Abstract
Insects have a large family of C‐type lectins involved in cell adhesion, pathogen recognition and activation of immune responses. In this study, 32 transcripts encoding C‐type lectin domain proteins (CTLDPs) were identified from the Thitarodes xiaojinensis transcriptome. According to their domain structures, six CTLDPs with one carbohydrate‐recognition domain (CRD) were classified into the CTL‐S subfamily. The other 23 CTLDPs with two CRDs were grouped into the immulectin (IML) subfamily. The remaining three with extra regulatory domains were sorted into the CTL‐X subfamily. Phylogenetic analysis showed that CTL‐S and CTL‐X members from different insects could form orthologous groups. In contrast, no T. xiaojinensis IML orthologues were found in other insects. Remarkable lineage‐specific expansion in this subfamily was observed reflecting that these CTLDPs, as important receptors, have evolved diversified members in response to a variety of microbes. Prediction of binding ligands revealed that T. xiaojinensis, a cold‐adapted species, conserved the ability of CRDs to combine with Ca2+ to keep its receptors from freezing. Comparative analysis of induction of CTLDP genes after different immune challenges indicated that IMLs might play critical roles in immune defenses. This study examined T. xiaojinensis CTLDPs and provides a basis for further studies of their characteristics.
Keywords: binding ligand, carbohydrate‐recognition domain, C‐type lectin domain protein, expression profiling, phylogenetic feature, Thitarodes xiaojinensis
Introduction
Insects live in a hostile environment and possess effective immune systems (Lemaitre & Hoffmann, 2007). The initial step in the initiation of immune responses relies on biosensor proteins called pattern recognition receptors (PRRs), which can detect and bind to certain pathogen‐associated molecular patterns (PAMPs) on the surface of invading microbes. PAMPs consist of lipopolysaccharides (LPS) or peptidoglycans (PGNs) in bacteria and β‐1, 3‐glucans in fungi (Pal & Wu, 2009; Wang et al., 2017). Insects have evolved a diverse group of PRRs corresponding to different PAMPs.
C‐type lectins constitute a large family of PRRs that have been identified in many organisms, ranging from plants, animals to viruses (Wang et al., 2013). In most cases, binding of the carbohydrate‐recognition domain (CRD) to sugars requires ligation to Ca2+. However, evidence shows that not all C‐type lectins have the ability to associate with carbohydrates or Ca2+ (Zelensky & Gready, 2005). We therefore employed annotation studies in Manduca sexta and Bombyx mori to use C‐type lectin domain proteins (CTLDPs) to refer to proteins harboring the domains (Rao et al., 2015a, b).
The genomes of many arthropods have been sequenced and a large number of CTLDPs have been identified. There are 34, 25, 39, 12, 16, 24, 34 and 23 genes encoding CTLDPs identified in the model insect species, Drosophila melanogaster, Anopheles gambiae, Aedes aegypti, Apis mellifera, Tribolium castaneum, Helicoverpa armigera, M. sexta and B. mori, respectively (Evans et al., 2006; Waterhouse et al., 2007; Zou et al., 2007; Xiong et al., 2015). Each CTLDP from the Dipteran and Hymenopteran species as listed contains a single CTLD and many of them are involved in immune responses (Table S1). For instance, D. melanogaster DL1, 2 and 3 with galactose specificity can agglutinate bacteria, and the latter two enhance melanization and activation of prophenoloxidase (PPO) (Tanji et al., 2006; Ao et al., 2007). AgCTL4 and AgCTLMA2 in the mosquito A. gambiae prevent ookinetes of Plasmodium berghei from melanization (Osta et al., 2004). A novel CTLDP from A. aegypti (CLSP2), composed of elastase‐like serine protein at the N‐terminus and CTLD at the C‐terminus, was shown to be a negative regulator in anti‐Beauveria bassiana infection (Wang et al., 2015). In Lepidoptera, the number of CTLDs in CTLDPs is distinctive. Besides the proteins present in one CTLD, there are also extraordinary CTLDPs possessing dual and triple CTLDs (Rao et al., 2015a, b). CTLDPs with two CTLDs are also found in T. castaneum (Zou et al., 2007). To date, Lepidoptera CTLDPs involved in immune responses were mainly found in proteins with two tandem CRDs (Table S2). For example, M. sexta immulectins (MsIML1–4) can be induced by bacteria and fungi, stimulating agglutination of the bacterial and fungal cells (MsIML1 and 4) and moderating PPO activation (MsIML1, 2 and 4) (Yu et al., 1999, 2005, 2006). In B. mori, the immune functions of IML1, 3, 4 and 5 have been investigated (Koizumi et al., 1999; Watanabe et al., 2006; Takase et al., 2009). In crustaceans, CTLDPs have also been identified and reported to be involved in immune defenses. For instance, a Pacifastacus leniusculus CTLDP with a single CRD plays a regulated role in prophenoloxidase activation (Wu et al., 2013).
Thitarodes xiaojinensis is a major host of Ophiocordyceps sisnensis (Zhang & Tu, 2015). Previously, 258 immunity‐related genes were identified in the transcriptome of T. xiaojinensis (Meng et al., 2015). The genus Hepialus is a phylogenetically primitive group of Lepidoptera (Nielsen et al., 2000). Therefore, analysis of the phylogenetic relationship between CTLDPs in T. xiaojinensis and those in other insects will help us understand if they share a common ancestor or emerged individually after divergence of phyla. To address this goal, transcriptome data were searched and we used recently released information on the CTLDPs of M. sexta and B. mori. A total of 32 transcripts encoding CTLDPs were annotated. These CTLDPs were classified into three groups based on domain structure. The lineage‐specific expansion in T. xiaojinensis CTLDPs is associated with expansion of proteins with tandem CTLDs. Additionally, expression profiling of the CTLDP genes after different immune challenges was used to explore their roles in T. xiaojinensis immunity.
Materials and methods
Identification and characteristic prediction of genes encoding CTLDPs in T. xiaojinensis
C‐type lectin domain protein amino acid sequences from M. sexta, B. mori and A. aegypti (Shin et al., 2011; Rao et al., 2015a, b; Wang et al., 2015) were accessed as queries to search the T. xiaojinensis unigene database (Meng et al., 2015). In total, 32 transcripts were confirmed by sequencing and deposited in the GenBank database (Table S3). The functional domain was detected using SMART (http://smart.embl.de/). Their signal peptides and transmembrane regions were predicted through CBS prediction server (http://www.cbs.dtu.dk/services/). Their domain architectures were visualized by software DOC (Ren et al., 2009).
Sequence alignments and analysis of phylogenesis
Subfamily‐specific CTLDPs from other insects were retrieved through BLASTP. Those sequences along with corresponding T. xiaojinensis CTLDPs were aligned by CLUSTALX 2.1 with the following settings: weight matrices of BLOSUM series, a gap penalty of 10 and an extension gap penalty of 0.1 (Thompson et al., 1997). According to alignment results, MEGA 6.0 was implemented to build unrooted neighbor‐joining trees using the bootstrap method (Tamura et al., 2013).
Prediction for ligand binding site and three‐dimensional (3D) model
Protein‐ligand binding sites in T. xiaojinensis CTLDs were predicted by the COACH (http://zhanglab.ccmb.med.umich.edu/COACH/) along with the I‐TASSER server (Roy et al., 2010; Yang et al., 2013) and denoted manually in the alignment of CTLDs. Putative 3D models were generated by I‐TASSER and shown using the PyMOL molecular graphic system.
Induction analysis of T. xiaojinensis CTLDPs after different microbial challenges
To investigate changes in transcriptional levels after immune stimulation, 10 complementary DNA (cDNA) libraries, referring to messenger RNA (mRNA) samples from the fat body of larvae without treatment (control group) and larvae exposed with Enterobacter cloacae for 6 h, Ringer's solution (8.05 g NaCl, 0.42 g KCl and 0.18 g CaCl2 per L) for 12 h, Ophiocordyceps sinensis for 12, 48, 72 h, 1 year, and Cordyceps militaris for 12, 48, 72 h, were constructed and separately sequenced on the Hiseq 2000 (Meng et al., 2015). The fragments per kilobase per million mapped reads (FPKM, representing transcript abundance) value of each gene encoding CTLDP was calculated and used to determine fold change by the DEGseq package in R https://www.r-project.org/about.html (P‐value < 0.001). Following this, values of log2‐fold change for 32 T. xiaojinensis CTLDP genes were used to draw a heatmap using heatmap.2 function in R environment. To validate the results of differentially expressed genes (DEGs), specific primers (Table S4) for six genes were designed to carry out quantitative real‐time polymerase chain reaction (qPCR). The cDNAs from the 10 libraries were used as the templates. The detailed procedure has been previously described (Meng et al., 2015). The acquired data were exported to Excel for computing ‐ΔΔCt, corresponding to log2‐fold change exhibiting in the heatmap.
Results
Features of T. xiaojinensis CTLDPs
There are 32 CTLDPs identified in the T. xiaojinensis transcriptome (Table S3) and these were divided into three subfamilies. Six proteins presenting a single CTLD were assigned to the CTL‐S subfamily, while 23 proteins containing two CTLDs were grouped into the immulectin (IML) subfamily; the remaining three proteins have more complex structures including both CTLDs and other conserved domains (Fig. 1). These were assigned to the CTL‐X subfamily. Initiation and/or stop codons could not be found in the coding sequences of CTL‐S5, IML13, 16–21, CTL‐X2, X4 and X6 (Table S3); therefore, the N‐ or/and C‐terminal regions in their architectures, as shown in Figure 1, were absent. Previous studies indicated that CTLs, possessing a signal peptide but not a transmembrane domain, execute their functions extracellularly (Table S1 and S2), and thus T. xiaojinensis CTL‐S1–S4, IML1–12, 14, 15, 17, 19, 21–23 and CTL‐X4 probably similarly work. Functions of CTLs with a transmembrane region were reported more in vertebrates. These receptors anchor to cell membrane and generally interact with ligands by their extracellular parts (Zelensky & Gready, 2005). T. xiaojinensis CTL‐X2 and X6 containing transmembrane regions are expected to work in a similar way. CTL‐S6 without the N‐terminal secretion signal or C‐terminal transmembrane region may be retained in the cytoplasma.
Figure 1.
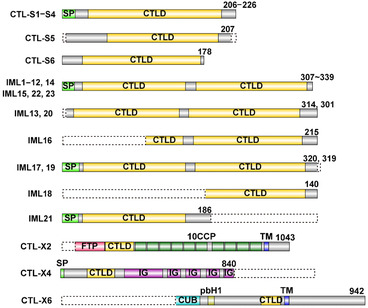
Schematic architectures of the 32 Thitarodes xiaojinensis C‐type lectin domain proteins (CTLDPs). The putative signal peptide (SP), CTLD, eel‐fucolectin tachylectin‐4 pentaxrin‐1 (FTP), complement control protein (CCP) (also known as sushi), immunoglobulin (Ig), complement C1r/C1s, Uegf, Bmp1 (CUB), parallel beta‐helix 1 (pbH1) and transmembrane domains (TM) are in different colored boxes. The sizes or size ranges of CTLDPs are indicated on the top right of each bar. The dashed boxes in the N‐ or C‐terminal ends represent the missing regions. The size of individual architectures is not proportional.
CTL‐S1, S2 and S3 contain the QPD motif, which may bind galactose (Table S3). In contrast, the CRD in CTL‐S6 is likely to interact with mannose, as it contained the EPN motif. The carbohydrate‐recognition motif in CTL‐S4 and CTL‐S5 presents an untypical signature (QRT and HPL, respectively), and thus their characteristics need further investigation. The EPN motifs are mostly localized in the first CRDs of T. xiaojinensis IML, namely IML1A, 2A, 4A, 7A, 8A, 10A, 15A–17A, 23A, while the QPD motifs are mostly localized in the second CRDs of T. xiaojinensis IML, namely IML1B, 3B–5B, 7B–11B, 13B–15B, 17B, 22B, 23B (Table S3). The two CRDs in IML6 are the same, namely EPN motifs, while the dual CTLDs in IML20 present two QPD motifs. IML12 and IML19 are exceptional; the motifs in the two CTLDs of the former are QPN and RRN; the motif in the first CRD of the latter is QPD (Table S3).We identified three proteins in T. xiaojinensis belonging to the CTL‐X group (Table S3). The conserved carbohydrate‐recognition motif, QPD, was only found in the CRD of T. xiaojinensis CTL‐X2 (Table S3). The CRD in CTL‐X4 contains a non‐canonical motif (DNH) and the motif for binding carbohydrate could not be identified in CTL‐X6 (Table S3). Even so, they might be involved in cell–cell interactions or complement activation due to the immunoglobulin (Ig) (in CTL‐X4) and CUB (complement C1r/C1s, Uegf, Bmp1) (in CTL‐X6) domain.
Evolutionary relationships among insect CTLDPs
Due to the remarkable differences in structures among the three CTLDP subfamilies, the phylogenetic relationships of CTLDPs were separately analyzed. CTL‐S subfamily members were divided into two clades (Fig. 2A). T. xiaojinensis CTL‐S1 through S5 formed clade A and might be closely related according to high bootstrap values and CTL‐S6 is located in clade B (Fig. 2A). In addition, T. xiaojinensis CTL‐S1 through S6 reveal monophyletic groups with their orthologues from other insects. In M. sexta and B. mori, three IMLs possess their own 1 : 1 orthologues (Rao et al., 2015a, b). However, orthologous pairs of all identified T. xiaojinensis IMLs could not be detected. A total of 23 T. xiaojinensis IMLs clustered together showing a clear species‐specific expansion (Fig. 2B). T. xiaojinensis IML1, 9, 18, IML3, 4, 15, 17, 20, 21, IML5, 10–14, 22, 23 and IML 6–8, 16, exhibited high bootstrap values among each other, and represent four groups of related IMLs that may have evolved through multiple gene duplications.
Figure 2.
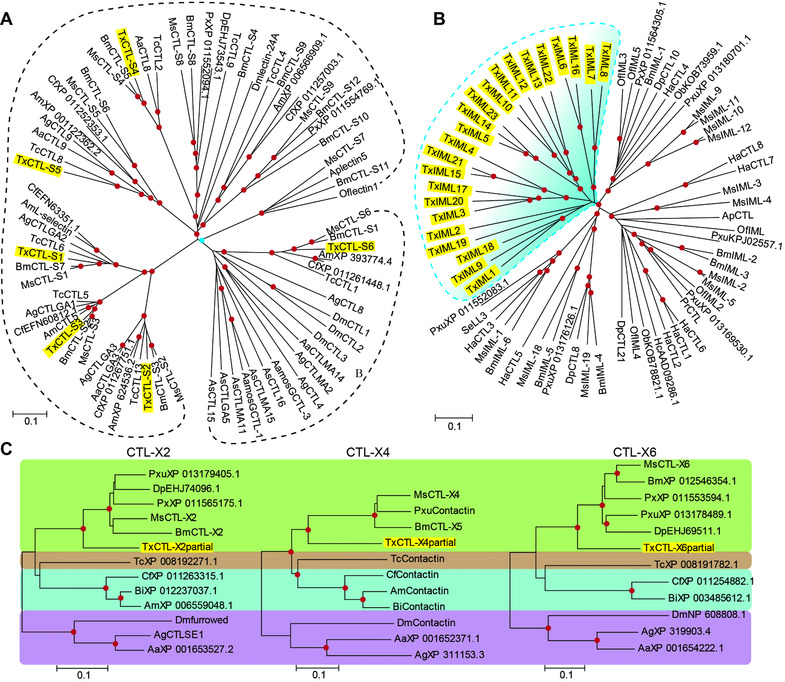
Phylogenetic relationships between C‐type lectin domain proteins (CTLDPs) of Thitarodes xiaojinensis (shaded yellow) and other insect species. Members belonging to T. xiaojinensis CTL‐S, immulectin (IML) and CTL‐X were separately analyzed with their orthologues, which present similar domain structures. (A) CTL‐S family. The evolutionary tree was separated into two clusters. Bootstrap value of the blue node is 963 (1000 trials). (B) IML family. Lineage‐specific expansion of T. xiaojinensis IMLs is denoted by blue shade. (C) CTL‐X family. Four divided subgroups are exhibited in all three CTL‐X trees and shaded in different colors. Each of green, pink, blue and purple clades only include Lepidoptera, Coleoptera, Diptera and Hymenoptera, respectively. Red dots at the nodes indicate bootstrap values >700 from 1000 trials. Amino acid sequences without names were represented by their Genbank accession numbers. Tx, T. xiaojinensis; Ms, Manduca sexta; Bm, Bombyx mori; Px, Plutella xylostella; Dp, Danaus plexippus; Of, Ostrinia furnacalis; Se, Spodoptera exigua; Pxu, Papilio xuthus; Ob, Operophtera brumata; Hc, Hyphantria cunea; Pr, Pieris rapae; Ap, Antheraea pernyi; Tc, Tribolium castaneum; Am, Apis mellifera; Cf, Camponotus floridanus; Dm, Drosophila melanogaster; Ag, Anopheles gambiae; Aa, Aedes aegypti; As, Armigeres subalbatus.
All three protein sequences of T. xiaojinensis CTL‐X were incomplete, but their CTLDs could be detected (Table S3). Considering that the domain architectures among CTL‐X2, X4 and X6 are variable, CTL‐X protein sequences from different insects were individually classified into CTL‐X2, X4 or X6 groups at first, and then members in the same classification were aligned with each other. The three CTL‐X groups revealed similar evolutionary relationships with those in CTL‐Ss. The CTL‐Xs formed strong monophyletic groups (Fig. 2C) suggesting a common ancestor before the divergence of these phyla.
Phylogenetic and structural characteristics of CTLDs
Research on the CTLDPs from T. xiaojinensis will aid understanding of the phylogenetic relationships among the Hepialidae. To achieve this goal, a neighbor‐joining tree was built based on sequence alignment of 53 T. xiaojinensis CTLDs and four clades were revealed (Fig. 3). All the IML‐As (the CTLDs near the N‐terminus) along with IML21 clustered into the blue clade and all the IML‐Bs (the CTLDs near C‐terminus) along with IML18 clustered into the green clade (Fig. 3). The pink clade contained members belonging to CTL‐S, while the yellow clade contained CTL‐X members (Fig. 3). These clustering relationships suggest that CTLDs from the same category evolved from a common ancestor, possessing similar composition of amino acid sequences, and were thus grouped together. The close relationship between IML‐A and IML‐B indicated that gene duplication probably gave rise to the emergence of the ancestor IML genes. The differentiated nodes denoted by blue arrows in Figure 3 suggest that the evolution of the CTL‐S group occurred before the variation pattern of the IML and CTL‐X groups. In addition, phylogenetic relationships of IML‐A members mostly corresponded to those in IML‐B and were also reflected in the IML evolutionary tree (Fig. 2B) derived from complete sequence alignments (i.e. IML3, 4, 15, 17, 20, IML5, 10–14, 22, 23 and IML1, 9, 18).
Figure 3.
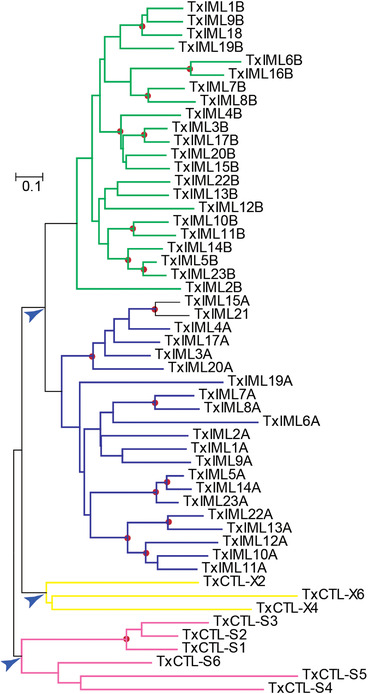
Evolutionary relationships of Thitarodes xiaojinensis C‐type lectin domains (CTLDs). A neighbor‐joining tree was built based on the alignment of CTLD sequences. CTLDs from the category of CTL‐S, CTL‐X, immulectin (IML)‐A and IML‐B formed four individual clades shaded pink, yellow, blue and green, respectively. Red dots at the nodes indicate bootstrap values >700 from 1000 trials. Blue arrows denote the key nodes at which the evolution has occurred. There were too few amino acid residues in the IML16A to be included.
To determine whether CTLDs from the same category have common features, the alignment of the 53 CTLD amino acid sequences was further analyzed. Six cysteine (Cys) residues, which may form three disulfide bridges for stability, are conserved in nearly all of the CTLDs belonging to the IML‐A group (Fig. 4). The disulfide linkages in CTL‐S and CTL‐X groups were variable. In the CTL‐S group, Cys‐3 and ‐6 as well as Cys‐4 and ‐5 are thought to build bridges in all members, while Cys‐2 is shifted in CTL‐S1–S3 resulting in its linkage with Cys‐1 being different from the others. All six Cys residues in conserved locations were identified in the CTLDs of CTL‐S5. In the CTL‐X group, CTL‐X2 and X6 maintain the three disulfide bridges in the conserved position. The residues around Cys‐4 revealed category‐specific features. All members in the IML‐B category contained a Gly residue at the first site after Cys‐4, while in members belonging to the IML‐A and CTL‐S categories, the residues at the same position were valine/isoleucine/leucine/alanine (Val/Ile/Leu/Ala).
Figure 4.
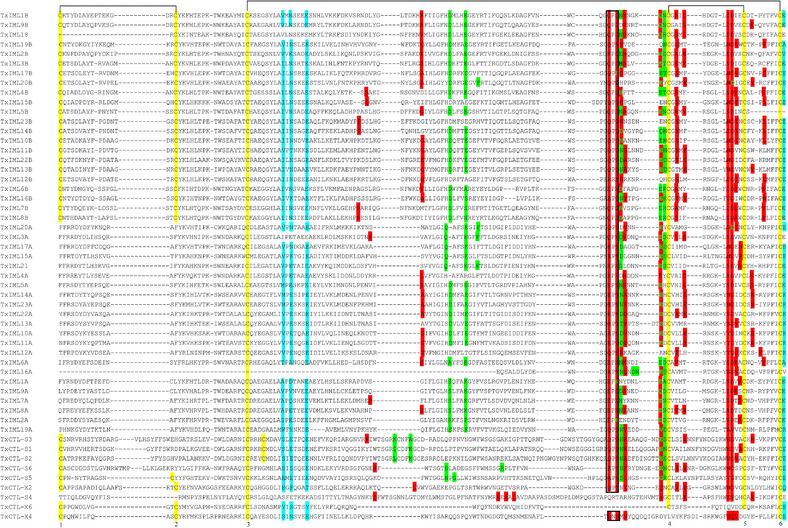
Multiple sequence alignments of C‐type lectin domains (CTLDs) in Thitarodes xiaojinensis. The conserved cysteine (Cys) residues are highlighted in yellow and marked with 1 to 6. The predicted disulfide linkages between Cys‐1 and ‐2, ‐3 and ‐6, ‐4 and ‐5 are shown by lines. The motifs (e.g. EPN and QPD) that usually participate in carbohydrate recognition in CTLDPs are indicated by the enclosed box. Residues involving in binding Ca2+ in site‐1 and site‐4 are shaded in green and cyan, respectively. Ligand binding sites in each CTLD were predicted through COACH combining with I‐TASSER server. The consensus binding residues are shaded in red, with those also ligating to Ca2+ in bold and green font.
To predict the functions of T. xiaojinensis CTLDPs, potential binding sites and corresponding ligands in the 53 CTLDs were detected by COACH server combining with I‐TASSER (Roy et al., 2010; Yang et al., 2013). In some CTLDs the residues (in bold) responsible for interaction with carbohydrates were presumed to associate with Ca2+ (Table S5). In particular, CTL‐S1–S3, S5, S6, IML1A–3A, 5A, 7A–11A, 13A–17A, 19A, 20A, 21, 22A, 23A, IML1B–11B, 13B, 14B, 16B, 17B, 18, IML20B and 22B might bind sugar ligands in a calcium‐dependent pattern. IML4A, 6A, 12A, 12B, 15B, 19B, 23B, CTL‐X2 and X4 were predicted to combine sugar as well as Ca2+, but their binding sites are not shared. Among the remaining CTLDs, one probably binds carbohydrates in the absence of Ca2+ and the other might only interact with Ca2+ (Table S5). CTLDs possessing EPN motifs usually have characteristics of mannose‐binding and those having QPD motifs usually reveal characteristics of galactose‐binding (van Vliet et al., 2008). In T. xiaojinensis, IML1A, 2A, 7A, 15A, 17A and 21 containing EPN motifs are likely to combine with mannose, as the predicted results are consistent with the rule. IML19A, 20A, 1B, 3B, 5B, 11B, 22B and CTL‐X2 probably have the ability to bind galactose, since their putative ligands are sugar derivatives of galactose (Table S5). In addition, secondary and tertiary structures of four CTLDs separately from IML‐A, IML‐B, CTL‐S and CTL‐X groups were compared with human dendritic cell‐specific intercellular adhesion molecule‐grabbing nonintegrin‐related (DC‐SIGNR) (1K9J), which is known to recognize mannose type ligands (Probert et al., 2013), to determine what properties in the structures determine their binding specificity (Figs. 5 and S1). The four representative T. xiaojinensis CTLDs (gray) had a typical CTLD structure model presenting double loops similar to those in DC‐SIGNR (black) (Fig. 5). The 2D structures of CTLDs generally have two β‐sheets at the N‐terminus followed by two α‐helices and two or three β‐sheets at the C‐terminus (Fig. S1). The N‐ and C‐terminal β stands come together to form an antiparallel β‐sheet located at the bottom of the putative crystal structures and the other two C‐terminal β strands (such as T. xiaojinensis IML2A β4 and β5) form the second β‐sheet located at the top of the structures (Fig. 5 and S1). The second loop is the primary region participating in binding carbohydrates and/or interaction with Ca2+ (Fig. 5). T. xiaojinensis IML2A, possesses the EPN motif and is predicted to bind mannose requiring Ca2+ coordination (Table S5). Its 3D structure aligned well with DC‐SIGNR (1K9J) (Fig. 5A), especially the region involved with mannose. However, for the other three CTLDPs, the areas involved in sugar binding in the second loops did not match those in DC‐SIGNR. These results were consistent with the prediction in Table S5 that the motifs in T. xiaojinensis IML2B, CTL‐S1 and CTL‐X2 are not EPN and their sugar ligands are N‐acetyl‐D‐glucosamine (NGA), alpha‐L‐fucopyranose and NGA, respectively. It indicates that the second loop is structurally flexible among CTLDPs and is the key component of binding specificity. Some T. xiaojinensis CTLDs (e.g. CTL‐S2, S3, S6, IML4A, 6A, etc.), had predictions for sugar ligands that were inconsistent with the rule referring to EPN and QPD motifs. Therefore, more evidence will be needed to confirm these predictions.
Figure 5.
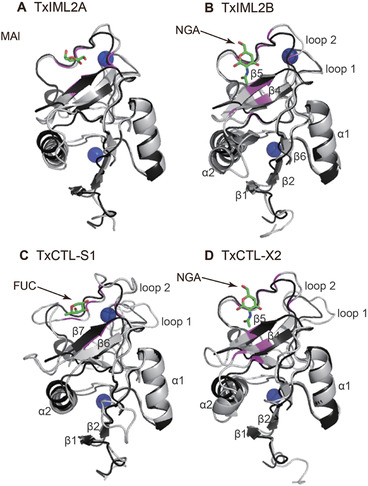
Structures of the C‐type lectin domains (CTLDs) in Thitarodes xiaojinensis immulectin (IML)2A (A), IML2B (B), CTL‐S1 (C) and CTL‐X2 (D) and structural comparison with the CTLD in human human dendritic cell‐specific intercellular adhesion molecule‐grabbing nonintegrin‐related (DC‐SIGNR) (1K9J). T. xiaojinensis and human CTLDs are colored gray and black, respectively. Secondary structure elements in Figure S1 are labeled in the corresponding models. The blue spheres indicate Ca2+ ions. The stick models represent the carbohydrate ligands (the C atoms are shaded in green and the O atoms are in pink). The residues involving in binding carbohydrate (Table S5) are highlighted in magenta. MAN, alpha‐D‐mannose; NGA, N‐acetyl‐D‐glucosamine; FUC, alpha‐L‐fucopyranose.
Expression profiles of CTLDPs in T. xiaojinensis fat body under various challenges
To investigate mRNA levels of T. xiaojinensis CTLDP genes, we searched their FPKM values in ten libraries (Meng et al., 2015). The results are listed in Table S6. Overall, genes encoding CTL‐S and CTL‐X expressed at low levels in all conditions. IML18 and IML19 presented similar mRNA levels with FPKM values less than 3.5 in untreated/treated groups. High transcript abundances in untreated larvae were observed in genes encoding IML7–9 (Table S6). Except for the five IMLs mentioned above, the remaining IMLs increase their mRNA levels to > 10 or even > 100 after at least one challenge. The maximum value (5768.7) was the IML1 expression in larvae exposed to C. militaris for 48 h (Table S6).
To compare induction of T. xiaojinensis CTLDP genes after Ringer's, O. sinensis, C. militaris and E. cloacae challenges, DEGs analysis was performed. For fungus O. sinensis and C. militaris infection, each induced two distinct genes (Fig. 6A). In particular, the mRNA amount of IML14 individually increased in fat body at 72 h after O. sinensis infection, while CTL‐S3 mRNA levels exhibited 3.1‐fold up‐regulation in the fat body exposed to O. sinensis for 48 h; IML8 and IML18 were greatly induced in C. militaris challenged larvae at 72 and 12 h post‐infection (hpi), respectively (Fig. 6B). After fungus O. sinensis or C. militaris infection, seven genes including IML3, 4, 16, 20, CTL‐S5, S6 and CTL‐X6 were commonly up‐regulated, and IML7, 9 and CTL‐S2 were commonly down‐regulated (Fig. 6A and 6B). Expression levels of the gene encoding CTL‐X2 generally showed a two‐fold up‐regulation after infection (Fig. 6A and 6B). There are nine genes including genes encoding IML1, 2, 5, 10–13, 15 and 22 that were all quickly activated in all conditions. No unique induction was seen in the Ringer's treated groups.
Figure 6.
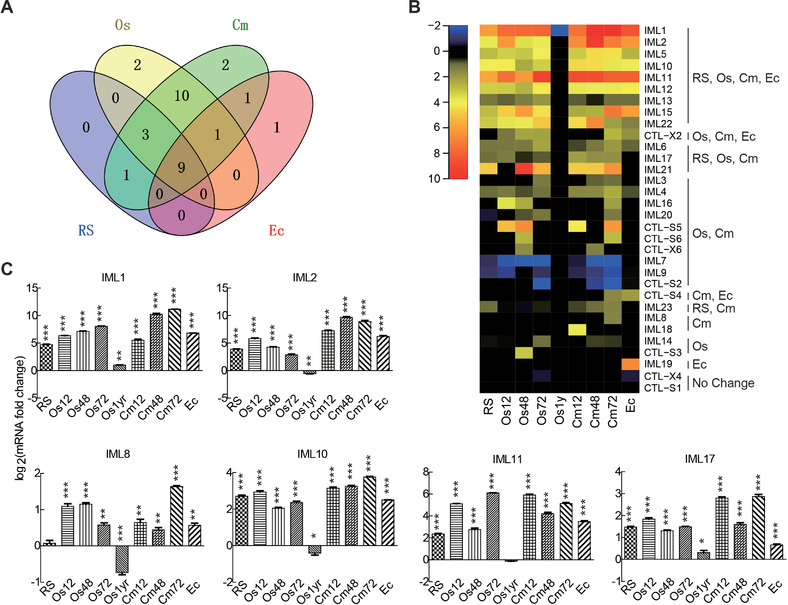
Induced expression of C‐type lectin domain protein (CTLDP) genes in Thitarodes xiaojinensis fat body by Ringer's (RS), Ophiocordyceps sinensis (Os), Cordyceps militaris (Cm) and Enterobacter cloacae (Ec) challenges. (A) Venn diagram demonstrating unique or common induction of a gene. The genes shown log2‐fold change > 1 or log2‐fold change < 0.5 after challenges were considered as differentially expressed genes and submitted to the Venny server (http://bioinfogp.cnb.csic.es/tools/venny/) for statistics. (B) Transcriptional changes of 32 candidate genes after different challenges. Gene boxes were colored based on the value of the log2‐fold change. Cm/Os12, Cm/Os48 and Cm/Os72 represent the larvae infected with C. militaris/O. sinensis for 12, 48 and 72 h. Os1yr represents the larvae infected with O. sinensis for a year. (C) Confirmation of differentially expressed genes by quantitative real‐time polymerase chain reaction. T. xiaojinensis ribosomal protein S3 (rpS3) was used as an internal control to normalize the templates. Messenger RNA levels are normalized to the control group and expressed as log2. Each untreated and treated group contained three larvae. The bars represent the means ± SEM (n = 3) from three replications and Student's t‐test was used to calculate the statistically significant difference among groups. * P < 0.05; ** P < 0.01; *** P < 0.001.
Discussion
In this study, 32 CTLDP genes were annotated from the T. xiaojinensis transcriptome. The orthologues of all T. xiaojinensis CTL‐Ss were found in other insects (Fig. 2A) indicating that the emergence of these six genes occurred earlier than species differentiation. T. xiaojinensis also lacked some genes, such as the orthologues to B. mori CTL‐S8–S12. The M. sexta genome encodes 19 IMLs, B. mori genome encodes six IMLs and the H. armigera genome encodes 11 IMLs (Rao et al., 2015a, b; Xiong et al., 2015). The number of IMLs (23) in T. xiaojinensis is relatively high and the phylogenetic tree shows a possible lineage‐specific expansion in Figure 2B. IMLs have probably experienced evolutionary selection pressure in the ghost moth. There are only three identified CTLDPs belonging to CTL‐X subfamily (Fig. 1 and Table S3). The neighbor‐joining tree in Figure 2C revealed that these three T. xiaojinensis CTL‐Xs individually formed orthologues to M. sexta CTL‐X2, X4 and X6. The absence of T. xiaojinensis orthologues to other M. sexta CTL‐X may be attributed to length limitations of the assembled transcripts. In addition, the distinct manner of evolution between the IML family (species‐specific expansion) and CTL‐S along with the CTL‐X family (monophyletic group) indirectly reflects their potential functions in the ghost moth. It seems that members in the IML family have undergone significant evolutionary pressure leading to duplication and divergence, indicating that they are probably targets for diverse foreigners. In contrast, conservation was observed in T. xiaojinensis CTL‐Ss and CTL‐Xs, the orthologues of which could be found in other species, indicating that they might deal with endogenous glycans or act in cell‐cell adhesion and tissue integration.
CTL‐S had sequences that were relatively simple with only one CRD and no other detectable domains (Table S3 and Fig. 1), and appears to be more primitive than the other two subfamilies. This was illustrated in the phylogenetic tree established by 53 CTLDs from T. xiaojinensis (Fig. 3). There were two reasons for building the tree in Figure 2A. One reason was to reveal the evolutionary relationships among the CTL‐Ss from various species. The close relation among CTL‐S1, S2 and S3 is consistent with the information in Figure 3. The second reason was to predict potential functions of T. xiaojinensis CTL‐S through their relationships with other characterized CTL‐S (Table S1). Two well‐known CTLDs (AgCTL4 and MA2) in A. gambiae have been reported to protect Plasmodium parasites from melanization (Osta et al., 2004). A. aegypti mosGCTL‐1 and ‐3 are used as receptors or attachment factors to facilitate West Nile virus (WNV) and dengue virus‐2 (DENV‐2) invasion, respectively (Cheng et al., 2010; Liu et al., 2014). Five CTL‐Ss from Armigeres subalbatus were tested for their roles in innate immunity and AsCTLGA5 was involved in resistance against Escherichia coli by rna interference (RNAi) experiments (Shi et al., 2014). However, none of these well‐studied CTL‐S exhibited close connections with T. xiaojinensis CTL‐S. They all clustered together reflecting mosquito‐specific expansion (Fig. 2A). These receptors have been exposed to significant evolutionary pressure and the distinctive extension in some members might be driven by particular microbes. In Lepidoptera, the characteristics of only one CTL‐S have been determined, namely B. mori CTL‐S4. This had no ability to bind microorganisms and its function is still unknown (Takase et al., 2009). Therefore, determining the roles of Lepidopteran CTL‐S will require more molecular and biochemical experiments.
Unlike mammalian or Dipteran members of the CTL‐S family, Lepidoptera have enlarged their own IML family during evolution. An apparent lineage‐specific expansion of IML is seen in many Lepidoptera. Based on the characteristics of IMLs from M. sexta, B. mori, Antheraea pernyi and H. armigera, these proteins are mainly involved in defense responses, such as binding to intruders and inducing phagocytosis, agglutination, encapsulation, PPO activation and melanization (Table S2). The evolved pattern of IMLs and known functions of some receptors suggest that IMLs may be primary targets for pathogens and parasites. Therefore, they may have evolved into tandem‐CRD forms to enhance binding affinity and extend their spectrum of recognition. B. mori IML6, M. sexta IML1, H. armigera CTL3, Spodoptera exigua LL3 and Papilio xuthus XP_011552083.1 (Genbank accession number) formed an orthologous group and so did B. mori IML4, M. sexta IML19, Danaus plexippus CTL18 and P. xuthus XP_013176126.1 (Fig. 2B). However, no orthologues to those IMLs were identified in T. xiaojinensis (Fig. 2B) indicating that these genes emerged after divergence of B. mori, M. sexta, H. armigera and butterflies from primitive insect groups. In the end of IML‐A in H. armigera there is a conserved PXXC motif, while in the end of IML‐B there is a conserved FXCE motif (Wang et al., 2012). Most dual CRDs in T. xiaojinensis IMLs are consistent with that rule, except IML2B, 5B, 10B, 19B and IML17A (Fig. 4). We found another interesting property in their compositions. There is a conserved glycine (Gly) behind the Cys‐4 of IML‐B and the residues at the first site after Cys‐4 of IML‐A are Val/Ile/Leu/Ala. This feature is not only seen in T. xiaojinensis IMLs but also in other identified IMLs (Rao et al., 2015a, b; Xiong et al., 2015). It is known that the four to six Cys residues in CTLDs are responsible for formation of disulfide bonds, and thus motif signatures around Cys, conserved in most IMLs, may have important effects on protein sorting. Uncovering the characteristics of IML sequences will help distinguish IML from CTL‐S and CTL‐X. Under some conditions there is an incomplete amino acid sequence with only one CTLD, but it probably belongs to the IML subfamily, like T. xiaojinensis IML18 and IML21 (Fig. 4).
CTL‐X represents a subfamily of CTLDPs with CTLDs and other identically conserved domains. In T. xiaojinensis, there were three transcripts encoding proteins belonging to this subfamily (Table S3 and Fig. 1). Although the N‐ or C‐terminus is absent in T. xiaojinensis CTL‐X2, 4 and 6, the matched parts of these CTLDPs showed high identity (77.85%, 58.25% and 70.87%, respectively) to CTL‐X2, 4 and 6 from M. sexta. The phylogenetic tree in Figure 2C revealed monophyletic groups among CTL‐X orthologues from other insects, indicating these receptors might have similar functions in different species. The insect CTL‐Xs are mainly associated with cell adhesion and developmental regulation. It is reported that the D. melanogaster furrowed gene in CTL‐X2 group encodes a protein, similar to vertebrate selectins, and mutation of complement control protein regions in the protein interrupted development of sensory organs (Leshko‐Lindsay & Corces, 1997). Genetic, molecular and biochemical features of D. melanogaster contactin (an orthologue to Thitarodes CTL‐X4) were studied by Faivre‐Sarrailh et al. (2004) who demonstrated that this cell adhesion molecule is required for organization of septate junctions involved in formation and maintenance of charge and size selective barriers. Although the functions of many CTL‐X group members have been reported in D. melanogaster, the roles of CTLDs are unknown. Characteristics of most identified CTL‐X from Lepidoptera have not been investigated, and their functions such as properties of their CTLDs are unknown.
In addition to phylogenetic analysis, binding ligands and structural models for T. xiaojinensis CTLDPs were also analyzed. Except for one CRD in CTL‐S4, the remaining CRDs in T. xiaojinensis were all identified as having at least one Ca2+‐association site (Table S5). This proportion is quite different from M. sexta and B. mori. A total of 56 CTLDs were investigated in M. sexta and only 18 models contained one or two Ca2+ ions (Rao et al., 2015a). In B. mori, nearly 70% of the CTLDs were expected to interact with Ca2+ (Rao et al., 2015b). This difference suggests that the sites for Ca2+ interaction in CTLDPs are variable in different Lepidoptera species. It is worth noting that a type II antifreeze protein from the liver of smelt (Osmerus mordax) is homologous to CTLDP and its antifreeze activity was responsive to Ca2+ (Ewart et al., 1992). T. xiaojinensis is adapted to the cold alpine meadows of Xiaojin County, where the mean annual temperature is –3.2°C (Zhu et al., 2016). B. mori and M. sexta, as model insects, are reared in the laboratory at 25°C. Therefore, the preservation of Ca2+‐binding sites for most CTLDPs may be an environmental adaptation of T. xiaojinensis. In asialoglycoprotein and macrophage mannose receptors, missing Ca2+ causes transformation of the CRD region and this results in release of the bound ligands (Loeb & Drickamer, 1988). There is evidence that Ca2+ also plays a negative role in association with ligands. Plasminogen cannot interact with tetranectin (a CTLDP in humans) until Ca2+ removes it from the binding site (Graversen et al., 1998). We believe that T. xiaojinensis uses Ca2+ ions as a mediator to flexibly control the recognition of CTLDPs to other ligands.
Expression profiles of CTLDPs in the fat body (a major immune tissue for insects) of T. xiaojinensis after different stimulations provide some indications about their functions. T. xiaojinensis CTL‐Xs are not expected to participate in immune defense, since their FPKM values are all very low (Table S6). Considering the characteristics of D. melanogaster CTL‐X, T. xiaojinensis CTL‐Xs may be involved in cell‐cell adhesion or tissue integration. T. xiaojinensis CTL‐S1–S6 are also expressed at low levels in all untreated and treated larvae (Table S6), indicating that their targets are probably endogenous sugars or sugar derivatives rather than microbes. To T. xiaojinensis, pathogenicity of O. sinensis is different from pathogenicity of C. militaris and E. cloacae. After infection with C. militaris or E. cloacae, the larvae die within several days. However, O. sinensis can infect larvae for a long time, ranging from several months to 1 year (Meng et al., 2015). As previously noted, IMLs probably play significant roles in binding a variety of microbes and activating immune responses in T. xiaojinensis. Therefore, the inducible ability of T. xiaojinensis IMLs could provide clues to decipher distinct reactions that are elicited by different challenges. IML8 is likely an important target for C. militaris, as its mRNA level was greatly up‐regulated in larvae infected by the fungus for 72 h (Fig. 6B). IML14 is likely a specific receptor for O. sinensis, as its significant up‐regulation was only detected in larvae infected by O. sinensis for 72 h (Fig. 6B). It would be useful to compare functional features of IML14 and 8 in T. xiaojinensis immunity.
In conclusion, 32 CTLDP genes identified from T. xiaojinensis were divided into three subfamilies and renamed. Phylogenetic analysis revealed evolutionary diversification of these genes. Gene duplication and merging may have resulted in IMLs possessing two CTLDs and natural selection‐based expansion of this subfamily in T. xiaojinensis. In addition, most of the identified CTLDPs in T. xiaojinensis had Ca2+ affinity possibly to regulate interactions of these receptors with other ligands and to protect them from freezing. Structural comparisons revealed that the structure of the second loop could also affect the specificity of binding ligands. CTLDP gene expression profiles in response to different immune challenges provide useful information for future functional studies in species of primitive Lepidoptera.
Disclosure
The authors declare no conflicts of interests.
Supporting information
Fig. S1. Structure‐based sequence alignment of Thitarodes xiaojinensis C‐type lectin domains (CTLDs) and the CTLD in human DC‐SIGNR. Multiple sequence alignment was carried out by MUSCLE, a module in MEGA 6.0, and depicted by ESPript. Identical residues are marked in red and similar residues are in yellow. The carbohydrate‐recognition motifs are indicated by the box enclosed with a red line.
Table S1. Functions of the C‐type lectin domain proteins (CTLDPs) with one carbohydrate‐recognition domain (CRD) in Dipteran.
Table S2. Functions of the C‐type lectin domain proteins (CTLDPs) with two carbohydrate‐recognition domains (CRDs) in Lepidoptera.
Table S3. Features of 32 Thitarodes xiaojinensis C‐type lectin domain proteins (CTLDPs).
Table S4. Primers for 6 genes encoding C‐type lectin domain proteins (CTLDPs).
Table S5. Structural characteristics of 53 C‐type lectin domains (CTLDs) in Thitarodes xiaojinensis.
Table S6. FPKM values of 32 Thitarodes xiaojinensis genes encoding C‐type lectin domain proteins (CTLDPs) in ten libraries.
Acknowledgements
This work was supported by National Key Plan for Scientific Research and Development of China (2017YFD0200400, 2016YFC1200603), National Natural Science Foundation of China (No. 31272366, 30900947, 31672291), and Open research Fund Program of State Key Laboratory of Integrated Pest Management (Chinese IPM1505, 1612).
The copyright line for this article was changed on 18 March 2019 after original online publication.
Contributor Information
Qi‐Lian Qin, Email: qinql@ioz.ac.cn.
Zhen Zou, Email: zouzhen@ioz.ac.cn.
References
- Ao, J. , Ling, E. and Yu, X.Q. (2007) Drosophila C‐type lectins enhance cellular encapsulation. Molecular Immunology, 44, 2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, G. , Cox, J. , Wang, P. , Krishnan, M.N. , Dai, J. , Qian, F. et al (2010) A C‐type lectin collaborates with a CD45 phosphatase homolog to facilitate west nile virus infection of mosquitoes. Cell, 142, 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao, J. , Ling, E. and Yu, X.Q. (2007) Drosophila C‐type lectins enhance cellular encapsulation. Molecular Immunology, 44, 2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, G. , Cox, J. , Wang, P. , Krishnan, M.N. , Dai, J. , Qian, F. et al (2010) A C‐type lectin collaborates with a CD45 phosphatase homolog to facilitate west nile virus infection of mosquitoes. Cell, 142, 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.D. , Aronstein, K. , Chen, Y.P. , Hetru, C. , Imler, J.L. , Jiang, H. et al (2006) Immune pathways and defence mechanisms in honey bees Apis mellifera . Insect Molecular Biology, 15, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart, K.V. , Rubinsky, B. and Fletcher, G.L. (1992) Structural and functional similarity between fish antifreeze proteins and calcium‐dependent lectins. Biochemical and Biophysical Research Communications, 185, 335–340. [DOI] [PubMed] [Google Scholar]
- Faivre‐Sarrailh, C. , Banerjee, S. , Li, J. , Hortsch, M. , Laval, M. and Bhat, M.A. (2004) Drosophila contactin, a homolog of vertebrate contactin, is required for septate junction organization and paracellular barrier function. Development, 131, 4931–4942. [DOI] [PubMed] [Google Scholar]
- Graversen, J.H. , Lorentsen, R.H. , Jacobsen, C. , Moestrup, S.K. , Sigurskjold, B.W. , Thogersen, H.C. et al (1998) The plasminogen binding site of the C‐type lectin tetranectin is located in the carbohydrate recognition domain, and binding is sensitive to both calcium and lysine. Journal of Biological Chemistry, 273, 29241–29246. [DOI] [PubMed] [Google Scholar]
- Koizumi, N. , Imai, Y. , Morozumi, A. , Imamura, M. , Kadotani, T. , Yaoi, K. et al (1999) Lipopolysaccharide‐binding protein of Bombyx mori participates in a hemocyte‐mediated defense reaction against gram‐negative bacteria. Journal of Insect Physiology, 45, 853–859. [DOI] [PubMed] [Google Scholar]
- Lemaitre, B. and Hoffmann, J.A. (2007) The host defense of Drosophila melanogaster . Annual Review of Immunology, 25, 697–743. [DOI] [PubMed] [Google Scholar]
- Leshko‐Lindsay, L.A. and Corces, V.G. (1997) The role of selectins in Drosophila eye and bristle development. Development, 124, 169–180. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Zhang, F.C. , Liu, J.Y. , Xiao, X.P. , Zhang, S.Y. , Qin, C.F. et al (2014) Transmission‐blocking antibodies against mosquito C‐type lectins for dengue prevention. PLoS Pathogens, 10, e1003931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J.A. and Drickamer, K. (1988) Conformational changes in the chicken receptor for endocytosis of glycoproteins. Modulation of ligand‐binding activity by Ca2+ and pH. Journal of Biological Chemistry, 263, 9752–9760. [PubMed] [Google Scholar]
- Meng, Q. , Yu, H.Y. , Zhang, H. , Zhu, W. , Wang, M.L. , Zhang, J.H. et al (2015) Transcriptomic insight into the immune defenses in the ghost moth, Hepialus xiaojinensis, during an Ophiocordyceps sinensis fungal infection. Insect Biochemistry and Molecular Biology, 64, 1–15. [DOI] [PubMed] [Google Scholar]
- Nielsen, E.S. , Robinson, G.S. and Wagner, D.L. (2000) Ghost‐moths of the world: a global inventory and bibliography of the Exoporia (Mnesarchaeoidea and Hepialoidea) (Lepidoptera). Journal of Natural History, 34, 823–878. [Google Scholar]
- Osta, M.A. , Christophides, G.K. and Kafatos, F.C. (2004) Effects of mosquito genes on Plasmodium development. Science, 303, 2030–2032. [DOI] [PubMed] [Google Scholar]
- Pal, S. and Wu, L.P. (2009) Lessons from the fly: pattern recognition in Drosophila melanogaster . Advances in Experimental Medicine and Biology, 653, 162–174. [DOI] [PubMed] [Google Scholar]
- Probert, F. , Whittaker, S.B. , Crispin, M. , Mitchell, D.A. and Dixon, A.M. (2013) Solution NMR analyses of the C‐type carbohydrate recognition domain of DC‐SIGNR protein reveal different binding modes for HIV‐derived oligosaccharides and smaller glycan fragments. Journal of Biological Chemistry, 288, 22745–22757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, X.J. , Cao, X. , He, Y. , Hu, Y. , Zhang, X. , Chen, Y.R. et al (2015a) Structural features, evolutionary relationships, and transcriptional regulation of C‐type lectin‐domain proteins in Manduca sexta . Insect Biochemistry and Molecular Biology, 62, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, X.J. , Shahzad, T. , Liu, S. , Wu, P. , He, Y.T. , Sun, W.J. et al (2015b) Identification of C‐type lectin‐domain proteins (CTLDPs) in silkworm Bombyx mori . Developmental & Comparative Immunology, 53, 328–338. [DOI] [PubMed] [Google Scholar]
- Ren, J. , Wen, L.P. , Gao, X.J. , Jin, C.J. , Xue, Y. and Yao, X.B. (2009) DOG 1.0: illustrator of protein domain structures. Cell Research, 19, 271–273. [DOI] [PubMed] [Google Scholar]
- Roy, A. , Kucukural, A. and Zhang, Y. (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X.Z. , Kang, C.J. , Wang, S.J. , Zhong, X. , Beerntsen, B.T. and Yu, X.Q. (2014) Functions of Armigeres subalbatus C‐type lectins in innate immunity. Insect Biochemistry and Molecular Biology, 52, 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S.W. , Zou, Z. and Raikhel, A.S. (2011) A new factor in the Aedes aegypti immune response: CLSP2 modulates melanization. EMBO Reports, 12, 938–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase, H. , Watanabe, A. , Yoshizawa, Y. , Kitami, M. and Sato, R. (2009) Identification and comparative analysis of three novel C‐type lectins from the silkworm with functional implications in pathogen recognition. Developmental & Comparative Immunology, 33, 789–800. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji, T. , Ohashi‐Kobayashi, A. and Natori, S. (2006) Participation of a galactose‐specific C‐type lectin in Drosophila immunity. Biochemical Journal, 396, 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet, S.J. , Saeland, E. and van Kooyk, Y. (2008) Sweet preferences of MGL: carbohydrate specificity and function. Trends in Immunology, 29, 83–90. [DOI] [PubMed] [Google Scholar]
- Wang, J.L. , Liu, X.S. , Zhang, Q. , Zhao, H.B. and Wang, Y.F. (2012) Expression profiles of six novel C‐type lectins in response to bacterial and 20E injection in the cotton bollworm (Helicoverpa armigera). Developmental & Comparative Immunology, 37, 221–232. [DOI] [PubMed] [Google Scholar]
- Wang, R.J. , Lin, Z. , Jiang, H. , Li, J.C. , Saha, T.T. , Lu, Z.Y. et al (2017) Comparative analysis of peptidoglycan recognition proteins in endoparasitoid wasp Microplitis mediator . Insect Science, 24, 2–16. [DOI] [PubMed] [Google Scholar]
- Wang, X.L. , Zhang, J.H. , Chen, Y. , Ma, Y.L. , Zou, W.J. , Ding, G.Y. et al (2013) A novel pattern recognition protein of the Chinese oak silkmoth, Antheraea pernyi, is involved in the pro‐PO activating system. Biochemistry and Molecular Biology Reports, 46, 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.H. , Hu, Y. , Xing, L.S. , Jiang, H. , Hu, S.N. , Raikhel, A.S. et al (2015) A critical role for CLSP2 in the modulation of antifungal immune response in mosquitoes. PLoS Pathogens, 11, e1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, A. , Miyazawa, S. , Kitami, M. , Tabunoki, H. , Ueda, K. and Sato, R. (2006) Characterization of a novel C‐type lectin, Bombyx mori multibinding protein, from the B. mori hemolymph: mechanism of wide‐range microorganism recognition and role in immunity. Journal of Immunology, 177, 4594–4604. [DOI] [PubMed] [Google Scholar]
- Waterhouse, R.M. , Kriventseva, E.V. , Meister, S. , Xi, Z. , Alvarez, K.S. , Bartholomay, L.C. et al (2007) Evolutionary dynamics of immune‐related genes and pathways in disease‐vector mosquitoes. Science, 316, 1738–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Charoensapsri, W. , Nakamura, S. , Tassanakajon, A. , Söderhäll, I. and Söderhäll, K. (2013) An MBL‐like protein may interfere with the activation of the proPO‐system, an important innate immune reaction in invertebrates. Immunobiology, 218, 159–168. [DOI] [PubMed] [Google Scholar]
- Xiong, G.H. , Xing, L.S. , Lin, Z. , Saha, T.T. , Wang, C. , Jiang, H. et al (2015) High throughput profiling of the cotton bollworm Helicoverpa armigera immunotranscriptome during the fungal and bacterial infections. BMC Genomics, 16, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Roy, A. and Zhang, Y. (2013) Protein‐ligand binding site recognition using complementary binding‐specific substructure comparison and sequence profile alignment. Bioinformatics, 29, 2588–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X.Q. , Tracy, M.E. , Ling, E. , Scholz, F.R. and Trenczek, T. (2005) A novel C‐type immulectin‐3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochemistry and Molecular Biology, 35, 285–295. [DOI] [PubMed] [Google Scholar]
- Yu, X.Q. , Gan, H. and Kanost, M.R. (1999) Immulectin, an inducible C‐type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochemistry and Molecular Biology, 29, 585–597. [DOI] [PubMed] [Google Scholar]
- Yu, X.Q. , Ling, E. , Tracy, M.E. and Zhu, Y. (2006) Immulectin‐4 from the tobacco hornworm Manduca sexta binds to lipopolysaccharide and lipoteichoic acid. Insect Molecular Biology, 15, 119–128. [DOI] [PubMed] [Google Scholar]
- Zelensky, A.N. and Gready, J.E. (2005) The C‐type lectin‐like domain superfamily. FEBS Journal, 272, 6179–6217. [DOI] [PubMed] [Google Scholar]
- Zhang, D.L. and Tu, Y.Q. (2015) The determination of Thitarodes xiaojinensis ecotype. Journal of Environmental Entomology, 37, 1055–1059. [Google Scholar]
- Zhu, W. , Zhang, H. , Li, X. , Meng, Q. , Shu, R.H. , Wang, M.L. et al (2016) Cold adaptation mechanisms in the ghost moth Hepialus xiaojinensis: Metabolic regulation and thermal compensation. Journal of Insect Physiology, 85, 76–85. [DOI] [PubMed] [Google Scholar]
- Zou, Z. , Evans, J.D. , Lu, Z. , Zhao, P. , Williams, M. , Sumathipala, N. et al (2007) Comparative genomic analysis of the Tribolium immune system. Genome Biology, 8, R177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Structure‐based sequence alignment of Thitarodes xiaojinensis C‐type lectin domains (CTLDs) and the CTLD in human DC‐SIGNR. Multiple sequence alignment was carried out by MUSCLE, a module in MEGA 6.0, and depicted by ESPript. Identical residues are marked in red and similar residues are in yellow. The carbohydrate‐recognition motifs are indicated by the box enclosed with a red line.
Table S1. Functions of the C‐type lectin domain proteins (CTLDPs) with one carbohydrate‐recognition domain (CRD) in Dipteran.
Table S2. Functions of the C‐type lectin domain proteins (CTLDPs) with two carbohydrate‐recognition domains (CRDs) in Lepidoptera.
Table S3. Features of 32 Thitarodes xiaojinensis C‐type lectin domain proteins (CTLDPs).
Table S4. Primers for 6 genes encoding C‐type lectin domain proteins (CTLDPs).
Table S5. Structural characteristics of 53 C‐type lectin domains (CTLDs) in Thitarodes xiaojinensis.
Table S6. FPKM values of 32 Thitarodes xiaojinensis genes encoding C‐type lectin domain proteins (CTLDPs) in ten libraries.


