Abstract
Understanding the mechanisms regulating recruitment of human skeletal (stromal or mesenchymal) stem cells (hMSC) to sites of tissue injury is a prerequisite for their successful use in cell replacement therapy. Chemokine‐like protein TAFA2 is a recently discovered neurokine involved in neuronal cell migration and neurite outgrowth. Here, we demonstrate a possible role for TAFA2 in regulating recruitment of hMSC to bone fracture sites. TAFA2 increased the in vitro trans‐well migration and motility of hMSC in a dose‐dependent fashion and induced significant morphological changes including formation of lamellipodia as revealed by high‐content‐image analysis at single‐cell level. Mechanistic studies revealed that TAFA2 enhanced hMSC migration through activation of the Rac1‐p38 pathway. In addition, TAFA2 enhanced hMSC proliferation, whereas differentiation of hMSC toward osteoblast and adipocyte lineages was not altered. in vivo studies demonstrated transient upregulation of TAFA2 gene expression during the inflammatory phase of fracture healing in a closed femoral fracture model in mice, and a similar pattern was observed in serum levels of TAFA2 in patients after hip fracture. Finally, interleukin‐1β was found as an upstream regulator of TAFA2 expression. Our findings demonstrate that TAFA2 enhances hMSC migration and recruitment and thus is relevant for regenerative medicine applications. Stem Cells 2019;37:407–416
Keywords: Mesenchymal stem (stromal) cell, Migration, Fracture healing, Regenerative medicine, TAFA2
During the inflammatory phase of fracture healing, elevated levels of interleukin‐1β induce TAFA2 production at the site of fracture, leading to recruitment of osteoprogenitor cells needed for subsequent bone formation.
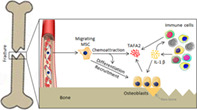
Significance Statement.
Skeletal (mesenchymal) stem cells (MSC) are being used in an increasing number of clinical trials for their therapeutic benefits in tissue regeneration. Understanding the mechanisms that regulate MSC migration and homing is a prerequisite for their successful use in cell therapy applications. Results show that TAFA2, a recently identified neurokine, is highly expressed at the site of skeletal fracture and induces MSC migration through activation of Rac1/p38 signaling.
Introduction
Skeletal (also known as bone marrow stromal or mesenchymal) stem cells (MSC) are nonhematopoietic cells residing in the bone marrow stroma and are capable of differentiation to mesodermal cell types such as osteoblasts, adipocytes, and chondrocytes 1, 2. MSC are recruited to the bone formation sites during bone remodeling and after bone fracture, where they differentiate into osteoblastic bone‐forming cells 3. Insufficient or defective recruitment of MSC to bone formation sites has been suggested as a mechanism mediating impaired bone formation in osteoporosis and in nonunion or poorly healing fractures 4, 5.
MSC are being tested in an increasing number of clinical trials for enhancing tissue regeneration, including bone tissue regeneration, where direct transplantation of MSC cultured on scaffolds has been tested with good preliminary results 6, 7, 8. Systemic (intravenous) infusion of MSC is an attractive route of administration because of its ease and clinical feasibility 6, 7, 8. Also hematopoietic stem cell (HSC) transplantation has demonstrated the ability of HSC to home to their bone marrow niche after intravenous infusion and to exert their biological functions 9, suggesting that MSC may be equipped with similar functional capacity.
Previous studies have demonstrated the presence of MSC in the circulation and that they can be recruited to injured tissues (for review, please see 10). MSC can adhere to endothelial cells and exhibit leukocyte‐like trans‐endothelial migration 11. However, few cells are recruited to fracture sites after systemic infusion of MSC 12, 13. Interestingly, some studies have reported enhanced tissue regeneration despite the presence of few engrafted MSC 14, 15, 16, 17. To increase the number of MSC homed to the bone, a number of approaches have been tried, for example, modifying CD44 expression by MSC 18 or surface conjugation of MSC with a synthetic peptidomimetic ligand with high affinity for activated α4β1 integrin to target MSC to the skeleton 19 or to precondition MSC with small molecules before systemic infusion in order to boost their skeletal homing properties 12.
In order to identify secreted factors with capacity for enhancing recruitment of MSC to the skeleton, we examined a number of factors known to regulate migration and motility of adult stem cells in a number of tissues. We identified TAFA2 in a screening experiment using chemoattraction of MSC as an endpoint. TAFA2 (also known as FAM19A2, family with sequence similarity 19 member A2) is a member of FAM19A family consisting of five highly homologous genes (TAFA1‐5) encoding small secreted proteins 20. TAFA proteins are broadly expressed with highest expression in the brain and function as neurokines 20. Despite lack of functional studies about biological roles of TAFA2, genome‐wide association studies have revealed an association between TAFA2 gene polymorphism and mental retardation and insulin sensitivity 21, 22. Here, we report a novel role for TAFA2 in recruiting human MSC (hMSC) to the site of skeletal fracture. Using cell‐based and in vivo studies, we show that TAFA2 stimulates hMSC migration through activation of Rac1‐p38 signaling. TAFA2 overexpression at the site of skeletal fracture increased the number of recruited hMSC to the injured tissue. TAFA2 levels were differentially regulated during the time course of fracture healing in a closed femoral fracture model in mice, and a similar pattern was observed in TAFA2 serum levels in patients with hip fractures.
Materials and Methods
MSC Culture
As a model for primary hMSC, we used our well‐characterized, immortalized human MSC line (hereafter referred to as hMSC), established by ectopic expression of the catalytic subunit of human telomerase as previously described 40. hMSC maintain all typical characteristics of primary hMSC 41. Primary bone marrow‐derived hMSC (hereafter referred to as hpMSC) were established from fresh bone marrow aspirates as previously described, from three young healthy donors after informed consent 42. The cells were cultured in a standard growth medium containing minimal essential medium (MEM; Gibco, Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Biochrom, Germany) and 1% penicillin/streptomycin (Gibco, UK) at 37°C in a humidified atmosphere containing 5% CO2. The medium was changed every third day until the cells became 90% confluent.
Boyden Chamber Chemotaxis Trans‐Well Migration Assay
The migration ability of hMSC toward a number of chemoattractant factors identified by NeuroSearch A/S, Copenhagen, Denmark, and provided by Dr. Per Horn was measured in a 48‐well microchemotaxis Boyden chamber‐based cell migration assay (8 μm pores, polycarbonate membrane, NeuroProbe, Inc., USA). The membrane was coated for 1 hour at 37°C with migration media (low glucose Dulbecco's MEM [LG‐DMEM, Gibco]), supplemented with 0.2% FBS (Gibco) containing 5 μg/ml fibronectin and 10 μg/ml rat tail collagen I. hMSC were serum starved in migration media 24 hours before migration assay. The chemotaxis and control media (27 μl) were placed in the lower chamber and covered by the coated membrane and fixed by the upper chamber. hMSC (1.25 × 104) suspended in 50 μl migration media were loaded into each well of the upper chamber. The migration was carried out for 4 hours in 37°C. Nonmigrated cells on the upper side of the membrane were scraped and rinsed with cold phosphate buffered saline (PBS). Migrated cells on the lower side of the membrane were fixed and stained with hemacolor staining kit (Merck, Germany). The migrated cells were scanned and photos were taken at ×20 magnification by a computer‐assisted cell‐counting system (Visiopharm, Denmark). Images were captured of the entire wells and counted by using ImagJ software. Results were expressed as mean number of cells per at least six replicates and three independent experiments. For inhibition of the signaling pathways, the cells were incubated 30 minutes before the seeding in the chambers, with p38 inhibitor SB203580 (Sigma) or dimethyl sulfoxide (DMSO) (Sigma, USA) used as vehicle.
Oris Cell Motility Assay
The assay was performed according to the manufacturer's instructions (Platypus, Madison, WI). Briefly, 96‐well cell culture plates were coated with fibronectin (10 μg/ml) in PBS for 2 hours at 37°C, rinsed twice with PBS. After washing, the Oris cell seeding stoppers were inserted in the wells. hMSC were trypsinized, washed twice with MEM, and resuspended in 0.2% serum medium, seeded and permitted to attach at a density of 2.5 × 103 cells in each well. Thereafter, the stoppers were gently lifted from all wells excluding the designated “reference” wells (n = 6). Then, cells were washed with migration media (LG‐DMEM, supplemented with 0.2% FBS) and permitted to migrate during 24 hours in fresh migration media supplemented with chemoattractant in a humidified chamber (37°C, 5% CO2). The cells were washed and fixed for 10 minutes in 4% paraformaldehyde/PBS, permeabilized with 0.1% Triton X‐100 in PBS and stained with tetramethylrhodamine (TRITC)‐conjugated phalloidin solution for 40 minutes followed by 4′,6‐diamidino‐2‐phenylindole (DAPI; 1 μg/ml). The mean fluorescence intensity of the migrated cells was analyzed by an enzyme‐linked immunosorbent assay (ELISA) microtiter plate reader (FLUOstar Omega, BMG LABTECH, Ortenberg, Germany).
Quantitation of Cell Spreading and Immunofluorescent Intensity
Ninety‐six‐well cell culture plates (Perkin Elmer, Denmark) were coated with fibronectin (10 μg/ml) in PBS for 2 hours at 37°C, rinsed twice with PBS, and blocked with 1% BSA for 1 hour. hMSC were trypsinized, washed twice with MEM media, and resuspended in serum‐free medium. Cells were stimulated with 10 μg/ml TAFA2 (R&D systems, Minneapolis, MN) for 30 minutes at 37°C and then replated onto fibronectin‐coated plates in standard culture medium. Cells were fixed in 4% paraformaldehyde for 10 minutes, washed with PBS, and stained for F‐actin with phalloidin (TRITC‐labeled, Sigma) and DAPI for nuclear staining. Fluorescent images were analyzed by Operetta high‐content‐imaging system (Perkin Elmer) at ×20 magnification using Harmony High Content Imaging and Analysis Software (PerkinElmer).
Western Blotting Analysis
hMSC were cultured until 80% confluent, then starved in migration media for 24 hours before treatment with TAFA2 at 10 μg/ml or 10% FBS. Protein samples were harvested at 0, 10, 30, 60, 120, and 240 minutes after treatment. Briefly, cells were washed in PBS and lysed in RIPA buffer (Sigma, USA) supplemented with protease and phosphatase inhibitors (Roche, Germany). After 30 minutes incubation at 4°C, samples were centrifuged for 10 minutes at 12,000 rpm, 4°C. Protein concentration was determined with Pierce Coomassie Plus Bradford assay (Thermo Fisher Scientific, USA), and equal amounts of proteins were loaded on a 10% polyacrylamide gel (Novex, Life technologies, USA). Blotted nitrocellulose membranes were incubated overnight with antibodies against P‐p38, p38, P‐AKT, AKT, P‐ERK, ERK2 (Cell Signaling, USA), and α‐tubulin (Sigma, USA), overnight at 4°C. Membranes were incubated with horseradish peroxidase (HRP)‐conjugated secondary antibody (Santa Cruz Biotechnology, USA) for 45 minutes at room temperature, and protein bands were visualized with Amersham ECL chemiluminescence detection system (GE Healthcare, UK).
In Vivo Studies of Fracture Healing
The ethical approval for femur fracture in mice was granted by the Danish National Animal Experiment Inspectorate (2012‐15‐2934‐00559). Female immunodeficient NOD.CB17‐Prkdcscid/J (NOD‐scid, 12 weeks old) and C57BL/6J (10 weeks old) mice were anesthetized by administration of intraperitoneal ketamine hydrochloride (100 mg/kg body weight) and xylazine hydrochloride (10 mg/kg). The closed femur fracture procedure was carried out as described 45. Briefly, an incision was made just medial to the patella, and the fracture was fixed by inserting a 0.5 mm needle between the condyles and drilled in a retrograde fashion into the medullary cavity of the femur. Afterward, a standardized reproducible closed fracture was induced using a blunt‐impact guillotine fracture apparatus. A radiograph was acquired after fracture induction (using Faxitron MX‐20; Faxitron, USA) to confirm the fracture induction and needle position.
Immunohistochemical Staining
Tissues were harvested at different time points and fixed in 4% formalin for 3 days. Bones were decalcified in formic acid. The tissue was paraffin embedded, sectioned, and stained for human TAFA2 protein expression. Briefly, the staining was performed using DAKO En Vision+ and PowerVision according to manufacturer's instructions. The tissue sections were demasked using TEG solution (Tris 10 mM, EGTA 0.5 mM, pH 9.0) (15 minutes) and incubated 1 hour with sheep anti‐human TAFA2 antibodies (R&D Systems, Abingdon, U.K.) diluted in ChemMate Antibody diluent (1:30 Dako, Glostrup, Denmark). After washing, the sections were incubated with secondary antibody (rabbit anti‐goat antibody, cell signaling, USA) for 20 minutes. The slides were processed on an automatic slide processor (Techmate500, Dako, Glostrup, Denmark). Analysis was carried out on an IX50 Olympus microscope using Olympus DP Software v3.1 (Olympus, Essex, U.K.) or a Leica DM4500 (Leica, Wetzlar, Germany) using the Surveyor Turboscan Mosaic acquisition imaging analysis system v5.04.01 (Objective Imaging Ltd, Cambridge, U.K.).
Antibody Labeling and Flow Cytometry Analysis
The cells were incubated with antibodies against cell surface proteins for 20 minutes on ice, and two washes with PBE (PBS containing 2 mM EDTA and 0.5% FBS) were done to remove unbound antibody. For intracellular staining of TAFA2, the cells were fixed and permeabilized with BD‐fixation and permeabilization kit (BD Biosciences, USA), and the cells were stained with 2 μg anti‐TAFA2 antibodies and incubated for 1 hour on ice in dark. Cells were washed, fixed in 2% paraformaldehyde, and cell acquisition was performed with flow cytometer BD FACS LSRII. Kaluza 1.2 software program was used for analysis of the data. Mean fluorescence intensity (MFI) value of a relevant isotype control staining was subtracted from the corresponding MFI of the specific staining. The flow cytometer was standardized by using positive and negative controls and flow check beads each time to reduce day‐to‐day variation.
RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction
RNA was isolated from organs or cultured cells according to previously published method. 43 After cDNA synthesis, quantitative reverse transcription polymerase chain reaction was performed using a comparative Ct method [(1/ (2ΔCt)] formula, and data were represented as relative expression to the HPRT1 reference gene.
Osteoblast Differentiation and ALP Activity Assay
Osteogenic induction was performed using MEM supplemented with 10 mM β‐glycerophosphate (Calbiochem‐Merck, Germany), 50 μg/ml l‐ascorbic acid‐2‐phosphate (Wako Chemicals GmbH, Germany), 10 nM dexamethasone (Sigma‐Aldrich, Denmark), and 10 nM calcitriol (1.25‐dihydroxy vitamin‐D3) kindly provided by Leo Pharma, Ballerup, Denmark. Medium was changed every third day during induction. The induction was performed with and without addition of different concentrations of rhTAFA2 protein. ALP activity was measured at day 6 of induction. Simultaneously, the numbers of viable cells were determined on the day of ALP activity measurement by adding the CellTiter‐Blue solution (Promega, USA) to the wells. The cells were then rinsed with TBS (20 mM Trizma base, 150 mM NaCl at pH = 7.5) and fixed in 3.7% formaldehyde‐90% ethanol for 30 seconds at room temperature. A reaction mixture containing 100 μl 50 mM NaHCO3, 1 mM MgCl2 (Sigma, USA), and 1 mg/ml of p‐nitrophenyl phosphate (Sigma) was added into each well and incubated 20 minutes at 37°C. The reaction was stopped by adding 50 μl of 3 M NaOH. The absorbance at 405 nm was determined using an ELISA microtiter plate reader (FLUOstar Omega, BMG LABTECH, Ortenberg, Germany). ALP enzymatic activity was normalized to the number of viable cells. After osteoblastic induction at day 15, assessment of hydroxyapatite deposition was performed using the OsteoImage Bone Mineralization Assay (Lonza, USA) according to manufacturer's instructions. Data were analyzed, and noninduced cells were compared with induced and induced cells supplemented with rhTAFA2 (10 μg/ml).
Adipogenic Differentiation
Adipogenic differentiation of hMSC was induced using MEM supplemented with 10% FBS (Sigma‐Aldrich, Brøndby, Denmark), 10% horse serum (Sigma‐Aldrich, USA), 100 nM dexamethasone (Sigma‐Aldrich, Denmark), 500 μM 1‐methyl‐3‐isobutylxanthine (IBMX, Sigma‐Aldrich, USA), 1 μM rosiglitazone (BRL49653, Cayman Chemical, Ann Arbor, MI), and 5 μg/ml insulin (Sigma‐Aldrich, USA) for 15 days. Media was changed every third day, and rhTAFA2 (10 μg/ml) was added to induced cells. Oil Red O staining and quantification was performed as previously described 43.
siRNA Transfection
siRNA transfection was performed using a reverse transfection protocol as previously described 42. Briefly, Lipofectamine 2000 was used as transfection reagent, and transfections were carried out according to the manufacturer's instructions (Invitrogen, USA). Nontargeting siRNA No.2 (Ambion, CA, USA) and siRNA against Rac1 (sense: CUACUGUCUUUGACAAUUAtt) (Ambion) were used at 25 nM.
ELISA Measurement of TAFA2 Concentrations in Clinical Samples
ELISA measurements of TAFA2 concentrations in human serum samples were performed using an established ELISA procedure (Mybiosource, San Diego, USA) according to the manufacturer's instructions. The human serum samples used in this study were from patients with traumatic hip fracture admitted to the Orthopaedic Department at Herlev Hospital. The experimental protocol was approved by the regional scientific ethical committee in Copenhagen (Number KA 05081). All patients received oral and written information and participated under signed informed consent. Collection and handling of the serum samples was previously published 44.
Results
TAFA2 Enhanced Migration and Motility of hMSC
To identify novel regulators of hMSC recruitment to injured tissues, we screened a number of newly identified chemoattractant neurokines, for their ability to enhance in vitro trans‐well migration of hMSC using a three‐dimensional‐Boyden chamber assay. The members of TAFA family, TAFA2, 4, and 5, exhibited the highest chemoattractant activity for hMSC (Fig. 1A). Next, we used the Oris assay to confirm the trans‐well migration‐stimulating activity of TAFA proteins and found that TAFA2 is the most potent stimulator of hMSC motility (Fig. 1B, 1C). TAFA2 also stimulated cell motility in cultures of human primary bone marrow‐derived MSC (hpMSC; Fig. 1D). To specify whether TAFA2‐induced cell migration is directional (chemotaxis) or random (chemokinesis), TAFA2 was added to both upper and lower compartments of the Boyden chamber and we observed a significant reduction in number of migrating hMSC (Fig. 1E) and hpMSC (Fig. 1F). TAFA2 exhibited a dose‐dependent chemoattractant effect on the trans‐well migration of hMSC (Fig. 1G), and a similar effect was observed in hpMSC (Fig. 1H, 1I). In addition, combining TAFA2 with different concentrations of anti‐human TAFA2 antibody reduced the chemotactic activity of TAFA2 (Fig. 1J).
Figure 1.
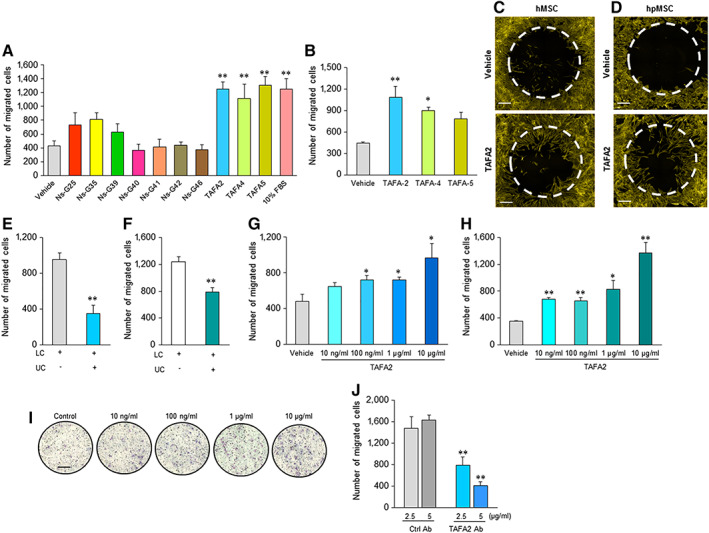
TAFA2‐enhanced migration of human skeletal (stromal, mesenchymal stem) cells (hMSC). (A): Screening of a number of secreted neurokines (25 μg/ml) for chemotactic activity in hMSC cultures using an in vitro three‐dimensional‐Boyden chamber trans‐well migration assay. Data represent mean ± SEM from six independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. (B, C): Measuring two‐dimensional (2D) migration of hMSC in the presence of TAFA2, TAFA4, and TAFA5 (10 μg/ml) using Oris cell migration assay. Scale bar: 50 μm. Data represent mean ± SEM from three independent experiments. *, p ≤ .05; **, p ≤ .01, two‐tailed unpaired Student t test. (D): 2D migration of primary hMSC (hpMSC) in the presence of TAFA2 (10 μg/ml) using Oris cell migration assay. Scale bar: 50 μm. Data represent three independent experiments. Quantitation of trans‐well migration of hMSC (E) and hpMSC (F) in the presence of recombinant human TAFA2 (10 μg/ml) in different compartments of Boyden chamber assay, LC and UC. Data represent mean ± SEM from three independent experiments **, p ≤ .01, two‐tailed unpaired Student t test. Quantitation of trans‐well migration of hMSC (G) and hpMSC (H) in the presence of different doses of TAFA2 (10 ng/ml, 100 ng/ml, 1 μg/ml, and 10 μg/ml). Data represent mean ± SEM from three independent experiments. *, p ≤ .05; **, p ≤ .01, two‐tailed unpaired Student t test. (I): Hemacolor staining of migrated cells toward different concentrations of TAFA2 (10 ng/ml, 100 ng/ml, 1 μg/ml, and 10 μg/ml) was present in the lower compartment of the Boyden chamber. Scale bar: 50 μm. Data are representative of three independent experiments. (J): Measuring hMSC trans‐well migration in the presence of TAFA2 (10 μg/ml) with and without anti‐human TAFA2 antibody (2.5 and 5 μg/ml). Data represent mean ± SEM from three independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. Abbreviations: LC, lower chamber; UC, upper chamber.
TAFA2 Stimulates hMSC Migration Through Activation of Rac1‐p38 Signaling
Formation of lamellipodia is an early event initiating migration of various cell types such as fibroblasts and leukocytes 23. We used the Operetta high‐content‐imaging system to perform quantitative analysis of TAFA2‐induced morphological changes in cultured hMSC at a single‐cell resolution. We noted that TAFA2 induced formation of lamellipodia (cell protrusions that are needed for cell movement) in cultured hMSC (Fig. 2A) that was associated with increased cell area (Fig. 2B). We then examined the activation status of Rac1 GTPase that is required for formation of lamellipodia and cell migration. Analysis of lysates from TAFA2‐stimulated hMSC showed significantly increased levels of Rac1 activity at 10, 15, and 30 minutes after TAFA2 stimulation, which returned to the initial levels after 60 minutes (Fig. 2C). To examine the role of Rac1 in TAFA2‐induced hMSC migration, we performed small interfering RNA (siRNA)‐mediated knockdown of Rac1, which decreased Rac1 expression in hMSC cultures (Fig. 2D) and abolished the TAFA2‐induced hMSC trans‐well migration (Fig. 2E). To identify the downstream signaling pathways involved in TAFA2‐mediated hMSC motility, we examined the activation status of several signaling factors involved in cell migration: p38, AKT, and ERK. We found increased levels of P‐p38 within the initial 2 hours after TAFA2 stimulation (Fig. 2F; Supporting Information Fig. S1). We did not detect changes in P‐Akt and P‐ERK levels in TAFA2‐stimulated hMSC cultures (Fig. 2F). Interestingly, inhibition of p38 signaling using SB203580 abolished TAFA2‐induced hMSC migration in a dose‐dependent manner (Fig. 2G, 2H).
Figure 2.
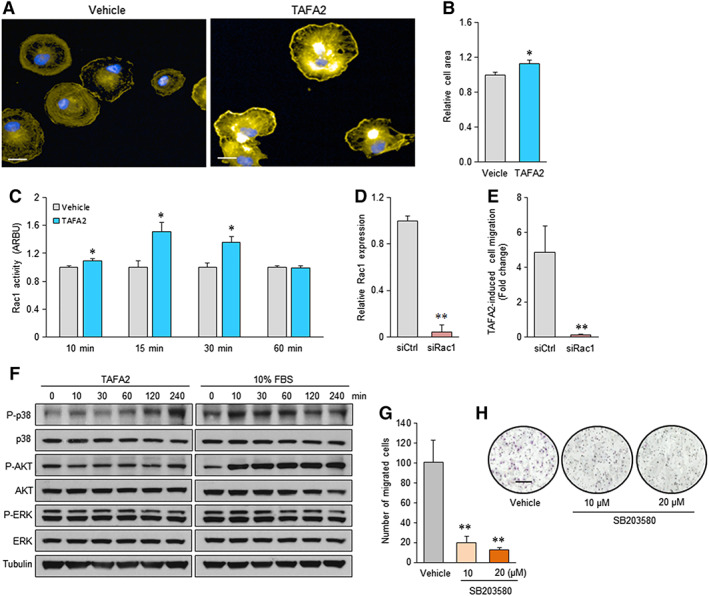
TAFA2‐induced lamellipodia formation and enhanced Rac1‐p38 signaling in cultures of human skeletal (mesenchymal, stromal) stem cells (hMSC). (A, B): Analysis of cell morphology of phalloidin‐stained hMSC cultures using Operetta high‐content imaging system and cell area quantitation at single‐cell level. Scale bar: 10 μm. Data represent mean ± SEM from three independent experiments. *, p ≤ .05, two‐tailed unpaired Student t test. (C): Quantitation of Rac1 activity in hMSC cultures, 10, 15, 30, and 60 minutes after stimulation with TAFA2 (10 μg/ml). (D): Quantitative reverse transcription polymerase chain reaction analysis of Rac1 expression in hMSC cultures transfected with siRac1 or siCtrl. Data represent mean ± SEM from three independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. (E): Quantitation of TAFA2‐induced trans‐well migration of hMSC transfected with siCtrl or siRac1. Data represent mean ± SEM from three independent experiments. *, p ≤ .05; **, p ≤ .01, two‐tailed unpaired Student t test. (F): Western blot analysis of P‐p38, p38, P‐Akt, Akt, P‐ERK, ERK, and tubulin in lysates from hMSC cultures at different time points after stimulation with TAFA2 (10 μg/ml) or 10% fetal bovine serum. Data are representative of two independent experiments. (G, H): Quantitation of TAFA2‐induced trans‐well migration in hMSC cultures pretreated with vehicle or p38 small molecule inhibitor (SB203580; 10 and 20 μM). Photomicrographs show Hemacolor staining of the migrated hMSC. Scale bar: 50 μm. Data represent mean ± SEM from three independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. Abbreviation: ARBU, arbitrary unit; siCtrl, control nontargeting small interfering RNA; siRac1, small interfering RNA against Rac1.
TAFA2 Regulates Proliferation, but Not Differentiation of hMSC
To investigate other possible functions of TAFA2 in hMSC biology, we tested whether TAFA2 could exert a regulatory effect on hMSC proliferation or differentiation. Adding TAFA2 to hMSC cultures in the presence of normal culture media increased cell proliferation as shown by increased cell number and Ki67 levels (Fig. 3A, 3B). Next, we examined TAFA2 expression during ex vivo differentiation of hMSC toward osteoblastic and adipocytic lineages. TAFA2 mRNA expression increased during osteoblast and adipocyte differentiation of hMSC (Fig. 3C, 3D). However, TAFA2 did not affect osteoblast differentiation as shown by quantitation of alkaline phosphatase (ALP) activity (Fig. 3E, 3F) and mineralized matrix formation assessed by measuring deposition of hydroxyapatite (Fig. 3G, 3H). Similarly, TAFA2 did not affect adipocyte differentiation of hMSC evidenced by measuring the number of mature lipid‐filled adipocytes (Fig. 3I, 3J).
Figure 3.
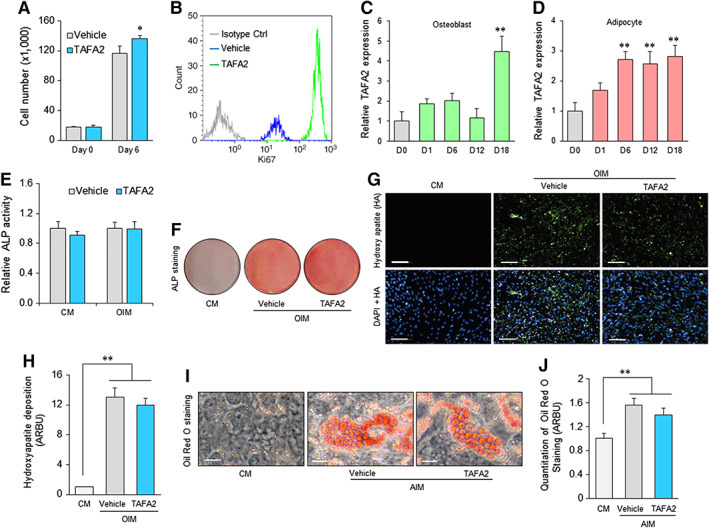
TAFA2‐enhanced proliferation, but not differentiation, of human skeletal (mesenchymal, stromal) stem cells (hMSC). (A): Cell number count in hMSC cultures at the baseline (day 0) or in the presence of vehicle or TAFA2 (10 μg/ml) for 6 days. Data represent mean ± SEM from three independent experiments. *, p ≤ .05, two‐tailed unpaired Student t test. (B): Flow cytometry analysis of Ki67 levels in hMSC cultures in the presence of vehicle or TAFA2 (10 μg/ml). TAFA2‐treated hMSC was also analyzed using isotype control antibody. (C, D): Quantitative reverse transcription polymerase chain reaction analysis of TAFA2 mRNA expression during osteoblast and adipocyte differentiation of hMSC. Data represent mean ± SEM from three independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. (E, F): Quantitation of ALP activity and ALP staining of osteogenic hMSC cultures 6 days after induction of differentiation. Data represent mean ± SEM from three independent experiments. (G, H): Analysis of mineralized matrix formation in osteogenic hMSC cultures by quantitation of hydroxyapatite deposition using Osteoimage bone mineralization assay on day 15 of differentiation. Scale bar: 100 μm. Data represent mean ± SEM from three independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. (I, J): Quantification of mature adipocytes by measuring accumulated lipid droplets in the presence of adipocyte induction medium using Oil Red O staining (day 15). Scale bar: 50 μm. Data represent mean ± SEM from three independent experiments. **, p ≤ .01, two‐tailed unpaired Student t test. Abbreviations: ALP, alkaline phosphatase; CM, culture media; DAPI, 4′,6‐diamidino‐2‐phenylindole; HA, hydroxy apatite; OIM, osteoblast induction media; AIM, adipocyte induction media.
TAFA2 Levels Are Differentially Regulated During Fracture Healing
To determine the relevance of TAFA2 to pathophysiology of fracture healing, we examined the pattern of TAFA2 expression at different phases of fracture healing in the mouse femoral fracture model. TAFA2 was highly expressed at the site of fracture 48 hours post induction. TAFA2 immunoreactivity was present in the fracture line‐adjacent bone marrow and in the injured muscle tissue above the fracture line, where the blunt impact from a dropped weight induced the mid‐diaphysis fracture (Fig. 4A). Interestingly, TAFA2 protein expression was already increased 2 hours after the fracture and expression peaked during the first week of fracture (Fig. 4B). TAFA2 mRNA and protein expression was also differentially regulated, with an initial increase in the early phase of fracture healing followed by downregulation (Fig. 4C, 4D). A similar pattern of expression was observed for the pro‐inflammatory cytokine interleukin‐1β (IL‐1β) (Fig. 4E). To investigate the role of IL‐1β as an upstream regulator of TAFA2 expression, we added different doses of IL‐1β to hMSC cultures and observed an enhancing effect on TAFA2 mRNA and protein levels (Fig. 4F, 4H). We then determined the cellular map of TAFA2 expression within the bone marrow during fracture healing. Interestingly, intracellular expression of TAFA2 in different leukocyte subpopulations was detectable with high levels of TAFA2 expression in monocytes and T cells at day 1 postfracture that was downregulated by day 3 (Fig. 4I), while monocyte cell frequency was not altered, and T‐cell frequency was increased on day 3 postfracture compared to day 1 (Fig. 4J). To extrapolate these findings to human physiology, we assessed the kinetics of TAFA2 levels in consecutive serum samples from patients after hip fracture. TAFA2 serum levels were high at the first day after hip fracture and decreased gradually in the subsequent measurements during a 21 day observation period (Fig. 4K).
Figure 4.
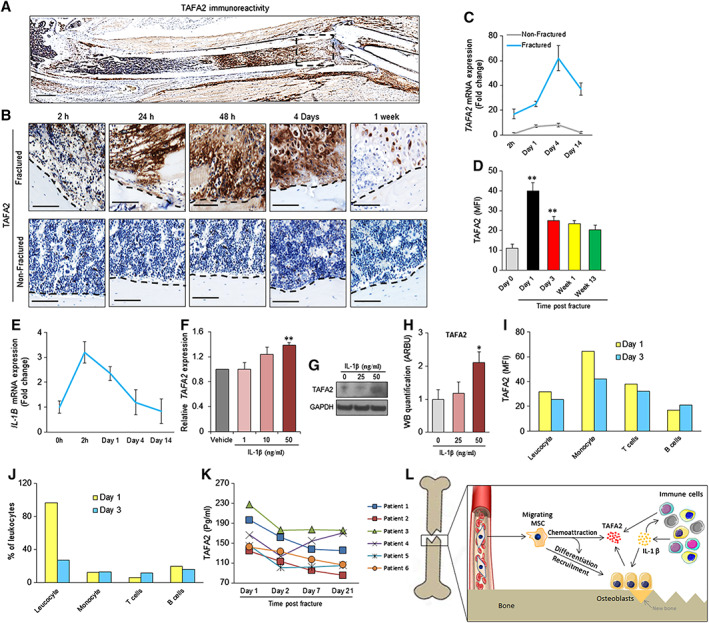
TAFA2 levels are regulated during the process of fracture healing. (A): Immunohistochemical analysis of TAFA2 protein expression in a fractured femur, 48 hours after induction of fracture. Scale bar: 1 mm. (B): Analysis of TAFA2 immunoreactivity at the fracture site (boxed area from A) at different time points (2, 24, and 48 hours, 4 days, and 1 week) after induction of femoral fracture (upper panel) and nonfractured femur (lower panel). Scale bar: 100 μm. (C): Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis of TAFA2 mRNA expression in nonfractured and fractured mouse femur at different time points after fracture induction (2 hours, day 1, day 4, and day 14). (D): Flow cytometry analysis of TAFA2 protein expression in bone marrow cells obtained from fractured femur at different time points. Data represent MFI ± SEM from three independent experiments. **, p ≤ .01; two‐tailed unpaired Student t test. (E): qRT‐PCR analysis of IL‐1β mRNA expression in mouse femur before fracture and at different time points after fracture induction (2 hours, day 1, day 4, and day 14). (F): qRT‐PCR analysis of TAFA2 mRNA expression in human skeletal (mesenchymal, stromal) stem cell (hMSC) cultures in the presence of different doses of IL‐1β (1, 10, and 50 ng/ml) for 48 hours. Data represent three independent experiments ± SEM; **, p ≤ .01, two‐tailed paired Student t test. (G, H): Western blot analysis and quantification of TAFA2 protein levels in hMSC cultures in the presence of different doses of IL‐1β. Data represent mean ± SEM from four independent experiments. *, p ≤ .05, two‐tailed unpaired Student t test. (I, J): Flow cytometry analysis of TAFA2 protein expression and percentage of leukocytes (CD45+), monocytes (CD14+), T cells (CD3+), and B cells (CD19+) at 1 and 3 days postfracture. Data are obtained from pooled bone marrow cells isolated from fractured femurs of three mice. (K): Enzyme‐linked immunosorbent assay measurement of TAFA2 levels in consecutive serum samples from patients with hip fracture. **, p ≤ .01, two‐tailed paired Student t test. (L): Proposed model for role of TAFA2 in recruiting MSC to the site of fracture. Abbreviations: IL, interleukins; MFI, mean fluorescence intensity; MSC, skeletal (mesenchymal) stem cell.
Discussion
In the present study, we identified the neurokine TAFA2 as a novel chemoattractant factor for hMSC that is expressed during the inflammatory phase of bone fracture healing and enhances in vitro transmigration of hMSC. Because of the limited number of factors known to exert specific effects on MSC migration and recruitment capacity, studying the role of TAFA2 in MSC biology is relevant for better understanding the biological processes of bone remodeling and fracture healing as well as possible use for enhancing MSC recruitment to injured sites in regenerative medicine protocols.
TAFA2 is a member of the TAFA gene family comprising newly described chemoattractant neurokines with high expression in the central nervous system. The TAFA family is composed of five highly homologous genes encoding small secreted proteins that contain conserved cysteine residues at fixed positions 20. The TAFA family is related to the chemokine, macrophage inflammatory protein‐1α (MIP1a/CCL3) that plays a role in granulocyte recruitment to sites of neuroinflammation 24. Several C‐C chemokines ligands, such as CCL3 24, CCL4/MIP‐β 25, and CCL2/Monocyte chemoattractant protein‐1 (MCP‐1) 26, are known for their modulatory functions on the migration of endogenous or transplanted neural progenitors toward damaged areas of the brain in ischemic stroke.
We observed that in vitro treatment of hMSC with TAFA2 resulted in enhanced trans‐well migration as well as significant changes in cell morphology and cytoskeletal remodeling as defined by rearrangement of F‐actin to form cell protrusions (lamellipodia) that are needed for cell movement 27. These responses are similar to changes in cell shape and cytoskeleton induced by chemokines known to affect cell motility 28. Also, previous studies have demonstrated that MSC can respond to chemoattractant proteins by reorganizing their actin cytoskeletons and become polarized in the direction of the chemoattractant gradient 29, 30. Indeed, a disorganization of cytoskeletal F‐actin has been associated with impaired migration of not only MSC but also epidermal cells and dendritic cells 31, 32, 33. Formation of lamellipodia and membrane polarization at the leading edge of a migrating cell enables directional cell movement 27. TAFA2 treatment of hMSC in two dimensional (2D) cultures resulted in formation of lamellipodia in a nondirectional fashion. It is plausible that restoration of Rac1 activity to initial levels at 60 minutes after treatment with TAFA2 represents a 2D‐culture phenomenon because of the absence of directional migration. In addition, the chemoattractant effect of TAFA2 was associated with increased activity of the p38 signaling which is known to mediate the migration of stem cells by several chemotactic factors including: IL‐33 34, stromal cell‐derived factor 1 alpha (SDF‐1α) 35, 36, and transforming growth factor beta (TGF‐β) 37. It is plausible that p38 signaling is induced after TAFA2 interaction with a cognate receptor that remains to be determined.
We observed that TAFA2 is constitutively expressed in the bone marrow by leukocyte and MSC, and its expression is upregulated after in vitro osteoblast differentiation of MSC. In contrast to several other factors, TAFA2 does not affect differentiation capacity of the cells but enhances their proliferation, which is consistent with a role in enhancing cell recruitment to sites where bone formation is needed. In vivo, TAFA2 expression was upregulated in the callus area during the inflammatory phase of fracture healing and also in the newly formed chondrocytes. This expression pattern is similar to what has been reported for other members of the C‐C family. For example, it has been reported that CCL3 and CCL4 (MIP1‐β) are expressed by the callus chondrocytes during fracture healing 38. The stimulating effects of TAFA2 on MSC in vitro transmigration, the increased expression of TAFA2 during fracture healing, and the absence of its direct effects of MSC differentiation suggest a possible role for in vivo chemotactic effects on MSC during fracture healing. Previous studies have shown that the number of osteoprogenitor cells in systemic circulation is increased after bone fracture 39. It is thus plausible that the presence of high levels of TAFA2 inside the bone fracture microenvironment during the inflammatory phase of fracture healing results in recruitment of osteoprogenitor cells needed for subsequent bone formation (Fig. 4L). Future studies are needed to determine whether local expression of TAFA2 at the fracture site could be used to increase recruitment of osteoprogenitors and to enhance the fracture healing.
Conclusion
We demonstrate that during the inflammatory phase of fracture healing, TAFA2 levels are increased at the site of fracture that leads to recruitment of osteogenic cells to the fracture site through activation of Rac1‐p38 signaling.
Author Contributions
A.J., A.I., L.C., N.D., W.Z., L.H., and C.C.: performed experiments, data analysis and interpretation; A.J., A.I., L.C., N.D., W.Z., L.H., H.E.J., B.M.A., C.C., and M.K.: conception/design, experimental design; H.E.J. provision of clinical samples; A.J., A.I., M.K.: manuscript writing; A.J., L.C., N.D., L.H., B.M.A., M.K.: manuscript editing. All authors: Final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Supplementary Figure 1: Quantitation of western blot analysis of P‐p38 levels in cell lysates at 10, 30, 60, 120, and 240 minutes after adding TAFA2 to hMSC cultures. Data represent mean ± s.e.m from three independent experiments. *P ≤ .05 (compared to 0 minutes control), two‐tailed unpaired Student t‐test.
Acknowledgments
This study was supported by a grant from the Innovation Foundations, the Novo Nordisk Foundation (NNF15OC0016284), Denmark, and KACST (Project Code: 10‐BIO1308‐02). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank Tina K. Nielsen, Bianca Jørgensen, Anita Friimose, and Charles E. Frary for excellent technical assistance. This article is dedicated to the loving memory of a wonderful physician, mentor, and scientist, Dr. Hans Johnsen.
The copyright line for this article was changed on 11 January 2020 after original online publication.
Contributor Information
Abbas Jafari, Email: ajafari@sund.ku.dk.
Moustapha Kassem, Email: mkassem@health.sdu.dk.
References
- 1. Abdallah BM, Kassem M. Human mesenchymal stem cells: From basic biology to clinical applications. Gene Ther 2008;15:109–116. [DOI] [PubMed] [Google Scholar]
- 2. Abdallah BM, Jafari A, Zaher W et al. Skeletal (stromal) stem cells: An update on intracellular signaling pathways controlling osteoblast differentiation. Bone 2015;70:28–36. [DOI] [PubMed] [Google Scholar]
- 3. Granero‐Molto F, Weis JA, Miga MI et al. Regenerative effects of transplanted mesenchymal stem cells in fracture healing. Stem Cells 2009;27:1887–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanghani‐Kerai A, Coathup M, Samazideh S et al. Osteoporosis and ageing affects the migration of stem cells and this is ameliorated by transfection with CXCR4. Bone Joint Res 2017;6:358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glass GE, Chan JK, Freidin A et al. TNF‐alpha promotes fracture repair by augmenting the recruitment and differentiation of muscle‐derived stromal cells. Proc Natl Acad Sci USA 2011;108:1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Pati S, Schreiber M. Cellular therapies and stem cell applications in trauma. Am J Surg 2018;215:963–972. [DOI] [PubMed] [Google Scholar]
- 7. Pearlin S, Nayak G, Manivasagam DS. Progress of regenerative therapy in orthopedics. Curr Osteoporos Rep 2018;16:169–181. [DOI] [PubMed] [Google Scholar]
- 8. Verboket R, Leiblein M, Seebach C et al. Autologous cell‐based therapy for treatment of large bone defects: from bench to bedside. Eur J Trauma Emerg Surg 2018;44:649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yin Y, Li X, He XT et al. Leveraging stem cell homing for therapeutic regeneration. J Dent Res 2017;96:601–609. [DOI] [PubMed] [Google Scholar]
- 10. Pignolo RJ, Kassem M. Circulating osteogenic cells: implications for injury, repair, and regeneration. J Bone Miner Res 2011;26:1685–1693. [DOI] [PubMed] [Google Scholar]
- 11. Teo GS, Ankrum JA, Martinelli R et al. Mesenchymal stem cells transmigrate between and directly through tumor necrosis factor‐alpha‐activated endothelial cells via both leukocyte‐like and novel mechanisms. Stem Cells 2012;30:2472–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levy O, Mortensen LJ, Boquet G et al. A small‐molecule screen for enhanced homing of systemically infused cells. Cell Rep 2015;10:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Becker A, Riet IV. Homing and migration of mesenchymal stromal cells: How to improve the efficacy of cell therapy? World J Stem Cells 2016;8:73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duffield JS, Park KM, Hsiao LL et al. Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow‐derived stem cells. J Clin Invest 2005;115:1743–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iso Y, Spees JL, Serrano C et al. Multipotent human stromal cells improve cardiac function after myocardial infarction in mice without long‐term engraftment. Biochem Biophys Res Commun 2007;354:700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Togel F, Hu Z, Weiss K et al. Administered mesenchymal stem cells protect against ischemic acute renal failure through differentiation‐independent mechanisms. Am J Physiol Renal Physiol 2005;289:F31–F42. [DOI] [PubMed] [Google Scholar]
- 17. Steiner B, Roch M, Holtkamp N et al. Systemically administered human bone marrow‐derived mesenchymal stem home into peripheral organs but do not induce neuroprotective effects in the MCAo‐mouse model for cerebral ischemia. Neurosci Lett 2012;513:25–30. [DOI] [PubMed] [Google Scholar]
- 18. Sackstein R, Merzaban JS, Cain DW et al. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat Med 2008;14:181–187. [DOI] [PubMed] [Google Scholar]
- 19. Guan M, Yao W, Liu R et al. Directing mesenchymal stem cells to bone to augment bone formation and increase bone mass. Nat Med 2012;18:456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tom Tang Y, Emtage P, Funk WD et al. TAFA: A novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics 2004;83:727–734. [DOI] [PubMed] [Google Scholar]
- 21. Borg K, Stankiewicz P, Bocian E et al. Molecular analysis of a constitutional complex genome rearrangement with 11 breakpoints involving chromosomes 3, 11, 12, and 21 and a approximately 0.5‐Mb submicroscopic deletion in a patient with mild mental retardation. Hum Genet 2005;118:267–275. [DOI] [PubMed] [Google Scholar]
- 22. Walford GA, Gustafsson S, Rybin D et al. Genome‐wide association study of the modified stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 2016;65:3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steffen A, Ladwein M, Dimchev GA et al. Rac function is crucial for cell migration but is not required for spreading and focal adhesion formation. J Cell Sci 2013;126:4572–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reichel CA, Rehberg M, Lerchenberger M et al. Ccl2 and Ccl3 mediate neutrophil recruitment via induction of protein synthesis and generation of lipid mediators. Arterioscler Thromb Vasc Biol 2009;29:1787–1793. [DOI] [PubMed] [Google Scholar]
- 25. Tatara Y, Ohishi M, Yamamoto K et al. Macrophage inflammatory protein‐1beta induced cell adhesion with increased intracellular reactive oxygen species. J Mol Cell Cardiol 2009;47:104–111. [DOI] [PubMed] [Google Scholar]
- 26. Yamagami S, Tamura M, Hayashi M et al. Differential production of MCP‐1 and cytokine‐induced neutrophil chemoattractant in the ischemic brain after transient focal ischemia in rats. J Leukoc Biol 1999;65:744–749. [DOI] [PubMed] [Google Scholar]
- 27. Krause M, Gautreau A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol 2014;15:577–590. [DOI] [PubMed] [Google Scholar]
- 28. Weber KS, Klickstein LB, Weber PC et al. Chemokine‐induced monocyte transmigration requires cdc42‐mediated cytoskeletal changes. Eur J Immunol 1998;28:2245–2251. [DOI] [PubMed] [Google Scholar]
- 29. Chamberlain G, Smith H, Rainger GE et al. Mesenchymal stem cells exhibit firm adhesion, crawling, spreading and transmigration across aortic endothelial cells: Effects of chemokines and shear. PLoS One 2011;6:e25663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belema‐Bedada F, Uchida S, Martire A et al. Efficient homing of multipotent adult mesenchymal stem cells depends on FROUNT‐mediated clustering of CCR2. Cell Stem Cell 2008;2:566–575. [DOI] [PubMed] [Google Scholar]
- 31. Reed MJ, Ferara NS, Vernon RB. Impaired migration, integrin function, and actin cytoskeletal organization in dermal fibroblasts from a subset of aged human donors. Mech Ageing Dev 2001;122:1203–1220. [DOI] [PubMed] [Google Scholar]
- 32. Shi D, Li X, Chen H et al. High level of reactive oxygen species impaired mesenchymal stem cell migration via overpolymerization of F‐actin cytoskeleton in systemic lupus erythematosus. Pathol Biol (Paris) 2014;62:382–390. [DOI] [PubMed] [Google Scholar]
- 33. Xu X, Liu X, Long J et al. Interleukin‐10 reorganizes the cytoskeleton of mature dendritic cells leading to their impaired biophysical properties and motilities. PLoS One 2017;12:e0172523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim J, Kim W, Le HT et al. IL‐33‐induced hematopoietic stem and progenitor cell mobilization depends upon CCR2. J Immunol 2014;193:3792–3802. [DOI] [PubMed] [Google Scholar]
- 35. Chen Y, Wei Y, Liu J et al. Chemotactic responses of neural stem cells to SDF‐1alpha correlate closely with their differentiation status. J Mol Neurosci 2014;54:219–233. [DOI] [PubMed] [Google Scholar]
- 36. Ryu CH, Park SA, Kim SM et al. Migration of human umbilical cord blood mesenchymal stem cells mediated by stromal cell‐derived factor‐1/CXCR4 axis via Akt, ERK, and p38 signal transduction pathways. Biochem Biophys Res Commun 2010;398:105–110. [DOI] [PubMed] [Google Scholar]
- 37. Saika S, Okada Y, Miyamoto T et al. Role of p38 MAP kinase in regulation of cell migration and proliferation in healing corneal epithelium. Invest Ophthalmol Vis Sci 2004;45:100–109. [DOI] [PubMed] [Google Scholar]
- 38. Alblowi J, Tian C, Siqueira MF et al. Chemokine expression is upregulated in chondrocytes in diabetic fracture healing. Bone 2013;53:294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eghbali‐Fatourechi GZ, Lamsam J, Fraser D et al. Circulating osteoblast‐lineage cells in humans. N Engl J Med 2005;352:1959–1966. [DOI] [PubMed] [Google Scholar]
- 40. Simonsen JL, Rosada C, Serakinci N et al. Telomerase expression extends the proliferative life‐span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol 2002;20:592–596. [DOI] [PubMed] [Google Scholar]
- 41. Abdallah BM, Haack‐Sorensen M, Burns JS et al. Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite [corrected] extensive proliferation. Biochem Biophys Res Commun 2005;326:527–538. [DOI] [PubMed] [Google Scholar]
- 42. Jafari A, Siersbaek MS, Chen L et al. Pharmacological Inhibition of protein kinase G1 enhances bone formation by human skeletal stem cells through activation of RhoA‐Akt signaling. Stem Cells 2015;33:2219–2231. [DOI] [PubMed] [Google Scholar]
- 43. Jafari A, Qanie D, Andersen TL et al. Legumain regulates differentiation fate of human bone marrow stromal cells and is altered in postmenopausal osteoporosis. Stem Cell Rep 2017;8:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hans Gottlieb TWK, Martin Boegsted BS, Olsen GS et al. Clinical study of circulating cellular and humoral biomarkers involved in bone regeneration following traumatic lesions. J Stem Cell Res Ther 2011;1:1–10. [Google Scholar]
- 45. Bonnarens F, Einhorn TA. Production of a standard closed fracture in laboratory animal bone. J Orthop Res 1984;2(1):97–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Quantitation of western blot analysis of P‐p38 levels in cell lysates at 10, 30, 60, 120, and 240 minutes after adding TAFA2 to hMSC cultures. Data represent mean ± s.e.m from three independent experiments. *P ≤ .05 (compared to 0 minutes control), two‐tailed unpaired Student t‐test.


