Summary
Thanks to the exponentially increasing number of publicly available bacterial genome sequences, one can now estimate the important contribution of integrated viral sequences to the diversity of bacterial genomes. Indeed, temperate bacteriophages are able to stably integrate the genome of their host through site‐specific recombination and transmit vertically to the host siblings. Lysogenic conversion has been long acknowledged to provide additional functions to the host, and particularly to bacterial pathogen genomes where prophages contribute important virulence factors. This review aims particularly at highlighting the current knowledge and questions about lysogeny in Salmonella genomes where functional prophages are abundant, and where genetic interactions between host and prophages are of particular importance for human health considerations.
Salmonella enterica genomes, as most other bacterial species, host multiple prophages whose genes can represent up to 5% of the genome content. In this review, we aim at putting into light the multiple genetic interactions that can be weaved between the host and its prophages and between prophages themselves within the host.
Glossary
Defective: a defective prophage has lost part of its genome and can no longer produce viral particles, but may still be able to excise from the host chromosome.
Functional: a prophage is considered functional if it can resume a lytic cycle and re‐infect naive cells.
Lysogeny: upon lysogenic infection by a temperate phage its genome is stably maintained as a prophage (most of the time integrated into the chromosome) in the host bacterium and vertically replicates with it. No progeny is produced until a lytic cycle resumes.
Lysogenic conversion: describes the phenotypic changes bacteria undergo upon infection by a temperate phage going through lysogeny.
Morons (“more on”): genes regulated independently from the rest of the prophage and conferring a fitness advantage under specific environmental conditions.
Polylysogeny: the hosting of multiple prophages in a single host genome.
Temperate bacteriophage: a phage that can multiply either lytically or through lysogeny.
Prophage/provirus: inherited form of a temperate bacteriophage; dormant form of the viral genome which replicates with the bacterial host genome.
Pseudolysogeny: a phage–host cell interaction in which the phage genome does not integrate into the host’s and forms an episomal form that can be still transmitted vertically. Pseudolysogeny applies to virulent and temperate phages and does not lead to the usual outcome lysis or lysogeny.
Pseudogenization: a gene becomes a pseudogene by accumulating mutations that hinder its correct transcription and/or translation.
Virulent or strictly lytic bacteriophage: a phage that multiplies exclusively through a lytic cycle.
Introduction
Bacteriophages are recognized as the most abundant biological entities on earth, participating to numerous biological cycles and constantly reshaping bacterial communities (Suttle, 2007; Brussaard et al., 2008). In all environments they outcompete the number of available hosts by one to several log10. Moreover, due to their propensity to lysogenize, i.e. become quiescent proviruses, temperate phages are recognized as essential drivers of bacterial genomes’ evolution (Roux et al., 2015a; 2015b; Casjens and Grose, 2016) (some detailed definitions can be found in the glossary).
This review aims at highlighting the contribution of prophage genes to the host physiology. To date, a lot of emphasis has been put on the identification and characterization of phage‐encoded virulence factors in various pathogenic bacteria (Brüssow et al., 2004; Dearborn and Dokland, 2012; Rabinovich et al., 2012; Busby et al., 2012; Fortier and Sekulovic, 2013; Davies et al., 2016; Kraushaar et al., 2017). However, as it becomes clearer that bacterial genomes contain large amounts of DNA from (pro)phage origin, we want to stress that these horizontally acquired genes are important contributors to the genomes evolution and provide discrete adaptive physiological contributions such as increasing fitness under certain environmental conditions or providing non‐obvious metabolic or signaling functions (D’Ari and Casadesús, 1998). We chose to focus on Salmonella enterica prophages for the following reasons: (i) it is a widespread enterobacteria displaying a broad host range, frequently carried by wild and domestic birds as well as rodents, and an animal and human pathogen, (ii) host‐prophages interactions have been studied for many years and still lead to amazing pieces of work encompassing many topics, such as host‐phage interactions, virulence, ecology and genome evolution.
Prophage abundance and integration sites in S. enterica genomes
The first prophages in Salmonella species have been identified in 1950, just before transduction has been discovered (Boyd, 1950; Zinder and Lederberg, 1952). Since the 1990s, the Bossi group has been a pioneer in S. enterica prophage research, which highlighted the diversity of the prophage repertoire of various strains (Figueroa‐Bossi et al., 1997; 2001; Figueroa‐Bossi and Bossi, 1999; Bossi et al., 2003). Since then many more prophages have been identified, every time a set of new Salmonella genomes is sequenced. Dormant prophages are transmitted vertically along with bacterial cell division and can be induced under stressful conditions, such as DNA damages or in animal guts (Kim et al., 2014). They can also undergo spontaneous induction, which can increase the fitness of a given strain whenever in competition when entering a new niche (Bossi et al., 2003).
Salmonella enterica genomes also carry defective prophages that are no longer able to form infectious particles, meanwhile being present – and perhaps maintained – in the host chromosome (Casjens, 2003; Bobay et al., 2013). Different events can lead to prophage degradation including large genomic reduction, targeting by insertion sequences (IS) as well as point mutations (Bobay et al., 2014). However, when prophages are not too degraded it is possible to “resuscitate” them into fully functional prophages, meaning inducible and able to form infectious particles. Such reactivations of defective prophages involve either a temporal complementation by an infecting phage that provides the missing function, or a recombination event that allows a permanent complementation (Figueroa‐Bossi and Bossi, 2004; De Paepe et al., 2014). The recombination events driven either by the host homologous recombinases or phage‐encoded recombinases inside the host cells are causing pervasive mosaicism in phage genomes (Lopes et al., 2010; De Paepe et al., 2014; Menouni et al., 2015). The best known and long studied temperate phage infecting S. enterica is P22 (the λ equivalent paradigm in S. enterica) that was a key model for transduction discovery (Boyd, 1950; Zinder and Lederberg, 1952). However, P22 itself is not a common prophage in S. enterica genomes that contain in average 5.29 prophages representing around 3.52% of the total gene content and close to 30% of the accessory genome (based on 21 S. enterica genomes analyzed) (Bobay et al., 2013). In other words, these numbers show that polylysogeny, i.e. the hosting of multiple prophages by a single genome, is a very frequent event. Another striking point is that integration sites are highly conserved between the two closely related Escherichia coli and S. enterica species and even beyond (Bobay et al., 2013; Oliveira et al., 2017). Among favored integration sites are found all categories of non‐translated RNA genes such as sRNA, tmRNA and tRNA, the latter being the most frequently targeted (Bobay et al., 2013). Other sites in the chromosome may be targeted as well, such as intergenic regions, while integration within protein‐encoding genes is much less frequent. Even when integrating at the 3′ end of genes, the site‐specific reaction involved in the integration process leads to the reconstitution of the targeted genes since the equivalent portion of the gene is provided on the phage genome, without affecting their function or expression (Argos et al., 1986). Alternatively, a prophage may disrupt a gene and therefore a cellular function. However, when the prophage excises, the interrupted gene can be reconstituted and the host regains the lost function, a process called phage‐driven regulatory switch or active lysogeny (Feiner et al., 2015). However, no such a switch has been experimentally described so far in S. enterica genomes.
The quasi‐weekly release of new draft genomes from S. enterica prevents an accurate description of the prophage content as prophage description and annotation are not so obvious, even though facilitated by various softwares (Clokie and Kropinski, 2009). Indeed, the presence of multiple contigs may hinder the correct description of prophages as they frequently co‐localize with contig borders, impairing correct genome assembly and are sometimes interrupted by insertion sequences (IS). As a result, prophage predictions need to undergo expert manual curation.
A recent study based on public health surveillance in the UK highlighted that S. enterica Typhimurium causing invasive non‐typhoidal salmonellosis in Africa carried a specific prophage as well as antibiotic resistance genes that are not found in the UK version of this lineage (Kintz et al., 2015; Owen et al., 2017; Ashton et al., 2017). As a consequence stably integrated prophages are useful tools as epidemiology markers in addition to CRISPR‐Cas typing. However, as the latter were found to be poorly active and show a very slow spacer turnover, such typing should be restricted to anciently diverged strains (Touchon and Rocha, 2010).
Lysogenic conversion
The notion of lysogenic conversion, meaning the propensity of temperate phage undergoing lysogeny to contribute to the host physiology has been described and admitted for numerous years. However, a strong bias is observed in the literature toward lysogenic conversion aspects that contribute the host virulence. As an example, the Gifsy‐2 encoded superoxide dismutase SodC that obviously contribute to the establishment of Salmonella cells into the macrophage (Figueroa‐Bossi and Bossi, 1999). Needless to say that S. enterica is an organism of choice for such contribution examples. However, one must consider that more subtle contributions do exist and pave the way for multiple interactions with the host genome as well as with the eukaryotic cells targeted by S. enterica or the microbiome encountered by the pathogen during its infectious journey in animals.
Prophage induction and prophage gene expression under lysogenic conditions
As in Escherichia coli, the repressor model is widespread in Salmonella’s Lambdoid prophages (Sauer et al., 1981; Campbell, 1994; Whipple et al., 1998). However, a striking and widely conserved feature is the involvement of antirepressor proteins in prophage induction. If most S. enterica prophages are induced by the activation of the SOS response due to DNA damaging factors (mitomycine C, UV or H2O2), the cleavage of the repressor is not the major outcome of the induction system. Indeed, it was shown for several S. enterica prophages that upon SOS response induction and LexA self‐cleavage, an antirepressor protein (Ant), homologous to the Tum one in phage 186, is being made that inhibits the lytic repressor through protein‐protein interaction (Shearwin et al., 1998; Lemire et al., 2011; Kim and Ryu, 2013). P22 also encodes such an antirepressor whose expression is negatively controlled by the Mnt repressor. Nevertheless, in this case, an ant mutant remains SOS inducible (Botstein et al., 1975; Levine et al., 1975). Interestingly, antirepressors are responsible for prophage induction crosstalk: a given prophage‐encoded antirepressor was shown to counteract the action of a repressor from another prophage (Lemire et al., 2011) (Fig. 1 and Table 1 subitem 2.1). This prophage crosstalk has probably a role in prophage dissemination, as non‐coordinated prophages could be lost upon massive host cell lysis provoked by a neighboring prophage undergoing induction. In contrast, a partial cell lysis is often provoked by spontaneous or uncompleted induction, which allows a non‐induced prophage to remain in the bacterial population.
Figure 1.
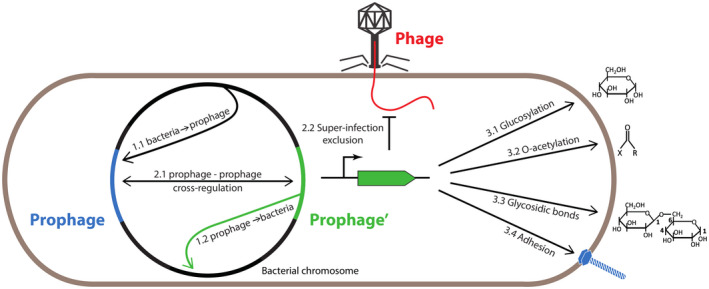
Multiple interactions between prophages and Salmonella hosts: The multiple host‐prophage and prophage‐prophage interactions depicted in the text are illustrated. For more details see Table 1.
Table 1.
Host‐prophage and prophage‐prophage interactions in S. enterica.
| 1. Interactions bacteria – prophage and prophage – bacteria |
| 1.1 Host factors |
|
| 1.2 Prophage factors |
|
| 2. Interactions prophage – (pro)phage |
| 2.1 Cross‐regulation |
| Repressor – Antirepressor (Fels‐2 and Gifsy prophages) |
| 2.2 Super‐infection exclusion |
|
| 3. Surface features encoded by prophages and phage remnants |
| 3.1 Glucosylation |
|
| 3.2 O‐acetylation |
|
| 3.3 Glycosidic bonds |
|
| 3.4 Adhesion |
|
A number of inducing conditions or chemicals have been described that induce prophages in Salmonella through the SOS‐response activation. However, induction driven by some of these should lead to preoccupation in the context of public health risks. Indeed, it has been shown recently that some prophages are induced by antibiotics widely used in agriculture, such as fluoroquinolones (Bearson and Brunelle, 2015) and carbadox (Bearson et al., 2014). As a result, these antibiotics that are most probably present in our daily food intake may have thus consequences on prophage induction in our gut. Also relevant to the physiology of Salmonella could be the induction by bile salts, which are encountered by this bacterium in its natural ecological niche (Hernández et al., 2012). In all cases, little is known about the in vivo consequences of these treatments. However, one should consider the demonstration of in vivo transfer of prophage‐encoded virulence genes at loci of inflammation (Diard et al., 2017), which raises the question of the consequences of inducing antibiotics on the transmission of prophage‐encoded genes.
A commonly admitted view is that prophage genes are under control either by the powerful phage repressor or by host factors that tend to limit the negative effects of horizontal gene acquisition. In the first case, thanks to the seminal work of Jacob and Monod “The operon model” (Lewis, 2011), the lysogeny repressor is known to control the expression of the lytic promoters (PL and PR) in the absence of induction. Host‐controlled expression of exogenic, and prophage genes in particular, has been described in several bacterial models (Navarre et al., 2006; Cardinale et al., 2008). However, it is in S. enterica that the role of the nucleoid associated protein H‐NS has been acknowledged as a genome sentinel (see below) (Navarre et al., 2006; Dorman, 2007; Navarre et al., 2007; Ali et al., 2013). Although there is no doubt about the extensive repression of prophage genes involved in lytic functions, we argue that it may have occulted the expression of those genes involved in lysogenic conversion and for which no obvious function in metabolic or pathogenic pathways have been identified. In order to find more of these genes whose expression remains significant under lysogenic conditions, one can look at the impressive amount of transcriptomic data available in the public databases and have a search in prophage‐related regions. One such very helpful database dedicated to S. enterica is the SalCom compendium developed by and hosted in Jay Hinton’s lab (Kröger et al., 2013; Colgan et al., 2016) (http://bioinf.gen.tcd.ie/cgi-bin/salcom.pl?_HL).
Contributions to phage resistance and physiology
Being extremely abundant on earth while presenting a gene content very different from that currently available in the genomic databases, bacteriophage genomes are believed to contribute extensively to the genetic “dark matter” (Rinke et al., 2013; Roux et al., 2015b; Nadeem and Wahl, 2017). Indeed, even when focusing on a single bacterial host the gene repertoire carried by its infecting phages is highly diverse (Hatfull et al., 2010; Cresawn et al., 2011; Hatfull, 2015), and a large proportion of these genes remain annotated with unknown functions. A number of contributions to the host physiology might also beneficiate to the prophages themselves; among them are those preventing phage superinfection and/or recognition by the immune system from mammals.
LPS modifications
One particularity of phages infecting Salmonella strains is that they often recognize the lipopolysaccharide (LPS) or endotoxin consisting of a lipid and a polysaccharide composed of O‐antigen, as receptor as opposed to E. coli bacteriophages that often target outer membrane proteins (Bertozzi Silva et al., 2016). LPS contributes to the structural integrity and negative charge of the outer membrane of Gram‐negative bacteria and protects the membrane from certain kinds of chemicals, such as anionic bile salts or lipophilic antibiotics. Interestingly, the cognate Salmonella prophages frequently carry genes encoding proteins involved in O‐antigen modifications (Fig. 1 and Table 1 subitem 3) (Duerr et al., 2009; Broadbent et al., 2010; Andres et al., 2013; Davies et al., 2013; Sun et al., 2014; Cota et al., 2015; Kintz et al., 2015).
One of the most prominent examples is bacteriophage P22 of S. enterica which recognizes O‐antigen polysaccharides with its tailspike protein and when integrated as a lysogen in the bacterial chromosome provides itself a gtrABC operon for O‐antigen glucosylation (Andres et al., 2010; 2013). Up to four gtr operons encoded on different prophages and responsible for distinct modifications can be found within one bacterial genome (Broadbent et al., 2010). The gtrA and gtrB genes encode conserved membrane proteins, the bactoprenol‐linked glucosyl translocase or “flippase” and the bactoprenol glucosyl transferase respectively. It is the third gene, gtrC, in the cluster that is variable and confers specificity since it encodes the glucosyl transferase attaching a glucose group at a distinct O‐antigen position. These gtr operons are regulated according to phase variation (see the “Negative host‐control of prophage genes” section). LPS is a potent activator of immune cells and therefore these temporarily changing surface modifications can shade the bacteria vis‐à‐vis the eukaryotic immune system (Duerr et al., 2009). Besides glucosylation, another possible LPS O‐antigen modification is acetylation. An example is the gene oafA which is located on a phage remnant (at ~2.33 Mb on the S. enterica 4/74 chromosome) and codes for an integral membrane transacylase conferring the specific serotype 05 (Slauch et al., 1996). More recently, a gene coding for a protein similar to the acyltransferase 3 of Pseudomonas syringae py. Syringae B728a has been identified on prophage SPC‐P1, which was associated to increased virulence in S. paratyphi C (Zou et al., 2010). Another intriguing example is the S. enterica serovar Anatum specific phage ε15 which modulates glycosidic linkage of O‐antigen by blocking the host α polymerase and producing its own β polymerase (changes from α‐1,6 to β‐1,6 glycosidic linkage). This modification restricts super‐infection by ε15 itself and in turn allows infection by phage ε34 (Kropinski et al., 2007).
All these genetic features responsible for O‐antigen modifications and prophage‐encoded factors without being “real” virulence factors, may affect the resistance to the intestinal environment as well as to sur‐infecting phages, and thus in sum the fitness and pathogenicity of the bacterial host.
Superinfection exclusion
Besides the LPS modifications, phages have acquired other ways to prevent super‐infection by themselves and other phages. As examples we will mention here the genes sieA and sieB and the so‐called phage carrier state of P22 (Fig. 1 and Table 1 subitem 2.2). The inner membrane protein SieA is seemingly responsible for blocking the phage DNA transfer across the membrane into the bacterial cytoplasm (Susskind et al., 1974; Susskind and Botstein, 1980). SieB, also encoded by E. coli phage λ, aborts the lytic development of other Salmonella phages ‐ such as P22‐like MG178 and MG40 ‐ by stopping RNA, DNA, and protein synthesis. P22 itself is not affected, since it produces an early uncharacterized escape factor (Susskind et al., 1974). Interestingly, both SieA and SieB inhibit infection of the same phages, including λ, although Salmonella is not a standard host for the latter (Ranade and Poteete, 1993).
Phage carrier cells were identified recently and are cells infected with P22 harboring an episomal form of P22 that is transmitted asymmetrically during division (Cenens et al., 2013a; 2013b). The daughter cell inheriting this episome enters lysogeny resulting in a chromosomally integrated prophage. The other daughter cell becomes P22‐free, but intriguingly stays resistant to P22 infection in a transient way. The immunity factors responsible for this resistance are GtrABC, SieA and the repressor C2, which have been constitutively produced by the phage carrier cell. The P22‐free daughter cells cytoplasmically inherit these immunity factors, which then dilute out upon subsequent cell divisions (Cenens et al., 2015). By conferring this temporary resistance to a bacterial subpopulation, phages might thus insure both vertical and horizontal transmission routes throughout an infected population while maintaining a bacterial population they can infect. This process, described as “host‐farming” by A. Aertsen, allows P22 to cultivate susceptible non‐immune cells as a prey reservoir. This is an example of how up‐to‐date single‐cell studies contribute to the field and also shows that even the extensively studied phages such as P22 still reserves surprising new features to uncover.
Contribution of prophage gene products to virulence
This section will be mainly dedicated to recent or not yet reviewed examples of prophage‐encoded factors that are involved in Salmonella virulence (Boyd and Brüssow, 2002; Boyd, 2012; Boyd et al., 2012). Salmonella displays full panoply of virulence factors permitting to adhere to and infect eukaryotic cells and survive within them, in particular the hostile microenvironment of macrophages.
Adhesion
Shah et al. have identified a prophage gene, gpE, coding for a putative tail‐spike protein in SopEΦ in S. enterica LT2 that increased binding to epithelial cells (specifically via Spectrin1, an eukaryotic surface protein) and increased cell invasion (Shah et al., 2014; Fig. 1, Table 1 subitem 3.4). However, this prophage‐encoded gene was only expressed when bacteria were exposed to a cold stress before the infection assay. This emphasizes the importance of storage conditions for Salmonella contaminated food (mostly eggs and poultry) that are stored at cold and then reheated exposing bacteria to a chain of stresses, which may in turn induce prophages genes and finally, increase bacterial virulence.
Host entry, manipulation and intracellular survival (virulence factors per se)
One of the most impressive features of S. enterica virulence is the secretion of multiple effectors involved in virulence. The effectors are secreted via two different Type Three Secretion Systems (T3SS) encoded by the pathogenicity islands 1 and 2 (SPI1 and 2). SPI1 is activated when S. enterica is in contact with eukaryotic host cells, whereas SPI2 is expressed during the phagocytosis step (Kaur and Jain, 2012). Bacterial effectors are able to interfere and hijack the host signaling pathways. In addition, other SPI‐encoded factors exist that facilitate bacterial survival, among which so‐called anti‐virulence factors, which deletions curiously confer more virulence to the bacteria.
Among the SPI1 secreted effectors, SopE is one of the best studied effector proteins and its secretion results in actin cytoskeleton rearrangements and stimulates membrane ruffling, promoting bacterial entry into non‐phagocytotic cells such as epithelial cells (reviewed in (Ehrbar and Hardt, 2005)). Its encoding gene has been originally identified on the SopEΦ prophage in S. enterica SL1344, but the sopE gene and a constant flanking sequence, called the SopE‐cassette, is sporadically distributed in other lambdoid prophages of the Gifsy family among several Salmonella serovars, as well as on a P2‐like prophage in S. typhi (Mirold et al., 2001; Bachmann et al., 2014). The SopE‐cassette has most probably been transferred and integrated among these prophages by homologous recombination resulting in multiple sopE copies present in a single bacterial genome (Hoffmann et al., 2014). In the context of evolution, such a modular exchange mechanism could enhance effector protein diversity, since genes may duplicate and then potentially evolve to other functions. However, even the well studied SopE virulence factor may still not have unveiled all its functions. Indeed, recently SopE has been found to be not only produced and secreted for entering the eukaryotic cell, but also during the intracellular state where it seems to participate to the formation of the early Salmonella‐containing vacuole (SCV) (Vonaesch et al., 2014). The SCV is formed in order to create a replicative niche for the bacteria within the host cell. This double function of SopE during cell entry as well as during intracellular survival suggests that other prophage‐encoded effectors may have additional functions for host‐cell manipulation.
Gifsy1 prophage has been found to encode three genes involved in intra‐cellular survival: gogB, sarA, and pagK2. The first gene, gogB, codes for an anti‐inflammatory effector, which inhibits NFκB activation by interaction with host factors Skp1 and FBX022. It is thought that this anti‐inflammatory effect limits tissue damage during longer term infection, while short‐term inflammation enhances colonization in the intestine (Pilar et al., 2012).
The second gene, sarA, has been identified only very recently. SarA is mainly secreted by SPI2‐encoded T3SS, although there is also some translocation by SPI1 T3SS. It activates the eukaryotic transcription factor STAT3, which induces the transcription of Il‐10 as well as of other anti‐inflammatory factors. SarA is thus the first example of an effector that activates an anti‐inflammatory pathway in the eukaryotic host cell (Jaslow et al., 2018). PagK2 is secreted in outer membrane vesicles and contributes to intracellular survival in macrophages through an unknown mechanism (Yoon et al., 2011). The anti‐inflammatory effects of the T3SS effectors seem to be crucial at systemic sites later in infection when S. enterica must evade immunity and promote intracellular growth. Apparently, there is an evolutionary advantage to maintain gogB and sarA on the same prophage and the recent identification of a new prophage ST‐1974 in S. enterica Enteritidis supports this idea (D’Alessandro et al., 2018). In this case, the two genes coding for anti‐inflammatory functions, gogB and ssek3, are present on a single prophage. As mentioned above, gogB is encoded on Gifsy1 but can be found elsewhere on the chromosome (see below), while sseK3 has been previously identified on prophage ST64B (Brown et al., 2011). So, it seems that recombination events, similar to the above‐mentioned modular exchange of the SopE‐cassette, have taken place between these prophages. Interestingly, both GogB and SseK3 act on the same anti‐inflammatory NFκB pathway. However, the SseK3 host targets remain to be identified (Yang et al., 2015).
Antivirulence
Some prophage‐encoded genes confer an intriguing phenotype termed anti‐virulence. The Gifsy2‐encoded grvA gene is such an anti‐virulence factor: in its absence, and in contrast to what one would expect with classical virulence genes, the bacterial host is more virulent than a wild type strain in competition assays in mice (Ho and Slauch, 2001). However, this phenotype is only observed when sodCI, a Gifsy2‐encoded superoxide dismutase, is present as well. Thus, it is hypothesized that in a wild type situation, GrvA decreases the pathogenicity of the host probably by affecting resistance to toxic oxygen species via SodCI through an unknown mechanism. Another peculiar example of a factor that can be a virulence or an anti‐virulence factor depending on the serovar type of its host, is bstA encoded on prophage BTP1. Indeed, it acts as virulence factor in S. enterica ST313, i.e. higher uptake in macrophages (Herrero‐Fresno et al., 2014), while it was described as an anti‐virulence factor (lower uptake) in S. enterica Dublin; however the molecular mechanisms underlying both phenotypes are not yet understood (Herrero‐Fresno et al., 2018). A potential reason for this difference may be that, similarly to GrvA, another virulence factor is affected by BstA and is present in only one serovar. This highlights the possibility of a different output of prophage genes depending of the pre‐existing bacterial regulatory networks. Currently, it is not fully understood why bacterial pathogens would possess these antivirulence genes and what the evolutionary advantage (for both the prophage and the host) might be. One may speculate that bacterial pathogens might evolve toward less virulence in order to ensure their own propagation by keeping the potential host in shape in a way resembling to the above described phage‐carrier state. The fact that these factors are prophage‐encoded might give an advantage to changing environmental niches of the mammal’s body, since prophages can be lost and acquired in only one recombination event in the gut and therefore provide a fast way of adaptation (Diard et al., 2017).
Host‐prophage regulatory networks
As mentioned earlier, genes from phage origin represent a large part of S. enterica accessory genome. Some of these genes contribute to the host physiology and therefore need to be expressed at the right time and the right place. To this end, they become part of the bacterial regulatory network. How does acquisition of these new genes not disturb the normal bacterial functioning? How are they integrated into the host regulatory network? What potential benefit do they provide to the bacterial host? How can bacterial regulators modulate prophage behavior by modulating gene expression?
Negative host‐control of prophage genes
Expression of new genes must not be detrimental for the bacterial host. Therefore, genes acquired by horizontal gene transfer (HGT) are generally first silenced before being integrated into the host regulatory network. The silencing of genes from foreign origin can, for example, occur via DNA modification or involve regulatory proteins that bind DNA to prevent transcription.
One of the most studied DNA modifications is responsible for an epigenetic regulation called phase variation and occurs only in a small fraction of the bacterial population. This regulation relies on the methylation of deoxyadenosines by the Dam methylase (Deoxyadenosine methyltransferase) (Casadesús, 2016). The Dam enzyme recognizes and specifically modifies the 5′‐GATC‐3′ sequences; when these sequences are localized in a promoter region, methylation events can block the binding of transcriptional regulators and consequently modify gene expression. DNA methylation is involved in the silencing of genes localized on the Gifsy1, Fels1 and ST64B prophages in S. enterica SL1344 (Balbontín et al., 2006). Strikingly, it negatively regulates most of the ST64B genes. This observation is in accordance with previous results showing that ST64B excision is inhibited by Dam regulation (Alonso et al., 2005). This has been attributed to the down regulation of two genes located on this prophage and coding for proteins involved in phage induction: the anti‐repressor Sb41 and the replication protein Sb42. The bacterial regulator involved in this regulation and hindered in its function by the Dam methylation has not been identified to date. While the Dam‐regulation observed for genes located on Gifsy1 and Fels1 prophages does not affect their excision, SopEΦ prophage excision is favored by Dam methylation. However, the transcriptional regulator as well as the target genes responsible for this phenotype has not been described (Alonso et al., 2005).
Epigenetic regulation is also involved in the regulation of O‐antigen glucosylation (see LPS modification section). Indeed, under lysogenic conditions expression of the P22 encoded gtrABC operon is regulated by Dam methylation and the bacterial regulator OxyR (Broadbent et al., 2010; Davies et al., 2013). The region upstream of the gtr operon contains several OxyR binding sites as well as several methylation sites. Depending on the methylation state, OxyR bind different sites and can act as an activator or a repressor of the system. OxyR binding to one site decreases the methylation by Dam on this site and thus increases its own binding. But this works also the other way around: increased methylation results in reduced OxyR binding, which favors methylation, etc. This confers heritability of the expression state to the system and only a part of the population is in an “ON” state, leading to an heterogeneous population (Broadbent et al., 2010; García‐Pastor et al., 2018). This regulatory mechanism is thought to be conserved among the P22 temperate phage family and can prevent superinfection by the same or other phages that use similar O‐antigen co‐receptor during a limited time (Davies et al., 2013).
Another factor involved in the silencing of genes acquired by HGT, in addition to core genes, is the DNA binding protein H‐NS (Lucchini et al., 2006; Navarre et al., 2006). By preferentially binding to AT rich sequences, this protein can discriminate between self and non‐self. Interestingly, several studies suggest that H‐NS dependent regulation would involve different mechanisms for ancestral genes or genes acquired by HGT (Vivero et al., 2008; Baños et al., 2009). It has been suggested that ancestral genes would be regulated directly by H‐NS binding whereas the genes acquired by HGT would require the formation of heterodimers between H‐NS and proteins belonging to the Hha family. What characteristics of the promoter are required to favor the binding of homodimers or heterodimers are not known. In the same order of idea, H‐NS proteins encoded by conjugative plasmids have evolved to specifically regulate foreign genes (Baños et al., 2009; 2011). This could be due to structural differences between plasmid‐encoded or chromosomally encoded H‐NS leading to different affinity for promoter regions. Indeed, although the N‐ and C‐terminal domains are conserved, the linker region presents some variability that could be responsible for this differential regulation. All these observations concern genes acquired by HGT in general, including genes from phage origin. However, it has been noticed that the GC content of prophages in the reference strain of S. enterica LT2 is similar to the average GC content of the genome (Navarre et al., 2007). Thus, we can wonder if the conclusions made above really apply to genes from prophage origin. Studies focusing specifically on the regulation of these genes are missing so far and need to be performed to answer this question. Ongoing work in our lab suggests however that H‐NS regulates prophage genes that have not been identified by global approaches in S. enterica ST4/74 and that these regulations have consequences on prophage maintenance in the host chromosome (Wahl et al., unpublished).
Positive host‐control of prophage genes
In addition to the negative regulation that we have just mentioned, some bacterial regulators also positively regulate genes from prophage origin. Surprisingly, if one looks at the different transcriptomic studies performed in S. enterica to define the targets of global regulator such as PhoP, SlyA, ArcA, FNR or RpoS, only a handful of prophage genes were identified (Navarre et al., 2005; Fink et al., 2007; Evans et al., 2011; Lévi‐Meyrueis et al., 2015). Furthermore, the molecular mechanism(s) leading to these regulations or the consequences on bacterial physiology are rarely looked at. Not surprisingly, what has been mostly studied is the regulation of genes coding for proteins involved in virulence and host colonization. These studies have shown that genes under the control of bacterial regulators are morons, which defines genes regulated independently from the rest of the prophage and conferring an advantage (fitness effect under specific conditions such as virulence) to the host. Among them, are several effectors proteins secreted by T3SS. As mentioned above, S. enterica possesses two T3SS encoded by the pathogenicity island 1 and 2 (SPI1 and 2). Among the regulators known to control the expression of genes located on the SPI are the InvF transcriptional regulator belonging to the AraC/XylS family for SPI1, itself regulated by the master regulator HilD, and the SsrAB two‐component system for SPI2. Although both regulators were initially thought to be only dedicated to the regulation of genes located on SPI1 and 2, they also regulate expression of prophage‐encoded effectors. Indeed, InvF regulates effectors from prophage origin secreted by SPI1, whereas SsrAB controls effectors that are SPI2‐dependent.
For example, SopE is a virulence factor encoded on the SopEΦ prophage in S. enterica SL1344 and secreted by the SPI1 system. Consequently, sopE expression is regulated by InvF, in association with the chaperon protein SicA (Darwin and Miller, 2000). The role of SicA is probably indirect, by stabilizing or allowing the function of a so far unidentified transcriptional regulator involved in sopE regulation (Tucker and Galán, 2000).
Other prophage‐encoded effectors are secreted by SPI2, and as a consequence, their expression depends on the SsrAB two‐component system. Among them, GogB is encoded by the first gene of the Gifsy1 prophage in S. enterica SL1344. Interestingly it has been shown that gogB expression is independent of Gifsy1 prophage factors since it can be transferred by itself in the enteropathogenic E. coli strain E2348/69, expressed from its own promoter and secreted via the T3SS of its new host. This shows that gogB can be integrated easily into the host regulatory network (Coombes et al., 2005). The GC content of gogB shows differences with the GC content of Gifsy1 suggesting that this gene has been recently acquired by the prophage. Moreover, gogB can be found outside of Gifsy1 and is not always prophage‐encoded, which further supports its transcriptional independence from the Gifsy1 prophage (Porwollik et al., 2002).
sseI is a gene located on the Gifsy2 prophage, encoding another T3SS effector. sseI expression is strongly activated by the direct binding of the phosphorylated form of SsrB in its promoter region (Worley et al., 2000; Feng et al., 2004). sseI expression is also regulated by the phosphorylated form of OmpR but it is not clear whether this regulation is direct or dependent on SsrB (Feng et al., 2004). Interestingly, the pseudogenization of sseI together with the higher expression of pgtE, encoding an outer membrane protein, allows S. enterica ST313 adaptation to cause systemic disease (Carden et al., 2017; Hammarlöf et al., 2018). The increase in pgtE transcription is due to a single SNP in its promoter region. Further studies are required to understand how these changes in gene expression modify S. enterica ST313 behavior (Hammarlöf et al., 2018).
SsrB also regulates genes in the phage remnant SPI12. Among the regulated genes STM2239 encodes a Q antiterminator protein that interacts with the RNA polymerase to facilitate the transcription of late promoters. The absence of STM2239 affects the fitness of the bacteria within the host. STM2239 allows the transcription of phage‐encoded genes but also of bacterial genes involved in metabolic pathways including ribose modification and transport, acetyl coenzyme A synthesis and recycling as well as galactose metabolism. None of these regulations have been characterized further, but it has been speculated that some of them may be important for S. enterica fitness within the host (Tomljenovic‐Berube et al., 2013).
Phage‐controlled bacterial genes
Except for virulence, examples of bacterial processes under prophage control are scarce (Fig. 1 and Table 1 subitem 1.2). However, P22 offers a nice illustration of bacterial genes encoding proteins involved in metabolism and under the control of a regulator from phage origin. The dgo operon is involved in the uptake and metabolism of D‐galactonate, an important carbon source during intracellular proliferation. In S. enterica LT2 strain, expression of the dgo operon is derepressed in the presence of Pid, a protein encoded on a moron locus in P22 (Cenens et al., 2013a; 2013b). This regulation only occurs when P22 undergoes pseudolysogeny, suggesting the existence of a dedicated genetic program in this condition.
Phage‐dependent regulation can be conserved among closely related bacteria. It is the case for the pckA gene encoding a phosphoenolpyruvate carboxykinase required for gluconeogenesis. In E. coli, this gene is under the control of the CI repressor of the λ phage (Chen et al., 2005). Interestingly, pckA regulatory region is conserved among Enterobacteriaceae and contains sequences homologs to several phage operators, one of them being the binding site for the C2 repressor of P22. This shared regulation between several prophages could be part of an adaptive strategy to increase lysogens fitness by lowering their growth rate under glucose‐limited conditions (Chen et al., 2005).
Prophages often integrate into tRNA encoding genes. One counter example is given by phage ΦW104 that integrates the host chromosome at a locus encoding the RyeA and RyeB sRNA located on the opposite DNA strand in S. enterica DT104 (Balbontín et al., 2008). The attachment site for ΦW104 is within the 23 last base pairs of ryeB and corresponds to an internal site in ryeA. Therefore, ΦW104 lysogenization modifies the 5′ portion of ryeA, leading to a decreased transcription of ryeA and an increased transcription of ryeB. This transcriptional regulation has probable physiological consequences on the bacterial host, by modifying the expression of RyeA and RyeB mRNA targets.
Finally, another example of host‐gene regulation involves a gene located on the Gifsy1 prophage, isrK (Hershko‐Shalev et al., 2016). This gene encodes an sRNA and a long polycistronic mRNA comprising isrK, orf45, anrP and isrJ coding sequences. However, there is no translation observed from this mRNA unless IsrK sRNA is present. Indeed, IrsK sRNA binds next to the orf45 ribosome binding site and facilitates the binding of the 30S ribosomal subunit to this site, leading to the translation of downstream sequences and the production of the AnrP protein. AnrP is an anti‐repressor activating the transcription of phage‐encoded genes. Among them, it activates the expression of the antQ gene coding for the AntQ anti‐terminator that interacts and forms a stable complex with RNA polymerase. This leads to an aberrant transcriptional elongation, DNA damage and ultimately cell death (Hershko‐Shalev et al., 2016).
All the above examples concern regulation of gene expression. However, phages can also influence bacterial physiology by other means. For example, the release of colicin 1b in S. enterica SL1344 depends on the lysis genes of the ST64B prophage (Nedialkova et al., 2015). Indeed, under specific conditions such as DNA damage or iron limitation, colicin 1b accumulates in the cell and needs the induction of ST64B lysis genes to be found in the extracellular medium. Interestingly, complex crosstalk between ST64B and other prophages present in that strain contributes to this regulation and need further characterization.
Conclusions
What is striking whenever considering and comparing different Salmonella genomes is the diversity of the prophage content as well as the diverse relationships these prophages engage with the host strains. As stated before, we think that contributions to virulence have been more studied and highlighted up to now than regulatory and metabolic interactions between the Salmonella host and its prophages. This bias seems largely due to the prevalent role of Salmonella species in public health threats. In addition, contrary to some previous statements, phage genomes rarely contain antibiotic resistance genes, and if antibioresistance transfer can be sometimes attributed to phages, it is more likely due to generalized transduction rather than lysogeny (Enault et al., 2017). Nevertheless, a new category of self transferable plasmid‐phages could change this view as some of them carry ATB resistance genes (SSU5 super‐cluster) (Gilcrease and Casjens, 2018).
Apart from actual contributions to virulence, we foresee that many more interactions do exist, and, it seems that microbiologists have only just began to explore the expense of prophage–host interactions and their short and long‐term effects on bacterial metabolism and evolution. More interactions certainly remain to be discovered such as the up‐to‐now neglected carrier state and its implications on the host metabolism (Cenens et al., 2015).
In a context of multidrug resistance spreading and recurrent warnings from WHO and other health authorities, the use of bacteriophages as therapeutic agents is coming back to the scene, not only as potent antimicrobials by themselves but also as synergistic or complementary agents in combination with antibiotics (Kamal and Dennis, 2015; Abedon, 2018). However, even though only virulent (or strictly lytic) phages are considered for therapeutic usage, one must be aware of the possible interference of prophages whenever considering phages as a treatment. As described above, prophages are important contributors of serotype conversion, and particularly in Salmonella species. The literature becomes quite abundant regarding Salmonella phage quests, but little is known about the consequences of poly‐lysogeny, which can rapidly modify the bacterial surface, and therefore, the resistance to surinfecting phages on the efficacy of phage cocktails, particularly for those developed to treat swine and poultry or in the case of adjuvant in food processing (Wernicki et al., 2017). We suggest to systematically assessing the prophage content of the targeted strains to evaluate and adapt the composition of phage cocktails.
Acknowledgements
Work in our group is supported by the French Research National Agency grants ANR‐16‐CE12‐0029‐01 and ANR‐17‐CE11‐0038‐02, and institutional funding from the Centre National de la Recherche Scientifique and the Aix‐Marseille Université. We are grateful to S. Champ, N. Ginet and the whole phage group at LCB for stimulating discussions and suggestions. We thank M. Touchon and E. Rocha for Salmonella genome analysis and to J.R. Fantino for his help in figure design. M.A. also thanks J. Casadesús who provided an inspiring environment while part of this review was written. Although literature citations have not been limited, we may have omitted some pieces of the abundant literature to be more focused, we apologize to those authors whose research is not included.
References
- Abedon, S.T. (2018) Phage therapy: various perspectives on how to improve the art. Methods in Molecular Biology (Clifton, NJ), 1734, 113–127. [DOI] [PubMed] [Google Scholar]
- Ali, S.S. , Whitney, J.C. , Stevenson, J. , Robinson, H. , Howell, P.L. and Navarre, W.W. (2013) Structural insights into the regulation of foreign genes in Salmonella by the Hha/H‐NS complex. The Journal of Biological Chemistry, 288, 13356–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, A. , Pucciarelli, M.G. , Figueroa‐Bossi, N. and García‐del Portillo, F. (2005) Increased excision of the Salmonella prophage ST64B caused by a deficiency in Dam methylase. Journal of Bacteriology, 187, 7901–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, D. , Gohlke, U. , Broeker, N.K. , Schulze, S. , Rabsch, W. , Heinemann, U. , et al. (2013) An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O‐antigen fragments to bind as nonsolution conformers. Glycobiology, 23, 486–494. [DOI] [PubMed] [Google Scholar]
- Andres, D. , Hanke, C. , Baxa, U. , Seul, A. , Barbirz, S. and Seckler, R. (2010) Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. The Journal of Biological Chemistry, 285, 36768–36775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos, P. , Landy, A. , Abremski, K. , Egan, J.B. , Haggard‐Ljungquist, E. , Hoess, R.H. et al (1986) The integrase family of site‐specific recombinases: regional similarities and global diversity. The EMBO Journal, 5, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton, P.M. , Owen, S.V. , Kaindama, L. , Rowe, W.P.M. , Lane, C.R. , Larkin, L. et al (2017) Public health surveillance in the UK revolutionises our understanding of the invasive Salmonella Typhimurium epidemic in Africa. Genome Medicine, 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann, N.L. , Petty, N.K. , Ben Zakour, N.L. , Szubert, J.M. , Savill, J. and Beatson, S.A. (2014) Genome analysis and CRISPR typing of Salmonella enterica serovar Virchow. BMC Genomics, 15, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontín, R. , Figueroa‐Bossi, N. , Casadesús, J. and Bossi, L. (2008) Insertion hot spot for horizontally acquired DNA within a bidirectional small‐RNA locus in Salmonella enterica . Journal of Bacteriology, 190, 4075–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontín, R. , Rowley, G. , Pucciarelli, M.G. , López‐Garrido, J. , Wormstone, Y. , Lucchini, S. et al (2006) DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. Journal of Bacteriology, 188, 8160–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baños, R.C. , Aznar, S. , Madrid, C. and Juárez, A. (2011) Differential functional properties of chromosomal‐ and plasmid‐encoded H‐NS proteins. Research in Microbiology, 162, 382–385. [DOI] [PubMed] [Google Scholar]
- Baños, R.C. , Vivero, A. , Aznar, S. , García, J. , Pons, M. , Madrid, C. et al (2009) Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H‐NS. PLoS Genetics, 5, e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson, B.L. , Allen, H.K. , Brunelle, B.W. , Lee, I.S. , Casjens, S.R. and Stanton, T.B. (2014) The agricultural antibiotic carbadox induces phage‐mediated gene transfer in Salmonella . Frontiers in Microbiology, 5, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson, B.L. and Brunelle, B.W. (2015) Fluoroquinolone induction of phage‐mediated gene transfer in multidrug‐resistant Salmonella . International Journal of Antimicrobial Agents, 46, 201–204. [DOI] [PubMed] [Google Scholar]
- Bertozzi Silva, J. , Storms, Z. and Sauvageau, D. (2016) Host receptors for bacteriophage adsorption. FEMS Microbiology Letters, 363. [DOI] [PubMed] [Google Scholar]
- Bobay, L.‐M. , Rocha, E.P.C. and Touchon, M. (2013) The adaptation of temperate bacteriophages to their host genomes. Molecular Biology and Evolution, 30, 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay, L.‐M. , Touchon, M. and Rocha, E.P.C. (2014) Pervasive domestication of defective prophages by bacteria. Proceedings of the National Academy of Sciences, 111, 12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi, L. , Fuentes, J.A. , Mora, G. and Figueroa‐Bossi, N. (2003) Prophage contribution to bacterial population dynamics. Journal of Bacteriology, 185, 6467–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein, K. , Lew, K.K. , Jarvik, V. and Swanson, C.A. (1975) Role of antirepressor in the bipartite control of repression and immunity by bacteriophage P22. Journal of Molecular Biology, 91, 439–462. [DOI] [PubMed] [Google Scholar]
- Boyd, E.F. (2012) Bacteriophage‐encoded bacterial virulence factors and phage‐pathogenicity island interactions. Advances in Virus Research, 82, 91–118. [DOI] [PubMed] [Google Scholar]
- Boyd, E.F. and Brüssow, H. (2002) Common themes among bacteriophage‐encoded virulence factors and diversity among the bacteriophages involved. Trends in Microbiology, 10, 521–529. [DOI] [PubMed] [Google Scholar]
- Boyd, E.F. , Carpenter, M.R. and Chowdhury, N. (2012) Mobile effector proteins on phage genomes. Bacteriophage, 2, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, J.S.K. (1950) The symbiotic bacteriophages of Salmonella typhimurium. The Journal of Pathology and Bacteriology, 62, 501–517. [DOI] [PubMed] [Google Scholar]
- Broadbent, S.E. , Davies, M.R. and Van Der Woude, M.W. (2010) Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation‐dependent mechanism. Molecular Microbiology, 77, 337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N.F. , Coombes, B.K. , Bishop, J.L. , Wickham, M.E. , Lowden, M.J. , Gal‐Mor, O. et al (2011) Salmonella phage ST64B encodes a member of the SseK/NleB effector family. PLoS One, 6, e17824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard, C.P.D. , Wilhelm, S.W. , Thingstad, F. , Weinbauer, M.G. , Bratbak, G. , Heldal, M. et al (2008) Global‐scale processes with a nanoscale drive: the role of marine viruses. The ISME Journal, 2, 575–578. [DOI] [PubMed] [Google Scholar]
- Brüssow, H. , Canchaya, C. and Hardt, W.‐D. (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiology and Molecular Biology Reviews, 68, 560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby, B. , Kristensen, D.M. and Koonin, E.V. (2012) Contribution of phage‐derived genomic islands to the virulence of facultative bacterial pathogens. Environmental Microbiology, 15, 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, A. (1994) Comparative molecular biology of lambdoid phages. Annual Review of Microbiology, 48, 193–222. [DOI] [PubMed] [Google Scholar]
- Carden, S.E. , Walker, G.T. , Honeycutt, J. , Lugo, K. , Pham, T. , Jacobson, A. et al (2017) Pseudogenization of the secreted effector gene ssei confers rapid systemic dissemination of S. Typhimurium ST313 within migratory dendritic cells. Cell Host & Microbe, 21, 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale, C.J. , Washburn, R.S. , Tadigotla, V.R. , Brown, L.M. , Gottesman, M.E. and Nudler, E. (2008) Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli . Science, 320, 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús, J. (2016) Bacterial DNA methylation and methylomes. Advances in Experimental Medicine and Biology, 945, 35–61. [DOI] [PubMed] [Google Scholar]
- Casjens, S. (2003) Prophages and bacterial genomics: what have we learned so far? Molecular Microbiology, 49, 277–300. [DOI] [PubMed] [Google Scholar]
- Casjens, S.R. and Grose, J.H. (2016) Contributions of P2‐ and P22‐like prophages to understanding the enormous diversity and abundance of tailed bacteriophages. Virology, 496, 255–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenens, W. , Makumi, A. , Govers, S.K. , Lavigne, R. and Aertsen, A. (2015) Viral transmission dynamics at single‐cell resolution reveal transiently immune subpopulations caused by a carrier state association. PLoS Genetics, 11, e1005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenens, W. , Makumi, A. , Mebrhatu, M.T. , Lavigne, R. and Aertsen, A. (2013a) Phage‐host interactions during pseudolysogeny: lessons from the Pid/dgo interaction. Bacteriophage, 3, e25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenens, W. , Mebrhatu, M.T. , Makumi, A. , Ceyssens, P.‐J. , Lavigne, R. , Van Houdt, R. et al (2013b) Expression of a novel P22 ORFan gene reveals the phage carrier state in Salmonella Typhimurium. PLoS Genetics, 9, e1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Golding, I. , Sawai, S. , Guo, L. and Cox, E.C. (2005) Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biology, 3, e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie M.R.J. and Kropinski A.M. (Eds.) (2009) Bacteriophages: Methods and Protocols. New York: Humana Press. [Google Scholar]
- Colgan, A.M. , Kröger, C. , Diard, M. , Hardt, W.‐D. , Puente, J.L. , Sivasankaran, S.K. et al (2016) The impact of 18 ancestral and horizontally‐acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genetics, 12, e1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes, B.K. , Wickham, M.E. , Brown, N.F. , Lemire, S. , Bossi, L. , Hsiao, W.W.L. et al (2005) Genetic and molecular analysis of GogB, a phage‐encoded type III‐secreted substrate in Salmonella enterica serovar typhimurium with autonomous expression from its associated phage. Journal of Molecular Biology, 348, 817–830. [DOI] [PubMed] [Google Scholar]
- Cota, I. , Sánchez‐Romero, M.A. , Hernández, S.B. , Pucciarelli, M.G. , García‐del Portillo, F. and Casadesús, J. (2015) Epigenetic control of Salmonella enterica O‐antigen chain length: a tradeoff between virulence and bacteriophage resistance. PLoS Genetics, 11(11), e1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn, S.G. , Bogel, M. , Day, N. , Jacobs‐Sera, D. , Hendrix, R.W. and Hatfull, G.F. (2011) Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics, 12, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro, B. , Pérez Escanda, V. , Balestrazzi, L. , Iriarte, A. , Pickard, D. , Yim, L. et al (2018) A novel prophage identified in strains from Salmonella enterica serovar enteritidis is a phylogenetic signature of the lineage ST‐1974. Microbial Genomics, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ari, R. and Casadesús, J. (1998) Underground metabolism. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 20, 181–186. [DOI] [PubMed] [Google Scholar]
- Darwin, K.H. and Miller, V.L. (2000) The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Molecular Microbiology, 35, 949–960. [DOI] [PubMed] [Google Scholar]
- Davies, E.V. , Winstanley, C. , Fothergill, J.L. and James, C.E. (2016) The role of temperate bacteriophages in bacterial infection. FEMS Microbiology Letters, 363, fnw015. [DOI] [PubMed] [Google Scholar]
- Davies, M.R. , Broadbent, S.E. , Harris, S.R. , Thomson, N.R. and van der Woude, M.W. (2013) Horizontally acquired glycosyltransferase operons drive Salmonellae lipopolysaccharide diversity. PLoS Genetics, 9, e1003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe, M. , Hutinet, G. , Son, O. , Amarir‐Bouhram, J. , Schbath, S. and Petit, M.‐A. (2014) Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52‐like recombinases. PLoS Genetics, 10, e1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn, A.D. and Dokland, T. (2012) Mobilization of pathogenicity islands by Staphylococcus aureus strain Newman bacteriophages. Bacteriophage, 2, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diard, M. , Bakkeren, E. , Cornuault, J.K. , Moor, K. , Hausmann, A. , Sellin, M.E. et al (2017) Inflammation boosts bacteriophage transfer between Salmonella spp. Science, 355, 1211–1215. [DOI] [PubMed] [Google Scholar]
- Dorman, C.J. (2007) H‐NS, the genome sentinel. Nature Reviews Microbiology, 5, 157–161. [DOI] [PubMed] [Google Scholar]
- Duerr, C.U. , Zenk, S.F. , Chassin, C. , Pott, J. , Gütle, D. , Hensel, M. et al (2009) O‐antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathogens, 5, e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrbar, K. and Hardt, W.‐D. (2005) Bacteriophage‐encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 5, 1–9. [DOI] [PubMed] [Google Scholar]
- Enault, F. , Briet, A. , Bouteille, L. , Roux, S. , Sullivan, M.B. and Petit, M.‐A. (2017) Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. The ISME Journal, 11, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M.R. , Fink, R.C. , Vazquez‐Torres, A. , Porwollik, S. , Jones‐Carson, J. , McClelland, M. et al (2011) Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiology, 11, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner, R. , Argov, T. , Rabinovich, L. , Sigal, N. , Borovok, I. and Herskovits, A.A. (2015) A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nature Reviews Microbiology, 13, 641–650. [DOI] [PubMed] [Google Scholar]
- Feng, X. , Walthers, D. , Oropeza, R. and Kenney, L.J. (2004) The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Molecular Microbiology, 54, 823–835. [DOI] [PubMed] [Google Scholar]
- Figueroa‐Bossi, N. and Bossi, L. (1999) Inducible prophages contribute to Salmonella virulence in mice. Molecular Microbiology, 33, 167–176. [DOI] [PubMed] [Google Scholar]
- Figueroa‐Bossi, N. and Bossi, L. (2004) Resuscitation of a defective prophage in Salmonella cocultures. Journal of Bacteriology, 186, 4038–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa‐Bossi, N. , Coissac, E. , Netter, P. and Bossi, L. (1997) Unsuspected prophage‐like elements in Salmonella typhimurium. Molecular Microbiology, 25, 161–173. [DOI] [PubMed] [Google Scholar]
- Figueroa‐Bossi, N. , Uzzau, S. , Maloriol, D. and Bossi, L. (2001) Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella . Molecular Microbiology, 39, 260–271. [DOI] [PubMed] [Google Scholar]
- Fink, R.C. , Evans, M.R. , Porwollik, S. , Vazquez‐Torres, A. , Jones‐Carson, J. , Troxell, B. et al (2007) FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica Serovar Typhimurium (ATCC 14028s). Journal of Bacteriology, 189, 2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier, L.‐C. and Sekulovic, O. (2013) Importance of prophages to evolution and virulence of bacterial pathogens. Virulence, 4, 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Pastor, L. , Puerta‐Fernández, E. and Casadesús, J. (2018) Bistability and phase variation in Salmonella enterica . Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms. [DOI] [PubMed] [Google Scholar]
- Gilcrease, E.B. and Casjens, S.R. (2018) The genome sequence of Escherichia coli tailed phage D6 and the diversity of Enterobacteriales circular plasmid prophages. Virology, 515, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlöf, D.L. , Kröger, C. , Owen, S.V. , Canals, R. , Lacharme‐Lora, L. , Wenner, N. et al (2018) Role of a single noncoding nucleotide in the evolution of an epidemic African clade of Salmonella . Proceedings of the National Academy of Sciences, 115, E2614–E2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull, G.F. (2015) Dark matter of the biosphere: the amazing world of bacteriophage diversity. Journal of Virology, 89, 8107–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull, G.F. , Jacobs‐Sera, D. , Lawrence, J.G. , Pope, W.H. , Russell, D.A. , Ko, C.‐C. et al (2010) Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. Journal of Molecular Biology, 397, 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, S.B. , Cota, I. , Ducret, A. , Aussel, L. and Casadesús, J. (2012) Adaptation and preadaptation of Salmonella enterica to Bile. PLoS Genetics, 8, e1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero‐Fresno, A. , Espinel, I. C. , Spiegelhauer, M. R. , Guerra, P. R. , Andersen, K. W. , and Olsen, J. E. (2018) The homolog of the gene bstA of the BTP1 Phage from Salmonella enterica Serovar Typhimurium ST313 is an antivirulence gene in Salmonella enterica Serovar Dublin. Infection and Immunity, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero‐Fresno, A. , Wallrodt, I. , Leekitcharoenphon, P. , Olsen, J.E. , Aarestrup, F.M. and Hendriksen, R.S. (2014) The role of the st313‐td Gene in virulence of Salmonella Typhimurium ST313. PLoS One, 9, e84566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko‐Shalev, T. , Odenheimer‐Bergman, A. , Elgrably‐Weiss, M. , Ben‐Zvi, T. , Govindarajan, S. , Seri, H. et al. (2016) Gifsy‐1 prophage IsrK with dual function as small and messenger RNA modulates vital bacterial machineries. PLoS Genetics, 12, e1005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T.D. and Slauch, J.M. (2001) Characterization of grvA, an antivirulence gene on the gifsy‐2 phage in Salmonella enterica serovar typhimurium. Journal of Bacteriology, 183, 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Zhao, S. , Pettengill, J. , Luo, Y. , Monday, S.R. , Abbott, J. et al (2014) Comparative genomic analysis and virulence differences in closely related Salmonella enterica serotype heidelberg isolates from humans, retail meats, and animals. Genome Biology and Evolution, 6, 1046–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaslow, S.L. , Gibbs, K.D. , Fricke, W.F. , Wang, L. , Pittman, K.J. , Mammel, M.K. et al (2018) Salmonella activation of STAT3 signaling by SarA effector promotes intracellular replication and production of IL‐10. Cell Reports, 23, 3525–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal, F. and Dennis, J.J. (2015) Burkholderia cepacia complex phage‐antibiotic synergy (PAS): antibiotics stimulate lytic phage activity. Applied and Environmental Microbiology, 81, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, J. and Jain, S.K. (2012) Role of antigens and virulence factors of Salmonella enterica serovar Typhi in its pathogenesis. Microbiological Research, 167, 199–210. [DOI] [PubMed] [Google Scholar]
- Kim, M. and Ryu, S. (2013) Antirepression system associated with the life cycle switch in the temperate podoviridae phage SPC32H. Journal of Virolog, 87, 11775–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Ryu, K. , Biswas, D. and Ahn, J. (2014) Survival, prophage induction, and invasive properties of lysogenic Salmonella Typhimurium exposed to simulated gastrointestinal conditions. Archives of Microbiology, 196, 655–659. [DOI] [PubMed] [Google Scholar]
- Kintz, E. , Davies, M.R. , Hammarlöf, D.L. , Canals, R. , Hinton, J.C.D. and van der Woude, M.W. (2015) A BTP1 prophage gene present in invasive non‐typhoidal Salmonella determines composition and length of the O‐antigen of the lipopolysaccharide. Molecular Microbiology, 96, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar, B. , Hammerl, J.A. , Kienöl, M. , Heinig, M.L. , Sperling, N. , Dinh Thanh, M. et al (2017) Acquisition of virulence factors in livestock‐associated MRSA: lysogenic conversion of CC398 strains by virulence gene‐containing phages. Scientific Reports, 7, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger, C. , Colgan, A. , Srikumar, S. , Händler, K. , Sivasankaran, S.K. , Hammarlöf, D.L. et al (2013) An infection‐relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host & Microbe, 14, 683–695. [DOI] [PubMed] [Google Scholar]
- Kropinski, A.M. , Kovalyova, I.V. , Billington, S.J. , Patrick, A.N. , Butts, B.D. , Guichard, J.A. et al (2007) The genome of epsilon15, a serotype‐converting, Group E1 Salmonella enterica‐specific bacteriophage. Virology, 369, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire, S. , Figueroa‐Bossi, N. and Bossi, L. (2011) Bacteriophage crosstalk: coordination of prophage induction by trans‐acting antirepressors. PLoS Genetics, 7, e1002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévi‐Meyrueis, C. , Monteil, V. , Sismeiro, O. , Dillies, M.‐A. , Kolb, A. , Monot, M. et al (2015) Repressor activity of the RpoS/σS‐dependent RNA polymerase requires DNA binding. Nucleic Acids Research, 43, 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M. , Truesdell, S. , Ramakrishnan, T. and Bronson, M.J. (1975) Dual control of lysogeny by bacteriophage P22: an antirepressor locus and its controlling elements. Journal of Molecular Biology, 91, 421–438. [DOI] [PubMed] [Google Scholar]
- Lewis, M. (2011) A tale of two repressors – a historical perspective. Journal of Molecular Biology, 409, 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, A. , Amarir‐Bouhram, J. , Faure, G. , Petit, M.‐A. and Guerois, R. (2010) Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Research, 38, 3952–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini, S. , Rowley, G. , Goldberg, M.D. , Hurd, D. , Harrison, M. and Hinton, J.C.D. (2006) H‐NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathogens, 2, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menouni, R. , Hutinet, G. , Petit, M.‐A. and Ansaldi, M. (2015) Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiology Letters, 362, 1–10. [DOI] [PubMed] [Google Scholar]
- Mirold, S. , Rabsch, W. , Tschäpe, H. and Hardt, W.D. (2001) Transfer of the Salmonella type III effector sopE between unrelated phage families. Journal of Molecular Biology, 312, 7–16. [DOI] [PubMed] [Google Scholar]
- Nadeem, A. and Wahl, L.M. (2017) Prophage as a genetic reservoir: promoting diversity and driving innovation in the host community. Evolution: International Journal of Organic Evolution, 71, 2080–2089. [DOI] [PubMed] [Google Scholar]
- Navarre, W.W. , Halsey, T.A. , Walthers, D. , Frye, J. , McClelland, M. , Potter, J.L. et al (2005) Co‐regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Molecular Microbiology, 56, 492–508. [DOI] [PubMed] [Google Scholar]
- Navarre, W.W. , McClelland, M. , Libby, S.J. and Fang, F.C. (2007) Silencing of xenogeneic DNA by H‐NS – facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes & Development, 21, 1456–1471. [DOI] [PubMed] [Google Scholar]
- Navarre, W.W. , Porwollik, S. , Wang, Y. , McClelland, M. , Rosen, H. , Libby, S.J. et al (2006) Selective silencing of foreign DNA with low GC content by the H‐NS protein in Salmonella . Science, 313, 236–238. [DOI] [PubMed] [Google Scholar]
- Nedialkova, L.P. , Sidstedt, M. , Koeppel, M.B. , Spriewald, S. , Ring, D. , Gerlach, R.G. et al (2015) Temperate phages promote colicin‐dependent fitness of Salmonella enterica serovar Typhimurium. Environmental Microbiology, 18, 1591–1603. [DOI] [PubMed] [Google Scholar]
- Oliveira, P.H. , Touchon, M. , Cury, J. and Rocha, E.P.C. (2017) The chromosomal organization of horizontal gene transfer in bacteria. Nature Communications, 8, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, S.V. , Wenner, N. , Canals, R. , Makumi, A. , Hammarlöf, D.L. , Gordon, M.A. et al (2017) Characterization of the prophage repertoire of African Salmonella Typhimurium ST313 reveals high levels of spontaneous induction of novel phage BTP1. Frontiers in Microbiology, 8, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilar, A.V.C. , Reid‐Yu, S.A. , Cooper, C.A. , Mulder, D.T. and Coombes, B.K. (2012) GogB is an anti‐inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLoS Pathogens, 8, e1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik, S. , Wong, R.M.‐Y. and McClelland, M. (2002) Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proceedings of the National Academy of Sciences, 99, 8956–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich, L. , Sigal, N. , Borovok, I. , Nir‐Paz, R. and Herskovits, A.A. (2012) Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell, 150, 792–802. [DOI] [PubMed] [Google Scholar]
- Ranade, K. and Poteete, A.R. (1993) Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. Journal of Bacteriology, 175, 4712–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke, C. , Schwientek, P. , Sczyrba, A. , Ivanova, N.N. , Anderson, I.J. , Cheng, J.‐F. et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature, 499, 431–437. [DOI] [PubMed] [Google Scholar]
- Roux, S. , Enault, F. , Hurwitz, B.L. and Sullivan, M.B. (2015a) VirSorter: mining viral signal from microbial genomic data. PeerJournal, 3, e985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, S. , Hallam, S.J. , Woyke, T. and Sullivan, M.B. (2015b) Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. eLife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, R.T. , Pan, J. , Hopper, P. , Hehir, K. , Brown, J. and Poteete, A.R. (1981) Primary structure of the phage P22 repressor and its gene c2. Biochemistry, 20, 3591–3598. [DOI] [PubMed] [Google Scholar]
- Shah, J. , Desai, P.T. and Weimer, B.C. (2014) Genetic mechanisms underlying the pathogenicity of cold‐stressed Salmonella enterica Serovar Typhimurium in cultured intestinal epithelial cells. Applied and Environmental Microbiology, 80, 6943–6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin, K.E. , Brumby, A.M. and Egan, J.B. (1998) The tum protein of coliphage 186 is an antirepressor. Journal of Biological Chemistry, 273, 5708–5715. [DOI] [PubMed] [Google Scholar]
- Slauch, J.M. , Lee, A.A. , Mahan, M.J. and Mekalanos, J.J. (1996) Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O‐antigen: oafA is a member of a family of integral membrane trans‐acylases. Journal of Bacteriology, 178, 5904–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Knirel, Y.A. , Wang, J. , Luo, X. , Senchenkova, S.N. , Lan, R. et al (2014) Serotype‐converting bacteriophage SfII encodes an acyltransferase protein that mediates 6‐O‐acetylation of GlcNAc in Shigella flexneri O‐antigens, conferring on the host a novel O‐antigen epitope. Journal of Bacteriology, 196, 3656–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind, M.M. and Botstein, D. (1980) Superinfection exclusion by lambda prophage in lysogens of Salmonella typhimurium. Virology, 100, 212–216. [DOI] [PubMed] [Google Scholar]
- Susskind, M.M. , Botstein, D. and Wright, A. (1974) Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium. III. Failure of superinfecting phage DNA to enter sieA+ lysogens. Virology, 62, 350–366. [DOI] [PubMed] [Google Scholar]
- Suttle, C.A. (2007) Marine viruses – major players in the global ecosystem. Nature Reviews Microbiology, 5, 801–812. [DOI] [PubMed] [Google Scholar]
- Tomljenovic‐Berube, A.M. , Henriksbo, B. , Porwollik, S. , Cooper, C.A. , Tuinema, B.R. , McClelland, M. et al (2013) Mapping and regulation of genes within Salmonella pathogenicity island 12 that contribute to in vivo fitness of Salmonella enterica Serovar Typhimurium. Infection and Immunity, 81, 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon, M. and Rocha, E.P.C. (2010) The small, slow and specialized CRISPR and anti‐CRISPR of Escherichia and Salmonella . PloS One, 5, e11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, S.C. and Galán, J.E. (2000) Complex function for SicA, a Salmonella enterica serovar typhimurium type III secretion‐associated chaperone. Journal of Bacteriology, 182, 2262–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivero, A. , Baños, R.C. , Mariscotti, J.F. , Oliveros, J.C. , García‐del Portillo, F. , Juárez, A. et al (2008) Modulation of horizontally acquired genes by the Hha‐YdgT proteins in Salmonella enterica serovar Typhimurium. Journal of Bacteriology, 190, 1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonaesch, P. , Sellin, M.E. , Cardini, S. , Singh, V. , Barthel, M. and Hardt, W.‐D. (2014) The Salmonella Typhimurium effector protein SopE transiently localizes to the early SCV and contributes to intracellular replication. Cellular Microbiology, 16, 1723–1735. [DOI] [PubMed] [Google Scholar]
- Wernicki, A. , Nowaczek, A. and Urban‐Chmiel, R. (2017) Bacteriophage therapy to combat bacterial infections in poultry. Virology Journal, 14, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple, F.W. , Hou, E.F. and Hochschild, A. (1998) Amino acid‐amino acid contacts at the cooperativity interface of the bacteriophage lambda and P22 repressors. Genes & Development, 12, 2791–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, M.J. , Ching, K.H. and Heffron, F. (2000) Salmonella SsrB activates a global regulon of horizontally acquired genes. Molecular Microbiology, 36, 749–761. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Soderholm, A. , Lung, T.W.F. , Giogha, C. , Hill, M.M. , Brown, N.F. et al (2015) SseK3 is a Salmonella effector that binds TRIM32 and modulates the host’s NF‐κB signalling activity. PLoS One, 10, e0138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H. , Ansong, C. , Adkins, J.N. and Heffron, F. (2011) Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infection and Immunity, 79, 2182–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder, N.D. and Lederberg, J. (1952) Genetic exchange in Salmonella . Journal of Bacteriology, 64, 679–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Q.‐H. , Li, Q.‐H. , Zhu, H.‐Y. , Feng, Y. , Li, Y.‐G. , Johnston, R.N. et al (2010) SPC‐P1: a pathogenicity‐associated prophage of Salmonella paratyphi C. BMC Genomics, 11, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]


