Abstract
Staphylococcus aureus is an important bacterial pathogen that can cause a wide spectrum of diseases in humans and other animals. S. aureus expresses a variety of virulence factors that promote infection with this pathogen. These include cell-surface proteins that mediate adherence of the bacterial cells to host extracellular matrix components, such as fibronectin and fibrinogen. Here, using immunoblotting, ELISA, and surface plasmon resonance analysis, we report that the iron-regulated surface determinant B (IsdB) protein, besides being involved in heme transport, plays a novel role as a receptor for the plasma and extracellular matrix protein vitronectin (Vn). Vn-binding activity was expressed by staphylococcal strains grown under iron starvation conditions when Isd proteins are expressed. Recombinant IsdB bound Vn dose dependently and specifically. Both near-iron transporter motifs NEAT1 and NEAT2 of IsdB individually bound Vn in a saturable manner, with KD values in the range of 16–18 nm. Binding of Vn to IsdB was specifically blocked by heparin and reduced at high ionic strength. Furthermore, IsdB-expressing bacterial cells bound significantly higher amounts of Vn from human plasma than did an isdB mutant. Adherence to and invasion of epithelial and endothelial cells by IsdB-expressing S. aureus cells was promoted by Vn, and an αvβ3 integrin-blocking mAb or cilengitide inhibited adherence and invasion by staphylococci, suggesting that Vn acts as a bridge between IsdB and host αvβ3 integrin.
Keywords: Staphylococcus aureus, plasma, extracellular matrix protein, adhesion, cell invasion, innate immunity, virulence factor, integrin, vitronectin (Vn), iron-regulated surface determinant B (IsdB), S. aureus
Staphylococcus aureus causes a wide range of opportunistic infections that range from superficial skin infections to life-threatening diseases, including endocarditis, pneumonia, and septicemia (1). Adherence of bacteria to host matrix components is the initial critical event in the pathogenesis of most infections. The extracellular matrix (ECM) essentially consists of macromolecules, such as collagens, proteoglycans, and glycoproteins, that serve as a substrate for the adhesion and migration of tissue cells. These processes involve integrins, a family of heterodimeric cell surface receptors that recognize specific ECM proteins (2, 3).
Bacteria, including S. aureus, also utilize the ECM as substrata for their adhesion through a family of cell wall-anchored (CWA) adhesins called MSCRAMMs (microbial surface component recognizing adhesive matrix molecules) that specifically recognize host matrix components (4, 5).
Vitronectin (Vn) is a glycoprotein that is synthesized in the liver and secreted into plasma (6) and is also an important component of the ECM (7). Vn is found at a high concentration in plasma (200–700 μg/ml) (8, 9) and is also present in different human tissues (10). The N-terminal portion of mature Vn (43 aa residues) consists of a somatomedin B (SMB) domain followed by the classical integrin-binding motif, Arg-Gly-Asp (RGD). Members of the integrin family that engage in Vn binding include integrin α3β1, αvβ3, αvβ5, and α5β1 (11). The next domain comprises four hemopexin-like domains with putative heme-binding motifs. In addition, Vn has three heparin-binding domains (HBD) spanning residues Vn82–137 (HBD-1), Vn175–219 (HBD-2), and Vn348–361 (HBD-3) (12–14) (Fig. S1A). Vn is present in the organism in different conformational states: as the native, folded monomer (65–75 kDa, the 65-kDa form being derived from the proteolytic cleavage in the C-terminal region of the protein) in plasma/serum and as a multimeric unfolded form in the ECM (6, 15, 16). Conformational change from the monomeric to the activated, multimeric state is promoted by exposure of Vn to agents, such as urea, or binding to physiological ligands, such as the thrombin-antithrombin complex and the membrane attack complex. Vn conformational activation reveals a number of cryptic sites, including the full exposure of the heparin-binding site at the C-terminal domain of the protein (17, 18) and cell-binding motif (RGD) (19, 20). Vn binds the terminal complement C5b-7 complex. It occupies the metastable membrane binding site and thereby inhibits membrane insertion of the complex (21). It also binds C9 and directly inhibits C9 polymerization (21, 22).
Several bacterial species interact with host cell-bound multimeric Vn, facilitating adherence to epithelial cells and artificial surfaces (23). Simultaneous interaction of Vn with an integrin and bacterial surface proteins results in the formation of a bridge between bacteria and host cells. This leads to internalization of bacteria, as exemplified by Streptococcus pneumoniae (24) or Pseudomonas aeruginosa (25), resulting in downstream signaling events (24).
Staphylococci contain several Vn-binding proteins, including the autolysins AtlE and Aae from S. epidermidis and the homologous proteins AtlA and Aaa from S. aureus (26, 27). Also, the multifunctional autolysin AtlL from Staphylococcus lugdunensis interacts with Vn (28).
Atl autolysins have a similar modular organization (signal peptide, propeptide, amidase activity, three major repeats, R1 to R3, and glucosaminidase activity), share a high degree of sequence similarity, and are functionally interchangeable (29). R1-R2 repeats are critical for autolysin binding to Vn (30). Moreover, the major autolysin, Atl, mediates S. aureus internalization via direct interaction with host heat shock protein Hsc70 (31).
Studies on S. aureus adhesion to and invasion of host cells have been performed with bacteria grown in rich medium containing iron (4). In contrast, in vivo S. aureus has restricted access to iron, and the lack of available iron leads to the upregulation of a number of genes, among which are those that encode surface determinant (Isd) proteins (32). The Isd system contains nine proteins whose expression is coordinately upregulated under iron-depleted conditions (33–36). The primary role of Isd proteins is to capture heme from hemoglobin (Hb) and transport it into the cell (32). These include IsdA, IsdB, IsdC, and IsdH, which are anchored to cell wall peptidoglycan by sortases and are exposed on the cell surface (37, 38). Each protein contains a structurally conserved near iron transporter (NEAT) motif(s) that binds Hb and heme. IsdA and IsdC contain one NEAT domain each, whereas IsdB and IsdH contain two and three NEAT domains, respectively. The NEAT domains adopt a beta sandwich fold that consists of two five-stranded antiparallel beta sheets (39).
Fig. S1B shows the organization and primary sequence comparisons between the seven known NEAT domains in S. aureus.
Sequence homologs of this class of proteins are found in a number of important human pathogens, such as S. lugdunensis, (40–42), Listeria monocytogenes (43), Bacillus anthracis (44), and Streptococcus pyogenes (45).
IsdA, IsdB, and IsdH of S. aureus are known to have other biological functions. IsdA interacts with an array of host proteins (36) and confers resistance to the innate defenses of the human skin (46). IsdH plays a role in the evasion of phagocytosis as a result of accelerated degradation of C3b (47). IsdB binds to platelets via direct interaction with the platelet integrin GPIIb/IIIa and also promotes S. aureus adherence to and internalization by nonphagocytic human cells (48). The objective of the current study was to investigate in more detail the binding of Vn to S. aureus cells. We show that cells expressing IsdB specifically bind to Vn and analyze the nature and the biological consequences of this interaction.
Results
Vn binding by S. aureus is promoted by growth under iron-restricted conditions
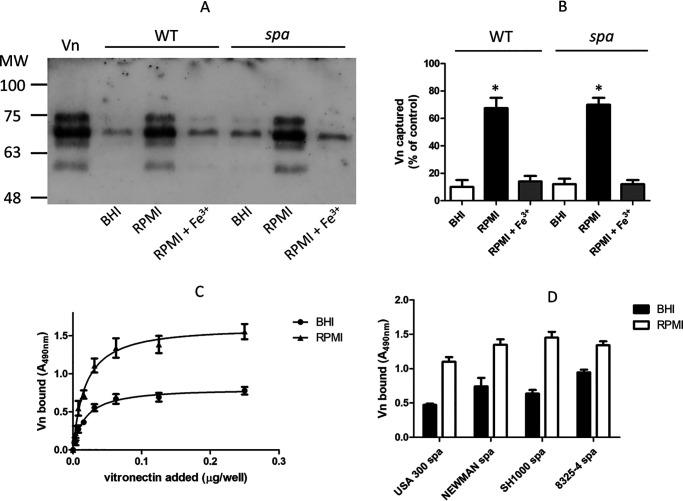
In preliminary experiments, we tested the capture of Vn by S. aureus strain SH1000 grown to stationary phase in rich brain heart infusion (BHI) or iron-restricted Roswell Park Memorial Institute 1640 (RPMI) medium with or without FeCl3. Bacteria grown in RPMI showed a higher ability to capture Vn than those grown in iron-rich medium or in RPMI supplemented with FeCl3, suggesting that binding of strain SH1000 depends on proteins induced by iron starvation. Interestingly, a protein A-deficient (spa) mutant showed a Vn-binding profile overlapping that of the WT strain, suggesting that Vn binding to the bacterial surface is not related to protein A expression (Fig. 1, A and B).
Figure 1.
Binding of purified Vn by S. aureus cells. A, S. aureus strain SH1000 and its spa mutant grown either in BHI or RPMI medium in the absence/presence of 1 mm FeCl3 were incubated with purified Vn. Bacterium-bound proteins were released by extraction buffer and separated by SDS-PAGE under reducing conditions and analyzed by far Western blotting. The membrane was probed with sheep anti-human Vn, followed by HRP-conjugated rabbit anti-sheep IgG. Molecular masses of standard proteins are indicated on the left. B, densitometric analysis of Vn bound to S. aureus SH1000 and the spa mutant as reported in panel A. The band intensity was quantified relative to a sample of purified human Vn (8 μg). The reported data are the mean values ± S.D. from three independent experiments. C, binding of increasing concentrations of Vn to immobilized S. aureus SH1000 spa cells grown in RPMI or BHI medium is shown. Bound Vn was detected as described above. The data points are the means ± S.D. from three independent experiments, each performed in triplicate. A statistically significant difference is indicated (Student's t test; *, p < 0.05). D, binding of Vn to strains of S. aureus spa. Microtiter wells coated with S. aureus spa strains grown in RPMI or BHI were incubated with Vn. Bound Vn was detected by addition of sheep anti-Vn polyclonal IgG and rabbit HRP-conjugated anti-sheep IgG.
To further analyze the interaction of Vn with S. aureus, SH1000 spa organisms grown in BHI and RPMI medium were immobilized onto microtiter wells and allowed to interact with increasing amounts of soluble Vn (Fig. 1C). Under both conditions, Vn bound to bacteria in a dose-dependent and saturable fashion. RPMI-grown bacteria captured significantly larger amounts of Vn, suggesting that S. aureus cells express a larger number of receptors on their surface or that novel receptors were induced when grown in iron starvation. To compare the Vn binding potential of different S. aureus strains, spa mutants of the S. aureus laboratory strains Newman, SH1000, and 8325-4 and the clinical strain USA300 grown in BHI or RPMI medium to the stationary phase were immobilized in microtiter wells and tested for binding to soluble Vn. All the strains grown in RPMI medium showed a higher ability to bind Vn when grown under iron starvation conditions than in iron-rich medium (Fig. 1D).
Identification of a Vn-binding protein
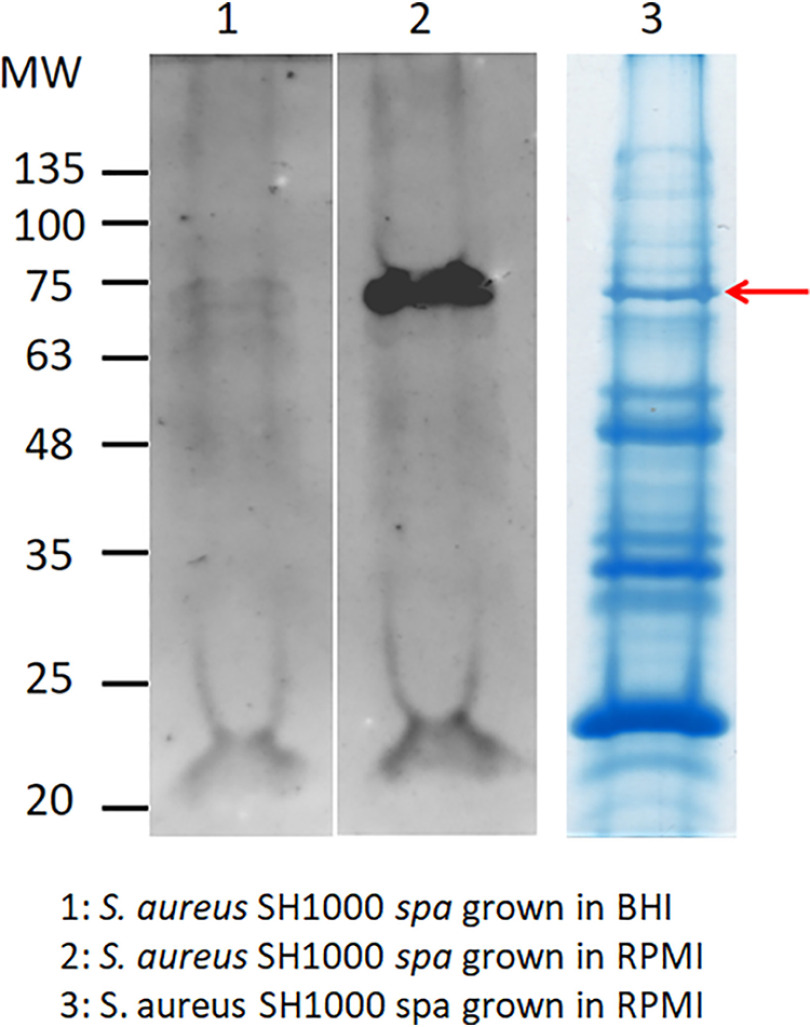
To identify the surface component(s) involved in Vn binding, SH1000 spa cells grown in BHI or RPMI medium were digested with lysostaphin and the released material subjected to SDS-PAGE under reducing conditions and far Western blotting. The nitrocellulose membrane was incubated with Vn, and the bound ligand was detected with anti-Vn IgG. A strong signal corresponding to a molecule of 75 kDa was noted in protein from cells grown in RPMI medium, whereas no significant signal was detected in material released from cells grown in BHI medium (Fig. 2, lanes 1 and 2).
Figure 2.
Far Western blotting of lysostaphin-released material from cells of SH1000 spa. Cells of SH1000 spa grown in BHI (lane 1) and RPMI (lane 2) were digested with lysostaphin and the released material subjected to far Western blotting. The nitrocellulose membrane was probed with Vn, followed by sheep anti-Vn and HRP-conjugated rabbit anti-sheep IgG. Molecular mass standards are indicated on the left. The lysostaphin-released material from cells of SH1000 spa grown in RPMI was also subjected to SDS-PAGE and stained with Coomassie blue (lane 3). The band corresponding to the 75-kDa bacterial component is marked by an arrow.
To further investigate this issue, proteins were separated by SDS-PAGE under reducing conditions and visualized by staining with Coomassie brilliant blue (Fig. 2, lane 3). A candidate protein of 75 kDa was excised from the stained gel, digested with trypsin, and analyzed by MS. The database search unequivocally identified IsdB as the potential Vn-binding protein of S. aureus SH1000 (Fig. S2A).
isdB gene expression is growth-phase dependent
Since S. aureus SH1000 spa cells grown to stationary phase in BHI medium bound Vn significantly less than when grown in RPMI medium (Fig. 1), we analyzed the expression of the isdB gene in cells grown both in BHI or RPMI medium to mid-exponential and stationary phases of growth by quantitative RT-PCR (qRT-PCR). Expression of isdB in cells grown to stationary phase in RPMI medium was about 5-fold higher than that in cells grown in BHI medium or cells grown to mid-exponential phase in either medium (Fig. S2B).
This finding was validated by comparing the expression levels of the IsdB protein by bacteria grown to mid-exponential and stationary phases in BHI and RPMI media by Western immunoblotting. While IsdB was virtually undetectable in material released from bacteria grown to both the mid-exponential and stationary phases in BHI medium, the protein was abundant in material released from bacteria grown to the stationary phase in RPMI medium (Fig. S2C).
Ectopic expression of IsdB by L. lactis and binding to Vn
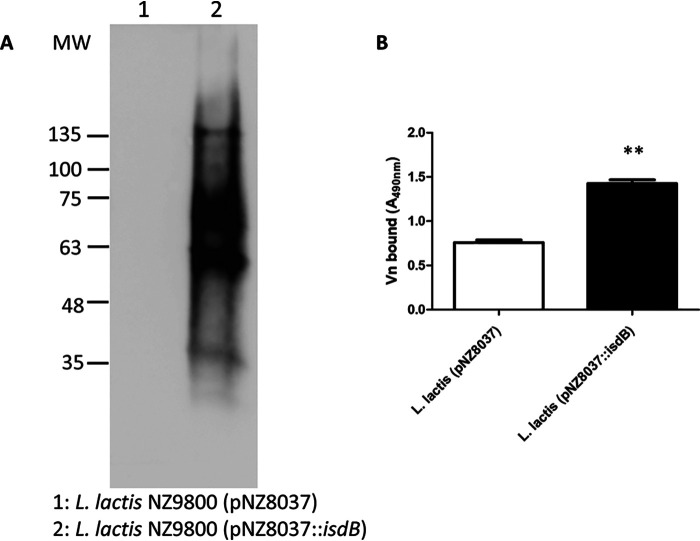
To study IsdB in isolation from other S. aureus CWA proteins, a strain of L. lactis expressing IsdB from a gene cloned into the plasmid vector pNZ8037 was used. To validate the expression of IsdB from the bacterium, L. lactis pNZ8037::isdB and the isogenic strain carrying the empty vector were treated with mutanolysin and lysozyme, and the released material subjected to far Western blotting. A Vn binding protein of 75 kDa was detected (along with an approximately 60-kDa protein, which may be a breakdown product), whereas the material released from L. lactis (pNZ8037) lacked reactivity (Fig. 3A). Moreover, the lactococci were immobilized onto microtiter wells, and binding of soluble Vn was examined by ELISA. Significantly higher binding of Vn to L. lactis (pNZ8037::isdB) was observed compared to that of L. lactis harboring the empty vector (Fig. 3B).
Figure 3.
Interaction of L. lactis-expressing IsdB with Vn. A, cell wall proteins were released by mutanolysin/lysozyme treatment from L. lactis expressing IsdB (pNZ8037::isdB) and control cells carrying the empty vector. The protein components in the mixture were separated by SDS-PAGE and subjected to far Western blotting. The membrane was probed with Vn, followed by sheep anti-Vn and HRP-conjugated rabbit anti-sheep IgG. The figure is representative of three independent experiments. Molecular mass standards are indicated on the left. B, interaction of Vn with L. lactis-expressing IsdB and cells carrying the empty vector assessed by ELISA. Bacteria were immobilized in microtiter plates and incubated with Vn, followed by addition to the wells of sheep anti-Vn polyclonal IgG and HRP-conjugated rabbit anti-sheep antibody. The data points are the means ± S.D. from three independent experiments, each performed in triplicate. Statistically significant differences are indicated (Student's t test; **, p < 0.01).
Specificity of Vn binding to IsdB
To investigate the specificity of Vn binding to IsdB, recombinant IsdB NEAT1-NEAT2 protein was immobilized onto microtiter wells and tested for binding to extracellular matrix proteins, including fibrinogen, fibronectin, collagen, and Vn. Only Vn bound to the surface-coated IsdB, whereas no binding of the other proteins was observed (Fig. 4A).
Figure 4.
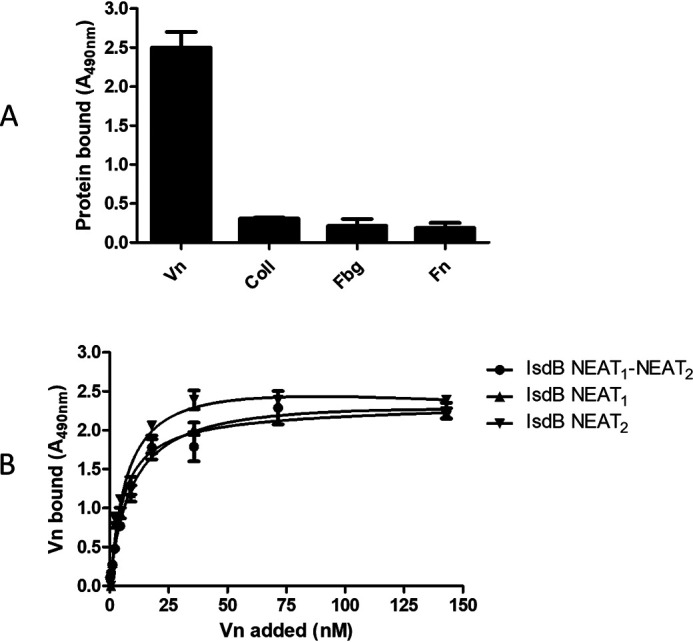
Characterization of Vn-binding activity of IsdB. A, specificity of recombinant IsdB interaction with extracellular matrix proteins. Microtiter wells were coated with IsdB NEAT1-NEAT2 and then incubated with the indicated extracellular matrix proteins Vn, collagen type I, fibrinogen, and fibronectin. Each bound protein was detected by the addition of ligand-specific polyclonal antibody and HRP-conjugated secondary IgG. The data points are the means ± S.D. from three independent experiments, each performed in triplicate. B, Vn binding to IsdB NEAT1-NEAT2 and its derivatives, NEAT1 and NEAT2. Recombinant IsdB NEAT1-NEAT2 and its derivatives, NEAT1 and NEAT2, were immobilized onto microtiter wells and incubated with increasing amounts of Vn. Bound Vn was detected by the addition of sheep anti-Vn polyclonal IgG and HRP-conjugated rabbit anti-sheep. The data points are the means ± S.D. from three independent experiments, each performed in triplicate.
Localization of the binding sites within IsdB
To localize the Vn-binding region within the IsdB protein, the NEAT1 and NEAT2 domains were expressed in E. coli and employed in binding studies. First, the binding of soluble Vn to immobilized recombinant NEAT1 and NEAT2 was determined by ELISA. Vn bound dose dependently and saturably to both NEAT1 and NEAT2 fragments and with a binding profile resembling that exhibited by intact full-length IsdB NEAT1-NEAT2 (Fig. 4B).
IsdB NEAT1-NEAT2 and single NEAT1 and NEAT2 domains bind to Vn with high affinity
Surface plasmon resonance (SPR) experiments were performed to compare the binding of Vn to those of immobilized IsdB NEAT1-NEAT2 and single NEAT1 and NEAT2 fragments. Vn displayed a high binding activity, as indicated by the high response values and the slow dissociation of the IsdB-Vn complexes upon removal of the ligand. The best fit of the data points was obtained with the Langmuir isotherm equation describing a one-site binding model. From this analysis, we obtained dissociation constant (KD) values of 17.85 ± 3.51 nm, 16.73 ± 3.02 nm, and 16.21 ± 2.79 nm for IsdB NEAT1-NEAT2/Vn, NEAT1/Vn, and NEAT2/Vn, respectively (Fig. 5). All these findings show that full-length IsdB contains two separate binding sites that interact with nearly identical high affinities for Vn.
Figure 5.
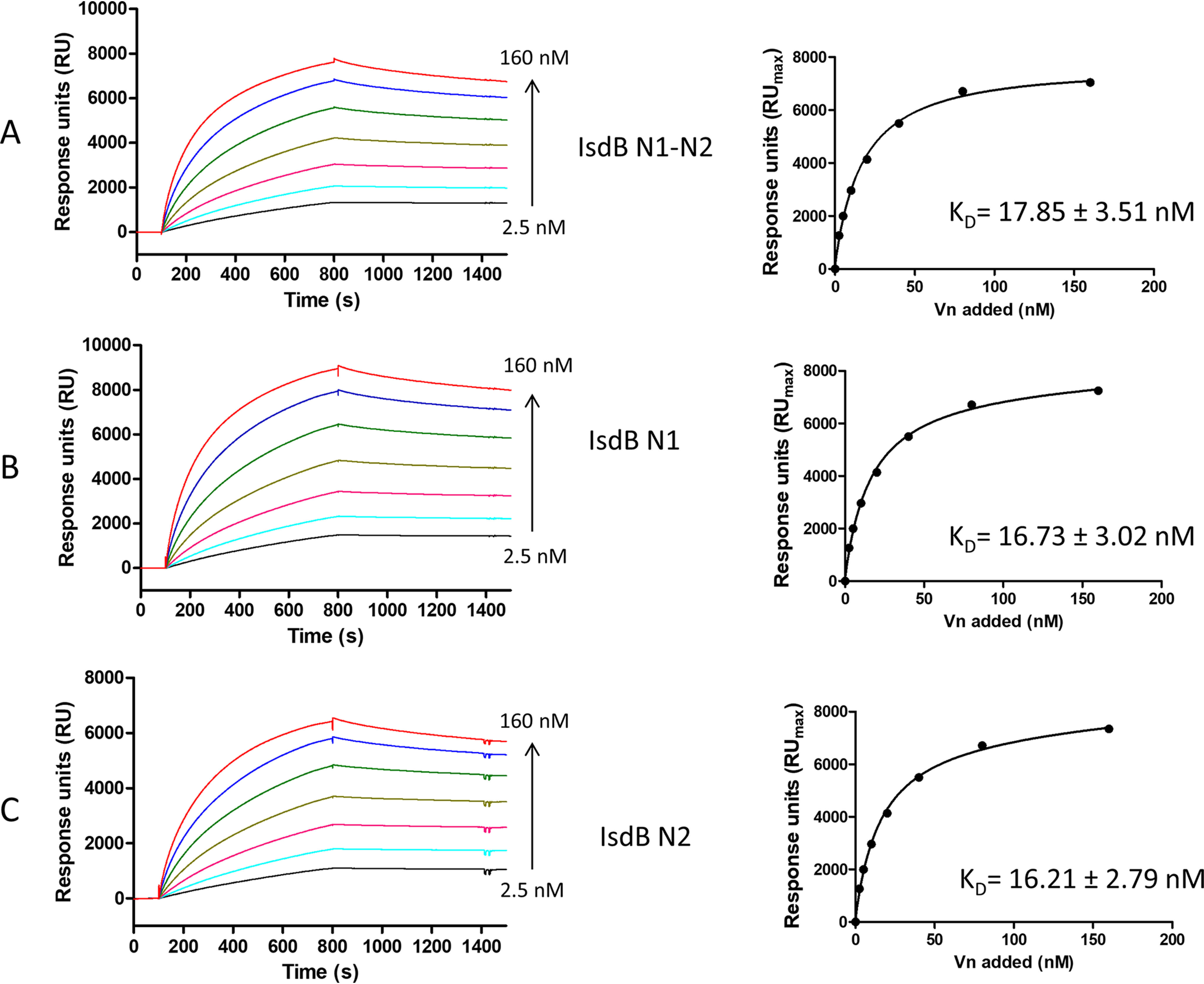
Analysis of the interaction between IsdB NEAT1-NEAT2 and its derivatives, NEAT1 and NEAT2, with Vn by SPR. Twofold linear dilution series (2.5–160 nm) of Vn were injected over the IsdB NEAT1-NEAT2 or its derivatives, NEAT1 and NEAT2, immobilized on the surface of a CM5 sensor chip. The sensorgrams obtained were normalized versus the response obtained when Vn was flowed over uncoated chips. The affinity was calculated from curve fitting to a plot of the response unit values at the steady state (RUmax) against increasing concentrations of Vn (left). Shown is one representative of three experiments.
Binding of Vn to IsdB is blocked by heparin and dependent on ionic strength
To investigate whether IsdB competes with glycosaminoglycans for binding to Vn, the effect of heparin, heparan sulfate, and chondroitin sulfate on Vn binding to immobilized IsdB NEAT1-NEAT2 was studied. In contrast to heparan sulfate and chondroitin sulfate, heparin dose dependently inhibited the IsdB-Vn interaction, suggesting that IsdB interacts with the heparin-binding domain(s) of the protein (Fig. S3A). To determine if ionic forces play a role in the interaction of IsdB with Vn, the effect of sodium chloride on Vn binding to IsdB was assessed. Addition of NaCl significantly reduced binding of Vn to immobilized IsdB, and at a concentration of 500 mm, NaCl reduced Vn binding to IsdB by almost 80% (Fig. S3B).
S. aureus IsdB captures Vn from plasma
To investigate if S. aureus cells expressing IsdB can recruit Vn from plasma, bacteria were incubated with human plasma and the amount of Vn captured was quantified by Western blotting and densitometry. S. aureus expressing IsdB captured approximately 4-fold more Vn than the isdB mutant. Thus, it can be concluded that IsdB expression is important for S. aureus to capture Vn from human plasma. However, it must be pointed out that a substantial amount of Vn bound to the isdB mutant, indicating that other Vn receptors are operational on the surface of S. aureus cells (Fig. 6).
Figure 6.
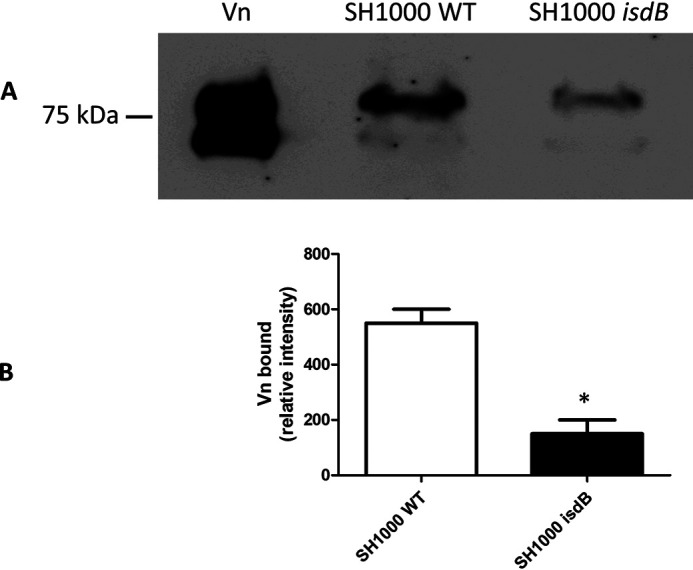
Capture of plasma vitronectin by S. aureus strain SH1000. A, S. aureus SH1000 and its isogenic isdB mutant were mixed with 1 ml of human plasma for 60 min. Proteins bound to the cell surface were released by extraction buffer, separated by SDS-PAGE under nonreducing conditions, and analyzed by Western blotting. The membrane was probed with sheep anti-Vn polyclonal IgG and HRP-conjugated rabbit anti-sheep and developed with the ECL Western blotting detection kit. The pure human Vn sample is shown on the left. B, densitometric analysis of Vn bound to and released from S. aureus SH1000 and its isogenic isdB mutant. The reported data are the mean values ± S.D. from three independent experiments. Statistically significant differences are indicated (Student's t test; *, p < 0.05).
Vn mediates adherence to and invasion of HeLa and HUVEC monolayers
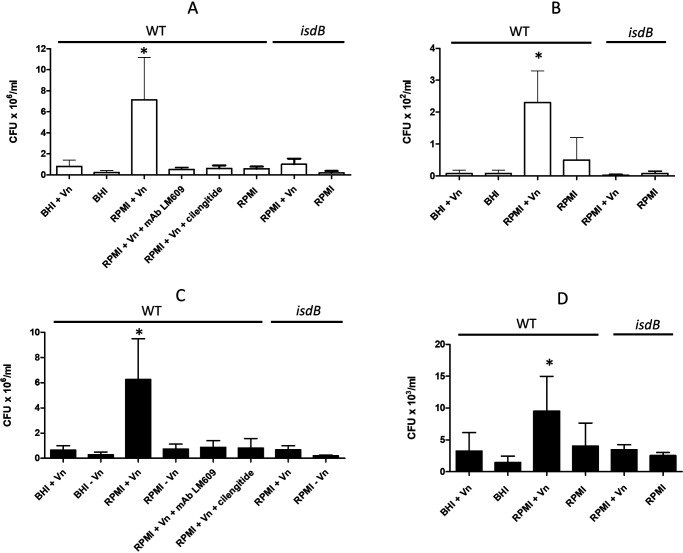
To investigate the role of the Vn-binding activity of IsdB in promoting adherence of S. aureus SH1000 to host cells, bacteria were grown to the stationary phase in BHI or RPMI medium and then tested for attachment to HeLa and human umbilical vein endothelial cell (HUVEC) monolayers (Fig. 7, A and C) in the absence of Vn. No adherence of the strain grown under either condition was observed. Conversely, the addition of exogenous human Vn to the cell monolayers promoted a high level of adhesion of the RPMI-grown but not the BHI-grown bacteria. Almost complete inhibition of adhesion was observed when the assays were performed in the presence of an integrin αvβ3 -binding mAb or the specific αvβ3 inhibitor cilengitide, suggesting the involvement of integrin αvβ3. To examine whether IsdB/Vn is also involved in the staphylococcal invasion of HeLa cells or HUVECs (Fig. 7, B and D), BHI- and RPMI-grown bacteria were incubated with cell monolayers in the presence or absence of Vn and then tested for internalization. We found a moderate level of invasion only by RPMI-grown bacteria in the presence of Vn, while no internalization was observed with BHI-grown bacteria even in the presence of Vn. In confirmation of this, neither adhesion nor invasion was observed with the isdB mutant (Fig. 7). Together, these data suggest that for promoting efficient adhesion and gaining entry of bacteria into HeLa cells or HUVECs, S. aureus can use Vn as a molecular bridge between surface-expressed IsdB and the αvβ3 integrin.
Figure 7.
Adhesion and internalization of S. aureus cells. Bacterial strains were grown overnight in BHI or RPMI medium, added to HeLa (A and B) or HUVEC (C and D) cell monolayers, and tested for adhesion (A and C) or internalization (B and D). The experiments were performed in DMEM for 2 h at 37 °C. The effects of αvβ3 mAb LM609 and cilengitide on bacterial adherence are also reported. The inocula and adherent and internalized bacteria were quantified by viable counting. Error bars show S.D. of the means from three independent determinations performed in triplicate with similar results (Student's t test; *, p < 0.05).
Staphylococcal adhesion to HeLa and HUVEC cell lines was similar, while bacterial invasion of HUVECs was 50-fold more effective than that of the HeLa cell line. Variation in expression levels of the αvβ3 integrin on the two cell types may contribute to the difference in pathogen invasiveness.
Notably, a persistent, low level of bacterial invasion was observed when bacteria grown in RPMI were incubated with HeLa and HUVECs in the absence of Vn, suggesting that other internalization mechanisms are operational.
Discussion
In our search for S. aureus MSCRAMMs with Vn-binding activity, here we show that S. aureus IsdB interacts with Vn. As previously demonstrated, IsdB is strongly upregulated under conditions of iron restriction and, in accordance with this property, Vn binding was strictly related to the expression of IsdB by bacterial strains. Bacteria bind Vn when grown under iron starvation conditions and not in an iron-rich medium. Moreover, binding of Vn by bacteria was mostly expressed by cells in the stationary phase of growth.
To characterize this binding to IsdB, we purified and used multimeric Vn, the conformational form of the protein found in the ECM. Notably, we found that bacteria expressing IsdB also captured Vn from plasma, where the monomeric form of the protein prevails. Thus, it seems that IsdB binds to both conformational states of Vn. This is reminiscent of the situation observed with Moraxella catarrhalis and Haemophilus influenzae, which interact with both isoforms of Vn (23).
The interaction of IsdB with Vn was confirmed using several approaches, including far Western blotting, ELISA, and SPR. Binding of IsdB to Vn was specific and saturable and exhibited a KD in the nanomolar range, which is comparable to that recorded for high-affinity CWA protein–host protein interactions (49–52).
To narrow down the Vn binding site(s) of IsdB, the NEAT1 and NEAT2 domains were recombinantly produced and tested for their ability to bind the ligand. The individual modules interacted with an affinity for Vn comparable to that of the full-length IsdB, suggesting that each module represents the minimal motif required for efficient interaction of IsdB with Vn and that IsdB potentially can bind two Vn molecules. The finding that both domains interact with Vn is particularly intriguing, in view of the fact that the two NEAT domains of IsdB have very low sequence identity (about 12%) and that they are functionally different. In fact, the NEAT1 domain is involved in Hb binding while NEAT2 plays a role in heme extraction from α and β chains of Hb (53). In view of the discovery that even NEAT motifs with low sequence identity show Vn-binding activity, other Isd proteins could exhibit Vn-binding activity. The presence of additional Vn receptors on the staphylococcal surface is consistent with the finding that the isdB mutant is capable of capturing Vn from human plasma and by the observation that other non-Isd surface proteins of S. aureus, such as Atl, have Vn-binding activity (26, 27). In apparent contradiction with these observations, IsdB seems to be the major Vn receptor when material released from bacterial cells was tested by far Western blotting (Fig. 2). This could be due to a higher level of expression of IsdB and/or to a higher affinity of IsdB for Vn compared with that of other potential Vn binders.
In this context, it will be worth investigating the activity of IsdB from Staphylococcus lugdunensis, which contains NEAT1 and NEAT2, 60 and 56% identical to S. aureus IsdB NEAT1 and NEAT2 domains, respectively (40), and the Vn-binding by other bacterial species expressing Isd proteins, such as Listeria monocytogenes and Bacillus anthracis.
Increasing the ionic strength had a dramatic effect on Vn binding to IsdB, indicating that Vn/IsdB complex formation is mainly driven by electrostatic interactions. Furthermore, the negatively charged heparin efficiently blocked the IsdB–Vn interaction, suggesting that the IsdB-binding region is located in the heparin-binding domain(s) of Vn. The inhibitory effect of heparin but not of heparan sulfate or chondroitin sulfate is in line with heparin possessing the highest negative charge density of any known biological macromolecule. In fact, although heparin and heparan sulfate are built up of linear chains of repeating disaccharide units, the average heparin disaccharide contains approximately 2.7 sulfate groups, whereas heparan sulfate contains ≥1 sulfate group per disaccharide (54).
Related to the finding that electrostatic bonds could be involved in Vn–IsdB interaction is the question of whether Vn and heme compete for the same binding site or have a different location on IsdB. The indication that IsdB uses only a hydrophobic pocket in the NEAT2 domain to bind and extract heme (38) and our observation that Vn binds to both NEAT1 and NEAT2 domains suggest that the binding sites in IsdB for heme and Vn are separate.
Binding of S. aureus to Vn contributes to bacterial adhesion to mammalian cells, as demonstrated by attachment of IsdB-expressing staphylococci to HeLa and HUVEC monolayers. The inhibition of the process by an αvβ3 mAb and cilengitide strongly suggests that adhesion involves the formation of a Vn bridged complex between IsdB and the integrin. Vn also mediated the internalization of staphylococci into epithelial and endothelial cells. However, due to the modest contribution of Vn to the process, it is possible that Vn-dependent invasion could act as a backup mechanism that becomes operational in the context of tissues where Fn is poorly expressed or under conditions (specific growth medium and/or phase of growth) where bacteria express reduced levels of Fn-binding proteins FnBPA and FnBPB (55, 56).
Binding to Fn by FnBPs allows the formation of a bridge between bacterial cells and the α5β1 integrin on host cells, and this is sufficient to trigger the bacterial uptake. This mechanism is widely acknowledged to be the main internalization process (57). Additional secondary invasion mechanisms have been reported, including, among others, the internalization mediated by EAP (58), Atl (31), and Lpl (59) proteins. Of note, the invasion assays performed in these studies have been carried out with experimental approaches and conditions (different cell lines and strains, absence/presence of serum) that significantly differ from the invasion pathway described in the present investigation. Thus, for an accurate analysis of the contribution of each mechanism to this multifactorial event, individual internalization processes should be evaluated and compared under identical standardized conditions.
It is unclear why the isdB deletion mutant, although it can express several Vn receptors (Fig. 6), does not adhere to and invade host cells (Fig. 7). If we hypothesize that non-IsdB receptors bind to Vn in a way that differs from IsdB, we can speculate that these receptors play a role other than adhesin/invasin in bacterial pathogenesis. For example, capture of Vn mediated by receptors could inhibit both assembly and deposition of the terminal complement complex on the bacterial surface. Alternatively, due to the plasminogen (PLG) binding ability of Vn (60), receptor-bound Vn could enhance plasminogen activation to plasmin, thereby contributing to the degradation of fibrin clots and bacterial spreading into the tissues.
IsdB binds directly to β3-containing integrins and promotes platelet activation and invasion of mammalian cells (48, 61). Consistent with this, we found a low level of invasion of both cell lines by bacteria grown in RPMI in the absence of Vn, suggesting that under these conditions the process can be driven by IsdB binding directly to integrins (48). As with many other MSCRAMMs, IsdB is a multivalent virulence factor and contributes in various ways to the pathogenesis of S. aureus. Examples of MSCRAMMs that bind to two or more ligands are clumping factor A (ClfA) that, in addition to binding to fibrinogen (62, 63), binds to complement factor I (64, 65), and clumping factor B (ClfB), another CWA protein that interacts with fibrinogen (66, 67), keratin 10 (68, 69), and loricrin (70, 71). These CWA proteins share similar mechanisms of binding (5). In parallel with this, the structural basis of Hb binding to IsdB NEAT motifs has been elucidated (72, 73). From this perspective, X-ray crystal structure analysis of the recombinant IsdB-binding domain of Vn in complex with NEAT motifs could provide clues about the mechanism and function of this important CWA protein in bacterial colonization of and survival within the host. Purified IsdB elicits antibodies that block heme iron scavenging, provide partial protection against S. aureus bacteremia in animal models (74–76), and promote opsonophagocytosis of S. aureus (75, 77). Although vaccination of humans with IsdB resulted in failure (78, 79), in light of these new findings, the use of IsdB as a vaccine component warrants further investigation.
Experimental procedures
Bacterial strains and culture conditions
All strains used in this study are listed in Table 1. S. aureus cells (48, 80–84) were grown overnight in BHI (VWR International Srl, Milan Italy) or in RPMI 1640 (Sigma-Aldrich, MO, USA) medium at 37 °C with shaking. L. lactis cells carrying the expression vector alone (pNZ8037) (85) or harboring the isdB gene (pNZ8037::isdB) (48) were grown overnight in BHI medium supplemented with chloramphenicol (10 μg/ml) at 30 °C without shaking. Cultures of L. lactis were diluted 1:100 in the same medium and allowed to reach exponential phase. Nisin (6.4 ng/ml) was added, and cultures were allowed to grow overnight as described above. In experiments where a defined number of cells were used, bacteria were harvested from the cultures by centrifugation, washed, and suspended in PBS at an optical density at 600 nm (OD600) of 1.0. Escherichia coli strain XL1-Blue (Agilent Technologies, CA, USA) transformed with vector pQE30 (Stratagene, La Jolla, CA) or derivatives were grown in Luria agar and Luria broth (VWR) containing 100 μg/ml ampicillin at 37 °C with shaking.
Table 1.
Bacterial strains
| Bacterial strain | Relevant propertiesa | Reference or source |
|---|---|---|
| S. aureus | ||
| LAC spa | Strain derivative of LAC* deficient in protein A; constructed by transduction of spa::Kanr by phage 85 into strain LAC* | 80 |
| Newman spa | Strain containing a null mutation in protein A obtained through transduction from S. aureus 8325-4 spa::Kanr (81) by generalized phage transduction using phage 85 | 82 |
| 8325-4 spa | spa gene inactivated by substituting part of the spa coding sequence for a DNA fragment specifying resistance to ethidium bromide. In vitro-constructed spa::EtBrr substitution mutation was introduced into the S. aureus chromosome by recombinational allele replacement | 81 |
| SH1000 | Laboratory strain. rsbU + derivative of S. aureus 8325‐4 rsbU + derivative of S. aureus 8325‐4 | 83 |
| SH1000 spa | spa::Tcr transduced from 8325-4 spa::Tcr | 84 |
| SH1000 isdB | isdB gene deleted by allelic exchange | 48 |
| L. lactis | ||
| NZ9800 (pNZ8037) | Expression vector with nisin-inducible promoter, Cmr | 85 |
| NZ9800 (pNZ8037::isdB) | isdB gene cloned in pNZ8037, Cmr | 48 |
| E. coli | ||
| XL1-Blue | E. coli cloning host | Stratagene |
a Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Tcr, tetracycline resistance.
Construction of S. aureus isdB deletion mutant and L. lactis expressing IsdB
Construction of Lactococcus lactis expressing IsdB and construction of the S. aureus isdB deletion mutant were performed as reported in Table 1.
Plasmid and DNA manipulation
Plasmid DNA (Table 2) was isolated using the WizardPlus SV miniprep kit (Promega, WI, USA), according to the manufacturer's instructions, and transformed into E. coli XL1-Blue cells using standard procedures (86). Transformants were screened by restriction analysis and verified by DNA sequencing (Eurofins Genomics, Milan, Italy). Chromosomal DNA was extracted using the bacterial genomic DNA purification kit (Edge Biosystems, MD, USA). Cloning of IsdB NEAT1-NEAT2 (aa residues 48–480) was performed as reported by Miajlovic et al. (61). Cloning of IsdB NEAT1 (aa residues 144–270) and IsdB NEAT2 (aa residues 334–458) domains was performed following the NEBuilder® HiFi DNA assembly according to the manufacturer's instructions (New England Biolabs, MA, USA). The primers used to amplify IsdB NEAT1 and IsdB NEAT2 domains and the pQE30 vector (Table S1) were purchased from Integrated DNA Technologies (Leuven, Belgium). DNA purification was carried out using the WizardSV gel and PCR clean-up system (Promega).
Table 2.
Plasmids
| Plasmid | Feature | Markera | Source or reference |
|---|---|---|---|
| pQE30 | E. coli vector for the expression of hexa-His-tagged recombinant proteins | Ampr | Qiagen |
| pQE30::isdB NEAT1-NEAT2 | pQE30 encoding residues 48–480 of IsdB NEAT1-NEAT2 protein | Ampr | 61 |
| pQE30::rIsdB-NEAT1 (144–270) | pQE30 derivative encoding the NEAT1 domain of IsdB NEAT1-NEAT2 protein from S. aureus SH1000 | Ampr | This study |
| pQE30::rIsdB-NEAT2 (334–458) | pQE30 derivative encoding the NEAT2 domain of IsdB protein from S. aureus SH1000 | Ampr | This study |
a Ampr, ampicillin resistance.
Expression and purification of recombinant proteins
Recombinant proteins IsdB NEAT1-NEAT2, IsdB-NEAT1, and IsdB-NEAT2 were expressed from pQE30 (Qiagen, Hilden, Germany) in E. coli XL1-Blue (Agilent Technologies, CA, USA). Overnight starter cultures were diluted 1:40 in Luria broth containing ampicillin (100 μg/ml) and incubated with shaking until the culture reached the exponential phase (optical density at 600 nm of 0.4–0.6). Recombinant protein expression was induced by the addition of 1 mm (final concentration) isopropyl 1-thio-β-D-galactopyranoside (IPTG) (Inalco, Milan, Italy) to the culture. After 4 h, bacterial cells were harvested by centrifugation and frozen at −80 °C. Cells were resuspended in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, pH 8.0) containing 1 mm PMSF (Sigma-Aldrich) as the protease inhibitor and 20 μg/ml DNase (Sigma-Aldrich) and lysed by freezing with liquid nitrogen and subsequent defrosting with warm water. The lysis procedure was repeated twice. The cell debris was removed by centrifugation, and proteins were purified from the supernatants by Ni+2-affinity chromatography on a HiTrap chelating column (GE Healthcare, Buckinghamshire, UK).
Protein purity was assessed by 15% SDS-PAGE and Coomassie brilliant blue staining (Fig. S4A). A bicinchoninic acid protein assay (Pierce, Rockford, IL, USA) was used to measure the concentration of purified proteins.
Reagents, proteins, and antibodies
BSA, hemoglobin, protease-free DNase I, skim milk, heparin, chondroitin sulfate, heparan sulfate, cilengitide, lysostaphin, trypsin, sheep anti-Vn polyclonal IgG, and HRP-conjugated rabbit anti-sheep IgG were purchased from Sigma-Aldrich. Human fibronectin (Fn) was purified from plasma by a combination of gelatin- and arginine-Sepharose affinity chromatography (87). The human Fn rabbit antibody was purchased from Pierce. Human fibrinogen (Fbg) was obtained from Calbiochem (Darmstadt, Germany). Collagen (Coll) type I was a generous gift of Professor R. Tenni (Department of Molecular Medicine, Pavia, Italy). Coll type I rabbit antibody was from Merck (Rome, Italy). αvβ3 integrin mAb LM609 was from Abcam (Cambridge, UK). Vn mAb 8E6 was purchased from Merck. IsdB and Fbg antibodies were raised in mice by a routine immunization procedure using purified IsdB NEAT1-NEAT2 and Fbg as the antigen, respectively. Rabbit anti-mouse HRP-conjugated secondary antibody was purchased from Dako Cytomation (Glostrup, Denmark).
Human plasma
Normal human plasma was prepared from freshly drawn blood obtained from healthy volunteers with informed consent and permission of the ethical board of the University of Pavia (permit no. 19092013). The study also abides by the Declaration of Helsinki principles. After centrifugation, the plasma fraction was frozen in aliquots and stored at −20 °C.
Isolation of Vn from human plasma
Human Vn was purified from human plasma on a heparin-Sepharose Hi-TrapTM column (GE Healthcare) by following the protocol described by Akiyama (88). The method takes advantage of the observation that the monomeric plasma Vn must be activated or “opened” with 8 M urea before it can bind well to heparin. Thus, the plasma is first depleted of compounds such as fibronectin that bind specifically or nonspecifically to heparin-Sepharose and then treated with 8 M urea to activate the heparin-binding activity of Vn, which is subsequently bound to a heparin affinity column and eluted in a multimeric conformation.
The purity of the isolated Vn was checked with 12.5% (w/v) SDS-PAGE under reducing conditions and Coomassie brilliant blue staining (Fig. S4B, lane 1) and its concentration quantified with the BCA assay. The multimeric conformation of the isolated Vn was assessed by 12.5% PAGE performed in the absence of SDS (Fig. S4B, lane 2) and by determination of the reactivity with the mAb 8E6, which preferentially recognizes the multimeric, activated form of Vn (data not shown).
Vn binding to S. aureus
To test binding of Vn, microtiter wells were coated overnight at 37 °C with 100 µl/well of stationary-phase S. aureus cells (OD600 of 1) in PBS. The plates were washed with PBS. To block free protein binding sites, the wells were treated for 1 h at 22 °C with BSA (2%, v/v) in PBS. The plates were then incubated for 1 h with increasing concentrations of Vn (up to 250 ng/well). After several washings with PBS, 0.5 μg of anti-Vn sheep IgG in BSA (1%, v/v) was added to the wells and incubated for 90 min.
The plates were washed and then incubated for 1 h with rabbit HRP-conjugated anti-sheep IgG diluted 1:1000. After washing, o-phenylenediamine dihydrochloride was added, and the absorbance at 490 nm was determined using an ELISA plate reader (Bio-Rad, CA USA).
Release of cell wall-anchored proteins from S. aureus and detection of Vn-binding activity
Lysostaphin digestion
To release cell wall-anchored proteins from S. aureus, bacteria were grown at 37 °C to exponential phase (OD600 of 0.4–0.6) or stationary phase, either in RPMI or BHI medium. Cells were harvested by centrifugation at 7000 × g at 4 °C for 15 min, washed three times with PBS, and resuspended to an A600 of 2.0 in lysis buffer (50 mm Tris-HCl, 20 mm MgCl2, pH 7.5) supplemented with 30% raffinose. Cell wall proteins were solubilized from S. aureus by incubation with lysostaphin (200 μg/ml) at 37 °C for 20 min in the presence of protease inhibitors (Complete Mini; Sigma-Aldrich). Protoplasts were recovered by centrifugation at 6000 × g for 20 min, and the supernatants were taken as the wall fraction. The supernatant was concentrated by treatment with 20% (v/v) TCA at 4 °C for 30 min. The precipitated proteins were washed twice with ice-cold acetone and dried overnight.
SDS-PAGE and far Western blotting
Proteins released by lysostaphin digestion were boiled for 10 min in sample buffer (0.125 M Tris-HCl, 4% [w/v] SDS, 20% [v/v] glycerol, 10% [v/v] β-mercaptoethanol, 0.002% [w/v] bromphenol blue) and separated by 12.5% (w/v) SDS-PAGE. The gels were stained with Coomassie brilliant blue (Bio-Rad). For Western blotting, proteins were subjected to SDS-PAGE and electroblotted onto a nitrocellulose membrane (GE Healthcare). After blocking with 5% (w/v) skim milk (Sigma-Aldrich) in PBS overnight at 4 °C, the membrane was probed with 2 μg/ml of Vn in PBS for 1 h at 22 °C followed by anti-Vn sheep IgG (1:10,000) and with rabbit HRP-conjugated anti-sheep IgG (1:100). Finally, blots were developed using the ECL Advance Western blotting detection kit (GE Healthcare), and images of the bands were captured by an ImageQuantTM LAS 4000 mini-biomolecular imager (GE Healthcare).
MS analysis
Proteins released by lysostaphin digestion of bacterial cells were separated by 12.5% SDS-PAGE and stained with GelCode blue stain reagent (Thermo Fisher Scientific, Waltham, MA, USA). The band marked by the red arrow (Fig. 2) was excised from the gel using a sterile scalpel and protein contained in the gel digested with sequencing grade trypsin (Sigma-Aldrich). Tryptic peptides were mixed with CHCA (alpha-cyano-4-hydroxycinnamic acid) and spotted onto the target plate. The spectra were acquired with a TOF/TOF 5800 system (AB SCIEX, Framingham, MT) using TOF/TOF Series Explorer acquisition software version 4.1.0. MS spectra were recorded in Reflecton positive mode (settings: fixed laser intensity, 3200 units; pulse rate, 400 Hz; total shots/spectrum, 1000; mass range (Da), 1000–4000 m/z). Calibration was performed using Peptide calibration Mix4 (LaserBio Laboratory, Sophia-Antipolis Cedex, France). MS-MS spectra of the most intensive MS signals were recorded in MS-MS positive mode (settings: fixed laser intensity, 3500 units; laser pulse rate, 1000 Hz; total shots/spectrum, 5000). MS-MS spectra were analyzed with Protein-PilotTM 5.0 software (AB SCIEX, Framingham, MT, USA) against the UniProtKB database updated 2 August 2016 using the following analysis parameters: (a) sample type, identification; (b) digestion, trypsin; (c) detected protein threshold [unused Protscore (Conf)], >0.05 (10%); (d) cysteine alkylation, iodoacetamide; (e) ID focus, biological modifications, amino acid substitutions; (f) FDR analysis, no; (g) taxonomy, no species.
Determination of isdB gene expression by qRT-PCR
Total RNA was extracted from S. aureus SH1000 spa cells during the exponential (OD600 of 0.4–0.6) and stationary phases of growth in BHI and RPMI media. Cells were harvested and total RNA was stabilized with RNeasy Protect bacterial reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total RNA was extracted using the Quick-RNA fungal/bacterial miniprep kit (Zymo Research, CA, USA) by following the manufacturer's recommendations. DNA was removed by DNase I treatment by using the TURBO DNA-free kit (Invitrogen, CA, USA). The RNA concentration was >100 ng/μl, and the A260/A280 ratio was >1.8. qRT-PCR was performed with an iTAq Universal SYBR Green one-step kit (Bio-Rad) using 4 ng of RNA in 20-µl volumes carried out on three replicates, and all reactions were performed in 20-μl volumes according to the manufacturer's protocol. The cDNA was analyzed using primers relative to the isdB coding sequence (Table S1). The conditions for thermal cycling were 50 °C for 10 min and 95 °C for 1 min, 35–40 cycles with 95 °C for 10 s, and then 60 °C for 10–30 s, followed by a slow increase of temperature by 0.5 °C per cycle to 95 °C, with a continuous measurement of the fluorescence. 4 ng of total RNA from each sample was used in a qRT-PCR experiment performed using a CFX Connect real-time PCR detection system (Bio-Rad) and an iTAq Universal SYBR Green one-step kit (Bio-Rad). Expression of isdB was analyzed using primers isdBF (GCAGGCGTTTTGTCTTTACC) and isdBR (GCCTAGCAAACCAACACCAT). Target gene expression was normalized using the reference gene rpoC, which was amplified using primers SAurpoCF (CCGCACCATCTGGTAAGATTAT) and SAurpoCR (GCTGTATCGGCAAGACCTTTA). No-template and no-RT controls were run for each assay, and the specificity of each amplification product was verified using dissociation curve analysis. Standard curves were generated from serial dilutions of chromosomal DNA spanning at least six orders of magnitude. All reactions proceeded with 90% to 110% efficiency. Gene expression analysis was performed using the CFX Manager software (Bio-Rad). Three technical replicates were performed for each experiment.
Determination of expression level of IsdB by Western immunoblotting
Material released by lysostaphin digestion of SH1000 spa grown either in BHI and RPMI media to both mid-exponential and stationary phases was subjected to SDS-PAGE and electroblotted onto nitrocellulose membrane, as reported above. After blocking with skim milk, the membrane was sequentially incubated with mouse IsdB antibody (1:5000) and HRP-conjugated rabbit anti-mouse IgG (1:10,000) and developed as described above.
Detection of the Vn-binding activity of IsdB-expressing L. lactis
Release of cell wall-anchored proteins from L. lactis transformants by mutanolysin/lysozyme digestion and far Western blotting
L. lactis was grown to late-exponential phase in BHI broth, washed twice in PBS, and concentrated to an A600 of 40 in 1 ml 26% raffinose (Sigma-Aldrich) in 20 mm Tris-HCl, pH 8, 10 mm MgCl2. Cell wall-associated proteins were released by incubation at 37 °C with occasional shaking with 500 U mutanolysin/ml and 200 µg lysozyme/ml in the presence of protease inhibitors (Complete Mini; Sigma-Aldrich). Protoplasts were removed by centrifugation at 6000 × g for 20 min, and the supernatant was concentrated as reported above. The interaction of Vn with IsdB was detected by SDS-PAGE followed by far Western blotting as reported above.
Vn binding to L. lactis by ELISA
To investigate the binding of Vn to L. lactis, microtiter wells were coated overnight at 37 °C with 100 µl/well of bacterial cell suspensions (OD600 of 1) in PBS. After treating with BSA, the plates were incubated with 10 µg/well Vn. Proteins bound to the bacterial cells were detected as described above.
Binding of Vn to IsdB by ELISA
The ability of immobilized recombinant IsdB proteins to bind to soluble Vn was determined using ELISA. Microtiter wells were coated overnight at 4 °C with 0.2 µg/well of the appropriate IsdB protein in 0.1 M sodium carbonate, pH 9.5. The plates were washed with 0.5% (v/v) Tween 20 in PBS (PBST) and then treated for 1 h at 22 °C with BSA (2%, v/v) in PBS. The plates were incubated for 1 h with increasing concentrations of soluble Vn in PBS. Bound Vn was detected by sheep anti-Vn IgG followed by rabbit HRP-conjugated anti-sheep IgG.
To determine the effect of ionic strength on the Vn-IsdB interaction, microtiter wells coated with 200 ng of IsdB NEAT1-NEAT2 were incubated with 500 ng/well of Vn in PBS containing increasing concentrations of NaCl. Complex formation was detected by incubation of the wells with antibodies as described above.
The effect of heparin, chondroitin sulfate, and heparan sulfate on the IsdB NEAT1-NEAT2–Vn interaction was examined by incubating IsdB NEAT1-NEAT2 immobilized onto microtiter plates (0.2 µg/well) with 0.5 µg/well of purified Vn in the presence of increasing concentrations of heparin, chondroitin sulfate, or heparan sulfate. Vn bound to IsdB NEAT1-NEAT2 was detected with antibodies as reported above.
SPR
To estimate the affinity of the interaction between Vn and IsdB NEAT1-NEAT2 or its domain NEAT1 or NEAT2, surface plasmon resonance (SPR) analyses were carried out using a multicycle injection strategy on a multiple flow cell Biacore X100 instrument (GE-Healthcare). IsdB NEAT1-NEAT2, NEAT1, or NEAT2 was covalently immobilized on a dextran matrix CM5 sensor chip surface in three different flow cells by using a protein solution (50 µg/ml in 50 mm sodium acetate buffer, pH 4.5) in a 1:1 dilution with N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride. The excess of the active groups on the dextran matrix was blocked using 1 M ethanolamine, pH 8.5. On the fourth flow cell, the dextran matrix was treated as described above but without any ligand to provide an uncoated reference flow cell. The running buffer used was PBS containing 0.005%, v/v, Tween 20. A 2-fold linear dilution series of Vn, in running buffer, was passed over the ligand at a flow rate of 30 µl/min, and all the sensorgrams were recorded at 22 °C. Assay channel data were subtracted from reference flow cell data. The response units at the steady state were plotted as a function of Vn concentration and fitted to the Langmuir equation to yield the KD values.
Capture of Vn by S. aureus cells
Stationary S. aureus strain SH1000 or the isdB mutant (108 cells/ml) were mixed with 1 ml of fresh human plasma for 30 min. Bacteria were then harvested by centrifugation, washed with PBS, and treated with the extraction buffer (125 mm Tris-HCl, pH 7.0, containing 2% SDS) for 3 min at 95 °C and then centrifuged at 10,000 × g for 3 min. The supernatants were subjected to SDS-PAGE under reducing conditions, and the proteins were transferred to a nitrocellulose membrane. The membrane was sequentially probed for bound Vn with antibodies as described above. The band intensities were quantified with Quantity One software (Bio-Rad).
To analyze the capture of purified Vn by S. aureus SH1000 and its spa mutant, bacteria (1 × 108cells/ml) were grown either in BHI or RPMI medium in the absence/presence of 1 mm FeCl3 and then incubated with purified Vn for 60 min. Bacterium-bound proteins were released from the bacterial surface by extraction buffer and subjected to SDS-PAGE and far Western blotting as reported above.
Cell culture infection with staphylococci and blocking experiments
HeLa cells were cultured in 24‐well plates until reaching confluence in DMEM supplemented with 10% FBS. HUVECs were cultured as previously reported (89).
24 h before adherence or invasion assays, the medium was removed from each culture and replaced with serum‐free medium. Overnight cultures of S. aureus SH1000 WT or the isdB mutant grown either in BHI or RPMI medium were suspended in DMEM without antibiotics and FBS to 1 × 107cells/ml and added to the monolayers for 2 h at 37 °C in 5% CO2. The effect of Vn on bacterial adhesion/invasion to monolayers was tested by pretreating the cells with 25 μg/ml of the protein 1 h prior to addition of the bacteria. To evaluate the role of αvβ3 integrin antibody and cilengitide on adhesion, monolayers were pretreated with Vn and then incubated with S. aureus SH1000 WT in the presence of 1 µg/ml of αvβ3 integrin LM609 mAb or 0.05 μm cilengitide. To determine bacterial adhesion, the infected cells were washed three times with Dulbecco's PBS, lysed, and plated on BHI agar for CFU counts. To enumerate internalized bacteria, the monolayers were further incubated for 2 h before cell lysis in medium supplemented with 100 μg/ml gentamicin to kill extracellular bacteria. Bacterial adherence and invasion are represented as recovered CFU/ml.
Statistical methods
Two group comparisons were performed by Student's t test. One-way analysis of variance, followed by Bonferroni's post hoc tests, was exploited for comparison of three or more groups. Analyses were performed using Prism 4.0 (GraphPad). Two-tailed P values of 0.05 were considered statistically significant.
Data availability
The MS proteomics data are available via ProteomeXchange with identifier PXD019371 (90). All other data supporting the findings of this study are available within the article and its supporting data.
Supplementary Material
Acknowledgments
We thank Giulia Barbieri, Department of Biology and Biotechnology, Pavia, Italy, for help with validation of isdB gene expression by qRT-PCR and cloning of recombinant fragments of IsdB. We thank PTS (Parco Tecnico Scientifico) of the University of Pavia and Polymerix srl, Pavia, Italy, for technical support with mass spectrometry.
This article contains supporting information.
Author contributions—G. P. conceptualization; G. P. resources; G. P., A. P., and M. J. A. data curation; G. P., A. P., and M. J. A. formal analysis; G. P., L. M., T. J. F., and P. S. supervision; G. P. funding acquisition; G. P., A. P., M. J. A., L. M., and P. S. validation; G. P., L. M., and P. S. visualization; G. P., A. P., M. J. A., L. M., and P. S. methodology; G. P., T. J. F., and P. S. writing-original draft; G. P. project administration; G. P., L. M., T. J. F., and P. S. writing-review and editing; A. P. and M. J. A. investigation; L. M. and P. S. software.
Funding and additional information—This research was funded by FFABR 2018, “Fondo di finanziamento per le attività base di ricerca,” Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) to G. P.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ECM
- extracellular matrix
- Aaa
- autolysin/adhesin from S. aureus
- Atl
- autolysin
- coll
- collagen
- EAP
- extracellular adherence protein
- Fn
- fibronectin
- Fbg
- fibrinogen
- Isd
- iron-regulated surface determinant
- Lpl
- lipoprotein-like lipoprotein
- NEAT
- near iron transporter
- MSCRAMM
- microbial surface component recognizing adhesive matrix molecule
- PLG
- plasminogen
- RU
- response unit
- SPR
- surface plasmon resonance
- Vn
- vitronectin
- OD
- optical density
- qRT-PCR
- quantitative RT-PCR
- CWA
- cell wall-anchored
- BHI
- brain heart infusion
- RPMI
- Roswell Park Memorial Institute
- HUVEC
- human umbilical vein endothelial cell.
References
- 1. Tong S. Y., Davis J. S., Eichenberger E., Holland T. L., and Fowler V. G. Jr. (2015) Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hynes R. O., and Naba A. (2012) Overview of the matrisome—an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 4, a004903 10.1101/cshperspect.a004903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naba A., Clauser K. R., Ding H., Whittaker C. A., Carr S. A., and Hynes R. O. (2016) The extracellular matrix: tools and insights for the “omics” era. Matrix Biol. 49, 10–24 10.1016/j.matbio.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foster T. J., Geoghegan J. A., Ganesh V. K., and Höök M. (2014) Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foster T. J. (2019) The MSCRAMM family of cell-wall-anchored surface proteins of gram-positive cocci. Trends Microbiol. 27, 927–941 10.1016/j.tim.2019.06.007 [DOI] [PubMed] [Google Scholar]
- 6. Preissner K. T., and Seiffert D. (1998) Role of vitronectin and its receptors in haemostasis and vascular remodeling. Thromb Res. 89, 1–21 10.1016/S0049-3848(97)00298-3 [DOI] [PubMed] [Google Scholar]
- 7. Leavesley D. I., Kashyap A. S., Croll T., Sivaramakrishnan M., Shokoohmand A., Hollier B. G., and Upton Z. (2013) Vitronectin—master controller or micromanager? IUBMB Life 65, 807–818 10.1002/iub.1203 [DOI] [PubMed] [Google Scholar]
- 8. Boyd N. A., Bradwell A. R., and Thompson R. A. (1993) Quantitation of vitronectin in serum: evaluation of its usefulness in routine clinical practice. J. Clin. Pathol. 46, 1042–1045 10.1136/jcp.46.11.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chauhan A. K., and Moore T. L. (2006) Presence of plasma complement regulatory proteins clusterin (Apo J) and vitronectin (S40) on circulating immune complexes (CIC). Clin. Exp. Immunol. 145, 398–406 10.1111/j.1365-2249.2006.03135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., et al. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 7, 2019–2027 10.1074/mcp.R800013-MCP200 [DOI] [PubMed] [Google Scholar]
- 11. Felding-Habermann B., and Cheresh D. A. (1993) Vitronectin and its receptors. Curr. Opin. Cell Biol. 5, 864–868 10.1016/0955-0674(93)90036-P [DOI] [PubMed] [Google Scholar]
- 12. Schvartz I., Seger D., and Shaltiel S. (1999) Vitronectin. Int. J. Biochem. Cell Biol. 31, 539–544 10.1016/S1357-2725(99)00005-9 [DOI] [PubMed] [Google Scholar]
- 13. Stanley K. K. (1986) Homology with hemopexin suggests a possible scavenging function for S-protein/vitronectin. FEBS Lett. 199, 249–253 10.1016/0014-5793(86)80489-6 [DOI] [PubMed] [Google Scholar]
- 14. Liang O. D., Rosenblatt S., Chhatwal G. S., and Preissner K. T. (1997) Identification of novel heparin-binding domains of vitronectin. FEBS Lett. 407, 169–172 10.1016/S0014-5793(97)00330-X [DOI] [PubMed] [Google Scholar]
- 15. Seiffert D. (1995) Evidence that conformational changes upon the transition of the native to the modified form of vitronectin are not limited to the heparin binding domain. FEBS Lett. 368, 155–159 10.1016/0014-5793(95)00630-R [DOI] [PubMed] [Google Scholar]
- 16. Izumi M., Yamada K. M., and Hayashi M. (1989) Vitronectin exists in two structurally and functionally distinct forms in human plasma. Biochim. Biophys. Acta 990, 101–108 10.1016/S0304-4165(89)80019-4 [DOI] [PubMed] [Google Scholar]
- 17. Stockmann A., Hess S., Declerck P., Timpl R., and Preissner K. T. (1993) Multimeric vitronectin. Identification and characterization of conformation-dependent self-association of the adhesive protein. J. Biol. Chem. 268, 22874–22882 [PubMed] [Google Scholar]
- 18. Zhuang P., Li H., Williams J. G., Wagner N. V., Seiffert D., and Peterson C. B. (1996) Characterization of the denaturation and renaturation of human plasma vitronectin. II. Investigation into the mechanism of formation of multimers. J. Biol. Chem. 271, 14333–14343 10.1074/jbc.271.24.14333 [DOI] [PubMed] [Google Scholar]
- 19. Lynn G. W., Heller W. T., Mayasundari A., Minor K. H., and Peterson C. B. (2005) A model for the three-dimensional structure of human plasma vitronectin from small-angle scattering measurements. Biochemistry 44, 565–574 10.1021/bi048347s [DOI] [PubMed] [Google Scholar]
- 20. Seiffert D., and Smith J. W. (1997) The cell adhesion domain in plasma vitronectin is cryptic. J. Biol. Chem. 272, 13705–13710 10.1074/jbc.272.21.13705 [DOI] [PubMed] [Google Scholar]
- 21. Milis L., Morris C. A., Sheehan M. C., Charlesworth J. A., and Pussell B. A. (1993) Vitronectin-mediated inhibition of complement: evidence for different binding sites for C5b-7 and C9. Clin. Exp. Immunol. 92, 114–119 10.1111/j.1365-2249.1993.tb05956.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Attia A. S., Ram S., Rice P. A., and Hansen E. J. (2006) Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 74, 1597–1611 10.1128/IAI.74.3.1597-1611.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh B., Su Y.-C., and Riesbeck K. (2010) Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol. Microbiol. 78, 545–560 10.1111/j.1365-2958.2010.07373.x [DOI] [PubMed] [Google Scholar]
- 24. Bergmann S., Lang A., Rohde M., Agarwal V., Rennemeier C., Grashoff C., Preissner K. T., and Hammerschmidt S. (2009) Integrin-linked kinase is required for vitronectin-mediated internalization of Streptococcus pneumoniae by host cells. J. Cell Sci. 122, 256–267 10.1242/jcs.035600 [DOI] [PubMed] [Google Scholar]
- 25. Buommino E., Di Domenico M., Paoletti I., Fusco A., De Gregorio V., Cozza V., Rizzo A., Tufano M. A., and Donnarumma G. (2014) AlphaVBeta5 integrins mediates Pseudomonas fluorescens interaction with A549 cells. Front. Biosci. 19, 408–415 10.2741/4215 [DOI] [PubMed] [Google Scholar]
- 26. Heilmann C., Hussain M., Peters G., and Götz F. (1997) Evidence for autolysin‐mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024 10.1046/j.1365-2958.1997.4101774.x [DOI] [PubMed] [Google Scholar]
- 27. Heilmann C., Thumm G., Chhatwal G. S., Hartleib J., Uekötter A., and Peters G. (2003) Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149, 2769–2778 10.1099/mic.0.26527-0 [DOI] [PubMed] [Google Scholar]
- 28. Hussain M., Steinbacher T., Peters G., Heilmann C., and Becker K. (2015) The adhesive properties of the Staphylococcus lugdunensis multifunctional autolysin AtlL and its role in biofilm formation and internalization. Int. J. Med. Microbiol. 305, 129–139 10.1016/j.ijmm.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 29. Zoll S., Schlag M., Shkumatov A. V., Rautenberg M., Svergun D. I., Götz F., and Stehle T. (2012) Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J. Bacteriol. 194, 3789–3802 10.1128/JB.00331-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kohler T. P., Gisch N., Binsker U., Schlag M., Darm K., Völker U., Zähringer U., and Hammerschmidt S. (2014) Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J. Biol. Chem. 289, 4070–4082 10.1074/jbc.M113.521229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirschhausen N., Schlesier T., Schmidt M. A., Götz F., Peters G., and Heilmann C. (2010) A novel staphylococcal internalization mechanism involves the major autolysin Atl and heat shock cognate protein Hsc70 as host cell receptor. Cell. Microbiol. 12, 1746–1764 10.1111/j.1462-5822.2010.01506.x [DOI] [PubMed] [Google Scholar]
- 32. Hammer N. D., and Skaar E. P. (2011) Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65, 129–147 10.1146/annurev-micro-090110-102851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. A., and Schneewind O. (2003) Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909 10.1126/science.1081147 [DOI] [PubMed] [Google Scholar]
- 34. Skaar E. P., and Schneewind O. (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6, 390–397 10.1016/j.micinf.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 35. Dryla A., Gelbmann D., Von Gabain A., and Nagy E. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin‐haemoglobin binding activity. Mol. Microbiol. 49, 37–53 10.1046/j.1365-2958.2003.03542.x [DOI] [PubMed] [Google Scholar]
- 36. Clarke S. R., Wiltshire M. D., and Foster S. J. (2004) IsdA of Staphylococcus aureus is a broad spectrum, iron‐regulated adhesin. Mol. Microbiol. 51, 1509–1519 10.1111/j.1365-2958.2003.03938.x [DOI] [PubMed] [Google Scholar]
- 37. Maresso A. W., and Schneewind O. (2006) Iron acquisition and transport in Staphylococcus aureus. Biometals 19, 193–203 10.1007/s10534-005-4863-7 [DOI] [PubMed] [Google Scholar]
- 38. Grigg J. C., Ukpabi G., Gaudin C. F., and Murphy M. E. (2010) Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 104, 341–348 10.1016/j.jinorgbio.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 39. Pilpa R. M., Fadeev E. A., Villareal V. A., Wong M. L., Phillips M., and Clubb R. T. (2006) Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J. Mol. Biol. 360, 435–447 10.1016/j.jmb.2006.05.019 [DOI] [PubMed] [Google Scholar]
- 40. Zapotoczna M., Heilbronner S., Speziale P., and Foster T. J. (2012) Iron-regulated surface determinant (Isd) proteins of Staphylococcus lugdunensis. J. Bacteriol. 194, 6453–6467 10.1128/JB.01195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farrand A. J., Haley K. P., Lareau N. M., Heilbronner S., McLean J. A., Foster T., and Skaar E. P. (2015) An iron-regulated autolysin remodels the cell wall to facilitate heme acquisition in Staphylococcus lugdunensis. Infect. Immun. 83, 3578–3589 10.1128/IAI.00397-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heilbronner S., Monk I. R., Brozyna J. R., Heinrichs D. E., Skaar E. P., Peschel A., and Foster T. J. (2016) Competing for iron: duplication and amplification of the isd locus in Staphylococcus lugdunensis HKU09-01 provides a competitive advantage to overcome nutritional limitation. PLoS Genet. 12, e1006246 10.1371/journal.pgen.1006246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Newton S. M., Klebba P. E., Raynaud C., Shao Y., Jiang X., Dubail I., Archer C., Frehel C., and Charbit A. (2005) The svpA‐srtB locus of Listeria monocytogenes: Fur‐mediated iron regulation and effect on virulence. Mol. Microbiol. 55, 927–940 10.1111/j.1365-2958.2004.04436.x [DOI] [PubMed] [Google Scholar]
- 44. Skaar E. P., Gaspar A. H., and Schneewind O. (2006) Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188, 1071–1080 10.1128/JB.188.3.1071-1080.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Andrade M. A., Ciccarelli F. D., Perez-Iratxeta C., and Bork P. (2002) NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 3, research0047-1 10.1186/gb-2002-3-9-research0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clarke S. R., Mohamed R., Bian L., Routh A. F., Kokai-Kun J. F., Mond J. J., Tarkowski A., and Foster S. J. (2007) The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell. Host Microbe 1, 199–212 10.1016/j.chom.2007.04.005 [DOI] [PubMed] [Google Scholar]
- 47. Visai L., Yanagisawa N., Josefsson E., Tarkowski A., Pezzali I., Rooijakkers S. H. M., Foster T. J., and Speziale P. (2009) Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology 155, 667–679 10.1099/mic.0.025684-0 [DOI] [PubMed] [Google Scholar]
- 48. Zapotoczna M., Jevnikar Z., Miajlovic H., Kos J., and Foster T. J. (2013) Iron-regulated surface determinant B (IsdB) promotes Staphylococcus aureus adherence to and internalization by non-phagocytic human cells. Cell Microbiol. 15, 1026–1041 10.1111/cmi.12097 [DOI] [PubMed] [Google Scholar]
- 49. Meenan N. A., Visai L., Valtulina V., Schwarz-Linek U., Norris N. C., Gurusiddappa S., Höök M., Speziale P., and Potts J. R. (2007) The tandem β-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 282, 25893–25902 10.1074/jbc.M703063200 [DOI] [PubMed] [Google Scholar]
- 50. Geoghegan J. A., Ganesh V. K., Smeds E., Liang X., Höök M., and Foster T. J. (2010) Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. J. Biol. Chem. 285, 6208–6216 10.1074/jbc.M109.062208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vazquez V., Liang X., Horndahl J. K., Ganesh V. K., Smeds E., Foster T. J., and Höök M. (2011) Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J. Biol. Chem. 286, 29797–29805 10.1074/jbc.M110.214981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ross C. L., Liang X., Liu Q., Murray B. E., Höök M., and Ganesh V. K. (2012) Targeted protein engineering provides insights into binding mechanism and affinities of bacterial collagen adhesins. J. Biol. Chem. 287, 34856–34865 10.1074/jbc.M112.371054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hare S. A. (2017) Diverse structural approaches to haem appropriation by pathogenic bacteria. Biochim. Biophys. Acta Proteins Proteom. 1865, 422–433 10.1016/j.bbapap.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 54. Toida T., Yoshida H., Toyoda H., Koshiishi I., Imanari T., Hileman R. E., Fromm J. R., and Linhardt R. J. (1997) Structural differences and the presence of unsubstituted amino groups in heparan sulphates from different tissues and species. Biochem. J. 322, 499–506 10.1042/bj3220499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johnson M. B., Pang B., Gardner D. J., Niknam-Benia S., Soundarajan V., Bramos A., Perrault D. P., Banks K., Lee G. K., Baker R. Y., Kim G. H., Lee S., Chai Y., Chen M., Li W., Young-Kwong H., et al. (2017) Topical fibronectin improves wound healing of irradiated skin. Sci. Rep. 7, 1–10 10.1038/s41598-017-03614-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saravia-Otten P., Müller H. P., and Arvidson S. (1997) Transcription of Staphylococcus aureus fibronectin-binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179, 5259–5263 10.1128/jb.179.17.5259-5263.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Josse J., Laurent F., and Diot A. (2017) Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front. Microbiol. 8, 2433 10.3389/fmicb.2017.02433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bur S., Preissner K. T., Herrmann M., and Bischoff M. (2013) The Staphylococcus aureus extracellular adherence protein promotes bacterial internalization by keratinocytes independent of fibronectin-binding proteins. J. Invest. Dermatol. 133, 2004–2012 10.1038/jid.2013.87 [DOI] [PubMed] [Google Scholar]
- 59. Tribelli P. M., Luqman A., Nguyen M. T., Madlung J., Fan S. H., Macek B., Sass P., Bitschar K., Schittek B., Kretschmer D., and Götz F. (2020) Staphylococcus aureus Lpl protein triggers human host cell invasion via activation of Hsp90 receptor. Cell. Microbiol. 22, e13111 10.1111/cmi.13111 [DOI] [PubMed] [Google Scholar]
- 60. Kost C., Stüber W., Ehrlich H. J., Pannekoek H., and Preissner K. T. (1992) Mapping of binding sites for heparin, plasminogen activator inhibitor-1, and plasminogen to vitronectin's heparin-binding region reveals a novel vitronectin-dependent feedback mechanism for the control of plasmin formation. J. Biol. Chem. 267, 12098–12105 [PubMed] [Google Scholar]
- 61. Miajlovic H., Zapotoczna M., Geoghegan J. A., Kerrigan S. W., Speziale P., and Foster T. J. (2010) Direct interaction of iron-regulated surface determinant IsdB of Staphylococcus aureus with the GPIIb/IIIa receptor on platelets. Microbiology 156, 920–928 10.1099/mic.0.036673-0 [DOI] [PubMed] [Google Scholar]
- 62. McDevitt D., Nanavaty T., House-Pompeo K., Bell E., Turner N., Mcintire L., Foster T., and Höök M. (1997) Characterization of the interaction between the Staphylococcus Aureus clumping factor (ClfA) and fibrinogen. Eur. J. Biochem. 247, 416–424 10.1111/j.1432-1033.1997.00416.x [DOI] [PubMed] [Google Scholar]
- 63. Ganesh V. K., Rivera J. J., Smeds E., Ko Y. P., Bowden M. G., Wann E. R., Gurusiddappa S., Fitzgerald J. R., and Höök M. (2008) A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 4, e1000226 10.1371/journal.ppat.1000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hair P. S., Ward M. D., Semmes O. J., Foster T. J., and Cunnion K. M. (2008) Staphylococcus aureus clumping factor A binds to complement regulator factor I and increases factor I cleavage of C3b. J. Infect. Dis. 198, 125–133 10.1086/588825 [DOI] [PubMed] [Google Scholar]
- 65. Hair P. S., Echague C. G., Sholl A. M., Watkins J. A., Geoghegan J. A., Foster T. J., and Cunnion K. M. (2010) Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infect. Immun. 78, 1717–1727 10.1128/IAI.01065-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eidhin D. Ní, Perkins S., Francois P., Vaudaux P., Höök M., and Foster T. J. (1998) Clumping factor B (ClfB), a new surface‐located fibrinogen‐binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30, 245–257 10.1046/j.1365-2958.1998.01050.x [DOI] [PubMed] [Google Scholar]
- 67. Perkins S., Walsh E. J., Deivanayagam C. C., Narayana S. V., Foster T. J., and Höök M. (2001) Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J. Biol. Chem. 276, 44721–44728 10.1074/jbc.M106741200 [DOI] [PubMed] [Google Scholar]
- 68. O'Brien L. M., Walsh E. J., Massey R. C., Peacock S. J., and Foster T. J. (2002) Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell. Microbiol. 4, 759–770 10.1046/j.1462-5822.2002.00231.x [DOI] [PubMed] [Google Scholar]
- 69. Walsh E. J., O'Brien L. M., Liang X., Hook M., and Foster T. J. (2004) Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J. Biol. Chem. 279, 50691–50699 10.1074/jbc.M408713200 [DOI] [PubMed] [Google Scholar]
- 70. Mulcahy M. E., Geoghegan J. A., Monk I. R., O'Keeffe K. M., Walsh E. J., Foster T. J., and McLoughlin R. M. (2012) Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog. 8, e1003092 10.1371/journal.ppat.1003092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vitry P., Valotteau C., Feuillie C., Bernard S., Alsteens D., Geoghegan J. A., and Dufrêne Y. F. (2017) Force-induced strengthening of the interaction between Staphylococcus aureus clumping factor B and loricrin. mBio 8, e01748–17 10.1128/mBio.01748-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bowden C., Chan A., Li E., Arrieta A. L., Eltis L. D., and Murphy M. (2018) Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin. J. Biol. Chem. 293, 177–190 10.1074/jbc.M117.806562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gianquinto E., Moscetti I., De Bei O., Campanini B., Marchetti M., Luque F. J., Cannistraro S., Ronda L., Bizzarri A. R., Spyrakis F., and Bettati S. (2019) Interaction of human hemoglobin and semi-hemoglobins with the Staphylococcus aureus hemophore IsdB: a kinetic and mechanistic insight. Sci. Rep. 9, 18629 10.1038/s41598-019-54970-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stranger-Jones Y. K., Bae T., and Schneewind O. (2006) Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U S A 103, 16942–16947 10.1073/pnas.0606863103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kuklin N. A., Clark D. J., Secore S., Cook J., Cope L. D., McNeely T., Noble L., Brown M. J., Zorman J. K., Wang X. M., Pancari G., Fan H., Isett K., Burgess B., Bryan J., et al. (2006) A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74, 2215–2223 10.1128/IAI.74.4.2215-2223.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim H. K., DeDent A., Cheng A. G., McAdow M., Bagnoli F., Missiakas D. M., and Schneewind O. (2010) IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28, 6382–6392 10.1016/j.vaccine.2010.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brown M., Kowalski R., Zorman J., Wang X. M., Towne V., Zhao Q., Secore S., Finnefrock A. C., Ebert T., Pancari G., Isett K., Zhang Y., Anderson A. S., Montgomery D., Cope L., et al. (2009) Selection and characterization of murine monoclonal antibodies to Staphylococcus aureus iron-regulated surface determinant B with functional activity in vitro and in vivo. Clin. Vaccine Immunol. 16, 1095–1104 10.1128/CVI.00085-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fowler V. G., Allen K. B., Moreira E. D., Moustafa M., Isgro F., Boucher H. W., Corey G. R., Carmeli Y., Betts R., Hartzel J. S., Chan I. S., McNeely T. B., Kartsonis N. A., Guris D., Onorato M. T., et al. (2013) Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 309, 1368–1378 10.1001/jama.2013.3010 [DOI] [PubMed] [Google Scholar]
- 79. Bagnoli F., Bertholet S., and Grandi G. (2012) Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Front. Cell. Infect. Microbiol. 2, 16 10.3389/fcimb.2012.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. O'Halloran D. P., Wynne K., and Geoghegan J. A. (2015) Protein A is released into the Staphylococcus aureus culture supernatant with an unprocessed sorting signal. Infect. Immun. 83, 1598–1609 10.1128/IAI.03122-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Patel A. H., Nowlan P., Weavers E. D., and Foster T. (1987) Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect. Immun. 55, 3103–3110 10.1128/IAI.55.12.3103-3110.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Higgins J., Loughman A., Van Kessel K. P., Van Strijp J. A., and Foster T. J. (2006) Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol. Lett. 258, 290–296 10.1111/j.1574-6968.2006.00229.x [DOI] [PubMed] [Google Scholar]
- 83. Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., and Foster S. J. (2002) σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184, 5457–5467 10.1128/jb.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Claro T., Widaa A., O'Seaghdha M., Miajlovic H., Foster T. J., O'Brien F. J., and Kerrigan S. W. (2011) Staphylococcus aureus Protein A Binds to Osteoblasts and Triggers Signals That Weaken Bone in Osteomyelitis. PLoS ONE 6, e18748 10.1371/journal.pone.0018748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. De Ruyter P. G., Kuipers O. P., and De Vos W. M. (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62, 3662–3667 10.1128/AEM.62.10.3662-3667.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 87. Speziale P., Visai L., Rindi S., and Di Poto A. (2008) Purification of human plasma fibronectin using immobilized gelatin and Arg affinity chromatography. Nat. Protoc. 3, 525–533 10.1038/nprot.2008.12 [DOI] [PubMed] [Google Scholar]
- 88. Akiyama S. K. (2013) Purification of vitronectin. Curr. Protoc. Cell Biol. 60, Unit 10.6 10.1002/0471143030.cb1006s60 [DOI] [PubMed] [Google Scholar]
- 89. Viela F., Prystopiuk V., Leprince A., Mahillon J., Speziale P., Pietrocola G., and Dufrêne Y. F. (2019) Binding of Staphylococcus aureus protein A to von Willebrand factor is regulated by mechanical force. mBio 10, e00555–19 10.1128/mBio.00555-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data are available via ProteomeXchange with identifier PXD019371 (90). All other data supporting the findings of this study are available within the article and its supporting data.