Abstract
Odorant binding proteins (OBPs) and chemosensory proteins (CSPs) play essential roles in insect chemosensory recognition. Here, we identified nine OBPs and nine CSPs from the Myzus persicae transcriptome and genome. Genomic structure analysis showed that the number and length of the introns are much higher, and this appears to be a unique feature of aphid OBP genes. Three M. persicae OBP genes (OBP3/7/8) as well as CSP1/4/6, CSP2/9 and CSP5/8 are tandem arrayed in the genome. Phylogenetic analyses of five different aphid species suggest that aphid OBPs and CSPs are conserved in single copy across all aphids (with occasional losses), indicating that each OBP and CSP class evolved from a single gene in the common ancestor of aphids without subsequent duplication. Motif pattern analysis revealed that aphid OBP and CSP motifs are highly conserved, and this could suggest the conserved functions of aphid OBPs and CSPs. Three OBPs (MperOBP6/7/10) are expressed antennae specifically, and five OBPs (MperOBP2/4/5/8/9) are expressed antennae enriched, consistent with their putative olfactory roles. M. persicae CSPs showed much broader expression profiles in nonsensory organs than OBPs. None of the nine MperCSPs were found to be antennae specific, but five of them (MperCSP1/2/4/5/6) showed higher expression levels in the legs than in other tissues. MperCSP10 mainly expressed in the antennae and legs. The broad and diverse expression patterns of M. persicae CSPs suggest their multifunctions in olfactory perception, development and other processes.
Keywords: Myzus persicae, odorant binding protein, chemosensory protein, genomic structure, motif‐pattern, tissue expression profiles
Introduction
The green peach aphid Myzus persicae (Hemiptera: Aphididae) is a major destructive polyphagous pest. They can use more than 400 plant species from more than 40 families for parthenogenetic reproduction, and use peach, its primary host, for sexual reproduction (Van Emden et al., 1969; Weber, 1986; Troncoso et al., 2005). Other than its direct feeding damage to plants, M. persicae can also transmit more than 100 plant viruses, including both persistent viruses such as potato leaf roll virus (Eskandari et al., 1979) and nonpersistent viruses such as cucumber mosaic virus (Bwye et al., 1997). In addition, M. persicae is a typical host‐alternating aphid species, and exhibits highly colour‐polymorphic association with different host plants. Green M. persicae aphids can produce pink offspring when they are fed on poor quality hosts (Williams et al., 2000), whereas red M. persicae aphids can produce green offspring when they are crowded on poor diets (Ueda and Takada, 1977). The ovipositing female aphids locate their hosts usually by chemical cues and antennal contacts (Read et al., 1970). The success of M. persicae in nature is due to its extremely high population adaptability to the environment, a wide genetic variability and a broad phenotypic plasticity for which chemical communications play a critical role (Weber, 1986).
Insects use their sensitive and selective olfactory organs, mainly the antennae, to perceive the surrounding world. Aphids, like other insects, rely heavily on chemical signals, including plant volatiles and species‐specific pheromones, to locate correct hosts, find mates and avoid predators or parasitoids (Pickett and Glinwood, 2007). Mature sexual females release sex pheromones from their tibiae of the hind legs to attract conspecific males (Marsh, 1972; Pickett and Glinwood, 2007). The sex pheromones of aphid species usually comprise (4aS,7S,7aR)‐nepetalactone and (1R,4aS,7S,7aR)‐nepetalactol, a mixture of two monoterpenoids (Pickett et al., 1992; Dewhirst et al., 2010). When attacked by predators or parasitoids, most aphid species, including M. persicae, emit chemical signals known as alarm pheromones that signal other individuals to escape and defend (Nault et al., 1973; Pickett and Griffiths, 1980), or even attack the predator (eg Ceratovacuna lanigera) (Arakaki, 1989). The alarm pheromones of most aphids are a mixture of (E)‐β‐farnesene (Eβf), α‐pinene, β‐pinene and β‐limonene (Pickett and Griffiths, 1980). Eβf, a sesquiterpene hydrocarbon, is the primary component of the alarm pheromones of many aphids, and thus is considered as a repellent to control aphids (Bowers et al., 1972). Chemoreception plays important roles in the complex life cycle of aphids. Studying the interaction of aphids with aphids, plants, natural enemies, competitors and mutualists will contribute to exploit novel environment‐friendly strategies for aphid control.
Odorant binding proteins (OBPs) and chemosensory proteins (CSPs) are two families of small water‐soluble proteins; they are highly concentrated (as high as 10 mm) in sensillum lymph of the antennal sensilla and believed to be involved in the initial chemosensory recognition of insects (Vogt and Riddiford, 1981; Calvello et al., 2003; Pelosi et al., 2006). Both OBPs and CSPs have small molecular weights, approximately 15 kDa for OBPs and 12 kDa for CSPs. The common feature of insect OBPs is that they have six highly conserved cysteines that are paired to form interlocked disulphide bridges (Scaloni et al., 1999; Northey et al., 2016). Insect CSPs, on other hand, have four conserved cysteines that are linked to form disulphide bridges (Angeli et al., 1999). So far, the genomic sequences of four aphid species have been published: the pea aphid Acyrthosiphon pisum (The International Aphid Genomics Consortium, 2010), the Russian wheat aphid Diuraphis noxia (Nicholson et al., 2015), the green peach aphid M. persicae (Mathers et al., 2017) and the soybean aphid Aphis glycines (Wenger et al., 2017). Meanwhile, the transcriptomic data of several aphid species are also available (Gu et al., 2013; Xue et al., 2016). Fifteen OBPs and 13 CSPs were identified from the A. pisum genomic data (Zhou et al., 2010). Nine OBPs and nine CSPs were identified in the cotton aphid Aphis gossypii (Gu et al., 2013), and 13 OBPs and five CSPs were identified in the grain aphid Sitobion avenae (Xue et al., 2016) from their transcriptomic data. For M. persicae, only four OBPs and five CSPs were identified from the expressed sequence tags (Xu et al., 2009). The functional studies of aphid OBPs and CSPs in recognizing and discriminating the sex pheromones, the alarm pheromone and the host plant volatiles, however, are still limited. The OBP3 of A. pisum (Qiao et al., 2009), the OBP7 of S. avenae (Vandermoten et al., 2011; Zhong et al., 2012), the OBP3 of Megoura viciae and Nasonovia ribisnigri (Northey et al., 2016) and the OBP3 and OBP7 of Rhopalosiphum padi (Fan et al., 2017) have high binding affinities with the alarm pheromone Eβf. However, the in vivo evidence of OBP3 and OBP7 participating in Eβf recognition is still not conclusive.
In this study, we identified nine OBP and nine CSP genes from the M. persicae transcriptomic and genomic sequence datasets and for the first time demonstrated their genomic structures, clusters and uniquely long introns. We further analysed their motif patterns and phylogenetic relationships with the OBPs and CSPs in five other aphid species and in other hemipterans. We also examined the tissue expression profiles of M. persicae OBP and CSP genes to infer their putative functions in aphid chemoreception of semiochemicals. The evolution and differentiation of Hemiptera OBPs and CSPs are also discussed.
Results
Sequencing, de novo assembly and functional annotation
A total of 110 059 204 raw reads were produced from the apterous M. persicae transcriptome sequencing project. After trimming adaptor sequences and removing low‐quality sequences, 107 199 360 clean reads were remained. After assembly, 50 352 unigenes were generated, with lengths ranging from 201 bp to 27.17 kb and an average length of 829 bp (N50 = 1745 bp, N90 = 294 bp).
The unigenes were searched with blastx and blastn programs against the sequences in the NCBI GenBank database. The results indicated that 15 881 of the 50 352 unigenes (31.54%) had blastx hits in the nonredundant protein (nr) databases, and 14 329 unigenes (28.46%) had blastn hits in the nonredundant nucleotide sequence (nt) databases. Some unigenes are homologous to more than one species, and most of the annotated unigenes have the best hit with Hemiptera insect genes.
The Gene Ontology (GO) category analysis revealed that only 12 521 of the 50 352 unigenes (24.87%) could be annotated into different functional groups (biological process, cellular components and molecular functions). The cellular process (7650 unigenes) and metabolic process (6913 unigenes) GO categories were most abundantly represented within the biological process GO. In the cellular components GO, the transcripts were mainly distributed in the cell (4832 unigenes) and cell part (4935 unigenes). The GO analysis also showed that the unigenes involving in binding (7306 unigenes) and catalytic activity (5696 unigenes) were most abundant in the molecular function ontology (Fig. 1).
Figure 1.
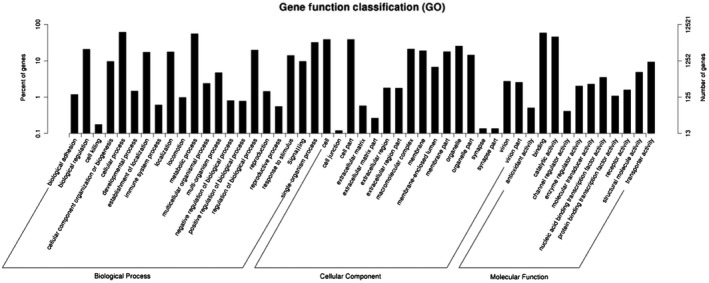
Gene Ontology (GO) classifications of the 50352 Myzus persicae unigenes.
Identification of OBP and CSP genes in M. persicae
A total of nine OBP and nine CSP genes were identified from the transcriptome and genome of M. persicae using motifsearch and tblastn program (Table 1). The identification of these OBPs and CSPs was confirmed by searching M. persicae genome sequences (https://bipaa.genouest.org/is/aphidbase/myzus_persicae). All M. persicae OBPs and CSPs are full‐lengths with open reading frames (ORFs) ranging from 396 to 858 bp. The nucleotide sequences of M. persicae OBP and CSP genes were verified by molecular cloning and sequencing. We name these M. persicae OBP genes as MperOBP2–10 and the CSP genes as MperCSP1–2 and MperCSP4–10 following the nomenclatures reported for the OBPs and CSPs of A. pisum (Zhou et al., 2010), A. gossypii (Gu et al., 2013) and S. avenae (Xue et al., 2016). Like in A. gossypii (Gu et al., 2013), both OBP1 and CSP3 identified in A. pisum are missing in the M. persicae transcriptome and genome dataset. Like AgosCSP1 (Gu et al., 2013), MperCSP1 does not have a signal peptide, a signature of secretory protein. The rest of the M. persicae OBP and CSP proteins all have a signal peptide at their N‐terminus (Table 1). The nine identified M. persicae OBPs can be divided into two distinct subfamilies: MperOBP2/3/4/7/8/9/10 belong to the classical OBP subfamily, which has the typical six conserved cysteines and fit the motif pattern of ‘C1‐X15–39‐C2‐X3‐C3‐X21–44‐C4‐X7–12‐C5‐X8‐C6’ (Fig. 2A, Zhou et al., 2004; Xu et al., 2009), MperOBP5 and MperOBP6 belong to the plus‐C OBP subfamily, which has one additional cysteine (C6a) and a conserved proline immediately after the sixth cysteine (Fig. 2A, Zhou et al., 2004). All of the MperCSPs have four conserved cysteines and fit the motif pattern of ‘C1‐X6–8‐C2‐X16–21‐C3‐X2‐C4’ (Fig. 2B, Zhou et al., 2004). The nucleotide sequences of MperOBPs and MperCSPs have been deposited in GenBank under the accession numbers MG356454 to MG356471.
Table 1.
List of odorant binding proteins and chemosensory proteins in Myzus persicae
| Gene name | RPKM | Signal peptide (aa) | Amino acids (aa) | ORF (bp) | Scaffold* | Accession no. | Gene annotation | Homology search with other aphids | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Score (bits) | E‐value | Identity (%) | ||||||||
| MperOBP2 | 383 | 1–19 | 244 | 732 | 105 (340085…345920,−) | MG356454 | Odorant‐binding protein 2 | A. cyrthosiphon pisum | NP 001153528 | 384 | 2e‐133 | 96 |
| MperOBP3 | 135 | 1–23 | 142 | 426 | 21 (798710…815519,+) | MG356455 | Odorant‐binding protein 3 | A. cyrthosiphon pisum | NP 001153529 | 281 | 6e‐96 | 96 |
| MperOBP4 | 21 | 1–22 | 200 | 600 | 300 (285472…288740,+) | MG356456 | Odorant‐binding protein 4 | S. avenae | APB03427 | 400 | 7e‐141 | 97 |
| MperOBP5 | 99 | 1–25 | 222 | 666 | 4 (1732552…1743198,−) | MG356457 | Odorant‐binding protein 5 | A. cyrthosiphon pisum | NP 001153531 | 450 | 4e‐160 | 97 |
| MperOBP6 | 73 | 1–19 | 216 | 648 | 77 (15902…21431,−) | MG356458 | Odorant‐binding protein 6 | S. avenae | APB03429 | 394 | 4e‐138 | 87 |
| MperOBP7 | 65 | 1–24 | 150 | 450 | 21 (790748…798189,+) | MG356459 | Odorant‐binding protein 7 | A. cyrthosiphon pisum | NP 001153533 | 251 | 1e‐83 | 87 |
| MperOBP8 | 33 | 1–18 | 162 | 486 | 21 (816539…821387,+) | MG356460 | Odorant‐binding protein 8 | A. cyrthosiphon pisum | NP 001153534 | 314 | 3e‐108 | 96 |
| MperOBP9 | 19 | 1–24 | 167 | 501 | 422 (21121…26183,+) | MG356461 | Odorant‐binding protein 9 | S. avenae | APB03432 | 265 | 2e‐88 | 90 |
| MperOBP10 | 8 | 1–24 | 144 | 432 | 47 (690675…693525,−) | MG356462 | Odorant‐binding protein 10 | S. avenae | APB03433 | 238 | 9e‐79 | 76 |
| MperCSP1 | 16 | No SP | 286 | 858 | 112 (593787…605068,−) | MG356463 | Chemosensory protein 1 | A. gossypii | AGE97641 | 273 | 1e‐89 | 87 |
| MperCSP2 | 330 | 1–20 | 132 | 396 | 55 (285362…286497,−) | MG356464 | Chemosensory protein 2 | A. gossypii | AGE97642 | 233 | 3e‐77 | 88 |
| MperCSP4 | 286 | 1–22 | 146 | 438 | 112 (578383…579415,−) | MG356465 | Chemosensory protein 4 | A. gossypii | AGE97643 | 256 | 6e‐86 | 94 |
| MperCSP5 | 609 | 1–19 | 140 | 420 | 10 (1364996…1366765,−) | MG356466 | Chemosensory protein 5 | A. gossypii | AGE97644 | 284 | 1e‐156 | 92 |
| MperCSP6 | 91 | 1–21 | 132 | 396 | 112 (615905…617608,−) | MG356467 | Chemosensory protein 6 | A. gossypii | AEG97645 | 208 | 3e‐67 | 87 |
| MperCSP7 | 40 | 1–24 | 156 | 468 | 910 (34801…42428,−) | MG356468 | Chemosensory protein 7 | A. gossypii | AGE97646 | 280 | 4e‐95 | 89 |
| MperCSP8 | 38 | 1–37 | 164 | 492 | 10 (1379554…1382877,−) | MG356469 | Chemosensory protein 8 | A. gossypii | AGE97647 | 240 | 9e‐79 | 75 |
| MperCSP9 | 330 | 1–22 | 163 | 489 | 55 (282954…285123,+) | MG356470 | Chemosensory protein 9 | A. gossypii | AGE97648 | 212 | 8e‐68 | 60 |
| MperCSP10 | 3 | 1–21 | 151 | 453 | 420 (101975…105884,−) | MG356471 | Chemosensory protein 10 | A. gossypii | AGE97649 | 177 | 2e‐54 | 83 |
aa, amino acids; ORF, open reading frame; RPKM, reads per kilobase per million; SP, signal peptide.
AphidBase scaffold ID (start…stop nt, orientation).
Figure 2.
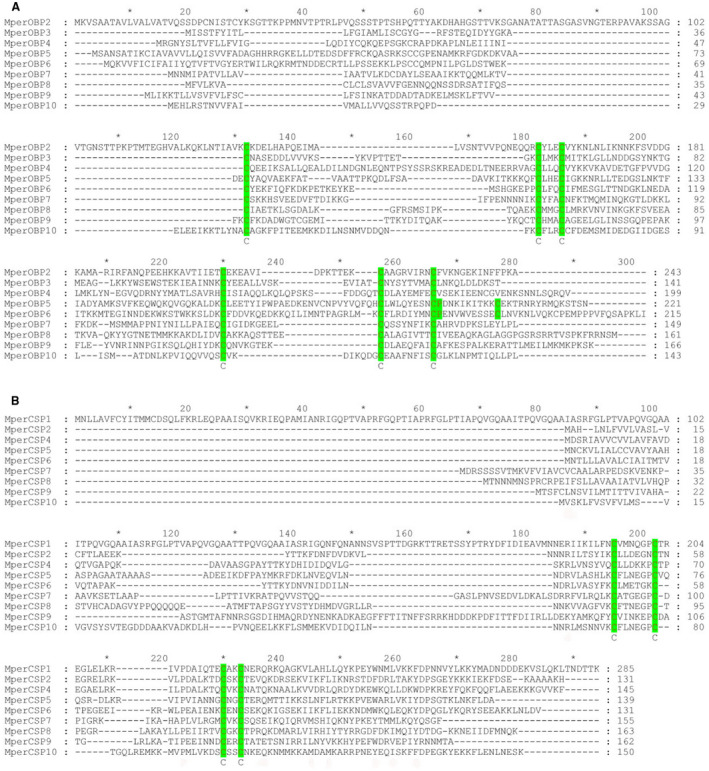
Alignment of the Mysuz persicae odorant binding proteins (OBPs) and chemosensory proteins (CSPs). The full‐length amino acid sequences are aligned by ClustalX 2.1 and then edited using GeneDoc. (A) Alignment of the M. persicae OBPs. The six conserved cysteine residues are indicated by the green shading. (B) Alignment of the M. persicae CSPs. The four conserved cysteine residues are shaded in green. [Colour figure can be viewed at wileyonlinelibrary.com]
Phylogenetic analyses of Hemiptera OBPs and CSPs
MperOBPs share relatively low amino acid identities (8–59%) with each other (Supporting Information Table S6), and MperCSPs show relatively high amino acid identities (10–71%) with each other (Supporting Information Table S7). But the amino acid identities of orthologous OBPs and CSPs in the five aphid species of the Aphididae family (M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae) are much higher (eg 93% among the OBP3 orthologues and 94% among the CSP4 orthologues). The phylogenetic tree of Hemiptera OBPs was built using 237 OBP sequences from 16 different aphid species (M. persicae, A. gossypii, A. pisum, A. glycines, S. avenae, Pterocomma salicis, Aphis fabae, Aphis craccivora, Tuberolachnus salignus, M. viciae, Metopolophium dirhodum, N. ribisnigri, R. padi, Lipaphis erysimi, Drepanosiphum platanoidis and Brevicoryne brassicae), four plant bug species (Adelphocoris suturalis, Apolygus lucorum, Adelphocoris lineolatus and Lygus lineolaris) and three plant hopper species (Nilaparvata lugens, Laodelphax striatellus and Sogatella furcifera) (Supporting Information Table S8; Fig. 3). Aphid OBPs are clearly separated from OBPs from plant bugs and plant hoppers and clustered together to form multiple homologous subgroups (lineages) named OBP1 to OBP10 subgroups supported by high bootstrap values of 89–100 (Fig. 3), indicating that these subgroups evolved from a single gene in the common ancestor of aphids. Furthermore, OBP1 subgroup and OBP8 subgroup are more closely related and clustered into a branch with an average amino acid identity of 61.54% and a bootstrap value of 99. The OBP1 and OBP8 subgroups contain OBPs from 11 aphid species (M. persicae, A. gossypii, A. pisum, A. glycines, S. avenae, M. dirhodum, A. fabae, N. ribisnigri, M. viciae, T. salignus and P. salicis), suggesting a more recent evolution into OBP1 subgroup and OBP8 subgroup from a common ancestor before aphid speciation (Zhou et al., 2010).
Figure 3.
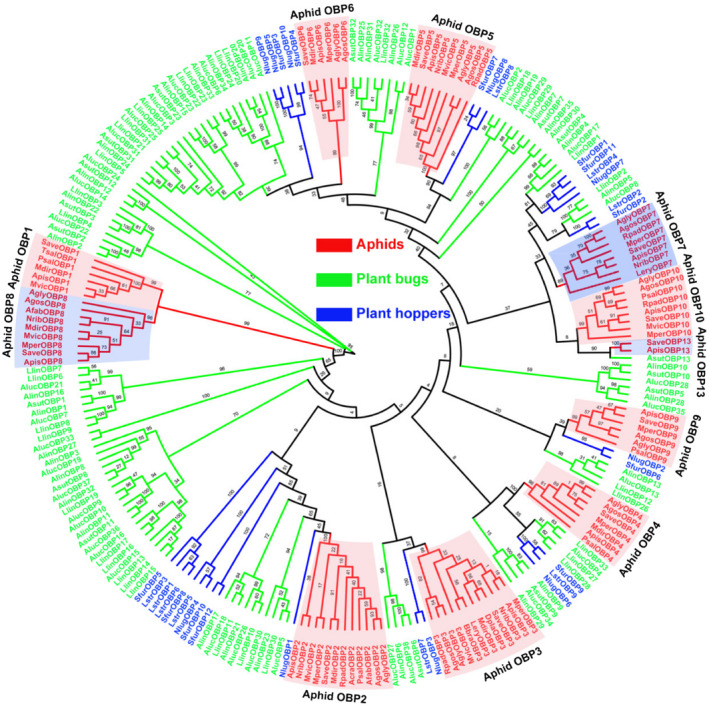
Phylogenetic tree of 237 odorant binding proteins (OBPs) from Hemiptera species. The phylogenetic tree of Hemiptera OBPs was built using 237 OBP sequences from 16 different aphid species (Myzus persicae, Aphis gossypii, Acyrthosiphon pisum, Aphis glycines, Sitobion avenae, Pterocomma salicis, Aphis fabae, Aphis craccivora, Tuberolachnus salignus, Megoura viciae, Metopolophium dirhodum, Nasonovia ribisnigri, Rhopalosiphum padi, Lipaphis erysimi, Drepanosiphum platanoidis and Brevicoryne brassicae), four plant bugs (Adelphocoris suturalis, Apolygus lucorum, Adelphocoris lineolatus and Lygus lineolaris) and three plant hoppers (Nilaparvata lugens, Laodelphax striatellus and Sogatella furcifera). The protein names and sequences that were used in phylogenetic analysis are listed in Supporting Information Table S8. [Colour figure can be viewed at wileyonlinelibrary.com]
The phylogenetic tree of Hemiptera CSPs was built using 110 CSP sequences from five different aphid species (M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae), three plant bugs (A. suturalis, A. lucorum and A. lineolatus) and three plant hoppers (N. lugens, L. striatellus and S. furcifera) (Fig. 4, Supporting Information Table S9). Like the OBPs shown in Fig. 3, the phylogenetic analysis clusters aphid CSPs into several clear clades of nine homologous subgroups (CSP1, CSP2, CSP4, CSP5, CSP6, CSP7, CSP8, CSP9 and CSP10). Interestingly, some aphid CSP subgroups are clustered with non‐aphid CSP genes in conserved clades. For example, the plant bug AsutCSP2 and the aphid CSP7 subgroup are clustered in the same clade with a bootstrap value as high as 95, and the plant hopper LstrCSP1 and the aphid CSP9 subgroup are clustered in the same branch with a bootstrap value as high as 93 (Fig. 4). This suggests that the AsutCSP2 and aphid CSP7s and the LstrCSP1 and aphid CSP9s evolved from the same ancestor and may play similar roles in these sucking insects.
Figure 4.
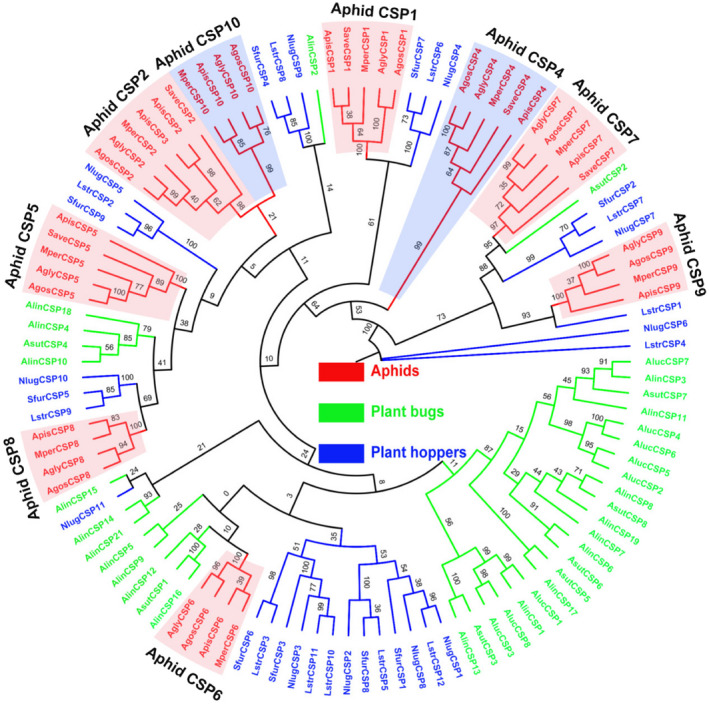
Phylogenetic tree of 110 chemosensory proteins (CSPs) from Hemiptera species. The phylogenetic tree of Hemiptera CSPs was built using 110 CSP sequences from five different aphid species (Myzus persicae, Aphis gossypii, Acyrthosiphon pisum, Aphis glycines and Sitobion avenae), three plant bugs (Adelphocoris suturalis, Apolygus lucorum and Adelphocoris lineolatus) and three plant hoppers (Nilaparvata lugens, Laodelphax striatellus and Sogatella furcifera). The protein names and sequences that were used in phylogenetic analysis are listed in Supporting Information Table S9. [Colour figure can be viewed at wileyonlinelibrary.com]
Genomic structure of M. persicae OBP and CSP genes
The genomic structures and the splice site of the intron–exon junctions of M. persicae OBP and CSP genes were analysed based on the M. persicae genome annotations and the GT–AG rules (Modrek and Lee, 2002). The results revealed that the sizes of the genomic sequences of MperOBP genes range from 2.85 to 16.81 kb with an average length of 6.92 kb. Five OBP genes (MperOBP4/7/8/9/10) have six introns and seven exons. The remaining four OBP genes (MperOBP2/3/5/6) have 4, 5, 8 and 7 introns and 5, 6, 9 and 8 exons, respectively (Fig. 5Table 5). The intron lengths of each MperOBP are variable; for example, the intron lengths of MperOBP3, MperOBP5 and MperOBP10 are 16.38 kb, 9.98 kb and 2.42 kb, respectively (Table 2). Cross‐species comparison reveals that the numbers of introns and exons of each OBP subgroup are the same among four different aphid species (M. persicae, A. gossypii, A. pisum and A. glycines), but the lengths of the introns and exons are different among the four species (Table 2).
Figure 5.
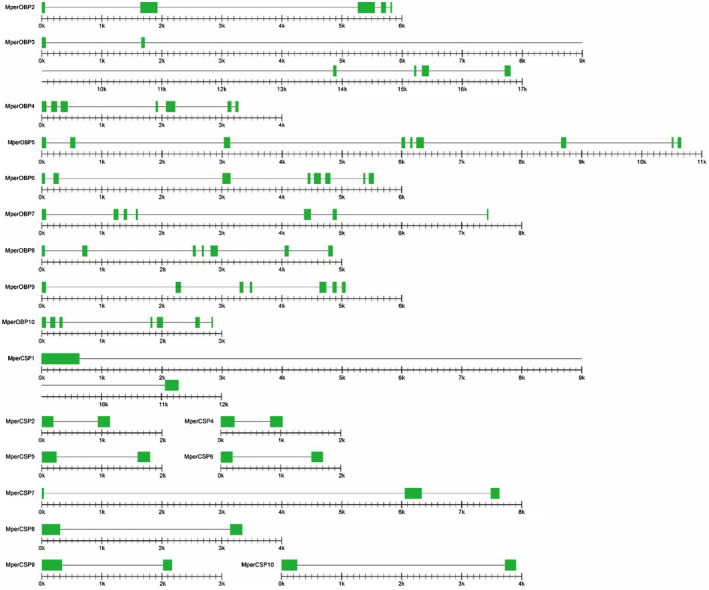
Genomic structure of Myzus persicae odorant binding protein (OBP) and chemosensory proteins (CSP) genes. The green rectangles and hairlines between two green rectangles represent the exons and introns, respectively. The length has been shown to scale with a scale bar under each M. persicae OBP and CSP gene; every minor mark represents 100 bp. [Colour figure can be viewed at wileyonlinelibrary.com]
Table 2.
The introns and exons of aphid odorant binding protein (OBP) and chemosensory protein (CSP) genes in Myzus persicae, Aphis gossypii, Acyrthosiphon pisum and Aphis glycines
| Gene name | Aphid species | Genomic DNA size (bp) | No. of introns | Total length of introns (bp) | Average intron size (bp) | No. of exons | Total length of exons (bp) | Average exon size (bp) | Scaffold | Strand |
|---|---|---|---|---|---|---|---|---|---|---|
| OBP2 | M. persicae | 5 836 | 4 | 5 104 | 1 276 | 5 | 732 | 146 | 105 | − |
| A. gossypii | 3 277 | 4 | 2 545 | 636 | 5 | 732 | 146 | x | x | |
| A. pisum | 3 885 | 4 | 3 153 | 788 | 5 | 732 | 146 | 748 | + | |
| A. glycines | 3 347 | 4 | 2 615 | 654 | 5 | 732 | 146 | 23 | + | |
| OBP3 | M. persicae | 16 810 | 5 | 16 384 | 3 277 | 6 | 426 | 71 | 21 | + |
| A. gossypii | 17 385 | 5 | 16 959 | 3 392 | 6 | 426 | 71 | x | x | |
| A. pisum | 17 976 | 5 | 17 550 | 3 510 | 6 | 426 | 71 | 195 | − | |
| A. glycines | 14 751 | 5 | 14 325 | 2 865 | 6 | 426 | 71 | 455 | − | |
| OBP4 | M. persicae | 3 269 | 6 | 2 669 | 445 | 7 | 600 | 86 | 300 | + |
| A. gossypii | 4 108 | 6 | 3 511 | 585 | 7 | 597 | 85 | x | x | |
| A. pisum | 3 250 | 6 | 2 668 | 445 | 7 | 582 | 83 | 116 | + | |
| A. glycines | 3 854 | 6 | 3 257 | 543 | 7 | 597 | 85 | 426 | + | |
| OBP5 | M. persicae | 10 647 | 8 | 9 981 | 1 248 | 9 | 666 | 74 | 4 | − |
| A. gossypii | 12 286 | 8 | 11 611 | 1 451 | 9 | 675 | 75 | x | x | |
| A. pisum | 10 371 | 8 | 9 705 | 1 213 | 9 | 666 | 74 | 838 | + | |
| A. glycines | 10 483 | 8 | 9 808 | 1 226 | 9 | 675 | 75 | 449 | + | |
| OBP6 | M. persicae | 5 530 | 7 | 4 882 | 697 | 8 | 648 | 81 | 77 | − |
| A. gossypii | 5 345 | 7 | 4 697 | 671 | 8 | 648 | 81 | x | x | |
| A. pisum | 5 349 | 7 | 4 701 | 672 | 8 | 648 | 81 | 211 | − | |
| A. glycines | 3 349 | 7 | 2 701 | 386 | 8 | 648 | 81 | 272 | − | |
| OBP7 | M. persicae | 7 442 | 6 | 6 992 | 1 165 | 7 | 450 | 64 | 21 | + |
| A. gossypii | 12 546 | 6 | 12 099 | 2 017 | 7 | 447 | 64 | x | x | |
| A. pisum | 8 526 | 6 | 8 058 | 1 343 | 7 | 468 | 67 | 195 | − | |
| A. glycines | 10 771 | 6 | 10 306 | 1 718 | 7 | 465 | 66 | 455 | − | |
| OBP8 | M. persicae | 4 849 | 6 | 4 363 | 727 | 7 | 486 | 69 | 21 | + |
| A. gossypii | 7 317 | 6 | 6 831 | 1 139 | 7 | 486 | 69 | x | x | |
| A. pisum | 6 425 | 6 | 5 939 | 990 | 7 | 486 | 69 | 195 | − | |
| A. glycines | 6 285 | 6 | 5 799 | 967 | 7 | 486 | 69 | 455 | − | |
| OBP9 | M. persicae | 5 063 | 6 | 4 562 | 760 | 7 | 501 | 72 | 422 | + |
| A. gossypii | 10 033 | 6 | 9 532 | 1 589 | 7 | 501 | 72 | x | x | |
| A. pisum | 5 803 | 6 | 5 302 | 884 | 7 | 501 | 72 | 754 | − | |
| A. glycines | 7 534 | 6 | 7 033 | 1 172 | 7 | 501 | 72 | 753 | − | |
| OBP10 | M. persicae | 2 851 | 6 | 2 419 | 403 | 7 | 432 | 62 | 47 | − |
| A. gossypii | 6 429 | 6 | 5 985 | 998 | 7 | 444 | 63 | x | x | |
| A. pisum | 2 839 | 6 | 2 404 | 401 | 7 | 435 | 62 | 244 | + | |
| A. glycines | 3 906 | 6 | 3 462 | 577 | 7 | 444 | 63 | Contig_3 | + | |
| CSP1 | M. persicae | 11 282 | 1 | 10 424 | 10 424 | 2 | 858 | 429 | 112 | − |
| A. gossypii | 6 763 | 1 | 6 250 | 6 250 | 2 | 513 | 257 | x | x | |
| A. pisum | 9 417 | 1 | 8 733 | 8 733 | 2 | 684 | 342 | 630 | − | |
| A. glycines | 6 077 | 1 | 5 564 | 5 564 | 2 | 513 | 257 | 331 | − | |
| CSP2 | M. persicae | 1 136 | 1 | 740 | 740 | 2 | 396 | 198 | 55 | − |
| A. gossypii | 1 110 | 1 | 705 | 705 | 2 | 405 | 203 | x | x | |
| A. pisum | 1 131 | 1 | 735 | 735 | 2 | 396 | 198 | 447 | + | |
| A. glycines | 1 118 | 1 | 713 | 713 | 2 | 405 | 203 | 391 | + | |
| CSP4 | M. persicae | 1 033 | 1 | 595 | 595 | 2 | 438 | 219 | 112 | − |
| A. gossypii | 1 024 | 1 | 586 | 586 | 2 | 438 | 219 | x | x | |
| A. pisum | 1 018 | 1 | 586 | 586 | 2 | 432 | 216 | 630 | − | |
| A. glycines | 1 017 | 1 | 579 | 579 | 2 | 438 | 219 | 331 | − | |
| CSP5 | M. persicae | 1 770 | 1 | 1 350 | 1 350 | 2 | 420 | 210 | 10 | − |
| A. gossypii | 1 770 | 1 | 1 350 | 1 350 | 2 | 420 | 210 | x | x | |
| A. pisum | 1 792 | 1 | 1 378 | 1 378 | 2 | 414 | 207 | 152 | − | |
| A. glycines | 1 771 | 1 | 1 351 | 1 351 | 2 | 420 | 210 | 192 | + | |
| CSP6 | M. persicae | 1 704 | 1 | 1 308 | 1 308 | 2 | 396 | 198 | 112 | − |
| A. gossypii | 1 818 | 1 | 1 422 | 1 422 | 2 | 396 | 198 | x | x | |
| A. pisum | 2 075 | 1 | 1 679 | 1 679 | 2 | 396 | 198 | 630 | − | |
| A. glycines | 1 802 | 1 | 1 406 | 1 406 | 2 | 396 | 198 | 331 | − | |
| CSP7 | M. persicae | 8 096 | 2 | 7 628 | 3 814 | 3 | 468 | 156 | 910 | − |
| A. gossypii | 7 760 | 2 | 7 301 | 3 651 | 3 | 459 | 153 | x | x | |
| A. pisum | 7 846 | 2 | 7 378 | 3 689 | 3 | 468 | 156 | 145 | − | |
| A. glycines | 7 718 | 2 | 7 259 | 3 630 | 3 | 459 | 153 | 1910 | − | |
| CSP8 | M. persicae | 3 324 | 1 | 2 832 | 2 832 | 2 | 492 | 246 | 10 | − |
| A. gossypii | 4 586 | 1 | 4 097 | 4 097 | 2 | 489 | 245 | x | x | |
| A. pisum | 5 252 | 1 | 4 760 | 4 760 | 2 | 492 | 246 | 152 | − | |
| A. glycines | 5 061 | 1 | 4 572 | 4 572 | 2 | 489 | 245 | 192 | + | |
| CSP9 | M. persicae | 2 170 | 1 | 1 681 | 1 681 | 2 | 489 | 245 | 55 | + |
| A. gossypii | 2 474 | 1 | 1 958 | 1 958 | 2 | 516 | 258 | x | x | |
| A. pisum | 2 532 | 1 | 2 034 | 2 034 | 2 | 498 | 249 | 447 | − | |
| A. glycines | 2 061 | 1 | 1 548 | 1 548 | 2 | 513 | 257 | 391 | − | |
| CSP10 | M. persicae | 3 910 | 1 | 3 457 | 3 457 | 2 | 453 | 227 | 420 | − |
| A. gossypii | 3701 | 2 | 3 251 | 1 626 | 3 | 450 | 150 | x | x | |
| A. pisum | 4 916 | 1 | 4 463 | 4 463 | 2 | 453 | 227 | 145 | + | |
| A. glycines | 3 811 | 1 | 3 355 | 3 355 | 2 | 456 | 228 | 550 | + |
‘x’ indicates the genomic location and transcriptional orientation of A. gossypii OBPs and CSPs cannot be investigated due to the A. gossypii genome not being available.
Eight of the nine MperCSPs (MperCSP1/2/4/5/6/8/9/10) have only one intron, and MperCSP7 has two introns (Fig. 5Table 5). The lengths of MperCSP genes are much shorter than those of MperOBP genes, ranging from 1.03 to 11.28 kb with an average length of 3.83 kb. All CSPs in the CSP subgroups (CSP1/2/4/5/6/8/9) from four different aphid species (M. persicae, A. gossypii, A. pisum and A. glycines) have one intron and two exons. The members in the CSP10 subgroup of M. persicae, A. pisum and A. glycines have only one intron, but A. gossypii CSP10 has two introns (Table 2, Zhou et al., 2010; Gu et al., 2013). These results suggest the formation of the intron–exon junctions in aphid CSP genes occurred at the early stages of aphid evolution before speciation.
We also analysed the genomic clustering of M. persicae OBPs and CSPs. The results indicated that the nine M. persicae OBPs are distributed among seven scaffolds (scaffolds 4/21/47/77/105/300/422) (Table 1), with MperOBP3/7/8 collocated in scaffold 21 (Fig. 6). The M. persicae CSPs are distributed across five scaffolds (scaffolds 10/55/112/420/910), with MperCSP1/4/6 collocated in scaffold 112, MperCSP2/9 located in scaffold 55, and MperCSP5/8 located in scaffold 10 (Table 1Fig. 1). Similar clusters of OBPs and CSPs were also found in A. pisum and A. glycines to form the OBP3/7/8 cluster, CSP1/4/6 cluster, CSP2/9 cluster and CSP5/8 cluster (Fig. 6). The phenomenon of gene cluster arrangement can also be found in other insects, such as in Bombyx mori, Drosophila melanogaster and Apis mellifera (Hekmat‐Scafe et al., 2002; Xu et al., 2003; Forêt and Maleszka, 2006; Gong et al., 2007, 2009 ).
Figure 6.
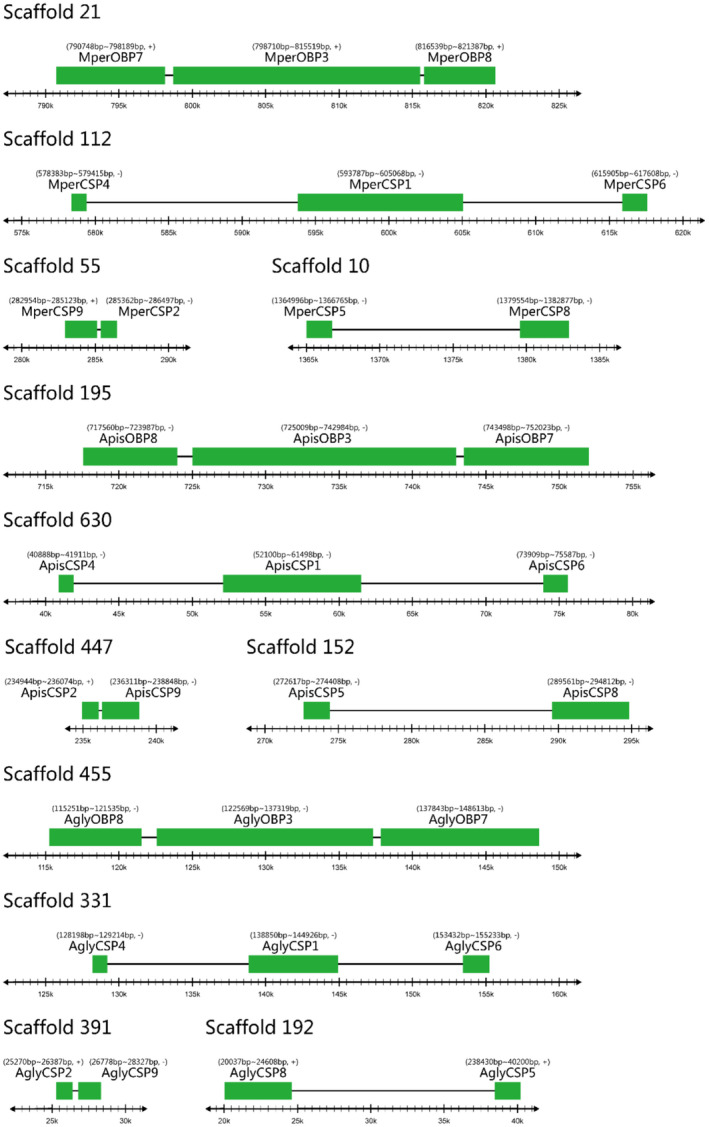
The tandem arrays of odorant binding protein (OBP) and chemosensory protein (CSP) genes in Myzus persicae, Acyrthosiphon pisum and Aphis glycines. The genomic sequences of the A. pisum, M. persicae and A. glycines were downloaded from the AphidBase at the BioInformatics Platform for Agroecosystem Arthropods (https://bipaa.genouest.org/is/). [Colour figure can be viewed at wileyonlinelibrary.com]
Motif pattern analysis
The conserved motifs are important elements of functional domains. In order to compare the amino acid motif patterns of aphid OBPs and CSPs, a total of 45 OBPs and 41 CSPs with intact ORFs from five aphid species (M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae) were combined into one set of sequences as a fasta format file and then submitted to the meme server (https://meme-suite.org/) to discover the conserved motifs within each aphid species and subgroups (Fig. 7). The meme program identified eight most conserved motifs (named motifs 1–8) from the 45 OBPs (Fig. 7A) and 41 CSPs (Fig. 7B). Based on the number, position and identity of motifs in the 45 OBPs, 13 motif patterns were identified in these 45 aphid OBP sequences. The most common eight motif patterns (with each motif pattern present in more than three OBPs) are shown as motifs 1/2/3/4/5/6/7/8 and their relative arrangements along each protein sequence are shown in Fig. 7A. Notably, all the paralogous OBPs present in a given aphid species have a unique motif pattern, while the homologous OBPs in each OBP subgroup from different aphid species often share one common motif pattern. The homologous OBP2, OBP3, OBP8, OBP9 and OBP10 subgroups, in the five aphid species M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae, display 4‐8‐5‐1‐7‐2‐6, 4‐6‐8‐1‐3‐7‐5‐2, 4‐5‐6‐1‐7‐2‐3, 4‐6‐5‐7‐1‐3‐2 and 4‐8‐5‐1‐3‐7‐2‐6 motif patterns, respectively. The OBP5 and OBP6 subgroups, in the four aphid species A. gossypii, A. pisum, A. glycines and S. avenae, have motif patterns of 4‐6‐7‐8‐1‐3‐5‐2 and 4‐6‐7‐1‐3‐5‐2, respectively. The OBP4 subgroup in the four aphid species M. persicae, A. gossypii, A. pisum and A. glycines has a motif pattern of 5‐6‐7‐1‐4‐2. Furthermore, 38 out of 45 aphid OBPs contain motifs 1, 2, 4, 5, 6 and 7. Motifs 1 and 2 are always located in the middle and at the C‐terminus in all 45 aphid OBPs sequences (Fig. 7A).
Figure 7.
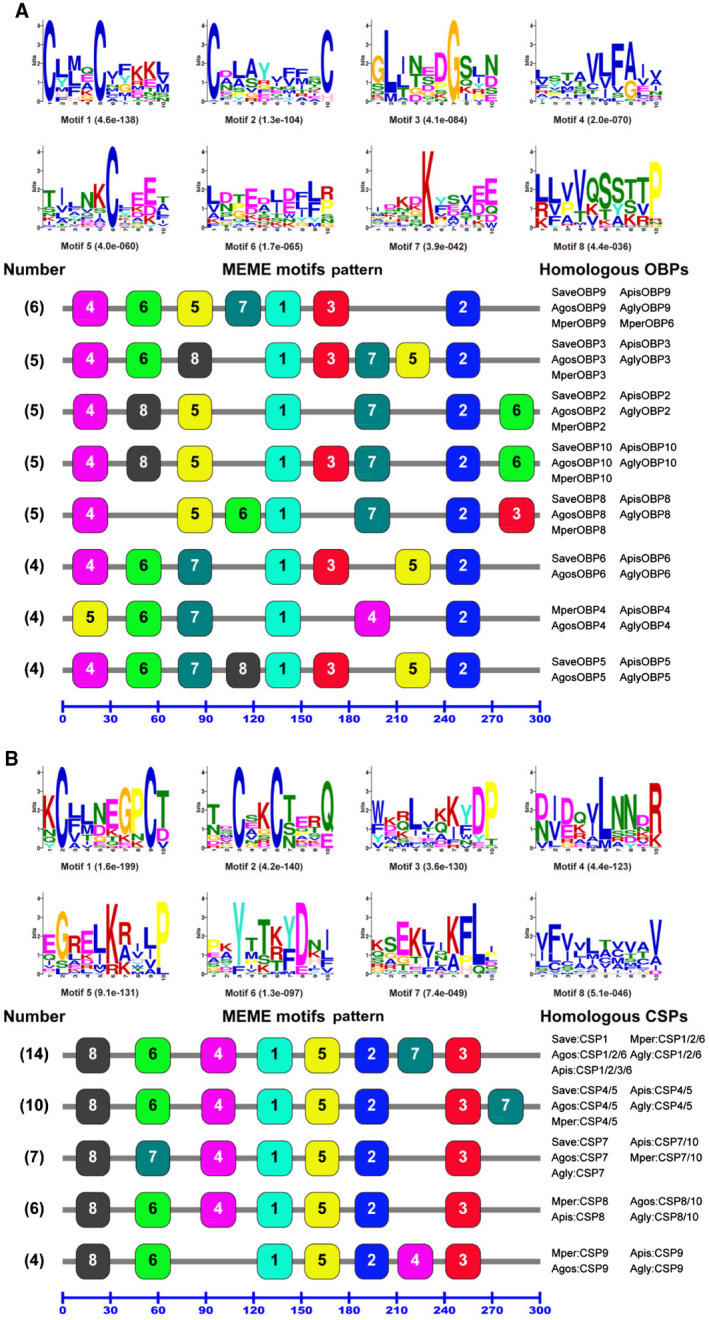
Motif analysis of odorant binding proteins (OBPs) and chemosensory proteins (CSPs) from five aphid species. The parameters used for motif discovery were minimum width = 6, maximum width = 10 and maximum number of motifs to find = 8. The upper parts in (A) and (B) list the eight motifs discovered in the five aphids’ OBPs and CSPs, respectively. All the motifs were discovered using meme (version 4.11.4; Bailey et al., 2009) online server (https://meme-suite.org/). The lower parts indicate approximate locations of each motif on the protein sequence. The numbers in the boxes correspond to the numbered motifs in the upper part of the figure, where small numbers indicate high conservation. The numbers on the bottom show the approximate locations of each motif on the protein sequence, starting from the N‐terminus. (A) The eight most common motif patterns which presented in 38 OBPs, with each motif pattern present in more than three OBPs; the remaining seven OBPs had five different motif patterns, with each of them presented in less than four OBPs. (B) All five motif patterns which presented in 41 CSPs, and each motif pattern presented in more than three CSPs. The protein names and sequences of the 45 OBPs and 41 CSPs from five different aphid species are listed in Supporting Information Table S2. [Colour figure can be viewed at wileyonlinelibrary.com]
The motif patterns of the aphid CSPs are much more conserved than the aphid OBPs, since there are only 5 motif patterns in the 41 aphid CSPs (Fig. 7B). Unlike OBP subgroups, paralogous CSPs in a given aphid species and different CSP subgroups may share a common motif pattern. For example, 14 CSPs in the CSP1, CSP2 and CSP6 subgroups (SaveCSP1, MperCSP1/2/6, AgosCSP1/2/6, AglyCSP1/2/6 and ApisCSP1/2/3/6) have the same motif pattern of 8‐6‐4‐1‐5‐2‐7‐3. Ten CSPs in the CSP4 and CSP5 subgroups (SaveCSP4/5, ApisCSP4/5, AgosCSP4/5, AglyCSP4/5 and MperCSP4/5) exhibit a motif pattern of 8‐6‐4‐1‐5‐2‐3‐7. ApisCSP10 and MperCSP10 in the CSP10 subgroup share the same motif pattern (8‐7‐4‐1‐5‐2‐3) with SaveCSP7, ApisCSP7, AgosCSP7, MperCSP7 and AglyCSP7 in the CSP7 subgroup. Similarly, two CSPs (AgosCSP10 and AglyCSP10) share one motif pattern (8‐6‐4‐1‐5‐2‐3) with four CSPs in the CSP8 subgroup (MperCSP8, AgosCSP8, ApisCSP8 and AglyCSP8). The four CSPs in the CSP9 subgroup (MperCSP9, ApisCSP9, AgosCSP9 and AglyCSP9), however, have a subgroup‐specific motif pattern of 8‐6‐1‐5‐2‐4‐3 (Fig. 7B). Meanwhile, motifs 1, 2, 3, 4, 5 and 8 are located at the same positions in all 41 aphid CSPs, except for the four CSP9 subgroup genes, in which motif 4 is located at the C‐terminus (Fig. 7B).
When the motif pattern of the 199 OBPs from 18 Hemiptera species were compared, we found a total of 51 different motif patterns (data not shown), but 51.3% of the 199 OBPs (102) are affiliated to the top six common motif patterns (Fig. 8A). These include 43 OBPs from the 5‐1‐3‐4‐2 pattern, 15 OBPs from the 1‐3‐4‐2 pattern, 14 OBPs from the 1‐3‐2 pattern, 10 OBPs from the 4‐5‐1‐3‐2 pattern, 10 OBPs from the 5‐1‐3‐8‐4‐2 pattern and another 10 OBPs from the 1‐2 pattern (Fig. 8A).
Figure 8.
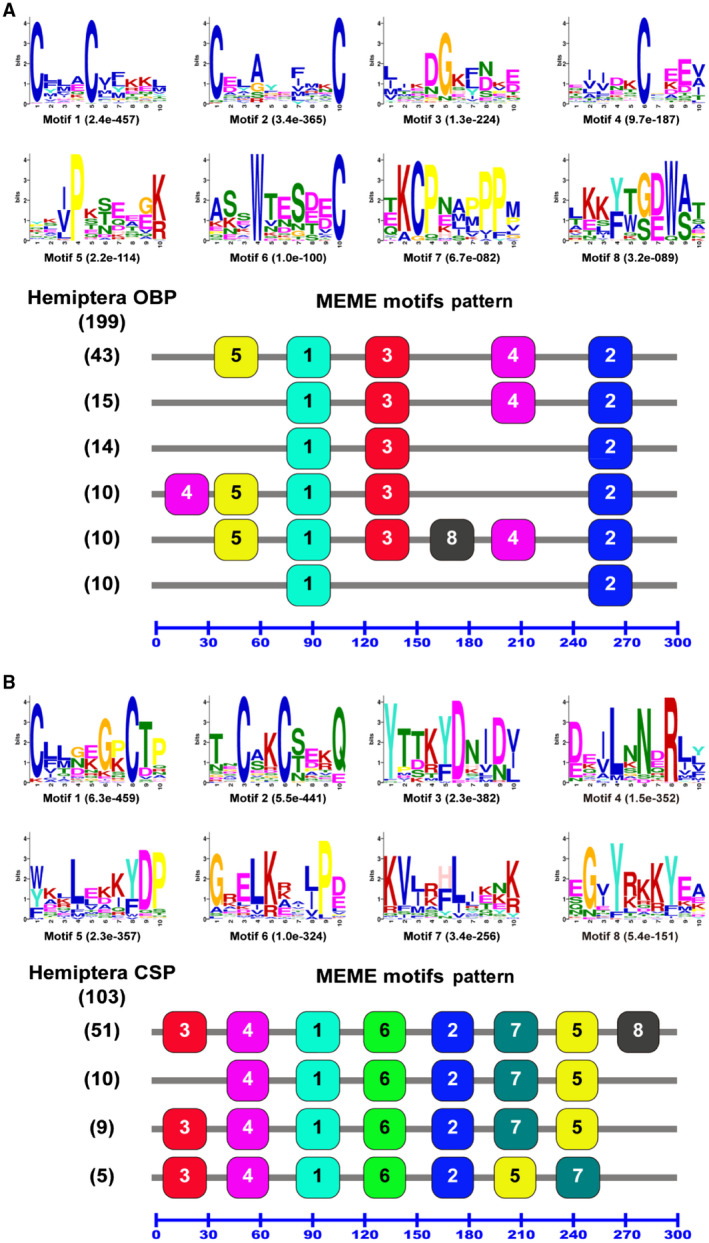
Motif analysis of Hemiptera odorant binding proteins (OBPs) and chemosensory proteins (CSPs). Parameters used for motif discovery were minimum width = 6, maximum width = 10 and maximum number of motif to find = 8. The upper parts in (A) and (B) list the eight motifs discovered in the Hemiptera OBPs and CSPs, respectively. All the motifs were discovered using meme (version 4.11.4; Bailey et al., 2009) online server (https://meme-suite.org/). The lower parts indicate approximate locations of each motif on the protein sequence. The numbers in the boxes correspond to the numbered motifs in the upper part of the figure, where small numbers indicate high conservation. The numbers on the bottom show the approximate locations of each motif on the protein sequence, starting from the N‐terminus. (A) The most common six motif patterns which presented in 102 OBPs, with each motif pattern present in more than nine OBPs; the remaining 97 OBPs had 45 different motif patterns, with each of them presented in less than 10 OBPs. (B) The four most common motif patterns which presented in 75 CSPs, with each motif pattern present in more than four CSPs; the remaining 28 CSPs had 13 different motif patterns, with each of them presented in less than five CSPs. The protein names and sequences of the 199 OBPs and 103 CSPs from 18 different Hemiptera species are listed in Supporting Information Table S3. [Colour figure can be viewed at wileyonlinelibrary.com]
Motifs 1 and 2 that contain four highly conserved cysteine residues (C2 and C3 in motif 1, C5 and C6 in motif 2) of the classic OBP subfamily in insects (Zhou et al., 2004, 2010 ) are present in all the 102 Hemiptera OBPs (Fig. 8A). Motif 3 with highly conserved glycine (G) and aspartic acid (D) is present in the top five common motif patterns (92 of the 102 OBPs). Motif 4, which contains another characteristically conserved cysteine residue (C4), was only mapped to some OBPs. However, motif 6, containing highly conserved tryptophan (W) and conserved cysteine 4 (C6a), and motif 7, containing highly conserved lysine (K), C6b and proline (P) of the plus‐C OBP subfamily, were not present in the 102 OBPs (Fig. 8A).
Motif analysis of a total of 103 Hemiptera CSPs revealed that 75 out of the 103 CSPs (72.8%) belong to the top four common motif patterns (Fig. 8B). These include 51 CSPs from the 3‐4‐1‐6‐2‐7‐5‐8 pattern, 10 CSPs from the 4‐1‐6‐2‐7‐5 pattern, 9 CSPs from the 3‐4‐1‐6‐2‐7‐5 pattern and 5 CSPs from the 3‐4‐1‐6‐2‐5‐7 pattern (Fig. 8B). Ten CSPs have lost motif 3, and 51 CSPs have motif 8. The motif patterns shown in Figs 7 and 8 further demonstrated that insect CSPs are more conversed than OBPs. It should be noticed that the motif patterns discovered by meme in Figs 7 and 8 are not comparable, because different input sequences were used in each analysis.
Expression profiles of M. persicae OBP and CSP genes in different tissues
The general and quantitative expression profiles of M. persicae OBP and CSP transcripts in different tissues (antennae, heads, legs and decapitated bodies) were examined using both semi‐quantitative reverse transcription PCR (RT‐PCR) and quantitative real‐time PCR (qRT‐PCR). Because equal amounts of cDNA (150 ng) were used in the RT‐PCR reactions, the intensity of the PCR bands of MperOBP2/3/6/9/10 and MperCSP2/4/5/9 was higher than the remaining MperOBPs and MperCSPs, which can reflect their relatively high expression levels in M. persicae. Actually, the reads per kilobase per million mapped reads (RPKM) analysis also showed their relative high abundances in the M. persicae transcriptome, with the RPKM values of MperOBP2/3 and MperCSP2/4/5/9 more than 100 (Table 1).
The absolute values of the slope of all lines from template dilution plots (log cDNA dilution vs. ΔC T) were <0.1 (Supporting Information Figs S2 and S3), and the real PCR amplification efficiencies of target and reference genes were more than 1.90 (calculated using the LinRegPCR program and listed in Supporting Information Table S5). Therefore, the efficiencies of the target and reference genes were similar in our analysis, and the ΔΔC T calculation method can be used for the relative quantification. Both RT‐PCR and qRT‐PCR results showed that three OBP genes (MperOBP6/7/10) were antennae specific, and their expression levels were approximately 101, 8.7 and 222 times higher in the antennae than in the body (p < 0.05), respectively (Fig. 9). MperOBP2/4/5/8/9 showed the highest expression in the antennae, and their expression levels were, respectively, 3.5, 3.8, 3.6, 6.5 and 63.9 times greater in the antennae than in the body (p < 0.05). MperOBP2/4/5/8/9 were also detectable in other tissues, such as head, leg and body (Fig. 9). MperOBP3 was the only OBP whose expression level was lower in the antennae than in the head and body.
Figure 9.
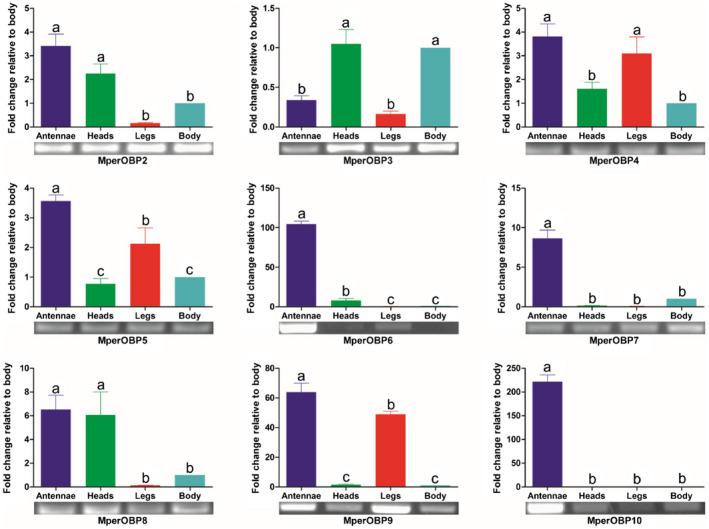
Myzus persicae odorant binding protein (OBP) transcript levels in different tissues assessed by reverse transcription PCR and quantitative real‐time PCR. The error bars present the standard error, and the different letters (a, b, c) indicate significant differences (p < 0.05). This figure was presented using GAPDH as reference gene to normalize the target gene expression and correct sample‐to‐sample variation; similar results were also obtained with β‐actin as reference gene. The standard error is represented by the error bar. [Colour figure can be viewed at wileyonlinelibrary.com]
None of the nine MperCSPs were found to be antennae specific, and five of them (MperCSP1/2/4/5/6) expressed at a higher expression level in the legs than the other three tissues (antennae, head and body) (Fig. 10), with the expression levels 20–50 times higher in the legs than in the decapitated bodies (p < 0.05). MperCSP10 expressed 41 and 39 times higher in the antenna and legs, respectively, than in the body (p < 0.05). MperCSP8 and MperCSP9 mainly expressed in the body. MperCSP7 exhibited a 2.2 times higher expression in the antennae than in the body (p < 0.05) (Fig. 10).
Figure 10.
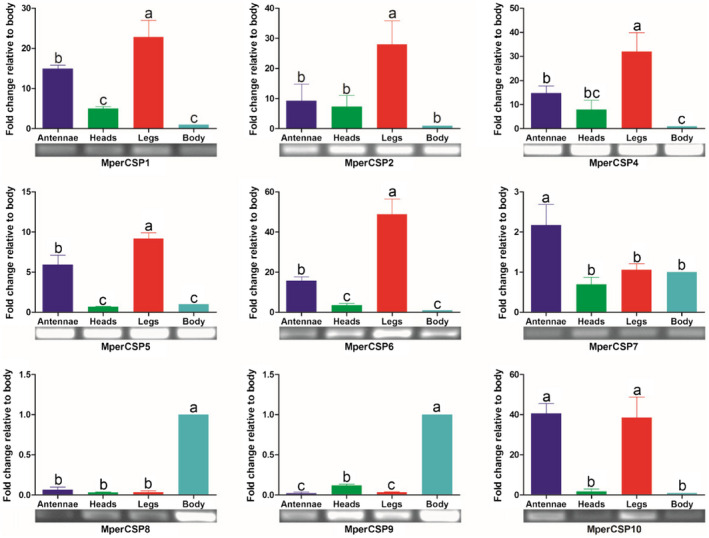
Myzus persicae chemosensory protein (CSP) transcript levels in different tissues assessed by reverse transcription PCR and quantitative real‐time PCR. The error bars present the standard error, and the different letters (a, b, c) indicate significant differences (p < 0.05). This figure was presented using GAPDH as reference gene to normalize the target gene expression and correct sample‐to‐sample variation; similar results were also obtained with β‐actin as reference gene. The standard error is represented by the error bar. [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
With the increase in insect genome projects and transcriptome sequencing projects, particularly in recent years, very large numbers of both OBPs and CSPs have been identified in different insect species. These studies have provided an excellent chance to comparatively study insect OBPs and CSPs. For the first time, we had identified nine OBPs and nine CSPs, both from the M. persicae transcriptomic and genomic data, including four OBPs and five CSPs reported from the expressed sequence tags in M. persicae (Xu et al., 2009). The number of M. persicae OBPs and CSPs identified here is less than the number of OBPs and CSPs identified in A. pisum (15 OBPs, 13 CSPs) (Zhou et al., 2010), the same as in A. gossypii (nine OBPs, nine CSPs) (Gu et al., 2013), and similar to those of other sucking insects such as the plant hoppers N. lugens (10 OBPs, 11 CSPs) (Zhou et al., 2014), S. furcifera (12 OBPs, nine CSPs) (He and He, 2014; Zhou et al., 2015) and the plant bug A. suturalis (16 OBPs, eight CSPs) (Cui et al., 2017). The high conservation in the sequence identities and genomic structures of the OBP and CSP orthologues among aphid species indicates that aphid OBPs and CSPs are conserved in a single copy across all aphids (with occasional losses), indicating that each OBP and CSP class evolved from a single gene in the common ancestor of aphids without subsequent duplication.
The absence of OBP1 and CSP3 homologous genes in M. persicae, as demonstrated in this study by transcriptome sequencing, searching the genome database and molecular cloning using gene‐specific primers of A. pisum OBP1 and CSP3 (data not shown here), is consistent with the same results reported in the cotton aphid A. gossypii (Gu et al., 2013) and recently in the A. glycines genome (Wenger et al., 2017). However, OBP1 homologous genes have been found in six other aphid species (A. pisum, S. avenae, M. viciae, M. dirhodum, T. salignu and P. salicis). The OBP1 orthologous genes are also not found in the plant hoppers and plant bugs in this study. It is possible that OBP1 and CSP3 genes might have been lost after aphid speciation of M. persicae, A. gossypii and A. glycines. Furthermore, there is a high amino acid identity between OBP1 and OBP8 subgroups in aphids, consistent with previous reports (Gu et al., 2013; He and He, 2014; Yuan et al., 2015; Xue et al., 2016).
The phylogenetic analyses clearly divided most of the aphid OBPs and CSPs into species‐specific homology subgroups. However, some aphid OBP and CSP subgroups are clustered with non‐aphid OBP and CSP genes in conserved clades. For example, aphid OBP4 and OBP5 subgroups are clustered with OBPs of plant hopper and plant bug with a bootstrap support of 100 and 84, respectively (Fig. 3), and aphid CSP7 and CSP9 subgroups are clustered with plant hopper and plant bug CSPs with a bootstrap support of 73 (Fig. 4), suggesting these genes may have a common function in these sucking insects.
The motif analysis contributes to the understanding of the functional domains of insect OBPs and CSPs. The members of each OBP subgroup have a common motif pattern, particularly for members of the aphid OBP family and members of the CSP9 subgroup (Fig. 7). Five motif patterns are shared by different CSP subgroups, further supporting previous reports of greater conservation between insect CSPs than between OBPs (Fig. 7). Interestingly, A. pisum ApisCSP3 shares the same motif pattern arrangement with members of the aphid CSP1 and CSP6 subgroups as well as ApisCSP2, despite the sequences between these three subgroups (CSP1, CSP2 and CSP6) being very different (Fig. 4). This result is not consistent with a previous report on the motif analysis of Hemiptera OBPs and CSPs (Wang et al., 2017). This could be because different sets of OBP and CSP protein sequences were used in these two studies. The comparative sequence analyses with other Hemiptera species confirmed the results from the previous studies that CSPs are more conserved than OBPs and some highly conserved cysteine residues (C2, C3, C5 and C6) in insect OBPs (Fig. 8), suggesting their involvement in protein structure and possible functions.
The intron numbers and lengths of M. persicae OBPs are nearly equal to other orthologous genes in A. pisum and A. gossypii (Zhou et al., 2010; Gu et al., 2013), but much greater than those in aphid CSPs; this appears to be a unique feature of aphid OBP genes in general. Four gene clusters (OBP3/7/8, CSP1/4/6, CSP2/9 and CSP5/8) are found in three aphid species (Fig. 6). The conserved genomic structure and same transcriptional orientation demonstrate that these aphid OBP and CSP genes were created by gene duplication, possibly through chromosome recombination, supporting the birth‐and‐death evolutionary model (Nei and Rooney, 2005; Zhou et al., 2010).
The major alarm pheromone component Eβf emitted from the cornicles of aphids (Edwards et al., 1973) is recognized by large placoid sensillum neurons, which express OR5 on the sixth antennal segment in A. pisum (Zhang et al., 2017); OBP3 and OBP7 are both expressed in the large placoid sensilla in M. persicae (Sun et al., 2013) and in the type II trichoid sensilla in A. pisum in the antennae (Biasio et al., 2015). OBP3 and/or OBP7 proteins in several aphid species (A. pisum, S. avenae, M. viciae, N. ribisnigri and R. padi) have high binding affinities with Eβf (Qiao et al., 2009; Vandermoten et al., 2011; Zhong et al., 2012; Fan et al., 2017), but the cellular location of all M. persicae OBPs in the antennal sensilla and their binding abilities with Eβf are still unknown. Our data reveal that MperOBP7 is antennae specific and expressed at a level that is 8.7 times higher in the antennae than in the body (p < 0.05) (Fig. 9), consistent with previous reports of the high binding affinity of this OBP to Eβf (Qiao et al., 2009; Vandermoten et al., 2011; Zhong et al., 2012; Fan et al., 2017) and its localization in antennal sensilla (Sun et al., 2013; Biasio et al., 2015). However, OBP3, which was also reported as the Eβf binding protein in several aphid species (A. pisum, S. avenae, M. viciae, N. ribisnigri and R. padi), is neither antennae specific nor antennae enriched, but is highly expressed in the heads and body in M. persicae (Fig. 9). This expression pattern is similar to that of A. pisum OBP3 (Biasio et al., 2015) and of S. avenae OBP3 (Xue et al., 2016), suggesting that aphid OBP3 proteins may have other functional roles in addition to alarm pheromone sensation. The fact that the A. pisum OBP3 was found in the type II trichoid sensilla in the antennae and in the terminal region of the body (Biasio et al., 2015) suggests that it may have dual functions of recognizing the Eβf from the environment and transporting the Eβf from the cornicles to the external environment.
Insect CSPs usually exhibit much broader expression profiles both in chemosensory organs and nonchemosensory organs than OBPs do, and play multiple roles in chemoreception, growth and development. For example, the American cockroach (Periplaneta americana) CSP gene P10 has a putative role in leg regeneration (Nomura et al., 1992), and the migratory locust (Locusta migratoria) antennae‐expressed CSP gene LmigCSP3 can regulate the rapid switch between attraction and repulsion behaviours in this species (Guo et al., 2011). In the present study, M. persicae CSPs also showed much broader expression profiles in nonsensory organs than M. persicae OBPs do (Fig. 10). None of the nine MperCSPs were found to be antennae‐specific. Five of them (MperCSP1/2/4/5/6) showed a higher expression level in the legs than the other three tissues (antennae, heads and body). MperCSP10 was mainly expressed in the antennae and legs. The broad and diverse expression patterns of M. persicae CSPs are consistent with their possible multifunctions in olfactory perception, development and other processes.
The sex pheromones from the Aphididae family are usually released from the tibiae of the hind legs of the mature sexual female aphids (Marsh, 1972; Pickett and Glinwood, 2007), and usually comprise of a mixture of the ubiquitous iridoids (4aS,7S,7aR)‐nepetalactone and (1R,4aS,7S,7aR)‐nepetalactol (Pickett et al., 1992; Dewhirst et al., 2010); however, in order to convey species integrity, the ratio of (4aS,7S,7aR)‐nepetalactone to (1R,4aS,7S,7aR)‐nepetalactol is species dependent, and this ratio is 1 : 1.5 for M. persicae (Dawson et al., 1990). The aphid OBPs and CSPs that can specially recognize the two ubiquitous sex pheromones are still unknown. It is reasonable to assume that the M. persicae OBPs and CSPs which are highly expressed in the antennae and legs may participate in the intersexual communication process. Till now, functional studies of the aphid OBPs and CSPs have narrowly focused on several OBPs (eg OBP3/7); the putative functional roles of other aphid OBPs and all the CSPs in the M. persicae and other aphid species still need to be elucidated. Future work towards the functions of these M. persicae OBPs and CSPs will enhance our understanding of the ecological context of aphid–aphid and aphid–plant interactions and facilitate design of novel sustainable strategies for aphid control.
Experimental procedures
Aphids, sample collection and RNA extraction
M. persicae aphids were collected from Chinese cabbage (Brassica rapa L. ssp. pekinensis) leaves at Langfang Experimental Station of the Chinese Academy of Agricultural Sciences, Hebei Province, China. Apterous adults were reared continuously as a parthenogenetic colony on tobacco (Nicotiana tabacum L.) seedlings in chambers, at 18–24 °C, 65–75% relative humidity, and a 16 h : 8 h light : dark photoperiod.
About 2000 apterous aphids were dissected with fine scissors to collect their antennae, heads, legs and decapitated bodies into separate tubes and stored in liquid nitrogen. Total RNAs from these samples were extracted using Trizol reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. The quantity and integrity of RNA samples were verified using 1.1% agarose gel electrophoresis and a NanoDrop ND‐1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
cDNA library construction and Illumina sequencing
About 50 mg of apterous aphids was collected and kept in a 1.5 ml centrifuge tube in liquid nitrogen for total RNA extraction using the Trizol reagent. The cDNA libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocol. Briefly, messenger RNAs (mRNAs) were isolated from 10 µg total RNAs from apterous aphids using oligo (dT) magnetic beads and fragmented into short nucleotides using the fragmentation buffer provided with the kit at 94 °C for 5 min. The cleaved mRNA was transcribed into the first‐strand cDNA using a random hexamer primer and M‐MuLV reverse transcriptase (RNaseH−). The second‐strand cDNA was subsequently synthesized using DNA polymerase I, deoxyribonucleotide triphosphate and ribonuclase H. After the end repair, dA‐tailing and adaptor ligation, the products were amplified by PCR and purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) to create the final sequencing library. The cDNA library was pair‐end sequenced using a PE150 strategy on an Illumina Hiseq2500 platform (Illumina, San Diego, CA, USA).
Bioinformatics analysis
The raw reads were cleaned by removing adaptor sequences and low‐quality sequences using Q20 with 99% accuracy using trimmomatic software (version 0.32) (Bolger et al., 2014). The clean reads were de novo assembled into unigenes with the short read assembly program trinity (version 20121005) with default parameters (Grabherr et al., 2011). After assembly, homology searches of all unigenes were performed using the blastx and blastn programs against the GenBank nonredundant protein (nr) and nucleotide sequence (nt) databases at the National Center for Biotechnology Information (NCBI). Matches with an E‐value less than 1.0 × 10−5 were considered to be significant and kept (Anderson and Brass, 1998). Gene names were assigned to each unigene based on the best blastx hit with the highest score value.
The BWA‐MEM alignment algorithm (Li, 2013) and htseq program (version 0.6.1) (Anders et al., 2015) were used for aligning the RNA reads and counting the read numbers mapped to each gene, respectively. The abundances of the unigenes in the transcriptome were calculated by the RPKM method (Mortazavi et al., 2008), using the formula
where RPKM(A) is the expression abundance of gene A, C is the number of reads that are uniquely mapped to gene A, N is the total number of reads that are uniquely mapped to all genes and L is the number of bases on gene A. The RPKM method is able to eliminate the influence of different gene lengths and sequencing discrepancies on the calculation of transcript abundance.
Annotation of putative OBP and CSP genes in M. persicae transcriptome and genome
The genomic sequences of M. persicae clone G006 assembly v2 were downloaded from AphidBase at the BioInformatics Platform for Agroecosystem Arthropods (https://bipaa.genouest.org/is/aphidbase/myzus_persicae/). Two methods were used to identify unigenes encoding putative OBPs and CSPs in the M. persicae transcriptome and genome. First, we ran the OBP motifsearch program on C1‐X15–39‐C2‐X3‐C3‐X21–44‐C4‐X7–12‐C5‐X8‐C6 (Zhou et al., 2008) and the CSP motifsearch program on C1‐X6‐8‐C2‐X16‐21‐C3‐X2‐C4 (Zhou et al., 2006) to identify the respective putative OBP and CSP genes from the M. persicae transcriptomic and genomic databases obtained earlier. Second, a tblastn search with known OBP and CSP sequences from other aphid species as the ‘query’ was performed to identify OBPs and CSPs from both the transcriptomic and genomic datasets. All candidate OBP and CSP genes were manually checked using the blastx program available at the NCBI.
Verification of OBP and CSP sequences by cloning and sequencing
ORFs of each identified OBP and CSP sequence were found by orf finder graphical analysis at NCBI (https://www.ncbi.nlm.nih.gov/gorf/gorf.html). Then, gene‐specific primers (Supporting Information Table S1) were designed using primer 5.0 software to clone the ORF of each M. persicae OBP and CSP gene. The template cDNA was synthesized using the Fast Quant RT Kit (TianGen, Beijing, China). PCR reactions were carried out with 200 ng antennal cDNAs with 0.5 units of Ex Taq DNA polymerase (TaKaRa, Dalian, China). The PCR amplification conditions were set as 95 °C for 2 min, followed by 36 cycles of 94 °C for 45 s, 56 °C for 1 min, 72 °C for 1 min and a final extension at 72 °C for 10 min. The PCR products were gel‐purified and subcloned into the pGEM‐T Easy vector (Promega, Madison, WI, USA). The inserts were confirmed by sequencing on the ABI3730XL automated sequencer (Applied Biosystems, Carlsbad, CA, USA) with standard M13 primers.
Genomic structure analysis of M. persicae OBPs and CSPs
The genomic DNA sequences of each M. persicae OBP and CSP gene were extracted using the blastn program, with the mRNA sequences of M. persicae OBPs and CSPs as ‘query’. The mRNA‐to‐genomic DNA alignment of each OBP and CSP gene was analysed using the splign online program (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi).
Motif analysis
A total of 45 OBPs and 41 CSPs from 5 different aphid species (M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae) (Supporting Information Table S2) were used for motif discovery and pattern analysis. A total of 199 OBPs and 103 CSPs from 18 different Hemiptera species (Supporting Information Table S3) were used for comparing the motif patterns between Hemiptera OBPs and CSPs. All OBP and CSP sequences used in this study have intact ORFs and similar lengths. The motif‐based sequence analysis tool meme (Bailey et al., 2009) (version 4.9.1; https://meme-suite.org/), which has been widely used for the discovery of DNA and protein motifs, was used to discover and analyse the motifs in the OBP and CSP sequences. The parameters used for motif discovery were as follows: minimum width = 6, maximum width = 10 and the maximum number of motifs to find = 8. This program analyses any multiple input sequences and predicts motifs and their arrangements along an individual sequence. The outputs are two graphic presentations: one contains the similarity plots for each identified motif and another contains a linear arrangement of each predicted motif on the sequences.
Sequence and phylogenetic analysis
The putative signal peptides of derived M. persicae OBP and CSP proteins were predicted using the SignalIP 4.1 Server (https://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011). The percentage identity matrix of each pair of OBPs was calculated using Vector NTI 11.5 (Invitrogen Corporation, Carlsbad, USA). Owing to the wide divergence of the 237 OBPs (12.63% amino acid identity) and 110 CSPs (16.48% amino acid identity) used in this study, sequence alignments were conducted with the prank alignment program (Löytynoja and Goldman, 2010), phylogenetic trees were established based on maximum likelihood by raxml version 8 (Stamatakis, 2014) with LG substitution matrix selected by the prottest 3 program (Darriba et al., 2011). Bootstrapping was performed to compute the confidence of the branches using 1000 replicates.
Expression profiles of M. persicae OBP and CSP genes in different tissues
The cDNAs from aphid antennae, heads, legs and the body parts were synthesized using the PrimeScript RT Reagent with gDNA Eraser (TaKaRa, Dalian, China). An equal amount of cDNA (150 ng) was used as the RT‐PCR and qRT‐PCR templates. Gene‐specific primer pairs for RT‐PCR analyses were designed with Primer 3 (https://frodo.wi.mit.edu/) or Primer Premier 5 (see Supporting Information Table S4). β‐Actin (GenBank Acc. XM_022309797) of M. persicae was used as the control gene to test the integrity of the cDNA. The PCR cycling profile was: 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 56 °C for 45 s, 72 °C for 1 min and a final extension for 10 min at 72 °C. The PCR products were separated on 1.2% agarose gels and stained with ethidium bromide. Each reaction was done at least six times with three biological replicates. In addition, the PCR products were randomly selected and verified by DNA sequencing.
qRT‐PCR was carried out on an ABI 7500 Real‐Time PCR system (Applied Biosystems, Carlsbad, CA, USA). The β‐actin and GAPDH (GenBank Acc. XM_022315441) genes are stably expressed in different tissues and developmental stages (Supporting Information Fig. S1), so were used as reference genes for normalizing the target gene expression and correcting the sample‐to‐sample variations. The primers used for qRT‐PCR were designed with beacon designer 7.90 (Supporting Information Table S5). Each qRT‐PCR reaction was conducted in a 25‐µl reaction mixture containing 12.5 µl of SuperReal PreMix Plus (TianGen, Beijing, China), 0.75 µl of each primer (10 µm), 1 µl of sample cDNA (150 ng/µl), 0.5 µl of Rox Reference Dye and 9.5 µl of sterilized water. The qRT‐PCR cycling parameters were 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 32 s. Then, the PCR products were heated to 95 °C for 15 s, cooled to 60 °C for 1 min, heated again to 95 °C for 30 s and cooled to 60 °C for 15 s to measure the melt curves. Negative controls without any template were included in each experiment. To ensure reproducibility, each qRT‐PCR reaction for each sample was performed in three technical replicates and three biological replicates. The comparative method (Livak and Schmittgen, 2001) was used to calculate the relative expressions between tissues. The comparative analyses of each target gene among various tissues were determined using a one‐way nested analysis of variance, followed by Tukey’s honest significance difference test using the spss statistics 18.0 software (SPSS Inc., Chicago, IL, USA). When applicable, the values were presented as the mean plus/minus the standard error.
An assumption in the ΔΔCT calculation for the comparative method for quantification is that the amplification efficiencies of the target and reference genes are approximately equal (Livak and Schmittgen, 2001). To confirm this, a pilot experiment was conducted to examine the variation of ΔC T (C T, Target − C T, β–actin/GAPDH) with template dilution. Briefly, five serial fivefold dilutions of cDNA from each sample were amplified. For each dilution, amplifications were performed in triplicate using primers for the target gene and β‐actin or GAPDH. Mean C T was calculated for both target gene and β‐actin or GAPDH, ΔC T was calculated, and log cDNA dilution vs. ΔC T was plotted. The real PCR amplification efficiencies of target and reference genes were calculated using the linregpcr program (version 11.0) (Ramakers et al., 2003).
Supporting information
FIGURE S1. The sTABLE expression of M. persicae β‐Actin and GAPDH in different development stages and tissues measured by RT‐PCR.
FIGURE S2. The standard curves of M. persicae OBPs and CSPs with GAPDH as reference gene.
FIGURE S3. The standard curves of M. persicae OBPs and CSPs with β‐actin as reference gene.
TABLE S1. Gene specific primers used for cloning of M. persicae OBP and CSP genes.
TABLE S2. The protein names and sequences of the 45 OBPs and 41 CSPs from M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae used in Fig. 7.
TABLE S3. The protein names and sequences of the 199 OBPs and 103 SPs from Hemiptera insects used in Fig. 8.
TABLE S4. Primers used in RT‐PCR for determination expression levels of M. persicae OBP and CSP genes.
TABLE S5. Primers used in real‐time PCR for determination expression level of M. persicae OBP and CSP genes. The real PCR amplification efficiencies of target and reference genes were calculated using LinRegPCR program (version 11.0) (Ramakers et al., 2003).
TABLE S6. A percent identity matrix of M. persicae OBPs.
TABLE S7. A percent identity matrix of M. persicae CSPs.
TABLE S8. The protein names and sequences of the Hemiptera 237 OBPs used in Fig. 3.
TABLE S9. The protein names and sequences of the Hemiptera 110 CSPs used in Fig. 4.
Acknowledgements
This work was supported by the Beijing Nova Programme (grant no. Z1511000003150118), the Beijing Talents Fund (grant no. 2015000021223ZK29), and the National Natural Science Foundation of China (grant no. 31772164 and 31401737) to Shao‐Hua Gu.
References
- Anders S., Pyl P.T. and Huber W. (2015) HTSeq – a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, I. and Brass, A. (1998) Searching DNA databases for similarities to DNA sequences: when is a match significant? Bioinformatics, 14, 349–356. [DOI] [PubMed] [Google Scholar]
- Angeli, S., Ceron, F., Scaloni, A., Monti, M., Monteforti, G., Minnocci, A. et al (1999) Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria . European Journal of Biochemistry, 262, 745–754. [DOI] [PubMed] [Google Scholar]
- Arakaki, N. (1989) Alarm pheromone eliciting attack and escape responses in the sugar cane woolly aphid, Ceratovacuna lanigera (Homoptera, Pemphigidae). Journal of Ethology, 7, 83–90. [Google Scholar]
- Bailey, T.L., Boden, M., Buske, F.A., Frith, M., Grant, C.E., Clementi, L. et al (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasio, F.D., Riviello, L., Bruno, D., Grimaldi, A., Congiu, T., Sun, Y.F. et al (2015) Expression pattern analysis of odorant‐binding proteins in the pea aphid Acyrthosiphon pisum . Insect Science, 22, 220–234. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M., Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, W.S., Nault, L.R., Webb, R.E. and Dutky, S.R. (1972) Aphid alarm pheromone: isolation, identification, synthesis. Science, 177, 1121–1122. [DOI] [PubMed] [Google Scholar]
- Bwye, A.M., Proudlove, W., Berlandier, F.A. and Jones, R.A.C. (1997) Effects of applying insecticides to control aphid vectors and cucumber mosaic virus in narrow‐leafed lupins (Lupinus angustifolius). Australian Journal of Experimental Agriculture, 37, 93–102. [Google Scholar]
- Calvello, M., Guerra, N., Brandazza, A., D’Ambrosio, C., Scaloni, A., Dani, F.R. et al (2003) Soluble proteins of chemical communication in the social wasp Polistes dominulus . Cellular and Molecular Life Sciences (CMLS), 60, 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H.H., Gu, S.H., Zhu, X.Q., Wei, Y., Liu, H.W., Khalid, H.D. et al (2017) Odorant‐binding and chemosensory proteins identified in the antennal transcriptome of Adelphocoris suturalis Jakovlev. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 24, 139–145. [DOI] [PubMed] [Google Scholar]
- Darriba, D., Taboada, G.L., Doallo, R. and Posada, D. (2011) ProtTest 3: fast selection of best‐fit models of protein evolution. Bioinformatics, 27, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, G.W., Griffiths, D.C., Merritt, L.A., Mudd, A., Pickett, J.A., Wadhams, L.J. et al (1990) Aphid semiochemicals – a review, and recent advances on the sex pheromone. Journal of Chemical Ecology, 16, 3019–3030. [DOI] [PubMed] [Google Scholar]
- Dewhirst, S.Y., Pickett, J.A. and Hardie, J. (2010) Aphid pheromones. Vitamins and Hormones, 83, 551–574. [DOI] [PubMed] [Google Scholar]
- Edwards, L.J., Siddall, J.B., Dunham, L.L., Uden, P. and Kislow, C.J. (1973) Trans‐β‐Farnesene, alarm pheromone of the green peach aphid, Myzus persicae (Sulzer). Nature, 241, 126–127.4121143 [Google Scholar]
- Eskandari, F., Sylvester, E.S. and Richardson J. (1979) Evidence for lack of propagation of potato leaf roll virus in its aphid vector, Myzus persicae . Phytopathology, 69, 45–47. [Google Scholar]
- Fan, J., Xue, W.X., Duan, H.X., Jiang, X., Zhang, Y., Yu, W.J. et al (2017) Identification of an intraspecific alarm pheromone and two conserved odorant‐binding proteins associated with (E)‐β‐farnesene perception in aphid Rhopalosiphum padi . Journal of Insect Physiology, 101, 151–160. [DOI] [PubMed] [Google Scholar]
- Forêt, S. and Maleszka, R. (2006) Function and evolution of a gene family encoding odorant binding‐like proteins in a social insect, the honey bee (Apis mellifera). Genome Research, 16, 1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, D.P., Zhang, H.J., Zhao, P., Lin, Y., Xia, Q.Y. and Xiang, Z.H. (2007) Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori . Insect Biochemistry and Molecular Biology, 37, 266–277. [DOI] [PubMed] [Google Scholar]
- Gong, D.P., Zhang, H.J., Zhao, P., Xia, Q.Y. and Xiang, Z.H. (2009) The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics, 10, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M.G., Haas, B.J., Yassour, M., Levin, J.Z., Thompson, D.A., Amit, I. et al (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology, 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, S.H., Wu, K.M., Guo, Y.Y., Field, L.M., Pickett, J.A., Zhang, Y.J. et al (2013) Identification and expression profiling of odorant binding proteins and chemosensory proteins between two wingless morphs and a winged morph of the cotton aphid Aphis gossypii Glover. PLoS ONE, 8, e73524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., Wang, X.H., Ma, Z.Y., Xue, L., Han, J.Y., Yu, D. et al (2011) CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genetics, 7, e1001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M. and He, P. (2014) Molecular characterization, expression profiling, and binding properties of odorant binding protein genes in the whitebacked planthopper, Sogatella furcifera . Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 174, 1–8. [DOI] [PubMed] [Google Scholar]
- Hekmat‐Scafe, D.S., Scafe, C.R., McKinney, A.J. and Tanouye, M.A. (2002) Genome‐wide analysis of the odorant‐binding protein gene family in Drosophila melanogaster . Genome Research, 12, 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA‐MEM.arXiv 1303.3997v2.
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2− ΔΔ Ct method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Löytynoja, A. and Goldman, N. (2010) webPRANK: a phylogeny‐aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics, 11, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, D. (1972) Sex pheromone in the aphid Megoura viciae . Nature New Biology, 238, 31–32. [DOI] [PubMed] [Google Scholar]
- Mathers, T.C., Chen, Y.Z., Kaithakottil, G., Legeai, F., Mugford, S.T., Baa‐Puyoulet, P. et al (2017) Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biology, 18, 27. Available at: doi: 10.1101/063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. (2002) A genomic view of alternative splicing. Nature Genetics, 30, 13–19. [DOI] [PubMed] [Google Scholar]
- Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nature Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Nault, L.R., Edwards, L.J. and Styer, W.E. (1973) Aphid alarm pheromones: secretion and reception. Environmental Entomology, 2, 101–105. [Google Scholar]
- Nei, M. and Rooney, A.P. (2005) Concerted and birth‐and‐death evolution of multigene families. Annual Review of Genetics, 39, 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, S.J., Nickerson, M.L., Dean, M., Song, Y., Hoyt, P.R., Rhee, H. et al. (2015) The genome of Diuraphis noxia, a global aphid pest of small grains. BMC Genomics, 16, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, A., Kawasaki, K., Kubo, T. and Natori, S. (1992) Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). The International Journal of Developmental Biology, 36, 391–398. [PubMed] [Google Scholar]
- Northey, T., Venthur, H., De Biasio, F., Chauviac, F.‐X., Cole, A., Ribeiro, K.A.L. et al (2016) Crystal structures and binding dynamics of odorant‐binding protein 3 from two aphid species Megoura viciae and Nasonovia ribisnigri . Scientific Reports, 6, 24739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, P., Zhou, J.J., Ban, L.P. and Calvello, M. (2006) Soluble proteins in insect chemical communication. Cellular and Molecular Life Sciences, 63, 1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, T.N., Brunak, S., Heijne, G.V. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Pickett, J.A. and Glinwood, R.T. (2007) Chemical ecology In: Van Emden, H.F. and Harrington, R. (Eds.) Aphids as Crop Pests. Wallingford, UK: CAB International Press, pp. 235–260. [Google Scholar]
- Pickett, J.A. and Griffiths, D.C. (1980) Composition of aphid alarm pheromones. Journal of Chemical Ecology, 6, 349–360. [Google Scholar]
- Pickett, J.A., Wadhams, L.J., Woodcock, C.M. and Hardie, J. (1992) The chemical ecology of aphids. Annual Review of Entomology, 37, 67–90. [Google Scholar]
- Qiao, H., Tuccori, E., He, X., Gazzano, A., Field, L., Zhou, J.‐J. et al (2009) Discrimination of alarm pheromone (E)‐β‐farnesene by aphid odorant‐binding proteins. Insect Biochemistry and Molecular Biology, 39, 414–419. [DOI] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H.L. and Moorman, A.F.M. (2003) Assumption‐free analysis of quantitative real‐time polymerase chain reaction (PCR) data. Neuroscience Letters, 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Read, D.P., Feeny, P.P. and Root, R.B. (1970) Habitat selection by the aphid parasite Diaeretiella rapae (Hymenoptera: Braconidae) and hyperparasite Charips brassicae (Hymenoptera: Cynipidae). The Canadian Entomologist, 102, 1567–1578. [Google Scholar]
- Scaloni, A., Monti, M., Angeli, S. and Pelosi, P. (1999) Structural analysis and disulfide‐bridge pairing of two odorant‐binding proteins from Bombyx mori . Biochemical and Biophysical Research Communications, 266, 386–391. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.P., Zhao, L.J., Sun, L., Zhang, S.G. and Ban, L.P. (2013) Immunolocalization of odorant‐binding proteins on antennal chemosensilla of the peach aphid Myzus persicae (Sulzer). Chemical Senses, 38, 129–136. [DOI] [PubMed] [Google Scholar]
- The International Aphid Genomics Consortium . (2010) Genome sequence of the pea aphid Acyrthosiphon pisum . PLOS Biology, 8, e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso, A.J., Vargas, R.R., Tapia, D.H., Olivares‐Donoso, R. and Niemeyer, H.M. (2005) Host selection by the generalist aphid Myzus persicae (Hemiptera: Aphididae) and its subspecies specialized on tobacco, after being reared on the same host. Bulletin of Entomological Research, 95, 23–28. [DOI] [PubMed] [Google Scholar]
- Ueda, N. and Takada, H. (1977) Differential relative abundance of green–yellow and red forms of Myzus persicae (Sulzer) (Homoptera: Aphididae) according to host plant and season. Applied Entomology and Zoology, 12, 124–133. [Google Scholar]
- Van Emden, H.F., Eastop, V.F., Hughes, R.D. and Way, M.J. (1969) The ecology of Myzus persicae . Annual Review of Entomology, 14, 197–270. [Google Scholar]
- Vandermoten, S., Francis, F., Haubruge, E. and Leal, W.S. (2011) Conserved odorant‐binding proteins from aphids and eavesdropping predators. PLoS ONE, 6, e23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, R.G. and Riddiford, L.M. (1981) Pheromone binding and inactivation by moth antennae. Nature, 293, 161–163. [DOI] [PubMed] [Google Scholar]
- Wang, R., Li, F., Zhang, W., Zhang, X., Qu, C., Tetreau, G. et al (2017) Identification and expression profile analysis of odorant binding protein and chemosensory protein genes in Bemisia tabaci MED by head transcriptome. PLoS ONE, 12, e0171739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, G. (1986) Ecological genetics of host plant exploitation in the green peach aphid, Myzus persicae . Entomologia Experimentalis et Applicata, 40, 161–168. [Google Scholar]
- Wenger, J.A. , Cassone, B.J., Legeai, F., Johnston, J.S., Bansal, R., Yates, A.D. et al (2017) Whole genome sequence of the soybean aphid. Insect Biochemistry and Molecular Biology. Available at: doi: 10.1016/j.ibmb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Williams, I.S., Dewar, A.M., Dixon, A.F.G. and Thornhill, W.A. (2000) Alate production by aphids on sugar beet: how likely is the evolution of sugar beet‐specific biotypes? Journal of Applied Ecology, 37, 40–51. [Google Scholar]
- Xu, P.X., Zwiebel, L.J. and Smith, D.P. (2003) Identification of a distinct family of genes encoding atypical odorant‐binding proteins in the malaria vector mosquito, Anopheles gambiae . Insect Molecular Biology, 12, 549–560. [DOI] [PubMed] [Google Scholar]
- Xu, Y.L., He, P., Zhang, L., Fang, S.Q., Dong, S.L., Zhang, Y.J. et al (2009) Large‐scale identification of odorant‐binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics, 10, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, W.X., Fan, J., Zhang, Y., Xu, Q.X., Han, Z.L., Sun, J.R. et al (2016) Identification and expression analysis of candidate odorant‐binding protein and chemosensory protein genes by antennal transcriptome of Sitobion avenae . PLoS ONE, 11, e0161839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H.B., Ding, Y.X., Gu, S.H., Sun, L., Zhu, X.Q., Liu, H.W. et al (2015) Molecular characterization and expression profiling of odorant‐binding proteins in Apolygus lucorum . PLoS ONE, 10, e0140562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R.B., Wang, B., Grossi, G., Falabella, P., Liu, Y., Yan, S.C. et al (2017) Molecular basis of alarm pheromone detection in aphids. Current Biology, 27, 55–57. [DOI] [PubMed] [Google Scholar]
- Zhong, T., Yin, J., Deng, S.S., Li, K.B. and Cao, Y.Z. (2012) Fluorescence competition assay for the assessment of green leaf volatiles and trans‐β‐farnesene bound to three odorant‐binding proteins in the wheat aphid Sitobion avenae (Fabricius). Journal of Insect Physiology, 58, 771–781. [DOI] [PubMed] [Google Scholar]
- Zhou, J.J., Huang, W.S., Zhang, G.A., Pickett, J.A. and Field, L.M. (2004) “Plus‐C” odorant‐binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae . Gene, 327, 117–129. [DOI] [PubMed] [Google Scholar]
- Zhou, J.J., Kan, Y.C., Antoniw, J., Pickett, J.A. and Field, L.M. (2006) Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chemical Senses, 31, 453–465. [DOI] [PubMed] [Google Scholar]
- Zhou, J.J., He, X.L., Pickett, J.A. and Field, L.M. (2008) Identification of odorant‐binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Molecular Biology, 17, 147–163. [DOI] [PubMed] [Google Scholar]
- Zhou, J.J., Vieira, F.G., He, X.L., Smadja, C., Liu, R., Rozas J. et al (2010) Genome annotation and comparative analyses of the odorant‐binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum . Insect Molecular Biology, 19, 113–122. [DOI] [PubMed] [Google Scholar]
- Zhou, S.S., Sun, Z., Ma, W.H., Chen, W. and Wang, M.Q. (2014) De novo analysis of the Nilaparvata lugens (Stål) antenna transcriptome and expression patterns of olfactory genes. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 9, 31–39. [DOI] [PubMed] [Google Scholar]
- Zhou, W.W., Yuan, X., Qian, P., Cheng, J.A., Zhang, C.X., Gurr, G. et al (2015) Identification and expression profiling of putative chemosensory protein genes in two rice planthoppers, Laodelphax striatellus (Fallén) and Sogatella furcifera (Horváth). Journal of Asia‐Pacific Entomology, 18, 771–778. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. The sTABLE expression of M. persicae β‐Actin and GAPDH in different development stages and tissues measured by RT‐PCR.
FIGURE S2. The standard curves of M. persicae OBPs and CSPs with GAPDH as reference gene.
FIGURE S3. The standard curves of M. persicae OBPs and CSPs with β‐actin as reference gene.
TABLE S1. Gene specific primers used for cloning of M. persicae OBP and CSP genes.
TABLE S2. The protein names and sequences of the 45 OBPs and 41 CSPs from M. persicae, A. gossypii, A. pisum, A. glycines and S. avenae used in Fig. 7.
TABLE S3. The protein names and sequences of the 199 OBPs and 103 SPs from Hemiptera insects used in Fig. 8.
TABLE S4. Primers used in RT‐PCR for determination expression levels of M. persicae OBP and CSP genes.
TABLE S5. Primers used in real‐time PCR for determination expression level of M. persicae OBP and CSP genes. The real PCR amplification efficiencies of target and reference genes were calculated using LinRegPCR program (version 11.0) (Ramakers et al., 2003).
TABLE S6. A percent identity matrix of M. persicae OBPs.
TABLE S7. A percent identity matrix of M. persicae CSPs.
TABLE S8. The protein names and sequences of the Hemiptera 237 OBPs used in Fig. 3.
TABLE S9. The protein names and sequences of the Hemiptera 110 CSPs used in Fig. 4.


