enhance the resistance and resilience of corals to changing ocean and reef conditions, are current challenges within this field.
Over the past 15 years, numerous review articles have reflected upon the relationship between corals and bacteria and archaea (Rosenberg and Ben‐Haim, 2002; Rohwer and Kelley, 2004; Mouchka et al., 2010; Bourne et al., 2016). These reviews repeatedly demonstrate that coral–microbes are taxonomically distinct from cells residing in the seawater, and there is general recognition of shared microbial groups over coral species and reefs (i.e., Roseobacter clade, Vibrio spp.). However, although most of the coral–microbial data are publically available, these data from multiple investigators and study environments data are not integrated. In other fields of study, combining existing data provides valuable resources for the community and especially for students and new investigators to these fields (e.g., Simister et al., 2012).
Integration of coral–microbial data is critical in the current framework of limited understanding about coral–microbial associations and during a time when many corals and reefs are experiencing disturbances from climate and anthropogenic sources (Hoegh‐Guldberg et al., 2007; Carpenter et al., 2008; Hughes et al., 2017, 2018). A growing community of scientists is pursuing studies aimed at understanding the resistance, resilience and impact of these events on coral microbiomes (e.g., Lee et al., 2015; Webster et al., 2016). For example, studies have shown that coral‐associated microbial communities demonstrate taxonomic shifts in composition in response to temperature, nutrient and pH‐based stressors (Littman et al., 2010; Meron et al., 2011; Shaver et al., 2017). The current research trend is to produce copious partial SSU rRNA gene sequences using next generation sequencing methods targeted at bacteria and archaea to inform questions about how microbial diversity, community structure and taxonomic composition changes under different environmental scenarios. However, sequences generated by these approaches are typically not long enough to allow for in‐depth phylogenetic analyses or primer and probe design. Further, taxonomic assignment is generally facilitated using reference databases such as Silva (Quast et al., 2013) or Greengenes (DeSantis et al., 2006), and these databases include sequences from biomedical‐based studies and it is often difficult to identify sequences originating from corals, thus hindering the development of coral‐specific analyses and resources, such as specific primers and probes.
In order to examine relatedness of microorganisms detected by sequencing approaches across numerous studies of corals, we followed an approach utilized by Simister et al. (2012) to examine the phylogeny of sponge‐associated microbes. Specifically, we utilized publically available Sanger produced sequence data from worldwide studies of corals to produce the Coral Microbiome Database (Coral MD), a coral‐specific SSU rRNA gene database hosting 21,100 coral–microbial sequences, housed within the ARB software (Ludwig et al., 2004). We utilized the Coral MD to examine the phylogenetic diversity of coral‐associated bacteria and archaea and these findings were confirmed in a subset of next‐generation sequencing data available for corals. Secondly, we identified putative monophyletic, coral‐specific sequence clusters and examined the distribution of these clusters among the Alcyonacea (soft corals) and the two main phylogenetic lineages of Scleractinia (robust/short and complex/long), which are separated by approximately 250 million years of divergence (Romano and Palumbi, 1996; Medina et al., 2006; Fukami et al., 2008). Our phylogenetic‐framed findings and the Coral MD provide a novel resource for the community of scientists studying coral‐associated microbes, and may advance investigations of the functional role of the coral microbiome and specific coral–microbial interactions.
Results and discussion
Overview of coral‐specific microbial sequence database
The Coral MD includes 21,100 sequences derived from coral‐associated bacteria and archaea which originated from public databases, and are compiled in the ARB software environment within the framework of the Silva reference database (Quast et al., 2013) (Supporting Information Methods). The database is available for download at BCO‐DMO (https://www.bco-dmo.org/dataset/724355). The mean and median lengths of sequences in the database are 893 and 750 bp respectively. There are 6833 nearly full‐length sequences greater than or equal to 1200 bp, and 5811 sequences are short reads, less than or equal to 600 bp. The sequences span 37 described and candidate phyla of Bacteria (Proteobacteria considered as single phylum) and 2 phyla of Archaea (Euryarchaeota and Thaumarchaeota) (Fig. 1). Of the bacteria, sequences are most commonly affiliated with the Proteobacteria (especially Gammaproteobacteria and Alphaproteobacteria), Bacteroidetes, Cyanobacteria and Firmicutes. Fewer sequences were associated with candidate divisions, including BRC1, OD1, OP11, OP3, OP8, SR1, TM7 and WS3.
Figure 1.
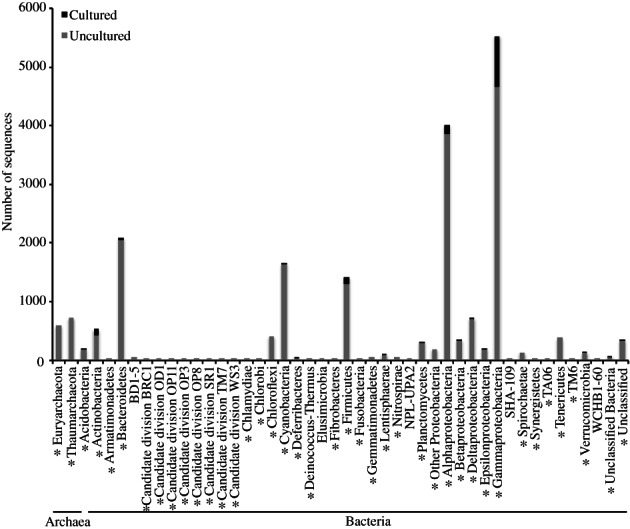
Sequences in the Coral Microbial Database. (21,100 Sanger‐produced sequences from archaeal and bacterial SSU rRNA genes) classified to phyla, candidate phyla and phyla‐level lineages from cultivated cells and environmental samples (uncultivated), based on the Silva reference taxonomy (Pruesse et al., 2007). Phyla that are also represented in next‐generation sequencing data available in the Coral Microbiome Portal are denoted (*).
In order to examine if the phyla‐based distribution of sequences in this database are representative of those available in next‐generation sequencing studies, we examined phyla‐based diversity of 715,114 coral–microbial sequences spanning nine studies and 23 coral species from the Alcyonacea as well as complex and robust Scleractinian lineages available within the Coral Microbiome Portal (CMP) (https://vamps2.mbl.edu/portals/CMP) (Sunagawa et al., 2010; Barott et al., 2011; Morrow et al., 2012; Apprill et al., 2013; Bayer et al., 2013a, b; Bourne et al., 2013; Šlapeta and Linares, 2013; Lesser and Jarett, 2014). We identified high consistency between the two types of sequence data, with three lineages represented in the next‐generation studies that were not present in the Sanger database (Candidate division OP10, TG‐1 and WS6), and five lineages in the Sanger sequencing based Coral MD database (BD1‐5, Elusimicrobia, NPL‐UPA2, SHA‐109 and WCHB1‐60) which were absent from the CMP. While examination of additional next‐generation sequencing based studies may further increase the phyla‐based diversity of coral microbes, these findings are, to our knowledge, the first to suggest such high (~ 39 Sanger from Coral MD; 42 from Coral MD + CMP database) phyla‐based microbial diversity associated with corals. This diversity is similar to the number of phyla observed in sponges, 41 (Thomas et al., 2016), but higher than many other host systems including the human skin, 19 phyla (Grice et al., 2009), termite, 10 phyla (Rahman et al., 2015) and honeybee, 5 phyla (Moran et al., 2012). It is interesting that such high similarity exists between the coral and sponge microbial phyla‐based diversity, as corals are host to more biochemical microenvironments of varying light, oxygen and pH, which, compared with sponges, which may provide numerous habitats for microorganisms (Kühl et al., 1995; Helmuth et al., 1997). Bacteria are known to reside in the tissues (Lesser et al., 2004; Ainsworth et al., 2006), outer protective mucus layer (Ducklow and Mitchell, 1979), and skeleton (Crossland and Barnes, 1976) of hard (scleractinian) corals, the major host type among the sequences examined. Scleractinian corals harbour several distinct tissue layers including an external protective ectoderm and an endodermal gastrovascular cavity that is used for digestion and also houses Symbiodinium, and these niches provide further habitat differentiation for microorganisms (Ainsworth et al., 2006; Rosenberg et al., 2007).
The Coral MD was queried to describe the most abundant coral–microbial lineages, represented by more than 100 sequences in the database, and these lineages spanned 14 phyla (Fig. 2, Supporting Information Table S1). The abundant groups included Vibrio (1217 sequences), found in 42 coral species, Endozoicomonas, with 943 sequences found in 35 coral species and an uncultured group of the Rhodobacteraceae family (Roseobacter clade) with 890 sequences from 37 coral species. Other lineages with large numbers of sequences included Marine Group II Euryarchaeota (493 sequences, 5 coral species), Marine Group I Thaumarchaeota (443 sequences, 7 coral species), and Ruegeria (356 sequences, 36 coral species) (Fig. 2). Lineages with over 200 sequences represented in the database include: Paenibacillus, Mycoplasma, Pseudoalteromonas, Fusibacter, Marinifilum, Bacillus, ‘Candidatus Nitrosopumilus’, “Candidatus Amoebophilus” and undescribed groups associated with Caldilineaceae, Flavobacteraceae and Alteromonadaceae families (Fig. 2), and sequences from most of these microbial groups were from less than 20 coral species. The abundant bacterial groups identified here corroborate with those identified by Mouchka et al. (2010), from 32 SSU rRNA gene studies with Sanger produced sequence data. Their dataset of 5711 sequences similarly found high observations of Rhodobacteraceae, Vibrio, Oceanospirillales, as well as Cyanobacteria, associated with corals of various health states, but only reported 11 bacterial phyla.
Figure 2.
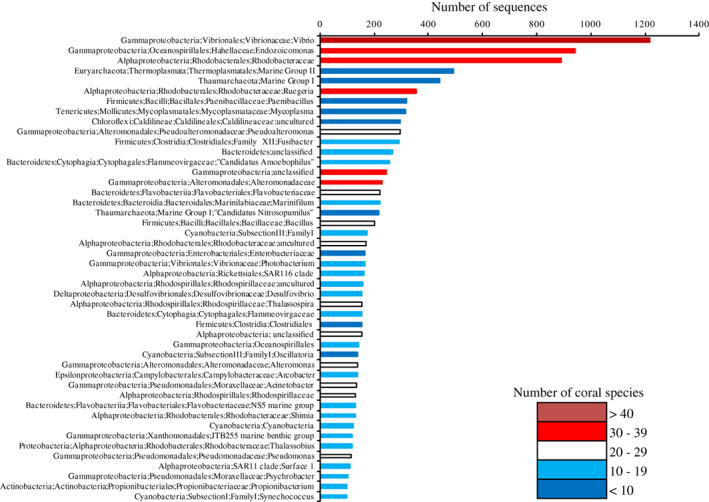
Abundances of archaeal and bacterial SSU rRNA gene sequences in the Coral Microbial Database according to most detailed taxonomic lineage. The colours reflect the representation of these sequences across different species of coral.
Coral‐specific microbial sequence clusters
In order to identify microbial groups that may be specific to the coral holobiont, we interrogated the most common taxonomic groups to determine the presence of monophyletic clusters originating only from corals. This approach follows that used by Simister et al. (2012) for sponge‐associated microorganisms. Clusters were assigned where three or more high‐quality sequences consistently formed a monophyly across neighbour joining, maximum likelihood and maximum parsimony methods of phylogenetic reconstruction (Supporting Information Methods). The taxa examined included those associated with the largest number of coral species, including Vibrio, Endozoicomonas and Ruegeria, as well as sequence clusters with relatives with known functions including the eukaryotic parasite “Candidatus Amoebophilus”, Thaumarchaeota including the ammonia oxidizing “Candidatus Nitrosopumilus” and under recognized Cyanobacterial lineages belonging to subsection III, Family I. We recognize that there may be next‐generation sequencing data available from other non‐coral environments which are associated with these lineages, but at this time that data is not compiled in a way that can be querried. Therefore, we identify these lineages as putatively monophyletic to corals.
Bacteria from the genus Vibrio have disparately been shown to be highly abundant (Bourne and Munn, 2005; Koren and Rosenberg, 2006) or only a small component (Apprill et al., 2013; Roder et al., 2014) of the coral associated microbial community. They are frequently associated with diseases including white syndrome, white band disease, yellow band disease and bleaching (see Tout et al., 2015 and reference therein). Our Vibrio phylogenetic analysis also included the closely related genera Photobacterium, Salinivibrio, Enterovibrio and Grimonita. Collectively there were 1461 coral associated sequences in the database that belonged to these genera, with 1217 assigned to Vibrio. Within the genus Vibrio, 37 putative monophyletic coral associated clusters (denoted VC1‐37) were identified, and 15 of these clusters were associated with both the complex and robust phylogenetic lineages of the Scleractinia (Fig. 3). Cluster VC31, associated with Dendronephthya, was exlusive to the Alcyonacea. The largest cluster (VC22) contained 37 sequences exclusively from Montastraea faveolata (now referred to as Orbicella faveolata) (Budd et al., 2012). Additional putative monophyletic coral associated clusters were associated with the genera Photobacterium (VC38‐VC40) and Salinivibrio (VC41) and were exclusive to the Scleractinia.
Figure 3.
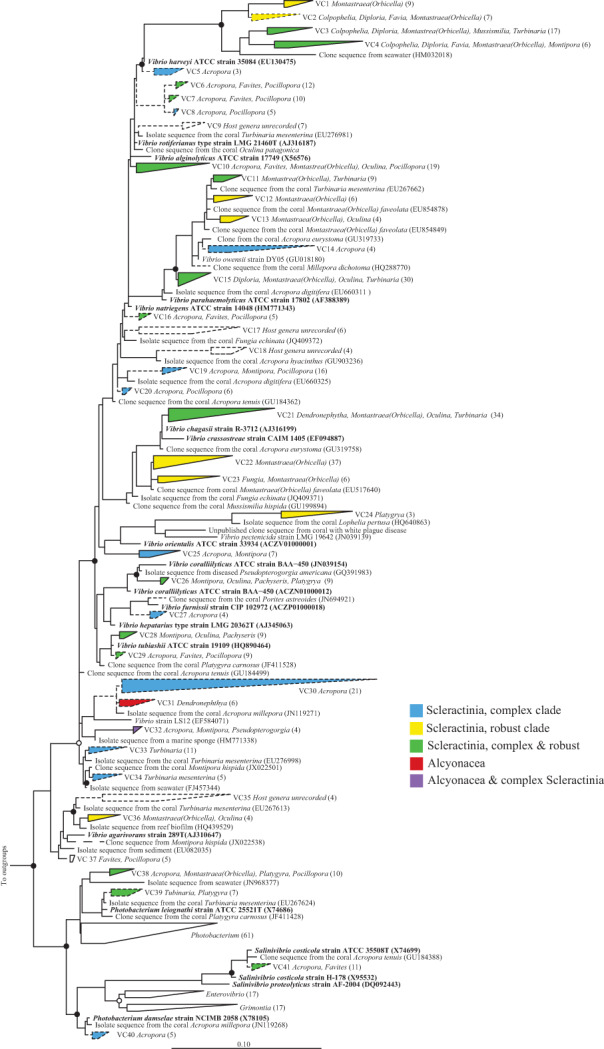
Phylogeny of coral‐specific clusters within the Gammaproteobacteria genus Vibrio and related sequences, based on 16S ribosomal RNA genes. The displayed tree is a maximum likelihood tree constructed based on long (> 1200 bp) sequences only; shorter coral‐associated sequences (indicated by dashed lines) were added using the ARB Parsimony (Quick add marked) tool in ARB. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of > 90%, and open circles represent > 75% support. Bar indicates 10% sequence divergence. The numbers of sequences in each cluster are indicated in brackets and the coral host genera for each cluster are listed adjacent to clusters, and microbial species name in bold and described or type species in culture. Accession numbers for specific sequences are shown in brackets, where accession numbers are absent none were available. ATCC, American Type Culture Collection.
The Gammaproteobacterial genus Endozoicomonas and closely related Endozoicomonas‐like sequences are one of the most recognized coral‐associated bacteria (Bayer et al., 2013b; Neave et al., 2016, 2017b) and abundances of Endozoicomonas sequences have been recorded at over 85% per colony (Morrow et al., 2012; Apprill et al., 2016). These cells are thought to play a role within host‐associated marine communities in protein and carbohydrate transport and cycling (Neave et al., 2017a) although their specific function is still undetermined. We identified 918 coral‐associated Endozoicomonas‐like sequences in the database and our phylogenetic analysis placed 798 of these into 10 monophyletic coral‐associated clusters, denoted EC1‐EC10 (Fig. 4). The largest of these (EC10) contained 357 sequences spanning both the robust and complex lineages of Scleractinia, and the most closely related sequences are from fish and sponges (Fig. 4). The second largest (EC1) contained 289 sequences and were widely associated with diverse hosts, including both clades of Scleractinia as well as the soft corals. Specific sequence clusters exclusive to the Alcyonacea include EC6, found in Gorgonia. Clusters exclusive to the robust clade of Scleractinia include EC4, EC8 and EC9, but no putative monophyletic Endozoicomonas sequence clusters were exclusive to the complex Scleractinia, which includes the corals with often the highest relative abundance of Endozoicomonas‐like sequences in their microbiome including Porites astreoides and Stylophora pistillata (Morrow et al., 2012; Bayer et al., 2013b; Apprill et al., 2016). This suggests that the complex clade corals may be colonized by Endozoicomonas‐like cells which are broad generalists for many distantly related corals, rather than specialists to related corals.
Figure 4.
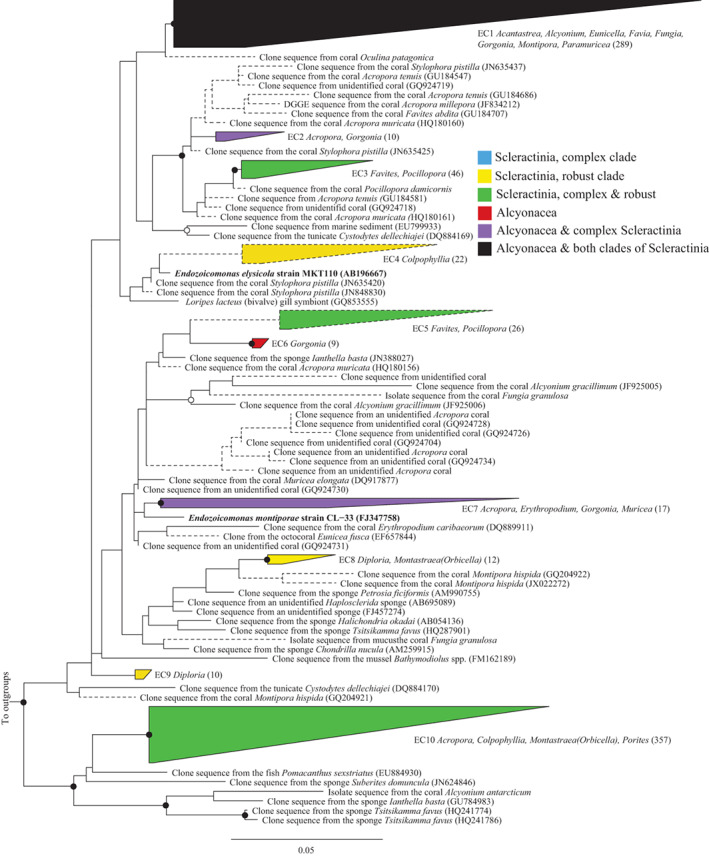
Phylogeny of coral‐specific clusters within the Gammaproteobacteria genus Endozoicomonas and related sequences, based on 16S ribosomal RNA genes. The displayed tree is a maximum likelihood tree constructed based on long (> 1200 bp) sequences only; shorter coral‐associated sequences (indicated by dashed lines) were added using the ARB Parsimony (Quick add marked) tool. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of > 90%, and open circles represent > 75% support. Bar indicates 5% sequence divergence. The numbers of sequences in each cluster are indicated in brackets and the coral host genera for each cluster are listed adjacent to clusters, and microbial species name in bold and described or type species in culture. Accession numbers for specific sequences are shown in brackets, where accession numbers are absent none were available.
The paraphyletic alphaproteobacterial genus Ruegeria has been identified as one of the more successful lineages of bacteria inhabiting a variety of marine environments, equipped with a large diversity of metabolic capabilities and regulatory pathways (Luo and Moran, 2014). We analysed 353 coral‐associated sequences identified taxonomically as Ruegeria and also found a number of paraphyletic groups, with some more phylogenetically related to sequences from the Roseobacter clade, including Phaeobacter, Marinovum and Thalassococcus (Fig. 5). We identified 9 small monophyletic coral associated clusters containing between 3 and 7 sequences, represented in either one or both clades of Scleractinia, and absent from soft corals. Eight of these clusters (denoted RC2‐RC10) were related to described Ruegeria species, with 1 cluster (denoted RC1) related to sequences from the genus Phaeobacter (Fig. 5).
Figure 5.
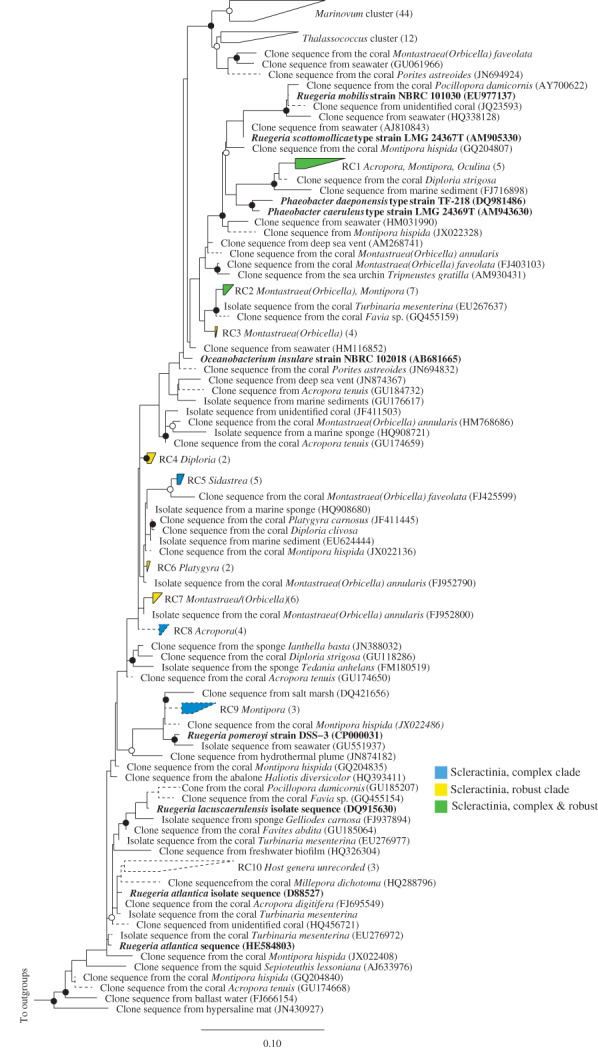
Phylogeny of coral‐specific clusters within the Alphaproteobacteria genus Ruegeria and related sequences, based on 16S ribosomal RNA genes. The displayed tree is a maximum likelihood tree constructed based on long (> 1200 bp) sequences only; shorter coral‐associated sequences (indicated by dashed lines) were added using the ARB Parsimony (Quick add marked) tool. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of > 90%, and open circles represent > 75% support. Bar indicates 10% sequence divergence. The numbers of sequences in each cluster are indicated in brackets and the coral host genera for each cluster are listed adjacent to clusters, and microbial species name in bold and described or type species in culture. Accession numbers for specific sequences are shown in brackets, where accession numbers are absent none were available. ATCC, American Type Culture Collection. NBRC, NTE (National Institute of Technology and Evaluation) Biological Resource Centre.
The Bacteroidetes genus “Candidatus Amoebophilus” was originally described as freshwater intracellular protozoan symbionts (Horn et al., 2001). Whole genome analysis has uncovered a suite of mechanisms by which “Ca. Amoebophilus” are thought to interact with their host, including a high number of eukaryotic protein domains (Schmitz‐Esser et al., 2010). Interestingly, members of this genus are dominant members of the microbiome of Caribbean corals, and are specifically associated with coral tissue (but not mucus) in relative abundances of up to 70% of affiliated bacterial and archaeal sequences (Apprill et al., 2016). Both the current phylogenetic analysis, and that conducted previously (Apprill et al., 2016) place the coral‐associated sequences in a separate lineage, adjacent to protozoan intracellular endosymbionts (Fig. 6), and into two monophyletic clusters, AC1 and AC2, which were exclusive to the Scleractinia and found across both robust and complex clades. Sequence cluster AC1 contained 232 sequences from various coral species including Montastraea/Orbicella faveolata, Montastraea/Orbicella annularis, M. franksi, Siderastrea stellata, Diploria strigosa, Porites astreoides and Montipora hispida. Cluster AC2 contained 13 sequences and was closely related to a seaweed endosymbiont (Fig. 6). It is unclear if these sequences are endosymbionts of the corals themselves or if instead they may be associated with other members of the holobiont such as a coral‐associated protozoan which is more prevalent in stony compared with soft corals.
Figure 6.
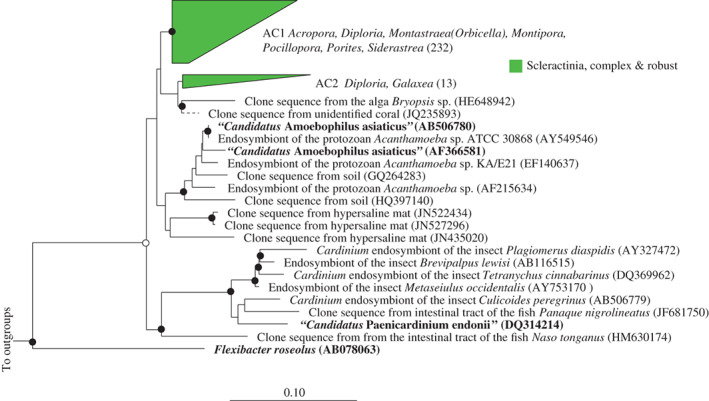
Phylogeny of coral‐specific clusters within the Bacteroidetes genus “Candidatus Amoebophilus” and related sequences, based on 16S ribosomal RNA genes. The displayed tree is a maximum likelihood tree constructed based on long (> 1200 bp) sequences only; shorter coral‐associated sequences (indicated by dashed lines) were added using the ARB Parsimony (Quick add marked) tool. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of > 90%, and open circles represent > 75% support. Bar indicates 10% sequence divergence. The numbers of sequences in each cluster are indicated in brackets and the coral host genera for each cluster are listed adjacent to clusters, and microbial species name in bold and described or type species in culture. Accession numbers for specific sequences are shown in brackets, where accession numbers are absent none were available.
The Thaumarchaeota Marine Group II, including the ammonia oxidizing “Candidatus Nitrosopumilus”, were represented in the database from scleractinian corals spanning both complex and robust clades, including Siderastrea stellata, Montipora hispida, Montastraea/Orbicella faveolata, Fungia sp., Favia sp. and Acantastrea sp. (Fig. 7). Previous studies show that the Archaeal portion of the coral‐associated microbial community can be dominated by sequences from this genus (Siboni et al., 2008) and suggest that these microbes may play a role in nitrogen cycling via ammonia oxidation due to the presence of archaeal ammonia monooxygenase genes (Beman et al., 2007; Siboni et al., 2012). Our phylogenetic analysis of sequences in this genus uncovered 12 monophyletic coral‐associated clusters that were consistent between all three treeing methods, denoted NC1‐NC12, with NC11 from the robust Scleractinia, including sequences from Acantastrea, Favia and Fungia most closely related to the ammonia oxidizing Nitrosopumilus maritimus (Könneke et al., 2005) (Fig. 7). Interestingly, the majority of the sequence clusters were exclusively associated with robust clade corals (NC1, NC6, NC7, NC10 and NC11), suggesting that these corals may harbour more specialized relationships with these microbes. Whole genome sequence analyses of marine Nitrosopumilus strains indicate that members of the genus are ammonia oxidizing autotrophs but vary in other metabolic traits, possibly promoting niche segregation and thus influencing nitrification rates in various environments (Bayer et al., 2015).
Figure 7.

Phylogeny of coral‐specific clusters within the Thaumarchaeota Marine Group II “Candidatus Nitrosopumilus” and related sequences, based on 16S ribosomal RNA genes. The displayed tree is a maximum likelihood tree constructed based on long (> 1200 bp) sequences only; shorter coral‐associated sequences (indicated by dashed lines) were added using the ARB Parsimony (Quick add marked) tool. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of > 90%, and open circles represent > 75% support. Bar indicates 10% sequence divergence. The numbers of sequences in each cluster are indicated in brackets and the coral host genera for each cluster are listed adjacent to clusters, and microbial species name in bold and described or type species in culture. Accession numbers for specific sequences are shown in brackets, where accession numbers are absent none were available.
Cyanobacteria are abundant primary producers in the waters surrounding corals (Sorokin, 1973), and have been shown to fix dinitrogen in some coral species (Lesser et al., 2004). Despite the potential functional importance of these cells, the vast majority of cyanobacterial sequences recovered from sequencing surveys of corals are not well‐resolved taxonomically. We examined the Cyanobacterial Family I lineage and identified 17 monophyletic coral‐specific sequence clusters spanning several described genera (Fig. 8). The largest cluster (denoted CC14) was most closely related to clusters CC13 and CC15, all from Montipora corals and related to Hydrocoleum lyngbyaceum and Trichodesmium erythraeum (Fig. 8). The filamentous Phormidium/Leptolyngbya genera contained 8 monophyletic clusters (denoted CC1–8), with up to 14 sequences in each cluster, and mostly spanning complex clade Scleractinia with CC8 present in both soft corals and both clades of stony corals. Leptolyngbya‐related sequences from A. eurystoma were found to persist even when exposed to reduced pH (Meron et al., 2011), suggesting that this group may be tolerant of a range of environmental conditions.
Figure 8.
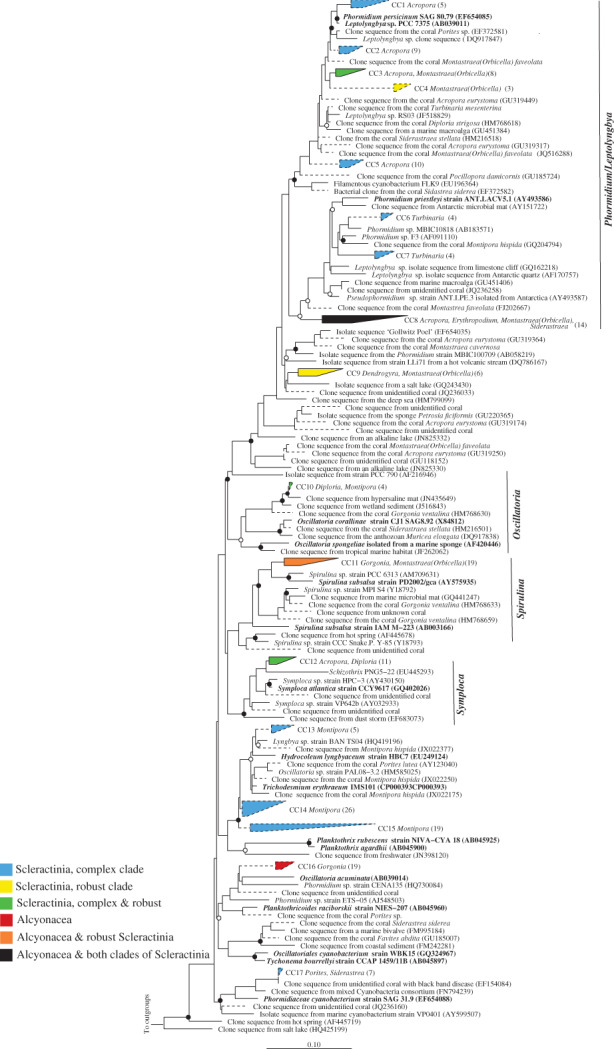
Phylogeny of coral‐specific clusters within the Cyanobacterial ‘Family I’ and related sequences, based on 16S ribosomal RNA genes. The displayed tree is a maximum likelihood tree constructed based on long (> 1200 bp) sequences only; shorter coral‐associated sequences (indicated by dashed lines) were added using the ARB Parsimony (Quick add marked) tool. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of > 90%, and open circles represent > 75% support. Bar indicates 10% sequence divergence. The numbers of sequences in each cluster are indicated in brackets and the coral host genera for each cluster are listed adjacent to clusters. Accession numbers for specific sequences are shown in brackets, where accession numbers are absent none were available.
Cultured isolates obtained from corals
Of the 21,100 sequences in the Coral MD, only 1378 (6.5%) were generated from cultured isolates (Supporting Information Table S2). A further 18,400 (87.4%) were identified as uncultured (either from clone libraries or other PCR based methods), and the remaining 1271 sequences (6.1%) did not have enough associated data to confirm their source (cultured or uncultured). Of the 41 identified taxonomic groups (which includes divisions of the Proteobacteria), 14 contained cultured representatives (Fig. 1). Gammaproteobacteria (870 sequences), dominated by sequences from the genera Vibrio (401 sequences), Pseudoalteromonas (149 sequences) and Photobacterium (110 sequences) were the most abundant cultured organisms in the database followed by Bacillus (Firmicutes, 71 sequences), Shewanella (Gammaproteobacteria, 27 sequences) and Ruegeria (Alphaproteobacteria, 25 sequences) (Fig. 1). A high number of Actinobacteria (114 sequences) were represented by cultures, primarily from the genera Streptomyces, Micromonospora, Kocuria, Micrococcus and Brachybacterium. A number of key taxa were dominant in the database – notably Euryarchaeota, Thaumarchaeota, Chloroflexi, Tenericutes, Spirochaetae and Gemmatimonadetes – yet contained no cultured representatives (Fig. 1). These groups denote important targets for future culture studies, along with highly abundant groups with minimal cultured representatives such as Endozoicomonas (4 cultured sequences) and Paenibacillus (1 cultured representative) (Fig. 1).
Utility of the coral MD
The Coral MD provides a number of support features that may advance coral microbiology studies. The sequences and their placement in ARB software are an ideal scenario for primer and probe design. The database provides a reference taxonomic framework for sequence classifications for amplicon sequence datasets produced from corals. Additionally, the database offers descriptive information about the coral‐associated microbes in a phylogenetic structure. The numerous taxonomic groups of bacteria and archaea associated with corals can be overwhelming to newcomers in the field, and the database provides a context to familiarize the user with phylogenetic groups that frequently associate with corals. Overall, the database has the potential to facilitate coral microbiology research toward the goal of understanding the role of coral‐associated microbes in the health and ecology of corals.
Supporting information
Appendix S1: Supporting Information Data 1. Summary of sequence data contained in the Coral MD and sorted by phyla.
Appendix S2: Supporting Information Methods
Supporting Information Table S1. Most abundant microbial taxa in the Coral MD, and their representation across coral hosts.
Supporting Information Table S2. Number of sequences in each microbial phylum identified from non‐cultured or cultured microorganisms.
Acknowledgements
This project was supported by NSF awards OCE‐1233612 and OCE‐1736288 to A.A and an Edith Cowan University Early Career Researcher award to M.J.H. We thank Evan Denmark for assisting with data analysis and Michael Rappé for providing the inspiration to develop this database. The authors have no conflicts of interest to report.
Contributor Information
Megan J. Huggett, Email: megan.huggett@newcastle.edu.au.
Amy Apprill, Email: aapprill@whoi.edu.
References
- Ainsworth, T.D. , Fine, M. , Blackall, L.L. , and Hoegh‐Guldberg, O. (2006) Fluorescence in situ hybridization and spectral imaging of coral‐associated bacterial communities. Appl Environ Microbiol 72: 3016–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth, T.D. , Fordyce, A.J. , and Camp, E.F. (2017) The other microeukaryotes of the coral reef microbiome. Trends Microbiol 25: 980–991. [DOI] [PubMed] [Google Scholar]
- Apprill, A. , Hughen, K. , and Mincer, T.J. (2013) Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ Microbiol 15: 2063–2072. [DOI] [PubMed] [Google Scholar]
- Apprill, A. , Weber, L. , and Santoro, A. (2016) Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1: e00143‐00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott, K.L. , Rodriguez‐Brito, B. , Janouškovec, J. , Marhaver, K.L. , Smith, J.E. , Keeling, P. , and Rohwer, F.L. (2011) Microbial diversity associated with four functional groups of benthic reef algae and the reef‐building coral Montastraea annularis . Environ Microbiol 13: 1192–1204. [DOI] [PubMed] [Google Scholar]
- Bayer, T. , Arif, C. , Ferrier‐Pagès, C. , Zoccola, D. , Aranda, M. , and Voolstra, C.R. (2013a) Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini . Mar Ecol Prog Ser 479: 75–84. [Google Scholar]
- Bayer, T. , Neave, M.J. , Alsheikh‐Hussain, A. , Aranda, M. , Yum, L.K. , Mincer, T. , et al (2013b) The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue‐associated Endozoicomonas bacteria. Appl Environ Microbiol 79: 4759–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, B. , Vojvoda, J. , Offre, P. , Alves, R.J. , Elisabeth, N.H. , Garcia, J.A. , et al (2015) Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J 10: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman, J.M. , Roberts, K.J. , Wegley, L. , Rohwer, F. , and Francis, C.A. (2007) Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Environ Microbiol 73: 5642–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D.G. , and Munn, C.B. (2005) Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol 7: 1162–1174. [DOI] [PubMed] [Google Scholar]
- Bourne, D.G. , Dennis, P.G. , Uthicke, S. , Soo, R.M. , Tyson, G.W. , and Webster, N. (2013) Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J 7: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D.G. , Morrow, K.M. , and Webster, N.S. (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70: 317–340. [DOI] [PubMed] [Google Scholar]
- Budd, A.F. , Fukami, H. , Smith, N.D. , and Knowlton, N. (2012) Taxonomic classification of the reef coral family Mussidae (Cnidaria: Anthozoa: Scleractinia). Zool J Linn Soc 166: 465–529. [Google Scholar]
- Carpenter, K.E. , Abrar, M. , Aeby, G. , Aronson, R.B. , Banks, S. , Bruckner, A. , et al (2008) One‐third of reef‐building corals face elevated extinction risk from climate change and local impacts. Science 321: 560–563. [DOI] [PubMed] [Google Scholar]
- Crossland, C.J. , and Barnes, D.J. (1976) Acetylene reduction by coral skeletons. Limnol Oceanogr 21: 153–156. [Google Scholar]
- DeSantis, T.Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E.L. , Keller, K. , et al (2006) Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducklow, H. , and Mitchell, R. (1979) Bacterial populations and adaptations in the mucus layers on living corals. Limnol Oceanogr 24: 715–725. [Google Scholar]
- Fukami, H. , Chen, C.A. , Budd, A.F. , Collins, A. , Wallace, C. , Chuang, Y.‐Y. , et al (2008) Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (order Scleractinia, class Anthozoa, phylum Cnidaria). PloS One 3: e3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice, E.A. , Kong, H.H. , Conlan, S. , Deming, C.B. , Davis, J. , Young, A.C. , et al (2009) Topographical and temporal diversity of the human skin microbiome. Science 324: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmuth, B.S.T. , Timmerman, B.E.H. , and Sebens, K.P. (1997) Interplay of host morphology and symbiont microhabitat in coral aggregations. Mar Biol 130: 1–10. [Google Scholar]
- Hoegh‐Guldberg, O. , Mumby, P.J. , Hooten, A.J. , Steneck, R.S. , Greenfield, P. , Gomez, E. , et al (2007) Coral reefs under rapid climate change and ocean acidification. Science 318: 1737–1742. [DOI] [PubMed] [Google Scholar]
- Horn, M. , Harzenetter, M.D. , Linner, T. , Schmid, E.N. , Müller, K.‐D. , Michel, R. , and Wagner, M. (2001) Members of the Cytophaga–Flavobacterium–Bacteroides phylum as intracellular bacteria of acanthamoebae: proposal of ‘Candidatus Amoebophilus asiaticus’. Environ Microbiol 3: 440–449. [DOI] [PubMed] [Google Scholar]
- Hughes, T.P. , Kerry, J.T. , Álvarez‐Noriega, M. , Álvarez‐Romero, J.G. , Anderson, K.D. , Baird, A.H. , et al (2017) Global warming and recurrent mass bleaching of corals. Nature 543: 373–377. [DOI] [PubMed] [Google Scholar]
- Hughes, T.P. , Anderson, K.D. , Connolly, S.R. , Heron, S.F. , Kerry, J.T. , Lough, J.M. , et al (2018) Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359: 80–83. [DOI] [PubMed] [Google Scholar]
- Kelly, L.W. , Williams, G.J. , Barott, K.L. , Carlson, C.A. , Dinsdale, E.A. , Edwards, R.A. , et al (2014) Local genomic adaptation of coral reef‐associated microbiomes to gradients of natural variability and anthropogenic stressors. PNAS 111: 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman, D. (2004) Antimicrobial activity of sponges and corals In Coral Health and Disease. Rosenberg E., and Loya Y. (eds). Berlin: Springer‐Verlag, pp. 243–258. [Google Scholar]
- Knowlton, N. , and Rohwer, F. (2003) Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am Nat 162: S51–S62. [DOI] [PubMed] [Google Scholar]
- Könneke, M. , Bernhard, A.E. , José, R. , Walker, C.B. , Waterbury, J.B. , and Stahl, D.A. (2005) Isolation of an autotrophic ammonia‐oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Koren, O. , and Rosenberg, E. (2006) Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol 72: 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl, M. , Cohen, Y. , Dalsgaaard, T. , Jørgensen, B.B. , and Revsbech, N.P. (1995) Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH, and light. Mar Ecol Prog Ser 117: 159–172. [Google Scholar]
- Lee, S.T.M. , Davy, S.K. , Tang, S.‐L. , Fan, T.‐Y. , and Kench, P.S. (2015) Successive shifts in the microbial community of the surface mucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbiol Ecol 91 10.1093/femsec/fiv142 [DOI] [PubMed] [Google Scholar]
- Lesser, M.P. , and Jarett, J.K. (2014) Culture‐dependent and culture‐independent analyses reveal no prokaryotic community shifts or recovery of Serratia marcescens in Acropora palmata with white pox disease. FEMS Microbiol Ecol 88: 457–467. [DOI] [PubMed] [Google Scholar]
- Lesser, M.P. , Mazel, C.H. , Gorbunov, M.Y. , and Falkowski, P.G. (2004) Discovery of symbiotic nitrogen‐fixing cyanobacteria in corals. Science 305: 997–1000. [DOI] [PubMed] [Google Scholar]
- Littman, R.A. , Bourne, D.G. , and Willis, B.L. (2010) Responses of coral‐associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol Ecol 19: 1978–1990. [DOI] [PubMed] [Google Scholar]
- Ludwig, W. , Strunk, O. , Westram, R. , Richter, L. , Meier, H. , Yadhukumar, A.B. , et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. , and Moran, M.A. (2014) Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, M. , Collins, A.G. , Takaoka, T.L. , Kuehl, J.V. , and Boore, J.L. (2006) Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci U S A 103: 9096–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron, D. , Atias, E. , Kruh, L.I. , Elifantz, H. , Minz, D. , Fine, M. , and Banin, E. (2011) The impact of reduced pH on the microbial community of the coral Acropora eurystoma . ISME J 5: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N.A. , Hansen, A.K. , Powell, J.E. , and Sabree, Z.L. (2012) Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7: e36393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, K.M. , Moss, A.G. , Chadwick, N.E. , and Liles, M.R. (2012) Bacterial associates of two Caribbean coral species reveal species‐specific distribution and geographic variability. Appl Environ Microbiol 78: 6438–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchka, M. , Hewson, I. , and Harvell, C.D. (2010) Coral‐associated bacterial assemblages: current knowledge and the potential for climate‐driven impacts. Integr Comp Biol 50: 662–674. [DOI] [PubMed] [Google Scholar]
- Neave, M.J. , Apprill, A. , Ferrier‐Pagès, C. , and Voolstra, C.R. (2016) Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas . Appl Microbiol Biotechnol 100: 8315–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave, M. , Mitchell, C. , Apprill, A. , and Voolstra, C. (2017a) Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep 7: 40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave, M.J. , Rachmawati, R. , Xun, L. , Michell, C.T. , Bourne, D.G. , Apprill, A. , and Voolstra, C.R. (2017b) Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J 11: 186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse, E. , Quast, C. , Knittel, K. , Fuchs, B. , Ludwig, W. , Peplies, J. , and Glöckner, F.O. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, N.A. , Parks, D.H. , Willner, D.L. , Engelbrektson, A.L. , Goffredi, S.K. , Warnecke, F. , et al (2015) A molecular survey of Australian and north American termite genera indicates that vertical inheritance is the primary force shaping termite gut microbiomes. Microbiome 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, J.‐B. , Tapiolas, D. , Willis, B.L. , and Bourne, D.G. (2009) Coral‐associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K.B. (2006) Regulation of microbial populations by coral surface mucus and mucus‐associated bacteria. Mar Ecol Prog Ser 322: 1–14. [Google Scholar]
- Roder, C. , Arif, C. , Bayer, T. , Aranda, M. , Daniels, C. , Shibl, A. , et al (2014) Bacterial profiling of white plague disease in a comparative coral species framework. ISME J 8: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer, F. , and Kelley, S. (2004) Culture‐independent analyses of coral‐associated microbes In Coral Health and Disease. Loya E.R.Y. (ed). Berlin, Heidelberg: Springer‐Verlag, pp. 265–277. [Google Scholar]
- Rohwer, F. , Breitbart, M. , Jara, J. , Azam, F. , and Knowlton, N. (2001) Diversity of bacteria associated with the Caribbean coral Montastraea franksi . Coral Reefs 20: 85–95. [Google Scholar]
- Rohwer, F. , Seguritan, V. , Azam, F. , and Knowlton, N. (2002) Diversity and distribution of coral‐associated bacteria. Mar Ecol Prog Ser 243: 1–10. [Google Scholar]
- Romano, S.L. , and Palumbi, S.R. (1996) Evolution of scleractinian corals inferred from molecular systematics. Science 271: 640–642. [Google Scholar]
- Rosenberg, E. , and Ben‐Haim, Y. (2002) Microbial diseases of corals and global warming. Environ Microbiol 4: 318–326. [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. , Koren, O. , Reshef, L. , Efrony, R. , and Zilber‐Rosenberg, I. (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5: 355–362. [DOI] [PubMed] [Google Scholar]
- Schmitz‐Esser, S. , Tischler, P. , Arnold, R. , Montanaro, J. , Wagner, M. , Rattei, T. , and Horn, M. (2010) The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba‐associated bacteria. J Bacteriol 192: 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver, E.C. , Shantz, A.A. , McMinds, R. , Burkepile, D.E. , Vega Thurber, R.L. , and Silliman, B.R. (2017) Effects of predation and nutrient enrichment on the success and microbiome of a foundational coral. Ecology 98: 830–839. [DOI] [PubMed] [Google Scholar]
- Siboni, N. , Ben‐Dov, E. , Sivan, A. , and Kushmaro, A. (2008) Global distribution and diversity of coral‐associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ Microbiol 10: 2979–2990. [DOI] [PubMed] [Google Scholar]
- Siboni, N. , Ben‐Dov, E. , Sivan, A. , and Kushmaro, A. (2012) Geographic specific coral‐associated ammonia‐oxidizing archaea in the northern gulf of Eilat (Red Sea). Microb Ecol 64: 18–24. [DOI] [PubMed] [Google Scholar]
- Simister, R.L. , Deines, P. , Botté, E.S. , Webster, N.S. , and Taylor, M.W. (2012) Sponge‐specific clusters revisited: a comprehensive phylogeny of sponge‐associated microorganisms. Environ Microbiol 14: 517–524. [DOI] [PubMed] [Google Scholar]
- Šlapeta, J. , and Linares, M.C. (2013) Combined amplicon pyrosequencing assays reveal presence of the apicomplexan ‘type‐N’ (cf. Gemmocystis cylindrus) and Chromera velia on the great barrier reef, Australia. PLoS ONE 8: e76095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin, Y.I. (1973) Trophical role of bacteria in the ecosystem of the coral reef. Nature 242: 415–417. [Google Scholar]
- Sunagawa, S. , Woodley, C.M. , and Medina, M. (2010) Threatened corals provide underexplored microbial habitats. PLoS ONE 5: e9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, T. , Moitinho‐Silva, L. , Lurgi, M. , Björk, J.R. , Easson, C. , Astudillo‐García, C. , et al (2016) Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7: 11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout, J. , Siboni, N. , Messer, L.F. , Garren, M. , Stocker, R. , Webster, N.S. , et al (2015) Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis . Front Microbiol 6: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trench, R.K. (1971) The physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates. II. Liberation of fixed 14C by zooxanthellae in vitro. Proc R Soc Lond B 177: 237–250. [Google Scholar]
- Webster, N. , Negri, A. , Botté, E. , Laffy, P. , Flores, F. , Noonan, S. , et al (2016) Host‐associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci Rep 6: 19324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information Data 1. Summary of sequence data contained in the Coral MD and sorted by phyla.
Appendix S2: Supporting Information Methods
Supporting Information Table S1. Most abundant microbial taxa in the Coral MD, and their representation across coral hosts.
Supporting Information Table S2. Number of sequences in each microbial phylum identified from non‐cultured or cultured microorganisms.


