Abstract
Human-specific pseudogenization of the CMAH gene eliminated the mammalian sialic acid (Sia) Neu5Gc (generating an excess of its precursor Neu5Ac), thus changing ubiquitous cell surface “self-associated molecular patterns” that modulate innate immunity via engagement of CD33-related-Siglec receptors. The Alu-fusion-mediated loss-of-function of CMAH fixed ∼2–3 Ma, possibly contributing to the origins of the genus Homo. The mutation likely altered human self-associated molecular patterns, triggering multiple events, including emergence of human-adapted pathogens with strong preference for Neu5Ac recognition and/or presenting Neu5Ac-containing molecular mimics of human glycans, which can suppress immune responses via CD33-related-Siglec engagement. Human-specific alterations reported in some gene-encoding Sia-sensing proteins suggested a “hotspot” in hominin evolution. The availability of more hominid genomes including those of two extinct hominins now allows full reanalysis and evolutionary timing. Functional changes occur in 8/13 members of the human genomic cluster encoding CD33-related Siglecs, all predating the human common ancestor. Comparisons with great ape genomes indicate that these changes are unique to hominins. We found no evidence for strong selection after the Human–Neanderthal/Denisovan common ancestor, and these extinct hominin genomes include almost all major changes found in humans, indicating that these changes in hominin sialobiology predate the Neanderthal–human divergence ∼0.6 Ma. Multiple changes in this genomic cluster may also explain human-specific expression of CD33rSiglecs in unexpected locations such as amnion, placental trophoblast, pancreatic islets, ovarian fibroblasts, microglia, Natural Killer(NK) cells, and epithelia. Taken together, our data suggest that innate immune interactions with pathogens markedly altered hominin Siglec biology between 0.6 and 2 Ma, potentially affecting human evolution.
Keywords: sialic acid, hominin, evolution, CD33rSiglecs, common ancestor, Neanderthal/Denisovan, great apes, archaic hominin
Introduction
Prior to the mid-1990s, the extreme similarity of human and chimpanzee protein sequences suggested that phenotypic differences were primarily due to differences in gene regulation (King and Wilson 1975). The first definitive exception was a fixed loss-of-function genomic mutation unique to the human lineage, in the gene-encoding CMP-Neu5Ac hydroxylase (CMAH) (Chou et al. 1998), an event mediated by an Alu–Alu fusion (Hayakawa et al. 2001). This mutation eliminated biosynthesis of the common mammalian sialic acid (Sia) N-glycolylneuraminic acid (Neu5Gc) and caused accumulation of its precursor N-acetylneuraminic acid (Neu5Ac), radically changing cell surface extracellular glycosylation throughout the body in the hominin lineage. The mutation was dated to >2 Ma by multiple methods (Chou et al. 2002; Hayakawa et al. 2006). Additional examples emerged a few years later, including the forkhead box protein P2 (FOXP2) (Enard et al. 2002, 2009), a myosin heavy chain family member (MHY16) (Currie 2004; Stedman et al. 2004), and examples of gene duplication followed by adaptive evolution (Johnson et al. 2001). Along with biomedical considerations (Varki 2000), such discoveries lead to sequencing of the chimpanzee genome (Chimpanzee Sequencing and Analysis Consortium 2005). There are now many more defined genetic differences between humans and our closest evolutionary cousins, involving not only gene expression (Enard et al. 2002; Fujiyama et al. 2002; Watanabe et al. 2004; Calarco et al. 2007; Kehrer-Sawatzki and Cooper 2007; Cruz-Gordillo et al. 2010; Otto et al. 2014; Atkinson et al. 2018) but also other distinct genomic changes ranging from massive (megabase) structural variation to differences in gene copy numbers, de novo genes, pseudogenization (O’Bleness et al. 2012; Ruiz-Orera et al. 2015), human-accelerated regions in noncoding regions, and microRNA genes (Eddy 2001; Mello and Conte 2004; Kim 2005; Ruiz-Orera et al. 2015).
This study focuses on human genes involved in the biology of sialic acids. All living cells are covered with complex arrays of glycoconjugates, with sialic acids occupying the majority of terminal positions on such glycan chains in animals of the Deuterostome lineage (including echinoderms and vertebrates) (Gagneux et al. 2015). These acidic, nine-carbon backbone amino-sugars play important roles in cell–cell and cell–matrix interactions, as well as in host–pathogen interactions (Varki et al. 2015). The hominin-specific loss-of-function mutation in CMAH mentioned above eliminated biosynthesis of Neu5Gc, which was a target for selective cell recognition by various nonhuman pathogens and toxins (Kyogashima et al. 1989; Martin et al. 2005; Campanero-Rhodes et al. 2007; Deng et al. 2014; Alisson-Silva et al. 2018). It is reasonable to speculate that exogenous pathogen recognition could have initially driven the CMAH loss-of-function, generating a polymorphism as the recessive loss-of-function mutant allele rose in frequency. However, this deletion was then fixed in an ancestral hominin population ∼2–3 Ma (Chou et al. 2002), possibly by directional selection against Neu5Gc via female immunity during reproduction, a mechanism demonstrated in vivo using transgenic mice that carry the same mutation in their Cmah gene as humans (Ghaderi et al. 2011). Given the likely timing of these events, we speculated that anti-Neu5Gc immunity of hominin females immunized against Neu5Gc by increased contact with Neu5Gc-rich vertebrate animal prey might have contributed to the origins of the genus Homo ∼2 Ma (Wood and Boyle 2016; Bergfeld et al. 2017).
Early discovery of some additional human-specific changes affecting sialic acid biology (Brinkman-Van der Linden et al. 2000; Angata, Varki, et al. 2001; Gagneux et al. 2003; Sonnenburg et al. 2004; Hayakawa et al. 2005; Nguyen et al. 2006) and system-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents (Altheide et al. 2006) suggested a possible “hotspot” in human sialic acid evolution (Varki 2009). With the availability of more human genomes (1000 Genomes Project Consortium et al. 2015), a recent study showed the patterns of genetic variation of 55 sialic acid biology-related genes in modern human populations did not significantly deviate from neutral expectations and were in fact not significantly different among genes belonging to different functional categories (Moon et al. 2018).
Sialic acid binding Ig-like lectin (Siglecs) are type I transmembrane proteins with an N-terminal immunoglobulin (Ig)-like-V-set domain that mediates sialic acid recognition, and a variable number of Ig-like-C-2 type domains (Angata, Hingorani, et al. 2001). Siglecs often have a cytoplasmic tail with one or more immunoreceptor tyrosine-based inhibitory motifs that can suppress immune cell activation. Alternatively, they can recruit adaptor proteins with immunoreceptor tyrosine-based activating motifs. Although Siglecs likely have multiple functions, one prominent role appears to be recognition of endogenous sialylated glycans as self-associated molecular patterns, suppressing reactions of innate immune cells against self (Varki 2011). Activating Siglecs have a positively charged arginine or lysine in their transmembrane domains that can recruit DAP12 and activate cellular immune responses against pathogens mimicking endogenous sialic acids (Schwarz et al. 2017). CD33-related Siglecs are rapidly evolving and among this family, nine inhibitory (hSiglec-3 and hSiglec-5 to hSiglec-12) and two activating members (hSiglec-14 and hSiglec-16) have been characterized in humans. Recently, an article published from our group showed variable presence or absence of functional changes in the CD33rSIGLEC cluster in 26 mammalian species including great apes (Khan et al. 2020).
With the availability of genomes from both living great apes (Prado-Martinez et al. 2013; Xue et al. 2015; Kronenberg et al. 2018) and extinct archaic hominins (Reich et al. 2010; Meyer et al. 2012; Prufer et al. 2014), we now systematically reassess the previous discoveries from multiple groups and also report on several new findings. Overall, we find that multiple complex changes in genes involving sialic acid biology beyond the initial CMAH mutation did indeed occur but are mostly confined to the CD33rSIGLEC gene cluster on chromosome 19 (Angata, Hingorani, et al. 2001), encoding CD33rSiglecs, prominent on innate immune cells (Varki and Angata 2006).
Overall, we found that multiple changes in the CD33rSIGLEC gene cluster are common in all human populations, postdate the common ancestor with the chimpanzee/bonobo lineage, but predate the common ancestor with Neanderthals and Denisovans. Such multiple complex changes in this gene cluster appear to be associated with altered expression of these genes, not just confined to innate immune cells, but also in other unexpected human cell types, some associated with diseases that appear to be uniquely human.
Materials and Methods
Genome Assembly and Identification of Variants across Humans and Great Apes
The great ape genome data (total number 147) for CMAH and CD33rSIGLECs were derived from three publications (Prado-Martinez et al. 2013; Xue et al. 2015; de Manuel et al. 2016; Kronenberg et al. 2018). These great ape genomes were mapped to human reference genome (GRCh37/hg19) retrieved from UCSC genome browser, using Burrows–Wheeler aligner and further processed with Picard tool to remove duplicates and variant were called using Genome-analysis toolkit. Moreover, the variants in variant call format of chimpanzee, bonobos, gorilla, and orangutan were visualized in integrative genome viewer with their respective reference genome (Pantro4, Gorgor3, and PonAbe2) and annotation file. All archaic hominins raw and processed files were obtained from the Max Planck Institute for Evolutionary Anthropology (Reich et al. 2010; Castellano et al. 2014; Prufer et al. 2014; Slon et al. 2018) (http://cdna.eva.mpg.de/neandertal). Bed coordinates of SIGLECs and CMAH genes were provided for each human and ape lineage as supplemental (chimpanzee, gorilla, and orangutan) (supplementary file 1, Supplementary Material online). Additionally, bed coordinates of additional polymorphism present in CD33rSIGLECs with allele frequency in great ape population were also provided as supplemental (supplementary file 2, Supplementary Material online).
Inference of Strong Archaic Natural Selection
The genes involved in sialic acid biology (67 genes) were overlapped with regions displaying signatures of ancient selective sweeps (Peyregne et al. 2017). The method used to identify these signatures of archaic selection relies on a hidden Markov model to detect extended regions in the genome where the Neanderthal and Denisovan lineages fall outside the human variation (Meyer et al. 2012; Prufer et al. 2014). This method can only detect events that occurred between the split of modern and archaic humans around 0.5 Ma (Prufer et al. 2017) and the split of modern human populations from each other around 0.2 Ma (Schiffels and Durbin 2014). For events with a selective advantage of 0.5% or larger, with an origin of the beneficial mutation as old as 600,000 years ago, the false positive rate of the method is lower than 0.1% and its true positive rate is larger than 65% (Peyregne et al. 2017). We also note that signals of positive selection are not detectable if the selection coefficient is smaller than 0.1%. The Ensembl database (release 82) (Aken et al. 2016) was used to annotate each gene with hg19 coordinates from transcription start to transcription end. Furthermore, L1CAM, PECAM, CMAH, SIGLEC13, SIGLEC16, and SIGLEC17 were excluded because they fall either in filtered regions not considered for the ancient sweep screen or could not be mapped to hg19 coordinates. As regulatory regions may also have been the target of positive selection, we extended the start and end coordinates 1 Mb upstream and downstream, respectively. If a neighboring gene was within 1 Mb, we only extended the coordinates until 5 or 1 kb from the transcription start or end of this neighboring gene. In order to test whether the lack of signatures of ancient selection is statistically significant, the candidate regions of ancient selection were randomly placed in the genome and the random placements of all regions were iterated 1,000 times, counting how often no overlap with genes involved in sialic acid biology was detected. The depletion in selection is not significant (356 sets never overlap; P value = 0.356).
Immunohistochemistry of Siglecs
Paraffin sections were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol and Tris-buffered saline–Tween (TBST). Following this, endogenous binding sites and peroxidases were blocked with 1% bovine serum albumin/TBST and 0.3% H2O2/TBST. This was followed by blocking of endogenous biotin and then heat-induced antigen retrieval was performed in citrate buffer pH 6. Primary antibodies (mouse anti-Siglec-7 and Rabbit anti-Siglec-13) or mouse IgG were then overlaid at optimal dilutions, and slides were incubated overnight at 4 °C in a humid chamber. Specific binding was detected using a biotinylated anti-mouse or anti-rabbit (for Siglec-13), followed by Horseradish Peroxidase (HRP) streptavidin and then biotinyl tyramide enhancement and then HRP streptavidin. Substrate color was developed, and nuclei were counterstained with hematoxylin and the slides were aqueous mounted for viewing and digital photomicrography using an Olympus BH2 microscope with an Olympus magnafire camera.
Results
Multiple Derived, Fixed, or Polymorphic Genomic SIGLEC Variants Are Present in All Modern Human Populations
Table 1 summarizes events and frequencies in human populations based on the 1000 Genomes Project data and compares these with archaic hominin and great ape genomes. All human genomic changes in CD33-related SIGLECs (affecting 8 out of 13 members of this class of genes) were present across modern human populations. Genome-wide association studies identified a derived CD33-linked allele (rs3865444(A), rs12459419(T)) that is protective against late onset Alzheimer’s disease (Schwarz et al. 2016). The linked single-nucleotide polymorphism was found to be variable across human populations with highest frequency in native American populations (48%) and lowest in African populations (5%). The previously reported Alu-mediated human SIGLEC13 deletion (Wang, Mitra, Secundino, et al. 2012) and SIGLEC17 (Siglec-P3) pseudogenization due to Open reading frame disruption and mutation of a critical arginine residue in its V-set domain (Wang, Mitra, Secundino, et al. 2012) appear to be fixed in all humans. SIGLEC12, SIGLEC14, and SIGLEC16 show polymorphic pseudogenization in variable frequencies across human populations, as detailed in table 1. SIGLEC12 also harbors a human-universal mutation of the critical arginine residues in the V-set domain (Arg -> Cys, first V-set domain) abrogating its ability to recognize sialic acid. An additional SIGLEC12 inactivation mutation (rs16982743) was found at an overall 18.6% frequency, highest in Africans (37%) and lowest in East Asians (5%). Another SIGLEC12 polymorphism, that is, frameshift (rs66949844) averages 59% (highest in native Americans and lowest in East Asians) in human population. Additionally, based on the two linked alleles present in SIGLEC16 (rs12611411(T) and rs12984584(C)), the pseudogene variant (SIGLEC16P) is present in higher frequency than the functional allele (SIGLEC16) in human populations (Wang, Mitra, Cruz, et al. 2012). Because these SIGLEC clusters are undergoing multiple gene conversions, some regions are highly prone to low mapping quality and hence some variants are not well defined, for example, SIGLEC14 fusion-deletion, which is highly prevalent in East Asian populations (Yamanaka et al. 2009; Ali et al. 2014). Due to the lack of high coverage sequence data, these regions have not been well genotyped in nonhuman genomes. The CMAH exon deletion is confirmed to be universal to humans.
Table 1.
Detailed Description of Genomic Variants and Their Percentage in Each Lineage Based on CD33rSIGLECs and CMAH Genes
| Gene | Genomic Changes | Changes | 1000 Genomes Data (%) |
Archaic Genomes (%) |
Great Apes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | America | E. Asia | Europe | S. Asia | Neanderthal | Denisovan | Chimpanzee | Bonobo | Gorilla | Orangutan | |||
| CD33m (SIGLEC3) | rs12459419(T) | TT | 5 | 48 | 19 | 31 | 16 | Ancestral | ND | Ancestral | Ancestral | Ancestral | Ancestral |
| rs3865444(A) | AA | 5 | 48 | 19 | 31 | 16 | Ancestral | Ancestral | Ancestral | Ancestral | Ancestral | Ancestral | |
| SIGLEC5 | Arg (R)/H | R | 100 | 100 | 100 | 100 | 100 | 100 | 100 | R/H | R | R/H | Low call |
| SIGLEC12 | Arg (R)/C | C | 100 | 100 | 100 | 100 | 100 | 100 | 100 | R | R | R | R |
| −/C(rs66949844) | C | 60 | 74 | 37 | 67 | 63 | 50 | 100 | — | — | — | — | |
| Q/*(rs16982743) | * | 37 | 13 | 4 | 20 | 11 | 60 | 50 | Q | Q | Q | Q | |
| SIGLEC13 | Deletion | Del | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 0 |
| SIGLEC14 | Arg (R) | R | 100 | 100 | 100 | 100 | 100 | 100 | 100 | R/H | R/H | R/H | Low call/Y |
| SIGLEC14 | Deletion | Fusion/deletion | 30 | 21 | 73 | 22 | 33 | ND | ND | ND | ND | ND | ND |
| SIGLEC16 | rs12611411 rs12984584 | 16P | 83 | 85 | 67 | 82 | 87 | 67 | 50 | Ancestral | Ancestral | Not well annotated | Ancestral |
| SIGLEC17 | Arg (R)/W | W | 100 | 100 | 100 | 100 | 100 | 100 | 100 | R | R | R | R |
| G/- | Disrupted ORF | 100 | 100 | 100 | 100 | 100 | 100 | 100 | G | G | G | G | |
| CMAH | Exon 6 deletion | CMAHP | 100 | 100 | 100 | 100 | 100 | 100 | 100 | Present | Present | Not well annotated | Present |
Note.—All the genomic changes were found in all existing human populations. CD33m (Siglec-3):allele increasing alternate splice form protective against Alzheimer’s disease; ND:not determined; Siglec-5: sialic acid binding arginine residue; Siglec-12: mutation of sialic acid binding arginine residue; Siglec-12: 1-bp insertion resulted in pseudogene (12P); Siglec-12: inactivation polymorphism at position 29 amino acid; Siglec-13: complete deletion by Alu–Alu fusion event; Siglec-14: sialic acid binding arginine residue; Siglec-14: 16-kb deletion/fusion, difficult to read from standard genomic data; Siglec-16:, pseudogene (16P) and gene (16) forms; Siglec-17: mutation of sialic acid binding arginine residue; Siglec-17: 1-bp deletion that disrupted the Open reading frame (ORF); CMAH: pseudogene due to 92-bp exon 6 deletion.
Great Ape Genomes Do Not Show a High Frequency of SIGLEC Gene Mutations
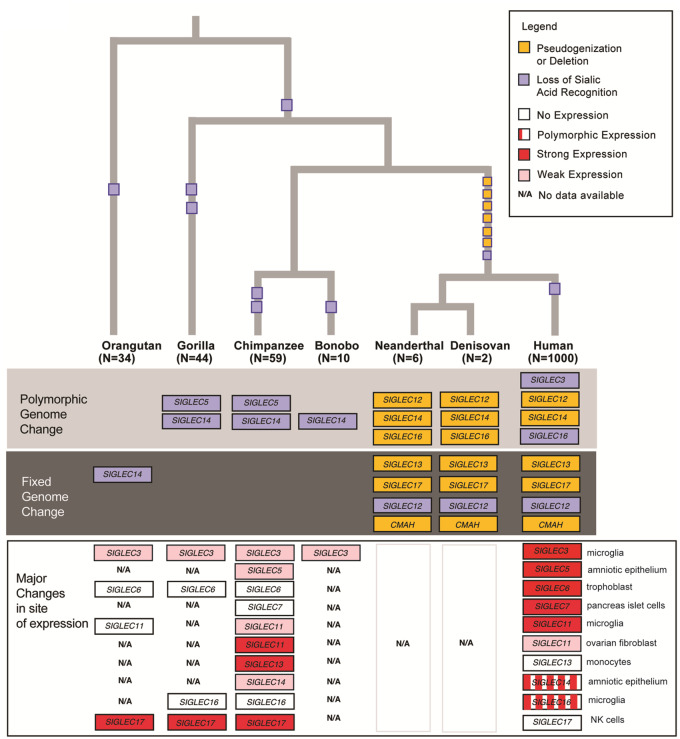
Our previous human–ape comparisons involved the incomplete draft of a single chimpanzee genome, a few additional incomplete great ape sequences and a small number of human genomes (Chimpanzee Sequencing and Analysis Consortium 2005). Thus, it remained possible that the genomic changes were actually a common feature of such genes across all these taxa. The availability of newly annotated versions of great ape genomes (Prado-Martinez et al. 2013; Xue et al. 2015; de Manuel et al. 2016; Kronenberg et al. 2018) now allows for much better control against ascertainment bias, by comparing human-specific polymorphisms with multiple different genomes for each ape species, capturing potential polymorphisms in the latter as well (Sullivan et al. 2017). We observed that great ape genomes share none of the mutations observed in human populations. Instead, we only observed independent changes in critical arginine residues in V-set domain of SIGLEC5/14, rendering them unable to recognize sialic acids (table 1). Although the essential arginine change (Arg -> His) in SIGLEC5 was found to be polymorphic in chimpanzee and gorilla, sequences in orangutan were not recognized due to low mappability. The SIGLEC14 essential arginine change (Arg -> His) was found to be polymorphic in chimpanzee, bonobo, and gorilla. However, sequences of SIGLEC14 in orangutan were not determined in most of the species due to low coverage, except for one that harbors an essential arginine change to tyrosine (Arg -> Tyr). The CMAH gene is functional in all great apes as expected, given that both types of sialic acids (Neu5Ac and Neu5Gc) are present in all these species (Muchmore et al. 1998). Overall, human genomes harbored a far greater number of structural/functional changes in CD33rSIGLEC genes (see fig. 1 and supplementary file 2, Supplementary Material online).
Fig. 1.
Evolutionary changes in hominid SIGLEC genes. The tree represents phylogenetic relatedness among hominids and different types of genomic events are depicted according to the color codes. Siglec expression differences are based on more limited comparisons (see table 1 and fig. 3 and see text for discussion).
Most Human Lineage-Specific Mutations Are Also Found in Archaic Hominin Genomes
To further understand the timing of these human genomic changes of these human genomic variants, we looked for their presence in archaic human genomes, that is, Neanderthal and Denisovan. The exon deletion leading to loss-of-function of the CMAH gene is shared by these two archaic genomes, as expected based on the existing estimates of fixation of the mutation over 2 Ma (Hayakawa et al. 2001). Notably, almost all other human CD33rSIGLECs variants (except for CD33-linked variants protective against late onset Alzheimer’s disease) were also present in these archaic hominin genomes in variable frequency (listed in table 1 and fig. 1) placing their origin before the human–Neanderthal common ancestor (∼0.6 Ma) (Prufer et al. 2017; Hajdinjak et al. 2018). Due to unavailability of well annotated sequences, the SIGLEC14 fusion/deletion could not be determined in archaic hominins.
Lack of Evidence for Strong Selection up to 500,000 Years Ago
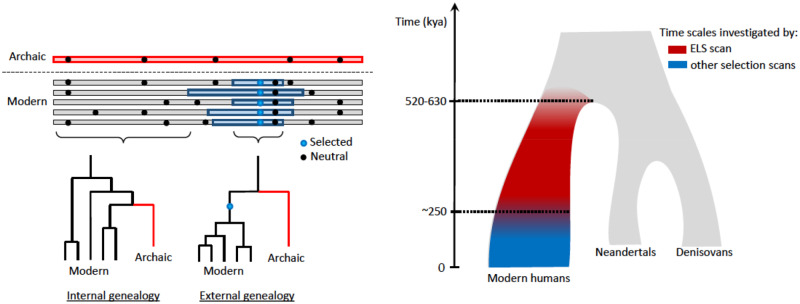
The presence of all genomic SIGLEC changes in human populations indicates that they predate the common ancestor of modern humans ∼0.2–0.3 Ma (Hublin et al. 2017). Consistent with this observation, the recent article by Moon et al. (2018) found that the patterns of genetic variation of most CD33rSIGLEC genes did not significantly deviate from neutral expectations, and the few that did significantly deviate from neutrality experienced either soft sweeps or population-specific hard sweeps (Moon et al. 2018). Using a method that allows detection of selection in deeper time (∼0.5 Ma) (Peyregne et al. 2017), we also found no evidence of strong selection after the ancestors of modern and archaic humans (Neanderthal–Denisovan) split from each other around 500,000 years ago. As selection may also target neighboring regulatory regions, we defined regulatory domains around each gene and looked for overlaps with candidate sweep regions. Using these extended gene coordinates, we again found no overlap with candidate regions for selective sweeps, suggesting that none of the genes involved in sialic acid biology exhibits strong signatures of selection more recently than 0.5 Myr in the common ancestral population of all modern humans. The description of extended lineage sorting (ELS) is provided in figure 2.
Fig. 2.
Description of the ELS scan. (A) Illustration of the selection signature. Patterns of shared mutations (dots) were used to reconstruct the genealogy that relate an archaic human Altai Neanderthal (Prufer et al. 2014); or Denisovan (Meyer et al. 2012); top rectangle to modern humans (bottom rectangles) at any given position along their genomes. There are two types of genealogies: the archaic falls either outside the modern human variation (external genealogy) or inside (internal genealogy). The internal genealogy is the most frequent in the genome. However, if a mutation spread among the ancestors of modern humans (blue dot) more recently than the population split with the ancestors of archaic humans, the archaic will be expected to fall outside the modern human variation in the genomic region around the mutation that has not been unlinked by recombination (blue rectangles). This region is expected to be large if the mutation was positively selected and spread quickly in the population, not leaving enough time for recombination to break its linkage with other mutations (black dots). (B) Time scales for the signatures of selection in humans. The tree represents the simplified population history of Neanderthals, Denisovans, and modern humans (Prufer et al. 2017). The colors indicate the time scales investigated by the ELS scan (red) and other methods (blue) used to detect events of positive selection on the modern human lineage.
Human-Specific Expression of CD33rSiglecs in Nonimmune Tissues
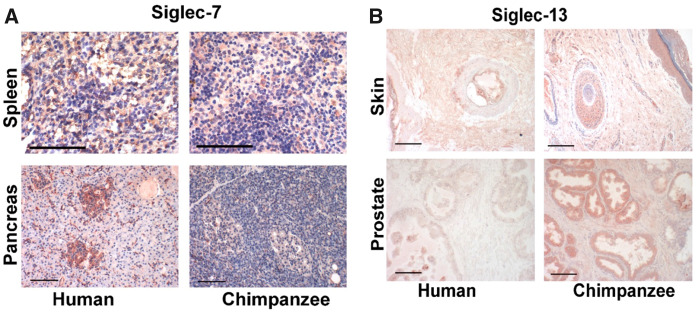
In addition to these multiple, polymorphic complex genomic changes in the human CD33rSIGLEC gene cluster, there appear to be unusual (derived) human-specific expression patterns of CD33rSiglecs (in nonhemopoietic cells) in locations such as placental trophoblast (SIGLEC6) (Kang et al. 2011), ovarian fibroblasts (SIGLEC11/16) (Wang et al. 2011), amniotic epithelium (SIGLEC5/14) (Ali et al. 2014), and microglia (SIGLEC11/16) (Hayakawa et al. 2005). In each of the above instances, we have previously reported human-specific expression and lack of expression in chimpanzee tissue samples (in some instances also other available great ape tissues). With regard to the recent report of SIGLEC7 expression in human pancreatic islets (Yamaguchi et al. 2017), we now show that such expression is missing in chimpanzee pancreatic islets (fig. 3). We also found that Siglec-13, which is deleted in hominins, is highly expressed in chimpanzee intestinal epithelium and skin (fig. 3). Current and previously published data are summarized in table 2. Although we obviously cannot detect tissue-specific expression in Neanderthals or Denisovans, it is reasonable to consider the possibility that these unusual human-specific expression patterns arose during the course of the large-scale changes affecting the CD33rSIGLECs gene cluster during the proposed “hotspot” and would thus have already existed in both of those extinct relatives.
Fig. 3.
Examples of immunohistochemistry using antibodies against Siglec-7 and Siglec-13 on human and chimpanzee tissues (see Materials and Methods for detail). (A) No difference in expression of Siglec-7 in splenic blood cells. There is markedly increased expression of Siglec-7 in human pancreatic islets. (B) Siglec-13 expression in chimpanzee skin and prostatic epithelium. The gene-encoding Siglec-13 is completely deleted in humans. Scale bars = 100 μm.
Table 2.
Differential Tissue and Cell Type Expression Profile of SIGLEC Genes among Humans and Great Apes
| Siglecs | Humans | “Great Apes” | Current Data and Literature Reference(s) | Potential Relevance to Diseases That Appear Unique to Humans |
|---|---|---|---|---|
| CD33 | High expression in human brain microglia | Low expression in chimpanzee brain microglia | Schwarz et al. (2016) | Alzheimer’s disease |
| Siglec-5 | Selectively expressed in amniotic epithelium | Not expressed in chimpanzee amniotic epithelium | Ali et al. (2014) | Intrauterine infections with group B Streptococcus |
| Siglec-6 | Selectively expressed in placental trophoblast | Not expressed in placental trophoblast chimpanzee, gorilla, and orangutan | Brinkman-Van der Linden et al. (2007) | Preeclampsia |
| Siglec-7 | Selectively expressed in pancreatic islet cells | Not expressed in chimpanzee | Present study (fig. 3A) | Type I diabetes |
| Siglec-11 | Selectively expressed in brain microglia | Less expression in chimpanzee, no expression in orangutan brain | Hayakawa et al. (2005) | Escherichia coli K1 meningitis in neonates and infants |
| Siglec-11 | Decreased expression in ovarian fibroblasts | Strong expression in chimpanzee ovarian fibroblasts | Wang et al. (2011) | Unknown |
| Siglec-13 | Deleted in humans | Expressed in chimpanzee monocytes | Wang, Mitra, Secundino, et al. (2012) | Unknown |
| Siglec-13 | Deleted in humans | Expressed in chimpanzee epithelium | Present study (fig. 3B) | Unknown |
| Siglec-14 | Polymorphic expression in amniotic epithelium | Not expressed in chimpanzee amniotic epithelium | Ali et al. (2014) | Intrauterine infections with Group B Streptococcus |
| Siglec-16 | Polymorphic expression in brain microglia | Not expressed in microglia of chimpanzee and gorilla | Hayakawa et al. (2005) | Escherichia coli K1 meningitis in neonates and infants |
| Siglec-17 | No protein expression | Expressed in natural killer cells of chimpanzee, gorilla, and orangutan | Wang, Mitra, Secundino, et al. (2012) | Unknown |
Discussion
Recent availability of sequences of many modern human genomes (1000 Genomes Project Consortium et al. 2015), archaic hominins (Green et al. 2010; Reich et al. 2010, 2011; Meyer et al. 2012, 2016; Castellano et al. 2014; Prufer et al. 2014, 2017; Sawyer et al. 2015; Kuhlwilm et al. 2016; Slon et al. 2018), and great apes (Prado-Martinez et al. 2013; Xue et al. 2015; de Manuel et al. 2016) provides a tool for understanding the role of evolution in shaping genetic architecture and allowing us to identify the genetic basis of phenotypic traits of various species (Haffter et al. 1996; Bryk and Tautz 2014; Valenzano et al. 2015; Castellano and Munch 2020). Our earlier studies noted multiple genomic changes affecting sialic acid biology, and we suggested the possibility of a hotspot in humans affecting these pathways compared with our closest evolutionary relatives (Altheide et al. 2006). However, the limited availability of annotated and high coverage genomes at the time left the question unresolved, and our suggestion has been recently tested by others (Moon et al. 2018), who could not find evidence for strong selective sweeps in current human populations. We herewith show that apart from the fixed CMAH null mutation and increased expression of ST6GAL1 (Gagneux et al. 2003), most of the human-specific changes affecting sialic acid biology are found in the SIGLEC gene cluster on chromosome 19, and that although great ape genomes do not show many changes in this cluster, almost all the human changes are also found in archaic genomes of Neanderthals and Denisovans (Reich et al. 2010; Castellano et al. 2014; Prufer et al. 2014; Slon et al. 2018; Bokelmann et al. 2019). In keeping with this overall conclusion, there was no evidence for strong selective pressure in this cluster after ∼0.6 Ma, when the human lineage diverged from Neanderthal–Denisovan common ancestor.
With regard to timing and order of changes, the most likely possibility is that the CMAH mutation first dramatically altered sialic acids throughout the body, possibly initiating a series of events that resulted in SIGLEC cluster changes. The complex and dynamic relationships between host and pathogens is often characterized by the Red Queen hypothesis, whereby slowly evolving hosts have to keep changing to keep pace with the pressure exerted by more rapidly evolving pathogens. Sialic acids form one primary interface, “the molecular frontier” in this evolutionary arms race. The loss-of-function of CMAH and the changes in ST6GAL1 expression undoubtedly led to multiple alterations of human–pathogen interactions. The direct consequence was the change in human cell surfaces, leading to overexpression of Neu5Ac, escaping Neu5Gc-specific pathogens, and favoring human pathogens that interact with Neu5Ac. There are many other known and possible biological consequences of human Neu5Gc loss (Varki 2009; Okerblom and Varki 2017), including protection from various Neu5Gc-recognizing pathogens such as malaria caused by Plasmodium reichenowi in African great apes (Martin et al. 2005), Escherichia coli K99 gastroenteritis (Kyogashima et al. 1989), transmissible gastroenteritis coronavirus (Schwegmann-Wessels and Herrler 2006), and simian virus 40 (Campanero-Rhodes et al. 2007). Conversely, CMAH loss likely made humans susceptible to Neu5Ac-preferring pathogens such as Vibrio cholerae (causative agent of the human-specific disease cholera) (Alisson-Silva et al. 2018) and typhoid fever caused by a secreted bacterial toxin that preferentially recognizes Neu5Ac (Deng et al. 2014). Another human-specific pathogen Streptococcus pneumoniae recognizes free Neu5Ac released from human cells by its secreted sialidase (Hentrich et al. 2016). Yet another secondary consequence appears to have been the evolution of pathogens that express Neu5Ac on their surfaces as a way to engage CD33-related Siglecs and downregulate innate immune responses in the human host. Again, the existence of many human-specific pathogens that display Neu5Ac supports this scenario (Angata 2018). For example, Group B Streptococcus type III, interacts with Siglec-9 through sialylated CPS and inhibits inflammatory response by neutrophils (Carlin et al. 2009). Likewise, nontypeable Haemophilus influenzae expressing sialic acid on its lipooligosaccharide binds to Siglec-5 and reduces cytokine production by myeloid cells (Angata et al. 2013), and the E. coli K1 strain that causes meningitis in neonates and infection in urinary tract, engages Siglec-11, and escapes killing (Hayakawa et al. 2017). Also, as expected, nonhuman primate Siglec-9 prefers binding to Neu5Gc, whereas human Siglec-9 prefers binding to Neu5Ac (Sonnenburg et al. 2004). Likewise, human Siglec-5 and CD33 prefer binding to Neu5Ac, compared with the baboon CD33, which strongly prefers Neu5Gc (Padler-Karavani et al. 2014). These findings imply that Siglecs underwent adaptation in new environments to “catch up” with the changes in human sialome caused by the hominin CMAH mutation. Some human Siglecs still prefer binding to Neu5Gc (Angata 2018), perhaps due to incomplete adaptation to the derived, human Neu5Ac-dominant sialome.
Although these genomic changes are ancient, evidence also suggests that existing human polymorphisms are associated with several diseases such as Chronic obstructive pulmonary disease, asthma, Alzheimer’s disease, and meningitis (Yamanaka et al. 2009; Gao et al. 2010; Ali et al. 2014; Schwarz et al. 2016). In addition, human Siglec-XII and chimpanzee Siglec12 are expressed on macrophages and luminal epithelia (Mitra et al. 2011). However, human Siglec-XII harbors a universal mutation (R122C) that makes the protein unable to recognize sialic acid. Interestingly, chimpanzee Siglec12 and human Siglec-XII with its arginine experimentally restored strongly prefer Neu5Gc (Mitra et al. 2011). These results suggest a scenario where Siglec-12 lost an endogenous ligand and is thus being eliminated from the population.
Prior studies have reported human-specific Siglec expression changes in placenta (Siglec-6) (Brinkman-Van der Linden et al. 2007), brain microglia (CD33/Siglec11/16) (Hayakawa et al. 2005; Schwarz et al. 2016), amniotic epithelium (Siglec-5/14) (Ali et al. 2014), ovarian fibroblasts (Siglec11/16) (Wang et al. 2011), and NK cells (Siglec-17) (Wang, Mitra, Secundino, et al. 2012). We here add to these expression differences by showing that Siglec-7 is upregulated in human but not chimpanzee pancreatic islets, and that the SIGLEC13 deletion resulted in a loss of Siglec-13 expression from human epithelia. Many of these expression changes could represent secondary consequences of multiple genomic changes that occurred in this gene cluster earlier in the hominin lineage. Overall, it is likely that the selective pressures driving all these changes were most prominent sometime after the split from the common ancestor of human and chimpanzee, but before the split from the Human–Neanderthal–Denisovan common ancestor.
Taken together, our data suggest that innate immune encounters with pathogens markedly altered hominin Siglec biology between 0.6 and 2 Ma, potentially affecting human evolution. Notably, this is the time period when a variety of changes were occurring in genus Homo, including exploration of new habitats (White et al. 1993; Semaw et al. 1997; deMenocal 2011), striding bipedalism and running (Bramble and Lieberman 2004; Lieberman 2015), and scavenging and hunting, involving butchery of animal carcasses with stone tools (Semaw et al. 1997; O’Connell et al. 2002; McPherron et al. 2010; Sayers and Lovejoy 2014; Harmand et al. 2015; Baird et al. 2016), activities that may have increased risk of injury, novel infections, and use of fire (Smith et al. 2015). In this regard, it is also interesting that human-like elimination of Cmah in mice enhances running ability (Okerblom et al. 2018) as well as macrophage activation (Okerblom et al. 2017).
Supplementary Material
Acknowledgments
We thank members of the Varki and Gagneux labs for valuable comments on this work. This research was supported by NIH grant R01GM32373 to A.V.. T.M.-B. is supported by BFU2017-86471-P (MINECO/FEDER, UE), U01 MH106874 grant, Howard Hughes International Early Career, Obra Social “La Caixa” and Secretaria d’Universitats i Recerca, and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880).
Literature Cited
- 1000 Genomes Project Consortium, et al. 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aken BL, et al. 2016. The Ensembl gene annotation system. Database (Oxford) 2016:baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SR, et al. 2014. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. 211(6):1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisson-Silva F, et al. 2018. Human evolutionary loss of epithelial Neu5Gc expression and species-specific susceptibility to cholera. PLoS Pathog. 14 (6):e1007133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altheide TK, et al. 2006. System-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents: evidence for two modes of rapid evolution. J Biol Chem. 281(35):25689–25702. [DOI] [PubMed] [Google Scholar]
- Angata T. 2018. Possible influences of endogenous and exogenous ligands on the evolution of human Siglecs. Front Immunol. 9:2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Hingorani R, Varki NM, Varki A.. 2001. Cloning and characterization of a novel mouse Siglec, mSiglec-F: differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem. 276(48):45128–45136. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki NM, Varki A.. 2001. A second uniquely human mutation affecting sialic acid biology. J Biol Chem. 276(43):40282–40287. [DOI] [PubMed] [Google Scholar]
- Angata T, et al. 2013. Loss of Siglec-14 reduces the risk of chronic obstructive pulmonary disease exacerbation. Cell Mol Life Sci. 70(17):3199–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson EG, et al. 2018. No evidence for recent selection at FOXP2 among diverse human populations. Cell 174(6):1424–1435.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird A, et al. 2016. Injury, inflammation and the emergence of human-specific genes. Wound Rep and Reg. 24(3):602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergfeld AK, et al. 2017. N-Glycolyl groups of nonhuman chondroitin sulfates survive in ancient fossils. Proc Natl Acad Sci U S A. 114 (39):E8155–E8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokelmann L, et al. 2019. A genetic analysis of the Gibraltar Neanderthals. Proc Natl Acad Sci U S A. 116(31):15610–15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramble DM, Lieberman DE.. 2004. Endurance running and the evolution of Homo. Nature 432(7015):345–352. [DOI] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, et al. 2000. Loss of N-glycolylneuraminic acid in human evolution. Implications for sialic acid recognition by siglecs. J Biol Chem. 275(12):8633–8640. [DOI] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, et al. 2007. Human-specific expression of Siglec-6 in the placenta. Glycobiology 17(9):922–931. [DOI] [PubMed] [Google Scholar]
- Bryk J, Tautz D.. 2014. Copy number variants and selective sweeps in natural populations of the house mouse (Mus musculus domesticus). Front Genet. 5:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, et al. 2007. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 21(22):2963–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero-Rhodes MA, et al. 2007. N-Glycolyl GM1 ganglioside as a receptor for simian virus 40. J Virol. 81(23):12846–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin AF, et al. 2009. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 206(8):1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano D, Munch K.. 2020. Population genomics in the great apes. Methods Mol Biol. 2090:453–463. [DOI] [PubMed] [Google Scholar]
- Castellano S, et al. 2014. Patterns of coding variation in the complete exomes of three Neandertals. Proc Natl Acad Sci U S A. 111(18):6666–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Chou HH, et al. 1998. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A. 95(20):11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, et al. 2002. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci U S A. 99(18):11736–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Gordillo P, et al. 2010. Extensive changes in the expression of the opioid genes between humans and chimpanzees. Brain Behav Evol. 76(2):154–162. [DOI] [PubMed] [Google Scholar]
- Currie P. 2004. Human genetics: muscling in on hominid evolution. Nature 428(6981):373–374. [DOI] [PubMed] [Google Scholar]
- de Manuel M, et al. 2016. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science 354(6311):477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deMenocal PB. 2011. Anthropology. Climate and human evolution. Science 331(6017):540–542. [DOI] [PubMed] [Google Scholar]
- Deng L, et al. 2014. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159(6):1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2001. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2(12):919–929. [DOI] [PubMed] [Google Scholar]
- Enard W, et al. 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418(6900):869–872. [DOI] [PubMed] [Google Scholar]
- Enard W, et al. 2009. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137(5):961–971. [DOI] [PubMed] [Google Scholar]
- Fujiyama A, et al. 2002. Construction and analysis of a human–chimpanzee comparative clone map. Science 295(5552):131–134. [DOI] [PubMed] [Google Scholar]
- Gagneux P, Aebi M, Varki A.. 2015. Evolution of glycan diversity In: Varki A, editor. Essentials of glycobiology. Cold Spring Harbor (NY: ): Cold Spring Harbor Laboratory Press. [Google Scholar]
- Gagneux P, et al. 2003. Human-specific regulation of alpha 2-6-linked sialic acids. J Biol Chem. 278(48):48245–48250. [DOI] [PubMed] [Google Scholar]
- Gao PS, et al. 2010. Polymorphisms in the sialic acid-binding immunoglobulin-like lectin-8 (Siglec-8) gene are associated with susceptibility to asthma. Eur J Hum Genet. 18(6):713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaderi D, et al. 2011. Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc Natl Acad Sci U S A. 108(43):17743–17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1–36. [DOI] [PubMed] [Google Scholar]
- Hajdinjak M, et al. 2018. Reconstructing the genetic history of late Neanderthals. Nature 555(7698):652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmand S, et al. 2015. 3.3-Million-year-old stone tools from Lomekwi 3, West Turkana, Kenya. Nature 521(7552):310–315. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, et al. 2001. Alu-mediated inactivation of the human CMP-N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci U S A. 98(20):11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, et al. 2005. A human-specific gene in microglia. Science 309(5741):1693. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, et al. 2006. Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics 172(2):1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, et al. 2017. Coevolution of Siglec-11 and Siglec-16 via gene conversion in primates. BMC Evol Biol. 17(1):228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich K, et al. 2016. Streptococcus pneumoniae senses a human-like sialic acid profile via the response regulator CiaR. Cell Host Microbe 20(3):307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hublin JJ, et al. 2017. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature 546(7657):289–292. [DOI] [PubMed] [Google Scholar]
- Johnson ME, et al. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413(6855):514–519. [DOI] [PubMed] [Google Scholar]
- Kang JH, et al. 2011. Preeclampsia leads to dysregulation of various signaling pathways in placenta. J Hypertens. 29(5):928–936. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN.. 2007. Understanding the recent evolution of the human genome: insights from human–chimpanzee genome comparisons. Hum Mutat. 28(2):99–130. [DOI] [PubMed] [Google Scholar]
- Khan N, et al. 2020. Maximum reproductive lifespan correlates with CD33rSIGLEC gene number: implications for NADPH oxidase-derived reactive oxygen species in aging. FASEB J. 34(2):1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 6(5):376–385. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC.. 1975. Evolution at two levels in humans and chimpanzees. Science 188(4184):107–116. [DOI] [PubMed] [Google Scholar]
- Kronenberg ZN, et al. 2018. High-resolution comparative analysis of great ape genomes. Science 360(6393):eaar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlwilm M, et al. 2016. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530(7591):429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogashima M, Ginsburg V, Krivan HC.. 1989. Escherichia coli K99 binds to N-glycolylsialoparagloboside and N-glycolyl-GM3 found in piglet small intestine. Arch Biochem Biophys. 270(1):391–397. [DOI] [PubMed] [Google Scholar]
- Lieberman DE. 2015. Human locomotion and heat loss: an evolutionary perspective. Compr Physiol. 5(1):99–117. [DOI] [PubMed] [Google Scholar]
- Martin MJ, et al. 2005. Evolution of human–chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci U S A. 102(36):12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron SP, et al. 2010. Evidence for stone-tool-assisted consumption of animal tissues before 3.39 million years ago at Dikika, Ethiopia. Nature 466(7308):857–860. [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D Jr. 2004. Revealing the world of RNA interference. Nature 431(7006):338–342. [DOI] [PubMed] [Google Scholar]
- Meyer M, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338(6104):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, et al. 2016. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531(7595):504–507. [DOI] [PubMed] [Google Scholar]
- Mitra N, et al. 2011. SIGLEC12, a human-specific segregating (pseudo)gene, encodes a signaling molecule expressed in prostate carcinomas. J Biol Chem. 286(26):23003–23011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JM, et al. 2018. Examination of signatures of recent positive selection on genes involved in human sialic acid biology. G3 (Bethesda) 8(4):1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchmore EA, Diaz S, Varki A.. 1998. A structural difference between the cell surfaces of humans and the great apes. Am J Phys Anthropol. 107(2):187–198. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, et al. 2006. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci U S A. 103(20):7765–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M, et al. 2012. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet. 13(12):853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JF, et al. 2002. Male strategies and Plio-Pleistocene archaeology. J Hum Evol. 43(6):831–872. [DOI] [PubMed] [Google Scholar]
- Okerblom J, Varki A.. 2017. Biochemical, cellular, physiological, and pathological consequences of human loss of N-glycolylneuraminic acid. ChemBioChem 18(13):1155–1171. [DOI] [PubMed] [Google Scholar]
- Okerblom J, et al. 2018. Human-like Cmah inactivation in mice increases running endurance and decreases muscle fatigability: implications for human evolution. Proc Biol Sci. 285(1886):20181656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okerblom JJ, et al. 2017. Loss of CMAH during human evolution primed the monocyte–macrophage lineage toward a more inflammatory and phagocytic state. J Immunol. 198(6):2366–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, et al. 2014. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nat Commun. 5(1):4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padler-Karavani V, et al. 2014. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. 28(3):1280–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyregne S, et al. 2017. Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 27(9):1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, et al. 2013. Great ape genetic diversity and population history. Nature 499(7459):471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, et al. 2017. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358(6363):655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468(7327):1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 89(4):516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, et al. 2015. Origins of de novo genes in human and chimpanzee. PLoS Genet. 11(12):e1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S, et al. 2015. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci U S A. 112(51):15696–15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers K, Lovejoy CO.. 2014. Blood, bulbs, and bunodonts: on evolutionary ecology and the diets of Ardipithecus, Australopithecus, and early Homo. Q Rev Biol. 89(4):319–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46(8):919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, et al. 2016. Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc Natl Acad Sci U S A. 113(1):74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, et al. 2017. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. 36(6):751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C, Herrler G.. 2006. Sialic acids as receptor determinants for coronaviruses. Glycoconj J. 23(1–2):51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaw S, et al. 1997. 2.5-million-year-old stone tools from Gona, Ethiopia. Nature 385(6614):333–336. [DOI] [PubMed] [Google Scholar]
- Slon V, et al. 2018. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561(7721):113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, et al. 2015. The significance of cooking for early hominin scavenging. J Hum Evol. 84:62–70. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Altheide TK, Varki A.. 2004. A uniquely human consequence of domain-specific functional adaptation in a sialic acid-binding receptor. Glycobiology 14(4):339–346. [DOI] [PubMed] [Google Scholar]
- Stedman HH, et al. 2004. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428(6981):415–418. [DOI] [PubMed] [Google Scholar]
- Sullivan AP, et al. 2017. An evolutionary medicine perspective on Neandertal extinction. J Hum Evol. 108:62–71. [DOI] [PubMed] [Google Scholar]
- Valenzano DR, et al. 2015. The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 163(6):1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 2000. A chimpanzee genome project is a biomedical imperative. Genome Res. 10(8):1065–1070. [DOI] [PubMed] [Google Scholar]
- Varki A. 2009. Multiple changes in sialic acid biology during human evolution. Glycoconj J. 26(3):231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 2011. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21(9):1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Angata T.. 2006. Siglecs—the major subfamily of I-type lectins. Glycobiology 16(1):1R–27R. [DOI] [PubMed] [Google Scholar]
- Varki A, et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology 25(12):1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mitra N, Cruz P, et al. 2012. Evolution of siglec-11 and siglec-16 genes in hominins. Mol Biol Evol. 29(8):2073–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mitra N, Secundino I, et al. 2012. Specific inactivation of two immunomodulatory SIGLEC genes during human evolution. Proc Natl Acad Sci U S A. 109(25):9935–9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. 2011. Expression of Siglec-11 by human and chimpanzee ovarian stromal cells, with uniquely human ligands: implications for human ovarian physiology and pathology. Glycobiology 21(8):1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, et al. 2004. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429(6990):382–388. [DOI] [PubMed] [Google Scholar]
- White TD, et al. 1993. New discoveries of Australopithecus at Maka in Ethiopia. Nature 366(6452):261–265. [DOI] [PubMed] [Google Scholar]
- Wood B, Boyle EK.. 2016. Hominin taxic diversity: fact or fantasy. Am J Phys Anthropol. 159(Suppl 61):37–78. [DOI] [PubMed] [Google Scholar]
- Xue Y, et al. 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348(6231):242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, et al. 2017. Chemical synthesis and evaluation of a disialic acid-containing dextran polymer as an inhibitor for the interaction between Siglec 7 and its ligand. ChemBioChem 18(13):1194–1203. [DOI] [PubMed] [Google Scholar]
- Yamanaka M, et al. 2009. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19(8):841–846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.