Abstract
NrCAM,a neuronal cell adhesion molecule in the L1 family of the immunoglobulin superfamily, is subjected to extensively alternative splicing and involved in neural development and some disorders. The aim of this study was to explore the role of Nrcam mRNA alternative splicing in neuropathic pain. A next generation RNA sequencing analysis of dorsal root ganglions (DRGs) showed the differential expression of two splicing variants of Nrcam, Nrcam+10 and Nrcam−10, in the injured DRG after the fourth lumbar spinal nerve ligation (SNL) in mice. SNL increased the exon 10 insertion, resulting in an increase in the amount of Nrcam+10 and a corresponding decrease in the level of Nrcam−10 in the injured DRGAn antisense oligonucleotide that specifically targeted exon 10 of Nrcam gene (Nrcam AON) repressed RNA expression of Nrcam+10 and increased RNA expression of Nrcam−10 in in vitro DRG cell culture. DRG microinjection or intrathecal injection of Nrcam AON attenuated SNL-induced mechanical allodynia, thermal hyperalgesia, or cold allodynia. Nrcam AON also relieved SNL- and chronic compression of DRG (CCD)-induced the maintenance if pain hypersensitivities in male and female mice.
Perspective: We conclude that the relative levels of alternatively spliced Nrcam variants are critical for neuropathic pain genesis. Targeting Nrcam alternative splicing via the antisense oligonucleotides may be a new potential avenue in neuropathic pain management.
Keywords: NrCAM, Alternative splicing, Dorsal root ganglion, Spinal nerve ligation, Neuropathic pain
Neuropathic pain is a very common clinical disorder, usually caused by nerve injury or accompanying some diseases (such as diabetes). It reduces the quality of life in patients due to a lack of effective analgesic strategies. The altered gene/protein expression in primary sensory neurons following peripheral nerve injury has been demonstrated as the key component of the pathophysiology of neuropathic pain16, 17, 29, 40, 41. A next generation RNA sequencing analysis revealed the differential expression of thousands of genes in the injured DRG caused by spinal nerve ligation (SNL)35. Further RNA alternative splicing analysis showed that many genes including Nrcam gene [encoding neuronal cell adhesion molecule (NrCAM)] underwent RNA alternative splicing changes in the injured DRG following SNL35. Whether these changes contribute to neuropathic pain is still elusive.
NrCAM is a neuronal cell adhesion molecule in the L1 family of the immunoglobulin superfamily. Full-length Nrcam cDNA encodes NrCAM, a single-transmembrane-domain protein, containing an N-terminal signal peptide, six immunoglobulin (Ig) domains, four to five fibronectin III repeats, a transmembrane domain, and a C-terminal cytoplasmic domain with tyrosine kinase phosphoacceptor sites8. Nrcam RNA is subject to extensive alternative splicing events. Multiple NrCAM isoforms, due to different RNA splicing variants, have been reported previously in chicken, rat, mouse, and human5, 7, 8, 32. These splicing processes include the 19 amino acid insertion located between Ig domains II and III encoded by exon 10, the 6 amino acid insertion located between the signal peptide and the first Ig domain encoded by exon 5, the 10 amino acid insertion located between the Ig domain VI and the first fibronectin III (FnIII) domain encoded by exon 198. NrCAM plays a wide variety of roles in neural development including cell proliferation and differentiation, axon growth and guidance, and synapse formation24. It was reported previously that alterations in NrCAM structure/expression, and its alternatively splicing variants, were associated with autism, drug addiction, and tumor progression8, 20, 28, 32. However, the function of the changed alternative splicing of NrCAM in the injured DRG under neuropathic pain conditions has not yet been explored.
In this study, we analyzed RNA sequence data from both sham and SNL DRGs using a probabilistic framework model implemented in the MISO (the mixture-of-isoforms) software. Our findings revealed that SNL led to a marked change in alternative splicing of NrCAM in the injured DRG. Suppression of exon 10 through DRG microinjection or intrathecal injection of specific splice-site antisense oligonucleotides (ASO) targeting exon 10 reversed the SNL-induced this change and attenuated SNL-induced pain hypersensitivities in both development and maintenance stage of neuropathic pain without sex-bias. Therefore, the alternative splicing of Nrcam exon 10 contributes to the peripheral mechanism of neuropathic pain. The ASO targeting Nrcam exon 10 alternative splicing may be beneficial for neuropathic pain patients.
Methods
Animal preparations
Adult male C57BL/6 mice were used in RNA sequencing experiments and adult male and female CD1 wild-type mice for remaining in vivo study. Three to four weeks old CD1 mice were used for DRG cell culture. All animals were kept in a standard 12-h light/dark cycle, with water and food available ad libitum. All procedures in this study were approved by the Animal Care and Use Committee at Rutgers New Jersey Medical School and are consistent with the ethical guidelines of the US National Institutes of Health and the International Association for the Study of Pain. All efforts were made to minimize animal suffering and to reduce the number of animals in this study. To eliminate bias, all of the experimenters were blind to treatment information.
RNA sequencing and splicing variants analysis
The information on RNA sequencing (Illumina, San Diego, CA) has been described previously35. Alternative splicing analysis was performed using MISO (mixture-of-isoforms) software9, 10, which used a probabilistic model to quantify the expression level of alternatively spliced genes from RNA-Seq data, and identified differentially regulated isoforms or exons across samples.
Design for Nrcam exon 10 antisense oligonucleotides (ASO)
Nrcam ASO was designed based on the previous reports4, 12. Nrcam ASO targeted the 5’ and 3’ splice-sites of exon 10, as well as the sequence adjacent to the 3’ splice-site of intron 9 −10, to block their recognition by the spliceosome. The designed Nrcam ASO sequence is UGUAACUCACCUGAAAUAAACAGAAUAUCAUGAAGAGGGCACAGAAGUAG. The ASO were 2′-O-methyl RNA oligonucleotides. The backbone of ASO was modified with five phosphorothioates on the 5′and 3′ends (Table 1). The scrambled ASO was designed based on the Nrcam ASO sequence without modifications of phosphorothioates. The designed sequences were sent to IDT (Integrated DNA Technologies, Inc. Coralville, Iowa) for production. The injected ASO solution was prepared through mixing 20% Glucose, TurboFect transfection reagent (ThermoFisher Scientific) and ASO by 15:5:1 in advance.
Table 1.
Sequences of Nrcam antisense oligonucleotide (ASO) or Scramble ASO.
| Name | Sequence |
|---|---|
| Scramble ASO | GCAUGUAAUUCCGGUAGCUACUCGAUUAGCCUAACAAUCC |
| Nrcam ASO | U*G*U*A*A*C*UCACCUGAAAUAAACAGAAUAUCAUGAAGAGGGCACAGA*A*G*U*A*G |
phosphorothioate modification.
Neuropathic pain models
Unilateral L4 spinal nerve ligation (SNL)16, 17 and chronic compression of DRG surgery (CCD)6, 34 were carried out as described previously. For SNL surgery, left L4 spinal nerve was exposed, ligated by 7–0 silk and transected at the distal site with a scissor under 2–3% isoflurane anesthesia. For CCD surgery, the intervertebral foramina of L3 and L4 were exposed and an L-shaped steel rod, 2 mm in length and 0.3 mm in diameter, was implanted into each foramen to compress corresponding DRGs.
Lumbar puncture
Lumbar puncture was performed as reported previously18. The mice were anesthetized with 2% isoflurane, and a 50 ml tube was placed under the belly of the mouse to elevate the spine. An insulin syringe (needle size: 0.3×8 mm) was injected into the subarachnoid space via L4-L5 interspace. A quick tail flick indicated that the needle was in the right place.
DRG microinjection
DRG microinjection was carried out as described with minor modification16, 41. The L4 DRG was exposed after removing the L4 articular processes. Nrcam ASO, scrambled ASO or vehicle solution (0.01 M phosphate buffered saline, 1 μl) was then injected into unilateral L4 DRG with a glass micropipette connected to a Hamilton syringe for 5 minutes. The surgical field was irrigated with sterile saline, and the skin incision closed with wound clips. Three days later, SNL or sham surgery was performed in the mice without signs of paresis or other abnormalities.
Behavioral tests
Mechanical behavioral test was carried out as described3, 16. Briefly, each mouse was placed in a Plexiglas chamber on an elevated mesh screen. Two calibrated von Frey filament (0.07g and 0.40 g; Stoelting Co., Wood Dale, IL, USA) was applied to the hind paw successively and each was applied 10 times. Each stimulation was sustained for approximately 1 s. The number of paw withdrawals in each of the 10 trials was expressed as a percent paw withdrawal frequency (PWF) [(number of paw withdrawals/10 trials) × 100 = % response frequency], and this percentage was used as an indication of the number of paw withdrawal.
Thermal behavioral test was carried out as described3, 16, 41. In brief, each mouse was placed in a Plexiglas chamber on a glass plate. A radiant heat from a Model 336 Analgesic Meter (IITC Inc./Life Science Instruments, Woodland Hills, CA, USA) was applied by aiming the beam of light through a hole in the light box through the glass plate to the middle of the plantar surface of each hind paw. When the animal lifted its foot, the light beam was turned off. The paw withdrawal latency was defined as the length of time between the start of the light beam and the foot lift. Each trial was repeated five times at 5-min intervals for each side. A cut-off time of 20 s was used to avoid tissue damage to the hind paw.
Cold behavioral test was carried out as reported previously 3,16,41. Each mouse was put on the cold aluminum plate on ice. The temperature was continuously kept at 0 °C. The paw withdrawal latencies to noxious cold (0 °C) were the length of time between the placement of the hind paw on the plate and the animal jumping, with or without paw licking or flinching16. Each trial was repeated three times at 15-min intervals for the ipsilateral paw. A cutoff time of 20 s was used to avoid tissue damage.
Locomotor function was tested as previously described27. Three reflex tests were carried out as follows. (1) Placing reflex: The mouse was held with the hind limbs slightly lower than the forelimbs, and the dorsal surfaces of the hind paws were brought into contact with the edge of a table. The experimenter recorded whether the hind paws were placed on the table surface reflexively; (2) Grasping reflex: The mouse was placed on a wire grid, and the experimenter recorded whether the hind paws grasped the wire on contact; (3) Righting reflex: The mouse was placed on its back on a flat surface, and the experimenter noted whether it immediately assumed the normal upright position. Scores for placing, grasping, and righting reflexes were based on counts of each normal reflex exhibited in five trials.
DRG cell culture and transfection of antisense oligonucleotides
Primary DRG cell cultures were carried out as described16, 17, 41. Briefly, 3–4 weeks old CD1 mice were euthanized after isoflurane anesthesia. All DRGs were collected in cold Neurobasal A Medium (Gibco/ThermoFisher Scientific, Grand Island, NY) with 10% fetal bovine serum (JR Scientific, Woodland, CA), 100 units/ml Penicillin, 100μg/ml Streptomycin (Quality Biological, Gaithersburg, MD) and then treated with enzyme solution (5 mg/ml dispase, 1 mg/ml collagenase type I in Hanks’ balanced salt solution (HBSS) without Ca2+ and Mg2+ (Gibco/ThermoFisher Scientific). After trituration and centrifugation, the dissociated cells were resuspended in mixed Neurobasal Medium and plated in a six-well plate coated with 100μg/ml poly-D-lysine (Sigma, St. Louis, MO). The cells were incubated at 95% O2, 5% CO2, and 37 °C. Nrcam exon 10 ASO or scrambled ASO was added to each 1 ml-well Opti-MEM Reduced Serum Medium by mixing with Lipofectamine 2000 Reagent (ThermoFisher Scientific). Cultured cells were collected 2 days later for RT-PCR analysis.
Reverse transcription (RT)-PCR
The injected DRGs from two mice were pooled together to obtain enough RNA. Total RNA from mouse DRG was extracted by an RNAeasy™ Animal RNA Isolation Kit with Spin Column (Beyotime, Shanghai, China). Total RNA was reverse-transcribed using the ThermoScript reverse transcriptase (Invitrogen/ThermoFisher Scientific) and oligo (dT) primer. Each sample was run in triplicate in a 10 μL reaction with 10 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), 20 ng of cDNA, and 250 nM of forward and reverse primers. Primers flanking alternatively spliced exon 10 were designed based on a previous report30. All primer sequences were listed in Table 2 and Fig. 3A. Total Nrcam mRNA was detected by forward (F) and reverse (R) pair primers (NrcamT), which amplified the fragment in exon 9. Nrcam+10 mRNA was amplified by forward (F) and reverse (R) pair primers (Nrcam+10), which amplified the fragment in exon 10. Nrcam−10 mRNA was detected by forward (F) and reverse (R) pair primers (Nrcam−10), which amplified the fragment including the junction between exon 9 and exon 11. To detect both Nrcam+10 and Nrcam−10 mRNAs through a 2% gel electrophoresis, forward (F) and reverse (R) pair primers (Nrcam+10&−10) were used, in which the reverse primer (Nrcam+10&−10) amplified the fragment in exon 11. All primers were purchased from Integrated DNA Technologies. For quantitative RT-PCR (qRT-PCR), the PCR amplification consisted of an initial 3-min incubation at 95°C, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. The procedure were set up and run in a BIO-RAD CFX96 real-time PCR system. All data were normalized to Tuba1a, an internal control. Ratios of ipsilateral-side mRNA levels to contralateral-side mRNA levels were calculated using the ΔCt method (2−ΔΔCt). For gel electrophoresis to visualize the size and intensity of the RT-PCR product, the PCR amplification cycles were reduced to 30 and a 2% agarose gel was used.
Table 2.
Primers.
| Species | mRNA name | Primer name | Primer sequence | Primer location | |
|---|---|---|---|---|---|
| Mouse | Total Nrcam | F | GTTTCCCAAGGCCTAAATGG | Exon 9 | |
| R(NrcamT) | AATAGGCTGCTTCTGCTGGA | Exon 9 | |||
| Nrcam+10 | F | GTTTCCCAAGGCCTAAATGG | Exon 9 | ||
| R(Nrcam+10) | CACCATAAAACTCAGTGTCACT | Exon 9 | |||
| Nrcam−10 | F | GTTTCCCAAGGCCTAAATGG | Exon9 | ||
| R(Nrcam−10) | ACTAGATTTAGCTGAAATCACC | The junction of exon 9 and 11 | |||
| Nrcam+10& Nrcam−10 | F | GTTTCCCAAGGCCTAAATGG | Exon 9 | ||
| R(Nrcam+10&−10) | TTTCATTGCCCTCTGGAGTT | Exon 11 | |||
| Tubala | F(Tubala) | GTGCATCTCCATCCATGTTG | |||
| R(Tubala) | GTGGGTTCCAGGTCTACGAA | ||||
| Human | Nrcam+10& Nrcam−10 | F(hNrcam+10&- 10) | TTTCCAATGTCCTCCCAGAG | ||
| R(hNrcam+10&- 10) | CTTGCATTGCCTTCTGGAGT | ||||
| Gapdh | F(hGapdh) | TCACCATCTTCCAGGAGCG | |||
| R(hGapdh) | CTGCTTCACCACCTTCTTGA | ||||
F: forward primer; R: Reverse primer.
Figure 3. SNL led to an increase in Nrcam+10 mRNA expression in the injured DRG.
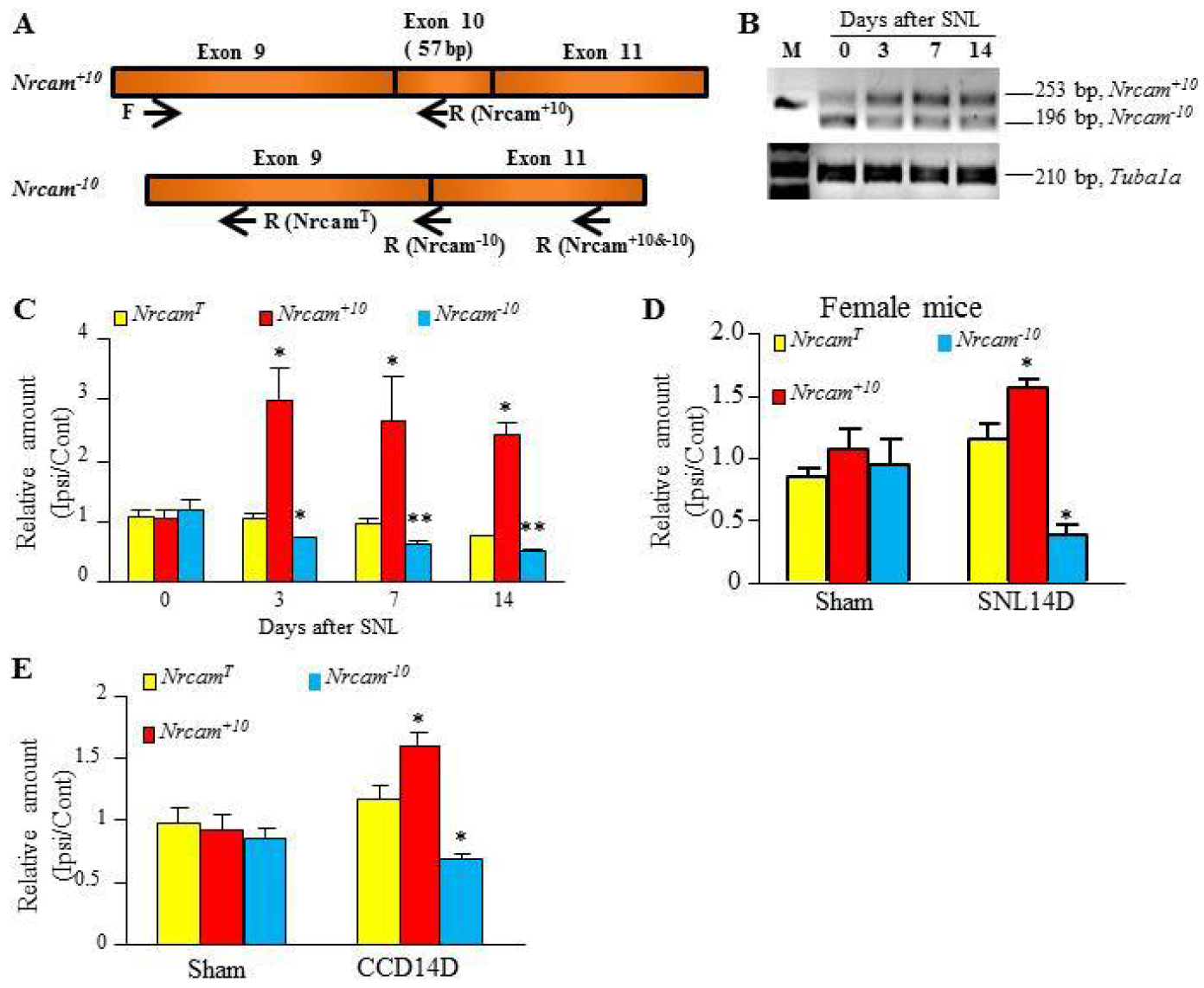
(A) Schematic graph showing Nrcam mRNA splicing variants Nrcam+10 and Nrcam−10 and the position of primers used in PCR reactions and quantitative reverse-transcriptase PCR (qRT-PCR) to examine total Nrcam mRNA and its variants. Their sequences are described in Table 2. (B) Typical photographs showing the increased Nrcam+10 and the decreased Nrcam−10 transcripts in the injured DRG after SNL or sham surgery by PCR with RT primer and gel electrophoresis. (C) The significant increases in the level of Nrcam+10 mRNA and the corresponding decreases in the amount of Nrcam−10 mRNA in the injured L4 DRG at 3, 7 and 14 days after SNL compared with sham group. N = 6 – 8 mice (3 – 4 biological repeats)/group. One way ANOVA followed by Tukey post hoc test. F(3,13) = 6.30 for Nrcam+10. F(3,13) = 9.27 for Nrcam−10. *P < 0.05, **P < 0.01 vs the corresponding control group (0 day). (D) The changes in the amounts of total Nrcam, Nrcam+10, and Nrcam−10 mRNA in the injured DRG after SNL or sham surgery in female mice. An increase in the level of Nrcam+10 mRNA, a decrease in the amount of Nrcam−10 mRNA, and no change in total Nrcam mRNA level on day 14 were seen in the injured DRG after SNL surgery. N = 10 mice (5 biological repeats)/group. *P < 0.05 vs the sham group by paired t-test. (E) The changes in the amounts of total Nrcam, Nrcam+10, and Nrcam−10 mRNA in mouse chronic compression of DRG (CCD) model. An increase in the level of Nrcam+10 mRNA, a decrease in the amount of Nrcam−10 mRNA, and no change in total Nrcam mRNA "level were seen in the injured DRG on day 14 after CCD surgery. N = 12 mice (6 biological repeats)/group. *P < 0.05 vs the sham group by paired t-test.
Total RNA from huam DRG was purchased from Clontech Laboratories (Mountain View, CA). According to the datasheet from the vendor, these DRGs came from 16–65 years old male/female Caucasians who died due to accident. The RNA was amplified by the PCR amplification procedure with an initial 3-min incubation at 95°C, followed by 30 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. A 2% agarose gel was used to visualize the size and intensity of the RT-PCR product. Gapdh was served as the internal control.
Statistical analysis
The animals were randomly distributed into various treatment groups. The number of mice per group for behavior tests is 8 to 9 and for other experiments 3 to 4. All of the results are expressed as means ± S.E.M. The data were statistically analyzed with two-tailed, paired Student’s t-test and a one-way or two-way repeated measure (RM) ANOVA. When ANOVA showed a significant difference, the post hoc Tukey test was performed to pairwise compare the difference between each mean (SigmaPlot 12.5, San Jose, CA). P values less than 0.05 were considered statistically significant.
Results
Spinal nerve ligation alters Nrcam RNA alternative splicing in the injured DRG
Our RNA sequencing data revealed that the Nrcam mRNA level decreased by 5.67% in the injured DRG from the SNL group compared to that from the sham group on day 7 after SNL [−1.11 folds; Sham (19.59) vs. SNL (17.64), P = 0.01]35. We applied the mixture-of-isoforms (MISO) software to estimate expression of alternatively spliced exons and isoforms, to assess the confidence in these estimates, and to detect differentially regulated exons or isoforms. There was a significant increase in the reading of a single exon of Nrcam in the injured DRG from the SNL group (Fig. 1). This exon is located at Chr12 (from 44545379 to 44545435) and includes 57 bases (5’-TGGATGAATTGAATGACACTATAGCTGCTAATTTGAGTGACACTGAGTTTTATGGTG-3’) encoding amino acids from 235 to 253 (VDELNDTIAANLSDTEFYGA) of NrCAM. By comparing the sequences reported in previous studies7, 8, 32, this alternative splicing occurs in exon 10 of Nrcam gene from human, mouse and rat.
Figure 1. RNA Seq data showed the changes in Nrcam mRNA and exon 10 reading in the injured DRG following spinal nerve ligation (SNL).

RNA-seq read densities in the sham-operated and SNL DRGs. Ipsilateral L4 DRGs from four mice were pooled together. N = 12 mice (3 biological repeats)/group. The exon 10 is framed in red.
The inclusion reads for exon 10 (X and Y) increased, while the exclusion reads for exon 10 (Z) decreased in the SNL group as compared with the sham group (P < 0.05, Figs. 2A, B). The Ψ value, which indicates ‘percentage spliced in’ denoting the fraction of mRNAs that represent the inclusion isoform9, increased significantly in the SNL group as compared with the sham group (P < 0.01, Figs. 2A, C). Thus, we concluded that there were at least two Nrcam RNA splicing variants in DRG, Nrcam RNA with exon 10 (Nrcam+10) and Nrcam RNA without exon 10 (Nrcam−10) (Fig. 3A). Nrcam−10 was dominant in naïve/sham DRGs, whereas Nrcam+10 was dominant in the SNL DRG.
Figure 2. SNL increased exon 10 insertion.
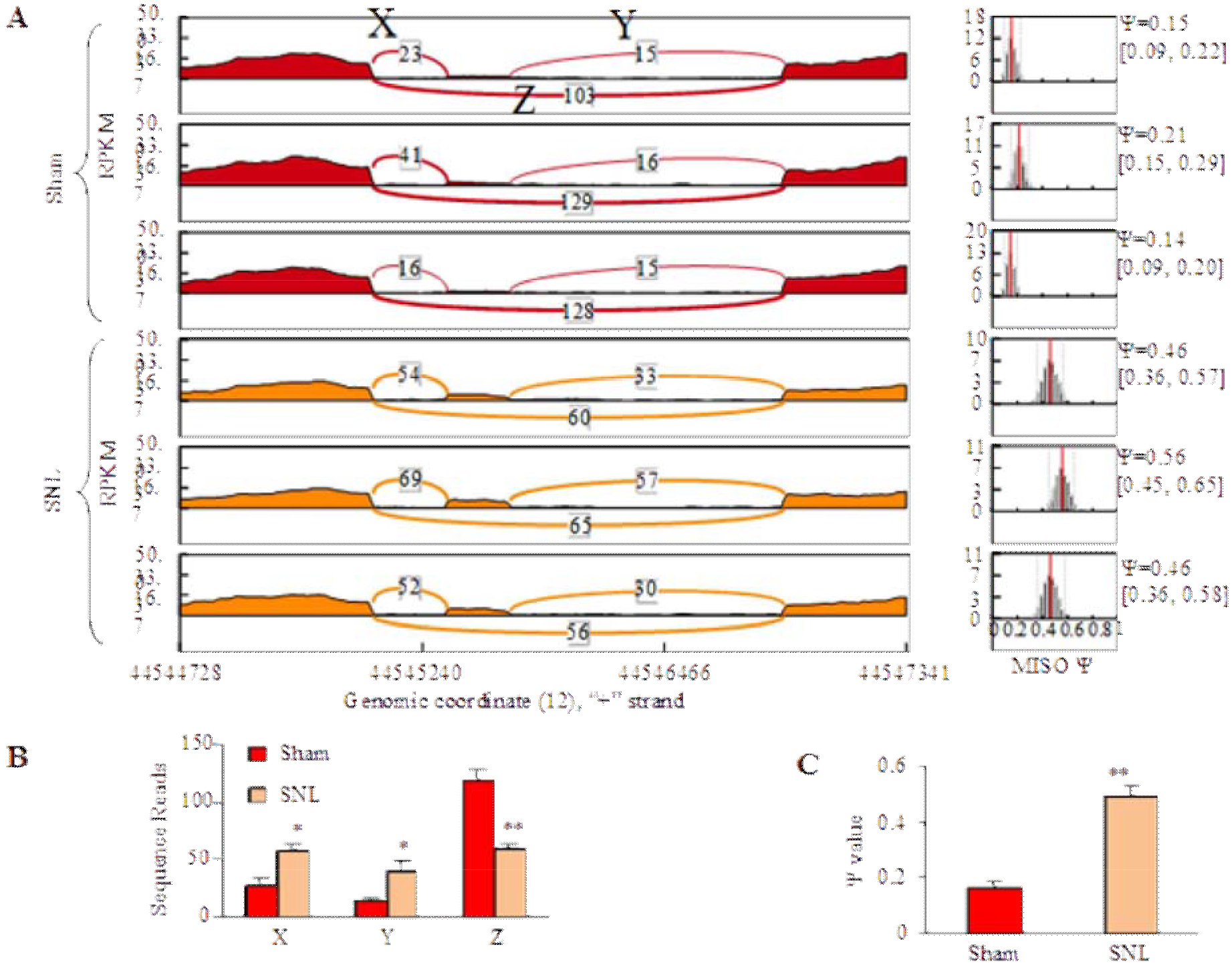
(A) MISO model showed inclusion reads (X and Y) and exclusion reads (Z) of Nrcam exon 10 for each sample from the sham group and the SNL group. (B) SNL significantly increased inclusion reads (X and Y) and decreased exclusion reads (Z) of Nrcam exon 10. N = 12 mice (3 biological repeats)/group. *P < 0.05, **P < 0.01 vs the sham group by paired t-test. (C) The Ψ value increased significantly in the SNL group. N = 12 mice (3 biological repeats)/group. **P < 0.01 vs the sham group by paired t-test.
We further carried out quantitative real-time PCR assay with specific primers for Nrcam+10 and Nrcam−10 variants, respectively, (Table 2 and Fig. 3A) and showed that the PCR product intensity of Nrcam+10 and Nrcam−10 mRNA displayed opposing changes after SNL (Fig. 3B). SNL significantly increased the level of Nrcam+10 mRNA and, correspondingly, reduced the amount of Nrcam−10 mRNA in the injured DRG on days 3, 7 and 14 after SNL (Figs. 3B, C). The levels of Nrcam+10 mRNA increased by 2.57-, 2.67-, and 2.44-fold on days 3, 7 and 14, respectively, after SNL compared to that in naïve mice (day 0. F (3, 13) = 6.30, P < 0.05). On the contrary, the levels of Nrcam−10 mRNA on days 3, 7, and 14 post-SNL decreased, respectively, by 31%, 38%, and 40% of the values from naïve mice (F (3, 13) = 9.27, P < 0.01) (Fig. 3B). Interestingly, the levels of total Nrcam mRNA showed a slight, but not significant, reduction after SNL (Fig. 3C, F(3, 13) = 2.18, P = 0.15). As showed in Fig. 3B, the fragment sizes of PCR product of Nrcam+10 and Nrcam−10 were 253 bp and 196 bp, respectively, as predicted (Fig. 3B). SNL-induced increase of Nrcam+10 mRNA level and decrease of Nrcam−10 mRNA were also observed in female mice (P < 0.05, Fig. 3D). Moreover, the changes in these two variants in the injured DRG were further confirmed in a chronic compression of DRG (CCD)-induced neuropathic pain model6, 36. CCD produced an increase in the level of Nrcam+10 mRNA and a corresponding reduction in the amount of Nrcam−10 mRNA in the compressed DRGs without a marked change in total Nrcam mRNA on day 14 after CCD (P < 0.05, Fig. 3E).
In addition, these two slicing variants were also detected in the trigeminal ganglion, heart and kidney (Fig. 4A). In the spinal cord, cortex, brainstem, hippocampus, cerebellum, and sciatic nerve, Nrcam+10, but not Nrcam−10, was detectable in mice (Figs. 4A, B). As expected, both variants were observed in human DRG (Fig. 4C).
Figure 4. Expression of Nrcam splicing variants in multiple tissues of mouse and in human dorsal root ganglion (DRG).
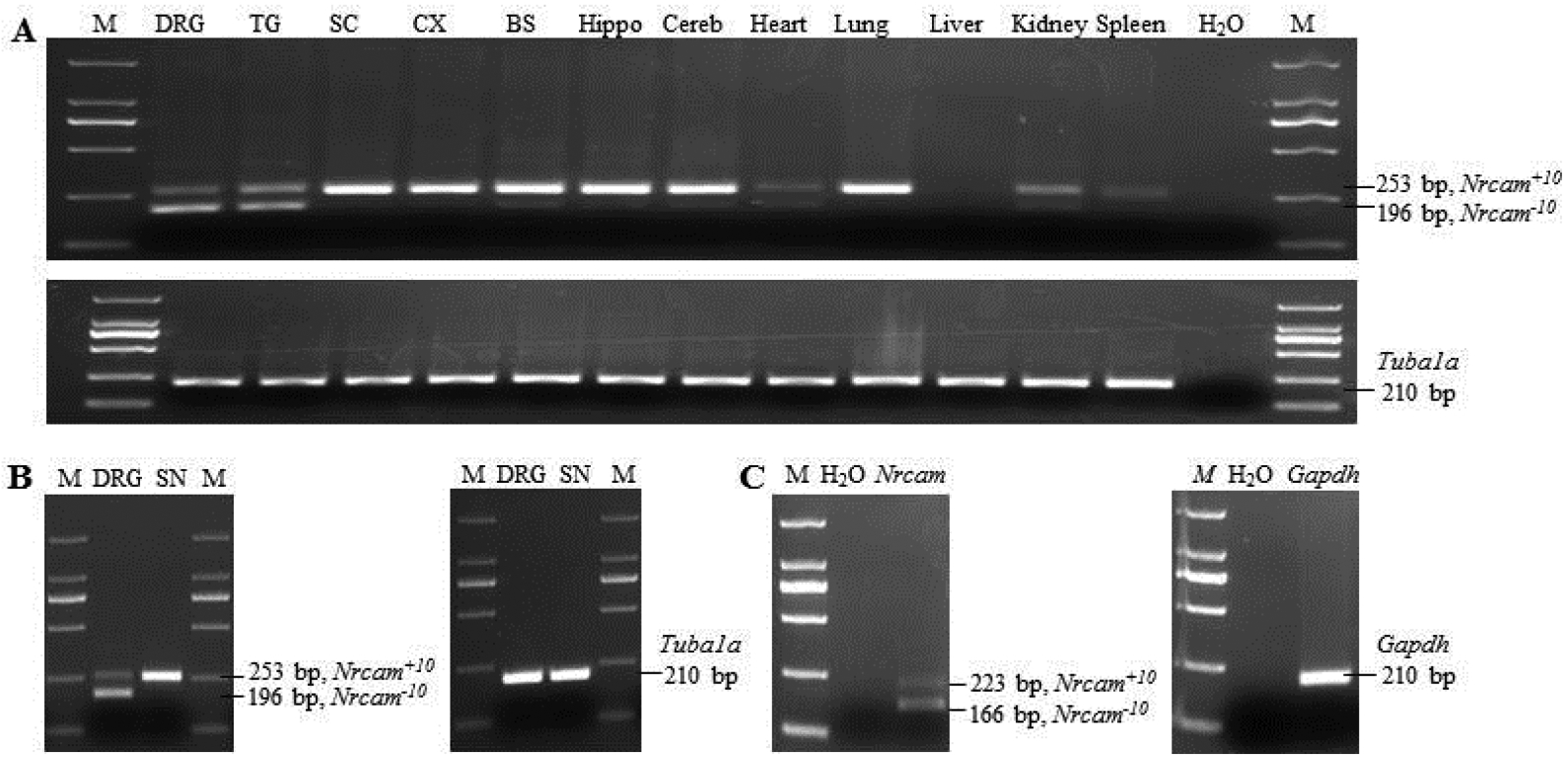
(A) Expression of Nrcam transcripts in mouse DRG, trigeminal ganglion (TG), spinal cord (SC), cortex (CX), brainstem (BS), hippocampus (Hippo), cerebellum (Cereb), heart, lung, liver, kidney, and spleen. Upper panel: Nrcam (Nrcam+10: 253 bp; Nrcam−10: 196 bp). Lower panel: Tubu1a (210 bp). (B) Expression of Nrcam transcripts in the DRG and sciatic nerve (SN). Left panel: Nrcam (long transcript: 253 bp; short transcript: 196 bp). Right panel: Tubu1a (210 bp). (C) Expression of Nrcam transcripts in human DRG. Left panel: human Nrcam (Nrcam+10: 223 bp; Nrcam−10: 166 bp). Right panel: human Gapdh (210 bp).
Antisense oligonucleotides (ASO) targeting the splice-sites of Nrcam exon 10 attenuates the development of SNL-induced pain hypersensitivities
In order to regulate exon 10 splicing expression, we designed splice-site 2’O-methyl ASO to specifically target the 5’ and 3’ splice-sites of exon 10, as well as the sequence adjacent to the 3’ splice-site of intron 9–10 (Table 1). Nrcam ASO could induce high levels of exon 10 skipping and expression of Nrcam−10. The scrambled ASO (Scr) was used as a control (Table 1). The effect of Nrcam ASO on exon 10 insertion was validated in the cultured DRG cells. As shown in Figure 5A, 50 nM Nrcam ASO did not affect total Nrcam RNA level (F(2, 8) = 3.26, P = 0.11), but significantly reduced Nrcam+10 expression level (F(2, 8) = 629.57, P < 0.001) and correspondingly increased the Nrcam−10 expression level (F(2, 8) = 191.11, P < 0.001). Compared to the naïve group, Nrcam ASO reduced the amount of Nrcam+10 RNA by 69% and increased the level of Nrcam−10 mRNA by 5.58-fold. This expressional shift indicates that Nrcam ASO blocks exon 10 insertion and consequently leads to more Nrcam−10 mRNA variant. As expected, the scrambled ASO had no such effect as compared to naïve group (Fig. 5A). Our findings indicate that the alternative splicing effect of Nrcam ASO is sequence-specific.
Figure 5. Intrathecal injection of Nrcam ASO relieved SNL-induced neuropathic pain.
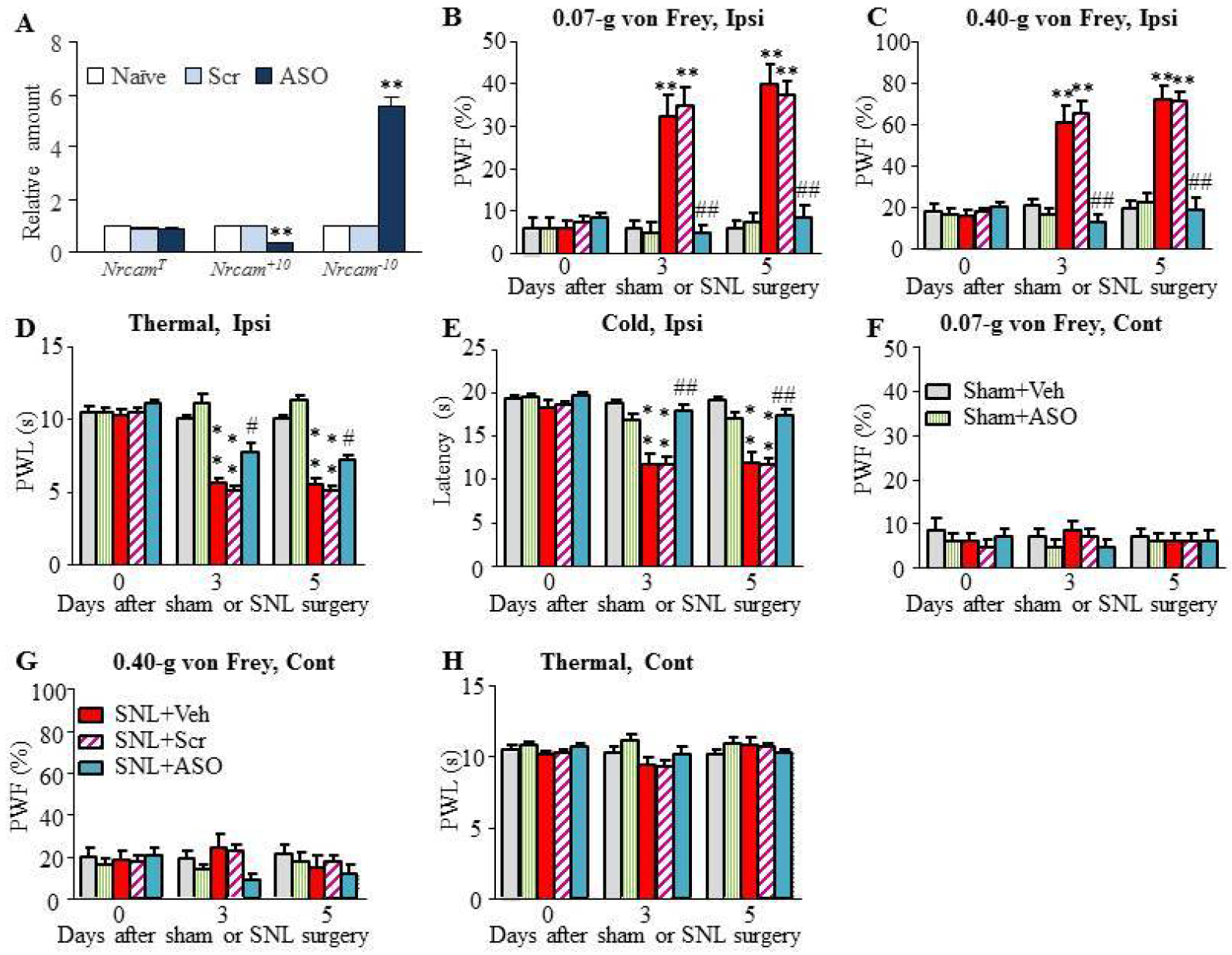
(A) Administration with Nrcam ASO had no effect on total Nrcam mRNA expression but decreased the level of Nrcam+10 mRNA and, correspondingly, increased the level of Nrcam−10 mRNA in the cultured DRG neurons. N = 6 mice (3 biological repeats)/group. One way ANOVA followed by Tukey post-hoc test. F = 629.57 for Nrcam+10, F (2, 8) (2, 8) = 191.11 for Nrcam−10. **P < 0.01 vs naïve or scramble (Scr) group. (B-H) Nrcam ASO (5 μM, 10 μl) were intrathecally injected through lumbar puncture 5 min before SNL or sham surgery and 1 day after SNL or sham surgery. The scrambled ASO (Scr) or normal saline (vehicle) served as the controls. Nrcam ASO, but not scrambled ASO and vehicle, attenuated SNL-induced decreases in paw withdrawal frequencies (PWFs) to 0.07-g (B) and 0.40-g (C) von Frey filaments stimuli and paw withdrawal latencies (PWLs) to thermal (D) or cold (E) stimulation on the ipsilateral side. Neither ASO nor vehicle had the effect on basal paw withdrawal responses to mechanical (F, G) and thermal (H) stimuli on the contralateral side. N = 8 mice/group. Two-way RM ANOVA (effect vs group × time interaction) followed by post hoc Tukey test. F (8, 119) = 9.78 (B), F (8, 119) = 14.84 (C), F (8, 119) = 22.19 (D), F (8, 119) = 9.08 (E), F (8, 119) = 0.39 (F), F (8, 119) = 1.59 (G), F (8, 119) = 1.35 (H), **P < 0.01 vs the sham + Veh group, #P < 0.05, ##P < 0.01 vs the SNL + Veh group.
Next, we examined if Nrcam ASO could relieve the SNL-induced mechanical allodynia, thermal hyperalgesia and cold allodynia during the development period. Given that lumbar puncture is used routinely for clinical analgesic strategy26, we intrathecally injected ASOs (5 μM, 5 μl) through lumbar puncture 5 min before SNL or sham surgery and 1 day after SNL or sham surgery. Scrambled ASO or normal saline (vehicle) served as the controls. As shown in Fig. 5B, Nrcam ASO significantly blocked the SNL-induced increases in ipsilateral paw withdrawal frequencies (PWFs) in response to 0.07-g (Fig. 5B) and 0.40-g (Fig. 5C) von Frey filament stimuli on days 3 and 5 after SNL. Consistently, Nrcam ASO significantly reversed the SNL-induced decreases in ipsilateral paw withdrawal latencies (PWLs) to thermal (Fig. 5D) and cold stimuli (Fig. 5E). Compared with the SNL plus vehicle group, scrambled ASO had no effect on SNL-induced mechanical allodynia, thermal hyperalgesia, or cold allodynia on the ipsilateral side (Figs. 5B–E). Neither Nrcam ASO nor scrambled ASO affected basal responses on the contralateral side (Figs. 5F–H).
Intrathecal injection may lack the site-specific effect of Nrcam ASO. To further confirm the role of DRG Nrcam gene alternative splicing in neuropathic pain genesis, we further microinjected ASO into the L4 DRG 3 days before SNL or sham surgery. The mechanical, thermal and cold tests were examined at days 3 and 5 after SNL or sham surgery. SNL led to mechanical allodynia, thermal hyperalgesia and cold allodynia on the ipsilateral (but not contralateral) side in the SNL plus vehicle-treated group on days 3 and 5 after SNL (Figs. 6A–D). These pain hypersensitivities were abolished or ameliorated on the ipsilateral side in the SNL plus Nrcam ASO-treated group on days 3 and 5 post-SNL (Figs. 6A–D). DRG microinjection of scrambled ASO had no effect on SNL-induced mechanical allodynia, thermal hyperalgesia and cold allodynia on the ipsilateral sides during observation period (Figs. 6A–D). As expected, neither Nrcam ASO nor scrambled ASO affected basal level of PWFs to mechanical stimulus or basal level of PWLs to thermal or cold stimulus on the contralateral side (Figs. 6A–G).
Figure 6. DRG microinjection of Nrcam ASO relieved SNL-induced neuropathic pain.
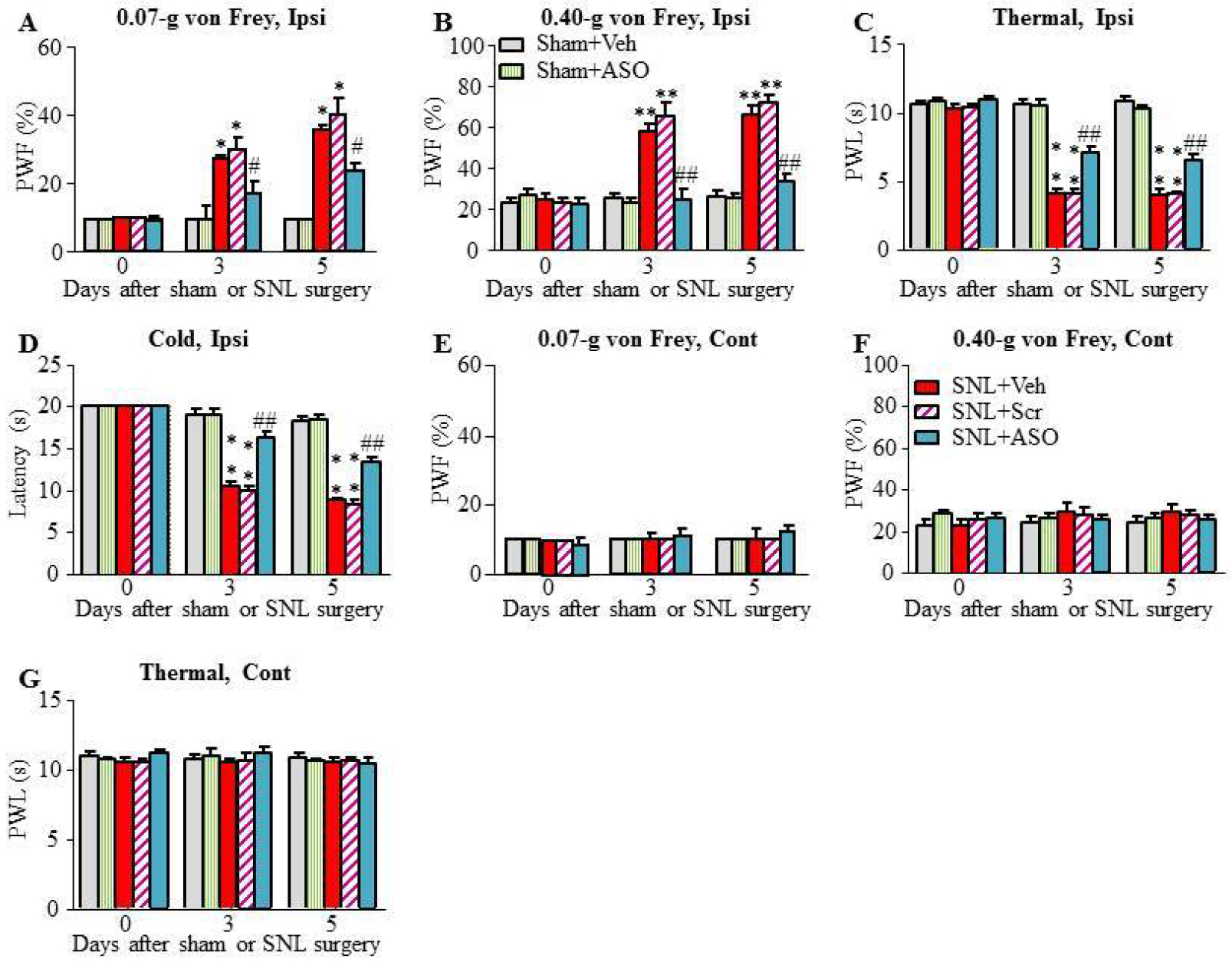
(A-E) ASOs or vehicle control were microinjected into the ipsilateral DRG before SNL or sham surgery at the same day. Nrcam ASO, but not scrambled ASO and vehicle, attenuated SNL-induced decreases in paw withdrawal frequencies (PWFs) to 0.07-g (A) and 0.40-g (B) von Frey filaments stimuli and paw withdrawal latencies (PWLs) to thermal (C) or cold (D) stimulation on the ipsilateral side. Neither ASO nor vehicle had the effect on basal paw withdrawal responses to mechanical (E, F) and thermal (G) stimuli on the contralateral side. N = 9 mice/group. Two-way RM ANOVA (effect vs group × time interaction) followed by post hoc Tukey test. F(8, 134) = 8.48 (A), F(8, 134) = 31.36 (B), F(8, 134) = 80.60 (C), F(8, 134) = 45.70 (D), F(8, 134)= 1.17 (E), F(8, 134)= 1.19 (F), F(8, 134)= 0.73 (G). **P < 0.01 vs the sham + Veh group. ##P < 0.01 vs the SNL + Veh group.
To exclude the possibility that the anti-nociceptive effects of Nrcam ASO were produced by impaired locomotor activities, the locomotor functions of all experimental mice were examined after the behavioral tests described above. None of the animals had locomotor deficits in placing, grasping, and righting reflex tests (Table 3). General behaviors, such as gait and spontaneous activity, were normal among the treatment groups. Hypermobility and convulsions were not seen in any animals.
Table 3.
Mean (Standard Error of the Mean) changes in the Locomotor function.
| Groups | Locomotor functional test | ||
|---|---|---|---|
| Placing | Grasping | Righting | |
| Sham + Veh | 5(0) | 5(0) | 5(0) |
| SNL + Veh | 5(0) | 5(0) | 5(0) |
| SNL + Scr | 5(0) | 5(0) | 5(0) |
| SNL + ASO | 5(0) | 5(0) | 5(0) |
| Sham + ASO | 5(0) | 5(0) | 5(0) |
N = 17 mice/group; 5 trials/test. SNL: spinal nerve ligation; Veh: Vehicle; Scr: scrambled antisense oligonucleotide; ASO: Nrcam antisense oligonucleotide.
Nrcam ASO attenuates the established pain hypersensitivities in mouse neuropathic pain models
We next observed the effect of intrathecal injection of Nrcam ASO on the maintenance of SNL-induced neuropathic pain. The ASOs and vehicle were administered on days 7, 9, and 11 when the pain hypersensitivities have been established in SNL-treated mice (Fig. 7A). As shown in Fig. 7, SNL led to the increase of PWFs to 0.07-g (Fig. 7B) and 0.40-g (Fig. 7C) von Frey filaments and the decrease of PWLs (Fig. 7D) to thermal stimulation on the ipsilateral, but not on the contralateral side, on day 7 through day 14 after SNL surgery in the scrambled ASO-treated group. These changes were reversed significantly by repeated Nrcam ASO treatment in male mice (Fig. 7B–D). Nrcam ASO treatment had no effect on the PWFs and PWLs on the contralateral side (Fig. 7B–D). To figure out the potential sex bias in the involvement of Nrcam mRNA variants in neuropathic pain, the effect of Nrcam ASO on SNL-induced pain hypersensitivities were further examined in female mice. As shown in Fig. 7, SNL produced mechanical and thermal hyperalgesia in female mice from day 7 until day 14 (Fig. 7E–G). Similar to the SNL male mice, Nrcam ASO treatment on days 7, 9, and 11 reversed the increase of PWFs to 0.07-g (Fig. 7E) and 0.40-g (Fig. 7F) von Frey filaments and the decrease of PWLs (Fig. 7G) to thermal stimulation on the ipsilateral side on all testing days and had no effect on basal responses on the contralateral side (Fig. 7E–G).
Figure 7. Intrathecal injection of Nrcam ASO relieved the maintenance of SNL-induced neuropathic pain in male and female mice.
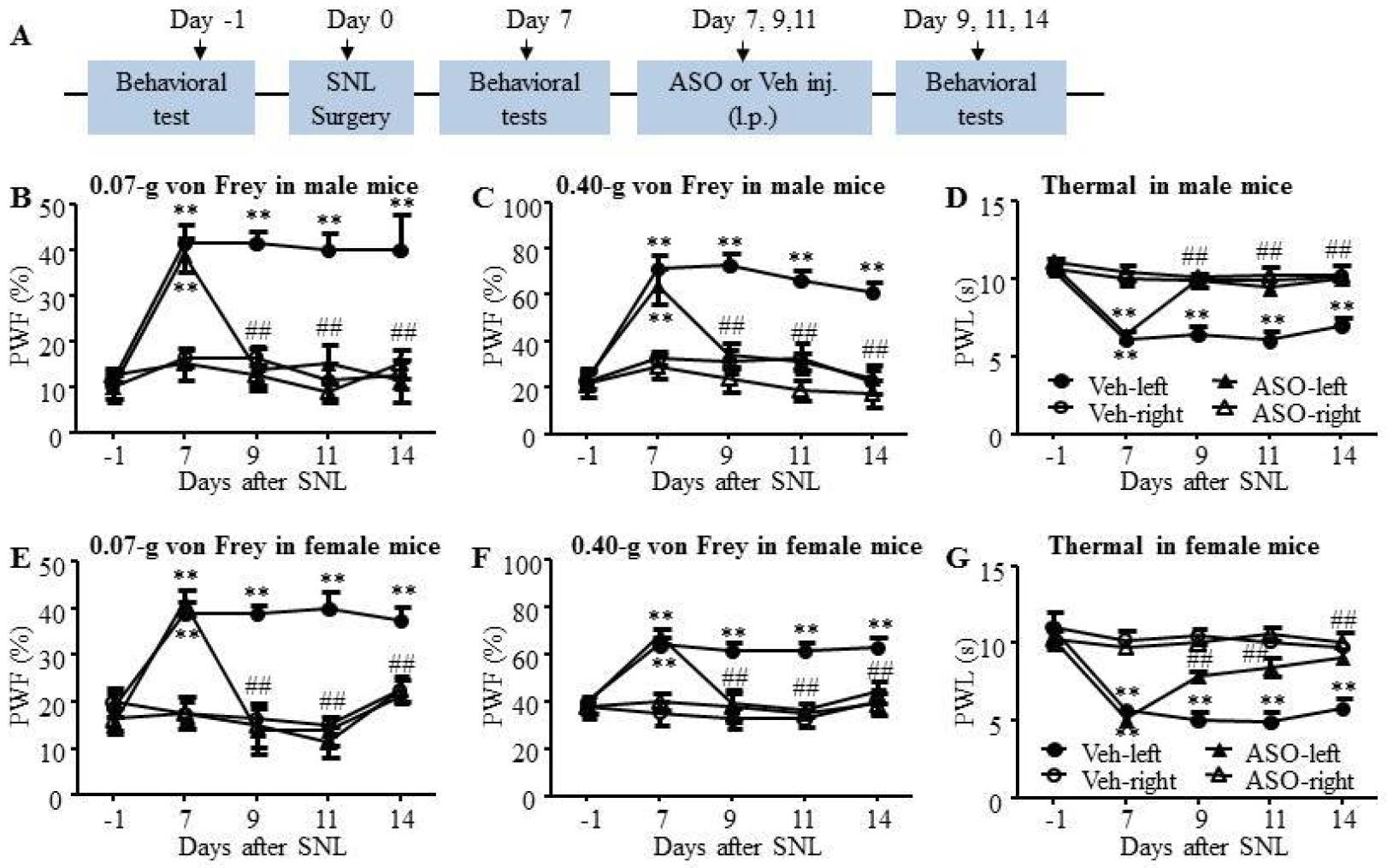
ASOs (5μM, 5μl) was injected intrathecally every other day starting from day 7 until day 14 after SNL. (A) The protocol of drug administration and behavioral tests. (B-D) Nrcam ASO reduced SNL-induced decreases in paw withdrawal frequencies (PWFs) to 0.07-g (B) and 0.40-g (C) von rey filaments stimuli and paw withdrawal latencies (PWLs) to thermal (D) stimulation on the ipsilateral side on day 9, 11 and 14 following SNL surgery in male mice, compared with vehicle treated-SNL male mice. N = 8 male mice/group. Two-way RM ANOVA (effect vs group × time interaction) followed by post hoc Tukey test. F(12, 159) = 4.99 (B), F(12, 159) = 5.98 (C), F(12, 159) = 6.83 (D), **P < 0.01 vs the values on day −1. ##P < 0.01 vs the SNL + Veh group. (E-G) Nrcam ASO reduced SNL-induced decreases in PWFs to 0.07-g (E) and 0.40-g (F) von Frey filaments stimuli and PWLs to thermal (G) stimulation on the ipsilateral side on day 9, 11 and 14 following SNL surgery in female mice, compared with vehicle treated-SNL female mice. The contralateral PWFs and PWLs were unaffected by ASO treatment in male and female mice (B-G). N = 8 female mice/group. Two-way RM ANOVA (effect vs group × time interaction) followed by post hoc Tukey test. F(12, 159) = 6.14 (E), F(12, 159) = 4.05 (F), F(12, 159) = 10.20 (G), **P < 0.01 vs the values on day −1. ##P < 0.01 vs the SNL + Veh group.
The shift of Nrcam+10 and Nrcam−10 mRNA in injured DRGs was also observed in CCD-induced neuropathic pain model (Fig. 3D). CCD model was used to mimic multilevel nerve root compression, intervertebral foramina stenosis-induced low back pain and radicular pain syndromes in human1, 6, 33, 37. Consistent with previous studies1, 6, 33, 37, CCD induced long term mechanical and thermal hyperalgesia in mice (Fig. 8A–C). Nrcam ASO was administered intrathecally every other day on days 7, 9, and 11 after CCD surgery. The CCD-induced increase in PWFs and decrease in PWLs were reversed gradually following repeated Nrcam ASO treatment (Fig. 8A–C). Moreover, the upregulation of Nrcam+10 mRNA and the downregulation of Nrcam−10 mRNA were abolished by repeated Nrcam ASO treatment (Fig. 8D). Our findings suggest that Nrcam ASO exerted the analgesic effect on neuropathic pain by normalizing mRNA levels of Nrcam+10 and Nrcam−10.
Figure 8. Intrathecal injection of Nrcam ASO relieved CCD-induced neuropathic pain.
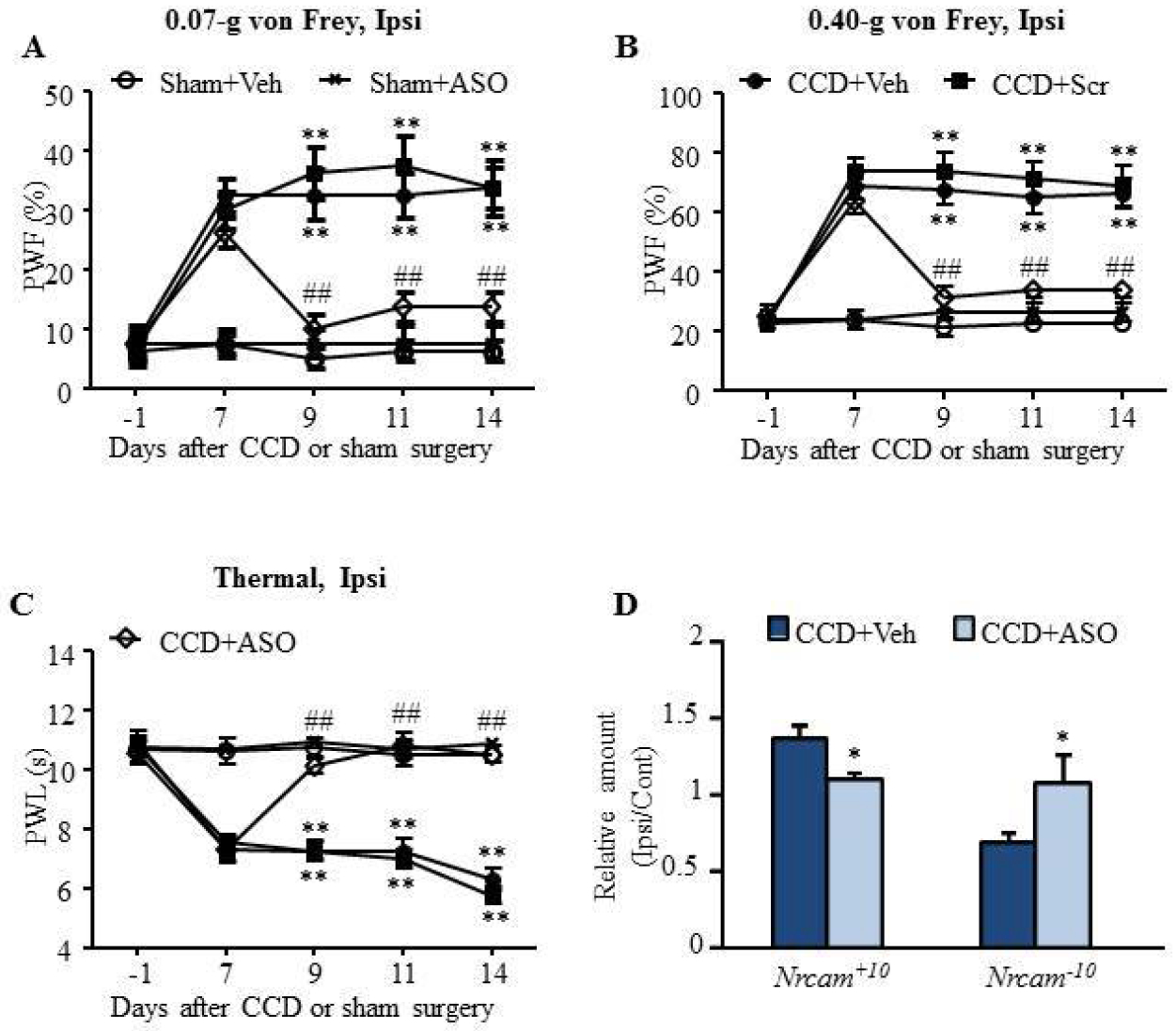
ASOs (5μM, 5μl) was injected intrathecally every other day starting from day 7 until day 14 after chronic compression of DRG (CCD) or sham surgery. (A-C) Nrcam ASO, but not scrambled ASO and vehicle, reduced CCD-induced decreases in paw withdrawal frequencies (PWFs) to 0.07-g (A) and 0.40-g (B) von Frey filaments stimuli and paw withdrawal latencies (PWLs) to thermal (C) stimulation on the ipsilateral side on day 9, 11 and 14 following CCD surgery. N = 8 male mice/group. Two-way RM ANOVA (effect vs group × time interaction) followed by post hoc Tukey test. F(16, 199) = 4.82 (A), F(16, 199) = 8.41 (B), F(16, 199) = 11.27 (C), **P < 0.01 vs the sham + Veh group. ##P < 0.01 vs the CCD + Veh group. (D) The increase of Nrcam+10 mRNA level and the decrease of Nrcam−10 mRNA level induced by CCD was abolished by Nrcam ASO treatment. *P < 0.05 vs the CCD + Veh group by paired t-test.
Discussion
In the present study, we demonstrated that SNL led to a marked change in Nrcam RNA alternative splicing, including an increase in the level of Nrcam+10 mRNA and a corresponding decrease in the amount of Nrcam−10 mRNA in the injured DRG. Blocking the increased Nrcam+10 mRNA through intrathecal administration or DRG microinjection of its specific ASO rescued DRG Nrcam−10 mRNA expression and prevented the development and maintenance of SNL-induced pain hypersensitivities. The analgesic effect of Nrcam ASO is observed in both sexes. Furthermore, the analgesic effect of Nrcam ASO was also observed in CCD-induced pain hypersensitivities, an animal model of neuropathic pain that highly mimics clinical conditions. Therefore, Nrcam+10 mRNA variant may be a potential target in the management of neuropathic pain.
NrCAM belongs to the L1 family of cell adhesion molecules, which contains six Ig-like domains and five Fn-type III repeats in its extracellular region, a transmembrane region, and a cytoplasmic region lacking any enzymatic activity24. NrCAM participates in neural development and some disorders, such as drug addiction and autism7, 24, 31. These functions may be related to extensively alternative processing of Nrcam RNA8, 32. Since Nrcam exon 10 alternative splicing was first reported in chickens in 19915, more Nrcam RNA variants have further been identified in humans 32. Nrcam exon 10 encodes 19 amino acids, which are located between immunoglobulin-like domains II and III (IgII and IgIII) of NrCAM protein32. Both Nrcam+10 and Nrcam−10 transcripts are abundant in adrenal glands, but Nrcam−10 is expressed predominately in the adult and fetal pancreas, whereas Nrcam+10 is expressed mainly in the placenta as well as adult and fetal brain7, 32. The present study reported that these two splicing variants were also detectable in the DRG, trigeminal ganglion, heart and kidney. As reported in humans32, Nrcam+10, but not Nrcam−10, was detected in the spinal cord, cortex, brainstem, hippocampus, cerebellum and sciatic nerve from mice. Since baseline levels of Nrcam+10 and Nrcam−10 transcripts in DRG were rather low, our pilot work showed that it was difficult to detect Nrcam+10 and Nrcam−10 hybridization signals in DRG cells. However, since previous studies reported both the Nrcam RNA hybridization signal and NrCAM immunoreactivity in brain neurons5, 8, we assume that Nrcam mRNA and NrCAM are neuronal-specific in the DRG although their detailed cellular distribution in the DRG is still unclear.
Targeting Nrcam RNA alternative splicing may be a potential avenue for neuropathic pain therapy. In the present study, we demonstrated a significant change in Nrcam exon 10 alternative splicing in the injured DRG of SNL mice without sex-bias. This change mainly included exon 10 insertion in Nrcam mRNA, resulting in an increase in Nrcam+10 mRNA variant expression with a corresponding decrease in short Nrcam−10 mRNA variant expression in the injured DRG on days 3 and 5 following SNL. ASO strategy has been developed for treatment of genetic diseases and certain cancers in clinic12, 13, 15. we carried out this strategy and reported that intrathecal administration or DRG microinjection of ASO specifically targeting Nrcam exon 10 relieved SNL or CCD-induced pain hypersensitivities in both development and maintenance stage in male and female mice. Our findings suggest that Nrcam+10 contributes to neuropathic pain genesis and may serve a potential target for neuropathic pain treatment.
The mechanism underlying the effect of Nrcam ASO is still unclear. The epidermal growth factor receptor (EGFR), a member of the ErbB family of receptor tyrosine kinases, regulates cellular growth, survival, proliferation, and differentiation of fibroblasts and hepatocytes2, 19, 23. Activation of DRG EGFR was involved in chronic pain induction19. Our recent study showed that blocking EGFR increase in the DRG attenuated CCD-induced pain hypersensitivities33. EGFR activation initiated the downstream intracellular signaling pathways including the mitogen-activated protein kinase (MAPK)/ extracellular signal-regulated kinase (ERK) and phosphoinositide 3 kinase (PI3K)/Protein kinase B (Akt)/ mammalian target of rapamycin (mTOR) pathways14, 25, 33. Given that NrCAM exerted its oncogenic function by binding to EGFR, subsequently activating downstream intracellular signaling pathways in thyroid cancer cells 39 and that both EGFR and its downstream cellular pathways (MAPK/ERK and PI3K/Akt/mTOR) contributed to neuropathic development11, 21, 22, 38, the anti-nociceptive effect of Nrcam ASO on neuropathic pain may be mediated through inhibition of EGFR-triggered activation of the intracellular MAPK/ERK and PI3K/Akt/mTOR pathway. This conclusion needs to be further confirmed in future studies.
Highlights.
Nrcam exon 10 splicing event was found in dorsal root ganglion (DRG).
SNL induced the increase in long Nrcam RNA variant with exon 10 in DRG.
Nrcam ASO attenuated pain hypersensitivities in mice neuropathic pain models
Funding:
The work was supported by the grant from the National Institutes of Health of USA (DA033390, NS094664, NS094224 and HL117684) to Y.X.T. .
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Briggs CA, Chandraraj S: Variations in the lumbosacral ligament and associated changes in the lumbosacral region resulting in compression of the fifth dorsal root ganglion and spinal nerve. Clin Anat 8:339–346,1995 [DOI] [PubMed] [Google Scholar]
- 2.Citri A, Yarden Y: EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 7:505–516,2006 [DOI] [PubMed] [Google Scholar]
- 3.Cui L, Miao X, Liang L, Abdus-Saboor I, Olson W, Fleming MS, Ma M, Tao YX, Luo W: Identification of Early RET+ Deep Dorsal Spinal Cord Interneurons in Gating Pain. Neuron 91:1137–1153,2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Disterer P, Al-Shawi R, Ellmerich S, Waddington SN, Owen JS, Simons JP, Khoo B: Exon skipping of hepatic APOB pre-mRNA with splice-switching oligonucleotides reduces LDL cholesterol in vivo. Mol Ther 21:602–609,2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grumet M, Mauro V, Burgoon MP, Edelman GM, Cunningham BA: Structure of a new nervous system glycoprotein, Nr-CAM, and its relationship to subgroups of neural cell adhesion molecules. J Cell Biol 113:1399–1412,1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu SJ, Xing JL: An experimental model for chronic compression of dorsal root ganglion produced by intervertebral foramen stenosis in the rat. Pain 77:15–23,1998 [DOI] [PubMed] [Google Scholar]
- 7.Ishiguro H, Hall FS, Horiuchi Y, Sakurai T, Hishimoto A, Grumet M, Uhl GR, Onaivi ES, Arinami T: NrCAM-regulating neural systems and addiction-related behaviors. Addict Biol 19:343–353,2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishiguro H, Liu QR, Gong JP, Hall FS, Ujike H, Morales M, Sakurai T, Grumet M, Uhl GR: NrCAM in addiction vulnerability: positional cloning, drug-regulation, haplotype-specific expression, and altered drug reward in knockout mice. Neuropsychopharmacology 31:572–584,2006 [DOI] [PubMed] [Google Scholar]
- 9.Katz Y, Wang ET, Airoldi EM, Burge CB: Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7:1009–1015,2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz Y, Wang ET, Silterra J, Schwartz S, Wong B, Thorvaldsdottir H, Robinson JT, Mesirov JP, Airoldi EM, Burge CB: Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics 31:2400–2402,2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kersten C, Cameron MG, Laird B, Mjaland S: Epidermal growth factor receptor-inhibition (EGFR-I) in the treatment of neuropathic pain. Br J Anaesth 115:761–767,2015 [DOI] [PubMed] [Google Scholar]
- 12.Khoo B, Roca X, Chew SL, Krainer AR: Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB. BMC Mol Biol 8:3,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khvorova A, Watts JK: The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35:238–248,2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolkova K, Novitskaya V, Pedersen N, Berezin V, Bock E: Neural cell adhesion molecule-stimulated neurite outgrowth depends on activation of protein kinase C and the Ras-mitogen-activated protein kinase pathway. J Neurosci 20:2238–2246,2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo T, Wood MJ: Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum Gene Ther 24:479–488,2013 [DOI] [PubMed] [Google Scholar]
- 16.Liang L, Gu X, Zhao JY, Wu S, Miao X, Xiao J, Mo K, Zhang J, Lutz BM, Bekker A, Tao YX: G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci Rep 6:37704,2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang L, Zhao JY, Gu X, Wu S, Mo K, Xiong M, Marie LB, Bekker A, Tao YX: G9a inhibits CREB-triggered expression of mu opioid receptor in primary sensory neurons following peripheral nerve injury. Mol Pain 12:2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang LL, Yang JL, Lu N, Gu XY, Zhang YQ, Zhao ZQ: Synergetic analgesia of propentofylline and electroacupuncture by interrupting spinal glial function in rats. Neurochem Res 35:1780–1786,2010 [DOI] [PubMed] [Google Scholar]
- 19.Martin LJ, Smith SB, Khoutorsky A, Magnussen CA, Samoshkin A, Sorge RE, Cho C, Yosefpour N, Sivaselvachandran S, Tohyama S, Cole T, Khuong TM, Mir E, Gibson DG, Wieskopf JS, Sotocinal SG, Austin JS, Meloto CB, Gitt JH, Gkogkas C, Sonenberg N, Greenspan JD, Fillingim RB, Ohrbach R, Slade GD, Knott C, Dubner R, Nackley AG, Ribeiro-da-Silva A, Neely GG, Maixner W, Zaykin DV, Mogil JS, Diatchenko L: Epiregulin and EGFR interactions are involved in pain processing. J Clin Invest 127:3353–3366,2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marui T, Funatogawa I, Koishi S, Yamamoto K, Matsumoto H, Hashimoto O, Nanba E, Nishida H, Sugiyama T, Kasai K, Watanabe K, Kano Y, Sasaki T, Kato N: Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. Int J Neuropsychopharmacol 12:1–10,2009 [DOI] [PubMed] [Google Scholar]
- 21.Obata K, Yamanaka H, Dai Y, Mizushima T, Fukuoka T, Tokunaga A, Noguchi K: Differential activation of MAPK in injured and uninjured DRG neurons following chronic constriction injury of the sciatic nerve in rats. Eur J Neurosci 20:2881–2895,2004 [DOI] [PubMed] [Google Scholar]
- 22.Obata K, Yamanaka H, Kobayashi K, Dai Y, Mizushima T, Katsura H, Fukuoka T, Tokunaga A, Noguchi K: Role of mitogen-activated protein kinase activation in injured and intact primary afferent neurons for mechanical and heat hypersensitivity after spinal nerve ligation. J Neurosci 24:10211–10222,2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastore S, Mascia F, Mariani V, Girolomoni G: The epidermal growth factor receptor system in skin repair and inflammation. J Invest Dermatol 128:1365–1374,2008 [DOI] [PubMed] [Google Scholar]
- 24.Sakurai T: The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Mol Cell Neurosci 49:351–363,2012 [DOI] [PubMed] [Google Scholar]
- 25.Schaefer AW, Kamiguchi H, Wong EV, Beach CM, Landreth G, Lemmon V: Activation of the MAPK signal cascade by the neural cell adhesion molecule L1 requires L1 internalization. J Biol Chem 274:37965–37973,1999 [DOI] [PubMed] [Google Scholar]
- 26.Storek B, Reinhardt M, Wang C, Janssen WG, Harder NM, Banck MS, Morrison JH, Beutler AS: Sensory neuron targeting by self-complementary AAV8 via lumbar puncture for chronic pain. Proc Natl Acad Sci U S A 105:1055–1060,2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao YX, Rumbaugh G, Wang GD, Petralia RS, Zhao C, Kauer FW, Tao F, Zhuo M, Wenthold RJ, Raja SN, Huganir RL, Bredt DS, Johns RA: Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J Neurosci 23:6703–6712,2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tollervey JR, Wang Z, Hortobagyi T, Witten JT, Zarnack K, Kayikci M, Clark TA, Schweitzer AC, Rot G, Curk T, Zupan B, Rogelj B, Shaw CE, Ule J: Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 21:1572–1582,2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda H: Molecular mechanisms of neuropathic pain-phenotypic switch and initiation mechanisms. Pharmacol Ther 109:57–77,2006 [DOI] [PubMed] [Google Scholar]
- 30.Vandenbroucke II, Vandesompele J, Paepe AD, Messiaen L: Quantification of splice variants using real-time PCR. Nucleic Acids Res 29:E68–E68,2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH: Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474:380–384,2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Williams H, Du JS, Terrett J, Kenwrick S: Alternative splicing of human NrCAM in neural and nonneural tissues. Mol Cell Neurosci 10:287–295,1998 [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Liu S, Xu L, Zhu X, Liu W, Tian L, Chen Y, Wang Y, Nagendra BVP, Jia S, Liang L, Huo FQ: The upregulation of EGFR in the dorsal root ganglion contributes to chronic compression of dorsal root ganglions-induced neuropathic pain in rats. Mol Pain 15:1744806919857297,2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Hurwitz O, Shimada SG, Qu L, Fu K, Zhang P, Ma C, LaMotte RH: Chronic Compression of the Dorsal Root Ganglion Enhances Mechanically Evoked Pain Behavior and the Activity of Cutaneous Nociceptors in Mice. PLoS One 10:e0137512,2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Marie LB, Miao X, Liang L, Mo K, Chang YJ, Du P, Soteropoulos P, Tian B, Kaufman AG, Bekker A, Hu Y, Tao YX: Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Mol Pain 12:2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie YB, Zhao H, Wang Y, Song K, Zhang M, Meng FC, Yang YJ, He YS, Kuang F, You SW, You HJ, Xu H: Bilateral Neuropathy of Primary Sensory Neurons by the Chronic Compression of Multiple Unilateral DRGs. Neural Plast 2016:2130901,2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie YB, Zhao H, Wang Y, Song K, Zhang M, Meng FC, Yang YJ, He YS, Kuang F, You SW, You HJ, Xu H: Bilateral Neuropathy of Primary Sensory Neurons by the Chronic Compression of Multiple Unilateral DRGs. Neural Plast 2016:2130901,2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu JT, Tu HY, Xin WJ, Liu XG, Zhang GH, Zhai CH: Activation of phosphatidylinositol 3-kinase and protein kinase B/Akt in dorsal root ganglia and spinal cord contributes to the neuropathic pain induced by spinal nerve ligation in rats. Exp Neurol 206:269–279,2007 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Sui F, Ma J, Ren X, Guan H, Yang Q, Shi J, Ji M, Shi B, Sun Y, Hou P: Positive Feedback Loops Between NrCAM and Major Signaling Pathways Contribute to Thyroid Tumorigenesis. J Clin Endocrinol Metab 102:613–624,2017 [DOI] [PubMed] [Google Scholar]
- 40.Zhao JY, Liang L, Gu X, Li Z, Wu S, Sun L, Atianjoh FE, Feng J, Mo K, Jia S, Lutz BM, Bekker A, Nestler EJ, Tao YX: DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun 8:14712,2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, Gao YJ, Hoffman PN, Cui H, Li M, Dong X, Tao YX: A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci 16:1024–1031,2013 [DOI] [PMC free article] [PubMed] [Google Scholar]


